Abstract
Caenorhabditis elegans is uniquely suited to the analysis of cell lineage patterns. C. elegans has a small number of somatic cells whose position and morphology are almost invariant from animal to animal. Because C. elegans is virtually transparent, cells can be identified in live animals using a simple bright-field microscopy technique, Nomarski differential interference contrast (DIC), or by expression of transgenic fluorescent reporter genes. The small size and rapid development of C. elegans mean that animals can develop while under continuous observation, allowing cell lineages to be analyzed throughout embronic and postembryonic development. Embryonic cell lineages can also be traced semiautomatically using timelapse imaging of GFP-labeled nuclei. Analysis of mutant cell lineages remains important for defining the roles of developmental control genes.
I. Introduction
Cell lineage analysis refers to the tracing of cellular genealogies by following cell divisions and migrations over time, beginning with specific progenitor cells and ending with their postmitotic descendants. The development of almost all metazoan animals can in principle be described as a lineage tree whose origin is the single-cell zygote. However, the variability of normal development means that cell lineage relationships can in general only be described in probabilistic terms. In contrast, for some animal groups, including nematodes, molluscs, and tunicates, the pattern of cell divisions throughout development is highly invariant between individuals. In such animals, the invariant lineage constitutes a complete fate map of development with single-cell resolution.
The first descriptions of nematode cell lineages began in the late 19th century and were based on a series of fixed specimens. These studies established that the pattern of embryonic cell divisions was virtually invariant from animal to animal. In some cases, the cell lineage was thought to generate a fixed number of cells in the adult (“cell constancy” or eutely), or at least in certain tissues (“partial constancy”) (van Cleave, 1932). However, it was not until the development of Nomarski DIC microscopy in the late 1960s (Allen et al., 1969; Padawer, 1968) that it became feasible to observe cell divisions in live animals.
Using Nomarski DIC microscopy of live animals the complete cell lineage of C. elegans from zygote to adult was delineated in a series of classic studies, culminating in the complete description of the embryonic cell lineage in 1983 (Sulston et al., 1983). All these descriptions were based on direct observation of live animals, without significant use of recording technology. Since then, cell lineages have been traced in over ten other nematode species (see Table I). The C. elegans “lineage papers” (Table II) remain an essential resource for learning cell identification and lineage analysis. For historical accounts of the early days of lineage analysis see Horvitz and Sulston (1990) and John Sulston’s Nobel Lecture (Sulston, 2003).
Table I.
Cell-lineage analysis in other nematode species
| Species | Lineages studied | Reference |
|---|---|---|
| Panagrellus redivivus | Postembryonic lineages | (Sternberg and Horvitz, 1981, 1982) |
| Turbatrix aceti | Early embryo | (Sulston et al., 1983) |
| Mesorhabditis etc. | Vulval lineages | (Sommer et al., 1994) |
| Pristionchus pacificus | Vulval lineages | (Sommer, 1997) |
| Acrobeloides | (Wiegner and Schierenberg, 1998) | |
| Oscheius tipulae | Vulval cell lineage | (Delattre and Felix, 2001) |
| Pellioditis marina | Complete embryonic lineage | (Houthoofd et al., 2003) |
| Halicephalobus gingivalis | (Houthoofd and Borgonie, 2007) | |
| Rhabditophanes | Embryonic lineage | (Houthoofd et al., 2008) |
| C. briggsae | Embryonic lineage; automated lineage tracing | (Zhao et al., 2008) |
| Diploscapter coronatus | Embryonic lineage | (Lahl et al., 2009) |
| Romanomermis culicivorax | Embryonic lineage | (Schulze and Schierenberg, 2008, 2009) |
Table II.
Key publications describing C. elegans lineages
| Lineages | Reference |
|---|---|
| Ventral nerve cord | (Sulston, 1976) |
| Postembryonic nongonadal lineages (hermaphrodite) | (Sulston and Horvitz, 1977) |
| Gonadal lineages (both sexes) | Kimble and Hirsh, 1979) |
| Postembryonic nongonadal lineages (male) | (Sulston et al., 1980) |
| Embryonic lineage | (Deppe et al., 1978; Sulston et al., 1983) |
With the advent of green fluorescent proteins (GFP) in the early 1990s (Chalfie et al., 1994), cell identification entered a new phase. Now specific cells or cell types could be identified more rapidly, without the need for meticulous drawing out of cells and their positions. Expression of a fluorescent marker provided an unambiguous measure of cell fate. For example, cells could be identified even when misplaced in aberrant locations as a result of a cell migration defect. Most importantly, whereas under DIC observation only cell nuclei are typically resolved, GFP markers can be used to visualize the entire cell, or specific subcellular compartments. The ability to see axons, muscle arms, and other structures in live animals opened up whole new areas of analysis.
GFP transgenic markers have in many cases replaced DIC for cell identification. Nevertheless there are still reasons to learn the DIC anatomy. First, transgenic markers are not available for all cell types or subsets of cell types. Second, care is needed to ensure that the GFP marker (often a high-copy number transgene) does not itself interfere with cell differentiation. Issues of photobleaching or phototoxicity often limit the amount of observation possible, although this has been to some extent overcome in the automated analysis of embryonic cell divisions. Finally, analyzing a number of markers can involve considerable strain construction, work that can be avoided if the cells can be identified by DIC.
There has been rapid recent progress in automation of cell identification and lineage analysis. Cell lineage analysis in embryos can be partly automated as discussed in detail below. Automatic cell lineage analysis in larval stages has so far not been possible, largely due to the difficulty in immobilizing larvae in a way that allows normal development. However, digital atlases of cell positions at defined stages can be generated, allowing gene expression patterns to be mapped semiautomatically (Long et al., 2009). At present such atlases represent ~65% of the nuclei in the L1 stage. However, many neuronal nuclei are too closely spaced to be reliably identified by automated analysis. Thus, to identify specific neuronal expression patterns a knowledge of the anatomy remains indispensable.
II. Rationale
A. Analysis of Mutant Phenotypes
One of the most frequent goals in cell-lineage analysis is to address the developmental basis of a specific phenotype, whether caused by mutation, RNAi, or some other perturbation. For example, using cell type specific GFP reporters it is straight-forward to screen for mutants affecting the number of cells that express a given reporter (Doitsidou et al., 2008; Kanamori et al., 2008). A change in the number of expressing cells could have a number of causes and lineage analysis can resolve these possibilities. For example, do excess GFP-expressing cells arise from ectopic expression of the reporter or from an overproliferation of specific precursors? Does failure to generate a given cell type reflect a cell fate transformation or an earlier defect in the lineage?
B. Cell Division Pattern as the Focus of Interest
In some cases, the pattern of cell divisions itself is the focus of interest, especially where no other molecular markers are available. For example, the role of the Wnt ligand LIN-44 in cell polarity was deduced from its effects on the polarity of certain cell divisions in the male tail (Herman and Horvitz, 1994). The stage specificity of cell-division patterns was critical in inferring the genetic control of developmental timing in larval development (Ambros and Horvitz, 1984). The regulative ability of certain tissues to undergo compensatory growth after damage was studied using cell-lineage analysis (Sulston and White, 1980).
Stem-cell-like division is inherently polarized. The stem-cell-like behavior of larval seam cells has been extensively analyzed by direct lineage analysis (e.g.Nimmo et al., 2005). Analysis of cell-lineage mutants has also been important in understanding the genetic basis of cell cycle control (e.g. Kostić and Roy, 2002; Fukuyama et al., 2003).
C. Cellular Patterns of Gene Expression
A common goal in cell identification is to define the cellular expression patterns of reporter genes. Some genes are expressed in relatively clear-cut patterns, for example, intestinal cells, all body wall muscles, all GABAergic neurons, and so on (cf. Chapter by Yan and JIn?). It is essential to learn to correlate such simple expression patterns first before attempting more complex patterns.
The nervous system poses the most daunting challenge for identification of gene-expression patterns. This is especially so in the large anterior ganglia, each of which contains 10–20 closely packed nuclei. However, the relative positions of most neuronal nuclei are fairly reproducible and can be learnt by reference to the maps in the L1 stage (Sulston et al., 1983). Maps of these nuclei in the adult are based on serial section EM reconstruction (White et al., 1986) and can be found in WormAtlas. Some neuronal nuclei display natural variability in location, and so cannot be conclusively identified based on position. Fortunately, identification of neurons is often made considerably easier by the distinctive disposition of the axons and dendrites of individual cell types.
D. Cell Killing by Laser Ablation
Cell ablation has been an important technique to define the developmental and physiological functions of cells (Bargmann and Avery, 1995). Individual cells can be killed with a laser microbeam focused on the cell nucleus. This depends on accurate cell identification, for which both DIC and GFP are now used. Ablation during development can be used to test the extent of replacement regulation by other cells. Physiological functions of cells can be addressed by ablation unless they are subject to replacement or compensation.
E. Genetic Mosaic Analysis
In C. elegans, genetic mosaic analysis relies on spontaneous loss of unstable extrachromosomal arrays or chromosomal duplications during development. By identifying the pattern of cells in which the array or duplication has been lost, the “loss point” in the early lineage can be deduced. Such patterns are generally examined in late larval stages, that is, after the cell divisions are largely complete.
F. Comparative Developmental Biology
Cell lineages can be traced using Nomarski DIC in any optically transparent organism that can develop under continuous observation. As a result, direct cell-lineage analysis has now been undertaken in over a dozen different nematode species (Table I). Embryonic lineages have now been traced in several species. Although initial studies suggested a high degree of conservation in early embryonic lineages (Sulston et al., 1983), subsequent studies of other species have revealed a remarkable diversity in the patterns of cell division within nematode embryos. Studies of vulval cell lineages in several species have been critical to our understanding of evolution of developmental mechanisms. As transgenic tools are now being developed in other nematode species, their use in automated analysis is likely to increase; the embryonic cell lineage of C. briggsae has already been followed using automated histone-GFP lineage tracing (Zhao et al., 2008).
III. Resources
The descriptions of cell lineages from the late 1970s remain the definitive descriptions of the cellular anatomy (Table II). In learning the anatomy an important initial goal is to compare one’s own drawings with the diagrams in the following papers. In particular, the description by Sulston et al. (1983) remains the best resource for learning embryonic anatomy; an “embryo” section of WormAtlas is currently under construction.
WormAtlas (www.wormatlas.org) and the C. elegans Atlas book (Hall and Altun, 2008) are invaluable for understanding adult anatomy and for correlating cellular anatomy with electron micrographs. The web site contains a small section on cell identification. A good online guide to cell identification is in Wormbook (Yochem, 2006), with plentiful Nomarski DIC images of “landmark” cells. This is an important addition to the original lineage papers. However, in our experience the only way to successfully learn cell identification is to sit at the microscope and draw what one sees.
IV. Nomenclature and Conventions
The nomenclature for cells was set out by Sulston and Horvitz (1977) and systematized by Sulston et al. (1983). Every cell in C. elegans can be named according to its ancestry, for example, ABpla. Terminally differentiated cells also have “functional” names that are either semiarbitrary (e.g., ASEL) or descriptive of terminal fate (hyp 7). For example, the cell ABalppppppaa is the neuron ASEL.
Embryonic cells are named beginning with one of the five early embryonic “founder cells”: AB, E, MS, C, D. The cells P0 through P4 denote the zygote and the precursors of the germ line, and should not be confused with the postembryonic blast cells P1–P12. Cells that go on to divide in postembryonic stages are renamed with a blast cell name (e.g., ABplapapaaa=QL), and their progeny named according to similar rules.
The suffixes in lineage names refer to the approximate orientation of the cell division relative to the overall axes of the embryo or larva: anterior/posterior, dorsal/ventral, left/right. Almost all cell divisions in C. elegans have a clear anterior–posterior orientation; indeed only ~8 embryonic cell divisions are predominantly in the transverse (left–right) axis. Cells are named according to the relative position of the daughters at the time of division, even if the daughters subsequently change relative position due to cell migration. In some places, such as at the anterior or posterior poles of the early embryo, steric constraints prevent the two daughters from remaining in strict anterior–posterior order, and their final positions are skewed relative to the initial orientation of the spindle.
A very small number of cells have variable ancestry. In several cases, a pair of cells constitutes an “equivalence group” in which each member of the pair can give rise to each fate. This is usually when pairs of cells formed on the left and right sides migrate to the ventral midline to form a single anterior–posterior series. For example, the cell ABplapaapp can become either of two ventral epidermal cells, P1 or P2, depending on whether it migrates to a midline position anterior or posterior to its contralateral homolog ABprapaapp. P1 is therefore denoted ABpl()apaapp. Such fate choices involve an interaction between the members of the cell pair.
V. Cell Identification and Lineage Analysis
A. Embryonic Cell Identification and Lineage Analysis
1. Manual Lineage Analysis
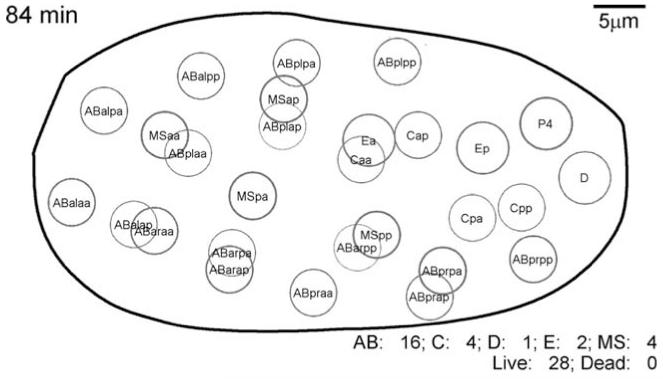
As all early embryonic blastomeres are very similar in morphology, early embryonic cell identification relies on the small number of cells involved and their invariant positions. Cell positions can be easily learnt up to the 28-cell stage (see Fig. 1). Between the 28-cell stage (100 min) and late gastrulation most cells can only be conclusively identified by following their lineage. After gastrulation (~240 min post first cleavage) cellular differentiation begins. Cells can be identified based on position relative to landmarks such as the first apoptotic cell deaths; maps of cell nuclei at 270 nuclei are in Figure 7 of Sulston et al. (1983).
Fig. 1.
Ventral view of wild-type embryo at 84 min/28-cell stage, redrawn from nuclear positions using NucleiTracker4D software (anterior is to left). Eggshell outline is an approximation based on nuclei filling in late embryogenesis. Depth is indicated by the thickness of the outline of each nucleus.
Cell and nuclear identification is more or less straightforward using the available maps up to the 1.75-fold stage when body wall muscle contractions begin. By the twofold stage the rolling activity of the embryo severely hampers attempts to identify cells in live specimens. To prevent this rapid movement the microscope objective can be transiently and reversibly cooled (Sulston et al., 1983). Overall the last 3 h of embryonic development remain relatively little studied.
The cell lineage reported in Sulston et al. (1983) was a composite built up from partial lineages of hundreds of embryos. With the improvement of imaging and storage technology by the late 1980s it became possible to record the complete development of single embryos using timelapse imaging in multiple focal planes. Such “four dimensional” (4D) microscopy was developed by J. G. White and used for studies of early C. elegans embryos (Hird and White, 1993). Several groups developed software for 4D acquisition and playback (Fire, 1994; Thomas et al., 1996). Commercial imaging software suites such as Improvision or Amira include options for automated 4D capture, as do the software suites for most commercial compound microscopes. Finally the Open Source software suite Micro-Manager (http://valelab.ucsf.edu/~MM/MMwiki/index.php/Micro-Manager) can be used for automated 4D acquisition.
Cell-lineage tracing from DIC 4D movies requires manual identification of nuclei at each time point. Software such as SIMI Biocell (Schnabel et al., 1997) (Table III) allows lineages to be generated in a straightforward manner through most of embryogenesis. The image degradation in deeper DIC focal planes limits complete lineaging of single embryos from DIC. Although computer image analysis can identify nuclei in DIC images (Hamahashi et al., 2005) the low contrast of DIC images makes this computationally challenging.
Table III.
Comparison of some methods for manual and semiautomatic embryonic lineage recording
| Software name | Data | Maximum nuclei tracked |
Method | Time required | Error rate | Reference |
|---|---|---|---|---|---|---|
| SIMI Biocell | DIC | 385 cells full/114 cells up to 1.5 fold |
Manual | N/A | N/A | (Schnabel et al., 1997) |
| Angler | DIC | end of gastrulation/ 300 cells |
Manual | N/A | N/A | (Martinelli et al., 1997) |
| N/A | DIC | 24 cells | Automated | N/A | N/A | (Hamahashi et al., 2005) |
| StarryNite | GFP | 350 cells | Automated | 8 h up to 350 cells |
1–3% up to 194 cells, larger up to 350 cells |
(Bao et al., 2006) |
| Endrov | DIC/GFP | 150 cells full/partial later stage |
Manual | N/A | N/A | (Hench et al., 2009) |
| NucleiTracker4D | GFP | 566 live/ 669 total cells |
Semiautomated | For GFP confocal = 4 h up to 350 cells 8–16 h to 525 cells |
Up to 3% to 350 cells, up to 15% to 525 cells (at each time-point). |
CAG & ADC, unpublished |
2. Semiautomated Lineage Analysis
With the advent of histone-GFP reporters marking all embryonic nuclei (Ooi et al., 2006), imaging and lineage analysis of embryo development could now be studied using the laser confocal microscope. The high fluorescence contrast of nuclear-localized GFP allows identification of nuclei by computer image segmentation in a more reliable fashion than from DIC data. Although photobleaching and phototoxicity limit the image quality, it is possible to image histone-GFP throughout embryonic development. The histone-GFP itself is presumably synthesized anew during each cell cycle, partly compensating for photobleaching effects. The Waterston lab implemented the first histone::GFP imaging platform for following embryo development through gastrulation and morphogenesis (Murray et al., 2006). To track moving and dividing nuclei the Starrynite software was developed (Bao et al., 2006). Additional visualization software allows curation of the tracking data and extraction of lineage information (Boyle et al., 2006).
The accuracy of the original Starrynite software declined in embryos with >350 nuclei. To allow nuclear tracking in later embryos several approaches are being tested. One approach is to optimize the segmentation algorithm for images of optically sectioned nuclei (Santella et al., 2010). Another approach, currently implemented in our laboratory (see below), is to curate nuclear identification at each time point, such that the automated tracking at the next time point t+1 starts with corrected information. A second modification to the search algorithm is to constrain the search for a particular nucleus (or its daughters) at t+1 only in the local neighborhood of its previous position at t. Rather than performing de novo segmentation, in this approach the maximum amount of information available at t is used for performing the segmentation and tracking for t+1 (Table III).
3. Extent of Variation in the Wild-Type Lineage
The ability to completely track all nuclei in individual embryos has prompted further examination of the degree of variability in wild-type development. Automated lineage tracing has confirmed the high degree of invariance in the assignment of cell fates, with the known exceptions of the midline cell pairs mentioned above. Some cell-division axes in the C lineage display variability. Cell division times can vary by a factor of 10% between embryos (Sulston et al., 1983), but within individual embryos the relative timing of cell divisions is highly consistent. The high degree of correlation of cell cycles within an individual embryo suggests the existence of a “developmental clock” controlling the rate of embryogenesis; the overall standard deviation in this developmental clock has been estimated at 4.5% (Bao et al., 2008).
Earlier lineage studies suggested that relative cell positions could show variability in midembryogenesis (Schnabel et al., 1997), although such variability decreases by the premorphogenetic stages. More recent studies have suggested that some of this variability may result from the slight compression introduced when an embryo is mounted on an agar pad or between slide and coverslip using beads (Hench et al., 2009). Compressed embryos display two stereotypical rotations. First, during gastrulation, the embryo turns from a left–right aspect to a dorsal–ventral aspect. Then, following epidermal enclosure, the embryo turns once again to display the left–right aspect and its comma shape. The increased variability in cell positions in such compressed embryos may reflect increased migration displacements in the flattened eggshell in conjunction with rotational movements. Unlike compressed embryos, freely mounted embryos attached to polylysine coated coverslips show less variability in cell positions (Hench et al., 2009; Schnabel et al., 2006) and do not display the typical left–right/dorsal–ventral rotations. However, the increased depth of the uncompressed embryo leads to a slight loss of optical quality, and many laboratories continue to use slightly compressed embryos for optimal imaging (see chapter by Hardin).
B. Postembryonic Cell Identification and Lineage Analysis
Cell identification in larval and adult stages is facilitated by the increased separation of nuclei and the differentiation of cell types. However as development proceeds nuclei tend to have slightly less stereotyped positions. Accurate identification of cells and nuclei is also complicated by the tendency of worms to move out of the field of view; at present there is no anesthetic or physical restraint that is compatible with long-term development.
Most cell types are readily identified by position and nuclear morphology. Complex cell groups such as the anterior ganglia require practice and tracing of cell positions from multiple animals. To begin identifying cells it is essential to start with simple easily recognized stages and tissues such as the 12-cell stage of vulval development. A novel approach to identifying new expression patterns is to analyze their intersection with previously characterized patterns using “split GFP” (Zhang et al., 2004).
Paralleling the automated lineaging efforts in the embryo, Long et al. (2009) have recently constructed a 3D atlas of nuclear positions in L1 larvae. Generating a standard 3D representation of the L1 larvae nuclei is instrumental for mapping gene expression patterns or high-throughput computer-controlled functional screens. Atlas building depends on (1) reliable identification of larval nuclei, (2) registration of multiple larval samples into the same standard representation, and (3) mapping of novel samples onto this standard representation. To achieve these goals Long et al. used DAPI stained worms to mark all cell nuclei, followed by several image processing steps. First, the images of larvae are straightened to a rod shape (Peng et al., 2008). Next, nuclei are automatically identified by adaptive thresholding and rule- and training-based segmentation. Nuclei can be validated and curated using the volume-object image annotation system (Peng et al., 2009). In this way, 357 out of the 558 L1 larval nuclei could be faithfully identified.
Using fiduciary muscle nuclei GFP markers, Long et al. registered as many as 40 L1 larvae samples into a 3D standard representation. However, as few as 15 samples were sufficient to infer correct nuclear positions along the body. A comparison of cell positions in the atlas samples along the three axes of the body showed that nucleus-to-nucleus spatial relationships are invariant, especially among cells belonging to the same tissue. After building the standard representation, Long et al. developed an automated procedure for mapping and annotating novel samples, such as expression patterns, onto this reference (Liu et al., 2009). Overall, their automated segmentation can identify nuclei in certain tissues with >80% accuracy. Improving accuracy and the ability to segment all of the 558 nuclei of larvae will entail using higher resolution microscopy methods like selective plane illumination or stimulated emission depletion.
VI. Materials, Methods, and Protocols
A general protocol for mounting C. elegans on agar pads for live analysis is provided in the chapter by Shaham in Worm Methods (http://www.wormbook.org/toc_wormmethods.html); see also the methods appendix to the C. elegans I book.
A. Protocol 1: Analysis of Embryonic Cell Lineages
1. Mounting
Detailed protocols for mounting C. elegans embryos are provided in the chapter by Hardin. Traditionally C. elegans embryos have been mounted on agar pads with buffer and a coverslip. Although embryos are completely viable under such conditions, it is clear that this method compresses the egg and eggshell. An uncompressed egg mounted in an aqueous medium is 50 μm long and 30 μm in diameter (Deppe et al., 1978). Also Ref. Blanchoud et al., 2010; Dev Dyn 239: 3285–96. Embryos mounted on agar pads are compressed to a thickness of ~20 μm (Schnabel et al., 1997). Mounting using spacer beads can also compress the embryo, depending on the bead size used. The lateral compression is helpful in reducing the number of optical sections needed for 4D lineage analysis and constraining the embryo to a fixed orientation for observation. However as mentioned in the text, compression may contribute to the variability in cell positions in early embryogenesis.
2. DIC Cell-Lineage Analysis and 4D Recording
Procedures for manual lineaging of embryos are described by Sulston et al. (1983). A number of software tools have been described over the years to allow time-lapse recording in multiple focal planes (4D recording) (see above). At present most commercial microscope vendors include 4D acquisition as an option. The minimal requirements are microscope equipped for Nomarski DIC optics, a motorized z-drive, a camera, and a computer workstation that controls the z-drive. A high-N.A. DIC objective (e.g., Zeiss Plan-Neofluar 100×) is essential for any lineage studies. Cell lineages can be traced manually from DIC 4D data sets. The software package SIMI Biocell is specifically designed to facilitate lineage construction from DIC 4D movies.
3. 4D Lineaging Using Histone-GFP
Procedures for 4D imaging of histone-GFP marked embryos are defined in Murray et al. (2006). Briefly, single transgenic HIS-72::GFP(zuIs178) embryos are mounted between coverslips in 8 μL of a mixture of 20 μm polystyrene beads (Polysciences Inc., Warrington, PA) in 1% methlycellulose in M9 (15% v/v beads, 85% v/v 1% methylcellulose in M9). The coverslip sandwich can be flipped to display the desired late development dorsal or ventral aspect and then attached to a slide and sealed.
We use a Zeiss LSM510 confocal with a 30 mWArgon laser. We acquire confocal z-stacks of size 64 × 35 × 30 μm3 with resolution of 0.125 × 0.125 × 0.85 μm3/voxel every minute for the first 300 min of development then every 2 min for the next 180 min. Two-color (GFP/mCherry) movies can be acquired to correlate cell-specific expression patterns with the ubiquitously expressed histone-GFP. Laser power, detector, and acquisition configurations are loaded through the MultiTime macro in the Zeiss LSM software.
Precise temperature control is extremely important to maintain embryo viability over prolonged periods of confocal imaging. Although embryos will survive 4D DIC imaging throughout embryogenesis at 25 °C, we find the upper limit for confocal imaging is 24 °C; the viability of imaged embryos should be checked whenever 4D imaging is being set up for the first time. There are several options for control of specimen temperature; we have used a custom-designed aluminum casing for the objective. The casing is cooled or heated by a small Peltier element and a liquid cooling system designed for computer chips.
The analysis of 4D LSM data sets by Starrynite and Acetree is described in detail by (Murray et al., 2006). We provide here a brief overview of our nuclear tracking approach (Giurumescu et al., in preparation). We analyze 4D LSM data sets with a user interface that combines the automated tracking and user-selected curation. At the first time point (usually 4–6 nuclei) the user manually identifies nuclei and names them according to the canonical wild-type lineage. For subsequent time z-stacks, the software first performs an automatic segmentation and tracking step using a minimal movement algorithm and local neighborhood search. Nuclei that do not satisfy the strict minimal movement condition (i.e., those that move less than their radius from t to t + 1, usually less than 5% of nuclei), are flagged for manual curation. Nuclear divisions are also curated manually. At each time point the correct set of nuclei is annotated, preventing the accumulation of annotation errors. Our software does not search for nuclear radii as an additional free parameter. Our initial manual lineaging confirmed the initial observation (Bao et al., 2006) that all nuclei in each of the major sublineages (AB, C, D, E, MS, and P4) show distinct radii values that linearly decrease with each round of division. Hence, our software prescribes nuclear radii values to all nuclei depending on their position in the lineage. Using this semiautomated approach it is possible to lineage essentially all nuclei up to the 1.5-fold stage (566 live nuclei, 103 cell deaths).
B. Protocol 2: Post-Embryonic Cell-Lineage Analysis
Worms to be lineaged must be in healthy, unstarved condition
Prepare a standard slide mount agar pad (cf. Sulston and Hodgkin methods appendix in C. elegans I). The agar should have been freshly prepared or melted.
Using a drawn-out capillary and mouth pipette pick up the worm(s) to be lineaged in a few microliters of M9 or S basal. Deposit the larva onto the agar pad together with a small volume of buffer. [If you are very dextrous, it is possible to do this with a worm pick, but small larvae are very easily injured. We recommend the mouth pipette.] Remove excess buffer by wicking with lens paper.
Using lens paper, wipe clean a small coverslip (18 × 18 mm OK, 12 × 12 mm best but can be hard to find). Using a worm pick, smear a small amount of OP50 E. coli onto the center of the coverslip. Place the coverslip gently over the buffer + worm so that the bacterial blob is within a couple of mm of the worm.
After 1–5 min the worm should become active and head toward the bacteria and start browsing contentedly. Under optimal circumstances, the worm will continue eating for hours, with occasional bouts of movement. If the worm does not move or begin eating within 10 min of mounting, it may be damaged.
If the worm appears healthy, trim the agar around the coverslip with a razor blade. Seal the edges of the coverslip with immersion oil or vaseline. Some brands of immersion oil are toxic and can interfere with long-term observation. Vaseline works fine unless your worm swims into it or you get some on the objective.
Find the area of interest in the animal and draw out everything you see as often as you can, identifying nuclei by reference to the standard maps in the papers in. With practice, multiple animals can be lineaged at a time, depending on the complexity of the lineage being traced. It is usually best to keep one animal per slide to avoid confusion.
1. Troubleshooting
The joy of observing a well-behaving worm is balanced by the frustration of a badly behaved worm that persistently heads for the edge of the agar pad, only to end up in the immersion oil or vaseline. To avoid such frustration it is important to check your worm frequently (every 10 min) and to learn how to recover the worm intact from the slide mount. Practice sliding off the coverslip and getting the worm into a buffer-drop from which you can suck it back into your capillary.
If the worm stops moving and cells lose contrast, the animal may be dying, or it may be entering lethargus, the 1–2 h period of inactivity that precedes each molt. If the developmental stage makes the latter explanation unlikely, there may be too much bacteria under the slide, leading to hypoxia. The worm can be revived by removing it from the slide (slide off the coverslip and use mouth pipette + drawn-out capillary to retrieve the worm). Place the worm on an NGM agar plate to recover for a few minutes, then remount.
The microscope DIC optics should be optimized (Köhler illumination). A heat filter must be used to prevent specimen heating under the prolonged observation. Immersion oil should be used between the objective and coverslip and between slide and condenser top lens. Ensure there are no bubbles or debris in the agar pad; once a worm crawls into a bubble, it will not come out again.
VII. Discussion
Cell-lineage analysis allows rigorous definition of cell ancestries and positions, mutant phenotypes, and gene expression patterns to single-cell resolution. The labor-intensive nature of lineage tracing from live samples has tended to limit its popularity. The recent development of automated lineage analysis promises to reduce the effort needed for early embryonic lineage studies, but lineage tracing in later embryos and in larvae remains a labor of love. Further computational advances may help to return lineage studies to the center of C. elegans developmental biology.
Acknowledgments
The authors thank members of their lab for comments and the C. elegans community for their development and sharing of the techniques mentioned here. Our work on embryonic development has been supported by an NIH NRSA Award to CAG and NIH R01 GM54657 to ADC.
References
- Allen RD, David GB, Nomarski G. The Zeiss-Nomarski differential interference equipment for transmitted-light microscopy. Z Wiss Mikrosk. 1969;69:193–221. [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Bao Z, Murray JI, Boyle T, Ooi SL, Sandel MJ, Waterston RH. Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2707–2712. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Zhao Z, Boyle TJ, Murray JI, Waterston RH. Control of cell cycle timing during C. elegans embryogenesis. Dev. Biol. 2008;318:65–72. doi: 10.1016/j.ydbio.2008.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Avery L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 1995;48:225–250. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle TJ, Bao Z, Murray JI, Araya CL, Waterston RH. AceTree: A tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinform. 2006;7:275. doi: 10.1186/1471-2105-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Delattre M, Felix MA. Polymorphism and evolution of vulval precursor cell lineages within two nematode genera, Caenorhabditis and Oscheius. Curr. Biol. 2001;11:631–643. doi: 10.1016/s0960-9822(01)00202-0. [DOI] [PubMed] [Google Scholar]
- Deppe U, Schierenberg E, Cole T, Krieg C, Schmitt D, Yoder B, von Ehrenstein G. Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 1978;75:376–380. doi: 10.1073/pnas.75.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Flames N, Lee AC, Boyanov A, Hobert O. Automated screening for mutants affecting dopaminergic-neuron specification in C. elegans. Nat. Meth. 2008;5:869–872. doi: 10.1038/nmeth.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. A four-dimensional digital image archiving system for cell lineage tracing and retrospective embryology. Comput. Appl. Biosci. 1994;10:443–447. doi: 10.1093/bioinformatics/10.4.443. [DOI] [PubMed] [Google Scholar]
- Fukuyama M, Gendreau SB, Derry WB, Rothman JH. Essential embryonic roles of the CKI-1 cyclin-dependent kinase inhibitor in cell-cycle exit and morphogenesis in C elegans. Dev. Biol. 2003;260(1):273–286. doi: 10.1016/s0012-1606(03)00239-2. [DOI] [PubMed] [Google Scholar]
- Hall DH, Altun ZF. C. elegans Atlas. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2008. [Google Scholar]
- Hamahashi S, Onami S, Kitano H. Detection of nuclei in 4D Nomarski DIC microscope images of early Caenorhabditis elegans embryos using local image entropy and object tracking. BMC Bioinform. 2005;6:125. doi: 10.1186/1471-2105-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hench J, Henriksson J, Luppert M, Burglin TR. Spatio-temporal reference model of Caenorhabditis elegans embryogenesis with cell contact maps. Dev. Biol. 2009;333:1–13. doi: 10.1016/j.ydbio.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Herman MA, Horvitz HR. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development. 1994;120:1035–1047. doi: 10.1242/dev.120.5.1035. [DOI] [PubMed] [Google Scholar]
- Hird SN, White JG. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Joy of the worm. Genetics. 1990;126:287–292. doi: 10.1093/genetics/126.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthoofd W, Borgonie G. The embryonic cell lineage of the nematode Halicephalobus gingivalis (Nematoda: Cephalobina: Panagrolaimoidea) Nematology. 2007;9:573–584. [Google Scholar]
- Houthoofd W, Jacobsen K, Mertens C, Vangestel S, Coomans A, Borgonie G. Embryonic cell lineage of the marine nematode Pellioditis marina. Dev. Biol. 2003;258:57–69. doi: 10.1016/s0012-1606(03)00101-5. [DOI] [PubMed] [Google Scholar]
- Houthoofd W, Willems M, Jacobsen K, Coomans A, Borgonie G. The embryonic cell lineage of the nematode Rhabditophanes sp. Int. J. Dev. Biol. 2008;52:963–967. doi: 10.1387/ijdb.072404wh. [DOI] [PubMed] [Google Scholar]
- Kanamori T, Inoue T, Sakamoto T, Gengyo-Ando K, Tsujimoto M, Mitani S, Sawa H, Aoki J, Arai H. Beta-catenin asymmetry is regulated by PLA1 and retrograde traffic in C. elegans stem cell divisions. EMBO J. 2008;27:1647–1657. doi: 10.1038/emboj.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kostić I, Roy R. Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans. Development. 2002;129(9):2155–2165. doi: 10.1242/dev.129.9.2155. [DOI] [PubMed] [Google Scholar]
- Lahl V, Schulze J, Schierenberg E. Differences in embryonic pattern formation between Caenorhabditis elegans and its close parthenogenetic relative Diploscapter coronatus. Int. J. Dev. Biol. 2009;53:507–515. doi: 10.1387/ijdb.082718vl. [DOI] [PubMed] [Google Scholar]
- Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sanchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, Myers EW, Kim SK. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell. 2009;139:623–633. doi: 10.1016/j.cell.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Peng H, Liu X, Kim SK, Myers E. A 3D digital atlas of C. elegans and its application to single-cell analyses. Nat. Meth. 2009;6:667–672. doi: 10.1038/nmeth.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli SD, Brown CG, Durbin R. Gene expression and development databases for C. elegans. Semin. Cell Dev. Biol. 1997;8:459–467. doi: 10.1006/scdb.1997.0171. [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Waterston RH. The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat. Protoc. 2006;1:1468–1476. doi: 10.1038/nprot.2006.222. [DOI] [PubMed] [Google Scholar]
- Nimmo R, Antebi A, Woollard A. mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development. 2005;132(22):5043–5054. [Google Scholar]
- Ooi SL, Priess JR, Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2006;2:e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padawer J. The Nomarski interference-contrast microscope. An experimental basis for image interpretation. J. R. Microsc. Soc. 1968;88:305–349. doi: 10.1111/j.1365-2818.1968.tb00616.x. [DOI] [PubMed] [Google Scholar]
- Peng H, Long F, Liu X, Kim SK, Myers EW. Straightening Caenorhabditis elegans images. Bioinformatics. 2008;24:234–242. doi: 10.1093/bioinformatics/btm569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Long F, Myers EW. VANO: A volume-object image annotation system. Bioinformatics. 2009;25:695–697. doi: 10.1093/bioinformatics/btp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella A, Du Z, Nowotschin S, Hadjantonakis AK, Bao Z. A hybrid blob-slice model for accurate and efficient detection of fluorescence labeled nuclei in 3D. BMC Bioinform. 2010;11:580. doi: 10.1186/1471-2105-11-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R, Bischoff M, Hintze A, Schulz AK, Hejnol A, Meinhardt H, Hutter H. Global cell sorting in the C. elegans embryo defines a new mechanism for pattern formation. Dev. Biol. 2006;294:418–431. doi: 10.1016/j.ydbio.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Schnabel R, Hutter H, Moerman D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: Variability of development and regional specification. Dev. Biol. 1997;184:234–265. doi: 10.1006/dbio.1997.8509. [DOI] [PubMed] [Google Scholar]
- Schulze J, Schierenberg E. Cellular pattern formation, establishment of polarity and segregation of colored cytoplasm in embryos of the nematode Romanomermis culicivorax. Dev. Biol. 2008;315:426–436. doi: 10.1016/j.ydbio.2007.12.043. [DOI] [PubMed] [Google Scholar]
- Schulze J, Schierenberg E. Embryogenesis of Romanomermis culicivorax: An alternative way to construct a nematode. Dev. Biol. 2009;334:10–21. doi: 10.1016/j.ydbio.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Sommer RJ. Evolutionary changes of developmental mechanisms in the absence of cell lineage alterations during vulva formation in the Diplogastridae (Nematoda) Development. 1997;124:243–251. doi: 10.1242/dev.124.1.243. [DOI] [PubMed] [Google Scholar]
- Sommer RJ, Carta LK, Sternberg PW. The evolution of cell lineage in nematodes. Dev. Suppl. 1994:85–95. [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. Gonadal cell lineages of the nematode Panagrellus redivivus and implications for evolution by the modification of cell lineage. Dev. Biol. 1981;88:147–166. doi: 10.1016/0012-1606(81)90226-8. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. Postembryonic nongonadal cell lineages of the nematode Panagrellus redivivus: Description and comparison with those of Caenorhabditis elegans. Dev. Biol. 1982;93:181–205. doi: 10.1016/0012-1606(82)90251-2. [DOI] [PubMed] [Google Scholar]
- Sulston JE. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976;275:287–297. doi: 10.1098/rstb.1976.0084. [DOI] [PubMed] [Google Scholar]
- Sulston JE. Caenorhabditis elegans: The cell lineage and beyond (Nobel lecture) Chembiochem. 2003;4:688–696. doi: 10.1002/cbic.200300577. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: Postembryonic development of nongonadal structures. Dev. Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sulston JE, White JG. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- Thomas C, DeVries P, Hardin J, White J. Four-dimensional imaging: Computer visualization of 3D movements in living specimens. Science. 1996;273:603–607. doi: 10.1126/science.273.5275.603. [DOI] [PubMed] [Google Scholar]
- van Cleave HJ. Eutely or cell constancy in its relation to body size. Q. Rev. Biol. 1932;7:59–67. [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wiegner O, Schierenberg E. Specification of gut cell fate differs significantly between the nematodes Acrobeloides nanus and Caenorhabditis elegans. Dev. Biol. 1998;204:3–14. doi: 10.1006/dbio.1998.9054. [DOI] [PubMed] [Google Scholar]
- Yochem J. WormBook. The C. elegans Research Community, WormBook; Jan 24, 2006. Nomarski images for learning the anatomy, with tips for mosaic analysis.http://www.wormbook.org doi/10.1895/wormbook.1.100.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Ma C, Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Boyle TJ, Bao Z, Murray JI, Mericle B, Waterston RH. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev. Biol. 2008;314:93–99. doi: 10.1016/j.ydbio.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]