Abstract
The arrival of multicellularity in evolution facilitated cell–cell signaling in conjunction with adhesion. As the ectodomains of cadherins interact with each other directly in trans (as well as in cis), spanning the plasma membrane and associating with multiple other entities, cadherins enable the transduction of “outside-in” or “inside-out” signals. We focus this review on signals that originate from the larger family of cadherins that are inwardly directed to the nucleus, and thus have roles in gene control or nuclear structure–function. The nature of cadherin complexes varies considerably depending on the type of cadherin and its context, and we will address some of these variables for classical cadherins versus other family members. Substantial but still fragmentary progress has been made in understanding the signaling mediators used by varied cadherin complexes to coordinate the state of cell–cell adhesion with gene expression. Evidence that cadherin intracellular binding partners also localize to the nucleus is a major point of interest. In some models, catenins show reduced binding to cadherin cytoplasmic tails favoring their engagement in gene control. When bound, cadherins may serve as stoichiometric competitors of nuclear signals. Cadherins also directly or indirectly affect numerous signaling pathways (e.g., Wnt, receptor tyrosine kinase, Hippo, NFκB, and JAK/STAT), enabling cell–cell contacts to touch upon multiple biological outcomes in embryonic development and tissue homeostasis.
The ability of cells to associate and organize themselves in response to one another the extracellular matrix or other cues to generate a variety of tissue shapes and forms has captivated biologists for centuries. Here, we discuss the broad principles of how the state of intercellular adhesion is communicated to a cell’s interior, with a focus on changes in nuclear gene activity. We will examine signals that originate from or involve the classic cadherin–catenin complex present at cell–cell contacts or more distantly related cadherin family members. Over 100 types of cadherins are present in the larger superfamily, with a variety of structural characteristics and functions (Hulpiau & van Roy, 2009; Oda & Takeichi, 2011).
Related aspects of this topic have been addressed in a number of excellent reviews, to which the reader is directed (Cavallaro & Christofori, 2004; Cavallaro & Dejana, 2011; Daugherty & Gottardi, 2007; Gavard, 2013; Geletu, Guy, Arulanandam, Feracci, & Raptis, 2013; Heuberger & Birchmeier, 2010; McEwen, Escobar, & Gottardi, 2012; Nelson & Nusse, 2004; Paulson, Prasad, Thuringer, & Manzerra, 2014; Philippova et al., 2009; Stepniak, Radice, & Vasioukhin, 2009; Sun, Parrish, Hill, & Meininger, 2014; van Roy, 2014). In addition to providing a current analysis, we aim here to provide a perspective on cadherins that addresses a cross-section of the superfamily, especially in vertebrates. Some recent findings include evidence, for example, of the intersection of classic cadherin– catenin complex components or functions with the Wnt or Hippo signaling pathways, each of which has powerful roles in development and human pathology. Signaling trajectories to the nucleus are initiated from additional types of cadherins as well, including desmosomal cadherins, or from cadherins that are more distantly related and comprise part of the planar cell polarity network, used in establishing proximal–distal identities within tissues. While less has been uncovered in these latter cases with regard to nuclear signaling, intriguing possibilities have arisen or are postulated.
Much of what we address here is within the sphere of “outside-in” signaling, that is, a form of signaling where a cue that originates from a cell’s exterior is propagated to the interior cytoplasm and nucleus. In the context of cadherin family members, this normally includes involvement in cytoskeletal control, such as the modulation of small GTPases. However, while such small GTPases and cytoskeletal effects are likely to contribute to down-stream effects upon gene activity, we will give this area relatively brief mention and refer the reader to other reviews (Anastasiadis, 2007; Hatzfeld, 2005; Kourtidis, Ngok, & Anastasiadis, 2013; Pieters, van Roy, & van Hengel, 2012; Ratheesh, Priya, & Yap, 2013). In a similar vein, cadherins can mechanically transduce outside pulling forces originating from adjacent cells into intracellular signals that regulate gene activity. Since the nuclear skeleton and the cytoskeletons are interconnected, mechanical changes at cell contacts can be rapidly propagated through the nuclear cytoskeleton with consequent but little understood effects upon the organization and activity of chromatin. Concerning this and other modes of mechanotransduction, we again direct the reader to other expert reviews (Brieher & Yap, 2013; Cavey & Lecuit, 2009; DuFort, Paszek, & Weaver, 2011; Gomez, McLachlan, & Yap, 2011; Huveneers & de Rooij, 2013; Maitre & Heisenberg, 2013; Smutny & Yap, 2010; Twiss & de Rooij, 2013).
1. β-CATENIN, A DUAL-FUNCTION ADHESION/TRANSCRIPTIONAL COACTIVATOR PROTEIN
β-Catenin was first isolated in association with the cytoplasmic domains of classical cadherins at cell–cell junctions and later was recognized to be the prime signal transducer of the canonical Wnt pathway, passing through nuclear pores en route to gene control regions (reviewed in Fagotto, 2013). In response to upstream Wnt pathway activation, β-catenin enters the nucleus to derepress (activate) genes that are otherwise repressed by T cell factor (TCF)/lymphoid enhancer factor (LEF). Since β-catenin is essential to most vertebrate developmental processes, with pathologic activity contributing to multiple human diseases, there have been innumerable attempts to unravel the highly involved set of molecular participants in what is commonly referred to as the “canonical Wnt pathway.”
How is the nuclear pool of β-catenin generated? Under normal cellular conditions, β-catenin that is not already participating in cadherin-based adhesions is rapidly destroyed. Wnt signals effectively block this elaborate destruction mechanism, which allows β-catenin protein to accumulate in the cytosol, translocate to the nucleus, bind TCF/LEF DNA-binding factors, and induce transcription. Although conceptually simple, a dizzying array of proteins contribute to the targeted destruction of β-catenin and its inhibition by Wnts, which implies that organisms go to great trouble to control the magnitude, duration, and cell-specific expression of genes imposed by Wnt signals in the name of normal development. Thus, rather than being comprehensive, we will summarize key steps in the Wnt pathway where cross talk with cadherins has been observed.
Briefly, the canonical Wnt pathway is initiated by an extracellular Wnt-ligand binding to a transmembrane coreceptor complex comprised of seven-membrane pass Frizzled (Fz) protein and Low-density lipoprotein Receptor-related Protein 5 or 6 (Lrp5/6). After engaging these receptors, a canonical Wnt acts to stabilize a pool of β-catenin that would otherwise be destroyed by a macromolecular assembly termed the “destruction complex.” This complex is organized by a key scaffold component, Axin. Through its ability to bind β-catenin, its regulatory kinases, and other modifiers, Axin favors sequential phosphorylations by casein kinase 1 alpha (CK1α) and glycogen synthase kinase 3β (GSK3β), events that ultimately “flag” β-catenin for E3-ligase (β-TrCP) phosphorecognition, ubiquitylation, and proteosomal destruction. The adenomatous polyposis coli (APC) tumor suppressor protein is a scaffold component with binding sites for β-catenin and Axin (among other proteins) and is particularly note-worthy because loss-of-function mutations in APC cause both heritable and sporadic forms of colon cancer through excessive β-catenin nuclear signaling (reviewed in Heinen, 2010; Polakis, 2000). Despite the clear importance of APC, molecular details as to how this protein leads to the destruction of β-catenin are just coming into view. APC may facilitate β-catenin “flux” through the destruction complex by its higher affinity binding to β-catenin than Axin (Ha, Tonozuka, Stamos, Choi, & Weis, 2004) as well as shield β-catenin’s N-terminal phosphate groups (required for β-TrCP-mediated ubiquitylation and destruction) from dephosphorylation by the phosphatase PP2 (Su et al., 2008), possibly through a region in APC that can bind the F-actin-binding protein α-catenin (Choi, Estaras, Moresco, Yates, & Jones, 2013). Wnt binding to cell surface Fz and Lrp5/6 receptors (Chu et al., 2013) favors phosphorylations along the cytoplasmic tail of Lrp5/6, leading to the recruitment of Axin and the inhibition of GSK-3β within the Axin complex (Cselenyi et al., 2008; Mi, Dolan, & Johnson, 2006; Piao et al., 2008; Stamos & Weis, 2013; Wu, Huang, Garcia Abreu, & He, 2009), in addition to β-catenin hypophosphorylation and its ultimate stabilization and nuclear signaling. Wnt signaling can also target GSK3 to multivesicular endosomes, which may be important for sustained GSK3 inhibition required for long-term β-catenin signaling activity (Taelman et al., 2010). While any point along this pathway may be subject to regulation, evidence (discussed below) indicates that cadherins largely affect β-catenin signaling at three levels: (1) direct, high-affinity binding to β-catenin that competes for its interactions with other binding partners (TCF, Axin, APC) and thereby prevents nuclear import (Fagotto, Gluck, & Gumbiner, 1998; Suh & Gumbiner, 2003) (Fig. 1); (2) increasing the rate of β-catenin flux through the destruction complex (Hay et al., 2009; Maher, Flozak, Stocker, Chenn, & Gottardi, 2009) (Figs. 2A and 3); and (3) facilitating activation of the Wnt receptor complex at the level of Lrp5/6 phosphorylation, endocytosis, and “signalosome” formation (Bilic et al., 2007; Blitzer & Nusse, 2006) (Figs. 2B and 3). We reason that these distinct inputs arose during the course of evolution so that the state of cadherin adhesive interactions could inform the magnitude and duration of the nuclear differentiation programs dictated by β-catenin/TCF and perhaps additional gene regulatory complexes.
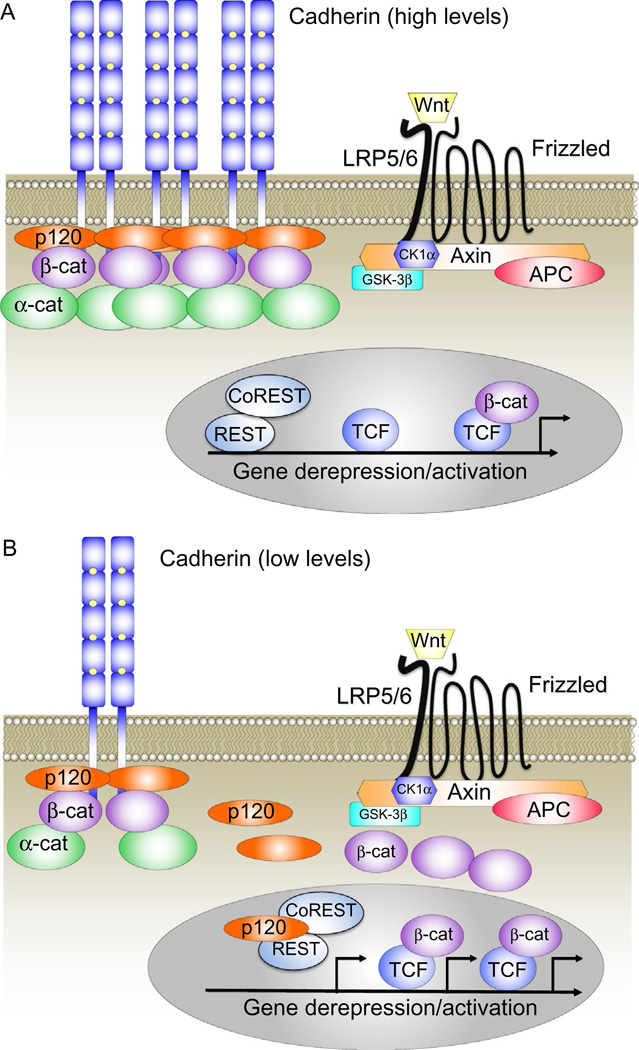
Figure 1.
Cadherins as competitive stoichiometric inhibitors of p120ctn and β-catenin signaling. The model presented in (A) and (B) reflects evidence that cells with greater cadherin abundance (A=high levels of cadherin expression; B=lower cadherin expression) can sequester and thereby inhibit the ability of p120ctn and β-catenin to derepress activities of their respective DNA-binding factors, Rest/CoRest and TCF/LEF.
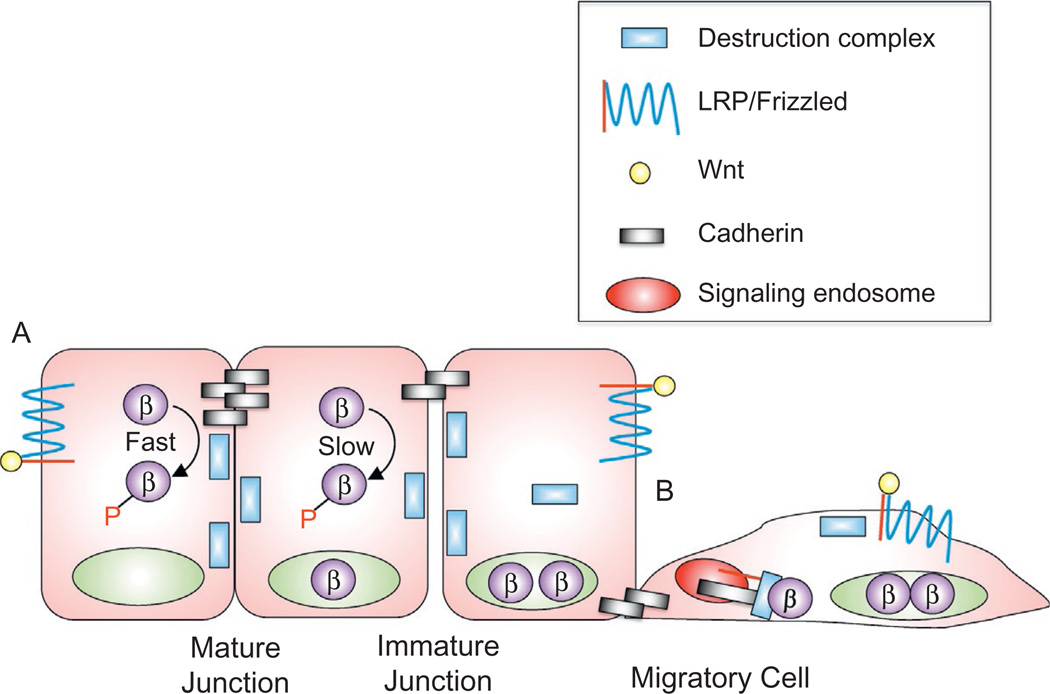
Figure 2.
Cadherin-based adhesion as an enhancer of β-catenin destruction. In densely confluent cells with mature junctions, cadherins promote a faster turnover of β-catenin than in less adhesive (subconfluent) cells with immature junctions (A). This may explain why cells migrating adjacent to a wound appear sensitized to Wnt signals (Howard, Deroo, Fujita, & Itasaki, 2011; Maher et al., 2009). Mechanistically, the latter study suggests that enhanced β-catenin signaling is due to an activation step that depends on cadherins and a signaling endosome (B), while the former suggests that these cells show reduced capacity to degrade β-catenin (A).
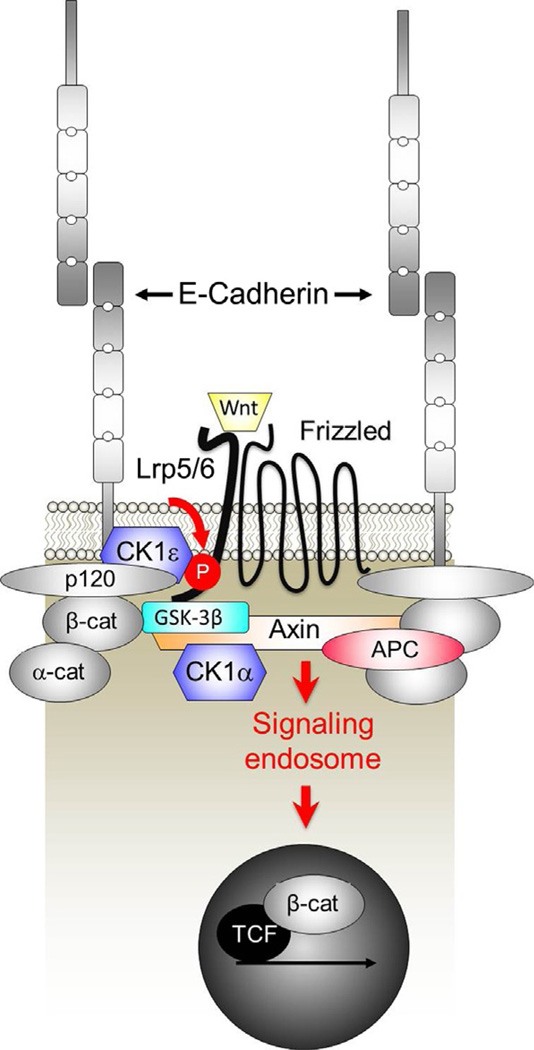
Figure 3.
Cadherins as facilitators of Wnt/β-catenin signaling. A number of studies show that E- and N-cadherins are required for efficient Wnt/β-catenin signaling, working at the level of the Wnt receptor complex. Work from the Dunach group shows that p120ctn facilitates Lrp5/6 phosphorylation and signaling by functioning as a proximal scaffold for CK1ε. p120ctn also contributes to the inhibition of GSK3 by multivesicular bodies leading to enhanced signaling from an endosome.
2. CADHERINS AS STOICHIOMETRIC INHIBITORS OF WNT/β-CATENIN SIGNALING
Evidence that β-catenin is both a critical component of the cadherin/catenin adhesive complex, serving as an important linker between the cadherin and the actin-binding protein, α-catenin (Dartsch, Schulte, Hagerling, Kiefer, & Vestweber, 2014), and an indispensible mediator of Wnt signaling raises intriguing questions as to whether or not adhesion and Wnt signaling are coordinated through the use of this common component, β-catenin. Indeed, experimental manipulations in cells and simple model organisms reveal that cadherin expression and β-catenin signaling are interrelated. For example, forced expression of the cadherin can antagonize β-catenin signaling activity in a number of systems (Fagotto, Funayama, Gluck, & Gumbiner, 1996; Heasman et al., 1994; Orsulic, Huber, Aberle, Arnold, & Kemler, 1999; Sanson, White, & Vincent, 1996). Conversely, lowering cadherin levels in developing tissues can lead to enhanced β-catenin signaling (Ciruna & Rossant, 2001; Cox, Kirkpatrick, & Peifer, 1996). Since biochemical and crystallographic evidence show that β-catenin binds cadherins or TCFs through an overlapping binding interface (Graham, Weaver, Mao, Kimelman, & Xu, 2000; Huber & Weis, 2001), where the cadherin:β-catenin interaction is of higher affinity than that of TCF:β-catenin (Choi, Huber, & Weis, 2006), these data collectively show how cadherins can, in principle, function as stoichiometric inhibitors of β-catenin/TCF (etc.) signaling. Importantly, the ability of lowered E-cadherin levels to lead to β-catenin signaling activation is not universal, as epithelial cancers that have lost E-cadherin expression by various means often fail to exhibit constitutive β-catenin signaling activation (Caca et al., 1999; Herzig, Savarese, Novatchkova, Semb, & Christofori, 2007; van de Wetering et al., 2001). Indeed, targeted loss of E-cadherin can be associated with downregulation of β-catenin (Hendriksen et al., 2008), presumably because there are no other β-catenin-binding cadherins in these systems (e.g., N-cadherin or P-cadherin), and loss of this major high-affinity β-catenin-binding partner leads to β-catenin elimination by the phosphodestruction complex. Thus, the ability of a cadherin to limit β-catenin signaling is contextual and appears to occur only when cells are actively engaged in Wnt signaling (Fig. 1).
While the aforementioned gain- and loss-of-function examples clearly show that cadherins levels can set thresholds for β-catenin signaling, we have few examples where cadherin levels are modulated in vivo to dampen or elevate β-catenin signaling. The best early example of this involves an FGF-induced epithelial–mesenchymal transition (EMT) that leads to an E-cadherin downregulation that is important for full β-catenin signaling required for primitive streak formation in the mouse (Ciruna & Rossant, 2001). It is also clear that stem cells downregulate E-cadherin as they begin transitioning away from pluripotency (discussed below) (Bao et al., 2009; del Valle et al., 2013; Lee, Ji, Furuta, Park, & McCrea, 2014; Li, Zhou, et al., 2010; Li, Wang, et al., 2010; reviewed in Pieters & van Roy, 2014). Conversely, while a few signals have been shown to increase E-cadherin transcription in both cell culture and developmental models (Hosono et al., 2000; Montell, Rorth, & Spradling, 1992; Niewiadomska, Godt, & Tepass, 1999; Ohira et al., 2003; Shimamura & Takeichi, 1992), it is unclear whether the observed increases in cadherin synthesis are used to dampen endogenous Wnt/β-catenin signals, in addition to enhancing cell–cell adhesiveness required in certain morphogenetic events. Interestingly, TCF-binding sites have been identified in the E-cadherin promoter (Huber et al., 1996), suggesting the existence of a negative feedback loop whereby Wnt/β-catenin signaling could increase E-cadherin levels, in turn dampening the signaling pool of β-catenin. While activation of β-catenin signaling has been associated with the upregulation of E-cadherin in mouse intestine (Wong, Rubinfeld, & Gordon, 1998) and a Drosophila cell line (Yanagawa et al., 1997), the universality of this feedback mechanism is unclear given evidence that a TCF site in the E-cadherin promoter interacts with other factors to inhibit E-cadherin transcription during hair follicle development (Jamora, DasGupta, Kocieniewski, & Fuchs, 2003).
3. THE CADHERIN–CATENIN COMPLEX AS BOTH A POTENTIATOR AND AN ATTENUATOR OF WNT/β-CATENIN SIGNALING
3.1. Cadherin-based adhesion and destruction complex activity
If it is clear that cadherin protein levels can impact the nuclear signaling capacity of β-catenin, it has been less clear whether changes in the state of cell–cell adhesion might impact β-catenin signaling. Intuitively, the pairing of a cell surface adhesion receptor (i.e., cadherin) with a transcriptional coactivator (i.e., β-catenin) for the purposes of cell–cell adhesion long suggested such a relationship. Recently, two labs independently uncovered proximal relationships between the cadherin/catenin and β-catenin destruction complexes (Hay et al., 2009; Maher et al., 2009). One group found that cadherin-based cell–cell adhesion across a number of epithelial cell lines could increase the amino-terminal phosphorylation of β-catenin and its subsequent rate of destruction (Maher et al., 2009). This ability of cadherins to promote β-catenin turnover was supported by evidence that the N-terminally phosphorylated forms of β-catenin colocalized at E-cadherin-based cell–cell contacts with its known destruction complex components, Axin, APC2, and GSK3β. Although this complex appeared to be molecularly distinct from the E-cadherin complex by sucrose density gradient analysis, the membrane proximity of the phosphodestruction complex suggests that it spatially poised to receive cadherin-contact-dependent signals. In osteoblasts, N-cadherin showed a similar capacity to promote the phosphorylation-dependent ubiquitylation (and ultimate destruction) of β-catenin (Hay et al., 2009). But in the latter study, the molecular relationship between N-cadherin, Lrp5/6, and Axin appears closer, as they were found to coassociate by immunoprecipitation (Hay et al., 2009). Differences aside, these results suggest a model where greater cadherin-mediated adhesion, as seen most in sedentary cells, disfavors canonical Wnt signaling by enhancing destruction complex activity (Fig. 2A). This model is consistent with earlier work demonstrating that less adhesive, motile cells display increased Wnt-reporter activity in Zebrafish embryos (Dorsky, Sheldahl, & Moon, 2002), as well as the more recent identification of a novel membrane-proximal inhibitor of β-catenin signaling, WTX/Amer1 (Major et al., 2007), which can impact the activity of the phosphodestruction complex (Tanneberger et al., 2011). An appealing feature of this model is that the activity of the β-catenin phosphodestruction complex can be “tuned” by the local adhesive environment, despite a uniform presence of Wnt ligand, which may be relevant to cell fate decisions that occur in various developmental contexts.
3.2. Cadherins as facilitators of Wnt/β-catenin signaling
For the two aforementioned examples where cadherin-based adhesion limits the activity of the phosphodestruction complex, recent studies highlight more numerous examples demonstrating a requirement of the cadherin complex for Wnt/β-catenin signaling. For example, N-cadherin:N-cadherin associations between cortical precursor cells in the ventricular neurogenic niche help to maintain β-catenin signaling in these cells, thus inhibiting their differentiation into mature neurons (Zhang et al., 2010). Later findings from the same group (Zhang et al., 2013) used conditional knockout approaches in mice together with in vivo electroporation of embryos in utero to induce focal loss of N-cadherin, as well as knockdown and antibody function-blocking approaches in cell lines to show that N-cadherin enhanced the phosphorylation of Lrp6 in the presence of Wnt signals in a cell-autonomous fashion (Zhang et al., 2013). Since Lrp5/6 phosphorylation leads to the recruitment of Axin and inhibition of GSK3 activity (Cselenyi et al., 2008; Mi et al., 2006; Piao et al., 2008; Stamos & Weis, 2013; Wu et al., 2009), these data suggest that N-cadherin:N-cadherin engagement facilitates an obligate early step in Wnt signal transduction (Fig. 3).
While the mechanism of N-cadherin’s requirement for Wnt signaling was not established in the above studies, it is important to bear in mind that, as the major cell-cell adhesion system in cells, the cadherin/β-catenin/α-catenin complex is critical not only for basic intercellular adhesion but also for establishing the close contacts that so many other juxtacrine signaling molecules depend upon, from membrane-anchored signaling pairs like Notch/Delta (Ferreira et al., 2011), Ephrins and Eph receptors (Zantek et al., 1999) to death receptor activation (Lu et al., 2014) and insulin secretion (Parnaud et al., 2014). Thus, via this fundamental mechanism, N-cadherin:N-cadherin engagement may enhance the interaction of Wnt ligands with Wnt receptor components to facilitate Wnt/β-catenin signaling. Relatedly, cadherin engagement is known to promote phosphatidylinositol 3-kinase activity (PI3K) (Tran, Adams, Vaillancourt, & Heimark, 2002), which through subsequent activation of AKT may facilitate β-catenin signaling through phosphoinhibition of GSK3β (via serine 9 phosphorylation; Fang et al., 2000), as well as phosphorylating a site in β-catenin that appears to enhance its transcriptional activity (Fang et al., 2007; Tian et al., 2004; Zhang et al., 2013).
Other evidence, however, suggests that the cadherin/catenin complex may play a more direct role in Wnt receptor complex activation. For example, the Dunach group has published a number of reports centered upon the role of the cadherin–catenin complex in recruiting kinases needed for the execution of canonical Wnt signals (Casagolda et al., 2010; Del Valle-Perez, Arques, Vinyoles, de Herreros, & Dunach, 2011; Del Valle-Perez, Casagolda, et al., 2011). The relationships uncovered are largely operating at the level of cadherin/catenin associations and their dissociations from Wnt–receptor complex components, Lrp5/6, CK1 family members, Dishevelled and Axin, as assessed by coimmunoprecipitation analysis in recombinant Wnt-treated cell lines. Indeed, a proximal relationship between a cadherin and Wnt receptor may be inferred from the existence of the Flamingo (Celsr in humans) planar cell polarity (PCP) protein, which appears to be a fusion between a nonclassical cadherin protein and a Frizzled receptor (Berger-Muller & Suzuki, 2011; Formstone, 2010).
As discussed above, Lrp5/6 phosphorylation is a key early step in Wnt signal transduction, leading to the recruitment of Dishevelled and Axin, inhibition of GSK3 activity, and β-catenin stabilization (MacDonald, Tamai, & He, 2009). While CK1γ and GSK3 can modify Lrp5/6 (Davidson et al., 2005; Mi et al., 2006; Zeng et al., 2005), what controls the local activation or accessibility of Lrp5/6 to these kinases has been unclear. Dunach and colleagues present evidence that p120-catenin (p120ctn) is required for Wnt-mediated phosphoactivation of Lrp6 (Casagolda et al., 2010; Del Valle-Perez, Arques, et al., 2011) (Fig. 3). A subsequent step involves the recruitment of Axin bound to another CK1 family member, CK1α. CK1α in turn phosphorylates p120ctn (Serines 268 and 269), dissociating it from the cadherin complex (Del Valle-Perez, Casagolda, et al., 2011; Vinyoles et al., 2014). Once dissociated from the cadherin, p120ctn is able to transduce signals to the nucleus in a manner that partially resembles β-catenin (discussed below) (Del Valle-Perez, Casagolda, et al., 2011). Cadherin-free p120ctn can also associate with the small-GTPase modulator Vav2 (Valls et al., 2012) and activate JNK kinases to promote β-catenin’s nuclear entry and activity (Wu et al., 2008). Lastly, evidence that endocytosis appears to be an obligate step for Wnt signal transduction (Bilic et al., 2007; Blitzer & Nusse, 2006), as is known for many other signaling pathways (Goh & Sorkin, 2013; Haglund & Dikic, 2012), raises the possibility that cadherins may impact Wnt signaling by controlling Wnt receptor complex internalization into signalosomes (Bilic et al., 2007). In this regard, recent studies show that Wnt receptor complex activation can promote the sequestration of GSK3 into multivesicular bodies (Taelman et al., 2010) (although perhaps not in flies; Gagliardi, Hernandez, McGough, & Vincent, 2014), leading to β-catenin stabilization and signaling, where the cadherin and p120ctn appear required for this process (Vinyoles et al., 2014). Given that the cadherin/catenin complex is fundamental not only for cell–cell adhesion but for basic membrane dynamics (Abreu-Blanco, Verboon, Liu, Watts, & Parkhurst, 2012; Schepis, Sepich, & Nelson, 2012), it may not be surprising to find these examples of the cadherin/catenin complex being both required for and inhibitory to Wnt/β-catenin signaling. A similar phenomenon has been described for the role of GSK3 in Wnt signaling, where it plays both activating and inhibitory roles at distinct steps of the pathway (Zeng et al., 2005). Nonetheless, we look forward to future studies that parse the activating and inhibitory contributions of the cadherin/catenin complex to Wnt signaling.
4. CADHERIN SIGNALING AND STEM CELL BEHAVIOR
One important context where cadherin levels are endogenously regulated during development occurs in mouse embryonic stem cells (mESCs), as they begin transitioning away from pluripotency (reviewed in Bhatt, Rizvi, Batta, Kataria, & Jamora, 2013; Farahani et al., 2014; Li, Bennett, & Wang, 2012; Pieters & van Roy, 2014). E-Cadherin levels are reduced as this transition begins, with cadherin levels being restored in subsequent differentiating lineages. The types of cadherin(s) that subsequently become expressed are lineage based. That is, cells moving toward a neural lineage will begin to express N-cadherin, and such alterations in E-, N-, or other cadherin levels likely impact β-catenin signaling as described above. For example, N-cadherin expression in cortical neural progenitors favors stemness through maintaining robust β-catenin signaling, while reductions in N-cadherin levels led to premature differentiation of neurons and their migration away from the stem cell niche (Woodhead, Mutch, Olson, & Chenn, 2006; Zhang et al., 2010). Other work suggests that N-cadherin’s ability to promote β-catenin signaling can favor differentiation of multipotential mesenchymal progenitor cells (murine C3H10T1/2 cells) toward the bone lineage, as opposed to maintaining stemness (Arnsdorf, Tummala, & Jacobs, 2009), fitting with long established evidence that Wnt signals can both promote stemness as well as differentiated states (Clevers, Loh, & Nusse, 2014; Lien & Fuchs, 2014; Serio, 2014).
5. β-CATENIN “RELEASE” FROM CORTICAL ENDOSOMES AND NUCLEAR SIGNALING
Two recent studies support the intriguing idea that a cadherin-dependent, membrane-proximal event may “prime” β-catenin for nuclear signaling. Similar to studies by the Chenn group (Zhang et al., 2013, 2010), Howard et al. found that N-cadherin is required for robust Wnt signaling in ingressing cells during mouse embryo gastrulation (Howard et al., 2011). Using a cell culture EMT model (meant to mimic aspects of ingression), this group demonstrated that an established phosphomimetic form of β-catenin previously shown to exhibit reduced binding to cadherin (Y654E; Roura, Miravet, Piedra, Garcia de Herreros, & Dunach, 1999; van Veelen et al., 2011) manifested reduced transcriptional activity compared to wild type or Y654F forms of β-catenin in the context of Wnt3 and HGF signals. Together with evidence that HGF-induced β-catenin signaling is sensitive to E-cadherin silencing and the endocytosis inhibitor dynasore, it appears that a cadherin-dependent priming event, from a signaling endosome, is required for efficient Wnt signaling. Additional evidence that HGF-induced β-catenin signaling is insensitive to the protein synthesis inhibitor, cycloheximide (despite an earlier report to the contrary; Willert, Epping, Pollack, Brown, & Nusse, 2002), raises the possibility that β-catenin signaling activity may be recruited from a preexisting cadherin-associated pool. Thus, mesenchymal cell types or shapes (e.g., even epithelial cells that are subconfluent) may be more sensitive to Wnt signals because cadherins are being more actively endocytosed, which appears important for β-catenin signaling. An attractive feature of this model is that it couples the dynamic state of cadherin cell–cell adhesive contacts with nuclear signaling functions that arise from those contacts, in this case taking place via β-catenin (Fig. 2B).
Remarkably, this “cortical release” model for β-catenin signaling activation may be evolutionarily conserved with a mechanism that controls cell division orientation in Caenorhabditis elegans (Kim et al., 2013). In worms, different forms of β-catenin-like proteins exist, with WRM1 being responsible for transducing noncanonical Wnt signals, as well as contributing to the orientation of cell division. While WRM1 does not bind cadherins or α-catenin to participate in cell adhesion (an activity carried out by HMP2), it is intriguing that WRM1’s nuclear activity is regulated in part by its release from the cell cortex in response to Wnt and CDK1 signals. Thus, in the context of the Howard et al. study discussed above, perhaps β-catenin release from cadherins shares features with the cortical release of C. elegans WRM1.
6. THE OTHER CATENINS
While β-catenin may be the best-known example of a dual-function adhesion-nuclear signaling protein, it is important to recognize that other catenins appear to follow the same paradigm. For example, plakoglobin (γ-catenin), which is within the β-catenin subfamily and is typically associated with desmosomal cadherins, can also interact with E-cadherin under conditions where β-catenin is limiting (Huelsken et al., 2000). Like β-catenin, plakoglobin contains an N-terminal phosphodestruction box, is stabilized by Wnts (more modestly than β-catenin), and interacts with TCF family DNA-binding proteins to modulate gene expression (Karnovsky & Klymkowsky, 1995; Klymkowsky, Williams, Barish, Varmus, & Vourgourakis, 1999; Kolligs, Hu, Dang, & Fearon, 1999; Simcha et al., 1998; Williams, Barish, Klymkowsky, & Varmus, 2000; Zhurinsky, Shtutman, & Ben-Ze’ev, 2000). This occurs in a manner that is less well understood than for β-catenin, as plakoglobin appears to bind a distinct region on TCF that has context-dependent positive and negative effects on gene expression (Miravet et al., 2002; Solanas et al., 2004).
Plakoglobin has been implicated in nuclear signaling by a number of groups (reviewed in Aktary & Pasdar, 2012; Swope, Li, & Radice, 2013), with some reports suggesting a signaling axis relating to the extent of its dissociation from classic or desmosomal cadherins present at desmosomal or adherens junctions (Hu, Berkowitz, O’Keefe, & Rubenstein, 2003; Hu, O’Keefe, & Rubenstein, 2001; Miravet et al., 2003; Williamson et al., 2006). In addition to TCF/LEF, plakoglobin is reported to associate with p53 and coregulate some p53 target genes (Aktary, Kulak, Mackey, Jahroudi, & Pasdar, 2013), as well as to associate with other proteins having nuclear activity (Aktary & Pasdar, 2013; Lam et al., 2012). Certain mutations in human desmosomal proteins, including desmosomal cadherins (e.g., desmoglein2 and desmocollin2), reduce plakoglobin’s junctional association, enhance its nuclear localization, and thereby contribute to the heart pathology arrhythmogenic right ventricular cardiomyopathy (ARVC) (Garcia-Gras et al., 2006). The condition can be mimicked by transgenic plakoglobin expression in mice, where plakoglobin shows enhanced nuclear localization and suppresses canonical Wnt signals (Lombardi et al., 2011). Conversely, the conditional removal of plakoglobin from the mouse heart leads to a decrease in desmosomal proteins and a coincident increase in β-catenin stabilization and Wnt signaling (Li et al., 2011). Possibly reflecting in part such a relationship of plakoglobin’s antagonism of Wnt/β-catenin signaling in some contexts, plakoglobin’s removal in keratinocytes protects keratinocytes from apoptosis (Dusek et al., 2007), whereas plakoglobin expression in mouse epidermis and hair follicles results in stunted proliferative potential and hair growth, and premature differentiation (Charpentier, Lavker, Acquista, & Cowin, 2000).
In addition to β-catenin and plakoglobin, at adherens junctions the cytodomains of classic cadherins associate with another armadillo-domain protein group known as the p120ctn subfamily. Members of the p120ctn family bind to the cadherin directly, but at a more membrane-proximal site than β-catenin or plakoglobin. The armadillo domains of the p120ctn-subfamily are similar in structure to β-catenin, even as their homology is not especially high. Members include p120ctn, ARVCF (armadillo-repeat gene deleted in velocardiofacial syndrome), p0071, and δ-catenin, and they bind to classic cadherins in a mutually exclusive manner. α-Catenin, on the other hand, is more structurally similar to vinculin, binds cadherin only indirectly (via β-catenin), and lacks an armadillo domain.
p120ctn, as well as other subfamily members, promotes cadherin stability, as best studied for E-, N-, and VE-cadherin (Chiasson, Wittich, Vincent, Faundez, & Kowalczyk, 2009; Davis, Ireton, & Reynolds, 2003; Ferreri, Minnear, Yin, Kowalczyk, & Vincent, 2008; Hatanaka, Simons, & Murakami, 2011; Iyer, Ferreri, DeCocco, Minnear, & Vincent, 2004; Mege, Gavard, & Lambert, 2006; Reynolds & Carnahan, 2004; Xiao et al., 2005). Additionally, p120ctn exhibits key roles in modulating the functions of small GTPases, as will be discussed later (reviewed in Anastasiadis, 2007; Kourtidis et al., 2013; Ratheesh et al., 2013). While the p120ctn subfamily is far less studied than β-catenin, each catenin also has nuclear roles. We will mention such functions briefly, while directing the reader to additional papers (reviewed in Bass-Zubek, Godsel, Delmar, & Green, 2009; Daniel, 2007; Hatzfeld, 2007; Kourtidis et al., 2013; McCrea & Park, 2007; Schackmann, Tenhagen, van de Ven, & Derksen, 2013; Schmidt & Jager, 2005; Stepniak et al., 2009).
p120ctn subfamily nuclear functions bear similar features to some β-catenin activities. For example, despite some interesting alternative views (Blattler et al., 2013; Ruzov, Hackett, et al., 2009; Ruzov, Savitskaya, et al., 2009), a number of p120ctn gene targets are shared with β-catenin. Further, the stability of p120ctn-isoform1, which determines the size of its signaling pool, is regulated by canonical Wnt signals in much the same way as β-catenin (Hong et al., 2010, 2012; Kim et al., 2004; Park et al., 2005; Spring et al., 2005). Even with a number of shared gene targets, each catenin is likely to have distinct nuclear roles. For example, in contrast to β-catenin, the expression of p120ctn or ARVCF alone cannot generate an ectopic dorsal axis when expressed on the ventral side of early Xenopus embryos (Geis, Aberle, Kuhl, Kemler, & Wedlich, 1998; Paulson, Fang, Ji, Reynolds, & McCrea, 1999). Future genome-wide analyses should assist in discerning the shared versus distinct gene targets and nuclear outputs of these catenins. At least three distinct transcription factors have thus far been found in direct protein–protein and functional association with p120ctn, with some evidence indicating that p120’s sequestration by cadherins or the cytoskeleton has an impact upon such nuclear signaling (Daniel & Reynolds, 1999; Hosking et al., 2007; Kim et al., 2004; Lee et al., 2014). Additional transcription factors bind to δ-catenin (Gu et al., 2011), ARVCF (unpublished), and plakophilin3-catenin (Munoz et al., 2014). Thus, just as β-catenin binds to multiple types of transcription factors (with the HMG-box containing TCF/LEF being the most prominent), each p120ctn subfamily member is likely to bind more than one DNA-binding factor, with some relationships being unique to that catenin. Future studies that uncover this larger network of catenin nuclear functions will likely provide additional opportunities to learn how the cadherin–catenin complex signals to the nucleus in varied contexts (Fig. 4).
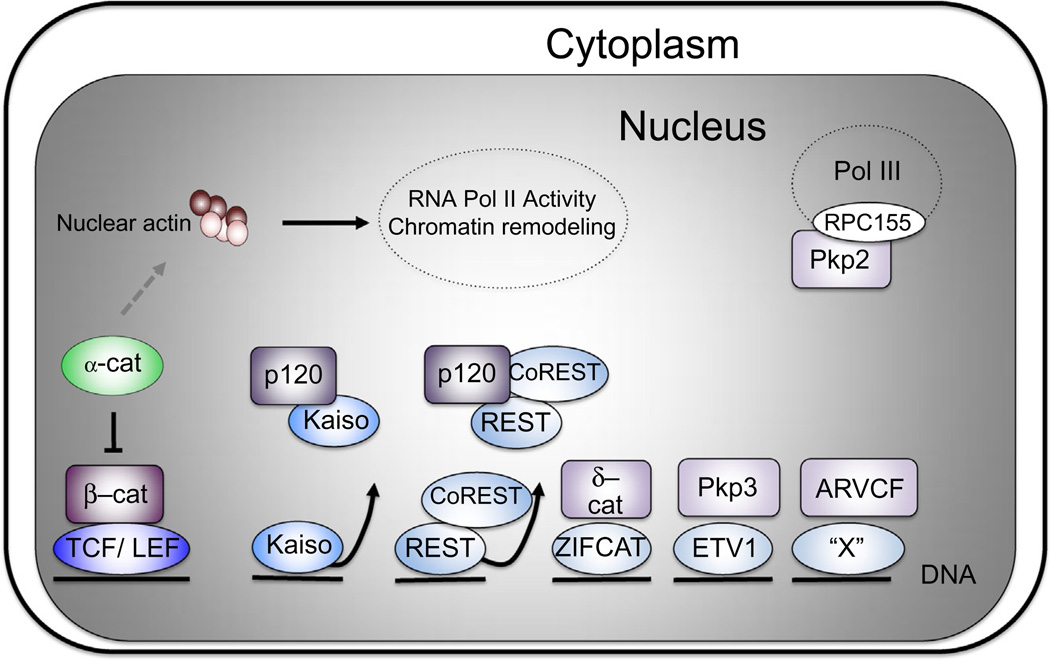
Figure 4.
Nuclear roles for catenins. The armadillo family of catenin proteins (β-catenin, p120ctn, δ-catenin, ARVCF, and plakophilins (PKP) 2 and 3 (shades of purple; light gray in the print version)) modulate gene expression via association with cognate DNA-binding factors (shades of blue; gray in the print version). In most instances characterized to date, derepression occurs to activate the target genes. However, Pkp3 appears to further activate gene targets already positively modulated by ETV1, and δ-catenin's action upon ZIFCAT is not yet known. Lacking an armadillo domain and homology to the other catenins, α-catenin (green; gray in the print version) is structurally related to vinculin and may impact transcription through its actin-binding function, inhibiting RNA polymerase II (RNA Pol II). PKP2 can interact with a component of the RNA Pol III complex RPC155.
Finally, it is worth noting that armadillo-repeat catenin proteins like β-catenin, plakoglobin, and p120ctn family members appear to have arisen from the same evolutionary predecessor as the importins (Andrade, Petosa, O’Donoghue, Muller, & Bork, 2001), which shuttle substrates across the nuclear membrane. Thus, in metazoans, catenin nuclear roles were likely selected for in concert with those relating to cadherins. In this scenario, catenins would have enhanced the functionality of cell–cell junctions on a structural level, and additionally on a signaling level, given their predisposed capacity to enter the nucleus (reviewed in (Fagotto, 2013; Hulpiau & van Roy, 2011)).
7. NUCLEAR SIGNALING FUNCTIONS OF THE ACTIN-BINDING PROTEIN, α-CATENIN
α-Catenin is an F-actin-binding protein and vinculin homologue and has been long appreciated to play an indispensible role in cell–cell adhesion by forming an essential link between the cadherin/β-catenin and the underlying actin cytoskeleton (Maiden & Hardin, 2011). As cells contain a substantial amount of cadherin-free α-catenin (Benjamin et al., 2010; Schneider, Herrenknecht, Butz, Kemler, & Hausen, 1993), there has been long-standing interest in the unique functions of this pool of α-catenin. Indeed, recent studies show that this pool of α-catenin can impact actin dynamics, such as limiting Arp2/3-based actin polymerization and severing by cofilin (Benjamin et al., 2010; Drees, Pokutta, Yamada, Nelson, & Weis, 2005; Hansen et al., 2013), which ultimately promotes the assembly and stabilization of unbranched filament bundles. This ability of α-catenin to impact F-actin dynamics might explain how α-catenin loss has been linked to the context-dependent activation of numerous pathways, such as Ras (Vasioukhin, Bauer, Degenstein, Wise, & Fuchs, 2001), NFκB (Kobielak & Fuchs, 2006; Piao et al., 2014), Hedgehog (Lien, Klezovitch, Fernandez, Delrow, & Vasioukhin, 2006; Rhee, Ryu, Kim, Chun, & Chun, 2012), and Hippo/Warts (Schlegelmilch et al., 2011; Silvis et al., 2011). While in some of these cases, α-catenin can be co-immunoprecipitated with various components of these pathways (e.g., the NFκB negative regulator, IκBα (Piao et al., 2014); the Hippo/Warts effector, Yap (Schlegelmilch et al., 2011; Silvis et al., 2011); and the Hedgehog nuclear effector, Gli3 (Rhee et al., 2012)), the molecular details of these interactions and the extent to which they are mediated by the cadherin-free pool of α-catenin remain unknown. In this regard, it is important to bear in mind that as the major cell–cell adhesion system in cells, the cadherin/β-catenin/α-catenin complex is critical not only for intercellular adhesion but also for establishing the close cell contacts that so many other junction and juxtacrine signaling molecules depend upon, from tight junctions in epithelia to gap junctions and desmosomes in cardiac tissue to membrane-anchored signaling pairs like Notch/Delta or Ephrins and Eph receptors, as mentioned above (Fagotto & Gumbiner, 1996; Ferreira et al., 2011; Zantek et al., 1999). Thus, it may be useful to consider some of the pleiotrophic signaling effects of α-catenin loss in the context of its broader role in cell–cell adhesion.
Regardless of this issue, it is clear that one important target of the cytosolic pool of α-catenin is its major stoichiometric binding partner, β-catenin. While a number of forced-expression studies showed that α-catenin could inhibit the nuclear accumulation (Takahashi, Ishihara, Takada, Tsukita, & Nagafuchi, 2000) and signaling of β-catenin (Giannini, Vivanco, & Kypta, 2000; Hwang et al., 2005; Merdek, Nguyen, & Toksoz, 2004; Sehgal, Gumbiner, & Reichardt, 1997), it was not until recently that α-catenin silencing studies were carried out in a way that revealed its capacity to function as a bona fide negative regulator of β-catenin signaling. In one study, α-catenin was found to limit β-catenin signaling by promoting its ubiquitylation and proteolysis by the “destruction complex” through an ability to bind a distinct region of APC (Choi et al., 2013). Interestingly, this function of α-catenin appears to occur at the level of β-catenin/TCF-occupied promoters and in the context of a histone H3 lysine K4 demethylase transcriptional repressor complex containing APC:CtBP: CoREST:LSD1 proteins. A second recent study confirmed earlier suggestions that endogenously expressed α-catenin can localize to the nucleus (El-Bahrawy, Talbot, Poulsom, & Alison, 2002; Giannini et al., 2000), but only under conditions where β-catenin is also nuclear, demonstrating that α-catenin nuclear function is largely β-catenin dependent (Daugherty et al., 2014). Like the study by Choi and colleagues, α-catenin can be found in a complex with both β-catenin and TCF on Wnt-responsive promoters, suggesting that α-catenin can limit β-catenin-mediated transcription at gene promoters, but mechanistically the studies diverge, probably because the latter study used APC mutant colon carcinoma cell lines. Indeed, Daugherty et al. found that the C-terminal actin-binding region of α-catenin is important for its capacity to inhibit β-catenin/TCF transcription, raising the possibility that α-catenin’s ability to impact actin organization might be related to its function as a transcriptional inhibitor. Indeed, cell nuclei contain a substantial amounts of G-actin and β-actin incorporates into all three RNA polymerase complexes and some chromatin-remodeling complexes required for transcription (Hofmann et al., 2004; Hu, Wu, & Hernandez, 2004; Kukalev, Nord, Palmberg, Bergman, & Percipalle, 2005; Philimonenko et al., 2004) suggesting that nuclear proteins with a capacity to bind actin may affect gene expression. Supporting this idea, a nuclear-targeted form of α-catenin could induce the formation of nuclear F-actin filaments, while cells lacking α-catenin showed greater nuclear actin mobility using a GFP-tagged, nuclear-localized actin. In addition, formation of nuclear actin filaments correlated with reduced RNA synthesis and a more compact chromatin organization, suggesting cross talk with components that drive gene repression (such as the CtBP:CoREST:LSD1 described above). While the studies by Choi et al. and Daugherty et al. present very different models of transcription inhibition by α-catenin, the models are not contradictory, and thus each set of observations likely inform different aspects of the transcription inhibitory mechanism. Moreover, evidence that the p120ctn family of proteins can interact with and negatively regulate the REST/CoREST repressor complex (Lee et al., 2014) suggests that nuclear-localized catenins may broadly affect gene expression and cell differentiation through regulation of this complex (Fig. 4).
Other proteins that associate with cadherins, such as the LIM-family members Ajuba and Zyxin, further exhibit cytoskeletal as well as nuclear localizations and functions (reviewed in Kadrmas & Beckerle, 2004; Smith, Hoffman, & Beckerle, 2014). The physical interactions of Ajuba include that with α-catenin, occurring at cadherin junctions (Marie et al., 2003). In the nucleus, Ajuba inhibits growth and promotes endodermal differentiation in P19 embryonal cells (Kanungo, Pratt, Marie, & Longmore, 2000), and it also modulates certain growth-hormone receptors, as well as an unrelated transcription factor (Montoya-Durango et al., 2008). Further, acting upstream of nuclear entry, Ajuba negatively regulates Hippo signaling through functional interactions with kinases that determine YAP’s nuclear transit (Das Thakur et al., 2010), while in Xenopus, Ajuba promotes meiotic maturation through ERK activation (Goyal et al., 1999). Thus, although additional insights are needed of how the functions of LIM-proteins at cadherin contacts are tied to those in the nucleus, a number of these proteins appear to be candidates in transducing cadherin-to-nuclear signals.
8. CADHERIN NUCLEAR SIGNALING VIA RTKs
In a number of systems, cadherins have been implicated in cross talk with receptor tyrosine kinases (RTKs), in some cases, through direct association with these receptors. Early interest in this area arose from findings that the time-course of contact-dependent inhibition of cell growth, mediated in part by cadherins, coincided with lessened RTK activity (Takahashi & Suzuki, 1996). There is also ample evidence that the cadherin/catenin complex is modified by RTKs (and their kinase effectors) (Daniel & Reynolds, 1997; Hoschuetzky, Aberle, & Kemler, 1994), which appears to change the affinity of the cadherin/β-catenin interface and thereby has consequences for both the adhesive and nuclear signaling function of β-catenin (reviewed in Daugherty & Gottardi, 2007). Both examples of cadherin nuclear signaling will be discussed below.
8.1. Cadherin-mediated inhibition of diverse RTKs
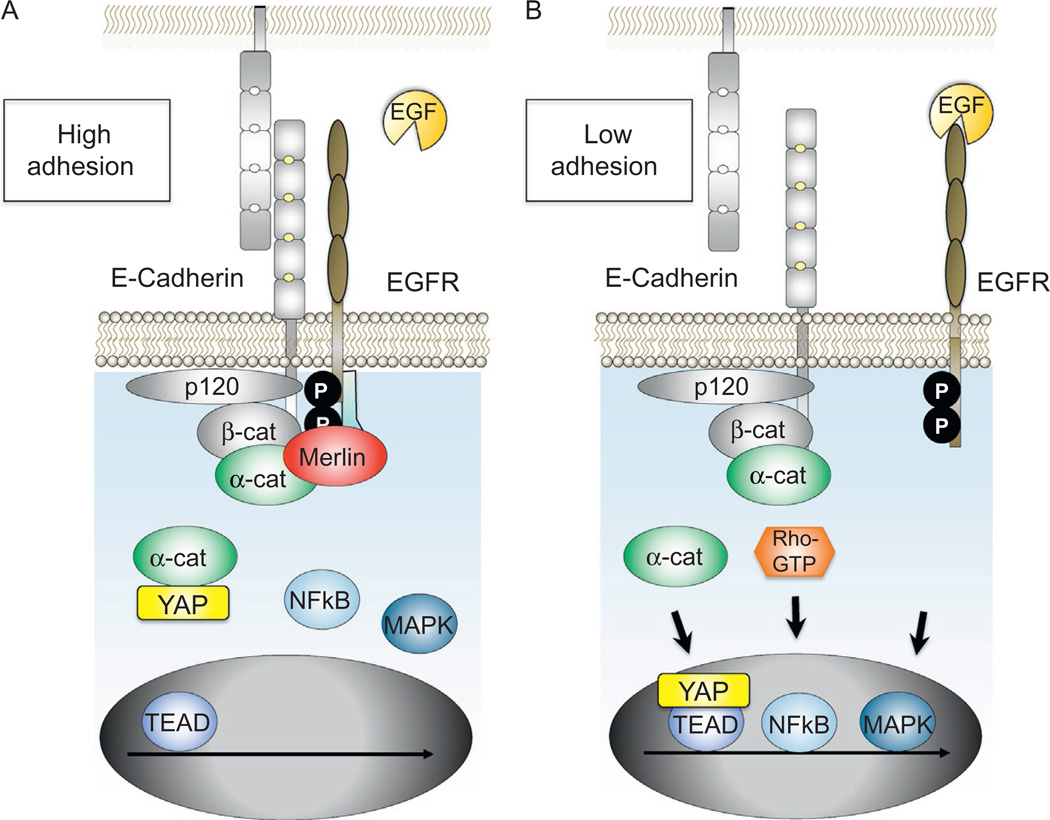
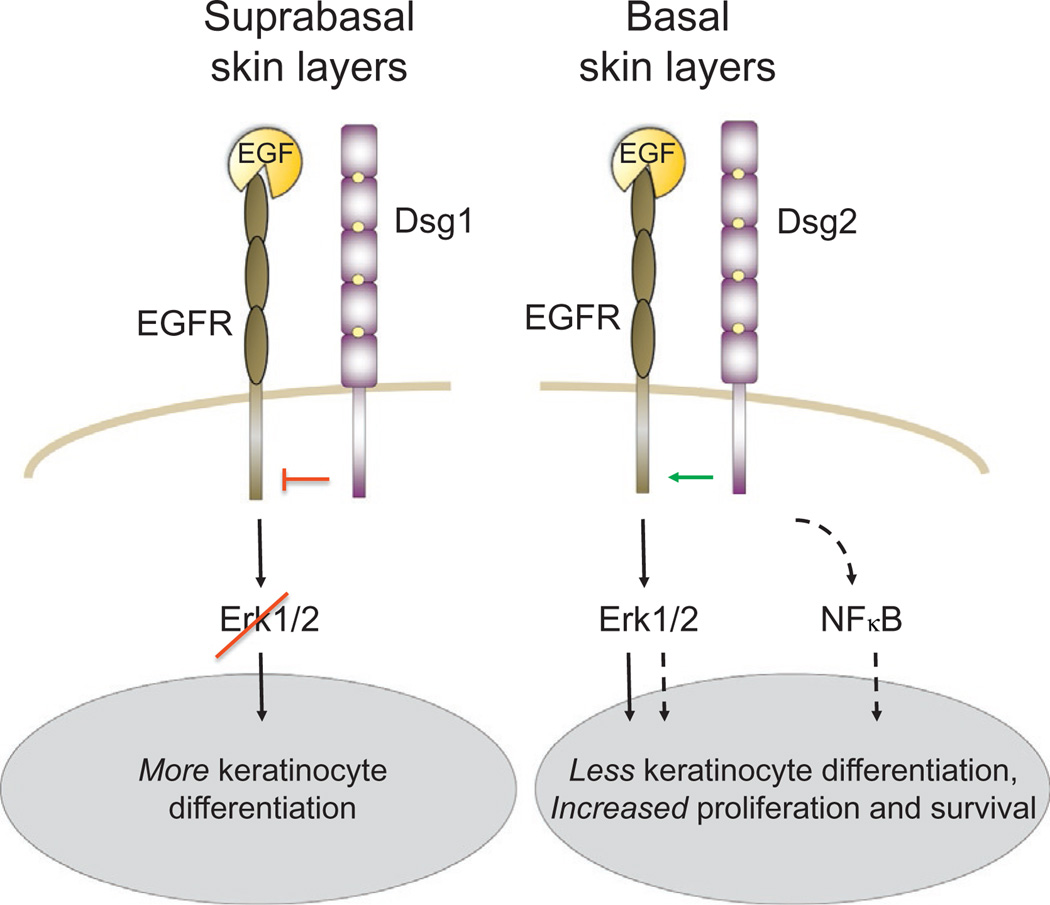
Early studies showed that the EGFR could colocalize with E-cadherin at apically localized adherens junctions (Chen, Solomon, Kui, & Soll, 2002), coassociate in immunoprecipitation assays (e.g., Hoschuetzky et al., 1994), and genetically interact in flies (Dumstrei, Wang, Shy, Tepass, & Hartenstein, 2002), suggesting a close relationship between these two proteins. Indeed, E-cadherin appears to limit EGFR activity at multiple levels. The cadherin ectodomain has been implicated in sterically preventing EGF-ligand binding (Qian, Karpova, Sheppard, McNally, & Lowy, 2004), while the cytodomain is required to suppress EGFR signaling downstream of its phosphoactivation (Perrais, Chen, Perez-Moreno, & Gumbiner, 2007). Other evidence suggests that physical interactions between the cadherin cytoplasmic domain, catenins, and the ERM protein, Merlin, are important for EGFR inhibition. Merlin associates with E-cadherin via α-catenin (Curto, Cole, Lallemand, Liu, & McClatchey, 2007; Gladden, Hebert, Schneeberger, & McClatchey, 2010) and acts as a link between cadherin-mediated contact maturation, the Par3 cell polarity complex, and the suppression of EGFR-mediated proliferation signals. Part of Merlin’s activity is to restrict EGFR to a membrane compartment incapable of passing on a signal (e.g., by preventing endocytosis). Merlin is thought to do this by binding the PDZ-domain containing protein NHE-RF1 (Curto et al., 2007; Gladden et al., 2010), which is in complex with EGFR. Since NHE-RF1 binds analogously to other RTKs, additional RTKs may also be subject to modulation by cadherins. While E-cadherin can suppress signaling from RTKs other than EGFR, including IGF-1R, c-Met, and ErB2–4 (Qian et al., 2004; Vermeer et al., 2003), Merlin is selective for the EGFR (Curto et al., 2007), suggesting that the cadherin/catenin complex uses molecules functionally analogous to Merlin to limit signaling from other types of RTKs (Fig. 5).
Figure 5.
Cadherin signaling via RTKs, NFκB, and Hippo pathways. E-Cadherin in densely packed epithelial monolayers can inhibit access of EGF to the EGFR as well as down-stream signaling from the EGFR via Merlin (A), as contrasted with less mature contacts characterized by lowered extents of trans-E-cadherin interactions, where the indicated nuclear signaling trajectories are enhanced (B). Cadherin engagement can also limit the nuclear accumulation of YAP and NFκB through α-catenin and Rho-dependent mechanisms, respectively.
Interestingly, cadherin homotypic adhesion between cells (i.e., in trans), often associated with cadherin clustering in cis, is not necessarily required for cadherins to influence RTKs (Cavallaro & Dejana, 2011). VE-cadherin’s cytodomain can interact with VEGFR2, preventing its endocytosis and facilitating its dephosphorylation via the junctional phosphatase, DEP1 (Grazia Lampugnani et al., 2003). This reduces MAPK signaling, leading to growth inhibition (Lampugnani, Orsenigo, Gagliani, Tacchetti, & Dejana, 2006). VE-cadherin’s association with VEGFR has also been tied to cell survival via the inhibition of apoptosis, as targeted truncation of VE-cadherin’s β-catenin-binding region led to mouse embryo lethality, in part, due to loss of responsiveness to survival signals from the VEGF-A ligand, leading to increased endothelial apoptosis (Carmeliet et al., 1999; Gavard, 2013). Thus, the catenin-binding domains of both E-cadherin and VE-cadherin are important for the regulation of their cognate RTKs. VE-cadherin has also been implicated in promoting or responding to signals from additional membrane receptors, including FGFR, TGFβ-R2, and the angiopoietin1 receptor, Tie2 (Fukuhara et al., 2008; Hayashi et al., 2013; Murakami et al., 2008; Rudini et al., 2008; Saharinen et al., 2008; Winderlich et al., 2009).
Another example where cadherins are involved in adhesion-independent signaling (observed even in single cells) is that of VE-cadherin bridging the mechanosensor PECAM to VEGFR2 (Shay-Salit et al., 2002; Tzima et al., 2005). Upon flow (shear-stress), PECAM activates Src-family kinases, while VE-cadherin assists PECAM in passing signals to VEGFR2, thereby enhancing PI3K activity and the NFκB pathway. The VE-cadherin enhancement of PI3K activity is also associated with cross talk between adherens and tight junctions through nuclear signaling by a FoxO family transcription factor (Taddei et al., 2008). In this study, VE-cadherin’s enhancement of PI3K and Akt activity led to phosphorylation of FoxO1 and the release of its repressive activity at the promoter of claudin1, a component of tight junctions. Once cells become fully confluent, VE-cadherin clustering activates PIP3K and Akt signaling more strongly in releasing FoxO1 from DNA. A VE-cadherin mutant unable to bind β-catenin or interact with PIP3K was unable to produce such effects. In addition, VE-cadherin limits the signaling pool of β-catenin, as noted earlier. Once sequestered, β-catenin can no longer form a repressive complex together with FoxO1 and TCF4 (Taddei et al., 2008).
Like VE-cadherin’s ability to facilitate VEGFR signaling discussed above, N-cadherin can stimulate FGFR signaling, even in single cells (Suyama, Shapiro, Guttman, & Hazan, 2002; reviewed in Radice, 2013). In this latter context, N-cadherin associates in cis with the extracellular domain of FGFR (within the same cell) (Utton, Eickholt, Howell, Wallis, & Doherty, 2001), preventing its ligand-mediated internalization and downregulation (Suyama et al., 2002). Lastly, E-cadherin-mediated cell–cell contact formation can lead to PI3K and Akt, activation, promoting cell survival in response to extracellular stimuli (McLachlan & Yap, 2007; Pece, Chiariello, Murga, & Gutkind, 1999), as well as EGFR-dependent, ligand-independent increases in MAPK activation (Pece & Gutkind, 2000). Taken together, the major varieties of classical cadherins (E-, Nand VE-types) play both facilitator and inhibitor roles toward their cognate RTKs (similar to E-cadherin’s contribution to Wnt signaling). While there appears to be no current unified mechanism for these regulatory roles, interactions mediated by both extracellular and intracellular catenin-binding domains are clearly important for RTK regulation by cadherins (Fig. 5).
8.2. β-Catenin as a key target of RTKs and other membrane-activated kinases
Given that β-catenin is a dual-function adhesion/transcriptional coactivator protein, it has long been speculated that modifications that reduce the affinity of β-catenin for the cadherin and α-catenin could lead to enhanced nuclear signaling activity of β-catenin. We will largely discuss the more recent evidence favoring this model, since this area has been generally addressed in reviews elsewhere (Cavallaro & Dejana, 2011; Daugherty & Gottardi, 2007; Harris & Tepass, 2010; Hartsock & Nelson, 2008; Heuberger & Birchmeier, 2010; Meng & Takeichi, 2009; Nelson & Nusse, 2004; Niessen, Leckband, & Yap, 2011).
The best evidence for the aforementioned model involves a tyrosine phosphorylation in β-catenin at residue 654, which leads to reduced E-cadherin binding both in vitro (Roura et al., 1999) and in vivo (van Veelen et al., 2011), and enhanced nuclear signaling activity (Jean et al., 2009; Kim et al., 2009; Piedra et al., 2001; van Veelen et al., 2011; Xi et al., 2013). Activation of a number of kinases is associated with phosphorylation of β-catenin at Y654, including EGFR (Jean et al., 2009), HGF (David et al., 2008), TGFβ (Kim et al., 2009), and Src downstream of a variety of signals (Condello, Cao, & Matei, 2013; Lee et al., 2013; Sumiyoshi, Takahashi, Obata, Sugimoto, & Kohara, 2011; Xi et al., 2013). In most of these cases, pY654 β-catenin shows reduced association with cadherin contacts as well as enhanced nuclear accumulation and target-gene activation. Often missing from the discussion of these data, however, is the extent to which reduced binding of β-catenin to a cadherin alone is sufficient for β-catenin signaling, given that cadherin-free β-catenin is thought to be rapidly degraded by the destruction complex. We speculate, therefore, that these kinases may in parallel generate “Wnt-like” signals that inhibit GSK3 and ultimately inhibit the N-terminal phosphorylation and degradation of β-catenin, resulting in a signaling form of β-catenin that is refractory to sequestration by cadherins.
Other sites in β-catenin have also been implicated in reduced adhesive activity and enhanced nuclear signaling. Tyrosine phosphorylation of β-catenin residue 142 reduces its association with α-catenin (Piedra et al., 2003). This would lead to a form of β-catenin that may be better able to signal because of its reduced association with α-catenin, which can inhibit β-catenin signaling (discussed above) and/or enhanced association with transcriptional coactivator protein Bcl9 (Brembeck et al., 2004; Kramps et al., 2002), although the role of Y142 may be less important in (Hoffmans & Basler, 2007) or indirect toward Bcl9 (Sampietro et al., 2006). Lastly, tyrosine 489 in β-catenin has also been associated with reduced cadherin binding and nuclear signaling activity downstream of a Slit-Robo abl tyrosine kinase mechanism (Rhee, Buchan, Zukerberg, Lilien, & Balsamo, 2007).
While the above examples focus on the impact of phosphorylation for just a few sites in β-catenin, we have many more historic examples that are discussed above where signaling is coassociated with changes in the cadherin–catenin complex along with elevated β-catenin signaling, but where precise mechanisms are lacking, such as following the activation of the FGFR (Pai et al., 2008), EGFR (Galbiati et al., 2000; Lu, Ghosh, Wang, & Hunter, 2003), IGF-2 (Morali et al., 2001), and IGFR-type1 (Playford, Bicknell, Bodmer, & Macaulay, 2000). Conversely, inhibition of certain RTKs, such as ErbB2, coincides with canonical Wnt signal suppression and greater cell–cell adhesive integrity (Bonvini et al., 2001). Not surprisingly, phosphatases appear to counter the effects of tyrosine kinase activation, resulting in enhanced integrity of the cadherin–catenin complex and cell–cell adhesion, and in some cases reduced β-catenin signaling (Balsamo, Arregui, Leung, & Lilien, 1998; Hellberg, Burden-Gulley, Pietz, & Brady-Kalnay, 2002; Nawroth et al., 2002; Novellino et al., 2008; Xu et al., 2004; Yan et al., 2006). Whether the above effects are largely mediated through modification of β-catenin at Y142, 489, or 654 or other presently uncharacterized sites is not known. Moreover, p120ctn is also targeted by many of these same kinases (reviewed in Alema & Salvatore, 2007; Daniel & Reynolds, 1997) and phosphatases, with contributions of individual phosphorylation sites to adhesive and signaling functions just emerging. For example, PKCα activation is associated with p120ctn’s phosphorylation at S879, resulting in its reduced association with VE-cadherin (Vandenbroucke St Amant et al., 2012), and conceivably increased p120ctn signaling. Likewise, as a consequence of ligand activation of the canonical Wnt pathway, CK1ε has been found to phosphorylate p120ctn at S268 and S269, resulting in its dissociation and enhanced nuclear activity in relieving Kaiso-mediated gene repression (Dann et al., 2014; Del Valle-Perez, Casagolda, et al., 2011). Other phosphorylation events are known to modulate p120ctn’s nuclear signaling (Hong et al., 2010, 2012), but are not (yet) linked with reduced binding to cadherins.
9. CADHERIN NUCLEAR SIGNALING BY SMALL GTPases AND NFκB
As alluded to above, the canonical Wnt pathway is defined as being mediated via β-catenin. However, there are multiple distinct signaling trajectories not involving β-catenin that may be activated following Wnt-ligand binding to Frizzled/Lrp5/6. To what extent these trajectories are activated in parallel or independently of β-catenin is a matter of context and uncertainty, with cross talk (much of it antagonistic) occurring between them. Relative to that of β-catenin, these trajectories have been more difficult to study, as we possess fewer available endogenous readouts, synthetic reporters, or protein-based assays. The most prominent “noncanonical” pathways involve activation of small GTPases such as RhoA, Rac1, and Cdc42, often monitored via pull-down of their active GTP-bound forms, or indirectly, by scoring for phenotypic effects upon the actin or microtubule cytoskeletons, cell motility, or invasion (reviewed in Gomez-Orte, Saenz-Narciso, Moreno, & Cabello, 2013). Trajectories based upon intracellular calcium release also reside within the collection of noncanonical Wnt pathways, and there are indications of the involvement of trimeric G-protein pathways as well (Kilander et al., 2014; Luchtenborg et al., 2014).
In vivo, one of the examples of cadherin involvement with small GTPases comes from Xenopus, where the cytoplasmic region of cadherin11 activates Rho-GTPases at membrane regions via its interaction with the Rho guanine nucleotide-exchange factor (GEF) Trio (Kashef et al., 2009; van Rijssel & van Buul, 2012). In neural crest migration, the cytodomain of cadherin11 positively regulates RhoA, Rac and Cdc42 and is required for cell migrations into regions such as the pharyngeal pouches for formation of craniofacial cartilage (Kashef et al., 2009). As opposed to being solely enriched only along cell–cell borders in cranial neural crest explants, cadherin11 is also present at cell–extracellular matrix interfaces and cell protrusions, such as filopodia and lamellipodia, whose formation cadherin11 promotes.
VE-cadherin and VE-cadherin trans-interactions have likewise been linked to recruiting the Rho GEF Tiam1 to activate Rac in endothelial cells (Birukova et al., 2011; Lampugnani et al., 2002). A variety of studies have linked VE-cadherin to the modulation of Rac, Rho, and/or Cdc42 (Gavard & Gutkind, 2006; Mehta & Malik, 2006; van Nieuw Amerongen, van Delft, Vermeer, Collard, & van Hinsbergh, 2000; Wojciak-Stothard, Potempa, Eichholtz, & Ridley, 2001; Wojciak-Stothard & Ridley, 2003). Cadherins in some cases interact indirectly with small GTPases via catenins such as p120ctn, which recruits the p190-RhoGEF to cadherins to enhance junctional integrity (Wildenberg et al., 2006). Through such effects, cadherins modulate small GTPases in manners that are relevant to cytoskeletal organization and force generation. Cadherin-dependent forces, for example, can include the generation of cortical tension that assists in driving many morphogenic events (reviewed in Nishimura & Takeichi, 2009; Niessen et al., 2011; Watanabe, Sato, & Kaibuchi, 2009).
In Xenopus, p120ctn and δ-catenin regulate the small GTPases Rac and Rho during gastrulation (Ciesiolka et al., 2004; Fang et al., 2004; Gu et al., 2009). In mice, multiple studies have likewise indicated that this functional link is important for development, as exemplified in forming neurons (Elia, Yamamoto, Zang, & Reichardt, 2006). Together, small-GTPase modulation appears to be a signaling feature of cadherins and the p120ctn subfamily (Anastasiadis, 2007; Hatzfeld, 2005; Kourtidis et al., 2013; Pieters, van Hengel, & van Roy, 2012; Ratheesh et al., 2013; Watanabe et al., 2009). While p120ctn’s cytoskeletal effects can be pronounced, the consequent small-GTPase impact upon nuclear events has yet to be determined. As noted earlier, such effects are likely, even if occurring indirectly via changes in cell tension and shape. The plakophilin-catenin subfamily may also prove to engage in such nuclear signaling via small-GTPase effects. Although we do not discuss the plakophilins in depth, they are structurally similar to the other armadillo-domain-containing catenins and were first found in association with desmosomal cadherins (desmogleins and desmocollins) (Heid et al., 1994; reviewed in Bass-Zubek et al., 2009).
p120ctn further engages in modulation of NFκB, a pathway having clear links to the nucleus. It is not currently known to what extent such regulation occurs only after p120ctn departs from the cadherin complex, perhaps in response to phosphorylation events, versus when p120ctn is still bound or conceivably oscillating on and off the cadherin (or newly synthesized). However, as for β-catenin, the signaling roles of p120ctn are usually thought to be higher when p120ctn is not cadherin bound. The knockout of p120ctn in mouse skin and certain other tissues leads to elevated NFκB activity (Davis & Reynolds, 2006; Kobielak & Fuchs, 2006; Perez-Moreno et al., 2006). It is believed that this effect, and another one upon mitotic chromosome segregation, comes about via p120ctn’s initial modulation of Rho, producing a downstream impact upon Rho-kinase and ultimately NFκB (Fig. 5).
In addition to cadherin modulation of NFκB signals via small GTPases such as Rho or Rac, more direct effects may be at play. In mammalian cultured cells, the exogenous expression of E-cadherin lowers β-catenin’s and NFκB’s association with gene targets such as fibronectin and Lef1 (Chen, Khan, Cukiernik, & Chakrabarti, 2003). Here, NFκB p65 appears to associate with the E-cadherin complex. In K-ras overexpressing cells, this association is lost, and such cells exhibit disrupted cell–cell junctions. Upon E-cadherin depletion, there is reduced NFκB association with cell–cell junctions as would be expected, and this is paralleled with an increase in NFκB transcriptional activity (under both basal or TNFα conditions). The concept is that the E-cadherin complex is not only relevant to signaling conferred via β-catenin but also that via NFκB, with each acting on certain shared gene promoters (e.g., fibronectin and Lef1). This relationship is proposed to tether E-cadherin levels to cell fate choices, such as in EMT (Solanas et al., 2008). A positive feedback loop may be involved, since β-catenin and NFκB also promote the expression of Slug, ZEB1, and ZEB2, which are repressors of E-cadherin transcription (Chua et al., 2007; Conacci-Sorrell et al., 2003).
We have briefly mentioned these less traditionally discussed catenin trajectories because they are apt to provide additional opportunities for the cadherin–catenin complex to engage in nuclear signaling. In the context of Xenopus development, a number of labs have examined the functional relationship of C-cadherin with the PAPC-cadherin (Chen & Gumbiner, 2006; Chen, Koh, Yoder, & Gumbiner, 2009) and of these cadherins with the Frizzled receptor (Kraft, Berger, Wallkamm, Steinbeisser, & Wedlich, 2012). Upon the binding of the Wnt11 ligand to the Frizzled7 receptor, these cadherins are reported to bind Frizzled7 directly and to partition into distinct complexes (Medina, Swain, Kuerner, & Steinbeisser, 2004). Adhesion normally conferred by C-cadherin is lowered via a block to C-cadherin’s lateral dimerization. As PAPC’s expression is restricted to regions undergoing morphogenic movements in Xenopus, such as during gastrulation (Medina et al., 2004; Winklbauer, Medina, Swain, & Steinbeisser, 2001), PAPC lowers C-cadherin’s adhesive function to enable needed cellular rearrangements. Further, PAPC’s cytoplasmic domain interacts with Sprouty and ANR5 (Chung, Yamamoto, & Ueno, 2007; Wang et al., 2008) permitting PAPC to participate in PCP signaling via the modulation of small GTPases.
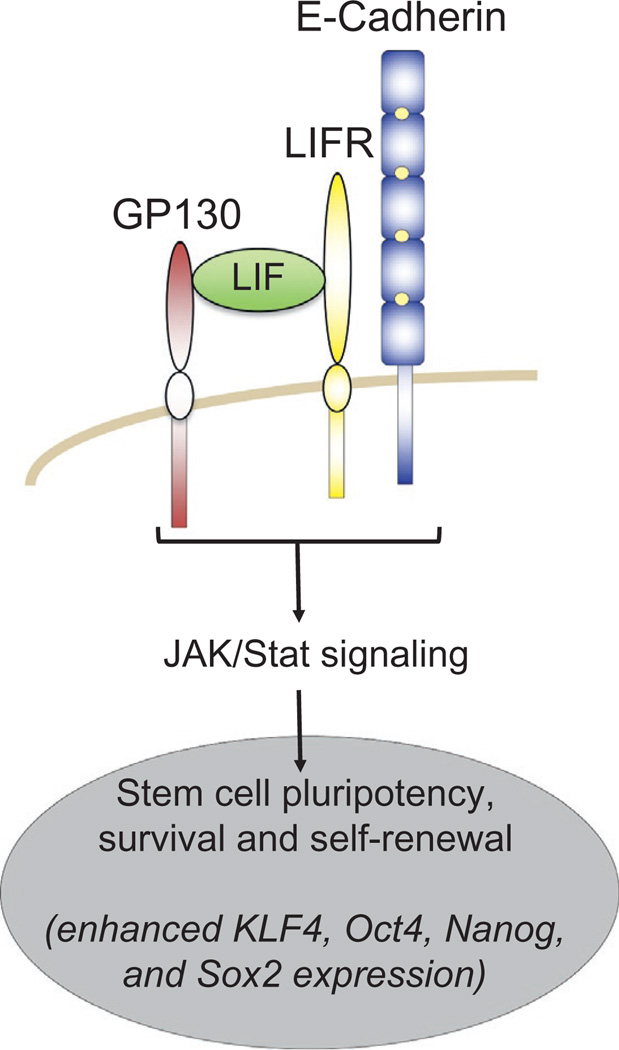
Other pathways are activated from cadherins that involve small GTPases. One relates to production of the IL6-cytokine, which binds cytokine receptors (e.g., gp130) on surrounding cells to initiate JAK-STAT signaling (Geletu et al., 2013). Cadherin ligation under high-density cell conditions, possibly mimicking cell-packing densities in vivo, affects Rho-family GTPase activity. The activation of Rac, for example, can lead to NFκB activation and thereby expression of the IL6-ligand. JAK-STAT signals often promote expression of cell survival gene products (e.g., Bcl-xL, survivin, Mcl1, myc, VEGF), and lower expression of p53. Such nuclear effects appear to be cadherin-dependent, since even a purified E-cadherin ectodomain fragment elicits such responses (Arulanandam et al., 2009). Further, this relationship may be quite general among cadherins, as it has been observed for C- and M-cadherin (Charrasse et al., 2007; Noren, Niessen, Gumbiner, & Burridge, 2001), as well as N-cadherin and cadherin11 (Geletu et al., 2013). Under the conditions examined, IL6 production following cadherin-mediated homophilic ligation did not activate ERK1/2 (Arulanandam et al., 2009; Geletu et al., 2013; Raptis et al., 2000). Thus, in the context of cadherin-mediated high cell densities, the STAT pathway appears to a major factor in conferring cell survival.
10. CADHERIN NUCLEAR SIGNALING VIA PROTEOLYSIS
A number of studies show production of cadherin proteolytic fragments promotes cadherin-to-nuclear signaling, and in some cancer contexts, the nuclear entry and staining of the cadherin cytodomain has already been used in diagnosis (reviewed in Chetty & Serra, 2008; David & Rajasekaran, 2012). There are reports of extracellular proteases such as ADAMs and MMPs being involved, as well as intramembrane or intracellular proteases whose activities can be coupled to the initial occurrence of extracellular cleavage events, for example, cleavages subsequently occurring via presenilins or caspases (Damsky, Richa, Solter, Knudsen, & Buck, 1983; De Wever et al., 2007; Dusek et al., 2006; Lochter et al., 1997; Marambaud et al., 2002; Maretzky, Reiss, et al., 2005; Maretzky, Schulte, et al., 2005; McCusker, Cousin, Neuner, & Alfandari, 2009; Noe et al., 2001; Steinhusen et al., 2001; Symowicz et al., 2007; Vallorosi et al., 2000; reviewed in David & Rajasekaran, 2012; McCusker & Alfandari, 2009; Niessen et al., 2011). We will focus upon the intracellular cadherin fragments, but some of the extracellular cleavage products also appear to have biological activity in both development and pathology, for example, in relation to RTK activation (e.g., ErbB/HER2:HER3) and the consequent upregulation of MMP activity or inhibition of apoptosis (Fedor-Chaiken, Hein, Stewart, Brackenbury, & Kinch, 2003; Najy, Day, & Day, 2008; Inge et al., 2011; Grabowska, Sandhu, & Day, 2012; Nawrocki-Raby et al., 2003; Zuo et al., 2011). Further, in vivo exogenous expression of the ectodomain fragment of C-cadherin, for example, disrupts early gastrulation movements in Xenopus, an effect that may take place via altered aPKC and Rac activity in the recipient cells expressing endogenous intact C-cadherin (Seifert, Ibrahim, Stodtmeister, Winklbauer, & Niessen, 2009). In MDCK cells, aPKC associates with the cytoplasmic tail of E-cadherin, an interaction partially dependent on β-catenin. Incubation with E-cadherin’s ectodomain increased the phosphorylation of aPKC and also its cytoplasmic association with native E-cadherin. aPKC and Rac are both involved in PCP pathways directing cell movements in development, so this work has suggested one means of cadherin outside-in signaling.
Intramembrane cleavage of cadherins has been evaluated for the release of active fragments to the cytoplasm or nucleus. One focus has been upon the presenilin protease, part of a membrane-associated complex that when misregulated promotes Alzheimer’s disease. In the context of cadherins, while reducing their adhesive function (Marambaud et al., 2002), presenilin generates a cytodomain fragment of N-cadherin with nuclear signaling activity (Marambaud et al., 2003). In particular, the fragment enters the nucleus to bind CBP (Creb-binding protein), leading to CBP’s degradation. This in turn lowers the ability of CBP’s binding partner, CREB (cAMP response element-binding protein), to transactivate gene targets. Thus, in cases where presenilin is inactive, the CBP/CREB complex is more active.
As additionally resolved in mice deficient for presenilin, β-catenin is more active (Kang et al., 2002). Conversely, presenilin expression reduces β-catenin activity. This effect may occur indirectly through promoting β-catenin’s phosphorylation via CK1 and GSK3β, and thereby reduced β-catenin stability (Kang et al., 1999, 2002; Soriano et al., 2001; Zhang et al., 1998). It is uncertain to what extent presenilin acts upon β-catenin that is associated with cadherin versus membrane localized via the destruction complex, as described (Maher et al., 2009).
Other proteases involved in the initiation of signals from the cadherin–catenin complex include the disintegrins and metalloproteases known as ADAMs. For example, ADAM10 colocalizes with N-cadherin in early mouse embryos, and they coassociate in cell line and in vitro contexts (Reiss et al., 2005). The extracellular cleavage of N-cadherin by ADAM10 first results in the generation of an extracellular fragment that retains some biological activities (Bixby & Zhang, 1990; Damsky et al., 1983; Paradies & Grunwald, 1993; Utton et al., 2001). The remaining membrane-bound fragment is then subject to an additional intramembrane cleavage step(s) by other proteases (Marambaud et al., 2003), such as gamma-secretase. In the case of N-cadherin, this results in lessened association with β-catenin and enhanced β-catenin cytoplasmic levels and nuclear activities (Fortini, 2002; Reiss et al., 2005; Shoval, Ludwig, & Kalcheim, 2007). The in vivo relevance was suggested in ADAM10-deficient mice embryos, where the loss of the N-cadherin C-terminal fragment dramatically reduced expression of an endogenous β-catenin reporter, cyclinD1. N-Cadherin proteolysis is initiated with the first extracellular cleavage step, which is itself responsive to the activation of PKC and calcium-mediated pathways. Thus, calcium influx during tissue remodeling or repair may act in part via shedding of the N-cadherin ectodomain in combination with cadherin cytodomain effects upon gene transcription. Such a scenario is likely to play out with additional cadherins as reported for E-cadherin in cancer cell-line contexts (Ito et al., 1999; Marambaud et al., 2002; Maretzky, Reiss, et al., 2005). Interestingly, p120ctn appears to enhance the nuclear entry of an E-cadherin cytodomain fragment, with the fragment enhancing p120ctn-facilitated relief of Kaiso-mediated target-gene repression and suppressing apoptosis (Ferber et al., 2008). Likewise, a fragment of E-cadherin arising as a consequence of presenilin 1 activity is reported to enhance β-catenin nuclear activity (Uemura et al., 2006). In the context of neurons, activation of the glutamate activated N-methyl-d-aspartate (NMDA) receptor, which contributes to long-term neuronal plasticity (as reviewed in Carroll & Zukin, 2002), leads to calcium influx and calpain activation, and the processing of both N-cadherin and β-catenin. Possibly initiated at the cadherin–catenin complex, the effects produced include enhanced β-catenin stability and signaling (Abe & Takeichi, 2007; reviewed in Arikkath & Reichardt, 2008).
In the developmental context of neural crest delamination in quail, bone morphogenic protein (BMP) signals result in N-cadherin fragmentation via ADAM10, leading to enhanced β-catenin expression and signaling, and thereby to enhanced cyclinD1 expression. Whereas such an N-cadherin cytodomain fragment might be expected to sequester β-catenin, another possibility in agreement with the observed downstream effects is that the fragment assists in protecting β-catenin from degradation until it reaches the nucleus to signal. Such a protection scenario is consistent with another study using the cytodomain E-cadherin (Simcha et al., 2001). In any case, interference with N-cadherin fragmentation compromised neural crest delamination. Prior to BMP signals, both the extracellular and intracellular domains of N-cadherin appear to be needed to maintain neural crest cells in a premigratory state (Shoval et al., 2007). Then, in initiating delamination and migration, BMP signals may act upon N-cadherin to both limit adhesive activity and coordinately enhance N-cadherin signaling output.
Desmosomal cadherins are essential in tissues that experience mechanical stress. In the intestine, cells differentiate as they progress along the crypt–villus axis. In the context of cadherin proteolytic fragments, the desmoglein2–cadherin was found to release a cytoplasmic fragment, likely resulting from a caspase, at the outset of apoptosis (Nava et al., 2007). Desmoglein’s ectodomain is additionally shed via ADAM10 and 17 cleavage (Bech-Serra et al., 2006). Further reports point to desmosomal cadherins being fragmented, such as desmoglein1 (Dusek et al., 2006; Weiske et al., 2001), in addition to classic cadherins (Rios-Doria & Day, 2005; Rios-Doria et al., 2003). In the case of desmoglein2, its cytoplasmic fragment promotes apoptosis, perhaps as proposed, in concert with the physiological extrusion of cells from the intestinal villus or during inflammation (Fig. 6).
Figure 6.
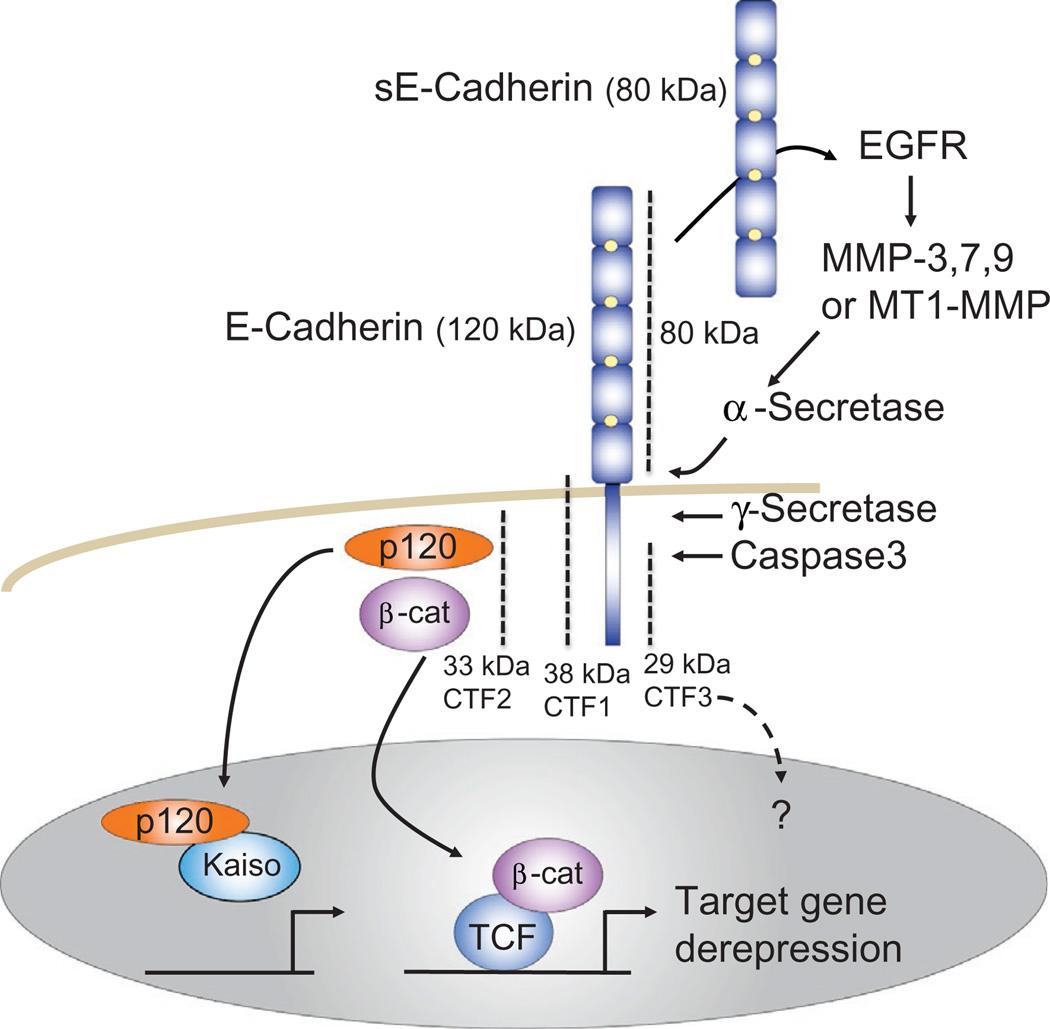
Cadherin signaling via proteolysis. Cadherins are cleaved at specific sites by proteases to generate fragments that are capable of transducing signals, either to the extracellular or to the intracellular space. The soluble extracellular E-cadherin fragment (sE-cadherin) can associate with intact E-cadherin present on other cells to alter cadherin-dependent cell properties inclusive of intracellular signaling (not shown). It also interacts with EGFR family members to activate MAPK signaling or metalloproteases, with the latter directly or indirectly resulting in the production of defined E-cadherin fragments. The intracellular C-terminal fragment number 2 (CTF2) is not membrane associated, so has been proposed to assist in the protection or translocation of p120ctn or β-catenin to the nucleus. Here, p120ctn displaces and thereby derepresses Kaiso-mediated repression of its gene targets, whereas β-catenin derepresses TCF/LEF-mediated gene repression via the recruitment of transcriptional coactivators. In the context of such selective proteolysis of E-cadherin, each catenin is thereby assisted in activating its respective gene targets.
11. PROTOCADHERIN SIGNALING TO THE NUCLEUS
The impact of the protocadherins on nuclear signaling is not as well studied as for the classical cadherins, even though the protocadherins constitute the largest group in the cadherin superfamily (reviewed in Kahr, Vandepoele, & van Roy, 2013;Weiner & Jontes, 2013). They typically have six to seven or more extracellular cadherin repeats, a single transmembrane pass, and interestingly, a cytoplasmic domain completely distinct from those of classical cadherins. Given that the cytodomains of protocadherins a and b are well conserved, their cytoplasmic domains likely execute conserved functions. They are not generally thought to interact with catenin proteins, although β-catenin has been reported to associate with protocadherin-PC (reviewed in Chen, Vacherot, et al., 2002; Vanhalst, Kools, Staes, van Roy, & Redies, 2005). Protocadherins are widely dispersed, with strong representation in neural tissues and those undergoing morphogenic movements (Chen & Gumbiner, 2006;Chen, Koh, et al., 2009;Nakao, Platek, Hirano,& Takeichi, 2008; Vanhalst et al., 2005; reviewed in Kahr et al., 2013). Some but not all have adhesive activity, possiblymodulated by cis-interactions occurring between different protocadherin members to provide combinatorial arrangements that assist in specifying cell–cell associations and identity (e.g., of neurons; reviewed in Chen & Maniatis, 2013). Protocadherins further interact with classic cadherins (Biswas, Emond, & Jontes, 2010; Chen, Koh, et al., 2009; Chen, Vacherot, et al., 2002; Emond, Biswas, Blevins, & Jontes, 2011; Emond, Biswas, & Jontes, 2009; Yasuda et al., 2007), with which they have been found in cis- or trans-associations. Via mechanisms noted above involving classic cadherins, nuclear signaling might thus occur for protocadherins indirectly as a consequence of these relationships.
Protocadherins also associate with Frizzled7 (Berndt et al., 2011; Kraft et al., 2012; Medina et al., 2004), implicated in noncanonical Wnt signaling, as well as the phosphatase PP1α (Chen, Vacherot, et al., 2002), which has been examined in contexts of learning and memory (Mulkey, Endo, Shenolikar, & Malenka, 1994). Neurobiologists are further interested in protocadherins given some of their interactions with proteins such as Dab1 (Homayouni, Rice, & Curran, 2001), which acts in the Reelin signaling pathway to influence neuronal positioning (reviewed in Bar, Lambert de Rouvroit, & Goffinet, 2000), or with the protocadherin arcadlin, which modulates N-cadherin cell surface levels and thereby dendritic spine densities (Yamagata et al., 1999; Yasuda et al., 2007).
Other associations include those with the tyrosine kinase Ret (Schalm, Ballif, Buchanan, Phillips, & Maniatis, 2010) or with CK2β, Sprouty, ANR5, or FLRT3 (Chen, Koh, et al., 2009; Chung et al., 2007; Kietzmann, Wang, Weber, & Steinbeisser, 2012; Wang et al., 2008; reviewed in Chen & Maniatis, 2013). Protocadherin interactions assist in keeping Ret stable and responsive to its GNDF ligand (Schalm et al., 2010), where they have been found to inhibit signaling from Pyk2 (prolinerich tyrosine kinase) and Fak (Chen, Lu, et al., 2009; Garrett, Schreiner, Lobas, & Weiner, 2012), kinases having roles in small-GTPase modulation (RhoA, Rac1), as well as signaling to the nucleus. Protocadherin PAPC’s interaction with Frizzled7 modulates noncanonical Wnt signaling (i.e., PCP signaling) as mediated via JNK, RhoA, and Rac, during tissue separation and elongation (i.e., convergence–extension movements) in Xenopus (Medina et al., 2004; Unterseher et al., 2004). While RhoA and Rac are often evaluated from the perspective of cytoskeletal control, they and JNK have the potential to pass signals directly to the nucleus. Likewise, interaction of the PAPC-protocadherin with CK2β facilitates the removal of β-catenin, making it more difficult for Wnt signals to inhibit β-catenin’s destruction complex. This alters the transcriptional activity of canonical Wnt gene targets and thereby gastrulation in Xenopus (Kietzmann et al., 2012).
Protocadherin nuclear signaling through the MAPK pathway has also been suggested with the involvement of p38 MAPK and MAPKKK (even as such studies have also focused also on MAPKKK in the context of neuronal junctions) (Yasuda et al., 2007). Protocadherin7 (NF-protocadherin) has likewise been implicated in nuclear signaling via interactions with chromatin as well as the transcriptional regulator TAF1/Set (Heggem & Bradley, 2003; Murakami, Hijikata, Matsukawa, Ishikawa, & Yorifuji, 2006). Interestingly, however, the TAF1 interaction appears to occur in the cytoplasm, with work in Xenopus including examinations of cell–cell adhesion outcomes and proper neural fold and tube formation (Rashid, Newell, Shama, & Bradley, 2006). Finally, there are indications of protocadherins being cleaved by γ-secretase as described above for classic cadherins, to generate fragments that may transduce information to the nucleus (Bonn, Seeburg, & Schwarz, 2007; Haas, Frank, Veron, & Kemler, 2005; Hambsch, Grinevich, Seeburg, & Schwarz, 2005). Such cleavage may be predominant during development, with less cleavage found to occur as neurons differentiate (Buchanan, Schalm, & Maniatis, 2010).
12. ATYPICAL CADHERIN NUCLEAR SIGNALING
“Atypical” cadherins refer to a number of cadherin superfamily members possessing primary and folding structures that distinguish them from classical cadherins. Those most reported are in the invertebrate literature, where they were first characterized and include members such as Fat, Daschous, and Flamingo (vertebrate Celsr). In common with classical cadherins, Fat and Daschous span the plasma membrane once, but as is typical to this cadherin subfamily have considerably larger extracellular domains with a greater number of cadherin repeats, as well as cytodomains that differ from each other and classical cadherins. For some of the atypical cadherins, such as Flamingo, evidence suggests that their roles center on signaling, and perhaps less so on adhesion (Shimada, Usui, Yanagawa, Takeichi, & Uemura, 2001; Usui et al., 1999; Zhou, Goffinet, & Tissir, 2008; reviewed in Wu & Mlodzik, 2009). In all cases, the extracellular domains of atypical cadherins are highly variable (Oda, Tagawa, & Akiyama-Oda, 2005; reviewed in Harris & Tepass, 2010; Oda & Takeichi, 2011), suggesting the potential to engage in a wide variety of interactions.
As noted, much of the work upon these cadherins was initially conducted in Drosophila, with both similarities and significant departures later found in vertebrates. While direct signaling to the nucleus may occur from atypical cadherins, there appears to be no uniform mode of signaling. Rather, considerable findings suggest their contributions to the modulation of cytoskeletal architecture and functions in epithelial tissue patterning (e.g., from proximal-to-distal), and directing the morphogenesis or generation of epithelial, neuroepithelial, and neuronal forms (e.g., proper formation of the neural tube or guidance of dendrites or axons). For example, interaction of Fat with Daschous promotes the activation of Frizzled and pathways thought to be largely distinct (despite cross talk) from β-catenin stabilization (Yang, Axelrod, & Simon, 2002). While small GTPases and other mediators of the observed cytoskeletal effects are likely to modulate downstream gene activity, this has not yet been the emphasis of recent studies.
In Drosophila, however, clear genetic evidence indicates that the atypical Fat cadherin controls tissue size through the nuclear-directed Hippo pathway (Bennett & Harvey, 2006; Cho et al., 2006; Hamaratoglu et al., 2006; Silva, Tsatskis, Gardano, Tapon, & McNeill, 2006; Willecke et al., 2006). While most components of this pathway are conserved in vertebrates, one recent report suggests that the Fat–Hippo relationship is not conserved (Bossuyt et al., 2014). Instead, it appears that differences arose in the Hippo pathway’s upstream modulators between arthropods and vertebrates. Nonetheless, in invertebrates at least, there is clear precedence for nuclear signaling being dependent upon atypical cadherins.
Fat’s putative connection to nuclear signaling was earlier made via the ERM-family protein Expanded, which together with Merlin modulates the EGFR and Hippo pathways (Bennett & Harvey, 2006; Curto et al., 2007; Gladden et al., 2010; Hamaratoglu et al., 2006; Silva et al., 2006; Willecke et al., 2006). In this nuclear-directed context, the definitive identity of upstream Drosophila ligands in relation to Fat remains in question. However, another Drosophila atypical cadherin, Daschous, is thought to functionally interact with Fat in certain contexts involving cell–cell interactions (Konsavage & Yochum, 2013; Rogulja, Rauskolb, & Irvine, 2008; reviewed in Matakatsu & Blair, 2006).
13. CADHERIN NUCLEAR SIGNALING VIA THE HIPPO PATHWAY
Evidence that cadherins functionally interact with the Hippo pathway has come from a variety of sources. In the context of a mammalian cancer cell-line deficient for E-cadherin, one of the key effectors of this pathway, Yap, is predominantly nuclear (Kim, Koh, Chen, & Gumbiner, 2011). When E-cadherin expression is restored, Yap moves to the cytoplasm in a cell density-dependent manner, and Yap target genes are no longer activated. The participation of the Hippo pathway in such cadherin and cell density-dependent effects upon cell growth was further made evident using beads coated with the E-cadherin ectodomain. Such beads normally have the capacity to inhibit cell growth, but not in the case where the Hippo pathway was inactivated through the removal of one of its components, or through the downstream expression of Yap (Kim et al., 2011). How cadherin mediates its effects upon Yap is not clear, but as noted below it may be via signals that involve α-catenin or other entities.
Even within a contiguous group of cells, the relationship of the cadherin complex to the Hippo pathway can vary according to cell density. For example, at the borders of a colony of cells grown in culture, cadherin-mediated cell–cell contacts and cell densities are often less mature than toward the center. This is reflected not only in the state of the contacts but also in the disposition of Hippo pathway components. Cells at the edge display a greater proportion of nuclear Yap and often display higher proliferative and migratory dispositions. Activation of the Hippo pathway, as seen in the phosphorylation of Yap by the Lats kinase as well as other measures, appears sensitive to the junctional status of cells. As noted, homophilic ligation of cadherins between cells (in trans) activates the Hippo kinase-mediated pathway to prevent Yap’s nuclear entry (Gumbiner & Kim, 2014). Despite the uncertainty regarding how this occurs, the association of β-catenin and α-catenin with cadherin appears to be needed for cadherin sensing to be integrated with the Hippo pathway. Other junction-associated proteins are also likely to be involved in some of these contexts, including, for example, Merlin, Ajuba, and Kibra (Baumgartner, Poernbacher, Buser, Hafen, & Stocker, 2010; Das Thakur et al., 2010; Genevet, Wehr, Brain, Thompson, & Tapon, 2010; Gladden et al., 2010; Huang, Wu, Barrera, Matthews, & Pan, 2005; Morrison et al., 2001).
α-Catenin has been proposed to limit Yap activity in a manner that bypasses some of the upstream Hippo pathway components. For example, by interacting with Yap indirectly via 14-3-3σ, α-catenin prevents the PP2A phosphatase from dephosphorylating Yap, so that Yap is kept out of the nucleus (Schlegelmilch et al., 2011). In this context, α-catenin might inhibit Yap nuclear function in ways that are independent of α-catenin being associated with cadherin (Fig. 5). In addition, N-terminally phosphorylated β-catenin may also cross-regulate and inhibit Hippo pathway effectors, Yap and Taz (Azzolin et al., 2012;Rosenbluh et al., 2012). Lastly, the cytoskeleton and cell shape have also been studied with regard to their impact upon Hippo signal transduction (Dupont et al., 2011; Sansores-Garcia et al., 2011). Since cadherins and cell–cell junctions are integral to larger cytoskeletal structures and cell-shape considerations, such mechanics reflect yet another manner that signals originating at cadherin-based structures may be moved to the nucleus, a process termed mechanotransduction. Whatever the mechanism, upon the lessening of cadherin-mediated contacts, there is often a corresponding lessening of upstream Hippo signaling, such that downstream Yap enters the nucleus to bind a variety of transcription factors (Basu, Totty, Irwin, Sudol, & Downward, 2003; Chen, Loh, & Song, 2010; Komuro, Nagai, Navin, & Sudol, 2003; Varelas et al., 2010; Yagi, Chen, Shigesada, Murakami, & Ito, 1999), with a strong proportion modulating growth.
14. STEM CELL MAINTENANCE VIA CADHERIN NUCLEAR SIGNALING
Despite early established relationships between E-cadherin and β-catenin signaling (McEwen et al., 2012), and β-catenin signaling and stem cell maintenance (reviewed in Miki, Yasuda, & Kahn, 2011), evidence clearly shows that maintaining mESCs in the pluripotent state requires E-cadherin, where its loss drives entry into what is referred to as a “primed” state, termed mEpiESCs (reviewed Li et al., 2012; Pieters & van Roy, 2014). Human ESCs likewise exhibit the requirement of E-cadherin for self-renewal, pluripotency, and long-term survival (Li, Wang, et al., 2010). Remarkably, the key progenitor-pathway downstream of E-cadherin involves LIF-STAT signaling. LIF is a secreted cytokine that promotes stemness in mESCs, and interestingly, the ecto- or transmembrane domain of E-cadherin forms a complex with the transmembrane LIF receptor and its coreceptor GP130 (del Valle et al., 2013). This stabilizes the receptor complex at the protein level to promote JAK/Stat signaling (Hirai, Karian, & Kikyo, 2011). In E-cadherin-deficient mESCs, a loss in LIF responsiveness is observed (del Valle et al., 2013; Hawkins, Mohamet, Ritson, Merry, & Ward, 2012), while downstream STAT activation in such cells is able to maintain the stemness state (Matsuda et al., 1999) (Fig. 7).
Figure 7.
Cadherin signaling promotes JAK/STAT signaling for stem cell maintenance. Mouse and human ESCs require E-cadherin for pluripotency, self-renewal, and long-term survival. The transmembrane and ectodomain of E-cadherin interact with the LIF receptor (LIFR) and GP130 coreceptor. This association stabilizes the complex and thereby promotes JAK/Stat signaling and consequent stemness properties to the stem cells.
In mEpiESCs, N-cadherin in combination with E-cadherin is present at cell–cell contacts (Bao et al., 2009). N-Cadherin assists in maintaining the primed state by enabling activin signaling (Soncin et al., 2009). N-Cadherin has further been found to assist in maintaining the neural progenitor pool, through the modulation of delamination and differentiation, as described, for one case, above (reviewed Paulson et al., 2014; Rousso et al., 2012; Seki, Namba, Mochizuki, & Onodera, 2007; Yagita et al., 2009; Zhang et al., 2010).
Cadherins also help to organize stem cells. In the Drosophila ovary, for example, DE-cadherin allows for germline stem cells to make contact with cap cells forming the niche; here, the stemness-promoting Wnt ligand is expressed (Gonzalez-Reyes, 2003; Song & Xie, 2003; Song, Zhu, Doan, & Xie, 2002). Maintaining a stem cell niche in bone marrow is like-wise dependent on stem cell attachment, but here the interaction occurs with osteoblast cells via N-cadherin (Zhang et al., 2003). Finally, cadherins play vital roles in stem cell differentiation (del Valle et al., 2013; Larue et al., 1996; Mohamet, Lea, & Ward, 2010; Moore, Radice, Dominis, & Kemler, 1999).
In stem cell contexts, cadherins might signal to the nucleus via catenins. However, thus far, the role of β-catenin has been discussed more in the context of the canonical Wnt pathway. How β-catenin operates in a signaling or other capacity is also under active discussion by stemness researchers (Anton, Kestler, & Kuhl, 2007; Lyashenko et al., 2011; Soncin et al., 2009). In the nucleus, events may involve β-catenin associating with transcription factors other than TCF/LEF. Some work suggests that β-catenin’s adhesive function is more pertinent to its role in stemness (del Valle et al., 2013; Lyashenko et al., 2011), even as other interpretations have been offered (Wray et al., 2011; reviewed in Pieters & van Roy, 2014).
Lastly, recent work in mESCs suggests that p120ctn might also require consideration. Roughly in line with known decreases in E-cadherin protein occurring in mESCs leaving their naive state, the signaling pool of p120ctn rises. This was seen to derepress (activate) gene targets of the transcriptional repressor REST, to which p120ctn binds, assisting in moving the mESCs toward a more differentiated state. While more work is required, E-cadherin depletion also resulted in elevation of these same gene targets, with p120ctn’s specific involvement suggested by a partial rescue of the observed effects upon the codepletion of p120ctn (but not β-catenin) (Lee et al., 2014).
15. DESMOSOMAL CADHERIN NUCLEAR SIGNALING
Desmoglein1–cadherin has been implicated in adhesion-independent signaling contributing to morphogenesis and differentiation of keratinocytes (Getsios et al., 2009). Here, via its cytoplasmic tail (when associated with the plasma membrane), Desmoglein1 suppressed EGFR activation and signaling, in particular the Erk1/2 trajectory, to facilitate differentiation (Getsios et al., 2009). Interestingly, the ectodomain of Desmoglein1 is not required for this activity, nor are Desmoglein1’s associations with Desmocollin1–cadherin nor γ-catenin (plakoglobin). Desmoglein1 was concluded to act within an early instructive pathway directing keratinocytes through terminal differentiation, preventing Erk1/2 signaling in the suprabasal layers. Conceivably, this occurs via the direct interaction of Desmoglein1 with the EGFR, as they colocalize. Interestingly, Desmoglein2 expression in the suprabasal skin layer acts in a contrary manner to Desmoglein1, where Desmoglein2 ectopic presence appears to be functionally coupled to enhanced cell survival via increased Erk1/2 and NFκB activity (Brennan et al., 2007). In the case of another type of desmosomal cadherin, Desmocollin3, its ectopic expression was tied to increased β-catenin levels and transcriptional activity (Hardman et al., 2005). Other work has likewise linked desmosomal cadherins to proliferation, differentiation, and morphogenesis, but not always with the same outcomes (Chidgey et al., 2001; Elias et al., 2001; Eshkind et al., 2002; Merritt et al., 2002). Given that these desmosomal cadherins are normally expressed in the basal versus suprabasal regions of skin, each may provide a respective homeostatic balance for the activities of the other (Fig. 8).
Figure 8.
Desmosomal cadherin signaling in skin. In the suprabasal skin layers, the cytoplasmic domain of Desmoglein1 inhibits EGFR signaling via the Erk1/2 signaling trajectory (Getsios et al., 2009). This might occur via a direct interaction of the Desmoglein1 cytodomain with EGFR. Ectopic Desmoglein2 expression (involucrin promoter) in the suprabasal keratinocyte layer increases proliferation and cell survival, apparently via effects upon EGFR and NFκB signaling (Brennan et al., 2007). Misexpression of desmocollin3a and 3b in subrabasal keratinocytes leads to proliferation and differentiation defects, with increased β-catenin signaling normally seen only in basal keratinocytes (Hardman et al., 2005) (not shown).
In particular, the ectopic in vivo expression of Desmocollin3a and 3b in suprabasal keratinocytes of transgenic mice increases β-catenin stability and signaling by an unknown mechanism (Hardman et al., 2005). Again while the responsible biochemical links are uncertain, the expression of desmoglein2 in the same region likewise increased proliferation and cell survival, but apparently through effects upon multiple signaling pathways including EGFR and NFκB (Brennan et al., 2007).
There are additional indications of desmosomal cadherins signal to the nucleus in development and disease. One interpretation for ARVCs in humans followed the use of a mouse model in which Desmoplakin, a protein linking desmosomal cadherins to the intermediate cytoskeleton, was ablated. γ-Catenin (plakoglobin) was released from the desmosomal adhesion complex and found to inhibit the expression of β-catenin gene targets, and thereby the canonical Wnt pathway. The loss of such essential Wnt signals is known to reduce the formation of muscle in favor of fatty tissue, fitting well with known features of the disease in humans (Ben-Ze’ev, Shtutman, & Zhurinsky, 2000; Garcia-Gras et al., 2006).
Work in intestinal cancer epithelial cells has likewise shown a functional interaction between Desmoglein2 and EGFR. The loss of Desmoglein2 showed reduced EGFR activation, and thereby reduced Erk and Src activity. Cell proliferation was concomitantly lessened, perhaps as a result of compensatory expression of Desmocollin2 followed by its association and inhibition of EGFR. Thus, Desmoglein2 and Desmocollin2 appear to have opposing roles in this cancer cell context, with Desmoglein2 promoting and Desmocollin2 inhibiting tumorigenesis (Kamekura et al., 2014).
Finally, it is interesting to consider that desmosomal cadherins also interact with catenin proteins, specifically plakophilin1–3. Additionally, a catenin found at both desmosomal and adherens junctions is γ-catenin (plakoglobin), with its signaling properties having been briefly mentioned earlier in this review. The plakophilin-catenins may possess roughly analogous roles to those of catenins present at the adherens junctions. That is, in addition to being at desmosomal junctions where they assist in linking Desmocollin- or Desmoglein-cadherins to the cytoskeleton (intermediate filaments), plakophilin1–3 are additionally present in the cytoplasm or nucleus of some cells (Mertens, Kuhn, & Franke, 1996). Plakophilin3 has been found in association with RNA-binding proteins such as G3BP, FXR1, and PABPC1 in cytoplasmic stress granules that arise and assist cells in response to adverse metabolic stimuli (Hofmann et al., 2006). Recently, plakophilin3 was found to bind the Ets-family transcription factor ETV1, with the functional interaction of these two proteins being supported in studies using Xenopus embryos (Munoz et al., 2014). Likewise, plakophilin2 interacts with a subunit of the RNA polymerase III complex (RPC155) (Mertens et al., 1996), possibly engaging in the modulation of ribosomal or tRNA transcription. Plakophilin2’s entry to the nucleus is negatively regulated via direct phosphorylation by the kinase C-TAK, which produces a binding site for 14-3-3 to retain it within the cytoplasm (Muller, Ritt, Copeland, & Morrison, 2003). Additionally, plakophilin2 has been reported in association with β-catenin and to modestly enhance β-catenin’s nuclear activity (Chen, Bonne, Hatzfeld, van Roy, & Green, 2002). These nuclear interactions of plakophilin2 or plakophilin3 leave open the possibility of cross talk between desmosomal cadherins and the nucleus, similar in concept to that discussed earlier for β-catenin and p120ctn.
16. CONCLUSIONS
The cadherin family, which includes classic, atypical, and protocadherin forms, comprises a diverse protein family. Most engage in cell– cell adhesion, but a long-standing question has been to what extent the cadherins also engage in signaling. This review has focused in particular upon the involvement of cadherins in signaling to the nucleus. It occurs via established pathways, such as those involving RTK or Wnt components, but we have also touched upon additional pathways such as Hippo, that while developmentally central are still emerging. We have concerned ourselves little with signals directed primarily to the cytoplasm or cytoskeleton, despite their importance, including signals that largely rely upon mechanical considerations (such as involving “tensegrity”). In these latter cases, we have tried to direct the reader to other papers or reviews.
Our review of evidence from many investigators indicates that cadherins are integrated into the larger fabric of cell and developmental decisions and functions through an array of direct or indirect protein–protein associations. A central concept for consideration is that organismal fitness benefited from mechanisms that reported cell–cell adhesive states to the nucleus— contributing to one of many forms of outside-in signaling. Indeed, such reporting may have been selected for over the course of evolution. For example, signals involving or originating from cadherins can lead to the activation or repression of gene targets that affect proliferation and differentiation, and in other cases motility or cell death. As cadherins are expressed and function in most tissues, and at most developmental time points, as well as in tissue regeneration and homeostasis, the phenotypes that result from their loss or gain of function are many. Along with integrins, that are best known to interact with the extracellular matrix (but also some neighboring-cell receptors), or with other classes of cell–cell adhesion proteins, cadherins comprise one class of the multiple transmembrane systems capable of integrating the cell’s outside world with that which lies within.
Since cadherins have adhesive as well as the noted signaling or additional functions, it can be challenging at the experimental level to discern if observed gain- or loss-of-function phenotypes are attributable mainly to altered adhesion-motility states or altered nuclear signaling (etc.). Indeed, since these functions engage each other in many cases, they are interdependent.
To recap, we have focused in this review on a few modalities of cadherin nuclear signal transduction. One involves the modulation by cadherins of other cell surface receptors, such as RTKs, or those that bind to Wnt ligands, such as Frizzled and Lrp5/6. Generally in this case, a series of indirect events transpire (such as promotion or inhibition of the MAPK cascade) that have an impact upon gene activity. A second type of cadherin-based mechanism involves events more proximal to the nucleus, such as direct interaction of cadherin cytodomains with proteins having the ability to enter the nucleus to alter gene activity. Examples here include the catenins and also DNA-binding transcription factors such NFκB. In some cases, we have seen that cadherin proteolytic fragments themselves exhibit the capacity to enter the nucleus to promote or inhibit gene programs.
Overall, our understanding of the larger picture of cadherin nuclear signaling is still quite modest. While many examples can be pointed out as summarized in this review, we lack an understanding at the systems level of the interrelationships that occur at the plasma membrane, cytoplasm, and nucleus. That is, while much information is known, we remain more in the position of imagining than comprehending how things fit together on a wider scale. This is of course a problem faced currently in all biological fields, and we thus look forward to the application of emerging or still-to-arise wet-bench technologies, as well as the development and wise application of more robust information processing and model testing.
ACKNOWLEDGMENTS
We regret not being able to discuss many studies relevant to this topic. P. D.Mand C. J. Gare supported by funds from the National Institutes of Health GM107079 and GM076561, respectively.
REFERENCES
- Abe K, Takeichi M. NMDA-receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron. 2007;53:387–397. doi: 10.1016/j.neuron.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Abreu-Blanco MT, Verboon JM, Liu R, Watts JJ, Parkhurst SM. Drosophila embryos close epithelial wounds using a combination of cellular protrusions and an actomyosin purse string. Journal of Cell Science. 2012;125:5984–5997. doi: 10.1242/jcs.109066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktary Z, Kulak S, Mackey J, Jahroudi N, Pasdar M. Plakoglobin interacts with the transcription factor p53 and regulates the expression of 14-3-3 sigma. Journal of Cell Science. 2013;126:3031–3042. doi: 10.1242/jcs.120642. [DOI] [PubMed] [Google Scholar]
- Aktary Z, Pasdar M. Plakoglobin: Role in tumorigenesis and metastasis. International Journal of Cell Biology. 2012;2012:189–521. doi: 10.1155/2012/189521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktary Z, Pasdar M. Plakoglobin represses SATB1 expression and decreases in vitro proliferation, migration and invasion. PLoS One. 2013;8:e78388. doi: 10.1371/journal.pone.0078388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alema S, Salvatore AM. p120 catenin and phosphorylation: Mechanisms and traits of an unresolved issue. Biochimica et Biophysica Acta. 2007;1773:47–58. doi: 10.1016/j.bbamcr.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ. p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochimica et Biophysica Acta. 2007;1773:34–46. doi: 10.1016/j.bbamcr.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. Journal of Molecular Biology. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Anton R, Kestler HA, Kuhl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Letters. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Arikkath J, Reichardt LF. Cadherins and catenins at synapses: Roles in synaptogenesis and synaptic plasticity. Trends in Neurosciences. 2008;31:487–494. doi: 10.1016/j.tins.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnsdorf EJ, Tummala P, Jacobs CR. Non-canonical Wnt signaling and N-cadherin related beta-catenin signaling play a role in mechanically induced osteogenic cell fate. PLoS One. 2009;4:e5388. doi: 10.1371/journal.pone.0005388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulanandam R, Vultur A, Cao J, Carefoot E, Elliott BE, Truesdell PF, et al. Cadherin-cadherin engagement promotes cell survival via Rac1/Cdc42 and signal transducer and activator of transcription-3. Molecular Cancer Research: MCR. 2009;7:1310–1327. doi: 10.1158/1541-7786.MCR-08-0469. [DOI] [PubMed] [Google Scholar]
- Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Balsamo J, Arregui C, Leung T, Lilien J. The nonreceptor protein tyrosine phosphatase PTP1B binds to the cytoplasmic domain of N-cadherin and regulates the cadherin-actin linkage. The Journal of Cell Biology. 1998;143:523–532. doi: 10.1083/jcb.143.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, et al. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar I, Lambert de Rouvroit C, Goffinet AM. The evolution of cortical development. An hypothesis based on the role of the Reelin signaling pathway. Trends in Neurosciences. 2000;23:633–638. doi: 10.1016/s0166-2236(00)01675-1. [DOI] [PubMed] [Google Scholar]
- Bass-Zubek AE, Godsel LM, Delmar M, Green KJ. Plakophilins: Multifunctional scaffolds for adhesion and signaling. Current Opinion in Cell Biology. 2009;21:708–716. doi: 10.1016/j.ceb.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Molecular Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Developmental Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Bech-Serra JJ, Santiago-Josefat B, Esselens C, Saftig P, Baselga J, Arribas J, et al. Proteomic identification of desmoglein 2 and activated leukocyte cell adhesion molecule as substrates of ADAM17 and ADAM10 by difference gel electrophoresis. Molecular and Cellular Biology. 2006;26:5086–5095. doi: 10.1128/MCB.02380-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JM, Kwiatkowski AV, Yang C, Korobova F, Pokutta S, Svitkina T, et al. AlphaE-catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. The Journal of Cell Biology. 2010;189:339–352. doi: 10.1083/jcb.200910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Current Biology. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Ben-Ze’ev A, Shtutman M, Zhurinsky J. The integration of cell adhesion with gene expression: The role of beta-catenin. Experimental Cell Research. 2000;261:75–82. doi: 10.1006/excr.2000.5045. [DOI] [PubMed] [Google Scholar]
- Berger-Muller S, Suzuki T. Seven-pass transmembrane cadherins: Roles and emerging mechanisms in axonal and dendritic patterning. Molecular Neurobiology. 2011;44:313–320. doi: 10.1007/s12035-011-8201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. Mind-bomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/beta-catenin signaling. The Journal of Cell Biology. 2011;194:737–750. doi: 10.1083/jcb.201107021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt T, Rizvi A, Batta SP, Kataria S, Jamora C. Signaling and mechanical roles of E-cadherin. Cell Communication & Adhesion. 2013;20:189–199. doi: 10.3109/15419061.2013.854778. [DOI] [PubMed] [Google Scholar]
- Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- Birukova AA, Zebda N, Fu P, Poroyko V, Cokic I, Birukov KG. Association between adherens junctions and tight junctions via Rap1 promotes barrier protective effects of oxidized phospholipids. Journal of Cellular Physiology. 2011;226:2052–2062. doi: 10.1002/jcp.22543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Emond MR, Jontes JD. Protocadherin-19 and N-cadherin interact to control cell movements during anterior neurulation. The Journal of Cell Biology. 2010;191:1029–1041. doi: 10.1083/jcb.201007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixby JL, Zhang R. Purified N-cadherin is a potent substrate for the rapid induction of neurite outgrowth. The Journal of Cell Biology. 1990;110:1253–1260. doi: 10.1083/jcb.110.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler A, Yao L, Wang Y, Ye Z, Jin VX, Farnham PJ. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin. 2013 doi: 10.1186/1756-8935-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzer JT, Nusse R. A critical role for endocytosis in Wnt signaling. BMC Cell Biology. 2006;7:28. doi: 10.1186/1471-2121-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Seeburg PH, Schwarz MK. Combinatorial expression of alpha- and gamma-protocadherins alters their presenilin-dependent processing. Molecular and Cellular Biology. 2007;27:4121–4132. doi: 10.1128/MCB.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvini P, An WG, Rosolen A, Nguyen P, Trepel J, Garcia de Herreros A, et al. Geldanamycin abrogates ErbB2 association with proteasome-resistant beta-catenin in melanoma cells, increases beta-catenin-E-cadherin association, and decreases beta-catenin-sensitive transcription. Cancer Research. 2001;61:1671–1677. [PubMed] [Google Scholar]
- Bossuyt W, Chen CL, Chen Q, Sudol M, McNeill H, Pan D, et al. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2014;33:1218–1228. doi: 10.1038/onc.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Essential role of BCL9-2 in the switch between beta-catenin’s adhesive and transcriptional functions. Genes & Development. 2004;18:2225–2230. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan D, Hu Y, Joubeh S, Choi YW, Whitaker-Menezes D, O’Brien T, et al. Suprabasal Dsg2 expression in transgenic mouse skin confers a hyperproliferative and apoptosis-resistant phenotype to keratinocytes. Journal of Cell Science. 2007;120:758–771. doi: 10.1242/jcs.03392. [DOI] [PubMed] [Google Scholar]
- Brieher WM, Yap AS. Cadherin junctions and their cytoskeleton(s) Current Opinion in Cell Biology. 2013;25:39–46. doi: 10.1016/j.ceb.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Buchanan SM, Schalm SS, Maniatis T. Proteolytic processing of protocadherin proteins requires endocytosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17774–17779. doi: 10.1073/pnas.1013105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caca K, Kolligs FT, Ji X, Hayes M, Qian J, Yahanda A, et al. Beta- and gamma-catenin mutations, but not E-cadherin inactivation, underlie T-cell factor/lymphoid enhancer factor transcriptional deregulation in gastric and pancreatic cancer. Cell Growth & Differentiation. 1999;10:369–376. [PubMed] [Google Scholar]
- Carmeliet P, Lampugnani MG, Moons L, Breviario F, Compernolle V, Bono F, et al. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Carroll RC, Zukin RS. NMDA-receptor trafficking and targeting: Implications for synaptic transmission and plasticity. Trends in Neurosciences. 2002;25:571–577. doi: 10.1016/s0166-2236(02)02272-5. [DOI] [PubMed] [Google Scholar]
- Casagolda D, Del Valle-Perez B, Valls G, Lugilde E, Vinyoles M, Casado-Vela J, et al. A p120-catenin-CK1epsilon complex regulates Wnt signaling. Journal of Cell Science. 2010;123:2621–2631. doi: 10.1242/jcs.067512. [DOI] [PubMed] [Google Scholar]
- Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nature Reviews. Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- Cavallaro U, Dejana E. Adhesion molecule signalling: Not always a sticky business. Nature Reviews. Molecular Cell Biology. 2011;12:189–197. doi: 10.1038/nrm3068. [DOI] [PubMed] [Google Scholar]
- Cavey M, Lecuit T. Molecular bases of cell-cell junctions stability and dynamics. Cold Spring Harbor Perspectives in Biology. 2009;1:a002998. doi: 10.1101/cshperspect.a002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Lavker RM, Acquista E, Cowin P. Plakoglobin suppresses epithelial proliferation and hair growth in vivo. The Journal of Cell Biology. 2000;149:503–520. doi: 10.1083/jcb.149.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S, Comunale F, Fortier M, Portales-Casamar E, Debant A, Gauthier-Rouviere C. M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Molecular Biology of the Cell. 2007;18:1734–1743. doi: 10.1091/mbc.E06-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Bonne S, Hatzfeld M, van Roy F, Green KJ. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta-catenin signaling. The Journal of Biological Chemistry. 2002;277:10512–10522. doi: 10.1074/jbc.M108765200. [DOI] [PubMed] [Google Scholar]
- Chen X, Gumbiner BM. Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. The Journal of Cell Biology. 2006;174:301–313. doi: 10.1083/jcb.200602062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Khan ZA, Cukiernik M, Chakrabarti S. Differential activation of NF-kappa B and AP-1 in increased fibronectin synthesis in target organs of diabetic complications. American Journal of Physiology. Endocrinology and Metabolism. 2003;284:E1089–E1097. doi: 10.1152/ajpendo.00540.2002. [DOI] [PubMed] [Google Scholar]
- Chen X, Koh E, Yoder M, Gumbiner BM. A protocadherin-cadherin-FLRT3 complex controls cell adhesion and morphogenesis. PLoS One. 2009;4:e8411. doi: 10.1371/journal.pone.0008411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Loh PG, Song H. Structural and functional insights into the TEAD-YAP complex in the Hippo signaling pathway. Protein & Cell. 2010;1:1073–1083. doi: 10.1007/s13238-010-0138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lu Y, Meng S, Han MH, Lin C, Wang X. Alpha- and gamma-protocadherins negatively regulate PYK2. The Journal of Biological Chemistry. 2009;284:2880–2890. doi: 10.1074/jbc.M807417200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Maniatis T. Clustered protocadherins. Development. 2013;140:3297–3302. doi: 10.1242/dev.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MC, Solomon TE, Kui R, Soll AH. Apical EGF receptors regulate epithelial barrier to gastric acid: Endogenous TGF-alpha is an essential facilitator. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2002;283:G1098–G1106. doi: 10.1152/ajpgi.00507.2001. [DOI] [PubMed] [Google Scholar]
- Chen MW, Vacherot F, De La Taille A, Gil-Diez-De-Medina S, Shen R, Friedman RA, et al. The emergence of protocadherin-PC expression during the acquisition of apoptosis-resistance by prostate cancer cells. Oncogene. 2002;21:7861–7871. doi: 10.1038/sj.onc.1205991. [DOI] [PubMed] [Google Scholar]
- Chetty R, Serra S. Nuclear E-cadherin immunoexpression: From biology to potential applications in diagnostic pathology. Advances in Anatomic Pathology. 2008;15:234–240. doi: 10.1097/PAP.0b013e31817bf566. [DOI] [PubMed] [Google Scholar]
- Chiasson CM, Wittich KB, Vincent PA, Faundez V, Kowalczyk AP. p120-catenin inhibits VE-cadherin internalization through a Rho-independent mechanism. Molecular Biology of the Cell. 2009;20:1970–1980. doi: 10.1091/mbc.E08-07-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey M, Brakebusch C, Gustafsson E, Cruchley A, Hail C, Kirk S, et al. Mice lacking desmocollin 1 show epidermal fragility accompanied by barrier defects and abnormal differentiation. The Journal of Cell Biology. 2001;155:821–832. doi: 10.1083/jcb.200105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nature Genetics. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Choi SH, Estaras C, Moresco JJ, Yates JR, 3rd, Jones KA. alpha-Catenin interacts with APC to regulate beta-catenin proteolysis and transcriptional repression of Wnt target genes. Genes & Development. 2013;27:2473–2488. doi: 10.1101/gad.229062.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Huber AH, Weis WI. Thermodynamics of beta-catenin-ligand interactions: The roles of the N- and C-terminal tails in modulating binding affinity. The Journal of Biological Chemistry. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- Chu ML, Ahn VE, Choi HJ, Daniels DL, Nusse R, Weis WI. Structural studies of Wnts and identification of an LRP6 binding site. Structure. 2013;21:1235–1242. doi: 10.1016/j.str.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: Potential involvement of ZEB-1 and ZEB-2. Oncogene. 2007;26:711–724. doi: 10.1038/sj.onc.1209808. [DOI] [PubMed] [Google Scholar]
- Chung HA, Yamamoto TS, Ueno N. ANR5, an FGF target gene product, regulates gastrulation in Xenopus. Current Biology. 2007;17:932–939. doi: 10.1016/j.cub.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Ciesiolka M, Delvaeye M, Van Imschoot G, Verschuere V, McCrea P, van Roy F. p120 catenin is required for morphogenetic movements involved in the formation of the eyes and the craniofacial skeleton in Xenopus. Journal of Cell Science. 2004;117:4325–4339. doi: 10.1242/jcs.01298. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Developmental Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: The roles of beta-catenin signaling, Slug, and MAPK. The Journal of Cell Biology. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello S, Cao L, Matei D. Tissue transglutaminase regulates beta-catenin signaling through a c-Src-dependent mechanism. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2013;27:3100–3112. doi: 10.1096/fj.12-222620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RT, Kirkpatrick C, Peifer M. Armadillo is required for adherens junction assembly, cell polarity, and morphogenesis during Drosophila embryogenesis. The Journal of Cell Biology. 1996;134:133–148. doi: 10.1083/jcb.134.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cselenyi CS, Jernigan KK, Tahinci E, Thorne CA, Lee LA, Lee E. LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3’s phosphorylation of beta-catenin. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8032–8037. doi: 10.1073/pnas.0803025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curto M, Cole BK, Lallemand D, Liu CH, McClatchey AI. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. The Journal of Cell Biology. 2007;177:893–903. doi: 10.1083/jcb.200703010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky CH, Richa J, Solter D, Knudsen K, Buck CA. Identification and purification of a cell surface glycoprotein mediating intercellular adhesion in embryonic and adult tissue. Cell. 1983;34:455–466. doi: 10.1016/0092-8674(83)90379-3. [DOI] [PubMed] [Google Scholar]
- Daniel JM. Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochimica et Biophysica Acta. 2007;1773:59–68. doi: 10.1016/j.bbamcr.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. Tyrosine phosphorylation and cadherin/catenin function. Bioessays. 1997;19:883–891. doi: 10.1002/bies.950191008. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Molecular and Cellular Biology. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann SG, Golas J, Miranda M, Shi C, Wu J, Jin G, et al. p120 catenin is a key effector of a Ras-PKCvarepsilon oncogenic signaling axis. Oncogene. 2014;33:1385–1394. doi: 10.1038/onc.2013.91. [DOI] [PubMed] [Google Scholar]
- Dartsch N, Schulte D, Hagerling R, Kiefer F, Vestweber D. Fusing VE-cadherin to alpha-catenin impairs fetal liver hematopoiesis and lymph but not blood vessel formation. Molecular and Cellular Biology. 2014;34:1634–1648. doi: 10.1128/MCB.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Current Biology. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty RL, Gottardi CJ. Phospho-regulation of beta-catenin adhesion and signaling functions. Physiology (Bethesda) 2007;22:303–309. doi: 10.1152/physiol.00020.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty RL, Serebryannyy L, Yemelyanov A, Flozak AS, Yu HJ, Kosak ST, et al. alpha-Catenin is an inhibitor of transcription. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5260–5265. doi: 10.1073/pnas.1308663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JM, Rajasekaran AK. Dishonorable discharge: The oncogenic roles of cleaved E-cadherin fragments. Cancer Research. 2012;72:2917–2923. doi: 10.1158/0008-5472.CAN-11-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David MD, Yeramian A, Dunach M, Llovera M, Canti C, de Herreros AG, et al. Signalling by neurotrophins and hepatocyte growth factor regulates axon morphogenesis by differential beta-catenin phosphorylation. Journal of Cell Science. 2008;121:2718–2730. doi: 10.1242/jcs.029660. [DOI] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, et al. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. The Journal of Cell Biology. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Reynolds AB. Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Developmental Cell. 2006;10:21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- De Wever O, Derycke L, Hendrix A, De Meerleer G, Godeau F, Depypere H, et al. Soluble cadherins as cancer biomarkers. Clinical & Experimental Metastasis. 2007;24:685–697. doi: 10.1007/s10585-007-9104-8. [DOI] [PubMed] [Google Scholar]
- del Valle I, Rudloff S, Carles A, Li Y, Liszewska E, Vogt R, et al. E-Cadherin is required for the proper activation of the Lifr/Gp130 signaling pathway in mouse embryonic stem cells. Development. 2013;140:1684–1692. doi: 10.1242/dev.088690. [DOI] [PubMed] [Google Scholar]
- Del Valle-Perez B, Arques O, Vinyoles M, de Herreros AG, Dunach M. Coordinated action of CK1 isoforms in canonical Wnt signaling. Molecular and Cellular Biology. 2011;31:2877–2888. doi: 10.1128/MCB.01466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle-Perez B, Casagolda D, Lugilde E, Valls G, Codina M, Dave N, et al. Wnt controls the transcriptional activity of Kaiso through CK1epsilondependent phosphorylation of p120-catenin. Journal of Cell Science. 2011;124:2298–2309. doi: 10.1242/jcs.082693. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Developmental Biology. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort CC, Paszek MJ, Weaver VM. Balancing forces: Architectural control of mechanotransduction. Nature Reviews. Molecular Cell Biology. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Shy D, Tepass U, Hartenstein V. Interaction between EGFR signaling and DE-cadherin during nervous system morphogenesis. Development. 2002;129(17):3983–3994. doi: 10.1242/dev.129.17.3983. [DOI] [PubMed] [Google Scholar]
- Dusek RL, Getsios S, Chen F, Park JK, Amargo EV, Cryns VL, et al. The differentiation-dependent desmosomal cadherin desmoglein 1 is a novel caspase-3 target that regulates apoptosis in keratinocytes. The Journal of Biological Chemistry. 2006;281:3614–3624. doi: 10.1074/jbc.M508258200. [DOI] [PubMed] [Google Scholar]
- Dusek RL, Godsel LM, Chen F, Strohecker AM, Getsios S, Harmon R, et al. Plakoglobin deficiency protects keratinocytes from apoptosis. The Journal of Investigative Dermatology. 2007;127:792–801. doi: 10.1038/sj.jid.5700615. [DOI] [PubMed] [Google Scholar]
- El-Bahrawy M, Talbot I, Poulsom R, Alison M. Variable nuclear localization of alpha-catenin in colorectal carcinoma. Laboratory Investigation. 2002;82:1167–1174. doi: 10.1097/01.lab.0000028821.41246.6a. [DOI] [PubMed] [Google Scholar]
- Elia LP, Yamamoto M, Zang K, Reichardt LF. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias PM, Matsuyoshi N, Wu H, Lin C, Wang ZH, Brown BE, et al. Desmoglein isoform distribution affects stratum corneum structure and function. The Journal of Cell Biology. 2001;153:243–249. doi: 10.1083/jcb.153.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond MR, Biswas S, Blevins CJ, Jontes JD. A complex of Protocadherin-19 and N-cadherin mediates a novel mechanism of cell adhesion. The Journal of Cell Biology. 2011;195:1115–1121. doi: 10.1083/jcb.201108115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond MR, Biswas S, Jontes JD. Protocadherin-19 is essential for early steps in brain morphogenesis. Developmental Biology. 2009;334:72–83. doi: 10.1016/j.ydbio.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Eshkind L, Tian Q, Schmidt A, Franke WW, Windoffer R, Leube RE. Loss of desmoglein 2 suggests essential functions for early embryonic development and proliferation of embryonal stem cells. European Journal of Cell Biology. 2002;81:592–598. doi: 10.1078/0171-9335-00278. [DOI] [PubMed] [Google Scholar]
- Fagotto F. Looking beyond the Wnt pathway for the deep nature of beta-catenin. EMBO Reports. 2013;14:422–433. doi: 10.1038/embor.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Funayama N, Gluck U, Gumbiner BM. Binding to cadherins antagonizes the signaling activity of beta-catenin during axis formation in Xenopus. The Journal of Cell Biology. 1996;132:1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Current Biology. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Gumbiner BM. Cell contact-dependent signaling. Developmental Biology. 1996;180:445–454. doi: 10.1006/dbio.1996.0318. [DOI] [PubMed] [Google Scholar]
- Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. The Journal of Biological Chemistry. 2007;282:11221–11229. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Ji H, Kim SW, Park JI, Vaught TG, Anastasiadis PZ, et al. Vertebrate development requires ARVCF and p120 catenins and their interplay with RhoA and Rac. The Journal of Cell Biology. 2004;165:87–98. doi: 10.1083/jcb.200307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Yu SX, Lu Y, Bast RC, Jr, Woodgett JR, Mills GB. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahani E, Patra HK, Jangamreddy JR, Rashedi I, Kawalec M, Rao Pariti RK, et al. Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis. 2014;35:747–759. doi: 10.1093/carcin/bgu045. [DOI] [PubMed] [Google Scholar]
- Fedor-Chaiken M, Hein PW, Stewart JC, Brackenbury R, Kinch MS. E-Cadherin binding modulates EGF receptor activation. Cell Communication & Adhesion. 2003;10:105–118. [PubMed] [Google Scholar]
- Ferber EC, Kajita M, Wadlow A, Tobiansky L, Niessen C, Ariga H, et al. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. The Journal of Biological Chemistry. 2008;283:12691–12700. doi: 10.1074/jbc.M708887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AC, Suriano G, Mendes N, Gomes B, Wen X, Carneiro F, et al. E-Cadherin impairment increases cell survival through Notch-dependent upregulation of Bcl-2. Human Molecular Genetics. 2011;21:334–343. doi: 10.1093/hmg/ddr469. [DOI] [PubMed] [Google Scholar]
- Ferreri DM, Minnear FL, Yin T, Kowalczyk AP, Vincent PA. N-cadherin levels in endothelial cells are regulated by monolayer maturity and p120 availability. Cell Communication & Adhesion. 2008;15:333–349. doi: 10.1080/15419060802440377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone CJ. 7TM-cadherins: Developmental roles and future challenges. Advances in Experimental Medicine and Biology. 2010;706:14–36. doi: 10.1007/978-1-4419-7913-1_2. [DOI] [PubMed] [Google Scholar]
- Fortini ME. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nature Reviews. Molecular Cell Biology. 2002;3:673–684. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Sako K, Minami T, Noda K, Kim HZ, Kodama T, et al. Differential function of Tie2 at cell-cell contacts and cell-substratum contacts regulated by angiopoietin-1. Nature Cell Biology. 2008;10:513–526. doi: 10.1038/ncb1714. [DOI] [PubMed] [Google Scholar]
- Gagliardi M, Hernandez A, McGough IJ, Vincent JP. Inhibitors of endocytosis prevent Wnt/Wingless signalling by reducing the level of basal beta-Catenin/Armadillo. Journal of Cell Science. 2014;127:4918–4926. doi: 10.1242/jcs.155424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Volonte D, Brown AM, Weinstein DE, Ben-Ze’ev A, Pestell RG, et al. Caveolin-1 expression inhibits Wnt/beta-catenin/Lef-1 signaling by recruiting beta-catenin to caveolae membrane domains. The Journal of Biological Chemistry. 2000;275:23368–23377. doi: 10.1074/jbc.M002020200. [DOI] [PubMed] [Google Scholar]
- Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. The Journal of Clinical Investigation. 2006;116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett AM, Schreiner D, Lobas MA, Weiner JA. Gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. 2012;74:269–276. doi: 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavard J. Endothelial permeability and VE-cadherin: A wacky comradeship. Cell Adhesion & Migration. 2013;7:455–461. doi: 10.4161/cam.27330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavard J, Gutkind JS. VEGF controls endothelial-cell permeability by promoting the beta-arrestin-dependent endocytosis of VE-cadherin. Nature Cell Biology. 2006;8:1223–1234. doi: 10.1038/ncb1486. [DOI] [PubMed] [Google Scholar]
- Geis K, Aberle H, Kuhl M, Kemler R, Wedlich D. Expression of the Armadillo family member p120cas1B in Xenopus embryos affects head differentiation but not axis formation. Development Genes and Evolution. 1998;207:471–481. doi: 10.1007/s004270050138. [DOI] [PubMed] [Google Scholar]
- Geletu M, Guy S, Arulanandam R, Feracci H, Raptis L. Engaged for survival: From cadherin ligation to STAT3 activation. JAK-STAT. 2013;2:e27363. doi: 10.4161/jkst.27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Developmental Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getsios S, Simpson CL, Kojima S, Harmon R, Sheu LJ, Dusek RL, et al. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. The Journal of Cell Biology. 2009;185:1243–1258. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini AL, Vivanco M, Kypta RM. Alpha-catenin inhibits beta-catenin signaling by preventing formation of a beta-catenin*T-cell factor*DNA complex. The Journal of Biological Chemistry. 2000;275:21883–21888. doi: 10.1074/jbc.M001929200. [DOI] [PubMed] [Google Scholar]
- Gladden AB, Hebert AM, Schneeberger EE, McClatchey AI. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Developmental Cell. 2010;19:727–739. doi: 10.1016/j.devcel.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harbor Perspectives in Biology. 2013;5:a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez GA, McLachlan RW, Yap AS. Productive tension: Force-sensing and homeostasis of cell-cell junctions. Trends in Cell Biology. 2011;21:499–505. doi: 10.1016/j.tcb.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Gomez-Orte E, Saenz-Narciso B, Moreno S, Cabello J. Multiple functions of the noncanonical Wnt pathway. Trends in Genetics: TIG. 2013;29:545–553. doi: 10.1016/j.tig.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A. Stem cells, niches and cadherins: A view from Drosophila. Journal of Cell Science. 2003;116:949–954. doi: 10.1242/jcs.00310. [DOI] [PubMed] [Google Scholar]
- Goyal RK, Lin P, Kanungo J, Payne AS, Muslin AJ, Longmore GD. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Molecular and Cellular Biology. 1999;19:4379–4389. doi: 10.1128/mcb.19.6.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska MM, Sandhu B, Day ML. EGF promotes the shedding of soluble E-cadherin in an ADAM10-dependent manner in prostate epithelial cells. Cellular Signalling. 2012;24:532–538. doi: 10.1016/j.cellsig.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- Grazia Lampugnani M, Zanetti A, Corada M, Takahashi T, Balconi G, Breviario F, et al. Contact inhibition of VEGF-induced proliferation requires vascular endothelial cadherin, beta-catenin, and the phosphatase DEP-1/CD148. The Journal of Cell Biology. 2003;161:793–804. doi: 10.1083/jcb.200209019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D, Sater AK, Ji H, Cho K, Clark M, Stratton SA, et al. Xenopus deltacatenin is essential in early embryogenesis and is functionally linked to cadherins and small GTPases. Journal of Cell Science. 2009;122:4049–4061. doi: 10.1242/jcs.031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D, Tonthat NK, Lee M, Ji H, Bhat KP, Hollingsworth F, et al. Caspase-3 cleavage links delta-catenin to the novel nuclear protein ZIFCAT. The Journal of Biological Chemistry. 2011;286:23178–23188. doi: 10.1074/jbc.M110.167544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM, Kim NG. The Hippo-YAP signaling pathway and contact inhibition of growth. Journal of Cell Science. 2014;127:709–717. doi: 10.1242/jcs.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Molecular Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Haas IG, Frank M, Veron N, Kemler R. Presenilin-dependent processing and nuclear function of gamma-protocadherins. The Journal of Biological Chemistry. 2005;280:9313–9319. doi: 10.1074/jbc.M412909200. [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. Journal of Cell Science. 2012;125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nature Cell Biology. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hambsch B, Grinevich V, Seeburg PH, Schwarz MK. {gamma}-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. The Journal of Biological Chemistry. 2005;280:15888–15897. doi: 10.1074/jbc.M414359200. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Kwiatkowski AV, Ouyang CY, Liu H, Pokutta S, Watkins SC, et al. alphaE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Molecular Biology of the Cell. 2013;24:3710–3720. doi: 10.1091/mbc.E13-07-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Liu K, Avilion AA, Merritt A, Brennan K, Garrod DR, et al. Desmosomal cadherin misexpression alters beta-catenin stability and epidermal differentiation. Molecular and Cellular Biology. 2005;25:969–978. doi: 10.1128/MCB.25.3.969-978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Tepass U. Adherens junctions: From molecules to morphogenesis. Nature Reviews. Molecular Cell Biology. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- Hartsock A, Nelson WJ. Adherens and tight junctions: Structure, function and connections to the actin cytoskeleton. Biochimica et Biophysica Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka K, Simons M, Murakami M. Phosphorylation of VE-cadherin controls endothelial phenotypes via p120-catenin coupling and Rac1 activation. American Journal of Physiology. Heart and Circulatory Physiology. 2011;300:H162–H172. doi: 10.1152/ajpheart.00650.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M. The p120 family of cell adhesion molecules. European Journal of Cell Biology. 2005;84:205–214. doi: 10.1016/j.ejcb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. Plakophilins: Multifunctional proteins or just regulators of desmosomal adhesion? Biochimica et Biophysica Acta. 2007;1773:69–77. doi: 10.1016/j.bbamcr.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Hawkins K, Mohamet L, Ritson S, Merry CL, Ward CM. E-Cadherin and, in its absence, N-cadherin promotes Nanog expression in mouse embryonic stem cells via STAT3 phosphorylation. Stem Cells. 2012;30:1842–1851. doi: 10.1002/stem.1148. [DOI] [PubMed] [Google Scholar]
- Hay E, Laplantine E, Geoffroy V, Frain M, Kohler T, Muller R, et al. N-cadherin interacts with axin and LRP5 to negatively regulate Wnt/beta-catenin signaling, osteoblast function, and bone formation. Molecular and Cellular Biology. 2009;29:953–964. doi: 10.1128/MCB.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Majumdar A, Li X, Adler J, Sun Z, Vertuani S, et al. VE-PTP regulates VEGFR2 activity in stalk cells to establish endothelial cell polarity and lumen formation. Nature Communications. 2013;4:1672. doi: 10.1038/ncomms2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, et al. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Heggem MA, Bradley RS. The cytoplasmic domain of Xenopus NF-protocadherin interacts with TAF1/set. Developmental Cell. 2003;4:419–429. doi: 10.1016/s1534-5807(03)00036-4. [DOI] [PubMed] [Google Scholar]
- Heid HW, Schmidt A, Zimbelmann R, Schafer S, Winter-Simanowski S, Stumpp S, et al. Cell type-specific desmosomal plaque proteins of the plakoglobin family: Plakophilin 1 (band 6 protein) Differentiation; Research in Biological Diversity. 1994;58:113–131. doi: 10.1046/j.1432-0436.1995.5820113.x. [DOI] [PubMed] [Google Scholar]
- Heinen CD. Genotype to phenotype: Analyzing the effects of inherited mutations in colorectal cancer families. Mutation Research. 2010;693:32–45. doi: 10.1016/j.mrfmmm.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellberg CB, Burden-Gulley SM, Pietz GE, Brady-Kalnay SM. Expression of the receptor protein-tyrosine phosphatase, PTPmu, restores E-cadherin-dependent adhesion in human prostate carcinoma cells. The Journal of Biological Chemistry. 2002;277:11165–11173. doi: 10.1074/jbc.M112157200. [DOI] [PubMed] [Google Scholar]
- Hendriksen J, Jansen M, Brown CM, van der Velde H, van Ham M, Galjart N, et al. Plasma membrane recruitment of dephosphorylated beta-catenin upon activation of the Wnt pathway. Journal of Cell Science. 2008;121:1793–1802. doi: 10.1242/jcs.025536. [DOI] [PubMed] [Google Scholar]
- Herzig M, Savarese F, Novatchkova M, Semb H, Christofori G. Tumor progression induced by the loss of E-cadherin independent of beta-catenin/Tcf-mediated Wnt signaling. Oncogene. 2007;26:2290–2298. doi: 10.1038/sj.onc.1210029. [DOI] [PubMed] [Google Scholar]
- Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harbor Perspectives in Biology. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Karian P, Kikyo N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. The Biochemical Journal. 2011;438:11–23. doi: 10.1042/BJ20102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmans R, Basler K. BCL9-2 binds Arm/beta-catenin in a Tyr142-independent manner and requires Pygopus for its function in Wg/Wnt signaling. Mechanisms of Development. 2007;124:59–67. doi: 10.1016/j.mod.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Hofmann I, Casella M, Schnolzer M, Schlechter T, Spring H, Franke WW. Identification of the junctional plaque protein plakophilin 3 in cytoplasmic particles containing RNA-binding proteins and the recruitment of plakophilins 1 and 3 to stress granules. Molecular Biology of the Cell. 2006;17:1388–1398. doi: 10.1091/mbc.E05-08-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nature Cell Biology. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- Homayouni R, Rice DS, Curran T. Disabled-1 interacts with a novel developmentally regulated protocadherin. Biochemical and Biophysical Research Communications. 2001;289:539–547. doi: 10.1006/bbrc.2001.5998. [DOI] [PubMed] [Google Scholar]
- Hong JY, Park JI, Cho K, Gu D, Ji H, Artandi SE, et al. Shared molecular mechanisms regulate multiple catenin proteins: Canonical Wnt signals and components modulate p120-catenin isoform-1 and additional p120 subfamily members. Journal of Cell Science. 2010;123:4351–4365. doi: 10.1242/jcs.067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JY, Park JI, Lee M, Munoz WA, Miller RK, Ji H, et al. Down’s-syndrome-related kinase Dyrk1A modulates the p120-catenin-Kaiso trajectory of the Wnt signaling pathway. Journal of Cell Science. 2012;125:561–569. doi: 10.1242/jcs.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoschuetzky H, Aberle H, Kemler R. Beta-catenin mediates the interaction of the cadherin-catenin complex with epidermal growth factor receptor. The Journal of Cell Biology. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosking CR, Ulloa F, Hogan C, Ferber EC, Figueroa A, Gevaert K, et al. The transcriptional repressor Glis2 is a novel binding partner for p120 catenin. Molecular Biology of the Cell. 2007;18:1918–1927. doi: 10.1091/mbc.E06-10-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono S, Gross I, English MA, Hajra KM, Fearon ER, Licht JD. E-cadherin is a WT1 target gene. The Journal of Biological Chemistry. 2000;275:10943–10953. doi: 10.1074/jbc.275.15.10943. [DOI] [PubMed] [Google Scholar]
- Howard S, Deroo T, Fujita Y, Itasaki N. A positive role of cadherin in Wnt/beta-catenin signalling during epithelial-mesenchymal transition. PLoS One. 2011;6:e23899. doi: 10.1371/journal.pone.0023899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Berkowitz P, O’Keefe EJ, Rubenstein DS. Keratinocyte adherens junctions initiate nuclear signaling by translocation of plakoglobin from the membrane to the nucleus. The Journal of Investigative Dermatology. 2003;121:242–251. doi: 10.1046/j.1523-1747.2003.12376.x. [DOI] [PubMed] [Google Scholar]
- Hu P, O’Keefe EJ, Rubenstein DS. Tyrosine phosphorylation of human keratinocyte beta-catenin and plakoglobin reversibly regulates their binding to E-cadherin and alpha-catenin. The Journal of Investigative Dermatology. 2001;117:1059–1067. doi: 10.1046/j.0022-202x.2001.01523.x. [DOI] [PubMed] [Google Scholar]
- Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes & Development. 2004;18:3010–3015. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mechanisms of Development. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. Requirement for beta-catenin in anterior-posterior axis formation in mice. The Journal of Cell Biology. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. The International Journal of Biochemistry & Cell Biology. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F. New insights into the evolution of metazoan cadherins. Molecular Biology and Evolution. 2011;28:647–657. doi: 10.1093/molbev/msq233. [DOI] [PubMed] [Google Scholar]
- Huveneers S, de Rooij J. Mechanosensitive systems at the cadherin-F-actin interface. Journal of Cell Science. 2013;126:403–413. doi: 10.1242/jcs.109447. [DOI] [PubMed] [Google Scholar]
- Hwang SG, Yu SS, Ryu JH, Jeon HB, Yoo YJ, Eom SH, et al. Regulation of beta-catenin signaling and maintenance of chondrocyte differentiation by ubiquitin-independent proteasomal degradation of alpha-catenin. The Journal of Biological Chemistry. 2005;280:12758–12765. doi: 10.1074/jbc.M413367200. [DOI] [PubMed] [Google Scholar]
- Inge LJ, Barwe SP, D’Ambrosio J, Gopal J, Lu K, Ryazantsev S, et al. Soluble E-cadherin promotes cell survival by activating epidermal growth factor receptor. Experimental Cell Research. 2011;317:838–848. doi: 10.1016/j.yexcr.2010.12.025. [DOI] [PubMed] [Google Scholar]
- Ito K, Okamoto I, Araki N, Kawano Y, Nakao M, Fujiyama S, et al. Calcium influx triggers the sequential proteolysis of extracellular and cytoplasmic domains of E-cadherin, leading to loss of beta-catenin from cell-cell contacts. Oncogene. 1999;18:7080–7090. doi: 10.1038/sj.onc.1203191. [DOI] [PubMed] [Google Scholar]
- Iyer S, Ferreri DM, DeCocco NC, Minnear FL, Vincent PA. VE-cadherin-p120 interaction is required for maintenance of endothelial barrier function. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2004;286:L1143–L1153. doi: 10.1152/ajplung.00305.2003. [DOI] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean C, Blanc A, Prade-Houdellier N, Ysebaert L, Hernandez-Pigeon H, Al Saati T, et al. Epidermal growth factor receptor/beta-catenin/T-cell factor 4/matrix metalloproteinase 1: A new pathway for regulating keratinocyte invasiveness after UVA irradiation. Cancer Research. 2009;69:3291–3299. doi: 10.1158/0008-5472.CAN-08-1909. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: From the cytoskeleton to the nucleus. Nature Reviews. Molecular Cell Biology. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kahr I, Vandepoele K, van Roy F. Delta-protocadherins in health and disease. Progress in Molecular Biology and Translational Science. 2013;116:169–192. doi: 10.1016/B978-0-12-394311-8.00008-X. [DOI] [PubMed] [Google Scholar]
- Kamekura R, Kolegraff KN, Nava P, Hilgarth RS, Feng M, Parkos CA, et al. Loss of the desmosomal cadherin desmoglein-2 suppresses colon cancer cell proliferation through EGFR signaling. Oncogene. 2014;33:4531–4536. doi: 10.1038/onc.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DE, Soriano S, Frosch MP, Collins T, Naruse S, Sisodia SS, et al. Presenilin 1 facilitates the constitutive turnover of beta-catenin: Differential activity of Alzheimer’s disease-linked PS1 mutants in the beta-catenin-signaling pathway. The Journal of Neuroscience. 1999;19:4229–4237. doi: 10.1523/JNEUROSCI.19-11-04229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DE, Soriano S, Xia X, Eberhart CG, De Strooper B, Zheng H, et al. Presenilin couples the paired phosphorylation of beta-catenin independent of axin: Implications for beta-catenin activation in tumorigenesis. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Molecular Biology of the Cell. 2000;11:3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A, Klymkowsky MW. Anterior axis duplication in Xenopus induced by the over-expression of the cadherin-binding protein plakoglobin. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4522–4526. doi: 10.1073/pnas.92.10.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashef J, Kohler A, Kuriyama S, Alfandari D, Mayor R, Wedlich D. Cadherin-11 regulates protrusive activity in Xenopus cranial neural crest cells upstream of Trio and the small GTPases. Genes & Development. 2009;23:1393–1398. doi: 10.1101/gad.519409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzmann A, Wang Y, Weber D, Steinbeisser H. Xenopus paraxial protocadherin inhibits Wnt/beta-catenin signalling via casein kinase 2beta. EMBO Reports. 2012;13:129–134. doi: 10.1038/embor.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilander MB, Petersen J, Andressen KW, Ganji RS, Levy FO, Schuster J, et al. Disheveled regulates precoupling of heterotrimeric G proteins to Frizzled 6. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2014;28:2293–2305. doi: 10.1096/fj.13-246363. [DOI] [PubMed] [Google Scholar]
- Kim S, Ishidate T, Sharma R, Soto MC, Conte D, Jr, Mello CC, et al. Wnt and CDK-1 regulate cortical release of WRM-1/beta-catenin to control cell division orientation in early Caenorhabditis elegans embryos. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E918–E927. doi: 10.1073/pnas.1300769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NG, Koh E, Chen X, Gumbiner BM. E-Cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kugler MC, Wei Y, Kim KK, Li X, Brumwell AN, et al. Integrin alpha3beta1-dependent beta-catenin phosphorylation links epithelial Smad signaling to cell contacts. The Journal of Cell Biology. 2009;184:309–322. doi: 10.1083/jcb.200806067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Park JI, Spring CM, Sater AK, Ji H, Otchere AA, et al. Noncanonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nature Cell Biology. 2004;6:1212–1220. doi: 10.1038/ncb1191. [DOI] [PubMed] [Google Scholar]
- Klymkowsky MW, Williams BO, Barish GD, Varmus HE, Vourgourakis YE. Membrane-anchored plakoglobins have multiple mechanisms of action in Wnt signaling. Molecular Biology of the Cell. 1999;10:3151–3169. doi: 10.1091/mbc.10.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak A, Fuchs E. Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2322–2327. doi: 10.1073/pnas.0510422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs FT, Hu G, Dang CV, Fearon ER. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Molecular and Cellular Biology. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. The Journal of Biological Chemistry. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Konsavage WM, Jr, Yochum GS. Intersection of Hippo/YAP and Wnt/beta-catenin signaling pathways. Acta Biochimica et Biophysica Sinica. 2013;45:71–79. doi: 10.1093/abbs/gms084. [DOI] [PubMed] [Google Scholar]
- Kourtidis A, Ngok SP, Anastasiadis PZ. p120 catenin: An essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Progress in Molecular Biology and Translational Science. 2013;116:409–432. doi: 10.1016/B978-0-12-394311-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft B, Berger CD, Wallkamm V, Steinbeisser H, Wedlich D. Wnt-11 and Fz7 reduce cell adhesion in convergent extension by sequestration of PAPC and C-cadherin. The Journal of Cell Biology. 2012;198:695–709. doi: 10.1083/jcb.201110076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, et al. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nature Structural & Molecular Biology. 2005;12:238–244. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- Lam L, Aktary Z, Bishay M, Werkman C, Kuo CY, Heacock M, et al. Regulation of subcellular distribution and oncogenic potential of nucleophosmin by plakoglobin. Oncogenesis. 2012;1:e4. doi: 10.1038/oncsis.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani MG, Orsenigo F, Gagliani MC, Tacchetti C, Dejana E. Vascular endothelial cadherin controls VEGFR-2 internalization and signaling from intracellular compartments. The Journal of Cell Biology. 2006;174:593–604. doi: 10.1083/jcb.200602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani MG, Zanetti A, Breviario F, Balconi G, Orsenigo F, Corada M, et al. VE-cadherin regulates endothelial actin activating Rac and increasing membrane association of Tiam. Molecular Biology of the Cell. 2002;13:1175–1189. doi: 10.1091/mbc.01-07-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Antos C, Butz S, Huber O, Delmas V, Dominis M, et al. A role for cadherins in tissue formation. Development. 1996;122:3185–3194. doi: 10.1242/dev.122.10.3185. [DOI] [PubMed] [Google Scholar]
- Lee M, Ji H, Furuta Y, Park JI, McCrea PD. p120-catenin regulates REST and CoREST, and modulates mouse embryonic stem cell differentiation. Journal of Cell Science. 2014;127:4037–4051. doi: 10.1242/jcs.151944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Kang DH, Choi M, Choi YJ, Lee JY, Park JH, et al. Peroxiredoxin-2 represses melanoma metastasis by increasing E-cadherin/beta-catenin complexes in adherens junctions. Cancer Research. 2013;73:4744–4757. doi: 10.1158/0008-5472.CAN-12-4226. [DOI] [PubMed] [Google Scholar]
- Li L, Bennett SA, Wang L. Role of E-cadherin and other cell adhesion molecules in survival and differentiation of human pluripotent stem cells. Cell Adhesion & Migration. 2012;6:59–70. doi: 10.4161/cam.19583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Swope D, Raess N, Cheng L, Muller EJ, Radice GL. Cardiac tissue-restricted deletion of plakoglobin results in progressive cardiomyopathy and activation of {beta}-catenin signaling. Molecular and Cellular Biology. 2011;31:1134–1144. doi: 10.1128/MCB.01025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang S, Jezierski A, Moalim-Nour L, Mohib K, Parks RJ, et al. A unique interplay between Rap1 and E-cadherin in the endocytic pathway regulates self-renewal of human embryonic stem cells. Stem Cells. 2010;28:247–257. doi: 10.1002/stem.289. [DOI] [PubMed] [Google Scholar]
- Li D, Zhou J, Wang L, Shin ME, Su P, Lei X, et al. Integrated biochemical and mechanical signals regulate multifaceted human embryonic stem cell functions. The Journal of Cell Biology. 2010;191:631–644. doi: 10.1083/jcb.201006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Fuchs E. Wnt some lose some: Transcriptional governance of stem cells by Wnt/beta-catenin signaling. Genes & Development. 2014;28:1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 2006;311:1609–1612. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell MJ. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. The Journal of Cell Biology. 1997;139:1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT, Marian AJ. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circulation Research. 2011;109:1342–1353. doi: 10.1161/CIRCRESAHA.111.255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4:499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- Lu M, Marsters S, Ye X, Luis E, Gonzalez L, Ashkenazi A. E-Cadherin couples death receptors to the cytoskeleton to regulate apoptosis. Molecular Cell. 2014;54:987–998. doi: 10.1016/j.molcel.2014.04.029. [DOI] [PubMed] [Google Scholar]
- Luchtenborg AM, Solis GP, Egger-Adam D, Koval A, Lin C, Blanchard MG, et al. Heterotrimeric Go protein links Wnt-Frizzled signaling with ankyrins to regulate the neuronal microtubule cytoskeleton. Development. 2014;141:3399–3409. doi: 10.1242/dev.106773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyashenko N, Winter M, Migliorini D, Biechele T, Moon RT, Hartmann C. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nature Cell Biology. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Developmental Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher MT, Flozak AS, Stocker A, Chenn A, Gottardi CJ. Activity of the beta-catenin phosphodestruction complex at cell-cell contacts is enhanced by cadherin-based adhesion. The Journal of Cell Biology. 2009;186:219–228. doi: 10.1083/jcb.200811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden SL, Hardin J. The secret life of alpha-catenin: Moonlighting in morphogenesis. The Journal of Cell Biology. 2011;195:543–552. doi: 10.1083/jcb.201103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre JL, Heisenberg CP. Three functions of cadherins in cell adhesion. Current Biology. 2013;23:R626–R633. doi: 10.1016/j.cub.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316:1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, et al. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. The EMBO Journal. 2002;21:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, et al. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, et al. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, Hartmann D, et al. L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Molecular and Cellular Biology. 2005;25:9040–9053. doi: 10.1128/MCB.25.20.9040-9053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. The Journal of Biological Chemistry. 2003;278:1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. The EMBO Journal. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea PD, Park JI. Developmental functions of the P120-catenin sub-family. Biochimica et Biophysica Acta. 2007;1773:17–33. doi: 10.1016/j.bbamcr.2006.06.009. [DOI] [PubMed] [Google Scholar]
- McCusker CD, Alfandari D. Life after proteolysis: Exploring the signaling capabilities of classical cadherin cleavage fragments. Communicative & Integrative Biology. 2009;2:155–157. doi: 10.4161/cib.7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C, Cousin H, Neuner R, Alfandari D. Extracellular cleavage of cadherin-11 by ADAM metalloproteases is essential for Xenopus cranial neural crest cell migration. Molecular Biology of the Cell. 2009;20:78–89. doi: 10.1091/mbc.E08-05-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen AE, Escobar DE, Gottardi CJ. Signaling from the adherens junction. Sub-Cellular Biochemistry. 2012;60:171–196. doi: 10.1007/978-94-007-4186-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan RW, Yap AS. Not so simple: The complexity of phosphotyrosine signaling at cadherin adhesive contacts. Journal of Molecular Medicine. 2007;85:545–554. doi: 10.1007/s00109-007-0198-x. [DOI] [PubMed] [Google Scholar]
- Medina A, Swain RK, Kuerner KM, Steinbeisser H. Xenopus paraxial protocadherin has signaling functions and is involved in tissue separation. The EMBO Journal. 2004;23:3249–3258. doi: 10.1038/sj.emboj.7600329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mege RM, Gavard J, Lambert M. Regulation of cell-cell junctions by the cytoskeleton. Current Opinion in Cell Biology. 2006;18:541–548. doi: 10.1016/j.ceb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Mehta D, Malik AB. Signaling mechanisms regulating endothelial permeability. Physiological Reviews. 2006;86:279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- Meng W, Takeichi M. Adherens junction: Molecular architecture and regulation. Cold Spring Harbor Perspectives in Biology. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdek KD, Nguyen NT, Toksoz D. Distinct activities of the alpha-catenin family, alpha-catulin and alpha-catenin, on beta-catenin-mediated signaling. Molecular and Cellular Biology. 2004;24:2410–2422. doi: 10.1128/MCB.24.6.2410-2422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt AJ, Berika MY, Zhai W, Kirk SE, Ji B, Hardman MJ, et al. Suprabasal desmoglein 3 expression in the epidermis of transgenic mice results in hyperproliferation and abnormal differentiation. Molecular and Cellular Biology. 2002;22:5846–5858. doi: 10.1128/MCB.22.16.5846-5858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens C, Kuhn C, Franke WW. Plakophilins 2a and 2b: Constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. The Journal of Cell Biology. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi K, Dolan PJ, Johnson GV. The low density lipoprotein receptor-related protein 6 interacts with glycogen synthase kinase 3 and attenuates activity. The Journal of Biological Chemistry. 2006;281:4787–4794. doi: 10.1074/jbc.M508657200. [DOI] [PubMed] [Google Scholar]
- Miki T, Yasuda SY, Kahn M. Wnt/beta-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Reviews. 2011;7:836–846. doi: 10.1007/s12015-011-9275-1. [DOI] [PubMed] [Google Scholar]
- Miravet S, Piedra J, Castano J, Raurell I, Franci C, Dunach M, et al. Tyrosine phosphorylation of plakoglobin causes contrary effects on its association with desmosomes and adherens junction components and modulates beta-catenin-mediated transcription. Molecular and Cellular Biology. 2003;23:7391–7402. doi: 10.1128/MCB.23.20.7391-7402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravet S, Piedra J, Miro F, Itarte E, Garcia de Herreros A, Dunach M. The transcriptional factor Tcf-4 contains different binding sites for beta-catenin and plakoglobin. The Journal of Biological Chemistry. 2002;277:1884–1891. doi: 10.1074/jbc.M110248200. [DOI] [PubMed] [Google Scholar]
- Mohamet L, Lea ML, Ward CM. Abrogation of E-cadherin-mediated cellular aggregation allows proliferation of pluripotent mouse embryonic stem cells in shake flask bioreactors. PLoS One. 2010;5:e12921. doi: 10.1371/journal.pone.0012921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell DJ, Rorth P, Spradling AC. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Montoya-Durango DE, Velu CS, Kazanjian A, Rojas ME, Jay CM, Longmore GD, et al. Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. The Journal of Biological Chemistry. 2008;283:32056–32065. doi: 10.1074/jbc.M802320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R, Radice GL, Dominis M, Kemler R. The generation and in vivo differentiation of murine embryonal stem cells genetically null for either N-cadherin or N- and P-cadherin. The International Journal of Developmental Biology. 1999;43:831–834. [PubMed] [Google Scholar]
- Morali OG, Delmas V, Moore R, Jeanney C, Thiery JP, Larue L. IGF-II induces rapid beta-catenin relocation to the nucleus during epithelium to mesenchyme transition. Oncogene. 2001;20:4942–4950. doi: 10.1038/sj.onc.1204660. [DOI] [PubMed] [Google Scholar]
- Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, et al. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes & Development. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkey RM, Endo S, Shenolikar S, Malenka RC. Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature. 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- Muller J, Ritt DA, Copeland TD, Morrison DK. Functional analysis of C-TAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate. The EMBO Journal. 2003;22:4431–4442. doi: 10.1093/emboj/cdg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz WA, Lee M, Miller RK, Ahmed Z, Ji H, Link TM, et al. Plakophilin-3 catenin associates with the ETV1/ER81 transcription factor to positively modulate gene activity. PLoS One. 2014;9:e86784. doi: 10.1371/journal.pone.0086784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Hijikata T, Matsukawa M, Ishikawa H, Yorifuji H. Zebrafish protocadherin 10 is involved in paraxial mesoderm development and somitogenesis. Developmental Dynamics. 2006;235:506–514. doi: 10.1002/dvdy.20622. [DOI] [PubMed] [Google Scholar]
- Murakami M, Nguyen LT, Zhuang ZW, Moodie KL, Carmeliet P, Stan RV, et al. The FGF system has a key role in regulating vascular integrity. The Journal of Clinical Investigation. 2008;118:3355–3366. doi: 10.1172/JCI35298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. The Journal of Biological Chemistry. 2008;283:18393–18401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao S, Platek A, Hirano S, Takeichi M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. The Journal of Cell Biology. 2008;182:395–410. doi: 10.1083/jcb.200802069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava P, Laukoetter MG, Hopkins AM, Laur O, Gerner-Smidt K, Green KJ, et al. Desmoglein-2: A novel regulator of apoptosis in the intestinal epithelium. Molecular Biology of the Cell. 2007;18:4565–4578. doi: 10.1091/mbc.E07-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki-Raby B, Gilles C, Polette M, Bruyneel E, Laronze JY, Bonnet N, et al. Upregulation of MMPs by soluble E-cadherin in human lung tumor cells. International Journal of Cancer. 2003;105:790–795. doi: 10.1002/ijc.11168. [DOI] [PubMed] [Google Scholar]
- Nawroth R, Poell G, Ranft A, Kloep S, Samulowitz U, Fachinger G, et al. VE-PTP and VE-cadherin ectodomains interact to facilitate regulation of phosphorylation and cell contacts. The EMBO Journal. 2002;21:4885–4895. doi: 10.1093/emboj/cdf497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: Dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiological Reviews. 2011;91:691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska P, Godt D, Tepass U. DE-Cadherin is required for intercellular motility during Drosophila oogenesis. The Journal of Cell Biology. 1999;144:533–547. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Takeichi M. Remodeling of the adherens junctions during morphogenesis. Current Topics in Developmental Biology. 2009;89:33–54. doi: 10.1016/S0070-2153(09)89002-9. [DOI] [PubMed] [Google Scholar]
- Noe V, Fingleton B, Jacobs K, Crawford HC, Vermeulen S, Steelant W, et al. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. Journal of Cell Science. 2001;114:111–118. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- Noren NK, Niessen CM, Gumbiner BM, Burridge K. Cadherin engagement regulates Rho family GTPases. The Journal of Biological Chemistry. 2001;276:33305–33308. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- Novellino L, De Filippo A, Deho P, Perrone F, Pilotti S, Parmiani G, et al. PTPRK negatively regulates transcriptional activity of wild type and mutated oncogenic beta-catenin and affects membrane distribution of beta-catenin/E-cadherin complexes in cancer cells. Cellular Signalling. 2008;20:872–883. doi: 10.1016/j.cellsig.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Oda H, Tagawa K, Akiyama-Oda Y. Diversification of epithelial adherens junctions with independent reductive changes in cadherin form: Identification of potential molecular synapomorphies among bilaterians. Evolution & Development. 2005;7:376–389. doi: 10.1111/j.1525-142X.2005.05043.x. [DOI] [PubMed] [Google Scholar]
- Oda H, Takeichi M. Evolution: Structural and functional diversity of cadherin at the adherens junction. The Journal of Cell Biology. 2011;193:1137–1146. doi: 10.1083/jcb.201008173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, et al. WNT7a induces E-cadherin in lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10429–10434. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-Cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. Journal of Cell Science. 1999;112(Pt 8):1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- Pai R, Dunlap D, Qing J, Mohtashemi I, Hotzel K, French DM. Inhibition of fibroblast growth factor 19 reduces tumor growth by modulating beta-catenin signaling. Cancer Research. 2008;68:5086–5095. doi: 10.1158/0008-5472.CAN-07-2325. [DOI] [PubMed] [Google Scholar]
- Paradies NE, Grunwald GB. Purification and characterization of NCAD90, a soluble endogenous form of N-cadherin, which is generated by proteolysis during retinal development and retains adhesive and neurite-promoting function. Journal of Neuroscience Research. 1993;36:33–45. doi: 10.1002/jnr.490360105. [DOI] [PubMed] [Google Scholar]
- Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Developmental Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Parnaud G, Lavallard V, Bedat B, Matthey-Doret D, Morel P, Berney T, et al. Cadherin engagement improves insulin secretion of single human beta-cells. Diabetes. 2014 Oct 2; doi: 10.2337/db14-0257. pii: DB_140257. [Epub ahead of print].. [DOI] [PubMed] [Google Scholar]
- Paulson AF, Fang X, Ji H, Reynolds AB, McCrea PD. Misexpression of the catenin p120(ctn)1A perturbs Xenopus gastrulation but does not elicit Wnt-directed axis specification. Developmental Biology. 1999;207:350–363. doi: 10.1006/dbio.1998.9158. [DOI] [PubMed] [Google Scholar]
- Paulson AF, Prasad MS, Thuringer AH, Manzerra P. Regulation of cadherin expression in nervous system development. Cell Adhesion & Migration. 2014;8:19–28. doi: 10.4161/cam.27839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Chiariello M, Murga C, Gutkind JS. Activation of the protein kinase Akt/PKB by the formation of E-cadherin-mediated cell-cell junctions. Evidence for the association of phosphatidylinositol 3-kinase with the E-cadherin adhesion complex. The Journal of Biological Chemistry. 1999;274:19347–19351. doi: 10.1074/jbc.274.27.19347. [DOI] [PubMed] [Google Scholar]
- Pece S, Gutkind JS. Signaling from E-cadherins to the MAPK pathway by the recruitment and activation of epidermal growth factor receptors upon cell-cell contact formation. The Journal of Biological Chemistry. 2000;275:41227–41233. doi: 10.1074/jbc.M006578200. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. p120-catenin mediates inflammatory responses in the skin. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrais M, Chen X, Perez-Moreno M, Gumbiner BM. E-Cadherin homophilic ligation inhibits cell growth and epidermal growth factor receptor signaling independently of other cell interactions. Molecular Biology of the Cell. 2007;18:2013–2025. doi: 10.1091/mbc.E06-04-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nature Cell Biology. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- Philippova M, Joshi MB, Kyriakakis E, Pfaff D, Erne P, Resink TJ. A guide and guard: The many faces of T-cadherin. Cellular Signalling. 2009;21:1035–1044. doi: 10.1016/j.cellsig.2009.01.035. [DOI] [PubMed] [Google Scholar]
- Piao S, Lee SH, Kim H, Yum S, Stamos JL, Xu Y, et al. Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS One. 2008;3:e4046. doi: 10.1371/journal.pone.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao HL, Yuan Y, Wang M, Sun Y, Liang H, Ma L. Alpha-catenin acts as a tumour suppressor in E-cadherin-negative basal-like breast cancer by inhibiting NF-kappaB signalling. Nature Cell Biology. 2014;16:245–254. doi: 10.1038/ncb2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra J, Martinez D, Castano J, Miravet S, Dunach M, de Herreros AG. Regulation of beta-catenin structure and activity by tyrosine phosphorylation. The Journal of Biological Chemistry. 2001;276:20436–20443. doi: 10.1074/jbc.M100194200. [DOI] [PubMed] [Google Scholar]
- Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia de Herreros A, et al. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Molecular and Cellular Biology. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters T, van Hengel J, van Roy F. Functions of p120ctn in development and disease. Frontiers in Bioscience. 2012;17:760–783. doi: 10.2741/3956. [DOI] [PubMed] [Google Scholar]
- Pieters T, van Roy F. Role of cell-cell adhesion complexes in embryonic stem cell biology. Journal of Cell Science. 2014;127:2603–2613. doi: 10.1242/jcs.146720. [DOI] [PubMed] [Google Scholar]
- Pieters T, van Roy F, van Hengel J. Functions of p120ctn isoforms in cell-cell adhesion and intracellular signaling. Frontiers in Bioscience. 2012;17:1669–1694. doi: 10.2741/4012. [DOI] [PubMed] [Google Scholar]
- Playford MP, Bicknell D, Bodmer WF, Macaulay VM. Insulin-like growth factor 1 regulates the location, stability, and transcriptional activity of beta-catenin. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12103–12108. doi: 10.1073/pnas.210394297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes & Development. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. The EMBO Journal. 2004;23:1739–1748. doi: 10.1038/sj.emboj.7600136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice GL. N-cadherin-mediated adhesion and signaling from development to disease: Lessons from mice. Progress in Molecular Biology and Translational Science. 2013;116:263–289. doi: 10.1016/B978-0-12-394311-8.00012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raptis L, Brownell HL, Vultur AM, Ross GM, Tremblay E, Elliott BE. Specific inhibition of growth factor-stimulated extracellular signal-regulated kinase 1 and 2 activation in intact cells by electroporation of a growth factor receptor-binding protein 2-Src homology 2 binding peptide. Cell Growth & Differentiation. 2000;11:293–303. [PubMed] [Google Scholar]
- Rashid D, Newell K, Shama L, Bradley R. A requirement for NF-protocadherin and TAF1/Set in cell adhesion and neural tube formation. Developmental Biology. 2006;291:170–181. doi: 10.1016/j.ydbio.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Ratheesh A, Priya R, Yap AS. Coordinating Rho and Rac: The regulation of Rho GTPase signaling and cadherin junctions. Progress in Molecular Biology and Translational Science. 2013;116:49–68. doi: 10.1016/B978-0-12-394311-8.00003-0. [DOI] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, et al. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. The EMBO Journal. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Carnahan RH. Regulation of cadherin stability and turnover by p120ctn: Implications in disease and cancer. Seminars in Cell & Developmental Biology. 2004;15:657–663. doi: 10.1016/j.semcdb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Rhee J, Buchan T, Zukerberg L, Lilien J, Balsamo J. Cables links Robo-bound Abl kinase to N-cadherin-bound beta-catenin to mediate Slit-induced modulation of adhesion and transcription. Nature Cell Biology. 2007;9:883–892. doi: 10.1038/ncb1614. [DOI] [PubMed] [Google Scholar]
- Rhee J, Ryu JH, Kim JH, Chun CH, Chun JS. alpha-Catenin inhibits beta-catenin-T-cell factor/lymphoid enhancing factor transcriptional activity and collagen type II expression in articular chondrocytes through formation of Gli3R.alpha-catenin. beta-catenin ternary complex. The Journal of Biological Chemistry. 2012;287:11751–11760. doi: 10.1074/jbc.M111.281014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Doria J, Day ML. Truncated E-cadherin potentiates cell death in prostate epithelial cells. The Prostate. 2005;63:259–268. doi: 10.1002/pros.20179. [DOI] [PubMed] [Google Scholar]
- Rios-Doria J, Day KC, Kuefer R, Rashid MG, Chinnaiyan AM, Rubin MA, et al. The role of calpain in the proteolytic cleavage of E-cadherin in prostate and mammary epithelial cells. The Journal of Biological Chemistry. 2003;278:1372–1379. doi: 10.1074/jbc.M208772200. [DOI] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Developmental Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura S, Miravet S, Piedra J, Garcia de Herreros A, Dunach M. Regulation of E-cadherin/catenin association by tyrosine phosphorylation. The Journal of Biological Chemistry. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, et al. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74:314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudini N, Felici A, Giampietro C, Lampugnani M, Corada M, Swirsding K, et al. VE-cadherin is a critical endothelial regulator of TGF-beta signalling. The EMBO Journal. 2008;27:993–1004. doi: 10.1038/emboj.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov A, Hackett JA, Prokhortchouk A, Reddington JP, Madej MJ, Dunican DS, et al. The interaction of xKaiso with xTcf3: A revised model for integration of epigenetic and Wnt signalling pathways. Development. 2009;136:723–727. doi: 10.1242/dev.025577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov A, Savitskaya E, Hackett JA, Reddington JP, Prokhortchouk A, Madej MJ, et al. The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development. 2009;136:729–738. doi: 10.1242/dev.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen P, Eklund L, Miettinen J, Wirkkala R, Anisimov A, Winderlich M, et al. Angiopoietins assemble distinct Tie2 signalling complexes in endothelial cell-cell and cell-matrix contacts. Nature Cell Biology. 2008;10:527–537. doi: 10.1038/ncb1715. [DOI] [PubMed] [Google Scholar]
- Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu W. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Molecular Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Sanson B, White P, Vincent JP. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. The EMBO Journal. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackmann RC, Tenhagen M, van de Ven RA, Derksen PW. p120-catenin in cancer—Mechanisms, models and opportunities for intervention. Journal of Cell Science. 2013;126:3515–3525. doi: 10.1242/jcs.134411. [DOI] [PubMed] [Google Scholar]
- Schalm SS, Ballif BA, Buchanan SM, Phillips GR, Maniatis T. Phosphorylation of protocadherin proteins by the receptor tyrosine kinase Ret. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13894–13899. doi: 10.1073/pnas.1007182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepis A, Sepich D, Nelson WJ. Alpha-E-catenin regulates cell-cell adhesion and membrane blebbing during zebrafish epiboly. Development. 2012;139:537–546. doi: 10.1242/dev.073932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Jager S. Plakophilins—Hard work in the desmosome, recreation in the nucleus? European Journal of Cell Biology. 2005;84:189–204. doi: 10.1016/j.ejcb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Schneider S, Herrenknecht K, Butz S, Kemler R, Hausen P. Catenins in Xenopus embryogenesis and their relation to the cadherin-mediated cell-cell adhesion system. Development. 1993;118:629–640. doi: 10.1242/dev.118.2.629. [DOI] [PubMed] [Google Scholar]
- Sehgal RN, Gumbiner BM, Reichardt LF. Antagonism of cell adhesion by an alpha-catenin mutant, and of the Wnt-signaling pathway by alpha-catenin in Xenopus embryos. The Journal of Cell Biology. 1997;139:1033–1046. doi: 10.1083/jcb.139.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K, Ibrahim H, Stodtmeister T, Winklbauer R, Niessen CM. An adhesion-independent, aPKC-dependent function for cadherins in morphogenetic movements. Journal of Cell Science. 2009;122:2514–2523. doi: 10.1242/jcs.042796. [DOI] [PubMed] [Google Scholar]
- Seki T, Namba T, Mochizuki H, Onodera M. Clustering, migration, and neurite formation of neural precursor cells in the adult rat hippocampus. The Journal of Comparative Neurology. 2007;502:275–290. doi: 10.1002/cne.21301. [DOI] [PubMed] [Google Scholar]
- Serio RN. Wnt of the two horizons: Putting stem cell self-renewal and cell fate determination into context. Stem Cells and Development. 2014;23:1975–1990. doi: 10.1089/scd.2014.0055. [DOI] [PubMed] [Google Scholar]
- Shay-Salit A, Shushy M, Wolfovitz E, Yahav H, Breviario F, Dejana E, et al. VEGF receptor 2 and the adherens junction as a mechanical transducer in vascular endothelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9462–9467. doi: 10.1073/pnas.142224299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Usui T, Yanagawa S, Takeichi M, Uemura T. Asymmetric colocalization of Flamingo, a seven-pass transmembrane cadherin, and Dishevelled in planar cell polarization. Current Biology. 2001;11:859–863. doi: 10.1016/s0960-9822(01)00233-0. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Takeichi M. Local and transient expression of E-cadherin involved in mouse embryonic brain morphogenesis. Development. 1992;116:1011–1019. doi: 10.1242/dev.116.4.1011. [DOI] [PubMed] [Google Scholar]
- Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Current Biology. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, et al. alpha-Catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Science Signaling. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcha I, Kirkpatrick C, Sadot E, Shtutman M, Polevoy G, Geiger B, et al. Cadherin sequences that inhibit beta-catenin signaling: A study in yeast and mammalian cells. Molecular Biology of the Cell. 2001;12:1177–1188. doi: 10.1091/mbc.12.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, et al. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. The Journal of Cell Biology. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Hoffman LM, Beckerle MC. LIM proteins in actin cytoskeleton mechanoresponse. Trends in Cell Biology. 2014;24:575–583. doi: 10.1016/j.tcb.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutny M, Yap AS. Neighborly relations: Cadherins and mechanotransduction. The Journal of Cell Biology. 2010;189:1075–1077. doi: 10.1083/jcb.201005151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G, Miravet S, Casagolda D, Castano J, Raurell I, Corrionero A, et al. beta-Catenin and plakoglobin N- and C-tails determine ligand specificity. The Journal of Biological Chemistry. 2004;279:49849–49856. doi: 10.1074/jbc.M408685200. [DOI] [PubMed] [Google Scholar]
- Solanas G, Porta-de-la-Riva M, Agusti C, Casagolda D, Sanchez-Aguilera F, Larriba MJ, et al. E-Cadherin controls beta-catenin and NF-kappaB transcriptional activity in mesenchymal gene expression. Journal of Cell Science. 2008;121:2224–2234. doi: 10.1242/jcs.021667. [DOI] [PubMed] [Google Scholar]
- Soncin F, Mohamet L, Eckardt D, Ritson S, Eastham AM, Bobola N, et al. Abrogation of E-cadherin-mediated cell-cell contact in mouse embryonic stem cells results in reversible LIF-independent self-renewal. Stem Cells. 2009;27:2069–2080. doi: 10.1002/stem.134. [DOI] [PubMed] [Google Scholar]
- Song X, Xie T. Wingless signaling regulates the maintenance of ovarian somatic stem cells in Drosophila. Development. 2003;130:3259–3268. doi: 10.1242/dev.00524. [DOI] [PubMed] [Google Scholar]
- Song X, Zhu CH, Doan C, Xie T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 2002;296:1855–1857. doi: 10.1126/science.1069871. [DOI] [PubMed] [Google Scholar]
- Soriano S, Kang DE, Fu M, Pestell R, Chevallier N, Zheng H, et al. Presenilin 1 negatively regulates beta-catenin/T cell factor/lymphoid enhancer factor-1 signaling independently of beta-amyloid precursor protein and notch processing. The Journal of Cell Biology. 2001;152:785–794. doi: 10.1083/jcb.152.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring CM, Kelly KF, O’Kelly I, Graham M, Crawford HC, Daniel JM. The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Experimental Cell Research. 2005;305:253–265. doi: 10.1016/j.yexcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harbor Perspectives in Biology. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. Cleavage and shedding of E-cadherin after induction of apoptosis. The Journal of Biological Chemistry. 2001;276:4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harbor Perspectives in Biology. 2009;1:a002949. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Fu C, Ishikawa S, Stella A, Kojima M, Shitoh K, et al. APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Molecular Cell. 2008;32:652–661. doi: 10.1016/j.molcel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Suh EK, Gumbiner BM. Translocation of beta-catenin into the nucleus independent of interactions with FG-rich nucleoporins. Experimental Cell Research. 2003;290:447–456. doi: 10.1016/s0014-4827(03)00370-7. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi E, Takahashi S, Obata H, Sugimoto A, Kohara Y. The beta-catenin HMP-2 functions downstream of Src in parallel with the Wnt pathway in early embryogenesis of C. elegans. Developmental Biology. 2011;355:302–312. doi: 10.1016/j.ydbio.2011.04.034. [DOI] [PubMed] [Google Scholar]
- Sun Z, Parrish AR, Hill MA, Meininger GA. N-cadherin, a vascular smooth muscle cell-cell adhesion molecule: Function and signaling for vasomotor control. Microcirculation. 2014;21:208–218. doi: 10.1111/micc.12123. [DOI] [PubMed] [Google Scholar]
- Suyama K, Shapiro I, Guttman M, Hazan RB. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2:301–314. doi: 10.1016/s1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- Swope D, Li J, Radice GL. Beyond cell adhesion: The role of armadillo proteins in the heart. Cellular Signalling. 2013;25:93–100. doi: 10.1016/j.cellsig.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symowicz J, Adley BP, Gleason KJ, Johnson JJ, Ghosh S, Fishman DA, et al. Engagement of collagen-binding integrins promotes matrix metalloproteinase-9-dependent E-cadherin ectodomain shedding in ovarian carcinoma cells. Cancer Research. 2007;67:2030–2039. doi: 10.1158/0008-5472.CAN-06-2808. [DOI] [PubMed] [Google Scholar]
- Taddei A, Giampietro C, Conti A, Orsenigo F, Breviario F, Pirazzoli V, et al. Endothelial adherens junctions control tight junctions by VE-cadherin-mediated upregulation of claudin-5. Nature Cell Biology. 2008;10:923–934. doi: 10.1038/ncb1752. [DOI] [PubMed] [Google Scholar]
- Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Ishihara S, Takada S, Tsukita S, Nagafuchi A. Posttranscriptional regulation of alpha-catenin expression is required for Wnt signaling in L cells. Biochemical and Biophysical Research Communications. 2000;277:691–698. doi: 10.1006/bbrc.2000.3748. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Suzuki K. Density-dependent inhibition of growth involves prevention of EGF receptor activation by E-cadherin-mediated cell-cell adhesion. Experimental Cell Research. 1996;226:214–222. doi: 10.1006/excr.1996.0221. [DOI] [PubMed] [Google Scholar]
- Tanneberger K, Pfister AS, Brauburger K, Schneikert J, Hadjihannas MV, Kriz V, et al. Amer1/WTX couples Wnt-induced formation of PtdIns(4,5)P2 to LRP6 phosphorylation. The EMBO Journal. 2011;30:1433–1443. doi: 10.1038/emboj.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Feetham MC, Tao WA, He XC, Li L, Aebersold R, et al. Proteomic analysis identifies that 14-3-3zeta interacts with beta-catenin and facilitates its activation by Akt. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15370–15375. doi: 10.1073/pnas.0406499101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NL, Adams DG, Vaillancourt RR, Heimark RL. Signal transduction from N-cadherin increases Bcl-2. Regulation of the phosphatidylinositol 3-kinase/Akt pathway by homophilic adhesion and actin cytoskeletal organization. The Journal of Biological Chemistry. 2002;277:32905–32914. doi: 10.1074/jbc.M200300200. [DOI] [PubMed] [Google Scholar]
- Twiss F, de Rooij J. Cadherin mechanotransduction in tissue remodeling. Cellular and Molecular Life Sciences. 2013;70:4101–4116. doi: 10.1007/s00018-013-1329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, et al. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- Uemura K, Kihara T, Kuzuya A, Okawa K, Nishimoto T, Bito H, et al. Activity-dependent regulation of beta-catenin via epsilon-cleavage of N-cadherin. Biochemical and Biophysical Research Communications. 2006;345:951–958. doi: 10.1016/j.bbrc.2006.04.157. [DOI] [PubMed] [Google Scholar]
- Unterseher F, Hefele JA, Giehl K, De Robertis EM, Wedlich D, Schambony A. Paraxial protocadherin coordinates cell polarity during convergent extension via Rho A and JNK. The EMBO Journal. 2004;23:3259–3269. doi: 10.1038/sj.emboj.7600332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, et al. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Utton MA, Eickholt B, Howell FV, Wallis J, Doherty P. Soluble N-cadherin stimulates fibroblast growth factor receptor dependent neurite outgrowth and N-cadherin and the fibroblast growth factor receptor co-cluster in cells. Journal of Neurochemistry. 2001;76:1421–1430. doi: 10.1046/j.1471-4159.2001.00140.x. [DOI] [PubMed] [Google Scholar]
- Vallorosi CJ, Day KC, Zhao X, Rashid MG, Rubin MA, Johnson KR, et al. Truncation of the beta-catenin binding domain of E-cadherin precedes epithelial apoptosis during prostate and mammary involution. The Journal of Biological Chemistry. 2000;275:3328–3334. doi: 10.1074/jbc.275.5.3328. [DOI] [PubMed] [Google Scholar]
- Valls G, Codina M, Miller RK, Del Valle-Perez B, Vinyoles M, Caelles C, et al. Upon Wnt stimulation, Rac1 activation requires Rac1 and Vav2 binding to p120-catenin. Journal of Cell Science. 2012;125:5288–5301. doi: 10.1242/jcs.101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Barker N, Harkes IC, van der Heyden M, Dijk NJ, Hollestelle A, et al. Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Research. 2001;61:278–284. [PubMed] [Google Scholar]
- van Nieuw Amerongen GP, van Delft S, Vermeer MA, Collard JG, van Hinsbergh VW. Activation of RhoA by thrombin in endothelial hyperpermeability: Role of Rho kinase and protein tyrosine kinases. Circulation Research. 2000;87:335–340. doi: 10.1161/01.res.87.4.335. [DOI] [PubMed] [Google Scholar]
- van Rijssel J, van Buul JD. The many faces of the guanine-nucleotide exchange factor trio. Cell Adhesion & Migration. 2012;6:482–487. doi: 10.4161/cam.21418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roy F. Beyond E-cadherin: Roles of other cadherin superfamily members in cancer. Nature Reviews. 2014;14:121–134. doi: 10.1038/nrc3647. [DOI] [PubMed] [Google Scholar]
- van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, et al. Beta-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011;60:1204–1212. doi: 10.1136/gut.2010.233460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke St Amant E, Tauseef M, Vogel SM, Gao XP, Mehta D, Komarova YA, et al. PKCalpha activation of p120-catenin serine 879 phospho-switch disassembles VE-cadherin junctions and disrupts vascular integrity. Circulation Research. 2012;111:739–749. doi: 10.1161/CIRCRESAHA.112.269654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhalst K, Kools P, Staes K, van Roy F, Redies C. delta-Protocadherins: A gene family expressed differentially in the mouse brain. Cellular and Molecular Life Sciences. 2005;62:1247–1259. doi: 10.1007/s00018-005-5021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Developmental Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- Vermeer PD, Einwalter LA, Moninger TO, Rokhlina T, Kern JA, Zabner J, et al. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature. 2003;422:322–326. doi: 10.1038/nature01440. [DOI] [PubMed] [Google Scholar]
- Vinyoles M, Del Valle-Perez B, Curto J, Vinas-Castells R, Alba-Castellon L, Garcia de Herreros A, et al. Multivesicular GSK3 sequestration upon Wnt signaling is controlled by p120-catenin/cadherin interaction with LRP5/6. Molecular Cell. 2014;53:444–457. doi: 10.1016/j.molcel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Wang Y, Janicki P, Koster I, Berger CD, Wenzl C, Grosshans J, et al. Xenopus paraxial protocadherin regulates morphogenesis by antagonizing Sprouty. Genes & Development. 2008;22:878–883. doi: 10.1101/gad.452908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Sato K, Kaibuchi K. Cadherin-mediated intercellular adhesion and signaling cascades involving small GTPases. Cold Spring Harbor Perspectives in Biology. 2009;1:a003020. doi: 10.1101/cshperspect.a003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner JA, Jontes JD. Protocadherins, not prototypical: A complex tale of their interactions, expression, and functions. Frontiers in Molecular Neuroscience. 2013;6:4. doi: 10.3389/fnmol.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiske J, Schoneberg T, Schroder W, Hatzfeld M, Tauber R, Huber O. The fate of desmosomal proteins in apoptotic cells. The Journal of Biological Chemistry. 2001;276:41175–41181. doi: 10.1074/jbc.M105769200. [DOI] [PubMed] [Google Scholar]
- Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, et al. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Current Biology. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Willert J, Epping M, Pollack JR, Brown PO, Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Developmental Biology. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BO, Barish GD, Klymkowsky MW, Varmus HE. A comparative evaluation of beta-catenin and plakoglobin signaling activity. Oncogene. 2000;19:5720–5728. doi: 10.1038/sj.onc.1203921. [DOI] [PubMed] [Google Scholar]
- Williamson L, Raess NA, Caldelari R, Zakher A, de Bruin A, Posthaus H, et al. Pemphigus vulgaris identifies plakoglobin as key suppressor of c-Myc in the skin. The EMBO Journal. 2006;25:3298–3309. doi: 10.1038/sj.emboj.7601224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winderlich M, Keller L, Cagna G, Broermann A, Kamenyeva O, Kiefer F, et al. VE-PTP controls blood vessel development by balancing Tie-2 activity. The Journal of Cell Biology. 2009;185:657–671. doi: 10.1083/jcb.200811159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklbauer R, Medina A, Swain RK, Steinbeisser H. Frizzled-7 signalling controls tissue separation during Xenopus gastrulation. Nature. 2001;413:856–860. doi: 10.1038/35101621. [DOI] [PubMed] [Google Scholar]
- Wojciak-Stothard B, Potempa S, Eichholtz T, Ridley AJ. Rho and Rac but not Cdc42 regulate endothelial cell permeability. Journal of Cell Science. 2001;114:1343–1355. doi: 10.1242/jcs.114.7.1343. [DOI] [PubMed] [Google Scholar]
- Wojciak-Stothard B, Ridley AJ. Shear stress-induced endothelial cell polarization is mediated by Rho and Rac but not Cdc42 or PI 3-kinases. The Journal of Cell Biology. 2003;161:429–439. doi: 10.1083/jcb.200210135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MH, Rubinfeld B, Gordon JI. Effects of forced expression of an NH2-terminal truncated beta-Catenin on mouse intestinal epithelial homeostasis. The Journal of Cell Biology. 1998;141:765–777. doi: 10.1083/jcb.141.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead GJ, Mutch CA, Olson EC, Chenn A. Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. The Journal of Neuroscience. 2006;26:12620–12630. doi: 10.1523/JNEUROSCI.3180-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nature Cell Biology. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Huang H, Garcia Abreu J, He X. Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One. 2009;4:e4926. doi: 10.1371/journal.pone.0004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Mlodzik M. A quest for the mechanism regulating global planar cell polarity of tissues. Trends in Cell Biology. 2009;19:295–305. doi: 10.1016/j.tcb.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Wei Y, Sennino B, Ulsamer A, Kwan I, Brumwell AN, et al. Identification of pY654-beta-catenin as a critical co-factor in hypoxia-inducible factor-1alpha signaling and tumor responses to hypoxia. Oncogene. 2013;32:5048–5057. doi: 10.1038/onc.2012.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Garner J, Buckley KM, Vincent PA, Chiasson CM, Dejana E, et al. p120-catenin regulates clathrin-dependent endocytosis of VE-cadherin. Molecular Biology of the Cell. 2005;16:5141–5151. doi: 10.1091/mbc.E05-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Craig AW, Greer P, Miller M, Anastasiadis PZ, Lilien J, et al. Continuous association of cadherin with beta-catenin requires the non-receptor tyrosinekinase Fer. Journal of Cell Science. 2004;117:3207–3219. doi: 10.1242/jcs.01174. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. The EMBO Journal. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y, Sakurai T, Tanaka H, Kitagawa K, Colman DR, Shan W. N-cadherin mediates interaction between precursor cells in the subventricular zone and regulates further differentiation. Journal of Neuroscience Research. 2009;87:3331–3342. doi: 10.1002/jnr.22044. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, Irie Y, et al. Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. The Journal of Biological Chemistry. 1999;274:19473–19479. doi: 10.1074/jbc.274.27.19473. [DOI] [PubMed] [Google Scholar]
- Yan HX, Yang W, Zhang R, Chen L, Tang L, Zhai B, et al. Proteintyrosine phosphatase PCP-2 inhibits beta-catenin signaling and increases E-cadherindependent cell adhesion. The Journal of Biological Chemistry. 2006;281:15423–15433. doi: 10.1074/jbc.M602607200. [DOI] [PubMed] [Google Scholar]
- Yanagawa S, Lee JS, Haruna T, Oda H, Uemura T, Takeichi M, et al. Accumulation of Armadillo induced by Wingless, Dishevelled, and dominant-negative Zeste-White 3 leads to elevated DE-cadherin in Drosophila clone 8 wing disc cells. The Journal of Biological Chemistry. 1997;272:25243–25251. doi: 10.1074/jbc.272.40.25243. [DOI] [PubMed] [Google Scholar]
- Yang CH, Axelrod JD, Simon MA. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell. 2002;108:675–688. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, et al. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zantek ND, Azimi M, Fedor-Chaiken M, Wang B, Brackenbury R, Kinch MS. E-Cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth & Differentiation. 1999;10:629–638. [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hartmann H, Do VM, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, et al. Destabilization of beta-catenin by mutations in presenilin-1 potentiates neuronal apoptosis. Nature. 1998;395:698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shemezis JR, McQuinn ER, Wang J, Sverdlov M, Chenn A. AKT activation by N-cadherin regulates beta-catenin signaling and neuronal differentiation during cortical development. Neural Development. 2013;8:7. doi: 10.1186/1749-8104-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Woodhead GJ, Swaminathan SK, Noles SR, McQuinn ER, Pisarek AJ, et al. Cortical neural precursors inhibit their own differentiation via N-cadherin maintenance of beta-catenin signaling. Developmental Cell. 2010;18:472–479. doi: 10.1016/j.devcel.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Goffinet AM, Tissir F. Role of the cadherin Celsr3 in the connectivity of the cerebral cortex. Médecine Sciences: M/S. 2008;24:1025–1027. doi: 10.1051/medsci/200824121025. [DOI] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze’ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Molecular and Cellular Biology. 2000;20:4238–4252. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng GQ, et al. Activation of EGFR promotes squamous carcinoma SCC10A cell migration and invasion via inducing EMT-like phenotype change and MMP-9-mediated degradation of E-cadherin. Journal of Cellular Biochemistry. 2011;112:2508–2517. doi: 10.1002/jcb.23175. [DOI] [PubMed] [Google Scholar]