Abstract
Gulf War illness (GWI) is a chronic disease of unknown etiology characterized by persistent symptoms such as cognitive impairment, unexplained fatigue, pervasive pain, headaches, and gastrointestinal abnormalities. Current reports suggest that as many as 200,000 veterans who served in the 1990–1991 Persian Gulf War were afflicted. Several potential triggers of GWI have been proposed including chemical exposure, toxins, vaccines, and unknown infectious agents. However, a definitive cause of GWI has not been identified and a specific biological marker that can consistently delineate the disease has not been defined. Myalgic encephalomyelitis (ME) is a disease with similar and overlapping symptomology, and subjects diagnosed with GWI typically fit the diagnostic criteria for ME. For these reasons, GWI is often considered a subgroup of ME. To explore this possibility and identify immune parameters that may help to understand GWI pathophysiology, we measured 77 serum cytokines in subjects with GWI and compared these data to that of subjects with ME as well as healthy controls. Our analysis identified a group of cytokines that identified ME and GWI cases with sensitivities of 92.5% and 64.9%, respectively. The five most significant cytokines in decreasing order of importance were IL-7, IL-4, TNF-α, IL-13, and IL-17F. When delineating GWI and ME cases from healthy controls, the observed specificity was only 33.3%, suggesting that with respect to cytokine expression, GWI cases resemble control subjects to a greater extent than ME cases across a number of parameters. These results imply that serum cytokines are representative of ME pathology to a greater extent than GWI and further suggest that the two diseases have distinct immune profiles despite their overlapping symptomology.
Keywords: cytokines, Gulf War illness, myalgic encephalomyelitis, cytokines, random forest: interleukin-7
1. Introduction
Gulf War illness (GWI) and myalgic encephalomyelitis (ME) are complex diseases of unknown etiology. They are often characterized by a constellation of unexplained and overlapping symptoms, which include widespread inflammation, fatigue, multisystemic neuropathology, joint and muscle pain and gastrointestinal pathology[1–3]. Although the two diseases are similar with overlapping symptoms, GWI is a specific term given to returning military veterans and civilian workers of the Persian Gulf War that took place from August 2, 1990 to February 28, 1991. ME is frequently associated with acute flu-like onset as well as noninfectious environmental triggers[4]; whereas, multiple factors including environmental exposure, toxins, vaccines, and unknown infectious agents have been evaluated as potential triggers for GWI[5, 6]. Indeed, GWI and ME have many clinical symptoms in common including long-term and severe fatigue that is not relieved by rest, gastrointestinal disorders, and neurological impairments[2]. Accordingly, it has been suggested that GWI cases meet the diagnostic criteria for ME and, therefore, represent discrete subsets of ME. Currently, there is no pathognomonic marker for either disease as well as no clinical diagnostic test available; for these reasons, diagnosis is mainly based on clinical observation, epidemiological evaluation, and medical anamnesis.
Immunological impairments in subjects with ME have been extensively documented. For example, several researchers have reported abnormalities in natural killer (NK) cell numbers and function[7, 8] as well as abnormalities in serum and plasma cytokine and chemokine levels[9–12]. Natelson et al. observed that levels of granulocyte-macrophage colony-stimulating factor (GM-CSF) in the cerebral spinal fluid of ME cases were lower than in controls and that levels of CXCL8 were elevated in cases with sudden, influenza-like onset when compared to cases with gradual onset or healthy controls[13]. In a study by Zhang and colleagues, two groups of cases who met the case definition for ME were compared to each other; Gulf War veterans who developed their malady after they had returned home from the Gulf and a group of nonveterans who developed the illness sporadically[14]. They reported that Gulf War veterans with ME had a statistically significant increase in total T cells and a lower percentage of NK cells when compared to respective controls. In addition, veterans with ME had higher levels of interleukin (IL)-2, IL-10, interferon (IFN)-γ, and tumor necrosis factor (TNF)-α over that of controls. However, they observed no difference in civilian veterans with ME when compared to controls across a number of immune parameters.
Unquestionably, serum or plasma inflammatory cytokine and chemokine levels are some of the most commonly reported differences between subjects with ME and healthy controls. For example, Maes et al. reported that subjects with ME have significantly higher levels of serum IL-1 and TNF-α when compared to controls[15, 16]. Also, Fletcher and coworkers observed increased levels of serum regulatory and proinflammatory cytokines such as lymphotoxin-α, IL-1α, IL-1β, IL-4, IL-5, IL-6, and IL-12 in ME cases when compared to controls[17]. Other studies have investigated cytokine profiles of subjects with GWI and ME[18]. A Th2 shift is commonly reported for subgroups of ME cases[11, 19], suggesting that those with ME may be hyper-responsive to allergens, toxins, extracellular bacteria, and parasites and hypo-responsive to viruses and intracellular bacteria. Smylie et al. reported a decreased Th2 polarity in females with ME as compared to GWI and suggested that an IL-23/Th17/IL-17 axis could be used to delineate GWI and ME[18]. Skowera and colleagues reported that, in contrast to asymptomatic Gulf War veterans, symptomatic veterans with “multisymptom illness” displayed an ongoing Th1-type immune activation with significantly elevated levels of IFN-γ and IL-2, in the absence of in vitro stimulation[20]. These studies suggest that although similar in clinical manifestations, GWI and ME potentially present discrete cytokine profiles, which may reflect differences in disease pathogenesis.
Cytokines orchestrate numerous immune functions including activating and prolonging leukocyte proliferation, directing migration, and influencing and shaping leukocyte functional activity. Abnormal leukocyte counts in subjects with GWI and ME may, in fact, be a consequence of dysregulated cytokine control. Subsequently, abnormal leukocyte counts may lead to a disturbed immune response, often manifesting with broad clinical presentations. It may also be suggested that cytokine profiles, which are reflective of the profound immune disturbances in subjects with GWI and ME, might potentially serve as useful biomarkers. A greater understanding regarding cytokine dysregulation in GWI and ME may also help to better understand the pathogenesis of these diseases, thus improving diagnosis, treatment efficacy, and prophylactic measures.
In the present study, we have conducted a comprehensive survey of 77 different cytokines and chemokines in an effort to better understand the immune responses associated with GWI and ME. Our results suggest that Th1 and Th17 cytokines underscore GWI cases, while Th1 and Th2 cytokines as well as a more diverse group of inflammatory cytokines and mononuclear chemoattractant cytokines characterize ME. Additionally, in order to identify the most important cytokines that distinguish these groups and potentially identify underlying pathology, we utilized the machine logic nearest neighbor predictor algorithm Random Forest to analyze these data. The five most significant cytokines identified by our model in decreasing order of importance were IL-7, IL-4, TNF-α, IL-13, and IL-17F. Although our Random Forest analysis produced a cytokine signature that identified ME cases with 92.5% sensitivity, only 64.9% sensitivity was achieved when delineating GWI cases. Further more, specificity was only 33.3%, suggesting that with respect to cytokine expression, GWI cases resemble control subjects to a greater extent than ME cases across a number of parameters. These results imply that serum cytokines are representative of ME pathology to a greater extent than GWI and further suggest that the two diseases have distinct immune profiles despite their overlapping symptomology.
2. Materials and methods
2.1 Study subjects
A total of 146 subjects were enrolled in these studies; 67 cases with a confirmed diagnosis of ME, 37 identified as having GWI, and 42 healthy controls. Informed consent was obtained from each participant according to human subjects protocols approved by the University of Nevada Biomedical Institutional Review Board (protocols B12-031 and B12-036). The cases identified as having ME were physician diagnosed and met the Carruthers et al. criteria for ME as well as the 1994 Fukuda et al. criteria[1, 21, 22]. ME subjects were recruited from across the United States and from individuals who sought treatment for ME at the Himmunitas ME/CFS clinic in Brussels Belgium. GWI subjects were recruited by the VA Sierra Nevada Health Care System Medical Center in Reno, Nevada, and were physician diagnosed satisfying the inclusion criteria of having been on active duty in the military during the Persian Gulf War (Operation Desert Storm: 1990–1991) and symptoms consistent with GWI as defined by the Centers for Disease Control and Prevention (CDC) and Kansas criteria for GWI[23, 24]. Cases were generally representative of the respective populations for each disease based on gender and age.
2.2. Serum samples
Our initial evaluation regarding the method of blood collection indicated that most anticoagulants we tested activated cytokine expression to some level over a 24-h time period (data not shown). The activation was the most pronounced with blood collected on heparin. Given that lymphocytes from ME subjects respond to a greater extent upon stimulation than controls (unpublished observation), this problem would not be normalized even when cases and controls are handled in an identical manner. Additionally, our study required some blood to be shipped overnight; therefore, we chose to conduct our analysis on serum rather than plasma. Whole blood was collected using serum-separator tubes, centrifuged immediately to isolate the serum, and aliquots were made at approximately 24 h post draw and stored at −80 °C until analyzed.
2.3. Cytokine analysis
Serum cytokine levels were analyzed on a Luminex 200 analyzer (Austin, TX) with Bio-Plex (Bio-Rad, Hercules, CA) multiplex magnetic bead-based antibody detection kits according to the manufacturer’s instructions. Bio-Plex Pro Human Chemokine panels (40-Plex), Bio-Plex Pro Human Th17 Cytokine panels, Bio-Plex Pro Human Cytokine 27-plex panels, and Bio-Plex Human Cytokine 21-plex panels were used to cover a total of 77 cytokines and chemokines (herein referred to as “cytokines”). For each subject, 50 μl of serum was analyzed and a minimum of 50 beads per cytokine was acquired. Data collected was analyzed using MasterPlex CT control software and MasterPlex QT analysis software (MiraiBio division of Hitachi Software, San Francisco, CA). Standard curves for each cytokine were generated using standards provided by the manufacturer and some samples were analyzed on multiple runs for quality control purposes and to normalize the collective runs.
2.4. Statistical analysis
In order to determine differences in cytokine values and distributions between GWI, ME, and control subjects, we initially performed Kolmogorov-Smirnov tests for normality, which revealed that the data were not normally distributed (data not shown). We therefore used the non-parametric Kruskal–Wallis (K.W.) one-way analysis of variance by ranks to confirm that the three populations did not originate from the same distribution. We then performed a Mann-Whittney (M.W.) analysis to identify differences in medians between GWI and ME cases as well as between GWI cases and controls and ME cases and controls. We additionally conducted Pearson correlation analysis, comparing cytokines to each other within each respective subject population. Finally, we performed classification analysis using the tree-based ensemble machine learning algorithm Random Forest[25]. For this analysis, 500 random trees were built using six predictors for each node, and auto-bootstrap out-of-bag sampling was used for testing the model.
3. Results
3.1. Differential expression of serum cytokines
In these studies, a total of 104 cases (67 ME and 37 GWI) and 42 controls were analyzed for 77 serum cytokines. Subjects classified as having ME were physician diagnosed and fulfilled the criteria described by Carruthers et al.[1, 21] as well as the Fukuda criteria[22]. GWI subjects were physician diagnosed at the VA Sierra Nevada Health Care System Medical Center in Reno, Nevada, and met the inclusion criteria as having been on active duty in the military during the Persian Gulf War (Desert Storm: 1990–1991) and symptoms consistent with GWI as defined by the Centers for Disease Control and Prevention (CDC) and Kansas criteria for GWI.[23, 24]. Subjects’ ages ranged from 23 to 81 years (mean age = 58.9 years). For ME cases, the ratio of females to males was approximately 2 to 1 (64% and 36%, respectively); for GWI cases the ratio of males to females was approximately 2 to 1 (64% and 36%, respectively). Controls were of approximately equal proportions (57% male and 43% female).
The K.W. test was initially utilized to compare the individual cytokines concurrently for GWI and ME cases as well as healthy controls. Of the 77 cytokines analyzed, 48 (63%) differed for at least one of the three groups (p ≤ 0.05), suggesting that the respective cytokine values did not originate from the same distribution (Supplemental data Table 1). We next utilized the M.W. test to compare the two groups of cases with each other and each group of cases with the control group (Supplemental data Table 1). When GWI and ME subjects were compared, 48 cytokines were observed to be significantly different (p ≤ 0.05). Additionally, when ME cases were compared to healthy controls, 42 cytokines were observed to be differentially expressed, 17 of which were upregulated and 26 were downregulated (Supplemental data Table 2). This is in contrast to only 14 cytokines that differed between GWI cases and controls, 7 of which were upregulated and 7 were downregulated (Table 1.). These observations suggest that, with respect to cytokines, GWI cases resemble healthy controls to a greater extent than they resemble ME cases.
Table 1.
Cytokines upregulated and downregulated in subjects with Gulf War illness (GWI) when compared to healthy controls
| Cytokines upregulated in GWI subjects | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Analyte | Group | Minimum | Maximum | Mean | Standard Deviation | P value by M.W. |
| CCL11 | CON | 20.7 | 90.9 | 47.1 | 16.4 | |
| GWI | 22.2 | 252.8 | 72 | 48.4 | 0.008 | |
| FGF | CON | 3 | 98.5 | 20.7 | 17 | |
| GWI | 9.2 | 57.7 | 23.9 | 11.1 | 0.026 | |
| IFN-γ | CON | 2.8 | 50.4 | 15.4 | 10.3 | |
| GWI | 7 | 47.5 | 22.4 | 10.6 | 0.003 | |
| IL-17A | CON | 2.5 | 10.5 | 7.5 | 1.5 | |
| GWI | 0.8 | 277.8 | 13.6 | 44.8 | 0.032 | |
| IL-17F | CON | 1 | 143.5 | 17.9 | 30.8 | |
| GWI | 9.4 | 678.8 | 47.8 | 113.5 | 0.001 | |
| IL-33 | CON | 8.2 | 2850.6 | 723.3 | 646 | |
| GWI | 56.5 | 4379.6 | 1112.2 | 883.4 | 0.011 | |
| IL-5 | CON | 2.4 | 6.4 | 5.9 | 0.9 | |
| GWI | 0.1 | 24.5 | 6.4 | 3.4 | 0.014 | |
|
| ||||||
| Cytokines downregulated in GWI subjects | ||||||
|
| ||||||
| Analyte | Group | Minimum | Maximum | Mean | Standard Deviation | P value by M.W. |
| CCL5 | CON | 300.4 | 27934.9 | 5520.3 | 4290.5 | |
| GWI | 497.4 | 10415.4 | 4059.3 | 2015 | 0.031 | |
| CXCL8 | CON | 6.4 | 3660.2 | 267.4 | 620.8 | |
| GWI | 2.2 | 1413.5 | 126.3 | 284 | 0.032 | |
| IL-13 | CON | 8.1 | 11.3 | 9 | 1.3 | |
| GWI | 8.1 | 13.9 | 8.2 | 1 | 0.001 | |
| IL-25 | CON | 0 | 4.7 | 2.4 | 2 | |
| GWI | 0 | 4.7 | 1.3 | 1.4 | 0.04 | |
| IL-4 | CON | 0.1 | 2 | 1.1 | 0.6 | |
| GWI | 0.1 | 4.6 | 0.9 | 0.7 | 0.041 | |
| IL-7 | CON | 1.4 | 11.8 | 5.5 | 3.2 | |
| GWI | 2.5 | 18.9 | 3.7 | 2.7 | 0.011 | |
| TNF-α | CON | 17.6 | 72.2 | 23.1 | 10.2 | |
| GWI | 5.7 | 20 | 19.3 | 3 | <0.0001 | |
3.2. Th1, Th2, and Th17 cytokine expression
Previous studies have reported that Th1, Th2, and Th17 cytokines, or combinations there of, characterize GWI and ME. To explore this possibility, we organized these cytokines into three groups (Table 2). For subjects with ME, the Th1 cytokines IFN-γ, IL-2, and IL-12(p75) were upregulated, while the Th2 cytokine IL-5 and IL-9 were downregulated. Paradoxically, the classical Th2 cytokines IL-4, IL-6, IL-10, IL-13 were also upregulated in subjects with ME. For subjects with GWI, the Th1 cytokine IFN-γ was upregulated; however, CXCL8 was slightly downregulated. Additionally, the Th2 cytokines IL-4, IL-13 and IL-25 were all significantly downregulated. For the Th17 cytokines, we observed IL-17F to be significantly downregulated in ME cases, while IL17A and IL17F were both significantly upregulated in GWI cases. Of the 48 cytokines that were differentially expressed in ME, 12 (25%) represented Th1, Th2, or Th17 cytokines. In contrast, of the 14 differentially expressed cytokines observed in GWI subjects (Table 2), 9 (64%) represented Th1, Th2, or Th17 cytokines.
Table 2.
Th1, Th2, and Th17 cytokines expression in subjects with GWI and ME
| Analyte | CON | ME | GWI | P value by Mann Whittney | |
|---|---|---|---|---|---|
| ME vs CON | GWI vs CON | ||||
| Th1 Cytokines | |||||
|
| |||||
| IFN-γ | 15.4 | 32.4 | 22.4 | 0.001 | 0.003 |
| IL-2 | 4.7 | 11.3 | 3.6 | 0.062 | 0.271 |
| TNF-α | 23.1 | 45.4 | 19.3 | <0.0001 | <0.0001 |
| IL-12(p75) | 20 | 32.7 | 21.9 | <0.0001 | 0.246 |
| CXCL8 | 267.4 | 191.9 | 126.3 | <0.0001 | 0.032 |
| IL-18 | 790.2 | 726.3 | 873.2 | 0.219 | 0.446 |
| IL-12(p40) | 290.1 | 238.3 | 309.9 | 0.089 | 0.914 |
|
| |||||
| Th2 cytokines | |||||
|
| |||||
| IL-4 | 1.1 | 1.7 | 0.9 | <0.0001 | 0.041 |
| IL-13 | 9 | 10.7 | 8.2 | <0.0001 | 0.001 |
| IL-1β | 7.9 | 14.4 | 6.6 | <0.0001 | 0.133 |
| IL-25 | 2.4 | 5 | 1.3 | <0.0001 | 0.04 |
| IL-10 | 10.2 | 31.7 | 10.8 | 0.017 | 0.223 |
| IL-5 | 5.9 | 5.3 | 6.4 | <0.0001 | 0.014 |
| IL-6 | 4.3 | 12.4 | 3.6 | <0.0001 | 0.202 |
| IL-9 | 31.7 | 8 | 8.7 | <0.0001 | 0.1 |
|
| |||||
| Th17 cytokines | |||||
|
| |||||
| IL-17A | 7.5 | 8.7 | 13.6 | 0.053 | 0.032 |
| IL-17F | 17.9 | 2.9 | 47.8 | <0.0001 | 0.001 |
| IL-21 | 30.1 | 44.5 | 36.2 | 0.918 | 0.081 |
3.3. Cytokine correlation analysis
In addition to T cells, other cells make many of the cytokines typically associated with a Th1, Th2 or Th17 shifts. For instance, the endogenous pyrogen TNF-α is primarily made by activated macrophages, but is also made by most nucleated cells including lymphocytes, fibroblasts and neurons[26, 27]. Likewise, IL-6 is produced by activated macrophages as well as T cells and can act in a proinflammatory or antiinflammatory capacity[28]. Therefore, we conducted correlation analysis, in order to provide additional clues as to which cells produce these cytokines in our study groups. Our results suggest the Th1 cytokines strongly correlate in the ME population but substantially less so for the Th2 cytokines (Table 3.). Additionally, a complete absence of correlation was observed for the Th17 cytokines in the ME group. In contrast to the ME group, only IL-2 and IL-12(p75) showed any significant correlation in GWI. Again, with respect to the GWI group, the Th1 cytokines showed a much weaker correlation and strikingly, TNF-α was negatively correlated with IFN-γ and IL-2, which is in contrast to the ME and control groups (Table 3.). Also, the Th17 cytokines showed a moderately positive correlation in the GWI population. Interestingly, the correlation between IL-17A and IL-21 showed a positive correlation in contrast to that of the control population, which showed a negative correlation.
Table 3.
Correlation of cytokines in ME, GWI and controls (values given as R-squared)
| Th1 Cytokines ME | Th1 Cytokines GWI | Th1 Cytokines CON | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-12p75 | TNFα | IL-2 | IL-12p75 | TNFα | IL-2 | IL-12p75 | TNFα | IL-2 | ||||||
| IFNγ | 0.944 | 0.797 | 0.934 | IFNγ | 0.408 | −0.495 | 0.424 | IFNγ | 0.305 | 0.346 | 0.339 | |||
| IL-2 | 0.929 | 0.562 | IL-2 | 0.724 | −0.166 | IL-2 | 0.837 | 0.802 | ||||||
| TNFα | 0.758 | TNFα | 0.012 | TNFα | 0.534 | |||||||||
|
| ||||||||||||||
| Th2 Cytokines ME | Th2 Cytokines GWI | Th2 Cytokines CON | ||||||||||||
| IL-9 | IL-6 | IL-5 | IL-13 | IL-9 | IL-6 | IL-5 | IL-13 | IL-9 | IL-6 | IL-5 | IL-13 | |||
|
| ||||||||||||||
| IL-4 | 0.065 | 0.282 | 0.561 | 0.679 | IL-4 | 0.354 | 0.152 | 0.674 | 0.872 | IL-4 | −0.011 | 0.660 | −0.742 | 0.878 |
| IL-13 | 0.074 | 0.068 | 0.946 | IL-13 | 0.249 | 0.16 | 0.896 | IL-13 | −0.121 | 0.680 | −0.633 | |||
| IL-5 | 0.056 | 0.019 | IL-5 | 0.306 | 0.117 | IL-5 | 0.113 | −0.462 | ||||||
| IL-6 | −0.017 | IL-6 | 0.013 | IL-6 | 0.041 | |||||||||
|
| ||||||||||||||
| Th17 Cytokines ME | Th17 Cytokines GWI | Th17 Cytokines CON | ||||||||||||
| IL-21 | IL-17F | IL-21 | IL-17F | IL-21 | IL-17F | |||||||||
|
| ||||||||||||||
| IL-17A | −0.016 | −0.022 | IL-17A | 0.349 | 0.102 | IL-17A | −0.544 | -0.337 | ||||||
| IL-17F | −0.036 | IL-17F | 0.592 | IL-17F | 0.815 | |||||||||
In addition to the Th1, Th2 and Th17 cytokines, we conducted correlation analysis on the remaining analytes and observed almost perfect correlation (R2≥0.90) between a number of cytokines in the ME group (26 cytokines) but fewer in the GWI group (14 cytokines) (Supplemental data Table 3). Of particular interest, we observed almost perfect correlation between IL-7/IL-13, IL-7/FGF, and IL-7/TNF-α and between IL-3/IFN-α in the ME group. We also observed near perfect correlation between IL-1α/IL-3, IL-1α/IL-12(p40), IL-1α/IL-2 and IL-1α/IL-22 and between IL-1/IL-7 and IL-1/IL-13 in the GWI group.
3.4. Classification of cytokines by importance
Both GWI and ME are diseases with undefined etiology and both are often characterized by aberrant cytokine expression; however, the patterns of cytokine expression appear to be more complex than can be described by a standard Th1, Th2 or Th1 7 shift. With this in mind, we utilized the machine logic algorithm Random Forest (RF) to analyze our data set and potentially identify the most important cytokines that define these diseases. For this analysis, 500 random decision trees were built using six predictors for each node, and auto bootstrap out-of-bag sampling was employed to text the model. The 20 most significant cytokines for delineation of each group of subject in order of decreasing importance were IL-7, IL-4, TNF-α, IL-13, IL-17F, IL-1, IL-5, IL-25, CXCL8, VEGF, CCL11, IL12(p75), IL-9, CFS3, IFN-γ, CCL4, IL-6, CCL2, CXCL1, and CXCL10 (Figure 1.). Using only serum cytokines, we were able to achieve sensitivity of 92.5% for delineating ME; however, only 64.9% sensitivity was achieved when delineating GWI with 33.3% overall specificity (Table 4). These data indicate that using serum cytokines alone may not yield an effective diagnostic tool; however, it may provide important clues regarding the underlying pathology of the disease.
Figure 1.
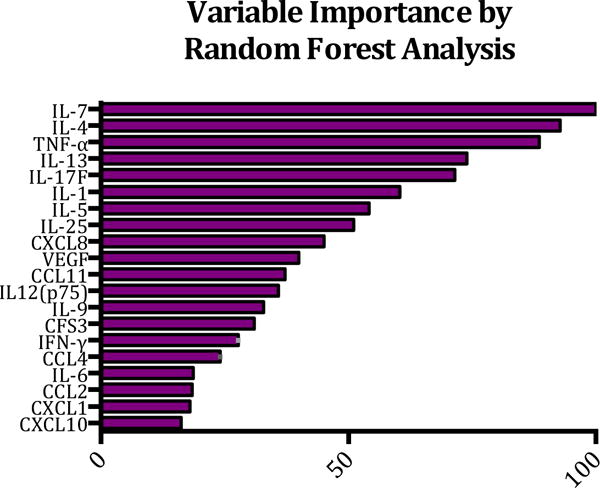
Classification analysis of cytokine data using Random Forest. In order to identify which cytokines most accurately predict disease status of subjects with GWI, ME or controls, Random Forest analysis was implemented whereby 500 random trees were built and six predictors were used at each node. Auto-bootstrap out-of-bag sampling was used for testing the model.
Table 4.
Random Forest model statistics
| Out-of-Bag Testing | |||||
|---|---|---|---|---|---|
| Misclassification | |||||
|
| |||||
| Class | N Cases | N Misclassified | Pct. Error | Cost | |
| ME | 67 | 5 | 7.46% | 0.0746 | |
| GWI | 37 | 13 | 35.14% | 0.3514 | |
| CON | 42 | 28 | 66.67% | 0.6667 | |
|
| |||||
| Out-of-Bag Testing | |||||
| Prediction Success | |||||
|
| |||||
| Actual Class | Total Class | Percent Correct |
ME N = 70 |
GWI N = 46 |
CON N = 30 |
| ME | 67 | 92.54% | 62 | 1 | 4 |
| GWI | 37 | 64.86% | 1 | 24 | 12 |
| CON | 42 | 33.33% | 7 | 21 | 14 |
| Total: | 146 | ||||
| Average: | 63.58% | ||||
| Overall% | |||||
| Correct: | 68.49% | ||||
4. Discussion
Previous studies of GWI and ME often report that study subjects are characterized by abnormal numerical and functional leukocyte parameters. For example, when compared to healthy controls, NK cell enumeration and functionality have been reported to be abnormal in both diseases[7, 29]. Additionally, atypical cytokine expression profiles are often reported in association with GWI and ME, although the results are often contradictory. For example, a distinct immune profile of attenuated Th1/Th17 and elevated Th2 responses was reported by Broderick et al., in subjects with ME[9]. However, in another study, Moss et al. observed upregulation of proinflammatory cytokines in the serum of ME cases, suggestive of a Th17 shift[30]. Likewise, cytokine profiling in GWI has been fraught by contradictory results, where predominant Th1 or Th2 immune responses have been reported[14, 20, 31]. Consequently, although differences in serum or plasma cytokines are well documented between cases and controls in both diseases, there is no consensus on a dominant cytokine expression profile for either disease. These conflicting findings may be a result of the heterogeneous nature of these diseases or perhaps a result of different methods of analysis or blood collection procedures. It is also likely that, at any given time, cytokine expression of an individual may change over time, complicating their use as a diagnostic marker.
In this report, we have presented a comprehensive analysis of 77 different cytokines, which to our knowledge represents the largest investigation of serum cytokines in GWI and ME subjects to date. Subjects’ blood was collected using serum separator tubes and centrifuged immediately in order to isolate serum cytokines without the use of anticoagulants. By using this method, we have eliminated the possibility that the observed results were subject to leukocyte activation associated with anti coagulants such as heparin or assay interference associated chelating agents like ethylenediaminetetraacetic acid (EDTA).
In this study, we observed differences between cases and controls for 48 of the 77 cytokines investigated, using a confidence interval of 95%. Of the cytokines analyzed, 42 (54.5%) were found to be significantly different between ME cases and healthy controls. In contrast, only 14 cytokines (17.7%) were found to be significantly different between GWI cases and controls. Additionally, when comparing GWI and ME cases, 48 of 77 cytokines were differentially expressed. These data suggest that subjects with GWI and ME are unlikely to represent the same population.
Previous studies have suggested that subjects with GWI and ME may be characterized through the expression of either Th1 or Th2 cytokines. Upon activation, proliferating helper T cells may develop into effector T cells that are often classified as either Th1 or Th2 cells. Th1 immunity is directed against intra cellular pathogens such as viruses and mycobacteria, whereas Th2 immunity is typically in response to extracellular pathogens such as fungi and helminths. For these reasons, the cytokines produced by these cells are also referred to as Th1- or Th2-type cytokines. The cytokine expression observed in this study with respect to ME cases, was largely inconsistent with a clear Th1- or Th2-type immune response. For instance, we observed an upregulation of IFN-γ (p≤0.001) and IL-12(p75)(p≤0.001) in the absence of an increase in IL-12(p40). These data are consistent with a classic Th1 response[32]. On the other hand, we also observed an upregulation of the IL-10 and IL-4, and when compared to healthy controls (p≤0.0001), suggestive of a Th2 response. These observations, in conjunction with our correlation analysis, suggest that the Th1 and Th2 cytokines observed in subjects with ME may originate from other immune cells in addition to T cells.
Our data more strongly supports a Th1/Th17 immune polarization in subjects with GWI. Serum cytokine analysis of these subjects showed an upregulation of the Th1 cytokine IFN-γ (p≤0.003) and the Th17 cytokines IL-17A (p≤0.032) and IL-17F (p≤0.001) and a concomitant downregulation of the Th2 cytokines IL-4 (p≤0.014) and IL-13 (p≤0.001) when compared to healthy controls. Exposures to such things as toxins, vaccines and unknown infectious agents have been suggested as potential triggers for GWI[5, 6]. Several such triggers have been associated with a Th1/Th17 cytokine shift. For instance, Robbe et al. reported that the occupational exposure to agricultural dust was associated with upregulation of IL-17 and IFN-γ[33], and Harris and coworkers reported that human DCs upregulate IL-17 and IFN-γ in response to the bacteria B. anthracis[34]. Additionally, the cationic liposome adjuvant system CAF01, which is commonly used in such vaccines as the trivalent influenza vaccine, is reported to promote a strong and sustained Th1 and Th17 response[35]. Although we cannot say that any of these triggers contribute to GWI, the observed Th1/Th17 shift would be consistent with such triggers.
Little is known regarding the pathophysiology of GWI and ME; nevertheless, the source or class of cytokines produced in subjects with these diseases may provide important clues. Indeed, cytokine profiling has provided valuable knowledge regarding the pathogenesis of other diseases. For example, De Furia et al. used cytokine profiling to identify the source of inflammatory cytokines associated with type 2 diabetes[36], and Swindle and coworkers utilized cytokine expression data to dissect the psoriatic transcriptome and identified the respective cellular contributions associated with this disease[37]. Additionally, Valeyev and colleagues showed that using a systems model approach; cytokine expression data could be used to provide a quantitative description of immune cell interactionsin subjects with psoriasis[38]. In order to identify potential cytokines that may provide information regarding the pathogenesis of GWI and ME, we implemented the machine logic algorithm Random Forest (RF) to analyze our data set. The RF algorithm uses an ensemble of unpruned classification or regression trees produced through bootstrap sampling of the training data set and random feature selection in tree generation. Prediction was made by a majority vote of the predictions of the ensemble. The strength of the analysis was evaluated by an out-of-bag sampling without replacement of the original data. The RF is an attractive method since it handles both discrete and continuous data, it accommodates and compensates for missing data, and it is invariant to monotonic transformations of the input variables. The RF algorithm is uniquely suited for cytokine analysis in that it can handle highly skewed values well and weighs the contribution of each cytokine according to its relatedness with others. Using cytokine expression as input variables and subject status (i.e., GWI case, ME case, and control) as the outcome variable, we identified a group of cytokines that associated with disease status and, therefore, may contribute to the pathogenesis of the disease.
The five most significant cytokines identified by our model in decreasing order of importance were IL-7, IL-4, TNF-α, IL-13, and IL-17F. These cytokines were also identified by significant correlations in our analysis. IL-7 is a hematopoietic growth factor and is important for development, maturation and homeostasis of B, T, and NK cells. Stromal cells of the bone marrow and thymus are the primary source of IL-7; however, is it is also produced to a lesser extent by DCs, hepatocytes, and neurons, but not by lymphocytes[39]. Our data suggest that IL-7 is over-expressed in ME (p≤0.001) and under-expressed in GWI (p≤0.01) when compared to healthy controls. Previous studies suggest that the administration of exogenous of IL-7 in humans leads to the expansion of CD4+ and CD8+ T cells with a concomitant decrease of CD4+ Tregs[40]. Other studies have shown that IL-7-treated animals have reduced numbers of T cells expressing the inhibitory molecules suppressor of cytokine signaling 3 (SOCS3) and programmed cell death protein 1 (PD-1)[41]. Several studies suggest that ME is an inflammatory disease, and multiple reports of individuals with ME expressing auto antibodies[42, 43] and the efficacious treatment of ME cases with the B-cell-depleting drug rituximab[44, 45], suggest that some components of ME pathology may also overlap with those of autoimmunity. Aberrant expression of IL-7 and its receptor has been associated with several autoimmune diseases including inflammatory bowel disease, multiple sclerosis, rheumatoid arthritis, and Sjögren’s syndrome (Reviewed in[46]). The upregulation of IL-7 may help explain some of the clinical observations associated with ME such as the inflammatory component or the presentation of autoimmune-like symptoms. In that IL-7 is primarily produced in the bone marrow, but not by lymphocytes, transcriptional studies of bone marrow would be prudent; however, given the difficulty in collecting bone marrow biopsies, deciphering its involvement in these diseases may prove difficult. It is also likely that transcriptional profiling studies that only utilize whole blood will fail to identify an important component to the pathophysiology of these diseases.
Our analysis identified IL-4 and TNF-α as the second and third most important cytokines when delineating ME, GWI, and controls. Both IL-4 and TNF-α were over-expressed in ME (p≤0.0001) and under-expressed in GWI (p≤0.04 and p≤0.0001). IL-4 is a classic Th2 cytokine and promotes the differentiation of naïve helper T cells into Th2 cells. Once differentiated, the Th2 cells can produce additional IL-4 in a positive feedback control loop[47]. The chronic nature of these diseases suggests that Th2 cells are a likely source of serum IL-4; however, our correlation analysis suggests that they may not be the only source. During an inflammatory response, IL-4 production is often accompanied with IL-10 production, which is also upregulated in our ME subjects; however, no statistical difference was observed for IL-10 in GWI subjects. Interestingly, IL-13 was the next most important cytokine in our model. Expression of IL-13 was slightly downregulated in GWI and upregulated in ME. Our analysis showed that IL-13 and TNF-α expression was almost perfectly correlated (R2 = 0.918). IL-13 is an anti inflammatory cytokine and its upregulation may be a response to counter the inflammatory effects of TNF-α. Lastly, our model identified IL-17F as the fifth most important cytokine in differentiating GWI cases, ME cases, and healthy controls. We observed IL-17F to be significantly downregulated in ME cases (p≤0.0001) and upregulated in GWI cases (p≤0.001). By increasing the production of inflammatory chemokines, IL-17 is a potent mediator of delayed-type responses and its expression promotes the recruitment of monocytes and neutrophils to the site of inflammation. IL-17F, in particular, is associated with respiratory pathology such as asthma[50].
Our analysis using RF suggests that any combination of the 77 cytokines analyzed in our study may not provide a stand-alone differential diagnosis of GWI and ME. Although the cytokine signature delineated ME cases with 92.5% efficiency, only 64.9% sensitivity was achieved when delineating GWI cases. Furthermore, specificity was 33.3% using cytokines only. Our ongoing research suggests that, by using a combination of cytokines and clinical parameters, we can far exceed the sensitivity and specificity of these results (data not shown). This observation further suggests that cytokines are useful when stratifying subjects into discrete subgroups. It also suggests that the “catch all” terms of GWI and ME may be overly broad. In light of the heterogeneous nature of these diseases, stratification into subgroups may be mandatory in order to make meaningful progress in understanding the pathophysiology of these diseases.
In conclusion, this study supports an involvement for Th1/Th17 cytokines in GWI and further identifies the cytokines IL-7, IL-4, TNF-α, IL-13, and IL-17F as potentially contributing to the pathogenesis of GWI and ME. This knowledge may provide direction in the development of therapeutic treatments for these diseases.
Supplementary Material
Highlights.
Gulf war illness (GWI) is characterized by a Th1/Th17 shift.
Th1, Th2 and inflammatory cytokines characterize myalgic encephalomyelitis (ME)
Cytokine importance by Random Forest were IL-7, IL-4, TNF-α, IL-13, and IL-17F.
GWI and ME have distinct cytokine profiles despite similar symptomology.
Acknowledgments
We are grateful to Nossa Van Den Vonder and Jan De Leersnijder at the Himmunit as ME/CFS clinic and Kellen Jones at WPI for their assistance in study subject recruitment. We also thank Dr. Doug Redelman for his insightful discussions regarding this work and Dr. Ken Hunter and Carli Kinnie for their critical proofreading of this manuscript.
Source of support
These studies were supported by awards from the Department of Defense (DOD) grant GW100091 and by the National Institutes of Health (NIH) grant R01 AI078234. A fellowship provided by the Program of Competitive Growth of Kazan Federal University supported the work of N. Blatt and A. Rizvanov. This material is, in part, the result of work supported with resources and the use of facilities in the VA Sierra Nevada Health Care System. The contents of this publication do not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Carruthers BM, van de Sande MI, De Meirleir KL, Klimas NG, Broderick G, Mitchell T, et al. Myalgic encephalomyelitis: International Consensus Criteria. J Intern Med. 2011;270:327–38. doi: 10.1111/j.1365-2796.2011.02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolson GL, Nicolson NL. Gulf War illnesses: complex medical, scientific and political paradox. Medicine, conflict, and survival. 1998;14:156–65. doi: 10.1080/13623699808409384. [DOI] [PubMed] [Google Scholar]
- 3.De Meirleir K, Khaiboullina S, Fremont M, Hulstaert J, Rizvanov A, Palotas A, et al. Plasmacytoid Dendritic Cells in the Duodenum of Individuals Diagnosed with Myalgic Encephalomyelitis Are Uniquely Immunoreactive to Antibodies to. Human Endogenous Retroviral ProteinsIn Vivo. 2013;27 [PMC free article] [PubMed] [Google Scholar]
- 4.Buchwald D, Umali J, Pearlman T, Kith P, Ashley R, Wener M. Post infectious chronic fatigue: a distinct syndrome? Clinial infectious diseases: an official publication of the Infectious Diseases Society of America. 1996;23:385–7. doi: 10.1093/clinids/23.2.385. [DOI] [PubMed] [Google Scholar]
- 5.Iversen A, Chalder T, Wessely S. Gulf War Illness: lessons from medically unexplained symptoms. Clinical psychology review. 2007;27:842–54. doi: 10.1016/j.cpr.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Fukuda K, Nisenbaum R, Stewart G, Thompson WW, Robin L, Washko RM, et al. Chronic multisymptom illness affecting Air Force veterans of the Gulf War. JAMA: the journal of the American Medical Association. 1998;280:981–8. doi: 10.1001/jama.280.11.981. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S, Vayuvegula B. A comprehensive immunological analysis in chronic fatigue syndrome. Scandinavian journal of immunology. 1991;33:319–27. doi: 10.1111/j.1365-3083.1991.tb01777.x. [DOI] [PubMed] [Google Scholar]
- 8.Whiteside TL, Friberg D. Natural killer cells and natural killer cell activity in chronic fatigue syndrome. The American journal of medicine. 1998;105:27S–34S. doi: 10.1016/s0002-9343(98)00155-7. [DOI] [PubMed] [Google Scholar]
- 9.Broderick G, Fuite J, Kreitz A, Vernon SD, Klimas N, Fletcher MA. A formal analysis of cytokine networks in chronic fatigue syndrome. Brain Behav Immun. 2010;24:1209–17. doi: 10.1016/j.bbi.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomoda A, Joudoi T, Rababel M, Matsumoto T, Park TH, Miike T. Cytokine production and modulation: comparison of patients with chronic fatigue syndrome and normal controls. Psychiatry research. 2005;134:101–4. doi: 10.1016/j.psychres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Patarca-Montero R, Antoni M, Fletcher MA, Klimas NG. Cytokine and other immunologic markers in chronic fatigue syndrome and their relation to neuro psychological factors. Applied neuro psychology. 2001;8:51–64. doi: 10.1207/S15324826AN0801_7. [DOI] [PubMed] [Google Scholar]
- 12.Chao CC, Janoff EN, Hu SX, Thomas K, Gallagher M, Tsang M, et al. Altered cytokine release in peripheral blood mononuclear cell cultures from patients with the chronic fatigue syndrome. Cytokine. 1991;3:292–8. doi: 10.1016/1043-4666(91)90497-2. [DOI] [PubMed] [Google Scholar]
- 13.Natelson BH, Weaver SA, Tseng CL, Ottenweller JE. Spinal fluid abnormalities in patients with chronic fatigue syndrome. Clinical and diagnostic laboratory immunology. 2005;12:52–5. doi: 10.1128/CDLI.12.1.52-55.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Zhou XD, Denny T, Ottenweller JE, Lange G, LaManca JJ, et al. Changes in immune parameters seen in Gulf War veterans but not in civilians with chronic fatigue syndrome. Clinical and diagnostic laboratory immunology. 1999;6:6–13. doi: 10.1128/cdli.6.1.6-13.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maes M, Twisk FN, Johnson C. Myalgic Encephalomyelitis (ME), Chronic Fatigue Syndrome (CFS), and Chronic Fatigue (CF) are distinguished accurately: results of supervised learning techniques applied on clinical and inflammatory data. Psychiatry research. 2012;200:754–60. doi: 10.1016/j.psychres.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 16.Maes M, Twisk FN, Kubera M, Ringel K. Evidence for inflammation and activation of cell-mediated immunity in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS): increased interleukin-1, tumor necrosis factor-alpha, PMN-elastase, lysozyme and neopterin. Journal of affective disorders. 2012;136:933–9. doi: 10.1016/j.jad.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher MA, Zeng XR, Barnes Z, Levis S, Klimas NG. Plasma cytokines in women with chronic fatigue syndrome. J Transl Med. 2009;7:96. doi: 10.1186/1479-5876-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smylie AL, Broderick G, Fernandes H, Razdan S, Barnes Z, Collado F, et al. A comparison of sex-specific immune signatures in Gulf War illness and chronic fatigue syndrome. BMC immunology. 2013;14:29. doi: 10.1186/1471-2172-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skowera A, Cleare A, Blair D, Bevis L, Wessely SC, Peakman M. High levels of type 2 cytokine-producing cells in chronic fatigue syndrome. Clinical and experimental immunology. 2004;135:294–302. doi: 10.1111/j.1365-2249.2004.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skowera A, Hotopf M, Sawicka E, Varela-Calvino R, Unwin C, Nikolaou V, et al. Cellular immune activation in Gulf War veterans. Journal of clinical immunology. 2004;24:66–73. doi: 10.1023/B:JOCI.0000018065.64685.82. [DOI] [PubMed] [Google Scholar]
- 21.Carruthers B, Jain AK, De Meirleir K, Peterson D, Klimas N, Lerner A, et al. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Clinical Working Case Definition, Diagnostic and Treatment Protocols. J Chronic Fatigue Syndrome. 2003;11:1–12. [Google Scholar]
- 22.Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Annals of internal medicine. 1994;121:953–9. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- 23.Centers for Disease C, Prevention. Unexplained illness among Persian Gulf War veterans in an Air National Guard Unit: preliminary report–August 1990-March 1995. MMWR Morbidity and mortality weekly report. 1995;44:443–7. [PubMed] [Google Scholar]
- 24.Steele L. Prevalence and patterns of Gulf War illness in Kansas veterans: association of symptoms with characteristics of person, place, and time of military service. American journal of epidemiology. 2000;152:992–1002. doi: 10.1093/aje/152.10.992. [DOI] [PubMed] [Google Scholar]
- 25.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 26.Ware CF, Crowe PD, Grayson MH, Androlewicz MJ, Browning JL. Expression of surface lymphotoxin and tumor necrosis factor on activated T, B, and natural killer cells. Journal of immunology. 1992;149:3881–8. [PubMed] [Google Scholar]
- 27.Grivennikov SI, Tumanov AV, Liepinsh DJ, Kruglov AA, Marakusha BI, Shakhov AN, et al. Distinct and nonredundant in vivo functions of TNF produced by t cells and macrophages/neutrophils: protective and deleterious effects. Immunity. 2005;22:93–104. doi: 10.1016/j.immuni.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochimica et biophysica acta. 2011;1813:878–88. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 29.Whistler T, Fletcher MA, Lonergan W, Zeng XR, Lin JM, Laperriere A, et al. Impaired immune function in Gulf War Illness. BMC Med Genomics. 2009;2:12. doi: 10.1186/1755-8794-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moss RB, Mercandetti A, Vojdani A. TNF-alpha and chronic fatigue syndrome. Journal of clinical immunology. 1999;19:314–6. doi: 10.1023/a:1020595709352. [DOI] [PubMed] [Google Scholar]
- 31.Rook GA, Zumla A. Gulf War syndrome: is it due to a systemic shift in cytokine balance towards a Th2 profile? Lancet. 1997;349:1831–3. doi: 10.1016/S0140-6736(97)01164-1. [DOI] [PubMed] [Google Scholar]
- 32.Abdi K. IL-12: the role of p40 versus p75. Scandinavian journal of immunology. 2002;56:1–11. doi: 10.1046/j.1365-3083.2002.01101.x. [DOI] [PubMed] [Google Scholar]
- 33.Robbe P, Spierenburg EA, Draijer C, Brandsma CA, Telenga E, van Oosterhout AJ, et al. Shifted T-cell polarisation after agricultural dust exposure in mice and men. Thorax. 2014;69:630–7. doi: 10.1136/thoraxjnl-2013-204295. [DOI] [PubMed] [Google Scholar]
- 34.Harris KM, Ramachandran G, Basu S, Rollins S, Mann D, Cross AS. The IL-23/Th17 axis is involved in the adaptive immune response to Bacillus anthracis in humans. European journal of immunology. 2014;44:752–62. doi: 10.1002/eji.201343784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenkrands I, Vingsbo-Lundberg C, Bundgaard TJ, Lindenstrom T, Enouf V, van der Werf S, et al. Enhanced humoral and cell-mediated immune responses after immunization with trivalent influenza vaccine adjuvanted with cationic liposomes. Vaccine. 2011;29:6283–91. doi: 10.1016/j.vaccine.2011.06.040. [DOI] [PubMed] [Google Scholar]
- 36.DeFuria J, Belkina AC, Jagannathan-Bogdan M, Snyder-Cappione J, Carr JD, Nersesova YR, et al. B cells promote inflammation in obesity and type 2 diabetes through regulation of T-cell function and an inflammatory cytokine profile. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5133–8. doi: 10.1073/pnas.1215840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swindell WR, Johnston A, Voorhees JJ, Elder JT, Gudjonsson JE. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC genomics. 2013;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valeyev NV, Hundhausen C, Umezawa Y, Kotov NV, Williams G, Clop A, et al. A systems model for immune cell interactions unravels the mechanism of inflammation in human skin. PLoS computational biology. 2010;6:e1001024. doi: 10.1371/journal.pcbi.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fry TJ, Mackall CL. Interleukin-7: from bench to clinic. Blood. 2002;99:3892–904. doi: 10.1182/blood.v99.11.3892. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg SA, Sportes C, Ahmadzadeh M, Fry TJ, Ngo LT, Schwarz SL, et al. IL-7 administration to humans leads to expansion of CD8+ and CD4+ cells but a relative decrease of CD4+ T-regulatory cells. Journal of immunotherapy. 2006;29:313–9. doi: 10.1097/01.cji.0000210386.55951.c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellegrini M, Calzascia T, Toe JG, Preston SP, Lin AE, Elford AR, et al. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell. 2011;144:601–13. doi: 10.1016/j.cell.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Konstantinov K, von Mikecz A, Buchwald D, Jones J, Gerace L, Tan EM. Autoantibodies to nuclear envelope antigens in chronic fatigue syndrome. The Journal of clinical investigation. 1996;98:1888–96. doi: 10.1172/JCI118990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Mikecz A, Konstantinov K, Buchwald DS, Gerace L, Tan EM. High frequency of auto antibodies to insoluble cellular antigens in patients with chronic fatigue syndrome. Arthritis and rheumatism. 1997;40:295–305. doi: 10.1002/art.1780400215. [DOI] [PubMed] [Google Scholar]
- 44.Fluge O, Bruland O, Risa K, Storstein A, Kristoffersen EK, Sapkota D, et al. Benefit from B-lymphocyte depletion using the anti-CD20 antibody rituximab in chronic fatigue syndrome. A double-blind and placebo-controlled study. PloS one. 2011;6:e26358. doi: 10.1371/journal.pone.0026358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fluge O, Mella O. Clinical impact of B-cell depletion with the anti-CD20 antibody rituximab in chronic fatigue syndrome: a preliminary case series. BMC neurology. 2009;9:28. doi: 10.1186/1471-2377-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dooms H. Interleukin-7: Fuel for the autoimmune attack. Journal of autoimmunity. 2013;45:40–8. doi: 10.1016/j.jaut.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Oliphant CJ, Barlow JL, McKenzie AN. Insights into the initiation of type 2 immune responses. Immunology. 2011;134:378–85. doi: 10.1111/j.1365-2567.2011.03499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lombardi VC, Khaiboullina SF. Plasmacytoid dendritic cells of the gut: Relevance to immunity and pathology. Clinical immunology. 2014;153:165–77. doi: 10.1016/j.clim.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munoz L, Nomdedeu JF, Lopez O, Carnicer MJ, Bellido M, Aventin A, et al. Interleukin-3 receptor alpha chain (CD123) is widely expressed in hematologic malignancies. Haematologica. 2001;86:1261–9. [PubMed] [Google Scholar]
- 50.Iwakura Y, Nakae S, Saijo S, Ishigame H. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunological reviews. 2008;226:57–79. doi: 10.1111/j.1600-065X.2008.00699.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


