Abstract
Age-related cataracts are a leading cause of blindness. Previously, we have demonstrated the association of unfolded protein response with various cataractogenic stressors. However, DNA methylation alterations leading to suppression of lenticular antioxidant protection remains unclear. Here, we report the methylglyoxal-mediated sequential events responsible for Keap1 promoter DNA demethylation in human lens epithelial cells, because Keap1 is a negative regulatory protein that regulates the Nrf2 antioxidant protein. Methylglyoxal induces the ER stress and activates the unfolded protein response leading to overproduction of ROS prior to human lens epithelial cells death. Methylglyoxal also suppresses the Nrf2 and DNA methyltransferases but activates the DNA demethylation pathway enzyme, TET1. Bisulfite genomic DNA sequencing confirms the methylglyoxal-mediated Keap1 promoter DNA demethylation leading to over-expression of Keap1 mRNA and protein. Similarly, bisulfite genomic DNA sequencing of human clear lenses (n=15) slowly lose 5-methylcytosine in the Keap1 promoter throughout life, at a rate of 1% per year. By contrast, diabetic cataractous lenses (n=21) lose an average of 90% of the 5-methylcytosine regardless of the age. Over-expressed Keap1 protein is responsible for decreasing the Nrf2 by proteasomal degradation, thereby suppressing the Nrf2 dependent stress protection. This study demonstrates for the first time about the associations of unfolded protein response activation, Nrf2 dependent antioxidant system failure and loss of Keap1 promoter methylation because of altered active and passive DNA demethylation pathway enzymes in human lens epithelial cells by methylglyoxal. As an outcome, cellular redox balance is altered towards lens oxidation and cataract formation.
Keywords: cataracts, methylglyoxal, ER stress, DNA methylation, unfolded protein response, Nrf2 dependent antioxidant protection, Keap1 promoter demethylation
Introduction
Age-related cataracts (ARCs) are a leading cause of visual impairments worldwide. The prevalence of ARCs escalates from 4% at age 52-64 to 50% at age 75-85. Diabetes is one of the potential risk factors with five-fold increases in the incidence and progression of early cataract formation [1, 2], influenced essentially by duration of the diabetes and the quality of glycemic control [1, 3]. Being a non-insulin dependent tissue, the lens is incapable of down-regulating glucose transport. The increase of extracellular glucose concentrations leads to hyperglycemic deleterious effects [4], which include hyperosmolarity combined with oxidation [5], increased cytosolic Ca2+ [6], proteolysis [7], lens epithelial cells (LECs) death [8], LECs migration [9], and aging [10].
Hyperglycemia results in abnormal cellular accumulation of reactive aldehydes, including methylglyoxal (MGO), which is generated as a consequence of both non-enzymatic reactions of glucose with proteins [11] and the enhanced formation and resultant breakdown of triose phosphates within cells [12]. MGO reacts with arginine and lysine residues of lens proteins, and generates protein adducts, and results in MGO-derived advanced glycation end-products (AGEs) thereby altering the functions and conformation of lens proteins by crystallin aggregations, a key contributor for increased light-scattering in lens opacity [13-15]. In addition, MGO reacts with DNA, forming major DNA adducts linked to increased DNA strand breaks and frameshift mutation [16], mitochondrial dysfunction and reactive oxygen species (ROS) formation [5, 17, 18].
Paradoxically, improper control of glycemia has been connected with activation of both oxidative and endoplasmic reticulum (ER) stress signaling pathways. A growing body of evidence suggests that ER stress triggers an evolutionarily conserved adaptive program known as the unfolded protein response (UPR), which combines the early inhibition of protein synthesis with a later upregulation of genes that stimulate protein folding or clearance [19, 20]. UPR is mediated by three ER transmembrane proteins: inositol-requiring enzyme 1α (IRE1α), PKR-like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6) thereby inducing overall gene expression changes to restore ER homeostasis [21, 22]. Further, if ER stress is not alleviated, the prolonged UPR activates apoptosis by up-regulating activating transcription factor 4 (ATF4), which promotes both the transcription of prosurvival genes and the expression of the proapoptotic transcription factor, C/EBP-homologous protein (CHOP) and caspases [23, 24]. Also, UPR upregulates intracellular ROS production and activates the transcriptional factor, nuclear factor-erythroid-2-related factor 2 (Nrf2), to maintain the cellular redox homeostasis from oxidative damage by controlling the inducible expression of many cytoprotective genes [25, 26].
Normally, Nrf2 is found in the cytosol by binding with its negative regulator, Kelch-like ECH-associated protein 1 (Keap1). Under basal conditions, Nrf2 is constantly ubiquitinated by the E3-ubiquitin ligase-like domain of Keap1, an oxygen free radical sensor protein [27], followed by 26S proteasomal degradation [28]. Upon oxidative/ER stress, Nrf2 dissociates from the Keap1, then translocates into the nucleus and activates the transcription of antioxidant response element (ARE)-containing genes [29]. Under terminal UPR, the level of Nrf2 decreases due to proteasomal degradation and proteolysis by m-calpain and caspase-3 and caspase-1 [30, 31]. Nrf2 also regulates the expression of the glyoxalase 1 (Glo-1) gene [32], which is a major MGO detoxification enzyme.
In addition, ER-induced oxidative stress targets Ca2+ release from ER calcium stores, which activates m-calpain in the lens [33]. Recent findings have suggested a direct link between the production of ROS and protein folding and oxidation, because oxidative stress and ROS production are integral components of ER stress and are not just consequences of ER stress induction [34]. The key enzymatic machinery of ROS production during UPR induction is driven by a protein relay involving a protein disulfide isomerase (PDI), ER oxidoreductin 1 (Ero1)-Lα and Ero1-Lβ, and molecular oxygen, a terminl electron acceptor [35, 36].
Moreover, ROS produced by oxidative/ER stress stimulates the alterations in DNA methylation patterns, without changing the DNA base sequence. Such alterations in DNA methylation patterns are known to strongly control the expression of several genes [37]. DNA methylation patterns are established during embryonic development and are then accurately inherited in the cells with a ‘maintenance’ mechanism, which involves the methylation of DNA daughter strand after replication by DNA methyltransferase 1 (Dnmt1). Failure of this ‘maintenance’ mechanism leads to DNA demethylation, which can result from passive demethylation in the absence of Dnmt1, or from the prevention of Dnmt1 action following DNA replication [38]. In contrast, active DNA demethylation occurs through the enzymatic substitution of 5-methylcytosine (5mC) with cytosine [39], where the ten-eleven-translocation 1 protein (TET1) and the activation-induced deaminase (AID) convert 5-mC to thymine followed by base-excision repair by thymine DNA glycosylase (TDG) in non-replicating cells [32, 40-42].
Here, we hypothesize that the MGO might induce overproduction of ROS, ER stress mediated activation of UPR, suppression of Nrf2 dependent antioxidant protection and alteration of passive and active DNA demethylation pathway enzymes, leading to reprogramming of gene expression. We further hypothesize that this sequence of events is responsible for Keap1 promoter DNA demethylation in diabetic ARC lenses. To test this hypothesis, we have studied the cell death, ROS overproduction, Ca2+ release, UPR induction, and evaluated the levels of Nrf2/Keap1, active and passive DNA demethylation pathway enzymes, and promoter DNA methylation status of the Keap1 gene in human lens epithelial cells (HLECs) treated with MGO. Our findings highlight the associations between induction of ER stress and UPR activation, ROS overproduction, Nrf2 dependent antioxidant system failure and loss of Keap1 promoter methylation because of altered active and passive DNA demethylation pathway enzymes in HLECs by MGO.
Materials and methods
Ethical statements
All animal experiments were approved by the University of Nebraska Animal Care and Use Committee and were in compliance with the Animal Welfare Act (Public Law 91–579) as mandated by the NIH Guide for Care and Use of Laboratory Animals and the procedures recommended by the Association for Research in Vision and Ophthalmology resolution on the use and treatment of animals in ophthalmic and vision research were followed. Further, the approval and oversight from the Institutional Review Board is not required to conduct research on human lens samples for bisulfite genomic sequencing of human Keap1 promoter DNA because this study did not obtain data through intervention or interaction with the human subjects as well as did not receive any identifiable private information from them. Therefore, Institutional Review Board approval for research involving leftover (excess) human biological material is not required. Human clear and diabetic cataractous lenses were obtained from National Disease Research Interchange (NDRI), Philadelphia, PA.
Cell culture
HLECs (SRA 01/04) [43] were cultured overnight in DMEM, High glucose (Life Technologies, Gaithersburg, MD) with 10% fetal bovine serum (Gemini Bio-Products, Calabasas, CA) under 20% atmospheric oxygen at 37°C. Cells were plated 24 h prior to experiment in DMEM, Low glucose (Life Technologies, Gaithersburg, MD) under 4% atmospheric oxygen and cultured with 10 μmoles/L 5-Aza-2′-deoxycytidine (5-Aza; Sigma-Aldrich, St. Louis, MO) for 7 days or with varying concentrations of MGO, a generous gift from Dr. Keshore R. Bidasee, UNMC, for 24 h. 4% atmospheric oxygen environment was generated in an O2/CO2 incubator (Sanyo, Osaka, Japan) with an attached 50 gallon liquid nitrogen gas tank, since human lens is located in a hypoxic environment [44]. At the end of the experiment, the cells were harvested and used for cell death and intracellular ROS production assays, bisulfite genomic DNA sequencing, real-time quantitative PCR (RT-qPCR), and Western blotting.
Induction of diabetes in Nrf2-/- and Nrf2+/+ C57BL/6 mice
Diabetes was induced in overnight fasted Nrf2-/- and Nrf2+/+ mice (8 weeks old; supplied from Jackson Laboratory, Bar Harbor, ME) by multiple i.p. injection of streptozotocin (50 mg/kg body weight; Sigma-Aldrich, St. Louis, MO) dissolved in 0.1 M cold citrate buffer, pH 4.5 for 5 consecutive days. After a week, mice having fasting blood glucose above 250 mg/dL were considered as diabetic and selected for the experiment. All animals received water and food ad libitum.
Cell viability/death and ROS staining
LECs isolated from diabetic Nrf2-/- and Nrf2+/+ C57BL/6 mice as well as HLECs were treated with MGO and were stained for cell viability and cell death using viability/cytotoxicity assay kits (Biotium Inc., Hayward, CA) for 30 min. After washing twice with PBS, they subjected to fluorescent microscopic imaging (Nikon, Eclipse TE2000-U) with a green filter (450-490 nm) for viable cells and with a red filter (510-560 nm) for dead cells, respectively. This viability/cytotoxicity assay kit is consisted of calcein AM and ethidium homodimer-III (EthD). Calcein AM is a non-fluorescent substrate for cell-permeate esterase and is cleaved by intracellular esterases to form highly fluorescent calcein, which is retained in the live cells and imparts intense green color fluorescence. EthD, a membrane-impermeable fluorescent dye, undergoes a fluorescence enhancement upon binding with nucleic acids and imparts red color fluorescence. EthD is only able to enter into the dead cells, because the plasma membranes of those cells are compromised. The cytosolic ROS level in LECs treated with MGO was also determined by 2′,7′-dichlorodihydrofluorescein diacetate (H2-DCFH-AD) (Invitrogen, Carlsbad, CA) for 30 min at 20°C. The cells were washed twice with PBS and subjected to fluorescent microscopic imaging (Nikon, Eclipse TE2000-U, Tokyo, Japan) with a green filter (450-490 nm) [45, 46].
Calcium imaging
HLECs were loaded with 5 μM Fluo-3, AM with Pluronic® F-127 (Invitrogen, Carlsbad, CA) in DMEM containing 1.8 mM CaCl2 for 30 min at 37°C. Cells were then washed twice with Ca2+-free DMEM before imaging. 25 μM MGO was added to the cells in Ca2+-free DMEM and time-lapse confocal live imaging was conducted to assess the intracellular Ca2+ changes. Cells earlier exposed to MGO were then challenged with 10 mM caffeine to assess ER-Ca2+ content changes, if any, for 2 min. A second bolus of MGO (25 μM) was also added to these cells to check, if any, the intracellular Ca2+ changes for an additional 3 min. Experiments were performed in line-scan mode by using a Zeiss 410 confocal microscope (Carl Zeiss Inc., Thornwood, NY) and field-stimulating cells at 0.25 Hz (10 V for 10 ms), as described previously [47]. Fluo-3, AM was excited by light at 488 nm, and fluorescence was measured at wavelengths of >515 nm.
Western blotting
Cells were lysed with RIPA buffer (Cell Signaling Technology, Inc. Beverly, MA). Western blotting was performed with antibodies specific to immunoglobulin heavy-chain binding protein (BiP; BD Biosciences, Franklin Lakes, NJ), β-actin, PERK, p-PERK, eukaryotic translation initiation factor 2α (eIF2α), p-eIF2α, IRE1α, ATF4, ATF6, CHOP, Ero1-α, Ero1-Lβ, Nrf2, glutathione reductase (GR), catalase, Glo-1, Dnmt1, Dnmt3a, Dnmt3b, TET1 (Santa Cruz Biotechnology, Santa Cruz, CA), and GAPDH (Novus Biological, Littleton, CO) as described elsewhere [45, 46]. The specificity of each antibodies were validated in HLECs and specificity of Nrf2 was validated in LECs isolated from Nrf2-/- and Nrf2+/+ mice prior to conduct experiments. The intensity of each band was normalized to that of GAPDH, or β-actin and the data were presented as a relative intensity using the ImageJ analysis software [48].
RT-qPCR
Total RNA was isolated from the HLECs with Quick-RNA™ MicroPrep solution (Zymo Research Corporation, Orange, CA). The column purified total RNA was reverse transcribed by iScript™ Reverse Transcription Supermix for real-time PCR (Bio-Rad, Hercules, CA) and was analyzed by MiniOpticon™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using the SsoFast™ EvaGreen® supermix (Bio-Rad, Hercules, CA) as described elsewhere [45, 46]. The primer sequences were designed using the ProbeFinder software (Roche, Indianapolis, IN) and were synthesized commercially. Primers used for RT-qPCR are listed in Table S1. In addition, the Validated All-in-One™ qPCR primers for Nrf2 target genes were purchased from GeneCopoeia and catalogs numbers were listed in the Table S2. Each reaction was carried out in triplicate and three independent experiments were run. Standard curves were prepared to determine individual PCR amplification efficiencies by using a serial dilution of a reference sample and was included in each real-time run to correct for possible variations in product amplification. The relative copy numbers were obtained from the standard curve and were normalized to the values obtained for Gapdh or β-actin, the internal control. Data acquisition, analysis and PCR efficiencies were done using Bio-Rad CFX manager software (version 3.1).
Bisulfite genomic DNA sequencing
The genomic DNA from LECs of human clear and diabetic cataractous lenses as well as HLECs treated with 100 μM MGO, 1 μg/μL tunicamycin, 1 μM thapsigargin, and 10 μmoles/L 5-Aza were subjected to bisulfite conversion by EZ DNA Methylation-Direct™ kit (Zymo Research Corporation, Orange, CA). The bisulfite-modified DNA was amplified by bisulfite sequencing PCR using Platinum® PCR SuperMix High Fidelity (Invitrogen, Carlsbad, CA) with primers specific to human Keap1 promoter (Table S3). Then, the amplified PCR products were cleaned by gel extraction with Zymoclean™ Gel DNA recovery kit (Zymo Research Corporation, Orange, CA), then cloned into pCR®4-TOPO vector using TOPO TA Cloning® kit (Invitrogen, Carlsbad, CA). The recombinant plasmids were transformed into One Shot® TOP10 chemically competent E. coli (Invitrogen, Carlsbad, CA) using the regular chemical transformation method. Plasmid DNA were isolated from about 10 independent clones of each amplicon with PureLink™ Quick Plasmid Miniprep kit (Invitrogen, Carlsbad, CA) and then sequenced (High-Throughput DNA Sequencing and Genotyping Core Facility, University of Nebraska Medical Center, Omaha, NE) to determine the status of CpG methylation. Clones with an insert with >99.5% bisulfite conversion, i.e., non-methylated cytosine residues to thymine were included in this study, and the remaining was excluded. Then the sequenced data of each clone was analyzed for DNA methylation in the Keap1 promoter by BISMA software (http://biochem.jacobs-university.de/BDPC/BISMA/) using default filtering threshold settings [49].
Proteasomal degradation studies
HLECs were precultured with 2.5 μM and 5 μM MG-132 (Selleck Chemicals, Houston, TX), a proteasome protease inhibitor for 4 h, then, followed by a washout in regular medium, with or without 100 μM MGO for 24 h. The harvested cells were lysed using RIPA buffer (Cell Signaling Technology, Inc. Beverly, MA) and the proteins were analyzed by Western blotting using the antibodies specific to Nrf2, Dnmt1, Dnmt3a and Dnmt3b (Santa Cruz Biotechnology, Santa Cruz, CA).
Statistical analysis
The results were expressed as mean ± SD. Statistical significance between two groups were determined by student's t test and comparisons among multiple groups were determined by one-way ANOVA followed by post hoc least significant difference test using the SPSS (version 15.0) software (SPSS Inc., Chicago, IL). Values were considered statistically significant when p < 0.05.
Results
MGO mobilizes Ca2+ from ER of HLECs
First, we examined the Ca2+ mobilization from the ER of HLECs treated with MGO by time-lapse confocal laser scanning microscopy, because disturbances in the regulation of ER Ca2+ in parallel disturb the cellular events that are vitally associated with cell life and death through ER stress mediated activation of UPR. The spontaneous release of Ca2+ at random locations within the HLECs is significantly increased 21 sec after the addition of 25 μM MGO (Fig. 1). The elevation in spontaneous Ca2+ release continues for 42 to 65 sec. A second dose of 25 μM MGO added after the first dose is incapable of eliciting Ca2+ release from the ER. Further, removal of Ca2+ from the culture medium did not alter the ability of MGO to change spontaneous Ca2+ release, which confirms that MGO is eliciting Ca2+ release from the ER.
Fig. 1.

Mobilization of Ca2+ from ER in HLECs treated with MGO. Time-lapse confocal images of HLECs showing the mobilization of Ca2+ from ER, when challenged with 25 μM MGO for about 5 min. Red arrows indicate spontaneous release of Ca2+ from ER. Three independent experiments were performed and all of which were exhibited similar pattern of Ca2+ release from ER when challenged with MGO. In the graph, “a” denotes the 1st dose of MGO and “b” denotes the 2nd dose of MGO.
MGO induces cell death through ROS production in HLECs
We next investigated the toxic nature of MGO by studying the cell death and ROS production in HLECs. The HLECs cultured with varying concentrations of MGO for 16 h shows increased cell death (Fig. 2A) with significant ROS production (Fig. 2B). In addition, the LECs isolated from Nrf2-/- mice as well as Nrf2+/+ mice cultured with 1 μM MGO for 20 h shows greater ROS production in Nrf2-/- mouse LECs than that of Nrf2+/+ mouse LECs (Fig. 3A). Further, the cultured LECs of the Nrf2-/- mouse treated with varying concentrations of MGO for 20 h shows more enhanced cell death than cultured LECs of the Nrf2+/+ mouse. These results confirm that MGO is nearly 10 times more able to induce cell death in cultured LECs of Nrf2-/- mouse than the cell death induced by MGO in cultured LECs of Nrf2+/+ mouse (Fig. 3A–B). Similarly, diabetic Nrf2-/- mice shows significant cell death with limited viable cells than diabetic Nrf2+/+ mice (Fig. 3C–D). Diabetic Nrf2-/- mice developed cataract 3 weeks earlier than diabetic Nrf2+/+ mice, but more than 60% of Nrf2-/- mice died prior to develop cataract (Fig. 3E-F). In addition, the lens enucleated from these animals exhibited superficial lens opacity on the lens surface and showed significant death of LECs.
Fig. 2.

Cell death and ROS production in HLECs treated with MGO. A. Photomicrographs of Ethidium homodimer-III staining for cell death in HLECs treated with 0, 25, 50, 100 and 200 μM MGO for 16 h. Graph shows the percentage of cell death measured from the images of the HLECs treated with varying concentrations of MGO. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group. B. The representative fluorescent photomicrographs of H2-DCFH-AD staining for ROS production in HLECs treated with 0, 25, 50, 100 and 200 μM MGO for 16 h. Graph shows the total fluorescence intensity measured from the images of the HLECs (n = 30 in each group) treated with varying concentrations of MGO. Background fluorescence level was corrected. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group.
Fig. 3.

ROS production and cell death in Nrf2-/- and Nrf2+/+ mouse LECs treated with MGO for 20 h as well as in the lenses of diabetic Nrf2-/- and diabetic Nrf2+/+ mice. A. Photomicrographs of H2-DCFH-AD staining for ROS production, and Ethidium homodimer-III staining for cell death in Nrf2-/- mouse LECs treated with 1 μM MGO for 20 h. B. Comparison of cell death between Nrf2-/- and Nrf2+/+ mouse LECs treated with 1, 3, 10, 30, and 100 μM MGO for 20 h. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group. C. Photomicrographs of Calcein AM staining for viable live cells in the lenses of diabetic Nrf2+/+ mice and diabetic Nrf2-/- mice. D. Photomicrographs of Ethidium homodimer-III staining for cell death in the lenses of diabetic Nrf2+/+ mice and diabetic Nrf2-/- mice. E. Photograph of diabetic Nrf2+/+ mouse showing clear eye. F. Photograph of diabetic Nrf2-/- mouse showing cataractous eye. Diabetic Nrf2-/- mice developed cataract 3 weeks earlier than diabetic Nrf2+/+ mice, but more than 60% of Nrf2-/- mice died prior to develop cataract.
MGO induces ER stress and activates the chronic UPR pathway in HLECs
We next investigated MGO mediated induction of ER stress and activation of UPR in HLECs. The expression levels of UPR specific proteins were analyzed in cultured HLECs treated with 100 μM MGO for different times (Fig. 4A). MGO activates ER stress by phosphorylating PERK and eIF2α, and the ratios between p-PERK/PERK and p-eIF2α/eIF2α are significantly increased in HLECs treated with 100 μM MGO for 2-8 h and then decreased after 12 to 24 h. The level of IRE1α is also increased at 4-8 h, but the level of ATF6 is not changed (Fig. 4A). These results confirm that MGO activates UPR by phosphorylating PERK in HLECs.
Fig. 4.

Activation of ER stress mediated UPR and suppression of Nrf2 dependent antioxidant protection in HLECs treated with MGO. A. Immunoblot of p-PERK, PERK, p-elF2α, elF2α, IRE1α, and ATF6 in HLECs treated with 100 μM MGO for 2, 4, 8, 12, and 24 h. GAPDH was probed as a loading control. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group. B. Immunoblot of p-PERK, PERK, p-elF2α, elF2α, ATF6, BiP, ATF4, and CHOP in HLECs treated with varying concentrations of MGO for 24 h. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group. C. Immunoblot of Keap1, Nrf2, GR, catalase, Glo-1, Ero1-Lα, Ero1-Lβ, and PDI in HLECs treated with varying concentrations of MGO for 24 h. GAPDH was probed as a loading control. GAPDH shown in Fig. 4C is common to Fig. 4B, because both experiments are same grouping. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group.
The activation of UPR solely depends on the concentration and exposure duration of the ER stressors. It is well known that low levels or short exposure of ER stressors result in adaptive UPR by decreasing the level of unfolded proteins in the ER. However, chronic UPR ultimately results in the inability to restore cellular homeostasis, eventually triggering apoptosis [21]. To determine the type of UPR activation, HLECs were cultured with varying concentrations of MGO for 24 h. The protein blot analyses of HLECs treated with 1, 3, and 10 μM MGO for 24 h do not show any significant changes in the levels of phosphorylation of PERK, and eIF2α, whereas, the HLECs treated with 30 and 100 μM MGO for 24 h reveal increased levels of phosphorylation of PERK and eIF2α when compared with control HLECs (Fig. 4B). In addition, HLECs treated with 100 αM MGO for 24 h show a higher ATF4 protein expression level than that at lower concentrations of MGO (Fig. 4B). By contrast, the protein expression levels of BiP and CHOP are notably decreased in HLECs treated with MGO for 24 h (Fig. 4B). These results confirm the induction of ER stress and strong activation of UPR within 24 h by MGO in HLECs.
MGO acts on ER localized oxidative enzymes and suppresses the Nrf2 dependent antioxidant protection in HLECs
Next, we set to study the ER localized oxidative enzyme levels and suppression of Nrf2 dependent stress/antioxidant protection in HLECs treated with varying concentrations of MGO for 24 h. The ER redox homeostasis is routinely accomplished by a protein relay between ER localized oxidative enzymes Ero1-Lα, and Ero1-Lβ, and PDI [35, 36]. The level of Ero1-Lα is increased in HLECs treated with 1, 3, 10, and 30 μM MGO for 24 h compared to that of control HLECs (Fig. 4C). However, the Ero1-Lα is notably decreased in HLECs treated with 100 μM MGO for 24 h (Fig. 4C). By contrast, the level of Ero1-Lβ in HLECs treated with 100 μM MGO is decreased, whereas those levels remain the same in HLECs treated with 1, 3, 10, and 30 μM MGO as in control HLECs (Fig. 4C). The level of PDI is not changed in HLECs treated with MGO for 24 h (Fig. 4C). Further, Ero1-Lα and Ero1-Lβ exert their function by forming complexes as Ero1-Lα/PDI or Ero1-Lβ/PDI.
Subsequently, the protein expression levels of the stress/antioxidant proteins such as, Nrf2, and its target gene product, GR, are decreased significantly in HLECs treated with 1, 3, 10, 30, and 100 μM MGO for 24h than that of control HLECs (Fig. 4C). But the level of catalase is significantly decreased in HLECs treated with 100 μM MGO compared with that of control HLECs as well as HLECs treated with 1, 3, 10, and 30 μM MGO for 24 h (Fig. 4C). By contrast, the Keap1, a negative regulator for Nrf2, protein level is increased significantly in HLECs treated with 30 and 100 μM MGO (Fig. 4C). The protein level of Glo-1 in HLECs treated with varying concentrations of MGO for 24 h is same as in control HLECs (Fig. 4C), because the half-life of Glo-1 is more than 6 days [50].
MGO decreases passive DNA demethylation pathway enzymes and increases active DNA demethylation pathway enzymes in HLECs
Because, overproduction of ROS from oxidative/ER stress triggers the changes in DNA methylation patterns [51, 52], we assessed the status of passive and active DNA demethylation pathway enzymes in HLECs treated with 100 μM MGO for 24 h and the results obtained were comparable with the standard ER stressors, such as tunicamycin and thapsigargin. The RT-qPCR reveals significant upregulation of passive DNA demethylation pathway enzymes, such as Dnmt1 (2.1 fold), Dnmt3a (1.1 fold), and Dnmt3b (1.7 fold), as well as active DNA demethylation pathway enzyme, Tet1 (2.3 fold), in HLECs treated with 100 μM MGO for 24 h (Fig. 5A). Similarly, the mRNA expression levels of Dnmt1, Dnmt3a, Dnmt3b and Tet1 are significantly upregulated in HLECs treated with the well-known ER stressors such as, tunicamycin (Fig. 5C), and thapsigargin (Fig. 5E), respectively.
Fig. 5.

Altered expressions of passive and active DNA demethylation pathway enzymes in HLECs treated with MGO, tunicamycin and thapsigargin. RT-qPCR analysis of HLECs treated with 100 μM MGO for 1 day (A), 1 μg/μL tunicamycin for 1 day (C), 1 μM thapsigargin for 3 days (E), showing the fold change in the mRNA expressions of passive DNA demethylation pathway enzymes, Dnmt1, Dnmt3a, and Dnmt3b, and active DNA demethylation pathway enzyme, Tet1. Fold change in the gene expressions were normalized with the internal control, Gapdh and the data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group. Immunoblot of passive DNA demethylation pathway enzymes, Dnmt1, Dnmt3a, and Dnmt3b, and active DNA demethylation pathway enzyme, TET1 in HLECs treated with 100 μM MGO for 1 day (B), 1 μg/μL tunicamycin for 1 day (D), 1 μM thapsigargin for 3 days (F). GAPDH was probed as a loading control. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group.
However, the protein blot analyses of the expression profiles for passive DNA demethylation pathway enzymes, such as Dnmt1, Dnmt3a, and Dnmt3b show significant decreases whereas the active DNA demethylation pathway enzymes, such as TET1 shows notable increases in HLECs cultured with 100 μM for 24 h (Fig. 5B). Similar results were obtained for the standard ER stressor, tunicamycin (Fig. 5D). However, another ER stressor, thapsigargin treated HLECs shows significantly decreased levels of Dnmt1, and Dnmt3a with notable increase in Dnmt3b, the TET1 protein level is not significantly altered (Fig. 5F). Hence, the decreased levels of passive DNA demethylation pathway enzymes and increased or constant levels of the active DNA demethylation pathway enzymes in HLECs by MGO could effect on the methylation status of a gene leading to loss of DNA methylation.
MGO induces loss of Keap1 promoter DNA methylation in HLECs
To further explore the relationship between ER stress-mediated suppression of Nrf2 dependent antioxidant protection and altered expressions of active and passive DNA demethylation pathway enzymes, we examined the promoter DNA methylation status of Keap1 in HLECs treated with 100 μM MGO for 24 h. The reason for studying the promoter DNA methylation status of the Keap1 gene is that Keap1 is a negative regulator of Nrf2 protein and also, the promoter of Keap1 gene has a predominant CpG island (between -460 and +341) with a total number of 68 CpG dinucleotides. Any changes occurring in the Keap1 promoter DNA methylation ultimately affect the expression profiles of Nrf2 dependent antioxidant protection. It has been previously shown that the CpG dinucleotides found in the Keap1 promoter region (20 CpGs between -433 and -96) have primarily undergone loss of methylation [45].
Similarly, we investigated the loss of Keap1 promoter DNA methylation in the HLECs treated with MGO. Surprisingly, bisulfite genomic DNA sequencing studies confirm 28% loss of 5-mC in the Keap1 promoter (between -433 and -96) of HLECs treated with 100 μM MGO for 24 h (Fig. 6B) compared to control HLECs (Fig. 6A). Interestingly, the well-known ER stressors, tunicamycin (1 μg/μL; 1 day) and thapsigargin (1 μM; 3 days) lose 22 and 10% of 5-mC in the Keap1 promoter of HLECs, respectively (Fig. 6C-D). Likewise, HLECs treated with 10 μmoles/L 5-Aza, an inhibitor of Dnmt1 for 7 days shows a 65% loss of 5-mC in the same region of the Keap1 promoter (Fig. 6E). These results further confirm that MGO induces loss of Keap1 promoter DNA methylation in HLECs by altered expressions of either passive [38] or active DNA demethylation pathway enzymes [39]. Being an inhibitor of Dnmt1 [53], 5-Aza induces loss of Keap1 promoter DNA methylation by the replication dependent, passive DNA demethylation pathway. However, we could not find any notable methylation/demethylation patterns in the Nrf2 promoter in HLECs treated with these chemicals as well as in clear and diabetic cataractous lenses (Results not shown).
Fig. 6.
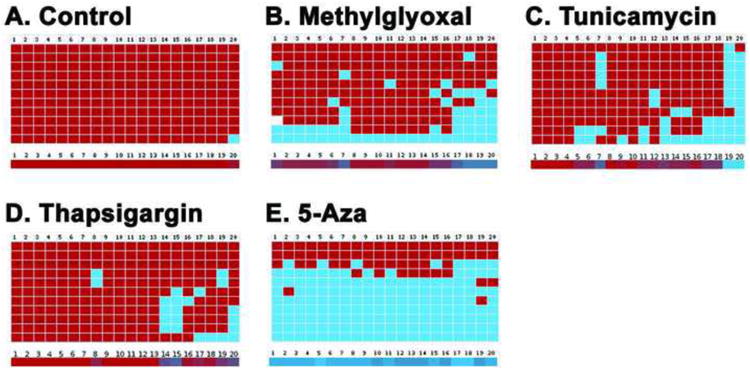
Loss of Keap1 promoter DNA methylation in HLECs treated with MGO, tunicamycin, thapsigargin, and 5-Aza. Bisulphite genomic DNA sequencing of control HLECs (A) showing highly methylated CpG dinucleotides whereas HLECs treated with 100 μM MGO for 1 day (B), 1 μg/μL tunicamycin for 1 day (C), 1 μM thapsigargin for 3 days (D), and 10 μmoles/L 5-Aza for 7 days (E) showing notably demethylated CpG dinucleotides in the region between -433 and -96 of Keap1 promoter. Each column represents a CpG dinucleotide site (1-20; there are 20 CpG dinucleotides) and each square indicates a CpG dinucleotide. Red squares represent methylated CpG dinucleotides. Blue squares represent unmethylated CpG dinucleotides, and white-square represents CpG status not determined. The color gradient bar (1-20) represents that red region contains more methylated CpG dinucleotides and blue region contains more unmethylated CpG dinucleotides.
MGO activates Keap1 overexpression by loss of Keap1 promoter DNA methylation, thereby suppressing the Nrf2 and its downstream target genes
To further confirm that the loss of Keap1 promoter DNA methylation results in the overexpression of Keap1, which eventually leads to the suppression of Nrf2 dependent antioxidant protection, we studied the mRNA expression profiles of Keap1 and Nrf2 in HLECs treated with 100 μM MGO for 24 h by RT-qPCR. Interestingly, the mRNA expression of Keap1 is increased 5 fold and Nrf2 is increased 2 fold in HLECs treated with 100 μM MGO for 24 h (Fig. 7A). Similarly, the mRNA expression of Keap1 and Nrf2 is increased in HLECs treated with the well-known ER stressors, tunicamycin (Fig. 7C) as well as thapsigargin (Fig. 7E), respectively. In the same way, the DNA demethylation agent, 5-Aza shows the increased expressions of Keap1 and Nrf2 mRNA in HLECs (Fig. 7G).
Fig. 7.

Over-expressed Keap1 protein suppresses the Nrf2 level in HLECs treated with MGO, tunicamycin, thapsigargin, and 5-Aza. RT-qPCR analyses of HLECs treated with 100 μM MGO for 1 day (A), 1 μg/μL tunicamycin for 1 day (C), 1 μM thapsigargin for 3 days (E), and 10 μmoles/L 5-Aza for 7 days (G) showing the increased expressions of Keap1 and Nrf2 mRNAs. Fold change in the gene expressions were normalized with the internal control, β-actin. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group. Immunoblot of Keap1 and Nrf2 in HLECs treated with 100 μM MGO for 1 day (B), 1 μg/μL tunicamycin for 1 day (D), 1 μM thapsigargin for 3 days (F), and 10 μmoles/L 5-Aza for 7 days (H) showing the increased level of Keap1 and decreased level of Nrf2. β-actin was probed as a loading control. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group.
We also examined the protein expression profiles of Keap1 and Nrf2 in HLECs treated with 100 μM MGO for 24 h as well as in HLECs treated with 1 μg/μL tunicamycin for 1 day, 1 μM thapsigargin for 3 days, and 10 μmoles/L 5-Aza for 7 days. The protein expression level of Keap1 is increased 1.8 fold and that of Nrf2 is decreased 0.4 fold in HLECs treated with MGO (Fig. 7B). Likewise, the protein expression level of Keap1 is increased and that of Nrf2 is decreased in HLECs treated with tunicamycin (Fig. 7D), thapsigargin (Fig. 7F), and 5-Aza (Fig. 7H). These results confirm that MGO mediated loss of Keap1 promoter DNA methylation significantly upregulates the transcription in HLECs. By contrast, the protein expression profile of Keap1 is increased, and Nrf2 is decreased in MGO as well as tunicamycin, thapsigargin, and 5-Aza treated HLECs. Assuming the rates of transcription and translation are approximately equal, the increased Keap1 protein must be responsible for decreasing the Nrf2 level by ubiquitin-mediated proteasomal degradation. Therefore, Nrf2 is degraded constantly and Keap1 is recycled or in part, taken up by autophagic clearance [54]. Further, decreased Nrf2 significantly downregulated mRNAs of its downstream target genes such as Gclc, Gclm, Nqo1, Srxn1, and TxrR1 in HLECs treated with 100 μM MGO for 24 h. But the mRNA expression of Glo-1 is unchanged (Fig. 8). This eventually results in the altered cellular redox balance, which ultimately causes lens oxidation and cataract formation.
Fig. 8.

Downregulation of Nrf2-target genes in HLECs treated with MGO. RT-qPCR analyses of HLECs treated with 100 μM MGO for 24 h showing the decreased expression of Nrf2-target genes, such as Gclc, Gclm, Nqo1, Srxn1, TxrR1, and Glo-1 mRNAs. Fold change in the gene expressions were normalized with the internal control, Gapdh. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group.
MGO augments the proteasomal degradation of Nrf2 protein and passive DNA demethylation pathway enzymes in HLECs
Similar to the binding of Nrf2 with Keap1 and Dnmt1 with acetylated histone, which result in ubiquitin-mediated proteasomal degradation, we expect that the protein degradation of Nrf2 (Fig. 7B), Dnmt1, Dnmt3a, and Dnmt3b (Fig. 5B) are due to proteasomal degradation, even though their mRNA levels are relatively high. To explore this, HLECs treated with 100 μM MGO for 24 h were pretreated with 2.5 and 5 μM MG-132, an inhibitor of proteasome protease, for 4 h. The protein expression profiles reveal that MGO treatment of HLECs enhances significant proteasomal degradation of Nrf2, Dnmt1, Dnmt3a, and Dnmt3b, but not the Keap1 and TET1 proteins (Fig. 9). However, MG-132 pretreated HLECs then cultured with 100 μM MGO for 24 h show significantly greater increases in the protein expressions of Nrf2, Keap1, Dnmt1, Dnmt3a, Dnmt3b, and TET1 than that of HLECs treated with MGO alone. These results further confirm that MGO stimulates degradation of Nrf2, Keap1, Dnmt1, Dnmt3a, and Dnmt3b proteins through activation of proteasomal degradation. Furthermore, MG-132 is reported to block the 5-Aza mediated promoter DNA demethylation by preventing the Dnmt1 degradation [55].
Fig. 9.

Prevention of MGO mediated protein degradation in HLECs by MG-132, a proteasome protease inhibitor. Immunoblot of Nrf2, Keap1, Dnmt1, Dnmt3a, Dnmt3b, and TET1 in HLECs precultured with 2.5 and 5 μM MG-132 for 4 h, followed by a washout in regular medium, with or without 100 μM MGO for 24 h. GAPDH was probed as a loading control. The data are presented as mean ± SD from three independent experiments. *p<0.05, vs control group.
ER stress not aging induces age-related cataract formation
Recently, we reported the loss of promoter DNA methylation in the Keap1 gene of human clear (n=15) and diabetic ARC (n=21) lenses [45]. Further, to analyze the methylation status of those promoter DNA sequences, we examined the association between the percentages of 5-mCs in the promoter of the Keap1 gene of human clear and diabetic ARC lenses as obtained from bisulfite genomic DNA sequencing with age (Fig. 10). This clearly shows linear age-related demethylation in the clear lenses, but drastic, clustered demethylation in the diabetic ARC lenses (Fig. 10). The Keap1 promoter DNA in clear lenses loses 5-mC at a rate of 1% per year i.e., the 99.5% of 5-mC in the Keap1 promoter at 17 years old is reduced to 60% at 86 years (Fig. 10). By contrast, the DNA of the Keap1 promoter in the diabetic ARC lenses has lost an average of 90% of the 5-mC methylation independent of age. These results suggest a notion that stressors are play a notable role in ARC formation, in addition with aging.
Fig. 10.

Association between age and Keap1 promoter DNA demethylation in human clear and diabetic cataractous lenses. The association between the percentages of methylated cytosines in the Keap1 gene of human clear (n=15) and diabetic cataractous (n=21) lenses as obtained from bisulfite genomic DNA sequencing (y-axis) with age (x-axis) is clearly showing linear age-related demethylation in the clear lenses, whereas drastic, clustered demethylation in the diabetic cataractous lenses. The diamond symbol indicates the clear lenses and the square symbol indicates the diabetic cataractous lenses. Horizontal black line is a borderline between clear and diabetic cataractous lenses.
Discussion
ARCs are multifactorial diseases induced by a variety of cataractogenic stresses with different pathogenesis. The consensus among lens researchers is that ARCs are closely associated with oxidation and aging of the lens [5, 17, 18]. Our studies have identified a general basic mechanism, which may be common among all types of cataract. We have previously reported that many cataractogenic stressors induce the ER stress mediated UPR in HLECs [46, 56]. In this report, we show that MGO mediated ER stress activates UPR, aggravates ROS production and cell death, suppresses the Nrf2 dependent antioxidant protection, and alters the active and passive DNA demethylation pathway enzymes. As an outcome, the Keap1 promoter DNA is demethylated in HLECs.
The concentration of MGO in the lenses is nearly 20-fold higher than that of blood samples from normal human subjects. Also, lenses are the prime site for protein modification by MGO. Further, the lenses of aged subjects have notably reduced capacity to detoxify MGO and are expected to be susceptible to the formation of MGO-modified proteins, especially lens crystallins and other proteins leading to formation of diabetic and senile cataract [57]. Moreover, the estimated rate of formation of methylglyoxal in the tissues of healthy individuals is 125 μM/day, whereas the serum MGO concentration is reported to be elevated up to several folds [58]. There are several reports about using supraphysiological concentrations of MGO to study its deleterious effects on various cells and tissues [59]. Further, ER stress is induced by a sum of concentration as well as duration of the stress. In our experiments, we used 100 μM MGO (supraphysiological level) with short duration (24 h), which is sufficient to induce deleterious effects. Further, the supraphysiological level of MGO (100 μM in HLECs) that we used in this study is significantly higher than that of MGO concentration (100nM) in the human lens [57]. Thus, the observed findings may be due to high concentration of MGO.
ER stress mediated activation of UPR by MGO
Hyperglycemia augments the generation of MGO in the lens, because the lens is located in a hypoxic environment [60-62]. This leads to accelerated formation and accumulation of MGO in the lens compared to other tissues and results in protein modification by glycoxidation. The MGO-modified proteins ultimately accumulate inside the cell thereby inducing ER stress in HLECs. Culturing the HLECs with 30 μM MGO for 24 h is sufficient to induce the ER stress, but 100 μM MGO is more optimal for studying the ER stress mediated activation of UPR and its consequent signalling relay. We found that MGO induces the ER stress by phosphorylating the ER transmembrane protein, PERK, into p-PERK, which is one of the strongest activations. Then, p-PERK phosphorylates eIF2α to form p-eIF2α as well as phosphorylating Nrf2 to p-Nrf2 [25, 26]. MGO also activates one of three unfolded protein sensors such as IRE-1α within 4 h, but these decreases-within 24 h. MGO does not activate the ATF6 protein, but induces Ca2+ release from the ER of HLECs
Loss of Nrf2 dependent antioxidant protection by MGO
Next, the p-Nrf2 dissociates from its negative regulatory protein, Keap1, and translocates into the nucleus, where it binds with the ARE elements in the promoter region of several antioxidant and stress associated genes to activate their transcription [29]. MGO also intensifies ROS production and cell death in HLECs, augmenting the Keap1 level. As a result, Nrf2, GR, and catalase levels are significantly decreased. Interestingly, the levels of ROS production associated enzymes, PDI, and Ero1-Lβ are constant in HLECs treated with varying concentrations of MGO for 24 h. This suggests that MGO mediated ROS production must be constant. However, the continued ROS production swamps the Nrf2 dependent antioxidant protection.
Further, lower doses of MGO significantly degrade the Nrf2 than those required to increase Keap1 protein expression or to induce PERK phosphorylation. However, this degradation is prevented by the presence of a proteasomal protease inhibitor, MG-132. These results strongly suggest that methylglyoxal induces the Nrf2 degradation. Currently, we don't know whether this degradation is induced by the modification of Nrf2 by MGO or translational suppression due to toxicity of MGO to the translational machinery. However, these effects appears likely that at least at lower doses of MGO is having either effects on Nrf2 degradation by the proteasome or Nrf2 translation, independent of either the ER stress or Keap1 protein.
ER-Ca2+ release and proteasomal degradation of Nrf2, Dnmt1, Dnmt3a, and Dnmt3b by MGO
The activation of UPR by MGO also induces Ca2+ release from the ER to the cytoplasm of HLECs, thereby increasing the cytosolic Ca2+ and up-regulating the m-calpain activity [63, 64]. These changes consecutively activate the multiple caspases including caspase-3 and caspase-1. m-calpain and caspases also known to cleave crystallin proteins [33, 46] leading to the formation of cataracts [65]. Nrf2 is one of the substrates for caspase-3 and has two caspase-3 cleavage consensus sites at D208 and D366 of its amino acid sequence [66]. This cleavage generates 30 and 50 kDa peptides, which are detected in HLECs treated with an ER stressor, homocysteine [46].
Increased accumulation of Ca2+ in the lens, and the influx of Ca2+ from the vitreous fluid due to the activation of Ca2+ dependent proteinases including m-calpain are reported in the majority of ARCs [63, 64]. Our results suggest that the cataractogenic stress induced ER stress activates the UPR, thereby increasing the release of Ca2+ from the ER, which is sufficient to activate m-calpain to cleave the lenticular proteins [46]. Further, Ca2+ release from the ER must be the initial event in the cascade, and repeated ER stress/UPR further releases the Ca2+ and induces damage in the lenses. These changes further upset the Ca2+ homeostasis and result in higher levels of Ca2+ and protein degradation in the lens. It is possible that high concentrations of MGO interact with N-terminal cysteines, which are contributing to ion channel activation as well as to the kinetic properties of ion channel responses [67].
Extensive studies on MGO-mediated modification of lens proteins in clear and cataractous lenses have revealed the formation of MGO adducts and AGEs, crystallin aggregation, functional alterations of chaperone proteins, and functional and conformational alterations of cellular proteins by proteasomal proteases [13, 14, 68]. Hyperglycemia-induced MGO formation also causes covalent modification of the 20S proteasome, which results in decreased chymotrypsin-like proteasome activity and reduced levels of the polyubiquitin receptor 19S-S5a [69]. Our results confirm that the proteasome protease inhibitor, MG-132, suppresses the MGO-induced loss of Keap1, Nrf2, Dnmt1, Dnmt3a, and Dnmt3b in HLECs by blocking their proteasomal degradation.
Altered levels of active and passive DNA demethylation pathway enzymes by MGO and Keap1 promoter DNA demethylation
DNA methylation in replicating LECs of the germinative zone can occur during lens differentiation under the action of DNA methyltransferases, Dnmt3a and Dnmt3b through de novo DNA methylation. This paternal methylation pattern in the daughter strand of hemi-methylated DNA is maintained by Dnmt1 [38]. Accordingly, the promoter CpG island of the Keap1 gene, that spans the region between -433 bp and -96 bp, is completely methylated in the HLEC line (SRA 01/04) as well as in a 17 year old clear human lens [45]. Being more susceptible to ER stressors, passive DNA demethylation in the Keap1 promoter can be initially induced in these replicating LECs by suppressing the Dnmts. Consequently, the lens fiber cells and LECs differentiated from these replicating LECs lose Keap1 promoter DNA methylation leading to lower levels of Nrf2. This is confirmed by culturing HLECs with 5-Aza, an inhibitor of Dnmt1, which significantly activates the Keap1 mRNA and protein by loss of Keap1 promoter DNA methylation. However, MGO suppresses the Dnmts and activates the TET1 and TDG in HLECs suggesting the involvement of active and passive DNA demethylation pathways in Keap1 promoter demethylation. These results further support an idea that loss of Keap1 promoter DNA methylation appears to be a primary event and then either translational arrest through UPR or proteasomal degradation can occur to lower the Nrf2 level. Consistent with our results, there is lot of literature correlating overexpression of Keap1 protein with decreased Nrf2. However, we are planning to conduct experiments using tagged Nrf2 and Keap1 in cell culture that MGO-mediated overexpression of Keap1 prevents Nrf2 from translocating to the nucleus in near future.
Loss of Keap1 promoter DNA methylation in human diabetic cataractous lenses
Our results demonstrate a unique feature in the loss of Keap1 promoter DNA methylation, which tends to be acquired in a gradual manner, i.e. loss of 1 % 5-mC per year, rather than an abrupt process in human clear lenses. By contrast, we found abrupt changes in the loss of Keap1 promoter DNA methylation, i.e. an average loss of 90% 5-mC in human diabetic cataractous lenses regardless of age, which results is aberrant gene modifications [37]. Similarly, cultured HLECs treated with MGO lose about 28% of the Keap1 promoter DNA methylation within 24 h. Persistent exposure of diabetic patients to MGO might also result in a loss of 5-mC in the Keap1 promoter within a short time. This may be associated with the severity of diabetic stress or the extent of tight control of the glycemic levels in each individual diabetic patient. Consequently, this may result in aberrant activation of Keap1 gene, which ultimately ends up with suppression of Nrf2 dependent cellular protection.
We found nearly 90% loss of Keap1 promoter DNA methylation in diabetic patients with all types of ARCs, which includes cortical, nuclear and posterior sub-capsular (PSC) cataracts. Diabetic ARCs are reported to be both cortical and PSC, however our results reveal that mixed types of cataracts are the most abundant with age above 70. Even though, these types of cataracts are thought to have different etiologies with different mechanism, our results reveal the same pattern of loss of Keap1 promoter DNA methylation (Table 1).
Table 1.
Different types diabetic cataracts and their Keap1 promoter methylation status.
| S. No. | Age | Gender | Type of cataract | % of Methylation | ||
|---|---|---|---|---|---|---|
|
| ||||||
| Cortical | Nuclear | PSC | ||||
| 1 | 48 | M | 2+ | 9.5 | ||
| 2 | 56 | F | 2+ | 12.0 | ||
| 3 | 59 | F | 2+ | 12.5 | ||
| 4 | 67 | M | 2+ | 7.5 | ||
| 5 | 68 | F | 2+ | 7.0 | ||
| 6 | 69 | M | 2+ | 2+ | 18.0 | |
| 7 | 70 | F | 2+ | 5.0 | ||
| 8 | 70 | M | 2+ | 6.0 | ||
| 9 | 72 | M | 2+ | 2+ | 12.5 | |
| 10 | 73 | M | 3+ | 3+ | 25.0 | |
| 11 | 73 | F | 2+ | 2+ | 2+ | 17.5 |
| 12 | 74 | F | 2+ | 2+ | 5.5 | |
| 13 | 76 | F | 2+ | 2+ | 2+ | 5.0 |
| 14 | 79 | F | 2+ | 2+ | 15.0 | |
| 15 | 79 | F | 2+ | 2+ | 5.5 | |
| 16 | 81 | M | 2+ | 21.5 | ||
| 17 | 81 | M | 3+ | 3+ | 4.5 | |
| 18 | 83 | F | 2+ | 20.5 | ||
| 19 | 83 | F | 2+ | 2+ | 15.5 | |
| 20 | 84 | M | 2+ | 2+ | 21.5 | |
| 21 | 86 | M | 3+ | 3+ | 7.5 | |
There are some reports demonstrating the promoter DNA hypermethylation of Keap1 gene in various cancerous cells and tissues [70-76], however, we found notable promoter DNA demethylation of Keap1 gene in human diabetic ARCs as well as HLECs treated with various ER stressors, including MGO. In addition, the human lens is the only organ in which cancer is never reported. Also, HLECs is considered to be a tumor cell line, because it is generated by transfection with plasmid vector DNA containing a large T antigen of SV40 [43]. Suitably, the promoter DNA of Keap1 gene in the HLECs is highly methylated, which is comparable to DNA methylation in various cancers [70-76]. Our next interest is to investigate how Keap1 promoter DNA methylation is induced in tumor cells and demethylation in diabetic cataractous lenses.
ARCs can be preventable
Currently ophthalmic researchers have little or no idea about the causes of ARCs in some individuals as young as age 40 while others have transparent lenses at their age 90. The prevalence of ARCs is most likely similar to our Keap1 promoter DNA demethylation results with human diabetic cataractous lenses. This suggests that lifestyle changes, systemic diseases, nutritional deficiencies, drugs, toxins aging may play a bigger role in the epigenetic changes through ROS production and UPR activation. The effort to prevent ARC formation has drifted towards prevention of secondary cataract.
Our bisulfite genomic DNA sequencing data on the human Keap1 gene suggest that if promoter DNA demethylation is avoided, ARCs can be preventable up to ages over 100. In addition, the Keap1-/- mouse dies postnatally [77], but the lung tissue specific Keap1-/- mouse has significantly enhanced antioxidant protection [78] suggesting that suppression of the Keap1 is potentially an excellent therapeutic approach for prevention of ARCs. In addition, avoidance of any cataractogenic stressors can be the best way of preventing ARCs in later life.
Overall, our studies suggest that MGO induces ER stress mediated activation of UPR, Ca2+ release from ER, overproduction of ROS and cell death, and loss of Keap1 promoter DNA methylation through altered levels of active and passive DNA demethylation pathway enzymes leading to failure of Nrf2 dependent antioxidant protection. These conditions result in a redox imbalance in the HLECs towards lens oxidation and cataract formation (Fig. 11).
Fig. 11.
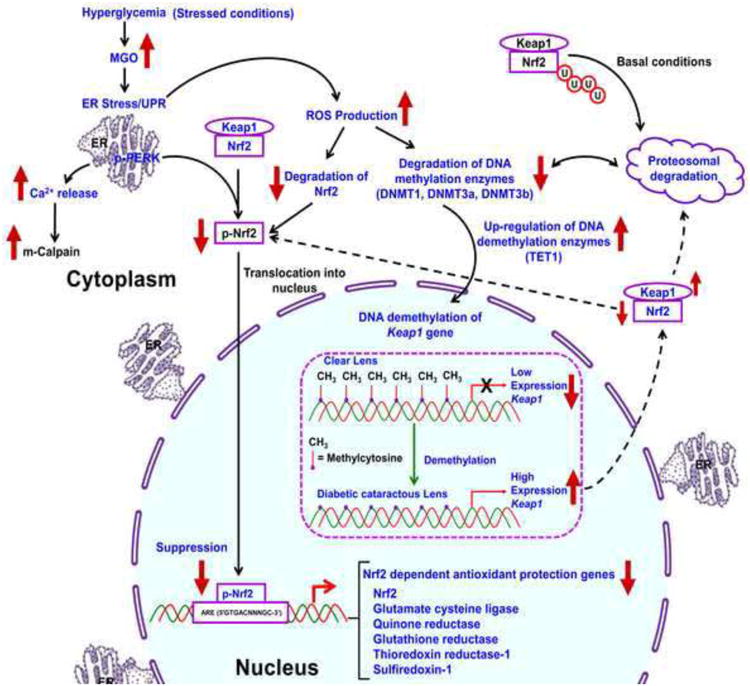
Schematic diagram for MGO mediated activation of ER stress, suppression of Nrf2 dependent antioxidant protection, demethylation of Keap1 promoter in LECs, and formation of cataract. Various age-dependent and environmental stresses induce ER stress. MGO is one of the diabetic stresses also induce ER stress. Severe or prolonged ER stress triggers an adaptive response known as UPR, which includes the early inhibition of protein synthesis with a later upregulation of genes that promote protein folding or disposal. Chronic UPR generates overproduction of ROS and results in oxidative stress. In response to oxidation, the Dnmts and Nrf2 are degraded by proteasomal degradation. Therefore, the decreased level of Dnmts can no longer methylate the DNA by maintenance mechanism and result in loss of DNA methylation. In addition, ER stressors stimulate the active DNA demethylation, by increasing the levels of TET1, AID, and TDG proteins, which further induce the loss of DNA methylation in the Keap1 promoter. Demethylated Keap1 promoter results in overexpression of the Keap1 gene and produces higher levels of Keap1 protein, which induces degradation of Nrf2 by ubiquitin-mediated proteasomal degradation. As a result, Nrf2 dependent stress protection is declined and the redox balance is altered towards lens oxidation leading to cataract formation. Our proven evidences are highlighted with blue color letters.
Supplementary Material
Highlights.
MGO induces ER stress and activates UPR in human lens epithelial cells
MGO demethylates the Keap1 promoter by altering DNA demethylation pathway enzymes
Over-expressed Keap1 suppresses Nrf2 dependent antioxidant protection
Loss of Keap1 promoter DNA methylation occurs in all type of diabetic ARCs
Acknowledgments
This work was supported in part by the RPB and EY0180172. We thank Janice A. Taylor and James R. Talaska of the Confocal Laser Scanning Microscope Core Facility at the University of Nebraska Medical Center for providing assistance with confocal microscopy and the Nebraska Research Initiative and the Eppley Cancer Center for their support of the Core Facility, and the UNMC DNA Sequencing Core Facility (supported by P20 RR016469 from the INBRE Program of the National Center for Research Resources) for sequencing analysis. We also thank the National Disease Research Interchange (NDRI), Philadelphia, PA for providing human lenses.
Abbreviations
- H2-DCFH-AD
2′,7′-dichlorodihydrofluorescein diacetate
- 5-Aza
5-Aza-2 -deoxycytidine
- 5mC
5-methylcytosine
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor 6
- AID
activation-induced deaminase
- AGEs
advanced glycation end-products
- ARCs
age-related cataracts
- ARE
antioxidant response element
- CHOP
C/EBP-homologous protein
- Dnmt1
DNA methyltransferase 1
- eIF2α
eukaryotic translation initiation factor 2α
- ER
endoplasmic reticulum
- Ero1
ER oxidoreductin 1
- GR
glutathione reductase
- Glo-1
glyoxalase 1
- HLECs
human lens epithelial cells
- IRE1α
inositol-requiring enzyme 1α
- Keap1
Kelch-like ECH-associated protein 1
- LECs
lens epithelial cells
- MGO
methylglyoxal
- Nrf2
nuclear factor-erythroid-2-related factor 2
- PERK
PKR-like endoplasmic reticulum kinase
- PSC
posterior sub-capsular cataract
- PDI
protein disulfide isomerase
- ROS
reactive oxygen species
- RT-qPCR
real-time quantitative PCR
- TET1
ten-eleven-translocation 1
- TDG
thymine DNA glycosylase
- UPR
unfolded protein response
Footnotes
Author Disclosure Statement: No competing financial interests exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pollreisz A, Schmidt-Erfurth U. Diabetic cataract-pathogenesis, epidemiology and treatment. J Ophthalmol. 2010;2010:608751. doi: 10.1155/2010/608751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falck A, Laatikainen L. Diabetic cataract in children. Acta Ophthalmol Scand. 1998;76:238–240. doi: 10.1034/j.1600-0420.1998.760223.x. [DOI] [PubMed] [Google Scholar]
- 3.Kim SI, Kim SJ. Prevalence and risk factors for cataracts in persons with type 2 diabetes mellitus. Korean J Ophthalmol. 2006;20:201–204. doi: 10.3341/kjo.2006.20.4.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyselova Z, Stefek M, Bauer V. Pharmacological prevention of diabetic cataract. J Diabetes Complications. 2004;18:129–140. doi: 10.1016/S1056-8727(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 5.Lou MF. Redox regulation in the lens. Prog Retin Eye Res. 2003;22:657–682. doi: 10.1016/s1350-9462(03)00050-8. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes JD, Russell SL, Illingworth CD, Duncan G, Wormstone IM. Regional differences in store-operated Ca2+ entry in the epithelium of the intact human lens. Invest Ophthalmol Vis Sci. 2009;50:4330–4336. doi: 10.1167/iovs.08-3222. [DOI] [PubMed] [Google Scholar]
- 7.Biswas S, Harris F, Dennison S, Singh JP, Phoenix D. Calpains: enzymes of vision? Med Sci Monit. 2005;11:RA301–310. [PubMed] [Google Scholar]
- 8.Li WC, Spector A. Lens epithelial cell apoptosis is an early event in the development of UVB-induced cataract. Free Radic Biol Med. 1996;20:301–311. doi: 10.1016/0891-5849(96)02050-3. [DOI] [PubMed] [Google Scholar]
- 9.Kohnen T, Koch DD, Font RL. Lensification of the posterior corneal surface. An unusual proliferation of lens epithelial cells. Ophthalmology. 1997;104:1343–1347. doi: 10.1016/s0161-6420(97)30137-7. [DOI] [PubMed] [Google Scholar]
- 10.Negahban K, Chern K. Cataracts associated with systemic disorders and syndromes. Curr Opin Ophthalmol. 2002;13:419–422. doi: 10.1097/00055735-200212000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Thornalley PJ, Langborg A, Minhas HS. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem J. 1999;344 Pt 1:109–116. [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips SA, Thornalley PJ. The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur J Biochem. 1993;212:101–105. doi: 10.1111/j.1432-1033.1993.tb17638.x. [DOI] [PubMed] [Google Scholar]
- 13.Ahmed N, Thornalley PJ, Dawczynski J, Franke S, Strobel J, Stein G, Haik GM. Methylglyoxal-derived hydroimidazolone advanced glycation end-products of human lens proteins. Invest Ophthalmol Vis Sci. 2003;44:5287–5292. doi: 10.1167/iovs.03-0573. [DOI] [PubMed] [Google Scholar]
- 14.Derham BK, Harding JJ. Effects of modifications of alpha-crystallin on its chaperone and other properties. Biochem J. 2002;364:711–717. doi: 10.1042/BJ20011512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seidler NW, Yeargans GS, Morgan TG. Carnosine disaggregates glycated alpha-crystallin: an in vitro study. Arch Biochem Biophys. 2004;427:110–115. doi: 10.1016/j.abb.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Thornalley PJ, Waris S, Fleming T, Santarius T, Larkin SJ, Winklhofer-Roob BM, Stratton MR, Rabbani N. Imidazopurinones are markers of physiological genomic damage linked to DNA instability and glyoxalase 1-associated tumour multidrug resistance. Nucleic Acids Res. 2010;38:5432–5442. doi: 10.1093/nar/gkq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennan LA, Kantorow M. Mitochondrial function and redox control in the aging eye: role of MsrA and other repair systems in cataract and macular degenerations. Exp Eye Res. 2009;88:195–203. doi: 10.1016/j.exer.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baynes JW, Thorpe SR. Role of oxidative stress in diabetic complications: a new perspective on an old paradigm. Diabetes. 1999;48:1–9. doi: 10.2337/diabetes.48.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146:743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Ishigaki Y, Yamada T, Kondo K, Yamaguchi S, Imai J, Uno K, Hasegawa Y, Sawada S, Ishihara H, Oyadomari S, Mori M, Oka Y, Katagiri H. Involvement of endoplasmic stress protein C/EBP homologous protein in arteriosclerosis acceleration with augmented biological stress responses. Circulation. 2011;124:830–839. doi: 10.1161/CIRCULATIONAHA.110.014050. [DOI] [PubMed] [Google Scholar]
- 21.Papa FR. Endoplasmic reticulum stress, pancreatic beta-cell degeneration, and diabetes. Cold Spring Harb Perspect Med. 2012;2:a007666. doi: 10.1101/cshperspect.a007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Back SH, Kaufman RJ. Endoplasmic reticulum stress and type 2 diabetes. Annu Rev Biochem. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tinhofer I, Anether G, Senfter M, Pfaller K, Bernhard D, Hara M, Greil R. Stressful death of T-ALL tumor cells after treatment with the anti-tumor agent Tetrocarcin-A. FASEB J. 2002;16:1295–1297. doi: 10.1096/fj.02-0020fje. [DOI] [PubMed] [Google Scholar]
- 24.Xie Q, Khaoustov VI, Chung CC, Sohn J, Krishnan B, Lewis DE, Yoffe B. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology. 2002;36:592–601. doi: 10.1053/jhep.2002.35441. [DOI] [PubMed] [Google Scholar]
- 25.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 26.Cullinan SB, Diehl JA. Coordination of ER and oxidative stress signaling: the PERK/Nrf2 signaling pathway. Int J Biochem Cell Biol. 2006;38:317–332. doi: 10.1016/j.biocel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- 28.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 29.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 30.So HS, Kim HJ, Lee JH, Park SY, Park C, Kim YH, Kim JK, Lee KM, Kim KS, Chung SY, Jang WC, Moon SK, Chung HT, Park RK. Flunarizine induces Nrf2-mediated transcriptional activation of heme oxygenase-1 in protection of auditory cells from cisplatin. Cell Death Differ. 2006;13:1763–1775. doi: 10.1038/sj.cdd.4401863. [DOI] [PubMed] [Google Scholar]
- 31.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Xue M, Rabbani N, Momiji H, Imbasi P, Anwar MM, Kitteringham N, Park BK, Souma T, Moriguchi T, Yamamoto M, Thornalley PJ. Transcriptional control of glyoxalase 1 by Nrf2 provides a stress-responsive defence against dicarbonyl glycation. Biochem J. 2012;443:213–222. doi: 10.1042/BJ20111648. [DOI] [PubMed] [Google Scholar]
- 33.Azuma M, Fukiage C, David LL, Shearer TR. Activation of calpain in lens: a review and proposed mechanism. Exp Eye Res. 1997;64:529–538. doi: 10.1006/exer.1996.0234. [DOI] [PubMed] [Google Scholar]
- 34.Higa A, Chevet E. Redox signaling loops in the unfolded protein response. Cell Signal. 2012;24:1548–1555. doi: 10.1016/j.cellsig.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Tu BP, Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 36.Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R. Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J Biol Chem. 2000;275:23685–23692. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 37.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132:2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- 38.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 39.Nabel CS, Kohli RM. Molecular biology. Demystifying DNA demethylation. Science. 2011;333:1229–1230. doi: 10.1126/science.1211917. [DOI] [PubMed] [Google Scholar]
- 40.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibaraki N, Chen SC, Lin LR, Okamoto H, Pipas JM, Reddy VN. Human lens epithelial cell line. Exp Eye Res. 1998;67:577–585. doi: 10.1006/exer.1998.0551. [DOI] [PubMed] [Google Scholar]
- 44.Elanchezhian R, Palsamy P, Madson CJ, Mulhern ML, Lynch DW, Troia AM, Usukura J, Shinohara T. Low glucose under hypoxic conditions induces unfolded protein response and produces reactive oxygen species in lens epithelial cells. Cell Death Dis. 2012;3:e301. doi: 10.1038/cddis.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palsamy P, Ayaki M, Elanchezhian R, Shinohara T. Promoter demethylation of Keap1 gene in human diabetic cataractous lenses. Biochem Biophys Res Commun. 2012;423:542–548. doi: 10.1016/j.bbrc.2012.05.164. [DOI] [PubMed] [Google Scholar]
- 46.Elanchezhian R, Palsamy P, Madson CJ, Lynch DW, Shinohara T. Age-related cataracts: homocysteine coupled endoplasmic reticulum stress and suppression of Nrf2-dependent antioxidant protection. Chem Biol Interact. 2012;200:1–10. doi: 10.1016/j.cbi.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao CH, Wehrens XH, Wyatt TA, Parbhu S, Rozanski GJ, Patel KP, Bidasee KR. Exercise training during diabetes attenuates cardiac ryanodine receptor dysregulation. J Appl Physiol (1985) 2009;106:1280–1292. doi: 10.1152/japplphysiol.91280.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 49.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA--fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frickel EM, Jemth P, Widersten M, Mannervik B. Yeast glyoxalase I is a monomeric enzyme with two active sites. J Biol Chem. 2001;276:1845–1849. doi: 10.1074/jbc.M005760200. [DOI] [PubMed] [Google Scholar]
- 51.Turk PW, Laayoun A, Smith SS, Weitzman SA. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis. 1995;16:1253–1255. doi: 10.1093/carcin/16.5.1253. [DOI] [PubMed] [Google Scholar]
- 52.Weitzman SA, Turk PW, Milkowski DH, Kozlowski K. Free radical adducts induce alterations in DNA cytosine methylation. Proc Natl Acad Sci U S A. 1994;91:1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 54.Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, Zhu M, Zhong Q. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6:614–621. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikesugi K, Yamamoto R, Mulhern ML, Shinohara T. Role of the unfolded protein response (UPR) in cataract formation. Exp Eye Res. 2006;83:508–516. doi: 10.1016/j.exer.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 57.Haik GM, Lo TW, Jr, Thornalley PJ. Methylglyoxal concentration and glyoxalase activities in the human lens. Exp Eye Res. 1994;59:497–500. doi: 10.1006/exer.1994.1135. [DOI] [PubMed] [Google Scholar]
- 58.McLellan AC, Thornalley PJ, Benn J, Sonksen PH. Glyoxalase system in clinical diabetes mellitus and correlation with diabetic complications. Clin Sci. 1994;87:21–29. doi: 10.1042/cs0870021. [DOI] [PubMed] [Google Scholar]
- 59.Chang KC, Paek KS, Kim HJ, Lee YS, Yabe-Nishimura C, Seo HG. Substrate-induced up-regulation of aldose reductase by methylglyoxal, a reactive oxoaldehyde elevated in diabetes. Mol Pharmacol. 2002;61:1184–1191. doi: 10.1124/mol.61.5.1184. [DOI] [PubMed] [Google Scholar]
- 60.Shui YB, Fu JJ, Garcia C, Dattilo LK, Rajagopal R, McMillan S, Mak G, Holekamp NM, Lewis A, Beebe DC. Oxygen distribution in the rabbit eye and oxygen consumption by the lens. Invest Ophthalmol Vis Sci. 2006;47:1571–1580. doi: 10.1167/iovs.05-1475. [DOI] [PubMed] [Google Scholar]
- 61.Holekamp NM, Shui YB, Beebe DC. Vitrectomy surgery increases oxygen exposure to the lens: a possible mechanism for nuclear cataract formation. Am J Ophthalmol. 2005;139:302–310. doi: 10.1016/j.ajo.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 62.Barbazetto IA, Liang J, Chang S, Zheng L, Spector A, Dillon JP. Oxygen tension in the rabbit lens and vitreous before and after vitrectomy. Exp Eye Res. 2004;78:917–924. doi: 10.1016/j.exer.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Morishima N, Nakanishi K, Takenouchi H, Shibata T, Yasuhiko Y. An endoplasmic reticulum stress-specific caspase cascade in apoptosis. Cytochrome c-independent activation of caspase-9 by caspase-12. J Biol Chem. 2002;277:34287–34294. doi: 10.1074/jbc.M204973200. [DOI] [PubMed] [Google Scholar]
- 64.Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 65.Shih M, David LL, Lampi KJ, Ma H, Fukiage C, Azuma M, Shearer TR. Proteolysis by m-calpain enhances in vitro light scattering by crystallins from human and bovine lenses. Curr Eye Res. 2001;22:458–469. doi: 10.1076/ceyr.22.6.458.5483. [DOI] [PubMed] [Google Scholar]
- 66.Ohtsubo T, Kamada S, Mikami T, Murakami H, Tsujimoto Y. Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases. Cell Death Differ. 1999;6:865–872. doi: 10.1038/sj.cdd.4400566. [DOI] [PubMed] [Google Scholar]
- 67.Eberhardt MJ, Filipovic MR, Leffler A, de la Roche J, Kistner K, Fischer MJ, Fleming T, Zimmermann K, Ivanovic-Burmazovic I, Nawroth PP, Bierhaus A, Reeh PW, Sauer SK. Methylglyoxal activates nociceptors through transient receptor potential channel A1 (TRPA1): a possible mechanism of metabolic neuropathies. J Biol Chem. 2012;287:28291–28306. doi: 10.1074/jbc.M111.328674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shamsi FA, Lin K, Sady C, Nagaraj RH. Methylglyoxal-derived modifications in lens aging and cataract formation. Invest Ophthalmol Vis Sci. 1998;39:2355–2364. [PubMed] [Google Scholar]
- 69.Queisser MA, Yao D, Geisler S, Hammes HP, Lochnit G, Schleicher ED, Brownlee M, Preissner KT. Hyperglycemia impairs proteasome function by methylglyoxal. Diabetes. 2010;59:670–678. doi: 10.2337/db08-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbano R, Muscarella LA, Pasculli B, Valori VM, Fontana A, Coco M, la Torre A, Balsamo T, Poeta ML, Marangi GF, Maiello E, Castelvetere M, Pellegrini F, Murgo R, Fazio VM, Parrella P. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics. 2013;8:105–112. doi: 10.4161/epi.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo D, Wu B, Yan J, Li X, Sun H, Zhou D. A possible gene silencing mechanism: hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem Biophys Res Commun. 2012;428:80–85. doi: 10.1016/j.bbrc.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 72.Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, Ishiguro A, Kijima H, Mimura J, Itoh K, Fukuda S, Saijo Y. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC cancer. 2012;12:66. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muscarella LA, Barbano R, D'Angelo V, Copetti M, Coco M, Balsamo T, la Torre A, Notarangelo A, Troiano M, Parisi S, Icolaro N, Catapano D, Valori VM, Pellegrini F, Merla G, Carella M, Fazio VM, Parrella P. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient's outcome. Epigenetics. 2011;6:317–325. doi: 10.4161/epi.6.3.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muscarella LA, Parrella P, D'Alessandro V, la Torre A, Barbano R, Fontana A, Tancredi A, Guarnieri V, Balsamo T, Coco M, Copetti M, Pellegrini F, De Bonis P, Bisceglia M, Scaramuzzi G, Maiello E, Valori VM, Merla G, Vendemiale G, Fazio VM. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics. 2011;6:710–719. doi: 10.4161/epi.6.6.15773. [DOI] [PubMed] [Google Scholar]
- 75.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 76.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 78.Blake DJ, Singh A, Kombairaju P, Malhotra D, Mariani TJ, Tuder RM, Gabrielson E, Biswal S. Deletion of Keap1 in the lung attenuates acute cigarette smoke-induced oxidative stress and inflammation. Am J Respir Cell Mol Biol. 2010;42:524–536. doi: 10.1165/rcmb.2009-0054OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


