Summary
Genome sequencing has revealed that bacteria contain many more biosynthetic gene clusters than predicted based on the number of secondary metabolites discovered to date. While this biosynthetic reservoir has fostered interest in new tools for natural product discovery, there remains a gap between gene cluster detection and compound discovery. Here we apply molecular networking and the new concept of pattern-based genome mining to 35 Salinispora strains including 30 for which draft genome sequences were either available or obtained for this study. The results provide a method to simultaneously compare large numbers of complex microbial extracts, which facilitated the identification of media components, known compounds and their derivatives, and new compounds that could be prioritized for structure elucidation. These efforts revealed considerable metabolite diversity and led to several molecular family-gene cluster pairings, of which the quinomycin-type depsipeptide retimycin A was characterized and linked to gene cluster NRPS40 using pattern-based bioinformatic approaches.
Introduction
The analysis of genome sequence data has revealed that even well studied bacteria can maintain the genetic potential to produce many more secondary or specialized metabolites than discoveries to date would suggest (Bentley, et al., 2002; Cimermancic, et al., 2014; Doroghazi, et al., 2014; Nett, et al., 2009). While this revelation has generated renewed interest in the field of natural product discovery, there remain inefficiencies in the processes by which known compounds are detected and new compounds prioritized for isolation and structure elucidation. Molecular networking is a tandem mass spectrometry (MS/MS) based computational approach that represents an important advance for the field of natural product research (Nguyen, et al., 2013; Winnikoff, et al., 2013; Yang, et al., 2013). This technique allows for high-throughput multi-strain comparisons and, with the integration of authentic standards, a rapid method to de-replicate (Koehn, 2008) and identify new compounds with known structural scaffolds (Krug and Müller, 2014; Yang, et al., 2013). Molecular networking also shows considerable potential as an aid to novel compound discovery, especially when complemented with genome sequence data and recently developed peptidogenomic and glycogenomic methods (Kersten, et al., 2011; Kersten, et al., 2013). Together, these approaches provide a rapid method to create bioinformatic links between parent ions and the pathways responsible for their biosynthesis. Furthermore, by analyzing large numbers of related strains, it becomes possible to address relationships between taxonomy and metabolome content, which can help resolve the ecological significance and evolutionary history of specific functional traits.
Molecular networks are used to organize MS/MS spectra into groups based upon similarities in their fragmentation patterns and the expectation that structurally related molecules will yield similar MS/MS spectra. In these networks, MS/MS spectra are represented as nodes, and the similarity between two spectra computed using a modified cosine score (Watrous, et al., 2012), which defines the edges connecting two nodes (Bandeira, 2007; Watrous, et al., 2012). A series of connected nodes generally indicates structurally related molecules or molecular families (MFs) (Nguyen, et al., 2013). Molecular networking provides a rapid and highly sensitive approach to compare metabolic profiles among strains without arduous data mining (Yang, et al., 2013). It has been used for the global visualization of the molecules produced by one (Liu, et al., 2014) or a large number of organisms (Nguyen, et al., 2013), to discover a new suite of natural products from S. coelicolor (Sidebottom, et al., 2013), to characterize small molecules dependent on the colibactin pathway in E. coli (Vizcaino, et al., 2014), to define the metabolomic potential of a new environmental taxon (Wilson, et al., 2014), and to study the chemical basis of microbial interactions on agar surfaces (Watrous, et al., 2012). However, molecular networking has not been used to simultaneously assess the pan-metabolome of a large number of closely related bacterial strains.
The marine actinomycete genus Salinispora consists of three closely related species (Freel, et al., 2013; Maldonado, et al., 2005) that share 99% 16S rRNA gene sequence identity (Jensen and Mafnas, 2006). They are the source of a wide range of structurally novel secondary metabolites (Jensen, et al., 2015) including salinosporamide A, which has undergone phase I clinical trials for the treatment of cancer (Feling, et al., 2003). Genome sequence data supports previous observations that some compounds are consistently produced by members of the same Salinispora species (Jensen, et al., 2007) while also revealing extraordinary levels of pathway diversity and considerable potential for new compound discovery (Penn, et al., 2009; Ziemert, et al., 2014). While genome mining has been used to target the products of individual Salinispora pathways (Udwary, et al., 2007), molecular networking provides the opportunity to simultaneously assess metabolite production in large numbers of strains. When coupled with genome sequence data, metabolite and biosynthetic gene cluster (BGC) distributions can be compared in a process we describe here as “pattern-based genome mining”, thus expanding on recent efforts to create bioinformatics links between BGCs and their small molecule products (Doroghazi, et al., 2014). Here we applied molecular networking to the analysis of 35 closely related Salinispora strains including 30 for which genome sequence data was available. This metabolomic approach compliments a previous bioinformatic study (Ziemert, et al., 2014) and further resolves the relationships among Salinispora species designations and secondary metabolite production (Jensen, et al., 2007). A de-replication strategy (Yang, et al., 2013) was used to populate the network with previously described Salinispora secondary metabolites and aid in the identification of known compounds and new derivatives. We then searched for “patterns” in an effort to create bioinformatic links between the presence of uncharacterized BGCs and the production of specific compounds. This combined approach, which has the potential to be automated, was then used to select compounds of interest for structure elucidation.
Results
Thirty-five Salinispora strains isolated from 10 global collection sites were analyzed (Table S1). These strains include a broad representation of the diversity within the three currently recognized species (Freel, et al., 2012) and 30 for which draft genome sequences are available, seven of which are new to this study. The fermentation conditions were standardized using the indicator phenol red such that all cultures were extracted upon entry into stationary phase (Figure S1), which occurred on day nine to 30 depending upon strain (Table S1) and is linked to a shift from primary to secondary metabolism (Nieselt, et al., 2010). This transition was associated with a pH shift from acidic (yellow) to basic (red), which corresponds to a change from rapid growth and net acetate excretion in the presence of abundant nutrients to slower growth and acetate assimilation following nutrient depletion (Wolfe, 2005). The extracts were analyzed by high-resolution tandem mass spectrometry (HR-MS/MS), the results from biological replicates combined for each strain, and m/z values <300 excluded, which resulted in the generation of over 200,000 MS1 HR-MS spectra over a mass range of 304.175–2485.4 m/z.
Molecular network
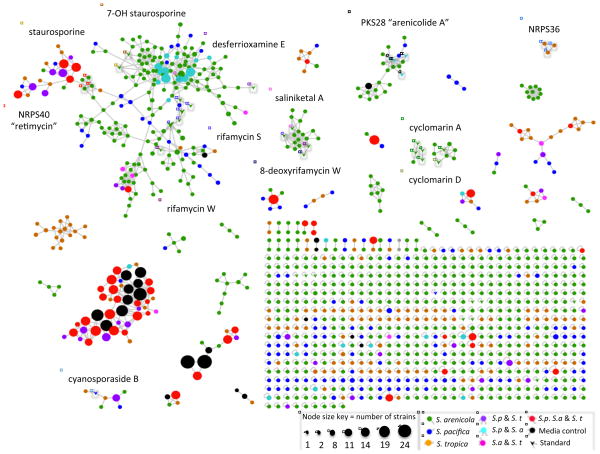
Analysis of the MS/MS data led to the identification of 1137 parent ions, which were visualized as nodes in a molecular network (Figure 1). The node size reflects the number of strains producing each parent ion and varies from strain-specific (one strain), which represents the vast majority of nodes, to the most ubiquitous metabolites, which were observed in a maximum of 24 of the 35 strains (Figure 1). When media components are excluded, the most common parent ion was observed in 15 strains (Figure S2). The network was screened against a spectral database generated from authentic standards, which led to the identification of seven compound classes previously described from Salinispora spp. (Table 1). The large number of nodes that networked with many of the standards suggests the presence of additional analogues in these compound classes. Examples include the cyclomarin (Renner, et al., 1999) and arenicolide (Williams, et al., 2007) molecular families, where networking reveals the production of both known compounds and what appear to be new analogs. For example, in addition to cyclomarin A and D, inspection of the parent mass and the MS/MS data suggests the molecular family contains putative demethylated, methylated, and hydrated cyclomarin A analogues (m/z: 1051.59, 1079.62 and 1083.61, respectively). Similarly, in the arenicolide cluster, putative dehydrogenated, methylated, and hydroxylated arenicolide A congeners were detected (m/z: 825.48, 841.47 and 843.49, respectively) (Figure 1).
Figure 1. Salinispora molecular network.
Related parent ions are networked based on similarities in MS/MS fragmentation patterns. Arrows indicate ions that matched known Salinispora secondary metabolites, with a representative of that compound molecular family named in a similarly colored box. Node size reflects the number of strains that produced each parent ion. Node color reflects the distribution of the parent ion among the three species.
Table 1. Linking pathway distributions to parent ion detection.
Biosynthetic pathways associated with 13 structurally characterized secondary metabolites were identified in the 30 Salinispora genome sequences analyzed. Products from eight of these pathways (including rtm, which is new to this study) were identified using HR-MS/MS data in comparison with authentic standards. All MS data, including raw data that did not appear in the network, were screened for parent ions associated with each compound class. Data are provided for only one representative of each structure class. ND = not detected, NA = not applicable.
| Biosynthetic gene cluster (BGC) | Product | Observed parent ion m/z | MF | MW | GnPS score | Expected parent ion m/z | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | # strains detected | Species detected | Name | # strains detected (%of total) | ||||||
| PKS28 | 2 | Sa, Sp | Arenicolide A* | 2 (100) | 827.49 [M+Na]+ | C45H72O12 | 804.50 | 0.62–0.76 [M+Na]+ | 827.492 [M+Na]+ | Williams (2007) |
| cya | 17 | Sp, St | Cyanosporaside B | 5 (29.4) | 440.26 [M+Na]+ | C21H20ClNO6 | 417.00 | 0.81 [M+Na]+ | 440.088 [M+Na]+ | Oh (2006) |
| cym | 1 | Sa | Cyclomarin A | 1 (100) | 1065.60 [M+Na]+ | C56H82N8O11 | 1042.61 | 0.97 [M+Na]+ | 1065.60 [M+Na]+ | Shultz (2008) |
| des | 21 | Sp | Desferioxamine E | 1 (4.7) | 601.21 [M+H]+ | C27H48N6O9 | 600.35 | 0.54 [M+H]+ | 601.36 [M+H]+ | Roberts (2012) |
| rif | 9 | Sa | Rifamycin S | 8 (88.9) | 718.29 [M+Na]+ | C37H45NO12 | 695.30 | 0.67 [M+Na]+ | 718.29 [M+Na]+ | Kim (2006) |
| sta | 15 | Sa, Sp, St | Staurosporine | 11 (73.3) | 467.22 [M+H]+ | C28H26N4O3 | 466.20 | 0.68 [M+H]+ | 467.208 [M+H]+ | Freel (2011) |
| lom | 16 | Sp, St | Lomaiviticin C | 5 (31.3) | 670.27 [M+2H]2+ | C68H82N4O24 | 1339.39 | NA | 670.277 [M+2H]2+ | He (2001) |
| slm | 13 | Sp, St | Salinilactam** | 0 (0) | ND | NA | NA | NA | NA | Udwary (2007) |
| sal | 13 | Sa, Sp, St | Salinosporamides | 0 (0) | ND | NA | NA | NA | NA | Fehling (2003) |
| spo | 4 | St | Sporolides** | 0 (0) | ND | NA | NA | NA | NA | Buchanan (2005) |
| lym | 28 | Sa, Sp, St | Lymphostins | 0 (0) | ND | NA | NA | NA | NA | Aotani (1997) |
| rtm | 1 | Sa | Retimycin A* | 1 (100) | 1185.44 [M+H]+ | C54H68N10O15S2 | 1184.33 | NA | 1185.439 [M+H]+ | this study |
Sa = S. arenicola, Sp = S. pacifica, St = S. tropica.
Links between BGC and product are bioinformatics-based.
Standards not available. MF = molecular formula, MW = molecular weight, GnPS = Global Natural Products Social Molecular Networking (http://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp).
Patterns visualized in the network include a large number of nodes and molecular families that are specific to S. arenicola. It is also clear that many of the ions observed from all three species networked with media components (shown as black nodes) suggesting these molecular families are of low interest in terms of secondary metabolite discovery. The exclusion of metabolites with m/z values <300 prevented the detection of some previously identified Salinispora metabolites such as salinosporamide K (274.1 m/z [M+Na]+) (Eustáquio, et al., 2011) and salinipyrone (293.17 m/z [M+H]+) (Oh, et al., 2008), however salinosporamide A (314.116 m/z [M+H]+) (Feling, et al., 2003), which has consistently been reported from S. tropica (Jensen, et al., 2007), was not observed in the molecular network or the raw experimental data. This may be due to feedback regulation of the biosynthetic pathway (Lechner, et al., 2011) the absence of adsorbent resins in the fermentation medium (Tsueng, et al., 2008), or the aqueous instability of salinosporamide A (Fenical, et al., 2009).
Parent ion distributions
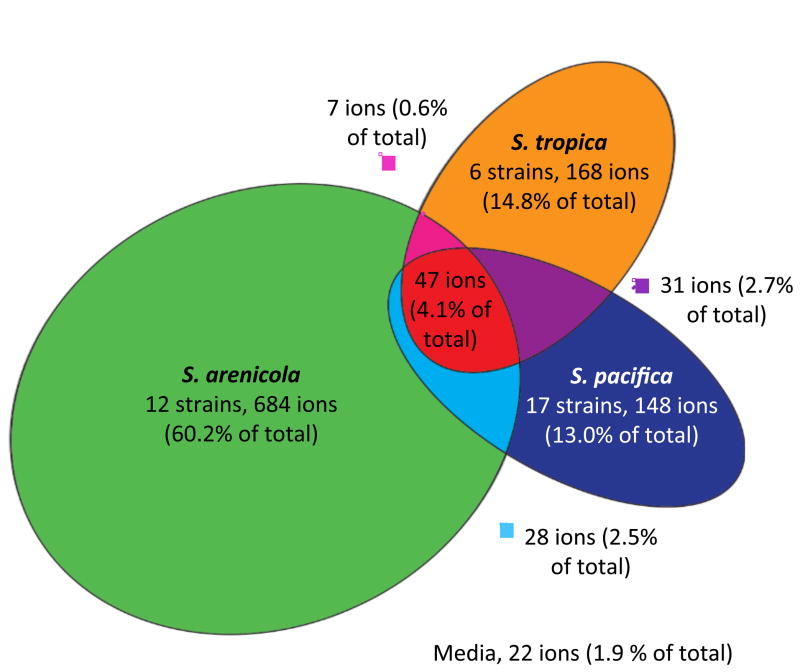
Only 22 of the 1137 parent ions (1.9%) were observed in the medium blank. The majority of ions (87.1%) were not produced across species boundaries, with only 5.8% of the total shared by two and 4.1% shared by all three species (Figure 2). This apparent species-specificity can be accounted for by the large number of ions that were observed in only one strain (Figure S2) thus indicating a high level of variability in secondary metabolite production among strains. These ions include a broad range of masses, suggesting there is no correlation between mass and the frequency with which a parent ion was detected. The results are in agreement with a bioinformatic analysis of 75 Salinispora genome sequences in which more than half of the polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) pathways were only observed in one or two strains (Ziemert, et al., 2014). There were no cases where all 35 strains analyzed, or all strains from the same species, produced the same parent ion (Figure S2). The number of unique parent ions was far greater for S. arenicola, which averaged 57.0 per strain, relative to S. tropica and S. pacifica, which averaged 28.0 and 8.7 per strain, respectively. Nodes not associated with edges (ions that are not part of a molecular family and potentially represent unique chemistry) were distributed among the species as follows, S. arenicola 371 (62.1%), S. pacifica 98 (16.4%) and S. tropica 98 (16.4%).
Figure 2. Parent ion distribution.
A proportional Euler diagram reveals that most ions were species-specific. Parent ions were considered shared when produced by at least one strain from two different species. Twenty-two ions (1.9% of total) assigned to the medium were not included in the analysis.
The phylogenetic relationships among the three Salinispora spp. are well resolved and reveal the basal position of S. arenicola relative to the more recently diverged sister taxa S. tropica and S. pacifica (Freel, et al., 2013). Based on the assumption that horizontal gene transfer has not ameliorated all evidence of vertical inheritance among the genes responsible for secondary metabolite production, it is expected that parent ion similarities would be greater between S. tropica and S. pacifica based on their relatively close evolutionary relationship than between either of these species and S. arenicola. A species-level pairwise comparison reveals that S. tropica and S. pacifica shared 5.6% and 7.9% of the parent ions they produced, respectively, with S. arenicola, while S. tropica and S. pacifica shared 18.1%. Thus, despite the high level of among strain variability in parent ion production, species relationships are reflected in the levels of shared metabolite production.
Linking compounds to gene clusters through molecular networking
The BCGs associated with the biosynthesis of 10 of the secondary metabolites reported from Salinispora spp. have been experimentally linked to their secondary metabolic products. These encode the production of the cyclomarins (cym) (Schultz, et al., 2008), cyanosporasides (cya) (Lane, et al., 2013), rifamycins (rif) (including saliniketal) (Wilson, et al., 2010; Ziemert, et al., 2014), lomaiviticins (lom) (Kersten, et al., 2013), desferrioxamines (des) (Roberts, et al., 2012), staurosporines (sta) (Onaka, et al., 2002), salinilactam (slm) (Udwary, et al., 2007), salinosporamides (sal) (Eustáquio, et al., 2009; Eustáquio, et al., 2011), sporolides (spo) (McGlinchey, et al., 2008), and lymphostin (lym) (Miyanaga, et al., 2011). Furthermore, the arn cluster has been bioinformatically linked to arenimycin biosynthesis (Asolkar, et al., 2010) via “glycogenomics” (Kersten, et al., 2013) while the rtm cluster (Table S2) has been bioinformatically linked to retimycin A biosynthesis (this study) using peptidogenomics (Kersten, et al., 2011). Overall, products associated with eight Salinispora BGCs were detected among the extracts analyzed (Table 1).
Molecular networking coupled with genome sequence data provides a rapid method to assess the relationships between the presence of a BCG and the detection of its secondary metabolic products. We refer to these correlative analyses as pattern-based genome mining, where the detection of a parent ion within a molecular family is used as a proxy for the expression of the associated BCG (Figure 3). In some cases, pattern-based genome mining revealed a perfect correlation between BCGs and products. This was observed for the arenicolides and cyclomarins, which were detected from all three of the strains that possessed the respective clusters (Table 1). In most cases however, the correlations were less perfect. For example, rifamycins were detected in eight of the nine strains that possessed the BCG while the desferioxamines were only observed in one of 21 strains. In the later case, it was surprising that even one strain produced these iron-chelating compounds, as the medium was not specifically designed to be iron limited, which is known to support production (Roberts, et al., 2012). While it remains unclear how much of the variability observed can be linked to the nuances associated with fermentation conditions, extraction protocols, and mass spectroscopy, pattern-based genome mining holds promise as a method to generate links between the presence of BCGs and the small molecules they ultimately produce. In total, products were detected in 34 of 140 cases (24%) in which the associated BGCs were detected. This suggests that many of the BGCs were not expressed under the culture conditions employed or that, if expressed, the products were either not extracted or went undetected in the MS analyses.
Figure 3. Pattern-based genome mining in 30 Salinispora strains.
Gene clusters are listed on the left, follow previous nomenclature (Ziemert at al., 2014), and include the addition of the staurosporine (sta) and desferrioxamine (des) pathways. Colored boxes indicate the presence of a biosynthetic gene cluster, vertical lines (shading) indicate the HR-MS detection of the products of that gene cluster. Colors indicate the distribution of the gene cluster across the three species: red (all three species), light blue (S. arenicola and S. pacifica), purple (S. tropica and S. pacifica), blue (S. pacifica only), orange (S. tropica only), and green (S. arenicola only). Three S. arenicola and two S. pacifica strains were excluded from the figure because genome sequences were not avialable.
Linking compounds to uncharacterized gene clusters
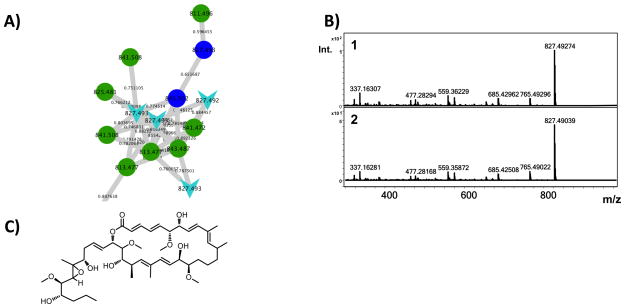
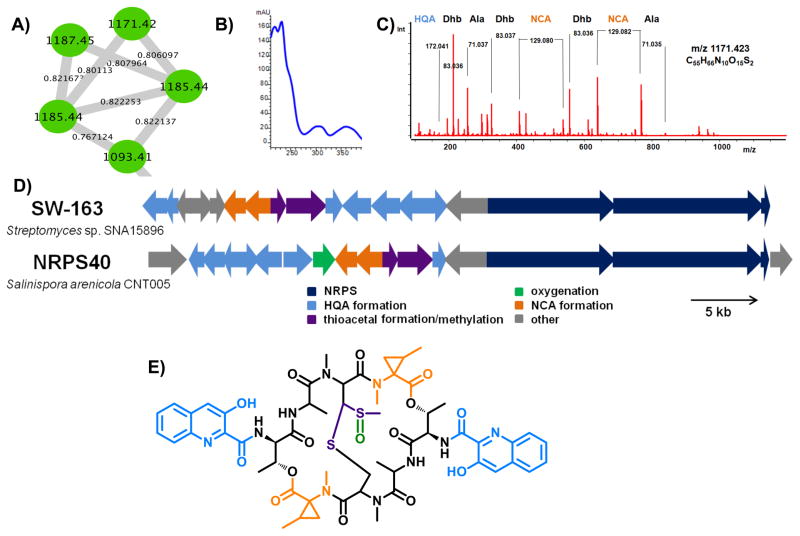
It was possible to link an ion that matched the previously described metabolite arenicolide A (Williams, et al., 2007) to an uncharacterized biosynthetic gene cluster (PKS28) based on a number of lines of evidence. First, pattern-based analysis revealed that the two strains producing the arenicolide A ion (827.492 m/z [M+Na]) were the only strains in which PKS28 was observed (S. arenicola strain CNQ-748 and S. pacifica strain CNT-138) (Figure 3). While incomplete genome assembly prevented a precise interpretation of this gene cluster, the KS sequences in PKS28 show a high level of sequence identity (92%) to KS sequences previously linked to arenicolide A production (Edlund, et al., 2011). The molecular network places arenicolide A within a larger molecular family of structurally related analogues (Figure 4), suggesting that additional diversity remains to be discovered in this class of compounds. This is the first evidence of arenicolide A production by S. pacifica. In a second example of pattern-based genome mining, the pathway NRPS40 was identified as unique to strain CNT-005 (Ziemert, et al., 2014) (Figure 3). In an effort to search for the products of this pathway, a series of nodes that were similarly unique to strain CNT-005 was explored in more detail (Figure 5). MS/MS analysis of the 1171.42 and 1185.43 m/z parent ions revealed peptide mass shifts while the characteristic UV profiles suggested the compounds may be related to the quinomycins, a group of highly cytotoxic and antibiotic dimeric depsipeptides (Dawson, et al., 2007; Zolova, et al., 2010). Analysis of the MS/MS spectra revealed alanine, several dehydrated threonine residues and unassigned mass shifts suggesting modified amino acid residues (Figure S3). An analysis of the NRPS gene clusters in strain CNT-005 led to NRPS40 as a candidate for the biogenesis of the 1171.42 and 1185.43 m/z parent ions.
Figure 4. Arenicolide A production.
A) An arenicolide A standard (expected 827.492 m/z) networked to 827.492 and 827.493 m/z ions (light blue arrows) produced by strains S. pacifica CNT-138 and S. arenicola CNQ-748. Structurally related analogues are produced by CNT-138 (dark blue nodes) and CNQ-748 (green nodes). B) MS/MS fragmentation pattern of the 827.492 m/z parent ion (1) in comparison with the arenicolide A standard (2). C) Structure of arenicolide A.
Figure 5. Identification of the novel non-ribosomal peptide retimycin A.
A) Analysis of the molecular network revealed a cluster of four parent ions produced only by S. arenicola strain CNT-005. B) UV spectra associated with parent ion 1171.423 m/z. C) MS/MS analysis of the 1171.423 m/z parent ion revealed amino acid shifts that corresponded to alanine, dehydrated threonine, and two unknown residues later assigned as HQA and NCA. D) Bioinformatic analysis of NRPS40 (rtm) in comparison to the related pathway responsible for the production of SW-163, a quinomycin-like depsipeptide. E) Chemical structure of retimycin A. The colors of the building blocks correspond to the colors of their respective biosynthetic genes in Figure 5D. See also Figures S3–S5 and Table S2.
Isolation and characterization of retimycin A
A detailed analysis of NRPS40 revealed considerable homology to the BGC responsible for the production of the quinomycin-like compound SW-163 (Watanabe, et al., 2009) including genes for the production of hydroxyquinaldic acid (HQA) and the cyclopropane-containing norcoronamic acid (NCA). However, the adenylation domain specificity of the second NRPS module differs from SW-163 and was not predictable by bioinformatic tools. Additionally, NRPS40 includes a CYP450 oxygenase, which collectively suggests the product differs from that of the previously characterized compound SW-163. Large-scale fermentation followed by the isolation of the 1171.42 m/z metabolite and subsequent 2D NMR characterization (Figures S4–S5, Table S3) confirmed the presence of HQA and NCA units. Assignment of the unknown amino acid revealed a threonine moiety, a previously unprecedented residue in the quinomycin family. The characterized members of this family contain D-serine (echinomycins), D-cysteine (thiocoralines), or D-diaminobutyric acid (quinaldopeptin) at this position (Fernández, et al., 2014). Marfey’s analysis of the hydrolyzed compound showed the stereochemistry of the threonine residue to be D-allo-Thr, which corresponds to the presence of an epimerase domain in the first module of RtmO. Assignment of the beta-protons of the cysteine residues revealed a thioacetal moiety, a common motif in the quinomycin family. MS/MS analysis showed a central fragment loss of m/z 63.99, which corresponds to a molecular formula of CH4SO (Figure S3). Taken together with the observation of a methyl singlet that shows correlation to the tertiary carbon of the thioacetal, we concluded that the central moiety is a methylated thioacetal that is oxidized at the rearranged sulfur, which is a novel feature in this family. Interestingly, in a recently described quinomycin, the non-rearranged sulfur was shown to be oxidized (Lim, et al., 2014). The 1171.42 m/z compound was named “retimycin A”, after the Latin word “reticulum” meaning network. It is the first fully characterized novel natural product discovered by molecular networking. For the analogue with m/z 1185.43, preliminary NMR data suggest that this molecule is not simply a methylated version of retimycin A, but lacks symmetry in the peptide backbone. A lack of material prevented the elucidation of this structure, which warrants further investigation. Bioactivity results revealed comparable cytotoxicity (IC50: <0.076 μg/mL) against an HCT-116 cell line for retimycin A and the structurally related compound echinomycin.
Discussion
The field of natural products chemistry was invigorated by the observation that even well studied bacteria can maintain the genetic potential to produce many new secondary or specialized metabolites (Bentley, et al., 2002). Spearheading this renewed interest is the application of genome mining techniques (Bachmann, et al., 2014) and focused efforts to develop new discovery platforms including heterologous expression (Bonet, et al., 2014; Yamanaka, et al., 2014), the activation of silent gene clusters (Seyedsayamdost, 2014), peptidogenomics (Kersten, et al., 2011), glycogenomics (Kersten, et al., 2013), and proteomics (Chen, et al., 2013), all of which have helped to realize this potential. While there have been efforts to automate the process of genome mining (Medema, et al., 2014; Mohimani, et al., 2014; Zhang, et al., 2014), there remain major gaps between the detection of BGCs in genome sequence data and the identification of the metabolites they produce (Jensen, et al., 2014). Bridging this gap presents considerable challenges due to our poor understanding of the relationships between BGC distributions, expression, and the successful biogenesis and detection of a secondary metabolite. Here we present pattern-based genome mining as a highly sensitive and scalable approach to link molecules detected by mass spectrometry to the BGCs responsible for their biogenesis. When applied to the marine actinomycete genus Salinispora, the results provided a rapid method to recognize previously described secondary metabolites, generate bioinformatic links between unidentified parent ions and their putative BGCs, and prioritize compounds for isolation and structure elucidation.
The global Salinispora molecular network revealed high levels of species and strain-specific production with only 15% of the parent ions crossing species boundaries. This variability is not entirely due to differences in genome content as the products of only 34 of 140 (24%) of the characterized BGCs were detected. Variables such as culture conditions likely contribute to this discrepancy, which was similarly reported in the analysis of 830 actinobacterial metabolomes (Doroghazi, et al., 2014). Considering the des pathway, it’s not surprising that the iron-chelating desferrioxamines were only detected in one of 21 cases given that the iron-replete growth medium was not designed to elicit production. The variables that affect the expression of other BGCs are less clear. Future transcriptome analyses will help define these variables and distinguish between “silent” BGCs and those for which the products are produced but remain undetected. An extended molecular network including different growth conditions and extraction methods is expected to provide better consistency between predicted compounds and detected ions. It is also anticipated that subjecting larger numbers of organisms to pattern-based genome mining will improve the correlative power of the approach.
It was interesting to note that S. arenicola produced the largest number of unique parent ions per strain. This was surprising considering that in a prior study S. pacifica was observed to maintain greater PKS and NRPS diversity (Ziemert, et al., 2014). None-the-less, this observation is supported by an independent LC-MS analysis in which twice the number of compounds were detected from S. arenicola (Bose, et al., 2014). A more comprehensive comparison of the BGCs and their levels of expression in these two species will provide a better understand of the relationships between BGC distributions and specialized metabolite production.
As has been shown previously with other bacteria (Yang, et al., 2013), seeding the Salinispora molecular network with previously identified secondary metabolites made it possible to rapidly identify known compounds and related molecular families among the 1137 nodes present in the network. It was also possible to identify common media components and what appear to be new derivatives of previously described molecules such as the cyclomarins and arenicolides, which in many cases (e.g. methylation, hydroxylation) could be readily identified by mass spectrometry. This study benefitted from the analysis of a large number of closely related strains and data from previously described Salinispora secondary metabolites. These methods will become increasingly useful for the analysis of diverse collections of bacteria as shared knowledge databases, such as that available though the Global Natural Products Social Molecular Networking site (http://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp), provide infrastructure to easily share MS/MS data with the larger community.
Using pattern-based genome mining in combination with peptidogenomics, NRPS40 was linked to a 1171.423 m/z parent ion, which was targeted for isolation and subsequently identified as retimycin A, a new quinomycin-like depsipeptide in the thiocoraline family. The detection of the retimycin gene cluster (rtm) in only one of the 30-genome sequences indicates the value of studying large numbers of closely related strains. Interestingly, the related metabolite thiocoraline has been reported from two Micromonospora strains isolated from marine invertebrates (Lombó, et al., 2006), suggesting that this pathway has been exchanged horizontally between these two closely related actinomycete genera. Retimycin A has now been added to the Salinispora MS/MS spectral library thus providing a useful reference to better assess the production of this and related compounds among other strains. This growing spectral library provides unique opportunities to obtain a more global view of the Salinispora secondary metabolome.
Significance
This study represents the first application of molecular networking to a large collection of environmental bacteria. It introduces the concept of pattern based genome mining, a scalable, mass spectrometry method with the potential to be automated. The molecular network made it possible to simultaneously visualize the molecular composition of organic extracts generated from 35 closely related strains belonging to the marine actinomycete genus Salinispora. Populating the network with standards facilitated the identification of known compounds and their derivatives and compounds of high priority for isolation and structure elucidation. A majority of parent ions were species or strain specific, resulting in a high degree of secondary metabolite diversity. When complemented with genome sequence data, pattern-based genome mining revealed non-perfect correlations between gene cluster distributions and the detection of the associated products, suggesting that gene expression may be highly variable among strains. Pattern based genome mining was used to identify NRPS40 as a candidate for natural product discovery while peptidogenomics was used to provide a bioinformatic link between this BGC and a parent ion observed in the molecular network. This compound was structurally characterized and named retimycin A, a new member of the quinomycin family of depsipeptide antitumor antibiotics. The growing MS/MS database of Salinispora natural products will continue to improve the effectiveness by which molecular networking can be used to rationalize chemical space and improve the effectiveness by which natural products are discovered from this genus.
Experimental Procedures
Strain culture conditions and extraction
Salinispora strains were selected based on 16S rRNA phylotype, isolation location, and the availability of genome sequence data (Table S1). Pre-cultures were grown in medium A1M1 [5 g/L soluble starch (Affymetrix), 2 g/L peptone (Fischer Scientific), 2 g/L yeast extract (Affymetrix), 22 g/L instant ocean (Marineland), 100 mL DI water, adjusted to pH 6.5 before autoclaving at 121°C for 45 minutes] for 7–9 days after which 5 mL was inoculated in duplicate into 100 mL A1M1 with 100 μL of 10 mg/mL filter sterilized phenol red solution (Sigma). Fermentations were performed in 500 mL Erlenmeyer flasks at 28°C and shaking at 160 rpm. Stainless steel springs were added to reduce cell clumping. The cultures were extracted with an equal volume of ethyl acetate (Fischer Scientific) when the indicator changed from yellow to red (corresponding to a pH of 8.0) and the ethyl acetate layers collected, dried in vacuo, and stored at -20°C.
Mass spectral data acquisition
Extracts were dissolved in MeOH at the final concentration of 0.1 mg/mL and injected onto a Phenomenex Kinetex (Torrance, CA, USA) C18 reversed-phase HPLC column (2.6 mm, 100 × 4.6 mm). Samples were analyzed using an Agilent 6530 Accurate-Mass Q-TOF spectrometer coupled to an Agilent 1260 LC system (Santa Clara, CA. USA) under the following LC conditions with 0.1 % TFA: 1–5 min (10% MeCN in H2O), 5–26 min (10–100% MeCN), 26–30 min (100% MeCN). The divert valve was set to waste for the first 5 min. Q-TOF MS settings during the LC gradient were as follows: positive ion mode mass range 300–2500 m/z, MS scan rate 1/s, MS/MS scan rate 5/s, fixed collision energy 20 eV; source gas temperature 300°C, gas flow 11 L/min, nebulizer 45 psig, scan source parameters: VCap 3000, fragmentor 100, skimmer1 65, octopoleRFPeak 750. The MS was auto-tuned using Agilent tuning solution in positive mode before each measurement. LC (DAD) data were analyzed with ChemStation software (Agilent) and MS data were analyzed with MassHunter software (Agilent). The spectral data generated from the biological replicates were combined for each strain prior to the network analyses.
Crude extract and standard compound analysis
HR-MS/MS fragmentation data was generated for the known Salinispora compounds listed in Table 1 under the acquisition conditions described above. Ions were selected via peak extraction using Masshunter software and the data converted to mzXML format using the Trans-Proteomic pipeline (Keller, et al., 2005). The data were uploaded to the molecular networking server as a Salinispora standards library. All HR-MS/MS raw data files were searched for high-resolution masses that matched the 12 classes of compounds previously reported from Salinispora species (Table 1) including those for which standards were not available. Methods include 1) the Mass Hunter ion extraction function 2) searching for HR masses in raw molecular networking files, and 3) searching for HR masses in the Cytoscape file. Searches were made for expected parent ions corresponding to multiple ions/analogues within each compound class.
Molecular networking
The MS/MS data of 35 Salinispora strains was converted from Mass Hunter data files (.d) to mzXML file format using the Trans-Proteomic pipeline (Institute for Systems Biology, Seattle) (Deutsch, et al., 2010) and clustered using structure-independent spectral alignment (MS-Cluster) (Frank, et al., 2007; Guthals, et al., 2012; Keller, et al., 2005). For network visualization, these were imported into Cytoscape 3.0 (Guthals, et al., 2012). Each node corresponds to a consensus spectrum and each edge represents a significant pairwise alignment.
Computationally, spectra were converted into unit vectors in n-dimensional space; pairs of vectors were compared with a dot product calculation, which includes the cosine of the angle between the two vectors, referred to as the cosine similarity score. Identical spectra were combined into consensus spectra that have a minimum of six ions that match. Cosine similarity scores range from 0–1, where identical spectra have a cosine score of 1. Two nodes are required to be in the top 10 cosine scores (K parameter) in both directions for an edge to connect them in Cytoscape. The pairs with a cosine score higher than 0.95 were combined into consensus spectra. The algorithm parameters include mass tolerance for fragment peaks (0.3 Da), parent mass tolerance (2.0 Da), a minimum number of matched peaks per spectral alignment (6), a maximum component size of 1 and a minimum cosine score of 0.5. This latter value was selected empirically to eliminate the clustering of different compound classes into the same molecular family (i.e., false positives). Cytoscape was used to visually display the data as a network of nodes and edges (Cline, et al., 2007) and organized with the edge-weighted force-directed layout plug-in.
Chemical isolation and structural characterization
For the isolation of retimycin A, the fermentation of strain CNT-005 was scaled up to a total volume of 7×1 L in medium A1M1 in 2.8 L Fernbach flasks. After seven days of cultivation, a 1:1 mixture of XAD7HP:XAD16 resin (Amberlite) was added and, after two hours, collected by filtration and eluted 2X with acetone. The extract was concentrated under vacuum and separated via preparative HPLC [Agilent Prostar, Synergi-10u Hydro-RP, 250×21.2 mm (Phenomenex)] using a gradient from 30% to 95% acetonitrile containing 0.1% TFA over 30 min (15 ml/min). A fraction eluting from 23–26 min was collected, dried, and subjected to semi-preparative HPLC purification [Agilent 1200, Luna 5u C18, 100A, 250×10 mm (Phenomenex)] with isocratic 55% acetonitrile (0.1% TFA), 2.5 ml/min) to yield pure retimycin (0.5 mg, tR= 36 min). Subsequent NMR characterization (1H, 1H-1H COSY, HSQC, HMBC, TOCSY) was carried out on a BRUKER Avance spectrometer (600 MHz).
Marfey’s analysis of retimycin
Retimycin (200 μg) was hydrolyzed (6 M HCl, 160°C, 5 min) and dried under nitrogen. The resulting solid was re-dissolved in 1 M sodium bicarbonate (200 μl) followed by addition of 1 mL of 1-fluoro-2,4-dinitrophenyl-D-alanine amide (D-FDAA) in acetone (1.5 mg/ml). The reaction was stirred at 50°C for 1 h, quenched with 1 M HCl (200 μL), and dried under nitrogen. The resulting solid was re-dissolved in 1:1 H2O:CH3CN (200 μl) and filtered. The resulting solution was analyzed (20 μL) by LC-MS (positive mode) on a Luna C18 column, 250×4.6 mm, 5u (Phenomenex) with a gradient from 0% CH3CN to 40% CH3CN in H2O over 95 minutes (0.4 ml/min). The mass for Thr-FDAA (m/z: 372.1) was extracted and determined to elute at 53.28 min. This was compared to the retention times for all 4 threonine isomers, derivatized with D-FDAA (D-Thr: 52.90 min, D-allo-Thr: 53.20 min, L-allo-Thr: 56.70 min, L-Thr: 61.50 min). The optical rotation was determined on a Jasco P200 polarimeter (c: 0.2, CHCl3).
Supplementary Material
Highlights.
Pattern-based genome mining was applied to 35 Salinispora strains
Molecular networking facilitated new compound discovery
The quinomycin-type depsipeptide retimycin A was characterized
Acknowledgments
This work was funded by the National Institutes of Health under grants RO1GM085770 (to PRJ and BSM) and R01GM097509 (to BSM, NB and PCD). MC was funded by a DFG postdoctoral fellowship. Genome sequencing was conducted by the U.S. Department of Energy Joint Genome Institute and supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. The authors thank W. Fenical for supplying previously identified Salinispora secondary metabolites as standards used to seed the molecular network. N. Millán-Aguiñaga is acknowledged for assistance with strain identification. We thank B.M. Duggan for assistance in NMR measurements and J.C. Busch for HCT cytotoxicity testing. D. A. Phillips is acknowledged for Figure 2 graphic design work.
Footnotes
Author Contributions: KD, AL, and PJ designed the study. KD and AL performed and analyzed the molecular networking experiments. KD and PJ wrote the manuscript. MC, BM, and JE isolated and solved he structure of retimycin. AS performed the fermentation studies. NZ and AL analyzed the gene clusters. MW and PD assisted with the networking analyses.
All authors declare no financial conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Bradley S. Moore, Email: bsmoore@ucsd.edu.
Pieter C. Dorrestein, Email: pdorrestein@ucsd.edu.
Paul R. Jensen, Email: pjensen@ucsd.edu.
References
- Asolkar RN, Kirkland TN, Jensen PR, Fenical W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J Antibiot. 2010;63:37–39. doi: 10.1038/ja.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J Ind Microbiol Biotechnol. 2014;41:175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandeira N. Spectral networks: a new approach to de novo discovery of protein sequences and posttranslational modifications. BioTechniques. 2007;42:687–695. doi: 10.2144/000112487. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Bonet B, Teufel R, Crüsemann M, Ziemert N, Moore BS. Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J Nat Prod. 2014 doi: 10.1021/np500664q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose U, Hodson MP, Shaw PN, Fuerst JA, Hewavitharana AK. Two peptides, cycloaspeptide A and nazumamide A from a sponge associated marine actinobacterium Salinispora sp. Nat Prod Comm. 2014;9:545–546. [PubMed] [Google Scholar]
- Chen Y, Unger M, Ntai I, McClure RA, Albright JC, Thomson RJ, Kelleher NL. Gobichelin A and B: mixed-ligand siderophores discovered using proteomics. MedChemComm. 2013;4:233–238. doi: 10.1039/C2MD20232H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P, Medema MH, Claesen J, Kurita K, Brown LCW, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S, Malkinson JP, Paumier D, Searcey M. Bisintercalator natural products with potential therapeutic applications: isolation, structure determination, synthetic and biological studies. Nat Prod Rep. 2007;24:109–126. doi: 10.1039/b516347c. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund A, Loesgen S, Fenical W, Jensen PR. Geographic distribution of secondary metabolite genes in the marine actinomycete Salinispora arenicola. Appl Environ Microbiol. 2011;77:5916–5925. doi: 10.1128/AEM.00611-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Nat Acad Sci. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustáquio AS, Nam SJ, Penn K, Lechner A, Wilson MC, Fenical W, Jensen PR, Moore BS. The discovery of salinosporamide K from the marine bacterium “Salinispora pacifica” by genome mining gives insight into pathway evolution. ChemBioChem. 2011;12:61–64. doi: 10.1002/cbic.201000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew Chem Int Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorg Med Chem. 2009;17:2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández J, Marín L, Álvarez-Alonso R, Redondo S, Carvajal J, Villamizar G, Villar CJ, Lombó F. Biosynthetic Modularity Rules in the Bisintercalator Family of Antitumor Compounds. Mar Drugs. 2014;12:2668–2699. doi: 10.3390/md12052668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, Pevzner PA. Clustering millions of tandem mass spectra. J Proteome Res. 2007;7:113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel KC, Edlund A, Jensen PR. Microdiversity and evidence for high dispersal rates in the marine actinomycete ‘Salinispora pacifica’. Environ Microbiol Rep. 2012;14:480–493. doi: 10.1111/j.1462-2920.2011.02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel KC, Millan-Aguinaga N, Jensen PR. Multilocus sequence typing reveals evidence of homologous recombination linked to antibiotic resistance in the genus Salinispora. Appl Environ Microbiol. 2013;79:5997–6005. doi: 10.1128/AEM.00880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthals A, Watrous JD, Dorrestein PC, Bandeira N. The spectral networks paradigm in high throughput mass spectrometry. Mol BioSystems. 2012;8:2535–2544. doi: 10.1039/c2mb25085c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Chavarria KL, Fenical W, Moore BS, Ziemert N. Challenges and triumphs to genomics-based natural product discovery. J Ind Microbiol Biotechnol. 2014;41:203–209. doi: 10.1007/s10295-013-1353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Mafnas C. Biogeography of the marine actinomycete Salinispora. Environ Microbiol. 2006;8:1881–1888. doi: 10.1111/j.1462-2920.2006.01093.x. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Moore BS, Fenical W. The marine actinomycete genus Salinispora: a model organism for secondary metabolite discovery. Nat Prod Rep. 2015 doi: 10.1039/c4np00167b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Eng J, Zhang N, Li Xj, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Sys Biol. 2005:1. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, Ziemert N, Gonzalez DJ, Duggan BM, Nizet V, Dorrestein PC, Moore BS. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc Natl Acad Sci. 2013;110:4407–4416. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn FE. Natural Compounds as Drugs. I. Springer; 2008. High impact technologies for natural products screening; pp. 175–210. [DOI] [PubMed] [Google Scholar]
- Krug D, Müller R. Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep. 2014;31:768–783. doi: 10.1039/c3np70127a. [DOI] [PubMed] [Google Scholar]
- Lane AL, Nam SJ, Fukuda T, Yamanaka K, Kauffman CA, Jensen PR, Fenical W, Moore BS. Structures and comparative characterization of biosynthetic gene clusters for cyanosporasides, enediyne-derived natural products from marine actinomycetes. J Am Chem Soc. 2013;135:4171–4174. doi: 10.1021/ja311065v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner A, Eustáquio A, Gulder TAM, Hafner M, Moore BS. Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem Biol. 2011;18:1527–1536. doi: 10.1016/j.chembiol.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CL, Nogawa T, Uramoto M, Okano A, Hongo Y, Nakamura T, Koshino H, Takahashi S, Ibrahim D, Osada H. RK-1355A and B, novel quinomycin derivatives isolated from a microbial metabolites fraction library based on NPPlot screening. J Antibiot. 2014;67:323–329. doi: 10.1038/ja.2013.144. [DOI] [PubMed] [Google Scholar]
- Liu WT, Lamsa A, Wong WR, Boudreau PD, Kersten R, Peng Y, Moree WJ, Duggan BM, Moore BS, Gerwick WH. MS/MS-based networking and peptidogenomics guided genome mining revealed the stenothricin gene cluster in Streptomyces roseosporus. J Antibiot. 2014;67:99–104. doi: 10.1038/ja.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombó F, Velasco A, Castro A, De la Calle F, Braña AF, Sánchez-Puelles JM, Méndez C, Salas JA. Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. ChemBioChem. 2006;7:366–376. doi: 10.1002/cbic.200500325. [DOI] [PubMed] [Google Scholar]
- Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M. Salinispora arenicola gen. nov., sp nov and Salinispora tropica sp nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005;55:1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- McGlinchey RP, Nett M, Moore BS. Unraveling the biosynthesis of the sporolide cyclohexenone building block. J Am Chem Soc. 2008;130:2406–2407. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- Medema MH, Paalvast Y, Nguyen DD, Melnik A, Dorrestein PC, Takano E, Breitling R. Pep2Path: Automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comp Biol. 2014;10:e1003822. doi: 10.1371/journal.pcbi.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A, Janso JE, McDonald L, He M, Liu H, Barbieri L, Eustáquio AS, Fielding EN, Carter GT, Jensen PR. Discovery and assembly-line biosynthesis of the lymphostin pyrroloquinoline alkaloid family of mTOR inhibitors in Salinispora bacteria. J Am Chem Soc. 2011;133:13311–13313. doi: 10.1021/ja205655w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H, Kersten RD, Liu WT, Wang M, Purvine SO, Wu S, Brewer HM, Pasa-Tolic L, Bandeira N, Moore BS, et al. Automated genome mining of ribosomal peptide natural products. ACS Chem Biol. 2014;9:1545–1551. doi: 10.1021/cb500199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DD, Wu CH, Moree WJ, Lamsa A, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci. 2013;110:E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieselt K, Battke F, Herbig A, Bruheim P, Wentzel A, Jakobsen ØM, Sletta H, Alam MT, Merlo ME, Moore J. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics. 2010;11:10. doi: 10.1186/1471-2164-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DC, Gontang EA, Kauffman CA, Jensen PR, Fenical W. Salinipyrones and pacificanones, mixed-precursor polyketides from the marine actinomycete Salinispora pacifica. J Nat Prod. 2008;71:570–575. doi: 10.1021/np0705155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H, Taniguchi S, Igarashi Y, Furumai T. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp TP-A0274 and its heterologous expression in Streptomyces lividans. J Antibiot. 2002;55:1063–1071. doi: 10.7164/antibiotics.55.1063. [DOI] [PubMed] [Google Scholar]
- Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009;3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner MK, Shen YC, Cheng XC, Jensen PR, Frankmoelle W, Kauffman CA, Fenical W, Lobkovsky E, Clardy J. Cyclomarins A-C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.) J Am Chem Soc. 1999;121:11273–11276. [Google Scholar]
- Roberts AA, Schultz AW, Kersten RD, Dorrestein PC, Moore BS. Iron acquisition in the marine actinomycete genus Salinispora is controlled by the desferrioxamine family of siderophores. FEMS Microbiol Lett. 2012;335:95–103. doi: 10.1111/j.1574-6968.2012.02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz AW, Oh DC, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J Am Chem Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidebottom AM, Johnson AR, Karty JA, Trader DJ, Carlson EE. Integrated metabolomics approach facilitates discovery of an unpredicted natural product suite from Streptomyces coelicolor M145. ACS Chem Biol. 2013;8:2009–2016. doi: 10.1021/cb4002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsueng G, Teisan S, Lam KS. Defined salt formulations for the growth of Salinispora tropica strain NPS21184 and the production of salinosporamide A (NPI-0052) and related analogs. Appl Microbiol Biotechnol. 2008;78:827–832. doi: 10.1007/s00253-008-1358-9. [DOI] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino MI, Engel P, Trautman E, Crawford JM. Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J Am Chem Soc. 2014;136:9244–9247. doi: 10.1021/ja503450q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Hotta K, Nakaya M, Praseuth AP, Wang CC, Inada D, Takahashi K, Fukushi E, Oguri H, Oikawa H. Escherichia coli allows efficient modular incorporation of newly isolated quinomycin biosynthetic enzyme into echinomycin biosynthetic pathway for rational design and synthesis of potent antibiotic unnatural natural product. J Amer Chem Soc. 2009;131:9347–9353. doi: 10.1021/ja902261a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci. 2012;109:1743–1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PG, Miller ED, Asolkar RN, Jensen PR, Fenical W. Arenicolides A-C, 26-membered ring macrolides from the marine actinomycete Salinispora arenicola. J Org Chem. 2007;72:5025–5034. doi: 10.1021/jo061878x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MC, Gulder TAM, Mahmud T, Moore BS. Shared biosynthesis of the saliniketals and rifamycins in Salinispora arenicola is controlled by the sare1259-encoded cytochrome P450. J Am Chem Soc. 2010;132:12757–12765. doi: 10.1021/ja105891a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MC, Mori T, Ruckert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UAE, Heycke N, Schmitt S, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- Winnikoff JR, Glukhov E, Watrous J, Dorrestein PC, Gerwick WH. Quantitative molecular networking to profile marine cyanobacterial metabolomes. J Antibiot. 2013;67:105–112. doi: 10.1038/ja.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, Wodtke A, de Felicio R, Fenner A, et al. Molecular networking as a dereplication strategy. J Nat Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ortega M, Shi Y, Wang H, Melby JO, Tang W, Mitchell DA, van der Donk WA. Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS. Proc Natl Acad Sci. 2014;111:12031–12036. doi: 10.1073/pnas.1406418111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz M, Millan-Aguinaga N, Chavarria KL, Jensen PR. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci. 2014;111:E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolova OE, Mady AS, Garneau-Tsodikova S. Recent developments in bisintercalator natural products. Biopolymers. 2010;93:777–790. doi: 10.1002/bip.21489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.