Abstract
Three dynamic models for the investigation of in vitro biofilm formation are described in this chapter. In the 6-well plate assay presented here, the placing of the plate on a rotating platform provides shear, thereby making the system dynamic with respect to the static microtiter assay.
The second reported model, especially suitable for harvesting high amounts of cells for transcriptomic or proteomic investigations, is based on numerous glass beads placed in a flask incubated with shaking on a rotating platform, thus increasing the surface area for biofilm formation. Finally, the flow-cell system, that is the driving model for elucidating the biofilm-forming process in vitro as well as the biofilm tolerance towards antibiotics and host defense components, is illustrated here.
Keywords: 6-Well plate assay, Glass beads, Flow-cell system, Confocal laser scanning microscopy
1 Introduction
Surface-attached biofilms can be studied using many different experimental setups. In this chapter we have chosen to describe three dynamic models for in vitro biofilm formation. All have advantages and disadvantages depending on the experiment and the desired outcome.
The first model is based on 6-well microtiter plates. Microtiter biofilm model was originally a static assay that was developed to study the ability of coagulase-negative staphylococci to adhere to surfaces [1]. At present time two versions of high-throughput screening, static microtiter plate assays exist, one where the biofilm is formed in the wells of the plate [2] and the Calgary Biofilm Device [3], which are 96 (or more) pegs which fits into microtiter plates. These assays can be used to test for biomass buildup, by staining the biomass using crystal violet. Crystal violet staining on the other hand does not discriminate between live and dead bacteria. To test whether the bacteria are being killed in these assays, the bacteria needs to be plated for CFU. These static assays are typically used for screening numerous mutants for biofilm forming capacity [4] or for the first screening of antibiofilm drugs both in terms of killing and dispersal [5]. The 6-well plate assays we present here are still very easy to use and control; however, the embedded cover slip can be removed and examined by microscopy, and placing the plate on a rotating platform provides shear, thereby making the system more dynamic. This is an advantage when investigating more than just biomass buildup. The second model we present is based on numerous glass beads in a flask incubated with shaking on a rotating platform. This increases the surface area for biofilm formation but still has the dynamic shear forces. This is especially important for harvesting high amounts of cells for transcriptomic or proteomic investigations.
To study biofilm development in real time and the different stages of formation and behavior of flow-cell systems [6], colony biofilms [7], drip flow reactors [8], or rotating disk reactors [9] can be used. The flow-cell system which we describe here was developed and described by Christensen et al. [6] and was based on the system described by Wolfaardt et al. [10]. The bacteria grow in small channels with a glass surface through which the biofilm can be monitored noninvasively and continuously using confocal laser scanning microscopy (CLSM). The flow-cell system has been the driving model system for elucidating biofilm-forming process [11–14] in vitro and the biofilm tolerance towards antibiotics [15, 16] and host defense components [15, 17–19]. The flow-cell system is superior for direct and noninvasive biofilm investigations; however, the price is that the number of samples to be tested in each experiment is limited and the method is time consuming compared to the microtiter assay.
The three methods we describe in this chapter can preferably be used in combination. The easier and fairly high-throughput 6-well method can be used to identify mutants, growth conditions, antibiofilm drugs of interest, or lead candidates from large libraries or collections. The bead method serves as a basis for much biofilm and subsequently transcriptomic and proteomic elaborations. The properties of biofilm formation gained in these two models can then be investigated in the flow cells.
2 Materials
2.1 Six-Well Microbial Biofilm Growth with Shear
Polystyrene 6-well plates.
22 × 22-1 cover slip.
Orbital shaker.
Sterile plastic container.
Ultrasonic bath.
Kinematica Polytron P1200E handheld homogenizer.
70 % ethanol.
Phosphate-buffered saline (PBS).
Epi-fluorescence microscope or confocal laser scanning microscope.
Appropriate PNA FISH probes such as Cy3-labeled C. albicans/FITC-labeled S. aureus PNA probe cocktail.
Vectashield Mounting Media.
Clear nail polish.
Trypticase soy broth (TSB).
Yeast peptone dextrose (YPD).
Sabouraud dextrose agar.
RPMI 1640 buffered with HEPES and supplemented with L -glutamine.
5 % heat-inactivated fetal bovine serum (RPMI-FBS).
YPD containing 5 % FBS medium.
2.2 Glass Bead Biofilms
In order to provide increased surface area for biofilm growth, glass beads are added to 500 ml flasks and then placed on an orbital shaker to provide shear.
Media: Brain–heart infusion (BHI) broth.
Solid soda lime.
Solid borosilicate glass balls.
TSB.
The protein preservation solution is composed of 10mM Tris•Cl, 1mM EDTA, 0.5 mg/ml PMSF (Phenylmethyl-sulphonylfluoride), and 10mM sodium azide.
RNAprotect reagent (Qiagen, Valencia, CA).
Homogenizer and conical tubes (one filled with ethanol, two filled with PBS).
Conical tubes (50 ml).
Glass 500 ml tissue culture bottles.
Plastic container.
Ultrasonic bath.
Kinematica Polytron P1200E handheld homogenizer.
70 % Ethanol.
2.3 Flow Cells
2.3.1 Components for Assembly of the Flow-Chamber System
Bubble traps.
Flow chambers.
Polycarbonate sheet plastic, 6 mm thick (optional, if flow chambers are to be made locally).
Substratum: 50 × 24-mm glass cover slips or other appropriate materials.
Marprene® tubing, 3 mm outer diameter, 1 mm inner diameter.
Silicone tubing, 3 mm outer diameter, 1 mm inner diameter.
Silicone tubing, 4 mm outer diameter, 2 mm inner diameter.
Silicone tubing, 7 mm outer diameter, 5 mm inner diameter.
Clear polypropylene plastic connectors and T-connectors (Cole Parmer), 1/8 in. (3.175 mm) and 1/16 in. (1.588 mm). Reduction connectors 1/8 to 1/16 in.
2-ml syringe.
Injection needles.
Medium bottles.
Waste container.
Silicone glue.
70 and 96 % (v/v) ethanol.
0.5 % (w/v) sodium hypochlorite.
H2O, sterile.
1 % hydrogen peroxide (optional).
Medium appropriate for organisms and type of biofilm being grown (e.g., biofilm minimal medium, FeEDTA-AB (FAB) [20]).
Peristaltic Pump (Watson-Marlow, 205S).
Microscope.
Rolling cart for flow systems and pumps (optional).
Computer Numerical Control (CNC) tooling machine or a drilling machine mounted on an upright stand and equipped with a milling drill tool (3 mm) (if flow chambers are to be made locally).
2.3.2 Components for Construction of the Bubble Trap (Advanced Version)
35 × 80 × 45-mm polycarbonate block.
CNC tooling machine.
5-ml syringes with inner diameter of 12.5 mm.
9 × 2-mm rubber gaskets (M-seals, 221355; http://www.m-seals.dk/cms.ashx).
Silicone glue.
Stoppers (e.g., http://www.nordson.com/en-us/divisions/efd) or use the leftover needle protective cover from the needles used for inoculating the flow cells (see above).
2.3.3 Components for Construction of the Bubble Trap (Simple Version)
A 10 mm thick polycarbonate block, 80 × 35 mm surface area.
Drilling machine mounted in a vertical stand.
An 8 and a 3 mm drill suitable for drilling in plastic.
2- or 5-ml syringes.
Silicone glue and stoppers as above.
2.3.4 Materials for Inoculation and Running of the Flow Cells
Inoculum, e.g., fresh overnight culture of the microorganisms under study.
70 and 96 % (v/v) ethanol.
Medium (e.g., FAB medium).
Silicone glue.
Flow-cell system (DTU Systems Biology, Technical University of Denmark, or see below).
Syringes and needles (0.4 × 12 mm, 0.5 ml).
Clamps.
2.3.5 Equipment for CLSM of Flow Cell-Grown Biofilms
Confocal laser scanning microscope (e.g., Zeiss LSM710).
Scalpels.
Computer software:
Imaris (Bitplane; http://www.bitplane.com).
ImageJ (http://rsb.info.nih.gov/ij).
Comstat version 2 (DTU Systems Biology, Technical University of Denmark, http://www.comstat.dk).
Java runtime environment (needed for Comstat v. 2, http://www.java.com).
3 Methods
3.1 Six-Well Microbial Biofilm Growth with Shear
3.1.1 Single-Species Biofilms (See Notes 1 and 2)
Starter cultures of bacteria (e.g., Staphylococcus aureus, Staphylococcus epider midis, Pseudomonas aer uginosa, Streptococcus pyogenes, Bacillus subtilis, Escherichia coli, Acinetobacter bau-mannii, Klebsiella pneumoniae, Salmonella typhimurium) are derived from frozen glycerol stocks that are streaked onto petri dishes in order to ensure a lack of contamination.
A minimum of ten colony-forming units (CFUs) should be picked and added to TSB or another appropriate growth medium and incubated overnight at 37 °C. By choosing ten CFUs, aberrant results due to clonal differences can be avoided.
Fresh log-phase bacterial starter cultures are grown by diluting the overnight culture 1:100 in fresh 37 °C pre-warmed TSB for 3 h. Bacterial cultures are then washed twice in sterile PBS.
Bacteria are then diluted to an OD of 0.1 at 600 nm, and 50 μl are added to each polystyrene 6-well plate with 5 ml of 37 °C pre-warmed TSB per well and each containing a sterile 22 × 22 × 1 cover slip.
Shear is provided by placing plates on a rotating platform during incubation at 100 rpm (ensure that this is consistent for all studies).
For growth longer than 24 h, the plate is removed from the incubator and immediately placed at an angle on another unused 6-well plate to allow solution to collect at the well sides.
Spent media is removed, and 5 ml of fresh, sterile 37 °C pre-warmed media is added carefully to the side of the well. The 6-well plate is returned to the incubator and allowed to rotate at 100 rpm for an additional 2 min.
The plate is removed from the incubator and again immediately placed at a slight angle to allow solution to collect at the well sides. Wash media is removed, and 5 ml of fresh, sterile 37 °C pre-warmed media is added carefully to the side of the well.
The 6-well plate is returned to the incubator and allowed to rotate at 100 rpm.
If multiple days are required for different biofilm growth stages (early, maturing, and fully mature as described below), repeat every 24 h (see Note 3).
3.1.2 Dual-Species Biofilm (Fungi and Bacteria) (See Notes 1 and 2)
Bacteria are grown, subcultured to log phase, washed, and diluted as described above.
An aliquot of a glycerol stock of Candida albicans is grown and maintained on Sabouraud dextrose agar. Cultures are grown overnight in YPD in an orbital shaker (100 rpm) at 37 °C under aerobic conditions. Yeast cells are harvested and washed twice in sterile PBS.
C. albicans overnight cultures are grown as described above and diluted to an OD of 1.0 at 540 nm. Aliquots of each species suspensions (50 μl) are added to each polystyrene 6-well plate with 5 ml of 37 °C pre-warmed broth (described below) per well and each containing a sterile cover slip [21].
Dual-species biofilms (C. albicans and bacterial species) are grown in RPMI 1640 buffered with HEPES and supplemented with L -glutamine and 5 % heat-inactivated fetal bovine serum (RPMI-FBS) when hyphal growth by C. albicans is needed or YPD containing 5 % FBS medium (YPDFBS) for experiments with yeast cells of C. albicans.
3.1.3 Single-or Dual-Species Coverslip Microscopy
PNA-FISH hybridization is performed as per the manufacturer's protocol (http://www.advandx.com/Technology/PNA-FISH-Technology.aspx). Use Cy3-labeled C. albicans/FITC-labeled S. aureus PNA probe cocktail for this cell combination. Non-adherent cells are removed by gently washing with PBS prior to imaging.
Add a single drop of Vectashield Mounting Media to the microscope slide, and lay cover slip face down. Seal edges between the cover slip and microscope slide using clear nail polish.
For all microbial microscopy counts, a minimum of ten random and blinded fields of view should be evaluated. In addition, each field of view must have a minimum of 20 cells per field of view. If less than 20 cells per field of view, increase the number of fields of view to attain 2,000 cells.
3.1.4 Biofilm Dispersal (See Note 4)
Biofilm dispersal method 1 [22, 23]: Each cover slip should be transferred into a sterile plastic container containing 5 ml PBS. The container is sealed and immersed in an ultrasonic bath. Sonication at 30 kHz with a power output of 300 W, as specified by the manufacturer, should be performed at 37 °C for 5 min.
Biofilm dispersal method 2 [23, 24]: Using a Kinematica Polytron P1200E handheld homogenizer at maximum speed (30,000 rpm), first disinfect with 70 % ethanol and wash twice in sterile PBS, followed by homogenization for 1 min on ice. Immediately serially dilute and plate for CFU determination.
3.2 Glass Bead Biofilms
3.2.1 Preparation of Glass Beads and Biofilm Culture Bottle
Wash beads with a detergent and dry.
Add beads to 500 ml glass bottles (approximately half full), add culture media (BHI or TSB) to 2–3 cm above beads so that all beads are submerged (e.g., BHI), and autoclave sterilize.
3.2.2 Protocol for Biofilm Growth
A minimum of ten colonies should be picked and added to TSB and incubated overnight at 37 °C. By choosing ten CFUs, aberrant results due to clonal differences can be avoided.
Fresh log-phase bacterial starter cultures are grown by diluting the overnight culture 1:100 in fresh 37 °C pre-warmed TSB for 3 h. Bacterial cultures are then washed twice in sterile PBS.
Bacteria are then diluted to an OD of 0.1 at 600 nm, and 500 μl are added to each bead and culture-containing 500 ml sterilized bottle.
Shear is provided by placing bottle in holding clamp on a rotating platform during incubation at 100 rpm (ensure that this is consistent for all studies) at 37 °C.
For growth longer than 24 h, the bottle is removed from the incubator and spent media is immediately removed via pipet. Fresh, sterile 37 °C pre-warmed media is added carefully to the side of the bottle.
The bottle is returned to the incubator and allowed to rotate at 100 rpm.
If multiple days are required for different biofilm growth stages (early, maturing, and fully mature as described below), repeat every 24 h (see Note 3).
3.2.3 Harvest
The bottle is removed from the incubator, and media containing planktonic bacteria is immediately removed via pipet. Fresh, sterile 37 °C pre-warmed media is added carefully to the side of the bottle, and it is then returned to the incubator and allowed to rotate at 100 rpm for an additional 2 min.
The bottle is removed from the incubator, and again wash media is immediately removed to remove all non-adherent bacteria.
3.2.4 Biofilm Dispersal (See Note 4)
Biofilm dispersal method 1 [22, 23]: Beads should be transferred into a sterile plastic container containing 50 ml PBS. The container is sealed and immersed in an ultrasonic bath. Sonication at 30 kHz with a power output of 300 W, as specified by the manufacturer, should be performed at 37 °C for 5 min. Immediately serially dilute and plate for CFU determination.
Biofilm dispersal method 2 [23, 24]: Sterile PBS is added to the washed beads in the 500 ml tissue culture bottle to a level just above the level of the beads. Using a Kinematica Polytron P1200E handheld homogenizer at maximum speed (30,000 rpm), first disinfect with 70 % ethanol and wash twice in sterile PBS, followed by homogenization for 1 min on ice. Immediately serially dilute and plate for CFU determination.
3.2.5 Preservation
For RNA or proteomic sample harvest, biofilms must also be dispersed. However, instead of using PBS as the solution to disperse the biofilm into, one should use RNA protect for RNA samples or PBS with protease inhibitor solution for proteomic samples. Following dispersion, transfer solution to centrifuge tubes and centrifuge to concentrate, and pour off supernatant. Freeze at −80 °C until ready for processing.
3.3 Flow Cells
Growing biofilms in flow-chamber devices allows for continuous, dynamic observation of living microbial communities and facilitates manipulations which consequences likewise can be monitored online. The requirements for such devices are that they should be mountable on a microscope without interfering with medium supply and that the growing biofilm should be easily observable while on the microscope. Furthermore, manipulations such as challenging the biofilm with antibiotics or changing nutrient availability should be easy to do.
The flow chambers used in this chapter are constructed from transparent, non-fluorescent plastic, polycarbonate (PC) which has the additional advantage of being tolerant to auto-claving at 115 °C. The flow chambers are essentially plastic blocks with milled or molded channels (40 × 4 × 1 mm) with an inlet and outlet connector in the ends. On the free side of the channels a piece of coverslip glass is glued with silicone glue. Nutrient medium is supplied to the flow chamber via silicone tubing, which is very flexible and also permeable to oxygen, ensuring oxygen saturation of the medium at the inlet to the flow chamber. Occasionally small air bubbles will form in the medium lines. These could be catastrophic to the biofilm structure, effectively acting like a razor blade when passing through the flow chamber. To remove such bubbles a device called a bubble trap is inserted in the supply line upstream of the flow chamber. The bubble trap is a small liquid-filled cylinder (a single-use syringe) sitting on top of the passing medium. If an air bubble is in the liquid while passing the trap, the bubble will flow to the top of the cylinder and stay there rather than moving on to the flow chamber. Medium is contained in a flask and led to the bubble traps and flow chambers via peristaltic pump. To prevent the tubing to break because of the wear applied by the pump special tubing is used in the pump. Marprene® tubing has proven to be a suitable material for this purpose—it can be autoclaved and is essentially inverted towards the medium. Marprene® can withstand pump wear for long periods of time and can be reused several times. Waste from the flow chambers is collected in suitable containers downstream of the flow chambers and can subsequently be disposed of by, e.g., autoclaving.
3.3.1 Fabrication of Flow Chambers
Flow chambers can be made by milling and moulding, or if the necessary tools are unavailable several sources for buying ready-made flow chambers exist. To construct flow chambers by milling a milling machine is required. While it is possible to control the milling process manually it is recommended to use a programmable machine (a CNC machine).
Use a sheet of polycarbonate, 6 × 76 × 26 mm, to mill the flow chamber shown in Fig. 1 . This will produce a flow chamber with dimension that fits on a standard microscope slide holder.
Start by making a flat flange in each end of the device 1 mm thick by milling away 8 mm into the plastic. The middle part is 6 mm thick and will accommodate the actual flow channels.
Mill the flow channels using a 4 mm flat head milling tool. The channels can be of any depth from 0.1 to 5 mm. The standard dimensions are 1 mm depth and 40 mm length. To allow for medium inflow the channels either drill a hole from each end of the flow chamber which allows insertion of a silicone tube or use an advanced milling machine to fabricate a stud in each end of the channels onto which the silicone medium line can be connected. Use a thin drill to open the tubes to connect to the flow channels.
Attach the selected substratum (usually a glass cover slip, 50 × 24 mm) to the flow chamber. Some silicone glues, in particular types intended for sealing sanitary installations, may contain bacteriostatic or bactericidal compounds and should be avoided. We found 3 M Super Sealant suitable for gluing flow chambers. Place thin strips of silicone glue along the edges of the channels, and take care not to leave gaps in the strips. A single-use 2 ml syringe with a cut pipette tip placed inside the syringe tip is a convenient tool for applying the glue: remove the piston, and insert the thin 1 cm of a 200 μl pipette tip in the syringe. Fill about 1 ml silicone glue in the open end of the syringe, and replace the piston. The filled syringe can now aid in applying the thin strips of glue (see Note 5).
Substrata other than glass can be used: Cut the desired substratum to cover the flow channels; i.e., it should have the same dimensions as a glass cover slip, approximately 50 × 24 mm.
Align the substratum to the part of the flow chamber with the applied glue. Gently press the substratum onto the chamber, and ensure that there are no visible leaks, i.e., areas where the glue seem to form gaps. If such gaps can be seen, use either the finger or the piston handle to press the substratum firmly to the chamber.
If it is not possible to completely seal the flow chamber, or if silicone glue has entered the channels, remove the substratum and clean the plastic base using 96 % ethanol. Dependent on the type of substratum either dispose and replace or clean using ethanol. Repeat steps 4–6 .
Allow at least several hours for the glue to solidify, preferably overnight before use.
If the chambers are fabricated using a machine that has made studs in the in- and outlet ends attach silicone tubing by twisting the tubing onto the studs. Alternatively, place cut tubing ends into the receiver holes and seal with silicone glue (manually fabricated chambers).
Fig. 1.
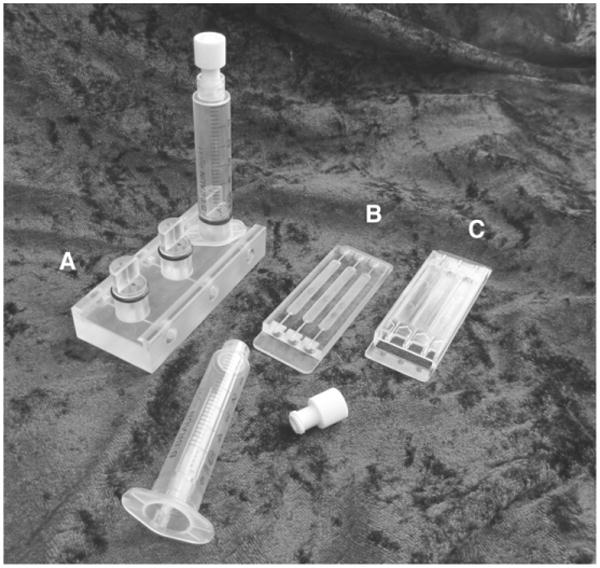
Bubble traps and flow cells (A–C)
3.3.2 Fabrication of Advanced Bubble Trap Using CNC Milling Equipment
The bubble traps in the system are required to remove small air bubbles from reaching the flow chambers, as described above (see Note 6). The simplest form of a bubble trap can be made from a single-use syringe, a cap, and two needles. A slightly more advanced version utilizes syringes glued to a plastic base with in- and outlet ports. More sophisticated versions must be fabricated using advanced milling tools. These are much easier to handle but also much more expensive. A milled version is shown in Fig. 1 .
Mill the base of the bubble trap from a single block of polycarbonate (35 × 80 × 45 mm).
Use the machine to carve rings to accommodate gaskets to prevent leakage when the syringes are mounted on the base. Also carve a rail along the sides of the base—this can be used to lock the syringes into place by twisting the syringe so that the handles will fit under the rails.
Place 5-ml syringes (inner diameter, 12.5 mm) as on each of the columns. Do not twist the syringe to fix it onto the base before the entire system has been tested. In case of a blocked line downstream of the bubble trap, the syringe can then pop off, relieving the built-up pressure. If the syringe is locked damage may occur to the flow chambers in this case.
Place a stopper on top of each syringe.
3.3.3 Fabrication of Simple Bubble Traps
Remove the piston from a 5 ml syringe, and detach the rubber seal from the plunger.
Put the rubber seal back into the end of the syringe.
Use silicone glue to attach this onto a flat base.
Penetrate two injection needles above the seal into the syringe, one needle 1 cm above the other. The lower needle is now the outlet port and the upper needle the inlet.
Place a stopper on top of the syringe.
3.3.4 Fabrication of Slightly More Advanced Bubble Traps
Prepare a base plate from a piece of polycarbonate (30 × 110 × 10 mm).
Drill three evenly spaced cavities on one side of the plastic (do not drill all the way through the plastic, ca. 8 mm) using an 8 mm drill.
Drill in- and outlet channels from each side. Use a 3 mm drill, and place the center 4 mm from the top of the base. Drill far enough that each cavity will have a connection on each side of the plastic.
Glue three single-use 2 ml syringes on top of each cavity using silicone glue.
Use 3 mm outer diameter silicone tubing in the in- and outlet ports.
Place a stopper on top of the syringes.
Allow solidifying for at least 24 h.
This construction is quite fragile, and great care should be taken to avoid breaking off the syringes from the base.
3.3.5 Assembly of the Flow-Cell System
Timing is crucial when setting up a flow-chamber experiment. The items that need to be glued should preferably be done so 1 day prior to use. After assembly and checking for leakages the system must be sterilized using hypochlorite for at least 4 h and washed with several passages of sterile water. Only then the system can be filled with medium and inoculated. The system is shown in Fig. 2 .
Fig. 2.
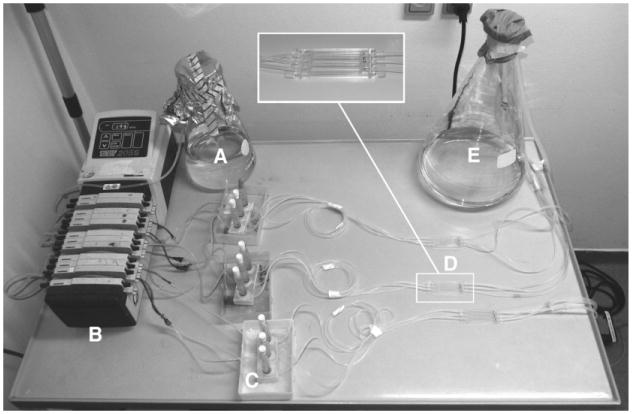
The assembled flow-cell system (A–E)
All tubing is of 1 mm inner diameter, except for the medium supply tubing and the waste tubing which have 2 mm inner diameter. All tubing is connected using polypropylene plastic connector. The entire system, except the pump, can be autoclaved before or after assembly.
Start by preparing the flow chambers and bubble traps (see Subheadings 3.3.1–3.3.4). Then prepare and autoclave the medium (remember to insert a medium-supply silicone tube into the medium and cap off the end with aluminum foil). The tubing system must then be prepared: a fan-out connector to split the medium supply so that each channel will have its own supply from the pump: this makes it much easier to, e.g., remove channels that are contaminated or broken from the system if needed. The remainders of the flow lines are individual for each channel.
Prepare flow chambers and bubble traps according to Subheadings 3.3.1–3.3.4.
Calculate the amount of medium you will need. To do this you can calibrate the usage by attaching a piece of Marprene® tubing to the pump and connect it to an inlet and outlet tubing. Set the pump to the desired velocity, and measure how much water is passed through the tubing for a given period, e.g., 1 h. Then you can easily calculate the medium requirement per day. Multiply by the number of channels to find the overall usage per 24 h. It is advisable to prepare more medium than needed to allow for unexpected delays and for having a buffer volume near the bottom of the supply flask.
Preparation of medium: Place a sufficient long silicone tube, 2 mm inner diameter, in the bottle. Fix the tubing outside of the bottle using autoclave tape. Attach a straight connector to the end, and protect the end against contamination with aluminum foil. It is essential to clamp off the tubing or siphoning may empty the bottle while autoclaving. Fill the bottle with medium, and cover the bottle opening with aluminum foil. Autoclave immediately.
Prepare all the tubing for the rest of the system:
Start by making the fan-out component: make one piece of silicone tubing (50 cm) that will attach to the silicone tubing from the medium bottle. Cover the end with aluminum foil. To the other end attach a T-connector (1/16 in.). To each end attach 5 cm of silicone tubing (1 mm inner diameter). To the two new ends add additional T-connector and 5 cm pieces of tubing. Continue until there is the same number of free ends as channels in the system.
Prepare short pieces of Marprene® tubing, just long enough to pass through the pump. In each end place a 1/16 in. straight connector. To prevent the pump from pulling off the connectors, wind four winds of autoclave tape around the Marprene® next to the connector on the inlet side of the pump.
On the outlet side of the pump attach a short (35 cm) piece of tubing and connect the other end to the bubble trap. On the outlet side of the bubble trap attach a long piece of tubing (150 cm, to allow the flow chamber to be taken from the experiment table to the microscope). Attach the other end to the flow chamber. At the outlet end of the flow chamber attach a very short (10 cm) piece of tubing and then a 1/16 to 1/8 straight reduction connector. Finally, attach the waste (effluent) tubing to the connector. The waste tubing is 2 mm inner diameter and should be sufficiently long to allow the flow chamber to be moved to the microscope while keeping the end in the waste container (120 cm is usually sufficient) (see Note 7).
If needed, autoclave the assembled flow system (recommended).
Attach all the pump tubing to the pump. It is highly recommended to keep all the tubing in order, such that the first flow channel in the first flow chamber is connected to the first channel in the first bubble trap and the first channel in the pump. This will make it much easier to find and fix problems such as leakages in the system later on.
Optionally place the entire setup on a rolling cart to make transport to the microscope and incubation room (if required) easy.
Place the waste lines into a suitable reception container.
3.3.6 Sterilization of the Flow-Cell System
Always use sterile water when operating the system, even before sterilization.
Remove all stoppers from the bubble traps, and store them in 70 % ethanol. Place all components on a flat surface, and make sure that the waste container is at the same level as the flow chambers.
Place the free end of the fan-out connector in 1 l of water. Fill the system by setting the pump to maximum speed. When the bubble traps are filled with water wait for about 15 s and then place the stopper on the syringe. Then observe that the rest of the system is filled. At this point it is important to detect and seal any leaks in the system.
Remove air bubbles (even tiny ones) in the flow chambers by tapping them firmly on the ends on a hard surface. This will release the bubbles, and they can flow away due to the high velocity of the liquid.
Lift the inlet tube into the air, and allow the system to be emptied completely.
Remove all the stoppers and fill the system as described above, with a solution of 0.5 % sodium hypochloride in water. When the system is completely filled and all air bubbles removed continue flow at maximum speed for another 5–10 min. Reduce flow rate to the minimum of the pump and sterilize for at least 1 h. It is not recommended to leave the sterilizing agent in the system for extended periods and never more than 24 h.
The system is now sterile, and handling should be done to prevent contamination.
Empty and fill the system three consecutive times with sterile water to remove all traces of the hypochloride. Optionally the second pass can be water with 1 % hydrogen peroxide.
If the system is to be used immediately, empty the system and fill with medium. The system is now ready for inoculation.
If the system is not used at once it is recommended to leave it with water, running at a low rate until use.
3.3.7 Inoculation of the Flow Cells (See Note 8)
The amount of cells to inoculate is very dependent on both the strain and the medium. As a starting point cells from an overnight culture must be diluted before inoculation. The exact dilution must be empirically determined. For Escherichia coli a dilution of 1:100 is suitable, while Acinetobacter spp. should only be diluted ten times. Pseudomonas aeruginosa is usually diluted 1:1,000.
Withdraw about 250 μl of the diluted culture into a syringe equipped with a thin needle, e.g., 27 G. Wipe the tubing on the inlet side of the flow chamber with 70 % ethanol. Stop the pump, and penetrate the silicone tubing as near the inlet of the channel as possible. Inject the inoculum into the channel, and observe that no air bubbles are injected. If that happens remove the air bubble using the syringe. Important: Never place fingers behind the tubing while inserting the needle—the risk of being stung by the needle is significant.
Seal the injection hole with silicone glue.
Leave the flow chamber with the substratum facing the table for 1 h without flow to allow initial adhesion of the cells to the substratum.
Reverse the flow chambers, and resume flow. In this system a flow rate of 1–3 ml per channel per hour is suitable for E. coli and P. aeruginosa . For other organisms other flow rates may be needed.
3.3.8 Running of the Flow-Cell System (See Note 9)
The system can run for several days or even weeks without interruption. However, check frequently that medium is available and the waste is not overflowing. Deal with any leaks. A broken flow chamber cannot be fixed and must be isolated from the system.
Take care to ensure uninterrupted flow as some strains react by detachment for even short periods of no-flow (see Note 10).
Dispose of waste according to local regulations.
3.3.9 Microscopic Inspection and Imaging of Flow Cell: Grown Biofilms (See Note 11)
The flow cells with the transparent glass cover slip which also serve as the substratum for the biofilm enable easy and non-invention visualization. All microscopes can basically be used; however, the optimal method of choice for visualization of flow chamber-grown biofilms is to use CLSM.
Practical information when using CLSM and flow cells:
The flow cell system should be close to the confocal microscope to avoid interruption of growth if the system has to be transported.
Make sure that the tubing in both ends on the flow cells are long enough for the flow cell to be placed under the microscope without interfering with the setup.
Make sure that the flow cell is firmly attached in the specimen holder. Adhesive tape to assist mounting the sample onto the microscope is a possibility.
WARNING: Before starting the microscopy be careful if the microscope is adjusted to automatically set parfocality as for normal microscope slides. The flow chambers are much thicker; as a consequence the microscope may position the lens wrongly and destroy the sample. An empty flow cell can be used to calibrate the microscope adjustments before mounting a real experiment.
3.3.10 End of Experiment
Many of the flow-cell components can be reused and should be thoroughly cleaned after each experiment. Other components such as the upstream silicone tubes should not be reused and discarded after each experiment.
Disassemble the inlet tube to allow emptying the remaining liquid from the still assembled system by filling with air. All waste should be collected and disposed of following local biohazard regulations.
Detach the flow chambers. The pump tubing and the downstream waste tubing can be reused if autoclaved.
The flow chambers are disassembled using a scalpel to remove the substratum. If the substratum is made of fragile material such as glass it will inevitably break in this process. Remember to use plastic gloves and protective eye wear.
Remove remains of silicone glue from the flow chamber using mechanical abrasion and 96 % ethanol.
The reusable tubing and the clean flow cells can be wrapped in metal foil, autoclaved, and stored for subsequent experiments.
3.3.11 Image Analysis
The CLSM imaging can be either used as qualitatively descriptive images or for quantitative measurements. In both instances the CLSM images obtained of the biofilm experiment by CLSM should most often be processed since most CLSM files are in a specific format.
For qualitative and descriptive analysis of the images, several software programs are available. We recommend the commercial package Imaris® software suite (http://www.bitplane.ch) which can create two- and three-dimensional visualizations with simple measurements, time-lapse movies, as well as animations. A freeware alternative is the program ImageJ [25] (http://rsb.info.nhi.gov/ij) which can be supplemented by user-written plug-ins to perform several graphical visualizations of CLSM images and extensive qualitative measurements. Using nonspecialized programs such as ImageJ does require more from the user to get full benefit of its capabilities than do commercial dedicated packages.
For quantitative analysis several programs have been developed, such as the ISA3D [26], Comstat [27] (http://www.comstat.dk), and Daime [28] (http://www.microbial-ecology.net/daime). Using these programs basic parameters from CLSM image stacks, such as biomass, roughness, and average thickness, can be calculated. The quantitative measurements are numbers, rather than images, and provide a way to directly evaluate both reproducibility of experiments and statistically compare different biofilms which qualitatively seem similar.
Notes
All experiments should be performed in triplicate (i.e., three true replicates).
Several strains of each species should be used including recent representative clinical isolates.
Each stage of biofilm growth is as different as biofilms are to planktonic cultures [29]. If multiple growth stages of biofilms are required, perform biofilm growth curves for each strain in which CFU/cm2 is determined at multiple time points following inoculation. The various stages include the following: (a) early-stage biofilms are those with monolayers soon after microbial attachment, (b) fully mature biofilms are those where the CFU/cm2 reaches a maximum and static level, and (c) maturing biofilms are those with CFU/cm2 at 50 % of fully mature biofilms.
In order to get representative CFU counts from biofilms, it is important to break up the biofilm conglomerates by either sonication or homogenization.
As with any liquid, even a small leak will render the entire flow cell useless and should be sealed before the experiment is initiated. Leaks are easy to spot when the setup is filled with water. Use normal water before the system is sterilized. If leaks are observed, either change the damaged part or seal with silicone glue; remember to allow for hardening. It is usually very difficult to seal leaks once the system and experiment are initiated since the flow should be turned off to allow for the silicone to dry. The glass cover slips on the flow cells are very fragile, so please be careful. If the flow cell leaks while experiment with bacteria is running the biohazard risk is great. Especially if the flow cell is mounted on the microscope, it can become harmful to the equipment and difficult to decontaminate. Either immediately attempt to reseal the leak or remove the flow cell from the microscope. While running an experiment, it is not possible to drain the system or even stop the flow temporarily. If leaks occur during an experiment clean and dry the area containing the leak, and keep a piece of paper towel at hand to repeatedly soak away leaking fluid. Apply excess of silicone glue to the leak, and continue to remove leaking liquid until it stops.
As with any liquid-changing temperature or flow rates, gas or air bubbles can form. These air bubbles can introduce artifacts in the developing biofilm eventually destroying or altering the three-dimensional structure of biofilms. Due to this it is recommended to use bubble traps to catch the gas bubbles before they reach the flow cells. Since the change in temperature, for example, when cooling the media from autoclaving to room temperature to heat it up to the experimental temperature, increases the bubble formation, it is recommended not to cool the medium after autoclaving, and to place it immediately at the correct temperature for the experiment. Furthermore, if running of the flow-cell system above room (e.g., 37 °C) the change of bubble formation increases. A recently described [30] setup with a modified medium container may be employed.
Place the end of the waste tubes downstream of the flow cell above the surface of the waste reservoir to avoid reverse siphoning from the waste container.
As of any other experimental setup, microbial growth in flow cells may be influenced by biological variation, such as selection of mutants. Due to this we recommend always to run at least two independent flow channels of each bacterium or strain, and as with any other biological setup experiments should always be performed in replicates.
Since many bacteria are motile and tend to migrate towards chemical gradients, such as the fresh medium supply, bacterial growth might occur in the tubing upstream of the flow cell also known as “backgrowth.” If such fouling occurs we recommend removing the contaminated part of the tubing since biofilm in the upstream tubing will use substrate and release waste products that may affect the biofilm in the flow cell. The contaminated tubing is removed by first clamping the tubing on the effluent side of the flow cells, so that waste medium does not flow backwards through the chamber, destroying the sample. Hence cut off the contaminated part, reconnect, and slowly remove clamping. It is in this case critical not to open clamps too abruptly as the concurrent liquid movement can disrupt the biofilm. The effluent tubes can also be changed if they become heavily fouled, but here clamping is not critical, but the biohazard of the contaminated waste should be avoided.
The biofilms formed by some microbial species will disperse in response to even short periods of change in flow rate. If the bacteria of a given experiment are expected to easily disperse it may be important to ensure constant running of the peristaltic pump.
Always perform the microscopy in the same area of the flow cell. The biofilm formation in the flow cell is very different in the inlet part compared to the outlet part, because the bacteria use substrate and release waste products. We recommend the microscopic inspection, and imaging of the biofilm should be done near the medium inlet to avoid uncontrolled conditions.
References
- 1.Christensen GD, Simpson WA, Younger JJ, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staph-ylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 3.Ceri H, Olson ME, Stremick C, et al. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee B, Haagensen JA, Ciofu O, et al. Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J Clin Microbiol. 2005;43:5247–5255. doi: 10.1128/JCM.43.10.5247-5255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hengzhuang W, Wu H, Ciofu O, et al. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2011;55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen BB, Sternberg C, Andersen JB, et al. Molecular tools for study of biofilm physiology. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- 7.Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goeres DM, Hamilton MA, Beck NA, et al. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc. 2009;4:783–788. doi: 10.1038/nprot.2009.59. [DOI] [PubMed] [Google Scholar]
- 9.Zelver N, Hamilton M, Pitts B, et al. Measuring antimicrobial effects on biofilm bacteria: from laboratory to field. Methods Enzymol. 1999;310:608–628. doi: 10.1016/s0076-6879(99)10047-8. [DOI] [PubMed] [Google Scholar]
- 10.Wolfaardt GM, Lawrence JR, Robarts RD, et al. Multicellular organization in a degradative biofilm community. Appl Environ Microbiol. 1994;60:434–446. doi: 10.1128/aem.60.2.434-446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klausen M, Heydorn A, Ragas P, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 12.Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 13.Sauer K, Camper AK, Ehrlich GD, et al. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 15.Bjarnsholt T, Jensen PØ, Burmølle M, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymor-phonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 16.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeru-ginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol. 2008;68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 17.Alhede M, Bjarnsholt T, Jensen PØ, et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155:3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 18.Jensen PO, Bjarnsholt T, Phipps R, et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology. 2007;153:1329–1338. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- 19.van Gennip M, Christensen LD, Alhede M, et al. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 2009;117:537–546. doi: 10.1111/j.1600-0463.2009.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2007;189:2531–2539. doi: 10.1128/JB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters BM, Ovchinnikova ES, Krom BP, et al. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology. 2012;158:2975–2986. doi: 10.1099/mic.0.062109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjerkan G, Witso E, Bergh K. Sonication is superior to scraping for retrieval of bacteria in biofilm on titanium and steel surfaces in vitro. Acta Orthop. 2009;80:245–250. doi: 10.3109/17453670902947457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol. 2004;186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady RA, O'May GA, Leid JG, et al. Resolution of Staphylococcus aureus biofilm infection using vaccination and antibiotic treatment. Infect Immun. 2011;79:1797–1803. doi: 10.1128/IAI.00451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abrámoff MD, Magalhâes PJ, Ram SJ. Image processing with ImageJ. Biophoton Int. 2004;11:36–41. [Google Scholar]
- 26.Beyenal H, Donovan C, Lewandowski Z, Harkin G. Three-dimensional biofilm structure quantification. J Microbiol Methods. 2004;59:395–413. doi: 10.1016/j.mimet.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Heydorn A, Nielsen AT, Hentzer M, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 28.Daims H, Lucker S, Wagner M. Daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 29.Southey-Pillig CJ, Davies DG, Sauer K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crusz SA, Popat R, Rybtke MT, et al. Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling. 2012;28:835–842. doi: 10.1080/08927014.2012.716044. [DOI] [PMC free article] [PubMed] [Google Scholar]


