Abstract
HIV-1 initiates infection by merging its envelope membrane with the target cell membrane, a process that is mediated by the viral Env glycoprotein following its sequential binding to CD4 and coreceptors, CXCR4 or CCR5. Although HIV-1 fusion has been a target for antiviral therapy, the virus has developed resistance to drugs blocking the CCR5 binding or Env refolding steps of this process. This highlights the need for novel inhibitors. Here, we adapted and optimized an enzymatic HIV–cell fusion assay, which reports the transfer of virus-encapsulated β-lactamase into the cytoplasm, to high-throughput screening (HTS) with a 384-well format. The assay was robustly performed in HTS format and was validated by the pilot screen of a small library of pharmacologically active compounds. Several hits identified by screening included a prominent cluster of purinergic receptor antagonists. Functional studies demonstrated that P2X1 receptor antagonists selectively inhibited HIV-1 fusion without affecting the fusion activity of an unrelated virus that enters cells through an endocytic route. The inhibition of HIV–cell fusion by P2X1 antagonists was not through downmodulation of the cell surface expression of CD4 or coreceptors, thus implicating P2X1 receptor in the HIV-1 fusion step. The ability of these antagonists to inhibit viruses regardless of their coreceptor (CXCR4 or CCR5) preference indicates that fusion is blocked at a late step downstream of coreceptor binding. A future large-scale screening campaign for HIV-1 fusion inhibitors, using the above functional readout, will likely reveal novel classes of inhibitors and suggest potential targets for antiviral therapy.
Introduction
The HIV-1 Env induces fusion between the viral and the host cell membranes through a multistep process that is initiated upon sequential engagement of CD4 and its coreceptors, CXCR4 or CCR5.1–4 The formation of ternary complexes between the gp120 subunit of Env, CD4, and coreceptors triggers the refolding of the transmembrane gp41 subunit, which promotes membrane merger.5,6 This refolding progresses through prehairpin intermediates, characterized by the formation/exposure of the N-proximal heptad repeat (HR1) regions and the membrane-proximal heptad repeat (HR2) regions (reviewed in Ref.7). In the final 6-helix bundle structure (6HB), three HR1 and three HR2 coalesce forming a highly stable antiparallel helical bundle. A number of small-molecule inhibitors of HIV-1 fusion that interfere with CD4-induced conformational changes in gp120,8,9 coreceptor binding,10–12 and the gp41 6HB formation13–24 have been identified by high-throughput screening (HTS). Currently, only two HIV-1 fusion inhibitors (enfuvirtide and maraviroc) have been approved for clinical use.25 The ease with which the virus develops resistance to these inhibitors highlights the need for novel therapeutic targets that could be blocked by small molecules.
The emergence of drug-resistant viruses has prompted a paradigm shift for prevention and therapy from targeting the HIV-1 proteins to much more conserved cellular proteins.26–35 Multiple screens for HIV-1 inhibitors relied on in vitro assays, which used viral proteins or their fragments, or on HIV-1 infections/replication as a readout. In vitro screening has identified competitive inhibitors of assembly of the gp41 HR1- and HR2-derived peptides into the 6HB.13–24 HTS for small-molecule inhibitors competing with the chemokine (RANTES) binding to CCR5 has led to the identification of identified coreceptor antagonists that effectively blocked fusion of CCR5-tropic viruses: maraviroc, Sch-C, and TAK-779.10–12 These narrowly focused readouts provide a powerful means to identify specific inhibitors of a given step of the virus entry, but exclude all other targets for inhibition of HIV-1 fusion. Infectivity-based HTS for fusion inhibitors identified BMS-806 that interferes with CD4-induced conformational changes in gp1208,9 as well as several postfusion inhibitors.36 An important caveat of infectivity-based screens when searching for viral fusion inhibitors is that they produce a large number of hits targeting postfusion steps of HIV-1 entry, thus complicating data analysis and hit validation.
Thus far, only one HTS campaign utilized an HIV Env-mediated cell–cell fusion assay.37 This screen identified new inhibitors of HIV-1 fusion, including the 18A compound (1-(2,1,3-benzothiadiazol-4-yl)-3-[(E)-(4-hydroxyphenyl)methyleneamino]urea), which blocks fusion of diverse HIV-1 isolates by interfering with CD4-induced conformational changes in Env glycoprotein.38 However, a cell–cell fusion-based screen should not detect inhibitors of virus endocytosis/trafficking steps of entry or virucidal compounds that selectively damaged the virus.39,40 These considerations warrant the implementation of virus–cell fusion assay for HTS, which appears particularly important in the light of our finding that HIV-1 enters cells through endocytosis and fusion with endosomal compartments.41,42 The results of such screen should reveal new targets for intervention—cellular factors involved in endocytosis and vesicular trafficking. Toward this goal, we adapted a direct virus–cell fusion assay42,43 for HTS and validated it by screening a small library of pharmacologically active compounds (LOPAC). The screen reveals promising hits that inhibit HIV-1 fusion without affecting cell viability, including several purinergic receptor antagonists. Functional characterization of these hits demonstrated that P2X1 receptor antagonists interfere with HIV-1 fusion by a yet unknown mechanism.
Materials and Methods
Cells and Reagents
HeLa-derived TZM-bl cells expressing CD4, CXCR4, and CCR5 (donated by Drs. J.C. Kappes and X. Wu, Ref.44) were obtained from the NIH AIDS Research and Reference Reagent Program. Human embryonic kidney HEK293T/17 cells were obtained from the ATCC (Manassas, VA). HeLa-ADA cells stably expressing Env and Tat from the HIV-1 ADA strain were a gift from Dr. Marc Alizon (Pasteur Institute, Paris, France).45 TZM-bl and HeLa-ADA cells were grown in Dulbecco's modified Eagle's medium (DMEM; Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO) and 100 U penicillin–streptomycin (Gemini Bio-Products, Sacramento, CA). HEK 293T/17 cells were maintained in DMEM supplemented with 10% FBS, 100 U penicillin/streptomycin, and 0.5 mg/mL G418 sulfate (Mediatech). Inhibitors NF279, NF449, NF340, A740003, MRS2500, PSB1114, and PPADS were purchased from Tocris Bioscience (Ellisville, MO). Bright-Glo™ Luciferase and CCF4-AM were obtained from Promega (Madison, WI) and Invitrogen (Carlsbad, CA), respectively. The C52L recombinant peptide derived from the HIV-1 gp41 glycoprotein46 was a gift from Dr. Min Lu (University of New Jersey, New Brunswick, NJ). The LOPAC was obtained from Sigma-Aldrich.
Virus Production
HIV-1 pseudoviruses bearing HXB2, Bal26, and R3A envelope glycoproteins and the β-lactamase-Vpr chimera (BlaM-Vpr) were produced by cotransfecting HEK293T/17 cells with the HIV-based pR8ΔEnv packaging vector, pMM310 plasmid expressing BlaM-Vpr,47 pcRev,48 and the vector encoding HXB2 Env49 or Bal26 (a gift from P. Clapham, University of Massachusetts, Worchester, MA) or R3A (a gift from Dr. J. Hoxie, University of Pennsylvania, Philadelphia, PA), using the jetPRIME® transfection reagent (VWR, Radnor, PA). The transfection medium was replaced with fresh DMEM/10% FBS after 12 h and cells were cultured for 36 h, after which time the virus-containing culture medium was collected, passed through a 0.45-μm filter, aliquoted, and stored at −80°C. Infectious titers were determined by a β-Gal assay,50 using TZM-bl cells.
Development of Virus–Cell Fusion Assay in a 384-Well HTS Format
HIV-1 fusion with target cells was measured using the BlaM assay,42,43 which measures the activity of β-lactamase incorporated into virions using the β-lactamase Vpr chimera. We optimized this assay for a 384-well format compatible with HTS (Table 1). Briefly, the day before the experiment, TZM-bl cells (2×104 cells/well in 25 μL culture medium) were seeded into a 384-well, flat, clear-bottom black wall plate (Cat. No. 3712; Corning, Corning, NY) using a 16-channel pipette. HIV-1 pseudoviruses (5 μL/well) were added to cells at multiplicity of infection (MOI) 1.0 and centrifuged at 2,095 g at 4°C for 30 min to facilitate virus binding to cells. Virus–cell fusion was initiated by incubating the plates at 37°C for 90 min in a cell culture incubator. The medium was then removed and 25 μL/well of 1.8 μM CCF4-AM BlaM substrate was added. The plates were incubated at 12°C overnight to allow for CCF4-AM cleavage by BlaM. The fluorescence intensity was measured using the Synergy HT fluorescence plate reader (Bio-Tek Instr., Bad Friedrichshall, Germany) with excitation at 400 nm and emissions at 460 and 528 nm for the blue and green signals of the substrate, respectively. The fusion signal was calculated and expressed as a ratio of blue over green signals after subtracting the blank fluorescence signal from wells with substrate but without virus, using the following equation: Fusion=(F460−F460 blank)/(F520−F520 blank). The performance of the virus–cell fusion assay for HTS was evaluated using Z′ and the signal-to-background (S/B) ratio, which were calculated using the following equations: Z′=1−(3·SDvirus+3·SDno-virus)/(Fusionvirus−Fusionno-virus) and S/B=Fusionvirus/Fusionno-virus, where SD is standard deviation.
Table 1.
Protocol for Virus–Cell Fusion High-Throughput Screening Assay in Regular 384-Well Cell Culture Plate
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Plate cells | 25 μL | 20,000 TZM-bl cells |
| 2 | Incubation time | 24 h | 37°C, 5% CO2 |
| 3 | Add virus | 5 μL | HIV-1 pseudoviruses at MOI 1.0 |
| 4 | Centrifuge | 30 min | 2,095 g at 4°C |
| 5 | Library compound | 0.1 μL | The final compound concentration is 10 μM |
| 6 | Add positive control | 0.5 μL | Positive control C52L peptide diluted in medium added at final concentration of 1 μM |
| 7 | Incubation time | 90 min | 37°C, 5% CO2 |
| 8 | Add CCF4-AM substrate | 25 μL | Remove the medium before adding substrate |
| 9 | Incubation time | 16 h | Overnight incubation at 12°C |
| 10 | Assay readout | Excitation at 400 nm, emissions at 460 nm and 520 nm | EnVision Multilabel plate reader |
Step Notes
1. Black wall clear-bottom cell culture plate was used. Cells were dispensed to all wells.
3. Virus was added from column 2 to 24. Equal volume of medium was added to column 1 as no virus control. MOI is multiplicity of infection.
5. Pintool was used for compound transfer.
6. Positive control C52L was added to column 24.
8. Follow manufacturer's instruction for preparing substrate. Substrate was added from column 3 to column 24.
10. Bottom read module was used for reading plate.
Pilot Screening in a 384-Well HTS Format
Pilot screening of the LOPAC library (Sigma-Aldrich) was carried out to validate the virus–cell fusion assay for HTS using a robotic platform. Twenty-five microliters of CD4- and CXCR4-expressing indicator TZM-bl cells (2×104/well) was dispensed into a 384-well cell culture plate, using Multidrop™ Combi (Thermo-Fisher Scientific, Waltham, MA), and cultured for 24 h. Concentrated suspension of HXB2 pseudoviruses (4×104 cfu/well, MOI=1) bearing BlaM-Vpr was dispensed into wells, and the plates were centrifuged at 2,095 g, 4°C for 30 min to facilitate virus binding. Next, 0.1 μL/well of 3 mM library compound dissolved in dimethyl sulfoxide (DMSO) was added to cells using the Beckman NX liquid handler with integrated Pintool. The final compound concentration in wells was 10 μM. Vehicle control wells in each plate received an equal volume of DMSO (final concentration 0.3% v/v) or 1 μM of the inhibitory gp41 HR2-derived peptide, C52L (positive control).46 Plates were incubated at 37°C, 5% CO2 for 90 min, after which time, the medium was removed using a BioTek plate washer, and the CCF4-AM substrate (25 μL/well) was dispensed into the wells. The resulting BlaM signal was measured after an overnight incubation at 12°C. Validation of selected hits was done manually, using TZM-bl cells (1×105/well) cultured in 96-well black clear-bottom plates (Corning).
Miniaturization of the Virus–Cell Fusion Assay to 384-Half-Well Plates
We further miniaturized the BlaM assay to reduce the consumption of the expensive CCF4-AM substrate and thereby minimize the cost of large-scale screening. Three thousand cells in 10 μL of culture medium were seeded into 384-half-well tissue culture plates (#3542; Corning), using the Multidrop Combi dispenser. After overnight incubation, 10 μL of virus was dispensed into the wells. The BlaM assay was performed, as described above, except that only 5 μL/well of the BlaM substrate was used. After incubating at 12°C for overnight, the fusion signal was measured using the EnVision Multilabel plate reader (PerkinElmer, Waltham, MA) with excitation at 400 nm and emissions at 460 and 520 nm with a dual mirror D425/D490 (in a bottom-reading mode). The dose–response curve was obtained, and IC50 was calculated using GraphPad Prism software.
Cell Viability Assay
After reading the BlaM signal and removing the CCF4-AM substrate, the MTS substrate (CellTiter Aqueous One; Promega) was added to wells. Plates were incubated for 90 min at 37°C, 5% CO2, and cell viability was measured by absorbance at 490 nm (Synergy HT plate reader). The MTS assay is highly reproducible and has excellent S/B ratio and Z′ values (data not shown). The effect of the compound on cell viability was normalized to vehicle (DMSO) control and expressed as % of Control. Overnight incubation of cells at 12°C did not reduce their viability or dampen their responses to cytotoxic compounds (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/adt).
Flow Cytometry
TZM-bl cells (1×106 cells) were incubated in the presence or absence of NF279 in complete media for 90 min at 37°C and detached with a nonenzymatic solution (Cellstripper™ solution; Mediatech). Harvested cells were washed thrice with cold phosphate-buffered saline (PBS) and incubated for 2 h at 4°C with antibodies diluted in PBS supplemented with 2% FBS. Cells were washed thrice with PBS before analysis. Cell surface CD4 and CXCR4 were detected using a 1/100 dilution of the mouse anti-hCD4 APC-conjugated SK3 clone antibody and the anti-CXCR4 APC-conjugated 12G5 antibody (both from eBioscience, San Diego, CA), respectively. CCR5 was detected using a mouse anti-CCR5 (BD Pharmingen, San Jose, CA) diluted to 1/200 and a sheep anti-mouse FITC-labeled secondary antibody (1/1,000 dilution; Sigma-Aldrich). Cells were analyzed using a BD LSR II FACS analyzer (BD Biosciences, San Jose, CA).
Cell–Cell Fusion Assay
Fusion between the indicator TZM-bl cells and HeLa-ADA cells expressing HIV-1 ADA Env and Tat was measured, as described in Miyauchi et al.42 Briefly, 0.5×105 TZM-bl cells were dispensed into each well of a 96-well Stripwell culture plate (Corning) and cultured for 24 h. HeLa-ADA cells were detached from culture dishes using a Cellstripper solution, and 0.5×105 cells per well were overlaid onto TZM-bl cells in the presence or absence of fusion inhibitors. Cell fusion was allowed to proceed for 60 min at 37°C, 5% CO2 and stopped by adding 5 μM of C52L. Cells were further incubated in the presence of C52L for 24 h at 37°C, 5% CO2. The extent of fusion was evaluated based upon the Tat-driven luciferase expression in TZM-bl cells, using Bright-Glo system (Promega).
Data Analysis
Screening results were analyzed using Cambridge Bioassay software (Cambridge, MA). Z′ and S/B were calculated for each plate to evaluate the performance and reproducibility of the assay. The effect of a compound on HIV-1 fusion was expressed as % Fusion=100×(Fcompound−Fbackground)/(Fcontrol−Fbackground)], where Fcontrol is the average fusion signal from wells containing virus and DMSO, Fcompound is the signal from wells with a compound, and Fbackground is the average signal from wells without virus.
Results
Direct Measurements of HIV-1 Fusion with Target Cells by the BlaM Assay
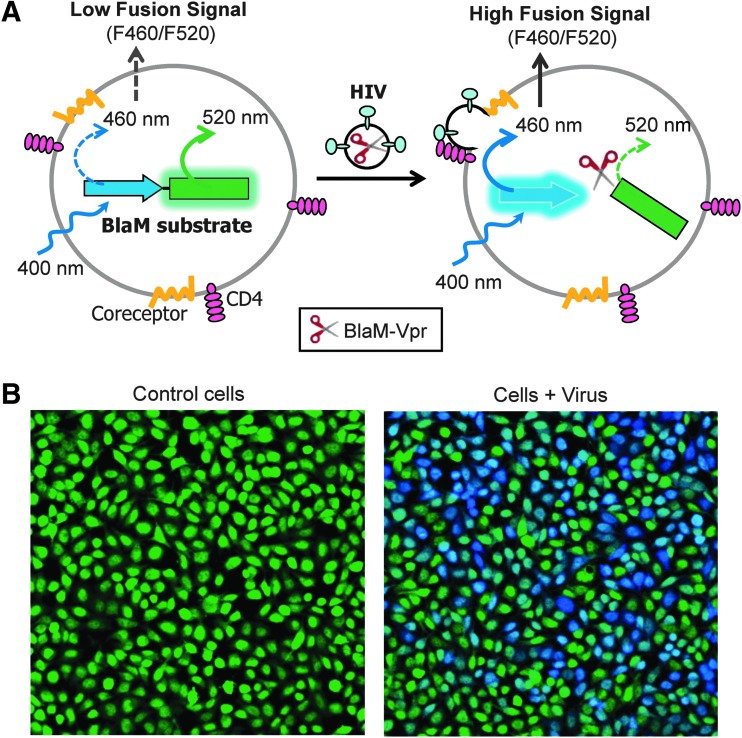
The BlaM assay is widely used to assess the extent and the kinetics of HIV-1 fusion with target cells.42,51,52 The principle of this assay is illustrated in Figure 1. HIV-1 pseudoviruses containing β-lactamase fused to the N-terminus of the HIV-1 Vpr (BlaM-Vpr) are produced by coexpressing the BlaM-Vpr and HIV-1 backbone in producer cells, as described in Miyauchi et al.42 HIV-1 Vpr is required to direct BlaM-Vpr incorporation into the viral core.53 The release of BlaM-Vpr into the cytoplasm as a result of virus–cell fusion (Fig. 1) is detected and quantified based on the extent of cleavage of a fluorescent BlaM substrate loaded into cells. The BlaM substrate consists of the fluorescence donor (coumarin, blue emission) and acceptor (fluorescein, green emission) moieties joined by the cephalosporin linker. Due to Förster resonance energy transfer (FRET) between the donor and acceptor, uncleaved substrate excited at 400 nm (to excite coumarin) emits primarily in green (528 nm). Cleavage of the cephalosporin linker by BlaM-Vpr released from the viral cores results in the loss of FRET and a blue shift (460 nm) in the emission spectrum (Fig. 1). The extent of fusion is thus readily measured as the ratio of fluorescence intensities at 460 nm over 528 nm, using a microplate reader or a flow cytometer.
Fig. 1.
Principle of the β-lactamase assay for quantifying virus–cell fusion. (A) A diagram of the Förster resonance energy transfer (FRET)-based BlaM assay. (B) Images of cells loaded with the CCF4-AM BlaM substrate (left panel) and loaded cells after inoculation with HXB2 pseudoviruses bearing the BlaM-Vpr chimera (right).
Implementation of the BlaM Assay in a 384-Well HTS Format
We have previously measured HIV–cell fusion by the BlaM assay in 96-well plates.41,42 Briefly, pseudoviruses containing the BlaM-Vpr chimera were prebound to target cells in the cold to prevent endocytosis and fusion, and virus entry/fusion was synchronously initiated by shifting to 37°C. Following the virus–cell incubation at 37°C, cells were loaded with a fluorescent BlaM substrate and incubated overnight at reduced temperature to allow for the substrate cleavage. Substrate cleavage by BlaM-Vpr is manifested in a blue shift in cell fluorescence.
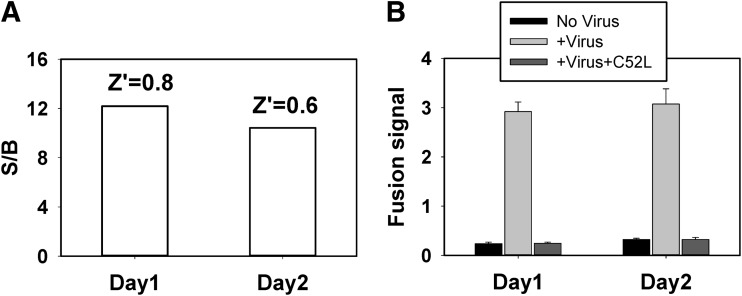
To adapt the BlaM assay for HTS, we modified it to a 384-well format (Table 1). After extensive optimization against a set of parameters such as cell number, the order of reagents addition, incubation time, and reader settings, we have simplified the assay to fewer steps for later automation adaptation. We evaluated the assay's performance parameters, Z′ and S/B ratio, to determine its compatibility with HTS. In general, a Z′ value of 0.5 or above (maximum=1) signifies an assay suitable for HTS,54 which requires a large dynamic signal range and low signal variability. As shown in Figure 2A, the BlaM assay performed in a 384-well format yielded an S/B ratio greater than 10 and Z′ above 0.6. The assay also exhibited minimal day-to-day variations. To determine the sensitivity and specificity of the virus–cell fusion assay in a 384-well format, we tested the effect of a known peptide inhibitor, C52L, derived from the HIV-1 gp41 glycoprotein.46 As shown in Figure 2B, the fusion signal was potently inhibited by C52L to the levels similar to the fusion signal from wells that did not receive the virus. These results demonstrate that the 384-well-based BlaM assay is compatible with screening for HIV-1 fusion inhibitors.
Fig. 2.
Assay development for a 384-well high-throughput screening (HTS) format. Fusion of HXB2.BlaM-Vpr pseudoviruses with TZM-bl cells cultured in 384-well plates. (A) The BlaM assay yields robust values for signal-to-background (S/B) and Z′ with minimal day-to-day variations. (B) The specificity of the BlaM signal was tested using a known inhibitor of HIV-1 fusion, C52L, added at 1 μM. The data are mean with standard deviation (SD) of four replicates.
Assay Validation for Small-Molecule Fusion Inhibitors Through Pilot Screening of LOPAC Library
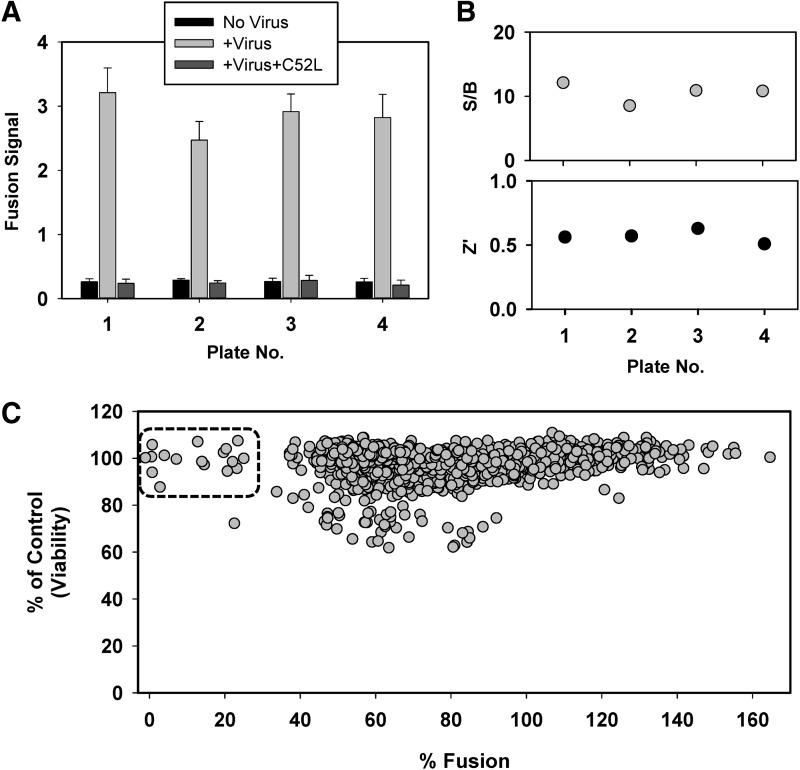
The virus–cell fusion assay was validated for use in a 384-well HTS format by a pilot screen of LOPAC library, which contains 1,280 pharmacologically active compounds frequently used for HTS assay validation.55 As shown in Figure 2, high fusion signals from LOPAC screening plates were observed for control virus-containing wells, with a minimal background signal in the absence of virus (Fig. 3A). The inhibitory effect of the C52L peptide on HIV-1 fusion was consistent across plates and similar with that observed in the assay validation experiment (Fig. 2). The S/B ratios were above 8 and Z′ above 0.5 across the four screening plates (Fig. 3B). These data further support the utility of our 384-well fusion assay for HTS for HIV-1 entry inhibitors.
Fig. 3.
Pilot screening to validate the BlaM assay for HTS. The BlaM virus–cell fusion assay in a 384-well format was validated for HTS, using the library of pharmacologically active compounds (LOPAC). (A) The fusion signal and the inhibitory effect of C52L on the fusion signal across four screening plates. The data shown are average with SD from 16 replicates. (B) S/B ratio (upper panel) and Z′ for four testing plates. (C) Scatter plot of pilot screening of 1,280 compounds using % Fusion (X-axis) against the % of Control (viability, Y-axis). The potential positives (rectangular region) are compounds with % Fusion <30 and % of Control (viability) >80.
To eliminate HTS false positives due to a cytotoxic effect, we coupled the primary BlaM assay with a standard MTS cell viability assay, similar to that described in Ref.41 The pairing of BlaM and MTS assays allows early identification and elimination of cytotoxic compounds in the primary screen format. The obtained screening results were plotted as the effect of compounds on HIV-1 fusion (expressed as % Fusion, Fig. 3C [X-axis]) against the effect on cell viability (% of Control [Y-axis]). Our screen identified 17 compounds as positive hits exhibiting more than 70% reduction of the HIV-1 fusion signal without significantly effecting cell viability (% of Control>80). The corresponding hit rate is 1.3%, which is within the expected range.
Positive Hits from the Pilot Screen
Seventeen compounds identified as positive hits with % Fusion <30% and cell viability >80% are shown in Figure 3 and Table 2. Two of these hits, cefsulodin and cephapirin, were false positive, since these compounds inhibit the enzymatic activity of β-lactamase.56,57 These two hits thus exemplify the compounds that interfere with the primary assay. Another hit, phorbol 12-myristate 13-acetate (PMA), likely inhibited HIV-1 fusion due to its ability to downregulate CD4 and CXCR4 expression.58,59 Interestingly, our screen identified a cluster of purinergic receptor inhibitors, suramin, NF023, NF449, PPNDS, and reactive blue 2 (Table 2). Suramin is a nonselective P2 purinergic antagonist60 and NF023 is a suramin analog that specifically antagonizes the P2X1 receptor activity.61 NF449 and PPNDS are also potent and selective P2X1 receptor inhibitors,62,63 whereas reactive blue 2 antagonizes the P2Y receptor activity.64 Other positive hits included cellular signaling and mitochondrial electron transport inhibitors, as well as antagonists of glutamate receptors (Table 2). As shown in Figure 3C, a number of components enhanced HIV-1 fusion with cells, perhaps by targeting cellular factors that restrict virus entry.
Table 2.
Positive Hits from Library of Pharmacologically Active Compounds Screen
| Target query of the PubChem Databaseb | ||||||||
|---|---|---|---|---|---|---|---|---|
| BlaM (% fusion) | MTS (% of control) | Hit No. | PubChem SID | Compound name | Targeta | No. active flags | No. total bioassays | % Activec |
| −1.1 | 100.2 | 1 | 17404637 | N-arachidonoyl glycine | FAAH | 40 | 581 | 6.9 |
| 0.5 | 100.6 | 2 | 17405836 | Aurintricarboxylic acid | TopoII | 133 | 501 | 26.5 |
| 0.7 | 105.7 | 3 | 17405738 | Suramin hexasodium | P2X, P2Y | 48 | 253 | 19.0 |
| 0.7 | 94 | 4 | 17405427 | NF449 octasodium salt | Gs-alpha | 33 | 180 | 18.3 |
| 2.8 | 87.7 | 5 | 17405495 | PPNDS tetrasodium | P2X1 | 26 | 169 | 15.4 |
| 3.9 | 101.2 | 6 | 17405842 | Reactive Blue 2 | P2Y | 33 | 111 | 29.7 |
| 7.2 | 99.6 | 7 | 17404937 | (2S,1′S,2′S)-2-(carboxycyclopropyl)glycine | mGluR2 | 54 | 301 | 17.9 |
| 12.8 | 107 | 8 | 17405664 | Sanguinarine chloride | Na+/K+ ATPase | 36 | 131 | 27.5 |
| 13.9 | 98.6 | 9 | 17405423 | Methoctramine tetrahydrochloride | M2 | 24 | 54 | 44.4 |
| 15.5 | 97.3 | 10 | 17405371 | NF023 | P2X1 | 22 | 176 | 12.5 |
| 19.7 | 102.5 | 11 | 17405743 | Theobromine | A1>A2 | 0 | 240 | 0.0 |
| 20.3 | 104 | 12 | 17405590 | Phorbol 12-myristate 13-acetate | PKC | 12 | 152 | 7.9 |
| 23.5 | 107.4 | 13 | 17405668 | Rotenone | Mitochondria | 209 | 2035 | 10.3 |
| 20.7 | 94.5 | 14 | 17404966 | Dephostatin | CD45 Tyrosine Kinase | 52 | 278 | 18.7 |
| 22.1 | 98.5 | 15 | 17405531 | Protoporphyrin IX disodium | Guanylyl cyclase | 137 | 822 | 16.7 |
| 23.2 | 95.7 | 16 | 17404822 | Cefsulodin sodium salt hydrate | Cell wall synthesis | 11 | 106 | 10.4 |
| 25.0 | 99.9 | 17 | 17404842 | Cephapirin sodium | Cell wall synthesis | 19 | 168 | 11.3 |
Target reported in LOPAC supporting information.
Based on the PubChem data from Emory, NCGC, and ChEMBL.
Active was calculated from (No. of active flags/No. of total bioassays)×100.
LOPAC, library of pharmacologically active compounds.
We have performed target query of PubChem database for promiscuous compounds for all 17 hits (Table 2). Compounds with a higher ratio of % active flags across the bioassays might be frequently revealed promiscuous compounds. For example, the percentage of active for aurintricarboxylic acid (hit No. 2) is 26.5%. This compound is actually a confirmed hit in the screening of inhibitors for AmpC β-lactamase (PubChem:AID 485341) and therefore might be a false positive in our assay.
Assay Miniaturization for 384-Half-Well Plates
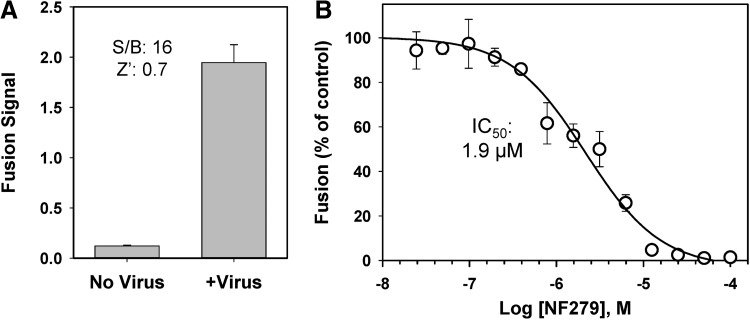
To reduce the cost of consumables for the future large-scale HTS campaign, we further optimized and miniaturized the BlaM assay for 384-half-well plates. The minimal volume for a 384-half-well is 5 μL, which is five times smaller than the minimal volume for a regular 384-well. After extensive optimization, we have obtained a robust signal for HTS in 384-half-well plates, as demonstrated by Z′ of 0.7 and an S/B ratio of 16 (Fig. 4A). The greater Z′ value compared to the standard 384-well format (Fig. 3B) could be due to the use of gas-permeable plate sealers to prevent medium evaporation in the miniaturized assay. To validate the miniaturized assay for HTS, we evaluated the effect of the highly selective P2X1 receptor inhibitor, NF279,65 on HIV-1 fusion. NF279 was chosen over NF023 and NF449 identified by our screen because of its greater potency (see section “Inhibitors of P2X1 Receptor Interfere with HIV-1 Fusion” below). NF279 inhibited the BlaM signal resulting from the fusion activity of the dual-tropic R3A Env,66 which can use either CXCR4 or CCR5 as a coreceptor (Fig. 4B). Inhibition occurred in a dose-dependent manner with IC50 at 1.9 μM. The above results validate the miniaturized BlaM assay format for screening inhibitors of HIV-1 entry/fusion.
Fig. 4.
Miniaturization of the BlaM assay for HTS. The BlaM assay was miniaturized and optimized for 384-half-well plates to achieve a fourfold reduction in the amount of CCF4-AM substrate required for the HTS campaign. (A) Performance of the miniaturized BlaM assay in 384-half-well plate format, using R3A Env-pseudotyped viruses. Robust fusion signal with S/B ratio of 15 and Z′ of 0.7 was obtained, indicating a high-quality assay for HTS. (B) Validation of the miniaturized BlaM assay for R3A Env-pseudotyped viruses, using an active compound, NF279, similar to those identified by a pilot screen. The effect of NF279 is expressed as % of control and the data shown are average and SD from triplicate samples. The IC50 was determined using GraphPad Prism.
Inhibitors of P2X1 Receptor Interfere with HIV-1 Fusion
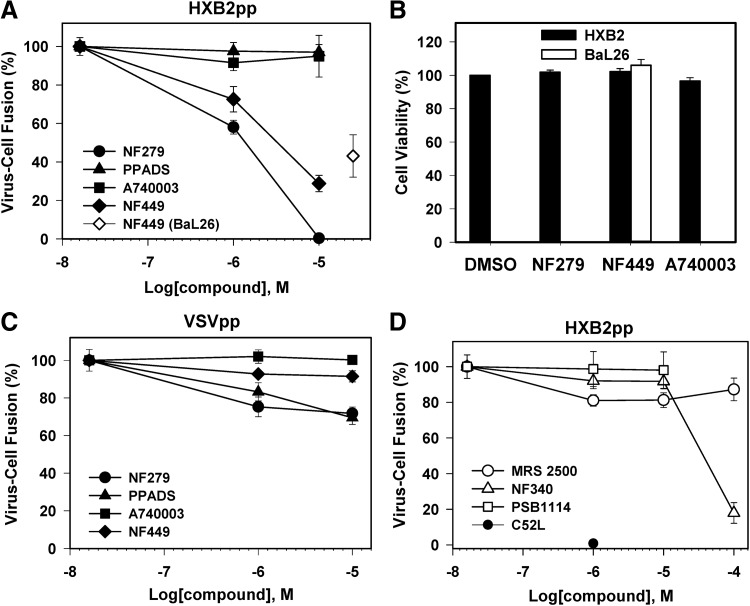
Because several purinergic receptor inhibitors were among the hits (Table 2), we further investigated the role of these compounds in HIV-1 fusion. Purinergic receptors have been previously reported to play a role in HIV-1 fusion and infection in macrophages and CD4+ T cells.67,68 The authors proposed that purinergic receptors are involved in a signaling cascade that leads to elevation of the cytosolic calcium, which in turn promotes HIV-1 fusion through an unknown mechanism. To determine the types of purinergic receptors required for HIV-1 fusion with target cells, we used a panel of specific inhibitors of P2X and P2Y receptors. When fusion experiments were performed in the presence of varied concentrations of these inhibitors, only P2X1 receptor antagonists, NF449 identified by the pilot screen and an additional compound NF279, diminished HIV-1 fusion (Fig. 5A). NF279 was more potent than NF449 (IC50 1.2 and 3.3 μM, respectively). Neither of these compounds affected cell viability within a concentration range tested (Fig. 5B). By contrast, the P2X7 antagonist A740003,69 a nonselective P2 inhibitor PPADS,70 and a panel of P2Y antagonists targeting P2Y1 (MRS250071), P2Y2 (PSB111472), and P2Y11 (NF34073) were without effect (Fig. 5D). Viral fusion was inhibited only by a very high concentration (100 μM) of NF340. Since this effect was not due to reduced cell viability (data not shown), it is possible that the P2Y11 receptor can also play a role in HIV-1 fusion.
Fig. 5.
The effects of P2 receptor inhibitors on HIV–cell fusion. Pseudoviruses encoding BlaM-Vpr (multiplicity of infection [MOI]=1) were bound to TZM-bl cells at 4°C. Viral fusion was initiated by shifting to 37°C for 90 min and measured by the BlaM assay in a 96-well format. (A) Fusion of HIV-1 pseudoviruses HXB2pp (CXCR4-tropic, filled symbols) or Bal26pp (CCR5-tropic, open diamond) with TZM-bl cells in the presence or absence of P2X receptor inhibitors: a nonselective P2 receptor inhibitor PPADS (▲), P2X1 inhibitors NF279 (●) and NF449 (♦ and ◊), and P2X7 inhibitor A740003 (■). (B) Effects of P2 receptor inhibitors on TZM-bl cell viability measured by the MTS assay. Cells infected with HXB2pp and BaL26pp were incubated with 10 and 25 μM of indicated inhibitors, respectively. (C) P2 receptor inhibitors do not considerably affect VSV-G pseudovirus (VSVpp) fusion (the symbol legend is the same as in A). (D) Effects of P2Y inhibitors MRS2500 (◯), PSB1114 (□), and NF340 (Δ) on HXB2pp fusion with TZM-bl cells. The extent of fusion in the presence of 1 μM C52L (●) is shown for comparison. Data points are mean and SEM of combined triplicate measurements from three independent experiments.
NF449 also inhibited fusion mediated by the CCR5-tropic HIV-1 BaL26 Env, although less efficiently than fusion of HXB2 particles (Fig. 5A). As shown above (Fig. 4A), this compound also interfered with fusion mediated by the dual-tropic R3A Env. To check for selectivity of the observed inhibition of HIV-1 fusion by purinergic receptor antagonists, we assessed their effects on low pH-dependent fusion mediated by the VSV G glycoprotein.74 This glycoprotein directs viral entry and fusion through an endosomal pathway. Fusion of pseudoviruses bearing VSV G (referred to as VSVpp) was largely unaffected by purinergic inhibitors, except for a modest (ca. 25%) reduction in the extent of fusion caused by NF279 and PPADS (Fig. 5C). In control experiments performed in the presence of NH4Cl, which raises endosomal pH, VSVpp fusion was nearly completely blocked (data not shown). These results demonstrate that the inhibitory effect of P2X1 antagonists is specific to HIV-1 Env-mediated fusion.
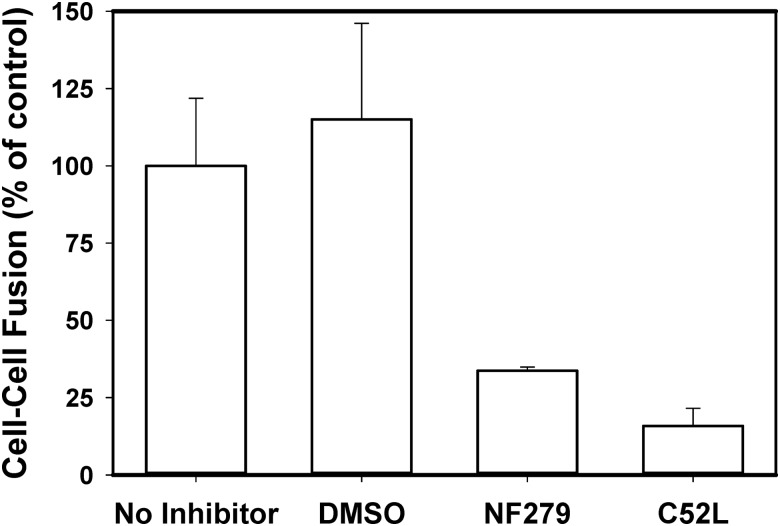
The inhibition of HIV-1 fusion by NF279 was not due to the diminished CD4 or coreceptor levels on the cell surface, as revealed by the flow cytometry experiments (Table 3). We also tested the possibility that this compound prevented viral fusion by blocking HIV-1 endocytosis. We and others have shown that inhibition of HIV-1 uptake by cells inhibits viral fusion and infection.42,52 Our functional studies imply that HIV-1 enters permissive cells through endocytosis and fusion with intracellular compartments.41,42 We indirectly tested the notion that NF279 can interfere with HIV-1 endocytosis and not with the downstream fusion step by measuring the effect of this drug on fusion between HIV-1 Env-expressing and target cells. Similar to virus–cell fusion, 10 μM NF279 effectively inhibited HIV-1 Env-mediated cell–cell fusion (Fig. 6A) to an extent similar to that observed in virus–cell fusion experiments (Fig. 5A). This result implies that NF279 targets the viral fusion step and not HIV-1 endocytosis.
Table 3.
Expression of CD4 and Coreceptors on TZM-bl Cells Measured by Flow Cytometry
| MFI (untreated) | MFI (50 μM NF279a) | Fold-change | |
|---|---|---|---|
| CD4 | 63.6 | 59.9 | 0.94 |
| CXCR4 | 66.0 | 67.7 | 1.03 |
| CCR5 | 192.2 | 180.2 | 0.94 |
This high concentration of NF279, which exceeds its fully inhibitory concentration for HXB2pp fusion by ∼5-fold, was selected to better illustrate the lack of effect on CD4 and coreceptor expression.
MFI, mean fluorescence intensity.
Fig. 6.
P2X1 receptor inhibitor blocks cell–cell fusion mediated by HIV-1 Env glycoprotein. HeLa cells constitutively expressing HIV-1 ADA Env and Tat were overlaid onto the indicator TZM-bl cells in the presence or absence of inhibitors, and cell–cell fusion was triggered by shifting to 37°C. The extent of cell–cell fusion was measured by the Tat-dependent luciferase expression in TZM-bl cells. Both NF279 (10 μM) and the fusion inhibitor C52L (1 μM) blocked HIV-1 Env-mediated fusion. The data are mean and SD from one experiment performed in triplicate.
Discussion
In this study, we adapted and optimized for HTS an enzymatic assay that directly measures the HIV-1 pseudovirus fusion with target cells. This assay performed robustly in regular 384-well and 384-half-well formats and was validated by screening a small library of bioactive compounds. The primary screen for HIV-1 fusion inhibitors combines the virus–cell fusion assay with a cell viability assay, thus helping to eliminate false-positive hits due to cell toxicity. The relatively low fraction of cytotoxic compounds in the LOPAC library indicates that cell viability assessment may not be necessary for the large-scale HTS campaign. Several hits that strongly inhibited HIV-1 fusion without affecting cell viability were identified. The results of the pilot screen strongly support the feasibility of a large-scale HTS campaign.
Unlike the cell–cell fusion-based HTS assay for screening for HIV-1 fusion inhibitors,37 our assay may identify endosomal trafficking factors involved in HIV-1 entry.41,42,75 This could be important, since several lines of evidence imply that productive HIV-1 entry into cells occurs through an endocytic route.41,42,52,76–79 Furthermore, cell fusion-based assays are generally not sensitive to inhibitors that inactivate diverse enveloped viruses through disrupting their membrane, while having no adverse effect on cell membranes.39,40
Our observation that P2X1 receptor antagonists inhibit HIV-1 fusion is in agreement with the literature. Published studies implicate the P2X1 receptor in HIV-1 entry/fusion and in cell–cell transmission.67,68,80 However, the mechanism by which these channels modulate HIV-1 fusion remains unclear. Purinergic receptors are widely distributed in the central nervous system and in many peripheral tissues and are classified into two main classes, P1 receptors that bind adenosine, and P2 receptors that are responsive to phosphorylated nucleosides, such as ATP, ADP, and related nucleotides.81–83 P2 receptors are further classified into two subfamilies, ionotropic P2X and metabotropic P2Y receptors. Metabotropic receptors are coupled to the intracellular signaling pathways through heterotrimeric G proteins, whereas ionotropic P2X receptors are ion channels that open upon ATP binding and allow Ca2+ influx. Seven members of the P2X family receptors (P2X1–7) and eight P2Y receptors (P2Y1, P2Y2, P2Y4, P2Y6, P2Y11, P2Y12, P2Y13, and P2Y14) have been identified. Upon activation, these receptors modulate diverse cellular functions, such as the plasma membrane permeabilization, elevation of intracellular calcium, cell death, as well as innate and/or adaptive immune responses against pathogens.82,83
Our results show that P2X1 receptor antagonists, NF279 and NF449, prevent HIV–cell and Env-mediated cell–cell fusion through a mechanism that does not involve downregulation of CD4 or coreceptors. This apparent requirement for P2X1 receptors is specific to HIV-1, since P2X1 antagonists do not interfere with VSV fusion. Our findings are thus consistent with the conclusion that purinergic antagonists inhibited HIV-1 fusion at a step downstream of CD4 engagement and virological synapse formation.80 The apparent reliance of the HIV-1 fusion process on P2X1 function indicates that these receptors can be targeted for antiviral therapy. Future validation of hits other than P2 receptor antagonists may reveal additional cellular targets that are essential for HIV-1 entry and fusion.
Supplementary Material
Abbreviations Used
- BlaM
β-lactamase
- C52L
HIV-1 gp41-derived recombinant peptide
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- FRET
Förster resonance energy transfer
- 6HB
six-helix bundle structure
- HR1 and HR2
HIV-1 gp41 heptad repeat 1 and 2 domains
- HTS
high-throughput screening
- LOPAC
library of pharmacologically active compounds
- MOI
multiplicity of infection
- PBS
phosphate-buffered saline
- S/B
signal/background
- SD
standard deviation
- TZM-bl
HIV-1 indicator cell line
Acknowledgments
We thank Dr. Min Lu for C52L peptide, Dr. P. Clapham for BaL26 Env, Dr. J. Hoxie for R3A Env, and Dr. J. Binley for HXB2 Env expression vectors. The use of the Emory+Children's Pediatric Research Flow Cytometry Core is gratefully acknowledged. This work was supported by the NIH R01 GM108480 grant to Y.D. and G.B.M., by the NIH R01 GM054787 grant to G.B.M., and by the Winship Cancer Institute to Y.D. (NIH 5P30CA138292).
Disclosure Statement
No competing financial interests exist.
References
- 1.Berger EA, Murphy PM, Farber JM: Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol 1999;17:657–700 [DOI] [PubMed] [Google Scholar]
- 2.Melikyan GB: Membrane fusion mediated by human immunodeficiency virus envelope glycoprotein. Curr Top Membr 2011;68:81–106 [DOI] [PubMed] [Google Scholar]
- 3.Doms RW: Chemokine receptors and HIV entry. AIDS 2001;15Suppl 1:S34–S35 [DOI] [PubMed] [Google Scholar]
- 4.Blumenthal R, Durell S, Viard M: HIV viral entry and envelope glycoprotein mediated fusion. J Biol Chem 2012;287:40841–40849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert DM, Kim PS: Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem 2001;70:777–810 [DOI] [PubMed] [Google Scholar]
- 6.Markosyan RM, Cohen FS, Melikyan GB: HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol Biol Cell 2003;14:926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melikyan GB: Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology 2008;5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin PF, Blair W, Wang T, et al. : A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci U S A 2003;100:11013–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Si Z, Madani N, Cox JM, et al. : Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A 2004;101:5036–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strizki JM, Xu S, Wagner NE, et al. : SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc Natl Acad Sci U S A 2001;98:12718–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baba M, Nishimura O, Kanzaki N, et al. : A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci U S A 1999;96:5698–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorr P, Westby M, Dobbs S, et al. : Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 2005;49:4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer M, Kapoor TM, Strassmaier T, et al. : Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat Struct Biol 1999;6:953–960 [DOI] [PubMed] [Google Scholar]
- 14.Liu S, Boyer-Chatenet L, Lu H, Jiang S: Rapid and automated fluorescence-linked immunosorbent assay for high-throughput screening of HIV-1 fusion inhibitors targeting gp41. J Biomol Screen 2003;8:685–693 [DOI] [PubMed] [Google Scholar]
- 15.Liu S, Jiang S, Wu Z, et al. : Identification of inhibitors of the HIV-1 gp41 six-helix bundle formation from extracts of Chinese medicinal herbs Prunella vulgaris and Rhizoma cibotte. Life Sci 2002;71:1779–1791 [DOI] [PubMed] [Google Scholar]
- 16.Jiang S, Lu H, Liu S, Zhao Q, He Y, Debnath AK: N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob Agents Chemother 2004;48:4349–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai L, Gochin M: A novel fluorescence intensity screening assay identifies new low-molecular-weight inhibitors of the gp41 coiled-coil domain of human immunodeficiency virus type 1. Antimicrob Agents Chemother 2007;51:2388–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC: Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc Natl Acad Sci U S A 2006;103:13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Hixon MS, Dawson PE, Janda KD: Development of a FRET assay for monitoring of HIV gp41 core disruption. J Org Chem 2007;72:6700–6707 [DOI] [PubMed] [Google Scholar]
- 20.Katritzky AR, Tala SR, Lu H, et al. : Design, synthesis, and structure-activity relationship of a novel series of 2-aryl 5-(4-oxo-3-phenethyl-2-thioxothiazolidinylidenemethyl)furans as HIV-1 entry inhibitors. J Med Chem 2009;52:7631–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Qi Z, Guo A, et al. : ADS-J1 inhibits human immunodeficiency virus type 1 entry by interacting with the gp41 pocket region and blocking fusion-active gp41 core formation. Antimicrob Agents Chemother 2009;53:4987–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray EJ, Leaman DP, Pawa N, et al. : A low-molecular-weight entry inhibitor of both CCR5- and CXCR4-tropic strains of human immunodeficiency virus type 1 targets a novel site on gp41. J Virol 2010;84:7288–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart KD, Huth JR, Ng TI, et al. : Non-peptide entry inhibitors of HIV-1 that target the gp41 coiled coil pocket. Bioorg Med Chem Lett 2010;20:612–617 [DOI] [PubMed] [Google Scholar]
- 24.Pang W, Wang RR, Gao YD, et al. : A novel enzyme-linked immunosorbent assay for screening HIV-1 fusion inhibitors targeting HIV-1 Gp41 core structure. J Biomol Screen 2011;16:221–229 [DOI] [PubMed] [Google Scholar]
- 25.Henrich TJ, Kuritzkes DR: HIV-1 entry inhibitors: recent development and clinical use. Curr Opin Virol 2013;3:51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brass AL, Dykxhoorn DM, Benita Y, et al. : Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008;319:921–926 [DOI] [PubMed] [Google Scholar]
- 27.Bushman FD, Malani N, Fernandes J, et al. : Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog 2009;5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konig R, Zhou Y, Elleder D, et al. : Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008;135:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H, Xu M, Huang Q, et al. : Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 2008;4:495–504 [DOI] [PubMed] [Google Scholar]
- 30.Khan MM, Simizu S, Lai NS, Kawatani M, Shimizu T, Osada H: Discovery of a small molecule PDI inhibitor that inhibits reduction of HIV-1 envelope glycoprotein gp120. ACS Chem Biol 2011;6:245–251 [DOI] [PubMed] [Google Scholar]
- 31.Freeman MM, Seaman MS, Rits-Volloch S, et al. : Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody. Structure 2010;18:1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stantchev TS, Markovic I, Telford WG, Clouse KA, Broder CC: The tyrosine kinase inhibitor genistein blocks HIV-1 infection in primary human macrophages. Virus Res 2007;123:178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmon B, Campbell N, Ratner L: Role of Abl kinase and the Wave2 signaling complex in HIV-1 entry at a post-hemifusion step. PLoS Pathog 2010;6:e1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Princen K, Schols D: HIV chemokine receptor inhibitors as novel anti-HIV drugs. Cytokine Growth Factor Rev 2005;16:659–677 [DOI] [PubMed] [Google Scholar]
- 35.Finnegan CM, Rawat SS, Puri A, Wang JM, Ruscetti FW, Blumenthal R: Ceramide, a target for antiretroviral therapy. Proc Natl Acad Sci U S A 2004;101:15452–15457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia JM, Gao A, He PL, et al. : High-throughput screening using pseudotyped lentiviral particles: a strategy for the identification of HIV-1 inhibitors in a cell-based assay. Antiviral Res 2009;81:239–247 [DOI] [PubMed] [Google Scholar]
- 37.Herschhorn A, Finzi A, Jones DM, et al. : An inducible cell-cell fusion system with integrated ability to measure the efficiency and specificity of HIV-1 entry inhibitors. PLoS One 2011;6:e26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herschhorn A, Gu C, Espy N, Richard J, Finzi A, Sodroski JG: A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat Chem Biol 2014;10:845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf MC, Freiberg AN, Zhang T, et al. : A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A 2010;107:3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St.Vincent MR, Colpitts CC, Ustinov AV, et al. : Rigid amphipathic fusion inhibitors, RAFIs, small molecule antiviral compounds against enveloped viruses. Proc Natl Acad Sci U S A 2010;107:17339–17344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de la Vega M, Marin M, Kondo N, et al. : Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 2011;8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB: HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 2009;137:433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavrois M, De Noronha C, Greene WC: A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol 2002;20:1151–1154 [DOI] [PubMed] [Google Scholar]
- 44.Wei X, Decker JM, Liu H, et al. : Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 2002;46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M: Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science 1997;276:1874–1878 [DOI] [PubMed] [Google Scholar]
- 46.Deng Y, Zheng Q, Ketas TJ, Moore JP, Lu M: Protein design of a bacterially expressed HIV-1 gp41 fusion inhibitor. Biochemistry 2007;46:4360–4369 [DOI] [PubMed] [Google Scholar]
- 47.Tobiume M, Lineberger JE, Lundquist CA, Miller MD, Aiken C: Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J Virol 2003;77:10645–10650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malim MH, Hauber J, Fenrick R, Cullen BR: Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 1988;335:181–183 [DOI] [PubMed] [Google Scholar]
- 49.Binley JM, Cayanan CS, Wiley C, Schulke N, Olson WC, Burton DR: Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J Virol 2003;77:5678–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimpton J, Emerman M: Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol 1992;66:2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavrois M, Neidleman J, Yonemoto W, Fenard D, Greene WC: HIV-1 virion fusion assay: uncoating not required and no effect of Nef on fusion. Virology 2004;328:36–44 [DOI] [PubMed] [Google Scholar]
- 52.Daecke J, Fackler OT, Dittmar MT, Krausslich HG: Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol 2005;79:1581–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selig L, Pages JC, Tanchou V, et al. : Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol 1999;73:592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang JH, Chung TD, Oldenburg KR: A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999;4:67–73 [DOI] [PubMed] [Google Scholar]
- 55.Li L, Du Y, Chen W, Fu H, Harrison DG: A novel high-throughput screening assay for discovery of molecules that increase cellular tetrahydrobiopterin. J Biomol Screen 2011;16:836–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones RN: A review of cephalosporin metabolism: a lesson to be learned for future chemotherapy. Diagn Microbiol Infect Dis 1989;12:25–31 [DOI] [PubMed] [Google Scholar]
- 57.Slack MP: Antipseudomonal beta-lactams. J Antimicrob Chemother 1981;8:165–170 [DOI] [PubMed] [Google Scholar]
- 58.Signoret N, Oldridge J, Pelchen-Matthews A, et al. : Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol 1997;139:651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelchen-Matthews A, Parsons IJ, Marsh M: Phorbol ester-induced downregulation of CD4 is a multistep process involving dissociation from p56lck, increased association with clathrin-coated pits, and altered endosomal sorting. J Exp Med 1993;178:1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Charlton SJ, Brown CA, Weisman GA, Turner JT, Erb L, Boarder MR: PPADS and suramin as antagonists at cloned P2Y- and P2U-purinoceptors. Br J Pharmacol 1996;118:704–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Rhee AM, van der Heijden MP, Beukers MW, AP IJ, Soudijn W, Nickel P: Novel competitive antagonists for P2 purinoceptors. Eur J Pharmacol 1994;268:1–7 [DOI] [PubMed] [Google Scholar]
- 62.Rettinger J, Braun K, Hochmann H, et al. : Profiling at recombinant homomeric and heteromeric rat P2X receptors identifies the suramin analogue NF449 as a highly potent P2X1 receptor antagonist. Neuropharmacology 2005;48:461–468 [DOI] [PubMed] [Google Scholar]
- 63.Lambrecht G, Rettinger J, Baumert HG, et al. : The novel pyridoxal-5′-phosphate derivative PPNDS potently antagonizes activation of P2X(1) receptors. Eur J Pharmacol 2000;387:R19–R21 [DOI] [PubMed] [Google Scholar]
- 64.Bean BP: Pharmacology and electrophysiology of ATP-activated ion channels. Trends Pharmacol Sci 1992;13:87–90 [DOI] [PubMed] [Google Scholar]
- 65.Damer S, Niebel B, Czeche S, et al. : NF279: a novel potent and selective antagonist of P2X receptor-mediated responses. Eur J Pharmacol 1998;350:R5–R6 [DOI] [PubMed] [Google Scholar]
- 66.Meissner EG, Duus KM, Gao F, Yu XF, Su L: Characterization of a thymus-tropic HIV-1 isolate from a rapid progressor: role of the envelope. Virology 2004;328:74–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hazleton JE, Berman JW, Eugenin EA: Purinergic receptors are required for HIV-1 infection of primary human macrophages. J Immunol 2012;188:4488–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seror C, Melki MT, Subra F, et al. : Extracellular ATP acts on P2Y2 purinergic receptors to facilitate HIV-1 infection. J Exp Med 2011;208:1823–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Honore P, Donnelly-Roberts D, Namovic MT, et al. : A-740003 [N-(1-{[(cyanoimino)(5-quinolinylamino) methyl]amino}-2,2-dimethylpropyl)-2-(3,4-dimethoxyphenyl)acetamide], a novel and selective P2X7 receptor antagonist, dose-dependently reduces neuropathic pain in the rat. J Pharmacol Exp Ther 2006;319:1376–1385 [DOI] [PubMed] [Google Scholar]
- 70.Lambrecht G, Friebe T, Grimm U, et al. : PPADS, a novel functionally selective antagonist of P2 purinoceptor-mediated responses. Eur J Pharmacol 1992;217:217–219 [DOI] [PubMed] [Google Scholar]
- 71.Kim HS, Ohno M, Xu B, et al. : 2-Substitution of adenine nucleotide analogues containing a bicyclo[3.1.0]hexane ring system locked in a northern conformation: enhanced potency as P2Y1 receptor antagonists. J Med Chem 2003;46:4974–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.El-Tayeb A, Qi A, Nicholas RA, Muller CE: Structural modifications of UMP, UDP, and UTP leading to subtype-selective agonists for P2Y2, P2Y4, and P2Y6 receptors. J Med Chem 2011;54:2878–2890 [DOI] [PubMed] [Google Scholar]
- 73.Meis S, Hamacher A, Hongwiset D, et al. : NF546 [4,4′-(carbonylbis(imino-3,1-phenylene-carbonylimino-3,1-(4-methyl-phenylene)-car bonylimino))-bis(1,3-xylene-alpha,alpha′-diphosphonic acid) tetrasodium salt] is a non-nucleotide P2Y11 agonist and stimulates release of interleukin-8 from human monocyte-derived dendritic cells. J Pharmacol Exp Ther 2010;332:238–247 [DOI] [PubMed] [Google Scholar]
- 74.Matlin KS, Reggio H, Helenius A, Simons K: Pathway of vesicular stomatitis virus entry leading to infection. J Mol Biol 1982;156:609–631 [DOI] [PubMed] [Google Scholar]
- 75.Demirkhanyan LH, Marin M, Padilla-Parra S, et al. : Multifaceted mechanisms of HIV-1 entry inhibition by human alpha-defensin. J Biol Chem 2012;287:28821–28838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carter GC, Bernstone L, Baskaran D, James W: HIV-1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology 2011;409:234–250 [DOI] [PubMed] [Google Scholar]
- 77.Kadiu I, Gendelman HE: Human immunodeficiency virus type 1 endocytic trafficking through macrophage bridging conduits facilitates spread of infection. J Neuroimmune Pharmacol 2011;6:658–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schaeffer E, Soros VB, Greene WC: Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J Virol 2004;78:1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.von Kleist L, Stahlschmidt W, Bulut H, et al. : Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 2011;146:471–484 [DOI] [PubMed] [Google Scholar]
- 80.Swartz TH, Esposito AM, Durham ND, Hartmann BM, Chen BK: P2X-selective purinergic antagonists are strong inhibitors of HIV-1 fusion during both cell-to-cell and cell-free infection. J Virol 2014;88:11504–11515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paoletti A, Raza SQ, Voisin L, et al. : Multifaceted roles of purinergic receptors in viral infection. Microbes Infect 2012;14:1278–1283 [DOI] [PubMed] [Google Scholar]
- 82.Miller CM, Zakrzewski AM, Ikin RJ, et al. : Dysregulation of the inflammatory response to the parasite, Toxoplasma gondii, in P2X7 receptor-deficient mice. Int J Parasitol 2011;41:301–308 [DOI] [PubMed] [Google Scholar]
- 83.Burnstock G, Kennedy C: P2X receptors in health and disease. Adv Pharmacol 2011;61:333–372 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.