Abstract
SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins are a highly regulated class of membrane proteins that drive the efficient merger of two distinct lipid bilayers into one interconnected structure. This protocol describes our fluorescence resonance energy transfer (FRET)-based single vesicle-vesicle fusion assays for SNAREs and accessory proteins. Both lipid-mixing (with FRET pairs acting as lipophilic dyes in the membranes) and content-mixing assays (with FRET pairs present on a DNA hairpin that becomes linear via hybridization to a complementary DNA) are described. These assays can be used to detect substages such as docking, hemifusion, and pore expansion and full fusion. The details of flow cell preparation, protein-reconstituted vesicle preparation, data acquisition and analysis are described. These assays can be used to study the roles of various SNARE proteins, accessory proteins and effects of different lipid compositions on specific fusion steps. The total time required to finish one round of this protocol is 3–6 d.
INTRODUCTION
Membrane fusion is an important cellular process by which two initially distinct lipid bilayers merge to form one interconnected structure. Intracellular trafficking, egg fertilization and communication between neurons are among the many processes that rely on some form of fusion1. Membrane fusion in eukaryotic cells is mediated by a conserved family of proteins called SNAREs2,3. All SNARE systems consist of two cognate pairs, v-and t-SNAREs (vesicular-and target-SNAREs), which are anchored in two fusing membranes. They can form a four-α-helix bundle via association of their ‘SNARE motifs’ that are composed of 15 hydrophobic core layers4. In particular, the neuronal SNAREs are responsible for the fast (submilliseconds) neurotransmitter release. The neurotransmitters encapsulated within synaptic vesicles in the axon bulb are released into the synaptic cleft in response to presynaptic depolarization. This fast calcium-triggered neurotransmitter release is the result of the fusion of synaptic vesicle membrane to the plasma membrane and is driven by the neuronal SNARE proteins, which are under tight regulation by a number of accessory proteins such as synaptotagmin, complexin and Munc18 (refs. 5 and 6).
Development of the protocol
Since 1998, ensemble bulk fluorescence (Förster) resonance energy transfer (FRET) has been used as a standard in vitro tool for studying SNARE-mediated membrane fusion (henceforth referred to as bulk lipid-mixing assay)7. In the most commonly used scheme, v-and t-SNARE proteins are reconstituted into two independent populations of vesicles, where one population of vesicles contains both acceptor and donor fluorophore-labeled lipophilic molecules and the other population contains no fluorophores. Upon fusion, lipid mixing results in an increase of the average distance between donors and acceptors, and the degree of fusion can be quantified from the recovery of the donor fluorescence emission. For SNARE protein reconstitution, both standard and direct methods have been widely used. The standard method is a co-micellization method whereby lipids and proteins are initially cosolubilized with detergent. The direct method refers to a preparation process whereby detergent solubilized proteins are incorporated into preformed vesicles. The diameter of protein-reconstituted vesicles may be obtained through electron microscopy or dynamic light scattering, and is ~50 nm for the standard method and ~90 nm for the direct method. The vesicle-incorporation rates of proteins can be assessed by a co-flotation assay. A detailed comparison between standard and direct methods regarding vesicle size, protein incorporation and fusion ability has been reported by the Rizo group8.
Despite its utilities, the bulk lipid-mixing assay cannot clearly distinguish different stages of the fusion process such as docking, hemifusion and full fusion because of ensemble averaging. Recognizing this limitation, many researchers developed fusion assays that can visualize the docking and fusion of individual vesicles to the planar bilayer9–11. Because having the SNARE proteins on the planar bilayer in close proximity to a glass surface may potentially influence the fusogenic function of the proteins9–11, we sought an alternative approach in which the reaction observed is the fusion between two single vesicles12. In this assay, one set of vesicles is surface-tethered and the other set of vesicles is free-floating above the surface. Unlike the bulk lipid-mixing assay in which both the donor and acceptor fluorophores are typically in the same vesicle before fusion, in the single-vesicle assay each vesicle pair is formed by one vesicle with donor fluorophore-conjugated lipids and another acceptor fluorophore-labeled vesicle. FRET between each pair of vesicles can be measured either after the samples have equilibrated and the unreacted vesicles have been washed away or in real-time as free-floating vesicles dock and fuse to the surface-tether vesicles. The data from the equilibrated samples can be used to differentiate between docked and lipid-mixed populations. Furthermore, through careful analysis of hundreds of real-time FRET efficiency trajectories (time resolution of 100–200 milliseconds), transient intermediate states between the docked (low FRET) and the fully lipid mixed (high FRET) state can be observed, strongly suggesting the presence of fusion intermediates, such as hemifusion. Using this assay, we have also shown evidence for distinct fusion pathways such as kiss-and-run events12 and full-collapse fusion13.
However, this single vesicle-vesicle lipid mixing is still blind to the fusion pore opening and expansion, the final crucial step of the full-collapse fusion pathway14. A fundamental assumption of using lipid mixing to study membrane fusion is that there is a direct correlation between lipid and content mixing. However, a recent study on DNA-mediated membrane fusion showed that this might not be the case14. By comparing results from separate sets of experiments, one using lipid-mixing indicators and the other using content-mixing indicators, that study showed that a high level of lipid mixing can occur with a limited degree of content mixing, thus demonstrating that lipid mixing alone may be an insufficient marker for fusion pore opening14. The next-generation liposome fusion assay would have to use content-mixing reporters in addition to lipid-mixing reporters. Attempts to use small molecules as indicators of membrane fusion15 have been hampered by leakage of the indicator molecules as a result of the transient destabilization of the membranes associated with the reconstitution of large numbers of SNARE proteins. For ensemble assays, this leakage problem makes the data analysis challenging, although in single-vesicle analysis, vesicles still containing the content-mixing probes can be selected and analyzed as long as the leakage problem is not too severe16,17. To circumvent this problem, Rothman’s group used radiolabeled DNA to report that SNARE proteins constitute the minimal machinery for membrane fusion18. A limitation of this approach is that the content-mixing signal was measured after vesicles were lysed by detergent, leaving open the possibility that docked (but not fused) vesicles could still contribute to the final readout.
In order to study the fusion pore expansion, inspired by the single-molecule vesicle encapsulation approach19, we developed a single-molecule FRET-based vesicle-vesicle fusion assay with fluorescent probes encapsulated inside SNARE-reconstituted vesicles13. The probe, often referred to as a molecular beacon, is a DNA molecule conjugated with a donor fluorophore (Cy3) and an acceptor fluorophore (Cy5). By itself, the probe forms a hairpin structure so that the two fluorophores are in close proximity, giving rise to high FRET. When a poly-A oligonucleotide is added, it hybridizes with the loop sequence (poly-T) of the hairpin probe, opening up the hairpin and reducing FRET. If the poly-A strand is encapsulated inside another vesicle, the FRET changes described here can only be seen when the two vesicles merge their content with a large enough pore for the DNA strand(s) to pass through.
Applications
By using our single vesicle-vesicle lipid-mixing assay on yeast SNAREs that function in membrane trafficking from endoplasmic reticulum to Golgi, we discovered the multiple intermediates12 and the role of the SN1 and SN2 helices of Sec9c (ref. 20). For the neuronal SNARE system, we found the dual function of complexin (namely, docking inhibition and enhancement of fusion after docking21), the rapid aggregation of vesicles induced by C2AB/Ca2+ (ref. 22), the fusion-promoting role of Munc18 binding to the SNARE core23 and fast lipid mixing induced by membrane-anchored synaptotagmin 1 (ref. 24). By using our vesicle-vesicle content-mixing assay, we detected the fusion pore expansion in SNARE-dependent membrane fusion13.
Other approaches
There are other researchers who pioneered single-molecule and single-vesicle approaches to study SNARE-mediated membrane fusion25–29. One such assay, developed by Rothman’s group28, which we call the single vesicle–bilayer fusion assay, detects individual fusion events between free-floating vesicles and the supported bilayer deposited on the surface. The probe may be a lipid-conjugated fluorophore, a soluble fluorophore within the vesicle or a combination of both. This single vesicle-bilayer fusion approach is described in the accompanying protocol30 and is attractive because the planar nature of the supported bilayer resembles the geometry of the neuronal plasma membrane. More recently, Brunger and co-workers developed an advanced single vesicle-vesicle fusion assay that allows the simultaneous monitoring of lipid and content mixing by using a small content-mixing probe17.
Limitations of our approach
The intermediate states in our lipid-mixing assay were mostly transient and are difficult to clearly demonstrate through the FRET histogram analysis because of lipid flip-flops and variations in the final FRET values, which can occur if there is a marked heterogeneity in vesicle sizes. Smith and Weisshaar29 found that identifying the hemifusion population solely from histogram peak fitting is often challenging unless a majority of the population are in that state. Instead, fluorescence intensity time-trace analysis is a more direct way to observe the hemifusion state. Within an individual pair of vesicles, we can identify the low FRET state (docking), intermediate FRET state(s) (hemifusion) and a high FRET state (lipid mixed). For our content-mixing assay, because of the large size of DNA content-mixing indicators, formation of small or transient pore formation will be missed.
Experimental design
Single-vesicle lipid-mixing assay
In our vesicle-vesicle lipid-mixing assay, the v-and t-SNARE proteins (or vice versa) are reconstituted into two different groups of vesicles labeled with acceptor (DiD) and donor (DiI) fluorophores, respectively (for more information, see the supplier’s website, e.g., http://www.invitrogen.com/). The acceptor-labeled vesicles are immobilized on the surface and then the donor-labeled vesicles are added in order to observe the formation of a single vesicle-vesicle complex through specific SNARE interactions12 (Fig. 1). A flow chamber is assembled on a quartz slide that has been coated with PEG cushions to eliminate nonspecific binding. This is a surface passivation technique31–33 that is an alternative to the use of a supported bilayer29. The passivation of the flow chamber surface is crucial to ensure (i) that tethered vesicles are free of interaction with the surface and (ii) that free-floating vesicles will not randomly adhere to the surface.
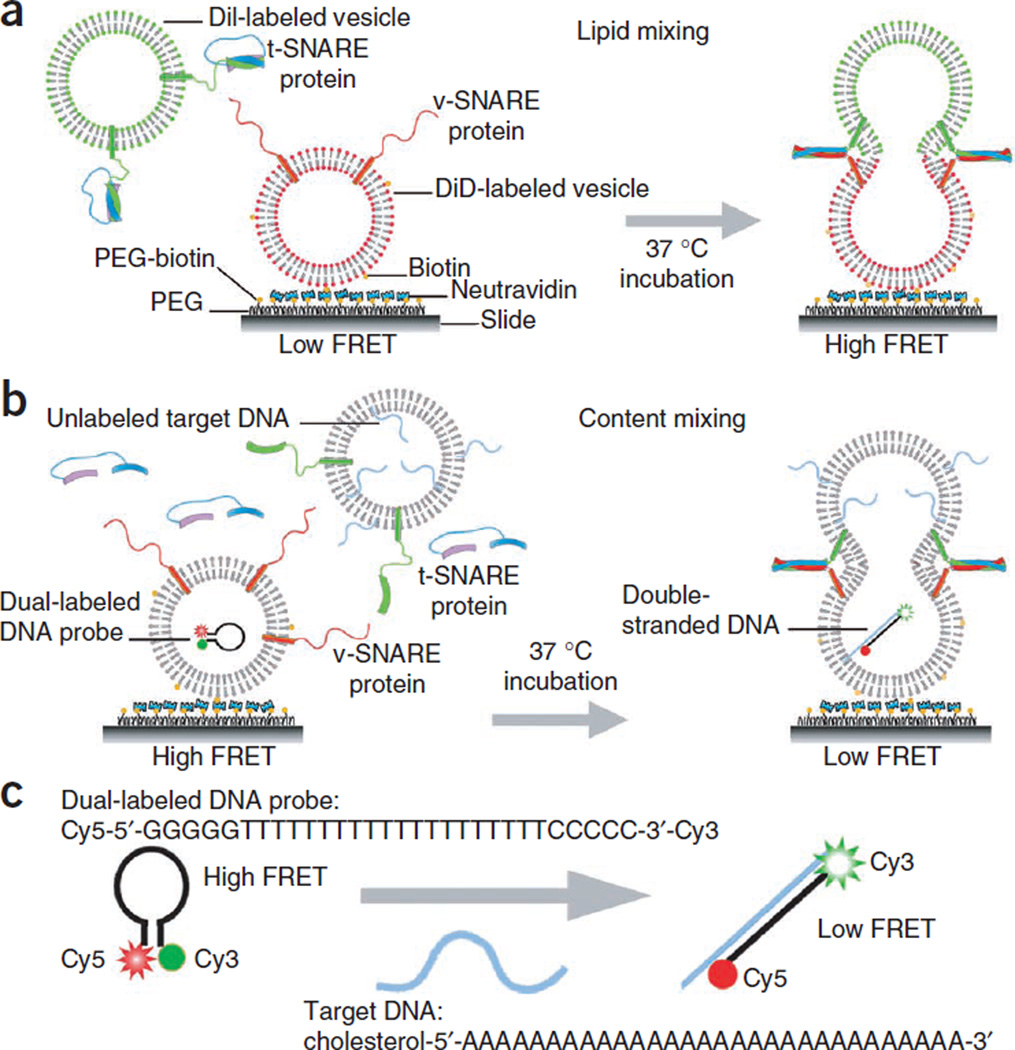
Figure 1.
Experimental scheme. (a) Single vesicle-vesicle lipid-mixing assay: DiD (acceptor)-labeled v-SNARE vesicles are immobilized on the bottom surface of a flow chamber, which is also the imaging surface of the TIR fluorescence microscope. DiI (donor)-labeled t-SNARE vesicles and accessory proteins are injected into the chamber space. After incubation at 37 °C, some t-SNARE vesicles dock to v-SNARE vesicles through the formation of trans-SNARE complexes, and accessory protein binds to the trans-SNARE complexes. When t- and v-SNARE vesicles proceed to fuse, resultant lipid mixing will entail an increase in the FRET efficiency. (b) Single vesicle-vesicle content-mixing assay: vesicles reconstituted with v-SNARE proteins and encapsulating dual-labeled DNA probes are immobilized on the surface of the flow cell. Vesicles reconstituted with t-SNARE proteins and encapsulating complementary poly-A DNA strands are flowed in, and the sample is incubated at 37 °C. The vesicle membrane is unlabeled for the content-mixing assay. (c) The Cy3/Cy5 dual-labeled DNA probe is a single-stranded oligonucleotide with a stem-and-loop hairpin structure. The loop contains a probe sequence (T20) that is complementary to a target DNA (A30), and the stem is formed by the annealing of complementary arm sequences that are located on either side of the probe sequence (GGGGG and CCCCC). When the hairpin probe is in the closed state, Cy3 and Cy5 fluorophores linked to the ends are in close proximity and show a high FRET value. When the hairpin probe hybridizes to a target DNA, it switches to the open state, increasing the distance between two dyes and showing a low FRET value. In order to achieve a high incorporation efficiency of target DNA in the t-SNARE vesicle, we included a cholesterol modification at the 5′ end of the target DNA strand. The size of both the probe and target DNA molecules is several nanometers in diameter, which is much larger than the size of normal neurotransmitters. We purchased DNA strands from Integrated DNA Technologies (http://www.idtdna.com/). Cholesterol is conjugated to DNA through a tetraethylene-glycol linker. Cyanine dye carrying an N-hydroxysuccinimidyl ester (NHS-ester) reactive group is linked to the aminoallyl nucleotide located at the end of DNA. Through the hydrophobic cholesterol molecule, each t-SNARE vesicle is anchored with 10–15 unlabeled target DNAs. By photobleaching Cy5, we determined that ~0.8 Cy3/Cy5 dual-labeled DNA probes are encapsulated in v-SNARE protein-reconstituted vesicles.
NeutrAvidin is then added to coat the surface. Through the specific interaction between NeutrAvidin and biotin-conjugated lipid molecule in the membrane, the v-SNARE vesicles are tethered to the bottom surface of the flow chamber. Nonspecific binding test of SNARE-reconstituted vesicles in the absence of NeutrAvidin or biotin-conjugated lipid can be performed to check the quality of the surface.
After washing out free v-SNARE vesicles, the t-SNARE vesicles are diluted with accessory proteins and injected into the flow chamber to induce assembly of SNARE complexes and membrane fusion between a single pair of t-and v-SNARE vesicles. The use of a low concentration of t-SNARE vesicles can dramatically reduce the probability of multiple vesicle interactions to guarantee an operation at the single vesicle-vesicle pair level. After incubation at 37 °C, free t-SNARE vesicles and accessory proteins are removed by washing, and FRET measurements are made using a total internal reflection (TIR) fluorescence microscope to reveal the final outcome of the fusion reaction. Docking and fusion reactions can also be observed in real time. From increasing values of the FRET efficiency, real-time fusion intermediates with different degrees of lipid mixing can be discerned. For example, an intermediate FRET is obtained for hemifusion, in which only the outer leaflets mix. From the hemifusion state, after merging inner leaflets, a fusion pore opens to mix content, which corresponds to the highest FRET value. During the method development process, we used a 1:100 protein-to-lipid ratio for both lipid and content-mixing assays of yeast SNAREs in order to maximize the reaction yield. The ratio may be reduced to a more biologically relevant value, as we did for our studies involving neuronal SNAREs21.
Single-vesicle content-mixing assay
Inspired by conventional molecular beacons which use a DNA hairpin labeled with a fluorophore and a quencher34, we designed a DNA hairpin composed of a 5-bp stem and a poly-thymidine loop (T20) labeled with a donor (Cy3) and an acceptor (Cy5) fluorophore at either end of the stem (Fig. 1c). When the loop region of this Cy3/Cy5 dual-labeled DNA probe hybridizes with a second DNA strand that has a complementary sequence (A30), the formation of a long double-strand breaks apart the stem region and separates the donor and the acceptor fluorophores from one another (Fig. 1b). The v-SNARE vesicles, with v-SNARE proteins reconstituted in the membrane and DNA hairpin encapsulated inside, are immobilized on a polymer-coated quartz surface via biotinylated lipids. The t-SNARE vesicles containing t-SNAREs and harboring multiple poly-A DNAs with a cholesterol modification are added together with soluble t-SNAREs. Cholesterol modification increases the incorporation efficiency of poly-A DNA in the t-SNARE vesicle membrane, thus increasing the probability that a fusion event results in the opening of DNA hairpin. When two vesicles dock and a large enough fusion pore forms between them, the two DNA molecules with a molecular weight of ~10 kDa, which is much larger than the size of normal neurotransmitters, should hybridize, thereby resulting in a change in the FRET efficieny (E). Because of the larger size of our probe, the assay is useful in detecting the pore expansion, but not the initial pore opening between fusing membranes.
MATERIALS
REAGENTS
1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate, DiIC18(5) (DiD; Invitrogen, cat. no. D307)
1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate, DiIC18(3) (DiI; Invitrogen, cat. no. D282)
1,2-Dioleoyl-sn-glycero-3-phospho-l-serine (sodium salt) (DOPS; Avanti Polar Lipids, cat. no. 840035C)
1,2-Dioleoyl-sn-glycerol-3-phosphoethanolamine (DOPE; Avanti Polar Lipids, cat. no. 850725C)
1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt) (Biotin-DPPE; Avanti Polar Lipids, cat. no. 870277X)
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; Avanti Polar Lipids, cat. no. 850457C)
Acetic acid (glacial; Fisher Scientific, cat. no. A38-500)
Acetone (Fisher Scientific, cat. no. A18-4)
Alconox (Lab Safety Supply, cat. no. 2494)
Biotin-polyethylene glycol-succinimidyl valerate (molecular weight 5,000, Biotin-PEG-SVA; Laysan Bio, cat. no. biotin-PEG-SVA-5000)
Bio-Beads SM-2 adsorbent (Bio-Rad, cat. no. 152-3920)
β-Mercaptoethanol (Sigma, cat. no. M7154)
Chloroform (Fisher Scientific, cat. no. C606-1) ! CAUTION Chloroform is a carcinogen. All sample preparations involving chloroform should be performed under a chemical hood.
Catalase (Sigma, cat. no. C3155)
Cholesterol (Avanti Polar Lipids, cat. no. 70000P)
Dextrose (Sigma, cat. no. D9559)
Glucose oxidase (Sigma, cat. no. G2133)
Glycerol (RPI, cat. no. G22020-4.0)
HEPES (RPI, cat. no. H75030-1000.0) ▲ CRITICAL Once any buffer including HEPES is prepared, it should be stored at 4 °C and used within 1 month.
Hydrochloric acid (HCl; Fisher Scientific, cat. no. A144c-212)
l-α-Phosphatidylinositol-4,5-bisphosphate (brain, porcine triammonium salt) (PIP2; Avanti Polar Lipid, cat. no. 8400046X)
Liquid nitrogen
Methanol (Fisher Scientific, cat. no. A412-4)
Methoxy-polyethylene glycol-succinimidyl valerate (molecular weight 5,000, mPEG-SVA; Laysan Bio, cat. no. mPEG-SVA-5000)
N-(2-aminoethyl)-3-aminopropyltrimethoxysilane, amino silane (United Chemical Technologies, cat. no. A0700)
NeutrAvidin (Thermo Scientific, cat. no. LF144746)
n-Octyl-β-d-glucopyranoside (OG; RPI, cat. no. N02007-10.0)
Potassium hydroxide (KOH; Fisher Scientific, cat. no. P250-3)
Potassium chloride (KCl; Fisher Scientific, cat. no. P217-3)
Sodium bicarbonate (Fisher Scientific, cat. no. S233-500)
Sodium chloride (NaCl; Fisher Scientific, cat. no. S271-3)
Tris(hydroxymethyl) aminomethane (RPI, cat. no. T60040-5000.0)
EQUIPMENT
Aluminum foil
Beakers
Coverslips (24 mm × 40 mm, no. 1 12, rectangular; VWR, cat. no. 48393230)
Diamond drill bits (3/4 mm; Kingsley North, cat. no. 1-0500-100)
Drilling set (Dremel 300-N/10 300; Dremel)
Double-sided tape (~100 µm thick; 3M, cat. no. 20014958)
Epoxy (5 min; Devcon)
Flask (Pyrex, Fisher Scientific, cat. no. 07-250-091)
Forceps (Fisher Scientific, cat. no. 13-812-8)
Glass staining dishes (Fisher Scientific, cat. no. 08-817)
Gastight syringes: 50–500 µl (Hamilton)
Glass tubes (13 mm × 100 mm; Fisher Scientific, cat. no. 14-958A)
GyroMini nutating mixer (RPI, cat. no. 246260)
Magnetic stirrer
Microfuge tubes (Eppendorf, cat. no. centrifuge 5415 D)
Mini extruder set: including one extruder, two syringes (250 µl), 100 PC membranes (0.1 µM), 100 filter supports and one holder/heating block (Avanti Polar Lipids, cat. no. 610023)
Needle (26 gauge, 3/8 inch; BD, cat. no. 305110)
Parafilm (Fisher Scientific, cat. no. 13-374-10)
Propane torch (Bernzomatic, Fisher Scientific, cat. no. 19-313-625)
PTFE tubing (Weico Wire, cat. no. ETT-28)
Slides (quartz microscope, 1 inch × 3 inch, 1 mm thick; G. Finkenbeiner, cat. no. 1×3×1)
Sterile disposable syringe (1 ml, BD, cat. no. 309602)
Vacuum desiccator
Vacuum pump
Vortex mixer (Fisher Scientific, cat. no. 02215365)
Milli-Q H2O (Millipore)
Water circulator (NESLAB RTE-7 Digital One refrigerated bath; Fisher Scientific)
Pellin-Broca prism (CVI Laser)
Quartz slide with a thin layer of immersion oil (Olympus)
Nd:YAG laser (Crystalaser)
HeNe laser (Melles Griot)
Water-immersion objective lens (Olympus)
Dual-band filter (Chroma Technology, 51007)
Dichroic mirror (Chroma Technology, 645DCXR)
Electron-multiplying charge-coupled device (EM-CCD) camera (Andor Technology, iXon DV 887-BI)
REAGENT SETUP
Biotin-DPPE stock
Prepare the stock in chloroform (0.1 mg ml− 1 or 0.5 mg ml− 1). The solution can be stored at −20 °C for up to 1 month.
Cholesterol stock
Prepare the stock in chloroform (20 mg ml− 1). The solution can be stored at −20 °C for up to 1 month.
Dialysis buffer
Dialysis buffer must be freshly prepared each time using HEPES buffer containing 5% (vol/vol) glycerol.
DiD working solution
DiD working solution is prepared using 100% ethanol (1 mg ml− 1). ▲ CRITICAL The solvent is subject to evaporation. An absorbance measurement may be done to verify the accuracy of the dye concentration. Store the solution at −20 °C for up to 1 month without recalibration.
DiI working solution
DiI working solution is prepared using 100% ethanol (1 mg ml− 1). ▲ CRITICAL The solvent is subject to evaporation. An absorbance measurement may be done to verify the accuracy of the dye concentration. Store the solution at −20 °C for up to 1 month without recalibration.
Fusion buffer
The solution consists of 25 mM HEPES, 100 mM KCl, and 5% (vol/vol) glycerol (pH 7.4, with KOH) and is filtered a through 0.2-µm filter. It can be stored at 4 °C for 1 month.
HEPES buffer
The solution consists of 25 mM HEPES and 100 mM KCl (pH 7.4, with KOH). Store HEPES buffer at 4 °C for up to 1 month.
HEPES buffer with the oxygen scavenging system
This buffer consists of HEPES buffer with 0.8% (wt/vol) dextrose, 165 U ml− 1 glucose oxidase, 2170 U ml− 1 catalase and 1% (vol/vol) β-mercaptoethanol. Store at 4 °C for up to 1 month.
PEG buffer
PEG buffer consists of 0.1 M sodium bicarbonate (pH 8.5). This solution must be freshly prepared.
OG stock, 10 wt%
Prepare the stock in fusion buffer. It can be stored at 4 °C for 1 month.
T50 buffer
The solution consists of 10 mM Tris-HCl and 50 mM NaCl (pH 8.0, with NaOH). Store T50 buffer at 4 °C for 1 month.
EQUIPMENT SETUP
TIR fluorescence microscope setup
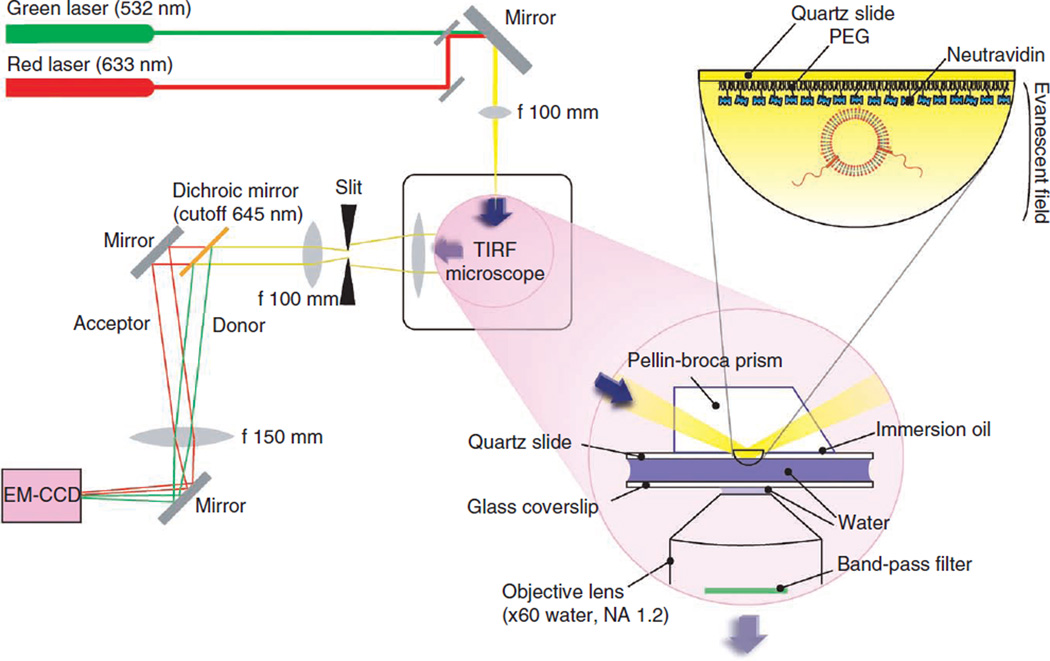
A schematic illustration of a prism-type TIR fluorescence microscopy is shown in Figure 2, and a photo of our homemade TIR fluorescence microscope setup is shown in Supplementary Figure 1a. The excitation beam is focused into a Pellin-Broca prism placed on top of a quartz slide with a thin layer of immersion oil (Supplementary Fig. 1b). The donor (DiI or Cy3) and acceptor (DiD or Cy5) dyes are excited by an Nd:YAG laser (532 nm) and a HeNe laser (633 nm), respectively (Supplementary Fig. 1c). The fluorescence emissions are collected by a water-immersion objective lens (×60, NA = 1.2). The excitation beam is blocked by a dual-band filter. The emission beam passes through a slit to adjust its total width (Supplementary Fig. 1d), and is split to donor and acceptor emission by a 645-nm dichroic mirror and detected simultaneously by the electron-multiplying charge-coupled device camera with a time resolution of 100 ms. The latest version of the data acquisition software is available for download from the group website of T.H. (http://bio.physics.illinois.edu/). The current procedure is written on the basis of the use of this software. Additional details of the wide-field TIR fluorescence microscope setup can be found in our previous review papers31–33.
Figure 2.
The schematic illustration of a prism-type TIR fluorescence (TIRF) microscopy with both excitation and emission pathways as well as the TIR occurring at the interface between a quartz slide and water. Photos of the homemade setup are shown in Supplementary Figure 1.
PROCEDURE
Preparation of PEG slides ● TIMING at least 7 h
-
1|
Take the amino silane out of the freezer and keep it in a dark place to equilibrate it to room temperature (21–25 °C).
-
2|
While wearing appropriate gloves, use acetone and tap water to rinse the slides with drilled holes for channels; scrub away any residues on their surfaces with your fingers.
-
3|
Sonicate the slides in a glass staining dish for 15 min in 10 wt% Alconox.
-
4|
Decant all the Alconox out of the glass staining jar and rinse it with excess tap water.
-
5|
Rinse the slides in the glass staining jar three times with deionized Milli-Q H2O.
-
6|
Sonicate the slides for 5 min in deionized Milli-Q H2O.
-
7|
7| Discard the Milli-Q H2O from the glass staining jar. Rinse the slides three times with deionized Milli-Q H2O.
-
8|
Sonicate the slides in the glass staining jar for 20 min in acetone.
-
9|
Discard the acetone from the glass staining jar. Rinse the slides three times in deionized Milli-Q H2O.
-
10|
Sonicate the slides, coverslips and a 250-ml Erlenmeyer flask for 1 h in 1 M KOH.
-
11|
Discard the KOH from the staining jars and flask. Rinse the slides and the coverslips in the glass staining jars and the flask three times in Milli-Q H2O.
-
12|
Fill the Erlenmeyer flask with methanol and sonicate it for 15 min.
-
13|
Burn the surface of each slide (hold it with forceps) using a propane torch to burn away any fluorescent organic molecules. Burn both sides of each slide for at least 30 s, then place it back in the glass staining jar. Burn each side of the coverslip (for no longer than 2 s to prevent it from breaking), then place it back in the glass staining jar.
-
14|
Discard the methanol in the flask. Prepare the amino silane solution by mixing 150 ml of methanol, 7.5 ml of acetic acid and 1.5 ml of amino silane in the flask. Promptly pour the solution into the emptied glass staining jars containing the slides and the coverslips. Before storing the amino silane, desiccate the amino silane for 10 min in a vacuum chamber and flow nitrogen for ~20 s into the chamber. Seal the cap with a strip of Parafilm for storage.
▲ CRITICAL STEP The solution should be poured into staining jars immediately after mixing.
-
15|
Incubate the slides and coverslips for 10 min, sonicate them for 1 min for a brief mixing and incubate them for additional 10 min in the amino silane–containing solution.
-
16|
Take out the mPEG and biotin-PEG from the freezer and equilibrate them to room temperature.
! CAUTION Avoid condensation by keeping caps closed during equilibration and minimize the exposure to the ambient light.
-
17|
Discard the amino silane solution from the glass staining jars.
-
18|
Use forceps to hold each slide and coverslip and rinse them with Milli-Q H2O followed by methanol.
-
19|
Dry each slide with nitrogen gas by blowing it from the edge while holding it with forceps.
-
20|
Place the slides on a flat surface (e.g., plastic container or a box).
-
21|
Prepare the reaction solution by dissolving 0.4 mg of biotin-PEG and 16 mg of mPEG in 64 µl of freshly made PEG buffer for each slide in a 1.5-ml Microfuge tube. Adjust the ratio of biotin-PEG and mPEG and sodium bicarbonate according to the number of slides.
▲ CRITICAL STEP This solution must be prepared just before applying to slide surfaces to minimize hydrolysis.
-
22|
Desiccate biotin-PEG and mPEG for 5 min in a vacuum chamber and flow nitrogen for ~20 s into the chamber before storage. Put Parafilm around the cap for storage.
-
23|
Dissolve the solution by inverting the tube several times. Centrifuge at 10,000g for 1 min at room temperature to remove air bubbles and to spin down any undissolved PEG.
-
24|
Immediately after the spin, add 70 µl of the reaction solution to the imaging area of each slide.
▲ CRITICAL STEP Make sure there are no air bubbles. To get rid of the bubbles, gently tap the bubble two or three times with a pipette tip.
-
25|
Place the coverslip over each slide to spread the solution evenly and to prevent drying.
-
26|
Incubate the slides in a dark and humid environment for at least 3 h. The incubation can also extend to 24 h.
▲ CRITICAL STEP Added humidity slows down the evaporation of the PEG solution and is crucial. H2O may be added to the bottom of a container (e.g., an empty pipette tip box) where the slides are kept.
-
27|
Disassemble the slides and coverslips by using forceps and rinse each of them with excess Milli-Q H2O and dry them with nitrogen. Store them at − 20 °C in 50-ml tubes or any appropriate containers until use (one slide and coverslip set per tube).
Vesicle preparation
-
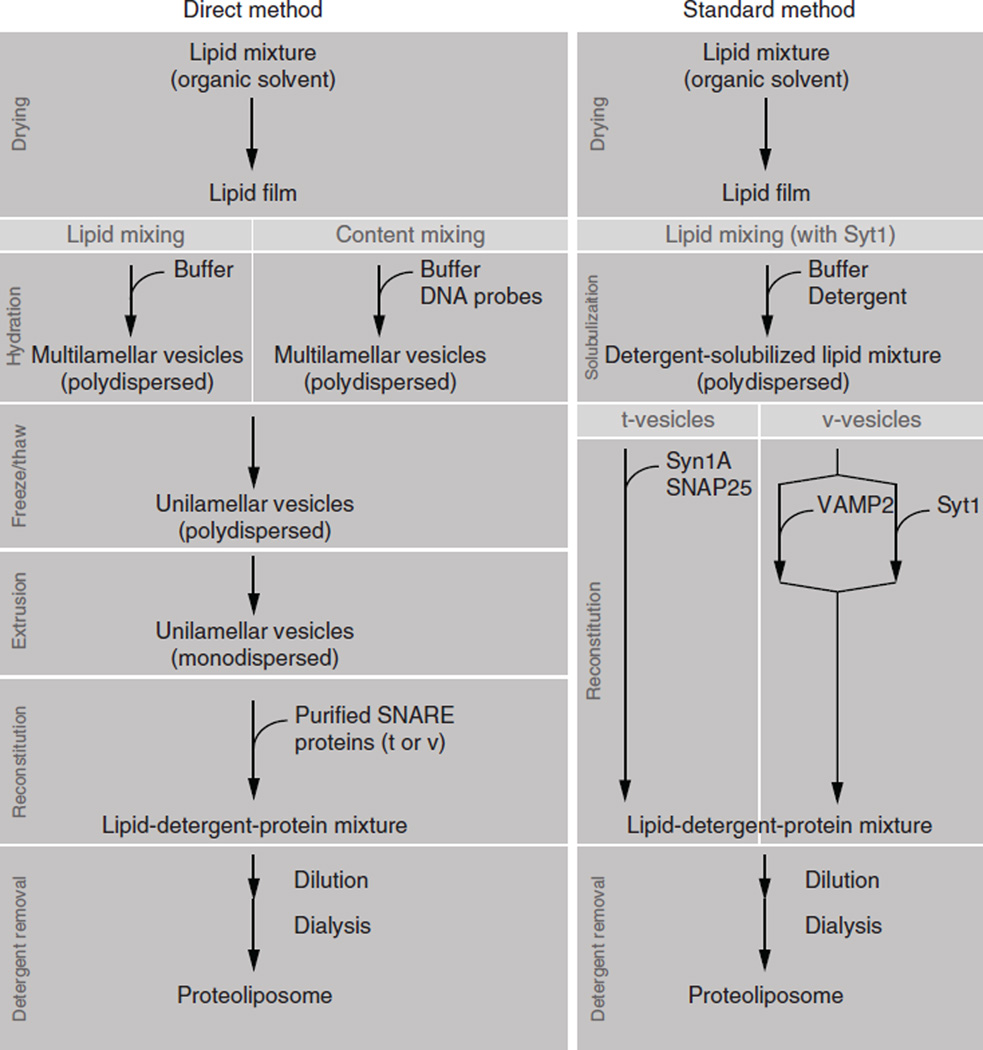
28|As summarized in Figure 3, vesicles prepared by both standard and direct methods8 can be used for the single vesicle-vesicle assay. The composition of the lipid solutions required are described in options A, B and C for the preparation of lipid-mixing vesicles (direct method), the preparation of content-mixing vesicles (direct method) and the preparation of the lipid mixture (standard method). The composition of the lipid can be changed according to the purpose of the experiment12,21. However, a verification of vesicle size and protein incorporation efficiency is suggested when using a different composition. Rizo, et al. reports the detailed procedures in ref. 8.
- Preparation of lipid-mixing vesicles (direct method) ● TIMING 5 h
-
Combine the following lipid solution in a glass tube for t- and v-vesicles, respectively (PC/PS/cholesterol = 43:15:40 mol%, 2 mol% dyes).
t-vesicle (10 mM) v-vesicle (10 mM) POPC (20 mg ml−1) 80 μl 80 µl DOPS (20 mg ml−1) 30.2 µl 30.2 µl Cholesterol (20 mg ml−1) 38.6 µl 38.6 µl DiI (1 mg ml−1) 100 µl — DiD (1 mg ml−1) — 98 µl Bio-DPPE (0.5 mg ml−1) — 6.4 µl ! CAUTION Chloroform is a carcinogen. The experiment should be performed in a chemical hood.
-
- Preparation of content-mixing vesicles (direct method) ● TIMING 5 h
-
Combine the following components in a glass tube for t- and v-vesicles, respectively (PC/PS/cholesterol = 45:15:40 mol%).
t-vesicle (10 mM) v-vesicle (10 mM) POPC (20 mg ml−1) 85.8 µl 85.8 µl DOPS (20 mg ml−1) 30.5 µl 30.5 µl Cholesterol (20 mg ml−1) 38.6 µl 38.6 µl Bio-DPPE (0.5 mg ml−1) 10.5 µl ! CAUTION Chloroform is a carcinogen. The experiment should be performed in a chemical hood.
-
- Preparation of lipid mixture (standard method) ● TIMING 4 h
-
Combine the following lipid components in a glass vial for making t-vesicles (PC/PS/cholesterol/PE/PIP2/DiI = 32:15:20:25:6:2 mol%) and v-vesicles (PC/PS/cholesterol/PE/DiD = 50:3:20:25:2 mol%), respectively.
t-vesicle (10 mM) v-vesicle (10 mM) DOPS (5 mg ml−1) 54 µl 11 µl POPC (10 mg ml−1) 54.5 µl 85.8 µl Cholesterol (5 mg ml−1) 34.5 µl 35 µl DOPE (10 mg ml−1) 41.5 µl 42 PIP2 (1 mg ml−1) 147 Biotin-DPPE (0.1 mg ml−1) 2.5 µl DiI (1 mg ml−1) 41.5 µl DiD (1 mg ml−1) 643 µl ! CAUTION Chloroform is a carcinogen. The experiment should be carried out in a chemical hood.
-
-
29|
Evaporate the organic solvent carefully under a gentle stream of nitrogen. Tilt and rotate the tube continuously until a thin film is formed at the bottom of the tube.
! CAUTION Adjust the air stream in an appropriate strength to prevent the lipid solution from spilling out of the glass tube.
-
30|
Wrap the tube with a piece of foil with a small hole on the top for chloroform evaporation and place the tube into a vacuum desiccator for at least 2 h for further drying.
▲ CRITICAL STEP Shield samples from exposure to the ambient light by aluminum foil or a box to minimize the photobleaching of dyes.
-
31|Hydrate the lipid film in 500 µl of buffer by vortexing until it is completely suspended (typically for 20 min). The buffer composition is different for the three options.
Option (from Step 28) Buffer Comment A HEPES Vortexing typically takes 20 min B HEPES buffer solution containing 10 µM target DNA and v-vesicle lipid film in 500 µl of HEPES buffer solution with 1 µM Cy3/Cy5 dual-labeled molecular beacon Vortex several times to completely dissolve the lipid C Fusion buffer containing 1.3 wt% OG -
32|Pipette the lipid solution into a 1.5-ml Microfuge tube. For the direct method (from Step 28A and Step 28B) and the standard method (from Step 28C) follow directions as given in the table below:
Step 28A, Step 28B Seal tightly with Parafilm and wrap in foil. Perform Steps 33–35 Step 28C Mix gently with a rotator for 1 h at room temperature, and proceed to Step 35 -
33|
Freeze and thaw the lipid solution using liquid nitrogen and warm water (30–40 °C) in the water bath for at least 5 cycles.
! CAUTION The Microfuge tube cap may pop open if it is not sealed tightly.
-
34|
Extrude the lipid solution through a 100-nm polycarbonate membrane (25 cycles) according to the manufacturer’s instructions. Detailed mini-extruder assembly and extrusion information are available from the Avanti Polar Lipids website (http://www.avantilipids.com/).
-
35|
Store the vesicle solution in a Microfuge tube at 4 °C for up to 1 week.
Figure 3.
Standard versus direct methods of preparing SNARE-reconstituted vesicles preparation. Direct method: Steps 28A/28B and 29–35 for vesicle preparation and Steps 36A and 37 for protein reconstitution. Standard method: Steps 28C–32 for lipid preparation and Steps 36B and 37 for protein reconstitution.
Reconstitution of SNAREs
-
36|Follow the steps in option A for the reconstitution of yeast SNAREs (direct method), and follow option B for the reconstitution of neuronal SNAREs (standard method).
- Reconstitution of yeast SNAREs (direct method) ● TIMING 1 h plus dialysis time (10–12 h)
-
Reconstitute the protein with lipid vesicles containing lipophilic or content dyes (from Step 35) at a 1:100 protein to lipid ratio, according to the table below, in a 1.5-ml Microfuge tube. Mix by inverting several times. Yeast t-SNARE protein Sso1pHT and v-SNARE protein Snc2p were dissolved in buffers containing 0.8 wt% OG detergent solution.
t-vesicle v-vesicle t-vesicles (10 mM) 10 µl v-vesicles (10 mM) 10 µl Sso1pHT proteins (20 µM) 50 µl Snc2p proteins (20 µM) 50 µl 10% OG 0.9 µl 0.9 µl ▲ CRITICAL STEP Keep proteins and vesicles on ice. Protect the vesicles from exposure to the light. 10 wt% OG is supplemented to keep the OG concentration of the mixture at ~0.8 wt% (> critical micelle concentration, CMC). - Wrap the tubes with foil and rock them in a cold room (4 °C) for 20 min.
-
Dilute the vesicle/protein mixture twofold with 61 µl of dialysis buffer. Mix by inverting four times.! CAUTION The vesicle harboring the SNARE protein is formed as the OG concentration is reduced below the CMC. Therefore, use a sufficient amount of fusion buffer to lower the OG concentration below the CMC.
- Centrifuge the tube at 1,000g for 2 min at room temperature using a bench-top centrifuge.
- Move the solutions into prehydrated dialysis tubes and dialyze them against 2 liters of dialysis buffer for 10–12 h at 4 °C.
-
- Reconstitution of neuronal SNAREs (standard method) ● TIMING 3 h plus dialysis time (10–12 h)
- For a t-SNARE vesicle sample, mix syntaxin-1A and SNAP-25 (molar ratio of 1:2 syntaxin-1A/SNAP-25) in fusion buffer containing 1.3 wt% OG by gentle agitation for 1 h at 4 °C to form a t-SNARE complex. The total volume of syntaxin-1A–SNAP-25 mixture is 40 µl.
-
Combine the DiI-containing lipid solution (from Step 32) and the t-SNARE precomplex in a 1.5-ml Microfuge tube by gentle agitation for 1 h at 4 °C.! CAUTION The protein-to-lipid ratio could be changed, depending on the purpose of the experiment. The volume of the mixture is generally 80 µl.
-
For the v-SNARE + synaptotagmin-1 (Syt1) vesicle sample, combine the DiD-containing lipid solution (from Step 32) and VAMP2 in fusion buffer containing 1.3 wt% OG and 1 mM DTT in a 1.5-ml Microfuge tube and gently agitate for 1 h at 4 °C. In a separate tube, similarly combine DiD-containing lipid solution (from Step 32) and Syt1.! CAUTION The protein-to-lipid ratio could be changed according to the purpose of the experiment, but the volume of each mixture is generally 40 µl.
-
Mix the lipid-VAMP2 and lipid-Syt1 solutions in a 1.5-ml Microfuge tube with gentle agitation for 1 h at 4 °C. Supplement with enough DTT so that its final concentration is 1 mM.! CAUTION The addition of DTT helps to prevent intermolecular interaction between VAMP2 and Syt1 and to reduce protein aggregates.
-
Dilute the both lipid/protein solutions by threefold with fusion buffer.! CAUTION Use a sufficient amount of fusion buffer to lower the OG concentration below the CMC.
- Transfer 2 g of Bio-beads SM-2 into a 50-ml plastic tube and wash the beads three times with excess distilled water.
- Hydrate the dialysis tubes with fusion buffer for 10 min at room temperature.
- Discard the buffer from dialysis tubes with a pipette.
- Transfer 1 liter of fusion buffer and prewashed SM-2 beads into a beaker and stir the buffer gently with a magnetic stir bar at 4 °C.
- Transfer each diluted sample into a separate prewetted dialysis tube and dialyze the samples for 2 h.
-
37|
Collect the dialyzed vesicles into Microfuge tubes. Measure the volume to calculate the lipid concentration.
▲ CRITICAL STEP The lipid concentration may change because of the change in the volume during dialysis. UV absorbance may also be used to compare the dye concentration before and after the dialysis. Although this is more quantitative, we have not seen a dramatic improvement over the volume-based concentration calculation approach.
■ PAUSEPOIN TThe protein-reconstituted vesicle samples may be stored at 4 °C, and should be used within 3 d.
Surface immobilization of v-vesicles ● TIMING 40 min
-
38|
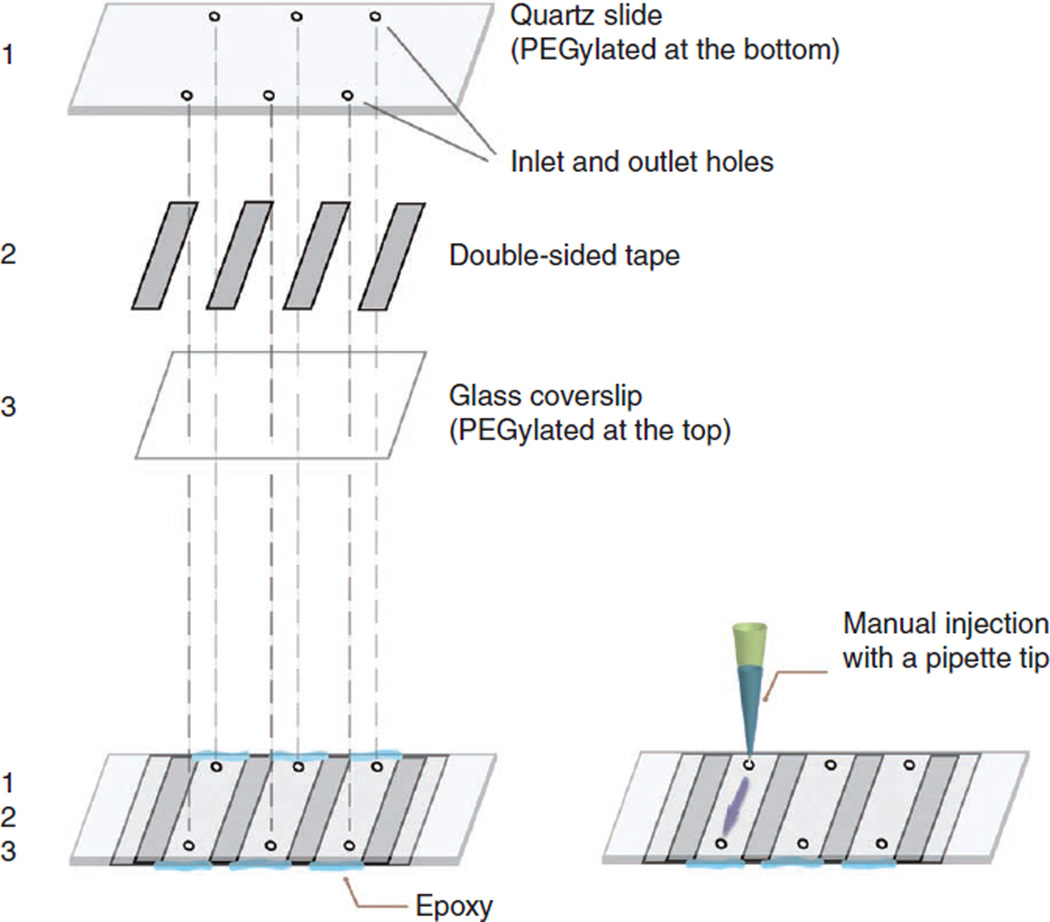
By following the steps shown in Figure 4, assemble the flow chamber using strips of double-sided tape and epoxy.
-
39|
Flow in 30 µl of 0.2 mg ml −1 NeutrAvidin in T50 into each empty flow channel and incubate for 5 min.
-
40|
Wash out the excess NeutrAvidin with HEPES buffer.
-
41|
Flow in 100 µl of 8 µM (lipid concentration) of v-vesicle and incubate for 15 min.
! CAUTION In general, minimize the exposure to ambient light to reduce photodamage.
-
42|
Check the surface density of v-vesicles by a TIR fluorescence microscope during the incubation using an appropriate laser excitation wavelength and intensity.
! CAUTION DiD dyes photobleach very quickly. Use the minimal sufficient laser intensity.
▲ CRITICAL STEP The entire surface should be covered by v-vesicles while individual vesicles may still be identified. There is batch-to-batch variation in the final vesicle concentration. The v-vesicle concentration should be adjusted to achieve the optimal vesicle density.
? TROUBLESHOOTING
-
43|
Wash out the excess v-vesicles with HEPES buffer (200–400 µl).
▲ CRITICAL STEP Avoid introduction of any air bubbles, as they will destroy vesicles on contact. Air bubbles may be minimized by first filling the flow chamber inlet with the buffer and making sure that there is no air in the tip of the pipette tip.
Figure 4.
Sample chambers are made by putting a quartz slide (1) and a glass coverslip (3) together with pieces of double-sided tape (2) and sealing with epoxy. The holes on the quartz slide are used for the inlet and outlet of solution exchange through each channel with a volume of ~10 µl.
Taking snapshots of single-vesicle fusion reaction ● TIMING up to 1 h
-
44|
Flow in 100 µl of solution containing 5 µM t-vesicles with the target concentration of Sec9c. Different solution conditions (e.g., protein concentrations, t-vesicle concentrations, lipid compositions) may be flowed into other channels for a comparison.
-
45|
Incubate the flow chamber at 37 °C inside a hydrated box (e.g., an empty pipette tip box with water).
▲ CRITICAL STEP The incubation time and t-vesicle concentration are both proportional to the docking/fusion. These parameters need to be optimized for each sample to show the best contrast from the control.
-
46|
Wash out the excess t-vesicles with 200–400 µl of HEPES buffer.
-
47|
Place the flow cell on the TIR fluorescence microscope setup and acquire short movies (ten frames with 532-nm excitation and five frames with 633-nm excitation, Fig. 5).
▲ CRITICAL STEP The excitation intensity needs to be adjusted such that single t-vesicles (the smallest fluorescent spots) emit a strong signal and a dim haze may be visible in the acceptor channel. With this intensity, fluorescent emission from aggregated vesicles will be saturated, but they will be filtered out during data analysis. Aggregation rarely occurs for fresh samples. We saw aggregations in aged samples and could remove them by centrifugation.
? TROUBLESHOOTING
-
48|
Acquire short movies from 20 to 30 random locations within the flow channel of interest. Select representative locations and avoid areas where large vesicle aggregates are present.
■ PAUSE POINT Movies from 20–30 random locations are enough to accurately determine the FRET distribution. A typical number of vesicle pairs we identify from these movies is ~4,000. No significant change was observed for imaging process involving ~100 random locations. A higher number of short movies may be necessary for samples with low docking efficiencies. Final saved data may be analyzed at any time, but data should be analyzed to check for an appropriate intensity distribution peaking between 300 and 1,000 (a.u.), which is instrument-dependent.
Figure 5.
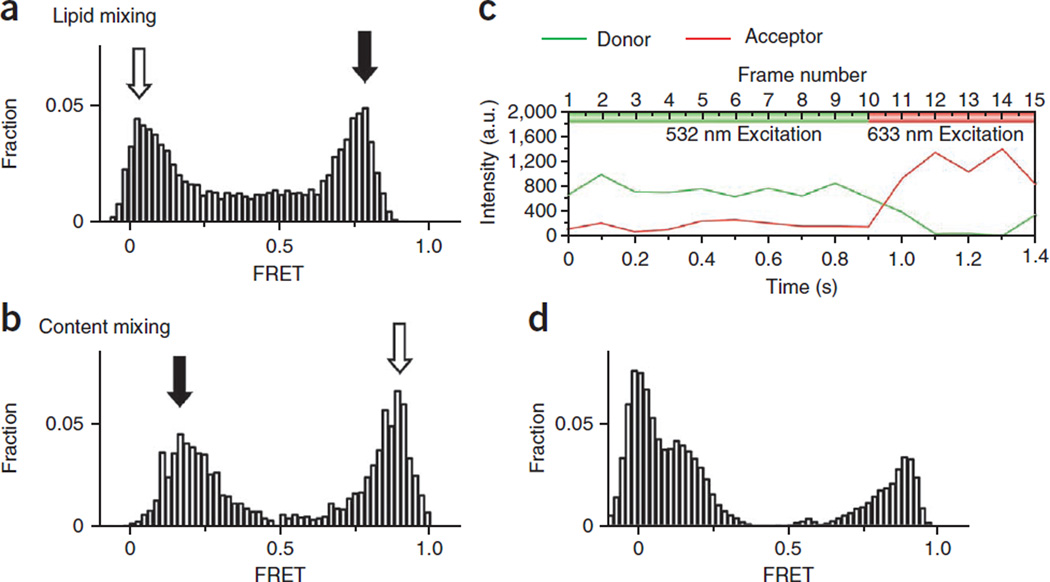
Experimental results. E distribution from a lipid mixing (a) is obtained by continuous 532-nm laser excitation. The y axis is the normalized population and the x axis is E. Open and solid arrows indicate nonfused and fused vesicle populations, respectively. Owing to background contribution in the acceptor channel from direct excitation, the FRET histogram is plotted on the basis of vesicles identified in the donor channel only. (b) The E distribution from a typical content-mixing assay is obtained from FRET beacon molecules containing both donor (Cy3) and acceptor (Cy5) dyes. Open and solid arrows indicate nonfused and fused vesicle populations, respectively. Unlike the lipid-mixing sample, in which labeling efficiency is ensured by the presence of multiple dyes in the vesicle membranes, some dual-labeled DNA probes may lack one or both dyes. (c) The presence of both dyes may be verified by short (5 frames) exposure of 633-nm illumination followed by 532-nm (10 frames) illumination. (d) With only 532-nm excitation, the E distribution shows an extra donor-only peak at E = 0. We selected only those DNA probes with both donor (532-nm excitation) and acceptor (633 nm excitation) fluorescence signals to construct the final histogram (b). a.u., arbitrary units.
Real-time single-vesicle fusion reaction ● TIMING 30 min
-
49|
Assemble the flow cell as shown in Figure 4.
-
50|
Turn on the temperature controller for the TIR fluorescence microscope setup to the target temperature (37 °C).
-
51|
Follow Steps 39–43 to immobilize v-vesicles on the flow chamber surface. For the final wash of the excess v-vesicles, use 200 µl of imaging buffer.
-
52|
Place the flow cell in the TIR fluorescence microscope setup, obtain the sharp focus and adjust the laser intensity.
▲ CRITICAL STEP The fluorescence signal will increase upon t-vesicle docking/fusion, so the laser intensity should be set lower to compensate for this. The flow channel used for the static measurement with the same condition will serve as a good reference point for setting this intensity.
-
53|
Load the t-vesicle and Sec9c mixture in the buffer reservoir.
-
54|
Quickly move to a new area of the flow chamber and start recording a movie file.
-
55|
Gently flow in the t-vesicle either manually or with a syringe pump by pulling the syringe plunger.
! CAUTION Flow rate is approximately 2 ml min −1. Because the flow is applied only for a few seconds and docking is allowed to occur over 10–15 min period for our snapshot-taking protocol, the flow rate should not influence the rate of docking.
-
56|
After 2 min, to avoid photobleaching, move to a new location within the flow channel and start acquiring another movie file.
-
57|
Repeat Step 56 two more times or until the frequency of docking dwindles.
Data analysis for static single-vesicle fusion ● TIMING 30 min
-
58|
Extract the donor and acceptor intensities from movie files for fitted molecules with acceptor intensity (on the basis of frames 12–14) using our custom software (available for download from our group website; http://bio.physics.illinois.edu/).
-
59|Calculate the relative FRET efficiency, taking into account the donor intensity bleed-through (BT) into the acceptor channel. The exact bleed-through factor (BF) needs to be measured for each instrument, but a typical range is 14–18%. represents the 10-frame-averaged intensity value of acceptor emission from excitation of the donor dye, and represents the 10-frame-averaged intensity value of donor emission from excitation of the donor dye.
-
60|Calculate the total intensity value for each molecule.
-
61|Calculate the stoichiometry value (S) for each molecule. IDexc represents the sum of emission intensities from donor-based excitation, IAexc represents the sum of emission intensities from acceptor-based excitation, and represents the 3-frame-averaged intensity (frames 12–14) value of acceptor emission from direct excitation of the acceptor dye.
-
62|
Select a range of total intensity (to avoid low-intensity background noise and high-intensity aggregates) and S (to select vesicle pairs with both donor and acceptor molecules) and plot the final FRET histogram on the basis of selected vesicle pairs.
Data analysis for real-time single-vesicle fusion ● TIMING up to 10 h
-
63|
Identify all molecule positions from series of 10-s averaged segments all through the duration of the experiment, and extract donor and acceptor intensities of these molecules from movie files using our custom software. This allows the identification of molecules docked to surface-bound vesicles at different time points during the experiment.
-
64|
Calculate the corrected acceptor intensity, total intensity and FRET values by following Steps 59 and 60.
-
65|
Analyze each time trace to extract out parameters such as docking dwell time, intermediate dwell time, intermediate FRET state and the final FRET state.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| Step | Problem | Reason | Solution |
|---|---|---|---|
| 42 | Too few vesicles | Insufficient biotins on the surface | Surface functionalization with the biotin-PEG may not have been done properly. To test for this, use other fluorescent markers such as dye-labeled streptavidin or oligonucleotides that have been dual-labeled with biotin and fluorescent dye. If the surface biotin-PEG is fine, there may be a problem with NeutrAvidin or the biotin-lipid |
| Large and bright vesicles | Aggregation of vesicles during reconstitution | Immobilize vesicles before reconstitution to identify the step at which the problem is caused. If the problem is during the reconstitution, the detergent concentration or the lipid composition may need to be optimized | |
| Too many vesicles | Incorrect vesicle concentration calculation | Use a fresh channel with a lower concentration of vesicles | |
| 47 | Too much donor bleed-through | Intensity is too high | Reduce the laser intensity with neutral density filter or a half-wave plate/beam splitter |
| Too many or too few docked vesicles | Unoptimized vesicle concentration or incubation time | Change the concentration or incubation time | |
| Too many docked vesicles | Nonspecific binding | Redo the PEGylation process. You may also run a control with a sample without Sec9c or SNAP-25 to check whether this is nonspecific binding |
● TIMING
Steps 1–27, preparation of PEG slides: at least 7 h
Step 28A (and 29–35), preparation of lipid-mixing vesicles for direct method: 5 h
Step 28B (and 29–35), preparation of content-mixing vesicles for direct method: 5 h
Steps 28C–32, preparation of lipid mixture for standard method: 4 h
Steps 36A and 37, reconstitution of yeast SNAREs: 1 h plus dialysis time (10–12 h)
Steps 36B and 37, reconstitution of neuronal SNAREs: 3 h plus dialysis time (10–12 h)
Steps 38–43, surface immobilization of v-vesicles: 40 min
Steps 44–48, static single-vesicle fusion reaction: up to 1 h
Steps 49–57, real-time single-vesicle fusion reaction: 30 min
Steps 58–62, data analysis for static single-vesicle fusion: 30 min
Steps 63–65, data analysis for real-time single-vesicle fusion: up to 10 h
ANTICIPATED RESULTS
FRET histograms are plotted from FRET signals collected from over one thousand vesicles before and after the fusion reaction. The y axis shows a normalized population in which we divided the distribution by the total number of vesicles measured, and the x axis is the FRET efficiency (E) value. For lipid mixing (Fig. 5a), the nonzero but low E value (open arrow) suggests close contact or docking between the donor and the acceptor vesicles without a high degree of lipid mixing, whereas the high E peak (solid arrow) indicates substantial lipid mixing. The relatively broad E distribution can be attributed to size variation of vesicles and measurement noise24. For content mixing (Fig. 5b), nonfused v-SNARE vesicles with dual-labeled probes in the closed state show a high E value (open arrow). In contrast, a nonzero low E peak (solid arrow) corresponds to the vesicle population with a DNA hybridized, opened probe induced by content mixing. E distribution of content mixing with the donor-only population subtracted can be obtained through an alternating laser excitation system of 532 and 633 nm (Fig. 5c). As shown in Figure 5d, a donor-only peak of zero E appears when we use 532-nm excitation only. As the nonzero low E peak is the positive sign of the content mixing, this zero E population complicates data analysis.
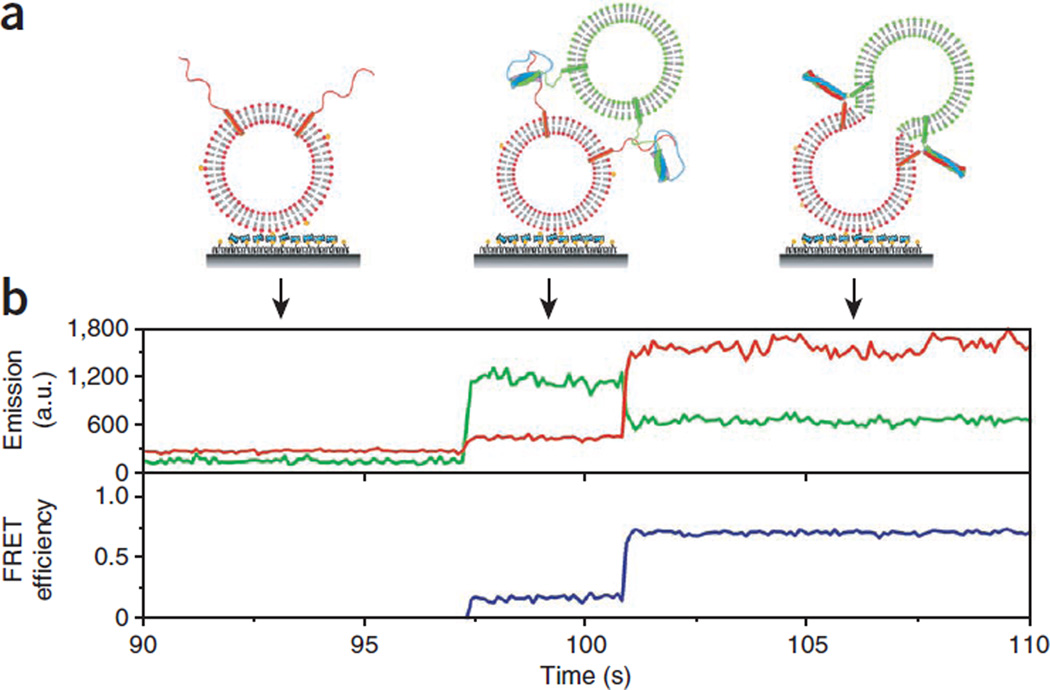
A typical single-vesicle lipid-mixing time trace is shown in Figure 6, demonstrating key advantages of our assay; fusion intermediates can be seen directly. Docking of a t-SNARE liposome to a surface-tethered v-SNARE vesicle is observed as an abrupt increase of the donor fluorescence intensity, and the FRET efficiency monotonically increases with increasing lipid mixing (observed as an increase in the acceptor intensity accompanied by an anticorrelated decrease in donor intensity) (Fig. 6). Additional docking by another t-SNARE vesicle can be ruled out, as such an event would show a further sharp increase in donor intensity, which was not observed at the low vesicle concentration used (~100 pM).
Figure 6.
Real-time lipid mixing for lipophilic dyes. (a) Cartoons showing different stages of fusion reaction between a vesicle pair. (b) Fluorescence intensity time traces of the donor (DiI, green) and the acceptor (DiD, red, top) and the corresponding trace of FRET efficiency, E (blue, bottom). No appreciable fluorescence signal change was observed until a t-SNARE liposome docked to a v-SNARE liposome. Rapid lipid mixing between the two vesicles caused by fusion leads to an increase in E. a.u., arbitrary units.
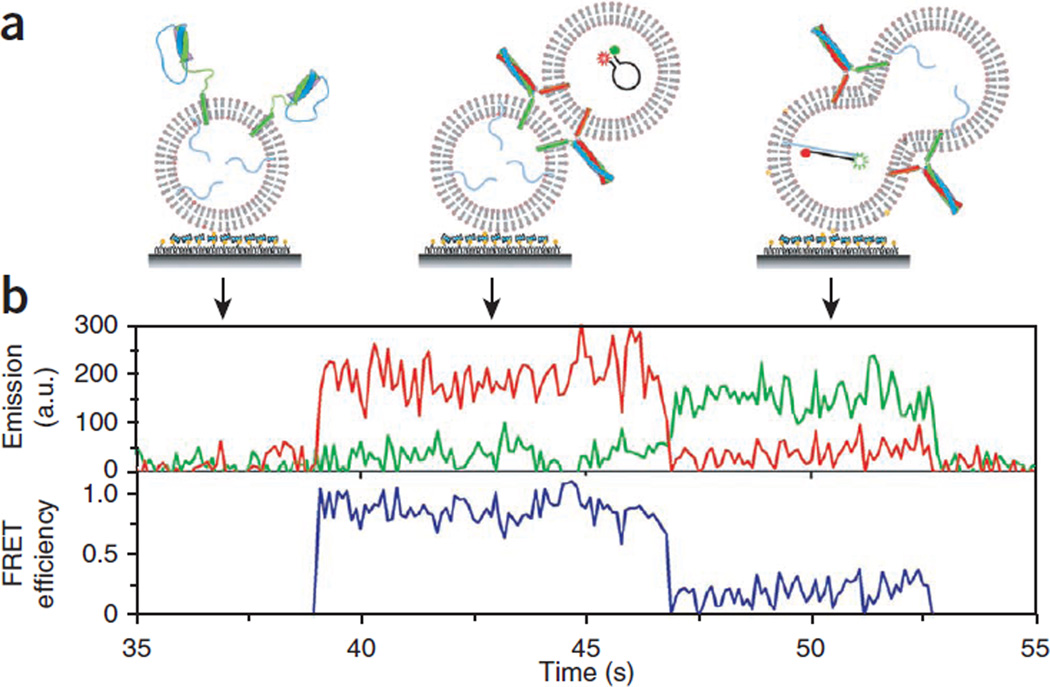
For the real-time content mixing (Fig. 7), the vesicle containing unlabeled target DNAs needs to be tethered to the surface. In this scheme, the fluorescence intensity time trace will initially show no FRET. The binding event of a v-vesicle that has the FRET beacon is seen as a sudden appearance of the fluorescence signal with a high E value. After a certain latent time, DNA hybridization caused by content mixing decreases E to a lower value, ~0.2.
Figure 7.
Real-time content mixing for content labeling only. (a) For this real-time experiment, we switched the surface immobilization configuration so that vesicle docking can be clearly marked as an abrupt increase in fluorescence signal. t-vesicles with the target DNAs (poly-A) are surface-immobilized and v-vesicles containing FRET beacons are flowed into the imaging chamber. To eliminate DNA molecules outside vesicles, DNase I treatment was applied before reactions. (b) A real-time content-mixing time trajectory of fluorescence intensities (green curve for Cy3, red curve for Cy5) and the corresponding E (blue curve). a.u., arbitrary unit.
Supplementary Material
Acknowledgments
This work was supported by the US National Institutes of Health Grants (R21 GM074526 to T.H. and R01 GM51290 to Y.-K.S.) and by the National Research Foundation of Korea grants funded by the Korean government (2009-0069857 and 2009-0090781 to T.-Y.Y.). T.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Note: Supplementary information is available via the HTML version of this article.
AUTHOR CONTRIBUTIONS J.D. and Y.I. performed experiments. J.D., Y.I. and C.J. drew figures. H.L. and T.-Y.Y. contributed the neuronal SNARE vesicle preparation protocols based on the standard method. Z.S. and Y.-K.S. developed the yeast SNARE reconstitution protocol based on the direct method. S.S. wrote the PEG slide preparation part. J.D., Y.I. and T.H. wrote the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 2.Wickner W, Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4-angstrom resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 5.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat. Struct. Mol. Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunger AT, Weninger K, Bowen M, Chu S. Single-molecule studies of the neuronal SNARE fusion machinery. Annu. Rev. Biochem. 2009;78:903–928. doi: 10.1146/annurev.biochem.77.070306.103621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 8.Chen XC, et al. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys. J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowen ME, Weninger K, Brunger AT, Chu S. Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs) Biophys. J. 2004;87:3569–3584. doi: 10.1529/biophysj.104.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fix M, et al. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl. Acad. Sci. USA. 2004;101:7311–7316. doi: 10.1073/pnas.0401779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu TT, Tucker WC, Bhalla A, Chapman ER, Weisshaar JC. SNARE-driven, 25-millisecond vesicle fusion in vitro. Biophys. J. 2005;89:2458–2472. doi: 10.1529/biophysj.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon TY, Okumus B, Zhang F, Shin YK, Ha T. Multiple intermediates in SNARE-induced membrane fusion. Proc. Natl. Acad. Sci. USA. 2006;103:19731–19736. doi: 10.1073/pnas.0606032103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diao JJ, et al. A single-vesicle content mixing assay for SNARE-mediated membrane fusion. Nat. Commun. 2010;1 doi: 10.1038/ncomms1054. Artn 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan YHM, van Lengerich B, Boxer SG. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc. Natl. Acad. Sci. USA. 2009;106:979–984. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerberg J, Cohen FS, Finkelstein A. Fusion of phospholipid-vesicles with planar phospholipid-bilayer membranes.1. Discharge of vesicular contents across the planar membrane. J. General Phys. 1980;75:241–250. doi: 10.1085/jgp.75.3.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JM, Ha T, Chu S, Boxer SG. Early steps of supported bilayer formation probed by single vesicle fluorescence assays. Biophys. J. 2002;83:3371–3379. doi: 10.1016/S0006-3495(02)75337-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyoung M, et al. In vitro system capable of differentiating fast Ca(2+)-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc. Natl. Acad. Sci. USA. 2011;108:E304–E313. doi: 10.1073/pnas.1107900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickel W, et al. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc. Natl. Acad. Sci. USA. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boukobza E, Sonnenfeld A, Haran G. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J. Phys. Chem. B. 2001;105:12165–12170. [Google Scholar]
- 20.Su ZL, Ishitsuka Y, Ha T, Shin YK. The SNARE complex from yeast is partially unstructured on the membrane. Structure. 2008;16:1138–1146. doi: 10.1016/j.str.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon TY, et al. Complexin and Ca2+ stimulate SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 2008;15:707–713. doi: 10.1038/nsmb.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diao JJ, Yoon TY, Su ZL, Shin YK, Ha T. C2AB: a molecular glue for lipid vesicles with a negatively charged surface. Langmuir. 2009;25:7177–7180. doi: 10.1021/la901676e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diao JJ, et al. Single-vesicle fusion assay reveals Munc18-1 binding to the SNARE core is sufficient for stimulating membrane fusion. ACS Chem. Neurosci. 2010;1:168–174. doi: 10.1021/cn900034p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HK, et al. Dynamic Ca2+-dependent stimulation of vesicle fusion by membrane-anchored synaptotagmin 1. Science. 2010;328:760–763. doi: 10.1126/science.1187722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen SM, Mortensen MW, Stamou DG. Single vesicle assaying of SNARE-synaptotagmin-driven fusion reveals fast and slow modes of both docking and fusion and Intrasample heterogeneity. Biophys. J. 2011;100:957–967. doi: 10.1016/j.bpj.2010.12.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domanska MK, Kiessling V, Stein A, Fasshauer D, Tamm LK. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J. Biol. Chem. 2009;284:32158–32166. doi: 10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. USA. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karatekin E, et al. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc. Natl. Acad. Sci. USA. 2010;107:3517–3521. doi: 10.1073/pnas.0914723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith EA, Weisshaar JC. Docking, not fusion, as the rate-limiting step in a SNARE-driven vesicle fusion assay. Biophys. J. 2011;100:2141–2150. doi: 10.1016/j.bpj.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karatekin E, Rothman JE. Fusion of single proteoliposomes with planar, supported bilayers in microfluidic flow cells. Nat. Protoc. 2012;7:903–920. doi: 10.1038/nprot.2012.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 32.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selvin PRH, Ha T. Single Molecule Techniques: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- 34.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.