Abstract
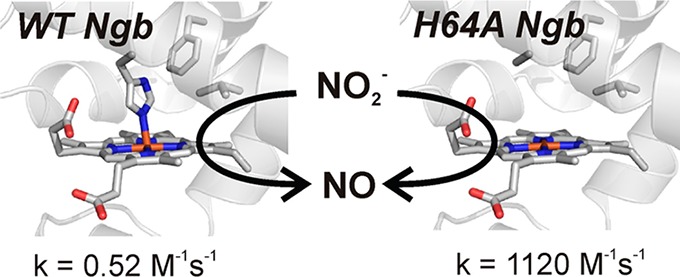
Neuroglobin (Ngb) is a six-coordinate globin that can catalyze the reduction of nitrite to nitric oxide. Although this reaction is common to heme proteins, the molecular interactions in the heme pocket that regulate this reaction are largely unknown. We have shown that the H64L Ngb mutation increases the rate of nitrite reduction by 2000-fold compared to that of wild-type Ngb [Tiso, M., et al. (2011) J. Biol. Chem. 286, 18277–18289]. Here we explore the effect of distal heme pocket mutations on nitrite reduction. For this purpose, we have generated mutations of Ngb residues Phe28(B10), His64(E7), and Val68(E11). Our results indicate a dichotomy in the reactivity of deoxy five- and six-coordinate globins toward nitrite. In hemoglobin and myoglobin, there is a correlation between faster rates and more negative potentials. However, in Ngb, reaction rates are apparently related to the distal pocket volume, and redox potential shows a poor relationship with the rate constants. This suggests a relationship between the nitrite reduction rate and heme accessibility in Ngb, particularly marked for His64(E7) mutants. In five-coordinate globins, His(E7) facilitates nitrite reduction, likely through proton donation. Conversely, in Ngb, the reduction mechanism does not rely on the delivery of a proton from the histidine side chain, as His64 mutants show the fastest reduction rates. In fact, the rate observed for H64A Ngb (1120 M–1 s–1) is to the best of our knowledge the fastest reported for a heme nitrite reductase. These differences may be related to a differential stabilization of the iron–nitrite complexes in five- and six-coordinate globins.
Six-coordinate globins make up a group of heme proteins that are structurally similar to five-coordinate globins like hemoglobin (Hb) and myoglobin (Mb). However, as a notable difference from five-coordinate globins, the distal histidine residue is bound to the heme iron in the ferrous (FeII) and usually in the ferric (FeIII) states, yielding a six-coordinate iron. In these proteins, distal histidine dissociation is required to allow binding of ligands to the heme iron. As mentioned, the distal histidine (E7 in general globin nomenclature) in five-coordinate globins is not in direct contact with the iron atom but plays a critical role in ligand stabilization. A vast literature has explored the importance of this and other residues in the heme distal pocket of Mb with regard to ligand binding, heme autoxidation, and other properties.1−3 Whereas we can expect six-coordinate globins to recapitulate some of the observed behaviors, there is limited information about the role of these residues in six-coordinate globins, and notable differences between both globin families may exist. As an example, we have observed very different behaviors in the nitrite reduction rates of His(E7) mutations.4 A better understanding of the structure–function relationships in six-coordinate globins is important for the elucidation of the often unknown function of these proteins.
The reduction of nitrite to nitric oxide (NO) is a reaction of important physiological consequences that can overcome the decrease in activity of NO synthases under hypoxic conditions.5 Nitrite reduction by hemoglobin has been proposed to regulate blood pressure, hypoxic vasodilation, platelet activation, and the cellular resilience to hypoxia.6−9 Among the proteins catalyzing this reaction, special emphasis has been placed on the role of heme proteins,10,11 with most studies involving the reaction of Hb.12−14 We have previously studied the nitrite reductase activity of other six-coordinate globins.4,15 Our results indicate that the reactivity of the proteins with nitrite spans at least 3 orders of magnitude and, at least in the case of neuroglobin, can increase up to 2500-fold after the removal of the His64(E7) side chain. The rates of nitrite reduction by wild-type six-coordinate globins appear to be higher than those of five-coordinate globins and vary over a wide range.4,15−17 Different hypotheses have been proposed to explain the faster rates in six-coordinate globins and distal histidine mutants. Factors like heme accessibility and redox potential have been related to the nitrite reductase rates.4,14 Studies of Ngb and other globins have shown the ability of heme distal pocket residues to modulate these and other heme properties.4,18−23 Here we present a detailed study of distal pocket mutations at positions Leu29(B10), His64(E7), and Val68(E11) (Figure 1) to elucidate the more relevant factors that modulate nitrite reduction. Our study aims to orient further work with six-coordinate globins to engineer proteins with tailored nitrite reductase activities.
Figure 1.
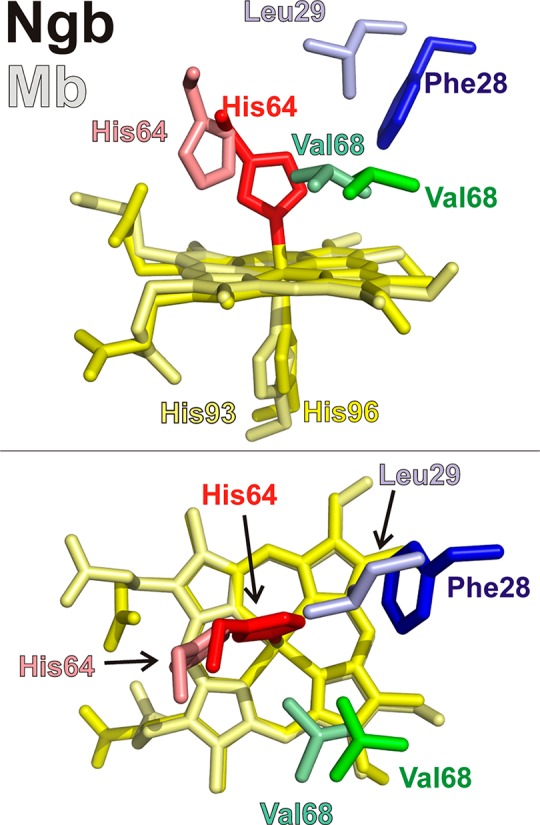
Location of selected heme pocket residues. The relative location of the neuroglobin residues studied in this work is shown. The top panel shows a side view and the bottom panel a top view. Heme moieties and side chains are shown as sticks (Ngb Phe28, blue; Ngb His64, red; Ngb Val68, green; Ngb His96 and Ngb heme, yellow; Mb Leu29, light blue; Mb His64, light red; Mb Val68, light green; Mb His93 and Ngb heme, pale yellow). The proximal histidines (Mb His93 and Ngb His96) have been omitted in the top view for the sake of clarity. Protein Data Bank entries 1OJ6 (Ngb) and 2W6W (Mb) were used.
Materials and Methods
Reagents and Protein Preparation
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. UV–visible spectra and kinetic data were recorded on an HP8453 UV–vis spectrophotometer (Agilent Technologies, Palo Alto, CA) or a Cary 50 spectrophotometer (Agilent Technologies).
Cloning, Expression, and Purification of Recombinant Ngb
Molecular biology was performed using standard techniques. The previously used pET28-Ngb plasmid encoding a His-tagged protein and a thrombin-cleaving site4 was modified so the Ngb coding sequence (originally inserted between the EcoRI and HindIII sites) was inserted between the NcoI and HindIII, sites and the protein was expressed without a His tag. The wild-type Ngb plasmid generated was used as a template for the site-directed mutagenesis reactions to produce the F28W, F28L, F28V, F28H, H64W, H64Q, H64A, H64L, V68A, V68F, and V68I mutants using the QuikChange II site-directed mutagenesis kit (Stratagene, Palo Alto, CA) with the adequate primers. The sequences were confirmed by DNA sequencing at the University of Pittsburgh Genomics and Proteomics Core. The plasmids were transformed into SoluBL21 Escherichia coli cells (Genlantis).
Purification of Ngb proteins was conducted using reported methods with a number of modifications.4,24 Initial cultures were grown overnight at 37 °C in LB broth containing 30 μg/mL kanamycin. The cultures were transferred to 4 L flasks containing 1 L of TB (50 mL of initial culture added per liter of TB) and grown at 37 °C until the OD600 reached 0.8. Then 0.4 mM δ-aminolevulinic acid was added to the medium, and protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside. Protein expression was continued for 20–24 h at 37 °C. Cells were harvested and either lysed or kept at −80 °C until they were processed. Cells were lysed in 50 mM MOPS buffer (pH 7.0) containing 1 mM EDTA, 1 mg/mL lysozyme, 1 mM phenylmethanesulfonyl fluoride, and 0.5 mM dithiothreitol. Cell lysis was accomplished by sonication (8–12 pulses of 30 s at 35% amplitude) using a Misonix S-4000 sonicator (Qsonica, Newtown, CT). The crude lysate was clarified by centrifugation at 20000g for 60 min. To remove nucleic acids from the sample, polyethylenimine was added to the clarified supernatant to a final concentration of 0.1% (v/v). The precipitated nucleic acids were removed by centrifugation at 8000g for 10 min. The supernatant was then loaded into a DE-32 column (Whatman) equilibrated with 50 mM MOPS (pH 7.0) and 10 mM NaCl. The protein was eluted with a gradient from 10 to 100 mM NaCl in 50 mM MOPS (pH 7.0). After the DE-32 anion exchange column, the pooled protein fractions were passed through an Amicon Ultra centrifugal filter (Millipore) with a 50 kDa cutoff to remove high-molecular weight contaminants. The flow-through was concentrated using a 10 kDa cutoff Amicon Ultra centrifugal filter (Millipore) and buffer exchanged to 100 mM phosphate buffer (pH 7.4). This procedure generally yielded >85% pure protein samples as assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Alternatively, instead of using the 50 kDa filters, some samples were further purified by gel chromatography. In these cases, after the DE-32 column samples were concentrated using a 10 kDa cutoff Amicon Ultra centrifugal filter (Millipore). The concentrated sample (2–3 mL) was loaded into a Sephacryl S200HR column (GE Healthcare) equilibrated with 100 mM phosphate buffer (pH 7.4).
Protein chromatography steps were conducted using an ÅKTA Purifier 10 FPLC system (GE Healthcare) with UNICORN software. Protein purity was assessed by SDS–PAGE and UV–visible spectroscopy.
Nitrite Reduction Experiments
The reactions were conducted anaerobically in 3.5 mL optical glass cuvettes (Starna Cells, Atascadero, CA) closed by a screw cap with a silicone septum. Reactions were followed at 37 °C in 100 mM sodium phosphate (pH 7.4). The experiments were conducted in the presence of 2.5 mM sodium dithionite. Under these conditions, the reduction of met-Ngb proceeds at rates of ∼20 s–1. The observed rate of Ngb-NO formation in the reactions is <0.2 s–1. Reactions were initiated by addition of sodium nitrite from an anaerobic stock solution (1–100 mM) to yield the desired final concentration of nitrite (50 μM to 10 mM).
Autoxidation Rates
The preparation of the oxy-Ngb (FeII-O2) species from the met-Ngb (FeIII) samples was as follows. Samples of wild-type or mutant neuroglobins (initial concentration of 10–50 μM) were reduced to the deoxy-Ngb form with excess dithionite at room temperature and then passed through a Sephadex G25 column (PD10, GE Healthcare) equilibrated with sodium phosphate buffer (100 mM, pH 7.4) to remove dithionite. Because of the high oxygen affinity of the protein, this step both removed the excess dithionite and produced, by reaction with oxygen in the buffer, quantitative formation of the oxy-Ngb (FeII-O2). The protein was collected after the sample had passed through the column and mixed with buffer at 37 °C in a 1:2 protein:buffer ratio to ensure an initial temperature of ∼37 °C. Because of the fast oxidation of some neuroglobins, significant oxidation of the oxy-Ngb during the column filtration step occurred in some cases. For the mutants with faster autoxidation rates, the deoxy-Ngb (FeII) species was prepared by dithionite reduction as described above, but the process was conducted in an anaerobic glovebox. The sample was then transferred to a sealed cuvette, and the reaction was started by addition of aerobic buffer. Reaction rates were followed at 37 °C in a Cary 50 spectrophotometer with a thermostated cell holder. Spectral changes were monitored between 450 and 700 nm for slower reactions or between 500 and 600 nm for fast reactions. Scans were taken every 12 or 6 s at a scan rate of 2400 nm/min. The wavelengths showing maximal absorbance changes between oxy-Ngb and met-Ngb species were used to determine the autoxidation rates. Spectral changes were fit to a single-exponential equation using Origin 8.0 (OriginLab Corp., Northampton, MA).
Redox Potentiometry
Redox titrations were performed inside a glovebox (Coy Laboratory Products, Grass Lake, MI) under a nitrogen atmosphere and 1–4% hydrogen to remove residual oxygen with a Pd catalyst. Neuroglobin samples (final concentration of ∼10 μM) were oxidized with potassium ferricyanide and then run through a Sephadex G25 column (PD10, GE Healthcare) equilibrated with anaerobic buffer to remove excess ferricyanide. Spectrophotometric measurements were taken at 25 °C in 100 mM sodium phosphate buffer (pH 7.0). Phenazine methosulfate (Em = 80 mV), 2-methyl-1,4-naphthoquinone (Em = 10 mV), Indigo tetrasulfonate (Em = −46 mV), 2-hydroxy-1,4-naphthoquinone (Em = −137 mV), and anthraquinone 2,6-sulfonate (Em = −184 mV) were used as redox mediators at concentrations between 1 and 5 μM. Potentials were determined with an MI-800/410 redox electrode (Microelectrodes Inc., Bedford, NH) coupled to an Accumet AB15 pH/mV meter. A correction factor of 199 mV at 25 °C was used for the electrode readings. The protein was titrated with sodium dithionite, and the spectra were monitored after each dithionite addition. The one-electron midpoint potentials were determined from the difference spectra. The fraction oxidized for each spectrum was calculated from the maximal difference between the oxidized and reduced spectra (around 430 and 390 nm). Using these data and the corresponding measured potentials (vs SHE) the midpoint potential of the half-reaction can be determined using the Nernst equation (eq 1):
| 1 |
where E is the measured equilibrium potential at each titration point, R is the gas constant (8.314 J mol–1 K–1), T is the experimental temperature in kelvin, n is the number of electrons in the half-reaction, F is the Faraday constant (96485 C/mol), and [oxidized]/[reduced] is the ratio of oxidized to reduced species.
Statistical Analysis
Data were analyzed using Origin 8.0 (OriginLab Corp.), and values are expressed as means ± the standard deviation of the mean.
Results
Protein Expression and Purification
The wild-type and mutant proteins were overexpressed in E. coli as described in Materials and Methods. We did not observe noticeable variations in the expression levels of the mutants compared to that of the wild-type protein. The protein is generally purified in the oxidized form; however, the His64 mutants (in particular H64Q and H64A) were purified mostly in the nitrosyl (Ngb FeII-NO) form as reported for other His64 mutants25 and consistent with intracellular scavenging of NO by these high-affinity mutants.
Spectral Properties
The mutations of neuroglobin residues Phe28(B10) and Val68(E11) did not produce significant changes in the spectral properties of the deoxy-Ngb (FeII) and met-Ngb (FeIII) species. As expected, mutation of the distal histidine residue His64 (E7) changes heme iron coordination form the wild-type six-coordinate form to a five-coordinate environment. This change is apparent in the spectra of the deoxy-Ngb (FeII) species, where the spectra of the six-coordinate heme with two peaks with maxima around 520 and 550 nm are shifted to a five-coordinate species with a single peak around 555 nm, as observed for H64L and H64Q.4,26,27 Remarkably, the spectrum of deoxy-Ngb H64A shows two peaks similar to those of the wild-type protein, and in the case of H64W, a shoulder around 520 nm is observed. The origin of this “residual” six-coordinate environment is unknown but may be due to the stabilization of a solvent molecule in the heme pocket. A similar phenomenon has been described in some cases for Ngb H64L.21,28
Autoxidation Rates of Mutant Neuroglobins
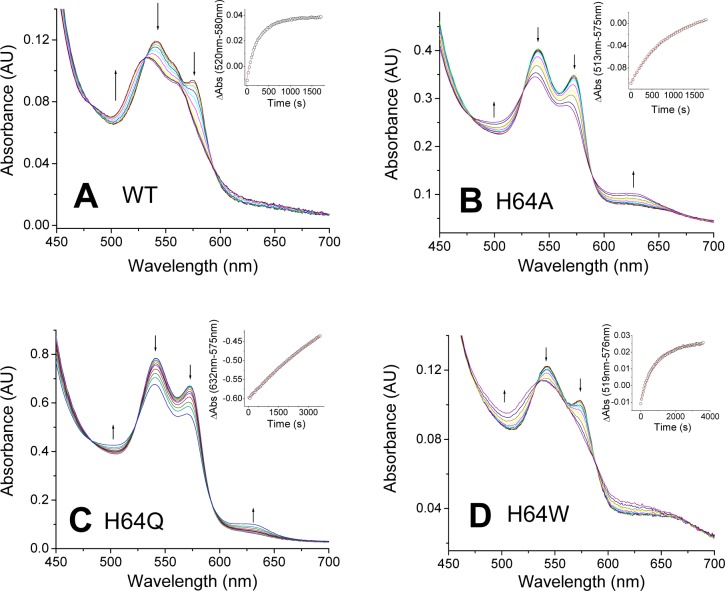
We studied the autoxidation of human wild-type and mutant neuroglobins at 37 °C in 100 mM sodium phosphate (pH 7.4). As previously shown, the oxygenated form of wild-type Ngb (oxy-Ngb, FeII-O2) is unstable and decays, in a single-exponential fashion, to form the oxidized, ferric form (FeIII, met-Ngb)26 (Figure 2A). For wild-type neuroglobin, the oxy-Ngb species decays at a rate of autoxidation of 0.23 min–1 at 37 °C (Table 1), not far from the value of 0.17 min–1 obtained at pH 7.5 and 25 °C by Fago et al.29
Figure 2.
Autoxidation of wild-type Ngb and its His64 mutants. The plots show selected spectra during the time course of autoxidation for each mutant: (A) wild type, (B) H64A, (C) H64Q, and (D) H64W. Arrows indicate the direction of the absorbance change. Insets show the fitting of the decay to a single-exponential equation.
Table 1. Autoxidation Rates of Wild-Type and Mutant Neuroglobins.
| Ngb | kautox (min–1) |
|---|---|
| wild-type | 0.23 ± 0.03 |
| F28W | 3.22 ± 0.42 |
| F28L | 1.76 ± 0.13 |
| F28V | 3.06 ± 0.22 |
| F28H | 11.3 ± 0.7 |
| H64W | 0.076 ± 0.006 |
| H64Q | 0.010 ± 0.002 |
| H64A | 0.066 ± 0.005 |
| V68A | 2.38 ± 0.21 |
| V68F | 0.039 ± 0.001 |
| V68I | 0.122 ± 0.005 |
All the His64 (E7) substitutions resulted in decreased autoxidation rates (Figure 2B–D and Table 1). H64W and H64A induced a 3-fold decrease in autoxidation rates, whereas H64Q was less susceptible to autoxidation, with a >20-fold decrease in autoxidation rate. This result is in agreement with previous observations of Ngb mutants mentioning that H64Q and H64L/K67L were “remarkably stable in the oxygenated form”.29
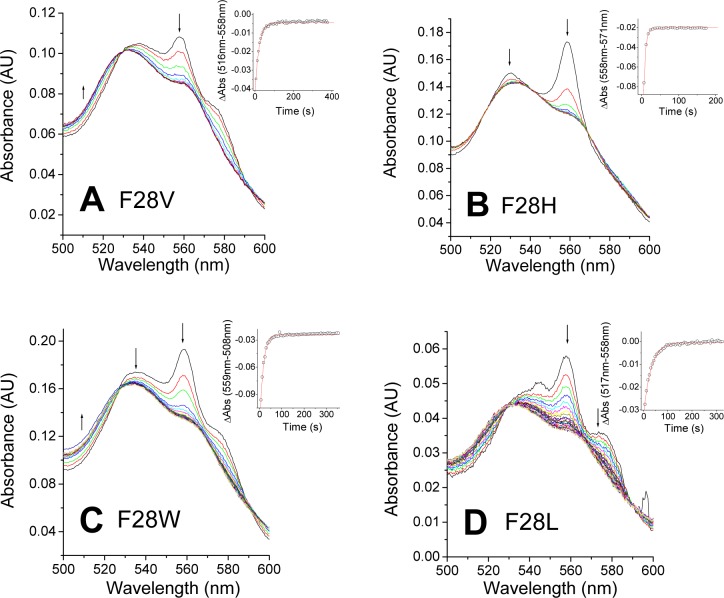
Mutation of the Phe28(B10) significantly decreases the stability of the oxy complex. As observed in Figure 3, the Phe28 mutants hardly form the FeII-O2 complex, with peaks around 545 and 575 nm, and instead, a mixture of deoxy-Ngb (with a peak around 560 nm) and oxy species is formed. The two species decay to the ferric form at apparently similar rates. Studies of myoglobin have shown that two mechanisms can be involved in heme autoxidation, an inner sphere, unimolecular autoxidation mechanism in which the FeO2 complex produces FeIII and superoxide and an outer sphere, bimolecular autoxidation mechanism in which the deoxy heme is oxidized by oxygen without formation of an FeII-O2 complex.18 Our results suggest that the autoxidation of the Phe28 mutants could involve both mechanisms, with a significant contribution of the outer sphere electron transfer mechanism, and no formation of the FeII-O2 complex. Partial formation of the oxy complex can be observed for F28V, F28W, and F28L (panels A, C, and D of Figure 3, respectively). In the case of F28H (Figure 3B), we do not observe significant buildup of oxy species and the deoxy decays to met-Ngb in what appears to be a purely outer sphere electron transfer reaction. Further work is required to clarify these autoxidation mechanisms (see also Figure 1 of the Supporting Information). In all cases, the rates of autoxidation are faster than that of the wild type (WT) (Table 1). The F28L mutant shows a 7-fold increase, whereas the F28W substitution causes a larger, 14-fold increase. The highest autoxidation rate is observed for the F28H mutant, showing a 50-fold increase versus that of the WT (Table 1). The large differences observed for F28H are unexpected given the aromatic character of both side chains. The His side chain may adopt a conformation different from that of the native Phe residue and can also interact with water molecules. In the absence of structural data, the true cause of this divergent behavior remains unclear. The ability of phenylalanine to stabilize the FeII-O2 complex has been shown in other studies of Mb and plant hemoglobins.18,20 In our case, the rate increase does not seem to relate to the size of the side chain as observed for sperm whale myoglobin.18 Our results are more comparable to those of the studies of the B10 residue in plant hemoglobins, where a phenylalanine was also the only residue providing stable oxygen binding.20 Similar results have been also reported in myoglobin, where a phenylalanine in position B10 yields the lower autoxidation rates; however, the reactions are several orders of magnitude slower than those seen in Ngb.18
Figure 3.
Autoxidation of Ngb Phe28 mutants. The plots show selected spectra during the time course of autoxidation for each mutant: (A) F28V, (B) F28H, (C) F28W, and (D) F28L. Arrows indicate the direction of the absorbance change. Insets show the fitting of the decay to a single-exponential equation.
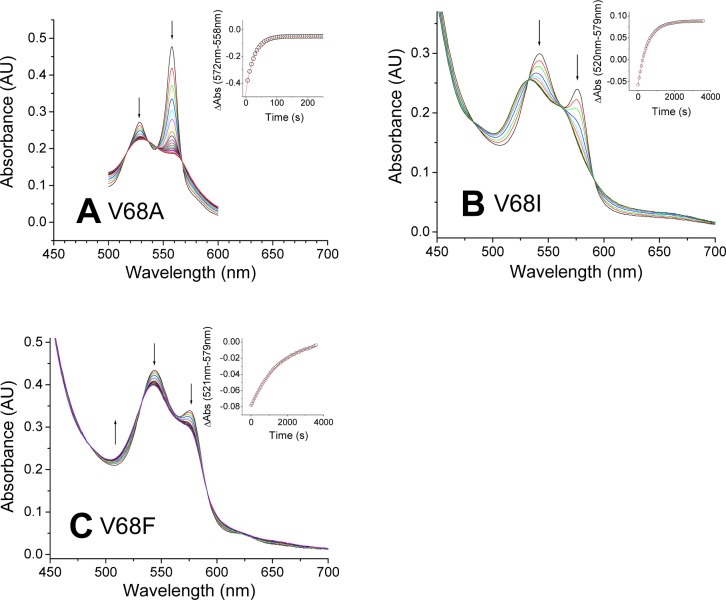
The changes in the Val68(E11) position modified the reactivity toward oxygen in opposite directions. The V68A mutant did not form a stable oxy complex, and as observed in the Phe28 mutants, a fast decay through an outer sphere mechanism is observed (Figure 4A). Conversely, replacement of Val68 with Phe and Ile (panels B and C of Figure 4, respectively) resulted in mutants showing nearly complete formation of an FeII-O2 species. These complexes decay at a rate 2–6-fold lower than those of the wild-type oxy complex (Table 1).
Figure 4.
Autoxidation of Ngb Val68 mutants. The plots show selected spectra during the time course of autoxidation for each mutant: (A) V68A, (B) V68I, and (C) V68F. Arrows indicate the direction of the absorbance change. Insets show the fitting of the decay to a single-exponential equation.
Nitrite Reduction by Mutant Neuroglobins
Neuroglobin, like other heme proteins, can catalyze the reduction of nitrite to form NO according to the scheme
| I |
| II |
As reaction II (rate constant of ∼2 × 108 M–1 s–1)25 is much faster than reaction I, reaction I is rate-limiting. It should be noted that in six-coordinate globins the binding of diatomic ligands is in practice limited by the dissociation rate of the distal histidine (∼0.4 s–1 for Ngb at 20 °C);30 however, the observed rates for Ngb mutants that retain the His64 residue are well below these values. In the presence of excess dithionite, the ferric Ngb formed is reduced back to ferrous Ngb, and the global process is as follows:
| III |
Removal of the distal histidine residue produces a five-coordinate heme that shows increased affinity for ligands.4,25,26 We have shown that the replacement of His64 with Gln or Leu leads to large increases in nitrite reduction rates.4 Here, we wanted to expand these studies by investigating the effect of other mutations of very different physicochemical properties. The observed spectral changes and calculated rate constants are listed in Table 2 and Figures 2–5 of the Supporting Information.
Table 2. Nitrite Reduction Rates of Wild-Type and Mutant Deoxyneuroglobins.
| Ngb | kNitrite (M–1 s–1) |
|---|---|
| wild-type | 0.52 ± 0.19 |
| F28W | 0.76 ± 0.24 |
| F28L | 2.70 ± 0.92 |
| F28V | 2.24 ± 0.33 |
| F28H | 0.37 ± 0.05 |
| H64W | 7.6 ± 1.3 |
| H64Q | 285 ± 23 |
| H64A | 1120 ± 140 |
| V68A | 0.090 ± 0.019 |
| V68F | 1.17 ± 0.34 |
| V68I | 5.07 ± 1.83 |
The H64W mutation retains the aromatic properties of the side chain and is theoretically able to form a hydrogen bond with the heme, but the larger size may cause conformational challenges for such interaction. The H64A mutation eliminates the ability to form hydrogen bonds and decreases the side chain size. The H64W mutation increases nitrite reduction rates by 15-fold (Table 2). Replacement of His64 with Gln leads to a 700-fold increase, and in the case of H64A, a rate more than 2000-fold higher is observed. These changes are consistent with a marked dependence on the size of the side chain at position E7 (Table 2).
F28 mutants show a modest effect in their nitrite reductase rates, with changes in nitrite reduction rates of 0.7–5-fold compared to that of WT Ngb. F28W and F28H mutations retain the aromatic character of the side chain and show the smaller changes. Replacement of the phenylalanine residue with a hydrophobic but not aromatic side chain (F28L and F28V) leads to 4–5-fold rate increases. These results suggest that the size of the side chain is the main factor in the rate.
In the case of the Val68(E11) position, we observe again different trends between the V68A mutation and V68F and V68I mutations. The V68A mutant reduced nitrite at a rate 6-fold slower than that of wild-type Ngb, whereas the other mutations cause rate increases of 2-fold (V68F) and 10-fold (V68I). We do not observe a direct effect of the side chain volume on the nitrite reduction rates as we see in Phe28 or His64; it is possible that significant rearrangements in the heme pocket are required to accommodate the side chain changes. In particular, the results observed for the V68A mutant suggest that some degree of collapse of the heme pocket might occur.
Redox Potential of Mutant Neuroglobins
To study the effect of the redox potential on the kinetic parameters, we determined the redox potential of wild-type neuroglobin and its mutants at 25 °C in 100 mM sodium phosphate (pH 7.0) (Table 3 and Figures 6–9 of the Supporting Information).
Table 3. Redox Potentials of Wild-Type Neuroglobin and Its Mutants.
| Ngb | Em (mV) |
|---|---|
| wild-type | –118 ± 4 |
| F28W | –136 ± 4 |
| F28L | –141 ± 4 |
| F28V | –146 ± 5 |
| F28H | –187 ± 2 |
| H64W | –53 ± 22 |
| H64Q | –62 ± 6 |
| H64A | –119 ± 2 |
| V68A | –107 ± 11 |
| V68F | –121 ± 3 |
| V68I | –126 ± 11 |
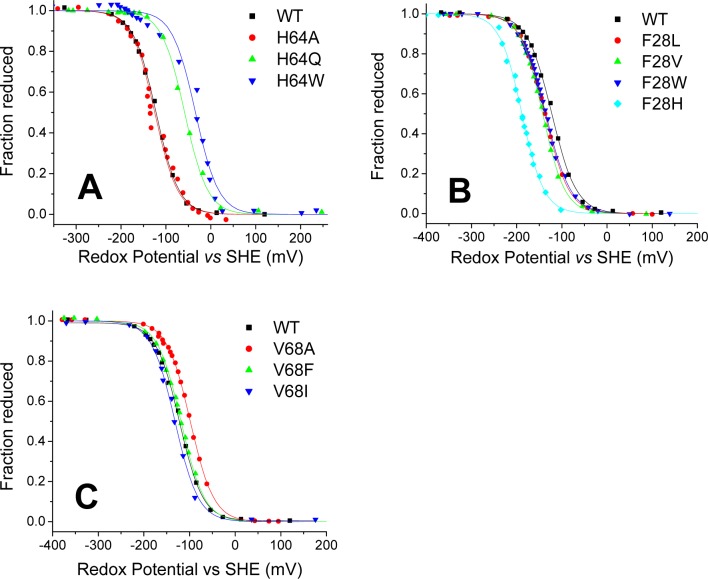
We first determined the redox potential of our wild-type Ngb (Figure 5A and Table 3). Our observed value of −118 ± 4 mV is in good agreement with the reported values of −129 mV26 and −115 mV.21
Figure 5.
Redox potentials of wild-type Ngb and its mutants: (A) His64 mutants, (B) Phe28 mutants, and (C) Val68 mutants. The plots show the fit of the fraction reduced (as determined from absorbance spectra) to the Nernst equation (solid lines). The titration of wild-type Ngb is included in all panels for reference.
Mutation of the distal histidine either had no effect on the redox potential or shifted the midpoint potential to more positive values (Figure 5A and Table 3). The H64A mutation showed little effect, whereas H64Q and H64W caused increases of 56 and 65 mV, respectively. The redox potential for the His(E7)Leu mutation in several six-coordinate globins has been studied by Halder et al.21 They observed a large increase in redox potential for the mutation in rHb1, Cgb, and SynHb, between 113 and 37 mV, but the potential of Ngb H64L was almost unchanged.21 We observe a similar phenomenon with H64A, where the potential is similar to that of wild-type Ngb. However, H64Q and H64W are more positive as expected. In terms of water coordination, we do not observe a correlation with the observed potentials. The ferric form of H64Q is clearly five-coordinate, whereas the spectra of the H64A and H64W ferric forms are consistent with a mainly water-bound heme (Figure 8 of the Supporting Information).
The presence of a water molecule bound to the ferrous heme has been shown to correlate with a more negative redox potential in myoglobin His64(E7) mutants.22 In our study, H64A retains the double peak of the ferrous form, whereas H64Q and H64W form a single-peak ferrous species. The trend in redox potential appears to match the observations in Mb, with H64Q and H64W showing a shift toward more positive values. Similar observations have arisen from the study of His(E7)Leu mutants in six-coordinate globins, where the mutations in Ngb and SynHb retain a six-coordinate character and show changes in redox potential smaller than those of the corresponding mutations in rHb1 or Cgb, where the mutant shows a five-coordinate ferrous spectrum.21
Replacement of the Phe28 residue caused in all cases a decrease in the redox potential of the heme. The shift was moderate for F28W, F28L, and F28V mutants [−18 to −28 mV (Figure 5B and Table 3)] and more pronounced for F28H [−69 mV (Table 3)]. Subtle structural changes may occur in the F28H mutant, as discussed above. The trend observed is similar to that reported in a study of another six-coordinate globin, rice nonsymbiotic hemoglobin 1 (rHb1). In rHb1, the replacement of the Phe(B10) residue with Trp or Leu also induced a decrease in the redox potential of 8 or 30 mV respectively.20
The mutation of the Val68(E11) residue induced a much smaller shift in the redox potential, as compared to the effect of mutation of Phe28(B10) or His64(E7). The values of V68A, V68F, and V68I are within ±11 mV of the wild-type value (Figure 5C and Table 3). Mutation of the homologous residue in myoglobin to polar residues can produce redox potential decreases of >180 mV.31 As the studied mutation retains the hydrophobicity of the wild-type residue, the possible effect of such polar side chains on the redox potential of six-coordinate globins remains uncertain.
Discussion
The mutation of the distal heme pocket residues in neuroglobin offers an opportunity to compare the properties of five-coordinate and six-coordinate heme globins. An extensive literature of protein engineering studies of five-coordinate globins exists. However, the applicability of these studies to six-coordinate globins is unknown. Here we show a systematic study of Ngb focusing on the effects on nitrite reductase rates, autoxidation kinetics, and redox potentials.
Nitrite Reductase Rates and Redox Potential
Previous studies of the nitrite reductase reaction of Hb and Mb showed a trend where the rate of the reaction was faster at more negative heme potentials.14 Subsequent work on six-coordinate globins does not seem to conform to this pattern. Whereas six-coordinate globins have in general redox potentials lower than those of five-coordinate globins,21,26 their nitrite reduction rates are not consistently faster. Ngb and Cgb appear to be slower nitrite reductases than Mb,4,17,32 and six-coordinate plant hemoglobins are faster nitrite reductases than Mb.15,16
The rates of nitrite reduction by Ngb mutants can be plotted versus their heme redox potential (Figure 6). There is no clear relationship between redox potential and nitrite reductase activity. The more striking feature of the plot is the fact that His64(E7) mutations greatly increase the nitrite reduction rate, in a manner independent of the redox potential of the mutant (Figure 6).
Figure 6.
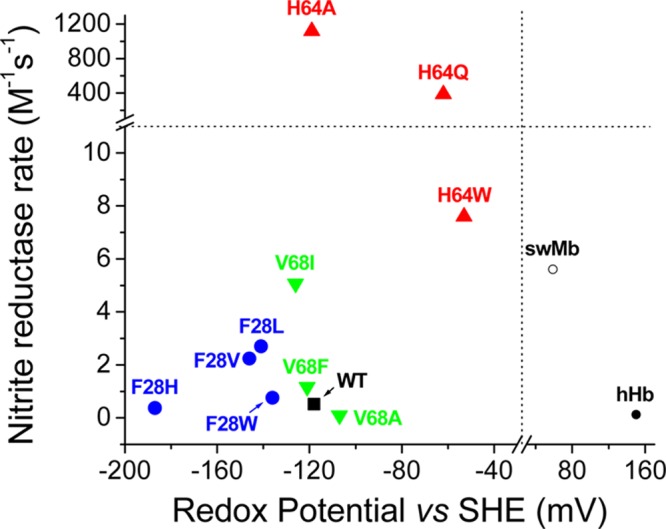
Relationship between the observed nitrite reductase rates and the redox potential. The different symbols denote different Ngb mutations as follows: filled square, wild-type Ngb; filled circles, Phe28 mutants; upward-pointing triangles, His64 mutants; downward-pointing triangles, Val68 mutants; empty circle, wild-type sperm whale myoglobin; filled circle, human hemoglobin. The dashed lines indicate the breaks in the X and Y axes.
In general, the nitrite reduction in Ngb appears to be limited by heme iron accessibility, and this limitation is partly contributed by the heme distal pocket but mainly depends on the ligation of His(E7) to the heme. In this context, it is tempting to speculate that the rate of nitrite reduction in six-coordinate globins will correlate with the distal histidine binding affinity. However, the relationship between the two factors appears to be more complex. A summary of histidine binding parameters for six-coordinate globins is shown in Table 4. As mentioned by Sturms et al.,16 tight binding of distal histidine (KHis > 100) can be observed in slow nitrite reductases, such as wild-type Ngb, but also in fast nitrite reductases, such as SynHb. Conversely, weaker distal histidine binding (KHis < 100) is observed in Cgb (slower reductase) and rHb1 (fast reductase). It is noteworthy that Ngb and Cgb have the slower histidine dissociation rates (Table 4). Actually, histidine dissociation rates are an imperfect yet better indicator of the nitrite reduction rates. Although the concentrations of nitrite used do not seem sufficiently high to overcome the fast histidine binding rates, it is possible that the presence of a nitrite in the heme pocket increases histidine dissociation rates by forming electrostatic and H-bonding interactions with the distal histidine side chain. Different reaction mechanisms for five- and six-coordinate globins may exist; this possibility is discussed below.
Table 4. Distal Histidine Binding Constants, Nitrite Reductase Rate Constants, and Redox Potentials for Selected Globinsa.
| distal
histidine binding |
|||||
|---|---|---|---|---|---|
| protein | kbinding (s–1) | kdissoc (s–1) | KHis | KNitrite (M–1 s–1) | Em (mV) |
| Ngb (human) | 2000b | 4.5b | 444b | 0.12c | –118d |
| Cgb (human) | 200e | 2e | 100e | 0.14f | –28g |
| SynHb | 4200h | 14h | 300h | 27i | –195g |
| RHb1 | 75h | 40h | 1.9h | 40i | –143g |
| AtHb1 | 230j | 110j | 2.1j | 19.8k | NDm |
| AtHb2 | 1600j | 38j | 42j | 4.9k | NDm |
| SwMb | NAn | NAn | NAn | 5.6c | –59l |
Histidine dissociation determined at 20 °C in 100 mM sodium phosphate buffer (pH 7.0). Nitrite reductase rates determined at 25 °C in 100 mM sodium phosphate buffer (pH 7.4), except for those Cgb, SynHb, and RHb1, which were determined at pH 7.0. All redox potentials determined at 25 °C in 100 mM sodium phosphate buffer (pH 7.0).
From ref (26).
From ref (4).
From this work.
From ref (45).
From ref (4).
From ref (21).
From ref (46).
From ref (16).
From ref (47).
From ref (15).
From ref (31).
Not determined.
Not applicable.
Autoxidation Rates and Redox Potential
We have investigated the relationship between the observed autoxidation rates for Ngb mutants and the redox potential of the heme group (Figure 7). In this case, we can observe a reasonable correlation between the autoxidation rates and the redox potential. The mechanisms of globin autoxidation have been discussed in detail by Shikama.33,34 The potential for formation of superoxide from oxygen is −330 mV; thus, theoretically a heme potential of −330 mV would be needed to provide the driving force necessary for the reaction to occur spontaneously.33,34 It follows that an efficient oxygen carrier protein will experience an evolutionary pressure to minimize this side reaction, and consequently, proteins like Hb and Mb have significantly positive reduction potentials (59 mV for Mb31 and 150 mV for Hb35). In general, lower redox potentials have been determined for Ngb [−118 mV (Table 3)] and other six-coordinate globins.21 In a hypothetical scenario in which the redox potential constitutes the sole driving force of the reaction, the more negative the heme redox potential, the faster the heme autoxidation (and concomitant oxygen reduction). Our results fit well into this overall scheme, with the proteins with more negative potential showing faster autoxidation rates (Figure 7). The inset of Figure 7 is consistent with a change in the activation energy of the reaction that depends linearly of the redox potential, as dictated by the Arrhenius equation. It is remarkable that this observation requires that all other factors (including but not limited to ligand accessibility and pH effects) have a limited influence on the rates. We have also included in the analysis a subset of four E7Mb mutants for which the redox potential and autoxidation rates are available (Table 1 of the Supporting Information).18,22 We do not observe a consistent behavior for the mutant proteins and wild-type Mb; however, a separate analysis of the mutants alone yields a linear fit, almost parallel to the neuroglobin data (Figure 7). This indicates that for some Mb mutants a situation similar to that of Ngb, with the redox potential dominating the rate, is plausible when the distal pocket is enlarged and does not provide much ligand stabilization. It is unlikely that this situation can be generalized to other Mb mutants, as the redox potential of the Mb mutants is not far from wild-type Mb values and most mutations do not yield deviations from wild-type Mb autoxidation rates as large as those of the E7 mutants (Table 5).
Figure 7.
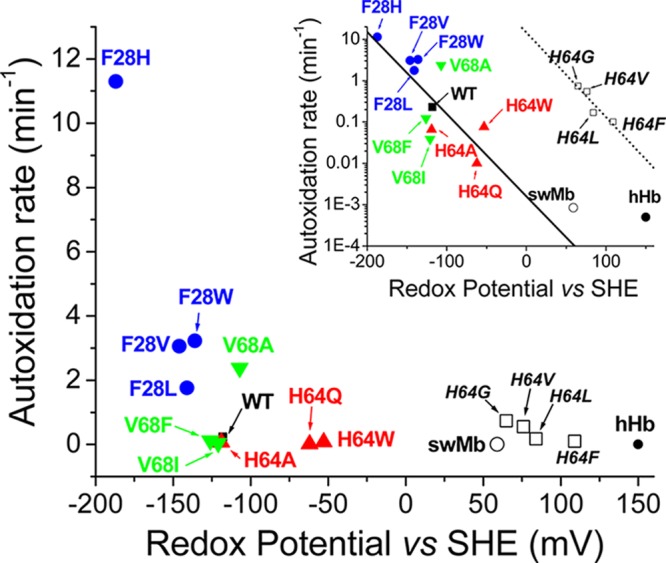
Relationship between the observed autoxidation rates and the redox potential. The different symbols denote different Ngb mutations as follows: filled square, wild-type Ngb; filled circles, Phe28 mutants; upward-pointing triangles, His64 mutants; downward-pointing triangles, Val68 mutants; empty circle, wild-type sperm whale myoglobin; filled circle, human hemoglobin. The empty squares denote data for sperm whale myoglobin mutants; mutations are indicated in italics. The inset shows the same points but using a logarithmic scale for the autoxidation rates. The solid line denotes the best fit of the neuroglobin data points to a linear equation; the dotted line denotes the best fit of the sperm whale myoglobin data points (except for that of the wild type) to a linear equation.
Table 5. Autoxidation Rates for Neuroglobin and Myoglobin Mutants.
| Ngb | kautoxa (min–1) | swMb equivalent | kautoxb (h–1) |
|---|---|---|---|
| wild-type | 0.23 | L29F | 0.005 |
| F28W | 3.22 | L29W | NDc |
| F28L | 1.76 | wild-type | 0.051 |
| F28V | 3.06 | L29V | 0.23 |
| F28H | 11.3 | L29H | NDc |
| F28A | NDc | L29A | 0.24 |
| H64W | 0.076 | H64W | NDc |
| H64Q | 0.010 | H64Q | 0.21 |
| H64A | 0.066 | H64A | 58 |
| H64L | NDc | H64L | 10 |
| H64G | NDc | H64G | 44 |
| H64V | NDc | H64V | 33 |
| H64T | NDc | H64T | 54 |
| H64F | NDc | H64F | 6 |
| V68A | 2.38 | V68A | 0.26 |
| V68F | 0.039 | V68F | 0.069 |
| V68I | 0.122 | V68I | 0.75 |
| V68L | NDc | V68L | 0.10 |
Values determined at 37 °C in 100 mM sodium phosphate buffer (pH 7.4) (this work).
Values determined at 37 °C in 100 mM potassium phosphate buffer (pH 7.0).18
Not determined.
Effects of Mutations on Ngb versus Mb
Autoxidation rates of a variety of Mb mutants are available.18 Notwithstanding the heterogeneity within five-coordinate globins,36 this offers a reasonable framework for comparison the effect of mutations on five- and six-coordinate globins. A comparison of the observed values is given in Table 5.
Mutations of the B10 residue (Phe28 in Ngb and Leu29 in swMb) indicate a common pattern for both globins. In both cases, the phenylalanine provides the maximal stability for the FeII-O2 complex (Table 5). Studies of the six-coordinate rHb1 have also indicated that a phenylalanine in position B10 is also the residue allowing for a slower autoxidation rate.20 The conservation of this residue in Ngb sequences can be related to a role of the protein in O2 transport and/or storage, but other functions such as NO scavenging would also benefit from a stable oxy species.20 In any case, we observe that other mutations can also increase FeII-O2 complex stability in Ngb, notably His64 substitutions (Tables 1 and 5). However, His64 and Val68 (and Phe28) are strictly conserved in the available Ngb sequences. This observation suggests that the O2 complex stability of wild-type Ngb is enough for its biological function, and other factors such as six-coordination are apparently more relevant to function. Remarkably, not all six-coordinate globins conserve a Phe residue at position B10 (for example, a leucine residue is conserved in Cgbs); this suggests that the stability of the FeII-O2 species has not been optimized during the evolution of six-coordinate globins.
Mutations of the residue in position E11 had an effect on Ngb autoxidation rates smaller than the effect of the equivalent mutations in swMb (Table 5). The Ngb V68A mutation was an exception to this trend, but on the basis of the slow nitrite reduction by this mutant, we hypothesize that substantial rearrangements of the heme environment, with a reduction of the accessible volume of the heme pocket, occur in this mutant.
Given the particular role of the E7 residue in six-coordinate globins, the fact that the differences between Ngb and Mb E7 mutants are maximal is not completely unexpected. Whereas this mutation increases autoxidation rates up to 1000-fold in Mb, all the Ngb mutants show improved FeII-O2 complex stability. The increase in autoxidation rates is not completely general to five-coordinate globins; in fact, the His(E7)Leu mutant of soybean leghemoglobin shows only a modest increase in its autoxidation rate.36 In Mb, the E7 histidine is both stabilizing bound O2 and preventing its protonation (which leads to fast autoxidation via the formation of FeII-O2H+ and fast dissociation to FeIII and HO2). This stabilized FeII-O2 species is reportedly inert toward autoxidation.18 The CO-bound Ngb structure shows the His side chain close to the CO molecule,37 and it is thus conceivable that the His E7 in Ngb could fulfill a similar role. However, our observed rates for His(E7) mutants indicate that a histidine is not necessary for the stabilization of the FeII-O2 species in Ngb. The differences between Ngb and Mb autoxidation reactions could also be mechanistic. Two different mechanisms (unimolecular vs bimolecular) can be involved in the autoxidation reaction.18 The swMb data show a reasonable correlation between autoxidation rates and oxygen dissociation constants; the correlation is consistent with a unimolecular autoxidation mechanism being predominant at higher oxygen concentrations.18 We investigated the nature of the autoxidation reaction mechanism in Ngb (Figure 1 of the Supporting Information). Our experiments indicate that wild-type Ngb and the F28W mutant show rates consistent with a unimolecular mechanism with little or no contribution from the bimolecular mechanism, unlike the mixed mechanism generally observed in swMbs.18 In the case of the H64A mutant, a behavior more consistent with a mixture of uni- and bimolecular mechanisms is observed. The contribution of a bimolecular autoxidation mechanism to the observed rate declines as the oxygen concentration increases and approaches zero at high O2 concentrations, whereas the unimolecular mechanism rate increases in a hyperbolic manner, reaching a saturation rate. As the autoxidation rates seem to reach a plateau at oxygen levels higher than 50 μM for the Ngbs studied, we expect the unimolecular mechanism to prevail under normoxic conditions for Ngb as it does for Mb. Therefore, a reasonable explanation for the opposite effect observed for the autoxidation rates of E7 mutants in Mb and Ngb can be traced to the opposite changes in oxygen affinity. The E7 mutants of swMb show a decreased affinity for oxygen,18 but the data for Ngb mutants H(E7)Q and H(E7)V indicate that the oxygen affinity is increased with respect to that of the wild-type protein.29 We speculate that oxygen affinities for the Ngb mutants will also show a correlation with their autoxidation rates, indicating that the dissociation of oxygen or superoxide from the heme is regulated by similar constraints.
Two Mechanisms of Nitrite Reduction?
Our results further indicate a dichotomy in the reaction of deoxy five- and six-coordinate globins toward nitrite. In Hb and Mb, there is a correlation between faster rates and more negative potentials.14 However, in Ngb, there is a marked increase in reaction rates as the distal pocket size increases, and the redox potential shows a poor relationship with reaction rate constants. These observations are puzzling; if the reaction of nitrite were limited in all cases by heme pocket size, Hb and Mb should show rates faster than those of six-coordinate globins. Differences in reaction mechanism may exist; Perissinotti et al. studied the nitrite reaction with deoxyhemoglobin by computational methods and showed that histidine protonation could be a limiting step in the reaction.38 Our studies showed that swMb mutants His(E7)Ala and His(E7)Leu are indeed poor nitrite reductases (Table 1 of the Supporting Information).4 In the case of horse Mb, the His(E7)Val mutant causes a 15-fold decrease in nitrite reduction rate.39 Conversely, our results indicate that for Ngb the reduction mechanism does not rely on the delivery of a proton from the His side chain, as His64 mutants show the fastest reduction rates. If the proton is delivered from a water molecule, we would expect that a mutant such as F28H or H64Q could help to keep that water molecule in place, but each of those is not faster than F28L or H64A, respectively.
The reaction of nitrite with deoxyglobins requires the formation of an elusive FeII-NO2– intermediate.40 The geometry of the intermediate is a matter of discussion, as the nitrite molecule can be bound through the nitrogen atom (FeII-N-nitro), one oxygen atom (FeII-O-nitrito), or even a bidentate (FeII-O,O-nitrito) species.41−44 Interestingly, the product of a nitrogen-bound nitrite will be FeIII-NO,4,38 whereas an oxygen-bound nitrite will yield an FeIII-OH intermediate.38 Recent studies by Silaghi-Dumitrescu et al.40 indicate that in the reaction of hemoglobin an FeIII-NO intermediate species can be detected, therefore implying an FeII-N-nitro intermediate. It is conceivable that the subtle structural changes between five- and six-coordinate globins can lead to a difference in the stabilization of the FeII-NO2– species. The possibility of five-coordinate globins favoring N-nitro binding modes and six-coordinate globins favoring a more reactive O-nitrito species deserves further study.
Glossary
Abbreviations
- Hb
red blood cell hemoglobin
- Mb
myoglobin
- Ngb
neuroglobin
- Cgb
cytoglobin
- rHb1
rice nonsymbiotic hemoglobin 1
- B10
10th amino acid of helix B
- E7
seventh amino acid of helix E
- E11
11th amino acid of helix E
- KHis
equilibrium constant for the distal histidine binding equilibrium (rate of His binding divided by rate of His dissociation)
- NO
nitric oxide
- SHE
standard hydrogen electrode
- SynHb
Synechocystis hemoglobin
- swMb
sperm whale myoglobin.
Supporting Information Available
Sample spectra and traces for the nitrite reduction reactions of wild-type neuroglobin and mutants, spectral data for the redox potential determinations of wild-type neuroglobin and mutants, autoxidation rates for WT Ngb, F28W, and H64A at several oxygen concentrations, and a compilation of autoxidation rates, nitrite reduction rates, and redox potential values determined for myoglobin mutants. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by National Institutes of Health Grants HL098032, HL096973, and DK085852 and funding from the Institute for Transfusion Medicine and the Hemophilia Center of Western Pennsylvania (to M.T.G.) and by funding from the Competitive Medical Research Fund of the University of Pittsburgh Medical Center Health System (to J.T.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Olson J. S.; Phillips G. N. (1996) Kinetic pathways and barriers for ligand binding to myoglobin. J. Biol. Chem. 271, 17593–17596. [DOI] [PubMed] [Google Scholar]
- Raven E. L.; Mauk A. G. (2001) Chemical reactivity of the active site of myoglobin. Adv. Inorg. Chem. 51, 1–49. [Google Scholar]
- Brunori M. (2010) Myoglobin strikes back. Protein Sci. 19, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso M.; Tejero J.; Basu S.; Azarov I.; Wang X.; Simplaceanu V.; Frizzell S.; Jayaraman T.; Geary L.; Shapiro C.; Ho C.; Shiva S.; Kim-Shapiro D. B.; Gladwin M. T. (2011) Human neuroglobin functions as a redox-regulated nitrite reductase. J. Biol. Chem. 286, 18277–18289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg J. O.; Weitzberg E.; Gladwin M. T. (2008) The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discovery 7, 156–167. [DOI] [PubMed] [Google Scholar]
- Cosby K.; Partovi K. S.; Crawford J. H.; Patel R. P.; Reiter C. D.; Martyr S.; Yang B. K.; Waclawiw M. A.; Zalos G.; Xu X.; Huang K. T.; Shields H.; Kim-Shapiro D. B.; Schechter A. N.; Cannon R. O. III; Gladwin M. T. (2003) Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat. Med. 9, 1498–1505. [DOI] [PubMed] [Google Scholar]
- Gladwin M. T.; Schechter A. N.; Kim-Shapiro D. B.; Patel R. P.; Hogg N.; Shiva S.; Cannon R. O. III; Kelm M.; Wink D. A.; Espey M. G.; Oldfield E. H.; Pluta R. M.; Freeman B. A.; Lancaster J. R. Jr.; Feelisch M.; Lundberg J. O. (2005) The emerging biology of the nitrite anion. Nat. Chem. Biol. 1, 308–314. [DOI] [PubMed] [Google Scholar]
- Velmurugan S.; Kapil V.; Ghosh S. M.; Davies S.; McKnight A.; Aboud Z.; Khambata R. S.; Webb A. J.; Poole A.; Ahluwalia A. (2013) Antiplatelet effects of dietary nitrate in healthy volunteers: Involvement of cGMP and influence of sex. Free Radical Biol. Med. 65, 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti P.; Tejero J.; Gladwin M. T. (2013) Evidence mounts that red cells and deoxyhemoglobin can reduce nitrite to bioactive NO to mediate intravascular endocrine NO signaling: Commentary on “Anti-platelet effects of dietary nitrate in healthy volunteers: Involvement of cGMP and influence of sex”. Free Radical Biol. Med. 65, 1518–1520. [DOI] [PubMed] [Google Scholar]
- Tejero J.; Gladwin M. T. (2014) The globin superfamily: Functions in nitric oxide formation and decay. Biol. Chem. 395, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladwin M. T.; Kim-Shapiro D. B. (2008) The functional nitrite reductase activity of the heme-globins. Blood 112, 2636–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M. P.; Pickering R. A.; DeWeert T. M.; Hoekstra J. W.; Pater D. (1981) Kinetics and mechanism of the oxidation of human deoxyhemoglobin by nitrites. J. Biol. Chem. 256, 12393–12398. [PubMed] [Google Scholar]
- Gladwin M. T.; Grubina R.; Doyle M. P. (2009) The new chemical biology of nitrite reactions with hemoglobin: R-state catalysis, oxidative denitrosylation, and nitrite reductase/anhydrase. Acc. Chem. Res. 42, 157–167. [DOI] [PubMed] [Google Scholar]
- Huang Z.; Shiva S.; Kim-Shapiro D. B.; Patel R. P.; Ringwood L. A.; Irby C. E.; Huang K. T.; Ho C.; Hogg N.; Schechter A. N.; Gladwin M. T. (2005) Enzymatic function of hemoglobin as a nitrite reductase that produces NO under allosteric control. J. Clin. Invest. 115, 2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso M.; Tejero J.; Kenney C.; Frizzell S.; Gladwin M. T. (2012) Nitrite reductase activity of nonsymbiotic hemoglobins from Arabidopsis thaliana. Biochemistry 51, 5285–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturms R.; DiSpirito A. A.; Hargrove M. S. (2011) Plant and cyanobacterial hemoglobins reduce nitrite to nitric oxide under anoxic conditions. Biochemistry 50, 3873–3878. [DOI] [PubMed] [Google Scholar]
- Petersen M. G.; Dewilde S.; Fago A. (2008) Reactions of ferrous neuroglobin and cytoglobin with nitrite under anaerobic conditions. J. Inorg. Biochem. 102, 1777–1782. [DOI] [PubMed] [Google Scholar]
- Brantley R. E. Jr.; Smerdon S. J.; Wilkinson A. J.; Singleton E. W.; Olson J. S. (1993) The mechanism of autooxidation of myoglobin. J. Biol. Chem. 268, 6995–7010. [PubMed] [Google Scholar]
- Dou Y.; Olson J. S.; Wilkinson A. J.; Ikeda-Saito M. (1996) Mechanism of hydrogen cyanide binding to myoglobin. Biochemistry 35, 7107–7113. [DOI] [PubMed] [Google Scholar]
- Smagghe B. J.; Kundu S.; Hoy J. A.; Halder P.; Weiland T. R.; Savage A.; Venugopal A.; Goodman M.; Premer S.; Hargrove M. S. (2006) Role of phenylalanine B10 in plant nonsymbiotic hemoglobins. Biochemistry 45, 9735–9745. [DOI] [PubMed] [Google Scholar]
- Halder P.; Trent J. T. III; Hargrove M. S. (2007) Influence of the protein matrix on intramolecular histidine ligation in ferric and ferrous hexacoordinate hemoglobins. Proteins 66, 172–182. [DOI] [PubMed] [Google Scholar]
- VanDyke B. R.; Saltman P.; Armstrong F. A. (1996) Control of myoglobin electron-transfer rates by the distal (nonbound) histidine residue. J. Am. Chem. Soc. 118, 3490–3492. [Google Scholar]
- Brancaccio A.; Cutruzzola F.; Allocatelli C. T.; Brunori M.; Smerdon S. J.; Wilkinson A. J.; Dou Y.; Keenan D.; Ikeda-Saito M.; Brantley R. E. Jr.; Olson J. S. (1994) Structural factors governing azide and cyanide binding to mammalian metmyoglobins. J. Biol. Chem. 269, 13843–13853. [PubMed] [Google Scholar]
- Dewilde S.; Mees K.; Kiger L.; Lechauve C.; Marden M. C.; Pesce A.; Bolognesi M.; Moens L. (2008) Expression, purification, and crystallization of neuro- and cytoglobin. Methods Enzymol. 436, 341–357. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer S.; Dewilde S.; Kiger L.; Nistor S. V.; Goovaerts E.; Marden M. C.; Moens L. (2003) Nitric oxide binding properties of neuroglobin. A characterization by EPR and flash photolysis. J. Biol. Chem. 278, 4919–4925. [DOI] [PubMed] [Google Scholar]
- Dewilde S.; Kiger L.; Burmester T.; Hankeln T.; Baudin-Creuza V.; Aerts T.; Marden M. C.; Caubergs R.; Moens L. (2001) Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J. Biol. Chem. 276, 38949–38955. [DOI] [PubMed] [Google Scholar]
- Astudillo L.; Bernad S.; Derrien V.; Sebban P.; Miksovska J. (2012) Conformational dynamics in human neuroglobin: Effect of His64, Val68, and Cys120 on ligand migration. Biochemistry 51, 9984–9994. [DOI] [PubMed] [Google Scholar]
- Nienhaus K.; Kriegl J. M.; Nienhaus G. U. (2004) Structural dynamics in the active site of murine neuroglobin and its effects on ligand binding. J. Biol. Chem. 279, 22944–22952. [DOI] [PubMed] [Google Scholar]
- Fago A.; Hundahl C.; Dewilde S.; Gilany K.; Moens L.; Weber R. E. (2004) Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J. Biol. Chem. 279, 44417–44426. [DOI] [PubMed] [Google Scholar]
- Brunori M.; Giuffre A.; Nienhaus K.; Nienhaus G. U.; Scandurra F. M.; Vallone B. (2005) Neuroglobin, nitric oxide, and oxygen: Functional pathways and conformational changes. Proc. Natl. Acad. Sci. U.S.A. 102, 8483–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan R.; Zewert T. E.; Gray H. B.; Boxer S. G. (1989) Effects of buried ionizable amino acids on the reduction potential of recombinant myoglobin. Science 243, 69–72. [DOI] [PubMed] [Google Scholar]
- Li H.; Hemann C.; Abdelghany T. M.; El-Mahdy M. A.; Zweier J. L. (2012) Characterization of the mechanism and magnitude of cytoglobin-mediated nitrite reduction and nitric oxide generation under anaerobic conditions. J. Biol. Chem. 287, 36623–36633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama K. (1998) The molecular mechanism of autoxidation for myoglobin and hemoglobin: A venerable puzzle. Chem. Rev. 98, 1357–1373. [DOI] [PubMed] [Google Scholar]
- Shikama K. (2006) Nature of the FeO2 bonding in myoglobin and hemoglobin: A new molecular paradigm. Prog. Biophys. Mol. Biol. 91, 83–162. [DOI] [PubMed] [Google Scholar]
- Antonini E.; Wyman J.; Brunori M.; Taylor J. F.; Rossi-Fanelli A.; Caputo A. (1964) Studies on the Oxidation-Reduction Potentials of Heme Proteins. I. Human Hemoglobin. J. Biol. Chem. 239, 907–912. [PubMed] [Google Scholar]
- Hargrove M. S.; Barry J. K.; Brucker E. A.; Berry M. B.; Phillips G. N.; Olson J. S.; Arredondo-Peter R.; Dean J. M.; Klucas R. V.; Sarath G. (1997) Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J. Mol. Biol. 266, 1032–1042. [DOI] [PubMed] [Google Scholar]
- Vallone B.; Nienhaus K.; Matthes A.; Brunori M.; Nienhaus G. U. (2004) The structure of carbonmonoxy neuroglobin reveals a heme-sliding mechanism for control of ligand affinity. Proc. Natl. Acad. Sci. U.S.A. 101, 17351–17356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissinotti L. L.; Marti M. A.; Doctorovich F.; Luque F. J.; Estrin D. A. (2008) A microscopic study of the deoxyhemoglobin-catalyzed generation of nitric oxide from nitrite anion. Biochemistry 47, 9793–9802. [DOI] [PubMed] [Google Scholar]
- Yi J.; Heinecke J.; Tan H.; Ford P. C.; Richter-Addo G. B. (2009) The distal pocket histidine residue in horse heart myoglobin directs the O-binding mode of nitrite to the heme iron. J. Am. Chem. Soc. 131, 18119–18128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silaghi-Dumitrescu R.; Svistunenko D. A.; Cioloboc D.; Bischin C.; Scurtu F.; Cooper C. E. (2014) Nitrite binding to globins: Linkage isomerism, EPR silence and reductive chemistry. Nitric Oxide 42, 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.; Safo M. K.; Richter-Addo G. B. (2008) The nitrite anion binds to human hemoglobin via the uncommon O-nitrito mode. Biochemistry 47, 8247–8249. [DOI] [PubMed] [Google Scholar]
- Copeland D. M.; Soares A. S.; West A. H.; Richter-Addo G. B. (2006) Crystal structures of the nitrite and nitric oxide complexes of horse heart myoglobin. J. Inorg. Biochem. 100, 1413–1425. [DOI] [PubMed] [Google Scholar]
- Ford P. C. (2010) Reactions of NO and nitrite with heme models and proteins. Inorg. Chem. 49, 6226–6239. [DOI] [PubMed] [Google Scholar]
- Tsou C. C.; Yang W. L.; Liaw W. F. (2013) Nitrite activation to nitric oxide via one-fold protonation of iron(II)-O,O-nitrito complex: Relevance to the nitrite reductase activity of deoxyhemoglobin and deoxyhemerythrin. J. Am. Chem. Soc. 135, 18758–18761. [DOI] [PubMed] [Google Scholar]
- Hamdane D.; Kiger L.; Dewilde S.; Green B. N.; Pesce A.; Uzan J.; Burmester T.; Hankeln T.; Bolognesi M.; Moens L.; Marden M. C. (2003) The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin. J. Biol. Chem. 278, 51713–51721. [DOI] [PubMed] [Google Scholar]
- Smagghe B. J.; Sarath G.; Ross E.; Hilbert J. L.; Hargrove M. S. (2006) Slow ligand binding kinetics dominate ferrous hexacoordinate hemoglobin reactivities and reveal differences between plants and other species. Biochemistry 45, 561–570. [DOI] [PubMed] [Google Scholar]
- Smagghe B. J.; Hoy J. A.; Percifield R.; Kundu S.; Hargrove M. S.; Sarath G.; Hilbert J. L.; Watts R. A.; Dennis E. S.; Peacock W. J.; Dewilde S.; Moens L.; Blouin G. C.; Olson J. S.; Appleby C. A. (2009) Correlations between Oxygen Affinity and Sequence Classifications of Plant Hemoglobins. Biopolymers 91, 1083–1096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.