Summary
Mycobacterium tuberculosis and Staphylococcus aureus secrete virulence factors via Type VII protein secretion (T7S), a system that intriguingly requires all of its secretion substrates for activity. To gain insights into T7S function, we used structural approaches to guide studies of the putative translocase EccC, a unique enzyme with three ATPase domains, and its secretion substrate EsxB. The crystal structure of EccC revealed that the ATPase domains are joined by linker/pocket interactions that modulate its enzymatic activity. EsxB binds via its signal sequence to an empty pocket on the C-terminal ATPase domain, which is accompanied by an increase in ATPase activity. Surprisingly, substrate binding does not activate EccC allosterically, but rather by stimulating its multimerization. Thus, the EsxB substrate is also an integral T7S component, illuminating a mechanism that helps explain interdependence of substrates and suggests a model in which binding of substrates modulates their coordinate release from the bacterium.
Introduction
While all cells secrete proteins through the conserved Sec system, bacteria also utilize specialized secretion systems to interact with their environment (Waksman, 2012). These systems are particularly important for bacterial pathogens, as they allow for regulated secretion of virulence factors into eukaryotic cells during infection. The Type VII secretion (T7S) system, the only specialized secretion system found exclusively in Gram-positive bacteria (Huppert et al., 2014; Waksman, 2012), is required for virulence of several bacterial pathogens, including Mycobacterium tuberculosis (Guinn et al., 2004; Houben et al., 2014; Hsu et al., 2003; Stanley et al., 2003), Mycobacterium marinum (Davis and Ramakrishnan, 2009; Gao et al., 2004), and Staphylococcus aureus (Burts et al., 2005). The significance of this secretion system is further highlighted by the fact that loss of the ESX-1 T7S system in M. tuberculosis is the most important genetic difference between virulent strains that cause tuberculosis and the live attenuated vaccine strain, BCG (Brodin et al., 2006; Mahairas et al., 1996; Pym et al., 2003). However, despite its medical importance and its broad evolutionary conservation, the molecular architecture, mechanism of secretion, and regulation of T7S are unknown.
T7S systems have been identified in many Gram-positive organisms and are defined by the presence of two conserved elements: EccC, a membrane-bound protein with three predicted ATPase domains, and EsxB, a small secretion substrate containing a WXG motif (Bitter et al., 2009; Pallen, 2002). Other components have been genetically linked to T7S, but these are not universally conserved (Abdallah et al., 2007). EccC and EsxB interact physically (Stanley et al., 2003) and the last seven amino acids of EsxB constitute a “signal-sequence” that is necessary and sufficient for secretion through the ESX-1 system (Champion et al., 2006), although additional signals adjacent to these sequences are also required for full secretion (Daleke et al., 2012; Sysoeva et al., 2014). The molecular basis of T7S substrate-targeting selection is not known, and our understanding of substrate recognition has been mostly limited to yeast two-hybrid and genetic studies. One interesting feature of T7S is that substrates are co-dependent for secretion (Fortune et al., 2005), in that genetic removal of one substrate abrogates secretion of all other substrates through a specific T7S system. This unique feature of T7S has complicated the study of individual virulence factors in the context of infection and has thwarted attempts to genetically engineer these systems to secrete heterologous proteins.
EccC has a unique multi-domain structure consisting of a two-pass transmembrane domain, a short domain of unknown function (DUF) and three P-loop NTPase domains that share approximately 20% identity to one another (Figure 1A). The ATPase domains are evolutionarily related to the ASCE (Additional Strand Conserved Glutamate) fold family that includes protein and DNA directed mechanoenzymes such as FtsK, VirD4 (TrwB) and VirB4 (TrwK) (Erzberger and Berger, 2006). These motor proteins generally assemble into hexameric rings with the ATPase activity dependent on “arginine finger” residues that extend into adjacent monomers to form the active site (Ahmadian et al., 1997). The individual ATPase domains of EccC are unique in that each has a long N-terminal linker that is of unknown function but contains several motifs that are highly conserved among all of the EccC proteins.
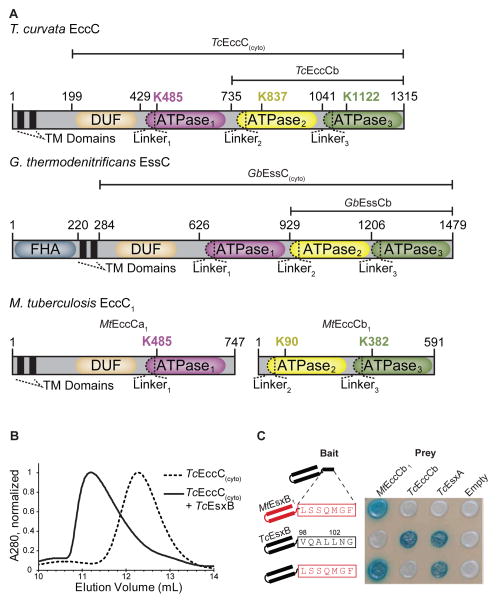
Figure 1. The EccC ATPase has a unique, conserved domain structure and binds to the EsxB signal sequence.
(A) Domain structure of the EccC and EssC ATPases. (B) Size exclusion chromatography showing that TcEsxB binds to TcEccC(cyto) and induces a large shift in elution volume. (C) Yeast-two-hybrid analysis of interactions between EccC and EsxB. Wild-type TcEsxB and MtEsxB1 are directed specifically to their cognate ATPase via the last seven amino acids (boxed), which are not required for interaction with EsxA. Also see Figure S1
We present here a series of structures of EccC, both with and without the EsxB signal sequence, which reveal that EccC exists in an autoinhibited state as a tightly integrated set of three ATPase domains joined to one another through specific linker/pocket interactions. We show that EccC activity is activated by disruption of one of these linker interactions and further activated through substrate-mediated multimerization of the enzyme. Our findings suggest that substrates, in addition to serving roles outside of the cell, are also necessary components of the secretion apparatus itself, and provide a mechanistic explanation for the unique interdependence of substrate secretion in T7S.
Results
The EsxB Signal Sequence Binds the EccC Translocase but does not Activate its ATPase Activity
To understand the nature of the interaction between EccC and EsxB using an in vitro system, we screened a panel of EccC/EsxB pairs from various bacterial species and found robust expression in E. coli of the cytoplasmic portion of EccC from the thermophilic actinobacterium Thermomonospora curvata (TcEccC(cyto)) and its cognate EsxB partner (TcEsxB). The T. curvata secretion system shares close homology with other actinomycete T7S systems and contains all of the conserved components identified in the M. tuberculosis Esx system including EsxA, EsxB, EccC, EccD, EccB, and MycP1 (Bitter et al., 2009) (Figure S1A). TcEccC(cyto), TcEsxA and TcEsxB were all stable in isolation and strongly bound one another to form an EccC:EsxB:EsxA complex (Figure 1B; Figure S1B and S1C). Similar to yeast two-hybrid studies with M. tuberculosis proteins (Champion et al., 2006), the last seven amino acids of TcEsxB specifically targeted the substrate to TcEccC, and swapping this sequence with the C-terminus of MtEsxB1 completely reversed the specificity (Figure 1C). Thus the known EccC interactions of the virulence associated ESX-1 system of M. tuberculosis are recapitulated in our model system.
In the ESX-1 system in M. tuberculosis, MtEccC is split into two polypeptides, MtEccCa (containing the trans- and juxtamembrane regions and ATPase1) and MtEccCb (containing ATPase2 and ATPase3), which interact with one another to form a complete MtEccCab complex (Stanley et al., 2003). The substrate MtEsxB interacts exclusively with MtEccCb and not with MtEccCa and we found this feature was conserved in our T. curvata system. When we artificially split TcEccC into “TcEccCa” and “TcEccCb” fragments orthologous to the tuberculosis ESX-1 proteins, these fragments interacted robustly in the two-hybrid assay (Figure S1B). Likewise, EsxB interacted directly with TcEccCb but not TcEccCa, which parallels the ESX-1 system (Figure S1B and S1D).
In analogy with other phylogenetically related translocases (Guglielmini et al., 2013), which are often strongly activated by their substrates (Massey et al., 2006), we hypothesized that binding of TcEccC(cyto) to the substrate TcEsxB would activate its ATPase domains. However we could not measure any ATPase activity in the TcEccC(cyto) or TcEccCb proteins, either in the presence or absence of TcEsxB (Figure S1E). Likewise, the nucleotide binding state of EccC had no effect on the apparent KD of signal sequence binding (Figure S1F). This unexpected result suggested that the binding to EccC does not immediately lead to work being done on the substrate.
The Structure of TcEccCb Bound to the Signal Sequence
In order to understand the interaction between EsxB and EccC, we solved the structure of TcEccCb (containing ATPase2 and ATPase3) using data to 3.24 Å resolution, in combination with a peptide containing the last 23 residues of the TcEsxB substrate, including the C-terminal signal sequence (Figure 2A; Figure S2A and Table S2). Both ATPase2 and ATPase3 are clearly bound to ATP in the structure (Figure 2B and 2C) suggesting that the ATPase activity of these domains is indeed extremely low, even in the presence of saturating amounts of EsxB signal sequence peptide. This ATPase-inactivated state appears to be evolutionarily conserved, as a high-resolution crystal structure of a fragment of the related EssC ATPase from Geobacillus thermodenitrificans (“GbEssCb”) has a very similar structure, with both domains bound to ATP (Figure S2B).
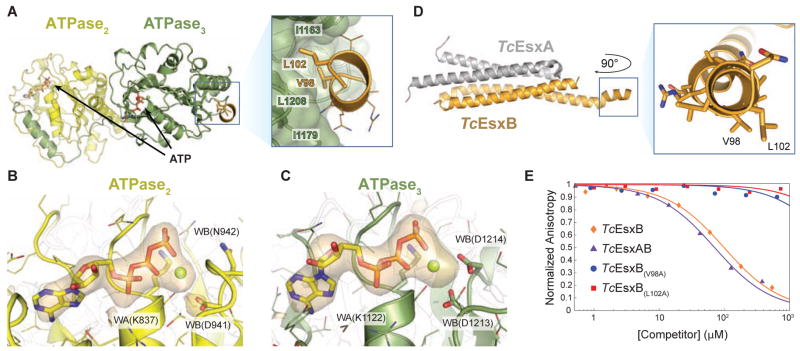
Figure 2. Co-crystal structure reveals signal sequence binding pocket in TcEccCb.
(A) The crystal structure of TcEccCb (ATPase domains colored as in Figure 1A) bound to the C-terminal signal sequence of TcEsxB (gold). Binding of the C-terminal amino acids of TcEsxB to ATPase3 is mediated by interactions with two conserved hydrophobic residues that bind in a hydrophobic binding pocket. Only the C-terminal signal sequence residues are interpretable in the electron density (Figure S2A), and the Y-X-X-X-D/E motif implicated in secretion (Daleke et al., 2012) appears disordered in the crystal. (B–C) The orange volume represents the simulated-annealing difference-density map calculated for ATPase2 ( B) and ATPase3 ( C) without nucleotide and contoured at 4 σ. (D) X-ray structure of Tc the EsxAB heterodimer with a close-up view of the C-terminal signal sequence helix. V98 and L102, which are necessary for binding to TcEccC, are labeled. (E) Binding of a fluorescently labeled signal sequence peptide (5-FAM-VNRVQALLNG) to TcEccC(cyto) monitored in the presence of increasing concentrations of unlabeled competing full length TcEsxB. Wild-type TcEsxB and TcEsxAB heterodimer compete with the peptide. Mutations in L102 or V98 prevent competition with the wild-type peptide, indicating that they do not bind. Presented data are representative experiments. Also see Figure S2.
The C-terminus of the signal sequence peptide, which was previously thought to be unstructured (Renshaw et al., 2005), forms a short amphipathic helix (residues 96–103) that interacts exclusively with the hydrophobic pocket on ATPase3 (pocket3) (Figure 2A). This C-terminal helix is likely a common feature of all EsxB homologs (Poulsen et al., 2014). The helix was also present in a higher resolution structure of the full length TcEsxBA complex in the absence of ATPase3 (Figure 2D and Table S2). Here, we observed the characteristic helical hairpin seen in all EsxB homolog proteins, however in our structure the chain makes a turn through a short extended region (residues 93–95) before ending in a helix that matches the length of helix observed in the ATPase complex structure. Of note, this helix is found in a crystal contact with an adjacent symmetry related molecule, which could artificially stabilize the helical structure. Although present in the EsxB fragment crystallized with EccC, the Y-X-X-X-D/E motif implicated in secretion (Daleke et al., 2012; Sysoeva et al., 2014) was disordered in our crystal, suggesting that it is not involved in recognition of the signal sequence motif. Importantly, pocket3 is distant from the ATP catalytic site and binding of the peptide does not appear to alter the ATP binding ability of ATPase3. Mutation of any of the EsxB interaction residues on either EsxB or EccC completely abrogated the interaction, demonstrating the specificity of its binding to pocket3 (Figure 2E and S2C). Together, these data show that TcEsxB is targeted to TcEccCb through specific binding to the hydrophobic pocket3 on ATPase3, but binding of the C-terminus of TcEsxB neither requires nor enhances nucleotide hydrolysis or exchange.
Because EccC lacked ATPase activity with or without substrate, we examined the evolutionary conservation of each ATPase domain amongst many unique EccC orthologs to determine if the residues required for ATPase activity are conserved. We found that the catalytic residues of ATPase2 and ATPase3 are highly degenerate with respect to other related ATPases, especially in the catalytic glutamate of the Walker B motif (Figure S2D). Such changes might be expected to greatly reduce or eliminate ATP hydrolysis (Wendler et al., 2012), which is consistent with the presence of ATP in these domains observed in our crystal structures. In contrast, ATPase1 is highly conserved with its closest known homolog, the motor protein FtsK, suggesting ATPase1 may serve as the active motor domain for EccC. Thus, ATPase2 and ATPase3 appear to be naturally suboptimal ATPases, similar to the catalytically inactive domains of other multimeric ATPases such as dynein (Carter et al., 2011) and the F1-ATPase (Walker, 2013).
ATPase1 is Inhibited by its Interaction with ATPase2
To understand the structure of ATPase1 and its relationship to ATPase2 and ATPase3, we solved the crystal structure of the full cytoplasmic domain of TcEccC, “TcEccC(cyto),” using data to 2.9 Å resolution (Figure 3A; Figure S3A and S3B; Table S3). Although the full protein is present in the crystal (Figure S3C), the N-terminal “DUF” domain and linker1 are disordered in the structure and could not be modeled. The structure is monomeric, as it is in solution (Figure S3D), and ATPase2 and ATPase3 are very similar in both their conformation and nucleotide binding state compared to the TcEccCb structure (RMSD 0.7 Å), showing that binding of the signal sequence does not alter EccC’s structure in these domains. The interface between ATPase1 and ATPase2 is remarkably similar to the interface between ATPase2 and ATPase3, joining together the three domains in a direct translation where the only interfaces between the domains are mediated by the inter-domain linkers. Highlighting the general importance of these linker interactions, removal of the N-terminal 34 amino acids homologous to linker2 on MtEccCb completely blocked binding to MtEccCa (Figure S3E and S3F). Single-particle electron microscopy and 3D reconstruction of TcEccC(cyto) and a related ATPase from Geobacillus thermodenitrificans revealed a similar monomeric structure that was remarkably rigid, as illustrated in the homogeneity of the class averages (Figure 3B and 3C; Figure S3G, S3H, S3I, and S3J). The DUF domain, which is required for secretion in vivo (Figure S3K), is also visible in these images, though its density is reduced, likely due to averaging of multiple flexible states.
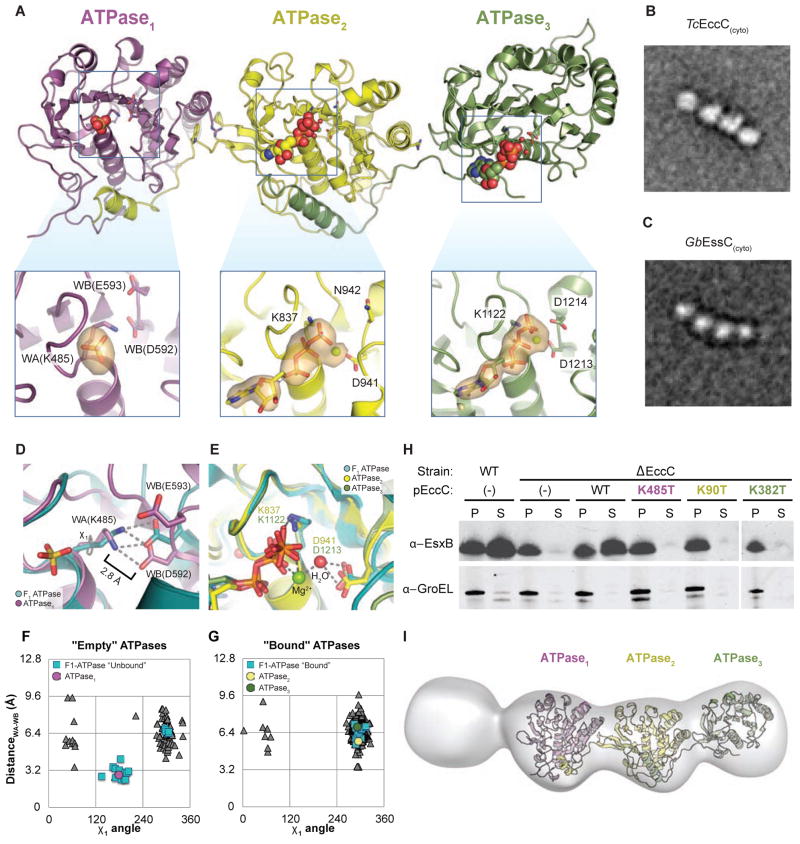
Figure 3. ATPase1 is autoinhibited and integrated into a ridged array of ATPase domains.
(A) The crystal structure of TcEccC(cyto) highlighting the differences between the ATP- bound catalytic sites of ATPase2 and ATPase3 and the nucleotide free site of ATPase1. The orange volume represents a simulated-annealing difference-density map calculated without nucleotide or sulfate and contoured at 3 σ. Note that the ATPase3 insert has been rotated slightly to allow for comparison between the ATPase active sites (B–C) Representative EM class average of (B) TcEccC(cyto) and ( GbC) EssC(cyto) showing the linear structure of EccC. Scale bars are 100 Å. (D) ATPase1 (purple), with the “empty” subunit of F1-ATPase (PDB 1H8H) overlaid in cyan and (E) ATPase2 (yellow) and ATPase3 (green) overlaid with the AMP-PNP-bound subunit of F1-ATPase (cyan) from 1H8H. (F) A graph representing the distance between the Walker A lysine amino group and the closest Walker B carboxylate oxygen, as a function of the rotameric position of the Walker A lysine. Each triangle represents one of 311 pdb chains of an ATP bound, P-loop ATPase identified by our protocol (see supplementary methods). (G) A similar graph to (F) except the triangle represents the residues of “empty” ATPases from PDB entries that contain both a bound and unbound P-loop ATPase domain in the same file. (H) Western blot detection of MtEsxB1 and GroEL from cell supernatants (S) and cell pellet lysate (P) fractions of MtEccC1 knockout and complemented cells. (I) Three- dimensional reconstruction at an estimated resolution of 23 Å, based on 1634 images in the presence of 1 mM ATP-γS and 10 mM MgCl2. The model has been contoured to fit the crystal structure. Though it is impossible to resolve the difference between the first and fourth domains, the electron density of one is much lower than the other three, suggesting this domain is the DUF domain, which is disordered in the crystal structure. Also see Figure S3.
In ATPase1 the nucleotide binding residues and nucleotide loading is strikingly different from the other two domains (Figure 3D and 3E). Despite the high ATP concentration in the crystallization solution (5 mM), ATPase1 contains a sulfate ion in the active site (Figure 3A) whereas ATPase2 and ATPase3 are bound to ATP as they were in the signal sequence-bound structure (Figure 2). Several structural features of the ATPase1 catalytic site are strongly reminiscent of the ATP “empty” (βE) subunit of the F1-ATPase (Figure 3D and 3E) which is known to have a very low affinity for nucleotide (Menz et al., 2001; Senior, 2012). In particular, the Walker A lysine is rotated into an unfavorable rotamer and bound to the Walker B aspartate, displacing the binding of magnesium in the active site and likely preventing binding of ATP. An analysis of all P-loop ATPases in the Protein Data Bank (Berman et al., 2002) that contain both ATP-bound and ATP-unbound subunits in the asymmetric unit found that this configuration of the enzymatic residues in the ATP binding site is very unusual under these conditions and is essentially restricted to structural models of the empty state of the F1-ATPase (Figure 3F and 3G).
Despite the low affinity state in the crystal, the ability of ATPase1 to bind ATP is required in vivo (Figure 3H and Figure S3L, S3M, and S3N) showing that cycling of ATPase1 into an ATP avid conformation is required for the function of the secretion system. We conclude that we have captured a low-nucleotide-affinity state of ATPase1 that must be reversed during the EccC catalytic cycle.
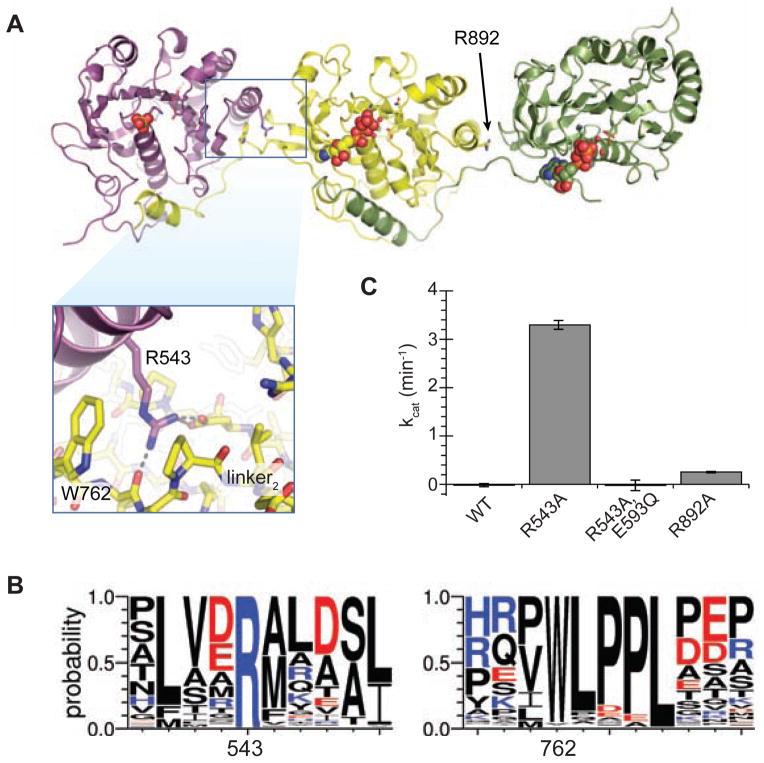
Overlaying the three ATPase domains revealed that the linker-pocket interactions of ATPase1 and ATPase2 are analogous to signal sequence binding of ATPase3 (Figure 4A). In particular, the important residues for signal sequence binding in pocket3 have clear homologs in the linker2-pocket1 interaction (Figure 4B). Since ATPases are often modulated by the effect of N- and C-terminal appendages (Besprozvannaya et al., 2013; Karamanou et al., 1999; Peña et al., 2011), we hypothesized that the attachment between pocket1 and linker2 might allosterically regulate ATPase1, locking it into the low affinity form seen in the crystal structure and leading to the low catalytic rate we observed in vitro. Much of the interface between ATPase1 and linker2 is mediated by a 100% conserved arginine in pocket1, R543, that interacts with W762 and L763 in linker2 (Figure 5A and 5B). We reasoned that loss of this interaction might mimic an allosteric effector binding in pocket1 in a manner analogous to signal sequence binding to ATPase3 and modulate the activity of ATPase1. Indeed, mutation of R543 to alanine resulted in a sharp increase in EccC ATPase activity (Figure 5C). Additional mutation of an ATPase1 catalytic residue (E593Q) completely inhibited this activation, suggesting that this increase in ATPase activity is dependent on the activity of ATPase1. Because R543 does not make any direct interactions with the ATP binding or catalytic residues, these results strongly suggest that the activity of ATPase1 is modulated allosterically by its interaction with the linker. Mutation of the equivalent residue at the ATPase2-ATPase3 interface had no significant effect on the activity of the enzyme. Therefore, we conclude that the activity of ATPase1 is controlled through its pocket-linker interaction with ATPase2. Importantly, mutation of R543 to alanine in MtEccCa1 severely reduced secretion of EsxB by M. tuberculosis (Figure S4A and S4B), indicating that the interface between ATPase1 and ATPase2 is critical for the secretion process. This is consistent with a model in which this interface couples substrate recognition in EccCb (ATPase2 and ATPase3) with EccCa (ATPase1) activity.
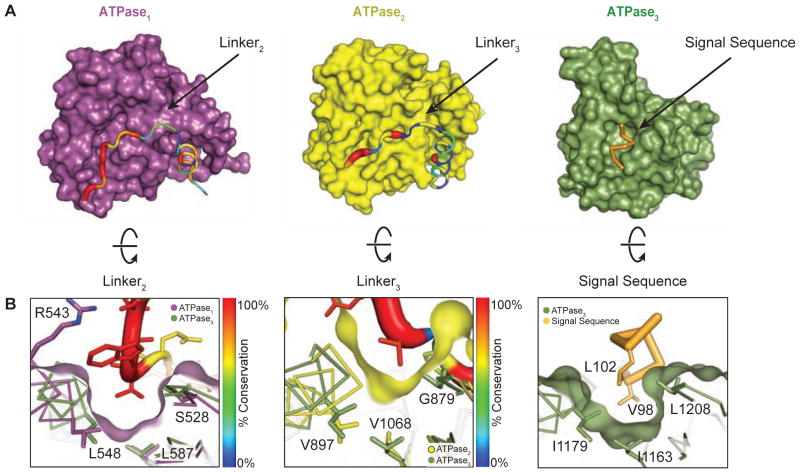
Figure 4. Residues in linker2 and linker3 mimic the substrate and bind to pocket1 and pocket2 on TcEccC.
(A) The individual ATPase domains are shown and have been rotated to reveal the path of the linker across the ATPase domain. The linker is colored and weighted in diameter according to the degree of conservation across 142 unique EccC sequences. (B) The surface has been rotated to highlight the linker groove. ATPase3 and the pocket residues (Figure S2C) overlay ATPase1 and ATPase2 to highlight the homologies in the linker binding and signal sequence binding pockets. The ATPase2 pocket is significantly shallower than ATPase1 and ATPase3.
Figure 5. ATPase1 is held in an autoinhibited state by inter-ATPase interactions.
(A) Crystal structure of TcEccC(cyto) with inset highlighting the interface between ATPase1 and ATPase2. (B) Logo diagram representing the alignment of 142 unique EccC sequences. (C) Disruption of ATPase1-ATPase2 interface by R543A mutation activates the ATPase activity of TcEccC, which requires the Walker B catalytic residue in ATPase1 (E593Q). An analogous mutation between ATPase2 and ATPase3, R892A, led to a small increase in activity. The graph represents three measurements of ATPase activity at an enzyme concentration of 1 μM ATPase with saturating ATP*MgCl2 (10 mM). Also see Figure S4.
EccC ATPase Activation by Substrate Binding
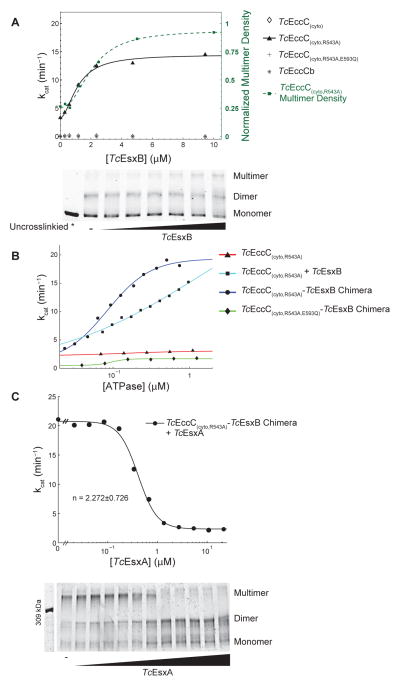
In contrast to the wild-type enzyme, addition of TcEsxB to the TcEccC(cyto,R543A) activated mutant led to a five-fold, saturating increase in the ATPase activity (Figure 6A) revealing that substrates can contribute to EccC activation if the autoinhibitory interaction between ATPase1 and ATPase2 is removed. Mutation of TcEsxB residues responsible for the interaction with ATPase3 abrogated stimulation, demonstrating that the effect is specific to signal sequence binding (Figure S4C). However, mutation of the mutation of the catalytic residues in ATPase2 and ATPase3 significantly reduced, but did not eliminate, overall ATPase activity (Figure S4E). In accord with this finding, ATP binding by ATPase2 and ATPase3 is also required for secretion in vivo (Figure 3H). In contrast, ATPase2 and ATPase3 alone had no activity and were not stimulated by TcEsxB (Figure 6A). Thus, although binding of ATP by ATPase2 and ATPase3 is required for full activity of EccC, these domains act to regulate the activity of ATPase1 rather than additively contribute to overall ATPase activity. This is consistent with recent genetic evidence that the different ATPase domains play distinct roles during secretion in vivo (Ramsdell et al., 2014).
Figure 6. EsxB and EsxA substrates control EccC activity via regulating enzyme multimerization.
(A) The ATPase activity of the indicated TcEccC proteins was measured at different concentrations of TcEsxB. Multimerization of TcEccC(cyto,R543A), detected by glutaraldehyde crosslinking (bottom panel), increases with addition of TcEsxB (0–10 μM). Quantification of the multimer band is also indicated on the ATPase activity graph (green dotted line with squares) to demonstrate correlation between multimer concentration and activity. (B) ATPase activity of the indicated proteins, either TcEccC(cyto,R543A) +/− TcETc sxB EccC or (cyto,R543A)-TcEsxB chimeras, was measured as a function of enzyme concentration. In (B) and (C) each point represents the mean of three independent measurements. (C) ATPase activity of the TcEccC(cyto,R543A)-TcEsxB chimera was measured at different concentrations of TcEsxA (top panel), and multimerization of the enzyme in these reactions was assessed by glutaraldehyde crosslinking followed by SDS-PAGE (bottom panel - top concentration of TcEsxA is 22 μM and concentrations are reduced 2-fold in each lane to the left). See also Figure S5.
EccC Activity is Controlled by Multimerization
Binding of the EsxB signal sequence to ATPase3 appears to be a simple molecular recognition event (Figure 2), and our results suggest that binding is unlikely to change the conformation of ATPase1. We thus reasoned that substrate binding could activate EccC via regulating multimerization. Indeed, the related FtsK and TrwB ATPases form multimers during their catalytic cycle in which arginine residues (“R-fingers”) complete the active site of neighboring subunits (Gomis-Rüth et al., 2001; Massey et al., 2006; Wendler et al., 2012). ATPase1 has a completely conserved R- finger (Figure S4G and S4H) that is required for secretion in vivo (Figure S4I), implying that formation of the active site of ATPase1 also involves multimerization. Furthermore, expression of ATPase-deficient versions of EccC in wild-type bacteria has a dominant- negative effect on secretion, consistent with this notion (Ramsdell et al., 2014).
In order to investigate the role of multimerization in the activation of EccC we measured the dependence of kcat on increasing concentrations of enzyme. In the absence of multimerization, the kcat should be a constant property of the enzyme, but if the catalytic pocket of one ATPase molecule is assembled in trans with an arginine donated by a different ATPase molecule, the kcat of the enzyme should increase as more arginine fingers become available with increasing concentration of enzyme. In the absence of TcEsxB, neither TcEccC(cyto) nor TcEccC(cyto,R543A) exhibited concentration- dependent ATPase activation (Figure 6B) suggesting their activity was not dependent on multimeriziation. In contrast, in the presence of a 10-fold excess of TcEsxB (TcEsxB+TcEccC(cyto,R543A)), the ATPase activity was strongly concentration dependent. To guarantee a one-to-one molar ratio between TcEsxB and TcEccC, we fused TcEsxB via a flexible 14 amino acid linker to the C-terminus of TcEccC(cyto,R543A). This protein was dimeric as determined by analytical ultracentrifugation (Figure S4F), and similarly to the TcEsxB+TcEccC(cyto,R543A) complex the kcat of this chimera was highly concentration dependent (Figure 6B), with a maximal activity similar to the saturated TcEsxB:TcEccC(cyto,R543A) complex (>100-fold over wild-type). Mutation of the R-finger residue in the activated, substrate-ATPase fusion protein (TcEsxB- TcEccC(cyto,R543A,R616Q)) reduced its activity down to the baseline activity level of the R543A mutant (Figure S4J). Thus the R543A mutant does not exhibit concentration dependent activation in the absence of the substrate and also does not require the R-finger residue for its baseline activity. These data strongly support the idea that the active form of EccC is multimeric, but this state is sparsely populated in the absence of the EsxB substrate. However, the activity of this multimeric form is only manifest in the setting of the permissive R543A mutation. These experiments define a hierarchy of activation where both the effect of the R543A mutation and the multimerization are required for appreciable ATPase activity.
EsxB, but not EsxA or EsxBA, Directly Multimerizes EccC Translocase
To probe the multimeric state of TcEccC(cyto) during active catalysis, we used glutaraldehyde crosslinking to capture higher-order multimers. Crosslinking of TcEccC(cyto,R543A) in the presence of native TcEsxB, or with the TcEsxB-TcEccC(cyto,R543A) fusion, revealed both dimer and higher-order oligomeric states that are strongly correlated with activity (Figure 6A). These oligomers were specific to EccC and did not form with the substrate alone (Figure S5A), or when ATPase was incubated with a signal sequence mutant, TcEsxB(V98A) (Figure S5C and S5D). Importantly, while TcEsxB binding was insufficient to activate ATPase activity of the wild-type enzyme (Figure 6A), it effectively drove TcEccC(cyto) into higher-order complexes (Figure S5B and S5D), indicating that substrate-mediated multimerization is not sufficient to override autoinhibition mediated by ATPase2. We found that EsxB exists as a homodimer in isolation (Figure S5E), and thus the substrate likely stabilizes multimers by first forming EccC:EsxB:EsxB:EccC complexes. Although the addition of EsxB leads to a clear increase in multimerization of the ATPase, it appears to stabilize a state that occurs in the absence of EsxB, as both crosslinking (Figure S5B) and analytical ultracentrifugation (Figure S4F) experiments revealed a low level of EccC multimerization without EsxB.
Surprisingly, addition of TcEsxA, another substrate that forms a tight 1:1 complex with TcEsxB (Figure 2D; Figure S5F, (Renshaw et al., 2002)), but does not directly bind to EccC (Figure S1B and S5G), strongly inhibited ATPase activity in a cooperative manner (Hill coefficient > 2, Figure 6C), suggesting that each TcEsxA molecule affects the activity of more than two TcEccC(cyto,R543A)-TcEsxB molecules. The higher-order multimer concentration decreased with increasing TcEsxA in a pattern that directly mirrored the cooperative decrease in ATPase activity (Figure 6C), and this was not affected by mutation of the arginine finger residue (Figure S5H and S5I). Furthermore, addition of TcEsxA to the TcEccC(cyto)-TcEsxB chimera led to loss of dimerization of this construct (Figure S5J). Thus, while EsxB homodimers promote assembly and activation of EccC, EsxA binding to EsxB-bound EccC leads to cooperative disassembly and inhibition of the multimeric ATPase.
To test whether EsxA-induced inhibition was due to disruption of the EsxB:EsxB dimerization event, we measured inhibition of ATPase activity in the TcEsxB- TcEccC(cyto,R543A) chimera with increasing concentrations of the signal sequence mutant TcEsxB(V98A), which can still form homodimers but cannot bind to EccC. We found that TcEsxB(V98A) also inhibits activity (Figure S5K), supporting the notion that EsxA inactivates EccC by removing the stabilizing effect of the EsxB:EsxB interaction, presumably by forming EccC:EsxB:EsxA trimers instead of EccC:EsxB:EsxB:EccC tetramers.
Discussion
In this work we have developed a thermophilic model system that allowed for the detailed dissection of the only two components of T7S conserved in all Gram-positive bacteria: EccC and EsxB. Based on our findings we posit a model in which secretory substrates play an active regulatory role in T7S by modulating the activity of EccC (Figure 7). In the absence of EsxB, EccC is monomeric and tightly inactivated via interactions between ATPase1 and ATPase2. EsxB binding to ATPase3, which is relatively weak (~10 μM), drives EccC multimerization, but is not sufficient for activation. Allosteric interactions through displacement of linker2 from pocket1, which relieves the inhibitory interaction with ATPase2, are also required to permit activation of EccC. While we do not yet know the nature of these activation signals, given the linker-pocket architecture found in each ATPase domain we suspect that other substrates and/or T7S components bind to these pockets to create an “AND” logic gate by which secretion of multiple substrates is coordinated, explaining the phenomenon of mutually dependent Type VII secretion (Fortune et al., 2005). The DUF and the transmembrane domains also play an important role in secretion and full delineation of their contribution to the process awaits further experimentation.
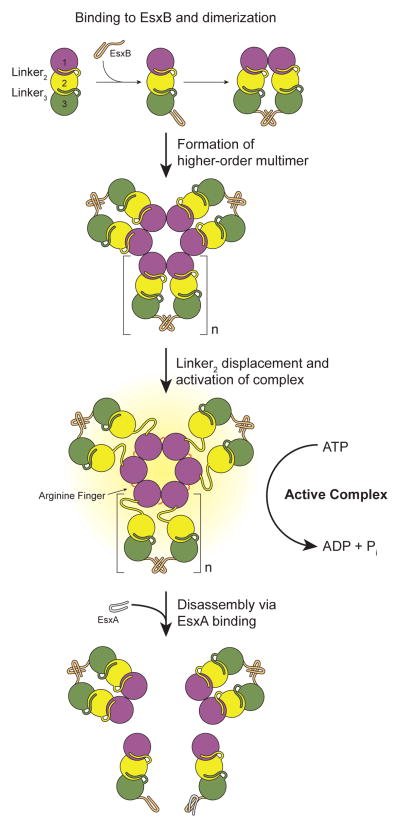
Figure 7. Model of substrate-mediated activation of EccC.
In the absence of substrates EccC is monomeric. Interaction with EsxB leads to dimerization of the ATPase and then higher order multimerization, but cannot activate the enzyme. In this study we used the R543A mutation to disrupt the interaction between ATPase1 and ATPase2 although in vivo this role may be played by other proteins that bind to ATPase1 analogously to the binding of EsxB to ATPase3, or other signals. Once ATPase1 is displaced, EccC is activated further by multimerization mediated by a conserved R-finger. EsxA can disrupt the EsxB:EsxB interaction and disassemble the multimer. We have no evidence for the structure of the EccC:EsxB dimer or stiochiomtry and structure of the multimeric form. Thus, both these aspects of the model are speculative, though based on prior structures of related substrate proteins (i.e. 3GVM) and the FtsK-like ATPases (i.e. 2IUU). We have indicated this ambiguity with the variable “n” for the number of subunits in the multimer.
This model suggests that T7S activity may be governed by a simple, “just-intime” post-translational control mechanism in which energy is expended only when key substrates are recognized by EccC (Bozdech et al., 2003). T7S may be poised for secretion under all conditions, with EccC waiting for delivery of complete sets of substrates, which would explain why removal of one substrate would inhibit secretion of others. In this way, T7S may not be directly regulated by environmental stimuli, but actuated by signal transduction pathways that regulate synthesis of substrates, such as PhoP/R (Ryndak et al., 2008) and EspR (Raghavan et al., 2008). Although other control mechanisms may be in play, this mode of regulation would not only conserve ATP consumption until it is needed, but would also allow for coordinate secretion of multiple substrates, a function that may be beneficial for the organism.
Our results also suggest that EsxB homodimers, in addition to EsxAB heterodimers (Renshaw et al., 2002), play an important role in Type VII secretion, a notion supported by the observation that ancestral T7S systems, such as those found in the phylum firmicutes, lack EsxA homologs and EsxB exists solely as a homodimer (Poulsen et al., 2014). Experimental evidence from the literature also supports the idea that EsxB dimers have an important physiological role. For example, recent work shows that EsxB in Bacillus subtilis is secreted as a dimer (Sysoeva et al., 2014). Likewise, at least four unique crystal structures of different EsxB homodimers from various bacterial species have been deposited in the Protein Data Bank including 2VRZ (Sundaramoorthy et al., 2008), 3GVM (Poulsen et al., 2014), 3ZBH (unpublished) and 3O9O (unpublished). Taken together, we believe that these results provide compelling evidence for the role of EsxB homodimers in vivo, and support a model that one role of EsxA is to antagonize the stimulatory effects of EsxB on EccC. How substrates are actually translocated out of the cell upon binding EccC, the oligomeric state of substrates during translocation, and how EccC and/or other T7S proteins modify substrates before export remain important questions that require further study.
Most motor proteins only display maximal ATPase activity in the presence of a mechanical load. Indeed, the activity of EccC, even when activated by the substrate and the R543A mutation, is relatively low. We believe the activity we measure likely represents a basal ATPase rate without the “load” of substrates to be translocated across a membrane. A graded activation of ATPase function is reminiscent of the activation of SecA translocase. In this case, SecA is nearly inactive when cytosolic (Lill et al., 1990), but is partially activated by its interaction with SecYEG, which releases an interdomain, allosteric inhibition in SecA leading to an increase in ATPase activity (Karamanou et al., 2007). The motor is thus primed for the translocation reaction, which is stimulated by its interaction with the signal sequence bearing protein (Chatzi et al., 2014).
Our structural analysis shows other intriguing similarities to the Sec translocation system. The Sec translocase binds to a similar, small helical peptide using mixed electrostatic and hydrophobic interactions. In both cases, the binding occurs in a specialized groove that is distant from the ATPase active site (Gelis et al., 2007) suggesting a role in targeting and orientation of substrates. There are also similarities to targeting of substrates in other secretion systems. For example, in the Type III secretion system a targeting sequence on a chaperone protein, CesAB, is required for interaction with the Type III ATPase, however the actual translocation is mediated by an entirely different signal (Chen et al., 2013). Given that several other regions of the EsxB and EsxA proteins have been implicated in translocation (Daleke et al., 2012; Sysoeva et al., 2014) we suspect a similar division between targeting and substrate orientation is also present in the Type VII system.
The EccC ATPase is phylogenetically related to the T4 secretion system coupling proteins, typified by the VirD4 ATPase in Agrobacterium tumefaciens (Guglielmini et al., 2013). These proteins also bind to a C-terminal sequence on substrate proteins that is necessary for secretion but the molecular interactions and biochemical effects of substrate binding in these systems is unknown (Trokter et al., 2014). In the A. tumefaciens system, three monomeric ATPases (VirB4, VirD4 and VirB11) are required for secretion of substrates by the system. It is intriguing to speculate that these three ATPases, which all appear to serve very different mechanistic purposes, may carry out functions analogous to the three ATPase domains of EccC, but this hypothesis awaits further structural information about the assembly and function of EccC and of the T4 secretion ATPases.
Targeting T7S for inhibition is an attractive antibacterial strategy, given the centrality of these systems to pathogenesis in M. tuberculosis and S. aureus, and their wide distribution among gram-positive bacteria (Chen et al., 2010). Our work suggests two unexpected targets for disruption of the function of T7 secretion. First, small molecules targeted to the inactive state of ATPase1 may stabilize its autoinhibition (Schindler et al., 2000). Secondly, the interaction pocket for the substrate is quite deep and may be amenable to small molecule targeting. Additionally, knowing the molecular determinants of signal sequence recognition may also allow us to design improved vaccine strains, which export subsets of immunodominant virulence factors but do not cause disease.
Experimental Procedures
A full description of the methods, reagents and crystallographic statistics is included in the supplemental materials.
Mycobacterial mutants and secretion assays
The ΔeccCa1Mt-ΔeccCb1Mt deletion strain was created by homologous recombination using specialized transducing phage as previously described (Glickman et al., 2000). Complementation of the eccC null mutant was carried out by cloning the entire M. tuberculosis rv3870-rv3871 locus into an integrating vector containing a C- terminal flag tag and under the control of the predicted native promoter. Secretion assays were performed as described previously (Ohol et al., 2010).
Protein expression and purification
Recombinant proteins were subcloned by PCR into a pET vector system and expressed and purified from C41(DE3) strain E. coli using standard techniques. Details are available in the Supplemental Experimental Procedures.
Electron microscopy
Single particles were picked from uranyl acetate-stained images and processed into classes containing approximately 60 images. Reconstructions were accomplished as described in the Supplemental Experimental Procedures.
Crystallization and structure solution
Crystallization and structure solution are described in detail in the Supplemental Experimental Procedures. For the EccCTc( 199-1315) structure, initial phases were determined with SAD phasing of a Ta6Br12 derivative at ~7.5 Å, then improved by MIR phasing with Pt and Hg derivatives. An anomalous difference map determined by comparison to a selenomethionine derivative assisted model building. The other structures were solved using standard methods.
Biochemical assays
Steady-state ATPase activity was analyzed using an assay that continuous coupled assay (Kornberg and Pricer, 1951) adapted to a 96 well format.
Fluorescence aniostropy was performed using a 5-FAM-VNRVQALLNG peptide interacting with the EccCTc( 199-1315) construct as described in the Supplemental Experimental Methods.
For crosslinking assays, 2-5 μg of total protein was incubated with 0.2% glutaraldehyde for 10 minutes and then quenched with 1 M Tris pH 8.0. Denaturing gels were stained with Coomassie or western blotted. A full description is available in the Supplemental Experimental Methods.
Bioinformatics
We attempted to identify all P-loop ATPases in the PDB on a per chain basis and then analyzed the position of the Walker A lysine side chain relative to the position of the Walker B aspartic acid using programs designed by the authors. The initial list and the sorted lists described in the text are available in Supplemental Table 4. Please see the Supplemental Experimental Methods for full details.
Genetic interaction studies
Directed yeast-two-hybrid studies were performed using a LacZ reporter system as described previously (Champion et al., 2006). Strain names and additional procedures are available in the Supplementary Experimental Procedures.
Supplementary Material
Highlights.
X-ray and EM structures reveal the structure the Type VII secretion ATPase EccC
EccC is a unique, linear array of three interlocking ATPase domains
The secretion substrate EsxB binds to an unexpected pocket on the third ATPase domain
Binding of substrates controls multimerization and activation of EccC
Acknowledgments
We acknowledge support from the NIH (K08AI091656 to OSR, R01AI081727 to JSC and U54GM094625 to RMS) and the NSF (Grant No. 1144247 to DD).
For technical assistance we thank Rebecca Robbins, Peter Bieling, Jenifer Du Mond, Lara Koehler, R. Stefan Isaac, Yaneth Robles, Diana Romero, Spenser Alexander and Andrew Rodriguez. For helpful discussions we thank Ron Vale, John Kuriyan, James Berger, Bennett Penn, Matt Lohse and Damian Ekiert.
We thank R. Rajashankar at the APS beamline NE-CAT 24ID, George Meigs and Jane Tanamachi at ALS BL 8.3.1 and Tzanko Doukov and Lisa Dunn at SSRL. APS is supported by the U.S. DOE Contract DE-AC02-06CH11357. ALS is supported by the U.S. DOE Contract DE-AC02-05CH11231. The SSRL Structural Molecular Biology Program is supported by the U.S. DOE Office of Biological and Environmental Research, and by the NIH-GMS, (including P41GM103393).
Footnotes
Author Contributions
O.S.R. and D.D. contributed equally to this work. O.S.R and J.S.C. conceived the research; O.S.R., D.D., X.L., Y.C., R.M.S., and J.S.C., designed experiments; O.S.R., D.D., X.L., L.C., and A.B. collected data; O.S.R., D.D., X.L., J.H., J.F.M., Y.C., R.M.S., and J.S.C. analyzed data; O.S.R., D.D., and J.S.C wrote the manuscript. All authors commented on the scientific content of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdallah AM, Gey van Pittius NC, Champion PAD, Cox J, Luirink J, Vanden-broucke-Grauls CMJE, Appelmelk BJ, Bitter W. Type VII secretion--mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated hydrolysis reaction of Ras. Nat Struct Mol Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystal-logr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- Besprozvannaya M, Pivorunas VL, Feldman Z, Burton BM. SpoIIIE achieves directional DNA translocation through allosteric regulation of ATPase activity by an accessory domain. J Biol Chem. 2013 doi: 10.1074/jbc.M113.484055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W, Houben ENG, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao L-Y, Liu J, et al. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009;5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD, Leclerc C, et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 2006;74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci USa. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion PAD, Stanley SA, Champion MM, Brown EJ, Cox JS. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- Chatzi KE, Sardis MF, Economou A, Karamanou S. SecA-mediated targeting and translocation of secretory proteins. Biochim Biophys Acta. 2014;1843:1466–1474. doi: 10.1016/j.bbamcr.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Chen JM, Pojer F, Blasco B, Cole ST. Towards anti-virulence drugs targeting ESX-1 mediated pathogenesis of Mycobacterium tuberculosis. Drug Discovery Today: Disease Mechanisms. 2010;7:e25–e31. [Google Scholar]
- Chen L, Ai X, Portaliou AG, Minetti CASA, Remeta DP, Economou A, Kalodimos CG. Substrate-Activated Conformational Switch on Chaperones Encodes a Targeting Signal in Type III Secretion. Celrep. 2013;3:709–715. doi: 10.1016/j.celrep.2013.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CMJE, Luirink J, Bitter W. General secretion signal for the mycobacterial type VII secretion pathway. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci USa. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L-Y, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Moncalián G, Pérez-Luque R, González A, Cabezón E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring hel-icases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- Guglielmini J, de la Cruz F, Rocha EPC. Evolution of Conjugation and Type IV Secretion Systems. Molecular Biology and Evolution. 2013;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben ENG, Korotkov KV, Bitter W. Take five - Type VII secretion systems of Mycobacteria. Biochim Biophys Acta. 2014;1843:1707–1716. doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci USa. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert LA, Ramsdell TL, Chase MR, Sarracino DA, Fortune SM, Burton BM. The ESX System in Bacillus subtilis Mediates Protein Secretion. PLoS ONE. 2014;9:e96267. doi: 10.1371/journal.pone.0096267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- Karamanou S, Gouridis G, Papanikou E, Sianidis G, Gelis I, Keramisanou D, Vrontou E, Kalodimos CG, Economou A. Preprotein-controlled catalysis in the helicase motor of SecA. Embo J. 2007;26:2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Pricer WE. Enzymatic phosphorylation of adenosine and 2,6-diaminopurine riboside. J Biol Chem. 1951;193:481–495. [PubMed] [Google Scholar]
- Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Löwe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. My-cobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ. The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- Peña A, Ripoll-Rozada J, Zunzunegui S, Cabezón E, la Cruz de F, Arechaga I. Autoinhibitory regulation of TrwK, an essential VirB4 ATPase in type IV secretion systems. J Biol Chem. 2011;286:17376–17382. doi: 10.1074/jbc.M110.208942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C, Panjikar S, Holton SJ, Wilmanns M, Song YH. WXG100 Protein Superfamily Consists of Three Subfamilies and Exhibits an α-Helical C-Terminal Conserved Residue Pattern. PLoS ONE. 2014;9:e89313. doi: 10.1371/journal.pone.0089313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, Griffiths KE, Marchal G, Leclerc C, Cole ST. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdell TL, Huppert LA, Sysoeva TA, Fortune SM, Burton BM. Linked Domain Architectures Allow for Specialization of Function in the FtsK/SpoIIIE ATPases of ESX Secretion Systems. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, Gordon SV, Hewinson RG, Burke B, Norman J, et al. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. Embo J. 2005;24:2491–2498. doi: 10.1038/sj.emboj.7600732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw PS, Panagiotidou P, Whelan A, Gordon SV, Hewinson RG, William-son RA, Carr MD. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J Biol Chem. 2002;277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- Ryndak M, Wang S, Smith I. PhoP, a key player in Mycobacterium tuberculosis virulence. Trends Microbiol. 2008;16:528–534. doi: 10.1016/j.tim.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Senior AE. Two ATPases. J Biol Chem. 2012;287:30049–30062. doi: 10.1074/jbc.X112.402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci USa. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy R, Fyfe PK, Hunter WN. Structure of Staphylococcus aureus EsxA suggests a contribution to virulence by action as a transport chaperone and/or adaptor protein. J Mol Biol. 2008;383:603–614. doi: 10.1016/j.jmb.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysoeva TA, Zepeda-Rivera MA, Huppert LA, Burton BM. Dimer recognition and secretion by the ESX secretion system in Bacillus subtilis. Proc Natl Acad Sci USa. 2014 doi: 10.1073/pnas.1322200111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G. ScienceDirectRecent advances in the structural and molecular biology of type IV secretion systems. Curr Opin Struct Biol. 2014;27:16–23. doi: 10.1016/j.sbi.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G. Bacterial secretion comes of age. Philos Trans R Soc Lond, B, Biol Sci. 2012;367:1014–1015. doi: 10.1098/rstb.2011.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- Wendler P, Ciniawsky S, Kock M, Kube S. Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.