Summary
Transcription is a highly dynamic process. Consequently, we have developed native elongating transcript sequencing technology for mammalian chromatin (mNET-seq), which generates single-nucleotide resolution, nascent transcription profiles. Nascent RNA was detected in the active site of RNA polymerase II (Pol II) along with associated RNA processing intermediates. In particular, we detected 5′splice site cleavage by the spliceosome, showing that cleaved upstream exon transcripts are associated with Pol II CTD phosphorylated on the serine 5 position (S5P), which is accumulated over downstream exons. Also, depletion of termination factors substantially reduces Pol II pausing at gene ends, leading to termination defects. Notably, termination factors play an additional promoter role by restricting non-productive RNA synthesis in a Pol II CTD S2P-specific manner. Our results suggest that CTD phosphorylation patterns established for yeast transcription are significantly different in mammals. Taken together, mNET-seq provides dynamic and detailed snapshots of the complex events underlying transcription in mammals.
Graphical Abstract
Highlights
-
•
Development of mammalian native elongating transcript sequencing (mNET-seq)
-
•
Dynamic Pol II CTD phosphorylation during transcription cycle
-
•
Co-transcriptional splicing and microprocessing detected by mNET-seq
-
•
Termination factors are associated with Pol II pausing at both TES and TSS
Sequencing nascent transcripts from the active site of mammalian RNA polymerase II by a technique called mNET-seq unravels dynamic insights into the transcription cycle, including co-transcriptional splicing and RNA microprocessing.
Introduction
Virtually all transcripts synthesized by RNA polymerase II (Pol II) from protein-coding genes are co-transcriptionally processed to generate the final functional mRNA (Moore and Proudfoot, 2009). First, a Cap structure (me7Gppp) is added to the transcript 5′ end soon after transcriptional initiation, which ultimately earmarks transcripts for efficient cytoplasmic translation. Then as the polymerase proceeds to elongate through the gene body (GB), intronic RNA, which often constitutes the majority of the primary transcript in mammalian genes, is removed by a splicing mechanism involving the stepwise assembly of a complex set of small RNA (snRNA) and associated proteins that together make up the spliceosome (Wahl et al., 2009). In outline, U1snRNA-protein complex (U1snRNP) identifies the intron 5′ splice site (SS) as soon as it is transcribed by Pol II, and then on reaching the 3′ end of the intron multiple snRNPs, U2, U4, U5, and U6 recognize the 3′SS and proximal intronic branch point on the nascent transcript. Following reorganization of snRNP/intron interactions, the branch point A nucleotide carries out a 2′OH nucleophilic attack on the 5′SS, resulting in cleavage of the intron from the upstream exon. The newly formed upstream exon 3′OH then undergoes a second nucleophilic attack on the 3′SS, resulting in precise fusion of adjacent exons and release of the intron. Prior to intron splicing, hairpin structures embedded within some introns are excised by the double-strand RNA-specific microprocessor complex. This comprises an RNA-binding protein DGCR8 together with the endonuclease Drosha, which facilitate release of pre-microRNA (miRNA) hairpins from the nascent transcript. These pre-miRNA go on to form cytoplasmic miRNA, which are critical for the translational regulation of many mRNA (Krol et al., 2010). Finally at gene 3′ ends, a further RNA-processing reaction involving cleavage of the nascent transcript at a specific poly(A) signal (PAS) occurs. This RNA cleavage reaction is mediated by an endonuclease (CPSF73) that is part of a large multimeric cleavage and polyadenylation complex. A poly(A) tail is then added to the mRNA 3′ end, promoting rapid release of mRNA from the chromatin template (Proudfoot, 2011). Although these individual RNA-processing mechanisms are well characterized, their interconnections with transcription remain enigmatic. We describe in this study a method to investigate these interconnections, genome wide.
The above outlined co-transcriptional pre-mRNA-processing reactions are precisely coordinated with the Pol II transcription cycle that proceeds from initiation at the transcription start site (TSS), leading on to elongation through the GB and ending with release of the mRNA at the PAS, also called the transcription end site (TES). Finally, termination occurs whereby Pol II separates from the DNA template. Both the Pol II transcription cycle and coupled pre-mRNA-processing reactions are orchestrated by a unique structural feature of Pol II. This comprises an extended C-terminal domain (CTD) of the large subunit (Rpb1) that has a heptad structure YSPTSPS repeated 52 times with some variation in mammals and 26 times in budding yeast. This CTD is separate from the main globular enzyme, being positioned close to the RNA exit channel. It is relatively unstructured (Meinhart and Cramer, 2004) and subject to extensive post-translational modification, especially phosphorylation of S2 and S5 but also Y1, T4, and S7 (Heidemann et al., 2013; Hsin and Manley, 2012). This combined but differential CTD phosphorylation is often considered to be a molecular code that acts to orchestrate transcription and coupled pre-mRNA processing. Especially in simpler eukaryotes, such as budding yeast, CTD S5P is correlated with TSS-associated events, whereas S2P is thought to correlate with TES events (Buratowski, 2009). However in the larger and more complex genes of mammals, this CTD code may be less clear-cut and vary between different gene classes.
To gain a more complete understanding of the Pol II transcription cycle and how this is coordinated with co-transcriptional pre-mRNA processing, genome-wide analysis of nascent RNA has been undertaken. For example, global nuclear run on-sequencing (GRO-seq) and precision nuclear run on-sequencing (PRO-seq) with modified nucleotides (Core et al., 2008; Kwak et al., 2013) provide a way to study Pol II profiles associated with nascent transcription. Similarly, 5′ capped nascent RNA isolated from insoluble chromatin can be sequenced at high resolution (3′NT-seq) (Weber et al., 2014). These approaches generated detailed maps of Pol II nascent transcription in mammals and flies, which accumulates at promoters, providing a regulated transition from initiation into productive elongation (Adelman and Lis, 2012; Core and Lis, 2008; Gilchrist et al., 2010; Rahl et al., 2010). Precise maps of PRO-seq reads identified two different types of Pol II pausing at the TSS, referred to as proximal and distal TSS pausing. PRO-seq additionally showed Pol II accumulation near 3′SS, possibly important for the selection of active exons (Kwak et al., 2013). GRO-seq has also shown a correlation between Pol II density and nucleosome occupancy as observed at the TES of many genes, suggesting a connection with transcription termination (Grosso et al., 2012). A significant limitation to these above nascent RNA-mapping techniques is that the relationship between Pol II CTD modification and nascent RNA was not established.
Precise maps of Pol II nascent RNA have also been generated by native elongating transcript sequencing (NET-seq) in yeast (Churchman and Weissman, 2011). Here, endogenous Pol II is flag tagged by genomic integration allowing immunoprecipitation (IP) of Pol II nascent RNA complexes. However, again connections between Pol II CTD modifications and nascent RNA could not be determined. In contrast, we establish mammalian NET-seq technology (mNET-seq) using a selection of CTD phosphorylation-specific Pol II antibodies to IP Pol II-associated transcripts. In detail, we have compared low or unphosphorylated (unph), S2P, S5P, and total (unph+ph) CTD mNET-seq profiles and show that unph CTD Pol II-nascent RNA are accumulated over the TSS, whereas S2P Pol II-nascent RNA are spread throughout the GB and TES. Remarkably S5P profiles precisely correlate with active splicing on protein-coding genes. An important feature of our analysis is that we are able to directly detect the initial 5′SS cleavage step in intron splicing and can also observe active Drosha cleavage of pre-miRNA hairpin structures present in gene introns. In effect, our extensive mNET-seq data sets provide a “treasure trove” of detailed information on nascent transcription and co-transcriptional RNA processing in mammalian cells.
Results
mNET-Seq Strategy
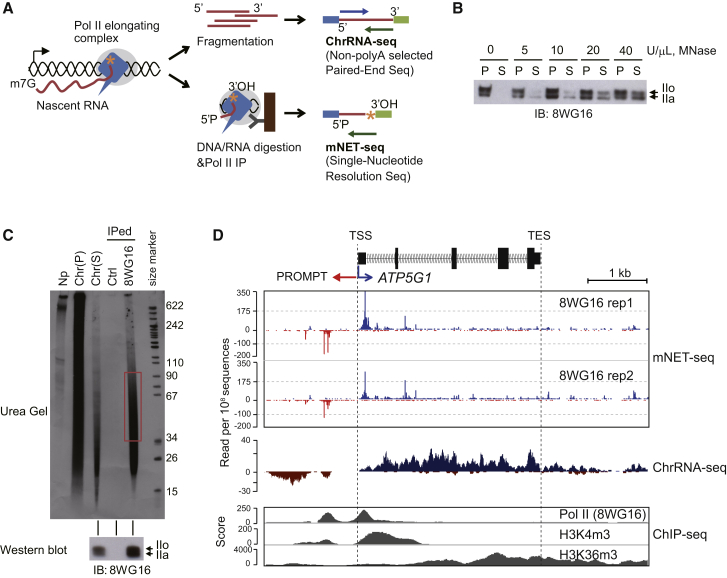
To detect unstable nascent RNA across the human genome, we isolated a nuclear chromatin fraction from HeLa cells enriched in transcriptionally active Pol II (Pol IIo) and associated nascent RNA (Figure 1A) (Nojima et al., 2013). This chromatin-bound RNA was fragmented to 150–200 nt and ligated to adaptors for strand-specific paired-end deep sequencing (Figure S1A, top; Experimental Procedures). ChrRNA-seq detects unstable RNA, such as promoter upstream transcripts (PROMPTs), introns, and read-through transcripts (Figures 1D and S1B). For mNET-seq, chromatin was digested with micrococcal nuclease (MNase) to release Pol II from insoluble chromatin. Note that accessible RNA will also be digested (Figure S1A, bottom; Experimental Procedures). Western blot analysis using Pol II 8WG16 antibody confirmed that both phosphorylated (Pol IIo) and unphosphorylated (Pol IIa) forms were released in a MNase dose-dependent manner (Figure 1B). Nascent RNA distribution was also tested after cell fractionation and MNase digestion, by using nuclear run on (NRO) nuclei, labeled with [α-32P] UTP (Figure 1C). The nucleoplasmic (Np) fraction contained long 32P-RNA (over 600 nt). However, after MNase digestion (40 U/μl), the residual chromatin pellet (P) contained RNA of 10–600 nt, whereas the chromatin supernatant (S) had shorter RNA of 10–200 nt. This supernatant fraction was then IPed using Pol II 8WG16 antibody, which efficiently precipitated this shorter RNA. Although the predominant size of the IPed RNA was 20–45 nt, we selected a longer RNA fraction (35–100 nt) to obtain unique alignment with the human genome after deep sequencing. In this method, the Pol II complex will protect nascent RNA from MNase digestion. The hydroxylated 3′ end (3′OH) of the nascent RNA corresponds to the terminal nucleotide synthesized by Pol II (Figure 1A, asterisk). The 5′ end of the cleaved Pol II-associated RNA is also hydroxylated after MNase digestion. To achieve strand-specific RNA sequencing, we carried out a kinase reaction on the IP beads to phosphorylate all nascent RNA 5′ ends but leave the Pol II-embedded 3′OH intact (Figure S1A). Illumina adapters were then ligated onto gel-purified RNA, and Illumina high-throughput paired-end sequencing was carried out and generated ∼108 reads for each mNET-seq sample. For library construction, we omitted the NRO step because the NRO reaction may perturb the native Pol II distribution. The above Pol II IP from MNase-treated chromatin coupled with isolation and sequencing of the associated RNA constitutes a refined mammalian NET-seq protocol.
Figure 1.
mNET-Seq Methodology
(A) ChrRNA-seq and mNET-seq strategies. Pol II (blue) elongating complex (gray circle) and associated nascent RNA (red line) in chromatin. Orange asterisk depicts the 3'OH of nascent RNA. For ChrRNA-seq (top), fragmented nascent RNA is subjected to directional paired-end deep sequencing. For mNET-seq (bottom), DNA and RNA are digested with MNase and Pol II-nascent RNA complex precipitated with Pol II antibody. Isolated RNA is deep sequenced, and the 3′ end nucleotide uniquely mapped on the human genome.
(B) Pol II release from insoluble chromatin DNA. Chromatin DNA was digested with increasing amounts of MNase. Western blot used 8WG16 Pol II antibody. P; pellet, S; supernatant. IIo and IIa indicate phosphorylated and unphosphorylated Pol II.
(C) Nascent RNA distribution in mNET-seq method. Nascent RNA was 32P-labeled by NRO reaction. Fractionated nascent RNA are nucleoplasm (Np), chromatin pellet (Chr (P)) and supernatant (Chr (S)). IP was with 8WG16 Pol II antibody. 35–100 nt RNA purified from gel (red box). IPed Pol II was detected by western blot (bottom).
(D) ATP5G1 mNET-seq. Two biological replicates of mNET-seq/unph using 8WG16 Pol II antibody. ChrRNA-seq shown as mNET-seq input. ChIP-seq (Pol II [8WG16], H3K4m3, and H3K36m3) data are from ENCODE project data sets (Consortium et al., 2012).
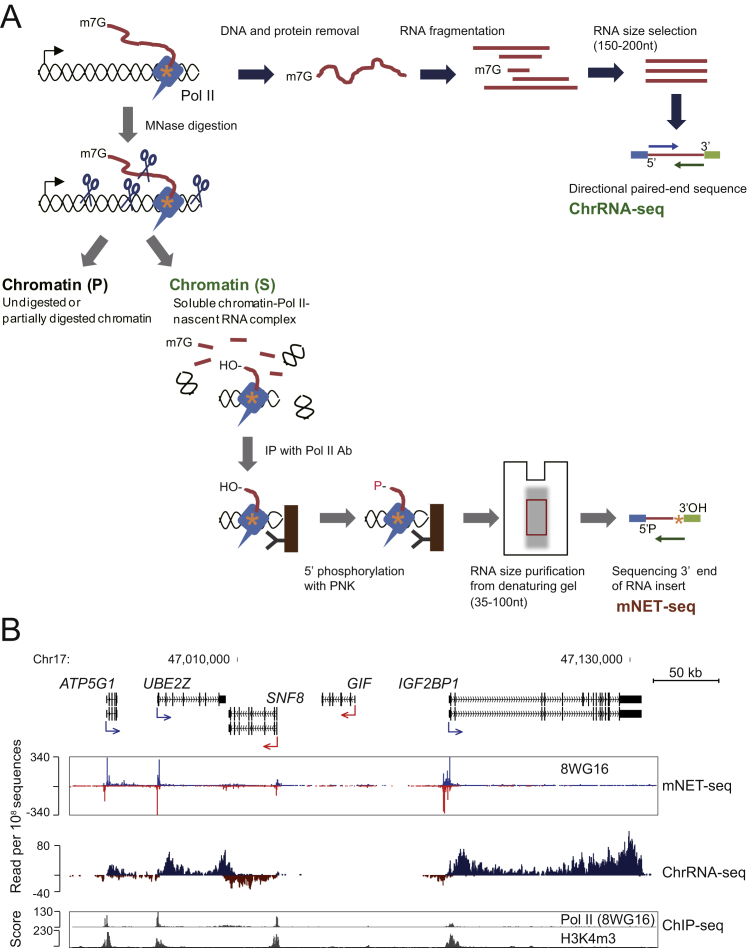
Figure S1.
Detailed ChrRNA-Seq and mNET-Seq Methods, Related to Figure 1
(A) (Above) ChrRNA-seq method. Pol II and RNA synthesis site are diagrammed as tailed blue box and orange asterisk, respectively. Chromatin-bound RNA (red line) is purified from isolated chromatin fraction by DNase and proteinase K treatments. RNA is fragmented to 150–200 nt and adapters ligated on both ends for paired-end 51bases directional deep sequencing (blue and green arrows).
(Below) mNET-seq method. Chromatin DNA and chromatin-bound RNA are digested with MNase I (light blue scissors). To separate insoluble pellet (P) and soluble chromatin supernatant (S), digested chromatin is centrifuged. Soluble Pol II-nascent RNA complex is immunoprecipitated (IP) with Pol II antibody. 5′ hydroxyl (OH) is then phosphorylated with PNK on beads and phenol extraction performed to remove DNA and proteins. IPed RNA is purified from denaturing gel (size range 35–100 nt). RNA adapters are added to both ends strand-specifically and deep sequencing is conducted from reverse sequence primer (green arrow) to read 3′ end of insert (orange asterisk).
(B) Example of mNET-seq and ChrRNA-seq data view on human chromosome 17. mNET-seq and ChrRNA-seq reads on the plus strand, blue and dark blue, respectively; mNET-seq and ChrRNA-seq reads on the minus strand, red and dark red, respectively. ChIP-seq (Pol II [8WG16] and H3K4m3) data are from the ENCODE project data sets (Consortium et al., 2012).
Finally, libraries were prepared from two biological replicates of HeLa native chromatin after Pol II 8WG16 IP. Deep sequencing was conducted using a reverse sequence primer to read the 3′ ends of the RNA insert, which corresponds to the RNA synthesis site in the Pol II active site (Figure 1A). mNET-seq data aligned to the human genome (hg19) was compared to 8WG16 chromatin IP (ChIP-seq) and ChrRNA-seq as shown for ATP5G1, a typical example of an actively expressed gene in HeLa cells (Figure 1D). A lower-resolution cluster of genes expressed at varying levels is also presented (Figure S1B). Note that as mNET-seq identifies the 3′ end of transcript within the Pol II active site, TSS-associated reads will only be detected 30 nt or beyond the exact TSS. Modifications of histone H3, H3K4m3 and H3K36m3, reflect active promoters and gene bodies, respectively. Strand-specific transcription activity was revealed by ChrRNA-seq. As expected, both replicates of mNET-seq/8WG16 (unph) display strong peaks at the active TSS, consistent with the previously described ChIP-seq/unph profiles. We therefore predict that this TSS-accumulated mNET-seq signal reflects Pol II pausing. Additionally, mNET-seq data revealed both sense and antisense transcription on active genes, as previously shown by GRO-seq and PRO-seq (Core et al., 2008; Kwak et al., 2013).
Pol II CTD Phosphorylation-Specific Nascent RNA Profiles at TSS and TES
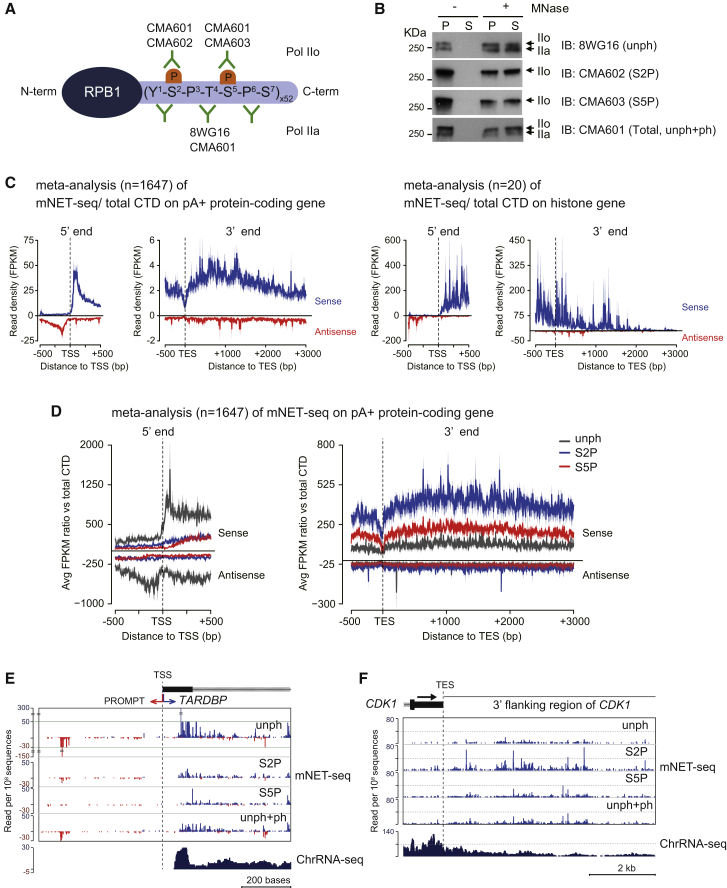
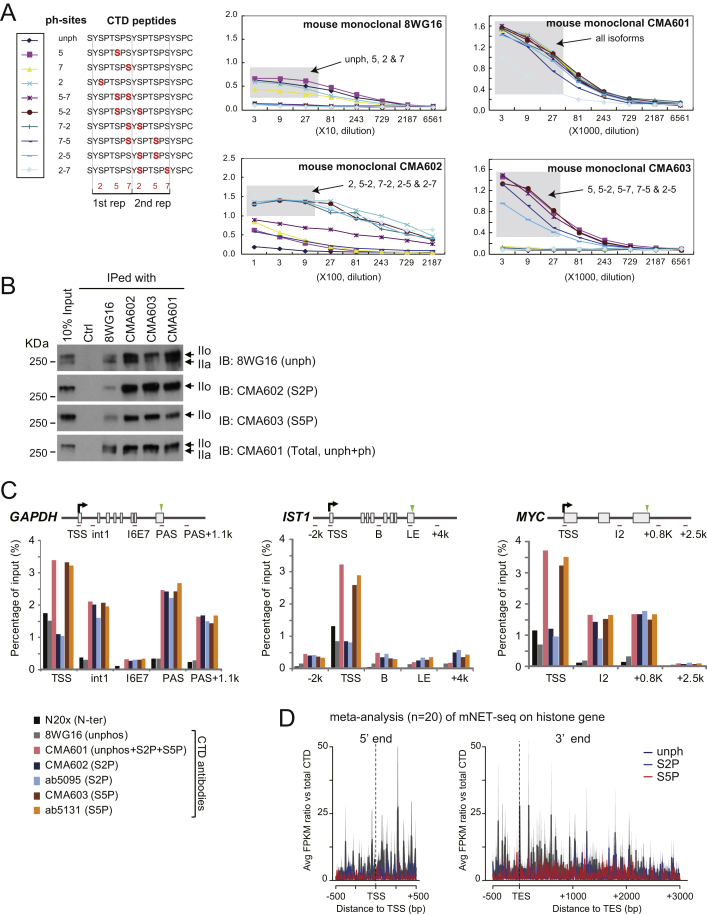
A major benefit of our mNET-seq procedure is that it allows the use of different Pol II antibodies to precipitate modified Pol II-associated nascent transcripts. We elected to employ newly described specific monoclonal antibodies to detect CTD phosphorylation-dependent nascent RNA profiles for S2P, S5P, and all CTD isoforms (Figure 2A) (Stasevich et al., 2014). We carried out further tests to confirm the specificity of these antibodies versus 8WG16. First we performed ELISA assays (Figure S2A) with synthetic peptides of 15 amino acids, containing two adjacent heptad repeats, either singly or doubly phosphorylated on S2P, S5P, and S7P. As expected, 8WG16 bound with relative specificity to unphosphorylated or singly phosphorylated CTD peptides. CMA601 bound all CTD peptides with or without serine phosphorylation, whereas CMA602 and CMA603 bound CTD peptides containing S2P and S5P, respectively. We also performed IP Pol II western blots (Figure S2B) with these four antibodies under mNET-seq conditions and confirmed their specificity and IP efficiency. We finally performed Pol II ChIP analysis on three specific genes comparing our monoclonal antibodies to commercial polyclonal antibodies (ab5095 [S2P] and ab5131 [S5P], respectively) that are widely used for ChIP-seq assays (Pérez-Lluch et al., 2011) (Figure S2C). Notably, matching ChIP profiles were observed for the different S2P- and S5P-specific antibodies. A potential concern with our mNET-seq protocol was that, as we only partially solubilize the chromatin pellet by MNase treatment, there may be selective release of different Pol II modifications. However, the chromatin pellet and supernatant following MNase treatment gave very similar patterns of Pol IIo and Pol IIa with all four antibodies arguing against selective release of differentially modified Pol II (Figure 2B).
Figure 2.
mNET-Seq with Different Phospho-CTD Modifications
(A) Diagram showing different Pol II antibody epitopes on CTD (Stasevich et al., 2014).
(B) Specificity of Pol II phosphorylation released from chromatin following MNase treatment with indicated Pol II antibodies.
(C) Meta-analyses of mNET-seq/unph+ph on TSS and TES of pA+ protein-coding genes (left) and histone genes (right). Read density (FPKM) of mNET-seq data were plotted around TSS (±0.5 kb) and TES (−0.5 k∼+3 kb). Data on pA+ and histone genes are represented as mean ± SEM. mNET-seq sense strand, blue; antisense strand, red.
(D) Meta-analyses of mNET-seq on TSS and TES of pA+ protein-coding genes. Ratio of read density (FPKM) of indicated mNET-seq data to mNET-seq/unph+ph data was plotted around TSS (±0.5 kb) and TES (−0.5 k∼+3 kb). unph, dark gray; S2P, blue; S5P, red. Line and shading represent mean ± SEM for each bin.
(E and F) mNET-seq profiles over TSS of TARDBP (E) and TES of CDK1 (F). Read density, read per 108 sequences.
Figure S2.
Specificity of Pol II Antibodies, Related to Figure 2
(A) ELISA assay (right) was performed with indicated CTD heptapeptides (left) and Pol II antibodies (right).
(B) Pol II precipitated from cell extracts with indicated antibodies detected by western blot using each antibody.
(C) Pol II ChIP was conducted with indicated Pol II antibodies on GAPDH, IST1, and MYC. Positions of primer sets and PAS are shown by red bars and green triangles, respectively. TSS denoted by black arrow.
(D) meta-analyses of mNET-seq on 5′ end (−0.5 k∼+0.5 kb from TSS) and 3′end (−0.5 k∼+3kb from TES) of histone gene. 8WG16 (unph), dark gray; CMA602 (S2P), blue; CMA603 (S5P), red; n = 20. Line and shading represent mean ± SEM for each bin.
Based on previously published RNA-seq data (Lacoste et al., 2014), we found 11,560 (45%) RefSeq genes actively transcribed in HeLa cells. However, to avoid over-represention of ncRNA (such as rRNA, tRNA, snoRNA, and snRNA) in the mNET-seq meta-analysis, we excluded genes overlapping these sequences. We also excluded overlapping transcription units as these might bias average profiles (see Extended Experimental Procedures). We initially looked at meta-profiles over TSS and TES regions for all Pol II isoforms (unph+ph antibody). As expected, bidirectional TSS mNET-seq peaks were detected and a wider, mainly sense peak beyond the TES. In contrast, the histone genes gave a flatter mNET-seq profile across these short poly(A) minus genes and diminished TSS antisense reads (Figure 2C). This clearly shows the specificity of our mNET-seq profiles. We next analyzed meta-profiles using the CTD phospho-specific Pol II antibodies (Figure 2D). To allow cross-comparison between the different antibodies, the data are presented as a ratio with mNET-seq reads obtained for total Pol II (unph+ph). Remarkably, only mNET-seq/unph gave a bidirectional TSS profile, whereas S2P and S5P show a gradual increase from low TSS signals to higher signals in the GB. The TES meta-profiles revealed the expected dominance of S2P. Single-gene TSS and TES mNET-seq profiles (TARDBP and CDK1, respectively) were consistent with the meta-profiles. The marked differences in mNET-seq profiles observed for specific CTD phosphorylation were not seen for histone genes, which showed little difference other than higher unph reads across the genes (Figure S2D). Overall, mNET-seq profiles reveal remarkable CTD phosphorylation specificity for poly(A)+ protein-coding genes.
Extended Experimental Procedures.
Antibodies
Pol II antibodies CMA601, CMA602, and CMA603 were generated by HK (Stasevich et al., 2014). 8WG16, Aly, and PTBP1 antibodies were purchased from Abcam. CPSF73, CstF-64, and CstF-64 tau antibodies were purchased from Bethyl laboratories. Tubulin antibody was purchased from Sigma. Xrn2 antibody was provided by Dr. N. Gromak.
siRNA Transfection
SMARTpool siRNA against human PTBP1, CPSF73 (CPSF3), and CstF64 (CSTF2) were purchased from Thermo scientific. ON-TARGET plus siRNA against Xrn2 was made by Thermo Scientific as following sequences.
Sense: AAGAGUACAGAUGAUCAUGUU;
Antisense: 5′-P CAUGAUCAUCUGUACUCUUUU.
Silencer select siRNA against CstF64 tau (CSTF2T) was designed by Life technologies.
Sense: CCAUUAUUGACUCACCCUAtt;
Antisense: UAGGGUGAGUCAAUAAUGGgc.
These siRNA (final concentration 30 nM) were transfected into HeLa cells using Lipofectamine RNAiMAX reagent (Life technologies) according to the manual and incubated for 60 to 72 hr.
RT-PCR Analysis
RNAs were isolated from HeLa cells that were transfected with siRNA against PTBP1 or treated with splicing inhibitor Pla-B. For reverse transcription, 1 μg of nuclear RNA was incubated with oligo(dT)20 and Superscript III reverse transcriptase (Life Technologies). PCR was performed using GO taq polymerase (Promega) and following primer set.
BRD2_ex4_FW: 5′-CAAAATTATAAAACAGCCTATGGACATG-3′
BRD2_ex5_RV: 5′-TTTTCCAGCGTTTGTGCCATTAGGA-3′
BZW1_ex3_FW: 5′-TACCGTCGATATGCAGAAACA-3′
BZW1_ex4_RV: 5′-GAGCAAATGCTTGCATGGTCT-3′
PKMex8_Fw: 5′-GATGGAGCCGACTGCATCATG-3′,
PKMex11_Rv: 5′-ATTCCGGGTCACAGCAATGAT-3′
For PKM, PCR products were digested by either NcoI (NEB) or PstI (NEB) for 6 hr. The PCR products were analyzed by 2% agarose gel electrophoresis, followed by ethidium bromide staining.
Purification of ChrRNA and Nucleoplasm RNA and RNA Library Preparation
Chromatin RNA fraction was prepared from ∼80% confluent HeLa cells in 100 mm dishes. Approximately 7 × 106 cells were washed with ice-cold PBS twice. The cells were lysed with ice-cold 4 ml of HLB/NP40 buffer (10 mM Tris-Hcl, pH 7.5, 10 mM NaCl, 0.5% NP40, and 2.5 mM MgCl2) and incubated on ice for 5 min. After the incubation, 1 ml of ice-cold HLB/NP40/Sucrose buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 0.5% NP40, 2.5 mM MgCl2, and 10% Sucrose) was under-laid and then the nuclei were collected under 1,400 rpm centrifuge at 4°C for 5 min. Isolated nuclei were resuspended in 125 μl of NUN1 solution (20 mM Tri-HCl, pH 8.0, 75mM NaCl, 0.5 mM EDTA, 50% Glycerol and proteinase inhibitor 1xComplete (Roche) followed by 1.2 ml NUN2 buffer (20 mM HEPES-KOH pH 7.6, 7.5 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 1 M Urea, 1% NP40, proteinase inhibitor 1xComplete and phosphatase inhibitor 1xPhosStop [Roche]). Fifteen minute incubation was carried out on ice with mixing by max speed vortex for 5 s every ∼4 min and then chromatin pellets were precipitated under 13,000 rpm centrifuge at 4°C for 10 min. The supernatant was collected for nucleoplasm RNA. Chromatin pellet was resuspended in 200 μl HSB (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, and 10 mM MgCl2) with 0.25 U/μl TURBO DNase (Life technologies) at 37°C for 10 min and then treated with Proteinase K for 10 min. Chromatin and nucleoplasm RNAs were extracted by Trizol reagent (Life technologies). The RNA extraction steps were repeated three times. Prior to RNA library preparations, rRNAs were depleted using Ribo-Zero rRNA removal kits (Epicenter) from 5 μg of Chromatin RNA and Nucleoplasm RNA. RNA was also fragmented 150–200 nt by heat treatment (94°C) for 15 min in 1xNEB first-strand synthesis buffer. Two hundred nanograms of RNA was used for RNA library preparations. These were carried out according to NEBNext Ultra Directional RNA Library Prep kit for Illumina (NEB). Deep sequencing using Hiseq2000 and Hiseq2500 were performed by the Wellcome Trust Centre for Human Genetics (WTCHG) Oxford UK.
mNET-Seq Method and RNA Library Preparation
Approximately 1.6 × 108 cells were used to generate nuclear and chromatin fractions. Isolated chromatin was washed in 100 μl of 1× Micrococcal Nuclease (MNase) buffer (NEB) and then incubated with MNase (40 u/μl) on Thermomixer (eppendorf, 1,400 rpm) at 37°C for 90 s. In order to inactivate MNase, EGTA (25 mM) was added immediately after the reaction and soluble-digested chromatin was collected by 13,000 rpm centrifuge for 5 min. The supernatant was diluted with 9 ml of NET-2 buffer and Pol II antibody-conjugated beads were added. 40 μg of Pol II antibody was used for each mNET-seq experiment. Immunoprecipitation was performed at 4°C for 1 hr. The beads were washed with 1 ml of NET-2 buffer six times and with 500 μl of 1×PNKT (1×PNK buffer and 0.1% Triton X-100) buffer once in the cold room. The washed beads were incubated in 100 μl of PNK reaction mix (1xPNKT, 1 mM ATP and 0.05 U/ml T4 PNK 3′phosphatase minus (NEB) on Thermomixer (1,400 rpm) at 37°C for 6 min. After, the reaction beads were washed with 1 ml of NET-2 buffer once and RNA was extracted with Trizol reagent. RNA was resolved on 8% denaturing acrylamide 7 M urea gels for size purification. 35–100 nt fragments were eluted from the gel using RNA elution buffer (1 M NaOAc and 1 mM EDTA) and RNA was precipitated in 75% Ethanol. RNA libraries were prepared according to the manual of Truseq small RNA library prep kit (Illumina). Deep sequencing was conducted by WTCHG in Oxford.
Analyses of In Vivo CLIP Assay for TSS
CLIP-sequencing data sets (Martin et al., 2012) were downloaded for the following transcription factors: CPSF-73, CstF-64, CstF-64tau, CPSF-160, CPSF-30 and CF-Im25. Normalized read counts were calculated for sense and antisense strands relative to the direction of gene transcription for a region of 3 kb upstream and downstream of annotated Refseq TSS and plotted for 10 bp bins (Table S1).
Data Pre-Processing
mNET-seq data adaptors were trimmed using Cutadapt (v1.1) (Martin, 2011), discarding reads with less than 10 bases. Then a Perl script was used to remove the reads left unpaired. The remaining reads were then aligned to the reference human genome (hg19) using TopHat (v2.0.9) (Kim et al., 2013) with a minimum anchor length of 5 bases, and only allowing for one alignment to the reference. It was then necessary to determine the last nucleotide incorporated by the polymerase and its directionality. This nucleotide was defined as the 5′ end of read two of the pair, with the directionality indicated by read one. Knowing this, the properly aligned read pairs were trimmed to solely keep the 5′ nucleotide of read two. This was done using SAMtools (Li et al., 2009) and a python script. SAMtools was also used to separate the reads by strand for further analysis.
ChrRNA-seq and nucleoplasm RNA-seq data were aligned using the same version of TopHat but allowing for the read pairs to be separated by 3 kb. For the metagene representation, SAMtools was used to separate the reads by strands.
ChIP-seq data for unphosphorylated Pol II, H3K4m3, and H3K36m3 (GEO accession numbers GSM935395, GSM945201, and GSM733711, respectively) were generated as part of the ENCODE Project (Consortium et al., 2012).
Determination of Expressed Genes
To determine genes expressed in HeLa S3 cells, strand-specific RNA-seq data from a previously published study (Lacoste et al., 2014) was used (GEO accession number GSM1155630). The data were aligned with TopHat and then Cufflinks (v2.1.1) (Trapnell et al., 2010) was used to acquire a FPKM value for each gene. These values were then converted to log2 and their distribution was plotted. The cut off value chosen to determine the expressed genes was the local minimum of the log2 (FPKM) distribution between the primary peak of high-expression genes and the long left shoulder of low-expression transcripts as previously reported (Hart et al., 2013). This defined 11,560 expressed genes, of which 10,473 were protein coding. From these genes a further selection of those where the GB and the adjacent regions do not intersect other genes was made. For most profiles the adjacent regions extended to TSS-1000 bp and TES+3 kb, which resulted in 1,647 genes used. For the profiles depicting the effect of CPA and termination factors at the TES (Figure 6), only the adjacent region was considered that extended to TES+7 kb in order to allow inclusion of more genes, while at the same time capturing more distal effects. In this case 1,586 genes were used. For the chosen replication-dependent histone genes adjacent regions (TSS-250 bp, TES+1 kb) also do not overlap other genes. Twenty in all were selected.
Meta-Profiles
Meta-profiles represent average Pol II or RNA abundance distribution across expressed genes. To generate these, genes were aligned by their annotated TSS and TES. The 5′ end, showing a span of 1 kb up and downstream of the TSS, and the 3′ end, showing the interval from TES-500 bp to TES+3 kb or TES+7 kb, were divided in 5 bp windows. Reads in each window were then counted and normalized for region and library size. For untreated mNET-seq data targeting different CTD modifications (unph, S2P, and S5P, see Figure 2A), a ratio against the corresponding total Pol II (unph+ph) mNET-seq value was calculated. Lastly, the obtained values were averaged for all genes under analysis to obtain the meta-profiles. Replication-dependent histone meta-profiles were generated using the same method, but the window around the TSS extended from TSS-250 bp to TSS+250 bp, and around the TES from TES-250 bp to TES+1 kb.
Splice junction average profiles, which show a region of 50 bases upstream and downstream of the splice sites, were made by first aligning all 5′ or 3′ splice junctions considered. Then for each base of each region, reads were counted and normalized, and then the average value for each position was plotted.
The individual profiles were plotted in single-base windows using a scale of reads per 108 sequences.
Determining Spliced Exons from mNET-Seq Data
Included exons co-transcriptionally spliced were identified by aligning gapped reads from Ser5 phosphorylation-targeting mNET-seq to exon pairs. Junctions were considered spliced if they had at least 10 reads supporting them and at least 3 reads per replicate when available.
Determination of Included and Excluded Exons
To determine whether alternative exons were included or excluded in the transcripts produced, previously described RNA-seq data used for determining expressed genes was analyzed with MISO (Katz et al., 2010). These results were compared to RefSeq exon reference data. Exons were then divided according to their Ψ value. Only exons with more than 0.9 or less than 0.1 were considered included or excluded, respectively.
Escaping and Read-Through Index
Escaping Index (EI) is defined as the proportion of Pol II from the TSS that proceeds to the elongation phase of transcription. It was calculated as follows:
where GB is the reads per kilobase per million reads (FPKM) of mNET-seq sense reads in the interval [TSS+500, TES], TSS is the FPKM of sense reads in [TSS-50, TSS+250]. The constant c was used to log the zeros in the data. The first 500 bases of each gene are excluded from the definition of the GB to prevent TSS polymerase accumulation from interfering with the counts for the GB. The Read-Through Index (RTI) was calculated using the same approach, but instead of considering the TSS interval, the FPKM of sense reads for [TES, TES+2000] was used to access the proportion of Pol II accumulating after the annotated transcript end. Normalized GB counts use the same formula but without dividing the FPKM from the GB region by any of the others.
Significance of the differences between control and knock-down for each index was calculated using a two-sided Mann-Whitney test. The p values were then adjusted using the Holm method.
PCR Primer Sequences for ChIP Assay
| GAPDH_TSS_F | 5′-CGGCTACTAGCGGTTTTACG-3′ |
| GAPDH_TSS_R | 5′-GCTGCGGGCTCAATTTATAG-3′ |
| GAPDH_int1_F | 5′-CCCCTTCATACCCTCACGTA-3′ |
| GAPDH_int1_R | 5′-GACAAGCTTCCCGTTCTCAG-3′ |
| GAPDH_I6E7_F | 5′-ACCCAGAAGACTGTGGATGG-3′ |
| GAPDH_I6E7_R | 5′-TTCAGCTCAGGGATGACCTT-3′ |
| GAPDH_PAS_F | 5′-CTGAATCTCCCCTCCTCACA-3′ |
| GAPDH_PAS_R | 5′-TGCCCCAGACCCTAGAATAA-3′ |
| GAPDH_PAS+1.1k_F | 5′-TCCAGCCTAGGCAACAGAGT-3′ |
| GAPDH_PAS+1.1k_R | 5′-TGTGCACTTTGGTGTCACTG-3′ |
| IST1_-2k_F | 5′-TGTTAGCCAGGGTGGTCTTC-3′ |
| IST1_-2k_R | 5′-GGTCAGGAGTTGGAGAGCAG-3′ |
| IST1_TSS_F | 5′-AACCCTGAAGTCGGTGTCTG-3′ |
| IST1_TSS_R | 5′-CTCCGAAGTCGTTTGAATCC-3′ |
| IST1_B_F | 5′-CACCATGCCCAGCTAATTTT-3′ |
| IST1_B_R | 5′-ACCCTCAGGTGGTTCTGATG-3′ |
| IST1_LE_F | 5′-TGAAGGCCTCGCTTAGTTGT-3′ |
| IST1_LE_R | 5′-GCACCTTGTCCTTTCTCTGC-3′ |
| IST1_+4k_F | 5′-TCCGCTGTCACTGCATAAAC-3′ |
| IST1_+4k_R | 5′-TTCCCATGGAGAGGAACATC-3′ |
| MYC_TSS_F | 5′-GGGATCGCGCTGAGTATAAA-3′ |
| MYC_TSS_R | 5′-CCTATTCGCTCCGGATCTC-3′ |
| MYC_I2_F | 5′-TGGCAGGGAGTGTATGAATG-3′ |
| MYC_I2_R | 5′-CACCCACTCTTGAGGCAGTT-3′ |
| MYC_+0.8K_F | 5′-ACATCAACCCCATGAAGGAG-3′ |
| MYC_+0.8K_R | 5′-GTGGCTTGGACAGGTTAGGA-3′ |
| MYC_+2.5k_F | 5′-GATGGAGACCATCCTGGCTA-3′ |
| MYC_+2.5k_R | 5′-ATGCAGTGGCACAATCTCAG-3′ |
Exon Tethering to Pol II with CTD S5P for Co-Transcriptional Splicing
The coupling of Pol II transcription to splicing is well established (Moore and Proudfoot, 2009). For example, altered Pol II elongation speed can affect alternative splicing patterns (Ip et al., 2011; Muñoz et al., 2009), indicating that Pol II slows down near splice sites to promote spliceosome assembly. In particular, genome-wide analysis of nascent RNA by high-resolution tiling arrays in yeast showed that Pol II is paused over terminal exons but only for co-transcriptionally spliced genes (Carrillo Oesterreich et al., 2010). Additionally, precisely timed ChIP analysis in yeast revealed that Pol II CTD S5P accumulates over the 3′SS of intron-containing genes (Alexander et al., 2010). Furthermore, this splicing-dependent Pol II pausing requires pre-spliceosome assembly (Chathoth et al., 2014).
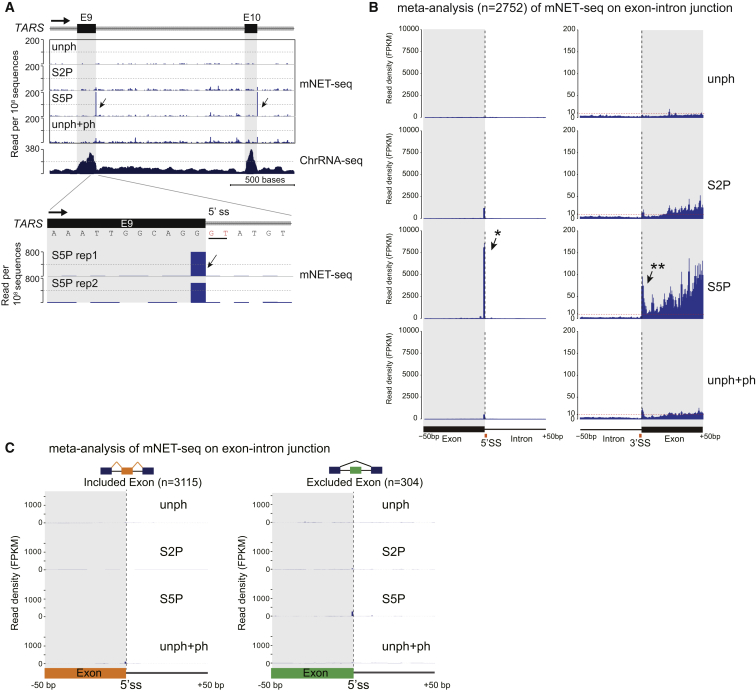
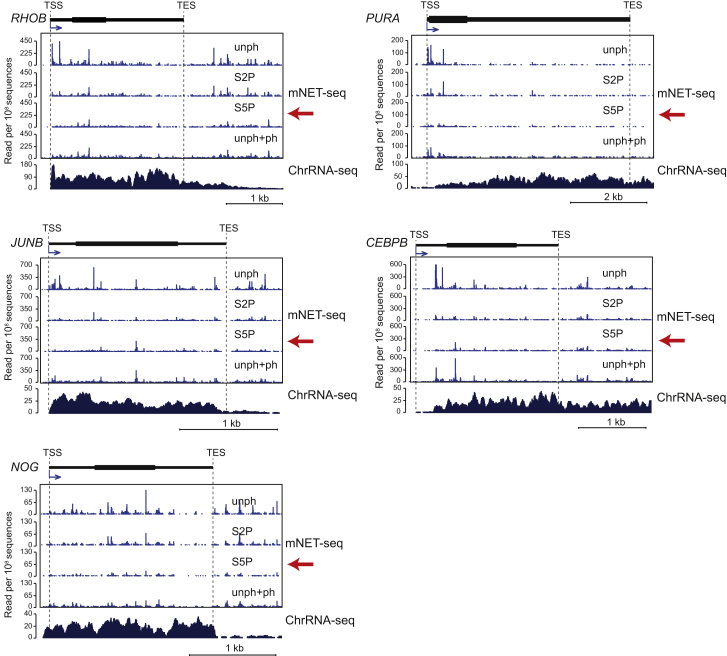
We were interested to determine whether our mNET-seq profiles reflect the co-transcriptionality of splicing, but we observed unexpected patterns. First, we present the mNET-seq profile of a specific gene, TARS, comparing the four different Pol II antibody profiles (Figure 3A). Surprisingly, mNET-seq/S5P selectively detected prominent exon peaks. We have reasoned that mNET-seq will specifically identify the nascent transcript 3′OH in the Pol II active site. However, as previously noted (Churchman and Weissman, 2011), co-precipitated spliceosomes contain 3′OH RNA derived from splicing intermediates that also yield NET-seq signal. Remarkably, single-nucleotide analysis of TARS exon 9 reveals that the major S5P peaks exactly match the 5′SS (Figure 3A, lower panel). These observations suggest that S5P detects the initial 5′SS cleavage intermediate, indicating that spliceosome complex C is associated with Pol II CTD S5P. We next performed meta-analysis of mNET-seq comparing all four antibodies over gene regions that are co-transcriptionally spliced as judged by fused exon reads (Figure 3B). As for TARS, these actively spliced introns give a strong 5′SS S5P-specific signal indicative of co-precipitated spliceosome C complex. Significantly, we also detect selective accumulation of S5P reads over the downstream exon. Apparently, Pol II CTD S5P pauses over exon sequences and so allows time for the spliceosome to perform the first catalytic step. This will generate intronic lariats and cleaved upstream exons, which remain tethered to the downstream positioned Pol II. To further substantiate this mechanism, we carried out additional meta-analysis of predicted included or excluded exons from final spliced mRNA in HeLa cells by analyzing total poly(A)+ RNA-seq data (Katz et al., 2010). Again, we demonstrate a strong 5′SS S5P signal for included but not excluded exons (Figure 3C). We finally present mNET-seq analysis for five intronless genes that show no clear S5P peaks (Figure S3).
Figure 3.
Exon Tethering to Ser5-Phosphorylated Pol II Complex
(A) TARS mNET-seq profile with different antibodies, followed by expanded view of exon 9 5′SS. S5P-dominant peaks are indicated by black arrows.
(B) Meta-analysis of mNET-seq profiles over 3′ ends (left) and 5′ ends (right) of co-transcriptionally spliced exons. Single asterisk, peak at 3′ end of spliced exon; double asterisk, accumulation of Pol II at 5′ end of spliced exon.
(C) Meta-analysis of mNET-seq data over 5′SS of included exons (orange) and excluded exons (green).
For (B) and (C), bars represent mean ± SEM for each base.
Figure S3.
Intronless Genes, Related to Figure 3
Example of intronless genes. mNET-seq with indicated Pol II antibodies and ChrRNA-seq data are shown on indicated intronless genes; RHOB, PURA, JUNB, CEBPB, and NOG. mNET-seq/S5P data set are indicated by red arrows. Read density, reads per 108 sequences.
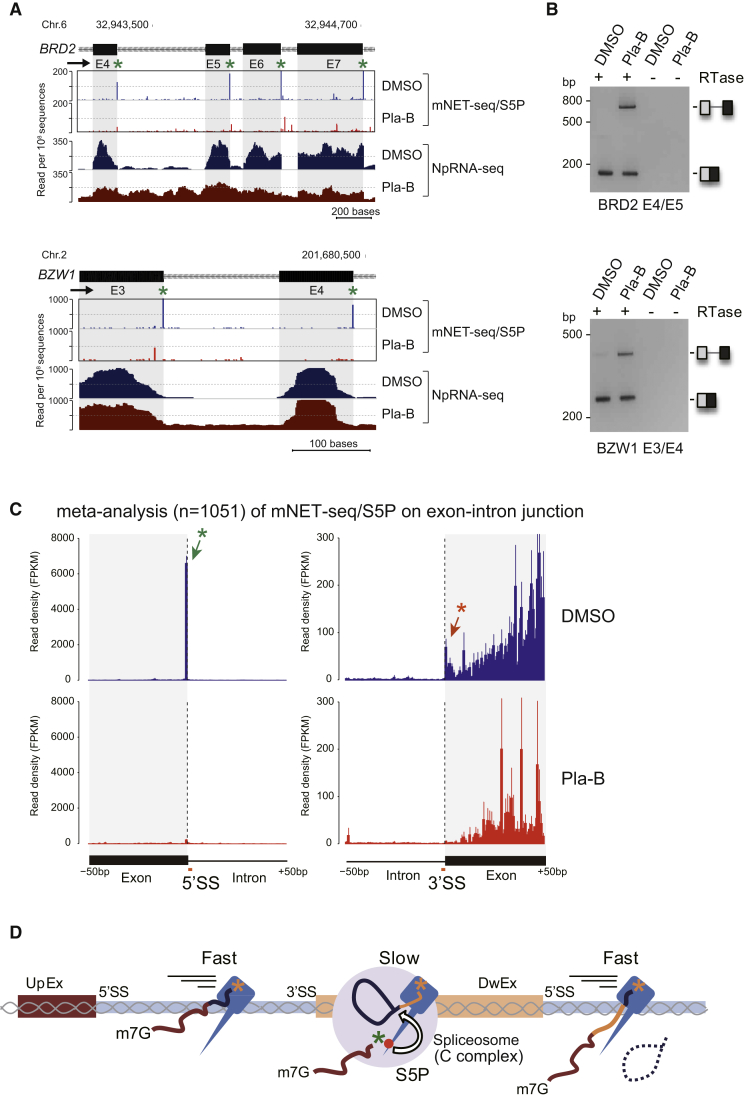
The surprising observation that mNET-seq/S5P profiles show a strong 5′SS signal merited further experimental validation. We therefore employed the splicing inhibitor pladienolide B (Pla-B), which is known to inactive the SF3b sub-complex of U2 snRNP (Kotake et al., 2007), required for intronic branch point recognition as a prelude to the first catalytic step of intron splicing. We initially confirmed the effect of Pla-B treatment on two specific genes (BRD2 and BZW1). First, nucleoplasmic RNA from control DMSO or Pla-B-treated cells was sequenced (NpRNA-seq), and the patterns obtained across these two genes showed a clear increase in intron retention (Figure 4A). This was confirmed by RT-PCR with specific exon primers (Figure 4B) where Pla-B treatment enhanced intron retention in both cases. Notably, mNET-seq/S5P analysis across these same two genes showed the usual high 5′SS peaks for the control but not Pla-B-treated cells (Figure 4A). To establish generality, we performed meta-analysis over 1,051 actively spliced introns (Figure 4C). As before, we saw the high 5′SS peak and enrichment of S5P reads over the downstream exon. Dramatically, Pla-B treatment eradicated the 5′SS signal and substantially reduced downstream exon pausing. These results confirm that the 5′SS mNET-seq/S5P signal that we detect genome wide for spliced exons is indeed a bona fide splicing intermediate.
Figure 4.
Effect of Splicing Inhibition on mNET-Seq and ChrRNA-Seq Profiles
(A) mNET-seq and NpRNA-seq on BRD2 and BZW1 from HeLa cell treated with DMSO (blue) or splicing inhibitor Pla-B (red). Green asterisks denote 5′SS peaks.
(B) RT-PCR analysis of indicated exon splicing showing unspliced and spliced RNA products.
(C) Meta-analysis of mNET-seq/S5P around exon 5′SS and 3′SS from DMSO (blue) and Pla-B (red) treated HeLa cells. S5P-peaks at 5′ and 3′ ends of spliced exons are shown by orange and green asterisks, respectively. Bars represent mean ± SEM for each base.
(D) Co-transcriptional splicing model. 3′OH of upstream exon (UpEx, dark red) and RNA in Pol II catalytic site are shown as green and orange asterisks, respectively. 3′OH of the UpEX RNA is protected in S5P Pol II-spliceosome C complex (gray circle). S5P Pol II pauses over DwEx.
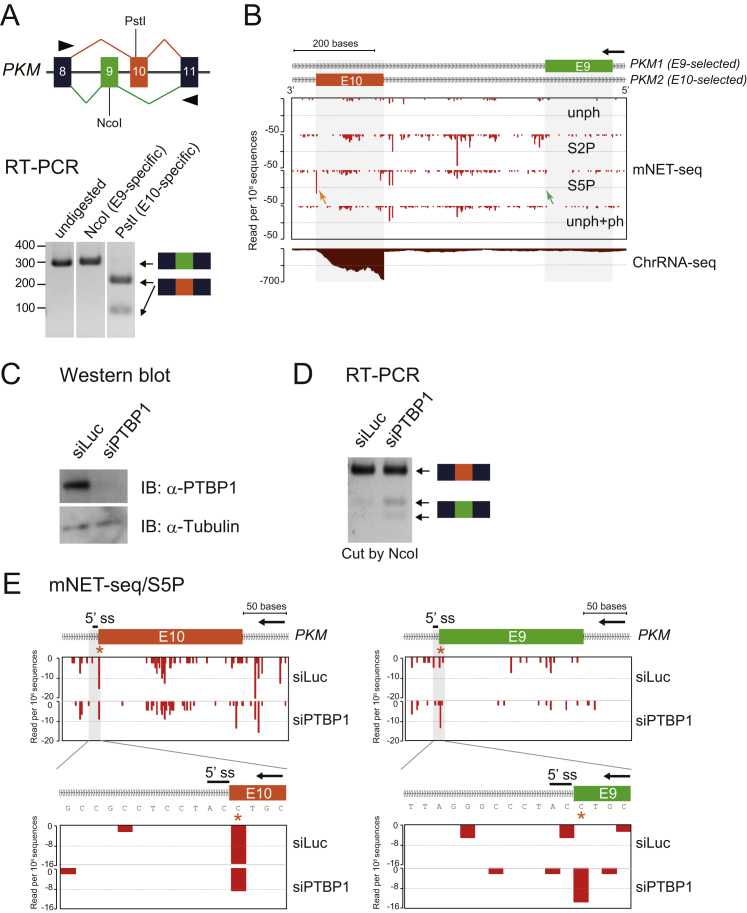
We also studied the mutually exclusive exons 9 and 10 of PKM. RT-PCR and ChrRNA-seq analyses show that exon 10 is predominantly included in mature PKM transcripts in HeLa cells (Figures S4A and S4B) (David et al., 2010). Furthermore the mNET-seq/S5P profile gave the characteristic 5′SS signal at the end of exon 10 but not exon 9 of PKM (Figure S4B). To experimentally manipulate this well-known case of alternative splicing, we performed S5P analysis on chromatin from cells with the splicing-regulatory protein PTBP1 depleted by siRNA treatment (Figure S4C), which is known to be required for the alternative splicing of PKM exon 10 (David et al., 2010). As shown by a lower-resolution and then single-nucleotide resolution mNET-seq profile, the 5′SS peak is reduced at the end of exon 10 but enhanced at the end of exon 9 after depletion of PTBP1 (Figure S4E). Again, this splice-site switch is confirmed by RT-PCR analysis (Figure S4D). Overall, these data on PKM exon 9 and 10 alternative splicing fully corroborate the general upstream exon-tethering pattern for actively spliced exons as demonstrated by our mNET-seq analysis.
Figure S4.
mNET-seq Profiles for PKM Alternative Splicing after PTBP1 Depletion, Related to Figure 4
(A) PKM exons 8–11 are illustrated. Exon 9 (green) and exon 10 (orange) are mutually exclusive. PCR primers indicated as black triangles. RT-PCR products were digested with indicated exon-specific restriction enzyme (NcoI or PstI).
(B) mNET-seq data around mutually exclusive exons 9 and 10 of PKM. mNET-seq/S5P signals at 3′ end of exon 9 and exon 10 are shown by green and orange arrows, respectively. Transcription direction, black arrow.
(C) Western blot of PTBP1 and tubulin from siPTBP1-treated HeLa cells.
(D) PKM RT-PCR products from PTBP1-depleted HeLa nuclear RNA were digested by NcoI.
(E) mNET-seq/S5P data over mutually exclusive exons 9 and 10 of PKM from siLuc and siPTBP1-treated HeLa cells (top), followed by expanded view around 5′SS of introns 10 and 11. S5P-peaks at 3′ ends of exons, orange asterisks. Transcription direction, black arrows.
Co-Transcriptional Pre-miRNA Biogenesis
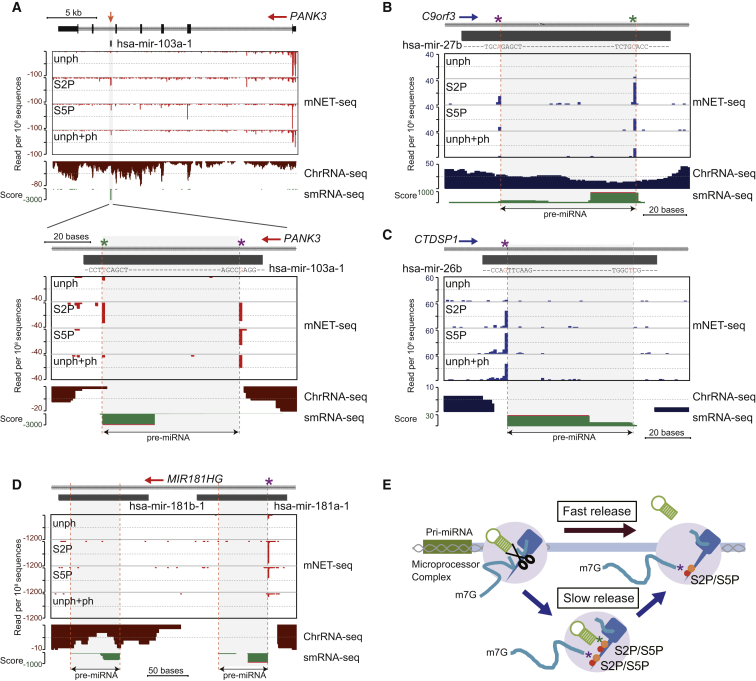
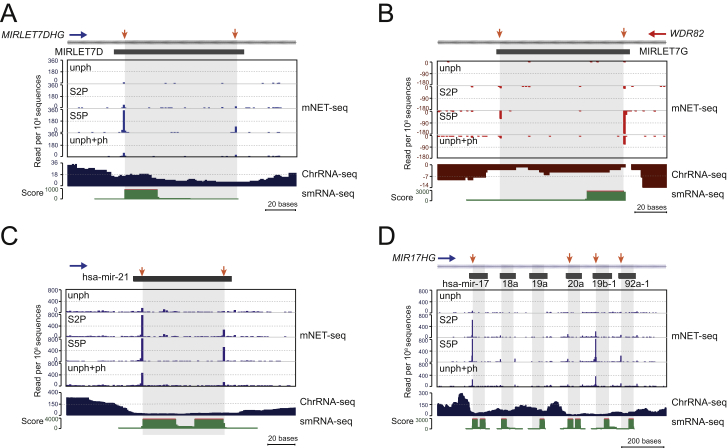
Most pre-miRNA are present within the introns of protein-coding genes and are excised co-transcriptionally by the microprocessor complex, containing Drosha and DGCR8 (Morlando et al., 2008; Pawlicki and Steitz, 2008). Drosha cleavage generates 3′OH ends that have the potential for mNET-seq detection. Because RNA cleavage sites on pre-miRNA generated by the microprocessor complex are quite variable, we individually checked the mNET-seq profiles for highly expressed pri-miRNA in HeLa cells. Our analysis began with PANK3, which harbors hsa-mir-103a-1 in its penultimate intron (Figure 5A). Its mNET-seq profiles show high S5P 5′SS peaks indicative of exon tethering for each exon except exon 5, before the pre-miRNA-containing intron 5. Instead a peak is detected with S5P- and S2P-specific antibodies over the pre-miRNA within this intron. The single-nucleotide resolution profile over hsa-mir-103a-1 (Figure 5A, bottom) shows two peaks by mNET-seq/S2P defining the pre-miRNA 5′ and 3′ ends. Notably, only the 5′ end is detected by mNET-seq/S5P. Similarly hsa-mir-27b in an intron of C9orf3 gives S5P and S2P peaks at both ends of the pre-miRNA (Figure 5B). In contrast, for intronic hsa-mir-26b (CTDSP1), only a 5′ peak is detectable (Figure 5C). A further three examples of intronic pre-miRNAs show both 5′ and 3′ pre-miRNA peaks detectable by either mNET-seq/S5P or S2P (Figures S5A–S5C). These specific 5′ and 3′ end pre-miRNA peaks correspond to the 3′ ends of the cleaved intron and the pre-miRNA, which reaffirms the co-transcriptionality of pre-miRNA processing. As with spliceosomes, we suggest that microprocessor is co-precipitated with Pol II so that 3′OH intermediates of Drosha cleavage are detected by mNET-seq. Two pre-miRNAs (hsa-mir181a-1 and hsa-mir181b-1) are located in the MIR181A1HG intron (Figure 5D). Although the ENCODE project data (Consortium et al., 2012) show that both mature miRNAs are expressed in HeLa cells, only hsa-mir181a-1 yields significant mNET-seq peaks. This correlates with ChrRNA-seq analysis showing a signal window over hsa-mir181a-1, but not hsa-mir181b-1. We infer that only hsa-mir181a-1 is co-transcriptionally processed. Evidently, mNET-seq distinguishes co-transcriptional and post-transcriptional pre-miRNA processing. We also note that the variable mNET-seq double peaks (i.e., hsa-mir-27b) and single peaks (i.e., hsa-mir-26b) suggest kinetic differences in pre-miRNA biogenesis. Some pre-miRNAs (such as pre-miRNA-26b and 181a-1) may be released immediately from the Pol II elongation complex after microprocessor cleavage. Other pre-miRNAs (such as pre-miRNA-27b and let-7g) may be more slowly released with the 3′ ends of the pre-miRNA still tethered to the Pol II elongation complex (Figure 5E, model). Significantly, S2P and S5P generally show larger peaks than unph for pre-miRNA processing, suggesting that CTD phosphorylation is important for co-transcriptional pre-miRNA biogenesis. For the MIR17HG locus containing six tandem pre-miRNA (Figure S5D), Drosha co-transcriptionally cleaves the outer pre-miRNA. However, more inner pre-miR18a and pre-miR19a appear to be processed post-transcriptionally, as judged by a lack of mNET-seq peaks and the absence of a hole in the ChrRNA-seq profile over these sequences (Conrad et al., 2014).
Figure 5.
Pre-miRNA Biogenesis from Protein-Coding Gene Introns
(A–D) mNET-seq with different Pol II antibodies versus ChrRNA-seq over intronic pre-miRNAs. (A) mNET-seq data on PANK3 with magnified view over hsa-mir-103a-1 denoted by a black rectangle. The pre-miRNA is indicated by an orange arrow (top). Three other pre-miRNA are also shown: hsa-mir-27b (B), hsa-mir-26b (C), and hsa-mir181a/b-1 (D). Drosha cleavage sites are identified by dashed orange lines, and asterisks indicate frequent cleavage sites (5′ end, purple; 3′end, green). Small RNA-seq data are shown below (green).
(E) Model of co-transcriptional pre-miRNA biogenesis. Pre-miRNA DNA and hairpin RNA are shown in green. Co-transcriptional Drosha cleavage (scissors) and spliceosome (gray) shown with 3′ ends of cleaved RNA (purple asterisk) and pre-miRNA (green asterisk) tethered to phosphorylated CTD. Pre-miRNA release may occur from the transcription complex, fast (dark red arrow) or slow (blue arrows).
Figure S5.
Further Examples of mNET-Seq and ChrRNA-Seq Profiles over Pre-miRNA, Related to Figure 5
mNET-seq analysis with unph, S2P, S5P, and unph+ph antibodies compared with ChrRNA-seq and small RNA-seq profiles for MIRLET7D (A), MIRLET7G (B), hsa-mir-21 (C), and MIR17HG (D). Note MIR17HG harbors polycistronic pre-miRNA. Frequent RNA cleavage sites (miRBase) are indicated by orange arrows.
Pol II Pausing Regulated by CPA Factors at TES
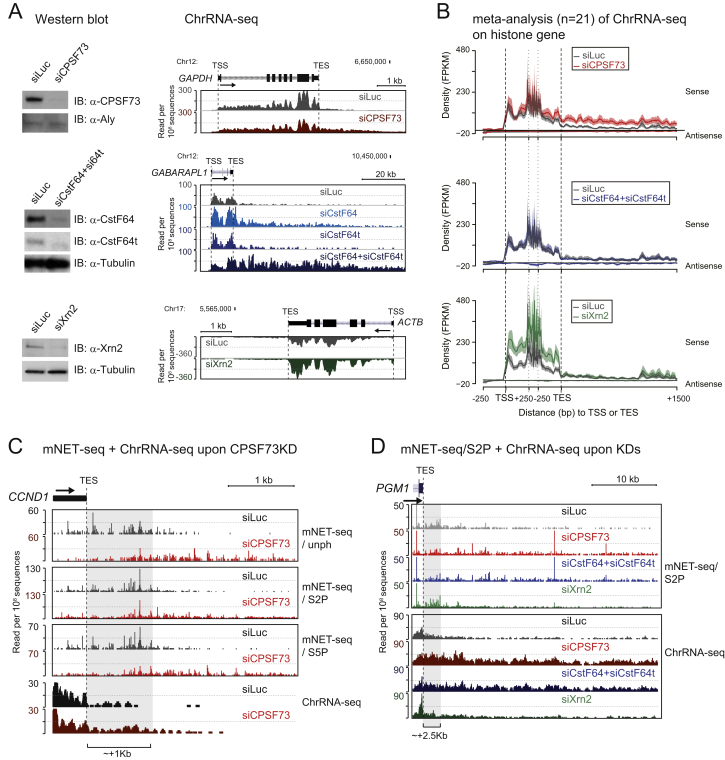
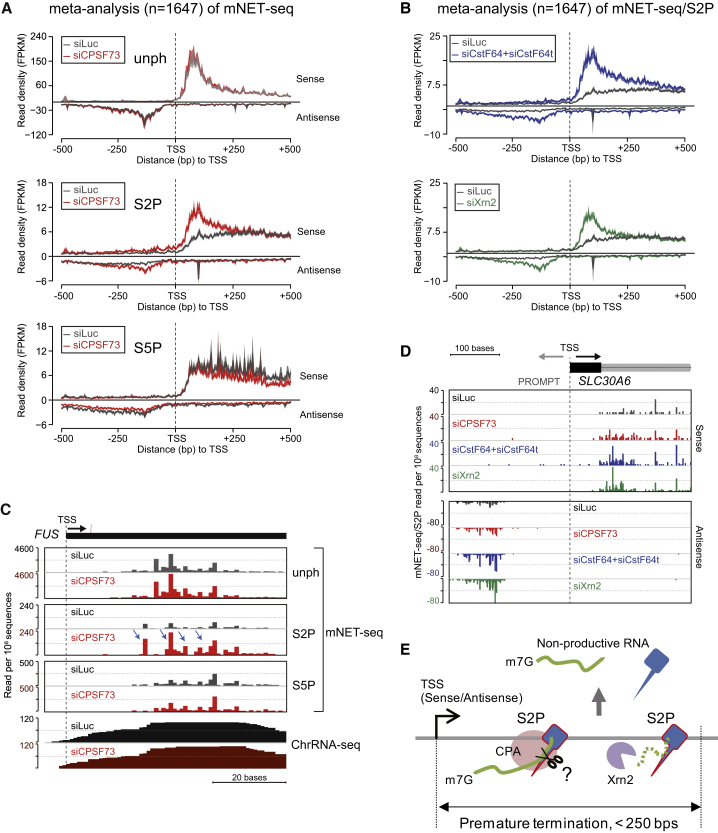
To establish the impact of CPA factors on mNET-seq profiles over and 3′ to TES, we depleted CPA (CPSF73 and CstF64+CstF64t) and Xrn2 by siRNA treatment (Figure S6A, left panels). ChrRNA-seq analyses for specific genes demonstrated clear Pol II termination defects after depletion of CPA factors (Figure S6A, right panels). Double-knockdown of CstF64+CstF64t proteins was necessary to see a full termination defect, presumably due to their functional redundancy in HeLa cells (Yao et al., 2012). Xrn2 knockdown showed no significant termination defect as suggested previously (Brannan et al., 2012). Possibly like CstF64, this factor acts redundantly with other termination factors. Interestingly, Xrn2 depletion increased transcript levels within the GB, suggesting a major role for Xrn2 in nuclear turnover (Davidson et al., 2012). We also performed ChrRNA-seq analysis for histone genes (Figure S6B). Here, CPSF73 still showed a clear termination defect consistent with the known association of CPSF with the histone 3′ processing machinery (Kolev and Steitz, 2005). In contrast, CstF64+CstF64t or Xrn2 knockdowns showed no termination defect. Notably, loss of Xrn2 significantly increased histone gene reads, again indicating a major role in histone mRNA turnover.
Figure S6.
mNET-Seq and ChrRNA-Seq Profiles upon CPA Factors and Xrn2 Knockdown, Related to Figure 6
(A) Termination defect on pA+ protein-coding genes. Western blots showing knockdown efficiencies of siRNA treatments for CPSF73, CstF64, CstF64t, and Xrn2. Aly and Tubulin proteins are loading controls (left). Termination defect detected following depletion of CPSF73 protein (red) on GAPDH (right top). Additive termination defect seen following double-knockdown of CstF64 and CstF64t (turquoise and blue and dark blue double) on GABARAPL1 (right middle). No termination defect detected following Xrn2 depletion (green) on ACTB (right bottom).
(B) mNET-seq meta-profile on histone gene upon CPA factors and Xrn2 knockdown. Effect of siCPSF73 (red, top), siCstF64+siCstF64t (blue, middle), and siXrn2 (green, bottom). n = 21. Line and shading represent mean ± SEM for each bin.
(C) TES of CCND1 as an example of mNET-seq with three different Pol II antibodies (top) and ChrRNA-seq (bottom) from siLuc (control siRNA, dark gray) and siCPSF73 (red) treated HeLa cells. Grey shaded region shows the decrease of Pol II density in siCPSF73-treated cells.
(D) TES of PGM1 as an example of mNET-seq/S2P (top) and ChrRNA-seq (bottom) from siLuc (control siRNA, dark gray), siCPSF73 (red), siCstF64+siCstF64t (blue), and siXrn2 (green) treated HeLa cells. Grey shaded region shows the decrease of Pol II density in siCPSF73 and siCstF64+siCstF64t-teated cells.
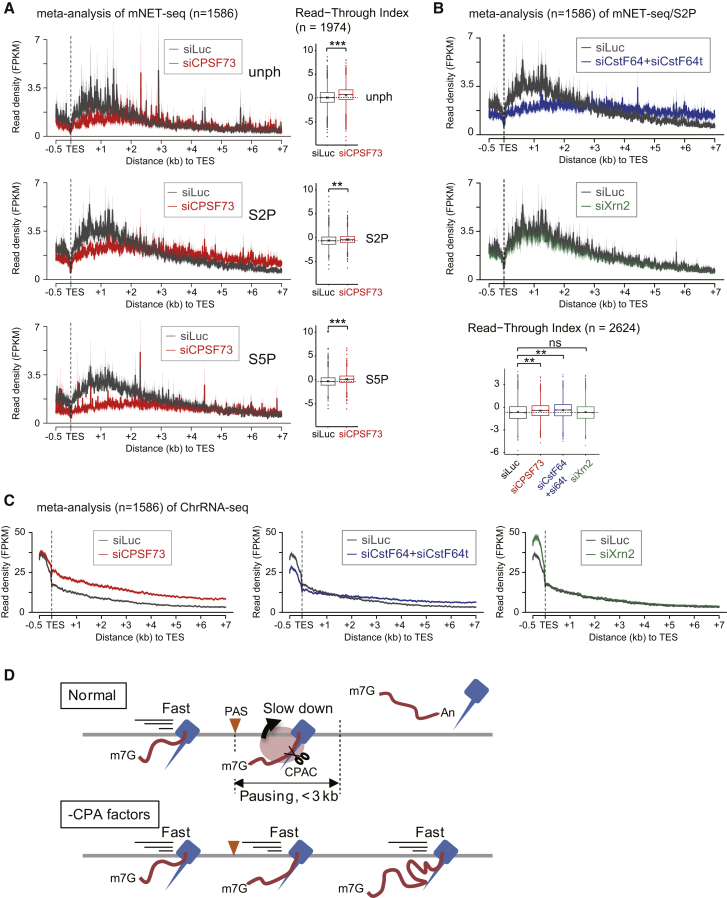
To extend our termination studies to mNET-seq, we principally analyzed CTD S2P profiles as these are most likely to show effects on 3′ end processing (Ahn et al., 2004; Hirose and Manley, 1998; McCracken et al., 1997). However, we also performed S5P and unph meta-analyses in CPSF73-depleted cells. Interestingly, depletion of CPSF73 substantially reduced Pol II unph, S2P, and S5P pausing over the TES (Figure 6A). Similarly CstF64+CstF64t double-knockdown reduced TES pausing. In contrast, Xrn2 knockdown showed no significant difference to the siLuc control (Figure 6B). We also observe that S2P profiles upon knockdown of CPA factors crossed over the siLuc control profile approximately 2.5 kb downstream of the TES, reflecting expected transcriptional termination defects (Figures 6A and 6B). These mNET-seq meta-analyses were complemented by ChrRNA-seq (Figure 6C) where meta-analysis of CPSF73 knockdown gave clear a termination defect immediately following the TES, whereas CstF64+CstF64t double-knockdown showed a termination defect further downstream. Again, specific genes are shown from both our mNET-seq and ChrRNA-seq data sets and show similar trends to those seen in meta-analyses after CPA knockdown (Figures S6C and S6D).
Figure 6.
Nascent RNA within Pol II Complex at TES
(A) Meta-analysis of mNET-seq with indicated Pol II antibodies over TES regions (−0.5 k∼+7kb) from siLuc (dark gray) and siCPSF73 (red) treated HeLa cells (left) is shown. Also shown are RTIs of mNET-seq following CPSF73 knockdown (right). GB signals were divided by signals in a 2 kb region from TES (TES+2k) for RTI (see Extended Experimental Procedures). Dashed line is median of siLuc. (∗∗) p value < 8.52 × 10−11, and (∗∗∗) p value < 2.17 × 10−35 by two-sided Mann-Whitney test.
(B) Meta-analysis of mNET-seq/S2P following termination factor knockdown over TES regions (top). siLuc (dark gray), siCstF64+siCstF64t (blue), and siXrn2 (green). RTI of mNET-seq following indicated knockdown (bottom) is shown. (∗∗) p value < 1.94 × 10−15 by two-sided Mann-Whitney test; ns indicates no difference between samples (p value = 0.9894 by two-sided Mann-Whitney test).
(C) Meta-profiles of ChrRNA-seq following indicated knockdown over TES. siLuc (dark gray), siCPSF73 (red), siCstF64+siCstF64t (blue), and siXrn2 (green).
(D) Model correlating Pol II pausing and PAS-dependent transcription termination at TES. RNA cleavage (scissors) by CPA complex (red circle) at PAS (orange triangle). Pol II elongation speed over 3′ flank region is regulated by PAS recognition on average over a 3 kb region from TES.
For (A)–(C), line and shading represent mean ± SEM for each bin.
3′ End Termination Machinery Regulates Levels of Promoter-Associated RNA
Although RNA cleavage sites have been previously identified near TSS (Almada et al., 2013), which factors are involved in this process has not been determined. Because CPSF73 contains the endonuclease activity, it could potentially cleave nascent RNA near the TSS by recognition of cryptic PAS. We therefore performed meta-analysis across TSS using the mNET-seq data obtained from knockdown of CPA factors and Xrn2. Interestingly, we observe an equivalent increase in TSS-associated S2P Pol II pausing on both mRNA and PROMPT strands after depletion of CPA factors and Xrn2 (Figures 7A and 7B). Notably, this effect is specific for S2P as S5P or unph meta-analysis following CPSF73 knockdown did not show a change in TSS pausing (Figure 7A). S2P meta-analysis of CstF64+CstF64t double-knockdown shows an average 3.6-fold increase as compared to siLuc (Figure 7B, top). Also, CPSF73 and Xrn2 knockdowns both show an average 2.3-fold increase in Pol II pausing (Figures 7A, middle and 7B, bottom). The extent of pausing varies with a more focused effect for CPSF73 and Xrn2 but more prolonged for CstF64+CstF64t on both mRNA and PROMPT strands (Figures 7A and 7B). We also present gene-specific examples to validate our TSS mNET-seq meta-analysis. FUS shows enhanced levels of TSS mNET-seq reads following CPSF73 knockdown, but only for S2P (Figure 7C). SLC30A6 also shows similar enhanced levels of TSS reads for S2P following each termination factor knockdown (Figure 7D).
Figure 7.
Promoter-Associated RNA Turnover Regulated by Termination Factors
(A) Meta-analysis of mNET-seq with indicated Pol II antibodies over TSS regions (−0.5k∼+0.5 kb) from siLuc (dark gray) and siCPSF73 (red) treated HeLa cells (left).
(B) Meta-analyses of mNET-seq/S2P following knockdown of CstF64+CstF64t (blue) and Xrn2 (green) at TSS (left).
(C) mNET-seq of FUS with indicated Pol II antibodies and ChrRNA-seq from siLuc (dark gray) and siCPSF73 (red) treated HeLa cells. Increased mNET-seq/S2P signals following depletion of CPSF73 are denoted by blue arrows.
(D) mNET-seq/S2P maps with indicated knockdowns around TSS of SLC30A6 on both mRNA and PROMPT strands.
(E) Model showing effects of CPA and Xrn2 at TSS. S2P Pol II-CPA complex (red circle) cleaves TSS-associated nascent RNA, and Xrn2 (purple) degrades cleaved RNA from 5′ end to 3′ end over a region of 250 bp from TSS.
For (A) and (B), line and shading represent mean ± SEM for each bin.
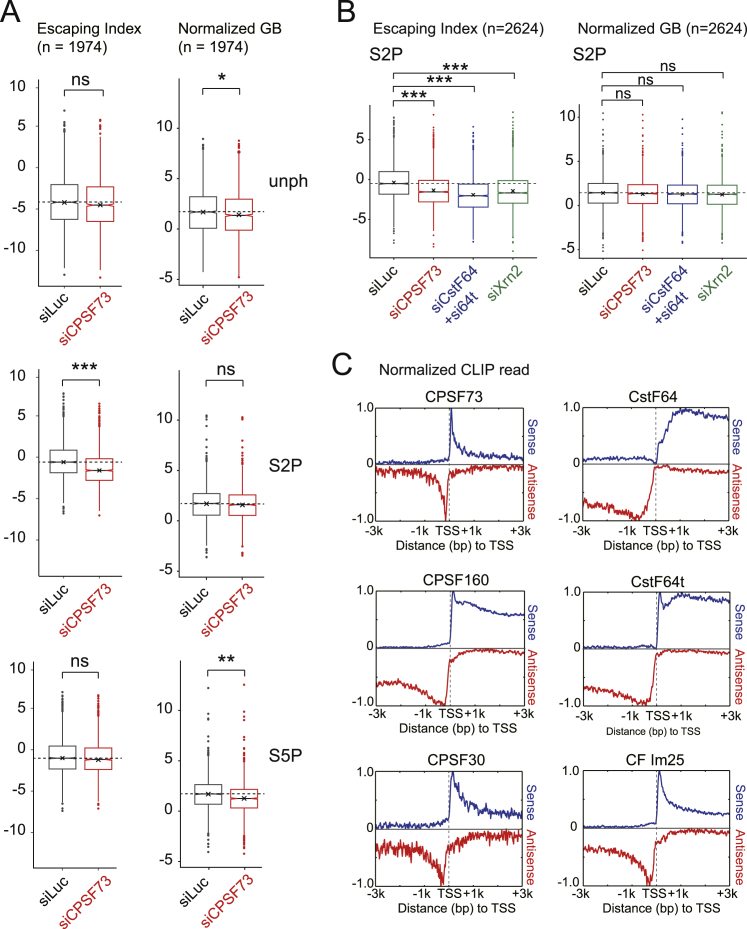
We quantitated the effects of termination factor knockdown by measuring the ratio change between mNET-seq reads over the TSS as compared to the GB; we refer to this as the Escaping Index (EI). We also calculated any changes in read values across the GB (Figure S7; Extended Experimental Procedures). The distribution of EI values clearly shows that depletion of all three factors increases promoter-associated S2P Pol II pausing but has no effect on S2P Pol II distribution across the GB. These results indicate that CPA factors and Xrn2 are involved in restricting the levels of promoter-associated non-productive transcripts.
Figure S7.
TSS Escaping Indexes and CLIP Analysis, Related to Figure 7
(A) EI and normalized GB profiles of each mNET-seq (right). GB signals were divided by signals in promoter region (PRO, −50 to +250 bp over TSS) for EI. The EI (n = 1,974) and normalized GB (n = 1,974) with indicated Pol II antibodies and siRNA treatments are shown below. (∗∗∗) p value < 8.4 × 10−48, (∗∗) p value < 6.7 × 10−20 and (∗) p value < 4.5 × 10−4 by two-sided Mann-Whitney test; (ns) indicates no difference between samples (p value > 0.001 by two-sided Mann-Whitney test).
(B) The EI (left, n = 2,106) and normalized GB (right, n = 1,974) with indicated siRNA treatments are shown below. (∗∗∗) p value < 1.3 × 10−63 by two-sided Mann-Whitney test; (ns) indicates no difference between samples (p value > 0.001 by two-sided Mann-Whitney test).
(C) CLIP analysis of TSS associated CPA factors (Martin et al., 2012). Normalized read counts and distance from TSS are shown at y and x axes. Sense, blue; antisense, red.
In order to examine whether CPA factors could directly bind to nascent RNA near TSS, we analyzed in vivo cross-linking and immunoprecipitation (CLIP) data for genome-wide alternative polyadenylation at TES (Martin et al., 2012). Surprisingly, all CPA factors, including CPSF73, CstF64, CstF64t, CPSF160, CPSF30, and CFIm25 proteins, are significantly detected on both strands within 500 nt of the TSS. Especially CPSF73 shows a substantial peak 160 nt upstream and 80 nt downstream of TSS (Figure S7C and Table S1). Together with our mNET-seq/S2P results, we conclude that the CPA complex cleaves not only pre-mRNA at the PAS to promote 3′ end termination but also promotes promoter-associated premature termination (Figure 7E). Notably, Xrn2 plays a unique role in TSS but not in TES termination.
Discussion
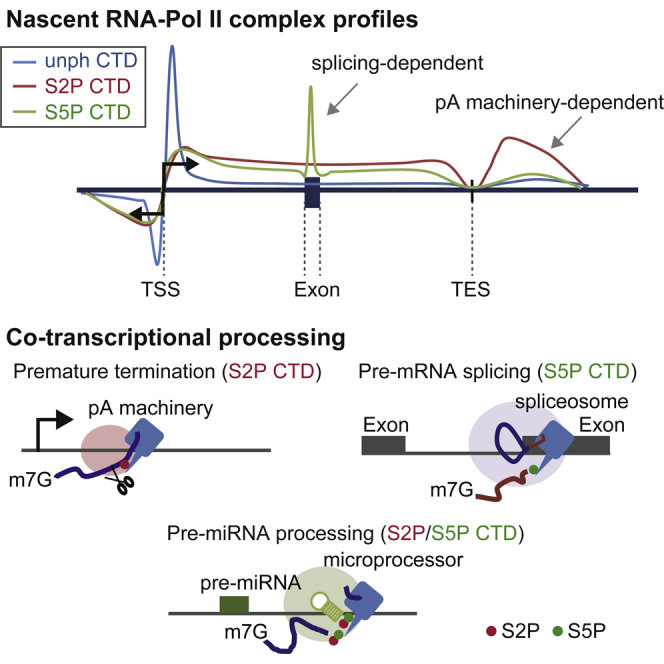
Our mNET-seq analysis reveals precise maps of both nascent RNA and the associated Pol II “CTD code.” We employed recently evaluated high-affinity and specificity monoclonal antibodies to Pol II CTD S5P, S2P, unph, and unph+ph (Figure S2; Stasevich et al., 2014) in our mNET-seq analysis. Interestingly, our mNET-seq data reveal significant differences in CTD modification profiles across mammalian protein-coding genes as compared to previous studies. In particular, we detect predominantly low or unphosphorylated CTD over the TSS region (at least lacking S5P and S2P modification). Furthermore, most detected S5P signal is found in the GBs, where it is particularly associated with actively spliced exons. Finally, although we find that S2P signal is more associated with TES regions (consistent with previous studies), we demonstrate a redistribution of this CTD mark to TSS following CPA depletion. Several explanations may account for the differences between our mNET-seq data and previous studies. Thus, mNET-seq does not involve cross-linking (by formaldehyde), which is by necessity used in ChIP analysis. The possibility that cross-linking distorts the native chromatin structure remains a concern. Similarly, mNET-seq detects nascent transcripts at single-nucleotide resolution, which cannot be achieved by GRO-seq analysis. Even though PRO-seq analysis does give single-nucleotide resolution, the act of isolating nuclei, treating with sarcosyl, and then carrying out an in vitro transcription reaction (using modified nucleotides) as in both GRO-seq and PRO-seq protocols may distort the native transcription profiles of genes. Clearly, in the future, we can extend our analysis to include other CTD phosphorylation marks using appropriate Pol II antibodies. For example, the CTD S7P mark is important to recruit Integrator complex to snRNA genes, which regulates 3′ end processing and termination (Egloff et al., 2007). Mutation of CTD T4 specifically represses histone gene expression by blocking 3′ end processing (Hsin et al., 2011). Another CTD modification, Y1P, stimulates the binding of elongation factor Spt6 and blocks recruitment of termination factors in yeast (Mayer et al., 2012). It remains a possibility that the mammalian CTD code may be significantly different than the likely simplified code for budding yeast. Notably, yeast CTD has only 26 heptad repeats in its CTD, and these are near identical, unlike the more variable mammalian heptad repeats. Possibly, the high S5P TSS signals observed in yeast are replaced by other CTD or indeed histone marks in higher eukaryotes. Furthermore, few yeast genes possess introns so that the dominant presence of S5P marks over mammalian exons would be less quantitatively significant in yeast. Even so, it has been shown that yeast introns display high S5P signals near their 3′ ends (Alexander et al., 2010), similarly to the mammalian S5P splicing association, described here.
A remarkable feature of our mNET-seq data is that we readily detect RNA 3′ ends formed as RNA-processing intermediates through co-association of RNA-processing complexes with elongating Pol II. In particular, the Pol II CTD S5P mark, previously thought to be mainly associated with TSS events such as co-transcriptional capping and early transcriptional elongation (Hsin and Manley, 2012), plays a major role in splicing. Thus, 5′SS peaks of mNET-seq/S5P are detected at the end of co-transcriptionally spliced exons (Figure 3), indicating that the 3′ cleaved upstream exon within the spliceosome is associated with Pol II elongation complexes in an S5P-dependent manner. We also note that the mNET-seq S5P reads are particularly high over spliced exons, suggesting that S5P Pol II pauses over functional exons allowing time for U2 snRNP-mediated activation of 5′SS cleavage. Indeed, we demonstrate this by directly inhibiting U2 snRNP function (Figures 4 and S4). Overall, we find that the 5′SS cleavage intermediate is retained within the spliceosome C complex associated with Pol II S5P until subsequent ligation with the downstream exon can occur (Figure 4D, model). In effect, we provide genome-wide support for exon tethering to Pol II as previously predicted from studies on transfected gene constructs wherein co-transcriptional intron cleavage did not prevent exon splicing across a discontinuous intron (Dye et al., 2006). We anticipate that our mNET-seq technology will provide new ways to unravel the complexity of the co-transcriptional splicing mechanism.
A surprising aspect of our mNET-seq analysis is that we do not detect a peak of signal associated with pre-mRNA 3′ end processing (TES meta-analysis in Figure 2C and Figures 6A and 6B). This contrasts the splicing-associated 5′SS and Drosha cleavage sites that are highly prevalent in our data (Figures 3, 4, and 5). We predict that 3′ end cleavage (coupled with polyadenylation) may cause rapid mRNA release from the Pol II complex and so escape mNET-seq detection, like in the pre-miRNA fast-release model (Figure 5E). Although 3′ end processing is known to be required for Pol II termination (Proudfoot, 2011), it is also thought that Pol II pausing at TES regulates both 3′ end processing and subsequent transcription termination (Gromak et al., 2006; Nag et al., 2007). We examined the effect on Pol II pausing at TES following depletion of CPA components. Consistent with previous reports, ChrRNA-seq reveals that CPSF73 and CstF64 depletion cause transcriptional termination defects on protein-coding genes (Figure 6). Interestingly, our mNET-seq data also reveal that depletion of CPA factors causes significantly less pausing immediately downstream of TES (<3 kb from TES) and then more Pol II occupancy at further downstream regions (>3 kb from TES) compared to control cells. This indicates that Pol II elongation speed is regulated by the CPA complex, which may be an important factor in mediating transcription termination (Figure 6D). We also demonstrate that no significant termination defect occurs following the TES upon knockdown of Xrn2 (Figure 6A, bottom). This observation contrasts our previous reports based on plasmid transfection studies (West et al., 2004). However, it has been shown more recently that Xrn2 has a required partner protein, TTF2, for transcription termination (Brannan et al., 2012). It seems likely that Xrn2-associated termination is redundant with other termination factors.
Unexpectedly, mNET-seq analysis showed a significant increase specifically in S2P Pol II pausing at the TSS (<250 base) for both mRNA and PROMPT transcription upon CPA factor and Xrn2 depletion. This suggests that CPA and Xrn2 are involved in premature termination at the TSS, consistent with a previous report (Brannan et al., 2012). Although CPA factors and Xrn2 affect S2P Pol II occupancy at TSS, they show no difference in S2P Pol II distribution across the GB. Recent studies have pointed toward differences between promoter-proximal termination for mRNA sense or antisense RNA (Almada et al., 2013; Ntini et al., 2013). Antisense TSS transcripts (PROMPTs) are thought to utilize cryptic PAS close to the TSS, whereas sense TSS transcripts may have reduced occurrence of cryptic PAS. Those that are present may be blocked by nearby 5′SS U1 snRNP recruitment (Kaida et al., 2010). These apparent differences in cryptic PAS usage between PROMPTs and sense TSS-associated transcripts may favor productive sense over non-productive antisense transcription. However, our mNET-seq data suggest that CPA factors and Xrn2 play equivalent roles in restricting sense and antisense TSS transcription. Thus, their depletion causes an equivalent increase in S2P Pol II pausing in both transcriptional directions. Also, we show by CLIP analysis that CPA factors are directly and equally associated with these two transcript classes (Martin et al., 2012). Our data suggest that transcriptional directionality at TSS is unlikely to be regulated by CPA-mediated termination. Rather both sense and antisense TSS-associated transcripts are restricted by normally TES-associated termination factors. Indeed, we observe a redistribution of S2P Pol II from the TES to the TSS following CPA factor and Xrn2 knockdown. This argues for close interconnections between both ends of the Pol II transcription unit, as previously demonstrated by 3C analysis (Ansari and Hampsey, 2005; O’Sullivan et al., 2004; Tan-Wong et al., 2012). Several gene-specific analyses in mammals have reported the co-association of CPA with transcription initiation factors. Thus, CPSF is a known component of some TFIID complexes (Dantonel et al., 1997), and CstF has been shown to associate with TFIIB (Wang et al., 2010). Also, mutating the PAS depleted promoter-associated transcription factors and increased promoter-associated Pol II CTD S2P (Mapendano et al., 2010). Finally, the elongation factor TFIIS has been shown to promote release of paused TSS transcripts in Drosophila (Adelman et al., 2005), and this may in turn relate to CPA promoter effects.
Overall, mNET-seq maps nascent transcription at single-nucleotide resolution, showing both Pol II pausing and associated co-transcriptional RNA cleavage. Importantly, this method can be applied genome wide to check for modified polymerase occupancy (even Pol I and Pol III) by selecting a range of different antibodies. We anticipate that mNET-seq will expand our knowledge of how different nascent RNA are associated with specific “CTD codes.”
Experimental Procedures
Antibodies and siRNA
Antibodies and siRNA information are available in the Extended Experimental Procedures. In outline, siRNA treatment was carried out for 3 days prior to cell harvesting. The efficiency of protein depletion was confirmed by western blot with appropriate antibodies.
Cell Culture, NRO Assay, and RT-PCR
Cell culture and NRO assay were as previously described (Nojima et al., 2013). RT-PCR and primers are described in the Extended Experimental Procedures.
In Vivo Splicing Inhibition
HeLa cells were treated with either DMSO (0.1%) or Pla-B (1 μM) for 4 hr. Pla-B was purchased from Santa Cruz (sc-391691).
RNA-Seq Methods
Preparation of chromatin and nucleoplasmic RNA was previously described (Nojima et al., 2013). For mNET-seq, isolated chromatin was incubated with MNase (40 u/μl). MNase was inactivated by EGTA, and the insoluble chromatin removed by centrifugation. IP was performed from the supernatant using specific Pol II antibody-conjugated beads for 1 hr. IPed RNA was 5′ end phosphorylated by polynucleotide kinase treatment of the washed beads. Purified RNA was fractionated on denaturing acrylamide gels, and a 35–100 nt fraction was isolated. RNA libraries were prepared according to the manual of Truseq small RNA library prep kit (Illumina). The reads were generated in Hiseq2000/2500 (Illumina). For full methods, see the Extended Experimental Procedures.
Data Pre-Processing
mNET-seq data adaptors were trimmed using Cutadapt (v1.1) (Martin, 2011). The remaining paired reads were aligned to the reference human genome (hg19) using TopHat (v2.0.9) (Kim et al., 2013) only allowing for one alignment to the reference. The last nucleotide incorporated by the polymerase was defined as the 5′ end of read two (green arrow, Figure 1A) of the pair, with the directionality indicated by read one (blue arrow, Figure 1A), and then the properly aligned read pairs were trimmed to solely keep the 5′ nucleotide of read two. ChrRNA-seq and nucleoplasm RNA-seq data were aligned using the same version of TopHat but allowing for the read pairs to be separated by 3 kb. Further details of data pre-processing and bioinfomatic analysis are available in the Extended Experimental Procedures.
Author Contributions
T.N. performed all molecular biology and genomic analyses, except that M.J.D. performed NRO. T.G. carried out all bioinformatic analyses aided by ARFG, except that S.D. analyzed CLIP-seq data. H.K. generated Pol II antibodies. T.N., M.C.-F., and N.J.P. designed the project and wrote the paper.
Acknowledgments
We thank the N.J.P. lab and Dr. M. Dienstbier for critical discussion. T.N. was supported by the KANAE foundation. H.K. was supported by JSPS KAKENHI and JST CREST. This work was supported by funding to N.J.P. (Wellcome Trust Programme [091805/Z/10/Z] and ERC Advanced [339270] Grants) and to M.C.-F. (Fundação Ciência e Tecnologia, Portugal).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
Maria Carmo-Fonseca, Email: carmo.fonseca@medicina.ulisboa.pt.
Nicholas J. Proudfoot, Email: nicholas.proudfoot@path.ox.ac.uk.
Accession Numbers
The data present in this work are deposited in NCBI’s Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under the accession number GSE60358.
Supplemental Information
References
- Adelman K., Lis J.T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K., Marr M.T., Werner J., Saunders A., Ni Z., Andrulis E.D., Lis J.T. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Ahn S.H., Kim M., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Alexander R.D., Innocente S.A., Barrass J.D., Beggs J.D. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almada A.E., Wu X., Kriz A.J., Burge C.B., Sharp P.A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K., Kim H., Erickson B., Glover-Cutter K., Kim S., Fong N., Kiemele L., Hansen K., Davis R., Lykke-Andersen J., Bentley D.L. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F., Preibisch S., Neugebauer K.M. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Chathoth K.T., Barrass J.D., Webb S., Beggs J.D. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol. Cell. 2014;53:779–790. doi: 10.1016/j.molcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman L.S., Weissman J.S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T., Marsico A., Gehre M., Orom U.A. Microprocessor activity controls differential miRNA biogenesis in vivo. Cell Rep. 2014;9:542–554. doi: 10.1016/j.celrep.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Consortium E.P., Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Lis J.T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel J.C., Murthy K.G., Manley J.L., Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- David C.J., Chen M., Assanah M., Canoll P., Manley J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L., Kerr A., West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31:2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye M.J., Gromak N., Proudfoot N.J. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Egloff S., O’Reilly D., Chapman R.D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist D.A., Dos Santos G., Fargo D.C., Xie B., Gao Y., Li L., Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N., West S., Proudfoot N.J. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso A.R., de Almeida S.F., Braga J., Carmo-Fonseca M. Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res. 2012;22:1447–1456. doi: 10.1101/gr.138057.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann M., Hintermair C., Voß K., Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Manley J.L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hsin J.P., Manley J.L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J.P., Sheth A., Manley J.L. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip J.Y., Schmidt D., Pan Q., Ramani A.K., Fraser A.G., Odom D.T., Blencowe B.J. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N.G., Steitz J.A. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kwak H., Fuda N.J., Core L.J., Lis J.T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., Almouzni G. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Mapendano C.K., Lykke-Andersen S., Kjems J., Bertrand E., Jensen T.H. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol. Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- Martin G., Gruber A.R., Keller W., Zavolan M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Mayer A., Heidemann M., Lidschreiber M., Schreieck A., Sun M., Hintermair C., Kremmer E., Eick D., Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Meinhart A., Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Proudfoot N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M.J., Pérez Santangelo M.S., Paronetto M.P., de la Mata M., Pelisch F., Boireau S., Glover-Cutter K., Ben-Dov C., Blaustein M., Lozano J.J. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Nag A., Narsinh K., Martinson H.G. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat. Struct. Mol. Biol. 2007;14:662–669. doi: 10.1038/nsmb1253. [DOI] [PubMed] [Google Scholar]
- Nojima T., Dienstbier M., Murphy S., Proudfoot N.J., Dye M.J. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Rep. 2013;3:1080–1092. doi: 10.1016/j.celrep.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E., Järvelin A.I., Bornholdt J., Chen Y., Boyd M., Jørgensen M., Andersson R., Hoof I., Schein A., Andersen P.R. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- O’Sullivan J.M., Tan-Wong S.M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N.J. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Pawlicki J.M., Steitz J.A. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Lluch S., Blanco E., Carbonell A., Raha D., Snyder M., Serras F., Corominas M. Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res. 2011;39:4628–4639. doi: 10.1093/nar/gkq1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich T.J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., Sakata-Sogawa K., Tokunaga M., Nagase T., Nozaki N., McNally J.G., Kimura H. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- Tan-Wong S.M., Zaugg J.B., Camblong J., Xu Z., Zhang D.W., Mischo H.E., Ansari A.Z., Luscombe N.M., Steinmetz L.M., Proudfoot N.J. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang Y., Fairley J.A., Roberts S.G. Phosphorylation of TFIIB links transcription initiation and termination. Curr. Biol. 2010;20:548–553. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C.M., Ramachandran S., Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. Human 5′ —> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Yao C., Biesinger J., Wan J., Weng L., Xing Y., Xie X., Shi Y. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc. Natl. Acad. Sci. USA. 2012;109:18773–18778. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Hart T., Komori H.K., LaMere S., Podshivalova K., Salomon D.R. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics. 2013;14:778. doi: 10.1186/1471-2164-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.