Abstract
Large collections of annotated cancer cell lines are powerful tools for precisely matching targeted drugs with genomic alterations that can be tested as biomarkers in the clinic. Whether these screening platforms, which utilize short-term cell survival to assess drug responses, can be applied to precision radiation medicine is not established. To this end, 32 cancer cell lines were screened using 18 targeted therapeutic agents with known or putative radiosensitizing properties (227 combinations). The cell number remaining after drug exposure with or without radiation was assessed by non-clonogenic assays. We derived short-term radiosensitization factors (SRF2Gy) and calculated clonogenic survival assay-based dose enhancement factors (DEFSF0.1). Radiosensitization was characterized by SRF2Gy values of mostly ~1.05–1.2 and significantly correlated with drug-induced changes in apoptosis and senescence frequencies. SRF2Gy was significantly correlated with DEFSF0.1, with a respective sensitivity and specificity of 91.7% and 81.5% for a 3-day endpoint, and 82.8% and 84.2% for a robotic 5-day assay. KRAS mutations (codons 12/13) were found to be a biomarker of radiosensitization by midostaurin in lung cancer, which was pronounced under conditions that enriched for stem cell-like cells. In conclusion, while short-term proliferation/survival assays cannot replace the gold standard clonogenic survival assay for measuring cellular radiosensitivity, they capture with high accuracy the relative change in radiosensitivity that is caused by a radiosensitzing targeted agent.
Keywords: precision radiation medicine, cancer cell lines, targeted drugs, biomarkers
Introduction
Large panels of annotated cancer cell lines provide useful preclinical models for identifying genotype-correlated drug sensitivities that can be clinically tested (1–5). The basic concept underlying the success of these analyses predicts that the cytostatic or cytotoxic effects of drugs in cultured cells translate into tumor regression, a standard criterion of efficacy in patients with metastatic cancer. However, regression is an insufficient surrogate endpoint for the outcome of radiation therapy with curative intent which requires eradication of all tumor cells that could give rise to a local recurrence (6). Traditionally these have been termed ‘clonogenic’ cells, i.e., cells that have the capacity to produce an expanding family of daughter cells and form colonies following irradiation in an in-vitro assay or give rise to a recurrent tumor in in-vivo models. To which extent clonogenic cells may represent cancer stem cells is unclear though more recently the terms have been used interchangeably (7, 8).
Because chromosomal damage caused by ionizing radiation (IR) may persist over several cell cycles before disrupting a cell’s ability to divide infinitely, colony formation or clonogenic survival assays (CSA) have been considered the ‘gold standard’ for assessing the cytotoxic effects of IR in cell culture, supporting the concept that cellular radiosensitivity is a major, though not the only, determinant of in-vivo radiosensitivity (9–14). In contrast, it is a long-held paradigm that radiosensitivity determined in short-term assays that measure cell proliferation or viability over a few days correlates poorly with radiosensitivity derived from CSA (15, 16).
The importance of pre-clinical and clinical drug development with IR and its challenges have been highlighted (17–20). Historically, the choice of radiosensitizers has conformed to a “one-size-fits-all” philosophy, but it has become increasingly apparent that radiosensitizing effects may be genotype-dependent, requiring predictive biomarkers for appropriate patient selection (21, 22). To this end, precision radiation medicine may leverage genomic information derived from human cancer cell lines or tissue samples. Unfortunately, CSA are not ideal for the large scale and high-throughput cell line screens that would be needed to identify tumor genotypes that correlate with sensitivity to IR/drug combinations owing to the often poor colony forming ability of human cancer cell lines and the time and resources it takes to conduct these assays. This is an important barrier to pre-clinical testing and clinical translation of novel IR/drug regimens.
We recently observed that the radiosensitizing effects of epidermal growth factor receptor (EGFR) inhibitors seen in a short-term viability assay correlated well with radiosensitization in a CSA because the premature senescence response underlying radiosensitization led to a proliferative delay that was captured in the 3-day assay (23). We, therefore, hypothesized that short-term assays can provide a measure of the change in cellular radiosensitivity that is caused by a targeted drug provided the drug alters the mode of cell inactivation observed within a few days following irradiation, such as senescence, apoptosis, or autophagy. Furthermore, we reasoned that robotic screening platforms can be adapted to capture the ultimately therapeutically significant but small magnitude effects of radiosensitizing drugs (~10% reduction in cell number) which stand in contrast to the typically large effects of targeted drugs alone in susceptible cell lines (>50% reduction) (23–25).
Methods
Cell Lines
Annotated cell lines were selected from previously published panels (1, 23–25). The identity of the cell lines had been tested as described (23), and additional authentication was performed by Bio-Synthesis, Inc. No cell line tested positive for mycoplasma (MycoAlert, Lonza). For 3D culture of tumor spheres, 5,000 cells/well were grown in low-binding 96-well plates (Thermo, 145399) using serum-free medium composed of DMEM (Sigma-Aldrich), basic fibroblast and epidermal growth factor (20 ng/mL each, Sigma-Aldrich), and B27 supplement (Life Technologies), followed by drug/IR treatments 3 days later.
Treatments
X-ray treatments were performed as described (23). Drugs were dissolved in Dimethyl Sulfoxide (DMSO) (Sigma-Aldrich), except chloroquine which was dissolved in deionized water. Drugs were aliquoted and stored according to manufacturers’ guidelines. Drugs were added to cells 1 hour before irradiation at appropriate concentrations (Supplementary Table 1).
Cell Survival Assays
Clonogenic cell survival was measured by seeding cells for colony formation at appropriate densities 16–18 hours prior to 2 Gy irradiation +/− drug pre-incubation as described (23, 25). Drugs were not washed out following irradiation except for NU7026 and olaparib after 24 hours. The syto60 assay has been described (23–25). The CellTiter-Glo® (CTG) luminescence (Promega, Madison, WI, USA) and MTT metabolic assays (Cayman Chemical, Ann Arbor, MI, USA) were performed following the manufacturer’s protocols. To adapt robotic screening (1), 96-well clear bottom black plates (Corning, NY, USA) with optimized cell density for each cell line (i.e. 70–80% confluence by end of the assay for control samples) were prepared. Cells were drugged by the liquid handling robot (Zephyr, Caliper Life Sciences, Hopkinton, MA) 1 hour pre-irradiation. CTG reagents were applied to cells 5 days later (EL406™, BioTek Instruments, Winooski, Vermont, USA). Signals were read by the MultiLabel reader, 2140 Envision (Perkin Elmer, Waltham, MA, USA).
Apoptosis and Senescence Assays
Seventy-two hours after irradiation, cells and media were collected, centrifuged, and resuspended in Annexin binding buffer with cell density adjusted to ~106/ml. Cells were stained with propidium iodide (Sigma-Aldrich) and Annexin V-Cy5 following the manufacturer’s protocol (BioVision, Milpitas, CA), and then analyzed by a LSRII flow cytometer (BD Biosciences, San Jose, CA). Senescence-associated β-galactosidase staining was performed using a commercial kit (Cell Signaling, #9860) as described (23).
Immunofluorescence Microscopy
Staining and visualization of γ-H2AX and 53BP1 foci were performed as previously described (23, 25).
Western Blotting
Whole cell lysates were prepared using standard methods. Specific antibodies against phospho-PKC (pan) (Cell Signaling, #9371) and total PKCα [Y124] (Abcam, ab32376), and horseradish peroxidase–conjugated secondary antibody (Santa Cruz) were used. Protein bands were visualized with enhanced chemiluminescence (Invitrogen) followed by autoradiography.
RNA Interference
PKCα siRNA transfections were carried out as described (25).
Flow Cytometry
Cells were labeled with CD133/1 (AC133) - PE antibody (Miltenyi Biotec), and high and low CD133 expressing cells were subjected to sterile sorting by flow cytometry.
Statistical Analysis
All data were analyzed by GraphPad Prism 6. Clonogenic survival data were fitted by the Linear-Quadratic formula. Statistical comparisons were carried out with the F-test. Unless noted all statistical tests were two-sided. Receiver-operating characteristic (ROC) curves were applied to evaluate the performance of the short-term screening platforms in comparison with the CSA to determine an appropriate cut-off for the testing assay.
Results
Non-Clonogenic Screening Platform
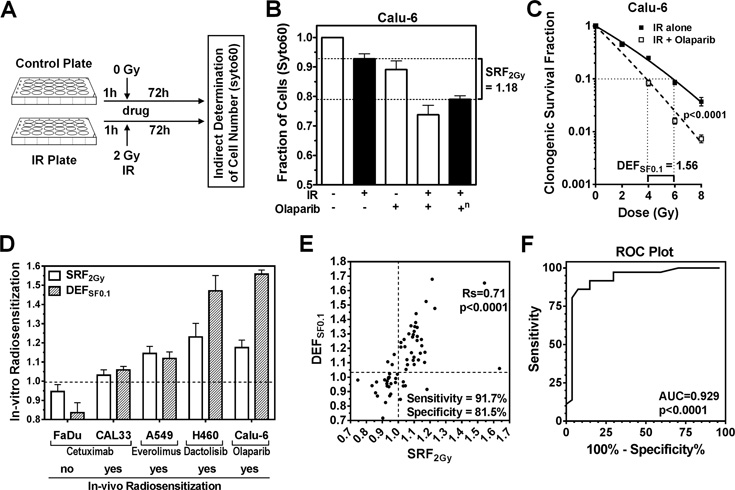
To determine the effects of irradiation +/− drug treatment on short-term survival of cancer cells, we initially employed a previously published assay that relies on fixation of persistent cells followed by incubation with a nucleic acids stain (syto60) for quantification (23, 24) (Fig. S1A–E), in addition to other common short-term assays (CTG, MTT) (Fig. S1F–I). We arbitrarily selected a pilot panel of 32 cell lines derived from lung, colorectal, genitourinary, and head and neck cancers as well as 18 molecular targeted drugs with known or putative radiosensitizing properties (1). Drug concentrations were selected to be minimally toxic for drug alone treatments, known to inhibit the target, and achievable in patients. Drugs were added to plates 1 hour before mock-treatment or irradiation with a clinically relevant dose of 2 Gy followed by incubation for ≥3 days depending on the particular experiment (Fig. 1A). In total, we assayed 227 cell line-drug combinations (Suppl. Tab. 1A,B). The effect of combined drug/IR relative to the effect of IR alone, and corrected for drug alone effect, was expressed as SRF2Gy (Short-term Radiosensitization Factor at 2 Gy) (Fig. S1J, 1B).
Figure 1. Correlation of radiosensitization in short-term syto60 and clonogenic assays.
A) Illustration of pilot set-up using a 24-well format and syto60 staining. IR, ionizing radiation; h, hours. B) Example of short-term radiosensitization using the PARP inhibitor olaparib in Calu-6 lung cancer cells. SRF2Gy, short-term radiosensitization factor for 2 Gy (for definition, see Fig. S1J). +, treatment with 2 Gy IR or/and 1 µM olaparib; −, no treatment; n, effect of combined IR and olaparib normalized for the effect of olaparib alone. C) Example of radiosensitizing drug effect using clonogenic survival as endpoint. DEFSF0.1, which is a standard descriptor of radiosensitization (14), represents the ratio of radiation doses required to achieve 0.1 clonogenic survival when given without and with drug. The drug + IR curve is corrected for the effect of drug alone. Statistical comparison by F-test. D) Illustration of the association of SRF2Gy with DEFSF0.1 values and previously reported radiosensitization of xenografts for various cell lines and targeted drugs (see text). E) Correlation of SRF2Gy with DEFSF0.1 for 25 cancer cell lines treated with up to 8 agents for a total of 63 comparisons. DEFSF0.1 values were derived from full clonogenic survival curves (Supplementary Table 1A). For sensitivity and specificity calculations, cut-offs of ≥1.01 for SRF2Gy and ≥1.04 for DEFSF0.1 were used to define a positive effect. Statistical comparison by Spearman rank correlation. Rs, Spearman rank coefficient. F) Receiver operating characteristic (ROC) curve using the data shown in panel E. AUC, area under the curve.
Correlation of Radiosensitization in Non-Clonogenic and Clonogenic Survival Assays
To correlate radiosensitization in the short-term syto60 assay (SRF2Gy) with radiosensitization using the CSA, standard Dose Enhancement Factors at 0.1 clonogenic survival fraction (DEFSF0.1) (14) were calculated (illustrated in Fig. 1C). Initial experiments using selected targeted drugs and cell lines suggested that radiosensitization described by SRF2Gy not only predicted drug effect in the CSA, but also correlated with the known ability of these drugs to enhance the effects of IR in-vivo (Fig. 1D) (26–29).
Next, we generated 63 comparisons of syto60-based SRF2Gy values and CSA-derived DEFSF0.1 values based on 25 cancer cell lines treated with up to 8 drugs (Fig. 1E, Suppl. Tab. 1A). There was a highly statistically significant correlation between SRF2Gy and DEFSF0.1 values (p<0.0001). Similarly, short-term and clonogenic SRF2Gy values were significantly correlated (p<0.0001) (Fig. S2A). A ROC plot confirmed the high accuracy of SRF2Gy values to predict radiosensitization (p<0.0001) (Fig. 1F). Notably, even small SRF2Gy values of 1.05 or less were often associated with radiosensitization in the CSA (Fig. 1E) so that we selected a cut-off of ≥1.01. For DEFSF0.1, we arbitrarily chose a cut-off of ≥1.04 due to data indicating that even DEF values this small could be clinically significant (Fig. 1D, S2B). With these cut-offs the overall sensitivity and specificity with regard to radiosensitization in the CSA was 91.7% and 81.5%, respectively.
We conclude that a short-term assay can capture the relative change in radiosensitivity caused by a radiosensitizing agent. Thus, specifically for radiosensitization short-term endpoints may be an appropriate surrogate of CSA. However, our data do not suggest that short-term assays should be generally substituted for CSA. In fact, we did not find any correlation between cellular radiosensitivity measured with the short-term assay and radiosensitivity determined using the CSA (Fig. S2C), which is consistent with historical data (15, 16).
Drug-Induced Changes in Apoptosis and Senescence Correlate with Radiosensitization
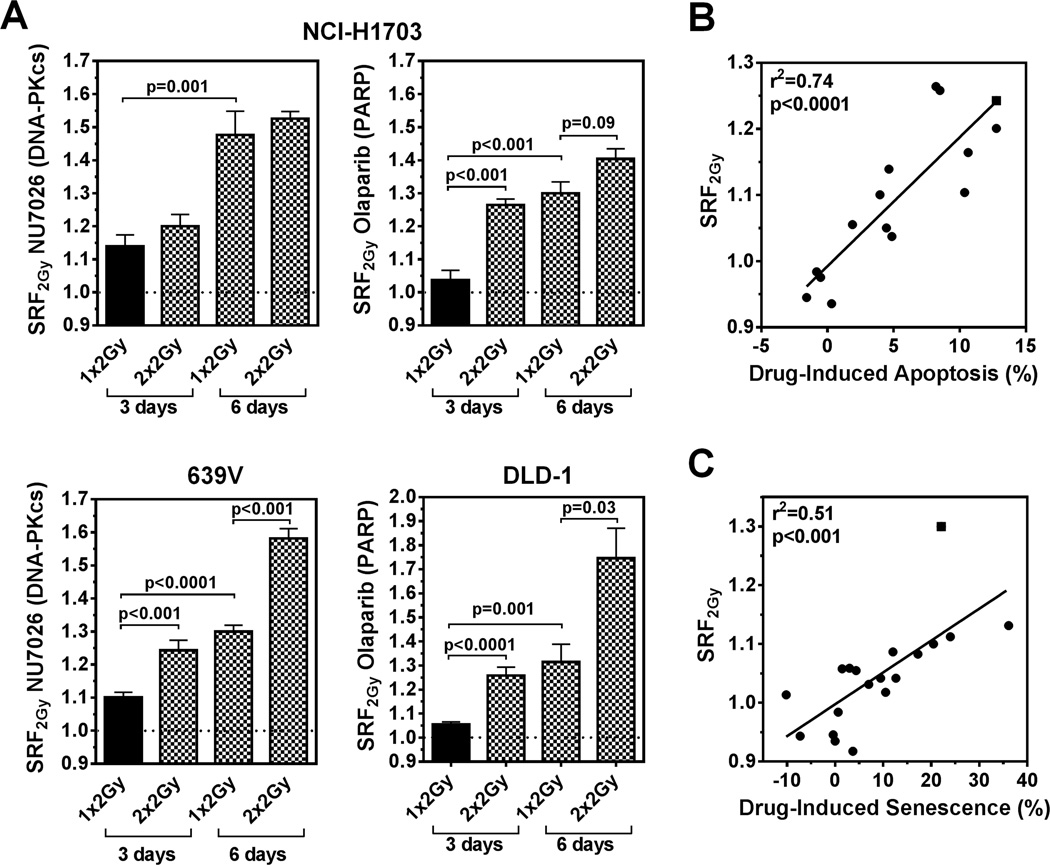
Notably, the SRF2Gy values that correlated with radiosensitization in the CSA were generally small, i.e., on average 1.12 (SD +/− 0.13) (Fig. 1E, and further illustrated in Fig. S2D). To increase our confidence that these small values represent true effects, we tested an alternate 2 × 2 Gy irradiation schedule because during a fractionated course of radiation therapy in the clinic the cytotoxic effect of a single dose is repeated. This schedule produced statistically significant increases in SRF2Gy for several cell-drug combinations (Fig. 2A). In addition, because IR-induced lethal chromosomal aberrations may inactivate cells only after a few cell divisions, we extended the incubation period from 3 to 6 days, which also yielded an often pronounced increase in SRF2Gy (Fig. 2A, S2E).
Figure 2. Factors that enhance short-term radiosensitization and correlation with apoptosis and premature senescence frequencies.
A) Examples of enhanced SRF2Gy when IR is repeated (2 Gy × 2, 24 hours apart) or when incubation times are extended to 6 days (incubation times counted from day of (first) irradiation). Data points shown represent mean (+/− standard error) based on at least three biological repeat experiments. Statistical comparisons by unpaired T-test, two-sided. B) Correlation of SRF2Gy values with relative change in the percentage of Annexin V positive cells upon adding drug to IR, normalized for drug alone effect. Data points represent differences between drug + 2 Gy versus 2 Gy alone effects in several cell lines, except for square symbol which indicates a 2×2 Gy treatment. Solid line, result of linear regression analysis. C) Analogous to panel B, correlation of SRF2Gy with relative change of SA-β-gal positive cells scored 3 days after irradiation, except for square symbol which re-presents a 6-day experiment.
Next, we investigated the cellular events underlying the observed radiosensitization by different drugs. A strong correlation between drug-induced apoptosis and SRF2Gy was found for several cell line-drug combinations (Fig. 2B, Fig. S3A–E). This is particularly well illustrated in NCI-H1703 cells, which are senescence-resistant due to non-functional p53/p16 (Fig. S3A–C). Drug-induced premature senescence could also be observed, as shown in Fig. S4, and correlated well with radiosensitization (Fig. 2C). Together, the data in Fig. 2 suggest that the observed SRF2Gy values (Fig. 1E) represent not only true effects that are based on drug-induced changes in apoptosis or senescence responses but also in many cases can be augmented by fractionation and/or prolongation of incubation times.
Implementing a Robotic High-Throughput Platform for Personalized Radiation Medicine
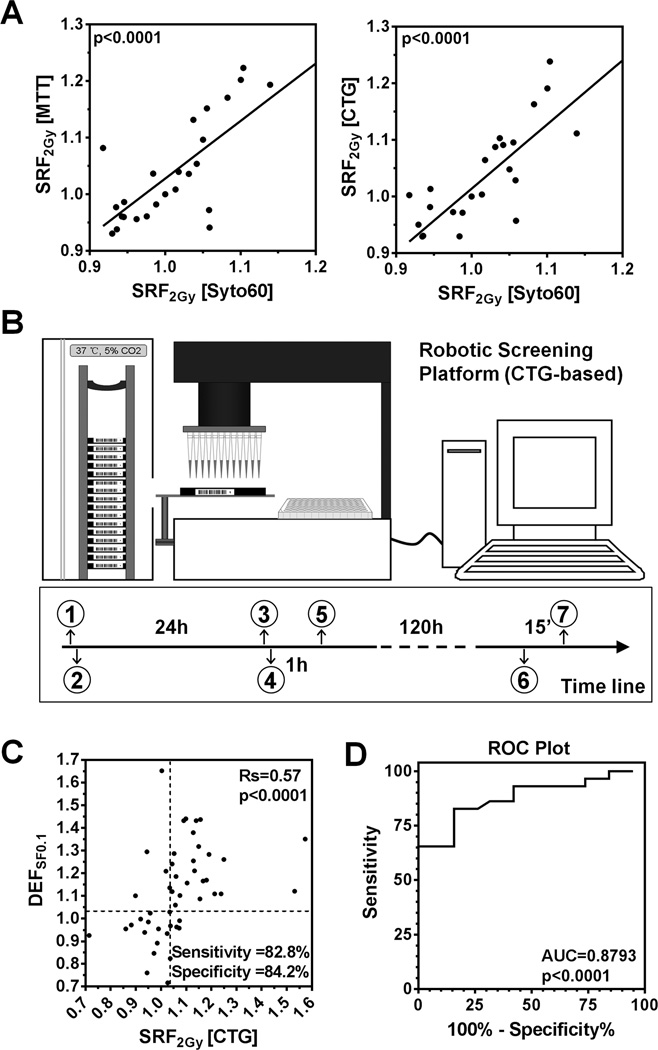
In order to adapt our approach for robotic high through-put screening (1), we confirmed that the observed radiosensitizing effects were not specific to the syto60 assay and could be detected with the commonly used MTT and CTG assays (p<0.0001) (Fig. 3A). Comparative analysis using a 96-well plate format indicated that the CTG assay was the most sensitive and robust of the three assays, and was thus selected for robotic platform testing (Fig. 3B, S1G–I). Ten cancer cell lines and 16 targeted drugs were chosen (Suppl. Tab. 1B). Clonogenic survival data were available for 48 cell line-drug combinations, and indicated a high accuracy of the CTG assay in terms of predicting radiosensitization, with a sensitivity of 82.8% and specificity 84.2% (Fig. 3C,D). A higher cut-off for SRF2Gy of ≥1.04 was chosen compared to the syto60 assay, given the tendency of the CTG assay to produce generally slightly higher SRF2Gy values.
Figure 3. Establishing a robotic cell line screening platform.
A) Comparison of the syto60 assay with the MTT or CTG assay. Solid lines and p-values represent results of linear regression analysis. Each data point is based on three biological repeats. B) Illustration of the robot-assisted screening process. (1) Prepare duplicate plates (mock-treatment, IR 2 Gy), (2) Seed one cell line per plate, (3) Prepare master drug plate, (4) Load plates, add drugs via robot, and incubate, (5) Treat plates and return to incubator, (6) Add CTG agents, (7) Read plates. C) Correlation of SRF2Gy values obtained with the 5-day robotic CTG assay and DEFSF0.1 values analogous to Fig. 1E. For sensitivity and specificity calculations, cut-offs of ≥1.04 for SRF2Gy and ≥1.03 for DEFSF0.1 were chosen to define a positive effect. Data points represent 48 comparisons based on 9 cell lines and 12 targeted agents. D) ROC plot analogous to Fig. 1F.
Genomic Biomarkers of Radiosensitization
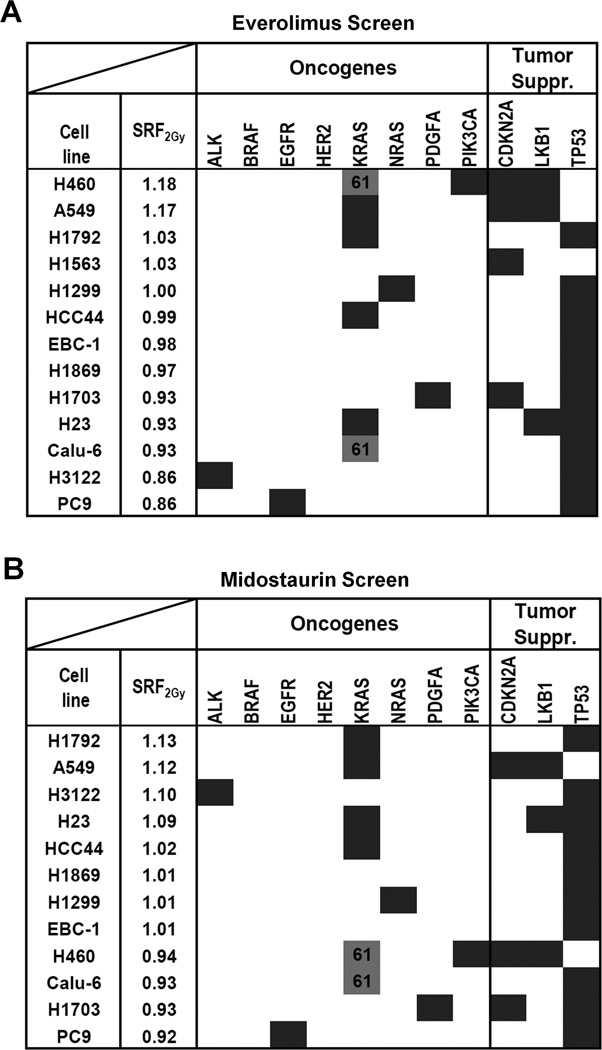
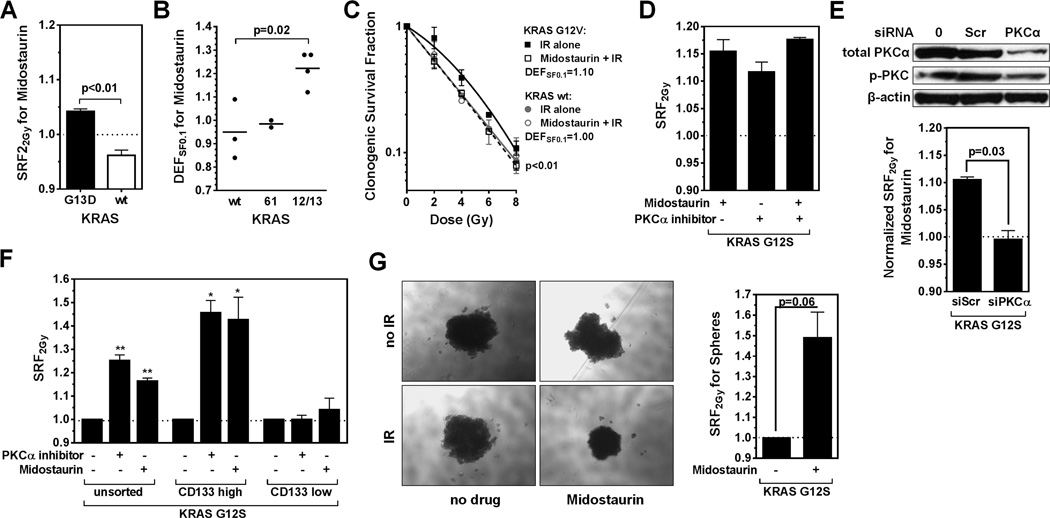
Next, we focused on a subset of lung cancer cell lines to determine if our screening platform can detect genetically defined mechanisms of radiosensitization. For this, we arbitrarily selected the mTOR inhibitor everolimus, a negative regulator of DNA damage-mediated autophagy, and the multi-kinase inhibitor midostaurin (30–33). For everolimus, radiosensitization was observed almost exclusively in cell lines with wild-type TP53 (p=0.001) (Fig. 4A), and this was confirmed in an isogenic cell pair (Fig. S5A,B). Consistent with a promoting role of p53 in autophagy induction and premature senescence (23, 34, 35), we observed everolimus-induced autophagy and senescence only in irradiated TP53 wild-type but not mutated cells (Fig. S5C,D). Of the top 5 cell lines radiosensitized by midostaurin (SRF2Gy of 1.02–1.13), four harbored KRAS mutations in codons 12 and 13 (Fig. 4B). In contrast, cells with wild-type KRAS or mutations in codons 61 did not show radiosensitization (p=0.01). KRAS codon 12/13 mutation-dependent radiosensitization was confirmed in isogenic cell pairs and the CSA (Fig. 5A–C). Midostaurin also increased the number of residual IR-induced DNA double-strand breaks and caused apoptosis and senescence in irradiated KRAS-mutant cells (Fig. S6A–C), in line with the correlations shown in Fig. 2B,C.
Figure 4. Identification of genomic biomarkers of radiosensitization by the mTOR inhibitor everolimus and the multi-kinase inhibitor midostaurin.
A) Results of IR/drug screen of 13 lung cancer cell lines with everolimus (20 nM) (Supplementary Table 1A). Cell lines are ranked by average SRF2Gy value. Dark fields indicate known mutations or other genomic alterations in common oncogenes and tumor suppressors (suppr.). KRAS codon 61 mutations (grey fill-in) are distinguished from codon 12/13 mutations (dark). SRF2Gy values are statistically significantly higher in p53 wild-type and than in mutated cell lines (p=0.001) (T-test). B) Analogous results for midostaurin (100 nM). SRF2Gy values are statistically significantly higher in cell lines with KRAS codon 12/13 mutations than in all other cell lines (p=0.01) and higher than in wild-type cell lines (p=0.04).
Figure 5. Follow-up analysis on the radiosensitizing effects of midostaurin.
A) SRF2Gy values for isogenic DLD-1 cells harboring a mutant KRAS or a deleted allele. B) DEFSF0.1 values derived from clonogenic survival curves according to KRAS status of non-isogenic lung cancer cell lines. Statistical comparison with t-test. C) Clonogenic survival of isogenic KRAS wild-type NCI-H1703 cells with or without stable expression of a mutant KRAS transgene. Data were fitted using the LQ formula, and statistical comparison was performed with the F-test. D) SRF2Gy values for KRAS-mutated A549 cells treated with midostaurin or the PKCα-specific Ro-32-0432 inhibitor (100 nM). E) Effect of PKCα depletion: Upper panel, Western blot of A549 cells transfected with no reagents (0), a scrambled control (scr), or siRNA against PKCα. p-PKC indicates a pan-antibody against phospho-PKC isoforms. Lower panel, SRF2Gy values for midostaurin with or without PKCα depletion. Statistical comparison with T-test. F) SRF2Gy values in A549 populations based on treatment with midostaurin or Ro-32-0432 following no sorting, sorting for high CD133 expressors, and sorting for low CD133 expressors. Statistical comparisons to IR alone with one-sample T-test, *, p≤0.05, **, p≤0.01. G) Left panel: representative images of A549 spheres following treatments as indicated. Right panel, quantification of results using the CTG assay. Statistical comparison with T-test. All data points shown represent mean +/− standard error based on at least three biological repeat experiments.
Interestingly, we recently found that PKCα, a known target of midostaurin, contributes to the radioresistance of KRAS-mutant cells (25). We, therefore, compared the radiosensitizing effect of a specific PKCα small molecular inhibitor to the effect of midostaurin and observed comparable results (Fig. 5D). Depletion of PKCα abrogated the radiosensitizing effect of midostaurin (Fig. 5, S6D). As PKCα was recently implicated in maintaining breast cancer stem cells (36), we asked whether midostaurin’s effect was more pronounced in a subpopulation of lung cancer cells. Strikingly, midostaurin poorly radiosensitized cells with low expression of the stem cell marker CD133 while a relatively large SRF2Gy of 1.43 was observed in a subpopulation of high CD133 expressors (Fig. 5F, S6E). Thus, a relatively small SRF2Gy seen in an unselected cell population, such as ~1.1 for midostaurin may be driven by the sensitivity of a stem cell-like subpopulation. Tumor spheres are thought to contain a higher fraction of stem cells compared to monolayer cultures (36). Again, the radiosensitizing effect of midostaurin was evident and enhanced in KRAS-mutant tumor spheres, i.e., SRF2Gy ~ 1.4 (Fig. 5G, S6F).
Discussion
Clonogenic survival assays have been considered the gold standard for assessing the cell-inactivating effects of IR in-vitro (37–39). Even though plate formats have been tested (38, 40, 41), CSA are not ideal for the high-throughput screens that are needed to match genomic tumor profiles with IR/drug sensitivities owing to the frequently poor colony forming ability of human cancer cell lines and the time it takes to conduct these assays. Short-term cell viability/survival assays, on the other hand, are generally not considered to provide appropriate surrogate endpoints of clonogenic survival (15, 16, 41). In individual cell lines, some short-term assays such as the MTT assay can capture radiosensitizing effects and correlate with CSA (42–44). However, to our knowledge, the utility of short-term assays as a surrogate for CSA for screening any larger number of cancer cell lines has never been validated.
Here, we establish a robust correlation between short-term and clonogenic radiosensitization for a variety of cell lines, drugs, and assay conditions (Fig. 1E, 3C). Short-term radiosensitization was measured for 2 Gy IR single doses which are clinically relevant and largely avoid cell cycle delay problems that can be caused by higher IR doses. The cumulative data suggest that changes in senescence, apoptosis, or/and autophagy caused by the radiosensitizing drug compared to cells treated with IR alone consistently affect the number of persisting cells that are measured by the short-term assays (Fig. 2, S3, S4). Because IR-induced lethal chromosomal aberrations typically abrogate cell proliferation after a few division cycles, for example by causing mitotic catastrophe, apoptosis, or senescence (45), we suggest that assessing cell numbers at ~5–6 days post-irradiation is the best compromise between giving enough time for drug effects to manifest (at least 2–3 days) and avoiding the lengthy incubation needed for selecting out the surviving cells as colonies (2–3 weeks). Our findings provide an important proof-of-principal regarding the close correlation between short-term and clonogenic assay results when assessing radiosensitizing effects. However, more data will be needed before we can establish robust cut-offs for predicting DEF values (Fig. 1E, 3C).
In contrast to studies on the anti-proliferative effects of anti-cancer drugs alone, which are typically pronounced in susceptible cell lines (24), the dynamic range of radiosensitizing effects when combining drugs with 2 Gy IR is much smaller though surprisingly robust. For example, in the 3-day syto60 assay the average SRF2Gy was only ~1.1 (Fig. 1E). A SRF2Gy as small as 1.03 for the combination of the EGFR-directed monoclonal antibody cetuximab and IR in the head and neck cancer cell line CAL33 appears to be clinically meaningful given the observed impact in-vivo (Fig. 1D, S2B) (26). That such small SRF2Gy values capture early cellular events corresponding to real radiosensitization that should translate into larger effects with prolonged radiation courses was further highlighted by several lines of experimentation which demonstrated an increase in SRF2Gy when: a) incubation time was prolonged (to allow for additional senescence and apoptosis events to occur) (Fig. 2A, S2E); b) repeat 2 Gy irradiation was performed (Fig. 2A, S3B,E); and c) drug effect was measured under stem cell-enriched culture conditions (Fig. 5F,G, S6F).
Our initial screens successfully established a genomic biomarker, KRAS mutation, for one of the targeted drugs, midostaurin (Fig. 4B, 5). Cells with codon 12/13 mutations were radiosensitized while those with codon 61 mutations were not, suggesting functional heterogeneity associated with different KRAS mutations although we did not pursue this further. Even though midostaurin is a “dirty” tyrosine kinase inhibitor with multiple targets (32, 33), our findings suggest that PKCα is a critical target for radiosensitization of KRAS-mutant cells (Figure 5D,E, S6D) (25). A phase I trial of midostaurin with radiation in rectal cancer is ongoing at our institution (NCT01282502, www.clinicaltrials.gov). The data highlight the potential clinical significance of this type of screening.
As KRAS mutations are present in ~30% of NSCLC (46), a relatively small cell line panel was sufficient to detect a potential association with drug effect (Fig. 4B). However, one can envision that drug/IR combinations exist that track with more uncommon genomic alterations, e.g., present in < 5–10% of tumors. To detect those associations, panels of ~50–100 cell lines will be needed. This represents a very different approach from traditional investigations of IR/drug combinations which have utilized only small numbers of in-vitro cell lines for a given cancer type (3, 17), consistent with the traditional “one size fits all” philosophy of combining IR with drugs in patients.
We believe that genomic biomarker discovery using established cancer cell lines has validity given the observed genotype and phenotype similarities with human cancers, though this is not undisputed (47, 48). It is also clear that in-vitro radiosensitization may not readily translate into in-vivo effects, and therefore a path to in-vivo validation of radiosensitizing effects remains a critical part of any pre-clinical investigation strategy (39, 49, 50).
In conclusion, while short-term assays cannot supplant the gold-standard CSA for measuring absolute radiosensitivity, screening platforms such as ours can capture with high accuracy the relative change in radiosensitivity that is caused by a targeted drug in an individual cell line. Genomic biomarkers identified through this type of screen may guide the identification of patients who would benefit from novel drug/IR combinations. We suggest that our data support a paradigm change regarding the utility of non-clonogenic survival assays in precision radiation medicine.
Supplementary Material
Implications.
This study supports a paradigm shift regarding the utility of short-term assays for precision radiation medicine, which should facilitate the identification of genomic biomarkers to guide the testing of novel drug/radiation combinations.
Acknowledgments
Funding: This work was supported by the Dana-Farber/Harvard Cancer Center Specialized Program Of Research Excellence in Lung Cancer (P50 CA090578) to BYY, JS, HW; American Cancer Society (123420RSG-12-224-01-DMC) to HW; UK Wellcome Trust (086357) to JS, CHB; a stipend from the Jinan Municipal Center for Disease Control and Prevention, Shandong, China to JH; and Federal Share of program income earned by Massachusetts General Hospital, Proton Therapy Research and Treatment Center (C06 CA059267) to BYY, JAE, KDH, HW.
Footnotes
Conflicts of Interest: Jeff Settleman, Ph.D., is employed by Genentech Inc., San Francisco, CA
References
- 1.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O'Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein JN. Drug discovery: Cell lines battle cancer. Nature. 2012;483:544–545. doi: 10.1038/483544a. [DOI] [PubMed] [Google Scholar]
- 5.Gazdar AF, Minna JD. Precision medicine for cancer patients: lessons learned and the path forward. J Natl Cancer Inst. 2013;105:1262–1263. doi: 10.1093/jnci/djt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willers H, Held KD. Introduction to clinical radiation biology. Hematol Oncol Clin North Am. 2006;20:1–24. doi: 10.1016/j.hoc.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Willers H, Azzoli CG, Santivasi WL, Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J. 2013;19:200–207. doi: 10.1097/PPO.0b013e318292e4e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause M, Yaromina A, Eicheler W, Koch U, Baumann M. Cancer stem cells: targets and potential biomarkers for radiotherapy. Clin Cancer Res. 2011;17:7224–7229. doi: 10.1158/1078-0432.CCR-10-2639. [DOI] [PubMed] [Google Scholar]
- 9.Fertil B, Malaise EP. Inherent cellular radiosensitivity as a basic concept for human tumor radiotherapy. Int J Radiat Oncol Biol Phys. 1981;7:621–629. doi: 10.1016/0360-3016(81)90377-1. [DOI] [PubMed] [Google Scholar]
- 10.Malaise EP, Fertil B, Chavaudra N, Guichard M. Distribution of radiation sensitivities for human tumor cells of specific histological types: comparison of in vitro to in vivo data. Int J Radiat Oncol Biol Phys. 1986;12:617–624. doi: 10.1016/0360-3016(86)90071-4. [DOI] [PubMed] [Google Scholar]
- 11.West CM, Davidson SE, Roberts SA, Hunter RD. The independence of intrinsic radiosensitivity as a prognostic factor for patient response to radiotherapy of carcinoma of the cervix. Br J Cancer. 1997;76:1184–1190. doi: 10.1038/bjc.1997.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjork-Eriksson T, West C, Karlsson E, Mercke C. Tumor radiosensitivity (SF2) is a prognostic factor for local control in head and neck cancers. Int J Radiat Oncol Biol Phys. 2000;46:13–19. doi: 10.1016/s0360-3016(99)00373-9. [DOI] [PubMed] [Google Scholar]
- 13.Yaromina A, Krause M, Thames H, Rosner A, Krause M, Hessel F, Grenman R, Zips D, Baumann M. Pre-treatment number of clonogenic cells and their radiosensitivity are major determinants of local tumour control after fractionated irradiation. Radiother Oncol. 2007;83:304–310. doi: 10.1016/j.radonc.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 14.Hall EJ, Giaccia AJ. Radiobiology for the Radiologist. 7th edition. Philadelphia, PA: Lippincott Williams & Wilkins; 2011. [Google Scholar]
- 15.Brown JM, Wouters BG. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 1999;59:1391–1399. [PubMed] [Google Scholar]
- 16.Brown JM, Wilson G. Apoptosis genes and resistance to cancer therapy: what does the experimental and clinical data tell us? Cancer Biol Ther. 2003;2:477–490. doi: 10.4161/cbt.2.5.450. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence YR, Vikram B, Dignam JJ, Chakravarti A, Machtay M, Freidlin B, Takebe N, Curran WJ, Jr, Bentzen SM, Okunieff P, Coleman CN, Dicker AP. NCI-RTOG translational program strategic guidelines for the early-stage development of radiosensitizers. J Natl Cancer Inst. 2013;105:11–24. doi: 10.1093/jnci/djs472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colevas AD, Brown JM, Hahn S, Mitchell J, Camphausen K, Coleman CN. Development of investigational radiation modifiers. J Natl Cancer Inst. 2003;95:646–651. doi: 10.1093/jnci/95.9.646. [DOI] [PubMed] [Google Scholar]
- 19.Harrington KJ, Billingham LJ, Brunner TB, Burnet NG, Chan CS, Hoskin P, Mackay RI, Maughan TS, Macdougall J, McKenna WG, Nutting CM, Oliver A, Plummer R, Stratford IJ, Illidge T. Guidelines for preclinical and early phase clinical assessment of novel radiosensitisers. Br J Cancer. 2011;105:628–639. doi: 10.1038/bjc.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz D, Ito E, Liu FF. On the path to seeking novel radiosensitizers. Int J Radiat Oncol Biol Phys. 2009;73:988–996. doi: 10.1016/j.ijrobp.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Lin SH, George TJ, Ben-Josef E, Bradley J, Choe KS, Edelman MJ, Guha C, Krishnan S, Lawrence TS, Le QT, Lu B, Mehta M, Peereboom D, Sarkaria J, Seong J, Wang D, Welliver MX, Coleman CN, Vikram B, Yoo S, Chung CH. Opportunities and challenges in the era of molecularly targeted agents and radiation therapy. J Natl Cancer Inst. 2013;105:686–693. doi: 10.1093/jnci/djt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das AK, Bell MH, Nirodi CS, Story MD, Minna JD. Radiogenomics predicting tumor responses to radiotherapy in lung cancer. Semin Radiat Oncol. 2010;20:149–155. doi: 10.1016/j.semradonc.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Morsbach F, Sander D, Gheorghiu L, Nanda A, Benes C, Kriegs M, Krause M, Dikomey E, Baumann M, Dahm-Daphi J, Settleman J, Willers H. EGF Receptor Inhibition Radiosensitizes NSCLC Cells by Inducing Senescence in Cells Sustaining DNA Double-Strand Breaks. Cancer Res. 2011;71:6261–6269. doi: 10.1158/0008-5472.CAN-11-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, Archibald H, Raudales R, Tam A, Lee D, Rothenberg SM, Supko JG, Sordella R, Ulkus LE, Iafrate AJ, Maheswaran S, Njauw CN, Tsao H, Drew L, Hanke JH, Ma XJ, Erlander MG, Gray NS, Haber DA, Settleman J. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M, Kern AM, Hülskötter M, Greninger P, Singh A, Pan Y, Chowdhury D, Krause M, Baumann M, Benes CH, Efstathiou JA, Settleman J, Willers H. EGFR-Mediated Chromatin Condensation Protects KRAS-Mutant Cancer Cells Against Ionizing Radiation. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-13-3157. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gurtner K, Deuse Y, Butof R, Schaal K, Eicheler W, Oertel R, Grenman R, Thames H, Yaromina A, Baumann M, Krause M. Diverse effects of combined radiotherapy and EGFR inhibition with antibodies or TK inhibitors on local tumour control and correlation with EGFR gene expression. Radiother Oncol. 2011;99:323–330. doi: 10.1016/j.radonc.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Senra JM, Telfer BA, Cherry KE, McCrudden CM, Hirst DG, O'Connor MJ, Wedge SR, Stratford IJ. Inhibition of PARP-1 by olaparib (AZD2281) increases the radiosensitivity of a lung tumor xenograft. Mol Cancer Ther. 2011;10:1949–1958. doi: 10.1158/1535-7163.MCT-11-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauceri HJ, Sutton HG, Darga TE, Kocherginsky M, Kochanski J, Weichselbaum RR, Vokes EE. Everolimus exhibits efficacy as a radiosensitizer in a model of non-small cell lung cancer. Oncol Rep. 2012;27:1625–1629. doi: 10.3892/or.2012.1666. [DOI] [PubMed] [Google Scholar]
- 29.Konstantinidou G, Bey EA, Rabellino A, Schuster K, Maira MS, Gazdar AF, Amici A, Boothman DA, Scaglioni PP. Dual phosphoinositide 3-kinase/mammalian target of rapamycin blockade is an effective radiosensitizing strategy for the treatment of non-small cell lung cancer harboring K-RAS mutations. Cancer Res. 2009;69:7644–7652. doi: 10.1158/0008-5472.CAN-09-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim KW, Mutter RW, Cao C, Albert JM, Freeman M, Hallahan DE, Lu B. Autophagy for cancer therapy through inhibition of pro-apoptotic proteins and mammalian target of rapamycin signaling. J Biol Chem. 2006;281:36883–36890. doi: 10.1074/jbc.M607094200. [DOI] [PubMed] [Google Scholar]
- 31.Zaugg K, Rocha S, Resch H, Hegyi I, Oehler C, Glanzmann C, Fabbro D, Bodis S, Pruschy M. Differential p53-dependent mechanism of radiosensitization in vitro and in vivo by the protein kinase C-specific inhibitor PKC412. Cancer Res. 2001;61:732–738. [PubMed] [Google Scholar]
- 32.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 33.Fabbro D, Ruetz S, Bodis S, Pruschy M, Csermak K, Man A, Campochiaro P, Wood J, O'Reilly T, Meyer T. PKC412--a protein kinase inhibitor with a broad therapeutic potential. Anticancer Drug Des. 2000;15:17–28. [PubMed] [Google Scholar]
- 34.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 35.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W, Chen X, Liu XS, Brown M, Lim B, Weinberg RA. Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puck TT, Marcus PI. Action of x-rays on mammalian cells. J Exp Med. 1956;103:653–666. doi: 10.1084/jem.103.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz D, Ito E, Lau KS, Mocanu JD, Bastianutto C, Schimmer AD, Liu FF. Increased efficiency for performing colony formation assays in 96-well plates: novel applications to combination therapies and high-throughput screening. Biotechniques. 2008;44:ix–xiv. doi: 10.2144/000112757. [DOI] [PubMed] [Google Scholar]
- 39.Kahn J, Tofilon PJ, Camphausen K. Preclinical models in radiation oncology. Radiat Oncol. 2012;7:223. doi: 10.1186/1748-717X-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abazeed ME, Adams DJ, Hurov KE, Tamayo P, Creighton CJ, Sonkin D, Giacomelli AO, Du C, Fries DF, Wong KK, Mesirov JP, Loeffler JS, Schreiber SL, Hammerman PS, Meyerson M. Integrative radiogenomic profiling of squamous cell lung cancer. Cancer Res. 2013;73:6289–6298. doi: 10.1158/0008-5472.CAN-13-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin SH, Zhang J, Giri U, Stephan C, Sobieski M, Zhong L, Mason KA, Molkentine J, Thames HD, Yoo SS, Heymach JV. A High Content Clonogenic Survival Drug Screen Identifies MEK Inhibitors as Potent Radiation Sensitizers for KRAS Mutant Non-Small-Cell Lung Cancer. J Thorac Oncol. 2014;9:965–973. doi: 10.1097/JTO.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of radiosensitivity. Cancer Res. 1987;47:943–946. [PubMed] [Google Scholar]
- 43.Dai XF, Ding J, Zhang RG, Ren JH, Ma CM, Wu G. Radiosensitivity enhancement of human hepatocellular carcinoma cell line SMMC-7721 by sorafenib through the MEK/ERK signal pathway. Int J Radiat Biol. 2013;89:724–731. doi: 10.3109/09553002.2013.791405. [DOI] [PubMed] [Google Scholar]
- 44.Xiao W, Graham PH, Hao J, Chang L, Ni J, Power CA, Dong Q, Kearsley JH, Li Y. Combination therapy with the histone deacetylase inhibitor LBH589 and radiation is an effective regimen for prostate cancer cells. PLoS One. 2013;8:e74253. doi: 10.1371/journal.pone.0074253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Ejeh F, Kumar R, Wiegmans A, Lakhani SR, Brown MP, Khanna KK. Harnessing the complexity of DNA-damage response pathways to improve cancer treatment outcomes. Oncogene. 2010;29:6085–6098. doi: 10.1038/onc.2010.407. [DOI] [PubMed] [Google Scholar]
- 46.Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, Pawlak A, Mino-Kenudson M, Yeap BY, Riely GJ, Iafrate AJ, Arcila ME, Ladanyi M, Engelman JA, Dias-Santagata D, Shaw AT. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19:4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105:452–458. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumann M, Krause M. Targeting the epidermal growth factor receptor in radiotherapy: radiobiological mechanisms, preclinical and clinical results. Radiother Oncol. 2004;72:257–266. doi: 10.1016/j.radonc.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Krause M, Zips D, Thames HD, Kummermehr J, Baumann M. Preclinical evaluation of molecular-targeted anticancer agents for radiotherapy. Radiother Oncol. 2006;80:112–122. doi: 10.1016/j.radonc.2006.07.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.