Abstract
Summary: Identifying, amongst millions of publications available in MEDLINE, those that are relevant to specific microRNAs (miRNAs) of interest based on keyword search faces major obstacles. References to miRNA names in the literature often deviate from standard nomenclature for various reasons, since even the official nomenclature evolves. For instance, a single miRNA name may identify two completely different molecules or two different names may refer to the same molecule. mirPub is a database with a powerful and intuitive interface, which facilitates searching for miRNA literature, addressing the aforementioned issues. To provide effective search services, mirPub applies text mining techniques on MEDLINE, integrates data from several curated databases and exploits data from its user community following a crowdsourcing approach. Other key features include an interactive visualization service that illustrates intuitively the evolution of miRNA data, tag clouds summarizing the relevance of publications to particular diseases, cell types or tissues and access to TarBase 6.0 data to oversee genes related to miRNA publications.
Availability and Implementation: mirPub is freely available at http://www.microrna.gr/mirpub/.
Contact: vergoulis@imis.athena-innovation.gr or dalamag@imis.athena-innovation.gr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
MicroRNAs (miRNAs) are small, single-stranded endogenous RNAs, which suppress the expression of protein coding genes. They are important regulators of many crucial cellular processes and they are implicated in a multitude of diseases such as rheumatoid arthritis, multiple sclerosis, Alzheimer, cancer, etc. (Soifer et al., 2007).
The number of miRNA-related publications is growing constantly every year, reaching more than 12,700 papers until 2013. Keyword search to retrieve publications relevant to particular miRNAs faces three obstacles: (i) publications may refer to the same miRNA using different names, (ii) publications may refer to miRNAs with old, obsolete names and (iii) results retrieved as relevant should be carefully examined, since, e.g. sequence changes might have happened in referenced miRNAs, after the publication date.
mirPub is a database with a powerful and intuitive interface that addresses the aforementioned issues. During miRNA literature search, mirPub takes into account both inconsistencies in naming miRNAs and the history of miRNA data as being recorded in all available miRBase (Griffiths-Jones et al., 2005) versions. mirPub considers miRNA-publication associations based on text mining techniques on MEDLINE articles, integrated data from several curated databases and data provided by its user community.
All stored associations are available through mirPub’s Web Interface (see Supplementary Fig. S1 of Supplementary Data). Users are able to search for publications related to particular miRNAs by providing keywords. mirPub matches the keywords to stored miRNA terms. In case of zero or multiple matches for a given keyword, mirPub provides a set of keyword suggestions (string similarity is used for the case of zero matches). Otherwise, the relevant publications are presented as a chronologically ordered list. Along with the article title and year of publication, each mirPub result provides access to TarBase 6.0 (Vergoulis et al., 2012) data to oversee gene targets mentioned in the article, as well as links to relevant external resources (e.g. articles abstract and full text) and MeSH metadata. mirPub expands the set of user keywords to also contain the families of the identified miRNAs and miRNA name variants as well (e.g. old or alternative names). The complete set of keywords used for each search is displayed and the user is able to keep only a subset of them filtering out the irrelevant publications (details in Section 1 of Supplementary Data). Finally, mirPub provides tag clouds summarizing MeSH diseases, tissues and cells that are relevant to the displayed publications, giving an insight about the role of the queried miRNAs.
Regarding the miRNA data history, mirPub also provides an interactive visualization service that intuitively illustrates the timeline of changes for any mature or hairpin miRNA (see Supplementary Fig. S2 in Supplementary Data). In particular, each mature or hairpin miRNA involved in a user query is accompanied by an information button, which activates a pop-up window that visualizes the name and sequence changes related to each particular molecule. This is a key feature of mirPub, since this information enables users to refine their search, excluding, for instance, publications related to a different molecule that had the same name in the past or including publications that refer to miRNAs of interest with different, older names. Preliminary experiments presented in Section 3 evaluate the usefulness of the aforementioned feature.
2 System implementation
The majority of miRNA-to-publication associations in mirPub database have been discovered by applying text mining techniques on titles, abstracts and full texts of all available MEDLINE publications. Full-text articles were obtained from PMC only for open-access papers. mirPub’s text mining method is presented in Figure 1. In brief, mirPub seeks appearances of terms that describe miRNA molecules or families in MEDLINE publications.
Fig. 1.
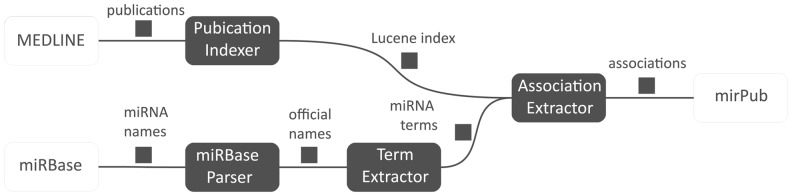
mirPub's text mining method. miRBase Parser produces the set of ‘official names’ consisting of all miRNA and family names recorded in miRBase. Term Extractor expands this set to produce the set of ‘miRNA terms’, which also contains variants of the official names based on predefined modification rules (see Section 2). Publication Indexer builds an inverted index on MEDLINE texts and association extractor utilizes this index to retrieve efficiently all appearances of miRNA terms in literature. The aforementioned appearances indicate miRNA publication associations
Many of the aforementioned terms are official miRNA molecule or family names collected from all available miRBase versions by mirPub’s ‘miRBase Parser’ component. During its execution, this component captures the whole miRBase history, relating each mature or hairpin name with particular nucleotide sequences, an information needed by mirPub’s miRNA data history visualization feature (for details see Section 2 of Supplementary Data). The rest terms are variants of the collected official names produced by mirPub’s ‘Term Extractor’ based on two predefined modification rules. The first rule consists of omitting the species prefix of the name (e.g. omit ‘hsa’ from ‘hsa-let-7a-5p’). In fact, it is very common in literature that a miRNA name appears without this prefix. The second rule employs replacing some tokens of miRNA names by others. For instance, token ‘mir’ is frequently replaced by tokens ‘mirna’ or ‘microrna’ and token ‘let’ is replaced by ‘mir-let’, ‘mirna-let’ or ‘microrna-let’. Similar rules have been also used in Xie et al. (2013).
To support efficient retrieval of publications that contain each miRNA term, mirPub builds a Lucene (http://lucene.apache.org/core/) inverted index for the titles, the abstracts and the full texts of MEDLINE articles. mirPub’s ‘Association Extractor’ probes this index for each miRNA term produced by Term Extractor and stores all discovered miRNA-to-publication associations in mirPub’s relational database.
Execution of the above-described text mining process will be performed in a regular basis on the most recent, hitherto, version of MEDLINE files. Nevertheless, this process fails to retrieve some miRNA-to-publication associations for various reasons. For instance, some associations are described in figures, thus optical character recognition must be used to retrieve them. Moreover, many articles are of restricted access, thus their full text is not available for text mining. To capture part of the previous cases, mirPub also incorporates data from several curated databases such as miRBase, TarBase and miR2Disease (Jiang et al., 2009). Additionally, mirPub incorporates a User Interface to report new miRNA-to-publication associations, as well as errors (see Supplementary Figs S1 and S3 of Supplementary Data). A specific protocol (for details refer to Section 1 of Supplementary Data) ensures that each newly reported association or error will be examined by a curator prior its inclusion in mirPub’s database. This supervised crowdsourcing approach is expected to guarantee that mirPub contains correct and up-to-date data.
3 Evaluating mirPub’s contribution
Currently, mirPub is the largest database that contains miRNA-to- publication associations counting >210,000 distinct associations (involving >19,800 articles). A comparison of mirPub to the most popular databases providing similar information can be found in Supplementary Table S1 of Supplementary Data. In brief, mirPub has been found to contain about 14-fold more publications than TarBase. Furthermore, preliminary experiments regarding mirPub’s effectiveness in retrieving miRNA literature are also presented in Supplementary Data. A first experiment indicates that mirPub has a recall that is more than 2-fold the recall of PubMed in retrieving miRNA-related publications (Supplementary Table S3 in Supplementary Data). This is possible as mirPub incorporates curated associations between miRNAs and publications. Another experiment concludes that taking into account miRNA data history improves search effectiveness (Supplementary Table S4 in Supplementary Data).
Supplementary Material
Acknowledgements
The authors acknowledge I. S. Vlachos for his substantial contribution in importing TarBase 6.0 data to mirPub.
Funding
This work was supported by the projects ‘MIKRORNA’, ‘TOM’ and ‘MEDA’ co-funded by ESF and national resources (actions ‘SYNERGASIA’, ‘ARISTEIA’ and ‘KRHPIS’, respectively).
Conflict of Interest: none declared.
References
- Griffiths-Jones S., et al. (2005) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res., 34(Suppl. 1), D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., et al. (2009) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res., 37(Database issue), D98–D104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer H., et al. (2007) MicroRNAs in disease and potential therpeutic applications. Mol. Ther., 15, 2070–2079. [DOI] [PubMed] [Google Scholar]
- Vergoulis T., et al. (2012) TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res., 40, D222–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B., et al. (2013) MirCancer: a microRNA-cancer association database constructed by text mining literature. Bioinformatics, 29,638–644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


