Abstract
Background and Purpose
Chronic pain is a serious worldwide health issue, with current analgesics having limited efficacy and dose-limiting side effects. Humans with loss-of-function mutations in the voltage-gated sodium channel NaV1.7 (hNaV1.7) are indifferent to pain, making hNaV1.7 a promising target for analgesic development. Since spider venoms are replete with NaV channel modulators, we examined their potential as a source of hNaV1.7 inhibitors.
Experimental Approach
We developed a high-throughput fluorescent-based assay to screen spider venoms against hNaV1.7 and isolate ‘hit’ peptides. To examine the binding site of these peptides, we constructed a panel of chimeric channels in which the S3b-S4 paddle motif from each voltage sensor domain of hNaV1.7 was transplanted into the homotetrameric KV2.1 channel.
Key Results
We screened 205 spider venoms and found that 40% contain at least one inhibitor of hNaV1.7. By deconvoluting ‘hit’ venoms, we discovered seven novel members of the NaSpTx family 1. One of these peptides, Hd1a (peptide μ-TRTX-Hd1a from venom of the spider Haplopelma doriae), inhibited hNaV1.7 with a high level of selectivity over all other subtypes, except hNaV1.1. We showed that Hd1a is a gating modifier that inhibits hNaV1.7 by interacting with the S3b-S4 paddle motif in channel domain II. The structure of Hd1a, determined using heteronuclear NMR, contains an inhibitor cystine knot motif that is likely to confer high levels of chemical, thermal and biological stability.
Conclusion and Implications
Our data indicate that spider venoms are a rich natural source of hNaV1.7 inhibitors that might be useful leads for the development of novel analgesics.
Tables of Links
| TARGETS | ||
|---|---|---|
| Cav channels | Nav1.2 channel | Nav1.6 channel |
| Kv2.1 channel | Nav1.3 channel | Nav1.7 channel |
| Kv11.1 (ERG) channel | Nav1.4 channel | Nav1.8 channel |
| Nav1.1 channel | Nav1.5 channel | Nav1.9 channel |
| LIGANDS |
|---|
| Glutathione |
| Tetrodotoxin (TTX) |
| Veratridine |
These Tables list key protein targets and ligands in this article which are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Pawson et al., 2014) and are permanently archived in the Concise Guide to PHARMACOLOGY 2013/14 (Alexander et al., 2013).
Introduction
Chronic pain is a major health issue worldwide that affects ∼15% of the adult population (Gaskin and Richard, 2012). The annual economic burden of chronic pain in the United States is ∼$600 billion, which exceeds the combined economic cost of cancer, diabetes and stroke (Gaskin and Richard, 2012). There is an unmet clinical need for more effective analgesics to treat chronic pain as most currently available drugs have limited efficacy and dose-limiting side effects.
Voltage-gated sodium (NaV) channels are transmembrane proteins that regulate the electrical properties of cells. There are nine mammalian subtypes denoted NaV1.1–NaV1.9 (Catterall et al., 2005). Several remarkable genetic studies led to the emergence of human NaV1.7 (hNaV1.7) as an analgesic target. Gain-of-function mutations in the SNC9A gene encoding hNaV1.7 cause painful inherited neuropathies (Yang et al., 2004; Fertleman et al., 2006; Estacion et al., 2008; Cheng et al., 2011; Theile et al., 2011), whereas loss-of-function mutations result in congenital indifference to all forms of pain (Cox et al., 2006). Moreover, single nucleotide polymorphisms in SCN9A are associated with differences in pain sensitivity (Reimann et al., 2010; Duan et al., 2013; Reeder et al., 2013). Thus, the combined genetic data suggest that subtype-selective inhibitors of hNaV1.7 are likely to be useful analgesics for treating a broad range of pain conditions (King and Vetter, 2014).
Spider venoms are a rich source of peptidic NaV channel modulators (Herzig et al., 2011; Gilchrist et al., 2014), which are classified into 12 families of NaV channel toxins (NaSpTxs) based upon their primary structure and disulfide framework (Klint et al., 2012). Most NaSpTxs are gating modifiers that perturb channel function by stabilizing one or more voltage sensors in a particular state (Gilchrist et al., 2014). Since the voltage sensors are less conserved than the pore region of NaV channels, these toxins have the potential to selectively modulate particular NaV channel subtypes.
Spiders are the most successful group of venomous animals, with >45 000 extant species. Their venoms contain hundreds to thousands of peptides (Escoubas et al., 2006), with even a conservative estimate of 200 peptides per venom, leading to a total of 9 million spider-venom peptides. Thus far, only ∼0.01% of this vast pharmacological landscape has been explored, providing massive scope for discovery of novel NaV channel modulators from these arachnids. Here, we describe a high-throughput pipeline for the discovery and characterization of hNaV1.7 modulators from spider venoms. We report seven novel peptides from NaSpTx family 1 (NaSpTx-F1) discovered using this approach, and use one of these peptides to exemplify high-throughput methods we developed for determining their structure and NaV channel binding site. We anticipate that development of detailed structure–function relationships for members of NaSpTx-F1, including the seven peptides described here, will facilitate the engineering of selective inhibitors of hNaV1.7 that might be useful analgesics.
Methods
High-throughput screen of spider venoms against hNaV1.7
Mild electrical currents were applied to the chelicerae of spiders to stimulate venom secretion. Venom was collected in polypropylene tubes, lyophilized and stored at −20°C until required. Venoms and venom fractions were assessed against hNaV1.7 endogenously expressed in SH-SY5Y neuroblastoma cells (Vetter et al., 2012). Briefly, SH-SY5Y cells were maintained at 37°C/5% CO2 in RPMI containing 15% FBS and 2 mM L-glutamine and seeded on black-walled 96- or 384-well imaging plates at a density of 30 000 or 150 000 cells per well. After 48 h, cells were loaded for 30 min at 37°C with Calcium 4 No-Wash dye (Molecular Devices, Sunnyvale, CA, USA) diluted in physiological salt solution [composition (in mM) NaCl 140, glucose 11.5, KCl 5.9, MgCl2 1.4, NaH2PO4 1.2, NaHCO3 5, CaCl2 1.8, HEPES 10]. Ca2+ responses following addition of veratridine (50 μM) were measured using a fluorescence imaging plate reader (FLIPRTETRA; Molecular Devices) (excitation 470–495 nm, emission 515–575 nm) after 5 min pre-incubation with venom or venom fractions. Under these conditions, most of the veratridine-induced response is mediated by hNaV1.7, with smaller contributions from hNaV1.2 and hNaV1.3 (Vetter et al., 2012).
Isolation of NaV1.7 inhibitors
Venom (1 mg) was diluted with H2O, filtered (Ultrafree-MC Centrifugal Filter, 0.22 μm; Merck Millipore, Bayswater, Vic, Australia) and then loaded onto an analytical C18 reverse-phase (RP) HPLC column (Vydac 4.6 × 250 mm, 5 μm; Grace, Columbia, MD, USA) attached to a Prominence HPLC system (Shimadzu, Rydalmere, NSW, Australia). Components were eluted at 1 mL·min−1 with solvent A [99.5% H2O, 0.05% trifluoroacetic acid (TFA)] and solvent B (90% CH3CN, 0.05% TFA in H2O) using isocratic elution at 5% solvent B for 5 min, followed by a gradient of 5–20% solvent B over 5 min, then 20–40% solvent B over 40 min and then 40–80% solvent B over 5 min (or minor variations of this gradient). hNaV1.7-active fractions were further fractionated using a polysulfoethyl cation exchange column (4.6 × 100 mm, 3 μm, 300 Å pore size) and eluted at 1 mL·min−1 with solvent A [KH2PO4, 20% (v/v) CH3CN, pH 2.7] and solvent B (1 M KCl in solvent A) using a gradient of 0–50% solvent B over 50 min. Absorbance was measured at 214 and 280 nm using a Shimadzu Prominence SPD-20A detector.
Sequencing of NaV1.7-active peptides
Peptide masses were determined using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS using a Model 4700 Proteomics Analyser (Applied Biosystems, Foster City, CA, USA). HPLC fractions were spotted with α-cyano-4-hydroxycinnamic acid (5 mg·mL−1 in 50% CH3CN). Peptides were reduced and alkylated before being sequenced. Disulfide bonds were reduced by incubating peptides for 15 min at 65oC in 150 mM Tris (pH 8), 1 mM EDTA and 5 mM DTT. Thiol groups were pyridylethylated using 5 μL of 95% 4-vinylpyridine and 20 μL of CH3CN; the reaction was allowed to proceed for 2 h in darkness at room temperature (RT). Both reactions were performed under N2. Samples were desalted using RP-HPLC (C18 column, 2.1 × 100 mm, 5 μm; Thermo Scientific, Waltham, MA, USA) and eluted in solvent B (5% for 15 min, 5–40% over 35 min, 40–80% over 10 min, 0.25 mL·min−1). Reaction progress was monitored using MS. N-terminal sequencing was performed by the Australian Proteome Analysis Facility.
Production of recombinant Hd1a
A synthetic gene encoding Hd1a (peptide μ-TRTX-Hd1a from the venom of the spider Haplopelma doriae), with codons optimized for Escherichia coli expression, was cloned into the pLic-MBP (maltose binding protein) vector (Cabrita et al., 2006). This plasmid, which enables periplasmic production of a His6-MBP-Hd1a fusion protein, was transformed into E. coli strain BL21(λDE3) for Hd1a production. Cultures were grown in Luria-Bertani medium at 37°C with shaking at 180 r.p.m. When the OD600 reached 0.8–1.0, the culture was cooled to 16°C and induced with 1 mM isopropyl b-d-1-thiogalactopyranoside (IPTG). Cells were harvested 12–14 h later by centrifugation for 15 min at 8000× g. For production of uniformly 13C/15N-labelled Hd1a, cultures were grown in minimal medium supplemented with 13C6-glucose and 15NH4Cl as the sole carbon and nitrogen sources respectively. Cells were disrupted under constant pressure at 30 kPa (TS Series Cell Disrupter, Constant Systems, Northants, UK), then the His6-MBP-toxin fusion protein was captured on Ni-NTA Superflow resin (Qiagen, Valencia, CA, USA). The resin was washed with 15 mM imidazole to remove non-specifically bound proteins and then fusion protein was eluted with 500 mM imidazole. After buffer exchange to remove imidazole, reduced and oxidized glutathione were added to 0.6 and 0.4 mM, respectively, to activate TEV protease and promote peptide folding. After the addition of His6-tagged TEV protease (∼40 μg·mg−1 Hd1a), cleavage was allowed to proceed at RT for 12 h. His6-MBP and His6-TEV protease were precipitated with TFA (1%), then Hd1a was purified from the supernatant using RP-HPLC (C4 column; 250 × 10 mm, 5 μm; flow rate 3 mL·min−1; Phenomenex, Torrance, CA, USA) with a gradient of 10–60% solvent B (0.43% TFA in 90% acetonitrile) in solvent A (0.5% TFA in water) over 50 min. If required, a final purification was performed (Aquasil C18 column, 150 × 4.6 mm, 5 μm; flow rate 1 mL·min−1) using a gradient of 25–30% solvent B over 20 min.
Construction of hNaV1.7/KV2.1 chimeras
Channel chimeras were generated using sequential PCR with KV2.1Δ7 (Frech et al., 1989; Swartz and MacKinnon, 1997) and hNaV1.7 (Origene Technologies, Rockville, MD, USA) as templates. The KV2.1Δ7 construct contains seven-point mutations in the outer vestibule that render the channel sensitive to agitoxin-2, a pore-blocking scorpion toxin (Garcia et al., 1994). cRNA was synthesized using T7 polymerase (mMessage mMachine kit; Life Technologies, Federick, MD, USA) after linearizing DNA with appropriate restriction enzymes.
Electrophysiology
Xenopus laevis oocytes were injected with cRNA encoding hNaV α and β1 subunits, KV2.1, KV2.1/hNaV1.7 chimeras, or KV2.1/hNaV1.9 chimeras. Two-electrode voltage-clamp electrophysiology (OC-725C, Warner Instruments, Hamden, CT, USA; 150 μL recording chamber) was used to measure currents 1–4 days after cRNA injection and incubation at 17°C in ND96 that contained (in mM) 96 NaCl, 2 KCl, 5 HEPES, 1 MgCl2, 1.8 CaCl2 and 50 μg·mL−1 gentamycin, pH 7.6. Data were filtered at 4 kHz and digitized at 20 kHz using pClamp software (Molecular Devices). Microelectrode resistances were 0.5–1 MΩ when filled with 3 M KCl. For KV channel experiments, the external recording solution contained (in mM) 50 KCl, 50 NaCl, 5 HEPES, 1 MgCl2, 0.3 CaCl2, pH 7.6 with NaOH. For NaV channel experiments, the external recording solution contained (in mM) 96 NaCl, 2 KCl, 5 HEPES, 1 MgCl2, 1.8 CaCl2, pH 7.6 with NaOH. All experiments were performed at RT (∼22 °C). Toxin samples were diluted in recording solution with 0.1% BSA. Leak and background conductance, identified by blocking channels with agitoxin-2 or tetrodotoxin (TTX), were subtracted for all KV or hNaV1.7 currents respectively.
Voltage–activation relationships were obtained by measuring tail currents for KV channels or by monitoring steady-state currents and calculating conductance for NaV channels. Occupancy of closed or resting channels by toxins was examined using negative holding voltages where the open probability was very low, and the fraction of unbound channels was estimated using depolarizations too weak to open toxin-bound channels. After the addition of toxin to the recording chamber, equilibration between toxin and channel was monitored using weak depolarizations elicited at 5–10 s intervals. For all channels, voltage–activation relationships were recorded in the absence and presence of toxin. Off-line data analysis was performed using Clampfit (Molecular Devices) and Origin 7.5 (Originlab, Northampton, MA, USA).
Determination of Hd1a structure
13C/15N-labelled Hd1a (500 μM in 20 mM sodium acetate buffer containing 5% D2O, pH 4.9) was filtered (Ultrafree-MC 0.22 μm centrifugal filter; Merck Millipore) and then 300 μL was added to a susceptibility-matched microtube (Shigemi, Allison Park, PA, USA). Data were acquired at 25°C on a 900 MHz NMR spectrometer (Bruker Biospin, Ettlingen, Germany) equipped with a cryogenic probe. Spectra used for resonance assignments were acquired using non-uniform sampling (NUS) (Mobli et al., 2010). NUS data were processed using the Rowland NMR toolkit with automatic selection of maximum entropy parameters (Mobli et al., 2007). 13C- and 15N-edited HSQC-NOESY experiments (mixing time 200 ms) were acquired with uniform sampling. Dihedral angles were derived using TALOS+ (Shen et al., 2009); restraint ranges for structure calculations were set to twice the estimated SD. NOESY spectra were manually peak picked and integrated, then peak lists were automatically assigned, distance restraints extracted and structures calculated using CYANA 3.0 (Güntert, 2004).
Results
Screening spider venoms for hNaV1.7 modulators
For this assay, we utilized a human cell line (SH-SY5Y) that expresses high levels of hNaV1.7 (Vetter et al., 2012). Activation of hNaV1.7 with veratridine leads to membrane depolarization, which activates endogenous voltage-gated calcium (CaV) channels (Sousa et al., 2013). The resultant influx of calcium ions induces a fluorescence response when they bind internalized Calcium 4 dye (see control wells 1A–1H and 12A–12H in Figure 1A). Due to the large cellular Ca2+ gradient and the comparatively high sensitivity of calcium dyes, this method yields better signal-to-noise ratio than alternative approaches, including membrane potential or Na+-specific dyes. We utilized a two-addition protocol in which spider venoms (0.4–20.0 μg) and subsequently veratridine were added to the wells of a 96-well plate, and used this assay to screen for hNaV1.7 modulators. Venoms with putative NaV1.7 inhibitors caused a loss of the veratridine-induced fluorescent signal (e.g., wells 2A, 3E, 5F in Figure 1A). In contrast, putative activators were characterized by a spontaneous increase in fluorescence prior to addition of veratridine (e.g., wells 2G and 7B). Some venoms appear to contain both activators and inhibitors of NaV1.7 as they induced a fluorescent signal prior to veratridine addition and also blocked the veratridine-induced response (e.g., well 3G). Since spider venoms are also a rich source of CaV channel modulators (King, 2007; King et al., 2008a), some venoms might contain molecules that abrogate the fluorescent response due block of CaV rather than NaV channels. Modulation of hNaV1.7 by crude venoms could also be due to the synergistic effect of multiple compounds, an effect that might be lost upon venom fractionation.
Figure 1.

High-throughput screen of spider venoms against hNaV1.7. (A) Example of a 96-well plate from a FLIPR-based screen of spider venoms against hNav1.7 endogenously expressed in SH-SY5Y cells. Wells 1A–1H and 12A–12H (pale green) were buffer controls showing the fluorescent response obtained when hNaV1.7 was activated by veratridine. All other wells contained spider venoms. Wells containing venoms that inhibited the veratridine-induced fluorescent response are highlighted in blue, whereas wells containing venoms that elicited a fluorescence response before the addition of veratridine and thus putatively contain activators of hNaV1.7 are highlighted in red. (B) Statistics of venom screen. Eighty-two of the 205 spider venoms inhibited the fluorescent response and therefore putatively contain inhibitors of hNaV1.7, whereas 30 venoms caused a spontaneous increase in fluorescence, suggesting that they contain activators of hNaV1.7. Some venoms caused a spontaneous increase in fluorescence as well as a subsequent reduction in the veratridine-induced fluorescent response (e.g. well G3), and hence they putatively contain both activators and inhibitors of hNaV1.7. For the statistical analysis, these venoms were counted only as inhibitors; hence, the number of activators might be higher than indicated here.
We used this assay to screen venoms from 205 spiders belonging to 13 different taxonomic families (Supporting Information Table S1). Eighty-two venoms (40%) inhibited the fluorescent response and therefore putatively contain at least one inhibitor of hNaV1.7, whereas 30 venoms (14.6%) caused a spontaneous increase in fluorescence and therefore putatively contain at least one activator of hNaV1.7 (Figure 1B). Crude venoms that potently inhibited hNaV1.7 were fractionated using RP-HPLC and then individual fractions were tested in the FLIPR assay. Active fractions were purified to single components (peptides) using an orthogonal ion-exchange chromatography step. These peptides were reduced, alkylated and sequenced using Edman degradation. Using this approach, 15 novel peptides were isolated from eight venoms. Seven peptides were found to be members of NaSpTx-F1 and they will be described here.
Novel members of NaSpTx-F1
As for all previously described NaSpTx-F1 members, the seven new peptides were isolated from theraphosid (tarantula) venoms. Toxins were named based upon the rational nomenclature proposed for spider-venom peptides (King et al., 2008b), with the μ prefix denoting inhibition of NaV channels. Although it is possible that some of these toxins might inhibit CaV rather than NaV channels, this seems unlikely since all NaSpTx-F1 toxins characterized to date exclusively target NaV channels (Klint et al., 2012).
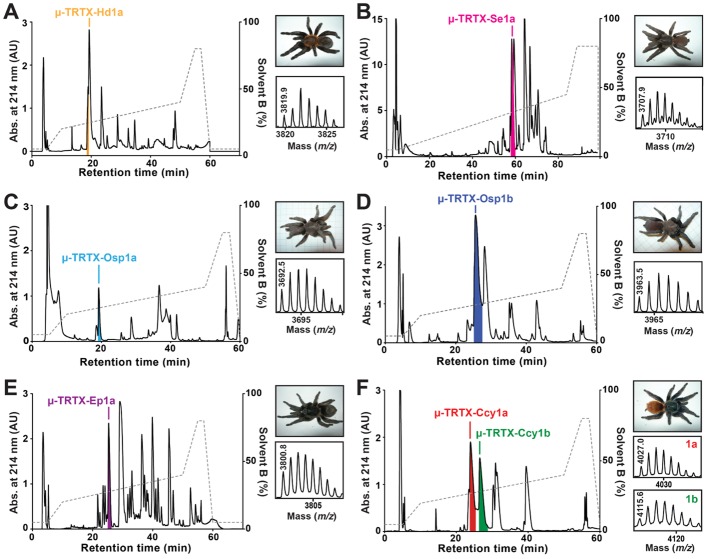
Figure 2 outlines purification of the seven peptides, while their amino acid sequences are provided in Table 1. Each peptide comprises 32–35 residues with six cysteine residues that form three disulfide bonds based upon mass of the reduced and oxidized peptides. We did not experimentally determine their disulfide connectivity since, based upon sequence homology, it is presumably the same as previously characterized NaSpTx-F1 members that all contain an inhibitor cystine knot (ICK) with cysteine connectivity C1–C4, C2–C5, C3–C6 (Pallaghy et al., 1994). As shown in Table 1, there is a good agreement between the molecular mass of the native toxins determined using MALDI-TOF MS and the mass calculated from the amino acid sequence. Thus, none of the peptides are C-terminally amidated or contain any other post-translational modification aside from disulfide bonds. Despite the fact that all seven peptides belong to NaSpTx-F1, their sequence similarity is only moderate (38–62% sequence identity with respect to Hd1a; see Table 1). μ-TRTX-Ccy1a and μ-TRTX-Ccy1b are paralogues isolated from the same venom that differ only at position 6 (Ile6 in Ccy1a, Phe6 in Ccy1b).
Figure 2.
Isolation of novel NaV1.7 inhibitors from spider venom. Each panel shows a chromatogram resulting from fractionation of crude venom using RP-HPLC; the acetonitrile gradient is indicated by the dotted line. Coloured fractions are those that putatively contain inhibitors of NaV1.7. These fractions were further purified using ion exchange chromatography to obtain pure, active peptides. The top inset in each panel shows a photo of the spider from which venom was obtained. The bottom inset in each panel is a MALDI-TOF MS spectrum showing the M + H+ ion for the purified active peptide. These six panels show isolation of (A) μ-TRTX-Hd1a from the venom of Haplopelma doriae; (B) μ-TRTX-Se1a from the venom of Selenocosmia effera; (C) μ-TRTX-Osp1a from the venom of Orphnaecus species 1; (D) μ-TRTX-Osp1b from the venom of Orphnaecus species 2; (E) μ-TRTX-Ep1a from the venom of Euathlus pulcherrimaklaasi; (F) μ-TRTX-Ccy1a and μ-TRTX-Ccy1b from the venom of Chromatopelma cyaneopubescens.
Table 1.
Amino acid sequences of hNaV1.7-active peptides discovered in this study
| Venom | Toxin name | Sequence | Observed mass (Da) | Calculated mass (Da) | Identity (%) |
|---|---|---|---|---|---|
| Haplopelma doriae | μ-TRTX-Hd1a | -ACLGFGKSCNPSNDQCCKSSSLACSTKHKWCKYEL | 3819.9 | 3819.7 | 100 |
| Chromatopelma cyaneopubescens | μ-TRTX-Ccy1a | DDCLGIFKSCNPDNDKCCES--YKCSRRDKWCKYVL | 4027.0 | 4027.8 | 62 |
| Chromatopelma cyaneopubescens | μ-TRTX-Ccy1b | DDCLGFFKSCNPDNDKCCEN--YKCNRRDKWCKYVL | 4115.6 | 4115.8 | 59 |
| Orphnaecus species 11 | μ-TRTX-Osp1a | -GCKGFGKACKYGADECCKN--LVCSKKHKWCKYTL | 3692.5 | 3692.8 | 60 |
| Orphnaecus species 21 | μ-TRTX-Osp1b | -ECLGWMKGCEPKNNKCC--SSYVCTYKYPWCRYDL | 3963.5 | 3963.7 | 56 |
| Euathlus pulcherrimaklaasi | μ-TRTX-Ep1a | -DCLKFGWKCNPRNDKCC--SGLKCGSNHNWCKLHL | 3800.8 | 3800.7 | 50 |
| Selenocosmia effera | μ-TRTX-Se1a | -DCLGWMAGCDFNDNKCC--AGYVCK-KHPWCRYDL | 3707.9 | 3707.5 | 38 |
Masses are monoisotopic masses, either determined experimentally using MALDI-TOF MS or calculated using PeptideMass (http://web.expasy.org/peptide_mass/). % identity was calculated relative to Hd1a.
Orphnaecus species 1 and 2 were collected from Sibaliw and Maanghit Caves, respectively, on Panai Islands in the Philippines. We have tentatively assumed that these are the same species, and the toxins have been named accordingly.
The seven spider-venom peptides are similar to several previously described members of NaSpTx-F1. μ-TRTX-Cc1a/b, μ-TRTX-Se1a and μ-TRTX-Osp1b are most similar to κ-TRTX-Gr4a (82, 55 and 55% identity, respectively) and β-TRTX-Cm1a (76, 52 and 58% identity, respectively). κ-TRTX-Gr4a (VSTX3) binds to the voltage sensor domain of the archaebacterial voltage-gated potassium channel KVAP (Ruta and MacKinnon, 2004) but it has not been tested against NaV channels. β-TRTX-Cm1a (ceratotoxin-1) was isolated based upon its ability to inhibit mammalian NaV channels; it potently inhibits NaV1.1, NaV1.2, NaV1.4 and NaV1.5 with IC50 values ranging from 3 to 890 nM (Bosmans et al., 2006). μ-TRTX-Hd1a (Hd1a), μ-TRTX-Osp1a and μ-TRTX-Ep1a are most similar to μ-TRTX-Hhn1b (hainantoxin-IV, Hhn1b) (82, 62 and 51% identity, respectively), a potent inhibitor of TTX-sensitive NaV currents in dorsal root ganglion (DRG) neurons (Liu et al., 2012).
Recombinant Hd1a preferentially inhibits hNaV1.7
In order to validate the ability of the FLIPR assay to identify inhibitors of hNaV1.7, we undertook detailed structure–function characterization of Hd1a. Hd1a was produced in the periplasm of E. coli to facilitate disulfide bond formation (Klint et al., 2013). The His6-MBP-Hd1a fusion protein, which was the major cellular protein produced after IPTG induction (Figure 3A, inset), was purified using nickel affinity chromatography, then Hd1a was liberated from the fusion protein with TEV protease and purified using RP-HPLC (Figure 3A). Hd1a was purified to >98% homogeneity as judged by RP-HPLC (Figure 3B) and MS (Figure 3C). In order to facilitate TEV protease cleavage of the His6-MBP-Hd1a fusion protein, Hd1a was produced with an additional N-terminal glycine residue, the preferred P1′ residue in the TEV protease recognition site (Kapust et al., 2002; Renicke et al., 2013); all remaining work was completed with this recombinant toxin (rHd1a).
Figure 3.

Expression and purification of recombinant Hd1a. (A) RP-HPLC chromatogram showing purification of recombinant Hd1a after cleavage from the His6-MBP fusion tag by TEV protease. The peak corresponding to Hd1a is highlighted with an asterisk. Inset: SDS-PAGE gel showing Escherichia coli cells before (lane 1) and after (lane 2) induction of Hd1a expression with IPTG. Lane 3 contains molecular mass standards, with masses indicated in kDa on the right of the gel. The arrow indicates the running position of the MBP-toxin fusion protein. (B) RP-HPLC chromatogram of recombinant Hd1a after purification, showing a single uniform peak. (C) MALDI-TOF MS spectrum showing the M + H+ ion for purified recombinant Hd1a (observed, 3877.77 Da; calculated, 3877.71 Da).
Application of 1 μM rHd1a caused almost complete inhibition of hNaV1.7-mediated currents recorded from oocytes (Figure 4A). Fitting the Hill equation to concentration–response data (Figure 4B) yielded an IC50 of 111 ± 7 nM and a Hill coefficient of 0.95 ± 0.06, indicative of a single toxin binding site. Conductance–voltage (G-V) relationships obtained before and after addition of a non-saturating concentration of rHd1a (50 nM) revealed no significant shift in activation (Figure 4C), and inhibition was not voltage-dependent over the range of −25 to 0 mV (Supporting Information Fig. S1A). While this suggests a pore-blocking mechanism of action, additional experiments designed to determine the Hd1a binding site (described below) indicate this is not the case. Hd1a had no effect on steady-state inactivation (Figure 4D) or recovery from inactivation (Figure 4E), and it caused only a minor increase in inactivation time constants (Supporting Information Fig. S2). Inhibition by Hd1a is reversible as hNaV1.7 currents recovered quickly following washout in toxin-free solution (Figure 4F).
Figure 4.
Effect of rHd1a on hNaV1.7/β1. Currents were recorded from Xenopus oocytes using the two-electrode voltage-clamp technique. (A) Sodium currents before (black) and after (red) addition of 1 μM rHd1a. (B) Current–voltage relationship before (black) and after (red) addition of 50 nM rHd1a. Currents were evoked from a holding potential of −90 mV, stepping from −70 to −10 mV in 5 mV increments. (C) I/I0 as a function of rHd1a concentration (n = 3–4; error bars represent SEM). Fitting the Hill equation to the data yielded an IC50 of 111 ± 7 nM and Hill coefficient of 0.95 ± 0.06. (D) Steady-state fast inactivation before (black) and after (red) addition of 1 μM rHd1a. Currents were evoked from a holding potential of −90 mV stepping to −15 mV in 5 mV increments. (E) Recovery from fast inactivation before (black) and after (red) addition of 1 μM rHd1a. Currents were evoked from a holding potential of −90 mV by applying a −15 mV pulse followed by repolarization to−90 mV. A second pulse to −15 mV was applied after a period ranging from 0 to 50 ms, increasing in 1 ms increments. (F) hNaV1.7 currents (I/Imax) following application of 1 μM rHd1a for 5 min followed by washout for 10 min. Depolarizing pulses were applied from −90 to −20 mV every 80 ms. (G) Selectivity of rHd1a. hNaV1 currents (I/I0) in the presence of 1 μM rHd1a (n = 3–4; error bars denote SEM). Orange and green bars denote TTX-sensitive and TTX-resistant hNaV subtypes respectively. All currents were evoked by a depolarization to −20 mV from a holding potential of −90 mV.
The subtype selectivity of rHd1a was assessed by examining its ability to inhibit a complete panel of hNaV channel subtypes expressed in oocytes, including the key off-target subtypes hNaV1.4, hNaV1.5 and hNaV1.6 (Figure 4G). At a concentration of 1 μM, Hd1a was highly selective with no inhibition of hNaV1.5 or hNaV1.8, 23–31% inhibition of hNaV1.3, hNaV1.4 and hNaV1.6, moderate inhibition of hNaV1.2 (55%), and robust inhibition of hNaV1.1 (87%) and hNaV1.7 (87%) (Figure 4D). Moreover, this selectivity pattern was maintained over a large voltage range (from −25 to 0 mV; Supporting Information Fig. S1B). Hd1a was inactive against hERG (Kv11.1 channels; Supporting Information Fig. S3).
Hd1a interacts with domain II voltage sensor of hNaV1.7
The NaV channel binding site has been examined for only one member of NaSpTx-F1, namely huwentoxin-IV (μ-TRTX-Hh2a, Hh2a), which binds to the S3b-S4 helix-turn-helix motif in the domain II voltage sensor (Xiao et al., 2008). This so-called paddle motif (Alabi et al., 2007; Bosmans et al., 2008) is targeted by a variety of animal toxins. In order to determine whether rHd1a also binds to the paddle region, we transplanted the S3b-S4 paddle motif from each of the four voltage-sensor domains of hNaV1.7 into the homotetrameric KV2.1 channel (Figure 5). For domains (D) I, III and IV of hNaV1.7, we generated functional chimeras using previously described paddle-motif boundaries (Bosmans et al., 2008; 2011) (Figure 5B and C). In the case of DII, we could only record ionic currents when four N-terminal residues were not transferred (Figure 5B and C). Nonetheless, the transferred region in all functional chimeras contained the crucial basic residues that contribute to gating charge movement in KV channels (Aggarwal and MacKinnon, 1996; Seoh et al., 1996), suggesting that the four voltage-sensing domains in hNaV1.7 contain paddle motifs that are involved in sensing membrane voltage changes.
Figure 5.

Sensitivity of hNaV1.7/rKV2.1 paddle chimeras to rHd1a. (A) Schematic of the hNaV1.7 channel showing the voltage-sensor paddle motifs (coloured segments) that were transferred from hNaV1.7 to rat KV2.1 (rKV2.1). The paddles from domains I, II, III and IV are coloured purple, red, cyan and green respectively. (B) Sequence alignment of the paddle region of rKV2.1 with the paddle motifs (S3b-S4 region) from each domain of hNaV1.7. For the domain II transplant, only the chimera highlighted with an asterisk was functional. (C) Families of potassium currents for each of the hNaV1.7/rKv2.1 chimeras. Scale bars: 1 μA (ordinate axis) and 50 ms (abscissa). (D) Tail current voltage-activation relationships for each of the hNaV1.7/rKV2.1 chimeras (n = 5; error bars denote SEM). The holding voltage was −90 mV (−120 mV for DIII) and the tail voltage was −60 mV (or −90 mV for DIII). (E) Mean time constants (τ) from single exponential fits to channel activation (filled circles) and deactivation (open circles) plotted as a function of the voltage at which the current was recorded (n = 3–5; error bars denote SEM). (F) Potassium currents elicited by depolarizations to 70 mV. Currents are shown before (black) and in the presence (coloured traces) of 1 μM rHd1a. Colours denote the domain from which the hNaV1.7 paddle motif was transferred: DI paddle (purple); DII paddle (red); DIII paddle (blue); DIV paddle (green). Scale bars: 50 ms (abscissa), 1 μA (DII, DIII) and 2 μA (DI, DIV, KV2.1) (ordinate axis).
Examination of the G-V relationships for the hNaV1.7/KV2.1 chimeras revealed that each voltage-sensor paddle has a distinct effect on KV2.1 gating; the midpoints of the G-V relations are ≥50, 44 ± 1, −16 ± 2 and 9 ± 1 mV for the DI, II, III and IV constructs respectively (Figure 5D). The voltage sensor in DIV plays a crucial role in fast inactivation, possibly due to its slower response to changes in membrane voltage compared with DI–III and its coupling to the inactivation gate (Sheets et al., 1999; Horn et al., 2000; Chanda and Bezanilla, 2002). The DIV paddle motifs within rNaV1.2 and rNaV1.4 delay channel opening when transplanted into KV channels, suggesting that this region helps determine the slower kinetics of DIV voltage-sensor activation (Bosmans et al., 2008). To explore whether the DIV paddle motif serves a similar role in hNaV1.7, we measured the kinetics of activation and deactivation of the four chimeras in response to membrane depolarization and repolarization respectively (Figure 5E). As observed for previously studied chimeric channels, the kinetics for the DIV hNaV1.7/KV2.1 chimera were slower compared with other hNaV1.7/KV2.1 chimeras, suggesting that the paddle motif in domain IV contributes to fast inactivation in hNaV1.7.
We next examined the effect of rHd1a on KV2.1 and the hNaV1.7/KV2.1 paddle chimeras. One micromolar rHd1a had no effect on wild-type KV2.1 or the DI, DIII and DIV chimeras, but it strongly inhibited K+ currents mediated by the DII hNaV1.7/KV2.1 chimera (Figure 5F). We conclude that rHd1a inhibits hNaV1.7 primarily by interacting with the S3b-S4 paddle in DII.
Since hNaV1.9 has proven recalcitrant to heterologous expression, we also examined the effect of rHd1a on a panel of previously described hNaV1.9/KV2.1 paddle chimeras in which the S3b-S4 paddle motifs of hNaV1.9 were transplanted into KV2.1 (Bosmans et al., 2011). rHd1a had no effect on currents mediated by any of these chimeras (Supporting Information Fig. S4). Since the transplanted S3b-S4 paddle region encompasses the binding site for rHd1a on hNaV1.7, we tentatively concluded that rHd1a has no effect on hNaV1.9.
Solution structure of rHd1a
The bacterial expression system described earlier allowed production of uniformly 13C/15N-labelled Hd1a for structure determination using NMR. 1HN, 15N, 13C′, 13Cα, 13Cβ resonance assignments for Hd1a were obtained by analysing amide-proton strips in three-dimensional (3D) HNCACB, CBCA(CO)NH and HNCO spectra. Additional side chain chemical shifts were obtained from 15N/13C-NOESY spectra and a four-dimensional (4D) HCC(CO)NH-TOCSY experiment that provides side chain 1H-13C connectivities (Mobli et al., 2010). An assigned two-dimensional (2D) 1H-15N HSQC spectrum of rHd1a is shown in Supporting Information Fig. S5, and complete chemical shifts are available from BioMagResBank (accession number 19998). CYANA 3.0 was used to automatically assign NOESY spectra and calculate 200 structures from random starting conformations; the 20 structures with best stereochemical quality as judged by MolProbity (Chen et al., 2010) were selected to represent the structure of rHd1a (PDB ID 2MPQ; Figure 6).
Figure 6.

Ensemble of rHd1a structures. Stereoview of the ensemble of 20 rHd1a structures (PDB 2MPQ) overlaid over the backbone atoms of residues 2–35. β strands are shown in blue and the three disulfide bonds are highlighted in yellow. The four inter-cystine loops and the N- and C-termini are labelled.
Statistics highlighting the high precision and stereochemical quality of the ensemble of rHd1a structures are shown in Table 2. The ensemble is highly precise with backbone and heavy-atom root-mean-square deviation values of 0.05 ± 0.01 and 0.41 ± 0.04 Å, respectively, and it ranks as ‘high resolution’ based upon measures of precision and stereochemical quality (Kwan et al., 2011).
Table 2.
Structural statistics for the ensemble of Hd1a structures
| Experimental restraints | |
| Interproton distance restraints | |
| Intra-residue | 178 |
| Sequential | 168 |
| Medium range (i–j < 5) | 142 |
| Long range (i–j ≥ 5) | 214 |
| Disulfide bond restraints | 9 |
| Dihedral angle restraints (ϕ, ψ) | 57 |
| Total number of restraints per residue | 21.3 |
| RMSD to mean coordinate structure (Å) | |
| Backbone atoms | 0.05 ± 0.01 |
| All heavy atoms | 0.41 ± 0.04 |
| Stereochemical quality | |
| Residues in most favoured Ramachandran region (%) | 87.9 ± 0.9 |
| Ramachandran outliers (%) | 0.0 ± 0.0 |
| Unfavourable side chain rotamers (%) | 11.1 ± 2.2 |
| Residues with bad angles | 0.0 ± 0.0 |
| Clashscore, all atoms | 0.0 ± 0.0 |
| Overall MolProbity score (percentile) | 1.89 ± 0.07 (81 ± 3) |
Root-mean-square deviation (RMSD) values were calculated over the well-defined regions of the structures (residues 2–35). Measures of stereochemical quality are from MolProbity (http://helix.research.duhs.duke.edu). Clashscore is the number of steric overlaps >0.4 Å per 103 atoms. All statistics are given as mean ± SD.
Figure 6 shows an overlay of the ensemble of 20 Hd1a structures. As expected, Hd1a contains an ICK motif in which the Cys17–Cys32 disulfide bond pierces a 12-residue ring formed by the other two disulfides (Cys3–Cys18, Cys10–Cys25) and the intervening sections of peptide backbone. Thus, Hd1a contains four inter-cystine loops (numbered 1–4 in Figure 6). The dominant secondary structure is a β hairpin comprising strands β1 (residues 23–26) and β2 (residues 31–34) connected by a short hairpin loop (residues 27–30; loop 4).
Discussion
High-throughput venom screens
In contrast to the diversity of NaV channels in mammals, insects contain a single isoform. Interference with this channel has lethal consequences, making it an ideal target for insect predators such as spiders, scorpions and centipedes (King et al., 2008a). As a consequence, the venoms of these arthropods are rich in NaV channel modulators (Rodriguez De La Vega and Possani, 2005; Gurevitz et al., 2007; King et al., 2008a; Yang et al., 2012; Gilchrist et al., 2014). Since there is no evolutionary selection pressure to prevent these toxins acting on vertebrates, many spider-venom peptides also modulate the activity of mammalian NaV channels (Nicholson and Little, 2005; Escoubas and Bosmans, 2007). However, since the level of sequence identity between insect and human NaV channels is only 55–60% (King et al., 2008a), it is difficult to predict the effect of these peptides on specific hNaV subtypes. We therefore developed a high-throughput approach for screening spider-venom peptides directly against hNaV1.7.
We took advantage of the observation that the neuroblastoma cell line SH-SY5Y expresses high levels of NaV1.7 to the exclusion of most other subtypes (Vetter et al., 2012). This formed the basis of a high-throughput FLIPR assay in which venoms were monitored for their ability to prevent a calcium-dependent fluorescent response induced by activation of the endogenous hNaV1.7 population with veratridine, a non-specific NaV channel agonist. This assay enabled ∼90 venoms to be screened in 96-well plate format; if we conservatively estimate that each venom contains 200 peptides, then this corresponds to a rapid screen of 18 000 peptides. Scaling to 384-well plates would enable rapid screening of >350 venoms (>70 000 peptides) and minimize the amount of venom required.
Using this high-throughput FLIPR assay, we demonstrated that spider venoms are an incredibly rich source of hNaV1.7 modulators with the potential to serve as leads for analgesic development. A remarkable 40% of all venoms screened putatively contain at least one inhibitor of hNaV1.7. Moreover, deconvolution of ‘hits’ revealed that some spider venoms contain more than one hNaV1.7 inhibitor (e.g. see Figure 2F).
High-throughput structure–function studies
Using our hNaV1.7 screen, we discovered seven novel members of NaSpTx-F1 and focused on one of these (Hd1a) in order to develop robust methods for structure–function characterization. rHd1a potently inhibited hNaV1.7 with an IC50 of ∼110 nM, with modest or zero effect on all other subtypes except NaV1.1. Since most spider-venom-derived NaV1.7 modulators are gating modifiers that bind to one or more of the voltage sensors, we developed an approach that allows rapid determination of which voltage-sensor is involved in toxin binding. Competition binding experiments can be difficult to interpret because some spider-venom peptides interact with more than one voltage sensor (Bosmans et al., 2008); therefore, we developed an alternative approach in which a panel of channel chimeras was created by transplanting the voltage-sensor paddles from each of the four domains of hNaV1.7 into rKV2.1 (Figure 5). This enabled us to ask whether transplantation of the hNaV1.7 paddles into rKV2.1 sensitizes this KV channel to the effects of a NaV channel toxin. Using this approach, we demonstrated that rHd1a binds exclusively to the voltage-sensor paddle from hNaV1.7 domain II (Figure 5F). Hd1a joins an extensive list of animal toxins that target this paddle motif to prevent Nav channel opening (Cestele et al., 1998; Catterall et al., 2007; Bosmans et al., 2008; Bosmans and Swartz, 2010; Xiao et al., 2011).
High-throughput screens such as the one described here are increasingly being used in venom-based drug discovery pipelines (Vetter et al., 2011). This results in more lead molecules becoming available very early in the discovery pipeline than in traditional discovery programmes focussed on one or several venoms (Bosmans et al., 2006). Hence, there is a need to rapidly determine the structure of several venom peptides to allow timely development of structure–activity relationships (SAR). Although NMR is the dominant approach for determining the structure of polypeptides <10 kDa (Kwan et al., 2011), it suffers from low-throughput. Thus, we integrated several approaches to speed up venom-peptide structure determination. Firstly, we acquired all scalar-coupling experiments using NUS in order to expedite data acquisition and increase resolution in the indirect frequency dimensions. This enables a complete 3D/4D data set to be acquired in <5 days, and the increased digital resolution makes the spectra highly amenable to automated assignment. Secondly, we used automated NOESY assignment and structure calculation (Güntert, 2004). Together, these approaches enable high-resolution structures of excellent stereochemical quality to be determined in a highly automated fashion in 1 week, as exemplified by the structure of rHd1a reported here (Figure 6).
The structure of rHd1a is similar to that of Hhn1b and Hh2a (Figure 7A). A positively charged patch on the surface of Hhn1b is important for its inhibition of TTX-sensitive NaV currents in DRG neurons, with mutation to alanine of residues K27, R29 and K32 resulting in >100-fold decrease in potency (Liu et al., 2012). These three cationic residues are conserved in Hd1a (corresponding residues are K28, K30 and K33; Figure 7B). Extensive mutagenesis of Hh2a, which is 68% identical to Hd1a, also revealed that these charged residues are important for hNaV1.7 inhibition, with a K32A mutation causing almost complete loss of activity (Minassian et al., 2013; Revell et al., 2013). Hydrophobic interactions with the channel are also important, as a W30A mutation caused complete loss of hNaV1.7 inhibition, whereas F6A and Y33A mutations triggered major reductions in potency (Minassian et al., 2013; Revell et al., 2013). Interestingly, W30 and K32 are two of the few residues that are completely conserved in NaSpTx-F1 (Klint et al., 2012). The mutational data are mapped onto the structure of rHd1a in Figure 7C. The functionally important hydrophobic and cationic residues form adjacent clusters that together create a contiguous surface epitope that likely represents the peptide's pharmacophore for hNaV1.7.
Figure 7.

Structural comparison of rHd1a with characterized NaSpTx-F1 members. (A) Ribbon representation of the 3D structures of Hd1a (green, PDB 2MPQ), μ-TRTX-Hhn1b (cyan, PDB 1NIY) and μ-TRTX-Hh2a (pink, PDB 1MB6). The C-termini are labelled. (B) Alignment of the amino sequences of the three peptides. Red denotes residues that cause a reduction in inhibition of NaV1.7 when mutated to alanine. Sequence numbering is with respect to μ-TRTX-Hh2a and μ-TRTX-Hhn1b. (C) Solvent accessible surface of Hd1a. Hydrophobic and basic residues are coloured green and blue, respectively, and are labelled. This view is related to that in panel (A) by a 180° rotation around the long axis of the molecule.
Most of the residues important for interaction of NaSpTx-F1 peptides with hNaV1.7 are located in the structurally well-conserved C-terminal region and loop 4 (Figure 7A). There are subtle sequence variations in these regions (see Table 1), and detailed SAR studies will be required to unravel the molecular determinants that underlie differences in subtype selectivity within NaSpTx-F1. The recently described tethered-toxin screening approach, which allows alanine scans to be performed without the need to produce and purify individual mutants (Gui et al., 2014), should expedite such studies.
In summary, we developed a high-throughput screen for discovery of hNaV1.7 modulators and used this screen to demonstrate that spider venoms are perhaps the best natural source of inhibitors of this target analgesic. We discovered seven novel members of NaSpTx-F1 using this screen. One of these toxins, rHd1a, potently inhibited hNaV1.7 with a high level of selectivity over other hNaV subtypes In addition, we engineered a panel of hNaV1.7/rKv2.1 channel chimeras that can be used to rapidly assess whether hNaV1.7 toxins target any of the channel's voltage-sensor paddles. The venom-based drug discovery pipeline described here provides a paradigm for high-throughput screening of animal venoms against other therapeutic ion channel targets.
Acknowledgments
We thank Kenton Swartz for the KV2.1 clone and agitoxin-2. Thanks to Jamie Vandenberg for the hERG clone and Ben Cristofori-Armstrong for helping with hERG experiments. Thanks to the members of the Deutsche Arachnologische Gesellschaft (DeArGe), particularly Arnd Schlosser, Gianni Sposato, Henrik Krehenwinkel, Reti Erler and Volker von Wirth, for providing spiders for milking. We acknowledge financial support from the Australian Research Council (Discovery Grant DP110103129 to G. F. K. and R. J. L.), the National Health & Medical Research Council (Principal Research Fellowships to G. F. K. and R. J. L.; Program Grant to R. J. L.), and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (Award R00NS073797 to F. B.). Access to the Australian Proteome Analysis Facility is facilitated by support from the Australian Government's National Collaborative Research Infrastructure Strategy.
Glossary
Abbreviations
- CaV channel
voltage-gated calcium channel
- FLIPR
fluorescence imaging plate reader
- Hd1a
peptide μ-TRTX-Hd1a from venom of the spider Haplopelma doriae
- hNaV1.7
human voltage-gated sodium channel subtype 1.7
- ICK
inhibitor cystine knot
- KV channel
voltage-gated potassium channel
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- MBP
maltose binding protein
- NaSpTx
spider toxin that targets voltage-gated sodium channels
- NaSpTx-F1
NaSpTx family 1
- NaV channel
voltage-gated sodium channel
- RMSD
root-mean-square deviation
- SAR
structure–activity relationship
- TTX
tetrodotoxin
Author contributions
G. F. K., R. J. L. and F. B. directed this program of research. G. F. K., R. L., I. V., V. H., J. K. K. and F. B. conceived and designed the experiments. J. K. K., J. J. S., D. R., S. Y. R., S. S., I. V., M. M., V. H. and F. B. performed the experiments: J. K. K., J. J. S., D. R., S. Y. R., S. S., M. M., F. B. and G. F. K. analysed the data. J. K. K., F. B. and G. F. K. wrote the paper.
Conflict of interest
The authors have no conflicts of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Figure S1 Voltage-dependence of NaV channel inhibition by rHd1a. (A) Inhibition of NaV1.7-mediated currents by rHd1a is not voltage-dependent from −25 to 0 mV (n = 3–4; error bars denote SEM). Currents were evoked from a holding potential of −90 mV, stepping from −70 to 0 mV in 5 mV increments. (B) Selectivity of rHd1a is not voltage-dependent from −25 to 0 mV. I/I0 in the presence of 1 μM rHd1a for hNaV1.2, hNaV1.3, hNaV1.5 and hNaV1.7 (n = 3–4; error bars denote SEM). Currents were evoked from a holding potential of −90 mV, stepping from −70 to 0 mV in 5 mV increments.
Figure S2 Effect of rHd1a on hNaV1.7 inactivation time constants. Time constant (|) for inactivation of hNaV1.7 in the absence (black) or presence (red) of 1 μM rHd1a (n = 5–7; error bars denote SEM). The rate of inactivation is slowed in the presence of rHd1a. Currents were evoked from a holding potential of −90 mV, stepping from −70 to +20 mV in 5 mV increments.
Figure S3 Effect of rHd1a on human ERG channels (hERG, Kv11.1). The current–voltage (I–V) relationship for hERG was measured by applying 2 s pulses to test potentials ranging from −70 to +40 mV in 10 mV increments and then by a repolarizing step to −70 mV for 1 s. Tail currents are plotted as a function of voltage and fitted to a Boltzmann equation (n = 6; error bars denote SEM). There was no significant difference between control currents (black) and those following addition of 1 μM rHd1a (red).
Figure S4 Effect of rHd1a on hNaV1.9/rKv2.1 chimeras. Tail current–voltage relationships for a panel of hNaV1.9/rKv2.1 chimeras obtained before (black) or after (red) addition of 1 μM rHd1a (n = 5–8; error bars denote SEM). These chimeras were constructed by transplanting the S3b-S4 paddle from each domain of hNaV1.9 (DI, DII, DIII and DIV, as indicated) into rKv2.1; construction and characterization of these chimeras was reported previously.1 The holding voltage was −90 mV (or −120 mV for DII) and the tail voltage was −70 mV for the DI (panel A), −90 mV for DII (panel B), −60 mV for DIII (panel C) and −80 mV for DIV (panel D) chimeras. rHd1a had no effect on any of the chimeric channels.
Figure S5 Fully assigned 2D 1H-15N-HSQC spectrum of Hd1a. Horizontal lines connect peaks corresponding to the two side chain NH groups of N11, N14, and Q16. The peak from the indole side chain of W31 is also labelled. All other peaks are from backbone amide groups. The 1H-15N-HSQC spectrum is missing resonances from the backbone amide groups of K30 and W31, but they were present in 3D 15N- and 13C-edited HSQC-NOESY spectra.
Table S1 List of species from which venom was obtained for this study.
References
- Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: Ion channels. Br J Pharmacol. 2013;170:1607–1651. doi: 10.1111/bph.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosmans F, Swartz KJ. Targeting voltage sensors in sodium channels with spider toxins. Trends Pharmacol Sci. 2010;31:175–182. doi: 10.1016/j.tips.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosmans F, Rash L, Zhu S, Diochot S, Lazdunski M, Escoubas P, et al. Four novel tarantula toxins as selective modulators of voltage-gated sodium channel subtypes. Mol Pharmacol. 2006;69:419–429. doi: 10.1124/mol.105.015941. [DOI] [PubMed] [Google Scholar]
- Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456:202–208. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosmans F, Puopolo M, Martin-Eauclaire MF, Bean BP, Swartz KJ. Functional properties and toxin pharmacology of a dorsal root ganglion sodium channel viewed through its voltage sensors. J Gen Physiol. 2011;138:59–72. doi: 10.1085/jgp.201110614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita LD, Dai W, Bottomley SP. A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotech. 2006;6:12. doi: 10.1186/1472-6750-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Cestele S, Yarov-Yarovoy V, Yu FH, Konoki K, Scheuer T. Voltage-gated ion channels and gating modifier toxins. Toxicon. 2007;49:124–141. doi: 10.1016/j.toxicon.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Cestele S, Qu Y, Rogers JC, Rochat H, Scheuer T, Catterall WA. Voltage sensor-trapping: enhanced activation of sodium channels by β-scorpion toxin bound to the S3-S4 loop in domain II. Neuron. 1998;21:919–931. doi: 10.1016/s0896-6273(00)80606-6. [DOI] [PubMed] [Google Scholar]
- Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol. 2002;120:629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Dib-Hajj SD, Tyrrell L, Te Morsche RH, Drenth JP, Waxman SG. Deletion mutation of sodium channel NaV1.7 in inherited erythromelalgia: enhanced slow inactivation modulates dorsal root ganglion neuron hyperexcitability. Brain. 2011;134:1972–1986. doi: 10.1093/brain/awr143. [DOI] [PubMed] [Google Scholar]
- Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan G, Xiang G, Zhang X, Yuan R, Zhan H, Qi D. A single-nucleotide polymorphism in SCN9A may decrease postoperative pain sensitivity in the general population. Anesthesiology. 2013;118:436–442. doi: 10.1097/ALN.0b013e31827dde74. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Bosmans F. Spider peptide toxins as leads for drug development. Expert Opin Drug Discov. 2007;2:823–835. doi: 10.1517/17460441.2.6.823. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Sollod B, King GF. Venom landscapes: mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47:650–663. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Estacion M, Dib-Hajj SD, Benke PJ, Te Morsche RH, Eastman EM, Macala LJ, et al. NaV1.7 gain-of-function mutations as a continuum: A1632E displays physiological changes associated with erythromelalgia and paroxysmal extreme pain disorder mutations and produces symptoms of both disorders. J Neurosci. 2008;28:11079–11088. doi: 10.1523/JNEUROSCI.3443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertleman CR, Baker MD, Parker KA, Moffatt S, Elmslie FV, Abrahamsen B, et al. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron. 2006;52:767–774. doi: 10.1016/j.neuron.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Frech GC, Vandongen AM, Schuster G, Brown AM, Joho RH. A novel potassium channel with delayed rectifier properties isolated from rat brain by expression cloning. Nature. 1989;340:642–645. doi: 10.1038/340642a0. [DOI] [PubMed] [Google Scholar]
- Garcia ML, Garcia-Calvo M, Hidalgo P, Lee A, Mackinnon R. Purification and characterization of three inhibitors of voltage-dependent K+ channels from Leiurus quinquestriatus var. hebraeus venom. Biochemistry. 1994;33:6834–6839. doi: 10.1021/bi00188a012. [DOI] [PubMed] [Google Scholar]
- Gaskin DJ, Richard P. The economic costs of pain in the United States. J Pain. 2012;13:715–724. doi: 10.1016/j.jpain.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Gilchrist J, Olivera BM, Bosmans F. Animal toxins influence voltage-gated sodium channel function. Handb Exp Pharmacol. 2014;221:203–229. doi: 10.1007/978-3-642-41588-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Liu B, Cao G, Lipchik AM, Perez M, Dekan Z, et al. A tarantula-venom peptide antagonizes the TRPA1 nociceptor ion channel by binding to the S1-S4 gating domain. Curr Biol. 2014;24:473–483. doi: 10.1016/j.cub.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitz M, Karbat I, Cohen L, Ilan N, Kahn R, Turkov M, et al. The insecticidal potential of scorpion β-toxins. Toxicon. 2007;49:473–489. doi: 10.1016/j.toxicon.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Herzig V, Wood DL, Newell F, Chaumeil PA, Kaas Q, Binford GJ, et al. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucl Acids Res. 2011;39:D653–D657. doi: 10.1093/nar/gkq1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn R, Ding S, Gruber HJ. Immobilizing the moving parts of voltage-gated ion channels. J Gen Physiol. 2000;116:461–476. doi: 10.1085/jgp.116.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Copeland TD, Waugh DS. The P1′ specificity of tobacco etch virus protease. Biochem Biophys Res Commun. 2002;294:949–955. doi: 10.1016/S0006-291X(02)00574-0. [DOI] [PubMed] [Google Scholar]
- King GF. Modulation of insect CaV channels by peptidic spider toxins. Toxicon. 2007;49:513–530. doi: 10.1016/j.toxicon.2006.11.012. [DOI] [PubMed] [Google Scholar]
- King GF, Vetter I. No gain, no pain: NaV1.7 as an analgesic target. ACS Chem Neurosci. 2014;5:749–751. doi: 10.1021/cn500171p. [DOI] [PubMed] [Google Scholar]
- King GF, Escoubas P, Nicholson GM. Peptide toxins that selectively target insect NaV and CaV channels. Channels. 2008a;2:100–116. doi: 10.4161/chan.2.2.6022. [DOI] [PubMed] [Google Scholar]
- King GF, Gentz MC, Escoubas P, Nicholson GM. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon. 2008b;52:264–276. doi: 10.1016/j.toxicon.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Klint JK, Senff S, Rupasinghe DB, Er SY, Herzig V, Nicholson GM, et al. Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon. 2012;60:478–491. doi: 10.1016/j.toxicon.2012.04.337. [DOI] [PubMed] [Google Scholar]
- Klint JK, Senff S, Saez NJ, Seshadri R, Lau HY, Bende NS, et al. Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of E. coli. PLoS ONE. 2013;8:e63865. doi: 10.1371/journal.pone.0063865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan AH, Mobli M, Gooley PR, King GF, Mackay JP. Macromolecular NMR spectroscopy for the non-spectroscopist. FEBS J. 2011;278:687–703. doi: 10.1111/j.1742-4658.2011.08004.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Li D, Wu Z, Li J, Nie D, Xiang Y, et al. A positively charged surface patch is important for hainantoxin-IV binding to voltage-gated sodium channels. J Pept Sci. 2012;18:643–649. doi: 10.1002/psc.2451. [DOI] [PubMed] [Google Scholar]
- Minassian NA, Gibbs A, Shih AY, Liu Y, Neff RA, Sutton SW, et al. Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin huwentoxin-IV (μ-TRTX-Hh2a) J Biol Chem. 2013;288:22707–22720. doi: 10.1074/jbc.M113.461392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobli M, Maciejewski MW, Gryk MR, Hoch JC. An automated tool for maximum entropy reconstruction of biomolecular NMR spectra. Nat Meth. 2007;4:467–468. doi: 10.1038/nmeth0607-467. [DOI] [PubMed] [Google Scholar]
- Mobli M, Stern AS, Bermel W, King GF, Hoch JC. A non-uniformly sampled 4D HCC(CO)NH-TOCSY experiment processed using maximum entropy for rapid protein sidechain assignment. J Magn Reson. 2010;204:160–164. doi: 10.1016/j.jmr.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson GM, Little MJ. Spider neurotoxins targeting voltage-gated sodium channels. Toxin Rev. 2005;24:315–345. [Google Scholar]
- Pallaghy PK, Nielsen KJ, Craik DJ, Norton RS. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson AJ, Sharman JL, Benson HE, Faccenda E, Alexander SP, Buneman OP, et al. NC-IUPHAR. The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledgebase of drug targets and their ligands. Nucl. Acids Res. 2014;42(Database Issue):D1098–1106. doi: 10.1093/nar/gkt1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder JE, Byler TK, Foster DC, Landas SK, Okafor H, Stearns G, et al. Polymorphism in the SCN9A voltage-gated sodium channel gene associated with interstitial cystitis/bladder pain syndrome. Urology. 2013;81:210. doi: 10.1016/j.urology.2012.07.072. , e1–210.e4. [DOI] [PubMed] [Google Scholar]
- Reimann F, Cox JJ, Belfer I, Diatchenko L, Zaykin DV, McHale DP, et al. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc Natl Acad Sci U S A. 2010;107:5148–5453. doi: 10.1073/pnas.0913181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renicke C, Spadaccini R, Taxis C. A tobacco etch virus protease with increased substrate tolerance at the P1′ position. PLoS ONE. 2013;8::e67915. doi: 10.1371/journal.pone.0067915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell JD, Lund PE, Linley JE, Metcalfe J, Burmeister N, Sridharan S, et al. Potency optimization of huwentoxin-IV on hNaV1.7: a neurotoxin TTX-S sodium-channel antagonist from the venom of the Chinese bird-eating spider Selenocosmia huwena. Peptides. 2013;44:40–46. doi: 10.1016/j.peptides.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Rodriguez De La Vega RC, Possani LD. Overview of scorpion toxins specific for Na+ channels and related peptides: biodiversity, structure-function relationships and evolution. Toxicon. 2005;46:831–844. doi: 10.1016/j.toxicon.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Ruta V, MacKinnon R. Localization of the voltage-sensor toxin receptor on KvAP. Biochemistry. 2004;43:10071–10079. doi: 10.1021/bi049463y. [DOI] [PubMed] [Google Scholar]
- Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- Sheets MF, Kyle JW, Kallen RG, Hanck DA. The Na channel voltage sensor associated with inactivation is localized to the external charged residues of domain IV, S4. Biophys J. 1999;77:747–757. doi: 10.1016/S0006-3495(99)76929-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SR, Vetter I, Ragnarsson L, Lewis RJ. Expression and pharmacology of endogenous CaV channels in SH-SY5Y human neuroblastoma cells. PLoS ONE. 2013;8:e59293. doi: 10.1371/journal.pone.0059293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz KJ, MacKinnon R. Hanatoxin modifies the gating of a voltage-dependent K+ channel through multiple binding sites. Neuron. 1997;18:665–673. doi: 10.1016/s0896-6273(00)80306-2. [DOI] [PubMed] [Google Scholar]
- Theile JW, Jarecki BW, Piekarz AD, Cummins TR. Nav1.7 mutations associated with paroxysmal extreme pain disorder, but not erythromelalgia, enhance Navβ4 peptide-mediated resurgent sodium currents. J Physiol. 2011;589:597–608. doi: 10.1113/jphysiol.2010.200915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I, Davis JL, Rash LD, Anangi R, Mobli M, Alewood PF, et al. Venomics: a new paradigm for natural products-based drug discovery. Amino Acids. 2011;40:15–28. doi: 10.1007/s00726-010-0516-4. [DOI] [PubMed] [Google Scholar]
- Vetter I, Mozar CA, Durek T, Wingerd JS, Alewood PF, Christie MJ, et al. Characterisation of NaV types endogenously expressed in human SH-SY5Y neuroblastoma cells. Biochem Pharmacol. 2012;83:1562–1571. doi: 10.1016/j.bcp.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Bingham JP, Zhu W, Moczydlowski E, Liang S, Cummins TR. Tarantula huwentoxin-IV inhibits neuronal sodium channels by binding to receptor site 4 and trapping the domain II voltage sensor in the closed configuration. J Biol Chem. 2008;283:27300–27313. doi: 10.1074/jbc.M708447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Jackson JO, 2nd, Liang S, Cummins TR. Common molecular determinants of tarantula huwentoxin-IV inhibition of Na+ channel voltage-sensors in domains II and IV. J Biol Chem. 2011;286:27301–27310. doi: 10.1074/jbc.M111.246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Liu Z, Xiao Y, Li Y, Rong M, Liang S, et al. Chemical punch packed in venoms makes centipedes excellent predators. Mol Cell Proteomics. 2012;11:640–650. doi: 10.1074/mcp.M112.018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Li S, Xu Z, Li H, Ma L, et al. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet. 2004;41:171–174. doi: 10.1136/jmg.2003.012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Voltage-dependence of NaV channel inhibition by rHd1a. (A) Inhibition of NaV1.7-mediated currents by rHd1a is not voltage-dependent from −25 to 0 mV (n = 3–4; error bars denote SEM). Currents were evoked from a holding potential of −90 mV, stepping from −70 to 0 mV in 5 mV increments. (B) Selectivity of rHd1a is not voltage-dependent from −25 to 0 mV. I/I0 in the presence of 1 μM rHd1a for hNaV1.2, hNaV1.3, hNaV1.5 and hNaV1.7 (n = 3–4; error bars denote SEM). Currents were evoked from a holding potential of −90 mV, stepping from −70 to 0 mV in 5 mV increments.
Figure S2 Effect of rHd1a on hNaV1.7 inactivation time constants. Time constant (|) for inactivation of hNaV1.7 in the absence (black) or presence (red) of 1 μM rHd1a (n = 5–7; error bars denote SEM). The rate of inactivation is slowed in the presence of rHd1a. Currents were evoked from a holding potential of −90 mV, stepping from −70 to +20 mV in 5 mV increments.
Figure S3 Effect of rHd1a on human ERG channels (hERG, Kv11.1). The current–voltage (I–V) relationship for hERG was measured by applying 2 s pulses to test potentials ranging from −70 to +40 mV in 10 mV increments and then by a repolarizing step to −70 mV for 1 s. Tail currents are plotted as a function of voltage and fitted to a Boltzmann equation (n = 6; error bars denote SEM). There was no significant difference between control currents (black) and those following addition of 1 μM rHd1a (red).
Figure S4 Effect of rHd1a on hNaV1.9/rKv2.1 chimeras. Tail current–voltage relationships for a panel of hNaV1.9/rKv2.1 chimeras obtained before (black) or after (red) addition of 1 μM rHd1a (n = 5–8; error bars denote SEM). These chimeras were constructed by transplanting the S3b-S4 paddle from each domain of hNaV1.9 (DI, DII, DIII and DIV, as indicated) into rKv2.1; construction and characterization of these chimeras was reported previously.1 The holding voltage was −90 mV (or −120 mV for DII) and the tail voltage was −70 mV for the DI (panel A), −90 mV for DII (panel B), −60 mV for DIII (panel C) and −80 mV for DIV (panel D) chimeras. rHd1a had no effect on any of the chimeric channels.
Figure S5 Fully assigned 2D 1H-15N-HSQC spectrum of Hd1a. Horizontal lines connect peaks corresponding to the two side chain NH groups of N11, N14, and Q16. The peak from the indole side chain of W31 is also labelled. All other peaks are from backbone amide groups. The 1H-15N-HSQC spectrum is missing resonances from the backbone amide groups of K30 and W31, but they were present in 3D 15N- and 13C-edited HSQC-NOESY spectra.
Table S1 List of species from which venom was obtained for this study.