Abstract
Ribosomally produced natural products, the RiPPs, exhibit features that are potentially useful in the creation of large chemical libraries using simple mutagenesis. RiPPs are encoded on ribosomal precursor peptides, but they are extensively posttranslationally modified, endowing them with properties that are useful in drug discovery and biotechnology. In order to determine which mutations are acceptable, strategies are required to determine sequence selectivity independently of the context of flanking amino acids. Here, we examined the absolute sequence selectivity of the trunkamide cyanobactin pathway, tru. A series of random double and quadruple simultaneous mutants were synthesized and produced in Escherichia coli. Out of a total of 763 mutated amino acids examined in 325 unique sequences, 323 amino acids were successfully incorporated in 159 sequences, leading to >300 new compounds. Rules for tru sequence selectivity were determined, which will be useful for the design and synthesis of combinatorial biosynthetic libraries. The results are also interpreted in comparison to the known natural products of tru and pat cyanobactin pathways.
Keywords: combinatorial biosynthesis, cyanobactin, trunkamide, patellamide
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) offer several advantages in the synthetic biology of complex materials and in drug discovery.1,2 RiPPs are universally synthesized on short precursor peptides, which encode a core peptide that is converted into a natural product. Enzymatic modifications to the core peptide, and usually proteolytic cleavage of the core from the precursor peptide, yield much smaller and highly chemically modified, bioactive compounds. These transformations mainly take place in vivo, meaning that in order to modify the structure of the resulting compound, it is sufficient to make mutations in the precursor peptide. Such mutagenesis has been applied to modify or improve initial biological activities of compounds, with the goal of increasing the activity of the natural product against its natural target.3−5 In a few cases, it has also been applied to create designer compounds to hit proteins not targeted by the initial natural product.6,7
Ideally, combinatorial RiPP libraries could be applied to any target, no matter what the initial activity of the compound might be. This could be achieved by, for example, creating a randomized library of core peptides. This method is analogous to that of phage display, intein cyclic peptide libraries, and related strategies.8,9 The major difference is that in the RiPPs, diverse posttranslational modifications impart further drug-like properties into the structures of the compounds.2 A major challenge has been in understanding the substrate selectivity of these modifying enzymes. If the enzymes are relatively selective for specific substrates, then the idea of using RiPPs for combinatorial biosynthesis would fail.
Three general strategies have been employed to address RiPP selectivity. The first involves making directed mutants, either via chemical or genetic methods, and determining which lead to functional products.3 A related second method involves synthesizing saturation libraries of single point mutants.5 Both of these methods are powerful, but they suffer from the fact that they do not distinguish the context dependence of the mutation. For example, an amino acid may be accepted in a certain position in a peptide sequence, but if a second position is simultaneously changed, that amino acid may no longer be accepted. This is because it is the overall molecular properties of a peptide, not its linear sequence, that determines whether or not it will be a substrate for enzymes. A third method involves synthesizing randomized libraries and looking for improved biological activity.4 This method relatively rapidly assesses large numbers of compounds, but it does not determine whether a compound can be made or not; instead, only active compounds are discovered. These methods have been widely and successfully used to create derivatives.3−5,10−14 However, in order to truly determine the sequence selectivity of a RiPP pathway, multiple amino acids must be simultaneously mutagenized, and the products must be chemically characterized.
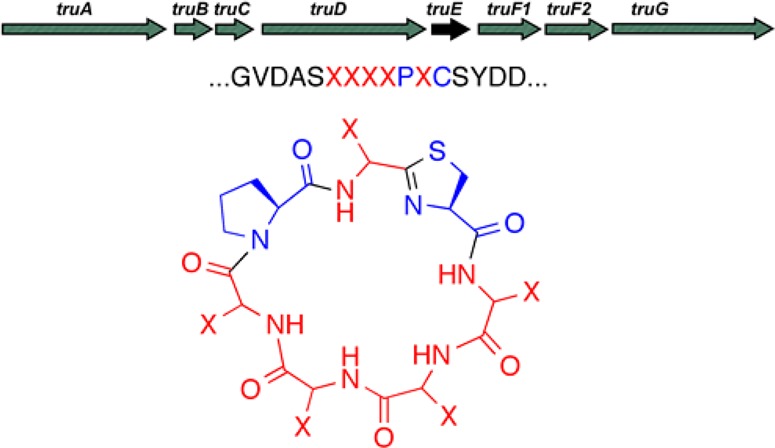
We sought to determine the sequence selectivity of the cyanobactin biosynthetic pathway, tru,15 independent of the surrounding sequence context and without dependence on a functional activity of the products. This pathway was already known in several studies to exhibit unusually relaxed sequence selectivity both in vivo and in vitro (Figure 1),6,15−25 but no methodical and unbiased studies have previously examined the absolute substrate selectivity of the pathway. The tru precursor peptide TruE is modified by at least four different enzymes, meaning that tru is a complex biosynthetic pathway. The products of the pathway are small, N–C macrocycles of 6–8 amino acids, which are highly decorated with posttranslational modifications. Thus, tru affords an opportunity to evaluate the use of RiPPs in combinatorial biosynthetic libraries, since the compounds are relatively small in size but are highly complex chemically. The tru pathway is also ideal because, although it originates in cyanobacteria, it can be functionally expressed in E. coli, making it experimentally tractable.15,17 Here, we report on our efforts to synthesize and characterize such libraries.
Figure 1.

Natural and genetically engineered cyanobactins. (A) TruE precursor peptides are processed by a series of enzymes to yield drug-like products such as trunkamide and patellin 3. (B) Natural and synthetic diversity of peptides previously known to be synthesized by pat (blue), tru (black), or both (green) pathways in vivo, emphasizing the large diversity synthesized by identical enzymes in E. coli. The known in vitro biosynthetic diversity is greater still. Numbering of substrate amino acids begins at the carboxyl terminal cysteine and proceeds backward to the amino terminus of the linear precursor.
Results and Discussion
Library Strategy
We designed a strategy focused on cyclic heptapeptides. Heptapeptides offered a representative sequence complexity in comparison to the hexa- and octa-peptide series, and the natural product trunkamide is a heptapeptide that was considered to be an anticancer lead at one time.26−28 Thus, we constructed and analyzed a relatively complex library while simultaneously building trunkamide variants for anticancer screening, the results of which will be reported elsewhere.
In synthesizing libraries, we desired to encode the entire tru operon on a single plasmid vector as previously described,17 including all modifying enzymes and the precursor peptide gene, truE. TruE consists of two core peptides that are modified enzymatically to produce two distinct natural products (Figure 2). We sought to keep intact the core peptide for the natural product, patellin 3,26,29 which would serve as an internal production control. The second core peptide sequence was modified to create libraries. The internal patellin 3 control is essential to our strategy. If patellin 3 is found, but the recombinant peptide is not, then the absence of that recombinant peptide is meaningful. However, if neither peptide is found, then there may be a technical failure in the recombinant expression run. In all of the results reported below, robust patellin 3 expression was observed from each reported recombinant E. coli strain.
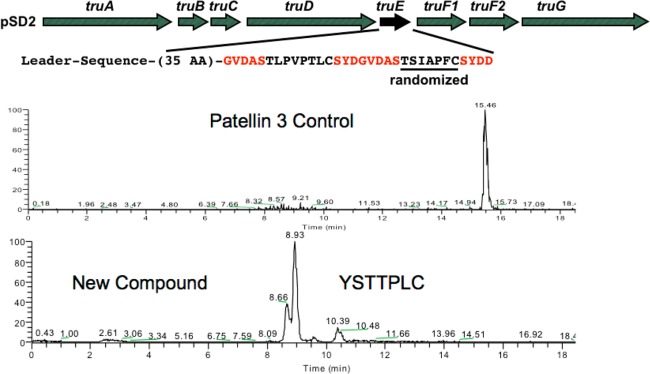
Figure 2.
Expression strategy for trunkamide derivatives. (Top) The full 11 kbp operon was expressed in E. coli, including a randomized truE gene sequence in cassette 2. (Bottom) Example of a randomly obtained clone and its production of a new cyanobactin derivative in E. coli. HPLC-MS run extracted ion chromatograms are shown.
In the design of the core peptide mutagenesis strategy, we selected NNK libraries (N, any nucleotide; K, guanine or thymidine). For the past two decades, this strategy has been the most common one used in creating genetically encoded libraries (such as phage display libraries), since it only encodes 32 codons with 1 stop sequence, rather than the 64 codons with 3 stop sequences encoded in NNN libraries.30
Previous studies have shown that a heterocycle (either proline or thiazol(in)e/oxazol(in)e) is required in position 1 in order for macrocyclase enzyme to be functional.22 Therefore, the thiazoline precursor Cys was fixed in position 1. We chose to fix Pro in position 3 in these studies because we reasoned that this position would be important in holding the resulting product in the correct orientation for trunkamide activity. Previously, NMR studies have shown that trunkamide adopts a specific shape, which might be essential for the anticancer mechanism of the natural product.31 A library with full 5-fold degeneracy contains 205 (3.2 million) possible sequences, meaning that even with Cys and Pro fixed, a large library would still be characterized, but the analysis is more chemically feasible than assessing 206 (64 million) sequences.
The challenge was to adequately characterize this enormous number of possible mutants, which requires expensive and time-consuming procedures. Briefly, in order to synthesize the reported libraries, randomized vectors were prepared in which the tru pathway was intact, but the second core peptide sequence of truE was mutagenized with the NNK strategy. The resulting library was transformed into E. coli, and individual colonies were picked from plates. The mutant truE gene was sequenced in each colony, and the colonies were arrayed into deep 24-well plates. After 5 days of incubation, the colonies were pelleted, extracted with organic solvent, and the resulting extracts were partially enriched on a solid phase resin. Production of cyclic peptides was analyzed by HPLC-mass spectrometry (Figure 2). Potential product masses were determined using the Sanger sequences of truE genes: a spreadsheet was produced in which the possible molecular weights of products in various states of posttranslational modification were listed.
For example, the spreadsheet contained the expected mass of the linear, unmodified product, and in addition, expected masses with 0–3 (or more if chemically possible) dehydration reactions to yield macrocycle and thiazoline or oxazoline, as well as with the addition of 0–3 isoprene units, which are added to the Ser or Thr side chains. These expected molecular weights were then sought in each HPLC-MS trace. If an expected peak was found, the presence of this peak was also sought in adjacent HPLC-MS runs using extracts of E. coli clones that contained other TruE2 derivative sequences, which thus served as negative controls. If a compound is prenylated, isoprene is partly eliminated in the MS, so that both the parent ion and deprenylated fragments are apparent in the MS.17,18 Each run was also assessed for the presence of the positive control, patellin 3, and its less prenylated derivatives. Taken together, the extensive positive and negative controls embedded in this library design allowed us to assign the identity of new compounds with high confidence. This general method was also validated previously on much smaller library sets.17
Finally, an additional complication of these libraries is that the alpha protons adjacent to thiazoline are readily epimerized.27 Indeed, we have previously described that the Phe adjacent to thiazoline in trunkamide is synthesized in the L-form and gradually epimerizes to the D-form.15 The metastable form of the compound seems to be determined by the preferred three-dimensional structure of the molecule, since only a subset of heptapeptides exhibits this trait. The practical outcome of epimerization is that there are often, but not always, two distinct isobaric peaks for each product formed (Figure 3).
Figure 3.
Example of peak doubling observed in libraries (top), compared to a similar compound that does not exhibit peak doubling (bottom). This phenomenon has previously been characterized with other derivatives to be caused by epimerization. Here, the possible positions that may be epimerized are shown with wavy lines. For each compound, the HPLC-MS derived extracted ion chromatogram is shown, along with integrated m/z for that peak area. Full spectra are shown in greater detail in Supporting Information Table S4.
While simpler chemical methods could perhaps be envisioned to analyze the libraries, we have tried many of them and found this procedure to be the most reproducible. In addition, the method provided very robust controls. In essence, each run served as a negative control for every other run. The presence of patellin 3 as a positive control, and the sequence-mass relationship of new compounds, enabled us to establish the presence or absence of each new compound with very high confidence. Another advantage was that the presence of patellin 3 enabled us to roughly estimate the amount of each mutant compound synthesized, which ranged from approximately 10–500 μg per liter in this library, using a previously described method.17 If a compound was not detected despite the positive controls working, we consider the sequence as not being a substrate for the tru pathway. However, there are many reasons that a compound might not be detected beyond it being a poor substrate (it might be lethal to E. coli, it may be secreted into the broth, etc.). It may as well simply not be detectable under the experimental conditions.
A strategy to elucidate RiPP libraries of 3.2 million members, with the complex chemical analysis problem described above, has not been previously reported and required careful design. In order to determine the allowed sequences, in the absence of a function-based assay, we chose to first simultaneously randomize two different amino acid positions. Since only 202 (400) double mutants are possible per pairwise sequence randomized, this strategy allowed us to perform an initial assessment of the absolute sequence selectivity at five different positions with a potential library size of 1200 sequences, rather than 3.2 million. In this design, we grouped positions 6 and 7 (XXIAPFC), 4 and 5 (TSXXPFC), and 2 and 4 (TSIXPXC). Subsequently, we performed a tandem quadruple mutation (XSXXPXC), which has a potential to accept a total of 204 (160 000) different sequences. These changes covered positions 2, 4, 5, and 6. Thus, taken together, positions 2, 4, 5, 6, and 7 were randomized in at least a subset of libraries.
Even these more limited libraries contained a greater sequence diversity than could be reasonably characterized. Therefore, we selected a strategy that enabled saturation coverage at each of five positions. Sanger sequencing of the resulting clones indicated that the positions were saturated with each of the 20 proteinogenic amino acids (Table 1). If each amino acid were represented once in each of the mutated positions, a total of 100 possible substitutions would be allowed, whereas we measured 763 individual amino acid substitutions. The only exceptions were the amino acids Met and Trp, which were absent from position 6, although overall they were encoded 19 and 25 times, respectively, in the libraries. Amino acids were incorporated into the library in their expected numbers based upon NNK codon frequency, with no undue bias by amino acid type (Figure 4).27 This showed that, overall, the libraries were of high quality.
Table 1. Number of Occurrences of Amino Acid Coding Mutations in the Librariesa.
| position | 7 | 6 | 5 | 4 | 2 | total | expect |
|---|---|---|---|---|---|---|---|
| A (Ala) | 9 | 3 | 8 | 12 | 5 | 37 | 49 |
| C (Cys) | 4 | 2 | 4 | 8 | 5 | 23 | 25 |
| D (Asp) | 5 | 3 | 8 | 11 | 5 | 32 | 25 |
| E (Glu) | 4 | 3 | 11 | 7 | 1 | 26 | 25 |
| F (Phe) | 2 | 2 | 7 | 8 | 1 | 20 | 25 |
| G (Gly) | 14 | 6 | 8 | 19 | 7 | 54 | 49 |
| H (His) | 6 | 2 | 9 | 16 | 4 | 37 | 25 |
| I (Ile) | 7 | 4 | 5 | 8 | 5 | 29 | 25 |
| K (Lys) | 7 | 5 | 11 | 12 | 2 | 37 | 25 |
| L (Leu) | 3 | 7 | 7 | 19 | 23 | 59 | 74 |
| M (Met) | 4 | 0 | 5 | 6 | 4 | 19 | 25 |
| N (Asn) | 10 | 4 | 11 | 7 | 3 | 35 | 25 |
| P (Pro) | 5 | 4 | 12 | 14 | 10 | 45 | 49 |
| Q (Gln) | 5 | 3 | 6 | 13 | 6 | 33 | 25 |
| R (Arg) | 10 | 6 | 26 | 17 | 3 | 62 | 74 |
| S (Ser) | 11 | 4 | 12 | 25 | 13 | 65 | 74 |
| T (Thr) | 9 | 3 | 15 | 14 | 10 | 51 | 49 |
| V (Val) | 5 | 3 | 9 | 20 | 7 | 44 | 49 |
| W (Trp) | 6 | 0 | 4 | 7 | 8 | 25 | 25 |
| Y (Tyr) | 8 | 5 | 3 | 9 | 5 | 30 | 25 |
| total | 134 | 69 | 181 | 252 | 127 | 763 |
Only full-length TruE2 mutants are considered in this analysis. Saturation was achieved for each amino acid at each site, with the exceptions of Met and Trp in position 6. Expect indicates the total number of times that the amino acid may be expected to occur in an NNK library with 763 mutants; observed values meet expectations for a library of this complexity, indicating no undue bias in the library preparation and analysis.
Figure 4.

Observed (x-axis) versus expected (y-axis) occurrence of each amino acid in the libraries, using the data shown in Table 1. Orange, hydrophobic; yellow, hydrophilic; light blue, aromatic; green, acidic; purple, Gly/Met/Pro/Cys; black, basic.
Double Mutant Libraries
The core peptide sequence of trunkamide, TSIAPFC, was mutated in NNK libraries encoding XXIAPFC, TSIXPXC, and TSXXPFC. The first of these was aimed at determining the importance of the posttranslational modification prenylation, which requires the presence of Ser or Thr in positions 6 and 7. The other libraries aimed to mutagenize the remaining three positions in the library.
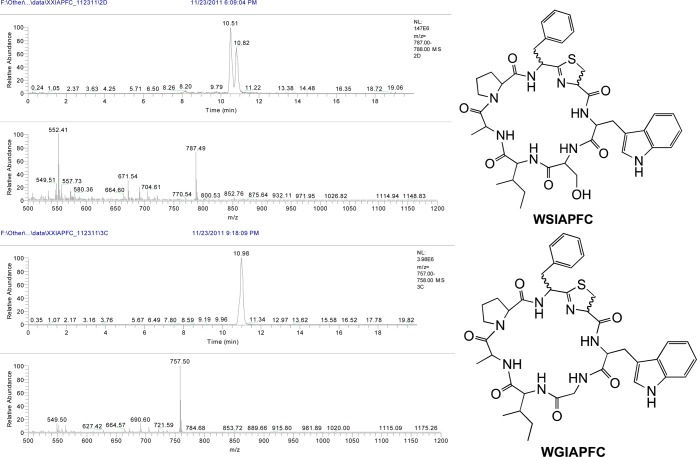
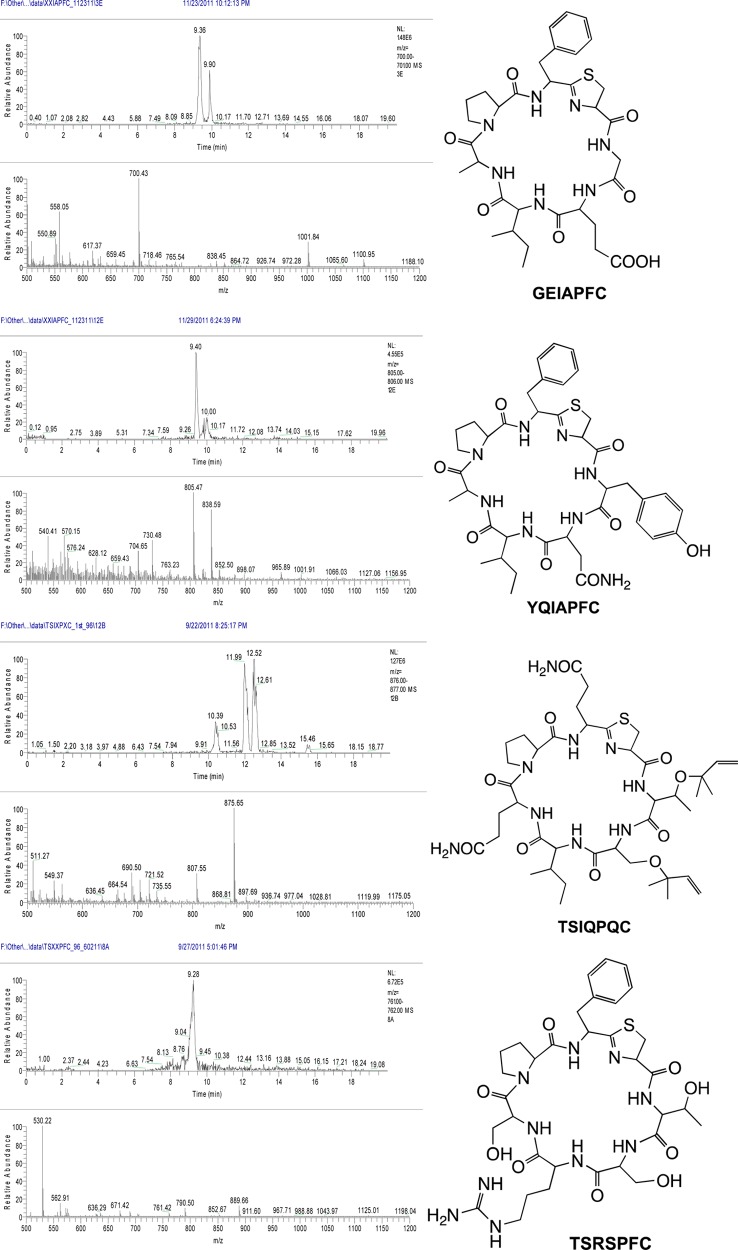
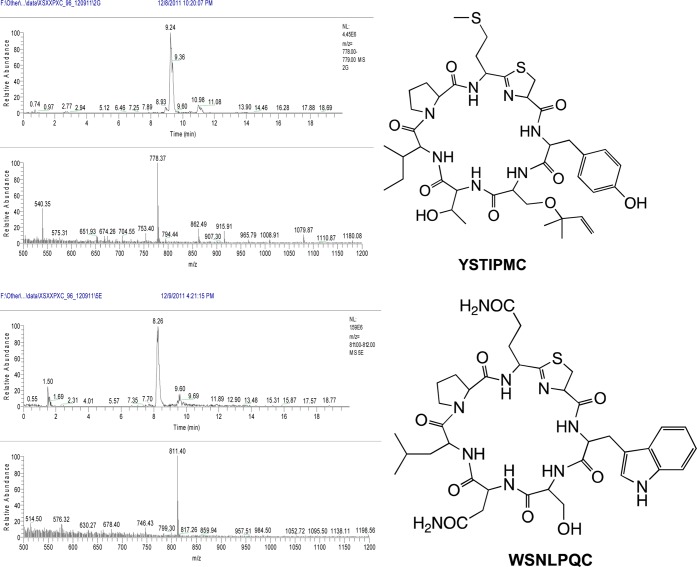
We screened a total of 460 random double mutant clones, out of which 260 encoded full-length TruE derivatives and the remainder contained stop codons or frame-shifts. Out of these 260, 150 mutant TruE products (142 unique) were detected as abundant components of these extracts for a ∼58% success rate (Figure 5). All three libraries exhibited an approximately equal success rate. Since 1200 possible double mutants might be created by this library, if the success rate could be extrapolated, then 675 compounds should be produced in these libraries. Details of which amino acids are found in which positions are described in ensuing sections below. However, a few examples of interesting sequences will be described. While the native sequence is TSIAPFC, we found that sequences such as PLIAPFC, HLIAPFC, GEIAPFC, TSIQPNC, TSISPTC, TSIFPWC, TSAEPFC, TSHPPFC, TSKQPFC, and many other highly unnatural sequences were competent for modification by the tru pathway and were found in whole cell extracts. These many promising and diverse sequences are listed in Supporting Information Table S1, and their raw MS data is shown in Supporting Information Table S4. Further examples are also shown in Figures 2 and 5. Many sequences also failed to be processed, as shown in Supporting Information Table S2. Supporting Information Table S3 summarizes observed mutations per position and reveals that every amino acid was incorporated into every position in the library, with the exception of Met and Trp in position 6, which were not encoded in the library.
Figure 5.
Examples of successfully produced double mutant derivatives. Larger spectra, as well as spectra for all produced compounds are provided in Supporting Information Table S4.
The double mutant libraries also clearly revealed the value and necessity of determining context-independent amino acid selectivity. As a concrete example in this library, the sequence TSRAPFC (versus the wild-type sequence, TSIAPFC) was not accepted; in a saturation library, it would appear that R is forbidden in position 5. However, in reality, R was relatively favorable in position 5, when position 4 was no longer the wild-type Ala sequence, and the following peptides were all successfully synthesized: TSRDPFC, TSRFPFC, TSRQPFC, TSRSPFC, TSRTPFC, TSRVPFC, TSRYPFC. Treating peptide substrates as linear sequences of letters is clearly inadequate to the task of determining the true substrate selectivity of RiPP pathways.
Quadruple Mutant Library
We designed the sequence, XSXXPXC, based upon the results above. We wished to preserve the Ser in position 6 so that this position would always be available for prenylation, allowing us to assess substrate selectivity of prenylation. In this event, 96 clones were screened, leading to 65 full-length TruE derivatives. Of these 65, nine led to full-length products, for a 14% success rate. These derivatives were highly sequence variable (Figure 6), reinforcing the value of the library. Results and data are provided in Supporting Information Tables S1–S4.
Figure 6.
Examples of successfully produced quadruple mutant derivatives. Larger spectra, as well as spectra for all produced compounds are provided in Supporting Information Table S4.
Sequence Selectivity Rules
We noticed that certain amino acids could never be found in certain positions, even if they were encoded in those positions in multiple independent events. This enabled us to form sequence selectivity rules that were useful in understanding library diversity. The formulation of rules is described in “Methods”, but briefly, we tabulated the amount of times an amino acid was found in each position in a mutant library, counting only mutated residues. If an amino acid was encoded in a specific position at least five times, but never led to a detectable product, that amino acid was considered to be “forbidden”.
In total, each of the 20 amino acids was present an average of 38.15 times, with 134, 69, 181, 251, and 127 point mutations in positions 7, 6, 5, 4, and 2, respectively (Table 1). This enabled the rules to be assigned with confidence. In particular, Cys was never found outside of position 1. Arg and Lys are unfavorable; they were never found in positions 2, 6, or 7, and were disfavored in positions 4 and 5. Asp, Gly, Glu, and Pro were never found in position 2, although Gly was only found in tandem with other relatively disfavored amino acids in this library and therefore was not considered as forbidden.
The rules are not absolute, and merely denote preferences that are reflected in the complexity of the resulting libraries. As a very concrete example, despite 23 codons encoding Cys in our libraries, we never saw Cys outside of position 1. However, in previous work Cys has been found in other positions in octapeptides produced by tru.17 Therefore, it is likely that Cys is acceptable in some contexts that were not fully explored by this library. Nonetheless, the sequence selectivity rules are useful in analyzing the experimental results. For example, if only rule-following sequences are considered, 80%, 84%, and 58% success rates are observed for the XXIAPFC, TSIXPXC, and TSXXPFC libraries, respectively. (The complication of the rules for the latter library, in which basic amino acids are sometimes accepted but greatly decrease the chances of production, explain the lower success rate. In fact, similar total numbers of products are accessible from each library.) Similarly, the quadruple library would have a 38% success rate following the rules. About the same total number of compounds would be anticipated in either event, but the background of unaccepted sequences is higher when the rules are not followed.
Additional variables increase the number of compounds produced (Figure 3 and Supporting Information Figure S4). In particular, both d- and l- amino acids are observed adjacent to thiazoline, which causes some peaks in the LC-MS traces to be doubled. Ser or Thr adjacent to thiazoline is partially dehydrated, leading to compounds that are 18 Da lighter than predicted. This dehydration is most probably due to the reverse Michael reaction, since it is observed to increase in extracts over time, whereas enzymatic reaction products are stable. Finally, mixtures of differentially prenylated products are observed, with 0, 1, 2, or even 3 prenylation events observed as mixtures of compounds in traces. It is easy to verify this as O-prenylation, since 68 Da (isoprene) is readily lost in these compounds, as previously described.17 Once these compound variants are taken into account, the 159 successfully produced sequences yielded >300 unique, new compounds.
Of note, prenylation was the most fastidious feature of the pathway. While all detected products were N–C macrocyclic and contained thiazoline, not all products were prenylated, even if prenylatable groups were present. We only observed prenylation when position 5 was comprised of Arg, Val, Leu, Ile, Met, Ser, or Thr.
Beyond the Rules: Relative Sequence Preferences
In nature, the tru and related pat pathways produce compounds that contain Ala, Cys (position 1 only in tru), Ile, Leu, Phe, Ser, Thr, and Val, or eight out of the 20 proteinogenic amino acids.33,34 There are no known tru or pat natural products containing Asp, Glu, Gly, His, Met, Asn, Gln, Trp, or Tyr. Nonetheless, from this library methodology, these amino acids are clearly highly favored in most positions, so that in this work we more than double the number of amino acids (to 17) that are known to be favored in tru and pat core sequences. In addition, Arg and Lys are never found in natural sequences. They are disfavored in most positions, but they can in fact be found with relatively high preference under some circumstances. Beyond these rules, there were minor variations in preference at individual amino acid positions, which are shown in Supporting Information Table S3.
The overall preference for each amino acid can be readily discerned from the available data (Figure 7 and Table 2). Models were created both with and without considering the codon bias of NNK libraries, yielding essentially identical results. Therefore, for simplicity, we chose to analyze the data as straight percentage of occurrence in compounds versus total appearances in the library. In so doing, it is apparent that amino acids behave relatively similarly as groups (Supporting Information Tables S3B and S3C). Preferred amino acids include the four hydrophobic amino acids, four hydrophilic amino acids, four aromatic amino acids, and Gly/Met, which were grouped. The two acidic amino acids and Pro are relatively favored except in specific positions. The two basic amino acids are highly disfavored. Although behavior is similar within groups, there are minor variations in incorporation efficiency between amino acids within groups; complete details are provided in Supporting Information Table S3.
Figure 7.
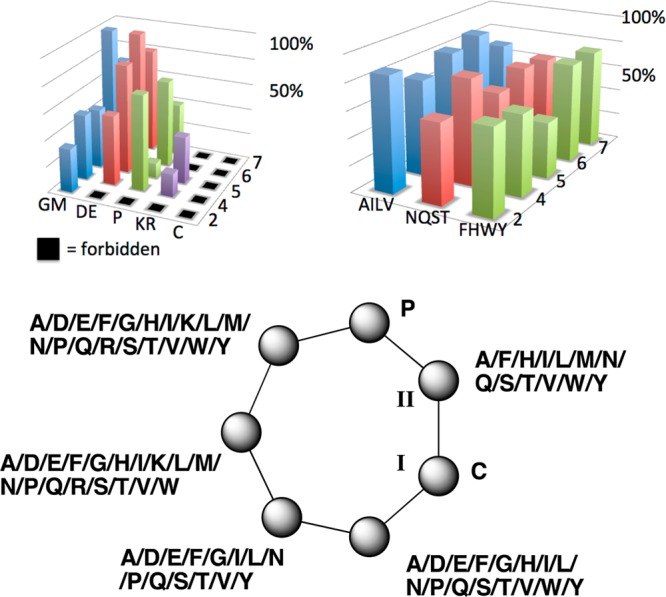
Graphical representation of results shown in Table 2. The vertical axis is the % chance that an amino acid will be found in a specified position in otherwise rule-following sequences (forbidden amino acids indicated by a black box). The horizontal axes represent the amino acids considered and the position in the peptide, with similar amino acids grouped and considered together. The circles indicate total accepted mutations at each position found in this study.
Table 2. Substitution Preference and Forbidden Positions in Heptapeptide Librariesa.
| 7 | 6 | 5 | 4 | 2 | ||
|---|---|---|---|---|---|---|
| pos. | T | S | I | A | F | total |
| AILV | 24, 70% | 16, 87% | 30, 85% | 60, 72% | 39, 86% | 171, 80% |
| NQST | 35, 64% | 14, 67% | 45, 56% | 64, 80% | 32, 60% | 189, 67% |
| DE | 10, 67% | 6, 100% | 19, 83% | 18, 53% | 10, 0% | 63, 70% |
| FHWY | 22, 75% | 9, 75% | 22, 44% | 39, 56% | 18, 64% | 110, 60% |
| GM | 18, 69% | 6, 00% | 13, 36% | 25, 50% | 11, 33% | 73, 53% |
| P | 4, 33% | 4, 67% | 12, 11% | 14, 73% | 10, 0% | 64, 32% |
| KR | 17, 0% | 11, 0% | 36, 36% | 29, 18% | 6, 0% | 99, 29% |
| C | 4, 0% | 2, 0% | 4, 0% | 8, 0% | 5, 0% | 23, 0% |
Given are the total number of appearances of amino acids in each position for the combined double and quadruple mutant libraries, then % success rate for rule-following sequences. Highlighted are positions of “forbidden” amino acid substitution. In the shaded series, the percentage value indicates that this amino acid is never observed in libraries, since they are never rule-followers. The wild type sequence is shown in row 2.
Interestingly, if one considers the eight amino acids naturally found in pat and tru products in the wild, there is only a slightly greater chance that these amino acids would be found in our library than the other 9 favored amino acids. Excluding Cys, the remaining 7 natural amino acids are successfully incorporated in degenerate positions 74%/55% of the time in double/double + quadruple mutant libraries. (These amino acids are encoded 307 times in the libraries.) In comparison, the 9 relatively preferred amino acids that are not found in nature are successfully incorporated 67%/47% of the time. (These amino acids are encoded 293 times in the libraries.) We hypothesize that the naturally observed amino acids are important for the phenotype of the compounds in their native environments. The pat and tru pathways are found so far only in symbiotic cyanobacteria that live in marine animals, and the resulting compounds are abundant components of the animals. It will thus be of interest to elucidate the natural roles of these compounds.
Library Quality and Statistics
The double mutant libraries allowed us to achieve confidence intervals of 11, 8, and 11 for the three libraries at the 95% confidence level. Overall, this indicates that together these 3 double mutant libraries would yield between 600 and 750 unique compounds. Similarly, although the quadruple mutant library is obviously undersampled (although it is still the largest of its kind reported to date), one could estimate that between 16 000 and 48 000 compounds could be produced using this methodology.
Outlook for Library Design
Here, we define the unbiased sequence selectivity of the tru pathway with the heptapeptide series. We show that virtually every amino acid is accepted at five separate positions within the heptapeptide series. By using tandem mutations, we were able to determine sequence selectivity regardless of the context of the amino acid. As the example of Arg in position 5 shows, context dependence is a critical but underappreciated aspect of substrate selectivity in RiPP libraries. Because every position was oversampled with the minor exceptions mentioned above, the results are obtained with high confidence. In studies of RiPP selectivity in vivo, these context-independent results are generally not obtained, or else they are usually constrained only to compounds that exhibited the sought biological activity. From this study, it seems likely that the breadth of products that could be synthesized by RiPP pathways has been underestimated.
These libraries have been used to screen for more active variants of the anticancer compound, trunkamide, which will be described in due course. This is not the ideal application of such libraries, since it is highly labor-intensive and leads to a relatively small compound diversity. We have created libraries containing >108 unique sequences; if even 1% of these are synthesized, the libraries should afford >106 unique compounds. In these types of libraries, in vivo screening and selection methods provide the most appropriate applications, since they require a minimum of manipulations and are much less expensive to conduct. These methods would be analogous to those used in phage display or intein circularization libraries.
The results of this study reveal several important points in the design and synthesis of randomized RiPP libraries. First, as found in other library-generating methods, we experienced a relatively high attrition rate of sequences even with standard methods such as NNK libraries employed. Therefore, in large RiPP libraries one has to consider that at minimum 30% of clones are nonfunctional because of stops, frameshifts, etc. Second, it appears that basic amino acids are relatively disfavored in E. coli (while they are seemingly not as unfavorable in vitro)22,35 in comparison to other amino acids. Other strategies, such as in vitro libraries, may serve to circumvent this problem, but they have disadvantages in some screening applications. Third, in RiPP libraries that are truly randomized, even with an enormous number of successfully produced new compounds, it is likely that most sequences will not be functionally produced. This requires a design of the screening strategy that minimizes the manipulation of individual colonies and that does not rely on chemical extraction methods. Finally, the majority of the limitations discussed above can be circumvented by producing libraries in which each position is randomized to contain only favored amino acids. This could be achieved by preparing libraries using trimer phosphoramidites, where each amino acid is encoded by a unique codon (Glen Research, Sterling, VA). Each randomized position could be produced using a trimer phosphoramidite mixture containing only the amino acids favored at each site. If these limitations are considered during the design of a screening campaign, RiPP libraries offer access to an unprecedented chemical diversity through simple mutagenesis.
In the case of the tru pathway, at least four individual enzymatic steps take place in the processing of the precursor peptide.15 Prior to the action of enzymes, a precursor peptide, TruE, is synthesized on the ribosome. This precursor peptide contains three distinct recognition sequences (RS) that attract modifying enzymes, as well as the amino acid sequence that encodes the natural (or unnatural) product.16 The first three enzymatic steps involve enzymes that target recognition elements in the core peptide. The first of these is heterocyclization of Cys residues by TruD, which binds to RSI in the leader peptide.16,21 Previous studies in vitro show that TruD and the related PatD are extremely broad-specificity enzymes,16,21,23,35,37 and thus it is likely that TruD is not a limiting enzyme in the synthesis of derivatives. In our study, we explicitly searched for derivatives lacking heterocyclization in Cys, but we did not find any such examples. Cys failed to be incorporated at any other position in the precursor peptide. Since it is known from in vitro studies that TruD cyclizes Cys in many different positions, it is likely that Cys in all positions is heterocyclized but that such products containing multiple heterocycles are not substrates for macrocyclization.
The next enzyme in the pathway is TruA, which cleaves the precursor peptide at RSII, releasing a free N-terminus that is the nucleophile for macrocyclization.16,24,38,39 Relatively less is known about the substrate selectivity of TruA (which is sequence identical to PatA), and our study was not designed to detect possible products in the case of TruA failure. Subsequently, TruG cleaves RSIII from the core peptide in tandem with macrocyclization.22,24,38,40 TruG and its close relative PatG have been extensively studied in vitro and exhibit very relaxed substrate selectivity. For example, in vitro substrates containing Arg or Lys are highly favorable,22 rather than being unfavorable as found in this study in vivo. Rarely, PatG macrocyclization leads to linear, rather than cyclic products in vitro. Our study was designed to detect short linear peptides resulting from core peptides, but these were not observed in any case.
In our study, when we observed products, they were universally macrocyclic and heterocyclic at Cys, indicating that the three enzymes TruD, TruA, and TruG were able to process all of the substrates and intermediates. In the last enzymatic step, one, two, or three units of isoprene are added by TruF.15,18,41 This enzyme was not always functional, and many products were observed in which prenylation was lacking, as discussed above. This is of significance because the cyanobactin prenyltransferases modify the cyclic peptide itself. No RS is present when TruF acts, and the precursor peptide has already been cleaved. Thus, this step would be expected to exhibit the most stringent substrate requirements. Even so, TruF can recognize a large number of different sequences and is quite substrate relaxed. It remains a mystery how an enzyme can recognize such a large variety of cyclic peptides, which to the best of our knowledge has not previously been observed for any other enzyme family.
Complicating the above discussion, nothing is known about the relative rates of the enzymes in vivo, or of the possible degradation or stability of proteins or intermediates in vivo. This means that, for example, a mutant may actually be a reasonable substrate for all of the enzymes, but the degradation rate of a short peptide intermediate may be faster than the relatively slow cyclization step. For reasons of in vivo kinetics, therefore, it is very difficult to firmly establish the reason for failure of individual intermediates. Perhaps conditions could be found that might affect these relative rates, broadening the potential substrates in E. coli.
How many total compounds might be made using the tru pathway? Our study was not aimed to answer this question, but an approximate estimate might be made. The lower-end estimate for the quadruple mutant library is that ∼20 000 compounds might be accessible. Position 6, not sampled in the quadruple mutant library, was very permissive in double mutant library, such that perhaps 100 000–200 000 compounds might be accessible in a pentuple mutant library based upon a linear extrapolation. If these numbers scale at all with larger libraries, and since multiple ring sizes are possible, a large number of derivatives might be anticipated from the tru pathway. It should be kept in mind that with the appropriate techniques, unnatural and nonproteinogenic motifs can also be incorporated by tru, increasing the biosynthetic potential of the pathway.17,22 While the estimates are merely suggestive, they receive some support in the very elaborate derivatives that have been synthesized to date in vivo and in vitro by the tru pathway, as well as by the fact that our hextuple mutant library afforded a ∼1% success rate in preliminary studies.
In summary, we show here specifically with the tru pathway example that RiPP libraries are promising sources of chemical diversity. These methods and libraries may find application in drug discovery, as well as in other problems in synthetic biology and biotechnology.
Methods
Library Design and Expression
The plasmid pSD-2 was synthesized containing the tru pathway genes: truA, truB, truC, truD, truF1, truF2, and truG. In place of native truE, a truE variant was constructed in which patellin 3 was present in the first core peptide cassette and a randomized NNK library was present in the second cassette. In this study, 4 libraries were synthesized based upon the heptameric trunkamide sequence: XXIAPFC, TSXXPFC, TSIXPXC, and XSXXPXC. An oligonucleotide duplex was prepared for each library, as follows. Each library template oligo was annealed separately with the Library Primer oligo and the duplexes were filled in using Taq DNA polymerase (NEB, Ipswich, MA) as per the manufacturer’s instructions. Each duplex was digested with MluI (NEB, Ipswich, MA), as per the manufacturer’s directions. The resulting MluI/BanII terminated oligo duplexes were ligated into BanII/MluI digested plasmid pSD-2. Each ligation mix was transformed by electroporation, using a BTX ECM600, into NEB10-β. Multiple transformations were performed to produce libraries of sufficient complexity. Plasmid purifications were performed using Qiagen mini-prep spin columns. The following oligonucleotides were used: library primer 5′-pCGCTCGGAGATTAGTCGTCATAGGA-3′ (p for 5′ phosphate); library template XXIAPFC 5′-CGTTGACGCGTCTNNKNNKATTGCGCCGTTCTGCTCCTATGACGACTAATCTCCGAGCGAGCC-3′; library template TSXXPFC 5′-CGTTGACGCGTCTACCAGCNNKNNKCCGTTCTGCTCCTATGACGACTAATCTCCGAGCGAGCC-3; library template TSIXPXC 5′-CGTTGACGCGTCTACCAGCATTNNKCCGNNKTGCTCCTATGACGACTAATCTCCGAGCGAGCC-3′; library template XSXXPXC 5′-CGTTGACGCGTCTNNKAGCNNKNNKCCGNNKTGCTCCTATGACGACTAATCTCCGAGCGAGCC-3′.
Library quality was verified by randomly picking and sequencing the truE mutants from individual E. coli clones and Sanger sequencing the truE gene. Titers of mutant libraries were verified by dilution transformation into E. coli, showing that >108 individual sequences were present in the 6-fold randomized library and that the other libraries contained enough diversity to fully sample the sequence space. The pathways containing mutant truE sequences were expressed in E. coli using 6 mL culture volumes in 24-well plates, using methods that were previously described.17 For each randomly picked expression clone, the truE mutant gene was sequenced, and the resulting sequence was used to predict potential molecular weights of the resulting cyanobactin products, as described in Results.
Chemical Analysis
After the growth period, cells were harvested by centrifugation, the supernatant was removed, and the pellet was extracted with acetone (3 × 1 mL, followed by centrifugation to pellet cells). The acetone was removed under vacuum, the extracts were passed through HP20 resin to eliminate salts and hydrophobic components, and the resulting methanol eluent was concentrated under vacuum. Samples were analyzed by HPLC-MS using a Thermo Finnigan TS Quantum mass spectrometer coupled to a Waters 2694 HPLC separations module, 2996 photodiode array detector, and 600 controller, using a Phenomenex C18 column (10 cm × 4.6 mm, 5 μm). Samples were analyzed for the presence or absence of the standard patellin 3 as a control for expression quality in each construct. This patellin 3 standard also verified that the total expression operon present in pSD-2 was functional and full-length. Compounds representing all masses calculated as described above were sought manually. Putative product peaks were verified by comparing their weights to control runs from other expression experiments in which that precise sequence was not present. When compounds are isoprenylated, the fragment isoprene loss can also be readily identified, further supporting the structure assignment. Thus, for each compound found, a large number of negative controls and significant MS data exist in support of the proposed identified compound. This method has been described in greater detail in previous publications in which cyanobactin derivatives were identified or synthesized from complex extracts.15,17−20,36 Raw data for each experiment is provided in Supporting Information Table S4.
Determination of Selectivity Rules
TruE mutant sequences that yielded products and those that failed to yield products were tabulated (Supporting Information Tables S1–S3). Mutants containing stop codons, frameshifts, or other errors within the cassette were not considered in this analysis, and only sequences expressing full-length TruE analogs were used. First, we analyzed which amino acids could possibly exist in each position. If a mutant residue was found at least 5 times in a specific position, and yet, the resulting predicted compound was never found, this amino acid was considered to be “forbidden” by the rules. For example, Arg/Lys was never found in positions 2, 6, or 7, despite occurring a total of 35 times in available sequences. In addition, Pro, Asp, and Glu were not allowed in position 2. Despite occurring 23 times in the library, Cys was never found in any variable position.
If an amino acid never appeared in a given position X, despite being present in the mutant library, it was given a score of −1. Sequences containing those forbidden amino acids (88 total sequences) were not considered in the remaining substrate selectivity analysis, since it was presumed that the effect of the forbidden amino acid dominated the nonproduction of compound from that sequence. The remaining 174 sequences were analyzed as follows. The percent of the time that an amino acid in an individual position led to successful production of compound was calculated (Supporting Information Figure S3). This value was taken to provide a measure of the relative favorability of that amino acid in a given position. Finally, the values and sequence selectivity rules were reevaluated with the complete set of data to ensure consistency.
These analyses were first performed using formulas that normalized to the expectation value given the NNK codon bias. However, since the observed sequences were truly random and representative, essentially identical results were obtained whether or not this bias was considered.
Acknowledgments
We thank Casey Van Wagoner and Zhidong Chen for technical assistance, Zhenjian Lin for help with NMR, and Symbion cofounder Sheryl Hohle.
Supporting Information Available
Tables of data showing which variants are synthesized and which are not detected and which amino acids are accepted in each position. All mass spectra used to assign compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was funded by National Institutes of Health (NIH) GM093790, AT006121, and GM071425.
The authors declare the following competing financial interest(s): DER and EWS are part-owners of Symbion Discovery.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K. D.; Fischbach M. A.; Garavelli J. S.; Goransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Muller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J.; Rebuffat S.; Ross R. P.; Sahl H. G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Sussmuth R. D.; Tagg J. R.; Tang G. L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. A.; Donia M. S.; Schmidt E. W. (2009) Ribosomal peptide natural products: Bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 26, 537–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdick D. A.; Dodd H. M.; Barraille P.; White J.; Stein T. H.; Chater K. F.; Gasson M. J.; Bibb M. J. (2003) Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus DSM 40005. Proc. Natl. Acad. Sci. U.S.A. 100, 4316–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S. J.; Link A. J. (2011) Sequence diversity in the lasso peptide framework: Discovery of functional microcin J25 variants with multiple amino acid substitutions. J. Am. Chem. Soc. 133, 5016–5023. [DOI] [PubMed] [Google Scholar]
- Young T. S.; Dorrestein P. C.; Walsh C. T. (2012) Codon randomization for rapid exploration of chemical space in thiopeptide antibiotic variants. Chem. Biol. 19, 1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M. S.; Hathaway B. J.; Sudek S.; Haygood M. G.; Rosovitz M. J.; Ravel J.; Schmidt E. W. (2006) Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2, 729–735. [DOI] [PubMed] [Google Scholar]
- Knappe T. A.; Manzenrieder F.; Mas-Moruno C.; Linne U.; Sasse F.; Kessler H.; Xie X.; Marahiel M. A. (2011) Introducing lasso peptides as molecular scaffolds for drug design: Engineering of an integrin antagonist. Angew. Chem., Int. Ed. Engl. 50, 8714–8717. [DOI] [PubMed] [Google Scholar]
- Scott J. K.; Smith G. P. (1990) Searching for peptide ligands with an epitope library. Science 249, 386–390. [DOI] [PubMed] [Google Scholar]
- Scott C. P.; Abel-Santos E.; Wall M.; Wahnon D. C.; Benkovic S. J. (1999) Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 96, 13638–13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierbaum G.; Reis M.; Szekat C.; Sahl H.-G. (1994) Construction of an expression system for engineering of the lantibiotic Pep5. Appl. Environ. Microbiol. 60, 4332–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D.; Connor P. M. O.; Cotter P. D.; Hill C.; Ross R. P. (2008) The generation of nisin variants with enhanced activity against specific Gram-positive bacteria. Mol. Microbiol. 69, 218–230. [DOI] [PubMed] [Google Scholar]
- Cotter P. D.; Deegan L. H.; Lawton E. M.; Draper L. A.; O’Connor P. M.; Hill C.; Ross R. P. (2006) Complete alanine scanning of the two-component lantibiotic lacticin 3147: Generating a blueprint for rational drug design. Mol. Microbiol. 62, 735–747. [DOI] [PubMed] [Google Scholar]
- Liu W.; Hansen J. N. (1992) Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J. Biol. Chem. 267, 25078–25085. [PubMed] [Google Scholar]
- Bierbaum G.; Sahl H.-G. (2009) Lantibiotics: Mode of action, biosynthesis, and bioengineering. Curr. Pharm. Biotechnol. 10, 2–18. [DOI] [PubMed] [Google Scholar]
- Donia M. S.; Ravel J.; Schmidt E. W. (2008) A global assembly line for cyanobactins. Nat. Chem. Biol. 4, 341–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar D.; Pierce E.; McIntosh J. A.; Schmidt E. W. (2014) Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth. Biol. 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tianero M. D.; Donia M. S.; Young T. S.; Schultz P. G.; Schmidt E. W. (2012) Ribosomal route to small-molecule diversity. J. Am. Chem. Soc. 134, 418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. A.; Donia M. S.; Nair S. K.; Schmidt E. W. (2011) Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J. Am. Chem. Soc. 133, 13698–13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M. S.; Fricke W. F.; Ravel J.; Schmidt E. W. (2011) Variation in tropical reef symbiont metagenomes defined by secondary metabolism. PloS One 6, e17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M. S.; Ruffner D. E.; Cao S.; Schmidt E. W. (2011) Accessing the hidden majority of marine natural products through metagenomics. ChemBioChem 12, 1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. A.; Donia M. S.; Schmidt E. W. (2010) Insights into heterocyclization from two highly similar enzymes. J. Am. Chem. Soc. 132, 4089–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. A.; Robertson C. R.; Agarwal V.; Nair S. K.; Bulaj G. W.; Schmidt E. W. (2010) Circular logic: Nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J. Am. Chem. Soc. 132, 15499–15501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. A.; Schmidt E. W. (2010) Marine molecular machines: Heterocyclization in cyanobactin biosynthesis. ChemBioChem. 11, 1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; McIntosh J.; Hathaway B. J.; Schmidt E. W. (2009) Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J. Am. Chem. Soc. 131, 2122–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. W.; Nelson J. T.; Rasko D. A.; Sudek S.; Eisen J. A.; Haygood M. G.; Ravel J. (2005) Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U.S.A. 102, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A. R.; Coll J. C.; Bourne D. J.; MacLeod J. K.; Zabriskie T. M.; Ireland C. M.; Bowden B. F. (1996) Patellins 1–6 and trunkamide A: Novel cyclic hexa-, hepta-, and octa-peptides from colonial ascidians Lissoclinum sp. Aust. J. Chem. 49, 659–667. [Google Scholar]
- Wipf P.; Uto Y. (2000) Total synthesis and revision of stereochemistry of the marine metabolite trunkamide A. J. Org. Chem. 65, 1037–1049. [DOI] [PubMed] [Google Scholar]
- Palomero F. A., Naudi J. M. C., Lledo E. G., Manzanares I., Rodriguez I. (2002) Process for producing trunkamide A compounds. WO2002081506.
- Zabriskie T. M.; Foster M. P.; Stout T. J.; Clardy J.; Ireland C. M. (1990) Studies on the solution- and solid-state structure of patellin 2. J. Am. Chem. Soc. 112, 8080–8084. [Google Scholar]
- Smith G. P.; Petrenko V. A. (1997) Phage display. Chem. Rev. 97, 391–410. [DOI] [PubMed] [Google Scholar]
- Salvatella X.; Caba J. M.; Albericio F.; Giralt E. (2003) Solution structure of the antitumor candidate trunkamide A by 2D NMR and restrained simulated annealing methods. J. Org. Chem. 68, 211–215. [DOI] [PubMed] [Google Scholar]
- Bosley A. D.; Ostermeier M. (2005) Mathematical expressions useful in the construction, description, and evaluation of protein libraries. Biomol. Eng. 22, 57–61. [DOI] [PubMed] [Google Scholar]
- Schmidt E. W., Donia M. S. (2009) Chapter 23. Cyanobactin ribosomally synthesized peptides—A case of deep metagenome mining, Methods Enzymol. 458, 575–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M. S., and Schmidt E. W. (2010) Cyanobactins—Ubiquitous cyanobacterial ribosomal peptide metabolites. In Comprehensive Natural Products II (Liu H.-W., and Mander L., Eds.), pp 539–558, Elsevier, Oxford. [Google Scholar]
- Goto Y.; Ito Y.; Kato Y.; Tsunoda S.; Suga H. (2014) One-pot synthesis of azoline-containing peptides in a cell-free translation system integrated with a posttranslational cyclodehydratase. Chem. Biol. 21, 766–774. [DOI] [PubMed] [Google Scholar]
- McIntosh J. A.; Lin Z.; Tianero M. D.; Schmidt E. W. (2013) Aestuaramides, a natural library of cyanobactin cyclic peptides resulting from isoprene-derived Claisen rearrangements. ACS Chem. Biol. 8, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J.; Bent A. F.; Zollman D.; Smith K.; Houssen W. E.; Zhu X.; Lebl T.; Scharff R.; Shirran S.; Botting C. H.; Jaspars M.; Schwarz-Linek U.; Naismith J. H. (2013) The cyanobactin heterocyclase enzyme: A processive adenylase that operates with a defined order of reaction. Angew. Chem. Int. Ed. 52, 13991–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V.; Pierce E.; McIntosh J.; Schmidt E. W.; Nair S. K. (2012) Structures of cyanobactin maturation enzymes define a family of transamidating proteases. Chem. Biol. 19, 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssen W. E.; Koehnke J.; Zollman D.; Vencome J.; Raab A.; Smith M. C. M.; Naismith J. H.; Jaspars M. (2012) The discovery of new cyanobactins from Cyanothece PCC 7425 defines a new signature for processing of patellamides. ChemBioChem. 13, 2683–2689. [DOI] [PubMed] [Google Scholar]
- Koehnke J.; Bent A.; Houssen W. E.; Zollman D.; Morawitz F.; Shirran S.; Vendome J.; Nneoyiegbe A. F.; Trembleau L.; Botting C. H.; Smith M. C. M.; Jaspars M.; Naismith J. H. (2012) The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Chem. Biol. 19, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A. F.; Koehnke J.; Houssen W. E.; Smith M. C. M.; Jaspars M.; Naismith J. H. (2013) Structure of PatF from Prochloron didemni. Acta Crystallogr. Sect. F 69, 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.