Abstract
Haplotype-resolved genomes equencing enables accurate interpretation of medically relevant genetic variation, deep inferences regarding population history, and the non-invasive prediction of fetal genomes. We describe an approach for genome-wide haplotyping based on contiguity preserving transposition (CPT-Seq) and combinatorial indexing. Tn5 transposition is used to modify DNA with adapter and index sequences while preserving contiguity. After dilution and compartmentalization, the transposase is removed, resolving the DNA into individually indexed libraries. The libraries in each compartment, enriched for neighboring genomic elements, are further indexed via PCR. Combinatorial 96-plex indexing at both the transposition and PCR stage enables the construction of phased synthetic reads from each of the nearly 10,000 “virtual compartments”. We demonstrate feasibility of this method by assembling >95% of heterozygous variants in a human genome into long, accurate haplotype blocks (N50 = 1.4–2.3 Mb). The rapid, scalable, and cost-effective workflow could enable haplotype resolution to become routine in human genome sequencing.
Most genomic studies to date ignore the diploid nature of the human genome1. However, the context in which variation occurs on each individual chromosome can have a significant impact on gene regulation and may have strong clinical significance1,2. Applications that can greatly benefit from phased genomes include medical genetics (e.g. detecting compound heterozygosity; non-invasive fetal genome sequencing3, 4), population genetics5–8, cancer genetics6, and HLA (Human Leukocyte Antigen) typing9. Thus, there is a strong need for cost-effective methods that support accurate and comprehensive haplotype-resolved sequencing of human genomes.
There are two general approaches for genome-wide haplotyping: computational and experimental phasing. Computational approaches in general pool information across multiple individuals, preferentially relatives, by using existing pedigree or population-level data1. Based on the quality and breadth of the reference genomes used, these methods cannot necessarily deliver phasing information across the whole genome. Because the performance of computational phasing is contingent upon multiple parameters including sample size, density of genetic markers, degree of relatedness, sample ethnicity, and allele frequency10, its performance for genome-wide phasing will inevitably be limited11–12. Importantly, rare and de novo variations, which are medically relevant but not observed at appreciable frequencies at the population level, may fail to phase accurately with computational methods. However, complementing the results of an experimental phasing platform with computational approaches, as long as data points can be traced to their experimental or computational origin, has the potential to improve the data, e.g. SNP coverage, and enable new discoveries, e.g. building a fine scale recombination map22.
Most experimental approaches to genome-wide haplotype-resolved sequencing take advantage of the concept of sub-haploid complexity reduction, thereby providing a direct and hypothesis-free approach to phasing13–19. In vitro implementations of complexity reduction separate the parental copies in compartments through sub-haploid dilution, amplify the individual copies using random primer amplification, and then derive haplotypes by inferring and genotyping the haploid molecules present in each compartment13, 15, 18. However, these methods suffer from several limitations. First, random primer amplification-based methods generate false variants through chimeric sequence formation15, can result in a biased representation of the genome with allelic drop-out in the diploid context13, and can yield underrepresentation of GC-rich sequences15. In part as a consequence, very deep sequencing, i.e., 200–500Gb, is required to obtain phasing information with N50 block sizes in the range of 700 kb to 1Mb (N50 is defined as the phased block length such that blocks of equal or longer lengths cover half the bases of the total phased portion of the genome). Second, the requirement of diluting to sub-haploid content and thus starting with minute amounts of DNA may put a burden on reproducibility, accuracy, and uniformity of amplification13, 15. The complexity of this step scales linearly with the number of compartments (usually between 96 and 384), in which each compartment represents an individual library preparation from a picogram-scale starting amount. Cloning-based approaches allow working with reasonable amounts of DNA, but require high-efficiency cloning which is time consuming and technically challenging and are also limited to the size of the cloning platform (fosmids/BACs)14, 17, 20. Finally, for some methods, there is a requirement for upfront size-selection of genomic DNA prior to sub-haploid complexity reduction. Since the reconstruction of long haplotypes is challenging, any limits on the length of input DNA molecules will fundamentally constrain the length of the resulting haplotypes20,21. Alternative approaches for obtaining long-range phasing information include long-read technologies, but these currently suffer from low accuracy and throughput22. Thus, despite advances in phasing methods, there remain major practical obstacles to their integration with routine human genome sequencing10.
We describe a novel approach to haplotype-resolved genome sequencing based on two technological advances: (a) transposition of specific adapters and index sequences into long DNA molecules at high frequency while preserving the contiguity and ordering information of the DNA, and (b) a combinatorial two-level indexing scheme of transposition and PCR, effectively enabling thousands of “virtual compartments”, from which very long, haploid reads can be constructed for phasing. The workflow of indexed transposition, dilution, and indexed PCR amplification contributes to the accuracy and robustness of the method and enables the generation of libraries ready for sequencing in under 3 hours. We present results demonstrating the feasibility of this approach by haplotype sequencing of a mother-father-child trio from the HapMap project18. The simplicity and power of this method for genome-wide haplotype resolution make it an effective complement to shotgun sequencing of individual human genomes.
RESULTS
Contiguity Preserving Transposition sequencing (CPT-seq)
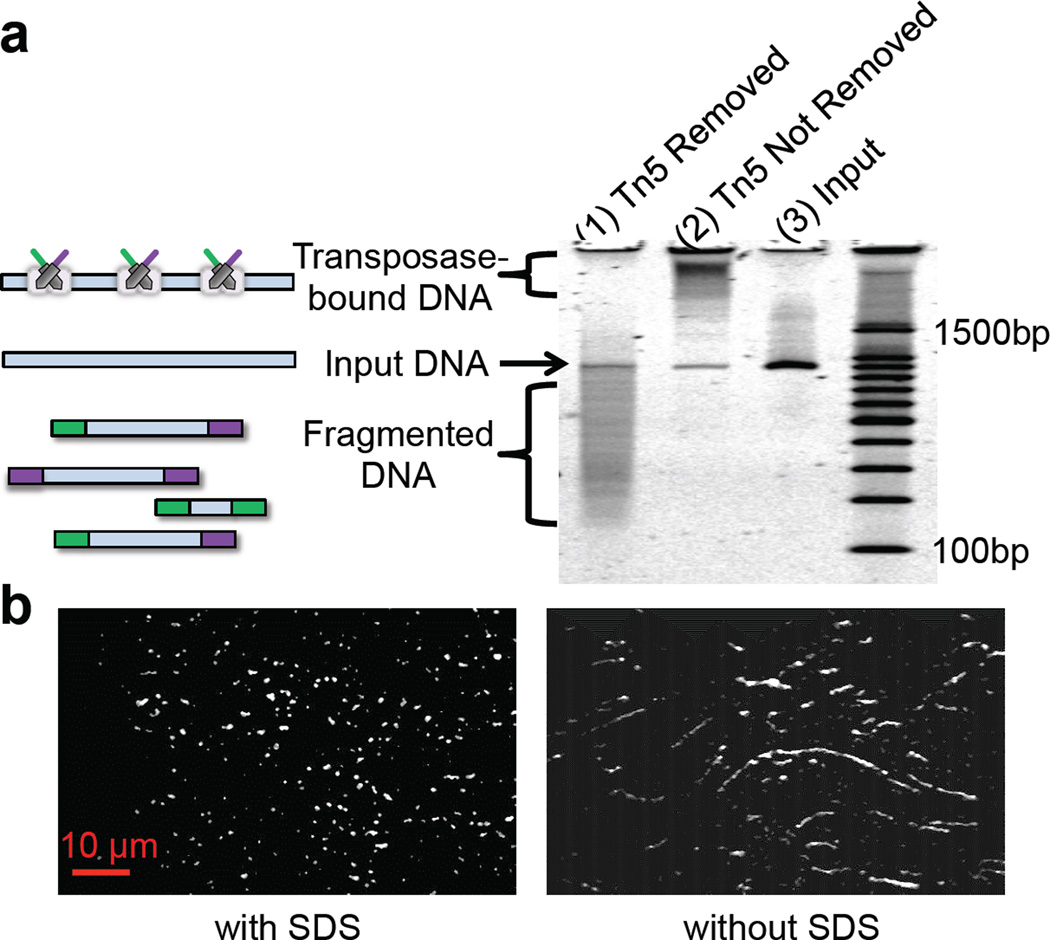
A hyperactive version of the Tn5 transposome was previously described to simultaneously fragment DNA and introduce adaptors at high-frequency (100–300bp intervals) in vitro, creating sequencing libraries for next generation DNA sequencing22, 23. This specific protocol removes any phasing or contiguity information due to the fragmentation of DNA. However, we observed from gel electrophoresis experiments that the Tn5 transposase enzyme stays bound to its DNA substrate post-transposition (Fig. 1a), and the protein-DNA complex only dissociates after removal of transposase by the addition of a protein denaturing agent such as SDS (Sodium Dodecyl Sulfate). This observation was independently validated by single molecule imaging experiments. DNA was labeled with YOYO-1 fluorescent dye and then subjected to Tn5 transposition. As shown in Fig. 1b, each transposed DNA molecule retains its high molecular weight, and is only fragmented when exposed to SDS. In a complementary experiment, transposomes were assembled with Cy5-labeled transposons and applied to high molecular weight(HMW) genomic DNA labeled with YOYO-1 (Supplementary Fig. 1). The “bead-on-a-string” configuration of transposomes on the substrate DNA post-transposition also indicates that target DNA is not fragmented after transposition. Transposed DNA undergoes fragmentation upon treatment with a protease, an alternative to SDS, which digests the transposase. These observations imply that Tn5 can be used for inserting custom-designed oligonucleotide sequences into target DNA while maintaining order and extensive contiguity of the original DNA molecule.
Figure 1.
Tn5 transposase maintains contiguity of target DNA post-transposition.(a) PAGE-analysis of transposase contiguity: Tn5 transposome was used to target a ~1kb PCR amplicon. Transposed DNA was either treated with SDS to remove the transposase enzyme (lane 1), or as a control without SDS treatment (lane 2). Lane 3 is the input DNA and lane 4 is a 100bp reference ladder. As shown here, Tn5 transposase enzyme stays bound to its substrate DNA post-transposition and the protein-DNA complex only dissociates after addition of the protein denaturing agent, i.e., SDS. (b) Single molecule imaging of Tn5transposed DNA: HMW DNA (see Online methods) labeled with YOYO-1 fluorescent dye was subjected to Tn5 transposition. SDS samples were treated with a final 0.05% SDS concentration and incubated at 55°C for 15 min.
In order to test whether we could leverage this unique characteristic of transposition for extracting haplotyping information, we designed a simple transposition and dilution experiment using HMW genomic DNA (see Online Methods). If DNA stays intact upon transposition, we should be able to transpose DNA first, and then dilute it to sub-haploid content. After sub-haploid compartmentalization, the transposase is removed from the target DNA (using SDS) and the library is PCR amplified with indexed primers and then sequenced. If DNA stays intact upon transposition, each index should be enriched for sequence reads that are in close proximity to one another in the genome. In contrast, enrichment of proximal fragments should be lost if the transposase is removed (i.e., SDS added) prior to dilution. Supplementary Fig. 2 shows the distribution of distances between tandem alignments(consecutive aligned reads) observed when SDS treatment was carried-out before (pre-dilution) or after (post-dilution) the dilution step. In both cases, 1.2 pg transposed DNA was used in the PCR step, translating to ~40% haploid content of the human genome. When SDS treatment occurs after dilution, enrichment is observed for reads that map to proximal regions (“islands”) of the genome(represented by the left peak in the bimodal distribution). When SDS treatment occurs prior to dilution, the proximal population is not observed. This demonstrates that genomic DNA largely stays intact during transposition and dilution, and therefore proximity information from each genomic DNA molecule can potentially be extracted.
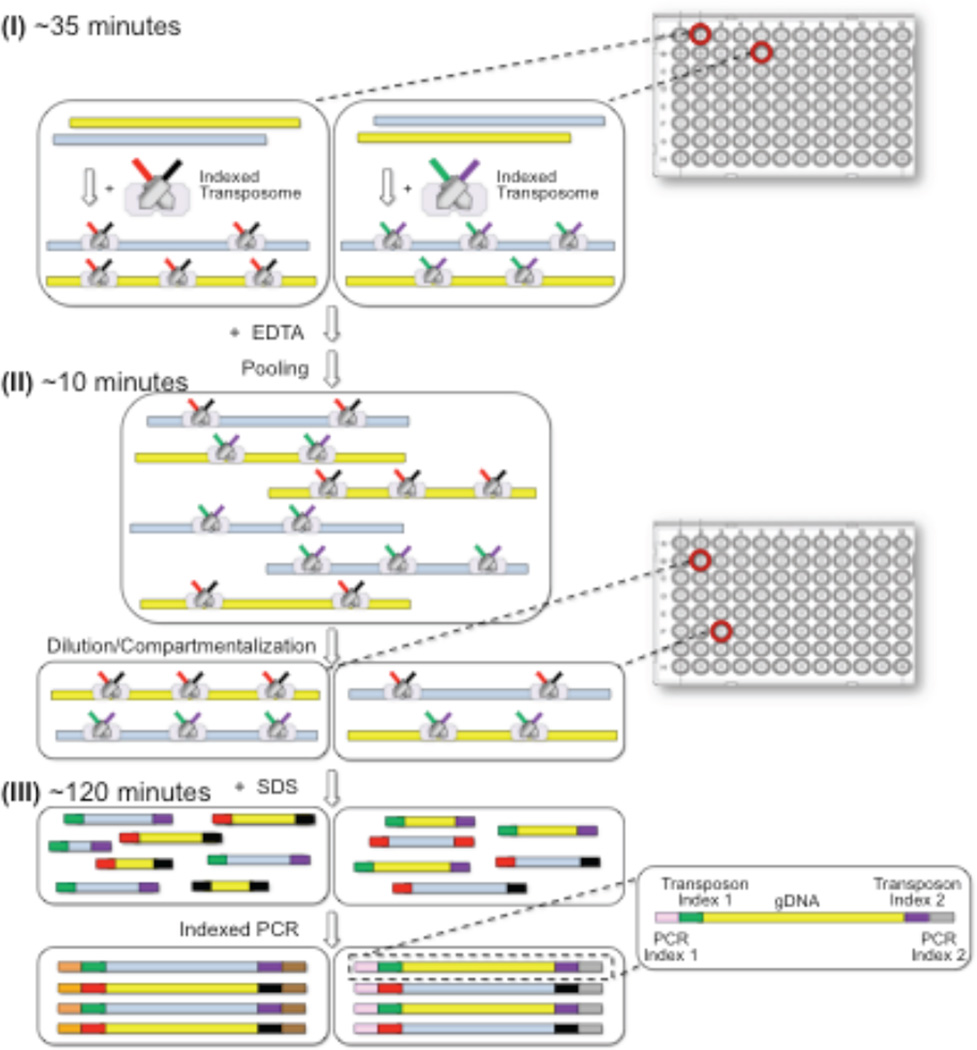
Effective dilution haplotyping requires sub-haploid dilution of the contiguity-preserved genomic libraries into multiple compartments followed by indexed PCR. To minimize the number of physical compartments required and to economize on reagents, we developed the concept of “virtual compartments”, i.e., virtual partitions within each physical compartment, using a combinatorial two-step indexing scheme (Fig. 2). The first index is incorporated into the gDNA library during transposition (defining the virtual partitions), and the second index is incorporated during indexed PCR of the physical compartment. In this manner, a set of virtual partitions, equal to the number of indexed transposition reactions in the first step, can be defined within each physical compartment of the subsequenct dilution step.
Figure 2.
Overview of the CPT-Seq workflow. There are three key steps: (I) indexed transposition, (II) pooling, diluting and compartmentalization, and (III) indexed PCR. A set of 96 different indexed transposome complexes are used to set up 96 independent transposition reactions to create separate genomic virtual partitions (step I). Transposition reactions are pooled together, diluted to sub-haploid DNA content, and split to 96 compartments (step II). Upon removal of the transposase with SDS, compartment-specific libraries are generated using indexed PCR (step III). All samples are pooled together after PCR, and prepared for sequencing.
As a concrete example, a single genomic sample is split in to M=96 independent transposition reactions, each employing unique indexed transposon adapters. These 96 separately indexed/adapterized genomic libraries (contiguity preserved), are then pooled, diluted, and redistributed into a set of N=96 discrete physical compartments (wells on a plate), with each physical compartment now having M=96virtual partitions. A second compartmental index (N=96) is incorporated during PCR. In this manner, this two-tier indexing process effectively creates a total of MxN = 96 × 96 = 9216 “virtual compartments” whose library elements are demultiplexed after sequencing by reading out the unique set of combinatorial indices. In addition to the two-tiers of indexing, indexes at a single tier are created from the combination of “left” and “right” index combinations (Supplementary Fig. 3) [1]. A set of only 8+12 oligonucleotides is used to make the 96 assymettrically indexed transposomes, and additional 8+12 oligonucleotide primers are used to create the 96 assymmetrically compartmental PCR indexes (see Online Methods).
Our implementation of this concept consists of taking a gDNA sample and aliquoting 1 ng into 96 different wells of a microtiter plate, each well containing a Tn5 transposome mix with a unique index (M = 96; “virtual partitions”). After tagmentation, the 96 indexed genomic libraries (contiguity preserved) are pooled, diluted, and redistributed into 96 separate wells of a microtiter plate (N = 96; “physical compartments”). The original pool is diluted such that there is approximately 3% haploid content per virtual partition or ~3 copies of genome per physical compartment (96 virtual partitions per physical compartment). Such low haploid content per virtual compartment is required to avoid collisions between maternal and paternal copies from the same genomic region. Importantly, given the virtual portioning, the amount of DNA per physical compartment is two orders of magnitude higher than other dilution-based haplotyping approaches contributing to the amplification robustness of our method. After physical compartmentalization, the DNA-transpososome complexes are denatured by addition of SDS detergent, and the content of each compartment is amplified with a pair of indexed PCR primers (96 different PCR indexes).
After PCR, combinatorially-indexed libraries from all wells are combined and sequenced using a dual indexing workflow to read all four indexes (two bipartite codes each consisting of left and right indices) and genomic DNA inserts (Supplementary Fig. 3). A standard IVC plot (sequencing Intensity Vs. Cycle number, Supplementary Fig. 4) confirms the four primer sequencing strategy; with sequence reads through genomic DNA (read 1), both bipartite indices (read 2, 3), and genomic DNA (read 4), respectively. In summary, genome-wide haplotype information can be captured with a simple workflow consisting of three steps: (I) parallelized and indexed transposition of HMW genomic DNA, (II) pooling, dilution, and physical compartmentalization of transposed libraries, and (III) parallelized and indexed PCR.
CPT-seq haplotyping results
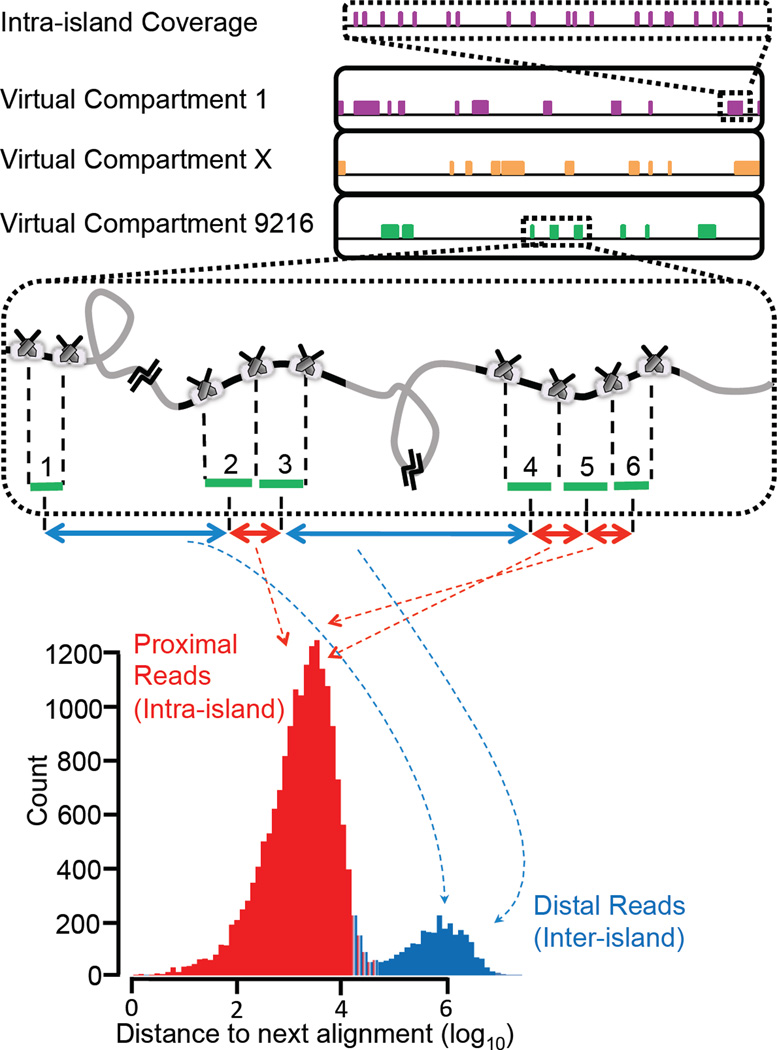
As a proof-of-concept, we applied this strategy to the haplotype-resolved whole genome sequencing of a classic HapMap trio18 (NA12878, NA12891, and NA12892). HMW input DNA for the trio samples was prepared using the Gentra™ Puregen kit (Qiagen) with 100–200 kb expected average size (Supplementary Fig. 5). For the NA12878 sample, we also used a commercially available genomic DNA source with low molecular weight (LMW, i.e., an average DNA size of 50 kb). All DNA samples were processed through the described workflow and sequenced on 4 lanes of a HiSeq 2000, generating between 80–130 Gb of data mapping to the genome (Table 1). Sequence reads were demultiplexed into 9216 distinct partitions based on their unique combinatorial index identity. Data from all partitions were mapped to the reference human genome (hg19). In Fig. 3 and Supplementary Fig. 6, schematic and representative genome coverage plots are shown for the mapped reads from three representative indexes. Clusters or “islands” of reads are observed scattered across the genome suggesting that these “island” originate from a single molecule. Furthermore, a plot of nearest neighbor distances between reads for a given index across a genomic region exhibits a bimodal distribution (Fig. 3, bottom panel and Supplementary Fig. 7). The reads from within an island (proximal reads) form one peak, and reads between islands or sparse singleton reads (distal reads) form the other peak. The coverage within each island (Fig. 3, top panel and Supplementary Fig. 8 for schematic and representative illustration, respectively) exhibits a “strobed” pattern with only 5 to 10% coverage. In spite of this low intra-island coverage per indexed partition, the combined genome-wide coverage from all 9216 partitions is between 97–99% (Table 1). Using HMW DNA, the N50 of informative islands is between 70–90 kb, and switching to LMW DNA decreases the island size significantly to 45 kb. As shown in Table 1, about 50–60 Mb of genome, or 2% haploid equivalents, is observed per virtual compartment.
Table 1.
Haplotyping results for the trio samples. Whole genome phasing data is provided for the samples NA12891, NA12892, and NA12878 prepared using the Gentra Puregene Cell kit (three columns on the right) or acquired from Coriell (for NA12878 only, left column; see Online Methods). The average DNA content of all partitions is shown in Mb and reflects the actual fraction of material added per virtual compartment after accounting for losses. In the ideal case, 3% haploid content (~90 Mb) was added per virtual partition. The analysis is carried out in three main steps: I (ReFHap phasing), II (removing single-linked and conflicting SNPs), and III(adding missing SNPs using 1000 Genomes). The fraction of SNPs phased and also point (PA) and long (LA) accuracies are reported for each step of the analysis. N50 values are reported for both haplotyping islands and haplotyping blocks obtained after phasing individual islands into longer contigs.
| DNA Source | NA12878 | NA12878 | NA12891 | NA12892 |
|---|---|---|---|---|
| DNA Prep | Coriell | Gentra | Gentra | Gentra |
| Read Length | 76 | 51 | 76 | 76 |
| Number of Reads (Millions) | 1749 | 1622 | 1395 | 1113 |
| Number of HiSeq Lanes | 4 | 4 | 4 | 4 |
| Mapped bases (Gb) | 133 | 83 | 106 | 85 |
| DNA /Partition | 21 Mb | 58 Mb | 62 Mb | 49 Mb |
| Informative Island N50 | 45 kb | 76 kb | 86 kb | 73 kb |
| %SNPs Covered | 93.15 | 98.46 | 98.53 | 96.8 |
| Phasing Block N50 (kb) | 490 | 1485 | 2286 | 1387 |
| Long Switches/Mb | 0.14 | 0.18 | 0.06 | 0.16 |
| Step I %Phased | 90.64 | 96.81 | 96.92 | 93.64 |
| Step I Accuracy | LA: 99.97 PA: 99.41 |
LA: 99.96 PA: 99.58 |
LA: 99.99 PA: 99.58 |
LA: 99.97 PA: 99.25 |
| Step II %Phased | 71.71 | 92.84 | 91.96 | 83.76 |
| Step II Accuracy | LA: 99.96 PA: 99.82 |
LA: 99.96 PA: 99.89 |
LA: 99.99 PA: 99.82 |
LA: 99.96 PA: 99.74 |
| Step III %Phased | 94.6 | 98.17 | 98.05 | 96.64 |
| Step III Accuracy | LA: 99.97 PA: 99.83 |
LA: 99.96 PA: 99.88 |
LA: 99.99 PA: 99.82 |
LA: 99.97 PA: 99.75 |
Figure 3.
Demonstration of haplotype read “islands”. Coverage plots are shown for three representative indexes across part of chromosome 22. Reads from the same contiguous molecule display as”islands” of read clusters across the genome (middle panel), or as one mode of a bimodal distribution from a nearest neighbor plot of mapped reads (bottom panel, a representative distance plot from one index). Grey and black regions in the middle panel represent regions of the chromosome that are, respectively, absent or present in a given PCR compartment. Only the black regions, i.e., haplotyping islands, are covered by sequencing reads that carry the index for that given physical compartment. Aligned reads are sorted based on their genomic coordinates and the distance between neighboring alignments from the same partition is recorded. A bimodal distribution is observed with grey regions represented by the distal, i.e., inter-island, subpopulation and the black regions or islands by the proximal, i.e., intra-island subpopulation. Breaks between the islands imply that two neighboring islands do not necessarily belong to the same haplotype. A high ratio of the intra-island to the inter-island peak indicates strong enrichment of the proximal regions of the genome that are in the same haplotype-phase. Representative intra-island coverage is shown in the top panel.
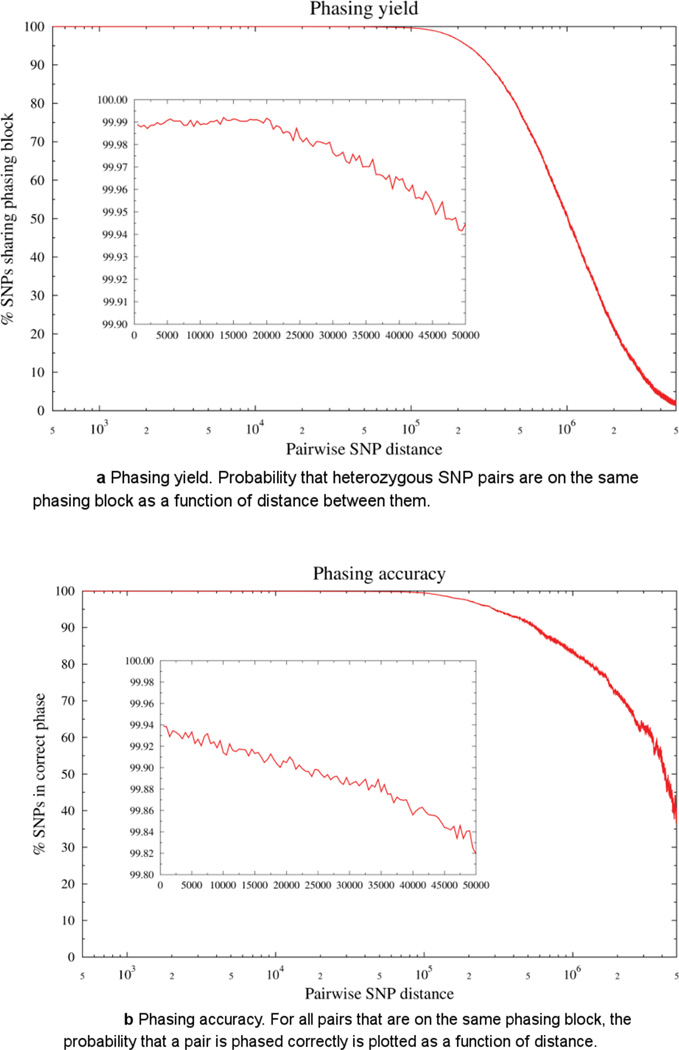
Assignment of SNPs to their respective haplotypes is performed in a multi-step process (see Online Methods and Supplementary Fig. 9)25. As shown in Table 1, 94–97% of SNPs from the HMW DNA samples are phased by ReFHap25. The N50 of assembled haplotyping blocks for HMW DNA samples ranges from 1.4 to 2.3 Mb (Supplementary Fig. 5). We evaluated phasing yield and accuracy of the current method by plotting the probability that heterozygous SNP pairs are on the same phasing block as a function of distance between them (Fig. 4a). For all pairs that are on the same phasing block, the probability that a pair is phased correctly, again as a function of distance is plotted (Fig. 4b). As shown in Figure 4a and 4b, phasing yield (i.e., average genomics distance with >80% SNPs sharing phasing block) is in Mb-scale blocks and minimum accuracy of 99.8% extends to 50 kb pairwise SNP distances. Analogous to other papers34 we have separated switch errors into two categories: long switches and point switches, with the total switch error defined as the sum of short and long switch error. Long switches are defined by large scale transition from one haplotype to another (for example, 00000001111111), whereas point switches are defined by local phasing errors that do not affect adjacent positions (for example, 000000100000). The ReFHap step has a long switch accuracy of 99.95–99.97% and point switch accuracy of 99.3–99.6% on the samples with four lanes of sequencing data. This level of coverage and accuracy can be achieved without any imputation and population-level data.
Figure 4.
a Phasing yield. Probability that heterozygous SNP pairs are on the same phasing block as a function of distance between them.
b Phasing accuracy. For all pairs that are on the same phasing block, the probability that a pair is phased correctly is plotted as a function of distance.
Phasing accuracy can be improved by removing conflicting SNPs and singletons, SNPs that are covered by only one data point (Supplementary Fig. 9 and Table 1). This filter enables creation of a high quality haplotyping backbone at the expected cost of lower coverage. Optionally, a subset of missing SNPs can be imputed using the 1000 Genomes panel26 data (Supplementary Fig. 9). The 1000 Genomes data is only used to perform imputation on SNPs that were not covered (“filling imputation”), but not for connecting haplotyping blocks (“stitching imputation”, as the latter results in much higher long switch error rate (Supplementary Fig. 10). Therefore, the N50 of assembled haplotyping blocks does not change after the imputation step, and there is minimal chance of introducing errors by making chimeric blocks from both parents at this step. As shown in Table 1, the final switch accuracy is very high (with point and long switch accuracies of 99.75% and 99.96%, respectively), with the major error mode being single point switches. Depending on the accuracy and coverage requirements, certain steps of the analysis pipeline can be either included or excluded.
We also analysed genome-wide phasing performance as a function of sequencing depth. Sequencing data was down-sampled and phased as previously described. As shown in Supplementary Table 1, even with as little as 40–60 Gb of sequencing data, an amount substantially less than previously reported studies15, about 95% of SNPs are covered and phased with a long switch accuracy of 99.96% or higher. Even with these very low long switch error rates, it is notable that an appreciable proportion of long haplotype blocks will still contain switch errors.
One key motivation for haplotype-resolved genome sequencing relates to the phasing of de novo mutations and compound heterozygous variations1. Based on our results (Supplementary Table 2), 45 out of the 48 de novo mutations previously described and validated for NA12878 cell line27 were successfully phased here, which further demonstrates the advantage of haplotype resolved whole genome sequencing. Additionally, 33 de novo SNPs observed both with CPT-seq and LFR (long Fragment Read)13, 15 were concordant with LFR data for NA12878 and verified with phasing data from the grandchildren NA12886 and NA12885. For each de-novo SNP, we located the phasing block of the SNP in CPT-seq and compared the phase of the ten neighboring SNPs to the LFR method. We found all of them to be concordant. Furthermore, to demonstrate phasing of compound heterozygous variants, we examined the SNPs in the genes described as putative compound heterozygotes in NA12878 by Kamphanset. al.28. From a total of 10 pairs and one trio (23 total SNPs), 19 of them are phased by CPT-seq, covering 6 pairs and the trio. All phased SNPs shared between both methods are concordant.
DISCUSSION
We present a simple and robust workflow for capturing genome-wide haplotype information in three basic steps: contiguity preserving transposition, dilution, and PCR (Fig. 2) with an overall process time of less than 3 hours. The strength of this platform relies on: (1) the ability to transpose DNA with universal primers and indexes while maintaining contiguity, taking full advantage of DNA quality without enforcing any size selection or physical constraint,(2) universal primer amplification, simplifying down-stream processes and contributing to a more uniform genome-wide representation as compared to random primer-based methods, and (3) combinatorial indexing enabling a parallelized highly-partitioned library construction process that creates thousands of indexed libraries useful in dilution haplotyping.
Another critical aspect of this platform is the scalability of the combinatorial indexing scheme. As we demonstrated here, 96 indexed transposition reactions and 96 indexed PCR reactions generated (using only 40 indexed oligonucleotides) 9216 “virtual” compartments. The combinatorial indexing scheme minimized both the number of physical compartments required and amount of reagents used. The method can easily be adapted to a different number of virtual compartments depending on application and available sequencing throughput. For example, 48×96 and 384×384 versions of our workflow are expected to generate approximately 4,600 and 147,000 compartments, respectively. Although the method is based on dilution, the concept of virtual compartments eliminates the need to dilute DNA to sub-haploid content per physical compartment, thereby minimizing the challenges of low-input amplification. This unique feature allows multiple parental copies of the same genomic region from a given sample to be present in each physical compartment as long as they are from different indexed transposition reactions.
Unlike other dilution haplotyping methods that build up redundancy by whole genome amplification (WGA), our approach transforms a set of sub-haploid genomic molecules directly into a library albeit with low coverage (i.e., strobed reads, Fig. 3, top panel) of any molecule within each partition. This low coverage is a consequence of several losses. First, a 50% loss arises by the fact that the initial transposition uses two different adapters (A and B) and only AB library elements amplify in PCR (50% loss from AA and BB elements)22, 23, 29. Second, non-uniform transposition creates a broad length distribution of library elements in which the shorter elements are preferrentally amplified in PCR. In spite of this shortcoming, the aggregate coverage across the thousands of virtual compartments more than compensates for the low coverage of strobed reads within any given haplotyping island leading to coverages of greater than 95% of all SNPs.
An important feature of CPT-Seq is the ability to create haplotype islands of strobed reads30 across long stretches of DNA, taking full advantage of highly intact starting gDNA. This is evident from the whole genome phasing data that was acquired for the NA12878 sample with high and low quality input DNA (Table 1, compare column 1 and 2). The more intact the DNA, the longer the island N50. As such, a haplotyping assay that uses HMW gDNA can phase more effectively across further distances than an assay using LMW gDNA. Long island N50s are especially important for studying information-poor regions of the genome, regions containing segmental duplications, or long stretches of homozygosity1. As such, a method that utilizes larger DNA fragments, ideally as long as intact chromosomes, will have the potential to retain and consequently deliver more genetic information and genomic context than methods that use fragmented DNA. Finally, the current workflow is simple and does not size select DNA making it more adaptable to automation.
For analysis, assignment of SNPs to their respective haplotypes was carried out in three major steps (See Online Methods and Supplementary Fig. 9). First, SNPs from all islands are phased using ReFHap25. Next, conflicting and singleton SNPs are removed to improve accuracy (Supplementary Fig. 9 and Table 1), with the tradeoff of reduced coverage. Finally, some of the missing SNPs can be imputed using 1000 Genomes26 or other population-level data (Supplementary Fig. 9 and Table 1). We note that imputation using 1000 Genomes data26 should be viewed with caution. One main motivation for haplotype-resolved genome sequencing relates to the phasing of rare or de novo variation1, which precisely are the sorts of variants that imputation fails to phase accurately unless trio data is available for the family. Based on our results (Supplementary Table 2), 45 out of 48 total de novo mutations previously described and validated for NA12878 cell line were successfully phased here, which is a clear advantage of haplotype-resolved whole genome sequencing. In our analysis pipeline, imputation is only used as an optional step to fill the gaps for SNPs that are missing from the experimental data, but not for connecting haplotyping blocks (Supplementary Fig. 10). This analysis approach avoids introducing long switch errors and ensures high quality phasing across the genome.
Whole genome phasing requires more sequencing in addition to the 30X coverage (i.e., ~100 Gb for human genome) which is routinely used for robust variant calling. The percentage of SNPs phased and the accuracy of phasing for our platform, which is a function of sequencing depth, is plotted in Supplementary Fig. 11. As shown in the figure, higher phasing output and accuracy require a higher sequencing depth (and consequently cost). Imputation can be leveraged to increase genome-wide coverage of phased SNPs at a lower sequencing depth, but cannot substitute for experimental phasing, particularly in clinical settings where accuracy is essential and where potentially compound heterozygous variants are usually rare and therefore poorly recovered by imputation. However, with lower sequencing cost over time, and with availability of sample preparation schemes like CPT-seq, which minimize the labor cost and turnaround time of the sample preparation, the monetary burden will be minimized to a level that, in the future, every genome can be individually phased for a nominal price.
In brief, we have introduced a simple, fast, and reliable approach for genome-wide, imputation-free haplotyping, based on sequencing of nearly 10,000 “virtual partitions” generated by contiguity preserving transposition and combinatorial indexing. With as little as 40–60 Gb of sequencing data, a small fraction compared to previously reported studies15, ~94–96% of SNPs in individual human genomes can be phased (Supplementary Table 3). Haplotyping blocks are in the megabase range with long switch error rate on the order of 1–2 per 10Mb, assuringaccurate phasing across the genome. The method uses readily available equipment, requires minimal hands-on time, and can potentially be integrated with microfluidic, droplet, and flow cell platforms31, 32. More generally, its scalability, speed, and cost-effectiveness may allow it to be adopted broadly, such that haplotype information can become a routine component of human genome sequencing in both research and clinical settings.
METHODS
Methods and any associated references are available in the online version of the paper.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper
ONLINE METHODS
Input DNA quality assessment
Genomic DNA was obtained from a commercial vendor (Coriell) as well as using the Gentra™ Puregene kit (Qiagen) on cultured lymphoblastoid cells. Size ranges were assessed via pulsed field gel electrophoresis on a BioRad Pulsed Field Gel Electrophoresis system using a 1% agarose TBE gel run at 120 V for 16 hours at 14°C with a switch time ranging from 1 to 6 seconds.
Single molecule imaging
Imaging was performed using an Olympus 100X, 1.49 NA oil immersion lens, on an Olympus CellTIRF microscope set-up outfitted with an AndoriXon EMCCD camera. Fiber coupled 488 nm and 640 nm Coherent Cube diode lasers were used to illuminate the YOYO-1 and Cy5 dyes, respectively. Multiline dichroic and emission filters (Semrock Di01-R405/488/561/635 and Semrock FF01-446/523/600/677) were used to image both dyes on the same filter cube. The lasers and the camera exposure were controlled by a custom Lab View code, such that 40 ms exposure images were collected from each laser consecutively. Data from 30 frames for each channel were averaged to get high quality images. Using the TIRF mode enabled efficient background subtraction. False color overlay of data from the YOYO and Cy5 channels are shown in Supplementary Figure 1.
10 ng of NA12878 human genomic DNA (Gentra™ protocol) was incubated with 2.5 pmoles of Cy5 labeled Tn5 transposome in the presence or absence of Mg2+ in a 20 µl reaction. After 5 min of incubation at 55°C, 0.5 µl of a 1:100 dilution of YOYO-1 DNA dye was added to the reaction. For the protease treated samples, 1 µl of protease (QIAGEN, Cat. No. 19157) was added and samples were incubated at 37°C for 20 min. Next, 2 µl of each protease treated sample was deposited onto a microscope slide, and a cover slip was applied to spread out the sample. For the other samples, 10 µl of the transposed DNA was added to a cover slip on a 2500 RPM spinning stage to get effective DNA stretching before imaging.
Assembly of 96 indexed transposome complexes
The 20 transposons were formed by annealing each individual indexed oligonucleotide (IDT, standard desalting, Supplementary Table 4), each containing the Tn5 Mosaic End (ME) sequence at their 3’-end, to a universal 5’-phosphorylated, 19 bp ME complimentary oligo in annealing buffer (10mM Tris-HCl, 1 mM EDTA, 25 mM NaCl; pH 8.0). Oligonucleotides were mixed in a 1:1 molar ratio at a final stock concentration of 100 µM and were annealed using the following thermocycling parameters: 95 °C for 5 min. followed by a slow ramp to 25 °C at 0.1 °C/sec. 8 of the oligonucleotides (i5 oligonucleotides) have adapter sequences to make them compatible with the P5 Illumina sequencing end, and the other 12 have the adapter for the P7 side (i7 oligonucleotides). The 20 unique annealed transposons were individually mixed on ice in a 1:1 molar ratio with EZ-Tn5 transposase (Epicentre) at a final stock concentration of 12.5 µM and incubated at 37 °C for 1 hour. Quality of complexes was assessed on an 8% TBE gel (Invitrogen). In order to make 96 unique transposome complexes, the 8 i5 and 12 i7 transposome complexes were aliquoted, with the same 1:1 ratio, into columns 1–12 and rows A-H, respectively, of a 96-well PCR plate and stored at −20 °C. A working stock of transposome complexes diluted to 2.5 µM was subsequently made and stored at −20 °C. Therefore, transposon indexing (which introduces a pair of indexes on either side of the genomic insert) is achieved by using only 20, i.e.,8 i5 (i.e. P5-side index) plus 12 i7 (i.e. P7-side index), index-containing oligonucleotides, creating 8×12=96 different index combinations.
Transposition of HMW gDNA
96 transposition reactions were set-up on ice in a low DNA-binding PCR plate (BioRad, Cat. No. HSS-9601). Each reaction mixture contained in a final volume of 20 uL: 1 ng of HMW gDNA, 10 µl of 2X Nextera Tagment DNA (TD) buffer from the Nextera DNA Sample Preparation Kit (Illumina, Cat. No. FC-121-1031), and 8 µl of water. 2.5 picomoles of transposome complex was added to its respective well and mixed gently by pipet. The transposition plate was incubated at 55 °C for 10 min in a thermocycler with a heated lid. The transposition was stopped by adding 20 µl of 40 mM EDTA (pH 8.0) to each reaction and incubated at 37 °C for 15 min. 20 µl from each well was then pooled into a plastic basin and gently rocked for 5 min at 2 rpm to mix well. The 25 pg/µl pool was then diluted to 1 pg/µl in 1X TE buffer in a PCR strip-tube. 10 pg of pooled dilution was added to each well of a low DNA-binding 96-well PCR plate containing 10 µl of 5 µM Nextera i7 primer (Illumina, Cat. No. FC-121-1012) and 200 ng BSA (New England Biolabs). In order to dissociate Tn5 from the transposed DNA, 2 µl of 1% SDS was added to each well, gently mixed by pipetting, and incubated at 55 °C for 15 min in a thermocycler. The i7 primer was used as a non-specific surface blocker, preventing any potential DNA loss post-SDS treatment.
PCR indexing
10 µl of 5 µM Nextera i5 primer (Illumina, Cat. No. FC-121-1012) was added to its respective well post-SDS treatment. 30 µl Nextera PCR Master Mix, (NPM, Illumina, Cat. No. FC-121-1031) and water were aliquoted into each well from a master mix to create a total PCR volume of 100 µl. Similar to transposon-level indexing, PCR-level indexing is also generated with 8 i5 and 12 i7 PCR primers, yielding 96 unique PCR index combinations. The following PCR parameters were used: initial extension at 72 °C for 3 min followed by an initial denaturation at 98 °C for 30 sec, 20 cycles of denaturation at 98 °C for 10 sec, annealing at 63 °C for 30 sec, and extension at 72 °C for 3 min. 96 PCR reactions were pooled and purified with Zymo Clean & Concentrator-500 (Zymo Research, Cat. No. D4031). The following procedure was used for purification: 50 µl from all 96 reactions were pooled into a plastic solution basin and combined with 9.6 ml Zymo binding buffer and added to a 500 µg Zymo-Spin column. Sample was washed two times with Zymo wash buffer and eluted in 2 ml Zymo elution buffer. The quality and insert size of the libraries were assessed using the High Sensitivity BioAnalyzer (Agilent).
Cluster generation and sequencing
Purified libraries were clustered on a PE HiSeq flow cell v3 using the Truseq v3 PE Cluster Kit (PE-401-3001) at a final concentration of 8 pM. The following procedure was used for clustering: libraries were diluted to 800 pM in RSB (Illumina, Cat. No. FC-121-1031), 10 µl of the diluted libraries were denatured with 10 µl of 0.1 N NaOH for 5 min at room temperature, and resuspended with 980 µl of chilled Hybridization buffer (HT1, Illumina). The denatured libraries were then aliquoted into an eight-well strip-tube. A custom Read-1 sequencing primer (IDT, standard desalting, Supplementary Table S4) was used at 0.5 µM final concentration for clustering on an Illumina cBot. The clustered flow cell was subjected to paired-end sequencing (51-bp reads) on an Illumina HiSeq 2000 using a custom sequencing recipe and two custom sequencing primers for index 1 and Read-2 (Table S4).
Data analysis
Source code files used for the analysis are available in Supplemental Materials (File name: CPT.tar.gz). VCF files of the genomes used in this study were obtained from previous whole genome sequencing studies (Eberle et al., unpublished results, manuscript in preparation). Raw sequencing reads were demultiplexed by unique indexes into 9126 separate fastq files. Reads for each partition were then aligned using bwa v 0.6.1 [33] to human genome reference hg19. Aligned reads from each index map to discrete islands along the genome. All cloud, VCF, fastq files for NA12878, NA12891, and NA12892 are located at bioproject PRJNA241346
Island boundaries were determined by finding clusters of reads such that the distance between any two consecutive reads does not exceed 15 kb and there are at least 5 unique read pairs in each cluster. Only islands that contained reads covering at least 2 heterozygous variants were retained. For each of the SNPs covered by an island, we recorded which variant was supported by the reads in the island.
Islands from all 9126 partitions were subsequently combined. We then split the genome into partitions of up to 3 Megabases in size such that no informative islands (i.e. islands covering more than 1 heterozygous SNP) overlapped with the partition boundaries. For each partition, we used ReFHap [25] to phase heterozygous SNPs and produce the initial phasing blocks. Subsequently, we removed phasing information for SNPs that are linked by only one data point because of the high switch error rate per island. We also removed SNPs showing conflicting calls by multiple islands. We then used 1000 Genomes phased panel to phase some of the remaining SNPs as follows: Each phasing block was divided into 100 kb windows and for each window we attempted to find the best matching phased genome from the 1000 Genomes reference panel using SNPs that were previously phased using ReFHap. This process was performed for both haplotypes reported by ReFHap.
For SNPs that are reported in the 1000 Genomes panel and not phased after the previous two steps, we assigned the phase from the 1000 Genomes panel if the haplotype call from 1000 Genomes panel of two best matching haplotypes was different. After 1000 Genomes phasing, we phased the remaining SNPs that were not phased by either ReFHap or 1000 Genomes but are connected to phased blocks of SNPs by a single island. The phasing assignment of the island to one of the haplotypes and the phase of the SNP was selected in a way that is best supported by this island.
Switch error definition and calculation: In order to assess phasing accuracy, “truth set” was built using phasing data obtained from sequencing the NA12878 trio (NA12878, NA12891, and NA12892). Next, for each phased block experimentally inferred phase was compared with the truth set. Each SNP found in both the phased block and the truth set was recoded as ‘0’ if reported phases matched and ‘1’ otherwise to obtain concordance strings (for example: 0000000000100000001110000000000011111111111111111111).
In the ideal case, the concordance string would consist of only ‘0’s or ‘1’s, however, switches may occur due to experimental or algorithmic errors. Switch errors were separated into two categories: long switches and point switches. Long switch is defined as a large scale transition from one haplotype to another (for example 00000001111111), whereas point switch is defined as a local phasing error that does not affect positions following that switch (for example 000000100000).
In order to decompose the concordance string into long and point switch instances, a Hidden Markov Model (HMM) was developed to establish underlying long range phase information according to the following scoring scheme: ‘-1’ for point mutation, and ‘-5’ for transition to another haplotype. The Viterbi algorithm was used to construct optimal underlying phase in the above example:
Actual concordance string: 0000000000100000001110000000000011111111111111111111
Phase computed by HMM: 0000000000000000000000000000000011111111111111111111
In the example above, the algorithm chose “1” and “111” to represent 1 and 3 point mutations, incurring penalties of −1 and −3, instead of calling them long switches and incurring extra penalty of −20. Therefore, the total score includes four point switches and one long switch: −1 + −3 + −5 = −9
The number of long switches is calculated by the number of transitions within the phase string from the HMM output (1 long switch which is underlined in the example above). The phase string from the HMM output was compared with the concordance string in order to compute the number of point switches (4 switches, underlined in the actual concordance string). Long switch error rate is defined as the number of long switches divided by the total number of SNPs shared between phased blocks and the truth set. Point switch error rate is defined similarly. Long and point switch accuracy is defined as 100*(1-error rate).
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Jocelyne Bruand, Fan Zhang, and AmirAli Kia for help with the data analysis. We also are thankful to Igor Goryshin, Nick Caruccio, and Ramesh Vaidyanathan for discussions at different stages of the project. We would also like to thank Steve Norberg, Joe Zhang, Josh Bernd, Tonya McSherry, Tony Le, Phyrup Diep, and Greg Roberts for performing sequencing, help with custom recipes, and supporting data transfer.
Footnotes
A AUTHOR CONTRIBUTIONS
F.J.S., S.A., and K.L.G. conceived the study; F.J.S. oversaw the technology development. S.A. led the assay development, performed the experiments, and analyzed the data. L.C., C.T., N.P., A.A., J.K. and I.G. performed experiments. T.R. and E.K. performed data analysis. D.P. developed the analysis pipeline. K.V. developed the single molecule imaging system and collected images for the single molecule experiments. S.A., L.C., D.P., M.R., K.L.G., J.S. and F.J.S.co-wrote the paper. All authors contributed to the revision and review of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests
Accession codes: bioproject PRJNA241346 (https://submit.ncbi.nlm.nih.gov/subs/bioproject/)
REFERENCES
- 1.Bansal V, et al. The next phase in human genetics. Nat. Biotechnol. 2011;29(1):38–39. doi: 10.1038/nbt.1757. [DOI] [PubMed] [Google Scholar]
- 2.Tewhey R, et al. The importance of phase information for human genomics. Nat Rev Genet. 2011;12(3):215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan HC, et al. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487(7407):320–324. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitzman JO, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4(137) doi: 10.1126/scitranslmed.3004323. 137ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 6.Adey A, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500(7461):207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tishkoff SA, et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271(5254):1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 8.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40(9):1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosomichi K, et al. Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics. 2013;14:355. doi: 10.1186/1471-2164-14-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 2011;12(10):703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal V, et al. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res. 2008;18(8):1336–1346. doi: 10.1101/gr.077065.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He D, et al. Optimal algorithms for haplotype assembly from whole-genome sequence data. Bioinformatics. 2010;26(12):183–190. doi: 10.1093/bioinformatics/btq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaper F, et al. Whole-genome haplotyping by dilution, amplification, and sequencing. Proc. Natl. Acad. Sci. U S A. 2013;110(14):5552–5557. doi: 10.1073/pnas.1218696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat. Biotechnol. 2011;29(1):59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487(7406):190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan HC, et al. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011;29(1):51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duitama J, et al. Fosmid-based whole genome haplotyping of a HapMap trio child: evaluation of Single Individual Haplotyping techniques. Nucleic Acids Res. 2012;40(5):2041–2053. doi: 10.1093/nar/gkr1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suk EK, et al. A comprehensively molecular haplotype-resolved genome of a European individual. Genome Res. 2011;21(10):1672–1685. doi: 10.1101/gr.125047.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo C, et al. On the design of clone-based haplotyping. Genome Biol. 2013;14(9):R100. doi: 10.1186/gb-2013-14-9-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geraci F. A comparison of several algorithms for the single individual SNP haplotyping reconstruction problem. Bioinformatics. 2010;26(18):2217–2225. doi: 10.1093/bioinformatics/btq411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol. Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- 23.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11(12):R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erlich Y, et al. DNA Sudoku--harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 2009;19(7):1243–1253. doi: 10.1101/gr.092957.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duitama J, et al. Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology. Niagara Falls, NY, USA: 2010. ReFHap: a reliable and fast algorithm for single individual haplotyping; pp. 160–169. [Google Scholar]
- 26.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43(7):712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamphans T, et al. Filtering for compound heterozygous sequence variants in non-consanguineous pedigrees. PLoS One. 2013;8(8):e70151. doi: 10.1371/journal.pone.0070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo C, et al. Strobe sequence design for haplotype assembly. BMC Bioinformatics. 2011;12(suppl. 1):S24. doi: 10.1186/1471-2105-12-S1-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu AY, et al. A microfabricated fluorescence-activated cell sorter. Nat. Biotechnol. 1999;17(11):1109–1111. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- 32.Hua Z, et al. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal Chem. 2010;82(6):2310–2316. doi: 10.1021/ac902510u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuleshov V, et al. Whole-genome haplotyping using long reads and statistical methods. Nat. Biotechnol. 2014;32:261–266. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.