Abstract
Background
Many papers have been published in biomedical journals reporting on the development of prognostic and therapy-guiding biomarkers or predictors developed from high-dimensional data generated by omics technologies. Few of these tests have advanced to routine clinical use.
Purpose
We discuss statistical issues in the development and evaluation of prognostic and therapy-guiding biomarkers and omics-based tests.
Methods
Concepts relevant to the development and evaluation of prognostic and therapy-guiding clinical tests are illustrated through discussion and examples. Some differences between statistical approaches for test evaluation and therapy evaluation are explained. The additional complexities introduced in the evaluation of omics-based tests are highlighted.
Results
Distinctions are made between clinical validity of a test and clinical utility. To establish clinical utility for prognostic tests it is explained why absolute risk should be evaluated in addition to relative risk measures. The critical role of an appropriate control group is emphasized for evaluation of therapy-guiding tests. Common pitfalls in the development and evaluation of tests generated from high-dimensional omics data such as model overfitting and inappropriate methods for test performance evaluation are explained, and proper approaches are suggested.
Limitations
The cited references do not comprise an exhaustive list of useful references on this topic, and a systematic review of the literature was not performed. Instead, a few key points were highlighted and illustrated with examples drawn from the oncology literature.
Conclusions
Approaches for the development and statistical evaluation of clinical tests useful for predicting prognosis and selecting therapy differ from standard approaches for therapy evaluation. Proper evaluation requires an understanding of the clinical setting and what information is likely to influence clinical decisions. Specialized expertise relevant to building mathematical predictor models from high-dimensional data is helpful to avoid common pitfalls in the development and evaluation of omics-based tests.
Keywords: biomarker, omics, clinical test, diagnostic test, therapy selection
Introduction
High-throughput omics technologies have generated much excitement for their potential to provide detailed biological characterizations of disease that can be used to optimize care for patients. Omics is defined as “study of related sets of biological molecules in a comprehensive fashion. Examples of omics disciplines include genomics, transcriptomics, proteomics, metabolomics, and epigenomics” [1]. When there is interest to develop a clinical test based on omics assays to aid in decision making for patient care, omics variables are typically combined by means of a computational model that produces a result that can direct clinical actions. We will use the term “omics predictor” to refer to this computational model, which usually takes the form of a risk score or classifier for predicting clinical outcome or benefit from a particular therapy or class of therapies. The omics assay in combination with a fully specified computational model (omics predictor) is referred to as an “omics test” [1], although we sometimes use the terms omics predictor and omics test interchangeably in the discussions in this paper.
Many papers published in biomedical journals have reported the development of omics predictors for clinical outcomes, but very few of these predictors have advanced to the point of incorporation into omics tests that are ready for clinical use. An important reason for the lack of advancement of omics predictors into clinically useful omics tests has been poor integration of statistical, computational, and laboratory expertise with clinical expertise. Our goal in this paper is to highlight issues to consider in the development of omics predictors and to provide guidance on how to properly evaluate published omics predictors for their potential as useful clinical tests.
The two types of omics tests we focus on here are tests that predict prognosis and tests that provide information useful for selecting therapy [2]. A prognostic test produces a measurement that is associated with clinical outcome in the absence of therapy (natural disease course), or sometimes the definition is broadened to mean a measurement that is associated with outcome in the context of a standard therapy that all patients are likely to receive. A therapy-guiding test produces a measurement that can identify subgroups of patients that differ in the benefit they receive from a particular therapy. For example if the test produces a binary result it could indicate that patients who have a negative test result do not benefit from a new therapy relative to some standard therapy (including possibly no therapy) while patients with a positive test result do receive clinically meaningful benefit from the new therapy. Clinical benefit might include outcomes such as shorter duration of disease symptoms or longer survival. These same principles apply even to tests based on a single biomarker. Alternative terms for biomarkers that are useful for guiding therapy include treatment-selection markers, treatment-stratification markers, treatment effect modifiers, or predictive markers (use of the term “predictive” is somewhat idiosyncratic to the medical subspecialty of oncology).
Our discussion of statistical issues relevant to omics test development begins with basic principles for evaluation of prognostic and therapy-guiding tests (applicable even to tests based on a single biomarker). We expand from that point to special additional considerations for omics tests related to the high-dimensional nature of omics data, where there are usually far more measured variables per patient than the total number of patients in the study. This leads to additional statistical challenges in the development of predictor models from high-dimensional omics data, as will be discussed.
A framework for evaluation of biomarker-based clinical tests
Several authors have discussed a framework for the evaluation of clinical tests [3–5] and pointed to the need to consider three important aspects of clinical tests: analytical validity, clinical validity, and clinical utility. Here we follow the definitions provided by McShane and Hayes [5]. Analytical validity refers to “analytical accuracy, reliability, and reproducibility of the test.” Clinical validity is the “demonstration that the test has a suitably strong association with a clinical outcome of interest.” Clinical utility implies that “use of the test to direct patient care has been shown to result in a favorable balance of benefits to harm leading to improved outcomes, compared to non-use” of the test. “Improvement in outcome may relate to overall survival, disease-free survival, quality of life, or cost of care.” All of these aspects must be considered in the statistical and clinical evaluations conducted during development of an omics predictor into an omics-based clinical test. For discussion of analytical validity, readers are referred to the paper by Pennello in this journal issue [6]. The discussions here will focus on clinical validity and clinical utility and the examples discussed are drawn from oncology.
Lack of statistical rigor in studies of prognostic and therapy-guiding omics tests
Statistically rigorous design and analysis methods have long been expected for clinical studies developing and evaluating new therapies, but many biomarker-based tests are not developed in a similarly rigorous environment. For omics tests the situation is perhaps even worse than for tests based on single biomarkers due to the need for new approaches for analysis of very high-dimensional data and the involvement of individuals with more varied types of scientific expertise, including laboratory, computational and bioinformatics expertise, where principles of clinical biostatistics are less familiar. Shortcomings in statistical and clinical rigor are frequently observed in published papers reporting development or evaluation of omics tests [7, 8]. Studies with poor designs (or perhaps more appropriately referred to as studies without a design) are common in the development and evaluation of both prognostic and predictive tests. Many studies aiming to evaluate a prognostic test fail to distinguish between a statistically significant association of an omics test result with a clinical outcome and a clinically useful association, and they fail to demonstrate that a test contributes clinically useful information after adjustment for standard clinical or pathological factors. For therapy-guiding tests, studies sometimes fail to include an appropriate control group, and this can lead to confusion between prognostic value of a test and its value for guiding therapy. These issues apply to tests based on single biomarkers as well as tests derived from high-dimensional omics data, but the latter present even more challenges. Specifically, flawed evaluations of predictor performance are frequently observed in studies that develop tests based on high-dimensional omics data due to the hazards of naively developing and evaluating models derived from large numbers of predictor variables. Each of these issues is now discussed in more depth with examples provided from oncology to reinforce the concepts.
General study design principles
In oncology, the vast majority of omics predictors have been developed retrospectively using tumor specimens that had been collected previously and stored. A major advantage of a retrospective approach is that follow-up for clinical outcome is already available for immediate use. Unfortunately, many omics predictor development studies have not been conducted under ideal conditions. Rather, the majority of omics predictors have been developed on specimen collections that are haphazardly assembled (i.e., “convenience samples”) and are often comprised of tumor specimens from heterogeneous groups of patients with many different clinical presentations (e.g., disease stages or subtypes) who received a variety of different treatments. Treatments received may be unknown and there may be confounding between treatment selection and clinical characteristics of the patient. The cases for which specimens are available might also represent a biased collection of cases, for example because there were certain requirements for the quantity and quality of specimens to ensure that they were amenable to omics assay. Studies based on convenience specimen sets represent particularly treacherous terrain for reliable predictor development, but chances for development of a useful predictor from a retrospective specimen collection can be increased if great care is taken to avoid excessive heterogeneity and bias.
After initial development of a predictor, it is important to validate its performance on additional data sets. The best type of retrospective study design to use for validation is one conducted using specimens that were collected from completed well-designed prospective cohort studies or randomized clinical trials. These types of specimen collection are preferable for predictor development as well, if multiple such collections are available to allow for both development and validation. The advantage of using specimen collections from clinical trials and prospective cohort studies are that typically patient inclusion and exclusion criteria were carefully specified, specimens were acquired and processed in a reasonably standardized way, and patients for whom specimens are available are representative of the full patient cohort. Prior to performing assays on the specimens for purposes for validating a predictor, there should be a rigorous study design in place that includes complete specification of the predictor and a statistical analysis plan that is prospectively defined and followed to assess the predictor performance. Under these conditions, a high level of evidence for clinical utility of a predictor can be obtained [4].
Additional principles specific to the development and evaluation of prognostic or predictive tests also apply. These additional principles are now discussed in detail, separately, for prognostic and therapy-guiding tests. Multiple examples from the published literature are cited to illustrate the points.
Principles for evaluation of prognostic tests
Statistical significance versus clinical utility for a prognostic test
To establish clinical validity of a claim that an omics predictor is prognostic requires one to show that the predictor results have a statistically significant association with a specified clinical outcome. In oncology, the usual endpoints of interest are disease recurrence, progression, survival, or composites of these endpoints. One might establish clinical validity, for example, by showing that a log-rank test comparing the clinical outcome of patients in the omics predictor-defined “favorable ” and “unfavorable” groups are statistically significantly different, but this would not necessarily establish clinical utility. To establish clinical utility, it would be necessary to show that the information provided by the test was not available from other standardly measured factors, that the additional information contributed could be acted upon clinically, and by so doing, the patient would be likely to have a better outcome as a result of the test-directed clinical management plan.
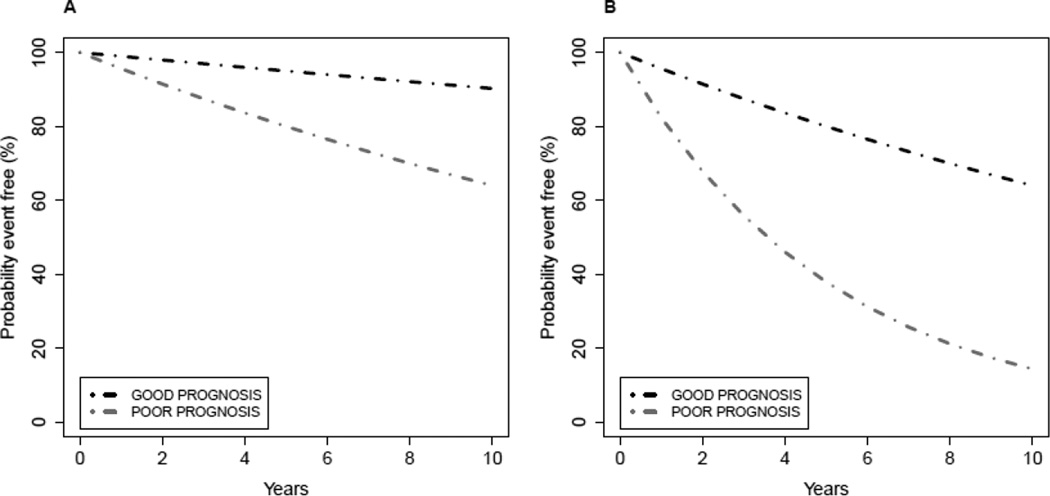
Figure 1 displays two example scenarios in which an omics test has prognostic value, but the potential for clinical utility for the omics test in represented in Figure 1A is stronger than for the test in Figure 1B. Suppose that the survival curves represent time from surgery for primary breast cancer to distant recurrence of breast cancer when the patients do not receive additional therapy following surgery. An important decision that early stage breast cancer patients and their physicians face following surgery is whether or not the patient should undergo systemic chemotherapy treatment (called “adjuvant” chemotherapy in this setting). In Figure 1A, the outcome for patients predicted to be in the favorable subgroup is so good that it is unlikely that many of those patients would want to undergo chemotherapy treatment with its associated toxicities, inconvenience, and costs given the very small potential benefits in terms of freedom from distant recurrence. Thus, assuming that the omics test provides information not readily or reliably available from other standard clinical or pathological factors, the omics test could provide information useful in clinical decision making (spare some patients chemotherapy), and those patients would likely have a more favorable benefit to risk ratio by foregoing chemotherapy. An example of this type of prognostic test is the Oncotype DX recurrence score [9], which has been established to have the ability to identify a group of node-negative hormone receptor-positive breast cancers that have such low risk of distant recurrence when treated with endocrine therapy alone that systemic chemotherapy would not usually be recommended. In contrast, the prognostic value of the test illustrated in Figure 1B may have limited clinical utility. With sufficient numbers of events (distant recurrences), statistical significance could be established for the difference in survival represented by the two curves in Figure 1B, but unless different clinical management strategies would be employed for the patients in those two subgroups (both have predicted poor outcome and would likely receive chemotherapy) and those differing strategies would benefit the patients, the omics test with characteristics as represented in Figure 1B does not have clinical utility.
Figure 1. Evaluation of a prognostic test.
Patients are uniformly treated with a standard therapy, and the test is performed on all patients. GOOD denotes a test result intended to indicate favorable prognosis, and POOR denotes a test result intended to indicate unfavorable prognosis.
A) The survival curves in plot A show a situation where event-free survival for patients in the GOOD prognosis group might be considered sufficiently favorable that no additional treatments would be recommended for that group.
B) The survival curves in plot B show a situation where event-free survival in both the GOOD and POOR prognosis groups might be considered sufficiently unfavorable that additional treatments would be recommended for both groups. In this setting the prognostic test results would not influence therapy decisions for the patients in either group.
Approaches for evaluation of clinical utility of a prognostic test compared to approaches for evaluation of a new therapeutic
The examples in Figure 1 suggest some important differences in the statistical approaches to evaluation of the clinical utility of a prognostic test compared to evaluation of the efficacy of a new therapy compared to a standard therapy (including possibly no therapy). To establish superiority of a new therapy in a randomized clinical trial one would power the trial to detect some prespecified magnitude of clinically meaningful improvement in outcome for the patients who receive the new therapy. In the prognostic marker setting in Figure 1A, it would be necessary to establish not only a difference in outcome between the two predictor-defined subgroups, but it would also have to be established that the survival probabilities in the predictor-defined favorable subgroup were suitably high, for example by showing that a lower confidence bound on survival probability exceeded some threshold value, so that the clinical management would be different (e.g., no or reduced chemotherapy recommended). (The test-directed change in clinical management would also have to be shown to provide clinical benefit for the patient.) Alternatively, one could show that a prognostic test was able to identify a subgroup within a larger group of risk patients normally considered to have very favorable (low) risk who do unusually poorly under standard care (e.g., no chemotherapy following surgery) and who would benefit from more aggressive therapy. In the context of lung cancer, Subramanian and Simon discuss two example scenarios in which prognostic tests could be useful [8]: 1) identify high-risk completely resected stage I patients who might benefit from chemotherapy following surgery (stage IA patients usually do not receive chemotherapy following surgery and use of chemotherapy in stage IB varies), or 2) identify stage II patients who have a low risk of recurrence in the absence of chemotherapy (usually chemotherapy is recommended for stage II patients).
Evaluation of a prognostic test in the context of standard clinical and pathological factors
Another difference between prognostic test evaluation and therapy evaluation is that in conventional randomized treatment trials, one can rely on randomization to approximately balance other standard prognostic factors between the two treatment arms (potentially enhanced in small trials by stratifying randomization on key prognostic variables), and usually studies are powered to globally assess the treatment benefit in the randomized eligible patients. In contrast, to establish the highest level of clinical utility for a prognostic test it should be demonstrated that the test provides clinically significant information beyond that provided by standard prognostic factors. It is a much more data intensive task to show value added beyond standard prognostic variables or to show prognostic value added in each of several finer subgroups defined by one or more standard prognostic variables. Further, the possibility exists that even if prognostic separation of predictor-defined subgroups is established in each of one or more subgroups, within certain subgroups the patients predicted to have favorable (unfavorable) outcome may not have sufficiently good (poor) outcome that a change in clinical management strategy would be considered.
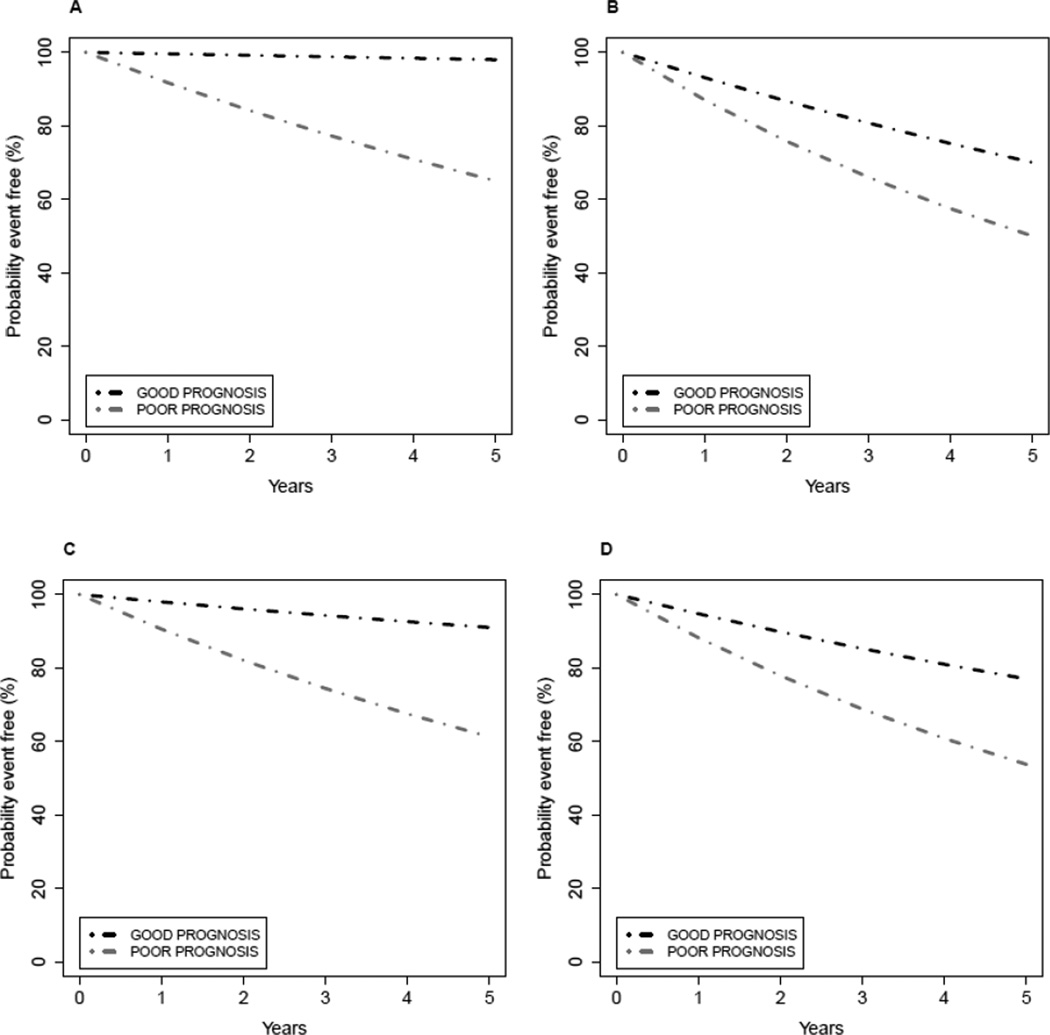
The paper by Hatzis and colleagues [10] illustrates the complexities of interpreting the value added by an omics predictor in the presence of heterogeneity in important standard factors. As seen in Figure 2 (middle) of that paper, the omics predictor described there separates patients (all of whom received a standard chemotherapy regimen plus endocrine therapy) into two groups designated as treatment sensitive and treatment insensitive, with the sensitive group having 3-year distant relapse-free survival = 92% and the insensitive group having 3-year distant relapse-free survival = 75%. (Although the names of the predictor-defined subgroups might seem to imply the predictor has value for selecting therapy, the design of the Hatzis study precludes one from drawing such a conclusion, a concept that will be further discussed under the topic of therapy-guiding tests.) Separating the patients into subgroups defined by estrogen receptor (ER) status (known to be a weak prognostic marker but a strong marker for guiding use of endocrine therapy in breast cancer) and examining the predictor performance in each of the ER subgroups separately suggests that the clinical utility of the prognostic information provided by the omics predictor may differ between the ER-defined subgroups (see Figure 3 in Hatzis et al [10]). The predictor-defined sensitive group in the ER-positive subgroup has 3-year distant relapse-free survival = 97%, whereas the sensitive group within the ER-negative subgroup has 3-year distant relapse-free survival = 83%. If these estimates represent the true underlying survival distributions, one would be unlikely to consider additional therapy for the predictor-defined sensitive ER-positive patients while alternative therapy (in addition to, or in place of standard therapy) might still be considered for the ER-negative patients called sensitive by the predictor. It is also possible that the ER-positive patients called sensitive by the predictor are even being overtreated and would do just as well with less therapy (e.g., endocrine therapy alone), although this cannot be established either way from the evidence supplied by the Hatzis study [10].
Figure 2. Effect of patient heterogeneity on assessment of clinical utility of a prognostic test.
Patients are uniformly treated with a standard therapy. GOOD denotes a test result intended to indicate favorable prognosis, and POOR denotes a test result intended to indicate unfavorable prognosis. Patients can also be segregated into two groups on the basis of other standard clinical and pathological factors, and these are designated as group1 and group 2.
A) Plot A shows the event-free survival curves for the subgroups identified by the prognostic test within patient group 1.
B) Plot B shows the event-free survival curves for the subgroups identified by the prognostic test within patient group 2.
C) Plot C shows the event-free survival curves for the subgroups identified by the prognostic test applied to groups 1 and 2 combined in proportion 75% (group 1) and 25% (group 2). For the setting depicted here, prognosis in the subgroup predicted to have GOOD prognosis by the test might be considered sufficiently favorable that no additional treatments would be recommended for that subgroup, whereas additional treatment might be recommended for the POOR subgroup.
D) Plot D shows the event-free survival curves for the subgroups identified by the prognostic test applied to groups 1 and 2 combined in proportion 25% (group 1) and 75% (group 2). For the setting depicted here, prognosis in both subgroups identified by the test might be considered sufficiently unfavorable that additional treatments would be recommended for both, and the prognostic test results would not influence therapy decisions for any patients.
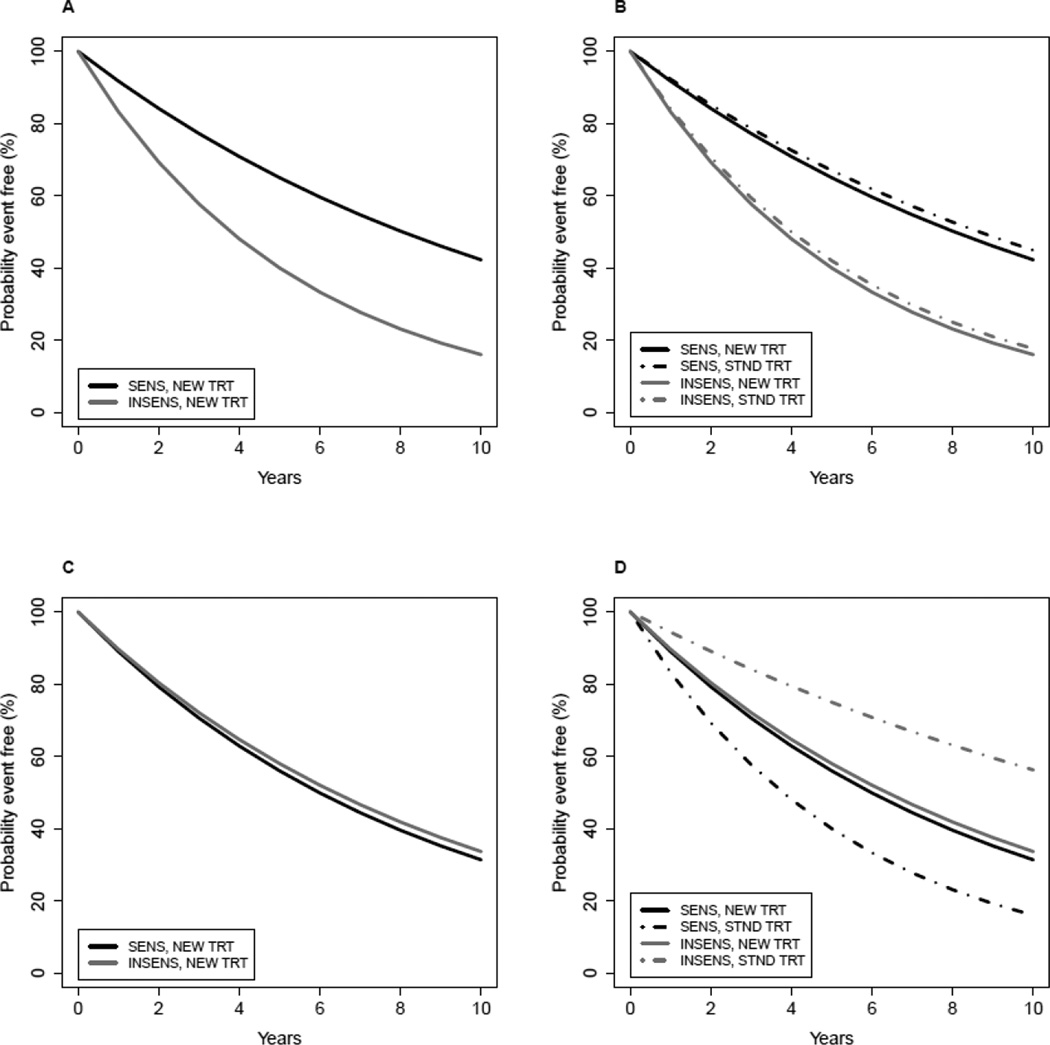
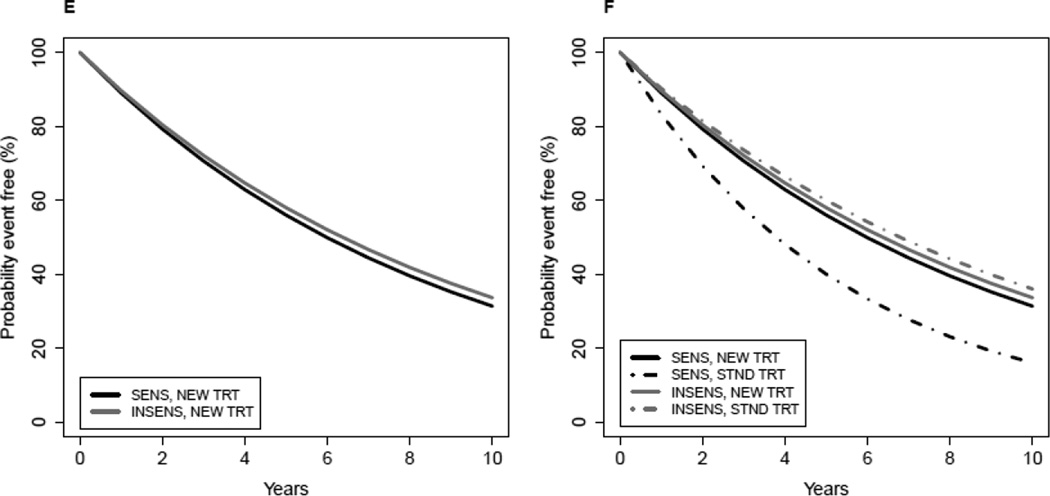
Figure 3. Evaluation of a therapy-guiding test.
A test is performed on all patients, and the patients are randomly assigned to treatment with the standard therapy (STND TRT) or the new therapy (NEW TRT). SENS denotes a test result that is intended to predict benefit (sensitivity) from NEW TRT relative to STND TRT. INSENS denotes a test result intended to predict lack of benefit (insensitivity) from NEW TRT compared to STND TRT.
A–B) The event-free survival curves in plots A–B show a situation in which the test is not useful for guiding therapy because within each category of test result the NEW and STND treatments result in the same event-free survival. If only plot A was examined, one might mistakenly conclude that the test identifies which patients benefit from NEW TRT.
C–D) The event-free survival curves in plots C–D show a situation in which the test is useful for guiding therapy because patients predicted by the test to be SENS have a better outcome when they receive NEW TRT compared to STND, whereas patients predicted by the test to be INSENS have a better outcome when they receive STND TRT compared to NEW.
E–F) The event-free survival curves in plots E-F show a situation in which the usefulness of the test for guiding therapy is not clear-cut. Patients predicted by the test to be SENS have a better outcome when they receive NEW TRT compared to STND; whereas, patients predicted by the test to be INSENS have the same event-free survival outcome regardless of treatment. (It is assumed the STND TRT has already been shown to offer benefit over no therapy.) In this setting, the utility of the omics test depends on whether one would prefer to give the NEW or STND TRT to patients whose test results are INSENS, and this may depend on other factors such as cost, convenience, and toxicity of NEW TRT compared to STND. If NEW therapy is preferred for all patients, then the omics test is not useful for therapy selection in this setting.
Figure 2 presents a hypothetical example to illustrate how mixing together, in different ratios, patient subgroups with distinct baseline prognostic characteristics can confuse the evaluation of clinical utility of a prognostic test. Patient groups 1 and 2 represented in Figures 2A–B have different baseline prognostic characteristics, although within each group the predictor is able to separate patients into subgroups with different prognosis. When combined in proportion 75% (group 1) and 25% (group 2) as in Figure 2C, one might conclude that survival for patients in the GOOD prognosis subgroup is sufficiently favorable that no additional treatments would be recommended for that subgroup, whereas additional treatment might be recommended for the POOR subgroup. When combined in proportion 25% (group 1) and 75% (group 2) as in Figure 2D, one might conclude that survival in both the GOOD and POOR prognosis subgroups is sufficiently unfavorable that additional treatments would be recommended for both subgroups. The test represented in Figure 2C has potential clinical utility, but the test in Figure 2D might not viewed as having clinical utility.
Heterogeneity in standard prognostic factors has implications for comparing predictor performance across different patient cohorts from different studies as well. The lung cancer predictor studied by Kratz and colleagues [11], for example, exhibited substantially different absolute levels of risk within each of the predictor-defined risk groups across the two different validation patient cohorts studied. One validation cohort was comprised of 433 patients with stage I non-squamous non-small-cell lung cancer who had surgery at hospitals in the Kaiser Permanente Northern California system. The second validation cohort was comprised of 1006 Chinese patients who had undergone surgery for early-stage non-small-cell lung cancer at one of several institutions participating in the China Clinical Trials Consortium. Among these 1006 Chinese patients, there were 471 patients who had stage I disease. For the Kaiser cohort (all stage I), the predictor divided patients into three risk groups with 5 year overall survival of 71.4% (95% CI 60.5–80.0) in the low-risk group, 58.3% (95% CI 48.9–66.6) in the intermediate-risk group, and 49.2% (42.2–55.8) in the high-risk group. For the Chinese cohort restricted to stage I, 5 year overall survival estimates in the three risk groups were 83.0% (95% CI 73.8–89.1) in the low-risk group, 67.7% (95% CI 54.8–77.7) in the intermediate-risk group, and 64.6% (57.9–70.5) in the high-risk group. The two cohorts overall (stage I only in the Kaiser cohort and stages I-III in the Chinese cohort) differed on the distribution of sex (55% female in Kaiser versus 38% female in Chinese), smoking history (85% positive history in Kaiser versus 49% positive in Chinese), and obviously with regard to ethnicity. (Covariate distributions were not reported separately for the stage I subgroup of the Chinese cohort.) The absolute levels of risk within each predictor-defined risk group differed between the two cohorts, although hazard ratios estimated from multivariable analyses (adjusting for slightly different standard prognostic factors) conducted on each overall cohort (n=433 for Kaiser and n=1006 for Chinese) were similar (hazard ratios relative to low risk group were 2.04 (95% CI 1.28–3.26) high risk and 1.66 (95% CI 1.00–2.74) intermediate risk for Kaiser; 2.37 (95% CI 1.63–3.43) high risk and 1.60 (95% CI 1.03–2.49) intermediate for Chinese). In the United States, patients with stage IA non-small-cell lung cancer typically do not receive adjuvant therapy. In contrast, there is more controversy surrounding the use of chemotherapy for treating stage IB patients, but many stage IB patients will receive chemotherapy. For determining clinical utility, it would have been useful to consider analyses of stage IA and IB separately because the absolute risk levels may differ and result in different treatment choices.
The preceding examples highlight how examination of only the hazard ratio associated with a prognostic test in a multivariable analysis adjusted for standard prognostic factors is generally not sufficient to interpret the potential clinical value of a prognostic test in the context of those known prognostic variables. In a poor risk category defined by some combination of standard prognostic variables, the outcome may be fairly poor in both the favorable and unfavorable subgroups defined by the new prognostic test and there would be no change in the clinical management of patients with poor risk standard variables on the basis of the results of the new prognostic test. Another possibility is that there is an interaction between the standard variables and the new prognostic test such that the new test is prognostic in some categories defined by standard variables but not in others. Unless these interactions are appropriately modeled and the study is sufficiently powered to detect such interactions (which is rare), the lack of prognostic value of the new test in certain standard prognostic groups could go unnoticed. In the situations just discussed the overall conclusion about the clinical utility of the new prognostic test would be driven by whichever prognostic categories based on standard variables were most abundantly represented in the study. These points all argue for avoiding studies that include patients representing an extremely heterogeneous mixture of standard clinical and pathological characteristics when trying to determine the clinical value added by a new test. It is usually more appropriate to consider patient subgroups segregated according to how they are currently managed clinically, and then evaluate what additional clinically useful information can be provided by the new test.
Principles for evaluation of therapy-guiding tests
Importance of an appropriate control group for evaluation of a therapy-guiding test
Putative therapy-guiding biomarker-based tests are most reliably evaluated in the context of randomized clinical trials. Randomized treatment is the best safeguard against confusion between test prognostic value and subgroup-specific benefit or lack of benefit from a particular therapy. It also avoids potential confounding between treatment choice and other patient characteristics. If the therapy is a new therapy, then the test might be co-developed with the therapy in the context of a prospective clinical trial. If the test is being developed to guide use of an existing therapy, then it might be possible to use retrospective specimen collections from completed clinical trials to develop and evaluate the test. To simplify discussion we refer to the situation of assessment of the value of a test for choosing between a new therapy and a standard therapy (which might be no therapy), although the basic principles apply for both new and existing therapies.
Figure 3 illustrates two possible scenarios to explain the main points. Figure 3A shows the association between the test result and outcome when all patients are treated with the new therapy. Sometimes investigators prematurely conclude that such an observation establishes that the test can be used to select patients who will benefit from the new therapy relative to standard clinical management, but this could be an erroneous conclusion as shown by Figure 3B. The situation depicted in Figures 3A–B represents a test that is prognostic but is not useful for guiding therapy because the patients who are positive on the test have better outcome regardless of whether they receive the new versus standard therapy. In addition for this example, there is no benefit of the new therapy relative to standard in either test-defined subgroup.
Figures 3C–D demonstrate a situation in which studying the test in patients receiving the new therapy only could lead one to erroneously discard a therapy that is useful for guiding therapy. The test depicted in Figures 3C–D is useful for guiding therapy because patients predicted to be sensitive to the new therapy by the test have a better outcome when they receive the new therapy compared to standard, whereas patients predicted by the test to be insensitive to the new therapy have a better outcome when they receive standard therapy. If the performance of the test had been examined only in patients who received the new therapy, in might have been incorrectly concluded that the test result has no relationship to benefit from the new therapy.
Figures 3E–F demonstrate a situation where interpretation of clinical value of the test is more complex and dependent on factors in addition to survival benefit. In this scenario the patients who are predicted to be sensitive by the test have inferior outcome compared to test-negative patients when all receive standard therapy, but the test-positive patients preferentially benefit from the new therapy. This preferential benefit from the new therapy improves their outcome to result in outcome similar to that for patients who are negative for the test and receive either the new therapy or standard therapy. The determination of which treatment to offer a patient who is predicted by the test to be insensitive to the new therapy would depend on additional factors such as toxicities, convenience, and cost of the different therapy options.
Many scenarios in addition to those presented in Figure 3 are possible. The test may or may not have prognostics value. Both patient subgroups might receive some benefit from the new therapy, but the magnitude of benefit might be different in the two test-defined subgroups. The statistical power for detecting benefit of the new treatment might also be different in the two test-defined subgroups due to differential magnitude of benefit and/or different samples sizes and numbers of events in the test-defined subgroups.
To move from establishing clinical validity of a putative therapy-guiding test to clinical utility ultimately requires consideration of a variety of factors in addition to survival benefit. These factors may include toxicities of the therapy options, potential risks associated with use of the test (e.g., if the test requires obtaining a specimen by a difficult invasive biopsy procedure), and costs associated both with the test and the therapy options. More detailed discussion and numerous examples of how to properly evaluate clinical utility of putative therapy-guiding tests can be found elsewhere [12]. Several prospective trial designs have been proposed for evaluation of clinical utility of therapy-guiding biomarker-based tests, but prospective trials are typically large, expensive, and difficult to conduct; therefore, retrospective evaluations are often attempted first [4]. Readers interested in prospective biomarker-based clinical trial designs are referred elsewhere for further discussion [2, 13–14].
Common pitfalls in the development of omics-based tests
Development and evaluation of prognostic and therapy-guiding tests from high-dimensional omics data adds substantial complexity to the difficulties already inherent in the evaluation of tests based on single biomarkers. A thorough discussion is beyond the scope of this paper, but it is worthwhile to briefly describe some of the common pitfalls encountered in the high-dimensional data setting. Readers interested in an extensive discussion of a multitude of considerations in the development of omics-based tests and evaluation of their readiness for use in clinical trials where they will be used to guide clinical management for patients are referred to McShane et al [15].
Predictor development process
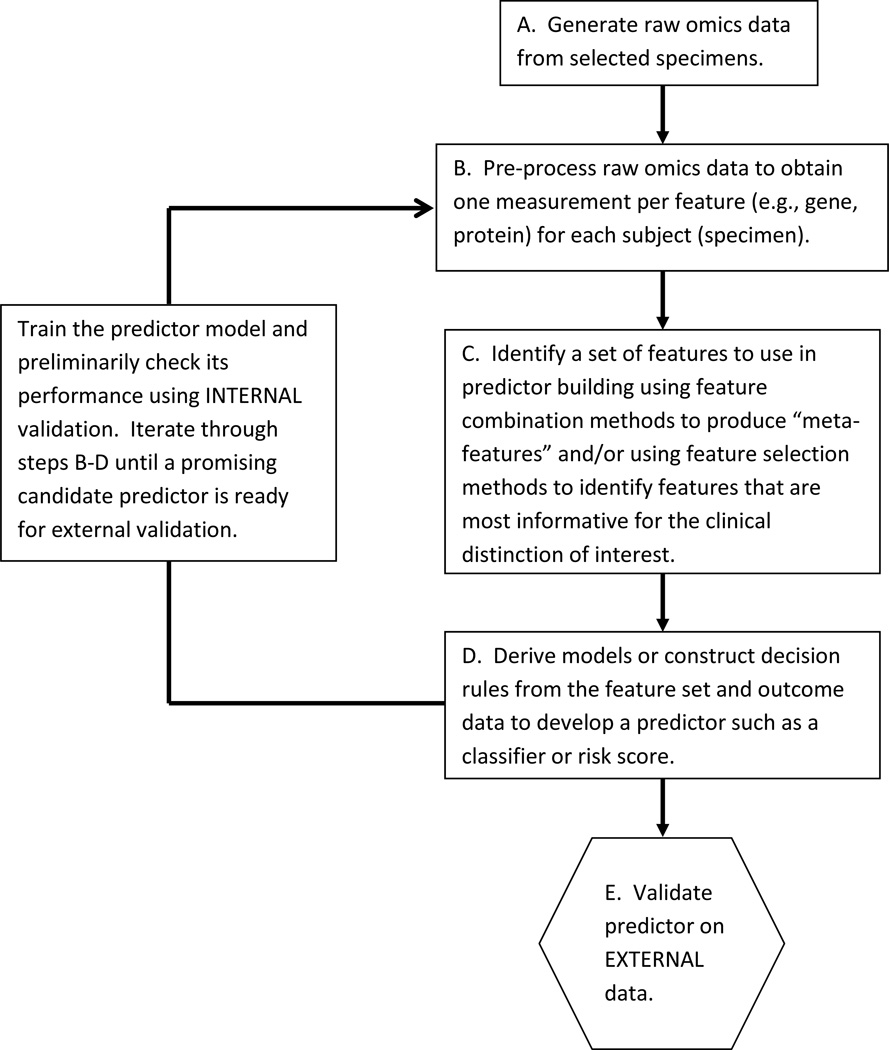
A schematic of the typical process followed to develop an omics predictor is represented in Figure 4. A number of initial data pre-processing steps are usually performed to transform raw omics data into summary measurements for each of many features per subject, for example the expression levels for 10,000 genes, abundance of thousands of proteins, and so forth, for each patient. Standard statistical modeling approaches cannot easily handle these large numbers of independent variables so some type of data reduction step is usually employed to either select subsets of variables for inclusion in the model (termed “feature selection”) or to derive “meta-variables”, where each meta-variable consists of combinations of original variables, for example principal components derived from the original variables. Some type of algorithm is then applied to develop a predictor model from that reduced set of variables into the form of a classifier or risk score. This entire process may occur seamlessly, or as two distinct steps (feature selection followed by model building). A variety of machine learning algorithms including random forests, support vector machines, and nearest neighbor classifiers are available [16]. Alternatively, more conventional statistical modeling techniques such as discriminant analysis (for binary endpoints) or Cox proportional hazards regression modeling (for time-to-event data) may be used once the number of variables has been reduced. Book-length treatments of model building approaches are available for interested readers [16, 17].
Figure 4. Schematic of omics predictor development process.
Step A: Specimens are gathered from potentially multiple sources and raw omics data are generated in possibly multiple laboratories. Some specimen eligibility criteria may have been applied, e.g., sufficient quality and quantity of biological material must be available.
Step B. Raw omics data undergo pre-processing to screen out poor quality or unreliable data. Data normalization, calibration, and other adjustment methods are typically applied in an effort to correct for artifacts such as laboratory equipment drift, and batch effects due to assay run or reagent lots. If the omics data originate from multiple sources, the pre-processing steps used may vary across different subsets of the data. Different data pre-processing steps applied to the same raw omics data will generally produce different results and may affect the performance of the predictor. Successful omics predictors must be robust to routine amounts of variation due to specimen handling or laboratory assay variation that cannot be controlled when the predictor is used in clinical practice.
Step C. The high dimension of typical omics data often requires that the number of features considered for use in predictor building be reduced. This can be accomplished by application of data reduction techniques that may or may not use outcome data. Examples of data reduction techniques that do not use outcome data include clustering to identify features contributing redundant information or principal components analysis to create “meta-features”, which are linear combinations of the original feature values. Data reduction approaches that use outcome data include univariate statistical analyses to identify features that individually have high correlation with outcome, for example using t-tests to identify all features that exhibit statistically significant mean differences between two outcome classes. Sometimes feature identification is incorporated seamlessly into the predictor building process and there is no distinct break between steps C and D.
Step D. A variety of regression modeling approaches, decision tree algorithms, or other machine learning techniques can be used to develop predictors from the feature data. It is common for there to be multiple iterations of predictor training steps B through D to make model adjustments until convergence on a predictor that looks promising.
Step E. Ideally, a predictor should be validated on a new data set that was in no way used to derive it. This external independent data should be obtained under specimen processing and handling conditions expected in routine clinical settings, from patients who are representative of the population in which the test is intended to be used. Internal validations, even if carefully conducted, always have potential limitations due to the possibility of biases affecting the entire data set.
Guard against overfitting complex models to high-dimensional data
Due to the high-dimensionality of omics data and complexity of many of the predictor models, there is substantial opportunity to inadvertently overfit models built from omics data. Overfitting refers to development of models that exaggerate minor fluctuations in the data due to fitting to random noise. A model that has been overfit to an initial data set (“training set”) will not exhibit good performance on a completely independent data set (“testing set”). The potential for overfitting can be reduced if constraints are placed on model complexity and one remains vigilant by monitoring model performance using internal validation strategies such as bootstrapping, cross-validation, or other resampling methods to compute preliminary estimates of model performance during the model building process [18]. Model complexity can be controlled using regularization techniques, which are methods to control smoothness of a fitted model or the number or magnitude of parameters in the model. Familiar examples of approaches incorporating regularization are penalized regression modeling methods such as ridge regression and lasso regression [16]. Internal validation, while advisable and helpful, is not foolproof. If the full training data set is subject to some major bias, for example bias due to some confounding with experimental artifacts, internal validation will not detect the problem. The most reliable way to assess model performance is to assess the model on a completely independent external testing data set.
Assessing predictor performance
Two key aspects of predictive model performance are discrimination and calibration [19]. To assess how well a predictor is calibrated, one examines the agreement between the probability of developing the outcome of interest (within a certain time period) or possessing the characteristic of interest and the observed frequencies of the same. Calibration is usually assessed graphically. Discrimination is the ability of a model to distinguish individuals who experience the outcome of interest or possess the characteristic of interest from individuals who don’t experience the event or do not possess the characteristic. An example of a widely used measure of discrimination is the C-index. For a simple binary prediction problem one might also examine positive and negative predictive value. Readers are referred to Moons et al [19–20] for a thorough discussion of development and validation of prediction models.
An unacceptable practice that is still found in some published articles presenting omics predictors is to assess predictor performance by plugging into the predictor the same data that were used to build it without use of proper resampling methods [7–8, 21]. Model performance estimates calculated this way (called “resubstitution estimates”) are highly biased in the direction of optimistic estimation of model performance. For example, Subramanian and Simon [8] showed through simulation that if one fits a model to data that are completely noise with no association between omics variables and survival outcome, a model built to predict into two prognostic classes will show dramatic separation of survival curves between the predicted classes on the training data; however the resulting model will have no ability to classify cases into subgroups having different prognosis on a completely independent data set (see Figure 2 in [8]).
A variation on resubstitution is the practice of using combined training and testing sets (“full” data) to identify informative features and then building the prediction model using those features to assess the model performance by an internal validation strategy on the training data. This approach, which we call “partial resubstitution”, allows information from the initial feature selection carried out on the full data set (which included the testing data) to leak into the predictor. As shown by Simon and colleagues, the bias in the accuracy estimate can be remarkably large even using partial resubstitution [21].
Bias can creep into model performance estimates in ways other than computing resubstitution estimates. One example of another flawed approach is to report model performance estimates on the combined training and testing sets, rather than on the testing set alone as would be appropriate. Even subtle decisions made when iteratively fitting models to the same training data can lead to a potential for bias in internally validated predictor performance metrics (see iterations in steps B–D in Figure 4). For example, if a decision is made to pre-process the raw omics data in a different way because it leads to a predictor with better performance on the testing (validation) data, then even a performance metric computed by internal validation on the training data could potentially be subject to some bias. The potential bias results from the look at the testing data to select pre-processing method, and from the fact that the final model was selected as the best performer among multiple models evaluated on the same testing data.
Bias also occurs when one builds a predictor model using a data set ignoring standard prognostic variables but then attempts to show on that same data set that the predictor contributes significant information beyond that provided by the standard variables by including calculated predictions into a multivariable model along with the standard variables and showing that the predictions make a statistically significant contribution to the model. This approach will lead to an exaggerated assessment of the ability of the predictor to contribute information over and above the information provided by the standard variables. This exaggeration occurs because the omics predictor has been specifically constructed to predict the outcome in the same data set, whereas the standard variables do not have a similar advantage.
Failure to maintain strict separation between training and testing sets can also introduce bias into assessments of putative therapy-guiding predictors. Suppose that one develops a prognostic predictor with omics data generated using specimens from patients accrued to the standard therapy arm (or arm receiving no therapy) of a clinical trial and then applies that predictor to data generated from patients on the experimental therapy arm. Simon and Freidlin [22] showed how this approach would lead to a biased evaluation of the treating-guiding ability of a predictor. The bias derives from the fact that the prognostic ability of the predictor would be exaggerated on the standard therapy arm (from which data were used to build the predictor) but would not have the same bias “advantage” when applied to the experimental therapy arm. The differential in apparent prognostic ability between arms could give rise to a spurious effect that could look like therapy-guiding ability of the predictor. Zhu et al [23] used an analysis approach like the one just described to conclude that the gene signature they developed had ability “to select patients with stage IB to II NSCLC most likely to benefit from adjuvant chemotherapy with cisplatin/vinorelbine.” Because their predictor was developed using the control arm (no chemotherapy) from the same trial, their results could be subject to the type of bias described by Simon and Freidlin [22]. The predictor must be evaluated on a completely independent data set in order to assess whether it has ability to accurately select patients who would benefit from chemotherapy.
Concluding remarks
Use of biomarker-based tests, including tests based on omics data, has steadily increased over the last several years in concert with efforts to refine treatment strategies to maximize chances patients will receive treatments that most benefit them. Proper development and evaluation approaches for tests that are useful in making treatment decisions for patients with diseases or other medical conditions is a relatively new topic for many biostatisticians and medical researchers compared to classical statistical methodology for clinical trials evaluating new therapies. We hope that the discussions presented here have highlighted some of the important statistical issues in this burgeoning area of biomedical research and will help to promote best practices for development and evaluation of prognostic and therapy-guiding clinical tests.
Acknowledgments
The views expressed in this article are the personal opinions of the authors and do not necessarily reflect policy of the U.S. National Cancer Institute.
References
- 1.Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington, DC: The National Academies Press; 2012. Institute of Medicine. [PubMed] [Google Scholar]
- 2.Clark GM, McShane LM. Biostatistical considerations in development of biomarker-based tests to guide treatment decisions. Stat Biopharm Res. 2011;3(4):549–560. [Google Scholar]
- 3.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: Methods of the EGAPP Working Group. Genet Med. 2009;11(1):3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McShane LM, Hayes DF. Publication of tumor marker research results: The necessity for complete and transparent reporting. J Clin Oncol. 2012;30(34):4223–4232. doi: 10.1200/JCO.2012.42.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennello G. (paper submitted for this special issue) [Google Scholar]
- 7.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99(2):147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 8.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: Ready for clinical use? J Natl Cancer Inst. 2010;102(7):464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 10.Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: Development and international validation studies. Lancet. 2012;379:823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polley MC, Freidlin B, Korn E, et al. Statistical and Practical Considerations for Clinical Evaluation of Predictive Biomarkers. J Natl Cancer Inst. 2013 doi: 10.1093/jnci/djt282. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sargent D, Conley BA, Allegra C, et al. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23(9):2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 14.Freidlin B, McShane LM, Korn EL. Randomized clinical trials with biomarkers: Design issues. J Natl Cancer Inst. 2010;102(3):152–160. doi: 10.1093/jnci/djp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McShane LM, Cavenagh MM, Lively TG, et al. Criteria for the use of omics-based predictors in clinical trials sponsored by the National Cancer Institute: Explanation & elaboration. BMC Medicine. 2013 doi: 10.1186/1741-7015-11-220. (in press; checklist available at http://www.cancerdiagnosis.nci.nih.gov/docs/checklist_draft_guidelines.pdf) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: data-mining, inference, and prediction. 2nd ed. New York: Springer; 2009. [Google Scholar]
- 17.Harrell FE. Regression Modeling Strategies: With applications to linear models, logistic regression, and survival analysis. New York: Springer; 2001. [Google Scholar]
- 18.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21(15):3301–3307. doi: 10.1093/bioinformatics/bti499. doi. [DOI] [PubMed] [Google Scholar]
- 19.Moons KGM, Kengne AP, Woodward M, et al. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 2012;98:683–690. doi: 10.1136/heartjnl-2011-301246. [DOI] [PubMed] [Google Scholar]
- 20.Moons KGM, Kengne AP, Grobbee DE, et al. Risk prediction models: II. External validation, model updating, and impact assessment. Heart. 2012;98:691–698. doi: 10.1136/heartjnl-2011-301247. [DOI] [PubMed] [Google Scholar]
- 21.Simon R, Radmacher MD, Dobbin K, et al. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 22.Simon RM, Freidlin B. Re: Designing a randomized clinical trial to evaluate personalized medicine: A new approach based on risk prediction [Correspondence] J Natl Cancer Inst. 2012;103(5):445. doi: 10.1093/jnci/djq552. (DOI:10.1093/jnci/djq552). [DOI] [PubMed] [Google Scholar]
- 23.Zhu C-Q, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28(29):4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]