Abstract
Motivation: Research on methods for the inference of networks from biological data is making significant advances, but the adoption of network inference in biomedical research practice is lagging behind. Here, we present Cyni, an open-source ‘fill-in-the-algorithm’ framework that provides common network inference functionality and user interface elements. Cyni allows the rapid transformation of Java-based network inference prototypes into apps of the popular open-source Cytoscape network analysis and visualization ecosystem. Merely placing the resulting app in the Cytoscape App Store makes the method accessible to a worldwide community of biomedical researchers by mouse click. In a case study, we illustrate the transformation of an ARACNE implementation into a Cytoscape app.
Availability and implementation: Cyni, its apps, user guides, documentation and sample code are available from the Cytoscape App Store http://apps.cytoscape.org/apps/cynitoolbox
Contact: benno.schwikowski@pasteur.fr
1 Introduction
The availability of large-scale experimental technologies has enabled the routine measurement of global abundances and states of molecular components in cellular systems. In contrast, the experimental measurement of component interactions—important for analyses in a network context—is still far more difficult. Perhaps for this reason the development of methods that infer networks from transcriptomics or other global molecular measurements has recently received much attention (Marbach et al., 2012; Poultney et al., 2012). However, the use and adoption of these methods in biomedical research has been slow.
To stimulate the exchange between the network inference and biomedical research communities, we have developed the open-source computational framework Cyni that allows to rapidly transform Java-based network inference core code into interactive components (apps) of the Cytoscape visualization and analysis platform (Cline et al., 2007; Shannon et al., 2003), and thereby to rapidly make novel methods accessible to a large community of biomedical researchers. Here, we describe the structure of Cyni, and illustrate the implementation of the ARACNE network inference method (Margolin et al., 2006), which has confirmed MYC as a major regulatory hub in human B cells, and identified a number of new MYC targets (Basso et al., 2005).
2 The Cyni framework
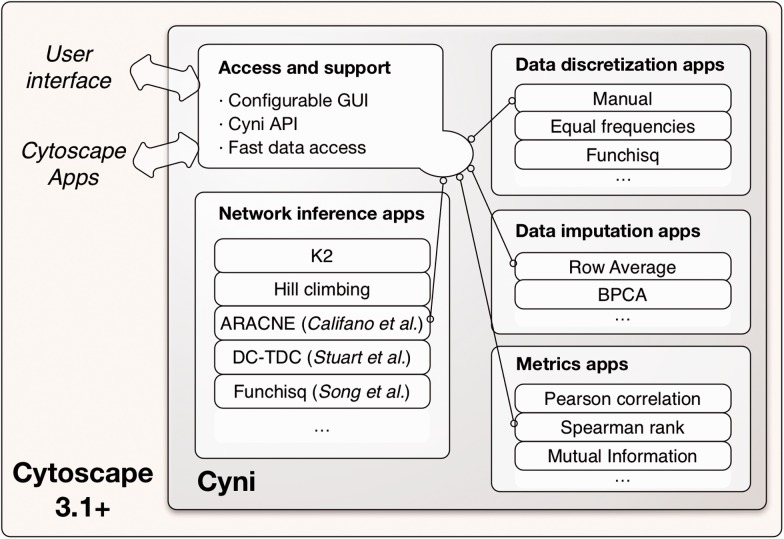
The Cyni framework provides support for network inference through an extensible system of Cytoscape apps that provide key functionality in and around network inference, while eliminating the need for interaction with the much more complex Cytoscape application programmer interface (API; Fig. 1). Cyni comprises app implementations of commonly used pairwise score functions (metrics), data discretization and data imputation, network inference, along with configurable graphical user interfaces (GUIs), API and common data structures. Like Cytoscape and its apps, Cyni and its apps are written in Java to allow easy portability to all common operating systems. For the user, Cyni apps and their parameters are accessible via the Cyni GUI panel within Cytoscape.
Fig. 1.
The Cyni framework for network inference
Network inference apps produce a new Cytoscape network from input data in a Cytoscape table. Apps that are built-in (using multi-threaded Java) include Bayesian algorithms, such as K2 (Cooper and Herskovits, 1992), hill climbing (Murphy, 2012), an information theory algorithm based on mutual information (Butte and Kohane, 2000) and a correlation-based algorithm, which employs any of the built-in (or additional) correlation metrics.
Data imputation apps aim at estimating missing values. There are different options for the user to indicate what constitutes missing data. Built-in apps are, besides row average and zero imputation, the Bayesian principle component analysis algorithm that has been tested favorably for bioinformatics applications (Aittokallio, 2010; Oba et al., 2003).
Data discretization apps allow users to discretize continuous input data, which is required by certain network inference algorithms. The discretized data are represented in newly generated Cytoscape data table columns. Two built-in discretization apps provide discretization of equal width or frequency, and manually controlled discretization through a GUI.
3 Case study: implementation of ARACNE using Cyni
Using the example of the ARACNE Java reference implementation (Margolin et al., 2006), we show how Cyni can be used to transform a pure network inference algorithm into a fully functional Cytoscape app in a few steps:
Step 1
Embed core implementation: Insert the existing code into the sample code of the CyniTask java class. Modify data access and network output to employ Cyni data structures. For ARACNE, we inserted the core algorithmic code into the CyniTask sample class and modified data access to read data from CyniTable. We added code to create a Cytoscape network from ARACNE’s result.
Step 2
Set up parameter handling: Extend the Cyni Context class, specify the user-controlled parameters in a predefined annotation and rename parameters to be class variables of the newly extended Cyni Context class.
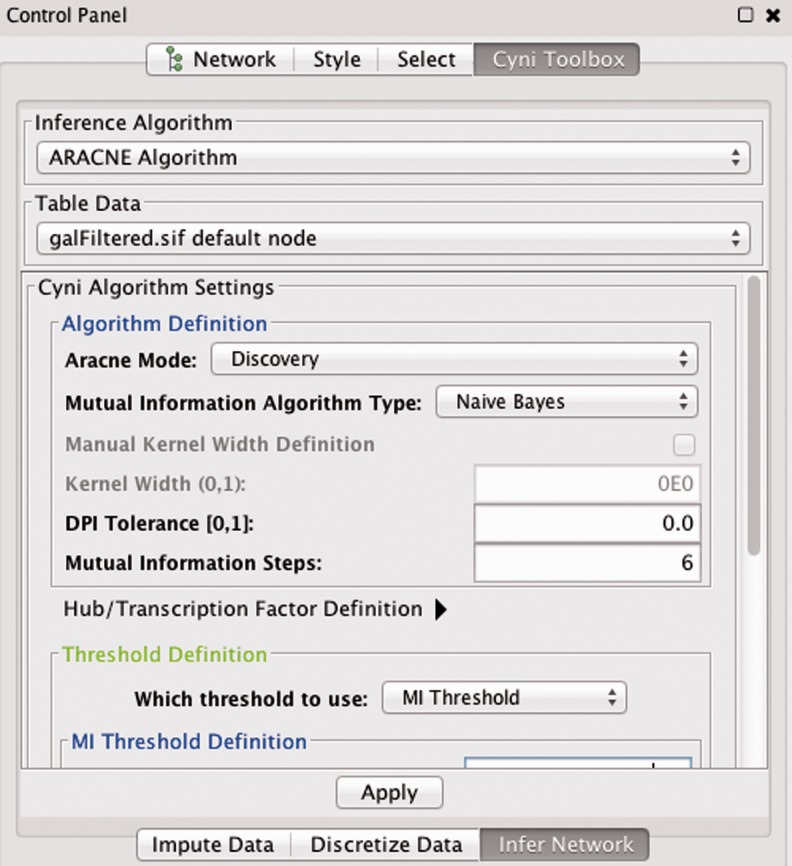
For ARACNE, we extended the Cyni Context class by specifying the 13 user-controllable ARACNE parameters of the reference implementation (see Fig. 2).
Fig. 2.
The ARACNE implementation using Cyni
Step 3
(Optional) Extract metric: If not yet available in Cyni, required mathematical scoring functions (metrics) can be made available to other inference methods by implementing them as a Cyni metric. This will also enable the user to choose from other Cyni metrics. The code for the continuous mutual information metric in ARACNE was extracted into a newly extended Cyni metric class and calls to this measurement into original code were replaced by calls to new Cyni metric.
Step 4
Submit the new app to the Cytoscape App Store: Connect to the Cytoscape App Store using a browser, create a free user account if necessary and upload the compiled .jar file. The ARACNE app is, together with its source code, available at http://apps.cytoscape.org/aracne, and can be installed into an open Cytoscape session through a mouse click.
To ∼90%, the ARACNE app source code consists of existing code (80% original Java code and 10% Cyni sample code). Only 10% had to be written to embed ARACNE into Cyni and Cytoscape. We note that similar steps can also be applied to bring novel data imputation and discretization methods to Cytoscape.
4 Conclusions
Cyni is a framework that allows the efficient embedding of Java-based network inference methods into the Cytoscape app ecosystem. A rich set of built-in network inference features, documentation, a tutorial and sample code help turning, with minor effort, existing algorithmic command line prototypes into interactive research tools, and to provide them to a large audience of biomedical researchers. The ability to mix and match components may also facilitate the systematic comparisons of different methods.
Funding
This project was funded by the National Institute of General Medical Sciences (NIGMS) under grant P41 GM103504.
Conflict of Interest: none declared.
References
- Aittokallio T. (2010) Dealing with missing values in large-scale studies: microarray data imputation and beyond. Brief. Bioinform., 11, 253–264. [DOI] [PubMed] [Google Scholar]
- Basso K., et al. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet., 4, 382–390. [DOI] [PubMed] [Google Scholar]
- Butte A.J., Kohane I.S. (2000) Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac. Symp. Biocomput., 5, 418–429. [DOI] [PubMed] [Google Scholar]
- Cline M.S., et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc., 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G., Herskovits E. (1992) A Bayesian method for the induction of probabilistic networks from data. Mach. Learn., 9, 309–347. [Google Scholar]
- Marbach D., et al. (2012) Wisdom of crowds for robust gene network inference. Nat. Methods, 9, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin A.A., et al. (2006) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics, 7 (Suppl. 1), S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.P. (2012) Machine Learning: A Probabilistic Perspective. MIT Press, Cambridge, MA, USA. [Google Scholar]
- Oba S., et al. (2003) A Bayesian missing value estimation method for gene expression profile data. Bioinformatics, 19, 2088–2096. [DOI] [PubMed] [Google Scholar]
- Poultney C.S., et al. (2012) Integrated inference and analysis of regulatory networks from multi-level measurements. Methods Cell Biol., 110, 19–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]