Abstract
Contractile motion is the simplest metric of cardiomyocyte health in vitro, but unbiased quantification is challenging. We describe a rapid automated method, requiring only standard video microscopy, to analyze the contractility of human-induced pluripotent stem cell-derived cardiomyocytes (iPS-CM). New algorithms for generating and filtering motion vectors combined with a newly developed isogenic iPSC line harboring genetically encoded calcium indicator, GCaMP6f, allow simultaneous user-independent measurement and analysis of the coupling between calcium flux and contractility. The relative performance of these algorithms, in terms of improving signal to noise, was tested. Applying these algorithms allowed analysis of contractility in iPS-CM cultured over multiple spatial scales from single cells to three-dimensional constructs. This open source software was validated with analysis of isoproterenol response in these cells, and can be applied in future studies comparing the drug responsiveness of iPS-CM cultured in different microenvironments in the context of tissue engineering.
Introduction
Human-induced pluripotent stem cell (iPSC) and genome engineering technologies now offer a means to model human disease, cardiotoxicity, and therapy in vitro, to overcome the limitations of animal models.1 However, robust unbiased methods to assess the physiology of cardiomyocytes derived from in vitro differentiation of pluripotent stem cells are lacking. Clinically, heart disease manifests through changes in physical characteristics of the tissue, leading to aberrations in mechanical strain, movement coordination, and beating frequency (e.g., arrhythmia). Based on the electrochemical activation underlying these changes, high-throughput in vitro assays are being developed to monitor the electrical behavior of either single cells or clusters of primary and pluripotent cell-derived cardiomyocytes.2–4 These high-throughput assays complement traditional lower-throughput patch-clamp electrophysiology, and are complemented by novel genetic tools for imaging calcium transients.5,6 However, these approaches either require that cardiomyocytes be plated onto specialized materials (e.g., multielectrode arrays, microelectrode arrays [MEAs], or synthetic hydrogels embedded with fluorescent particles to track strain7) or involve genetic manipulations.5 In contrast, qualitative aspects of cardiomyocyte physiology can be easily monitored with standard optical microscopy, which does not require genetic engineering or specialized substrates and can be performed continuously throughout the time course of differentiation or drug evaluation. Hence, image-processing tools have been applied in attempts to turn such qualitative observations into quantitative data.3,8–10
Popular image-processing techniques include edge detection and simple pixel intensity monitoring.8,10 These techniques require the user to specify regions of interest to monitor, which introduces inherent bias.8,11 To avoid this bias, a series of more robust methods to detect motion, collectively referred to as optical flow,12 are now used to analyze primary13,14 and pluripotent stem cell-derived9,15 cardiomyocytes. In the field of computer vision, optical flow describes the motion of pixels from one frame to the next in a two-dimensional (2D) video.12 One of the simplest means of calculating optical flow is through block matching, in which macroblocks of pixels within an image are used as fiduciary markers to estimate movement from one still frame to the next (Fig. 1).
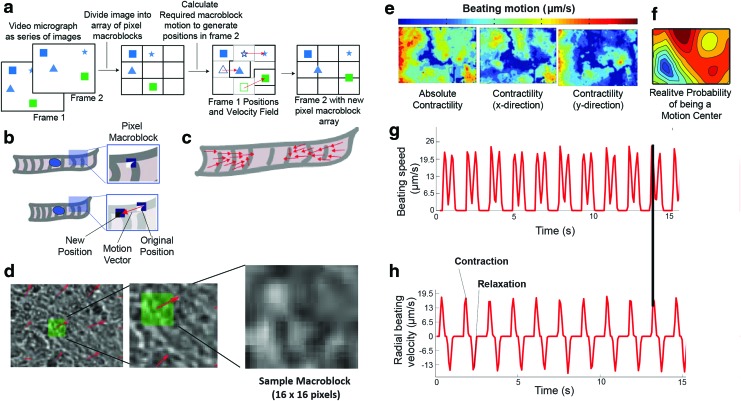
FIG. 1.
Block-matching and cleaning algorithms to analyze cardiomyocyte contractility. (a) A block-matching optical flow algorithm estimates motion in arbitrary sets of images. First, a still frame (Frame 1; left) is divided up into a grid of macroblocks. In a subsequent frame (Frame 2; right), the position of the macroblock is identified, and the motion vector (red) connecting the original and new macroblock positions is calculated (center). Finally, the image in the new frame is divided into a new set of macroblocks, and the process is repeated. Note: Macroblocks are placed based on overall image boundaries, irrespective of the position of individual objects. (b) Schematic depicting the application of block matching to cardiomyocyte motion analysis. (c) By tracking movement of all macroblocks from Frame 1 to Frame 2, a field of vectors indicating motion velocity is generated, as shown in schematic form (top) and within a beating sheet of induced pluripotent stem-derived cardiomyocytes (iPS-CM, bottom). (d) Example of a typical macroblock from iPS-CM motion analysis; A close-up image within an iPS-CM sheet, indicating a macroblock (green) that is shown at progressively higher magnification. (e) Heat maps depicting the time-averaged magnitude of all motion (left), horizontal motion only (center), or vertical motion only (right) for the images used to generate the data in Figure 1. (f) Heat map depicting the likelihood that a specific region within the image is a center of contraction. A contractile center is defined as a region from which contractions (inward motion) are maximized over the time-averaged course of the movie. Note: Regions of maximum contraction tend to be outside of motion centers. (g) Representative tracing of the average speed versus time for a cardiomyocyte sheet. Contraction and relaxation appear as doublet peaks, and beat rate is calculated from the time shift between contractions. (h) Tracings obtained for the same region as in (g), but depicting average velocity versus time. Black drop line denotes a contraction, as defined by both methods. Color images available online at www.liebertpub.com/tec
Analysis of motion vector fields generated by block matching yields several parameters related to the health of cardiac tissue that cannot be identified with simple edge detection, such as the robustness of mechanical coupling across tissue, which can be assessed through coordination of motion from different parts of the tissue.12–15 However, because of technical obstacles, many users have not adopted this unbiased approach for analyzing cardiomyocyte contractility. First, analyzing videos of single or sparse cardiomyocytes is difficult because their contraction speed is often obscured by nearby Brownian motion or camera noise. This prevents direct comparisons of the drug IC50 and other relevant parameters between cardiomyocytes cultured in 2D versus three-dimensional (3D), a topic of interest to tissue engineers. Second, optical flow analysis has been applied only to bright-field videos of cardiomyocytes,9,13,14 in which motion cannot be tracked simultaneously with voltage or calcium flux using fluorescent reporters (e.g., to assess electromechanical coupling). Finally, optical flow analysis typically requires significant experience in computer programming.
Cardiomyocytes are derived from in vitro differentiation of pluripotent stem cells, because these cells do not have the characteristic rod shape or contractile motion of adult cardiomyocytes. Thus, to quantify drug response, disease physiology, and maturation characteristics of this cell population, we developed new algorithms for optical flow analysis that allow greater sensitivity to contractile motion and improved ability to reject noise from Brownian motion and other background sources, to monitor the physiology of the contractility of iPS-derived cardiomyocytes (iPS-CM) independent of the culture platform. By applying an improved single-sweep exhaustive search block-matching algorithm, we were able to rapidly determine contractile motion in sheets of iPS-CM derived from high-efficiency differentiations. New algorithms were applied to detect the center of contractile motion and to measure cardiomyocyte contractility even in sparse cultures or singularized cells. These algorithms also allowed optical flow analysis of movies taken with the genetic calcium reporter, GCaMP6f, thus allowing, for the first time, simultaneous measurements of motion and calcium flux. This may be applied in future studies to detect the effects of drugs or genetic mutations on calcium–contraction coupling over long-term culture. To demonstrate the power of these tools, we applied them toward analyzing the dose and temporal response of iPS-CM to isoproterenol. The code required to implement this analysis is also provided in this study with a user-friendly interface, to obviate the need for programming ability. This user-friendly tool requires only standard video microscopy, and even relatively low frame rates (<30 frames per second [fps]) can be used to acquire most of the parameters relevant to assessing iPS-CM behavior and drug response, including beat rate. It can be a widely used tool as an alternative to methods that require more expertise and costly equipment.
Materials and Methods
iPSC culture and cardiomyocyte differentiation
Wild-type (WT) human iPSCs were reprogrammed using nonintegrating vectors16 from dermal fibroblasts obtained from a healthy volunteer who had a normal electrocardiogram and no known family history of cardiac disease at the time of donation. The cells are hereafter referred to as WTC. Before cardiac differentiation, iPSCs were passaged without feeder layers in the Essential 8 medium (Life Technologies) at a constant density of 8000 cells/cm2 on substrates coated with growth factor-reduced Matrigel (BD Biosciences) for at least three passages.
Cardiomyocyte differentiation was achieved by modifying the Wnt GiWi method developed by Lian et al.17,18 Briefly, human iPSCs were dissociated with Accutase (Life Technologies) and seeded onto Matrigel-coated cell culture plates at 25,000 cells/cm2 in E8 media supplemented with 10 μM Y-27632 (Sigma). The medium was changed 24 h later to E8 without Y27632, and cells were expanded in E8 for 2 additional days. On day 0, iPSCs were treated with 12 μM of the GSK3-β inhibitor, CHIR99021 (CHIR; Tocris), in the RPMI 1640 medium containing B27 supplement without insulin (RPMI/B27-I; Life Technologies). Exactly 24 h after adding CHIR (day 1), the medium was changed to RPMI/B27-I and cells were incubated for another 48 h. On day 3, cells were exposed to a 50:50 mixture of their own conditioned media and fresh RPMI/B27-I. The combined media were supplemented with the Wnt inhibitor IWP-2 (Tocris) at a final concentration of 5 μM. On day 5, 48 h after adding IWP-2, media were changed to RPMI/B27-I for 2 days, and then changed to RPMI 1640 containing B27 complete supplement (RPMI/B27C; Life Technologies) on day 7. Thereafter, cells were fed every 3 days with RPMI/B27C.
For motion tracking studies, iPS-CM were analyzed either within the wells where they were originally differentiated or they were singularized and replated onto a second set of substrates. Using the modified GiWi differentiation protocols (above), we achieved differentiation efficiencies of above 70% (based on flow cytometry analysis of cardiac Troponin T15), and only wells that had visibly beating sheets were used for tracking analysis. For replating, iPS-CM on days 15–18 of differentiation were retrieved by digestion for 10–15 min in 0.25% trypsin, which was quenched in embryoid body media 20 (EB20; Knockout Dulbecco's Modified Eagle's Medium containing 20% characterized fetal bovine serum, 1 mM nonessential amino acids, 1 mM l-glutamine, 0.1 mM β-mercaptoethanol). Cells were carefully singularized to prevent mechanical shear19 before they were counted. To seed single cells, iPS-CM were replated at 10,000 cells/cm2, whereas for clusters, cells were replated at 50,000 cells/cm2. For 3D scaffolds, cells were seeded at a density of 107 cells/mL. Cells were replated onto Matrigel-coated substrates, or 3D scaffolds, in EB20 supplemented with 10 μM Y27632.20
Analysis of iPS-CM spontaneous beating and drug response with video microscopy
iPS-CM were imaged on a microscope at 37°C and 5% CO2 to maintain physiologic conditions. The reproducibility of the approach was verified by performing the analysis using two different systems: (1) Molecular Devices ImageXpress Micro and (2) Zeiss Z1 AxioObserver Microscope equipped with a Hamamatsu ORCA-Flash 4.0 sCMOS camera. The x, y, and z positions of each region of interest (ROI) were preselected at random and programmed using Zeiss Zen software. Plates containing samples were incubated on the microscope for 30 min to equilibrate conditions before analyzing baseline beating of the cells. Videos of cells were taken at 14 fps over 10–20 s for each position. At least 30 positions were used to analyze baseline beating. For drug response studies, media containing 10×stock solutions of isoproterenol were equilibrated to the same conditions within a separate multiwell plate, and immediately following the baseline imaging, the drug was rapidly added. The final concentration of isoproterenol ranged from 100 nM to 10 μM after diluting into culture media. Bright-field videos (14 fps) were taken at 0.5-, 2-, 8-, and 24-h intervals after adding the drug. To verify that a 14 fps capture rate was sufficient to capture dynamic cell responses, we also performed analysis of baseline iPS-CM beating at 100 fps using a Hamamatsu ORCA-Flash4.0 camera.
Software availability
All image analyses tools described in this study were developed in Matlab (Mathworks). The implementation of these algorithms requires the Image Processing and Parallel Computing Toolboxes, in addition to the base Matlab computing environment. Although the code is distributed in this study for users to further modify, under GNU license, users who simply wish to apply the code can do so without programming knowledge using the supplied user interfaces (Supplementary Figs. S1 and S2; Supplementary Data are available online at www.liebertpub.com/tec). The software is available at http://gladstone.ucsf.edu/46749d811/.
Optical flow analysis of spontaneous cardiomyocyte beating
Videos of spontaneously beating iPS-CM (no exogenous electrical stimulation) were exported as a series of single-frame image (.tiff) files. The block-matching method used for motion tracking employs an exhaustive step search. Movement of a given pixel macroblock at the ith frame (Fig. 1a–c) is calculated by matching a block of pixels (typically 16×16; Fig. 1d) to an identically-sized block of pixels in the i+dth frame, where d is a delay in frames that the user selects. Similar to previous analyses of confluent monolayers of rodent primary cardiomyocytes,13 we empirically found an optimal delay of two frames. Before performing this analysis, the user inputs a maximum pixel shift parameter that describes the largest possible movement, p, in pixels that a given macroblock can make. We empirically set p to 7 for images taken with 10×or 20×objectives, 1.02 and 0.512 μm per pixel, respectively. For block matching, a given pixel macroblock of n×n pixels from time i is first set to the exact same spatial position at time i+d. The entire macroblock is then subjected to rigid vertical and horizontal motion, one pixel at a time, until the maximum displacement p has been reached in each of the four cardinal directions. This sweeps the macroblock across p×p+1 different positions (the extra position is the origin) to create p×p+1 temporary new macroblocks. For each new temporary macroblock from time i, the quality of registration between the temporary macroblock and that set at the origin for time i+d is calculated by measuring the mean absolute difference (MAD) between each pixel. The MAD is then used to assign a cost to each possible position for macroblock movement. Although more complex methods for image registration are available, MAD can be measured very rapidly. Within the p×p+1 array of possible new macroblock positions, the position that yields the minimum cost is selected as the new position.
By repeating the above procedure for each macroblock in the image, the computer generates an array of motion vectors that represent the motion of cells and tissue for each time frame (Fig. 1a–c, g). Taking all frames together, a time series of motion vectors that display the kinetics of the beating motion is obtained, which visually aids in interpreting videos of beating cardiomyocytes (Supplementary Movie S1).
Computational analysis of beating centers
To identify beating centers—defined as regions from which motion originates—new algorithms were developed. First, the user selects a ROI and then a subregion that contains the origin of contraction. The software then calculates, within the larger ROI, the net movement toward and away from each macroblock contained within the smaller ROI. The center of motion is identified as the pixel from which the difference between inward motion (toward the pixel; contraction, blue) and outward motion (away from the pixel; relaxation, red) is maximized (Fig. 1f; Supplementary Movie S2). The software further produces an image, depicting the likelihood that a given location is a beating center, and corresponding plots of beating toward and away from the motion center (Fig. 1h).
MEA analysis
On day 18, iPS-CM were plated onto Matrigel-coated MEA chips (MED-P545A; Alpha MED Scientific, Inc.). Field potential waveforms were recorded using a MED64 system (Alpha MED Scientific, Inc.). Bright-field videos were recorded using a CCD camera (QICAM; QImaging) on an Eclipse TE-300 microscope (Nikon) equipped with a heated stage (TP-SQ05; Tokai Hit Co.). During MEA recording and imaging, the cells were maintained at 37°C.
Patch clamp analysis
Patch clamp was performed as previously described.21 Briefly, 30 days after initiating differentiation with CHIR, visibly beating myocytes were dispersed by trypsinization onto fibronectin-coated coverslips (no. 1, CS15R; Warner Instruments). Populated coverslips were incubated in the RPMI complete medium (changed every 3 days) and were transferred to the superfusion bath (Warner RC-26GLP) on a Nikon TiS inverted microscope equipped with a photomultiplier (PMT) microfluorometer system (IonOptix; PMT400). Extracellular solutions, delivered locally near the patch-clamp electrode, were warmed to 30°C with a superfusion system (AutoMate Scientific). One myocyte of a synchronously beating microcluster was patch clamped through an Axopatch 200B amplifier (Molecular Devices) coupled through pClamp software (v10) to patch electrodes of 2–3 MΩ (1B-150F; WPI). The patch pipettes were filled with a solution containing (mM) 120 KCl, 20 NaHEPES, 10 MgATP, 5.0 K2EGTA, and 2 MgCl2, set to pH 7.1 with KOH, and supplemented with 240 μg/mL of amphotericin B.22 In the recording chamber, myocytes were superfused at constant flow (W2-64; Warner Instruments) with modified Tyrode's solution containing (mM) 137 NaCl, 10 NaHEPES, 10 dextrose, 5 KCl, 2 CaCl2, and 1 MgCl2, set to pH 7.4 with NaOH. With the Axopatch amplifier in current clamp mode at zero applied current, trains of spontaneous action potentials were digitized at 5 kHz with low-pass filtering at 2 kHz, recorded for 30 s per data file.
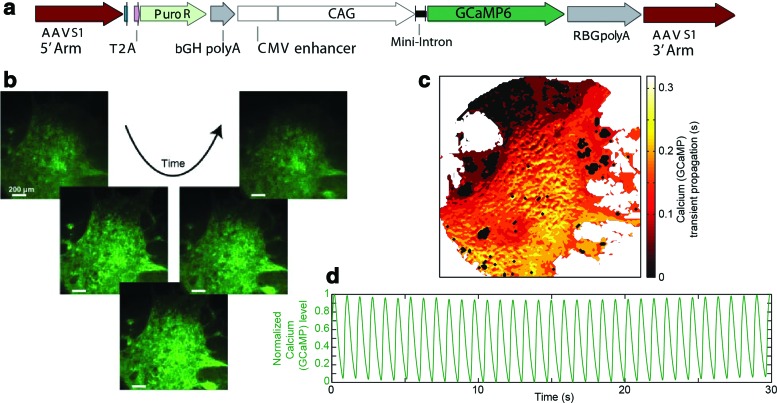
Insertion of the genetically encoded calcium indicator GCaMP6f into the AAVS1 safe harbor locus
The fast green fluorescent calcium indicator GCaMP6f open reading frame5 was placed under the control of the CAG promoter, and a puromycin resistance gene was genetically engineered into the AAVS1 locus of WTC iPSCs, through transcription activator-like effector nuclease (TALEN)-mediated genome editing.23 Note, puromycin is driven by the endogenous AAVS1 promoter, to improve the fidelity of targeting.21 iPSC lines containing the GCaMP6f cassette (GCaMP-WTC) were selected using puromycin (0.5 μg/mL). A heterozygous knockin clone was generated and differentiated into iPS-CM to check GCaMP6f functionality.
Batch analysis
To enable batch analysis of large datasets, several features were added to the software. First, a guided user interface specific to batch generation of motion vectors was created. Through this interface, users select any number of image series, for which motion tracking analysis will be performed, to generate motion vectors. For each image series, the user defines the delay, frame rate, and macroblock size, along with method (if any) to be used for segmenting regions containing cardiomyocytes.
Statistical analyses
The significance of differences between data points was assessed with a two-tailed student's t-test. To test the correlation between isoproterenol concentration and initial beat rate and the chronotropic response of iPS-CM, a Pearson correlation analysis was used (preceded by a Shapiro–Will test). Analyses were performed with GraphPad Prism 6 software.
Results
Optical flow analysis of cardiomyocytes differentiated from human iPSCs
As in previous studies, efficient differentiations yielded sheets of cardiomyocytes wherein at least 70% of cells were positive for cardiac troponin T by FACS analysis15 (Data not shown). Optical flow analysis applied to these sheets yielded similar results to those previously reported for sheets of neonatal or human embryonic stem cell-derived cardiomyocytes.9,13 One obvious benefit of optical flow is that it can summarize temporal information about beating through heat maps of the absolute motion and motion along specific axes (Fig. 1e). These maps provide a rapid overview of the relative movement in different parts of the field; acquiring similar information would take considerably longer with manual edge detection methods. Furthermore, with parallel computing, the computation time was significantly decreased depending on the number of available computing cores; typically, the time required to process 150 frames at 1024×1024 pixel resolution and a macroblock size of 16 decreases from 45 min to a maximum of 1.5 min. By selecting only a subregion of an image series for analysis, computing time could be further reduced to 30 s or less.
Within a differentiated sheet, some regions are responsible for generating the contractile motion and this may reflect differences in the transcriptome or strength of substrate adhesion of these regions compared to their surroundings. Our algorithms were able to identify these contractile centers in an unbiased manner (Fig. 1f). Often, though, a robust estimate of beating intensity (e.g., amplitude of motion vectors) and periodicity of contraction (e.g., beat rate) is sufficient to gauge cardiomyocyte behavior. These data can be extracted from motion vector maps simply by plotting the magnitude of vectors in a given region of interest. For cardiomyocytes, performing this analysis on a specific ROI produces a tracing somewhat reminiscent of action potential measurements (Fig. 1g).
Because this simple analysis does not discriminate based on the direction of motion vectors, it yields positive peaks both for contraction and relaxation. Simple peak identification tools can then be used to find contraction and relaxation peaks, in an automated manner, to measure maximum contraction velocity (a surrogate for beating intensity) and beat rate. A user interface allows peaks to be discriminated from background noise based on their magnitude and temporal separation. For a more robust analysis of peaks—or when the magnitude of motion varies over time—the autocorrelation analysis can be used to estimate beat rate. Both types of analyses can be performed within the DataEvaluation guided user interface (Supplementary Fig. S2).
To further discriminate between tachycardia and more pathologic beating aberrations (e.g., delayed after depolarizations, and early after depolarizations) either at a high or low frame rate, it is useful to calculate contractile motion toward a center of movement and relaxation away from the same center (Supplementary Fig. S3). Depending on cellular organization, a given field of view may have several contractile centers (Fig. 1f). For a given center of beating and an area immediately surrounding it, one can then generate a one-dimensional (1D) plot of contraction toward and relaxation away from the beating center (Fig. 1h).
Effects of camera frame rate on optical flow analysis
Although low frame rate cameras (those typically used on video microscopy systems) are capable of estimating beat rate regularity and direction in cells beating up to 3 Hz (a reasonable limit for human cardiomyocytes), robust estimation of maximum contraction velocity and overall shape of the contraction–relaxation waveforms requires videos taken at a high frame rate (Fig. 2).
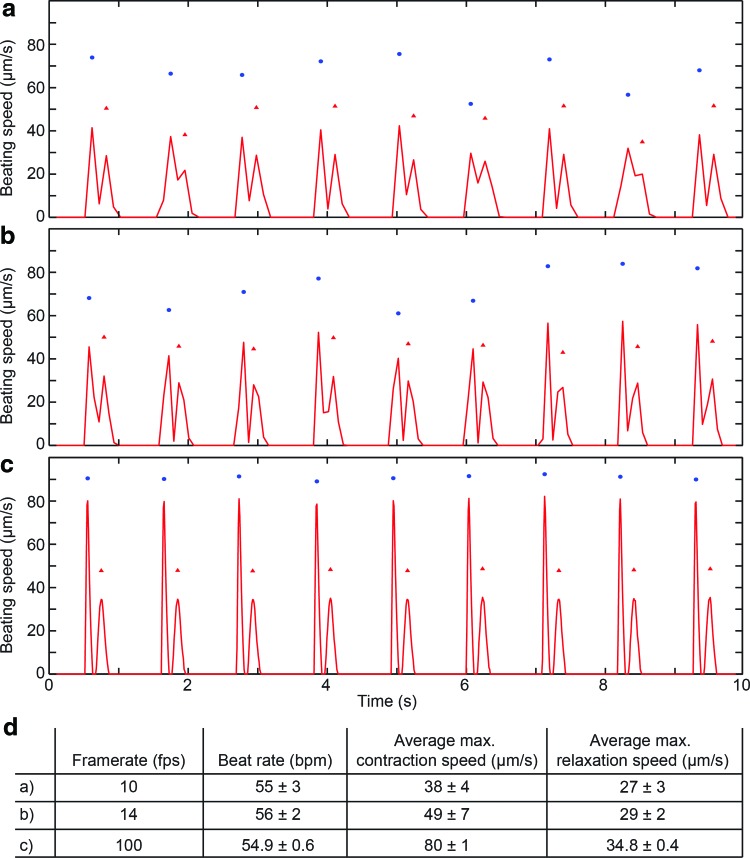
FIG. 2.
Effects of camera frame rate on accuracy of block-matching optical flow analysis of iPS-CM sheet beating. (a–c) Capture of beating motion for a sheet of iPS-CM either at (a) 10 frames per second (fps) and (b)14 fps or (c) 100 fps. Contractions (maximum contraction velocity for each beat, represented by a doublet of peaks) are denoted with blue dots and relaxation cycles (maximum relaxation velocity for each beat represented by a doublet of peaks). (d) Table of calculated values for maximum contraction velocity, maximum relaxation velocity, and beat rate. Note: The beat rate calculated based on the distance between contraction peaks or the periodicity of doublets is the same, but sampling at a lower frame rate typically causes an underestimate of the maximum contraction velocity and overestimate of the variability in waveform shape. Color images available online at www.liebertpub.com/tec
Verification of optical flow software with standard electrophysiology assays
We verified that motion tracking provides a surrogate for direct electrical measurements by sequentially characterizing movement and field potential waveforms of iPS-CM cultured on an MEA (Fig. 3a). Measurements of the same sample revealed no significant deviation between these measurements, in terms of the measured beat rate and regularity, suggesting that motion tracking software provides a surrogate for MEA action potential measurements, which require special equipment (Fig. 3b–d). To further verify computational motion tracking methodology with a gold standard method, video microscopy was performed on iPS-CM during a patch-clamp experiment, in which triggered activity was observed in a cell cluster (Fig. 3e; Supplementary Movie S3). Because of the high level of noise within bright-field video microscopy of patch-clamped iPS-CM in the apparatus (due to fluid flow and intensity variation of the light source), additional measures were taken while processing. Briefly, the time series was treated as a 3D volume, and a 3D Gaussian blur was applied. Analysis of tracings obtained for contraction speed versus time in this cell cluster suggested that as with MEA, electrical action potentials corresponded to contractile motion (Fig. 3f, g). Furthermore, motion tracking analysis successfully detected the aforementioned contractile abnormality.
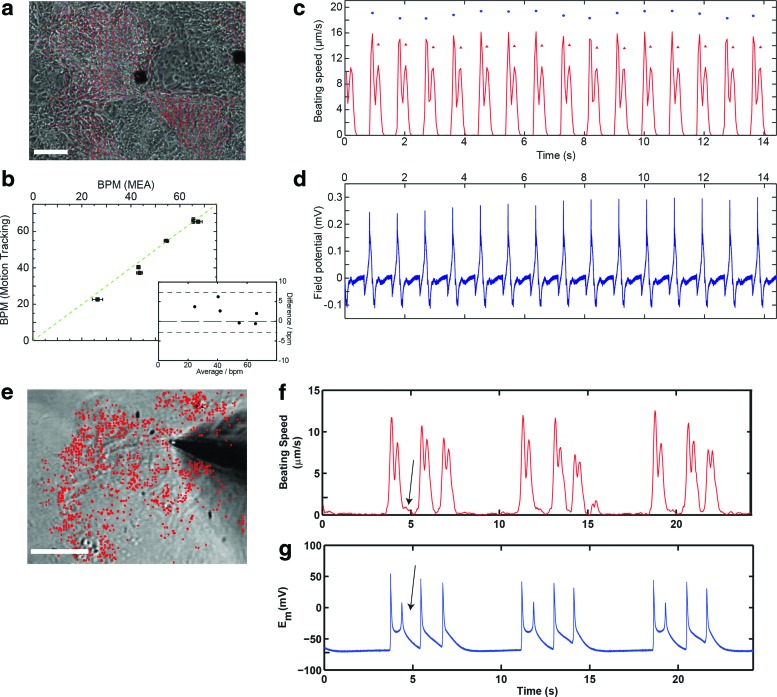
FIG. 3.
Validation of motion tracking software with standard electrophysiologic methods. (a–d) Comparison of motion tracking software to microelectrode array (MEA). (a) Bright-field image of replated iPS-CM on an MEA chip overlayed with the respective motion vectors for this still frame from a movie (Scale bar 150 μm). (b) Measurements on the same sample reveal that there are no significant differences in the beat frequency determined by electrophysiological MEA and motion tracking analysis (highlighted by the Bland–Altman plot in the inset). (c) Representative motion tracking characterization: Tracing of the average motion featuring the typical contraction and relaxation peaks. (d) Representative electrophysiologic MEA characterization: Time course of the field potential. (e–g) Comparison of motion tracking software to patch clamp. (e) Bright-field image of a patch of iPS-CM, which were analyzed simultaneously by video microscopy and patch clamp (Scale bar 100 μm). Triggered activity, as detected by both techniques, is denoted with black arrows. (f, g) Quantitative, (f) motion tracking analysis, and (g) patch-clamp measurements of contraction speed versus time and transmembrane potential and Em versus time, respectively. Color images available online at www.liebertpub.com/tec
Optical flow analysis of sparse cultures and single cells
Previously developed block-matching algorithms can monitor the behavior of confluent cardiomyocyte sheets9,13,14 or cells in 3D constructs such as collagen gels (Fig. 4a), but these algorithms fail when applied to sparse or singularized cardiomyocytes because motion caused by nonspecific signals (e.g., Brownian motion), obscures the signal from cardiomyocyte motion (Fig. 4b, c; Supplementary Movies S4 and S5). The magnitude of motion vectors associated with noise was very high for acellular regions, and there was little to no correlation between the magnitude of motion from one macroblock to the next (Supplementary Movies S4 and S5; Fig. 4b, c). In contrast, motion associated with clusters of cardiomyocytes was relatively well coordinated from one macroblock to the next (Fig. 4b), and motion occurred at regular intervals (e.g., during beating). Based on these observations, two preprocessing algorithms were tested: (1) global edge detection was on the image at the earliest time point to identify cellular regions or (2) a moving foreground detector was used to identify connected groups of pixels, which underwent repetitive motion. The moving foreground detector assesses whether or not a single pixel has changed from one frame to the next, and then calculates the cumulative movement for that pixel over the entire time series of moves.
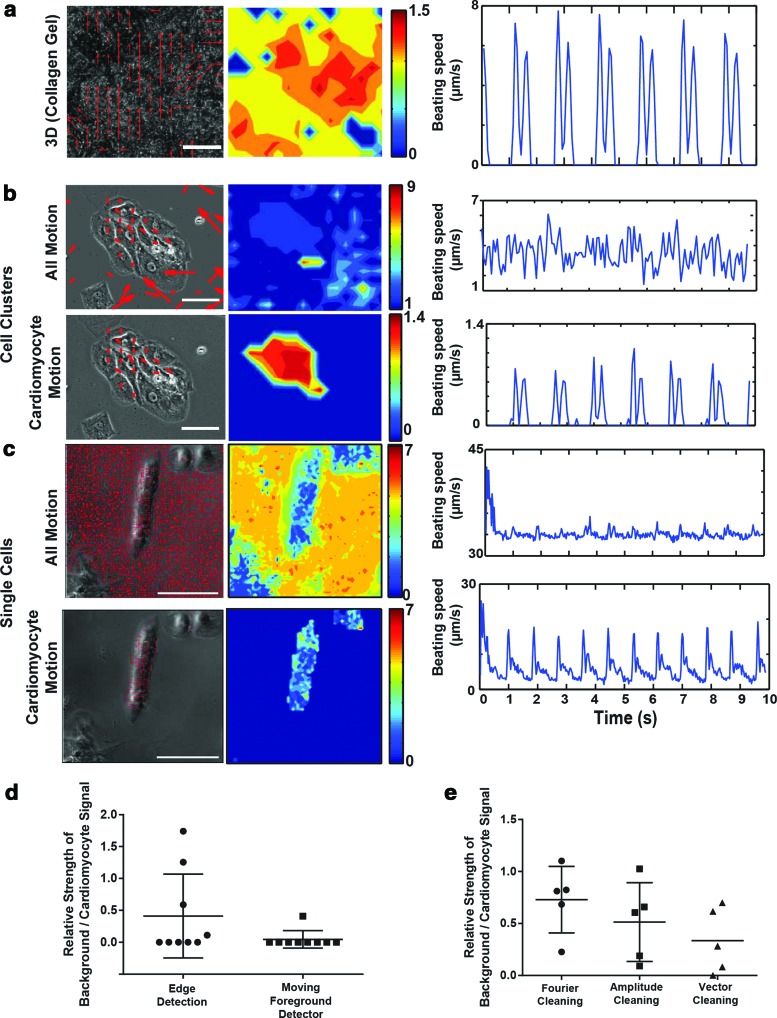
FIG. 4.
Filtering algorithms allow motion tracking over multiple spatial scales. (a) Representative frame (left), heat map of time-averaged motion (center), and one-dimensional (1D) tracing of motion vector magnitude (right) from motion capture analysis of a beating three-dimensional construct with iPS-CM encapsulated into collagen gel. Note that motion is substantial and coordinated in space, resulting in an easily interpretable 1D motion tracing. (b, c) Representative frames from motion capture (left), time-averaged heat maps of motion magnitude (center), and 1D tracings (right) for either (b) cardiomycotye clusters or (c) singularized cardiomycotyes. Note that without preprocessing (All Motion), there is substantial noise, which is uncoordinated in space and tends to result in very large amplitude vectors (compare size and magnitude of motion vectors for motion generated outside of cell boundaries to motion generated within cell boundaries). This resulted in 1D tracings, in which cardiomyocyte motion was difficult to distinguish. (d) Quantitative comparison of improvement in ratio of intensity of background noise (from user-defined cell-free background regions) to intensity of signal from user-defined beating cardiomyocytes, when singularized cardiomyocytes and clusters were preprocessed with moving foreground or edge detectors. (e) Effects of postcleaning algorithms on the signal-to-noise ratio, in which preprocessing algorithms did not completely eliminate background noise. Scale bars: 100 μm. Color images available online at www.liebertpub.com/tec
To further concentrate on cardiomyocyte motion, we applied one of three different clean-up algorithms to the motion vectors: amplitude-based cleaning, vector-based cleaning, and frequency domain cleaning. For amplitude-based cleaning, the amplitude of each motion vector at a given time point was compared with the eight surrounding motion vectors at time i and times i−1 and i+1. If the difference between this vector and the surrounding vectors exceeds a user-determined threshold, the motion vector in question is assumed to be generated due to noise and is thereafter set to zero. Vector-based cleaning was performed in a manner similar to amplitude-based cleaning, with the addition that differences in directional information were taken into account. For frequency domain cleaning, it was assumed that the beating motion of human cardiomyocytes has a maximum frequency (user definable; default value 4 Hz) and that motion at higher frequencies is noise. Typically, amplitude-based cleaning was used in concert with global edge detection and pixel intensity normalization described above. To analyze single cells, smaller macroblocks (4×4 pixels) were used.
To assess the robustness of these preprocessing and postcleaning methods, we compared the ability to distinguish cardiomyocyte motion signal from background noise signal with user-identified cellular and background regions within the same image. Signal and noise were detected by measuring the area under curves of time versus contraction speed for cardiomyocyte regions versus background regions, respectively. Data are reported as the relative improvement in the ratio of noise/signal, normalized to the noise/signal ratio obtained for the same cell, without preprocessing or postprocessing (Fig. 4d). To compare the ability of different postcleaning techniques to allow automated peak identification, we measured the percentage of peaks that were identified manually by a trained user to the peaks identified within the program, after analyzing preprocessed cells with different postcleaning techniques (Fig. 4e).
Qualitatively, time-averaged contraction heat maps and 1D tracings of contraction speed versus time, both suggest that preprocessing either with edge detection (Fig. 4b) or moving foreground detection (Fig. 4c) was sufficient to identify beating regions. Quantitative analysis of the diminishing ratio of noise (area under curve for contraction speed vs. time detected in user-identified background regions) to cardiomyocyte signal (area under curve for contraction speed vs. time detected in user-defined beating cardiomyocyte regions) confirmed that either preprocessing algorithm could be used in the majority of cases, though the moving foreground detector outperformed edge detection (Fig. 4d). Importantly, because block matching is not performed in regions that are not identified by initial segmentation, both preprocessing algorithms could decrease computation time by as much as 90% when smaller macroblocks were used to assess single cell motion (data not shown). When preprocessing alone was insufficient to yield negligible levels of background noise, postprocessing techniques were effective in further reducing background noise levels (Fig. 4e). Among the three postprocessing methods tested, vector cleaning (e.g., penalizing motion vectors, which had significantly different magnitude and direction from their neighbors in adjacent macroblocks) performed slightly better than either frequency domain (Fourier) cleaning or amplitude cleaning (e.g., penalizing motion vectors, which had a significantly different magnitude from their neighbors in adjacent macroblocks regardless of direction). However, when we measured the percentage of beats (contraction–relaxation cycles) that were identified manually by a trained user and compared this with the number of contraction–relaxation cycles identified within the program, all three postprocessing methods yielded a similar ability to identify beat rate and regularity in an automated manner (data not shown).
Analysis of calcium flux and electromechanical coupling with optical flow analysis on fluorescent videos
iPSCs with a constitutively expressed GCaMP6f inserted into the AAVS1 locus (Fig. 5a) were generated, and cardiac differentiation was performed to yield a beating sheet of cells (Supplementary Movie S6). High-speed imaging captured waves of calcium flux traveling through these sheets (Fig. 5b, c) with the intensity of the GCaMP signal following the expected sinusoid pattern (Fig. 5d). These results are consistent with previous work indicating that integration of the GCaMP vector in the defined AAVS locus drives sufficient expression levels to perform quantitative physiologic analysis if optimal promoters are used.5
FIG. 5.
Cardiac tissue derived from a genetically engineered iPSC line harboring a GCaMP Ca reporter. (a) Vector design for introducing GCaMP6f into the AAVS1 safe harbor locus of iPSCs. Note: Puromycin resistance (Puro R) is driven by the endogenous promoter, while a CAG promoter drives GCaMP6f. (b) Representative time series of images taken on sheets of iPS-CM harboring the GCaMP6f reporter, which were tracked for calcium transients (fluorescence intensity). Note: Images are contrast enhanced to aide in visualization. (c) Isochronal activation heat map depicting the propagation of the peak in the calcium transient through the tissue. (d) Tracing of the GCaMP signal featuring the beating region of the cell sheet. Scale bars: 200 μm. Color images available online at www.liebertpub.com/tec
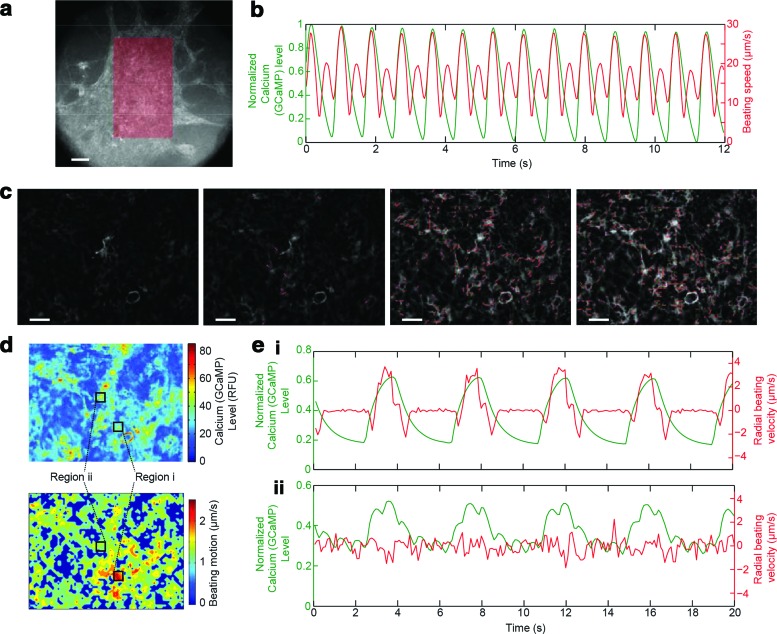
Although calcium flux is often used as a surrogate marker for cardiomyocyte electrical responses,21 there are disease conditions and drugs that cause decoupling between calcium influx and mechanical contraction. Hence, optical flow algorithms were modified to allow direct analysis of contractile motion from GCaMP movies, to facilitate future measurements of electromechanical coupling. Typically, an assumption of block matching is that the intensity of images is relatively constant over time,12 which is clearly not the case for video micrographs taken with fluorescent reporters like GCaMP6f. Thus, we modified the block-matching process to include local intensity normalization steps so that the texture rather than the intensity of the image was used to calculate macroblock motion. First, to identify cellular versus acellular ROI, a minimum fluorescence intensity threshold corresponding to the baseline fluorescence intensity of GCaMP6f in nonexcited cells was set. This allowed cells to be identified from acellular regions by edge detection. To sharpen the image features/edges, the background was subtracted and a minimum filter (radius of 2 pixels) was applied. Additionally, the block-matching algorithm was upgraded with an intensity normalization step; The intensity of pixels within each macroblock (at time i) was normalized to the maximum pixel intensity within the same macroblock (at time i), so that any differences between images would be due to changes in the location of macroblocks rather than absolute pixel intensity (i.e., the texture rather than the intensity of the image was used to calculate macroblock motion).
Simultaneous analysis of these two signals indicated that grossly, for homogenous cell sheets, Ca2+ flux and contractile motion were spatially and temporally correlated (Fig. 6a, b, e[i]; Supplementary Movie S6) and contraction followed calcium flux after a slight delay, as described in previous work.24 However, less connected or branch-type tissues, however, can show different types of electromechanical coupling and this was apparent by directly comparing heat maps for time-averaged motion and GCaMP signals (Fig. 6c–e; Supplementary Movie S7). Region [i] (Fig. 6e), for instance, features a slight millisecond-scale delay between the calcium transient and mechanical motion. Several other regions, for example, region [ii] (Fig. 6e) exhibited calcium flux, but no contractile motion likely reflecting noncontractile cells.
FIG. 6.
Combining motion tracking analysis with a genetically encoded calcium indicator to assess electromechanical coupling. (a) Analyzed region of interest (ROI) of the dense and homogeneous, high-density cardiac tissue from Figure 5. (b) Combined tracings of the GCaMP signal and the motion tracking revealing spatially and temporally correlated Ca2+ flux and contractile motion. (c) GFP channel (pseudo colored in grayscale) overlayed with the derived motion vectors of a low-density branch-type tissue. (d) Heat maps of the tissue in (c) depicting time-averaged calcium (GCaMP6f intensity) levels and beating motion. (e) Motion tracings associated with two ROIs [(i) and (ii)] within (d): the GFP channel was used to generate motion vectors to simultaneously monitor calcium levels (GCaMP6f intensity; green) and radial beating velocity (red). These ROIs show GCaMP-iPS-CM that either (i) generated contractile motion (i.e., each calcium transient is tightly coupled to beating, and the motion lags behind the rise in calcium by several milliseconds) or (ii) exhibited rapid, low-intensity calcium fluctuations that do not generate mechanical motion, indicating poor electromechanical coupling or contractility. Color images available online at www.liebertpub.com/tec
Application examples of block-matching analysis
Baseline beat rate and drug response of iPS-CM analyzed by optical flow
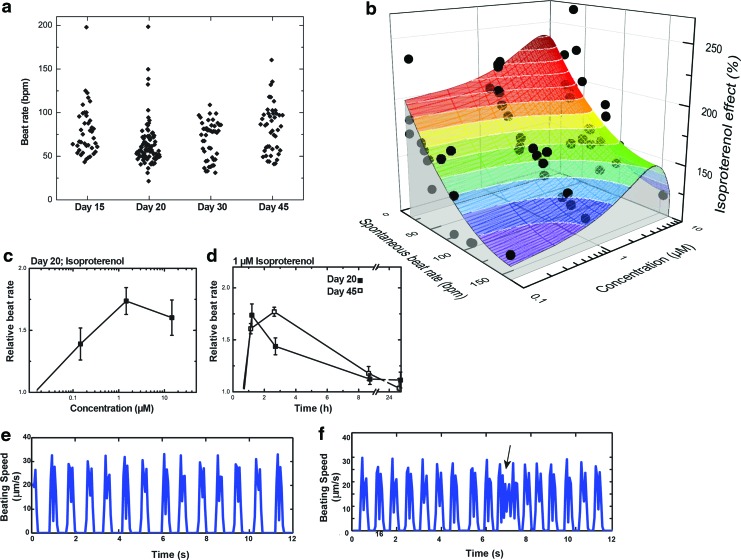
The motion tracking software was applied to assess the baseline behavior and drug response of iPS-CM. To evaluate the high-throughput and noninvasive nature of the software, we initially tracked beat rate over the course of development of iPS-CM. This revealed marked heterogeneity in the spontaneous beat rate of these cell preparations, which was not otherwise apparent (Fig. 7a–e). Interestingly, the cells' chronotropic response to this drug depended on their initial beating rate (Fig. 7b) with the greatest fold change when basal beat rates were between 30 and 50 beats per minute (bpm). In fact, the correlation between initial beat rate and the fold isoproterenol-induced increase in beat rate was much stronger (Pearson correlation coefficient −0.749) than the correlation between the isoproterenol dose (within 100 nM and 10 μM) and the fold increase in beat rate (Pearson correlation coefficient 0.064). Hence, further pharmacologic analysis was performed only on iPS-CM with an initial beat rate between 30 and 90 bpm. These studies showed that on day 45, cells exhibited a longer-lived chronotropic response than on day 20, though the fold increase in beat rate was constant (1.5-fold; Fig. 7c, d).
FIG. 7.
Analyzing iPS-CM pharmacology with motion tracking software. (a) Baseline beat rate of iPS-CM within the original wells where differentiation was performed. (b) Response surface analysis of the combined effects of isoproterenol dose and spontaneous (initial) beat rate on iPS-CM chronotropic (beat rate) response to isoproterenol dosing. Only the spontaneous initial beat rate had significant effects on the fold increase in beat rate (Pearson correlation analysis). (c, d) Isoproterenol response, as assessed by the chronotropic dose–response (c) and the time course of the chronotropic response in iPS-CMs (d). (e, f) Usage examples for analysis of isoproterenol-induced arrhythmia. Motion tracings of iPS-CM either (e) before or (f) 5 min after adding 1 μM isoproterenol to cells. Note: Arrhythmia occurred at 7 s (black arrow), as detected by tracking beating speed. Error bars: standard error of the mean, n=6–8. Color images available online at www.liebertpub.com/tec
Discussion
The computational motion tracking software provided in this study is a robust tool for unbiased analysis of cardiomyocyte contractile behavior on multiple spatial scales. Block matching provides information not only on beat rate but also yields vector fields that can be used to quantify the spatial distribution of beating of tissue constructs. These vector fields can potentially also be used to quantify temporal correlations in motion within different parts of tissues as a means of gauging the success of tissue assembly and the ability to generate physiologically relevant tissues with uniaxial contraction. Generation of the vectors is user independent and robust, and the process of generating vectors and subsequently assessing beat rate regularity and abnormalities is highly amenable to automation. Using modern architecture for parallel computing, this procedure can rapidly analyze many samples. Although more complex assays, including patch-clamp electrophysiology, can provide additional information, computational motion tracking is noninvasive and can be performed on cardiomyocytes in any culture format with minimal additional expertise.
As suggested by the differences in beat rate and regularity between cell sheets used for MEA analysis and cell clusters used for patch clamp, plating conditions can have a significant effect on the electrophysiologic behavior of iPS-CM, emphasizing the need for analytical tools that do not require specific culture conditions. Therefore, this system may be suitable for comparing the behavior, drug response, and maturation of isolated iPS-CM and iPS-CM grown in more in vivo-like microenvironments.15,25,26 To this end, the enabling nature of this software was demonstrated in a previous study, in which long QT syndrome type 3 patient-specific iPSCs were differentiated into iPS-CM and cultured on synthetic biomaterials to generate a physiologically relevant in vitro model of this disease.15 Direct analysis of cells in these 3D environments would present significant challenges with patch-clamp methodology and would not be possible with MEA.
The ability to specifically detect the cells that initiate beating in cardiomyocyte sheets or cells that couple electrically, but not mechanically, to their neighbors may aid in subsequent isolation and transcriptional analysis of these potentially distinct populations. Another new feature introduced in this study—the ability to measure electromechanical coupling through optical flow analysis of fluorescent images—may be especially useful for in vitro disease modeling. For example, abnormal calcium flux has been reported in iPS-CM derived from patients with hypertrophic cardiomyopathy who harbor mutations in MYH7, a component of the cell contraction apparatus.27 This aberration may be explained by stress-induced decoupling between calcium flux and mechanical contractility, and this cardiomyopathy could be tracked noninvasively with genetic calcium sensors combined with computational motion tracking. The same features could be used to test drugs that desensitize sarcomeres to calcium.
In addition to the aforementioned features, with batch analysis, the software is well suited to assess larger sample numbers to detect effects of subtle epidemiologically common mutations with in vitro models. The ability to share motion vector fields generated by this software provides an opportunity for data mining in the rapidly growing area of genetic and other disease models based on human cardiomyocytes. Most importantly, we provide the software as an open source resource, so that it can be easily used—and improved upon—by the biomedical and tissue engineering communities.
Supplementary Material
Acknowledgments
The authors thank Douglas Kim, Janelia Farm, HHMI, for providing the original GCaMP6f plasmid and Loren Looger, Janelia Farm, HHMI, for GCaMP information and advice. The authors thank Jidong Fu, Gladstone Institute of Cardiovascular Disease, for advice on electrophysiology. This work was funded, in part, from the NIH, NHLBI R01HL096525, R01HL108677, UH2TR000487, U01HL100406, U01HL098179, U01GM09614, P01HL089707, and the Gladstone Institutes. N.H. and A.M. are postdoctoral fellows of CIRM training programs TG2-01160 (N.H.) and TG2-01164 (A.M.). P.L. is supported by a postdoctoral fellowship from the German Research Foundation, Lo 2081/1-1. L.M.J. is a clinical fellow of CIRM training program TG2-01160. M.A.M. is supported by a postdoctoral fellowship from the Canadian Institutes of Health Research, 129844. The authors acknowledge assistance from the Roddenberry Center for Stem Cell Biology at Gladstone and Berkeley Stem Cell Shared Facility and Biological Imaging Facility. All the software developed in this work is available under a GNU license at http://gladstone.ucsf.edu/46749d811/.
Disclosure Statement
No competing financial interests exist.
References
- 1.Mathur A., et al. Human induced pluripotent stem cell-based microphysiological tissue models of myocardium and liver for drug development. Stem Cell Res Ther 4Suppl 1,S14, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burridge P.W., Keller G., Gold J.D., and Wu J.C.Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 10,16, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirenko O., Cromwell E.F., Crittenden C., Wignall J.A., Wright F.A., and Rusyn I.Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity. Toxicol Appl Pharmacol 273,500, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson C., et al. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J Biomol Screen 18,1203, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Chen T.W., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499,295, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiba Y., et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 489,322, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engler A.J., et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci 121,3794, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barzan C., Barba D.T., Blomgren P., and Paolini P.Image processing techniques for assessing contractility in isolated adult cardiac myocytes. Int J Biomed Imaging 2009,352954, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen A., et al. Integrated platform for functional monitoring of biomimetic heart sheets derived from human pluripotent stem cells. Biomaterials 35,675, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hossain M.M., Shimizu E., Saito M., Rao S.R., Yamaguchi Y., and Tamiya E.Non-invasive characterization of mouse embryonic stem cell derived cardiomyocytes based on the intensity variation in digital beating video. Analyst 135,1624, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Delbridge L.M., and Roos K.P.Optical methods to evaluate the contractile function of unloaded isolated cardiac myocytes. J Mol Cell Cardiol 29,11, 1997 [DOI] [PubMed] [Google Scholar]
- 12.Horn B.K.P., and Schunkck B.G.Determining optical flow. Artif Intell 17,185, 1981 [Google Scholar]
- 13.Hayakawa T., et al. Noninvasive evaluation of contractile behavior of cardiomyocyte monolayers based on motion vector analysis. Tissue Eng C 18,21, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Shapira-Schweitzer K., and Seliktar D.Matrix stiffness affects spontaneous contraction of cardiomyocytes cultured within a PEGylated fibrinogen biomaterial. Acta Biomater 3,33, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Ma Z., et al. Three-dimensional filamentous human diseased cardiac tissue model. Biomaterials 35,1367, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J., et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324,797, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lian X., et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109,e1848, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lian X., et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 8,162, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu W.Z., Van Biber B., and LaFlamme M.A.Methods for the derivation and use of cardiomyocytes from human pluripotent stem cells. Methods Mol Biol 767,419, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braam S.R., Nauw R., Ward-van Oostwaard D., Mummery C., and Passier R.Inhibition of ROCK improves survival of human embryonic stem cell-derived cardiomyocytes after dissociation. Ann N Y Acad Sci 1188,52, 2010 [DOI] [PubMed] [Google Scholar]
- 21.Spencer C.I., Baba S., Nakamura K., et al. Calcium transients closely reflect prolonged action potentials in iPSC models of inherited cardiac arrhythmia. Stem Cell Reports 3,1, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rae J., Cooper K., Gates P., Watsky M., et al. Low access resistance perforated patch recordings using amphotericin B. J Neurosci Methods 37,1, 1991 [DOI] [PubMed] [Google Scholar]
- 23.Hockemeyer D., Wang H., Kiani S., et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29,731, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wehrens X.H.T., Lehnart S.E., and Marks A.R.Intracellular calcium release and cardiac disease. Annu Rev Physiol 67,69, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Nunes S.S., et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods 10,781, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiburcy M., et al. Terminal differentiation, advanced organotypic maturation, and modeling of hypertrophic growth in engineered heart tissue. Circ Res 109,1105, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Lan F., Lee A.S., Liang P., et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 12,101, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.