Abstract
Exopolysaccharides contribute significantly to attachment and biofilm formation in the opportunisitc pathogen Pseudomonas aeruginosa. The Psl polysaccharide, which is synthesized by the polysaccharide synthesis locus (psl), is required for biofilm formation in nonmucoid strains that do not rely on alginate as the principal biofilm polysaccharide. In-frame deletion and complementation studies of individual psl genes revealed that eleven psl genes, pslACDEFGHIJKL, are required for Psl production and surface attachment. We also present the first structural analysis of the psl-dependent polysaccharide, which consists of a repeating pentasaccharide containing d-mannose, d-glucose, and l-rhamnose:
In addition we identified the sugar nucleotide precursors involved in Psl generation and demonstrated the requirement for GDP-d-mannose, UDP-d-glucose, and dTDP-l-rhamnose in Psl production and surface attachment. Finally, genetic analyses revealed that wbpW restored Psl production in a pslB mutant and pslB promoted A-band LPS synthesis in a wbpW mutant, indicating functional redundancy and overlapping roles for these two enzymes. The structural and genetic data presented here provide a basis for further investigation of the Psl proteins and potential roles for Psl in the biology and pathogenesis of P. aeruginosa.
Introduction
Pseudomonas aeruginosa is a Gram-negative pathogen responsible for a variety of opportunistic infections, including chronic airway infections in patients with cystic fibrosis (CF) (Driscoll et al., 2007; Govan and Deretic, 1996). During the course of chronic infection, P. aeruginosa forms matrix-encased, surface-associated communities called biofilms. Biofilms are thought to contribute to persistence in the CF airway by contributing to evasion of the host immune response and antimicrobial therapy (Parsek and Singh, 2003; Ramsey and Wozniak, 2005; Stewart and Costerton, 2001). Polysaccharide has been identified as a significant component of the P. aeruginosa biofilm matrix (Branda et al., 2005) with alginate, an unbranched heteropolymer of β-1,4-linked d-mannuronic and d-guluronic acids, as the defining polysaccharide of clinical mucoid isolates (Govan and Deretic, 1996; Ramsey and Wozniak, 2005). Nonmucoid P. aeruginosa strains do not rely on alginate for production of the biofilm matrix, but instead utilize the exopolysaccharides Pel and/or Psl (Branda et al., 2005; Ryder et al., 2007; Wozniak et al., 2003). The pel locus contains seven genes involved in the production of a glucose-rich polysaccharide important in pellicle formation in standing liquid cultures and maintenance of biofilm structure (Friedman and Kolter, 2004a; Vasseur et al., 2005). The polysaccharide synthesis locus (psl) is a group of fifteen co-transcribed genes proposed to encode the necessary enzymes for Psl biosynthesis (Friedman and Kolter, 2004b; Jackson et al., 2004; Matsukawa and Greenberg, 2004). It is predicted by us and others that the resulting polysaccharide contains d-mannose (d-Man), d-glucose (d-Glc), and l-rhamnose (l-Rha), and possibly galactose, although the structure of this key biofilm matrix component is unknown (Friedman and Kolter, 2004b; Jackson et al., 2004; Ma et al., 2007; Matsukawa and Greenberg, 2004; Rocchetta et al., 1998; Wozniak et al., 2003).
P. aeruginosa has the potential to produce several polysaccharides or polysaccharide-containing compounds. Besides alginate, Pel, and Psl, P. aeruginosa can make two other important polysaccharide-containing compounds, rhamnolipid and lipopolysaccharide (LPS). Although their structures are radically different, two or more of these compounds shares some of the same basic building blocks. Polysaccharide synthesis commonly relies on a pool of sugar nucleotide precursors which are modified or directly incorporated into the elongating polysaccharide chain (Rocchetta et al., 1999; Samuel and Reeves, 2003; Whitfield and Roberts, 1999). In addition to the proposed composition of Psl, one or more of the sugar nucleotide precursors GDP-d-Man, UDP-d-Glc, and dTDP-l-Rha is also critical for the production of alginate, rhamnolipid, both types of P. aeruginosa LPS, and potentially Pel. AlgD uses GDP-d-Man as a substrate for the first step in alginate biosynthesis (Govan and Deretic, 1996; Ramsey and Wozniak, 2005). Rhamnolipid synthesis requires dTDP-l-Rha, which is linked by a β-glycosidic bond to a 3-hydroxyfatty acid in mono- or di-rhamnose configurations (Soberon-Chavez et al., 2005). A-band LPS is a homopolymer of α-1,2, α-1,3, α-1,3-linked d-Rha trisaccharides attached to core oligosaccharide-lipid A (Rivera et al., 1988; Rivera and McGroarty, 1989; Rocchetta et al., 1998). B-band LPS is a serotype-specific heteropolymer. The common laboratory wild-type strain, PAO1, belongs to serotype O5, and produces B-band LPS containing an O-antigen trisaccharide repeating unit of 2-acetamido-3-acetamidino-2,3-dideoxy-d-mannuronic acid [Man(2NAc3N)A], 2,3-diacetamido-d-mannuronic acid [Man(NAc)2A], and 2-acetamido-2,6-dideoxy-d-galactose (Fuc2NAc) (Rocchetta et al., 1998; Rocchetta et al., 1999). Synthesis of A-band LPS relies on GDP-d-Man as a precursor to d-Rha (Rocchetta et al., 1998), and both A-band and B-band LPS require dTDP-l-Rha as l-Rha is the linking sugar for B-band and presumably A-band attachment to the outer core. Although B-band LPS is typically reduced in P. aeruginosa CF isolates, nonmucoid PAO1 expresses both A-band and B-band LPS (Hancock et al., 1983; Rocchetta et al., 1999).
In this study we show that eleven of the fifteen psl genes are necessary for Psl synthesis, while four psl genes, pslB, pslM, pslN, and pslO, are not required. Prior to this study there had been no structural investigation of the Psl polysaccharide. Using an inducible Psl overexpression strain to generate polysaccharide, we determined that a fraction of Psl is released from the cell in the form of a branched pentasaccharide repeating unit containing d-Man, d-Glc, and l-Rha. This was predicted by prior analyses (Friedman and Kolter, 2004b; Jackson et al., 2004; Kocharova et al., 1988; Matsukawa and Greenberg, 2004; Rocchetta et al., 1998; Wozniak et al., 2003) and is identical to that described previously by Kocharova et al. (1988). We inactivated one or more critical genes in each sugar nucleotide precursor pathway and show that GDP-d-Man, UDP-d-Glc, and dTDP-l-Rha are each required for Psl production. We also discovered that the A-band LPS gene product, WbpW, functions in the synthesis of both Psl and A-band LPS.
Results and Discussion
Anti-Psl antiserum specifically detects Psl
To assist in detecting Psl in polysaccharide extracts, we had antiserum (α-Psl) generated to the extracellular material from the arabinose-inducible Psl overexpression strain WFPA801 (Ma et al., 2006). The antiserum was then adsorbed multiple times against WFPA800, a strain engineered that lacks Psl due to a deletion in the promoter region upstream of pslA (Ma et al., 2006). The specificity of α-Psl was first determined by immunoblotting polysaccharide extracts from wild type and three psl mutants. The psl operon structures of these four strains are shown in Fig. 1A, along with the putative functions of psl-encoded gene products as annotated previously (Friedman and Kolter, 2004b; Jackson et al., 2004; Stover et al., 2000). WFPA826 contains a deletion from upstream of pslA to downstream of pslO, removing all psl open reading frames. As shown in Fig. 1B, when immunoblotted, extracts from these strains revealed the predicted interactions with the α-Psl antiserum. Extracts from WFPA800 and WFPA826 did not react with α-Psl, while WFPA801 extracts reacted strongly with α-Psl, requiring a 1:50 dilution to be within the range of chemiluminescent detection. The fact that WFPA800 has the same loss of α-Psl reactivity as WFPA826 confirms that the psl promoter deletion is a valid negative control for Psl synthesis. The intermediate reactivity of PAO1 extracts with α-Psl demonstrates a lower level of Psl being produced by the wild type strain as compared to the level produced by induced strain WFPA801.
Figure 1.
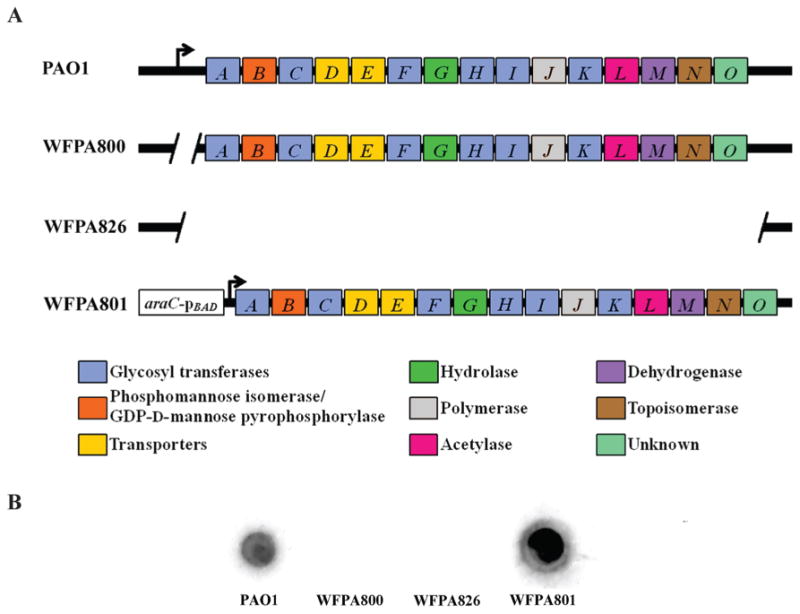
Structure of the psl operon and confirmation of Psl-specific antisera.
A. Map of the psl operon in PAO1 and selected mutants used in this study. Genes pslA-O are shown in boxes (not to scale), with the color corresponding to the putative functions assigned to the gene products, shown in the lower panel. Functions were assigned using bioinformatics by three independent sources (Friedman and Kolter, 2004b; Jackson et al., 2004; Stover et al., 2000). Angled lines represent the extent of deleted sequence, and black arrows indicate transcriptional start sites (not to scale).
B. Crude polysaccharide extracts from PAO1, WFPA800, WFPA826, and WFPA801 were blotted on nitrocellulose, probed with α-Psl, and detected by chemiluminescence using a HRP-conjugated secondary antibody. Extract from WFPA801 was diluted 1:50 in order to remain within range of detection.
Mutagenesis reveals that all psl genes except pslBMNO are necessary for Psl synthesis and P. aeruginosa attachment
When initially studied, the psl locus was found to encode fifteen co-transcribed open reading frames predicted to be involved in polysaccharide synthesis (Friedman and Kolter, 2004b; Jackson et al., 2004; Matsukawa and Greenberg, 2004). The phenotype of a psl promoter deletion mutant is a severe attenuation of surface attachment and biofilm forming ability under both static and continuous flow conditions (Ma et al., 2006). Few studies have been carried out on deletion mutants of individual psl genes (Campisano et al., 2006; Friedman and Kolter, 2004b; Jackson et al., 2004; Ma et al., 2007), but of those, all show essential roles in biofilm formation. To investigate the contribution of each psl gene to generation of Psl and the attachment phenotype, in-frame, non-polar deletions were made in each psl gene following a similar overlapping PCR strategy to that which was used to create WFPA800 (Table 1) (Ma et al., 2006). The ability of these fifteen mutants to attach to a solid surface was first assessed by crystal violet staining (Fig. 2A), with values normalized to the PAO1 background strain. Attachment of WFPA800 and eleven psl mutants (pslACDEFGHIJKL) was significantly reduced (p<0.001, Dunnett's Test following one-way ANOVA) compared to that of PAO1. Interestingly, deletion of pslB, pslM, pslN, or pslO did not result in an observable attachment defect compared to PAO1. Polysaccharide extracts from each of the psl mutants were blotted on nitrocellulose and probed with α-Psl to directly evaluate Psl production (Fig. 2A). The same eleven strains that were deficient in attachment likewise did not produce detectable levels of Psl. Mutants that had comparable attachment to PAO1, however, did produce Psl. The correlation between Psl production and solid surface attachment has been previously reported (Friedman and Kolter, 2004b; Jackson et al., 2004; Ma et al., 2006; Ma et al., 2007; Ma et al., 2009), and this analysis of individual psl mutants confirms these observations.
Table 1.
Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| PAO1 | Prototroph | (Holloway, 1955) |
| WFPA800 | psl promoter deletion | Ma, et al. (2006) |
| WFPA801 | psl-araC-pBAD promoter replacement | Ma, et al. (2006) |
| WFPA811 | In-frame pslA deletion | This work |
| WFPA812 | In-frame pslB deletion | This work |
| WFPA813 | In-frame pslC deletion | This work |
| WFPA814 | In-frame pslD deletion | This work |
| WFPA815 | In-frame pslE deletion | This work |
| WFPA816 | In-frame pslF deletion | This work |
| WFPA817 | In-frame pslG deletion | This work |
| WFPA818 | In-frame pslH deletion | Ma, et al. (2007) |
| WFPA819 | In-frame pslI deletion | Ma, et al. (2007) |
| WFPA820 | In-frame pslJ deletion | This work |
| WFPA821 | In-frame pslK deletion | This work |
| WFPA822 | In-frame pslL deletion | This work |
| WFPA823 | In-frame pslM deletion | This work |
| WFPA824 | In-frame pslN deletion | This work |
| WFPA825 | In-frame pslO deletion | This work |
| WFPA841 | In-frame wbpW deletion | This work |
| WFPA842 | In-frame pslB, wbpW deletion | This work |
| RML-ΔC | Non-polar interruption of rmlC by FRT sequences | Rahim, et al. (2000) |
| R1O5 | rmd::GmR in PAO1 | Rocchetta, et al. (1998) |
| PAO1waaL | waaL::GmR in PAO1 | Abeyrathne, et al. (2005) |
| PAO1-ΔgalU | galU::GmR in PAO1 | R. E. Hancock Lab |
Figure 2.
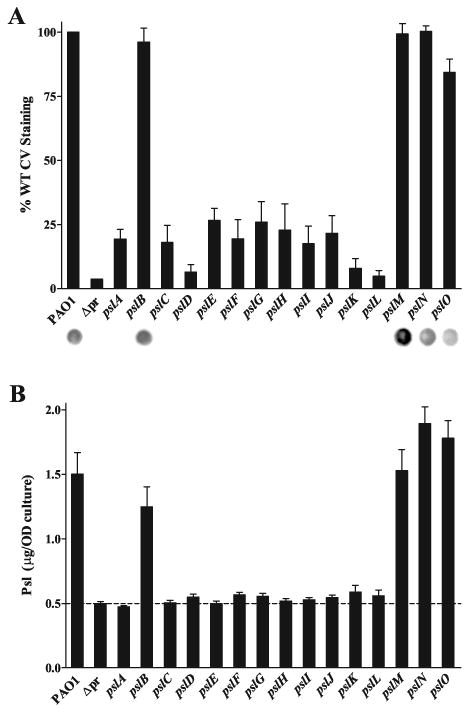
Mutagenesis of psl genes reveals that pslACDEFGHIJKL are necessary for Psl synthesis and P. aeruginosa attachment.
A. Solid surface attachment of psl single mutants correlates with Psl production. Crystal violet staining, read at 540 nm, shown as a percentage of PAO1. The promoter deletion strain WFPA800 is notated as Δpr in both A and B. Values are mean ± SEM from two experiments each in triplicate. Psl production by α-Psl immunoblot is shown below each bar.
B. Psl is produced by eleven of fifteen psl single mutants. ELISA performed using plates coated with crude polysaccharide extracts and probed with α-Psl, read at 450 nm. Values are mean ± SEM from two experiments each in triplicate; dashed line shows background signal of extracts derived from the Δpr strain, WFPA800. The mass of Psl is normalized to OD600 equivalents used to generate the extracts.
We were able to quantify Psl production by individual psl mutants using an ELISA assay, in which Psl-specific antiserum was used to detect Psl in polysaccharide extracts bound to 96-well microtiter plates. A standard curve using purified Psl allowed us to quantify the amount of Psl (Fig. 2B). Similar to both the crystal violet staining and immunoblot, the ELISA revealed that eleven psl genes, pslACDEFGHIJKL, are essential for Psl production. The differences in Psl produced by these eleven Psl-deficient mutants were highly significant by Dunnett's Test (p<0.001) compared to PAO1. When complemented by plasmids expressing the relevant psl gene, each of these mutants produced Psl at levels comparable to or greater than wild type (data not shown). This provides convincing genetic evidence that most but not all of the psl genes within the operon are required for Psl synthesis and the attachment phenotype linked to Psl.
The lack of an attachment phenotype or effect on Psl production in pslM, pslN, and pslO mutants (pslB is discussed below) does not exclude these genes as members of the psl operon. However, recent transcriptional profiling data reveals that pslM, pslN, and pslO are not co-transcribed with pslA-pslL (Goodman et al., 2004; Hickman et al., 2005; Starkey et al., 2009). It is conceivable that these three gene products may have regulatory functions in the Psl pathway, or might modify the polysaccharide without altering its attachment properties or epitopes recognized by α-Psl. Further work will need to be done to determine the inclusion and precise function of the last three genes in the psl operon.
Psl is a pentasaccharide containing d-Man, d-Glc, and l-Rha
As no polysaccharide structure has been definitively associated with the psl locus, we first sought to determine Psl structure using biochemical approaches. P. aeruginosa WFPA801 cells were grown statically in a chemically defined medium (Vasseur et al., 2007) supplemented with arabinose (0.4% final concentration) in order to induce the production of Psl. Upon growth, the bacteria produced thick transparent extracellular material, composed mostly of nucleic acids and proteins, which is in good agreement with literature data (Matsukawa and Greenberg, 2004). Crude carbohydrate extracts were obtained from both cell-associated matrix and from the growth medium, then further treated to remove nucleic acids and proteins. These methods of purification of carbohydrate polymers, as described in the Experimental Procedures, have been previously employed for the isolation of extracellular teichoic acids from staphylococcal biofilms (Sadovskaya et al., 2004; Vinogradov et al., 2006).
The total carbohydrate extract was fractionated on a Sephadex G-50 column. A typical elution profile of the crude carbohydrate of the growth medium (1 L) is shown in Fig. 3A. Fractionation of the carbohydrate extract of the cell-surface-associated matrix gave similar profiles (data not shown). The majority of carbohydrate material eluted in lower MW fractions, in the range of 3000-6000. Yields of this material reached 10 mg/L of culture from the growth medium, while an additional 2-3 mg was obtained from the cell-associated matrix. The material was separated in three fractions, A, B, and C, according to the MW (Fig. 3A). Monosaccharide analysis of fraction C indicated the presence of Man, Glc, and Rha in the approximate ratios of 3:1:1. Monosaccharide analysis and NMR spectra of fraction B indicated a complex mixture, containing β-(1,3)-linked glycerophosphorylated glucan and LPS, but also Man, Glc, and Rha (Vinogradov et al., unpublished data). Polysaccharide from fraction C was chosen for the detailed NMR analysis due to better quality of its NMR spectra. Its structure was analyzed by 2D NMR spectroscopy, and was found to have a pentasaccharide repeating unit structure I (Table 2 and Fig. 4).
Figure 3.
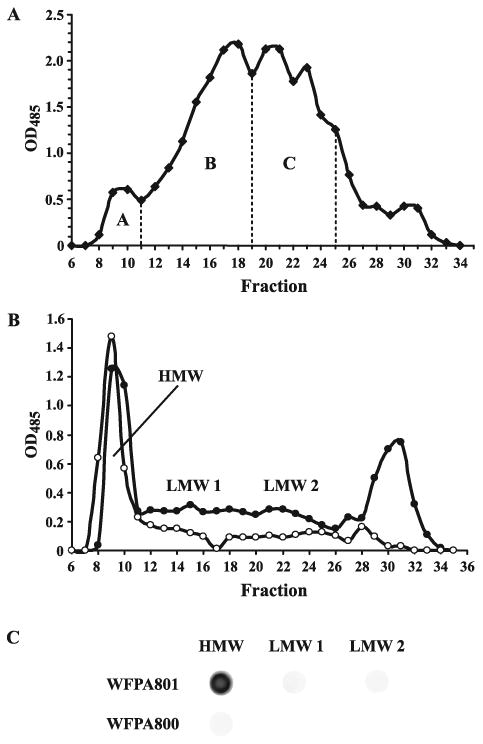
Fractionation of carbohydrate extracts reveals a psl-dependent polysaccharide.
A. Typical elution profile of crude carbohydrate extract of the growth medium (1 L) of P. aeruginosa psl-inducible strain WFPA801 on a Sephadex G-50 column. Cells were grown in M63 medium (1 L) with addition of l-arabinose (0.4 %). Fractions A, B, and C are as indicated. Fractions B and C contain polysaccharide with ∼3-5 and ∼1-2 pentasaccharide repeating units, respectively. Fraction C was used for detailed NMR analysis.
B. Elution profile of crude carbohydrate extracts of the growth medium of P. aeruginosa psl-inducible strain WFPA801 (closed circles) and psl promoter deletion strain WFPA800 (open circles) on a Sephadex G-50 column. Cells were grown in 0.5 L LBNS with addition of 2.0% l-arabinose. A high MW fraction (HMW) and two low MW fractions (LMW 1 and LMW 2) are as indicated.
C. Antiserum reactivity with WFPA801 and WFPA800 cell-associated polysaccharide fractions from B screened by immunoblot analysis.
Table 2.
NMR data. Polysaccharide spectra were recorded at 30°C in D2O with acetone standard.
| Unit | Nucleus | δ (ppm) | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| H/C 1 | H/C 2 | H/C 3 | H/C 4 | H/C 5 | H/C 6a | H/C 6b | ||
| α-Man A | H | 5.38 | 4.06 | 3.97 | 3.70 | 4.20 | 3.79 | 3.89 |
| C | 101.5 | 71.3 | 71.5 | 67.8 | 73.5 | 61.5 | ||
| α -Man A′ | H | 5.21 | 4.10 | 3.97 | 3.70 | 4.20 | 3.79 | 3.89 |
| C | 102.3 | 71.3 | 71.5 | 67.8 | 73.5 | 61.5 | ||
| α -Rha B | H | 5.14 | 4.30 | 3.94 | 3.59 | 4.08 | 1.27 | |
| C | 102.1 | 71.5 | 80.6 | 72.3 | 70.0 | 17.7 | ||
| β-Man C | H | 4.87 | 4.39 | 4.00 | 3.75 | 3.44 | 3.79 | 3.89 |
| C | 97.8 | 74.4 | 82.4 | 66.8 | 77.7 | 61.5 | ||
| β-Man C′ | H | 4.88 | 4.23 | 3.80 | 3.63 | 3.41 | 3.79 | 3.89 |
| C | 97.8 | 76.8 | 74.9 | 68.1 | 77.9 | 61.5 | ||
| β-Man D | H | 4.88 | 4.31 | 3.90 | 3.66 | 3.40 | 3.79 | 3.89 |
| C | 102.4 | 68.9 | 80.3 | 66.3 | 77.4 | 61.5 | ||
| β-Glc E | H | 4.65 | 3.47 | 3.63 | 3.48 | 3.48 | 3.73 | 3.93 |
| C | 101.6 | 74.9 | 83.8 | 69.4 | 77.2 | 62.1 | ||
| α -Glc E′ | H | 5.20 | 3.63 | 3.80 | 3.48 | 3.47 | 3.72 | 3.90 |
| C | 93.4 | 73.2 | 80.9 | 69.4 | 77.2 | 62.1 | ||
| β-Glc E″ | H | 4.65 | 3.35 | 3.61 | 3.47 | 3.47 | 3.72 | 3.90 |
| C | 96.9 | 75.9 | 83.4 | 69.4 | 77.2 | 62.1 | ||
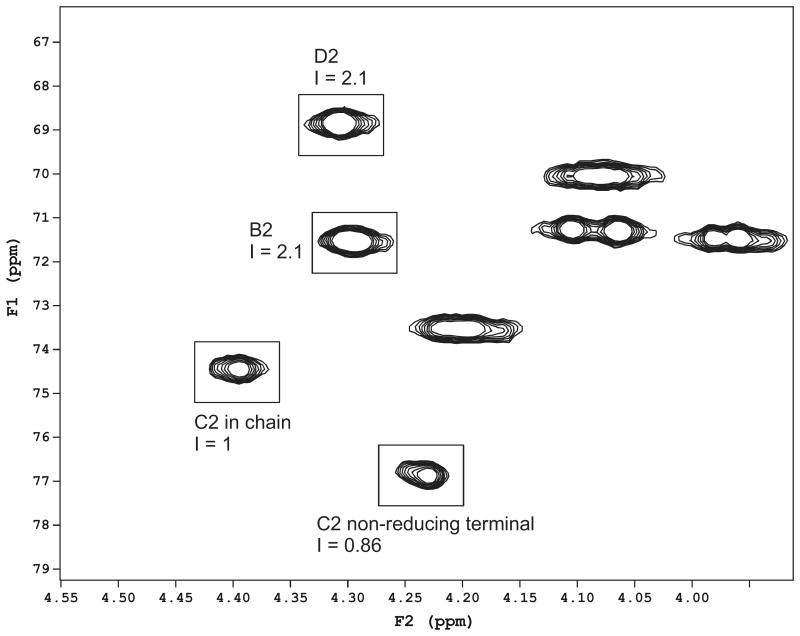
Figure 4.
Partial 2D HSQC spectrum of Psl and the integration data of signals, characteristic for a single pentasaccharide repeating unit (structure I) and its oligomers. I, integral value.
| I |
The structure of this repeating unit is identical to the one of the low molecular weight neutral extracellular polysaccharide of a clinical isolate of P. aeruginosa immunotype 4 and has been characterized earlier (Kocharova et al., 1988).
In addition to the monosaccharides of the polymeric chain, signals of the reducing terminal α- and β-Glc, substituted by variants of α-Rha were present. Signals of the residue C, not substituted at O-3 by Glc E, (C′) were also identified (Table 2), thus indicating the presence of a pentasaccharide, corresponding to a single repeating unit with the Glc residue E at the reducing end (II):
| II |
Integration of the H/C-2 signals of the residues B, C, D (Fig. 4) shows that the overall degree of polymerization is very small, and the mixture may contain an equimolar amount of components with n = 1, 2, and 3, with very little amount of longer chains.
In agreement with the NMR data, methylation analysis of the polysaccharides of fractions B and C indicated the presence of terminal Manp, 3-O-substituted Manp, 2,3-O-substituted Manp, 3-O-substituted Glcp and 3-O-substituted Rhap; as well as 2-O-substituted Manp characteristic for structure II. Methylation analysis indicated an average length of 1-2 repeating units in polysaccharide from fraction C and 4-5 repeating units in polysaccharide from fraction B.
Wild type P. aeruginosa PAO1 were grown in identical conditions but without the addition of arabinose, and the crude polysaccharide extract was fractionated on a Sephadex G-50 column. A low MW polysaccharide containing Man, Glc, and Rha eluted at ∼4000. In a separate experiment, we assessed relative quantities of the Man-containing polysaccharide in the wild type and WFPA801 strains and found it to be ∼4 times higher in the WFPA801 strain with Psl synthesis induced with 0.4% arabinose (data not shown), which is in a good agreement with the transcription data (Ma et al., 2006).
Importantly, no Man-containing carbohydrates were detected in the cell-associated matrix or growth media of the psl promoter deletion mutant WFPA800 (data not shown). From these analyses we can affirm that a Man-rich polysaccharide is indeed the product of the psl locus. The chemical structure of the repeating unit of this polysaccharide has been determined and was found to be identical to one described in the literature (Kocharova et al., 1988). We have found that the polysaccharides produced by the psl-inducible strain WFPA801 and the wild-type strain PAO1 had a slightly lower average MW than the polysaccharide described by Kocharova et al. In addition, we were able to identify a biological repeating unit of this polysaccharide (Structure II).
To determine α-Psl antiserum reactivity with fractionated polysaccharide, WFPA800 and WFPA801 cells were grown in LBNS supplemented with 2% l-arabinose, then treated with ethanol/CaCl2 and proteinase K as described in Experimental Procedures. The crude cell-associated polysaccharide was fractionated on a Sephadex G-50 column (Fig. 3B) and three fractions corresponding to fractions A-C in Fig. 3A were evaluated for Psl by immunoblot analysis. The high MW fraction from WFPA801 reacted strongly with α-Psl, while the two low MW fractions did not show detectable reactivity with the antiserum (Fig. 3C). The monosaccharide composition of this high MW fraction is consistent with the structure of Psl (data not shown). The lack of reactivity with either low MW fraction is currently under investigation. However, high MW cell-associated polysaccharide of WFPA800 did not react with α-Psl, confirming that the α-Psl antiserum is Psl-specific but is likely directed primarily towards a high MW form of Psl (Fig. 3C). Quantities of low MW cell-associated polysaccharide from WFPA800 sufficient for immunoblotting could not be obtained.
A study performed over two decades ago identified a neutral, low molecular weight (∼6500) polysaccharide with a pentasaccharide repeating unit in a clinical P. aeruginosa isolate. The pentasaccharide repeating unit consisted of d-Glc, d-Man, and l-Rha:
Antibodies to this pentasaccharide were cross-reactive with LPS from several Fisher immunotype strains of P. aeruginosa, suggesting that the structure may be a conserved component of the LPS core or O-antigen (Kocharova et al., 1988). However, the large proportion of d-Man in this structure is not consistent with the lack of such residues in the inner core or either of the two outer core glycoforms (Bystrova et al., 2006; Komarova et al., 2008; Poon et al., 2008). Additionally, no structures identical or highly similar to this pentasaccharide have been reported in P. aeruginosa O-antigen polysaccharides to date (Rocchetta et al., 1999). A later study investigating the synthesis of the A-band LPS precursor d-Rha identified a cluster of three open reading frames predicted to encode enzymes involved in production of a polysaccharide containing d-Man, d-Glc, and l-Rha (Rocchetta et al., 1998). The authors speculated that the gene cluster which they identified might be responsible for a polysaccharide distinct from LPS and alginate, and could account for the pentasaccharide repeating unit published by Kocharova et al. (Rocchetta et al., 1998). Following the publication of the psl locus by three individual groups (Friedman and Kolter, 2004b; Jackson et al., 2004; Matsukawa and Greenberg, 2004), it was clear that the three ORFs found by Rocchetta et al. were the first three genes in the psl cluster, pslA, pslB, and pslC. This observation raised the hypothesis that the newly annotated psl locus encoded enzymes necessary for constructing the polysaccharide isolated by Kocharova et al., but the hypothesis was not followed up until this investigation of the structure of Psl polysaccharide.
Previous reports are conflicting as to the presence of galactose in Psl (Friedman and Kolter, 2004b; Ma et al., 2007; Matsukawa and Greenberg, 2004; Wozniak et al., 2003). Binding of a galactose-specific lectin to intact P. aeruginosa was shown to be psl-dependent, and monosaccharide analysis revealed galactose and mannose as predominant extracellular components of WFPA801 induced with 2% arabinose (Ma et al., 2007). However, we did not detect significant levels of galactose in any carbohydrate preparation used for structural or antiserum reactivity experiments. In the present study, NMR was utilized as a more accurate and direct method of structural determination compared with lectin binding assays and compositional analyses used previously. It is also possible that variations in growth conditions used in these studies could account for the differences in monosaccharide composition.
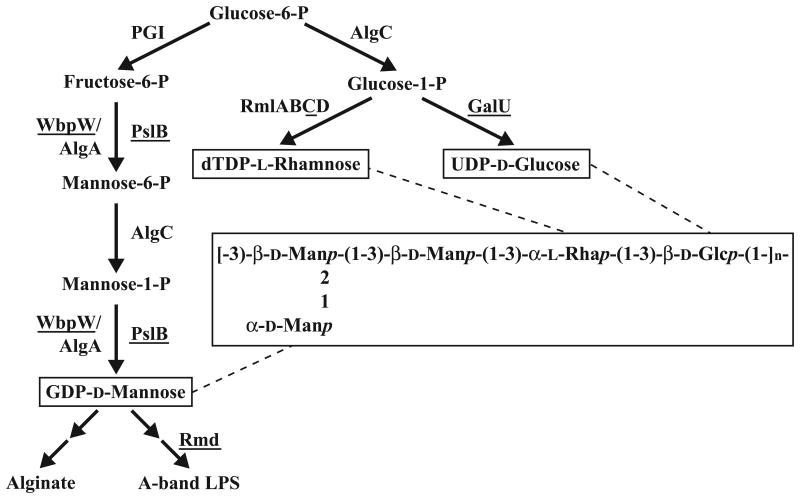
Shared sugar nucleotide precursors involved in Psl synthesis
Knowing the structure of Psl allowed us to then identify the pathways involved in production of precursor sugars used by Psl enzymes, and determine if Psl production is affected when these pathways are disturbed. The pathways and the Psl structure indicating the proposed location of incorporated sugars are summarized in Fig. 5. The sugar nucleotide precursors GDP-d-Man, UDP-d-Glc, and dTDP-l-Rha are synthesized in three distinct pathways, making removal of one sugar at a time possible (Rahim et al., 2000; Rocchetta et al., 1998; Rocchetta et al., 1999). It is important to note that while the biosynthetic gene clusters for the previously mentioned P. aeruginosa polysaccharides are located at sites distinct from the psl locus (Friedman and Kolter, 2004b; Jackson et al., 2004; Matsukawa and Greenberg, 2004), the use of common precursors, as discussed in the introduction, suggests functional overlap of known polysaccharide synthesis enzymes with gene products of the psl locus.
Figure 5.
Pathways involved in synthesis of Psl sugar nucleotide precursors.
Terminal steps of GDP-d-Man, UDP-d-Glc, and dTDP-l-Rha synthesis pathways shown. Sugar nucleotides are shown in boxes and enzymes deleted in this study are underlined. In the Psl repeating unit s`, larger box, p indicates the pyranosyl form of the monosaccharide residue. PGI, phosphoglucose isomerase.
dTDP-l-Rha is necessary for Psl synthesis
Consistent with the presence of l-Rha in the Psl structure (I), and to provide an independent validation of the Psl structure, we reasoned that disrupting the l-Rha pathway would result in a loss of Psl synthesis and a defect in surface attachment. The rml locus is responsible for the generation of dTDP-l-Rha, the sugar-nucleotide precursor allowing l-Rha to be incorporated in the LPS outer core and rhamnolipid, as well as the O polysaccharide of certain P. aeruginosa serotypes (Rahim et al., 2000). RmlC is a dTDP-4-keto-6-deoxy-d-glucose-3,5-epimerase that catalyzes the penultimate step in the dTDP-l-Rha pathway, and when deleted, results in a defect in LPS outer core synthesis leading to a loss of both A-band polysaccharide and B-band O-antigen on the cell surface (Dong et al., 2007; Rahim et al., 2000). The O-antigen defect in an rmlC mutant is in agreement with results obtained by previous studies demonstrating that l-Rha is the acceptor residue for O-antigen addition to the outer core (Sadovskaya et al., 1998; Sadovskaya et al., 2000). It is likely that terminal l-Rha on the outer core is also the site of attachment for A-band polysaccharide, but this has not been definitively established (Rahim et al., 2000) and work is underway by two of the authors, Lam and Vinogradov, to address this issue.
An rmlC mutant was evaluated for surface attachment and Psl production by immunoblot (Fig. 6). There was a significant decrease in the attachment of the rmlC mutant compared to the parental wild type PAO1 strain in which this mutation was generated (p<0.001, Dunnett's Test), but not as severe a defect as in WFPA800 (Δpr, Fig. 6). Immunoblotting polysaccharide extract from the rmlC mutant with α-Psl antiserum did not reveal any detectable Psl, implicating the rml locus in its production by providing the l-Rha precursor dTDP-l-Rha. The fact that the rmlC mutant shows greater attachment than WFPA800 might be attributed to the altered surface physiology of the rmlC mutant compared to WFPA800, due to a loss of both A-band polysaccharide and B-band O-antigen from the cell surface. Previous reports have indicated an increase in surface adherence in certain P. aeruginosa strains lacking both A-band and B-band LPS, but this has not been confirmed for all serotypes (Rocchetta et al., 1999).
Figure 6.
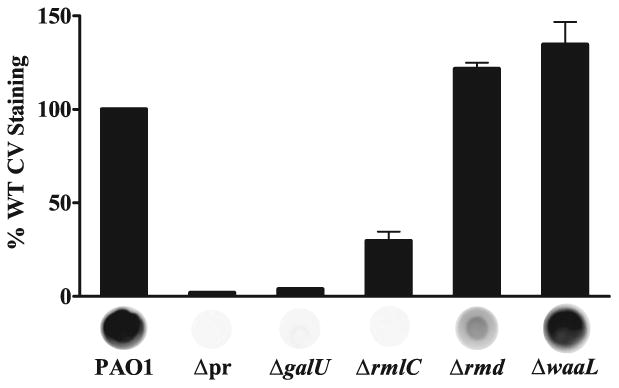
dTDP-l-Rha and UDP-d-Glc are essential for Psl production.
Deletion of rmlC or galU results in defective surface attachment due to loss of Psl, while loss of rmd does not affect attachment despite a reduction in Psl. Crystal violet staining, read at 540 nm, shown as a percentage of PAO1. The promoter deletion strain WFPA800 is notated as Δpr. Values are mean ± SEM from two or three experiments. Psl production by α-Psl immunoblot shown below each bar.
To determine if the attachment defect in the rmlC mutant might be due to effects on LPS, a waaL mutant was examined (Fig. 6). WaaL is the O-antigen ligase responsible for addition of both A-band polysaccharide and B-band O-antigen to the LPS core (Abeyrathne and Lam, 2007). The LPS phenotypes of rmlC and waaL mutants are similar in that A- and B-band polysaccharides are not displayed on the cell surface but instead remain linked to the lipid carrier undecaprenol-phosphate (Und-P) (Abeyrathne et al., 2005; Rahim et al., 2000). Interestingly, attachment of the waaL mutant was significantly greater than that of PAO1 (0.001<p<0.01, Dunnett's Test), suggesting that the lack of surface-associated A-band and B-band polysaccharide facilitates attachment in the waaL mutant but that the loss of Psl takes precedence over the LPS alteration in defining the rmlC attachment phenotype. The increase in attachment of the waaL strain is not due to an increase in Psl expression, as the waaL mutant produced Psl at a level comparable to that of PAO1.
To demonstrate the specificity of Psl biosynthetic enzymes for the l-isoform of rhamnose, an rmd mutant was evaluated for surface attachment and Psl production (Fig. 6). Rmd is a GDP-4-keto-6-deoxy-d-mannose reductase and performs the final step in conversion of GDP-d-Man into GDP-d-Rha, the substrate for A-band polysaccharide production (Maki et al., 2002; Rocchetta et al., 1998). Since GDP-d-Rha synthesis occurs downstream of GDP-d-Man synthesis, we predicted that deletion of rmd would result in a loss of only A-band LPS. The rmd mutant was not defective in attachment but in fact attached to a greater degree than wild type (p<0.05, Dunnett's Test). This is similar to the waaL-deficient strain and suggests that the loss of A-band LPS increases attachment, perhaps by alteration of the surface properties of the cell. Psl production in the rmd mutant was detectable by immunoblot, but rmd extracts reacted substantially weaker with α-Psl compared to wild type. Deletion of rmd may result in accumulation of GDP-d-Man that inhibits the phosphomannose isomerase (PMI) activity of PslB, resulting in a reduction in Psl (Lee et al., 2008). This explanation, however, does not take into account the compensatory PMI function of WbpW (see discussion below, Fig. 7), an enzyme for which GDP-d-Man feedback inhibition has yet to be demonstrated. It is unlikely that α-Psl is cross-reactive with A-band LPS since WFPA800 produces A-band LPS at a level comparable to that of wild type (see Fig. 9A), but displays no detectable reactivity with α-Psl. The observed reactivity of extracts from the rmd but not the rmlC mutant with α-Psl confirms that Psl enzymes rely on the precursor sugar nucleotide dTDP-l-Rha in Psl production and cannot use GDP-d-Rha exclusively. The intermediate Psl phenotype of the rmd mutant, however, does not rule out a contribution by GDP-d-Rha.
Figure 7.
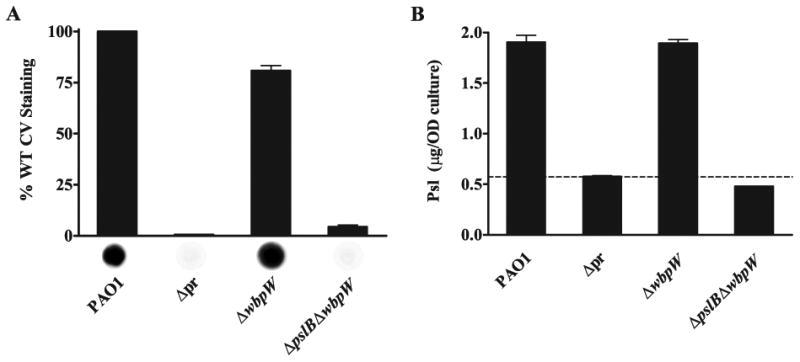
WbpW is the PMI/GMP in Psl synthesis in the absence of PslB.
A. Deletion of pslB and wbpW results in an attachment defect and loss of Psl. Crystal violet staining, read at 540 nm, shown as a percentage of PAO1. The promoter deletion strain WFPA800 is notated as Δpr in both A and B. Values are mean ± SEM from three experiments. Psl production by α-Psl immunoblot shown below each bar.
B. Psl is produced by a wbpW mutant but not by a pslB wbpW double mutant. ELISA performed using plates coated with crude polysaccharide extracts and probed with α-Psl, read at 450 nm. Values are mean ± SEM from two experiments each in triplicate; dashed line shows background signal of extracts derived from the Δpr strain, WFPA800. The mass of Psl is normalized to OD600 equivalents used to generate the extracts.
Figure 9.
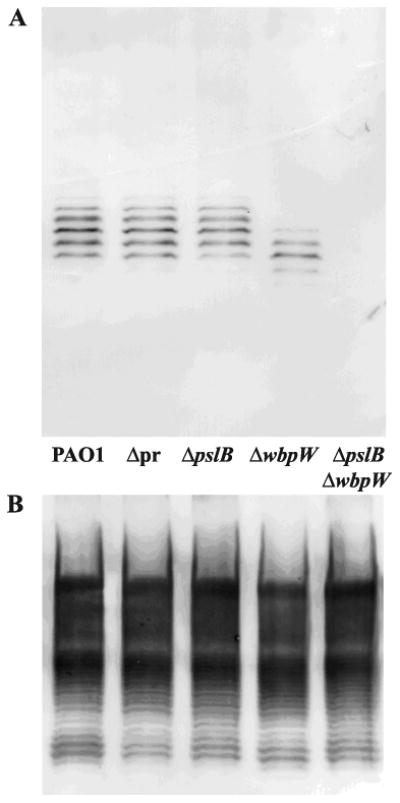
PslB is the PMI/GMP in A-band LPS synthesis in the absence of WbpW.
A. A-band LPS is detectable in a wbpW mutant but not in a pslB wbpW double mutant. LPS was purified from P. aeruginosa grown on plates 12 h at 37°C. Western blot analysis of PAO1, WFPA800 (notated as Δpr), pslB, wbpW, and pslB wbpW, using the A-band-specific mAb N1F10.
B. B-band LPS is unaffected in mutant P. aeruginosa strains. Western blot analysis of strains from A, using the B-band-specific mAb MF15-4.
UDP-d-Glc is required for Psl synthesis
Continuing with the analysis of sugar precursors incorporated into the Psl structure, we predicted that interfering with UDP-d-Glc generation would likewise prevent synthesis of Psl and result in defective surface attachment characteristic of the Psl-null phenotype. In P. aeruginosa, galU encodes the UDP-d-Glc pyrophosphorylase which joins Glc-1-P to its nucleotide carrier in the final step of the UDP-d-Glc pathway (Rocchetta et al., 1999). Similar to deletion of rmlC, interruption of galU results in truncation of the LPS outer core preventing surface expression of both A-band and B-band LPS (Choudhury et al., 2005). Unlike the rmlC mutant, attachment of which is somewhat greater than WFPA800 (p<0.05, Tukey's Test), the galU mutant displays a significant (p<0.001, Dunnett's Test) defect in surface attachment comparable to that of WFPA800 (Fig. 6). Given that the loss of surface-associated A-band LPS appears to increase surface attachment, as in rmlC, rmd, and waaL mutants, resolving the lack of such an increase in the galU mutant will require further investigation. Immunoblotting extracts from the galU mutant revealed a complete loss of Psl as seen in the rmlC-deficient strain, confirming that in addition to dTDP-l-Rha, UDP-d-Glc is essential for Psl production. When compared to the waaL mutant lacking both A- and B-band LPS on the surface, the attachment phenotype of the galU mutant, similar to the rmlC strain, is dominated by the loss of Psl and not the loss of surface-associated A- and B-band LPS.
WbpW allows for Psl production in a pslB mutant
The pslM, pslN, and pslO gene products have no homologs in P. aeruginosa and do not appear to have annotated functions associated with synthesis of a polysaccharide (Friedman and Kolter, 2004b; Jackson et al., 2004; Stover et al., 2000). PslB, however, is a paralog of WbpW and AlgA, being 60% identical at the amino acid level with WbpW (Jackson et al., 2004; Rocchetta et al., 1998). WbpW and AlgA have been shown to be bifunctional enzymes with PMI and GDP-d-Man pyrophosphorylase (GMP) activities and function in the A-band LPS and alginate pathways, respectively (Rocchetta et al., 1998; Shinabarger et al., 1991). It has been recently reported that PslB also possesses both PMI and GMP activities as predicted by homology to WbpW and AlgA (Lee et al., 2008). In fact, prior to the psl nomenclature, an open reading frame designated orf488 (pslB) was identified as coding for a putative PMI/GMP, and was positioned between two other orfs (pslA and pslC) collectively implicated in the synthesis of a polysaccharide containing d-Man, d-Glc, and l-Rha (Rocchetta et al., 1998). In that same report, A-band LPS was produced at a low level in a wbpW algA double mutant, suggesting that ORF488/PslB was able to partially compensate for the loss of PMI/GMP activity in the synthesis of A-band LPS (Rocchetta et al., 1998); however, this was not directly tested. Since PslB appears to be functional in the A-band LPS pathway, we hypothesized that WbpW compensates for PslB to allow for the production of GDP-d-Man and Psl in the pslB mutant. To test this we created an in-frame, non-polar wbpW deletion in the pslB background. A single wbpW mutant was also created to confirm that PslB is indeed functional in Psl synthesis. We did not delete algA in this study because the algD promoter responsible for transcription of algA is not significantly activated in nonmucoid strains, resulting in no detectable alginate (Wozniak and Ohman, 1994; Wozniak et al., 2003).
Solid surface attachment for the pslB wbpW mutant was assessed, and polysaccharide extracts were used to quantify Psl production by immunoblot and ELISA (Fig. 7). Crystal violet staining revealed a significant (p<0.001) defect in attachment for the pslB wbpW mutant similar to that seen in the psl promoter deletion strain WFPA800 (Fig. 7A). The wbpW mutant exhibited a reduction in attachment (p<0.001) compared to wild type, yet still attached at approximately 80% of wild type. Immunoblot analysis of the pslB wbpW mutant showed no detectable Psl, while the wbpW mutant produced Psl at a level similar to that of wild type (Fig. 7A).
To directly visualize attachment of the pslB wbpW mutant, we allowed mid-log cells to attach to vertically oriented glass coverslips for 4 h, then analyzed early biofilm formation at the air-liquid interface by scanning electron microscopy (Parise et al., 2007). Wild type cells attached in a well-defined zone at the air-liquid interface, and upon higher magnification appeared tightly clustered and in contact with the glass along the long axes of the cells (Fig. 8A). In agreement with what is observed in the microtiter dish attachment assay, attachment of WFPA800 was greatly reduced compared to wild type; attaching only in isolated clusters of a few dozen cells each (Fig. 8B). The pslB wbpW double mutant attached similarly to WFPA800, confirming the attachment defect observed in the microtiter plate assay (Fig. 8E). The wbpW mutant displayed a well-defined line at the air-liquid interface similar to wild type, but had a web-like appearance in the submerged region of the coverslip (Fig. 8D). This may be related to the altered A-band phenotype (see Fig. 9A). Lastly, the pslB mutant showed abundant attachment both at the air-liquid interface and submerged regions of the coverslip (Fig. 8C). However, several of the pslB mutant cells were attached at the pole, in contrast to wild type or the other mutants (Fig. 8C). The cause of this altered attachment phenotype is not yet understood and will require further investigation of the surface properties of pslB-deficient cells compared to wild type and wbpW mutant cells.
Figure 8.
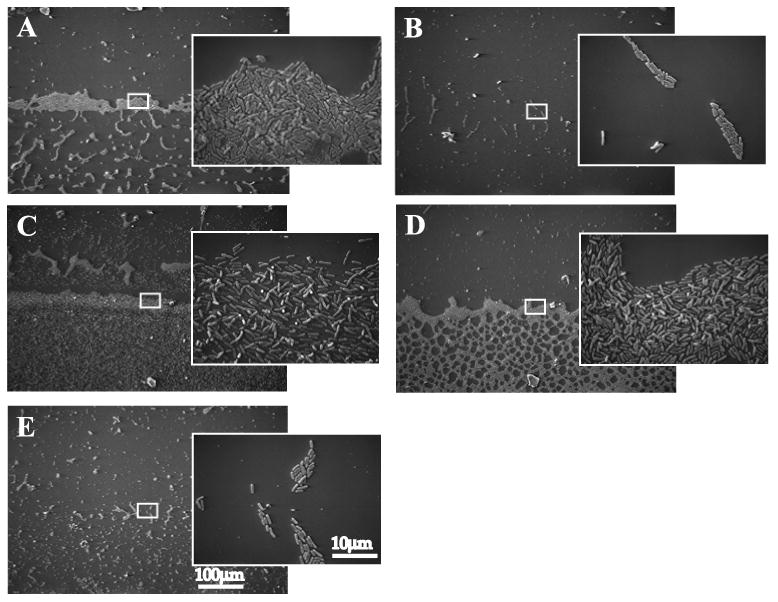
Attachment phenotypes of pslB and wbpW single and double mutants.
Cells were allowed to attach to vertically oriented glass coverslips for 4 h, then fixed and visualized by SEM. A, PAO1; B, WFPA800; C, pslB; D, wbpW; E, pslB wbpW. Larger images taken at 170×, inset images taken at 2500×. White boxes indicate the magnified region of the air-liquid interface shown in inset.
An ELISA quantifying Psl production by these same strains revealed a complete loss of Psl in extracts from the pslB wbpW double mutant but no significant decrease for extracts derived from the wbpW mutant (Fig. 7B). Together, the attachment, immunoblot, and ELISA data confirm that, in a pslB mutant, WbpW is the PMI/GMP responsible for generating the GDP-d-Man essential in the production of Psl. The lack of a Psl phenotype in the wbpW mutant demonstrates that PslB does not require WbpW for Psl synthesis and is capable of functioning as a PMI/GMP, in agreement with recently published PslB enzymatic data (Lee et al., 2008).
PslB is the PMI/GMP that promotes A-band LPS production in a wbpW mutant
In a previous study, it was proposed that ORF488 (PslB) encoded a PMI/GMP responsible for the low level of A-band LPS observed in a wbpW algA mutant; however, pslB was not deleted in that study and therefore its role in A-band LPS synthesis could not be confirmed (Rocchetta et al., 1998). The pslB wbpW double mutant generated in this study provides a means of addressing this unresolved question, namely, that PslB can serve as the PMI/GMP in the A-band LPS pathway much as WbpW can function as the PMI/GMP in the Psl pathway. A western blot analysis of A-band LPS using the A-band-specific monoclonal antibody N1F10 (Lam et al., 1989) in pslB, wbpW, and pslB wbpW mutants was performed (Fig. 9). A-band LPS production in the pslB mutant and WFPA800 was similar to wild type, suggesting that no psl genes are needed for A-band synthesis and more specifically that WbpW functions as a PMI/GMP without PslB (Fig. 9A). Compared with the wild type strain, WFPA800, and the pslB mutant, the wbpW mutant produced reduced, but detectable A-band LPS in a faster migrating cluster of bands (Fig. 9A). A-band production in this mutant is presumably due to the PMI/GMP activity of PslB. In contrast to the other strains, the pslB wbpW double mutant failed to produce detectable A-band polysaccharide (Fig. 9A), indicating that A-band LPS synthesis requires the redundant PMI/GMP activity of either PslB or WbpW to generate product.
The unexpected faster migration of N1F10-reactive bands in the wbpW mutant may be due to PslB functioning in an atypical pathway for a Psl enzyme, resulting in a shorter A-band polymer, but this explanation lacks a molecular basis since generation of GDP-d-Man by PslB occurs upstream of A-band assembly and should not interfere with this process. Although in-frame mutagenesis was performed, it is possible that deletion of wbpW had a polar effect on wzm and/or wzt, which are directly downstream of wbpW and encode the ABC transporter responsible for transport of A-band to the periplasm for attachment to the LPS core (Rocchetta and Lam, 1997). Deleting either of these genes results in A-band polysaccharide remaining in the cytoplasm attached to the lipid carrier Und-P, which runs slightly faster on a PAGE gel similar to what was seen in the wbpW mutant (Rocchetta and Lam, 1997). To address this, samples were prepared using the hot aqueous phenol method that results in cleavage of the highly labile A-band-Und-P linkage, preventing detection by western blotting (Rocchetta and Lam, 1997). Both A-band and B-band LPS were detectable for all strains except the pslB wbpW double mutant, indicating that the faster migrating bands in the wbpW mutant are not due to lipid linkage, but to another cause that remains to be determined (data not shown). The lack of A-band in the double mutant, however, indicates that in spite of possible polar effects on wzm and/or wzt, wbpW was indeed deleted and is not able to contribute to GDP-d-Man synthesis. As a control B-band LPS was analyzed by Western blot using the B-band-specific monoclonal antibody MF15-4 (Lam et al., 1987). There were no differences in B-band LPS between the strains shown in Fig. 9A, confirming that neither WbpW nor PslB is involved in production of the sero-specific O-antigen (Fig. 9B).
Conclusions and perspectives
In this study we have demonstrated that eleven of the fifteen psl genes, pslACDEFGHIJKL, are required for Psl synthesis and solid surface attachment. Prior to this work the structure of the Psl polysaccharide, while elucidated biochemically over two decades ago, had not been firmly linked to the psl operon. Here we show that the structure of Psl from cell-associated and growth medium fractions contains d-Man, d-Glc, and l-Rha in a pentasaccharide repeating unit distinct from other known polysaccharide moieties. Data presented in this study reveal a role for the sugar nucleotide precursors GDP-d-Man, UDP-d-Glc, and dTDP-l-Rha, all with previously identified functions in polysaccharide synthesis, in Psl polysaccharide production. Following up on the lack of a Psl phenotype in the pslB mutant, we have shown that the overlapping functions of PslB and WbpW permit Psl synthesis in a pslB mutant and A-band LPS synthesis in a wbpW mutant. The use of common precursors and similarly functioning enzymes in generating multiple polysaccharides demonstrates metabolic efficiency and contributes to the characteristic adaptability of P. aeruginosa as an opportunistic pathogen. For this overlap to be productive, however, it is important that products from all required pathways be expressed under the same conditions. Overlap among polysaccharide synthesis pathways is likewise apparent in other bacterial pathogens. In E. coli O9:K30, the O-antigen GMP RfbM produces sufficient GDP-Man for both O-antigen and capsular polysaccharide production (Jayaratne et al., 1994). The sugar nucleotide UDP-GlcNAc and its derivatives are also commonly shared among surface polysaccharides; PGA, peptidoglycan, and LPS in E. coli (Wang et al., 2004), and LOS, capsule, and Pgl heptasaccharide in Campylobacter jejuni (Bernatchez et al., 2005). A recent report identified redundant paralogs, at least one of which having a putative function in an LPS pathway, at both the initiating glycosyltransferase and polymerase steps in synthesis of the Caulobacter crescentus holdfast polysaccharide (Toh et al., 2008). Although our understanding of the structure of Psl and the contribution of individual psl genes to Psl production has been extended by this study, much remains unknown about the functions of essential Psl proteins, regulation of the polysaccharide, and bacterial-host interactions mediated by Psl. It is also unclear how the Psl pentasaccharide repeating unit assembles into a biofilm matrix material. Future studies requiring biochemical, genetic, and immunological approaches will be necessary to fully understand the larger role of Psl in P. aeruginosa biology and pathogenesis.
Experimental Procedures
Strains and growth conditions
Escherichia coli strains were grown in Luria Bertani (LB; 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) broth, and P. aeruginosa were routinely grown in LB or LB without NaCl (LBNS) at 37°C. Solid media with (LA) or without (LANS) NaCl were prepared with the addition of 1.5% agar. Sucrose was added at 5% for counterselection experiments. Antibiotics were used at the following concentrations: gentamicin (Gen) 15 and 100 μg/mL; ampicillin (Amp) 100 μg/mL; carbenicillin (Crb) 300 μg/mL; irgasan (Irg) 25 μg/mL. E. coli JM109 was used for cloning and SM10 for biparental mating with P. aeruginosa. Strains used in this study are presented in Table 1, and all plasmids and oligonucleotides are shown in Tables S1 and S2. Addition of l-arabinose at 2.0% was used for induction of Psl expression in WFPA801. For structural studies, P. aeruginosa were grown statically in 140–mm diameter Petri dishes in humidified chambers at 37°C for 24 hrs in M63 minimal medium ((NH4)2SO4, 2g/L; KH2PO4, 13.6 g/L; FeCl3, 0.5 mg/L); pH 7; supplemented with 0.5% Casaminoacids (Difco), 1 mM MgCl2, and 0.2% glucose (Vasseur et al., 2007). For growth of the psl-inducible WFPA801, medium was additionally supplemented with 0.4% l-arabinose (Acros Organic).
Preparation of Psl for structural analysis
P. aeruginosa strains PAO1, WFPA801, and WFPA800 were grown statically in 140–mm diameter Petri dishes in humidified chambers at 37°C as outlined above. The transparent gelatinous material loosely attached to the bottom of the Petri dishes was collected together with the cells and growth medium. Cells and cell-associated matrix material were separated from the viscous growth media by centrifugation (6000 × g, 4°C, 30 min.). The pellet was suspended in 0.9% NaCl and the cell-surface-associated polymers were detached by mild sonication (IKA Labotecknik sonicator, 3 × 30 sec, 50% cycle, intensity 0.5). The pellet was removed by centrifugation and treated with a fresh portion of saline; this procedure was repeated twice more to give crude cell-associated matrix extract.
This extract and the growth media were further treated in the same way. TCA was added to a final concentration of 5% (wt/vol) in order to precipitate extracellular DNA and proteins. After centrifugation (10000 × g, 10 min.) the clear supernatant was extensively dialyzed and lyophilized. The residue was resuspended in water, the insoluble material removed by centrifugation (10000 × g, 10 min.), and the supernatant deproteinated by extraction with phenol (2×) and phenol-chloroform. The obtained crude carbohydrate extract was fractionated on a Sepahdex G-50 column.
Alternatively, WFPA801 and WFPA800 were grown in LBNS (0.5 L) with addition of 2% l-arabinose. Biomass was collected by centrifugation, re-suspended in 0.9% NaCl, vortexed, and pelleted. From the supernatant, extracellular DNA was removed by precipitation with 25% ethanol and 0.1 M CaCl2. The supernatant was dialyzed, lyophilized, resuspended in water, and treated with proteinase K (final concentration 0.5 mg/mL) for 60 min at 60°C, followed by proteinase K inactivation for 30 min at 80°C. This crude polysaccharide extract was fractionated on a Sephadex G-50 column and carbohydrate-containing fractions were screened by Western immunoblotting.
Analytical methods
Methylation analysis was carried out by the Ciucanu and Kerek method after reduction of polysaccharides with NaBH4 (Ciucanu and Kerek, 1984).
Gas-liquid chromatography
Monosaccharides were identified by GLC on a Shimatzu GC-14 gas chromatograph equipped with flame ionization detector and Zebron ZB-5 capillary column (30 m × 0.25 mm), with hydrogen as carrier gas, using a temperature gradient 170°C (3 min.), 260°C at 5°C/min. Prior to analysis, polysaccharides were hydrolyzed with 4 M TFA (110°C, 3 h) and converted to alditol acetates by conventional methods.
Gel-permeation chromatography
Gel-permeation chromatography was carried out on a Sephadex G-50 column (1.6 × 95 cm; Pharmacia), irrigated with water. Fractions (5 mL) were assayed colorimetrically for aldose (Dubois, 1956). Dextrans with average molecular masses 1000, 5, and 1 KDa (Fluka) were used as MW markers.
NMR spectroscopy
Two-dimensional 1H and 13C NMR spectra were recorded using a Varian Inova 500 MHz and 400 MHz spectrometers for samples in D2O solutions at 25°C with acetone internal reference (2.23 ppm for 1H and 31.5 ppm for 13C) using standard pulse sequences DQCOSY, TOCSY (mixing time 120 ms), NOESY (mixing time 400 ms), HSQC and HMBC (100 ms long range transfer delay).
Manipulations and mutagenesis of P. aeruginosa
In-frame, non-polar deletions were made in each psl gene and wbpW using overlap extension PCR. EcoRI and HindIII cut sites were engineered into the outer pair of primers for cloning, except for pslM and pslN for which BamHI was used in place of HindIII. Amplicons containing each deletion were digested with EcoRI and HindIII or BamHI and cloned into pEX18Gm for psl genes or pEX18Ap for wbpW. Integrants in P. aeruginosa were selected with Gen 100/Irg 25 for pEX18Gm or Crb 300/Irg 25 for pEX18Ap. Counterselection with sucrose was performed, and clones were confirmed by Gen 100 or Crb 300 sensitivity for pEX18Gm and pEX18Ap, respectively. Deletions were verified by sequencing of PCR amplicons, as well as complementation in trans for the psl mutants displaying a Psl-null phenotype.
Generation of Psl antiserum
WFPA801 was grown on cellophane over LANS + 2% arabinose at 37°C overnight. Biomass was removed and resuspended in 250mL of 0.9% NaCl, vortexed and pelleted. The supernatant was precipitated with 95% ethanol at 4°C overnight and washed twice with 100% ethanol. The subsequent pellet was lyophilized and used for antisera generation by New England Peptide. New Zealand white rabbits were immunized with 400 μg of the crude Psl material followed by three subsequent boosts of 200 μg each. Serum from the final bleed was used for subsequent adsorptions. For adsorptions, WFPA800 cells were cultured on LANS plates, removed with LBNS, and pelleted. Intact bacteria (∼ 105 CFU) were resuspended in 5 mL final bleed serum and incubated with rocking at 4°C for 1 h. Cells were pelleted and supernatant was adsorbed repeatedly with fresh cells (total of four rounds). Antiserum enriched against Psl (α-Psl) was filtered through a 0.22 μm acrodisc filter, and aliquots were stored at -20°C.
Western immunoblotting and ELISA of polysaccharide extracts
Crude polysaccharide extracts were obtained by resuspending approximately 10 OD equivalents (vol. [mL] = 10/culture OD600) of overnight culture (∼16 h) in 100 μL 0.5M EDTA and boiling 5 min. at 100°C (Parise et al., 2007). The supernatant was collected and treated with proteinase K for 60 min. at 60°C (final concentration 0.5 mg/mL), followed by proteinase K inactivation for 30 min. at 80°C. Samples were stored at 4°C for immunoblotting and ELISA. For immunoblotting 1-5 μL of polysaccharide extract was spotted on a nitrocellulose membrane and allowed to air dry. The membrane was blocked with 10% skim milk in TBST (20 mM Tris, 137 mM NaCl, 0.1% Tween 20, pH 7.6) then probed with α-Psl (1:25000 in TBST) for 45 min. at 25°C with agitation. After washing, the secondary horseradish peroxidase-conjugated donkey anti-rabbit whole IgG (1:10000 in TBST, GE Healthcare) was applied and the blot incubated 45 min. at 25°C. The membrane was washed again and detected using SuperSignal West Dura Extended Duration Substrate per manufacturer's instructions (Pierce). Images were recorded using a Kodak Image Station 2000RT System and analyzed with Kodak Molecular Imaging Software.
In order to quantify Psl production an ELISA was modified from an existing protocol (Honko et al., 2006). Flat-bottom 96-well MaxiSorp plates (Nunc) were coated in triplicate overnight at 4°C with 100 μL of a 1:100 dilution of the crude polysaccharide extracts also used for immunoblotting. Plates were washed 3× with wash buffer (PBS + 0.05% Tween 20) and blocked for 2 h at 25°C with 10% newborn calf serum in PBS. Plates were washed 3× and incubated with 100 μL α-Psl (1:25000 in PBS) for 2 h at 25°C. Following five washes, the secondary horseradish peroxidase-conjugated donkey anti-rabbit whole IgG (1:10000 in PBS, GE Healthcare) was applied and the plates incubated for 1 h at 25°C. The plates were washed 7× and 100 μL of 3,3′,5,5′-Tetramethylbenzidine (TMB, Sigma) substrate was applied and incubated 30 min. at 25°C. To stop development 50 μL of a 2N solution of H2SO4 was added and the plates were read at 450 nm. Psl concentration was determined from absorbance values using a Psl standard curve then normalized to the optical density of cells used to make the polysaccharide extract. The Psl standard was the lyophilized extract used in antibody generation resuspended at a known concentration, typically 2 mg/mL, in pyrogen free distilled water.
Microtiter dish attachment assay
Attachment of P. aeruginosa cells to an abiotic surface was evaluated by crystal violet staining essentially as described (Ma et al., 2006). A volume of 100 μL of mid-log culture (OD600 ∼0.5) was added to the wells of a 96-well PVC microtiter plate (BD Falcon) and allowed to attach for 30 min. at 25°C. Nonattached cells were removed and the plate rinsed thoroughly with water. For staining, 100 μL of a 0.1% crystal violet solution was added to each well and the plate incubated for 30 min. at 25°C. After washing as above, the crystal violet bound to cells was solubilized in 200 μL of 95% ethanol for 30 min. at 25°C, then 100 μL of the solution was transferred to a new polystyrene microtiter plate to measure absorbance at 540 nm.
LPS preparation and Western blot analysis
LPS was prepared by the proteinase K method (Hitchcock and Brown, 1983) from P. aeruginosa grown overnight on plates at 37°C. Cells (1 mL OD600 ∼0.5) were collected and treated with proteinase K overnight at 55°C. LPS was separated on a 12.5% SDS-PAGE gel, transferred to a BioTrace172 NT nitrocellulose membrane (Pall) and analyzed by Western immunoblotting as described previously (Rocchetta et al., 1998). Mouse-derived monoclonal antibodies N1F10 (Lam et al., 1989) and MF15-4 (Lam et al., 1987) specific for A-band and B-band LPS, respectively, were used to probe membranes. Blots were developed using goat anti-mouse alkaline phosphatase-conjugated secondary antibody (Jackson ImmunoResearch) and BCIP/NBT (Sigma).
Scanning electron microscopy
Biofilm formation was visualized essentially as described (Parise et al., 2007). Cells grown to mid-log (OD600 ∼0.5) were allowed to attach 4 h at 25°C on vertically oriented 12 mm coverslips partially submerged in culture to create an air-liquid interface. Coverslips were gently rinsed in PBS, fixed with 2.5% glutaraldehyde for 1 h, and routinely processed for SEM. Cells were visualized using a Philips 515 SEM.
Statistical calculations and image processing
Graphing and statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, Inc.). Images were processed using Adobe Photoshop 7.0 (Adobe Systems, Inc.). Figures were assembled using Adobe Illustrator 10.0 (Adobe Systems, Inc.).
Supplementary Material
Acknowledgments
The authors wish to acknowledge Ken Grant and MicroMed for assistance with microscopy, Cynthia Ryder, Jordi Torrelles, John Gunn, and Elizabeth Waligora for scientific and manuscript advice. The galU mutant strain was kindly provided by Bob Hancock and Irith Wiegand of the University of British Columbia. This work was supported by Public Health Service grants AI061396 and HL058334 (D.J.W) and NRSA fellowship AI07870002 (M.S.B) and by an operating grant from the Canadian Cystic Fibrosis Foundation (J.S.L.). J.S.L. holds a Canadian Research Chair in Cystic Fibrosis and Microbial Glycobiology.
References
- Abeyrathne PD, Daniels C, Poon KK, Matewish MJ, Lam JS. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J Bacteriol. 2005;187:3002–3012. doi: 10.1128/JB.187.9.3002-3012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeyrathne PD, Lam JS. WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol Microbiol. 2007;65:1345–1359. doi: 10.1111/j.1365-2958.2007.05875.x. [DOI] [PubMed] [Google Scholar]
- Bernatchez S, Szymanski CM, Ishiyama N, Li J, Jarrell HC, Lau PC, Berghuis AM, Young NM, Wakarchuk WW. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J Biol Chem. 2005;280:4792–4802. doi: 10.1074/jbc.M407767200. [DOI] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Bystrova OV, Knirel YA, Lindner B, Kocharova NA, Kondakova AN, Zahringer U, Pier GB. Structures of the core oligosaccharide and O-units in the R- and SR-type lipopolysaccharides of reference strains of Pseudomonas aeruginosa O-serogroups. FEMS Immunol Med Microbiol. 2006;46:85–99. doi: 10.1111/j.1574-695X.2005.00004.x. [DOI] [PubMed] [Google Scholar]
- Campisano A, Schroeder C, Schemionek M, Overhage J, Rehm BH. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl Environ Microbiol. 2006;72:3066–3068. doi: 10.1128/AEM.72.4.3066-3068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury B, Carlson RW, Goldberg JB. The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohydr Res. 2005;340:2761–2772. doi: 10.1016/j.carres.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohyrdate Research. 1984;131:209–217. [Google Scholar]
- Dong C, Major LL, Srikannathasan V, Errey JC, Giraud MF, Lam JS, Graninger M, Messner P, McNeil MR, Field RA, Whitfield C, Naismith JH. RmlC, a C3′ and C5′ carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation. J Mol Biol. 2007;365:146–159. doi: 10.1016/j.jmb.2006.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- Dubois M. Colorimetric methods for determination of sugars and related substances. Anal Biochem. 1956;28:350–356. [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004a;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004b;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE, Mutharia LM, Chan L, Darveau RP, Speert DP, Pier GB. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BW. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- Honko AN, Sriranganathan N, Lees CJ, Mizel SB. Flagellin is an effective adjuvant for immunization against lethal respiratory challenge with Yersinia pestis. Infect Immun. 2006;74:1113–1120. doi: 10.1128/IAI.74.2.1113-1120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaratne P, Bronner D, MacLachlan PR, Dodgson C, Kido N, Whitfield C. Cloning and analysis of duplicated rfbM and rfbK genes involved in the formation of GDP-mannose in Escherichia coli O9:K30 and participation of rfb genes in the synthesis of the group I K30 capsular polysaccharide. J Bacteriol. 1994;176:3126–3139. doi: 10.1128/jb.176.11.3126-3139.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocharova NA, Knirel YA, Shashkov AS, Kochetkov NK, Pier GB. Structure of an extracellular cross-reactive polysaccharide from Pseudomonas aeruginosa immunotype 4. J Biol Chem. 1988;263:11291–11295. [PubMed] [Google Scholar]
- Komarova BS, Tsvetkov YE, Pier GB, Nifantiev NE. First Synthesis of Pentasaccharide Glycoform I of the Outer Core Region of the Pseudomonas aeruginosa Lipopolysaccharide. J Org Chem. 2008 doi: 10.1021/jo801561p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam JS, MacDonald LA, Lam MY, Duchesne LG, Southam GG. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect Immun. 1987;55:1051–1057. doi: 10.1128/iai.55.5.1051-1057.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MY, McGroarty EJ, Kropinski AM, MacDonald LA, Pedersen SS, Hoiby N, Lam JS. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:962–967. doi: 10.1128/jcm.27.5.962-967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Chang HY, Venkatesan N, Peng HL. Identification of amino acid residues important for the phosphomannose isomerase activity of PslB in Pseudomonas aeruginosa PAO1. FEBS Lett. 2008;582:3479–3483. doi: 10.1016/j.febslet.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak DJ. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007;189:8353–8356. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki M, Jarvinen N, Rabina J, Roos C, Maaheimo H, Renkonen R. Functional expression of Pseudomonas aeruginosa GDP-4-keto-6-deoxy-d-mannose reductase which synthesizes GDP-rhamnose. Eur J Biochem. 2002;269:593–601. doi: 10.1046/j.0014-2956.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parise G, Mishra M, Itoh Y, Romeo T, Deora R. Role of a putative polysaccharide locus in Bordetella biofilm development. J Bacteriol. 2007;189:750–760. doi: 10.1128/JB.00953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Poon KK, Westman EL, Vinogradov E, Jin S, Lam JS. Functional characterization of MigA and WapR: putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J Bacteriol. 2008;190:1857–1865. doi: 10.1128/JB.01546-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology. 2000;146(Pt 11):2803–2814. doi: 10.1099/00221287-146-11-2803. [DOI] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Rivera M, Bryan LE, Hancock RE, McGroarty EJ. Heterogeneity of lipopolysaccharides from Pseudomonas aeruginosa: analysis of lipopolysaccharide chain length. J Bacteriol. 1988;170:512–521. doi: 10.1128/jb.170.2.512-521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M, McGroarty EJ. Analysis of a common-antigen lipopolysaccharide from Pseudomonas aeruginosa. J Bacteriol. 1989;171:2244–2248. doi: 10.1128/jb.171.4.2244-2248.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchetta HL, Lam JS. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchetta HL, Pacan JC, Lam JS. Synthesis of the A-band polysaccharide sugar d-rhamnose requires Rmd and WbpW: identification of multiple AlgA homologues, WbpW and ORF488, in Pseudomonas aeruginosa. Mol Microbiol. 1998;29:1419–1434. doi: 10.1046/j.1365-2958.1998.01024.x. [DOI] [PubMed] [Google Scholar]
- Rocchetta HL, Burrows LL, Lam JS. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovskaya I, Brisson JR, Lam JS, Richards JC, Altman E. Structural elucidation of the lipopolysaccharide core regions of the wild-type strain PAO1 and O-chain-deficient mutant strains AK1401 and AK1012 from Pseudomonas aeruginosa serotype O5. Eur J Biochem. 1998;255:673–684. doi: 10.1046/j.1432-1327.1998.2550673.x. [DOI] [PubMed] [Google Scholar]
- Sadovskaya I, Brisson JR, Thibault P, Richards JC, Lam JS, Altman E. Structural characterization of the outer core and the O-chain linkage region of lipopolysaccharide from Pseudomonas aeruginosa serotype O5. Eur J Biochem. 2000;267:1640–1650. doi: 10.1046/j.1432-1327.2000.01156.x. [DOI] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Li J, Jabbouri S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus epidermidis RP62A, a reference biofilm-positive strain. Carbohydr Res. 2004;339:1467–1473. doi: 10.1016/j.carres.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Samuel G, Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res. 2003;338:2503–2519. doi: 10.1016/j.carres.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Shinabarger D, Berry A, May TB, Rothmel R, Fialho A, Chakrabarty AM. Purification and characterization of phosphomannose isomerase-guanosine diphospho-d-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1991;266:2080–2088. [PubMed] [Google Scholar]
- Soberon-Chavez G, Lepine F, Deziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Toh E, Kurtz HD, Jr, Brun YV. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol. 2008;190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151:985–997. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- Vasseur P, Soscia C, Voulhoux R, Filloux A. PelC is a Pseudomonas aeruginosa outer membrane lipoprotein of the OMA family of proteins involved in exopolysaccharide transport. Biochimie. 2007;89:903–915. doi: 10.1016/j.biochi.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Vinogradov E, Sadovskaya I, Li J, Jabbouri S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus aureus MN8m, a biofilm forming strain. Carbohydr Res. 2006;341:738–743. doi: 10.1016/j.carres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- Wozniak DJ, Ohman DE. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak DJ, Wyckoff TJ, Starkey M, Keyser R, Azadi P, O′Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.