Abstract
PKD-mediated phosphorylation of class IIa HDACs frees the MEF2 transcription factor to activate genes that govern muscle differentiation and growth. Studies of the regulation and function of this signaling axis have involved MC1568 and Gö-6976, which are small molecule inhibitors of class IIa HDAC and PKD catalytic activity, respectively. We describe unanticipated effects of these compounds. MC1568 failed to inhibit class IIa HDAC catalytic activity in vitro, and exerted divergent effects on skeletal muscle differentiation compared to a bona fide inhibitor of these HDACs. In cardiomyocytes, Gö-6976 triggered calcium signaling and activated stress-inducible kinases. Based on these findings, caution is warranted when employing MC1568 and Gö-6976 as pharmacological tool compounds to assess functions of class IIa HDACs and PKD.
Keywords: class IIa histone deacetylase, protein kinase D, small molecule inhibitors, muscle
1. Introduction
The 18 mammalian histone deacetylases (HDACs) are grouped into four classes (I, II, III and IV) [1]. Class II HDACs are further divided into two subclasses, IIa (HDACs 4, 5, 7 and 9) and IIb (HDACs 6 and 10). Class IIa HDACs serve critical roles in the control of muscle gene expression [2]. These HDACs have long (~500 amino acid) amino-terminal extensions that harbor binding sites for pro-myogenic transcription factors such as myocyte enhancer factor 2 (MEF2) [3–7]. In skeletal muscle cells, binding of class IIa HDACs to MEF2 results in suppression of MEF2 target genes that govern differentiation of myoblasts into multi-nucleated myotubes [8;9]. In cardiac muscle, interaction of class II HDACs with MEF2 blocks hypertrophic growth of the heart [10].
The amino-terminal extensions of class IIa HDACs also harbor two serine residues that confer signal-responsiveness to the enzymes. These sites are phosphorylated by members of the calcium/calmodulin-dependent kinase (CaM kinase) superfamily, which includes CaM kinase II and protein kinase D (PKD) [11–14]. Phosphorylation of class IIa HDACs creates docking sites for the 14-3-3 chaperone protein, which promotes nuclear export of the transcriptional repressors. Thus, in response to CaMK/PKD signaling, class IIa HDACs dissociate from MEF2, freeing the transcription factor to activate downstream target genes. The identification of PKD as a class IIa HDAC kinase was aided by the use of a pharmacological tool compound, Gö-6976 [15], which functions as an ATP-competitive inhibitor of the three PKD isoforms, PKD-1, -2 and -3 [16].
Despite having conserved catalytic domains, class IIa HDACs are unable to deacetylate histones [17]. In fact, bona fide substrates of class IIa HDACs have yet to be identified, and class IIa HDAC catalytic activity can only be monitored using an artificial substrate [18]. Class IIa HDAC catalytic activity is largely resistant to commonly used HDAC inhibitors, such as suberanilohydroxamic acid (SAHA) [19]. Several years ago, MC1568 (originally referred to as compound 2f) was described as the first class IIa HDAC-selective inhibitor [20]. MC1568 has been used in a multitude of studies as a chemical biological probe to assess the function of class IIa HDAC catalytic activity in various physiological and pathophysiological processes. For example, MC1568 was recently found to block pulmonary artery endothelial cell proliferation and rescue experimental pulmonary hypertension [21], as well as block hepatic stellate cell activation and liver fibrosis in mice [22]. Paradoxically, MC1568 was shown to block skeletal muscle differentiation by stabilizing MEF2:HDAC4 complexes and preventing MEF2 acetylation [23].
In the current studies, we confirm that MC1568 blocks differentiation of C2C12 myoblasts into multinucleate myotubes. However, the anti-myogenic activity of this compound appears to be due to off-target action(s), since enzymatic assays revealed an inability of MC1568 to inhibit class IIa HDAC catalytic activity. Furthermore, we validate that Gö-6976 is a PKD inhibitor. Surprisingly, however, exposure of primary cardiomyocytes to Gö-6976 leads to enhanced calcium signaling and activation of stress-inducible kinases such as p38 mitogen-activated protein kinase and c-Jun N-terminal kinase (JNK). These findings highlight the need to employ multiple, structurally distinct compounds when assessing the functions of class IIa HDACs and PKD using pharmacological tools.
2. Materials and methods
2.1. Inhibitors, antibodies and adenoviruses
Gö-6976 was obtained from EMD Millipore and A.G. Scientific. MC1568 was purchased from both Sigma-Aldrich and Selleck Chemicals. SAHA, MGCD0103 and Tubastatin A were purchased from Selleck Chemicals, whereas diphenylacetohydroxamic acid (DPAH) [24] was obtained from Sigma-Aldrich (D6071). DPAH was also synthesized in-house, and its purity was confirmed to be greater than 95%. U-73122 was procured from Tocris Bioscience. For all studies, DMSO was used as the compound vehicle (final DMSO concentration = 0.1%). For immunoblotting, cells were homogenized in PBS (pH 7.4) containing 0.5% Triton X-100, 300 mM NaCl and protease/phosphatase inhibitor cocktail (Thermo Fisher) using a Bullet Blender homogenizer (Next Advance). The following antibodies were used for immunoblotting: anti-calnexin (Santa Cruz Biotechnology; sc-11397), anti-PKD1 (Cell Signaling Technology; #2052), anti-phospho-PKD1 Ser-738/742 (Cell Signaling Technology; #2054), anti-phospho-PKD1 Ser-910 (Cell Signaling Technology; #2051), anti-phospho-ERK1/2 (Cell Signaling Technology; 4370), anti-phospho-p38 (Cell Signaling Technology; 4631) and anti-phospho-JNK (Cell Signaling Technology; 4668). Adenoviruses for wild-type PKD1 and catalytically inactive PKD1 harboring a K612W substitution (K/W) [25] were generated using pShuttle-CMV, as previously described [26].
2.2. Primary cardiac myocyte preparation
Neonatal rat ventricular myocytes (NRVMs) were prepared from hearts of 1- to 3-day-old Sprague–Dawley rats, as previously described [27]. Cells were cultured overnight on 10-cm plates coated with gelatin (0.2%; Sigma-Aldrich) in Dulbecco’s Modified Eagle’s Medium (DMEM) containing calf serum (10%), L-glutamine (2 mM), and penicillin–streptomycin. After overnight culture, cells were washed with serum-free medium and maintained in DMEM supplemented with L-glutamine, penicillin–streptomycin, and Neutridoma-SP (0.1%; Roche Applied Science), which contains albumin, insulin, transferrin, and other defined organic and inorganic compounds. Adult rat ventricular myocytes (ARVMs) were obtained from female Sprague–Dawley rats, as described previously [28]. ARVMs were plated at a density of 100–150 cells/ mm2 on laminin-coated 60-mm plates and maintained in serum-free DMEM supplemented with albumin (2 mg/mL), 2,3-butanedione monoxime (1 mg/mL), L-carnitine (2 mM), creatine (5 mM), penicillin–streptomycin (100 μg/mL), triiodothyronine (1 pM), and taurine (5 mM).
2.3. C2C12 culture and transfection
C2C12 myoblasts were obtained from ATCC and were maintained in DMEM containing 10% FBS, L-glutamine (2 mM), and penicillin–streptomycin. C2C12 cells were induced to differentiate by replacing FBS in the culture medium with 2% horse serum. C2C12 cells were transfected with a 3× MEF2-luciferase plasmid, which contains three tandem copies of the mouse muscle creatine kinase enhancer MEF2 site upstream of the thymidine kinase promoter [29], using FuGENE 6 (Promega). To control for non-specific effects of compounds on luciferase expression, parallel plates of C2C12 cells were transfected with a TATA-luciferase plasmid lacking MEF2 binding sites. Luciferase activity is expressed as a ratio of MEF2-luciferase:TATA-luciferase.
2.4. In vitro kinase assays
NRVMs on 10-cm plates (2 × 106 cells/plate) were serum starved for 4 hours and subsequently exposed to phenylephrine (10 μM) for 1 hour. Following treatment, total protein lysates were prepared, as described above for immunoblotting. Total protein (200 μg) was incubated in buffer (500 μl) containing antibodies specific for PKD1 (Santa Cruz Biotechnology; sc-639), PKD2 (Bethyl Laboratories; BL844) or PKD3 (Bethyl Laboratories; BL1282), overnight at 4°C with rocking. Antibody-protein complexes were captured with Protein G-Sepharose beads (GE Healthcare Life Sciences), with incubation for 1 hour at 4°C. Immunoprecipitates were equilibrated with kinase buffer (20 mM HEPES, pH 7.5, 20 mM MgCl2), and kinase reactions were initiated upon addition of ATP (0.1 mM), 10 μCi [γ-32P]-ATP, and 2 μg Syntide-2 substrate (Anaspec). Kinase reactions were carried out at 30°C for 30 minutes, and terminated with 2× SDS-PAGE loading dye. Phospho-Syntide-2 peptide was resolved by SDS-PAGE and visualized by autoradiography.
2.5. Calcium measurements
NRVMs were loaded with 5 μM Fluo 3-AM and 0.05% pluronic acid (both from Invitrogen) dissolved in Tyrode’s solution (in mM: 140 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES, 5 Glucose, pH adjusted to 7.4 with NaOH) for twenty minutes. NRVMs were then washed three times in Tyrode’s solution with gentle agitation. After a ten minute de-esterification period, Fluo-3 AM-loaded cells, bathed in Tyrode’s solution, were placed on the stage of an LSM 510 META scanning laser confocal microscope, and viewed with a Zeiss Plan-Neofluar 40× objective (Zeiss). Fluo-3 AM was excited with the 488-nm line of an argon laser (30-milliwatt maximum output, operated at 50% or 6.3A, attenuated to 5%). The emitted fluorescence was directed through a dual 488/543 dichroic mirror to a photomultiplier equipped with a 505 nm long-pass filter. Caffeine (Sigma-Aldrich) or Gö-6976 (Sigma-Aldrich) were diluted to working concentrations (5 mM and 1 μM, respectively) in Tyrode’s solution, and were delivered via a manually-operated, gravity-driven global perfusion system. Spontaneous Ca2+ fluctuations were analyzed qualitatively using NIH Image J software. Fluorescence amplitude data are expressed as ΔF/F, where F represents baseline fluorescence prior to application of caffeine or Gö-6976, and ΔF represents the change in peak fluorescence during the application of either compound. All Ca2+ imaging experiments were performed at room temperature (~25°C) in the University of Colorado Denver Advanced Light Microscopy Core.
2.6. Immunostaining
Following experimental treatments, NRVMs were washed twice in PBS (pH 7.4), fixed with formalin (10%) in PBS, permeabilized and blocked with PBS containing NP-40 (0.1%) and bovine serum albumin (BSA; 3%), and then incubated in the same solution containing primary antibody specific for PKD1 phosphorylated on serine-916 (1:1,000 dilution; Cell Signaling Technology; #2051) overnight at 4°C. Cells were washed four times in PBS and incubated in PBS-NP-40-BSA containing a Cy3-conjugated secondary antibody (1:200 dilution; Jackson Laboratories) for 30 minutes at room temperature. Cells were washed four times in PBS and then covered with mounting solution (SlowFade; Molecular Probes) and glass coverslips. For myosin staining of skeletal muscle cells, immunohistochemical analysis was performed using an anti-sarcomeric myosin (MF20; Iowa Hybridoma Bank) primary antibody, a secondary antibody (biotinylated anti-mouse IgG; Vector Labs), and Elite ABC and VECTOR NovaRED Substrate Kits (Vector Labs). The percentage of the plate containing myosin-positive myotubes was quantified in a blinded manner using an Axiovert 200 inverted microscope with a digital camera equipped with AxioVision imaging software (Zeiss). Values were averaged from five independent plates of cells per condition.
2.7. HDAC activity assays
HDAC activity assays were completed as previously reported [30]. Each HDAC substrate was based on ε-N-acylated lysine, derivatized on the carboxyl group with amino-methylcoumarin (AMC) [18]. Subsequent to deacylation by HDACs, trypsin was used to release AMC, resulting in a significant increase in fluorescence. Rat heart homogenates were employed as a source of HDAC activity. Homogenates were prepared in PBS (pH 7.4) containing 0.5% Triton X-100, 300 mM NaCl and protease/phosphatase inhibitor cocktail (Thermo Fisher), using a Bullet Blender homogenizer (Next Advance). Tissue extracts were diluted into PBS in 100 μl total volume in a 96-well plate (60 μg ventricular protein/well). Tissue extracts were incubated with increasing concentrations of HDAC inhibitors for 1hr. Substrates were added (5 μl of 1 mM DMSO stock solutions), and the plates were placed at 37 °C for 2–3 hours. Finally, developer/stop solution was added (50 μl per well of PBS with 1.5% Triton X-100, 3 μM TSA, and 0.75 mg/ml trypsin), followed by an additional 20 minute incubation at 37 °C. AMC fluorescence was measured using a BioTek Synergy 2 plate reader, with excitation and emission filters of 360 nm and 460 nm, respectively (each with 40 nm bandwidth), along with a 400 nm dichroic top mirror. As a control, HDAC inhibitors were incubated with AMC without the appended substrate. The importance of this step is illustrated by the fact that MC1568 quenches AMC-generated fluorescence (sFig. 1). Background signals from buffer blanks were subtracted, and the data were normalized to appropriate controls. To monitor recombinant HDAC4 activity, 15ng of recombinant human HDAC4 (BPS Bioscience; #50004) or PBS blank was added to a 96-well plate. Wells were incubated for 1hr with increasing concentrations of DPAH or MC1568 (Sigma-Aldrich), prior to addition of the cell-permeable class IIa fluorescent HDAC substrate. After two hours, stop buffer was added to each well and incubated for 20 min at 37 °C, and deacetylase activity was quantified as described above.
3. Results
3.1 Differential effects of class IIa HDAC inhibitors on skeletal muscle differentiation
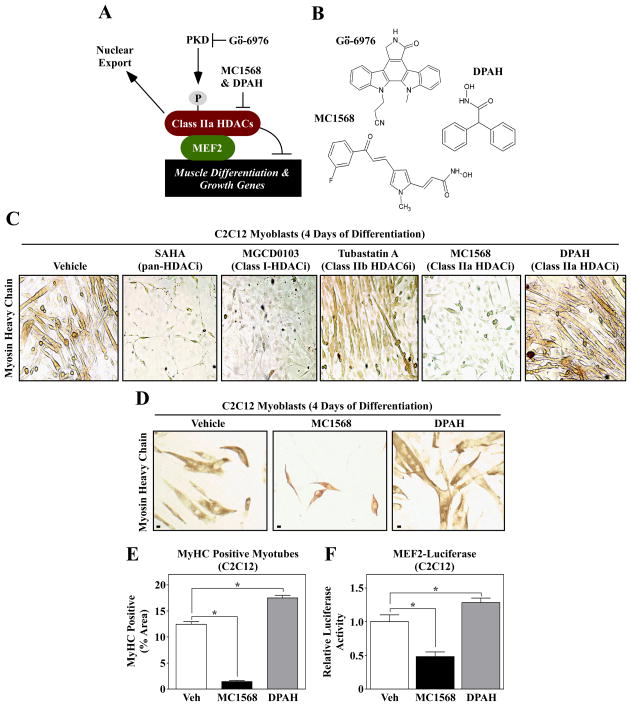
Association of MEF2 with class IIa HDACs leads to repression of downstream target genes that promote skeletal muscle differentiation (Fig. 1A). When class IIa HDACs are phosphorylated by kinases such as PKD, these HDACs are exported to the cytoplasm, and MEF2 becomes transcriptionally active. Gö-6976 inhibits the catalytic activity of PKD, while MC1568 and DPAH are small molecule inhibitors of class IIa HDAC catalytic activity (Fig. 1A and B).
Fig. 1.
Differential effects of isoform-selective HDAC inhibitors on skeletal muscle differentation. (A) A schematic representation of the PKD-class IIa HDAC axis. Gö-6976 blocks the catalytic activity of PKD, while MC1568 and DPAH inhibit the enzymatic activity of class IIa HDACs. (B) The molecular structures of the compounds used in this study are shown. (C) C2C12 myoblasts were induced to differentiate in the absence (vehicle) or presence of the indicated HDAC inhibitors; SAHA (1 μM), MGCD0103 (500 nM), Tubastatin A (1 μM), MC1568 (10 μM) and DPAH (10 μM). After four days of differentiation, cells were fixed and stained with an anti-myosin antibody to reveal myosin heavy chain (MyHC)-positive skeletal myotubes. (D) C2C12 cells were induced to differentiate in the absence or presence of the class IIa HDAC inhibitors MC1568 and DPAH, stained as described for (C), and differentiation was quantified (E); Scale bar = 10 μm. (F) C2C12 myoblasts were transfected with a MEF2-dependent luciferase reporter gene, and the cells were subsequently induced to differentiate for three days in the absence or presence of MC1568 or DPAH. For (E) and (F), five independent plates of cells were analyzed per condition. Numbers represent averages +SEM; *P < 0.05 vs. vehicle-treated cells.
To assess the impact of isoform-selective HDAC inhibition on skeletal muscle differentiation, C2C12 myoblasts were differentiated in the absence or presence SAHA (pan-HDAC inhibitor), MGCD0103 (class I HDAC inhibitor), Tubastatin A (HDAC6 inhibitor), MC1568 or DPAH. Pre-treatment with SAHA and MGCD0103 led to complete inhibition of C2C12 cell differentiation, as indicated by the absence of myosin-positive myotubes in these cultures (Fig. 1C). Consistent with prior findings, HDAC6 inhibition with Tubastatin A moderately impaired myogenesis [31], while the class IIa HDAC inhibitor MC1568 completely abolished formation of multinucleated myotubes [23]. Surprisingly, however, the structurally distinct class IIa HDAC inhibitor, DPAH, failed to block differentiation of C2C12 cells. Higher magnification images of cells from a second experiment with MC1568 and DPAH revealed that DPAH actually enhanced C2C12 myotube formation (Fig. 1D and E). The differential actions of the two compounds on muscle differentiation correlated with their effects on MEF2 transcriptional activity (Fig. 1F), with MC1568 and DPAH repressing and stimulating a MEF2-dependent reporter gene, respectively.
3.2. Commercially available MC1568 does not inhibit class IIa HDAC catalytic activity
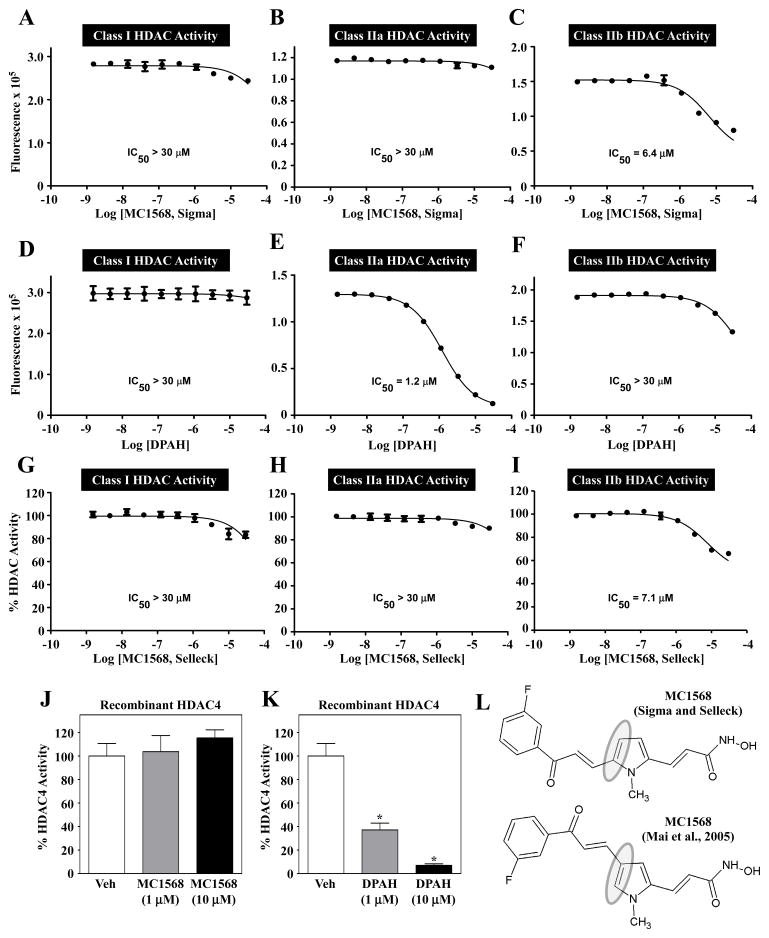
To begin to address the discrepant results with MC1568 and DPAH, the compounds were tested for their ability to block HDAC catalytic activity in vitro. Dose-response studies were performed with substrates that enable specific assessment of class I, IIa or IIb HDAC activity. Rat left ventricular homogenates were used as a rich source of endogenous HDACs [30]. Consistent with prior findings [20], MC1568 had no observable effect on class I HDAC activity (Fig. 2A). Surprisingly, however, MC1568 also failed to inhibit class IIa HDAC activity and, instead, modestly suppressed class IIb HDAC catalytic activity (Fig. 2B and C); HDAC6 is the predominant class IIb HDAC in these lysates [30]. In contrast, DPAH dose-dependently inhibited class IIa HDACs (IC50 = 1.2 μM), while having no effect on class I or IIb HDACs (Fig. 2D – F). Similar results were obtained using independent sources of MC1568 and DPAH (Fig. 2G–I and data not shown). To confirm these findings, recombinant HDAC4 was used as a representative class IIa HDAC. Consistent with the results obtained using endogenous HDACs, MC1568 failed to block the activity of recombinant HDAC4 (Fig. 2J), while DPAH efficiently inhibited this class IIa HDAC (Fig. 2K). Quantification of class IIa HDAC activity was enabled by the use of a substrate having a trifluoroacetyl group on the ε-amine, which is deacetylated by class IIa HDACs but not class I or IIb HDACs [32].
Fig. 2.
MC1568 does not block class IIa HDAC catalytic activity. Dose-response studies were performed to assess the ability of MC1568 and DPAH to inhibit HDAC catalytic activity in vitro. (A – C) MC1568 (from Sigma-Aldrich) failed to inhibit class I and class IIa HDAC activity, but did block class IIb activity at higher concentrations. (D – F) DPAH dose-dependently, and selectively, inhibited class IIa HDAC catalytic activity. MC1568 from Selleck Chemicals also failed to inhibit endogenous class IIa HDAC activity (G – I). MC1568 was unable to inhibit the catalytic activity of a recombinant form of a prototypical class IIa HDAC, HDAC4, while DPAH efficiently inhibited recombinant HDAC4 (J and K). (L) Analysis of NMR data from Sigma and Selleck revealed the commercial MC1568 to be a 2,5-disubstituted pyrrole, as opposed to the 2,4-disubstituted isomer shown in Fig. 1B.
The mechanism underlying the inability of MC1568 to inhibit class IIa HDACs could be related to compound structure. Indeed, nuclear magnetic resonance (NMR) data revealed the commercial compound to be an isomer of the previously published structure (Fig. 2L).
3.3. Gö-6976 activates stress-inducible kinases in cardiac myocytes
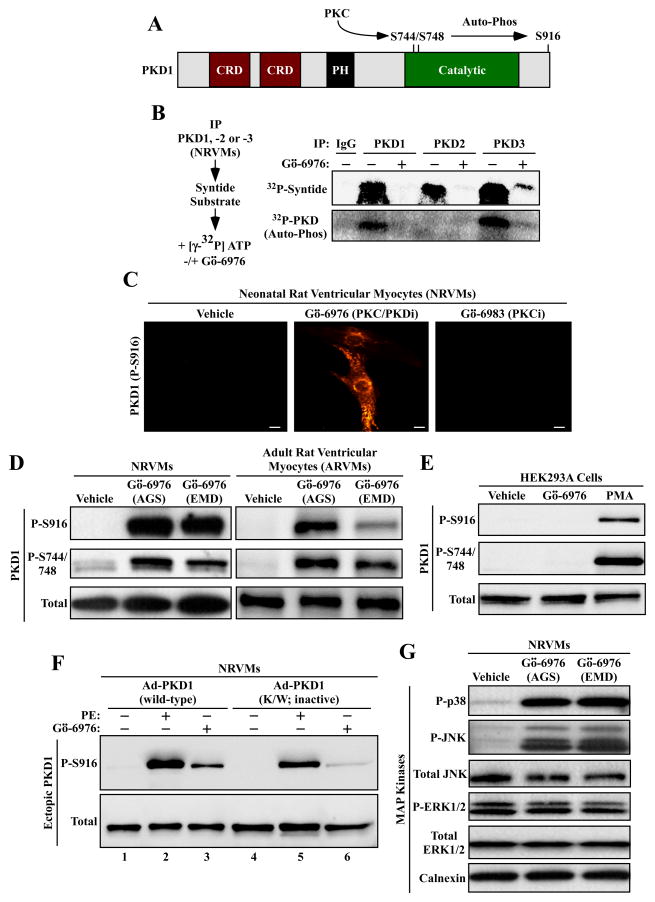
Among the downstream effectors of Gαq-coupled receptors in cardiac myocytes are members of the PKD family [26;33]. The three PKD isoforms exhibit a high degree of sequence similarity, consisting of 900 amino acids, with two amino-terminal cysteine-rich domains (CRDs), an internal pleckstrin homology domain (PH), and carboxy-terminal catalytic domains [34;35]. PKD can be activated by protein kinase C (PKC)-mediated phosphorylation of activation loop sites within the catalytic domain, with subsequent auto-phosphorylation of PKD on serine-916 (Fig. 3A).
Fig. 3.
Gö-6976 activates stress-dependent kinases in cardiomyocytes. (A) Schematic representation of PKD1, with cysteine-rich domains (CRD) and a pleckstrin homology (PH) domain indicated. PKC phosphorylates activation loop sites in the catalytic domain of PKD1 (serines-744 and -748), resulting in stimulation of the kinase. Serine-916 can undergo auto-phosphorylation. (B) NRVMs were stimulated with phenylephrine (PE; 10 μM) for 1 hour. Whole cell lysates were immunoprecipitated (IP) with antibodies to PKD1, -2 or -3 and incorporated into in vitro kinase reactions in the absence or presence of Gö-6976 (10 μM). Gö-6976 inhibited the ability of each PKD isoform to phosphorylate the synthetic syntide substrate, and blocked auto-phosphorylation of PKD1 and PKD3. (C) NRVMs were treated with Gö-6976 (10 μM) or Gö-6983 (10 μM) for 1 hour, and PKD serine-916 phosphorylation was assessed by indirect immunofluorescence. Scale bar = 10 μm. (D) NRVMs (left panel) or adult rat ventricular myocytes (right panel) were treated for 1 hour with vehicle control or Gö-6976 (10 μM) from two independent commercial sources, as described in the Materials and methods. Protein homogenates were immunoblotted with antibodies to the indicated forms of PKD1. (E) HEK293A fibroblasts were treated for 1 hour with Gö-6976 (10 μM) or phorbol-12-myristate-13-acetate (PMA; 50 nM), which served as a positive control for PKC and PKD activation. Immunblotting was performed as in (D). (F) NRVMs were infected with adenoviruses encoding wild-type PKD1 or PKD1 K/W, which is catalytically inactive. Cells were treated for 1 hour with PE (10 μM) or Gö-6976 (10 μM), lysed, and immunoblotted with the indicated antibodies. (G) NRVMs were treated for 1 hour with Gö-6976 (10 μM) from two different vendors, and protein homogenates were immunoblotted with antibodies to the indicated proteins.
PKD1, -2 and -3 were immunoprecipitated from NRVMs that had been stimulated with the α1-adrenergic receptor agonist phenylephrine (PE), which potently activates PKD in cardiac myocytes. Immunoprecipitates were incorporated into in vitro kinase reactions containing syntide-2 substrate and [γ-32P]-ATP, in the absence or presence of Gö-6976. As shown in Fig. 3B, Gö-6976 effectively inhibited the ability of PKD1, -2 and -3 to phosphorylate the synthetic substrate, and also blocked auto-phosphorylation of PKD1 and PKD3; PKD2 auto-phosphorylation was not detected in the assay.
Next, the impact of Gö-6976 on auto-phosphorylation of PKD1 on serine-916 in intact, unstimulated cardiomyocytes was assessed. Paradoxically, treatment of NRVMs with this compound led to enhanced serine-916 phosphorylation, while a related compound, Gö-6983, which targets PKC but not PKD, had no effect on PKD1 phosphorylation (Fig. 3C). Stimulation of PKD1 serine-916 phosphorylation by Gö-6976 in NRVMs was confirmed by immunoblotting, and was shown to occur with compounds obtained from two independent vendors (Fig. 3D; left-hand panels). Gö-6976 similarly promoted phosphorylation of the activation loop sites on PKD1 (serines-744 and -748) in NRVMs. Furthermore, Gö-6976 was also able to increase PKD1 phosphorylation in adult rat ventricular myocytes (ARVMs) (Fig. 3D; right-hand panels), but not in HEK293 fibroblasts (Fig. 3E), suggesting cell type-specificity of the effects.
Stimulation of PKD1 serine-916 phosphorylation by Gö-6976 is paradoxical, since serine-916 is thought to be an auto-phosphorylation site, and Gö-6976 is an ATP-competitive inhibitor of PKD [16]. To determine if Gö-6976 triggers PKD auto-phosphorylation, experiments were performed with ectopic PKD1 expressed in NRVMs via adenoviruses. Consistent with the results with endogenous PKD1, ectopically expressed wild-type PKD1 was efficiently phosphorylated on serine-916 upon treatment with PE or Gö-6976 (Fig. 3F). When normalized to total PKD1 expression, serine-916 phosphorylation of catalytically inactive PKD1 (K/W) was attenuated in PE-treated cells (Fig. 3F, lane 5), consistent with α1-adrenergic receptor signaling causing PKD1 auto-phosphorylation [26;33]. Remarkably, however, serine-916 was still phosphorylated following treatment with Gö-6976 (Fig. 3F, lane 6), suggesting that the compound stimulates PKD1 serine-916 phosphorylation through a PKD-independent mechanism governed by a distinct kinase(s). This explains how Gö-6976 can inhibit PKD catalytic activity (Fig. 3B), and still trigger PKD Ser-916 phosphorylation. Additional immunoblotting studies revealed that Gö-6976 also stimulated phosphorylation of the stress-inducible MAP kinases p38 and JNK in cardiomyocytes, but not extracellular signal-regulated kinase (ERK1/2) (Fig. 3G).
3.4. Gö-6976 alters calcium signaling in cardiac myocytes
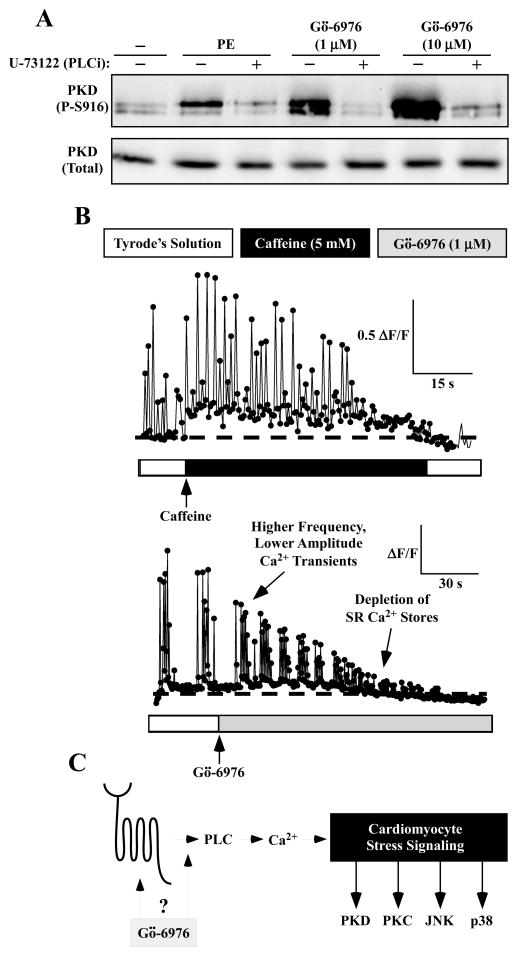
Given the ability of Gö-6976 to activate multiple kinases (PKC, p38 and JNK) in cultured cardiomyocytes, experiments were next performed to address the possibility that the compound functions proximally, via a general signaling mediator(s), to stimulate diverse downstream events. Consistent with this notion, the phospholipase C (PLC) inhibitor U-73122 blocked Gö-6976-mediated PKD phosphorylation as efficiently as it blocked PE-induced phosphorylation of the kinase (Fig. 4A). Subsequently, effects of Gö-6976 and caffeine on calcium signaling in cardiomyocytes were assessed by imaging cells that were loaded with a calcium-sensitive dye, Fluo-3 AM. Spontaneous calcium transients were detected in NRVMs prior to application of either caffeine or Gö-6976 (Fig. 4B). Caffeine treatment of NRVMs led to induction of sarcoplasmic reticulum (SR) calcium release (Fig. 4B, top panel). Strikingly, Gö-6976 also triggered myoplasmic calcium fluctuations within seconds after addition to NRVMs (Fig. 4B, bottom panel). Effects of caffeine and Gö-6976 on NRVM calcium were distinct, with Gö-6976-induced transients having lesser amplitude than spontaneous transients. Furthermore, Gö-6976 treatment caused a rapid increase in myoplasmic calcium concentration, which ultimately led to depletion of SR calcium stores.
Fig. 4.
Gö-6976 alters calcium signaling in cardiac myocytes. (A) NRVMs were pre-treated with the phospholipase C (PLC) inhibitor U-73122 (1 μM) for 20 minutes prior to addition of phenylephrine (PE; 10 μM) or Gö-6976 (1 or 10 μM) for 1 hour. Immunoblotting was performed to assess levels of phospho- and total PKD1. (B) NRVMs were loaded with the calcium-sensitive dye, Fluo-3 AM. Calcium fluctuations prior to and after treatment with caffeine (5 mM) or Gö-6976 (1 μM) were measured using a confocal microscope. Fluorescence amplitude data are expressed as ΔF/F, where F represents baseline fluorescence prior to application of caffeine or Gö-6976, and ΔF represents the change in peak fluorescence during the application of either compound.
4. Discussion
The current study demonstrates that MC1568 and Gö-6976 exert paradoxical effects when delivered to striated muscle cell cultures. Consistent with prior results, MC1568 inhibited differentiation of C2C12 myoblasts into multinucleate myotubes. However, this compound failed to block the enzymatic activity of class IIa HDACs in in vitro assays. Identical findings were made with MC1568 acquired from two different vendors. In contrast to the data obtained with MC1568, a structurally distinct compound, DPAH, dose-dependently inhibited class IIa HDAC catalytic activity and enhanced, rather than inhibited, C2C12 differentiation. The contrapositive effects of the compounds on muscle differentiation correlated with effects on MEF2 transcriptional activity, with MC1568 and DPAH inhibiting and activating a MEF2-driven reporter gene, respectively. These results suggest that MC1568 inhibits skeletal myogenesis through a mechanism that is independent of suppression of class IIa HDAC catalytic activity, perhaps through an off-target action. Furthermore, the findings with DPAH illustrate that inhibition of class IIa HDAC catalytic activity is not sufficient to block myogenesis, as previously suggested, and, conversely, may enhance muscle differentiation.
At least three explanations could account for our inability to replicate the previous finding that MC1568 inhibits class IIa HDAC catalytic activity. First, analysis of NMR spectra revealed that commercially available MC1568 is an isomer of the published structure. Of note, a recent medicinal chemistry report of the re-synthesis of MC1568 defined the major synthetic product to be the same isomer sold by Sigma and Selleck [36]. As such, we cannot rule out the possibility that the differential effects on class IIa HDAC activity are due to distinct properties of the isomers. A second possibility is related to the assays that were used to define class IIa HDAC catalytic activity. We monitored catalytic activity of the HDACs with a synthetic substrate that is specifically deacetylated by class IIa HDACs; this substrate is not deacetylated by other HDAC isoforms. In contrast, Mai et al. used a histone H4-3H-acetyl-peptide substrate [20]. However, it should be noted that follow-up studies by Mai and others did employ the same AMC-based fluorescent class IIa substrate used in our work [23], and we have found that the AMC fluorophore is non-specifically quenched by MC1568 (sFig. 1). Thus, a third explanation for our disparate findings is that MC1568 creates a false positive reading by quenching AMC-generated fluorescence of non-histone synthetic substrates.
We also show that the staurosporine analog, Gö-6976, stimulates stress-dependent kinases in culture neonatal and adult cardiac myocytes, but not in fibroblasts. This surprising action of Gö-6976 is blocked by U-73122, which is a PLC inhibitor. Furthermore, we show that Gö-6976 alters calcium signaling in NRVMs within seconds after treatment. These findings suggest the possibility that Gö-6976 has an off-target action that serves to activate a cell surface receptor(s) to stimulate PLC-mediated calcium release (Fig. 4C).
Activation of cardiac stress signaling by Gö-6976 was initially indicated by induction of PKD phosphorylation on serine-916, which is unexpected since this is an auto-phosphorylation site, and Gö-6976 is an ATP-competitive inhibitor of PKD [16]. In this regard, it is important to stress the distinction between auto-phosphorylation of PKD and phosphorylation of downstream substrates by PKD. Rybin et al. showed that PKD1 serine-916 auto-phosphorylation does not correlate with the ability of the kinase to phosphorylate downstream substrates such as CREB [37]. Furthermore, several studies have shown that PKD-mediated phosphorylation of HDAC5 can be blocked under conditions where serine-916 phosphorylation persists [15;26;38]. Our results with catalytically inactive PKD1 (Fig. 3F) suggest that another kinase(s) is phosphorylating serine-916 in Gö-6976-treated cells.
Chemical biological probes have the potential to provide key insights into the roles of specific targets in the control of various biological processes and disease pathways. Ideally, results should be confirmed with at least two independent, structurally distinct compounds that target the same protein, thereby reducing the possibility that observed effects are governed by “off-target” actions of a given small molecule. In this regard, several novel and highly selectively small molecule inhibitors of class IIa HDACs and PKD have recently been described [24;38–46]. These powerful pharmacological tools should help clarify the roles of class IIa HDAC and PKD catalytic activity in striated muscle cell differentiation and growth.
Supplementary Material
Highlights.
Commercially available MC1568 does not block class IIa HDAC catalytic activity
Inhibition of class IIa HDAC catalytic activity enhances skeletal myogenesis
Gö-6976 alters calcium signaling in primary cardiomyocytes
Gö-6976 stimulates stress-inducible kinases in primary cardiac myocytes
Acknowledgments
This work is dedicated to the loving memory of Dr. Douglas Lemon, an amazing scientist, colleague, friend and family man. We thank R.A. Bannister for calcium imaging, P.J. Papst for assistance with in vitro kinase assays, and J. H. Mahaffey and M.Y. Jeong for ARVMs. T.A. McKinsey was supported by NIH grants (HL116848, AG043822 and HL114887) and the American Heart Association (Grant-in-Aid, 14510001). M.F.W. received funding to support the Medicinal Chemistry Core Facility via Colorado Clinical and Translational Sciences Institute grant NIH-NCATS, UL1TR001082. M.S. Stratton received funding from T32 training grants from the NIH (HL007822 and HL007171).
Abbreviations
- HDAC
histone deacetylase
- PKD
protein kinase D
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 2.McKinsey TA, Zhang CL, Olson EN. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 3.Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J Biol Chem. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparrow DB, Miska EA, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, Spohr G, Kouzarides T, Mohun TJ. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang AH, Bertos NR, Vezmar M, Pelletier N, Crosato M, Heng HH, Th’ng J, Han J, Yang XJ. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 9.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossuyt J, Helmstadter K, Wu X, Clements-Jewery H, Haworth RS, Avkiran M, Martin JL, Pogwizd SM, Bers DM. Ca2+/calmodulin-dependent protein kinase IIdelta and protein kinase D overexpression reinforce the histone deacetylase 5 redistribution in heart failure. Circ Res. 2008;102:695–702. doi: 10.1161/CIRCRESAHA.107.169755. [DOI] [PubMed] [Google Scholar]
- 12.Kreusser MM, Backs J. Integrated mechanisms of CaMKII-dependent ventricular remodeling. Front Pharmacol. 2014;5:36. doi: 10.3389/fphar.2014.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinsey TA. Derepression of pathological cardiac genes by members of the CaM kinase superfamily. Cardiovasc Res. 2007;73:667–677. doi: 10.1016/j.cardiores.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 14.Weeks KL, Avkiran M. Roles and post-translational regulation of cardiac class IIa histone deacetylase isoforms. J Physiol. 2014:1–13. doi: 10.1113/jphysiol.2014.282442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gschwendt M, Dieterich S, Rennecke J, Kittstein W, Mueller HJ, Johannes FJ. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 1996;392:77–80. doi: 10.1016/0014-5793(96)00785-5. [DOI] [PubMed] [Google Scholar]
- 17.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo SP, Carfi A, Koch U, De FR, Steinkuhler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heltweg B, Dequiedt F, Marshall BL, Brauch C, Yoshida M, Nishino N, Verdin E, Jung M. Subtype selective substrates for histone deacetylases. J Med Chem. 2004;47:5235–5243. doi: 10.1021/jm0497592. [DOI] [PubMed] [Google Scholar]
- 19.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai A, Massa S, Pezzi R, Simeoni S, Rotili D, Nebbioso A, Scognamiglio A, Altucci L, Loidl P, Brosch G. Class II (IIa)-selective histone deacetylase inhibitors. 1 Synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J Med Chem. 2005;48:3344–3353. doi: 10.1021/jm049002a. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Hwangbo C, Hu X, Kang Y, Papangeli I, Mehrotra D, Park H, McLean DL, Ju H, Comhair SA, Erzurum SC, Chun HJ. Restoration of Impaired Endothelial MEF2 Function Rescues Pulmonary Arterial Hypertension. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.114.013339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannaerts I, Eysackers N, Onyema OO, Van BK, Valente S, Mai A, Odenthal M, van Grunsven LA. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS One. 2013;8:e55786. doi: 10.1371/journal.pone.0055786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nebbioso A, Manzo F, Miceli M, Conte M, Manente L, Baldi A, De LA, Rotili D, Valente S, Mai A, Usiello A, Gronemeyer H, Altucci L. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009;10:776–782. doi: 10.1038/embor.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tessier P, Smil DV, Wahhab A, Leit S, Rahil J, Li Z, Deziel R, Besterman JM. Diphenylmethylene hydroxamic acids as selective class IIa histone deacetylase inhibitors. Bioorg Med Chem Lett. 2009;19:5684–5688. doi: 10.1016/j.bmcl.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Storz P, Doppler H, Toker A. Protein kinase Cdelta selectively regulates protein kinase D-dependent activation of NF-kappaB in oxidative stress signaling. Mol Cell Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison BC, Kim MS, van RE, Plato CF, Papst PJ, Vega RB, McAnally JA, Richardson JA, Bassel-Duby R, Olson EN, McKinsey TA. Regulation of cardiac stress signaling by protein kinase d1. Mol Cell Biol. 2006;26:3875–3888. doi: 10.1128/MCB.26.10.3875-3888.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer JN, Hartogensis WE, Patten M, Fortuin FD, Long CS. Interleukin-1 beta induces cardiac myocyte growth but inhibits cardiac fibroblast proliferation in culture. J Clin Invest. 1995;95:2555–2564. doi: 10.1172/JCI117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satoh N, Suter TM, Liao R, Colucci WS. Chronic alpha-adrenergic receptor stimulation modulates the contractile phenotype of cardiac myocytes in vitro. Circulation. 2000;102:2249–2254. doi: 10.1161/01.cir.102.18.2249. [DOI] [PubMed] [Google Scholar]
- 29.Molkentin JD, Black BL, Martin JF, Olson EN. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol Cell Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemon DD, Horn TR, Cavasin MA, Jeong MY, Haubold KW, Long CS, Irwin DC, McCune SA, Chung E, Leinwand LA, McKinsey TA. Cardiac HDAC6 catalytic activity is induced in response to chronic hypertension. J Mol Cell Cardiol. 2011;51:41–50. doi: 10.1016/j.yjmcc.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di FS, Azakir BA, Therrien C, Sinnreich M. Dysferlin interacts with histone deacetylase 6 and increases alpha-tubulin acetylation. PLoS One. 2011;6:e28563. doi: 10.1371/journal.pone.0028563. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Jones P, Altamura S, De FR, Gallinari P, Lahm A, Neddermann P, Rowley M, Serafini S, Steinkuhler C. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008;18:1814–1819. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Haworth RS, Goss MW, Rozengurt E, Avkiran M. Expression and activity of protein kinase D/protein kinase C mu in myocardium: evidence for alpha1-adrenergic receptor- and protein kinase C-mediated regulation. J Mol Cell Cardiol. 2000;32:1013–1023. doi: 10.1006/jmcc.2000.1143. [DOI] [PubMed] [Google Scholar]
- 34.Fu Y, Rubin CS. Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 2011;12:785–796. doi: 10.1038/embor.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology (Bethesda ) 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleming CL, Ashton TD, Gaur V, McGee SL, Pfeffer FM. Improved synthesis and structural reassignment of MC1568: a class IIa selective HDAC inhibitor. J Med Chem. 2014;57:1132–1135. doi: 10.1021/jm401945k. [DOI] [PubMed] [Google Scholar]
- 37.Rybin VO, Guo J, Steinberg SF. Protein kinase D1 autophosphorylation via distinct mechanisms at Ser744/Ser748 and Ser916. J Biol Chem. 2009;284:2332–2343. doi: 10.1074/jbc.M806381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meredith EL, Ardayfio O, Beattie K, Dobler MR, Enyedy I, Gaul C, Hosagrahara V, Jewell C, Koch K, Lee W, Lehmann H, McKinsey TA, Miranda K, Pagratis N, Pancost M, Patnaik A, Phan D, Plato C, Qian M, Rajaraman V, Rao C, Rozhitskaya O, Ruppen T, Shi J, Siska SJ, Springer C, van EM, Vega RB, von MA, Yang L, Yoon T, Zhang JH, Zhu N, Monovich LG. Identification of orally available naphthyridine protein kinase D inhibitors. J Med Chem. 2010;53:5400–5421. doi: 10.1021/jm100075z. [DOI] [PubMed] [Google Scholar]
- 39.Di GE, Gagliostro E, Brancolini C. Selective class IIa HDAC inhibitors: myth or reality. Cell Mol Life Sci. 2014;72:73–86. doi: 10.1007/s00018-014-1727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gamber GG, Meredith E, Zhu Q, Yan W, Rao C, Capparelli M, Burgis R, Enyedy I, Zhang JH, Soldermann N, Beattie K, Rozhitskaya O, Koch KA, Pagratis N, Hosagrahara V, Vega RB, McKinsey TA, Monovich L. 3,5-diarylazoles as novel and selective inhibitors of protein kinase D. Bioorg Med Chem Lett. 2011;21:1447–1451. doi: 10.1016/j.bmcl.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 41.George KM, Frantz MC, Bravo-Altamirano K, Lavalle CR, Tandon M, Leimgruber S, Sharlow ER, Lazo JS, Wang QJ, Wipf P. Design, Synthesis, and Biological Evaluation of PKD Inhibitors. Pharmaceutics. 2011;3:186–228. doi: 10.3390/pharmaceutics3020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, Murray-Thompson M, Schwartz B, Chakravorty S, Wu Z, Mander PK, Kruidenier L, Reid RA, Burkhart W, Turunen BJ, Rong JX, Wagner C, Moyer MB, Wells C, Hong X, Moore JT, Williams JD, Soler D, Ghosh S, Nolan MA. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol. 2013;9:319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 43.Meredith EL, Beattie K, Burgis R, Capparelli M, Chapo J, Dipietro L, Gamber G, Enyedy I, Hood DB, Hosagrahara V, Jewell C, Koch KA, Lee W, Lemon DD, McKinsey TA, Miranda K, Pagratis N, Phan D, Plato C, Rao C, Rozhitskaya O, Soldermann N, Springer C, van EM, Vega RB, Yan W, Zhu Q, Monovich LG. Identification of potent and selective amidobipyridyl inhibitors of protein kinase D. J Med Chem. 2010;53:5422–5438. doi: 10.1021/jm100076w. [DOI] [PubMed] [Google Scholar]
- 44.Monovich L, Vega RB, Meredith E, Miranda K, Rao C, Capparelli M, Lemon DD, Phan D, Koch KA, Chapo JA, Hood DB, McKinsey TA. A novel kinase inhibitor establishes a predominant role for protein kinase D as a cardiac class IIa histone deacetylase kinase. FEBS Lett. 2010;584:631–637. doi: 10.1016/j.febslet.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Muraglia E, Altamura S, Branca D, Cecchetti O, Ferrigno F, Orsale MV, Palumbi MC, Rowley M, Scarpelli R, Steinkuhler C, Jones P. 2-Trifluoroacetylthiophene oxadiazoles as potent and selective class II human histone deacetylase inhibitors. Bioorg Med Chem Lett. 2008;18:6083–6087. doi: 10.1016/j.bmcl.2008.09.076. [DOI] [PubMed] [Google Scholar]
- 46.Sharlow ER, Giridhar KV, Lavalle CR, Chen J, Leimgruber S, Barrett R, Bravo-Altamirano K, Wipf P, Lazo JS, Wang QJ. Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem. 2008;283:33516–33526. doi: 10.1074/jbc.M805358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.