Abstract
When the first version of this unit was written in 1995 protein purification of recombinant proteins was based on a variety of standard chromatographic methods and approaches many of which were described and mentioned in this unit and elsewhere in the book. In the interim there has been a shift towards an almost universal usage of the affinity or fusion tag. This may not be the case for biotechnology manufacture where affinity tags can complicate producing proteins under regulatory conditions. Regardless of the protein expression system, questions are asked as to which and how many affinity tags to use, where to attach them in the protein and whether to engineer a self cleavage system or simply leave them on. We will briefly address some of these issues. Also although this overview focuses on E.coli, protein expression and purification from the other commonly used expression systems are mentioned and apart from cell breakage methods, the protein purification methods and strategies are essentially the same.
Protein Expression
The expression of recombinant proteins, especially using bacterial vectors and hosts, is a mature technology. With the appropriate cDNA and PCR methods, expression plasmids can be rapidly produced. Following sequence determination of the constructs, plasmids are transformed into expression hosts, single colonies picked, and fermentation performed. With E. coli, a 2-liter fermentation using complex media will generate ~ 50 to 80 g (wet weight of cells). Assuming modest protein expression (2% to 5% of the total cellular protein), between 100 and 300 mg of recombinant protein is available in the cells. The problem is, of course, how to isolate it in an active form. Soluble proteins can be recovered with good yields (>50%), and insoluble proteins, which must undergo a denaturation and folding cycle, can be recovered with more modest yields (5% to 20%). Hence, using small-scale fermentations and laboratory-scale processing equipment, proteins (or subdomains thereof) can usually be produced in sufficient quantities (10 to 100 mg) to initiate most studies including detailed structural determinations. Some strategies for achieving high-level expression of genes in E. coli have been reviewed by Markrides (1996) and Baneyx (1999) and are also discussed in Unit 5.24.
Some of the above characteristics also hold true for the production of proteins using yeast and baculovirus eukaryotic expression systems, although more effort and expertise is required to construct the vectors and, with the baculovirus system, produce cells for processing. A yeast expression system may be a wise choice for proteins that form insoluble inclusions in bacteria, and for the production of membrane-associated proteins (Cereghino and Clegg, 1999; UNITS 5.6–5.8). The baculovirus system has proven very useful for producing phosphorylated proteins and glycoproteins (Kost, 1999; UNITS 5.4–5.5) and for the co-expression of interacting proteins. The construction of stable mammalian protein expression vectors requires considerably more time and effort but may be the only approach for producing complex multidomain proteins (UNITS 5.9–5.10). Cells growing to cell densities of 1–5 ×109 cells/ml can be expected to typically secrete >10 mg/liter of product. Alternatively, transient gene expression systems using various viral vectors (e.g., vaccinia virus; UNITS 5.12–5.15), can be used to produce lesser amounts of protein, which is useful for feasibility studies. It is of interest to note that the large-scale transient expression systems in mammalian cells are being actively developed by biotechnology companies (Wurm and Bernard, 1999).
The choice of a host system for the production of recombinant proteins is discussed in unit 5.16 and is also concisely summarized by Brondyke (2009). Also, there is a special issue on the production of recombinant proteins in the journal Biotechnology Advances (Sanchez and Demin, 2012). In this issue there are excellent overviews of protein expression and production using E.coli (Chen, 2012); yeast (Celik and Calik, 2012); insect cell and the baculovirus system (Drugmand et al 2012); mammalian cells (Zhu, 2012); cell free systems (Carlson et al., 2012) and plant cells (Xu et al., 2012).
As mentioned by Chen (2012), for many investigators the initial choice is often Escherichia coli which remains the preferred system for laboratory investigations and initial development in commercial activities and is a benchmark for comparison among the other various expression platforms. This is due to such factors as ease of genetic manipulation, availability of optimized expression plasmids, and ease of growth. This unit presents an overview of recombinant protein purification with special emphasis on proteins expressed in E. coli. Practical aspects and strategies are stressed throughout, and wherever possible, the discussion is cross-referenced to the example protocols described in the rest of Chapter 6.
The first section deals with information pertinent to protein purification that can be derived from translation of the cDNA sequence. This is followed by a brief discussion of some of the common problems associated with bacterial protein expression (see also UNIT 5.1). Planning a protein purification strategy requires that the solubility of the expression product be determined; it is also useful to establish the location of the protein in the cell—e.g., cytoplasm or periplasm. This unit includes flow charts that summarize approaches for establishing solubility and localization of bacterially produced proteins (see also UNIT 5.2).
Purification strategies for both soluble and insoluble proteins are reviewed and summarized in flow charts (see also Chapter 1). Many of the individual purification steps, especially those involving chromatography, are covered in detail in Chapters 8 and 9, and elsewhere (Scopes, 1994; Janson, 2011). The methodologies and approaches described here are essentially suitable for laboratory-scale operations. Large-scale methodologies have been previously reviewed (Asenjo and Patrick 1990; Thatcher, 1996; Sofer and Hagel, 1997).
A section on glycoproteins produced in bacteria in the nonglycosylated state is included to emphasize that, although they may not be useful for in vivo studies, such proteins are well suited for structural studies. The final sections deal with protein handling, scale and aims of purification, and specialized equipment needed for recombinant protein purification and characterization.
An Overall Summary of Protein Production and Characterization
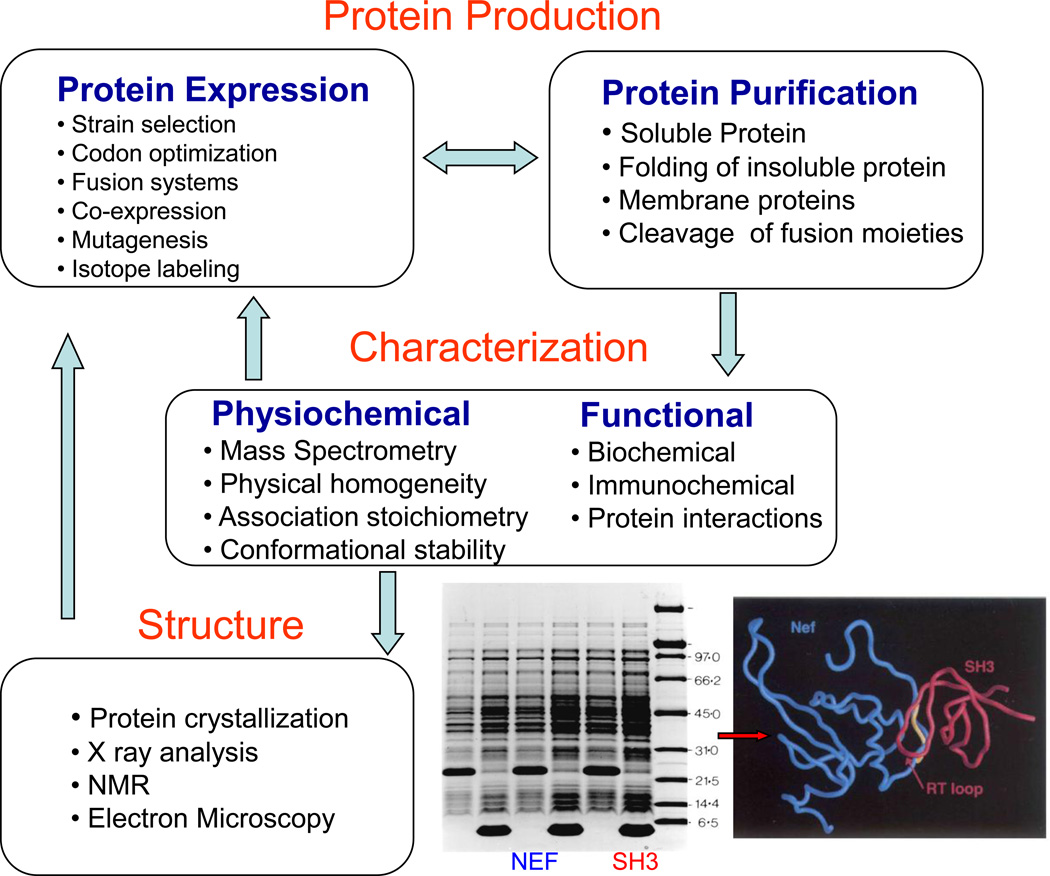
Protein expression is achieved by a recombinant expression system often E.coli as summarized in Figure 6.1.1. The expression system is optimized for protein expression of wild type sequence or a fusion tagged version. Stable isotopes C-14, N-15, H-2 can be incorporated into the growth media for labeling protein to be studied by NMR. Protein purification proceeds following the approaches and methods discussed in the unit and elsewhere (Chapter 9). The protein is characterized using various biophysical and biochemical methods which have also been detailed in the various Chapters of the book. The level of characterization depends on the final usage of the protein. It can be argued that characterization for structural determination requires the most rigorous approach, as micro chemical and physical heterogeneities can, for example, prevent protein crystallization. Characterization of therapeutic protein will also require rigor but more emphases will be placed on biochemical, immunological and functional testing. As the direction of arrows indicates (Figure 6.1.1) all the various stages are interdependent and there are always adjustments to be made based on the accumulation of information on the protein system being studied. Under ideal conditions, there is very high expression of biomedically important proteins such as HIV-1 Nef (NEF) and a Src homology-3 (SH3) domain of tyrosine kinase. These purified proteins form a complex the structure of which was solved by NMR (Grzesiek, et.al., 1996a).
Figure 6.1.1.
Overview of Protein Production and Characterization. See text for details
PROTEIN SEQUENCE AND COMPOSITIONAL ANALYSIS
Analyzing the Protein Sequence
The protein sequence translated from the DNA coding sequence is usually available, and before attempting any laboratory work, it is useful to carry out a literature survey and basic computer analyses (see Chapter 2). First, if the natural protein has been isolated and characterized, reviewing the physicochemical properties of the protein and the established purification techniques used may aid in planning a strategy for isolation from the recombinant host. Recombinant proteins that accumulate as insoluble aggregates or inclusion bodies, require folding into native-like conformations (Lilie et al., 1998; De Bernardez Clark et al., 1999; Shing and Panda, 2005; UNIT 6.4). The amyloid-like nature of inclusion body protein which appears to co-exist with the presence of protein with native-like structure has been reviewed (Ventura and Villaverde, 2006). Recently, the potential biotechnological potential of inclusion body is under exploration as functional, protein-releasing materials in regenerative medicine and protein replacement therapies (Villaverde, et al 2012).
Second, for uncharacterized proteins, analyses of related proteins with sequence similarities or known motifs may provide useful clues for selecting purification steps (UNIT 2.1; see also the PROSITE database of protein families and domains at the ExPASy Molecular Biology Server at http://ca.expasy.org/prosite). For example, if the protein contains the well-known kringle domain, lysine affinity chromatography might be a successful purification technique (Cleary et al., 1989). On the other hand, if the protein contains no recognizable motifs and has no similarity to other proteins, yet contains many cysteine residues, other strategies and precautions would be warranted as described in UNITS 6.3–6.5.
The amino acid sequence can be used to direct the synthesis of peptides corresponding to potential epitopes (e.g., 10 to 20 residues; UNIT 2.2). Polyclonal antibodies raised against the peptides may be suitable for detecting the protein of interest by immunoblotting. This approach may be especially valuable for monitoring proteins expressed at low levels— e.g., when E. coli secretion vectors are used. The antibodies may also be useful for immunoaffinity chromatography.
Analyzing the Amino Acid Composition
The amino acid composition (UNIT 3.2) of the protein will also allow calculation of some basic physicochemical parameters. Using average pKa values for ionizable side chains in proteins (Matthew et al., 1978), the isoelectric point (pI) can be estimated by applying the well-known Henderson-Hasselbach relationship. The calculations can be performed using an electronic spreadsheet such as Excel or via the internet using one of the many molecular biology servers, e.g., ExPASy (http://www.expasy.ch/tools/pi_tool.html). The values obtained, although only approximate, are useful for guiding the initial selection of ion-exchange resins and the pH of column buffers. When eukaryotic hosts are selected for protein expression, it should be noted that post-translational modifications such as phosphorylation and glycosylation will affect the pI.
Another parameter that can be estimated from the amino acid composition is the extinction coefficient (ε), usually at 280 nm (Pace et al., 1995). Although this information will be more useful when the protein has been purified, as most columns are monitored by UV absorption, proteins with an unusually low ε (no tryptophan and little or no tyrosine) may be difficult to detect during the early stages of purification.
Other physicochemical parameters that can be calculated include hydrodynamic parameters such as molecular radii and sedimentation coefficients, the program SEDNTERP is especially useful (http://www.jphilo.mailway.com/download.htm). These parameters may help in interpreting results of gel-filtration and centrifugational separations.
CHARACTERISTICS OF THE HOST-VECTOR SYSTEM
Choosing an Expression System
Popular protein expression systems include E. coli, yeast, baculovirus-infected insect cells, and cultured mammalian cell lines (see Chapter 5). If the requirement is to obtain a protein post-translationally modified via glycosylation (see Chapter 12) or phosphorylation (see Chapter 13), then a eukaryotic expression system must be used. Stable mammalian expression systems are the most time-consuming to establish and require the most expertise; however, they may be the only successful system for certain requirements including, e.g., proteins with authentic glycosylation patterns; large multidomain and multisubunit proteins, and especially proteins that are insoluble in E. coli. Post-translational modifications may aid purification (e.g., lectin affinity chromatography can be used for glycoproteins; UNIT 9.1). On the other hand, these modifications may introduce charge heterogeneity—as is commonly observed with glycosylation due to loss of sialic acid— which may then complicate purification, especially with methods such as ion-exchange chromatography (UNIT 8.2). Specific modification of proteins expressed in E. coli can be achieved by the co-expression of modifying enzymes, such as phosphorylation of tryrosyl residues by tyrosine kinase (Ren and Schaefer, 2001; Agilent: http://www.chem.agilent.com). However, most of the post-translational modifications observed in E. coli are nonspecific, such as deamidation (Wingfield et al., 1987a) and proteolytic clipping (Nagata et al., 1986). Other less common sources of protein heterogeneity arising from E. coli expression are: (1) internal starts in translation (Dale et al., 1994); (2) partial readthroughs of the termination codon (Danley et al., 1991), and (3) translation errors (Lu et al., 1993).
The initial choice for protein expression is often E. coli but if direct expression of a protein of interest fails or yields an insoluble product, there are many other options available including generating fusion proteins and many other approaches discussed elsewhere in this overview. If other expression hosts are to be screened, there are universal cloning systems commercially available (e.g., Gateway cloning system at http://www.lifetechnologies.com; Unit 5.17) that allow the rapid transfer of the gene of interest into multivector systems including yeast, baculovirus, and mammalian cells.
Minimizing Proteolysis
If the protein is expressed in the cytoplasm in a soluble state, the purification can be carried out directly after cell lysis. Soluble recombinant proteins are, however, susceptible to proteolysis, which can occur before or after extraction from the cell (Maurizi, 1992). Choosing protease-deficient E. coli host strains (Goff and Goldberg, 1985), manipulating growth conditions, especially the time of induction for inducible promoters (Allet et al., 1988), and using exogenous protease inhibitors can minimize this problem. Nevertheless, more extreme steps may be required, such as inducing the expressed protein to form insoluble inclusion bodies, using a secretion vector to locate the protein to the periplasm or medium, and changing to a eukaryotic expression system. In addition, there is a protein engineering approach that requires knowledge of the proteolytic cleavage site(s) to stabilize the protein. It requires alteration of one or both of the residues forming the scissile bond by site-directed mutagenesis (Mildner et al., 1994). For discussion on strategies to minimize proteolytic degradation, see reviews by Murby et al. (1996) and Makrides (1996). UNIT 5.25 also reviews approaches to preventing and avoiding proteolysis during expression and purification of proteins.
Removing the Amino-Terminal Methionine
Another common problem with proteins expressed directly in E. coli is retention of the N-terminal methionine derived from the initiating N-formylmethionine (the formyl group is almost always removed). The N-terminal methionine is generally removed when the second amino acid is alanine, glycine, proline, serine, threonine, or valine (cleavable residues), but not when it is arginine, asparagine, aspartic acid, glutamic acid, glutamine, isoleucine, leucine, lysine, or methionine (noncleavable residues (Sherman et al., 1985; Giglione, et al., 2004). When recombinant proteins are expressed at very high levels, the N-terminal methionine can be retained regardless of the nature of the second amino acid, presumably due to saturation of the processing enzymes or depletion of required metal cofactors. Removal of cleavable N-terminal methionine can be carried out in vitro by digestion with purified methionine aminopeptidase (Miller et al., 1987) or by co-expression of the processing enzyme (Ben-Bassat et al., 1987; Hwang, et al., 1999).
The need to remove noncleavable methionines can be circumvented by incorporating an N-terminal secretion leader sequence that localizes the protein, minus the leader, to the periplasmic space (Holland et al., 1990). Other approaches utilize the incorporation of N-terminal fusions with, e.g., ubiquitin, which can be cleaved in vitro or in vivo with a processing enzyme (ubiquitin hydrolase). This approach involves co-expression of the hydrolase (Miller et al., 1989). In an novel approach Liao et al., 2004, used an engineered E. coli methionine aminopeptidase which was able to remove bulky penultimate residues not cleaved by the wild type enzyme. Finally, it should be noted that it is sometimes possible to resolve proteins containing N-terminal Met from those lacking it by chromatographic methods (Wingfield et al., 1987b).
Dealing with Inclusion Bodies
The expression of eukaryotic proteins in E. coli often leads to the accumulation of insoluble protein called inclusion bodies (UNITS 6.3 & 6.5). Inclusion bodies can be easily observed by phase-contrast microscopy as dense bodies, usually located at the polar extremities of the cell and they can be isolated by centrifugation (Georgiou and Valax, 1999).
The rate of protein biosynthesis in prokaryotes is about ten times faster than in eukaryotes. Comparison of the rates of in vitro refolding of orthologous prokaryotic and eukaryotic proteins indicates that the former refolds six times faster. This suggests that the rate of folding correlates with the rate of elongation of polypeptide chains. Hence, part of the problem in expressing eukaryotic proteins in bacteria might be due to combination of fast synthesis and slow folding, which favors aggregation (Widmann and Christen, 2000). Proteins in the unfolded state at high concentration, even small rapidly folding proteins, are prone to aggregation due to exposure of hydrophobic surfaces that are normally buried in the native state (see Fersht, 1998, for further discussion). Some proteins are helped to fold in vivo by binding to accessory proteins called chaperones (see below for additional comments).
The formation of inclusion bodies can occasionally protect proteins against proteolysis and can also allow accumulation of proteins normally toxic to the cell; some examples include proteases (HIV-1 protease; Cheng et al., 1990) and membrane-spanning domains (Jones et al., 2000). The formation of inclusion bodies also simplifies purification of the protein, albeit in a denatured/aggregated state (see below). The main disadvantage is that the protein must be extracted with protein denaturants and then folded into a native-like conformation. For small (10-to 20-kDa) single-domain proteins, this is usually not problematic, although the overall recoveries may only be 5% to 20% of those of similar or identical proteins expressed in a soluble state. For large (40- to 70-kDa), multiple-domain proteins, recoveries may be negligible, although there have been a number of successful cases, such as the 69-kDa tissue plasminogen activator (Grunfeld et al., 1992).
The formation of inclusion bodies can sometimes be prevented by changing the promoter, host strain, and combinations thereof; controlling the growth conditions (especially the pH of the culture); adding nonmetabolizable sugars such as sucrose and sorbitol to the fermentation medium; and changing the temperature of induction, usually by lowering it (for reviews, see Schein, 1989; Wetzel, 1992; Baneyx, 1999). In an opposite approach to this, the incorporation of the ketosteroid isomerase tag usually results in inclusion body formation which can be useful for the production of peptides etc., which otherwise would be susceptible to proteolysis (see for example: Jaroniec et al., 2005).
The recombinant protein may be located in both the insoluble and soluble fractions of the cell (mixed-phase expression), and in these cases, better yields may be obtained by processing the soluble material (discarding the insoluble) even though it might constitute only a minor portion of the total expressed protein (Thatcher and Panayotatos, 1986; Wingfield et al., 1987c). Soluble protein purified from mixed-phase expressions should be carefully analyzed to check its authenticity (e.g., by mass spectrometry) as the solubility may have resulted from minor modifications such as deamidation or proteolysis of a few residues from either the N or C terminus (P.T. Wingfield unpub. observ.).
A successful approach for avoiding inclusion body formation is the use of an appropriate secretion vector (Guisez et al., 1998; Cornelius, 2001) The N-terminal secretion signal directs protein to the periplasmic space (see Localizing Protein), and translocation across the plasma membrane results in cleavage of the secretion leader sequence. The periplasm contains enzymes that accelerate folding and formation of disulfide bonds (for reviews, see Missiakas and Raina 1997). Purification is also simplified as the protein content of the periplasmic space constitutes only 4% of the total E. coli protein (Beacham, 1979).
Affinity Tag and Fusion Proteins
Apart from direct expression, there are many examples of fusion protein expression. Fusion proteins consist of the protein of interest partnered or tagged with proteins or protein domains appended to either the N-or C-terminus (or both) (UNIT 5.1; Uhlen et al., 1992; also see Table 3 in Makrides, 1996). The appended moieties are commonly called “tags” and are often linked to the host protein by a short linker sequence containing a specific chemical (e.g., Met or Asp-Pro) or protease cleavage site (e.g., thrombin). One of the main purposes of constructing fusion proteins is to facilitate the recovery and purification of the recombinant protein. The most popular fusion partners are the poly-histidine tag (His-tag) and the glutathione-S-transferase (GST-tag), these are discussed in more detail in UNITS 6.5 & 6.6. A tag may help maintain the solubility of a protein that is normally expressed in an insoluble form (LaVallie et al., 1993; Zhang et al., 1998). Alternatively, as mentioned above, the tags may promote insolubility, especially useful for protecting short, partially structured polypeptides, and for expressing proteins that are normally toxic to E. coli (e.g., proteins with membrane-associating or -spanning regions). The Gateway universal cloning system, previously mentioned, has been used to screen for improved solubility by comparing the effects of six different N-terminal fusion proteins and the His-tag (Hammarstrom et al., 2002; Unit 5.17). This type of study using conventional cloning and expression would represent a major undertaking.
The expression of fusion proteins with affinity handles, such as those containing stretches of polyhistidine (His-tagged; see also UNIT 6.5), has become extremely popular due to the ease of protein purification under both nondenaturing and denaturing solvent conditions (for more details, see discussion on Purifying Denatured Proteins). The soluble fusion proteins often have native-like conformations and are biologically active. It cannot be assumed, however, that a tag will have no effect on the protein’s function or activity. From a protein purification viewpoint, the main advantage of affinity-tagged proteins is realized when they are combined with a secretion vector (Skerra et al., 1991); in such a case, the protein will be translocated to the periplasm or the medium, though often at low concentrations, and the tagged protein can be readily purified from the culture medium after osmotically shocking the cells
Some of currently used tags are summarized by Lichty et al (2005); Esposito and Chatterjee (2006) and Unit 9.9. Waugh (2005) describes combinational tagging and some methods to remove the tags. There is also a useful discussion and recommendations for the use and sequence positioning of His-tags by Graslund et al., (2008). Although the incorporation of tags has become the de facto standard for recombinant protein production, the additional use of conventional protein purification methods such as ion-exchange and gel filtration chromatographies cannot be ignored to resolve, for example, charge and especially size heterogeneities.
SOLUBILITY AND LOCATION OF THE PROTEIN
Determining Solubility
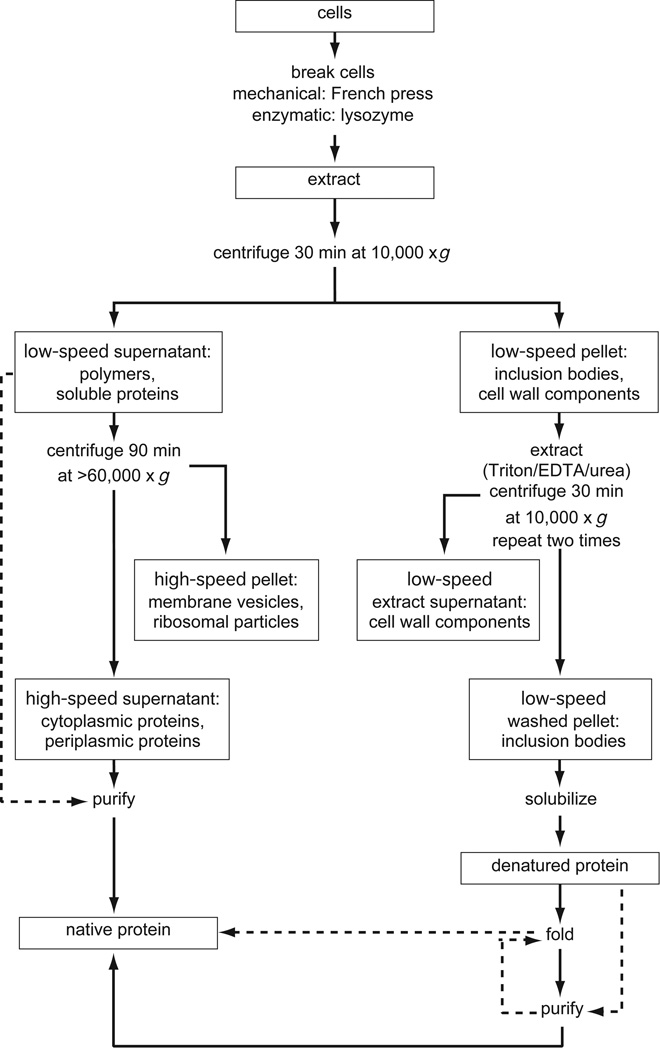
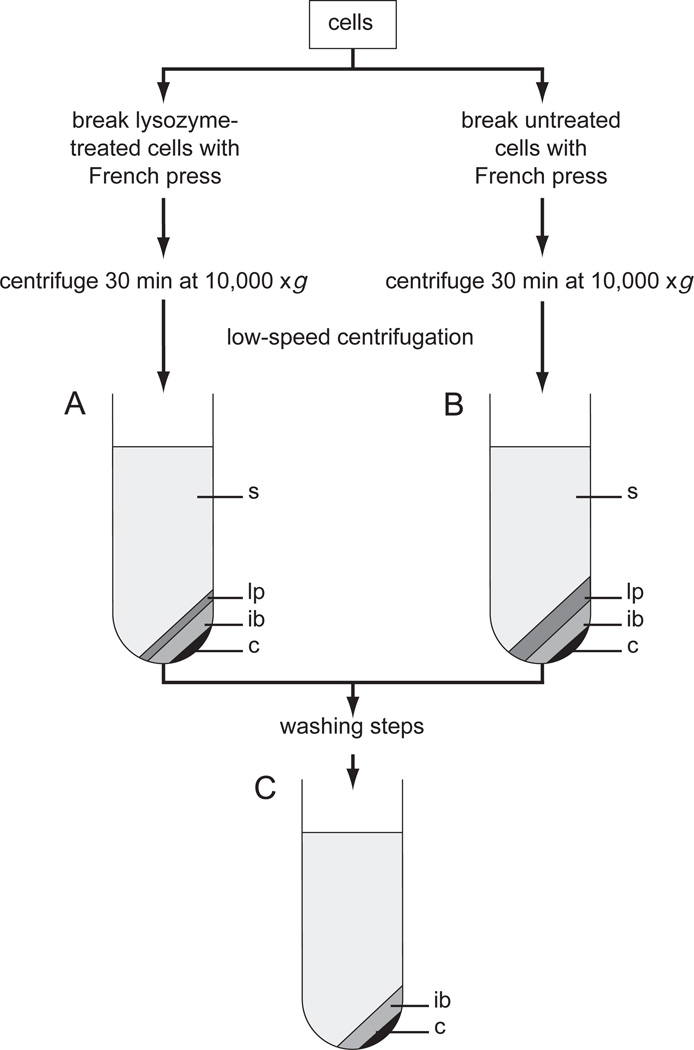
Figure 6.1.2 shows a simple centrifugation scheme that indicates how to determine the solubility of a protein expressed in E. coli (see also UNIT 5.2). The recombinant protein in the various fractions is assayed by SDS-PAGE (UNIT 10.1); if more sensitive methods are required, immunoblotting or biological assays may be used.
Figure 6.1.2.
Differential centrifugation of E. coli cell lysates. Cells are broken with a French press or by lysozyme treatment. Insoluble (inclusion body) proteins, from either the cytoplasm or periplasm, are located in the low-speed pellet, which is subjected to preextraction to remove outer membrane and peptidoglycan material. Inclusion bodies are extracted from washed pellets with strong protein denaturants such as guanidine·HCl. The solubilized protein, which is denatured and reduced (free sulfhydryl residues), is either directly folded and oxidized (disulfide bonds formed) or purified before folding. Soluble proteins (from the periplasm and cytoplasm) are located in the low-speed and high-speed supernatants. The latter can be used directly for chromatography, whereas the former requires clarification by other techniques such as ammonium sulfate fractionation or membrane filtration.
Cell breakage carried out with a French press (UNIT 6.2) will disrupt both the outer and inner membranes. The peptidoglycan layer, which lies underneath the outer membrane in Gram-negative bacteria such as E. coli, will be fragmented into sheets. Low-speed centrifugation (30 min at 10,000 × g) separates unbroken cells, bacterial outer membrane, and peptidoglycan components, and highly aggregated inclusion body proteins (pellet fraction) from soluble bacterial proteins, soluble recombinant proteins, and polymeric materials, including ribosomal protein complexes and inner membrane vesicles (supernatant fraction). High-speed centrifugation (90 min at 100,000 × g) of the low-speed supernatant will pellet polymers. Soluble proteins, derived mainly from the cytoplasm and periplasmic space, can then be recovered from the clarified supernatant. Soluble proteins in the low-speed or high-speed supernatant are purified directly using conventional methods (UNIT 6.2).
It should be noted that a working definition of solubility is the presence of protein in the supernatant after centrifugation for 100 min at 100,000 × g. This definition applies to solvents of viscosity or density close to that of water.
Occasionally, recombinant protein will be found in both the pellet and supernatant fractions after low-speed centrifugation due to the accumulation of both soluble and inclusion body proteins. If partitioning is observed only following high-speed centrifugation, then specific self-association or nonspecific association involving E. coli proteins and nucleic acid may be suspected. Recombinant proteins that normally bind RNA or DNA often bind nonspecifically to bacterial nucleic acid (Sherman and Fyfe, 1990; Wingfield et al., 1990). Lindwall et al. (2000) have developed a sparse screen approach to optimizing the buffer composition for extracting and solubilizing folded (non-aggregated) proteins.
Inclusion body proteins, which are located in the low-speed pellet fraction, can be partially purified by extracting with a mixture of detergent [usually 1% to 5% (v/v) Triton X-100] and denaturant, either urea or guanidine·HCl. The concentration of denaturant used for pellet washing is determined empirically and should be below the concentration required for solubilization of the recombinant protein; the usual ranges are 1 to 4 M urea and 0.5 to 1.5 M guanidine·HCl. The cloudy extract will consist of complex carbohydrate from the fragmented peptidoglycan layer, lipopolysaccharide, and outer membrane proteins. The inclusion body proteins in the washed pellets are then extracted with solvents that disrupt protein-protein interactions (e.g., 6 to 8 M urea or guanidine·HCl) and processed as described below.
Localizing Protein
When proteins incorporating a secretion vector are expressed in E. coli, advantage can be taken of the fact that the recombinant proteins will be located in the periplasmic space and/or the culture medium. Secretion into the medium is due to “leakage” from the periplasm and appears to depend on the level of accumulation and the fermentation conditions. Figure 6.1.3 summarizes approaches used to recover proteins selectively from the periplasmic space or the medium (see also UNIT 5.2). High-level secretion into the periplasm sometimes results in the formation of aggregates, analogous to cytoplasmic inclusion bodies (Bowden et al., 1991). Periplasmic inclusion bodies can be extracted from the low-speed pellet fraction following normal cell breakage (see Fig. 6.1.2).
Figure 6.1.3.
Localization of secreted and periplasmic proteins in E. coli. Periplasmic protein produced via a secretion vector can leak into the medium and be recovered by centrifugation (supernatant, S1) or filtration. Washing cells with an isotonic solution such as lightly buffered 0.15 M NaCl or 0.25 M sucrose can also release protein (S2). The compartmentalized periplasmic proteins are released by isotonic shock treatment by directly suspending normal cell paste or plasmolyzed cell paste into hypotonic medium. Plasmolyzed cell paste is derived by suspending cells in hypertonic medium and then pelleting. (In hypertonic medium the cell contracts, separating the inner membrane from the cell wall, and is said to be osmotically sensitized.) The hypertonic wash often releases protein (P1). The supernatant from shocked cells (P2) will contain constitutive E. coli proteins and the recombinant product. Osmotically sensitized cells can also be treated with lysozyme to fragment the outer membrane, thus releasing periplasmic proteins (P3).The pellet from the lysozyme treatment contains spheroplasts (cells with fragmented outer membranes), which are easily disrupted by detergents, sonication, or hypotonic shock to release cytoplasmic proteins.
Proteins in the medium can be recovered by subjecting the culture medium to centrifugation or filtration, steps that remove intact cells and large debris. The clarified protein is usually dilute and is often concentrated prior to purification by affinity or conventional chromatography. Periplasmic proteins can be selectively released by osmotic shock (preferred method) or by selective disruption of the outer membrane and peptidoglycan layer using lysozyme.
Apart from its use in dissecting the bacterial compartments, lysozyme is often employed to prepare complete cell lysates, especially in laboratories that do not have access to a French press. Cells treated with lysozyme can be disrupted with detergents or by brief sonication (UNIT 6.5).
Useful microscale (<1 ml) E. coli cell fractionation schemes have been based on osmotic shock treatment (Yarranton and Mountain, 1992) or repeated freezing and thawing of cells (Johnson and Hecht, 1994). UNIT 5.2 describes small-scale (1-to 25-ml) procedures for preparing samples of periplasmic extracts and extracellular media for analysis by SDS-PAGE (UNIT 10.1).
STRATEGIES FOR ISOLATION OF SOLUBLE PROTEINS
The isolation methods used to isolate soluble recombinant or non-recombinant proteins depend on the intrinsic physiochemical properties of the proteins unless they are tagged. A flow chart summarizing some of the methods commonly used for bacterial and other recombinant produced protein is shown in Figure 6.1.3 (see also Chapter 1). In Figure 6.1.3, the step, Perform Affinity Methods, refers not only to conventional affinity purification methods (see Chapter 9), but also to affinity methods based on the use of fusion proteins. A specific protocol detailing the purification of the soluble protein interleukin-1β (IL-1β) is presented in UNIT 6.2 and two more recent examples are discussed below. Comments on the various stages are given in order of their application. As mentioned earlier, affinity methods such as nickel-chelate chromatography (for hist-tag proteins) will be most commonly used following clarification of the cell lysate. It is worth mentioning that GE Heathcare Streamline Chelating (6% cross-linked agarose containing a quartz core) is extremely robust and can be added directly to crude extracts then collected on a sintered glass funnel and the hist-tagged protein eluted by step-wise elation.
Determining the Isoelectric Point
The section on Analyzing the Amino Acid Composition mentions how the isoelectric point of a protein can be estimated from the pKa values for ionizable side-chain groups. The pI can also be determined experimentally by subjecting the soluble protein extract to 1-D isoelectric focusing (UNIT 10.2) or 2-D titration curve analysis (Watanabe et al., 1994; UNIT 7.3). If the recombinant protein is not a major component in the cell extract, specific detection on the 2-D gel by immunoblotting will be required. The calculated pI can be used to optimize the buffer pH in subsequent ion-exchange steps.
Breaking Cells
Cells are efficiently broken by high-pressure homogenization using a continuous-fill French press, which is suitable for processing volumes of 40 to 250 ml (reviewed by Hopkins, 1991; see UNIT 6.2). (Yeast cells can also be conveniently broken with the French press, although least two passes are required). For volumes exceeding 500 ml, the MantonGaulin-APV homogenizer (APV Gaulin) should be used. Sonication is also useful for breaking cells but is best suited for volumes <100 ml. Alternatively, the outer cell wall can be enzymatically digested with lysozyme (200 µg/ml) and the cells broken by detergents, sonication, or both (Kaback, 1971; Burgess and Jendrisak, 1975; UNIT 6.5). Proteins that are secreted into the periplasmic space can be selectively released by hypotonic (osmotic) shock (Heppel, 1967).
The viscosity of the cell lysate may be high due to released nucleic acid. Before centrifugation, the viscosity must be reduced either by sonicating or by adding DNase (25 to 50 µg/ml plus 5 to 10 mM Mg2+) and RNase (50 µg/ml; no Mg2+ requirement). A standard protease inhibitor mixture should be included in the buffer—containing, for example, 2 to 5 mM EDTA, 0.5 to 1.0 mM phenylmethylsulfonyl fluoride (PMSF) or 5 mM benzamidine, and 1 µM pepstatin A. The serine protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) is a water-soluble substitute for PMSF with a much longer half-life in aqueous solution and is used at ~ 50 µM. (Roche Applied Science: http://www.roche-applied-science.com. Go to the Biochemistry section to download the booklet: “The complete guide for protease inhibition” that lists properties for most commercially available reagents). The addition of α2-macroglobulin (1 µg/mg recombinant protein) before the final purification step(s) can protect protease-sensitive proteins (Ultsch et al., 1991; see also section on MAP30 purification below). The crude extracts should be kept cold and the recombinant protein taken rapidly to a stage of the purification process where it is stable against contaminating proteases (e.g., as an ammonium sulfate precipitate).
Clarifying Cell Extract by Centrifugation or Selective Precipitation
The lysate is subjected first to low-speed centrifugation to remove unbroken cells and large cellular debris, then to high-speed centrifugation to remove ribosomal material and other particulates (see Fig. 6.1.1). If an ultracentrifuge is not available, the extract can be clarified by the following techniques: ammonium sulfate or polyethylene glycol fractionation (reviewed by Scopes, 1994), phase partitioning (reviewed by Walter and Johansson, 1986), and membrane filtration (van Reis and Zydney, 2001; useful guides on filtration technology are available on-line from Millipore and GE Heathcare Lifesciences. Expanded bed adsorption is a chromatographic technique where crude extracts can be directly applied to adsorbents, for example ion exchangers, without initial clarification (see GE Heathcare Life Sciences for literature http://www.gelifesciences.com).
Proteins that bind tightly to nucleic acid can be selectively precipitated with polyethyleneimine and resolubilized by salt extraction (Burgess and Jendrisak, 1975). In practice, particular properties of the protein can be exploited at this stage; for example, the protein of interest may be soluble under conditions where most E. coli proteins are insoluble, such as acidic pH or high temperature.
Applying Clarified Extract to a Weak Anion Exchanger
Fractionating the extract with an anion-exchange resin is a useful first step as it removes host E. coli proteins, many of which have pI values in the range 5.0 to 7.0 and will thus bind to a column equilibrated in 50 to 100 mM Tris·Cl, pH 7.5 to 8.0. The positively charged matrix will also tightly bind non-proteinaceous materials such as nucleic acid and other polyanionic species (e.g., lipopolysaccharide derived from the bacterial outer membrane). A useful cleanup of the protein will take place whether or not the protein of interest binds to the column (see UNITS 6.2 & 6.5). The following column sizes are recommended for processing extracts: for 5 g cells, 2.5-cm diameter packed to a height of 10 to 15 cm; for 50 g cells, 5.0-cm diameter packed to a height of 20 cm.
Preparing for Repeat Ion-Exchange Step
Before repeating ion exchange, the solvent pH and ionic strength usually need adjustment. This can be carried out by dialysis (UNIT 6.2) or by gel filtration on a desalting column using, for example, Sephadex G-25 or G-50 (UNIT 8.3). In preparation for cation-exchange chromatography, dialysis against slightly acidic buffers (pH 5.0 to 6.0) will result in the helpful precipitation of some E. coli proteins. It may also be advisable to include a relatively low concentration of urea (0.5 to 2 M) or a nonionic or zwitterionic detergent in the dialysis buffer to minimize coprecipitation with contaminants (for listings of detergents and properties, see Anatrace http://www.anatrace.com; le Maire et.al., 2000). Basic proteins (pI >9.0), which do not bind to the DEAE column, can be applied after dilution to a cation exchanger equilibrated at pH 7.0 to 7.5 without careful buffer exchange (Allet et al., 1988).
Repeating Ion-Exchange Chromatography
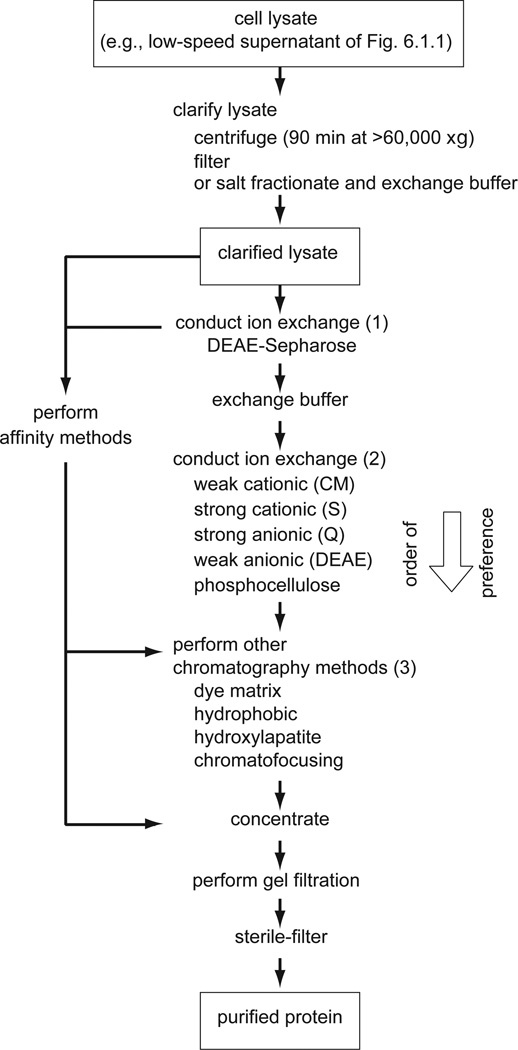
For a second round of ion-exchange chromatography, one of the ion-exchange resins indicated in Figure 6.1.4 should be used. Selection kits are available for rapidly screening and selecting the most suitable ion-exchanger (GE Heathcare Life Sciences and others). For cation-exchange chromatography, phosphate buffer (10 to 50 mM) between pH 5.0 and 7.5 should be tried first. Cellulose Phosphate (a bifunctional cation exchanger manufactured by Whatman (now part of GE Heathcare: resin may no longer be available but supply from other suppliers can be checked) is effective for nucleic acid–binding proteins (Kelley and Stump, 1979). Protein is usually eluted from cellulose phosphate columns using phosphate gradients.
Figure 6.1.4.
Purification of soluble proteins from bacterial cell and other cell lysates. Abbreviations for ion-exchange resins are as follows: CM, carboxymethyl; DEAE, diethylaminoethyl; Q, quaternary ammonium; S, methyl sulfonate. The order of preference for the stages of ion-exchange (2) and other methods (3) is based on the author’s opinion and does not necessarily represent a consensus view. On the other hand, the use of a DEAE-based matrix at an early stage (1) is common practice. Affinity methods (see text and Chapter 9) can be performed at any stage following clarification of the lysate.
After two stages of ion exchange, many proteins will be pure enough for the final gel-filtration step (see Performing Gel Filtration). However, if the sample contains contaminants close in size to the protein of interest, then further purification is required. Some of the frequently used methods are listed in Figure 6.1.4. Hydrophobic-interaction chromatography (UNIT 8.4) is especially useful following ammonium sulfate fractionation or salt elution from an ion-exchange resin. Screening kits are also available for rapidly checking protein binding on several different agarose-dye matrices (Sigma at http://www.sigmaaldrich.com).
Performing Gel Filtration
The final purification step of gel filtration (using a column 1.5 to 5.0 cm in diameter and 60 to 100 cm in length) will provide good separation of the recombinant protein from higher- and lower-molecular-weight E. coli protein contaminants. Gel filtration will also separate aggregated or highly associated recombinant protein from the physically stable form of the protein (e.g., monomer or dimer). Finally, gel filtration chromatography allows for easy exchange of the buffer. The protein solution is usually concentrated before being applied to the column. After chromatography, the protein will be diluted three- to five-fold (or more) and may therefore require repeat concentration.
Other Methods
In addition to the generalized approach described, affinity methods can be applied at any stage following clarification of the extract. Biospecific affinity can be exploited with immobilized natural ligands such as antibodies, substrates, and receptor ligands. Affinity chromatography, which selects for particular classes of proteins, is carried out with immobilized lectins (for glycoproteins), dyes (for nucleotide-binding proteins), and nucleic acids or heparin (for RNA-and DNA-binding proteins). Commercially available antibodies against post-translationally modified residues (e.g., phosphotyrosine) are also useful. The application of affinity tags or fusions has been previously described. Affinity methods are most useful when high degrees of purification are required—e.g., for proteins secreted into the medium, for small-scale isolations, or for rapid purification requirements.
The most commonly used affinity method is immunoaffinity chromatography. The ideal reagent is a monoclonal antibody that has been specifically selected to have a moderate-to-low affinity for the ligand in question, thus allowing elution under nondenaturing conditions. Antibodies raised against peptides often have lower affinities for the native protein than antibodies raised against the intact protein. Elution from peptide-antibody immunoaffinity columns can be achieved using the competing immunizing peptide (reviewed by Sutcliffe et al., 1983). Directed immobilization of the antibody, where only the Fc domain is bound to the column matrix and the antigen binding site (Fab domain) is thus oriented away from the matrix, results in higher binding efficiencies. An oriented antibody matrix can be made by binding antibody to immobilized protein A-Sepharose (or protein G-Sepharose) and fixing it in position with a covalent cross-linking reagent (Schneider et al., 1982; commercial kits can be obtained from GE Heathcare, Thermo Scientific and others.
Compilations of standard chromatographic fractionation media (and related supplies) are available on the various manufacturers’ web sites (GE Heathcare, BioRad etc). Companies which make chromatography matrices etc., are constantly being shuffled and rebranded; therefore, the supply of your favorite media may not be guaranteed in the future so be aware of alternate supplies and suppliers.
STRATEGIES FOR ISOLATION OF INSOLUBLE PROTEINS
Recombinant proteins expressed in E. coli that are located in the low-speed pellet fraction (see Fig. 6.1.2) following cell lysis are highly aggregated (i.e., inclusion bodies). Inclusion bodies are normally derived from protein aggregation in the cytoplasm, or in the periplasm if a secretion vector was used. As mentioned above, protein can also be located in either the low-or high-speed pellet fractions because of interaction with bacterial nucleic acids. Furthermore, if the protein is known to undergo polymerization in vitro (e.g., viral nucleocapsid subunits), expression in E. coli can also be expected to lead to polymerization in vivo to varying degrees, and such proteins will be partitioned in both the supernatant and pellet fractions (Wingfield et al., 1995). There are also examples of membrane proteins that, when expressed in E. coli, associate with the inner cytoplasmic membrane and can be extracted with nondenaturing detergents (Bibi and Beja, 1994, and references cited therein).
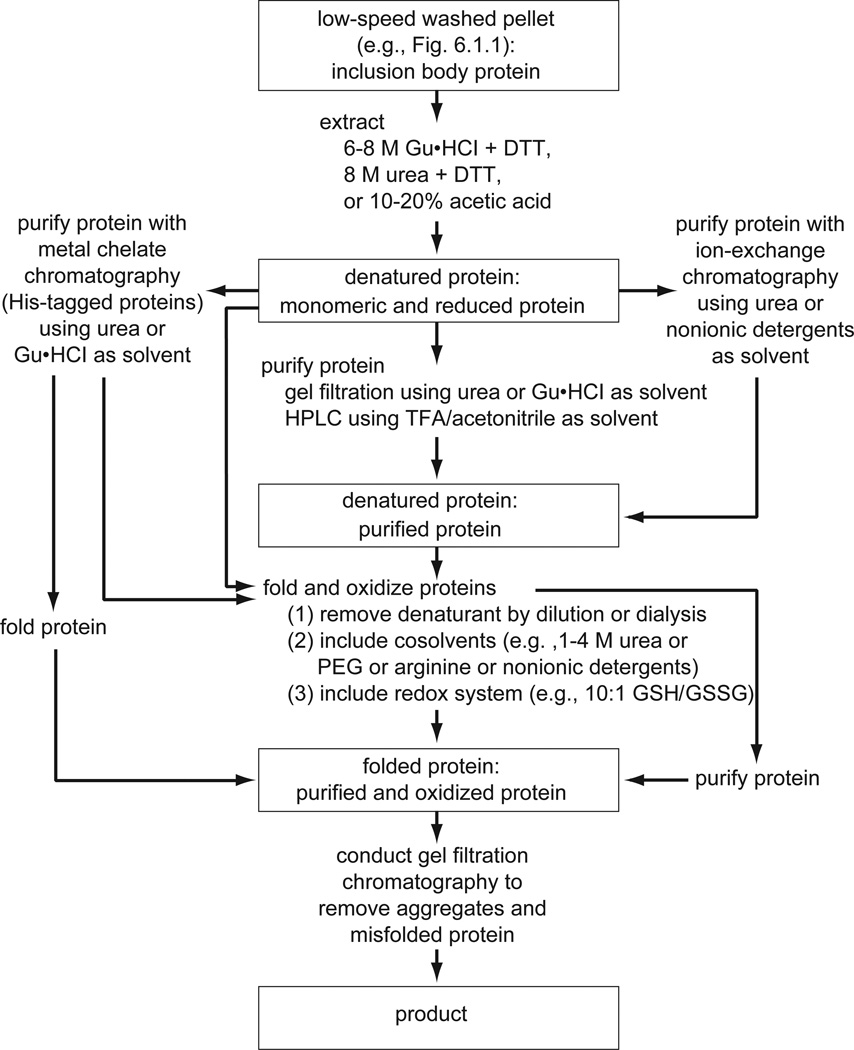
When apparent insolubility is due to interactions involving folded protein as described above, extraction under nondenaturing conditions should be attempted, for example, using various pH buffers containing salt (e.g., 0.25 to 1.0 M NaCl) and nondenaturing detergents (e.g., 10 mM CHAPS or 2% Triton X-100). Insolubility due to classic inclusion body formation requires extraction with denaturing solvents, and the remainder of this section deals with this subject. The flow chart in Figure 6.1.5 illustrates some of the approaches possible for processing protein extracted from inclusion bodies.
Figure 6.1.4.
Folding and purification of inclusion body proteins from E. coli. The protein is extracted with protein denaturants such as guanidine·HCl (Gu·HCl), urea, or an organic acid. The reductant dithiothreitol (DTT) is included to prevent artificial disulfide bond formation (especially intermolecular bonds). The denatured protein can be purified by various methods and then folded, or it can be directly folded. Typically, some purification (e.g., gel filtration in Gu·HCl) prior to folding is recommended as it often results in higher folding yields. Protein folding and oxidation are carried out concurrently. Disulfide bond formation is catalyzed by low-molecular-weight thiol/disulfide pairs such as reduced (GSH) and oxidized (GSSG) glutathione. GSH/GSSG ratios of 5:1 to 10:1 are normally used, which are similar to those found in vivo in the endoplasmic reticulum (Hwang et al., 1992). A cosolvent is included to maintain solubility during folding. Folded protein is purified if necessary (purification is usually needed if the protein is directly folded). Gel filtration is a useful final step for removing aggregated and or misfolded protein.
Breaking Cells
Cells can be broken by mechanical means (UNIT 6.2), enzymatically with lysozyme (UNIT 6.5), or by a combination of methods (UNIT 6.5). It is advantageous to break cells as completely as possible, as any unbroken cells will be located in the low-speed pellet fraction from which the recombinant, insoluble protein will be extracted.
Preparing Washed Pellets
The object of the initial low-speed centrifugation and pellet “washing” is to extract as many E. coli contaminants as possible without solubilizing the recombinant protein. This is usually carried out as described in the section on Determining Solubility (see also Fig. 6.1.1).
When a fixed-angle rotor is used, pellets from the low-speed centrifugation consist of at least two light-colored layers and a darker, hard-packed pellet at the bottom of the tube (Fig. 6.1.6). The hard-packed material is probably a small amount of unbroken cells. The next layer is inclusion body protein, and the top layer (least dense and lightest in color) is outer membrane and peptidoglycan fragments. Analysis of the top layer by SDS-PAGE (after heating proteins in SDS sample buffer at >80°C) will reveal two strong bands at ~ 35 and 38 kDa representing OmpA and the matrix proteins OmpC and OmpF, respec-tively, from the outer membrane (DiRienzo et al., 1978; see also Fig. 6.3.2). The outer membrane/peptidoglycan layer can be partially removed by resuspending and centrifuging at reduced speed (or time) or by diluting the suspension. Alternatively, the cells can be pretreated with lysozyme prior to the French-press cell breakage as described in UNIT 6.5. The lysozyme treatment reduces the size of the loosely pelleted outer membrane/peptidoglycan material so it locates predominately in the low-speed supernatant (Fig. 6.1.6). The recombinant protein in a well-prepared washed pellet will typically be >60% pure when analyzed by SDS-PAGE (UNIT 6.3).
Figure 6.1.5.
Preparation of washed pellets using lysozyme and the French press. Cells are broken with the French press with or without prior treatment with lysozyme. After low–speed centrifugation using a fixed-angle rotor, the contents of the centrifuge tubes have the characteristics shown. The contents of tubes A and B are labeled: s, supernatant; lp, loose pellet; ib, inclusion body protein; and c, unbroken cells and large cellular debris. The loose pellet material is derived from the outer cell wall and outer membrane (see text for further details). After washing the insoluble material (UNIT 6.3), the pellet should consist mainly of the inclusion body layer (tube C), and the supernatant should be fairly clear.
Extracting Protein
The washed pellets are extracted with high concentrations of protein denaturants such as 6 to 8 M guanidine·HCl or urea. It should be noted that some proteins are resistant to denaturation with high concentrations of these reagents, especially urea. Some washed pellets extracted with 8 M guanidine·HCl can be viscous and unsuitable for direct chromatography. In these cases, pre extraction of the washed pellets with a limiting concentration (0.5 to 2.0 M) of guanidine·HCl can often overcome this problem.
Solubilization with the anionic detergent N-lauroylsarcosine (Nguyen et al., 1993; Burgess and Knuth, 1996) and with 10% to 20% acetic acid has also been useful (UNIT 6.5); other denaturants for extracting inclusion bodies are described elsewhere (UNIT 6.3; Marston and Hartley, 1990). For background information on the mode of action of protein denaturants, readers should consult the reviews of Tanford (1968) and Creighton (1993). If the protein contains cysteine residues, it is essential to include a reducing agent, preferably 5 to 10 mM dithiothreitol (DTT). Even in the presence of strong protein denaturants, it may be necessary to sonicate or heat samples briefly to completely disperse and solubilize the protein.
The extraction process should completely disaggregate and denature the protein into unfolded monomers. Urea is not recommended for the initial extraction. For example, even if it is known that a native version of protein can be unfolded with 4 M urea, the same protein in an E. coli inclusion body will almost certainly not be completely extracted as unfolded monomers with that same concentration of urea (or in most cases, even with 8 M urea). Initial extraction trials should be carried out with guanidine·HCl, which is more effective than urea. Most proteins will be extracted with 6 to 8 M guanidine·HCl. There should be adequate reductant present to maintain sulfhydryl groups in the reduced state, and thus prevent artificial disulfide bond formation. The presence of EDTA and a slightly acidic pH of 6.0 to 6.5 will help minimize cysteine oxidation. The extract may require clarification by filtration or centrifugation.
Choosing Purification or Folding
The extracted protein can be further purified, or it can be directly folded and then purified. Protein folding appears to be unaffected by the protein background in bacterial extracts (London et al., 1974), however, removal of nonproteinaceous material prior to folding has been reported to be beneficial (Darby and Creighton, 1990). It is worth considering that high concentrations of background bacterial protein may promote aggregation of the unfolded recombinant protein by macromolecular crowding effects (Ellis, 2001). If purification of protein in the denatured state is possible, use the purified material to develop a folding protocol. Then use this protocol with clarified protein extracts, or better still with protein partially purified by DEAE-Sepharose, to observe if the presence of contaminants has any effect on the yield of folded protein.
Finally, there may be specific reasons for purifying proteins in the denatured state. For example, some proteolytic enzymes, such as HIV-1 protease, self-digest (undergo auto- proteolysis) in the uninhibited state (Mildner et al., 1994, and references cited therein) but can be purified intact in the denatured (inactive) state, then refolded when required. Other proteins once folded may have low solubilities and be especially susceptible to aggregation, resulting in poor behavior on column matrices (see VP26 purification below). However, in general, unfolded proteins are more susceptible to chemical and proteolytic modifications.
Purifying Denatured Proteins
If the protein is extracted with guanidine·HCl, gel filtration is a useful first purification method; often protein >80% pure can be obtained (UNIT 6.3; Wingfield et al., 1997). The proteins exist as random coils in the denaturant and their elution from the column should be in order of their molecular weight and not be influenced by shape. If the protein is located in several peaks there may have been incomplete solubilization during the extraction. In this case, 8 M guanidine·HCl should be used for the extraction and the protein dispersed by sonication or by heating if necessary. Another possibility is intermolecular disulfide bond formation, in which case the DTT concentration in the sample and column buffers should be increased. The column can often be equilibrated and eluted with lower guanidine·HCl concentrations (e.g., 4 M) than those used for the actual extraction process. Only monomeric protein should be selected for further processing. The protein at this stage can be stored frozen, ideally at −80°C.
The partially purified protein in guanidine·HCl can be directly folded (see Performing Protein Folding), or the denaturant can be exchanged by dialysis or gel filtration for 1% to 5% (v/v) acetic or formic acids (acetonitrile at 5% to 10% v/v can also be included) and then lyophilized. Alternatively, the protein can be acidified with trifluoroacetic acid (TFA; ≤0.1% v/v) and further purified by reversed-phase chromatography (Wingfield, 1997; Wingfield et al., 1999). Useful high-flow matrices (Source 15RPC from GE Heathcare) can be purchased as bulk media. These matrices may not have the resolution of traditional prepackaged silica-based reversed-phase columns, but they have high capacity, can be eluted at higher flow rates, and are stable over a wide range of pH. Proteins eluted with acetonitrile/TFA are also suitable for lyophilization.
Proteins tagged with histidine residues can be purified in guanidine·HCl, urea, or even SDS containing buffers, using metal chelate chromatography (UNIT 6.5). There are many reports of “on-column protein folding” by binding the unfolded protein in guanidine·HCl or urea and then accomplishing folding using a reverse urea gradient (e.g., Gulnik et al., 2001).
Proteins in urea and non-ionic or zwitterionic detergents (e.g., CHAPS) can be purified by ion-exchange chromatography (e.g., Wingfield et al., 1990). For ion-exchange chromatography, better results have been reported using protein that has been first extracted with guanidine·HCl, and then exchanged into urea (Shire et al., 1984).
If urea is used either for extraction or for maintaining solubility during refolding, a cyanate scavenger such as a glycine or Tris-based buffer should be included to prevent carbamylation of the protein (Stark et al., 1960). For critical work, urea can be deionized with a mixed bed ion-exchange resin (see discussion of Protein Folding Reagents in APPENDIX 3A).
Performing Protein Folding
Protocols for folding proteins basically involve controlled removal of the denaturant under conditions that minimize aggregation and allow correct formation of disulfide bonds. For overviews of the practical aspects of protein folding, see UNIT 6.4; Wetzel (1992); Thatcher et al. (1996); Rudolph et al. (1997); Lilie et al. (1998); De Bernardez Clark et al. (1999); and De Bernardez Clark (2001); Vallejo and Rinas (2004); Shing and Panda (2005) Yang et al., (2011);Yamaguchi and Miyazaki (2014).
To minimize nonproductive aggregation, folding is normally carried out at low protein concentrations (e.g., 0.01 to 0.10 mg/ml); for small, single-domain proteins, higher concentrations (e.g., 0.1 to 1.0 mg/ml) can often be tolerated. Dilution and dialysis are the most common methods for removing the denaturant. Solubility during folding can be maintained with co-solvents such as nondenaturing concentrations of urea (1 to 4 M; London et al., 1974; UNIT 6.5) or guanidine·HCl (0.1 to 1.5 M; Orsini and Goldberg, 1978), arginine (0.4 to 0.8 M; De Bernardez Clark et al., 1999), nonionic detergents and lipids (Zardeneta and Horowitz, 1994), cationic detergents (Puri et al., 1992), and polyethylene glycol (PEG; Cleland et al., 1992). These various additives function by minimizing intermolecular associations between “sticky” hydrophobic surfaces present in folding intermediates. For further discussion of aggregation versus folding, see Goldberg et al. (1991) and Kiefhaber et al. (1991). Additives such as ammonium sulfate, glycerol, sucrose, enzyme substrates or inhibitors, and ligands have also been used to improve protein folding (see Table 1 in De Bernardez Clark et al., 1999, for a useful list of additives used in folding).
Protein expressed in the cytoplasm of E. coli is in the reduced state; this is true for both soluble and insoluble proteins. Once insoluble protein is solubilized, it needs to be maintained in a reduced state by the presence of reductant until protein folding is initiated. The oxidative formation of disulfide bonds (one of the rate-limiting steps in protein folding) can be catalyzed by low-molecular-weight thiol and disulfide pairs such as reduced and oxidized glutathione (GSH/GSSG). Redox buffers facilitate oxidation through thiol/disulfide exchange reactions (reviewed by Wetlaufer, 1984; Creighton, 1984; Gilbert, 1995). Normally GSH/GSSG ratios of 5 to 10 are used with a total glutathione concentration of 1 to 5 mM (Wetlaufer, 1984). To reduce the rate of GSH loss due to air oxidation, 1 mM EDTA should be included in the buffer (Wetlaufer et al., 1987). The optimal concentrations and ratios of reagents must be established in an empirical manner. Folding and oxidation are normally carried out concurrently (for further details, see Rudolph et al., 1997). Analogous to the approach commonly used to optimize conditions for protein crystallization, various screens have been developed to establish initial conditions for protein renaturation and oxidation (Hofmann et al., 1995; Armstrong et al., 1999) and kits are commercially available (FoldIt Screen from Hampton Research at http://www.hamptonresearch.com).
For examples of preparative protein folding, see UNIT 6.5. In addition, some recent examples from the author’s laboratory are given below. The refolding of Fab fragments expressed in E. coli (Buchner and Rudolph, 1992) is illustrative of the systematic and empirical approach used to optimize folding conditions. Other examples of interest are described by Kohno et al. (1990) and Grunfeld et al. (1992).
Protein-assisted folding and oxidation
Protein folding in vivo is assisted in both eukaryotes and prokaryotes by two classes of accessory proteins: folding catalysts (for a review, see Schiene and Fischer, 2000) and molecular chaperones (Eisenberg, 1999; Feldman and Frydman, 2000; Saibil, 2013). Folding catalysts accelerate rate-limiting steps in protein folding such as disulfide bond formation (Narayan, 2012) and the rotation of X-Pro bonds (peptidyl prolyl cis-trans isomerase) during protein folding. Chaperones bind denatured or unfolded proteins thus preventing misfolding and aggregation. The cytoplasm of E. coli is maintained in the reduced state by thioredoxin and the glutathione/glutaredoxin pathways. In hosts where the reduction of thioredoxin and glutathione is impaired by mutations to the thioredoxin reductases and glutathione reductase genes, the resultant oxidizing conditions allow the formation of disulfide bonds in expressed proteins located in bacterial cytoplasm (Bessette et al., 1999). Expression kits are commercially available with Origami host strains which are K-12 derivatives that have mutations in both the thioredoxin reductase (trxB) and glutathione reductase (gor) genes, (Novagen: http://www.emdmillipore.com). The periplasm of E. coli also contains protein disulfide isomerases, the Dsb enzymes, which have thioredoxin-like folds and act as strong thiol:disulfide oxidants (Missiakkas and Raina, 1997; Braun et al., 1999). Secretion of proteins into the periplasmic space has been the traditional approach for producing oxidized proteins in vivo and is well suited for proteins that are toxic to the cell when expressed in the cytoplasm (Cornelis, 2000). In unit 6.10 there is an example of the production of antibody fragments by independent secretion of heavy chain fragment and light chain to the periplasm where they form disulfide bridges and associate. To direct secretion to the periplasm, the heavy chain fragment has the N-terminal signal sequence from the periplasmic protein pectate lyase B (pelB) of Gram-negative bacteria and light chain, the signal sequence from the major outer membrane protein (ompA). In addition to providing an oxidizing milieu, the periplasm of E. coli also contains thiol-disulfide oxidoreductases that catalyze the formation of disulfide bridges, as well as other enzymes that promote protein folding such as peptidyl-prolyl cis/trans isomerases.
As mentioned, molecular chaperones prevent aggregation by interacting transiently with hydrophobic patches on unfolded proteins and suppressing aggregation and promoting folding (UNIT 6.4; reviewed by Jaenicke, 1993; Ellis and Hart, 1999; Feldman and Frydman, 2000). There are now many examples of chaperone-assisted protein expression in which the endogenous levels of the bacterial chaperones GroES and GroEL (~1%) are increased up to ten-fold by co-expression with a target protein (Cole, 1996; Goenka and Rao, 2001). Often, increases in soluble protein expression are observed, but this is not always the case.
Chaperones have also been used in vitro as protein folding reagents and some examples of folding in the presence of protein disulfide isomerase, peptidyl prolyl cis-trans isomerase and GroES/GroEL are given in Rudolph et al. (1997). Protocols for the high-level expression and rapid purification of E. coli GroEL and GroES are described by Kamireddi et al. (1997). Clontect Laboratories (http://www.clontech.com) produce a chaperone plasmid kit consisting of five different plasmids, each of which is designed to express multiple molecular chaperones that function together as a "chaperone team" to enable optimal protein expression and folding and to reduce protein misfolding. Co-expression of a target protein with one of these five chaperone plasmids could increase the recovery of soluble protein. Clontect also market the Takara-brand pCold Expression vectors which contain the cold shock protein A (cspA) promoter for expression of high purity and high yield recombinant protein in E. coli (Quing et al., 2004) These vectors selectively induce target protein synthesis at low temperatures (15°C) while the bacterial cell is in a state of suppressed host protein expression and decreased protease activity. The pCold vectors can be used in conjunction with the chaperone plasmid set. A recent and interesting application of the cold shock vector system is the bacterial expression of the most abundant protein in human cytoplasm, namely, beta-actin (Tamura et al, 2011).
Purifying Folded Protein
Once the protein has been folded, any of the purification methods discussed in Chapters 8 and 9 can be used. The number of purification steps required should be fewer than those for a protein expressed in a soluble state because of the purification factor obtained by preparation of washed inclusion bodies (UNIT 6.3). One of the purification methods that should be included is gel filtration, which may be the only one required. A correctly selected matrix should remove any remaining E. coli proteins and separate aggregated and misfolded protein from the native folded protein. Misfolded protein may be expected to have a larger molecular radius (higher apparent mass) than the corresponding native protein.
Monitoring Protein Folding
The restoration of function (e.g., enzymatic or biological activity) is perhaps the best criterion for detecting successful folding. However, it is not always practical to use activity measurements to monitor folding. It is also worth mentioning that an unfolded protein may become activated following the dilution required for many activity measurements. Conversely, native proteins can be denatured or inactivated during prolonged incubation at 37°C or by adsorption to microtiter plates. The use of antibodies to monitor protein folding is briefly reviewed by Goldberg (1991), and reviews of common spectroscopic methods, such as circular dichroism and fluorescence, are provided in Chapter 7 and by Schmid (1997); Creighton (2010).
EXPRESSION OF GLYCOPROTEINS
Because E. coli lacks glycosylation machinery, expression of glycoproteins in E. coli systems results in the synthesis of nonglycosylated variants. Glycoproteins expressed in E. coli are often, but not always, insoluble. In vitro folding studies with glycosylated and nonglycosylated forms of proteins indicate that the carbohydrate can stabilize folding intermediates, and thus enhance folding, while not necessarily affecting the stability of the native state (Kern et al., 1993, and references cited therein). In eukaryotic cells, interference with protein glycosylation can lead to the formation of misfolded, aggregated, and degraded protein. This indicates that in vivo glycosylation (N-linked) may also prevent the aggregation of folding intermediates (reviewed by Helenius, 1994). Detailed NMR studies on glycoproteins have clearly shown that carbohydrates stabilize folded proteins and even prevent marginally stable proteins from unfolding (for a review, see Wyss and Wagner, 1996).
Despite potential pitfalls, many nonglycosylated protein variants have been successfully folded from E. coli inclusion bodies. Examples include cytokines of biomedical importance such as granulocyte/macrophage colony-stimulating factor (GM-CSF; Diederichs et al., 1991) and interleukin 5 (IL-5; Milburn et al., 1993). Inclusion body formation was avoided in some studies by using secretion vectors; examples include GM-CSF (Walter et al., 1992) and the extracellular domain of the human growth hormone receptor (deVos et al., 1992). The aforementioned proteins have been crystallized and their structures determined by X-ray crystallography, supporting the view that the structural integrity and conformation of the proteins were not affected by the lack of glycosylation and their respective preparative histories.
If a glycoprotein of interest is available from a eukaryotic recombinant expression system or if the natural protein is available, then before investing time with E. coli expression, it may be worthwhile to determine whether the protein can be denatured and refolded in vitro. Pilot experiments can be carried out on intact protein and on protein enzymatically deglycosylated with glycosidases and, if disulfides are present, with and without reduction. Of course, if the protein can be secreted to the periplasm, aggregation and the necessity for in vitro folding may be avoided.
The production of deglycosylated proteins in E. coli expression systems for in vitro biochemical and structural studies is obviously of great value; however, the proteins may not always be suitable for in vivo studies due to low biological activity. Compared to authentic proteins, nonglycosylated variants can have a reduced circulatory lifetime and can exhibit increased immunogenicity and protease sensitivity (Rasmussen, 1992).
For the production of glycoproteins for therapeutic applications (Ghanderi et al, 2012) mammalian cell expression is the system of choice. Furthermore, mammalian expression has enabled the structural investigation of a whole new set of targets including large, multi-domain and highly glycosylated eukaryotic cell surface receptors and their supra-molecular assemblies (Aricescu and Owens, 2013).
STRATEGIES FOR ISOLATION OF MEMBRANE PROTEINS
Membrane proteins contain one or more regions which are anchored or inserted in lipid bilayers. These membrane associated domains are often helical and amphiphatic and are released or solubilized using detergents. Detergents associate to form micelles and the membrane associated regions insert into these micelles which now act as surrogate membranes. It is only the original membrane associated region that binds detergent and as a rule of thumb one micelle per protein (this would have to verified by direct determination of the protein: detergent ratio). Commonly used detergents are listed in Appendix 1B and the company Antarace (http://www.anatrace.com) provides lots useful on-line information. Also, Chapter 29 deals specifically with membrane proteins and describes some of the recombinant systems used to express membrane proteins.
In the following section a few strategic decisions for isolating and characterizing membrane proteins will be briefly discussed.
Firstly, there is the decision as whether to truncate the membrane associated regions and express the ectodomain. This removes the problem of requiring detergents and assumes that the functionality of the ectodomain is independent or unaffected by the truncation. There are numerous examples of this in the literature including the HIV-1 gp41 ectodomain work described below.
Secondly, the membranes hosting the recombinant proteins can be isolated by subcellular fractionation taking advantage of the density differences between lipoprotein, protein and protein – nucleic acid complexes. This results in enrichment of the target protein as opposed to direct detergent extraction from the cells. Small scale extractions should be used to select the best detergent: use, for example, 5- to 10-fold higher than the detergent critical micelle concentration (cmc), mix (homogenize, sonicate etc) and then spin at 100,000 × g for 1–2 h (or perform small scale gel filtration chromatography). Make a judgment on most effective solubilization conditions and remember that the detergent can always be switched once the protein purified. Once solubilized, the protein can be isolated using standard purification techniques but you have be aware that you are dealing with a binary system, protein plus detergent, and the detergent will effect the physiochemical properties of the protein, examples, higher mass during gel filtration and charge properties shifts depending on whether the bound detergent is anionic, cationic or zwitterionic (no change). Also, with tagged proteins, the detergent may affect binding to affinity matrices.
The concentration of proteins in detergents is often not straightforward, especially with membrane filtration, and the comments in unit 17.9 (critical parameter and troubleshooting section) should be consulted for details.
If membrane proteins are produced in E.coli and appear aggregated they can be extracted under denaturing conditions and folded using various detergent combinations (see for example, Lakamek et al., 2014). There is an example of the purification of a membrane protein receptor expressed in E. coli in Chapter 6 (unit 6.8).
SOME EXAMPLES OF PROTEIN EXPRESSION AND PURIFICATION
Examples of protein expression and purification can be found in most biochemical journals, two which may be especially useful: Protein Expression and Purification (http://www.academicpress.com/pep), which covers advances in the expression and purification of recombinant proteins mainly from E. coli although other expression systems are often included; and Current Opinion in Biotechnology, which regularly provides updates on various aspects of recombinant protein production as well as useful reference lists. Detailed protocols are also given in the units of this Chapter and a few recent examples of protein expression and purification are discussed below to illustrate some of the general approaches used to deal with soluble and insoluble E. coli protein expression.
Soluble Proteins
HIV Nef
Nef is a 205-residue myristolylated protein expressed at high levels in the early stages of HIV infection. The protein is important for the induction of AIDS and is being actively researched as a potential drug target. Unlike most HIV-1 and related proteins expressed in bacteria, Nef is recovered from the soluble fraction of E. coli extracts. The purification protocol adopted following cell breakage and low-speed centrifugation is fairly straightforward comprising two stages of ion-exchange chromatography using DEAE-Sepharose (weak exchanger) followed by Q Sepharose (strong exchanger) and finally gel filtration using Superdex 75. Characterization of the purified protein yielded the following information.
Nef has a maximum solubility of ~ 0.5 to 0.6 mM (<10 mg/ml) in low-ionic strength buffers at pH 7.5 to 8.0, (e.g., 5 mM Tris·Cl). The solubility can be increased by the inclusion of nondenaturing concentrations (2 M) of urea, as established by titration studies monitored by far-UV circular dichroism. Acetonitrile (5% to 10%) also increases the solubility of protein.
The protein contains three cysteines (positions 54, 141, and 205), none of which are involved in native disulfide bond formation. The cysteines at positions 54 and 205 are solvent-exposed.
Digestion of the purified protein with proteases indicated rapid digestion of the N-terminal region (residues 1–38). For example, digestion was complete with a few minutes using relatively low concentration of trypsin (1% w/w).
The above information was exploited to increase the robustness of the purification protocol. Low solubility was a major issue during purification and this was improved by including 4 M urea in the extraction buffer and 2 M urea in the two anion exchange column buffers. For the final gel filtration step, 10% acetonitrile was included to help maintain both the solubility of Nef and fortuitously cause aggregation of some E. coli contaminants that eluted in the void volume. Neither the urea nor the acetonitrile at the concentrations used resulted in Nef denaturation. The problem of cysteine oxidations was circumvented by mutating cysteines 54 and 205 to alanines. Mutation of cysteine 205 alone and including 5 mM DTT in all the column buffers was also a satisfactory solution. The high susceptibly of the N-terminal region to proteolytic processing indicates that it is solvent-accessible and likely to be unstructured. In the case of Nef, this region can be deleted without affecting the folding of the protein and removes the potential for heterogeneity due to partial processing by E. coli proteases. The NMR structure of HIV Nef was determined with protein prepared as described above (Grzesiek et al., 1997).
MAP30
MAP30 is a plant protein obtained from bitter melon that has anti-HIV and anti-tumor activities. The 30-kDa protein is well expressed in E. coli as a soluble protein and is purified by two stages of exchange chromatography followed by gel filtration. The clarified extract is first applied to a DEAE-Sepharose column at pH 8.0; the majority of MAP30 does not bind or weakly binds the exchange resin. The column flow-through and early eluting fractions are dialyzed against pH 6.5 buffer then fractionated using SP-Sepharose (strong cation exchanger). The final step is gel filtration using a Superdex 200 column at pH 8.0.
There are clear similarities between the MAP30 purification scheme and the one developed for the Nef protein; both utilize an initial clean-up step using DEAE-Sepharose followed by a second more discriminating ion-exchange step and finally a “polishing” step using gel filtration. For Nef, the second ion-exchange step employs an anion-exchange resin while the MAP30 method uses a cation-exchange resin. The choice of resin for the second step reflects the difference in the isoelectric points of these proteins. Nef has a calculated pI of ~ 5.95 and is positively charged at pH values greater than this. MAP30 has a slightly basic pI of 9.00 and is negatively charged at pH values below this. Thus, Nef binds to DEAE-Sepharose and Q-Sepharose at pH 7.4 and 8.0, respectively.
On the other hand, MAP30 does not bind to DEAE-Sepharose at pH 7.4 but binds strongly to a cation exchanger at pH 6.5.
Apart from purification, there is also another similarity between Nef and MAP30, namely susceptibility to proteolytic processing during purification. As previously mentioned, the N-terminal region (residues 1–38) of Nef is at risk for proteolysis, and to maintain the structural integrity, especially during cell breakage and the initial processing, protease inhibitor cocktails must be included in the buffers. MAP30 also has a region susceptible to processing, namely, the ~ 20 residues at the C-terminal end of the protein. Again, this is due to the fact that this region is largely unstructured in an otherwise folded and stable molecule (Wang et al., 1999). When purifying MAP30, standard protease inhibitors are included during the early stages of purification and, in addition, α-macroglobulin (15 to 2.0 µg/mg protein) is added to the protein prior to the gel-filtration step. The macroglobulin inhibits a wide range of proteases by a trapping mechanism (Sottrup-Jensen, 1989). If proteins are to be used for structural studies, deletion mutants can eliminate unstructured regions at the N- and C- terminal regions. Deletions of such regions from either Nef or MAP30 do not significantly change the pI of either protein, so the same purification procedures can be applied to the deletion mutants. Although incremental structural determination is an important strategy in structural biology, one should always be aware that regions deleted, even those that appear unstructured, may have important functional roles. There are many examples of disordered proteins and protein domains that adopt folded structures upon binding to their biological targets (for a review, see Dyson and Wright, 2001), and in the case of Nef, it appears that the apparently unstructured N-terminal region (residues 1–57) mediates binding to the tumor suppressor protein p53, possibly enhancing HIV-1 replication (Greenway et al., 2002). Multiple vector co-expression systems for producing heteromeric complexes in E. coli (Johnson et al., 2000; Tolia and Joshua-To, 2006) may be useful for producing proteins requiring binding partners for folding and stability.
Insoluble proteins
HIV-1 gp41 ectodomain
The membrane-associated glycoproteins of HIV-1 include gp120 and gp41, the latter mediating membrane fusion with the host cell. These viral envelope proteins have been the subject of intense structural analysis over the last several years as inhibition of membrane fusion, hence viral entry, is a potential drug target in the development of therapeutics for AIDS. A basic strategy in tackling membrane-associated proteins is to remove the membrane-spanning region by expressing the non-membrane-associated region or ectodomain.
The gp41 ectodomain is a 150-residue protein that is expressed in E. coli as an insoluble protein. The protein can be extracted from inclusion bodies with 8 M guanidine·HCl and purified by one step of gel filtration in the presence of 4 M guanidine·HCl. The guanidine is removed by preparative reversed-phase HPLC and the protein folded by dialysis against 50 mM sodium formate at pH 3.0. The yield of folded protein is >90%. Characterization of the protein indicates that its solubility decreases dramatically below pH 4.0. Between pH 3.0 and 4.0, the protein has an all α-helical secondary structure with a trimeric subunit structure. The protein was demonstrated to have folded by determining its full structure at pH 3.5 using multidimensional NMR (Caffrey et al., 1998). The protein was also crystallized from a buffer at pH 3.5 and its structure determined by X-ray crystallography (Yang et al, 1999).
Other insoluble proteins expressed in E. coli that exhibit acid stability similar to the gp41 ectodomain can be processed and folded using a similar scheme as described above. For example, the HIV protease can be purified and folded with this method. The HIV protease, after folding at pH 3.5, exhibits fair solubility up to pH 5.0, with solubility decreasing at higher pH values. Other proteins may only be partially folded or unfolded at acidic pH values; in these cases, the reversed-phase HPLC step could be used to simply remove the denaturant, then the protein can be freeze dried from TFA-acetonitrile solvent and used for folding trials.
The gp41 ectodomain contains two cysteine residues in a loop region connecting N- and C-terminal helical domains. These cysteines do not form intramolecular disulfides and can be substituted by alanine residues. This is a common theme. If a protein contains free solvent-accessible cysteines that play no structural or functional role, it is often a good idea to substitute them (usually with Ala), especially if structural studies are planned.
Human Tissue Inhibitor of Metalloprotease-2 (TIMP-2) and Hepatocyte Growth Factor isoforms (NK1 and NK2)
The TIMP families of proteins are inhibitors of the matrix metalloproteases and are critical effectors of extracellular matrix turnover. The hepatocyte growth factor (HGF) is a multifunctional protein stimulating a wide range of cellular targets. The HGF gene codes for three distinct proteins: the full-length form and two truncated isoforms that include an N-terminal domain (N) and one-kringle (NK1) or two-kringle domains (NK2). TIMP-2 (21 kDa), NK1 (21 kDa), and NK2 (30 kDa) contain multiple disulfides that stabilize the folded conformations. For example, TIMP-2, apart from having 12 cysteines that form 6 disulfides, contains a cysteine as the N-terminal residue. All three proteins were expressed in E. coli as insoluble proteins, extracted with guanidine·HCl and reductant, and the unfolded protein separated by gel filtration in a similar manner to that previously discussed. The partially purified proteins can be conveniently stored frozen in guanidine·HCl at −80°C for several years without deleterious effects on folding or recovery of active protein. The folding and oxidation of the proteins are detailed in the respective publications, Stahl et al. (1997) and Wingfield et al. (1999), but briefly, the protocols involve equilibrium dialysis incorporating urea as a co-solvent to maintain solubility during folding, and a glutathione-based oxido-shuffling system (redox buffer) to promote formation of disulfide bonds (this approach is also detailed in Basic Protocol 1 in UNIT 6.5). The final stage of the purification process is gel filtration of the folded proteins, which, apart from removing host contaminates, separates folded monomers from any misfolded and aggregated protein.
When recombinant expressed proteins are insoluble in E. coli, the purification scheme can be very simple as illustrated above where one or two steps of gel filtration may be all that is required; the challenge is determining a method to fold and oxidize the protein. In all three examples discussed above, the key to efficient folding is maintaining solubility, whether by taking advantage of the acid stability of the protein and working at pH 3.5, or by including the co-solvent urea. As mentioned above, TIMP-2 has an N-terminal cysteine residue. When this protein was originally expressed, an alanine was appended to the N-terminus since it had been observed that partial N-terminal processing occurred when cysteine was the terminal residue. The alanine residue was added in an effort to produce homogeneous protein for structural studies. The purified Ala+ TIMP-2 appeared monomeric and folded, yet was devoid of its normal inhibitory activity (Wingfield et al., 1999). It was determined that the coordination of a zinc atom by the N-terminal cysteine stabilized substrate binding and required a free amino terminal group. This was demonstrated by exopeptidase digestion (using aminopeptidase 1) of Ala+ TIMP-2, which removed the N-terminal alanine making cysteine the N-terminal residue and, thus, restoring biological activity.
A GST fusion protein
The protein VP26 is a 12-kDa capsid protein of the herpes simplex virus and initial attempts to directly express this protein in E. coli failed. It was possible, however, to produce this protein at fairly high levels in E. coli as a GST fusion (Wingfield et al., 1997a). The insoluble protein was treated in the usual manner: solubilized with guanidine·HCl and partially purified by gel filtration also in guanidine·HCl. The usual purification for GST fusion proteins is affinity chromatography using immobilized glutathione, which requires that the GST moiety be folded (UNIT 6.6). Due to the low solubility of VP26 and its high propensity for aggregation, the following approach was used. First, the VP26-GST fusion was folded from the guanidine·HCl solution by equilibrium dialysis against buffer containing 2.5 M urea, 10 mM CHAPS, and 0.25 M NaCl, and then against the same buffer lacking the urea. The buffer additives were included to maintain protein solubility (solubility is improved with >0.25 M NaCl, but the cleavage of the GST moiety by thrombin is inhibited by high salt concentrations). Following cleavage of GST and VP26, the proteins were denatured again with guanidine·HCl, separated by gel filtration and the purified VP26 refolded from urea and CHAPS as described above. As an aside, the GST moiety is readily refolded from guanidine·HCl and does not require high salt or CHAPS to maintain solubility during the dialysis steps. The purification approach used here may appear inelegant, but the fusion system was used not to facilitate purification, but to facilitate expression of the protein.
PROTEIN HANDLING
Storing Purified Proteins
Purified protein should be filter-sterilized prior to storage. Millex-GV 0.22-µm filters (http://www.emdmillipore.com) employ hydrophilic membranes with low binding capacities and are recommended for most proteins. Proteins are best stored at −80°C or may be stored on ice; freezing at −20°C is not recommended. Rapid freezing in small aliquots using dry ice/ethanol mixtures is preferred to slow freezing at −20°C. The addition of sucrose or glycerol often increases protein stability during storage and during freezing and thawing cycles (Arakawa and Timasheff, 1985; Timasheff and Arakawa, 1997). Lyophilization is best for long-term storage; however, care should be taken in choosing the protein solvent (Franks, 1993). Trehalose can stabilize protein molecules for storage and help them retain their functional activity (Unit 4.9).
Promoting Protein Solubility and Stability
If the recombinant protein contains reactive unpaired sulfhydryl groups in the native conformation, 1 to 5 mM DTT should be included in the column buffers during purification. However, reductant should not be used gratuitously, as the native protein may contain intra- or intermolecular disulfide bonds, disruption of which can reduce the stability and solubility of the protein. Reductants should be included, for example, during gel filtration if dimers or higher aggregates need to be converted to active monomeric protein. The presence of intermolecular (and occasionally intramolecular) disulfide bonds can be determined analytically by SDS-PAGE under nonreducing conditions (UNIT 6.5) by pretreating proteins sequentially with iodoacetamide (to prevent artificial disulfide exchange) and then with SDS in the absence of reductant. The use of reductants can best be rationalized once the native protein has been characterized.
EDTA (1 to 5 mM) is often included in buffers to remove heavy metals that can catalyze oxidative processes and inhibit certain proteases. It should be noted that EDTA will bind to anion exchange resins (Scopes, 1994).
Other components often added to buffers to promote protein solubility during purification include nonionic or zwitterionic detergents, low concentrations of urea (1 to 2 M), and salt (0.5 to 1 M NaCl). These additives are compatible with ion-exchange chromatography, except for high-salt concentrations, which are compatible with hydrophobic-interaction chromatography (UNIT 8.4), affinity chromatography (Chapter 9), and gel-filtration chromatography (UNIT 8.3). Solvent pH is one of the most important variables for maintaining protein solubility; in general, proteins are least soluble at or near their isoelectric points.
Preventing Contamination
Precautions to prevent contamination of the protein of interest are as follows:
To avoid cross-contamination, especially from other recombinant proteins, dedicate one set of chromatography resins for the purification of each protein. If this is not possible, or if expensive prepackaged matrices are used, be sure to clean resins thoroughly after each use. Check the manufacturer’s recommendations and be aware of the chemical stability of the resin, especially for extremes of pH.
Store resins with preservatives (e.g., 1 mM sodium azide) and avoid storage in phosphate buffers, which provide a good medium for bacterial growth.
To generate reproducible protocols using ion-exchange methods, monitor the pH and conductivity of all buffers and column effluents (the latter ideally in-line).
Avoid protein cross-contamination in concentration equipment such as stirred ultrafiltration cells with ultrafiltration membranes.
Keep pH and conductivity probes scrupulously clean, especially when used with solutions containing proteases. Likewise, use care when using cuvettes for UV measurements.
Avoid vigorous stirring of protein solutions to prevent shear denaturation, and handle soft agarose-based column matrices carefully to prevent bead fragmentation
Removing Pyrogens
Recombinant proteins used for in vivo studies should be free of endotoxins (pyrogenic lipopolysaccharide derived from the bacterial outer membrane of Gram-negative bacteria). Yeast and mammalian cell hosts do not contain endotoxins; however, exogenous contamination from water and others must be avoided. Pyrogens can be detected using the sensitive Limulus amoebocyte lysate (LAL) assay kits available from Sigma and other suppliers. As endotoxins are negatively charged, they will be removed by anion-exchange chromatography. Other methods are reviewed in detail by Petsch and Anspach (2000).
SCALE OF OPERATIONS AND AIMS OF PURIFICATION
Determining Scale
The amount of protein required and the level of purity will vary dramatically from laboratory to laboratory and study to study. The following guidelines will help in planning a strategy.
If a Coomassie blue–stained band corresponding to the expressed protein is observed on one-dimensional SDS-PAGE analysis of a whole-cell extract, then the protein constitutes at least 0.5% to 1% of the total protein. Wet E. coli cell paste contains ~ 10% to 15% protein by weight (reviewed by Neidhardt, 1987). If the level of expression is low to average (e.g., 5%), then 1 g wet weight of cells will contain ~ 5 mg recombinant protein. Hence, a cell paste of 20 to 50 g (a typical yield from 1-to 2- liter benchtop fermentation) will contain 100 to 250 mg recombinant protein, and often two-to five-fold more. Shaker-flask fermentations of equivalent volumes might yield 5% or 10% of these amounts. Thus, for soluble proteins, or insoluble ones that can be refolded (with ≥5% yield), significant amounts of protein can be obtained from relatively small fermentations. For proteins secreted into the periplasm or medium, fermentations on larger scales may be required, as expression levels are usually considerably lower than that for direct expression.
Deciding the Aims of the Purification
There are many reasons, both scientific and commercial, for producing purified recombinant proteins. The development of laboratory-scale purification schemes that produce pure protein (a single band on SDS-PAGE) should be relatively straightforward given the relatively high abundance of recombinant proteins in cell extracts. Protein present at 1% of the total cell extract requires only a 100-fold purification compared to the several thousand-fold sometimes required for the purification of non-recombinant proteins (reviewed by Stein, 1991). The widely used method of affinity tagging proteins allows the non-specialist to rapidly purify protein for biochemical and activity studies without investment in some of the specialized equipment mentioned below. However, far more time and expertise is required to develop protocols that produce purified recombinant proteins having the physical and chemical homogeneity required for clinical use and for structural determinations. Furthermore, only after detailed characterization of the isolated protein will chemical and physical heterogeneities be revealed in enough detail for steps to be taken to either prevent their occurrence or rationalize fractionation of modified species.
Therapeutic proteins
Mammalian cells are the production host for many current protein therapeutics, however, E. coli, is also used to produce major biotechnological products including insulin and bovine growth hormone. Some advances in E. coli production of therapeutic proteins and methods used to fold solubilized protein for industrial processes have been recently reviewed (De Bernardez Clark, 2001; Swartz, 2001). Proteins used for clinical studies must be manufactured according to applicable FDA guidelines that include Good Manufacturing Procedures (GMP). Sofer and Hagel (1997) provide practical coverage of modern process development, including process chromatography and its scale-up. The physiochemical characterization of protein pharmaceuticals can be especially challenging and many of the methods and approaches used rely on mass spectrometry (see Chapter 16).
Structure Determination
For many investigators, a primary goal is to correlate the structure of a protein with its function (and vice versa). Many proteins produced by recombinant DNA technology are present only in trace amounts in nature (e.g., interferons and other cytokines; Ealick et al., 1991), and authentic material is not available for detailed molecular characterization. Knowledge of the 3-D structure allows a rational approach to protein engineering and the design of drugs that modulate the biological activity of the protein. The substitution, deletion, and insertion of residues allow a structure-function hypothesis to be tested and new, sometimes improved protein variants (or mutants) to be produced.
NMR Spectroscopy
It is a major challenge to produce proteins suitable for structural determination, not only in terms of quality, but in terms of the quantity which may be required, especially for NMR (UNIT 17.5). Many proteins, although they have native-like structure and biological activity, are not suitable for structural determination due to, e.g., limited solubility, conformational flexibility (floppy regions/domains), and heterogeneity of posttranslational modifications (especially carbohydrate). Often, these problems can be resolved by a combination of protein biochemistry and protein engineering approaches and requires a close collaboration between the structural biologist and the molecular/protein chemist. The NMR determination of the HIV-1 Nef structure is an example of this integrated approach (see above).
Structural determination in solution by multidimensional NMR is presently limited to proteins 30 to 40 kDa (reviewed by Clore and Gronenborn, 1994; Unit 17.5). One of the largest proteins solved to date is the 44-kDa trimeric SIV gp41 ectodomain (Caffrey et al., 1998). Larger proteins can be studied incrementally (using the dissection approach) if information on the domain boundaries is known (Campbell and Downing, 1998). The sample demands can be as high as several hundred milligrams, and larger proteins (>10 kDa) must be uniformly labeled with various combinations of 2H, 13C, and 15N. Some of the labeling scenarios required to solve the HIV Nef structure are presented in Table 1 of Grzesiek et al. (1997). New developments in isotope labeling strategies are reviewed by Goto and Kay (2000). Labeling in E. coli is achieved by growing the bacteria in minimal medium containing one or more of the following stable (nonradioactive) isotopes: 15NH4Cl (sole nitrogen source), [13C] glucose (sole carbon source), and 2H2O (UNIT 5.3). The 15N and 13C labeling of the HIV protease using a 2-liter fermentor is detailed by Yamazaki et al. (1996). Label incorporation is conveniently monitored by mass spectrometry of the purified protein.
Over the lifetime of the structural study (4 to 12 months), because of the multisample requirements, a reliable and robust purification method is essential. It should also be noted that the labeling requirement usually dictates that the protein be produced in bacteria, although labeled proteins have been produced in yeast and insect cells (Goto and Kay, 2000) and cell free systems (Takeda and Kainosho, 2012).
For NMR purposes, the recombinant protein must be homogeneous and soluble at 1 to 3 mM concentrations, preferably with solvents below pH 7.0 and at temperatures >30°C. As measurements take many hours to complete, the presence of trace amounts of proteases can ruin the experiment. In addition, particular attention must be paid to maintaining solvent-accessible and reactive cysteines (unpaired) in the reduced state (usually by including DTT or TCEP) and often cysteines are mutated to alanine residues (Wingfield et al., 1997).
Protein crystallization and X-ray crystallography
The rate-limiting step in structure determination using X-ray crystallography is production of crystals that diffract to high resolution (UNIT 17.4). The scientists involved in the production and characterization of the protein are often best situated to crystallize the protein. Furthermore, once crystallization conditions have been optimized, it can be quite easy to interest structural groups in collaboration.
Determining optimal crystallization conditions may require as little as a few milligrams or as much as 100 mg of pure protein. The protein itself must usually be physically and chemically homogeneous; small amounts of protein impurities may significantly interfere with crystallization. In general, physical homogeneity is more critical than chemical homogeneity. Some of the methods used to establish physical and chemical homogeneity are discussed elsewhere (Chapter 7; Jones et al., 1994). Many investigators use the sparse matrix sampling technology to screen for initial crystallization conditions and commercial kits are available for this purpose (e.g., Hampton Research: http://www.hamptonresearch.com). The company site also has useful tips and protocols for protein crystallization in general.
The phase problem in crystallographic analysis has traditionally been solved by isomorphous replacement with heavy atoms, now multiwavelengh anomalous diffraction (MAD) is often used (Hendrickson et al., 1990). For this approach, selenium is incorporated into recombinant proteins via selenomethionine (seleno-L-methionine; available from Sigma and others) using a methionine-requiring auxotroph. In studies of the gp41 protein (mentioned above), the T7 expression system (Novagen, http://www.emdmillipore.com) and the host strain B834/DE3 (Novagen) were used. Briefly, transformed cells were grown overnight in a 0.5-liter shaker flask containing minimal media plus 1 mM methionine. Cells were collected and resuspended in 0.5 liters of media minus methionine. The cells were grown at 37°C in a small fermentor and fed 5 ml of 10 mg/ml selenomethionine. The cells were induced for 3 hr with IPTG and fed an additional 50 mg of selenomethionine (total feed: 100 mg). Cells were collected (~ 7.0 g wet weight) and 130 mg of pure gp41 ectodomain was isolated as described above. Mass spectrometry indicated that the single methionine was >98% labeled. The protein was then crystallized as previously described (Wingfield et al., 1997). For more details on labeling using E. coli, see Chapter 5 (UNIT 5.3). Selenomethionine incorporation into eukaryotic systems is not as successful as in E. coli; incorporation can be as high as 90% in baculovirus, but only ~ 60% in yeast.
The production of well-defined protein complexes for structural studies can be straightforward. For example, monomeric proteins can be expressed in bacteria which self-associate into stable complexes ranging from simple dimers (e.g., γ-IFN) and trimers (e.g., α-TNF) to complex structures such as viral nucleocapsids (e.g., Hepatitis B Virus core antigen, 180-mer). These stable (tightly associated) homopolymers are well suited for structural studies. Heteroprotein complexes can be made by either co-expression of protein subunits (Johnson et al., 2000; Kholod and Mustelin, 2001) or by in vitro assembly of individual components. The former approach may be required in the case where individual subunits are unstable (Nash et al., 1987). In contrast to stable complexes, there are many biologically significant complexes characterized by weak association (see UNIT 20.13). Many protein-protein interactions of interest, e.g., signal transduction pathways, may be somewhat transitory and involve weak interactions. In these complexes, the dissociation constants (Kd) between proteins are <10−6 M, and therefore, tend to exhibit concentration dependent reversible self-association. This behavior results in physical heterogeneity, thus, complicating crystallization attempts. NMR has been used to examine weakly associating systems, e.g., the binding of the CD4 determinant to HIV-1 Nef (Grzesiek et al., 1996b). To study protein-protein complexes characterized by low Kd, it may be necessary to use protein engineering and other approaches to generate more stable and tighter interactions.
It has been mentioned that one of the reasons for fusion tagging proteins is to increase their potential for stable expression and accumulation. The enhancement of a protein’s physical properties, especially solubility, by appending protein (e.g., GST) or peptide (e.g., FLAG) tags makes these fusion proteins good candidates for crystallization trails.
Some investigators have reported problems with the crystallization of His-tagged proteins; in our own work we have not removed the tag without problems, for example, Dimattia et al (2010).
For crystallization studies, acquisition of a robot system for rapid screening, good quality microscopes, including a UV-microscope (budget allowing), and incubation cabinets are suggested.
Biophysical Studies
Information on the conformational properties, including denaturation/folding curves, can help rationalize the development of preparative protein folding processes. Low-resolution structural studies using various biophysical methodologies (Jones et al., 1994) can be made with less material (<1 to 10 mg). Proteins for spectroscopic studies should be >95% pure and previously fractionated on a gel-filtration column to remove aggregated and possibly misfolded variants. The removal of aggregates is especially important for spectroscopic studies including UV/Vis, fluorescence and circular dichroism where excessive light scattering must be avoided (see Chapter 7; Colon, 1999).
Various labeling and tagging strategies can be used to aid both structural and functional studies. The most common approach is to append affinity tags that can then be used to immobilize the protein in a directed manner (Nilsson et al., 1997). This approach is especially useful for studying protein interactions. Also, analogous to the in vivo protein labeling scenarios as described above for selenomethionine, specific residues can be modified. For example, tryptophan in recombinant proteins can be replaced by 5-hydroxytryptophan by using an E. coli Trp auxotroph. Protein thus labeled has a strong absorbance at 310 nm that can be exploited in structure-function studies (Laue et al., 1993).
SPECIALIZED EQUIPMENT
Breaking and Fractionating Cells
For small- to medium-scale work on a regular basis, a French press (Thermo Scientific, http://www.thermoscientific.com) with a continuous-fill cell is recommended (UNIT 6.2). It is also useful for breaking yeast cells. It should be mentioned that Thermo no longer sell the French press, used equipment can still be found for sale at various on-line sites. For large-scale work (>500 ml), the Manton-Gaulin-APV homogenizer (http://www.spx.com) is recommended. For further processing of cells and cell lysates (e.g., UNITS 6.2 & 6.3), an ultrasonic homogenizer is required. An instrument with a 400-W (or higher) capacity is recommended, for example from Branson (http://www.emersonindustrial.com). After low-speed centrifugation using standard preparative centrifuges (Beckman Coulter “Avanti Series” can be found at http://www.beckmancoulter.com), high-speed centrifugation is a convenient and rapid cleanup step before column chromatography (Fig. 6.1.4). With Beckman ultracentrifuges, the 45 Ti rotor is recommended. This six-place rotor has a maximum speed of 235,000 × g; with thick-walled polycarbonate tubes, its capacity is ~ 400 ml.
Chromatographing Proteins
Most chromatography is carried out at 4°C either in a cold room or, more conveniently, in a cold cabinet in the laboratory. The basic components of a chromatography system are as follows: column, column matrix, pumps, a gradient-making device, UV/visible or other detection system, and a fraction collector. These components can be bought as units such as the AKTA Explorer chromatograph systems (GE Heathcare), which can be used for laboratory-scale to large-scale work. Column matrices can be purchased prepacked or as bulk media that are packed in columns by the user. Ion-exchange separations, using standard low- to medium-pressure resins (agarose/dextran/cellulose-based), require at least one narrow (2.5-cm) and one wide (5.0-cm) column with adjustable flow adapters so that the resin height can be varied between 5 and 30 cm. Gel filtration requires columns with diameters of 1.25 and 2.5 cm (5 cm for larger-scale work) and lengths of 60 to 100 cm. Simple gradient makers with capacities of 150 ml to 2 liters are generally available.
Concentrating Proteins
Stirred ultrafiltration cells are recommended for laboratory-scale work. The cells range in size from 3 ml to 2 liters and are used in conjunction with variable molecular weight cutoff membranes (EM Millipore, http://www.emdmillipore.com). For larger volumes, Millipore also sells various systems. For smaller volumes (0.5 to 15 ml), centrifugational concentrators are available (EM Millipore and others). For a review of the equipment used for protein concentration, see Harris (1989).
Making Analytical Measurements
A protein purification laboratory should have a dependable scanning UV/visible spectrophotometer, ideally an instrument with computerized data collection and analysis. Hewlett Packard (Agilent) instruments with diode array detectors are recommended for most routine work (http://www.agilent.com). For laboratories specializing in purifying recombinant proteins from E. coli, access to a spectropolarimeter (e.g., Jasco J-815, http://www.jascoinc.com) will be helpful for monitoring and developing folding protocols. For rapid chemical characterization and identity check of proteins, access to a mass spectrometer is also desired (Chapter 16). Most of the companies mentioned have excellent Web sites where technical information is posted. There are also companies which can perform pay for services including analytical ultracentrifugation (AUC), circular dichroism, light scattering, DSC, fluorescence etc., for example, Alliance Protein Laboratories, Inc. Finally, there are an excellent series of handbooks on chromatographic separations and protein analytical techniques published by GE Heathcare Lifesciences and which can be conveniently downloaded as free pdf files.
Literature Cited
- 1.Allet B, Payton M, Mattaliano RJ, Gronenborn AM, Clore GM, Wingfield PT. Purification and characterization of the DNA-binding protein Ner of bacteriophage Mu. Gene. 1988;65:259–268. doi: 10.1016/0378-1119(88)90462-3. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa T, Timasheff SN. Theory of protein solubility. Methods Enzymol. 1985;114:49–77. doi: 10.1016/0076-6879(85)14005-x. [DOI] [PubMed] [Google Scholar]
- 3.Aricescu AR, Owens RJ. Expression of recombinant glycoproteins in mammalian cells: towards an integrative approach to structural biology. Current Opinion in Structural Biology. 2013;23:345–356. doi: 10.1016/j.sbi.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong N, De Lencastre A, Gouaux E. A new protein folding screen: Application to the ligand binding domain of a glutamate and kainite receptor and to a lysozyme and carbonic anhydrase. Protein Sci. 1999;8:1475–1483. doi: 10.1110/ps.8.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asenjo JA, Patrick I. In: Large-scale protein purification. In Protein Purification Applications: A Practical Approach. Harris ELV, Angal S, editors. Oxford: IRL Press; 1990. pp. 1–27. [Google Scholar]
- 6.Baneyx F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 7.Beacham IR. Periplasmic enzymes in Gram-negative bacteria. Int. Biochem. 1979;10:877–883. doi: 10.1016/0020-711x(79)90117-4. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Bassat A, Bauer K, Chang S-Y, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: Properties of the E. coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessette PH, Aslund F, Beckwick J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibi E, Beja O. Membrane topology of multidrug resistant protein expressed in E. coli. J. Biol. Chem. 1994;31:19910–19915. [PubMed] [Google Scholar]
- 11.Bowden GA, Paredes AM, Georgiou G. Structure and morphology of protein inclusion bodies in E. coli. Biotechnology. 1991;9:725–730. doi: 10.1038/nbt0891-725. [DOI] [PubMed] [Google Scholar]
- 12.Braun P, Gerritse G, van Dijl J-M, Quax WJ. Improving protein secretion by engineering components of the bacterial translocation machinery. Curr. Opin. Biotechnol. 1999;10:376–381. doi: 10.1016/s0958-1669(99)80068-8. [DOI] [PubMed] [Google Scholar]
- 13.Brondyke WH. Selecting and appropriate method for expressing a recombinant protein. Methods in Enzymology. 2009;463:131–144. doi: 10.1016/S0076-6879(09)63011-1. [DOI] [PubMed] [Google Scholar]
- 14.Buchner J, Rudolph R. Renaturation and characterization of recombinant Fab fragments produced in Escherichia coli. Biotechnology. 1992;9:157–162. doi: 10.1038/nbt0291-157. [DOI] [PubMed] [Google Scholar]
- 15.Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of E. coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA-cellulose chromatography. J. Biol. Chem. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 16.Burgess RR, Knuth MW. Purification of a recombinant protein overproduced in Escherichia coli. In: Marshak DR, Kadonaga JT, Burgess RR, Knuth MW, Brennan WA, Lin S-H, editors. Strategies for Protein Purification and Characterization: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 205–217.pp. 245–262. [Google Scholar]
- 17.Caffrey M, Cai M, Kaufman J, Stahl S, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. Three dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell I, Downing AK. NMR of modular proteins. Nat. Struct. Biol. 1998;5:496–499. doi: 10.1038/733. [DOI] [PubMed] [Google Scholar]
- 19.Carlson ED, Gan R, Hodgman EC, Jewett MC. Cell-free protein synthesis: Applications come of age. Biotechnol. Advances. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celik E, Calik P. Production of recombinant proteins by yeast cells. Biotechnol. Advances. 30:1108–1118. doi: 10.1016/j.biotechadv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Cereghino GPL, Clegg JM. Applications of yeast in biotechnology: Protein production and genetic analysis. Curr. Opin. Biotechnol. 1999;10:422–427. doi: 10.1016/s0958-1669(99)00004-x. [DOI] [PubMed] [Google Scholar]
- 22.Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol. Advances. 30:1102–1107. doi: 10.1016/j.biotechadv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Y-SE, McGowan MH, Kettner CA, Schloss JV, Erickson-Viitanen S, Yin FH. High synthesis of recombinant HIV-1 protease and the recovery of active enzyme from inclusion bodies. Gene. 1990;87:243–248. doi: 10.1016/0378-1119(90)90308-e. [DOI] [PubMed] [Google Scholar]
- 24.Cleary S, Mulkerrin MG, Kelley RF. Purification and characterization of tissue plasminogen activator kringle-2 domain expressed in E. coli. J. Biol. Chem. 1989;28:1884–1891. doi: 10.1021/bi00430a068. [DOI] [PubMed] [Google Scholar]
- 25.Cleland JL, Builder SE, Swartz JR, Winkler M, Chang JY, Wang DIC. Poly-ethylene glycol enhanced protein refolding. Biotechnology. 1992;10:1013–1019. doi: 10.1038/nbt0992-1013. [DOI] [PubMed] [Google Scholar]
- 26.Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:249–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 27.Cole PA. Chaperone-assisted protein expression. Structure. 1996;4:239–242. doi: 10.1016/s0969-2126(96)00028-7. [DOI] [PubMed] [Google Scholar]
- 28.Colon W. Analysis of protein structure by solution optical spectroscopy. Methods Enzymol. 1999;309:605, 632. doi: 10.1016/s0076-6879(99)09041-2. [DOI] [PubMed] [Google Scholar]
- 29.Cornelis P. Expressing genes in different Escherichia coli compartments. Curr. Opin. Biotechnol. 2000;11:450–454. doi: 10.1016/s0958-1669(00)00131-2. [DOI] [PubMed] [Google Scholar]
- 30.Creighton TE. Physical and Chemical Basis of Molecular Biology. Helvetian Press; 2010. [Google Scholar]
- 31.Creighton TE. Disulfide bond formation in proteins. Methods Enzymol. 1984;107:305–329. doi: 10.1016/0076-6879(84)07021-x. [DOI] [PubMed] [Google Scholar]
- 32.Creighton TE. Proteins: Structures and Molecular Properties. 2nd ed. New York: Freeman; 1993. pp. 292–296. [Google Scholar]
- 33.Dale GE, Broger C, Langen H, D’Arcy A, Struber D. Improving protein stability through rationally designed amino acid replacements: Solubilization of the trimethoprim-resistant type S1 dihydrofolate reductase. Protein Eng. 1994;7:933–939. doi: 10.1093/protein/7.7.933. [DOI] [PubMed] [Google Scholar]
- 34.Danley DE, Strick CA, James LC, Lanzetti AJ, Otterness IG, Grenett HE, Fuller GM. Identification and characterization of a C-terminally extended form of recombinant murine IL-6. FEBS Lett. 1991;283:135–139. doi: 10.1016/0014-5793(91)80571-j. [DOI] [PubMed] [Google Scholar]
- 35.Darby NJ, Creighton TE. Folding proteins. Nature. 1990;344:715–716. doi: 10.1038/344715b0. [DOI] [PubMed] [Google Scholar]
- 36.De Bernardez Clark E. Protein folding for industrial processes. Curr. Opin. Biotechnol. 2001;12:202–207. doi: 10.1016/s0958-1669(00)00200-7. [DOI] [PubMed] [Google Scholar]
- 37.De Bernardez Clark E, Schwartz E, Rudolph R. Inhibition of aggregation side reactions during in-vitro protein folding. Methods Enzymol. 1999;309:217–236. doi: 10.1016/s0076-6879(99)09017-5. [DOI] [PubMed] [Google Scholar]
- 38.deVos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: Crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 39.Diederichs K, Boone T, Karplus A. Novel fold and putative receptor binding site of granulocyte-macrophage colony stimulating factor. Science. 1991;254:1779–1782. doi: 10.1126/science.1837174. [DOI] [PubMed] [Google Scholar]
- 40.DiMattia M, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. HIV-1 Rev dimer structure at 3.2Å resolution: Implications oligomerization and binding to the Rev Response Element”. Proc. Natl. Acad. Sci. USA. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiRienzo JM, Nakamura K, Inouye M. The outer membrane proteins of Gram-negative bacteria: Biosynthesis, assembly and function. Annu. Rev. Biochem. 1978;47:481–532. doi: 10.1146/annurev.bi.47.070178.002405. [DOI] [PubMed] [Google Scholar]
- 42.Drugmand J-C, Schneider Y-J, Agathos SN. Insect cells as factories for biomanufacturing. Biotechnol. Advances. 2012;30:1140–1157. doi: 10.1016/j.biotechadv.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 43.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2001;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 44.Ealick SE, Cook WJ, Vijay-Kumar S, Carson M, Nagabhushan TL, Trotta PP, Bugg CE. Three-dimensional structure of recombinant human interferon-gamma. Science. 1991;252:698–702. doi: 10.1126/science.1902591. [DOI] [PubMed] [Google Scholar]
- 45.Eisenberg D. How chaperones protect virgin proteins. Science. 1999;285:1021–1022. doi: 10.1126/science.285.5430.1021. [DOI] [PubMed] [Google Scholar]
- 46.Ellis RJ. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 47.Ellis RJ, Hart FU. Principles of protein folding in the cellular environment. Curr. Opin. Struct. Biol. 1999;9:102–110. doi: 10.1016/s0959-440x(99)80013-x. [DOI] [PubMed] [Google Scholar]
- 48.Esposito D, Chatterjee DK. Enhancement of soluble expression through the use of fusion tags. Current Opinion in Biotechnology. 2006;17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Feldman DE, Frydman J. Protein folding in vivo: The importance of molecular chaperones. Curr. Opin. Struct. Biol. 2000;1026 doi: 10.1016/s0959-440x(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 50.Fersh A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: W.H. Freeman and Company; 1998. [Google Scholar]
- 51.Franks F. Storage stabilization of proteins. In: Franks F, editor. Protein Biotechnology. Totowa, NJ: Humana Press; 1993. pp. 486–531. [Google Scholar]
- 52.Georgiou G, Valax P. Isolating inclusion bodies from bacteria. Methods Enzymol. 1999;309:48–58. doi: 10.1016/s0076-6879(99)09005-9. [DOI] [PubMed] [Google Scholar]
- 53.Ghanderi D, Zhang M, Hurtado-Ziola N, Varki A. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnology and Genetic Engineering Reviews. 2012;28:147–176. doi: 10.5661/bger-28-147. [DOI] [PubMed] [Google Scholar]
- 54.Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell Mol Life Sci. 2004;61:1455–1474. doi: 10.1007/s00018-004-3466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 56.Goenka S, Rao CM. Expression of recombinant ζ-crystallin in Escherichia coli with the help of GroEL/ES and its purification. Protein Expr. Purif. 2001;21:260–267. doi: 10.1006/prep.2000.1359. [DOI] [PubMed] [Google Scholar]
- 57.Goff SA, Goldberg ME. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 58.Goldberg ME. Investigating protein conformation dynamics and folding with monoclonal antibodies. Trends Biochem. Sci. 1991;16:358–362. doi: 10.1016/0968-0004(91)90148-o. [DOI] [PubMed] [Google Scholar]
- 59.Goldberg ME, Rudolph R, Jaenicke R. A kinetic study of the competition between renaturation and aggregation during the refolding of denatured-reduced egg white lysozyme. Biochemistry. 1991;30:2790–2797. doi: 10.1021/bi00225a008. [DOI] [PubMed] [Google Scholar]
- 60.Goto NK, Kay LE. New developments in isotope labeling strategies for protein solution NMR spectroscopy. Curr. Opin. Struct. Biol. 2000;10:585–592. doi: 10.1016/s0959-440x(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 61.Greenway AL, McPhee DA, Allen K, Johnson R, Holloway G, Mills J, Azad A, Sankovich S, Lambert P. Human immunodeficiency virus type 1 Nef binds to tumor suppressor p53 and protects cells against p53-mediated apoptosis. J. Virol. 2002;76:2692–2702. doi: 10.1128/JVI.76.6.2692-2702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grunfeld H, Patel A, Shatzman A, Nishikawa AH. Effector-assisted refolding of recombinant tissue-plasminogen activator produced in Escherichia coli. Appl. Biochem. Biotechnol. 1992;33:117–138. doi: 10.1007/BF02950781. [DOI] [PubMed] [Google Scholar]
- 63.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu J-S, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine kinase. Nature Struct. Biol. 1996a;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 64.Grzesiek S, Bax A, Hu J-S, Kaufman JD, Palmer I, Stahl SJ, Tjandra N, Wingfield PT. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Science. 1997;6:1248–1263. doi: 10.1002/pro.5560060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef: Mapping of the Nef binding surface by NMR. Biochemistry. 1996b;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 66.Gulnik SV, Afonina EI, Gustchina E, Yu B, Silva AM, Kim Y, Erickson JW. Utility of (His)6 Tag for purification and refolding of proplasmepsin-2 and mutants with altered activation properties. Protein Expr. Purif. 2001;24:412–419. doi: 10.1006/prep.2001.1590. [DOI] [PubMed] [Google Scholar]
- 67.Hammarstrom M, Hellgren N, Van Den Berg S, Berglund H, Hard T. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 2002;11:313–321. doi: 10.1110/ps.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harris EL. Concentration of the extract. In: Harris ELV, Angal S, editors. Protein Purification Methods: A Practical Approach. Oxford: IRL Press; 1989. pp. 125–172. [Google Scholar]
- 69.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hendrickson WA, Horton JR, LeMaster DM. Selenenomethionyl proteins for analysis by multiwavelengh anomalous diffraction (MAD): A vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heppel LA. Selective release of enzymes from bacteria. Science. 1967;156:1451–1455. doi: 10.1126/science.156.3781.1451. [DOI] [PubMed] [Google Scholar]
- 72.Hofmann A, Tai M, Wong W, Glabe CG. A sparse matrix screen to establish initial conditions for protein renaturation. Anal. Biochem. 1995;230:8–15. doi: 10.1006/abio.1995.1429. [DOI] [PubMed] [Google Scholar]
- 73.Holland IB, Kenny B, Steipe B, Pluckthun A. Secretion of proteins in E. coli. Methods Enzymol. 1990;182:132–143. doi: 10.1016/0076-6879(90)82013-r. [DOI] [PubMed] [Google Scholar]
- 74.Hopkins TR. Physical and chemical cell disruption for the recovery of intracellular proteins. In: Seetharam R, Sharma SK, editors. Purification and Analysis of Recombinant Proteins. New York: Marcel Dekker; 1991. pp. 57–83. [PubMed] [Google Scholar]
- 75.Hwang DDW, Liu L-F, Kuan I-C, Lin L-Y, Tam T-CS, Tam MF. Co-expression of glutathione S-transferase with methionine aminopeptidase: A system of producing enriched N-terminal processed proteins in E.coli. Biochem J. 1999;338:335–342. [PMC free article] [PubMed] [Google Scholar]
- 76.Jaenicke R. Role of accessory proteins in protein folding. Curr. Opin. Struct. Biol. 1993;3:104–112. [Google Scholar]
- 77.Janson J-C. Protein purification: Principles, high resolution methods, and applications: Volume 54. 3rd ed. New York: Wiley; 2011. [Google Scholar]
- 78.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 79.Johnson BH, Hecht MH. Recombinant proteins can be isolated from E. coli by repeated cycles of freezing and thawing. Biotechnology. 1994;12:1357–1360. doi: 10.1038/nbt1294-1357. [DOI] [PubMed] [Google Scholar]
- 80.Johnson K, Clements A, Venkataramani RN, Trievel RC, Marmorstein R. Coexpression of proteins in bacteria using a T7-based expression plasmid: Expression of heteromeric cell cycle and transcriptional regulatory complexes. Protein Expr. Purif. 2000;20:435–443. doi: 10.1006/prep.2000.1313. [DOI] [PubMed] [Google Scholar]
- 81.Jones C, Mulloy B, Thomas AH. Microscopy, optical spectroscopy, and macroscopic techniques. Methods Mol. Biol. 1994;22:1–245. [Google Scholar]
- 82.Jones DH, Ball EH, Sharpe S, Barber KR, Grant CWM. Expression and membrane assembly of a transmembrane region from Neu. Biochemistry. 2000;39:1878-1878. doi: 10.1021/bi992495e. [DOI] [PubMed] [Google Scholar]
- 83.Kaback HR. Bacterial membranes. Methods Enzymol. 1971;22:99–120. [Google Scholar]
- 84.Kamireddi M, Eisenstein E, Reddy P. Stable expression and rapid purification of Escherichia coli GroEL and GroES chaperones. Protein Expr. Purif. 1997;11:47–52. doi: 10.1006/prep.1997.0764. [DOI] [PubMed] [Google Scholar]
- 85.Kelley WS, Stump KH. A rapid procedure for isolation of large quantities of E. coli DNA polymerase 1 utilizing a λ polA transducing phage. J. Biol. Chem. 1979;254:3206–3210. [PubMed] [Google Scholar]
- 86.Kern G, Kern D, Jaenicke R, Seckler RL. Kinetics of folding and association of differentially glycosylated variants of invertase from Saccharomyces cerevisiae. Protein Sci. 1993;2:1862–1868. doi: 10.1002/pro.5560021108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kholod N, Mustelin T. Novel vectors for co-expression of two proteins in E.coli. Biotechniques. 2001;31:322–328. doi: 10.2144/01312st03. [DOI] [PubMed] [Google Scholar]
- 88.Kiefhaber T, Rudolph R, Kohler H-H, Buchner J. Protein aggregation in vitro and in vivo: A quantitative model of the kinetic competition between folding and aggregation. Biotechnology. 1991;9:825–829. doi: 10.1038/nbt0991-825. [DOI] [PubMed] [Google Scholar]
- 89.Kohno T, Carmichael DF, Sommer A, Thompson RC. Refolding of recombinant proteins. Methods Enzymol. 1990;185:187–195. doi: 10.1016/0076-6879(90)85018-j. [DOI] [PubMed] [Google Scholar]
- 90.Kost TA, Condreay JP. Recombinant baculovirus as expression vectors for insect and mammalian cells. Curr. Opin. Biotechnol. 1999;10:428–433. doi: 10.1016/s0958-1669(99)00005-1. [DOI] [PubMed] [Google Scholar]
- 91.Lakomek N-A, Kaufman JA, Stahl SJ, Wingfield PT. HIV-1 envelope protein gp41: An NMR study of dodecyl phosphocholine embedded gp41 reveals a dynamic prefusion intermediate conformation. Structure. 2014;22:1311–1321. doi: 10.1016/j.str.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laue TM, Senear DF, Eaton S, Ross JBA. 5-Hydroxytryptophan as a new intrinsic probe for investigating protein-DNA interactions by analytical ultracentrifugation. Study of the effect of DNA on the self-assembly of the bacteriophage λ cI repressor. J. Biol. Chem. 1993;32:2469–2472. doi: 10.1021/bi00061a003. [DOI] [PubMed] [Google Scholar]
- 93.LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- le Maire M, Champeil P, Moller JV. Interaction of membrane proteins and lipids with solubilizing detergents. Biochimica et Biophysica Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Liao Y-D, Jeng J-C, Wang C-F, Wang S-C, Chang S-T. Removal of N-terminal methionine from recombinant proteins by engineered E. coli methionine aminopeptidase. Protein Science. 2004;13:1802–1810. doi: 10.1110/ps.04679104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty JJ, Malecki JL, Agnew HD, Michelson-Horowitz DJ, Tan S. Comparison of affinity tags for protein production. Protein Expression and Purification. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Lilie H, Schwartz E, Rudolph R. Advances in refolding of proteins produced in E.coli. Curr. Opin. Biotechnol. 1998;9:497–501. doi: 10.1016/s0958-1669(98)80035-9. [DOI] [PubMed] [Google Scholar]
- Lindwall G, Chau M-F, Gardner SR, Kohlstaedt LA. A sparse matrix approach to the solubilization of overexpression proteins. Protein Eng. 2000;13:67–71. doi: 10.1093/protein/13.1.67. [DOI] [PubMed] [Google Scholar]
- London J, Skrzynia C, Goldberg ME. Renaturation of Escherichia coli tryptophanase after exposure to 8 M urea. Eur. J. Biochem. 1974;47:409–415. doi: 10.1111/j.1432-1033.1974.tb03707.x. [DOI] [PubMed] [Google Scholar]
- Lu HS, Fausset PR, Sotos LS, Clogston CL, Rohde MF, Stoney KS, Herman AC. Isolation and characterization of three recombinant human granulocyte colony stimulating factor His to Gln isoforms produced in E. coli. Protein Expr. Purif. 1993;4:465–472. doi: 10.1006/prep.1993.1061. [DOI] [PubMed] [Google Scholar]
- Markrides S. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston FAO, Hartley DL. Solubilization of protein aggregates. Methods Enzymol. 1990;182:264–276. doi: 10.1016/0076-6879(90)82022-t. [DOI] [PubMed] [Google Scholar]
- Matthew JB, Friend SH, Botelho LD, Lehman LD, Hanania GI, Gurd FRH. Discrete charge calculations of potentiometric titrations for globular proteins. Biochem. Biophys. Res. Commun. 1978;81:416–421. doi: 10.1016/0006-291x(78)91549-8. [DOI] [PubMed] [Google Scholar]
- Maurizi MR. Proteases and protein degradation in Escherichia coli. Experientia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Hassel AM, Lambert MH, Jordon SR, Proudfoot AEI, Graber P, Wells TNC. A novel dimer configuration revealed by the crystal structure at 2.4 angstrom resolution of human interleukin-5. Nature. 1993;363:172–176. doi: 10.1038/363172a0. [DOI] [PubMed] [Google Scholar]
- Mildner AM, Rothrock DJ, Leone JW, Bannow CA, Lull JM, Reardon IM, Sarcich JL, Howe WJ, Tomich C-SC, Smith CW, Heinrikson RL, Tomasselli AG. The HIV-1 protease as enzyme and substrate: Mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry. 1994;33:9405–9413. doi: 10.1021/bi00198a005. [DOI] [PubMed] [Google Scholar]
- Miller CG, Strauch KL, Kurral AM, Miller JL, Wingfield PT, Mazzei GJ, Werlen RC, Graber P, Movva NR. N-Terminal methionine-specific peptidase in Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 1987;84:2718–2772. doi: 10.1073/pnas.84.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller HI, Henzel WJ, Ridgway JB, Kuang W-J, Chrisholm V, Liu C-C. Cloning and expression of a yeast ubiquitin-protein cleaving activity in E. coli. Biotechnology. 1989;7:698–704. [Google Scholar]
- Missiakas D, Raina S. Protein folding in the bacterial periplasm. J. Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murby M, Uhlen M, Stahl S. Upstream strategies to minimize proteolytic degradation upon recombinant protein in Escherichia coli. Protein Expr. Purif. 1996;7:129–136. doi: 10.1006/prep.1996.0018. [DOI] [PubMed] [Google Scholar]
- Nagata K, Kikuchi N, Ohara O, Teraoka H, Yoshida N, Kawade Y. Purification and characterization of recombinant murine immune interferon. FEBS Lett. 1986;205:200–204. doi: 10.1016/0014-5793(86)80897-3. [DOI] [PubMed] [Google Scholar]
- Narayan M. Disulfide bonds: protein folding and subcellular protein trafficking. FEBS J. 2012;279:2272–2282. doi: 10.1111/j.1742-4658.2012.08636.x. [DOI] [PubMed] [Google Scholar]
- Nash HA, Robertson CA, Flamm E, Weisberg RA, Miller H. Overproduction of Escherichia coli integration host factor, a protein with noidentical subunits. J. Bacteriol. 1987;169:4124. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC. Chemical composition of Escherichia coli. In: Neidhardt FC, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 3–6. [Google Scholar]
- Nguyen LH, Jenson DB, Burgess RR. Overproduction and purification of sigma-32, the Escherichia coli heat shock transcription factor. Protein Expr. Purif. 1993;4:425–433. doi: 10.1006/prep.1993.1056. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Stahl S, Lundeberg J, Uhlen M, Nygren P-A. Affinity fusion strategies for detection, purification, and immobilization of recombinant proteins. Protein Expr. Purif. 1997;11:1–16. doi: 10.1006/prep.1997.0767. [DOI] [PubMed] [Google Scholar]
- Orsini G, Goldberg ME. The renaturation of reduced chymotrypsin A in guanidine·HCl. J. Biol. Chem. 1978;253:3453–3458. [PubMed] [Google Scholar]
- Pace NC, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsch D, Anspach FB. Endotoxin removal from protein solutions. J. Biotechnol. 2000;76:97–119. doi: 10.1016/s0168-1656(99)00185-6. [DOI] [PubMed] [Google Scholar]
- Puri NK, Crivelli E, Cardamome M, Fiddes R, Bertoloini J, Ninham B, Brandon MR. Solubilization of growth hormone and other recombinant proteins from Escherichia coli by using a cationic surfactant. Biochem. J. 1992;285:871–879. doi: 10.1042/bj2850871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quing G, Ma LC, Korchid A, Swapna GV, Mal TK, Takayama MM, Xia B, Phadtare S, Ke H, Acton T, Montelione GT, Ikura M, Inouye M. Cold Shock Induced high-yield protein production in Escherichia coli. Nat Biotechnol. 2004;22:877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- Rasmussen JR. Effect of glycosylation on protein function. Curr. Opin. Struct. Biol. 1992;2:682–686. [Google Scholar]
- Ren Z, Schaefer TS. Isopropyl-b-D-thiogalactosidase (IPTG)-inducible tyrosine phosphorylation of protein in E.coli. Biotechniques. 2001;31:1254–1258. doi: 10.2144/01316bm05. [DOI] [PubMed] [Google Scholar]
- Rudolph R, Bohm G, Lilie H, Jaenicke R. Folding proteins. In: Creighton TE, editor. Protein Function: A Practical Approach. 2nd Edition. Oxford: IRL Press; 1997. pp. 57–99. [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nature Reviews Molecular Cell Biology. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S, Demain D. Special issue on the production of recombinant proteins: editorial comments. Biotechnol. Advances. 2012;30:1100–1101. doi: 10.1016/j.biotechadv.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Schein CH. Production of soluble recombinant proteins in bacteria. Biotechnology. 1989;7:1141–1147. [Google Scholar]
- Schiene C, Fisher G. Enzymes that catalyze the restructuring of proteins. Curr. Opin. Struct. Biol. 2000;10:40–45. doi: 10.1016/s0959-440x(99)00046-9. [DOI] [PubMed] [Google Scholar]
- Schmid FX. Optical spectroscopy to characterize protein conformation and conformational changes. In: Creighton TE, editor. Protein Structure: A Practical Approach. Second Edition. Oxford: IRL Press; 1997. pp. 261–296. [Google Scholar]
- Schneider C, Newman RA, Sutherland DR, Asser U, Greaves MF. A one step purification of membrane proteins using a high affinity immunomatrix. J. Biol. Chem. 1982;257:10766–10769. [PubMed] [Google Scholar]
- Scopes RK. Protein Purification: Principles and Practice. 3rd ed. New York and Heidelberg: Springer-Verlag; 1994. [Google Scholar]
- Sherman F, Stewart JW, Tsunasawa S. Methionine or not methionine at the beginning of a protein. Bioessays. 1985;3:27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in E. coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins. J. Bioscience and Bioengineering. 2005;99:303–310. doi: 10.1263/jbb.99.303. [DOI] [PubMed] [Google Scholar]
- Shire SJ, Bock L, Ogez J, Builder S, Kleid D, Moore DM. Purification and immunogenicity of fusion VP1 protein of foot and mouth disease virus. Biochemistry. 1984;23:6474–6480. doi: 10.1021/bi00321a031. [DOI] [PubMed] [Google Scholar]
- Skerra A, Pfitzinger I, Pluckthun A. The functional expression of antibody Fv fragments in Escherichia coli: Improved vectors and a generally applicable purification technique. Biotechnology. 1991;9:273–278. doi: 10.1038/nbt0391-273. [DOI] [PubMed] [Google Scholar]
- Sofer G, Hagel L. Handbook of process chromatography: A guide to optimization, scale-up, and validation. San Diego, Calif: Academic Press; 1997. [Google Scholar]
- Sottrup-Jensen L. Alpha-macroglobulins: Structure, shape, and mechanism of proteinase complex formation. J. Biol. Chem. 1989;264:11539–11542. [PubMed] [Google Scholar]
- Stahl SJ, Wingfield PT, Kaufman JD, Pannell LK, Cioce V, Sakata H, Taylor WG, Rubin JS, Bottaro DP. Functional and biophysical characterization of recombinant human growth factor isoforms produced in. Escherichia coli. Biochem. J. 1997;326:763–772. doi: 10.1042/bj3260763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Stein WH, Moore S. Reactions of cyanate present in aqueous urea with amino acids and proteins. J. Biol. Chem. 1960;235:3177–3181. [Google Scholar]
- Stein S. Isolation of natural proteins. In: Stein S, editor. Fundamentals of Protein Biotechnology. New York: Marcel Dekker; 1991. pp. 137–160. [Google Scholar]
- Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JG, Shinnick TM, Green N, Lerner RA. Antibodies that react with predetermined sites on proteins. Science. 1983;219:660–665. doi: 10.1126/science.6186024. [DOI] [PubMed] [Google Scholar]
- Swartz JP. Advances in Escherichia coli production of therapeutic proteins. Curr. Opin. Biotechnol. 2001;12:195–201. doi: 10.1016/s0958-1669(00)00199-3. [DOI] [PubMed] [Google Scholar]
- Takeda M, Kainosho M. Cell-free protein production for NMR studies. Methods Mol Biol. 2012;831:71–84. doi: 10.1007/978-1-61779-480-3_5. [DOI] [PubMed] [Google Scholar]
- Tamura M, Ito K, Kunihiro S, Yamasaki C, Haragauchi M. Production of human beta-actin and a mutant using a bacterial expression with a cold shock vector. Protein Expression and Purification. 2011;78:1–5. doi: 10.1016/j.pep.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv. Protein Chem. 1968;23:122–275. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Thatcher DR. Industrial scale purification of proteins. In: Price NC, editor. Proteins LabFax. Oxford: BIOS Scientific Publishers; 1996. pp. 131–137. [Google Scholar]
- Thatcher DR, Panayotatos N. Purification of recombinant IFN-2. Methods Enzymol. 1986;119:166–177. doi: 10.1016/0076-6879(86)19026-4. [DOI] [PubMed] [Google Scholar]
- Thatcher DR, Wilks P, Chaudhuri J. Inclusion bodies and refolding. In: Price NC, editor. Proteins LabFax. Oxford: BIOS Scientific Publishers; 1996. pp. 119–130. [Google Scholar]
- Timasheff SN, Arakawa T. Stabilization of protein structure by solvents. In: Creighton TE, editor. Protein Structure: A Practical Approach. Second Edition. Oxford: IRL Press; 1997. pp. 349–363. [Google Scholar]
- Tolia NH, Joshua-To L. Strategies for protein coexpression in Escherichia coli. Nature Methods. 2006;3:55–64. doi: 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Forberg G, Moks T, Hartmanis M, Nilsson B. Fusion proteins in biotechnology. Curr. Opin. Biotechnol. 1992;3:363–369. doi: 10.1016/0958-1669(92)90164-e. [DOI] [PubMed] [Google Scholar]
- Ultsch M, deVos AM, Kossiakoff AA. Crystals of the complex between human growth hormone and the extracellular domain of its receptor. J. Mol. Biol. 1991;222:865–868. doi: 10.1016/0022-2836(91)90578-t. [DOI] [PubMed] [Google Scholar]
- Vallejo LF, Rinas U. Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microbial Cell Factories. 2004;3:11. doi: 10.1186/1475-2859-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reis R, Zydney A. Membrane separations in biotechnology. Curr. Opin. Biotechnol. 2001;12:208–211. doi: 10.1016/s0958-1669(00)00201-9. [DOI] [PubMed] [Google Scholar]
- Ventura S, Villaverde A. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 2006;24:179–185. doi: 10.1016/j.tibtech.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Villaverde A, García-Fruitós E, Rinas U, Seras-Franzoso J, Kosoy A, Luis Corchero J, Vazquez E. Packaging protein drugs as bacterial inclusion bodies for therapeutic applications. Microbial Cell Factories. 2012;11:76. doi: 10.1186/1475-2859-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter H, Johansson G. Partitioning in aqueous two-phase systems: An overview. Anal. Biochem. 1986;155:215–242. doi: 10.1016/0003-2697(86)90431-8. [DOI] [PubMed] [Google Scholar]
- Walter MR, Cook WJ, Ealick SE, Nagabhushan TL, Trotta PP, Bugg CE. Three-dimensional structure of human recombinant granulocyte-macrophage colony stimulating factor. J. Mol. Biol. 1992;224:1075–1085. doi: 10.1016/0022-2836(92)90470-5. [DOI] [PubMed] [Google Scholar]
- Wang Y-X, Neamati N, Jacob J, Palmer I, Stahl SJ, Kaufman JD, Huang PL, Huang PL, Winslow HE, Pommier Y, Wingfield PT, Lee-Huang S, Bax A, Torchia DA. Solution structure of anti-HIV-1 and anti-tumor protein MAP30: Structural insights into its multiple functions. Cell. 1999;99:433–442. doi: 10.1016/s0092-8674(00)81529-9. [DOI] [PubMed] [Google Scholar]
- Watanabe E, Tsoka S, Asenjo JA. Selection of chromatographic protein purification operations based on physicochemical properties. Ann. N.Y. Acad. Sci. 1994;721:348–364. doi: 10.1111/j.1749-6632.1994.tb47407.x. [DOI] [PubMed] [Google Scholar]
- Waugh DS. Making the most of affinity tags. Trends Biotechnology. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Wetlaufer DB. Nonenzymatic formation and isomerization of protein disulfides. Methods Enzymol. 1984;107:301–304. doi: 10.1016/0076-6879(84)07020-8. [DOI] [PubMed] [Google Scholar]
- Wetlaufer DB, Branca PA, Chen G-X. The oxidative folding of proteins by disulfides plus thiol does not correlate with redox potential. Protein Eng. 1987;1:141–146. doi: 10.1093/protein/1.2.141. [DOI] [PubMed] [Google Scholar]
- Wetzel R. Principles of protein stability. Part 2—Enhanced folding and stabilization of proteins by suppression of aggregation in vitro and in vivo. In: Rees AR, Sternberg MJE, Wetzel R, editors. Protein Engineering: A Practical Approach. Oxford: IRL Press; 1992. pp. 191–216. [Google Scholar]
- Widmann M, Christen P. Comparison of folding rates of homologous prokaryotic and eukaryotic proteins. J. Biol. Chem. 2000;275:18619–18622. doi: 10.1074/jbc.C000156200. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Graber P, Rose K, Simona MG, Hughes GJ. Chromatofocusing of N-terminally processed forms of proteins: Isolation and characterization of two forms of interleukin 1β and bovine growth hormone. J. Chromatogr. 1987b;387:291–300. doi: 10.1016/s0021-9673(01)94532-7. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Mattaliano RJ, MacDonald HR, Craig S, Clore GM, Gronenborn AM, Schmeissner U. Recombinant-derived interleukin 1α stabilized against specific deamidation. Protein Eng. 1987a;1:413–417. doi: 10.1093/protein/1.5.413. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Payton M, Graber P, Rose K, Dayer J-M, Shaw AR, Schmeissner U. Purification and characterization of human interleukin 1α produced in Escherichia coli. Eur. J. Biochem. 1987c;165:537–541. doi: 10.1111/j.1432-1033.1987.tb11472.x. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Kaufman J, Palmer I, Chung V, Sax JK, Kleiner DE, Stetler-Stevenson GW. Functional and biophysical characterization of full length, recombinant human TIMP-2 produced in Escherichia coli: Comparison of wild type and N-terminal alanine substituted variant. J. Biol. Chem. 1999;274:21362–21368. doi: 10.1074/jbc.274.30.21362. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Kaufman J, Zlotnick A, Hyde CC, Gronenborn AM, Clore GM. The extracellular domain of immunodeficiency virus gp41 protein: Expression in Escherichia coli, purification and crystallization. Protein Sci. 1997;6:1653–1660. doi: 10.1002/pro.5560060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Payton MA, Venkatesan S, Misra M, Steven A. HIV-1 Rev expressed in recombinant Escherichia coli: Purification polymerization and conformational properties. Biochemistry. 1990;30:7527–7534. doi: 10.1021/bi00244a023. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Thomsen DR, Homa FL, Booy FP, Trus BL, Steven AC. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of the VP5, the major capsid protein of Herpes Simplex Virus. J. Virology. 1997a;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Williams RW, Steven AC. Hepatitis core antigen produced in E. coli: Conformational analysis, and in vitro assembly. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- Wurm F, Bernard A. Large–scale transient expression in mammalian cells for recombinant protein production. Curr. Opin. Biotechnol. 1999;10:156–159. doi: 10.1016/s0958-1669(99)80027-5. [DOI] [PubMed] [Google Scholar]
- Wyss DF, Wagner G. The structure of sugars in glycoproteins. Curr. Opin. Struct. Biol. 1996;7:409–416. doi: 10.1016/s0958-1669(96)80116-9. [DOI] [PubMed] [Google Scholar]
- Xu J, Dolan MC, Medrano G, Cramer CL, Weathers PJ. Green factory: Plants as bioproduction platforms for recombinant proteins. Biotechnol. Advances. 2012;30:1171–1184. doi: 10.1016/j.biotechadv.2011.08.020. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Miyazaki M. Refolding Techniques for Recovering Biologically Active Recombinant Proteins from Inclusion Bodies. Biomolecules. 2014;4:235–251. doi: 10.3390/biom4010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Hinck AP, Wang Y-X, Nicholson LK, Torchia DA, Wingfield PT, Stahl SJ, Kaufman JD, Chang C-H, Domaille PJ, Lam PYS. Three dimensional solution structure of the HIV-1 protease complexed with DMP 323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy. Protein Sci. 1996;5:495–506. doi: 10.1002/pro.5560050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang L, Zhang T, Feng Y, Lu W, Wang J, Wu H, Cao C, Wang X. Highly Efficient Production of Soluble Proteins from Insoluble Inclusion Bodies by a Two-Step-Denaturing and Refolding Method. PLOS ONE. 2011 doi: 10.1371/journal.pone.0022981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z-N, Mueser TC, Kaufman J, Stahl SJ, Wingfield PT, Hyde C. The structure of the SIV gp41 ectodomain at 1.47 A. J. Struct. Biol. 1999;126:133–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- Yarranton GT, Mountain A. Expression of proteins in prokaryotic systems—Principles and case studies. In: Rees AR, Sternberg MJE, Wetzel R, editors. Protein Engineering: A Practical Approach. Oxford: IRL Press; 1992. pp. 303–324. [Google Scholar]
- Zardeneta G, Horowitz PM. Detergent, liposome, and micelle-assisted protein refolding. Anal. Biochem. 1994;223:1–6. doi: 10.1006/abio.1994.1537. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Olsen DR, Nguyen KB, Olson PS, Rhodes ET, Mascarenhas D. Expressoin of eukaryotic proteins in soluble form in Escherichia coli. Protein Expr. Purif. 1998;12:159–165. doi: 10.1006/prep.1997.0834. [DOI] [PubMed] [Google Scholar]
- Zhu J. Mammalian cell protein expression for biopharmaceutical production. Biotechnol. Advances. 2012;30:1158–1170. doi: 10.1016/j.biotechadv.2011.08.022. [DOI] [PubMed] [Google Scholar]