Abstract
Alzheimer’s disease (AD) is a chronic neurodegenerative disorder characterized by progressive neuropathology and cognitive decline. We describe a cross-tissue analysis of methylomic variation in AD using samples from three independent human post-mortem brain cohorts. We identify a differentially methylated region in the ankyrin 1 (ANK1) gene that is associated with neuropathology in the entorhinal cortex, a primary site of AD manifestation. This region was confirmed as significantly hypermethylated in two other cortical regions (superior temporal gyrus and prefrontal cortex) but not in the cerebellum, a region largely protected from neurodegeneration in AD, nor whole blood obtained pre-mortem, from the same individuals. Neuropathology-associated ANK1 hypermethylation was subsequently confirmed in cortical samples from three independent brain cohorts. This study represents the first epigenome-wide association study (EWAS) of AD employing a sequential replication design across multiple tissues, and highlights the power of this approach for identifying methylomic variation associated with complex disease.
Alzheimer’s disease (AD) contributes significantly to the global burden of disease affecting in excess of 26 million people worldwide1,2. The pathogenesis associated with AD is characterized by the accumulation of amyloid plaques, tangles of intracellular hyperphosphorylated tau, gliosis, synaptic dysfunction and eventually cell death3,4. Although the neuropathological manifestation of AD is well characterized in post-mortem brain, little is known about the underlying risk factors or mechanism(s) involved in disease progression. Of note, different parts of the brain show differential vulnerability to AD; although there is progressive neurodegeneration across the cortex with areas such as the entorhinal cortex (EC) being characterized by considerable and early neuropathology, regions such as the cerebellum (CER) are relatively resistant to neuronal damage, with little or no plaque or neurofibrillary tangle pathology5.
Contemporary research aimed at exploring the etiology of AD has focused primarily on DNA sequence variation, with some notable success6. Increasing knowledge about the biology of the genome7 also implicates an important role for epigenetic variation in human health and disease, and recent methodological advances mean that epigenome-wide association studies (EWAS) are now feasible for complex disease phenotypes including AD8. Epigenetic epidemiology is a relatively new endeavor, however, and there are important considerations regarding study design, tissue-type, analysis strategy and data interpretation9,10. Here we describe the first systematic cross-tissue EWAS analysis of DNA methylation in AD using a powerful sequential replication design, with the goal of identifying disease-associated methylomic variation across pathologically-relevant regions of the brain.
The first (‘discovery’) stage of our analysis utilized multiple tissues from donors (N = 117) archived in the MRC London Brainbank for Neurodegenerative Disease. From each donor, genomic DNA was isolated from four brain regions (EC, superior temporal gyrus (STG), prefrontal cortex (PFC) and CER) and, where available, whole blood obtained pre-mortem (Supplementary Table S1 and Supplementary Table S2). DNA methylation was quantified using the Illumina 450K HumanMethylation array, with pre-processing, normalization and stringent quality control undertaken as previously described11 (see Online Methods). Our analyses focussed on identifying differentially-methylated positions (DMPs) associated with Braak staging, a standardized measure of neurofibrillary tangle burden determined at autopsy12, with all analyses controlling for age and sex.
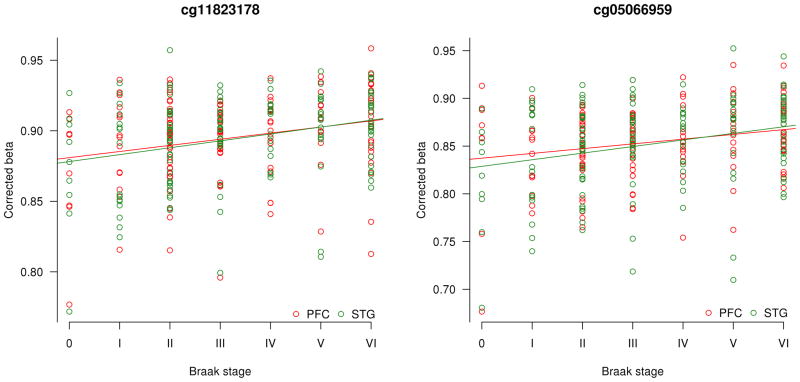
We first assessed DNA methylation differences identified in the EC, given that it is a primary and early site of neuropathology in AD5. The top-ranked Braak-associated EC DMPs are shown in Table 1 and Supplementary Table S3, with results for the other brain regions profiled (STG, PFC, and CER) shown in Supplementary Tables S4–S6. Two of the top-ranked EC DMPs (cg11823178, the top-ranked EC DMP, and cg05066959, the fourth-ranked EC DMP) are located just 91bp away from each other within the ankyrin 1 (ANK1) gene on chromosome 8, encoding a brain-expressed protein13 involved in compartmentalization of the neuronal plasma membrane14 (Fig. 1a). These DMPs are also located proximal to the NKX6-3 gene, encoding a homeodomain transcription factor involved in development of the brain15,16. Increased EC DNA methylation at both CpG sites is associated with Braak stage (cg11823178: r = 0.47, t(102) = 5.39, P = 4.59E–7; cg05066959: r = 0.41, t(102) = 5.37, P = 1.34E–5) (Fig. 1b). As AD is characterized by significant neuronal loss we used an in silico algorithm to confirm that the observed association is not confounded by differences in neuronal proportions between individuals17; both CpG sites remain significantly associated with Braak score after correction for estimated cellular heterogeneity (cg11823178: P = 7.09E–7; cg05066959: P = 6.20E–6) (Table 1). We used comb-p18 to identify spatially-correlated regions of differential DNA methylation, highlighting a Braak-associated DMR spanning these CpG sites (P = 6.04E–7) (Supplementary Table S7). Hypermethylation at both DMPs is significantly associated with Braak score in the STG (cg11823178: r = 0.37, t(111) = 4.15, P = 6.51E–5; cg05066959: r = 0.33, t(111) = 3.67, P = 3.78E–4) and the PFC (cg11823178: r = 0.29, t(108) = 3.12, P = 2.33E–3; cg05066959: r = 0.32, t(108) = 3.52, P = 6.48E–4) (Fig. 1c). In contrast, no significant neuropathology-associated hypermethylation is detected at either CpG site in the CER (cg11823178: r = 0.01, t(106) = 0.082, P = 0.935; cg05066959: r = −0.08, t(106) = 0.085, P = 0.395) (Fig. 1d), a region largely protected from neurodegeneration in AD, nor is elevated DNA methylation at either site associated with AD diagnosis in whole blood collected pre-mortem (data not shown).
Table 1. The ten top-ranked Braak-associated DMPs in EC.
Shown for each DMP are chromosomal location (hg19), up/downstream genes, P value from our quantitative model (see Online Methods), difference (Δ) in corrected DNA methylation (%) between individuals with the lowest (score 0) and highest (score VI) Braak score, and CETS p-value17 to highlight whether these top-ranked DMPs are mediated by the effect of differential neuronal cell proportions across samples. Also shown are the corresponding statistics across the three other matched brain regions (STG, PFC, CER) in the London cohort and the STG and PFC in the Mount Sinai ‘replication’ cohort for CpG sites showing a nominally significant difference in the same direction. The 100 top-ranked EC DMPs are given in Supplementary Table S3, with data for other brain regions given in Supplementary Tables S4–S6.
| London Cohort | Mount Sinai Cohort | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC | STG | PFC | CER | STG | PFC | ||||||||||||
| Probe | Position | Illumina Gene Annotation |
Genes with TSS within 5Kb upstream |
Genes with TSS within 1Kb downstream |
P value | Δ | CETS- adjusted P |
P value | Δ | P value | Δ | P value | Δ | P value | Δ | P value | Δ |
| cg11823178 | 8:41519399 | ANK1; MIR486 | NKX6-3 | ANK1 | 4.59E–07 | 4.62 | 7.09E–07 | 6.51E–05 | 3.12 | 2.33E–03 | 2.05 | - | - | 1.63E–04 | 3.36 | 1.07E–03 | 2.97 |
| cg22997194 | 10:72647819 | PCBD1 | PCBD1 | - | 9.99E–07 | −3.89 | 0.05 | - | - | - | - | - | - | - | - | - | - |
| cg06653632 | 12:129281444 | SLC15A4 | SLC15A4 | TMEM132C | 1.98E–06 | 3.51 | 5.74E–07 | 5.02E–03 | 2.01 | 0.0381 | 1.42 | - | - | 9.21E–04 | 2.40 | 0.0124 | 1.89 |
| cg05066959 | 8:41519308 | ANK1; MIR486 | NKX6-3 | ANK1 | 1.35E–05 | 5.45 | 6.20E–06 | 3.78E–04 | 4.03 | 6.48E–04 | 3.45 | - | - | 5.78E–04 | 4.75 | 4.00E–03 | 3.41 |
| cg24152732 | 19:4180820 | SIRT6 | SIRT6 | CREB3L3 | 1.37E–05 | −3.11 | 2.45E–06 | - | - | - | - | 0.0429 | −1.86 | - | - | - | - |
| cg14972141 | 13:100217995 | - | CLYBL | TM9SF2 | 1.48E–05 | −4.01 | 0.0025 | 3.41E–04 | −2.25 | 0.0179 | −2.23 | 7.86E–04 | −2.30 | - | - | - | - |
| cg04029027 | 7:130125811 | MEST | CEP41 | MEST | 2.29E–05 | 3.41 | 2.06E–05 | 0.0292 | 1.92 | - | - | - | - | - | - | - | - |
| cg05030077 | 16:2255199 | MLST8 | MLST8 | - | 2.69E–05 | −1.89 | 0.12 | - | - | - | - | - | - | - | - | - | - |
| cg04151012 | 2:27806529 | ZNF512 | ZNF512 | - | 2.88E–05 | 3.21 | 1.39E–05 | - | - | - | - | - | - | - | - | - | - |
| cg18522315 | 20:8000623 | TMX4 | TMX4 | - | 3.27E–05 | 1.75 | 9.18E–05 | 0.0422 | 0.74 | - | - | - | - | - | - | - | - |
FIGURE 1. Cortex-specific hypermethylation of ANK1 is correlated with AD-associated neuropathology in the brain.


Linear regression models demonstrated that a) cg11823178 in ANK1 is the top-ranked neuropathology-associated differentially methylated position (DMP) in the EC in the London discovery cohort (N = 104). The adjacent probe, cg05066959, is also significantly associated with neuropathology. Green bars denote the location of annotated CpG islands. b) EC DNA methylation at both CpG sites is strongly associated with Braak score (cg11823178: r = 0.47, t(102) = 5.39, P = 4.59E–7; cg05066959: r = 0.41, t(102) = 5.37, P = 1.34E–5). c) Both probes are also associated with neuropathology in the other cortical regions assessed in the same individuals, being significantly correlated with Braak score in the STG (N = 113) (cg11823178: r = 0.37, t(111) = 4.15, P = 6.51E–5; cg05066959: r = 0.33, t(111) = 3.67, P = 3.78E–4) and the PFC (N = 110) (cg11823178: r = 0.29, t(108) = 3.12, P = 2.33E–3; cg05066959: r = 0.32, t(108) = 3.52, P = 6.48E–4). d) There is no association between DNA methylation and Braak score at either ANK1 probe in the CER (N = 108) (cg11823178: r = 0.01, t(106) = 0.082, P = 0.935; cg05066959: r = −0.08, t(106) = 0.085, P = 0.395), a region largely protected against AD-related neuropathology. e) cg11823178 is the top-ranked cross-cortex DMP (Fisher’s χ2(6) = 60.6, P = 3.42E–11), with cg05066959 also strongly associated with Braak score (Fisher’s χ2(6) = 52.9, P = 1.24E–9).
Interestingly, we observe significant overlap in Braak-associated DMPs across the three cortical regions profiled in the London ‘discovery’ cohort; 38 (permuted P-value < 0.005) and 30 (permuted P-value <0.005) of the 100 top-ranked EC probes are significantly differentially methylated in the same direction in the STG and PFC, respectively (Supplementary Table S8), with a highly significant correlation of top-ranked Braak-associated DNA methylation scores across these sites (EC vs STG: r = 0.88, P = 6.73E–14; EC vs PFC: r = 0.83, P = 8.77E–13). There is, however, a clear distinction between cortical regions and CER, with the top-ranked CER DMPs appearing to be more tissue–specific and not differentially methylated in cortical regions (permuted P-values for enrichment all > 0.05), although ~15% of the top-ranked cortical DMPs are differentially methylated in CER (permuted P-values all ≤ 0.01), indicating that these represent relatively pervasive AD-associated changes that are observed across multiple tissues. We subsequently used a meta-analysis method (see Online Methods) to highlight consistent Braak-associated DNA methylation differences across all three cortical regions in the ‘discovery’ cohort. The top-ranked cross-cortex DMPs are shown in Table 2 and Supplementary Table S9, with DMRs identified using comb-p listed in Supplementary Table S10. Of note, cg11823178 is the most significant cross-cortex DMP (Δ = 3.20, Fisher’s P = 3.42E–11, Brown’s P = 1.00E–6), with cg05066959 again ranked fourth (Δ = 4.26, Fisher’s P = 1.24E–9, Brown’s P = 6.24E–6) (Fig. 1e) and a DMR spanning these probes being associated with neuropathology (Sidak-corrected P = 3.39E–4) (Supplementary Table S10). Together, these data suggest that cortical DNA hypermethylation at the ANK1 locus is robustly associated with AD-related neuropathology.
Table 2. The ten top-ranked cross-cortex Braak-associated DMPs.
Shown for each DMP are chromosomal location (hg19), up/downstream genes, Fisher’s P-value, Brown’s P-value, and cross-cortex estimate (Δ) of methylation difference between individuals with the lowest (score 0) and highest (score VI) Braak stage. Also shown are differences (Δ) between Braak 0 and Braak IV and corresponding P values from our individual cortex models in the London discovery cohort (see Online Methods) and the STG and PFC in the Mount Sinai replication cohort for CpG sites showing a nominally significant difference in the same direction. The 100 top-ranked cross-cortex Braak-associated DMPs are given in Supplementary Table S9.
| Probe Information | London Cohort | Mount Sinai Cohort | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cross-cortex model | EC | STG | PFC | CER | STG | PFC | |||||||||||||
| Probe | Position | Illumina Annotation | Genes with TSS within 5Kb upstream |
Genes with TSS within 1Kb downstream |
Fisher’s P Value |
Brown’s P Value |
Δ | P value | Δ | P value | Δ |
P value |
Δ | P value | Δ | P value | Δ | P value | Δ |
| cg11823178 | 8:41519399 | ANK1;MIR486 | NKX6-3 | ANK1 | 3.42E–11 | 1.00E–06 | 3.20 | 4.59E–07 | 4.62 | 6.51E–05 | 3.12 | 2.33E–03 | 2.05 | - | - | 1.63E–04 | 3.36 | 1.07E–03 | 2.97 |
| cg25018458 | 17:980014 | ABR | TIMM22 | ABR | 1.89E–10 | 9.91E–07 | 1.34 | 1.05E–04 | 1.29 | 4.36E–06 | 1.59 | 9.48E–04 | 1.12 | - | - | 4.46E–05 | 1.82 | 2.14E–04 | 1.93 |
| cg05810363 | 17:74475270 | RHBDF2 | RHBDF2 | AANAT | 9.42E–10 | 9.15E–06 | 3.36 | 1.37E–04 | 3.79 | 2.26E–05 | 3.58 | 7.93E–04 | 2.76 | - | - | 3.55E–03 | 3.65 | 5.54E–03 | 3.19 |
| cg05066959 | 8:41519308 | ANK1;MIR486 | NKX6-3 | ANK1 | 1.24E–09 | 6.24E–06 | 4.26 | 1.35E–05 | 5.45 | 3.78E–04 | 4.03 | 6.48E–04 | 3.45 | - | - | 5.78E–04 | 4.75 | 4.00E–03 | 3.41 |
| cg18428542 | 2:91787196 | - | ACTR3BP2 | - | 1.94E–09 | 2.53E–05 | −4.00 | 2.09E–04 | −4.05 | 1.73E–04 | −3.93 | 1.47E–04 | −4.04 | 7.46E–05 | −4.43 | - | - | - | - |
| cg16665310 | 13:100218136 | - | CLYBL | TM9SF2 | 6.11E–09 | 1.11E–04 | −5.73 | 1.39E–04 | −6.71 | 1.17E–04 | −5.15 | 1.13E–03 | −5.50 | 2.32E–04 | −3.84 | - | - | - | - |
| cg05912299 | 13:100217962 | - | CLYBL | TM9SF2 | 7.17E–09 | 1.38E–04 | −6.03 | 6.76E–05 | −7.46 | 3.67E–03 | −4.64 | 8.85E–05 | −6.29 | 9.86E–04 | −5.63 | - | - | - | - |
| cg03169557 | 16:89598950 | SPG7 | RPL13 | SPG7 | 7.95E–09 | 7.91E–06 | 1.63 | 3.01E–04 | 1.31 | 1.35E–04 | 1.97 | 6.05E–04 | 1.52 | - | - | 2.85E–03 | 2.11 | 7.98E–04 | 2.34 |
| cg23968456 | 10:73521631 | CDH23;C10orf54 | C10orf105 | C10orf54 | 1.09E–08 | 1.09E–05 | 1.22 | 1.89E–03 | 0.87 | 2.21E–03 | 1.13 | 8.27E–06 | 1.62 | - | - | 3.07E–03 | 1.72 | 0.0416 | 1.05 |
| cg02672452 | 2:91818189 | - | ACTR3BP2 | - | 1.16E–08 | 1.27E–04 | −4.42 | 4.71E–05 | −5.29 | 2.35E–03 | −3.64 | 3.33E–04 | −4.49 | 6.75E–04 | −5.87 | - | - | - | - |
A cortical ‘replication’ dataset was generated using DNA isolated from two regions (STG and PFC) obtained from a cohort of brains archived in the Mount Sinai Alzheimer’s Disease and Schizophrenia Brain Bank (N = 144) with detailed neuropathology data including Braak staging and amyloid burden (see Online Methods)19. Strikingly, Braak-associated DNA methylation scores for the 100 top-ranked cross-cortex DMPs identified in the London discovery cohort (listed in Supplementary Table 9) are strongly correlated with neuropathology-associated differences at the same probes in both cortical regions profiled in the Mount Sinai replication cohort (STG Braak score: r = 0.63, P = 2.66E–12; PFC Braak score: r = 0.64, P = 6.03E–13; STG amyloid burden: r = 0.46, P = 1.09E–6; PFC amyloid burden: r = 0.65, P = 2.87E–13) (see Fig. 2a). Furthermore, increased DNA methylation at each of the two ANK1 CpG sites is significantly associated with elevated Braak staging (Table 1, Fig. 2b) and amyloid burden (Fig. 2c) in both cortical regions. To further confirm the association between cortical ANK1 hypermethylation and neuropathology we used bisulfite-pyrosequencing to quantify DNA methylation across an extended region spanning eight CpG sites, including cg11823178 and cg05066959 in DNA extracted from a third independent collection of matched EC, STG and PFC tissue (N = 62) obtained from the Thomas Willis Oxford Brain Collection20 (see Online Methods and Supplementary Table S11a). Average DNA methylation across this region was significantly elevated in all three cortical regions tested (EC: P = 0.0004; STG: P = 0.0008; PFC: P = 0.014) in affected individuals (Supplementary Fig. S1), most notably in the EC where six of the eight CpG sites assessed are characterized by significant AD-associated hypermethylation (Fig. 2d). A meta-analysis of cg11823178 and cg05066959 across all three independent cohorts confirms consistent neuropathology-associated hypermethylation in each of the cortical regions assessed (Fig. 2e and Fig. 2f). Further evidence to support our conclusions comes from an independent EWAS of AD pathology in 708 cortical samples (De Jager et al., co-submitted article21). There is a significant correlation (r = 0.57, P = 1.55E–9) between the DNA methylation changes identified in our cross-cortex analyses and neuropathology-associated differences at the same probes in the study by De Jager et al. (Fig. 2g)21. Conversely, neuropathology-associated DNA methylation scores for top-ranked DMPs in the study of De Jager and colleagues are strongly correlated (r = 0.49, P = 7.8E–10) with those observed using the cross-cortex model for the same probes in our discovery cohort (Supplementary Fig. S2). In particular, De Jager et al. also identify a highly significant association between elevated DNA methylation at cg11823178 and cg05066959 and AD-related neuropathology. Together, these data provide compelling evidence for an association between ANK1 hypermethylation and the neuropathological features of AD, specifically in the cortical regions associated with disease manifestation. Although not previously implicated in dementia, genetic variation in ANK1 is associated with diabetic phenotypes22–24, an interesting observation given the established links between type 2 diabetes and AD25.
FIGURE 2. Neuropathology-associated DMPs are consistent across sample cohorts, with replicated evidence for ANK1 hypermethylation.
a) Braak-associated DNA methylation scores for the top-ranked cross-cortex DMPs identified using linear regression models in the London discovery cohort (listed in Supplementary Table S9) are significantly correlated with neuropathology-associated differences at the same probes in both cortical regions profiled in the Mount Sinai replication cohort using linear regression models (PFC (N =142) Braak score: r = 0.64, P = 6.03E–13; STG (N = 144) Braak score: r = 0.63, P = 2.66E–12; PFC amyloid burden: r = 0.65, P = 2.87E–13; STG amyloid burden: r = 0.46, P = 1.09E–6). Shown is data for Mount Sinai PFC Braak score analysis, with the two ANK1 probes (cg11823178 and cg05066959) highlighted in red. cg11823178 and cg05066959 are significantly associated with b) Braak score in the STG (cg11823178: r = 0.28, t(142) = 3.62, P = 1.63E–04; cg05066959: r = 0.25, t(142) = 3.29, P = 5.78E–04) and PFC (cg11823178: r = 0.24, t(140) = 3.14, P = 1.07E–03; cg05066959: r = 0.21, t(140) = 2.75, P = 4.00E–03) and also c) amyloid pathology in the STG (cg11823178: r = 0.21, t(142) = 2.81, P = 4.99E–04; cg05066959: r = 0.27, t(142) = 3.47, P =5.65E–04) and PFC (cg11823178: r = 0.29, t(140) = 3.69, P = 2.35E–04; cg05066959: r = 0.19, t(140) = 2.56, P = 9.93E–03). In the Oxford replication cohort, bisulfite–pyrosequencing was used to quantify DNA methylation across eight CpG sites spanning an extended ANK1 region. Linear models, adjusting for age and gender, confirmed significant neuropathology-associated hypermethylation in all three cortical regions assessed (see Supplementary Fig. S1), d) most notably in the EC (N=51), where six of the eight CpG sites showed a significant (amplicon average P = 0.0004) neuropathology-associated increase in DNA methylation (data is represented as mean +/− SEM, with *=p<0.05, **=p<0.01, and ***=p<0.005). Meta-analyses across the three sample cohorts (London, Mount Sinai and Oxford) confirms Braak-associated cortex-specific hypermethylation for both e) cg11823178 and f) cg05066959. Finally, there is striking consistency in neuropathology-associated DMPs identified in our discovery cohort and those identified in the co-submitted study by De Jager and colleagues. g) Braak-associated DNA methylation scores for the 100 top-ranked cross-cortex DMPs identified in the London discovery cohort are significantly correlated with neuropathology-associated differences (neuritic-plaque load) at the same probes in the dorsolateral prefrontal cortex (DLPFC) identified by De Jager and colleagues in 708 individuals (r = 0.57, P = 1.55E–9)21. The two ANK1 probes (cg11823178 and cg05066959) are highlighted in red.
ANK1 is a transcriptionally complex gene, with multiple isoforms and several alternative promoters identified (Supplementary Fig. S3). Given the established role of DNA methylation in regulating isoform-specific gene expression we examined whether AD neuropathology was associated with the differential abundance of various ANK1 isoforms in the EC using qPCR (see Online Methods). Briefly, three assays with specificity to ANK1 isoforms i) 1,2,3 and 4, ii) 9, and iii) 5,7 and 10 (Supplementary Table S11b) were used to profile 36 EC samples from whom high quality RNA was available (see Supplementary Table S2). Our linear model highlighted a significant association (P = 0.04) between the abundance of isoforms 5,7 and 10 transcripts and AD-associated neuropathology (Supplementary Fig. S4). No significant differences in transcript levels were observed for the other two isoform-specific assays (data not shown).
As a definitive diagnosis of AD can only be made via neuropathological examination at autopsy, there is considerable interest in the identification of clinical biomarkers that may have both diagnostic and prognostic utility during the early stages of the disorder26,27. Recent work has identified several transcriptomic blood biomarkers for AD with potential clinical utility for the early diagnosis of the disease28–32. In this study we had access to matched pre-mortem whole blood DNA for methylomic profiling from a subset of samples in the London ‘discovery’ cohort (N=93). Because of the duration elapsed between blood sampling and mortality (average = 4.15 +/− 3.00 years), analyses on these data were restricted to the identification of DMPs associated with a clinical diagnosis of AD, rather than Braak score. We identified a number of AD-associated DMPs (Supplementary Table S12) in pre-mortem blood, many in the vicinity of genes of relevance to AD including DAPK1 (cg14067233), previously implicated in genetic studies33,34; GAS1 (cg14067233), an APP-interacting protein involved in the control of APP maturation and processing35; and NDUFS5 (cg17074958), a mitochondrial gene previously shown to be differentially expressed in AD blood36. Our data suggest, however, that the top-ranked DMPs in blood are distinct to those identified in the brain; there is no significant overlap with either cortex or CER (permuted P-values for enrichment in EC = 0.89, PFC = 0.40, STG = 0.45, and CER = 0.41) suggesting that AD-associated DMPs in blood are unlikely to be directly related to the actual neurodegenerative process itself. Using data from our previous independent blood-based transcriptomic analyses of both AD and mild cognitive impairment (MCI)36, however, we observe that 18 of our top-ranked blood DMPs are located in the vicinity of known differentially expressed transcripts (Supplementary Table S13). These data suggest that, although distinct from AD-associated changes occurring in the brain, many of the AD-associated DMPs identified in blood prior to death may mediate detectable transcriptomic changes and, given the relative stability and ease of profiling DNA modifications compared to RNA, have potential utility as diagnostic biomarkers of the disorder.
Definitively distinguishing cause from effect in epigenetic epidemiology is difficult, especially for disorders like AD that are manifest in inaccessible tissues such as the brain and not amenable to longitudinal study9,10. However, our observation of highly consistent changes across multiple regions of the cortex in several independent sample cohorts suggests that the identified loci are directly relevant to the pathogenesis of AD. In this regard, the ANK1 DMR reported here, subsequently confirmed in the study by De Jager and colleagues21, represents one of the most robust molecular associations with AD yet identified. One issue in EWAS analyses using platforms such as the Illumina 450K array relates to potential technical artifacts caused by genetic variation, although we are confident that the DMPs identified in this study do not result from polymorphisms in (or flanking) the assayed CG dinucleotides. We used a stringent two-pronged strategy to exclude these effects: i) the direct exclusion of probes known to be affected by common single nucleotide polymorphisms (SNPs) and ii) the statistical filtering of extreme sample outliers within individual probe data that are frequently caused by rare SNPs (see Online Methods). Although this study was unable to explore the extent to which AD-associated variation is driven by genetic variation, the role of genetic-epigenetic interactions in complex disease represents an important area for further study37. Finally, power calculations for EWAS analyses are difficult, especially given the paucity of existing data for brain DNA methylation and limited information about the extent of inter-individual variation occurring at individual CpG sites. Conventional methods for multiple-test correction such as those used in genome-wide association studies (GWAS) are likely to be overly stringent given the non-independence of DNA methylation across multiple CpG sites9,10,38. Studies investigating the role of epigenetic dysfunction in complex brain diseases such as AD are in their infancy, and no real precedents have yet been set about the optimal sample-sizes needed to detect them9. Our conservative power calculation (see Online Methods), suggests we are well-powered to identify relatively small (~5%) DNA methylation differences between groups for the majority of probes on the Illumina 450K array. More importantly, our study represents the largest cross-tissue study of AD using DNA from both affected and unaffected brain regions, and the first to employ a sequential replication design incorporating independent study cohorts and two independent technologies (Illumina 450K array and bisulfite–pyrosequencing). The striking overlap between DMPs identified across our sample cohorts (Fig. 2a), and with those identified by De Jager et al. (Fig. 2g and Supplementary Fig. S2), suggests our study is adequately powered to detect robust AD-associated differences that can be replicated in other studies.
In summary, our data provide evidence for extensive differences in DNA methylation across brain regions in AD. Our analyses of multiple brain regions obtained from three independent cohorts implicates a role for cortex-specific hypermethylation across a region within ANK1 in AD-associated neuropathology, with methylomic changes mirroring known patterns of neuropathology and being most significant in the EC. This finding is strengthened by the independent identification of the same DMR in another large EWAS of AD21. Finally, although most brain-identified DMPs, including ANK1, are not detected in blood, we do identify multiple AD-associated DNA methylation differences in pre-mortem blood samples, many located in the vicinity of genes previously found to be transcriptionally altered even in patients with MCI during the early stages of cognitive decline. Our study represents the first EWAS of AD employing a sequential replication design across multiple tissues, and highlights the power of this approach more broadly for the identification of disease-associated DMRs.
ONLINE METHODS
Subjects and samples
Brain tissue was obtained from three independent sample cohorts, enabling us to take a powerful cross-tissue sequential-replication approach to identifying DNA methylation differences in AD. Our discovery cohort comprised of entorhinal cortex (EC), superior temporal gyrus (STG), prefrontal cortex (PFC), and cerebellum (CER) tissue obtained from 117 individuals archived in the MRC London Neurodegenerative Disease Brain Bank (http://www.kcl.ac.uk/iop/depts/cn/research/MRC-London-Neurodegenerative-Diseases-Brain-Bank/MRC-London-Neurodegenerative-Diseases-Brain-Bank.aspx). Ethical approval for the study was provided by the NHS South East London REC 3. Matched blood samples collected prior to death were available for a subset of individuals (Supplementary Tables S1 and S2) as part of the Alzheimer’s Research UK funded study “Biomarkers of AD Neurodegeneration”, with informed consent according to the Declaration of Helsinki (1991). For validation purposes STG and PFC tissue was obtained from 144 individuals archived in the Mount Sinai Alzheimer’s Disease and Schizophrenia Brain Bank (http://icahn.mssm.edu/research/labs/neuropathology-and-brain-banking)19 and EC, STG and PFC samples from an additional 62 individuals archived in the Thomas Willis Oxford Brain Collection (http://www.medsci.ox.ac.uk/optima/information-for-patients-and-the-public/the-thomas-willis-oxford-brain-collection)20. All samples were dissected by trained specialists, snap-frozen and stored at −80°C. Further information about the samples is given in Supplementary Table S1 and Supplementary Table S2. Genomic DNA was isolated from ~100mg of each dissected brain region or whole blood stored in EDTA collection tubes using a standard phenol-chloroform extraction method, and tested for degradation and purity prior to analysis.
Power
Power calculations for EWAS analyses are difficult given the paucity of existing data for brain DNA methylation and limited information about the extent of inter-individual variation occurring at individual CpG sites9. As we have previously discussed, studies investigating the role of epigenetic dysfunction in complex brain diseases such as AD are in their infancy, and no real precedents have yet been set about the optimal sample-sizes needed to detect them9. A conservative power calculation using methylome data from this and other ongoing studies in our lab11,39–41, suggests we are well-powered to identify DNA methylation differences of ~5% between groups for the majority of probes on the Illumina 450K array based conservatively on a case-control t-test with an array-wide Bonferroni threshold and the observed distribution of beta-value variances for the entorhinal cortex data set. More importantly, our study represents the largest cross-tissue study of AD using DNA from both affected and unaffected brain regions, and the first to employ a sequential replication design incorporating three independent study cohorts and two independent technologies (Illumina 450K array and bisulfite-pyrosequencing). The striking overlap between DMPs identified across our sample cohorts (Fig. 2a), and with those identified by De Jager et al.21 (Fig. 2g and Supplementary Fig. S2), suggests our study was adequately powered to detect robust AD-associated differences.
Methylomic profiling
500ng DNA from each sample was sodium bisulfite-treated using the Zymo EZ 96 DNA methylation kit (Zymo Research, CA, USA) according to the manufacturer’s standard protocol. Samples were assessed using the Illumina Infinium HumanMethylation450K BeadChip (Illumina Inc, CA, USA) using a Illumina HiScan System (Illumina, CA, USA). All samples were assigned a unique code for the purpose of the experiment and grouped by tissue and randomized with respect to sex and disease status to avoid batch effects, and processed in batches of four BeadChips. Illumina Genome Studio software was used to extract the raw signal intensities of each probe (without background correction or normalization).
Data analysis
All computations and statistical analyses were performed using R 3.0.242 and Bioconductor 2.1343. Signal intensities were imported into R using the methylumi package44 as a methylumi object. Initial quality control checks were performed using functions in the methylumi package to assess concordance between reported and genotyped gender. Non-CpG SNP probes on the array were also used to confirm that all four brain regions and matched bloods were sourced from the same individual in the London Cohort and two brain regions in the Mount Sinai cohort where expected. Data was pre-processed in the R package wateRmelon using the dasen function as previously described11. Array data for each of the tissues was normalized separately and initial analyses were performed separately by tissue. The effects of age and sex were regressed out before subsequent analysis. For identification of DMPs specifically altered with respect to neuropathological measures of AD, we performed a quantitative analysis where samples were analyzed using linear regression models in respect to Braak stage (London N = 117, Mount Sinai N = 144) and amyloid burden (Mount Sinai N = 144). We used a two-level strategy for avoiding spurious signals due to SNPs rather than DNA methylation differences. Probes with common (MAF > 5%) SNPs in the CG or single base extension position or probes that are nonspecific or mismapped were flagged and disregarded in the evaluation of our results45. In order to also clean up rarer SNPs whilst discarding minimum data, within each tissue, and for each probe, we discarded beta values lying more than four times the interquartile range from the mean; these extreme outliers are generally the result of polymorphisms. Data was analyzed separately in each brain region using linear regression with probes ranked according to P value, and Q-Q plots assessed to check for P value inflation (see Supplementary Fig. S5 for example). To identify differentially methylated regions (DMRs), we identified spatially correlated P values within our data using the Python module comb-p18 to group ≥4 spatially correlated CpGs within a 500bp sliding window. The CETS package in R17 was used to check whether our top-ranked DMPs were mediated by the effect of differential neuronal cell proportions across samples. To identify probes with consistent associations between Braak stage and methylation across the three cortical regions, we employed a meta-analysis of EC, STG and PFC. P values from the individual region results for each site were generated using Fisher’s method and (as a way of controlling for the covariance of the samples which come from the same individuals) Brown’s method. Raw data has been deposited in GEO under accession number GSE43414.
Targeted replication using bisulfite pyrosequencing
Bisulfite pyrosequencing was used to quantify DNA methylation across eight individual ANK1 CpG sites, including cg05066959 and cg11823178, spanning from 41519302 to 41519420 within chromosome 8 (hg19). A single amplicon (246bp) was amplified using primers designed using the PyroMark Assay Design software 2.0 (Qiagen, UK) (Supplementary Table 11a), and sequenced using two sequencing primers to maximize coverage across eight CpG sites. DNA methylation was quantified in 62 samples within the Oxford replication cohort using the Pyromark Q24 system (Qiagen, UK) following the manufacturer’s standard instructions and the Pyro Q24 CpG 2.0.6 software. Data was adjusted for the effects of age and sex. An analysis was performed to compare samples with Braak scores 0-II to samples with Braak scores V-VI at a) individual CpGs and b) amplicon-averaged DNA methylation.
Transcript variant analysis
A subset of samples from the London cohort was selected for RNA analyses. RNA was extracted from 30mg brain tissue using the Qiagen RNeasy mini kit and those with a concentration >90ng/ul and a RNA integrity number (RIN) >7 (N = 36) were used for subsequent qRT-PCR (see Supplementary Table S2 for specific samples used in this analysis). 20μl cDNA was synthesized from 1300ng total RNA using the SuperScript® VILO™ cDNA Synthesis Kit according to the manufacturer’s protocol and diluted five to tenfold for PCR, depending on the downstream assay. Off the shelf TaqMan® Gene Expression assays (Life Technologies) were purchased for the five housekeeping genes (EIF4A2, GAPDH, ACTB, SF3A1, UBC) identified as being most stably expressed in the brain using GeNORM (Primer Design, Southampton, UK). At least ten known protein coding splice variants for ANK1 have been characterized (Supplementary Fig. S3), and we were able to design three custom TaqMan® Gene Expression assays to target (a) variants 1,2,3 and 4, (b) variants 5, 7 and 10, and (c) variant 9 (Supplementary Table S11b). qRT-PCR was performed using TaqMan® Gene Expression Mastermix (Life Technologies) for each sample in duplicate on an ABI7900HT according to the manufacturer’s protocol. The abundance of ANK1 transcript variants was determined by relative quantification to the geometric mean of the five housekeeping genes. Data was adjusted for the effect of age and sex and linear models used to analyze variant levels with respect to Braak score.
Supplementary Material
Acknowledgments
This work was funded by NIH grant R01 AG036039 to JM and an Equipment Grant from Alzheimer’s Research UK. We thank Carolyn Sloan for technical support and Istvan Bodi and Andrew King for neuropathological diagnosis of cases. We also thank the Oxford Project to Investigate Memory and Ageing (OPTIMA), the National Institute for Health (NIHR) Biomedical Research Unit in Dementia in the South London and Maudsley NHS Foundation Trust (SLaM), Brains for Dementia Research (Alzheimer Brain Bank UK) and the donors and families who made this research possible. Blood samples from the London cohort were collected as part of the Alzheimer’s Research UK funded study “Biomarkers of AD Neurodegeneration”. The Oxford Brain Bank is supported in part by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust and University of Oxford. Brain banking and neuropathology assessments for the Mount Sinai cohort was supported by NIH grants AG02219, AG05138 and MH064673 and the Department of Veterans Affairs VISN3 MIRECC. Replication work in Boston was supported by the National Institutes of Health grants: R01 AG036042, R01AG036836, R01 AG17917, R01 AG15819, R01 AG032990, R01 AG18023, RC2 AG036547, P30 AG10161, P50 AG016574, U01 ES017155, KL2 RR024151, K25 AG041906-01.
Footnotes
AUTHOR CONTRIBUTIONS
KL, RS, RM, MV, DC, and JB conducted laboratory experiments. JM conceived and supervised the project. EH, LS RS and KL undertook data analyses and bioinformatics. CT, SL, SAS, PK, VH, and CJ provided brain tissue for analysis. PDJ, GS and DB provided replication data. ZK provided help with cellular heterogeneity correction. LH provided help with the alternative splicing assays. JM, KL and LS drafted the manuscript. All authors read and approved the final submission.
A Supplementary Methods Checklist is available.
References
- 1.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007;3:186–91. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 2.Knapp M, Prince M, Dementia UK. The Full report. Alzheimer’s Society. 2007
- 3.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–5. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 4.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 5.Wenk GL. Neuropathologic changes in Alzheimer’s disease. J Clin Psychiatry. 2003;64 (Suppl 9):7–10. [PubMed] [Google Scholar]
- 6.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Encode Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lunnon K, Mill J. Epigenetic studies in Alzheimer’s disease: current findings, caveats, and considerations for future studies. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:789–99. doi: 10.1002/ajmg.b.32201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet. 2013;14:585–94. doi: 10.1038/nrg3405. [DOI] [PubMed] [Google Scholar]
- 10.Murphy TM, Mill J. Epigenetics in health and disease: heralding the EWAS era. Lancet. 2014;383:1952–4. doi: 10.1016/S0140-6736(14)60269-5. [DOI] [PubMed] [Google Scholar]
- 11.Pidsley R, et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 13.Kordeli E, Bennett V. Distinct ankyrin isoforms at neuron cell bodies and nodes of Ranvier resolved using erythrocyte ankyrin-deficient mice. J Cell Biol. 1991;114:1243–59. doi: 10.1083/jcb.114.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boiko T, et al. Ankyrin-dependent and -independent mechanisms orchestrate axonal compartmentalization of L1 family members neurofascin and L1/neuron-glia cell adhesion molecule. J Neurosci. 2007;27:590–603. doi: 10.1523/JNEUROSCI.4302-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson SB, Janiesch C, Sander M. Expression of Nkx6 genes in the hindbrain and gut of the developing mouse. J Histochem Cytochem. 2005;53:787–90. doi: 10.1369/jhc.5B6619.2005. [DOI] [PubMed] [Google Scholar]
- 16.Alanentalo T, et al. Cloning and analysis of Nkx6.3 during CNS and gastrointestinal development. Gene Expr Patterns. 2006;6:162–70. doi: 10.1016/j.modgep.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Guintivano J, Aryee M, Kaminsky Z. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8 doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28:2986–8. doi: 10.1093/bioinformatics/bts545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haroutunian V, et al. Regional distribution of neuritic plaques in the nondemented elderly and subjects with very mild Alzheimer disease. Arch Neurol. 1998;55:1185–91. doi: 10.1001/archneur.55.9.1185. [DOI] [PubMed] [Google Scholar]
- 20.Esiri MM. Brain banks: the Oxford experience. J Neural Transm Suppl. 1993;39:25–30. [PubMed] [Google Scholar]
- 21.De Jager PL, et al. Alzheimer’s disease pathology is associated with early alterations in brain DNA methylation at ANK1, BIN1 and other loci. Nat Neurosci. 2014 doi: 10.1038/nn.3786. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soranzo N, et al. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–39. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura M, et al. A single-nucleotide polymorphism in ANK1 is associated with susceptibility to type 2 diabetes in Japanese populations. Hum Mol Genet. 2012;21:3042–9. doi: 10.1093/hmg/dds113. [DOI] [PubMed] [Google Scholar]
- 24.Harder MN, et al. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased beta-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J Clin Endocrinol Metab. 2013;98:E801–6. doi: 10.1210/jc.2012-4169. [DOI] [PubMed] [Google Scholar]
- 25.Barnes DE, Yaffe K. The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol. 2011;10:819–28. doi: 10.1016/S1474-4422(11)70072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovestone S. Searching for biomarkers in neurodegeneration. Nat Med. 2010;16:1371–2. doi: 10.1038/nm1210-1371b. [DOI] [PubMed] [Google Scholar]
- 27.Hooper C, Lovestone S, Sainz-Fuertes R. Alzheimer’s Disease, Diagnosis and the Need for Biomarkers. Biomark Insights. 2008;3:317–323. doi: 10.4137/bmi.s682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fehlbaum-Beurdeley P, et al. Toward an Alzheimer’s disease diagnosis via high-resolution blood gene expression. Alzheimers Dement. 2010;6:25–38. doi: 10.1016/j.jalz.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Rye PD, et al. A novel blood test for the early detection of Alzheimer’s disease. J Alzheimers Dis. 2011;23:121–9. doi: 10.3233/JAD-2010-101521. [DOI] [PubMed] [Google Scholar]
- 30.Booij BB, et al. A gene expression pattern in blood for the early detection of Alzheimer’s disease. J Alzheimers Dis. 2011;23:109–19. doi: 10.3233/JAD-2010-101518. [DOI] [PubMed] [Google Scholar]
- 31.Lunnon K, et al. A blood gene expression marker of early Alzheimer’s disease. J Alzheimers Dis. 2013;33:737–53. doi: 10.3233/JAD-2012-121363. [DOI] [PubMed] [Google Scholar]
- 32.Fehlbaum-Beurdeley P, et al. Validation of AclarusDx, a Blood-Based Transcriptomic Signature for the Diagnosis of Alzheimer’s Disease. J Alzheimers Dis. 2012;32:169–81. doi: 10.3233/JAD-2012-120637. [DOI] [PubMed] [Google Scholar]
- 33.Wu ZC, et al. Association of DAPK1 genetic variations with Alzheimer’s disease in Han Chinese. Brain Res. 2011;1374:129–33. doi: 10.1016/j.brainres.2010.12.036. [DOI] [PubMed] [Google Scholar]
- 34.Laumet G, et al. Systematic analysis of candidate genes for Alzheimer’s disease in a French, genome-wide association study. J Alzheimers Dis. 2010;20:1181–8. doi: 10.3233/JAD-2010-100126. [DOI] [PubMed] [Google Scholar]
- 35.Chapuis J, Vingtdeux V, Campagne F, Davies P, Marambaud P. Growth arrest-specific 1 binds to and controls the maturation and processing of the amyloid-beta precursor protein. Hum Mol Genet. 2011;20:2026–36. doi: 10.1093/hmg/ddr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lunnon K, et al. Mitochondrial dysfunction and immune activation are detectable in early Alzheimer’s disease blood. J Alzheimers Dis. 2012;30:685–710. doi: 10.3233/JAD-2012-111592. [DOI] [PubMed] [Google Scholar]
- 37.Meaburn EL, Schalkwyk LC, Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics. 2010;5:578–82. doi: 10.4161/epi.5.7.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–9. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies MN, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dempster EL, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–96. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong CC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2014;19:495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 43.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis S, Du P, Bilke S, Triche J, Bootwalla M. Methylumi: Handle Illumina Methylation Data. R package version 2.10.0. 2014 [Google Scholar]
- 45.Chen YA, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.