Abstract
Recent evidence reveals a crucial role for acetylcholine and its receptors in the regulation of inflammation, particularly of nicotinic acetylcholine receptor α7 (Chrna7) and muscarinic acetylcholine receptor 3 (Chrm3). Immunohistochemistry is a key tool for their cellular localization in functional tissues. We evaluated nine different commercially available antibodies on back skin tissue from wild-type (Wt) and gene-deficient (KO) mice. In the immunohistochemical analysis, we focused on key AChR-ligand sensitive skin cells (mast cells, nerve fibers and keratinocytes). All five antibodies tested for Chrm3 and the first three Chrna7 antibodies stained positive in both Wt and respective KO skin. With the 4th antibody (ab23832) nerve fibers were unlabeled in the KO mice. By western blot analysis, this antibody detected bands in both Wt and Chrna7 KO skin and brain. qRT-PCR revealed mRNA amplification with a primer set for the undeleted region in both Wt and KO mice, but none with a primer set for the deleted region in KO mice. By 2D electrophoresis, we found β-actin and β-enolase cross reactivity, which was confirmed by double immunolabeling. In view of the present results, the tested antibodies are not suitable for immunolocalization in skin and suggest thorough control of antibody specificity is required if histomorphometry is intended.
Keywords: atopic dermatitis-like allergic dermatitis (AlD), muscarinic acetylcholine receptor 3 (Chrm3), non-neuronal cholinergic system (NNCS), nicotinic acetylcholine receptor α7 (Chrna7), stress
Introduction
Non-neuronal cholinergic expression has gained significant attention in the past two decades. Apart from its classical neurotransmission function, acetylcholine (ACh) has been shown to have important roles in non-neuronal tissues, particularly in inflammatory reactions in organs at the self-environment border (Roosterman et al. 2006). The skin is one such organ, targeted by many challenges that invoke an inflammatory response to maintain homeostasis and integrity. In the skin, the expression of ACh and its receptors has been found in nerve fibers, keratinocytes, melanocytes, and in cells of the immune system, such as mast cells and lymphocytes (Bering et al. 1987; Grando et al. 1993; Iyengar 1989; Lammerding-Koppel et al. 1997; Richman and Arnason 1979; Schmelz et al. 2000). Furthermore, human keratinocytes have been shown to express ACh synthesizing and hydrolyzing enzymes, namely choline acetyltransferase (ChAT) and acetylcholinesterase (AChE) (Grando 1997). The skin can therefore be considered as a prime source as well as target for non-neuronal ACh signaling (Kurzen et al. 2007).
Binding of ACh results in the activation of two different classes of receptors, namely the muscarinic and nicotinic acetylcholine receptors (AChRs). In the central nervous system, functional characterization of these AChRs has been extensively reported (Hogg et al. 2003). Functional data on the role of ACh in the homeostasis of skin cell populations largely relies on cell culture experiments. Lately, however, it has become evident that, in a living organism, the functional outcome of ACh signaling depends on a complex interplay between various cell populations and signaling cascades (Kawashima et al. 2012; Picciotto et al. 2012). This makes in vivo studies essential, especially for those studies involving environmental stressors that affect the non-neuronal cholinergic system (NNCS).
To now better understand the multiple roles of peripheral AChRs, an extensive assessment of their cellular and subcellular localizations in intact, whole organisms is essential. This is especially important to investigate the real life interactions of these proteins in complex organisms, such as animals under stress. The use of anti-receptor subunit antibodies to localize AChRs in situ, however, is commonly controlled only by standard negative staining procedures, such as omission of the primary antibody or incubation with species-specific IgG1 instead of the primary antibody. Data obtained by immunohistochemistry (IHC) is usually trusted if they are in accordance with other methods that approve protein localization, such as western blot analysis (WB) or ligand binding studies. However, reported staining results are often unable to be reproduced independently. Also, substantial discrepancies have been reported (Gotti and Clementi 2004) as to the localization and the functional relevance of AChR activation in the inflammatory responses of the skin. Reliable and reproducible methods for their in situ detection and more elaborate methods to control for label-specificity in IHC are therefore urgently required.
In this study, we evaluated nine different antibodies for Chrm3 and Chrna7 by IHC. To reliably prove specificity, we employed AChR subunit-deficient mouse tissue for negative controls, which, in our opinion, provides an ideal tool for testing the specificity of subunit-directed antibodies. Additionally, we quantitatively analyzed the mRNA expression of Chrna7 using qRT-PCR analysis and protein expression was evaluated by western blotting and 2D gel electrophoresis. With this approach, we aim to reliably localize and characterize AChRs in the murine skin.
Materials & Methods
Animals
The present study was performed with permission of the state of Hessen, Regierungspräsidium Giessen, according to section 8 of the German Law for the Protection of Animals, and conforms to the NIH guidelines for the care and use of laboratory animals. Young, female wild-type (Wt), Chrm3 knock-out (KO) and Chrna7 KO mice (7–8 weeks of age) were used in this study. Their generation and characterization have been described in detail in earlier studies (Orr-Urtreger et al. 1997; Yamada et al. 2001). Mice were housed under a 12/12 hr light/dark cycle in temperature-regulated rooms (22–24°C), with food and water ad libitum. The genotype of each mouse was confirmed by PCR in all experiments. The corresponding background strains for the knockout mice (C57Bl/6J for Chrna7 and B6NTac for Chrm3) were used as Wt mice.
Genotyping
Genotyping of all mice used in this study was performed as described elsewhere (Moser et al. 2007). PCR was carried out on approximately 0.5 µg DNA using Kapa Mouse Genotyping Kit (Peqlab; Erlangen, Germany), according to manufacturer’s protocol. The thermal cycler profile was as follows: 95°C for 3 min; followed by 35 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec); and then 72°C for 7 min.
Antibodies
In this study, five different antibodies with presumed specificity for Chrm3 and four different antibodies for Chrna7 were tested in parallel by IHC and WB on skin and brain biopsies from Wt and KO mice. A detailed description of the antibodies applied is provided in Table 1.
Table 1.
Antibodies Tested for Specificity in Wild Type and Corresponding Knock-out Mice Skin.
| AChR subunit | Company | Catalogue Number | Dilutions Tested | Epitope |
|---|---|---|---|---|
| Rabbit anti-Chrm3 | Abcam (Cambridge, UK) | ab41169 | 1:50; 1:100; 1:200; 1:400 | C-terminal region |
| Goat anti-Chrm3 | Santa Cruz Biotechnology (Dallas, TX) | sc-31486 | 1:50;1:100;1:200; 1:250; 1:400 | N-terminal extracellular domain |
| Goat anti-Chrm3 | Santa Cruz Biotechnology | sc-31487 | 1:50;1:100;1:200; 1:250; 1:400 | Cytoplasmic domain |
| Rabbit anti-Chrm3 | Sigma-Aldrich (Steinheim, Germany) | M0194 | 1:100; 1:200; 1:400 | Amino acids 461–479 (3rd intra cellular loop) |
| Rabbit anti- Chrm3 | Genetex (Irvine, CA) | GTX111637 | 1:100; 1:200 | Amino acids 286–501 |
| Rabbit anti-Chrna7 | Abcam | ab28741 | 1:100; 1:200 | Amino acids 398–447 (exon 10, within deleted region in Chrna7 KO mice) |
| Rabbit anti-Chrna7 | Santa Cruz Biotechnology | sc-5544 | 1:100; 1:200; 1:400 | Amino acids 367–502 (exon 10, within deleted region in Chrna7 KO mice) |
| Rabbit anti-Chrna7 | R&D Abs (Las Vegas, NV) | AS-5631S | 1:100; 1:200 | Amino acids 493–502 (exon 10, within deleted region in Chrna7 KO mice) |
| Rabbit anti-Chrna7 | Abcam | ab23832 | 1:100; 1:200 | Amino acids 1–100 (exons 1–4, not in deleted region in Chrna7 KO mice) |
| Rabbit anti β-enolase | LSBio (Eching, Germany) | LS-C81180 | 1:200; 1:400; 1:800 | N-terminal region |
| Rabbit anti β-actin | Abcam | a8227 | 1:100; 1:200; 1:400 | Amino acids 1–100 |
AChR, acetylcholine receptor.
Preparation of Tissue Samples
Mice were deeply anesthetized with a mixture of 2% xylazin (CEVA, Ceva Sante Animale, Libourne, France; 12 mg/kg body weight) and 25 mg/ml ketanest S (Pfizer, Berlin, Germany; 100 mg/kg body weight). The hair on the back skin was then gently shaved with a beard and hair trimmer (Aesculap Isis GT420; Suhl, Germany). After cervical dislocation, the skin on the back was dissected under sterile conditions using fine laboratory scissors and scalpel, and quickly shock-frozen in liquid nitrogen. Whole brains were also removed aseptically from the mice and frozen in liquid nitrogen. All samples were finally stored at -80°C until further processing. For skin biopsies intended for immunofluorescence, the mice were administered with a lethal dose of anesthesia. The abdominal cavity and rib cage were opened and the fixative was injected using a butterfly syringe inserted through the left ventricle of the heart. For optimal circulation of the fixative throughout the body, the right ventricle was cut open (Hendrix et al. 2008; Peters et al. 2011). LANA solution, composed of paraformaldehyde, NaOH, saturated picric acid and distilled water, was used as the fixative agent. In addition, frozen tissues from all of the aforementioned organs were used for protein extraction and subsequent immunoblotting and for RNA isolation, as described below in qRT-PCR section.
Immunohistochemistry
For IHC, 14-µm-thick mouse back skin cryostat sections were used. Frozen sections were air-dried for 30 min, and washed in Tris-buffered saline (TBS) (3×5 min). Blocking was performed for 30 min with 5% normal serum (NS) of the host species in which the secondary antibody was generated. Primary antibodies were diluted in TBS with 0.3% Triton X-100 and 2% NS, and incubated for 24 hr at 4°C. In addition to this standard protocol, all antibodies were tested with antigen retrieval using 1% sodium dodecyl sulfate (SDS) (Brown et al. 1996) for 5 min with no effect on the below-described results. After the primary antibody incubation, the secondary antibody (diluted 1:200 in TBS) was incubated for 1 hr at 37°C with Cyanine (Cy3)-conjugated F(ab)2 fragments of donkey anti-rabbit or donkey anti-goat IgG (Dianova, Hamburg, Germany). After washing with TBS (3×5 min), mast cell staining was performed with fluorescein isothiocyanate (FITC)-Avidin (Botchkarev et al. 1997; Tharp et al. 1985) (diluted 1:5000 in TBS) for 20 min at 37°C followed by counterstaining of nuclei with DAPI. The staining patterns were reproduced at least three times per staining protocol. Standard controls were performed by omitting the primary antibodies and by incubation with mouse IgG1 instead of primary antibodies. Images of the observed positively stained sections were taken using a Leica DMI5000 M microscope with DFC345 FX camera and Leica Application Suite Advanced Fluorescence (LAS-AF) 2.7 software (Leica Microsystems; Wetzlar, Germany).
Western Blotting
For analysis of Chrna7 protein from skin and brain tissues of Chrna7 KO mouse and Wt mouse, total protein was isolated using a urea extraction method (Paddenberg et al. 2012). Protein homogenates (40 μg) were loaded into each well of 12% gels. SDS-PAGE was followed by a semi-dry transfer onto PVDF membranes. Rabbit anti-Chrna7 (ab23832; Abcam, Cambridge, UK) and mouse anti-β-actin (ab8227; Abcam) antibodies were used as primary antibodies followed by anti-rabbit or -mouse HRP-conjugated secondary antibodies (Dako, Hamburg, Germany). The epitope recognition region spans the first 100 amino acids of the Chrna7 protein. β-actin served as a loading control.
Quantitative Real Time PCR (qRT-PCR)
Mouse skin or brain biopsies were homogenized to obtain a fine powder using a metal ball mill (MM2000; Retsch, Haan, Germany). This step was necessary for the efficient disintegration of biopsy specimens. Powdered biopsies were then completely dissolved in RLT buffer and subjected to total RNA isolation using the RNeasy Mini Kit (Qiagen; Hilden, Germany) according to the manufacturer’s protocol. Contaminating DNA was removed using DNase (1 U/µl; Life Technologies, Darmstadt, Germany) in the presence of 20 mM Tris-HCl (pH 8.4), 2 mM MgCl2, 50 mM KCl for 15 min at 25°C. Equal amounts of RNA were reverse transcribed with Superscript II reverse transcriptase (Life Technologies) for 50 min at 42°C. cDNA was then subjected to qRT-PCR analysis (StepOne Real-time PCR system, Life Technologies) using QuantiTect Probe PCR kit (Qiagen) and two sets of specific forward and reverse primers for Chrna7 spanning exons 1–4 and exons 8–10. The primers used in the study are listed in Table 2. Gene expression was normalized to HPRT calculating the difference between the cycle time (CT) for HPRT and the CT for the respective transcript.
Table 2.
List of Primers and Probes Used for qRT-PCR.
| Gene | Primer Sequence |
|---|---|
| Chrna7 (exons 1–4) | Forward: CCTGCAAGGCGAGTTCC |
| Reverse: CTCAGGGAGAAGTACACGGTGA | |
| Taqman probe: 6FAM-CTGGTCAAGAACTACAACCCGCTGGA–BBQ | |
| Chrna7 (exons 8–10) | Forward: GCCCTTGATAGCACAGTACTTCG |
| Reverse: GATCCTGGTCCACTTAGGCATTT | |
| Taqman probe: 6FAM-CAGTGGTCGTGACAGTGATTGTGCTGC-BBQ | |
| HPRT | Forward: GTTGGATACAGGCCAGACTTTGT |
| Reverse: CACAGGACTAGAACACCTGC | |
| Taqman probe: 6FAM-TTGCAGATTCAACTTGCGCTCATCTT–BBQ |
2D Gel Electrophoresis
Protein Isolation
The tissue was cut into pieces. Samples were homogenized in 250 µl lysis buffer consisting of 6 M urea (Sigma-Aldrich; Taufkirchen, Germany), 2 M thiourea (Sigma-Aldrich), 4% 3-3′-(cholamidopropyl)-3,3-dimethylammoniumpropylsulfate (CHAPS; Roth, Karlsruhe, Germany), 30 mM dithiothreitol (DTT; Fluka, Seelze, Germany), 2% IPG-buffer pH 3–10 (GE Healthcare; Freiburg, Germany) for 3 min at intensity 8 in a bullet blender using 0.5 mm glass beads (Next Advance; New York, USA). Samples were then centrifuged at 20,000 × g, for 1 hr at 4°C.
Two-dimensional Gel Electrophoresis
Isoelectric focusing (IEF)-strips (pH 3–10 NL, 13 cm; GE Healthcare, Freiburg, Germany) were loaded with samples containing 268 µg (for preparative gels) or 100 µg (for WB) of protein by rehydration for 24 hr. For isoelectric focusing, a Multiphor II system (GE Healthcare) operating in gradient mode was used. Focusing was performed with voltage gradients from 0–100 V, 1 mA, 2 W for 5 hr; from 100–3500 V, 2 mA, 5 W for 6 hr, and at 3500 V, 2 mA, 5 W for 6 hr. After focusing, the IEF-strips were either stored at -80°C or immediately equilibrated for 15 min in 5 ml equilibration stock solution (ESS; 6 M urea, 4% SDS, 0.01% bromophenol blue, 50 mM Tris-HCl pH 8.8, 30% glycerol (v/v), 65 mM DTT), followed by 15 min in ESS containing 216 mM iodacetamide. Protein separation in 2D was performed by electrophoresis using 12.5% SDS polyacrylamide gels, according to the protocol of Laemmli (Laemmli 1970). Electrophoresis was carried out in a Hoefer600 cell (GE Healthcare, Freiburg, Germany) with the following program: 15 min at 15 mA/gel (300 V and 25 W limits, and 2 hr at 110 mA (400 V and 25 W limits). Gels were stained with Flamingo (Bio-Rad) or electroblotted.
Western Blotting
For WB, 2D-separated proteins were transferred to PVDF membranes by tank blotting. After blocking with Roti-Block (Roth; Karlsruhe, Germany), the membranes were incubated with the Chrna7 antibody (ab23832, Abcam, 1:1000 dilution) overnight at 4°C in Roti-Block. Afterwards, the membranes were washed 5 times for 3 min with PBS-T (PBS containing 0.05 % Tween 20), and then incubated with HRP-conjugated anti-rabbit Ig (P0217, Dako Cytomation, Hamburg, Germany; 1: 3000 dilution) for 1 hr at room temperature. Membranes were again washed 5 times for 3 min with Roti-Block. PC-modified proteins were visualized by enhanced chemiluminescence using the ECL SuperSignal kit (GE Healthcare). The corresponding protein spots were excised from preparative gels with a ExQuest™ Spot Cutter (Bio-Rad) and transferred into 96-well plates (Greiner Bio-One; Frickenhausen, Germany).
Tryptic In-gel Digestion of Proteins
The simultaneous digestion of the samples was performed by a pipetting robot (MicroStarlet; Hamilton Robotics, Munich, Germany). The excised gel plugs were destained with 25 mM ammonium hydrogen carbonate containing 50% (v/v) acetonitrile, and then dehydrated with 100% acetonitrile and re-swelled in 50 mM ammonium hydrogen carbonate before again being dehydrated with 100% acetonitrile. Samples were finally dried at 56°C. For tryptic digestion, gel plugs were rehydrated with a solution of 17 µl of 25 mM ammonium hydrogen carbonate containing 4.5 ng/µl trypsin (sequencing grade; Promega, Mannheim, Germany) and 0.02% Proteasemax (Promega) and incubated at 45°C for 2 hr. Peptides were recovered by extraction with 15 µl of 1% trifluoroacetic acid (TFA; Applied Biosystems, Warrington, UK) and stored at 4°C until further use.
Matrix-Assisted Laser-Desorption Ionization Time-Of-Flight Mass Spectrometry (MALDI-TOF-MS)
MALDI-TOF-MS was performed on an Ultraflex I TOF/TOF mass spectrometer (Bruker Daltonics; Bremen, Germany) equipped with a nitrogen laser and a LIFT-MS/MS facility. The instrument was operated in the positive-ion reflectron mode using 2,5-dihydroxybenzoic acid (5 mg/ml; Sigma-Aldrich), and methylendiphosphonic acid (5 mg/ml; Fluka) in 0.1% TFA as matrix. Sum spectra, consisting of 200–400 single spectra, were acquired. For data processing and instrument control, we used the Compass 1.3 software package consisting of FlexControl 2.4, FlexAnalysis 3.0, BioTools 3.0. Data storage and database searching were performed using ProteinScape 3.1 (Bruker Daltonics).
Database Search
Proteins were identified by MASCOT peptide mass fingerprint search (Matrix Science; London, UK) using the Uniprot Mouse database (20140219; 51373 sequences, 24629748 residues). For the search, the allowed criteria were: a mass tolerance of 75 ppm, carbamidomethylation of cysteine as a global modification, oxidation of methionine as a variable modification, and one missed cleavage. Identifications with a Mascot Score higher than 60 were considered as statistically significant (p<0.05).
Results
Chrm3 Antibodies Stain Positively in Both Wild Type and Chrm3 KO Mice
In the case of Chrm3, for each of the five antibodies tested, staining in Wt animals rendered staining patterns that suggested successful positive staining. Most antibodies showed clear staining of the epidermis, nerve fibers and cells of the immune system, such as mast cells. Staining was negative when standard control staining methods were employed (omission of primary antibody). Different dilutions of each Chrm3 antibody were tested (Table 1). There was no significant effect of the dilution in terms of antibody binding specificity. A lower concentration of primary antibody resulted in lower staining intensity but the same positive structures, or complete negative staining.
However, staining in the skin from the KO animal was not negative and largely resembled those staining patterns observed in Wt samples. In detail, the Chrm3 antibody ab41169 (Abcam) showed staining in the epidermis, mast cells and nerve fibers in both Wt as well as KO skin, with the only difference that staining in the Wt skin was more intense. The Chrm3 antibody sc-31486 (Santa Cruz Biotechnology) showed staining in the epidermis, mast cells and skin fibroblasts in both Wt and KO samples. The Chrm3 antibody sc-31487 (Santa Cruz Biotechnology) labelled venules located next to hair follicles in Wt and KO samples. In addition, this antibody labelled nerve fibers exclusively in the KO skin. The Chrm3 antibodies GTX111637 (Genetex) and M0194 (Sigma-Aldrich) paradoxically showed more intense staining in all the analyzed cell populations of the KO tissue as compared with that in the Wt skin (Fig. 1 and Table 3).
Figure 1.
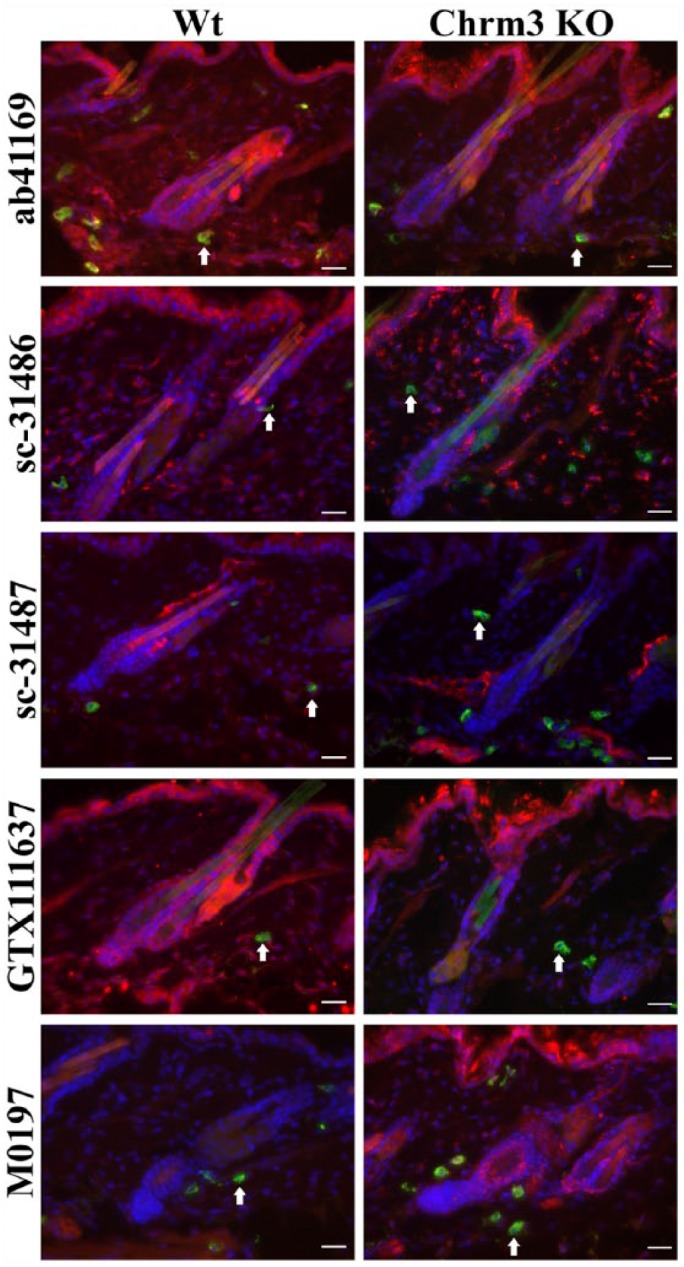
Representative photomicrographs of an immunohistochemical triple staining as described in the Materials & Methods section. The stained antigen is Chrm3 (red), with staining performed using ab41169, sc-31486, sc-31487, GTX 111637 or M0194 antibodies. Mast cells (FITC-labeled avidin, green; white arrows) and cell nuclei (DAPI, blue) are counterstained. Double labelling results in a yellow signal. The left panel shows an example of the staining pattern obtained in a skin biopsy in a wild type (Wt) animal. Right panel shows the staining of skin from a Chrm3 knock-out (KO) animal. Scale, 25 μm.
Table 3.
Differences in the Staining Intensities of Different Chrna7 Antibodies.
| Epidermis |
Mast Cells |
Single Nerve Fibers |
||||
|---|---|---|---|---|---|---|
| Wt | KO | Wt | KO | Wt | KO | |
| Exon 10 (Abcam; ab28741) | ++ | + | ++ | + | ++ | + |
| Exon 10 (Santa Cruz Biotechnology; sc-5544) | +++ | ++ | +++ | +++ | ++ | + |
| Exon 10 (R&D Abs; AS-5631S) | +++ | + | +++ | + | +++ | + |
| Exons 1-4 (Abcam; ab23832) | +++ | + | +++ | +++ | +++ | - |
“-” = no staining; “+ - +++” = positive staining of differing intensities. Wt, wild type; KO, knock-out.
Differential Staining Pattern of Different Chrna7 Antibodies in Skin from KO Mice
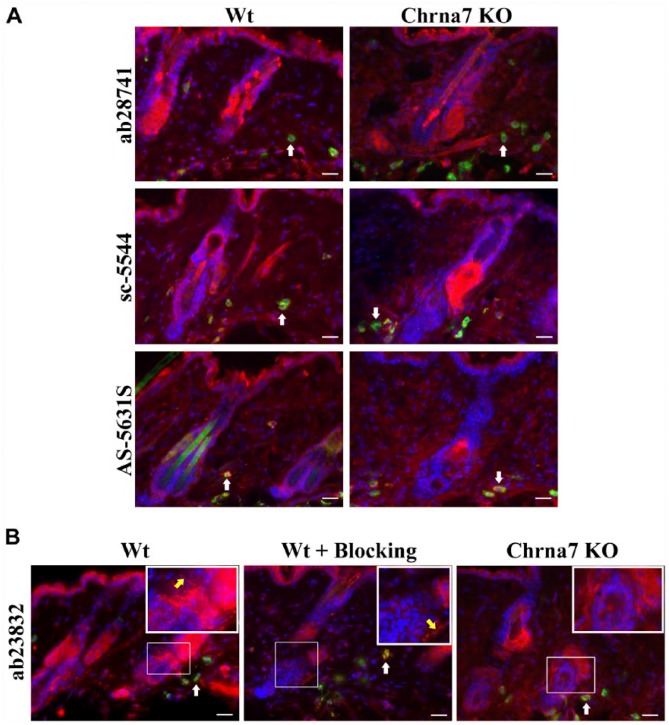
The three antibodies used for Chrna7—ab28741 (Abcam), sc-5544 (Santa Cruz Biotechnology) and AS-5631S (R&D Abs)—were raised against the C-terminal end of Chrna7 (spanning exon 10) and showed staining in keratinocytes, mast cells and nerve fibers in Wt mouse skin. In Chrna7 KO mouse skin, we observed reduced staining intensity, but all structures labelled in the Wt skin were positive (Fig. 2A). The Abcam antibody, ab23832 (raised against protein coded in exons 1–4, which is not deleted in Chrna7 KO), showed clear labeling of keratinocytes, mast cells and nerve fibers in Wt mouse skin. In KO skin, the staining intensity in mast cells and keratinocytes was reduced and, interestingly, nerve fibers were exclusively unlabelled in the KO mouse skin with ab23832 (Table 3, Fig. 2B). Hence, the staining appears to be partially detecting Chrna7 antigen. Pre-incubation of ab23832 with its specific blocking peptide ab24285 (Abcam) at different concentrations (10 µg/ml, 100 µg/ml, 200 µg/ml), different temperatures (room temperature or 4°C), and for different incubation times (30 min or 1 hr or overnight) with or without agitation resulted in less staining intensity, but the positive signal remained (Fig. 2B). The antibody dilution did not produce any significant change in the staining pattern of all four of the Chrna7 antibodies tested (Table 1).
Figure 2.
Representative photomicrographs of an immunohistochemical triple staining, as described in the Materials & Methods section. (A) Stained antigen is Chrna7 (red) (ab28741, sc-5544, AS-5631S). Mast cells are FITC-labeled avidin (green, white arrows) and cell nuclei are stained with DAPI (blue). Double labelling results in a yellow signal. The left panel shows an example of the staining pattern obtained in a skin biopsy of a wild type (Wt) animal. The right panel shows the staining of skin from a Chrna7 knock-out (KO) animal. (B) Stained antigen is Chrna7 (red) (ab23832), mast cells (FITC-labeled avidin - green, pointed at by white arrows) and cell nuclei (DAPI – blue). Double labelling results in yellow signal. Regions of interest are indicated by thin white squares and shown as blow-ups in thicker white squares. Blow-ups show positive staining of nerve fibers (pointed at by yellow arrow) with ab23832 in Wt skin with or without blocking peptide (ab24285), but no positive nerve fibers in Chrna7 KO animal. Scale, 25 μm.
Western Blotting Detects Expected Double Bands Only With One Chrna7 Antibody
By WB, the first three antibodies for Chrna7 produced either multiple unspecific bands or no bands at all at the region corresponding to Chrna7 (40–60 kDa; data not shown). Only when using the ab23832 (exons 1–4) antibody did we detect a clear double band at the expected 45 and 56 kDa (Fig. 3). Membranes tested without the primary antibody or non-specific IgG1 did not reveal any signals.
Figure 3.
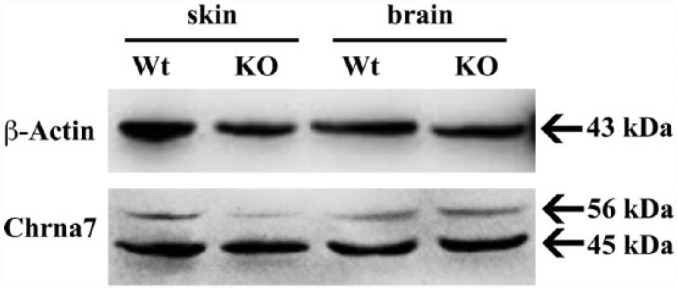
Analysis of brain and skin biopsy lysates from wild type (Wt) and Chrna7 knock-out (KO) mice by western blotting using the Chrna7 (ab23832) antibody. Lanes 1 and 2 show skin lysates, 3 and 4 show brain lysates. The immunoblot is representative of three independent experiments. Equal loading was checked using β-actin.
qRT-PCR Demonstrates the Presence of a Transcript for the Undeleted Chrna7 Region in KO Mice
In Wt mice skin biopsies, we could detect the mRNA expression of Chrna7 by qRT-PCR. This was accomplished using primers for Chrna7 exons 1–4 as well as exons 8–10. We also detected the presence of a variant Chrna7 mRNA, which lacked exons 8–10, in Chrna7 KO mice using the primers for exons 1–4 (Fig. 4A and B).
Figure 4.
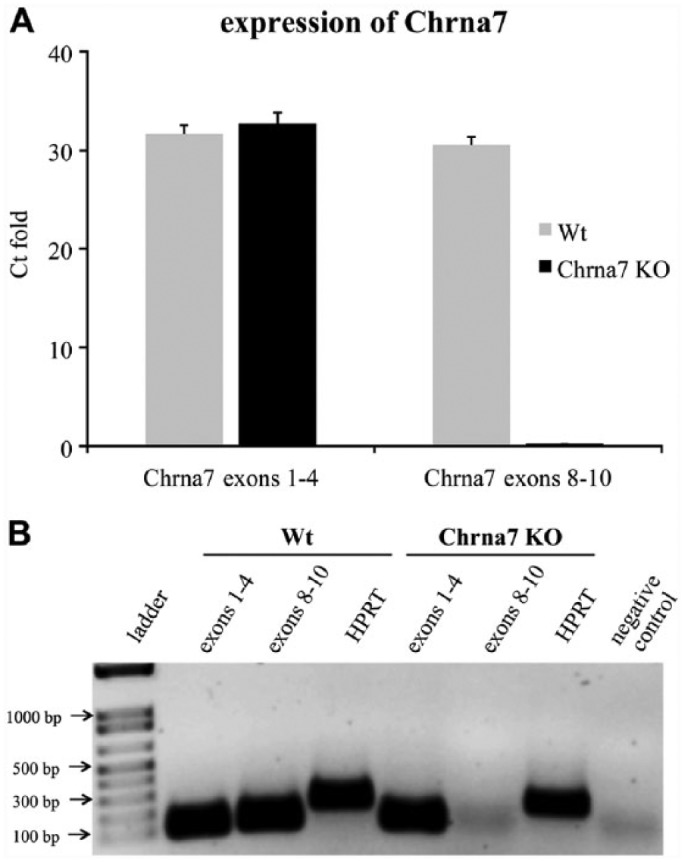
Expression of Chrna7 mRNA. (A) qRT-PCR analysis of Chrna7 mRNA expression in Chrna7 knock out (KO) mice as compared to wild type (Wt) mice (C57Bl/6). Data represent the mean values of three independent experiments ± SD. (B) Ethidium bromide-stained agarose gel electrophoresis shows the qRT-PCR products. Lane 1 depicts the DNA ladder, lanes 2–3 and lanes 5–6 show the mRNA expression of Chrna7 in Wt and Chrna7 KO skin biopsies, respectively. The house keeping gene HPRT was used as an internal control (lanes 4 and 7). PCR quality control sterile water (without DNA) served as a negative control (lane 8).
Ab23832 Antibody Identifies β-actin and β-enolase with Immunoblotting
The extracts of whole skin homogenates of Wt mice were electrophoretically separated in a 2D SDS-PAGE followed by immunoblotting using ab23832. The analysis revealed a distinct spot pattern in Wt (C57Bl6 J) mice skin. By MALDI-TOF-MS, the molecular range of the spots of interest was determined and revealed that ab23832 labeled a series of spots of approximately 40–55 kDa with a pI of approximately 5.5 (Fig. 5). However, the MASCOT peptide mass fingerprint search did not confirm any of these spots to be Chrna7. Instead of labelling Chrna7 protein, these spots corresponded to that of β-actin and β-enolase. Hence, no Chrna7 spot could be detected, questioning the above described staining result in nerve fibers.
Figure 5.
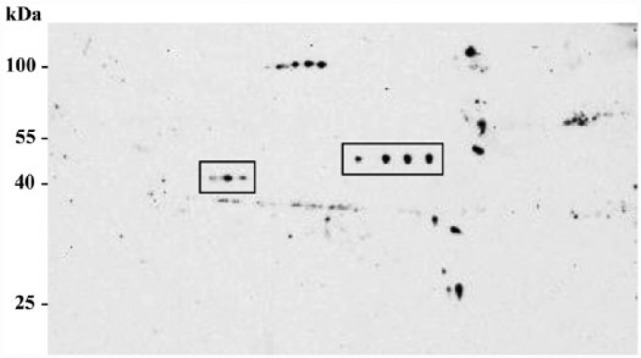
Western blotting of whole skin biopsy lysates after 2D SDS-PAGE detected with the Chrna7 ab23832 antibody. The protein lysates were pooled from each of three wild type (Wt) mice. Approximately 40 µg of protein were resolved on 2D gels and analyzed by western blotting for ab23832 (size, ~50 kDa; pI, ~5.5).
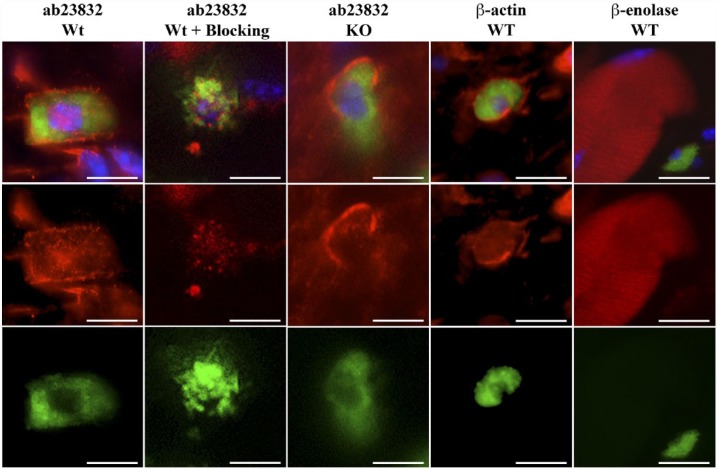
Chrna7 (ab23832) Antibody Shows the Same Staining Pattern as That for β-actin, but Not for β-enolase, in Mast Cells
To confirm that the Chrna7 antibody, ab23832, stains β-actin or β-enolase, we performed IHC triple staining with DAPI for the nuclei, FITC-labeled avidin for the mast cells and Chrna7 (ab23832) or β-actin or β-enolase. The staining pattern of ab23832 in Wt and Chrna7 KO skin as exactly the same pattern as that for β-actin. As β-enolase is muscle specific, we detected it only in muscle fibers of the Wt skin. In mast cell staining, the β-actin antibody showed a similar staining pattern to that of ab23832 in Wt and Chrna7 KO skin (Fig. 6).
Figure 6.
Representative photomicrographs of an immunohistochemical triple staining as described in the Materials & Methods section. Stained antigens are Chrna7, β-actin or β-enolase (red) (Abcam, ab23832; Abcam; ab8227; LSBio, LS-C81180, respectively), mast cells (FITC-labeled avidin; green) and cell nuclei (DAPI; blue). First and second column panels show an example of the staining pattern obtained in the mast cells of wild type (Wt) skin without or with blocking peptide (ab24285) and the third column panels show that of skin from Chrna7 KO animals with ab23832. The fourth column panels show an example of the staining pattern of β-actin in mast cells of the Wt animal. The fifth column panels show an example of the staining pattern of β-enolase in muscle fibers of the Wt animal. Scale, 10 μm.
Discussion
We here report the unspecific labeling that is obtained using nine different commercial antibodies directed against Chrm3 and Chrna7. All produced similar staining patterns in skin from Wt and KO mice. Only one, from the nine antibodies tested, appeared to give specific staining in skin mast cells, keratinocytes and nerve fibers; albeit, this was disguised by the additional labeling of unrelated proteins.
The Chrm3 KO mice strain used in this study was generated and characterized by Yamada and colleagues, wherein the Chrm3 gene was deleted via targeted disruption (Yamada et al. 2001). A small part of the C-terminus is not knocked out. However, since the start codon and the following exons are deleted, there is no possibility of Chrm3 gene transcription that would result in protein detection. Chrna7 is a membrane-bound ion channel consisting of pentameric subunits. Each subunit is composed of 502 amino acids [1506 base pairs (bp)]. The Chrna7 gene is encoded by 10 exons (2091 bp) and the coding sequence lies between 51–1559 bp. The Chrna7 KO animal strain used in this study was designed by Orr-Urtreger and colleagues (Orr-Urtreger et al. 1997). The functional part of the protein located in the C-terminal end, coded by exons 8–10, is deleted in this Chrna7 KO mouse. As the start codon and initial exons coding the extracellular, ligand binding domains of Chrna7 subunit are not deleted, there is a possibility of an mRNA coding exons 1 through 7 and therefore a partial protein.
Chrm3 expression has been previously shown in mast cells (Sawada et al. 2010) and epithelial cells, such as keratinocytes (Arredondo et al. 2003; Ndoye et al. 1998). Similarly, the expression of Chrna7 has been reported in mast cells (Aung et al. 2011), keratinocytes (Arredondo et al. 2006), and nerve fibers (Downs et al. 2014). Here, we used standard protocols for IHC. To examine the specificity of antibodies raised against Chrm3 and Chrna7 subunits in skin samples, we employed WB, KO mouse tissues, and protein expression in brain sections, as well as standard negative controls for Wt mouse tissues.
Unexpectedly, in the case of Chrm3, each of the five antibodies tested showed indecisive immunoreactivity and a staining pattern that was similar between Wt and Chrm3 KO samples. This positive staining in KO tissue strongly suggests that the antibodies are not suitable for immunolocalization (Fig. 1). Hence, we decided not to investigate further the unsuitability of these Chrm3 antibodies.
For Chrna7, the antibody ab23832, raised against the N-terminus of Chrna7 (exons 1-4), showed positive staining in all of the investigated cell populations in the skin of Wt and Chrna7 KO mice. The use of the blocking peptide did not obliterate the staining, which also suggested non-specificity of the antibody generated against a synthetic peptide within residues 1–100 of human Chrna7 and conjugated to KLH. Interestingly, nerve fibers were exclusively unlabeled in the KO mice (Fig. 2B). On the basis of these staining results in mouse skin, we initially surmised that perhaps there may be a truncated Chrna7 protein coded between exons 1–7. Similar evidence of a truncated protein was shown, for example, for tyrosine kinase B (TrkB) in keratinocytes (Kryl and Barker 2000; Marconi et al. 2003), and this triggered our curiosity. Furthermore, reports of alternatively spliced variants of mouse Chrna7 have been published (Saragoza et al. 2003). Our qRT-PCR analysis revealed an mRNA amplification with a primer set for the undeleted region (exons 1–4) in both Wt and Chrna7 KO animal skin and brain, but none with a primer set for the deleted region (exons 8–10) in KO mice (Fig. 4). If the remaining mRNA (Fig. 4) is translated into a protein, this would be around 40 kDa or less. By WB, the Chrna7 antibody (ab23832) produced a clear double band showing its protein expression in skin and brain of Wt and Chrna7 KO animals at 45 and 56 kDa (Fig. 3). The predicted band size for full-length Chrna7 matches with the 56 kDa band. The additional band at 45 kDa is not predicted by any reported variants of Chrna7. qRT-PCR of Wt and Chrna7 KO thus suggested the possibility of a variant Chrna7 mRNA in Wt and Chrna7 KO mice. By WB, however, we could not detect a truncated protein exactly matching the predicted size.
To better understand the specificity of ab23832, we performed 2D electrophoresis and MALDI-TOF-MS, which clearly showed a series of spots at around 40–50 kDa (Fig. 5). The lower set of spots (~40 kDa) were confirmed to be that of β-actin, whereas the higher set spots (~50 kDa) was that of β-enolase. In concordance, the IHC of β-actin and β-enolase antibodies in Wt mouse skin clearly showed a similar staining pattern between β-actin and Chrna7 (ab23832) antibodies, whereas β-enolase, as a muscle-specific protein, was only detected in the muscle fibers of the skin. Staining of ab23832 in Chrna7 KO tissue shows exactly the same pattern, especially in the mast cells (Fig. 6). Using BLAST protein sequence alignment, we determined a 39% similarity for the first 100 amino acids (region detected by ab23832) of mouse Chrna7 and β-actin and a similarity of 35% for Chrna7 and β-enolase, which could be the potential reason for the positive signal detected in KO tissue. Hence, the presence of a truncated protein can, by large, be ruled out.
After 2D gel electrophoresis followed by MALDI-TOF-MS and double immunostaining with β-actin and β-enolase, it is very clear that ab23832 is not detecting Chrna7; instead, it is cross reacting with β-actin and β-enolase in murine skin. Therefore, we conclude that this antibody is also unsuitable for IHC.
The other three Chrna7 antibodies raised against the C-terminal end of the protein were used in this study to circumvent false positive staining of a not yet excluded variant of Chrna7 (Fig. 2A). However, all three antibodies showed staining in Wt as well as in Chrna7 KO mouse skin.
Our results clearly show that it is necessary to do further controls in terms of antibody specificity even if the expected staining patterns and routine controls suggest specificity. Since reported by Herber and colleagues 11 years ago (Herber et al. 2004), several other groups have also pointed out unspecific staining patterns of AChR antibodies in IHC (Jositsch et al. 2009; Moser et al. 2007; Pradidarcheep et al. 2009). With rising interest in the NNCS, as evident from recent literature, it is inevitable to localize the expression of AChRs in intact functional tissues for exact biological characterization (Di Angelantonio et al. 2003; Gotti and Clementi 2004; Wevers et al. 1999). Unfortunately, the availability of specific antibodies does not seem to have improved in the past decade. Researchers who try to identify expression patterns and functions of AChR in unreported tissues, species and experimental paradigms, such as the skins’ response to stress, have to spend huge amounts of time and energy on multiple techniques to confirm the correct localization of these receptors by IHC.
In our opinion, IHC could be the most instructive method to localize AChR expression in functional tissues under defined experimental conditions. However, this remains hampered by the lack of reliable commercial antibodies for immune detection. A thorough evaluation of AChR antibody specificity with functional, molecular and quantitative techniques is desirable. The use of KO animal tissue to control the suitability of antibodies appears to be one holistic approach available. We are very much aware that extraction of a full-length protein in its native state for the generation of highly specific antibodies is sometimes impossible or complicated. However, synthetic peptides apparently are not often sufficient templates for antibody generation because physiological folding of a protein cannot be mimicked artificially.
With these results it is clearly evident that all of the nine tested AChR antibodies are not suitable for the immunolocalization of AChR in skin samples. If histomorphometry is to be used, we recommend a complete validation of antibody specificity. Unfortunately, we have to report that there is no good antibody as yet available for these AChRs since the report of Herber et al. (2004), and that most of these antibodies are still commercially available.
Acknowledgments
We wish to thank Sandra Laux, Barbara Gries, Lianne Renno, Michael Dreisbach, Christina Schmidt and Hans-Günter Welker for their excellent technical assistance.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work was supported by LOEWE grants to EP, UG, GL and WK.
References
- Arredondo J, Chernyavsky AI, Jolkovsky DL, Pinkerton KE, Grando SA. (2006). Receptor-mediated tobacco toxicity: cooperation of the Ras/Raf-1/MEK1/ERK and JAK-2/STAT-3 pathways downstream of alpha7 nicotinic receptor in oral keratinocytes. FASEB J 20:2093-2101. [DOI] [PubMed] [Google Scholar]
- Arredondo J, Hall LL, Ndoye A, Chernyavsky AI, Jolkovsky DL, Grando SA. (2003). Muscarinic acetylcholine receptors regulating cell cycle progression are expressed in human gingival keratinocytes. J Periodontal Res 38:79-89. [DOI] [PubMed] [Google Scholar]
- Aung G, Niyonsaba F, Ushio H, Kajiwara N, Saito H, Ikeda S, Ogawa H, Okumura K. (2011). Catestatin, a neuroendocrine antimicrobial peptide, induces human mast cell migration, degranulation and production of cytokines and chemokines. Immunology 132:527-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bering B, Moises HW, Muller WE. (1987). Muscarinic cholinergic receptors on intact human lymphocytes. Properties and subclass characterization. Biol Psychiatry 22:1451-1458. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Eichmuller S, Peters EM, Pietsch P, Johansson O, Maurer M, Paus R. (1997). A simple immunofluorescence technique for simultaneous visualization of mast cells and nerve fibers reveals selectivity and hair cycle–dependent changes in mast cell–nerve fiber contacts in murine skin. Arch Dermatol Res 289:292-302. [DOI] [PubMed] [Google Scholar]
- Brown D, Lydon J, McLaughlin M, Stuart-Tilley A, Tyszkowski R, Alper S. (1996). Antigen retrieval in cryostat tissue sections and cultured cells by treatment with sodium dodecyl sulfate (SDS). Histochem Cell Biol 105:261-267. [DOI] [PubMed] [Google Scholar]
- Di Angelantonio S, Matteoni C, Fabbretti E, Nistri A. (2003). Molecular biology and electrophysiology of neuronal nicotinic receptors of rat chromaffin cells. Eur J Neurosci 17:2313-2322. [DOI] [PubMed] [Google Scholar]
- Downs AM, Bond CE, Hoover DB. (2014). Localization of alpha7 nicotinic acetylcholine receptor mRNA and protein within the cholinergic anti-inflammatory pathway. Neuroscience 266:178-185. [DOI] [PubMed] [Google Scholar]
- Gotti C, Clementi F. (2004). Neuronal nicotinic receptors: from structure to pathology. Prog Neurobiol 74:363-396. [DOI] [PubMed] [Google Scholar]
- Grando SA. (1997). Biological functions of keratinocyte cholinergic receptors. J Investig Dermatol Symp Proc 2:41-48. [DOI] [PubMed] [Google Scholar]
- Grando SA, Kist DA, Qi M, Dahl MV. (1993). Human keratinocytes synthesize, secrete, and degrade acetylcholine. J Invest Dermatol 101:32-36. [DOI] [PubMed] [Google Scholar]
- Hendrix S, Picker B, Liezmann C, Peters EM. (2008). Skin and hair follicle innervation in experimental models: a guide for the exact and reproducible evaluation of neuronal plasticity. Exp Dermatol 17:214-227. [DOI] [PubMed] [Google Scholar]
- Herber DL, Severance EG, Cuevas J, Morgan D, Gordon MN. (2004). Biochemical and histochemical evidence of nonspecific binding of alpha7nAChR antibodies to mouse brain tissue. J Histochem Cytochem 52:1367-1376. [DOI] [PubMed] [Google Scholar]
- Hogg RC, Raggenbass M, Bertrand D. (2003). Nicotinic acetylcholine receptors: from structure to brain function. Rev Physiol Biochem Pharmacol 147:1-46. [DOI] [PubMed] [Google Scholar]
- Iyengar B. (1989). Modulation of melanocytic activity by acetylcholine. Acta Anat (Basel) 136:139-141. [DOI] [PubMed] [Google Scholar]
- Jositsch G, Papadakis T, Haberberger RV, Wolff M, Wess J, Kummer W. (2009). Suitability of muscarinic acetylcholine receptor antibodies for immunohistochemistry evaluated on tissue sections of receptor gene-deficient mice. Naunyn Schmiedebergs Arch Pharmacol 379:389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima K, Fujii T, Moriwaki Y, Misawa H. (2012). Critical roles of acetylcholine and the muscarinic and nicotinic acetylcholine receptors in the regulation of immune function. Life Sci 91:1027-1032. [DOI] [PubMed] [Google Scholar]
- Kryl D, Barker PA. (2000). TTIP is a novel protein that interacts with the truncated T1 TrkB neurotrophin receptor. Biochem Biophys Res Commun 279:925-930. [DOI] [PubMed] [Google Scholar]
- Kurzen H, Wessler I, Kirkpatrick CJ, Kawashima K, Grando SA. (2007). The non-neuronal cholinergic system of human skin. Horm Metab Res 39:125-135. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- Lammerding-Koppel M, Noda S, Blum A, Schaumburg-Lever G, Rassner G, Drews U. (1997). Immunohistochemical localization of muscarinic acetylcholine receptors in primary and metastatic malignant melanomas. J Cutan Pathol 24:137-144. [DOI] [PubMed] [Google Scholar]
- Marconi A, Terracina M, Fila C, Franchi J, Bonte F, Romagnoli G, Maurelli R, Failla CM, Dumas M, Pincelli C. (2003). Expression and function of neurotrophins and their receptors in cultured human keratinocytes. J Invest Dermatol 121:1515-1521. [DOI] [PubMed] [Google Scholar]
- Moser N, Mechawar N, Jones I, Gochberg-Sarver A, Orr-Urtreger A, Plomann M, Salas R, Molles B, Marubio L, Roth U, Maskos U, Winzer-Serhan U, Bourgeois JP, Le Sourd AM, De Biasi M, Schroder H, Lindstrom J, Maelicke A, Changeux JP, Wevers A. (2007). Evaluating the suitability of nicotinic acetylcholine receptor antibodies for standard immunodetection procedures. J Neurochem 102:479-492. [DOI] [PubMed] [Google Scholar]
- Ndoye A, Buchli R, Greenberg B, Nguyen VT, Zia S, Rodriguez JG, Webber RJ, Lawry MA, Grando SA. (1998). Identification and mapping of keratinocyte muscarinic acetylcholine receptor subtypes in human epidermis. J Invest Dermatol 111:410-416. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Goldner FM, Saeki M, Lorenzo I, Goldberg L, De Biasi M, Dani JA, Patrick JW, Beaudet AL. (1997). Mice deficient in the alpha7 neuronal nicotinic acetylcholine receptor lack alpha-bungarotoxin binding sites and hippocampal fast nicotinic currents. J Neurosci 17:9165-9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddenberg R, Tiefenbach M, Faulhammer P, Goldenberg A, Gries B, Pfeil U, Lips KS, Piruat JI, Lopez-Barneo J, Schermuly RT, Weissmann N, Kummer W. (2012). Mitochondrial complex II is essential for hypoxia-induced pulmonary vasoconstriction of intra- but not of pre-acinar arteries. Cardiovasc Res 93:702-710. [DOI] [PubMed] [Google Scholar]
- Peters EM, Liezmann C, Spatz K, Daniltchenko M, Joachim R, Gimenez-Rivera A, Hendrix S, Botchkarev VA, Brandner JM, Klapp BF. (2011). Nerve growth factor partially recovers inflamed skin from stress-induced worsening in allergic inflammation. J Invest Dermatol 131:735-743. [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Higley MJ, Mineur YS. (2012). Acetylcholine as a neuromodulator: cholinergic signaling shapes nervous system function and behavior. Neuron 76:116-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradidarcheep W, Stallen J, Labruyere WT, Dabhoiwala NF, Michel MC, Lamers WH. (2009). Lack of specificity of commercially available antisera against muscarinergic and adrenergic receptors. Naunyn Schmiedebergs Arch Pharmacol 379:397-402. [DOI] [PubMed] [Google Scholar]
- Richman DP, Arnason BG. (1979). Nicotinic acetylcholine receptor: evidence for a functionally distinct receptor on human lymphocytes. Proc Natl Acad Sci U S A 76:4632-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosterman D, Goerge T, Schneider SW, Bunnett NW, Steinhoff M. (2006). Neuronal control of skin function: the skin as a neuroimmunoendocrine organ. Physiol Rev 86:1309-1379. [DOI] [PubMed] [Google Scholar]
- Saragoza PA, Modir JG, Goel N, French KL, Li L, Nowak MW, Stitzel JA. (2003). Identification of an alternatively processed nicotinic receptor alpha7 subunit RNA in mouse brain. Brain Res Mol Brain Res 117:15-26. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Nakamura M, Bito T, Fukamachi S, Kabashima R, Sugita K, Hino R, Tokura Y. (2010). Cholinergic urticaria: studies on the muscarinic cholinergic receptor M3 in anhidrotic and hypohidrotic skin. J Invest Dermatol 130:2683-2686. [DOI] [PubMed] [Google Scholar]
- Schmelz M, Michael K, Weidner C, Schmidt R, Torebjork HE, Handwerker HO. (2000). Which nerve fibers mediate the axon reflex flare in human skin? Neuroreport 11:645-648. [DOI] [PubMed] [Google Scholar]
- Tharp MD, Seelig LL, Jr., Tigelaar RE, Bergstresser PR. (1985). Conjugated avidin binds to mast cell granules. J Histochem Cytochem 33:27-32. [DOI] [PubMed] [Google Scholar]
- Wevers A, Monteggia L, Nowacki S, Bloch W, Schutz U, Lindstrom J, Pereira EF, Eisenberg H, Giacobini E, de Vos RA, Steur EN, Maelicke A, Albuquerque EX, Schroder H. (1999). Expression of nicotinic acetylcholine receptor subunits in the cerebral cortex in Alzheimer’s disease: histotopographical correlation with amyloid plaques and hyperphosphorylated-tau protein. Eur J Neurosci 11:2551-2565. [DOI] [PubMed] [Google Scholar]
- Yamada M, Miyakawa T, Duttaroy A, Yamanaka A, Moriguchi T, Makita R, Ogawa M, Chou CJ, Xia B, Crawley JN, Felder CC, Deng CX, Wess J. (2001). Mice lacking the M3 muscarinic acetylcholine receptor are hypophagic and lean. Nature 410:207-212. [DOI] [PubMed] [Google Scholar]