SUMMARY
Squamous cell carcinoma (SCC) of the lung is the second most common subtype of lung cancer. With limited treatment options, the 5-year survival rate of SCC is only 15%. Although genomic alterations in SCC have been characterized, identifying the alterations that drive SCC is critical for improving treatment strategies. Mouse models of SCC are currently limited. Using lentiviral delivery of Sox2 specifically to the mouse lung, we tested the ability of Sox2 to promote tumorigenesis in multiple tumor suppressor backgrounds. Expression of Sox2, frequently amplified in human SCC, specifically cooperates with loss of Lkb1 to promote squamous lung tumors. Mouse tumors exhibit characteristic histopathology and biomarker expression similar to human SCC. They also mimic human SCCs by activation of therapeutically relevant pathways including STAT and mTOR. This model may be utilized to test the contribution of additional driver alterations in SCC, as well as for preclinical drug discovery.
INTRODUCTION
Lung cancer is a leading cause of cancer-related deaths in the United States and worldwide. Squamous cell carcinoma (SCC) is the second most common type of lung cancer, representing ~30% of cases and over 400,000 deaths worldwide each year (Cancer Genome Atlas Research Network, 2012; Jemal et al., 2011). Currently, the 5-year survival rate for SCC is approximately 15%. One of the major problems contributing to treatment failure is a lack of targeted therapies. Targeted therapies that are effective against adenocarcinoma, the other major subtype of non-small-cell lung cancer (NSCLC), are ineffective or contraindicated for SCC. The current standard of care for SCC involves surgery when operable, or combination chemotherapy, usually a platinum doublet, which has a poor response rate (Oliver et al., 2013). Identification of new therapeutic targets for SCC requires elucidation of the critical genes and pathways driving this disease.
Cancer Genome Atlas Research Network (2012) recently sequenced 178 human SCCs, identifying numerous genetic alterations that may serve as valuable therapeutic targets. However, SCC has one of the highest mutation rates of all tumor types, making it difficult to distinguish “driver” from “passenger” mutations. Some genetic alterations in SCC occur in proteins for which targeted therapies are available, such as fibroblast growth factor receptor (FGFR) and phosphatidyl inositol 3-kinase (PI3K)/ AKT. Other potential oncogenic drivers were discovered for which there are currently no available therapies, such as the transcription factors SOX2, P63, and NRF2 (Cancer Genome Atlas Research Network, 2012).
Mouse models of adenocarcinoma and small cell lung cancer (SCLC) have been utilized to understand mechanisms of tumor initiation, progression, and therapeutic response (Kwon and Berns, 2013). Genetic models of SCC have only recently been generated (Ji et al., 2007; Xiao et al., 2013). The combination of KrasG12D expression and Lkb1 loss in the mouse lung leads to lung tumors of multiple lineages (adenocarcinoma, mixed adenosquamous, squamous, and large cell) (Jackson et al., 2001; Ji et al., 2007). While KRAS is commonly mutated in human lung adenocarcinomas (~21%), it is rarely altered in SCC (~6%) (Perez-Moreno et al., 2012). In the mouse lung, KrasG12D expression alone promotes the exclusive development of lung adenocarcinomas (Jackson et al., 2001). LKB1 (also known as serine/threonine kinase 11; STK11), a tumor suppressor implicated in metabolism, cell polarity, and growth control, is lost or mutated in 5%–19% of human SCCs (Ji et al., 2007; Perez-Moreno et al., 2012; Shackelford and Shaw, 2009). This suggests that Lkb1 loss in the KrasG12D/+Lkb1fl/fl model contributes to the altered spectrum of lung tumor types, including squamous tumors.
Recently, a kinase-dead IKKα knockin mouse was reported to develop lung SCCs (Xiao et al., 2013). This discovery followed other mouse models with loss-of-function IKKα alleles that developed papillomas and skin SCC (Liu et al., 2012). The kinase-dead IKKα knockin mice develop spontaneous SCCs of the lung but also develop tumors in the skin, forestomach, and esophagus, contributing to early mortality (Xiao et al., 2013). When wild-type IKKα expression is restored using a skin promoter, mice survive longer and develop lung SCCs but still have defects in the forestomach and esophagus. These phenotypes are consistent with the observation that IKKα is downregulated in human skin SCC and in head and neck SCC (Liu et al., 2012), but the role of IKKα in lung SCC is less clear. Copy number losses and genomic mutations in IKKα are rare in human lung SCCs, and other mechanisms of altering IKKα and its related pathways are not well defined (Cancer Genome Atlas Research Network, 2012). The broad spectrum of lung tumor types in KrasG12D/+Lkb1fl/fl mice and the extrapulmonary phenotypes in the kinase-dead IKKα mice make the study of biomarkers, mechanisms of progression, and squamous-specific therapies difficult.
SOX2 is one of the most frequently altered genes in human SCC, amplified in ~21% and overexpressed in 60%–90% of tumors (Bass et al., 2009; Brcic et al., 2012; Cancer Genome Atlas Research Network, 2012; Hussenet et al., 2010). SOX2 is also frequently expressed in early-stage SCC, suggesting that SOX2 expression may be an initiating event in SCC development (Bass et al., 2009; Brcic et al., 2012; Hussenet et al., 2010). Gain-of-function and loss-of-function studies have previously underscored a critical role for Sox2 in lung development, specifically its importance in the proliferation and differentiation of basal and neuroendocrine cells (Gontan et al., 2008; Lu et al., 2010; Que et al., 2009). Consistent with this observation, SOX2 amplification is common in SCC and SCLC, lung tumors that are thought to originate from basal and neuroendocrine cells, respectively (Bass et al., 2009; Hussenet et al., 2010; Rudin et al., 2012). However, expression of Sox2 alone in the lung promotes hyperplasia and tumors of the adenocarcinoma lineage with aberrant basal cell marker expression (Lu et al., 2010). Although SOX2 is one of the most frequent genetic alterations associated with human SCC, it has not yet been shown to promote squamous lung tumors in vivo. Here, we sought to identify combinations of gene drivers that promote lung SCC.
RESULTS AND DISCUSSION
Lentiviral Approach to Identify Combinatorial Drivers of Lung SCC
To identify genetic combinations that facilitate Sox2-driven SCC, we used a lentiviral approach to combine Sox2 expression with loss of distinct tumor suppressor genes. Bicistronic lentiviruses expressing Sox2 and Cre recombinase are delivered specifically to the mouse lung using intranasal inhalation, allowing constitutive expression of two genes driven by β-Actin and Pgk promoters, respectively. Lentiviruses are administered to mice harboring conditional LoxP-flanked (“floxed”) tumor suppressor alleles to simultaneously create two genetic “hits”: expression of Sox2 via the lentivirus and deletion of a tumor suppressor gene using the Cre/LoxP system (Figure S1A). Control cDNA (GFP) or murine Sox2 were cloned into bicistronic lentiviral vectors under control of the β-Actin promoter (referred to as Lenti-GFP-Cre or Lenti-Sox2-Cre, respectively). Sox2 expression was validated by immunoblot analysis and Cre expression was verified using a human embryonic kidney 293T (HEK293T) reporter system that was used to calculate viral titer (Figures S1B and S1C).
Mice harboring conditional LoxP-flanked tumor suppressor alleles (Lkb1fl/fl, p53fl/fl, or p53fl/flRbfl/fl) were infected with Lenti-GFP-Cre or Lenti-Sox2-Cre. LKB1 is lost or mutated in 5%–19% of human SCCs, but loss of Lkb1 alone in the mouse lung does not lead to tumor formation (Ji et al., 2007). TP53 is one of the most common genetic alterations in SCC (mutated in ~81% of tumors), but loss of p53 alone in the mouse lung leads to rare adenocarcinomas after long latencies (Cancer Genome Atlas Research Network, 2012; Jackson et al., 2005; Meuwissen et al., 2003). Both TP53 and RB1 alterations are common in human SCLC (in which SOX2 is genomically amplified in ~27% of cases), and loss of p53/Rb in the mouse lung leads to SCLC after long latencies, as well as rare adenocarcinomas (Cancer Genome Atlas Research Network, 2012; Meuwissen et al., 2003; Rudin et al., 2012). We reasoned that these tumor suppressor genes may cooperate with Sox2 expression to promote lung tumor development. Beginning at 3 months postinfection, mice were monitored for tumor formation every month by low-dose microcomputed tomography (microCT) imaging for 7.5–12 months or until tumors were detected. The Sox2-Lkb1 combination had the greatest reduction in tumor-free survival compared to other genetic combinations tested (Figure S1D). Individual tumors were observed by microCT in all three conditional genetic backgrounds and analyzed by histopathology (Figures 1A and 1B; Table 1). It is striking that most of the tumors in the Lenti-Sox2-Cre Lkb1fl/fl combination (n = 7 of 9 tumors in 17 mice) were classified as squamous tumors, as determined by analysis of hematoxylin and eosin (H&E)-stained sections by two independent pathologists (Figure 1B; Table 1). Sox2-Lkb1 squamous tumors exhibited characteristic SCC features, including keratin pearls and desmoplastic stroma (Brcic et al., 2012) (Figure 1B). Lkb1fl/fl mice receiving Lenti-GFP-Cre did not develop tumors (n = 0 of 14 mice) as expected (Table 1). Sox2 expression in the context of p53 loss or loss of p53/Rb did not lead to squamous lung tumors, but it did lead to a few adenocarcinomas at latencies of 5–12 months (Table 1). The Sox2-Lkb1 combination exhibited significantly increased tumor incidence (Fisher’s exact test, two sided, p = 0.009) and a significant enrichment in squamous tumors (89% versus 0%, Fisher’s exact test, two sided, p = 0.001), compared to all other genetic combinations combined (Table 1). Together, these findings suggest that Sox2 expression and Lkb1 loss specifically cooperate to promote squamous tumorigenesis.
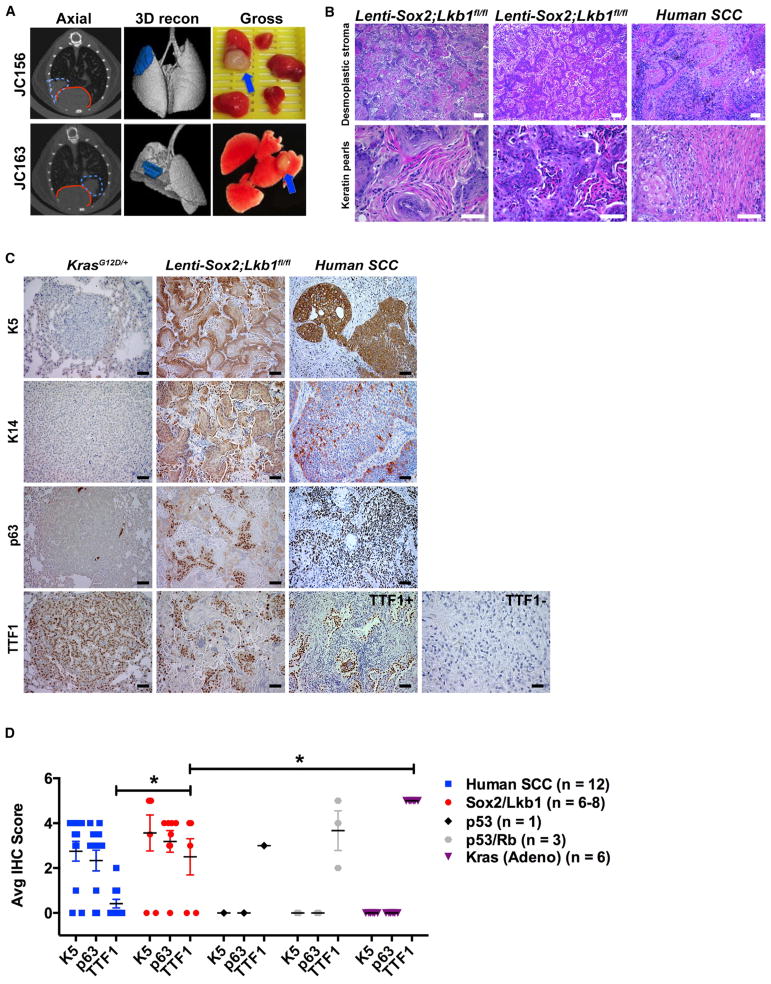
Figure 1. Lenti-Sox2;Lkb1fl/fl Tumors Express Biomarkers of Human SCC.
(A) Left panels: two representative microCT images with tumor outlined in dashed blue lines and heart outlined in solid red lines in axial view. Middle panels: three-dimensional microCT reconstructions (3D recon) with lung tumors in blue. Right panels: gross morphology of dissected lungs with tumors indicated by blue arrows. Mouse IDs (JC156 and JC163) correspond to tumors in Table 1.
(B) Lenti-Sox2;Lkb1fl/fl tumors and human SCC stained with H&E. Top scale bars represent 100 μM; bottom scale bars represent, 50 μM.
(C) Representative KrasG12D/+ mouse lung adenocarcinomas, Lenti-Sox2;Lkb1fl/fl tumors, or human lung SCCs stained with K5, K14, p63, or TTF1. Scale bar represents 50 μM.
(D) Average IHC score based on 0–5 scoring system where 5 indicates >90% positive; 4 indicates >75%; 3 indicates >50%; 2 indicates >25%; 1 indicates >10%; and 0 indicates negative. Human SCCs (n = 12), tumors from Table 1, and KrasG12D/+ adenocarcinomas were compared for K5, p63, and TTF1 IHC. Number of tumors analyzed is indicated in the color key. Error bars represent mean ± SEM. Student’s unpaired t test, *p = 0.02 for Sox2/Lkb1 versus Kras, and p = 0.04 for Sox2/Lkb1 versus human SCC.
See also Figure S1.
Table 1.
Lenti-Sox2;Lkb1fl/fl Mice Develop Squamous Lung Tumors
| Genotype | Virus delivered | No. of mice with tumors/ total mice | p value for tumor incidence Sox2 versus GFP | Mouse ID with tumor | Pathological review based on H&E (subtype after biomarker staining) | Biomarker staining | No. of squamous tumors/total tumors (based on biomarker staining) | Lkb1 recombination by PCR | Latency |
|---|---|---|---|---|---|---|---|---|---|
| Lkb1fl/fl | GFP | 0/14 (0%) | 0/0 (0%) | 6–10 months | |||||
| Sox2 | 7/17 (41%) | p = 0.009 | 8/9 (89%) | 6–10 months | |||||
| JC156 | squamous | K5+, K14+, p63+, Sox2+, TTF1 low | Yes | 6 months, 5 days | |||||
| JC156b | squamous | n/a | n/a | 6 months, 5 days | |||||
| JC163 | squamous | K5+, K14+, p63+, Sox2+, TTF1+ | Yes | 7 months, 15 days | |||||
| JC163b | squamous | K5+, K14+, p63+, Sox2+, TTF1+ | n/a | 7 months, 15 days | |||||
| JC191 | squamous | K5+, p63+, Sox2+, TTF1− | Yes | 7 months, 9 days | |||||
| JC205 | well-differentiated adenocarcinoma (squamous) | K5+, p63+, Sox2+ | Yes | 7 months, 5 days | |||||
| JC215 | well-differentiated adenocarcinoma | K5−, p63−, Sox2+ | Yes | 9 months, 8 days | |||||
| JC217b | squamous | K5−, p63+, Sox2+, TTF1− | n/a | 9 months, 12 days | |||||
| JC229 | squamous | K5+, p63+, Sox2+, TTF1− | n/a | 9 months, 18 days | |||||
| p53fl/fl | GFP | 0/10 (0%) | 0/0 (0%) | 5.5–7.5 months | |||||
| Sox2 | 1/8 (11.1%) | p = 0.44 | 0/1 (0%) | 5.5–7.5 months | |||||
| JC147 | well-differentiated adenocarcinoma | K5−, p63−, Sox2−, TTF1+ | 5 months, 19 days | ||||||
| Rbfl/flp53fl/fl | GFP | 2/28 (7%) | 0/2(0%) | 6–12 months | |||||
| JC118 | well-differentiated adenocarcinoma | n/a | 6 months, 6 days | ||||||
| JC204 | poorly differentiated NSCLC (Adeno) | K5−, p63−, Sox2−, TTF1+ | 8 months, 12 days | ||||||
| Sox2 | 2/14 (14%) | p = 0.59 | 0/3 (0%) | 6–12 months | |||||
| JC189 | well-differentiated adenocarcinoma | K5−, p63−, Sox2−, TTF1+ | 10 months, 27 days | ||||||
| JC216 | poorly differentiated NSCLC (Adeno) | K5−, p63−, Sox2−, TTF1+ | 12 months | ||||||
| JC216b | poorly differentiated NSCLC (Adeno) | n/a | 12 months |
Detailed histopathology, biomarker staining, Lkb1 recombination results, and latency of tumors are identified in mice receiving GFP or Sox2 lentiviruses in indicated conditional genetic backgrounds. n/a indicates insufficient tissue available for analysis. See also Figure S1.
Lenti-Sox2-Cre Lkb1fl/fl Tumors Express Biomarkers of Human SCC
Expression of basal cell markers such as cytokeratin-5 (K5), -14 (K14), and P63 distinguish human SCC from adenocarcinoma (Reis-Filho et al., 2003; Terry et al., 2010). We analyzed tumors from each genetic combination, as well as 12 human SCCs for squamous biomarker expression. Sox2-Lkb1 squamous tumors consistently expressed all of the basal cell markers examined (K5, K14, and p63) and were similar to human SCC, whereas adenocarcinomas in mice from other genetic combinations including KrasG12D did not (Figures 1C and 1D; Figure S2; Table 1). Most human lung tumors express TTF1 (also known as Nkx2.1), but mean expression of TTF1 is significantly reduced in SCCs compared to adenocarcinomas and is reportedly expressed in only ~10% of SCCs (Perner et al., 2009). Compared to KrasG12D-driven mouse adenocarcinomas that were uniformly TTF1 positive, TTF1 levels were significantly reduced in Sox2-Lkb1 tumors (Student’s unpaired t test, p = 0.02) (Figures 1C and 1D). Sox2-Lkb1 tumors exhibited a broad spectrum of TTF1 expression, ranging from no TTF1 expression (similar to human SCCs) to higher levels of expression. Sox2-Lkb1 tumors expressed significantly less TTF1 than mouse adenocarcinomas but significantly more TTF1 than human SCCs (Student’s unpaired t test, p = 0.04) (Figures 1C and 1D). Taken together, these results demonstrate that Sox2-Lkb1 tumors highly resemble human SCC at the level of histopathology and squamous biomarker expression.
Next, we sought to validate Sox2 expression and Lkb1 loss in mouse squamous tumors. All Sox2-Lkb1 tumors were strongly positive for nuclear Sox2 expression, similar to 75% of human SCCs (n = 9 of 12) (Figures 2A and 2B). KrasG12D-driven adenocarcinomas and adenocarcinomas arising from other genetic combinations (p53 or p53/Rb loss) were negative for Sox2 (Figures 2A and 2B; Figure S2). Given that the same lentiviruses were used in all experiments, this suggests that there may be a selective advantage for high Sox2 expression in the context of Lkb1 loss. It is interesting that KrasG12D/+Lkb1fl/fl squamous tumors had little or no Sox2 expression, suggesting that there may be Sox2-independent mechanisms of squamous tumorigenesis (Figures 2A and 2B). Recombination of the Lkb1 allele was determined by PCR analysis on DNA from macrodissected Sox2-Lkb1 tumors. All tumors examined (n = 5 of 5) demonstrated the presence of the recombined Lkb1 floxed allele as well as Sox2 expression (Figure 2C; Table 1). Two of nine Sox2-Lkb1 tumors appeared as adenocarcinomas by H&E, and both tumors exhibited Sox2 expression and Lkb1 recombination; one tumor expressed basal cell markers and, as such, was reclassified by pathologists as squamous, whereas the other tumor did not express K5 or p63 (Table 1). This suggests that Sox2 expression and Lkb1 loss may not be sufficient for squamous tumorigenesis in all contexts and/or that this adenocarcinoma represents an intermediate state that has the potential to trans-differentiate to a squamous tumor under certain conditions (Han et al., 2014).
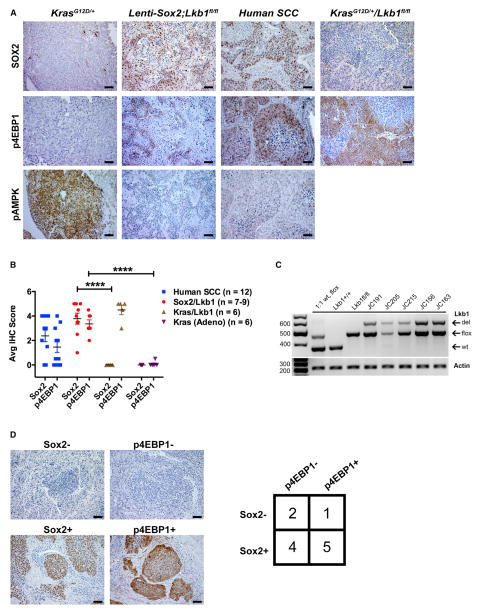
Figure 2. Lenti-Sox2;Lkb1fl/fl Tumors Express Sox2 and Exhibit Activation of the mTOR Pathway Similar to Human SCCs.
(A) Representative IHC of Sox2, p4EBP1, and pAMPK in KrasG12D/+ mouse lung adenocarcinomas, Lenti-Sox2;Lkb1fl/fl tumors, human lung SCCs, and KrasG12D/+Lkb1fl/fl squamous tumors. Brown/red staining is positive. Scale bar represents 50 μm.
(B) Average IHC scores for Sox2 and p4EBP1 from individual tumors indicated in Table 1 based on 0–5 scoring system where 5 indicates >90% positive; 4 indicates >75%; 3 indicates >50%; 2 indicates >25%; 1 indicates >10%; and 0 indicates negative. Number of tumors analyzed is indicated in color key at right. Error bars represent mean ± SEM. Student’s unpaired t test, ****p < 0.001.
(C) PCR validation of Lkb1 recombination in tumor samples from mice indicated in Table 1. Wild-type (WT), floxed (flox), and recombined (del) Lkb1 alleles indicated. Actin serves as input control. Mixed 1:1 Lkb1 WT and floxed DNA, Lkb1+/+, and Lkb1fl/fl normal lung DNA serve as controls.
(D) Left panels: representative images of Sox2− and p4EBP1-positive and -negative IHC. Right panels: contingency table of human SCCs (n = 12) stained with antibodies to SOX2 and p4EBP1.
See also Figure S2.
LKB1 phosphorylates and activates adenosine monophosphate-activated protein kinase (AMPK), which negatively regulates the mammalian target of rapamycin (mTOR) pathway. Immunohistochemical (IHC) analysis of Sox2-Lkb1 tumors demonstrated an absence of phosphorylated AMPK (pAMPK) and enhanced expression of phosphorylated eukaryotic translation initiation factor 4E-binding protein 1 (p4EBP1), an mTOR substrate (Hay and Sonenberg, 2004; Perner et al., 2009), con-firming activation of the mTOR pathway (Figures 2A and 2B). The mTOR pathway is frequently activated in human lung SCCs and represents a potential therapeutic target (Mantripragada and Khurshid, 2013). Indeed, we observed that, of the human SCCs with strong SOX2 expression (9 of 12), five demonstrated evidence of mTOR pathway activation as well (55% of Sox2+ SCCs) (Figure 2D). Altogether, these data reveal that SOX2 expression and mTOR activity frequently co-occur in human SCC and that Sox2-Lkb1 tumors strongly resemble human SCCs at the level of pathway activation.
Sox2-Driven SCCs Exhibit Expression or Activation of Potential Therapeutic Targets
To identify pathways dysregulated in Sox2-driven SCC, we performed IHC analysis for therapeutically relevant pathways in Sox2-Lkb1 tumors, KrasG12DLkb1fl/fl squamous tumors identified by K5 staining, and human SCCs. FGFR family members are frequently amplified and/or harbor activating point mutations in human SCC, and FGFR inhibitors are currently in clinical trials (Cancer Genome Atlas Research Network, 2012; Dutt et al., 2011; Liao et al., 2013). Notably, SOX2 and FGFR2 expression are highly correlated in human SCC, and SOX2 can bind the FGFR2 promoter (Bass et al., 2009; Boyer et al., 2005; Fang et al., 2011), suggesting that FGFR2 may be a direct target of SOX2 in lung cancer. We examined expression of FGFR2 in each tumor group and found that Fgfr2 expression tended to be higher in Sox2-Lkb1 tumors compared to KrasG12DLkb1fl/fl squamous tumors, but these data did not reach statistical significance (Figures 3A and 3B). FGFR2 was highly induced by SOX2 expression in human A549 lung cancer cells harboring LKB1 mutations (Figure S3) but not in two other cell lines with LKB1 mutations (H23 and H157) (Mahoney et al., 2009; Figure S3). We were not able to detect phospho-Frs or pFGFR2 in the Sox2-Lkb1 tumors, so it remains to be determined whether pathways downstream of FGFR2 are activated.
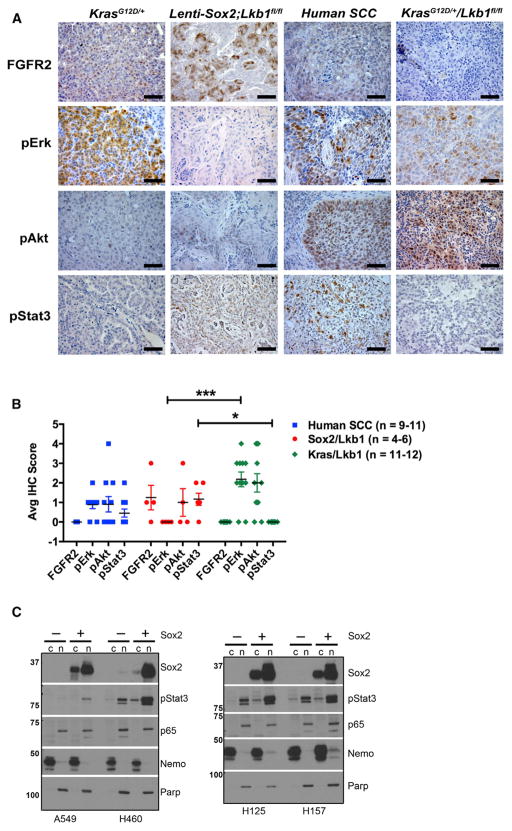
Figure 3. Murine SCCs Exhibit Expression of Potential Therapeutic Targets.
(A) Representative IHC of KrasG12D/+ mouse lung adenocarcinomas, mouse Lenti-Sox2;Lkb1fl/fl tumors, human lung SCCs, and KrasG12D/+Lkb1fl/fl squamous tumors stained with indicated antibodies. Brown/red staining is positive. Scale bar represents 50 μm.
(B) Average IHC score of FGFR2, pErk, pAkt, and pStat3 stains in individual tumors from Table 1, human SCCs, and KrasG12D/+Lkb1fl/fl squamous tumors. Number of tumors analyzed is indicated in color key at right. IHC scoring system is based on scoring system where 5 indicates >75% positive; 4 indicates >50%; 3 indicates >25%; 2 indicates >10%; 1 indicates >2.5%; and 0 indicates negative. Error bars represent mean ± SEM. For Student’s unpaired t test, *** indicates p < 0.0002 and *p < 0.01.
(C) Immunoblot of cytoplasmic (c) and nuclear (n) protein extracts for SOX2, pStat3, and NF-κB p65 from human lung cancer cell lines under control (−) or 48 hr SOX2 induction (+) in stable Tet-On cells. Nemo and Parp serve as loading controls for cytoplasmic and nuclear fractions, respectively.
See also Figure S3.
The nuclear factor κB (NF-κB) pathway is activated and implicated in various cancers, including lung adenocarcinoma (Meylan et al., 2009; Xue et al., 2011) and lung SCC (Xiao et al., 2013). Sox2-Lkb1 tumors demonstrated evidence of NF-κB pathway activity as indicated by nuclear p65, similar to Kras-driven adenocarcinomas (Figure S3). Although, the role of NF-κB signaling in SCC development remains to be determined, activation of this pathway in multiple mouse models of SCC warrants further study of NF-κB as a potential therapeutic target. Inducible SOX2 expression alone was not sufficient to increase nuclear p65 in human cell lines regardless of LKB1 status, suggesting that other factors are likely driving NF-κB signaling in these cells (Figure 3C).
The RAS/MAPK/PI3K pathways are frequently activated in human squamous tumors via different mechanisms (Cancer Genome Atlas Research Network, 2012). To examine the status of these pathways, we stained tumors for pErk and pAkt. Notably, pErk was significantly reduced in Sox2-Lkb1 tumors compared to KrasG12DLkb1fl/fl squamous tumors and human SCCs (Figures 3A and 3B). We speculate that Kras activation in KrasG12DLkb1fl/fl squamous tumors maintains pErk signaling, which is absent in Sox2-driven tumors. We observed variable levels of pAkt in Sox2-Lkb1 tumors, ranging from little or no pAkt to high levels similar to those in KrasG12DLkb1fl/fl tumors (Figures 3A and 3B). Together, Sox2-Lkb1 tumors had reduced MAPK pathway activity compared to KrasG12DLkb1fl/fl squamous tumors, suggesting that Sox2-Lkb1 tumors may rely more heavily on other growth factor signaling pathways for proliferation, including mTOR. A recent study published while this article was under consideration demonstrated that Pten loss cooperates with Lkb1 loss to promote SCC (Xu et al., 2014). Notably, Pten/Lkb1-null tumors also exhibited low MAPK pathway activation.
Stat3 has been shown to cooperate with Sox2 to promote tumor progression in esophageal and forestomach tumors, cancers in which SOX2 is genomically amplified (Liu et al., 2013). Preclinical studies have identified STAT3 inhibitors, and STAT3 decoys are now being tested in patients with head and neck SCC (Sen et al., 2012). Compared to KrasG12D adenocarcinomas and KrasG12DLkb1fl/fl squamous tumors, the Jak-Stat pathway was significantly activated in Sox2-Lkb1 tumors (indicated by phospho-Stat3) with clear nuclear pStat3 in some cells within the squamous tumors (Figures 3A and 3B). Human SCCs had variable levels of pStat3 similar to those in Sox2-Lkb1 tumors (Figures 3A and 3B). It is striking that, in multiple human lung cancer cell lines of adenocarcinoma and squamous lineage, inducible expression of SOX2 consistently promoted nuclear pStat3 accumulation (Figure 3C). Altogether, these data suggest that Sox2 contributes to Stat signaling in vivo and thus represents a therapeutic target warranting further investigation for SCC treatment.
To summarize, we have used a lentiviral-based approach to generate a mouse model of SCC based on Sox2 expression and Lkb1 loss, highlighting their cooperation in lung SCC development. Both SOX2 expression and mTOR pathway activity frequently co-occur in human SCC, making this model particularly relevant to the human disease. Mouse tumors recapitulate the human disease at the level of histopathology, biomarker expression, and activation of potential therapeutic targets that have been largely unexplored in SCC treatment. It is important to note that Stat signaling was induced by SOX2 in vitro and enriched in Sox2-driven squamous tumors. This model thus serves as a preclinical tool to test whether STAT inhibition is therapeutically effective against Sox2-driven SCCs.
The low penetrance of tumor formation in this model may be due to low lentiviral titer and/or the underlying cell of origin. Although we recognize that tumor numbers in this study are low, the paucity of other squamous lung tumor models suggests that this particular genetic combination is highly relevant. While this article was under consideration, Xu et al. demonstrated that Pten/Lkb1 loss promotes squamous lung tumors exclusively, in contrast to Kras/Lkb1 mice, which develop multiple tumor types (Ji et al., 2007; Xu et al., 2014). It remains unknown why PI3K activation, as opposed to MAPK, would lead exclusively to squamous tumors in the context of Lkb1 loss, especially since PI3K pathway alterations also occur in adenocarcinoma (Ding et al., 2008). We speculate that, since Pten/Lkb1-null tumors express Sox2, Sox2 may be a critical target of the PI3K pathway in these tumors.
In both Pten/Lkb1 and Sox2/Lkb1 mouse models, inflammatory microenvironment and activation of immune signaling pathways appear to distinguish SCC from adenocarcinoma (Xu et al., 2014). A notable difference between the two models is that adenoviruses used in Pten/Lkb1 and Kras/Lkb1 models are thought to enhance inflammation compared to lentiviruses used here. These models will serve as preclinical tools for further investigation of immunotherapy efficacy and function.
It is unclear whether SCCs arise from basal cells or whether the transforming events promote basal cell differentiation. The identity of the cell of origin for SCC remains an important unanswered question. One model suggests that basal cells serve as the cells of origin because SCCs express basal cell markers and Sox2 is expressed in basal cells (Que et al., 2009; Sutherland and Berns, 2010). However, other studies suggest that Sox2 promotes basal cell fate (Han et al., 2014; Lu et al., 2010). Recent studies in KrasG12D/+Lkb1fl/fl mice suggest that type II pneumocyte-derived lung adenocarcinomas can transdifferentiate into SCCs (Han et al., 2014). Given that lentiviruses in our model may target multiple cell types in the mouse lung, it is possible that Sox2 expression and Lkb1 loss are capable of altering cell fate to a squamous-like identity. Given that we also detected one tumor that appeared as an adenocarcinoma expressing basal cell markers, we speculate that transdifferentiation may also occur in this model. The lentiviral approach used here could be adapted to investigate the cell of origin for Sox2-driven SCCs.
Bicistronic lentiviruses may also be used to test other candidate SCC drivers, such as P63 and NRF2, as well as to identify cooperating genetic events that promote SCC. We expect that murine squamous tumors will have fewer “passenger” genetic alterations than human SCC due to lack of carcinogen exposure, thereby providing a biological filter to identify “driver” genetic changes in SCC. Beyond lung cancer, findings from this model may have important clinical implications for other squamous or SOX2-driven malignancies such as SCLC and brain, esophageal, and oral cancers.
EXPERIMENTAL PROCEDURES
Mouse Breeding and Lentiviral Infections
Mice were housed in an environmentally controlled room according to the Committee of Animal Care. Lkb1fl/fl mice were purchased from The Jackson Laboratory. LSL-KrasG12D/+ and p53fl/fl animals were kindly provided by Tyler Jacks. p53fl/flRbfl/fl mice were generated by Anton Berns (Meuwissen et al., 2003) and obtained from T. Jacks. Lung tumor tissue from KrasG12D/+Lkb1fl/fl mice was kindly provided by Reuben Shaw. At 6–8 weeks of age, anesthetized mice were infected with ~107 infectious units/ml of Lenti-Sox2-Cre or GFP-Cre lentiviruses or 6.47 ~107 plaque-forming units of AdCre (University of Iowa) by nasal instillation as described elsewhere (Jackson et al., 2001). Viruses were administered in a Biosafety Level 2+ room according to Institutional Biosafety Committee guidelines.
Lentivirus Production
GFP and murine Sox2 cDNAs were cloned into bicistronic lentiviral vectors expressing Cre to generate Lenti-Sox2-Cre and Lenti-GFP-Cre plasmids and confirmed by direct sequencing. 293T cells were transfected with a three-plasmid transfection system including the lentiviral vector, pCMV-delta-8.2 (Addgene) and pCMV-VSV-G (Addgene). Virus was harvested posttransfection, concentrated by ultracentrifugation (24,000 × g), and titered using HEK293T reporter cells stably expressing a Lox-DsRed2(Stop)-Lox-GFP cassette. Number of cells expressing green fluorescent protein was measured by flow cytometry and used to calculate titer (infectious U/ml).
MicroCT Imaging
At indicated time points, mice were scanned for 30 s to 2 min under isoflurane anesthesia using a small animal Quantum FX microCT (PerkinElmer) at 45 υm resolution, 90 kV, with 160 υA current. Images were acquired using PerkinElmer Quantum FX software and processed with Analyze 11.0 (AnalyzeDirect).
Human Tissue
Excess deidentified fresh tissue was obtained with prior patient consent under approved protocol by the institutional review board (#10924). Institutional guidelines regarding specimen use were followed.
Supplementary Material
Acknowledgments
We thank Drs. W. Akerley, S. Slomowitz, and A.J. Bhutkar for valuable advice and Dr. K. Jones for critical reading of the manuscript. We are especially grateful to Dr. R. Shaw for providing Kras/Lkb1 tissue. Thanks to members of the T.G.O. Lab for technical support, especially M. Terry, R. Arya, and J. Clegg. We thank Y. Derose, B. Anderson, and K. Gligorich for histological services; M. DuPage, K. Lane, and the T. Jacks laboratory for lentiviral constructs; and M. Van Brocklin for HEK293T reporter cells. T.G.O. was supported in part by a Department of Defense Lung Concept Award (DoD USAMRAA W81XWH-12-1-0211) and a V Scholar award from The V Foundation for Cancer Research. T.G.O. is a Damon Runyon-Rachleff Innovation Awardee supported in part by the Damon Runyon Cancer Research Foundation (DRR-26-13).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and three figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.05.036.
AUTHOR CONTRIBUTIONS
A.M. designed and performed the majority of experiments, acquired and analyzed data, and wrote the manuscript. K.B., U.K., P.C., and S.P. performed experiments. S.C provided human tissue. B.W. and S.C. analyzed histopathology and provided clinical expertise. T.O. conceived of the project, analyzed the data, and wrote the paper.
References
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brcic L, Sherer CK, Shuai Y, Hornick JL, Chirieac LR, Dacic S. Morphologic and clinicopathologic features of lung squamous cell carcinomas expressing Sox2. Am J Clin Pathol. 2012;138:712–718. doi: 10.1309/AJCP05TTWQTWNLTN. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS ONE. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G, Lin B. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev Biol. 2008;317:296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Han X, Li F, Fang Z, Gao Y, Li F, Fang R, Yao S, Sun Y, Li L, Zhang W, et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat Commun. 2014;5:3261. doi: 10.1038/ncomms4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembelé D, Martinet N, Thibault C, Huelsken J, Brambilla E, du Manoir S. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS ONE. 2010;5:e8960. doi: 10.1371/journal.pone.0008960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Kwon MC, Berns A. Mouse models for lung cancer. Mol Oncol. 2013;7:165–177. doi: 10.1016/j.molonc.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, Liu Q, Pugh TJ, Pedamallu CS, Hayes DN, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen Z, Zhu F, Hu Y. IκB kinase alpha and cancer. J Interferon Cytokine Res. 2012;32:152–158. doi: 10.1089/jir.2011.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, Onaitis MW. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney CL, Choudhury B, Davies H, Edkins S, Greenman C, Haaften Gv, Mironenko T, Santarius T, Stevens C, Stratton MR, Futreal PA. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer. 2009;100:370–375. doi: 10.1038/sj.bjc.6604886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantripragada K, Khurshid H. Targeting genomic alterations in squamous cell lung cancer. Front Oncol. 2013;3:195. doi: 10.3389/fonc.2013.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Patel J, Akerley W. Squamous non-small cell lung cancer as a distinct clinical entity. Am J Clin Oncol. 2013 doi: 10.1097/COC.0b013e3182a0e850. Published online July 24, 2013 http://dx.doi.org/10.1097/COC.0b013e3182a0e850. [DOI] [PubMed]
- Perez-Moreno P, Brambilla E, Thomas R, Soria JC. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012;18:2443–2451. doi: 10.1158/1078-0432.CCR-11-2370. [DOI] [PubMed] [Google Scholar]
- Perner S, Wagner PL, Soltermann A, LaFargue C, Tischler V, Weir BA, Weder W, Meyerson M, Giordano TJ, Moch H, Rubin MA. TTF1 expression in non-small cell lung carcinoma: association with TTF1 gene amplification and improved survival. J Pathol. 2009;217:65–72. doi: 10.1002/path.2443. [DOI] [PubMed] [Google Scholar]
- Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136:1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Simpson PT, Martins A, Preto A, Gärtner F, Schmitt FC. Distribution of p63, cytokeratins 5/6 and cytokeratin 14 in 51 normal and 400 neoplastic human tissue samples using TARP-4 multi-tumor tissue microarray. Virchows Arch. 2003;443:122–132. doi: 10.1007/s00428-003-0859-2. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, Johnson JT, Duvvuri U, Lee J, Sahu N, Joyce S, et al. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2012;2:694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KD, Berns A. Cell of origin of lung cancer. Mol Oncol. 2010;4:397–403. doi: 10.1016/j.molonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry J, Leung S, Laskin J, Leslie KO, Gown AM, Ionescu DN. Optimal immunohistochemical markers for distinguishing lung adenocarcinomas from squamous cell carcinomas in small tumor samples. Am J Surg Pathol. 2010;34:1805–1811. doi: 10.1097/PAS.0b013e3181f7dae3. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Jiang Q, Willette-Brown J, Xi S, Zhu F, Burkett S, Back T, Song NY, Datla M, Sun Z, et al. The pivotal role of IKKα in the development of spontaneous lung squamous cell carcinomas. Cancer Cell. 2013;23:527–540. doi: 10.1016/j.ccr.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y, Chen Z, Herter-Sprie GS, Akbay EA, Tchaicha JH, Altabef A, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Meylan E, Oliver TG, Feldser DM, Winslow MM, Bronson R, Jacks T. Response and resistance to NF-κB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 2011;1:236–247. doi: 10.1158/2159-8290.CD-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.