Abstract
Telomerase is a large ribonucleoprotein complex minimally composed of a catalytic telomerase reverse transcriptase (TERT) and an RNA component (TR) that provides the template for telomeric DNA synthesis. However, it remains unclear how TERT and TR assemble into a functional telomerase. Here we report the crystal structure of the conserved regions 4 and 5 (CR4/5) of TR in complex with the TR-binding domain (TRBD) of TERT from the teleost fish Oryzias latipes. The structure shows that CR4/5 adopts an L-shaped three-way-junction conformation with its two arms clamping onto TRBD. Both the sequence and conformation of CR4/5 are required for the interaction. Our structural and mutational analyses suggest that the observed CR4/5-TRBD recognition is common to most eukaryotes, and CR4/5 in vertebrate TR might have a similar role in telomerase regulation as that of stem-loop IV in Tetrahymena TR.
Telomeres, the ends of linear eukaryotic chromosomes, are highly specialized structures and are essential for genome integrity and stability. In most eukaryotes, the length of the telomere is replenished by telomerase, a specialized reverse transcriptase that iteratively adds telomeric repeats at the chromosome ends1. Telomerase is strongly repressed in most normal somatic cells with the exception of several highly proliferative cells such as germline cells and hematopoietic cells2,3. In contrast, telomerase is reactivated in more than 90% of all cancer types, and this accounts for the telomere maintenance and proliferative immortality of cancer cells during tumor progression; thus, inhibition of telomerase may be a promising approach for cancer therapies4.
Telomerase is a ribonucleoprotein complex minimally composed of a catalytic TERT and a TR that provides the template for telomeric DNA synthesis1. TERT is highly conserved across eukaryotes, and it can be divided into four consecutive structural domains: the telomerase essential N-terminus domain (TEN), the TRBD, the reverse transcriptase domain (RT) and the C-terminal extension domain (CTE)5. The crystal structure of the Tribolium castaneum TERT (TcTERT), which lacks the TEN domain, reveals a ring-like organization with the TRBD and CTE domains in close contact6. The interior cavity of the TERT ring could accommodate a synthetic RNA–DNA duplex that resembles the hybrid of the RNA template and the telomeric DNA7. In contrast to TERT, the telomerase RNA from different organisms varies substantially in both length and sequence8. However, several conserved secondary structures of vertebrate TRs, which include the template-pseudoknot (T-PK) structure, CR4/5, box H/AC and conserved region 7 (CR7), have been identified on the basis of phylogenetic comparative analyses8,9. The T-PK and CR4/5 domains bind to TERT independently and are sufficient to reconstitute telomerase activity in trans with TERT, whereas the box H/ACA and CR7 domains are essential for the biogenesis and stability of TR10–12. Mutations in human telomerase RNA may cause several inherited disorders, such as autosomal dominant dyskeratosis congenita and aplastic anemia13,14.
The CR4/5 domain of vertebrate TRs specifically binds to the TRBD domain of TERT and is essential to the assembly and the enzymatic activity of telomerase10,15. The CR4/5 RNA comprises three conserved base-paired regions—stems P5, P6 and P6.1—and potentially forms a three-way-junction (TWJ) structure8,16. The stem-loop P6.1 is the most conserved element and is critical to the telomerase activity16. In yeast telomerase RNA, a conserved TWJ element similar to the vertebrate CR4/5 domain has also been identified17. In Tetrahymena telomerase, a conserved stem-loop IV (SL4) domain instead of the CR4/5 domain binds to TRBD and is required for telomerase activity18,19.
In this study, we set out to understand the molecular mechanism of how TERT and TR assemble into a functional telomerase. We determined the crystal structure of the TRBD–CR4/5 complex—the first, to our knowledge—from the teleost fish O. latipes (Japanese medaka) and revealed an important TERT-TR recognition interface in telomerase. Our structure together with mutational data suggests that the observed mechanism for the TRBD-CR4/5 recognition is common to most eukaryotes. By using information from related structures, we provide structural evidence that CR4/5 in vertebrate TRs might have a similar role in telomerase catalysis as that of SL4 in Tetrahymena TR. In addition, we have also modeled a minimum telomerase catalytic core including the TRBD, RT and CTE domains of TERT and the CR4/5 and T-PK RNAs, on the basis of our results.
RESULTS
Structure of the telomerase TRBD–CR4/5 complex
In a previous study, we mapped the binding interface between TRBD and CR4/5 of the medaka telomerase through photoreagent-mediated cross-linking analyses and identified three cross-linked pairs between the CR4/5 nucleotides and the TRBD residues (U182-Tyr503, U187-Phe355 and U205-Trp477)20. Isothermal titration calorimetry (ITC) measurements showed that the purified TRBD formed a stable binary complex with CR4/5 with an equilibrium dissociation constant of 0.76 μM (Fig. 1a and Supplementary Fig. 1a). To reveal the mechanism of how CR4/5 is recognized by TRBD, we crystallized the TRBD–CR4/5 complex and determined its structure by single-wavelength anomalous dispersion (SAD) at a resolution of 3 Å (Table 1). The calculated electron density map allowed unambiguous tracing of most of the complex except for a few loop regions in TRBD and loop L6.1 in CR4/5 (Supplementary Fig. 1b). There were two TRBD–CR4/5 complexes in one asymmetric unit, and they exhibited almost identical conformations except for the orientations of helices α4 and α9 of TRBD, owing to the crystal packing (Supplementary Fig. 1c). Helices α4 and α9 of TRBD are not involved in CR4/5 recognition (Fig. 1b and Supplementary Fig. 1c).
Figure 1.
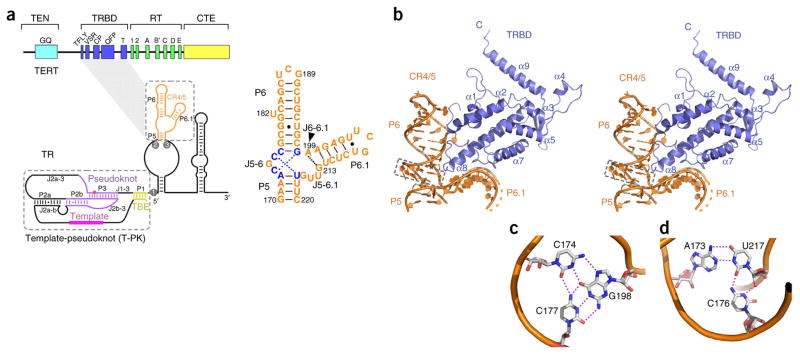
Overview of the CR4/5–TRBD complex structure. (a) Domain organization of the vertebrate telomerase TERT–TR complex. Top, domain organization of TERT. Domains are indicated above brackets and motifs below brackets. Bottom left, predicted secondary structures of TR. Conserved domains and motifs are denoted. The TRBD-CR4/5 interaction is indicated with a connecting gray shadow. Bottom right, secondary structure of CR4/5. (b) Stereo view of the overall structure of the CR4/5–TRBD complex. TRBD and CR4/5 are colored in blue and orange, respectively. The base triples at junction J5-6 are denoted with a dashed box. (c) Base-triple interactions among C174, G198 and C177. Hydrogen bonds are indicated with magenta dotted lines. (d) Base-triple interactions among C176, U217 and A173.
Table 1.
Data collection and refinement statistics
| TRBD–CR4/5 (Native) | TRBD–CR4/5 (Hg-SAD) | |
|---|---|---|
| Data collection | ||
| Space group | P 6222 | P 6222 |
| Cell dimensions | ||
| a, b, c (Å) | 118.1, 118.1, 358.0 | 117.7, 117.7, 354.7 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 3.00 (3.11–3.00)a | 3.30 (3.42–3.30) |
| Rmerge | 0.096 (0.568) | 0.113 (0.800) |
| I / σI | 30.3 (3.9) | 33.3 (3.0) |
| Completeness (%) | 99.9 (100.0) | 94.7 (94.3) |
| Redundancy | 8.2 (8.4) | 13.3 (13.8) |
| Refinement | ||
| Resolution (Å) | 41.97–3.00 | |
| No. of reflections | 30,523 | |
| Rwork / Rfree (%) | 22.1 / 26.1 | |
| No. of atoms | ||
| TRBD | 3,849 | |
| CR4/5 | 2,000 | |
| Sulfate | 60 | |
| Water | 31 | |
| B factors (Å2) | ||
| TRBD | 77.06 | |
| CR4/5 | 148.03 | |
| Sulfate | 101.64 | |
| Water | 66.54 | |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | |
| Bond angles (°) | 1.018 | |
Values in parentheses are for highest-resolution shell. Three crystals were used for the native data set, and one crystal was used for the SAD data set on the mercury derivative (Hg-SAD).
The TRBD–CR4/5 complex exhibits a 1:1 stoichiometry between TRBD and CR4/5. TRBD adopts a mostly helical and globular conformation that closely resembles the recently reported TRBD structure from the Fugu fish Takifugu rubripes21 (Fig. 1b and Supplementary Fig. 1d). The CR4/5-binding site on TRBD partially overlaps with the conserved QFP motif of TRBD and also involves the N-terminal region of helix α2 (Fig. 1b and Supplementary Fig. 2a). Other conserved motifs in TRBD—TFLY, VSR, T and CP—do not participate in CR4/5 binding and presumably have roles in other functions of TERT (Supplementary Fig. 2a). A recent report has linked the TFLY, T and CP motifs to the binding of the template boundary element (TBE) of TR21.
The TRBD–CR4/5 complex structure reveals that CR4/5 forms an L-shaped TWJ structure (Fig. 1a,b). Stems P5 and P6 stack coaxially to form the longer arm of the L-shaped RNA (Fig. 1a,b). There are two base triples stacking at the junction between stems P5 and P6 (Fig. 1b). C174 at the junction J5-6, instead of forming a Watson-Crick base pair with nucleotide G216 as previously predicted8,16, inserts its base into the major groove of stem P6 and pairs with the Hoogsteen face of base pair C177-G198 (Fig. 1c). Similarly, the base of C176 penetrates into the A173-U217 minor groove at the end of stem P5, coordinating two hydrogen-bonding interactions with the base and sugar edge of U217 (Fig. 1d). The resulting base triples, C174•G198-C177 and C176•U217-A173, adopt nearly planar conformations that allow the coaxial and direct stacking of stems P5 and P6 (Fig. 1b). CR4/5 makes a sharp turn at A199, the only nucleotide of junction J6-6.1 (Fig. 1a). Consequently, stem P6.1 branches out from the long axis of stems P5 and P6 by almost 90° to form the short arm of the L-shaped RNA (Fig. 1a,b). The length and sequence of stem P6.1 are invariant in all vertebrate CR4/5 domains and are critical for telomerase activity (Supplementary Fig. 2b)8,16. Consistently with this notion, in the crystal structure of the TRBD–CR4/5 complex the conformation of the four-base-pair stem of medaka P6.1 is almost identical to those of the NMR structures of human wild-type and pseudouridylated stem P6.1 (Supplementary Fig. 3a)22,23. The electron density of loop L6.1 in the TRBD–CR4/5 complex structure is poorly defined, presumably because L6.1 adopts a flexible conformation in the crystal.
The NMR structure of the medaka CR4/5 has recently been reported24. Comparison of the CR4/5 structure in the TRBD–CR4/5 complex with the free CR4/5 NMR structure reveals that TRBD binding results in a dramatic conformational change in CR4/5. In the free CR4/5, stems P5, P6 and P6.1 are arranged into a Y-shaped conformation, in which P6 and P6.1 are aligned closely to each other, with an interhelical angle of ~35° (Fig. 2a)24. However, when CR4/5 is in complex with TRBD, stem P6.1 rotates away from stem P6 over 180° around the junction region, thus allowing the clamping of P6 and P6.1 on TRBD (Fig. 2a). The global conformational change of CR4/5 is also accompanied by an extensive rearrangement of the base-pairings within and around the junctions J5-6, J6-6.1 and J5-6.1, including the formation of the base triples C174•G198-C177 and C176•U217-A173 (Fig. 2b).
Figure 2.
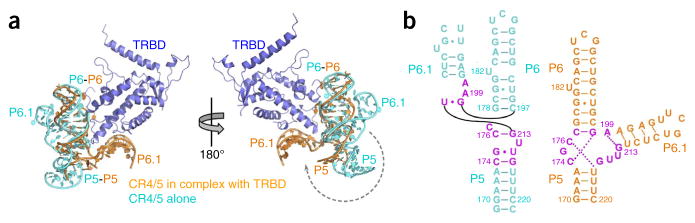
Comparison of CR4/5 conformations in free form and in the CR4/5–TRBD complex. (a) Superposition of the crystal structure of TRBD–CR4/5 with the NMR structure of the medaka CR4/5 (PDB 2MHI24), on the basis of stem P6 of CR4/5 in the two structures. The superposed structures are shown in two opposite views. TRBD is colored in blue, the free CR4/5 in cyan and the CR4/5 in the complex in orange. (b) Secondary structures of the free (cyan) and complexed CR4/5 (orange). The nucleotides that are differently base-paired in the two structures are colored in magenta.
The protein-RNA interaction
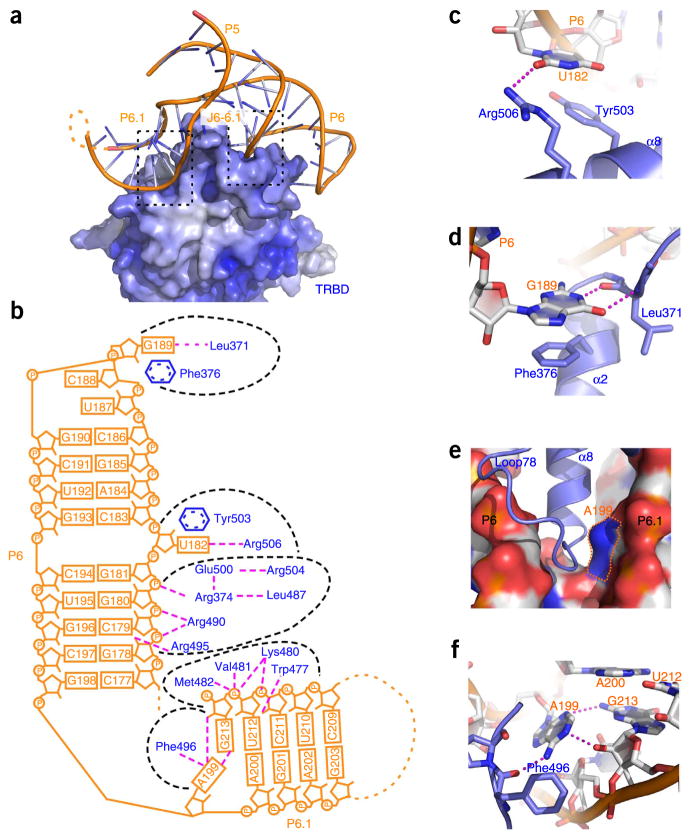
A detailed analysis of the contact between CR4/5 and TRBD suggests that the specific recognition of the RNA by TRBD relies on both the conformation and the sequence of CR4/5. Stems P6 and P6.1 clamp onto a triangular protrusion of TRBD, causing the burial of ~1,040 Å2 of surface area at the interface (Figs. 1b and 3a). On the P6 arm of CR4/5, four continuous nucleotides, G178, C179, G180 and G181, are embedded in a concave groove formed by loop L78 (the loop between helices α7 and α8) and helices α8 and α2 of TRBD (Fig. 3a and Supplementary Fig. 3b). This interface is stabilized by seven hydrogen bonds between the RNA backbone and residues Arg374, Arg490 and Arg495 of TRBD (Fig. 3b and Supplementary Fig. 3b). Similarly, on the other arm of CR4/5 the backbone phosphate groups of C211, U212 and G213 in stem P6.1 adhere to a curved surface of TRBD via five hydrogen-bonding interactions with the side chain of Lys480 as well as the main chain amino groups of Trp477, Val481 and Met482 of TRBD (Fig. 3b and Supplementary Fig. 3c). These interactions involve only the backbone of CR4/5 and presumably mainly contribute to the binding affinity with TRBD. Notably, TRBD has no direct contact with stem P5 and junctions J5-6 and J5-6.1 of CR4/5, in accordance with those regions of CR4/5 being dispensable for TRBD binding20.
Figure 3.
Structural and mutational analysis of the CR4/5-TRBD interaction. (a) RNA backbone–mediated interactions between CR4/5 and TRBD. TRBD is shown as electrostatic-potential surface. The two black dashed boxes denote the regions where the backbone of CR4/5 contacts the TRBD. The detailed interactions are shown in Supplementary Figure 3b,c. (b) Schematic diagram of the CR4/5-TRBD interactions. Stems P6 and P6.1 of CR4/5 are shown in orange, and their interacting TRBD residues are shown in blue. Separate CR4/5-TRBD contact patches are delineated with black dashed lines. Hydrogen bonds are shown with magenta dashed lines. (c) Detailed interactions mediated by the bulge U182 in stem P6 of CR4/5. (d) Detailed interactions of the nucleotide G189 with its surrounding TRBD residues. (e) Outline of the bottom of junction J6-6.1. TRBD is in cartoon representation, and CR4/5 is in surface representation. The base of A199 is denoted with dotted lines. (f) Hydrogen-bonding interactions between nucleotides A199 and G213 as well as the adjacent carbonyl group of TRBD residue Phe496.
The specific recognition of CR4/5 by TRBD relies on three nucleotides: U182, G189 and A199. In the middle of stem P6, the base of nucleotide U182 flips out of the stem and forms a π-stacking interaction with the aromatic side chain of TRBD Tyr503 (Fig. 3b,c). This is consistent with our previous data indicating that the 5-iodouridine–labeled U182 cross-linked efficiently to Tyr503 (ref. 20). Similarly, G189 in the loop region of stem P6 packs against the phenol ring of Phe376 from helix α2 of TRBD (Fig. 3b,d). Removal of these interactions by alanine substitution of Tyr503 and Phe376 either greatly weakened or completely disrupted the TRBD-CR4/5 interaction (Table 2 and Supplementary Fig. 3d). In addition to the stacking contacts, the bases of U182 and G189 are also specifically recognized by multiple hydrogen bonds with the peripheral TRBD residues Arg506 and Leu371, respectively (Fig. 3c,d). These electrostatic interactions provide a strong sequence constraint to CR4/5 because replacement of U182 and G189 with any other nucleotide abolished the association between CR4/5 and TRBD (Table 2 and Supplementary Fig. 3d).
Table 2.
ITC data of the wild-type and mutant medaka CR4/5-TRBD interactionsa
| TRBD | CR4/5 | Kd (μM) |
|---|---|---|
| WT | WT | 0.76 ± 0.21 |
| Y503A | WT | 10 ± 2.2 |
| WT | U182A | >1,000 |
| WT | U182C | >1,000 |
| WT | U182G | >1,000 |
| F376A | WT | >1,000 |
| WT | G189C | >1,000 |
| WT | G189U | >1,000 |
| WT | G189A | 175 ± 47 |
| WT | A199G | >1,000 |
| WT | A199U | >1,000 |
| WT | A199C | >1,000 |
Additional data in Supplementary Figure 3d. WT, wild type.
A199 is the single nucleotide that connects stems P6 and P6.1. This junction nucleotide anchors TRBD into the cleft between the two arms of the L-shaped CR4/5 (Fig. 3e). The edge of the base of A199 is surrounded with three hydrogen-bonding interactions, one with the carbonyl group of TRBD Phe496 and two with nucleotide G213 (Fig. 3f). The latter two allow A199 and G213 to form an unusual base pair that stabilizes the bent backbone of the RNA and makes a sharp turn between stems P6 and P6.1 (Fig. 3f). The conformation of A199 shapes the bottom outline of junction J6-6.1, which snugly accommodates the complementary surface of the N terminus of helix α8 of TRBD (Fig. 3e). Consistently with this observation, substitution of A199 with any other nucleotide (C, U or G) led to a complete loss of the TRBD-CR4/5 interaction (Table 2 and Supplementary Fig. 3d).
Close inspection of the NMR structure of the free CR4/5 reveals that in the absence of TRBD, the TRBD-binding nucleotides on stem P6 are accessible to their complementary surface on TRBD, whereas those on stem P6.1 are obscured from TRBD binding by the adjacent stem P5 (Supplementary Fig. 4a). We speculate that TRBD might initially bind to stem P6 and then trigger the conformational transition of CR4/5 to favor the attachment of P6.1 to TRBD.
A common TRBD-CR4/5 recognition mechanism across species
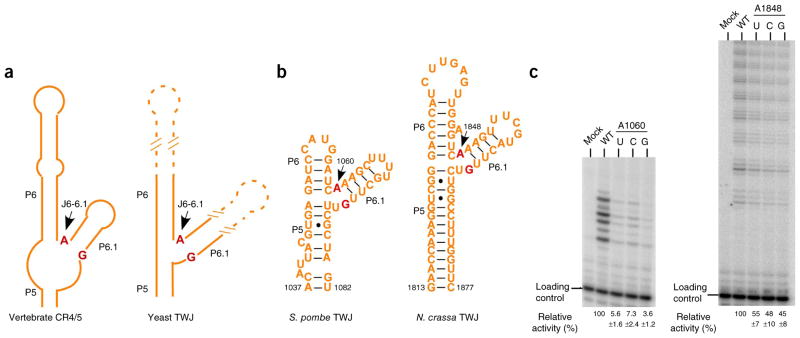
Structure-based sequence analysis of all vertebrate CR4/5 and yeast TWJ domains clearly shows that junction J6-6.1 invariably contains only a single nucleotide, A (A199 in medaka CR4/5) (Fig. 4a and Supplementary Fig. 2b). Furthermore, the G residue at the 5′ terminus of junction J5-6.1 (G213 in medaka CR4/5) is also highly conserved across eukaryotes (Fig. 4a and Supplementary Fig. 2b). This observation suggests that formation of an unusual A•G pair at the junction between stems P6 and P6.1 is a conserved feature of all CR4/5 and TWJ of TRs and that the TRBD-CR4/5 recognition mechanism revealed in the medaka crystal structure is very likely to be common to all species whose TR contains a CR4/5 or TWJ structure. To test this hypothesis, we examined the interaction between TRBD and TWJ from the telomerase of fission yeast Schizosaccharomyces pombe (Fig. 4b). Wild-type S. pombe TRBD bound to S. pombe TWJ with an affinity of 0.57 μM, a value comparable to that of the medaka TRBD-CR4/5 interaction (Table 3 and Supplementary Fig. 4b). In contrast, substitution of the junction nucleotide A1060 of S. pombe TWJ with any other nucleotide either severely impaired its binding to S. pombe TRBD or disrupted the folding of S. pombe TWJ (Table 3 and Supplementary Fig. 4b). In addition, we reconstituted S. pombe telomerase from in vitro–generated TERT protein and TR fragments (T-PK and TWJ) and examined telomerase activities with a direct primer-extension assay. We found that the mutations of A1060 also caused greatly impaired in vitro telomerase activities, thus suggesting that the junction A nucleotide is important for the proper assembly of an active yeast telomerase ribonucleoprotein (RNP) complex (Fig. 4c and Supplementary Fig. 4c). In the telomerase of the filamentous ascomycete Neurospora crassa, substitution of the junction nucleotide A1848 with any other nucleotide also led to decreased telomerase activity in vitro, although to a lesser extent compared to the decrease in activity of the S. pombe telomerase after similar nucleotide substitution (Fig. 4b,c and Supplementary Fig. 4c). Notably, an A-to-G mutation of the junction nucleotide A1130 in TR from the budding yeast Kluyveromyces lactis resulted in growth defects and shortened telomere length17, thus indicating that this linker nucleotide is also crucial for telomere function in cells.
Figure 4.
Phylogenetic and functional analysis of the invariant junction J6-6.1 nucleotide A. (a) Schematic diagram of the secondary structures of vertebrate CR4/5 and yeast TWJ. The structure-based sequence alignment is shown in Supplementary Figure 2b. The invariant nucleotide A at the junction J6-6.1 and its pairwise nucleotide G are colored in red. (b) Secondary structures of S. pombe and N. crassa TWJ domains. (c) Telomerase primer-extension assays of wild-type and mutant S. pombe and N. crassa telomerases (additional data in Supplementary Fig. 4c). Activities of mutant telomerases (A1060U, A1060C and A1060G substitutions for S. pombe and A1848U, A1848C and A1848G substitutions for N. crassa) are normalized to the wild type (WT) and shown below the gel as relative activities ± s.d. from three independent assays.
Table 3.
ITC data of the wild-type and mutant TWJ-TRBD interactions in S. pombe telomerasea
| TRBD | TWJ | Kd (μM) |
|---|---|---|
| WT | WT | 0.57 ± 0.07 |
| WT | A1060G | NAb |
| WT | A1060U | >1,000 |
| WT | A1060C | 39 ± 14 |
Additional data in Supplementary Figure 4b.
NA, not available for ITC measurement because the mutant RNA was misfolded.
Stem P6 varies substantially in both length and sequence in different species (Supplementary Fig. 2b)8,25. Nucleotides U182 and G189, which mediate the specific interaction between medaka CR4/5 and TRBD, are conserved between only closely related species, such as the fugu fish Takifugu rubripe (Supplementary Fig. 2b)25,26. Consistently with this, we found that fugu CR4/5 could efficiently bind to the medaka TRBD domain (Supplementary Fig. 4d). This is also consistent with our previous observation that fugu T-PK and CR4/5 can reconstitute telomerase activity with medaka TERT25. In contrast, S. pombe TWJ, which contains a much shorter stem P6 with no bulged nucleotide, could not bind to medaka TRBD (Fig. 4b and Supplementary Fig. 3d), thus suggesting that S. pombe TRBD must recognize TWJ through some features that are different from U182 and G189 of medaka CR4/5. Collectively, our structural and biochemical analyses suggest that CR4/5 (TWJ) is bound by TRBD through two types of interactions. The helical conformation of stems P6 and P6.1 and the conserved A nucleotide at the junction J6-6.1 define the overall shape of CR4/5 (TWJ) essential for the TRBD-CR4/5 binding in all vertebrates and yeasts. The species incompatibility of the TRBD-CR4/5 (TWJ) interaction presumably results from the coevolution of the protein and RNA components of telomerase along different eukaryotic lineages.
Implications for the functions of stem-loop P6.1
Stem-loop P6.1, the most conserved element in CR4/5, is essential for the catalytic activity of telomerase8,16. Overlay of the medaka CR4/5–TRBD complex structure with the structure of TcTERT in complex with an RNA–DNA hairpin generates a homology model of the CR4/5–TcTERT complex (Fig. 5a). TcTERT lacks the N-terminal TEN domain, which is conserved in other species6. For simplicity, hereafter we will refer to the TEN-less TcTERT as TERT. The homology model of the CR4/5–TERT complex reveals that P6.1 is not in the vicinity of the catalytic center of TERT7 (Fig. 5a), thus suggesting that P6.1 might allosterically participate in the catalytic process20. Stem-loop P6.1 extends to the surface of the CTE of TERT, and nucleotides G203 and U204 spatially collide with the loop region N terminal to helix α18 as well as the loop between helixes α21 and α22 of TERT (Fig. 5b). This implies that the CTE of TERT and P6.1 of CR4/5 might undergo local conformational rearrangement when TERT and TR assemble into a functional telomerase RNP complex, although further investigations are required to test this hypothesis.
Figure 5.
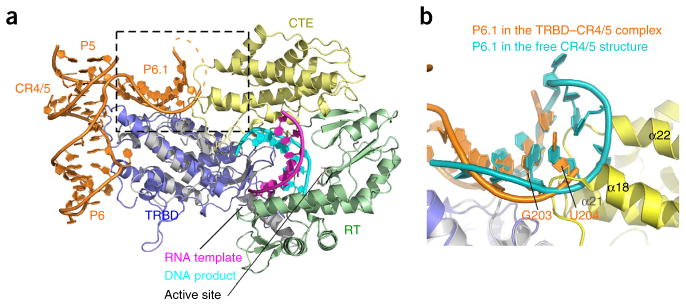
Homology model of the CR4/5–TERT complex. (a) Structural superposition of the CR4/5–TRBD crystal structure with the T. castaneum TERT–DNA–RNA complex structure (PDB 3KYL7). The missing L6.1 region in the TRBD–CR4/5 complex is shown as dotted lines. The spatial collision between P6.1 and the CTE domain of TERT is denoted with a dashed box. (b) Enlargement of the boxed region in a. Stem P6.1 (cyan) in the NMR structure of the medaka CR4/5 is superposed on the stem P6.1 (orange) of the crystal structure of the medaka TRBD–CR4/5 complex.
Tetrahymena telomerase RNA lacks the CR4/5 or TWJ domains but instead has a unique SL4 region that is required for telomerase activity18,19. The low-resolution architecture of the Tetrahymena telomerase holoenzyme, which was reported recently, has indicated that the distal SL4 region might be flanked by the CTE and TRBD domains of TERT27. Comparison of the medaka CR4/5–TRBD structure with the proposed model of the Tetrahymena telomerase holoenzyme reveals that both P6.1 of CR4/5 and distal SL4 of Tetrahymena TR point to the same interface between TRBD and CTE of their respective TERT proteins (Supplementary Fig. 5a), thus suggesting that both loops might have similar structural roles in telomerase RNP assembly.
DISCUSSION
The crystal structure of the CR4/5–TRBD complex provides clues for how the T-PK domain, the other essential element of TR, might interact with TERT. Vertebrate T-PK consists of the template, the TBE (stem P1 in vertebrate TR) and an arc-shaped helical region that includes the triple-helical PK structure28–30 (Fig. 1a). The model of the CR4/5–TERT complex reveals a large shallow concaved surface on one side of TERT that probably accommodates the arc-shaped PK (Supplementary Fig. 5b). This arrangement of PK could bring its conserved triple-helix region close to the template-primer in agreement with previous findings that the triple helix contributes to telomerase activity, and its nucleotide A131 (A176 in human TR) participates in telomerase catalysis31 (Supplementary Fig. 5b). In addition, Qiao et al.32 previously reported that an engineered human cis-telomerase that linked the 3′ end of the telomerase PK region to the 5′ end of the DNA primer with a five-nucleotide linker could generate a near-optimal telomerase activity, thus indicating that the distance between the 3′ end of PK and the 5′ end of DNA primer is ~33 Å. In keeping with this observation, the orientation of PK in our proposed model could allow the 3′ end of PK to be adjacent to the 5′ end of the DNA (Supplementary Fig. 5b). In this model, the TBE-proximal end of the PK is immediately adjacent to the T-CP motif of TRBD so that the TBE stem P1, which is only a few nucleotides 3′ to the PK, could directly contact the T-CP motif, as suggested by a recent study21 (Supplementary Fig. 5b). In addition, this architecture also allows the 5′ end of the template to be connected back to stem P1 through a groove between the RT and TRBD domains of TERT7 (Supplementary Fig. 5b). Future studies are required to determine the precise location of the T-PK domain of TR in the telomerase RNP complex.
In summary, we report the crystal structure of the TRBD domain of TERT in complex with the CR4/5 domain of TR. The CR4/5–TRBD structure offers a starting point for further investigation into the structure and function of the telomerase RNP complex. Moreover, because telomerase is strongly upregulated in most cancer cells and has been studied as a plausible anticancer target, the crystal structure of the CR4/5–TRBD complex offers a new opportunity for the development of therapeutics that modulate telomerase activity for the treatment of human diseases.
ONLINE METHODS
Protein expression and purification
Medaka TRBD (residues 318–579) was expressed in Escherichia coli strain ScarabXpress T7lac cells (Scarab Genomics) with a modified pMAL-2CX vector with an MBP protein fused at its N terminus. After induction for 16 h with 0.1 mM IPTG at 20 °C, the cells were harvested by centrifugation, and the pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), 1 mM PMSF, 5 mM benzamidine, 1 μg/ml leupeptin and 1 μg/ml pepstatin). The cells were then lysed by sonication, and the debris was removed by ultracentrifugation. The supernatant was subjected to 55% ammonium sulfate precipitation. The precipitated proteins were resuspended in lysis buffer and were mixed with amylose resin (New England BioLabs) for 2 h, and then the MBP-TRBD proteins were eluted with 10 mM maltose. After purification by gel-filtration chromatography on HiLoad Superdex200 (GE Healthcare), the MBP-TRBD proteins were treated with protease 3C at 4 °C overnight and were then rebound to the amylose resin to remove the MBP proteins. After a final step of gel-filtration chromatography on HiLoad Superdex200, the purified proteins were concentrated to 10 mg/ml and stored at −80 °C. Wild-type and mutant TRBD proteins used for ITC measurements were purified similarly.
RNA preparation and purification
CR4/5 RNAs (nucleotides 170–220), with a glucosamine-6-phosphate–activated ribozyme (GlmS ribozyme) fused at the 3′ terminus, were generated by in vitro transcription with T7 RNA polymerase. CR4/5-GlmS RNAs were purified by gel-filtration chromatography with the buffer 50 mM HEPES-KOH, pH 7.5, 150 mM NaCl and 10 mM MgCl2 to remove the DNA template and NTPs. The purified RNAs were then treated with 1 mM glucosamine-6-phosphate at 25 °C for 30 min to cleave the GlmS ribozyme from the 3′ site of CR4/5. Then the CR4/5 RNAs were purified by gel-filtration chromatography on HiLoad Superdex200 and were concentrated to 10 mg/ml.
Crystallization, data collection and structure determination
The TRBD proteins were mixed with the CR4/5 RNAs at a molar ratio of 1:1.2. After 1 h incubation on ice, the CR4/5–TRBD complex was purified by gel-filtration chromatography and then concentrated to 12 mg/ml. The TRBD–CR4/5 complex was crystallized by the sitting-drop vapor diffusion method at 4 °C. The precipitant solution contained 100 mM sodium acetate, pH 4.5, 0.9–1 M ammonium sulfate and 0.2–0.3 M NaCl. The TRBD–CR4/5 crystals were then dehydrated with the solution containing 100 mM sodium acetate, pH 4.5, 2 M ammonium sulfate and 0.3 M NaCl. For preparation of a mercury derivative, the dehydrated crystals were soaked in the dehydration buffer with 0.1 mM HgBr2 for 1 h. All of the crystals were gradually transferred into a harvesting solution containing the precipitants plus 5 M sodium formate before being flash frozen in liquid nitrogen for storage. Diffraction data were collected under cryogenic conditions (100 K) at Advanced Photon Source beamline 21-ID-D and processed with HKL2000 (ref. 33). SAD data from the mercury derivative were used to obtain the initial phases with SHARP34. The initial SAD map was substantially improved by solvent flattening. A model of the TRBD–CR4/5 complex was built into the density with Coot35 and refined in Phenix36. Native data sets of the TRBD–CR4/5 complex were solved by molecular replacement with Phaser37 in the CCP4i suite, and the structure was further refined in Phenix. The majority (~92%) of the residues lie in the most favored region in the Ramachandran plot, and the remaining ones lie in the additionally stereochemically allowed regions in the Ramachandran plot.
Isothermal titration calorimetry
The equilibrium dissociation constants of the wild-type and mutant TRBD-CR4/5 and TRBD-TWJ interactions were determined with an ITC200 calorimeter (MicroCal). The binding enthalpies were measured at 20 °C in 50 mM Tris-HCl, pH 7.5, 500 mM NaCl and 1 mM MgCl2. Two independent experiments were performed for every interaction described here. ITC data were subsequently analyzed and fitted with Origin 7 (OriginLab).
Telomerase in vitro reconstitution and telomerase activity assay
The S. pombe and N. crassa telomerases were in vitro reconstituted as previously described38. The TERT protein was synthesized in rabbit reticulocyte lysate (RRL) with the TnT Quick Coupled Transcription/Translation System (Promega) and then assembled with 1 μM of in vitro–transcribed RNA fragments in the RRL. S. pombe TERT was assembled with S. pombe TR fragments T-PK (nt 83–957) and TWJ (nt1037–1082), whereas N. crassa TERT was assembled with N. crassa TR RNA fragments T-PK (nt 225–1515, Δ463–1288, Δ256–433) and TWJ (nt 1813–1877). The telomerase assay was carried out with 2 μl of in vitro–reconstituted telomerase in a 10-μl reaction. For S. pombe telomerase assays, the reaction contained reaction buffer (50 mM Tris-HCl, pH 8.0, 0.5 mM MgCl2, 1 mM spermidine and 2 mM DTT) and 100 μM dATP, 100 μM dCTP, 100 μM dTTP, 5 μM dGTP, 0.165 μM [α-32P]dGTP (3,000 Ci/mmol, 10 mCi/ml, PerkinElmer) and 1 μM DNA primer 5′-GTTACGGTTACAGGTTACG-3′. For N. crassa telomerase assays, the reaction contained 1× reaction buffer, 1 mM dATP, 1 mM dTTP, 5 μM dGTP, 0.165 μM [α-32P]dGTP (3,000 Ci/mmol, 10 mCi/ml, PerkinElmer) and 1 μM DNA primer 5′-(TTAGGG)3-3′. The reactions were incubated at 30 °C for 60 min and terminated by phenol/chloroform extraction and subsequent ethanol precipitation. Telomerase-extended products were resolved on a denaturing 8 M urea/10% polyacrylamide gel. The dried gel was exposed to a phosphor storage screen and analyzed with a Bio-Rad FX Pro Molecular Imager. Original images of autoradiographs can be found in Supplementary Figure 4c.
Supplementary Material
Acknowledgments
We thank Y. Li (University of Michigan) and K. Wan (University of Michigan) for technical support and F. Guo (University of California, Los Angeles) for suggestions. M.L. is supported as a Howard Hughes Medical Institute Early Career Scientist. This work was supported by grants from the Ministry of Science and Technology of China (2013CB910400 to M.L.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010201 to M.L.) and the US National Institutes of Health (RO1GM094450 to J.J.-L.C.).
Footnotes
Accession codes. Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 4O26.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.H. carried out the bulk of the experiments; C.J.B. and D.P.R. established constructs and protocols for expression and purification of the soluble telomerase monomeric protein–RNA core complex; A.F.B. performed the telomerase assays; J.W., L.W. and R.Z. determined the CR4/5–TRBD structure; J.X. purified the S. pombe TRBD protein; J.J.-L.C. and M.L. designed the project and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 2.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 3.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 4.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 5.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 6.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 8.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tesmer VM, et al. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol. 1999;19:6207–6216. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vulliamy T, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 14.Vulliamy T, Marrone A, Dokal I, Mason PJ. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359:2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 15.Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4–CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown Y, et al. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007;35:6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason DX, Goneska E, Greider CW. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol. 2003;23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards RJ, et al. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA. 2006;12:1475–1485. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bley CJ, et al. RNA-protein binding interface in the telomerase ribonucleoprotein. Proc Natl Acad Sci USA. 2011;108:20333–20338. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure. 2013;21:1870–1878. doi: 10.1016/j.str.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim NK, Theimer CA, Mitchell JR, Collins K, Feigon J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res. 2010;38:6746–6756. doi: 10.1093/nar/gkq525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leeper T, Leulliot N, Varani G. The solution structure of an essential stem-loop of human telomerase RNA. Nucleic Acids Res. 2003;31:2614–2621. doi: 10.1093/nar/gkg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim NK, Zhang Q, Feigon J. Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function. Nucleic Acids Res. 2013;42:3395–3408. doi: 10.1093/nar/gkt1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie M, et al. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem. 2008;283:2049–2059. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- 26.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J, et al. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci USA. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cash DD, et al. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci USA. 2013;110:10970–10975. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao F, Goodrich KJ, Cech TR. Engineering cis-telomerase RNAs that add telomeric repeats to themselves. Proc Natl Acad Sci USA. 2010;107:4914–4918. doi: 10.1073/pnas.0909366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.De La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi X, et al. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res. 2013;41:450–462. doi: 10.1093/nar/gks980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.