Abstract
A recent hydrogen-deuterium exchange study of folding of the GroEL/GroES-dependent bacterial enzyme DapA has suggested that the DapA folding pathway when free in solution may differ from the folding pathway used in the presence of the GroEL/GroES chaperonin. Here, we have investigated whether DapA aggregation might be occurring in free solution under the conditions of the exchange experiment, as this would confound interpretation of the pathway predictions. Dynamic light scattering data, sedimentation analysis and refolding yield indicate that significant aggregation occurs upon dilution of DapA from denaturant, bringing into question the earlier conclusion that different folding pathways occur in the absence and presence of the chaperonin system.
Keywords: Dap, GroEL, aggregation, light scattering, protein folding
A recent study of the GroEL/GroES-dependent substrate protein DapA has suggested that its folding pathway inside the GroEL/GroES chaperonin cavity may differ from that taken in free solution [1] under so-called “permissive conditions” of low temperature and concentration [2], where the protein can reach the native form either inside the encapsulated chaperonin cavity or free in solution [3]. This was based on pulsed hydrogen-deuterium exchange experiments carried out at various times after initiating folding, producing distinct patterns of deuterium incorporation in the chaperonin reaction vs under solution conditions. A conclusion was drawn that chaperonin “confinement” of the encapsulated DapA mediates a different folding trajectory. Here we have studied DapA under the conditions employed for folding in the published study, 10°C and 2.4 μM final concentration of DapA monomer, and find that the behavior of DapA in free solution under these conditions is complicated by aggregation, as contrasted with absence of aggregation in the presence of chaperonin. We conclude that the exchange experiment cannot be interpreted as indicating distinct folding pathways, because folding in solution under the conditions of the study is a convolution of both folding to reach the native state and misfolding that produces aggregation.
Materials and Methods
Proteins
DapA was cloned into the NdeI and BamHI sites of pET 17b following PCR amplification of E. coli total DNA using PrimeStar Max DNA Polymerase (Takara). Clones were sequenced, and one with the correct sequence was transformed into BL21(DE3) cells, grown in large volume at 22°C to an OD650 = ~0.8, induced with 0.5 mM IPTG, and grown further overnight at 22°C. Cells were harvested, resuspended in 20 mM Tris-HCl, pH 7.5, 1 mM DTT, and disrupted by passage through a MicroFluidizer (Microfluidics). After centrifugation to remove debris, the supernatant was applied to a Fast Flow Q column (GE Lifesciences) in 20 mM Tris-HCl, pH 7.5, 1 mM DTT. The column was eluted with a gradient from 0 to 1 M NaCl in the same buffer. DapA-containing fractions were pooled, adjusted to 2 M (NH4)2SO4, and applied to a phenyl-Sepharose column (GE Lifesciences). The column was eluted with a reverse gradient from 2 M to 0 M (NH4)2SO4, and the DapA-containing fractions were pooled, concentrated, and dialyzed against 20 mM Tris-HCl, pH 7.5, 100 mM KCl, 1 mM DTT (buffer A). Aliquots were chromatographed on a 1.6 × 100 cm Sephacryl S-200 column (GE Lifesciences) in buffer A. DapA-containing fractions showed no contamination on SDS-PAGE. They were pooled and concentrated using a Centriprep 30 ultrafiltration device (Millipore).
Wild-type GroEL, the GroEL trap mutant D87K, and GroES were purified as previously described [4].
Dynamic light scattering (DLS)
DLS experiments were carried out on a DynaPro Titan temperature-controlled instrument (Wyatt Technologies), fitted with a 12 μl observation cuvette and controlled by Dynamics 6.7.3 software. Laser power was set at 15%. The instrument and cuvette were allowed to equilibrate at 10°C for at least 30 min before adding samples and acquiring data. DapA refolding in solution was typically carried out by rapidly diluting 2 μl of DapA, denatured in 7.2 M GuHCl, 10 mM DTT, into 198 μl of refolding buffer (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 10 mM MgCl2, 30 mM sodium pyruvate) at 10°C. An aliquot of the refolding reaction solution was pipetted into the observation cuvette immediately after mixing. Data acquisition was initiated immediately, so that the first data point was acquired at ~5 sec after mixing. For all DLS experiments, the buffers and protein solutions, except the actual refolding reactions, were filtered through 0.22 μm centrifugal or syringe filters (Millipore) before use, and all were examined in the DLS instrument to ensure that no large scattering particles were present. For the GroEL-bound DapA sample, denatured DapA was diluted into buffer containing 4.8 μM GroEL, and an aliquot of the sample was examined after equilibration at 10°C. Both average intensity of scattered light and autocorrelation data were generated at each acquisition time, and the Dynamics software automatically fitted the autocorrelation data to produce estimates of the hydrodynamic radii of the scattering particles at each data point.
Sedimentation analysis
Twenty μl DapA, denatured in 7.2 M GuHCl/10 mM DTT, as above, was directly diluted 100-fold into 2 ml refolding buffer equilibrated at 10°C to a final concentration of 2.4 μM, as in the light scattering experiment, incubated for 30 min at 10°C, then centrifuged at 10°C for 10 min at ~250,000 × g (Beckman Type 70.1 Ti rotor). The protein concentrations of aliquots of the supernatant were estimated using the Bio-Rad Protein Assay reagent. These values were compared to those produced by identically sized aliquots of 2.4 μM native DapA.
Results and Discussion
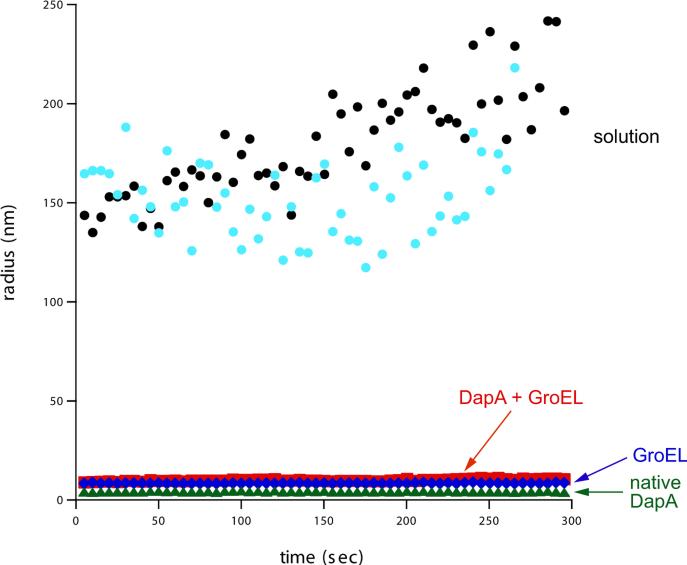
When we diluted DapA from GuHCl to 2.4 μM in free solution at 23°C, we immediately observed visible cloudiness in the solution. We therefore carried out the same dilution at 10°C, the conditions of refolding in free solution used in the hydrogen-deuterium exchange experiment [1], and monitored the solution using dynamic light scattering. In particular, DapA was diluted from GuHCl to 2.4 μM final concentration in either free solution at 10°C or into the same solution containing a 2-fold molar excess of GroEL (Fig. 1). At the earliest measurable time following dilution of unfolded DapA directly into solution (~5 sec), in two independent tests, dynamic light scattering revealed the accumulation of particles of ~150 nm diameter (Fig. 1). By contrast, no significant light scattering beyond that produced by the GroEL tetradecamer was observed when DapA was diluted into a buffer with GroEL, consistent with the chaperonin's ability to efficiently bind the non-native form(s) of substrate proteins and prevent multimolecular aggregation (ref.5 and Fig.1). Additional control measurements revealed that GroEL alone and native DapA tetramer exhibited only the scattering appropriate for their molecular sizes (Fig. 1). We examined light scattering of the solution mixture at later times after dilution of DapA from denaturant and observed the same light scattering as during the first 5 min. We conclude that aggregation occurs under the conditions of folding in free solution at 10°C and 2.4 μM, employed in the hydrogen exchange study.
Fig. 1. DapA aggregates upon dilution from denaturant directly into solution.
DapA, denatured in guanidine HCl, was rapidly diluted into refolding buffer at 10°C, and this solution was pipetted into a cuvette equilibrated at 10°C in the sample compartment of a dynamic light scattering (DLS) instrument. Measurements were taken at 5 sec intervals for 5 min. The instrument software was used to estimate the size (hydrodynamic radius) of the scattering particles from the autocorrelation function at each acquisition point. These data are plotted vs. the acquisition time, starting at 5 sec after mixing. Data from two independent experiments are displayed (black and turquoise circles). In another experiment, denatured DapA was rapidly diluted (2.4 μM final concentration) into a solution containing 4.8 μM GroEL at 23°C. After 5 min, the solution was equilibrated at 10°C and placed in the DLS instrument. Measurements were taken and analyzed as for the dilution directly into solution (red squares). As controls, similar measurements were carried out with native DapA tetramer (24 μM; green triangles) and GroEL alone (4.8 μM; blue diamonds). Note that the estimated radii are averages calculated from the data at each acquisition time. The scattering particles in the direct dilution experiments were highly polydisperse, with average radii of 150-180 nm. Those in the GroEL-DapA complex experiment were somewhat polydisperse, and those in the control experiments were effectively monodisperse. Average hydrodynamic radii for these latter samples were: GroEL complex with DapA, 7.9 nm; GroEL, 6.8 nm; DapA tetramer, 3.3 nm.
To measure the fraction of DapA that aggregates in the light scattering study, the refolding mixtures were sedimented after 30 min, maintaining the temperature at 10°C for both incubation and centrifugation. Comparison of protein concentration of the solution before and after centrifugation revealed that only 77±3.4% (n=4) of the DapA was recovered in the soluble fraction, reflecting that ~25% of the protein had aggregated. Such aggregation offers explanation for why recovery of native enzyme in free solution is uniformly reduced relative to GroEL/GroES mediated folding ([1] and Fig.S1).
The lack of concentration dependence of the rates of refolding, as reported in [1], suggests that the non-native monomer of DapA in solution is in equilibrium with an off-pathway monomeric state (or states) that aggregates irreversibly. Such an off-pathway state would slow folding relative to that at GroEL/GroES. Importantly, the presence of such off-pathway states would also complicate any attempt to use hydrogen exchange to resolve a folding trajectory to the native state because they would be subject to exchange and be analyzed along with the folding-competent states. The earlier study sought to directly address whether any aggregation might be occurring by carrying out an FCS study at 100 pM concentration. No self-association of denatured DapA monomers in solution was observed [1], but we note that this experiment was carried out at approximately four orders of magnitude lower concentration than the conditions of the folding experiment that were used for the hydrogen-deuterium exchange experiment and thus would not be able to inform about the possible occurrence of aggregation at the higher concentration (see Supplementary Discussion concerning behavior of DapA monomer at low concentration).
In sum, then, it seems impossible to interpret the hydrogen-deuterium exchange data from the earlier study as providing evidence for a “confinement” role of the GroEL/GroES cavity that would alter the folding trajectory of the DapA protein. By contrast, another study showed that the GroEL substrate, double mutant MBP, which, like DapA, folds more slowly in solution than at GroEL/GroES but fully reaches the native state, also aggregates, and that when such aggregation is relieved, the protein now folds in free solution at the same rate as at GroEL/GroES [6]. As concerns folding in the GroEL/GroES cavity, we note that a recent study of wild-type MBP shows that it rapidly collapses upon dilution from denaturant, within the dead time of mixing [7]. Such collapse likely also occurs for substrates released from the GroEL cavity wall into the GroEL/GroES chamber, and this would seem to obviate a need for “confinement”. By contrast, the encapsulation of a collapsed monomer inside the GroEL/GroES complex, an “Anfinsen cage” [8], ensures that, during folding, the protein simply cannot aggregate with any other protein.
Supplementary Material
Highlights.
DapA diluted into solution produces large light scattering signals
Sedimentation indicates precipitation of ~25% of DapA when refolded in solution
Yield of DapA refolded in solution is reduced compared to that from GroEL/GroES
Aggregation of DapA confounds interpretation of refolding pathway data
Acknowledgements
This work was supported by a grant (RO1 ES 023875) from the NIEHS (E.C.) and by the Howard Hughes Medical Institute (A.H.).
Abbreviations
- DLS
dynamic light scattering
- ASA
aspartate semialdehyde
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- FCS
fluorescence correlation spectroscopy
- MBP
maltose binding protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Georgescauld F, Popova K, Gupta AJ, Bracher A, Engen JR, Hayer-Hartl M, Hartl FU. GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding. Cell. 2014;157:922–934. doi: 10.1016/j.cell.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viitanen PV, Lubben TH, Reed J, Goloubinoff P, O'Keefe DP, Lorimer GH. Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (GroEL) are K+ dependent. Biochemistry. 1990;29:5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- 3.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 4.Weissman JS, Kashi Y, Fenton WA, Horwich AL. GroEL-mediated protein folding proceeds by multiple rounds of release and rebinding of non-native forms. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 5.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and MgATP. Nature. 1989;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 6.Tyagi NK, Fenton WA, Deniz AA, Horwich AL. Double mutant MBP refolds at same rate in free solution as inside the GroEL/GroES chaperonin chamber when aggregation in free solution is prevented. FEBS Lett. 2011;585:1969–1972. doi: 10.1016/j.febslet.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters BT, Mayne L, Hinshaw JR, Sosnick TR, Englander SW. Folding of a large protein at high structural resolution. Proc. Natl. Acad. Sci. USA. 2013;110:18898–18903. doi: 10.1073/pnas.1319482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saibil HR, Zheng D, Roseman AM, Hunter AS, Watson GM, Chen S, auf der Mauer A, O'Hara BP, Wood SP, Mann NH, et al. ATP induces large quaternary rearrangements in a cage-like chaperonin structure. Current Biol. 1993;3:265–273. doi: 10.1016/0960-9822(93)90176-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.