Abstract
Although technically a member of the Inhibitor of Apoptosis (IAP) gene family, survivin has consistently defied assumptions, refuted predictions and challenged paradigms. Despite its more than 5,500 citations currently in Medline, the biology of survivin has remained fascinatingly complex, its exploitation in human disease, most notably cancer, tantalizing, and its regulation of cellular homeostasis unexpectedly far-reaching. An inconvenient outsider that resists schemes and dogmas, survivin continues to hold great promise to unlock fundamental circuitries of cellular functions in health and disease.
Keywords: Survivin, IAP, cell division, apoptosis, cancer, spindle assembly, mitotic catastrophe
INTRODUCTION
Inhibitor-of-Apoptosis (IAP) proteins are multifunctional molecules structurally identified by the presence of a ~70 amino acid Baculovirus IAP Repeat (BIR), a zinc finger fold coordinated by histidine and cysteine residues present at least once in each family member [1]. The eight IAP family members in the human genome contain one to three BIRs, typically arranged in the protein’s amino-terminus. Additional protein domains found in IAPs include a carboxyl-terminus RING, which functions as an E3 ubiquitin ligase, a ubiquitin-associated domain implicated in binding to ubiquitinated proteins, and a caspase-recruitment domain (CARD, in c-IAP1 and c-IAP2), of less clear function.
At 142 amino acids, survivin [2-6] is the smallest member of the IAP family [1], containing a single BIR and a carboxyl-terminus α-helix, but no other identifiable protein domain (Figure 1). X-ray crystallography data have shown that survivin forms a stable homodimer in solution [7], but definitive evidence that this structure is actually required for function(s), in vivo, is still lacking. To the contrary, there is evidence that some key protein-protein interactions, for instance the recognition of the chromosomal passenger protein, Borealin [8, 9], as well as mechanisms of subcellular localization [9], in particular nucleo-cytoplasmic trafficking [10], or apoptosis inhibition [11], require a monomeric survivin protein.
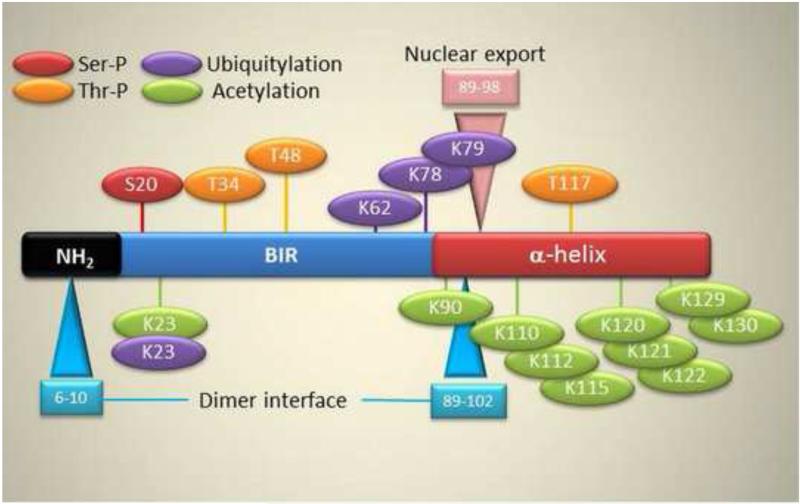
Figure 1. Post-translational modifications in survivin.
Amino acids undergoing Ser/Thr phosphorylation, acetylation or ubiquitylation are indicated. The positions of the proposed dimer interface and nuclear export sequence (NES) are shown.
Located at the tip of chromosome 17 (17q25) in the human genome, a TATA-less survivin locus has been the subject of considerable studies [12]. This is more than an exercise since agents that shut down survivin gene expression have successfully entered the clinic as anticancer strategies [13]. Transcription of the survivin gene relies on three essential Sp1 sites [14], and canonical CDE/CHR boxes that impart sharp cell cycle periodicity of survivin gene expression at mitosis [15]. A CG-rich CpG island also in the proximal survivin promoter has been investigated for potential epigenetic modifications, but clear-cut implications for this mechanism are still lacking [12]. The regulation of the survivin locus is highly complex, and a plethora of transcription factors, many of which are intercalated in cell proliferation, cell survival or developmental pathways upregulate survivin gene expression [12]. An equally long list of molecules has been associated with actively repressing survivin gene transcription, in particular, p53 [16, 17], Pten [18], and BRCA1-SIRT1 [19]. This suggests that transcription of the survivin gene is a finely tuned process, carefully balanced between activators and inhibitors, and with potential unique cell type-specificities and developmental regulation. The fact that many of these transcriptional inhibitors are bona fide tumor suppressors, mutated or otherwise lost in cancer, suggests that repression of survivin gene expression is an important barrier against malignant transformation, and, by extension, that successful tumor suppression depends on ablation of this pathway [20].
A mature survivin protein is extensively post-translationally modified (Figure 1), and phosphorylation plays an important role in survivin functions [21]. Best studied are the mitotic kinases Cdk1 [22-24], Aurora B [25, 26], casein kinase [27], and Polo-like kinase 1 [27] that phosphorylate survivin at different residues, regulating protein stability, subcellular localization, association with protein partners, various aspects of mitosis, and apoptosis inhibition. There may be potentially important roles for other kinases, for instance PKA [28] regulation of survivin cytoprotection, and even more phosphorylation events are predicted by consensus algorithms [21] (Figure 1). An additional post-translational modification of survivin involves acetylation of many lysine residues predominantly clustered in the -COOH terminus of the protein (Figure 1). Mostly mediated by Creb-binding protein (CBP) [21], cycles of survivin acetylation and deacetylation have been implicated in nucleo-cytoplasmic shuttling and a potential new mechanism of downstream regulation of gene expression [29] (Figure 1).
NEW TRICKS FOR OLD SURVIVIN: CELL DIVISION AND CELL SURVIVAL
That survivin is essential for cell division is established, inferred from its sharp cell cycle-dependent expression at G2/M, localization to various aspects of the mitotic apparatus and the plethora of mitotic defects ensuing from targeting survivin by many approaches and in disparate cell types [30, 31]. The phenotype of survivin-knockout embryos, which do not remain viable past blastocyst state [32], the extensive mitotic defects in adult tissues where survivin has been conditionally ablated [33, 34], and the cell division defects caused by loss of survivin-like molecules in C.elegans [35, 36], and yeast [37], all support a model where a survivin pathway is indispensable at cell division.
Together with Aurora B, Borealin and INCENP, survivin is the fourth member of the chromosomal passenger complex (CPC) [38], a regulator of chromosome-microtubule attachment, spindle assembly checkpoint, and cytokinesis at cell division (Figure 2). Structurally, survivin recognizes the CPC through its dimerization domain, suggesting that it functions as a monomer in the complex [9]. Functionally, the CPC ensures a fidelity checkpoint that destabilizes errors in microtubule-kinetochore attachment via Aurora B-dependent phosphorylation of target substrates [38]. It derives from this model that a precise but dynamic localization of Aurora B to the inner centromere is paramount to ensure CPC checkpoint function. This centromere targeting activity is provided by survivin (Figure 2). It was first suggested that this could be accomplished by reversible mono-ubiquitylation of survivin. In this model, Ufd1 (ubiquitin fusion degradation protein 1) ubiquitylation of survivin promoted CPC centromere binding, whereas deubiquitylation by hFAM mediated its dissociation [39].
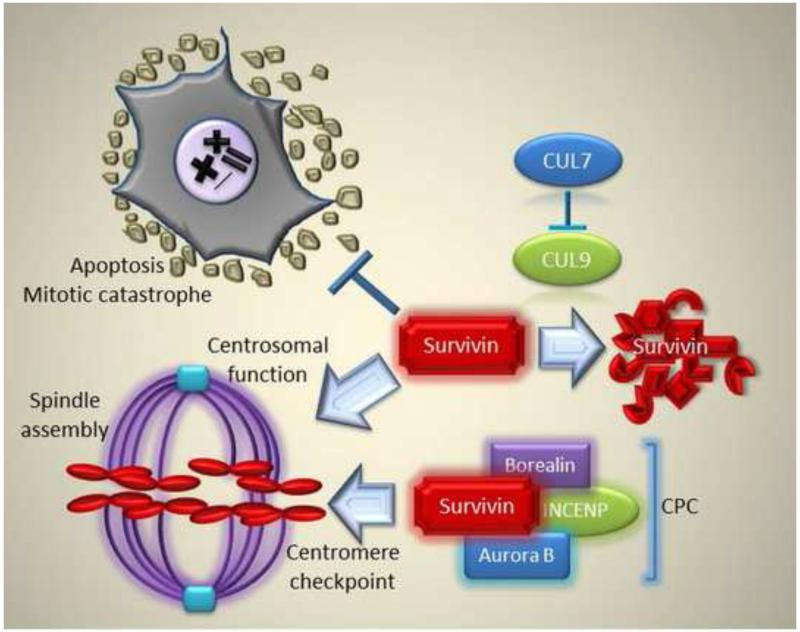
Figure 2. Tripartite role of survivin at mitosis.
A proposed role of survivin in inhibiting apoptosis/mitotic catastrophe (i), regulating spindle assembly (ii) and CPC targeting for a centromere checkpoint (iii) are indicated. See text for additional details.
More recent studies, however, pointed to a very different scenario, in which survivin “reads” a mitotic histone mark introduced by Haspin phosphorylation of Thr3 on histone 3 (H3) [40-42]. By binding to phosphorylated H3, survivin then localizes the entire CPC to inner centromeres, precisely where is needed to correct errors in kinetochore-microtubule attachment [40-42]. Interference with the survivin-H3 recognition mislocalizes Aurora B, and causes many, but not all of the mitotic defects associated with survivin loss [40-42].
Although attractive, there may be more layers of complexity to this model. First, there is an additional mitotic histone mark created by Bub1 phosphorylation of histone H2A on Thr120 that may also promote CPC recruitment to centromeres through the adaptor protein Shugoshin (Sgo2) [40], itself another survivin-interacting molecule [43]. Second, at least in budding yeast, CPC targeting by survivin does not seem required for chromosomal bi-orientation, a function that may be accomplished just by clustering of Aurora B on chromatin or microtubules [44]. And, finally, the binding requirements of survivin to H3 may be exceedingly intricate, in vivo. In fact, the survivin region centered on Asp70 and Asp71 [42], which recognizes phosphorylated H3 [45, 46], is also the binding interface for the pro-apoptotic mediator, Smac [46]. It is still unclear how much of the anti-apoptotic function of survivin depends on sequestering Smac away from the caspase inhibitor, XIAP [1]. However, this mechanism has been repeatedly proposed [47], either involving monomeric survivin [11], thus the same structure recruited to the CPC [9], or mitochondria-localized survivin [48]. There may be differences in the affinity with which survivin binds H3 versus Smac [46]. However, this is controversial, as the dissociation constants for the survivin-Smac complex obtained from isothermal calorimetry [46] or NMR [49] do not agree. And to add another layer of complexity, Glu65 in survivin, which has also been implicated in binding phosphorylated H3 [46], is essential for the recognition of yet another survivin partner at mitosis, the GTPase Ran [50]. Functionally, this protein complex is important to deliver the Ran effector, TPX2 to microtubules, thus supporting bipolar spindle assembly [50], and, potentially, a scaffold function for CPC activation [51].
A CPC BAND OR A SURVIVIN ORCHESTRA?
Although dubbed as controversial [30], a potential mitotic role of survivin in spindle assembly has been proposed [31]. This was linked to a pool of survivin localized to centrosomes and microtubules [52], and which regulates bipolar spindle assembly [53] via active repression of microtubule dynamics [54]. This model is consistent with a large body of literature, where loss of survivin has been invariably associated with a plethora of microtubule defects, including centrosomal abnormalities, formation of multipolar spindles, misaligned spindles and, ultimately, polyploidy [4, 55] (Figure 2). Such “microtubule” phenotype is also seen in adult tissues after conditional ablation of survivin, in vivo [56, 57].
Fresh evidence has now uncovered a new pathway for how survivin may regulate microtubule integrity and cell viability at cell division (Figure 2). This involved a centrosomal localization of CUL7. Together with OBSL1 and CCDC8, CUL7 is a component of the so-called 3M complex [58], molecules that are mutated in rare growth retardation syndromes [59]. It turns out that centrosomal CUL7 is essential for microtubule dynamics, as its depletion causes a microtubule phenotype not dissimilar to what observed after survivin loss, with prometaphase arrest, tetraploidy, and, ultimately, death of cells attempting to traverse mitosis [58]. In further dissecting the pathway, it was shown that CUL7 inhibits the E3 ligase activity of its associated molecule, CUL9 [60], and this was functionally important because depletion of CUL9 rescued the microtubule phenotype induced by CUL7 silencing [61]. The third component of this response, and the actual effector of microtubule dynamics, was identified as survivin, in agreement with earlier data [54], which is destroyed by CUL9-mediated ubiquitylation [61]. Accordingly, loss of CUL9, dubbed as a tumor suppressor based on the cancer-prone phenotype of knockout mice, was sufficient to increase survivin levels, in vivo [61], and, conversely, re-expression of survivin in CUL7-silenced cells was sufficient not only to restore microtubule dynamics but also to rescue cell viability from apoptosis at cell division [61] (Figure 2).
That in addition to controlling CPC targeting (via H3 binding) and now microtubule/spindle dynamics (via TPX2 delivery and 3M complex regulation), survivin may have a third function at mitosis in countering cell death has long been proposed [15]. This idea has been vigorously challenged, and, at times, openly derided [62]. And yet, it is well known that when cells approach mitosis abnormally, whether because of a damaged mitotic apparatus, or under stress conditions, as the norm for tumor cells, they activate a dual necrotic and apoptotic suicidal pathway loosely defined as mitotic catastrophe [63]. This is likely a fail-safe mechanism to prevent the accumulation of an aneuploidy progeny, and survivin has long been recognized as an important antagonist of this process [3]. The reverse may also be true, and silencing of survivin has been associated with mitotic catastrophe [64] (Figure 2). Mechanistically, it may not be unexpected that survivin inhibits caspase-9 in concert with XIAP [28], and antagonizes p53-dependent cell death [65], two effector pathways of mitotic catastrophe [63].
Recent data further support this model, suggesting a unifying context for an integrated, tripartite role of survivin at mitosis. It turns out that stressed tumor cells exposed to low level, non-cytotoxic DNA damage recruit the essential autophagy regulator, ATG5 to the nucleus [66]. Here, ATG5 forms a complex with nuclear survivin, and sequesters it, triggering hallmarks of mitotic catastrophe, including multipolar spindles, missegregated chromosomes, and cell death [66]. It is intriguing that mislocalization of the CPC, potentially contributed by ATG5 sequestration of survivin in the nucleus [66], has also been implicated in the activation of mitotic catastrophe [63], and this is the type of cell death seen with small molecule CPC antagonists pursued as anticancer agents [67] (Figure 2).
Taken together, these results point to a complex model for a simultaneous tripartite role of survivin at cell division, overseeing faithful chromosomal segregation by CPC targeting (i), bipolar spindle formation via centrosomal regulation of microtubule dynamics (ii), and inhibition of mitotic catastrophe through suppression of caspase 9 and sequestration of Smac (iii) (Figure 2). Whether this integrated survivin pathway is uniquely exploited in tumor cells, compared to the normal tissues, remains to be demonstrated. It could be argued that aneuploid and genetically deranged tumor cells, constantly exposed to disparate stress stimuli in their microenvironment, may become especially dependent or“ addicted” to the anti-apoptotic function of survivin at mitosis, a prediction consistent with the phenotype of transgenic mouse models [68]. On the other hand, survivin is also essential roles to maintain the viability of specialized compartments of normal cells (see below), potentially through the same integrated, tripartite pathway (Figure 2).
NEW TRICKS FOR NEW SURVIVIN
For its role at the crossroads of multiple signaling pathways [4], it is not surprising that fresh experimental evidence continues to uncover novel roles for survivin in cellular homeostasis. One of these emerging trends involves an essential developmental role in stem cell maintenance.
STEM CELL MAINTENANCE
Stemming from its abundant expression during embryonic and fetal development [69], and the panoply of developmental signaling pathways that target survivin, in particular Wnt/β-catenin [70], Hedgehog[71], Hippo [72], and Notch [73], it may not be surprising that human [74, 75], and mouse [76] embryonic stem cells contain high levels of survivin. Although the biochemical wiring of this pathway is not completely defined, survivin is clearly indispensable for the integrity of stem/progenitor cells. This has been demonstrated for pluripotent stem cells, where expression and subcellular localization of survivin and its splice variants [77] may be regulated by microRNAs [78], as well as neural [79], hematopoietic [80, 81], epidermal [82], and intestinal [83] stem cells. Other data point to a pivotal role of survivin in stem cell-driven malignancy, initiating hematopoietic transformation in transgenic mice [84], conferring a drug-resistant phenotype in acute lymphoblastic leukemia [85], cooperating with Myc-driven signaling in mesenchymal transformation [86], and enabling survival of stem/progenitor-like cells during gliomagenesis [87]. Consistent with these observations, survivin is a reliable indicator of poor prognosis in stem cell-derived tumors, including acute leukemia [88], and gliomas [89]. Based on these results, targeting survivin may provide a viable strategy to deplete a potential cancer stem cell reservoir, often linked to disease recurrence and drug resistance. Accordingly, interference with survivin expression or function inhibited pluripotent stem cell-derived teratomas [90], eradicated acute leukemia [85], or suppressed gliomagenesis using small molecule [87] or oncolytic viral [91] approaches.
More work is required to dissect how survivin maintains a stem cell compartment. However, stem cells seem to need survivin predominantly, if not exclusively, for protection against apoptosis [84, 87, 90, 92]. Second, there may be an unexpected upstream role of survivin in controlling pluripotency at the level of gene expression. Although this is a novel field of investigation, it is intriguing that survivin expression in CD34+38- myeloblastic stem cells is associated with extensive transcriptional reprogramming and upregulation of regulators of cell proliferation, PI3K, and cell migration/invasion [88]. Conversely, deletion of survivin in hematopoietic stem cells disrupts an Evi-1-dependent transcriptional program, resulting in loss of downstream target genes, Gata2, Pbx1 and Sall2 [81]. A potential upstream role of survivin in controlling gene expression, including in stem cells, clearly remains to be fully elucidated. However, earlier studies have shown that modulation of survivin acetylation by CBP suppresses STAT3-dependent gene transcription [93], and transgenic expression of survivin in the urothelium was sufficient to produce a transcriptional gene signature of increased inflammation and heightened cell motility [94].
CELL MOTILITY AND METASTASIS
A fairly unanimous consensus is that expression of survivin in virtually every human tumor is associated with poor prognosis [95]. There could be various mechanisms for this correlation, and some have been documented, including inhibition of apoptosis, including mitotic catastrophe [96], enhanced drug resistance [6], and maintenance of cancer stem cells [88]. However, the main cause of cancer death comes from disease dissemination to distant organs, i.e. metastasis. Retrospective studies and meta-analyses have consistently shown that survivin expression correlates with metastatic disease in breast [97], colorectal [98], gastric [99], thyroid [100], esophageal [101], and non-small cell lung [102] cancer. Even assuming that this is a direct effect, the mechanistic underpinning of how survivin may influence metastatic competency is not firmly established. In fact, a role of IAPs in general in cell motility has been intensely debated [103]. Although these molecules may inhibit cell migration under certain conditions, potentially via ubiquitination of Rac1 [104], or regulation of RAF destruction [105], most data suggest that IAPs [106, 107] function as potentially evolutionary-conserved [108] enablers of cell motility [103]. Data on how survivin may participate in this response are still scant, but emerging evidence also points to survivin as an important mediator of tumor cell motility and invasion, potentially affecting metastatic competency, in vivo [109].
A survivin pathway in controlling cell motility may be entirely separate from other functions in this molecule in apoptosis inhibition and mitosis. Instead, experimental data have mechanistically linked survivin expression in tumors to increased production of matrix metalloproteinase(s) for digestion of the extracellular matrix [110], upregulation of α5 integrin combined with Akt activation [111, 112], or NFκB-dependent paracrine release of fibronectin by a survivin-XIAP complex, which resulted in a highly metastatic phenotype, in vivo [113]. Intriguingly, a new function of survivin in regulating cell motility may not be restricted to tumors. Accordingly, silencing of survivin in vascular smooth muscle cells did not affect cell proliferation or cell viability, but caused disorganized actin filaments, changes in cell shape, and impaired chemotaxis [114].
CONCLUDING REMARKS AND THE NEXT SURVIVIN WAVE(S)
If nothing else, the remarks in this contribution should highlight the humbling complexity of survivin biology. Despite considerable efforts over almost two decades, the last chapter of how survivin works in normal or tumor cells has not yet been written. New, unexpected, and sometimes bewildering findings continuously emerging in the literature make us rethink what we really know about this unique signaling network in health and disease. Attempts to (over)simplify the biology of survivin, discount its diversity and artificially categorize its consequences have created biased perceptions. Just because survivin-like molecules in lower organisms do not affect cell death, this does not mean that survivin is not an apoptosis inhibitor, but simply that it targets a cell death pathway not operative in those models. And just because cell division is a carefully orchestrated cascade, it does not mean that it is error-free, especially under stress, and that does not couple to a fail-safe mechanism of mitotic cell death, of which survivin is an integral part.
Perhaps an unwelcomed consequence of these preconceived ideas has been the paucity of therapeutic strategies to disable the survivin pathway in cancer. This is a curious deficiency, especially when one considers the urgency for new, more effective treatments in oncology, the undisputable importance of survivin as a disease driver in virtually every human tumor, and our better understanding of some of its functions. Although the lack of progress in this area may reflect a broader risk-avulsion and redundancy of current oncology drug discovery efforts, there is confidence that the pace of survivin discovery will continue unabated in the years to come, refining its biochemical pathways, uncovering new functions, and opening innovative prospects for personalized therapeutics.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grants P01 CA140043 and R01 CA78810 and CA190027.
Footnotes
COMPETING FINANCIAL INTEREST
The author declares that no competing financial interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Srinivasula SM, Ashwell JD. IAPs: what’s in a name? Mol Cell. 2008;30:123–35. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stauber RH, Mann W, Knauer SK. Nuclear and cytoplasmic survivin: molecular mechanism, prognostic, and therapeutic potential. Cancer Res. 2007;67:5999–6002. doi: 10.1158/0008-5472.CAN-07-0494. [DOI] [PubMed] [Google Scholar]
- [3].Ryan BM, O’Donovan N, Duffy MJ. Survivin: A new target for anti-cancer therapy. Cancer Treat Rev. 2009 doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- [4].Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- [5].Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- [6].Singh N, Subramanian K, Kanwar RK, Cheung CH, Kanwar JR. Clinical aspects for survivin: a crucial molecule for targeting drug-resistant cancers. Drug Discov Today. 2014 doi: 10.1016/j.drudis.2014.11.013. [DOI] [PubMed] [Google Scholar]
- [7].Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7:602–8. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- [8].Bourhis E, Hymowitz SG, Cochran AG. The mitotic regulator Survivin binds as a monomer to its functional interactor Borealin. J Biol Chem. 2007;282:35018–23. doi: 10.1074/jbc.M706233200. [DOI] [PubMed] [Google Scholar]
- [9].Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin-Borealin-INCENP Core Complex Reveals How Chromosomal Passengers Travel Together. Cell. 2007;131:271–85. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- [10].Engelsma D, Rodriguez JA, Fish A, Giaccone G, Fornerod M. Homodimerization antagonizes nuclear export of survivin. Traffic. 2007;8:1495–502. doi: 10.1111/j.1600-0854.2007.00629.x. [DOI] [PubMed] [Google Scholar]
- [11].Pavlyukov MS, Antipova NV, Balashova MV, Vinogradova TV, Kopantzev EP, Shakhparonov MI. Survivin monomer plays an essential role in apoptosis regulation. J Biol Chem. 2011;286:23296–307. doi: 10.1074/jbc.M111.237586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Boidot R, Vegran F, Lizard-Nacol S. Transcriptional regulation of the survivin gene. Mol Biol Rep. 2014;41:233–40. doi: 10.1007/s11033-013-2856-0. [DOI] [PubMed] [Google Scholar]
- [13].Rauch A, Hennig D, Schafer C, Wirth M, Marx C, Heinzel T, et al. Survivin and YM155: how faithful is the liaison? Biochim Biophys Acta. 2014;1845:202–20. doi: 10.1016/j.bbcan.2014.01.003. [DOI] [PubMed] [Google Scholar]
- [14].Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J. 1999;344(Pt 2):305–11. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–4. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- [16].Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–22. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- [17].Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–57. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- [18].Guha M, Plescia J, Leav I, Li J, Languino LR, Altieri DC. Endogenous tumor suppression mediated by PTEN involves survivin gene silencing. Cancer Res. 2009;69:4954–8. doi: 10.1158/0008-5472.CAN-09-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guha M, Altieri DC. Survivin as a global target of intrinsic tumor suppression networks. Cell Cycle. 2009;8:2708–10. doi: 10.4161/cc.8.17.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nogueira-Ferreira R, Vitorino R, Ferreira-Pinto MJ, Ferreira R, Henriques-Coelho T. Exploring the role of post-translational modifications on protein-protein interactions with survivin. Arch Biochem Biophys. 2013;538:64–70. doi: 10.1016/j.abb.2013.07.027. [DOI] [PubMed] [Google Scholar]
- [22].O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, et al. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci U S A. 2000;97:13103–7. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13:820–7. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- [24].Barrett RM, Osborne TP, Wheatley SP. Phosphorylation of survivin at threonine 34 inhibits its mitotic function and enhances its cytoprotective activity. Cell Cycle. 2009;8:278–83. doi: 10.4161/cc.8.2.7587. [DOI] [PubMed] [Google Scholar]
- [25].Wheatley SP, Barrett RM, Andrews PD, Medema RH, Morley SJ, Swedlow JR, et al. Phosphorylation by Aurora-B Negatively Regulates Survivin Function During Mitosis. Cell Cycle. 2007:6. doi: 10.4161/cc.6.10.4179. [DOI] [PubMed] [Google Scholar]
- [26].Wheatley SP, Henzing AJ, Dodson H, Khaled W, Earnshaw WC. Aurora-B Phosphorylation in Vitro Identifies a Residue of Survivin That Is Essential for Its Localization and Binding to Inner Centromere Protein (INCENP) in Vivo. J Biol Chem. 2004;279:5655–60. doi: 10.1074/jbc.M311299200. [DOI] [PubMed] [Google Scholar]
- [27].Barrett RM, Colnaghi R, Wheatley SP. Threonine 48 in the BIR domain of survivin is critical to its mitotic and anti-apoptotic activities and can be phosphorylated by CK2 in vitro. Cell Cycle. 2011;10:538–48. doi: 10.4161/cc.10.3.14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dohi T, Xia F, Altieri DC. Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection. Mol Cell. 2007;27:17–28. doi: 10.1016/j.molcel.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Riolo MT, Cooper ZA, Holloway MP, Cheng Y, Bianchi C, Yakirevich E, et al. Histone deacetylase 6 (HDAC6) deacetylates survivin for its nuclear export in breast cancer. J Biol Chem. 2012;287:10885–93. doi: 10.1074/jbc.M111.308791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lens SM, Vader G, Medema RH. The case for Survivin as mitotic regulator. Curr Opin Cell Biol. 2006;18:616–22. doi: 10.1016/j.ceb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- [31].Altieri DC. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr Opin Cell Biol. 2006;18:609–15. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- [32].Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–28. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- [33].Okada H, Mak TW. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat Rev Cancer. 2004;4:592–603. doi: 10.1038/nrc1412. [DOI] [PubMed] [Google Scholar]
- [34].Gurbuxani S, Xu Y, Keerthivasan G, Wickrema A, Crispino JD. Differential requirements for survivin in hematopoietic cell development. Proc Natl Acad Sci U S A. 2005;102:11480–5. doi: 10.1073/pnas.0500303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fraser AG, James C, Evan GI, Hengartner MO. Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr Biol. 1999;9:292–301. doi: 10.1016/s0960-9822(99)80137-7. [DOI] [PubMed] [Google Scholar]
- [36].Speliotes EK, Uren A, Vaux D, Horvitz HR. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol Cell. 2000;6:211–23. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- [37].Huang HK, Bailis JM, Leverson JD, Gomez EB, Forsburg SL, Hunter T. Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol Cell Biol. 2005;25:9000–15. doi: 10.1128/MCB.25.20.9000-9015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- [40].Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–43. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- [41].Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–9. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–5. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–7. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- [44].Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–21. doi: 10.1038/nature12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jeyaprakash AA, Basquin C, Jayachandran U, Conti E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 2011;19:1625–34. doi: 10.1016/j.str.2011.09.002. [DOI] [PubMed] [Google Scholar]
- [46].Du J, Kelly AE, Funabiki H, Patel DJ. Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human Survivin. Structure. 2012;20:185–95. doi: 10.1016/j.str.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Song Z, Yao X, Wu M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J Biol Chem. 2003;278:23130–40. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- [48].Ceballos-Cancino G, Espinosa M, Maldonado V, Melendez-Zajgla J. Regulation of mitochondrial Smac/DIABLO-selective release by survivin. Oncogene. 2007;26:7569–75. doi: 10.1038/sj.onc.1210560. [DOI] [PubMed] [Google Scholar]
- [49].Sun C, Nettesheim D, Liu Z, Olejniczak ET. Solution structure of human survivin and its binding interface with Smac/Diablo. Biochemistry. 2005;44:11–7. doi: 10.1021/bi0485171. [DOI] [PubMed] [Google Scholar]
- [50].Xia F, Canovas PM, Guadagno TM, Altieri DC. A survivin-ran complex regulates spindle formation in tumor cells. Mol Cell Biol. 2008;28:5299–311. doi: 10.1128/MCB.02039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Iyer J, Tsai MY. A novel role for TPX2 as a scaffold and co-activator protein of the Chromosomal Passenger Complex. Cell Signal. 2012;24:1677–89. doi: 10.1016/j.cellsig.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–6. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- [53].Giodini A, Kallio MJ, Wall NR, Gorbsky GJ, Tognin S, Marchisio PC, et al. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002;62:2462–7. [PubMed] [Google Scholar]
- [54].Rosa J, Canovas P, Islam A, Altieri DC, Doxsey SJ. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol Biol Cell. 2006;17:1483–93. doi: 10.1091/mbc.E05-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Saito T, Hama S, Izumi H, Yamasaki F, Kajiwara Y, Matsuura S, et al. Centrosome amplification induced by survivin suppression enhances both chromosome instability and radiosensitivity in glioma cells. Br J Cancer. 2008;98:345–55. doi: 10.1038/sj.bjc.6604160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Okada H, Bakal C, Shahinian A, Elia A, Wakeham A, Suh WK, et al. Survivin Loss in Thymocytes Triggers p53-mediated Growth Arrest and p53-independent Cell Death. J Exp Med. 2004;199:399–410. doi: 10.1084/jem.20032092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Aboualaiwi WA, Muntean BS, Ratnam S, Joe B, Liu L, Booth RL, et al. Survivin-induced abnormal ploidy contributes to cystic kidney and aneurysm formation. Circulation. 2014;129:660–72. doi: 10.1161/CIRCULATIONAHA.113.005746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yan J, Yan F, Li Z, Sinnott B, Cappell KM, Yu Y, et al. The 3M complex maintains microtubule and genome integrity. Mol Cell. 2014;54:791–804. doi: 10.1016/j.molcel.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huber C, Dias-Santagata D, Glaser A, O’Sullivan J, Brauner R, Wu K, et al. Identification of mutations in CUL7 in 3-M syndrome. Nat Genet. 2005;37:1119–24. doi: 10.1038/ng1628. [DOI] [PubMed] [Google Scholar]
- [60].Skaar JR, Florens L, Tsutsumi T, Arai T, Tron A, Swanson SK, et al. PARC and CUL7 form atypical cullin RING ligase complexes. Cancer Res. 2007;67:2006–14. doi: 10.1158/0008-5472.CAN-06-3241. [DOI] [PubMed] [Google Scholar]
- [61].Li Z, Pei XH, Yan J, Yan F, Cappell KM, Whitehurst AW, et al. CUL9 mediates the functions of the 3M complex and ubiquitylates survivin to maintain genome integrity. Mol Cell. 2014;54:805–19. doi: 10.1016/j.molcel.2014.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- [63].Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12:385–92. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- [64].Lamers F, van der Ploeg I, Schild L, Ebus ME, Koster J, Hansen BR, et al. Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocr Relat Cancer. 2011;18:657–68. doi: 10.1530/ERC-11-0207. [DOI] [PubMed] [Google Scholar]
- [65].Beltrami E, Plescia J, Wilkinson JC, Duckett CS, Altieri DC. Acute ablation of survivin uncovers p53-dependent mitotic checkpoint functions and control of mitochondrial apoptosis. J Biol Chem. 2004;279:2077–84. doi: 10.1074/jbc.M309479200. [DOI] [PubMed] [Google Scholar]
- [66].Maskey D, Yousefi S, Schmid I, Zlobec I, Perren A, Friis R, et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. doi: 10.1038/ncomms3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–7. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- [68].Xia F, Altieri DC. Mitosis-independent survivin gene expression in vivo and regulation by p53. Cancer Res. 2006;66:3392–5. doi: 10.1158/0008-5472.CAN-05-4537. [DOI] [PubMed] [Google Scholar]
- [69].Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43–9. [PMC free article] [PubMed] [Google Scholar]
- [70].Kim PJ, Plescia J, Clevers H, Fearon ER, Altieri DC. Survivin and molecular pathogenesis of colorectal cancer. Lancet. 2003;362:205–9. doi: 10.1016/S0140-6736(03)13910-4. [DOI] [PubMed] [Google Scholar]
- [71].Brun SN, Markant SL, Esparza LA, Garcia G, Terry D, Huang JM, et al. Survivin as a therapeutic target in Sonic hedgehog-driven medulloblastoma. Oncogene. 2014 doi: 10.1038/onc.2014.304. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tsuneki M, Madri JA. Adhesion molecule-mediated hippo pathway modulates hemangioendothelioma cell behavior. Mol Cell Biol. 2014;34:4485–99. doi: 10.1128/MCB.00671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lee CW, Raskett CM, Prudovsky I, Altieri DC. Molecular Dependence of Estrogen Receptor-Negative Breast Cancer on a Notch-Survivin Signaling Axis. Cancer Res. 2008;68:5273–81. doi: 10.1158/0008-5472.CAN-07-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Blum B, Bar-Nur O, Golan-Lev T, Benvenisty N. The anti-apoptotic gene survivin contributes to teratoma formation by human embryonic stem cells. Nat Biotechnol. 2009;27:281–7. doi: 10.1038/nbt.1527. [DOI] [PubMed] [Google Scholar]
- [75].Filion TM, Qiao M, Ghule PN, Mandeville M, van Wijnen AJ, Stein JL, et al. Survival responses of human embryonic stem cells to DNA damage. J Cell Physiol. 2009;220:586–92. doi: 10.1002/jcp.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Coumoul X, Li W, Wang RH, Deng C. Inducible suppression of Fgfr2 and Survivin in ES cells using a combination of the RNA interference (RNAi) and the Cre-LoxP system. Nucleic Acids Res. 2004;32:e85. doi: 10.1093/nar/gnh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mull AN, Klar A, Navara CS. Differential localization and high expression of SURVIVIN splice variants in human embryonic stem cells but not in differentiated cells implicate a role for SURVIVIN in pluripotency. Stem Cell Res. 2014;12:539–49. doi: 10.1016/j.scr.2014.01.002. [DOI] [PubMed] [Google Scholar]
- [78].Kapinas K, Kim H, Mandeville M, Martin-Buley LA, Croce CM, Lian JB, et al. microRNA-mediated survivin control of pluripotency. J Cell Physiol. 2015;230:63–70. doi: 10.1002/jcp.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Miranda CJ, Braun L, Jiang Y, Hester ME, Zhang L, Riolo M, et al. Aging brain microenvironment decreases hippocampal neurogenesis through Wnt-mediated survivin signaling. Aging Cell. 2012;11:542–52. doi: 10.1111/j.1474-9726.2012.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Leung CG, Xu Y, Mularski B, Liu H, Gurbuxani S, Crispino JD. Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J Exp Med. 2007;204:1603–11. doi: 10.1084/jem.20062395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Fukuda S, Hoggatt J, Singh P, Abe M, Speth JM, Hu P, et al. Survivin modulates genes with divergent molecular functions and regulates proliferation of hematopoietic stem cells through Evi-1. Leukemia. 2014 doi: 10.1038/leu.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Marconi A, Dallaglio K, Lotti R, Vaschieri C, Truzzi F, Fantini F, et al. Survivin identifies keratinocyte stem cells and is downregulated by anti-beta1 integrin during anoikis. Stem Cells. 2007;25:149–55. doi: 10.1634/stemcells.2006-0165. [DOI] [PubMed] [Google Scholar]
- [83].George RJ, Sturmoski MA, May R, Sureban SM, Dieckgraefe BK, Anant S, et al. Loss of p21Waf1/Cip1/Sdi1 enhances intestinal stem cell survival following radiation injury. Am J Physiol Gastrointest Liver Physiol. 2009;296:G245–54. doi: 10.1152/ajpgi.00021.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Small S, Keerthivasan G, Huang Z, Gurbuxani S, Crispino JD. Overexpression of survivin initiates hematologic malignancies in vivo. Leukemia. 2010;24:1920–6. doi: 10.1038/leu.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Park E, Gang EJ, Hsieh YT, Schaefer P, Chae S, Klemm L, et al. Targeting survivin overcomes drug resistance in acute lymphoblastic leukemia. Blood. 2011;118:2191–9. doi: 10.1182/blood-2011-04-351239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hipp NI, Christner L, Wirth T, Mueller-Klieser W, Walenta S, Schrock E, et al. MYCN and survivin cooperatively contribute to malignant transformation of fibroblasts. Carcinogenesis. 2014;35:479–88. doi: 10.1093/carcin/bgt341. [DOI] [PubMed] [Google Scholar]
- [87].Guvenc H, Pavlyukov MS, Joshi K, Kurt H, Banasavadi-Siddegowda YK, Mao P, et al. Impairment of glioma stem cell survival and growth by a novel inhibitor for Survivin-Ran protein complex. Clin Cancer Res. 2013;19:631–42. doi: 10.1158/1078-0432.CCR-12-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Carter BZ, Qiu Y, Huang X, Diao L, Zhang N, Coombes KR, et al. Survivin is highly expressed in CD34(+)38(-) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML. Blood. 2012;120:173–80. doi: 10.1182/blood-2012-02-409888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lv S, Dai C, Liu Y, Shi R, Tang Z, Han M, et al. The Impact of Survivin on Prognosis and Clinicopathology of Glioma Patients: A Systematic Meta-Analysis. Mol Neurobiol. 2014:1–6. doi: 10.1007/s12035-014-8823-5. [DOI] [PubMed] [Google Scholar]
- [90].Lee MO, Moon SH, Jeong HC, Yi JY, Lee TH, Shim SH, et al. Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc Natl Acad Sci U S A. 2013;110:E3281–90. doi: 10.1073/pnas.1303669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ahmed AU, Thaci B, Tobias AL, Auffinger B, Zhang L, Cheng Y, et al. A preclinical evaluation of neural stem cell-based cell carrier for targeted antiglioma oncolytic virotherapy. J Natl Cancer Inst. 2013;105:968–77. doi: 10.1093/jnci/djt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Feng R, Zhou S, Liu Y, Song D, Luan Z, Dai X, et al. Sox2 protects neural stem cells from apoptosis via up-regulating survivin expression. Biochem J. 2013;450:459–68. doi: 10.1042/BJ20120924. [DOI] [PubMed] [Google Scholar]
- [93].Wang H, Holloway MP, Ma L, Cooper ZA, Riolo M, Samkari A, et al. Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J Biol Chem. 2010;285:36129–37. doi: 10.1074/jbc.M110.152777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Salz W, Eisenberg D, Plescia J, Garlick DS, Weiss RM, Wu XR, et al. A survivin gene signature predicts aggressive tumor behavior. Cancer Res. 2005;65:3531–4. doi: 10.1158/0008-5472.CAN-04-4284. [DOI] [PubMed] [Google Scholar]
- [95].Rodel F, Sprenger T, Kaina B, Liersch T, Rodel C, Fulda S, et al. Survivin as a prognostic/predictive marker and molecular target in cancer therapy. Curr Med Chem. 2012;19:3679–88. doi: 10.2174/092986712801661040. [DOI] [PubMed] [Google Scholar]
- [96].Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol. 2012;14:1203–11. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- [97].Li Y, Ma X, Wu X, Liu X, Liu L. Prognostic significance of survivin in breast cancer: meta-analysis. Breast J. 2014;20:514–24. doi: 10.1111/tbj.12303. [DOI] [PubMed] [Google Scholar]
- [98].Goossens-Beumer IJ, Zeestraten EC, Benard A, Christen T, Reimers MS, Keijzer R, et al. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br J Cancer. 2014;110:2935–44. doi: 10.1038/bjc.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Chen J, Li T, Liu Q, Jiao H, Yang W, Liu X, et al. Clinical and prognostic significance of HIF-1alpha, PTEN, CD44v6, and survivin for gastric cancer: a meta-analysis. PLoS One. 2014;9:e91842. doi: 10.1371/journal.pone.0091842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Selemetjev S, Dencic TI, Marecko I, Jankovic J, Paunovic I, Savin S, et al. Evaluation of survivin expression and its prognostic value in papillary thyroid carcinoma. Pathol Res Pract. 2014;210:30–4. doi: 10.1016/j.prp.2013.09.014. [DOI] [PubMed] [Google Scholar]
- [101].Cao M, Yie SM, Wu SM, Chen S, Lou B, He X, et al. Detection of survivin-expressing circulating cancer cells in the peripheral blood of patients with esophageal squamous cell carcinoma and its clinical significance. Clin Exp Metastasis. 2009;26:751–8. doi: 10.1007/s10585-009-9274-7. [DOI] [PubMed] [Google Scholar]
- [102].Yie SM, Lou B, Ye SR, He X, Cao M, Xie K, et al. Clinical significance of detecting survivin-expressing circulating cancer cells in patients with non-small cell lung cancer. Lung Cancer. 2009;63:284–90. doi: 10.1016/j.lungcan.2008.05.024. [DOI] [PubMed] [Google Scholar]
- [103].Fulda S. Regulation of cell migration, invasion and metastasis by IAP proteins and their antagonists. Oncogene. 2014;33:671–6. doi: 10.1038/onc.2013.63. [DOI] [PubMed] [Google Scholar]
- [104].Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012;31:14–28. doi: 10.1038/emboj.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Dogan T, Harms GS, Hekman M, Karreman C, Oberoi TK, Alnemri ES, et al. X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat Cell Biol. 2008;10:1447–55. doi: 10.1038/ncb1804. [DOI] [PubMed] [Google Scholar]
- [106].Liu J, Zhang D, Luo W, Yu Y, Yu J, Li J, et al. X-linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)-dependent regulation of the cytoskeleton. J Biol Chem. 2011;286:15630–40. doi: 10.1074/jbc.M110.176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yu J, Zhang D, Liu J, Li J, Yu Y, Wu XR, et al. RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J Biol Chem. 2012;287:13752–60. doi: 10.1074/jbc.M111.337469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Geisbrecht ER, Montell DJ. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 2004;118:111–25. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- [109].Ye Q, Cai W, Zheng Y, Evers BM, She QB. ERK and AKT signaling cooperate to translationally regulate survivin expression for metastatic progression of colorectal cancer. Oncogene. 2014;33:1828–39. doi: 10.1038/onc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gao F, Zhang Y, Yang F, Wang P, Wang W, Su Y, et al. Survivin promotes the invasion of human colon carcinoma cells by regulating the expression of MMP7. Mol Med Rep. 2014;9:825–30. doi: 10.3892/mmr.2014.1897. [DOI] [PubMed] [Google Scholar]
- [111].McKenzie JA, Liu T, Goodson AG, Grossman D. Survivin enhances motility of melanoma cells by supporting Akt activation and {alpha}5 integrin upregulation. Cancer Res. 2010;70:7927–37. doi: 10.1158/0008-5472.CAN-10-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].McKenzie JA, Liu T, Jung JY, Jones BB, Ekiz HA, Welm AL, et al. Survivin promotion of melanoma metastasis requires upregulation of alpha5 integrin. Carcinogenesis. 2013;34:2137–44. doi: 10.1093/carcin/bgt155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Nabzdyk CS, Lancero H, Nguyen KP, Salek S, Conte MS. RNA interference-mediated survivin gene knockdown induces growth arrest and reduced migration of vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2011;301:H1841–9. doi: 10.1152/ajpheart.00089.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]