Abstract
Gene expression changes during aging are partly conserved across species, and suggest that oxidative stress, inflammation and proteotoxicity result from mitochondrial malfunction and abnormal mitochondrial-nuclear signaling. Mitochondrial maintenance failure may result from trade-offs between mitochondrial turnover versus growth and reproduction, sexual antagonistic pleiotropy and genetic conflicts resulting from uni-parental mitochondrial transmission, as well as mitochondrial and nuclear mutations and loss of epigenetic regulation. Aging phenotypes and interventions are often sex-specific, indicating that both male and female sexual differentiation promote mitochondrial failure and aging. Studies in mammals and invertebrates implicate autophagy, apoptosis, AKT, PARP, p53 and FOXO in mediating sex-specific differences in stress resistance and aging. The data support a model where the genes Sxl in Drosophila, sdc-2 in C. elegans, and Xist in mammals regulate mitochondrial maintenance across generations and in aging. Several interventions that increase life span cause a mitochondrial unfolded protein response (UPRmt), and UPRmt is also observed during normal aging, indicating hormesis. The UPRmt may increase life span by stimulating mitochondrial turnover through autophagy, and/or by inhibiting the production of hormones and toxic metabolites. The data suggest that metazoan life span interventions may act through a common hormesis mechanism involving liver UPRmt, mitochondrial maintenance and sexual differentiation.
Keywords: UPRmt, sexual antagonistic pleiotropy, sexual conflict, mother’s curse, heteroplasmy, dosage compensation
Mitochondrial maintenance failure and aging
Mitochondrial malfunction is implicated in aging across species, including yeast [1–5] C. elegans [6, 7], Drosophila [8–10] and mammals [11, 12], indicating possible conservation of basic mechanisms. Several non-exclusive and potentially synergistic mechanisms may contribute to the observed mitochondrial failure during aging (Figure 1). Evolutionary theory predicts trade-offs between reproduction and somatic maintenance required for optimal life span [13]. Increasing evidence suggests that growth and reproduction may occur at the expense of mitochondrial turnover, leading to longer-lived and more damage-prone mitochondria. For example, down-regulation of mitochondrial gene expression is observed in several species at the end of developmental growth and during adult aging [14–17]. Similarly, sex-specific selective pressures, including ones resulting from uni-parental inheritance of the mitochondria, may lead to sexual antagonistic pleiotropy (SAP) of genes with mitochondrial functions [18]. Finally, inherited mitochondrial mutations (heteroplasmy) and new mitochondrial mutations arising during development and aging may synergize with these effects to cause mitochondrial maintenance failure during aging.
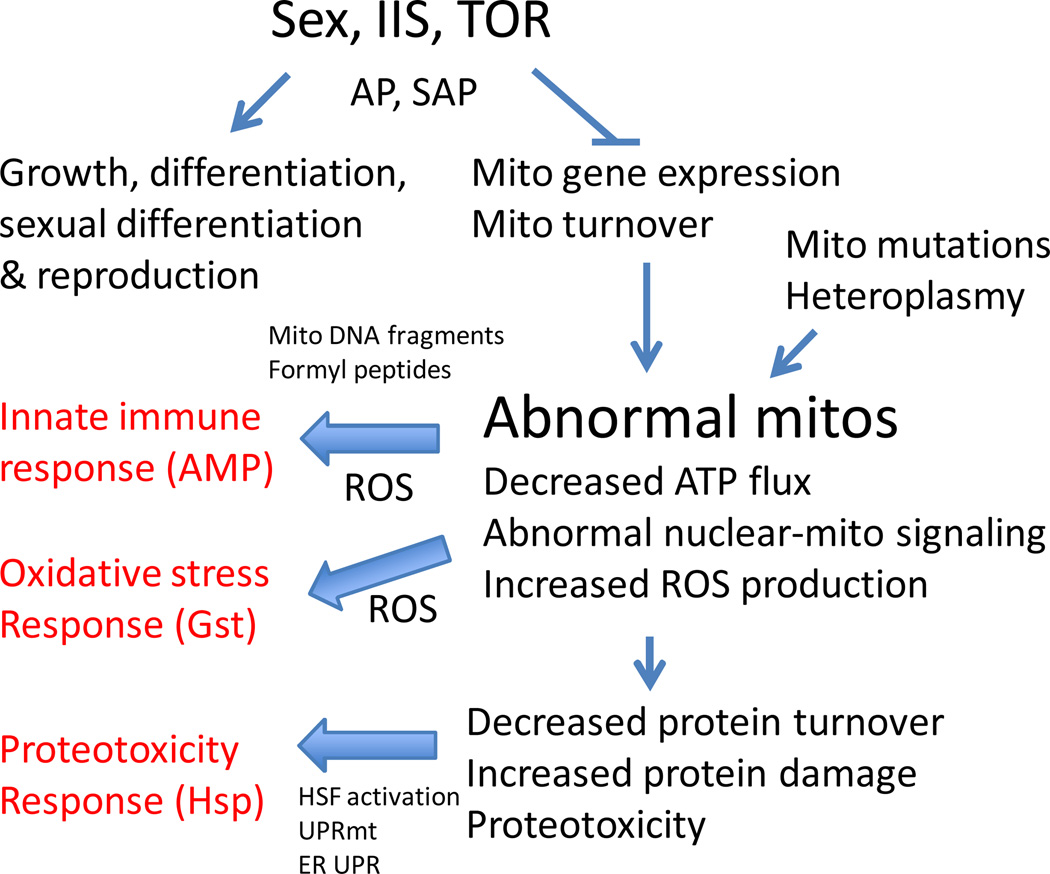
Figure 1.
Model for aging gene expression patterns. Chromosomal sex and sexual differentiation pathways (Sex), in concert with insulin/IGF1-like signaling (IIS) and Target-of-Rapamycin (TOR) pathways, promote growth, sexual differentiation and reproduction at the expense of costly mitochondrial gene expression and turnover. AP, antagonistic pleiotropy; SAP, sexual antagonistic pleiotropy. Reduced mitochondrial turnover leads to abnormal mitochondria, the UPRmt, and the stress-response gene expression patterns that characterize aging (indicated in red). Mitochondrial mutations and heteroplasmy synergize with these effects to produce abnormal mitochondria during aging. Mito, mitochondria. AMP, anti-microbial peptide. Gst, Glutathione-S-transferase. Hsp, heat shock protein. HSF, heat shock transcription factor. UPRmt, mitochondrial unfolded protein response. ER UPR, endoplasmic reticulum unfolded protein response.
Structural and functional abnormalities of mitochondria with age
Pioneering studies beginning in the 1970’s described the accumulation of mitochondria with abnormal structure in various tissues of Drosophila and other dipterans, including gut, flight muscle and fat-body [19–24]. Electron microscopy revealed abnormalities including a swollen appearance, inclusions, and disordered membrane structures. The abnormal mitochondria of flight muscle often have a characteristic rearrangement of the internal membrane described as a “whorl” or “swirl” [25, 26]. When mitochondria are isolated from tissues of aged flies, they exhibit functional abnormalities including decreased electron transport chain (ETC) enzyme activity and increased production of reactive oxygen species (ROS) [8, 27–30]. Mitochondria in tissues of mammals [31–33] and C. elegans [34, 35] show a similar range of structural and functional abnormalities with age. Consistent with a loss of normal mitochondrial function, human aging is associated with decreased metabolic rate and often with a disruption of energy homeostasis called metabolic syndrome [36, 37].
Mitochondrial dynamics and mitochondrial maintenance
Mitochondria are normally degraded in cells through selective macroautophagy (also called autophagy or mitophagy), involving engulfment by the autophagosome followed by fusion with the lysosome and degradation of the mitochondrial material (diagrammed in Figure 2) [38]. Decreased membrane potential may be one signal that marks mitochondria for degradation [39]. A decline in this process with age and the accumulation of partly-degraded mitochondrial material is implicated in the production of age pigment, or lipofuscin [40]. Mitochondria normally undergo dynamic changes in structure mediated by fission and fusion events [41], and a decrease in fission has been suggested as one mechanism for increased mitochondrial size with age in certain tissues. Fission is also implicated in normal mitophagy, in part by generating mitochondria of appropriate size for engulfment by the autophagosome. The importance of fission and fusion events in mitochondrial maintenance during aging is underscored by the identification of mutations in genes that control these pathways, including PARKIN, that predispose human patients to age-related neurodegenerative disease [39]. The PARKIN pathway promotes mitochondrial turnover by autophagy, and in Drosophila this pathway has been shown to also promote selective turnover of ETC components [42]. Notably, over-expression of Parkin in adult female Drosophila is reported to alter mitochondrial dynamics during aging and to increase life span [43].
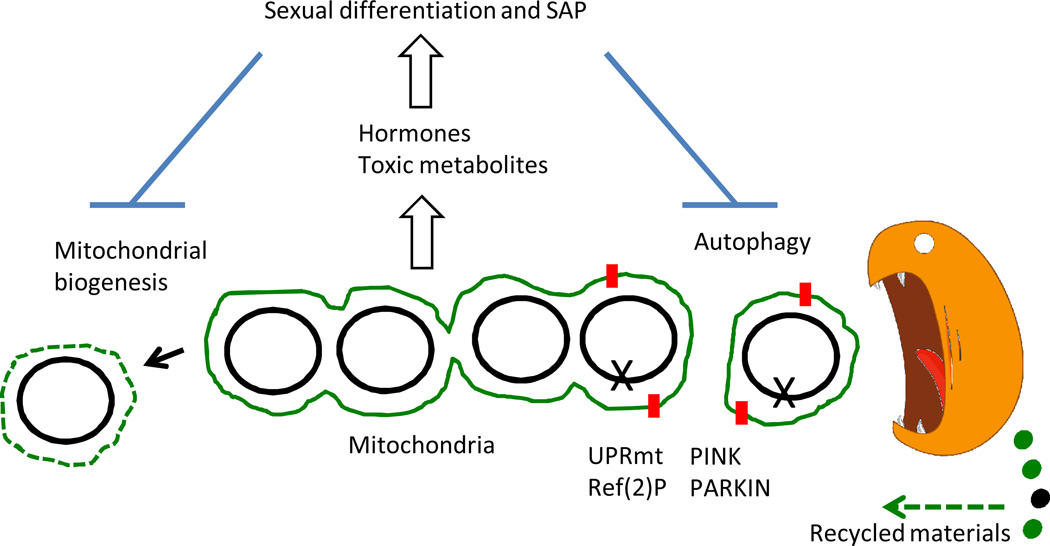
Figure 2.
Models for life span extension by UPRmt and hormesis. Mitochondria (indicated in green) contain multiple mitochondrial genomes (black circles). A mitochondrial genome mutation (indicated by X) causes UPRmt and loss of membrane potential. These changes signal marking by Ref(2)P (indicated with red squares) and activation of the PINK/PARKIN pathway for fission of mitochondria and destruction by the autophagy pathway (cartooned in orange). Degradation products are recycled for use in biogenesis of new mitochondria. Induction of the UPRmt in young animals (hormesis) would inhibit the production of toxic metabolites including hormones and age pigment. Hormones promote sexual differentiation and the deleterious effects of many genes (through sexual antagonistic plieotropy, SAP). Sexual differentiation and SAP in turn inhibit mitochondrial turnover and maintenance, resulting in aging, oxidative stress and a toxic aging-associated UPRmt.
Gene expression changes during aging indicate mitochondrial maintenance failure
The patterns of gene expression observed during aging can vary with species, tissue and sex, however several conserved themes have emerged that are each consistent with a failure in mitochondrial maintenance (Figure 1). Genome-wide analysis of gene expression patterns in adult male Drosophila revealed that aging is characterized by down-regulation of mitochondrial genes and up-regulation of genes associated with innate-immune response, oxidative stress response, proteotoxicity response, and purine biosynthesis [14, 17]. These same patterns have been found in aging of one or more mammalian tissues [16, 44, 45]. Up-regulated stress response and down-regulated metabolism genes have also been identified in certain studies of C. elegans aging [46]. The down-regulation of mitochondrial genes is expected to reduce mitochondrial turnover, resulting in longer-lived and more damage-prone mitochondria, consistent with the structural and functional abnormalities discussed above. Mitochondria are the main source of ROS in the cell, and compromised mitochondrial function during aging is associated with increased production of ROS [47]. The up-regulation of innate immune response genes may result from the pro-inflammatory effects of mitochondrial DNA fragments, mitochondrial formyl peptides and ROS [14, 48, 49]; in Drosophila, increased microbial load also contributes to this up-regulation [50]. Reduced mitochondrial ATP production is expected to result in decreased rates of cellular protein synthesis and turnover, and the longer-lived proteins will be more susceptible to damage, in particular due to increased production of ROS. Consistent with this scenario is the up-regulated basal expression of the proteotoxicity response, including heat shock proteins (Hsps) targeted to the cytoplasm and mitochondria [51–56]. Gene expression changes during aging are also sexually-dimorphic, for example, in the vertebrate liver [57], brain [45, 58] and heart [59, 60], where males tend to show relatively greater reduction in mitochondrial gene expression.
Mis-regulated apoptosis during aging
Apoptosis (programmed cell death) mechanisms involve regulation by the mitochondria in both mammals and invertebrates [61, 62]. Mis-regulated apoptosis is observed during aging and is consistent with tissue-specific outcomes for mitochondrial maintenance failure [63]. For example, mitochondrial malfunction and apoptotic-like events are implicated in age-related muscle-wasting (sarcopenia) and neurodegenerative disease in mammals [33, 64]. Apoptotic-like events are also associated with aging in Drosophila muscle and fat cells [65]. In contrast, a down-regulation of apoptosis is associated with both cell senescence and cancer in mammals [66–68] and with tissue over-growth in the aging C. elegans gonad [69].
Sexual antagonistic pleiotropy (SAP) and consequences of mitochondrial uniparental transmission
Aging and aging-associated diseases are hypothesized to result from antagonistic pleiotropy of gene function between developmental stages and the sexes [3, 6, 18, 68, 70–77]. Antagonistic pleiotropy is when a gene has a beneficial effect during the growth and reproductive period, such as increasing reproductive fitness, but is detrimental at later ages and contributes to aging. For example, the target-of-rapamycin (TOR) pathway promotes growth and can also inhibit autophagy [5, 78–80], making it a candidate for mediating a trade-off between growth and mitochondrial maintenance required for longevity (Figure 1). Sexual antagonist pleiotropy (SAP) is when a gene responds to sex-specific selective pressures, resulting in a benefit for one sex and a detriment for the other sex, or even a detriment for both sexes [18, 74, 81–83]. Likely examples of sexual antagonistic pleiotropy are the Drosophila sex peptide [84, 85], and pheromones of both Drosophila [86] and C. elegans [87, 88], that are produced in one sex but act to reduce life span in the other sex.
Sexual differentiation is controlled by environmental signals and the chromosomal sex of the animal, for example, X/X genotype for Drosophila and mammalian females and the C. elegans hermaphrodite, hereafter referred to as the C. elegans female (Figure 3) [89–91]. The presence of two X chromosomes sets the master regulatory gene for dosage compensation (DC) to the “on” state (Sxl in Drosophila, sdc-2 in C. elegans, and Xist in mammals) also called the binary “Switch-Gene” [18, 92]. In males, where only one X chromosome is present, the Switch-Gene is in the “off” state. Maternal factors provided to the zygote from the mother, including mitochondria, combine with chromosomal sex and the Switch-Gene on/off state to control sexual differentiation (Figure 3). Female sexual differentiation enables preferential transmission of mitochondria to the offspring through the oocyte. The extreme bias [93, 94] towards uni-parental transmission of mitochondrial genomes may be one force maintaining deleterious alleles in the population that contribute to aging, because it creates potentially powerful sex-specific selective pressures (Figure 4) [95–97]. Non-exclusive explanations for why mitochondria are preferentially inherited through the mother include the avoidance of conflicts between different mitochondrial genome alleles in the zygote [98], avoiding damage to the mitochondrial genome that might be greater in the more metabolically active sperm [99–101], and the potential to create the sexes and promote evolution [18, 102, 103].
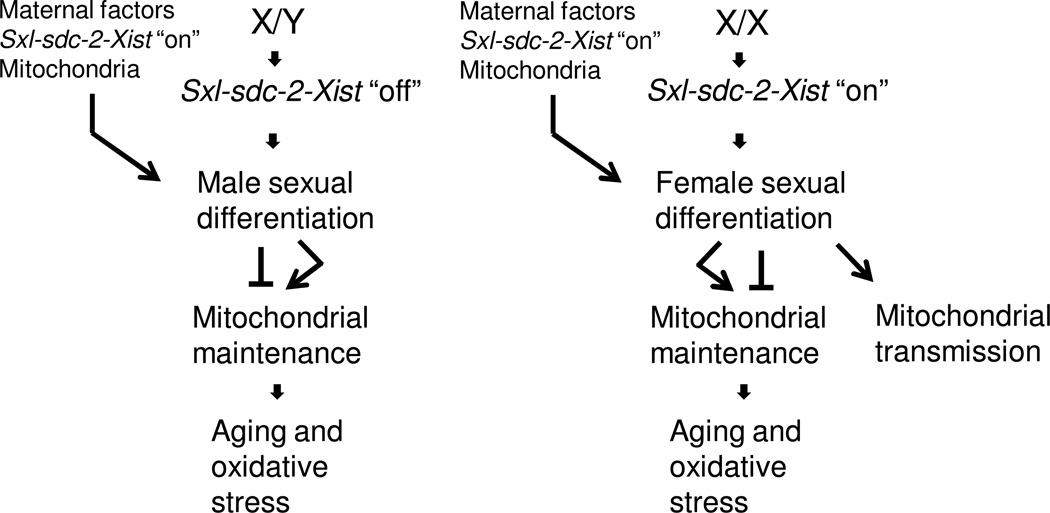
Figure 3.
Sex-specific regulation of aging and oxidative stress in Drosophila, C. elegans and mammals. In females (X/X) the master regulatory gene (or “Switch-Gene”) for Dosage Compensation (DC) is in the “on” state: Sxl in Drosophila, sdc-2 in C. elegans, and Xist in mammals. In males (X/Y) these genes are in the “off” state. Chromosomal sex, the Switch-Gene on/off state, and DC regulate mitochondrial maintenance as follows: Sexual differentiation, in particular DC, is required for animal viability including mitochondrial maintenance during development. In the adult, sexual differentiation mediates trade-offs between growth and reproduction and long-term mitochondrial maintenance that leads to aging and oxidative stress (see also Figures 1, 2). Female sexual differentiation mediates the preferential transmission of the mitochondria to offspring. Maternal factors, including mitochondria, are provided to the egg from the mother and are required for viability and sexual differentiation. In C. elegans (X/X) is the hermaphrodite, and the Y chromosome is absent in males (genotype X/O).
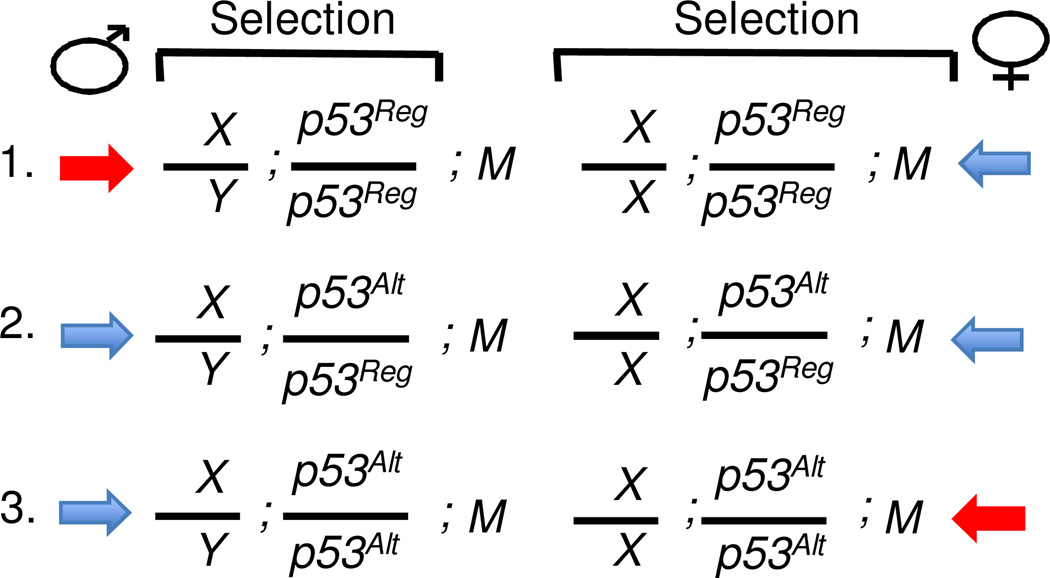
Figure 4.
Model for the genetic interaction between the mitochondrial genotype (M) and an autosomal gene (p53). Three possible autosomal genotypes are presented: (1.) Male and female homozygous for p53[Regular]. (2.) Male and female heterozygous for p53[Regular] and p53[Altered]. (3.) Male and female homozygous for p53[Altered]. Blue-color arrows indicate genotypes potentially beneficial for the indicated sex; red-color arrow indicates genotype potentially detrimental for that sex. For example, arrows might relate to larval survival and/or adult survival and reproduction. Details: (1.) In females natural selection acts to optimize the fit of both nuclear and mitochondrial alleles (p53Reg and M). (2. & 3.) In males natural selection can only act to optimize how nuclear genes cope with the mitochondrial genome (M), leading to selection for p53Alt. (3.) In females, p53Alt tends to be non-optimal. Reg, Regular. Alt, Altered.
The presence of more than one inherited mitochondrial allele (heteroplasmy) has been found to be common in Drosophila [94, 104, 105] mouse [98, 106] and humans [107]. Heteroplasmy has significant implications for aging, as studies in mouse show that the presence of more than one mitochondrial genotype can cause tissue-specific conflicts in metabolic regulation that result in deleterious phenotypes [98]. The female germ line has been confirmed to be acting as a selective sieve that reduces the transmission of non-optimal mitochondrial genomes in both Drosophila [108–110] and vertebrates, including mammals [11, 111, 112]. The data from Drosophila suggest that the mitochondria are being selected for their relative replication ability within the female germ line cells [108, 113]. Therefore natural selection is acting to optimize mitochondrial function both in the female germ-line, where the selective sieve operates, as well as in the female soma, because only viable and reproductively successful females will be able to pass on their germ-line mitochondria. The female-biased action of mitochondrial selection is expected to allow for accumulation of mitochondrial mutations that are relatively deleterious to the male.
Because mitochondrial genes are inherited almost exclusively from the mother, natural selection can only act to optimize mitochondrial gene function and nuclear-mitochondrial gene interactions in females (Figure 4). This is expected to lead to mitochondrial genome function that is optimized for the female and less optimal for the male; a situation sometimes called “mother’s curse” [76, 93, 114]. Indeed, recent experiments in Drosophila support the existence of a load of mitochondrial mutations that preferentially promote aging in males [109]. Because natural selection cannot act to optimize mitochondrial gene function for the male, the expectation is that natural selection will act on nuclear genes in the male, in particular nuclear mitochondrial genes, to select for alleles that can compensate for the non-optimal mitochondrial function (Figure 4). In turn, in the next generation, the female will inherit these nuclear alleles that are likely to be non-optimal for female physiology and female nuclear-mitochondrial genetic interactions. This ongoing battle between male and female is similar to a “Red Queen” situation [115–117] and may maintain deleterious alleles in the population that contribute to aging (sexual antagonistic pleiotropy, or SAP), in particular alleles affecting mitochondrial maintenance [18, 118]. In turn this mechanism may be beneficial for driving evolution and creating the sexes [18]. These models suggest that genes with sex-specific effects on aging should be common, and their functions should center on the mitochondria. One consequence of gene alleles exhibiting such SAP may be the failure in mitochondrial maintenance discussed above.
Genes with sexual antagonistic pleiotropy (SAP)
The evolutionary models predict that the deleterious effects of many genes will be sex-biased or sex-specific, and these deleterious effects will be regulated by chromosomal sex and sexual differentiation pathways (Figure 1). This prediction is supported by the fact that the onset of senescence often correlates with the sexual and reproductive maturation of the animal [18, 115, 119, 120]. ROS signaling [121] can promote mammalian cell differentiation [122, 123], sexual differentiation in yeasts [124, 125] and reproduction in humans [126, 127]. In Drosophila, hydrogen peroxide induces the expression of numerous developmental and signaling genes [17]. It is tempting to speculate that reduced mitochondrial turnover and moderately increased basal ROS levels could be selected for in part because of a benefit for sexual differentiation and reproductive fitness, despite the negative consequences for aging.
Genes in several species have been identified that have sex-specific effects on life span and/or mitochondrial function indicative of SAP. For example, the Drosophila sex peptide and the Drosophila and C. elegans pheromones mentioned above are sex-specific and dramatically shorten life span, indicating that the genes that encode these factors exhibit SAP. In humans both p53 and MDM2 alleles are reported to have sex-specific effects on longevity and cancer rates [128, 129]. Human genome-side association studies have revealed sex-specific quantitative trait loci (QTL) that regulate mitochondrial content of blood tissue, including male-specific effects of an allele of mitochondrial ribosomal protein gene MRPL37 [130]. In mice with an ETC complex III gene mutation, males have reduced life span, whereas females show a subset with increased life span [131]. Drosophila studies have identified many QTLs and genes with sex-specific and sexually-antagonistic effects on aging and mitochondrial function [73, 110, 132–136]. For example, Drosophila p53 exhibits developmental stage-specific and sex-specific effects on adult life span indicative of SAP [137], and these effects are modulated in a sex-specific way by foxo [82, 138]. Finally, Drosophila p53 dominant mutants have sex-specific effects on life span that are dependent upon the environment [137, 139], and recent studies show nucleus-by-mitochondria-by-environment genetic effects on Drosophila life span [140].
Sex-specific regulation of p53 and foxo
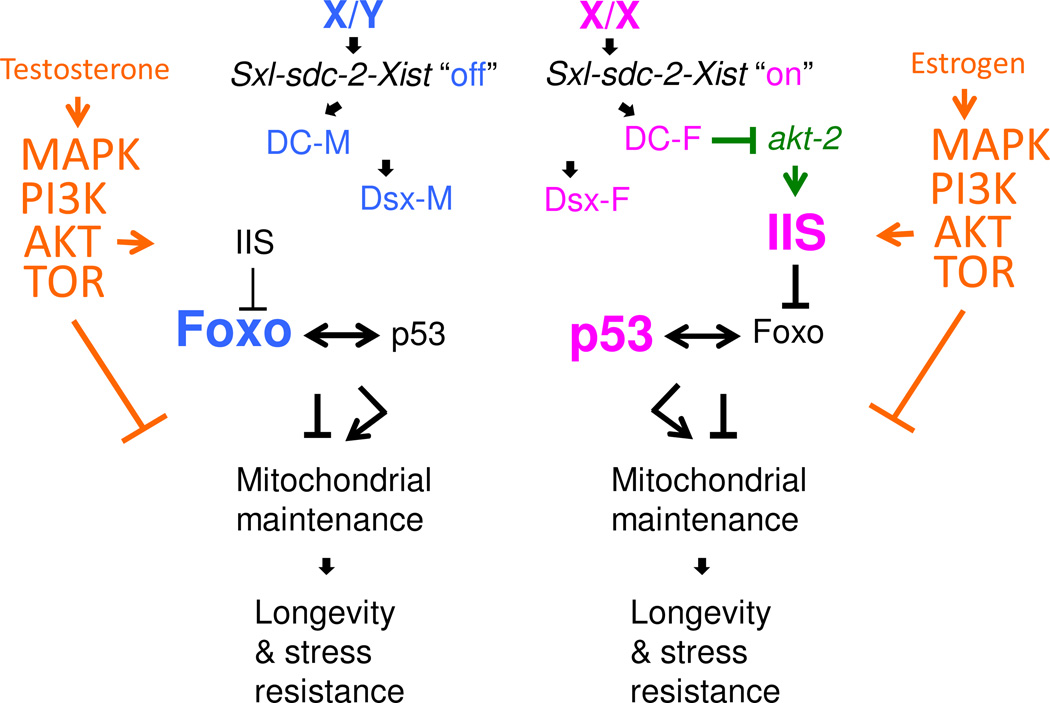
Several lines of evidence indicate sex-specific activity of the conserved mitochondrial and life span regulators p53 and foxo. It is likely the details of their regulation will differ by species, tissue, and environmental condition, however several themes have emerged (Figure 5). In humans, women are more sensitive to insulin than men with regards to glucose metabolism in the muscle and the liver, suggesting that insulin signaling may be greater in women than in men [141]. In Drosophila and mice, mutations that disrupt insulin/insulin-like growth factor 1-like signaling (IIS) increase life span in females to a greater extent than in males, suggesting relatively greater activity of IIS in females than in males [18, 39, 142, 143]. IIS negatively regulates the activity of the conserved transcription factor encoded by the foxo gene [144]. Consistent with the idea of relatively lower IIS in Drosophila males, several lines of evidence indicate relatively greater foxo protein activity in males (Figure 5). Several foxo protein transcriptional targets are expressed at higher levels in Drosophila males than in females [138, 145], in a foxo-dependent manner [82, 138]. Recent studies in Drosophila and beetles reveal the role of foxo in regulating sex-specific tissue growth and plasticity, with relatively greater activity observed in male tissues [146, 147]. In mammals, foxo gene family members and p53 interact genetically and regulate common target genes [148]. In Drosophila, the foxo gene was found to act preferentially in males to alter the effects of p53 on life span [82, 138], consistent with greater foxo activity in males.
Figure 5.
Model for sex-specific regulation of mitochondrial maintenance pathways. Chromosomal sex determines the on/off state of the master regulator of dosage compensation (DC) the “Switch-Gene”: Sxl in Drosophila, sdc-2 in C. elegans, and Xist in humans. This sets DC to either the male (M) or female (F) state. Chromosomal sex also directs the expression the dsx-like gene in either the male or female state, which in turn regulates somatic sexual differentiation. Females exhibit relatively greater IIS and p53 activity (indicated in pink). Males exhibit relatively greater Foxo activity (indicated in blue). In C. elegans, DC negatively regulates expression of the X-linked gene akt-2, which is a positive regulator of IIS (indicated in green). In mammals, the gonadal hormones testosterone and estrogen activate a MAPK/PI3K/AKT/TOR signaling pathway that can activate IIS and potentially inhibit autophagy (indicated in orange).
Several lines of evidence suggest relatively greater p53 activity in females (Figure 5). In Drosophila, p53 limited life span to a greater extent in adult females than in adult males, suggesting greater p53 activity in females [137]. Interestingly, p53 also appears to be more active in human females as compared to males with regard to tumor suppression [149], and to have greater developmental phenotypes in female mice relative to male mice [150]. Studies of Drosophila life span reveal gene-by-sex-by-environment interactions for p53 that are opposite in male and female [137, 139]. In C. elegans females, p53 can have either positive or negative effects on life span depending upon the nature of the intervention [151] and the degree of stress [152]. Males were not analyzed, however it seems likely that the threshold for p53 positive versus p53 negative effects could differ between the C. elegans sexes under appropriate conditions. These studies indicate that SAP is common, including conserved regulators of mitochondrial function and life span such as p53 and foxo, and suggest that greater p53 activity in females may be common to Drosophila, mice and humans.
In C. elegans, the DC pathway was found to negatively regulate IIS signaling during development in females, in part by reducing expression of the X-linked gene akt-2 (Figure 5, indicated in green). This result is consistent with the idea of the Switch-Gene on/off state regulating DC and IIS, however, it would seem to be more consistent with decreased IIS in females relative to males. One possible explanation might be that a partial repression of akt-2 and other X-linked genes in adults (i.e., escape from X-inactivation) might contribute to relatively greater AKT and IIS activity in females. Alternatively it might be that dimorphism in IIS signaling has a different pattern in C. elegans. Consistent with this latter possibility, in C. elegans, males are the longer-lived sex, and this was dependent upon foxo but not on daf-2 (the insulin-like receptor homolog) [153, 154]. In either event, the studies indicate that sexual dimorphism and SAP in life span regulation is common across species, including conserved regulators of mitochondrial function such as IIS, foxo and p53.
Mitochondrial mutations during development and aging
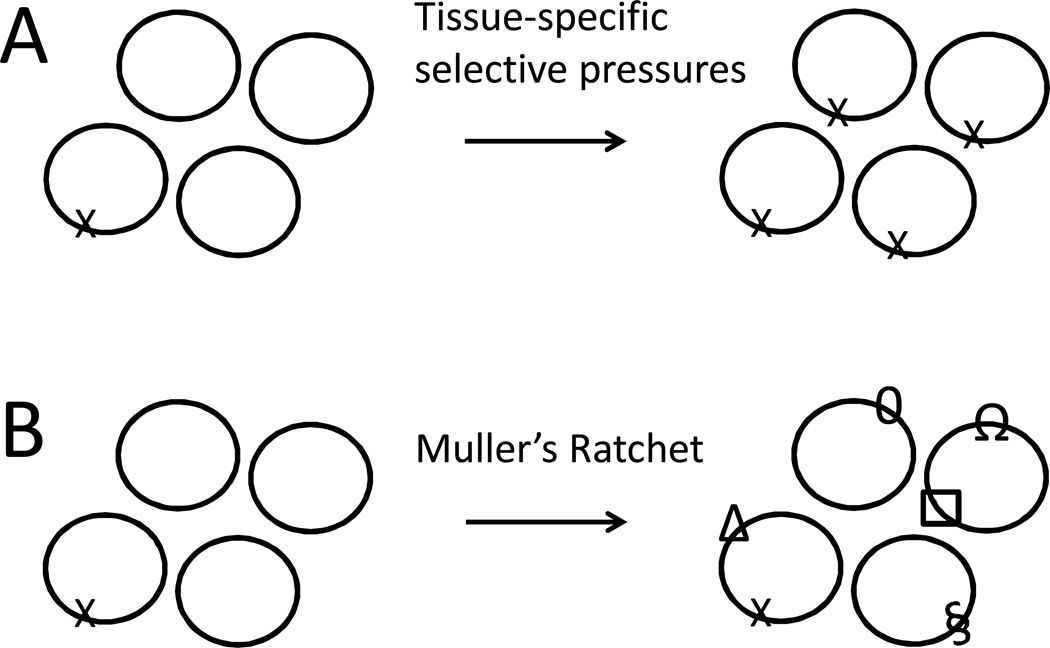
Mitochondrial mutations have long been hypothesized to contribute to aging. Because there are many copies of the mitochondrial genome per cell, one long-standing question is whether a mutation can become sufficiently abundant to have a deleterious effect. Mounting evidence indicates that this is often the case. Inherited mitochondrial heteroplasmy and new mitochondrial mutations arising during development and aging are implicated in a variety of human aging-related diseases [155–157], including Parkinson’s Disease [158], age-related macular degeneration [159], and cancer [155, 160]. Tissue-specific selective pressures acting on either heteroplasmy or new mutations are hypothesized to cause increased abundance of a particular mitochondrial genotype in a cell or tissue relative to surrounding cells or tissues, through at least three non-exclusive mechanisms (Figure 6A). (i) Cells can favor the replication and/or transmission of one mitochondrial genotype over another during cell division, through mechanisms that are not yet entirely clear [41, 157, 161]. (ii) One mitochondrial genotype may better favor survival of cells, as is suggested by studies of cancer [155, 160, 162]. (iii) One mitochondrial genotype may have an inherent replication advantage over another, such as a smaller genome or more active DNA replication origin [113, 163]. These mechanisms may cause deleterious mitochondrial alleles to accumulate in the cell to the point that they compromise normal cell function and promote aging.
Figure 6.
Potential mechanisms for mitochondrial mutation load increase within a cell. Two non-exclusive mechanisms are diagrammed. A. Tissue-specific selective pressures can promote the replication/survival of one mitochondrial allele over another. B. Muller’s ratchet. Because mitochondrial genomes do not recombine there is no mechanism to remove deleterious mutations from a mitochondrial genome lineage. Eventually all mitochondrial genomes may accumulate one ore more detrimental mutations (indicated by symbols).
Mitochondrial genomes do not recombine, or recombine at extremely low levels [164]. As a consequence, when a mitochondrial genome lineage acquires a new mutation there is no way for this mutation to be lost, a phenomenon called “Muller’s ratchet” [116]. Muller’s ratchet provides another mechanism for how mitochondrial DNA mutations might accumulate to levels sufficient to compromise cell function. Potentially each mitochondrial DNA lineage in the cell could accumulate a unique spectrum of mutations, such that no one mutation is present at high frequency, but most genomes have at least one or more detrimental mutations (Figure 6B). Because the mitochondrial genome contains a small number of genes encoding the ETC and translation components, these different mutations might often affect the same gene and/or the same process.
An increased load of mitochondrial mutations with age has been reported for Drosophila [165, 166], C. elegans [167], and mammals [12]. Experiments in mice suggest that most of this load results from expansion of mutations that arose during development, with a more minor contribution by new mutations arising during aging [106, 156]. The mitochondrial DNA mutator mouse has been particularly useful in investigating these relationships [168]. In this mouse the proofreading ability of the mitochondrial DNA polymerase is crippled, leading to greatly increased rates of mitochondrial mutation. Using this mouse both inherited mitochondrial mutations (heteroplasmy) and somatic mitochondrial mutations have been shown to contribute to aging phenotypes [106].
The total abundance of mitochondrial DNA in human tissues is also emerging as a marker for metabolic disease [169, 170] and cancer [171, 172], including sex-specific associations [173–175]. While there are some conflicting reports and technical issues associated with measurement of mitochondrial DNA, the data suggest that mitochondrial DNA copy number is mis-regulated in human metabolic disease and cancer, involving increases or decreases in abundance of intact genomes, as well as accumulation of damaged and deleted molecules [176]. In addition to promoting inflammation [111], mitochondrial DNA fragments are emerging as potential mediators of nuclear DNA damage during aging [155, 177, 178].
Sex-specific mitochondrial maintenance and aging
Additional observations support a role for sexual differentiation in contributing to mitochondrial maintenance failure during aging. In particular, mitochondrial function and mitochondrial regulatory pathways, including autophagy and apoptosis, show sex-specificity at the level of animals, tissues and cells. Life span typically differs between males and females, for example, human females have greater life expectancy than males [179]. Metabolic regulation also differs, for example, men have greater basal metabolic rate, whereas women are more sensitive to insulin with regard to glucose metabolism in muscle and liver [141]. Aging-associated diseases also show sex bias. For example, men have greater cardiovascular disease, cancer and stroke, whereas women have greater autoimmune disease and osteoporosis [180–185]. Genetic and environmental life span interventions are usually sex-biased in their effects in both invertebrates and mammals [18, 131, 135, 186–188]. Notably, both dietary restriction and reduced insulin-like signaling cause greater life span increase in females in both Drosophila and mice. Experimental studies have begun to hint at possible cellular mechanisms underlying sex-dimorphism in stress resistance and aging, including sex-specific regulation of autophagy and apoptosis pathways.
Estrogen and testosterone are sex-specific regulators of mammalian stress response
Both estrogen receptor (ER) and androgen receptor (AR) are found associated with the mammalian mitochondria. Estrogen is most often reported to be anti-apoptotic in both muscle and neural tissues upon stress [180, 189]. For example, estrogen is cardioprotective in mouse ischemia-reperfusion model in ovariectimized females [190], and can also reduce infarct in males [191]. In mouse skeletal muscle cell line subjected to oxidative stress, the estrogen receptor is implicated in inhibiting CASPASE 3, as well as activating a p38 MAPK/PI3K/AKT signaling cascade that prevents BAD activation and apoptosis [189]. In neonatal rat cardiomyocytes the protective effect of estrogen was associated with p38beta MAPK activation and phosphorylation of mitochondrial MnSOD [192]. In mice, estrogen was found to be protective for female cardiomyocytes in vivo and in vitro through estrogen receptor and PI3K-dependent pathways [193]. In cultured adult mouse cardiomyocytes, female cells had greater levels of phosphorylated AKT at baseline and in response to stress, with consequently reduced apoptosis, perhaps to due to greater abundance of ER-alpha [194]. The results for testosterone in heart and muscle cells are more mixed [180]. For example, testosterone could decrease infarct in male mice subjected to ischemia-reperfusion [195]. Testosterone was also antiapoptotic in mouse skeletal muscle cell line subjected to oxidative stress, and the targets for testosterone benefit appeared different from estrogen, implicating HSP90 translocation and reduced BAX [196]. In contrast, testosterone was pro-apoptotic in assays of cultured rat myocytes [180]. One explanation may be that the beneficial effects of testosterone are observed preferentially in male cells, such as recently shown for rat pancreatic cells [197]. In nervous tissue the effects of testosterone are more consistently positive, with implication of MAPK and AKT signaling [189].
Taken together, the data demonstrate that mammalian sex-steroids are powerful regulators of stress response at the level of tissues and individual cells. Both estrogen and testosterone are implicated in activation of MAPK/PI3K/AKT/TOR signaling pathways (Figure 5, indicated in orange) [198–201]. One interpretation is that male and female cells are adapted to physiological levels of the corresponding sex-specific hormones for near-optimal stress response in the intact animal, and that under specific conditions these hormones can have either positive or negative effects [202]. For example, through their ability to promote IIS and TOR signaling these mammalian steroids may promote growth and sexual differentiation at the expense of mitochondrial turnover and maintenance important for longevity (Figures 1, 2, 5) [199, 200]. Notably, circulating steroid hormones are also implicated in regulating sex differences in aging in C. elegans and Drosophila [203–205].
Sex-specific autophagy and apoptosis in mammalian cells and tissues
One theme that has emerged is that in mammals female cells are generally more resistant to stress than are male cells. This might be related to the better regulation of mitochondrial functions predicted by the evolutionary theories discussed above. In vivo and in vitro studies have characterized sex-specific differences in stress resistance of specific tissues, including the heart and the brain.
The heart and cultured cardiomyocytes show better stress response in mammalian females. In humans, women show less apoptosis and less maladaptive remodeling relative to men in response to acute coronary ischemia [206]. Under normal conditions men’s hearts have increased expression of CARBONIC ANHYDRASE 3, a gene associated with hypertrophy and heart failure, and decreased expression of APOJ/CLUSTERIN, an autophagy regulator thought to be protective upon inflammatory injury [207]. Studies of human cells and rodent models indicate that estrogen is generally protective but may not be responsible for all the sex differences in heart stress responses.
Autophagy appears to be important in mediating heart stress response. Most studies indicate that autophagy limits myocyte death during acute ischemia-reperfusion and improves subsequent heart function, as shown for mouse myocyctes in vitro and in vivo using inhibitors of autophagy pathways [208]. However, certain studies have reported negative effects of autophagy [180]. For example, upon ischemia-reperfusion, inhibition of Beclin1 reduced rat cardiomyocyte cell death [209], and Beclin1 mutant mice had reduced infarct [210]. One suggestion is that autophagy may be protective during ischemia but detrimental during reperfusion [210]. Autophagy also appears to be important in mediating sex difference in heart stress response. In adult mouse heart, fasting caused greater activation of AKT and AMPK signaling and greater glycogen accumulation in females than in males, consistent with greater autophagy pathway activation upon stress in females [211]. In rat, greater basal autophagy markers were observed in male heart, associated with greater protein carbonyl content and possibly indicating greater baseline oxidative stress [212]. However, in response to acute ischemia-reperfusion, female rat heart showed greater autophagy markers than male, associated with smaller infarcts and fewer apoptotic cells [213].
Additional tissues show greater resistance to stress in mammalian females. In humans, peripheral blood mononuclear cells isolated from women were more resistant to radiation-induced apoptosis than were those from men [214]. In mice, females were more resistant to ischemia in the liver than were males [215], and similarly, rat females were more resistant to post-ischemia renal failure than were males [216]. Also, vascular smooth muscle cells isolated from male rats preferentially underwent apoptosis in response to UVB stress, whereas cells from females exhibited greater autophagy markers and increased survival [217].
The brain and cultured neurons show striking differences in stress response between males and females (summarized in Figure 7). In rodents, nutrient starvation killed neurons and fibroblasts isolated from males to a greater extent than those from females, and the increased survival of female neurons was associated with accumulation of triglycerides and lipid droplets [218]. Interestingly, inhibiting autophagy rescued male neurons but increased the death of male fibroblasts. In mice subjected to moderate hypoxia-ischemia, females were more resistant to brain injury, and in neonates, males showed preferential AIF translocation whereas the females showed preferential CASPASE 3 activation [219]. Similarly, in rat neonates, ischemia caused greater CASPASE 3 activation in female brain relative to males, and females had greater basal levels of autophagy markers [220]. Consistent with greater stress sensitivity in male brain, a mutation of the mouse Dual endothelin-1/VEGF signal peptide-activated receptor (DEspR) caused autophagic neuronal cell death in vivo specifically in males, perhaps through altered TOR pathway signaling [221].
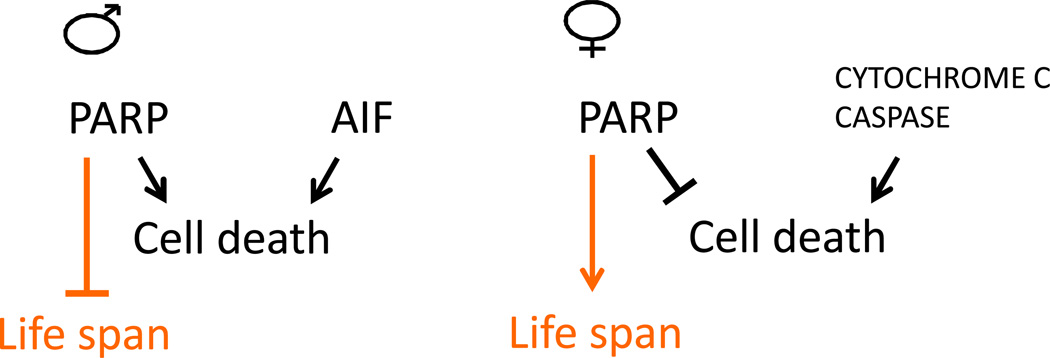
Figure 7.
Sex-specific regulation of cell death in response to stress in neurons. In male mouse neurons, cell death is promoted by PARP and AIF, and is independent of caspase activity (indicated in black). In female mouse neurons, cell death is promoted by CYTOCHROME C and caspase, and is inhibited by PARP activity (indicated in black). In Drosophila, over-expression of PARP in nervous tissue increases life span in females, and decreases life span in males (indicated in orange).
Studies of mouse brain ischemia reveal mechanistic differences in neuronal cell death between males and females. In females cell death is associated with CYTOCHROME c release from mitochondria, and is caspase-dependent, whereas death of male cells is associated with AIF release from mitochondria, and is caspase-independent (Figure 7, indicated in black) [222]. Moreover, in females PARP is protective, whereas in males cell death is induced by PARP [223, 224]. Consistent with this conclusion, in Parp-1 knockout mice the infarct in males is reduced whereas in females the infarct is more severe [225–227]. Both NAD+ depletion and PAR polymer formation may be toxic events during mouse ischemia [228]. Nicotinamide supplementation blunted the loss of NAD+ levels upon stress, and reduced infarct in wild type males, and Parp-1 knockout mice of both sexes, but had no effect in wild type females [226], consistent with the negative effect of PARP in males. Intriguingly, over-expression of Parp in adult Drosophila nervous tissue decreased life span in males and increased life span in females [229], suggesting a possible conservation of mechanisms (Figure 7, indicated in orange).
The data indicate that mammalian gonadal sex hormones such as estrogen and testosterone play an important role in mediating sex-specific differences in stress resistance and disease phenotypes. However, gonadal sex hormones are not required for all the sex differences in mammalian cellular stress resistance. In the female mouse brain the X-linked inhibitor of apoptosis protein (XIAP) gene showed greater expression at baseline, and greater reduction upon ischemia than in males, consistent with preferential activation of CASPASE 3, and these differences were independent of the presence of gonads and estrogen supplementation [222]. In addition, the differences in rodent brain stress resistance are observed in neonates, as well as in cultured cells [230–234]. Sex-specific differences in susceptibility to apoptosis were apparent in mouse embryonic cells where female cells were more sensitive to apoptosis induced by ethanol and camptothecin [235, 236]. Moreover, in cultured mammalian embryos females had better survival in response to heat and oxidative stress, and this correlated with increased expression of several X-linked genes. These genes encode GLUCOSE-6-PHOSPHATE DEHYDROGENASE (G6PDH), which generates reducing equivalents critical for oxidative stress resistance, and XIAP, which inhibits caspase activity [237]. These results suggest that there are inherent differences in stress resistance of male and female mammalian cells independent of gonadal sex hormones, and that these differences correlate with differences in X-linked gene expression. Studies of mice with altered sex chromosome content further support this conclusion.
Sex chromosome effects and escape from X-inactivation
Sophisticated manipulation of mouse sex chromosomes combined with optional removal of gonads reveals effects of the sex chromosome complement (i.e., number of X chromosomes and presence/absence of Y) independent of the gonads and gonadal sex hormones [238]. These phenotypes may or may not involve sex-specific hormones produced by tissues other than the gonads, such as fat or liver. For example, the X chromosome complement but not the Y chromosome had an effect on body weight [238]. Notably, X/X cells had greater expression of Akt1, Akt3 and several other genes relative to X/Y cells, and these changes were hypothesized to result from escape from X-inactivation and/or effects of sex chromosomes titrating chromatin factors away from the autosomes [239]. Strikingly, X/X mice were more sensitive to heart ischemia-reperfusion and apoptosis than were X/Y mice, (i.e., opposite to the observations in gonad-intact mice). The effect was due to the presence of two X chromosomes as opposed to the absence of the Y, and was associated with greater expression in the female heart of several genes that escape X-inactivation, including Eif2s3x, Kdm6a, Kdm5c, and Usp9x [240]. Eif2s3x encodes a translation initiation factor and Kdm5c and Kdm6a encode histone demethylases that could potentially affect expression of numerous autosomal genes. Interestingly, Usp9x encodes a ubiquitin-specific protease implicated in cell death pathways and negative regulation of mTOR. The different effect of sex chromosome composition in the presence/absence of gonadal sex hormones underscores the complexity and compensatory nature of sex-specific factors, consistent with the idea that males and females have evolved distinct regulatory networks [238].
Many genes escape from X-inactivation in humans and mice during development and in the adult, and the degree of expression depends upon the tissue, the stage of development and the age of the animal [241, 242]. Escape from X-inactivation is implicated in disease, including cancer and autoimmune disease. In mice, inactivation of Xist in the adult hematopoietic system caused cancer and lethality [243]. In humans, escape from X-inactivation is implicated in increasing expression of O-LINKED N-ACETYLGLUCOSAMINE TRANSFERASE (OGT) and the CD40 ligand CD40LG. OGT is a nutrient-sensitive chromatin modifier with effects on immune function and metabolism [244] and CD40LG is an immune signaling molecule [245], and increased expression of these genes could conceivably predispose women to autoimmune disorders. Numerous genes that regulate autophagy, apoptosis and metabolism are located on the human X chromosome and escape from X-inactivation to differing degrees, including G6PD, XIAP and LAMP2 [246]. Interestingly, in C. elegans, dosage compensation is required for regulation of X-linked regulators of IIS including pdk-1 and akt-2, and for normal dauer arrest and longevity [247–249]. The data from mammals indicate that the degree of escape from X-inactivation varies between tissues and between individuals for each gene [246].
In summary, the preferential resistance of female mammals to several types of stress is recapitulated at the level of cells and tissues, and correlates with sex-specific regulation of mitochondrial pathways including autophagy and apoptosis. These results might be compatible with the evidence for increased p53 activity in females, in that p53 can positively regulate autophagy through transcriptional activation, and can negatively regulate autophagy through direct effects [250]. One general theme that emerges is that female cells show relatively greater resistance to stress mediated by female-specific hormones and cell-autonomous effects including X-linked gene activation. The data suggest that females take advantage of the presence of two copies of critical X-linked genes, and a female state of dynamic DC to achieve expression of these genes across a greater dynamic range than is possible in males. This additional level of dynamic X-linked gene expression and regulation may allow females to more effectively regulate mitochondrial function and stress resistance. At the same time, the variable escape of genes from X-inactivation is implicated in disease, including aging-related disease [244].
Hormesis and mitochondrial maintenance failure in aging
Conditioning hormesis refers to the phenomenon where a mild stress treatment protects against a subsequent and more severe stress treatment [251]. For example, pre-treatment of mammalian cells, Drosophila flies and C. elegans worms with a mild oxidative stress protects the cells and animals from the lethal effects of a subsequent and more severe oxidative stress [252, 253]. Mild stress treatments including oxidative stress, heat and ionizing radiation can also sometimes lead to increased animal life span, a phenomenon also referred to as hormesis [254–256]. Hormesis is generally thought to result from the up-regulation of stress response genes, including proteolytic systems and Hsps [54, 257–260]. Several interventions that can increase life span have been found to involve mitochondrial stress. Because mitochondrial malfunction and mitochondrial stress are observed during normal aging, these observations suggest a hormesis-type mechanism where mitochondrial stress applied early in life protects against mitochondrial stress and failure during aging [9, 261, 262].
The autophagy/life span paradox
When autophagy is inhibited during animal development there are negative consequences for tissue structure and function and the viability of the adult animal [263–265]. In contrast the role of autophagy in longevity of the adult animal is less clear. Several interventions that can increase life span in adult C. elegans, including dietary restriction, p53 mutation, TOR pathway inhibition, germ-line ablation, and mitochondrial gene mutants have been found to be dependent upon autophagy, in that coincident inhibition of autophagy prevents the life span increase [266–270]. Similarly, interventions that can extend life span in adult Drosophila including the TOR pathway inhibitor rapamycin [271] and spermidine [272] were reported to require autophagy. These observations indicate that increased autophagy is part of the mechanism(s) for increased life span, and by implication, that autophagy might be a rate-limiting process for adult longevity. However, when conditional RNAi was used to inhibit autophagy in otherwise normal adult C. elegans there was no reduction in life span, indicating that autophagy is not normally rate-limiting for adult C. elegans life span [268, 270]. Similarly, when conditional RNAi was used to inhibit autophagy in adult Drosophila, both starvation resistance and immune function were reduced, yet there was no reduction in life span, indicating that autophagy is not normally rate-limiting for adult Drosophila life span [271, 273]. Over-expression of the Drosophila Atg8a gene in nervous tissue [274] and muscle tissue [275] has been reported to increase adult autophagy levels and increase life span, however these interventions were not specific to the adult stage, and the life span changes might be due to effects in addition to altered adult autophagy. Therefore, the paradox is that autophagy is required for many (and possibly all) interventions that increase life span in the adult [268], yet autophagy is not normally rate-limiting for adult life span.
One suggestion for how to reconcile these seemingly conflicting observations is that increased adult autophagy is not sufficient for life span extension unless accompanied by up-regulation of biosynthetic pathway(s) to direct the appropriate utilization of the liberated materials [270]. This would imply that under normal conditions some pathway other than autophagy is rate-limiting for adult life span, possibly a biosynthetic pathway such as purine biosynthesis or mitochondrial biogenesis. An alterative explanation is that autophagy might be toxic in the oldest animals, such as part of an autophagic cell death pathway. Intriguingly, inhibition of Atg7 by RNAi in adult fly epidermis delayed aging-related changes [276], suggesting that autophagy might contribute to aging of this tissue. Autophagy is also implicated in neuronal cell death during aging [277]. In this scenario the acute activation of autophagy earlier in adult life is beneficial through a hormesis-type mechanism that dis-favors subsequent autophagic cell death during aging, perhaps through a mechanism such as selective degradation of non-optimal mitochondrial genomes [278] (Figure 2), even though these genomes are not yet life span-limiting. Finally, it is conceivable that inducing autophagy in a tissue where it is not life span-limiting and where RNAi is effective, could send a signal to induce autophagy in a tissue where autophagy is life span-limiting but RNAi is not effective, such as certain neurons [279, 280]. Continued investigation of the relationship between tissue-specific autophagy regulation, hormesis and life span will be an important area for future research.
The mitochondrial stress response and unfolded protein response (UPRmt)
A stress response in the mitochondria involving a retrograde signal to the nucleus was characterized in yeast involving activation of the RTG transcription factors and induction of genes in the glutamate biosynthetic pathway [4, 5]. As the source of nitrogen for biosynthetic pathways, maintenance of glutamate levels appears critical to maintain viability in respiration-deficient cells. Retrograde stress responses have since been characterized for Drosophila, mammals and C. elegans [281, 282]. The metazoan mitochondrial stress response is characterized by induction of specific Hsp genes in the nucleus and the targeting of the Hsps to the mitochondria, and has several links to mitochondrial maintenance failure during aging. For simplicity all mitochondrial stress responses resulting in the induction of nuclear Hsp genes and the targeting of the Hsps to the mitochondria are referred to here as the mitochondrial unfolded protein response (UPRmt), however, as discussed below, there appears to be more than one type of UPRmt.
The Drosophila small Hsp called Hsp22 is a member of the alpha-crystallin family found in all metazoans [54, 55]. Drosophila Hsp22 is induced in response to heat stress and oxidative stress [14], and is targeted to the mitochondrial matrix [283], indicating a UPRmt. Hsp22 is also up-regulated during normal adult aging, and shows one of the largest aging-related increases known for a eukaryotic protein (>150 fold) [51, 52]. When Drosophila strains were genetically selected for increased life span, they were found to show increased Hsp22 expression during the first half of adult life, suggesting that Hsp22 in young flies might be beneficial for life span [284], possibly through a hormesis mechanism. Consistent with this idea, experimentally up-regulated expression of Hsp22 is reported to increase life span [285]. In contrast, high-level over-expression of Hsp22, particularly at late ages, may be toxic [286], and the time course of Hsp22 induction in aging flies is a biomarker of remaining life span [287].
The Drosophila mitochondrial ribosomal protein S12 is encoded by the gene tko, and is required for translation in the mitochondria [288]. A partial-loss-function mutation (tko[25t]) causes disrupted mitochondrial ribosomal structure and ETC deficiency, and a dramatic up-regulation of Hsp22 [289]. These studies identify Hsp22 as a robust marker of mitochondrial stress and aging in Drosophila, and suggest that Hsp22 is induced in response to UPRmt. A UPRmt has been characterized in mammalian cells [290], C. elegans [291, 292] and Drosophila [278, 293] involving up-regulated expression of mitochondrial Hsp60 and mitochondrial Hsp70, and in C. elegans the retrograde signal was shown to require the signaling gene ubl-5. Based on the similarities, the induction of Drosophila Hsp22 by mitochondrial protein folding disruption and by aging is hereafter referred to as the UPRmt, similar to the UPRmt characterized by induction of mitochondrial Hsp60 and mitochondrial Hsp70, however it remains possible there is more than one UPRmt pathway for mitochondrial Hsp induction.
The Drosophila protein Ref(2)P is the homolog of mammalian p62, a conserved protein implicated in marking mitochondria with UPRmt for autophagy [278, 294–297]. Drosophila Ref(2)P is induced in response to stresses that cause UPRmt, and is also induced during normal aging, consistent with an aging-associated UPRmt [9, 14, 17, 294, 298, 299].
In Drosophila muscle tissue, the over-expression of foxo, the IIS inhibitor Pten, and the foxo target 4E-BP could each stimulate autophagy, suggesting that reduced IIS and consequent foxo protein activation may normally stimulate autophagy [298]. In C. elegans, reduced IIS and FOXO activation cause up-regulated expression of mitochondrial MnSOD, and a retrograde ROS signal is implicated in this response, sometimes called “mitohormesis” [262, 300]. Some evidence indicates that mitohormesis does not involve the UPRmt [301]. However, given that the mitohormesis response induces mitochondrial MnSOD, and that over-expression of mitochondrial MnSOD can induce the UPRmt and increase life span in both Drosophila [302, 303] and C. elegans [304], it is tempting to speculate that these mitochondrial stress responses may be related. Life span extension in C. elegans can also be induced by paraquat and this involves a retrograde ROS signal that requires a mitochondrial MnSOD as well as the conserved caspase-dependent intrinsic apoptosis pathway [305]. This caspase-dependent ROS signaling is also proposed to be distinct from the UPRmt [306]. Therefore there may be a life span-extending mechanism involving a retrograde ROS signal that is distinct from the UPRmt, however, because both ROS and the UPRmt are up-regulated during normal aging, each of these life span-extending interventions may be examples of hormesis [9].
In support of the idea of more than one mitochondrial stress response pathway that can increase life span, certain factors required for induction of the UPRmt in C. elegans are specific for the particular type of mitochondrial stress [307–311]. Moreover, induction of a C. elegans UPRmt using constitutively active alleles of the transcription factor ATFS-1 failed to increase life span, indicating that not all UPRmt can increase life span [312]. These results support the idea that there is more than one type of retrograde mitochondrial stress signal that can increase life span, and the beneficial effects may be limited to specific conditions such as the nature of the stress, and the age and sex of the animal.
Recently numerous life-span extending interventions have been found to be associated with and/or require the UPRmt. In both Drosophila and C. elegans, RNAi inhibition of certain genes encoding ETC components can increase life span [7, 313–315]. This result was initially interpreted to suggest that life span increase might be due to decreased metabolic activity. However, subsequent analysis in C. elegans revealed that this intervention induces the UPRmt and requires the activity of ubl-5 for life span increase [313]. In both Drosophila and C. elegans, over-expression of the mitochondrial enzyme MnSOD can increase life span, and this was found to cause up-regulated expression of mitochondrial Hsps associated with the UPRmt including Hsp22 and Hsp60 [303, 304, 316]. RNAi knockdown of ETC components in the Drosophila muscle tissue induced the UPRmt and increased life span and muscle function with age, and coincident knockdown of UPRmt genes (Hsp60C or Clpx) or autophagy genes blocked the life span increase [317]. Notably, the UPRmt in the Drosophila muscle caused expression and secretion of ImpL2 (an insulin-binding protein homolog), and this was required for life span extension [317], indicating that signaling to some other tissue is required for increased life span. ImpL2 overexpression increases life span suggesting a mechanism of reduced IIS [318]. In mice a genome-wide association study of life span implicated the mitochondrial ribosomal protein S5 gene (Mrps5), and Mrps5 gene expression levels were negatively correlated with mouse strain life span [261]. Moreover, analysis in C. elegans confirmed that knockdown of the homologous mitochondrial ribosomal subunit gene caused the UPRmt and a life span increase dependent upon ubl-5. Experiments in C. elegans also implicate the UPRmt in the life span extending effects of the drugs rapamycin and resveratrol as well as manipulation of NAD(+) levels and SIRTUIN activity [261, 319]. Finally, several additional interventions reported to increase mouse life span, including mutations of ETC components p66SHC [320] and SURF1 [321, 322], and ectopic targeting of cytoplasmic catalase to the mitochondria [323], may be causing protein folding stress in the mitochondria and a beneficial UPRmt.
Dynamic interactions between mitochondria and nuclear chromatin state
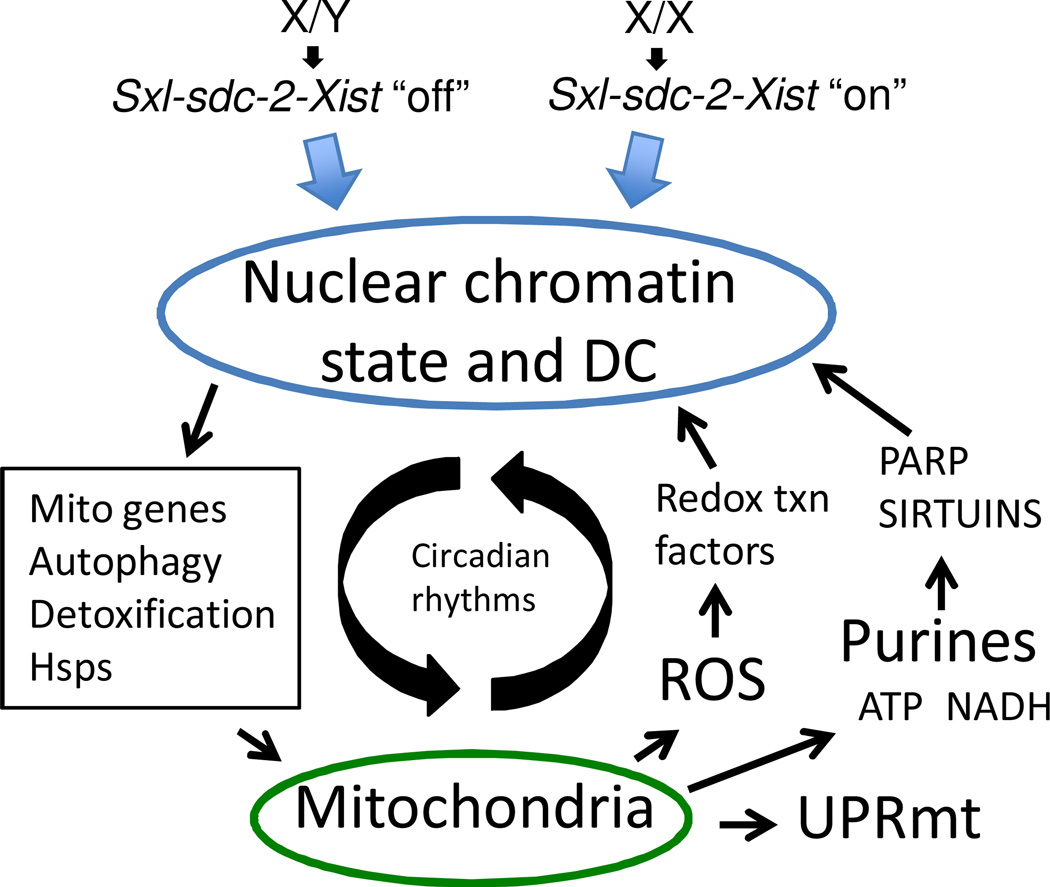
In further support of a hormesis model, it is noteworthy that most life span interventions involve a retrograde mitochondrial-to-nuclear signal that is up-regulated or altered during normal aging: UPRmt, ROS, and purine metabolism (Figure 8). Basal levels of UPRmt signaling may be part of the normal regulation of mitochondrial turnover [41, 317], and studies indicate increased UPRmt during normal aging [9, 14, 17, 298, 316]. Similarly, retrograde ROS signaling is implicated in regulating normal metabolic cycles [324] and mitochondrial turnover [41], and extensive literature documents increased ROS with aging [47]. Finally, purines including ATP and NADH are important retrograde signals for circadian gene expression [324, 325]; Drosophila studies implicate nucleotide levels in compensating for mitochondrial malfunction [293], and all the genes of the purine biosynthetic pathway are up-regulated during normal aging [14, 17]. These cyclical retrograde signals generated by mitochondrial metabolism are thought to be involved in the regulation of dynamic chromatin states in the nucleus [324–326], and this may include DC and X-linked gene expression (Figure 8). Consistent with this idea, in C. elegans, TOR signaling has recently been found to regulate DC [249], and in turn DC has been found to regulate IIS through alterations in X-linked gene expression [247].
Figure 8.
Nuclear chromatin state and metabolic circadian rhythms in mitochondrial maintenance. Chromosomal sex and the Switch-Gene on/off state regulate nuclear chromatin state, dosage compensation (DC) and gene expression. The nucleus regulates mitochondrial maintenance through expression of genes for mitochondrial biosynthesis and turnover (autophagy/mitophagy), detoxification and Hsps. The mitochondria send retrograde signals to the nucleus including ROS, UPRmt (including peptides, not shown), and purines, including ATP and NADH. Redox-sensitive transcription factors, PARP and SIRTUINS regulate nuclear chromatin state in response to mitochondrial ROS and purines.
Autophagy gene expression and the autophagy pathway are circadian-regulated in human heart [327], liver [328], skeletal muscle [329], and brain [330], and in the metabolic rhythms of yeast [331]. The Sirtuins are conserved NAD-dependent protein deacetylases that act in the nucleus to regulate chromatin state and circadian gene expression [325]. Interestingly, both ER-alpha and the Sirtuin SIRT3 have been implicated in mediating a retrograde UPRmt signal in human breast cancer cells [332]. In C. elegans, the Sirtuin SIR-2.1 and NAD were found to promote longevity by inducing a favorable UPRmt [319]. By responding to mitochondrial signals including NAD, ROS and UPRmt the Sirtuins may regulate circadian rhythms of autophagy to promote optimal mitochondrial turnover and homeostasis, to the benefit of longevity [333].
Tissue-specific and cell-specific consequences of mitochondrial failure in aging
Recent studies support the conclusion that mitochondrial maintenance failure is a common feature of aging, and has tissue-specific and cell-specific phenotypes. Aging phenotypes vary between individuals and between different tissues in the same animal in Drosophila, mammals and C. elegans with regard to tissue deterioration and changes in gene expression [51, 334–337]. Recently the induction of Hsp22 during aging in Drosophila oenocytes was found to vary dramatically between different cells, and between different developmental cell lineages. These variegated patterns suggest a heritable event during developmental cell divisions that predisposes a particular cell lineage to more rapid aging-associated UPRmt, such as possibly a mitochondrial mutation [316]. Oxidative stress is associated with aging in multiple tissues [47], for example mammalian skeletal muscle [338], and flight muscle in Drosophila [51, 339], whereas reductive stress is implicated in inherited cardiomyopathies involving mutations in small Hsps and potentially in ischemia [340]. The aging-like phenotypes of the mitochondrial mutator mouse support a causative role for mitochondrial mutations in aging, however previously the phenotypes identified did not always indicate increased oxidative stress [341–343], which is typically associated with mammalian aging. Recent results suggest that the consequences of the mitochondrial mutations are tissue-specific and that indeed the mutator mouse has increased markers of oxidative damage in muscle tissue [344, 345].
The liver may be a particularly important target tissue for life span interventions, in particular the UPRmt [9, 346]. In Drosophila both MnSOD over-expression and Hsp22 over-expression caused up-regulation of Hsp22 preferentially in the oenocytes [316], which are the Drosophila liver-like cells [347–349]. In C. elegans life span extension required UPRmt in the gut [313], which is thought to be a liver-like tissue. Finally, in mammals, the liver mitochondria exhibit characteristic changes in response to DR [350], and a hepatocyte cell line was particularly sensitive to induction of UPRmt [261]. As a central regulator of lipid metabolism, liver may favor life span by generating and mobilizing fat reserves that favor long-term survival [269]. A related possibility is that liver limits life span through the production of hormones and toxic metabolites (Figure 2). Finally, liver may be especially sensitive to mis-regulation of mitochondria and autophagy during aging [328, 346]. For example, accumulation of age pigment is observed in the mammalian liver as well as in the Drosophila oenocytes [316, 351–353]. Further investigation of tissue-specific, cell-lineage specific, and cell-specific causes and outcomes for mitochondrial failure during aging should be an important area for future research.
A common mechanism for metazoan life span interventions?
Because the UPRmt and autophagy are implicated in the mechanism of several genetic and pharmacologic life span interventions, and because the UPRmt is also observed during normal aging, it suggests a possible conserved hormesis mechanism. The common result may be increased autophagy and the breakdown and replacement of abnormal mitochondria, including the preferential destruction of mutated mitochondrial genomes. The related possibility is that the UPRmt reduces the production of one or more hormones or toxic metabolites by the liver mitochondria. The hormones might promote sexual differentiation and reproduction at the expense of longevity (Figures 1, 2).
Extensive data indicates that hormones that promote sexual differentiation can decrease life span. IIS shortens life span and promotes sexual differentiation in both invertebrates and mammals [144, 354–356] [120, 357–360], including the production of sex-specific hydrocarbons by the Drosophila liver-like cells [361]. Ample precedent for the negative effects of hormones comes from the fact that factors produced by the gonads can shorten life span in both C. elegans [362] and Drosophila [363, 364]. The Drosophila steroid hormone ecdysone and the terpene hydrocarbon hormone juvenile-hormone promote growth and sexual differentiation and also decrease life span [203, 365–367]. The Drosophila gene takeout encodes a lipophilic hormone-binding protein, and overexpression of takeout was reported to reduce sex-specific behaviors and increase life span [368], consistent with a negative effect of hormones on life span.
Not all hormones decrease life span. Hormones can sometimes inhibit sexual differentiation and promote life span, typically by opposing the effects of other hormones. For example, in C. elegans, the bile-acid-like steroid hormone dafachronic acid can either positively or negatively regulate life span depending on genetic background, dietary environment and signaling state of the animal [369–373]. In mice, fasting causes the liver to produce Fibroblast growth factor-21, which in turn can inhibit GH/IIS signaling, decrease reproduction [374] and increase life span [375]. In Drosophila, the neuropeptide adipokinetic hormone is a Drosophila equivalent of glucagon that acts in opposition to IIS. Adipokinetic hormone promotes mobilization of lipid stores and is reported to increase life span [376–378].
Several observations are consistent with a model where metazoan life span interventions act by reducing the production of sexual-differentiation hormones and toxic sex-specific metabolites by the mitochondria. The mitochondria are essential for the production of steroid hormones [379]. By producing hormones that promote sexual differentiation, the mitochondria in the liver-like cells and gonads could ultimately lead to their own demise by causing trade-offs and SAP throughout the tissues of the body. Notably, in C. elegans, inhibiting mitochondrial function early in life was beneficial [380, 381], whereas inhibiting mitochondrial function later in life was not, consistent with the possibility that mitochondria produce compounds that ultimately lead to their own demise. The Drosophila mitochondrial ribosomal protein S12 gene mutation tko not only causes UPRmt, but also reduces expression of sex-specific genes and sex-specific behaviors [289], consistent with a role for the mitochondria in promoting adult sexual differentiation.
Increasing evidence implicates liver-like cells as an important source of life-span limiting sexual-differentiation hormones and toxic sex-specific metabolites. Increased life span in the Ames dwarf mouse strain is associated with loss of sex-dimorphic gene expression in the liver [382], indicating reduced sex-specific liver metabolism. In Drosophila, up-regulation of MnSOD in young animals can extend fly life span, and also induces UPRmt markers and reduces the accumulation of age pigment in the liver-like oenocytes, suggesting reduced or altered oenocyte metabolism [316]. The oenocytes also produce sex-specific hydrocarbons that mark fly sexual identity [348, 349, 361] and pheromones that reduce fly life span [86], and it is likely that production of these compounds is also reduced by the UPRmt.
In conclusion, it can be argued that most of the life span-extending interventions in metazoans can be interpreted in terms of a mechanism involving hormesis, and in particular the liver UPRmt. Non-exclusive possibilities for the mechanism of increased life span include production of new and better-functioning mitochondria, altered DC, and the reduced production of hormones and toxic metabolites.
Implications for sex-specific interventions in human aging and disease
The increasing understanding of mechanisms for sex-dimorphic stress responses and mitochondrial maintenance may lead to improved interventions for human age-related diseases based on sex. For example, diabetes and metabolic syndrome increase the risk for heart attack to a greater extent in women than in men [383], underscoring the importance of dealing with these issues in female heart disease [384]. Sex-specific steroids may decrease stroke incidence in the corresponding sex, and possible sex-specific interventions are currently under study [185, 385]. Sex-dimorphic regulation of human immune response includes greater regulation by the IFNy cytokine in males and greater regulation by the IL6 cytokine in females [386–388], suggesting possible sex-specific targets for interventions in immune disorders and the inflammation associated with aging [389, 390]. Drugs currently used to treat heart disease have different efficacy and side effects in women compared to men, perhaps related to the sex-dimorphism in regulation of autophagy and apoptosis discussed above, and this makes sex an important consideration when designing treatment regimens [391]. The NIH has recently strengthened rules for including both sexes in biomedical research [392]. It appears likely that our increasing understanding of sex-specific regulation of stress responses and mitochondrial maintenance will continue to lead to improved interventions in human aging and disease.
Highlights.
Gene expression changes during aging indicate mitochondrial maintenance failure
Sexual differentiation may promote mitochondrial maintenance failure during aging
Sexual differentiation, autophagy and dosage compensation regulate stress resistance, mitochondrial maintenance and aging
Life span interventions in metazoans may involve hormesis and inhibition of sexual differentiation by liver UPRmt
Acknowledgements
Thank you to Laura Corrales-Diaz Pomatto and the anonymous reviewers for comments on the manuscript. This work was supported by a grant from the Department of Health and Human Services, National Institute on Aging (AG011833) and by pilot project funding from the Southern California Environmental Health Sciences Center (5P30ES007048).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parrella E, Longo VD. The chronological life span of Saccharomyces cerevisiae to study mitochondrial dysfunction and disease. Methods. 2008;46:256–262. doi: 10.1016/j.ymeth.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Osiewacz HD, Brust D, Hamann A, Kunstmann B, Luce K, Muller-Ohldach M, Scheckhuber CQ, Servos J, Strobel I. Mitochondrial pathways governing stress resistance, life, and death in the fungal aging model Podospora anserina. Ann N Y Acad Sci. 2010;1197:54–66. doi: 10.1111/j.1749-6632.2010.05190.x. [DOI] [PubMed] [Google Scholar]
- 3.Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and Chronological Aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jazwinski SM. The retrograde response: a conserved compensatory reaction to damage from within and from without. Progress in molecular biology and translational science. 2014;127:133–154. doi: 10.1016/B978-0-12-394625-6.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 6.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 7.Munkacsy E, Rea SL. The paradox of mitochondrial dysfunction and extended longevity. Exp Gerontol. 2014;56:221–233. doi: 10.1016/j.exger.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho J, Hur JH, Walker DW. The role of mitochondria in Drosophila aging. Exp Gerontol. 2011;46:331–334. doi: 10.1016/j.exger.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Tower J. Aging, MnSOD, and hormesis mechanisms converge on liver mUPR. Cell Cycle. 2013;12:3237–3238. doi: 10.4161/cc.26354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskalev AA, Aliper AM, Smit-McBride Z, Buzdin A, Zhavoronkov A. Genetics and epigenetics of aging and longevity. Cell Cycle. 2014;13:1063–1077. doi: 10.4161/cc.28433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environmental and molecular mutagenesis. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 12.Sevini F, Giuliani C, Vianello D, Giampieri E, Santoro A, Biondi F, Garagnani P, Passarino G, Luiselli D, Capri M, Franceschi C, Salvioli S. mtDNA mutations in human aging and longevity: Controversies and new perspectives opened by high-throughput technologies. Exp Gerontol. 2014 doi: 10.1016/j.exger.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 14.Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, Tavare S, Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 16.Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landis G, Shen J, Tower J. Gene expression changes in response to aging compared to heat stress, oxidative stress and ionizing radiation in Drosophila melanogaster. Aging (Albany NY) 2012;4:768–789. doi: 10.18632/aging.100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tower J. Sex-specific regulation of aging and apoptosis. Mech Ageing Dev. 2006;127:705–718. doi: 10.1016/j.mad.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi A, Philpott DE, Miquel J. Electron microscope studies on aging Drosophila melanogaster. 3. Flight muscle. J Gerontol. 1970;25:222–228. doi: 10.1093/geronj/25.3.222. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi A, Philpott DE, Miquel J. Electron microscope studies on aging Drosophila melanogaster. II. Intramitochondrial crystalloid in fat body cells. J Gerontol. 1970;25:218–221. doi: 10.1093/geronj/25.3.218. [DOI] [PubMed] [Google Scholar]
- 21.Miquel J, Economos AC, Bensch KG, Atlan H, Johnson JJE. Review of cell aging in Drosophila and mouse. Age. 1979;2:78–88. [Google Scholar]
- 22.Anton-Erxleben F, Miquel J, Philpott DE. Fine-structural changes in the midgut of old Drosophila melanogaster. Mech Ageing Dev. 1983;23:265–276. doi: 10.1016/0047-6374(83)90027-1. [DOI] [PubMed] [Google Scholar]
- 23.Sohal RS. Mitochondrial changes in the heart of Drosophila repleta, Wollaston with age. Exp Gerontol. 1970;5:213–216. doi: 10.1016/0531-5565(70)90040-9. [DOI] [PubMed] [Google Scholar]
- 24.Sohal RD. Mitochondrial changes in flight muscles of normal and flightless Drosophila melanogaster with age. J Morphol. 1975;145:337–353. doi: 10.1002/jmor.1051450307. [DOI] [PubMed] [Google Scholar]
- 25.Walker DW, Benzer S. Mitochondrial "swirls" induced by oxygen stress and in the Drosophila mutant hyperswirl. Proc Natl Acad Sci U S A. 2004;101:10290–10295. doi: 10.1073/pnas.0403767101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacktor B, Shimada Y. Degenerative changes in the mitochondria of flight muscle from aging blowflies. J Cell Biol. 1972;52:465–477. doi: 10.1083/jcb.52.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarze SR, Weindruch R, Aiken JM. Oxidative stress and aging reduce COX I RNA and cytochrome oxidase activity in Drosophila. Free Radic Biol Med. 1998;25:740–747. doi: 10.1016/s0891-5849(98)00153-1. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson M, Mockett RJ, Shen Y, Orr WC, Sohal RS. Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem J. 2005;390:501–511. doi: 10.1042/BJ20042130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubessay P, Garreau-Balandier I, Jarrousse AS, Fleuriet A, Sion B, Debise R, Alziari S. Aging impact on biochemical activities and gene expression of Drosophila melanogaster mitochondria. Biochimie. 2007;89:988–1001. doi: 10.1016/j.biochi.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Morrow G, Tanguay RM. Mitochondria and ageing in Drosophila. Biotechnol J. 2008;3:728–739. doi: 10.1002/biot.200800015. [DOI] [PubMed] [Google Scholar]
- 31.Hwang ES, Yoon G, Kang HT. A comparative analysis of the cell biology of senescence and aging. Cell Mol Life Sci. 2009;66:2503–2524. doi: 10.1007/s00018-009-0034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzetti E, Csiszar A, Dutta D, Balagopal G, Calvani R, Leeuwenburgh C. Role of mitochondrial dysfunction and altered autophagy in cardiovascular aging and disease: from mechanisms to therapeutics. American journal of physiology Heart and circulatory physiology. 2013;305:H459–H476. doi: 10.1152/ajpheart.00936.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzetti E, Calvani R, Cesari M, Buford TW, Lorenzi M, Behnke BJ, Leeuwenburgh C. Mitochondrial dysfunction and sarcopenia of aging: from signaling pathways to clinical trials. Int J Biochem Cell Biol. 2013;45:2288–2301. doi: 10.1016/j.biocel.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasuda K, Ishii T, Suda H, Akatsuka A, Hartman PS, Goto S, Miyazawa M, Ishii N. Age-related changes of mitochondrial structure and function in Caenorhabditis elegans. Mech Ageing Dev. 2006;127:763–770. doi: 10.1016/j.mad.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Regmi SG, Rolland SG, Conradt B. Age-dependent changes in mitochondrial morphology and volume are not predictors of lifespan. Aging (Albany NY) 2014 doi: 10.18632/aging.100639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Extermann M. Metabolic syndrome, aging, and cancer. Critical reviews in oncogenesis. 2013;18:515–529. doi: 10.1615/critrevoncog.2014010612. [DOI] [PubMed] [Google Scholar]
- 37.Guarner-Lans V, Rubio-Ruiz ME, Perez-Torres I, Banos de MacCarthy G. Relation of aging and sex hormones to metabolic syndrome and cardiovascular disease. Exp Gerontol. 2011;46:517–523. doi: 10.1016/j.exger.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amadoro G, Corsetti V, Florenzano F, Atlante A, Bobba A, Nicolin V, Nori SL, Calissano P. Morphological and bioenergetic demands underlying the mitophagy in post-mitotic neurons: the pink-parkin pathway. Frontiers in aging neuroscience. 2014;6:18. doi: 10.3389/fnagi.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 41.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golden TR, Hubbard A, Dando C, Herren MA, Melov S. Age-related behaviors have distinct transcriptional profiles in Caenorhabditis elegans. Aging Cell. 2008;7:850–865. doi: 10.1111/j.1474-9726.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmon AB, Richardson A, Perez VI. Update on the oxidative stress theory of aging: does oxidative stress play a role in aging or healthy aging? Free Radic Biol Med. 2010;48:642–655. doi: 10.1016/j.freeradbiomed.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 50.Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler JC, Bieschke ET, Tower J. Muscle-specific expression of Drosophila hsp70 in response to aging and oxidative stress. Proc Natl Acad Sci U S A. 1995;92:10408–10412. doi: 10.1073/pnas.92.22.10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King V, Tower J. Aging-specific expression of Drosophila hsp22. Dev Biol. 1999;207:107–118. doi: 10.1006/dbio.1998.9147. [DOI] [PubMed] [Google Scholar]
- 53.Calabrese V, Scapagnini G, Ravagna A, Colombrita C, Spadaro F, Butterfield DA, Giuffrida Stella AM. Increased expression of heat shock proteins in rat brain during aging: relationship with mitochondrial function and glutathione redox state. Mech Ageing Dev. 2004;125:325–335. doi: 10.1016/j.mad.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 54.Tower J. Hsps and aging. Trends Endocrinol Metab. 2009;20:216–222. doi: 10.1016/j.tem.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tower J. Heat shock proteins and Drosophila aging. Exp Gerontol. 2011;46:355–362. doi: 10.1016/j.exger.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maiello M, Boeri D, Sampietro L, Pronzato MA, Odetti P, Marinari UM. Basal synthesis of heat shock protein 70 increases with age in rat kidneys. Gerontology. 1998;44:15–20. doi: 10.1159/000021977. [DOI] [PubMed] [Google Scholar]
- 57.Roy AK, Chatterjee B. Sexual dimorphism in the liver. Annu Rev Physiol. 1983;45:37–50. doi: 10.1146/annurev.ph.45.030183.000345. [DOI] [PubMed] [Google Scholar]
- 58.Arslan-Ergul A, Adams MM. Gene expression changes in aging zebrafish (Danio rerio) brains are sexually dimorphic. BMC neuroscience. 2014;15:29. doi: 10.1186/1471-2202-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan L, Ge H, Li H, Lieber SC, Natividad F, Resuello RR, Kim SJ, Akeju S, Sun A, Loo K, Peppas AP, Rossi F, Lewandowski ED, Thomas AP, Vatner SF, Vatner DE. Gender-specific proteomic alterations in glycolytic and mitochondrial pathways in aging monkey hearts. J Mol Cell Cardiol. 2004;37:921–929. doi: 10.1016/j.yjmcc.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 60.Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, Noppinger PR. Sexually dimorphic gene expression in the heart of mice and men. J Mol Med (Berl) 2008;86:61–74. doi: 10.1007/s00109-007-0240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yacobi-Sharon K, Namdar Y, Arama E. Alternative germ cell death pathway in Drosophila involves HtrA2/Omi, lysosomes, and a caspase-9 counterpart. Dev Cell. 2013;25:29–42. doi: 10.1016/j.devcel.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Estaquier J, Vallette F, Vayssiere JL, Mignotte B. The mitochondrial pathways of apoptosis. Adv Exp Med Biol. 2012;942:157–183. doi: 10.1007/978-94-007-2869-1_7. [DOI] [PubMed] [Google Scholar]
- 63.Shen J, Tower J. Programmed cell death and apoptosis in aging and life span regulation. Discov Med. 2009;8:223–226. [PubMed] [Google Scholar]
- 64.Ghavami S, Shojaei S, Yeganeh B, Ande SR, Jangamreddy JR, Mehrpour M, Christoffersson J, Chaabane W, Moghadam AR, Kashani HH, Hashemi M, Owji AA, Los MJ. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Zheng J, Edelman SW, Tharmarajah G, Walker DW, Pletcher SD, Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proc Natl Acad Sci U S A. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marcotte R, Lacelle C, Wang E. Senescent fibroblasts resist apoptosis by downregulating caspase-3. Mech Ageing Dev. 2004;125:777–783. doi: 10.1016/j.mad.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 67.King KL, Cidlowski JA. Cell cycle and apoptosis: common pathways to life and death. J Cell Biochem. 1995;58:175–180. doi: 10.1002/jcb.240580206. [DOI] [PubMed] [Google Scholar]
- 68.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGee MD, Day N, Graham J, Melov S. cep-1/p53-dependent dysplastic pathology of the aging C. elegans gonad. Aging (Albany NY) 2012;4:256–269. doi: 10.18632/aging.100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 71.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- 73.Magwire MM, Yamamoto A, Carbone MA, Roshina NV, Symonenko AV, Pasyukova EG, Morozova TV, Mackay TF. Quantitative and molecular genetic analyses of mutations increasing Drosophila life span. PLoS Genet. 2010;6:e1001037. doi: 10.1371/journal.pgen.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pennell TM, Morrow EH. Two sexes, one genome: the evolutionary dynamics of intralocus sexual conflict. Ecology and evolution. 2013;3:1819–1834. doi: 10.1002/ece3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Connallon T, Clark AG. Evolutionary inevitability of sexual antagonism. Proc Biol Sci. 2014;281:20132123. doi: 10.1098/rspb.2013.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maklakov AA, Lummaa V. Evolution of sex differences in lifespan and aging: causes and constraints. Bioessays. 2013;35:717–724. doi: 10.1002/bies.201300021. [DOI] [PubMed] [Google Scholar]
- 77.Partridge L, Tower J. Yeast, a Feast: The Fruit Fly Drosophila as a Model Organism for Research into Aging. In: Guarente LP, Partridge L, Wallace DC, editors. Molecular Biology of Aging. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2008. pp. 267–308. [Google Scholar]
- 78.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katewa SD, Kapahi P. Role of TOR signaling in aging and related biological processes in Drosophila melanogaster. Exp Gerontol. 2011;46:382–390. doi: 10.1016/j.exger.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCormick MA, Tsai SY, Kennedy BK. TOR and ageing: a complex pathway for a complex process. Philos Trans R Soc Lond B Biol Sci. 2011;366:17–27. doi: 10.1098/rstb.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bonduriansky R. Reappraising sexual coevolution and the sex roles. PLoS Biol. 2009;7:e1000255. doi: 10.1371/journal.pbio.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tower J. The genetic architecture of aging: sexual antagonistic pleiotropy of p53 and foxo. Cell Cycle. 2010;9:3840–3841. doi: 10.4161/cc.9.19.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Promislow D. Mate choice, sexual conflict, and evolution of senescence. Behav Genet. 2003;33:191–201. doi: 10.1023/a:1022562103669. [DOI] [PubMed] [Google Scholar]
- 84.Gioti A, Wigby S, Wertheim B, Schuster E, Martinez P, Pennington CJ, Partridge L, Chapman T. Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proc Biol Sci. 2012;279:4423–4432. doi: 10.1098/rspb.2012.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 2014;10:e1004108. doi: 10.1371/journal.pgen.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gendron CM, Kuo TH, Harvanek ZM, Chung BY, Yew JY, Dierick HA, Pletcher SD. Drosophila life span and physiology are modulated by sexual perception and reward. Science. 2014;343:544–548. doi: 10.1126/science.1243339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, Brunet A. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science. 2014;343:541–544. doi: 10.1126/science.1244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi C, Murphy CT. Mating induces shrinking and death in Caenorhabditis mothers. Science. 2014;343:536–540. doi: 10.1126/science.1242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat Rev Genet. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salz HK. Sex determination in insects: a binary decision based on alternative splicing. Curr Opin Genet Dev. 2011;21:395–400. doi: 10.1016/j.gde.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murray SM, Yang SY, Van Doren M. Germ cell sex determination: a collaboration between soma and germline. Curr Opin Cell Biol. 2010;22:722–729. doi: 10.1016/j.ceb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolff JN, Gemmell NJ. Mitochondria, maternal inheritance, and asymmetric fitness: why males die younger. Bioessays. 2013;35:93–99. doi: 10.1002/bies.201200141. [DOI] [PubMed] [Google Scholar]
- 94.Wolff JN, Nafisinia M, Sutovsky P, Ballard JW. Paternal transmission of mitochondrial DNA as an integral part of mitochondrial inheritance in metapopulations of Drosophila simulans. Heredity (Edinb) 2013;110:57–62. doi: 10.1038/hdy.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Babcock CS, Asmussen MA. Effects of differential selection in the sexes on cytonuclear dynamics. Life stages with sex differences. Genetics. 1998;149:2063–2077. doi: 10.1093/genetics/149.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Babcock CS, Asmussen MA. Effects of differential selection in the sexes on cytonuclear polymorphism and disequilibria. Genetics. 1996;144:839–853. doi: 10.1093/genetics/144.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rand DM, Clark AG, Kann LM. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics. 2001;159:173–187. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, Crimi M, Waymire K, Lin CS, Masubuchi S, Friend N, Koike M, Chalkia D, MacGregor G, Sassone-Corsi P, Wallace DC. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151:333–343. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Paula WB, Lucas CH, Agip AN, Vizcay-Barrena G, Allen JF. Energy, ageing, fidelity and sex: oocyte mitochondrial DNA as a protected genetic template. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120263. doi: 10.1098/rstb.2012.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de Paula WB, Agip AN, Missirlis F, Ashworth R, Vizcay-Barrena G, Lucas CH, Allen JF. Female and male gamete mitochondria are distinct and complementary in transcription, structure, and genome function. Genome biology and evolution. 2013;5:1969–1977. doi: 10.1093/gbe/evt147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tilly JL, Sinclair DA. Germline energetics, aging, and female infertility. Cell Metab. 2013;17:838–850. doi: 10.1016/j.cmet.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allen JF. Separate sexes and the mitochondrial theory of ageing. Journal of theoretical biology. 1996;180:135–140. doi: 10.1006/jtbi.1996.0089. [DOI] [PubMed] [Google Scholar]
- 103.Lane N. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb Perspect Biol. 2014;6:a015982. doi: 10.1101/cshperspect.a015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rand DM. Population genetics of the cytoplasm and the units of selection on mitochondrial DNA in Drosophila melanogaster. Genetica. 2011;139:685–697. doi: 10.1007/s10709-011-9576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nunes MD, Dolezal M, Schlotterer C. Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster. Molecular ecology. 2013;22:2106–2117. doi: 10.1111/mec.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ross JM, Stewart JB, Hagstrom E, Brene S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L, Larsson NG. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Payne BA, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22:384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma H, Xu H, O'Farrell PH. Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat Genet. 2014;46:393–397. doi: 10.1038/ng.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Camus MF, Clancy DJ, Dowling DK. Mitochondria, maternal inheritance, and male aging. Curr Biol. 2012;22:1717–1721. doi: 10.1016/j.cub.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 110.Innocenti P, Morrow EH. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 2010;8:e1000335. doi: 10.1371/journal.pbio.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Soares P, Abrantes D, Rito T, Thomson N, Radivojac P, Li B, Macaulay V, Samuels DC, Pereira L. Evaluating purifying selection in the mitochondrial DNA of various mammalian species. PLoS One. 2013;8:e58993. doi: 10.1371/journal.pone.0058993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wolff JN, White DJ, Woodhams M, White HE, Gemmell NJ. The strength and timing of the mitochondrial bottleneck in salmon suggests a conserved mechanism in vertebrates. PLoS One. 2011;6:e20522. doi: 10.1371/journal.pone.0020522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet. 2014;46:389–392. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gemmell NJ, Metcalf VJ, Allendorf FW. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 2004;19:238–244. doi: 10.1016/j.tree.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 115.Bell G. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. Berkeley: University of California Press; 1982. [Google Scholar]
- 116.Hartfield M, Keightley PD. Current hypotheses for the evolution of sex and recombination. Integrative zoology. 2012;7:192–209. doi: 10.1111/j.1749-4877.2012.00284.x. [DOI] [PubMed] [Google Scholar]
- 117.Lively CM, Morran LT. The ecology of sexual reproduction. J Evol Biol. 2014;27:1292–1303. doi: 10.1111/jeb.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Morrow EH, Connallon T. Implications of sex-specific selection for the genetic basis of disease. Evolutionary applications. 2013;6:1208–1217. doi: 10.1111/eva.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yoshida K, Fujisawa T, Hwang JS, Ikeo K, Gojobori T. Degeneration after sexual differentiation in hydra and its relevance to the evolution of aging. Gene. 2006;385:64–70. doi: 10.1016/j.gene.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 120.Yuan R, Gatti DM, Krier R, Malay E, Schultz D, Peters LL, Churchill GA, Harrison DE, Paigen B. Genetic Regulation of Female Sexual Maturation and Longevity Through Circulating IGF1. J Gerontol A Biol Sci Med Sci. 2014 doi: 10.1093/gerona/glu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Forman HJ. Reactive oxygen species and alpha, beta-unsaturated aldehydes as second messengers in signal transduction. Ann N Y Acad Sci. 2010;1203:35–44. doi: 10.1111/j.1749-6632.2010.05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Oh J, Lee YD, Wagers AJ. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat Med. 2014;20:870–880. doi: 10.1038/nm.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tower J. Stress and stem cells. Wiley interdisciplinary reviews Developmental biology. 2012;1:789–802. doi: 10.1002/wdev.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lara-Ortiz T, Riveros-Rosas H, Aguirre J. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol Microbiol. 2003;50:1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x. [DOI] [PubMed] [Google Scholar]
- 125.Takemoto D, Tanaka A, Scott B. NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal genetics and biology : FG & B. 2007;44:1065–1076. doi: 10.1016/j.fgb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 126.Agarwal A, Gupta S, Sharma RK. Role of oxidative stress in female reproduction. Reproductive biology and endocrinology : RB&E. 2005;3:28. doi: 10.1186/1477-7827-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Courtois F, Carrier S, Charvier K, Guertin PA, Journel NM. The control of male sexual responses. Curr Pharm Des. 2013;19:4341–4356. doi: 10.2174/13816128113199990333. [DOI] [PubMed] [Google Scholar]
- 128.Di Pietro F, Dato S, Carpi FM, Corneveaux JJ, Serfaustini S, Maoloni S, Mignini F, Huentelman MJ, Passarino G, Napolioni V. TP53*P72 Allele Influences Negatively Female Life Expectancy in a Population of Central Italy: Cross-Sectional Study and Genetic-Demographic Approach Analysis. J Gerontol A Biol Sci Med Sci. 2012 doi: 10.1093/gerona/gls223. [DOI] [PubMed] [Google Scholar]
- 129.Bojesen SE, Nordestgaard BG. The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell Cycle. 2008;7:158–163. doi: 10.4161/cc.7.2.5249. [DOI] [PubMed] [Google Scholar]
- 130.Lopez S, Buil A, Souto JC, Casademont J, Blangero J, Martinez-Perez A, Fontcuberta J, Lathrop M, Almasy L, Soria JM. Sex-specific regulation of mitochondrial DNA levels: genome-wide linkage analysis to identify quantitative trait loci. PLoS One. 2012;7:e42711. doi: 10.1371/journal.pone.0042711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hughes BG, Hekimi S. A mild impairment of mitochondrial electron transport has sex-specific effects on lifespan and aging in mice. PLoS One. 2011;6:e26116. doi: 10.1371/journal.pone.0026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jumbo-Lucioni P, Bu S, Harbison ST, Slaughter JC, Mackay TF, Moellering DR, De Luca M. Nuclear genomic control of naturally occurring variation in mitochondrial function in Drosophila melanogaster. BMC Genomics. 2012;13:659. doi: 10.1186/1471-2164-13-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morrow EH, Stewart AD, Rice WR. Assessing the extent of genome-wide intralocus sexual conflict via experimentally enforced gender-limited selection. J Evol Biol. 2008;21:1046–1054. doi: 10.1111/j.1420-9101.2008.01542.x. [DOI] [PubMed] [Google Scholar]
- 134.Ingleby FC, Innocenti P, Rundle HD, Morrow EH. Between-sex genetic covariance constrains the evolution of sexual dimorphism in Drosophila melanogaster. J Evol Biol. 2014;27:1721–1732. doi: 10.1111/jeb.12429. [DOI] [PubMed] [Google Scholar]
- 135.Burger JM, Promislow DE. Sex-specific effects of interventions that extend fly life span. Sci Aging Knowledge Environ. 2004;2004:pe30. doi: 10.1126/sageke.2004.28.pe30. [DOI] [PubMed] [Google Scholar]
- 136.Sun X, Wheeler CT, Yolitz J, Laslo M, Alberico T, Sun Y, Song Q, Zou S. A Mitochondrial ATP Synthase Subunit Interacts with TOR Signaling to Modulate Protein Homeostasis and Lifespan in Drosophila. Cell reports. 2014;8:1781–1792. doi: 10.1016/j.celrep.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Waskar M, Landis GN, Shen J, Curtis C, Tozer K, Abdueva D, Skvortsov D, Tavare S, Tower J. Drosophila melanogaster p53 has developmental stage-specific and sex-specific effects on adult life span indicative of sexual antagonistic pleiotropy. Aging (Albany NY) 2009;1:903–936. doi: 10.18632/aging.100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shen J, Tower J. Drosophila foxo acts in males to cause sexual-dimorphism in tissue-specific p53 life span effects. Exp Gerontol. 2010;45:97–105. doi: 10.1016/j.exger.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Finch CE, Tower J. Sex-specific aging in flies, worms, and missing great-granddads. Cell. 2014;156:398–399. doi: 10.1016/j.cell.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhu CT, Ingelmo P, Rand DM. GxGxE for lifespan in Drosophila: mitochondrial, nuclear, and dietary interactions that modify longevity. PLoS Genet. 2014;10:e1004354. doi: 10.1371/journal.pgen.1004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Magkos F, Wang X, Mittendorfer B. Metabolic actions of insulin in men and women. Nutrition. 2010;26:686–693. doi: 10.1016/j.nut.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 143.Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, Piper MD, Al-Qassab H, Speakman JR, Carmignac D, Robinson IC, Thornton JM, Gems D, Partridge L, Withers DJ. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. Faseb J. 2008;22:807–818. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 144.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 145.Mattila J, Bremer A, Ahonen L, Kostiainen R, Puig O. Drosophila FoxO regulates organism size and stress resistance through an adenylate cyclase. Mol Cell Biol. 2009;29:5357–5365. doi: 10.1128/MCB.00302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Snell-Rood EC, Moczek AP. Insulin signaling as a mechanism underlying developmental plasticity: the role of FOXO in a nutritional polyphenism. PLoS One. 2012;7:e34857. doi: 10.1371/journal.pone.0034857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tang HY, Smith-Caldas MS, Driscoll MV, Salhadar S, Shingleton AW. FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet. 2011;7:e1002373. doi: 10.1371/journal.pgen.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochimica et biophysica acta. 2011;1813:1978–1986. doi: 10.1016/j.bbamcr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 149.Wu CC, Shete S, Amos CI, Strong LC. Joint effects of germ-line p53 mutation and sex on cancer risk in Li-Fraumeni syndrome. Cancer research. 2006;66:8287–8292. doi: 10.1158/0008-5472.CAN-05-4247. [DOI] [PubMed] [Google Scholar]
- 150.Chen X, Watkins R, Delot E, Reliene R, Schiestl RH, Burgoyne PS, Arnold AP. Sex difference in neural tube defects in p53-null mice is caused by differences in the complement of X not Y genes. Dev Neurobiol. 2008;68:265–273. doi: 10.1002/dneu.20581. [DOI] [PubMed] [Google Scholar]
- 151.Baruah A, Chang H, Hall M, Yuan J, Gordon S, Johnson E, Shtessel LL, Yee C, Hekimi S, Derry WB, Lee SS. CEP-1, the Caenorhabditis elegans p53 homolog, mediates opposing longevity outcomes in mitochondrial electron transport chain mutants. PLoS Genet. 2014;10:e1004097. doi: 10.1371/journal.pgen.1004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, Johnson TE. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell. 2009;8:380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McCulloch D, Gems D. Evolution of male longevity bias in nematodes. Aging Cell. 2003;2:165–173. doi: 10.1046/j.1474-9728.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- 154.Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ameur A, Stewart JB, Freyer C, Hagstrom E, Ingman M, Larsson NG, Gyllensten U. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 2011;7:e1002028. doi: 10.1371/journal.pgen.1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jokinen R, Junnila H, Battersby BJ. Gimap3: A foot-in-the-door to tissue-specific regulation of mitochondrial DNA genetics. Small GTPases. 2011;2:31–35. doi: 10.4161/sgtp.2.1.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rochet JC, Hay BA, Guo M. Molecular insights into Parkinson's disease. Progress in molecular biology and translational science. 2012;107:125–188. doi: 10.1016/B978-0-12-385883-2.00011-4. [DOI] [PubMed] [Google Scholar]
- 159.Jarrett SG, Lin H, Godley BF, Boulton ME. Mitochondrial DNA damage and its potential role in retinal degeneration. Progress in retinal and eye research. 2008;27:596–607. doi: 10.1016/j.preteyeres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 160.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Carling PJ, Cree LM, Chinnery PF. The implications of mitochondrial DNA copy number regulation during embryogenesis. Mitochondrion. 2011;11:686–692. doi: 10.1016/j.mito.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 162.Gasparre G, Hervouet E, de Laplanche E, Demont J, Pennisi LF, Colombel M, Mege-Lechevallier F, Scoazec JY, Bonora E, Smeets R, Smeitink J, Lazar V, Lespinasse J, Giraud S, Godinot C, Romeo G, Simonnet H. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet. 2008;17:986–995. doi: 10.1093/hmg/ddm371. [DOI] [PubMed] [Google Scholar]
- 163.McKinney EA, Oliveira MT. Replicating animal mitochondrial DNA. Genetics and molecular biology. 2013;36:308–315. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hagstrom E, Freyer C, Battersby BJ, Stewart JB, Larsson NG. No recombination of mtDNA after heteroplasmy for 50 generations in the mouse maternal germline. Nucleic Acids Res. 2014;42:1111–1116. doi: 10.1093/nar/gkt969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yui R, Ohno Y, Matsuura ET. Accumulation of deleted mitochondrial DNA in aging Drosophila melanogaster. Genes Genet Syst. 2003;78:245–251. doi: 10.1266/ggs.78.245. [DOI] [PubMed] [Google Scholar]
- 166.Yui R, Matsuura ET. Detection of deletions flanked by short direct repeats in mitochondrial DNA of aging Drosophila. Mutat Res. 2006;594:155–161. doi: 10.1016/j.mrfmmm.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 167.Golden TR, Melov S. Mitochondrial DNA mutations, oxidative stress, and aging. Mech Ageing Dev. 2001;122:1577–1589. doi: 10.1016/s0047-6374(01)00288-3. [DOI] [PubMed] [Google Scholar]
- 168.Kujoth GC, Prolla TA. Evolving insight into the role of mitochondrial DNA mutations in aging. Exp Gerontol. 2008;43:20–23. doi: 10.1016/j.exger.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 169.Weng SW, Lin TK, Liou CW, Chen SD, Wei YH, Lee HC, Chen IY, Hsieh CJ, Wang PW. Peripheral blood mitochondrial DNA content and dysregulation of glucose metabolism. Diabetes research and clinical practice. 2009;83:94–99. doi: 10.1016/j.diabres.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 170.Liu CS, Kuo CL, Cheng WL, Huang CS, Lee CF, Wei YH. Alteration of the copy number of mitochondrial DNA in leukocytes of patients with hyperlipidemia. Ann N Y Acad Sci. 2005;1042:70–75. doi: 10.1196/annals.1338.008. [DOI] [PubMed] [Google Scholar]
- 171.Purdue MP, Hofmann JN, Colt JS, Hoxha M, Ruterbusch JJ, Davis FG, Rothman N, Wacholder S, Schwartz KL, Baccarelli A, Chow WH. A case-control study of peripheral blood mitochondrial DNA copy number and risk of renal cell carcinoma. PLoS One. 2012;7:e43149. doi: 10.1371/journal.pone.0043149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xie H, Lev D, Gong Y, Wang S, Pollock RE, Wu X, Gu J. Reduced mitochondrial DNA copy number in peripheral blood leukocytes increases the risk of soft tissue sarcoma. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt023. [DOI] [PubMed] [Google Scholar]
- 173.Lee SH, Chung DJ, Lee HS, Kim TJ, Kim MH, Jeong HJ, Im JA, Lee DC, Lee JW. Mitochondrial DNA copy number in peripheral blood in polycystic ovary syndrome. Metabolism: clinical and experimental. 2011;60:1677–1682. doi: 10.1016/j.metabol.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 174.Kim JH, Lee DC. Mitochondrial DNA copy number in peripheral blood is associated with femoral neck bone mineral density in postmenopausal women. The Journal of rheumatology. 2012;39:1465–1472. doi: 10.3899/jrheum.111444. [DOI] [PubMed] [Google Scholar]
- 175.Zhao S, Yang Y, Liu J, Liu H, Ge N, Yang H, Zhang H, Xing J. Association of mitochondrial DNA content in peripheral blood leukocyte with hepatitis B virus-related hepatocellular carcinoma in a Chinese Han population. Cancer Sci. 2011;102:1553–1558. doi: 10.1111/j.1349-7006.2011.01968.x. [DOI] [PubMed] [Google Scholar]
- 176.Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2012 doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 177.Cheng X, Ivessa AS. The migration of mitochondrial DNA fragments to the nucleus affects the chronological aging process of Saccharomyces cerevisiae. Aging Cell. 2010;9:919–923. doi: 10.1111/j.1474-9726.2010.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Caro P, Gomez J, Arduini A, Gonzalez-Sanchez M, Gonzalez-Garcia M, Borras C, Vina J, Puertas MJ, Sastre J, Barja G. Mitochondrial DNA sequences are present inside nuclear DNA in rat tissues and increase with age. Mitochondrion. 2010;10:479–486. doi: 10.1016/j.mito.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 179.Barford A, Dorling D, Davey Smith G, Shaw M. Life expectancy: women now on top everywhere. BMJ. 2006;332:808. doi: 10.1136/bmj.332.7545.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pierdominici M, Ortona E, Franconi F, Caprio M, Straface E, Malorni W. Gender specific aspects of cell death in the cardiovascular system. Curr Pharm Des. 2011;17:1046–1055. doi: 10.2174/138161211795656891. [DOI] [PubMed] [Google Scholar]
- 181.Laurent M, Antonio L, Sinnesael M, Dubois V, Gielen E, Classens F, Vanderschueren D. Androgens and estrogens in skeletal sexual dimorphism. Asian journal of andrology. 2014;16:213–222. doi: 10.4103/1008-682X.122356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bubb KJ, Khambata RS, Ahluwalia A. Sexual dimorphism in rodent models of hypertension and atherosclerosis. British journal of pharmacology. 2012;167:298–312. doi: 10.1111/j.1476-5381.2012.02036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Frontiers in neuroendocrinology. 2014;35:347–369. doi: 10.1016/j.yfrne.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 184.Baggio G, Corsini A, Floreani A, Giannini S, Zagonel V. Gender medicine: a task for the third millennium. Clinical chemistry and laboratory medicine : CCLM / FESCC. 2013;51:713–727. doi: 10.1515/cclm-2012-0849. [DOI] [PubMed] [Google Scholar]
- 185.Zuo W, Zhang W, Chen NH. Sexual dimorphism in cerebral ischemia injury. European journal of pharmacology. 2013;711:73–79. doi: 10.1016/j.ejphar.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 186.Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, Lago CU, Zhang S, DuBois W, Ward T, deCabo R, Gavrilova O, Mock B, Finkel T. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell reports. 2013;4:913–920. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Harrison DE, Strong R, Allison DB, Ames BN, Astle CM, Atamna H, Fernandez E, Flurkey K, Javors MA, Nadon NL, Nelson JF, Pletcher S, Simpkins JW, Smith D, Wilkinson JE, Miller RA. Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell. 2014;13:273–282. doi: 10.1111/acel.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, Javors MA, Li X, Nadon NL, Nelson JF, Pletcher S, Salmon AB, Sharp ZD, Van Roekel S, Winkleman L, Strong R. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:468–477. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Vasconsuelo A, Pronsato L, Ronda AC, Boland R, Milanesi L. Role of 17-betaestradiol and testosterone in apoptosis. Steroids. 2011;76:1223–1231. doi: 10.1016/j.steroids.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 190.Lin J, Steenbergen C, Murphy E, Sun J. Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation. 2009;120:245–254. doi: 10.1161/CIRCULATIONAHA.109.868729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lagranha CJ, Deschamps A, Aponte A, Steenbergen C, Murphy E. Sex differences in the phosphorylation of mitochondrial proteins result in reduced production of reactive oxygen species and cardioprotection in females. Circ Res. 2010;106:1681–1691. doi: 10.1161/CIRCRESAHA.109.213645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu H, Yanamandala M, Lee TC, Kim JK. Mitochondrial p38beta and manganese superoxide dismutase interaction mediated by estrogen in cardiomyocytes. PLoS One. 2014;9:e85272. doi: 10.1371/journal.pone.0085272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Patten RD, Pourati I, Aronovitz MJ, Baur J, Celestin F, Chen X, Michael A, Haq S, Nuedling S, Grohe C, Force T, Mendelsohn ME, Karas RH. 17beta-estradiol reduces cardiomyocyte apoptosis in vivo and in vitro via activation of phosphoinositide-3 kinase/Akt signaling. Circulation research. 2004;95:692–699. doi: 10.1161/01.RES.0000144126.57786.89. [DOI] [PubMed] [Google Scholar]
- 194.Wang F, He Q, Sun Y, Dai X, Yang XP. Female adult mouse cardiomyocytes are protected against oxidative stress. Hypertension. 2010;55:1172–1178. doi: 10.1161/HYPERTENSIONAHA.110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Callies F, Stromer H, Schwinger RH, Bolck B, Hu K, Frantz S, Leupold A, Beer S, Allolio B, Bonz AW. Administration of testosterone is associated with a reduced susceptibility to myocardial ischemia. Endocrinology. 2003;144:4478–4483. doi: 10.1210/en.2003-0058. [DOI] [PubMed] [Google Scholar]
- 196.Pronsato L, Boland R, Milanesi L. Testosterone exerts antiapoptotic effects against H2O2 in C2C12 skeletal muscle cells through the apoptotic intrinsic pathway. The Journal of endocrinology. 2012;212:371–381. doi: 10.1530/JOE-11-0234. [DOI] [PubMed] [Google Scholar]
- 197.Palomar-Morales M, Morimoto S, Mendoza-Rodriguez CA, Cerbon MA. The protective effect of testosterone on streptozotocin-induced apoptosis in beta cells is sex specific. Pancreas. 2010;39:193–200. doi: 10.1097/MPA.0b013e3181c156d9. [DOI] [PubMed] [Google Scholar]
- 198.Hadji P, Coleman R, Gnant M. Bone effects of mammalian target of rapamycin (mTOR) inhibition with everolimus. Critical reviews in oncology/hematology. 2013;87:101–111. doi: 10.1016/j.critrevonc.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 199.Ciruelos Gil EM. Targeting the PI3K/AKT/mTOR pathway in estrogen receptorpositive breast cancer. Cancer treatment reviews. 2014;40:862–871. doi: 10.1016/j.ctrv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 200.Serra C, Sandor NL, Jang H, Lee D, Toraldo G, Guarneri T, Wong S, Zhang A, Guo W, Jasuja R, Bhasin S. The effects of testosterone deprivation and supplementation on proteasomal and autophagy activity in the skeletal muscle of the male mouse: differential effects on high-androgen responder and low-androgen responder muscle groups. Endocrinology. 2013;154:4594–4606. doi: 10.1210/en.2013-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Basualto-Alarcon C, Jorquera G, Altamirano F, Jaimovich E, Estrada M. Testosterone signals through mTOR and androgen receptor to induce muscle hypertrophy. Medicine and science in sports and exercise. 2013;45:1712–1720. doi: 10.1249/MSS.0b013e31828cf5f3. [DOI] [PubMed] [Google Scholar]
- 202.Brinton RD. The healthy cell bias of estrogen action: mitochondrial bioenergetics and neurological implications. Trends in neurosciences. 2008;31:529–537. doi: 10.1016/j.tins.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fagegaltier D, Konig A, Gordon A, Lai EC, Gingeras TR, Hannon GJ, Shcherbata HR. A Genome-Wide Survey of Sexually Dimorphic Expression of Drosophila miRNAs Identifies the Steroid Hormone-Induced miRNA let-7 as a Regulator of Sexual Identity. Genetics. 2014 doi: 10.1534/genetics.114.169268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Galikova M, Klepsatel P, Senti G, Flatt T. Steroid hormone regulation of C. elegans and Drosophila aging and life history. Exp Gerontol. 2011;46:141–147. doi: 10.1016/j.exger.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 205.McCulloch D, Gems D. Sex-specific effects of the DAF-12 steroid receptor on aging in Caenorhabditis elegans. Ann N Y Acad Sci. 2007;1119:253–259. doi: 10.1196/annals.1404.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dunlay SM, Roger VL. Gender differences in the pathophysiology, clinical presentation, and outcomes of ischemic heart failure. Current heart failure reports. 2012;9:267–276. doi: 10.1007/s11897-012-0107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Koenig A, Sateriale A, Budd RC, Huber SA, Buskiewicz IA. The role of sex differences in autophagy in the heart during coxsackievirus B3-induced myocarditis. J Cardiovasc Transl Res. 2014;7:182–191. doi: 10.1007/s12265-013-9525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kanamori H, Takemura G, Goto K, Maruyama R, Ono K, Nagao K, Tsujimoto A, Ogino A, Takeyama T, Kawaguchi T, Watanabe T, Kawasaki M, Fujiwara T, Fujiwara H, Seishima M, Minatoguchi S. Autophagy limits acute myocardial infarction induced by permanent coronary artery occlusion. American journal of physiology Heart and circulatory physiology. 2011;300:H2261–H2271. doi: 10.1152/ajpheart.01056.2010. [DOI] [PubMed] [Google Scholar]
- 209.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 210.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 211.Reichelt ME, Mellor KM, Curl CL, Stapleton D, Delbridge LM. Myocardial glycophagy - a specific glycogen handling response to metabolic stress is accentuated in the female heart. J Mol Cell Cardiol. 2013;65:67–75. doi: 10.1016/j.yjmcc.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 212.Campesi I, Straface E, Occhioni S, Montella A, Franconi F. Protein oxidation seems to be linked to constitutive autophagy: a sex study. Life Sci. 2013;93:145–152. doi: 10.1016/j.lfs.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 213.Chen C, Hu LX, Dong T, Wang GQ, Wang LH, Zhou XP, Jiang Y, Murao K, Lu SQ, Chen JW, Zhang GX. Apoptosis and autophagy contribute to gender difference in cardiac ischemia-reperfusion induced injury in rats. Life Sci. 2013;93:265–270. doi: 10.1016/j.lfs.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 214.Applebaum MA, Skol AD, Bond EE, Overholtzer M, Bond GL, Onel K. Radiation-induced apoptosis varies among individuals and is modified by sex and age. International journal of radiation biology. 2014:1–6. doi: 10.3109/09553002.2014.925603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Harada H, Pavlick KP, Hines IN, Lefer DJ, Hoffman JM, Bharwani S, Wolf RE, Grisham MB. Sexual dimorphism in reduced-size liver ischemia and reperfusion injury in mice: role of endothelial cell nitric oxide synthase. Proc Natl Acad Sci U S A. 2003;100:739–744. doi: 10.1073/pnas.0235680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fekete A, Vannay A, Ver A, Vasarhelyi B, Muller V, Ouyang N, Reusz G, Tulassay T, Szabo AJ. Sex differences in the alterations of Na(+), K(+)-ATPase following ischaemia-reperfusion injury in the rat kidney. J Physiol. 2004;555:471–480. doi: 10.1113/jphysiol.2003.054825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Straface E, Vona R, Gambardella L, Ascione B, Marino M, Bulzomi P, Canu S, Coinu R, Rosano G, Malorni W, Franconi F. Cell sex determines anoikis resistance in vascular smooth muscle cells. FEBS Lett. 2009;583:3448–3454. doi: 10.1016/j.febslet.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 218.Du L, Hickey RW, Bayir H, Watkins SC, Tyurin VA, Guo F, Kochanek PM, Jenkins LW, Ren J, Gibson G, Chu CT, Kagan VE, Clark RS. Starving neurons show sex difference in autophagy. J Biol Chem. 2009;284:2383–2396. doi: 10.1074/jbc.M804396200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhu C, Xu F, Wang X, Shibata M, Uchiyama Y, Blomgren K, Hagberg H. Different apoptotic mechanisms are activated in male and female brains after neonatal hypoxia-ischaemia. Journal of neurochemistry. 2006;96:1016–1027. doi: 10.1111/j.1471-4159.2005.03639.x. [DOI] [PubMed] [Google Scholar]
- 220.Weis SN, Toniazzo AP, Ander BP, Zhan X, Careaga M, Ashwood P, Wyse AT, Netto CA, Sharp FR. Autophagy in the brain of neonates following hypoxiaischemia shows sex- and region-specific effects. Neuroscience. 2014;256:201–209. doi: 10.1016/j.neuroscience.2013.10.046. [DOI] [PubMed] [Google Scholar]
- 221.Herrera VL, Decano JL, Bagamasbad P, Kufahl T, Steffen M, Ruiz-Opazo N. Sex-specific hippocampus-dependent cognitive deficits and increased neuronal autophagy in DEspR haploinsufficiency in mice. Physiol Genomics. 2008;35:316–329. doi: 10.1152/physiolgenomics.00044.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Siegel C, Li J, Liu F, Benashski SE, McCullough LD. miR-23a regulation of X-linked inhibitor of apoptosis (XIAP) contributes to sex differences in the response to cerebral ischemia. Proc Natl Acad Sci U S A. 2011;108:11662–11667. doi: 10.1073/pnas.1102635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lang JT, McCullough LD. Pathways to ischemic neuronal cell death: are sex differences relevant? Journal of translational medicine. 2008;6:33. doi: 10.1186/1479-5876-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Renolleau S, Fau S, Charriaut-Marlangue C. Gender-related differences in apoptotic pathways after neonatal cerebral ischemia. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2008;14:46–52. doi: 10.1177/1073858407308889. [DOI] [PubMed] [Google Scholar]
- 225.Liu F, Lang J, Li J, Benashski SE, Siegel M, Xu Y, McCullough LD. Sex differences in the response to poly(ADP-ribose) polymerase-1 deletion and caspase inhibition after stroke. Stroke. 2011;42:1090–1096. doi: 10.1161/STROKEAHA.110.594861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Siegel CS, McCullough LD. NAD+ and nicotinamide: sex differences in cerebral ischemia. Neuroscience. 2013;237:223–231. doi: 10.1016/j.neuroscience.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Siegel C, Turtzo C, McCullough LD. Sex differences in cerebral ischemia: possible molecular mechanisms. Journal of neuroscience research. 2010;88:2765–2774. doi: 10.1002/jnr.22406. [DOI] [PubMed] [Google Scholar]
- 228.Siegel C, McCullough LD. NAD+ depletion or PAR polymer formation: which plays the role of executioner in ischaemic cell death? Acta Physiol (Oxf) 2011;203:225–234. doi: 10.1111/j.1748-1716.2010.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shaposhnikov MV, Moskalev AA, Plyusnina EN. Effect of PARP-1 overexpression and pharmacological inhibition of NF-kB on the lifespan of Drosophila melanogaster. Advances in gerontology = Uspekhi gerontologii / Rossiiskaia akademiia nauk, Gerontologicheskoe obshchestvo. 2011;24:405–419. [PubMed] [Google Scholar]
- 230.Hagberg H, Wilson MA, Matsushita H, Zhu C, Lange M, Gustavsson M, Poitras MF, Dawson TM, Dawson VL, Northington F, Johnston MV. PARP-1 gene disruption in mice preferentially protects males from perinatal brain injury. Journal of neurochemistry. 2004;90:1068–1075. doi: 10.1111/j.1471-4159.2004.02547.x. [DOI] [PubMed] [Google Scholar]
- 231.Yuan M, Siegel C, Zeng Z, Li J, Liu F, McCullough LD. Sex differences in the response to activation of the poly (ADP-ribose) polymerase pathway after experimental stroke. Exp Neurol. 2009;217:210–218. doi: 10.1016/j.expneurol.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Du L, Bayir H, Lai Y, Zhang X, Kochanek PM, Watkins SC, Graham SH, Clark RS. Innate gender-based proclivity in response to cytotoxicity and programmed cell death pathway. J Biol Chem. 2004;279:38563–38570. doi: 10.1074/jbc.M405461200. [DOI] [PubMed] [Google Scholar]
- 233.Li H, Pin S, Zeng Z, Wang MM, Andreasson KA, McCullough LD. Sex differences in cell death. Annals of neurology. 2005;58:317–321. doi: 10.1002/ana.20538. [DOI] [PubMed] [Google Scholar]
- 234.Li R, Strykowski R, Meyer M, Mulcrone P, Krakora D, Suzuki M. Male-specific differences in proliferation, neurogenesis, and sensitivity to oxidative stress in neural progenitor cells derived from a rat model of ALS. PLoS One. 2012;7:e48581. doi: 10.1371/journal.pone.0048581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Penaloza C, Estevez B, Orlanski S, Sikorska M, Walker R, Smith C, Smith B, Lockshin RA, Zakeri Z. Sex of the cell dictates its response: differential gene expression and sensitivity to cell death inducing stress in male and female cells. FASEB J. 2009;23:1869–1879. doi: 10.1096/fj.08-119388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Giampietri C, Petrungaro S, Filippini A, Ziparo E. Sex-related differences in death control of somatic cells. J Cell Mol Med. 2013;17:550–551. doi: 10.1111/jcmm.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Perez-Crespo M, Ramirez MA, Fernandez-Gonzalez R, Rizos D, Lonergan P, Pintado B, Gutierrez-Adan A. Differential sensitivity of male and female mouse embryos to oxidative induced heat-stress is mediated by glucose-6-phosphate dehydrogenase gene expression. Mol Reprod Dev. 2005;72:502–510. doi: 10.1002/mrd.20366. [DOI] [PubMed] [Google Scholar]
- 238.Arnold AP. Conceptual frameworks and mouse models for studying sex differences in physiology and disease: Why compensation changes the game. Exp Neurol. 2014 doi: 10.1016/j.expneurol.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Seney ML, Ekong KI, Ding Y, Tseng GC, Sibille E. Sex chromosome complement regulates expression of mood-related genes. Biology of sex differences. 2013;4:20. doi: 10.1186/2042-6410-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li J, Chen X, McClusky R, Ruiz-Sundstrom M, Itoh Y, Umar S, Arnold AP, Eghbali M. The number of X chromosomes influences protection from cardiac ischaemia/reperfusion injury in mice: one X is better than two. Cardiovasc Res. 2014;102:375–384. doi: 10.1093/cvr/cvu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Abramowitz LK, Olivier-Van Stichelen S, Hanover JA. Chromosome imbalance as a driver of sex disparity in disease. Journal of genomics. 2014;2:77–88. doi: 10.7150/jgen.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 246.Zhang Y, Castillo-Morales A, Jiang M, Zhu Y, Hu L, Urrutia AO, Kong X, Hurst LD. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol Biol Evol. 2013;30:2588–2601. doi: 10.1093/molbev/mst148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Dumas KJ, Delaney CE, Flibotte S, Moerman DG, Csankovszki G, Hu PJ. Unexpected role for dosage compensation in the control of dauer arrest, insulinlike signaling, and FoxO transcription factor activity in Caenorhabditis elegans. Genetics. 2013;194:619–629. doi: 10.1534/genetics.113.149948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hartman PS, Ishii N. Chromosome dosage as a life span determinant in Caenorhabiditis elegans. Mech Ageing Dev. 2007;128:437–443. doi: 10.1016/j.mad.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 249.Webster CM, Wu L, Douglas D, Soukas AA. A non-canonical role for the C. elegans dosage compensation complex in growth and metabolic regulation downstream of TOR complex 2. Development. 2013;140:3601–3612. doi: 10.1242/dev.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Calabrese EJ, Bachmann KA, Bailer AJ, Bolger PM, Borak J, Cai L, Cedergreen N, Cherian MG, Chiueh CC, Clarkson TW, Cook RR, Diamond DM, Doolittle DJ, Dorato MA, Duke SO, Feinendegen L, Gardner DE, Hart RW, Hastings KL, Hayes AW, Hoffmann GR, Ives JA, Jaworowski Z, Johnson TE, Jonas WB, Kaminski NE, Keller JG, Klaunig JE, Knudsen TB, Kozumbo WJ, Lettieri T, Liu SZ, Maisseu A, Maynard KI, Masoro EJ, McClellan RO, Mehendale HM, Mothersill C, Newlin DB, Nigg HN, Oehme FW, Phalen RF, Philbert MA, Rattan SI, Riviere JE, Rodricks J, Sapolsky RM, Scott BR, Seymour C, Sinclair DA, Smith-Sonneborn J, Snow ET, Spear L, Stevenson DE, Thomas Y, Tubiana M, Williams GM, Mattson MP. Biological stress response terminology: Integrating the concepts of adaptive response and preconditioning stress within a hormetic dose-response framework. Toxicol Appl Pharmacol. 2007;222:122–128. doi: 10.1016/j.taap.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 252.Pickering AM, Staab TA, Tower J, Sieburth DS, Davies KJ. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative-stress adaptation in mammals C. elegans and D. melanogaster. J Exp Biol. 2012 doi: 10.1242/jeb.074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Cypser JR, Johnson TE. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci. 2002;57:B109–B114. doi: 10.1093/gerona/57.3.b109. [DOI] [PubMed] [Google Scholar]
- 254.Rattan SI. Hormesis in aging. Ageing research reviews. 2008;7:63–78. doi: 10.1016/j.arr.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 255.Epel ES, Lithgow GJ. Stress biology and aging mechanisms: toward understanding the deep connection between adaptation to stress and longevity. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S10–S16. doi: 10.1093/gerona/glu055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Moskalev AA, Plyusnina EN, Shaposhnikov MV. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: the role of cellular stress-resistance mechanisms. Biogerontology. 2011;12:253–263. doi: 10.1007/s10522-011-9320-0. [DOI] [PubMed] [Google Scholar]
- 257.Salminen A, Kaarniranta K. ER stress and hormetic regulation of the aging process. Ageing Res Rev. 2010;9:211–217. doi: 10.1016/j.arr.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 258.Moskalev A, Shaposhnikov M, Turysheva E. Life span alteration after irradiation in Drosophila melanogaster strains with mutations of Hsf and Hsps. Biogerontology. 2009;10:3–11. doi: 10.1007/s10522-008-9147-5. [DOI] [PubMed] [Google Scholar]
- 259.Sorensen JG, Kristensen TN, Kristensen KV, Loeschcke V. Sex specific effects of heat induced hormesis in Hsf-deficient Drosophila melanogaster. Exp Gerontol. 2007;42:1123–1129. doi: 10.1016/j.exger.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 260.Cypser JR, Tedesco P, Johnson TE. Hormesis and aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:935–939. doi: 10.1016/j.exger.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, Hoch M, Hohfeld J. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 264.Juhasz G, Neufeld TP. Drosophila Atg7: Required for stress resistance, longevity and neuronal homeostasis, but not for metamorphosis. Autophagy. 2008;4 doi: 10.4161/auto.5572. [DOI] [PubMed] [Google Scholar]
- 265.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 267.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 268.Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nature communications. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Hansen M, Flatt T, Aguilaniu H. Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 2013;17:10–19. doi: 10.1016/j.cmet.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Frohlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 273.Ren C, Finkel SE, Tower J. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp Gerontol. 2009;44:228–235. doi: 10.1016/j.exger.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 275.Bai H, Kang P, Hernandez AM, Tatar M. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet. 2013;9:e1003941. doi: 10.1371/journal.pgen.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Scherfer C, Han VC, Wang Y, Anderson AE, Galko MJ. Autophagy drives epidermal deterioration in a Drosophila model of tissue aging. Aging (Albany NY) 2013;5:276–287. doi: 10.18632/aging.100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Takacs-Vellai K, Bayci A, Vellai T. Autophagy in neuronal cell loss: a road to death. Bioessays. 2006;28:1126–1131. doi: 10.1002/bies.20489. [DOI] [PubMed] [Google Scholar]
- 278.de Castro IP, Costa AC, Celardo I, Tufi R, Dinsdale D, Loh SH, Martins LM. Drosophila ref(2)P is required for the parkin-mediated suppression of mitochondrial dysfunction in pink1 mutants. Cell death & disease. 2013;4:e873. doi: 10.1038/cddis.2013.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Maglioni S, Schiavi A, Runci A, Shaik A, Ventura N. Mitochondrial stress extends lifespan in C. elegans through neuronal hormesis. Exp Gerontol. 2014;56:89–98. doi: 10.1016/j.exger.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 280.Zarkower D. Somatic sex determination. WormBook : the online review of C elegans biology. 2006:1–12. doi: 10.1895/wormbook.1.84.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends in cell biology. 2013;23:311–318. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Jensen MB, Jasper H. Mitochondrial Proteostasis in the Control of Aging and Longevity. Cell Metab. 2014;20:214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Morrow G, Inaguma Y, Kato K, Tanguay RM. The small heat shock protein Hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J Biol Chem. 2000;275:31204–31210. doi: 10.1074/jbc.M002960200. [DOI] [PubMed] [Google Scholar]
- 284.Kurapati R, Passananti HB, Rose MR, Tower J. Increased hsp22 RNA levels in Drosophila lines genetically selected for increased longevity. J Gerontol A Biol Sci Med Sci. 2000;55:B552–B559. doi: 10.1093/gerona/55.11.b552. [DOI] [PubMed] [Google Scholar]
- 285.Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. Faseb J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 286.Bhole D, Allikian MJ, Tower J. Doxycycline-regulated over-expression of hsp22 has negative effects on stress resistance and life span in adult Drosophilamelanogaster. Mech Ageing Dev. 2004;125:651–663. doi: 10.1016/j.mad.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 287.Yang J, Tower J. Expression of hsp22 and hsp70 transgenes is partially predictive of drosophila survival under normal and stress conditions. J Gerontol A Biol Sci Med Sci. 2009;64:828–838. doi: 10.1093/gerona/glp054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Jacobs HT, Fernandez-Ayala DJ, Manjiry S, Kemppainen E, Toivonen JM, O'Dell KM. Mitochondrial disease in flies. Biochimica et biophysica acta. 2004;1659:190–196. doi: 10.1016/j.bbabio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 289.Fernandez-Ayala DJ, Chen S, Kemppainen E, O'Dell KM, Jacobs HT. Gene expression in a Drosophila model of mitochondrial disease. PLoS One. 2010;5:e8549. doi: 10.1371/journal.pone.0008549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 292.Baker BM, Haynes CM. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci. 2011;36:254–261. doi: 10.1016/j.tibs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 293.Tufi R, Gandhi S, de Castro IP, Lehmann S, Angelova PR, Dinsdale D, Deas E, Plun-Favreau H, Nicotera P, Abramov AY, Willis AE, Mallucci GR, Loh SH, Martins LM. Enhancing nucleotide metabolism protects against mitochondrial dysfunction and neurodegeneration in a PINK1 model of Parkinson's disease. Nat Cell Biol. 2014;16:157–166. doi: 10.1038/ncb2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Pimenta de Castro I, Costa AC, Lam D, Tufi R, Fedele V, Moisoi N, Dinsdale D, Deas E, Loh SH, Martins LM. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ. 2012;19:1308–1316. doi: 10.1038/cdd.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C. P62/SQSTM1 at the interface of aging, autophagy, and disease. Age (Dordr) 2014;36:9626. doi: 10.1007/s11357-014-9626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Lippai M, Low P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. BioMed research international. 2014;2014:832704. doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Manley S, Williams JA, Ding WX. Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp Biol Med (Maywood) 2013;238:525–538. doi: 10.1177/1535370213489446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Kuttner V, Bhukel A, Marino G, Pietrocola F, Harger A, Zimmermann A, Moustafa T, Sprenger A, Jany E, Buttner S, Carmona-Gutierrez D, Ruckenstuhl C, Ring J, Reichelt W, Schimmel K, Leeb T, Moser C, Schatz S, Kamolz LP, Magnes C, Sinner F, Sedej S, Frohlich KU, Juhasz G, Pieber TR, Dengjel J, Sigrist SJ, Kroemer G, Madeo F. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19:431–444. doi: 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB Journal. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 301.De Haes W, Frooninckx L, Van Assche R, Smolders A, Depuydt G, Billen J, Braeckman BP, Schoofs L, Temmerman L. Metformin promotes lifespan through mitohormesis via the peroxiredoxin PRDX-2. Proc Natl Acad Sci U S A. 2014;111:E2501–E2509. doi: 10.1073/pnas.1321776111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–672. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavare S, Tower J. Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 2007;8:R262. doi: 10.1186/gb-2007-8-12-r262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Cabreiro F, Ackerman D, Doonan R, Araiz C, Back P, Papp D, Braeckman BP, Gems D. Increased life span from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage. Free Radic Biol Med. 2011;51:1575–1582. doi: 10.1016/j.freeradbiomed.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157:897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- 307.Bennett CF, Kaeberlein M. The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation? Exp Gerontol. 2014;56:142–146. doi: 10.1016/j.exger.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Runkel ED, Liu S, Baumeister R, Schulze E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 2013;9:e1003346. doi: 10.1371/journal.pgen.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Runkel ED, Baumeister R, Schulze E. Mitochondrial stress: balancing friend and foe. Exp Gerontol. 2014;56:194–201. doi: 10.1016/j.exger.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 310.Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nature communications. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 315.Hur JH, Stork DA, Walker DW. Complex-I-ty in aging. Journal of bioenergetics and biomembranes. 2014;46:329–335. doi: 10.1007/s10863-014-9553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Tower J, Landis G, Gao R, Luan A, Lee J, Sun Y. Variegated Expression of Hsp22 Transgenic Reporters Indicates Cell-specific Patterns of Aging in Drosophila Oenocytes. J Gerontol A Biol Sci Med Sci. 2013 doi: 10.1093/gerona/glt078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Paik D, Jang YG, Lee YE, Lee YN, Yamamoto R, Gee HY, Yoo S, Bae E, Min KJ, Tatar M, Park JJ. Misexpression screen delineates novel genes controlling Drosophila lifespan. Mech Ageing Dev. 2012;133:234–245. doi: 10.1016/j.mad.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, Auwerx J. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 321.Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 322.Pulliam DA, Deepa SS, Liu Y, Hill S, Lin AL, Bhattacharya A, Shi Y, Sloane L, Viscomi C, Zeviani M, Van Remmen H. Complex IV-deficient Surf1(−/−) mice initiate mitochondrial stress responses. Biochem J. 2014;462:359–371. doi: 10.1042/BJ20140291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 324.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Imai SI, Guarente L. NAD and sirtuins in aging and disease. Trends in cell biology. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Orozco-Solis R, Sassone-Corsi P. Circadian clock: linking epigenetics to aging. Curr Opin Genet Dev. 2014;26C:66–72. doi: 10.1016/j.gde.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Rotter D, Rothermel BA. Targets, trafficking, and timing of cardiac autophagy. Pharmacological research : the official journal of the Italian Pharmacological Society. 2012;66:494–504. doi: 10.1016/j.phrs.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Czaja MJ, Ding WX, Donohue TM, Jr, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH, Perlmutter DH, Randall G, Ray RB, Tsung A, Yin XM. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131–1158. doi: 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ma D, Li S, Molusky MM, Lin JD. Circadian autophagy rhythm: a link between clock and metabolism? Trends Endocrinol Metab. 2012;23:319–325. doi: 10.1016/j.tem.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nature reviews Neuroscience. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Sachdeva UM, Thompson CB. Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy. 2008;4:581–589. doi: 10.4161/auto.6141. [DOI] [PubMed] [Google Scholar]
- 332.Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci. 2011;124:1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Hwang JW, Yao H, Caito S, Sundar IK, Rahman I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic Biol Med. 2013;61C:95–110. doi: 10.1016/j.freeradbiomed.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 335.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Girardot F, Lasbleiz C, Monnier V, Tricoire H. Specific age-related signatures in Drosophila body parts transcriptome. BMC Genomics. 2006;7:69. doi: 10.1186/1471-2164-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Glass D, Vinuela A, Davies MN, Ramasamy A, Parts L, Knowles D, Brown AA, Hedman AK, Small KS, Buil A, Grundberg E, Nica AC, Di Meglio P, Nestle FO, Ryten M, Durbin R, McCarthy MI, Deloukas P, Dermitzakis ET, Weale ME, Bataille V, Spector TD. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 2013;14:R75. doi: 10.1186/gb-2013-14-7-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Calvani R, Joseph AM, Adhihetty PJ, Miccheli A, Bossola M, Leeuwenburgh C, Bernabei R, Marzetti E. Mitochondrial pathways in sarcopenia of aging and disuse muscle atrophy. Biological chemistry. 2013;394:393–414. doi: 10.1515/hsz-2012-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Das N, Levine RL, Orr WC, Sohal RS. Selectivity of protein oxidative damage during aging in Drosophila melanogaster. Biochem J. 2001;360:209–216. doi: 10.1042/0264-6021:3600209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Brewer AC, Mustafi SB, Murray TV, Rajasekaran NS, Benjamin IJ. Reductive stress linked to small HSPs, G6PD, and Nrf2 pathways in heart disease. Antioxid Redox Signal. 2013;18:1114–1127. doi: 10.1089/ars.2012.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 342.Hiona A, Leeuwenburgh C. The role of mitochondrial DNA mutations in aging and sarcopenia: implications for the mitochondrial vicious cycle theory of aging. Exp Gerontol. 2008;43:24–33. doi: 10.1016/j.exger.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Hiona A, Sanz A, Kujoth GC, Pamplona R, Seo AY, Hofer T, Someya S, Miyakawa T, Nakayama C, Samhan-Arias AK, Servais S, Barger JL, Portero-Otin M, Tanokura M, Prolla TA, Leeuwenburgh C. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One. 2010;5:e11468. doi: 10.1371/journal.pone.0011468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Kolesar JE, Safdar A, Abadi A, MacNeil LG, Crane JD, Tarnopolsky MA, Kaufman BA. Defects in mitochondrial DNA replication and oxidative damage in muscle of mtDNA mutator mice. Free Radic Biol Med. 2014 doi: 10.1016/j.freeradbiomed.2014.07.038. [DOI] [PubMed] [Google Scholar]
- 345.Ahlqvist KJ, Hamalainen RH, Yatsuga S, Uutela M, Terzioglu M, Gotz A, Forsstrom S, Salven P, Angers-Loustau A, Kopra OH, Tyynismaa H, Larsson NG, Wartiovaara K, Prolla T, Trifunovic A, Suomalainen A. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 346.Schneider JL, Cuervo AM. Liver autophagy: much more than just taking out the trash. Nature reviews Gastroenterology & hepatology. 2014;11:187–200. doi: 10.1038/nrgastro.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- 348.Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461:987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- 349.Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7:e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Khraiwesh H, Lopez-Dominguez JA, Lopez-Lluch G, Navas P, de Cabo R, Ramsey JJ, Villalba JM, Gonzalez-Reyes JA. Alterations of Ultrastructural and Fission/Fusion Markers in Hepatocyte Mitochondria From Mice Following Calorie Restriction With Different Dietary Fats. J Gerontol A Biol Sci Med Sci. 2013 doi: 10.1093/gerona/glt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Jung T, Hohn A, Grune T. Lipofuscin: detection and quantification by microscopic techniques. Methods Mol Biol. 2010;594:173–193. doi: 10.1007/978-1-60761-411-1_13. [DOI] [PubMed] [Google Scholar]
- 352.Yin D. Biochemical basis of lipofuscin, ceroid, and age pigment-like fluorophores. Free Radic Biol Med. 1996;21:871–888. doi: 10.1016/0891-5849(96)00175-x. [DOI] [PubMed] [Google Scholar]
- 353.Schmucker DL, Sachs H. Quantifying dense bodies and lipofuscin during aging: a morphologist's perspective. Archives of gerontology and geriatrics. 2002;34:249–261. doi: 10.1016/s0167-4943(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 354.Brown-Borg HM, Bartke A. GH and IGF1: roles in energy metabolism of long-living GH mutant mice. J Gerontol A Biol Sci Med Sci. 2012;67:652–660. doi: 10.1093/gerona/gls086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Nef S, Verma-Kurvari S, Merenmies J, Vassalli JD, Efstratiadis A, Accili D, Parada LF. Testis determination requires insulin receptor family function in mice. Nature. 2003;426:291–295. doi: 10.1038/nature02059. [DOI] [PubMed] [Google Scholar]
- 356.Adham IM, Agoulnik AI. Insulin-like 3 signalling in testicular descent. International journal of andrology. 2004;27:257–265. doi: 10.1111/j.1365-2605.2004.00481.x. [DOI] [PubMed] [Google Scholar]
- 357.Yuan R, Meng Q, Nautiyal J, Flurkey K, Tsaih SW, Krier R, Parker MG, Harrison DE, Paigen B. Genetic coregulation of age of female sexual maturation and lifespan through circulating IGF1 among inbred mouse strains. Proc Natl Acad Sci U S A. 2012;109:8224–8229. doi: 10.1073/pnas.1121113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Vellai T, McCulloch D, Gems D, Kovacs AL. Effects of sex and insulin/insulin-like growth factor-1 signaling on performance in an associative learning paradigm in Caenorhabditis elegans. Genetics. 2006;174:309–316. doi: 10.1534/genetics.106.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Pitetti JL, Calvel P, Romero Y, Conne B, Truong V, Papaioannou MD, Schaad O, Docquier M, Herrera PL, Wilhelm D, Nef S. Insulin and IGF1 receptors are essential for XX and XY gonadal differentiation and adrenal development in mice. PLoS Genet. 2013;9:e1003160. doi: 10.1371/journal.pgen.1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Belgacem YH, Martin JR. Disruption of insulin pathways alters trehalose level and abolishes sexual dimorphism in locomotor activity in Drosophila. J Neurobiol. 2006;66:19–32. doi: 10.1002/neu.20193. [DOI] [PubMed] [Google Scholar]
- 361.Kuo TH, Fedina TY, Hansen I, Dreisewerd K, Dierick HA, Yew JY, Pletcher SD. Insulin signaling mediates sexual attractiveness in Drosophila. PLoS Genet. 2012;8:e1002684. doi: 10.1371/journal.pgen.1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Antebi A. Steroid regulation of C. elegans diapause, developmental timing, and longevity. Curr Top Dev Biol. 2013;105:181–212. doi: 10.1016/B978-0-12-396968-2.00007-5. [DOI] [PubMed] [Google Scholar]
- 363.Shen J, Ford D, Landis GN, Tower J. Identifying sexual differentiation genes that affect Drosophila life span. BMC Geriatr. 2009;9:56. doi: 10.1186/1471-2318-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci U S A. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Tricoire H, Battisti V, Trannoy S, Lasbleiz C, Pret AM, Monnier V. The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mech Ageing Dev. 2009;130:547–552. doi: 10.1016/j.mad.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 366.Simon AF, Shih C, Mack A, Benzer S. Steroid control of longevity in Drosophila melanogaster. Science. 2003;299:1407–1410. doi: 10.1126/science.1080539. [DOI] [PubMed] [Google Scholar]
- 367.Yamamoto R, Bai H, Dolezal AG, Amdam G, Tatar M. Juvenile hormone regulation of Drosophila aging. BMC Biol. 2013;11:85. doi: 10.1186/1741-7007-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Chamseddin KH, Khan SQ, Nguyen ML, Antosh M, Morris SN, Kolli S, Neretti N, Helfand SL, Bauer JH. takeout-dependent longevity is associated with altered Juvenile Hormone signaling. Mech Ageing Dev. 2012;133:637–646. doi: 10.1016/j.mad.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Magner DB, Wollam J, Shen Y, Hoppe C, Li D, Latza C, Rottiers V, Hutter H, Antebi A. The NHR-8 nuclear receptor regulates cholesterol and bile acid homeostasis in C. elegans. Cell Metab. 2013;18:212–224. doi: 10.1016/j.cmet.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Thondamal M, Witting M, Schmitt-Kopplin P, Aguilaniu H. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nature communications. 2014;5:4879. doi: 10.1038/ncomms5879. [DOI] [PubMed] [Google Scholar]
- 372.Dumas KJ, Guo C, Shih HJ, Hu PJ. Influence of steroid hormone signaling on life span control by Caenorhabditis elegans insulin-like signaling. G3 (Bethesda) 2013;3:841–850. doi: 10.1534/g3.112.005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Broue F, Liere P, Kenyon C, Baulieu EE. A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell. 2007;6:87–94. doi: 10.1111/j.1474-9726.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- 374.Owen BM, Bookout AL, Ding X, Lin VY, Atkin SD, Gautron L, Kliewer SA, Mangelsdorf DJ. FGF21 contributes to neuroendocrine control of female reproduction. Nat Med. 2013;19:1153–1156. doi: 10.1038/nm.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Zhang Y, Xie Y, Berglund ED, Coate KC, He TT, Katafuchi T, Xiao G, Potthoff MJ, Wei W, Wan Y, Yu RT, Evans RM, Kliewer SA, Mangelsdorf DJ. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. eLife. 2012;1:e00065. doi: 10.7554/eLife.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, Kapahi P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Hauser F, Grimmelikhuijzen CJ. Evolution of the AKH/corazonin/ACP/GnRH receptor superfamily and their ligands in the Protostomia. General and comparative endocrinology. 2014 doi: 10.1016/j.ygcen.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 378.Waterson MJ, Chung BY, Harvanek ZM, Ostojic I, Alcedo J, Pletcher SD. Water sensor ppk28 modulates Drosophila lifespan and physiology through AKH signaling. Proc Natl Acad Sci U S A. 2014;111:8137–8142. doi: 10.1073/pnas.1315461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Miller WL. Steroid hormone synthesis in mitochondria. Mol Cell Endocrinol. 2013;379:62–73. doi: 10.1016/j.mce.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 380.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 381.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 382.Amador-Noguez D, Zimmerman J, Venable S, Darlington G. Gender-specific alterations in gene expression and loss of liver sexual dimorphism in the long-lived Ames dwarf mice. Biochem Biophys Res Commun. 2005;332:1086–1100. doi: 10.1016/j.bbrc.2005.05.063. [DOI] [PubMed] [Google Scholar]
- 383.Regitz-Zagrosek V, Lehmkuhl E, Mahmoodzadeh S. Gender aspects of the role of the metabolic syndrome as a risk factor for cardiovascular disease. Gend Med. 2007;4(Suppl B):S162–S177. doi: 10.1016/s1550-8579(07)80056-8. [DOI] [PubMed] [Google Scholar]
- 384.Oertelt-Prigione S, Regitz-Zagrosek V. Women's cardiovascular health: prevention is key. Archives of internal medicine. 2009;169:1740–1741. doi: 10.1001/archinternmed.2009.353. [DOI] [PubMed] [Google Scholar]
- 385.Ritzel RM, Capozzi LA, McCullough LD. Sex, stroke, and inflammation: the potential for estrogen-mediated immunoprotection in stroke. Hormones and behavior. 2013;63:238–253. doi: 10.1016/j.yhbeh.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 387.Pellegrini P, Contasta I, Del Beato T, Ciccone F, Berghella AM. Gender-specific cytokine pathways, targets, and biomarkers for the switch from health to adenoma and colorectal cancer. Clinical & developmental immunology. 2011;2011:819724. doi: 10.1155/2011/819724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Oukka M. Th17 cells in immunity and autoimmunity. Annals of the rheumatic diseases. 2008;67(Suppl 3):iii26–iii29. doi: 10.1136/ard.2008.098004. [DOI] [PubMed] [Google Scholar]
- 389.Contasta I, Totaro R, Pellegrini P, Del Beato T, Carolei A, Berghella AM. A gender-related action of IFNbeta-therapy was found in multiple sclerosis. Journal of translational medicine. 2012;10:223. doi: 10.1186/1479-5876-10-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Berghella AM, Contasta I, Marulli G, D'Innocenzo C, Garofalo F, Gizzi F, Bartolomucci M, Laglia G, Valeri M, Gizzi M, Friscioni M, Barone M, Del Beato T, Secinaro E, Pellegrini P. Ageing gender-specific "Biomarkers of Homeostasis", to protect ourselves against the diseases of the old age. Immunity & ageing : I & A. 2014;11:3. doi: 10.1186/1742-4933-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Regitz-Zagrosek V. Therapeutic implications of the gender-specific aspects of cardiovascular disease. Nature reviews Drug discovery. 2006;5:425–438. doi: 10.1038/nrd2032. [DOI] [PubMed] [Google Scholar]
- 392.Clayton JA, Collins FS. Policy: NIH to balance sex in cell and animal studies. Nature. 2014;509:282–283. doi: 10.1038/509282a. [DOI] [PMC free article] [PubMed] [Google Scholar]