Abstract
Objectives
To investigate the role of genetic variation in influencing the risk of metabolic complications associated with highly active anti-retroviral therapy (HAART).
Methods
Cluster analysis of metabolic traits of 189 patients enrolled in ACTG5005s, the metabolic sub-study of ACTG384, a clinical trial of HAART, was performed to identify a sub-group of subjects with increased risk of developing a cluster of metabolic abnormalities after exposure to HAART. Almost 300 single nucleotide polymorphisms (SNPs) in 135 candidate genes were evaluated for their association with this sub-group.
Results
A sub-group of patients was identified that had a normal metabolic profile at baseline but developed significantly elevated lipids and insulin resistance on HAART. This high-risk sub-group of patients also experienced significant body composition changes, particularly limb fat loss. Candidate gene analysis revealed that a single nucleotide polymorphism in resistin, a gene previously implicated in obesity and insulin resistance, was associated with this high-risk group (P = 0.0003).
Conclusions
Genetic variation in resistin is associated with metabolic complications caused by highly active anti-retroviral therapy.
Keywords: highly active antiretroviral therapy, HAART, dyslipidemia, lipoatrophy, pharmacogenetics, single nucleotide polymorphism, resistin
Introduction
Highly active anti retroviral (HAART) regimens can be associated with multiple metabolic abnormalities, including elevated lipids, glucose and insulin resistance, which are accompanied by changes in body composition such as visceral obesity and limb fat loss. Typically these abnormalities cluster together in a syndrome called human immunodeficiency virus (HIV) lipodystrophy [1,2]. With the reduction in morbidity and mortality resulting from HAART, these metabolic side-effects are of concern as they are well-known risk factors for future cardiovascular disease [3]. The etiology of HIV lipodystrophy is poorly understood, although some PIs may be more likely to cause metabolic abnormalities [2,4,5]. The genetic basis of HIV lipodystrophy is also unclear. Some studies have implicated nucleotide variation in ApoCIII, the β3 adrenergic receptor or TNF-alpha [6,7,8]. However, these studies focused only on one trait (e.g. triglycerides or insulin resistance) and examined selected SNPs in a single gene.
In this study, we employed a new approach to analyze metabolic data collected in a clinical trial of HAART to identify a sub-group of patients who developed marked lipid elevations and insulin resistance after exposure to HAART. The clustering approach we used to identify this sub-group is a powerful method for finding patterns in multi-variable data, and has been successfully employed to detect sub-groups of cancer patients based on gene expression profiles [9, 10]. We reasoned that this approach was particularly suited to the metabolic complications caused by HAART because of the apparently syndromic nature of the side-effects [1,2]. Having identified this sub-group of patients, we performed a comprehensive candidate gene analysis to implicate resistin, a gene previously associated with obesity and insulin resistance [11].
Methods
Subjects, genotyping and sequencing
189 individuals who participated in ACTG5005s [12] and who provided written informed consent for genetic analysis were analyzed in this study. Briefly, drug naive HIV+ patients were randomized to a backbone of NRTIs comprised of didanosine/stavudine or zidovudine/lamivudine [13,14]. In addition, subjects were randomized to receive a PI (nelfinavir), an NNRTI (efavirenz) or the combination. Metabolic traits were measured at baseline and then at 16 week intervals up to 64 weeks. Body fat was measured in all consenting patients using dual-energy x-ray absorptiometry at baseline and every 16 weeks on HAART. Genotyping was done using the TaqMan method [15]. The sequence of the entire resistin gene was determined in 47 high-risk individuals, and for comparison, up to 32 individuals from the diversity panel (M44PDR, Coriell repository) to identify SNPs.
Statistical analysis
Insulin resistance by homeostasis-model assessment (HOMA-IR) was calculated using the formula: fasting insulin (μU/mL) x fasting glucose (mmol/L)/22.5. Traits measured after 32 weeks of HAART were standardized to have a mean of 0 and variance of 1 and then used in the clustering. Traits measured at 32 weeks were selected for clustering because metabolic side-effects become apparent by this time-point and because later time-points had substantial numbers of missing values. For each individual a profile was made of body mass index, total, LDL, HDL and non-HDL cholesterol, triglyceride, glucose, and HOMA-IR. These profiles were clustered together using “two-step” clustering [16] as implemented in SPSS version 12 (Chicago, Illinois, US). In this clustering method, data are first grouped into sub-clusters using agglomerative clustering and then cluster assignment is refined to determine the optimal number of clusters. The optimal number of clusters was determined in two steps. First, Bayesian information criterion was calculated for the specified number of clusters to obtain an initial estimate of cluster number. Second, this estimate was refined by finding the greatest change in distance between two clusters in each stage of hierarchical clustering. The distance between two clusters was defined as the decrease in log-likelihood resulting from the two clusters being combined into a single cluster. Up to 20 clusters were permitted, but the algorithm repeatedly determined that a two cluster solution was optimal. To assess stability of the cluster solution, multiple runs of clustering (N = 20) were performed using randomly sorted data. All runs produced clusters identical to those described in this study. Differences in means between clusters for metabolic traits and changes in body fat over time were evaluated using Kruskal-Wallis and repeated measures ANOVA, respectively. Total, trunk and limb fat values were natural log transformed to approximate normality and then used in the repeated measures ANOVA. Genetic association between SNPs and clusters was assessed using Fisher’s exact test. Linkage disequilibrium between SNPs and haplotypes were analyzed using Haploview [17].
Results
Clustering of metabolic profiles to identify a sub-group of patients at increased risk of developing metabolic abnormalities after exposure to HAART
We analyzed 189 subjects, enrolled in a HAART clinical trial [13,14], who had longitudinal metabolic measurements, including lipids, glucose and insulin, at baseline and up to 64 weeks of treatment [12]. We applied a clustering approach to the analysis of metabolic traits to identify a sub-group of patients with pronounced changes in their metabolic profiles after treatment with HAART. Because of the stronger phenotypic differences displayed by such subgroups, we reasoned that these subgroups might show greater contrast, and thus increase the likelihood of finding a genetic association [18,19]. In addition, because HIV+ patients on HAART typically present with a cluster of metabolic abnormalities [1,2], it was logical to analyze these traits in unison as opposed to sequential genetic analysis of individual metabolic traits. Furthermore, this approach has the potential to reduce noise, as patients with multiple abnormalities are more likely to be truly affected than those with merely sporadic elevations of a single metabolic trait. Finally, since we analyzed all the metabolic traits at once in this multivariable analytic approach we reduced the number of statistical tests by an order of magnitude, thereby significantly increasing the power of the study, an important consideration given the limited number of samples that were available for genetic analysis.
For each patient, we created a metabolic profile comprised of body mass index, total, LDL, HDL and non-HDL cholesterol, triglycerides, glucose and insulin resistance by homeostasis model assessment (HOMA-IR), all measured after thirty-two weeks of HAART. We selected this time-point because metabolic abnormalities become apparent at this level of exposure and because later time-points had significant numbers of missing values. We then clustered these metabolic profiles such that individuals with similar metabolic profiles were grouped together. This process was repeated iteratively until all individuals were clustered into two groups. This approach is analogous to the widely used method of clustering gene expression profiles to identify subgroups of tumors or cell-types [9,10].
Examination of mean values of metabolic traits led to labeling one cluster “normal” (N = 142) and the other “high-risk” (N = 47; Table 1). At baseline, lipids and insulin resistance index by HOMA-IR were comparable between the two clusters, although the high-risk group was a little heavier and had higher mean lipid levels. After 32 weeks of HAART, the normal cluster (N = 142) continued to exhibit normal mean total cholesterol (180 ± 29 mg/dl), LDL (109 ± 28 mg/dl), triglycerides (142 ± 77 mg/dl), glucose (90 ± 9 mgdL) and insulin resistance by HOMA-IR (1.5 ± 0.9). Relative to this subgroup, the high-risk cluster (N = 47) experienced significant increases in mean total cholesterol (261 ± 45 mg/dl, P< 0.001), LDL (178 ± 30 mg/dl, P<0.001), triglycerides (310 ± 236 mg/dl, P< 0.001), glucose (102 ± 23 mg/dL, P<0.001) and insulin resistance by HOMA-IR (4.3 ± 5.6, P < 0.001). These mean lipid levels and HOMA-IR are considered clinically relevant hyperlipidemia and insulin resistance, respectively [20]. Both clusters experienced only modest and statistically insignificant increases in mean HDL levels after exposure to HAART.
Table 1.
Metabolic traits at baseline and after 32 weeks of HAART by cluster
| Trait | Mean ± SD | P value* | |
|---|---|---|---|
| Normal cluster N = 142 | High-risk cluster N = 47 | ||
| Total cholesterol, mg/dL | 159 ± 31 | 182 ± 33 | <0.001 |
| 180 ± 29 | 261 ± 45 | <0.001 | |
| LDL cholesterol, mg/dL | 98 ± 28 | 120 ± 30 | <0.001 |
| 109 ± 28 | 178 ± 30 | <0.001 | |
| Non-HDL cholesterol, mg/dL | 125 ± 30 | 148 ± 30 | <0.001 |
| 138 ± 28 | 221 ± 44 | <0.001 | |
| HDL cholesterol, mg/dL | 34 ± 10 | 34 ± 9 | 0.98 |
| 42 ± 14 | 40 ± 13 | 0.34 | |
| Triglycerides, mg/dL | 145 ± 121 | 176 ± 128 | 0.04 |
| 142 ± 77 | 310 ± 236 | <0.001 | |
| HOMA-IR | 2.1 ± 2.7 | 2.0 ± 1.5 | 0.52 |
| 1.5 ± 0.9 | 4.3 ± 5.6 | <0.001 | |
| Glucose, mg/dL | 85 ± 11 | 89 ± 32 | 0.94 |
| 90 ± 9 | 102 ± 23 | <0.001 | |
| Body mass index, kg/m2 | 24.4 ± 4.9 | 26.3 ± 5.1 | 0.01 |
| 25.3 ± 5.5 | 27.3 ± 5.5 | 0.01 | |
For each trait the baseline and week 32 values are indicated in the first and second row, respectively. Univariate P value from Kruskal-Wallis test
Body composition measurements as determined by dual energy X-ray absorptiometry (DEXA) were available for a subset (N =76) of the patients evaluated above. DEXA measurements were made at 16 week intervals up to 64 weeks because these time-points were consistent with patient visits planned in the trial. Moreover, at the time of this clinical trial prospective data about DEXA measurements and knowledge of how these measurements change over time were unavailable. Since then however, another study [21] employing only 40 subjects found significant changes in both lean and fat masses as measured by DEXA at 12 week intervals, thereby providing reassurance that the changes in DEXA measurements we note below are clinically relevant and unlikely to be simply fluctuations in DEXA values.
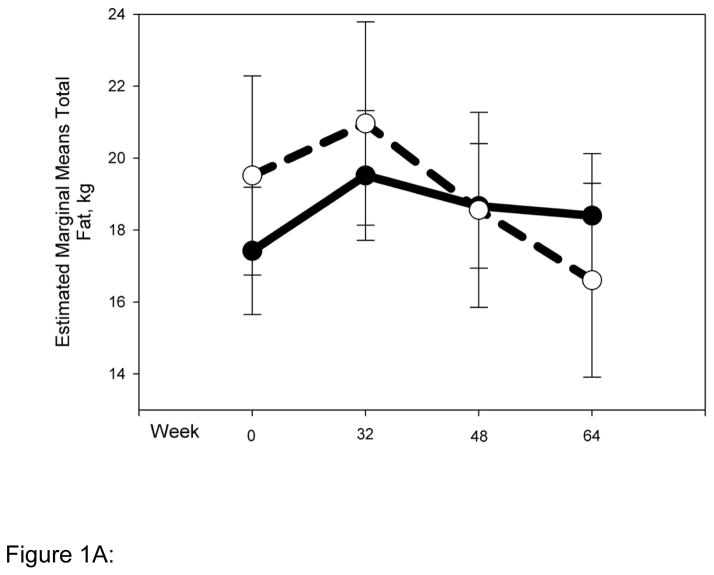
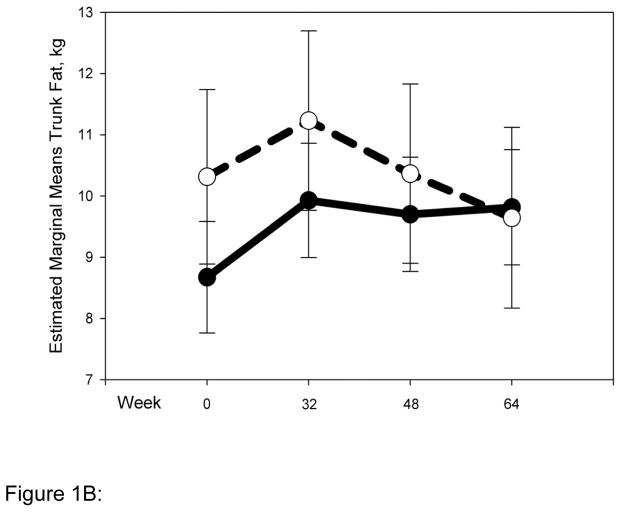
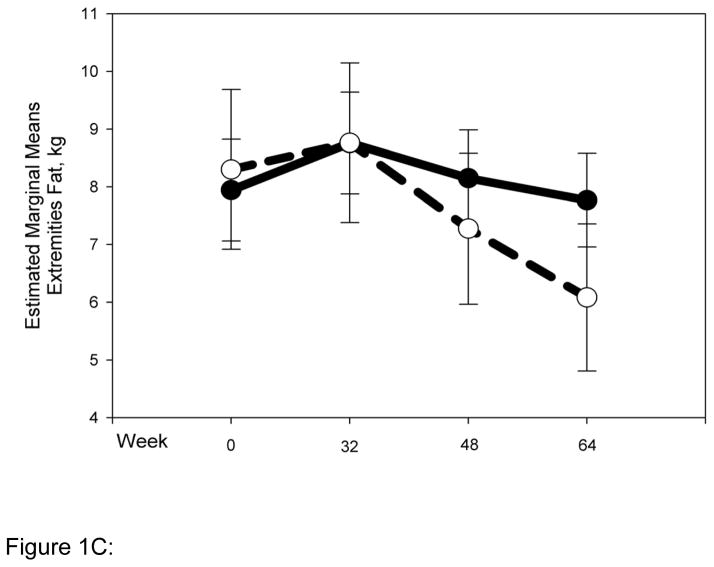
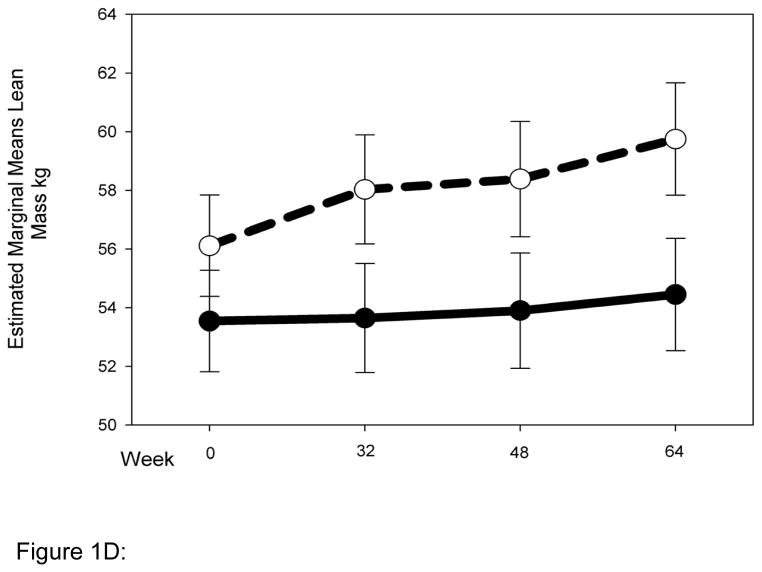
Striking differences in body composition over 64 weeks of HAART (Fig. 1 A-D) were observed between the normal (N = 54) and high-risk (N = 22) clusters described above. The high-risk group had higher mean total and trunk fat at baseline (Fig. 1 A, B), and both groups gained a little over 1 kg of fat up to 32 weeks. Beyond 32 weeks however, the normal cluster stabilized, whereas the high-risk cluster experienced reductions of ~5 kg and ~2.5 kg in mean total (P for interaction with cluster 0.01) and trunk fat (P for interaction with cluster 0.1), respectively. Changes in limb fat over time were even more striking (Fig. 1C). Both clusters had similar mean limb fat levels at baseline, and gained approximately 0.5 kg up to 32 weeks. The high-risk group then experienced a precipitous decline in mean limb fat or lipoatrophy of approximately 2.5 kg, but the normal cluster lost only approximately 0.5 kg of mean limb fat more gradually and returned to baseline by 64 weeks (P for interaction with cluster 0.001). The high-risk cluster had greater mean lean mass at baseline than the normal cluster. This mean lean mass increased by ~4 kg up to 64 weeks of HAART. In contrast, the normal cluster experienced little gain (0.5 kg) in mean lean mass for the duration of the study (P for interaction with cluster 0.009). This study does not address the mechanistic basis of this interesting difference in lean mass, and future studies will be needed to address this issue.
Figure 1.
Normal (N = 54) and high-risk (N = 22) clusters show markedly different body composition responses to HAART. Changes in body composition over time and interaction with clusters were analyzed using repeated measures ANOVA. Solid and dashed lines indicate values for the normal and high-risk clusters respectively. Error bars represent standard error of the mean. Although natural log transformed total, trunk and limb fat values were used in the repeated measures ANOVA to determine significance, for clarity untransformed values are provided in the graph (A) Total fat (P value for interaction with clusters = 0.01) (B) Trunk fat (P = 0.11) (C) Limb fat (P = 0.001) (D) Lean mass (P = 0.009).
Figure 1A: Change in total fat by cluster
Figure 1B: Change in trunk fat by cluster
Figure 1C: Change in limb fat by cluster
Figure 1D: Change in lean mass by cluster
Taken together, these results demonstrate that the clustering approach successfully classified patients into subgroups that displayed markedly different and clinically relevant metabolic and body composition responses to HAART.
Genetic association with cluster membership
We evaluated age, sex, race, HIV disease severity (baseline HIV copy number and CD4 cell-count) and individual drug treatment arm as potential predictors of high-risk cluster membership. None was significantly predictive (Table 2). Hepatitis C co-infection status or plasma markers of inflammation were not available for most patients, so were not evaluated as potential predictors.
Table 2.
No association between demographics, HIV disease severity, drug treatment arm and cluster membership
| Trait | Mean ± SD (Percent) | P value | |
|---|---|---|---|
| Normal cluster N = 142 | High-risk cluster N = 47 | ||
| Age, years | 38 ± 9 | 38 ± 10 | 0.9 |
| Sex, % men | 87 | 92 | 0.6 |
| Race, N (%) | 0.4 | ||
| White | 81 (57) | 24 (51) | |
| Black | 37 (26) | 16 (34) | |
| Hispanic | 20 (14) | 7 (15) | |
| Asian/PI | 4 (3) | 0 | |
| CD4 count, cells/mm3 | 257 ± 206 | 289 ± 208 | 0.4 |
| HIV copy number, log copies/mL | 5.1 ± 0.8 | 5.0 ± 0.8 | 0.6 |
| Intravenous drug use, never N (%) | 129 (91) | 44 (94) | 0.5 |
| Lipid-lowering treatment, N (%) | 1 (0.7) | 0 | 0.5 |
| Diabetes, N (%) | 1 (0.7) | 0 | 0.5 |
| Hypertension, N (%) | 8 (6) | 3 (6) | 0.9 |
| Anti-retroviral treatment arm, N (%) | 0.3 | ||
| Efavirenz + Didanosine + Stavudine | 30 (21) | 7 (15) | |
| Efavirenz + Zidovudine + Lamivudine | 24 (17) | 4 (9) | |
| Nelfinavir + Didanosine + Stavudine | 26 (18) | 6 (13) | |
| Nelfinavir + Zidovudine + Lamivudine | 17 (12) | 9 (19) | |
| Nelfinavir + Efavirenz + Didanosine + Stavudine | 26 (18) | 14 (30) | |
| Nelfinavir + Efavirenz + Zidovudine + Lamivudine | 19 (13) | 7 (15) | |
| Didanosine + Stavudine vs. Zidovudine + Lamivudine | 1 | ||
| Didanosine + Stavudine | 82 (58) | 27 (57) | |
| Zidovudine + Lamivudine | 60 (42) | 20 (43) | |
| Efavirenz vs. Nelfinavir vs. Efavirenz + Nelfinavir | 0.1 | ||
| Efavirenz | 54 (38) | 11 (23) | |
| Nelfinavir | 43 (30) | 15 (32) | |
| Efavirenz + Nelfinavir | 45 (32) | 21 (45) | |
We next examined if nucleotide variation in candidate genes was associated with cluster membership. We genotyped these 189 individuals for 299 SNPs in 135 candidate genes. We selected genes based on their likely involvement in regulating lipid and glucose metabolism, cytokines, drug metabolizing enzymes and transcription factors that regulate expression of these genes. We also selected genes whose expression in cell-culture was significantly perturbed after exposure to protease inhibitors [22]. To reduce genotyping effort and because the HapMap project [23] was not complete when this work was initiated, we concentrated our attention on SNPs that are more likely to affect the activity (e.g. coding sequence changes) or expression (promoter or near splice-sites) of the gene [24].
A SNP (dbSNP ID rs3219177) thirty-nine base-pairs downstream of the second exon of the resistin gene was highly significantly associated with cluster membership (Table 3). The frequency of this SNP was 0.16 in the normal cluster and 0.33 in the high-risk group (P = 0.0003; P = 0.04 after adjusting for 135 genes that were tested). Heterozygotes and homozygotes were 3 (95% C.I. 1.3–5.3) and 19 (95% C.I. 2–183) times more likely than wild-type to be classified in the high-risk cluster. The association was consistently observed in whites (P = 0.002) and blacks (P = 0.049), but because of the small number of hispanics (N = 27), the association was not significant in this race (Table 4); nonetheless heterozygotes were at consistently increased risk in whites (odds ratio, OR = 2.3), blacks (OR = 3.3) and hispanics (OR = 2.3) and there was no evidence of heterogeneity among the three races (P = 0.9). As noted below in the Discussion, resistin is a strong candidate for the metabolic changes observed in the high-risk cluster. For this reason, we decided to further investigate genetic variation at this locus.
Table 3.
Association between Resistin SNPs and Cluster Membership
| Resistin SNP genotypes | SNP Locationa | Number, (%) | Genotype frequency difference Pb | Allele frequency difference Pb | |
|---|---|---|---|---|---|
| Normal Cluster | High-risk Cluster | ||||
| rs3760678 | 7639120 | 0.03 | 0.01 | ||
| A/A | 114 (81) | 29 (64) | |||
| A/G | 23 (16) | 13 (29) | |||
| G/G | 3 (2) | 3 (7) | |||
| rs1862513 | 7639793 | 0.5 | 0.6 | ||
| C/C | 59 (42) | 16 (34) | |||
| C/G | 63 (45) | 26 (55) | |||
| G/G | 17 (12) | 5 (11) | |||
| rs3219175 | 7639855 | 0.09 | 0.03 | ||
| G/G | 122 (87) | 46 (98) | |||
| G/A | 15 (11) | 1 (2) | |||
| A/A | 3 (2) | 0 (0) | |||
| 5’UTR | 7640204 | 0.06 | 0.01 | ||
| G/G | 139 (100) | 45 (96) | |||
| G/A | 0 (0) | 2 (4) | |||
| rs3219177 | 7640369 | 0.001 | 0.0003 | ||
| C/C | 97 (69) | 20 (43) | |||
| C/T | 42 (30) | 23 (49) | |||
| T/T | 1 (1) | 4 (9) | |||
| INT1_NEW | 7640573 | 0.3 | 0.08 | ||
| A/A | 141 (100) | 45 (98) | |||
| A/G | 0 (0.8) | 1 (2) | |||
| IVS3+30C/T | 7640814 | 0.4 | 0.3 | ||
| C/C | 119 (87) | 36 (80) | |||
| C/T | 17 (12) | 9 (20) | |||
| T/T | 1 (1) | 0 (0) | |||
| rs3219178 | 7640951 | 0.7 | 0.6 | ||
| C/C | 63 (47) | 25 (53) | |||
| C/G | 61 (45) | 18 (38) | |||
| G/G | 11 (8) | 4 (9) | |||
Number indicates genomic nucleotide location of SNP based on reference sequence NT077812.
P values were determined from Fisher’s exact tests.
Table 4.
Association between resistin SNP rs3219177 and cluster membership by ethnicity
| Ethnicity | Number, (%) | P valuea | |
|---|---|---|---|
| Normal Cluster | High-risk Cluster | ||
| Caucasians | 0.002 | ||
| C/C | 55 (69) | 10 (42) | |
| C/T | 24 (30) | 10 (42) | |
| T/T | 1 (1) | 4 (16) | |
| African-American | 0.049 | ||
| C/C | 24 (67) | 6 (38) | |
| C/T | 12 (33) | 10 (62) | |
| T/T | 0 (0) | 0 (0) | |
| Hispanics | 0.332 | ||
| C/C | 15 (75) | 4 (57) | |
| C?T | 5 (25) | 3 (43) | |
| T/T | 0 (0) | 0 (0) | |
P value from Fisher’s exact test
To examine if other SNPs in the resistin gene were also associated with cluster membership we determined the sequence of the entire resistin gene in all 47 individuals classified as high-risk and, for comparison, up to 32 individuals from a human diversity panel. In addition, from dbSNP we selected SNPs that are located in phylogenetically conserved regions in the vicinity of the resistin gene. We genotyped the entire cohort for eight SNPs in resistin, including four newly identified by sequencing and two from phylogenetically conserved regions. Although three other SNPs were also associated with cluster membership (P < 0.05), none was as significantly associated as the SNP in intron 2 described above (Table 3) Furthermore, haplotype analysis revealed that only haplotypes bearing the intron 2 SNP, rs3219177, were significantly associated with cluster membership (data not shown). Taken together these results indicate that this SNP is potentially causative.
Discussion
By grouping patients with similar metabolic profiles after exposure to HAART using a powerful clustering approach, we identified a sub-group of patients that was especially vulnerable to the metabolic side-effects caused by HAART. This sub-group of patients had slightly elevated lipids at baseline, which were made worse after exposure to HAART, suggesting that HAART exacerbated underlying pre-disposition to dyslipidemia and insulin resistance.
As has been noted by others [25], “The idea that complex data can be grouped into clusters or categories is central to our understanding of the world, and this structure arises in many diverse contexts.” Similar clustering approaches have been employed widely in the analysis of whole genome expression arrays [9,10] to identify patterns in data that would not be apparent in univariate analysis. We believe such an approach is particularly suited to complex datasets with limited number of patients, such as the current study, to increase statistical power.
By performing a comprehensive candidate gene analysis we implicated resistin in the susceptibility to HIV lipodystrophy. Resistin was cloned as an adipocyte hormone that links obesity and insulin resistance [11]. Increased resistin resulted in mice with abnormal glucose tolerance [11]. Conversely, resistin knockout mice had lower fasting blood glucose [26]. SNPs in resistin have also been variably associated with metabolic traits captured by the clusters described above including body mass index, fat mass, insulin resistance and diabetes [27–31]. Engert et al. [27] examined SNPs in the 5’ flanking region of the resistin gene and found significant association between SNP rs1862513 and increased body mass index in non-diabetic obese subjects. Although this particular SNP was not significantly associated with cluster membership in this study, two flanking SNPs, rs321975 and rs3760678, which are 62 and 673 basepairs respectively from rs1862513 were significantly associated with high-risk cluster (Table 3). Wang et al. [28] demonstrated significant interaction between rs3219177 and BMI and insulin sensitivity, with the common CC genotype demonstrating greater mean insulin sensitivity. The association between this SNP and clusters which show significant differences in mean insulin resistance is consistent with this observation (Tables 1, 3). Sentinelli [29] did not find significant association between one SNP that they tested and diabetes status in a small case-control study. We did not detect this SNP in our population. Coneely et al. [30] found significant association between SNP rs3219177 and increased weight in elderly non-diabetic subjects, an observation that is completely consistent with the highly significant association we found between the same SNP and high-risk cluster, which also had increased mean BMI (Table 1). Osawa et al. [31] found significant association between a SNP in the promoter, rs1862513, and diabetes status. As noted above, we did not find signficant association with this particular SNP but two SNPs in proximity to this one were significantly associated with cluster membership in this study. More recently, the same group also found significant association between this SNP and plasma resistin levels, which in turn, were correlated with C-reactive protein in a Japanese population [32]. Taken together with the highly significant genetic association that we found, these genetic studies in non-HIV subjects support a strong link between genetic variation in resistin and the metabolic side-effects observed in the high-risk cluster. Because this SNP is located in non-coding sequence we hypothesize that it could influence expression of resistin, perhaps by affecting its splicing or transcription. Sequence comparison revealed that this SNP is located in a consensus binding-site for the T-cell transcription factor, TCF7L2, which has been previously implicated in susceptibility to Type II diabetes [33].
Additional work will be needed to understand how genetic variation in resistin might predispose individuals to HIV lipodystrophy. One possibility is that HIV+ patients on HAART who are in a chronic inflammatory state [34], might have increased resistin, as resistin gene expression can be induced by the pro-inflammatory molecules lipopolysaccharide and tumor necrosis factor alpha [35,36]. Individuals carrying the associated SNP in resistin might be particularly vulnerable to such changes in resistin gene expression. Given the link between inflammation and resistin levels and genotypes, it will be important in future studies of patients treated with HAART to determine circulating levels of inflammatory molecules including TNF-α, its soluble receptor and C-reactive protein in addition to resistin.
Our work suggests a potential basis for managing lipodystrophy in high-risk individuals. Since increased resistin is associated with obesity and insulin resistance, agents that reduce resistin including the marketed peroxisome proliferator-activated receptor agonists pioglitazone and rosiglitazone could potentially have a beneficial impact on HAART-induced lipodystrophy. In fact, consistent with this idea, rosiglitazone can reduce elevated circulating resistin levels in subjects with HIV lipodystrophy [37]. Another study did not find correlation between circulating serum resistin and lipodystrophy [38]; possibly because the different diagnostic criteria used to define lipodystrophy in that study led to subjects with normal lipid levels being classified as lipodystrophic. It is also possible that the paracrine effects of resistin are crucial to HIV lipodystrophy, and circulating resistin levels as measured in plasma in these studies do not accurately capture resistin levels at the site of its action.
In conclusion, we have used a clustering approach to identify a sub-group of patients with increased susceptibility to the metabolic side-effects cause by HAART. Genetic analysis revealed highly significant association between SNPs in the resistin gene and this subgroup of patients. Additional studies with larger cohorts exposed to other HAART regimens will be needed to determine the general applicability of these findings.
Acknowledgments
We thank patients for participating in the study. We thank David Haas and Cara-Ballard Sutcliffe at the Vanderbilt DNA repository for providing high-quality DNA. We thank T. Delmonte, L. Hui and E. Emison for help with genotyping and haplotype analyses and N. Dracopoli and R. Parker for helpful discussion.
Funding
This work was supported by grants to the Adult AIDS Clinical Trials Group from the National Institute of Allergy and Infectious Diseases (AI38558 and AI068636). The parent protocol (ACTG 384) was supported in part by Agouron Pharmaceuticals, Inc., Bristol-Myers Squibb Company, DuPont Pharmaceutical Company, GlaxoSmithKline, Inc., and Merck and Co., Inc. The genetic analysis was funded by Bristol-Myers Squibb Co.
Footnotes
Author contributions: PT, KM, WP, SG and MD ran the clinical trial and collected patients. KR, WG, OF, MN designed and executed the genetic analysis. KR, WG, OF, MN, PT, SG, MD drafted the manuscript.
Competing interests
KR, OF, WG are employees of Bristol-Myers Squibb Co. MN is currently employed by GlaxoSmithKline.
References
- 1.Carr A, Samaras K, Burton S, Law M, Freund J, Chisholm DJ, et al. A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance in patients receiving HIV protease inhibitors. AIDS. 1998;12:F51–8. doi: 10.1097/00002030-199807000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Grinspoon S, Carr A. Cardiovascular risk and body fat abnormalities in HIV infected individuals. N Engl J Med. 2005;352:48–62. doi: 10.1056/NEJMra041811. [DOI] [PubMed] [Google Scholar]
- 3.Friis-Moller N, Sabin CA, Weber R, d’Arminio Monforte A, El-Sadr WM, Reiss P, et al. Combination anti-retroviral therapy and the risk of myocardial infarction. N Engl J Med. 2003;349:1993–2003. doi: 10.1056/NEJMoa030218. [DOI] [PubMed] [Google Scholar]
- 4.Purnell JQ, Zambon A, Knopp RH, Pizzuti DJ, Achari R, Leonard JM, et al. Effect of ritonavir on lipids and post-heparin lipase activities in normal subjects. AIDS. 2000;14:51–7. doi: 10.1097/00002030-200001070-00006. [DOI] [PubMed] [Google Scholar]
- 5.Jemsek JG, Arathoon E, Arlotti M, Perez C, Sosa N, Pokrovskiy V, et al. Body fat and other metabolic effects of atazanavir and efavirenz, each administered in combination with zidovudine plus lamivudine, in antiretroviral-naive HIV-infected patients. Clin Infect Dis. 2006;42:273–80. doi: 10.1086/498505. [DOI] [PubMed] [Google Scholar]
- 6.Vonkeman HE, ten Napel CH, van Oeveren-Dybicz AM, Vermes I. Beta3-adrenergic receptor polymorphism and the antiretroviral therapy-related lipodystrophy syndrome. AIDS. 2000;14:1463–4. doi: 10.1097/00002030-200007070-00027. [DOI] [PubMed] [Google Scholar]
- 7.Maher B, Alfirevic A, Vilar FJ, Wilkins EG, Park BK, Pirmohamed M. TNF-alpha promoter region gene polymorphisms in HIV-positive patients with lipodystrophy. AIDS. 2002;15:2013–8. doi: 10.1097/00002030-200210180-00005. [DOI] [PubMed] [Google Scholar]
- 8.Foulkes AS, Wohl DA, Frank I, Puleo E, Restine S, Wolfe ML, et al. Associations among Race/Ethnicity, ApoC-III Genotypes, and Lipids in HIV-1-Infected Individuals on Antiretroviral Therapy. PLoS Med. 2006;3:e52. doi: 10.1371/journal.pmed.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–5. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 10.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 11.Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–12. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 12.Dube MP, Parker RA, Tebas P, Grinspoon SK, Zackin RA, Robbins GK, et al. Glucose metabolism, lipid, and body fat changes in antiretroviral-naive subjects randomized to nelfinavir or efavirenz plus dual nucleosides. AIDS. 2005;19:1807–18. doi: 10.1097/01.aids.0000183629.20041.bb. [DOI] [PubMed] [Google Scholar]
- 13.Robbins GK, De Gruttola V, Shafer RW, Smeaton LM, Snyder SW, Pettinelli C, et al. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349:2293–2303. doi: 10.1056/NEJMoa030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shafer RW, Smeaton LM, Robbins GK, De Gruttola V, Snyder SW, D’Aquila RT, et al. Comparison of four-drug regimens and pairs of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349:2304–2315. doi: 10.1056/NEJMoa030265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranade K, Chang MS, Ting CT, Pei D, Hsiao CF, Olivier M, et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–8. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, Ramakrishnan R, Livny M. BIRCH: A new data clustering algorithm and its applications. Data Mining and Knowledge Discovery. 1997;1:141–182. [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Risch N, Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science. 1995;268:1584–1589. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- 19.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 20.Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults. Executive Summary of the Third Report of the NCEP Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) JAMA. 2001;19:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 21.Mallon PWG, Miller J, Cooper DA, Carr A. Prospective evaluation of the effects of anti-retroviral therapy on body composition in HIV-1 infected men starting therapy. AIDS. 2003;17:971–979. doi: 10.1097/00002030-200305020-00005. [DOI] [PubMed] [Google Scholar]
- 22.Parker RA, Flint OP, Mulvey R, Elosua C, Wang F, Fenderson W, et al. Endoplasmic reticulum stress links dyslipidemia to inhibition of proteasome activity and glucose transport by HIV protease inhibitors. Mol Pharmacol. 2005;67:1909–19. doi: 10.1124/mol.104.010165. [DOI] [PubMed] [Google Scholar]
- 23.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33 (Suppl):228–37. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 25.Slonim N, Atwal GS, Tkacik G, Bialek W. Information based clustering. Proc Natl Acad Sci USA. 2005;51:18297–302. doi: 10.1073/pnas.0507432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee RR, Rangwala SM, Shapiro JS, Rich AS, Rhoades B, Qi Y, et al. Regulation of fasted blood glucose by resistin. Science. 2004;203:1195–98. doi: 10.1126/science.1092341. [DOI] [PubMed] [Google Scholar]
- 27.Engert JC, Vohl MC, Williams SM, Lepage P, Loredo-Osti JC, Faith J, et al. 5’ Flanking variants of resistin are associated with obesity. Diabetes. 2002;51:1629–34. doi: 10.2337/diabetes.51.5.1629. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Chu W, Hemphill C, Elbein SC. Human resistin gene: molecular scanning and evaluation of association with insulin sensitivity and type 2 diabetes in Caucasians. J Clin Endo Metab. 2002;87:2520–24. doi: 10.1210/jcem.87.6.8528. [DOI] [PubMed] [Google Scholar]
- 29.Sentinelli F, Romeo S, Arca M, Filippi E, Leonetti F, Banchieri M, et al. Human resistin gene, obesity, and type 2 diabetes mutational analysis and population study. Diabetes. 2002;51:860–62. doi: 10.2337/diabetes.51.3.860. [DOI] [PubMed] [Google Scholar]
- 30.Conneely KN, Silander K, Scott LJ, Mohlke KL, Lazaridis KN, Valle TT, et al. Variation in the resistin gene is associated with obesity and insulin-related phenotypes in Finnish subjects. Diabetologia. 2004;47:1782–88. doi: 10.1007/s00125-004-1537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osawa H, Yamada K, Onuma H, Murakami A, Ochi M, Kawata H, et al. The G/G genotype of resistin single-nucleotide polymorphism at -420 increases type 2 diabetes mellitus susceptibility by inducing promoter activity through specific binding of Sp1/3. Am J Hum Genet. 2004;75:678–86. doi: 10.1086/424761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osawa H, Tabara Y, Kawamoto R, Ohashi J, Ochi M, Onuma H, et al. Plasma resistin, associated with single nucleotide polymorphism -420, is correlated with insulin resistance, lower HDL cholesterol, and high-sensitivity C-reactive protein in the Japanese general population. Diabetes Care. 2007;30:1501–6. doi: 10.2337/dc06-1936. [DOI] [PubMed] [Google Scholar]
- 33.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 34.Ledru E, Christeff N, Patey O, de Truchis P, Melchior JC, Gougeon ML. Alteration of tumor necrosis factor-a T cell homeostasis following potent anti-retroviral therapy: contribution to the development of human immunodeficiency virus-associated lipodystrophy syndrome. Blood. 2000;95:3191–3198. [PubMed] [Google Scholar]
- 35.Lehrke M, Reilly MP, Millington SC, Iqbal N, Rader DJ, Lazar MA. An inflammatory cascade leading to hyperresistinemia in humans. PLoS Med. 2004;2:e45. doi: 10.1371/journal.pmed.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reilly MP, Lehrke M, Wolfe ML, Rohatgi A, Lazar MA, Rader DJ. Resistin is an inflammatory marker of atherosclerosis in humans. Circulation. 2005;111:932–9. doi: 10.1161/01.CIR.0000155620.10387.43. [DOI] [PubMed] [Google Scholar]
- 37.Kamin D, Hadigan C, Lehrke M, Mazza S, Lazar MA, Grinspoon S. Resistin levels in human immunodeficiency virus-infected patients with lipoatrophy decrease in response to rosiglitazone. J Clin Endocrinol Metab. 2005;90:3423–6. doi: 10.1210/jc.2005-0287. [DOI] [PubMed] [Google Scholar]
- 38.Barb D, Wadhwa SG, Kratzsch J, Gavrila A, Chan JL, Williams CJ, et al. Circulating resistin levels are not associated with fat redistribution, insulin resistance or metabolic profile in patients with the highly active anti-retroviral therapy-induced metabolic syndrome. J Clin Endocrinol Metab. 2005;90:5324–5328. doi: 10.1210/jc.2005-0742. [DOI] [PubMed] [Google Scholar]