Abstract
The ultimate goal of regenerative medicine is to replace damaged tissues with new functioning ones. This can potentially be accomplished by stem cell transplantation. While stem cell transplantation for blood diseases has been increasingly successful, widespread application of stem cell therapy in the clinic has shown limited results. Despite successful efforts to refine existing methodologies and to develop better ones for reprogramming, clinical application of stem cell therapy suffers from issues related to the safety of the transplanted cells, as well as the low efficiency of reprogramming technology. Better understanding of the underlying mechanism(s) involved in pluripotency should accelerate the clinical application of stem cell transplantation for regenerative purposes. This review outlines the main decision-making factors involved in pluripotency, focusing on the role of microRNAs, epigenetic modification, signaling pathways, and toll-like receptors. Of special interest is the role of toll-like receptors in pluripotency, where emerging data indicate that the innate immune system plays a vital role in reprogramming. Based on these data, we propose that nongenetic mechanisms for reprogramming provide a novel and perhaps an essential strategy to accelerate application of regenerative medicine in the clinic.
Keywords: dedifferentiation, transdifferentiation, reprogramming, pluripotency, microRNAs, epigenetic modifications, signaling pathways, toll-like receptors
Current status of regenerative medicine
Humans have a constrained ability to regenerate and restore their tissues and organs. Some organs have a higher regenerative capacity, such as the blood and the liver, while other organs have very limited ability to self-renew, such as the heart and the brain. Some tissues, such as the liver, regenerate by proliferation, while others undergo self-renewal, such as hematopoietic stem cells. In contrast with humans, some vertebrates have far-reaching regenerative capacities that, in specific cases, extend as far as replacing complete limbs.1
The field of regenerative medicine aims to cure intractable illness by replacing damaged tissues and failing organs, and/or fortifying the body’s own repair mechanisms. Regenerative medicine also incorporates tissue engineering, through which scaffolds, cells, and biologically active molecules are combined into functional tissues. This powerful capacity can conceivably overcome the hurdles of organ transplantation, including the shortage of organs available for donation and the severe problems associated with graft rejection or graft versus host disease.2
Stem cell therapy aims to regenerate malfunctioning tissues via several mechanisms.3 Injectable stem cells may replace defective cells and differentiate into functioning ones. Alternatively, stem and progenitor cells can induce regeneration by secreting biologically active molecules. Furthermore, stem cells can be differentiated in vitro into functioning cells and grown on scaffolds into functioning organs prior to transplantation.4
By taking advantage of biotechnological advances in scaffolding material, tissue engineering appears to be today’s promising treatment for replacing damaged tissues. The principle is straightforward: cells are gathered and introduced with or without alteration of their biological properties directly into the harmed tissue or into a permeable three-dimensional matrix. In these carefully designed scaffolds, stem cells are manipulated in a controlled environment in which physicochemical and mechanical parameters are carefully monitored. After reaching the desired differentiation, these cells or tissues could be grafted.
Dedifferentiation, transdifferentiation, and reprogramming; the three processes compared
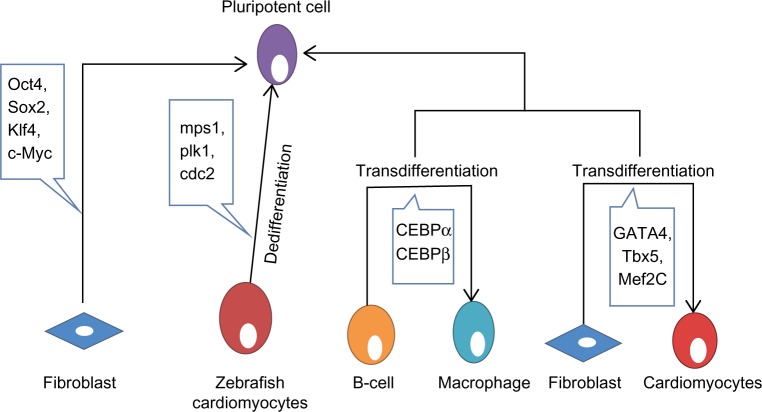
Dedifferentiation is one of the mechanisms linked to natural regeneration, in which a terminally differentiated cell returns back to a less differentiated stage from within its own particular genealogy. This procedure permits the cell to proliferate again before redifferentiating, prompting the substitution of those cells that have been lost. A few nonmammalian vertebrates have a remarkable ability to regenerate. As a rule, this process includes the dedifferentiation of mature cells followed by redifferentiation. In nonmammalian vertebrates, there are several illustrations of dedifferentiation, such as heart regeneration in zebrafish. Zebrafish can completely regenerate their heart following amputation of up to 20% of the ventricle.5 Throughout this process, differentiated cardiomyocytes that are still present in the heart dedifferentiate and proliferate to recover the missing tissue.6 As the cardiomyocytes dismantle their contractile apparatus, they additionally detach from each other and begin to express the positive cell cycle regulators monopolar spindle 1, polo-like kinase 1, and cdc2 (Figure 1).6 At present, little is known about the signaling pathways involved, despite the fact that fibroblast growth factor and platelet-derived growth factor signaling have been specifically linked to this regeneration. Hindrance of fibroblast growth factor or platelet-derived growth factor signaling prompts incomplete regeneration and formation of scar tissue.7 In this case, dedifferentiation seems to be unpredictably linked to the cell cycle, as re-entry into the cell cycle does not appear to be vital following dedifferentiation.8 Recent evidence suggests that the tumor suppressor retinoblastoma protein not only controls cell cycle arrest but might also play an active role in maintaining the differentiated status of a cell. Interestingly, blocking proliferation by blocking retinoblastoma protein does not prevent dedifferentiation from happening. It appears that dedifferentiation is independent of the cell cycle, and that retinoblastoma protein likewise has an important role in keeping up the differentiated status of a cell.9
Figure 1.
Review of reprogramming, dedifferentiation, and transdifferentiation. Pluripotent cells have the potential to give rise to any lineage of cells. Fibroblasts can be reprogrammed back to pluripotent cells by inducing expression of Oct4, Sox2, Klf4, and c-Myc.24 Zebrafish cardiomyocytes provide a good example of dedifferentiation by expressing mps1, plk1, and cdc2.6 B-cells can be transdifferentiated into macrophages utilizing CEBPα and CEBPβ.14 Also, fibroblasts can be transdifferentiated into cardiomyocytes utilizing GATA4, Tbx5, and Mef2c.17
Abbreviations: CEBP, CCAAT-enhancer-binding protein; mps1, monopolar spindle 1; plk1, polo-like kinase 1.
Transdifferentiation involves the irreversible change of cells from one differentiated cell type to another. Biological transdifferentiation takes place in two stages: first, the cell dedifferentiates, then the natural developmental program is stimulated, permitting the cell to differentiate into the new lineage.10 This sequence, however, may not be mandatory in all cases. The regenerating lens of the newt and salamander perfectly depicts the naturally occurring transdifferentiation process.11 In these species, after evacuation of the lens from the eye, new pigmented epithelial cells of the lens regenerate and further differentiate to structure a new lens.11 This regeneration involves inactivation of retinoblastoma protein, permitting the cells to re-emerge the cell cycle.12 Recently, Day and Beck showed that Wnt and bone morphogenetic protein signaling pathways are required for transdifferentiation from cornea to lens, a process in which both the PITX and Wnt genes are essential.13 The experimental transdifferentiation of B-cells into macrophages utilizing the transcription factors CCAAT-enhancer-binding protein-α (CEBPα) and CEBPβ provides a good illustration of in vitro transdifferentiation (Figure 1).14 Forced expression of CEBPβ and CEBPα in differentiated B-cells prompts reprogramming them into macrophages without notable changes in DNA methylation.15 Following a comparable strategy, it is conceivable to transdifferentiate murine fibroblasts into functional neurons using three transcription factors (Brn2, Ascl1, and Mytl1).16 Fibroblasts have been transdifferentiated into cardiomyocytes utilizing GATA4, Tbx5, and Mef2c (Figure 1).17 GATA4 activates the process by opening chromatin, permitting access to the other transcription factors.18 This offers a new strategy to replace the lost or damaged cardiomyocytes by inducing fibroblast transdifferentiation in vivo. Human dermal fibroblasts have additionally been transdifferentiated into multipotent progenitors that have the potential to differentiate into monocytic, granulocytic, erythroid, and megakaryocytic lineages with in vivo engraftment capacity utilizing the ectopic expression of Oct4 in the presence of particular cytokine treatment.19 A therapeutic example of transdifferentiation has been shown by Sapir et al, when human liver cells were induced to transdifferentiate into functional insulin-producing β-cells by transduction with PDX-1 virus.20 In an effort to determine the underlying mechanisms of transdifferentiation, a recent report showed that stepwise epigenetic modifications stabilize the changes caused by transcriptional factors to ensure transdifferentiation.21 Zuryn et al have further shown that there is parallelism between epigenetic modifications underlying transdifferentiation in vivo and efficient cell reprogramming in vitro.22
Reprogramming occurs naturally during fertilization to produce totipotent cells that can differentiate into any cell type.23 Reprogramming can also be induced artificially, whereby a fully differentiated cell is induced to regress into its pluripotent origin, with the possibility of differentiating into almost any other cell type. In 2006, Takahashi and Yamanaka induced pluripotency of a differentiated somatic cell through overexpression of only four transcription factors (Oct4, Sox2, c-Myc, and Klf4, Figure 1).24 These cells were called induced pluripotent stem cells (iPSCs). Oct4 has a fundamental role in the development and maintenance of pluripotency by activating or repressing specific genes,25 such as Nanog,26 Fgf4,27 Utf1,28 and Zfp206.29 Indeed, cells lacking functional Oct4 can no longer support this key property.30 In addition to activating pluripotency, Oct4 prevents the differentiation of pluripotent cells by acting as a depressor of lineage-specific transcription factors. Oct4 forms a repressive complex with Cdx2 (a transcription factor important for trophectoderm specification), and hinders its transcription.31 Moreover, Oct4 has been shown to suppress the transcription of the epithelial mesenchymal transition mediator Snail to encourage reprogramming.32 Recently, Oct4 was found to interact with nuclear β-catenin and to enhance its proteasomal degradation, thus maintaining the undifferentiated state of embryonic stem cells (ESCs).33 Despite the fact that Sox2 is involved in the self-renewal of stem cells, one of its essential capacities is to maintain Oct4 expression at the levels required to maintain pluripotency.34 It has been shown that Sox2 may act with Oct4 to activate Oct-Sox enhancers that in turn upregulate the expression of many pluripotency genes, including Nanog, Oct4, and Sox2 itself.34 During development, Nanog is expressed by the pluripotent cells of the inner cell mass, and appropriately, embryos that fail to express this gene fail to develop the inner cell mass.35 Nonetheless, if Nanog is removed from cells that have effectively accomplished pluripotency, they still hold many stem cell features.36 Subsequently, it appears that the major role of Nanog is in sustaining pluripotency. Interestingly, the cells most adjustable to reprogramming have been neural stem cells, which only require Oct4.37 Why certain cell types require fewer factors for reprogramming remains debatable. One possibility is that less differentiated cells are closer to pluripotency and subsequently need fewer factors. Another possibility is that certain cell types already express some of these factors. However, the strongest evidence points to the fundamental role of Oct4 in reprogramming. A recent study showed that Tet-1, a 5-methylcytosine hydroxylase, could replace Oct4 in reprogramming somatic cells and generating fully pluripotent iPSCs,38 which indicates that DNA methylation and hydroxymethylation play important roles in epigenetic remodeling in pluripotency.
All of the three processes discussed so far can prompt terminally differentiated cells to become highly plastic and suitable for therapeutic purposes However, one of the major differences between reprogramming and both transdifferentiation and dedifferentiation is their current potential for in vivo therapy. Although both transdifferentiation and dedifferentiation can be successfully accomplished in vivo, directing pluripotent cells into a new lineage is a complex process, that has so far been successful only in vitro. Although the three processes can induce considerable changes in differentiated cells, each has definite advantages in terms of regenerative medicine. If the aim is to replace cells lost as a consequence of injury or disease, this could be accomplished by reprogramming cells taken from the patient in vitro and afterward differentiating them into the desired cell type, followed by engraftment back into the patient. A simpler in vivo approach would be either to induce cells to dedifferentiate and then proliferate or to induce a more abundant or less specialized cell type to transdifferentiate into the desired cell types. If, however, the objective is to correct a genetic mutation, attempting to transdifferentiate or dedifferentiate any of the patient’s cells would not cure the disease, as the new cells would still contain the mutation. In this situation, the more feasible option would be to reprogram the patient’s own cells in vitro, followed by correcting the damaged gene before differentiating the cells into the desired lineage and injecting them back to the patient.39
MicroRNAs involved in pluripotency
The role of microRNAs (miRNAs) in pluripotency has been investigated by several laboratories. MiRNAs are small noncoding RNAs, typically comprising 19–25 nucleotides, that regulate gene expression post transcriptionally by translational silencing of their targeted messenger RNAs (mRNAs).40 In most cases, the effect of miRNAs in ESC physiology remains unknown. However, the presence of a subset of miRNAs that are only expressed in ESCs (Table 1) and, most importantly, the failure to create viable ESCs from mice deficient in Dicer,41 the enzyme needed for miRNA processing, suggest an essential role for miRNAs in ESC self-renewal.
Table 1.
MicroRNAs involved in pluripotency, self-renewal, differentiation, and reprogramming
| Stem cell process | MicroRNAs involved | References |
|---|---|---|
| Pluripotency | miR-290, miR-291, miR-292, miR-293, miR-294, miR-295, miR-302, miR-317, miR-327, miR-373, miR-367 | 44,43 |
| Self-renewal | miR-21, miR-141, miR-200, miR-429 | 119,120 |
| Differentiation | miR-134, miR-296, miR-470, miR-145 | 45,46 |
| Reprogramming | miR-302–367 cluster | 121 |
DGCR8 is a nuclear protein, essential for miRNA processing. Like the Dicer-null ESCs, DGCR8-deficient ESCs show either delayed or decreased expression of differentiation markers, in addition to delayed kinetics of cell cycle progression.42 Most DGCR8-deficient ESCs are arrested in the G1 phase. This indicates that the fundamental role of the miRNA pathway is to regulate the ESC cycle at the G1-S phase transition. In addition, DGCR8-null ESCs display differentiation defects as they fail to steadily silence the expression of self-renewal markers, including OcT4, Sox2, Nanog, and Rex1.42
The pattern of expression of miRNA in human ESCs seems to vary extraordinarily from that of differentiated cells.43 Studies showed that the miR-290–295 cluster and miR-296 are peculiar to ESCs and play specific roles in maintaining pluripotency44 and that their levels are reduced as the stem cells differentiate (Figure 2).44 In contrast, levels of miR-21 and miR-22 increase significantly after induction of differentiation, indicating that these miRNAs may play a critical role in stem cell differentiation (Figure 2).43 Other miRNAs, including miR-134, miR-296, and miR-470, were recently found to target coding regions of Oct4, Sox2 and Nanog to promote differentiation.45 Suh et al reported a group of miRNAs that are abundantly expressed in human ESCs. These miRNAs, termed ESC-specific miRNAs, incorporate miR-302a, miR-302b, miR-302c, miR-302d, miR-367, and miR-371–373 (Table 1).43 Xu et al showed a critical role for miR-145 in repression of the 3′-untranslated regions of Oct4, Sox2, and Klf4. Loss of miR-145 ameliorated differentiation and increased the expression of the most essential reprogramming factors, indicating its potential role in the creation of iPSCs.46
Figure 2.
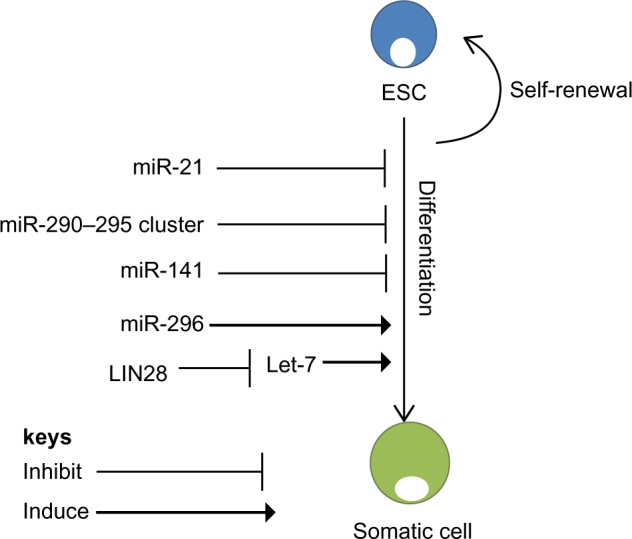
microRNA regulation of self-renewal and differentiation. ESCs have the potential to self-renew or differentiate into somatic cells. MicroRNAs regulate both self-renewal and differentiation pathways of ESCs. miR-21 and the miR-290–295 cluster are essential for self-renewal of ESCs.43 Another regulator of ESCs is Lin28, which binds to Let-7, promoting self-renewal.51 However, miR-296 promotes differentiation of ESCs.45
Abbreviation: ESCs, embryonic stem cells.
Another seemingly important regulator of pluripotency is survivin. Kapinas et al showed that survivin is selectively expressed under the control of miRNAs.47 They showed that miR-203 expression and activity is harmonious with the expression of survivin isoforms and subsequently controls expression of the Oct4 and Nanog transcription factors to maintain pluripotency.47
Efforts to identify the mRNA targets that underlie the capacity of ESC-specific miRNAs have been undertaken and reported by several groups.48 Subramanyam et al demonstrated that miRNAs stimulate reprogramming by simultaneously targeting several discriminating cellular pathways involving regulators of the G1-S cell cycle checkpoint, the mesenchymal-to-epithelial transition, and DNA methyl binding proteins.49 These cellular processes interact with a subset of the mRNAs targeted by these miRNAs. The ESC-specific miRNAs likewise target mRNAs that control apoptosis and bone morphogenetic protein signaling, although the role of these targets in reprogramming has not yet been elucidated.50
Despite these reports, regulators of miRNA biogenesis have been demonstrated to be among the handful of factors that can change differentiated cells into iPSCs. This is the case with the RNA-binding protein Lin28, which can proficiently change fibroblasts into iPSCs when transduced together with Oct4, Sox2, and Nanog.51 Evidently, Lin28 is an ESC-specific RNA-binding protein that interferes with and suppresses the action of let-7 miRNAs.52 let-7 miRNA is expressed at low levels in ESCs, and is rapidly induced upon differentiation.53 Additionally, Lin28 expression is activated by c-Myc in numerous human and mouse tumor models (Figure 2).54 These data provide evidence that Lin28 has a central role in blocking miRNA-mediated differentiation in stem cells and in substitution of c-Myc throughout iPSC formation. Evidently, using miRNAs to control the differentiation of stem cells can prompt diverse tissues to be generated from stem cells for the purpose of cell therapy. For instance, expression of miR-181 results in an increase in the number of cells entering the B-lymphoid lineage, but not the number of cells entering the T-lymphoid lineage, while ectopic expression of miR-142 or miR-223 has the opposite effect.55
Epigenetic modifications in pluripotency
Pluripotency of ESCs is maintained by epigenetic factors closely associated with the pluripotency transcription factor network (Table 2).56 Epigenetic modifications of gene expression include DNA methylation and histone modification, each of which adjusts how genes are expressed without modifying the underlying DNA sequence. Epigenetic factors involved in maintaining the pluripotency of ESCs must be activated throughout the reprogramming process,57 as during reprogramming, pluripotency genes that are hypermethylated in somatic cells must be demethylated and activated. The genomes of ESCs express an open chromatin state that is highly accessible to transcription factors.58 The open chromatin status is locally stimulated by chromatin-remodeling factors, such as the SWI-SNF complex and Chd1. Knockdown of Chd1 blocks the iPSC, while overexpression of the SWI-SNF complex enhances its formation.59,60
Table 2.
List of epigenetic markers and their role in reprogramming
| Epigenetic marker | Reprogramming effect | Epigenetic regulators mechanisms | References |
|---|---|---|---|
| H3K4 | Required for self-renewal, and is needed during the initial phase of reprogramming | Wdr5 catalyzes H3K4 methylation | 122 |
| H3K9 | Required for differentiation, and its removal results in efficient conversion of partially reprogrammed cells to iPSCs | By depletion of Suv39h1, Suv39h2, Setdb1, or Ehmt2 | 64 |
| H3K27 | Required for differentiation, and its removal is also involved in reprogramming | PRC2 and Utx interacts with reprogramming factors and impairs reactivation of pluripotency genes | 123,124 |
| H3K36 | Knockdown decreases reprogramming, while overexpression improves reprogramming | Kdm2a and Kdm2b facilitates tearly transcriptional response to the reprogramming factors | 125 |
| H3K79 | Depletion in the early phase enhances reprogramming | Kmt4 inhibition results in upregulation of pluripotency genes, such as Nanog and Lin28 | 126 |
| Histone acetylation | HDAC2 knockout allows reprogramming to be driven by overexpression of only microRNAs | Small-molecule inhibitors of histone deacetylases (such as valproic acid, trichostatin A, and butyrate) enhance reprogramming and replace ectopic c-Myc or Klf4 | 121,127,128 |
Abbreviation: iPSCs, induced pluripotent stem cells.
Histone modifications
During iPSC generation, somatic cell chromatin requires reorganization to an ESC-like state.61 It seems that the chromatin reorganization event occurs in a coordinated and sequential manner. Rearrangement of heterochromatin, distinguished by the presence of histone H3 lysine 9 trimethylation (H3K9me3) and heterochromatin protein 1, occurs before activation of Nanog, while enrichment of euchromatin markers happens simultaneously with activation of Nanog.62 Pluripotent ESCs are known for active histone markers, including H3K4me3, but as ESCs differentiate, the regions marked by repressive H3K9me3 and H3K27me3 extend and spread (Table 2).63 H3K9 methylation is a tangible barrier to reprogramming, so decreasing the levels of H3K9 methyltransferases or overexpressing H3K9 demethylases enhances the efficiency of creating iPSCs (Table 2).64,65 ESCs thus express bivalent histone modifications, ie, H3K4me3 and H3K27me3, which hinder many differentiation genes.66
DNA methylation
Like histone modifications, DNA methylations are well established in ESCs and iPSCs.67 Methylation is mediated by DNA methyltransferases, which stimulate the addition of Met groups to Cys residues in CpG islands. In general, regions rich in CpG that overlap gene promoters are unmethylated and their expression is decreased.68 Notwithstanding this, many promoters in CpG islands undergo methylation changes during differentiation, like promoters of pluripotency genes, which are free from methylation in undifferentiated ESCs and iPSCs, but become methylated at differentiation.69 Indeed, for ESCs to differentiate, DNA demethylation is needed as DNA methyltransferase-depleted cells conserve their stem cell characteristics, indicating that DNA methylation is essential for the maintenance of stemness.70
Chromatin remodeling
DNA is packaged into chromatin by associations with histone and non-histone proteins. The main unit of chromatin is the nucleosome, structured by the wrapping of 146 base pairs of DNA around a core of two copies, each composed of four histone proteins (H2A, H2B, H3, and H4).71 Pluripotent ESCs are characterized by a global chromatin structure that is generally dynamic and specific to the transcriptional machinery. Upon differentiation, chromatin is changed into a more repressive state.58 Components of the chromatin structure also play a key role in controlling gene expression profiles in pluripotent ESCs. Direct interaction between reprogramming factors and chromatin regulators may be essential for pluripotency. For example, Oct4 can interact with subunits of the BAF chromatin-remodeling complex,25,72 which improves reprogramming and could stimulate the binding of transcription factors to nucleosomal sites.59 Also, it has been shown that overexpression of Baf155/Brg1 (an ATP-dependent chromatin-remodeling complex) enhances reprogramming by enhancing the binding of Oct4 to its pluripotency targets.59 Additionally, Chd1, the chromatin-remodeling factor, is required to maintain the open chromatin state of pluripotent stem cells and is also required for efficient reprogramming of fibroblasts to the pluripotent stem cell state.60 Thus, somatic cell reprogramming using chromatin-remodeling molecules represents an efficient method of generating reprogrammed cells.
Recent work in iPSC programming showed that if cells are deficient in Mbd3, a nuclear protein of the Mbd3/NuRD (nucleosome remodeling and acetylation) complex, the efficiency of reprogramming induced by Oct4, Sox2, Klf4, and c-Myc increases to nearly 100%.73 It is also worth pointing out that the peremptory reprogramming of Mbd3-depleted somatic cells reported by Brumbaugh et al has been challenged by a more recent study showing a need for functioning of Mbd3/NuRD for efficient reprogramming.73 Santos et al have showed that depletion of Mbd3 led to reduction in the efficiency of reprogramming and that overexpression of Mbd3 facilities reprogramming of neural stem cells when combined with expression of Nanog, but not with other tested reprogramming factors.74 It seems that a number of differences, including choice of reprogramming cassettes and reprogramming culture conditions, have influenced the differences between these results. Also, 5-azacytidine, a DNA methyltransferase inhibitor, encourages transformation of partially reprogrammed cells to iPSCs, and upgrades overall reprogramming efficiency when used at the late stage of reprogramming.75
The ability to establish patient-matched ESC lines is currently challenged by the fact that there are abnormal epigenetic modifications during the reprogramming process. The above data thus provide strong evidence that epigenetic modifications have significant control over pluripotency, and should be considered when using iPSCs for cell therapy.
Signaling pathways involved in pluripotency
Wnt/β-catenin signaling pathway
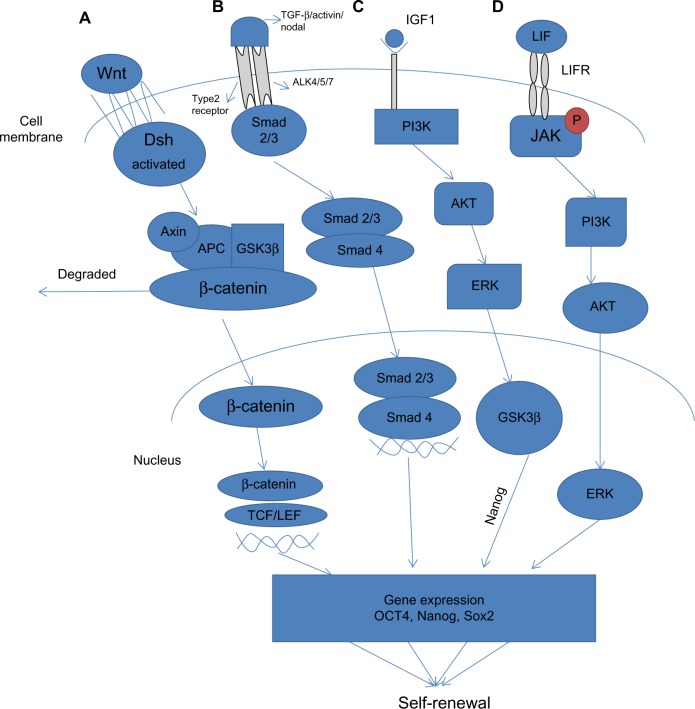
The Wnt signaling pathways function through groups of surface proteins, such as Wnt, which has been linked to embryonic development. β-catenin, a part of the cadherin cell adhesion complex, assumes an essential role in E-cadherin-mediated cell-cell adhesion and is an essential intermediate in the Wnt signaling pathway. It is well established that human and murine ESCs can be maintained in an undifferentiated state by the activation of Wnt signaling, which upregulates the expression of Oct4, Nanog, and Rex1 through the action of β-catenin (Figure 3).76 The role of Wnt signaling in the reprogramming processes is confounding since high levels of Wnt activity in glycogen synthase kinase (GSK)-3 double-knockout mice were shown to prompt differentiation of ESCs.77 (GSK-3 is a serine/threonine protein kinase that has been demonstrated to phosphorylate β-catenin, thus targeting it for recession).
Figure 3.
Signaling pathways involved in pluripotency. (A) Wnt signaling pathway is stimulated via binding of Wnt proteins with frizzled receptors. This subsequently activates Dsh, which replaces GSK-3β from the Axin-APC complex, preventing degradation of β-catenin. Subsequently, β-catenin translocates into the nucleus where it associates with TCF/LEF proteins to activate transcription of Wnt target genes that cause self-renewal. (B) TGF-β signals through two main receptors, ie, the type II receptor and the Activin receptor. Upon activation, ligand-specific Smad proteins (Smad2/3) are phosphorylated and form a complex with Smad4, which translocates to the nucleus to activate gene expression. (C) The PI3K/Akt pathway is activated by IGF-1. Activation of PI3K activates Akt, which modulates the threshold of Smad2/3 activity and inhibits Erk and maintains GSK-3β activity, which in turn induces expression of Nanog. (D) The Jak/Stat signal pathway is principally initiated by LIF, which likewise initiates PI3K through which Akt and Erk are activated, ultimately resulting in upregulation of expression of Klf4 and Myc.
Abbreviations: Dsh, Dishevelled; LIF, leukemia inhibitory factor; LIFR, leukemia inhibitory factor receptor; TGF-β, transforming growth factor-beta; PI3K, phosphoinositide-3-kinase; GSK-3β, glycogen synthase kinase 3-beta; IGF-1, insulin-like growth factor-1.
Wnt has been definitively shown to regulate dedifferentiation. Without Wnt signaling, β-catenin is phosphorylated by functional interaction with GSK-3β and hence targeted to degradation by the ubiquitin-proteasome system.78 Another putative “reprogrammer” enacted by Wnt signaling might be T-cell factor/lymphocyte enhancer factor, which indicates a high level of colocalization with Oct4, Sox2, and Nanog (Figure 3).79 Additionally, Wnt could activate endogenous c-Myc directly, substituting the need for exogenous c-Myc. This supports the evidence that Wnt signaling can promote reprogramming of somatic cells to pluripotency.80
TGF-β signaling pathway
Transforming growth factor-beta (TGF-β) is a protein that controls proliferation and cellular differentiation in different capacities in most cells. The signaling role of TGF-β in stem cell reprogramming has been investigated by Ichida et al, who showed that inhibiting TGF-β signaling replaces Sox2 in reprogramming by inducing the transcription of Nanog (Figure 3).81 It is well established that bone morphogenic protein 4, a member of the TGF-β superfamily, participates with leukemia inhibitory factor in maintaining pluripotency in murine ESCs. Under serum-free culture conditions, leukemia inhibitory factor alone prompted neural differentiation of murine ESCs. This effect was annulled by treatment with bone morphogenic protein 4, which maintained the undifferentiated state of ESCs, and without serum.82
In 2009, Maherali et al demonstrated that hindrance of TGF-β receptor I kinase with an Activin-like kinase 5 inhibitor upgraded both the proficiency and kinetics of murine embryonic fibroblasts reprogrammed by Oct4, Sox2, Klf4, and c-Myc. Activation of the TGF-β signaling pathway on the other hand blocked reprogramming.83 This discrepancy suggests that hindrance of TGF-β acts only on direct reprogramming pathways that are already working in ESCs. Inhibition of TGF-β could replace Sox2 and c-Myc in induction of iPSCs. This highlights the dual role of TGF-β as an inducer of pluripotency by replacing Sox2 and c-Myc.83 Inhibition of TGF-β signaling also promotes the mesenchymal-to-epithelial transition process, which is a critical step in somatic reprogramming.32 TGF-β prevents mesenchymal-to-epithelial transition by inhibiting both upregulation of epithelial markers and downregulation of the mesenchymal transcriptional repressor Snail.32
PI3K/Akt signaling pathway
Phosphoinositide-3-kinases (PI3Ks) are lipid kinases that induce generation of phosphatidylinositol trisphosphate, a signaling lipid, upon stimulation by numerous growth factor receptor tyrosine kinases, such as epidermal growth factor, fibroblast growth factor, and platelet-derived growth factor. Akt1, a serine/threonine kinase, is one of the fundamental players in this pathway. It modulates the functions of different cellular responses, including cell proliferation, growth, adhesion, and death, in addition to inducing tumorigenesis.84 Activation of Akt1 signaling is sufficient to maintain the pluripotency of murine ESCs without leukemia inhibitory factor (Figure 3).85 However, despite what might be expected, treatment of murine ESCs with the PI3K inhibitor LY294002 promoted a loss of ESC characteristics, even in the presence of leukemia inhibitory factor.86 Further, initiation of PI3K signaling is critical to the advancement of dedifferentiation in embryonic germ cells from primordial germ cells.87 The role of the PI3K pathway in ESC biology and in reprogramming remains largely unknown. Nakamura et al showed that activation of Akt signaling stimulated reprogramming after fusion of ESCs with thymocytes or murine embryonic fibroblasts, which prompted the formation of ESC-like hybrid cells.88 Supporting these results, it has been shown that ERas, an ESC-specific Ras gene, is closely associated with Akt in enhancing reprogramming89 and modulation of Akt signaling by genetic or chemical means.89
Jak-Stat signaling pathway
The Jak-Stat signaling pathway transmits information from chemical signals outside the cell through the cell membrane and into gene promoters in the DNA in the nucleus, causing DNA transcription and activity. Activation of the Janus kinase/signal transducer and activator of transcription 3 (Jak/Stat3) signaling pathway maintains murine ESC pluripotency.90 The Jak/Stat3 signaling pathway is principally initiated by leukemia inhibitory factor, which likewise initiates the PI3K/protein kinase B (also known as Akt) and extracellular signal-regulated kinase 1/2 (Erk1/2) pathways in ESCs (Figure 3).91 Artificially activated Stat3 sustains self-renewal of murine ESCs in the absence of leukemia inhibitory factor.92 Little is known about the downstream mechanism of Stat3 in maintenance of pluripotency, despite reports that leukemia inhibitory factor/Stat3 upregulates expression of Klf4 and Myc in murine ESCs.93
Activin/Nodal pathway
Nodal, a member of the TGF-β superfamily, acts by binding to heteromeric complexes between type I and type II Activin receptors, which in turn act through the Smad2/Smad3 signaling pathway.94 Recent data indicates that overexpression of the Nodal growth factor in human ESCs can obstruct their default neuroectoderm differentiation by formation of embryoid bodies.95 Nodal itself is expressed in human ESCs and disappears rapidly upon differentiation, indicating that Nodal signaling could be included in the maintenance of pluripotency. Restraint of Nodal does not induce differentiation of human ESCs. However, hindrance of the Activin/Nodal/TGF-β signaling pathway induces differentiation of human ESCs, showing that the Activin/Nodal pathway is fundamental for support of pluripotency.96 Evidently, understanding the molecular mechanisms and signaling pathways that regulate stemness is a mandatory step toward the design of rational clinical treatments.
Toll-like receptors involved in pluripotency
Toll-like receptors (TLRs) are a class of proteins with a key role in the innate immune response. TLR signaling comprises two different pathways, ie, a myeloid differentiation primary response gene (MyD 88)-dependent pathway and a MyD88-independent pathway.97 The MyD88-dependent pathway is common to all TLRs, except for TLR3.98 The TLR3 pathway is initiated by viral double-stranded RNA and is independent of MyD88. To investigate the role of this pathway in reprogramming using retroviral plasmid-containing stem cell factors (Oct4, Sox2, Klf4, and c-Myc), knockdown of TLR3 signaling diminished the pluripotency gene expression induced by Oct4. Similarly, short hairpin RNA knockdown of TLR3 diminished the ability of Oct4 to induce expression of the target pluripotent genes.99 This indicates that TLR3 knockdown inhibits activation of downstream pluripotency genes when utilizing retroviral vectors or mRNA to overexpress the reprogramming factors and reduces the efficiency and yield of human iPSCs. Taken together, stimulation of TLR3 produces substantial changes in the expression of epigenetic modifiers to enhance chromatin rebuilding and nuclear reprogramming and stimulates pluripotency genes and generation of human iPSC colonies.99
Emerging data suggest that TLRs may play a role in regulation of stem cell fate. TLR ligands have been shown to contribute to differentiation of mesenchymal stem cells (MSCs). Supplementation of adipogenic differentiation medium with TLR7 and TLR2 agonists induced MSCs to produce a significantly high percentage of adipose cells, with more promising results achieved with TLR7 (Figure 4).100 Also, TLR9 increased the osteogenic differentiation of MSCs (Figure 4).100 Human bone marrow hematopoietic stem cells could be differentiated into myeloid cells just by stimulation with TLR7/8 agonists.101 TLR2 agonists have been shown to have similar effects in hematopoietic stem cell differentiation into the myeloid lineage (Figure 4).101 Interestingly, TLR7/8 agonists were more effective at inducing CD11+CD14− cells, whereas TLR2 was more effective at inducing CD11+CD14+ cells.101 On the basis of these findings, it can be suggested that TLR signaling can influence the outcome of stem cell differentiation, and that differentiation is not produced through the same signaling pathways.
Figure 4.
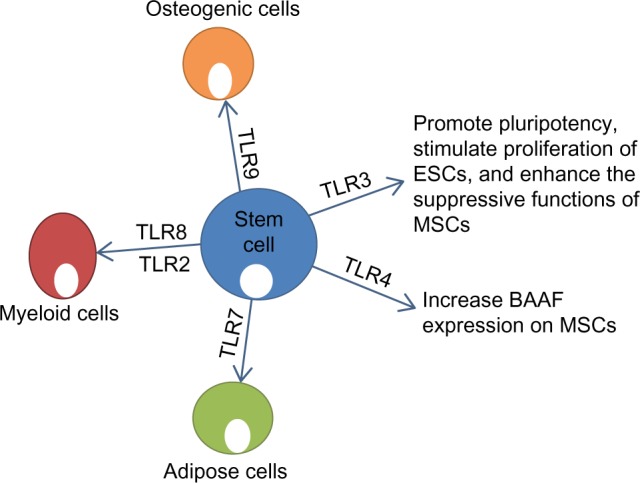
Effect of stimulation of TLRs on function and differentiation of stem cells. TLR3 plays a role in maintaining pluripotency and enhancing the function of MSCs.99 TLR4 increases expression of BAAF on MSCs. TLR7 induces adipogenic differentiation of MSCs.98 TLR8/2 induces production of myeloid cells from HSCs.100 TLR9 induces osteogenic differentiation of MSCs.98
Abbreviations: TLR, Toll-like receptor; ESCs, embryonic stem cells; HSCs, hematopoietic stem cells; MSCs, mesenchymal stem cells.
Human MSCs have been shown to produce proinflammatory cytokines, such as interleukin (IL)-1β, IL-6, IL-8, CCL5, and tumor necrosis factor-alpha, in response to TLR agonists.102 TLR2 agonists stimulated CD34+ progenitors to secrete granulocyte-macrophage colony-stimulating factor.101 It has been shown that production of cytokines in response to TLR stimulation occurs through nuclear factor kappa B signaling.103 MSCs from diverse sources express TLRs at the mRNA level, although expression at a protein level appears to be low (ie, compared with monocytes). Detection of TLRs in MSCs by flow cytometry has also been troublesome.104 Pevsner-Fischer et al demonstrated that murine MSCs express many TLRs, and specifically TLR2, which was essential for their differentiation capacity.105 Like murine MSCs, human MSCs also express many TLRs.106 In murine MSCs, TLR signaling may be connected to the multipotency of MSCs, as MyD88-deficient bone marrow MSCs failed to efficiently differentiate into chondrogenic and osteogenic lineages.105 In human MSCs, only activation of TLR9 has been shown to affect adipogenic differentiation.100
Giuliani et al have shown that TLRs can modulate the function of MSCs.107 TLR-primed adult and ESCs were more resistant than unprimed stem cells to natural killer-induced killing.107 In addition, TLR3-primed but not TLR4-primed MSCs showed enhanced suppressive functions against natural killer cells.107 On the other hand, Yan et al showed that TLR4 priming of MSCs increased the expression of B-cell activating factor,108 which indicates that TLR4 has an important role in B-lymphocyte immune regulation of MSCs (Figure 4).
Lee et al observed that TLR ligands such as lipopolysaccharide, a ligand of TLR4, and Poly (I:C), a ligand of TLR3, stimulated the proliferation of murine ESCs (ESD3) developed on gelatin-coated dishes in standard ESC culture medium containing leukemia inhibitory factor (Figure 4).99 The TLR ligands, lipopolysaccharide and Poly (I:C), were likewise found to induce ESCs to form primary embryoid bodies.109 Evidently, the cells from the lipopolysaccharide-stimulated primary embryoid bodies were still capable of forming the secondary embryoid bodies more efficiently in the presence of lipopolysaccharide, suggesting that lipopolysaccharide-stimulated ESCs retain competence for self-renewal.110
TLR signaling plays a similarly significant role in promoting differentiation of murine ESCs into hematopoietic progenitors.109 Adding lipopolysaccharide to the ESC hematopoietic differentiation culture favorably increased the differentiation of myeloid lineage progenitors.109 Also, treatment with the TLR4 ligand stimulated hematopoietic progenitors to differentiate from Twist-2-deficient ESCs,111 indicating the important role of the transcriptional repressor Twist-2 in controlling hematopoietic differentiation from ESCs.109
It appears that TLRs and their ligands can serve as regulators of stem cell proliferation and differentiation and might affect the maintenance of MSC multipotency. Our understanding of the biological significance of functional TLRs in stem cells and their role in dedifferentiation, transdifferentiation, and reprogramming seem to be evolving, and further investigation is warranted.
Conclusion and perspectives
One of the most significant challenges in regenerative medicine is being able to provide transplantable cells.112 Stem cell transplantation has shown impressive results in the treatment of a number of intractable diseases, such as leukemia, lymphoma, and immune deficiency disorders. However, use of stem cells for solid tissue transplantation has not been met with similar success.113 Issues with safety and production efficiency have hampered the progress and clinical applications of stem cell therapy, especially utilizing iPSCs.114 Evidently, the stress generated during reprogramming prompts the selection of iPSCs in which stress regulatory genes are mutated, rendering iPSCs more prone to tumor development.115 In this review, we have attempted to explain the role of miRNAs, epigenetic modifications, signaling pathways, and TLRs in pluripotency and the potential application of this knowledge in facilitating stem cell therapy.
The role of ESC-specific miRNAs in effective reprogramming of ESCs looks very promising. ESC-specific miRNAs alone can accomplish successful reprogramming, leading to the conclusion that these miRNAs have an important role in the process. The primary advantage of miRNAs is that, unlike transcription factors, they are more effective and take less time to induce cell redirection. However, the mechanism by which ESC-specific miRNAs redirect somatic cells to assume pluripotency remains unknown. miRNA-based reprogramming may herald a new era in regenerative medicine, and the introduction of miRNAs directly into patients to change the fate of certain cells does seem highly promising.
Epigenetic modifications have been shown to be closely connected to the pluripotent state of ESCs, as in the case of H3K4me3, which was shown to support the self-renewal capacity of ESCs.116 Further understanding of the regulatory connections between epigenetic modifications and pluripotency will aid in the generation of “high-quality” iPSCs suitable for therapeutic application.
Although the signaling pathways involved in dedifferentiation, transdifferentiation, and reprogramming have not been completely elucidated, recent evidence suggests that these pathways may have a critical role in application of stem cell therapy.117 The Wnt/β-catenin, TGF-β, PI3K/Akt, Activin/Nodal, and Jak-Stat signaling pathways all have an essential role in pluripotency.118 Recognizing which signaling pathways play a key role during dedifferentiation, transdifferentiation, and reprogramming could conceivably allow development of new approaches to control these processes through better control of the process of pluripotency. By activating or inhibiting certain signaling pathways, pluripotency can be turned on and off.
Understanding of the molecular mechanisms that control pluripotency or stimulate differentiation will enhance application of stem cell therapy in the clinical setting. Of special importance is the role of TLRs in embryonic and adult stem cells, with new reports suggesting that TLRs may play a dual activation and regulatory role in the immune system. This role may be more pronounced in regulation of mammalian stem cells, and highlights a previously unrecognized role for activation of innate immunity in pluripotency. These emerging data should have significant implications for understanding stem cell biology and applying stem cell therapy in the clinical setting.
Despite the large volume of research in the field, regenerative medicine has shown slow progress at the bedside, largely because of issues related to the safety of ESCs and iPSCs and the limited efficiency of reprogramming technology. Most efforts to reprogram somatic cells to behave like stem cells have focused on genetic modifications. Data presented in this review highlight important, novel, and nongenetic factors that can significantly alter somatic cells, enhance pluripotency, and provide an efficient means for reprogramming and stem cell therapy.
Acknowledgments
The Center of Excellence for Stem Cells and Regenerative Medicine is funded by a grant (number 5300) from The Science and Technology Development Fund in Egypt.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Brockes JP, Kumar A. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat Rev Mol Cell Biol. 2002;3(8):566–574. doi: 10.1038/nrm881. [DOI] [PubMed] [Google Scholar]
- 2.Mason C, Dunnill P. A brief definition of regenerative medicine. Regen Med. 2008;3(1):1–5. doi: 10.2217/17460751.3.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Riazi AM, Kwon SY, Stanford WL. Stem cell sources for regenerative medicine. Methods Mol Biol. 2009;482:55–90. doi: 10.1007/978-1-59745-060-7_5. [DOI] [PubMed] [Google Scholar]
- 4.Muneoka K, Allan CH, Yang X, Lee J, Han M. Mammalian regeneration and regenerative medicine. Birth Defects Res C Embryo Today. 2008;84(4):265–280. doi: 10.1002/bdrc.20137. [DOI] [PubMed] [Google Scholar]
- 5.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 6.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4(8):e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odelberg SJ. Inducing cellular dedifferentiation: a potential method for enhancing endogenous regeneration in mammals. Semin Cell Dev Biol. 2002;13(5):335–343. doi: 10.1016/s1084952102000897. [DOI] [PubMed] [Google Scholar]
- 9.Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, Frolov MV. Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet. 2010;6(4):e1000918. doi: 10.1371/journal.pgen.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 11.Tsonis PA, Madhavan M, Tancous EE, Del Rio-Tsonis K. A newt’s eye view of lens regeneration. Int J Dev Biol. 2004;48(8–9):975–980. doi: 10.1387/ijdb.041867pt. [DOI] [PubMed] [Google Scholar]
- 12.Thitoff AR, Call MK, Del Rio-Tsonis K, Tsonis PA. Unique expression patterns of the retinoblastoma (Rb) gene in intact and lens regeneration-undergoing newt eyes. Anat Rec A Discov Mol Cell Evol Biol. 2003;271(1):185–188. doi: 10.1002/ar.a.10023. [DOI] [PubMed] [Google Scholar]
- 13.Day RC, Beck CW. Transdifferentiation from cornea to lens in Xenopus laevis depends on BMP signalling and involves upregulation of Wnt signalling. BMC Dev Biol. 2011;11:54. doi: 10.1186/1471-213X-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Ubreva J, Ciudad L, Gomez-Cabrero D, et al. Pre-B cell to macrophage transdifferentiation without significant promoter DNA methylation changes. Nucleic Acids Res. 2011;40(5):1954–1968. doi: 10.1093/nar/gkr1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Efe JA, Zhu S, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011;108(19):7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 19.Szabo E, Rampalli S, Risueno RM, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468(7323):521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 20.Sapir T, Shternhall K, Meivar-Levy I, et al. Cell-replacement therapy for diabetes: generating functional insulin-producing tissue from adult human liver cells. Proc Natl Acad Sci USA. 2005;102(22):7964–7969. doi: 10.1073/pnas.0405277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyret E, Izpisua Belmonte JC. Worming toward transdifferentiation, one (epigenetic) step at a time. Dev Cell. 2014;30(6):641–642. doi: 10.1016/j.devcel.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Zuryn S, Ahier A, Portoso M, et al. Transdifferentiation. Sequential histone-modifying activities determine the robustness of transdifferentiation. Science. 2014;345(6198):826–829. doi: 10.1126/science.1255885. [DOI] [PubMed] [Google Scholar]
- 23.de Vries WN, Evsikov AV, Brogan LJ, et al. Reprogramming and differentiation in mammals: motifs and mechanisms. Cold Spring Harb Symp Quant Biol. 2008;73:33–38. doi: 10.1101/sqb.2008.73.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Pardo M, Lang B, Yu L, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6(4):382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodda DJ, Chew JL, Lim LH, et al. Transcriptional regulation of Nanog by OCT4 and SOX2. J Biol Chem. 2005;280(26):24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 27.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17(11):6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19(8):5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang ZX, Teh CH, Kueh JL, Lufkin T, Robson P, Stanton LW. Oct4 and Sox2 directly regulate expression of another pluripotency transcription factor, Zfp206, in embryonic stem cells. J Biol Chem. 2007;282(17):12822–12830. doi: 10.1074/jbc.M611814200. [DOI] [PubMed] [Google Scholar]
- 30.Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 31.Niwa H, Toyooka Y, Shimosato D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123(5):917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 32.Li R, Liang J, Ni S, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Abu-Remaileh M, Gerson A, Farago M, et al. Oct-3/4 regulates stem cell identity and cell fate decisions by modulating Wnt/beta-catenin signalling. EMBO J. 2002;29(19):3236–3248. doi: 10.1038/emboj.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masui S, Nakatake Y, Toyooka Y, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9(6):625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 35.Silva J, Nichols J, Theunissen TW, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138(4):722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 37.Kim JB, Greber B, Arauzo-Bravo MJ, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461(7264):649–643. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y, Chen J, Li K, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12(4):453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2010;12(2):79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 40.Ying SY, Chang DC, Miller JD, Lin SL. The microRNA: overview of the RNA gene that modulates gene functions. Methods Mol Biol. 2006;342:1–18. doi: 10.1385/1-59745-123-1:1. [DOI] [PubMed] [Google Scholar]
- 41.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270(2):488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5(2):351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 45.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 46.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 47.Kapinas K, Kim H, Mandeville M, et al. microRNA-mediated survivin control of pluripotency. J Cell Physiol. 2015;230(1):63–70. doi: 10.1002/jcp.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27(5):459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanyam D, Lamouille S, Judson RL, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipchina I, Elkabetz Y, Hafner M, et al. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25(20):2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 52.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14(8):1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang TC, Zeitels LR, Hwang HW, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA. 2009;106(9):3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 56.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145(6):835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7(7):540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 59.Singhal N, Graumann J, Wu G, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141(6):943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 60.Gaspar-Maia A, Alajem A, Polesso F, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460(7257):863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fussner E, Djuric U, Strauss M, et al. Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 2011;30(9):1778–1789. doi: 10.1038/emboj.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattout A, Biran A, Meshorer E. Global epigenetic changes during somatic cell reprogramming to iPS cells. J Mol Cell Biol. 2011;3(6):341–350. doi: 10.1093/jmcb/mjr028. [DOI] [PubMed] [Google Scholar]
- 63.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41(2):246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Liu H, Liu J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45(1):34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 65.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151(5):994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao XD, Han X, Chew JL, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1(3):286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 67.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 68.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143(3):470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farthing CR, Ficz G, Ng RK, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4(6):e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jackson M, Krassowska A, Gilbert N, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24(20):8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 72.van den Berg DL, Snoek T, Mullin NP, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6(4):369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brumbaugh J, Hochedlinger K. Removing reprogramming roadblocks: Mbd3 depletion allows deterministic iPSC generation. Cell Stem Cell. 2013;13(4):379–381. doi: 10.1016/j.stem.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 74.dos Santos RL, Tosti L, Radzisheuskaya A, et al. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15(1):102–110. doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10(1):55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 77.Ying J, Li H, Yu J, et al. WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/beta-catenin signaling, and is frequently methylated in colorectal cancer. Clin Cancer Res. 2008;14(1):55–61. doi: 10.1158/1078-0432.CCR-07-1644. [DOI] [PubMed] [Google Scholar]
- 78.Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta. 1997;1332(3):F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 79.Tam WL, Lim CY, Han J, et al. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26(8):2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marson A, Foreman R, Chevalier B, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3(2):132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ichida JK, Blanchard J, Lam K, et al. A small-molecule inhibitor of TGF-beta signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell. 2009;5(5):491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115(3):281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 83.Maherali N, Hochedlinger K. TGF-beta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol. 2009;19(20):1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 85.Watanabe S, Umehara H, Murayama K, Okabe M, Kimura T, Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25(19):2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 86.Liu N, Lu M, Feng XM, et al. Exogenous Nanog alleviates but is insufficient to reverse embryonic stem cells differentiation induced by PI3K signaling inhibition. J Cell Biochem. 2009;106(6):1041–1047. doi: 10.1002/jcb.22082. [DOI] [PubMed] [Google Scholar]
- 87.Kimura T, Tomooka M, Yamano N, et al. AKT signaling promotes derivation of embryonic germ cells from primordial germ cells. Development. 2008;135(5):869–879. doi: 10.1242/dev.013474. [DOI] [PubMed] [Google Scholar]
- 88.Nakamura T, Inoue K, Ogawa S, et al. Effects of Akt signaling on nuclear reprogramming. Genes Cells. 2008;13(12):1269–1277. doi: 10.1111/j.1365-2443.2008.01243.x. [DOI] [PubMed] [Google Scholar]
- 89.Yu Y, Liang D, Tian Q, et al. Stimulation of somatic cell reprogramming by ERas-Akt-FoxO1 signaling axis. Stem Cells. 2014;32(2):349–363. doi: 10.1002/stem.1447. [DOI] [PubMed] [Google Scholar]
- 90.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12(13):2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460(7251):118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 92.Matsuda T, Nakamura T, Nakao K, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18(15):4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hall J, Guo G, Wray J, et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5(6):597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 94.Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 95.Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol. 2004;275(2):403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 96.Inman GJ, Nicolas FJ, Callahan JF, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62(1):65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 97.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 98.Kaisho T, Akira S. Toll-like receptor function and signaling. J Allergy Clin Immunol. 2006;117(5):979–987. doi: 10.1016/j.jaci.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 99.Lee J, Sayed N, Hunter A, et al. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151(3):547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hwa Cho H, Bae YC, Jung JS. Role of toll-like receptors on human adipose-derived stromal cells. Stem Cells. 2006;24(12):2744–2752. doi: 10.1634/stemcells.2006-0189. [DOI] [PubMed] [Google Scholar]
- 101.Romieu-Mourez R, Francois M, Boivin MN, Bouchentouf M, Spaner DE, Galipeau J. Cytokine modulation of TLR expression and activation in mesenchymal stromal cells leads to a proinflammatory phenotype. J Immunol. 2009;182(12):7963–7973. doi: 10.4049/jimmunol.0803864. [DOI] [PubMed] [Google Scholar]
- 102.Sioud M, Floisand Y. TLR agonists induce the differentiation of human bone marrow CD34+ progenitors into CD11c+ CD80/86+ DC capable of inducing a Th1-type response. Eur J Immunol. 2007;37(10):2834–2846. doi: 10.1002/eji.200737112. [DOI] [PubMed] [Google Scholar]
- 103.Zhao JL, Ma C, O’Connell RM, et al. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell. 2014;14(4):445–459. doi: 10.1016/j.stem.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van den Berk LC, Jansen BJ, Siebers-Vermeulen KG, et al. Toll-like receptor triggering in cord blood mesenchymal stem cells. J Cell Mol Med. 2009;13(9B):3415–3426. doi: 10.1111/j.1582-4934.2009.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pevsner-Fischer M, Morad V, Cohen-Sfady M, et al. Toll-like receptors and their ligands control mesenchymal stem cell functions. Blood. 2007;109(4):1422–1432. doi: 10.1182/blood-2006-06-028704. [DOI] [PubMed] [Google Scholar]
- 106.Tomchuck SL, Zwezdaryk KJ, Coffelt SB, Waterman RS, Danka ES, Scandurro AB. Toll-like receptors on human mesenchymal stem cells drive their migration and immunomodulating responses. Stem Cells. 2008;26(1):99–107. doi: 10.1634/stemcells.2007-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Giuliani M, Bennaceur-Griscelli A, Nanbakhsh A, et al. TLR ligands stimulation protects MSC from NK killing. Stem Cells. 2014;32(1):290–300. doi: 10.1002/stem.1563. [DOI] [PubMed] [Google Scholar]
- 108.Yan H, Wu M, Yuan Y, Wang ZZ, Jiang H, Chen T. Priming of Toll-like receptor 4 pathway in mesenchymal stem cells increases expression of B cell activating factor. Biochem Biophys Res Commun. 2014;448(2):212–217. doi: 10.1016/j.bbrc.2014.04.097. [DOI] [PubMed] [Google Scholar]
- 109.Lee SH, Hong B, Sharabi A, Huang XF, Chen SY. Embryonic stem cells and mammary luminal progenitors directly sense and respond to microbial products. Stem Cells. 2009;27(7):1604–1615. doi: 10.1002/stem.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chan RJ, Johnson SA, Li Y, Yoder MC, Feng GS. A definitive role of Shp-2 tyrosine phosphatase in mediating embryonic stem cell differentiation and hematopoiesis. Blood. 2003;102(6):2074–2080. doi: 10.1182/blood-2003-04-1171. [DOI] [PubMed] [Google Scholar]
- 111.Sharabi AB, Lee SH, Goodell MA, Huang XF, Chen SY. Enhanced generation of myeloid lineages in hematopoietic differentiation from embryonic stem cells by silencing transcriptional repressor Twist-2. Cloning Stem Cells. 2009;11(4):523–533. doi: 10.1089/clo.2009.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhagavati S. Stem cell therapy: challenges ahead. Indian J Pediatr. 2015;82(3):286–291. doi: 10.1007/s12098-014-1521-5. [DOI] [PubMed] [Google Scholar]
- 113.Owaki T, Shimizu T, Yamato M, Okano T. Cell sheet engineering for regenerative medicine: current challenges and strategies. Biotechnol J. 2014;9(7):904–914. doi: 10.1002/biot.201300432. [DOI] [PubMed] [Google Scholar]
- 114.Medvedev SP, Shevchenko AI, Zakian SM. Induced pluripotent stem cells: problems and advantages when applying them in regenerative medicine. Acta Naturae. 2010;2(2):18–28. [PMC free article] [PubMed] [Google Scholar]
- 115.Duinsbergen D, Salvatori D, Eriksson M, Mikkers H. Tumors originating from induced pluripotent stem cells and methods for their prevention. Ann N Y Acad Sci. 2009;1176:197–204. doi: 10.1111/j.1749-6632.2009.04563.x. [DOI] [PubMed] [Google Scholar]
- 116.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 117.Shoni M, Lui KO, Vavvas DG, et al. Protein kinases and associated pathways in pluripotent state and lineage differentiation. Curr Stem Cell Res Ther. 2014;9(5):366–387. doi: 10.2174/1574888x09666140616130217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu K, Song Y, Yu H, Zhao T. Understanding the roadmaps to induced pluripotency. Cell Death Dis. 2014;5:e1232. doi: 10.1038/cddis.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463(7281):621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Melton C, Blelloch R. MicroRNA regulation of embryonic stem cell self-renewal and differentiation. Adv Exp Med Biol. 2011;695:105–117. doi: 10.1007/978-1-4419-7037-4_8. [DOI] [PubMed] [Google Scholar]
- 121.Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ang YS, Tsai SY, Lee DF, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Buganim Y, Faddah DA, Cheng AW, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150(6):1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mansour AA, Gafni O, Weinberger L, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488(7411):409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 125.Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat Cell Biol. 2012;14(5):457–466. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Onder TT, Kara N, Cherry A, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483(7391):598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285(33):25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]