Abstract
The 300 kDa cation-independent mannose 6-phosphate receptor (CI-MPR) plays an essential role in lysosome biogenesis by targeting ∼60 different phosphomannosyl-containing acid hydrolases to the lysosome. This type I membrane glycoprotein has a large extracellular region comprised of 15 homologous domains. Two mannose 6-phosphate (M6P) binding sites have been mapped to domains 3 and 9, whereas domain 5 binds preferentially to the phosphodiester, M6P-N-acetylglucosamine (GlcNAc). A structure-based sequence alignment predicts that the C-terminal domain 15 contains three out of the four conserved residues identified as essential for carbohydrate recognition by domains 3, 5 and 9 of the CI-MPR, but lacks two cysteine residues that are predicted to form a disulfide bond. To determine whether domain 15 of the CI-MPR has lectin activity and to probe its carbohydrate-binding specificity, truncated forms of the CI-MPR were tested for binding to acid hydrolases with defined N-glycans in surface plasmon resonance analyses, and used to interrogate a phosphorylated glycan microarray. The results show that a construct encoding domains 14–15 binds both M6P and M6P-GlcNAc with similar affinity (Kd = 13 and 17 μM, respectively). Site-directed mutagenesis studies demonstrate the essential role of the conserved Tyr residue in domain 15 for phosphomannosyl binding. A structural model of domain 15 was generated that predicted an Arg residue to be in the binding pocket and mutagenesis studies confirmed its important role in carbohydrate binding. Together, these results show that the CI-MPR contains a fourth carbohydrate-recognition site capable of binding both phosphomonoesters and phosphodiesters.
Keywords: glycan microarray, lectin, lysosome, lysosomal enzymes, mannose 6-phosphate receptor
Introduction
The 300 kDa cation-independent mannose 6-phosphate receptor (CI-MPR) and the 46 kDa cation-dependent MPR (CD-MPR) play critical roles in phagocytic and autophagic degradative pathways by supplying the lysosome with its repertoire of ∼60 different hydrolytic enzymes (Kornfeld and Sly 2001; Braulke and Bonifacino 2009). Newly synthesized soluble acid hydrolases are marked for recognition by the MPRs in a two-step process that generates phosphomannosyl residues and facilitates their delivery to lysosomes. The first step occurs in the cis Golgi and is carried out by uridine diphosphate (UDP)-N-acetylglucosamine:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc- 1-phosphotransferase, EC 2.7.8.17) that transfers GlcNAc-1-phosphate to the C-6 hydroxyl group of an N-glycan mannose within acid hydrolases to form a phosphodiester (Hasilik et al. 1980; Varki and Kornfeld 1980; Bao et al. 1996; Kudo et al. 2005). The second step, which may not always occur, takes place in the trans Golgi network (TGN) where N-acetylglucosamine-1-phosphodiester α-N-acetylglucosaminidase (EC 3.1.4.45; also known as “uncovering enzyme”) removes the GlcNAc to form a phosphomonoester, mannose 6-phosphate (M6P) (Waheed et al. 1981; Varki et al. 1983; Kornfeld et al. 1999; Rohrer and Kornfeld 2001; Do et al. 2002). The heterogeneity of the resulting phosphorylated N-glycans is high due to several factors including the number and location of the phosphate in the glycan chain: five out of the nine mannose residues of high mannose-type N-glycans can be modified by GlcNAc-1-phosphotransferase and each N-glycan chain can contain one or two phosphomannosyl residues, which can be phosphomonoesters and/or phosphodiesters (Varki and Kornfeld 1980). Therefore, the MPRs must interact with a diverse set of glycans in order to effectively target acid hydrolases to the lysosome.
The CI-MPR has a large extracellular region comprised of 15 domains, referred to as Mannose 6-phosphate Receptor Homology (MRH) domains (Munro 2001; Castonguay et al. 2011). Each ∼150-residue domain has three or four disulfides and share 15–38% sequence identity when compared with each other or to the CD-MPR [reviewed in references Brown et al. (2009) and Kim et al. (2009)] (Figure 1). Some, but not all domains of the CI-MPR have known functions. Residues important for plasminogen and urokinase-type plasminogen activator receptor binding are found in the N-terminal domain 1 (Leksa et al. 2002). The single insulin-like growth factor II binding site resides in domain 11, with residues in domain 13 increasing the affinity of the interaction by ∼10-fold and was the first demonstration of interplay between domains (Devi et al. 1998; Linnell et al. 2001). Carbohydrate-binding activity has been mapped to individual MRH domains: three phosphomannosyl-binding sites are localized to domains 3, 5 and 9 and differ in their preference for phosphomonoester- and phosphodiester-containing glycans (Hancock, Yammani et al. 2002; Reddy et al. 2004; Bohnsack et al. 2009) (Figure 1). Similar to domain 11, the binding affinity exhibited by the carbohydrate recognition domains (CRDs) 3 and 5 is enhanced by the presence of additional regions of the receptor. A construct encoding domain 3 alone exhibits ∼1000-fold lower affinity for a lysosomal enzyme than domains 1–3 (Hancock, Yammani et al. 2002), and the crystal structure of domains 1–3 shows that residues in domain 1 and domain 2 do not directly contact M6P, but act to stabilize interactions to the loops of the binding pocket located within domain 3 (Olson, Dahms et al. 2004). Similar results were observed for the phosphodiester-specific-binding site within domain 5: a construct encoding domains 5–9 binds with ∼60-fold higher affinity to the acid α-glucosidase (GAA) diester than a construct encoding domain 5 alone (Bohnsack et al. 2009). These results suggest that, like domain 3, domain 5 utilizes other domains of the receptor to stabilize its carbohydrate-binding pocket.
Fig. 1.
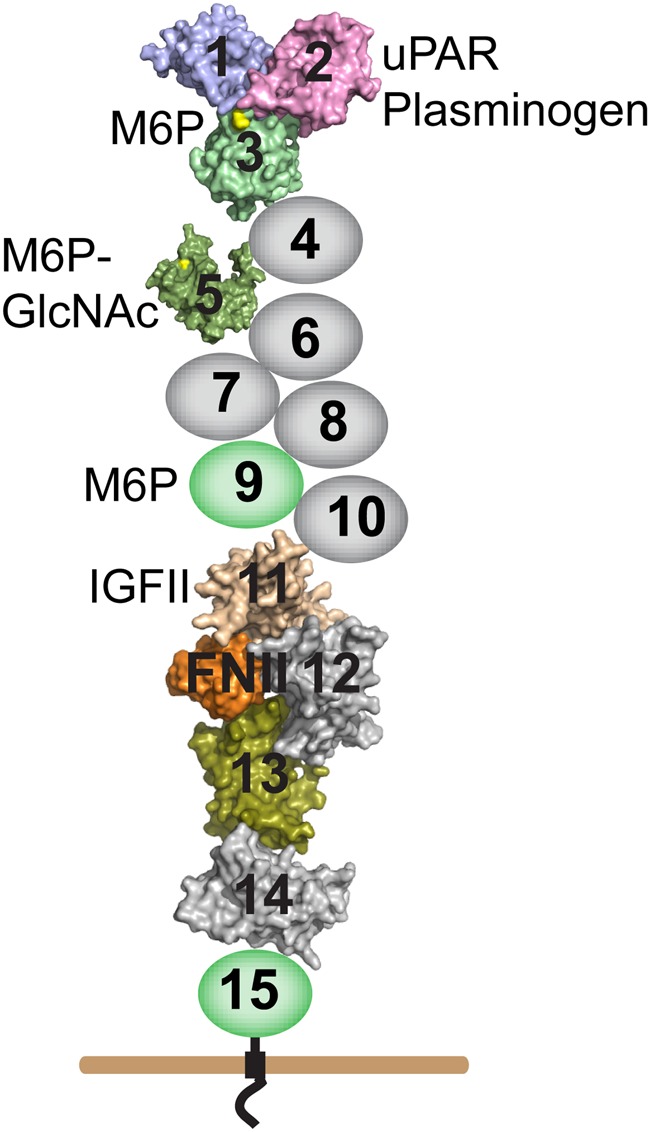
Schematic diagram of the CI-MPR. The CI-MPR is a type I transmembrane glycoprotein with an ∼2300-residue extracellular region. The locations of the ligand-binding sites are indicated. The fibronectin type II insert present in domain 13 (orange) interacts with domain 11 to increase the CI-MPR's affinity for IGF-II. Domains with solved structures are represented by their molecular surface generated in PyMol from RCSB deposited coordinates (domains 1–3, 1Q25; domain 5, 2KVB and domains 11–14, 2V5O). Domains with no known function are colored grey while domains with known function are colored (carbohydrate-binding domains are colored in green, with the M6P-binding site highlighted in yellow). This figure is available in black and white in print and in colour at Glycobiology online.
In addition to the MPRs, a few other proteins in the secretory pathway have been identified that contain MRH domains (Munro 2001): Three resident proteins of the endoplasmic reticulum (ER) involved in the quality control of glycoprotein folding that recognize mannose residues of high mannose-type N-glycans on newly synthesized glycoproteins [OS-9, XTP3-B and the β-subunit of the heterodimeric glucosidase II (GIIβ)] and the γ-subunit of the Golgi-localized GlcNAc-1-phosphotransferase [reviewed in ref. D'Alessio and Dahms (2015)]. The three-dimensional structure of MRH domains are known for eight (i.e. domains 1–3, domain 5, domains 11–14) out of the 15 domains of CI-MPR (Uson et al. 2003; Olson, Dahms et al. 2004; Olson, Yammani et al. 2004; Brown et al. 2008; Olson et al. 2010; Brown et al. 2008), CD-MPR (Roberts et al. 1998; Olson et al. 1999), OS-9 (Satoh et al. 2010) and GIIβ (Olson et al. 2013). The overall fold of an MRH domain comprises a flattened β-barrel structure consisting of nine β-strands organized into two orthogonally oriented antiparallel β-sheets, β1–β4 and β5–β9, with β9 interjecting between β7 and β8 (Figure 2A). Differences between MRH domains are due predominantly to differences in the length and positioning of loops that connect the β-strands. A comparison of the MRH domains that bind carbohydrate reveal that the binding pocket surrounding mannose is highly conserved and shows that the positions of the four residues critical for interacting with the mannose ring are nearly identical: Gln, Arg and Tyr are found on strands β3, β7 and β9, respectively, with Glu located in loop D between strands β8 and β9 (Kim et al. 2009; Olson et al. 2013) (Figure 2B and C). Together, these four residues serve as a signature motif for mannose recognition by MRH domains.
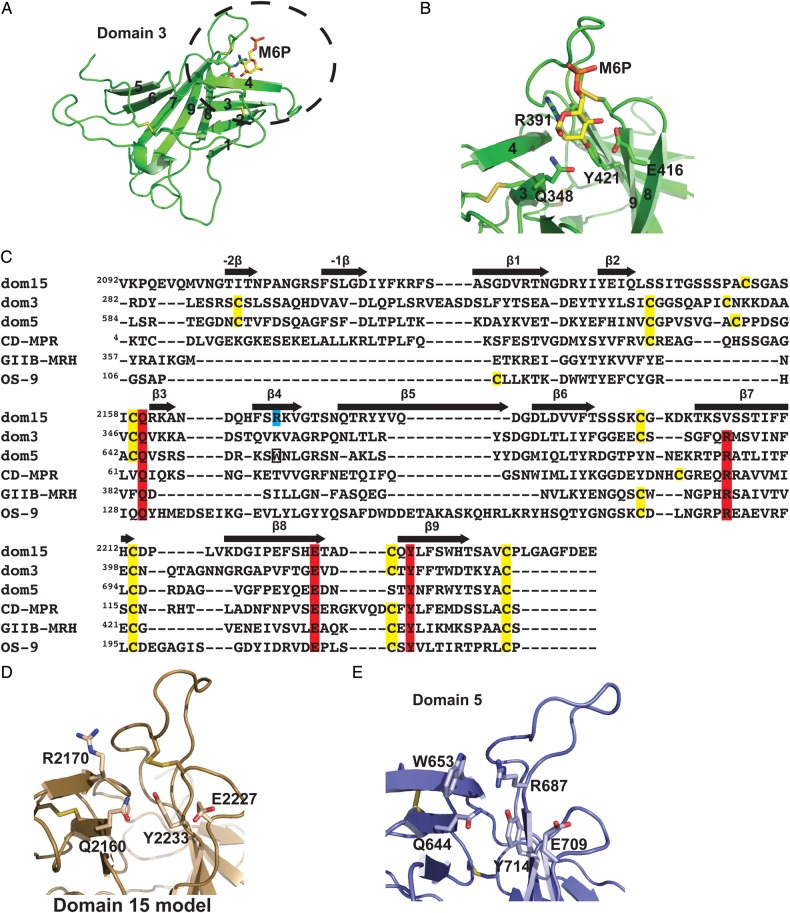
Fig. 2.
Comparison of carbohydrate-binding MRH domains. (A) Cartoon representation of the overall fold of an MRH domain using the structure of domain 3 (PDB entry 1Q25) as a model. The β-strands 1–9 are labeled along with the M6P ligand, which is present in the crystal structure. (B) The close-up view of domain 3's binding pocket with the four conserved signature motif residues labeled that interact with M6P (gold ball-and-stick). (C) Structure-based sequence alignment of MRH domains. The essential residues for carbohydrate binding are shaded in red while the critical Trp residue in domain 5 is boxed. The newly identified essential Arg residue in domain 15 is shaded in cyan. The locations of β-strands of CI-MPR domain 3 (PDB entry 1Q25) are indicated with arrows above the sequence. (D) The model of the binding pocket of domain 15. Residues predicted to be in the binding pocket (Gln, Glu, Tyr and newly identified Arg on β4-strand) are labeled. (E) The close-up view of the domain 5 (PDB entry 2KVB) binding pocket showing the four essential residues along with W653. This figure is available in black and white in print and in colour at Glycobiology online.
The presence of three distinct CRDs in the CI-MPR that recognize phosphomonoesters (domains 3 and 9) or phosphodiesters (domain 5) (Marron-Terada et al. 2000; Chavez et al. 2007; Bohnsack et al. 2009) explains, in part, why the CI-MPR is more efficient than the single CRD-containing, phosphomonoester-specific CD-MPR in targeting acid hydrolases to the lysosome (Hoflack et al. 1987; Ludwig et al. 1994; Pohlmann et al. 1995; Kasper et al. 1996; Sleat and Lobel 1997; Sohar et al. 1998). To further probe CI-MPR's ability to interact with a heterogeneous population of acid hydrolases, we performed a structure-based sequence alignment using the MRH domain structures of domains 3 (Olson Dahms et al. 2004; Olson, Yammani et al. 2004) and 5 (Olson et al. 2010) of CI-MPR, CD-MPR (Roberts et al. 1998; Olson et al. 1999), OS-9 (Satoh et al. 2010) and GIIβ (Olson et al. 2013) (Figure 2C). This analysis revealed that only domains 3, 5 and 9 of CI-MPR contain the four residues of the signature motif; however, domain 15 contains three of these four residues with Val at position 2205 instead of the conserved Arg residue on strand β7 (Figure 2C). To determine whether domain 15 of the CI-MPR contains a functional carbohydrate-binding site, we conducted several different experiments including surface plasmon resonance (SPR) analyses using immobilized acid hydrolases and probed a glycan microarray containing purified phosphorylated glycans. The results demonstrate that domain 15 of CI-MPR binds both phosphomonoester- and phosphodiester-containing high mannose-type glycans; thus, CI-MPR contains four carbohydrate-binding sites.
Results
Modeling of the C-terminal region of the CI-MPR predicts conserved residues form a binding pocket
Using the crystal structure of bovine CI-MPR domain 3 as a template (Figure 2A), a structural model of bovine domain 15 was generated, which predicts the three conserved residues (Gln, Glu and Tyr) are in close proximity. Although domain 15 lacks the conserved Arg residue on strand β4 (Figure 2C), the model predicts that an Arg residue located on a different β-strand, β4, is present in the binding pocket (Figure 2D). This Arg residue at position 2170 corresponds to a conserved Trp residue, W653, of domain 5 (Figure 2E). W653 is found in the binding pocket (Olson et al. 2010) and mutagenesis studies have shown that W653 is important for phosphodiester binding by domain 5 of the CI-MPR (Castonguay et al. 2011). A comparison of the amino acid sequence of CI-MPR domain 15 from various species reveals that the three residues (Gln2160, Glu2227 and Tyr2233) plus Arg2170 on β4 are absolutely conserved, along with the four Cys residues (Supplementary data, Figure S1). Taken together, these results suggest that domain 15 harbors a carbohydrate-binding site.
The C-terminal region of the CI-MPR binds acid hydrolases
To test the hypothesis that domain 15 of the CI-MPR can interact with mannose-containing ligands, constructs encompassing the C-terminal region of the receptor were generated as C-terminal His-tagged proteins in Sf9 cells. The 15 domains of the CI-MPR are joined together by short linkers of 5–12 amino acids that span the conserved cysteine residues marking the N- and C-terminus of each domain. However, domain 15, which is the C-terminal MRH domain adjacent to the membrane (Figure 1), is tethered to the transmembrane region by a linker of 26 amino acids. Initial experiments utilized two constructs, either terminating one amino acid after the final cysteine of the domain or extended by an additional eight residues. The presence of the additional eight residues at the C-terminus resulted in a higher yield of recombinant protein and these residues are present in constructs used in subsequent studies (Dom15, Dom14–15, Dom7–15 and Dom1–15).
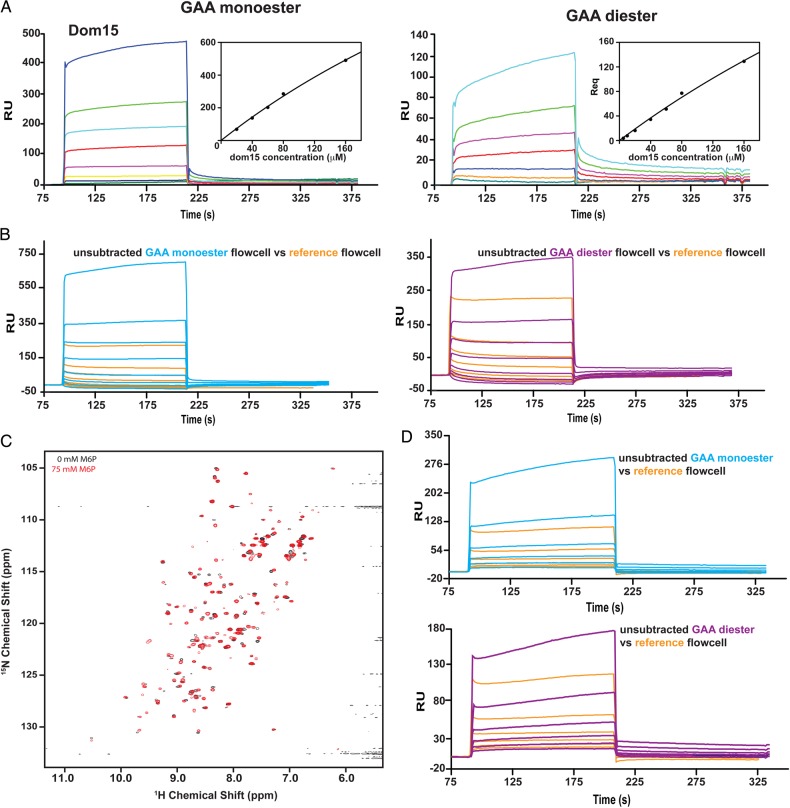
We have used GAA, a lysosomal enzyme that has been enzymatically modified to contain only phosphomonoesters (GAA monoester) or phosphodiesters (GAA diester), to determine the carbohydrate specificity and binding affinity of various truncated forms of the MPRs (Chavez et al. 2007; Bohnsack et al. 2009; Castonguay et al. 2012). SPR studies were undertaken in which GAA monoester and GAA diester were immobilized on separate flow cells of a CM5 sensor chip and various concentrations of Dom15 were flowed over the surface. Although Dom15 demonstrated measureable binding to both surfaces (KD = 700 and 900 μM, respectively: Table I and Figure 3A), the protein displayed significant interaction with the unmodified reference surface (Figure 3B). In an attempt to rectify this issue, Dom15 was expressed in Escherichia coli, which produces nonglycosylated proteins, and compared with the glycosylated protein isolated from Sf9 insect cells. The structural integrity of the Dom15 protein, which is refolded following its purification from inclusion bodies, was evaluated by nuclear magnetic resonance (NMR) spectroscopy. The 15N-1H-heteronuclear single quantum coherence (HSQC) spectrum displays a peak dispersion indicative of a folded protein, although the nonuniform peak intensities could indicate either some element of disorder or protein dynamics (Figure 3C). Addition of 75 mM M6P to the sample resulted in chemical shift perturbations in a select group of backbone NH groups suggestive of ligand binding, and other peaks appearing, which could indicate a change in the dynamics of the protein (Figure 3C). Similar to the glycosylated Dom15 isolated from Sf9 cells (Figure 3A and B), SPR analyses show Dom15 interacts with both GAA mono- and GAA diester surfaces, plus significant interaction with the reference flow cell was observed making it unsuitable for further SPR studies (Figure 3D).
Table I.
Kinetic data obtained from SPR analyses
| Construct |
KD (nM) |
|||||||
|---|---|---|---|---|---|---|---|---|
|
ka1 (M−1s−1 × 103) |
kd1 (s−1 × 10−3) |
Monoester |
Diester |
|||||
| Monoester | Diester | Monoester | Diester | kd1/ka1 | Steadystate | kd1/ka1 | Steadystate | |
| D1–15 | 184 ± 20 | 137 ± 30 | 0.8 ± 0.1 | 6.7 ± 0.6 | 4.5 ± 0.7 | 71 ± 10 | 51 ± 1 | 148 ± 20 |
| D1–14 | 65 ± 9 | 60 ± 9 | 2.0 ± 0.5 | 25 ± 3 | 31 ± 7 | 254 ± 55 | 436 ± 99 | 700 ± 270 |
| D7–15 | 122 ± 9 | 59 ± 5 | 10 ± 1 | 87 ± 3 | 80 ± 11 | 123 ± 15 | 1500 ± 163 | 1600 ± 390 |
| D7–15m15YF | 84 ± 30 | 21 ± 4 | 45 ± 3 | 6 ± 1 | 574 ± 166 | 418 ± 106 | DNB | DNB |
| D7–15m15RA | 36 ± 7 | DNB | 45 ± 2 | DNB | 1300 ± 303 | 1070 ± 236 | DNB | DNB |
| D7–15m9 | 1.6 ± 0.3 | 1.6 ± 0.3 | 112 ± 44 | 7 ± 3 | 12700 ± 2000 | 9340 ± 1120 | 4000 ± 2000 | 16300 ± 7800 |
| D7–15m9m15 | DNB | DNB | DNB | DNB | DNB | DNB | DNB | DNB |
| D14–15 | 20 ± 3 | 5 ± 3 | 274 ± 31 | 106 ± 82 | 13000 ± 3000 | 8200 ± 1250 | 17000 ± 7000 | 21300 ± 2980 |
| D15 (sf9) | 700000 | 900000 | ||||||
| D14–15m15YF | DNB | DNB | DNB | DNB | DNB | DNB | DNB | DNB |
| D14–15m15RA | DNB | DNB | DNB | DNB | DNB | DNB | DNB | DNB |
|
KD2(mM) |
||||||||
|
ka2 (M−1s−1 × 10−3) |
kd2 (s−1 × 10−3) |
kd2/ka2 |
||||||
| Construct | Monoester | Diester | Monoester | Diester | Monoester | Diester | ||
| D1–15 | 3 ± 2 | 3.9 ± 0.1 | 1.1 ± 0.9 | 0.06 ± 0.06 | 300 ± 150 | 16 ± 14 | ||
| D1–14 | 2 ± 2 | 9.0 ± 0.6 | 1 ± 2 | 2.5 ± 0.1 | 400 ± 500 | 279 ± 19 | ||
| D7–15 | 0.9 ± 0.6 | 8 ± 1 | 0.6 ± 0.9 | 6 ± 1 | 200 ± 400 | 750 ± 44 | ||
| D7–15m15YF | 10 ± 1 | 3 ± 2 | 4.0 ± 0.2 | 1 ± 2 | 410 ± 43 | DNB | ||
| D7–15m15RA | 11 ± 3 | DNB | 6 ± 0.7 | DNB | 545 ± 62 | DNB | ||
| D7–15m9 | 15.4 ± 0.6 | 5 ± 2 | 6.4 ± 0.4 | 0.2 ± 0.3 | 259 ± 117 | 0.4 ± 0.6 | ||
| D7–15m9m15 | DNB | DNB | DNB | DNB | DNB | DNB | ||
| D14–15 | 18 ± 10 | 8 ± 7 | 8 ± 5 | 8 ± 7 | 500 ± 500 | 750 ± 670 | ||
| D15 (sf9) | 9 | 11 | ||||||
| D14–15m15YF | DNB | DNB | DNB | DNB | DNB | DNB | ||
| D14–15m15RA | DNB | DNB | DNB | DNB | DNB | DNB | ||
DNB denotes did not bind. Kinetic and steady state affinity data calculated from SPR sensorgrams obtained with GAA mono and GAA diester immobilized on the sensor chip surface. Purified recombinant protein was injected as the free analyte. Upper table reports kinetic parameters of the major component obtained from fitting sensorgrams to a two-state model with conformational change. The lower table reports kinetic parameters for the minor component with KD values in the millimolar range. All values were determined from at least three independent trials.
Fig. 3.
Characterization of domain 15 construct by SPR and NMR. (A) Increasing concentrations of domain 15 (1, 5, 10, 20, 40, 60, 80 and 100 μM) produced in Sf9 cells were flowed over either a reference flow cell or GAA mono- or diester coupled to CM5 surface and the response was measured. Insets depict the response at equilibrium (Req) plotted against receptor concentration; data were fitted to a steadystate model (BIAevaluation software) and KD values were determined. (B) Unsubtracted responses for flow cells 1 (reference, orange) and GAA monoester (cyan) and diester (magenta) coupled flow cells illustrating the significant interaction of domain 15 protein produced in Sf9 cells with the reference, uncoupled dextran surface of the CM5 sensor chip. (C) 1H-15N HSQC spectra of E. coli expressed and refolded domain 15 protein in the absence (black) and presence of 75 mM M6P (red). (D) Sensorgrams of response units (RU) versus increasing concentrations of E. coli expressed domain 15 protein (1, 5, 10, 20, 40 and 80 μM) comparing the reference flow cell (orange) with the unsubtracted GAA monoester (cyan) or GAA diester (magenta) flow cells. This figure is available in black and white in print and in colour at Glycobiology online.
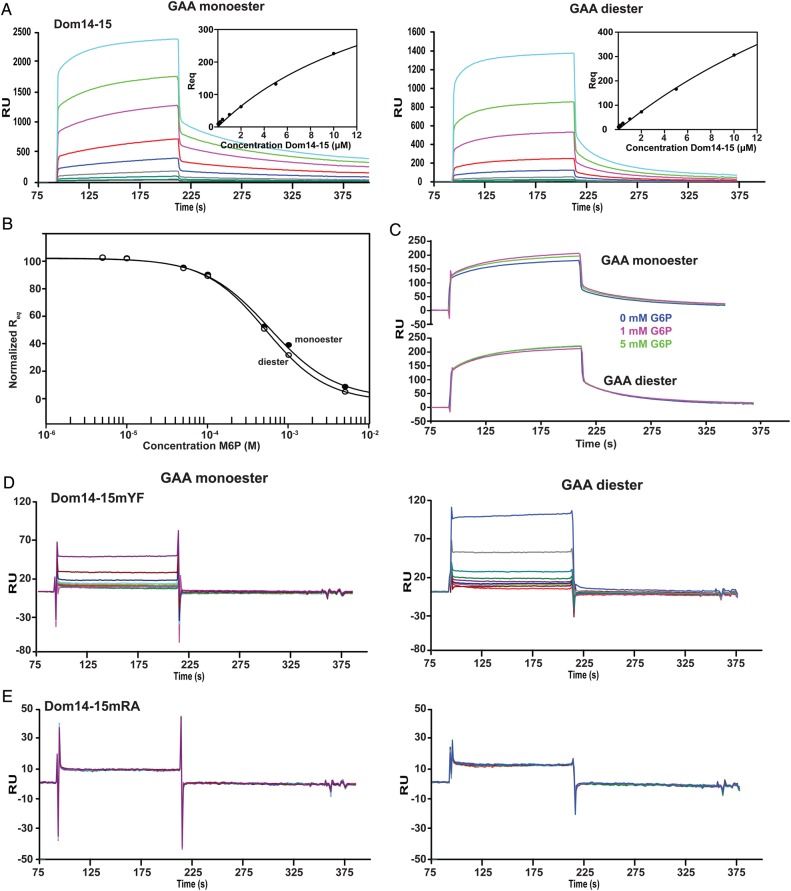
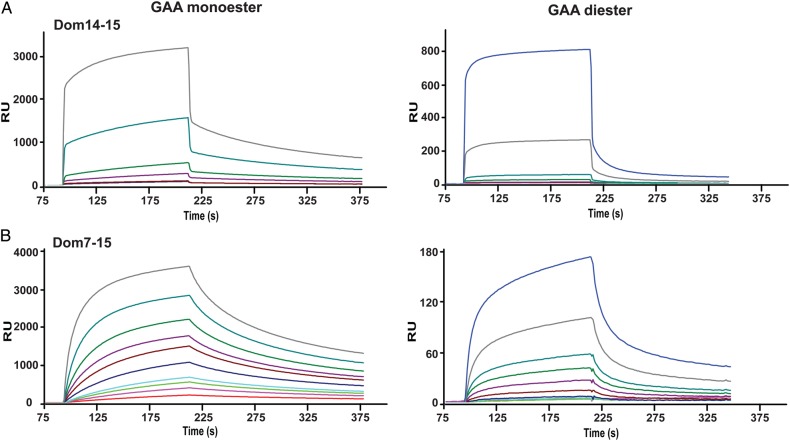
We next asked whether domains adjacent to domain 15 enhance carbohydrate-binding affinity, as we have shown for domains 3 and 5 of the CI-MPR (Hancock, Yammani et al. 2002; Bohnsack et al. 2009). A construct expressing two domains was identified that satisfies this criterion: domains 14 and 15 (Dom14–15). In contrast to the single domain 15 construct produced in Sf9 insect cells (Figure 3A and B) or E. coli (Figure 3D) in which significant binding to the unmodified, reference surface was detected, SPR analyses of the two domain construct, Dom14–15, revealed minimal interaction with the reference surface that accounted for <2% (20RU or less) of the response detected with the GAA monoester- or GAA diester-modified surfaces (Figure 4A). SPR studies show that Dom14–15 binds GAA monoester and GAA diester with similar affinities (KD1 = 13 ± 3 μM and KD1 = 17 ± 7 μM respectively; Figure 4A, Table I) that is ∼50-fold higher affinity than observed by Dom15. This interaction is inhibited by M6P (GAA monoester Ki = 580 ± 70 μM, GAA diester Ki = 520 ± 31 μM; Figure 4B), but not G6P at concentrations up to and including 5 mM (Figure 4C), demonstrating that the interaction between Dom14–15 and these lysosomal enzymes is dependent upon the presence of phosphomannosyl residues. Furthermore, substitution of the conserved signature motif Tyr2233 residue with Phe in domain 15 (Dom14–15mYF) resulted in a dramatic loss of binding activity with both GAA monoester- or GAA diester-coupled surfaces (Figure 4D).
Fig. 4.
SPR studies characterizing the interaction between truncated MPRs (Dom14–15, Dom14–15m15YF, Dom14–15m15RA and lysosomal enzyme GAA. (A) Increasing concentrations (40, 80, 120, 200, 400, 1000, 2000, 5000 and 10000 nM) of the Dom14–15 protein were flowed over the reference surface or GAA monoester or GAA diester coupled surfaces for 2 min followed by buffer and regeneration with 10 mM HCl. Representative sensorgrams are shown. Insets depict the response at equilibrium (Req) plotted against receptor concentration and data are fitted by nonlinear regression to calculate the KD under steadystate conditions. (B) Dom14–15 binding to GAA monoester (closed circles) and GAA diester (open circles) surfaces is inhibited by increasing concentrations of M6P. The equilibrium response (Req), normalized to the highest response, was plotted against the concentration of M6P. The Ki was calculated using nonlinear regression (SigmaPlot, version 10.0). (C) Dom14–15 was flowed over either the reference surface or GAA mono- or diester-coupled surface in the absence or presence of 1 or 5 mM G6P. The resulting sensorgrams are shown depicting the inability of G6P to inhibit receptor binding to the GAA-coupled surfaces. Sensorgrams of increasing concentrations [same as (A)] of either Dom14–15YF (D) or Dom14–15RA (E) flowed over either the reference surface or GAA mono- or diester-coupled surface. In both cases there is at least a 20-fold difference in RU response at the highest concentrations (increase in bulk refractive index due to high protein concentration). This figure is available in black and white in print and in colour at Glycobiology online.
Taken together, these results show that domain 15 can interact with both phosphomonoester- and phosphodiester-containing lysosomal enzymes and that Tyr2233 in domain 15 is essential for the interaction. Furthermore, the affinity of the interaction between Dom14–15 and the phosphomannosyl-containing GAA is comparable with domain 5 of the CI-MPR that we have shown to contain a low affinity site for M6P. In contrast to Dom14–15, Dom5 binds preferentially to glycans with phosphodiesters (Reddy et al. 2004; Chavez et al. 2007; Bohnsack et al. 2009).
Conserved arginine in domain 15 is a key determinant for phosphomannosyl binding
A structural model of domain 15 predicts that Arg2170 on β4 is in close proximity to the other three conserved residues (Gln2160, Glu2227 and Tyr2233) in the binding pocket (Figure 2D). We hypothesized that this Arg residue compensates for the missing signature motif Arg at position 2205 (Figure 2C). To test whether Arg2170 is important for carbohydrate recognition, Arg2170 was replaced with Ala in Dom14–15 (Dom14–15mRA). The resulting mutant was evaluated for its ability to bind GAA monoester and GAA diester. The SPR analyses showed that Dom14–15mRA exhibits no significant binding above background to either GAA monoester- or GAA diester-coupled surfaces (Figure 4E). These results demonstrate that Arg2170 of domain 15 is essential for phosphomannosyl recognition by Dom14–15.
Glycan microarray analyses
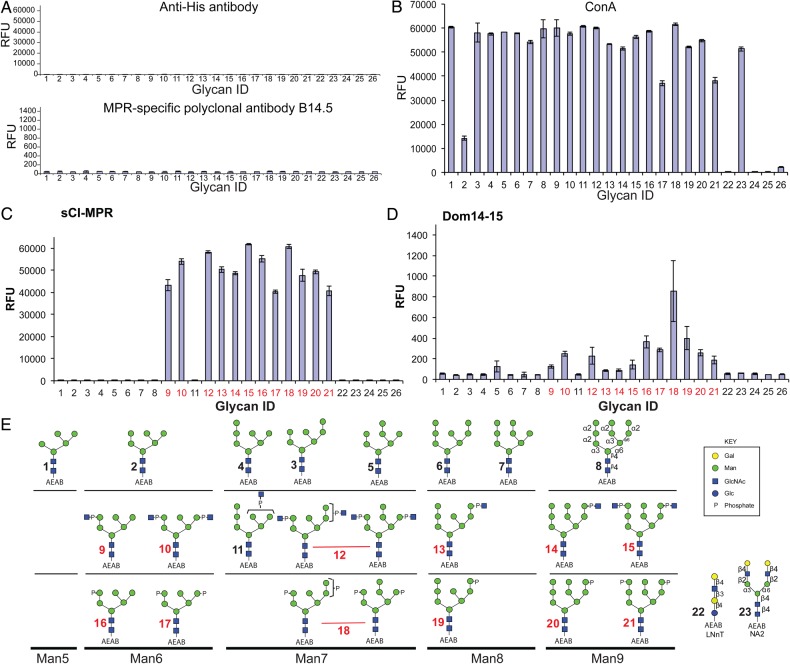
We have previously generated a novel glycan microarray that contains high mannose-type glycans with or without phosphomonoesters or phosphodiesters (Song et al. 2009, 2012). The purified glycans containing a bifunctional fluorescent linker, 2-amino-N-(2-aminoethyl)-benzamide (AEAB), are covalently immobilized onto N-hydroxysuccinimide (NHS)-activated glass slides. The slides were incubated with MPR proteins and their binding interactions were detected using MPR-specific antibodies or antibodies directed against the His-tag, which were not bound by the glycan microarray (Figure 5A). As a control to demonstrate the presence of glycans printed on the array, we used the plant lectin concanavalin A (ConA), which specifically binds high mannose-, hybrid- and complex-type biantennary glycans (Baenziger and Fiete 1979; Brenckle and Kornfeld 1980; Brewer and Bhattacharyya 1986). Consistent with its known specificity, biotinylated ConA bound to the expected glycans on the array (glycans 1–21 and 23) (Figure 5B). In addition, the similar intensity of binding by biotinylated ConA indicated that all glycans, except glycans 2, 17 and 21, were immobilized on the slides at comparable levels. As expected, little or no interaction was observed to the control glycan LNnT-AEAB (glycan 22) or the phosphate-buffered saline control (positions 24 and 25). The positive signal for biotin [position 26, ∼2000 relative fluorescence unit (RFU)] detected with the Cy5-labeled streptavidin is used for aligning the grid for analysis.
Fig. 5.
Interaction of CI-MPR constructs with phosphorylated and nonphosphorylated, high mannose glycans. The glycan microarray was printed as described in the text and the individual glycans are identified by their glycan number as indicated in (E). Glycans 1–8 are nonphosphorylated high mannose-type oligosaccharides; 9–15, phosphodiesters of high mannose-type oligosaccharides and 16–21, phosphomonoesters of high mannose-type oligosaccharides. (A) Controls showing background binding of rabbit antibody B14.5 and anti-penta His. MPR-specific rabbit antibody B14.5 at a 1:250 dilution followed by Cy5-labeled goat anti-rabbit immunoglobulin G (IgG) (5 μg/mL). Anti-penta His (5 μg/mL) followed by Alexa633 goat anti-mouse IgG (5 μg/mL). (B) Biotinylated ConA (0.5 μg/mL) detected with 0.5 μg/mL of Cy5-labeled streptavidin. (C) Soluble CI-MPR (sCI-MPR) isolated from bovine serum (5 μg/mL) detected with rabbit antibody B14.5 (1:250) and Cy5-labeled goat anti-rabbit IgG (5 μg/mL), (D) Dom14–15 (150 μg/mL) detected with anti-penta His (5 μg/mL) and Alexa633 goat anti-mouse IgG (5 μg/mL). The assays are single experiments where each glycan is printed in replicates of four, and the RFU value is the mean of the four. The error bars represent the mean ± S.D. The glycan ID numbers highlighted in red represent glycans that exhibit significant interaction with the receptor. This figure is available in black and white in print and in colour at Glycobiology online.
A soluble form of the CI-MPR (sCI-MPR) was purified from bovine serum; as we previously reported (Bohnsack et al. 2009; Song et al. 2009), it binds to all phosphomonoester- and phosphodiester-containing glycans on the array with the exception of glycan 11 (GPM7(3) or Man7-P-GlcNAc-b), but not high mannose-type glycans (glycans 1, 3–8; binding to glycan 2 could not be fully evaluated due to the reduced level (∼25%) of glycan 2 immobilized on the slides, see Figure 5B) (Figure 5C). Dom14–15 binding was significantly weaker but nevertheless exhibited a similar profile as the sCI-MPR in that it interacts with all phosphomonoester- and phosphodiester-containing glycans with the exception of glycan 11. However, at the single concentration of Dom14–15 used, a more robust interaction was observed with phosphomonoester-containing glycans than with the phosphodiester-containing glycans (Figure 5D; glycan structures are shown in Figure 5E). Dom14–15 did not bind to high mannose-type glycans, except for glycan 5 (Man7-c) (Figure 5D). In addition, no interaction was observed between Dom14–15 and the nonphosphorylated, galactose-terminated glycans LNnT-AEAB (glycan 22) and NA2-AEAB (glycan 23), further demonstrating Dom14–15′s requirement for phosphomannosyl residues as recognition determinants. Although sCI-MPR and Dom14–15 bound both glycans 17 and 21 (Figure 5C and D), it is likely that the observed response underrepresents the binding capacity of the receptors given that glycans 17 and 21 are immobilized at a lower level (∼65%) compared with the other glycans (Figure 5B).
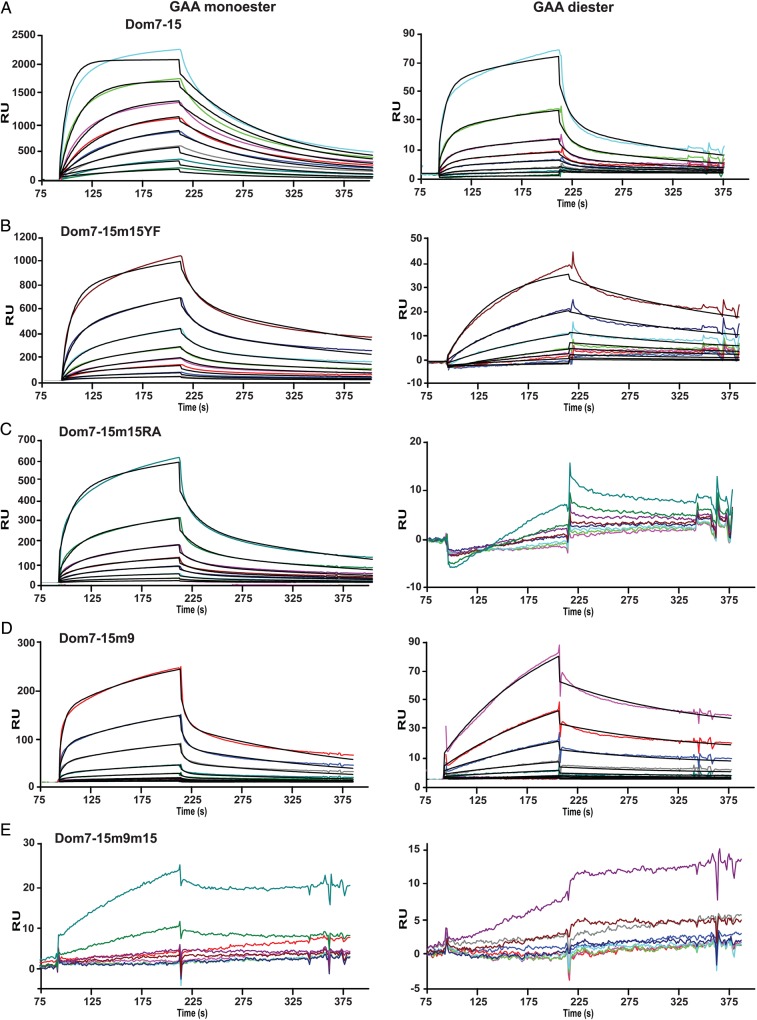
Influence of additional domains on the binding affinity of domain 15
To assess whether additional domains influence the binding affinity of domain 15, a construct encoding domains 7–15 (Dom7–15) was generated which contains the phosphomonoester-specific CRD, domain 9 (Bohnsack et al. 2009), along with the newly identified binding site in domain 15 (Figure 1). SPR analyses show that Dom7–15 binds GAA monoester with a higher affinity than GAA diester (KD1 = 80 ± 11 nM versus KD1 = 1500 ± 163 nM; Figure 6A, Table I). However, Dom7–15 exhibits an ∼11-fold higher affinity for GAA diester than Dom14–15 (KD1 = 17000 ± 7000 nM). Substitution of the conserved signature motif Tyr2233 residue with Phe in domain 15 (Dom7–15m15YF) or Arg2170, which is predicted to be in the binding pocket (Figure 2), with Ala (Dom7–15m15RA) resulted in the loss of interaction with the GAA diester-coupled surface (Figure 6B and C), similar to that observed when these mutations where placed in the context of Dom14–15 (Figure 4). Because the carbohydrate-binding site of domain 9 is specific for phosphomonoesters (Marron-Terada et al. 2000; Bohnsack et al. 2009), these data support the hypothesis that residues in domain 15 (Arg2170 and Tyr2233) are required for the ability of Dom7–15 to recognize phosphodiesters.
Fig. 6.
Interaction between Dom7–15 constructs and the lysosomal enzyme GAA modified with either M6P (monoester) or M6P-GlcNAc (diester). Representative sensorgrams from SPR analyses showing response units (RU) with increasing concentrations (10, 20, 40, 80, 120, 200 and 400 nM) of (A) Dom7–15, (B) Dom7–15m9, (C) Dom7–15m15YF, (D) Dom7–15RA or (E) Dom7–15m9m15. This figure is available in black and white in print and in colour at Glycobiology online.
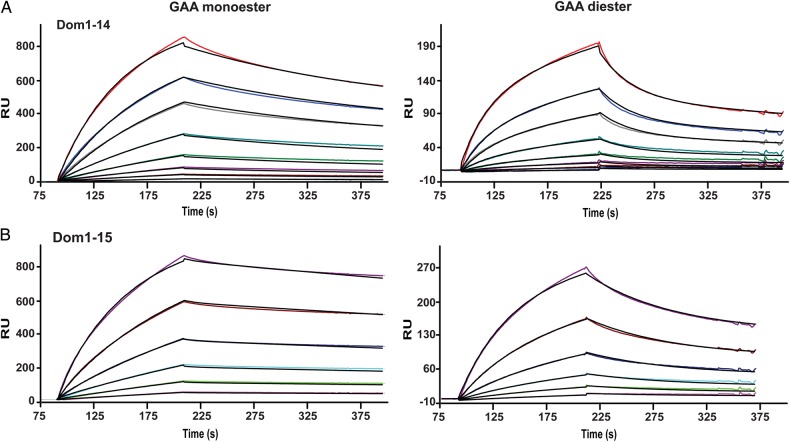
To evaluate the role of the presence of domains 7–14 on the carbohydrate-binding ability of domain 15, the essential Arg1290 residue of domain 9 was mutated to an alanine residue in the Dom7–15 construct (Dom7–15m9) and the ability of this construct to bind phosphomono- and diesters was evaluated by SPR. This mutation produced protein with similar affinities for the GAA mono and GAA diester surfaces (KD1 = 12700 ± 2000 nM and 4000 ± 2000 nM, respectively) as Dom14–15 (KD1 = 13000 ± 3000 nM and 17000 ± 7000 nM, respectively), indicating the inclusion of the seven additional domains did not result in additional enhancement of carbohydrate binding above that obtained by the presence of domain 14 (Figure 6D, Table I). To confirm there were no other regions in the Dom7–15 construct capable of binding these surfaces, we performed SPR analyses of a construct containing two amino acid substitutions to eliminate the carbohydrate-binding activity in domains 9 and 15; this construct (Dom7–15m9m15) demonstrated no measurable response to GAA monoester or GAA diester surfaces except at the highest concentration (1000 nM) tested (Figure 6E). As a control, a construct encoding the entire extracytoplasmic region of the CI-MPR (Dom1–15), terminating at the identical C-terminal residue as Dom14–15, was generated and flowed over the GAA coupled sensor chip. As shown in Figure 7, the binding of Dom1–15 (and Dom1–14) do not reach equilibrium within the time frame of the association phase, resulting in an underestimate of the binding affinity calculated by steady state methods. Thus, the KD values calculated from the ratio of the dissociation and association rate constants (kd/ka) are more accurate as this kinetic method does not rely on the achievement of steady state equilibrium conditions. Additionally, the KD values obtained for Dom1–15 as determined by (kd/ka) are similar to previously published values obtained for the CI-MPR isolated from bovine serum (Bohnsack et al. 2009). Evaluation of the kinetic data show that Dom1–15 binds with high affinity to GAA monoester (KD1 = 4.5 ± 0.7 nM) and GAA diester (KD1 = 51 ± 1 nM) (Figure 7B, Table I), A construct encoding domains 1–14 (Dom1–14) was also subjected to SPR analyses. Dom1–14, which lacks the CRD in domain 15, exhibits ∼10-fold decrease in affinity for both GAA mono and GAA diester surfaces (KD1 = 31 ± 7 nM and KD1 = 436 ± 99 nM) (Figure 7A, Table I) compared with Dom1–15. The presence of the M6P-GlcNAc-specific domain 5 in Dom1–14 results in binding to the diester surface, although with an ∼9-fold lower affinity compared with Dom1–15. Taken together, these data support the hypothesis that, similar to domains 3 and 5, additional domains interact with domain 15 to enhance the carbohydrate-binding affinity of domain 15.
Fig. 7.
Influence of the presence of domain 14 on receptor interaction with the lysosomal enzyme GAA modified with either M6P (monoester) or M6P-GlcNAc (diester). Representative sensorgrams from SPR analyses showing response units (RU) with increasing concentrations of (A) Dom1–14 (2, 5, 10, 20, 40, 80, 120 and 200 nM) and (B) Dom1–15 (2, 5, 10, 20, 40 and 80 nM). This figure is available in black and white in print and in colour at Glycobiology online.
Discussion
In the current study, a new phosphomannosyl-binding site in the CI-MPR that is located in domain 15 has been identified and characterized. We show that this domain is capable of recognizing both phosphodiester-containing glycans as well as phosphomonoesters, thereby distinguishing it from domain 5 as well as from the phosphomonoester-specific domains 3 and 9.
What is the functional significance of having multiple carbohydrate-binding sites with differing specificities? The CI-MPR, unlike the CD-MPR, is capable of recognizing the products of both enzymes that modify N-glycans on acid hydrolases: M6P-GlcNAc generated by GlcNAc-1-phosphotransferase and M6P produced by the action of the uncovering enzyme. The combination of the different glycan specificities of these four glycan-binding CRDs (domains 3, 5, 9 and 15) allows the CI-MPR to interact with the structurally diverse repertoire of phosphorylated N-glycans present on acid hydrolases, and supports previous in vitro (Hoflack et al. 1987; Sleat and Lobel 1997) and in vivo (Ludwig et al. 1994; Pohlmann et al. 1995; Kasper et al. 1996; Sohar et al. 1998) studies showing that the CI-MPR is more efficient in delivering acid hydrolases to lysosomes than the CD-MPR, which has a single CRD and cannot bind phosphodiesters (Hoflack and Kornfeld 1985; Tong and Kornfeld 1989; Roberts et al. 1998; Olson et al. 1999; Song et al. 2009). In addition, the uncovering enzyme is synthesized as a proenzyme that must undergo proteolytic processing by the endoprotease furin in the TGN in order to become active (Do et al. 2002). The CI-MPR may have evolved to contain phosphodiester-binding sites, as their presence would be advantageous to the organism in circumstances where the uncovering enzyme fails to undergo proteolytic activation or is missing, and thus only phosphodiesters would be present on acid hydrolases. In support of this hypothesis are studies showing that transgenic mice expressing only covered, M6P-GlcNAc-containing acid hydrolases have a normal phenotype (Boonen et al. 2009). This is in sharp contrast to patients who are deficient in GlcNAc-1-phosphotransferase activity and are unable to tag their acid hydrolases with phosphomannosyl residues needed for their delivery to lysosomes. Genetic mutations in GlcNAc-1-phosphotransferase cause mucolipidosis II (I-cell disease), a severe lysosomal storage disorder characterized by aberrant mistargeting and secretion of lysosomal enzymes that results in cardiomegaly, hepatomegaly, and death in the first decade of life (Kornfeld and Sly 2001; Tiede et al. 2005; Kudo et al. 2006). Mice lacking GlcNAc-1-phosphotransferase activity exhibit a similar phenotype as patients with mucolipidosis II (Gelfman et al. 2007). Together these studies suggests that the CI-MPR, due to its ability to bind phosphodiesters via its CRDs 5 and 15, rescues these mice from the severe mucolipidosis II-like phenotype observed in mice or patients lacking GlcNAc-1-phosphotransferase activity.
The presence of four different phosphomannosyl-binding sites in the CI-MPR not only provides the possibility of multivalent interactions and increased avidity with newly synthesized acid hydrolases to enhance the efficiency of their transport from the TGN to endosomal compartments, but also with intravenously administered recombinant acid hydrolases used in enzyme replacement treatment (ERT) of patients with lysosomal storage disorders. The rationale behind ERT for lysosomal storage diseases is based on providing cells with a functional enzyme that is lacking due to a genetic defect in these patients. The glycobiology surrounding the targeted delivery of acid hydrolases to the lysosome is well understood and has been instrumental in the development of Food and Drug Administration approved ERT for six lysosomal storage diseases, five of which target cell surface CI-MPR for the internalization of the intravenously administered recombinant enzyme (Brady 2006). The in vivo production of acid hydrolases with phosphomannosyl-containing glycans remains a challenge, and studies show that increasing the M6P content on recombinant human GAA improves the efficacy of ERT in Pompe disease (Zhu et al. 2005; McVie-Wylie et al. 2008; Zhu et al. 2009). In order to improve the design of high affinity acid hydrolases that will interact with the CI-MPR in a multivalent fashion, the spatial orientation of the CI-MPR's four carbohydrate-binding sites is needed. To date, the three-dimensional structures of eight (domains 1–3, domain 5, domains 11–14) out of the 15 MRH domains of the CI-MPR have been determined (Brown et al. 2002; Uson et al. 2003; Olson Dahms et al. 2004; Olson, Yammani et al. 2004; Brown et al. 2008; Olson et al. 2010), with structures of two (domains 9 and 15) out of the four carbohydrate-binding sites remaining to be determined and none of the existing multidomain structures (domains 1–3, domains 11–14) contain two carbohydrate-binding sites. Combining the two existing multidomain structures, and along with the prediction that the remaining two carbohydrate-binding sites will each occur in a tri-domain compact structure like domains 1–3 as opposed to the elongated structure of domains 11–14, we (Dahms et al. 2008) and others (Brown et al. 2008) have proposed simplified models of the CI-MPR in which the ligand-binding sites are oriented on the same face of the receptor, similar to that depicted in Figure 1. However, there are no data to support the positioning of all ligand-binding sites on one face of the CI-MPR, and additional structural studies are needed to test this model. Therefore, the structure of the entire extracytoplasmic region of the receptor is needed to determine how the four carbohydrate-binding sites are oriented spatially with respect to each other. This information will be critical for the design of high affinity, multivalent ligands used in ERT in which M6P and/or M6P-GlcNAc residues are properly spaced in order to align and engage with the corresponding carbohydrate-binding sites of the CI-MPR.
It is intriguing that all the known ligand-binding sites of the CI-MPR are localized to the odd-numbered domains of the receptor (Figure 1). Whether other domains will be identified as binding novel ligands remains to be determined. However, it is unlikely that additional phosphomannosyl-binding sites will be identified in the CI-MPR. Our prediction is based on structural studies of several MRH domain-containing proteins (Roberts et al. 1998; Olson et al. 1999; Olson, Dahms et al. 2004; Olson, Yammani et al. 2004; Olson et al. 2010; Satoh et al. 2010; Olson et al. 2013) and our use of a structure-based sequence alignment coupled with experimental data that together have identified a spatially conserved signature motif of four residues (Gln, Arg, Glu and Tyr) within the binding pocket that are essential for mannose recognition. However, structural studies of the phosphomannosyl-binding sites in domain 9 and domain 15 of the CI-MPR are needed to confirm the mutagenesis studies (Hancock, Haskins et al. 2002) (and this report) that the signature motif residues including the newly identified Arg2170 predicted to be on β-strand 4 of domain 15 are localized in the binding pocket and directly interact with the mannose ring. Our analyses of the remaining domains of the CI-MPR reveal that no other domain of the bovine or human CI-MPR has more than one of these four conserved residues, and notably domain 7 does not contain any of these four positionally conserved residues (data not shown). In support of this prediction, we have shown that constructs encoding: (i) Domain 1 or domains 1–2 do not interact with a phosphomonoester-containing affinity column (Westlund et al. 1991) or the lysosomal enzyme, β-glucuronidase, as assessed by SPR analyses (Bohnsack et al. 2010), (ii) domains 1–8 containing a point mutation in the binding pocket of domain 3 that renders it nonfunctional does not interact with a phosphomonoester-containing affinity column (Dahms et al. 1993) and (iii) domains 7–15 in which the binding sites in domains 9 and 15 were inactivated by single amino acid substitution of a residue in their binding pocket exhibits no appreciable binding to GAA monoester or GAA diester as assessed by SPR analyses (Figure 6). Together, these data are consistent with only domains 3, 5, 9 and 15 of the CI-MPR having phosphomannosyl-binding activity.
In addition to acid hydrolases, either newly synthesized or exogenously administered as in the case of ERT, the CI-MPR may interact with other phosphomannosyl-containing ligands at the cell surface or in extracellular fluids. For example, the CI-MPR has been reported to interact with glycosylphosphatidyl inositol (GPI)-linked proteins (Green et al. 1995), and the newly identified binding site in domain 15, which is juxtaposed to the membrane, may be ideally positioned to interact with GPI-linked proteins on the cell surface. Supporting its role both intracellularly and at the cell surface, we have shown that domain 15 is capable of binding GAA monoester and GAA diester at pH values found in the Golgi (pH 6.5) (Figure 8, Table II) and at the cell surface (pH 7.4) (Figures 3–7). Furthermore, a soluble form of the CI-MPR, which contains most of the extracellular region of the receptor, is present in serum and extracellular fluids of several species, including human, and its expression is developmentally regulated (Kiess et al. 1987; Causin et al. 1988; MacDonald et al. 1989; Valenzano et al. 1995; Xu et al. 1998). The presence of a carbohydrate-binding site exposed at the C-terminal end of the soluble CI-MPR (domain 15) may allow this soluble form of the receptor to engage in multivalent interactions with phosphomannosyl-containing ligands in the circulation. Leukemia inhibitory factor, a secreted cytokine that stimulates cell proliferation and differentiation (Mathieu et al. 2012), is another example of a nonacid hydrolase that bears phosphomannosyl residues and its bioavailability has been shown to be regulated by the CI-MPR (Barnes et al. 2011) [see Dahms and Hancock (2002) and Sleat et al. (2013, 2006)] for list of additional M6P-containing proteins]. Therefore, additional studies are needed to understand the molecular basis of CI-MPR's ability to modulate the cellular location and activity of a diverse set of ligands.
Fig. 8.
Interaction between Dom7–15 and Dom14–15 constructs and the lysosomal enzyme GAA modified with either M6P (monoester) or M6P-GlcNAc (diester) at the pH of the Golgi. Representative sensorgrams collected at pH 6.5 showing the interaction of increasing concentrations of (A) Dom14–15 (120, 200, 400, 1000, 5000 and 20000 nM) and (B) Dom7–15 (5, 10, 15, 20, 40, 80, 120, 200, 400, and 1000 nM) with GAA monoester and GAA diester coupled CM5 surfaces. Response units (RU) are shown. This figure is available in black and white in print and in colour at Glycobiology online.
Table II.
Kinetic data obtained from SPR analyses at pH 6.5
| Construct |
KD (nM) |
|||||||
|---|---|---|---|---|---|---|---|---|
|
ka1 (M−1s−1 × 103) |
kd1 (s−1 × 10−3) |
Monoester |
Diester |
|||||
| Monoester | Diester | Monoester | Diester | kd1/ka1 | Steadystate | kd1/ka1 | Steadystate | |
| D7–15 | 101 | 53 | 7.42 | 0.0706 | 74 | 97 | 1300 | 926 |
| D14–15 | 6.4 | 4.6 | 0.11 | 0.14 | 17000 | 9600 | 29000 | 45000 |
|
KD2 (mM) |
||||||||
|
ka2 (M−1s−1 × 10−3) |
kd2 (s−1 × 10−3) |
kd2/ka2 |
||||||
| Construct | Monoester | Diester | Monoester | Diester | Monoester | Diester | ||
| D7–15 | 0.69 | 7.1 | 0.13 | 5.7 | 0.19 | 0.81 | ||
| D14–15 | 28 | 3.8 | 17.4 | 9.8 | 0.62 | 2.6 | ||
Kinetic and steady state affinity values determined from SPR sensorgrams obtained with GAA monoester and GAA diester immobilized on sensor chip surfaces. Purified recombinant protein was injected as the free analyte. Upper table reports kinetic parameters of the major component obtained from fitting sensorgrams to a two-state model with conformational change. The lower table reports kinetic parameters for the minor component with KD values reported in mM.
Materials and methods
Generation, expression and purification of CI-MPR constructs
All constructs were cloned from the bovine CI-MPR cDNA (residue numbers correspond to amino acid position in the mature CI-MPR protein): Domain 15 (Dom15, residues 2092–2296), domains 14–15 (Dom14–15, residues 1955–2296), domains 7–15, (Dom7–15, residues 888–2296), domains 1–14 (Dom1–14, residues 1–1953) and domains 1–15 (Dom1–15, residues 1–2296). Constructs were cloned into the pVL1392/3 baculovirus transfer vector. All constructs, unless noted, were generated with a C-terminal tag of six histidine residues that was preceded by a thrombin cleavage site (LVPRGSHHHHHH) to facilitate purification by nickel nitrilotriacetic acid (Ni-NTA) affinity chromatography. Mutant cDNAs were generated using DpnI mediated site-directed mutagenesis and confirmed by DNA sequencing. The constructs were expressed in the fall army worm ovarian cell line Spodoptera frugiperda (Sf9, Expression Systems, Davis, CA) and the cells grown in shaker flasks using ESF921 serum-free medium (Expression Systems) as described previously (Bohnsack et al. 2009). These constructs, which use the promoter from the polyhedrin gene for constitutive protein expression, were engineered in-frame with the native bovine CI-MPR signal sequence, resulting in proteins that are secreted from Sf9 cells. Recombinant baculovirus was added to serum-free medium at a multiplicity of infection of two and cultures were harvested after 4 days of growth at 27°C. Following removal of the cells by centrifugation, the medium was concentrated by filtration using Amicon stirred cells and dialyzed against Ni-NTA binding buffer (20 mM Tris, 300 mM NaCl, pH 8.0). The dialyzed medium was passed over a Ni-NTA agarose (Clontech, Mountain View, CA) column, washed and then eluted with Ni-NTA binding buffer containing 20–400 mM imidazole. The proteins were subjected to gel filtration using a Superose 12 column (10 × 300 mm) to isolate a single species. Dom15 also was expressed in E. coli using a pET28/sumo vector. Cells were grown to an OD of 0.8 at 37°C before induction with 0.5 mM IPTG. Cultures were allowed to grow overnight at 16°C before harvesting. Isotopically labeled protein was generated by substituting [15N] ammonium sulfate for the unlabeled compound in the growth medium. Protein was isolated and refolded from inclusion bodies as previously described (Olson et al. 2013). The Bradford protein assay (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard was used to determine protein concentrations.
Purification of soluble CI-MPR from fetal bovine serum
A soluble form of the CI-MPR (sCI-MPR) was purified from fetal bovine serum by phosphomannan affinity chromatography as described previously (Valenzano et al. 1995).
Biosensor studies
All SPR measurements were performed at 25°C using a Biacore 3000 instrument (BIAcore, GE Healthcare, Piscataway, NJ) as described previously (Bohnsack et al. 2009) and using the lysosomal enzyme GAA with defined N-glycans (Chavez et al. 2007). Purified proteins (GAA monoester and GAA diester) were immobilized on CM5 sensor chips following activation of the surface using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and NHS as recommended by the manufacturer. After coupling, unreacted NHS was blocked with ethanolamine. The reference surface was treated in the same way except that protein was omitted. Samples of purified truncated CI-MPR proteins (Dom15, Dom14–15, Dom14–15mYF, Dom14–15mRA, Dom7–15, Dom7–15m9, Dom7–15m15YF, Dom7–15m15RA, Dom1–15 and Dom1–14) were prepared in 50 mM imidazole, 150 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 5 mM CaCl2 pH 7.4 supplemented with 0.005% (v/v) P20 and were injected in a volume of 80 µl over the coupled and reference flow cells at a flow rate of 40 µl/min. After 2 min, the solutions containing the purified proteins were replaced with buffer and the complexes were allowed to dissociate for 2 min. The sensor chip surface was regenerated with a 20 µl injection 10 mM HCl at a flow rate of 10 µl/min. The surface was allowed to re-equilibrate in running buffer for 1 min prior to subsequent injections. The response at equilibrium (Req) for each concentration of protein was determined by averaging the response over a 10 s period within the steady state region of the sensorgram using the BIAevaluation software package (version 4.0.1). The response at equilibrium (Req) was plotted versus the concentration of protein and fit to a 1 : 1 binding isotherm. Because steady state was not achieved in the cases of Dom1–14 and Dom1–15, data were also analyzed using a two-state reaction with conformational change-binding (BIAevaluation software package version 4.0.1). In all cases, the minor component made an insignificant contribution to the overall affinity and as such only the kinetic components of the major binding component are used. All response data were double-referenced (Myszka 2000), where controls for the contribution of the change in refractive index were performed in parallel with flow cells derivatized in the absence of protein and subtracted from all binding sensorgrams. Inhibition studies were performed using increasing concentrations of M6P or 1 and 5 mM glucose 6-phosphate (G6P). G6P, which is the 2-epimer of M6P, is used as a control because it had been shown previously to bind with low affinity (KD = 10–80 mM) to the full-length CI-MPR (Tong and Kornfeld 1989).
Interrogation of glycan microarray with MPRs
The phosphorylated glycan microarray used in these studies was prepared using a selection of the glycans (printed in replicates of n = 4) prepared as described previously; however, some glycans were no longer available (Song et al. 2009). Interaction between MPRs and purified glycans was determined as described previously (Bohnsack et al. 2009; Song et al. 2009) using 50 mM imidazole, pH 6.5, 150 mM NaCl, 10 mM MnCl2. To demonstrate the presence of the high mannose N-glycans and their phosphorylated derivatives, the array was interrogated with biotin-ConA (0.5 µg/mL) in 50 mM imidazole, pH 6.5 containing, 150 mM NaCl, 10 mM MnCl2, 1% BSA and 0.05% Tween 20 (binding buffer), and washes were with the same buffer without BSA at room temperature for 1 h. After washing, the array was incubated with Cy5-labeled Streptavidin (0.5 µg/mL) in binding buffer; and after washing, slides were scanned for fluorescence in a microarray scanner (PerkinElmer, Waltham, MA). The signals were processed as previously described to generate histograms showing average RFU associated with each printed glycan (n = 4). Structures associated with the Glycan ID are shown in Figure 5E. The soluble CI-MPR and Dom14–15 were similarly analyzed at 5 and 150 µg/mL, respectively. CI-MPR binding was detected using a 1:250 dilution of the specific polyclonal antiserum B14.5 followed by C5-labeled goat anti-rabbit IgG (5 µg/mL). Dom14–15 binding was detected using 5 µg/mL of an anti-penta-His monoclonal antibody (Life Technologies, Grand Island, NY) followed by Alexa633-labeled goat anti-mouse IgG (5 µg/mL).
1H-15N HSQC of E. coli-expressed domain 15 protein
NMR spectra were acquired at 25°C on a Bruker 600-MHz spectrometer equipped with a triple resonance CryoProbe™. Spectral data were processed using NMRPipe (Delaglio et al. 1995) and visualized using XEASY (Bartels et al. 1995).
Supplementary data
Supplementary data for this article are available online at http://glyco.oxfordjournals.org.
Funding
This work was supported in part by the National Institutes of Health grants (R01DK042667 to N.M.D. and P41GM10394 to R.D.C.). The National Center for Functional Glycomics is supported by National Institutes of Health grant P41GM103694 to R.D.C.
Conflict of interest statement
None declared.
Abbreviations
AEAB, 2-amino-N-(2-aminoethyl)-benzamide; CD-MPR, cation-dependent mannose 6-phosphate receptor; CI-MPR, cation-independent mannose 6-phosphate receptor; ConA, concanavalin A; CRD, carbohydrate recognition domain; ER, endoplasmic reticulum; ERT, enzyme replacement treatment; G6p, glucose 6-phosphate; GAA, acid α-glucosidase; GIIβ, β-subunit of the heterodimeric glucosidase II; GlcNAc, N-acetylglucosamine; GPI, glycosylphosphatidyl inositol; HSQC, heteronuclear single quantum coherence; IgG, immunoglobulin G; IGF-II, insulin-like growth factor II; M6P, mannose 6-phosphate; MRH, Mannose 6-phosphate Receptor Homology; NHS, N-hydroxysuccinimide; Ni-NTA, nickel nitriloacetic acid; NMR, nuclear magnetic resonance; SPR, surface plasmon resonance; TGN, trans Golgi network; UDP, uridine diphosphate.
Supplementary Material
Acknowledgements
We thank Dr. Jung-Ja Kim for critical reading of the manuscript, and Richard Bohnsack for technical assistance. The BIAcore 3000 instrument was purchased through a grant from the Advancing a Healthier Wisconsin program.
References
- Baenziger JU, Fiete D. 1979. Structural determinants of concanavalin A specificity for oligosaccharides. J Biol Chem. 254:2400–2407. [PubMed] [Google Scholar]
- Bao M, Booth JL, Elmendorf BJ, Canfield WM. 1996. Bovine UDP-N-acetylglucosamine:lysosomal-enzyme N-acetylglucosamine-1-phosphotransferase. I. Purification and subunit structure. J Biol Chem. 271:31437–31445. [DOI] [PubMed] [Google Scholar]
- Barnes J, Lim JM, Godard A, Blanchard F, Wells L, Steet R. 2011. Extensive mannose phosphorylation on leukemia inhibitory factor (LIF) controls its extracellular levels by multiple mechanisms. J Biol Chem. 286:24855–24864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 6:1–10. [DOI] [PubMed] [Google Scholar]
- Bohnsack RN, Patel M, Olson LJ, Twining SS, Dahms NM. 2010. Residues essential for plasminogen binding by the cation-independent mannose 6-phosphate receptor. Biochemistry. 49:635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack RN, Song X, Olson LJ, Kudo M, Gotschall RR, Canfield WM, Cummings RD, Smith DF, Dahms NM. 2009. Cation-independent mannose 6-phosphate receptor: A composite of distinct phosphomannosyl binding sites. J Biol Chem. 284:35215–35226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen M, Vogel P, Platt KA, Dahms N, Kornfeld S. 2009. Mice lacking mannose 6-phosphate uncovering enzyme activity have a milder phenotype than mice deficient for N-acetylglucosamine-1-phosphotransferase activity. Mol Biol Cell. 20:4381–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady RO. 2006. Enzyme replacement for lysosomal diseases. Annu Rev Med. 57:283–296. [DOI] [PubMed] [Google Scholar]
- Braulke T, Bonifacino JS. 2009. Sorting of lysosomal proteins. Biochim Biophys Acta. 1793:605–614. [DOI] [PubMed] [Google Scholar]
- Brenckle R, Kornfeld R. 1980. Structure of the oligosaccharides of mouse immunoglobulin M secreted by the MOPC 104E plasmacytoma. Arch Biochem Biophys. 201:160–173. [DOI] [PubMed] [Google Scholar]
- Brewer CF, Bhattacharyya L. 1986. Specificity of concanavalin A binding to asparagine-linked glycopeptides. A nuclear magnetic relaxation dispersion study. J Biol Chem. 261:7306–7310. [PubMed] [Google Scholar]
- Brown J, Delaine C, Zaccheo OJ, Siebold C, Gilbert RJ, van Boxel G, Denley A, Wallace JC, Hassan AB, Forbes BE, et al. 2008. Structure and functional analysis of the IGF-II/IGF2R interaction. EMBO J. 27:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Esnouf RM, Jones MA, Linnell J, Harlos K, Hassan AB, Jones EY. 2002. Structure of a functional IGF2R fragment determined from the anomalous scattering of sulfur. EMBO J. 21:1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Jones EY, Forbes BE. 2009. Keeping IGF-II under control: Lessons from the IGF-II-IGF2R crystal structure. Trends Biochem Sci. 34:612–619. [DOI] [PubMed] [Google Scholar]
- Castonguay AC, Lasanajak Y, Song X, Olson LJ, Cummings RD, Smith DF, Dahms NM. 2012. The glycan-binding properties of the cation-independent mannose 6-phosphate receptor are evolutionary conserved in vertebrates. Glycobiology. 22:983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castonguay AC, Olson LJ, Dahms NM. 2011. Mannose 6-phosphate receptor homology (MRH) domain-containing lectins in the secretory pathway. Biochim Biophys Acta. 1810:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causin C, Waheed A, Braulke T, Junghans U, Maly P, Humbel RE, von Figura K. 1988. Mannose 6-phosphate/insulin-like growth factor II-binding proteins in human serum and urine. Their relation to the mannose 6-phosphate/insulin-like growth factor II receptor. Biochem J. 252:795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez CA, Bohnsack RN, Kudo M, Gotschall RR, Canfield WM, Dahms NM. 2007. Domain 5 of the cation-independent mannose 6-phosphate receptor preferentially binds phosphodiesters (mannose 6-phosphate N-acetylglucosamine ester). Biochemistry. 46:12604–12617. [DOI] [PubMed] [Google Scholar]
- D'Alessio C, Dahms NM. 2015. Glucosidase II and MRH domain-containing proteins in the secretory pathway. Curr Protein Pept Sci. 16:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms NM, Hancock MK. 2002. P-type lectins. Biochim Biophys Acta. 1572:317–340. [DOI] [PubMed] [Google Scholar]
- Dahms NM, Olson LJ, Kim JJ. 2008. Strategies for carbohydrate recognition by the mannose 6-phosphate receptors. Glycobiology. 18:664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms NM, Rose PA, Molkentin JD, Zhang Y, Brzycki MA. 1993. The bovine mannose 6-phosphate/insulin-like growth factor II receptor. The role of arginine residues in mannose 6-phosphate binding. J Biol Chem. 268:5457–5463. [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Vax A. 1995. NMRPipe: A multidemensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- Devi GR, Byrd JC, Slentz DH, MacDonald RG. 1998. An insulin-like growth factor II (IGF-II) affinity-enhancing domain localized within extracytoplasmic repeat 13 of the IGF-II/mannose 6-phosphate receptor. Mol Endocrinol. 12:1661–1672. [DOI] [PubMed] [Google Scholar]
- Do H, Lee WS, Ghosh P, Hollowell T, Canfield W, Kornfeld S. 2002. Human mannose 6-phosphate-uncovering enzyme is synthesized as a proenzyme that is activated by the endoprotease furin. J Biol Chem. 277:29737–29744. [DOI] [PubMed] [Google Scholar]
- Gelfman CM, Vogel P, Issa TM, Turner CA, Lee WS, Kornfeld S, Rice DS. 2007. Mice lacking alpha/beta subunits of GlcNAc-1-phosphotransferase exhibit growth retardation, retinal degeneration, and secretory cell lesions. Invest Ophthalmol Vis Sci. 48:5221–5228. [DOI] [PubMed] [Google Scholar]
- Green PJ, Ferguson MA, Robinson PJ, Feizi T. 1995. The cation-independent mannose-6-phosphate receptor binds to soluble GPI-linked proteins via mannose-6-phosphate. FEBS Lett. 360:34–38. [PubMed] [Google Scholar]
- Hancock MK, Haskins DJ, Sun G, Dahms NM. 2002. Identification of residues essential for carbohydrate recognition by the insulin-like growth factor II/mannose 6-phosphate receptor. J Biol Chem. 277:11255–11264. [DOI] [PubMed] [Google Scholar]
- Hancock MK, Yammani RD, Dahms NM. 2002. Localization of the carbohydrate recognition sites of the insulin-like growth factor II/mannose 6-phosphate receptor to domains 3 and 9 of the extracytoplasmic region. J Biol Chem. 277:47205–47212. [DOI] [PubMed] [Google Scholar]
- Hasilik A, Klein U, Waheed A, Strecker G, von Figura K. 1980. Phosphorylated oligosaccharides in lysosomal enzymes: Identification of alpha-N-acetylglucosamine(1)phospho(6)mannose diester groups. Proc Natl Acad Sci USA. 77:7074–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoflack B, Fujimoto K, Kornfeld S. 1987. The interaction of phosphorylated oligosaccharides and lysosomal enzymes with bovine liver cation-dependent mannose 6-phosphate receptor. J Biol Chem. 262:123–129. [PubMed] [Google Scholar]
- Hoflack B, Kornfeld S. 1985. Purification and characterization of a cation-dependent mannose 6-phosphate receptor from murine P388D1 macrophages and bovine liver. J Biol Chem. 260:12008–12014. [PubMed] [Google Scholar]
- Kasper D, Dittmer F, von Figura K, Pohlmann R. 1996. Neither type of mannose 6-phosphate receptor is sufficient for targeting of lysosomal enzymes along intracellular routes. J Cell Biol. 134:615–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiess W, Greenstein LA, White RM, Lee L, Rechler MM, Nissley SP. 1987. Type II insulin-like growth factor receptor is present in rat serum. Proc Natl Acad Sci USA. 84:7720–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Olson LJ, Dahms NM. 2009. Carbohydrate recognition by the mannose-6-phosphate receptors. Curr Opin Struc Biol. 19:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R, Bao M, Brewer K, Noll C, Canfield W. 1999. Molecular cloning and functional expression of two splice forms of human N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase. J Biol Chem. 274:32778–32785. [DOI] [PubMed] [Google Scholar]
- Kornfeld S, Sly WS. 2001. I cell disease and pseudo-Hurler polydystrophy: Disorders of lysosomal enzyme phosphorylation and localization. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Metabolic and Molecular Bases of Inherited Diseases. New York: McGraw Hill; p. 3469–3482. [Google Scholar]
- Kudo M, Bao M, D'Souza A, Ying F, Pan H, Roe BA, Canfield WM. 2005. The alpha- and beta-subunits of the human UDP-N-acetylglucosamine:lysosomal enzyme phosphotransferase are encoded by a single cDNA. J Biol Chem. 280:36141–36149. [DOI] [PubMed] [Google Scholar]
- Kudo M, Brem MS, Canfield WM. 2006. Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase alpha/beta -subunits precursor gene. Am J Hum Genet. 78:451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leksa V, Godar S, Cebecauer M, Hilgert I, Breuss J, Weidle UH, Horejsi V, Binder BR, Stockinger H. 2002. The N terminus of mannose 6-phosphate/insulin-like growth factor 2 receptor in regulation of fibrinolysis and cell migration. J Biol Chem. 277:40575–40582. [DOI] [PubMed] [Google Scholar]
- Linnell J, Groeger G, Hassan AB. 2001. Real time kinetics of insulin-like growth factor II (IGF-II) interaction with the IGF-II/mannose 6-phosphate receptor. The effects of domain 13 and pH. J Biol Chem. 276:23986–23991. [DOI] [PubMed] [Google Scholar]
- Ludwig T, Munier-Lehmann H, Bauer U, Hollinshead M, Ovitt C, Lobel P, Hoflack B. 1994. Differential sorting of lysosomal enzymes in mannose 6-phosphate receptor-deficient fibroblasts. EMBO J. 13:3430–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald RG, Tepper MA, Clairmont KB, Perregaux SB, Czech MP. 1989. Serum form of the rat insulin-like growth factor II/mannose 6-phosphate receptor is truncated in the carboxyl-terminal domain. J Biol Chem. 264:3256–3261. [PubMed] [Google Scholar]
- Marron-Terada PG, Hancock MK, Haskins DJ, Dahms NM. 2000. Recognition of Dictyostelium discoideum lysosomal enzymes is conferred by the amino-terminal carbohydrate binding site of the insulin-like growth factor II/mannose 6-phosphate receptor. Biochemistry. 39:2243–2253. [DOI] [PubMed] [Google Scholar]
- Mathieu ME, Saucourt C, Mournetas V, Gauthereau X, Theze N, Praloran V, Thiebaud P, Boeuf H. 2012. LIF-dependent signaling: New pieces in the Lego. Stem Cell Rev. 8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVie-Wylie AJ, Lee KL, Qiu H, Jin X, Do H, Gotschall R, Thurberg BL, Rogers C, Raben N, O'Callaghan M, et al. 2008. Biochemical and pharmacological characterization of different recombinant acid alpha-glucosidase preparations evaluated for the treatment of Pompe disease. Mol Genet Metab. 94:448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. 2001. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr Biol. 11:R499–R501. [DOI] [PubMed] [Google Scholar]
- Myszka DG. 2000. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 323:325–340. [DOI] [PubMed] [Google Scholar]
- Olson LJ, Dahms NM, Kim JJ. 2004. The N-terminal carbohydrate recognition site of the cation-independent mannose 6-phosphate receptor. J Biol Chem. 279:34000–34009. [DOI] [PubMed] [Google Scholar]
- Olson LJ, Orsi R, Alculumbre SG, Peterson FC, Stigliano ID, Parodi AJ, D'Alessio C, Dahms NM. 2013. Structure of the lectin mannose 6-phosphate receptor homology (MRH) domain of glucosidase II, an enzyme that regulates glycoprotein folding quality control in the endoplasmic reticulum. J Biol Chem. 288:16460–16475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LJ, Peterson FC, Castonguay A, Bohnsack RN, Kudo M, Gotschall RR, Canfield WM, Volkman BF, Dahms NM. 2010. Structural basis for recognition of phosphodiester-containing lysosomal enzymes by the cation-independent mannose 6-phosphate receptor. Proc Natl Acad Sci USA. 107:12493–12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LJ, Yammani RD, Dahms NM, Kim JJ. 2004. Structure of uPAR, plasminogen, and sugar-binding sites of the 300 kDa mannose 6-phosphate receptor. EMBO J. 23:2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LJ, Zhang J, Lee YC, Dahms NM, Kim J-JP. 1999. Structural basis for recognition of phosphorylated high mannose oligosaccharides by the cation-dependent mannose 6-phosphate receptor. J Biol Chem. 274:29889–29896. [DOI] [PubMed] [Google Scholar]
- Pohlmann R, Boeker MW, von Figura K. 1995. The two mannose 6-phosphate receptors transport distinct complements of lysosomal proteins. J Biol Chem. 270:27311–27318. [DOI] [PubMed] [Google Scholar]
- Reddy ST, Chai W, Childs RA, Page JD, Feizi T, Dahms NM. 2004. Identification of a low affinity mannose 6-phosphate-binding site in domain 5 of the cation-independent mannose 6-phosphate receptor. J Biol Chem. 279:38658–38667. [DOI] [PubMed] [Google Scholar]
- Roberts DL, Weix DJ, Dahms NM, Kim J-JP. 1998. Molecular basis of lysosomal enzyme recognition: Three-dimensional structure of the cation-dependent mannose 6-phosphate receptor. Cell. 93:639–648. [DOI] [PubMed] [Google Scholar]
- Rohrer J, Kornfeld R. 2001. Lysosomal hydrolase mannose 6-phosphate uncovering enzyme resides in the trans-Golgi network. Mol Biol Cell. 12:1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y. 2010. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell. 40:905–916. [DOI] [PubMed] [Google Scholar]
- Sleat DE, Lobel P. 1997. Ligand binding specificities of the two mannose 6-phosphate receptors. J Biol Chem. 272:731–738. [DOI] [PubMed] [Google Scholar]
- Sleat DE, Sun P, Wiseman JA, Huang L, El-Banna M, Zheng H, Moore DF, Lobel P. 2013. Extending the mannose 6-phosphate glycoproteome by high resolution/accuracy mass spectrometry analysis of control and acid phosphatase 5-deficient mice. Mol Cell Prot. 12:1806–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat DE, Wang Y, Sohar I, Lackland H, Li Y, Li H, Zheng H, Lobel P. 2006. Identification and validation of mannose 6-phosphate glycoproteins in human plasma reveal a wide range of lysosomal and non-lysosomal proteins. Mol Cell Prot. 5:1942–1956. [DOI] [PubMed] [Google Scholar]
- Sohar I, Sleat D, Gong Liu C, Ludwig T, Lobel P. 1998. Mouse mutants lacking the cation-independent mannose 6- phosphate/insulin-like growth factor II receptor are impaired in lysosomal enzyme transport: Comparison of cation-independent and cation-dependent mannose 6-phosphate receptor-deficient mice. Biochem J. 330:903–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Heimburg-Molinaro J, Dahms NM, Smith DF, Cummings RD. 2012. Preparation of a mannose- 6-phosphate glycan microarray through fluorescent derivatization, phosphorylation, and immobilization of natural high-mannose N-glycans and application in ligand identification of P-type lectins. Methods Mol Biol. 808:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lasanajak Y, Olson LJ, Boonen M, Dahms NM, Kornfeld S, Cummings RD, Smith DF. 2009. Glycan microarray analysis of P-type lectins reveals distinct phosphomannose glycan recognition. J Biol Chem. 284:35201–35214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede S, Storch S, Lubke T, Henrissat B, Bargal R, Raas-Rothschild A, Braulke T. 2005. Mucolipidosis II is caused by mutations in GNPTA encoding the alpha/beta GlcNAc-1-phosphotransferase. Nature Med. 11:1109–1112. [DOI] [PubMed] [Google Scholar]
- Tong PY, Kornfeld S. 1989. Ligand interactions of the cation-dependent mannose 6-phosphate receptor. Comparison with the cation-independent mannose 6-phosphate receptor. J Biol Chem. 264:7970–7975. [PubMed] [Google Scholar]
- Uson I, Schmidt B, von Bulow R, Grimme S, von Figura K, Dauter M, Rajashankar KR, Dauter Z, Sheldrick GM. 2003. Locating the anomalous scatterer substructures in halide and sulfur phasing. Acta Crystallog Section D. 59:57–66. [DOI] [PubMed] [Google Scholar]
- Valenzano KJ, Remmler J, Lobel P. 1995. Soluble insulin-like growth factor II/mannose 6-phosphate receptor carries multiple high molecular weight forms of insulin-like growth factor II in fetal bovine serum. J Biol Chem. 270:16441–16448. [DOI] [PubMed] [Google Scholar]
- Varki A, Kornfeld S. 1980. Structural studies of phosphorylated high mannose-type oligosaccharides. J Biol Chem. 255:10847–10858. [PubMed] [Google Scholar]
- Varki A, Sherman W, Kornfeld S. 1983. Demonstration of the enzymatic mechanisms of alpha-N-acetyl-D-glucosamine-1-phosphodiester N-acetylglucosaminidase (formerly called alpha-N-acetylglucosaminylphosphodiesterase) and lysosomal alpha-N-acetylglucosaminidase. Arch Biochem Biophys. 222:145–149. [DOI] [PubMed] [Google Scholar]
- Waheed A, Hasilik A, von Figura K. 1981. Processing of the phosphorylated recognition marker in lysosomal enzymes. Characterization and partial purification of a microsomal alpha-N-acetylglucosaminyl phosphodiesterase. J Biol Chem. 256:5717–5721. [PubMed] [Google Scholar]
- Westlund B, Dahms NM, Kornfeld S. 1991. The bovine mannose 6-phosphate/insulin-like growth factor II receptor. Localization of mannose 6-phosphate binding sites to domains 1–3 and 7–11 of the extracytoplasmic region. J Biol Chem. 266:23233–23239. [PubMed] [Google Scholar]
- Xu Y, Papageorgiou A, Polychronakos C. 1998. Developmental regulation of the soluble form of insulin-like growth factor-II/mannose 6-phosphate receptor in human serum and amniotic fluid. J Clin Endocrinol Metab. 83:437–442. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Jiang JL, Gumlaw NK, Zhang J, Bercury SD, Ziegler RJ, Lee K, Kudo M, Canfield WM, Edmunds T, et al. 2009. Glycoengineered acid alpha-glucosidase with improved efficacy at correcting the metabolic aberrations and motor function deficits in a mouse model of Pompe disease. Mol Therapy. 17:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li X, McVie-Wylie A, Jiang C, Thurberg BL, Raben N, Mattaliano RJ, Cheng SH. 2005. Carbohydrate-remodelled acid alpha-glucosidase with higher affinity for the cation-independent mannose 6-phosphate receptor demonstrates improved delivery to muscles of Pompe mice. Biochem J. 389:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.