Abstract
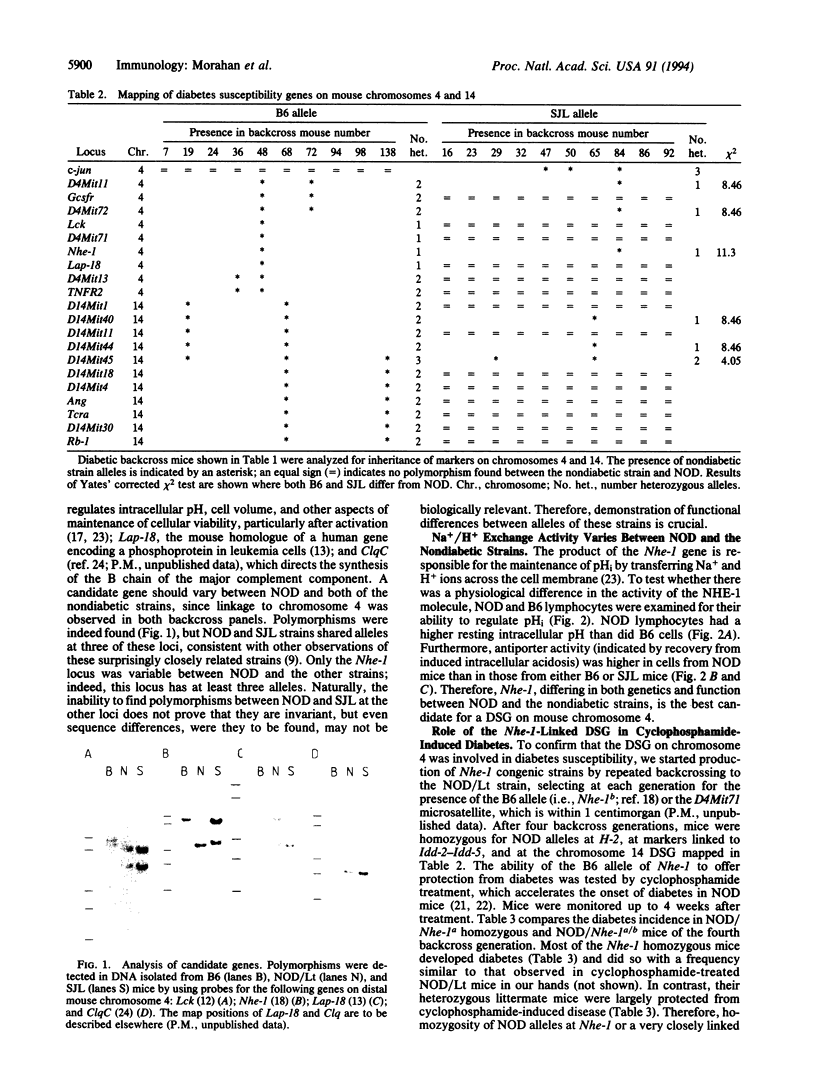
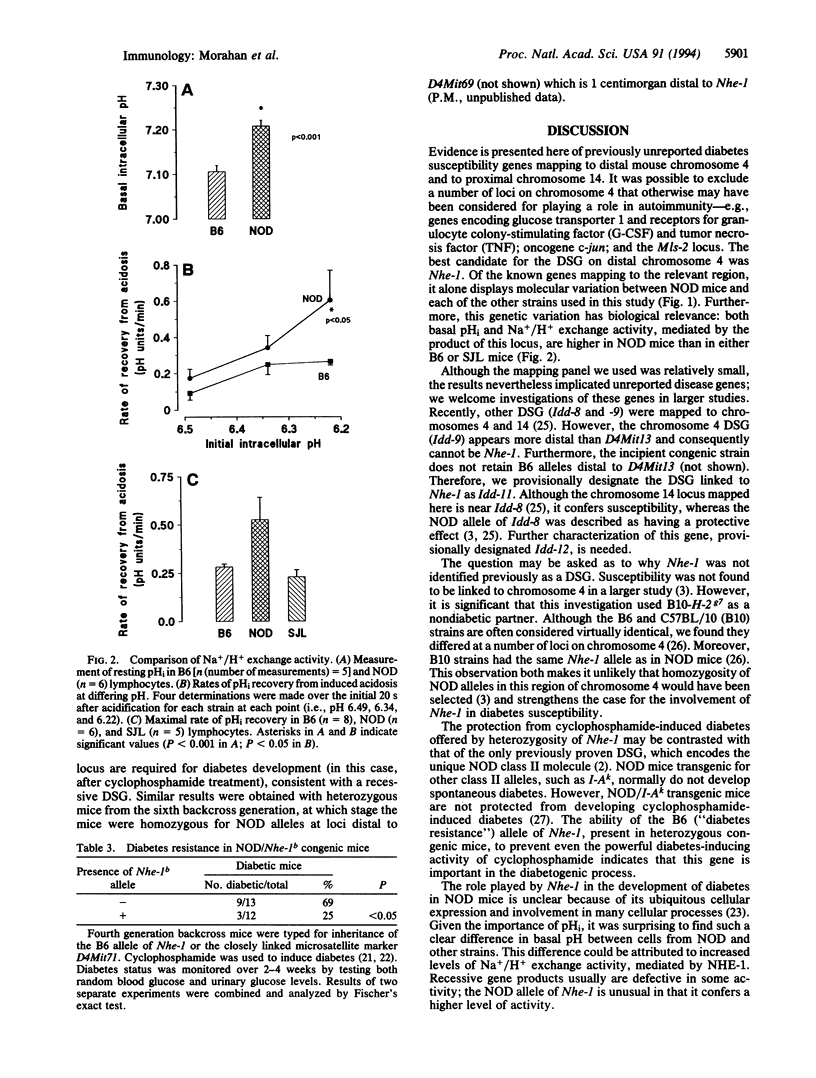
Insulin-dependent diabetes mellitus is a multigenic autoimmune disease, for which one of the best animal models is the nonobese diabetic (NOD) mouse strain. In both humans and NOD mice, major histocompatibility complex genes are implicated as risk factors in the disease process. Other susceptibility genes are also involved, and a number have been mapped in the mouse to specific chromosomal locations. To identify further susceptibility genes, diabetic backcross mice, produced after crossing NOD/Lt to the nondiabetic strains SJL and C57BL/6 (B6), were examined for markers not previously associated with disease susceptibility. Linkage was found to loci on chromosomes 4 and 14. Of the candidate loci on chromosome 4, the gene encoding the Na+/H+ exchanger-1, Nhe-1, was the most likely, since the NOD allele was different from that of both nondiabetic strains. NOD lymphocytes were found to have a higher level of Na+/H+ exchange activity than lymphocytes from either B6 or SJL mice. Since the chromosome 4 susceptibility gene is recessive, the B6 allele should prevent diabetes. This prediction was tested in fourth-generation backcross mice, selected for retention of the B6 allele at Nhe-1. Mice homozygous for Nhe-1 developed diabetes after cyclophosphamide treatment, but heterozygotes were largely protected from disease. These results implicate the Na+/H+ exchanger (antiporter) in the development of type 1 diabetes and may provide a screening test for at-risk individuals as well as offering prospects for disease prevention.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyarsky G., Rosenthal N., Barrett E., Boron W. F. Effect of diabetes on Na(+)-H+ exchange by single isolated hepatocytes. Am J Physiol. 1991 Jan;260(1 Pt 1):C167–C175. doi: 10.1152/ajpcell.1991.260.1.C167. [DOI] [PubMed] [Google Scholar]
- Carr P., Taub N. A., Watts G. F., Poston L. Human lymphocyte sodium-hydrogen exchange. The influences of lipids, membrane fluidity, and insulin. Hypertension. 1993 Mar;21(3):344–352. doi: 10.1161/01.hyp.21.3.344. [DOI] [PubMed] [Google Scholar]
- Charlton B., Bacelj A., Slattery R. M., Mandel T. E. Cyclophosphamide-induced diabetes in NOD/WEHI mice. Evidence for suppression in spontaneous autoimmune diabetes mellitus. Diabetes. 1989 Apr;38(4):441–447. doi: 10.2337/diab.38.4.441. [DOI] [PubMed] [Google Scholar]
- Cornall R. J., Prins J. B., Todd J. A., Pressey A., DeLarato N. H., Wicker L. S., Peterson L. B. Type 1 diabetes in mice is linked to the interleukin-1 receptor and Lsh/Ity/Bcg genes on chromosome 1. Nature. 1991 Sep 19;353(6341):262–265. doi: 10.1038/353262a0. [DOI] [PubMed] [Google Scholar]
- Davies J. E., Ng L. L., Kofoed-Enevoldsen A., Li L. K., Earle K. A., Trevisan R., Viberti G. Intracellular pH and Na+/H+ antiport activity of cultured skin fibroblasts from diabetics. Kidney Int. 1992 Nov;42(5):1184–1190. doi: 10.1038/ki.1992.403. [DOI] [PubMed] [Google Scholar]
- Dietrich W., Katz H., Lincoln S. E., Shin H. S., Friedman J., Dracopoli N. C., Lander E. S. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992 Jun;131(2):423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A. C., Seuanez H. N., Hanash S. M., Atweh G. F. A gene that encodes for a leukemia-associated phosphoprotein (p18) maps to chromosome bands 1p35-36.1. Genes Chromosomes Cancer. 1990 Jul;2(2):125–129. doi: 10.1002/gcc.2870020208. [DOI] [PubMed] [Google Scholar]
- Garchon H. J., Bedossa P., Eloy L., Bach J. F. Identification and mapping to chromosome 1 of a susceptibility locus for periinsulitis in non-obese diabetic mice. Nature. 1991 Sep 19;353(6341):260–262. doi: 10.1038/353260a0. [DOI] [PubMed] [Google Scholar]
- Garchon H. J. Non-MHC-linked genes in autoimmune diseases. Curr Opin Immunol. 1992 Dec;4(6):716–722. doi: 10.1016/0952-7915(92)90051-f. [DOI] [PubMed] [Google Scholar]
- Genes V. S. Simple methods of cybernetic processing of medical diagnosis data. Biomed Eng (NY) 1968 May-Jun;2(3):129–133. doi: 10.1007/BF00568813. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Palmer S. M., Rodrigues N. R., Cordell H. J., Hearne C. M., Cornall R. J., Prins J. B., McShane P., Lathrop G. M., Peterson L. B. Polygenic control of autoimmune diabetes in nonobese diabetic mice. Nat Genet. 1993 Aug;4(4):404–409. doi: 10.1038/ng0893-404. [DOI] [PubMed] [Google Scholar]
- Halkin A., Benjamin N., Doktor H. S., Todd S. D., Viberti G., Ritter J. M. Vascular responsiveness and cation exchange in insulin-dependent diabetes. Clin Sci (Lond) 1991 Aug;81(2):223–232. doi: 10.1042/cs0810223. [DOI] [PubMed] [Google Scholar]
- Harada M., Makino S. Promotion of spontaneous diabetes in non-obese diabetes-prone mice by cyclophosphamide. Diabetologia. 1984 Dec;27(6):604–606. doi: 10.1007/BF00276978. [DOI] [PubMed] [Google Scholar]
- Huppi K., Mock B. A., Schricker P., D'Hoostelaere L. A., Potter M. Organization of the distal end of mouse chromosome 4. Curr Top Microbiol Immunol. 1988;137:276–288. doi: 10.1007/978-3-642-50059-6_42. [DOI] [PubMed] [Google Scholar]
- Makino S., Kunimoto K., Muraoka Y., Mizushima Y., Katagiri K., Tochino Y. Breeding of a non-obese, diabetic strain of mice. Jikken Dobutsu. 1980 Jan;29(1):1–13. doi: 10.1538/expanim1978.29.1_1. [DOI] [PubMed] [Google Scholar]
- McClive P. J., Baxter A. G., Morahan G. Genetic polymorphisms of the non-obese diabetic (NOD) mouse. Immunol Cell Biol. 1994 Apr;72(2):137–142. doi: 10.1038/icb.1994.21. [DOI] [PubMed] [Google Scholar]
- McClive P. J., Huang D., Morahan G. C57BL/6 and C57BL/10 inbred mouse strains differ at multiple loci on chromosome 4. Immunogenetics. 1994;39(4):286–288. doi: 10.1007/BF00188793. [DOI] [PubMed] [Google Scholar]
- Morahan G., Hoffmann M. W., Miller J. F. A nondeletional mechanism of peripheral tolerance in T-cell receptor transgenic mice. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11421–11425. doi: 10.1073/pnas.88.24.11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan G., Rakar S. Localization of the mouse Na+/H+ exchanger gene on distal chromosome 4. Genomics. 1993 Jan;15(1):231–232. doi: 10.1006/geno.1993.1044. [DOI] [PubMed] [Google Scholar]
- Ng L. L., Davies J. E. Lipids and cellular Na+/H+ antiport activity in diabetic nephropathy. Kidney Int. 1992 Apr;41(4):872–876. doi: 10.1038/ki.1992.133. [DOI] [PubMed] [Google Scholar]
- Petry F., Reid K. B., Loos M. Gene expression of the A- and B-chain of mouse C1q in different tissues and the characterization of the recombinant A-chain. J Immunol. 1991 Dec 1;147(11):3988–3993. [PubMed] [Google Scholar]
- Prochazka M., Leiter E. H., Serreze D. V., Coleman D. L. Three recessive loci required for insulin-dependent diabetes in nonobese diabetic mice. Science. 1987 Jul 17;237(4812):286–289. doi: 10.1126/science.2885918. [DOI] [PubMed] [Google Scholar]
- Roos A., Boron W. F. Intracellular pH. Physiol Rev. 1981 Apr;61(2):296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- Salles J. P., Ser N., Fauvel J., Couvaras O., Bouissou F., Ghisolfi J., Barthe P., Chap H. Platelet Na(+)-H+ exchange in juvenile diabetes mellitus. J Hypertens Suppl. 1991 Dec;9(6):S222–S223. [PubMed] [Google Scholar]
- Sardet C., Franchi A., Pouysségur J. Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell. 1989 Jan 27;56(2):271–280. doi: 10.1016/0092-8674(89)90901-x. [DOI] [PubMed] [Google Scholar]
- Slattery R. M., Miller J. F., Heath W. R., Charlton B. Failure of a protective major histocompatibility complex class II molecule to delete autoreactive T cells in autoimmune diabetes. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10808–10810. doi: 10.1073/pnas.90.22.10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhelper M. E., Field L. J. Assignment of the angiogenin gene to mouse chromosome 14 using a rapid PCR-RFLP mapping technique. Genomics. 1992 Jan;12(1):177–179. doi: 10.1016/0888-7543(92)90427-t. [DOI] [PubMed] [Google Scholar]
- Stone J. C., Crosby J. L., Kozak C. A., Schievella A. R., Bernards R., Nadeau J. H. The murine retinoblastoma homolog maps to chromosome 14 near Es-10. Genomics. 1989 Jul;5(1):70–75. doi: 10.1016/0888-7543(89)90088-8. [DOI] [PubMed] [Google Scholar]
- Todd J. A., Aitman T. J., Cornall R. J., Ghosh S., Hall J. R., Hearne C. M., Knight A. M., Love J. M., McAleer M. A., Prins J. B. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991 Jun 13;351(6327):542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- Todd J. A. Genetic control of autoimmunity in type 1 diabetes. Immunol Today. 1990 Apr;11(4):122–129. doi: 10.1016/0167-5699(90)90049-f. [DOI] [PubMed] [Google Scholar]
- Wakabayashi S., Sardet C., Fafournoux P., Counillon L., Meloche S., Pagés G., Pouysségur J. Structure function of the growth factor-activatable Na+/H+ exchanger (NHE1). Rev Physiol Biochem Pharmacol. 1992;119:157–186. doi: 10.1007/3540551921_6. [DOI] [PubMed] [Google Scholar]
- de Gouyon B., Melanitou E., Richard M. F., Requarth M., Hahn I. H., Guenet J. L., Demenais F., Julier C., Lathrop G. M., Boitard C. Genetic analysis of diabetes and insulitis in an interspecific cross of the nonobese diabetic mouse with Mus spretus. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1877–1881. doi: 10.1073/pnas.90.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]




