Abstract
A series of eleven conformationally restricted, 4-substituted 2,6-dimethylfuro[2,3-d]pyrimidines was designed to explore the bioactive conformation required for dual inhibition of microtubule assembly and receptor tyrosine kinases (RTKs), and their biological activities are reported. All three rotatable single bonds in the lead compound 1 were sequentially restricted to address the role of each in SAR for microtubule and RTK inhibitory effects. Compounds 2, 3, 7 and 10 showed microtubule depolymerizing activity comparable to or better than the lead 1, some with nanomolar EC50 values. While compound 8 had no effect on microtubules, 8 and 10 both showed potent RTK inhibition with nanomolar IC50s. These compounds confirm that the bioactive conformation for RTK inhibition is different from that for tubulin inhibition. The tetrahydroquinoline analog 10 showed the most potent dual tubulin and RTK inhibitory activities (low nanomolar inhibition of EGFR, VEGFR2 and PDGFR-β). Compound 10 is highly potent activity against many NCI cancer cell lines, including several chemo-resistant cell lines, and could serve as a lead for further preclinical studies.
Introduction
Angiogenesis is required to provide the “life blood” of tumors, and, without a new blood supply, tumors cannot grow and metastasize.1 During carcinogenesis, an angiogenic switch occurs, and several angiogenic growth factors stimulate their receptor tyrosine kinases (RTKs) to initiate multiple pro-angiogenic events.2 A therapeutic strategy to inhibit these key angiogenic proteins or their RTKs was envisioned, 3–5 and multiple inhibitors targeting EGFR, VEGF and/or PDGFR-β among others are now used clinically. These RTKs are noted to have multi-kinase effects,6 and this appears to be important for optimal activities. Antiangiogenic therapies have proven to be useful clinically in combination with other approaches, and new agents continue to be developed.6
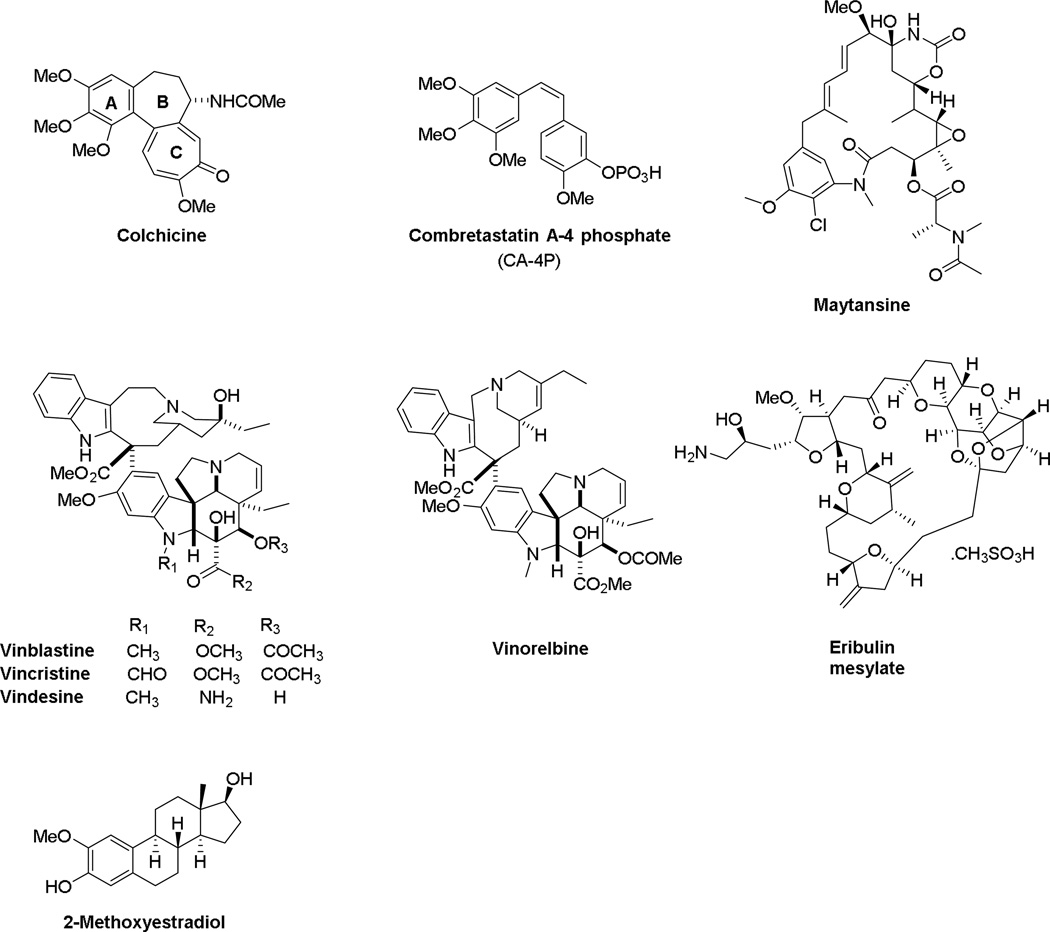
Turning to cytotoxic chemotherapy, tubulin binding agents are among the most successful anticancer drugs in clinical use (Figure 1).7 These compounds can be classified as microtubule stabilizers that stimulate tubulin polymerization or destabilizers that inhibit tubulin polymerization. The destabilizing agents bind to tubulin at different binding sites, including the vinca domain and the colchicine site.7 The vinca alkaloids, including vincristine, vinblastine, and vindesine (Figure 1), bind competitively within the vinca site. These vinca alkaloids have utility in the treatment of both solid and liquid tumors and are used in cancer therapy in both adults and children. Structurally diverse natural products and their analogs, including eribulin mesylate and maytansine (Figure 1) displace the vincas in a noncompetitive manner and they were assumed to bind within the vinca domain and initiate allosteric effects.8, 9 Very recent studies by the Steinmetz laboratory demonstrated that maytansine binds to a distinct microtubule depolymerizer site on β-tubulin that they have designated as the maytansine10 site. Occupancy of this site inhibits tubulin polymerization by preventing the addition of new subunits at the plus ends of the microtubule, a mechanism different from vinca site agents.10 These microtubule targeting agents have clinical utility, since a maytansine derivative is the payload in the antibody drug conjugate T-DM1 (ado-trastuzumab emtansine, Kadcyla®) and eribulin is used in the treatment of breast cancer. The colchicine site is a third non-overlapping microtubule destabilizer binding site on β-tubulin and is located at its interface with α-tubulin. While colchicine is too toxic for use in cancer therapy, a number of colchicine site agents have been evaluated for clinical activity, including 2-methoxyestradiol (2ME2), combretastatin A-4 phosphate (CA-4P) (fosbretastatin), the combretastatin CA-1P prodrug (OXi4503) and other closely related compounds.7, 11, 12 The interaction of colchicine site agents with tubulin is intriguing in that some colchicine site agents, notably 2ME2, were developed based on antiangiogenic effects, while others, including CA-4P, have antivascular effects that initiate rapid destruction of the tumor vasculature. Thus, while these colchicine site agents inhibit tubulin polymerization and cause microtubule depolymerization in cells, there are other mechanistic differences that are not fully understood.
Figure 1.
Structures of chemically diverse microtubule depolymerizing agents.
Multidrug resistance remains a major challenge in the curative treatment of cancer, and colchicine site agents have advantages over other microtubule targeting agents because most of them circumvent the P-glycoprotein (Pgp) and βIII tubulin-mediated resistance that have been implicated in limiting the clinical efficacy of other microtubule targeting agents.7, 13 In spite of this, no colchicine site agents have yet achieved FDA approval for anticancer therapy. This site has excellent potential for new drug discovery.
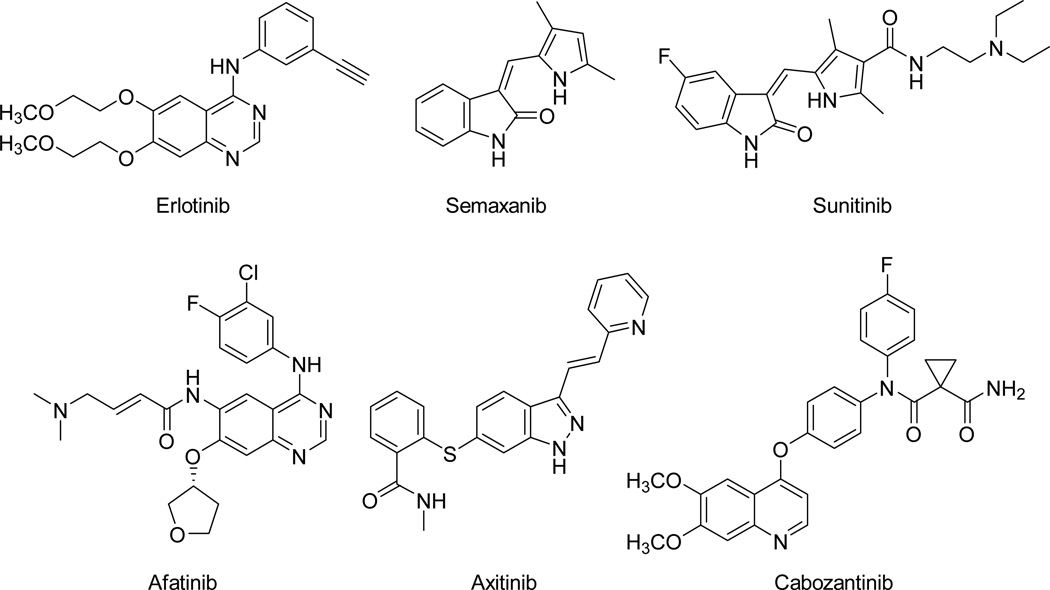
Returning to RTKs, vascular endothelial growth factor receptor (VEGFR), platelet derived growth factor receptor (PDGFR), epidermal growth factor receptor (EGFR) and related enzymes are inhibited by small molecules, such as erlotinib, semaxanib, sunitinib, afatinib, axitinib and cabozanitinb, that have considerable utility as targeted cancer chemotherapeutic agents (Figure 2).14–20 It is well established in the literature that these RTK inhibitors are cytostatic.14, 18, 19 Combination chemotherapy with RTK inhibitors as the antiangiogenic component along with cytotoxic chemotherapeutic agents are in clinical trials.15, 17, 20 Examples of such combinations currently in clinical trials include the combination of lapatinib with carboplatin, paclitaxel and trastuzumab in metastatic breast cancer21 and docetaxel, gemcitabine and pazopanib as treatment for soft tissue sarcoma22 among others.23 The advantages of combination chemotherapy, particularly with RTK inhibitors, addresses pathway redundancy,17 as well as tumor heterogeneity among other resistance mechanisms, and is beneficial when RTK inhibitors are combined with conventional cancer therapeutics.15, 17, 20
Figure 2.
Selected RTK inhibitors
We sought to combine RTK inhibitory and cytotoxic activities in single molecules to afford combination chemotherapeutics via single agents.24, 25 In keeping with the principles of combination chemotherapy25, 26 such single entities would act simultaneously at two or more distinct targets and prevent or delay the emergence of resistance, avoid drug-drug interactions, circumvent pharmacokinetic problems and overlapping toxicities that plague combination chemotherapy with two or more separate agents.26 In this study, we report the design, synthesis and biological activities of novel conformationally restricted bicyclic furo[2,3-d]pyrimidines, some of which possess potent activities against both tubulin and RTKs and potently inhibit cancer cells in culture.
Rationale
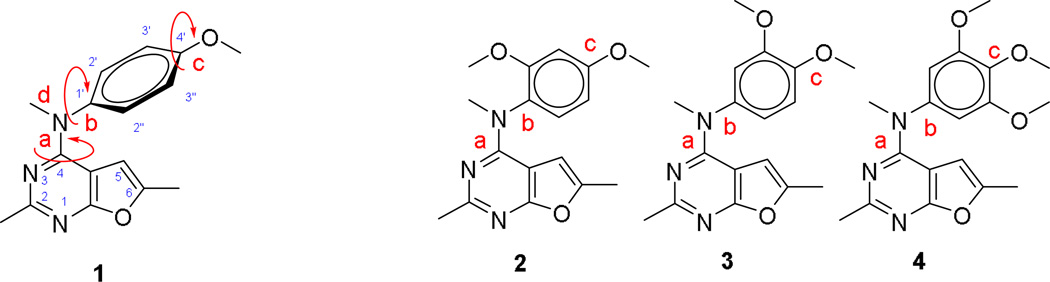
We25 recently reported the design and discovery of a series of novel bicyclic furo[2,3-d]pyrimidines, some of which possess both RTK and tubulin inhibitory properties and display potent in vivo antitumor activities. In this reported series of compounds, compound 1 (Figure 3) showed the most potent activities against both RTKs and tubulin. As shown in Figure 3, the bioactive conformation of 1 is determined by three rotatable single bonds: 4-position C-N bond (bond a), 1’-position C-N bond (bond b) and 4’-position C-O bond (bond c). Conformational analysis via molecular modeling and 1H NMR studies suggested that the methyl group on the aniline nitrogen in 1 restricted the free rotation of bond a as well as bond b (Figure 3) and thus restricts the conformation of the anilino ring.25 The potent activities of compound 1 against RTKs and tubulin were thought to be correlated with the conformational restriction in the molecule.25
Figure 3.
Structure and conformational analysis of compounds 1 – 4.
To further explore the bioactive conformation and improve the biological activity of 1, compounds 2–4 (Figure 3) were designed, in which the free rotation of the bonds a, b and c were sequentially restricted by replacement of a proton by a methoxy group. Our initial strategy to explore the bioactive conformation was to increase the energy barrier between the preferred conformation and non-preferred conformation by introducing bulky groups adjacent to the rotatable σ bonds… In compound 2 the additional 2’-methoxy group restricts the rotation of both b while rotation around bonds a and c are mostly unaffected. Compound 3 (Figure 3) was designed to restrict the free rotation of bond c by introducing an additional methoxy group at the 3’-position, . Using molecular modeling, we determined that compared to the 3’-proton in 1, the bulky 3’-methoxy group forces the methyl in the 4’-methoxy group to point away from the 3’-methoxy group. Introduction of methoxy groups at both the 3’ and 5’-positions in compound 4 (Figure 3), both of which are ortho to the 4’-position, severely restricts the rotation of bond c and forces the 4’-methoxy group to adopt a conformation that minimizes repulsive interactions with the ortho substitutions. Similar to 3, the rotation of bonds a and b are not affected in 4. We were aware that the additional methoxy groups in 2–4 may not only restrict the free rotation of single bonds but also provide extra binding or hindrance to binding due to steric and/or electronic properties with the targets. The choice of the methoxy moieties was based on the molecular modeling docked poses of our previous analog 1 and its 4’-methoxy group overlap with the methoxy groups of colchicine in its binding pocket in tubulin.25
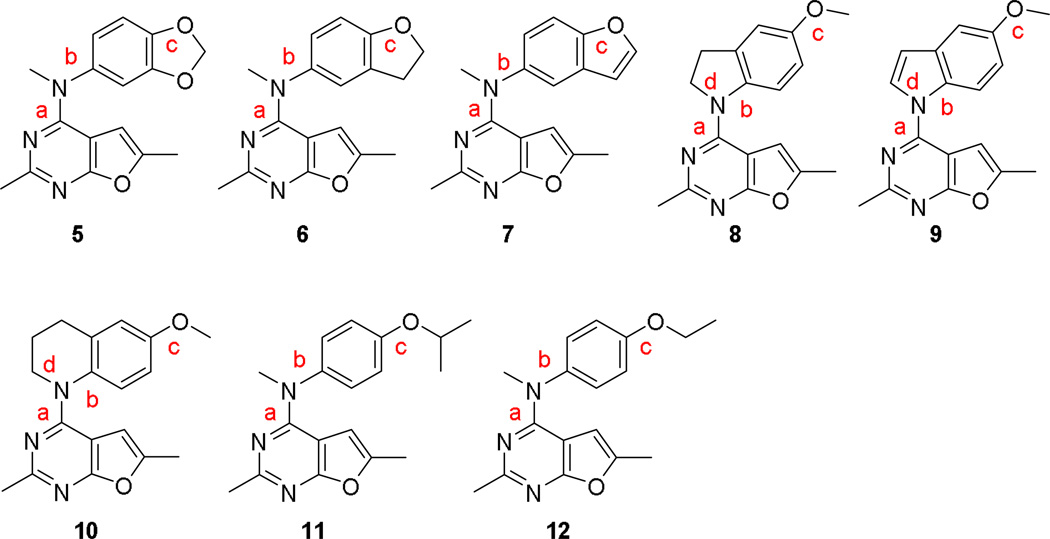
A second strategy to explore the influence of conformational restriction was to restrict the free rotation of single bonds by incorporating the bond into a ring. By changing the ring size and bond order, the conformation of the compounds can be further manipulated. With optimal ring size and bond order, the preferred conformation of the compound for activity could be defined. Compounds 5–10 (Figure 4) were designed using this strategy to explore the bioactive conformation.
Figure 4.
Structures of compounds 5–12.
In compounds 5–7, fused bicyclic systems were designed to replace the monocyclic 4’-methoxyphenyl group of 1. The 4’-methoxy group in 1 was converted to a methylenedioxy moiety as part of a fused bicyclic ring system. In compounds 5–7, bond c is locked into a fixed conformation, while rotation of bonds a and b are mostly unaffected. The methylenedioxy ring in 5, the dihydrobenzofuran in 6 and the benzofuran in 7 mimic the function of the 4’-methoxy group in 1 and could also provide additional binding interactions or steric hindrance with the target proteins.
Compounds 8–10 were designed to restrict the rotation of bonds a, b and d with free rotation of bond c unaffected. In 8, the N-methyl moiety and the phenyl ring of 1 is connected through an additional carbon providing a dihydroindole ring. The N-methyl group and the phenyl ring in 1 were incorporated into an indole ring to give compound 9. Connecting the N-methyl moiety and the phenyl ring via two carbon atoms affords the tetrahydroquinoline analog 10. Bond b in compounds 8–10 is restricted by incorporation into a fused bicyclic ring system and is no longer freely rotatable. The bulk of the 4-N-substitutions also restrict the rotation of bond a in 8–10. The dihydroindole (8), the tetrahydroquinoline (9), and indole (10) analogs have different ring size and increased ring rigidity, thus the fused phenyl ring in 8–10 was locked into somewhat different conformations, which may translate into increased potency and/or selectivity.
Compounds 11 and 12 introduce bulk at the 4’-position of the aniline moiety and were designed to explore the effect of bulk at that position. In addition, the 4’-isopropyl (11) and 4’-ethyl (12) could also affect rotation around bond c.
A systematic conformational search around the rotatable bonds of 1 (bonds a, b and c, Figure 3, 5° increments) and 10 (bonds a and c, Figure 4) using Sybyl X 2.1.127 showed a dramatic reduction in the number of conformations for 10 (1525 conformations)as compared to 1 (74,400 conformations), evidence of the expected conformational restriction due to the tetrahydroquinoline moiety. Similar reductions in the number of conformations were observed with the other restricted analogs (5 – 9, results not shown).
Molecular Modeling
Docking studies were performed for compounds 2 – 12 in the colchicine site of tubulin (PDB: 1SA028, 3.58Å), in VEGFR2 (PDB: 1YWN29, 1.71Å), in EGFR (PDB: 1M1730) and in a homology model of PDGFR-β24 using LeadIT 2.1.6 31 (validated by re-docking the crystal structure ligands) using previously reported methods.24, 25 Docking studies of the lead 125 and standard compounds (semaxanib for VEGFR2, erlotinib for EGFR and sunitinib for PDGFR-β) were performed using the previous methods. The docked poses were visualized using the software CCP4mg.32
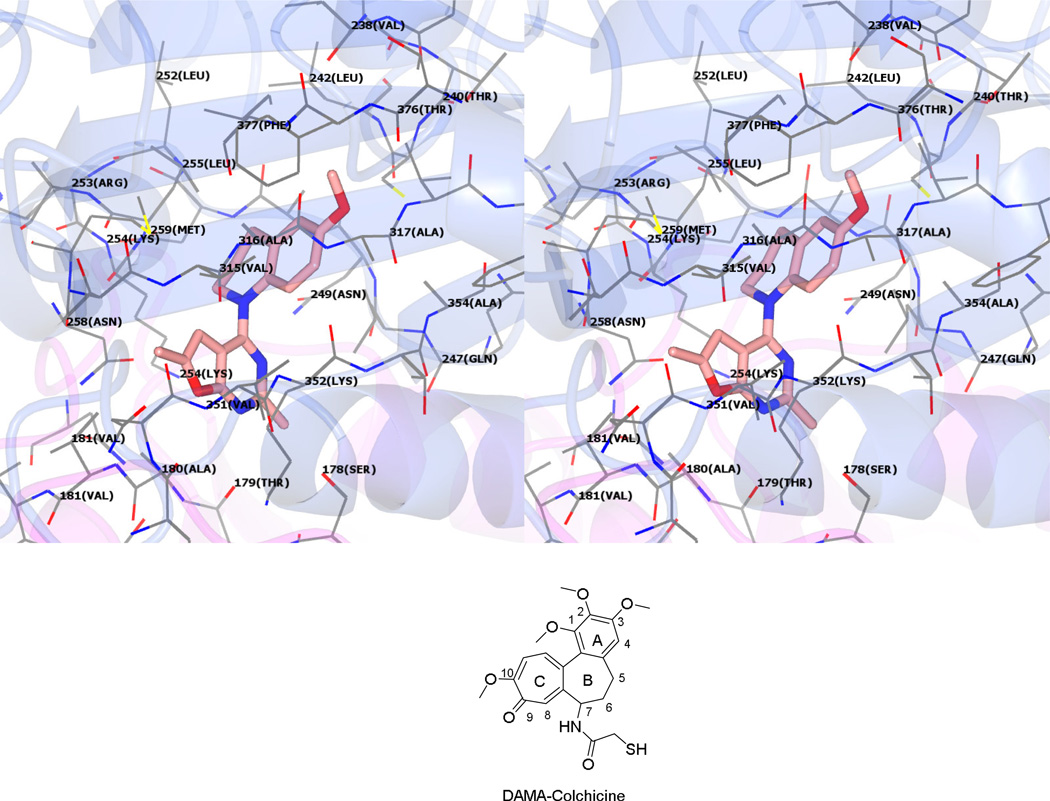
Multiple low-energy conformations (within 1 kcal/mol of the best pose) were obtained on docking 2 – 12 in the ATP site of the three target kinases and the colchicine site of tubulin. The docked conformation of 10 in the colchicine site of tubulin (Figure 5) is presented as a working model for compounds 2 – 12 based on their similarity to the bound conformation of N-deacetyl-N-(2-mercaptoacetyl)colchicine28 (DAMA-colchicine) (Figure 5, not shown in the model). The 6’-OMe phenyl group of 10 is oriented towards the triOMe containing A-ring of DAMA-colchicine and interacts with Leuβ246, Alaβ248, Leuβ253, Alaβ314, Ileβ376 and Valβ316. The 4’-OMe of 10 forms a hydrogen bond with Cysβ241, analogous to the hydrogen bond formed by the 3’-OMe group of DAMA-colchicine with Cysβ241. The tetrahydroquinoline ring of 10 occupies a region in space in proximity to the C5 and C6 of the B-ring of colchicine and is involved in hydrophobic interactions with Lysβ252, Alaβ248 and Leuβ246. The furo[2,3-d]pyrimidine scaffold of 10 partly overlaps with the C-ring of DAMA colchicine and forms hydrophobic interactions with Leuβ253, Asnβ256 and Lysβ250. Compound 10 had a docked score of −25 kJ/mol, which was better than the docked score of 1 (−23 kJ/mol). These results are in accord with results from recently published molecular modeling studies at the colchicine site of tubulin of our lead compound 1.25
Figure 5.
Stereoview. Docked pose of 10 in the colchicine site of tubulin. PDB: 1SA0.28
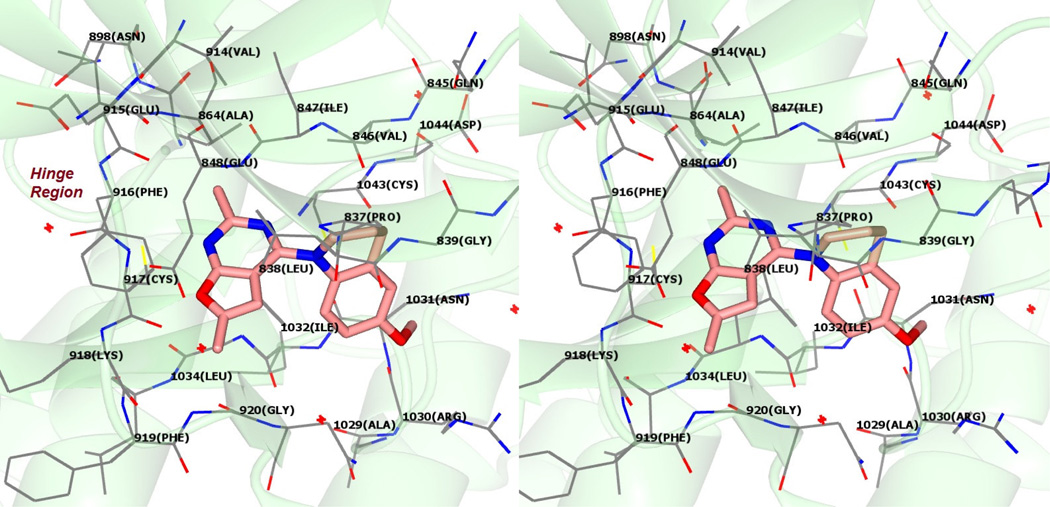
Figure 6 shows the docked pose of 10 in the ATP site of VEGFR2 (PDB: 1YWN29). The furo[2,3-d]pyrimidine scaffold of 10 is oriented parallel to the Hinge region amino acids (Figure 6) and occupies the adenine binding region of the ATP site of VEGFR2. The N1 nitrogen of 10 forms a hydrogen bond with the backbone NH of Cys917 in the Hinge region. Additionally, the scaffold is stabilized by hydrophobic interactions with Leu838 and Leu1033. The bulk of the tetrahydroquinoline moiety forces a vertical orientation of the molecule in the pocket and additionally interacts with the side chain carbon atoms of Ile1042 (not shown) and Cys1043. This vertical conformation orients the 6’-OMe moiety towards the sugar binding pocket in the ATP site of VEGFR2.29 The vertical orientation of the furo[2,3-d]pyrimidine scaffold of 10 is in contrast to the horizontal orientation of the purine ring of ATP in the ATP site of VEGFR2.25, 33 Compound 10 showed an improved docking score of −25 kJ/mol compared to 1 (−24 kJ/mol) in VEGFR2.
Figure 6.
Stereoview. Docked pose of 10 in VEGFR2. PDB: 1YWN29
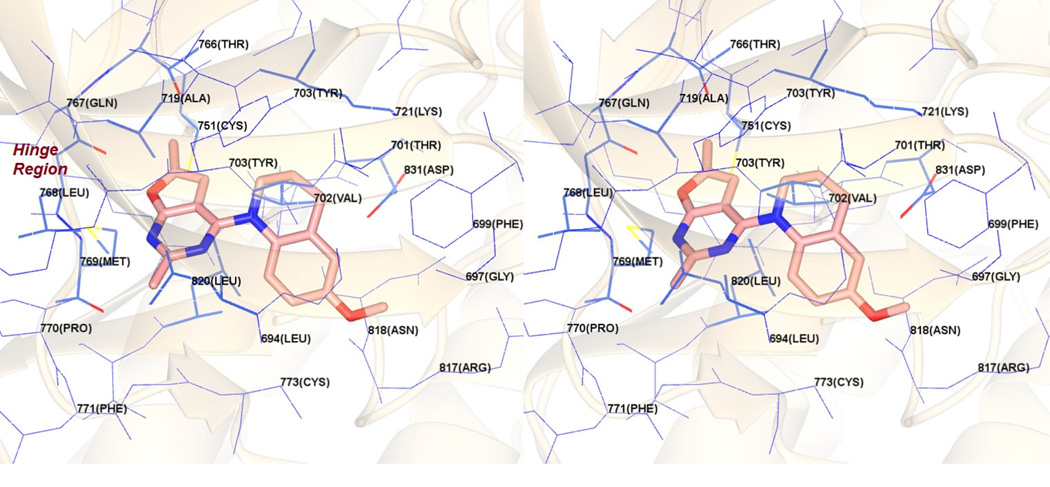
Figure 7 shows the docked pose of 10 in the ATP site of EGFR (PDB: 1M1730). As with the docked pose of 10 in VEGFR2, the furo[2,3-d]pyrimidine scaffold of 10 is oriented parallel to the Hinge region (Figure 7) due to the bulky tetrahydroquinoline ring and lies in the adenine binding region of the ATP binding site. The N1 nitrogen of 10 forms a hydrogen bond with the backbone NH of Met769 in the Hinge region. Additionally, the bicyclic scaffold forms hydrophobic interactions with Leu694, Met769, Leu820 and Met769. The 2-Me moiety interacts with Leu694 and Leu768 while the 6-Me moiety interacts with Cys751, Thr766, Leu768 and Met769. The tetrahydroquinoline moiety provides hydrophobic interactions with Phe699, Val702 and Lys721. In this conformation the 6’-OMe moiety lies in a polar pocket formed by the salt bridge between the side chains of Lys721 and Asp831. The vertical orientation of 10 is similar to that seen with the docked pose of 10 in VEGFR2 (Figure 6) and is a consequence of the steric hindrance afforded by the tetrahydroquinoline with the Hinge region amino acids. The docked score of 10 was −26 kJ/mol, better than the docking score of −24 kJ/mol for 1 in these studies.
Figure 7.
Stereoview. Docked pose of 10 in EGFR. PDB: 1M1730
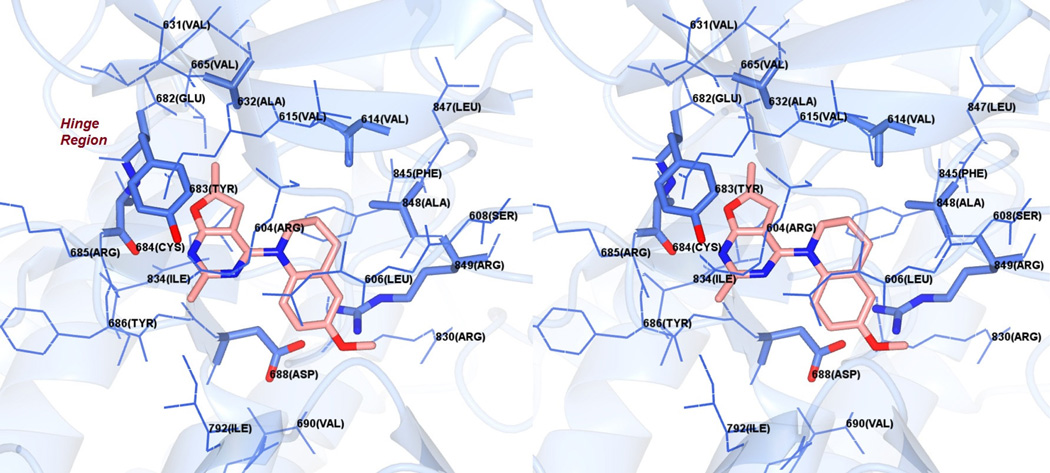
Figure 8 shows the docked pose of 10 in the ATP binding site of a PDGFR-β homology model.24 The furo[2,3-d]pyrimidine scaffold of 10 is once again oriented parallel to the Hinge region (Figure 8) and occupies the adenine region of the ATP site. The furo oxygen of 10 forms a hydrogen bond with the backbone NH of Cys684 in the Hinge region. The bicyclic scaffold forms hydrophobic interactions with Tyr683, Tyr686, Gly687 and Leu833 in the binding pocket. The 2-Me moiety interacts with Gly687, while the 6-Me moiety interacts with Ala632, Tyr683 and Leu833. The tetrahydroquinoline group interacts with Leu606 (not labeled), Val614 and Ala848. As is seen with the docked conformation of 10 in EGFR (Figure 7), the 6’-OMe moiety lies in a polar pocket formed by the salt bridge formed between the side chains of Arg849 and Asp688. The docked score of 10 was −20 kJ/mol, better than the docked score of −19 kJ/mol for 1 in these studies.
Figure 8.
Stereoview. Docked pose of 10 in a PDGFR-β homology model.24
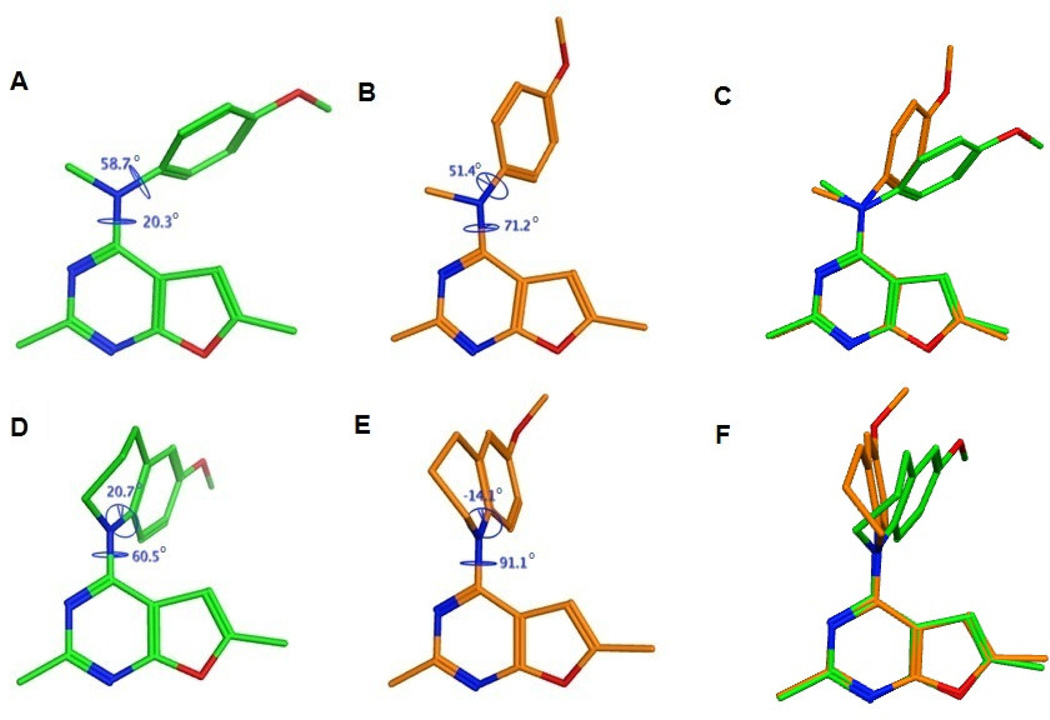
The conformations and energies of 1 and 10 in their lowest energy (Figure 9 A and D respectively) and the docked conformations in the colchicine site of tubulin (Figure 9 B and E respectively) are presented as representative examples of compounds 1 – 12. Superimposition of the docked and the energy minimum conformations of 1 and 10 (Figures 9C and 9F respectively) are also presented. Analysis of these conformations indicates that the docked conformation of 10 is energetically (ΔE between conformations D and E in Figure 9 is 0.42 kcal/mol) and conformationally closer to its lowest energy conformation compared to 1 (ΔE between conformations A and B in Figure 9 is 3.7 kcal/mol). Thus, compound 10 could be anticipated to bind more efficiently to the colchicine site of tubulin compared to 1 and was expected to exhibit increased potency. Similar results were predicted for the other conformationally restricted analogs 2 – 9 compared to 1 (modeling not shown).
Figure 9.
Comparison of 1 and 10 in the colchicine site of tubulin. Lowest energy conformations of 1 (A) and 10 (D) predicted by the systematic conformational search. Docked conformations of 1 (B) and 10 (E) in the colchicine binding site of tubulin. The poses were generated by superimposition of the furo[2,3-d]pyrimidine scaffolds of the docked and energy minimum poses of 1 (C) and 10 (F) and highlight differences in the orientations of the 4-position substituents.
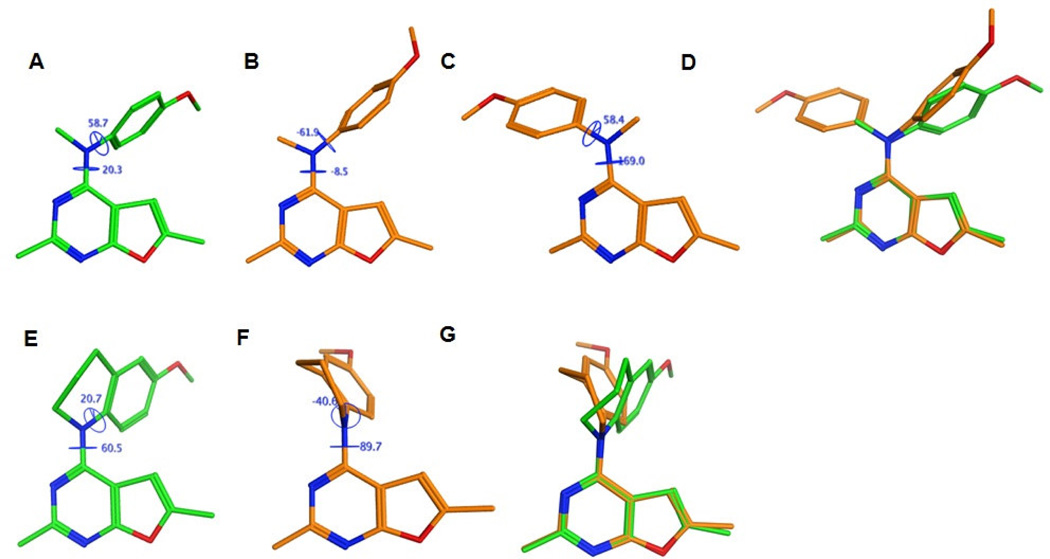
We previously reported two possible binding modes for 1 that varied in the orientation of the furo[2,3-d]pyrimidine scaffold relative to the Hinge (either parallel to the Hinge, termed ‘vertical’ binding mode, analogous to that seen in the docked conformation of 10 in Figure 6, or perpendicular to the Hinge, termed ‘horizontal’ binding mode) in the ATP site of VEGFR2.25 On comparing the conformations and energies of the docked poses of 1 and 10 to the conformations obtained by the systematic search using Sybyl-X 2.1, it was observed that the docked conformations of 1 in its horizontal binding mode and 10 in its vertical binding mode were conformationally and energetically closest to the lowest energy conformations of 1 and 10 (for 1, ΔE between conformations A and B in Figure 10 = 0.81 kcal/mol and for 10, ΔE between conformations E and F in Figure 10 = 1.03 kcal/mol), respectively. In contrast, the predicted vertical binding mode for 1 was conformationally and energetically distant from the lowest energy conformation of 1 from the systematic search study (ΔE between conformations A and C in Figure 10 = 1.78 kcal/mol). Similar results were obtained for the docked poses of 10 and 1 in EGFR and PDGFR-β and their respective low energy conformations (data not shown). These studies indicate that some flexibility in the molecule is necessary and is an important determining factor for the bioactive conformations of these compounds in RTKs and tubulin to allow adaptation of different conformations in order to function as multi-targeted inhibitors at different sites.
Figure 10.
Comparison of 1 and 10 in the ATP site of VEGFR2. Lowest energy conformations of 1 (A) and 10 (E) predicted by the systematic conformational search. Docked conformations of 125 in the vertical (B) and horizontal (C) binding modes and 10 (F) in the vertical binding mode in the ATP site of VEGFR2. The poses were generated by superimposition of the furo[2,3-d]pyrimidine scaffolds of the docked and energy minimum poses of 1 (D) and 10 (G) and highlight differences in the orientations of the 4-position substituents.
Chemistry
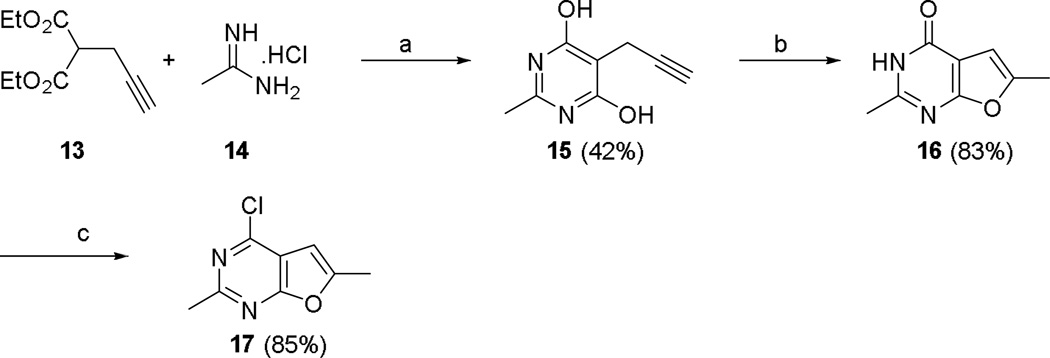
The synthesis of the key intermediate, compound 17, is shown in Scheme 1. As we recently reported, 25 a three step reaction, starting from diethyl propargyl malonate 13, was successfully employed in the synthesis of 4-chloro-2,6-dimethylfuro[2,3-d]pyrimidine 17 (Scheme 1). The condensation of diethyl propargyl malonate 13 and acetamidine hydrochloride 14 was carried out as a route to pyrimidine 15. Intramolecular cyclization of 15 to the furo[2,3-d]pyrimidine 16 (87%) proceeded under H2SO4 (conc.) at rt.
Scheme 1.
The synthesis of 4-chloro-2,6-dimethylfuro[2,3-d]pyrimidine 17.
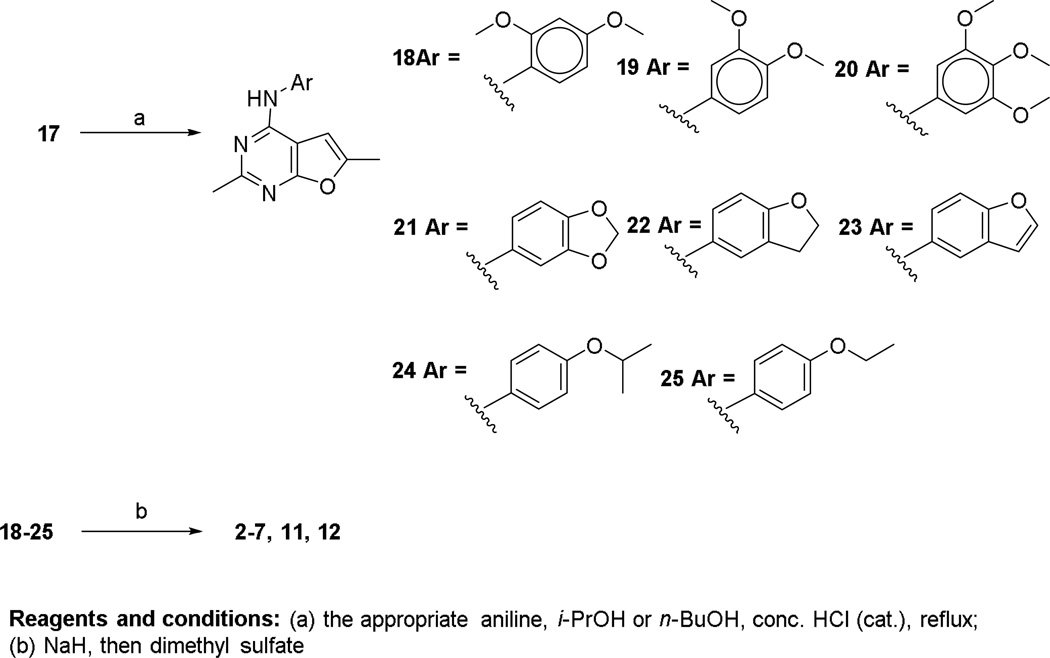
Chlorination of 16 with POCl3 afforded the 4-chloro-2,6-dimethylfuro[2,3-d]pyrimidine 17. This key intermediate was reacted with the appropriate anilines (Scheme 2), in nBuOH at reflux in the presence of a catalytic amount of HCl, to give intermediate compounds 18–25. Compounds 2–7, 11 and 12 were synthesized via N-methylation of 18–25 with dimethyl sulfate in the presence of NaH (Scheme 2).
Scheme 2.
The synthesis of N-aryl-2,6-dimethylfuro[2,3-d]pyrimidin-4-amines 2–7, 11 and 12.
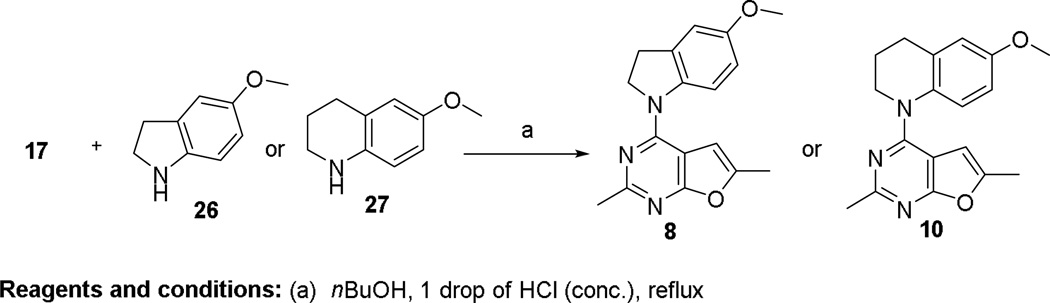
Under nucleophilic displacement condition, intermediate 17 (Scheme 3) reacted with 26 or 27 at reflux in nBuOH and a catalytic amount of HCl to provide 8 or 10, respectively.
Scheme 3.
The synthesis of 8 and 10.
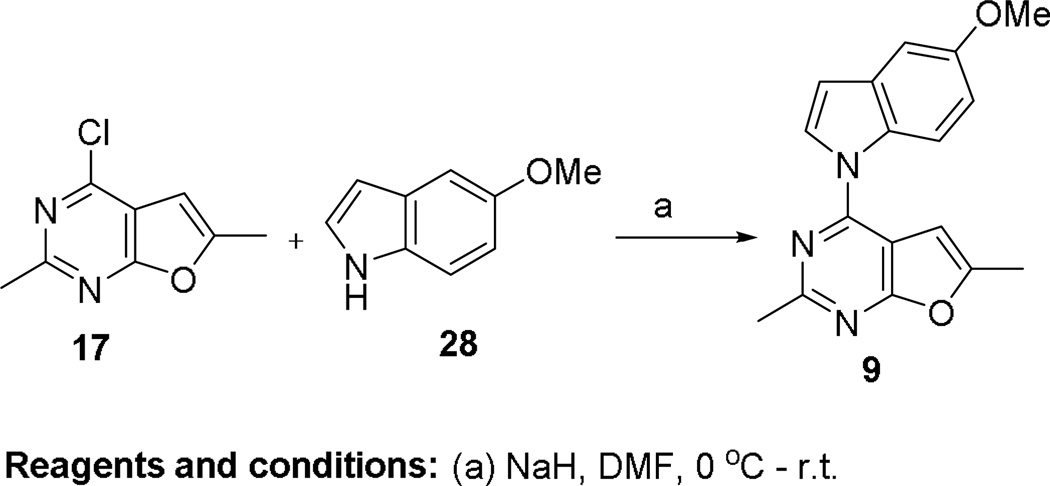
Direct aromatization of 8 to 9 was attempted. The conversion of 8 to 9 under different oxidation conditions including the use of MnO2,34 Pd/C,35 SeO236–38 and DDQ36–38 were unfruitful. This inability to convert 8 to 9 attests to the stability of 8 to aromatization. The failure of the above strategies prompted us to develop an alternate procedure for the synthesis of 9, as shown in Scheme 4. The deprotonation of 5-methoxyindoline by NaH in DMF resulted in the formation of a pyrrole anion, which was then reacted with 17 via SNAr displacement to afford 9 in 41% yield. The structure of the target 9 was confirmed by 1H NMR and elemental analysis.
Scheme 4.
The synthesis of compound 9.
Biological Evaluations and Discussion
RTK inhibitory activity of the compounds 2 – 12 was evaluated using human cancer cells known to express high levels of EGFR, VEGFR-2 or PDFGR-β using a phosphotyrosine ELISA cytoblot, and the data obtained are summarized in Table 1. Compound 1 and compounds known to inhibit a particular RTK were used as positive controls for these assays. The effects of the compounds on cell proliferation were measured in A431 cancer cells known to overexpress EGFR (Table 1). Finally, the effects of 2 – 12 on blood vessel formation was assessed using the chicken embryo chorioallantoic membrane (CAM) assay, a standard test for angiogenesis, and the results are presented in Table 1. In the CAM assay, purified angiogenic growth factors were placed locally on a vascularized membrane of a developing chicken embryo together with compounds of interest. Digitized images of the vasculature were taken 48 h after growth factor administration and the number of vessels per unit area evaluated as a measure of vascular density.
Table 1.
RTK, A431 and CAM inhibitory activities (IC50 ± SD) of compounds 1 – 12.
| Compound | EGFR kinase inhibition (nM) |
VEGFR-2 kinase inhibition (nM) |
PDGFR-β kinase inhibition (nM) |
A431 Cytotoxicity (nM) |
CAM angiogenesis inhibition (µM) |
|---|---|---|---|---|---|
| 1 | 68. 2 ± 6.2 | 19.1 ± 3.0 | 22.8 ± 4.9 | 20.8 ± 4.2 | 3.9 ± 0.4 |
| 2 | 162.3 ± 22.2 | 32.3 ± 5.0 | 40.4 ± 5.3 | 17.1 ± 2.0 | 53.6 ± 8.4 |
| 3 | 90.3 ± 10.2 | 18.6 ± 3.0 | 66.1 ± 8.1 | 9.7 ± 1.2 | 34.8 ± 5.1 |
| 4 | > 500 | > 300 | 166.3 ± 20.2 | > 300 | > 300 |
| 5 | 298.2 ± 38.1 | 19.9 ± 2.1 | 50.1 ± 7.8 | 48.9 ± 5.1 | 38.2 ± 5.1 |
| 6 | 226.4 ± 50.2 | 6.7 ± 0.81 | 24.6 ± 4.3 | 38.3 ± 4.2 | 14.0 ± 1.7 |
| 7 | 306.6 ± 42.1 | 5.8 ± 0.73 | 25.0 ± 4.8 | 73.2 ± 10.1 | 15.2 ± 0.81 |
| 8 | 103.2 ± 18.2 | 20.9 ± 3.7 | 73.2 ± 4.9 | 42.2 ± 5.0 | 38.2 ± 4.2 |
| 9 | > 500 | > 300 | 198.0 ± 22.5 | > 300 | > 300 |
| 10 | 3.4 ± 0.8 | 33.2 ± 5.0 | 58.2 ± 8.0 | 18.0 ± 2.9 | 104.2 ± 13.2 |
| 11 | 102.2 ± 20.1 | 38.5 ± 4.7 | 84.0 ± 10.0 | 11.6 ± 2.1 | 50.1 ± 8.2 |
| 12 | > 500 | 121.3 ± 18.6 | 132.0 ± 14.4 | 92.9 ± 10.1 | 201.0 ± 34.9 |
| semaxanib | n.d. | 12.9 ± 2.7 | n.d. | n.d. | 60.0 ± 10.1 |
| cisplatin | n.d. | n.d. | n.d. | 10.6 ± 2.9 | n.d. |
| sunitinib | 172.1 ± 19.4 | 18.9 ± 2.7 | 83.1 ± 10.1 | n.d. | 1.3 ± 0.07 |
| erlotinib | 1.2 ± 0.2 | 124.7 ± 18.2 | n.d. | n.d. | 29.1 ± 1.9 |
n.d. not determined
In the cellular assays using cells with high expression of EGFR, compound 10 showed potency similar to that of the standard erlotinib and was about 30-fold more potent than 1. All the other compounds (2 – 9, 11 and 12) were less potent than erlotinib and 1 in EGFR expressing cells. This result suggests that the tetrahydroquinoline ring system in 10 is highly favorable for EGFR inhibition and also indicates that conformational restriction by other structural constraints described above, besides the tetrahydroquinoline, were detrimental to inhibition of EGFR overexpressing cancer cells as compared to 1.
Against VEGFR-2 expressing cells, both 6 and 7 with the conformationally restricted 4’-OMe of 1 afforded excellent inhibitory activity, better than 1 and the standard semaxanib and 3fold better than sunitinib. Conformationally restricted analogs 3 and 5, with restrictions at the 4’-OMe of 1, were equipotent with 1 and sunitinib, as is 8 with a 5’-OMe dihydroindole substituent at the 4-position. Compounds 2, 10, and 11 maintain two-digit nanomolar potency but were less potent than 1 and sunitinib. Compounds 4 and 9 were relatively inactive, with 12 six-fold less active than 1. With the exception of 4, 9 and 12, conformational restriction affords good potency against VEGFR-2 overexpressing cells, with some analogs better than 1 and the standard sunitinib.
In the PDGFR-β expressing cells, as in VEGFR-2 expressing cells, 6 and 7 were the most potent and were similar to 1 and about 4-fold better than the standard sunitinib. Compounds 2, 5 and 10 were less potent than 1 but better than sunitinib. Compounds 3, 8, and 11 were similar to sunitinib but less potent than 1. Finally, 4, 9 and 12 were the least potent, but possessed three-digit nanomolar IC50 values. Thus, like VEGFR-2, with the exception of 4, 9 and 12, conformational restriction of 1 was conducive to PDGFR-β inhibition.
Against A431 cells all of the analogs (with 4 as an exception) had good activity as compared to 1 and were 2- to 3-fold less potent than 1. In the CAM angiogenesis assay, all of the compounds showed activity, with 6 and 7 being most potent and 4, 10, and 12 least potent.
The ability of compounds 2–12 to cause dose-dependent loss of cellular microtubules was evaluated and the data summarized in Table 2 in comparison with the data obtained with 1, CA-4 and paclitaxel as controls. Compounds 2, 3, 6, 7, 10 and 12 showed dose-dependent loss of cellular microtubules. Among these compounds, 10 had an EC50 of 21 nM, which is 5- to 10-fold more potent than all the other analogs, including 1, and comparable to the value obtained with CA-4. The second most potent new compound was 12 and the third most active was 6. Compounds 2, 3 and 7 were significantly less potent. With 10, it appears that restriction in conformation can lead to an increased effect on cellular microtubules, while with 4, 5, 8, 9 and 11 perhaps the bulk and/or conformational rigidity prevented optimal interactions with tubulin. Although 8 showed good RTK inhibition, it had no effect on cellular microtubules. Structure– activity analyses of all the members of this series identified that the conformation of the compounds did affect potency as disruptors of cellular microtubules (Table 2), and this activity was independent of the bioactive conformation required for RTK inhibition.
Table 2.
Microtubule depolymerizing and antiproliferative activities (IC50 ± SD) of 2 – 12.
| Compound | EC50 for MT depolymerization in A·10 cells (nM) |
IC50 ± SD in MDA-MB-435 cells (nM) |
EC50/IC50 ratio |
IC50 ± SD in HeLa cells (nM) |
IC50 ± SD in WTβIII (nM) |
Rr value | IC50 ± SD in SK-OV- 3 cells (nM) |
IC50 ± SD in SK- OV-3 MDR1- M6/6 cells (nM) |
Rr value |
|---|---|---|---|---|---|---|---|---|---|
| 1* | 103 | 17.1 ± 1.5 | 6 | 33.1 ±1.6 | 27.2 | 0.8 | 36.7 ±2.3 | 63.3 ±4.8 | 1.7 |
| 2 | 2,994 | 139.5 ± 8.8 | 22 | 321.4 ± 71.3 | 235.7 ± 10.6 | 0.7 | 372.3 ± 26.2 | 482.9 ± 18.5 | 1.3 |
| 3 | 2,719 | 50.6 ± 7.6 | 54 | 122.7 ± 10.1 | 113.1 ± 26.4 | 0.9 | 129.0 ± 14.2 | 203.3 ± 0.6 | 1.6 |
| 4 | > 10,000 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 5 | > 10,000 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 6 | 606 | 96.2 ± 13 | 6 | 104.6 ± 5.8 | 94.0 ± 7.0 | 0.9 | 120.5 ± 5.0 | 155.2 ± 17.2 | 1.3 |
| 7 | 3,786 | 133 ± 1.7 | 28 | 301.8 ± 23.4 | 242.7 ± 15.1 | 0.8 | 425.3 ± 53.1 | 509.0 ± 4.8 | 1.2 |
| 8 | > 10,000 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 9 | > 10,000 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 10 | 21 | 9.8 ± 0.9 | 2 | 13.5 ± 0.5 | 9.1 ± 1.8 | 0.7 | 12.9 ± 1.2 | 19.4 ± 2.9 | 1.5 |
| 11 | > 10,000 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 12 | 182 | 13.4 ± 1.6 | 14 | 28.2 ± 0.7 | 19.4 ± 0.7 | 0.7 | 24.6 ± 5.1 | 30.8 ± 1.7 | 1.3 |
| CA-4 | 9.8 | 5.0 ± 0.3 | 2 | 5.6 ± 0.3 | 5.7 ± 0.4 | 1.0 | 6.9 ± 0.1 | 3.0 ± 0.3 | 0.4 |
| Paclitaxel | n.d. | 3.3 ± 0.3 | n.d. | 1.9 ± 0.3 | 32.9 ± 5.8 | 17.3 | 6.3 ± 0.4 | 1,187.2 ± 265.2 | 188 |
n.d. not determined
Comparing the biological activities of 2 – 4 against RTKs and tubulin (Table 1 and Table 2) suggests that increasing the energy barrier for rotation by introducing bulky groups adjacent to the rotatable σ bonds was detrimental to biological activity in RTKs and tubulin compared to 1, except in A431 cells, where 2 and 3 show improved activities compared to 1. However, the extent of the loss of activity of 2 – 4 against RTKs was less than that against tubulin. This indicates that the ability to rotate bonds a – c to achieve the bioactive conformations of these compounds is of greater importance against tubulin than against RTKs.
Compounds 5 – 7, where the design strategy was to restrict rotation of bond c by incorporation into a ring, showed improved activities against VEGFR2 (compounds 6 and 7), but these compounds lost activity against EGFR, PDGFR-β and tubulin. Comparing 8 – 10, in which both bonds a and b are restricted by incorporation in a ring, shows that complete restriction of rotation in 9 eliminated RTK as well as tubulin inhibitory activities. Increasing flexibility around bond a in 8 restored some of the lost inhibitory activity against both RTKs and tubulin. The most flexible analog of the 8 – 10 compounds, 10, showed improved activities in inhibition of EGFR and tubulin compared to 1, once again indicating that some flexibility is necessary for dual RTK/tubulin inhibitory effects.
Compounds 11 and 12, which were designed to explore the effect of bulk at the 4’-position of the aniline moiety, showed loss of activities against RTKs as compared to 1. However, 12 showed improved potency in most of the tubulin assays (Table 2) as compared to 1, indicating that bulk at the 4’-position is beneficial for antitubulin activity.
The ability to circumvent drug resistance mechanisms is a notable advantage of most colchicine site agents, and the effects of compounds 2–12 were evaluated in two pairs of multidrug resistant cell lines (Table 2). The microtubule active compounds were evaluated in the parental HeLa cells and in an engineered cell line overexpressing βIII tubulin (WTβIII). The cells were equally sensitive to each of the compounds, with relative resistance values of 0.7–0.9. In addition, the compounds were evaluated for their ability to inhibit the growth of SK-OV-3 cells overexpressing Pgp in comparison to the parental line (Table 2). The relative resistance values range from 1.2–1.6, while the relative resistance of the Pgp substrate paclitaxel was 188. These results demonstrated that compounds 2, 3, 6, 7, 10 and 12 overcome both βIII and Pgp mediated drug resistance mechanisms.
To our knowledge, of all the compounds having dual RTK inhibition and microtubule disrupting activity, compound 10 showed the best EGFR inhibition and anti-microtubule activity. Studies were conducted to determine if 10 interacted directly with purified tubulin. We determined the IC50 for inhibition of tubulin polymerization of 10 as compared to CA-4 and 1. In this assay, 10 inhibited tubulin assembly about as well as CA-4 (Table 3) and was better than 1. The data in Table 4 also showed that 10 binds at the colchicine site on tubulin, since it inhibited [3H]colchicine binding to the protein as potently as did CA-4.
Table 3.
Effects of 1 and 10 on tubulin polymerization and inhibition of colchicine binding.
| Compound | Inhibition tubulin assembly IC50(µM) ± SD |
Inhibition of colchicine binding % inhibition ± SD |
|
|---|---|---|---|
| 1 µM | 5 µM | ||
| CA-4 | 1.0 ± 0.09 | 88 ± 2 | 99 ± 0.2 |
| 1 | 2.4 ± 0.01 | 63 ± 5 | 88 ± 3 |
| 10 | 1.1 ± 0.1 | 82 ± 3 | 96 ± 1 |
Table 4.
Cancer cell inhibitory activity (GI50, nM) of 1, 6, 7 and 10 (NCI 60 cell line panel).
| Panel/Cell line |
GI50 (nM) | Panel/Cell line |
GI50 (nM) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 6 | 7 | 10 | 1 | 6 | 7 | 10 | ||
| NSCLC | Renal Cancer | ||||||||
| A549/ATCC | 37.6 | n.d. | n.d. | <10 | 786 – 0 | 43.6 | 80.4 | 1040 | <10 |
| EKVX | 64.8 | 70 | 396 | <10 | A498 | 19.5 | n.d. | n.d. | <10 |
| HOP-62 | 32.2 | 39.6 | 75.2 | <10 | ACHN | 55.9 | 64.2 | 265 | <10 |
| HOP-92 | n.d. | 45.8 | 226 | 58.6 | CAKI-1 | 16.3 | 40.1 | 57.9 | <10 |
| NCI-H226 | 84.1 | 162 | 217 | 13 | RXF 393 | n.d. | 21.2 | 57.2 | <10 |
| NCI-H23 | 40.7 | 43.9 | 206 | <10 | SN 12C | 56.7 | 97.2 | 611 | <10 |
| NCI-H322M | n.d. | n.d. | n.d. | <10 | TK-10 | n.d. | 50.2 | 382 | <10 |
| NCI-H460 | 33.1 | 42.0 | 138 | <10 | UO-31 | 75.2 | n.d. | 278 | <10 |
| NCI-H522 | <10 | 12.9 | 29.9 | <10 | Colon Cancer | ||||
| CNS Cancer | COLO 205 | 20 | 40.3 | 139 | <10 | ||||
| SF-268 | 33.6 | 13.9 | 527 | 16.8 | HCC-2998 | 28.5 | 54.4 | 185 | 12.0 |
| SF-295 | 13.8 | 30.5 | 41.7 | <10 | HCT-116 | 34 | 45.1 | 77.1 | <10 |
| SF-539 | 20.0 | 31.6 | 60.9 | <10 | HCT-15 | 25.3 | 38.9 | 51.9 | <10 |
| SNB-19 | 33.3 | 56.5 | 93.6 | <10 | HT29 | 32.5 | 32.4 | 51.9 | <10 |
| SNB-75 | 53.2 | 30.4 | 103 | <10 | KM12 | 21.1 | 40.1 | 58.4 | <10 |
| U251 | 30.8 | 39.7 | 84 | <10 | SW-620 | 29.8 | 41.6 | 58.8 | <10 |
| Ovarian cancer | Prostate Cancer | ||||||||
| IGROV1 | n.d. | 62.9 | 142 | <10 | PC-3 | n.d. | 52.2 | 183 | <10 |
| OVCAR-3 | 29.1 | 24.5 | 30 | <10 | DU-145 | 26.2 | 44.1 | 198 | <10 |
| OVCAR-4 | 60.4 | 83.4 | 553 | 51 | Breast Cancer | ||||
| OVCAR-5 | 55.8 | 262 | 531 | 43 | MCF7 | 42.2 | 40.2 | 43.8 | <10 |
| OVCAR-8 | 36.6 | 63 | 330 | <10 | MDA-MB-231/ATCC | 44.3 | 148 | 223 | 11.4 |
| NCI/ADR-RES | n.d. | 26.5 | 37.6 | <10 | HS 578T | 15.3 | 31.0 | 51.7 | <10 |
| SK-OV-3 | 25.2 | 37.6 | 98.0 | <10 | BT-549 | 48.0 | 77.2 | 432 | <10 |
| Melanoma | MDA-MB-468 | 34.9 | 22.1 | 31.5 | <10 | ||||
| LOX IMVI | 54.9 | 45.6 | 162 | <10 | Leukemia | ||||
| MALME-3M | 42.3 | n.d. | n.d. | <10 | CCRF-CEM | 25.3 | 58.1 | 121 | <10 |
| M14 | 23.3 | 34.8 | 47.3 | <10 | HL-60(TB) | 17.3 | 27.6 | 41.2 | <10 |
| MDA-MB-435 | n.d. | 14.0 | 21.1 | <10 | K-562 | 13.2 | 31.7 | 40.4 | <10 |
| SK-MEL-2 | 33.9 | 26.4 | 82.4 | <10 | MOLT-4 | 67.9 | 76.7 | 211 | 15.0 |
| SK-MEL-28 | 37.7 | n.d. | n.d. | <10 | RPMI-8226 | 42.8 | 54.2 | 389 | <10 |
| SK-MEL-5 | 22.9 | 31.4 | 50.4 | <10 | SR | 32.0 | 36.6 | 62.4 | n.d. |
| UACC-62 | 14.6 | 61.3 | 57.1 | <10 | |||||
n.d. not determined
Compounds 6 and 10, the most potent microtubule depolymerization agents in the current study, and compound 7 for comparison, were selected by the National Cancer Institute for evaluation in the NCI 60 cell line panel and compared with 1. All three compounds (6, 7 and 10) showed potent GI50s against most of the NCI 60 cancer cell lines (Table 4). Although 6 and 7 were less potent than 1 in most cancer cell lines, 10 showed a significantly improved potency against all of the cancer cells when compared to 1.
Summary
A series of eleven furo[2,3-d]pyrimidine compounds was designed and evaluated as dual multi-targeted RTK inhibitors and antimitotic agents, using a conformational restriction strategy. Compounds 8 and 10 showed EGFR inhibitory activity similar to or better than the parent analog 1, while inhibitory effects on VEGFR2 and PDGFR-β were less than those of 1. SAR analysis indicates that restriction of only bond b (2) leads to improvement in A431 cytotoxicity compared to 1. For bond c, complete restriction (6 and 7) results in an increase in VEGFR2 inhibitory activity but a decrease in all other biological tests compared to 1. Partial restriction of bond c with an ortho substitution (3) leads to an improvement in A431 cytotoxicity compared to 1 but is detrimental to all other activities (Table 1). Further restriction with a di-ortho substitution (4) or as a methylene dioxy (5) of bond c is detrimental to activity across the board. Severe restriction of bonds b and d (9) results in loss of activity for both RTKs and tubulin. However, partial restriction (8) of bonds b and d restores RTK activity. Further relaxation of restriction of bonds b and d provides optimal RTK and tubulin activity in 10. The lack of tubulin activity for 11 is attributed more to bulk than restriction of bond c. This is apparent on comparison of 11 and 12. Remarkably, six of the compounds, 2, 3, 6, 7, 10 and 12 disrupted cellular microtubules and had significant cytotoxic activities indicating that restriction of bonds a, b and d are conducive to dual RTK and tubulin inhibitory properties. Compound 10 has the best balance of RTK and tubulin inhibitory activities resulting from the conformational restriction of bonds a, b and d.
However, as appears to be the case with compound 8, the bioactive conformation for RTK inhibition is predicted to be independent of the bioactive conformation required for tubulin inhibition. The preferred conformation of the tetrahydroquinoline substituted analog 10 is optimal for EGFR as well as for tubulin inhibition. Compound 10 is more potent than the lead compound 1 for both EGFR (20-fold) and tubulin (5-fold) inhibition, but not for all RTKs nor for antiangiogenic activities, indicating that the bioactive conformation in furo[2,3-d]pyrimidine analogs for dual RTK and tubulin inhibitory activities are different. This indicates that suitable conformational restriction increases potency for both tubulin and EGFR without loss of activity against VEGFR2 and PDGFR-β. Among multi-targeted inhibitors, the best analog is selected on the basis of the compound that has the best balance of inhibitory activities against the various targets. For this series, 10 was the best analog as evidenced by the activities in the NCI-60 cancer cell line panel, where it had significantly greater activity than the parent 1. Compound 10 is currently in further preclinical evaluations, the results of which will be the subject of future communications.
Experimental section
All evaporations were carried out in vacuo with a rotary evaporator. Analytical samples were dried in vacuo (0.2 mmHg) in a CHEM-DRY drying apparatus over P2O5 at 55 °C. Melting points were determined on a MEL-TEMP II melting point apparatus with FLUKE 51 K/J electronic thermometer and are uncorrected. Proton nuclear magnetic resonance (1H NMR) spectra were recorded on a Bruker WH-300 (300 MHz) or a Bruker 400MHz/52 MM (400 MHz) spectrometer. The chemical shift values are expressed in ppm (parts per million) relative to tetramethylsilane as internal standard: s ) singlet, d ) doublet, t ) triplet, q ) quartet, m ) mutiplet, br ) broad singlet. The relative integrals of peak areas agreed with those expected for the assigned structures. Thin-layer chromatography (TLC) was performed on Whatman Sil G/UV254 silica gel plates with fluorescent indicator, and the spots were visualized under 254 and 366 nm illumination. Proportions of solvents used for TLC are by volume. Column chromatography was performed on a 230–400 mesh silica gel (Fisher, Somerville, NJ) column. Elemental analyses were performed by Atlantic Microlab, Inc., Norcross, GA. Element compositions were within ± 0.4% of the calculated values. Fractional moles of water or organic solvents found in some analytical samples could not be prevented despite 24–48 h of drying in vacuo and were confirmed where possible by their presence in the 1H NMR spectra. All solvents and chemicals were purchased from Aldrich Chemical Co. or Fisher Scientific and were used as received. Purities of the final compounds 2–10 were > 95% by elemental analysis.
2-Methyl-6-hydroxy-5-prop-2-yn-1-ylpyrimidin-4(3H)-one (15)
To a mixture of diethyl prop-2-yn-1-ylmalonate 13 (11.9 g, 60 mmol) and acetamidine hydrochloride 14 (5.68 g, 60 mmol) in MeOH (100 mL) was added sodium metal (1.38 g, 60 mmol) portion wise. The resulted mixture was heated to reflux for 24 h. The suspension was then cooled in an ice-bath and the precipitate formed was collected by filtration and dissolved in 40 mL of water. This solution was adjusted to pH 3–4 with 1 N HCl, whereupon a thick precipitate formed, which was collected through filtration and washed with a small amount of water and acetone and dried over P2O5 to afford 4.1 g (42%) of 15 as a white solid; mp >300 °C; Rf 0.11 (CHCl3/MeOH 6:1); 1H NMR (DMSO-d6) δ 2.23 (s, 3 H), 3.05 (s, 2 H), 3.32 (s, 1 H), 11.92 (s, 2 H).
2,6-Dimethylfuro[2,3-d]pyrimidin-4(3H)-one (16)
To a 25 mL round-bottomed flask were added 15 (1.64 g, 10 mmol) and concentrated sulfuric acid (15 mL). The resulting solution was stirred overnight at ambient temperature and poured into 100 mL distilled water and extracted with chloroform (3 x 30 mL). The organic layers were pooled, dried over Na2SO4 and concentrated to afford 16 (1.36g, 83%) as a yellow powder; mp >300 °C; Rf 0.35 (CHCl3/MeOH 6:1); 1H NMR (DMSO-d6) δ 2.42 (s, 3 H, CH3), 2.44 (s, 3 H, CH3), 6.63 (s, 1 H, CH), 12.50 (s, 1 H, 3-NH exch).
4-Chloro-2,6-dimethylfuro[2,3-d]pyrimidine (17)
To a 50 mL flask were added 16 (1.64 g, 1 mmol) and 10 mL POCl3. The resulting mixture was maintained at reflux for 2 h, and the solvent was removed under reduced pressure to afford a dark residue. The crude mixture was purified by silica gel column chromatography using EtOAc/Hexane = 20:1 as the eluent. Fractions containing the product (TLC) were combined and evaporated to afford 1.55 g (85%) 17 as a yellow solid; mp 47.6–48.1 °C; Rf 0.26 (EtOAc/Hexane 15:1); 1H NMR (DMSO-d6) δ 2.48 (s, 3 H), 2.63 (s, 3 H), 6.77 (s, 1 H).
General procedure for the synthesis of 18–25
To a 100-mL round-bottomed flask, flushed with nitrogen, were added 17 (127 mg, 0.7 mmol), the appropriate aniline (1.05 mmol), BuOH (20 mL), and 2–3 drops of conc. HCl. The reaction mixture was heated at reflux with stirring for 2 h until the starting material 17 disappeared (TLC). The reaction solution was allowed to cool to room temperature; the solvent was removed under reduced pressure, and the residue was purified by column chromatography on silica gel with 10% EtOAc/Hexane as the eluent. Fractions containing the product (TLC) were combined and evaporated to afford target compounds.
N-(2,4-dimethoxyphenyl)-2,6-dimethylfuro[2,3-d]pyrimidin-4-amine (18)
Using the general procedure described above, compound 18 (77%) was obtained as an off-white powder; mp 97.7–97.9 °C; Rf 0.62 (EtOAc/Hexane 1:1); 1H NMR (DMSO-d6) δ 2.31 (s, 3 H), 2.37 (s, 3 H), 3.73 (s, 3 H, OCH3), 3.81 (s, 3 H, OCH3), 5.93 (s, 1 H, 5-CH), 6.55–6.58 (dd, 1 H, J = 6.8 Hz, J = 2 Hz C6H3), 7.33–7.35 (d, 2 H, J = 6.8 Hz, C6H3), 8.78 (s, 1 H, NH exch), Anal. (C16H17N3O3) C, H, N.
N-(3,4-dimethoxyphenyl)-2,6-dimethylfuro[2,3-d]pyrimidin-4-amine (19)
Using the general procedure described above, compound 19 (82%) was obtained as a gray solid; mp 150.8–151.4 °C; Rf 0.50 (EtOAc/Hexane 1:1); 1H NMR (DMSO-d6) δ 2.41 (s, 3 H), 2.48 (s, 3 H), 3.75 (s, 3 H, OCH3), 3.78 (s, 3 H, OCH3), 6.57 (s, 1 H, 5-CH), 6.94–6.96 (d, 1 H, J = 7.2 Hz, C6H3), 7.24–7.26 (dd, 1 H, J = 7.2 Hz, C6H3), 7.54 (s, 1 H), 9.36 (s, 1 H, NH exch), Anal. (C16H17N3O3·H2O) C, H, N.
2,6-Dimethyl-N-(3,4,5-trimethoxyphenyl)furo[2,3-d]pyrimidin-4-amine (20)
Using the general procedure described above, compound 20 (83%) was obtained as a yellow powder; mp 173.5–173.7 °C; Rf 0.3 (EtOAc/Hexane 3:1); 1H NMR (DMSO-d6) δ 2.43 (s, 3 H), 2.50 (s, 3 H), 3.65 (s, 3 H, OCH3), 3.80 (s, 6 H, 2 OCH3), 6.71 (s, 1 H, 5-CH), 7.29 (s, 2 H, C6H2), 9.43 (s, 1 H, NH exch), Anal. (C17H19N3O4·0.2H2O) C, H, N.
N-1,3-benzodioxol-5-yl-2,6-dimethylfuro[2,3-d]pyrimidin-4-amine (21)
Using the general procedure described above, compound 21 (72%) was obtained as a brown solid; mp 185.5–187.1 °C; Rf 0.10 (EtOAc/Hexane 1:3); 1H NMR (DMSO-d6) δ 2.40 (s, 3 H, CH3), 2.46 (s, 3 H, CH3), 6.01 (s, 2 H, OCH2O), 6.59 (br, 1 H, NH exch), 6.89–6.91 (d, 1 H, J = 6.8 Hz), 7.09–7.11 (dd, 1 H, J = 8.4 Hz ), 7.51 (s, 1 H), 9.37 (s, 1 H), Anal. (C15H13N3O2) C, H, N.
N-(2,3-dihydro-1-benzofuran-5-yl)-2,6-dimethylfuro[2,3-d]pyrimidin-4-amine (22)
Using the general procedure described above, compound 22 (70%) was obtained as a brown solid; mp 193.7–195.2 °C; Rf 0.10 (EtOAc/Hexane 1:3); 1H NMR (DMSO-d6) δ 2.38 (s, 3 H, CH3), 2.44 (s, 3 H, CH3), 3.18–3.23 (t, 2 H, J = 8.4 Hz, CH2CH2), 4.52–4.56 (t, 2 H, J = 8.4 Hz, CH2CH2), 6.43 (br, 1 H, NH exch), 6.74–6.77 (d, 1 H, J = 8.4 Hz), 7.33–7.35 (d, 1 H, J = 8.4 Hz), 7.57 (s, 1 H), 9.50 (s, 1 H), Anal. (C16H15N3O2) C, H, N.
N-1-benzofuran-5-yl-2,6-dimethylfuro[2,3-d]pyrimidin-4-amine (23)
Using the general procedure described above, compound 23 (66%) was obtained as colorless crystals; mp 173.4–175.0 °C; Rf 0.10 (EtOAc/Hexane 1:3); 1H NMR (DMSO-d6) δ 2.40 (s, 3 H, 6-CH3), 2.49 (s, 3 H, 2-CH3), 6.55 (br, 1 H, NH exch), 6.98 (d, 1 H, J = 1.6 Hz, 5-CH), 7.58–7.60 (d, 2 H, C8H5), 7.99 (d, 1 H, C8H5), 8.11–8.13 (t, 1 H, C8H5), 9.50 (s, 1 H, C8H5), Anal. (C16H13N3O2) C, H, N.
N-(4-propoxyphenyl)furo[2,3-d]pyrimidin-4-amine (24)
Using the general procedure described above, compound 24 (67%) was obtained as colorless crystals; mp 136.3–137.1°C; Rf 0.51 (EtOAc/Hexane 1:3); 1H NMR (DMSO-d6) δ 0.98–1.00 (t, 3 H, OCH2CH2CH3), 1.70–1.77 (m, 2 H, OCH2CH2CH3), 2.40 (s, 3 H, 6-CH3), 2.45 (s, 3 H, 2-CH3), 3.91–3.94 (t, 2 H, OCH2CH2CH3), 6.53 (s, 1 H, 5-CH), 6.93–6.94 (d, 2 H, J = 7.2 Hz, C6H4), 7.61–7.62 (d, 2 H, J = 7.2 Hz, C6H4), 9.33 (s, 1 H, NH exch); Anal. (C17H19N3O2) C, H, N.
N-(4-ethoxyphenyl)-2,6-dimethylfuro[2,3-d]pyrimidin-4-amine (25)
Using the general procedure described above, compound 25 (80%) was obtained as an off-white powder; mp 160.6–162.1 °C; Rf 0.51 (EtOAc/Hexane 1:3); 1H NMR (DMSO-d6) δ 1.32–1.35 (t, 3 H, J = 5.6 Hz, OCH2CH3), 2.40 (s, 3 H, 6-CH3), 2.45 (s, 3 H, 2-CH3), 4.00–4.04 (q, 2 H, J = 5.6 Hz, OCH2CH3), 6.50 (s, 1 H, 5-CH), 6.93–6.94 (d, 2 H, J = 6.8 Hz, C6H4), 7.61 (d, 2 H, J = 6.8 Hz, C6H4), 9.34 (s, 1 H, NH exch); Anal. (C16H17N3O2) C, H, N.
General procedure for the synthesis of 2–7, 11 and 12
To a 25 mL round bottomed flask was added the appropriate amount of 18–25 (0.5 mmol), which was dissolved in DMF (2 mL). The flask was purged with argon for five min, followed by cooling to 0 °C using an ice bath. Sodium hydride (36 mg, 1.5 mmol) was added to the solution at 0 °C. The solution was stirred for 30 min at 0 °C under an argon atmosphere. Dimethyl sulfate (150 mg, 1.2 mmol) was introduced to the reaction mixture with the help of a syringe, and the flask was warmed to room temperature. The mixture was stirred at room temperature for another 3 h, and 5 mL of 1 N HCl was added carefully to quench the reaction. The reaction solvent was removed under reduced pressure, and the residue was suspended in water (20 mL). The suspension was extracted using EtOAc (2 x 10 mL). The combined organic extracts were washed with brine (10 mL), dried over anhydrous sodium sulfate and removed by evaporation under reduced pressure. The residue was purified by column chromatography on silica gel with 20% EtOAc/Hexane as the eluent. Fractions containing the product (TLC) were combined and evaporated to afford target compounds.
N-(2,4-dimethoxyphenyl)-N,2,6-trimethylfuro[2,3-d]pyrimidin-4-amine (2)
Using the general procedure described above, compound 2 (74%) was obtained as orange crystals: mp 166.1–166.4 °C; Rf 0.38 (EtOAc/Hexane, 1:1); 1H NMR (DMSO-d6) δ 2.16 (s, 3 H, CH3), 2.47 (s, 3 H, CH3), 3.41 (s, 3 H, NCH3), 3.70 (s, 3 H, OCH3), 3.85 (s, 3 H, OCH3), 4.57 (s, 1 H, 5-CH), 6.62–6.64 (dd, 1 H, J = 6.8 Hz, J = 2.0 Hz, C6H3), 6.75–6.76 (d, 1 H, J = 6.8 Hz, C6H3), 7.24–7.22 (d, 1 H, J = 2.0 Hz, C6H3), Anal. (C17H19N3O) C, H, N.
N-(3,4-dimethoxyphenyl)-N,2,6-trimethylfuro[2,3-d]pyrimidin-4-amine (3)
Using the general procedure described above, compound 3 (50%) was obtained as orange crystals: mp 114.2–116.6 °C; Rf 0.28 (EtOAc/Hexane, 1:1); 1H NMR (DMSO-d6) δ 2.17 (s, 3 H, CH3), 2.48 (s, 3 H, CH3), 3.47 (s, 3 H, NCH3), 3.73 (s, 3 H, OCH3), 3.82 (s, 3 H, OCH3), 4.64 (s, 1 H, 5-CH), 6.87–6.89 (dd, 1 H, J = 6.8 Hz, J = 2.0 Hz, C6H3), 7.02–7.07 (d, 1 H, J = 2.0 Hz, C6H3), 7.04–7.06 (d, 1 H, J = 6.8 Hz, C6H3), Anal. (C17H19N3O3) C, H, N.
N,2,6-trimethyl-N-(3,4,5-trimethoxyphenyl)furo[2,3-d]pyrimidin-4-amine (4)
Using the general procedure described above, compound 4 (63%) was obtained as light yellow crystals: mp 176.5–178.1 °C; Rf 0.19 (EtOAc/Hexane, 2:1); 1H NMR (DMSO-d6) δ 2.20 (s, 3 H, CH3), 2.49 (s, 3 H, CH3), 3.50 (s, 3 H, NCH3), 3.74(s, 9 H, 3 OCH3), 4.73 (s, 1 H, 5-CH), 6.73 (s, 2 H, C6H2), Anal. (C18H21N3O4) C, H, N.
N-1,3-benzodioxol-5-yl-N,2,6-trimethylfuro[2,3-d]pyrimidin-4-amine (5)
Using the general procedure described above, compound 5 (67%) was obtained as colorless crystals; mp 200.0–200.7 °C; Rf 0.48 (EtOAc/Hexane 1:1); 1H NMR (DMSO-d6) δ 2.21 (s, 3 H, CH3), 2.48 (s, 3 H, CH3), 3.44 (s, 3 H, OCH3), 4.76 (s, 1 H, 5-CH), 6.14 (s, 2 H, OCH2O), 6.82–6.84 (m, 1 H), 7.01–7.04 (m, 2 H), Anal. (C16H15N3O3) C, H, N.
N-(2,3-dihydro-1-benzofuran-5-yl)-N,2,6-trimethylfuro[2,3-d]pyrimidin-4-amine (6)
Using the general procedure described above, compound 6 (59%) was obtained as colorless crystals; mp 167.2–168.4 °C; Rf 0.16 (EtOAc/Hexane 3:1); 1H NMR (DMSO-d6) δ 2.18 (s, 3 H, CH3), 2.48 (s, 3 H, CH3), 3.21–3.25 (t, 2 H, J = 6.8 Hz,CH2CH2), 3.45 (s, 3 H, OCH3), 4.61–4.64 (t, 2 H, J = 6.8 Hz, CH2CH2), 4.65 (s, 1 H, 5-CH), 6.86–6.88 (d, 1 H, J = 6.8 Hz), 7.05–7.07 (dd, 1 H, J = 8.4 Hz ), 7.24 (s, 1 H), Anal. (C17H17N3O2) C, H, N.
N-1-benzofuran-5-yl-N,2,6-trimethylfuro[2,3-d]pyrimidin-4-amine (7)
Using the general procedure described above, compound 7 (63%) was obtained as colorless crystals; mp 193.0–194.2 °C; Rf 0.22 (EtOAc/Hexane 3:1); 1H NMR (DMSO-d6) δ 2.10 (s, 3 H, CH3), δ 2.49 (s, 3 H, CH3), 3.38 (s, 3 H, OCH3), 4.37 (s, 1 H, 5-CH), 7.03 (s, 1 H), 7.30–7.32 (d, 1 H, J = 6.8 Hz), 7.69 (s, 1 H), 7.74–7.76 (d, 1 H, J = 6.8 Hz), 8.13 (s, 1 H), Anal. (C17H15N3O2), C, H, N.
N-2,6-trimethyl-N-(4-propoxyphenyl)furo[2,3-d]pyrimidin-4-amine (11)
Using the general procedure described above, compound 11 (61%) was obtained as a light brown solid: mp 100.7–100.8 °C; Rf 0.7 (EtOAc/Hexane, 1:3); 1H NMR (DMSO-d6) δ 0.99–1.03 (t, 3 H, J = 5.6 Hz, OCH2CH2CH3), 1.75–1.79 (m, 2 H, J = 5.6 Hz, OCH2CH2CH3), 2.17 (s, 3 H, CH3), 2.48 (s, 3 H, CH3), 3.45 (s, 3 H, NCH3), 3.99–4.02 (t, 2 H, J = 5.6 Hz, OCH2CH2CH3), 4.62 (s, 1 H, 5-CH), 7.05–7.07 (d, 2 H, J = 7.2 Hz, C6H4), 7.27–7.28 (d, 2 H, J = 7.2 Hz, C6H4); Anal. (C18H21N3O2)C, H, N..
N-(4-ethoxyphenyl)-N,2,6-trimethylfuro[2,3-d]pyrimidin-4-amine (12)
Using the general procedure described above, compound 12 (49%) was obtained as colorless crystals: mp 107.6–108.2 °C; Rf 0.64 (EtOAc/Hexane, 1:3); 1H NMR (DMSO-d6) δ 1.36–1.38 (t, 3 H, J = 5.6 Hz, OCH2CH3), 2.16 (s, 3 H, CH3),2.48 (s, 3 H, CH3), 3.45 (s, 3 H, NCH3), 4.85–4.10 (q, 2 H, OCH2CH3), 4.58 (s, 1 H, 5-CH), 7.04–7.06 (d, 2 H, J = 7.2 Hz, C6H4), 7.26–7.28 (d, 2 H, J = 7.2 Hz, C6H4); Anal. (C17H19N3O2), C, H, N.
4-(5-methoxyindolin-1-yl)-2,6-dimethylfuro[2,3-d]pyrimidine (8)
To a 100 mL round-bottomed flask, flushed with nitrogen, were added 17 (91 mg, 0.5 mmol), 5-methoxyindoline (82 mg, 0.55 mmol), n-BuOH (10 mL), and 2–3 drops of conc. HCl. The reaction mixture was heated at reflux with stirring for 12 h until the starting material 17 disappeared (TLC). The reaction solution was allowed to cool to room temperature; the solvent was removed under reduced pressure, and the residue was purified by column chromatography on silica gel with hexane: acetyl acetate = 20:1 as the eluent. Fractions containing the product (TLC) were combined and evaporated to afford 93 mg (63%) 8 as a white powder: mp 201.1–202.3 °C; Rf 0.5 (EtOAc/Hexane 3:1); 1H NMR (DMSO-d6) δ 2.42 (s, 3 H), 2.53 (s, 3 H), 3.25 (t, 2 H), 3.75 (s, 3 H), 4.39 (t, 2 H), 6.79 (t, 2 H), 6.89 (d, 1 H), 8.48 (d, 1 H), Anal. (C17H17N3O2), C, H, N.
4-(5-methoxy-1H-indol-1-yl)-2,6-dimethylfuro[2,3-d]pyrimidine (9)
To a solution of 5-methoxy-1H-indole (74 mg, 0.5 mmol) in 5 mL DMF was added NaH (13 mg, 0.55 mmol) at 0 °C and stirred for 30 min at the same temperature. To the solution was added 17 (273 mg, 1.5 mmol), and the mixture was stirred for another 2 h at ambient temperature. After adding 1 mL 1 N HCl to terminate the reaction, the solvent was removed under reduced pressure. The crude mixture was purified by silica gel column chromatography using hexane: acetyl acetate=20:1 as the eluent. Fractions containing the product (TLC) were combined and evaporated to afford 66 mg (41%) 9 as colorless crystals: mp 131.6–133.2 °C; Rf 0.28 (EtOAc/Hexane 3:1); 1H NMR (DMSO-d6) δ 2.51 (s, 3 H), 2.71 (s, 3 H), 3.81 (s, 3 H), 6.84 (d, 1 H, J = 2.8 Hz ), 6.95 (dd, 1 H, J1 = 7.2 Hz, J2 = 2.0 Hz), 7.09 (s, 1 H), 7.20 (d, 1 H, J = 2.0 Hz ), 8.04 (d, 1 H, J = 2.8 Hz), 8.57 (d, 1 H, J = 7.2 Hz ), Anal. (C17H15N3O2) C, H, N.
4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2,6-dimethylfuro[2,3-d]pyrimidine (10)
To a 100 mL round-bottomed flask flushed with nitrogen were added 17 (91 mg, 0.5 mmol), 6-methoxy-1,2,3,4-tetrahydroquinoline (90 mg, 0.55 mmol), BuOH (10 mL), and 2–3 drops of conc. HCl. The reaction mixture was heated at reflux with stirring for 12 h until the starting material 17 disappeared (TLC). The reaction solution was allowed to cool to room temperature; the solvent was removed under reduced pressure, and the residue was purified by column chromatography on silica gel with hexane: acetyl acetate = 20:1 as the eluent. Fractions containing the product (TLC) were combined and evaporated to afford 74 mg (48%) of 10 as a pink powder: mp 108.9–109.6 °C; Rf 0.5 (EtOAc/Hexane 3:1); 1H NMR (DMSO-d6) δ 1.90 (m, 2 H), 2.28 (s, 3 H), 2.48 (s, 3 H), 2.73 (t, 2 H), 3.75 (s, 3 H), 3.94 (t, 2 H), 5.53 (s, 1 H), 6.76 (dd, 1 H, J1 = 7.2 Hz, J2 = 2.0 Hz), 6.85 (d, 1 H, J = 2.0 Hz), 6.05 (d, 1 H, J = 7.2 Hz), Anal. (C18H19N3O2) C, H, N.
Molecular modeling
The X-ray crystal structures of tubulin co-crystallized with DAMA-colchicine, a close structural analog of colchicine (PDB: 1SA028, 3.58 Å resolution) and VEGFR-2 co-crystallized with a furo[2,3-d]pyrimidine inhibitor (PDB: 1YWN,29 1.71 Å resolution) were obtained from the protein database. The preparation and validation of the PDGFR-β homology model has been previously reported.24
Preparation of receptor and ligands for docking
The crystal structures of tubulin and VEGFR-2 and the homology model for PDGFR-β were imported into MOE 2013.08.39 After addition of hydrogen atoms, the protein was then “prepared” using the LigX function in MOE 2013.08, which is a collection of procedures that conducts interactive ligand modification and energy minimization in the active site of a flexible receptor. The procedure was performed with the default settings.
Ligands were built using the molecule builder function in MOE, energy minimized to local minima using the MMF94X forcefield to a constant (0.05 kcal/mol). Ligands were docked into the active site of the prepared protein using LeadIT 2.1.6.31 The docking site was restricted to the active site pocket residues defined as residues within 6.5 Å of the bound crystal structure ligand. Docking was performed using a hybrid (enthalpy and entropy) approach as the placement method. Docked poses were scored using the default threshold (full score contribution threshold of 0.3; no score contribution threshold of 0.7). Clash handling (protein-ligand and intra-ligand were set at default values of 2.9 Å3 and 0.6 Å3, respectively. The maximum number of solutions per iteration was set at 300, and the maximum number of solutions per fragment was set at 300. The best docked poses were exported as sdf files and visualized using the software CCP4mg.32
Docking Procedure
Colchicine site on tubulin
The A and B subunits of the protein, along with the crystallized ligand, were retained, while the C, D, and E subunits, GTP, GDP, and Mg ions were deleted. After addition of hydrogen atoms, the protein was then “prepared” using the LigX function in MOE as described above.
To validate the utility of LeadIT 2.1.6 for docking ligands into the active site, DAMA-colchicine, the co-crystallized ligand in the crystal structure (PDB: 1SA0),28 was built using the molecule builder, energy minimized, and docked into the active site using the above parameters. The best docked pose of DAMA-colchicine displayed an rmsd of 1.10 Å compared with the crystal structure pose of DAMA-colchicine. LeadIT 2.1.6 was thus validated for our docking studies. Docking studies were performed for 1–12 and the standard compounds using the same settings. Poses from the docking experiment were visualized using MOE and CCP4mg.
Kinase docking
After addition of hydrogen atoms, the proteins were then “prepared” using the LigX function in MOE as described above. To validate the utility of LeadIT 2.1.6 for docking ligands into the active site of the RTKs, the co-crystallized ligands in the crystal structures (N-{4-[4-amino-6-(4-methoxyphenyl)furo[2,3-d]pyrimidin-5-yl]phenyl}-N'-[2-fluoro-5-(trifluoromethyl)phenyl]urea from PDB 1YWN29 for VEGFR2, [6,7-Bis(2-methoxy-ethoxy)quinazoline-4-yl]-(3-ethynylphenyl)amine from PDB 1M17 for EGFR30) were built using the molecule builder, energy minimized using MMFF94x forcefield to a gradient of 0.05 kcal/mol and docked into the active site using the above parameters. The best docked poses of the original ligands displayed RMSD values of 0.9 Å and 0.91 Å, respectively, compared to the crystal structure poses, thus validating our docking procedure with LeadIT 2.1.6
Systematic conformational search using Sybyl-X 2.1.1
A systematic conformational search was carried out for 1 – 12 using Sybyl-X 2.1.127 by importing the energy minimized conformations of these compounds used for the docking studies. All applicable rotatable bonds (bonds a–c, see Figures 3 & 4) were selected for each compound and the search was carried out using 5° increments, and the energy of the ensuing poses was recorded. The poses were exported to MOE 2013.08,39 and RMSD calculations with the docked poses were performed in MOE 2013.0839 using an SVL code (mol_rmsd) obtained from the SVL exchange website.40
Biological evaluation methods
Antibodies
The PY-HRP antibody was from BD Transduction Laboratories (Franklin Lakes, NJ). Antibodies against EGFR, PDGFR-β, and VEGFR-2 were purchased from Cell Signaling Technology (Danvers, MA).
Phosphotyrosine ELISA
Cells used were cancer cell lines naturally expressing high levels of EGFR (A431), VEGFR-2 (U251), and PDGFR-β (SH-SY5Y). Expression levels at the RNA level were derived from the NCI Developmental Therapeutics Program (NCI-DTP) web site public molecular target information. Briefly, cells at 60–75% confluence were placed in serum-free medium for 18 h to reduce the background of phosphorylation. Cells were always >98% viable by Trypan blue exclusion. Cells were then pretreated for 60 min with a range of concentrations of 10,000-0.17 nM compound, in ⅓ log increments. The cells were then stimulated by the addition of 100 ng/mL EGF, VEGF, or PDGF-BB for 10 min. The reaction was stopped, and cells were permeabilized by quickly removing the media and adding ice-cold Tris-buffered saline (TBS) containing 0.05% Triton X-100, protease inhibitor cocktail and tyrosine phosphatase inhibitor cocktail. The TBS solution was then removed and cells fixed to the plate for 30 min at 60 °C, with a further incubation in 70% ethanol for 30 min. Cells were exposed to a blocking solution (TBS with 1% bovine serum albumin) for 1 h, washed, and then a horseradish peroxidase (HRP)-conjugated phosphotyrosine (PY) antibody was added overnight. The antibody was removed, cells were washed again in TBS, exposed to an enhanced luminol ELISA substrate (Pierce Chemical EMD, Rockford, IL) and light emission was measured using a Biotek (Winooski, VT) microplate reader. Data were graphed as a percent of the control (cells receiving growth factor alone), and IC50 values were calculated from two to three separate experiments with 6–8 replicates for each experiment using non-linear regression dose-response relation analysis with Prism 6.0 software (GraphPad, San Diego CA).
CAM assay of angiogenesis
The CAM assay is a standard assay for testing antiangiogenic agents. The CAM assay used in these studies was performed as described previously.22 Briefly, fertile leghorn chicken eggs (Ideal Poultry, Cameron, TX) were incubated for 10 days. The proangiogenic factors, human VEGF-165 and bFGF (100 ng each) were then added at saturation to a 6 mm microbial testing disk (BBL, Cockeysville, MD) and placed onto the CAM by breaking a small hole in the superior surface of the egg. Antiangiogenic compounds, at various doses, were added 8 h after the VEGF/bFGF at saturation to the same microbial testing disk, and the embryos were incubated for an additional 40 h. After 48 h, the CAMs were perfused with 2% paraformaldehyde containing 0.025% Triton X-100 for 20 sec, excised around the area of treatment, fixed again in 4% paraformaldehyde for 30 min, placed on Petri dishes, and a digitized image was taken using a dissecting microscope (Wild M400; Bannockburn, IL) at 7.5X and SPOT enhanced digital imaging system (Diagnostic Instruments, Sterling Heights, MI). A grid was then added to the digital CAM images, and the average number of vessels within 5–7 grids counted as a measure of vascularity. Sunitinib and semaxanib were used as positive controls for antiangiogenic activity. Data were graphed as a percent of CAMs receiving bFGF/VEGF only and IC50 values calculated from two to three separate experiments with 2–5 replicates per experiment using non-linear regression dose-response relation analysis.
Evaluation of Microtubule Effects
Indirect immunofluorescence was used to evaluate the effects of the compounds on cellular microtubules as described previously.41 Briefly, A-10 cells were treated for 18 h with vehicle (DMSO) or a compound, and microtubule effects were evaluated microscopically using a β-tubulin antibody. The EC50 was calculated from 3 independent experiments.
Antiproliferative Effects
The antiproliferative effects of compounds were evaluated using the sulforhodamine B (SRB) assay as described previously.41 The cells were treated with the compounds, vehicle (DMSO) or positive controls, (paclitaxel, CA-4) for 48 h. The IC50 values represent the mean of 3 independent experiments each conducted with triplicate wells ± SD.
Tubulin studies
Bovine brain tubulin was purified as described previously.42 The tubulin assembly assay43 and the [3H]colchicine binding assay44 were performed as described previously.
Supplementary Material
Acknowledgements
We gratefully acknowledge the support of the NCI for grant CA142868 (AG, SLM) and for conducting the 60 cell line screen, support by the Duquesne University Adrian Van Kaam Chair in Scholarly Excellence (AG), the CTRC Cancer Center Support Grant, P30 CA054174 and an NSF equipment grant for NMR instrumentation (NMR: CHE 0614785).
Abbreviations
- CA-4P
combretastatin A-4 phosphate
- VEGFR-2
vascular endothelial growth factor receptor-2
- RTKs
receptor tyrosine kinases
- 2ME2
2-methoxyestradiol
- PDGFR-β
platelet-derived growth factor receptor β
- CAM
chorioallantoic membrane
- DDQ
2,3-dichloro-5,6-dicyano-1,4-benzoquinone
- DAMA-colchicine
N-deacetyl-N-(2-mercaptoacetyl)colchicine
- Pgp
P-glycoprotein
- TBS
Tris-buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available: Elemental analysis is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Folkman J. Anti-Angiogenesis: New Concept for Therapy of Solid Tumors. Annals of Surgery. 1972;175:409–416. doi: 10.1097/00000658-197203000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Folkman J. Patterns and Emerging Mechanisms of the Angiogenic Switch during Tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 3.Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nat Rev Cancer. 2002;2:727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain RK, Carmeliet P. SnapShot: Tumor Angiogenesis. Cell. 2012;149:1408–1408.e1. doi: 10.1016/j.cell.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 7.Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai R, Nguyen TL, Burnett JC, Atasoylu O, Munro MHG, Pettit GR, Smith AB, Gussio R, Hamel E. Interactions of Halichondrin B and Eribulin with Tubulin. Journal of Chemical Information and Modeling. 2011;51:1393–1404. doi: 10.1021/ci200077t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamel E. Natural products which interact with tubulin in the vinca domain: Maytansine, rhizoxin, phomopsin a, dolastatins 10 and 15 and halichondrin B. Pharmacology & Therapeutics. 1992;55:31–51. doi: 10.1016/0163-7258(92)90028-x. [DOI] [PubMed] [Google Scholar]
- 10.Prota AE, Bargsten K, Diaz JF, Marsh M, Cuevas C, Liniger M, Neuhaus C, Andreu JM, Altmann K-H, Steinmetz MO. A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs. Proceedings of the National Academy of Sciences. 2014;111:13817–13821. doi: 10.1073/pnas.1408124111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nathan P, Zweifel M, Padhani AR, Koh D-M, Ng M, Collins DJ, Harris A, Carden C, Smythe J, Fisher N, Taylor NJ, Stirling JJ, Lu S-P, Leach MO, Rustin GJS, Judson I. Phase I Trial of Combretastatin A4 Phosphate (CA4P) in Combination with Bevacizumab in Patients with Advanced Cancer. Clinical Cancer Research. 2012;18:3428–3439. doi: 10.1158/1078-0432.CCR-11-3376. [DOI] [PubMed] [Google Scholar]
- 12.Patterson DM, Zweifel M, Middleton MR, Price PM, Folkes LK, Stratford MRL, Ross P, Halford S, Peters J, Balkissoon J, Chaplin DJ, Padhani AR, Rustin GJS. Phase I Clinical and Pharmacokinetic Evaluation of the Vascular-Disrupting Agent OXi4503 in Patients with Advanced Solid Tumors. Clinical Cancer Research. 2012;18:1415–1425. doi: 10.1158/1078-0432.CCR-11-2414. [DOI] [PubMed] [Google Scholar]
- 13.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10:194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 14.Cesca M, Bizzaro F, Zucchetti M, Giavazzi R. Tumor delivery of chemotherapy combined with inhibitors of angiogenesis and vascular targeting agents. Front Oncol. 2013;3:259. doi: 10.3389/fonc.2013.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall M, Gourley C, McNeish I, Ledermann J, Gore M, Jayson G, Perren T, Rustin G, Kaye S. Targeted anti-vascular therapies for ovarian cancer: current evidence. Br J Cancer. 2013;108:250–258. doi: 10.1038/bjc.2012.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heath VL, Bicknell R. Anticancer strategies involving the vasculature. Nat Rev Clin Oncol. 2009;6:395–404. doi: 10.1038/nrclinonc.2009.52. [DOI] [PubMed] [Google Scholar]
- 17.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 18.Ma J, Waxman DJ. Combination of antiangiogenesis with chemotherapy for more effective cancer treatment. Mol Cancer Ther. 2008;7:3670–84. doi: 10.1158/1535-7163.MCT-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews DJGME. Current Challenges and Future Directions. In: Matthews DJGME, editor. Targeting Protein Kinases for Cancer Therapy. Hoboken, NJ: Wiley & Sons Inc; 2010. pp. 623–663. [Google Scholar]
- 20.Postel-Vinay S, Armand J-P. Targeting Angiogenesis. In: Giaccone G, Soria J-C, editors. Targeted Therapies in Oncology. 2nd ed. Boca Raton, FL: CRC Press; 2013. pp. 283–314. [Google Scholar]
- 21.GlaxoSmithKline. Combination Of Lapatinib With Carboplatin, Paclitaxel and Trastuzumab In Metastatic Breast Cancer. In: ClinicalTrials.gov [Internet] Bethesda (MD): National Library of Medicine (US); [[cited 2015 Mar 06]]. 2000. Available from: http://clinicaltrials.gov/show/NCT00367471 NLM Identifier: NCT00367471. [Google Scholar]
- 22.Vermont Uo. ClinicalTrials.gov [Internet] Bethesda (MD): National Library of Medicine (US); 2000. [cited 2015 Mar 06]. Docetaxel, Gemcitabine and Pazopanib as Treatment for Soft Tissue Sarcoma. Available from: http://clinicaltrials.gov/show/NCT01719302 NLM Identifier: NCT01719302. [Google Scholar]
- 23.Twenty eight clinical trials were found on clinicaltrials.gov (accessed in March 2015) site in which RTK inhibitors were being used in combination with anti-tubulins and other chemotherapeutic agents. Of these, 16 trials are currently in progress and the identification numbers in clinicaltrials.gov are provided below: NCT01855750; NCT01804530; NCT01606878; NCT01746277; NCT02326285; NCT01620190; NCT01974440; NCT01683994; NCT02378389; NCT01939054; NCT01719302; NCT02191059; NCT02191059; NCT01876082; NCT00567554; NCT00367471. https://clinicaltrials.gov/ct2/results?term=receptor+tyrosine+kinase+AND+tubulin&Search=Search (03/06/2015)
- 24.Gangjee A, Zaware N, Raghavan S, Ihnat M, Shenoy S, Kisliuk RL. Single Agents with Designed Combination Chemotherapy Potential: Synthesis and Evaluation of Substituted Pyrimido[4,5-b]indoles as Receptor Tyrosine Kinase and Thymidylate Synthase Inhibitors and as Antitumor Agents. Journal of Medicinal Chemistry. 2010;53:1563–1578. doi: 10.1021/jm9011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Raghavan S, Ihnat M, Thorpe JE, Disch BC, Bastian A, Bailey-Downs LC, Dybdal-Hargreaves NF, Rohena CC, Hamel E, Mooberry SL, Gangjee A. The design and discovery of water soluble 4-substituted-2,6-dimethylfuro[2,3-d]pyrimidines as multitargeted receptor tyrosine kinase inhibitors and microtubule targeting antitumor agents. Bioorg Med Chem. 2014;22:3753–72. doi: 10.1016/j.bmc.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anighoro A, Bajorath J, Rastelli G. Polypharmacology: Challenges and Opportunities in Drug Discovery. Journal of Medicinal Chemistry. 2014 doi: 10.1021/jm5006463. [DOI] [PubMed] [Google Scholar]
- 27.Certera LP. 210 N. Tucker Blvd, Suite 350, St. Louis MO 63101. Sybyl X 2.1.1. 2013 www.certara.com.
- 28.Ravelli RBG, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki Y, Matsunaga S, Tang J, Maeda Y, Nakano M, Philippe RJ, Shibahara M, Liu W, Sato H, Wang L, Nolte RT. Novel 4-amino-furo[2,3-d]pyrimidines as Tie-2 and VEGFR2 dual inhibitors. Bioorganic & Medicinal Chemistry Letters. 2005;15:2203–2207. doi: 10.1016/j.bmcl.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 30.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–72. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 31.LeadIT 2.1.6. St. Augustin, Germany: BioSolveIT GmbH, An der Ziegelei 79, 53757; [Google Scholar]
- 32.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallographica Section D. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornev AP, Haste NM, Taylor SS, Ten Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proceedings of the National Academy of Sciences. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gangjee A, Yu J, Copper JE, Smith CD. Discovery of novel antitumor antimitotic agents that also reverse tumor resistance. J Med Chem. 2007;50:3290–301. doi: 10.1021/jm070194u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Zhou X, Kisliuk RL, Piraino J, Cody V, Gangjee A. Design, synthesis, biological evaluation and X-ray crystal structure of novel classical 6,5,6-tricyclic benzo[4,5]thieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors. Bioorg Med Chem. 2011;19:3585–94. doi: 10.1016/j.bmc.2011.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosowsky A, Chen KK, Lin M. 2,4-Diaminothieno(2,3-d)pyrimidines as antifolates and antimalarials. 3. Synthesis of 5,6-disubstituted derivatives and related tetracyclic analogs. J Med Chem. 1973;16:191–4. doi: 10.1021/jm00261a004. [DOI] [PubMed] [Google Scholar]
- 37.Wartenberg FHKT, Wetzel W, Wydra M, Benz A. Method for Producing Benzo Annelated Heterocycles. 2001 [Google Scholar]
- 38.Yadav PP, Gupta P, Chaturvedi AK, Shukla PK, Maurya R. Synthesis of 4-hydroxy-1-methylindole and benzo[b]thiophen-4-ol based unnatural flavonoids as new class of antimicrobial agents. Bioorganic & Medicinal Chemistry. 2005;13:1497–1505. doi: 10.1016/j.bmc.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molecular Operating Environment (MOE), 2013.08. 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: Chemical Computing Group Inc.; 2014. [Google Scholar]
- 40.mol_rmsd, Scientific Vector Language (SVL) source code provided by Chemical Computing Group Inc. 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: 2013. [Google Scholar]
- 41.Risinger AL, Jackson EM, Polin LA, Helms GL, LeBoeuf DA, Joe PA, Hopper-Borge E, Ludueña RF, Kruh GD, Mooberry SL. The Taccalonolides: Microtubule Stabilizers That Circumvent Clinically Relevant Taxane Resistance Mechanisms. Cancer Research. 2008;68:8881–8888. doi: 10.1158/0008-5472.CAN-08-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamel E, Lin CM. Separation of active tubulin and microtubule-associated proteins by ultracentrifugation and isolation of a component causing the formation of microtubule bundles. Biochemistry. 1984;23:4173–4184. doi: 10.1021/bi00313a026. [DOI] [PubMed] [Google Scholar]
- 43.Hamel E. Evaluation of antimitotic agents by quantitative comparisons of their effects on the polymerization of purified tubulin. Cell Biochemistry and Biophysics. 2003;38:1–21. doi: 10.1385/CBB:38:1:1. [DOI] [PubMed] [Google Scholar]
- 44.Verdier-Pinard P, Lai J-Y, Yoo H-D, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Structure-Activity Analysis of the Interaction of Curacin A, the Potent Colchicine Site Antimitotic Agent, with Tubulin and Effects of Analogs on the Growth of MCF-7 Breast Cancer Cells. Molecular Pharmacology. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.