Abstract
Motivation: Classical nuclear export signals (NESs) are short cognate peptides that direct proteins out of the nucleus via the CRM1-mediated export pathway. CRM1 regulates the localization of hundreds of macromolecules involved in various cellular functions and diseases. Due to the diverse and complex nature of NESs, reliable prediction of the signal remains a challenge despite several attempts made in the last decade.
Results: We present a new NES predictor, LocNES. LocNES scans query proteins for NES consensus-fitting peptides and assigns these peptides probability scores using Support Vector Machine model, whose feature set includes amino acid sequence, disorder propensity, and the rank of position-specific scoring matrix score. LocNES demonstrates both higher sensitivity and precision over existing NES prediction tools upon comparative analysis using experimentally identified NESs.
Availability and implementation: LocNES is freely available at http://prodata.swmed.edu/LocNES
Contact: yuhmin.chook@utsouthwestern.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Active transport of macromolecules between the nucleus and the cytoplasm controls the localization and functions of many proteins and RNAs. The majority of nuclear-cytoplasmic transport of macromolecules is mediated by nuclear transport receptors of the Karyopherin β (Kap) family, which bind nuclear localization or export signals (NLSs or NESs) in their cargoes (Conti and Izaurralde, 2001; Görlich and Kutay, 1999; Tran et al., 2007; Weis, 2003; Xu et al., 2010). CRM1 (for Chromosome Region Maintenance 1, also known as Exportin-1 or XPO1) is the best characterized export-Kap or exportin (Fornerod et al., 1997; Fukuda et al., 1997; Neville et al., 1997; Noske et al., 2008; Ossareh-Nazari et al., 1997; Stade et al., 1997). Approximately 300 broadly functioning protein cargoes for CRM1 have been experimentally identified and compiled into three separate databases NESbase, NESdb, and ValidNESs (la Cour et al., 2003; Xu et al., 2012; Fu et al., 2013). Many CRM1 cargoes participate in important cellular processes such as gene expression, signal transduction, immune response, and cell differentiation. Aberrant CRM1-mediated nuclear export causes diseases such as cancer, viral and inflammatory diseases (Etchin et al., 2013; Fung and Chook, 2014; Lapalombella et al., 2012; Turner et al., 2012; Zhou et al., 2013).
Cognate peptide segments in protein cargoes that bind CRM1 are known as classical nuclear export signals or NESs (previously also known as leucine-rich NESs). NES peptides are usually 8–15 amino acids long with regularly spaced conserved hydrophobic residues. The first NES consensus of L-X2,3-[LIVFM]-X2,3-L-X-[LI] was established from results of in vivo NES randomization-selection assays (Bogerd et al., 1996). Subsequently, La Cour et al. re-defined the NES consensus as Φ1-X2,3-Φ2-X2,3-Φ3-X-Φ4 (Φn represents Leu, Val, Ile, Phe or Met; X stands for any amino acid) according to alignment of 80 experimentally defined NESs (thereafter shortened as experimental NESs) collected in NESbase (la Cour et al., 2004). Inclusion of additional hydrophobic residues significantly increased the NES consensus coverage. Kosugi et al. further expanded the NES consensus through analysis of 101 distinct NES peptides obtained from a random peptide library screen (Kosugi et al., 2008). The Kosugi set of consensus sequences included four previously defined patterns, termed NES Classes 1a-d (Class 1a: Φ1-X3-Φ2-X2-Φ3-X-Φ4; Class 1b: Φ1-X2-Φ2-X2-Φ3-X-Φ4; Class 1c: Φ1-X3-Φ2-X3-Φ3-X-Φ4; Class 1d: Φ1-X2-Φ2-X3-Φ3-X-Φ4;) and two new patterns (Class 2: Φ1-X-Φ2-X2-Φ3-X-Φ4; Class 3: Φ1-X2-Φ2-X3-Φ3-X2-Φ4), to describe additional spacings between key hydrophobic residues of their NES peptides. The set of consensus sequences also allowed Ala, Thr, Cys or Trp to occur only once at hydrophobic positions of an NES.
Crystal structures of CRM1 bound to NESs from PKIα, Snurportin-1 and the HIV-1 Rev protein revealed that NESs bind directly to a groove on the convex surface of CRM1. The NES groove contains five hydrophobic pockets that accommodate conserved hydrophobic NES residues (Dong et al., 2009; Güttler et al., 2010; Monecke et al., 2009). The PKIα (LALKLAGLDI, Class 1a) and Snurportin (MEELSQALASSFSV, Class 1c) NESs adopt α-helical conformations at their N-termini and transition to loops at the C-termini when bound to CRM1. In contrast, the CRM1-bound Rev NES (LPPLERLTL, Class 2) adopts an extended conformation. Examination of CRM1-NES interactions led to a structure-based consensus, which adds a fifth hydrophobic position at the N-terminus of the NES (Φ0) as modulator of CRM1-NES binding affinities (Güttler et al., 2010).
Of the different proposed NES consensus patterns, the Kosugi set of consensus sequences has the highest sensitivity, with 89% coverage of experimental NESs versus 65% for the la Cour consensus. However, the precision rate of the Kosugi consensus is low (4% compared to 12% for the la Cour consensus). The generally low precision rates of the different consensus are probably due to degeneracy of the consensus patterns, which describe the 2-turn amphipathic helix that is ubiquitous in the proteome. We refined the Kosugi consensus sequences based on analysis of 234 experimental NESs collected in NESdb (Xu et al., 2012). The refined consensus only marginally improved the prediction precision (6%), suggesting that NES consensus sequences alone are insufficient to accurately locate NESs in CRM1 cargoes.
Several computational tools have been developed to predict classical NESs. A predictor named NES-Finder (http://research.nki.nl/fornerodlab/NES-Finder.htm) was the first available web-server to identify sequence motifs that fit a subset of NES consensus (Classes 1a, 1b, and 1d). ELM is another pattern matching method although it uses different regular expressions to define NESs (Gould et al., 2010). NetNES, developed in 2004, is the first NES predictor that does not explicitly use consensus patterns (la Cour et al., 2004). Instead, it employs machine-learning algorithms like Neural Networks (NN) and Hidden Markov Models (HMM) and relies only on protein sequence as features. NetNES integrates the outputs of NN and HMM trained with experimental NESs in NESbase to assign a score to each residue in the input protein. NetNES increased prediction precision to ∼30% at the cost of lowering the maximum recall rate to ∼40% (Fu et al., 2011). More recently, the NES predictor NESsential applied simplified consensus patterns to the query sequence as a pre-filter followed by Support Vector Machine (SVM) classification that incorporates both sequence and biophysical features such as predicted intrinsic disorder, secondary structure and solvent accessibilities (Fu et al., 2011). NESsential achieved better precision at lower recall levels when tested with 85 experimental NESs collected in ValidNESs. For example, at 20% recall level, NESsential increased precision by 17% compared with NetNES (Fu et al., 2011). In 2014, a NES prediction tool named Wregex was published (Prieto et al., 2014). Like NESsential, Wregex scans the query with regular expressions to generate a list of NES candidates. A position-specific scoring matrix (PSSM) is then used to compute a score for these candidates. Comparison between NESsential and Wregex using NES motifs in human deubiquitinases (DUBs) showed that Wregex produces fewer NES candidates than NESsential. However, since Wregex does not incorporate predicted biophysical features, it takes less time to predict NESs.
In this study, we present a new computational tool named LocNES to locate classical NESs in CRM1 cargoes. LocNES first ranks the NES consensus fitting peptides according to its PSSM score. The PSSM score rank, protein sequence, consensus pattern, and disorder propensity are used as feature set of a SVM model. LocNES was tested with a large set of experimental NESs and showed improved performance over existing NES prediction methods.
2 Methods
2.1 NES datasets
Entries in two of the most recent NES databases, NESdb and ValidNESs, were examined and compared. At the time of manuscript preparation, NESdb and ValidNESs contain 253 and 221 experimental CRM1 cargoes, respectively. Entries in both databases were combined and 36 entries with contradicting experimental evidence (listed as ‘NESs in doubt’ in NESdb) were removed. Sequence similarities among the remaining proteins were detected using programs CD-HIT (Li and Godzik, 2006; clustering threshold 40%) and BLASTClust (Altschul et al., 1997; similarity threshold 10%) and identified homologs were removed. 246 non-redundant proteins containing 290 experimental NESs were compiled in a dataset named the Dbase dataset and listed in Supplementary Table S1.
Thirty-two functional NES motifs and 78 non-functional NES motifs from 56 DUBs were identified using a nuclear export assay (Garcia-Santisteban et al., 2012). A second dataset (DUB dataset) was constructed with these 110 DUB NES motifs (functional and non-functional). The DUB NES motifs used in the export assay are peptides with 19–22 amino acids.
2.2 NES candidates in the Dbase dataset
LocNES scans CRM1 cargoes in both the Dbase and DUB datasets with a sliding window protocol to retrieve NES candidates (peptides that conform to NES consensus patterns). The NES consensus sequences used by LocNES are a modified version of the Kosugi consensus sequences (Xu et al., 2012): Φ1-X1,2,3-Φ2-[^W]2-Φ3-[^W]-Φ4; Φ1-X2,3-Φ2-[^W]3-Φ3-[^W]-Φ4; or Φ1-X2-Φ2-X[^W]2-Φ3-[^W]2-Φ4 ([^W] is any of the 20 amino acids except Trp; Ala or Thr can be used once at Φ1 or Φ2; X stands for any amino acid). Each NES candidate consists of 15 amino acids (shorter if located at the protein N-terminus). The C-terminal amino acid of each peptide is Φ4 in the NES consensus. If the Φ2-Φ4 portion of an NES candidate overlaps with an experimental NES, it is deemed as a real NES. Otherwise, the NES candidate is defined as a negative NES. By these criteria, LocNES located 4201 NES candidates in the Dbase dataset. Among them, 493 NES candidates are real NESs and the remaining 3708 are negative NESs. LocNES found no NES candidate for 42 of the 290 experimental NESs in the Dbase dataset. LocNES assigns a zero probability score to these 42 NESs.
2.3 NES candidates in the DUB dataset
NES motifs in the DUB dataset are longer than existing NES consensus patterns. Consequently, LocNES typically retrieves 2–3 NES candidates from each DUB NES motif. Since experimental testing of the DUB dataset was conducted at the motif level, LocNES assigns a probability score to each motif in DUB dataset in the following manner. First, LocNES calculates the probability score for each NES candidate in the DUB dataset. Then the highest probability score among NES candidates from the same DUB NES motif is designated as the probability score of the motif.
Both NESsential and Wregex (the two most recent NES predictors used in our performance comparison) retrieve NES candidates from query proteins in a similar manner as LocNES. However, since the three predictors use different NES consensus patterns, the number of NES candidates retrieved varies among them.
2.4 Calculation of PSSM score
The PSSM used to score peptides in each testing set was constructed with the NES peptides (aligned at the Φ4 position) in its corresponding training set. The element in the matrix is calculated as log likelihood ratio of the position-specific probability and uniform background probability (0.05). A pseudocount of 1 is added for each position. The PSSM score for a given sequence is calculated as the sum of log likelihood ratio of every residue.
2.5 Prediction pipeline of LocNES
LocNES first retrieves NES candidates from a query protein (see the previous section for details). Next, the PSSM score is computed for each NES candidate and all candidates are ranked according to its PSSM score. The feature set for SVM model is then constructed, which includes PSSM score rank, peptide sequence represented by a vector containing twenty-one indicator variables (one for each amino acid plus a blank position) for each residue, and the types of consensus (Classes 1a, 1b, 1c, 1d, 2, or 3) for the NES candidate. The feature set also contains disorder propensities, computed with DISOPRED (Ward et al., 2004) for every residue of the NES candidate and for the 15 residues of both N- and C-terminal peptides flanking the NES candidate. In addition, LocNES includes an indicator variable that is set to 1 if the NES candidate is located within six residues of another NES candidate with higher PSSM score rank (0 if otherwise). To ensure a fair comparison between LocNES and other NES predictors, our SVM model was trained with 124 NESs, which are a subset of the 154 experimental NESs that trained NESsential and Wregex. SVM algorithm was implemented by LIBSVM integrated in Scikit-learn python package (Chang and Lin, 2011; Pedregosa et al., 2011). The output of the SVM model is the probability score of the NES candidate.
2.6 Performance evaluation
Both Dbase and DUB datasets were used to compare LocNES with NESsential and Wregex. If the probability score of a real NES is above a pre-defined threshold, it is counted as a true positive. Otherwise, it is a false negative. If the probability score of a negative NES is below the pre-defined threshold, it will be counted as a true negative. Otherwise, it is a false positive. Receiver Operating Characteristic (ROC) curve and its area under the curve (AUC) are computed to evaluate NES predictors’ performance. Precision-recall (PR) curves are also generated. Recall is defined as the fraction of real NESs whose probability score is higher than a threshold value. Precision measures the percentage of real NESs among NES candidates with probability score higher than a threshold value.
2.7 Availability of LocNES
LocNES is freely available at http://prodata.swmed.edu/LocNES. The web interface was developed using PHP. The only required input is the query protein sequence in FASTA format. A standalone Linux version of LocNES is available upon request.
2.8 In vitro CRM1-NES binding assays
Expression constructs for 11 different Class 3 NES peptides were generated by ligation of annealed oligonucleotides into the pGEX-Tev vector and verified by sequencing. GST-NESs were expressed and purified as previously reported (Dong et al., 2009). Approximately 60 µg of GST-NESs were immobilized on glutathione sepharose beads and incubated with excess human CRM1 in a total volume of 100 µl for 30 min at 4°C in the presence or absence of RanGTP. After extensive washing with buffer containing 20 mM Hepes, pH 7.3, 110 mM potassium acetate, 15% glycerol, 2 mM magnesium acetate, and 2 mM DTT, bound proteins were separated by SDS/PAGE and visualized by Coomassie staining. GST-NESs were analysed by mass spectroscopy to detect potential protease degradation or truncation of the fusion peptides. GST-NESs were also subjected to gel filtration analysis using the Superdex 200 column (GE Healthcare) to detect potential aggregation.
2.9 Nuclear-cytoplasmic cellular localization assay
Nuclear-cytoplasmic distribution of EYFP2-NLS-NES fusion proteins was observed in HeLa cells. Expresion constructs for EYFP2-NLS-NES fusion proteins were generated by ligation of annealed oligonucleotides into a pEYFP2-NLS(SV40) vector and verified by sequencing 11 different Class 3 NES peptides. The plasmids were transfected using Lipofectamine 2000 (Invitrogen, Life Technologies) into HeLa cells that were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin and amphotericin B, seeded onto glass-bottom 24-well culture plates (MatTek) and grown to 50–70% confluency. Live cells were imaged 24 h after transfection. To determine whether CRM1 mediates the nuclear export of EYFP2-NLS-NES fusion proteins, transfected cells were also incubated with 2 nM leptomycin B (LMB) for 16 h. Live cell image acquisition was performed at 37°C in a 5% CO2 atmosphere using a spinning disk confocal microscope system (Nikon-Andor), the MetaMorph software and analysed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
3 Results
3.1 The Dbase and DUB datasets
Two datasets, named Dbase and DUB, were compiled for training and evaluating LocNES. The Dbase dataset contains 246 non-redundant CRM1 cargoes with 290 experimental NESs culled from the NESdb and ValidNESs databases (Fu et al., 2013; Xu et al., 2012). LocNES retrieved 4201 NES candidates from the Dbase dataset (see Section 2.2 for details). Among them, 493 are real NESs and 3708 are negative NESs.
The DUB dataset was constructed using 32 functional and 78 nonfunctional NES motifs (each 19–22 amino acids long) in 56 human DUBs proteins. Performance comparison using the DUB dataset was conducted at the motif level with prediction scores determined as described in Section 2.3.
3.2 Cross-validation of LocNES
NES candidates retrieved by LocNES from the Dbase dataset were partitioned into a five-fold cross-validation dataset to search for the optimal parameters for SVM models. Each training set includes 378–409 real NESs and 2828–3076 negative NESs. Each test set contains 84–115 real NESs and 728–986 negative NESs. Each test set also includes 6–11 experimental NESs that do not match the refined Kosgui consensus (details in Section 2.2).
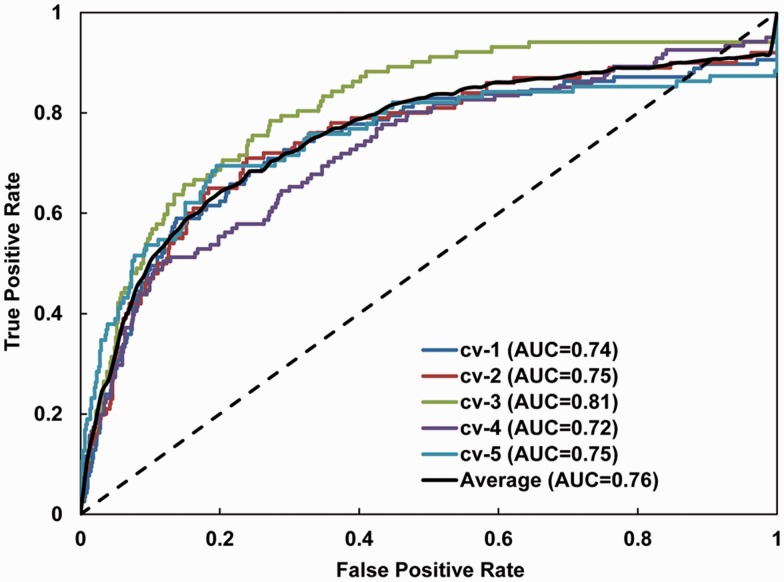
Extensive search of parameter space for both linear and RBF kernel of SVM models were conducted. The penalty parameter (C) for the linear kernel was sampled from 2−15 to 210. The penalty parameter and radius (C, γ) for the RBF kernel were sampled from 2−4 to 215 and from 2−15 to 210, respectively. We identified that a linear SVM model with C = 0.01 produced the largest average AUC value (0.76) and was used in subsequent performance comparisons. ROC curves for cross-validation using the linear SVM classifier are shown in Figure 1.
Fig. 1.
Receiver operating characteristic (ROC) curves of LocNES. ROC curves were generated using the five-way cross-validation (cv) set of Dbase. Each cv test set includes approximately 90 real and 900 negative NESs. The black dotted line represents random guessing
3.3 Features in the SVM model of LocNES
LocNES incorporates four categories of features in its SVM feature space to capture both the biophysical and sequence properties of NES candidate: PSSM score rank, peptide sequence, disorder propensity and the types of consensus pattern (Classes 1a, 1b, 1c, 1d, 2, or 3) for the NES candidate. In addition to these four feature categories, LocNES also includes a feature that indicates whether the NES candidate is located within six residues of another NES candidate with higher PSSM score rank. We name this feature the neighboring feature. The neighboring feature is introduced based on the observation that occasionally several NES candidates overlap with a real NES. We computed the F-scores for LocNES features using LIBSVM’s feature selection tool on the combined Dbase dataset (Chen and Lin, 2006). Results showed that among all the features tested, PSSM score rank has the highest discriminative power and the neighboring feature ranks the second among all the single features by F-score (Supplementary Table S2). An SVM model whose feature set includes just PSSM score rank and the neighboring feature produced an average AUC value of 0.71 when tested with Dbase cross-validation sets. PSSM score is less effective than the rank of PSSM score (AUC = 0.65). Peptide sequence has a slightly lower discriminative power than PSSM score rank (AUC = 0.70). Although disorder propensity is less discriminative than PSSM score rank or peptide sequence, it is still much better than random guessing as an SVM model with disorder propensity as its only feature gave an AUC value of 0.60 (versus AUC = 0.5). This is consistent with previous findings that NES regions tend to have higher disorder propensity than false positive matches (Fu et al., 2011; Xu et al., 2012). Information on NES consensus sequence types is the least discriminative feature (AUC = 0.55). Finally, although NESs found in available 3D structures tend to adopt α-helix-loop conformations and disfavor β-sheet conformations (Xu et al., 2012), an SVM model incorporating predicted secondary structure provided no performance improvement beyond an SVM model using disorder propensity (AUC = 0.58). It is likely that other features in our SVM model already implicitly represent secondary structural information.
3.4 Comparison of LocNES with NESsential and Wregex using the Dbase dataset
In order to compare the performance of LocNES with NES predictors NESsential and Wregex, we divided the Dbase dataset into training and test sets. The Dbase training set contains 124 experimental NESs from 103 CRM1 cargoes. The Dbase training set is a subset of the NESsential training set, which contains 154 NESs. Wregex used the same training set as NESsential. The remaining 143 CRM1 cargoes (containing 166 experimental NESs) in Dbase formed the test set. LocNES retrieved 1847 NES candidates from Dbase training set: 226 are real NESs (consensus matching peptides overlapping with experimental NESs) and 1621 are negative NESs (consensus matching peptides not overlapping with experimental NESs). A LocNES model was first trained using the parameters identified in cross-validation with this training set and then the model was used to locate NESs in the test set.
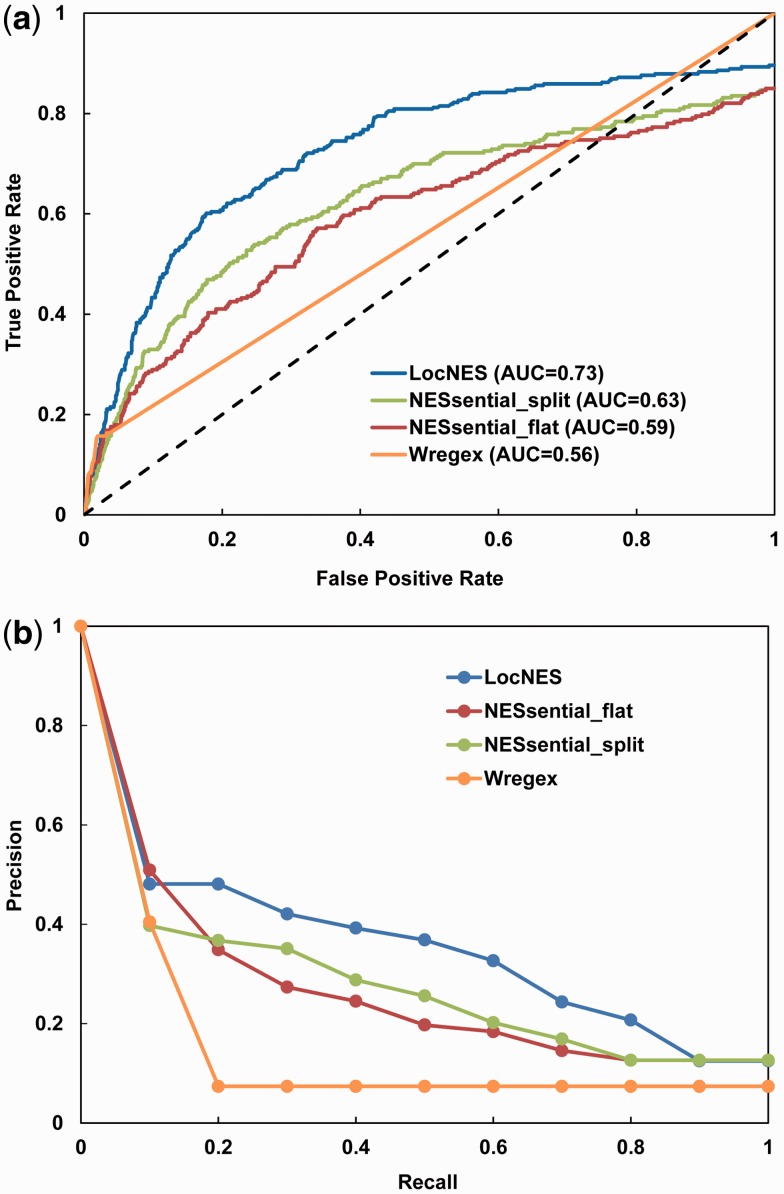
LocNES retrieved 267 real NESs and 2087 negative NESs from the Dbase test set. 31 experimental NESs in the test set do not match the refined Kosugi consensus patterns and hence were not retrieved by LocNES. Like LocNES, NESsential employs a pre-filter to identify NES candidates. NESsential found 232 real NESs and 1888 negative NESs from the Dbase test set. NESsential also failed to retrieve 41 experimental NESs in the Dbase test set. LocNES is able to retrieve a few more NES candidates than NESsential because the consensus sequences used by LocNES is more tolerant than the pre-filter used by NESsential (Φ-X2,3-Φ-X-Φ). Performances of LocNES and NESsential were evaluated with both the ROC and PR curves. As shown in Figure 2a, LocNES increases AUC values by 0.1 and 0.14 compared to NESsential’s flat and split modes, respectively. The PR curves in Figure 2b show that LocNES achieves higher precision than NESsential at most recall levels.
Fig. 2.
Performance of LocNES compared with NESsential and Wregex, using the Dbase test set. (a) ROC curve. The black dotted line represents random guessing. (b) Precision-Recall (PR) curve
Wregex is the most recently published NES predictor. The program combines regular expression matching and PSSM score. Several configurations are available in Wregex depending on the choice of regular expression and PSSM. Here, we used the Wregex A configuration, which was trained with the NESsential training set. Since Wregex uses a more restrictive regular expression ([DEQ].{0,1})([LIM])(.{2,3})([LIVMF])([^P]{2,3})([LMVF])([^P])([LMIV])(.{0,3}[DEQ]) than both LocNES and NESsential, it retrieved only 27 real NESs and 78 negative NESs from the Dbase test set. To compensate for the large discrepancy of real negative NESs between Wregex and LocNES, we added 2009 true negative NES candidates when we evaluate the performance of Wregex (i.e. Wregex always correctly predicts these 2009 NES candidates; 2087-78 = 2009). However, since Wregex found no matches for 139 experimental NESs in the test set, its maximum recall rate is only 16% compared to 90% for LocNES and 85% for NESsential. Consequently, the AUC value of Wregex is considerably smaller than that of LocNES and NESsential (Fig. 2a).
3.5 Comparison of LocNES with NESsential and Wregex using the DUB dataset
The DUB dataset includes 32 functional and 78 non-functional NES motifs (Garcia-Santisteban et al., 2012). Each NES motif in the DUB dataset is 19–22 amino acids long. Since NESs of the DUB dataset were tested at the motif level, performance comparison was conducted only at the same motif level (described in Section 2.3) to ensure that all predictors have the same numbers of real positives and real negatives.
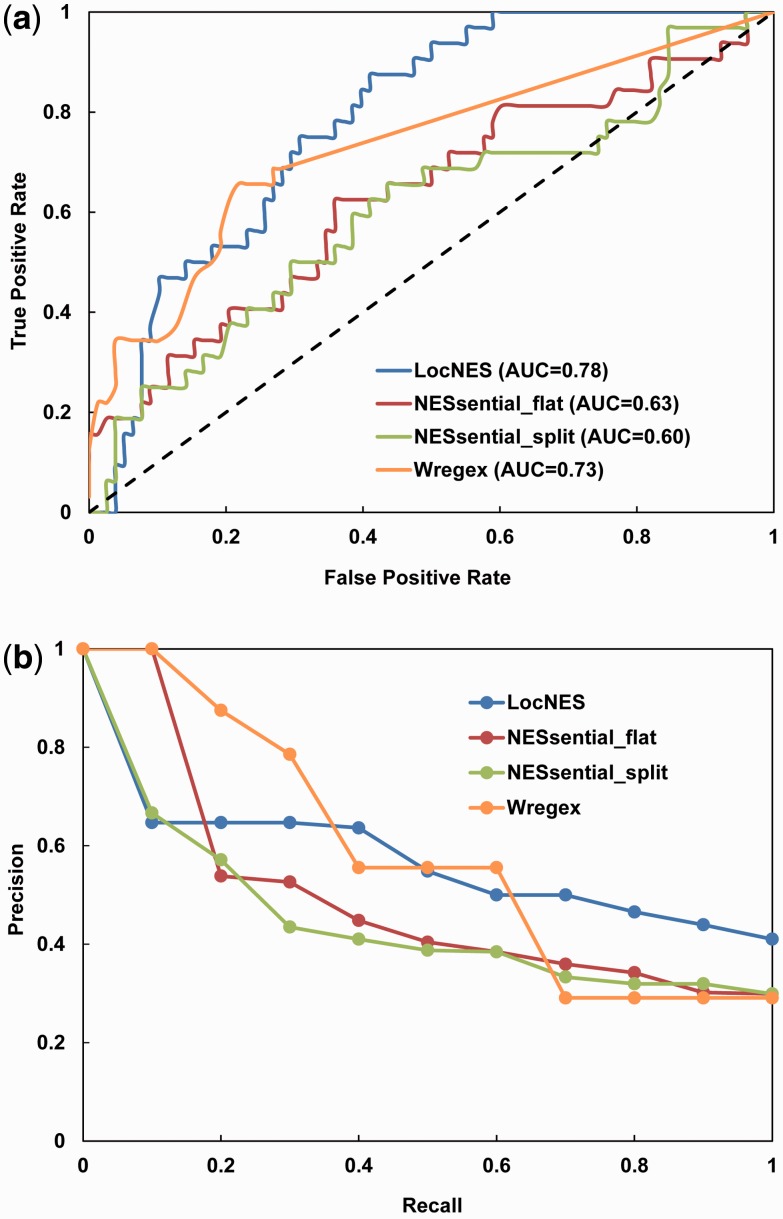
LocNES was able to retrieve at least one NES candidate for each of the 32 functional and 71 nonfunctional DUB motifs. NESsential found matches for 32 functional motifs and 75 nonfunctional motifs. LocNES and NESsential found no matches for 7 and 3 nonfunctional DUB motifs, respectively, and the prediction score of these motifs were set to zero. Wregex failed to find NES candidates for 10 functional and 56 nonfunctional DUB motifs. Thus, the maximum recall rate for Wregex is only 69% compared to 100% for LocNES and NESsential. Figure 3a shows that LocNES gives the best AUC value among the three predictors. On the other hand, Wregex achieves the best precision when the recall rate is lower than 40% (Fig. 3b). A meta-predictor that combines LocNES with Wregex improves the prediction performance on the DUB dataset compared with individual predictors (see Supplementary Figure S1). However, such a meta-predictor shows no performance improvement on the Dbase dataset.
Fig. 3.
Performance of LocNES compared with NESsential and Wregex using the DUB data set. (a) ROC curve. The black dotted line represents random guessing. (b) PR curve. The DUB test set includes 32 functional and 78 nonfunctional NES motifs
3.6 Prediction of Class 3 NESs by LocNES
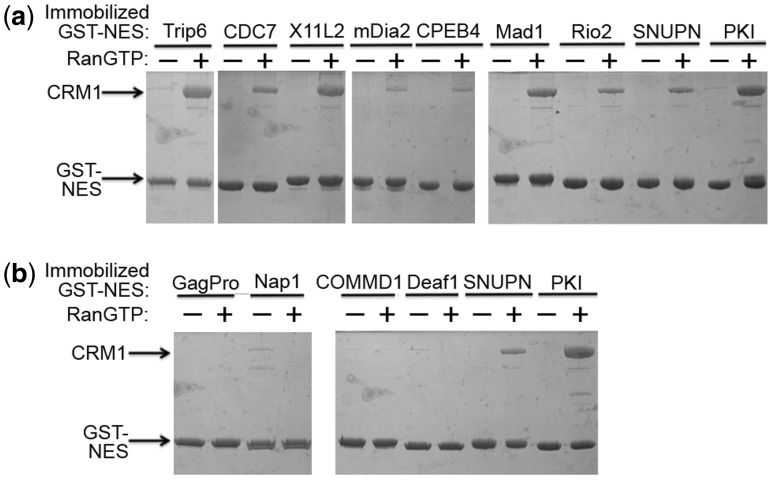
The NES consensus pattern Φ1-X2-Φ2-X3-Φ3-X2-Φ4 for Class 3 NESs was introduced in 2008 (Kosugi et al., 2008). Of the six NES patterns, only the Class 3 pattern contains two residues between Φ3 and Φ4. ∼10% of experimental NESs match the Class 3 pattern. Signals classified as Class 1b, 1c, 1d, and 2 NESs also have similar levels of occurrences (Kosugi et al., 2008). Among the 166 experimental NESs in the Dbase test set, 11 contain exclusively Class 3 NES candidates (Table 1). None of the class 3 NESs had been tested for CRM1 binding. Therefore, we tested these Class 3 NESs for direct CRM1 binding in pull-down binding assays using recombinant GST-NESs, CRM1 and RanGTP. Figure 4 shows that seven Class 3 NESs (Rio2, Mad1, CDC7, X11L2, CPEB4, mDia2, and Trip6) bind CRM1 in stoichiometric manner, further validating them as real NESs.
Table 1.
Results of CRM1-NES pull down binding assays, NES activity assay, and LocNES prediction scores for Class 3 NESs in Dbase test set.
| NES ID | Protein Name | Uniprot ID | NES sequence | Bind CRM1 | NES activity | Score |
|---|---|---|---|---|---|---|
| 77 | mDia2 | Q9Z207 | 1157SVPEVEALLARLRAL1171 | Yes | Yes | 0.38 |
| 104 | Mad1 | P40957 | 558AQTTIQLLQEKLEKL572 | Yes | Yes | 0.07 |
| 117 | COMMD1 | Q8N668 | 171ILKTLSEVEESISTL185 | No | No | 0.36 |
| 137 | Trip6 | Q9Z1Y4 | 93LDAEIDSLTSMLADL107 | Yes | Yes | 0.21 |
| 141 | X11L2 | O96018 | 53DESSLQELVQQFEAL67 | Yes | Yes | 0.28 |
| 153 | Rio2 | Q9BVS4 | 389RSFEMTEFNQALEEI403 | Yes | Yes | 0.04 |
| 197 | CDC7 | O00311 | 454PAQDLRKLCERLRGM468 | Yes | Yes | 0.09 |
| 198 | CPEB4 | Q17RY0 | 379RTFDMHSLESSLIDI393 | Yes | Yes | 0.14 |
| 254 | Nap1 | P25293 | 95KLLSLKTLQSELFEV109 | No | Yes | 0.18 |
| P147a | Deaf1 | O75398 | 459MVNSLLNTAQQLKTL473 | No | No | 0.55 |
| P148a | GagPro | P03322 | 225VREELASTGPPVVAM239 | No | No | 0.01 |
aCargoes collected in ValidNESs.
Fig. 4.
Interactions of Class 3 NESs with CRM1. Binding between recombinant purified GST-NESs and CRM1 is shown by pull-down binding assays. Eleven GST-Class 3 NESs were immobilized and incubated with CRM1 in the presence and absence of RanGTP. Bound proteins were resolved with SDS-PAGE and visualized by Coomassie staining. All 11 NESs conform only to Class 3 patterns. (a) Class 3 NESs that bind CRM1. (b) Class 3 NESs that do not bind CRM1
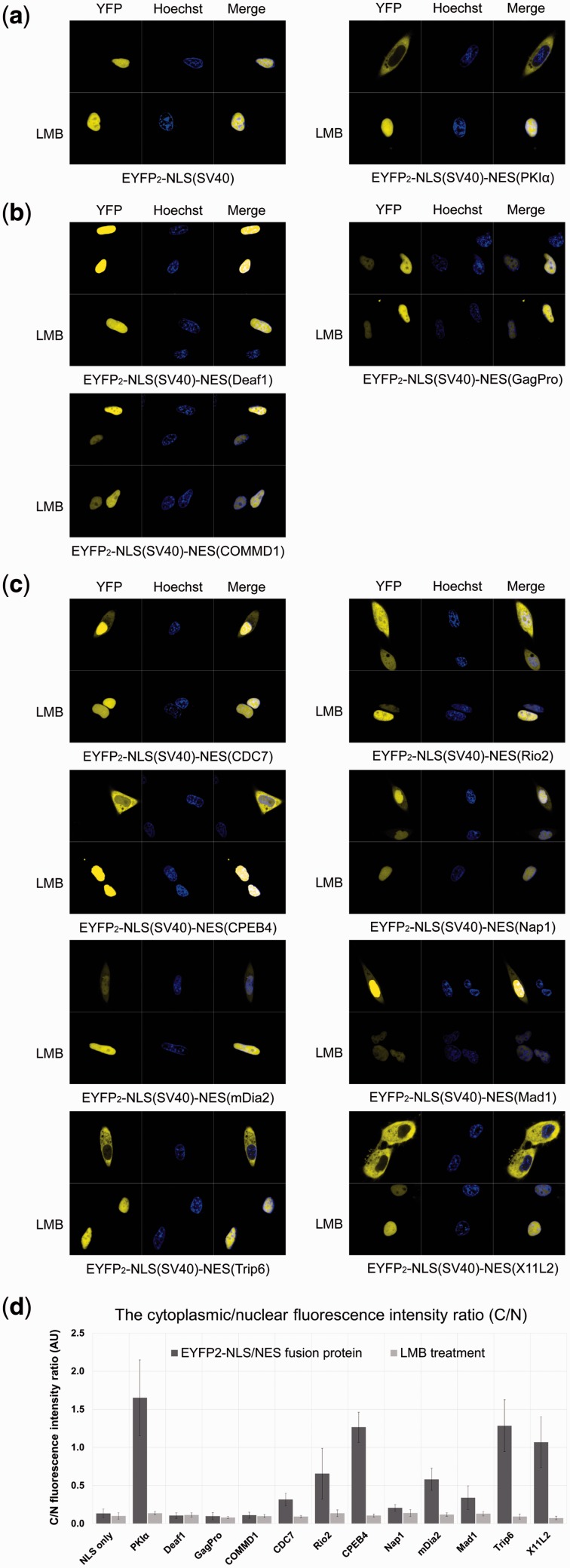
We also performed nuclear–cytoplasmic localization assays in HeLa cells to probe NES activities of the same eleven Class 3 NESs. As shown in Figure 5 and Supplementary Figure S2, eight of the eleven NESs (Rio2, Mad1, CDC7, X11L2, CPEB4, mDia2, Trip6 and Nap1) were able to target the EYFP2-NLS reporter to the cytoplasm. Among these Class 3 NESs, which show positive nuclear export activity, the Nap1 NES is the only one that did not bind CRM1. Interestingly, we observed cytoplasmic localization of EYFP2-NLS-NES(Nap1) in only 55% of HeLa cells which were transfected with the plasmid as compared to 100% of cells transfected with the seven CRM1 binders (see Supplementary Table S3). These results suggest that the Nap1 NES is a very weak NES. The remaining three Class 3 NESs (COMMD1, GagPro, and Deaf1) showed no detectable CRM1 binding nor cytoplasmic localization in HeLa cells, suggesting that they may have been incorrectly identified and are likely negative NESs.
Fig. 5.
Nuclear-cytoplasmic distribution of EYFP2-NLS/NES fusion proteins in HeLa cells. DIC, YFP (pseudocolored in yellow), Hoechst (pseudocolored in blue) and YFP merged with Hoechst images were captured using a spinning disk confocal microscope (60x). DIC images are shown in Supplementary Figure S2. All 11 NESs conform only to Class 3 pattern. CRM1-dependence is demonstrated by the nuclear accumulation after treatment with 2 nM leptomycin B for 16 h. (a) Controls with a classical monopartite NLS (SV40) and a classical NES (PKIα). (b) Class 3 NESs that presented positive in nuclear export activity. (c) Class 3 NESs that presented negative in nuclear export activity. Images shown here are representatives of at least three independent experiments and over a total of 350 transfected cells. The percentage of transfected cells showing cytoplasmic localization is listed in Supplementary Table S3. (d) The average ratio of cytoplasmic/nuclear fluorescence intensity (C/N ratio). Error bars indicate the standard deviations. For each EYFP2-NLS/NES fusion protein, fluorescence intensities were measured in 10 independent cells from different experiments using ImageJ software. AU, arbitrary units
NESsential and Wregex were able to identify all four Class 3 non-binders as negative NESs. However, both predictors have 0% recall rate for the seven Class 3 CRM1 binders since they filter out peptides with Φ3-X2-Φ4 spacing at the initial stage of prediction. NES-Finder also does not predict any Class 3 NESs binders since it only finds NES candidates that fit Class 1a, 1b or 1d patterns. LocNES is able to retrieve four Class 3 CRM1 binders (mDia2, Trip6, X11L2, and CPEB4; recall rate 57%) at the default threshold value of 0.1 (Table 1). LocNES also correctly identified one non-binder (GagPro) but mislabeled the remaining three non-binders as NESs. The older NetNES predictor does not employ pre-filtering and therefore is also able to find some Class 3 NESs. Of the seven Class 3 CRM1 binders, NetNES identified Class 3 NESs only in Mad1 and CDC7 (recall rate 28%) using its default cutoff of 0.5. All four non-binders were correctly identified as such at the same cutoff. NetNES may have a low recall rate for Class 3 NESs because few Class 3 NES peptides were identified before 2004. Class 3 NESs do not fit the traditional NES consensus patterns and were probably overlooked during experimental searches for NESs. Therefore, the machine learning algorithms in NetNES (and to a lesser extent LocNES) may not be optimally trained to recognize Class 3 NESs. Recall rate for Class 3 NESs will likely increase as more Class 3 NESs are identified and reported.
4 Discussion
4.1 Features that improved LocNES performance
Among all the features employed by the LocNES SVM model, the rank of PSSM score has the highest discriminative power. Interestingly, PSSM score is less effective than PSSM score rank (AUC 0.65 versus 0.71). The enhanced effectiveness of PSSM score rank may be due to over-representation of Class 1a NESs among experimental NESs (Xu et al., 2012). Since the PSSM score of a sequence measures how closely it resembles known NESs, a real NES that doesn’t fit class 1a spacing may have a low PSSM score. For example, the zebrafish protein Vsx1, a paired-like subclass of homeodomain protein, was shown to have a highly conserved Class 1b NES (31GFRSKGFAITDLLGL45; Knauer et al., 2005). LocNES gave the Vsx1 NES a rather low PSSM score of 0.56 but ranks it the highest among all NES candidates within Vsx1. As expected, a SVM model using PSSM score rank as a feature produced a higher prediction score for the Vsx1 NES than a SVM model using the absolute PSSM score as a feature (0.26 versus 0.09). The PSSM currently used by LocNES may not be large or diverse enough to completely sample the entire range of experimental NESs. It will be important to update the PSSM as more NESs are identified experimentally, as prediction performance will likely improve with a more balanced and representative PSSM.
Although LocNES uses PSSM to represent sequence information, incorporation of sequence into SVM model did produce a small but obvious improvement to the prediction performance (AUC increased by 0.04). There appears to be additional implicit information encoded in the sequence that enhance CRM1 binding or export activity of NESs but yet not detected by PSSM. Sequence analysis of CRM1 cargoes collected in NESdb revealed that real NESs exhibit distinct amino acid composition from negative NES candidates. In particular, acidic residues such as Asp and Glu are more prevalent in the non-conserved positions in real NESs (Xu et al., 2012). These negatively charged residues may form electrostatic contact with the basic amino acids that lined the NES binding groove as observed in structures of CRM1-NES complexes, thus increasing the CRM1-NES binding affinity. In addition, the presence of bulky amino acids like tryptophan in negative NES candidates, which is absent from the C-terminus of real NESs, can potentially cause steric clash with the narrow groove at the C-terminal end of NES binding groove and prevent binding of negative NES candidates.
Incorporation of disorder propensity noticeably increases LocNES prediction performance. In fact, the disorder status of the sequence on the C-terminal side of NESs is among the top-10 ranking features (Supplementary Table 2). The developers of NESsential also reported similar findings (Fu et al., 2011). It has been noticed that intrinsically disordered region of a protein often contain functionally important sites due to its flexibility and modularity. Comparison of the disorder scores between real NESs and negative NES candidates revealed that sequences flanking real NESs have significantly higher disorder scores than sequences surrounding negative NES candidates. In addition, examination of existing three-dimensional structures containing real NESs or negative NES candidates showed that real NESs are more likely to locate at the termini of protein domains or flanked by long loops (Xu et al., 2012). The location of real NESs near protein termini and within loops or disordered regions may increase their accessibility to CRM1 or enhance their adaptability for suitable CRM1-binding conformations.
LocNES outperforms existing NES predictors in Class 3 NES prediction. To explore which features of LocNES enhance Class 3 NES prediction, we computed the F-scores on Dbase dataset with the Class 3 NESs removed (Supplementary Table S4). Comparison between Supplementary Tables 2 and 4 showed that the neighboring feature, which indicates if another NES candidate is in close proximity (details in Section 3.3), is not among the top-10 ranking features when Class 3 NESs are not included in the dataset. Instead, amino acid identity at positions 6 and 14 become top ranking features, as well as the NES consensus type (if the NES candidate belongs to Class 1a). These feature changes when Class 3 NESs are removed from dataset demonstrate the prevalence of Class 1a among all experimental NESs. Furthermore, it indicates that the neighboring feature boosts Class 3 NES prediction performance. If the weight of Class 3 NESs is increased in the dataset (doubled or tripled), the F-score of the neighboring feature becomes slightly higher than the corresponding value calculated with the original Dbase dataset. Since the mean value of neighboring feature of real Class 3 NESs is higher than that of the negative Class 3 NES candidates, it seems that real Class 3 NESs tend to locate close to other NESs of higher PSSM score rank. Incorporation of neighboring feature likely helps to raise the prediction score for these Class 3 NESs. One interesting feature of Class 3 NES is that the spacing between its Φ2 and Φ4 (Φ2-X3-Φ3-X2-Φ4) is the same as the spacing between Φ1 and Φ3 of a Class 1a NES (Φ1-X3-Φ2-X2-Φ3). Therefore, there are some instances where a Class 3 NES and a Class 1a NES overlap the same experimental NES. For example, it was shown that human CPEB1 protein (Uniprot ID: Q9BZB8) harbors an experimental NES 92ANDLCLGLQSLSL104 (Ernoult-Lange et al., 2009). Two NES candidates can be found within this experimental NES: ANDLCLGLQSL (Class 3) and LCLGLQSLSL (Class 1a). We assume both NES candidates are real NESs since it is impossible to pinpoint which NES candidate is functional without elaborate point mutation studies. It is likely that both NES candidates are indeed functional. They may possess different binding affinities to CRM1 or the two NESs may act concertedly as a high-affinity NES with five hydrophobic residues (Φ1 in Class 3 NES acts as Φ0) as described by the structural-based consensus (Güttler et al., 2010). In the case of the CPEB1 protein, the prediction score of the Class 3 NES is almost the same as the Class 1a NES despite of the lower PSSM score rank of the Class 3 NES compared with the Class 1a NES.
4.2 Machine learning algorithms in NES prediction
Among the three NES predictors that we compared in performance analysis, Wregex is the only predictor that does not rely on a supervised machine-learning algorithm. Instead, it relies on PSSM to capture the intrinsic contribution of each residue to NES activity. While PSSM based methods have proven a useful tool for annotating functional sites, the low recall rate of Wregex indicates that PSSM by itself is insufficient in predicting NES. The major advantage of Wregex is its speed. It only takes seconds for Wregex to process a large number of proteins. Both NESsential and LocNES need time to generate features (such as disorder propensity or solvent accessibility) for machine-learning methods. Typically, it takes two minutes for LocNES to process a protein with ∼600 amino acids. However, since machine-learning methods are especially suited for pattern recognition problems when the patterns are not easily described by a well-defined set of rules, the potential improvement in prediction performance outweighs the extra cost in time. In NES prediction, machine-learning methods may be a contributing factor to the significantly higher recall rate of LocNES and NESsential. Furthermore, incorporation of PSSM together with sequence and structural properties of NESs into the feature set of machine-learning algorithm may further boost LocNES performance. Both NESsential and LocNES implemented machine-learning algorithm using SVM. We also experimented with Random forests, another popular machine learning classifier. When the same feature sets were used, Random Forests performed similarly as SVM models (data not shown).
4.3 Prediction at the protein level
In addition to determining NES locations within CRM1 cargoes, NESsential also attempted classification of NES-containing proteins versus non-NES-containing proteins. LocNES, on the other hand, assumes that the query protein is a CRM1 cargo and focuses on locating NES sites within the query. The lack of a large and reliable negative training/testing data is the major reason for this decision. NESsential selected 541 proteins annotated in Uniprot as located in only one compartment, either the nucleus or the cytosol, as non-NES containing proteins. There is concern that cellular localization information in Uniprot may not be up-to-date due to the rapid discovery of CRM1 cargoes. A quick search showed that among the 13 CRM1 cargoes from Saccharomyces cerevisiae collected in NESdb, 20% or three proteins (Hsp70, MCM3 and Map1), are annotated as localized to either the nucleus or the cytosol thus fitting the NESsential criteria of non-NES containing protein (Liku et al., 2005; Scott et al., 2009; Shulga et al., 1999). Furthermore, nuclear import and export processes are highly regulated. Many CRM1 cargos are modified and accessible to CRM1 only in specific cellular or signaling states. Therefore, steady state subcellular localization of protein may not reflect their status as CRM1 cargoes or inform on their localization upon stimulation from external cues or change of cellular states.
Enhanced performance of LocNES compared with NESsential and Wregex resulted from a more tolerant pre-filter, a more representative feature set for machine-learning models, a more accurate training dataset, and the combined use of machine-learning method, position specific scoring matrix and biophysical properties of NESs. As more CRM1 cargoes/NESs are discovered, increased size, diversity and accuracy of experimental NES databases will continue to improve training/testing datasets for future NES predictors. The rapid growth of high-resolution protein structures will also guide identification of NES candidates that are inaccessible to CRM1 and thus increase precision of NES prediction. Finally, advances in modeling conformational changes induced by post-transcriptional modification or binding partners may facilitate the discovery of highly regulated NESs and similarly increase the sensitivity of NES prediction.
5 Conclusion
LocNES is a supervised machine-learning algorithm to predict NESs in potential CRM1 cargoes. LocNES integrates the rank of PSSM score, peptide sequence, disorder propensity and NES consensus type into an SVM model. Performance comparison between LocNES and two latest NES predictors, NESsential and Wregex using two separate test datasets showed that LocNES achieved higher precision at most recall levels with more than 0.1 increase of AUC value than NESsential. LocNES produced at least 30% higher maximum recall rate than Wregex. In addition, LocNES is the only tool that can predict Class 3 NESs with over 60% recall rate at the default threshold.
Supplementary Material
Acknowledgements
While this paper was under review, another NES predictor, NESmapper (Kosugi et al. PLoS Comput. Biol. 2014), was published.
Funding
This work was funded by Cancer Prevention Research Institute of Texas (CPRIT) Grant RP120352 (to Y.M.C.), National Institutes of Health Grants [F32GM093493 to D.X., GM094575 to N.V.G, and R01 GM069909 to Y.M.C.], the University of Texas Southwestern Endowed Scholars Program (Y.M.C.), Welch Foundation Grant [I-1505 to N.V.G and I-1532 to Y.M.C.], and a Leukemia and Lymphoma Society Scholar Award (to Y.M.C.).
Conflict of Interest: none declared.
References
- Altschul S.F., et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., et al. (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell Biol., 16, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-C., Lin C.-J. (2011) LIBSVM : a library for support vector machines. ACM Transactions on Intelligent Systems and Technology, 2, 27:21–27:27. [Google Scholar]
- Chen Y.-W., Lin C.-J. (2006) Combining SVMs with various feature selection strategies. In I. Guyon, et al. (eds), Feature Extraction, Foundations and Applications, Springer, Berlin Heidelberg, pp. 315–324. [Google Scholar]
- Conti E., Izaurralde E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Dong X., et al. (2009) Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature, 458, 1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernoult-Lange M., et al. (2009) Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol. Biol Cell., 20, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchin J., et al. (2013) Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia, 27, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., et al. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fu S.C., et al. (2013) ValidNESs: a database of validated leucine-rich nuclear export signals. Nucleic Acids Res., 41, D338–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S.C., et al. (2011) Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res., 39, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., et al. (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature, 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Fung H.Y., Chook Y.M. (2014) Atomic basis of CRM1-cargo recognition, release and inhibition. Semin. Cancer Biol., 27, 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Santisteban I., et al. (2012) A global survey of CRM1-dependent nuclear export sequences in the human deubiquitinase family. Biochem. J., 441, 209–217. [DOI] [PubMed] [Google Scholar]
- Görlich D., Kutay U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Gould C.M., et al. (2010) ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res., 38, D167–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttler T., et al. (2010) NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol., 17, 1367–1376. [DOI] [PubMed] [Google Scholar]
- Knauer S.K., et al. (2005) Nuclear export is evolutionarily conserved in CVC paired-like homeobox proteins and influences protein stability, transcriptional activation, and extracellular secretion. Mol. Cell Biol., 25, 2573–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., et al. (2008) Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic, 9,2053–2062. [DOI] [PubMed] [Google Scholar]
- la Cour T., et al. (2003) NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res., 31, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T., et al. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel., 17, 527–536. [DOI] [PubMed] [Google Scholar]
- Lapalombella R., et al. (2012) Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood, 120, 4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22, 1658–1659. [DOI] [PubMed] [Google Scholar]
- Liku M.E., et al. (2005) CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2-7 complex prevents chromosomal rereplication. Mol. Biol. Cell, 16, 5026–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke T., et al. (2009) Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science, 324, 1087–1091. [DOI] [PubMed] [Google Scholar]
- Neville M., et al. (1997) The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol., 7, 767–775. [DOI] [PubMed] [Google Scholar]
- Noske A., et al. (2008) Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer, 112, 1733–1743. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B., et al. (1997) Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science, 278, 141–144. [DOI] [PubMed] [Google Scholar]
- Pedregosa F.V.G., et al. (2011) Scikit-learn: machine learning in python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Prieto G., et al. (2014) Prediction of nuclear export signals using weighted regular expressions (Wregex). Bioinformatics, 30, 1220–1227. [DOI] [PubMed] [Google Scholar]
- Scott R.J., et al. (2009) The nuclear export factor Xpo1p targets Mad1p to kinetochores in yeast. J. Cell Biol., 184, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., et al. (1999) A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J. Biol. Chem., 274, 16501–16507. [DOI] [PubMed] [Google Scholar]
- Stade K., et al. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Tran E.J., et al. (2007) SnapShot: nuclear transport. Cell, 131, 420. [DOI] [PubMed] [Google Scholar]
- Turner J.G., et al. (2012) Nuclear export of proteins and drug resistance in cancer. Biochem. Pharmacol., 83, 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.J., et al. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol., 337,635–645. [DOI] [PubMed] [Google Scholar]
- Weis K. (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell, 112, 441–451. [DOI] [PubMed] [Google Scholar]
- Xu D., et al. (2010) Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr. Opin. Struct. Biol., 20, 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., et al. (2012) Sequence and structural analyses of nuclear export signals in the NESdb database. Mol. Biol. Cell, 23, 3677–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., et al. (2012) NESdb: a database of NES-containing CRM1 cargoes. Mol. Biol. Cell, 23, 3673–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., et al. (2013) CRM1 is a novel independent prognostic factor for the poor prognosis of gastric carcinomas. Med. Oncol., 30, 726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.