Abstract
Background
Hepatocellular carcinoma (HCC) is the most common liver cancer, leading to many cancer-related deaths worldwide. Several studies have shown an association between pre-S deletion mutation of hepatitis B virus (HBV) and HCC risk, but the results remain conflicting. We aimed to verify HBV pre-S deletion mutations in relation to the risk of HCC.
Material/Methods
We searched the commonly used electronic databases for relevant studies of this association among the Asian population until September 30th, 2014. Odds ratios (ORs) with 95% confidence intervals (CIs) were employed to calculate the association.
Results
A total of 17 case-control studies were screened out, including 2837 HBV-infected patients, of whom 1246 had HCC. The results showed that the frequency of pre-S deletion of HBV in patients with HCC was higher than that in patients without HCC (35.7% vs. 11.5%), indicating the prevalence of this mutation in patients with HCC. Statistically significant correlations were observed for pre-S deletion mutation and risk of HCC in a random-effects model (OR=3.90, 95% CI=2.80–5.44, P<0.00001). This association was also found in Chinese populations (OR=4.84, 95% CI=2.86–8.20, P<0.00001).
Conclusions
Our data indicate that HBV pre-S deletion mutations might be associated with HCC risk. Their oncogenic role may be important in studying the potential mechanism of HBV hepatocarcinogenesis.
MeSH Keywords: Carcinoma, Hepatocellular; Hepatitis B virus; Meta-Analysis
Background
Hepatocellular carcinoma (HCC) is the most common solid tumor worldwide, and is the leading factor responsible for cancer mortality [1]. Most HCCs occur in China, Southeast Asia, and in parts of Africa [2]. The main risk factors contributing to the occurrence of HCC are virus-induced chronic infection caused by hepatitis C virus (HCV) or hepatitis B virus (HBV), alcohol abuse, liver cirrhosis, exposure to aflatoxin B1, and diabetes [3]. Among these, HBV is a major etiologic agent and is causes 80% of HCC cases. Furthermore, HBV-related liver disease is the major reason for liver transplantation [4,5]. Research has estimated that both genetic and viral factors are crucial for the development of HCC; however, the exact mechanism leading to HBV tumorigenesis has not yet been fully elucidated [6–8].
HBV is a small, enveloped, double-stranded DNA virus. It contains 4 main regions: the pre-S/S region, the enhancer II region, basal core promoter region, and the precore region [9]. Several HBV mutant strains in these regions were reported to contribute to the progression of liver disease [10]. One of the most commonly reported HBV mutations is the pre-S region deletion mutation. The pre-S proteins are produced from the surface gene (S gene) region, including 3 different translation sites: pre-S1, pre-S2, and S [11]. The pre-S protein is reported to be involved in hepatocyte attachment of the virus [12] and contains B cell and T cell epitopes [13]. The B cell and T cell epitopes are binding sites for neutralizing anti-pre-S2 antibody. An S promoter has been identified in pre-S that is associated with the production of middle and small HBs [11]. The pre-S1 and pre-S2 regions are crucial for the interplay within the host immune responses [14]. Numerous reports of mechanisms suggested that deletion mutations in pre-S in the integrated HBV DNA may impair the secretion of HBsAg, leading to increased endoplasmic reticulum and oxidative stress in hepatocytes [15].
Recent studies have demonstrated that pre-S mutations affect the severity of liver disease; its mutation generally presents in the form of deletions [16,17]. Studies have determined that mutation of the promoter sites of pre-S1 and pre-S2 significantly increase the risk of HCC development [18,19]; however, a clinical study [20] study has shown that the pre-S internal deletion mutants are unlikely to play crucial roles in hepatocarcinogenesis. These discrepancies may be due to genetic trait differences and different linkage disequilibrium. Therefore, in order to derive a more comprehensive estimation of the associations between pre-S deletion mutation and HCC risk, we conducted a meta-analysis to assess the relationship only among Asians.
Material and Methods
Literature search
An electronic literature search was conducted using PubMed, Embase, and China National Knowledge Infrastructure (CNKI) databases until September 30th, 2013. The Medical Subject Heading (MeSH) terms “hepatitis B virus,” “HBV,” “pre-S deletion,” “mutation,” “hepatocellular carcinoma”, and the individual corresponding free terms were employed as the searching words. In addition, the citations in the retrieved articles were reviewed to search for relevant studies.
Criteria for article screening
The studies included met the following criteria: 1) assessed the association between pre-S mutations of HBV and HCC risk; 2) case-control or cohort study; 3) the diagnosis of liver disease followed the guidelines of the American Association for the Study of Liver Diseases [14,21], and matched HBV-infected non-HCC cases as controls; 4) odds ratio with the 95% confidence interval (CI) was reported or could be determined from the available data; and 5) Asian population.
Data extraction
Two investigators independently assessed the extracted data from the included studies and reached a consensus on final results. Any disagreement was resolved by discussing with a third expert. The following information was extracted from all obtained publications: first author’s name, publication year, country or area, the number of included patients, and the number of deletion mutations. We extracted data on the participants whose mutation status had been determined. Our evaluation included deletions in pre-S1 and/or pre-S2 that were predicted or known to decrease pre-S or S expression.
Statistical analysis
Statistical analyses were carried out using Review Manager 5.2 (The Cochrane Collaboration). The crude odds ratios (ORs) and their corresponding 95% CI were used as a measure of the associations between HBV pre-S deletion mutation and risk of HCC. The overall effect was evaluated through the Z test. P value less than 0.05 was deemed to be statistically significant. The heterogeneity between all the included articles was evaluated by the Q test and the I2 statistics. P value less than 0.1 and I2 less than 50% was considered to be statistically significant. A fixed- or random-effects model was used to estimate the heterogeneity. A fixed-effects model was used when the effects were assumed to be homogenous, and a random-effects model was used when they were heterogenous. A funnel plot test was performed to examine publication bias.
Results
Study selection
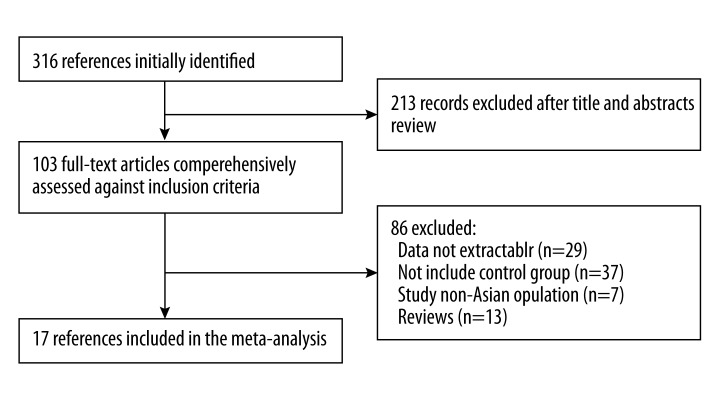
Our search strategy yielded 316 papers. We reviewed the titles, abstracts, and full texts of all retrieved articles using the defined criteria. Finally, 17 studies evaluating the relationship between pre-S deletion mutation of HBV and risk of HCC were selected in our meta-analysis. Figure 1 shows the flow diagram. Characteristics of the included studies are shown in Table 1. Of the 17 included studies, 8 were from mainland China [17,22–29], 3 were from Taiwan [16,30,31], 1 was from Thailand [32], 4 were from South Korea [33–36], and 1 was from mixed Asian countries [37]. These studies included 2837 HBV-infected participants, of whom 1246 had HCC.
Figure 1.
Studies identified with criteria for inclusion and exclusion.
Table 1.
Characteristics of studies included in this meta-analysis.
| First author | Year | Country | Mean age | Control type | Sample size | No. of deletion mutations | ||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases/controls | Cases | Controls | ||||
| Choi MS | 2007 | Korean | 51.9±7.8 | 39.4±12.2 | AC+CH+LC | 72/228 | 11 | 16 |
| Gao ZY | 2007 | China | 55.56±12.28 | 20.04±0.89 | AC+CHB | 26/53 | 10 | 3 |
| Mun H | 2008 | Korean | 47.9±17.3 | 47.9±17.3 | AC+CH+LC | 40/80 | 21 | 16 |
| Fang ZL | 2008 | China | 30–55 | 30–55 | Non-HCC | 33/33 | 15 | 6 |
| Chen C | 2008 | Taiwan | 50.7±11.3 | 48.9±10.6 | Non-HCC | 80/160 | 28 | 27 |
| Cao ZG | 2008 | China | 42.1±8.8 | 39.9±12.4 | CH | 47/50 | 24 | 9 |
| Jang JS | 2009 | South Korea | – | – | Non-HCC | 48/71 | 17 | 13 |
| Abe K | 2009 | Four countries | 11±3 | 6±5 | CH | 30/10 | 23 | 0 |
| Wu XD | 2010 | China | 46.7±10.7 | 45.8±13.3 | AC | 44/43 | 21 | 6 |
| Yin JH | 2010 | China | 49.9±11.0 | 30.0±16.9 | AC | 231/603 | 94 | 43 |
| Huang H | 2010 | Taiwan | 12.7±6.6 | 12.3±0.8 | CH | 19/19 | 9 | 1 |
| Yeung P | 2011 | Taiwan | 56.9±11.5 | 56.8±11.5 | Non-HCC | 96/96 | 28 | 14 |
| Qiang FL | 2011 | China | 42–67 | 21–39 | AC | 26/25 | 6 | 0 |
| Lee MH | 2011 | Korean | 44.3±7.8 | 44.3±8.0 | Non-HCC | 135/135 | 25 | 6 |
| Thongbai C | 2013 | Thailand | 58.1±12.0 | 41.3±8.8 | Non-HCC | 65/89 | 11 | 9 |
| Zhao ZM | 2014 | China | – | – | Non-HCC | 157/160 | 74 | 45 |
| Qu LS | 2014 | China | 44.2±9.5 | 45.0±8.3 | Non-HCC | 97/96 | 28 | 11 |
AC – asymptomatic carrier; CH – chronic hepatitis; LC – liver cirrhosis; HCC – hepatocellular carcinoma; ‘–’ – not applicable.
Pre-S deletions and the risk of HCC
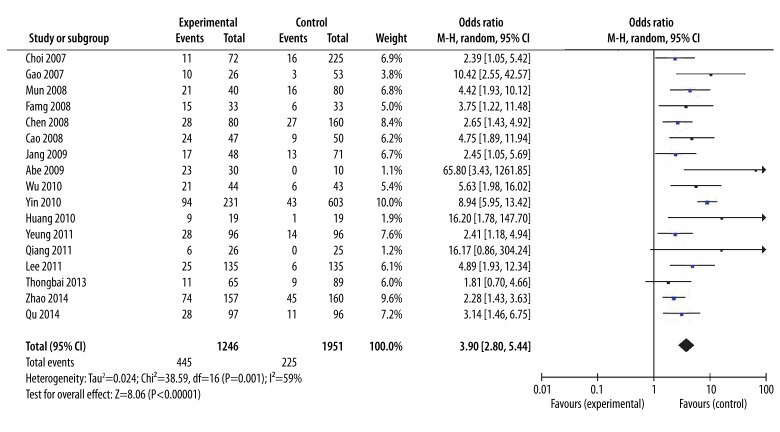
We mainly focused on the pre-S deletion mutation in this meta-analysis. As shown in Figure 2, we observed a statistically significant association between pre-S deletion mutation and HCC risk. The data show that patients with HCC had more HBV with pre-S deletions than patients without HCC (35.7% vs. 11.5%). The overall risk estimates OR for HCC of pre-S deletion was 3.90 (95% CI=2.80–5.44, P<0.00001) in a random-effects model. When only considering the Chinese population, our results found that the pre-S deletions were significantly associated with HCC risk (OR=4.84, 95% CI=2.86–8.20, P<0.00001).
Figure 2.
Forest plot for the correlation between pre-S deletion and HCC risk. Pooled odds ratio for the association of pre-S deletion mutation and HCC risk using random-effects model.
Sensitivity analysis
For this meta-analysis, the ORs were not significantly changed by deleting any single study, indicating that no individual study affected the statistical significance of the overall results.
Publication bias
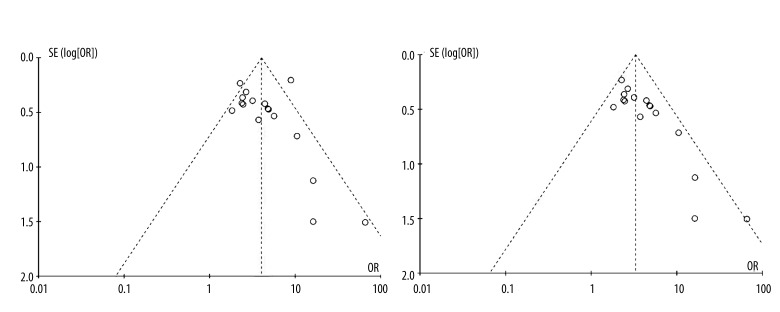
Publication bias was found in pre-S deletion mutation studies and funnel plot analysis showed that the study by Yin et al. was the outlier. After removing this study, the between-study heterogeneity decreased from 59% to 20%. Figure 3A, 3B display the funnel plots for the associations between pre-S deletion and HCC risk before and after omitting this report.
Figure 3.
Funnel plots analysis to detect publication bias of pre-S deletion mutation before (A) and after (B) three studies omitting. Each point represents an independent study for the indicated association.
Discussion
The pre-S region of HBV is located at 5′ of the S gene, and pre-S is composed of pre-S1 and pre-S2. Natural pre-S mutations of HBV have been frequently observed in chronic HBV infections [38]. Pre-S region deletion mutation is known to be a risk factor for hepatocarcinogenesis and often occurs in patients treated with interferon. This suggests attempts by the virus to evade host immune response [39]. Pre-S1 region deletion and pre-S2 region deletion are 2 common variants in the pre-S region. The deletion in the pre-S region results in the dysfunctional immune response and hepatocyte pathogenesis [40], finally leading to the occurrence of HCC. Thus, these mutations might be used as biomarkers in predicting the development of HCC.
We identified an association of pre-S deletion mutation with HCC risk in this meta-analysis. The results showed that pre-S mutation statistically correlates with the significantly increased risk of HCC. The pooled crude ORs from the included studies suggest that pre-S mutation is associated with 4.64-fold increased risk of HCC compared with wild-type HBV (without mutations) (95% CI= 2.92–8.12, P<0.00001) in a random-effects model.
To determine if specific pre-S mutations correlate with increased risk of HCC occurrence, we compared and analyzed the deleted portions of pre-S region in HBV-infected patients with or without HCC. We found that the 5′-terminal half of pre-S2 (nucleotides 3206-60) was the most commonly deleted portion of the pre-S region but that the frequency of this deletion in patients with and without HCC showed no significant statistical difference.
Although many studies have suggested the close relationship of pre-S deletions with the prevalence of HCC, they were mainly based on cross-sectional studies without thoroughly considering various possible confounding factors. Male sex, older age, and infection with HBV are the main risk factors for HCC occurrence [41,42]. Older age and HBV genotype C are closely linked to the presence of pre-S deletions [18,43]. The potential effect of the interaction of these factors should be considered in studying the role of pre-S deletions in HCC development in further research.
Several potential limitations exist in this meta-analysis. First, the observed association is substantially based on cross-sectional studies without considering other possible confounding factors. Second, publication biases cannot be completely excluded because all of the included studies were mainly relying on observation. Third, our study only focused on pre-S deletion mutation and we needed to also identify single-base substitutions at the pre-S1 and pre-S2 start codon. Fourth, all studies utilized in this meta-analysis were performed in eastern Asia, where HBV is endemic. Fifth, the control cases were not consistent; therefore, our results may not be generalizable to populations infected by other HBV genotypes. Furthermore, articles published in other languages were not incorporated into this analysis due to the anticipated difficulties in acquiring accurate medical translation.
Conclusions
Despite the above limitations, our results show that pre-S mutations are associated with the increased risk of HCC development. However, large-scale, well-designed, case-control studies concentrating on the epigenetic influence and the genetic variations are still needed to prove this association.
Footnotes
Conflict of interests
None.
Source of support: This study is supported by Jinan science and technology plan projects, No: 201408020
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Tsukuma H, Tanaka H, Ajiki W, Oshima A. Liver cancer and its prevention. Asian Pac J Cancer Prev. 2005;6(3):244–50. [PubMed] [Google Scholar]
- 3.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 4.Yu MC, Yuan JM, Govindarajan S, Ross RK. Epidemiology of hepatocellular carcinoma. Can J Gastroenterol. 2000;14(8):703–9. doi: 10.1155/2000/371801. [DOI] [PubMed] [Google Scholar]
- 5.Yuen MF, Hou JL, Chutaputti A. Hepatocellular carcinoma in the Asia pacific region. J Gastroenterol Hepatol. 2009;24(3):346–53. doi: 10.1111/j.1440-1746.2009.05784.x. [DOI] [PubMed] [Google Scholar]
- 6.Chan HL, Sung JJ. Hepatocellular carcinoma and hepatitis B virus. Semin Liver Dis. 2006;26(2):153–61. doi: 10.1055/s-2006-939753. [DOI] [PubMed] [Google Scholar]
- 7.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 8.Taura N, Ichikawa T, Miyaaki H, et al. Frequency of elevated biomarkers in patients with cryptogenic hepatocellular carcinoma. Med Sci Monit. 2013;19:742–50. doi: 10.12659/MSM.889361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shouval D, Samuel D. Hepatitis B immune globulin to prevent hepatitis B virus graft reinfection following liver transplantation: a concise review. Hepatology. 2000;32(6):1189–95. doi: 10.1053/jhep.2000.19789. [DOI] [PubMed] [Google Scholar]
- 10.Kao JH. Role of viral factors in the natural course and therapy of chronic hepatitis B. Hepatol Int. 2007;1(4):415–30. doi: 10.1007/s12072-007-9033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milich DR, Thornton GB, Neurath AR, et al. Enhanced immunogenicity of the pre-S region of hepatitis B surface antigen. Science. 1985;228(4704):1195–99. doi: 10.1126/science.2408336. [DOI] [PubMed] [Google Scholar]
- 12.Berthelot P, Neurath R, Courouce AM, et al. Hepatitis B vaccines with pre-S gene product. Lancet. 1986;1(8490):1150. doi: 10.1016/s0140-6736(86)91860-x. [DOI] [PubMed] [Google Scholar]
- 13.Milich DR, McNamara MK, McLachlan A, et al. Distinct H-2-linked regulation of T-cell responses to the pre-S and S regions of the same hepatitis B surface antigen polypeptide allows circumvention of nonresponsiveness to the S region. Proc Natl Acad Sci USA. 1985;82(23):8168–72. doi: 10.1073/pnas.82.23.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42(5):1208–36. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 15.Fang ZL, Sabin CA, Dong BQ, et al. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103(9):2254–62. doi: 10.1111/j.1572-0241.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeung P, Wong DK, Lai CL, et al. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in chronic hepatitis B. J Infect Dis. 2011;203(5):646–54. doi: 10.1093/infdis/jiq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin J, Xie J, Zhang H, et al. Significant association of different preS mutations with hepatitis B-related cirrhosis or hepatocellular carcinoma. J Gastroenterol. 2010;45(10):1063–71. doi: 10.1007/s00535-010-0253-1. [DOI] [PubMed] [Google Scholar]
- 18.Lin CL, Liu CH, Chen W, et al. Association of pre-S deletion mutant of hepatitis B virus with risk of hepatocellular carcinoma. J Gastroenterol Hepatol. 2007;22(7):1098–103. doi: 10.1111/j.1440-1746.2006.04515.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, Zhang H, Gu C, et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101(15):1066–82. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatsukawa M, Takaki A, Shiraha H, et al. Hepatitis B virus core promoter mutations G1613A and C1653T are significantly associated with hepatocellular carcinoma in genotype C HBV-infected patients. BMC Cancer. 2011;11:458. doi: 10.1186/1471-2407-11-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45(2):507–39. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 22.Fang ZL, Sabin CA, Dong BQ, et al. Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: a matched nested case-control study. J Gen Virol. 2008;89(Pt 11):2882–90. doi: 10.1099/vir.0.2008/002824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Z, Bai X, Guo X, et al. High prevalence of hepatitis B virus pre-S mutation and its association with hepatocellular carcinoma in Qidong, China. Arch Virol. 2008;153(10):1807–12. doi: 10.1007/s00705-008-0176-9. [DOI] [PubMed] [Google Scholar]
- 24.Qu L, Kuai X, Liu T, et al. Pre-S deletion and complex mutations of hepatitis B virus related to young age hepatocellular carcinoma in Qidong, China. PLoS One. 2013;8(3):e59583. doi: 10.1371/journal.pone.0059583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao ZY, Li T, Wang J, et al. Mutations in preS genes of genotype C hepatitis B virus in patients with chronic hepatitis B and hepatocellular carcinoma. J Gastroenterol. 2007;42(9):761–68. doi: 10.1007/s00535-007-2085-1. [DOI] [PubMed] [Google Scholar]
- 26.Qiang F, Wu X, Yang Z. Associations between hepatitis B virus preS2 deletion mutation and hepatocarcinoma. The Practical Journal of Cancer. 2011;20:21–26. [Google Scholar]
- 27.Wu X. Pre-S/S gene and Pre-C/C gene mutation in Guangdong Province of China. Lanzhou University; 2010. Study on the relationship between HBV-related HCC and HBV genotype; p. 42. [Google Scholar]
- 28.Zhao ZM, Jin Y, Gan Y, et al. Novel approach to identifying the hepatitis B virus pre-S deletions associated with hepatocellular carcinoma. World J Gastroenterol. 2014;20(37):13573–81. doi: 10.3748/wjg.v20.i37.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu LS, Liu JX, Liu TT, et al. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in Qidong, China. PLoS One. 2014;9(5):e98257. doi: 10.1371/journal.pone.0098257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CH, Changchien CS, Lee CM, et al. Combined mutations in pre-s/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: a case-control study. J Infect Dis. 2008;198(11):1634–42. doi: 10.1086/592990. [DOI] [PubMed] [Google Scholar]
- 31.Huang HP, Hsu HY, Chen CL, et al. Pre-S2 deletions of hepatitis B virus and hepatocellular carcinoma in children. Pediatr Res. 2010;67(1):90–94. doi: 10.1203/PDR.0b013e3181c1b0b7. [DOI] [PubMed] [Google Scholar]
- 32.Thongbai C, Sa-nguanmoo P, Kranokpiruk P, et al. Hepatitis B virus genetic variation and TP53 R249S mutation in patients with hepatocellular carcinoma in Thailand. Asian Pac J Cancer Prev. 2013;14(6):3555–59. doi: 10.7314/apjcp.2013.14.6.3555. [DOI] [PubMed] [Google Scholar]
- 33.Lee MH, Kim do Y, Kim JK, et al. Combination of preS deletions and A1762T/G1764A mutations in HBV subgenotype C2 increases the risk of developing HCC. Intervirology. 2012;55(4):296–302. doi: 10.1159/000329941. [DOI] [PubMed] [Google Scholar]
- 34.Choi MS, Kim DY, Lee DH, et al. Clinical significance of pre-S mutations in patients with genotype C hepatitis B virus infection. J Viral Hepat. 2007;14(3):161–68. doi: 10.1111/j.1365-2893.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- 35.Mun HS, Lee SA, Jee Y, et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008;80(7):1189–94. doi: 10.1002/jmv.21208. [DOI] [PubMed] [Google Scholar]
- 36.Jang JS, Kim HS, Kim HJ, et al. Association of concurrent hepatitis B surface antigen and antibody to hepatitis B surface antigen with hepatocellular carcinoma in chronic hepatitis B virus infection. J Med Virol. 2009;81(9):1531–38. doi: 10.1002/jmv.21577. [DOI] [PubMed] [Google Scholar]
- 37.Huy TT, Ushijima H, Win KM, et al. High prevalence of hepatitis B virus pre-s mutant in countries where it is endemic and its relationship with genotype and chronicity. J Clin Microbiol. 2003;41(12):5449–55. doi: 10.1128/JCM.41.12.5449-5455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan YF, Lu CC, Chen WC, et al. Prevalence and significance of hepatitis B virus (HBV) pre-S mutants in serum and liver at different replicative stages of chronic HBV infection. Hepatology. 2001;33(1):277–86. doi: 10.1053/jhep.2001.21163. [DOI] [PubMed] [Google Scholar]
- 39.Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127(2):164–76. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 40.Brunetto MR, Stemler M, Bonino F, et al. A new hepatitis B virus strain in patients with severe anti-HBe positive chronic hepatitis B. J Hepatol. 1990;10(2):258–61. doi: 10.1016/0168-8278(90)90062-v. [DOI] [PubMed] [Google Scholar]
- 41.Yuen MF, Tanaka Y, Shinkai N, et al. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut. 2008;57(1):98–102. doi: 10.1136/gut.2007.119859. [DOI] [PubMed] [Google Scholar]
- 42.Yuen MF, Tanaka Y, Fong DY, et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol. 2009;50(1):80–88. doi: 10.1016/j.jhep.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 43.Sugauchi F, Ohno T, Orito E, et al. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol. 2003;70(4):537–44. doi: 10.1002/jmv.10428. [DOI] [PubMed] [Google Scholar]