Abstract
Titanium (Ti) and its alloys are among the most successful implantable materials for dental and orthopedic applications. The combination of excellent mechanical and corrosion resistance properties makes them highly desirable as endosseous implants that can withstand a demanding biomechanical environment. Yet, the success of the implant depends on its osteointegration, which is modulated by the biological reactions occurring at the interface of the implant. A recent development for improving biological responses on the Ti-implant surface has been the realization that bifunctional peptides can impart material binding specificity not only because of their molecular recognition of the inorganic material surface, but also through their self-assembly and ease of biological conjugation properties. To assess peptide-based functionalization on bioactivity, the present authors generated a set of peptides for implant-grade Ti, using cell surface display methods. Out of 60 unique peptides selected by this method, two of the strongest titanium binding peptides, TiBP1 and TiBP2, were further characterized for molecular structure and adsorption properties. These two peptides demonstrated unique, but similar molecular conformations different from that of a weak binder peptide, TiBP60. Adsorption measurements on a Ti surface revealed that their disassociation constants were 15-fold less than TiBP60. Their flexible and modular use in biological surface functionalization were demonstrated by conjugating them with an integrin recognizing peptide motif, RGDS. The functionalization of the Ti surface by the selected peptides significantly enhanced the bioactivity of osteoblast and fibroblast cells on implant-grade materials.
Keywords: Implants, Titanium binding peptide, Molecular recognition, Biomaterial interface, Bioenabled surface modification
1. Introduction
Titanium (Ti) and Ti alloys have become some of the most widely used medical materials, particularly as orthopedic and dental implants. The combined properties of relatively low density, high toughness, strength, elastic modulus and excellent corrosion resistance make Ti alloys good candidates as a biomaterial to replace bone tissue or the roots of teeth without compromising functional performance. Over the past decades, there has been much improvement in medical-grade Ti and its alloys, with a focus on the metal surface intended to increase their biocompatibility and service life [1–3]. The surface topography and chemistry-related surface properties have been investigated frequently in order to enhance bioactivity [1,2]. Modifying the surface topography through roughening by means of grinding, sand blasting, etching or some combination thereof to a root mean squared (rms) roughness, measured in micrometers and nanometers, have generally resulted in surfaces that are more favorable for cell adhesion [2,4–26]. Also, coating the implant surfaces with calcium phosphate, in the form of tricalcium phosphate or hydroxyapatite (HAP) has been widely used [7–12]. A major concern for these ceramic coatings has been that the bonding strength between the coating and the metal is typically not strong and often results in cracking and spallation, therefore limiting their broad use in clinical settings [13,14].
Immobilization of bioactive molecules on implant surfaces has also been widely studied as a means of enhancing surface biocompatibility. Among bioactive molecules, extracellular matrix (ECM) proteins or synthetic peptides derived from native matrix proteins have been demonstrated as potential molecules to control cell response on implant surfaces. A short peptide fragment derived from the ECM protein fibronectin, arginine–glycine–aspartic acid (RGD) has been used widely, owing to its capacity to stimulate cell adhesion through activation of the integrin receptors. The first examples of synthetic peptide immobilization on Ti surfaces using silane chemistry were investigated by Xiao and colleagues [15,16]. The peptide composed of Arg–Gly–Asp–Cys (RGDC) sequence was covalently attached to the Ti surface using 3-amino-propyltriethoxylsilane and N-succinimidyl-3-maleimidopropionate [16,17]. Although these strategies improved the biocompatibility of Ti implant surfaces at various levels, they have limitations, owing to their need for an extensive chemical process or their use of organic solvents, as well as their low coupling efficiencies with bioactive molecules, which led to diminished activity and stability of the bioactive moiety. Moreover, the low material selectivity properties of these coupling agents restrict their use to a limited variety of materials. Furthermore, the behavior and stability of the modified surface under physiological conditions are not well understood [18,19]. Therefore, controlling the inorganic surface properties remains a challenge to induce enhanced interaction at the biomaterial interface, despite the availability of established biofunctional molecules, e.g., arginine–glycine–aspartic acid (RGD), polyethylene glycol (PEG) and bone morphogenetic protein 2 [20–25]. As a result, in order to advance the surface functionalization, techniques targeting the controlled biomaterial interface with material selectivity and coupling flexibility will positively impact the functional performance of implant materials.
During the last decade, inorganic binding peptides possessing affinity and specificity to select inorganic surfaces have been generated using phage and cell surface display techniques [26,27]. Their potential use was increasingly demonstrated in various scientific disciplines including surface functionalization [28–32], biomineralization [33,34], tissue engineering [31] and regenerative medicine [35,36]. Application of surface-binding peptides to hard tissue regeneration and restorative medicine is particularly intriguing and, consequently, there has been a great deal of interest in identifying and characterizing peptides that bind to various materials, such as TiO2 [37], Au [38,39], SiO2 [40,41], HAP [33,42,43] and select polymers [44]. Recently, several research groups selected peptides that are specific to Ti/TiOx surfaces, using phage display techniques. Among them, Sano et al. reported their experience with Ti binding peptides (TiBP) and investigated the effect of specific residues to contribute to binding by mutating amino acids within the oligopeptide [37]. Vreuls et al. identified selected peptide binding to amorphous TiO2 plasma-vapor-deposition coating on a stainless steel substrate using phage display technology [45]. The potential of peptide-based functionalization is undoubtedly appealing for implant materials because of the added return on investment provided by the biological relevance of the peptide to contribute to molecular recognition, self-organization and ease of conjugation with other biomolecules for enhanced functionality. Commercially pure (cp) Ti implants have been commonly used in restorative dentistry and orthopedics. These are available in four different grades, depending on the amount of interstitial elements in the Ti lattice and, hence, the compression of the Ti structure [46]. Among them, cp Grade 4 Ti, with the most interstitial concentration, exhibits the highest strength (~550 MPa tensile strength, ~250 MPa Vickers hardness). However cp Grade 1 Ti with the lowest yield strength (240 MPa tensile strength, ~120 MPa Vickers hardness) has better formability compared with those with higher interstitial content [46].
In the present study, it was hypothesized that TiBP generated by cell surface display on implant-grade Ti are capable of binding to Ti with high affinity, and they can be conjugated to other bioactive molecules without losing their surface-related function, resulting in the enhancement of the bioactivity obtained on the Ti surface. To test this hypothesis, peptides for cp Grade 4 Ti were selected using a FliTrx cell surface display library. Each of the selected TiBP was then characterized semi-quantitatively via fluorescence microscopy (FM) for affinity to their respective Ti surface. Peptides chosen because of their strong (TiBP1 and TiBP2) or weak (TiBP60) affinities were further subjected to quantitative binding characterization via quartz crystal microbalance (QCM), as well as analysis of their molecular conformation properties by protein structure interrogation techniques. The selected subset of TiBP was then examined for cytotoxicity. Building upon their affinity for the implant-grade Ti surface and their favorable cytotoxicity analysis, the TiBP modularity was evaluated by conjugating them with an integrin recognition sequence, Arg–Gly–Asp–Ser (RGDS). It was demonstrated that the conjugated TiBP–RGDS not only retain their surface binding affinity, but also gain enhanced bioactivity to osteoblast and fibroblast cells. Bioenabled implant surface functionalization using peptide-based linkers provides a platform for integrating the modular domains for multifunctional probe attachment on the surfaces in a single-step process under biologically relevant environments.
2. Materials and methods
2.1. Implant-grade Ti preparation
Implant-grade Ti sheets of cp Grade 1 and cp Grade 4 (A.D. Mac-Kay, Red Hook, New York) were prepared as disks 19 mm in diameter. These disks were used to mimic an implant surface. Surface roughness was achieved through hand polishing with a 600-grit finish silicon carbide metallurgical paper [47]. Disks were cleaned by first soaking in 1% sodium dodecyl sulfate (SDS) buffer overnight, followed by sonication for 1 h. Disks were washed in double-distilled water four times before and after passivation in 30% nitric acid for 1 h. Disks were sterilized by 15 min irradiation to each surface with UV light prior to cell culture experiments [47].
2.2. Surface characterization
Surface properties of cp Grades 1 and 4 Ti disks were determined using atomic force microscopy (AFM) and scanning electron microscopy (SEM). The rms of the prepared Ti disks was examined using a Nanoscope III/MMAFM system (Veeco Instruments, Santa Barbara, CA). Scans of size 30 × 30 μm were made in contact mode using standard SiN AFM tips with a nominal tip radius of 20 nm at a scan resolution of 512 × 512 pixels, or 60 nm pixel−1. The rms values were calculated from these scans using software provided by the manufacturer. The gross surface morphology of the prepared disks was examined using a JSM 7000 SEM (JEOL USA, Peabody, MA) at 10 keV by a LaB6 filament.
2.3. Cell surface display
The FliTrx bacterial cell surface display system [48] (Invitrogen, Carlsbad, CA) was used to select peptide sequences against the cp Grade 4 Ti surface [49,50]. Four selection rounds were applied in the biopanning process for Ti binding clone enrichment, and the DNA nucleotide sequence analysis of each of the 60 clones was performed (Fig. S1). The binding affinities of the isolated clones were further characterized by quantitative fluorescent microscopy employing a Nikon Eclipse TE-2000U fluorescent microscope (Melville, NY) equipped with a Hamamatsu ORCA-ER cooled CCD camera (Bridgewater, NJ), imaged using a FITC filter (exciter 460–500 nm, dichroic 505 nm, emitter 510–560 nm) and METAMORPH software (Universal Imaging, USA). The affinity level for each clone was established by calculating the average number of adherent cells on the Ti surface in triplicate samples. Consequently, the TiBP were grouped as strong, moderate or weak binders, according to their binding level.
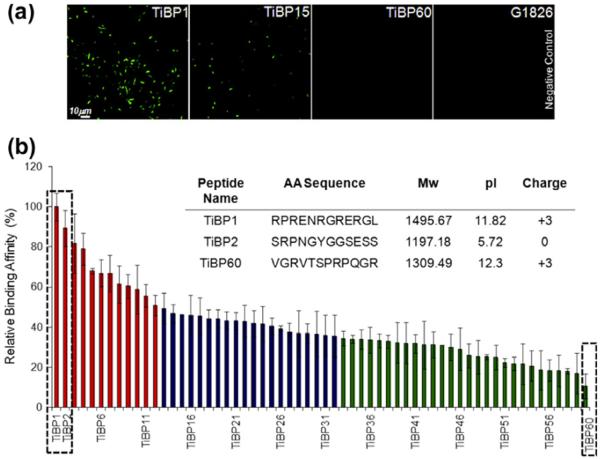
Fig. 1.
Ti binding peptides (TiBP) selected by cell surface display: (a) examples of FM images of TiBP with various binding affinities; (b) categorization of the Ti binding clones based on relative binding affinity analysis via FM. (b inset) Amino acid (AA) sequence and physicochemical properties of TiBP1, TiBP2 and TiBP60 (Mw: Molecular Weight, pI: Isoelectric point).
2.4. Peptide synthesis
A standard solid phase peptide synthesis technique was performed on Wang resin (Novabiochem, San Diego, CA) using F-moc chemistry. A CSBio 336S automated peptide synthesizer (C S Bio Co., Menlo Park, CA) with HBTU activation was used for the synthesis. The resulting resin-bound peptides were cleaved and side chain de-protected using Reagent K (TFA/thioanisole/H2O/phenol/ethanedithiol, relative ratio: 87.5:5:5:2.5) and precipitated by cold ether. The crude peptides obtained were purified by reversed-phase high-performance liquid chromatography at > 98% purity (Gemini 10u C18 110A column). The purified peptides were checked by mass spectroscopy, using a MALDI-TOF mass spectrometer (Bruker Daltonics, Billerica, MA) A representative chromatogram and the mass spectrum of the resultant pure peptide can be found in the Supplementary Data (Figs. S3–S5).
Bifunctional peptides were designed by conjugating the TiBP with the integrin binding domain, RGDS, via a three peptide flexible linker consisting of the amino acid glycine.
2.5. Circular dichroism spectroscopy
A solution containing 30 μM peptide, 100 mM Tris–HCl at pH 7.4 and varying volumes of 2,2,2-trifluoroethanol (TFE) (99.8% purity, Acros Organics, Pittsburgh, PA) was prepared for circular dichroic analysis. The spectrum, an average of eight scans from 185 to 260 nm, with a scan rate of 0.5 nm s−1, was collected at 20 °C for each sample, using an AVIV Stopped Flow 202SF circular dichroism (CD) Spectropolarimeter (Lakewood, NJ, USA). Appropriate background buffer subtraction was performed on each sample. The CD instrument was carefully calibrated using (1S)-(+)-10-camphorsulfonic acid (99%, Sigma–Aldrich, St. Louis, MO). Ellipticity is reported as mean residue ellipticity (M (deg cm2 dmol−1). All spectral smoothing (Savitzky–Golay algorithm) was executed using commercial graphing software (IGOR Pro. 6.0). For secondary structure estimation, a section of the smoothed spectrum (from 190 to 240 nm) was compared with a five-component reference spectra database [(1) alpha helix, (2) beta sheet, (3) beta turn, Type-I, (4) beta turn, Type-II, and (5) random coil] using a constrained least-squares fit [51]. The standard spectra do not consider any aromatic or disulfide dichroic contributions, since the peptides analyzed do not contain significant non-structural features (TiBP2 is the only peptide with an aromatic residue, Y). The secondary structure estimates are reported as the fractional weight ± the standard deviation.
2.6. Molecular modeling studies
Three different TiBP were modeled, TiBP1 and TiBP2 as strong binders and TiBP60 as a weak binder. To model these three peptides, linear forms were built using molecular modeling software (Hyperchem 7.5, Gainesville, FL). The energy minimizations of these peptides were carried out under implicit solvent conditions to reduce the otherwise huge computation time required to model individual water molecules. To analyze the energy landscape and thus to detect the minimum-energy structures, the chosen dihedral angles were first varied randomly, and then these newly formed initial peptide structures were minimized using the Polak–Ribiere conjugate gradient method. This process was reiterated until convergence of the gradient (0.01 kJ mol−1) was achieved using the CHARMM 27 force field [52]. In each round of energy minimization, unique low-energy conformations were stored, and high-energy and duplicate structures were discarded. Using the conformational search module, 1000 different local minima on the potential energy surface were found, and the lowest one was chosen as the global minimum or the lowest-energy conformation [53,54]. The lowest energy conformations were next solvated with TiP3P water explicitly and, finally, the overall system was energy minimized using the Polak–Ribiere conjugate gradient method by means of the convergence criterion and the force field parameters, as previously provided [52]. The final configurations and the corresponding Ramachandran plots were generated using VMD (Visual Molecular Dynamics) software (NIH) [55].
2.7. Binding affinity measurement
A QCM system was used to quantify the binding strength of the three Ti-binding peptides (TiBP1, TiBP2 and TiBP60) on the Ti surface. Five-megahertz quartz crystals (Q-Sense, Linthicum, MD) were coated with 25 nm of Ti film formed, via physical vapor deposition, and the coated crystals were used in a KSV QCM-D Z500 parallel flow system (Instrument Ltd, Helsinki, Finland), which monitors frequency change over time. Phosphate buffered saline (PBS) at various concentrations was used to dilute the peptides and to introduce the peptide to the crystal surface within a flow cell. Fluid flow was stopped to allow the peptide in solution to bind to the surface until equilibrium was reached. Binding was observed via the frequency shift, a parameter that is related to the mass of the adsorbed peptide. To determine the dissociation constant (Kd) of each peptide on Ti, the equilibrium frequency shift caused by peptide binding was measured at several concentrations. The resulting values were fit using the Langmuir adsorption model. Peptide concentrations of 0.1–2 μM and 2–15 μM were used for all binders initially; they were then adjusted to be in a similar range according to the Kd value for each peptide. After testing, each Ti surface was cleaned using a surfactant solution (1% SDS, 1 N NaOH) and all the surfaces were ozone cleaned for 15 min before their reuse.
2.8. Cell culture
MC3T3-E1 preosteoblast (CRL-2593™, ATCC, Manassas, VA) and NIH3T3 (CRL-1658™, ATCC, Manassas, VA) were cultured in alpha minimum essential medium (MEM) supplemented with 10% fetal bovine serum, 2 mM glutamine and 1% antibiotic solution (all media ingredients used were from Gibco, Carlsbad, CA). Cells were incubated at 37 °C in air supplemented with 5% CO2 for 1 week. The cells were then enzymatically detached from the surface of the cell culture dishes using 0.25% trypsin–EDTA solution (Gibco, Carlsbad, CA) and collected by centrifugation (2000 rpm for 5 min).
2.9. Cytotoxicity assay (MTT assay)
The peptides were dissolved and diluted to 200 μM in 1 × PBS (Gibco, Carlsbad, CA), and 750 μl of each peptide solution was incubated on sterilized cp Grades 1 and 4, implant-grade Ti surfaces. As a control, each Ti surface was also incubated with 1X PBS for 4 h at 37 °C in 5% CO2. Following incubation, all Ti specimens were washed twice in 1X PBS to remove unbound peptides. A total of 2 ml of 8 × 105 cells ml−1 MC3T3-E1 cells were plated in each well of the 12-well plates with the Ti disks functionalized with either the TiBP or PBS (negative control). To avoid fibronectin arising from animal serum sources, the test cultures were maintained in serum-free alpha-MEM with 1% antibiotic solution at 37 °C in 5% CO2 for 24 h. Following incubation, 200 μl media was removed from each well, 200 μl of an MTT (Sigma–Aldrich, St. Louis, MO) reagent (5 mg ml−1) (1:10 ratio) was added to each well and incubated for 3 h. At the end of incubation time, purple formazan crystals were observed using an inverted light microscope with Kohler illumination. Each Ti surface was transferred to a clean well, and the formazan crystals were dissolved and incubated with 1.5 ml DMSO and 0.1 M glycine, NaCl pH 10.5 (10:1) ratio for 5 min. Absorbance of the converted formazan dye was measured at a wavelength of 570 nm with background subtraction. Relative cell viability (%) was calculated and compared based on the PBS control bare Ti surface area for each group. Experiments were repeated in triplicate using three different Ti disk samples.
2.10. Cell adhesion and spreading
The MC3T3-E1 cells (104 cells ml−1) in serum-free media were inoculated on cp Grade 1 and 4 Ti surfaces at 37 °C and maintained for 2 h, as described above. Following the incubation period, the cells were fixed in 500 ll of 2% glutaraldehyde (Ted Pella, Redding CA) solution in PBS for 20 min at room temperature and dehydrated with a series of increasing ethanol solutions (10–30–60–90–100%). The Ti disks with cells were rinsed twice with PBS and treated with a stock solution of Alexa Fluor488-Phalloidin (Invitrogen Co.) diluted prior to use to obtain a working solution of ~33 nM. Then, 500 μl ofthe final working solution was added to each sample and kept at room temperature away from light for 20 min. The samples were then rinsed twice with distilled water, dried under N2, and observed using a TE 300L microscope. Metamorph and Image J Software (NIH) was used to analyze cell number and cell spreading. Similar procedures were used for NIH3T3 cells plated on Ti-coated glass surfaces. All experiments were repeated with three unique samples, with data determined in triplicate for each sample.
3. Results
The cell surface display of peptides that bind to implant-grade Ti surfaces resulted in the selection of two high-affinity binding sequences, TiBP1 and TiBP2. These high-affinity peptides and a weak binding sequence, TiBP60, were analyzed in detail for subsequent structural properties, binding kinetics parameters and bioactivities. The results of the findings are discussed as follows.
3.1. Selection and characterization of TiBP
A cell surface display technique [48] was applied on cp Grade 4 Ti surfaces [47] to select peptides that could serve as potential biomolecular surface linkers to improve cell responses to these implant materials (Supplementary Data, Fig. S1a). Throughout the cell surface display selection, four successive rounds of biopanning were performed, resulting in 60 unique clones, which were then subjected to DNA sequence analysis (Fig. 1). Semi-quantitative initial binding analysis based on the FM technique was applied to assess the affinity level among the selected clones. The binding affinity level for each of the individual clones was obtained by incubating them with the Ti surface, staining them with Syto 9 dye, which binds to nucleic acids, and evaluating their numbers by imaging with fluorescent microscopy. The bound bacterial cells expressing Ti-binding sequences were visualized as uniformly distributed bright green rods on a dark background, as opposed to GI826 plasmid-free control cells (Fig. 1a), which fail to bind. Binding affinity of the selected peptide clones was estimated by enumerating the adhered cells over three random regions in triplicate experiments. Based on these results, all the identified peptides were successfully categorized as strong, moderate and weak binders (Fig. 1b). The role of amino acid distribution on binding affinity was explored next. To calculate the relative abundances of amino acids, the observed amino acid distribution among strong and weak binder groups was compared with respect to the native (unpanned) library peptide sequences [39]. Based on the amino acid distribution analysis, the presence of polar (Ser, Asn) and basic (Arg) amino acids were evident (Supplementary Data, Fig. S2).
3.2. Conformational properties of TiBP
TiBP were synthesized by solid phase synthesis, and their conformational properties were investigated. First, the presence of ordered or disordered structure was assayed within each sequence and, second, the folding propensity of each sequence in the presence and absence of the structure-stabilizing solvent TFE was determined. As shown in Fig. 2a, at 0% TFE in aqueous media, each peptide exhibits a strong negative ellipticity band representing the pi–pi* transition [51,56]. For TiBP1 and TiBP2, this band is centered near 198 nm and is characteristic of random coil conformation in equilibrium with other secondary structures such as alpha-helix and beta turn that are featured in these sequences (Table S1) [33,39,51,56,57]. The presence of beta turn structures may arise from the Arg, Gly and Ser residues in TiBP1 and TiBP2 that would promote turn or loop-like regions in either sequence [51,56,58,59]. In addition, TiBP2 possesses a second, slightly positive ellipticity band centered near 218 nm, representing the n–pi* transition. Under the same conditions, the weak binding TiBP60 peptide sample features a pi–pi* band centered near 200 nm, and this reflects a shift away from random coil conformation towards other secondary structures. As shown in Table S1, TiBP60 possess more beta strand structure and reduced beta type II content compared with either TiBP1 or TiBP2, and this may be due to the presence of an extended beta strand forming a tetramer amino acid sequence cluster, –PRPQ–[60], located near the middle of the TiBP60 sequence. Thus, the –PRPQ tetramer sequence found in TiBP60 is not found in the collapsed random coil/beta turn structures of TiBP1 or TiBP2. From these data, it is concluded that each TiBP exhibits intrinsic disorder, with TiBP1 and TiBP2 adopting a combination of coil, turn or loop structures in solution, whereas TiBP60 adopts a more extended conformation under the same conditions. With the addition of TFE, it is noted that, for each peptide, the pi–pi* transition ellipticity band is reduced in intensity as a function of TFE addition and undergoes a 7–8 nm red shift in absorption wavelength. This red shift is indicative of a shift in secondary structure population away from a random coil towards other secondary structures. Thus, like other intrinsically disordered inorganic binding sequences, each TiBP undergoes some degree of conformational reordering in the presence of TFE. This fact is reflected in Table S1, where the percentage of random coil structure is observed to decrease for each peptide at 75% v/v TFE compared with 0% v/v TFE. In addition, it is noted that TiBP60 exhibits an isochromic transition point [51] centered at 215 nm, and this is not observed for either TiBP1 or TiBP2. From this transition point, it is inferred that the conformational transition for TiBP60 differs from that of TiBP1 and TiBP2, and this difference may arise from the presence of the extended –PRPQ– sequence in TiBP60.
Fig. 2.
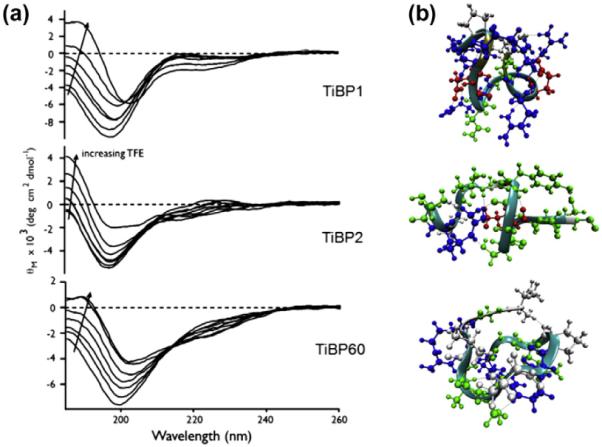
(a) CD spectra of 30 μM TiBP (TiBP1, TiBP2, TiBP60) in the presence of varying volume percentages of TFE in 100 mM Tris–HCl, pH 7.4. Arrow indicates increasing TFE concentration (0, 10, 20, 30, 40, 50, 75%). (b) Overlapped ribbon and CPK models of the predicted structures of the TiBP1, TiBP2 and TiBP60 (from top to bottom respectively). The residues are colored according to residue type (basic, blue; acidic, red; polar, green; non-polar, gray). The backbone is colored according to secondary structure (turn, cyan; random coil, gray; isolated bridge, tan). Water molecules are omitted for clarity.
The structural models of the three different Ti-binding peptides, two of them strong (TiBP1 and TiBP2) and one of them weak (TiBP60), were obtained using CHARMM force field parameters for the peptide and TIP3P parameters for the solvent. All three peptides exhibit mainly turn and random coil conformations, lacking regular secondary structure elements such as an alpha helix or a beta sheet (Fig. 2b).
3.3. Adsorption behavior of TiBP on Ti surfaces
The adsorption behavior of TiBP1, TiBP2 and TiBP60 was quantified via QCM by measuring the maximum vibration frequency shift (△Fmax) as a consequence of peptide binding. By measuring △Fmax at several peptide concentrations, two types of binding data, the disassociation constant (Kd) and free energy of adsorption (△Gads), were obtained by the following relationships based on the Langmuir adsorption model:
| (1) |
where △Fmax is the frequency shift when the surface is saturated, and C is the concentration of the bulk solution. The unknown constants, △Fmax and Kd, were fit to the data using a least squares regression. The dissociation constant can then be related to the free energy of adsorption by the following relationship:
| (2) |
where Kd values represent the concentration necessary to achieve 50% surface coverage on a given surface. While not intended to account for the absolute absorbed dry mass [61–66], these calculated Kd and △Gads values serve the purpose of comparison, e.g., the lower the Kd value, the more strongly the peptide binds to that surface. It was also assumed that the peptide films were rigid, resulting in a proportional frequency shift to the overall film mass, and that there was no significant difference between the wet and dry film masses, which were reasonable, given the small size of the peptide [66]. TiBP1, TiBP2 and TiBP60 showed vast differences in their binding affinity to Ti-coated quartz crystals. In the case of TiBP1 and TiBP2, both of which showed strong binding by FM analysis, the peptide saturated the surface at relatively low concentrations (Fig. 3), yielding Kd values of 0.90 μM and 018 μM, respectively, as shown in the inset in Fig. 3. Both these peptides can be characterized as strong binders from these Kd values compared with other TiBP in the literature [37]. However, TiBP60 showed much weaker binding, resulting in a Kd value of 14.76 μM (Fig. 3 inset), confirming the results obtained from FM analysis (Fig. 1). TiBP60 requires a ~100-fold greater concentration in solution to achieve the same surface coverage as TiBP2 (Fig. 3). There is a significant difference in the binding strengths between the two strong binding peptides. TiBP2 showed a fivefold lower Kd value than TiBP1. The free energy of adsorption (△Gads) values (Fig. 3 inset), however, did not display such significant differences. Here, TiBP60 yielded greater binding energy (–6.59 ± 0.12 kcal mol−1) compared with the value calculated for TiBP1 or TiBP2 (−8.25 ± 0.08,−9.19 ± 0.11 kcal mol−1, respectively).
Fig. 3.
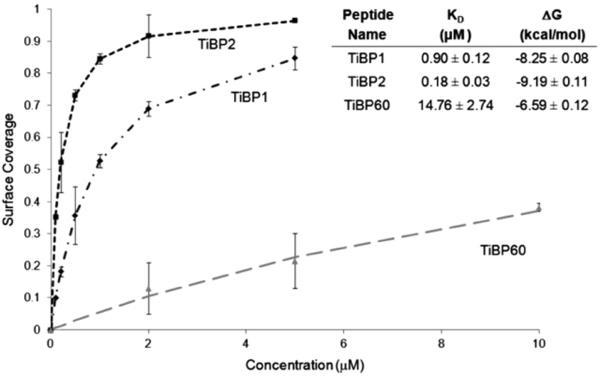
Surface coverage and observed Kd and ΔG values (inset) of TiBP1, TiBP2 and TiBP60 determined using the QCM with on Ti-coated quartz crystal.
3.4. Surface topography of Ti surfaces
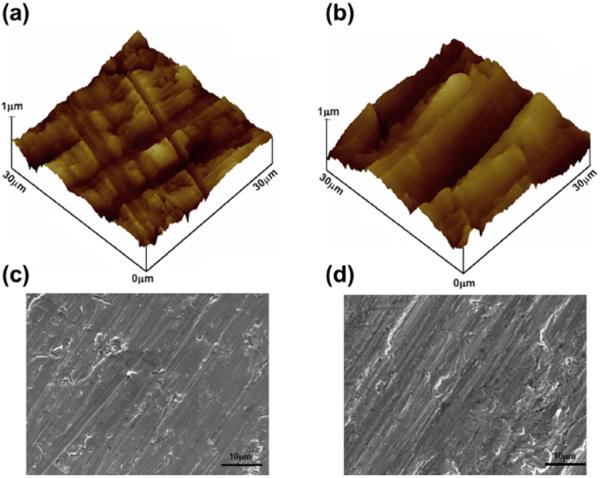
The biocompatibility of a biomaterial is closely related to the cellular functions it elicits, particularly cell viability and adhesion, i.e., cell properties that are important during the early stage of cell-surface interactions [67]. This interaction is limited and dependent on the surface characteristics of the materials, such as topography, chemistry and surface energy. Both material composition and surface morphology influence protein adsorption and, therefore, the resulting adhesion and proliferation of cells. First, the surface roughness of implant-grade Ti (cp Grades 1 and 4) was examined by AFM imaging. The data revealed that the Grade 4 surface was relatively smoother than the Grade 1 surface, the rms roughness value being 170 nm with a peak to valley range of 1.6 μm, and 300 nm with peak to valley range of 2.6μm for cp Grade 4 and 1 surfaces, respectively. While both cp Grades 1 and 4 Ti were chemically similar and both were polished with 600 grit SiC paper, the roughness difference is attributed to the result of the hardness difference between the two grades. A depiction of the different surface groove depths is shown by the 30×30 μm cut-out of the AFM scans in three-dimensional (3D) view shown in Fig. 4a and b. From the analyses of SEM images, the general view of the surface properties of these materials is evident. Grade 1 clearly reveals deeper grooves than those of Grade 4 material. Larger SEM scans revealed that the groove patterns are similar between the two grades of Ti surfaces, with Grade 4 having a smoother surface (Fig. 4c and d).
Fig. 4.
AFM scans in 3D view over a 30 × 30 μm region: (a) cp Grade 4 Ti; (b) cp Grade 1 Ti. SEM scans of Ti implant surfaces: (c) cp Grade 4 Ti; (d) cp Grade 1 Ti.
3.5. Potential cytotoxicity of the peptides under in vitro conditions
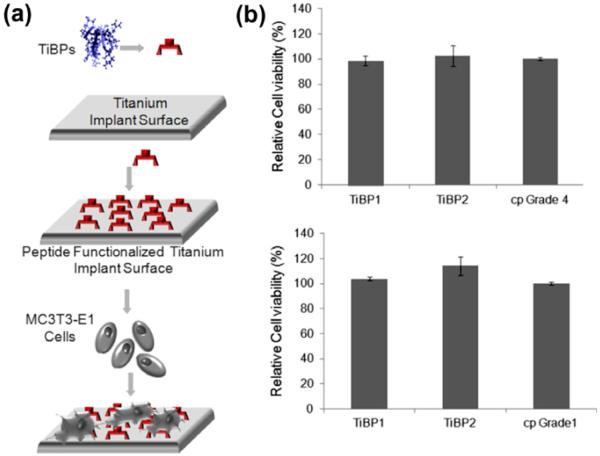
It was next evaluated whether there is any growth inhibition effect caused by the peptides under in vitro conditions. Cell viability was first tested on peptide-functionalized implant-grade Ti surface using an MTT assay. This is a colorimetric assay that measures the reduction of a yellow 3-(4,5-dimethythiazol-2-yl)-2,5-diphenly tetrazolium bromide enzymatically to dark purple colored formazan product by the metabolically active cells. Following peptide self-assembly on either of the two Ti surfaces, MC3T3-E1 cells were exposed to peptide modified surfaces to carry out the MTT assay (Fig. 5a). The results of the MTT assay demonstrate that cell viability and growth have not been affected by the presence of either TiBP1 or TiBP2 modified surfaces, compared with values obtained from control groups (Fig. 5b and c).
Fig. 5.
(a) Schematic representation of peptide functionalized implant surface in the presence of MC3T3-E1 cells for MTT assay. (b) TiBP1 and TiBP2 functionalized cp Grade 4 and (c) Grade 1 implant-grade Ti surfaces.
Next, assays were targeted to showing the effect that peptide-functionalized Ti surfaces exerted on cell adhesion. To test the versatility of the TiBP, the MC3T3-E1 osteoblast-like cells were exposed to peptide functionalized cp Grade 4 and 1 test surfaces including positive control groups. After 2 h of incubation, the cytoskeleton of adherent MC3T3-E1 was visualized with phalloidin labeling under FM. At 2 h, in the control and experimental groups the number of cells on both surfaces did not differ significantly (Fig. 6a and b), but the cells were observed to spread more on the peptide-coated Ti surfaces (Fig. 6a). Significantly more cells had extended their surface contacts on cp Grade 4 than on Grade 1. However, the cells on cp Grade 1 were more elongated compared with Grade 4 surfaces. In the case of TiBP functionalized cp Grade 4 surfaces, the number of cells on control and peptide-functionalized surfaces was very similar. In contrast, the spreading of cells on peptide functionalized surfaces was significantly different from the control group. A slight increase in cell spreading was observed on the cp Grade 4 Ti surfaces modified with TiBP1 and TiBP2 (Fig. 6a and c, respectively). In case of TiBP functionalized cp Grade 1 surfaces, there was no significant change in cell number or spreading among control and peptide-functionalized surfaces.
Fig. 6.
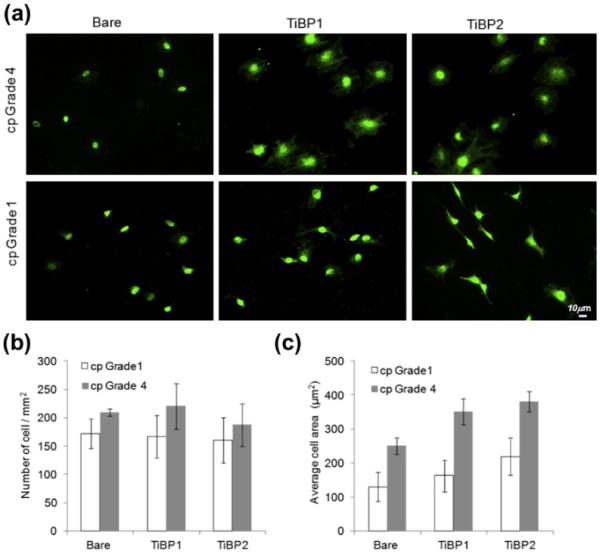
(a) FM of phalloidin stained MC3T3-E1 cells attached to cp Grade 1 and Grade 4 Ti surfaces. (b) The number of adherent MC3T3-E1 cells per mm2 in serum-free conditions on cp Grade 1 and Grade 4 Ti surfaces. (c) Average cell spread on cp Grade 1 and Grade 4 Ti surfaces.
3.6. TiBP–RGDS mediated bioactive surface modifications
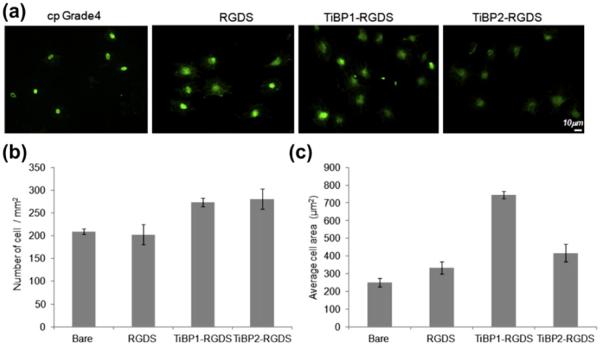
To demonstrate the modularity and flexibility of TiBP as molecular surface linkers, a bifunctional peptide was designed by conjugating TiBP with an oligopeptide constituting an integrin binding RGDS domain via a peptide linker consisting of three repeated glycine residues. The effect of the combined TiBP–RGDS on cell viability, adhesion and spreading was examined on cp Grade 4 Ti surfaces as well as Ti-coated glass surfaces in the presence of MC3T3-E1 osteoblast-like and fibroblast cells, respectively. First, the efficiency TiBP1–RGDS and TiBP2–RGDS on cell adhesion and spreading was explored on the implant-grade Ti surfaces that included negative (bare surface) and positive control (RGDS) in the presence of MC3T3-E1 (Fig. 7). A 1.5-fold difference was observed in the adherent cell number among the two bifunctional peptides functionalized surfaces (TiBP1–RGDS and TiBP2–RGDS), with TiBP1–RGDS showing the enhanced binding. Negative (bare) and positive control groups (RGDS) are shown in FM micrographs, and their response is plotted in graphic form (Fig. 7a, b). A significant difference in cell spreading behavior was also observed among TiBP1–RGDS vs. TiBP2–RGDS with respect to all control groups, as shown in Fig. 7c. Although both of the peptides resulted in better cell spreading compared with controls, TiBP1–RGDS resulted in an approximately threefold greater cell-spreading compared with TiBP2–RGDS on the surface (Fig. 7c).
Fig. 7.
(a) FM micrographs of phalloidin stained MC3T3-E1 cells on RGDS, TiBP1–RGDS and TiBP2–RGDS-treated cp Grade 4 Ti surfaces. (b) The number of adhered MC3T3-E1 cells mm−2 in serum-free conditions on control and peptide-treated surfaces. (c) Average cell spreading on control and peptide-treated surfaces.
Next, a Ti-coated glass substrate was used as the test surface to be functionalized with TiBP1–RGDS. The peptide's effect on cell adhesion and spreading in the presence of NIH3T3 mouse fibroblast was examined, compared with both negative control (bare surface) and positive control (RGDS) surfaces. The adhesion and spreading of the NIH3T3 cells on Ti surfaces were increased 3.5- 5-fold for TiBP1–RGDS functionalized surfaces compared with unmodified negative controls (Fig. 8a, b). No significant change was observed for cell adhesion on the surfaces incubated with RGDS alone or with TiBP1 peptide alone. This outcome was expected, because the role of TiBP alone on promoting cell adhesion and spreading is minor, since the residues promoting specific and avid binding to a material surface are predicted to provide little or no improvement in cell binding. Similarly, the integrin binding domain, i.e., RGDS alone, is not competent to provide an interface that promotes binding and spreading, since the integrin receptor ligand is not immobilized to the surface, but rather it is only passively absorbed. Cell adhesion and spreading was significantly enhanced on the Ti surface coated with the bifunctional peptide relative to surfaces of cp Grade 4 Ti and Ti thin-film treated with RGDS alone. These results suggest that the structural conformation of the peptide may allow the RGDS domain to be freely exposed, an outcome resulting in better cell adhesion and spreading. Also peptide charge and hydrophilicity may affect surface chemistry that can alter cell behavior. Surface micro- and nanotopography may have an effect not only on cell attachment and spreading, but also on peptide binding and availability of RGDS domain. Cell adhesion and spreading assays demonstrated that TiBP1–RGDS preserved its dual functionality effectively, with one peptide domain facilitating binding to the Ti surface, while the other peptide domain retained its function serving as a recognition site for cells.
Fig. 8.
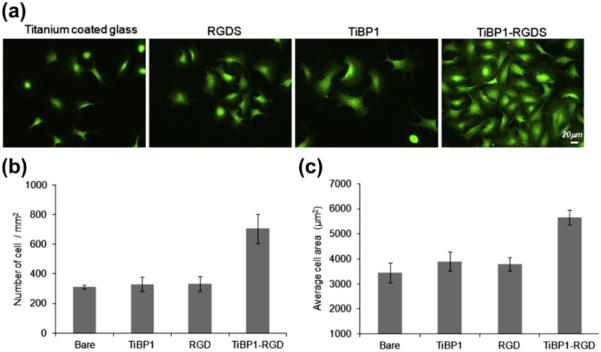
(a) FM micrographs of phalloidin stained NIH3T3 cells on RGDS, TiBP1–RGDS-treated glass-coated Ti surfaces. (b) The number of adhered NIH3T3 cells mm−2 in serum-free conditions on control and peptide-treated surfaces. (c) Average cell spreading mm−2 on control and peptide-treated surfaces.
4. Discussion
In this study, a series of Ti binding peptides was developed by a combinatorial cell surface display library. It demonstrated how these peptides could be converted to a bifunctional peptide by linking it to a bioactive molecule, RGDS, to enhance biocompatibility on the Ti surface for potential implant applications. In addition to selecting the Ti binding peptides and demonstrating their biocompatibility, extensive binding and structural/conformational characterizations were performed to elucidate the binding mechanisms operating for these peptides.
4.1. Peptide structure and binding affinity
Following the peptide selection for Ti surfaces, the physicochemical properties of the peptides were analyzed further, to gain additional insight into the mechanism of peptide binding on Ti surfaces. It was found that the presence of polar (Ser, Asn) and basic (Arg) amino acids were prominent in the selected peptides. The role of amino acid composition vs. amino acid positioning in the sequence and the role of peptide net charge vs. local charge distribution have been studied by different research groups, including the present authors, to investigate the binding mechanism of peptides to the desired surfaces [32,37,42,56,57,68,69]. A number of related probing studies in the literature focused on the interactions of peptides with metal, metal-oxide and mineral surfaces based on amino acid type and position relations [37,68,69]. For example, the HAP mineral binding sequences are considered to be rich in acidic residues (aspartate, glutamate and serine) resulting in a net negative charge that promotes binding to positively charged calcium atoms in the apatite crystal faces [32,42,43]. In the case of metal-oxide (e.g., Ti with native oxide surface and silica), peptides enriched with charged and polar amino acid residues (Lys, Arg, His, Asp and Glu) have been claimed to have high affinity with the negatively charged metal-oxide surfaces, probably because of their positive charge in an aqueous environment at neutral pH [37,68,69]. The observed abundance of Ser is remarkable among the strong binders of the selected peptides compared with His, Lys and Arg, which were considered as playing a role for peptide binding to metal-oxide surfaces in earlier studies. However, the abundance of Asp and Glu residues among the peptides is similar to the distribution described in previous studies (SupplementaryData, Fig. S2). While there are various peptide sequences with affinity for metal, metal-oxide and mineral surfaces, it remains a challenge to identify a clear contribution for each residue alone [37,40,68–70]. However, the overall folding on the solids is starting to be considered one of the key factors playing a crucial role in binding and self-assembly. The target surface and solution properties as well as the biopanning technique applied, including the design of this process, are the additionally important key factors.
In the literature, peptide sequences were reported as being selected using phage display against various size particles of Ti oxide powders, bulk Ti oxide (anatase) and even metallic Ti particles with native surface oxide [37,68,70]. In general, during the selection of peptides, features of the phage or cell surface library, the choice of biopanning protocol and strategy may collectively contribute to the differences observed in amino acid distribution, peptide generation efficiency and random sequence diversity. As unique peptides are identified not only by binding different materials, but also by affinity to different types of similar materials, it may be possible that amino acid sequence information may be deciphered, and this knowledge could lead to the prediction of the molecular recognition ability of a given peptide towards a given inorganic surface. In this study, three different peptides, specifically TiBP1, TiBP2 and TiBP60, were characterized in detail. TiBP1 and TiBP2 have a similar high affinity, despite their distinctive physicochemical properties, but TiBP60 has the lowest affinity, despite exhibiting physicochemical properties similar to those of the higher affinity TiBP1 (Fig. 1b inset). This apparent paradox may arise from a lack of knowledge regarding the possible conformation of the molecular structure of the peptide on the given solid surface which then dictates the specific molecular interactions with that surface, and has yet to be fully deciphered. Indeed, several material-specific polypeptides exhibit some degree of intrinsic disorder or unfolded structure [33,39,56,57,71,72]. In addition, it has been shown that target- or environment-induced folding propensity [60,71] is another molecular characteristic that functionally distinguishes different inorganic binding protein sequences from one another [33,56]. In the study described here, this issue was further investigated by studying the peptide conformational properties.
The CD studies revealed a significant conformational differences between strong binding peptides (TiBP1 and TiBP2) and weak binding peptides (TiBP60), and this difference may be a result of the presence of the –PRPQ– sequence in TiBP60. These differences in folding propensity may reflect the ability of each disordered TiBP sequence to adapt to Ti surfaces and significantly contribute to their observed binding propensities. Additional structural analyses were also in agreement with the CD analysis and indicated that all three peptides exhibited some degree of intrinsic disorder or unfolded structure [33,39,56,57,71,72]. The plots shown in Fig. 2b reveal the structure of the peptides in water, which is omitted for clarity, and reflects the CD experiments in the absence of the structure-stabilizing solvent (TFE). These plots indicate that, TiBP1 and TiBP2 have slightly more compact structures than TiBP60, a finding which may explain the differences in conformational transition studied via CD experiments in TFE conditions.
Understanding the peptide binding mechanism on solid surfaces has been elusive for practical applications [29]. As a prelude, selected peptides that can mediate cell adhesion were tested as adsorbed or covalently attached domains on various biomaterial surfaces. However, these cell adhesive peptide sequences were not designed with preferential affinity towards a specific material surface. Weakly attached molecules can be displaced and redistributed by cells attempting to attach to the same surface. In this case, formation of focal adhesion that influences cell attachment and behavior will not occur. This demonstrates the need to design a peptide including both material surface binding specificity and cell adhesive domains, both acting at the interface cooperatively. To understand better the behavior of peptide assembly on implant material surfaces requires additional knowledge. Therefore, the present authors further explored the binding affinities of the selected peptides on implant-grade Ti surfaces, while also intending to identify the means of increasing their cell adhesion behavior.
Correlating with the FM, CD and structural analyses, the QCM data revealed that TiBP1 and TiBP2 displayed stronger binding affinities (lower Kd and ΔGads values) than TiBP60. While the Ti surface used for QCM experiments was not the same one used in peptide selection, i.e., cp Grade 4 Ti vs. >99.9% pure Ti, the aim here is to quantitatively measure the relative binding differences among the TiBP and elucidate their binding mechanisms in the light of their structural conformation. Since both surfaces contain amorphous oxides, it is not anticipated that the resultant native oxide would differ. The nature of the native Ti oxide is an underappreciated and understudied issue. However, the parameter that would influence the native oxide is the electrolyte. Since the same buffer was used in the present studies, one expects the native oxide to be the same for both Ti film and cp Ti. In the literature, the assumption is that peptide binding adsorption relies on the formation of a passivated oxide layer and charged group interactions contributed from amino acid residues. Identified peptides tend to exhibit significant enrichment in histidine residues and hydroxyl-containing residues with high cationic charge [40,70]. However, the trends observed by the present authors regarding the adsorption behavior of TiBP indicate that it is not only the consequence of electrostatic, hydrogen, hydrophobic and dipole interactions, or the combination of these forces, but rather a more complex mechanism for binding that lies beyond simple binding affinity, amino acid content and peptide net charge correlation properties that normally specify peptide inorganic interactions. The observed differences in binding affinity, as revealed by QCM measurements, may be attributed to the similarities in their molecular architecture, allowing conformational transition to play a critical role in the ability of these sequences to adapt to the Ti implant material surfaces.
4.2. Bioactivity assays
Osteoblast-like MC3T3-E1 cells were used to study cell activity on the surface of implant-grade TiBP modified surfaces. There was a difference in cell spreading on TiBP modified cp Grade 1 and cp Grade 4 Ti surfaces. With respect to cell behavior, the parameter of surface roughness is among the important factors to consider, in addition to the chemical and wetting properties of the surfaces [67,73]. In the present case, the peptides were selected on cp Grade 4 Ti only, although there are only minute differences in chemical impurity content between the cp Grade 1 and cp Grade 4 Ti materials. However, the influences of these other factors were probably small compared with a nearly twofold difference in roughness between the two grades of Ti. The less rough cp Grade 4 Ti surface revealed a moderate increase in cell attachment number and cell spreading when functionalized with TiBP peptides. However, the more deeply grooved cp Grade 1 Ti revealed little difference in cellular behavior between TiBP-coated surfaces and non-treated surfaces This was probably a result of the depths of the grooves in the present cp Grade 1 Ti substrates being too severe to realize the benefit of functionalization by the TiBP. Because the cp Grade 4 Ti revealed improvement in cellular activity when coated with TiBP1 and TiBP2, the experiments with cp Grade 4 surfaces were continued, where bifunctional TiBP was used for surface functionalization to determine whether the cellular bioactivity could be further enhanced. After conjugating the RGDS peptide domain with either the TiBP1 or TiBP2, it was found that the TiBP1–RGDS functionalized cp Grade 4 Ti surface outperformed that of the TiBP2–RGDS functionalized surface. Altogether, the results indicated that cell–implant material interactions can be enhanced using the simple, flexible and modular dual-functional peptide-based single-step procedure for effective surface functionalization.
5. Conclusion
Deciphering the code for the physicochemical and structural properties that permit short peptides to interact with affinity and specificity with various inorganic surfaces is an important research avenue requiring improvement so that the host response to various biomedical device surfaces, whether they are used for soft or hard tissue implantable devices, can be optimized. Here, the present authors demonstrated an empirical approach to identifying such peptides, using a strategy of biopanning for random peptides expressed by phage or bacteria to bind to a specific inorganic solid material surface. Specifically, cell surface display was used to screen a set of peptides that bind to implant-grade Ti, biomaterial in frequent use for dental and osseous implantable devices. Detailed quantitative analysis on the subset of the two strong binding peptides provided the affinity and concentration range for the peptides to be used as novel implant-grade surface functionalization molecules. While peptides were demonstrated as effective surface linkers under the biological activity, their use as modular domains in creating multifunctional peptides was also tested. Conjugating these peptides to integrin recognition peptide sequence RGDS resulted in a single molecule when coated on Ti surfaces in a single step, and it enhanced the bioactivity of RGDS in the presence of pre-osseoblast and fibroblast cells. This combination of techniques can be easily manipulated to provide high-throughput screening for a variety of biomaterial surfaces, and the resulting binding peptides can be coupled to domains active in directing cell behavior that can be used to restore the function of tissues lost from disease, injury and congenital anomaly.
Understanding the first principles that regulate short peptide binding specificity and affinity responses to implant materials leverages the approach of using surface functionalization through single-step applications of biologically active coating under environmentally friendly conditions. Such implant binding peptides are candidates to serve as biomolecular linkers to functionalize implant surfaces by providing modular domains that ease their conjugation to biologically active signaling molecules, while retaining their remarkable binding and selectivity to the solid substrate in the absence of cytotoxicity properties. Since material surface binding peptides can be conjugated with a variety of bioactive molecules that enhance cell attachment, cell proliferation, cellular spreading and other fundamental properties of cell behavior, the TiBP studied here offer a unique utility for tissue engineering studies. The proposed peptide-based surface coating is universal and can be applied to induce various desired biological activity on any implant material from dental to orthopedic applications, using an easily adaptable single-step biologically relevant set of conditions.
Supplementary Material
Acknowledgements
This research is supported by NIH 1R21AR062249–01, NSF-MRSEC Program through the University of Washington GEMSEC DMR 0520567, NSF-BioMaterial Program, NSF-IRES and TUBITAK/NSF joint project (TBAG 107T250), and Institute of Science and Technology via Graduate Thesis Supporting Fund at Istanbul Technical University (Project Number 34009), National Institute for Dental & Craniofacial Research Grant # DE 130045 (MLS). Portions of this work (CD) represent contribution number 68 from the Laboratory of Chemical Physics, New York University.
Appendix A. Figures with essential colour discrimination
Certain figures in this article, particularly Figs. 1, 2 and 4–8, are difficult to interpret in black and white. The full colour images can be found in the on-line version, at http://dx.doi.org/10.1016/j.actbio.2012.11.004.
Appendix B. Supplementary data
Detailed experimental procedures are provided in Supplementary Data on the peptide selection, binding characterization, amino acid distribution of all generated strong and weak TiBP, secondary structure distribution of TiBP1, 2 and 60 and the MALDI/TOF mass spectrum of synthesized Ti binding peptides.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.actbio.2012.11.004.
Footnotes
Part of the Special Issue “TMS 2012 Biological Materials”, guest-edited by Nima Rahbar.
References
- [1].Hanawa T. An overview of biofunctionalization of metals in Japan. J R Soc Interface. 2009;6(Suppl 3):S361–369. doi: 10.1098/rsif.2008.0427.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Variola F, Brunski JB, Orsini G, Tambasco de Oliveira P, Wazen R, Nanci A. Nanoscale surface modifications of medically relevant metals: state-of-the art and perspectives. Nanoscale. 2011;3:335–53. doi: 10.1039/c0nr00485e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Long M, Rack HJ. Titanium alloys in total joint replacement - a materials science perspective. Biomaterials. 1998;19:1621–39. doi: 10.1016/s0142-9612(97)00146-4. [DOI] [PubMed] [Google Scholar]
- [4].Bagno A, Di Bello C. Surface treatments and roughness properties of Ti-based biomaterials. J Mater Sci Mater Med. 2004;15:935–49. doi: 10.1023/B:JMSM.0000042679.28493.7f. [DOI] [PubMed] [Google Scholar]
- [5].Lincks J, Boyan BD, Blanchard CR, Lohmann CH, Liu Y, Cochran DL, et al. Response of MG63 osteoblast-like cells to titanium and titanium alloy is dependent on surface roughness and composition. Biomaterials. 1998;19:2219–32. doi: 10.1016/s0142-9612(98)00144-6. [DOI] [PubMed] [Google Scholar]
- [6].Martin JY, Schwartz Z, Hummert TW, Schraub DM, Simpson J, Lankford J, et al. Effect of titanium surface roughness on proliferation, differentiation, and protein synthesis of human osteoblast-like cells (MG63) J Biomed Mater Res. 1995;29:389–401. doi: 10.1002/jbm.820290314. [DOI] [PubMed] [Google Scholar]
- [7].Heimann RB, Kurzweg H, Ivey DG, Wayman ML. Microstructural and in vitro chemical investigations into plasma-sprayed bioceramic coatings. J Biomed Mater Res. 1998;43:441–50. doi: 10.1002/(sici)1097-4636(199824)43:4<441::aid-jbm12>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- [8].Kurzweg H, Heimann RB, Troczynski T, Wayman ML. Development of plasma-sprayed bioceramic coatings with bond coats based on titania and zirconia. Biomaterials. 1998;19:1507–11. doi: 10.1016/s0142-9612(98)00067-2. [DOI] [PubMed] [Google Scholar]
- [9].Bigi A, Fini M, Bracci B, Boanini E, Torricelli P, Giavaresi G, et al. The response of bone to nanocrystalline hydroxyapatite-coated Ti13Nb11Zr alloy in an animal model. Biomaterials. 2008;29:1730–6. doi: 10.1016/j.biomaterials.2007.12.011. [DOI] [PubMed] [Google Scholar]
- [10].Hanawa T, Kamiura Y, Yamamoto S, Kohgo T, Amemiya A, Ukai H, et al. Early bone formation around calcium-ion-implanted titanium inserted into rat tibia. J Biomed Mater Res. 1997;36:131–6. doi: 10.1002/(sici)1097-4636(199707)36:1<131::aid-jbm16>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- [11].Rautray TR, Narayanan R, Kwon TY, Kim KH. Surface modification of titanium and titanium alloys by ion implantation. J Biomed Mater Res B Appl Biomater. 2010;93:581–91. doi: 10.1002/jbm.b.31596. [DOI] [PubMed] [Google Scholar]
- [12].Ogawa T, Saruwatari L, Takeuchi K, Aita H, Ohno N. Ti nano-nodular structuring for bone integration and regeneration. J Dent Res. 2008;87:751–6. doi: 10.1177/154405910808700809. [DOI] [PubMed] [Google Scholar]
- [13].Chang CK, Wu JS, Mao DL, Ding CX. Mechanical and histological evaluations of hydroxyapatite-coated and noncoated Ti6Al4V implants in tibia bone. J Biomed Mater Res. 2001;56:17–23. doi: 10.1002/1097-4636(200107)56:1<17::aid-jbm1063>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- [14].Rakngarm A, Mutoh Y. Characterization and fatigue damage of plasma sprayed HAp top coat with Ti and HAp/Ti bond coat layers on commercially pure titanium substrate. J Mech Behav Biomed Mater. 2009;2:444–53. doi: 10.1016/j.jmbbm.2008.11.001. [DOI] [PubMed] [Google Scholar]
- [15].Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- [16].Xiao SJ, Textor M, Spencer ND, Wieland M, Keller B, Sigrist H. Immobilization of the cell-adhesive peptide Arg-Gly-Asp-Cys (RGDC) on titanium surfaces by covalent chemical attachment. J Mater Sci-Mater Med. 1997;8:867–72. doi: 10.1023/a:1018501804943. [DOI] [PubMed] [Google Scholar]
- [17].Morra M. Biochemical modification of titanium surfaces: peptides and ECM proteins. Euro Cells Mater. 2006;12:1–15. doi: 10.22203/ecm.v012a01. [DOI] [PubMed] [Google Scholar]
- [18].Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem Rev. 2005;105(4):1103–69. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- [19].Mrksich M, Whitesides GM. Using self-assembled monolayers to understand the interactions of man-made surfaces with proteins and cells. Ann Rev Biophys Biomol Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- [20].Bierbaum S, Beutner R, Hanke T, Scharnweber D, Hempel U, Worch H. Modification of Ti6Al4V surfaces using collagen I, III, and fibronectin. I. Biochemical and morphological characteristics of the adsorbed matrix. J Biomed Mater Res A. 2003 Nov 1;67:421–30. doi: 10.1002/jbm.a.10080. [DOI] [PubMed] [Google Scholar]
- [21].Bierbaum S, Hempel U, Geissler U, Hanke T, Scharnweber D, Wenzel KW, et al. Modification of Ti6AL4V surfaces using collagen I, III, and fibronectin. II. Influence on osteoblast responses. J Biomed Mater Res A. 2003;67:431–8. doi: 10.1002/jbm.a.10084. [DOI] [PubMed] [Google Scholar]
- [22].Ku Y, Chung CP, Jang JH. The effect of the surface modification of titanium using a recombinant fragment of fibronectin and vitronectin on cell behavior. Biomaterials. 2005;26:5153–7. doi: 10.1016/j.biomaterials.2005.01.060. [DOI] [PubMed] [Google Scholar]
- [23].Martin HJ, Schulz KH, Bumgardner JD, Walters KB. XPS study on the use of 3-aminopropyltriethoxysilane to bond chitosan to a titanium surface. Langmuir. 2007;23:6645–51. doi: 10.1021/la063284v. [DOI] [PubMed] [Google Scholar]
- [24].Darouiche RO, Mansouri MD, Zakarevicz D, Alsharif A, Landon GC. In vivo efficacy of antimicrobial-coated devices. J Bone Joint Surg Am. 2007;89:792–7. doi: 10.2106/JBJS.F.00414. [DOI] [PubMed] [Google Scholar]
- [25].Balasundaram G, Webster TJ. Increased osteoblast adhesion on nanograined Ti modified with KRSR. J Biomed Mater Res A. 2007;80:602–11. doi: 10.1002/jbm.a.30954. [DOI] [PubMed] [Google Scholar]
- [26].Sarikaya M, Tamerler C, Schwartz DT, Baneyx FO. Materials assembly and formation using engineered polypeptides. Ann Rev Mater Res. 2004;34:373–408. [Google Scholar]
- [27].Kriplani U, Kay BK. Selecting peptides for use in nanoscale materials using phagedisplayed combinatorial peptide libraries. Curr Opin Biotech. 2005;16:470–5. doi: 10.1016/j.copbio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- [28].Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Molecular biomimetics: nanotechnology through biology. Nat Mater. 2003;2:577–85. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- [29].Shiba K. Exploitation of peptide motif sequences and their use in nanobiotechnology. Curr Opin Biotech. 2010;21:412–425. doi: 10.1016/j.copbio.2010.07.008. [DOI] [PubMed] [Google Scholar]
- [30].Khatayevich D, Gungormus M, Yazici H, So C, Cetinel S, Ma H, et al. Biofunctionalization of materials for implants using engineered peptides. Acta Biomater. 2010;6:4634–4641. doi: 10.1016/j.actbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- [31].Tamerler C, Khatayevich D, Gungormus M, Kacar T, Oren EE, Hnilova M, et al. Molecular biomimetics: GEPI-based biological routes to technology. Biopolymers. 2010;94:78–94. doi: 10.1002/bip.21368. [DOI] [PubMed] [Google Scholar]
- [32].Meyers SR, Khoo XJ, Huang X, Walsh EB, Grinstaff MW, Kenan DJ. The development of peptide-based interfacial biomaterials for generating biological functionality on the surface of bioinert materials. Biomaterials. 2009;30:277–86. doi: 10.1016/j.biomaterials.2008.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gungormus M, Fong H, Kim IW, Evans JS, Tamerler C, Sarikaya M. Regulation of in vitro calcium phosphate mineralization by combinatorially selected hydroxyapatite-binding peptides. Biomacromolecules. 2008;9:966–73. doi: 10.1021/bm701037x. [DOI] [PubMed] [Google Scholar]
- [34].Sano KI, Sasaki H, Shiba K. Specificity and biomineralization activities of Ti-binding peptide-1 (TBP-1) Langmuir. 2005;21:3090–5. doi: 10.1021/la047428m. [DOI] [PubMed] [Google Scholar]
- [35].Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled ss-hairpin peptide hydrogel surfaces. Biomaterials. 2005 Sep;26(25):5177–86. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- [36].Kashiwagi K, Tsuji T, Shiba K. Directional BMP-2 for functionalization of titanium surfaces. Biomaterials. 2009;30:1166–75. doi: 10.1016/j.biomaterials.2008.10.040. [DOI] [PubMed] [Google Scholar]
- [37].Sano KI, Shiba K. A hexapeptide motif that electrostatically binds to the surface of titanium. J Am Chem Soc. 2003;125:14234–5. doi: 10.1021/ja038414q. [DOI] [PubMed] [Google Scholar]
- [38].Brown S. Metal-recognition by repeating polypeptides. Nat Biotechnol. 1997;15:269–72. doi: 10.1038/nbt0397-269. [DOI] [PubMed] [Google Scholar]
- [39].Hnilova M, Oren EE, Seker UOS, Wilson BR, Collino S, Evans JS, et al. Effect of molecular conformations on the adsorption behavior of gold-binding peptides. Langmuir. 2008;24:12440–5. doi: 10.1021/la801468c. [DOI] [PubMed] [Google Scholar]
- [40].Naik RR, Brott LL, Clarson SJ, Stone MO. Silica-precipitating peptides isolated from a combinatorial phage display peptide library. J Nanosci Nanotech. 2002;2:95–100. doi: 10.1166/jnn.2002.074. [DOI] [PubMed] [Google Scholar]
- [41].Seker UOS, Wilson B, Sahin D, Tamerler C, Sarikaya M. Quantitative affinity of genetically engineered repeating polypeptides to inorganic surfaces. Biomacromolecules. 2009;10:250–7. doi: 10.1021/bm8009895. [DOI] [PubMed] [Google Scholar]
- [42].Segvich SJ, Smith HC, Kohn DH. The adsorption of preferential binding peptides to apatite-based materials. Biomaterials. 2009;30:1287–98. doi: 10.1016/j.biomaterials.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Weiger MC, Park JJ, Roy MD, Stafford CM, Karim A, Becker ML. Quantification of the binding affinity of a specific hydroxyapatite binding peptide. Biomaterials. 2010;31:2955–63. doi: 10.1016/j.biomaterials.2010.01.012. [DOI] [PubMed] [Google Scholar]
- [44].Sanghvi AB, Miller KPH, Belcher AM, Schmidt CE. Biomaterials functionalization using a novel peptide that selectively binds to a conducting polymer. Nat Mater. 2005;4:496–502. doi: 10.1038/nmat1397. [DOI] [PubMed] [Google Scholar]
- [45].Vreuls C, Zocchi G, Genin A, Archambeau C, Martial J, Van de Weerdt C. Inorganic-binding peptides as tools for surface quality control. J Inorg Biochem. 2010;104:1013–21. doi: 10.1016/j.jinorgbio.2010.05.008. [DOI] [PubMed] [Google Scholar]
- [46].Boyer R, Welsch G, Collings EW. Materials properties handbook - titanium alloys. ASM International; Materials Park, OH: 1994. [Google Scholar]
- [47].Ahmad M, Gawronski D, Blum J, Goldberg J, Gronowicz G. Differential response of human osteoblast-like cells to commercially pure (cp) titanium grades 1 and 4. J Biomed Mater Res. 1999;46:121–31. doi: 10.1002/(sici)1097-4636(199907)46:1<121::aid-jbm14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- [48].Lu ZJ, Murray KS, Vancleave V, Lavallie ER, Stahl ML, Mccoy JM. Expression of thioredoxin random peptide libraries on the Escherichia coli cell-surface as functional fusions to flagellin - a system designed for exploring protein-protein interactions. Bio-Technology. 1995;13:366–72. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- [49].Tamerler C, Khatayevich D, Gungormus M, Kacar T, Oren EE, Hnilova M, et al. Molecular biomimetics: GEPI-based biological routes to technology. Biopolymers. 2010;94:78–94. doi: 10.1002/bip.21368. [DOI] [PubMed] [Google Scholar]
- [50].Tamerler C, Sarikaya M. Molecular biomimetics: utilizing nature's molecular ways in practical engineering. Acta Biomater. 2007;3:289–99. doi: 10.1016/j.actbio.2006.10.009. [DOI] [PubMed] [Google Scholar]
- [51].Reed J, Reed TA. A set of constructed type spectra for the practical estimation of peptide secondary structure from circular dichroism. Anal Biochem. 1997;254:36–40. doi: 10.1006/abio.1997.2355. [DOI] [PubMed] [Google Scholar]
- [52].MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- [53].Cubellis MV, Caillez F, Blundell TL, Lovell SC. Properties of polyproline II, a secondary structure element implicated in protein-protein interactions. Protein Struct Funct Bioinformatics. 2005;58:880–92. doi: 10.1002/prot.20327. [DOI] [PubMed] [Google Scholar]
- [54].Oren EE, Tamerler C, Sarikaya M. Metal recognition of septapeptides via polypod molecular architecture. Nano Lett. 2005;5:415–9. doi: 10.1021/nl048425x. [DOI] [PubMed] [Google Scholar]
- [55].Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [56].Amos FF, Evans JS. AP7, a partially disordered pseudo C-ring protein, is capable of forming stabilized aragonite in vitro. Biochemistry. 2009;48:1332–9. doi: 10.1021/bi802148r. [DOI] [PubMed] [Google Scholar]
- [57].Delak K, Giocondi J, Orme C, Evans JS. Modulation of crystal growth by the terminal sequences of the prismatic-associated asprich protein. Cryst Growth Design. 2008;8:4481–6. [Google Scholar]
- [58].Wustman BA, Santos R, Zhang B, Evans JS. Identification of a “glycine-loop”-like coiled structure in the 34 AA Pro, Gly, Met repeat domain of the biomineral-associated protein, PM27. Biopolymers. 2002;65(5):362–72. doi: 10.1002/bip.10274. [DOI] [PubMed] [Google Scholar]
- [59].Xu GZ, Evans JS. Model peptide studies of sequence repeats derived from the intracrystalline biomineralization protein, SM50. I. GVGGR and GMGGQ repeats. Biopolymers. 1999;49(4):303–12. doi: 10.1002/(SICI)1097-0282(19990405)49:4<303::AID-BIP5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- [60].Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan YW, Moradian-Oldak J, et al. The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48(10):2272–81. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Limson J, Odunuga OO, Green H, Hook F, Blatch GL. The use of a quartz crystal microbalance with dissipation for the measurement of protein–protein interactions: a qualitative and quantitative analysis of the interactions between molecular chaperones. S Afr J Sci. 2004;100(11–12):678–82. [Google Scholar]
- [62].Pesquero NC, Pedroso MM, Watanabe AM, Goldman MHS, Faria RC, Roque-Barreira MC, et al. Real-time monitoring and kinetic parameter estimation of the affinity interaction of jArtinM and rArtinM with peroxidase glycoprotein by the electrogravimetric technique. Biosens Bioelectron. 2010;26(1):36–42. doi: 10.1016/j.bios.2010.04.047. [DOI] [PubMed] [Google Scholar]
- [63].Pfeiffer I, Hook F. Bivalent cholesterol-based coupling of oligonucletides to lipid membrane assemblies. J Am Chem Soc. 2004;126(33):10224–5. doi: 10.1021/ja048514b. [DOI] [PubMed] [Google Scholar]
- [64].Su XD, Wu YJ, Knoll W. Comparison of surface plasmon resonance spectroscopy and quartz crystal microbalance techniques for studying DNA assembly and hybridization. Biosens Bioelectron. 2005;21(5):719–26. doi: 10.1016/j.bios.2005.01.006. [DOI] [PubMed] [Google Scholar]
- [65].Su XD, Zhang H. Comparison of surface plasmon resonance spectroscopy and quartz crystal microbalance for human IgE quantification. Sensors Actuators BChem. 2004;100(3):309–14. [Google Scholar]
- [66].Tamerler C, Oren EE, Duman M, Venkatasubramanian E, Sarikaya M. Adsorption kinetics of an engineered gold binding Peptide by surface plasmon resonance spectroscopy and a quartz crystal microbalance. Langmuir. 2006;22(18):7712–8. doi: 10.1021/la0606897. [DOI] [PubMed] [Google Scholar]
- [67].Anselme K. Osteoblast adhesion on biomaterials. Biomaterials. 2000 Apr;21(7):667–81. doi: 10.1016/s0142-9612(99)00242-2. [DOI] [PubMed] [Google Scholar]
- [68].Chen HB, Su XD, Neoh KG, Choe WS. QCM-D analysis of binding mechanism of phage particles displaying a constrained heptapeptide with specific affinity to SiO2 and TiO2. Anal Chem. 2006;78(14):4872–9. doi: 10.1021/ac0603025. [DOI] [PubMed] [Google Scholar]
- [69].Chen HB, Su XD, Neoh KG, Choe WS. Probing the interaction between peptides and metal oxides using point mutants of a TiO2-binding peptide. Langmuir. 2008;24(13):6852–7. doi: 10.1021/la800314p. [DOI] [PubMed] [Google Scholar]
- [70].Fang Y, Poulsen N, Dickerson MB, Cai Y, Jones SE, Naik RR, et al. Identification of peptides capable of inducing the formation of titania but not silica via a subtractive bacteriophage display approach. J Mater Chem. 2008;18(32):3871–5. [Google Scholar]
- [71].Evans JS, Samudrala R, Walsh TR, Oren EE, Tamerler C. Molecular design of inorganic-binding polypeptides. MRS Bull. 2008;33(5):514–8. [Google Scholar]
- [72].Evans JS. “Tuning in” to mollusk shell nacre- and prismatic-associated protein terminal sequences. implications for biomineralization and the construction of high performance inorganic–organic composites. Chem Rev. 2008;108(11):4455–62. doi: 10.1021/cr078251e. [DOI] [PubMed] [Google Scholar]
- [73].Jager M, Zilkens C, Zanger K, Krauspe R. Significance of nano- and microtopography for cell–surface interactions in orthopaedic implants. J Biomed Biotech. 2007;2007:1–19. doi: 10.1155/2007/69036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.