Abstract
Inherited arrhythmia syndromes are collectively associated with substantial morbidity, yet our understanding of the genetic architecture of these conditions remains limited. Recent technological advances in DNA sequencing have led to the commercialization of genetic testing now widely available in clinical practice. In particular, next generation sequencing allows the large-scale and rapid assessment of entire genomes. Although next generation sequencing represents a major technological advance, it has introduced numerous challenges with respect to the interpretation of genetic variation, and has opened a veritable floodgate of biological data of unknown clinical significance to practitioners. In this review, we discuss current genetic testing indications for inherited arrhythmia syndromes, broadly outline characteristics of next generation sequencing techniques, and highlight challenges associated with such testing. We further summarize future directions that will be necessary to address to enable the widespread adoption of next generation sequencing in the routine management of patients with inherited arrhythmia syndromes.
Keywords: Sequencing, mutation, arrhythmia, genetics
Inherited arrhythmia syndromes (Table 1) are collectively associated with substantial risks of morbidity and sudden cardiac death. Over the past three decades, recognized familial aggregation and traditional genetic mapping efforts have substantiated the genetic basis of these arrhythmia syndromes. Yet for many inherited arrhythmia syndromes, our understanding of the genetic architecture and causal mechanisms remains limited.
Table 1.
Inherited arrhythmia syndromes and their predominant genetic causes.
| Condition | Predominant gene(s)* |
|---|---|
| Long QT syndrome | KCNQ1, KCNH2, SCN5A |
| Catecholaminergic polymorphic ventricular tachycardia | RYR2, CASQ |
| Brugada syndrome | SCN5A |
| Cardiac conduction system disease | SCN5A |
| Short QT syndrome | KCNH2, KCNQ1, KCNJ2 |
| Hypertrophic cardiomyopathy | MYBPC3, MYH7, TNNI3, TNNT2, TPM1 |
| Arrhythmogenic ventricular cardiomyopathy | DSC2, DSG2, DSP, JUP, PKP2, TMEM43 |
| Dilated cardiomyopathy (with conduction system disease) | TTN, (LMNA, SCN5A) |
| Left ventricular noncompaction cardiomyopathy | LBD3 |
| Restrictive cardiomyopathy | MYH7, TNNI3 |
| Sudden unexplained death & sudden infant death syndrome | KCNQ1, KCNH2, SCN5A, RYR2 |
| Atrial fibrillation | Predominant forms are polygenic |
The reader is referred to other sources for comprehensive reviews on the genetic bases of these conditions. 1-8
Recent technological advances in DNA sequencing have enabled insights into human biology and disease that were not previously feasible, and have led to the commercialization of genetic testing now widely available in clinical practice. In particular, high-throughput sequencing techniques (also referred to as next generation sequencing) have been developed that allow large-scale and rapid assessment of entire genomes. Although next generation sequencing represents a major technological advance, it has introduced numerous challenges with respect to the interpretation of genetic variation, and has unleashed a flood of biological data of unknown clinical significance to practitioners.
In this review, we discuss current genetic testing indications for inherited arrhythmia syndromes, broadly outline characteristics of next generation sequencing techniques, and highlight challenges associated with such testing. We also discuss future directions relevant to the application of genetic testing in the clinical management of patients with inherited arrhythmia syndromes.
Genetic basis of arrhythmia syndromes
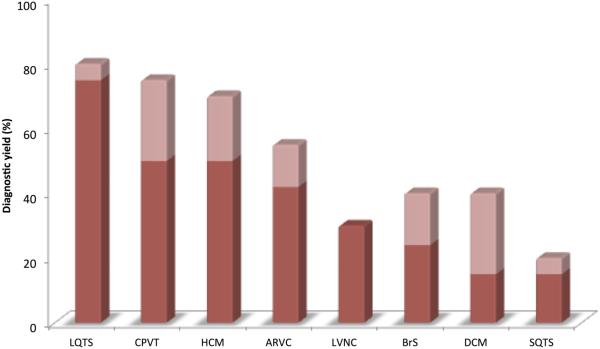
Over the past three decades, causal genes have been identified for most recognized inherited arrhythmia syndromes (Table 1). Discovered mutations in ion channel subunits governing cardiac electrical function, in sarcomeric proteins critical for cardiac contractile and structural integrity, and in other genes have informed our understanding of cardiac development and physiology. Yet currently recognized disease susceptibility genes, summarized elsewhere,1-8 represent merely a partial list of causal genes underlying these conditions. Indeed, conventional genetic testing for known genes underlying these conditions has been associated with incomplete diagnostic yield (Figure 1).9
Figure 1. The diagnostic yield of genetic testing varies by condition.
Percentage yields are drawn from Sturm and Hershberger.9 Dark red represents the lower bounds of diagnostic yield estimates, and the light red represents upper bounds of estimates. The potential yield of genetic testing is expected to rise with genetic testing approaches that encompass more genes.
LQTS = long QT syndrome; CPVT = catecholaminergic polymorphic ventricular tachycardia; HCM = hypertrophic cardiomyopathy; ARVC = arrhythmogenic ventricular cardiomyopathy; LVNC = left ventricular noncompaction cardiomyopathy; BrS = Brugada Syndrome; DCM = dilated cardiomyopathy; SQTS = short QT syndrome
Genetic mapping of inherited arrhythmia syndromes is associated with limited power owing to the incomplete penetrance, variable expressivity, and locus heterogeneity characteristic of these conditions. For example, in Brugada syndrome, not all SCN5A mutation carriers in an affected family manifest the disorder (incomplete penetrance), and often the disorder does not manifest until adulthood (age-dependent penetrance),10 making it difficult to understand whether a genetic variant is truly disease-causing or not. Mutations in some genes, such as those in SCN5A, may manifest with a wide array of phenotypes (variable expressivity) such as long QT syndrome, Brugada syndrome, cardiomyopathy, atrial fibrillation, and conduction system disease,11 which similarly complicates efforts to discern whether a variant is truly pathogenic. Some inherited conditions may be caused by genetic variation in any number of genes (locus heterogeneity), as is the case with dilated cardiomyopathy.12 Furthermore, large and multigenerational families with inherited arrhythmia syndromes affecting multiple individuals are rare, making it challenging to robustly associate a given genetic locus segregating with disease.
As such, knowledge of the genetic underpinnings of many arrhythmia syndromes remains incomplete. Genetic testing for inherited arrhythmia syndromes is therefore imperfect, with current tests generally having limited sensitivity for the detection of disease causing variants underlying these conditions.
Establishing the rationale for performing genetic testing
Broadly, genetic testing is used for diagnostic, predictive, therapeutic, pharmacogenetic, preimplant testing, newborn screening, and forensic applications. In adult cardiovascular medicine, genetic testing for inherited arrhythmia syndromes is most often reserved for diagnostic, predictive, or therapeutic applications.
The clinical utility of genetic testing for inherited arrhythmia syndromes is most relevant for confirmation of a suspected condition, or cascade screening in relatives of probands (index affected family members) that carry a disease-causing mutation. Some examples exist whereby mutation characteristics may influence prognosis, as in the case of mutations in pore forming (KCNH2)13 and transmembrane spanning regions (KCNQ1)14 in long QT syndrome. However, in aggregate, the prognostic features of specific genotypes currently remain limited. There are some data that suggest treatment may differ based on a particular genotype,15-18 most notably in the context of long QT syndrome type 3, where mutations leading to persistent late inward sodium current (INa) may warrant treatment with a sodium channel blocker.15 Future research may expand our understanding of the prognostic and therapeutic implications of specific genetic testing findings.
Inherited arrhythmia conditions for which genetic testing is indicated
In recent years consensus guidelines have emerged that provide recommendations for genetic testing for suspected inherited arrhythmia syndromes.1,2 Whereas these guidelines provide general consensus as to when testing may or may not be medically indicated, it is acknowledged that a variety of disease-specific, patient-level, and external factors must be weighed to determine the appropriateness of genetic testing in any given individual.9,19 In Table 2, we have summarized current indications for genetic testing in the context of inherited arrhythmia syndromes.
Table 2.
Aggregated consensus guideline indications for genetic testing in inherited arrhythmia syndromes.
| Condition | Diagnosis / Strong suspicion |
Relative is index case with known mutation |
Reference |
|---|---|---|---|
| Long QT syndrome | I1 / IIb2 | I | 1 |
| Catecholaminergic polymorphic ventricular
tachycardia |
I | I | 1 |
| Brugada syndrome | IIa | I | 1 |
| Cardiac conduction system disease | IIb3 | I | 1 |
| Short QT syndrome | IIb | I | 1 |
| Hypertrophic cardiomyopathy | I | I | 1,2 |
| Arrhythmogenic ventricular cardiomyopathy | IIa4 / IIb5 | I | 1 |
| Dilated cardiomyopathy | I6 / IIa7 | I | 1 |
| Left ventricular noncompaction cardiomyopathy | IIa | I | 1 |
| Restrictive cardiomyopathy | IIb | I | 1 |
| Out of hospital cardiac arrest | I8 | – | 1 |
| Sudden unexplained death & sudden infant death
syndrome |
IIb9 | I | 1 |
| Atrial fibrillation | – | – | 1 |
Strong clinical suspicion, or asymptomatic with QTc > 500 (adult) or > 480 (prepuberty) on repeated occasions
Asymptomatic with QTc > 480 (adult) or >460 (prepuberty)
Isolated CCD or with concomitant congenital heart disease, especially with family history of cardiac conduction system disease
Task force clinical diagnosis of arrhythmogenic ventricular cardiomyopathy
Task force possible arrhythmogenic ventricular cardiomyopathy
With conduction disease or family history of sudden death
To confirm diagnosis with familial dilated cardiomyopathy
If exam suggests channelopathy or cardiomyopathy
If evaluation suggests long QT syndrome or catecholaminergic polymorphic ventricular tachcyardia
These indications represent a summary of class I and II indications based on consensus guideline statements. 1,2
Overview of genetic sequencing techniques
The human genome is comprised of about 3 billion nucleotide pairs, and contains about 20,000 known genes.20-22 Each gene is comprised of both protein coding (exons) and noncoding (e.g., introns, promoter region, etc.) sequences. Protein-coding regions comprise about 1% of the human genome. Despite widespread and increasing recognition that non-coding portions of the genome have important regulatory properties and may participate in disease pathogenesis,23,24 clinical genetic testing for inherited arrhythmia syndromes classically is targeted and focuses on protein-coding regions.
Commercial genetic sequencing techniques have changed in recent years as a result of high-throughput technology. These changes have impacted the numbers of genes available for interrogation when ordering genetic testing. To provide the reader with an understanding of the scope of sequencing available when ordering tests today, we briefly discuss the technical properties of traditional sequencing and contrast it with high-throughput sequencing. Although additional methods for genetic variant detection exist, we will focus our discussion by contrasting Sanger and next generation sequencing techniques, which are most commonly employed for genetic testing in inherited arrhythmia syndromes.
Sanger sequencing
Causal gene discovery was traditionally performed using a combination of linkage mapping in affected families followed by candidate gene sequencing within an identified disease susceptibility locus. This approach has been a successful mainstay of causal variant and gene discovery for inherited arrhythmia syndromes for the past several decades. The ability to accurately decipher the coding sequences of genes within a genomic locus of interest has largely relied on advances described in 1977 by Allan Maxam and Walter Gilbert,25 and by Frederick Sanger.26 These methods were described about 25 years after Watson and Crick reported on the double-stranded helical structure of DNA.27
Sanger sequencing involves the competitive process of both synthesis and termination of DNA templates of interest using 2’deoxynucleotides (dNTPs) and 2’3’-dideoxynucleotides (ddNTPs) using DNA polymerase, respectively. The process of synthesis and termination creates a population of sequences of different lengths, which can be separated by size using gel electrophoresis. Typically, the ddNTPs are tagged with a fluorescent dye specific to that nucleotide, which produce detectable and specific emissions in response to excitation by a laser within the sequencer. The specific emissions thus reflect the individual nucelotides in the sequenced templates, and after transformation and processing, yield the sequences of the DNA templates.
Desirable qualities of Sanger sequencing include the high quality and accuracy of generated reads. Indeed, Sanger sequencing was used to create the initial genome in the Human Genome Project20,21 and remains the standard for definitive confirmation of single genetic variants. Nevertheless, Sanger sequencing has several limitations, including those related to inefficient excitation of the ddNTP fluorescent dyes, and ineffective detection or discrimination of the emitted signals.28 Sanger sequencing is time-consuming and expensive, which has traditionally relegated its application to targeted sequencing, rather than to high-throughput whole exome or whole genome sequencing. While Sanger sequencing is reasonably priced when performed at a small scale such as screening a limited set of genes, it remains far too slow and expensive when scaled to exomes or genomes.
Next generation sequencing
Recent technological advances have enhanced the scale of sequencing, allowing for the assessment of nucleotide variants across the genome with rapidity and relatively low cost. Next generation sequencing describes a number of different techniques, which collectively generate massive amounts of sequence reads that are aligned to a backbone reference genome for subsequent variant assessment. A full review of the chemistry and sequencing approaches is beyond the scope of this review, but excellent resources describing various sequencing approaches have been summarized elsewhere.29,30 Below, we have described one specific next generation sequencing technology as an illustration.
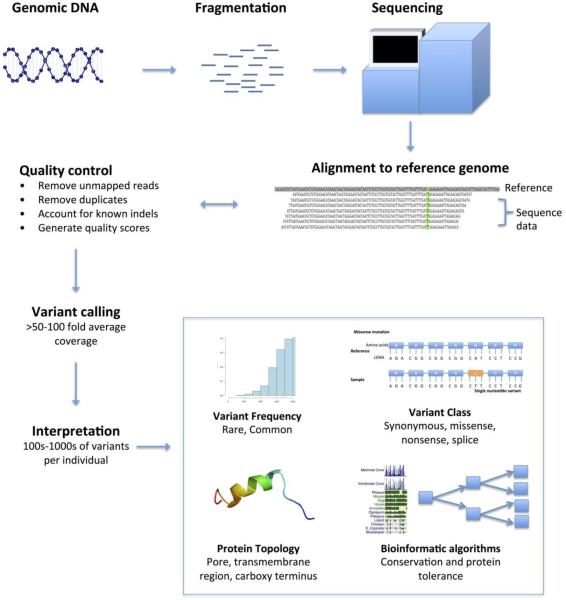
Next generation sequencing (Figure 2) approaches involve a combination of DNA template preparation, sequencing, and data analysis. First, DNA template preparation involves the fragmentation of genomic DNA into small segments, in contrast to Sanger sequencing in which specific templates of interest are often amplified using the polymerase chain reaction. Second, after fragmentation of genomic DNA, DNA templates are immobilized to a solid-state medium to enable multiple simultaneous sequencing reactions. In some cases, the DNA templates are clonally amplified, whereas other approaches rely on single molecule templates generated from the fragmentation step. Sequencing technologies vary substantially, but broadly involve the extension of a DNA template with detection of the added nucleotides through an imaging mechanism that is specific to the individual nucleotides. These imaging mechanisms may rely on the detection of dye or fluorescently-labeled nucleotides, detection of light energy released from pyrophosphates during nucleotide incorporation, or other approaches. Reactions are often repeated 50 or more times to generate sufficient coverage to provide confidence in the observed alignments and reads later performed. Third, data analysis involves the computational assembly of the many short DNA sequence reads by aligning them to a reference genome backbone, and subsequently determining the presence of genomic variation. This latter step requires considerable computational effort.31
Figure 2. Illustration depicting the flow of next generation sequencing data.
The figure displays the basic processing steps involved in the performance and interpretation of next generation sequencing for an isolated individual undergoing exome sequencing. In addition to using bioinformatic annotations, pathogenicity of identified variants can be inferred by using information on functional characterization information and family segregation, if available.
Major advantages of next generation sequencing include the capacity and efficiency of the approach, which has enabled the application of the technology to families and large study samples at a massive scale. Next generation sequencing can be applied to targeted panels of genes (e.g., 5-15 genes for conditions such as arrhythmogenic cardiomyopathy or long QT syndrome), comprehensive panels (e.g., 20-80 genes for comprehensive arrhythmia or cardiomyopathy tests), whole exomes, and even whole genomes.
Despite the promise, numerous limitations of the next generation sequencing exist. For example, exome sequencing for genetic diagnostics faces several challenges,32 some of which are summarized here. By definition, many genomic regions of interest are not included in commercially available capture methods (e.g., promoter regions, intronic enhancers, other functional noncoding regions). False negative findings may also occur because of technical limitations resulting in errant alignment of reads, insertions or deletions, or poor capture or sequencing over a particular gene or region. Indeed, relatively lower depth of coverage, or the number of times that a particular region of interest is interrogated, has been observed in genes such as KCNH2 with exome sequencing.33 Higher coverage results in more reliable discovery of genetic variation. Variability in sequencing coverage throughout the genome results from the facts that probes designed to amplify or immobilize specific exons do not have uniform specificity, and sequencing efficiency is not uniform across all DNA templates. Furthermore, capture probes are designed only for regions of the genome for which we have adequate templates, yet many segments of the genome remain difficult to ascertain.
Interpretation of results
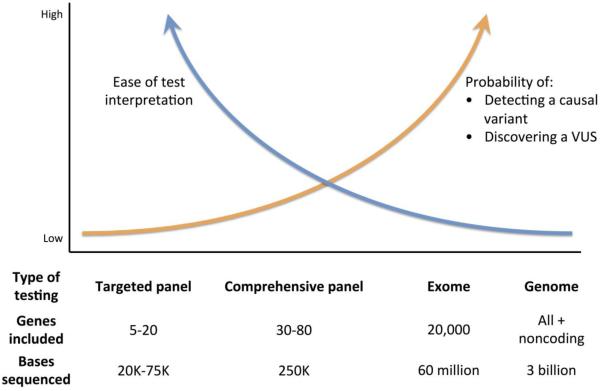
Although interpretation of sequencing results is challenging irrespective of the approach chosen, the massive amount of data generated from next generation sequencing poses a specific challenge unto itself. Sequencing of an exome or the protein coding region of the genome typically identifies ~200 novel protein altering, single nucleotide variants per individual,34 which leads to major challenges with respect to data manipulation and interpretation. Many of these variants are of unknown significance, underscoring the tradeoff between test sensitivity and intepretability that occurs when more comprehensive sequencing panels, such as whole exomes, are employed (Figure 3).
Figure 3. Schematic displaying the tradeoffs between increasingly comprehensive sequencing panels available with next generation sequencing.
The numbers of bases sequenced using each sequencing approach are estimates. VUS = variant of unknown significance
A full-length review of variant interpretation is beyond the scope of this article but can be found elsewhere.35 Here we briefly summarize general features of variants that suggest pathogenicity. Specific features utilized to infer deleteriousness include rarity, occurrence of variants at nucleotide or amino acid positions that are conserved across species, deleterious classifications (as in the case of nonsense mutations), and location within functional domains of proteins. In addition to these and many other bioinformatic considerations, pathogenicity is supported by the presence of segregation with disease within a family, and by evidence of a functional effect in model systems (although functional characterization of variants is not feasible in the clinical diagnostic settings). Inferring pathogenicity based on the integration of these diverse data elements is a somewhat subjective process,36 which introduces potential bias in the interpretation of observed variants. The extent to which subjectivity contributes to differences in interpretation of variants as pathogenic between research or clinical laboratories is unknown.
Defining and annotating variant pathogenicity is a major problem for inherited arrhythmia syndromes. Whereas repositories of patients with inherited arrhythmia syndromes have contributed to our understanding of the distribution of variation in these conditions,37-39 recent publications have called into question the pathogenicity of variants that previously were thought to be causally related to arrhythmia syndromes.40-42 As such, pathogenicity is subject to changes and modifications over time as more knowledge is gained. How that information will be communicated to clinicians, and what the clinician’s responsibility is with respect to periodically updating the annotation information, is unclear at the moment.
In summary, determining pathogenicity of identified variants is best performed in the context of information demonstrating reliable co-segregation of a particular variant with disease in a family. Furthermore, robust functional data supporting the pathogenicity of specific variants, while rare and often infeasible in clinical settings, lends credence to the deleterious nature of a variant. In the future, large and well-annotated databases of deleterious genetic variation will facilitate the interpretation of observed genetic variation generated from sequencing, whether performed in the clinical setting or research laboratory.
Practical genetic testing in the clinic
The advent of high-throughput sequencing has produced a number of commercially available sequencing options that include a) targeted panels that range from a handful or more genes, b) broad panels containing dozens of genes for a class of traits such as pan-cardiomyopathy or pan-arrhythmia, c) whole exomes, and d) whole genomes. Additional commercially available options for testing include insertion or deletion tests, as well as targeted single variant testing (which is typically reserved for testing whether a relative of an affected carrier also carries a variant of interest). The advantage of the more comprehensive tests is the potential for increased sensitivity of variant detection. The major drawbacks of more comprehensive testing include potentially increased costs, increased detection of genetic variants of unknown clinical significance, and decreased interpretability (Figure 3). It is also worth mentioning that comprehensive testing may be associated with a variety of incidental findings, ranging from variants that potentially affect drug response to variants that increase risk of non-cardiovascular diseases (e.g., dementia, breast cancer).43-45 As such, targeted testing currently remains the cornerstone of testing in the clinical setting.46
Determining which of the many commercially available targeted testing options to choose from depends on the purpose of the test, pretest probability of disease, and the specific test characteristics. Since the yield of genetic testing varies by the suspected inherited arrhythmia syndrome (Figure 1), having a high pretest probability for a specific condition and understanding the genetic basis of that particular condition are essential. For example, initial testing for conditions with relatively high diagnostic yields such as long QT syndrome, catecholaminergic polymorphic ventricular tachycardia, and hypertrophic cardiomyopathy need not include a broad sequencing panel, considering that most affected individuals have variation in only one of a small number of known genes. In contrast, more comprehensive testing may be appropriate for diseases with substantial locus heterogeneity, such as dilated cardiomyopathy, though this approach requires sufficient expertise to interpret variants of unknown significance and provide proper counseling to patients. In some cases, the specific genes included in a panel may vary from one laboratory to another, and may influence decisions about which test to order (Table 3).
Table 3.
Comparison of genetic testing panels between select companies for several different phenotypes, excluding whole exomes and genomes.
|
Transgenomic
www.transgenomic.com |
GeneDx
www.genedx.com |
Correlagan
www.correlagen.com |
Partners
Laboratory for Molecular Medicine www.personalizedmedicine.partners.org/Laboratory-For-Molecular-Medicine/Default.aspx |
Genes in
common |
|
|---|---|---|---|---|---|
|
| |||||
| Number of genes tested in panel | |||||
| Long QT syndrome | 13 | 12 | – | – | 12 |
| Brugada syndrome | 9 | 7 | – | – | 7 |
| Hypertrophic cardiomyopathy |
12 | 18 | – | 18 | 12 |
| Dilated cardiomyopathy | 13 | 38 | 6 | 27 | 6 |
| Arrhythmogenic ventricular cardiomyopathy |
5 | 7 | 5 | 8 | 5 |
| Pan-cardiomyopathy panel | – | 76 | – | 51 | 45 |
| Pan-arrhythmia panel | 25 | 30 | – | – | 23 |
Here we would like to emphasize that the decision to perform genetic testing can be complex, and extends beyond the technical aspects of different sequencing panels. Prior to performing testing, it is advised that pre-test counseling be performed to articulate such issues as the goals, potential consequences, and limitations of genetic testing, as well as address how the results will specifically alter each individual’s care. Similarly, after test results have been returned, it is important to summarize with patients, and often their relatives, how the findings may impact them and what the next steps are in light of the knowledge gained. Given these complexities, it is recommended that genetic testing be performed in centers with experience in the counseling of patients and interpretation of genetic testing results.
Future directions / challenges
Technological innovations have improved the extent to which we can now probe the genome, both in research and clinical settings. However several obstacles remain with respect to the appropriate use of next generation sequencing and other technologies in clinical settings. Among these challenges are cost, technological hurdles, determination of variant pathogenicity, and legal and ethical conundrums. The utility of genetic sequencing in the clinic, particularly in the area of complex or polygenic diseases, is of unclear utility. Existing challenges and potential future directions are summarized in Table 4.
Table 4.
Challenges and future directions in the application of next generation and other sequencing technologies in practice.
| Challenge | Problem | Potential solutions / future directions |
|---|---|---|
| Costs |
|
|
| Genome coverage |
|
|
| Determining variant pathogenicity |
|
|
| Legal and ethical considerations |
|
|
| Role of genetic variation in complex (polygenic) diseases |
|
|
Conclusions
The efficiency of next generation sequencing promises to elucidate the genetic contributions to many diseases. Next generation sequencing has facilitated the development of various different sequencing options to choose from when testing patients with inherited arrhythmia syndromes. Yet the implementation of next generation sequencing into clinical practice carries unique challenges. The data generated from this tool requires thoughtful analysis in order to be of practical use. With improvements in cost, further advances in sequencing technology, larger reference genome data banks for comparison, and improved functional assessment of identified variants in coming years, next generation sequencing is likely to become a familiar instrument in the routine management of patients with suspected inherited arrhythmia syndromes.
Acknowledgments
Funding: This work was supported by grants from the National Institutes of Health (1RO1HL092577, 1RO1HL104156, 1K24HL105780, HL065962). Dr. Lubitz is also supported by NIH/NHLBI K23HL114724 and a Doris Duke Charitable Foundation Clinical Scientist Development Award 2014105. Dr. Ellinor is also supported by an Established Investigator Award from the American Heart Association (13EIA14220013) and by support from the Fondation Leducq (14CVD01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: None
Contributor Information
Steven A. Lubitz, Cardiac Arrhythmia Service and Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA.
Patrick T. Ellinor, Medical and Population Genetics Program, The Broad Institute, Cambridge, MA.
References
- 1.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart rhythm : the official journal of the Heart Rhythm Society. 2011 Aug;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 2.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011 Dec 13;124:2761–2796. doi: 10.1161/CIR.0b013e318223e230. [DOI] [PubMed] [Google Scholar]
- 3.Hofman N, Tan HL, Alders M, Kolder I, de Haij S, Mannens MM, Lombardi MP, Dit Deprez RH, van Langen I, Wilde AA. Yield of molecular and clinical testing for arrhythmia syndromes: report of 15 years' experience. Circulation. 2013 Oct 1;128:1513–1521. doi: 10.1161/CIRCULATIONAHA.112.000091. [DOI] [PubMed] [Google Scholar]
- 4.Wilde AA, Behr ER. Genetic testing for inherited cardiac disease. Nat Rev Cardiol. 2013 Oct;10:571–583. doi: 10.1038/nrcardio.2013.108. [DOI] [PubMed] [Google Scholar]
- 5.Crotti L, Marcou CA, Tester DJ, Castelletti S, Giudicessi JR, Torchio M, Medeiros-Domingo A, Simone S, Will ML, Dagradi F, Schwartz PJ, Ackerman MJ. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012 Oct 9;60:1410–1418. doi: 10.1016/j.jacc.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Priori SG, Napolitano C, Memmi M, et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002 Jul 2;106:69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 7.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012 Aug 21;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 8.Marcus FI, Edson S, Towbin JA. Genetics of arrhythmogenic right ventricular cardiomyopathy: a practical guide for physicians. J Am Coll Cardiol. 2013 May 14;61:1945–1948. doi: 10.1016/j.jacc.2013.01.073. [DOI] [PubMed] [Google Scholar]
- 9.Sturm AC, Hershberger RE. Genetic testing in cardiovascular medicine: current landscape and future horizons. Curr Opin Cardiol. 2013 May;28:317–325. doi: 10.1097/HCO.0b013e32835fb728. [DOI] [PubMed] [Google Scholar]
- 10.Nannenberg EA, Sijbrands EJG, Dijksman LM, Alders M, van Tintelen JP, Birnie M, van Langen IM, Wilde AAM. Mortality of Inherited Arrhythmia Syndromes; Insight into Their Natural History. Circulation: Cardiovascular Genetics. 2012 Mar 28; doi: 10.1161/CIRCGENETICS.111.961102. [DOI] [PubMed] [Google Scholar]
- 11.Wilde AA, Brugada R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ Res. 2011 Apr 1;108:884–897. doi: 10.1161/CIRCRESAHA.110.238469. [DOI] [PubMed] [Google Scholar]
- 12.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014 Aug;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 13.Moss AJ, Zareba W, Kaufman ES, et al. Increased risk of arrhythmic events in long-QT syndrome with mutations in the pore region of the human ether-a-go-go-related gene potassium channel. Circulation. 2002 Feb 19;105:794–799. doi: 10.1161/hc0702.105124. [DOI] [PubMed] [Google Scholar]
- 14.Moss AJ, Shimizu W, Wilde AA, et al. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007 May 15;115:2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Priori SG, Wilde AA, Horie M, et al. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013 Dec;10:1932–1963. doi: 10.1016/j.hrthm.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Laurent G, Saal S, Amarouch MY, et al. Multifocal ectopic Purkinje-related premature contractions: a new SCN5A-related cardiac channelopathy. J Am Coll Cardiol. 2012 Jul 10;60:144–156. doi: 10.1016/j.jacc.2012.02.052. [DOI] [PubMed] [Google Scholar]
- 17.Mann SA, Castro ML, Ohanian M, et al. R222Q SCN5A mutation is associated with reversible ventricular ectopy and dilated cardiomyopathy. J Am Coll Cardiol. 2012 Oct 16;60:1566–1573. doi: 10.1016/j.jacc.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 18.Nair K, Pekhletski R, Harris L, Care M, Morel C, Farid T, Backx PH, Szabo E, Nanthakumar K. Escape capture bigeminy: phenotypic marker of cardiac sodium channel voltage sensor mutation R222Q. Heart Rhythm. 2012 Oct;9:1681–1688. doi: 10.1016/j.hrthm.2012.06.029. e1681. [DOI] [PubMed] [Google Scholar]
- 19.Ashley EA, Hershberger RE, Caleshu C, et al. Genetics and cardiovascular disease: a policy statement from the American Heart Association. Circulation. 2012 Jul 3;126:142–157. doi: 10.1161/CIR.0b013e31825b07f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001 Feb 16;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 21.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 22.International Human Genome Sequencing C Finishing the euchromatic sequence of the human genome. Nature. 2004 Oct 21;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 23.Consortium EP, Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007 Jun 14;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012 Sep 6;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 28.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005 Dec;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 29.Hutchison CA., 3rd DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 2007;35:6227–6237. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010 Jan;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 31.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 May;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011 Nov;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 33.Bagnall RD, Das KJ, Duflou J, Semsarian C. Exome analysis-based molecular autopsy in cases of sudden unexplained death in the young. Heart Rhythm. 2014 Apr;11:655–662. doi: 10.1016/j.hrthm.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 34.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011 Sep;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 36.Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am J Hum Genet. 2013 Oct 3;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapplinger JD, Landstrom AP, Salisbury BA, et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011 Jun 7;57:2317–2327. doi: 10.1016/j.jacc.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010 Jan;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapplinger JD, Tester DJ, Salisbury BA, Carr JL, Harris-Kerr C, Pollevick GD, Wilde AA, Ackerman MJ. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009 Sep;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Refsgaard L, Holst AG, Sadjadieh G, Haunso S, Nielsen JB, Olesen MS. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur J Hum Genet. 2012 Aug;20:905–908. doi: 10.1038/ejhg.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreasen C, Nielsen JB, Refsgaard L, Holst AG, Christensen AH, Andreasen L, Sajadieh A, Haunso S, Svendsen JH, Olesen MS. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013 Sep;21:918–928. doi: 10.1038/ejhg.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jabbari J, Jabbari R, Nielsen MW, Holst AG, Nielsen JB, Haunso S, Tfelt-Hansen J, Svendsen JH, Olesen MS. New exome data question the pathogenicity of genetic variants previously associated with catecholaminergic polymorphic ventricular tachycardia. Circ Cardiovasc Genet. 2013 Oct;6:481–489. doi: 10.1161/CIRCGENETICS.113.000118. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014 Nov 12;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabor HK, Auer PL, Jamal SM, Chong JX, Yu JH, Gordon AS, Graubert TA, O'Donnell CJ, Rich SS, Nickerson DA, Project NES, Bamshad MJ. Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am J Hum Genet. 2014 Aug 7;95:183–193. doi: 10.1016/j.ajhg.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013 Jul;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013 Apr;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]