Abstract
Zinc finger protein, FOG2 family member 2 (ZFPM2) (previously named FOG2) gene defects result in the highly morbid congenital diaphragmatic hernia (CDH) in humans and animal models. In a cohort of 275 CDH patient exomes, we estimated the prevalence of damaging ZFPM2 mutations to be almost 5%. Genetic analysis of a multigenerational family identified a heritable intragenic ZFPM2 deletion with an estimated penetrance of 37.5%, which has important implications for genetic counseling. Similarly, a low penetrance ZFPM2 frameshift mutation was observed in a second multiplex family. Isolated CDH was the predominant phenotype observed in our ZFPM2 mutation patients. Findings from the patients described herein indicate that ZFPM2 point mutations or deletions are a recurring cause of CDH.
Keywords: congenital diaphragmatic hernia, DNA copy number variation, exome, FOG2, penetrance, ZFPM2
Congenital diaphragmatic hernia (CDH) is a common birth defect (1 in 3000 live births) associated with significant morbidity and mortality. Most CDH patients have a diaphragmatic defect as an isolated, left posterolateral defect, which often includes lung hypoplasia. The remainder have a complex phenotype with additional malformations, at times as part of a recognizable syndrome (1). Most CDH cases are sporadic, with a single affected individual in the family. However, rare CDH kindreds have proven instructive for gene discovery (1–3).
Point mutations and deletions in ZFPM2 (zinc finger protein, FOG family member 2) have been reported in patients with diaphragmatic defects. ZFPM2 was identified as a candidate based on a mouse model and a patient with a de novo non-sense mutation, and it is expressed diffusely in the developing mouse diaphragm before and after muscularization and in the pulmonary mesenchyme during branching morphogenesis (4). Subsequently, two isolated CDH patients with missense variants in conserved residues were reported; because parental samples were not available it was not possible to assess whether they were inherited (5). Mutations in ZFPM2 have been identified in patients with tetralogy of fallot (TOF) or double outlet right ventricle (DORV), without diaphragmatic involvement (6).
More recently, ZFPM2 deletions were identified in two unrelated patients with isolated CDH; in both cases the deletion was inherited from an unaffected parent, suggesting reduced penetrance (7). However, the prevalence of ZFPM2 mutations and the degree of penetrance have never been systematically determined in CDH patients. We address the former by exome sequencing analysis in a cohort of sporadic unrelated CDH cases, while the latter is estimated by ZFPM2 findings in familial cases.
Materials and methods
Patient recruitment
Informed consent was obtained according to Partners Human Research Committee and Children’s Hospital Boston Clinical Investigation standards (Protocol 2000P000372 and 05-07-105R, respectively). All consented individuals underwent examination by a geneticist and/or review of medical records.
Sample collection and processing
Whole blood samples were collected for direct extraction (QIAamp DNA Blood Maxi kit, Qiagen, Valencia, CA) and Epstein-Barr virus (EBV) transformation (8). Primary fibroblast cultures were established from mechanically dissociated skin biopsies, plated in Dulbecco’s Modified Eagle’s medium (DMEM) with 10% fetal bovine serum (Gibco®|Life Technologies™, Grand Island, NY).
Whole exome sequencing
Whole exome sequencing was performed on 93 CDH patients at the Northwest Genomics Center (Seattle, WA) (9); and on 182 patients at Yale Center for Genomic Analysis (New Haven, CT), using the Illumina Genome Analyzer II (San Diego, CA). Reads were aligned using MAQ (sourceforge.net) and variant calling was performed by Genome Analysis Toolkit (GATK). ZFPM2 variants were reviewed by Ingenuity Variant Analysis™ (reference: NM_012082.3). 2-II-3 underwent clinical exome using Illumina HiSeq platform. The data were converted to FastQ by Illumina CASAVA 1.7 and mapped to BWA. Variant calling was performed using Atlas-SNP and Atlasindel (sourceforge.net).
Sanger sequencing
Primers were designed using PrimerBLAST (NCBI). Polymerase chain reaction (PCR) was performed using Qiagen Taq PCR Master Mix (Qiagen, Valencia, CA), sequenced by Taq DyeDeoxy Terminator cycle sequencing kit and resolved on the ABI 3730XL DNA analyzer (Applied Biosystems, Grand Island, NY).
Copy number variant (CNV) analysis
In Family 1, Affymetrix 6.0 chips and Agilent 1 M microarrays were hybridized as previously described (10) (Santa Clara, CA). Agilent 244 k arrays were performed on individual CDH12. Birdsuite (broadinstitute.org) and Agilent Feature Extraction (10.7.3.1), respectively, were used to generate CNV calls. Clinical microarrays were obtained on 2-II-1 [ClariSure CGH, Quest Diagnostics (Madison, NJ)], 2-II-2 and 2-II-3 [GenomeDx v5, GeneDx (Gaithersburg, MD)].
Expression of mutant ZFPM2 allele
Lymphoblastoid lines were treated with retinoic acid (1 × 10−7 M) and dibutyryl-cyclic AMP (1 mM) (Sigma, St Louis, MO) for 24 h before RNA extraction, and retrotranscribed with SuperScript® III Reverse Transcriptase (Invitrogen Life Technologies, Grand Island, NY). PCR was performed with GoTaq® Green Master Mix (Promega Corporation, Madison, WI).
Results
Sporadic CDH cohort
From 275 CDH patient exomes, we identified 14 potentially damaging heterozygous ZFPM2 sequence mutations in 13 unrelated CDH patients (5%) (Table 1; patients CDH1-CDH13). The majority of ZFPM2 mutations were missense and mapped to highly conserved nucleotides (phyloP p-value <10−4) (11) or known functional domains; p.E58X (CDH4) was a pre-mature stop codon, and p.N1062fs*23 (CDH13) was a frameshift expected to cause the loss of the fifth ZFPM2 zinc finger domain. ZFPM2 mutations were paternally or maternally inherited, except one, which arose de novo (Table 1). Because of lack of parental samples, inheritance could not be determined for every mutation.
Table 1.
Exonic ZFPM2 variants in 13 CDH study cohort patients. Reported associations, phenotypes associated to this mutation in the medical literature
| Sample ID |
hg19 location |
Mutation nomenclature
|
Prediction algorithms
|
dbSNP ID | Frequency (%) in unaffected individuals
|
phyloP (×10−4) |
Inheritance | Reported associations |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cDNA | Protein | SIfT | PolyPhen | 1000 genomes |
CG public genomes |
NHLBIESP | ||||||
| CDH1 | 106431420 | c.89A > G | p.E30G | Damaging | Possibly damaging | 121908601 | 0.26 | 0.92 | 0.45 | 0.23 | de novo | TOF |
| CDH2 | 106431420 | c.89A > G | p.E30G | Damaging | Possibly damaging | 121908601 | 0.26 | 0.92 | 0.45 | 0.23 | Unknown | TOF |
| CDH3 | 106431420 | c.89A > G | p.E30G | Damaging | Possibly damaging | 121908601 | 0.26 | 0.92 | 0.45 | 0.23 | Maternal | TOF |
| CDH4 | 106431503 | c.172G > T | p.E58* | NA | NA | – | 0 | 0 | 0 | 0.02 | Paternal | – |
| CDH5 | 106456600 | c.292G > A | p.D98N | Tolerated | Possibly damaging | 202217256 | 0.14 | 0 | 0.37 | 1.22 | Unknown | – |
| CDH6 | 106456600 | c.292G > A | p.D98N | Tolerated | Possibly damaging | 202217256 | 0.14 | 0 | 0.37 | 1.22 | Maternal | – |
| CDH7 | 106801092 | c.679A > G | p.I227V | Tolerated | Probably damaging | 202204708 | 0 | 0 | 0.08 | 0.13 | Paternal | DORV |
| CDH7 | 106813942 | c.1632G > A | p.M544I | Tolerated | Benign | 187043152 | 0.24 | 0 | 0.35 | 0.32 | Maternal | TOF |
| CDH8 | 106801092 | c.679A > G | p.I227V | Tolerated | Probably damaging | 202204708 | 0 | 0 | 0.08 | 0.13 | Paternal | DORV |
| CDH9 | 106811172 | c.960C > G | p.H320Q | Tolerated | Possibly damaging | – | 0 | 0 | 0 | 0.63 | Paternal | - |
| CDH10 | 106814811 | c.2501A > G | p.K834R | Damaging | Probably damaging | 113289249 | 0.05 | 0 | 0.25 | 0.66 | Unknown | - |
| CDH11 | 106814903 | c.2593A > G | p.K865E | Tolerated | Probably damaging | 367893066 | 0 | 0 | 0.02 | 2.6 | Unknown | - |
| CDH12 | 106815396 | c.3086A > T | p.K1029I | Tolerated | Probably damaging | 201729935 | 0 | 0 | 0.02 | 0.13 | Unknown | - |
| CDH13 | 106815496 | c.3186delC | p.N1062fs*23 | NA | NA | – | 0 | 0 | 0 | NA | Unknown | - |
CDH, congenital diaphragmatic hernia; NA, not applicable.
Patient CDH13 had a potential second hit with a p.R213C variant (phyloP p-value 7.228 × 10−7) in NR2F2, also implicated in CDH (1). The NR2F2 variant maps to the protein domain thought to bind ZFPM2 (UniProtKB, uniprot.org).
Every patient presented with isolated posterolateral CDH, except CDH12, who also had craniofacial abnormalities, TOF with an overriding aorta, a ventricular septal defect, and a narrow right ventricular outflow tract. It should be noted that CDH12 had also a 250 kb copy number gain of unknown significance in 8q24.21 (chr8:130,867,815-131,117,179; NCBI36/hg18), detected by Agilent 244 K aCGH and containing the FAM49B and ASAP1 genes. Parental samples were not available, precluding determination whether this CNV was de novo. Additional possibly pathogenic mutations or CNVs were not detected in these 13 patients.
Family 1
We characterized a multigenerational family of European ancestry, in which four relatives (1-II-6; 1-III-1; 1-III-2; 1-III-4) displayed isolated diaphragmatic defects, inherited in an autosomal dominant manner with incomplete penetrance (Fig. 1a). Clinical descriptions are available in the Supporting Information.
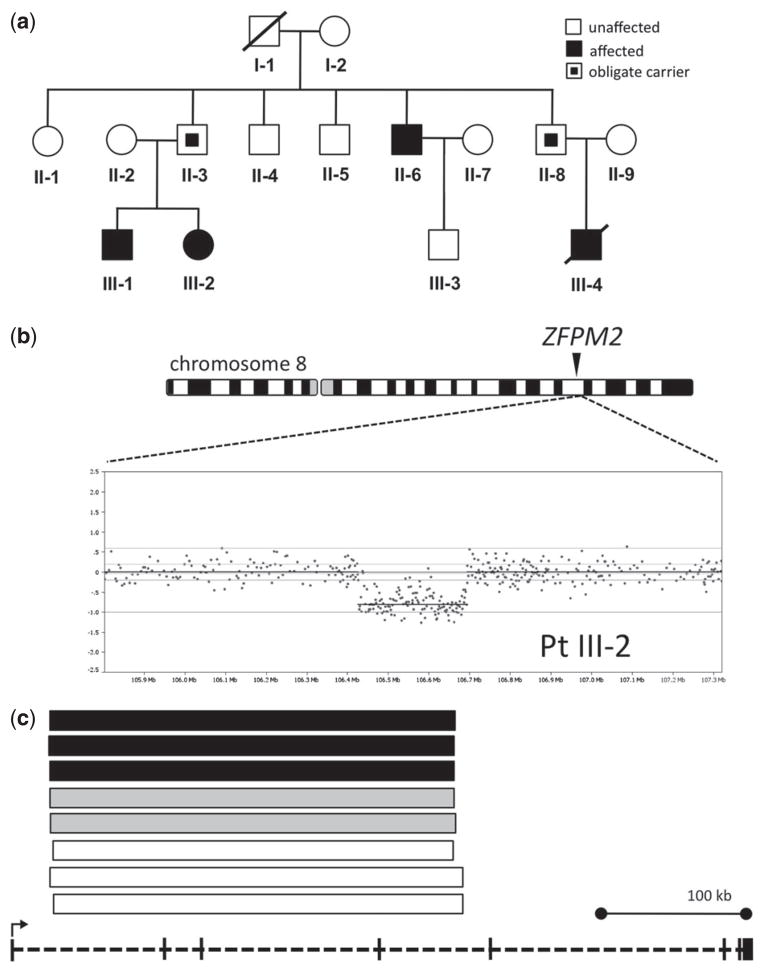
Fig. 1.
ZFPM2 deletion in Family 1. (A) Pedigree. Black inner square, unaffected carrier. (B) Chromosome 8 (schematic) and Agilent 1 M aCGH findings in III-2. (C) ZFPM2 deletion in other family members: affected (black), obligate carriers (gray), and unaffected individuals (white). Deletion intervals plotted against a diagram of the ZFPM2 gene (bottom). ZFPM2 deletions were not present in 7000 neuropsychiatric patients and 6000 controls.
A single copy number loss within the ZFPM2 gene was detected in all three individuals with CDH from whom a DNA sample was available (Fig. 1a–c). This intragenic deletion, spanning chr8:106,417,969-106,704,253 (NCBI36/hg18), was also present in both unaffected obligate carriers (1-II-3, 1-II-8), and as well as in three additional unaffected family members (Fig. 1c). The mutated transcript is predicted to be 3076 nucleotides long (380 bases shorter than the normal transcript) because of a frameshift introducing a pre-mature stop codon. In the theoretical resulting peptide, only 63 amino acids would be the same as normal ZFPM2.
To determine if the mutant allele was transcribed, immortalized lymphoblastoid cell lines from two affected individuals (1-III-2, and 1-II-6), one unaffected deletion carrier (1-II-3), and one family member without the deletion (1-II-4) were studied. In all samples, we detected only a 672 bp band, corresponding to the wild type allele, suggesting the predicted mutant transcript was unstable and/or underwent non-sense mediated decay (Fig. 2).
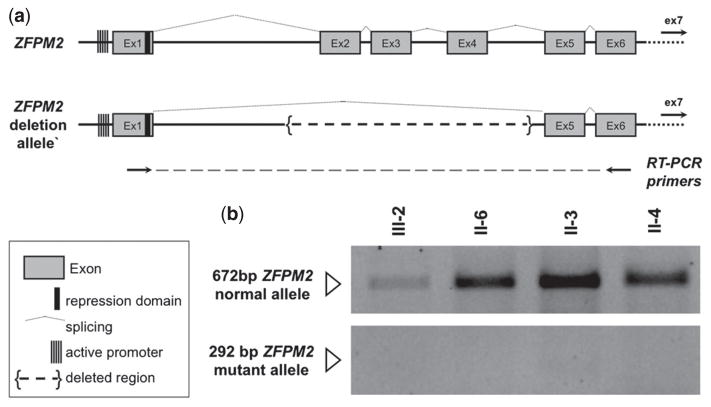
Fig. 2.
Expression of ZFPM2 mutant transcript. (a) Predicted splicing of normal and mutant ZFPM2 allele. Retro-transcription (RT) PCR primers were designed in exons 1 and 6 to amplify and discriminate full length and mutant transcripts. (b) RT-PCR on lymphoblastoid line cDNAs from III-2, II-6 (deletion carriers, affected), II-3 (deletion carrier, unaffected), and II-4 (non-carrier, unaffected). ZPMF2 promoter methylation was not different in affected or unaffected deletion carriers.
Family 2
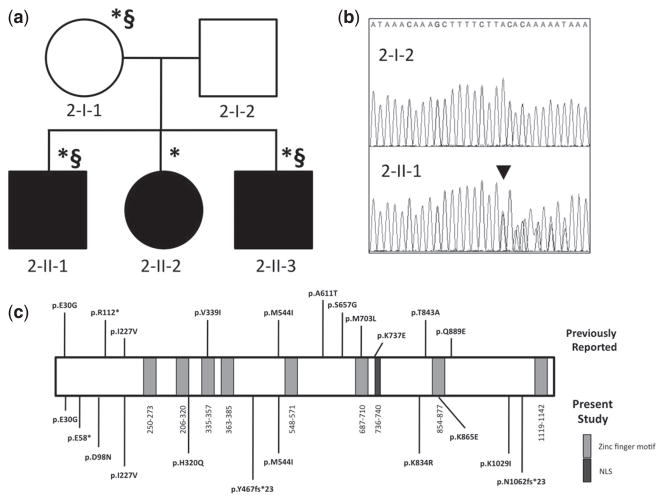
Phenotypically normal parents in a second multiplex family, also of European ancestry, had three children with left posterolateral CDH (2-II-1; 2-II-2; 2-II-3) (Fig. 3a). All siblings carried a p.Y467fs*23 mutation in ZFPM2, inherited from their mother (2-I-1), who did not have a clinical diagnosis of CDH (Fig. 3b). The p.Y467fs*23 mutation results in the truncation of over 50% of the protein, including the zinc finger domains (Fig. 3c). In addition to CDH, individual 2-II-1 had mild facial dysmorphism, hydrocephalus, and autistic behaviors, and individual 2-II-3 had reported developmental delays. Their mother (2-I-1) also had self-reported history of learning disabilities. These three individuals, but not 2-II-2, also harbor a chromosomal anomaly del(1)(q21.1q21.2)/dup(1)(q21.1), which appeared to segregate with the neurodevelopmental phenotype.
Fig. 3.
ZFPM2 mutation in Family 2. (a) Pedigree. *, p.Y467fs*23. §, 1q21 del/dup. (b) ZFPM2 reference and c.1396_1399dup (p.Y467fs*23, black arrowhead) in 2-II-1. (c) Familial mutation compared to sporadic cases (below) and previously reported ZFPM2 variants (above).
Discussion
In this study, we identified ZFPM2 mutations in a cohort of CDH patients, as well as investigated two families with inherited ZFPM2 mutations showing reduced penetrance. ZFPM2 mutations, present in ~5% of our CDH cohort, were considered pathogenic either on the basis of in silico algorithms or because they were reported to cause heart defects (6, 12, 13), a common CDH comorbidity (14). Seven mutations were familial, while one was de novo [p.E30G (CDH1)]. Interestingly, CDH7 carried two ZFPM2 missense variants, each inherited from a different parent, indicating compound heterozygosity. Although both had been previously associated with conotruncal defects (13), functional studies were never carried out to determine their pathogenicity, which raises the possibility that one is a benign variant. Even though p.E30G mutations have a frequency of up to 1% in individuals without a history of CDH (Table 1), this observation does not discount a role in CDH pathogenesis, as this and other studies have shown reduced penetrance or subclinical diaphragmatic defects in carriers of ZFPM2 mutations (7).
Mutations predicted to cause ZFPM2 because of pre mature stop codons leading to non-sense mediated decay constituted a major risk factor for CDH in two kindreds with apparent autosomal dominant inheritance and reduced penetrance. Because diaphragm morphology and function could not be studied in our phenotypically normal carriers, it is possible that these individuals could have a subclinical diaphragm abnormality. In Family 1, we estimate the penetrance for clinically relevant diaphragmatic defects to be around 37.5% of carriers.
Our case series indicates that patients with ZFPM2 mutations most often present with an isolated CDH phenotype. ZFPM2 mutations have been previously reported in patients with conotruncal heart defects, specifically TOF and DORV (6, 12, 13). Among the few reports to date, mutations associated with DORV were frequently de novo and occurred in exon 8 (13), but reduced penetrance was observed in at least two TOF patients (12). Interestingly, ZFPM2 mutation M703L has been associated with DORV (13) and CDH (5), but not in the same individual. Only patient CDH12 displayed a more complex phenotype. In addition to CDH and TOF with a ventricular septal defect, he displayed left anophthalmos and cleft palate, which sometimes co-occur with CDH (1). We cannot determine whether these anomalies are because of his ZFPM2 mutation or to the 8q24.21 copy number gain.
Several individuals from Family 2 were found to carry a chromosome 1q21 deletion/duplication in addition to the p.Y467fs*23 ZFPM2 mutation. 1q21 abnormalities have been associated with a variable phenotype, from developmental delay, mental retardation, learning disability, microcephaly and autism to no clinically relevant findings (15). Therefore, it is possible that the mild learning impairments in 2-I-1 and 2-II-3, and hydrocephalus and autism in 2-II-1 are because of this chromosomal rearrangement. Sibling 2-II-2, on the other hand, had only the p.Y467fs*23 ZFPM2 mutation and displayed isolated CDH with no apparent neurodevelopmental findings.
Despite reported monogenic forms, isolated CDH is believed to be mostly multifactorial/polygenic. There is precedence for non-Mendelian genetics in disorders such as cleft lip and palate and congenital heart defects, where the phenotype is affected by unknown risk factors, modifier genes, and environmental triggers, all of which must be kept in mind during genetic counseling (16). We propose that the ZFPM2 mutations described above, in light of their reduced penetrance, should be considered risk factors. Our findings imply that ZFPM2 may require additional hits, absence of a protective gene, or environmental factor to cause CDH with all of its clinical implications.
Supplementary Material
Acknowledgments
Funding provided by NIH/NICHD 1 R01-HD055150 and 1 P01-HD068250. Resequencing was partially provided through the RS&G Service by the Northwest Genomics Center at the University of Washington, under U.S. Federal Government contract number HHSN268201100037C from the NHLBI. We thank the patients and their families for their support; Alison J. Pfeister, JD, for logistical assistance; Steve DePalma, PhD and Michael E. Talkowski, PhD, for bioinformatics guidance.
Footnotes
Conflict of interest
The authors have declared no conflicting interests.
The following Supporting information is available for this article:
Appendix S1. Clinical reports.
Additional Supporting information may be found in the online version of this article.
References
- 1.Pober BR, Russell MK, Ackerman KG. Congenital diaphragmatic hernia overview. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. SourceGeneReviewsTM. Seattle, WA: University of Washington; 2010. [Google Scholar]
- 2.Czeizel A, Kovács M. A family study of congenital diaphragmatic defects. Am J Med Genet. 1985;21 (1):105–117. doi: 10.1002/ajmg.1320210115. [DOI] [PubMed] [Google Scholar]
- 3.David TJ, Illingworth CA. Diaphragmatic hernia in the south-west of England. J Med Genet. 13(4):253–262. doi: 10.1136/jmg.13.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackerman KG, Herron BJ, Vargas SO, et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1 (1):58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleyl SB, Moshrefi A, Shaw GM, et al. Candidate genes for congenital diaphragmatic hernia from animal models: sequencing of FOG2 and PDGFRalpha reveals rare variants in diaphragmatic hernia patients. Eur J Hum Genet. 2007;15 (9):950–958. doi: 10.1038/sj.ejhg.5201872. [DOI] [PubMed] [Google Scholar]
- 6.De Luca A, Sarkozy A, Ferese R, et al. New mutations in ZFPM2/FOG2 gene in tetralogy of Fallot and double outlet right ventricle. Clin Genet. 2011;80 (2):184–190. doi: 10.1111/j.1399-0004.2010.01523.x. [DOI] [PubMed] [Google Scholar]
- 7.Wat MJ, Veenma D, Hogue J, et al. Genomic alterations that contribute to the development of isolated and non-isolated congenital diaphragmatic hernia. J Med Genet. 2011;48 (5):299–307. doi: 10.1136/jmg.2011.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson MA, Gusella JF. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro. 1984;20 (11):856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 9.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461 (7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinto D, Darvishi K, Shi X, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29 (6):512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15 (7):901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pizzuti A, Sarkozy A, Newton AL, et al. Mutations of ZFPM2/FOG2 gene in sporadic cases of tetralogy of Fallot. Hum Mutat. 2003;22 (5):372–377. doi: 10.1002/humu.10261. [DOI] [PubMed] [Google Scholar]
- 13.Tan Z-P, Huang C, Xu Z-B, Yang J-F, Yang Y-F. Novel ZFPM2/FOG2 variants in patients with double outlet right ventricle. Clin Genet. 2012;82(5):466–471. doi: 10.1111/j.1399-0004.2011.01787.x. [DOI] [PubMed] [Google Scholar]
- 14.Menon SC, Tani LY, Weng HY, Lally PA, Lally KP, Yoder BA. Clinical characteristics and outcomes of patients with cardiac defects and congenital diaphragmatic hernia. J Pediatr. 2013;162 (1):114–119. doi: 10.1016/j.jpeds.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 15.Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21. 1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40 (12):1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12 (3):167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.