Abstract
The Systems Biology for Infectious Diseases Research program was established by the U.S. National Institute of Allergy and Infectious Diseases to investigate host-pathogen interactions at a systems level. This program generated 47 transcriptomic and proteomic datasets from 30 studies that investigate in vivo and in vitro host responses to viral infections. Human pathogens in the Orthomyxoviridae and Coronaviridae families, especially pandemic H1N1 and avian H5N1 influenza A viruses and severe acute respiratory syndrome coronavirus (SARS-CoV), were investigated. Study validation was demonstrated via experimental quality control measures and meta-analysis of independent experiments performed under similar conditions. Primary assay results are archived at the GEO and PeptideAtlas public repositories, while processed statistical results together with standardized metadata are publically available at the Influenza Research Database (www.fludb.org) and the Virus Pathogen Resource (www.viprbrc.org). By comparing data from mutant versus wild-type virus and host strains, RNA versus protein differential expression, and infection with genetically similar strains, these data can be used to further investigate genetic and physiological determinants of host responses to viral infection.
Background & Summary
With the recognition that host responses to pathogen infection play key roles in disease severity and mortality, virologists have shifted toward integrated systems biology approaches to identify therapeutics that target host pathways1,2. To support the cross-disciplinary approaches necessary to address a systems-level analysis, the Division of Microbiology and Infectious Diseases (DMID) at the National Institute of Allergy and Infectious Disease (NIAID) established the Systems Biology for Infectious Diseases Research (SysBio) program that provided support for four centers from 2008–2013: Systems Influenza, Systems Virology, Systems Biology of Enteropathogens, and Mycobacterium tuberculosis Systems Biology3. The Systems Influenza and Systems Virology centers focused on elucidating the mechanisms of how viral regulation of the host cellular circuitry contributes to viral replication and disease severity, thereby elucidating host pathways that could serve as potential new therapeutic targets.
Several publications have reported the findings from these virology-focused SysBio centers. Overall characterization of the host responses demonstrated that pro-inflammatory interferon (IFN) signalling pathways were enriched following infection with either influenza A or Betacoronavirus infection in vitro4,5 and in vivo6,7. These studies characterized several aspects of viral-host interactions that influence disease severity. Experiments designed to tease out the contribution of the virus and the host to the severity of host response reported that specific mutations in the H5N1 viral genome affected the kinetics and magnitude of the host response, rather than changes in specific host response factors8. Another study found that early response signatures to influenza virus infection correlated with the severity of late disease9. Lipidomics analysis of mice infected with various influenza A virus strains of varying pathogenicity have also been reported10. Similarly, the disease severity of SARS-CoV correlated with the dysregulation of the urokinase pathway and an increase in fibrinolysin pathway activity7. An integrated network interrogation approach using both transcriptomic and proteomic data sets predicted a subset of key regulator molecules responding to SARS-CoV and influenza infections that could be used as host targets for therapeutic intervention11,12. Genetic modification of non-structural protein 16 in SARS-CoV showed enhanced susceptibility to type I and III interferon responses, making it a good vaccine strain candidate13.
To support the broad dissemination of research datasets from these SysBio centers and other supported programs, DMID established the Bioinformatics Resource Centers for Infectious Diseases (BRC) program to provide public database and analysis resources for the infectious disease research community. A data dissemination working group, consisting of representatives from NIAID, the SysBio centers, and the BRCs was established to develop a data management and release approach that would maximize the reuse potential of the generated data. The working group leveraged existing public archiving strategies to store the primary assay results and devised metadata standards to represent the experiment descriptions and interpreted results in a consistent and standardized way. The final derived host response data and associated structured standardized metadata were made publically available at the BRC appropriate for the system under study. As the virus-centric BRCs, the Influenza Research Database (IRD, www.fludb.org)14 and the Virus Pathogen Database and Analysis Resource (ViPR, www.viprbrc.org)15 host data produced by the Systems Influenza and Systems Virology centers.
Here we describe 47 transcriptomic and proteomic datasets from 30 studies generated by these SysBio centers and made available through public resources. These studies characterized the host response to infection by members of the Orthomyxoviridae and Coronaviridae virus families, including pandemic influenza A H1N1 virus, highly pathogenic H5N1 avian influenza (HPAI) virus, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV); all of which are capable of causing severe respiratory infections and pose significant threats to humans on a global scale4,16. Host responses were evaluated in either in vivo (mouse) or in vitro (human cell line) model systems using three major experimental designs: longitudinal time-course, dose response, and genetic modification (involving both genetically manipulated viruses and hosts) comparisons. Host responses to these manipulated variables were assessed using transcriptomic (gene expression microarray) and proteomic (mass spectrometry) assays. Together, this collection represents the first coordinated effort to create a systems level description of host-pathogen interactions using multiple viral strains, host models, and ‘-omics’ technologies.
Methods
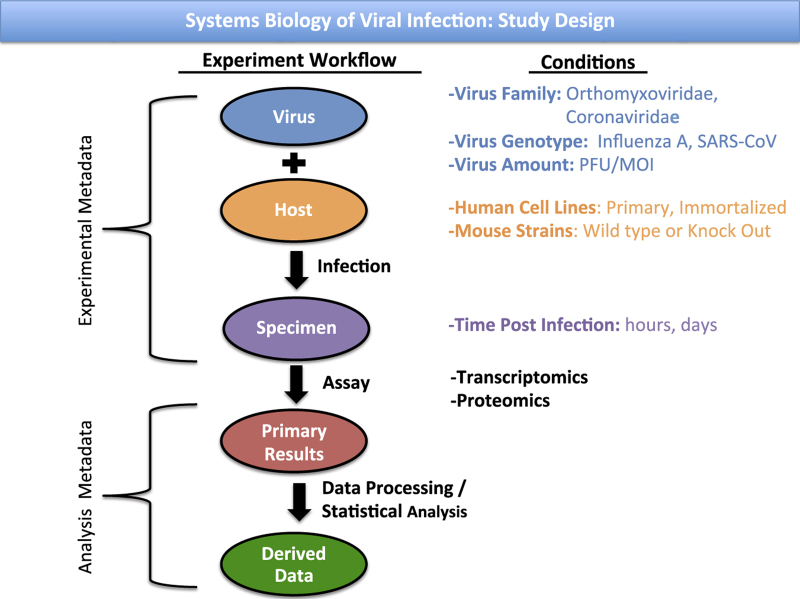
The experimental designs reported here include longitudinal (i.e. time course), dose response, and genetic modification of both virus and host strain comparisons. The intent of these designs was to assemble a unified view of the virulence mechanisms and replication strategies resulting from viral regulation of host cellular processes. For the present experiments, either transcriptomic (using gene expression microarrays) or proteomic (using liquid chromatography coupled with mass spectrometry) methodologies were used to collect the reported data. The materials used, sample preparation protocols, validation procedures, data processing, and hypothesis testing that were performed are described below. Throughout the Methods section, Study IDs are appended to indicate which studies used a given method. A summary of the overall experiment workflow and the experiment factor conditions that were compared is presented in Fig. 1. The relationship between the experiment workflow and the experimental metadata, analysis metadata, primary assay results, and derived data is also presented. Study designs for all experiments, including relevant repository identifiers, are summarized in Table 1 (available online only), with individual RNA experiment samples detailed in Table 2 (available online only) and individual protein experiment samples detailed in Table 3 (available online only). Sample tracking from animal subjects to experiment samples is given in Table 4 (available online only).
Figure 1. Overall study design for the study of the systems biology of viral infection.
The ovals down the center represent steps in the experimental workflow common to all datasets. Text on the left gives the type of metadata required to describe the components of the workflow. On the right lists the experiment conditions investigated at each step throughout these datasets.
Table 1. Study and experiment level metadata.
| Study | Study Name | Experiment ID | Analyte Type | Treatment Agent | Dose | Time Post Administration | Study Design | Host Species | Strain/Line | Sample Source | Institution | Principle Investigator | Primary Public Repository | Primary Archive Accession | BRC | BRC ACCESSION |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA04M001 | CA04M001 :A/CA/04/09 (H1N1) infection in C57BL6 mice with variable doses and times post infection. | CA04M001-P | Protein Quantification | A/California/04/2009 (H1N1) | 1.0E04, 1.0E05 PFU | 1,2,4,7 days | Dose Response, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00423 | IRD/ViPR | IRD_SV_CA04M001-P |
| CA04M001 | CA04M001 :A/CA/04/09 (H1N1) infection in C57BL6 mice with variable doses and times post infection. | CA04M001-R | Transcript Quantification | A/California/04/2009 (H1N1) | 1.0E03, 1.0E04, 1.0E05, 1.0E06 PFU | 1,2,4,7 days | Dose Response, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE37569 | IRD/ViPR | IRD_SV_CA04M001-R |
| ECL001 | ECL001: MERS-CoV infection in Calu3 cells: A time course | ECL001-R | Transcript Quantification | MERS-CoV | 5 MOI | 0,3,7,12,18,24 h | Longitudinal | Human | Calu3 cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE45042 | IRD/ViPR | IRD_SV_ECL001-R |
| ICL004 | ICL004: A/Vietnam/1203/2004 (H5N1) infection in Calu3 cell: A time course | ICL004-P | Protein Quantification | A/Vietnam/1203/2004 (H5N1) | 1 MOI | 0/3,7,12,18/24 h | Longitudinal | Human | Calu3 cells | Cell Line | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00424 | IRD/ViPR | IRD_SV_ICL004-P |
| ICL004 | ICL004: A/Vietnam/1203/2004 (H5N1) infection in Calu3 cell: A time course | ICL004-R | Transcript Quantification | A/Vietnam/1203/2004 (H5N1) | 1 MOI | 0,12,18,24,3,7 h | Longitudinal | Human | Calu3 cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE28166 | IRD/ViPR | IRD_SV_ICL004-R |
| ICL006 | ICL006: A/CA/04/09 (H1N1) infection in Calu3 cell: A time course | ICL006-P | Protein Quantification | A/California/04/2009 (H1N1) | 3 MOI | 0,3,7,12,18,24,30,36,48 h | Longitudinal | Human | Calu3 cells | Cell Line | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00425 | IRD/ViPR | IRD_SV_ICL006-P |
| ICL006 | ICL006: A/CA/04/09 (H1N1) infection in Calu3 cell: A time course | ICL006-R | Transcript Quantification | A/California/04/2009 (H1N1) | 3 MOI | 0,12,18,24,3,30,36,48,7 h | Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE37571 | IRD/ViPR | IRD_SV_ICL006-R |
| ICL010 | ICL010: A/Netherlands/602/2009 (H1N1) and A/CA/04/2009 (H1N1) infection in Calu3 cells: A time course | ICL010-P | Protein Quantification | A/Netherlands/602/2009 (H1N1) | 3 MOI | 0,3,7,12,18,24,30,36,48 h | Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00426 | IRD/ViPR | IRD_SV_ICL010-P |
| ICL010 | ICL010: A/Netherlands/602/2009 (H1N1) and A/CA/04/2009 (H1N1) infection in Calu3 cells: A time course | ICL010-R | Transcript Quantification | A/California/04/2009 (H1N1),NL602 (H1N1) | 3 MOI | 0,12,24,48,3,7,18,30,36 h | Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE40844 | IRD/ViPR | IRD_SV_ICL010-R |
| ICL011 | ICL011: VN1203 PB2-627E and PB1-F2del infection in Calu3 cells: A time course | ICL011-P | Protein Quantification | A/Vietnam/1203/2004 (H5N1), A/Vietnam/1203-CIP048_RG2 (H5N1), A/Vietnam/1203-CIP048_RG3 (H5N1) | 1 MOI | 7,24,0,3,12,18 h | Genetic, Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00427 | IRD/ViPR | IRD_SV_ICL011-P |
| ICL011 | ICL011: VN1203 PB2-627E and PB1-F2del infection in Calu3 cells: A time course | ICL011-R | Transcript Quantification | A/Vietnam/1203/2004 (H5N1), A/Vietnam/1203-CIP048_RG2 (H5N1), A/Vietnam/1203-CIP048_RG3 (H5N1) | 1 MOI | 7,12,18,24,0,3 h | Genetic, Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE43203 | IRD/ViPR | IRD_SV_ICL011-R |
| ICL012 | ICL012: Vietnam/1203/2004 (H5N1) and Vietnam/1203-NS1-trunc124/2004 (H5N1) infection in Calu3 cells: A time course | ICL012-P | Protein Quantification | A/Vietnam/1203/2004 (H5N1), A/Vietnam/1203-CIP048_RG4 (H5N1) | 1 MOI | 24,7,0,3,12,18 h | Genetic, Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00428 | IRD/ViPR | IRD_SV_ICL012-P |
| ICL012 | ICL012: Vietnam/1203/2004 (H5N1) and Vietnam/1203-NS1-trunc124/2004 (H5N1) infection in Calu3 cells: A time course | ICL012-R | Transcript Quantification | A/Vietnam/1203/2004 (H5N1), A/Vietnam/1203-CIP048_RG4 (H5N1) | 1 MOI | 7,24,0,3,12,18 h | Genetic, Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE43204 | IRD/ViPR | IRD_SV_ICL012-R |
| IM001 | IM001:A/Vietnam/1203/2004(H5N1) infection in C57BL6 mice with variable doses and times post infection. | IM001-P | Protein Quantification | A/Vietnam/1203/2004 (H5N1) | 1.0E02, 1.0E03, 1.0E04 PFU | 1,2,4,7 days | Dose Response, Longitudinal | Mouse | C57BL6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00418 | IRD/ViPR | IRD_SV_IM001-P |
| IM001 | IM001:A/Vietnam/1203/2004(H5N1) infection in C57BL6 mice with variable doses and times post infection. | IM001-R | Transcript Quantification | A/Vietnam/1203/2004 (H5N1) | 1.0E02, 1.0E03, 1.0E04 PFU | 1,2,4,7 days | Dose Response, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE33263 | IRD/ViPR | IRD_SV_IM001-R |
| IM002 | IM002: Influenza A/NJ/8/76 (H1N1), A/Mexico/4482/2009 (H1N1), Brisbane/59/2007 (H1N1) or reconstructed 1918 (H1N1) infection in BALB/c mice: A time course | IM002-R | Transcript Quantification | A/New Jersey/8/76 (H1N1), A/Mexico/4482/2009 (H1N1), A/Brisbane/59/2007 (H1N1), Reconstructed 1918 (H1N1) | 1.0E06 PFU | 1,3,5 days | Genetic, Longitudinal | Mouse | BALB/c | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE36328 | IRD/ViPR | IRD_SV_IM002-R |
| IM004 | IM004:VN1203 HA avirluent mutant virus ( A/Vietnam/1203-CIP048_RG1/2004 (H5N1)) infection in C57BL6 mice: A time course | IM004-P | Protein Quantification | A/Vietnam/1203-CIP048_RG1 (H5N1) | 1.0E04 PFU | 4,7,1,2 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00419 | IRD/ViPR | IRD_SV_IM004-P |
| IM004 | IM004:VN1203 HA avirluent mutant virus ( A/Vietnam/1203-CIP048_RG1/2004 (H5N1)) infection in C57BL6 mice: A time course | IM004-R | Transcript Quantification | A/Vietnam/1203-CIP048_RG1 (H5N1) | 1.0E04 PFU | 1,2,4,7 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE37572 | IRD/ViPR | IRD_SV_IM004-R |
| IM005 | IM005: Vietnam/1203-PB2-627E/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM005-P | Protein Quantification | A/Vietnam/1203-CIP048_RG2 (H5N1) | 1.0E04 PFU | 1,2,4,7 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00420 | IRD/ViPR | IRD_SV_IM005-P |
| IM005 | IM005: Vietnam/1203-PB2-627E/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM005-R | Transcript Quantification | A/Vietnam/1203-CIP048_RG2 (H5N1) | 1.0E04 PFU | 4,1,2,7 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE43301 | IRD/ViPR | IRD_SV_IM005-R |
| IM006A | IM006A: Vietnam/1203-PB1-F2del/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM006A-P | Protein Quantification | A/Vietnam/1203-CIP048_RG3 (H5N1) | 1.0E03 PFU | 1,2,4,7 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00422 | IRD/ViPR | IRD_SV_IM006A-P |
| IM006A | IM006A: Vietnam/1203-PB1-F2del/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM006A-R | Transcript Quantification | A/Vietnam/1203-CIP048_RG3 (H5N1) | 1.0E03 PFU | 1,2,4,7 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE43302 | IRD/ViPR | IRD_SV_IM006A-R |
| IM006B | IM006B: Vietnam/1203-PB1-F2del/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM006B-P | Protein Quantification | A/Vietnam/1203-CIP048_RG3 (H5N1) | 1.0E04 PFU | 1,2,4 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00422 | IRD/ViPR | IRD_SV_IM006B-P |
| IM006B | IM006B: Vietnam/1203-PB1-F2del/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM006B-R | Transcript Quantification | A/Vietnam/1203-CIP048_RG3 (H5N1) | 1.0E04 PFU | 1,2,7,4 days | Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE44441 | IRD/ViPR | IRD_SV_IM006B-R |
| IM007 | IM007: Vietnam/1203-NS1trunc-124/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM007-P | Protein Quantification | A/Vietnam/1203-CIP048_RG4 (H5N1) | 1.0E03, 1.0E04 PFU | 2,4,7,1 days | Dose Response, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00421 | IRD/ViPR | IRD_SV_IM007-P |
| IM007 | IM007: Vietnam/1203-NS1trunc-124/2004 (H5N1) infection in C57 Black 6 Mice: A time course | IM007-R | Transcript Quantification | A/Vietnam/1203-CIP048_RG4 (H5N1) | 1.0E03, 1.0E04 PFU | 1,2,4,7 days | Dose Response, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE44445 | IRD/ViPR | IRD_SV_IM007-R |
| IM009 | IM009: Influenza A/CA/04/2009 (H1N1) or, mouse-adapted A/CA/04/2009 (H1N1) infection in BALB/c mice: A time course | IM009-R | Transcript Quantification | A/California/04/2009 (H1N1), MA1-A/ California/04/2009 (H1N1) | 1.0E06 PFU | 1,3,5 days | Genetic, Longitudinal | Mouse | BALB/c | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE36328 | IRD/ViPR | IRD_SV_IM009-R |
| IM010 | IM010: Vietnam/1203/2004 (H5N1) infection in knockout mutants of C57 Black 6 Mice | IM010-R | Transcript Quantification | A/Vietnam/1203/2004 (H5N1) | 1.0E03 PFU | 2,6 days | Genetic, Longitudinal | Mouse | C57BL/6,IDO1,Tnfrsf1b | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE40792 | IRD/ViPR | IRD_SV_IM010-R |
| SBRI_AA_E1 | SBRI_AA: PR8, X31, VN6+2 12hrs infection in whole lung tissue. 10^5 pfu. | SBRI_AA_E1 | Transcript Quantification | A/Aichi/02/68 (HA, NA) x A/Puerto Rico/8/34 (X31), A/Puerto Rico/8/34 (H1N1),A/Vietnam/1203/04 (HA Vn1203, NA) x A/Puerto Rico/8/34 (VN6+2) | 1.0E05 PFU | 12 h | Genetic | Mouse | C57BL/6J | Lung Tissue | Systems Influenza | Alan Aderem | GEO | GSE37245 | IRD/ViPR | IRD_SI_SBRI_AA_E1 |
| SCL005 | SCL005: SARS Urbani or delta ORF6 Urbani infection in Calu3 cells with variable doses and times post infection. | SCL005-P | Protein Quantification | icSARS CoV Urbani,icSARS dORF6 | 5 MOI | 0,3,7,12,60,72,24,30,36,48,54 h | Genetic, Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00430 | IRD/ViPR | IRD_SV_SCL005-P |
| SCL005 | SCL005: SARS Urbani or delta ORF6 Urbani infection in Calu3 cells with variable doses and times post infection. | SCL005-R | Transcript Quantification | icSARS CoV Urbani,icSARS dORF6 | 5 MOI | 0,12,24,3,30,36,48,54,60,7,72 h | Genetic, Longitudinal | Human | Calu-3 cells (clone 2B5) | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE33267 | IRD/ViPR | IRD_SV_SCL005-R |
| SCL006 | SCL006: icSARS urbani vs icSARS BatSRBD mutant infection in Calu3 cell: A time course | SCL006-P | Protein Quantification | icSARS Bat SRBD,icSARS CoV Urbani | 1 MOI | 0,3,7,12,24,30,36,48,54,60,72 h | Genetic, Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00431 | IRD/ViPR | IRD_SV_SCL006-P |
| SCL006 | SCL006: icSARS urbani vs icSARS BatSRBD mutant infection in Calu3 cell: A time course | SCL006-R | Transcript Quantification | icSARS Bat SRBD,icSARS CoV Urbani | 1 MOI | 0,12,24,30,36,48,54,60,7,72 h | Genetic, Longitudinal | Human | Calu-3 cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE37827 | IRD/ViPR | IRD_SV_SCL006-R |
| SHAE002 | SHAE005: SARS-CoV, deltaORF6, BatSRBD mutants and A/CA/04/2009 (H1N1) infection of HAE cells: A time course | SHAE002-R | Transcript Quantification | A/California/04/2009 (H1N1),icSARS Bat SRBD,icSARS CoV Urbani,icSARS dORF6 | 2 MOI | 96,0,12,24,36,48,60,72,84,6,18 h | Genetic, Longitudinal | Human | HAE cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE47960 | IRD/ViPR | IRD_SV_SHAE002-R |
| SHAE003 | SHAE005: SARS-CoV, deltaORF6, BatSRBD mutants and A/CA/04/2009 (H1N1) infection of HAE cells: A time course | SHAE003-R | Transcript Quantification | A/California/04/2009 (H1N1),icSARS Bat SRBD,icSARS CoV Urbani,icSARS dORF6 | 2 MOI | 24,48,60,72,84,96,0,6,12,18,36 h | Genetic, Longitudinal | Human | HAE cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE47961 | IRD/ViPR | IRD_SV_SHAE003-R |
| SHAE004 | SHAE005: SARS-CoV, deltaORF6, BatSRBD mutants and A/CA/04/2009 (H1N1) infection of HAE cells: A time course | SHAE004-R | Transcript Quantification | A/California/04/2009 (H1N1),icSARS Bat SRBD,icSARS CoV Urbani,icSARS dORF6 | 1,2 MOI | 0,12,24,36,48,60,72,84,96,6,18 h | Genetic, Longitudinal | Human | HAE cells | Cell Line | Systems Virology | Michael G. Katze | GEO | GSE47962 | IRD/ViPR | IRD_SV_SHAE004-R |
| SM001 | SM001: SARS MA15 infection in C57BL6 mice with variable doses and times post infection. | SM001-P | Protein Quantification | SARS CoV MA15 | 1.0E02, 1.0E03, 1.0E04, 1.0E05 PFU | 1,2,4,7 days | Dose Response, Longitudinal | Mouse | C57BL6 | Lung Tissue | Systems Virology | Michael G. Katze | PeptideAtlas | PASS00433 | IRD/ViPR | IRD_SV_SM001-P |
| SM001 | SM001: SARS MA15 infection in C57BL6 mice with variable doses and times post infection. | SM001-R | Transcript Quantification | SARS CoV MA15 | 1.0E02, 1.0E03, 1.0E04, 1.0E05 PFU | 1,2,4,7 days | Dose Response, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE33266 | IRD/ViPR | IRD_SV_SM001-R |
| SM003 | SM003: icSARS, SARS MA15 or SARS BatSRBD mutant virus infection of C57 Black 6 mice: A time course | SM003-R | Transcript Quantification | SARS CoV MA15,icSARS Bat SRBD,icSARS CoV Urbani | 1.0E04, 1.0E05 PFU | 1,2,4,7 days | Genetic, Dose Response, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE50000 | IRD/ViPR | IRD_SV_SM003-R |
| SM004 | SM004: SARS MA15 infection in C57 Black 6, TIMP1 and Serpine1 (PAI1) knock-out mice: A time course | SM004-R | Transcript Quantification | SARS CoV MA15 | 1.0E04 PFU | 4,7 days | Dose Response, Longitudinal | Mouse | C57BL/6,Serpine1 (PAI1),TIMP1 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE51386 | IRD/ViPR | IRD_SV_SM004-R |
| SM007 | SM007: SARS MA15 infection in CXCR3 knockout mutant of C57 Black 6 Mice. | SM007-R | Transcript Quantification | SARS CoV MA15 | 1.0E04 PFU | 7,1,2,4 days | Dose Response, Longitudinal | Mouse | CXCR3,C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE50878 | IRD/ViPR | IRD_SV_SM007-R |
| SM009 | SM009: SARS MA15 infection in C57 Black 6 and PLAT knock-out mice: A time course | SM009-R | Transcript Quantification | SARS CoV MA15 | 1.0E05 PFU | 4,7 days | Dose Response, Longitudinal | Mouse | PLAT,C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE51387 | IRD/ViPR | IRD_SV_SM009-R |
| SM012 | SM012: SARS deltaORF6 mutant infection in C57 Black 6 Mice: A time course | SM012-R | Transcript Quantification | SARS CoV MA15,icSARS dORF6 | 1.0E06 PFU | 4,1,2,7 days | Genetic, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE49262 | IRD/ViPR | IRD_SV_SM012-R |
| SM014 | SM012: SARS nsp16 mutant infection in C57 Black 6 Mice: A time course | SM014-R | Transcript Quantification | SARS CoV MA15,icSARS dNSP16 | 1.0E06 PFU | 1,2,4,7 days | Genetic, Longitudinal | Mouse | C57BL/6 | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE49263 | IRD/ViPR | IRD_SV_SM014-R |
| SM015 | SM015: SARS MA15 infection in Tnfrsf1a/1b knockout mutant of C57 Black 6 Mice. | SM015-R | Transcript Quantification | SARS CoV MA15 | 1.0E05 PFU | 7,4 days | Longitudinal | Mouse | C57BL/6,Tnfrsf1a/1b | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE43301 | IRD/ViPR | IRD_SV_SM015-R |
| SM019 | SM019: SARS MA15 infection in Tnfrsf1b knockout mutant of C57 Black 6 Mice. | SM019-R | Transcript Quantification | SARS CoV MA15 | 1.0E05 PFU | 7,4 days | Longitudinal | Mouse | C57BL/6,Tnfrsf1b | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE40824 | IRD/ViPR | IRD_SV_SM019-R |
| SM020 | SM020: SARS MA15 infection in ppp1r14c knockout mutant of C57 Black 6 Mice. | SM020-R | Transcript Quantification | SARS CoV MA15 | 1.0E05 PFU | 7,4 days | Longitudinal | Mouse | C57BL/6,ppp1r14c | Lung Tissue | Systems Virology | Michael G. Katze | GEO | GSE40827 | IRD/ViPR | IRD_SV_SM020-R |
Table 2. Experiment samples from transcriptomic assays.
| Experiment ID | Experiment Sample ID | Biological Sample Type | PUBLIC REPOSITORY NAME | REPOSITORY ID |
|---|---|---|---|---|
| CA04M001-R | CA04M001_10^3_1d_2_RNA_ExpSam | RNA | GEO | GSM921810 |
| CA04M001-R | CA04M001_10^3_1d_3_RNA_ExpSam | RNA | GEO | GSM921811 |
| CA04M001-R | CA04M001_10^3_1d_4_RNA_ExpSam | RNA | GEO | GSM921812 |
| CA04M001-R | CA04M001_10^3_2d_1_RNA_ExpSam | RNA | GEO | GSM921813 |
| CA04M001-R | CA04M001_10^3_2d_2_RNA_ExpSam | RNA | GEO | GSM921814 |
| CA04M001-R | CA04M001_10^3_2d_3_RNA_ExpSam | RNA | GEO | GSM921815 |
| CA04M001-R | CA04M001_10^3_4d_1_RNA_ExpSam | RNA | GEO | GSM921816 |
| CA04M001-R | CA04M001_10^3_4d_2_RNA_ExpSam | RNA | GEO | GSM921817 |
| CA04M001-R | CA04M001_10^3_4d_3_RNA_ExpSam | RNA | GEO | GSM921818 |
| CA04M001-R | CA04M001_10^3_4d_4_RNA_ExpSam | RNA | GEO | GSM921819 |
| CA04M001-R | CA04M001_10^3_7d_2_RNA_ExpSam | RNA | GEO | GSM921820 |
| CA04M001-R | CA04M001_10^3_7d_3_RNA_ExpSam | RNA | GEO | GSM921821 |
| CA04M001-R | CA04M001_10^3_7d_4_RNA_ExpSam | RNA | GEO | GSM921822 |
| CA04M001-R | CA04M001_10^4_1d_1_RNA_ExpSam | RNA | GEO | GSM921823 |
| CA04M001-R | CA04M001_10^4_1d_2_RNA_ExpSam | RNA | GEO | GSM921824 |
| CA04M001-R | CA04M001_10^4_1d_3_RNA_ExpSam | RNA | GEO | GSM921825 |
| CA04M001-R | CA04M001_10^4_1d_4_RNA_ExpSam | RNA | GEO | GSM921826 |
| CA04M001-R | CA04M001_10^4_2d_1_RNA_ExpSam | RNA | GEO | GSM921827 |
| CA04M001-R | CA04M001_10^4_2d_2_RNA_ExpSam | RNA | GEO | GSM921828 |
| CA04M001-R | CA04M001_10^4_2d_3_RNA_ExpSam | RNA | GEO | GSM921829 |
| CA04M001-R | CA04M001_10^4_2d_4_RNA_ExpSam | RNA | GEO | GSM921830 |
| CA04M001-R | CA04M001_10^4_4d_1_RNA_ExpSam | RNA | GEO | GSM921831 |
| CA04M001-R | CA04M001_10^4_4d_2_RNA_ExpSam | RNA | GEO | GSM921832 |
| CA04M001-R | CA04M001_10^4_4d_3_RNA_ExpSam | RNA | GEO | GSM921833 |
| CA04M001-R | CA04M001_10^4_7d_1_RNA_ExpSam | RNA | GEO | GSM921834 |
| CA04M001-R | CA04M001_10^4_7d_2_RNA_ExpSam | RNA | GEO | GSM921835 |
| CA04M001-R | CA04M001_10^4_7d_3_RNA_ExpSam | RNA | GEO | GSM921836 |
| CA04M001-R | CA04M001_10^5_1d_1_RNA_ExpSam | RNA | GEO | GSM921837 |
| CA04M001-R | CA04M001_10^5_1d_2_RNA_ExpSam | RNA | GEO | GSM921838 |
| CA04M001-R | CA04M001_10^5_1d_3_RNA_ExpSam | RNA | GEO | GSM921839 |
| CA04M001-R | CA04M001_10^5_1d_4_RNA_ExpSam | RNA | GEO | GSM921840 |
| CA04M001-R | CA04M001_10^5_2d_1_RNA_ExpSam | RNA | GEO | GSM921841 |
| CA04M001-R | CA04M001_10^5_2d_2_RNA_ExpSam | RNA | GEO | GSM921842 |
| CA04M001-R | CA04M001_10^5_2d_3_RNA_ExpSam | RNA | GEO | GSM921843 |
| CA04M001-R | CA04M001_10^5_2d_4_RNA_ExpSam | RNA | GEO | GSM921844 |
| CA04M001-R | CA04M001_10^5_4d_1_RNA_ExpSam | RNA | GEO | GSM921845 |
| CA04M001-R | CA04M001_10^5_4d_2_RNA_ExpSam | RNA | GEO | GSM921846 |
| CA04M001-R | CA04M001_10^5_4d_3_RNA_ExpSam | RNA | GEO | GSM921847 |
| CA04M001-R | CA04M001_10^5_4d_4_RNA_ExpSam | RNA | GEO | GSM921848 |
| CA04M001-R | CA04M001_10^5_7d_1_RNA_ExpSam | RNA | GEO | GSM921849 |
| CA04M001-R | CA04M001_10^5_7d_2_RNA_ExpSam | RNA | GEO | GSM921850 |
| CA04M001-R | CA04M001_10^5_7d_3_RNA_ExpSam | RNA | GEO | GSM921851 |
| CA04M001-R | CA04M001_10^5_7d_4_RNA_ExpSam | RNA | GEO | GSM921852 |
| CA04M001-R | CA04M001_10^5_7d_5_RNA_ExpSam | RNA | GEO | GSM921853 |
| CA04M001-R | CA04M001_10^6_1d_1_RNA_ExpSam | RNA | GEO | GSM921854 |
| CA04M001-R | CA04M001_10^6_1d_3_RNA_ExpSam | RNA | GEO | GSM921855 |
| CA04M001-R | CA04M001_10^6_1d_4_RNA_ExpSam | RNA | GEO | GSM921856 |
| CA04M001-R | CA04M001_10^6_2d_1_RNA_ExpSam | RNA | GEO | GSM921857 |
| CA04M001-R | CA04M001_10^6_2d_2_RNA_ExpSam | RNA | GEO | GSM921858 |
| CA04M001-R | CA04M001_10^6_2d_3_RNA_ExpSam | RNA | GEO | GSM921859 |
| CA04M001-R | CA04M001_10^6_2d_4_RNA_ExpSam | RNA | GEO | GSM921860 |
| CA04M001-R | CA04M001_10^6_4d_1_RNA_ExpSam | RNA | GEO | GSM921861 |
| CA04M001-R | CA04M001_10^6_4d_2_RNA_ExpSam | RNA | GEO | GSM921862 |
| CA04M001-R | CA04M001_10^6_4d_3_RNA_ExpSam | RNA | GEO | GSM921863 |
| CA04M001-R | CA04M001_10^6_7d_1_RNA_ExpSam | RNA | GEO | GSM921864 |
| CA04M001-R | CA04M001_10^6_7d_2_RNA_ExpSam | RNA | GEO | GSM921865 |
| CA04M001-R | CA04M001_10^6_7d_3_RNA_ExpSam | RNA | GEO | GSM921866 |
| CA04M001-R | CA04M001_10^6_7d_4_RNA_ExpSam | RNA | GEO | GSM921867 |
| CA04M001-R | CA04M001_10^6_7d_5_RNA_ExpSam | RNA | GEO | GSM921868 |
| CA04M001-R | CA04M001_mock_1d_1_RNA_ExpSam | RNA | GEO | GSM921869 |
| CA04M001-R | CA04M001_mock_1d_2_RNA_ExpSam | RNA | GEO | GSM921870 |
| CA04M001-R | CA04M001_mock_2d_1_RNA_ExpSam | RNA | GEO | GSM921871 |
| CA04M001-R | CA04M001_mock_2d_2_RNA_ExpSam | RNA | GEO | GSM921872 |
| CA04M001-R | CA04M001_mock_4d_1_RNA_ExpSam | RNA | GEO | GSM921873 |
| CA04M001-R | CA04M001_mock_4d_2_RNA_ExpSam | RNA | GEO | GSM921874 |
| CA04M001-R | CA04M001_mock_4d_3_RNA_ExpSam | RNA | GEO | GSM921875 |
| CA04M001-R | CA04M001_mock_7d_1_RNA_ExpSam | RNA | GEO | GSM921876 |
| CA04M001-R | CA04M001_mock_7d_2_RNA_ExpSam | RNA | GEO | GSM921877 |
| CA04M001-R | CA04M001_mock_7d_3_RNA_ExpSam | RNA | GEO | GSM921878 |
| ECL001-R | ECL001_EMC_0h_1_array | RNA | GEO | GSM1096530 |
| ECL001-R | ECL001_EMC_0h_2_array | RNA | GEO | GSM1096515 |
| ECL001-R | ECL001_EMC_0h_3_array | RNA | GEO | GSM1096512 |
| ECL001-R | ECL001_EMC_12h_2_array | RNA | GEO | GSM1096529 |
| ECL001-R | ECL001_EMC_12h_3_array | RNA | GEO | GSM1096526 |
| ECL001-R | ECL001_EMC_18h_1_array | RNA | GEO | GSM1096540 |
| ECL001-R | ECL001_EMC_18h_2_array | RNA | GEO | GSM1096536 |
| ECL001-R | ECL001_EMC_18h_3_array | RNA | GEO | GSM1096532 |
| ECL001-R | ECL001_EMC_24h_1_array | RNA | GEO | GSM1096528 |
| ECL001-R | ECL001_EMC_24h_2_array | RNA | GEO | GSM1096513 |
| ECL001-R | ECL001_EMC_24h_3_array | RNA | GEO | GSM1096519 |
| ECL001-R | ECL001_EMC_3h_1_array | RNA | GEO | GSM1096521 |
| ECL001-R | ECL001_EMC_3h_2_array | RNA | GEO | GSM1096517 |
| ECL001-R | ECL001_EMC_3h_3_array | RNA | GEO | GSM1096542 |
| ECL001-R | ECL001_EMC_7h_1_array | RNA | GEO | GSM1096541 |
| ECL001-R | ECL001_EMC_7h_2_array | RNA | GEO | GSM1096518 |
| ECL001-R | ECL001_EMC_7h_3_array | RNA | GEO | GSM1096535 |
| ECL001-R | ECL001_Mock_0h_1_array | RNA | GEO | GSM1096531 |
| ECL001-R | ECL001_Mock_0h_2_array | RNA | GEO | GSM1096514 |
| ECL001-R | ECL001_Mock_0h_3_array | RNA | GEO | GSM1096534 |
| ECL001-R | ECL001_Mock_12h_1_array | RNA | GEO | GSM1096511 |
| ECL001-R | ECL001_Mock_12h_2_array | RNA | GEO | GSM1096533 |
| ECL001-R | ECL001_Mock_12h_3_array | RNA | GEO | GSM1096522 |
| ECL001-R | ECL001_Mock_24h_1_array | RNA | GEO | GSM1096525 |
| ECL001-R | ECL001_Mock_24h_2_array | RNA | GEO | GSM1096524 |
| ECL001-R | ECL001_Mock_24h_3_array | RNA | GEO | GSM1096520 |
| ECL001-R | ECL001_Mock_3h_1_array | RNA | GEO | GSM1096538 |
| ECL001-R | ECL001_Mock_3h_2_array | RNA | GEO | GSM1096516 |
| ECL001-R | ECL001_Mock_3h_3_array | RNA | GEO | GSM1096539 |
| ECL001-R | ECL001_Mock_7h_1_array | RNA | GEO | GSM1096537 |
| ECL001-R | ECL001_Mock_7h_2_array | RNA | GEO | GSM1096523 |
| ECL001-R | ECL001_Mock_7h_3_array | RNA | GEO | GSM1096527 |
| ICL004-R | ICL004_Mock_0H_1_RNA_ExpSam | RNA | GEO | GSM697564 |
| ICL004-R | ICL004_Mock_0H_2_RNA_ExpSam | RNA | GEO | GSM697565 |
| ICL004-R | ICL004_Mock_0H_3_RNA_ExpSam | RNA | GEO | GSM697566 |
| ICL004-R | ICL004_Mock_12H_1_RNA_ExpSam | RNA | GEO | GSM697573 |
| ICL004-R | ICL004_Mock_12H_2_RNA_ExpSam | RNA | GEO | GSM697574 |
| ICL004-R | ICL004_Mock_12H_3_RNA_ExpSam | RNA | GEO | GSM697575 |
| ICL004-R | ICL004_Mock_18H_1_RNA_ExpSam | RNA | GEO | GSM697576 |
| ICL004-R | ICL004_Mock_18H_2_RNA_ExpSam | RNA | GEO | GSM697577 |
| ICL004-R | ICL004_Mock_18H_3_RNA_ExpSam | RNA | GEO | GSM697578 |
| ICL004-R | ICL004_Mock_24H_1_RNA_ExpSam | RNA | GEO | GSM697579 |
| ICL004-R | ICL004_Mock_24H_2_RNA_ExpSam | RNA | GEO | GSM697580 |
| ICL004-R | ICL004_Mock_24H_3_RNA_ExpSam | RNA | GEO | GSM697581 |
| ICL004-R | ICL004_Mock_3H_1_RNA_ExpSam | RNA | GEO | GSM697567 |
| ICL004-R | ICL004_Mock_3H_2_RNA_ExpSam | RNA | GEO | GSM697568 |
| ICL004-R | ICL004_Mock_3H_3_RNA_ExpSam | RNA | GEO | GSM697569 |
| ICL004-R | ICL004_Mock_7H_1_RNA_ExpSam | RNA | GEO | GSM697570 |
| ICL004-R | ICL004_Mock_7H_2_RNA_ExpSam | RNA | GEO | GSM697571 |
| ICL004-R | ICL004_Mock_7H_3_RNA_ExpSam | RNA | GEO | GSM697572 |
| ICL004-R | ICL004_VN1203_0H_1_RNA_ExpSam | RNA | GEO | GSM697582 |
| ICL004-R | ICL004_VN1203_0H_2_RNA_ExpSam | RNA | GEO | GSM697583 |
| ICL004-R | ICL004_VN1203_0H_3_RNA_ExpSam | RNA | GEO | GSM697584 |
| ICL004-R | ICL004_VN1203_12H_1_RNA_ExpSam | RNA | GEO | GSM697591 |
| ICL004-R | ICL004_VN1203_12H_2_RNA_ExpSam | RNA | GEO | GSM697592 |
| ICL004-R | ICL004_VN1203_12H_3_RNA_ExpSam | RNA | GEO | GSM697593 |
| ICL004-R | ICL004_VN1203_18H_1_RNA_ExpSam | RNA | GEO | GSM697594 |
| ICL004-R | ICL004_VN1203_18H_2_RNA_ExpSam | RNA | GEO | GSM697595 |
| ICL004-R | ICL004_VN1203_18H_3_RNA_ExpSam | RNA | GEO | GSM697596 |
| ICL004-R | ICL004_VN1203_24H_1_RNA_ExpSam | RNA | GEO | GSM697597 |
| ICL004-R | ICL004_VN1203_24H_2_RNA_ExpSam | RNA | GEO | GSM697598 |
| ICL004-R | ICL004_VN1203_24H_3_RNA_ExpSam | RNA | GEO | GSM697599 |
| ICL004-R | ICL004_VN1203_3H_1_RNA_ExpSam | RNA | GEO | GSM697585 |
| ICL004-R | ICL004_VN1203_3H_2_RNA_ExpSam | RNA | GEO | GSM697586 |
| ICL004-R | ICL004_VN1203_3H_3_RNA_ExpSam | RNA | GEO | GSM697587 |
| ICL004-R | ICL004_VN1203_7H_1_RNA_ExpSam | RNA | GEO | GSM697588 |
| ICL004-R | ICL004_VN1203_7H_2_RNA_ExpSam | RNA | GEO | GSM697589 |
| ICL004-R | ICL004_VN1203_7H_3_RNA_ExpSam | RNA | GEO | GSM697590 |
| ICL006-R | ICL006_CA04_0h_1_RNA_ExpSam | RNA | GEO | GSM921945 |
| ICL006-R | ICL006_CA04_0h_2_RNA_ExpSam | RNA | GEO | GSM921944 |
| ICL006-R | ICL006_CA04_0h_3_RNA_ExpSam | RNA | GEO | GSM921943 |
| ICL006-R | ICL006_CA04_12h_1_RNA_ExpSam | RNA | GEO | GSM921942 |
| ICL006-R | ICL006_CA04_12h_2_RNA_ExpSam | RNA | GEO | GSM921941 |
| ICL006-R | ICL006_CA04_12h_3_RNA_ExpSam | RNA | GEO | GSM921940 |
| ICL006-R | ICL006_CA04_18h_1_RNA_ExpSam | RNA | GEO | GSM921939 |
| ICL006-R | ICL006_CA04_18h_2_RNA_ExpSam | RNA | GEO | GSM921938 |
| ICL006-R | ICL006_CA04_18h_3_RNA_ExpSam | RNA | GEO | GSM921937 |
| ICL006-R | ICL006_CA04_24h_1_RNA_ExpSam | RNA | GEO | GSM921936 |
| ICL006-R | ICL006_CA04_24h_2_RNA_ExpSam | RNA | GEO | GSM921935 |
| ICL006-R | ICL006_CA04_24h_3_RNA_ExpSam | RNA | GEO | GSM921934 |
| ICL006-R | ICL006_CA04_30h_1_RNA_ExpSam | RNA | GEO | GSM921933 |
| ICL006-R | ICL006_CA04_30h_2_RNA_ExpSam | RNA | GEO | GSM921932 |
| ICL006-R | ICL006_CA04_30h_3_RNA_ExpSam | RNA | GEO | GSM921931 |
| ICL006-R | ICL006_CA04_36h_1_RNA_ExpSam | RNA | GEO | GSM921930 |
| ICL006-R | ICL006_CA04_36h_2_RNA_ExpSam | RNA | GEO | GSM921929 |
| ICL006-R | ICL006_CA04_36h_3_RNA_ExpSam | RNA | GEO | GSM921928 |
| ICL006-R | ICL006_CA04_3h_1_RNA_ExpSam | RNA | GEO | GSM921927 |
| ICL006-R | ICL006_CA04_3h_2_RNA_ExpSam | RNA | GEO | GSM921926 |
| ICL006-R | ICL006_CA04_3h_3_RNA_ExpSam | RNA | GEO | GSM921925 |
| ICL006-R | ICL006_CA04_48h_1_RNA_ExpSam | RNA | GEO | GSM921924 |
| ICL006-R | ICL006_CA04_48h_2_RNA_ExpSam | RNA | GEO | GSM921923 |
| ICL006-R | ICL006_CA04_48h_3_RNA_ExpSam | RNA | GEO | GSM921922 |
| ICL006-R | ICL006_CA04_7h_1_RNA_ExpSam | RNA | GEO | GSM921921 |
| ICL006-R | ICL006_CA04_7h_2_RNA_ExpSam | RNA | GEO | GSM921920 |
| ICL006-R | ICL006_CA04_7h_3_RNA_ExpSam | RNA | GEO | GSM921919 |
| ICL006-R | ICL006_mock_0h_1_RNA_ExpSam | RNA | GEO | GSM921918 |
| ICL006-R | ICL006_mock_0h_2_RNA_ExpSam | RNA | GEO | GSM921917 |
| ICL006-R | ICL006_mock_0h_3_RNA_ExpSam | RNA | GEO | GSM921916 |
| ICL006-R | ICL006_mock_12h_1_RNA_ExpSam | RNA | GEO | GSM921915 |
| ICL006-R | ICL006_mock_12h_2_RNA_ExpSam | RNA | GEO | GSM921914 |
| ICL006-R | ICL006_mock_12h_3_RNA_ExpSam | RNA | GEO | GSM921913 |
| ICL006-R | ICL006_mock_18h_1_RNA_ExpSam | RNA | GEO | GSM921912 |
| ICL006-R | ICL006_mock_18h_2_RNA_ExpSam | RNA | GEO | GSM921911 |
| ICL006-R | ICL006_mock_18h_3_RNA_ExpSam | RNA | GEO | GSM921910 |
| ICL006-R | ICL006_mock_24h_1_RNA_ExpSam | RNA | GEO | GSM921909 |
| ICL006-R | ICL006_mock_24h_2_RNA_ExpSam | RNA | GEO | GSM921908 |
| ICL006-R | ICL006_mock_24h_3_RNA_ExpSam | RNA | GEO | GSM921907 |
| ICL006-R | ICL006_mock_30h_1_RNA_ExpSam | RNA | GEO | GSM921906 |
| ICL006-R | ICL006_mock_30h_2_RNA_ExpSam | RNA | GEO | GSM921905 |
| ICL006-R | ICL006_mock_30h_3_RNA_ExpSam | RNA | GEO | GSM921904 |
| ICL006-R | ICL006_mock_36h_1_RNA_ExpSam | RNA | GEO | GSM921903 |
| ICL006-R | ICL006_mock_36h_2_RNA_ExpSam | RNA | GEO | GSM921902 |
| ICL006-R | ICL006_mock_36h_3_RNA_ExpSam | RNA | GEO | GSM921901 |
| ICL006-R | ICL006_mock_3h_1_RNA_ExpSam | RNA | GEO | GSM921900 |
| ICL006-R | ICL006_mock_3h_2_RNA_ExpSam | RNA | GEO | GSM921899 |
| ICL006-R | ICL006_mock_3h_3_RNA_ExpSam | RNA | GEO | GSM921898 |
| ICL006-R | ICL006_mock_48h_1_RNA_ExpSam | RNA | GEO | GSM921897 |
| ICL006-R | ICL006_mock_48h_2_RNA_ExpSam | RNA | GEO | GSM921896 |
| ICL006-R | ICL006_mock_48h_3_RNA_ExpSam | RNA | GEO | GSM921895 |
| ICL006-R | ICL006_mock_7h_1_RNA_ExpSam | RNA | GEO | GSM921894 |
| ICL006-R | ICL006_mock_7h_2_RNA_ExpSam | RNA | GEO | GSM921893 |
| ICL006-R | ICL006_mock_7h_3_RNA_ExpSam | RNA | GEO | GSM921892 |
| ICL010-R | ICL010_Cal_0h_1_array | RNA | GEO | GSM1003148 |
| ICL010-R | ICL010_Cal_0h_2_array | RNA | GEO | GSM1003149 |
| ICL010-R | ICL010_Cal_0h_3_array | RNA | GEO | GSM1003150 |
| ICL010-R | ICL010_Cal_12h_1_array | RNA | GEO | GSM1003151 |
| ICL010-R | ICL010_Cal_12h_2_array | RNA | GEO | GSM1003152 |
| ICL010-R | ICL010_Cal_12h_3_array | RNA | GEO | GSM1003153 |
| ICL010-R | ICL010_Cal_24h_1_array | RNA | GEO | GSM1003154 |
| ICL010-R | ICL010_Cal_24h_2_array | RNA | GEO | GSM1003155 |
| ICL010-R | ICL010_Cal_24h_3_array | RNA | GEO | GSM1003156 |
| ICL010-R | ICL010_Cal_48h_1_array | RNA | GEO | GSM1003157 |
| ICL010-R | ICL010_Cal_48h_2_array | RNA | GEO | GSM1003158 |
| ICL010-R | ICL010_Cal_48h_3_array | RNA | GEO | GSM1003159 |
| ICL010-R | ICL010_mock_0h_1_array | RNA | GEO | GSM1003160 |
| ICL010-R | ICL010_mock_0h_2_array | RNA | GEO | GSM1003161 |
| ICL010-R | ICL010_mock_0h_3_array | RNA | GEO | GSM1003162 |
| ICL010-R | ICL010_mock_12h_1_array | RNA | GEO | GSM1003163 |
| ICL010-R | ICL010_mock_12h_2_array | RNA | GEO | GSM1003164 |
| ICL010-R | ICL010_mock_12h_3_array | RNA | GEO | GSM1003165 |
| ICL010-R | ICL010_mock_18h_1_array | RNA | GEO | GSM1003166 |
| ICL010-R | ICL010_mock_18h_2_array | RNA | GEO | GSM1003167 |
| ICL010-R | ICL010_mock_18h_3_array | RNA | GEO | GSM1003168 |
| ICL010-R | ICL010_mock_24h_1_array | RNA | GEO | GSM1003169 |
| ICL010-R | ICL010_mock_24h_2_array | RNA | GEO | GSM1003170 |
| ICL010-R | ICL010_mock_24h_3_array | RNA | GEO | GSM1003171 |
| ICL010-R | ICL010_mock_30h_1_array | RNA | GEO | GSM1003172 |
| ICL010-R | ICL010_mock_30h_2_array | RNA | GEO | GSM1003173 |
| ICL010-R | ICL010_mock_30h_3_array | RNA | GEO | GSM1003174 |
| ICL010-R | ICL010_mock_36h_1_array | RNA | GEO | GSM1003175 |
| ICL010-R | ICL010_mock_36h_2_array | RNA | GEO | GSM1003176 |
| ICL010-R | ICL010_mock_36h_3_array | RNA | GEO | GSM1003177 |
| ICL010-R | ICL010_mock_3h_1_array | RNA | GEO | GSM1003178 |
| ICL010-R | ICL010_mock_3h_2_array | RNA | GEO | GSM1003179 |
| ICL010-R | ICL010_mock_3h_3_array | RNA | GEO | GSM1003180 |
| ICL010-R | ICL010_mock_48h_1_array | RNA | GEO | GSM1003181 |
| ICL010-R | ICL010_mock_48h_2_array | RNA | GEO | GSM1003182 |
| ICL010-R | ICL010_mock_48h_3_array | RNA | GEO | GSM1003183 |
| ICL010-R | ICL010_mock_7h_1_array | RNA | GEO | GSM1003184 |
| ICL010-R | ICL010_mock_7h_2_array | RNA | GEO | GSM1003185 |
| ICL010-R | ICL010_mock_7h_3_array | RNA | GEO | GSM1003186 |
| ICL010-R | ICL010_NL_0h_1_array | RNA | GEO | GSM1003187 |
| ICL010-R | ICL010_NL_0h_2_array | RNA | GEO | GSM1003188 |
| ICL010-R | ICL010_NL_0h_3_array | RNA | GEO | GSM1003189 |
| ICL010-R | ICL010_NL_12h_1_array | RNA | GEO | GSM1003190 |
| ICL010-R | ICL010_NL_12h_2_array | RNA | GEO | GSM1003191 |
| ICL010-R | ICL010_NL_12h_3_array | RNA | GEO | GSM1003192 |
| ICL010-R | ICL010_NL_18h_1_array | RNA | GEO | GSM1003193 |
| ICL010-R | ICL010_NL_18h_2_array | RNA | GEO | GSM1003194 |
| ICL010-R | ICL010_NL_18h_3_array | RNA | GEO | GSM1003195 |
| ICL010-R | ICL010_NL_24h_1_array | RNA | GEO | GSM1003196 |
| ICL010-R | ICL010_NL_24h_2_array | RNA | GEO | GSM1003197 |
| ICL010-R | ICL010_NL_24h_3_array | RNA | GEO | GSM1003198 |
| ICL010-R | ICL010_NL_30h_1_array | RNA | GEO | GSM1003199 |
| ICL010-R | ICL010_NL_30h_2_array | RNA | GEO | GSM1003200 |
| ICL010-R | ICL010_NL_30h_3_array | RNA | GEO | GSM1003201 |
| ICL010-R | ICL010_NL_36h_1_array | RNA | GEO | GSM1003202 |
| ICL010-R | ICL010_NL_36h_2_array | RNA | GEO | GSM1003203 |
| ICL010-R | ICL010_NL_36h_3_array | RNA | GEO | GSM1003204 |
| ICL010-R | ICL010_NL_3h_1_array | RNA | GEO | GSM1003205 |
| ICL010-R | ICL010_NL_3h_2_array | RNA | GEO | GSM1003206 |
| ICL010-R | ICL010_NL_3h_3_array | RNA | GEO | GSM1003207 |
| ICL010-R | ICL010_NL_48h_1_array | RNA | GEO | GSM1003208 |
| ICL010-R | ICL010_NL_48h_2_array | RNA | GEO | GSM1003209 |
| ICL010-R | ICL010_NL_48h_3_array | RNA | GEO | GSM1003210 |
| ICL010-R | ICL010_NL_7h_1_array | RNA | GEO | GSM1003211 |
| ICL010-R | ICL010_NL_7h_2_array | RNA | GEO | GSM1003212 |
| ICL010-R | ICL010_NL_7h_3_array | RNA | GEO | GSM1003213 |
| ICL011-R | ICL011_mock_0h_1_array | RNA | GEO | GSM1058227 |
| ICL011-R | ICL011_mock_0h_2_array | RNA | GEO | GSM1058200 |
| ICL011-R | ICL011_mock_0h_3_array | RNA | GEO | GSM1058202 |
| ICL011-R | ICL011_mock_12h_1_array | RNA | GEO | GSM1058190 |
| ICL011-R | ICL011_mock_12h_2_array | RNA | GEO | GSM1058217 |
| ICL011-R | ICL011_mock_12h_3_array | RNA | GEO | GSM1058234 |
| ICL011-R | ICL011_mock_18h_1_array | RNA | GEO | GSM1058207 |
| ICL011-R | ICL011_mock_18h_2_array | RNA | GEO | GSM1058220 |
| ICL011-R | ICL011_mock_18h_3_array | RNA | GEO | GSM1058193 |
| ICL011-R | ICL011_mock_24h_1_array | RNA | GEO | GSM1058243 |
| ICL011-R | ICL011_mock_24h_2_array | RNA | GEO | GSM1058203 |
| ICL011-R | ICL011_mock_24h_3_array | RNA | GEO | GSM1058236 |
| ICL011-R | ICL011_mock_3h_1_array | RNA | GEO | GSM1058232 |
| ICL011-R | ICL011_mock_3h_2_array | RNA | GEO | GSM1058206 |
| ICL011-R | ICL011_mock_3h_3_array | RNA | GEO | GSM1058228 |
| ICL011-R | ICL011_mock_7h_1_array | RNA | GEO | GSM1058226 |
| ICL011-R | ICL011_mock_7h_2_array | RNA | GEO | GSM1058197 |
| ICL011-R | ICL011_mock_7h_3_array | RNA | GEO | GSM1058222 |
| ICL011-R | ICL011_VN-PB1-F2-del_0h_1_array | RNA | GEO | GSM1058219 |
| ICL011-R | ICL011_VN-PB1-F2-del_0h_2_array | RNA | GEO | GSM1058210 |
| ICL011-R | ICL011_VN-PB1-F2-del_0h_3_array | RNA | GEO | GSM1058216 |
| ICL011-R | ICL011_VN-PB1-F2-del_12h_1_array | RNA | GEO | GSM1058208 |
| ICL011-R | ICL011_VN-PB1-F2-del_12h_2_array | RNA | GEO | GSM1058241 |
| ICL011-R | ICL011_VN-PB1-F2-del_12h_3_array | RNA | GEO | GSM1058198 |
| ICL011-R | ICL011_VN-PB1-F2-del_18h_1_array | RNA | GEO | GSM1058215 |
| ICL011-R | ICL011_VN-PB1-F2-del_18h_2_array | RNA | GEO | GSM1058201 |
| ICL011-R | ICL011_VN-PB1-F2-del_18h_3_array | RNA | GEO | GSM1058246 |
| ICL011-R | ICL011_VN-PB1-F2-del_24h_1_array | RNA | GEO | GSM1058240 |
| ICL011-R | ICL011_VN-PB1-F2-del_24h_2_array | RNA | GEO | GSM1058230 |
| ICL011-R | ICL011_VN-PB1-F2-del_24h_3_array | RNA | GEO | GSM1058209 |
| ICL011-R | ICL011_VN-PB1-F2-del_3h_1_array | RNA | GEO | GSM1058229 |
| ICL011-R | ICL011_VN-PB1-F2-del_3h_2_array | RNA | GEO | GSM1058245 |
| ICL011-R | ICL011_VN-PB1-F2-del_3h_3_array | RNA | GEO | GSM1058213 |
| ICL011-R | ICL011_VN-PB1-F2-del_7h_1_array | RNA | GEO | GSM1058242 |
| ICL011-R | ICL011_VN-PB1-F2-del_7h_2_array | RNA | GEO | GSM1058199 |
| ICL011-R | ICL011_VN-PB1-F2-del_7h_3_array | RNA | GEO | GSM1058225 |
| ICL011-R | ICL011_VN-PB2-627E_0h_1_array | RNA | GEO | GSM1058189 |
| ICL011-R | ICL011_VN-PB2-627E_0h_2_array | RNA | GEO | GSM1058223 |
| ICL011-R | ICL011_VN-PB2-627E_0h_3_array | RNA | GEO | GSM1058237 |
| ICL011-R | ICL011_VN-PB2-627E_12h_1_array | RNA | GEO | GSM1058211 |
| ICL011-R | ICL011_VN-PB2-627E_12h_2_array | RNA | GEO | GSM1058191 |
| ICL011-R | ICL011_VN-PB2-627E_12h_3_array | RNA | GEO | GSM1058187 |
| ICL011-R | ICL011_VN-PB2-627E_18h_1_array | RNA | GEO | GSM1058224 |
| ICL011-R | ICL011_VN-PB2-627E_18h_2_array | RNA | GEO | GSM1058188 |
| ICL011-R | ICL011_VN-PB2-627E_18h_3_array | RNA | GEO | GSM1058212 |
| ICL011-R | ICL011_VN-PB2-627E_24h_1_array | RNA | GEO | GSM1058218 |
| ICL011-R | ICL011_VN-PB2-627E_24h_2_array | RNA | GEO | GSM1058233 |
| ICL011-R | ICL011_VN-PB2-627E_24h_3_array | RNA | GEO | GSM1058214 |
| ICL011-R | ICL011_VN-PB2-627E_3h_1_array | RNA | GEO | GSM1058195 |
| ICL011-R | ICL011_VN-PB2-627E_3h_2_array | RNA | GEO | GSM1058205 |
| ICL011-R | ICL011_VN-PB2-627E_3h_3_array | RNA | GEO | GSM1058221 |
| ICL011-R | ICL011_VN-PB2-627E_7h_1_array | RNA | GEO | GSM1058196 |
| ICL011-R | ICL011_VN-PB2-627E_7h_2_array | RNA | GEO | GSM1058231 |
| ICL011-R | ICL011_VN-PB2-627E_7h_3_array | RNA | GEO | GSM1058204 |
| ICL011-R | ICL011_VN1203_24h_1_array | RNA | GEO | GSM1058244 |
| ICL011-R | ICL011_VN1203_24h_2_array | RNA | GEO | GSM1058235 |
| ICL011-R | ICL011_VN1203_24h_3_array | RNA | GEO | GSM1058192 |
| ICL011-R | ICL011_VN1203_7h_1_array | RNA | GEO | GSM1058194 |
| ICL011-R | ICL011_VN1203_7h_2_array | RNA | GEO | GSM1058238 |
| ICL011-R | ICL011_VN1203_7h_3_array | RNA | GEO | GSM1058239 |
| ICL012-R | ICL012_mock_0h_2_array | RNA | GEO | GSM1058271 |
| ICL012-R | ICL012_mock_0h_3_array | RNA | GEO | GSM1058285 |
| ICL012-R | ICL012_mock_12h_1_array | RNA | GEO | GSM1058274 |
| ICL012-R | ICL012_mock_12h_2_array | RNA | GEO | GSM1058269 |
| ICL012-R | ICL012_mock_12h_3_array | RNA | GEO | GSM1058251 |
| ICL012-R | ICL012_mock_18h_1_array | RNA | GEO | GSM1058255 |
| ICL012-R | ICL012_mock_18h_2_array | RNA | GEO | GSM1058257 |
| ICL012-R | ICL012_mock_18h_3_array | RNA | GEO | GSM1058260 |
| ICL012-R | ICL012_mock_24h_1_array | RNA | GEO | GSM1058256 |
| ICL012-R | ICL012_mock_24h_2_array | RNA | GEO | GSM1058283 |
| ICL012-R | ICL012_mock_24h_3_array | RNA | GEO | GSM1058266 |
| ICL012-R | ICL012_mock_3h_1_array | RNA | GEO | GSM1058263 |
| ICL012-R | ICL012_mock_3h_2_array | RNA | GEO | GSM1058253 |
| ICL012-R | ICL012_mock_3h_3_array | RNA | GEO | GSM1058259 |
| ICL012-R | ICL012_mock_7h_1_array | RNA | GEO | GSM1058279 |
| ICL012-R | ICL012_mock_7h_2_array | RNA | GEO | GSM1058262 |
| ICL012-R | ICL012_mock_7h_3_array | RNA | GEO | GSM1058254 |
| ICL012-R | ICL012_NS1trunc_0h_1_array | RNA | GEO | GSM1058280 |
| ICL012-R | ICL012_NS1trunc_0h_2_array | RNA | GEO | GSM1058258 |
| ICL012-R | ICL012_NS1trunc_0h_3_array | RNA | GEO | GSM1058273 |
| ICL012-R | ICL012_NS1trunc_12h_1_array | RNA | GEO | GSM1058284 |
| ICL012-R | ICL012_NS1trunc_12h_2_array | RNA | GEO | GSM1058265 |
| ICL012-R | ICL012_NS1trunc_12h_3_array | RNA | GEO | GSM1058261 |
| ICL012-R | ICL012_NS1trunc_18h_1_array | RNA | GEO | GSM1058282 |
| ICL012-R | ICL012_NS1trunc_18h_2_array | RNA | GEO | GSM1058286 |
| ICL012-R | ICL012_NS1trunc_18h_3_array | RNA | GEO | GSM1058278 |
| ICL012-R | ICL012_NS1trunc_24h_1_array | RNA | GEO | GSM1058275 |
| ICL012-R | ICL012_NS1trunc_24h_2_array | RNA | GEO | GSM1058270 |
| ICL012-R | ICL012_NS1trunc_24h_3_array | RNA | GEO | GSM1058267 |
| ICL012-R | ICL012_NS1trunc_3h_1_array | RNA | GEO | GSM1058268 |
| ICL012-R | ICL012_NS1trunc_3h_2_array | RNA | GEO | GSM1058272 |
| ICL012-R | ICL012_NS1trunc_3h_3_array | RNA | GEO | GSM1058248 |
| ICL012-R | ICL012_NS1trunc_7h_1_array | RNA | GEO | GSM1058276 |
| ICL012-R | ICL012_NS1trunc_7h_2_array | RNA | GEO | GSM1058264 |
| ICL012-R | ICL012_NS1trunc_7h_3_array | RNA | GEO | GSM1058249 |
| ICL012-R | ICL012_VN1203_24h_1_array | RNA | GEO | GSM1058252 |
| ICL012-R | ICL012_VN1203_24h_2_array | RNA | GEO | GSM1058250 |
| ICL012-R | ICL012_VN1203_24h_3_array | RNA | GEO | GSM1058277 |
| ICL012-R | ICL012_VN1203_7h_1_array | RNA | GEO | GSM1058281 |
| ICL012-R | ICL012_VN1203_7h_2_array | RNA | GEO | GSM1058247 |
| ICL012-R | ICL012_VN1203_7h_3_array | RNA | GEO | GSM1058287 |
| IM001-R | IM001_Mock_D1_1_RNA_ExpSam | RNA | GEO | GSM822968 |
| IM001-R | IM001_Mock_D1_2_RNA_ExpSam | RNA | GEO | GSM822969 |
| IM001-R | IM001_Mock_D1_3_RNA_ExpSam | RNA | GEO | GSM822970 |
| IM001-R | IM001_Mock_D2_1_RNA_ExpSam | RNA | GEO | GSM822971 |
| IM001-R | IM001_Mock_D2_2_RNA_ExpSam | RNA | GEO | GSM822972 |
| IM001-R | IM001_Mock_D2_3_RNA_ExpSam | RNA | GEO | GSM822973 |
| IM001-R | IM001_Mock_D4_1_RNA_ExpSam | RNA | GEO | GSM822974 |
| IM001-R | IM001_Mock_D4_2_RNA_ExpSam | RNA | GEO | GSM822975 |
| IM001-R | IM001_Mock_D4_3_RNA_ExpSam | RNA | GEO | GSM822976 |
| IM001-R | IM001_Mock_D7_1_RNA_ExpSam | RNA | GEO | GSM822977 |
| IM001-R | IM001_Mock_D7_2_RNA_ExpSam | RNA | GEO | GSM822978 |
| IM001-R | IM001_Mock_D7_3_RNA_ExpSam | RNA | GEO | GSM822979 |
| IM001-R | IM001_VN1203_10^2pfu_D1_1_RNA_ExpSam | RNA | GEO | GSM822980 |
| IM001-R | IM001_VN1203_10^2pfu_D1_2_RNA_ExpSam | RNA | GEO | GSM822981 |
| IM001-R | IM001_VN1203_10^2pfu_D1_3_RNA_ExpSam | RNA | GEO | GSM822982 |
| IM001-R | IM001_VN1203_10^2pfu_D1_4_RNA_ExpSam | RNA | GEO | GSM822983 |
| IM001-R | IM001_VN1203_10^2pfu_D1_5_RNA_ExpSam | RNA | GEO | GSM822984 |
| IM001-R | IM001_VN1203_10^2pfu_D2_1_RNA_ExpSam | RNA | GEO | GSM822985 |
| IM001-R | IM001_VN1203_10^2pfu_D2_2_RNA_ExpSam | RNA | GEO | GSM822986 |
| IM001-R | IM001_VN1203_10^2pfu_D2_3_RNA_ExpSam | RNA | GEO | GSM822987 |
| IM001-R | IM001_VN1203_10^2pfu_D2_4_RNA_ExpSam | RNA | GEO | GSM822988 |
| IM001-R | IM001_VN1203_10^2pfu_D2_5_RNA_ExpSam | RNA | GEO | GSM822989 |
| IM001-R | IM001_VN1203_10^2pfu_D4_1_RNA_ExpSam | RNA | GEO | GSM822990 |
| IM001-R | IM001_VN1203_10^2pfu_D4_2_RNA_ExpSam | RNA | GEO | GSM822991 |
| IM001-R | IM001_VN1203_10^2pfu_D4_3_RNA_ExpSam | RNA | GEO | GSM822992 |
| IM001-R | IM001_VN1203_10^2pfu_D4_4_RNA_ExpSam | RNA | GEO | GSM822993 |
| IM001-R | IM001_VN1203_10^2pfu_D4_5_RNA_ExpSam | RNA | GEO | GSM822994 |
| IM001-R | IM001_VN1203_10^2pfu_D7_1_RNA_ExpSam | RNA | GEO | GSM822995 |
| IM001-R | IM001_VN1203_10^2pfu_D7_2_RNA_ExpSam | RNA | GEO | GSM822996 |
| IM001-R | IM001_VN1203_10^2pfu_D7_3_RNA_ExpSam | RNA | GEO | GSM822997 |
| IM001-R | IM001_VN1203_10^2pfu_D7_4_RNA_ExpSam | RNA | GEO | GSM822998 |
| IM001-R | IM001_VN1203_10^3pfu_D1_1_RNA_ExpSam | RNA | GEO | GSM822999 |
| IM001-R | IM001_VN1203_10^3pfu_D1_2_RNA_ExpSam | RNA | GEO | GSM823000 |
| IM001-R | IM001_VN1203_10^3pfu_D1_3_RNA_ExpSam | RNA | GEO | GSM823001 |
| IM001-R | IM001_VN1203_10^3pfu_D1_4_RNA_ExpSam | RNA | GEO | GSM823002 |
| IM001-R | IM001_VN1203_10^3pfu_D1_5_RNA_ExpSam | RNA | GEO | GSM823003 |
| IM001-R | IM001_VN1203_10^3pfu_D2_1_RNA_ExpSam | RNA | GEO | GSM823004 |
| IM001-R | IM001_VN1203_10^3pfu_D2_2_RNA_ExpSam | RNA | GEO | GSM823005 |
| IM001-R | IM001_VN1203_10^3pfu_D2_3_RNA_ExpSam | RNA | GEO | GSM823006 |
| IM001-R | IM001_VN1203_10^3pfu_D2_4_RNA_ExpSam | RNA | GEO | GSM823007 |
| IM001-R | IM001_VN1203_10^3pfu_D2_5_RNA_ExpSam | RNA | GEO | GSM823008 |
| IM001-R | IM001_VN1203_10^3pfu_D4_1_RNA_ExpSam | RNA | GEO | GSM823009 |
| IM001-R | IM001_VN1203_10^3pfu_D4_2_RNA_ExpSam | RNA | GEO | GSM823010 |
| IM001-R | IM001_VN1203_10^3pfu_D4_3_RNA_ExpSam | RNA | GEO | GSM823011 |
| IM001-R | IM001_VN1203_10^3pfu_D4_4_RNA_ExpSam | RNA | GEO | GSM823012 |
| IM001-R | IM001_VN1203_10^3pfu_D4_5_RNA_ExpSam | RNA | GEO | GSM823013 |
| IM001-R | IM001_VN1203_10^3pfu_D7_1_RNA_ExpSam | RNA | GEO | GSM823014 |
| IM001-R | IM001_VN1203_10^3pfu_D7_2_RNA_ExpSam | RNA | GEO | GSM823015 |
| IM001-R | IM001_VN1203_10^3pfu_D7_3_RNA_ExpSam | RNA | GEO | GSM823016 |
| IM001-R | IM001_VN1203_10^4pfu_D1_1_RNA_ExpSam | RNA | GEO | GSM823017 |
| IM001-R | IM001_VN1203_10^4pfu_D1_2_RNA_ExpSam | RNA | GEO | GSM823018 |
| IM001-R | IM001_VN1203_10^4pfu_D1_3_RNA_ExpSam | RNA | GEO | GSM823019 |
| IM001-R | IM001_VN1203_10^4pfu_D1_4_RNA_ExpSam | RNA | GEO | GSM823020 |
| IM001-R | IM001_VN1203_10^4pfu_D1_5_RNA_ExpSam | RNA | GEO | GSM823021 |
| IM001-R | IM001_VN1203_10^4pfu_D2_1_RNA_ExpSam | RNA | GEO | GSM823022 |
| IM001-R | IM001_VN1203_10^4pfu_D2_2_RNA_ExpSam | RNA | GEO | GSM823023 |
| IM001-R | IM001_VN1203_10^4pfu_D2_3_RNA_ExpSam | RNA | GEO | GSM823024 |
| IM001-R | IM001_VN1203_10^4pfu_D2_4_RNA_ExpSam | RNA | GEO | GSM823025 |
| IM001-R | IM001_VN1203_10^4pfu_D2_5_RNA_ExpSam | RNA | GEO | GSM823026 |
| IM001-R | IM001_VN1203_10^4pfu_D4_1_RNA_ExpSam | RNA | GEO | GSM823027 |
| IM001-R | IM001_VN1203_10^4pfu_D4_2_RNA_ExpSam | RNA | GEO | GSM823028 |
| IM001-R | IM001_VN1203_10^4pfu_D4_3_RNA_ExpSam | RNA | GEO | GSM823029 |
| IM001-R | IM001_VN1203_10^4pfu_D4_4_RNA_ExpSam | RNA | GEO | GSM823030 |
| IM001-R | IM001_VN1203_10^4pfu_D4_5_RNA_ExpSam | RNA | GEO | GSM823031 |
| IM002-R | IM002_Brisbane_10^6pfu_D1_2_array | RNA | GEO | GSM888961 |
| IM002-R | IM002_Brisbane_10^6pfu_D1_3_array | RNA | GEO | GSM888973 |
| IM002-R | IM002_Brisbane_10^6pfu_D3_2_array | RNA | GEO | GSM888958 |
| IM002-R | IM002_Brisbane_10^6pfu_D3_3_array | RNA | GEO | GSM888965 |
| IM002-R | IM002_Brisbane_10^6pfu_D5_1_array | RNA | GEO | GSM888966 |
| IM002-R | IM002_Brisbane_10^6pfu_D5_2_array | RNA | GEO | GSM888968 |
| IM002-R | IM002_Brisbane_10^6pfu_D5_3_array | RNA | GEO | GSM888975 |
| IM002-R | IM002_Mex_10^6pfu_D1_1_array | RNA | GEO | GSM888960 |
| IM002-R | IM002_Mex_10^6pfu_D1_2_array | RNA | GEO | GSM888982 |
| IM002-R | IM002_Mex_10^6pfu_D1_3_array | RNA | GEO | GSM888978 |
| IM002-R | IM002_Mex_10^6pfu_D3_1_array | RNA | GEO | GSM888992 |
| IM002-R | IM002_Mex_10^6pfu_D3_2_array | RNA | GEO | GSM888989 |
| IM002-R | IM002_Mex_10^6pfu_D3_3_array | RNA | GEO | GSM888988 |
| IM002-R | IM002_Mex_10^6pfu_D5_1_array | RNA | GEO | GSM888972 |
| IM002-R | IM002_Mex_10^6pfu_D5_2_array | RNA | GEO | GSM888964 |
| IM002-R | IM002_Mex_10^6pfu_D5_3_array | RNA | GEO | GSM888983 |
| IM002-R | IM002_Mock_D1_1_array | RNA | GEO | GSM888979 |
| IM002-R | IM002_Mock_D1_2_array | RNA | GEO | GSM888976 |
| IM002-R | IM002_Mock_D3_1_array | RNA | GEO | GSM888956 |
| IM002-R | IM002_Mock_D3_2_array | RNA | GEO | GSM888981 |
| IM002-R | IM002_Mock_D5_2_array | RNA | GEO | GSM888991 |
| IM002-R | IM002_NJ_10^6pfu_D1_1_array | RNA | GEO | GSM888962 |
| IM002-R | IM002_NJ_10^6pfu_D1_3_array | RNA | GEO | GSM888990 |
| IM002-R | IM002_NJ_10^6pfu_D3_1_array | RNA | GEO | GSM888984 |
| IM002-R | IM002_NJ_10^6pfu_D3_2_array | RNA | GEO | GSM888959 |
| IM002-R | IM002_NJ_10^6pfu_D3_3_array | RNA | GEO | GSM888969 |
| IM002-R | IM002_NJ_10^6pfu_D5_2_array | RNA | GEO | GSM888967 |
| IM002-R | IM002_NJ_10^6pfu_D5_3_array | RNA | GEO | GSM888971 |
| IM002-R | IM002_r1918_10^6pfu_D1_1_array | RNA | GEO | GSM888974 |
| IM002-R | IM002_r1918_10^6pfu_D1_2_array | RNA | GEO | GSM888986 |
| IM002-R | IM002_r1918_10^6pfu_D1_3_array | RNA | GEO | GSM888957 |
| IM002-R | IM002_r1918_10^6pfu_D3_1_array | RNA | GEO | GSM888985 |
| IM002-R | IM002_r1918_10^6pfu_D3_2_array | RNA | GEO | GSM888980 |
| IM002-R | IM002_r1918_10^6pfu_D3_3_array | RNA | GEO | GSM888955 |
| IM002-R | IM002_r1918_10^6pfu_D5_1_array | RNA | GEO | GSM888963 |
| IM002-R | IM002_r1918_10^6pfu_D5_2_array | RNA | GEO | GSM888970 |
| IM002-R | IM002_r1918_10^6pfu_D5_3_array | RNA | GEO | GSM888977 |
| IM004-R | IM004_HAavirulent_10^4pfu_1d_1_RNA_ExpSam | RNA | GEO | GSM921978 |
| IM004-R | IM004_HAavirulent_10^4pfu_1d_2_RNA_ExpSam | RNA | GEO | GSM921977 |
| IM004-R | IM004_HAavirulent_10^4pfu_1d_3_RNA_ExpSam | RNA | GEO | GSM921976 |
| IM004-R | IM004_HAavirulent_10^4pfu_1d_4_RNA_ExpSam | RNA | GEO | GSM921975 |
| IM004-R | IM004_HAavirulent_10^4pfu_1d_5_RNA_ExpSam | RNA | GEO | GSM921974 |
| IM004-R | IM004_HAavirulent_10^4pfu_2d_1_RNA_ExpSam | RNA | GEO | GSM921973 |
| IM004-R | IM004_HAavirulent_10^4pfu_2d_2_RNA_ExpSam | RNA | GEO | GSM921972 |
| IM004-R | IM004_HAavirulent_10^4pfu_2d_3_RNA_ExpSam | RNA | GEO | GSM921971 |
| IM004-R | IM004_HAavirulent_10^4pfu_2d_4_RNA_ExpSam | RNA | GEO | GSM921970 |
| IM004-R | IM004_HAavirulent_10^4pfu_2d_5_RNA_ExpSam | RNA | GEO | GSM921969 |
| IM004-R | IM004_HAavirulent_10^4pfu_4d_1_RNA_ExpSam | RNA | GEO | GSM921968 |
| IM004-R | IM004_HAavirulent_10^4pfu_4d_2_RNA_ExpSam | RNA | GEO | GSM921967 |
| IM004-R | IM004_HAavirulent_10^4pfu_4d_3_RNA_ExpSam | RNA | GEO | GSM921966 |
| IM004-R | IM004_HAavirulent_10^4pfu_4d_4_RNA_ExpSam | RNA | GEO | GSM921965 |
| IM004-R | IM004_HAavirulent_10^4pfu_4d_5_RNA_ExpSam | RNA | GEO | GSM921964 |
| IM004-R | IM004_HAavirulent_10^4pfu_7d_1_RNA_ExpSam | RNA | GEO | GSM921963 |
| IM004-R | IM004_HAavirulent_10^4pfu_7d_2_RNA_ExpSam | RNA | GEO | GSM921962 |
| IM004-R | IM004_HAavirulent_10^4pfu_7d_3_RNA_ExpSam | RNA | GEO | GSM921961 |
| IM004-R | IM004_HAavirulent_10^4pfu_7d_4_RNA_ExpSam | RNA | GEO | GSM921960 |
| IM004-R | IM004_HAavirulent_10^4pfu_7d_5_RNA_ExpSam | RNA | GEO | GSM921959 |
| IM004-R | IM004_mock_1d_1_RNA_ExpSam | RNA | GEO | GSM921958 |
| IM004-R | IM004_mock_1d_2_RNA_ExpSam | RNA | GEO | GSM921957 |
| IM004-R | IM004_mock_1d_3_RNA_ExpSam | RNA | GEO | GSM921956 |
| IM004-R | IM004_mock_2d_1_RNA_ExpSam | RNA | GEO | GSM921955 |
| IM004-R | IM004_mock_2d_2_RNA_ExpSam | RNA | GEO | GSM921954 |
| IM004-R | IM004_mock_2d_3_RNA_ExpSam | RNA | GEO | GSM921953 |
| IM004-R | IM004_mock_4d_1_RNA_ExpSam | RNA | GEO | GSM921952 |
| IM004-R | IM004_mock_4d_2_RNA_ExpSam | RNA | GEO | GSM921951 |
| IM004-R | IM004_mock_4d_3_RNA_ExpSam | RNA | GEO | GSM921950 |
| IM004-R | IM004_mock_7d_1_RNA_ExpSam | RNA | GEO | GSM921949 |
| IM004-R | IM004_mock_7d_2_RNA_ExpSam | RNA | GEO | GSM921948 |
| IM004-R | IM004_mock_7d_3_RNA_ExpSam | RNA | GEO | GSM921947 |
| IM005-R | IM005_Mock_1d_1_array | RNA | GEO | GSM1060268 |
| IM005-R | IM005_Mock_1d_2_array | RNA | GEO | GSM1060285 |
| IM005-R | IM005_Mock_1d_3_array | RNA | GEO | GSM1060278 |
| IM005-R | IM005_Mock_2d_1_array | RNA | GEO | GSM1060271 |
| IM005-R | IM005_Mock_2d_2_array | RNA | GEO | GSM1060261 |
| IM005-R | IM005_Mock_2d_3_array | RNA | GEO | GSM1060267 |
| IM005-R | IM005_Mock_4d_1_array | RNA | GEO | GSM1060281 |
| IM005-R | IM005_Mock_4d_2_array | RNA | GEO | GSM1060275 |
| IM005-R | IM005_Mock_4d_3_array | RNA | GEO | GSM1060273 |
| IM005-R | IM005_Mock_7d_1_array | RNA | GEO | GSM1060260 |
| IM005-R | IM005_Mock_7d_2_array | RNA | GEO | GSM1060287 |
| IM005-R | IM005_Mock_7d_3_array | RNA | GEO | GSM1060265 |
| IM005-R | IM005_PB2_627E_10^4pfu_1d_1_array | RNA | GEO | GSM1060288 |
| IM005-R | IM005_PB2_627E_10^4pfu_1d_2_array | RNA | GEO | GSM1060263 |
| IM005-R | IM005_PB2_627E_10^4pfu_1d_3_array | RNA | GEO | GSM1060269 |
| IM005-R | IM005_PB2_627E_10^4pfu_1d_4_array | RNA | GEO | GSM1060274 |
| IM005-R | IM005_PB2_627E_10^4pfu_1d_5_array | RNA | GEO | GSM1060258 |
| IM005-R | IM005_PB2_627E_10^4pfu_2d_1_array | RNA | GEO | GSM1060282 |
| IM005-R | IM005_PB2_627E_10^4pfu_2d_2_array | RNA | GEO | GSM1060259 |
| IM005-R | IM005_PB2_627E_10^4pfu_2d_3_array | RNA | GEO | GSM1060276 |
| IM005-R | IM005_PB2_627E_10^4pfu_2d_4_array | RNA | GEO | GSM1060272 |
| IM005-R | IM005_PB2_627E_10^4pfu_2d_5_array | RNA | GEO | GSM1060280 |
| IM005-R | IM005_PB2_627E_10^4pfu_4d_1_array | RNA | GEO | GSM1060266 |
| IM005-R | IM005_PB2_627E_10^4pfu_4d_2_array | RNA | GEO | GSM1060262 |
| IM005-R | IM005_PB2_627E_10^4pfu_4d_3_array | RNA | GEO | GSM1060257 |
| IM005-R | IM005_PB2_627E_10^4pfu_4d_4_array | RNA | GEO | GSM1060286 |
| IM005-R | IM005_PB2_627E_10^4pfu_4d_5_array | RNA | GEO | GSM1060284 |
| IM005-R | IM005_PB2_627E_10^4pfu_7d_1_array | RNA | GEO | GSM1060283 |
| IM005-R | IM005_PB2_627E_10^4pfu_7d_2_array | RNA | GEO | GSM1060277 |
| IM005-R | IM005_PB2_627E_10^4pfu_7d_3_array | RNA | GEO | GSM1060270 |
| IM005-R | IM005_PB2_627E_10^4pfu_7d_4_array | RNA | GEO | GSM1060279 |
| IM005-R | IM005_PB2_627E_10^4pfu_7d_5_array | RNA | GEO | GSM1060264 |
| IM006A-R | IM006A_Mock_1d_1_array | RNA | GEO | GSM1060316 |
| IM006A-R | IM006A_Mock_1d_2_array | RNA | GEO | GSM1060297 |
| IM006A-R | IM006A_Mock_1d_3_array | RNA | GEO | GSM1060304 |
| IM006A-R | IM006A_Mock_2d_1_array | RNA | GEO | GSM1060307 |
| IM006A-R | IM006A_Mock_2d_2_array | RNA | GEO | GSM1060289 |
| IM006A-R | IM006A_Mock_2d_3_array | RNA | GEO | GSM1060303 |
| IM006A-R | IM006A_Mock_4d_1_array | RNA | GEO | GSM1060295 |
| IM006A-R | IM006A_Mock_4d_2_array | RNA | GEO | GSM1060313 |
| IM006A-R | IM006A_Mock_4d_3_array | RNA | GEO | GSM1060319 |
| IM006A-R | IM006A_Mock_7d_1_array | RNA | GEO | GSM1060296 |
| IM006A-R | IM006A_Mock_7d_2_array | RNA | GEO | GSM1060306 |
| IM006A-R | IM006A_Mock_7d_3_array | RNA | GEO | GSM1060301 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_1_array | RNA | GEO | GSM1060299 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_2_array | RNA | GEO | GSM1060308 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_3_array | RNA | GEO | GSM1060291 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_4_array | RNA | GEO | GSM1060293 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_5_array | RNA | GEO | GSM1060310 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_1_array | RNA | GEO | GSM1060318 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_2_array | RNA | GEO | GSM1060312 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_3_array | RNA | GEO | GSM1060298 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_4_array | RNA | GEO | GSM1060294 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_5_array | RNA | GEO | GSM1060315 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_1_array | RNA | GEO | GSM1060300 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_2_array | RNA | GEO | GSM1060302 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_3_array | RNA | GEO | GSM1060314 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_4_array | RNA | GEO | GSM1060309 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_5_array | RNA | GEO | GSM1060290 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_7d_1_array | RNA | GEO | GSM1060311 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_7d_2_array | RNA | GEO | GSM1060305 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_7d_3_array | RNA | GEO | GSM1060292 |
| IM006A-R | IM006A_PB1_F2del_10^3pfu_7d_5_array | RNA | GEO | GSM1060317 |
| IM006B-R | IM006B_Mock_1d_1_array | RNA | GEO | GSM1085373 |
| IM006B-R | IM006B_Mock_1d_2_array | RNA | GEO | GSM1085385 |
| IM006B-R | IM006B_Mock_1d_3_array | RNA | GEO | GSM1085396 |
| IM006B-R | IM006B_Mock_2d_1_array | RNA | GEO | GSM1085387 |
| IM006B-R | IM006B_Mock_2d_2_array | RNA | GEO | GSM1085379 |
| IM006B-R | IM006B_Mock_2d_3_array | RNA | GEO | GSM1085389 |
| IM006B-R | IM006B_Mock_4d_1_array | RNA | GEO | GSM1085384 |
| IM006B-R | IM006B_Mock_4d_2_array | RNA | GEO | GSM1085376 |
| IM006B-R | IM006B_Mock_4d_3_array | RNA | GEO | GSM1085391 |
| IM006B-R | IM006B_Mock_7d_1_array | RNA | GEO | GSM1085393 |
| IM006B-R | IM006B_Mock_7d_2_array | RNA | GEO | GSM1085382 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_1_array | RNA | GEO | GSM1085383 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_2_array | RNA | GEO | GSM1085397 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_3_array | RNA | GEO | GSM1085378 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_4_array | RNA | GEO | GSM1085380 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_5_array | RNA | GEO | GSM1085386 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_1_array | RNA | GEO | GSM1085392 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_2_array | RNA | GEO | GSM1085377 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_3_array | RNA | GEO | GSM1085374 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_4_array | RNA | GEO | GSM1085388 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_5_array | RNA | GEO | GSM1085381 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_1_array | RNA | GEO | GSM1085398 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_2_array | RNA | GEO | GSM1085395 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_3_array | RNA | GEO | GSM1085375 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_4_array | RNA | GEO | GSM1085394 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_5_array | RNA | GEO | GSM1085390 |
| IM006B-R | IM006B_PB1_F2del_10^4pfu_7d_1_array | RNA | GEO | GSM1085372 |
| IM007-R | IM007_Mock_1d_1_array | RNA | GEO | GSM1085555 |
| IM007-R | IM007_Mock_1d_2_array | RNA | GEO | GSM1085571 |
| IM007-R | IM007_Mock_1d_3_array | RNA | GEO | GSM1085545 |
| IM007-R | IM007_Mock_2d_1_array | RNA | GEO | GSM1085564 |
| IM007-R | IM007_Mock_2d_2_array | RNA | GEO | GSM1085558 |
| IM007-R | IM007_Mock_2d_3_array | RNA | GEO | GSM1085547 |
| IM007-R | IM007_Mock_4d_1_array | RNA | GEO | GSM1085590 |
| IM007-R | IM007_Mock_4d_2_array | RNA | GEO | GSM1085551 |
| IM007-R | IM007_Mock_4d_3_array | RNA | GEO | GSM1085562 |
| IM007-R | IM007_Mock_7d_1_array | RNA | GEO | GSM1085567 |
| IM007-R | IM007_Mock_7d_2_array | RNA | GEO | GSM1085539 |
| IM007-R | IM007_Mock_7d_3_array | RNA | GEO | GSM1085542 |
| IM007-R | IM007_NS1trunc124_10^3pfu_1d_1_array | RNA | GEO | GSM1085575 |
| IM007-R | IM007_NS1trunc124_10^3pfu_1d_2_array | RNA | GEO | GSM1085568 |
| IM007-R | IM007_NS1trunc124_10^3pfu_1d_3_array | RNA | GEO | GSM1085577 |
| IM007-R | IM007_NS1trunc124_10^3pfu_1d_4_array | RNA | GEO | GSM1085559 |
| IM007-R | IM007_NS1trunc124_10^3pfu_1d_5_array | RNA | GEO | GSM1085579 |
| IM007-R | IM007_NS1trunc124_10^3pfu_2d_1_array | RNA | GEO | GSM1085580 |
| IM007-R | IM007_NS1trunc124_10^3pfu_2d_2_array | RNA | GEO | GSM1085581 |
| IM007-R | IM007_NS1trunc124_10^3pfu_2d_3_array | RNA | GEO | GSM1085572 |
| IM007-R | IM007_NS1trunc124_10^3pfu_2d_4_array | RNA | GEO | GSM1085560 |
| IM007-R | IM007_NS1trunc124_10^3pfu_2d_5_array | RNA | GEO | GSM1085552 |
| IM007-R | IM007_NS1trunc124_10^3pfu_4d_1_array | RNA | GEO | GSM1085543 |
| IM007-R | IM007_NS1trunc124_10^3pfu_4d_2_array | RNA | GEO | GSM1085553 |
| IM007-R | IM007_NS1trunc124_10^3pfu_4d_3_array | RNA | GEO | GSM1085582 |
| IM007-R | IM007_NS1trunc124_10^3pfu_4d_4_array | RNA | GEO | GSM1085583 |
| IM007-R | IM007_NS1trunc124_10^3pfu_4d_5_array | RNA | GEO | GSM1085584 |
| IM007-R | IM007_NS1trunc124_10^3pfu_7d_1_array | RNA | GEO | GSM1085574 |
| IM007-R | IM007_NS1trunc124_10^3pfu_7d_2_array | RNA | GEO | GSM1085548 |
| IM007-R | IM007_NS1trunc124_10^3pfu_7d_3_array | RNA | GEO | GSM1085566 |
| IM007-R | IM007_NS1trunc124_10^3pfu_7d_4_array | RNA | GEO | GSM1085546 |
| IM007-R | IM007_NS1trunc124_10^3pfu_7d_5_array | RNA | GEO | GSM1085563 |
| IM007-R | IM007_NS1trunc124_10^4pfu_1d_1_array | RNA | GEO | GSM1085578 |
| IM007-R | IM007_NS1trunc124_10^4pfu_1d_2_array | RNA | GEO | GSM1085550 |
| IM007-R | IM007_NS1trunc124_10^4pfu_1d_3_array | RNA | GEO | GSM1085540 |
| IM007-R | IM007_NS1trunc124_10^4pfu_1d_4_array | RNA | GEO | GSM1085557 |
| IM007-R | IM007_NS1trunc124_10^4pfu_1d_5_array | RNA | GEO | GSM1085570 |
| IM007-R | IM007_NS1trunc124_10^4pfu_2d_1_array | RNA | GEO | GSM1085549 |
| IM007-R | IM007_NS1trunc124_10^4pfu_2d_2_array | RNA | GEO | GSM1085561 |
| IM007-R | IM007_NS1trunc124_10^4pfu_2d_3_array | RNA | GEO | GSM1085554 |
| IM007-R | IM007_NS1trunc124_10^4pfu_2d_4_array | RNA | GEO | GSM1085585 |
| IM007-R | IM007_NS1trunc124_10^4pfu_2d_5_array | RNA | GEO | GSM1085586 |
| IM007-R | IM007_NS1trunc124_10^4pfu_4d_1_array | RNA | GEO | GSM1085544 |
| IM007-R | IM007_NS1trunc124_10^4pfu_4d_2_array | RNA | GEO | GSM1085556 |
| IM007-R | IM007_NS1trunc124_10^4pfu_4d_3_array | RNA | GEO | GSM1085569 |
| IM007-R | IM007_NS1trunc124_10^4pfu_4d_4_array | RNA | GEO | GSM1085541 |
| IM007-R | IM007_NS1trunc124_10^4pfu_4d_5_array | RNA | GEO | GSM1085573 |
| IM007-R | IM007_NS1trunc124_10^4pfu_7d_1_array | RNA | GEO | GSM1085565 |
| IM007-R | IM007_NS1trunc124_10^4pfu_7d_2_array | RNA | GEO | GSM1085587 |
| IM007-R | IM007_NS1trunc124_10^4pfu_7d_3_array | RNA | GEO | GSM1085588 |
| IM007-R | IM007_NS1trunc124_10^4pfu_7d_4_array | RNA | GEO | GSM1085589 |
| IM007-R | IM007_NS1trunc124_10^4pfu_7d_5_array | RNA | GEO | GSM1085576 |
| IM009-R | IM009_CA04_10^6pfu_D1_1_array | RNA | GEO | GSM888953 |
| IM009-R | IM009_CA04_10^6pfu_D1_2_array | RNA | GEO | GSM888936 |
| IM009-R | IM009_CA04_10^6pfu_D1_3_array | RNA | GEO | GSM888950 |
| IM009-R | IM009_CA04_10^6pfu_D3_1_array | RNA | GEO | GSM888947 |
| IM009-R | IM009_CA04_10^6pfu_D3_2_array | RNA | GEO | GSM888940 |
| IM009-R | IM009_CA04_10^6pfu_D3_3_array | RNA | GEO | GSM888942 |
| IM009-R | IM009_CA04_10^6pfu_D5_1_array | RNA | GEO | GSM888937 |
| IM009-R | IM009_CA04_10^6pfu_D5_2_array | RNA | GEO | GSM888929 |
| IM009-R | IM009_CA04_10^6pfu_D5_3_array | RNA | GEO | GSM888931 |
| IM009-R | IM009_MA-CA04_10^6pfu_D1_1_array | RNA | GEO | GSM888952 |
| IM009-R | IM009_MA-CA04_10^6pfu_D1_2_array | RNA | GEO | GSM888943 |
| IM009-R | IM009_MA-CA04_10^6pfu_D1_3_array | RNA | GEO | GSM888935 |
| IM009-R | IM009_MA-CA04_10^6pfu_D3_1_array | RNA | GEO | GSM888946 |
| IM009-R | IM009_MA-CA04_10^6pfu_D3_2_array | RNA | GEO | GSM888930 |
| IM009-R | IM009_MA-CA04_10^6pfu_D3_3_array | RNA | GEO | GSM888945 |
| IM009-R | IM009_MA-CA04_10^6pfu_D5_1_array | RNA | GEO | GSM888954 |
| IM009-R | IM009_MA-CA04_10^6pfu_D5_2_array | RNA | GEO | GSM888939 |
| IM009-R | IM009_MA-CA04_10^6pfu_D5_3_array | RNA | GEO | GSM888934 |
| IM009-R | IM009_Mock_D1_1_array | RNA | GEO | GSM888948 |
| IM009-R | IM009_Mock_D1_2_array | RNA | GEO | GSM888941 |
| IM009-R | IM009_Mock_D1_3_array | RNA | GEO | GSM888933 |
| IM009-R | IM009_Mock_D3_1_array | RNA | GEO | GSM888932 |
| IM009-R | IM009_Mock_D3_2_array | RNA | GEO | GSM888951 |
| IM009-R | IM009_Mock_D3_3_array | RNA | GEO | GSM888938 |
| IM009-R | IM009_Mock_D5_1_array | RNA | GEO | GSM888944 |
| IM009-R | IM009_Mock_D5_2_array | RNA | GEO | GSM888949 |
| IM010-R | IM010_B6_Mock_2d_1_array | RNA | GEO | GSM1001726 |
| IM010-R | IM010_B6_Mock_2d_2_array | RNA | GEO | GSM1001727 |
| IM010-R | IM010_B6_Mock_2d_3_array | RNA | GEO | GSM1001728 |
| IM010-R | IM010_B6_VN1203_2d_1_array | RNA | GEO | GSM1001729 |
| IM010-R | IM010_B6_VN1203_2d_2_array | RNA | GEO | GSM1001730 |
| IM010-R | IM010_B6_VN1203_2d_3_array | RNA | GEO | GSM1001731 |
| IM010-R | IM010_IDO1_Mock_2d_1_array | RNA | GEO | GSM1001704 |
| IM010-R | IM010_IDO1_Mock_2d_2_array | RNA | GEO | GSM1001705 |
| IM010-R | IM010_IDO1_Mock_2d_3_array | RNA | GEO | GSM1001706 |
| IM010-R | IM010_IDO1_Mock_6d_1_array | RNA | GEO | GSM1001707 |
| IM010-R | IM010_IDO1_Mock_6d_2_array | RNA | GEO | GSM1001708 |
| IM010-R | IM010_IDO1_Mock_6d_3_array | RNA | GEO | GSM1001709 |
| IM010-R | IM010_IDO1_VN1203_2d_1_array | RNA | GEO | GSM1001710 |
| IM010-R | IM010_IDO1_VN1203_2d_2_array | RNA | GEO | GSM1001711 |
| IM010-R | IM010_IDO1_VN1203_2d_3_array | RNA | GEO | GSM1001712 |
| IM010-R | IM010_IDO1_VN1203_6d_1_array | RNA | GEO | GSM1001713 |
| IM010-R | IM010_IDO1_VN1203_6d_2_array | RNA | GEO | GSM1001714 |
| IM010-R | IM010_Tnfrsf1b_Mock_2d_1_array | RNA | GEO | GSM1001715 |
| IM010-R | IM010_Tnfrsf1b_Mock_2d_2_array | RNA | GEO | GSM1001716 |
| IM010-R | IM010_Tnfrsf1b_Mock_2d_3_array | RNA | GEO | GSM1001717 |
| IM010-R | IM010_Tnfrsf1b_Mock_6d_1_array | RNA | GEO | GSM1001718 |
| IM010-R | IM010_Tnfrsf1b_Mock_6d_2_array | RNA | GEO | GSM1001719 |
| IM010-R | IM010_Tnfrsf1b_Mock_6d_3_array | RNA | GEO | GSM1001720 |
| IM010-R | IM010_Tnfrsf1b_VN1203_2d_1_array | RNA | GEO | GSM1001721 |
| IM010-R | IM010_Tnfrsf1b_VN1203_2d_2_array | RNA | GEO | GSM1001722 |
| IM010-R | IM010_Tnfrsf1b_VN1203_2d_3_array | RNA | GEO | GSM1001723 |
| IM010-R | IM010_Tnfrsf1b_VN1203_6d_1_array | RNA | GEO | GSM1001724 |
| IM010-R | IM010_Tnfrsf1b_VN1203_6d_2_array | RNA | GEO | GSM1001725 |
| SBRI_AA_E1 | AA_Mock_001 | RNA | GEO | GSM914274 |
| SBRI_AA_E1 | AA_Mock_002 | RNA | GEO | GSM914275 |
| SBRI_AA_E1 | AA_Mock_003 | RNA | GEO | GSM914276 |
| SBRI_AA_E1 | AA_Mock_005 | RNA | GEO | GSM914277 |
| SBRI_AA_E1 | AA_PR8_006 | RNA | GEO | GSM914259 |
| SBRI_AA_E1 | AA_PR8_007 | RNA | GEO | GSM914260 |
| SBRI_AA_E1 | AA_PR8_008 | RNA | GEO | GSM914261 |
| SBRI_AA_E1 | AA_PR8_009 | RNA | GEO | GSM914262 |
| SBRI_AA_E1 | AA_PR8_010 | RNA | GEO | GSM914263 |
| SBRI_AA_E1 | AA_VN6+2_016 | RNA | GEO | GSM914264 |
| SBRI_AA_E1 | AA_VN6+2_017 | RNA | GEO | GSM914265 |
| SBRI_AA_E1 | AA_VN6+2_018 | RNA | GEO | GSM914266 |
| SBRI_AA_E1 | AA_VN6+2_019 | RNA | GEO | GSM914267 |
| SBRI_AA_E1 | AA_VN6+2_020 | RNA | GEO | GSM914268 |
| SBRI_AA_E1 | AA_X31_011 | RNA | GEO | GSM914269 |
| SBRI_AA_E1 | AA_X31_012 | RNA | GEO | GSM914270 |
| SBRI_AA_E1 | AA_X31_013 | RNA | GEO | GSM914271 |
| SBRI_AA_E1 | AA_X31_014 | RNA | GEO | GSM914272 |
| SBRI_AA_E1 | AA_X31_015 | RNA | GEO | GSM914273 |
| SCL005-R | SCL005_DORF6_0H_1_RNA_ExpSam | RNA | GEO | GSM823243 |
| SCL005-R | SCL005_DORF6_0H_2_RNA_ExpSam | RNA | GEO | GSM823244 |
| SCL005-R | SCL005_DORF6_0H_3_RNA_ExpSam | RNA | GEO | GSM823245 |
| SCL005-R | SCL005_DORF6_12H_1_RNA_ExpSam | RNA | GEO | GSM823252 |
| SCL005-R | SCL005_DORF6_12H_2_RNA_ExpSam | RNA | GEO | GSM823253 |
| SCL005-R | SCL005_DORF6_12H_3_RNA_ExpSam | RNA | GEO | GSM823254 |
| SCL005-R | SCL005_DORF6_24H_1_RNA_ExpSam | RNA | GEO | GSM823255 |
| SCL005-R | SCL005_DORF6_24H_2_RNA_ExpSam | RNA | GEO | GSM823256 |
| SCL005-R | SCL005_DORF6_24H_3_RNA_ExpSam | RNA | GEO | GSM823257 |
| SCL005-R | SCL005_DORF6_30H_1_RNA_ExpSam | RNA | GEO | GSM823258 |
| SCL005-R | SCL005_DORF6_30H_2_RNA_ExpSam | RNA | GEO | GSM823259 |
| SCL005-R | SCL005_DORF6_30H_3_RNA_ExpSam | RNA | GEO | GSM823260 |
| SCL005-R | SCL005_DORF6_36H_1_RNA_ExpSam | RNA | GEO | GSM823261 |
| SCL005-R | SCL005_DORF6_36H_2_RNA_ExpSam | RNA | GEO | GSM823262 |
| SCL005-R | SCL005_DORF6_36H_3_RNA_ExpSam | RNA | GEO | GSM823263 |
| SCL005-R | SCL005_DORF6_3H_1_RNA_ExpSam | RNA | GEO | GSM823246 |
| SCL005-R | SCL005_DORF6_3H_2_RNA_ExpSam | RNA | GEO | GSM823247 |
| SCL005-R | SCL005_DORF6_3H_3_RNA_ExpSam | RNA | GEO | GSM823248 |
| SCL005-R | SCL005_DORF6_48H_1_RNA_ExpSam | RNA | GEO | GSM823264 |
| SCL005-R | SCL005_DORF6_48H_2_RNA_ExpSam | RNA | GEO | GSM823265 |
| SCL005-R | SCL005_DORF6_48H_3_RNA_ExpSam | RNA | GEO | GSM823266 |
| SCL005-R | SCL005_DORF6_54H_1_RNA_ExpSam | RNA | GEO | GSM823267 |
| SCL005-R | SCL005_DORF6_54H_2_RNA_ExpSam | RNA | GEO | GSM823268 |
| SCL005-R | SCL005_DORF6_54H_3_RNA_ExpSam | RNA | GEO | GSM823269 |
| SCL005-R | SCL005_DORF6_60H_1_RNA_ExpSam | RNA | GEO | GSM823270 |
| SCL005-R | SCL005_DORF6_60H_2_RNA_ExpSam | RNA | GEO | GSM823271 |
| SCL005-R | SCL005_DORF6_60H_3_RNA_ExpSam | RNA | GEO | GSM823272 |
| SCL005-R | SCL005_DORF6_72H_1_RNA_ExpSam | RNA | GEO | GSM823273 |
| SCL005-R | SCL005_DORF6_72H_2_RNA_ExpSam | RNA | GEO | GSM823274 |
| SCL005-R | SCL005_DORF6_72H_3_RNA_ExpSam | RNA | GEO | GSM823275 |
| SCL005-R | SCL005_DORF6_7H_1_RNA_ExpSam | RNA | GEO | GSM823249 |
| SCL005-R | SCL005_DORF6_7H_2_RNA_ExpSam | RNA | GEO | GSM823250 |
| SCL005-R | SCL005_DORF6_7H_3_RNA_ExpSam | RNA | GEO | GSM823251 |
| SCL005-R | SCL005_mock_0H_1_RNA_ExpSam | RNA | GEO | GSM823177 |
| SCL005-R | SCL005_mock_0H_2_RNA_ExpSam | RNA | GEO | GSM823178 |
| SCL005-R | SCL005_mock_0H_3_RNA_ExpSam | RNA | GEO | GSM823179 |
| SCL005-R | SCL005_mock_12H_1_RNA_ExpSam | RNA | GEO | GSM823186 |
| SCL005-R | SCL005_mock_12H_2_RNA_ExpSam | RNA | GEO | GSM823187 |
| SCL005-R | SCL005_mock_12H_3_RNA_ExpSam | RNA | GEO | GSM823188 |
| SCL005-R | SCL005_mock_24H_1_RNA_ExpSam | RNA | GEO | GSM823189 |
| SCL005-R | SCL005_mock_24H_2_RNA_ExpSam | RNA | GEO | GSM823190 |
| SCL005-R | SCL005_mock_24H_3_RNA_ExpSam | RNA | GEO | GSM823191 |
| SCL005-R | SCL005_mock_30H_1_RNA_ExpSam | RNA | GEO | GSM823192 |
| SCL005-R | SCL005_mock_30H_2_RNA_ExpSam | RNA | GEO | GSM823193 |
| SCL005-R | SCL005_mock_30H_3_RNA_ExpSam | RNA | GEO | GSM823194 |
| SCL005-R | SCL005_mock_36H_1_RNA_ExpSam | RNA | GEO | GSM823195 |
| SCL005-R | SCL005_mock_36H_2_RNA_ExpSam | RNA | GEO | GSM823196 |
| SCL005-R | SCL005_mock_36H_3_RNA_ExpSam | RNA | GEO | GSM823197 |
| SCL005-R | SCL005_mock_3H_1_RNA_ExpSam | RNA | GEO | GSM823180 |
| SCL005-R | SCL005_mock_3H_2_RNA_ExpSam | RNA | GEO | GSM823181 |
| SCL005-R | SCL005_mock_3H_3_RNA_ExpSam | RNA | GEO | GSM823182 |
| SCL005-R | SCL005_mock_48H_1_RNA_ExpSam | RNA | GEO | GSM823198 |
| SCL005-R | SCL005_mock_48H_2_RNA_ExpSam | RNA | GEO | GSM823199 |
| SCL005-R | SCL005_mock_48H_3_RNA_ExpSam | RNA | GEO | GSM823200 |
| SCL005-R | SCL005_mock_54H_1_RNA_ExpSam | RNA | GEO | GSM823201 |
| SCL005-R | SCL005_mock_54H_2_RNA_ExpSam | RNA | GEO | GSM823202 |
| SCL005-R | SCL005_mock_54H_3_RNA_ExpSam | RNA | GEO | GSM823203 |
| SCL005-R | SCL005_mock_60H_1_RNA_ExpSam | RNA | GEO | GSM823204 |
| SCL005-R | SCL005_mock_60H_2_RNA_ExpSam | RNA | GEO | GSM823205 |
| SCL005-R | SCL005_mock_60H_3_RNA_ExpSam | RNA | GEO | GSM823206 |
| SCL005-R | SCL005_mock_72H_1_RNA_ExpSam | RNA | GEO | GSM823207 |
| SCL005-R | SCL005_mock_72H_2_RNA_ExpSam | RNA | GEO | GSM823208 |
| SCL005-R | SCL005_mock_72H_3_RNA_ExpSam | RNA | GEO | GSM823209 |
| SCL005-R | SCL005_mock_7H_1_RNA_ExpSam | RNA | GEO | GSM823183 |
| SCL005-R | SCL005_mock_7H_2_RNA_ExpSam | RNA | GEO | GSM823184 |
| SCL005-R | SCL005_mock_7H_3_RNA_ExpSam | RNA | GEO | GSM823185 |
| SCL005-R | SCL005_WT_0H_1_RNA_ExpSam | RNA | GEO | GSM823210 |
| SCL005-R | SCL005_WT_0H_2_RNA_ExpSam | RNA | GEO | GSM823211 |
| SCL005-R | SCL005_WT_0H_3_RNA_ExpSam | RNA | GEO | GSM823212 |
| SCL005-R | SCL005_WT_12H_1_RNA_ExpSam | RNA | GEO | GSM823219 |
| SCL005-R | SCL005_WT_12H_2_RNA_ExpSam | RNA | GEO | GSM823220 |
| SCL005-R | SCL005_WT_12H_3_RNA_ExpSam | RNA | GEO | GSM823221 |
| SCL005-R | SCL005_WT_24H_1_RNA_ExpSam | RNA | GEO | GSM823222 |
| SCL005-R | SCL005_WT_24H_2_RNA_ExpSam | RNA | GEO | GSM823223 |
| SCL005-R | SCL005_WT_24H_3_RNA_ExpSam | RNA | GEO | GSM823224 |
| SCL005-R | SCL005_WT_30H_1_RNA_ExpSam | RNA | GEO | GSM823225 |
| SCL005-R | SCL005_WT_30H_2_RNA_ExpSam | RNA | GEO | GSM823226 |
| SCL005-R | SCL005_WT_30H_3_RNA_ExpSam | RNA | GEO | GSM823227 |
| SCL005-R | SCL005_WT_36H_1_RNA_ExpSam | RNA | GEO | GSM823228 |
| SCL005-R | SCL005_WT_36H_2_RNA_ExpSam | RNA | GEO | GSM823229 |
| SCL005-R | SCL005_WT_36H_3_RNA_ExpSam | RNA | GEO | GSM823230 |
| SCL005-R | SCL005_WT_3H_1_RNA_ExpSam | RNA | GEO | GSM823213 |
| SCL005-R | SCL005_WT_3H_2_RNA_ExpSam | RNA | GEO | GSM823214 |
| SCL005-R | SCL005_WT_3H_3_RNA_ExpSam | RNA | GEO | GSM823215 |
| SCL005-R | SCL005_WT_48H_1_RNA_ExpSam | RNA | GEO | GSM823231 |
| SCL005-R | SCL005_WT_48H_2_RNA_ExpSam | RNA | GEO | GSM823232 |
| SCL005-R | SCL005_WT_48H_3_RNA_ExpSam | RNA | GEO | GSM823233 |
| SCL005-R | SCL005_WT_54H_1_RNA_ExpSam | RNA | GEO | GSM823234 |
| SCL005-R | SCL005_WT_54H_2_RNA_ExpSam | RNA | GEO | GSM823235 |
| SCL005-R | SCL005_WT_54H_3_RNA_ExpSam | RNA | GEO | GSM823236 |
| SCL005-R | SCL005_WT_60H_1_RNA_ExpSam | RNA | GEO | GSM823237 |
| SCL005-R | SCL005_WT_60H_2_RNA_ExpSam | RNA | GEO | GSM823238 |
| SCL005-R | SCL005_WT_60H_3_RNA_ExpSam | RNA | GEO | GSM823239 |
| SCL005-R | SCL005_WT_72H_1_RNA_ExpSam | RNA | GEO | GSM823240 |
| SCL005-R | SCL005_WT_72H_2_RNA_ExpSam | RNA | GEO | GSM823241 |
| SCL005-R | SCL005_WT_72H_3_RNA_ExpSam | RNA | GEO | GSM823242 |
| SCL005-R | SCL005_WT_7H_1_RNA_ExpSam | RNA | GEO | GSM823216 |
| SCL005-R | SCL005_WT_7H_2_RNA_ExpSam | RNA | GEO | GSM823217 |
| SCL005-R | SCL005_WT_7H_3_RNA_ExpSam | RNA | GEO | GSM823218 |
| SCL006-R | SCL006_BatSRBD_0h_1_RNA_ExpSam | RNA | GEO | GSM928559 |
| SCL006-R | SCL006_BatSRBD_0h_2_RNA_ExpSam | RNA | GEO | GSM928560 |
| SCL006-R | SCL006_BatSRBD_0h_3_RNA_ExpSam | RNA | GEO | GSM928561 |
| SCL006-R | SCL006_BatSRBD_12h_1_RNA_ExpSam | RNA | GEO | GSM928562 |
| SCL006-R | SCL006_BatSRBD_12h_2_RNA_ExpSam | RNA | GEO | GSM928563 |
| SCL006-R | SCL006_BatSRBD_12h_3_RNA_ExpSam | RNA | GEO | GSM928564 |
| SCL006-R | SCL006_BatSRBD_24h_1_RNA_ExpSam | RNA | GEO | GSM928565 |
| SCL006-R | SCL006_BatSRBD_24h_2_RNA_ExpSam | RNA | GEO | GSM928566 |
| SCL006-R | SCL006_BatSRBD_24h_3_RNA_ExpSam | RNA | GEO | GSM928567 |
| SCL006-R | SCL006_BatSRBD_30h_1_RNA_ExpSam | RNA | GEO | GSM928568 |
| SCL006-R | SCL006_BatSRBD_30h_2_RNA_ExpSam | RNA | GEO | GSM928569 |
| SCL006-R | SCL006_BatSRBD_30h_3_RNA_ExpSam | RNA | GEO | GSM928570 |
| SCL006-R | SCL006_BatSRBD_36h_1_RNA_ExpSam | RNA | GEO | GSM928571 |
| SCL006-R | SCL006_BatSRBD_36h_2_RNA_ExpSam | RNA | GEO | GSM928572 |
| SCL006-R | SCL006_BatSRBD_36h_3_RNA_ExpSam | RNA | GEO | GSM928573 |
| SCL006-R | SCL006_BatSRBD_48h_1_B_RNA_ExpSam | RNA | GEO | GSM928574 |
| SCL006-R | SCL006_BatSRBD_48h_2_RNA_ExpSam | RNA | GEO | GSM928575 |
| SCL006-R | SCL006_BatSRBD_48h_3_RNA_ExpSam | RNA | GEO | GSM928576 |
| SCL006-R | SCL006_BatSRBD_54h_1_B_RNA_ExpSam | RNA | GEO | GSM928577 |
| SCL006-R | SCL006_BatSRBD_54h_2_RNA_ExpSam | RNA | GEO | GSM928578 |
| SCL006-R | SCL006_BatSRBD_54h_3_RNA_ExpSam | RNA | GEO | GSM928579 |
| SCL006-R | SCL006_BatSRBD_60h_1_RNA_ExpSam | RNA | GEO | GSM928580 |
| SCL006-R | SCL006_BatSRBD_60h_2_RNA_ExpSam | RNA | GEO | GSM928581 |
| SCL006-R | SCL006_BatSRBD_60h_3_RNA_ExpSam | RNA | GEO | GSM928582 |
| SCL006-R | SCL006_BatSRBD_72h_1_RNA_ExpSam | RNA | GEO | GSM928583 |
| SCL006-R | SCL006_BatSRBD_72h_2_RNA_ExpSam | RNA | GEO | GSM928584 |
| SCL006-R | SCL006_BatSRBD_72h_3_RNA_ExpSam | RNA | GEO | GSM928585 |
| SCL006-R | SCL006_icSARSCoV_0h_1_RNA_ExpSam | RNA | GEO | GSM928586 |
| SCL006-R | SCL006_icSARSCoV_0h_2_RNA_ExpSam | RNA | GEO | GSM928587 |
| SCL006-R | SCL006_icSARSCoV_0h_3_RNA_ExpSam | RNA | GEO | GSM928588 |
| SCL006-R | SCL006_icSARSCoV_12h_1_RNA_ExpSam | RNA | GEO | GSM928592 |
| SCL006-R | SCL006_icSARSCoV_12h_2_RNA_ExpSam | RNA | GEO | GSM928593 |
| SCL006-R | SCL006_icSARSCoV_12h_3_B_RNA_ExpSam | RNA | GEO | GSM928594 |
| SCL006-R | SCL006_icSARSCoV_24h_1_RNA_ExpSam | RNA | GEO | GSM928595 |
| SCL006-R | SCL006_icSARSCoV_24h_2_RNA_ExpSam | RNA | GEO | GSM928596 |
| SCL006-R | SCL006_icSARSCoV_24h_3_RNA_ExpSam | RNA | GEO | GSM928597 |
| SCL006-R | SCL006_icSARSCoV_30h_1_RNA_ExpSam | RNA | GEO | GSM928598 |
| SCL006-R | SCL006_icSARSCoV_30h_2_RNA_ExpSam | RNA | GEO | GSM928599 |
| SCL006-R | SCL006_icSARSCoV_30h_3_RNA_ExpSam | RNA | GEO | GSM928600 |
| SCL006-R | SCL006_icSARSCoV_36h_1_RNA_ExpSam | RNA | GEO | GSM928601 |
| SCL006-R | SCL006_icSARSCoV_36h_2_RNA_ExpSam | RNA | GEO | GSM928602 |
| SCL006-R | SCL006_icSARSCoV_36h_3_RNA_ExpSam | RNA | GEO | GSM928603 |
| SCL006-R | SCL006_icSARSCoV_48h_1_RNA_ExpSam | RNA | GEO | GSM928604 |
| SCL006-R | SCL006_icSARSCoV_48h_2_RNA_ExpSam | RNA | GEO | GSM928605 |
| SCL006-R | SCL006_icSARSCoV_48h_3_RNA_ExpSam | RNA | GEO | GSM928606 |
| SCL006-R | SCL006_icSARSCoV_54h_1_RNA_ExpSam | RNA | GEO | GSM928607 |
| SCL006-R | SCL006_icSARSCoV_54h_2_RNA_ExpSam | RNA | GEO | GSM928608 |
| SCL006-R | SCL006_icSARSCoV_54h_3_RNA_ExpSam | RNA | GEO | GSM928609 |
| SCL006-R | SCL006_icSARSCoV_60h_1_RNA_ExpSam | RNA | GEO | GSM928610 |
| SCL006-R | SCL006_icSARSCoV_60h_2_RNA_ExpSam | RNA | GEO | GSM928611 |
| SCL006-R | SCL006_icSARSCoV_60h_3_B_RNA_ExpSam | RNA | GEO | GSM928612 |
| SCL006-R | SCL006_icSARSCoV_72h_1_RNA_ExpSam | RNA | GEO | GSM928613 |
| SCL006-R | SCL006_icSARSCoV_72h_2_RNA_ExpSam | RNA | GEO | GSM928614 |
| SCL006-R | SCL006_icSARSCoV_72h_3_RNA_ExpSam | RNA | GEO | GSM928615 |
| SCL006-R | SCL006_icSARSCoV_7h_1_RNA_ExpSam | RNA | GEO | GSM928589 |
| SCL006-R | SCL006_icSARSCoV_7h_2_RNA_ExpSam | RNA | GEO | GSM928590 |
| SCL006-R | SCL006_icSARSCoV_7h_3_RNA_ExpSam | RNA | GEO | GSM928591 |
| SCL006-R | SCL006_mock_0h_1_RNA_ExpSam | RNA | GEO | GSM928529 |
| SCL006-R | SCL006_mock_0h_2_RNA_ExpSam | RNA | GEO | GSM928530 |
| SCL006-R | SCL006_mock_0h_3_RNA_ExpSam | RNA | GEO | GSM928531 |
| SCL006-R | SCL006_mock_12h_1_RNA_ExpSam | RNA | GEO | GSM928535 |
| SCL006-R | SCL006_mock_12h_2_RNA_ExpSam | RNA | GEO | GSM928536 |
| SCL006-R | SCL006_mock_12h_3_RNA_ExpSam | RNA | GEO | GSM928537 |
| SCL006-R | SCL006_mock_24h_1_RNA_ExpSam | RNA | GEO | GSM928538 |
| SCL006-R | SCL006_mock_24h_2_RNA_ExpSam | RNA | GEO | GSM928539 |
| SCL006-R | SCL006_mock_24h_3_RNA_ExpSam | RNA | GEO | GSM928540 |
| SCL006-R | SCL006_mock_30h_1_RNA_ExpSam | RNA | GEO | GSM928541 |
| SCL006-R | SCL006_mock_30h_2_RNA_ExpSam | RNA | GEO | GSM928542 |
| SCL006-R | SCL006_mock_30h_3_RNA_ExpSam | RNA | GEO | GSM928543 |
| SCL006-R | SCL006_mock_36h_1_RNA_ExpSam | RNA | GEO | GSM928544 |
| SCL006-R | SCL006_mock_36h_2_RNA_ExpSam | RNA | GEO | GSM928545 |
| SCL006-R | SCL006_mock_36h_3_RNA_ExpSam | RNA | GEO | GSM928546 |
| SCL006-R | SCL006_mock_48h_1_RNA_ExpSam | RNA | GEO | GSM928547 |
| SCL006-R | SCL006_mock_48h_2_RNA_ExpSam | RNA | GEO | GSM928548 |
| SCL006-R | SCL006_mock_48h_3_RNA_ExpSam | RNA | GEO | GSM928549 |
| SCL006-R | SCL006_mock_54h_1_RNA_ExpSam | RNA | GEO | GSM928550 |
| SCL006-R | SCL006_mock_54h_2_RNA_ExpSam | RNA | GEO | GSM928551 |
| SCL006-R | SCL006_mock_54h_3_RNA_ExpSam | RNA | GEO | GSM928552 |
| SCL006-R | SCL006_mock_60h_1_RNA_ExpSam | RNA | GEO | GSM928553 |
| SCL006-R | SCL006_mock_60h_2_RNA_ExpSam | RNA | GEO | GSM928554 |
| SCL006-R | SCL006_mock_60h_3_RNA_ExpSam | RNA | GEO | GSM928555 |
| SCL006-R | SCL006_mock_72h_1_RNA_ExpSam | RNA | GEO | GSM928556 |
| SCL006-R | SCL006_mock_72h_2_RNA_ExpSam | RNA | GEO | GSM928557 |
| SCL006-R | SCL006_mock_72h_3_RNA_ExpSam | RNA | GEO | GSM928558 |
| SCL006-R | SCL006_mock_7h_1_RNA_ExpSam | RNA | GEO | GSM928532 |
| SCL006-R | SCL006_mock_7h_2_RNA_ExpSam | RNA | GEO | GSM928533 |
| SCL006-R | SCL006_mock_7h_3_RNA_ExpSam | RNA | GEO | GSM928534 |
| SHAE002-R | SHAE002_BAT_0h_1_array | RNA | GEO | GSM1163233 |
| SHAE002-R | SHAE002_BAT_0h_2_array | RNA | GEO | GSM1163234 |
| SHAE002-R | SHAE002_BAT_0h_3_array | RNA | GEO | GSM1163235 |
| SHAE002-R | SHAE002_BAT_0h_4_array | RNA | GEO | GSM1163236 |
| SHAE002-R | SHAE002_BAT_12h_1_array | RNA | GEO | GSM1163237 |
| SHAE002-R | SHAE002_BAT_12h_2_array | RNA | GEO | GSM1163238 |
| SHAE002-R | SHAE002_BAT_12h_3_array | RNA | GEO | GSM1163239 |
| SHAE002-R | SHAE002_BAT_12h_4_array | RNA | GEO | GSM1163240 |
| SHAE002-R | SHAE002_BAT_24h_1_array | RNA | GEO | GSM1163241 |
| SHAE002-R | SHAE002_BAT_24h_2_array | RNA | GEO | GSM1163242 |
| SHAE002-R | SHAE002_BAT_24h_3_array | RNA | GEO | GSM1163243 |
| SHAE002-R | SHAE002_BAT_24h_4_array | RNA | GEO | GSM1163244 |
| SHAE002-R | SHAE002_BAT_36h_1_array | RNA | GEO | GSM1163245 |
| SHAE002-R | SHAE002_BAT_36h_2_array | RNA | GEO | GSM1163246 |
| SHAE002-R | SHAE002_BAT_36h_3_array | RNA | GEO | GSM1163247 |
| SHAE002-R | SHAE002_BAT_36h_4_array | RNA | GEO | GSM1163248 |
| SHAE002-R | SHAE002_BAT_48h_1_array | RNA | GEO | GSM1163249 |
| SHAE002-R | SHAE002_BAT_48h_3_array | RNA | GEO | GSM1163250 |
| SHAE002-R | SHAE002_BAT_48h_4_array | RNA | GEO | GSM1163251 |
| SHAE002-R | SHAE002_BAT_60h_1_array | RNA | GEO | GSM1163252 |
| SHAE002-R | SHAE002_BAT_60h_2_array | RNA | GEO | GSM1163390 |
| SHAE002-R | SHAE002_BAT_60h_3_array | RNA | GEO | GSM1163253 |
| SHAE002-R | SHAE002_BAT_60h_4_array | RNA | GEO | GSM1163254 |
| SHAE002-R | SHAE002_BAT_72h_1_array | RNA | GEO | GSM1163255 |
| SHAE002-R | SHAE002_BAT_72h_2_array | RNA | GEO | GSM1163256 |
| SHAE002-R | SHAE002_BAT_72h_3_array | RNA | GEO | GSM1163257 |
| SHAE002-R | SHAE002_BAT_72h_4_array | RNA | GEO | GSM1163258 |
| SHAE002-R | SHAE002_BAT_84h_1_array | RNA | GEO | GSM1163392 |
| SHAE002-R | SHAE002_BAT_84h_2_array | RNA | GEO | GSM1163259 |
| SHAE002-R | SHAE002_BAT_84h_3_array | RNA | GEO | GSM1163260 |
| SHAE002-R | SHAE002_BAT_84h_4_array | RNA | GEO | GSM1163261 |
| SHAE002-R | SHAE002_BAT_96h_1_array | RNA | GEO | GSM1163262 |
| SHAE002-R | SHAE002_BAT_96h_2_array | RNA | GEO | GSM1163263 |
| SHAE002-R | SHAE002_BAT_96h_3_array | RNA | GEO | GSM1163264 |
| SHAE002-R | SHAE002_BAT_96h_4_array | RNA | GEO | GSM1163265 |
| SHAE002-R | SHAE002_dORF6_0h_1_array | RNA | GEO | GSM1163322 |
| SHAE002-R | SHAE002_dORF6_0h_2_array | RNA | GEO | GSM1163323 |
| SHAE002-R | SHAE002_dORF6_0h_3_array | RNA | GEO | GSM1163324 |
| SHAE002-R | SHAE002_dORF6_0h_4_array | RNA | GEO | GSM1163325 |
| SHAE002-R | SHAE002_dORF6_12h_1_array | RNA | GEO | GSM1163326 |
| SHAE002-R | SHAE002_dORF6_12h_2_array | RNA | GEO | GSM1163327 |
| SHAE002-R | SHAE002_dORF6_12h_3_array | RNA | GEO | GSM1163328 |
| SHAE002-R | SHAE002_dORF6_12h_4_array | RNA | GEO | GSM1163329 |
| SHAE002-R | SHAE002_dORF6_24h_1_array | RNA | GEO | GSM1163330 |
| SHAE002-R | SHAE002_dORF6_24h_2_array | RNA | GEO | GSM1163331 |
| SHAE002-R | SHAE002_dORF6_24h_4_array | RNA | GEO | GSM1163332 |
| SHAE002-R | SHAE002_dORF6_36h_1_array | RNA | GEO | GSM1163333 |
| SHAE002-R | SHAE002_dORF6_36h_2_array | RNA | GEO | GSM1163334 |
| SHAE002-R | SHAE002_dORF6_36h_3_array | RNA | GEO | GSM1163335 |
| SHAE002-R | SHAE002_dORF6_36h_4_array | RNA | GEO | GSM1163336 |
| SHAE002-R | SHAE002_dORF6_48h_1_array | RNA | GEO | GSM1163337 |
| SHAE002-R | SHAE002_dORF6_48h_2_array | RNA | GEO | GSM1163338 |
| SHAE002-R | SHAE002_dORF6_48h_3_array | RNA | GEO | GSM1163339 |
| SHAE002-R | SHAE002_dORF6_48h_4_array | RNA | GEO | GSM1163340 |
| SHAE002-R | SHAE002_dORF6_60h_1_array | RNA | GEO | GSM1163341 |
| SHAE002-R | SHAE002_dORF6_60h_2_array | RNA | GEO | GSM1163342 |
| SHAE002-R | SHAE002_dORF6_60h_3_array | RNA | GEO | GSM1163343 |
| SHAE002-R | SHAE002_dORF6_60h_4_array | RNA | GEO | GSM1163344 |
| SHAE002-R | SHAE002_dORF6_72h_1_array | RNA | GEO | GSM1163345 |
| SHAE002-R | SHAE002_dORF6_72h_2_array | RNA | GEO | GSM1163346 |
| SHAE002-R | SHAE002_dORF6_72h_3_array | RNA | GEO | GSM1163347 |
| SHAE002-R | SHAE002_dORF6_72h_4_array | RNA | GEO | GSM1163348 |
| SHAE002-R | SHAE002_dORF6_84h_1_array | RNA | GEO | GSM1163349 |
| SHAE002-R | SHAE002_dORF6_84h_2_array | RNA | GEO | GSM1163350 |
| SHAE002-R | SHAE002_dORF6_84h_3_array | RNA | GEO | GSM1163351 |
| SHAE002-R | SHAE002_dORF6_84h_4_array | RNA | GEO | GSM1163352 |
| SHAE002-R | SHAE002_dORF6_96h_1_array | RNA | GEO | GSM1163353 |
| SHAE002-R | SHAE002_dORF6_96h_2_array | RNA | GEO | GSM1163354 |
| SHAE002-R | SHAE002_dORF6_96h_3_array | RNA | GEO | GSM1163355 |
| SHAE002-R | SHAE002_dORF6_96h_4_array | RNA | GEO | GSM1163356 |
| SHAE002-R | SHAE002_H1N1_0h_1_array | RNA | GEO | GSM1163266 |
| SHAE002-R | SHAE002_H1N1_0h_2_array | RNA | GEO | GSM1163267 |
| SHAE002-R | SHAE002_H1N1_0h_3_array | RNA | GEO | GSM1163268 |
| SHAE002-R | SHAE002_H1N1_12h_1_array | RNA | GEO | GSM1163269 |
| SHAE002-R | SHAE002_H1N1_12h_2_array | RNA | GEO | GSM1163270 |
| SHAE002-R | SHAE002_H1N1_12h_3_array | RNA | GEO | GSM1163271 |
| SHAE002-R | SHAE002_H1N1_18h_1_array | RNA | GEO | GSM1163272 |
| SHAE002-R | SHAE002_H1N1_18h_2_array | RNA | GEO | GSM1163273 |
| SHAE002-R | SHAE002_H1N1_18h_3_array | RNA | GEO | GSM1163395 |
| SHAE002-R | SHAE002_H1N1_24h_1_array | RNA | GEO | GSM1163274 |
| SHAE002-R | SHAE002_H1N1_24h_2_array | RNA | GEO | GSM1163275 |
| SHAE002-R | SHAE002_H1N1_24h_3_array | RNA | GEO | GSM1163276 |
| SHAE002-R | SHAE002_H1N1_36h_1_array | RNA | GEO | GSM1163277 |
| SHAE002-R | SHAE002_H1N1_36h_2_array | RNA | GEO | GSM1163278 |
| SHAE002-R | SHAE002_H1N1_36h_3_array | RNA | GEO | GSM1163394 |
| SHAE002-R | SHAE002_H1N1_48h_1_array | RNA | GEO | GSM1163279 |
| SHAE002-R | SHAE002_H1N1_48h_2_array | RNA | GEO | GSM1163280 |
| SHAE002-R | SHAE002_H1N1_48h_3_array | RNA | GEO | GSM1163281 |
| SHAE002-R | SHAE002_H1N1_48h_4_array | RNA | GEO | GSM1163282 |
| SHAE002-R | SHAE002_H1N1_6h_1_array | RNA | GEO | GSM1163283 |
| SHAE002-R | SHAE002_H1N1_6h_2_array | RNA | GEO | GSM1163284 |
| SHAE002-R | SHAE002_H1N1_6h_3_array | RNA | GEO | GSM1163285 |
| SHAE002-R | SHAE002_Mock_0h_1_array | RNA | GEO | GSM1163286 |
| SHAE002-R | SHAE002_Mock_0h_2_array | RNA | GEO | GSM1163287 |
| SHAE002-R | SHAE002_Mock_0h_3_array | RNA | GEO | GSM1163288 |
| SHAE002-R | SHAE002_Mock_12h_1_array | RNA | GEO | GSM1163289 |
| SHAE002-R | SHAE002_Mock_12h_2_array | RNA | GEO | GSM1163290 |
| SHAE002-R | SHAE002_Mock_12h_3_array | RNA | GEO | GSM1163291 |
| SHAE002-R | SHAE002_Mock_18h_1_array | RNA | GEO | GSM1163292 |
| SHAE002-R | SHAE002_Mock_18h_2_array | RNA | GEO | GSM1163293 |
| SHAE002-R | SHAE002_Mock_18h_3_array | RNA | GEO | GSM1163294 |
| SHAE002-R | SHAE002_Mock_24h_1_array | RNA | GEO | GSM1163295 |
| SHAE002-R | SHAE002_Mock_24h_2_array | RNA | GEO | GSM1163389 |
| SHAE002-R | SHAE002_Mock_24h_3_array | RNA | GEO | GSM1163296 |
| SHAE002-R | SHAE002_Mock_36h_1_array | RNA | GEO | GSM1163297 |
| SHAE002-R | SHAE002_Mock_36h_2_array | RNA | GEO | GSM1163298 |
| SHAE002-R | SHAE002_Mock_36h_3_array | RNA | GEO | GSM1163299 |
| SHAE002-R | SHAE002_Mock_48h_1_array | RNA | GEO | GSM1163300 |
| SHAE002-R | SHAE002_Mock_48h_3_array | RNA | GEO | GSM1163301 |
| SHAE002-R | SHAE002_Mock_48h_4_array | RNA | GEO | GSM1163302 |
| SHAE002-R | SHAE002_Mock_60h_1_array | RNA | GEO | GSM1163303 |
| SHAE002-R | SHAE002_Mock_60h_2_array | RNA | GEO | GSM1163304 |
| SHAE002-R | SHAE002_Mock_60h_3_array | RNA | GEO | GSM1163305 |
| SHAE002-R | SHAE002_Mock_60h_4_array | RNA | GEO | GSM1163306 |
| SHAE002-R | SHAE002_Mock_6h_1_array | RNA | GEO | GSM1163307 |
| SHAE002-R | SHAE002_Mock_6h_2_array | RNA | GEO | GSM1163308 |
| SHAE002-R | SHAE002_Mock_6h_3_array | RNA | GEO | GSM1163309 |
| SHAE002-R | SHAE002_Mock_72h_1_array | RNA | GEO | GSM1163310 |
| SHAE002-R | SHAE002_Mock_72h_2_array | RNA | GEO | GSM1163311 |
| SHAE002-R | SHAE002_Mock_72h_3_array | RNA | GEO | GSM1163312 |
| SHAE002-R | SHAE002_Mock_72h_4_array | RNA | GEO | GSM1163313 |
| SHAE002-R | SHAE002_Mock_84h_1_array | RNA | GEO | GSM1163314 |
| SHAE002-R | SHAE002_Mock_84h_2_array | RNA | GEO | GSM1163315 |
| SHAE002-R | SHAE002_Mock_84h_3_array | RNA | GEO | GSM1163316 |
| SHAE002-R | SHAE002_Mock_84h_4_array | RNA | GEO | GSM1163317 |
| SHAE002-R | SHAE002_Mock_96h_1_array | RNA | GEO | GSM1163318 |
| SHAE002-R | SHAE002_Mock_96h_2_array | RNA | GEO | GSM1163319 |
| SHAE002-R | SHAE002_Mock_96h_3_array | RNA | GEO | GSM1163320 |
| SHAE002-R | SHAE002_Mock_96h_4_array | RNA | GEO | GSM1163321 |
| SHAE002-R | SHAE002_SARS_0h_1_array | RNA | GEO | GSM1163357 |
| SHAE002-R | SHAE002_SARS_0h_2_array | RNA | GEO | GSM1163358 |
| SHAE002-R | SHAE002_SARS_0h_3_array | RNA | GEO | GSM1163359 |
| SHAE002-R | SHAE002_SARS_0h_4_array | RNA | GEO | GSM1163360 |
| SHAE002-R | SHAE002_SARS_12h_1_array | RNA | GEO | GSM1163361 |
| SHAE002-R | SHAE002_SARS_12h_2_array | RNA | GEO | GSM1163362 |
| SHAE002-R | SHAE002_SARS_12h_3_array | RNA | GEO | GSM1163391 |
| SHAE002-R | SHAE002_SARS_12h_4_array | RNA | GEO | GSM1163363 |
| SHAE002-R | SHAE002_SARS_24h_1_array | RNA | GEO | GSM1163364 |
| SHAE002-R | SHAE002_SARS_24h_2_array | RNA | GEO | GSM1163365 |
| SHAE002-R | SHAE002_SARS_24h_3_array | RNA | GEO | GSM1163366 |
| SHAE002-R | SHAE002_SARS_24h_4_array | RNA | GEO | GSM1163367 |
| SHAE002-R | SHAE002_SARS_36h_1_array | RNA | GEO | GSM1163368 |
| SHAE002-R | SHAE002_SARS_36h_2_array | RNA | GEO | GSM1163369 |
| SHAE002-R | SHAE002_SARS_48h_1_array | RNA | GEO | GSM1163370 |
| SHAE002-R | SHAE002_SARS_48h_2_array | RNA | GEO | GSM1163371 |
| SHAE002-R | SHAE002_SARS_48h_3_array | RNA | GEO | GSM1163372 |
| SHAE002-R | SHAE002_SARS_48h_4_array | RNA | GEO | GSM1163373 |
| SHAE002-R | SHAE002_SARS_60h_1_array | RNA | GEO | GSM1163374 |
| SHAE002-R | SHAE002_SARS_60h_2_array | RNA | GEO | GSM1163375 |
| SHAE002-R | SHAE002_SARS_60h_3_array | RNA | GEO | GSM1163393 |
| SHAE002-R | SHAE002_SARS_60h_4_array | RNA | GEO | GSM1163376 |
| SHAE002-R | SHAE002_SARS_72h_1_array | RNA | GEO | GSM1163377 |
| SHAE002-R | SHAE002_SARS_72h_2_array | RNA | GEO | GSM1163378 |
| SHAE002-R | SHAE002_SARS_72h_3_array | RNA | GEO | GSM1163379 |
| SHAE002-R | SHAE002_SARS_72h_4_array | RNA | GEO | GSM1163380 |
| SHAE002-R | SHAE002_SARS_84h_1_array | RNA | GEO | GSM1163381 |
| SHAE002-R | SHAE002_SARS_84h_2_array | RNA | GEO | GSM1163382 |
| SHAE002-R | SHAE002_SARS_84h_3_array | RNA | GEO | GSM1163383 |
| SHAE002-R | SHAE002_SARS_84h_4_array | RNA | GEO | GSM1163384 |
| SHAE002-R | SHAE002_SARS_96h_1_array | RNA | GEO | GSM1163385 |
| SHAE002-R | SHAE002_SARS_96h_2_array | RNA | GEO | GSM1163386 |
| SHAE002-R | SHAE002_SARS_96h_3_array | RNA | GEO | GSM1163387 |
| SHAE002-R | SHAE002_SARS_96h_4_array | RNA | GEO | GSM1163388 |
| SHAE003-R | SHAE003_BAT_0h_1_array | RNA | GEO | GSM1163396 |
| SHAE003-R | SHAE003_BAT_0h_2_array | RNA | GEO | GSM1163397 |
| SHAE003-R | SHAE003_BAT_0h_3_array | RNA | GEO | GSM1163398 |
| SHAE003-R | SHAE003_BAT_0h_4_array | RNA | GEO | GSM1163399 |
| SHAE003-R | SHAE003_BAT_24h_3_array | RNA | GEO | GSM1163400 |
| SHAE003-R | SHAE003_BAT_24h_4_array | RNA | GEO | GSM1163401 |
| SHAE003-R | SHAE003_BAT_48h_1_array | RNA | GEO | GSM1163402 |
| SHAE003-R | SHAE003_BAT_48h_2_array | RNA | GEO | GSM1163403 |
| SHAE003-R | SHAE003_BAT_48h_3_array | RNA | GEO | GSM1163404 |
| SHAE003-R | SHAE003_BAT_48h_4_array | RNA | GEO | GSM1163405 |
| SHAE003-R | SHAE003_BAT_60h_1_array | RNA | GEO | GSM1163406 |
| SHAE003-R | SHAE003_BAT_60h_2_array | RNA | GEO | GSM1163407 |
| SHAE003-R | SHAE003_BAT_60h_3_array | RNA | GEO | GSM1163408 |
| SHAE003-R | SHAE003_BAT_60h_4_array | RNA | GEO | GSM1163409 |
| SHAE003-R | SHAE003_BAT_72h_1_array | RNA | GEO | GSM1163410 |
| SHAE003-R | SHAE003_BAT_72h_2_array | RNA | GEO | GSM1163411 |
| SHAE003-R | SHAE003_BAT_72h_3_array | RNA | GEO | GSM1163412 |
| SHAE003-R | SHAE003_BAT_72h_4_array | RNA | GEO | GSM1163413 |
| SHAE003-R | SHAE003_BAT_84h_1_array | RNA | GEO | GSM1163414 |
| SHAE003-R | SHAE003_BAT_84h_2_array | RNA | GEO | GSM1163415 |
| SHAE003-R | SHAE003_BAT_84h_3_array | RNA | GEO | GSM1163416 |
| SHAE003-R | SHAE003_BAT_84h_4_array | RNA | GEO | GSM1163417 |
| SHAE003-R | SHAE003_BAT_96h_1_array | RNA | GEO | GSM1163418 |
| SHAE003-R | SHAE003_BAT_96h_2_array | RNA | GEO | GSM1163419 |
| SHAE003-R | SHAE003_BAT_96h_3_array | RNA | GEO | GSM1163420 |
| SHAE003-R | SHAE003_BAT_96h_4_array | RNA | GEO | GSM1163421 |
| SHAE003-R | SHAE003_dORF6_0h_1_array | RNA | GEO | GSM1163510 |
| SHAE003-R | SHAE003_dORF6_0h_2_array | RNA | GEO | GSM1163511 |
| SHAE003-R | SHAE003_dORF6_0h_3_array | RNA | GEO | GSM1163512 |
| SHAE003-R | SHAE003_dORF6_0h_4_array | RNA | GEO | GSM1163513 |
| SHAE003-R | SHAE003_dORF6_24h_1_array | RNA | GEO | GSM1163514 |
| SHAE003-R | SHAE003_dORF6_24h_2_array | RNA | GEO | GSM1163515 |
| SHAE003-R | SHAE003_dORF6_24h_3_array | RNA | GEO | GSM1163516 |
| SHAE003-R | SHAE003_dORF6_48h_1_array | RNA | GEO | GSM1163517 |
| SHAE003-R | SHAE003_dORF6_48h_2_array | RNA | GEO | GSM1163518 |
| SHAE003-R | SHAE003_dORF6_48h_3_array | RNA | GEO | GSM1163519 |
| SHAE003-R | SHAE003_dORF6_48h_4_array | RNA | GEO | GSM1163520 |
| SHAE003-R | SHAE003_dORF6_60h_1_array | RNA | GEO | GSM1163521 |
| SHAE003-R | SHAE003_dORF6_60h_2_array | RNA | GEO | GSM1163522 |
| SHAE003-R | SHAE003_dORF6_60h_3_array | RNA | GEO | GSM1163523 |
| SHAE003-R | SHAE003_dORF6_60h_4_array | RNA | GEO | GSM1163524 |
| SHAE003-R | SHAE003_dORF6_72h_1_array | RNA | GEO | GSM1163525 |
| SHAE003-R | SHAE003_dORF6_72h_2_array | RNA | GEO | GSM1163526 |
| SHAE003-R | SHAE003_dORF6_72h_3_array | RNA | GEO | GSM1163527 |
| SHAE003-R | SHAE003_dORF6_72h_4_array | RNA | GEO | GSM1163528 |
| SHAE003-R | SHAE003_dORF6_84h_1_array | RNA | GEO | GSM1163529 |
| SHAE003-R | SHAE003_dORF6_84h_2_array | RNA | GEO | GSM1163530 |
| SHAE003-R | SHAE003_dORF6_84h_3_array | RNA | GEO | GSM1163531 |
| SHAE003-R | SHAE003_dORF6_84h_4_array | RNA | GEO | GSM1163532 |
| SHAE003-R | SHAE003_dORF6_96h_1_array | RNA | GEO | GSM1163533 |
| SHAE003-R | SHAE003_dORF6_96h_2_array | RNA | GEO | GSM1163534 |
| SHAE003-R | SHAE003_dORF6_96h_3_array | RNA | GEO | GSM1163535 |
| SHAE003-R | SHAE003_dORF6_96h_4_array | RNA | GEO | GSM1163536 |
| SHAE003-R | SHAE003_H1N1_0h_1_array | RNA | GEO | GSM1163422 |
| SHAE003-R | SHAE003_H1N1_0h_2_array | RNA | GEO | GSM1163423 |
| SHAE003-R | SHAE003_H1N1_0h_3_array | RNA | GEO | GSM1163424 |
| SHAE003-R | SHAE003_H1N1_12h_1_array | RNA | GEO | GSM1163430 |
| SHAE003-R | SHAE003_H1N1_12h_2_array | RNA | GEO | GSM1163431 |
| SHAE003-R | SHAE003_H1N1_12h_3_array | RNA | GEO | GSM1163432 |
| SHAE003-R | SHAE003_H1N1_18h_1_array | RNA | GEO | GSM1163434 |
| SHAE003-R | SHAE003_H1N1_18h_2_array | RNA | GEO | GSM1163435 |
| SHAE003-R | SHAE003_H1N1_18h_3_array | RNA | GEO | GSM1163436 |
| SHAE003-R | SHAE003_H1N1_24h_1_array | RNA | GEO | GSM1163438 |
| SHAE003-R | SHAE003_H1N1_24h_2_array | RNA | GEO | GSM1163439 |
| SHAE003-R | SHAE003_H1N1_24h_3_array | RNA | GEO | GSM1163440 |
| SHAE003-R | SHAE003_H1N1_36h_1_array | RNA | GEO | GSM1163442 |
| SHAE003-R | SHAE003_H1N1_36h_2_array | RNA | GEO | GSM1163443 |
| SHAE003-R | SHAE003_H1N1_36h_3_array | RNA | GEO | GSM1163444 |
| SHAE003-R | SHAE003_H1N1_48h_1_array | RNA | GEO | GSM1163446 |
| SHAE003-R | SHAE003_H1N1_48h_2_array | RNA | GEO | GSM1163447 |
| SHAE003-R | SHAE003_H1N1_48h_3_array | RNA | GEO | GSM1163448 |
| SHAE003-R | SHAE003_H1N1_48h_4_array | RNA | GEO | GSM1163449 |
| SHAE003-R | SHAE003_H1N1_6h_1_array | RNA | GEO | GSM1163426 |
| SHAE003-R | SHAE003_H1N1_6h_2_array | RNA | GEO | GSM1163427 |
| SHAE003-R | SHAE003_H1N1_6h_3_array | RNA | GEO | GSM1163428 |
| SHAE003-R | SHAE003_Mock_0h_1_array | RNA | GEO | GSM1163478 |
| SHAE003-R | SHAE003_Mock_0h_2_array | RNA | GEO | GSM1163479 |
| SHAE003-R | SHAE003_Mock_0h_3_array | RNA | GEO | GSM1163480 |
| SHAE003-R | SHAE003_Mock_12h_1_array | RNA | GEO | GSM1163484 |
| SHAE003-R | SHAE003_Mock_12h_2_array | RNA | GEO | GSM1163485 |
| SHAE003-R | SHAE003_Mock_12h_3_array | RNA | GEO | GSM1163486 |
| SHAE003-R | SHAE003_Mock_18h_1_array | RNA | GEO | GSM1163487 |
| SHAE003-R | SHAE003_Mock_18h_3_array | RNA | GEO | GSM1163488 |
| SHAE003-R | SHAE003_Mock_24h_1_array | RNA | GEO | GSM1163489 |
| SHAE003-R | SHAE003_Mock_24h_2_array | RNA | GEO | GSM1163490 |
| SHAE003-R | SHAE003_Mock_24h_3_array | RNA | GEO | GSM1163491 |
| SHAE003-R | SHAE003_Mock_36h_1_array | RNA | GEO | GSM1163492 |
| SHAE003-R | SHAE003_Mock_36h_2_array | RNA | GEO | GSM1163493 |
| SHAE003-R | SHAE003_Mock_36h_3_array | RNA | GEO | GSM1163494 |
| SHAE003-R | SHAE003_Mock_48h_1_array | RNA | GEO | GSM1163495 |
| SHAE003-R | SHAE003_Mock_48h_2_array | RNA | GEO | GSM1163496 |
| SHAE003-R | SHAE003_Mock_48h_3_array | RNA | GEO | GSM1163497 |
| SHAE003-R | SHAE003_Mock_60h_1_array | RNA | GEO | GSM1163498 |
| SHAE003-R | SHAE003_Mock_60h_2_array | RNA | GEO | GSM1163499 |
| SHAE003-R | SHAE003_Mock_60h_3_array | RNA | GEO | GSM1163500 |
| SHAE003-R | SHAE003_Mock_6h_1_array | RNA | GEO | GSM1163481 |
| SHAE003-R | SHAE003_Mock_6h_2_array | RNA | GEO | GSM1163482 |
| SHAE003-R | SHAE003_Mock_6h_3_array | RNA | GEO | GSM1163483 |
| SHAE003-R | SHAE003_Mock_72h_1_array | RNA | GEO | GSM1163501 |
| SHAE003-R | SHAE003_Mock_72h_2_array | RNA | GEO | GSM1163502 |
| SHAE003-R | SHAE003_Mock_72h_3_array | RNA | GEO | GSM1163503 |
| SHAE003-R | SHAE003_Mock_84h_1_array | RNA | GEO | GSM1163504 |
| SHAE003-R | SHAE003_Mock_84h_2_array | RNA | GEO | GSM1163505 |
| SHAE003-R | SHAE003_Mock_84h_3_array | RNA | GEO | GSM1163506 |
| SHAE003-R | SHAE003_Mock_96h_1_array | RNA | GEO | GSM1163507 |
| SHAE003-R | SHAE003_Mock_96h_2_array | RNA | GEO | GSM1163508 |
| SHAE003-R | SHAE003_Mock_96h_3_array | RNA | GEO | GSM1163509 |
| SHAE003-R | SHAE003_SARS_0h_1_array | RNA | GEO | GSM1163450 |
| SHAE003-R | SHAE003_SARS_0h_2_array | RNA | GEO | GSM1163451 |
| SHAE003-R | SHAE003_SARS_0h_3_array | RNA | GEO | GSM1163452 |
| SHAE003-R | SHAE003_SARS_0h_4_array | RNA | GEO | GSM1163453 |
| SHAE003-R | SHAE003_SARS_24h_1_array | RNA | GEO | GSM1163454 |
| SHAE003-R | SHAE003_SARS_24h_2_array | RNA | GEO | GSM1163455 |
| SHAE003-R | SHAE003_SARS_24h_3_array | RNA | GEO | GSM1163456 |
| SHAE003-R | SHAE003_SARS_24h_4_array | RNA | GEO | GSM1163457 |
| SHAE003-R | SHAE003_SARS_48h_1_array | RNA | GEO | GSM1163458 |
| SHAE003-R | SHAE003_SARS_48h_2_array | RNA | GEO | GSM1163459 |
| SHAE003-R | SHAE003_SARS_48h_3_array | RNA | GEO | GSM1163460 |
| SHAE003-R | SHAE003_SARS_60h_1_array | RNA | GEO | GSM1163462 |
| SHAE003-R | SHAE003_SARS_60h_2_array | RNA | GEO | GSM1163463 |
| SHAE003-R | SHAE003_SARS_60h_3_array | RNA | GEO | GSM1163464 |
| SHAE003-R | SHAE003_SARS_60h_4_array | RNA | GEO | GSM1163465 |
| SHAE003-R | SHAE003_SARS_72h_1_array | RNA | GEO | GSM1163466 |
| SHAE003-R | SHAE003_SARS_72h_2_array | RNA | GEO | GSM1163467 |
| SHAE003-R | SHAE003_SARS_72h_3_array | RNA | GEO | GSM1163468 |
| SHAE003-R | SHAE003_SARS_72h_4_array | RNA | GEO | GSM1163469 |
| SHAE003-R | SHAE003_SARS_84h_1_array | RNA | GEO | GSM1163470 |
| SHAE003-R | SHAE003_SARS_84h_2_array | RNA | GEO | GSM1163471 |
| SHAE003-R | SHAE003_SARS_84h_3_array | RNA | GEO | GSM1163472 |
| SHAE003-R | SHAE003_SARS_84h_4_array | RNA | GEO | GSM1163473 |
| SHAE003-R | SHAE003_SARS_96h_1_array | RNA | GEO | GSM1163474 |
| SHAE003-R | SHAE003_SARS_96h_2_array | RNA | GEO | GSM1163475 |
| SHAE003-R | SHAE003_SARS_96h_3_array | RNA | GEO | GSM1163476 |
| SHAE003-R | SHAE003_SARS_96h_4_array | RNA | GEO | GSM1163477 |
| SHAE004-R | SHAE004_BAT_0h_1_array | RNA | GEO | GSM1163537 |
| SHAE004-R | SHAE004_BAT_0h_2_array | RNA | GEO | GSM1163538 |
| SHAE004-R | SHAE004_BAT_0h_3_array | RNA | GEO | GSM1163539 |
| SHAE004-R | SHAE004_BAT_12h_1_array | RNA | GEO | GSM1163540 |
| SHAE004-R | SHAE004_BAT_12h_2_array | RNA | GEO | GSM1163541 |
| SHAE004-R | SHAE004_BAT_12h_3_array | RNA | GEO | GSM1163542 |
| SHAE004-R | SHAE004_BAT_24h_1_array | RNA | GEO | GSM1163543 |
| SHAE004-R | SHAE004_BAT_24h_2_array | RNA | GEO | GSM1163544 |
| SHAE004-R | SHAE004_BAT_36h_1_array | RNA | GEO | GSM1163545 |
| SHAE004-R | SHAE004_BAT_36h_2_array | RNA | GEO | GSM1163546 |
| SHAE004-R | SHAE004_BAT_36h_3_array | RNA | GEO | GSM1163547 |
| SHAE004-R | SHAE004_BAT_48h_1_array | RNA | GEO | GSM1163548 |
| SHAE004-R | SHAE004_BAT_48h_2_array | RNA | GEO | GSM1163549 |
| SHAE004-R | SHAE004_BAT_48h_3_array | RNA | GEO | GSM1163550 |
| SHAE004-R | SHAE004_BAT_60h_1_array | RNA | GEO | GSM1163551 |
| SHAE004-R | SHAE004_BAT_60h_2_array | RNA | GEO | GSM1163552 |
| SHAE004-R | SHAE004_BAT_60h_3_array | RNA | GEO | GSM1163553 |
| SHAE004-R | SHAE004_BAT_72h_1_array | RNA | GEO | GSM1163554 |
| SHAE004-R | SHAE004_BAT_72h_2_array | RNA | GEO | GSM1163555 |
| SHAE004-R | SHAE004_BAT_72h_3_array | RNA | GEO | GSM1163556 |
| SHAE004-R | SHAE004_BAT_84h_1_array | RNA | GEO | GSM1163557 |
| SHAE004-R | SHAE004_BAT_84h_2_array | RNA | GEO | GSM1163558 |
| SHAE004-R | SHAE004_BAT_84h_3_array | RNA | GEO | GSM1163559 |
| SHAE004-R | SHAE004_BAT_96h_1_array | RNA | GEO | GSM1163560 |
| SHAE004-R | SHAE004_BAT_96h_2_array | RNA | GEO | GSM1163561 |
| SHAE004-R | SHAE004_BAT_96h_3_array | RNA | GEO | GSM1163562 |
| SHAE004-R | SHAE004_dORF6_0h_1_array | RNA | GEO | GSM1163563 |
| SHAE004-R | SHAE004_dORF6_0h_2_array | RNA | GEO | GSM1163564 |
| SHAE004-R | SHAE004_dORF6_0h_3_array | RNA | GEO | GSM1163565 |
| SHAE004-R | SHAE004_dORF6_12h_1_array | RNA | GEO | GSM1163566 |
| SHAE004-R | SHAE004_dORF6_12h_2_array | RNA | GEO | GSM1163567 |
| SHAE004-R | SHAE004_dORF6_12h_3_array | RNA | GEO | GSM1163568 |
| SHAE004-R | SHAE004_dORF6_24h_1_array | RNA | GEO | GSM1163569 |
| SHAE004-R | SHAE004_dORF6_24h_2_array | RNA | GEO | GSM1163570 |
| SHAE004-R | SHAE004_dORF6_24h_3_array | RNA | GEO | GSM1163571 |
| SHAE004-R | SHAE004_dORF6_36h_1_array | RNA | GEO | GSM1163572 |
| SHAE004-R | SHAE004_dORF6_36h_2_array | RNA | GEO | GSM1163573 |
| SHAE004-R | SHAE004_dORF6_36h_3_array | RNA | GEO | GSM1163574 |
| SHAE004-R | SHAE004_dORF6_48h_1_array | RNA | GEO | GSM1163575 |
| SHAE004-R | SHAE004_dORF6_48h_2_array | RNA | GEO | GSM1163576 |
| SHAE004-R | SHAE004_dORF6_48h_3_array | RNA | GEO | GSM1163577 |
| SHAE004-R | SHAE004_dORF6_60h_1_array | RNA | GEO | GSM1163578 |
| SHAE004-R | SHAE004_dORF6_60h_2_array | RNA | GEO | GSM1163579 |
| SHAE004-R | SHAE004_dORF6_60h_3_array | RNA | GEO | GSM1163580 |
| SHAE004-R | SHAE004_dORF6_72h_1_array | RNA | GEO | GSM1163581 |
| SHAE004-R | SHAE004_dORF6_72h_2_array | RNA | GEO | GSM1163582 |
| SHAE004-R | SHAE004_dORF6_72h_3_array | RNA | GEO | GSM1163583 |
| SHAE004-R | SHAE004_dORF6_84h_1_array | RNA | GEO | GSM1163584 |
| SHAE004-R | SHAE004_dORF6_84h_2_array | RNA | GEO | GSM1163585 |
| SHAE004-R | SHAE004_dORF6_84h_3_array | RNA | GEO | GSM1163586 |
| SHAE004-R | SHAE004_dORF6_96h_1_array | RNA | GEO | GSM1163587 |
| SHAE004-R | SHAE004_dORF6_96h_2_array | RNA | GEO | GSM1163588 |
| SHAE004-R | SHAE004_dORF6_96h_3_array | RNA | GEO | GSM1163589 |
| SHAE004-R | SHAE004_H1N1_0h_1_array | RNA | GEO | GSM1163590 |
| SHAE004-R | SHAE004_H1N1_0h_2_array | RNA | GEO | GSM1163591 |
| SHAE004-R | SHAE004_H1N1_0h_3_array | RNA | GEO | GSM1163592 |
| SHAE004-R | SHAE004_H1N1_12h_1_array | RNA | GEO | GSM1163596 |
| SHAE004-R | SHAE004_H1N1_12h_2_array | RNA | GEO | GSM1163597 |
| SHAE004-R | SHAE004_H1N1_12h_3_array | RNA | GEO | GSM1163598 |
| SHAE004-R | SHAE004_H1N1_18h_1_array | RNA | GEO | GSM1163599 |
| SHAE004-R | SHAE004_H1N1_18h_2_array | RNA | GEO | GSM1163600 |
| SHAE004-R | SHAE004_H1N1_18h_3_array | RNA | GEO | GSM1163601 |
| SHAE004-R | SHAE004_H1N1_24h_1_array | RNA | GEO | GSM1163602 |
| SHAE004-R | SHAE004_H1N1_24h_2_array | RNA | GEO | GSM1163603 |
| SHAE004-R | SHAE004_H1N1_24h_3_array | RNA | GEO | GSM1163604 |
| SHAE004-R | SHAE004_H1N1_36h_1_array | RNA | GEO | GSM1163605 |
| SHAE004-R | SHAE004_H1N1_36h_2_array | RNA | GEO | GSM1163606 |
| SHAE004-R | SHAE004_H1N1_36h_3_array | RNA | GEO | GSM1163607 |
| SHAE004-R | SHAE004_H1N1_48h_1_array | RNA | GEO | GSM1163608 |
| SHAE004-R | SHAE004_H1N1_48h_2_array | RNA | GEO | GSM1163609 |
| SHAE004-R | SHAE004_H1N1_48h_3_array | RNA | GEO | GSM1163610 |
| SHAE004-R | SHAE004_H1N1_6h_1_array | RNA | GEO | GSM1163593 |
| SHAE004-R | SHAE004_H1N1_6h_2_array | RNA | GEO | GSM1163594 |
| SHAE004-R | SHAE004_H1N1_6h_3_array | RNA | GEO | GSM1163595 |
| SHAE004-R | SHAE004_Mock_0h_1_array | RNA | GEO | GSM1163638 |
| SHAE004-R | SHAE004_Mock_0h_2_array | RNA | GEO | GSM1163639 |
| SHAE004-R | SHAE004_Mock_0h_3_array | RNA | GEO | GSM1163640 |
| SHAE004-R | SHAE004_Mock_12h_1_array | RNA | GEO | GSM1163644 |
| SHAE004-R | SHAE004_Mock_12h_2_array | RNA | GEO | GSM1163645 |
| SHAE004-R | SHAE004_Mock_12h_3_array | RNA | GEO | GSM1163646 |
| SHAE004-R | SHAE004_Mock_18h_1_array | RNA | GEO | GSM1163647 |
| SHAE004-R | SHAE004_Mock_18h_2_array | RNA | GEO | GSM1163648 |
| SHAE004-R | SHAE004_Mock_18h_3_array | RNA | GEO | GSM1163649 |
| SHAE004-R | SHAE004_Mock_24h_1_array | RNA | GEO | GSM1163650 |
| SHAE004-R | SHAE004_Mock_24h_2_array | RNA | GEO | GSM1163651 |
| SHAE004-R | SHAE004_Mock_24h_3_array | RNA | GEO | GSM1163652 |
| SHAE004-R | SHAE004_Mock_36h_1_array | RNA | GEO | GSM1163653 |
| SHAE004-R | SHAE004_Mock_36h_2_array | RNA | GEO | GSM1163654 |
| SHAE004-R | SHAE004_Mock_36h_3_array | RNA | GEO | GSM1163655 |
| SHAE004-R | SHAE004_Mock_48h_1_array | RNA | GEO | GSM1163656 |
| SHAE004-R | SHAE004_Mock_48h_2_array | RNA | GEO | GSM1163657 |
| SHAE004-R | SHAE004_Mock_48h_3_array | RNA | GEO | GSM1163658 |
| SHAE004-R | SHAE004_Mock_60h_1_array | RNA | GEO | GSM1163659 |
| SHAE004-R | SHAE004_Mock_60h_2_array | RNA | GEO | GSM1163660 |
| SHAE004-R | SHAE004_Mock_60h_3_array | RNA | GEO | GSM1163661 |
| SHAE004-R | SHAE004_Mock_6h_1_array | RNA | GEO | GSM1163641 |
| SHAE004-R | SHAE004_Mock_6h_2_array | RNA | GEO | GSM1163642 |
| SHAE004-R | SHAE004_Mock_6h_3_array | RNA | GEO | GSM1163643 |
| SHAE004-R | SHAE004_Mock_72h_1_array | RNA | GEO | GSM1163662 |
| SHAE004-R | SHAE004_Mock_72h_2_array | RNA | GEO | GSM1163663 |
| SHAE004-R | SHAE004_Mock_72h_3_array | RNA | GEO | GSM1163664 |
| SHAE004-R | SHAE004_Mock_84h_1_array | RNA | GEO | GSM1163665 |
| SHAE004-R | SHAE004_Mock_84h_2_array | RNA | GEO | GSM1163666 |
| SHAE004-R | SHAE004_Mock_84h_3_array | RNA | GEO | GSM1163667 |
| SHAE004-R | SHAE004_Mock_96h_1_array | RNA | GEO | GSM1163668 |
| SHAE004-R | SHAE004_Mock_96h_2_array | RNA | GEO | GSM1163669 |
| SHAE004-R | SHAE004_Mock_96h_3_array | RNA | GEO | GSM1163670 |
| SHAE004-R | SHAE004_SARS_0h_1_array | RNA | GEO | GSM1163611 |
| SHAE004-R | SHAE004_SARS_0h_2_array | RNA | GEO | GSM1163612 |
| SHAE004-R | SHAE004_SARS_0h_3_array | RNA | GEO | GSM1163613 |
| SHAE004-R | SHAE004_SARS_12h_1_array | RNA | GEO | GSM1163614 |
| SHAE004-R | SHAE004_SARS_12h_2_array | RNA | GEO | GSM1163615 |
| SHAE004-R | SHAE004_SARS_12h_3_array | RNA | GEO | GSM1163616 |
| SHAE004-R | SHAE004_SARS_24h_1_array | RNA | GEO | GSM1163617 |
| SHAE004-R | SHAE004_SARS_24h_2_array | RNA | GEO | GSM1163618 |
| SHAE004-R | SHAE004_SARS_24h_3_array | RNA | GEO | GSM1163619 |
| SHAE004-R | SHAE004_SARS_36h_1_array | RNA | GEO | GSM1163620 |
| SHAE004-R | SHAE004_SARS_36h_2_array | RNA | GEO | GSM1163621 |
| SHAE004-R | SHAE004_SARS_36h_3_array | RNA | GEO | GSM1163622 |
| SHAE004-R | SHAE004_SARS_48h_1_array | RNA | GEO | GSM1163623 |
| SHAE004-R | SHAE004_SARS_48h_2_array | RNA | GEO | GSM1163624 |
| SHAE004-R | SHAE004_SARS_48h_3_array | RNA | GEO | GSM1163625 |
| SHAE004-R | SHAE004_SARS_60h_1_array | RNA | GEO | GSM1163626 |
| SHAE004-R | SHAE004_SARS_60h_2_array | RNA | GEO | GSM1163627 |
| SHAE004-R | SHAE004_SARS_60h_3_array | RNA | GEO | GSM1163628 |
| SHAE004-R | SHAE004_SARS_72h_1_array | RNA | GEO | GSM1163629 |
| SHAE004-R | SHAE004_SARS_72h_2_array | RNA | GEO | GSM1163630 |
| SHAE004-R | SHAE004_SARS_72h_3_array | RNA | GEO | GSM1163631 |
| SHAE004-R | SHAE004_SARS_84h_1_array | RNA | GEO | GSM1163632 |
| SHAE004-R | SHAE004_SARS_84h_2_array | RNA | GEO | GSM1163633 |
| SHAE004-R | SHAE004_SARS_84h_3_array | RNA | GEO | GSM1163634 |
| SHAE004-R | SHAE004_SARS_96h_1_array | RNA | GEO | GSM1163635 |
| SHAE004-R | SHAE004_SARS_96h_2_array | RNA | GEO | GSM1163636 |
| SHAE004-R | SHAE004_SARS_96h_3_array | RNA | GEO | GSM1163637 |
| SM001-R | SM001_Mock_D1_1_RNA_ExpSam | RNA | GEO | GSM823085 |
| SM001-R | SM001_Mock_D1_2_RNA_ExpSam | RNA | GEO | GSM823086 |
| SM001-R | SM001_Mock_D1_3_RNA_ExpSam | RNA | GEO | GSM823087 |
| SM001-R | SM001_Mock_D2_1_RNA_ExpSam | RNA | GEO | GSM823088 |
| SM001-R | SM001_Mock_D2_2_RNA_ExpSam | RNA | GEO | GSM823089 |
| SM001-R | SM001_Mock_D2_3_RNA_ExpSam | RNA | GEO | GSM823090 |
| SM001-R | SM001_Mock_D4_1_RNA_ExpSam | RNA | GEO | GSM823091 |
| SM001-R | SM001_Mock_D4_2_RNA_ExpSam | RNA | GEO | GSM823092 |
| SM001-R | SM001_Mock_D4_3_RNA_ExpSam | RNA | GEO | GSM823093 |
| SM001-R | SM001_Mock_D7_1_RNA_ExpSam | RNA | GEO | GSM823094 |
| SM001-R | SM001_Mock_D7_2_RNA_ExpSam | RNA | GEO | GSM823095 |
| SM001-R | SM001_Mock_D7_3_RNA_ExpSam | RNA | GEO | GSM823096 |
| SM001-R | SM001_SARS_10^2pfu_D1_1_RNA_ExpSam | RNA | GEO | GSM823097 |
| SM001-R | SM001_SARS_10^2pfu_D1_2_RNA_ExpSam | RNA | GEO | GSM823098 |
| SM001-R | SM001_SARS_10^2pfu_D1_3_RNA_ExpSam | RNA | GEO | GSM823099 |
| SM001-R | SM001_SARS_10^2pfu_D1_4_RNA_ExpSam | RNA | GEO | GSM823100 |
| SM001-R | SM001_SARS_10^2pfu_D1_5_RNA_ExpSam | RNA | GEO | GSM823101 |
| SM001-R | SM001_SARS_10^2pfu_D2_1_RNA_ExpSam | RNA | GEO | GSM823102 |
| SM001-R | SM001_SARS_10^2pfu_D2_2_RNA_ExpSam | RNA | GEO | GSM823103 |
| SM001-R | SM001_SARS_10^2pfu_D2_3_RNA_ExpSam | RNA | GEO | GSM823104 |
| SM001-R | SM001_SARS_10^2pfu_D2_4_RNA_ExpSam | RNA | GEO | GSM823105 |
| SM001-R | SM001_SARS_10^2pfu_D2_5_RNA_ExpSam | RNA | GEO | GSM823106 |
| SM001-R | SM001_SARS_10^2pfu_D4_1_RNA_ExpSam | RNA | GEO | GSM823107 |
| SM001-R | SM001_SARS_10^2pfu_D4_2_RNA_ExpSam | RNA | GEO | GSM823108 |
| SM001-R | SM001_SARS_10^2pfu_D4_3_RNA_ExpSam | RNA | GEO | GSM823109 |
| SM001-R | SM001_SARS_10^2pfu_D4_4_RNA_ExpSam | RNA | GEO | GSM823110 |
| SM001-R | SM001_SARS_10^2pfu_D4_5_RNA_ExpSam | RNA | GEO | GSM823111 |
| SM001-R | SM001_SARS_10^2pfu_D7_1_RNA_ExpSam | RNA | GEO | GSM823112 |
| SM001-R | SM001_SARS_10^2pfu_D7_2_RNA_ExpSam | RNA | GEO | GSM823113 |
| SM001-R | SM001_SARS_10^2pfu_D7_3_RNA_ExpSam | RNA | GEO | GSM823114 |
| SM001-R | SM001_SARS_10^2pfu_D7_4_RNA_ExpSam | RNA | GEO | GSM823115 |
| SM001-R | SM001_SARS_10^2pfu_D7_5_RNA_ExpSam | RNA | GEO | GSM823116 |
| SM001-R | SM001_SARS_10^3pfu_D1_1_RNA_ExpSam | RNA | GEO | GSM823117 |
| SM001-R | SM001_SARS_10^3pfu_D1_2_RNA_ExpSam | RNA | GEO | GSM823118 |
| SM001-R | SM001_SARS_10^3pfu_D1_3_RNA_ExpSam | RNA | GEO | GSM823119 |
| SM001-R | SM001_SARS_10^3pfu_D1_4_RNA_ExpSam | RNA | GEO | GSM823120 |
| SM001-R | SM001_SARS_10^3pfu_D1_5_RNA_ExpSam | RNA | GEO | GSM823121 |
| SM001-R | SM001_SARS_10^3pfu_D2_1_RNA_ExpSam | RNA | GEO | GSM823122 |
| SM001-R | SM001_SARS_10^3pfu_D2_2_RNA_ExpSam | RNA | GEO | GSM823123 |
| SM001-R | SM001_SARS_10^3pfu_D2_3_RNA_ExpSam | RNA | GEO | GSM823124 |
| SM001-R | SM001_SARS_10^3pfu_D2_4_RNA_ExpSam | RNA | GEO | GSM823125 |
| SM001-R | SM001_SARS_10^3pfu_D2_5_RNA_ExpSam | RNA | GEO | GSM823126 |
| SM001-R | SM001_SARS_10^3pfu_D4_1_RNA_ExpSam | RNA | GEO | GSM823127 |
| SM001-R | SM001_SARS_10^3pfu_D4_2_RNA_ExpSam | RNA | GEO | GSM823128 |
| SM001-R | SM001_SARS_10^3pfu_D4_3_RNA_ExpSam | RNA | GEO | GSM823129 |
| SM001-R | SM001_SARS_10^3pfu_D4_4_RNA_ExpSam | RNA | GEO | GSM823130 |
| SM001-R | SM001_SARS_10^3pfu_D4_5_RNA_ExpSam | RNA | GEO | GSM823131 |
| SM001-R | SM001_SARS_10^3pfu_D7_1_RNA_ExpSam | RNA | GEO | GSM823132 |
| SM001-R | SM001_SARS_10^3pfu_D7_2_RNA_ExpSam | RNA | GEO | GSM823133 |
| SM001-R | SM001_SARS_10^3pfu_D7_3_RNA_ExpSam | RNA | GEO | GSM823134 |
| SM001-R | SM001_SARS_10^3pfu_D7_4_RNA_ExpSam | RNA | GEO | GSM823135 |
| SM001-R | SM001_SARS_10^3pfu_D7_5_RNA_ExpSam | RNA | GEO | GSM823136 |
| SM001-R | SM001_SARS_10^4pfu_D1_1_RNA_ExpSam | RNA | GEO | GSM823137 |
| SM001-R | SM001_SARS_10^4pfu_D1_2_RNA_ExpSam | RNA | GEO | GSM823138 |
| SM001-R | SM001_SARS_10^4pfu_D1_3_RNA_ExpSam | RNA | GEO | GSM823139 |
| SM001-R | SM001_SARS_10^4pfu_D1_4_RNA_ExpSam | RNA | GEO | GSM823140 |
| SM001-R | SM001_SARS_10^4pfu_D1_5_RNA_ExpSam | RNA | GEO | GSM823141 |
| SM001-R | SM001_SARS_10^4pfu_D2_1_RNA_ExpSam | RNA | GEO | GSM823142 |
| SM001-R | SM001_SARS_10^4pfu_D2_2_RNA_ExpSam | RNA | GEO | GSM823143 |
| SM001-R | SM001_SARS_10^4pfu_D2_3_RNA_ExpSam | RNA | GEO | GSM823144 |
| SM001-R | SM001_SARS_10^4pfu_D2_4_RNA_ExpSam | RNA | GEO | GSM823145 |
| SM001-R | SM001_SARS_10^4pfu_D2_5_RNA_ExpSam | RNA | GEO | GSM823146 |
| SM001-R | SM001_SARS_10^4pfu_D4_1_RNA_ExpSam | RNA | GEO | GSM823147 |
| SM001-R | SM001_SARS_10^4pfu_D4_2_RNA_ExpSam | RNA | GEO | GSM823148 |
| SM001-R | SM001_SARS_10^4pfu_D4_3_RNA_ExpSam | RNA | GEO | GSM823149 |
| SM001-R | SM001_SARS_10^4pfu_D4_4_RNA_ExpSam | RNA | GEO | GSM823150 |
| SM001-R | SM001_SARS_10^4pfu_D4_5_RNA_ExpSam | RNA | GEO | GSM823151 |
| SM001-R | SM001_SARS_10^4pfu_D7_1_RNA_ExpSam | RNA | GEO | GSM823152 |
| SM001-R | SM001_SARS_10^4pfu_D7_2_RNA_ExpSam | RNA | GEO | GSM823153 |
| SM001-R | SM001_SARS_10^4pfu_D7_3_RNA_ExpSam | RNA | GEO | GSM823154 |
| SM001-R | SM001_SARS_10^4pfu_D7_4_RNA_ExpSam | RNA | GEO | GSM823155 |
| SM001-R | SM001_SARS_10^4pfu_D7_5_RNA_ExpSam | RNA | GEO | GSM823156 |
| SM001-R | SM001_SARS_10^5pfu_D1_1_RNA_ExpSam | RNA | GEO | GSM823157 |
| SM001-R | SM001_SARS_10^5pfu_D1_2_RNA_ExpSam | RNA | GEO | GSM823158 |
| SM001-R | SM001_SARS_10^5pfu_D1_3_RNA_ExpSam | RNA | GEO | GSM823159 |
| SM001-R | SM001_SARS_10^5pfu_D1_4_RNA_ExpSam | RNA | GEO | GSM823160 |
| SM001-R | SM001_SARS_10^5pfu_D1_5_RNA_ExpSam | RNA | GEO | GSM823161 |
| SM001-R | SM001_SARS_10^5pfu_D2_1_RNA_ExpSam | RNA | GEO | GSM823162 |
| SM001-R | SM001_SARS_10^5pfu_D2_2_RNA_ExpSam | RNA | GEO | GSM823163 |
| SM001-R | SM001_SARS_10^5pfu_D2_3_RNA_ExpSam | RNA | GEO | GSM823164 |
| SM001-R | SM001_SARS_10^5pfu_D2_4_RNA_ExpSam | RNA | GEO | GSM823165 |
| SM001-R | SM001_SARS_10^5pfu_D2_5_RNA_ExpSam | RNA | GEO | GSM823166 |
| SM001-R | SM001_SARS_10^5pfu_D4_1_RNA_ExpSam | RNA | GEO | GSM823167 |
| SM001-R | SM001_SARS_10^5pfu_D4_2_RNA_ExpSam | RNA | GEO | GSM823168 |
| SM001-R | SM001_SARS_10^5pfu_D4_3_RNA_ExpSam | RNA | GEO | GSM823169 |
| SM001-R | SM001_SARS_10^5pfu_D4_4_RNA_ExpSam | RNA | GEO | GSM823170 |
| SM001-R | SM001_SARS_10^5pfu_D4_5_RNA_ExpSam | RNA | GEO | GSM823171 |
| SM001-R | SM001_SARS_10^5pfu_D7_1_RNA_ExpSam | RNA | GEO | GSM823172 |
| SM001-R | SM001_SARS_10^5pfu_D7_2_RNA_ExpSam | RNA | GEO | GSM823173 |
| SM001-R | SM001_SARS_10^5pfu_D7_3_RNA_ExpSam | RNA | GEO | GSM823174 |
| SM001-R | SM001_SARS_10^5pfu_D7_4_RNA_ExpSam | RNA | GEO | GSM823175 |
| SM001-R | SM001_SARS_10^5pfu_D7_5_RNA_ExpSam | RNA | GEO | GSM823176 |
| SM003-R | SM003_10^4_MA15_d1_1_array | RNA | GEO | GSM1211755 |
| SM003-R | SM003_10^4_MA15_d1_2_array | RNA | GEO | GSM1211756 |
| SM003-R | SM003_10^4_MA15_d1_3_array | RNA | GEO | GSM1211757 |
| SM003-R | SM003_10^4_MA15_d1_4_array | RNA | GEO | GSM1211758 |
| SM003-R | SM003_10^4_MA15_d2_1_array | RNA | GEO | GSM1211759 |
| SM003-R | SM003_10^4_MA15_d2_2_array | RNA | GEO | GSM1211760 |
| SM003-R | SM003_10^4_MA15_d2_3_array | RNA | GEO | GSM1211761 |
| SM003-R | SM003_10^4_MA15_d2_4_array | RNA | GEO | GSM1211762 |
| SM003-R | SM003_10^4_MA15_d4_1_array | RNA | GEO | GSM1211763 |
| SM003-R | SM003_10^4_MA15_d4_2_array | RNA | GEO | GSM1211764 |
| SM003-R | SM003_10^4_MA15_d4_3_array | RNA | GEO | GSM1211765 |
| SM003-R | SM003_10^4_MA15_d4_4_array | RNA | GEO | GSM1211766 |
| SM003-R | SM003_10^4_MA15_d7_1_array | RNA | GEO | GSM1211767 |
| SM003-R | SM003_10^4_MA15_d7_2_array | RNA | GEO | GSM1211768 |
| SM003-R | SM003_10^4_MA15_d7_3_array | RNA | GEO | GSM1211769 |
| SM003-R | SM003_10^5_MA15_d1_1_array | RNA | GEO | GSM1211771 |
| SM003-R | SM003_10^5_MA15_d1_3_array | RNA | GEO | GSM1211773 |
| SM003-R | SM003_10^5_MA15_d1_4_array | RNA | GEO | GSM1211774 |
| SM003-R | SM003_10^5_MA15_d2_2_array | RNA | GEO | GSM1211776 |
| SM003-R | SM003_10^5_MA15_d2_3_array | RNA | GEO | GSM1211777 |
| SM003-R | SM003_10^5_MA15_d2_4_array | RNA | GEO | GSM1211778 |
| SM003-R | SM003_10^5_MA15_d4_1_array | RNA | GEO | GSM1211779 |
| SM003-R | SM003_10^5_MA15_d4_2_array | RNA | GEO | GSM1211780 |
| SM003-R | SM003_10^5_MA15_d4_3_array | RNA | GEO | GSM1211781 |
| SM003-R | SM003_10^5_MA15_d4_4_array | RNA | GEO | GSM1211782 |
| SM003-R | SM003_10^5_MA15_d4_5_array | RNA | GEO | GSM1211783 |
| SM003-R | SM003_10^5_MA15_d7_1_array | RNA | GEO | GSM1211784 |
| SM003-R | SM003_10^5_MA15_d7_2_array | RNA | GEO | GSM1211785 |
| SM003-R | SM003_10^5_MA15_d7_3_array | RNA | GEO | GSM1211786 |
| SM003-R | SM003_BAT_d1_1_array | RNA | GEO | GSM1211787 |
| SM003-R | SM003_BAT_d1_2_array | RNA | GEO | GSM1211788 |
| SM003-R | SM003_BAT_d1_3_array | RNA | GEO | GSM1211789 |
| SM003-R | SM003_BAT_d1_4_array | RNA | GEO | GSM1211790 |
| SM003-R | SM003_BAT_d1_5_array | RNA | GEO | GSM1211791 |
| SM003-R | SM003_BAT_d2_1_array | RNA | GEO | GSM1211792 |
| SM003-R | SM003_BAT_d2_2_array | RNA | GEO | GSM1211793 |
| SM003-R | SM003_BAT_d2_3_array | RNA | GEO | GSM1211794 |
| SM003-R | SM003_BAT_d2_4_array | RNA | GEO | GSM1211795 |
| SM003-R | SM003_BAT_d2_5_array | RNA | GEO | GSM1211796 |
| SM003-R | SM003_BAT_d4_1_array | RNA | GEO | GSM1211797 |
| SM003-R | SM003_BAT_d4_2_array | RNA | GEO | GSM1211798 |
| SM003-R | SM003_BAT_d4_3_array | RNA | GEO | GSM1211799 |
| SM003-R | SM003_BAT_d4_4_array | RNA | GEO | GSM1211800 |
| SM003-R | SM003_BAT_d4_5_array | RNA | GEO | GSM1211801 |
| SM003-R | SM003_BAT_d7_1_array | RNA | GEO | GSM1211802 |
| SM003-R | SM003_BAT_d7_3_array | RNA | GEO | GSM1211803 |
| SM003-R | SM003_BAT_d7_4_array | RNA | GEO | GSM1211804 |
| SM003-R | SM003_BAT_d7_5_array | RNA | GEO | GSM1211805 |
| SM003-R | SM003_Mock_d1_1_array | RNA | GEO | GSM1211825 |
| SM003-R | SM003_Mock_d1_2_array | RNA | GEO | GSM1211826 |
| SM003-R | SM003_Mock_d1_3_array | RNA | GEO | GSM1211827 |
| SM003-R | SM003_Mock_d1_4_array | RNA | GEO | GSM1211828 |
| SM003-R | SM003_Mock_d2_1_array | RNA | GEO | GSM1211829 |
| SM003-R | SM003_Mock_d2_2_array | RNA | GEO | GSM1211830 |
| SM003-R | SM003_Mock_d2_3_array | RNA | GEO | GSM1211831 |
| SM003-R | SM003_Mock_d2_4_array | RNA | GEO | GSM1211832 |
| SM003-R | SM003_Mock_d4_1_array | RNA | GEO | GSM1211833 |
| SM003-R | SM003_Mock_d4_2_array | RNA | GEO | GSM1211834 |
| SM003-R | SM003_Mock_d4_3_array | RNA | GEO | GSM1211835 |
| SM003-R | SM003_Mock_d4_4_array | RNA | GEO | GSM1211836 |
| SM003-R | SM003_Mock_d7_1_array | RNA | GEO | GSM1211837 |
| SM003-R | SM003_Mock_d7_2_array | RNA | GEO | GSM1211838 |
| SM003-R | SM003_Mock_d7_3_array | RNA | GEO | GSM1211839 |
| SM003-R | SM003_Mock_d7_4_array | RNA | GEO | GSM1211840 |
| SM003-R | SM003_SARS_d1_1_array | RNA | GEO | GSM1211806 |
| SM003-R | SM003_SARS_d1_2_array | RNA | GEO | GSM1211807 |
| SM003-R | SM003_SARS_d1_3_array | RNA | GEO | GSM1211808 |
| SM003-R | SM003_SARS_d1_4_array | RNA | GEO | GSM1211809 |
| SM003-R | SM003_SARS_d2_1_array | RNA | GEO | GSM1211810 |
| SM003-R | SM003_SARS_d2_2_array | RNA | GEO | GSM1211811 |
| SM003-R | SM003_SARS_d2_3_array | RNA | GEO | GSM1211812 |
| SM003-R | SM003_SARS_d2_4_array | RNA | GEO | GSM1211813 |
| SM003-R | SM003_SARS_d2_5_array | RNA | GEO | GSM1211814 |
| SM003-R | SM003_SARS_d4_1_array | RNA | GEO | GSM1211815 |
| SM003-R | SM003_SARS_d4_2_array | RNA | GEO | GSM1211816 |
| SM003-R | SM003_SARS_d4_3_array | RNA | GEO | GSM1211817 |
| SM003-R | SM003_SARS_d4_4_array | RNA | GEO | GSM1211818 |
| SM003-R | SM003_SARS_d4_5_array | RNA | GEO | GSM1211819 |
| SM003-R | SM003_SARS_d7_1_array | RNA | GEO | GSM1211820 |
| SM003-R | SM003_SARS_d7_2_array | RNA | GEO | GSM1211821 |
| SM003-R | SM003_SARS_d7_3_array | RNA | GEO | GSM1211822 |
| SM003-R | SM003_SARS_d7_4_array | RNA | GEO | GSM1211823 |
| SM003-R | SM003_SARS_d7_5_array | RNA | GEO | GSM1211824 |
| SM004-R | SM004_B6_MA15_d4_1_array | RNA | GEO | GSM1244476 |
| SM004-R | SM004_B6_MA15_d4_2_array | RNA | GEO | GSM1244477 |
| SM004-R | SM004_B6_MA15_d4_3_array | RNA | GEO | GSM1244478 |
| SM004-R | SM004_B6_MA15_d4_4_array | RNA | GEO | GSM1244479 |
| SM004-R | SM004_B6_MA15_d7_1_array | RNA | GEO | GSM1244480 |
| SM004-R | SM004_B6_MA15_d7_3_array | RNA | GEO | GSM1244448 |
| SM004-R | SM004_B6_MA15_d7_4_array | RNA | GEO | GSM1244449 |
| SM004-R | SM004_B6_Mock_d4_1_array | RNA | GEO | GSM1244450 |
| SM004-R | SM004_B6_Mock_d4_2_array | RNA | GEO | GSM1244451 |
| SM004-R | SM004_B6_Mock_d4_3_array | RNA | GEO | GSM1244452 |
| SM004-R | SM004_B6_Mock_d4_4_array | RNA | GEO | GSM1244453 |
| SM004-R | SM004_B6_Mock_d7_1_array | RNA | GEO | GSM1244454 |
| SM004-R | SM004_B6_Mock_d7_2_array | RNA | GEO | GSM1244455 |
| SM004-R | SM004_B6_Mock_d7_3_array | RNA | GEO | GSM1244456 |
| SM004-R | SM004_B6_Mock_d7_4_array | RNA | GEO | GSM1244457 |
| SM004-R | SM004_PAI1_MA15_d4_1_array | RNA | GEO | GSM1244465 |
| SM004-R | SM004_PAI1_MA15_d4_2_array | RNA | GEO | GSM1244466 |
| SM004-R | SM004_PAI1_MA15_d4_3_array | RNA | GEO | GSM1244467 |
| SM004-R | SM004_PAI1_MA15_d4_4_array | RNA | GEO | GSM1244468 |
| SM004-R | SM004_PAI1_MA15_d7_1_array | RNA | GEO | GSM1244469 |
| SM004-R | SM004_PAI1_MA15_d7_2_array | RNA | GEO | GSM1244470 |
| SM004-R | SM004_PAI1_MA15_d7_3_array | RNA | GEO | GSM1244471 |
| SM004-R | SM004_PAI1_Mock_d4_1_array | RNA | GEO | GSM1244472 |
| SM004-R | SM004_PAI1_Mock_d4_2_array | RNA | GEO | GSM1244473 |
| SM004-R | SM004_PAI1_Mock_d7_1_array | RNA | GEO | GSM1244474 |
| SM004-R | SM004_PAI1_Mock_d7_2_array | RNA | GEO | GSM1244475 |
| SM004-R | SM004_TIMP1_MA15_d4_1_array | RNA | GEO | GSM1244443 |
| SM004-R | SM004_TIMP1_MA15_d4_2_array | RNA | GEO | GSM1244444 |
| SM004-R | SM004_TIMP1_MA15_d4_3_array | RNA | GEO | GSM1244445 |
| SM004-R | SM004_TIMP1_MA15_d4_4_array | RNA | GEO | GSM1244446 |
| SM004-R | SM004_TIMP1_MA15_d7_1_array | RNA | GEO | GSM1244447 |
| SM004-R | SM004_TIMP1_MA15_d7_2_array | RNA | GEO | GSM1244458 |
| SM004-R | SM004_TIMP1_MA15_d7_3_array | RNA | GEO | GSM1244459 |
| SM004-R | SM004_TIMP1_MA15_d7_4_array | RNA | GEO | GSM1244460 |
| SM004-R | SM004_TIMP1_Mock_d4_1_array | RNA | GEO | GSM1244461 |
| SM004-R | SM004_TIMP1_Mock_d4_2_array | RNA | GEO | GSM1244462 |
| SM004-R | SM004_TIMP1_Mock_d7_1_array | RNA | GEO | GSM1244463 |
| SM004-R | SM004_TIMP1_Mock_d7_2_array | RNA | GEO | GSM1244464 |
| SM007-R | SM007_B6_MA15_d2_1_array | RNA | GEO | GSM1231651 |
| SM007-R | SM007_B6_MA15_d2_2_array | RNA | GEO | GSM1231652 |
| SM007-R | SM007_B6_MA15_d2_3_array | RNA | GEO | GSM1231653 |
| SM007-R | SM007_B6_MA15_d4_1_array | RNA | GEO | GSM1231654 |
| SM007-R | SM007_B6_MA15_d4_2_array | RNA | GEO | GSM1231655 |
| SM007-R | SM007_B6_MA15_d7_1_array | RNA | GEO | GSM1231656 |
| SM007-R | SM007_B6_MA15_d7_2_array | RNA | GEO | GSM1231657 |
| SM007-R | SM007_B6_MA15_d7_3_array | RNA | GEO | GSM1231658 |
| SM007-R | SM007_B6_Mock_d2_1_array | RNA | GEO | GSM1231659 |
| SM007-R | SM007_B6_Mock_d2_2_array | RNA | GEO | GSM1231660 |
| SM007-R | SM007_B6_Mock_d2_3_array | RNA | GEO | GSM1231661 |
| SM007-R | SM007_B6_Mock_d4_1_array | RNA | GEO | GSM1231662 |
| SM007-R | SM007_B6_Mock_d4_2_array | RNA | GEO | GSM1231663 |
| SM007-R | SM007_B6_Mock_d4_3_array | RNA | GEO | GSM1231664 |
| SM007-R | SM007_B6_Mock_d7_1_array | RNA | GEO | GSM1231665 |
| SM007-R | SM007_B6_Mock_d7_2_array | RNA | GEO | GSM1231666 |
| SM007-R | SM007_B6_Mock_d7_3_array | RNA | GEO | GSM1231667 |
| SM007-R | SM007_CXCR3_MA15_d2_1_array | RNA | GEO | GSM1231668 |
| SM007-R | SM007_CXCR3_MA15_d2_2_array | RNA | GEO | GSM1231669 |
| SM007-R | SM007_CXCR3_MA15_d2_3_array | RNA | GEO | GSM1231670 |
| SM007-R | SM007_CXCR3_MA15_d4_2_array | RNA | GEO | GSM1231671 |
| SM007-R | SM007_CXCR3_MA15_d4_3_array | RNA | GEO | GSM1231672 |
| SM007-R | SM007_CXCR3_MA15_d7_1_array | RNA | GEO | GSM1231673 |
| SM007-R | SM007_CXCR3_MA15_d7_2_array | RNA | GEO | GSM1231674 |
| SM007-R | SM007_CXCR3_MA15_d7_3_array | RNA | GEO | GSM1231675 |
| SM007-R | SM007_CXCR3_MA15_d7_4_array | RNA | GEO | GSM1231676 |
| SM007-R | SM007_CXCR3_Mock_d2_2_array | RNA | GEO | GSM1231677 |
| SM007-R | SM007_CXCR3_Mock_d2_3_array | RNA | GEO | GSM1231678 |
| SM007-R | SM007_CXCR3_Mock_d4_1_array | RNA | GEO | GSM1231679 |
| SM007-R | SM007_CXCR3_Mock_d4_2_array | RNA | GEO | GSM1231680 |
| SM007-R | SM007_CXCR3_Mock_d7_1_array | RNA | GEO | GSM1231681 |
| SM007-R | SM007_CXCR3_Mock_d7_2_array | RNA | GEO | GSM1231682 |
| SM007-R | SM007_CXCR3_Mock_d7_3_array | RNA | GEO | GSM1231683 |
| SM009-R | SM009_B6_MA15_d4_2_array | RNA | GEO | GSM1244490 |
| SM009-R | SM009_B6_MA15_d4_3_array | RNA | GEO | GSM1244491 |
| SM009-R | SM009_B6_MA15_d4_4_array | RNA | GEO | GSM1244492 |
| SM009-R | SM009_B6_MA15_d7_2_array | RNA | GEO | GSM1244493 |
| SM009-R | SM009_B6_MA15_d7_4_array | RNA | GEO | GSM1244494 |
| SM009-R | SM009_B6_Mock_d4_1_array | RNA | GEO | GSM1244495 |
| SM009-R | SM009_B6_Mock_d4_2_array | RNA | GEO | GSM1244496 |
| SM009-R | SM009_B6_Mock_d7_1_array | RNA | GEO | GSM1244497 |
| SM009-R | SM009_B6_Mock_d7_2_array | RNA | GEO | GSM1244498 |
| SM009-R | SM009_PLAT_MA15_d4_3_array | RNA | GEO | GSM1244481 |
| SM009-R | SM009_PLAT_MA15_d4_4_array | RNA | GEO | GSM1244482 |
| SM009-R | SM009_PLAT_MA15_d7_1_array | RNA | GEO | GSM1244483 |
| SM009-R | SM009_PLAT_MA15_d7_2_array | RNA | GEO | GSM1244484 |
| SM009-R | SM009_PLAT_MA15_d7_4_array | RNA | GEO | GSM1244485 |
| SM009-R | SM009_PLAT_Mock_d4_1_array | RNA | GEO | GSM1244486 |
| SM009-R | SM009_PLAT_Mock_d4_2_array | RNA | GEO | GSM1244487 |
| SM009-R | SM009_PLAT_Mock_d7_1_array | RNA | GEO | GSM1244488 |
| SM009-R | SM009_PLAT_Mock_d7_2_array | RNA | GEO | GSM1244489 |
| SM012-R | SM012_dORF6_10^5pfu_1d_1_array | RNA | GEO | GSM1196066 |
| SM012-R | SM012_dORF6_10^5pfu_1d_3_array | RNA | GEO | GSM1196067 |
| SM012-R | SM012_dORF6_10^5pfu_1d_5_array | RNA | GEO | GSM1196068 |
| SM012-R | SM012_dORF6_10^5pfu_2d_1_array | RNA | GEO | GSM1196069 |
| SM012-R | SM012_dORF6_10^5pfu_2d_3_array | RNA | GEO | GSM1196070 |
| SM012-R | SM012_dORF6_10^5pfu_2d_4_array | RNA | GEO | GSM1196071 |
| SM012-R | SM012_dORF6_10^5pfu_4d_1_array | RNA | GEO | GSM1196072 |
| SM012-R | SM012_dORF6_10^5pfu_4d_2_array | RNA | GEO | GSM1196073 |
| SM012-R | SM012_dORF6_10^5pfu_4d_3_array | RNA | GEO | GSM1196074 |
| SM012-R | SM012_dORF6_10^5pfu_7d_1_array | RNA | GEO | GSM1196075 |
| SM012-R | SM012_dORF6_10^5pfu_7d_2_array | RNA | GEO | GSM1196076 |
| SM012-R | SM012_dORF6_10^5pfu_7d_3_array | RNA | GEO | GSM1196077 |
| SM012-R | SM012_MA15_10^5pfu_1d_2_array | RNA | GEO | GSM1196078 |
| SM012-R | SM012_MA15_10^5pfu_1d_3_array | RNA | GEO | GSM1196079 |
| SM012-R | SM012_MA15_10^5pfu_1d_4_array | RNA | GEO | GSM1196080 |
| SM012-R | SM012_MA15_10^5pfu_2d_1_array | RNA | GEO | GSM1196081 |
| SM012-R | SM012_MA15_10^5pfu_2d_3_array | RNA | GEO | GSM1196082 |
| SM012-R | SM012_MA15_10^5pfu_2d_5_array | RNA | GEO | GSM1196083 |
| SM012-R | SM012_MA15_10^5pfu_4d_1_array | RNA | GEO | GSM1196084 |
| SM012-R | SM012_MA15_10^5pfu_4d_2_array | RNA | GEO | GSM1196085 |
| SM012-R | SM012_MA15_10^5pfu_4d_3_array | RNA | GEO | GSM1196086 |
| SM012-R | SM012_MA15_10^5pfu_7d_1_array | RNA | GEO | GSM1196087 |
| SM012-R | SM012_MA15_10^5pfu_7d_2_array | RNA | GEO | GSM1196088 |
| SM012-R | SM012_MA15_10^5pfu_7d_3_array | RNA | GEO | GSM1196089 |
| SM012-R | SM012_Mock_1d_1_array | RNA | GEO | GSM1196090 |
| SM012-R | SM012_Mock_1d_4_array | RNA | GEO | GSM1196091 |
| SM012-R | SM012_Mock_1d_5_array | RNA | GEO | GSM1196092 |
| SM012-R | SM012_Mock_2d_2_array | RNA | GEO | GSM1196093 |
| SM012-R | SM012_Mock_2d_3_array | RNA | GEO | GSM1196094 |
| SM012-R | SM012_Mock_2d_4_array | RNA | GEO | GSM1196095 |
| SM012-R | SM012_Mock_4d_1_array | RNA | GEO | GSM1196096 |
| SM012-R | SM012_Mock_4d_2_array | RNA | GEO | GSM1196097 |
| SM012-R | SM012_Mock_4d_3_array | RNA | GEO | GSM1196098 |
| SM012-R | SM012_Mock_7d_1_array | RNA | GEO | GSM1196099 |
| SM012-R | SM012_Mock_7d_3_array | RNA | GEO | GSM1196100 |
| SM014-R | SM014_MA15_10^5pfu_1d_1_array | RNA | GEO | GSM1196101 |
| SM014-R | SM014_MA15_10^5pfu_1d_2_array | RNA | GEO | GSM1196102 |
| SM014-R | SM014_MA15_10^5pfu_1d_3_array | RNA | GEO | GSM1196103 |
| SM014-R | SM014_MA15_10^5pfu_1d_4_array | RNA | GEO | GSM1196104 |
| SM014-R | SM014_MA15_10^5pfu_2d_1_array | RNA | GEO | GSM1196105 |
| SM014-R | SM014_MA15_10^5pfu_2d_2_array | RNA | GEO | GSM1196106 |
| SM014-R | SM014_MA15_10^5pfu_2d_3_array | RNA | GEO | GSM1196107 |
| SM014-R | SM014_MA15_10^5pfu_2d_4_array | RNA | GEO | GSM1196108 |
| SM014-R | SM014_MA15_10^5pfu_4d_1_array | RNA | GEO | GSM1196109 |
| SM014-R | SM014_MA15_10^5pfu_4d_2_array | RNA | GEO | GSM1196110 |
| SM014-R | SM014_MA15_10^5pfu_4d_3_array | RNA | GEO | GSM1196111 |
| SM014-R | SM014_MA15_10^5pfu_4d_4_array | RNA | GEO | GSM1196112 |
| SM014-R | SM014_MA15_10^5pfu_7d_1_array | RNA | GEO | GSM1196113 |
| SM014-R | SM014_MA15_10^5pfu_7d_2_array | RNA | GEO | GSM1196114 |
| SM014-R | SM014_MA15_10^5pfu_7d_4_array | RNA | GEO | GSM1196115 |
| SM014-R | SM014_Mock_1d_2_array | RNA | GEO | GSM1196116 |
| SM014-R | SM014_Mock_1d_3_array | RNA | GEO | GSM1196117 |
| SM014-R | SM014_Mock_2d_1_array | RNA | GEO | GSM1196118 |
| SM014-R | SM014_Mock_2d_2_array | RNA | GEO | GSM1196119 |
| SM014-R | SM014_Mock_2d_3_array | RNA | GEO | GSM1196120 |
| SM014-R | SM014_Mock_4d_1_array | RNA | GEO | GSM1196121 |
| SM014-R | SM014_Mock_4d_2_array | RNA | GEO | GSM1196122 |
| SM014-R | SM014_Mock_4d_3_array | RNA | GEO | GSM1196123 |
| SM014-R | SM014_Mock_7d_1_array | RNA | GEO | GSM1196124 |
| SM014-R | SM014_Mock_7d_2_array | RNA | GEO | GSM1196125 |
| SM014-R | SM014_Mock_7d_3_array | RNA | GEO | GSM1196126 |
| SM014-R | SM014_nsp16_10^5pfu_1d_2_array | RNA | GEO | GSM1196127 |
| SM014-R | SM014_nsp16_10^5pfu_1d_3_array | RNA | GEO | GSM1196128 |
| SM014-R | SM014_nsp16_10^5pfu_1d_4_array | RNA | GEO | GSM1196129 |
| SM014-R | SM014_nsp16_10^5pfu_2d_1_array | RNA | GEO | GSM1196130 |
| SM014-R | SM014_nsp16_10^5pfu_2d_2_array | RNA | GEO | GSM1196131 |
| SM014-R | SM014_nsp16_10^5pfu_2d_3_array | RNA | GEO | GSM1196132 |
| SM014-R | SM014_nsp16_10^5pfu_2d_4_array | RNA | GEO | GSM1196133 |
| SM014-R | SM014_nsp16_10^5pfu_4d_1_array | RNA | GEO | GSM1196134 |
| SM014-R | SM014_nsp16_10^5pfu_4d_3_array | RNA | GEO | GSM1196135 |
| SM014-R | SM014_nsp16_10^5pfu_4d_4_array | RNA | GEO | GSM1196136 |
| SM014-R | SM014_nsp16_10^5pfu_7d_1_array | RNA | GEO | GSM1196137 |
| SM014-R | SM014_nsp16_10^5pfu_7d_2_array | RNA | GEO | GSM1196138 |
| SM014-R | SM014_nsp16_10^5pfu_7d_3_array | RNA | GEO | GSM1196139 |
| SM014-R | SM014_nsp16_10^5pfu_7d_4_array | RNA | GEO | GSM1196140 |
| SM015-R | SM015_B6_MA15_d4_2_array | RNA | GEO | GSM1003080 |
| SM015-R | SM015_B6_MA15_d7_1_array | RNA | GEO | GSM1003081 |
| SM015-R | SM015_B6_MA15_d7_2_array | RNA | GEO | GSM1003082 |
| SM015-R | SM015_B6_MA15_d7_3_array | RNA | GEO | GSM1003083 |
| SM015-R | SM015_B6_Mock_d4_1_array | RNA | GEO | GSM1003084 |
| SM015-R | SM015_B6_Mock_d4_2_array | RNA | GEO | GSM1003085 |
| SM015-R | SM015_B6_Mock_d7_1_array | RNA | GEO | GSM1003086 |
| SM015-R | SM015_B6_Mock_d7_2_array | RNA | GEO | GSM1003087 |
| SM015-R | SM015_B6_Mock_d7_3_array | RNA | GEO | GSM1003088 |
| SM015-R | SM015_Tnfrsf1a/1b_MA15_d4_1_array | RNA | GEO | GSM1003089 |
| SM015-R | SM015_Tnfrsf1a/1b_MA15_d4_2_array | RNA | GEO | GSM1003090 |
| SM015-R | SM015_Tnfrsf1a/1b_MA15_d7_1_array | RNA | GEO | GSM1003091 |
| SM015-R | SM015_Tnfrsf1a/1b_MA15_d7_2_array | RNA | GEO | GSM1003092 |
| SM015-R | SM015_Tnfrsf1a/1b_MA15_d7_3_array | RNA | GEO | GSM1003093 |
| SM015-R | SM015_Tnfrsf1a/1b_Mock_d4_1_array | RNA | GEO | GSM1003094 |
| SM015-R | SM015_Tnfrsf1a/1b_Mock_d4_2_array | RNA | GEO | GSM1003095 |
| SM015-R | SM015_Tnfrsf1a/1b_Mock_d7_1_array | RNA | GEO | GSM1003096 |
| SM015-R | SM015_Tnfrsf1a/1b_Mock_d7_2_array | RNA | GEO | GSM1003097 |
| SM015-R | SM015_Tnfrsf1a/1b_Mock_d7_3_array | RNA | GEO | GSM1003098 |
| SM019-R | SM019_B6_MA15_d4_1_array | RNA | GEO | GSM1002567 |
| SM019-R | SM019_B6_MA15_d4_2_array | RNA | GEO | GSM1002568 |
| SM019-R | SM019_B6_MA15_d4_3_array | RNA | GEO | GSM1002569 |
| SM019-R | SM019_B6_MA15_d7_1_array | RNA | GEO | GSM1002570 |
| SM019-R | SM019_B6_MA15_d7_2_array | RNA | GEO | GSM1002571 |
| SM019-R | SM019_B6_MA15_d7_3_array | RNA | GEO | GSM1002572 |
| SM019-R | SM019_B6_Mock_d4_1_array | RNA | GEO | GSM1002573 |
| SM019-R | SM019_B6_Mock_d4_2_array | RNA | GEO | GSM1002574 |
| SM019-R | SM019_B6_Mock_d4_3_array | RNA | GEO | GSM1002575 |
| SM019-R | SM019_B6_Mock_d7_1_array | RNA | GEO | GSM1002576 |
| SM019-R | SM019_B6_Mock_d7_2_array | RNA | GEO | GSM1002577 |
| SM019-R | SM019_B6_Mock_d7_3_array | RNA | GEO | GSM1002578 |
| SM019-R | SM019_Tnfrsf1b_MA15_d4_1_array | RNA | GEO | GSM1002579 |
| SM019-R | SM019_Tnfrsf1b_MA15_d4_2_array | RNA | GEO | GSM1002580 |
| SM019-R | SM019_Tnfrsf1b_MA15_d4_3_array | RNA | GEO | GSM1002581 |
| SM019-R | SM019_Tnfrsf1b_MA15_d7_2_array | RNA | GEO | GSM1002582 |
| SM019-R | SM019_Tnfrsf1b_MA15_d7_3_array | RNA | GEO | GSM1002583 |
| SM019-R | SM019_Tnfrsf1b_Mock_d4_1_array | RNA | GEO | GSM1002584 |
| SM019-R | SM019_Tnfrsf1b_Mock_d4_2_array | RNA | GEO | GSM1002585 |
| SM019-R | SM019_Tnfrsf1b_Mock_d4_3_array | RNA | GEO | GSM1002586 |
| SM019-R | SM019_Tnfrsf1b_Mock_d7_2_array | RNA | GEO | GSM1002587 |
| SM019-R | SM019_Tnfrsf1b_Mock_d7_3_array | RNA | GEO | GSM1002588 |
| SM020-R | SM020_B6_MA15_d4_1_array | RNA | GEO | GSM1002603 |
| SM020-R | SM020_B6_MA15_d4_2_array | RNA | GEO | GSM1002604 |
| SM020-R | SM020_B6_MA15_d7_1_array | RNA | GEO | GSM1002605 |
| SM020-R | SM020_B6_MA15_d7_2_array | RNA | GEO | GSM1002606 |
| SM020-R | SM020_B6_MA15_d7_3_array | RNA | GEO | GSM1002607 |
| SM020-R | SM020_B6_Mock_d4_1_array | RNA | GEO | GSM1002608 |
| SM020-R | SM020_B6_Mock_d4_2_array | RNA | GEO | GSM1002609 |
| SM020-R | SM020_B6_Mock_d7_1_array | RNA | GEO | GSM1002610 |
| SM020-R | SM020_B6_Mock_d7_2_array | RNA | GEO | GSM1002611 |
| SM020-R | SM020_B6_Mock_d7_3_array | RNA | GEO | GSM1002612 |
| SM020-R | SM020_ppp1r14c_MA15_d4_1_array | RNA | GEO | GSM1002613 |
| SM020-R | SM020_ppp1r14c_MA15_d4_2_array | RNA | GEO | GSM1002614 |
| SM020-R | SM020_ppp1r14c_MA15_d7_2_array | RNA | GEO | GSM1002615 |
| SM020-R | SM020_ppp1r14c_MA15_d7_3_array | RNA | GEO | GSM1002616 |
| SM020-R | SM020_ppp1r14c_Mock_d4_1_array | RNA | GEO | GSM1002617 |
| SM020-R | SM020_ppp1r14c_Mock_d4_2_array | RNA | GEO | GSM1002618 |
| SM020-R | SM020_ppp1r14c_Mock_d7_1_array | RNA | GEO | GSM1002619 |
| SM020-R | SM020_ppp1r14c_Mock_d7_2_array | RNA | GEO | GSM1002620 |
| SM020-R | SM020_ppp1r14c_Mock_d7_3_array | RNA | GEO | GSM1002621 |
Table 3. Experiment samples from proteomic assays.
| Experiment ID | Experiment Sample ID | Biological Sample Type | Public Repository Name |
|---|---|---|---|
| CA04M001-P | CA04M001_CA04_10^4_1d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_1d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_1d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_1d_4_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_2d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_2d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_2d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_2d_4_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_4d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_4d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_4d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_7d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_7d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^4_7d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_1d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_1d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_1d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_1d_4_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_2d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_2d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_2d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_2d_4_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_4d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_4d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_4d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_4d_4_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_7d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_7d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_7d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_7d_4_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_CA04_10^5_7d_5_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_7d_1_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_7d_2_proteomics | Protein | PeptideAtlas |
| CA04M001-P | CA04M001_Mock_7d_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_0h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_0h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_0h_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_12h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_12h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_12h_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_18h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_18h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_18h_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_24h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_24h_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_3h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_7h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_7h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_Mock_7h_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_0h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_0h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_12h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_12h_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_18h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_18h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_18h_3_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_24h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_24h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_3h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_3h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_7h_1_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_7h_2_proteomics | Protein | PeptideAtlas |
| ICL004-P | ICL004_VN1203_7h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_0h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_0h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_0h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_12h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_12h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_12h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_18h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_18h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_18h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_24h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_24h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_24h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_30h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_30h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_30h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_36h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_36h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_36h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_3h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_3h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_3h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_48h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_48h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_48h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_7h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_7h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_CA04_7h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_0h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_0h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_0h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_12h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_12h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_12h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_18h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_18h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_18h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_24h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_24h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_24h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_30h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_30h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_30h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_36h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_36h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_36h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_3h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_3h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_3h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_48h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_48h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_48h_3_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_7h_1_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_7h_2_proteomics | Protein | PeptideAtlas |
| ICL006-P | ICL006_Mock_7h_3_proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_0h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_0h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_0h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_12h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_12h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_12h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_18h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_18h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_18h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_24h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_24h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_24h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_30h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_30h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_30h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_36h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_36h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_36h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_3h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_3h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_3h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_48h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_48h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_48h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_7h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_7h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_mock_7h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_0h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_0h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_0h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_12h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_12h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_12h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_18h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_18h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_18h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_24h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_24h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_24h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_30h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_30h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_30h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_36h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_36h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_36h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_3h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_3h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_3h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_48h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_48h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_48h_3_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_7h_1_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_7h_2_Proteomics | Protein | PeptideAtlas |
| ICL010-P | ICL010_NL_7h_3_Proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_0h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_0h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_0h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_12h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_12h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_12h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_18h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_18h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_18h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_24h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_24h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_24h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_3h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_3h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_3h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_7h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_7h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_mock_7h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_0h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_0h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_0h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_12h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_12h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_12h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_18h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_18h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_18h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_24h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_24h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_24h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_3h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_3h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_3h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_7h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_7h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB1-F2-del_7h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_0h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_0h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_0h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_12h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_12h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_12h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_18h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_18h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_18h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_24h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_24h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_24h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_3h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_3h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_3h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_7h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_7h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN-PB2-627E_7h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN1203_24h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN1203_24h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN1203_24h_3_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN1203_7h_1_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN1203_7h_2_proteomics | Protein | PeptideAtlas |
| ICL011-P | ICL011_VN1203_7h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_0h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_0h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_0h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_12h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_12h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_12h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_18h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_18h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_18h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_24h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_24h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_24h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_3h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_3h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_3h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_7h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_7h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_mock_7h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_0h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_0h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_0h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_12h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_12h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_12h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_18h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_18h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_18h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_24h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_24h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_24h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_3h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_3h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_3h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_7h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_7h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_NS1trunc_7h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_VN1203_24h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_VN1203_24h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_VN1203_24h_3_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_VN1203_7h_1_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_VN1203_7h_2_proteomics | Protein | PeptideAtlas |
| ICL012-P | ICL012_VN1203_7h_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_1d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_2d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_2d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_7d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_7d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_Mock_7d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_1d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_1d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_1d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_1d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_1d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_2d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_2d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_2d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_2d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_2d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_4d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_4d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_4d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_4d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_4d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_7d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_7d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_7d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^2_7d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_1d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_1d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_1d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_1d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_1d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_2d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_2d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_2d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_2d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_2d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_4d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_4d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_4d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_4d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_4d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_7d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_7d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^3_7d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_1d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_1d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_1d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_1d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_1d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_2d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_2d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_2d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_2d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_2d_5_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_4d_1_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_4d_2_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_4d_3_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_4d_4_proteomics | Protein | PeptideAtlas |
| IM001-P | IM001_VN1203_10^4_4d_5_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_1d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_1d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_1d_3_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_1d_4_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_1d_5_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_2d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_2d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_2d_3_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_2d_4_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_2d_5_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_4d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_4d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_4d_3_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_4d_4_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_4d_5_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_7d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_7d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_7d_3_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_7d_4_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_HAavir_10^4_7d_5_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_1d_3_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_2d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_2d_3_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_7d_1_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_7d_2_proteomics | Protein | PeptideAtlas |
| IM004-P | IM004_Mock_7d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_1d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_2d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_2d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_7d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_7d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_Mock_7d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_1d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_1d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_1d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_1d_4_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_1d_5_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_2d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_2d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_2d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_2d_4_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_2d_5_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_4d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_4d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_4d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_4d_4_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_4d_5_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_7d_1_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_7d_2_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_7d_3_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_7d_4_proteomics | Protein | PeptideAtlas |
| IM005-P | IM005_PB2_627E_10^4pfu_7d_5_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_1d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_2d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_2d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_7d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_7d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_Mock_7d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_4_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_5_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_4_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_5_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_4_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_5_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_1_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_2_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_3_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_4_proteomics | Protein | PeptideAtlas |
| IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_5_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_1d_3_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_2d_2_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_2d_3_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_1_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_2_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_3_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_4_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_5_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_1_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_2_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_3_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_4_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_5_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_1_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_2_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_3_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_4_proteomics | Protein | PeptideAtlas |
| IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_1d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_2d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_2d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_7d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_7d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_Mock_7d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_1d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_1d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_1d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_1d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_1d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_2d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_2d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_2d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_2d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_2d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_4d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_4d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_4d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_4d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_4d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_7d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_7d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_7d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_7d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^3pfu_7d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_1d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_1d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_1d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_1d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_1d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_2d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_2d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_2d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_2d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_2d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_4d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_4d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_4d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_4d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_4d_5_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_7d_1_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_7d_2_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_7d_3_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_7d_4_proteomics | Protein | PeptideAtlas |
| IM007-P | IM007_NS1trunc124_10^4pfu_7d_5_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_0h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_0h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_0h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_12h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_12h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_12h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_24h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_24h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_24h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_30h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_30h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_30h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_36h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_36h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_36h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_3h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_3h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_3h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_48h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_48h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_48h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_54h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_54h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_60h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_60h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_60h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_72h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_72h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_72h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_7h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_7h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_dORF6_7h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_0h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_0h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_0h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_12h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_12h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_24h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_24h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_24h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_30h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_30h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_30h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_36h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_36h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_36h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_3h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_3h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_3h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_48h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_48h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_48h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_54h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_54h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_54h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_60h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_60h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_60h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_72h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_72h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_72h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_7h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_icSARS_7h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_0h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_0h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_0h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_12h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_12h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_12h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_24h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_24h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_24h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_30h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_30h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_30h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_36h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_36h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_36h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_3h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_3h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_3h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_48h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_48h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_48h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_54h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_54h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_54h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_60h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_60h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_60h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_72h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_72h_2_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_72h_3_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_7h_1_proteomics | Protein | PeptideAtlas |
| SCL005-P | SCL005_Mock_7h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_0h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_0h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_0h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_12h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_12h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_12h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_24h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_24h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_24h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_30h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_30h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_36h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_36h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_36h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_3h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_3h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_3h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_48h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_48h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_54h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_54h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_60h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_60h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_60h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_72h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_72h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_72h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_7h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_7h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_BatSRBD_7h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_0h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_0h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_0h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_12h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_12h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_12h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_24h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_24h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_24h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_30h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_30h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_36h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_36h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_36h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_48h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_48h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_48h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_54h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_54h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_54h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_60h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_60h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_60h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_72h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_72h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_72h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_7h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_7h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_icSARS_7h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_0h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_0h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_12h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_12h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_12h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_24h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_24h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_24h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_30h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_30h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_30h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_36h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_36h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_36h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_3h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_3h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_3h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_48h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_48h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_48h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_54h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_54h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_54h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_60h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_60h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_72h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_72h_3_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_7h_1_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_7h_2_proteomics | Protein | PeptideAtlas |
| SCL006-P | SCL006_Mock_7h_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_1d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_1d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_1d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_2d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_2d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_2d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_4d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_4d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_4d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_7d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_7d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_Mock_7d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_1d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_1d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_1d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_1d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_1d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_2d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_2d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_2d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_2d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_2d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_4d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_4d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_4d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_4d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_4d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_7d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_7d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_7d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_7d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^2_7d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_1d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_1d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_1d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_1d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_1d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_2d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_2d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_2d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_2d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_2d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_4d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_4d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_4d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_4d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_4d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_7d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_7d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_7d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_7d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^3_7d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_1d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_1d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_1d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_1d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_1d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_2d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_2d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_2d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_2d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_2d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_4d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_4d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_4d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_4d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_4d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_7d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_7d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_7d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_7d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^4_7d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_1d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_1d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_1d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_1d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_1d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_2d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_2d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_2d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_2d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_2d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_4d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_4d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_4d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_4d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_4d_5_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_7d_1_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_7d_2_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_7d_3_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_7d_4_proteomics | Protein | PeptideAtlas |
| SM001-P | SM001_SARS_10^5_7d_5_proteomics | Protein | PeptideAtlas |
Table 4. Sample tracking from the subject to the experiment sample level.
| STUDY ID | Subject ID | Species Name | Biological Sample ID | Sample Type | Experiment ID | Experiment Sample ID | Biological Sample Type | Experiment ID | Experiment Sample ID | Biological Sample Type |
|---|---|---|---|---|---|---|---|---|---|---|
| CA04M001 | CA04M001_10^3_1d_2 | Mus musculus | CA04M001_10^3_1d_2_lung | Tissue | CA04M001-R | CA04M001_10^3_1d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_1d_3 | Mus musculus | CA04M001_10^3_1d_3_lung | Tissue | CA04M001-R | CA04M001_10^3_1d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_1d_4 | Mus musculus | CA04M001_10^3_1d_4_lung | Tissue | CA04M001-R | CA04M001_10^3_1d_4_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_2d_1 | Mus musculus | CA04M001_10^3_2d_1_lung | Tissue | CA04M001-R | CA04M001_10^3_2d_1_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_2d_2 | Mus musculus | CA04M001_10^3_2d_2_lung | Tissue | CA04M001-R | CA04M001_10^3_2d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_2d_3 | Mus musculus | CA04M001_10^3_2d_3_lung | Tissue | CA04M001-R | CA04M001_10^3_2d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_4d_1 | Mus musculus | CA04M001_10^3_4d_1_lung | Tissue | CA04M001-R | CA04M001_10^3_4d_1_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_4d_2 | Mus musculus | CA04M001_10^3_4d_2_lung | Tissue | CA04M001-R | CA04M001_10^3_4d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_4d_3 | Mus musculus | CA04M001_10^3_4d_3_lung | Tissue | CA04M001-R | CA04M001_10^3_4d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_4d_4 | Mus musculus | CA04M001_10^3_4d_4_lung | Tissue | CA04M001-R | CA04M001_10^3_4d_4_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_7d_2 | Mus musculus | CA04M001_10^3_7d_2_lung | Tissue | CA04M001-R | CA04M001_10^3_7d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_7d_3 | Mus musculus | CA04M001_10^3_7d_3_lung | Tissue | CA04M001-R | CA04M001_10^3_7d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^3_7d_4 | Mus musculus | CA04M001_10^3_7d_4_lung | Tissue | CA04M001-R | CA04M001_10^3_7d_4_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^4_1d_1 | Mus musculus | CA04M001_10^4_1d_1_lung | Tissue | CA04M001-R | CA04M001_10^4_1d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_1d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^4_1d_2 | Mus musculus | CA04M001_10^4_1d_2_lung | Tissue | CA04M001-R | CA04M001_10^4_1d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_1d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^4_1d_3 | Mus musculus | CA04M001_10^4_1d_3_lung | Tissue | CA04M001-R | CA04M001_10^4_1d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_1d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^4_1d_4 | Mus musculus | CA04M001_10^4_1d_4_lung | Tissue | CA04M001-R | CA04M001_10^4_1d_4_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_1d_4_proteomics | Protein |
| CA04M001 | CA04M001_10^4_2d_1 | Mus musculus | CA04M001_10^4_2d_1_lung | Tissue | CA04M001-R | CA04M001_10^4_2d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_2d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^4_2d_2 | Mus musculus | CA04M001_10^4_2d_2_lung | Tissue | CA04M001-R | CA04M001_10^4_2d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_2d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^4_2d_3 | Mus musculus | CA04M001_10^4_2d_3_lung | Tissue | CA04M001-R | CA04M001_10^4_2d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_2d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^4_2d_4 | Mus musculus | CA04M001_10^4_2d_4_lung | Tissue | CA04M001-R | CA04M001_10^4_2d_4_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_2d_4_proteomics | Protein |
| CA04M001 | CA04M001_10^4_4d_1 | Mus musculus | CA04M001_10^4_4d_1_lung | Tissue | CA04M001-R | CA04M001_10^4_4d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_4d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^4_4d_2 | Mus musculus | CA04M001_10^4_4d_2_lung | Tissue | CA04M001-R | CA04M001_10^4_4d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_4d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^4_4d_3 | Mus musculus | CA04M001_10^4_4d_3_lung | Tissue | CA04M001-R | CA04M001_10^4_4d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_4d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^4_7d_1 | Mus musculus | CA04M001_10^4_7d_1_lung | Tissue | CA04M001-R | CA04M001_10^4_7d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_7d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^4_7d_2 | Mus musculus | CA04M001_10^4_7d_2_lung | Tissue | CA04M001-R | CA04M001_10^4_7d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_7d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^4_7d_3 | Mus musculus | CA04M001_10^4_7d_3_lung | Tissue | CA04M001-R | CA04M001_10^4_7d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^4_7d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^5_1d_1 | Mus musculus | CA04M001_10^5_1d_1_lung | Tissue | CA04M001-R | CA04M001_10^5_1d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_1d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^5_1d_2 | Mus musculus | CA04M001_10^5_1d_2_lung | Tissue | CA04M001-R | CA04M001_10^5_1d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_1d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^5_1d_3 | Mus musculus | CA04M001_10^5_1d_3_lung | Tissue | CA04M001-R | CA04M001_10^5_1d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_1d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^5_1d_4 | Mus musculus | CA04M001_10^5_1d_4_lung | Tissue | CA04M001-R | CA04M001_10^5_1d_4_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_1d_4_proteomics | Protein |
| CA04M001 | CA04M001_10^5_2d_1 | Mus musculus | CA04M001_10^5_2d_1_lung | Tissue | CA04M001-R | CA04M001_10^5_2d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_2d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^5_2d_2 | Mus musculus | CA04M001_10^5_2d_2_lung | Tissue | CA04M001-R | CA04M001_10^5_2d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_2d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^5_2d_3 | Mus musculus | CA04M001_10^5_2d_3_lung | Tissue | CA04M001-R | CA04M001_10^5_2d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_2d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^5_2d_4 | Mus musculus | CA04M001_10^5_2d_4_lung | Tissue | CA04M001-R | CA04M001_10^5_2d_4_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_2d_4_proteomics | Protein |
| CA04M001 | CA04M001_10^5_4d_1 | Mus musculus | CA04M001_10^5_4d_1_lung | Tissue | CA04M001-R | CA04M001_10^5_4d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_4d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^5_4d_2 | Mus musculus | CA04M001_10^5_4d_2_lung | Tissue | CA04M001-R | CA04M001_10^5_4d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_4d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^5_4d_3 | Mus musculus | CA04M001_10^5_4d_3_lung | Tissue | CA04M001-R | CA04M001_10^5_4d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_4d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^5_4d_4 | Mus musculus | CA04M001_10^5_4d_4_lung | Tissue | CA04M001-R | CA04M001_10^5_4d_4_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_4d_4_proteomics | Protein |
| CA04M001 | CA04M001_10^5_7d_1 | Mus musculus | CA04M001_10^5_7d_1_lung | Tissue | CA04M001-R | CA04M001_10^5_7d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_7d_1_proteomics | Protein |
| CA04M001 | CA04M001_10^5_7d_2 | Mus musculus | CA04M001_10^5_7d_2_lung | Tissue | CA04M001-R | CA04M001_10^5_7d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_7d_2_proteomics | Protein |
| CA04M001 | CA04M001_10^5_7d_3 | Mus musculus | CA04M001_10^5_7d_3_lung | Tissue | CA04M001-R | CA04M001_10^5_7d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_7d_3_proteomics | Protein |
| CA04M001 | CA04M001_10^5_7d_4 | Mus musculus | CA04M001_10^5_7d_4_lung | Tissue | CA04M001-R | CA04M001_10^5_7d_4_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_7d_4_proteomics | Protein |
| CA04M001 | CA04M001_10^5_7d_5 | Mus musculus | CA04M001_10^5_7d_5_lung | Tissue | CA04M001-R | CA04M001_10^5_7d_5_RNA_ExpSam | RNA | CA04M001-P | CA04M001_CA04_10^5_7d_5_proteomics | Protein |
| CA04M001 | CA04M001_10^6_1d_1 | Mus musculus | CA04M001_10^6_1d_1_lung | Tissue | CA04M001-R | CA04M001_10^6_1d_1_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_1d_3 | Mus musculus | CA04M001_10^6_1d_3_lung | Tissue | CA04M001-R | CA04M001_10^6_1d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_1d_4 | Mus musculus | CA04M001_10^6_1d_4_lung | Tissue | CA04M001-R | CA04M001_10^6_1d_4_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_2d_1 | Mus musculus | CA04M001_10^6_2d_1_lung | Tissue | CA04M001-R | CA04M001_10^6_2d_1_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_2d_2 | Mus musculus | CA04M001_10^6_2d_2_lung | Tissue | CA04M001-R | CA04M001_10^6_2d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_2d_3 | Mus musculus | CA04M001_10^6_2d_3_lung | Tissue | CA04M001-R | CA04M001_10^6_2d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_2d_4 | Mus musculus | CA04M001_10^6_2d_4_lung | Tissue | CA04M001-R | CA04M001_10^6_2d_4_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_4d_1 | Mus musculus | CA04M001_10^6_4d_1_lung | Tissue | CA04M001-R | CA04M001_10^6_4d_1_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_4d_2 | Mus musculus | CA04M001_10^6_4d_2_lung | Tissue | CA04M001-R | CA04M001_10^6_4d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_4d_3 | Mus musculus | CA04M001_10^6_4d_3_lung | Tissue | CA04M001-R | CA04M001_10^6_4d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_7d_1 | Mus musculus | CA04M001_10^6_7d_1_lung | Tissue | CA04M001-R | CA04M001_10^6_7d_1_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_7d_2 | Mus musculus | CA04M001_10^6_7d_2_lung | Tissue | CA04M001-R | CA04M001_10^6_7d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_7d_3 | Mus musculus | CA04M001_10^6_7d_3_lung | Tissue | CA04M001-R | CA04M001_10^6_7d_3_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_7d_4 | Mus musculus | CA04M001_10^6_7d_4_lung | Tissue | CA04M001-R | CA04M001_10^6_7d_4_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_10^6_7d_5 | Mus musculus | CA04M001_10^6_7d_5_lung | Tissue | CA04M001-R | CA04M001_10^6_7d_5_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_mock_1d_1 | Mus musculus | CA04M001_mock_1d_1_lung | Tissue | CA04M001-R | CA04M001_mock_1d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_1d_1_proteomics | Protein |
| CA04M001 | CA04M001_mock_1d_2 | Mus musculus | CA04M001_mock_1d_2_lung | Tissue | CA04M001-R | CA04M001_mock_1d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_1d_2_proteomics | Protein |
| CA04M001 | CA04M001_mock_2d_1 | Mus musculus | CA04M001_mock_2d_1_lung | Tissue | CA04M001-R | CA04M001_mock_2d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_2d_1_proteomics | Protein |
| CA04M001 | CA04M001_mock_2d_2 | Mus musculus | CA04M001_mock_2d_2_lung | Tissue | CA04M001-R | CA04M001_mock_2d_2_RNA_ExpSam | RNA | |||
| CA04M001 | CA04M001_mock_4d_1 | Mus musculus | CA04M001_mock_4d_1_lung | Tissue | CA04M001-R | CA04M001_mock_4d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_4d_1_proteomics | Protein |
| CA04M001 | CA04M001_mock_4d_2 | Mus musculus | CA04M001_mock_4d_2_lung | Tissue | CA04M001-R | CA04M001_mock_4d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_4d_2_proteomics | Protein |
| CA04M001 | CA04M001_mock_4d_3 | Mus musculus | CA04M001_mock_4d_3_lung | Tissue | CA04M001-R | CA04M001_mock_4d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_4d_3_proteomics | Protein |
| CA04M001 | CA04M001_mock_7d_1 | Mus musculus | CA04M001_mock_7d_1_lung | Tissue | CA04M001-R | CA04M001_mock_7d_1_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_7d_1_proteomics | Protein |
| CA04M001 | CA04M001_mock_7d_2 | Mus musculus | CA04M001_mock_7d_2_lung | Tissue | CA04M001-R | CA04M001_mock_7d_2_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_7d_2_proteomics | Protein |
| CA04M001 | CA04M001_mock_7d_3 | Mus musculus | CA04M001_mock_7d_3_lung | Tissue | CA04M001-R | CA04M001_mock_7d_3_RNA_ExpSam | RNA | CA04M001-P | CA04M001_Mock_7d_3_proteomics | Protein |
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_0hr_1_Cells | Cells | ECL001-R | ECL001_Mock_0h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_0hr_2_Cells | Cells | ECL001-R | ECL001_Mock_0h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_0hr_3_Cells | Cells | ECL001-R | ECL001_Mock_0h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_3hr_1_Cells | Cells | ECL001-R | ECL001_Mock_3h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_3hr_2_Cells | Cells | ECL001-R | ECL001_Mock_3h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_3hr_3_Cells | Cells | ECL001-R | ECL001_Mock_3h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_7hr_1_Cells | Cells | ECL001-R | ECL001_Mock_7h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_7hr_2_Cells | Cells | ECL001-R | ECL001_Mock_7h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_7hr_3_Cells | Cells | ECL001-R | ECL001_Mock_7h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_12hr_1_Cells | Cells | ECL001-R | ECL001_Mock_12h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_12hr_2_Cells | Cells | ECL001-R | ECL001_Mock_12h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_12hr_3_Cells | Cells | ECL001-R | ECL001_Mock_12h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_18hr_1_Cells | Cells | ECL001-R | RNA | ||||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_18hr_2_Cells | Cells | ECL001-R | RNA | ||||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_18hr_3_Cells | Cells | ECL001-R | RNA | ||||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_24hr_1_Cells | Cells | ECL001-R | ECL001_Mock_24h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_24hr_2_Cells | Cells | ECL001-R | ECL001_Mock_24h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_Mock_24hr_3_Cells | Cells | ECL001-R | ECL001_Mock_24h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_0hr_1_Cells | Cells | ECL001-R | ECL001_EMC_0h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_0hr_2_Cells | Cells | ECL001-R | ECL001_EMC_0h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_0hr_3_Cells | Cells | ECL001-R | ECL001_EMC_0h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_3hr_1_Cells | Cells | ECL001-R | ECL001_EMC_3h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_3hr_2_Cells | Cells | ECL001-R | ECL001_EMC_3h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_3hr_3_Cells | Cells | ECL001-R | ECL001_EMC_3h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_7hr_1_Cells | Cells | ECL001-R | ECL001_EMC_7h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_7hr_2_Cells | Cells | ECL001-R | ECL001_EMC_7h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_7hr_3_Cells | Cells | ECL001-R | ECL001_EMC_7h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_12hr_1_Cells | Cells | ECL001-R | RNA | ||||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_12hr_2_Cells | Cells | ECL001-R | ECL001_EMC_12h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_12hr_3_Cells | Cells | ECL001-R | ECL001_EMC_12h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_18hr_1_Cells | Cells | ECL001-R | ECL001_EMC_18h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_18hr_2_Cells | Cells | ECL001-R | ECL001_EMC_18h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_18hr_3_Cells | Cells | ECL001-R | ECL001_EMC_18h_3_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_24hr_1_Cells | Cells | ECL001-R | ECL001_EMC_24h_1_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_24hr_2_Cells | Cells | ECL001-R | ECL001_EMC_24h_2_array | RNA | |||
| ECL001 | ECL001 2B-4 Cells | Homo sapiens | ECL001_EMC_24hr_3_Cells | Cells | ECL001-R | ECL001_EMC_24h_3_array | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_0H_1_cell | Cells | ICL004-R | ICL004_Mock_0H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_0h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_0H_2_cell | Cells | ICL004-R | ICL004_Mock_0H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_0h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_0H_3_cell | Cells | ICL004-R | ICL004_Mock_0H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_0h_3_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_12H_1_cell | Cells | ICL004-R | ICL004_Mock_12H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_12h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_12H_2_cell | Cells | ICL004-R | ICL004_Mock_12H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_12h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_12H_3_cell | Cells | ICL004-R | ICL004_Mock_12H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_12h_3_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_18H_1_cell | Cells | ICL004-R | ICL004_Mock_18H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_18h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_18H_2_cell | Cells | ICL004-R | ICL004_Mock_18H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_18h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_18H_3_cell | Cells | ICL004-R | ICL004_Mock_18H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_18h_3_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_24H_1_cell | Cells | ICL004-R | ICL004_Mock_24H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_24h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_24H_2_cell | Cells | ICL004-R | ICL004_Mock_24H_2_RNA_ExpSam | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_24H_3_cell | Cells | ICL004-R | ICL004_Mock_24H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_24h_3_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_3H_1_cell | Cells | ICL004-R | ICL004_Mock_3H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_3h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_3H_2_cell | Cells | ICL004-R | ICL004_Mock_3H_2_RNA_ExpSam | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_3H_3_cell | Cells | ICL004-R | ICL004_Mock_3H_3_RNA_ExpSam | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_7H_1_cell | Cells | ICL004-R | ICL004_Mock_7H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_7h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_7H_2_cell | Cells | ICL004-R | ICL004_Mock_7H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_7h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_Mock_7H_3_cell | Cells | ICL004-R | ICL004_Mock_7H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_Mock_7h_3_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_0H_1_cell | Cells | ICL004-R | ICL004_VN1203_0H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_0h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_0H_2_cell | Cells | ICL004-R | ICL004_VN1203_0H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_0h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_0H_3_cell | Cells | ICL004-R | ICL004_VN1203_0H_3_RNA_ExpSam | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_12H_1_cell | Cells | ICL004-R | ICL004_VN1203_12H_1_RNA_ExpSam | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_12H_2_cell | Cells | ICL004-R | ICL004_VN1203_12H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_12h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_12H_3_cell | Cells | ICL004-R | ICL004_VN1203_12H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_12h_3_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_18H_1_cell | Cells | ICL004-R | ICL004_VN1203_18H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_18h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_18H_2_cell | Cells | ICL004-R | ICL004_VN1203_18H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_18h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_18H_3_cell | Cells | ICL004-R | ICL004_VN1203_18H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_18h_3_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_24H_1_cell | Cells | ICL004-R | ICL004_VN1203_24H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_24h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_24H_2_cell | Cells | ICL004-R | ICL004_VN1203_24H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_24h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_24H_3_cell | Cells | ICL004-R | ICL004_VN1203_24H_3_RNA_ExpSam | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_3H_1_cell | Cells | ICL004-R | ICL004_VN1203_3H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_3h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_3H_2_cell | Cells | ICL004-R | ICL004_VN1203_3H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_3h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_3H_3_cell | Cells | ICL004-R | ICL004_VN1203_3H_3_RNA_ExpSam | RNA | |||
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_7H_1_cell | Cells | ICL004-R | ICL004_VN1203_7H_1_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_7h_1_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_7H_2_cell | Cells | ICL004-R | ICL004_VN1203_7H_2_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_7h_2_proteomics | Protein |
| ICL004 | ICL004 Calu-3 Cells | Homo sapiens | ICL004_VN1203_7H_3_cell | Cells | ICL004-R | ICL004_VN1203_7H_3_RNA_ExpSam | RNA | ICL004-P | ICL004_VN1203_7h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_0h_1_cell | Cells | ICL006-R | ICL006_CA04_0h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_0h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_0h_2_cell | Cells | ICL006-R | ICL006_CA04_0h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_0h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_0h_3_cell | Cells | ICL006-R | ICL006_CA04_0h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_0h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_12h_1_cell | Cells | ICL006-R | ICL006_CA04_12h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_12h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_12h_2_cell | Cells | ICL006-R | ICL006_CA04_12h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_12h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_12h_3_cell | Cells | ICL006-R | ICL006_CA04_12h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_12h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_18h_1_cell | Cells | ICL006-R | ICL006_CA04_18h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_18h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_18h_2_cell | Cells | ICL006-R | ICL006_CA04_18h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_18h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_18h_3_cell | Cells | ICL006-R | ICL006_CA04_18h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_18h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_24h_1_cell | Cells | ICL006-R | ICL006_CA04_24h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_24h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_24h_2_cell | Cells | ICL006-R | ICL006_CA04_24h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_24h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_24h_3_cell | Cells | ICL006-R | ICL006_CA04_24h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_24h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_30h_1_cell | Cells | ICL006-R | ICL006_CA04_30h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_30h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_30h_2_cell | Cells | ICL006-R | ICL006_CA04_30h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_30h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_30h_3_cell | Cells | ICL006-R | ICL006_CA04_30h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_30h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_36h_1_cell | Cells | ICL006-R | ICL006_CA04_36h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_36h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_36h_2_cell | Cells | ICL006-R | ICL006_CA04_36h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_36h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_36h_3_cell | Cells | ICL006-R | ICL006_CA04_36h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_36h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_3h_1_cell | Cells | ICL006-R | ICL006_CA04_3h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_3h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_3h_2_cell | Cells | ICL006-R | ICL006_CA04_3h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_3h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_3h_3_cell | Cells | ICL006-R | ICL006_CA04_3h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_3h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_48h_1_cell | Cells | ICL006-R | ICL006_CA04_48h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_48h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_48h_2_cell | Cells | ICL006-R | ICL006_CA04_48h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_48h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_48h_3_cell | Cells | ICL006-R | ICL006_CA04_48h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_48h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_7h_1_cell | Cells | ICL006-R | ICL006_CA04_7h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_7h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_7h_2_cell | Cells | ICL006-R | ICL006_CA04_7h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_7h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_CA04_7h_3_cell | Cells | ICL006-R | ICL006_CA04_7h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_CA04_7h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_0h_1_cell | Cells | ICL006-R | ICL006_mock_0h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_0h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_0h_2_cell | Cells | ICL006-R | ICL006_mock_0h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_0h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_0h_3_cell | Cells | ICL006-R | ICL006_mock_0h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_0h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_12h_1_cell | Cells | ICL006-R | ICL006_mock_12h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_12h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_12h_2_cell | Cells | ICL006-R | ICL006_mock_12h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_12h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_12h_3_cell | Cells | ICL006-R | ICL006_mock_12h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_12h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_18h_1_cell | Cells | ICL006-R | ICL006_mock_18h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_18h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_18h_2_cell | Cells | ICL006-R | ICL006_mock_18h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_18h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_18h_3_cell | Cells | ICL006-R | ICL006_mock_18h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_18h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_24h_1_cell | Cells | ICL006-R | ICL006_mock_24h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_24h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_24h_2_cell | Cells | ICL006-R | ICL006_mock_24h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_24h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_24h_3_cell | Cells | ICL006-R | ICL006_mock_24h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_24h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_30h_1_cell | Cells | ICL006-R | ICL006_mock_30h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_30h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_30h_2_cell | Cells | ICL006-R | ICL006_mock_30h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_30h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_30h_3_cell | Cells | ICL006-R | ICL006_mock_30h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_30h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_36h_1_cell | Cells | ICL006-R | ICL006_mock_36h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_36h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_36h_2_cell | Cells | ICL006-R | ICL006_mock_36h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_36h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_36h_3_cell | Cells | ICL006-R | ICL006_mock_36h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_36h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_3h_1_cell | Cells | ICL006-R | ICL006_mock_3h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_3h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_3h_2_cell | Cells | ICL006-R | ICL006_mock_3h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_3h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_3h_3_cell | Cells | ICL006-R | ICL006_mock_3h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_3h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_48h_1_cell | Cells | ICL006-R | ICL006_mock_48h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_48h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_48h_2_cell | Cells | ICL006-R | ICL006_mock_48h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_48h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_48h_3_cell | Cells | ICL006-R | ICL006_mock_48h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_48h_3_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_7h_1_cell | Cells | ICL006-R | ICL006_mock_7h_1_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_7h_1_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_7h_2_cell | Cells | ICL006-R | ICL006_mock_7h_2_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_7h_2_proteomics | Protein |
| ICL006 | ICL006 Calu-3 Cells | Homo sapiens | ICL006_mock_7h_3_cell | Cells | ICL006-R | ICL006_mock_7h_3_RNA_ExpSam | RNA | ICL006-P | ICL006_Mock_7h_3_proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_0h_1_Cells | Cells | ICL010-R | ICL010_NL_0h_1_array | RNA | ICL010-P | ICL010_NL_0h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_0h_2_Cells | Cells | ICL010-R | ICL010_NL_0h_2_array | RNA | ICL010-P | ICL010_NL_0h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_0h_3_Cells | Cells | ICL010-R | ICL010_NL_0h_3_array | RNA | ICL010-P | ICL010_NL_0h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_3h_1_Cells | Cells | ICL010-R | ICL010_NL_12h_1_array | RNA | ICL010-P | ICL010_NL_12h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_3h_2_Cells | Cells | ICL010-R | ICL010_NL_12h_2_array | RNA | ICL010-P | ICL010_NL_12h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_3h_3_Cells | Cells | ICL010-R | ICL010_NL_12h_3_array | RNA | ICL010-P | ICL010_NL_12h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_7h_1_Cells | Cells | ICL010-R | ICL010_NL_18h_1_array | RNA | ICL010-P | ICL010_NL_18h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_7h_2_Cells | Cells | ICL010-R | ICL010_NL_18h_2_array | RNA | ICL010-P | ICL010_NL_18h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_7h_3_Cells | Cells | ICL010-R | ICL010_NL_18h_3_array | RNA | ICL010-P | ICL010_NL_18h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_12h_1_Cells | Cells | ICL010-R | ICL010_NL_24h_1_array | RNA | ICL010-P | ICL010_NL_24h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_12h_2_Cells | Cells | ICL010-R | ICL010_NL_24h_2_array | RNA | ICL010-P | ICL010_NL_24h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_12h_3_Cells | Cells | ICL010-R | ICL010_NL_24h_3_array | RNA | ICL010-P | ICL010_NL_24h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_18h_1_Cells | Cells | ICL010-R | ICL010_NL_30h_1_array | RNA | ICL010-P | ICL010_NL_30h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_18h_2_Cells | Cells | ICL010-R | ICL010_NL_30h_2_array | RNA | ICL010-P | ICL010_NL_30h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_18h_3_Cells | Cells | ICL010-R | ICL010_NL_30h_3_array | RNA | ICL010-P | ICL010_NL_30h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_24h_1_Cells | Cells | ICL010-R | ICL010_NL_36h_1_array | RNA | ICL010-P | ICL010_NL_36h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_24h_2_Cells | Cells | ICL010-R | ICL010_NL_36h_2_array | RNA | ICL010-P | ICL010_NL_36h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_24h_3_Cells | Cells | ICL010-R | ICL010_NL_36h_3_array | RNA | ICL010-P | ICL010_NL_36h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_30h_1_Cells | Cells | ICL010-R | ICL010_NL_3h_1_array | RNA | ICL010-P | ICL010_NL_3h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_30h_2_Cells | Cells | ICL010-R | ICL010_NL_3h_2_array | RNA | ICL010-P | ICL010_NL_3h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_30h_3_Cells | Cells | ICL010-R | ICL010_NL_3h_3_array | RNA | ICL010-P | ICL010_NL_3h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_36h_1_Cells | Cells | ICL010-R | ICL010_NL_48h_1_array | RNA | ICL010-P | ICL010_NL_48h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_36h_2_Cells | Cells | ICL010-R | ICL010_NL_48h_2_array | RNA | ICL010-P | ICL010_NL_48h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_36h_3_Cells | Cells | ICL010-R | ICL010_NL_48h_3_array | RNA | ICL010-P | ICL010_NL_48h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_48h_1_Cells | Cells | ICL010-R | ICL010_NL_7h_1_array | RNA | ICL010-P | ICL010_NL_7h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_48h_2_Cells | Cells | ICL010-R | ICL010_NL_7h_2_array | RNA | ICL010-P | ICL010_NL_7h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_NL_48h_3_Cells | Cells | ICL010-R | ICL010_NL_7h_3_array | RNA | ICL010-P | ICL010_NL_7h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_0h_1_Cells | Cells | ICL010-R | ICL010_mock_0h_1_array | RNA | ICL010-P | ICL010_mock_0h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_0h_2_Cells | Cells | ICL010-R | ICL010_mock_0h_2_array | RNA | ICL010-P | ICL010_mock_0h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_0h_3_Cells | Cells | ICL010-R | ICL010_mock_0h_3_array | RNA | ICL010-P | ICL010_mock_0h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_3h_1_Cells | Cells | ICL010-R | ICL010_mock_12h_1_array | RNA | ICL010-P | ICL010_mock_12h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_3h_2_Cells | Cells | ICL010-R | ICL010_mock_12h_2_array | RNA | ICL010-P | ICL010_mock_12h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_3h_3_Cells | Cells | ICL010-R | ICL010_mock_12h_3_array | RNA | ICL010-P | ICL010_mock_12h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_7h_1_Cells | Cells | ICL010-R | ICL010_mock_18h_1_array | RNA | ICL010-P | ICL010_mock_18h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_7h_2_Cells | Cells | ICL010-R | ICL010_mock_18h_2_array | RNA | ICL010-P | ICL010_mock_18h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_7h_3_Cells | Cells | ICL010-R | ICL010_mock_18h_3_array | RNA | ICL010-P | ICL010_mock_18h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_12h_1_Cells | Cells | ICL010-R | ICL010_mock_24h_1_array | RNA | ICL010-P | ICL010_mock_24h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_12h_2_Cells | Cells | ICL010-R | ICL010_mock_24h_2_array | RNA | ICL010-P | ICL010_mock_24h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_12h_3_Cells | Cells | ICL010-R | ICL010_mock_24h_3_array | RNA | ICL010-P | ICL010_mock_24h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_18h_1_Cells | Cells | ICL010-R | ICL010_mock_30h_1_array | RNA | ICL010-P | ICL010_mock_30h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_18h_2_Cells | Cells | ICL010-R | ICL010_mock_30h_2_array | RNA | ICL010-P | ICL010_mock_30h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_18h_3_Cells | Cells | ICL010-R | ICL010_mock_30h_3_array | RNA | ICL010-P | ICL010_mock_30h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_24h_1_Cells | Cells | ICL010-R | ICL010_mock_36h_1_array | RNA | ICL010-P | ICL010_mock_36h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_24h_2_Cells | Cells | ICL010-R | ICL010_mock_36h_2_array | RNA | ICL010-P | ICL010_mock_36h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_24h_3_Cells | Cells | ICL010-R | ICL010_mock_36h_3_array | RNA | ICL010-P | ICL010_mock_36h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_30h_1_Cells | Cells | ICL010-R | ICL010_mock_3h_1_array | RNA | ICL010-P | ICL010_mock_3h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_30h_2_Cells | Cells | ICL010-R | ICL010_mock_3h_2_array | RNA | ICL010-P | ICL010_mock_3h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_30h_3_Cells | Cells | ICL010-R | ICL010_mock_3h_3_array | RNA | ICL010-P | ICL010_mock_3h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_36h_1_Cells | Cells | ICL010-R | ICL010_mock_48h_1_array | RNA | ICL010-P | ICL010_mock_48h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_36h_2_Cells | Cells | ICL010-R | ICL010_mock_48h_2_array | RNA | ICL010-P | ICL010_mock_48h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_36h_3_Cells | Cells | ICL010-R | ICL010_mock_48h_3_array | RNA | ICL010-P | ICL010_mock_48h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_48h_1_Cells | Cells | ICL010-R | ICL010_mock_7h_1_array | RNA | ICL010-P | ICL010_mock_7h_1_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_48h_2_Cells | Cells | ICL010-R | ICL010_mock_7h_2_array | RNA | ICL010-P | ICL010_mock_7h_2_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_mock_48h_3_Cells | Cells | ICL010-R | ICL010_mock_7h_3_array | RNA | ICL010-P | ICL010_mock_7h_3_Proteomics | Protein |
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_0h_1_Cells | Cells | ICL010-R | ICL010_Cal_0h_1_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_0h_2_Cells | Cells | ICL010-R | ICL010_Cal_0h_2_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_0h_3_Cells | Cells | ICL010-R | ICL010_Cal_0h_3_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_12h_1_Cells | Cells | ICL010-R | ICL010_Cal_12h_1_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_12h_2_Cells | Cells | ICL010-R | ICL010_Cal_12h_2_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_12h_3_Cells | Cells | ICL010-R | ICL010_Cal_12h_3_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_24h_1_Cells | Cells | ICL010-R | ICL010_Cal_24h_1_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_24h_2_Cells | Cells | ICL010-R | ICL010_Cal_24h_2_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_24h_3_Cells | Cells | ICL010-R | ICL010_Cal_24h_3_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_48h_1_Cells | Cells | ICL010-R | ICL010_Cal_48h_1_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_48h_2_Cells | Cells | ICL010-R | ICL010_Cal_48h_2_array | RNA | |||
| ICL010 | ICL010 Calu-3 cells | Homo sapiens | ICL010_Cal_48h_3_Cells | Cells | ICL010-R | ICL010_Cal_48h_3_array | RNA | |||
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_0h_1_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_0h_1_array | RNA | ICL011-P | ICL011_VN-PB2-627E_0h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_0h_2_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_0h_2_array | RNA | ICL011-P | ICL011_VN-PB2-627E_0h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_0h_3_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_0h_3_array | RNA | ICL011-P | ICL011_VN-PB2-627E_0h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_0h_1_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_0h_1_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_0h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_0h_2_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_0h_2_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_0h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_0h_3_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_0h_3_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_0h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_0h_1_Cells | Cells | ICL011-R | ICL011_mock_0h_1_array | RNA | ICL011-P | ICL011_mock_0h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_0h_2_Cells | Cells | ICL011-R | ICL011_mock_0h_2_array | RNA | ICL011-P | ICL011_mock_0h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_0h_3_Cells | Cells | ICL011-R | ICL011_mock_0h_3_array | RNA | ICL011-P | ICL011_mock_0h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_3h_1_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_3h_1_array | RNA | ICL011-P | ICL011_VN-PB2-627E_3h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_3h_2_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_3h_2_array | RNA | ICL011-P | ICL011_VN-PB2-627E_3h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_3h_3_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_3h_3_array | RNA | ICL011-P | ICL011_VN-PB2-627E_3h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_3h_1_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_3h_1_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_3h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_3h_2_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_3h_2_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_3h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_3h_3_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_3h_3_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_3h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_3h_1_Cells | Cells | ICL011-R | ICL011_mock_3h_1_array | RNA | ICL011-P | ICL011_mock_3h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_3h_2_Cells | Cells | ICL011-R | ICL011_mock_3h_2_array | RNA | ICL011-P | ICL011_mock_3h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_3h_3_Cells | Cells | ICL011-R | ICL011_mock_3h_3_array | RNA | ICL011-P | ICL011_mock_3h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_7h_1_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_7h_1_array | RNA | ICL011-P | ICL011_VN-PB2-627E_7h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_7h_2_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_7h_2_array | RNA | ICL011-P | ICL011_VN-PB2-627E_7h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_7h_3_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_7h_3_array | RNA | ICL011-P | ICL011_VN-PB2-627E_7h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_7h_1_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_7h_1_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_7h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_7h_2_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_7h_2_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_7h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_7h_3_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_7h_3_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_7h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN1203_7h_1_Cells | Cells | ICL011-R | ICL011_VN1203_7h_1_array | RNA | ICL011-P | ICL011_VN1203_7h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN1203_7h_2_Cells | Cells | ICL011-R | ICL011_VN1203_7h_2_array | RNA | ICL011-P | ICL011_VN1203_7h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN1203_7h_3_Cells | Cells | ICL011-R | ICL011_VN1203_7h_3_array | RNA | ICL011-P | ICL011_VN1203_7h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_7h_1_Cells | Cells | ICL011-R | ICL011_mock_7h_1_array | RNA | ICL011-P | ICL011_mock_7h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_7h_2_Cells | Cells | ICL011-R | ICL011_mock_7h_2_array | RNA | ICL011-P | ICL011_mock_7h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_7h_3_Cells | Cells | ICL011-R | ICL011_mock_7h_3_array | RNA | ICL011-P | ICL011_mock_7h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_12h_1_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_12h_1_array | RNA | ICL011-P | ICL011_VN-PB2-627E_12h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_12h_2_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_12h_2_array | RNA | ICL011-P | ICL011_VN-PB2-627E_12h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_12h_3_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_12h_3_array | RNA | ICL011-P | ICL011_VN-PB2-627E_12h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_12h_1_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_12h_1_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_12h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_12h_2_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_12h_2_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_12h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_12h_3_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_12h_3_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_12h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_12h_1_Cells | Cells | ICL011-R | ICL011_mock_12h_1_array | RNA | ICL011-P | ICL011_mock_12h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_12h_2_Cells | Cells | ICL011-R | ICL011_mock_12h_2_array | RNA | ICL011-P | ICL011_mock_12h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_12h_3_Cells | Cells | ICL011-R | ICL011_mock_12h_3_array | RNA | ICL011-P | ICL011_mock_12h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_18h_1_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_18h_1_array | RNA | ICL011-P | ICL011_VN-PB2-627E_18h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_18h_2_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_18h_2_array | RNA | ICL011-P | ICL011_VN-PB2-627E_18h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_18h_3_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_18h_3_array | RNA | ICL011-P | ICL011_VN-PB2-627E_18h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_18h_1_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_18h_1_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_18h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_18h_2_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_18h_2_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_18h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_18h_3_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_18h_3_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_18h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_18h_1_Cells | Cells | ICL011-R | ICL011_mock_18h_1_array | RNA | ICL011-P | ICL011_mock_18h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_18h_2_Cells | Cells | ICL011-R | ICL011_mock_18h_2_array | RNA | ICL011-P | ICL011_mock_18h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_18h_3_Cells | Cells | ICL011-R | ICL011_mock_18h_3_array | RNA | ICL011-P | ICL011_mock_18h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_24h_1_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_24h_1_array | RNA | ICL011-P | ICL011_VN-PB2-627E_24h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_24h_2_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_24h_2_array | RNA | ICL011-P | ICL011_VN-PB2-627E_24h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB2-627E_24h_3_Cells | Cells | ICL011-R | ICL011_VN-PB2-627E_24h_3_array | RNA | ICL011-P | ICL011_VN-PB2-627E_24h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_24h_1_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_24h_1_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_24h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_24h_2_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_24h_2_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_24h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN-PB1-F2-del_24h_3_Cells | Cells | ICL011-R | ICL011_VN-PB1-F2-del_24h_3_array | RNA | ICL011-P | ICL011_VN-PB1-F2-del_24h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN1203_24h_1_Cells | Cells | ICL011-R | ICL011_VN1203_24h_1_array | RNA | ICL011-P | ICL011_VN1203_24h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN1203_24h_2_Cells | Cells | ICL011-R | ICL011_VN1203_24h_2_array | RNA | ICL011-P | ICL011_VN1203_24h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_VN1203_24h_3_Cells | Cells | ICL011-R | ICL011_VN1203_24h_3_array | RNA | ICL011-P | ICL011_VN1203_24h_3_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_24h_1_Cells | Cells | ICL011-R | ICL011_mock_24h_1_array | RNA | ICL011-P | ICL011_mock_24h_1_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_24h_2_Cells | Cells | ICL011-R | ICL011_mock_24h_2_array | RNA | ICL011-P | ICL011_mock_24h_2_proteomics | Protein |
| ICL011 | ICL011 Calu-3 Cells | Homo sapiens | ICL011_Mock_24h_3_Cells | Cells | ICL011-R | ICL011_mock_24h_3_array | RNA | ICL011-P | ICL011_mock_24h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_0h_1_Cells | Cells | ICL012-R | ICL012_NS1trunc_0h_1_array | RNA | ICL012-P | ICL012_NS1trunc_0h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_0h_2_Cells | Cells | ICL012-R | ICL012_NS1trunc_0h_2_array | RNA | ICL012-P | ICL012_NS1trunc_0h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_0h_3_Cells | Cells | ICL012-R | ICL012_NS1trunc_0h_3_array | RNA | ICL012-P | ICL012_NS1trunc_0h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_0h_1_Cells | Cells | ICL012-R | RNA | ICL012-P | ICL012_mock_0h_1_proteomics | Protein | |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_0h_2_Cells | Cells | ICL012-R | ICL012_mock_0h_2_array | RNA | ICL012-P | ICL012_mock_0h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_0h_3_Cells | Cells | ICL012-R | ICL012_mock_0h_3_array | RNA | ICL012-P | ICL012_mock_0h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_3h_1_Cells | Cells | ICL012-R | ICL012_NS1trunc_3h_1_array | RNA | ICL012-P | ICL012_NS1trunc_3h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_3h_2_Cells | Cells | ICL012-R | ICL012_NS1trunc_3h_2_array | RNA | ICL012-P | ICL012_NS1trunc_3h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_3h_3_Cells | Cells | ICL012-R | ICL012_NS1trunc_3h_3_array | RNA | ICL012-P | ICL012_NS1trunc_3h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_3h_1_Cells | Cells | ICL012-R | ICL012_mock_3h_1_array | RNA | ICL012-P | ICL012_mock_3h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_3h_2_Cells | Cells | ICL012-R | ICL012_mock_3h_2_array | RNA | ICL012-P | ICL012_mock_3h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_3h_3_Cells | Cells | ICL012-R | ICL012_mock_3h_3_array | RNA | ICL012-P | ICL012_mock_3h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_7h_1_Cells | Cells | ICL012-R | ICL012_NS1trunc_7h_1_array | RNA | ICL012-P | ICL012_NS1trunc_7h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_7h_2_Cells | Cells | ICL012-R | ICL012_NS1trunc_7h_2_array | RNA | ICL012-P | ICL012_NS1trunc_7h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_7h_3_Cells | Cells | ICL012-R | ICL012_NS1trunc_7h_3_array | RNA | ICL012-P | ICL012_NS1trunc_7h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_7h_1_Cells | Cells | ICL012-R | ICL012_mock_7h_1_array | RNA | ICL012-P | ICL012_mock_7h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_7h_2_Cells | Cells | ICL012-R | ICL012_mock_7h_2_array | RNA | ICL012-P | ICL012_mock_7h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_7h_3_Cells | Cells | ICL012-R | ICL012_mock_7h_3_array | RNA | ICL012-P | ICL012_mock_7h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_VN1203_7h_1_Cells | Cells | ICL012-R | ICL012_VN1203_7h_1_array | RNA | ICL012-P | ICL012_VN1203_7h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_VN1203_7h_2_Cells | Cells | ICL012-R | ICL012_VN1203_7h_2_array | RNA | ICL012-P | ICL012_VN1203_7h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_VN1203_7h_3_Cells | Cells | ICL012-R | ICL012_VN1203_7h_3_array | RNA | ICL012-P | ICL012_VN1203_7h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_12h_1_Cells | Cells | ICL012-R | ICL012_NS1trunc_12h_1_array | RNA | ICL012-P | ICL012_NS1trunc_12h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_12h_2_Cells | Cells | ICL012-R | ICL012_NS1trunc_12h_2_array | RNA | ICL012-P | ICL012_NS1trunc_12h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_12h_3_Cells | Cells | ICL012-R | ICL012_NS1trunc_12h_3_array | RNA | ICL012-P | ICL012_NS1trunc_12h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_12h_1_Cells | Cells | ICL012-R | ICL012_mock_12h_1_array | RNA | ICL012-P | ICL012_mock_12h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_12h_2_Cells | Cells | ICL012-R | ICL012_mock_12h_2_array | RNA | ICL012-P | ICL012_mock_12h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_12h_3_Cells | Cells | ICL012-R | ICL012_mock_12h_3_array | RNA | ICL012-P | ICL012_mock_12h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_18h_1_Cells | Cells | ICL012-R | ICL012_NS1trunc_18h_1_array | RNA | ICL012-P | ICL012_NS1trunc_18h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_18h_2_Cells | Cells | ICL012-R | ICL012_NS1trunc_18h_2_array | RNA | ICL012-P | ICL012_NS1trunc_18h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_18h_3_Cells | Cells | ICL012-R | ICL012_NS1trunc_18h_3_array | RNA | ICL012-P | ICL012_NS1trunc_18h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_18h_1_Cells | Cells | ICL012-R | ICL012_mock_18h_1_array | RNA | ICL012-P | ICL012_mock_18h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_18h_2_Cells | Cells | ICL012-R | ICL012_mock_18h_2_array | RNA | ICL012-P | ICL012_mock_18h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_18h_3_Cells | Cells | ICL012-R | ICL012_mock_18h_3_array | RNA | ICL012-P | ICL012_mock_18h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_24h_1_Cells | Cells | ICL012-R | ICL012_NS1trunc_24h_1_array | RNA | ICL012-P | ICL012_NS1trunc_24h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_24h_2_Cells | Cells | ICL012-R | ICL012_NS1trunc_24h_2_array | RNA | ICL012-P | ICL012_NS1trunc_24h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_NS1trunc_24h_3_Cells | Cells | ICL012-R | ICL012_NS1trunc_24h_3_array | RNA | ICL012-P | ICL012_NS1trunc_24h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_24h_1_Cells | Cells | ICL012-R | ICL012_mock_24h_1_array | RNA | ICL012-P | ICL012_mock_24h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_24h_2_Cells | Cells | ICL012-R | ICL012_mock_24h_2_array | RNA | ICL012-P | ICL012_mock_24h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_Mock_24h_3_Cells | Cells | ICL012-R | ICL012_mock_24h_3_array | RNA | ICL012-P | ICL012_mock_24h_3_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_VN1203_24h_1_Cells | Cells | ICL012-R | ICL012_VN1203_24h_1_array | RNA | ICL012-P | ICL012_VN1203_24h_1_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_VN1203_24h_2_Cells | Cells | ICL012-R | ICL012_VN1203_24h_2_array | RNA | ICL012-P | ICL012_VN1203_24h_2_proteomics | Protein |
| ICL012 | ICL012 Calu-3 Cells | Homo sapiens | ICL012_VN1203_24h_3_Cells | Cells | ICL012-R | ICL012_VN1203_24h_3_array | RNA | ICL012-P | ICL012_VN1203_24h_3_proteomics | Protein |
| IM001 | IM001_Mock_1d_1 | Mus musculus | IM001_Mock_D1_1_lung | Tissue | IM001-R | IM001_Mock_D1_1_RNA_ExpSam | RNA | IM001-P | IM001_Mock_1d_1_proteomics | Protein |
| IM001 | IM001_Mock_1d_2 | Mus musculus | IM001_Mock_D1_2_lung | Tissue | IM001-R | IM001_Mock_D1_2_RNA_ExpSam | RNA | IM001-P | IM001_Mock_1d_2_proteomics | Protein |
| IM001 | IM001_Mock_1d_3 | Mus musculus | IM001_Mock_D1_3_lung | Tissue | IM001-R | IM001_Mock_D1_3_RNA_ExpSam | RNA | IM001-P | IM001_Mock_1d_3_proteomics | Protein |
| IM001 | IM001_Mock_2d_1 | Mus musculus | IM001_Mock_D2_1_lung | Tissue | IM001-R | IM001_Mock_D2_1_RNA_ExpSam | RNA | IM001-P | IM001_Mock_2d_1_proteomics | Protein |
| IM001 | IM001_Mock_2d_2 | Mus musculus | IM001_Mock_D2_2_lung | Tissue | IM001-R | IM001_Mock_D2_2_RNA_ExpSam | RNA | IM001-P | IM001_Mock_2d_2_proteomics | Protein |
| IM001 | IM001_Mock_2d_3 | Mus musculus | IM001_Mock_D2_3_lung | Tissue | IM001-R | IM001_Mock_D2_3_RNA_ExpSam | RNA | IM001-P | IM001_Mock_2d_3_proteomics | Protein |
| IM001 | IM001_Mock_4d_1 | Mus musculus | IM001_Mock_D4_1_lung | Tissue | IM001-R | IM001_Mock_D4_1_RNA_ExpSam | RNA | IM001-P | IM001_Mock_4d_1_proteomics | Protein |
| IM001 | IM001_Mock_4d_2 | Mus musculus | IM001_Mock_D4_2_lung | Tissue | IM001-R | IM001_Mock_D4_2_RNA_ExpSam | RNA | IM001-P | IM001_Mock_4d_2_proteomics | Protein |
| IM001 | IM001_Mock_4d_3 | Mus musculus | IM001_Mock_D4_3_lung | Tissue | IM001-R | IM001_Mock_D4_3_RNA_ExpSam | RNA | IM001-P | IM001_Mock_4d_3_proteomics | Protein |
| IM001 | IM001_Mock_7d_1 | Mus musculus | IM001_Mock_D7_1_lung | Tissue | IM001-R | IM001_Mock_D7_1_RNA_ExpSam | RNA | IM001-P | IM001_Mock_7d_1_proteomics | Protein |
| IM001 | IM001_Mock_7d_2 | Mus musculus | IM001_Mock_D7_2_lung | Tissue | IM001-R | IM001_Mock_D7_2_RNA_ExpSam | RNA | IM001-P | IM001_Mock_7d_2_proteomics | Protein |
| IM001 | IM001_Mock_7d_3 | Mus musculus | IM001_Mock_D7_3_lung | Tissue | IM001-R | IM001_Mock_D7_3_RNA_ExpSam | RNA | IM001-P | IM001_Mock_7d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D1_1 | Mus musculus | IM001_VN1203_10^2pfu_D1_1_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D1_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_1d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D1_2 | Mus musculus | IM001_VN1203_10^2pfu_D1_2_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D1_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_1d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D1_3 | Mus musculus | IM001_VN1203_10^2pfu_D1_3_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D1_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_1d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D1_4 | Mus musculus | IM001_VN1203_10^2pfu_D1_4_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D1_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_1d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D1_5 | Mus musculus | IM001_VN1203_10^2pfu_D1_5_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D1_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_1d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D2_1 | Mus musculus | IM001_VN1203_10^2pfu_D2_1_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D2_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_2d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D2_2 | Mus musculus | IM001_VN1203_10^2pfu_D2_2_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D2_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_2d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D2_3 | Mus musculus | IM001_VN1203_10^2pfu_D2_3_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D2_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_2d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D2_4 | Mus musculus | IM001_VN1203_10^2pfu_D2_4_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D2_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_2d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D2_5 | Mus musculus | IM001_VN1203_10^2pfu_D2_5_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D2_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_2d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D4_1 | Mus musculus | IM001_VN1203_10^2pfu_D4_1_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D4_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_4d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D4_2 | Mus musculus | IM001_VN1203_10^2pfu_D4_2_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D4_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_4d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D4_3 | Mus musculus | IM001_VN1203_10^2pfu_D4_3_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D4_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_4d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D4_4 | Mus musculus | IM001_VN1203_10^2pfu_D4_4_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D4_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_4d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D4_5 | Mus musculus | IM001_VN1203_10^2pfu_D4_5_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D4_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_4d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D7_1 | Mus musculus | IM001_VN1203_10^2pfu_D7_1_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D7_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_7d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D7_2 | Mus musculus | IM001_VN1203_10^2pfu_D7_2_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D7_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_7d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D7_3 | Mus musculus | IM001_VN1203_10^2pfu_D7_3_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D7_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_7d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^2pfu_D7_4 | Mus musculus | IM001_VN1203_10^2pfu_D7_4_lung | Tissue | IM001-R | IM001_VN1203_10^2pfu_D7_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^2_7d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D1_1 | Mus musculus | IM001_VN1203_10^3pfu_D1_1_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D1_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_1d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D1_2 | Mus musculus | IM001_VN1203_10^3pfu_D1_2_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D1_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_1d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D1_3 | Mus musculus | IM001_VN1203_10^3pfu_D1_3_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D1_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_1d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D1_4 | Mus musculus | IM001_VN1203_10^3pfu_D1_4_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D1_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_1d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D1_5 | Mus musculus | IM001_VN1203_10^3pfu_D1_5_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D1_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_1d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D2_1 | Mus musculus | IM001_VN1203_10^3pfu_D2_1_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D2_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_2d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D2_2 | Mus musculus | IM001_VN1203_10^3pfu_D2_2_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D2_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_2d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D2_3 | Mus musculus | IM001_VN1203_10^3pfu_D2_3_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D2_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_2d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D2_4 | Mus musculus | IM001_VN1203_10^3pfu_D2_4_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D2_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_2d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D2_5 | Mus musculus | IM001_VN1203_10^3pfu_D2_5_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D2_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_2d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D4_1 | Mus musculus | IM001_VN1203_10^3pfu_D4_1_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D4_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_4d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D4_2 | Mus musculus | IM001_VN1203_10^3pfu_D4_2_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D4_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_4d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D4_3 | Mus musculus | IM001_VN1203_10^3pfu_D4_3_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D4_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_4d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D4_4 | Mus musculus | IM001_VN1203_10^3pfu_D4_4_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D4_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_4d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D4_5 | Mus musculus | IM001_VN1203_10^3pfu_D4_5_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D4_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_4d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D7_1 | Mus musculus | IM001_VN1203_10^3pfu_D7_1_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D7_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_7d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D7_2 | Mus musculus | IM001_VN1203_10^3pfu_D7_2_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D7_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_7d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^3pfu_D7_3 | Mus musculus | IM001_VN1203_10^3pfu_D7_3_lung | Tissue | IM001-R | IM001_VN1203_10^3pfu_D7_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^3_7d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D1_1 | Mus musculus | IM001_VN1203_10^4pfu_D1_1_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D1_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_1d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D1_2 | Mus musculus | IM001_VN1203_10^4pfu_D1_2_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D1_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_1d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D1_3 | Mus musculus | IM001_VN1203_10^4pfu_D1_3_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D1_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_1d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D1_4 | Mus musculus | IM001_VN1203_10^4pfu_D1_4_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D1_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_1d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D1_5 | Mus musculus | IM001_VN1203_10^4pfu_D1_5_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D1_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_1d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D2_1 | Mus musculus | IM001_VN1203_10^4pfu_D2_1_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D2_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_2d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D2_2 | Mus musculus | IM001_VN1203_10^4pfu_D2_2_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D2_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_2d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D2_3 | Mus musculus | IM001_VN1203_10^4pfu_D2_3_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D2_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_2d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D2_4 | Mus musculus | IM001_VN1203_10^4pfu_D2_4_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D2_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_2d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D2_5 | Mus musculus | IM001_VN1203_10^4pfu_D2_5_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D2_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_2d_5_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D4_1 | Mus musculus | IM001_VN1203_10^4pfu_D4_1_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D4_1_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_4d_1_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D4_2 | Mus musculus | IM001_VN1203_10^4pfu_D4_2_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D4_2_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_4d_2_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D4_3 | Mus musculus | IM001_VN1203_10^4pfu_D4_3_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D4_3_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_4d_3_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D4_4 | Mus musculus | IM001_VN1203_10^4pfu_D4_4_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D4_4_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_4d_4_proteomics | Protein |
| IM001 | IM001_VN1203_10^4pfu_D4_5 | Mus musculus | IM001_VN1203_10^4pfu_D4_5_lung | Tissue | IM001-R | IM001_VN1203_10^4pfu_D4_5_RNA_ExpSam | RNA | IM001-P | IM001_VN1203_10^4_4d_5_proteomics | Protein |
| IM002 | IM002_Brisbane_10^6pfu_D1_2 | Mus musculus | IM002_Brisbane_10^6pfu_D1_2_Lung | Tissue | IM002-R | IM002_Brisbane_10^6pfu_D1_2_array | RNA | |||
| IM002 | IM002_Brisbane_10^6pfu_D1_3 | Mus musculus | IM002_Brisbane_10^6pfu_D1_3_Lung | Tissue | IM002-R | IM002_Brisbane_10^6pfu_D1_3_array | RNA | |||
| IM002 | IM002_Brisbane_10^6pfu_D3_2 | Mus musculus | IM002_Brisbane_10^6pfu_D3_2_Lung | Tissue | IM002-R | IM002_Brisbane_10^6pfu_D3_2_array | RNA | |||
| IM002 | IM002_Brisbane_10^6pfu_D3_3 | Mus musculus | IM002_Brisbane_10^6pfu_D3_3_Lung | Tissue | IM002-R | IM002_Brisbane_10^6pfu_D3_3_array | RNA | |||
| IM002 | IM002_Brisbane_10^6pfu_D5_1 | Mus musculus | IM002_Brisbane_10^6pfu_D5_1_Lung | Tissue | IM002-R | IM002_Brisbane_10^6pfu_D5_1_array | RNA | |||
| IM002 | IM002_Brisbane_10^6pfu_D5_2 | Mus musculus | IM002_Brisbane_10^6pfu_D5_2_Lung | Tissue | IM002-R | IM002_Brisbane_10^6pfu_D5_2_array | RNA | |||
| IM002 | IM002_Brisbane_10^6pfu_D5_3 | Mus musculus | IM002_Brisbane_10^6pfu_D5_3_Lung | Tissue | IM002-R | IM002_Brisbane_10^6pfu_D5_3_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D1_1 | Mus musculus | IM002_Mex_10^6pfu_D1_1_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D1_1_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D1_2 | Mus musculus | IM002_Mex_10^6pfu_D1_2_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D1_2_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D1_3 | Mus musculus | IM002_Mex_10^6pfu_D1_3_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D1_3_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D3_1 | Mus musculus | IM002_Mex_10^6pfu_D3_1_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D3_1_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D3_2 | Mus musculus | IM002_Mex_10^6pfu_D3_2_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D3_2_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D3_3 | Mus musculus | IM002_Mex_10^6pfu_D3_3_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D3_3_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D5_1 | Mus musculus | IM002_Mex_10^6pfu_D5_1_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D5_1_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D5_2 | Mus musculus | IM002_Mex_10^6pfu_D5_2_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D5_2_array | RNA | |||
| IM002 | IM002_Mex_10^6pfu_D5_3 | Mus musculus | IM002_Mex_10^6pfu_D5_3_Lung | Tissue | IM002-R | IM002_Mex_10^6pfu_D5_3_array | RNA | |||
| IM002 | IM002_Mock_D1_1 | Mus musculus | IM002_Mock_D1_1_Lung | Tissue | IM002-R | IM002_Mock_D1_1_array | RNA | |||
| IM002 | IM002_Mock_D1_2 | Mus musculus | IM002_Mock_D1_2_Lung | Tissue | IM002-R | IM002_Mock_D1_2_array | RNA | |||
| IM002 | IM002_Mock_D3_1 | Mus musculus | IM002_Mock_D3_1_Lung | Tissue | IM002-R | IM002_Mock_D3_1_array | RNA | |||
| IM002 | IM002_Mock_D3_2 | Mus musculus | IM002_Mock_D3_2_Lung | Tissue | IM002-R | IM002_Mock_D3_2_array | RNA | |||
| IM002 | IM002_Mock_D5_2 | Mus musculus | IM002_Mock_D5_2_Lung | Tissue | IM002-R | IM002_Mock_D5_2_array | RNA | |||
| IM002 | IM002_NJ_10^6pfu_D1_1 | Mus musculus | IM002_NJ_10^6pfu_D1_1_Lung | Tissue | IM002-R | IM002_NJ_10^6pfu_D1_1_array | RNA | |||
| IM002 | IM002_NJ_10^6pfu_D1_3 | Mus musculus | IM002_NJ_10^6pfu_D1_3_Lung | Tissue | IM002-R | IM002_NJ_10^6pfu_D1_3_array | RNA | |||
| IM002 | IM002_NJ_10^6pfu_D3_1 | Mus musculus | IM002_NJ_10^6pfu_D3_1_Lung | Tissue | IM002-R | IM002_NJ_10^6pfu_D3_1_array | RNA | |||
| IM002 | IM002_NJ_10^6pfu_D3_2 | Mus musculus | IM002_NJ_10^6pfu_D3_2_Lung | Tissue | IM002-R | IM002_NJ_10^6pfu_D3_2_array | RNA | |||
| IM002 | IM002_NJ_10^6pfu_D3_3 | Mus musculus | IM002_NJ_10^6pfu_D3_3_Lung | Tissue | IM002-R | IM002_NJ_10^6pfu_D3_3_array | RNA | |||
| IM002 | IM002_NJ_10^6pfu_D5_2 | Mus musculus | IM002_NJ_10^6pfu_D5_2_Lung | Tissue | IM002-R | IM002_NJ_10^6pfu_D5_2_array | RNA | |||
| IM002 | IM002_NJ_10^6pfu_D5_3 | Mus musculus | IM002_NJ_10^6pfu_D5_3_Lung | Tissue | IM002-R | IM002_NJ_10^6pfu_D5_3_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D1_1 | Mus musculus | IM002_r1918_10^6pfu_D1_1_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D1_1_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D1_2 | Mus musculus | IM002_r1918_10^6pfu_D1_2_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D1_2_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D1_3 | Mus musculus | IM002_r1918_10^6pfu_D1_3_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D1_3_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D3_1 | Mus musculus | IM002_r1918_10^6pfu_D3_1_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D3_1_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D3_2 | Mus musculus | IM002_r1918_10^6pfu_D3_2_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D3_2_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D3_3 | Mus musculus | IM002_r1918_10^6pfu_D3_3_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D3_3_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D5_1 | Mus musculus | IM002_r1918_10^6pfu_D5_1_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D5_1_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D5_2 | Mus musculus | IM002_r1918_10^6pfu_D5_2_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D5_2_array | RNA | |||
| IM002 | IM002_r1918_10^6pfu_D5_3 | Mus musculus | IM002_r1918_10^6pfu_D5_3_Lung | Tissue | IM002-R | IM002_r1918_10^6pfu_D5_3_array | RNA | |||
| IM004 | IM004_HAavirulent_10^4pfu_1d_1 | Mus musculus | IM004_HAavirulent_10^4pfu_1d_1_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_1d_1_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_1d_1_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_1d_2 | Mus musculus | IM004_HAavirulent_10^4pfu_1d_2_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_1d_2_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_1d_2_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_1d_3 | Mus musculus | IM004_HAavirulent_10^4pfu_1d_3_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_1d_3_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_1d_3_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_1d_4 | Mus musculus | IM004_HAavirulent_10^4pfu_1d_4_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_1d_4_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_1d_4_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_1d_5 | Mus musculus | IM004_HAavirulent_10^4pfu_1d_5_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_1d_5_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_1d_5_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_2d_1 | Mus musculus | IM004_HAavirulent_10^4pfu_2d_1_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_2d_1_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_2d_1_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_2d_2 | Mus musculus | IM004_HAavirulent_10^4pfu_2d_2_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_2d_2_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_2d_2_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_2d_3 | Mus musculus | IM004_HAavirulent_10^4pfu_2d_3_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_2d_3_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_2d_3_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_2d_4 | Mus musculus | IM004_HAavirulent_10^4pfu_2d_4_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_2d_4_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_2d_4_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_2d_5 | Mus musculus | IM004_HAavirulent_10^4pfu_2d_5_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_2d_5_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_2d_5_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_4d_1 | Mus musculus | IM004_HAavirulent_10^4pfu_4d_1_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_4d_1_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_4d_1_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_4d_2 | Mus musculus | IM004_HAavirulent_10^4pfu_4d_2_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_4d_2_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_4d_2_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_4d_3 | Mus musculus | IM004_HAavirulent_10^4pfu_4d_3_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_4d_3_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_4d_3_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_4d_4 | Mus musculus | IM004_HAavirulent_10^4pfu_4d_4_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_4d_4_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_4d_4_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_4d_5 | Mus musculus | IM004_HAavirulent_10^4pfu_4d_5_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_4d_5_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_4d_5_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_7d_1 | Mus musculus | IM004_HAavirulent_10^4pfu_7d_1_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_7d_1_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_7d_1_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_7d_2 | Mus musculus | IM004_HAavirulent_10^4pfu_7d_2_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_7d_2_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_7d_2_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_7d_3 | Mus musculus | IM004_HAavirulent_10^4pfu_7d_3_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_7d_3_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_7d_3_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_7d_4 | Mus musculus | IM004_HAavirulent_10^4pfu_7d_4_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_7d_4_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_7d_4_proteomics | Protein |
| IM004 | IM004_HAavirulent_10^4pfu_7d_5 | Mus musculus | IM004_HAavirulent_10^4pfu_7d_5_lung | Tissue | IM004-R | IM004_HAavirulent_10^4pfu_7d_5_RNA_ExpSam | RNA | IM004-P | IM004_HAavir_10^4_7d_5_proteomics | Protein |
| IM004 | IM004_mock_1d_1 | Mus musculus | IM004_mock_1d_1_lung | Tissue | IM004-R | IM004_mock_1d_1_RNA_ExpSam | RNA | IM004-P | IM004_Mock_1d_1_proteomics | Protein |
| IM004 | IM004_mock_1d_2 | Mus musculus | IM004_mock_1d_2_lung | Tissue | IM004-R | IM004_mock_1d_2_RNA_ExpSam | RNA | IM004-P | IM004_Mock_1d_2_proteomics | Protein |
| IM004 | IM004_mock_1d_3 | Mus musculus | IM004_mock_1d_3_lung | Tissue | IM004-R | IM004_mock_1d_3_RNA_ExpSam | RNA | IM004-P | IM004_Mock_1d_3_proteomics | Protein |
| IM004 | IM004_mock_2d_1 | Mus musculus | IM004_mock_2d_1_lung | Tissue | IM004-R | IM004_mock_2d_1_RNA_ExpSam | RNA | IM004-P | IM004_Mock_2d_1_proteomics | Protein |
| IM004 | IM004_mock_2d_2 | Mus musculus | IM004_mock_2d_2_lung | Tissue | IM004-R | IM004_mock_2d_2_RNA_ExpSam | RNA | IM004-P | IM004_Mock_2d_2_proteomics | Protein |
| IM004 | IM004_mock_2d_3 | Mus musculus | IM004_mock_2d_3_lung | Tissue | IM004-R | IM004_mock_2d_3_RNA_ExpSam | RNA | IM004-P | IM004_Mock_2d_3_proteomics | Protein |
| IM004 | IM004_mock_4d_1 | Mus musculus | IM004_mock_4d_1_lung | Tissue | IM004-R | IM004_mock_4d_1_RNA_ExpSam | RNA | IM004-P | IM004_Mock_4d_1_proteomics | Protein |
| IM004 | IM004_mock_4d_2 | Mus musculus | IM004_mock_4d_2_lung | Tissue | IM004-R | IM004_mock_4d_2_RNA_ExpSam | RNA | IM004-P | IM004_Mock_4d_2_proteomics | Protein |
| IM004 | IM004_mock_4d_3 | Mus musculus | IM004_mock_4d_3_lung | Tissue | IM004-R | IM004_mock_4d_3_RNA_ExpSam | RNA | IM004-P | IM004_Mock_4d_3_proteomics | Protein |
| IM004 | IM004_mock_7d_1 | Mus musculus | IM004_mock_7d_1_lung | Tissue | IM004-R | IM004_mock_7d_1_RNA_ExpSam | RNA | IM004-P | IM004_Mock_7d_1_proteomics | Protein |
| IM004 | IM004_mock_7d_2 | Mus musculus | IM004_mock_7d_2_lung | Tissue | IM004-R | IM004_mock_7d_2_RNA_ExpSam | RNA | IM004-P | IM004_Mock_7d_2_proteomics | Protein |
| IM004 | IM004_mock_7d_3 | Mus musculus | IM004_mock_7d_3_lung | Tissue | IM004-R | IM004_mock_7d_3_RNA_ExpSam | RNA | IM004-P | IM004_Mock_7d_3_proteomics | Protein |
| IM005 | IM005_mock_1d_1 | Mus musculus | IM005_mock_1d_1_Lung | Tissue | IM005-R | IM005_Mock_1d_1_array | RNA | IM005-P | IM005_Mock_1d_1_proteomics | Protein |
| IM005 | IM005_mock_1d_2 | Mus musculus | IM005_mock_1d_2_Lung | Tissue | IM005-R | IM005_Mock_1d_2_array | RNA | IM005-P | IM005_Mock_1d_2_proteomics | Protein |
| IM005 | IM005_mock_1d_3 | Mus musculus | IM005_mock_1d_3_Lung | Tissue | IM005-R | IM005_Mock_1d_3_array | RNA | IM005-P | IM005_Mock_1d_3_proteomics | Protein |
| IM005 | IM005_mock_2d_1 | Mus musculus | IM005_mock_2d_1_Lung | Tissue | IM005-R | IM005_Mock_2d_1_array | RNA | IM005-P | IM005_Mock_2d_1_proteomics | Protein |
| IM005 | IM005_mock_2d_2 | Mus musculus | IM005_mock_2d_2_Lung | Tissue | IM005-R | IM005_Mock_2d_2_array | RNA | IM005-P | IM005_Mock_2d_2_proteomics | Protein |
| IM005 | IM005_mock_2d_3 | Mus musculus | IM005_mock_2d_3_Lung | Tissue | IM005-R | IM005_Mock_2d_3_array | RNA | IM005-P | IM005_Mock_2d_3_proteomics | Protein |
| IM005 | IM005_mock_4d_1 | Mus musculus | IM005_mock_4d_1_Lung | Tissue | IM005-R | IM005_Mock_4d_1_array | RNA | IM005-P | IM005_Mock_4d_1_proteomics | Protein |
| IM005 | IM005_mock_4d_2 | Mus musculus | IM005_mock_4d_2_Lung | Tissue | IM005-R | IM005_Mock_4d_2_array | RNA | IM005-P | IM005_Mock_4d_2_proteomics | Protein |
| IM005 | IM005_mock_4d_3 | Mus musculus | IM005_mock_4d_3_Lung | Tissue | IM005-R | IM005_Mock_4d_3_array | RNA | IM005-P | IM005_Mock_4d_3_proteomics | Protein |
| IM005 | IM005_mock_7d_1 | Mus musculus | IM005_mock_7d_1_Lung | Tissue | IM005-R | IM005_Mock_7d_1_array | RNA | IM005-P | IM005_Mock_7d_1_proteomics | Protein |
| IM005 | IM005_mock_7d_2 | Mus musculus | IM005_mock_7d_2_Lung | Tissue | IM005-R | IM005_Mock_7d_2_array | RNA | IM005-P | IM005_Mock_7d_2_proteomics | Protein |
| IM005 | IM005_mock_7d_3 | Mus musculus | IM005_mock_7d_3_Lung | Tissue | IM005-R | IM005_Mock_7d_3_array | RNA | IM005-P | IM005_Mock_7d_3_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_1d_1 | Mus musculus | IM005_PB2_627E_10^4pfu_1d_1_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_1d_1_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_1d_1_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_1d_2 | Mus musculus | IM005_PB2_627E_10^4pfu_1d_2_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_1d_2_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_1d_2_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_1d_3 | Mus musculus | IM005_PB2_627E_10^4pfu_1d_3_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_1d_3_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_1d_3_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_1d_4 | Mus musculus | IM005_PB2_627E_10^4pfu_1d_4_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_1d_4_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_1d_4_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_1d_5 | Mus musculus | IM005_PB2_627E_10^4pfu_1d_5_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_1d_5_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_1d_5_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_2d_1 | Mus musculus | IM005_PB2_627E_10^4pfu_2d_1_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_2d_1_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_2d_1_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_2d_2 | Mus musculus | IM005_PB2_627E_10^4pfu_2d_2_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_2d_2_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_2d_2_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_2d_3 | Mus musculus | IM005_PB2_627E_10^4pfu_2d_3_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_2d_3_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_2d_3_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_2d_4 | Mus musculus | IM005_PB2_627E_10^4pfu_2d_4_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_2d_4_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_2d_4_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_2d_5 | Mus musculus | IM005_PB2_627E_10^4pfu_2d_5_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_2d_5_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_2d_5_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_4d_1 | Mus musculus | IM005_PB2_627E_10^4pfu_4d_1_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_4d_1_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_4d_1_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_4d_2 | Mus musculus | IM005_PB2_627E_10^4pfu_4d_2_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_4d_2_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_4d_2_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_4d_3 | Mus musculus | IM005_PB2_627E_10^4pfu_4d_3_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_4d_3_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_4d_3_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_4d_4 | Mus musculus | IM005_PB2_627E_10^4pfu_4d_4_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_4d_4_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_4d_4_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_4d_5 | Mus musculus | IM005_PB2_627E_10^4pfu_4d_5_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_4d_5_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_4d_5_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_7d_1 | Mus musculus | IM005_PB2_627E_10^4pfu_7d_1_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_7d_1_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_7d_1_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_7d_2 | Mus musculus | IM005_PB2_627E_10^4pfu_7d_2_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_7d_2_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_7d_2_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_7d_3 | Mus musculus | IM005_PB2_627E_10^4pfu_7d_3_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_7d_3_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_7d_3_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_7d_4 | Mus musculus | IM005_PB2_627E_10^4pfu_7d_4_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_7d_4_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_7d_4_proteomics | Protein |
| IM005 | IM005_PB2_627E_10^4pfu_7d_5 | Mus musculus | IM005_PB2_627E_10^4pfu_7d_5_Lung | Tissue | IM005-R | IM005_PB2_627E_10^4pfu_7d_5_array | RNA | IM005-P | IM005_PB2_627E_10^4pfu_7d_5_proteomics | Protein |
| IM006A | IM006A_mock_1d_1 | Mus musculus | IM006A_mock_1d_1_Lung | Tissue | IM006A-R | IM006A_Mock_1d_1_array | RNA | IM006A-P | IM006A_Mock_1d_1_proteomics | Protein |
| IM006A | IM006A_mock_1d_2 | Mus musculus | IM006A_mock_1d_2_Lung | Tissue | IM006A-R | IM006A_Mock_1d_2_array | RNA | IM006A-P | IM006A_Mock_1d_2_proteomics | Protein |
| IM006A | IM006A_mock_1d_3 | Mus musculus | IM006A_mock_1d_3_Lung | Tissue | IM006A-R | IM006A_Mock_1d_3_array | RNA | IM006A-P | IM006A_Mock_1d_3_proteomics | Protein |
| IM006A | IM006A_mock_2d_1 | Mus musculus | IM006A_mock_2d_1_Lung | Tissue | IM006A-R | IM006A_Mock_2d_1_array | RNA | IM006A-P | IM006A_Mock_2d_1_proteomics | Protein |
| IM006A | IM006A_mock_2d_2 | Mus musculus | IM006A_mock_2d_2_Lung | Tissue | IM006A-R | IM006A_Mock_2d_2_array | RNA | IM006A-P | IM006A_Mock_2d_2_proteomics | Protein |
| IM006A | IM006A_mock_2d_3 | Mus musculus | IM006A_mock_2d_3_Lung | Tissue | IM006A-R | IM006A_Mock_2d_3_array | RNA | IM006A-P | IM006A_Mock_2d_3_proteomics | Protein |
| IM006A | IM006A_mock_4d_1 | Mus musculus | IM006A_mock_4d_1_Lung | Tissue | IM006A-R | IM006A_Mock_4d_1_array | RNA | IM006A-P | IM006A_Mock_4d_1_proteomics | Protein |
| IM006A | IM006A_mock_4d_2 | Mus musculus | IM006A_mock_4d_2_Lung | Tissue | IM006A-R | IM006A_Mock_4d_2_array | RNA | IM006A-P | IM006A_Mock_4d_2_proteomics | Protein |
| IM006A | IM006A_mock_4d_3 | Mus musculus | IM006A_mock_4d_3_Lung | Tissue | IM006A-R | IM006A_Mock_4d_3_array | RNA | IM006A-P | IM006A_Mock_4d_3_proteomics | Protein |
| IM006A | IM006A_mock_7d_1 | Mus musculus | IM006A_mock_7d_1_Lung | Tissue | IM006A-R | IM006A_Mock_7d_1_array | RNA | IM006A-P | IM006A_Mock_7d_1_proteomics | Protein |
| IM006A | IM006A_mock_7d_2 | Mus musculus | IM006A_mock_7d_2_Lung | Tissue | IM006A-R | IM006A_Mock_7d_2_array | RNA | IM006A-P | IM006A_Mock_7d_2_proteomics | Protein |
| IM006A | IM006A_mock_7d_3 | Mus musculus | IM006A_mock_7d_3_Lung | Tissue | IM006A-R | IM006A_Mock_7d_3_array | RNA | IM006A-P | IM006A_Mock_7d_3_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_1d_1 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_1d_1_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_1_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_1_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_1d_2 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_1d_2_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_2_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_2_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_1d_3 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_1d_3_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_3_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_3_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_1d_4 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_1d_4_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_4_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_4_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_1d_5 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_1d_5_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_1d_5_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_1d_5_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_2d_1 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_2d_1_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_1_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_1_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_2d_2 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_2d_2_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_2_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_2_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_2d_3 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_2d_3_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_3_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_3_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_2d_4 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_2d_4_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_4_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_4_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_2d_5 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_2d_5_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_2d_5_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_2d_5_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_4d_1 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_4d_1_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_1_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_1_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_4d_2 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_4d_2_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_2_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_2_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_4d_3 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_4d_3_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_3_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_3_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_4d_4 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_4d_4_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_4_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_4_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_4d_5 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_4d_5_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_4d_5_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_4d_5_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_7d_1 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_7d_1_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_7d_1_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_1_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_7d_2 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_7d_2_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_7d_2_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_2_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_7d_3 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_7d_3_Lung | Tissue | IM006A-R | IM006A_PB1_F2del_10^3pfu_7d_3_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_3_proteomics | Protein |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_7d_4 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_7d_4_Lung | Tissue | IM006A-R | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_4_proteomics | Protein | |
| IM006A | IM006A_PB1-F2 Del_10^3pfu_7d_5 | Mus musculus | IM006A_PB1-F2 Del_10^3pfu_7d_5_Lung | Tissue | IM006B-R | IM006A_PB1_F2del_10^3pfu_7d_5_array | RNA | IM006A-P | IM006A_PB1_F2del_10^3pfu_7d_5_proteomics | Protein |
| IM006B | IM006B_mock_1d_1 | Mus musculus | IM006B_mock_1d_1_Lung | Tissue | IM006B-R | IM006B_Mock_1d_1_array | RNA | IM006B-P | IM006B_Mock_1d_1_proteomics | Protein |
| IM006B | IM006B_mock_1d_2 | Mus musculus | IM006B_mock_1d_2_Lung | Tissue | IM006B-R | IM006B_Mock_1d_2_array | RNA | IM006B-P | IM006B_Mock_1d_2_proteomics | Protein |
| IM006B | IM006B_mock_1d_3 | Mus musculus | IM006B_mock_1d_3_Lung | Tissue | IM006B-R | IM006B_Mock_1d_3_array | RNA | IM006B-P | IM006B_Mock_1d_3_proteomics | Protein |
| IM006B | IM006B_mock_2d_1 | Mus musculus | IM006B_mock_2d_1_Lung | Tissue | IM006B-R | IM006B_Mock_2d_1_array | RNA | IM006B-P | IM006B_Mock_2d_1_proteomics | Protein |
| IM006B | IM006B_mock_2d_2 | Mus musculus | IM006B_mock_2d_2_Lung | Tissue | IM006B-R | IM006B_Mock_2d_2_array | RNA | IM006B-P | IM006B_Mock_2d_2_proteomics | Protein |
| IM006B | IM006B_mock_2d_3 | Mus musculus | IM006B_mock_2d_3_Lung | Tissue | IM006B-R | IM006B_Mock_2d_3_array | RNA | IM006B-P | IM006B_Mock_2d_3_proteomics | Protein |
| IM006B | IM006B_mock_4d_1 | Mus musculus | IM006B_mock_4d_1_Lung | Tissue | IM006B-R | IM006B_Mock_4d_1_array | RNA | IM006B-P | IM006B_Mock_4d_1_proteomics | Protein |
| IM006B | IM006B_mock_4d_2 | Mus musculus | IM006B_mock_4d_2_Lung | Tissue | IM006B-R | IM006B_Mock_4d_2_array | RNA | IM006B-P | IM006B_Mock_4d_2_proteomics | Protein |
| IM006B | IM006B_mock_4d_3 | Mus musculus | IM006B_mock_4d_3_Lung | Tissue | IM006B-R | IM006B_Mock_4d_3_array | RNA | IM006B-P | IM006B_Mock_4d_3_proteomics | Protein |
| IM006B | IM006B_mock_7d_1 | Mus musculus | IM006B_mock_7d_1_Lung | Tissue | IM006B-R | IM006B_Mock_7d_1_array | RNA | |||
| IM006B | IM006B_mock_7d_2 | Mus musculus | IM006B_mock_7d_2_Lung | Tissue | IM006B-R | IM006B_Mock_7d_2_array | RNA | |||
| IM006B | IM006B_mock_7d_3 | Mus musculus | IM006B_mock_7d_3_Lung | Tissue | IM006B-R | RNA | ||||
| IM006B | IM006B_PB1-F2 Del_10^4pfu_1d_1 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_1d_1_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_1_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_1_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_1d_2 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_1d_2_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_2_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_2_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_1d_3 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_1d_3_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_3_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_3_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_1d_4 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_1d_4_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_4_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_4_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_1d_5 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_1d_5_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_1d_5_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_1d_5_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_2d_1 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_2d_1_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_1_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_1_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_2d_2 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_2d_2_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_2_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_2_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_2d_3 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_2d_3_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_3_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_3_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_2d_4 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_2d_4_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_4_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_4_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_2d_5 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_2d_5_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_2d_5_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_2d_5_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_4d_1 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_4d_1_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_1_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_1_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_4d_2 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_4d_2_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_2_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_2_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_4d_3 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_4d_3_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_3_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_3_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_4d_4 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_4d_4_Lung | Tissue | IM006B-R | IM006B_PB1_F2del_10^4pfu_4d_4_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_4_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_4d_5 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_4d_5_Lung | Tissue | IM007-R | IM006B_PB1_F2del_10^4pfu_4d_5_array | RNA | IM006B-P | IM006B_PB1_F2del_10^4pfu_4d_5_proteomics | Protein |
| IM006B | IM006B_PB1-F2 Del_10^4pfu_7d_1 | Mus musculus | IM006B_PB1-F2 Del_10^4pfu_7d_1_Lung | Tissue | IM007-R | IM006B_PB1_F2del_10^4pfu_7d_1_array | RNA | |||
| IM007 | IM007_mock_1d_1 | Mus musculus | IM007_mock_1d_1_Lung | Tissue | IM007-R | IM007_Mock_1d_1_array | RNA | IM007-P | IM007_Mock_1d_1_proteomics | Protein |
| IM007 | IM007_mock_1d_2 | Mus musculus | IM007_mock_1d_2_Lung | Tissue | IM007-R | IM007_Mock_1d_2_array | RNA | IM007-P | IM007_Mock_1d_2_proteomics | Protein |
| IM007 | IM007_mock_1d_3 | Mus musculus | IM007_mock_1d_3_Lung | Tissue | IM007-R | IM007_Mock_1d_3_array | RNA | IM007-P | IM007_Mock_1d_3_proteomics | Protein |
| IM007 | IM007_mock_2d_1 | Mus musculus | IM007_mock_2d_1_Lung | Tissue | IM007-R | IM007_Mock_2d_1_array | RNA | IM007-P | IM007_Mock_2d_1_proteomics | Protein |
| IM007 | IM007_mock_2d_2 | Mus musculus | IM007_mock_2d_2_Lung | Tissue | IM007-R | IM007_Mock_2d_2_array | RNA | IM007-P | IM007_Mock_2d_2_proteomics | Protein |
| IM007 | IM007_mock_2d_3 | Mus musculus | IM007_mock_2d_3_Lung | Tissue | IM007-R | IM007_Mock_2d_3_array | RNA | IM007-P | IM007_Mock_2d_3_proteomics | Protein |
| IM007 | IM007_mock_4d_1 | Mus musculus | IM007_mock_4d_1_Lung | Tissue | IM007-R | IM007_Mock_4d_1_array | RNA | IM007-P | IM007_Mock_4d_1_proteomics | Protein |
| IM007 | IM007_mock_4d_2 | Mus musculus | IM007_mock_4d_2_Lung | Tissue | IM007-R | IM007_Mock_4d_2_array | RNA | IM007-P | IM007_Mock_4d_2_proteomics | Protein |
| IM007 | IM007_mock_4d_3 | Mus musculus | IM007_mock_4d_3_Lung | Tissue | IM007-R | IM007_Mock_4d_3_array | RNA | IM007-P | IM007_Mock_4d_3_proteomics | Protein |
| IM007 | IM007_mock_7d_1 | Mus musculus | IM007_mock_7d_1_Lung | Tissue | IM007-R | IM007_Mock_7d_1_array | RNA | IM007-P | IM007_Mock_7d_1_proteomics | Protein |
| IM007 | IM007_mock_7d_2 | Mus musculus | IM007_mock_7d_2_Lung | Tissue | IM007-R | IM007_Mock_7d_2_array | RNA | IM007-P | IM007_Mock_7d_2_proteomics | Protein |
| IM007 | IM007_mock_7d_3 | Mus musculus | IM007_mock_7d_3_Lung | Tissue | IM007-R | IM007_Mock_7d_3_array | RNA | IM007-P | IM007_Mock_7d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_1d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_1d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_1d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_1d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_1d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_1d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_1d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_1d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_1d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_1d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_1d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_1d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_1d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_1d_4_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_1d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_1d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_1d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_1d_5_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_1d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_1d_5_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_2d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_2d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_2d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_2d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_2d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_2d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_2d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_2d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_2d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_2d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_2d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_2d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_2d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_2d_4_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_2d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_2d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_2d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_2d_5_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_2d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_2d_5_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_4d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_4d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_4d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_4d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_4d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_4d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_4d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_4d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_4d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_4d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_4d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_4d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_4d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_4d_4_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_4d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_4d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_4d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_4d_5_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_4d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_4d_5_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_7d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_7d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_7d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_7d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_7d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_7d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_7d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_7d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_7d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_7d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_7d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_7d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_7d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_7d_4_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_7d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_7d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^3_7d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^3_7d_5_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^3pfu_7d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^3pfu_7d_5_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_1d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_1d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_1d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_1d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_1d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_1d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_1d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_1d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_1d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_1d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_1d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_1d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_1d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_1d_4_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_1d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_1d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_1d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_1d_5_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_1d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_1d_5_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_2d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_2d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_2d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_2d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_2d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_2d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_2d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_2d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_2d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_2d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_2d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_2d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_2d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_2d_4_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_2d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_2d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_2d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_2d_5_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_2d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_2d_5_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_4d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_4d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_4d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_4d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_4d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_4d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_4d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_4d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_4d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_4d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_4d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_4d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_4d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_4d_4_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_4d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_4d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_4d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_4d_5_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_4d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_4d_5_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_7d_1 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_7d_1_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_7d_1_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_7d_1_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_7d_2 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_7d_2_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_7d_2_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_7d_2_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_7d_3 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_7d_3_Lung | Tissue | IM007-R | IM007_NS1trunc124_10^4pfu_7d_3_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_7d_3_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_7d_4 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_7d_4_Lung | Tissue | IM009-R | IM007_NS1trunc124_10^4pfu_7d_4_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_7d_4_proteomics | Protein |
| IM007 | IM007_VN1203-NS1trunc-124_10^4_7d_5 | Mus musculus | IM007_VN1203-NS1trunc-124_10^4_7d_5_Lung | Tissue | IM009-R | IM007_NS1trunc124_10^4pfu_7d_5_array | RNA | IM007-P | IM007_NS1trunc124_10^4pfu_7d_5_proteomics | Protein |
| IM009 | IM009_CA04_10^6pfu_D1_1 | Mus musculus | IM009_CA04_10^6pfu_D1_1_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D1_1_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D1_2 | Mus musculus | IM009_CA04_10^6pfu_D1_2_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D1_2_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D1_3 | Mus musculus | IM009_CA04_10^6pfu_D1_3_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D1_3_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D3_1 | Mus musculus | IM009_CA04_10^6pfu_D3_1_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D3_1_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D3_2 | Mus musculus | IM009_CA04_10^6pfu_D3_2_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D3_2_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D3_3 | Mus musculus | IM009_CA04_10^6pfu_D3_3_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D3_3_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D5_1 | Mus musculus | IM009_CA04_10^6pfu_D5_1_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D5_1_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D5_2 | Mus musculus | IM009_CA04_10^6pfu_D5_2_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D5_2_array | RNA | |||
| IM009 | IM009_CA04_10^6pfu_D5_3 | Mus musculus | IM009_CA04_10^6pfu_D5_3_Lung | Tissue | IM009-R | IM009_CA04_10^6pfu_D5_3_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D1_1 | Mus musculus | IM009_MA-CA04_10^6pfu_D1_1_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D1_1_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D1_2 | Mus musculus | IM009_MA-CA04_10^6pfu_D1_2_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D1_2_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D1_3 | Mus musculus | IM009_MA-CA04_10^6pfu_D1_3_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D1_3_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D3_1 | Mus musculus | IM009_MA-CA04_10^6pfu_D3_1_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D3_1_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D3_2 | Mus musculus | IM009_MA-CA04_10^6pfu_D3_2_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D3_2_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D3_3 | Mus musculus | IM009_MA-CA04_10^6pfu_D3_3_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D3_3_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D5_1 | Mus musculus | IM009_MA-CA04_10^6pfu_D5_1_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D5_1_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D5_2 | Mus musculus | IM009_MA-CA04_10^6pfu_D5_2_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D5_2_array | RNA | |||
| IM009 | IM009_MA-CA04_10^6pfu_D5_3 | Mus musculus | IM009_MA-CA04_10^6pfu_D5_3_Lung | Tissue | IM009-R | IM009_MA-CA04_10^6pfu_D5_3_array | RNA | |||
| IM009 | IM009_Mock_D1_1 | Mus musculus | IM009_Mock_D1_1_Lung | Tissue | IM009-R | IM009_Mock_D1_1_array | RNA | |||
| IM009 | IM009_Mock_D1_2 | Mus musculus | IM009_Mock_D1_2_Lung | Tissue | IM009-R | IM009_Mock_D1_2_array | RNA | |||
| IM009 | IM009_Mock_D1_3 | Mus musculus | IM009_Mock_D1_3_Lung | Tissue | IM009-R | IM009_Mock_D1_3_array | RNA | |||
| IM009 | IM009_Mock_D3_1 | Mus musculus | IM009_Mock_D3_1_Lung | Tissue | IM009-R | IM009_Mock_D3_1_array | RNA | |||
| IM009 | IM009_Mock_D3_2 | Mus musculus | IM009_Mock_D3_2_Lung | Tissue | IM009-R | IM009_Mock_D3_2_array | RNA | |||
| IM009 | IM009_Mock_D3_3 | Mus musculus | IM009_Mock_D3_3_Lung | Tissue | IM009-R | IM009_Mock_D3_3_array | RNA | |||
| IM009 | IM009_Mock_D5_1 | Mus musculus | IM009_Mock_D5_1_Lung | Tissue | IM010-R | IM009_Mock_D5_1_array | RNA | |||
| IM009 | IM009_Mock_D5_2 | Mus musculus | IM009_Mock_D5_2_Lung | Tissue | IM010-R | IM009_Mock_D5_2_array | RNA | |||
| IM010 | IM010_B6_Mock_2d_1 | Mus musculus | IM010_B6_Mock_2d_1_Lung | Tissue | IM010-R | IM010_B6_Mock_2d_1_array | RNA | |||
| IM010 | IM010_B6_Mock_2d_2 | Mus musculus | IM010_B6_Mock_2d_2_Lung | Tissue | IM010-R | IM010_B6_Mock_2d_2_array | RNA | |||
| IM010 | IM010_B6_Mock_2d_3 | Mus musculus | IM010_B6_Mock_2d_3_Lung | Tissue | IM010-R | IM010_B6_Mock_2d_3_array | RNA | |||
| IM010 | IM010_B6_VN1203_2d_1 | Mus musculus | IM010_B6_VN1203_2d_1_Lung | Tissue | IM010-R | IM010_B6_VN1203_2d_1_array | RNA | |||
| IM010 | IM010_B6_VN1203_2d_2 | Mus musculus | IM010_B6_VN1203_2d_2_Lung | Tissue | IM010-R | IM010_B6_VN1203_2d_2_array | RNA | |||
| IM010 | IM010_B6_VN1203_2d_3 | Mus musculus | IM010_B6_VN1203_2d_3_Lung | Tissue | IM010-R | IM010_B6_VN1203_2d_3_array | RNA | |||
| IM010 | IM010_IDO1_Mock_2d_1 | Mus musculus | IM010_IDO1_Mock_2d_1_Lung | Tissue | IM010-R | IM010_IDO1_Mock_2d_1_array | RNA | |||
| IM010 | IM010_IDO1_Mock_2d_2 | Mus musculus | IM010_IDO1_Mock_2d_2_Lung | Tissue | IM010-R | IM010_IDO1_Mock_2d_2_array | RNA | |||
| IM010 | IM010_IDO1_Mock_2d_3 | Mus musculus | IM010_IDO1_Mock_2d_3_Lung | Tissue | IM010-R | IM010_IDO1_Mock_2d_3_array | RNA | |||
| IM010 | IM010_IDO1_Mock_6d_1 | Mus musculus | IM010_IDO1_Mock_6d_1_Lung | Tissue | IM010-R | IM010_IDO1_Mock_6d_1_array | RNA | |||
| IM010 | IM010_IDO1_Mock_6d_2 | Mus musculus | IM010_IDO1_Mock_6d_2_Lung | Tissue | IM010-R | IM010_IDO1_Mock_6d_2_array | RNA | |||
| IM010 | IM010_IDO1_Mock_6d_3 | Mus musculus | IM010_IDO1_Mock_6d_3_Lung | Tissue | IM010-R | IM010_IDO1_Mock_6d_3_array | RNA | |||
| IM010 | IM010_IDO1_VN1203_2d_1 | Mus musculus | IM010_IDO1_VN1203_2d_1_Lung | Tissue | IM010-R | IM010_IDO1_VN1203_2d_1_array | RNA | |||
| IM010 | IM010_IDO1_VN1203_2d_2 | Mus musculus | IM010_IDO1_VN1203_2d_2_Lung | Tissue | IM010-R | IM010_IDO1_VN1203_2d_2_array | RNA | |||
| IM010 | IM010_IDO1_VN1203_2d_3 | Mus musculus | IM010_IDO1_VN1203_2d_3_Lung | Tissue | IM010-R | IM010_IDO1_VN1203_2d_3_array | RNA | |||
| IM010 | IM010_IDO1_VN1203_6d_1 | Mus musculus | IM010_IDO1_VN1203_6d_1_Lung | Tissue | IM010-R | IM010_IDO1_VN1203_6d_1_array | RNA | |||
| IM010 | IM010_IDO1_VN1203_6d_2 | Mus musculus | IM010_IDO1_VN1203_6d_2_Lung | Tissue | IM010-R | IM010_IDO1_VN1203_6d_2_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_Mock_2d_1 | Mus musculus | IM010_Tnfrsf1b_Mock_2d_1_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_Mock_2d_1_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_Mock_2d_2 | Mus musculus | IM010_Tnfrsf1b_Mock_2d_2_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_Mock_2d_2_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_Mock_2d_3 | Mus musculus | IM010_Tnfrsf1b_Mock_2d_3_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_Mock_2d_3_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_Mock_6d_1 | Mus musculus | IM010_Tnfrsf1b_Mock_6d_1_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_Mock_6d_1_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_Mock_6d_2 | Mus musculus | IM010_Tnfrsf1b_Mock_6d_2_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_Mock_6d_2_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_Mock_6d_3 | Mus musculus | IM010_Tnfrsf1b_Mock_6d_3_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_Mock_6d_3_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_VN1203_2d_1 | Mus musculus | IM010_Tnfrsf1b_VN1203_2d_1_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_VN1203_2d_1_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_VN1203_2d_2 | Mus musculus | IM010_Tnfrsf1b_VN1203_2d_2_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_VN1203_2d_2_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_VN1203_2d_3 | Mus musculus | IM010_Tnfrsf1b_VN1203_2d_3_Lung | Tissue | IM010-R | IM010_Tnfrsf1b_VN1203_2d_3_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_VN1203_6d_1 | Mus musculus | IM010_Tnfrsf1b_VN1203_6d_1_Lung | Tissue | SBRI_AA_E1 | IM010_Tnfrsf1b_VN1203_6d_1_array | RNA | |||
| IM010 | IM010_Tnfrsf1b_VN1203_6d_2 | Mus musculus | IM010_Tnfrsf1b_VN1203_6d_2_Lung | Tissue | SBRI_AA_E1 | IM010_Tnfrsf1b_VN1203_6d_2_array | RNA | |||
| SBRI_AA | AA001_Mock | Mus musculus | AA001_Mock_lung | Tissue | SBRI_AA_E1 | AA_Mock_001 | RNA | |||
| SBRI_AA | AA002_Mock | Mus musculus | AA002_Mock_lung | Tissue | SBRI_AA_E1 | AA_Mock_002 | RNA | |||
| SBRI_AA | AA003_Mock | Mus musculus | AA003_Mock_lung | Tissue | SBRI_AA_E1 | AA_Mock_003 | RNA | |||
| SBRI_AA | AA005_Mock | Mus musculus | AA005_Mock_lung | Tissue | SBRI_AA_E1 | AA_Mock_005 | RNA | |||
| SBRI_AA | AA006_PR8 | Mus musculus | AA006_PR8_lung | Tissue | SBRI_AA_E1 | AA_PR8_006 | RNA | |||
| SBRI_AA | AA007_PR8 | Mus musculus | AA007_PR8_lung | Tissue | SBRI_AA_E1 | AA_PR8_007 | RNA | |||
| SBRI_AA | AA008_PR8 | Mus musculus | AA008_PR8_lung | Tissue | SBRI_AA_E1 | AA_PR8_008 | RNA | |||
| SBRI_AA | AA009_PR8 | Mus musculus | AA009_PR8_lung | Tissue | SBRI_AA_E1 | AA_PR8_009 | RNA | |||
| SBRI_AA | AA010_PR8 | Mus musculus | AA010_PR8_lung | Tissue | SBRI_AA_E1 | AA_PR8_010 | RNA | |||
| SBRI_AA | AA011_X31 | Mus musculus | AA011_X31_lung | Tissue | SBRI_AA_E1 | AA_VN6+2_016 | RNA | |||
| SBRI_AA | AA012_X31 | Mus musculus | AA012_X31_lung | Tissue | SBRI_AA_E1 | AA_VN6+2_017 | RNA | |||
| SBRI_AA | AA013_X31 | Mus musculus | AA013_X31_lung | Tissue | SBRI_AA_E1 | AA_VN6+2_018 | RNA | |||
| SBRI_AA | AA014_X31 | Mus musculus | AA014_X31_lung | Tissue | SBRI_AA_E1 | AA_VN6+2_019 | RNA | |||
| SBRI_AA | AA015_X31 | Mus musculus | AA015_X31_lung | Tissue | SBRI_AA_E1 | AA_VN6+2_020 | RNA | |||
| SBRI_AA | AA016_VN6+2 | Mus musculus | AA016_VN6+2_lung | Tissue | SBRI_AA_E1 | AA_X31_011 | RNA | |||
| SBRI_AA | AA017_VN6+2 | Mus musculus | AA017_VN6+2_lung | Tissue | SBRI_AA_E1 | AA_X31_012 | RNA | |||
| SBRI_AA | AA018_VN6+2 | Mus musculus | AA018_VN6+2_lung | Tissue | SBRI_AA_E1 | AA_X31_013 | RNA | |||
| SBRI_AA | AA019_VN6+2 | Mus musculus | AA019_VN6+2_lung | Tissue | SCL005-R | AA_X31_014 | RNA | |||
| SBRI_AA | AA020_VN6+2 | Mus musculus | AA020_VN6+2_lung | Tissue | SCL005-R | AA_X31_015 | RNA | |||
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_0H_1_cell | Cells | SCL005-R | SCL005_DORF6_0H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_0h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_0H_2_cell | Cells | SCL005-R | SCL005_DORF6_0H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_0h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_0H_3_cell | Cells | SCL005-R | SCL005_DORF6_0H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_0h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_12H_1_cell | Cells | SCL005-R | SCL005_DORF6_12H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_12h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_12H_2_cell | Cells | SCL005-R | SCL005_DORF6_12H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_12h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_12H_3_cell | Cells | SCL005-R | SCL005_DORF6_12H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_12h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_24H_1_cell | Cells | SCL005-R | SCL005_DORF6_24H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_24h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_24H_2_cell | Cells | SCL005-R | SCL005_DORF6_24H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_24h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_24H_3_cell | Cells | SCL005-R | SCL005_DORF6_24H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_24h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_30H_1_cell | Cells | SCL005-R | SCL005_DORF6_30H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_30h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_30H_2_cell | Cells | SCL005-R | SCL005_DORF6_30H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_30h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_30H_3_cell | Cells | SCL005-R | SCL005_DORF6_30H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_30h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_36H_1_cell | Cells | SCL005-R | SCL005_DORF6_36H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_36h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_36H_2_cell | Cells | SCL005-R | SCL005_DORF6_36H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_36h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_36H_3_cell | Cells | SCL005-R | SCL005_DORF6_36H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_36h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_3H_1_cell | Cells | SCL005-R | SCL005_DORF6_3H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_3h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_3H_2_cell | Cells | SCL005-R | SCL005_DORF6_3H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_3h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_3H_3_cell | Cells | SCL005-R | SCL005_DORF6_3H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_3h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_48H_1_cell | Cells | SCL005-R | SCL005_DORF6_48H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_48h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_48H_2_cell | Cells | SCL005-R | SCL005_DORF6_48H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_48h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_48H_3_cell | Cells | SCL005-R | SCL005_DORF6_48H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_48h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_54H_1_cell | Cells | SCL005-R | SCL005_DORF6_54H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_54h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_54H_2_cell | Cells | SCL005-R | SCL005_DORF6_54H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_54h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_54H_3_cell | Cells | SCL005-R | SCL005_DORF6_54H_3_RNA_ExpSam | RNA | |||
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_60H_1_cell | Cells | SCL005-R | SCL005_DORF6_60H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_60h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_60H_2_cell | Cells | SCL005-R | SCL005_DORF6_60H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_60h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_60H_3_cell | Cells | SCL005-R | SCL005_DORF6_60H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_60h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_72H_1_cell | Cells | SCL005-R | SCL005_DORF6_72H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_72h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_72H_2_cell | Cells | SCL005-R | SCL005_DORF6_72H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_72h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_72H_3_cell | Cells | SCL005-R | SCL005_DORF6_72H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_72h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_7H_1_cell | Cells | SCL005-R | SCL005_DORF6_7H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_7h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_7H_2_cell | Cells | SCL005-R | SCL005_DORF6_7H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_7h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_DORF6_7H_3_cell | Cells | SCL005-R | SCL005_DORF6_7H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_dORF6_7h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_0H_1_cell | Cells | SCL005-R | SCL005_mock_0H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_0h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_0H_2_cell | Cells | SCL005-R | SCL005_mock_0H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_0h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_0H_3_cell | Cells | SCL005-R | SCL005_mock_0H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_0h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_12H_1_cell | Cells | SCL005-R | SCL005_mock_12H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_12h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_12H_2_cell | Cells | SCL005-R | SCL005_mock_12H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_12h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_12H_3_cell | Cells | SCL005-R | SCL005_mock_12H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_12h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_24H_1_cell | Cells | SCL005-R | SCL005_mock_24H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_24h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_24H_2_cell | Cells | SCL005-R | SCL005_mock_24H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_24h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_24H_3_cell | Cells | SCL005-R | SCL005_mock_24H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_24h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_30H_1_cell | Cells | SCL005-R | SCL005_mock_30H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_30h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_30H_2_cell | Cells | SCL005-R | SCL005_mock_30H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_30h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_30H_3_cell | Cells | SCL005-R | SCL005_mock_30H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_30h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_36H_1_cell | Cells | SCL005-R | SCL005_mock_36H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_36h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_36H_2_cell | Cells | SCL005-R | SCL005_mock_36H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_36h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_36H_3_cell | Cells | SCL005-R | SCL005_mock_36H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_36h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_3H_1_cell | Cells | SCL005-R | SCL005_mock_3H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_3h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_3H_2_cell | Cells | SCL005-R | SCL005_mock_3H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_3h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_3H_3_cell | Cells | SCL005-R | SCL005_mock_3H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_3h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_48H_1_cell | Cells | SCL005-R | SCL005_mock_48H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_48h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_48H_2_cell | Cells | SCL005-R | SCL005_mock_48H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_48h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_48H_3_cell | Cells | SCL005-R | SCL005_mock_48H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_48h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_54H_1_cell | Cells | SCL005-R | SCL005_mock_54H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_54h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_54H_2_cell | Cells | SCL005-R | SCL005_mock_54H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_54h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_54H_3_cell | Cells | SCL005-R | SCL005_mock_54H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_54h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_60H_1_cell | Cells | SCL005-R | SCL005_mock_60H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_60h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_60H_2_cell | Cells | SCL005-R | SCL005_mock_60H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_60h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_60H_3_cell | Cells | SCL005-R | SCL005_mock_60H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_60h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_72H_1_cell | Cells | SCL005-R | SCL005_mock_72H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_72h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_72H_2_cell | Cells | SCL005-R | SCL005_mock_72H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_72h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_72H_3_cell | Cells | SCL005-R | SCL005_mock_72H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_72h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_7H_1_cell | Cells | SCL005-R | SCL005_mock_7H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_7h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_7H_2_cell | Cells | SCL005-R | SCL005_mock_7H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_Mock_7h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_mock_7H_3_cell | Cells | SCL005-R | SCL005_mock_7H_3_RNA_ExpSam | RNA | Protein | ||
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_0H_1_cell | Cells | SCL005-R | SCL005_WT_0H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_0h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_0H_2_cell | Cells | SCL005-R | SCL005_WT_0H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_0h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_0H_3_cell | Cells | SCL005-R | SCL005_WT_0H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_0h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_12H_1_cell | Cells | SCL005-R | SCL005_WT_12H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_12h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_12H_2_cell | Cells | SCL005-R | SCL005_WT_12H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_12h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_12H_3_cell | Cells | SCL005-R | SCL005_WT_12H_3_RNA_ExpSam | RNA | |||
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_24H_1_cell | Cells | SCL005-R | SCL005_WT_24H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_24h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_24H_2_cell | Cells | SCL005-R | SCL005_WT_24H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_24h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_24H_3_cell | Cells | SCL005-R | SCL005_WT_24H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_24h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_30H_1_cell | Cells | SCL005-R | SCL005_WT_30H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_30h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_30H_2_cell | Cells | SCL005-R | SCL005_WT_30H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_30h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_30H_3_cell | Cells | SCL005-R | SCL005_WT_30H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_30h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_36H_1_cell | Cells | SCL005-R | SCL005_WT_36H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_36h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_36H_2_cell | Cells | SCL005-R | SCL005_WT_36H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_36h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_36H_3_cell | Cells | SCL005-R | SCL005_WT_36H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_36h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_3H_1_cell | Cells | SCL005-R | SCL005_WT_3H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_3h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_3H_2_cell | Cells | SCL005-R | SCL005_WT_3H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_3h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_3H_3_cell | Cells | SCL005-R | SCL005_WT_3H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_3h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_48H_1_cell | Cells | SCL005-R | SCL005_WT_48H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_48h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_48H_2_cell | Cells | SCL005-R | SCL005_WT_48H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_48h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_48H_3_cell | Cells | SCL005-R | SCL005_WT_48H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_48h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_54H_1_cell | Cells | SCL005-R | SCL005_WT_54H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_54h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_54H_2_cell | Cells | SCL005-R | SCL005_WT_54H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_54h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_54H_3_cell | Cells | SCL005-R | SCL005_WT_54H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_54h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_60H_1_cell | Cells | SCL005-R | SCL005_WT_60H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_60h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_60H_2_cell | Cells | SCL005-R | SCL005_WT_60H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_60h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_60H_3_cell | Cells | SCL005-R | SCL005_WT_60H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_60h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_72H_1_cell | Cells | SCL005-R | SCL005_WT_72H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_72h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_72H_2_cell | Cells | SCL005-R | SCL005_WT_72H_2_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_72h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_72H_3_cell | Cells | SCL005-R | SCL005_WT_72H_3_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_72h_3_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_7H_1_cell | Cells | SCL005-R | SCL005_WT_7H_1_RNA_ExpSam | RNA | SCL005-P | SCL005_icSARS_7h_1_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_7H_2_cell | Cells | SCL006-R | SCL005_WT_7H_2_RNA_ExpSam | RNA | SCL006-P | SCL005_icSARS_7h_2_proteomics | Protein |
| SCL005 | SCL005 2B-4 Cells | Homo sapiens | SCL005_WT_7H_3_cell | Cells | SCL006-R | SCL005_WT_7H_3_RNA_ExpSam | RNA | Protein | ||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_0h_1_cell | Cells | SCL006-R | SCL006_BatSRBD_0h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_0h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_0h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_0h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_0h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_0h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_0h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_0h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_3h_1_Cells | Cells | SCL006-P | SCL006_BatSRBD_7h_1_proteomics | Protein | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_3h_2_Cells | Cells | SCL006-P | SCL006_BatSRBD_7h_2_proteomics | Protein | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_3h_3_Cells | Cells | SCL006-P | SCL006_BatSRBD_7h_3_proteomics | Protein | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_7h_1_Cells | Cells | SCL006-P | SCL006_BatSRBD_3h_1_proteomics | Protein | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_7h_2_Cells | Cells | SCL006-P | SCL006_BatSRBD_3h_2_proteomics | Protein | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_7h_3_Cells | Cells | SCL006-P | SCL006_BatSRBD_3h_3_proteomics | Protein | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_12h_1_cell | Cells | SCL006-R | SCL006_BatSRBD_12h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_12h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_12h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_12h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_12h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_12h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_12h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_12h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_24h_1_cell | Cells | SCL006-R | SCL006_BatSRBD_24h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_24h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_24h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_24h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_24h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_24h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_24h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_24h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_30h_1_cell | Cells | SCL006-R | SCL006_BatSRBD_30h_1_RNA_ExpSam | RNA | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_30h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_30h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_30h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_30h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_30h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_30h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_36h_1_cell | Cells | SCL006-R | SCL006_BatSRBD_36h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_36h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_36h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_36h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_36h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_36h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_36h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_36h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_48h_1_B_cell | Cells | SCL006-R | SCL006_BatSRBD_48h_1_B_RNA_ExpSam | RNA | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_48h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_48h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_48h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_48h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_48h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_48h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_54h_1_B_cell | Cells | SCL006-R | SCL006_BatSRBD_54h_1_B_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_54h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_54h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_54h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_54h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_54h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_54h_3_RNA_ExpSam | RNA | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_60h_1_cell | Cells | SCL006-R | SCL006_BatSRBD_60h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_60h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_60h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_60h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_60h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_60h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_60h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_60h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_72h_1_cell | Cells | SCL006-R | SCL006_BatSRBD_72h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_72h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_72h_2_cell | Cells | SCL006-R | SCL006_BatSRBD_72h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_72h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_BatSRBD_72h_3_cell | Cells | SCL006-R | SCL006_BatSRBD_72h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_BatSRBD_72h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_0h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_0h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_0h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_0h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_0h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_0h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_0h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_0h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_0h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_12h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_12h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_12h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_12h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_12h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_12h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_12h_3_B_cell | Cells | SCL006-R | SCL006_icSARSCoV_12h_3_B_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_12h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_24h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_24h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_24h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_24h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_24h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_24h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_24h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_24h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_24h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_30h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_30h_1_RNA_ExpSam | RNA | |||
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_30h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_30h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_30h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_30h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_30h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_30h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_36h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_36h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_36h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_36h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_36h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_36h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_36h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_36h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_36h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_48h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_48h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_48h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_48h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_48h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_48h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_48h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_48h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_48h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_54h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_54h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_54h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_54h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_54h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_54h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_54h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_54h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_54h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_60h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_60h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_60h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_60h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_60h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_60h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_60h_3_B_cell | Cells | SCL006-R | SCL006_icSARSCoV_60h_3_B_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_60h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_72h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_72h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_72h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_72h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_72h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_72h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_72h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_72h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_72h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_7h_1_cell | Cells | SCL006-R | SCL006_icSARSCoV_7h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_7h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_7h_2_cell | Cells | SCL006-R | SCL006_icSARSCoV_7h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_7h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_icSARSCoV_7h_3_cell | Cells | SCL006-R | SCL006_icSARSCoV_7h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_icSARS_7h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_0h_1_cell | Cells | SCL006-R | SCL006_mock_0h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_0h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_0h_2_cell | Cells | SCL006-R | SCL006_mock_0h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_0h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_0h_3_cell | Cells | SCL006-R | SCL006_mock_0h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_12h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_12h_1_cell | Cells | SCL006-R | SCL006_mock_12h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_12h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_12h_2_cell | Cells | SCL006-R | SCL006_mock_12h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_12h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_12h_3_cell | Cells | SCL006-R | SCL006_mock_12h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_24h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_24h_1_cell | Cells | SCL006-R | SCL006_mock_24h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_24h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_24h_2_cell | Cells | SCL006-R | SCL006_mock_24h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_24h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_24h_3_cell | Cells | SCL006-R | SCL006_mock_24h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_30h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_30h_1_cell | Cells | SCL006-R | SCL006_mock_30h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_30h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_30h_2_cell | Cells | SCL006-R | SCL006_mock_30h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_30h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_30h_3_cell | Cells | SCL006-R | SCL006_mock_30h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_36h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_36h_1_cell | Cells | SCL006-R | SCL006_mock_36h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_36h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_36h_2_cell | Cells | SCL006-R | SCL006_mock_36h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_36h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_36h_3_cell | Cells | SCL006-R | SCL006_mock_36h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_3h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_48h_1_cell | Cells | SCL006-R | SCL006_mock_48h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_3h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_48h_2_cell | Cells | SCL006-R | SCL006_mock_48h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_3h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_48h_3_cell | Cells | SCL006-R | SCL006_mock_48h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_48h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_54h_1_cell | Cells | SCL006-R | SCL006_mock_54h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_48h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_54h_2_cell | Cells | SCL006-R | SCL006_mock_54h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_48h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_54h_3_cell | Cells | SCL006-R | SCL006_mock_54h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_54h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_60h_1_cell | Cells | SCL006-R | SCL006_mock_60h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_54h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_60h_2_cell | Cells | SCL006-R | SCL006_mock_60h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_54h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_60h_3_cell | Cells | SCL006-R | SCL006_mock_60h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_60h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_72h_1_cell | Cells | SCL006-R | SCL006_mock_72h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_60h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_72h_2_cell | Cells | SCL006-R | SCL006_mock_72h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_72h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_72h_3_cell | Cells | SCL006-R | SCL006_mock_72h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_72h_3_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_7h_1_cell | Cells | SCL006-R | SCL006_mock_7h_1_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_7h_1_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_7h_2_cell | Cells | SHAE002-R | SCL006_mock_7h_2_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_7h_2_proteomics | Protein |
| SCL006 | SCL006 2B-4 Cells | Homo sapiens | SCL006_mock_7h_3_cell | Cells | SHAE002-R | SCL006_mock_7h_3_RNA_ExpSam | RNA | SCL006-P | SCL006_Mock_7h_3_proteomics | Protein |
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_0h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_0h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_0h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_0h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_0h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_0h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_0h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_0h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_12h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_12h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_12h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_12h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_12h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_12h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_12h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_12h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_24h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_24h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_24h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_24h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_24h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_24h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_24h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_24h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_36h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_36h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_36h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_36h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_36h_3_Cells | Cells | SHAE002-R | RNA | ||||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_36h_4_Cells | Cells | SHAE002-R | RNA | ||||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_48h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_48h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_48h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_48h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_48h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_48h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_48h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_48h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_60h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_60h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_60h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_60h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_60h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_60h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_60h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_60h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_72h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_72h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_72h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_72h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_72h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_72h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_72h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_72h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_84h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_84h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_84h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_84h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_84h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_84h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_84h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_84h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_96h_1_Cells | Cells | SHAE002-R | SHAE002_SARS_96h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_96h_2_Cells | Cells | SHAE002-R | SHAE002_SARS_96h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_96h_3_Cells | Cells | SHAE002-R | SHAE002_SARS_96h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_SARS_96h_4_Cells | Cells | SHAE002-R | SHAE002_SARS_96h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_0h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_0h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_0h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_0h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_0h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_0h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_0h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_0h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_12h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_12h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_12h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_12h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_12h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_12h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_12h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_12h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_24h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_24h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_24h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_24h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_24h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_24h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_24h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_24h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_36h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_36h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_36h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_36h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_36h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_36h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_36h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_36h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_48h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_48h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_48h_2_Cells | Cells | SHAE002-R | RNA | ||||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_48h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_48h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_48h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_48h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_60h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_60h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_60h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_60h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_60h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_60h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_60h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_60h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_72h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_72h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_72h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_72h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_72h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_72h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_72h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_72h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_84h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_84h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_84h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_84h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_84h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_84h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_84h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_84h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_96h_1_Cells | Cells | SHAE002-R | SHAE002_BAT_96h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_96h_2_Cells | Cells | SHAE002-R | SHAE002_BAT_96h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_96h_3_Cells | Cells | SHAE002-R | SHAE002_BAT_96h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_BAT_96h_4_Cells | Cells | SHAE002-R | SHAE002_BAT_96h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_0h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_0h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_0h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_0h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_0h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_0h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_0h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_0h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_12h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_12h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_12h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_12h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_12h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_12h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_12h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_12h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_24h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_24h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_24h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_24h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_24h_3_Cells | Cells | SHAE002-R | RNA | ||||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_24h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_24h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_36h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_36h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_36h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_36h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_36h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_36h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_36h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_36h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_48h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_48h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_48h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_48h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_48h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_48h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_48h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_48h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_60h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_60h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_60h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_60h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_60h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_60h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_60h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_60h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_72h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_72h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_72h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_72h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_72h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_72h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_72h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_72h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_84h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_84h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_84h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_84h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_84h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_84h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_84h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_84h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_96h_1_Cells | Cells | SHAE002-R | SHAE002_dORF6_96h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_96h_2_Cells | Cells | SHAE002-R | SHAE002_dORF6_96h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_96h_3_Cells | Cells | SHAE002-R | SHAE002_dORF6_96h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_dORF6_96h_4_Cells | Cells | SHAE002-R | SHAE002_dORF6_96h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_0h_1_Cells | Cells | SHAE002-R | SHAE002_H1N1_0h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_0h_2_Cells | Cells | SHAE002-R | SHAE002_H1N1_0h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_0h_3_Cells | Cells | SHAE002-R | SHAE002_H1N1_0h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_6h_1_Cells | Cells | SHAE002-R | SHAE002_H1N1_12h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_6h_2_Cells | Cells | SHAE002-R | SHAE002_H1N1_12h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_6h_3_Cells | Cells | SHAE002-R | SHAE002_H1N1_12h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_12h_1_Cells | Cells | SHAE002-R | SHAE002_H1N1_18h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_12h_2_Cells | Cells | SHAE002-R | SHAE002_H1N1_18h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_12h_3_Cells | Cells | SHAE002-R | SHAE002_H1N1_18h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_18h_1_Cells | Cells | SHAE002-R | SHAE002_H1N1_24h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_18h_2_Cells | Cells | SHAE002-R | SHAE002_H1N1_24h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_18h_3_Cells | Cells | SHAE002-R | SHAE002_H1N1_24h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_24h_1_Cells | Cells | SHAE002-R | SHAE002_H1N1_36h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_24h_2_Cells | Cells | SHAE002-R | SHAE002_H1N1_36h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_24h_3_Cells | Cells | SHAE002-R | SHAE002_H1N1_36h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_36h_1_Cells | Cells | SHAE002-R | SHAE002_H1N1_48h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_36h_2_Cells | Cells | SHAE002-R | SHAE002_H1N1_48h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_36h_3_Cells | Cells | SHAE002-R | SHAE002_H1N1_48h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_48h_1_Cells | Cells | SHAE002-R | SHAE002_H1N1_48h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_48h_2_Cells | Cells | SHAE002-R | SHAE002_H1N1_6h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_48h_3_Cells | Cells | SHAE002-R | SHAE002_H1N1_6h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_H1N1_48h_4_Cells | Cells | SHAE002-R | SHAE002_H1N1_6h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_0h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_0h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_0h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_0h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_0h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_0h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_6h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_12h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_6h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_12h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_6h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_12h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_12h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_18h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_12h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_18h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_12h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_18h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_18h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_24h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_18h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_24h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_18h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_24h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_24h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_36h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_24h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_36h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_24h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_36h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_36h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_48h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_36h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_48h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_36h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_48h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_48h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_60h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_48h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_60h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_48h_4_Cells | Cells | SHAE002-R | SHAE002_Mock_60h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_60h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_60h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_60h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_6h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_60h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_6h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_60h_4_Cells | Cells | SHAE002-R | SHAE002_Mock_6h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_72h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_72h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_72h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_72h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_72h_3_Cells | Cells | SHAE002-R | SHAE002_Mock_72h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_72h_4_Cells | Cells | SHAE002-R | SHAE002_Mock_72h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_84h_1_Cells | Cells | SHAE002-R | SHAE002_Mock_84h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_84h_2_Cells | Cells | SHAE002-R | SHAE002_Mock_84h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_84h_3_Cells | Cells | SHAE003-R | SHAE002_Mock_84h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_84h_4_Cells | Cells | SHAE003-R | SHAE002_Mock_84h_4_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_96h_1_Cells | Cells | SHAE003-R | SHAE002_Mock_96h_1_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_96h_2_Cells | Cells | SHAE003-R | SHAE002_Mock_96h_2_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_96h_3_Cells | Cells | SHAE003-R | SHAE002_Mock_96h_3_array | RNA | |||
| SHAE005 | SHAE002 HAE cells | Homo sapiens | SHAE002_Mock_96h_4_Cells | Cells | SHAE003-R | SHAE002_Mock_96h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_0h_1_Cells | Cells | SHAE003-R | SHAE003_SARS_0h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_0h_2_Cells | Cells | SHAE003-R | SHAE003_SARS_0h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_0h_3_Cells | Cells | SHAE003-R | SHAE003_SARS_0h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_0h_4_Cells | Cells | SHAE003-R | SHAE003_SARS_0h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_24h_1_Cells | Cells | SHAE003-R | SHAE003_SARS_24h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_24h_2_Cells | Cells | SHAE003-R | SHAE003_SARS_24h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_24h_3_Cells | Cells | SHAE003-R | SHAE003_SARS_24h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_24h_4_Cells | Cells | SHAE003-R | SHAE003_SARS_24h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_48h_1_Cells | Cells | SHAE003-R | SHAE003_SARS_48h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_48h_2_Cells | Cells | SHAE003-R | SHAE003_SARS_48h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_48h_3_Cells | Cells | SHAE003-R | SHAE003_SARS_48h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_48h_4_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_60h_1_Cells | Cells | SHAE003-R | SHAE003_SARS_60h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_60h_2_Cells | Cells | SHAE003-R | SHAE003_SARS_60h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_60h_3_Cells | Cells | SHAE003-R | SHAE003_SARS_60h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_60h_4_Cells | Cells | SHAE003-R | SHAE003_SARS_60h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_72h_1_Cells | Cells | SHAE003-R | SHAE003_SARS_72h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_72h_2_Cells | Cells | SHAE003-R | SHAE003_SARS_72h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_72h_3_Cells | Cells | SHAE003-R | SHAE003_SARS_72h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_72h_4_Cells | Cells | SHAE003-R | SHAE003_SARS_72h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_84h_1_Cells | Cells | SHAE003-R | SHAE003_SARS_84h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_84h_2_Cells | Cells | SHAE003-R | SHAE003_SARS_84h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_84h_3_Cells | Cells | SHAE003-R | SHAE003_SARS_84h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_84h_4_Cells | Cells | SHAE003-R | SHAE003_SARS_84h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_96h_1_Cells | Cells | SHAE003-R | SHAE003_SARS_96h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_96h_2_Cells | Cells | SHAE003-R | SHAE003_SARS_96h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_96h_3_Cells | Cells | SHAE003-R | SHAE003_SARS_96h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_SARS_96h_4_Cells | Cells | SHAE003-R | SHAE003_SARS_96h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_0h_1_Cells | Cells | SHAE003-R | SHAE003_BAT_0h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_0h_2_Cells | Cells | SHAE003-R | SHAE003_BAT_0h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_0h_3_Cells | Cells | SHAE003-R | SHAE003_BAT_0h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_0h_4_Cells | Cells | SHAE003-R | SHAE003_BAT_0h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_24h_3_Cells | Cells | SHAE003-R | SHAE003_BAT_24h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_24h_4_Cells | Cells | SHAE003-R | SHAE003_BAT_24h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_48h_1_Cells | Cells | SHAE003-R | SHAE003_BAT_48h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_48h_2_Cells | Cells | SHAE003-R | SHAE003_BAT_48h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_48h_3_Cells | Cells | SHAE003-R | SHAE003_BAT_48h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_48h_4_Cells | Cells | SHAE003-R | SHAE003_BAT_48h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_60h_1_Cells | Cells | SHAE003-R | SHAE003_BAT_60h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_60h_2_Cells | Cells | SHAE003-R | SHAE003_BAT_60h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_60h_3_Cells | Cells | SHAE003-R | SHAE003_BAT_60h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_60h_4_Cells | Cells | SHAE003-R | SHAE003_BAT_60h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_72h_1_Cells | Cells | SHAE003-R | SHAE003_BAT_72h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_72h_2_Cells | Cells | SHAE003-R | SHAE003_BAT_72h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_72h_3_Cells | Cells | SHAE003-R | SHAE003_BAT_72h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_72h_4_Cells | Cells | SHAE003-R | SHAE003_BAT_72h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_84h_1_Cells | Cells | SHAE003-R | SHAE003_BAT_84h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_84h_2_Cells | Cells | SHAE003-R | SHAE003_BAT_84h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_84h_3_Cells | Cells | SHAE003-R | SHAE003_BAT_84h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_84h_4_Cells | Cells | SHAE003-R | SHAE003_BAT_84h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_96h_1_Cells | Cells | SHAE003-R | SHAE003_BAT_96h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_96h_2_Cells | Cells | SHAE003-R | SHAE003_BAT_96h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_96h_3_Cells | Cells | SHAE003-R | SHAE003_BAT_96h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_BAT_96h_4_Cells | Cells | SHAE003-R | SHAE003_BAT_96h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_0h_1_Cells | Cells | SHAE003-R | SHAE003_dORF6_0h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_0h_2_Cells | Cells | SHAE003-R | SHAE003_dORF6_0h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_0h_3_Cells | Cells | SHAE003-R | SHAE003_dORF6_0h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_0h_4_Cells | Cells | SHAE003-R | SHAE003_dORF6_0h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_24h_1_Cells | Cells | SHAE003-R | SHAE003_dORF6_24h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_24h_2_Cells | Cells | SHAE003-R | SHAE003_dORF6_24h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_24h_3_Cells | Cells | SHAE003-R | SHAE003_dORF6_24h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_48h_1_Cells | Cells | SHAE003-R | SHAE003_dORF6_48h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_48h_2_Cells | Cells | SHAE003-R | SHAE003_dORF6_48h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_48h_3_Cells | Cells | SHAE003-R | SHAE003_dORF6_48h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_48h_4_Cells | Cells | SHAE003-R | SHAE003_dORF6_48h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_60h_1_Cells | Cells | SHAE003-R | SHAE003_dORF6_60h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_60h_2_Cells | Cells | SHAE003-R | SHAE003_dORF6_60h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_60h_3_Cells | Cells | SHAE003-R | SHAE003_dORF6_60h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_60h_4_Cells | Cells | SHAE003-R | SHAE003_dORF6_60h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_72h_1_Cells | Cells | SHAE003-R | SHAE003_dORF6_72h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_72h_2_Cells | Cells | SHAE003-R | SHAE003_dORF6_72h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_72h_3_Cells | Cells | SHAE003-R | SHAE003_dORF6_72h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_72h_4_Cells | Cells | SHAE003-R | SHAE003_dORF6_72h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_84h_1_Cells | Cells | SHAE003-R | SHAE003_dORF6_84h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_84h_2_Cells | Cells | SHAE003-R | SHAE003_dORF6_84h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_84h_3_Cells | Cells | SHAE003-R | SHAE003_dORF6_84h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_84h_4_Cells | Cells | SHAE003-R | SHAE003_dORF6_84h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_96h_1_Cells | Cells | SHAE003-R | SHAE003_dORF6_96h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_96h_2_Cells | Cells | SHAE003-R | SHAE003_dORF6_96h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_96h_3_Cells | Cells | SHAE003-R | SHAE003_dORF6_96h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_dORF6_96h_4_Cells | Cells | SHAE003-R | SHAE003_dORF6_96h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_0h_1_Cells | Cells | SHAE003-R | SHAE003_H1N1_0h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_0h_2_Cells | Cells | SHAE003-R | SHAE003_H1N1_0h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_0h_3_Cells | Cells | SHAE003-R | SHAE003_H1N1_0h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_0h_4_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_6h_1_Cells | Cells | SHAE003-R | SHAE003_H1N1_6h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_6h_2_Cells | Cells | SHAE003-R | SHAE003_H1N1_6h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_6h_3_Cells | Cells | SHAE003-R | SHAE003_H1N1_6h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_6h_4_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_12h_1_Cells | Cells | SHAE003-R | SHAE003_H1N1_12h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_12h_2_Cells | Cells | SHAE003-R | SHAE003_H1N1_12h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_12h_3_Cells | Cells | SHAE003-R | SHAE003_H1N1_12h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_12h_4_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_18h_1_Cells | Cells | SHAE003-R | SHAE003_H1N1_18h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_18h_2_Cells | Cells | SHAE003-R | SHAE003_H1N1_18h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_18h_3_Cells | Cells | SHAE003-R | SHAE003_H1N1_18h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_18h_4_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_24h_1_Cells | Cells | SHAE003-R | SHAE003_H1N1_24h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_24h_2_Cells | Cells | SHAE003-R | SHAE003_H1N1_24h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_24h_3_Cells | Cells | SHAE003-R | SHAE003_H1N1_24h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_24h_4_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_36h_1_Cells | Cells | SHAE003-R | SHAE003_H1N1_36h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_36h_2_Cells | Cells | SHAE003-R | SHAE003_H1N1_36h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_36h_3_Cells | Cells | SHAE003-R | SHAE003_H1N1_36h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_36h_4_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_48h_1_Cells | Cells | SHAE003-R | SHAE003_H1N1_48h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_48h_2_Cells | Cells | SHAE003-R | SHAE003_H1N1_48h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_48h_3_Cells | Cells | SHAE003-R | SHAE003_H1N1_48h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_H1N1_48h_4_Cells | Cells | SHAE003-R | SHAE003_H1N1_48h_4_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_0h_1_Cells | Cells | SHAE003-R | SHAE003_Mock_0h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_0h_2_Cells | Cells | SHAE003-R | SHAE003_Mock_0h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_0h_3_Cells | Cells | SHAE003-R | SHAE003_Mock_0h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_6h_1_Cells | Cells | SHAE003-R | SHAE003_Mock_6h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_6h_2_Cells | Cells | SHAE003-R | SHAE003_Mock_6h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_6h_3_Cells | Cells | SHAE003-R | SHAE003_Mock_6h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_12h_1_Cells | Cells | SHAE003-R | SHAE003_Mock_12h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_12h_2_Cells | Cells | SHAE003-R | SHAE003_Mock_12h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_12h_3_Cells | Cells | SHAE003-R | SHAE003_Mock_12h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_18h_1_Cells | Cells | SHAE003-R | SHAE003_Mock_18h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_18h_2_Cells | Cells | SHAE003-R | RNA | ||||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_18h_3_Cells | Cells | SHAE003-R | SHAE003_Mock_18h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_24h_1_Cells | Cells | SHAE003-R | SHAE003_Mock_24h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_24h_2_Cells | Cells | SHAE003-R | SHAE003_Mock_24h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_24h_3_Cells | Cells | SHAE003-R | SHAE003_Mock_24h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_36h_1_Cells | Cells | SHAE003-R | SHAE003_Mock_36h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_36h_2_Cells | Cells | SHAE003-R | SHAE003_Mock_36h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_36h_3_Cells | Cells | SHAE003-R | SHAE003_Mock_36h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_48h_1_Cells | Cells | SHAE003-R | SHAE003_Mock_48h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_48h_2_Cells | Cells | SHAE004-R | SHAE003_Mock_48h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_48h_3_Cells | Cells | SHAE004-R | SHAE003_Mock_48h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_60h_1_Cells | Cells | SHAE004-R | SHAE003_Mock_60h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_60h_2_Cells | Cells | SHAE004-R | SHAE003_Mock_60h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_60h_3_Cells | Cells | SHAE004-R | SHAE003_Mock_60h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_72h_1_Cells | Cells | SHAE004-R | SHAE003_Mock_72h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_72h_2_Cells | Cells | SHAE004-R | SHAE003_Mock_72h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_72h_3_Cells | Cells | SHAE004-R | SHAE003_Mock_72h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_84h_1_Cells | Cells | SHAE004-R | SHAE003_Mock_84h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_84h_2_Cells | Cells | SHAE004-R | SHAE003_Mock_84h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_84h_3_Cells | Cells | SHAE004-R | SHAE003_Mock_84h_3_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_96h_1_Cells | Cells | SHAE004-R | SHAE003_Mock_96h_1_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_96h_2_Cells | Cells | SHAE004-R | SHAE003_Mock_96h_2_array | RNA | |||
| SHAE005 | SHAE003 HAE cells | Homo sapiens | SHAE003_Mock_96h_3_Cells | Cells | SHAE004-R | SHAE003_Mock_96h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_18h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_18h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_24h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_24h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_24h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_24h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_24h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_24h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_36h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_36h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_36h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_36h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_36h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_36h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_48h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_48h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_48h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_48h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_48h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_48h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_60h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_60h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_60h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_60h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_60h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_60h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_72h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_72h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_72h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_72h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_72h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_72h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_84h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_84h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_84h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_84h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_84h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_84h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_96h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_96h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_96h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_96h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_96h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_96h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_0h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_0h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_0h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_0h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_0h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_0h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_12h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_12h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_12h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_12h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_12h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_12h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_24h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_24h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_24h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_24h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_24h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_24h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_36h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_36h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_36h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_36h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_36h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_36h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_48h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_48h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_48h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_48h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_48h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_48h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_60h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_60h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_60h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_60h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_60h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_60h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_72h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_72h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_72h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_72h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_72h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_72h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_84h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_84h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_84h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_84h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_84h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_84h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_96h_1_Cells | Cells | SHAE004-R | SHAE004_SARS_96h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_96h_2_Cells | Cells | SHAE004-R | SHAE004_SARS_96h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_SARS_96h_3_Cells | Cells | SHAE004-R | SHAE004_SARS_96h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_0h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_0h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_0h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_0h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_0h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_0h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_12h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_12h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_12h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_12h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_12h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_12h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_24h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_24h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_24h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_24h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_36h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_36h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_36h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_36h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_36h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_36h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_48h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_48h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_48h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_48h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_48h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_48h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_60h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_60h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_60h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_60h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_60h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_60h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_72h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_72h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_72h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_72h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_72h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_72h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_84h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_84h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_84h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_84h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_84h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_84h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_96h_1_Cells | Cells | SHAE004-R | SHAE004_BAT_96h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_96h_2_Cells | Cells | SHAE004-R | SHAE004_BAT_96h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_BAT_96h_3_Cells | Cells | SHAE004-R | SHAE004_BAT_96h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_0h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_0h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_0h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_0h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_0h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_0h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_12h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_12h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_12h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_12h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_12h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_12h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_24h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_24h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_24h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_24h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_24h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_24h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_36h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_36h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_36h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_36h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_36h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_36h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_48h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_48h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_48h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_48h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_48h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_48h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_60h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_60h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_60h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_60h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_60h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_60h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_72h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_72h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_72h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_72h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_72h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_72h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_84h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_84h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_84h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_84h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_84h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_84h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_96h_1_Cells | Cells | SHAE004-R | SHAE004_dORF6_96h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_96h_2_Cells | Cells | SHAE004-R | SHAE004_dORF6_96h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_dORF6_96h_3_Cells | Cells | SHAE004-R | SHAE004_dORF6_96h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_0h_1_Cells | Cells | SHAE004-R | SHAE004_H1N1_0h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_0h_2_Cells | Cells | SHAE004-R | SHAE004_H1N1_0h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_0h_3_Cells | Cells | SHAE004-R | SHAE004_H1N1_0h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_6h_1_Cells | Cells | SHAE004-R | SHAE004_H1N1_12h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_6h_2_Cells | Cells | SHAE004-R | SHAE004_H1N1_12h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_6h_3_Cells | Cells | SHAE004-R | SHAE004_H1N1_12h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_12h_1_Cells | Cells | SHAE004-R | SHAE004_H1N1_18h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_12h_2_Cells | Cells | SHAE004-R | SHAE004_H1N1_18h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_12h_3_Cells | Cells | SHAE004-R | SHAE004_H1N1_18h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_18h_1_Cells | Cells | SHAE004-R | SHAE004_H1N1_24h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_18h_2_Cells | Cells | SHAE004-R | SHAE004_H1N1_24h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_18h_3_Cells | Cells | SHAE004-R | SHAE004_H1N1_24h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_24h_1_Cells | Cells | SHAE004-R | SHAE004_H1N1_36h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_24h_2_Cells | Cells | SHAE004-R | SHAE004_H1N1_36h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_24h_3_Cells | Cells | SHAE004-R | SHAE004_H1N1_36h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_36h_1_Cells | Cells | SHAE004-R | SHAE004_H1N1_48h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_36h_2_Cells | Cells | SHAE004-R | SHAE004_H1N1_48h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_36h_3_Cells | Cells | SHAE004-R | SHAE004_H1N1_48h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_48h_1_Cells | Cells | SHAE004-R | SHAE004_H1N1_6h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_48h_2_Cells | Cells | SHAE004-R | SHAE004_H1N1_6h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_H1N1_48h_3_Cells | Cells | SHAE004-R | SHAE004_H1N1_6h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_0h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_0h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_0h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_0h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_0h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_0h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_6h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_6h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_6h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_6h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_6h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_6h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_12h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_12h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_12h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_12h_2_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_12h_3_Cells | Cells | SHAE004-R | SHAE004_Mock_12h_3_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_18h_1_Cells | Cells | SHAE004-R | SHAE004_Mock_18h_1_array | RNA | |||
| SHAE005 | SHAE004 HAE cells | Homo sapiens | SHAE004_Mock_18h_2_Cells | Cells | SHAE004-R | SHAE004_Mock_18h_2_array | RNA | |||
| SM001 | SM001_Mock_D1_1 | Mus musculus | SM001_Mock_D1_1_lung | Cells | SM001-R | SM001_Mock_D1_1_RNA_ExpSam | RNA | SM001-P | SM001_Mock_1d_1_proteomics | Protein |
| SM001 | SM001_Mock_D1_2 | Mus musculus | SM001_Mock_D1_2_lung | Cells | SM001-R | SM001_Mock_D1_2_RNA_ExpSam | RNA | SM001-P | SM001_Mock_1d_2_proteomics | Protein |
| SM001 | SM001_Mock_D1_3 | Mus musculus | SM001_Mock_D1_3_lung | Cells | SM001-R | SM001_Mock_D1_3_RNA_ExpSam | RNA | SM001-P | SM001_Mock_1d_3_proteomics | Protein |
| SM001 | SM001_Mock_D2_1 | Mus musculus | SM001_Mock_D2_1_lung | Cells | SM001-R | SM001_Mock_D2_1_RNA_ExpSam | RNA | SM001-P | SM001_Mock_2d_1_proteomics | Protein |
| SM001 | SM001_Mock_D2_2 | Mus musculus | SM001_Mock_D2_2_lung | Cells | SM001-R | SM001_Mock_D2_2_RNA_ExpSam | RNA | SM001-P | SM001_Mock_2d_2_proteomics | Protein |
| SM001 | SM001_Mock_D2_3 | Mus musculus | SM001_Mock_D2_3_lung | Cells | SM001-R | SM001_Mock_D2_3_RNA_ExpSam | RNA | SM001-P | SM001_Mock_2d_3_proteomics | Protein |
| SM001 | SM001_Mock_D4_1 | Mus musculus | SM001_Mock_D4_1_lung | Cells | SM001-R | SM001_Mock_D4_1_RNA_ExpSam | RNA | SM001-P | SM001_Mock_4d_1_proteomics | Protein |
| SM001 | SM001_Mock_D4_2 | Mus musculus | SM001_Mock_D4_2_lung | Cells | SM001-R | SM001_Mock_D4_2_RNA_ExpSam | RNA | SM001-P | SM001_Mock_4d_2_proteomics | Protein |
| SM001 | SM001_Mock_D4_3 | Mus musculus | SM001_Mock_D4_3_lung | Cells | SM001-R | SM001_Mock_D4_3_RNA_ExpSam | RNA | SM001-P | SM001_Mock_4d_3_proteomics | Protein |
| SM001 | SM001_Mock_D7_1 | Mus musculus | SM001_Mock_D7_1_lung | Cells | SM001-R | SM001_Mock_D7_1_RNA_ExpSam | RNA | SM001-P | SM001_Mock_7d_1_proteomics | Protein |
| SM001 | SM001_Mock_D7_2 | Mus musculus | SM001_Mock_D7_2_lung | Cells | SM001-R | SM001_Mock_D7_2_RNA_ExpSam | RNA | SM001-P | SM001_Mock_7d_2_proteomics | Protein |
| SM001 | SM001_Mock_D7_3 | Mus musculus | SM001_Mock_D7_3_lung | Cells | SM001-R | SM001_Mock_D7_3_RNA_ExpSam | RNA | SM001-P | SM001_Mock_7d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D1_1 | Mus musculus | SM001_SARS_10^2pfu_D1_1_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D1_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_1d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D1_2 | Mus musculus | SM001_SARS_10^2pfu_D1_2_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D1_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_1d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D1_3 | Mus musculus | SM001_SARS_10^2pfu_D1_3_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D1_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_1d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D1_4 | Mus musculus | SM001_SARS_10^2pfu_D1_4_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D1_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_1d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D1_5 | Mus musculus | SM001_SARS_10^2pfu_D1_5_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D1_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_1d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D2_1 | Mus musculus | SM001_SARS_10^2pfu_D2_1_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D2_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_2d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D2_2 | Mus musculus | SM001_SARS_10^2pfu_D2_2_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D2_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_2d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D2_3 | Mus musculus | SM001_SARS_10^2pfu_D2_3_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D2_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_2d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D2_4 | Mus musculus | SM001_SARS_10^2pfu_D2_4_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D2_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_2d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D2_5 | Mus musculus | SM001_SARS_10^2pfu_D2_5_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D2_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_2d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D4_1 | Mus musculus | SM001_SARS_10^2pfu_D4_1_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D4_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_4d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D4_2 | Mus musculus | SM001_SARS_10^2pfu_D4_2_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D4_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_4d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D4_3 | Mus musculus | SM001_SARS_10^2pfu_D4_3_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D4_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_4d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D4_4 | Mus musculus | SM001_SARS_10^2pfu_D4_4_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D4_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_4d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D4_5 | Mus musculus | SM001_SARS_10^2pfu_D4_5_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D4_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_4d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D7_1 | Mus musculus | SM001_SARS_10^2pfu_D7_1_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D7_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_7d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D7_2 | Mus musculus | SM001_SARS_10^2pfu_D7_2_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D7_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_7d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D7_3 | Mus musculus | SM001_SARS_10^2pfu_D7_3_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D7_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_7d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D7_4 | Mus musculus | SM001_SARS_10^2pfu_D7_4_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D7_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_7d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^2pfu_D7_5 | Mus musculus | SM001_SARS_10^2pfu_D7_5_lung | Cells | SM001-R | SM001_SARS_10^2pfu_D7_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^2_7d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D1_1 | Mus musculus | SM001_SARS_10^3pfu_D1_1_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D1_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_1d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D1_2 | Mus musculus | SM001_SARS_10^3pfu_D1_2_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D1_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_1d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D1_3 | Mus musculus | SM001_SARS_10^3pfu_D1_3_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D1_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_1d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D1_4 | Mus musculus | SM001_SARS_10^3pfu_D1_4_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D1_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_1d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D1_5 | Mus musculus | SM001_SARS_10^3pfu_D1_5_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D1_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_1d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D2_1 | Mus musculus | SM001_SARS_10^3pfu_D2_1_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D2_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_2d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D2_2 | Mus musculus | SM001_SARS_10^3pfu_D2_2_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D2_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_2d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D2_3 | Mus musculus | SM001_SARS_10^3pfu_D2_3_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D2_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_2d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D2_4 | Mus musculus | SM001_SARS_10^3pfu_D2_4_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D2_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_2d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D2_5 | Mus musculus | SM001_SARS_10^3pfu_D2_5_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D2_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_2d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D4_1 | Mus musculus | SM001_SARS_10^3pfu_D4_1_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D4_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_4d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D4_2 | Mus musculus | SM001_SARS_10^3pfu_D4_2_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D4_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_4d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D4_3 | Mus musculus | SM001_SARS_10^3pfu_D4_3_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D4_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_4d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D4_4 | Mus musculus | SM001_SARS_10^3pfu_D4_4_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D4_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_4d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D4_5 | Mus musculus | SM001_SARS_10^3pfu_D4_5_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D4_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_4d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D7_1 | Mus musculus | SM001_SARS_10^3pfu_D7_1_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D7_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_7d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D7_2 | Mus musculus | SM001_SARS_10^3pfu_D7_2_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D7_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_7d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D7_3 | Mus musculus | SM001_SARS_10^3pfu_D7_3_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D7_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_7d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D7_4 | Mus musculus | SM001_SARS_10^3pfu_D7_4_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D7_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_7d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^3pfu_D7_5 | Mus musculus | SM001_SARS_10^3pfu_D7_5_lung | Cells | SM001-R | SM001_SARS_10^3pfu_D7_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^3_7d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D1_1 | Mus musculus | SM001_SARS_10^4pfu_D1_1_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D1_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_1d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D1_2 | Mus musculus | SM001_SARS_10^4pfu_D1_2_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D1_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_1d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D1_3 | Mus musculus | SM001_SARS_10^4pfu_D1_3_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D1_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_1d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D1_4 | Mus musculus | SM001_SARS_10^4pfu_D1_4_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D1_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_1d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D1_5 | Mus musculus | SM001_SARS_10^4pfu_D1_5_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D1_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_1d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D2_1 | Mus musculus | SM001_SARS_10^4pfu_D2_1_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D2_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_2d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D2_2 | Mus musculus | SM001_SARS_10^4pfu_D2_2_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D2_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_2d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D2_3 | Mus musculus | SM001_SARS_10^4pfu_D2_3_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D2_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_2d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D2_4 | Mus musculus | SM001_SARS_10^4pfu_D2_4_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D2_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_2d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D2_5 | Mus musculus | SM001_SARS_10^4pfu_D2_5_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D2_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_2d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D4_1 | Mus musculus | SM001_SARS_10^4pfu_D4_1_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D4_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_4d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D4_2 | Mus musculus | SM001_SARS_10^4pfu_D4_2_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D4_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_4d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D4_3 | Mus musculus | SM001_SARS_10^4pfu_D4_3_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D4_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_4d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D4_4 | Mus musculus | SM001_SARS_10^4pfu_D4_4_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D4_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_4d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D4_5 | Mus musculus | SM001_SARS_10^4pfu_D4_5_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D4_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_4d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D7_1 | Mus musculus | SM001_SARS_10^4pfu_D7_1_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D7_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_7d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D7_2 | Mus musculus | SM001_SARS_10^4pfu_D7_2_lung | Cells | SM001-R | SM001_SARS_10^4pfu_D7_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_7d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D7_3 | Mus musculus | SM001_SARS_10^4pfu_D7_3_lung | Tissue | SM001-R | SM001_SARS_10^4pfu_D7_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_7d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D7_4 | Mus musculus | SM001_SARS_10^4pfu_D7_4_lung | Tissue | SM001-R | SM001_SARS_10^4pfu_D7_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_7d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^4pfu_D7_5 | Mus musculus | SM001_SARS_10^4pfu_D7_5_lung | Tissue | SM001-R | SM001_SARS_10^4pfu_D7_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^4_7d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D1_1 | Mus musculus | SM001_SARS_10^5pfu_D1_1_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D1_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_1d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D1_2 | Mus musculus | SM001_SARS_10^5pfu_D1_2_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D1_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_1d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D1_3 | Mus musculus | SM001_SARS_10^5pfu_D1_3_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D1_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_1d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D1_4 | Mus musculus | SM001_SARS_10^5pfu_D1_4_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D1_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_1d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D1_5 | Mus musculus | SM001_SARS_10^5pfu_D1_5_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D1_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_1d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D2_1 | Mus musculus | SM001_SARS_10^5pfu_D2_1_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D2_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_2d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D2_2 | Mus musculus | SM001_SARS_10^5pfu_D2_2_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D2_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_2d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D2_3 | Mus musculus | SM001_SARS_10^5pfu_D2_3_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D2_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_2d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D2_4 | Mus musculus | SM001_SARS_10^5pfu_D2_4_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D2_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_2d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D2_5 | Mus musculus | SM001_SARS_10^5pfu_D2_5_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D2_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_2d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D4_1 | Mus musculus | SM001_SARS_10^5pfu_D4_1_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D4_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_4d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D4_2 | Mus musculus | SM001_SARS_10^5pfu_D4_2_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D4_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_4d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D4_3 | Mus musculus | SM001_SARS_10^5pfu_D4_3_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D4_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_4d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D4_4 | Mus musculus | SM001_SARS_10^5pfu_D4_4_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D4_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_4d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D4_5 | Mus musculus | SM001_SARS_10^5pfu_D4_5_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D4_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_4d_5_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D7_1 | Mus musculus | SM001_SARS_10^5pfu_D7_1_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D7_1_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_7d_1_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D7_2 | Mus musculus | SM001_SARS_10^5pfu_D7_2_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D7_2_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_7d_2_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D7_3 | Mus musculus | SM001_SARS_10^5pfu_D7_3_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D7_3_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_7d_3_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D7_4 | Mus musculus | SM001_SARS_10^5pfu_D7_4_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D7_4_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_7d_4_proteomics | Protein |
| SM001 | SM001_SARS_10^5pfu_D7_5 | Mus musculus | SM001_SARS_10^5pfu_D7_5_lung | Tissue | SM001-R | SM001_SARS_10^5pfu_D7_5_RNA_ExpSam | RNA | SM001-P | SM001_SARS_10^5_7d_5_proteomics | Protein |
| SM003 | SM003_10^4_MA15_d1_1 | Mus musculus | SM003_10^4_MA15_d1_1_Lung | Tissue | SM003-R | SM003_10^4_MA15_d1_1_array | RNA | |||
| SM003 | SM003_10^4_MA15_d1_2 | Mus musculus | SM003_10^4_MA15_d1_2_Lung | Tissue | SM003-R | SM003_10^4_MA15_d1_2_array | RNA | |||
| SM003 | SM003_10^4_MA15_d1_3 | Mus musculus | SM003_10^4_MA15_d1_3_Lung | Tissue | SM003-R | SM003_10^4_MA15_d1_3_array | RNA | |||
| SM003 | SM003_10^4_MA15_d1_4 | Mus musculus | SM003_10^4_MA15_d1_4_Lung | Tissue | SM003-R | SM003_10^4_MA15_d1_4_array | RNA | |||
| SM003 | SM003_10^4_MA15_d2_1 | Mus musculus | SM003_10^4_MA15_d2_1_Lung | Tissue | SM003-R | SM003_10^4_MA15_d2_1_array | RNA | |||
| SM003 | SM003_10^4_MA15_d2_2 | Mus musculus | SM003_10^4_MA15_d2_2_Lung | Tissue | SM003-R | SM003_10^4_MA15_d2_2_array | RNA | |||
| SM003 | SM003_10^4_MA15_d2_3 | Mus musculus | SM003_10^4_MA15_d2_3_Lung | Tissue | SM003-R | SM003_10^4_MA15_d2_3_array | RNA | |||
| SM003 | SM003_10^4_MA15_d2_4 | Mus musculus | SM003_10^4_MA15_d2_4_Lung | Tissue | SM003-R | SM003_10^4_MA15_d2_4_array | RNA | |||
| SM003 | SM003_10^4_MA15_d4_1 | Mus musculus | SM003_10^4_MA15_d4_1_Lung | Tissue | SM003-R | SM003_10^4_MA15_d4_1_array | RNA | |||
| SM003 | SM003_10^4_MA15_d4_2 | Mus musculus | SM003_10^4_MA15_d4_2_Lung | Tissue | SM003-R | SM003_10^4_MA15_d4_2_array | RNA | |||
| SM003 | SM003_10^4_MA15_d4_3 | Mus musculus | SM003_10^4_MA15_d4_3_Lung | Tissue | SM003-R | SM003_10^4_MA15_d4_3_array | RNA | |||
| SM003 | SM003_10^4_MA15_d4_4 | Mus musculus | SM003_10^4_MA15_d4_4_Lung | Tissue | SM003-R | SM003_10^4_MA15_d4_4_array | RNA | |||
| SM003 | SM003_10^4_MA15_d7_1 | Mus musculus | SM003_10^4_MA15_d7_1_Lung | Tissue | SM003-R | SM003_10^4_MA15_d7_1_array | RNA | |||
| SM003 | SM003_10^4_MA15_d7_2 | Mus musculus | SM003_10^4_MA15_d7_2_Lung | Tissue | SM003-R | SM003_10^4_MA15_d7_2_array | RNA | |||
| SM003 | SM003_10^4_MA15_d7_3 | Mus musculus | SM003_10^4_MA15_d7_3_Lung | Tissue | SM003-R | SM003_10^4_MA15_d7_3_array | RNA | |||
| SM003 | SM003_10^4_MA15_d7_4 | Mus musculus | SM003_10^4_MA15_d7_4_Lung | Tissue | SM003-R | RNA | ||||
| SM003 | SM003_10^5_MA15_d1_1 | Mus musculus | SM003_10^5_MA15_d1_1_Lung | Tissue | SM003-R | SM003_10^5_MA15_d1_1_array | RNA | |||
| SM003 | SM003_10^5_MA15_d1_3 | Mus musculus | SM003_10^5_MA15_d1_3_Lung | Tissue | SM003-R | SM003_10^5_MA15_d1_3_array | RNA | |||
| SM003 | SM003_10^5_MA15_d1_4 | Mus musculus | SM003_10^5_MA15_d1_4_Lung | Tissue | SM003-R | SM003_10^5_MA15_d1_4_array | RNA | |||
| SM003 | SM003_10^5_MA15_d2_1 | Mus musculus | SM003_10^5_MA15_d2_1_Lung | Tissue | SM003-R | RNA | ||||
| SM003 | SM003_10^5_MA15_d2_2 | Mus musculus | SM003_10^5_MA15_d2_2_Lung | Tissue | SM003-R | SM003_10^5_MA15_d2_2_array | RNA | |||
| SM003 | SM003_10^5_MA15_d2_3 | Mus musculus | SM003_10^5_MA15_d2_3_Lung | Tissue | SM003-R | SM003_10^5_MA15_d2_3_array | RNA | |||
| SM003 | SM003_10^5_MA15_d2_4 | Mus musculus | SM003_10^5_MA15_d2_4_Lung | Tissue | SM003-R | SM003_10^5_MA15_d2_4_array | RNA | |||
| SM003 | SM003_10^5_MA15_d4_1 | Mus musculus | SM003_10^5_MA15_d4_1_Lung | Tissue | SM003-R | SM003_10^5_MA15_d4_1_array | RNA | |||
| SM003 | SM003_10^5_MA15_d4_2 | Mus musculus | SM003_10^5_MA15_d4_2_Lung | Tissue | SM003-R | SM003_10^5_MA15_d4_2_array | RNA | |||
| SM003 | SM003_10^5_MA15_d4_3 | Mus musculus | SM003_10^5_MA15_d4_3_Lung | Tissue | SM003-R | SM003_10^5_MA15_d4_3_array | RNA | |||
| SM003 | SM003_10^5_MA15_d4_4 | Mus musculus | SM003_10^5_MA15_d4_4_Lung | Tissue | SM003-R | SM003_10^5_MA15_d4_4_array | RNA | |||
| SM003 | SM003_10^5_MA15_d4_5 | Mus musculus | SM003_10^5_MA15_d4_5_Lung | Tissue | SM003-R | SM003_10^5_MA15_d4_5_array | RNA | |||
| SM003 | SM003_10^5_MA15_d7_1 | Mus musculus | SM003_10^5_MA15_d7_1_Lung | Tissue | SM003-R | SM003_10^5_MA15_d7_1_array | RNA | |||
| SM003 | SM003_10^5_MA15_d7_2 | Mus musculus | SM003_10^5_MA15_d7_2_Lung | Tissue | SM003-R | SM003_10^5_MA15_d7_2_array | RNA | |||
| SM003 | SM003_10^5_MA15_d7_3 | Mus musculus | SM003_10^5_MA15_d7_3_Lung | Tissue | SM003-R | SM003_10^5_MA15_d7_3_array | RNA | |||
| SM003 | SM003_BAT_d1_1 | Mus musculus | SM003_BAT_d1_1_Lung | Tissue | SM003-R | SM003_BAT_d1_1_array | RNA | |||
| SM003 | SM003_BAT_d1_2 | Mus musculus | SM003_BAT_d1_2_Lung | Tissue | SM003-R | SM003_BAT_d1_2_array | RNA | |||
| SM003 | SM003_BAT_d1_3 | Mus musculus | SM003_BAT_d1_3_Lung | Tissue | SM003-R | SM003_BAT_d1_3_array | RNA | |||
| SM003 | SM003_BAT_d1_4 | Mus musculus | SM003_BAT_d1_4_Lung | Tissue | SM003-R | SM003_BAT_d1_4_array | RNA | |||
| SM003 | SM003_BAT_d1_5 | Mus musculus | SM003_BAT_d1_5_Lung | Tissue | SM003-R | SM003_BAT_d1_5_array | RNA | |||
| SM003 | SM003_BAT_d2_1 | Mus musculus | SM003_BAT_d2_1_Lung | Tissue | SM003-R | SM003_BAT_d2_1_array | RNA | |||
| SM003 | SM003_BAT_d2_2 | Mus musculus | SM003_BAT_d2_2_Lung | Tissue | SM003-R | SM003_BAT_d2_2_array | RNA | |||
| SM003 | SM003_BAT_d2_3 | Mus musculus | SM003_BAT_d2_3_Lung | Tissue | SM003-R | SM003_BAT_d2_3_array | RNA | |||
| SM003 | SM003_BAT_d2_4 | Mus musculus | SM003_BAT_d2_4_Lung | Tissue | SM003-R | SM003_BAT_d2_4_array | RNA | |||
| SM003 | SM003_BAT_d2_5 | Mus musculus | SM003_BAT_d2_5_Lung | Tissue | SM003-R | SM003_BAT_d2_5_array | RNA | |||
| SM003 | SM003_BAT_d4_1 | Mus musculus | SM003_BAT_d4_1_Lung | Tissue | SM003-R | SM003_BAT_d4_1_array | RNA | |||
| SM003 | SM003_BAT_d4_2 | Mus musculus | SM003_BAT_d4_2_Lung | Tissue | SM003-R | SM003_BAT_d4_2_array | RNA | |||
| SM003 | SM003_BAT_d4_3 | Mus musculus | SM003_BAT_d4_3_Lung | Tissue | SM003-R | SM003_BAT_d4_3_array | RNA | |||
| SM003 | SM003_BAT_d4_4 | Mus musculus | SM003_BAT_d4_4_Lung | Tissue | SM003-R | SM003_BAT_d4_4_array | RNA | |||
| SM003 | SM003_BAT_d4_5 | Mus musculus | SM003_BAT_d4_5_Lung | Tissue | SM003-R | SM003_BAT_d4_5_array | RNA | |||
| SM003 | SM003_BAT_d7_1 | Mus musculus | SM003_BAT_d7_1_Lung | Tissue | SM003-R | SM003_BAT_d7_1_array | RNA | |||
| SM003 | SM003_BAT_d7_3 | Mus musculus | SM003_BAT_d7_3_Lung | Tissue | SM003-R | SM003_BAT_d7_3_array | RNA | |||
| SM003 | SM003_BAT_d7_4 | Mus musculus | SM003_BAT_d7_4_Lung | Tissue | SM003-R | SM003_BAT_d7_4_array | RNA | |||
| SM003 | SM003_BAT_d7_5 | Mus musculus | SM003_BAT_d7_5_Lung | Tissue | SM003-R | SM003_BAT_d7_5_array | RNA | |||
| SM003 | SM003_Mock_d1_1 | Mus musculus | SM003_Mock_d1_1_Lung | Tissue | SM003-R | SM003_Mock_d1_1_array | RNA | |||
| SM003 | SM003_Mock_d1_2 | Mus musculus | SM003_Mock_d1_2_Lung | Tissue | SM003-R | SM003_Mock_d1_2_array | RNA | |||
| SM003 | SM003_Mock_d1_3 | Mus musculus | SM003_Mock_d1_3_Lung | Tissue | SM003-R | SM003_Mock_d1_3_array | RNA | |||
| SM003 | SM003_Mock_d1_4 | Mus musculus | SM003_Mock_d1_4_Lung | Tissue | SM003-R | SM003_Mock_d1_4_array | RNA | |||
| SM003 | SM003_Mock_d2_1 | Mus musculus | SM003_Mock_d2_1_Lung | Tissue | SM003-R | SM003_Mock_d2_1_array | RNA | |||
| SM003 | SM003_Mock_d2_2 | Mus musculus | SM003_Mock_d2_2_Lung | Tissue | SM003-R | SM003_Mock_d2_2_array | RNA | |||
| SM003 | SM003_Mock_d2_3 | Mus musculus | SM003_Mock_d2_3_Lung | Tissue | SM003-R | SM003_Mock_d2_3_array | RNA | |||
| SM003 | SM003_Mock_d2_4 | Mus musculus | SM003_Mock_d2_4_Lung | Tissue | SM003-R | SM003_Mock_d2_4_array | RNA | |||
| SM003 | SM003_Mock_d4_1 | Mus musculus | SM003_Mock_d4_1_Lung | Tissue | SM003-R | SM003_Mock_d4_1_array | RNA | |||
| SM003 | SM003_Mock_d4_2 | Mus musculus | SM003_Mock_d4_2_Lung | Tissue | SM003-R | SM003_Mock_d4_2_array | RNA | |||
| SM003 | SM003_Mock_d4_3 | Mus musculus | SM003_Mock_d4_3_Lung | Tissue | SM003-R | SM003_Mock_d4_3_array | RNA | |||
| SM003 | SM003_Mock_d4_4 | Mus musculus | SM003_Mock_d4_4_Lung | Tissue | SM003-R | SM003_Mock_d4_4_array | RNA | |||
| SM003 | SM003_Mock_d7_1 | Mus musculus | SM003_Mock_d7_1_Lung | Tissue | SM003-R | SM003_Mock_d7_1_array | RNA | |||
| SM003 | SM003_Mock_d7_2 | Mus musculus | SM003_Mock_d7_2_Lung | Tissue | SM003-R | SM003_Mock_d7_2_array | RNA | |||
| SM003 | SM003_Mock_d7_3 | Mus musculus | SM003_Mock_d7_3_Lung | Tissue | SM003-R | SM003_Mock_d7_3_array | RNA | |||
| SM003 | SM003_Mock_d7_4 | Mus musculus | SM003_Mock_d7_4_Lung | Tissue | SM003-R | SM003_Mock_d7_4_array | RNA | |||
| SM003 | SM003_SARS_d1_1 | Mus musculus | SM003_SARS_d1_1_Lung | Tissue | SM003-R | SM003_SARS_d1_1_array | RNA | |||
| SM003 | SM003_SARS_d1_2 | Mus musculus | SM003_SARS_d1_2_Lung | Tissue | SM003-R | SM003_SARS_d1_2_array | RNA | |||
| SM003 | SM003_SARS_d1_3 | Mus musculus | SM003_SARS_d1_3_Lung | Tissue | SM003-R | SM003_SARS_d1_3_array | RNA | |||
| SM003 | SM003_SARS_d1_4 | Mus musculus | SM003_SARS_d1_4_Lung | Tissue | SM003-R | SM003_SARS_d1_4_array | RNA | |||
| SM003 | SM003_SARS_d2_1 | Mus musculus | SM003_SARS_d2_1_Lung | Tissue | SM003-R | SM003_SARS_d2_1_array | RNA | |||
| SM003 | SM003_SARS_d2_2 | Mus musculus | SM003_SARS_d2_2_Lung | Tissue | SM003-R | SM003_SARS_d2_2_array | RNA | |||
| SM003 | SM003_SARS_d2_3 | Mus musculus | SM003_SARS_d2_3_Lung | Tissue | SM003-R | SM003_SARS_d2_3_array | RNA | |||
| SM003 | SM003_SARS_d2_4 | Mus musculus | SM003_SARS_d2_4_Lung | Tissue | SM003-R | SM003_SARS_d2_4_array | RNA | |||
| SM003 | SM003_SARS_d2_5 | Mus musculus | SM003_SARS_d2_5_Lung | Tissue | SM003-R | SM003_SARS_d2_5_array | RNA | |||
| SM003 | SM003_SARS_d4_1 | Mus musculus | SM003_SARS_d4_1_Lung | Tissue | SM003-R | SM003_SARS_d4_1_array | RNA | |||
| SM003 | SM003_SARS_d4_2 | Mus musculus | SM003_SARS_d4_2_Lung | Tissue | SM003-R | SM003_SARS_d4_2_array | RNA | |||
| SM003 | SM003_SARS_d4_3 | Mus musculus | SM003_SARS_d4_3_Lung | Tissue | SM003-R | SM003_SARS_d4_3_array | RNA | |||
| SM003 | SM003_SARS_d4_4 | Mus musculus | SM003_SARS_d4_4_Lung | Tissue | SM003-R | SM003_SARS_d4_4_array | RNA | |||
| SM003 | SM003_SARS_d4_5 | Mus musculus | SM003_SARS_d4_5_Lung | Tissue | SM003-R | SM003_SARS_d4_5_array | RNA | |||
| SM003 | SM003_SARS_d7_1 | Mus musculus | SM003_SARS_d7_1_Lung | Tissue | SM003-R | SM003_SARS_d7_1_array | RNA | |||
| SM003 | SM003_SARS_d7_2 | Mus musculus | SM003_SARS_d7_2_Lung | Tissue | SM003-R | SM003_SARS_d7_2_array | RNA | |||
| SM003 | SM003_SARS_d7_3 | Mus musculus | SM003_SARS_d7_3_Lung | Tissue | SM003-R | SM003_SARS_d7_3_array | RNA | |||
| SM003 | SM003_SARS_d7_4 | Mus musculus | SM003_SARS_d7_4_Lung | Tissue | SM003-R | SM003_SARS_d7_4_array | RNA | |||
| SM003 | SM003_SARS_d7_5 | Mus musculus | SM003_SARS_d7_5_Lung | Tissue | SM003-R | SM003_SARS_d7_5_array | RNA | |||
| SM004 | SM004_B6_MA15_d4_1 | Mus musculus | SM004_B6_MA15_d4_1_Lung | Tissue | SM004-R | SM004_B6_MA15_d4_1_array | RNA | |||
| SM004 | SM004_B6_MA15_d4_2 | Mus musculus | SM004_B6_MA15_d4_2_Lung | Tissue | SM004-R | SM004_B6_MA15_d4_2_array | RNA | |||
| SM004 | SM004_B6_MA15_d4_3 | Mus musculus | SM004_B6_MA15_d4_3_Lung | Tissue | SM004-R | SM004_B6_MA15_d4_3_array | RNA | |||
| SM004 | SM004_B6_MA15_d4_4 | Mus musculus | SM004_B6_MA15_d4_4_Lung | Tissue | SM004-R | SM004_B6_MA15_d4_4_array | RNA | |||
| SM004 | SM004_B6_MA15_d7_1 | Mus musculus | SM004_B6_MA15_d7_1_Lung | Tissue | SM004-R | SM004_B6_MA15_d7_1_array | RNA | |||
| SM004 | SM004_B6_MA15_d7_3 | Mus musculus | SM004_B6_MA15_d7_3_Lung | Tissue | SM004-R | SM004_B6_MA15_d7_3_array | RNA | |||
| SM004 | SM004_B6_MA15_d7_4 | Mus musculus | SM004_B6_MA15_d7_4_Lung | Tissue | SM004-R | SM004_B6_MA15_d7_4_array | RNA | |||
| SM004 | SM004_B6_Mock_d4_1 | Mus musculus | SM004_B6_Mock_d4_1_Lung | Tissue | SM004-R | SM004_B6_Mock_d4_1_array | RNA | |||
| SM004 | SM004_B6_Mock_d4_2 | Mus musculus | SM004_B6_Mock_d4_2_Lung | Tissue | SM004-R | SM004_B6_Mock_d4_2_array | RNA | |||
| SM004 | SM004_B6_Mock_d4_3 | Mus musculus | SM004_B6_Mock_d4_3_Lung | Tissue | SM004-R | SM004_B6_Mock_d4_3_array | RNA | |||
| SM004 | SM004_B6_Mock_d4_4 | Mus musculus | SM004_B6_Mock_d4_4_Lung | Tissue | SM004-R | SM004_B6_Mock_d4_4_array | RNA | |||
| SM004 | SM004_B6_Mock_d7_1 | Mus musculus | SM004_B6_Mock_d7_1_Lung | Tissue | SM004-R | SM004_B6_Mock_d7_1_array | RNA | |||
| SM004 | SM004_B6_Mock_d7_2 | Mus musculus | SM004_B6_Mock_d7_2_Lung | Tissue | SM004-R | SM004_B6_Mock_d7_2_array | RNA | |||
| SM004 | SM004_B6_Mock_d7_3 | Mus musculus | SM004_B6_Mock_d7_3_Lung | Tissue | SM004-R | SM004_B6_Mock_d7_3_array | RNA | |||
| SM004 | SM004_B6_Mock_d7_4 | Mus musculus | SM004_B6_Mock_d7_4_Lung | Tissue | SM004-R | SM004_B6_Mock_d7_4_array | RNA | |||
| SM004 | SM004_PAI1_MA15_d4_1 | Mus musculus | SM004_PAI1_MA15_d4_1_Lung | Tissue | SM004-R | SM004_PAI1_MA15_d4_1_array | RNA | |||
| SM004 | SM004_PAI1_MA15_d4_2 | Mus musculus | SM004_PAI1_MA15_d4_2_Lung | Tissue | SM004-R | SM004_PAI1_MA15_d4_2_array | RNA | |||
| SM004 | SM004_PAI1_MA15_d4_3 | Mus musculus | SM004_PAI1_MA15_d4_3_Lung | Tissue | SM004-R | SM004_PAI1_MA15_d4_3_array | RNA | |||
| SM004 | SM004_PAI1_MA15_d4_4 | Mus musculus | SM004_PAI1_MA15_d4_4_Lung | Tissue | SM004-R | SM004_PAI1_MA15_d4_4_array | RNA | |||
| SM004 | SM004_PAI1_MA15_d7_1 | Mus musculus | SM004_PAI1_MA15_d7_1_Lung | Tissue | SM004-R | SM004_PAI1_MA15_d7_1_array | RNA | |||
| SM004 | SM004_PAI1_MA15_d7_2 | Mus musculus | SM004_PAI1_MA15_d7_2_Lung | Tissue | SM004-R | SM004_PAI1_MA15_d7_2_array | RNA | |||
| SM004 | SM004_PAI1_MA15_d7_3 | Mus musculus | SM004_PAI1_MA15_d7_3_Lung | Tissue | SM004-R | SM004_PAI1_MA15_d7_3_array | RNA | |||
| SM004 | SM004_PAI1_Mock_d4_1 | Mus musculus | SM004_PAI1_Mock_d4_1_Lung | Tissue | SM004-R | SM004_PAI1_Mock_d4_1_array | RNA | |||
| SM004 | SM004_PAI1_Mock_d4_2 | Mus musculus | SM004_PAI1_Mock_d4_2_Lung | Tissue | SM004-R | SM004_PAI1_Mock_d4_2_array | RNA | |||
| SM004 | SM004_PAI1_Mock_d7_1 | Mus musculus | SM004_PAI1_Mock_d7_1_Lung | Tissue | SM004-R | SM004_PAI1_Mock_d7_1_array | RNA | |||
| SM004 | SM004_PAI1_Mock_d7_2 | Mus musculus | SM004_PAI1_Mock_d7_2_Lung | Tissue | SM004-R | SM004_PAI1_Mock_d7_2_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d4_1 | Mus musculus | SM004_TIMP1_MA15_d4_1_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d4_1_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d4_2 | Mus musculus | SM004_TIMP1_MA15_d4_2_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d4_2_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d4_3 | Mus musculus | SM004_TIMP1_MA15_d4_3_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d4_3_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d4_4 | Mus musculus | SM004_TIMP1_MA15_d4_4_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d4_4_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d7_1 | Mus musculus | SM004_TIMP1_MA15_d7_1_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d7_1_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d7_2 | Mus musculus | SM004_TIMP1_MA15_d7_2_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d7_2_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d7_3 | Mus musculus | SM004_TIMP1_MA15_d7_3_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d7_3_array | RNA | |||
| SM004 | SM004_TIMP1_MA15_d7_4 | Mus musculus | SM004_TIMP1_MA15_d7_4_Lung | Tissue | SM004-R | SM004_TIMP1_MA15_d7_4_array | RNA | |||
| SM004 | SM004_TIMP1_Mock_d4_1 | Mus musculus | SM004_TIMP1_Mock_d4_1_Lung | Tissue | SM004-R | SM004_TIMP1_Mock_d4_1_array | RNA | |||
| SM004 | SM004_TIMP1_Mock_d4_2 | Mus musculus | SM004_TIMP1_Mock_d4_2_Lung | Tissue | SM004-R | SM004_TIMP1_Mock_d4_2_array | RNA | |||
| SM004 | SM004_TIMP1_Mock_d7_1 | Mus musculus | SM004_TIMP1_Mock_d7_1_Lung | Tissue | SM004-R | SM004_TIMP1_Mock_d7_1_array | RNA | |||
| SM004 | SM004_TIMP1_Mock_d7_2 | Mus musculus | SM004_TIMP1_Mock_d7_2_Lung | Tissue | SM004-R | SM004_TIMP1_Mock_d7_2_array | RNA | |||
| SM007 | SM007_B6_MA15_d2_1 | Mus musculus | SM007_B6_MA15_d2_1_Lung | Tissue | SM007-R | SM007_B6_MA15_d2_1_array | RNA | |||
| SM007 | SM007_B6_MA15_d2_2 | Mus musculus | SM007_B6_MA15_d2_2_Lung | Tissue | SM007-R | SM007_B6_MA15_d2_2_array | RNA | |||
| SM007 | SM007_B6_MA15_d2_3 | Mus musculus | SM007_B6_MA15_d2_3_Lung | Tissue | SM007-R | SM007_B6_MA15_d2_3_array | RNA | |||
| SM007 | SM007_B6_MA15_d4_1 | Mus musculus | SM007_B6_MA15_d4_1_Lung | Tissue | SM007-R | SM007_B6_MA15_d4_1_array | RNA | |||
| SM007 | SM007_B6_MA15_d4_2 | Mus musculus | SM007_B6_MA15_d4_2_Lung | Tissue | SM007-R | SM007_B6_MA15_d4_2_array | RNA | |||
| SM007 | SM007_B6_MA15_d7_1 | Mus musculus | SM007_B6_MA15_d7_1_Lung | Tissue | SM007-R | SM007_B6_MA15_d7_1_array | RNA | |||
| SM007 | SM007_B6_MA15_d7_2 | Mus musculus | SM007_B6_MA15_d7_2_Lung | Tissue | SM007-R | SM007_B6_MA15_d7_2_array | RNA | |||
| SM007 | SM007_B6_MA15_d7_3 | Mus musculus | SM007_B6_MA15_d7_3_Lung | Tissue | SM007-R | SM007_B6_MA15_d7_3_array | RNA | |||
| SM007 | SM007_B6_Mock_d2_1 | Mus musculus | SM007_B6_Mock_d2_1_Lung | Tissue | SM007-R | SM007_B6_Mock_d2_1_array | RNA | |||
| SM007 | SM007_B6_Mock_d2_2 | Mus musculus | SM007_B6_Mock_d2_2_Lung | Tissue | SM007-R | SM007_B6_Mock_d2_2_array | RNA | |||
| SM007 | SM007_B6_Mock_d2_3 | Mus musculus | SM007_B6_Mock_d2_3_Lung | Tissue | SM007-R | SM007_B6_Mock_d2_3_array | RNA | |||
| SM007 | SM007_B6_Mock_d4_1 | Mus musculus | SM007_B6_Mock_d4_1_Lung | Tissue | SM007-R | SM007_B6_Mock_d4_1_array | RNA | |||
| SM007 | SM007_B6_Mock_d4_2 | Mus musculus | SM007_B6_Mock_d4_2_Lung | Tissue | SM007-R | SM007_B6_Mock_d4_2_array | RNA | |||
| SM007 | SM007_B6_Mock_d4_3 | Mus musculus | SM007_B6_Mock_d4_3_Lung | Tissue | SM007-R | SM007_B6_Mock_d4_3_array | RNA | |||
| SM007 | SM007_B6_Mock_d7_1 | Mus musculus | SM007_B6_Mock_d7_1_Lung | Tissue | SM007-R | SM007_B6_Mock_d7_1_array | RNA | |||
| SM007 | SM007_B6_Mock_d7_2 | Mus musculus | SM007_B6_Mock_d7_2_Lung | Tissue | SM007-R | SM007_B6_Mock_d7_2_array | RNA | |||
| SM007 | SM007_B6_Mock_d7_3 | Mus musculus | SM007_B6_Mock_d7_3_Lung | Tissue | SM007-R | SM007_B6_Mock_d7_3_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d2_1 | Mus musculus | SM007_CXCR3_MA15_d2_1_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d2_1_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d2_2 | Mus musculus | SM007_CXCR3_MA15_d2_2_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d2_2_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d2_3 | Mus musculus | SM007_CXCR3_MA15_d2_3_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d2_3_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d4_2 | Mus musculus | SM007_CXCR3_MA15_d4_2_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d4_2_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d4_3 | Mus musculus | SM007_CXCR3_MA15_d4_3_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d4_3_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d7_1 | Mus musculus | SM007_CXCR3_MA15_d7_1_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d7_1_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d7_2 | Mus musculus | SM007_CXCR3_MA15_d7_2_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d7_2_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d7_3 | Mus musculus | SM007_CXCR3_MA15_d7_3_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d7_3_array | RNA | |||
| SM007 | SM007_CXCR3_MA15_d7_4 | Mus musculus | SM007_CXCR3_MA15_d7_4_Lung | Tissue | SM007-R | SM007_CXCR3_MA15_d7_4_array | RNA | |||
| SM007 | SM007_CXCR3_Mock_d2_2 | Mus musculus | SM007_CXCR3_Mock_d2_2_Lung | Tissue | SM007-R | SM007_CXCR3_Mock_d2_2_array | RNA | |||
| SM007 | SM007_CXCR3_Mock_d2_3 | Mus musculus | SM007_CXCR3_Mock_d2_3_Lung | Tissue | SM007-R | SM007_CXCR3_Mock_d2_3_array | RNA | |||
| SM007 | SM007_CXCR3_Mock_d4_1 | Mus musculus | SM007_CXCR3_Mock_d4_1_Lung | Tissue | SM007-R | SM007_CXCR3_Mock_d4_1_array | RNA | |||
| SM007 | SM007_CXCR3_Mock_d4_2 | Mus musculus | SM007_CXCR3_Mock_d4_2_Lung | Tissue | SM007-R | SM007_CXCR3_Mock_d4_2_array | RNA | |||
| SM007 | SM007_CXCR3_Mock_d7_1 | Mus musculus | SM007_CXCR3_Mock_d7_1_Lung | Tissue | SM007-R | SM007_CXCR3_Mock_d7_1_array | RNA | |||
| SM007 | SM007_CXCR3_Mock_d7_2 | Mus musculus | SM007_CXCR3_Mock_d7_2_Lung | Tissue | SM007-R | SM007_CXCR3_Mock_d7_2_array | RNA | |||
| SM007 | SM007_CXCR3_Mock_d7_3 | Mus musculus | SM007_CXCR3_Mock_d7_3_Lung | Tissue | SM007-R | SM007_CXCR3_Mock_d7_3_array | RNA | |||
| SM009 | SM009_B6_MA15_d4_2 | Mus musculus | SM009_B6_MA15_d4_2_Lung | Tissue | SM009-R | SM009_B6_MA15_d4_2_array | RNA | |||
| SM009 | SM009_B6_MA15_d4_3 | Mus musculus | SM009_B6_MA15_d4_3_Lung | Tissue | SM009-R | SM009_B6_MA15_d4_3_array | RNA | |||
| SM009 | SM009_B6_MA15_d4_4 | Mus musculus | SM009_B6_MA15_d4_4_Lung | Tissue | SM009-R | SM009_B6_MA15_d4_4_array | RNA | |||
| SM009 | SM009_B6_MA15_d7_2 | Mus musculus | SM009_B6_MA15_d7_2_Lung | Tissue | SM009-R | SM009_B6_MA15_d7_2_array | RNA | |||
| SM009 | SM009_B6_MA15_d7_4 | Mus musculus | SM009_B6_MA15_d7_4_Lung | Tissue | SM009-R | SM009_B6_MA15_d7_4_array | RNA | |||
| SM009 | SM009_B6_Mock_d4_1 | Mus musculus | SM009_B6_Mock_d4_1_Lung | Tissue | SM009-R | SM009_B6_Mock_d4_1_array | RNA | |||
| SM009 | SM009_B6_Mock_d4_2 | Mus musculus | SM009_B6_Mock_d4_2_Lung | Tissue | SM009-R | SM009_B6_Mock_d4_2_array | RNA | |||
| SM009 | SM009_B6_Mock_d7_1 | Mus musculus | SM009_B6_Mock_d7_1_Lung | Tissue | SM009-R | SM009_B6_Mock_d7_1_array | RNA | |||
| SM009 | SM009_B6_Mock_d7_2 | Mus musculus | SM009_B6_Mock_d7_2_Lung | Tissue | SM009-R | SM009_B6_Mock_d7_2_array | RNA | |||
| SM009 | SM009_PLAT_MA15_d4_3 | Mus musculus | SM009_PLAT_MA15_d4_3_Lung | Tissue | SM009-R | SM009_PLAT_MA15_d4_3_array | RNA | |||
| SM009 | SM009_PLAT_MA15_d4_4 | Mus musculus | SM009_PLAT_MA15_d4_4_Lung | Tissue | SM009-R | SM009_PLAT_MA15_d4_4_array | RNA | |||
| SM009 | SM009_PLAT_MA15_d7_1 | Mus musculus | SM009_PLAT_MA15_d7_1_Lung | Tissue | SM009-R | SM009_PLAT_MA15_d7_1_array | RNA | |||
| SM009 | SM009_PLAT_MA15_d7_2 | Mus musculus | SM009_PLAT_MA15_d7_2_Lung | Tissue | SM009-R | SM009_PLAT_MA15_d7_2_array | RNA | |||
| SM009 | SM009_PLAT_MA15_d7_4 | Mus musculus | SM009_PLAT_MA15_d7_4_Lung | Tissue | SM009-R | SM009_PLAT_MA15_d7_4_array | RNA | |||
| SM009 | SM009_PLAT_Mock_d4_1 | Mus musculus | SM009_PLAT_Mock_d4_1_Lung | Tissue | SM009-R | SM009_PLAT_Mock_d4_1_array | RNA | |||
| SM009 | SM009_PLAT_Mock_d4_2 | Mus musculus | SM009_PLAT_Mock_d4_2_Lung | Tissue | SM009-R | SM009_PLAT_Mock_d4_2_array | RNA | |||
| SM009 | SM009_PLAT_Mock_d7_1 | Mus musculus | SM009_PLAT_Mock_d7_1_Lung | Tissue | SM009-R | SM009_PLAT_Mock_d7_1_array | RNA | |||
| SM009 | SM009_PLAT_Mock_d7_2 | Mus musculus | SM009_PLAT_Mock_d7_2_Lung | Tissue | SM009-R | SM009_PLAT_Mock_d7_2_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d1_1 | Mus musculus | SM012_dORF6_10^5_d1_1_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_1d_1_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d1_3 | Mus musculus | SM012_dORF6_10^5_d1_3_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_1d_3_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d1_5 | Mus musculus | SM012_dORF6_10^5_d1_5_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_1d_5_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d2_1 | Mus musculus | SM012_dORF6_10^5_d2_1_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_2d_1_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d2_3 | Mus musculus | SM012_dORF6_10^5_d2_3_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_2d_3_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d2_4 | Mus musculus | SM012_dORF6_10^5_d2_4_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_2d_4_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d4_1 | Mus musculus | SM012_dORF6_10^5_d4_1_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_4d_1_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d4_2 | Mus musculus | SM012_dORF6_10^5_d4_2_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_4d_2_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d4_3 | Mus musculus | SM012_dORF6_10^5_d4_3_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_4d_3_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d7_1 | Mus musculus | SM012_dORF6_10^5_d7_1_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_7d_1_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d7_2 | Mus musculus | SM012_dORF6_10^5_d7_2_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_7d_2_array | RNA | |||
| SM012 | SM012_dORF6_10^5_d7_3 | Mus musculus | SM012_dORF6_10^5_d7_3_Lung | Tissue | SM012-R | SM012_dORF6_10^5pfu_7d_3_array | RNA | |||
| SM012 | SM012_MA15_10^5_d1_2 | Mus musculus | SM012_MA15_10^5_d1_2_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_1d_2_array | RNA | |||
| SM012 | SM012_MA15_10^5_d1_3 | Mus musculus | SM012_MA15_10^5_d1_3_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_1d_3_array | RNA | |||
| SM012 | SM012_MA15_10^5_d1_4 | Mus musculus | SM012_MA15_10^5_d1_4_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_1d_4_array | RNA | |||
| SM012 | SM012_MA15_10^5_d2_1 | Mus musculus | SM012_MA15_10^5_d2_1_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_2d_1_array | RNA | |||
| SM012 | SM012_MA15_10^5_d2_3 | Mus musculus | SM012_MA15_10^5_d2_3_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_2d_3_array | RNA | |||
| SM012 | SM012_MA15_10^5_d2_5 | Mus musculus | SM012_MA15_10^5_d2_5_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_2d_5_array | RNA | |||
| SM012 | SM012_MA15_10^5_d4_1 | Mus musculus | SM012_MA15_10^5_d4_1_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_4d_1_array | RNA | |||
| SM012 | SM012_MA15_10^5_d4_2 | Mus musculus | SM012_MA15_10^5_d4_2_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_4d_2_array | RNA | |||
| SM012 | SM012_MA15_10^5_d4_3 | Mus musculus | SM012_MA15_10^5_d4_3_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_4d_3_array | RNA | |||
| SM012 | SM012_MA15_10^5_d7_1 | Mus musculus | SM012_MA15_10^5_d7_1_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_7d_1_array | RNA | |||
| SM012 | SM012_MA15_10^5_d7_2 | Mus musculus | SM012_MA15_10^5_d7_2_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_7d_2_array | RNA | |||
| SM012 | SM012_MA15_10^5_d7_3 | Mus musculus | SM012_MA15_10^5_d7_3_Lung | Tissue | SM012-R | SM012_MA15_10^5pfu_7d_3_array | RNA | |||
| SM012 | SM012_Mock_d1_1 | Mus musculus | SM012_Mock_d1_1_Lung | Tissue | SM012-R | SM012_Mock_1d_1_array | RNA | |||
| SM012 | SM012_Mock_d1_4 | Mus musculus | SM012_Mock_d1_4_Lung | Tissue | SM012-R | SM012_Mock_1d_4_array | RNA | |||
| SM012 | SM012_Mock_d1_5 | Mus musculus | SM012_Mock_d1_5_Lung | Tissue | SM012-R | SM012_Mock_1d_5_array | RNA | |||
| SM012 | SM012_Mock_d2_2 | Mus musculus | SM012_Mock_d2_2_Lung | Tissue | SM012-R | SM012_Mock_2d_2_array | RNA | |||
| SM012 | SM012_Mock_d2_3 | Mus musculus | SM012_Mock_d2_3_Lung | Tissue | SM012-R | SM012_Mock_2d_3_array | RNA | |||
| SM012 | SM012_Mock_d2_4 | Mus musculus | SM012_Mock_d2_4_Lung | Tissue | SM012-R | SM012_Mock_2d_4_array | RNA | |||
| SM012 | SM012_Mock_d4_1 | Mus musculus | SM012_Mock_d4_1_Lung | Tissue | SM012-R | SM012_Mock_4d_1_array | RNA | |||
| SM012 | SM012_Mock_d4_2 | Mus musculus | SM012_Mock_d4_2_Lung | Tissue | SM012-R | SM012_Mock_4d_2_array | RNA | |||
| SM012 | SM012_Mock_d4_3 | Mus musculus | SM012_Mock_d4_3_Lung | Tissue | SM012-R | SM012_Mock_4d_3_array | RNA | |||
| SM012 | SM012_Mock_d7_1 | Mus musculus | SM012_Mock_d7_1_Lung | Tissue | SM012-R | SM012_Mock_7d_1_array | RNA | |||
| SM012 | SM012_Mock_d7_3 | Mus musculus | SM012_Mock_d7_3_Lung | Tissue | SM012-R | SM012_Mock_7d_3_array | RNA | |||
| SM014 | SM014_MA15_10^5_d1_1 | Mus musculus | SM014_MA15_10^5_d1_1_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_1d_1_array | RNA | |||
| SM014 | SM014_MA15_10^5_d1_2 | Mus musculus | SM014_MA15_10^5_d1_2_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_1d_2_array | RNA | |||
| SM014 | SM014_MA15_10^5_d1_3 | Mus musculus | SM014_MA15_10^5_d1_3_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_1d_3_array | RNA | |||
| SM014 | SM014_MA15_10^5_d1_4 | Mus musculus | SM014_MA15_10^5_d1_4_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_1d_4_array | RNA | |||
| SM014 | SM014_MA15_10^5_d2_1 | Mus musculus | SM014_MA15_10^5_d2_1_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_2d_1_array | RNA | |||
| SM014 | SM014_MA15_10^5_d2_2 | Mus musculus | SM014_MA15_10^5_d2_2_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_2d_2_array | RNA | |||
| SM014 | SM014_MA15_10^5_d2_3 | Mus musculus | SM014_MA15_10^5_d2_3_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_2d_3_array | RNA | |||
| SM014 | SM014_MA15_10^5_d2_4 | Mus musculus | SM014_MA15_10^5_d2_4_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_2d_4_array | RNA | |||
| SM014 | SM014_MA15_10^5_d4_1 | Mus musculus | SM014_MA15_10^5_d4_1_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_4d_1_array | RNA | |||
| SM014 | SM014_MA15_10^5_d4_2 | Mus musculus | SM014_MA15_10^5_d4_2_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_4d_2_array | RNA | |||
| SM014 | SM014_MA15_10^5_d4_3 | Mus musculus | SM014_MA15_10^5_d4_3_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_4d_3_array | RNA | |||
| SM014 | SM014_MA15_10^5_d4_4 | Mus musculus | SM014_MA15_10^5_d4_4_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_4d_4_array | RNA | |||
| SM014 | SM014_MA15_10^5_d7_1 | Mus musculus | SM014_MA15_10^5_d7_1_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_7d_1_array | RNA | |||
| SM014 | SM014_MA15_10^5_d7_2 | Mus musculus | SM014_MA15_10^5_d7_2_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_7d_2_array | RNA | |||
| SM014 | SM014_MA15_10^5_d7_4 | Mus musculus | SM014_MA15_10^5_d7_4_Lung | Tissue | SM014-R | SM014_MA15_10^5pfu_7d_4_array | RNA | |||
| SM014 | SM014_MA15_10^5_d7_5 | Mus musculus | SM014_MA15_10^5_d7_5_Lung | Tissue | SM014-R | RNA | ||||
| SM014 | SM014_Mock_d1_1 | Mus musculus | SM014_Mock_d1_1_Lung | Tissue | SM014-R | RNA | ||||
| SM014 | SM014_Mock_d1_2 | Mus musculus | SM014_Mock_d1_2_Lung | Tissue | SM014-R | SM014_Mock_1d_2_array | RNA | |||
| SM014 | SM014_Mock_d1_3 | Mus musculus | SM014_Mock_d1_3_Lung | Tissue | SM014-R | SM014_Mock_1d_3_array | RNA | |||
| SM014 | SM014_Mock_d2_1 | Mus musculus | SM014_Mock_d2_1_Lung | Tissue | SM014-R | SM014_Mock_2d_1_array | RNA | |||
| SM014 | SM014_Mock_d2_2 | Mus musculus | SM014_Mock_d2_2_Lung | Tissue | SM014-R | SM014_Mock_2d_2_array | RNA | |||
| SM014 | SM014_Mock_d2_3 | Mus musculus | SM014_Mock_d2_3_Lung | Tissue | SM014-R | SM014_Mock_2d_3_array | RNA | |||
| SM014 | SM014_Mock_d4_1 | Mus musculus | SM014_Mock_d4_1_Lung | Tissue | SM014-R | SM014_Mock_4d_1_array | RNA | |||
| SM014 | SM014_Mock_d4_2 | Mus musculus | SM014_Mock_d4_2_Lung | Tissue | SM014-R | SM014_Mock_4d_2_array | RNA | |||
| SM014 | SM014_Mock_d4_3 | Mus musculus | SM014_Mock_d4_3_Lung | Tissue | SM014-R | SM014_Mock_4d_3_array | RNA | |||
| SM014 | SM014_Mock_d7_1 | Mus musculus | SM014_Mock_d7_1_Lung | Tissue | SM014-R | SM014_Mock_7d_1_array | RNA | |||
| SM014 | SM014_Mock_d7_2 | Mus musculus | SM014_Mock_d7_2_Lung | Tissue | SM014-R | SM014_Mock_7d_2_array | RNA | |||
| SM014 | SM014_Mock_d7_3 | Mus musculus | SM014_Mock_d7_3_Lung | Tissue | SM014-R | SM014_Mock_7d_3_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d1_2 | Mus musculus | SM014_nsp16_10^5_d1_2_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_1d_2_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d1_3 | Mus musculus | SM014_nsp16_10^5_d1_3_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_1d_3_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d1_4 | Mus musculus | SM014_nsp16_10^5_d1_4_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_1d_4_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d2_1 | Mus musculus | SM014_nsp16_10^5_d2_1_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_2d_1_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d2_2 | Mus musculus | SM014_nsp16_10^5_d2_2_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_2d_2_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d2_3 | Mus musculus | SM014_nsp16_10^5_d2_3_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_2d_3_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d2_4 | Mus musculus | SM014_nsp16_10^5_d2_4_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_2d_4_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d4_1 | Mus musculus | SM014_nsp16_10^5_d4_1_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_4d_1_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d4_3 | Mus musculus | SM014_nsp16_10^5_d4_3_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_4d_3_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d4_4 | Mus musculus | SM014_nsp16_10^5_d4_4_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_4d_4_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d7_1 | Mus musculus | SM014_nsp16_10^5_d7_1_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_7d_1_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d7_2 | Mus musculus | SM014_nsp16_10^5_d7_2_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_7d_2_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d7_3 | Mus musculus | SM014_nsp16_10^5_d7_3_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_7d_3_array | RNA | |||
| SM014 | SM014_nsp16_10^5_d7_4 | Mus musculus | SM014_nsp16_10^5_d7_4_Lung | Tissue | SM014-R | SM014_nsp16_10^5pfu_7d_4_array | RNA | |||
| SM015 | SM015_B6_MA15_d4_1 | Mus musculus | SM015_B6_MA15_d4_1_Lung | Tissue | SM015-R | RNA | ||||
| SM015 | SM015_B6_MA15_d4_2 | Mus musculus | SM015_B6_MA15_d4_2_Lung | Tissue | SM015-R | SM015_B6_MA15_d4_2_array | RNA | |||
| SM015 | SM015_B6_MA15_d7_1 | Mus musculus | SM015_B6_MA15_d7_1_Lung | Tissue | SM015-R | SM015_B6_MA15_d7_1_array | RNA | |||
| SM015 | SM015_B6_MA15_d7_2 | Mus musculus | SM015_B6_MA15_d7_2_Lung | Tissue | SM015-R | SM015_B6_MA15_d7_2_array | RNA | |||
| SM015 | SM015_B6_MA15_d7_3 | Mus musculus | SM015_B6_MA15_d7_3_Lung | Tissue | SM015-R | SM015_B6_MA15_d7_3_array | RNA | |||
| SM015 | SM015_B6_Mock_d4_1 | Mus musculus | SM015_B6_Mock_d4_1_Lung | Tissue | SM015-R | SM015_B6_Mock_d4_1_array | RNA | |||
| SM015 | SM015_B6_Mock_d4_2 | Mus musculus | SM015_B6_Mock_d4_2_Lung | Tissue | SM015-R | SM015_B6_Mock_d4_2_array | RNA | |||
| SM015 | SM015_B6_Mock_d7_1 | Mus musculus | SM015_B6_Mock_d7_1_Lung | Tissue | SM015-R | SM015_B6_Mock_d7_1_array | RNA | |||
| SM015 | SM015_B6_Mock_d7_2 | Mus musculus | SM015_B6_Mock_d7_2_Lung | Tissue | SM015-R | SM015_B6_Mock_d7_2_array | RNA | |||
| SM015 | SM015_B6_Mock_d7_3 | Mus musculus | SM015_B6_Mock_d7_3_Lung | Tissue | SM015-R | SM015_B6_Mock_d7_3_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_MA15_d4_1 | Mus musculus | SM015_Tnfrsf1a/1b_MA15_d4_1_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_MA15_d4_1_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_MA15_d4_2 | Mus musculus | SM015_Tnfrsf1a/1b_MA15_d4_2_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_MA15_d4_2_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_MA15_d7_1 | Mus musculus | SM015_Tnfrsf1a/1b_MA15_d7_1_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_MA15_d7_1_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_MA15_d7_2 | Mus musculus | SM015_Tnfrsf1a/1b_MA15_d7_2_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_MA15_d7_2_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_MA15_d7_3 | Mus musculus | SM015_Tnfrsf1a/1b_MA15_d7_3_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_MA15_d7_3_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_Mock_d4_1 | Mus musculus | SM015_Tnfrsf1a/1b_Mock_d4_1_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_Mock_d4_1_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_Mock_d4_2 | Mus musculus | SM015_Tnfrsf1a/1b_Mock_d4_2_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_Mock_d4_2_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_Mock_d7_1 | Mus musculus | SM015_Tnfrsf1a/1b_Mock_d7_1_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_Mock_d7_1_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_Mock_d7_2 | Mus musculus | SM015_Tnfrsf1a/1b_Mock_d7_2_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_Mock_d7_2_array | RNA | |||
| SM015 | SM015_Tnfrsf1a/1b_Mock_d7_3 | Mus musculus | SM015_Tnfrsf1a/1b_Mock_d7_3_Lung | Tissue | SM015-R | SM015_Tnfrsf1a/1b_Mock_d7_3_array | RNA | |||
| SM019 | SM019_B6_MA15_d4_1 | Mus musculus | SM019_B6_MA15_d4_1_Lung | Tissue | SM019-R | SM019_B6_MA15_d4_1_array | RNA | |||
| SM019 | SM019_B6_MA15_d4_2 | Mus musculus | SM019_B6_MA15_d4_2_Lung | Tissue | SM019-R | SM019_B6_MA15_d4_2_array | RNA | |||
| SM019 | SM019_B6_MA15_d4_3 | Mus musculus | SM019_B6_MA15_d4_3_Lung | Tissue | SM019-R | SM019_B6_MA15_d4_3_array | RNA | |||
| SM019 | SM019_B6_MA15_d7_1 | Mus musculus | SM019_B6_MA15_d7_1_Lung | Tissue | SM019-R | SM019_B6_MA15_d7_1_array | RNA | |||
| SM019 | SM019_B6_MA15_d7_2 | Mus musculus | SM019_B6_MA15_d7_2_Lung | Tissue | SM019-R | SM019_B6_MA15_d7_2_array | RNA | |||
| SM019 | SM019_B6_MA15_d7_3 | Mus musculus | SM019_B6_MA15_d7_3_Lung | Tissue | SM019-R | SM019_B6_MA15_d7_3_array | RNA | |||
| SM019 | SM019_B6_Mock_d4_1 | Mus musculus | SM019_B6_Mock_d4_1_Lung | Tissue | SM019-R | SM019_B6_Mock_d4_1_array | RNA | |||
| SM019 | SM019_B6_Mock_d4_2 | Mus musculus | SM019_B6_Mock_d4_2_Lung | Tissue | SM019-R | SM019_B6_Mock_d4_2_array | RNA | |||
| SM019 | SM019_B6_Mock_d4_3 | Mus musculus | SM019_B6_Mock_d4_3_Lung | Tissue | SM019-R | SM019_B6_Mock_d4_3_array | RNA | |||
| SM019 | SM019_B6_Mock_d7_1 | Mus musculus | SM019_B6_Mock_d7_1_Lung | Tissue | SM019-R | SM019_B6_Mock_d7_1_array | RNA | |||
| SM019 | SM019_B6_Mock_d7_2 | Mus musculus | SM019_B6_Mock_d7_2_Lung | Tissue | SM019-R | SM019_B6_Mock_d7_2_array | RNA | |||
| SM019 | SM019_B6_Mock_d7_3 | Mus musculus | SM019_B6_Mock_d7_3_Lung | Tissue | SM019-R | SM019_B6_Mock_d7_3_array | RNA | |||
| SM019 | SM019_Tnfrsf1b_MA15_d4_1 | Mus musculus | SM019_Tnfrsf1b_MA15_d4_1_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_MA15_d4_1_array | RNA | |||
| SM019 | SM019_Tnfrsf1b_MA15_d4_2 | Mus musculus | SM019_Tnfrsf1b_MA15_d4_2_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_MA15_d4_2_array | RNA | |||
| SM019 | SM019_Tnfrsf1b_MA15_d4_3 | Mus musculus | SM019_Tnfrsf1b_MA15_d4_3_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_MA15_d4_3_array | RNA | |||
| SM019 | SM019_Tnfrsf1b_MA15_d7_2 | Mus musculus | SM019_Tnfrsf1b_MA15_d7_2_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_MA15_d7_2_array | RNA | |||
| SM019 | SM019_Tnfrsf1b_MA15_d7_3 | Mus musculus | SM019_Tnfrsf1b_MA15_d7_3_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_MA15_d7_3_array | RNA | |||
| SM019 | SM019_Tnfrsf1b_Mock_d4_1 | Mus musculus | SM019_Tnfrsf1b_Mock_d4_1_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_Mock_d4_1_array | ||||
| SM019 | SM019_Tnfrsf1b_Mock_d4_2 | Mus musculus | SM019_Tnfrsf1b_Mock_d4_2_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_Mock_d4_2_array | ||||
| SM019 | SM019_Tnfrsf1b_Mock_d4_3 | Mus musculus | SM019_Tnfrsf1b_Mock_d4_3_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_Mock_d4_3_array | ||||
| SM019 | SM019_Tnfrsf1b_Mock_d7_2 | Mus musculus | SM019_Tnfrsf1b_Mock_d7_2_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_Mock_d7_2_array | ||||
| SM019 | SM019_Tnfrsf1b_Mock_d7_3 | Mus musculus | SM019_Tnfrsf1b_Mock_d7_3_Lung | Tissue | SM019-R | SM019_Tnfrsf1b_Mock_d7_3_array | ||||
| SM020 | SM020_B6_MA15_d4_1 | Mus musculus | SM020_B6_MA15_d4_1_Lung | Tissue | SM020-R | SM020_B6_MA15_d4_1_array | ||||
| SM020 | SM020_B6_MA15_d4_2 | Mus musculus | SM020_B6_MA15_d4_2_Lung | Tissue | SM020-R | SM020_B6_MA15_d4_2_array | ||||
| SM020 | SM020_B6_MA15_d7_1 | Mus musculus | SM020_B6_MA15_d7_1_Lung | Tissue | SM020-R | SM020_B6_MA15_d7_1_array | ||||
| SM020 | SM020_B6_MA15_d7_2 | Mus musculus | SM020_B6_MA15_d7_2_Lung | Tissue | SM020-R | SM020_B6_MA15_d7_2_array | ||||
| SM020 | SM020_B6_MA15_d7_3 | Mus musculus | SM020_B6_MA15_d7_3_Lung | Tissue | SM020-R | SM020_B6_MA15_d7_3_array | ||||
| SM020 | SM020_B6_Mock_d4_1 | Mus musculus | SM020_B6_Mock_d4_1_Lung | Tissue | SM020-R | SM020_B6_Mock_d4_1_array | ||||
| SM020 | SM020_B6_Mock_d4_2 | Mus musculus | SM020_B6_Mock_d4_2_Lung | Tissue | SM020-R | SM020_B6_Mock_d4_2_array | ||||
| SM020 | SM020_B6_Mock_d7_1 | Mus musculus | SM020_B6_Mock_d7_1_Lung | Tissue | SM020-R | SM020_B6_Mock_d7_1_array | ||||
| SM020 | SM020_B6_Mock_d7_2 | Mus musculus | SM020_B6_Mock_d7_2_Lung | Tissue | SM020-R | SM020_B6_Mock_d7_2_array | ||||
| SM020 | SM020_B6_Mock_d7_3 | Mus musculus | SM020_B6_Mock_d7_3_Lung | Tissue | SM020-R | SM020_B6_Mock_d7_3_array | ||||
| SM020 | SM020_ppp1r14c_MA15_d4_1 | Mus musculus | SM020_ppp1r14c_MA15_d4_1_Lung | Tissue | SM020-R | SM020_ppp1r14c_MA15_d4_1_array | ||||
| SM020 | SM020_ppp1r14c_MA15_d4_2 | Mus musculus | SM020_ppp1r14c_MA15_d4_2_Lung | Tissue | SM020-R | SM020_ppp1r14c_MA15_d4_2_array | ||||
| SM020 | SM020_ppp1r14c_MA15_d7_2 | Mus musculus | SM020_ppp1r14c_MA15_d7_2_Lung | Tissue | SM020-R | SM020_ppp1r14c_MA15_d7_2_array | ||||
| SM020 | SM020_ppp1r14c_MA15_d7_3 | Mus musculus | SM020_ppp1r14c_MA15_d7_3_Lung | Tissue | SM020-R | SM020_ppp1r14c_MA15_d7_3_array | ||||
| SM020 | SM020_ppp1r14c_Mock_d4_1 | Mus musculus | SM020_ppp1r14c_Mock_d4_1_Lung | Tissue | SM020-R | SM020_ppp1r14c_Mock_d4_1_array | ||||
| SM020 | SM020_ppp1r14c_Mock_d4_2 | Mus musculus | SM020_ppp1r14c_Mock_d4_2_Lung | Tissue | SM020-R | SM020_ppp1r14c_Mock_d4_2_array | ||||
| SM020 | SM020_ppp1r14c_Mock_d7_1 | Mus musculus | SM020_ppp1r14c_Mock_d7_1_Lung | Tissue | SM020-R | SM020_ppp1r14c_Mock_d7_1_array | ||||
| SM020 | SM020_ppp1r14c_Mock_d7_2 | Mus musculus | SM020_ppp1r14c_Mock_d7_2_Lung | Tissue | SM020-R | SM020_ppp1r14c_Mock_d7_2_array | ||||
| SM020 | SM020_ppp1r14c_Mock_d7_3 | Mus musculus | SM020_ppp1r14c_Mock_d7_3_Lung | Tissue | SM020-R | SM020_ppp1r14c_Mock_d7_3_array |
Viruses
Within the studies reported here, the host response to viral agents from two major viral families, Orthomyxoviridae and Coronaviridae, are characterized. From Orthomyxoviridae, two influenza A subtypes H5N1 and H1N1 were chosen in order to investigate influenza viruses with higher and lower reported pathogenicity, respectively. Highly Pathogenic Avian Influenza H5N1 (HPAI) has been commonly referred to as ‘Avian Flu’ as it is endemic in bird populations. HPAI is known to exhibit ‘stuttering transmission’ in humans, with a relatively high mortality rates (>50%), but to-date has not acquired efficient human-to-human transmission. This high mortality rate together with its pathological similarities to the 1918 H1N1 ‘Spanish Flu’ make it an important model to study and prevent future lethal pandemics. On the other hand, the more recent seasonal and pandemic H1N1 strains are considered to have lower pathogenicity, rarely causing severe disease or death. Since its emergence, pandemic H1N1 has been found to recur in the human population as a seasonal influenza. H1N1 influenza isolates used in these studies include recent pandemic strains, previous seasonal strains, and a reconstructed 1918 strain.
Virus preparations for the Influenza A/Vietnam/1203/2004 (H5N1) (abbreviated as VN1203) isolate were derived from a plasmid-based reverse-genetic system as previously described17,18. Four A/Viet Nam/1203/2004 mutant viruses that contain modified pathogenicity determinants were similarly generated by reverse genetics coupled with site-directed mutagenesis17–19, including H5N1 VN1203-HAavir in which the hemagglutinin (HA) surface protein poly-basic cleavage site was mutated20; H5N1 VN1203-PB2627E which possesses a K to E amino acid substitution at residue 627 of the PB2 protein21; H5N1 VN1203-NS1trunc, in which a stop codon was introduced at amino acid 124 of the NS1 protein open reading frame8; and H5N1 VN1203-PB1F2del, in which the PB1-F2 open reading frame was eliminated8. Additional natural influenza strains used in these studies include: A/California/04/2009 (H1N1) (abbreviated as CA04), A/Netherlands/602/2009 (H1N1) (abbreviated as NL602; kindly provided by Ron Fouchier, Erasmus Medical Center, Rotterdam, Netherlands)22, A/Mexico/4482/09 (H1N1) (Mex/4482), A/New Jersey/8/1976 (H1N1) (NJ/8/76), and A/Brisbane/59/07 (H1N1) (Brisbane/59/07)6. The reconstructed r1918 strain was also included in one study23. All virus strains were propagated and amplified in Madin-Darby canine kidney (MDCK) cells at 37° C in an atmosphere of 5% CO2, except NJ/8/76, which was propagated in 10-day-old embryonated hens’ eggs6. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, IM009, IM010, SBRI_AA_E1).
The PR8, X31, and VN1203(6+2) influenza A viruses were generated using the 8-plasmid influenza reverse genetics system24. All three viruses are based on PR8 (A/Puerto Rico/8/34), a mouse-adapted H1N1 strain originally derived from a human isolate. X31 is a mouse adapted H3N2 strain with the 6 internal genes of PR8 and the HA and NA derived genes from A/Aichi/2/1968. rgVN1203(6+2) (VN) contains the 6 internal genes of PR8 and the H5N1 HA and NA genes from A/Vietnam/1203/2004 with the polybasic cleavage site of the HA modified to restrict its replication to the airway25,26. (SBRI_AA_E1).
In addition to the influenza A strains, the studies reported here investigate recently characterized Coronaviridae strains. These strains are all members of the betacoronavirus genus including Severe Acute Respiratory Syndrome (SARS-CoV) and Middle Eastern Respiratory Syndrome (MERS-CoV) Coronaviruses. Both viruses cause severe acute respiratory illness, with SARS-CoV and MERS-CoV infection resulting in an overall mortality rate of ~10% and 38%, respectively. SARS-CoV was marked by its ease of transmissibility between humans, while MERS-CoV appears to be more limited in its human transmissibility. These viruses represent emerging pathogens not formerly recognized as a threat to human health, which adds an aspect of urgency to their research and characterization.
The SARS-CoV virus preparations used in these studies include a wild-type infectious clone derived from SARS-CoV (abbreviated icSARS-CoV), icSARS-CoV dORF6 in which accessory ORF6 was deleted, and Bat-SRBD, a reconstruction from a Bat-SCoV consensus genome with SARS-CoV receptor-binding domain (RBD)27. These strains were obtained from the Baric laboratory’s existing infectious clone constructs as previously described28–30. Recombinant mouse-adapted SARS-CoV (MA15)31, SARS-CoV dNSP16 (ref. 13), and human coronavirus Middle East Respiratory Syndrome Coronavirus strain EMC 2012 (abbreviated MERS-CoV) (from Bart L. Haagmans Erasmus Medical Center, Rotterdam, Netherlands) were also evaluated. All strains belonging to the Betacoronavirus genus were propagated in Vero E6 cells. For more details, see virus strain descriptions given in Table 5 (available online only). (ECL001, SCL005, SCL006, SHAE002, SHAE003, SHAE004, SM001, SM003, SM004, SM007, SM009, SM012, SM014, SM015, SM019, SM020).
Table 5. Description of viral strains.
| Virus Strain | Virus Description | Representative Accessions * | Sequence Alterations | Description Source |
|---|---|---|---|---|
| A/California/04/2009 (H1N1) | A/California/04/2009 (H1N1) - An isolate of the 2009 influenza pandemic | 1=FJ966079, 2=FJ966080, 3=FJ966081, 4=FJ966082, 5=FJ966083, 6=FJ966084, 7=FJ969513, 8=FJ966086 | None Specified | Josset et al., 2012 6 |
| MERS-CoV | MERS-CoV - A member of the betacoronavirus genus isolated from patients in the Middle East that causes severe acute respitory syndrome (SARS) fomerly known as vHuman coronavirus EMC 2012 (HCoV-EMC) [PMID: 23631916] | NC_019843 | None Specified | Josset et al., 2013 5 |
| A/Vietnam/1203/2004 (H5N1) | A/Vietnam/1203/2004 (H5N1)–Highly pathogenic avian influenza (HPAI) H5N1 | 1=AY651719, 2=AY651665, 3=AY651611, 4=AY651334, 5=AY651499, 6=AY651447, 7=AY651388, 8=AY651553, | None Specified | Li et al., 2011 4 |
| A/Netherlands/602/2009 (H1N1) | A/Netherlands/602/2009 (H1N1) - An isolate of the 2009 influenza pandemic | 1=CY046940, 2=CY046941, 3=CY046942, 4=CY039527, 5=CY046943, 6=CY039528, 7=CY046944, 8=CY046945 | None Specified | Mitchell et al., 2013 11 |
| A/New Jersey/8/1976 (H1N1) | A/New Jersey/8/76 (H1N1) - Isolated from throat swab material of a young military recruit during an outbreak at Fort Dix, NJ, in 1976. | 1=CY130125, 2=CY130124, 3=CY130123, 4=CY130118, 5=CY130121, 6=CY130120, 7=CY130119, 8=CY130122 | None Specified | Josset et al., 2012 6 |
| A/Mexico/4482/2009 (H1N1) | A/Mexico/4482/2009 (H1N1) - An Isolate from the 2009 H1N1 pandemic that caused severe respiratory illness in humans | 1=CY098505, 2=GQ149675, 3=GQ149676, 4=GQ149671, 5=GQ149673, 6=GQ149675, 7=GQ149669, 8=GQ379818, | None Specified | Josset et al., 2012 6 |
| Reconstructed 1918 (H1N1) | Reconstructed 1918 (H1N1) - The r1918 influenza virus segments were derived from the published sequences of strains A/South Carolina/1/18 (H1N1) HA, and A/Brevig Mission/1/1918 (H1N1) NA, M, NS, NP, PA, PB1 and PB2. The 1918 recombinant viruses also contained the 5′ and 3′ non-coding regions from strain A/WSN/1933 (H1N1). | 1=DQ208309, 2=DQ208310 3=DQ208311, 4=AF117241, 5=AY744935, 6=AF250356, 7=AY130766, 8=AF333238 | 5′ and 3′ non-coding regions were supplied by strain A/WSN/1933 (H1N1) | Kash et al., 2006 23 |
| A/Brisbane/59/2007 (H1N1) | A/Brisbane/59/2007 (H1N1) - H1N1 used in trivalent vaccine from 2008-2010 in Northern Hemisphere | 1=CY058484, 2=CY058485, 3=CY058486, 4=CY058487, 5=CY058488, 6=CY058489, 7=CY058490, 8=CY058491 | None Specified | Josset et al., 2012 6 |
| A/Vietnam/1203-CIP048_RG1 (H5N1) | A/Vietnam/1203-CIP048_RG1 (H5N1) - The H5N1 VN1203-HAavir mutant virus harbors an altered multi-basic cleavage site – a virulence factor important for expanded tissue range – and exhibits restricted systemic viral spread due to limited HA susceptibility to furin protease activity. | Parent Strain: A/Vietnam/1203/2004 (H5N1) | hemagglutinin (HA) surface protein poly-basic cleavage site (RERRRKKR↓G) was mutated to (RETR↓G) | Tchitchek et al., 2013 8 |
| A/Vietnam/1203-CIP048_RG2 (H5N1) | A/Vietnam/1203-CIP048_RG2 (H5N1) - The H5N1 VN1203-PB2627E mutant possesses an amino acid substitution (Lys-to-Glu) at position 627 in the PB2 polymerase subunit. This mutation is known to confer increased polymerase activity in mammalian cells, and also modulates anti-viral activity, apoptosis, and viral clearance. | Parent Strain: A/Vietnam/1203/2004 (H5N1) | PB2: K627E | Tchitchek et al., 2013 8 |
| A/Vietnam/1203-CIP048_RG3 (H5N1) | A/Vietnam/1203-CIP048_RG3 (H5N1) - H5N1 VN1203-PB1F2del mutant lacks expression of the PB1-F2 protein, potentially impacting an array of functions. PB1-F2 is a viral pathogenicity factor in mammals and birds, and has been shown to modulate viral polymerase activity, enhance lung inflammation, modulate innate immune responses, and demonstrate pro-apoptotic activity. | Parent Strain: A/Vietnam/1203/2004 (H5N1) | PB1 gene segment: T120C, C153G, G291A (all stops) | Tchitchek et al., 2013 8 |
| A/Vietnam/1203-CIP048_RG4 (H5N1) | A/Vietnam/1203-CIP048_RG4 (H5N1) - H5N1 VN1203-NS1trunc mutant virus produces a 91 amino acid C-terminal truncation in the effector domain of the NS1 host response antagonist protein. The NS1 protein inhibits RIG-I activation and cellular mRNA processing, and also promotes PI3K activation. The truncation results in the loss of the NS1 nuclear localization signal, a PI3K-binding motif, and binding domains. | Parent Strain: A/Vietnam/1203/2004 (H5N1) | NS1: 124(Stop), NS Gene segment: T400A | Tchitchek et al., 2013 8 |
| MA1-A/ California/04/2009 (H1N1) | MA1-A/ California/04/2009 (H1N1) - Mouse adapted MA-CA/04 contains five genetic mutations (in HA, G155E, S183P, and D222G; in PB2, E158G; and in NP, D101G) and is associated with a dramatic increase in virulence in mice | Parent Strain: A/California/04/2009 (H1N1) | PB2: E158GPA: L295PHA: G155E S183P R221R D222GNP: D101G | Ilyushina NA et al., 2010 27 |
| A/Aichi/02/68 (HA, NA) x A/Puerto Rico/8/34 (X31) | A/Aichi/02/68 (HA, NA) x A/Puerto Rico/8/34 (HKx31) - A reassortant virus that combines the surface HA and NA from an H3N2 virus (A/HK/x-31) with the 6 internal genes of PR8. Despite containing the same replication proteins as PR8, it is significantly less pathogenic in mice. | 1=CY009451, 2=CY009450, 3=CY009449, 4=AB295605, 5=CY009447, 6=AB295606, 7=CY009445, 8=CY009448 | None Specified | Askovich et al. 9 |
| A/Puerto Rico/8/1934 (H1N1) | A/Puerto Rico/8/34 (H1N1) - The H1N1 influenza virus A/Puerto Rico/34 (PR8) has been adapted to grow efficiently in the airways of mice, and is lethal at relatively low doses. | 1=CY009451, 2=CY009450, 3=CY009449, 4=CY009444, 5=CY009447, 6=CY009446, 7=CY009445, 8=CY009448 | None Specified | Askovich et al., 2013 9 |
| A/Vietnam/1203/04 (HA Vn1203, NA) x A/Puerto Rico/8/34 (VN6+2) | A/Vietnam/1203/04 (HA Vn1203, NA) x A/Puerto Rico/8/34 (VN6+2) - This H5N1 reassortant virus, designated rg/VN1203(6+2) contains the 6 internal genes from PR8 and the HA and NA from VN1203. The HA of this virus has had its polybasic cleavage site removed to restrict replication to the lung and airway epithelium. This virus is more pathogenic than PR8 and serves as a murine model for infection with a highly pathogenic strain. | 1=CY009451, 2=CY009450, 3=CY009449, 4=AY651334, 5=CY009447, 6=AY651447, 7=CY009445, 8=CY009448 | None Specified | Askovich et al., 2013 9 |
| icSARS Bat SRBD | SARS Bat SRBD - A SARS-CoV like virus isolated from bats that contains the receptor binding domain RBD from wild type SARS-CoV Urbani | FJ211859 | Bat-SCoV RBD (amino acid 323–505) was replaced with the SARS-CoV RBD (amino acid 319–518): FJ211860 | Becker, M.M., 2008 28 |
| icSARS CoV Urbani | icSARS CoV Urbani - Wild type infectious clone of SARS-CoV derived from genome fragments that were amplified in E. coli, ligated, and purified prior to in vitro transcription reactions to synthesize full length genomic RNA which were transfected into VeroE6 cells. | NC_004718* | Numerous alterations were made, see publications for details. Breifly, alterations included removal of one endogenous BglI site and the addition of three. Nucleotide alterations included an A to G at position 6460, T to C at position 14178, T to C at position 15740, C to T at position 19814, A to G at position 20528, and a T to C at position 20555. | Yount, B. et al., 2003 30 |
| icSARS dORF6 | icSARS dORF6 - infectious clone of a mutant Urbani SARS CoV in which the ORF6 protein is not expressed | Parent Strain icSARS CoV Urbani | Entire ORF6 deleted (nucleotides 27,074-27265) | Sims et al., 2013 12 |
| SARS CoV MA15 | SARS CoV MA15 - The mouse-adapted SARS-CoV (MA15) | FJ882957 | Gralinski et al., 2013 7 | |
| icSARS dNSP16 | icSARS dNSP16 - Genetic modification of non-structural protein 16 in SARS-CoV was shown to give rise to enhanced susceptibility to type I and III interferon responses | Parent Strain icSARS CoV Urbani | NSP16: K46A, K170A, D130A | Menachery et al., 2014 13 |
*Strains used were not directly sequenced. Accessions given are representative sequences with necessary details for recreating the strain used given in the ‘Sequence Alterations’ column.
Human cell lines, infections, and extractions
Most cell line studies involving infection with influenza A viruses were conducted in Calu-3 cells, a human lung adenocarcinoma cell line (kindly provided by Raymond Pickles; University of North Carolina, Chapel Hill, NC, USA). Cells were maintained in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 nutrient medium (DF12; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. For infection, Calu-3 cells were plated in 6 well plates at 1x106 cells per well. Cells were washed after 24 hours and infected 48 hours after plating. Infections were performed by removing culture medium, washing cells twice with 1x PBS, and inoculating each well with 300 μl of DF12 supplemented with 0.3% bovine serum albumin (DF12-BSA) containing virus at a multiplicity of infection of 1 to 3 plaque forming units per cell. Cultures were mock-infected by inoculating with DF12-BSA lacking virus. Cells were then incubated for 40 min at 37° C, and after the inoculum was removed, cells were washed with 1x PBS and cultured in 2 mL of DF12-BSA containing L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin. At each sample collection time point, aliquots of medium were reserved for plaque assays (see below). Time zero samples were immediately harvested using the appropriate procedure depending on the assay to be performed.
Studies examining members of the Betacoronavirus genus were performed in a clonal population of Calu-3 cells sorted for high levels of expression of the SARS-CoV cellular receptor angiotensin-converting enzyme 2 (ACE2), referred to as Calu-3 2B4 cells (kindly provided by Chien K. Tseng, University of Texas Medical Branch, Galveston, TX)32. Cells were maintained in 1x MEM (Gibco catalog number 11095) supplemented with defined Fetal Bovine Serum (HyClone catalog number SH30070.03) and antibiotic/antimycotic (100x Gibco catalog number 15240). Infections of Calu-3 2B4 cells were performed in a similar manner to the above Calu-3 cells. In the case of Calu-3 2B4 cells, 1.5×106 cells per well were seeded with 3 mL of complete culture medium. Culture medium was refreshed after 24 hours and infections administered after 48 hours as above. (SCL005, SCL006).
A subset of studies involving infection with influenza A virus and members of the Betacoronavirus genus were performed in human airway epithelial (HAE) cells, which were obtained from airway specimens resected from patients undergoing surgery under University of North Carolina Institutional Review Board, with approved protocols by the Cystic Fibrosis Center Tissue Culture Core. Primary cells were plated at a density of 2.5x105 cells per well on permeable Transwell-COL (12-mm-diameter) supports. HAE cultures were generated over 6 to 8 weeks, forming well-differentiated polarized cultures that resembled in vivo pseudostratified mucociliary epithelium33. Infection of HAE cultures began by washing with 1x PBS followed by challenge with 200 μL of either viral agent or mock to the apical surface. Cultures were incubated at 37° C for 2 hours and the inoculum and unbound viruses were removed by washing three times with 1x PBS. Apical wash samples were harvested to analyze viral growth kinetics and titers by plaque assay in Vero E6 cells (see below). (SHAE002, SHAE003, SHAE004).
Isolation of RNA from cell cultures was performed as follows: cell monolayers were first washed with 5 mL of cold PBS and then lysed directly in the culture dish by adding 1 mL of TRIZOL reagent. Cells were pipetted up and down until cells had a uniform color and consistency. They were then transferred to a 2 ml polypropylene tube (O-ring, screw cap), vortexed thoroughly, and incubated for 5 minutes at room temperature. Lysates were flash frozen on dry ice and stored at −80° C until shipping on dry ice. (ICL004, ICL006, ICL010, ICL011, ICL012, ECL001, SCL005, SCL006, SHAE002, SHAE003, SHAE004).
Isolation of protein from cell cultures was performed as follows: culture media was removed and cells were washed with 1 mL cold 150 mM (pH 8.5) ammonium bicarbonate buffer. This process was repeated for a total of 3 washes. 300 μL of cold 8 M urea dissolved in 50 mM ammonium bicarbonate buffer [pH 7.8] was then added directly to the well. Cells were scraped into a pre-sterilized 2 ml siliconized tube and incubated at room temperature for 15 minutes minutes to inactive viruses. Samples were then frozen in liquid nitrogen or dry ice/ethanol bath and stored at −80 °C until further processing. (ICL004, ICL006, ICL010, ICL011, ICL012, SCL005, SCL006).
Animal strains, infections, and extractions
The following mouse strains were obtained from the Jackson Laboratory (Bar Harbor, ME, USA): C57BL/6 J (stock no. 000664), Serpine1−/− (stock no. 002507), PLAT−/− (stock no. 002508), B6;129S6-Ppp1r14ctm1Uhl/J (stock no. 013041), B6.129S2-Tnfrsf1btm1Mwm/J (stock no. 002620), B6.129S7-Tnfrsf1btm1Imx/J (stock no. 003243), B6.129S4-Timp1tm1Pds/J (stock no. 006243), B6.129P2-Cxcr3tm1Dgen/J (stock no. 005796), and B6.129-Ido1tm1Alm/J (stock no. 005867). Young female BALB/c (Six-to eight-week-old) mice were purchased from Charles River Laboratories (Wilmington, MA, USA). All animal housing, care and treatment were conducted in accordance with the University of Wisconsin-Madison School of Veterinary Medicine Animal Care and Use Committee, or by the University of North Carolina Institutional Animal Care and Use Committee or under the guidance of the CDC's Institutional Animal Care and Use Committee in an animal facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. For virus infections, mice were anesthetized by isofluorane inhalation (VN1203) or lightly anesthetized with ketamine/xylazine (SARS-CoV and CA04) and intranasally inoculated with either 50 μL of phosphate-buffered saline (PBS) alone (mock infection), or PBS containing a viral agent. Individual or group mouse body weights were collected on a daily basis to monitor the disease course, and mice were humanely euthanized upon reaching the experimental endpoint (i.e. sample collection or severe clinical symptoms). Weight loss significance was determined by Student’s t test (Microsoft Excel), and significance in survival data was determined by the Mantel-Cox test (GraphPad). Both viral titers and qPCR assays were used to confirm the infection status of all mice (see below). For more details, see animal strain and cell line descriptions given in Table 6 (available online only).
Table 6. Description of host animal strains and cell lines.
| Host Species (Common) | Strain/Line (full name) | Description |
|---|---|---|
| Mouse | C57BL/6J (stock no. 000664) | C57BL/6 is the most widely used inbred strain. It is commonly used as a general purpose strain and background strain for the generation of congenics carrying both spontaneous and induced mutations. |
| Mouse | IDO1: B6.129-Ido1tm1Alm/J (stock no. 005867) | In contrast to wild-type, anti-proliferative treatments (CTLA4-Ig, IFNalpha, or CpG-ODN) do not suppress T cell expansion both in vivo and in vitro. |
| Mouse | Tnfrsf1b: B6.129S2-Tnfrsf1btm1Mwm/J (stock no. 002620) | Homozygous mutant mice show normal T-cell development and activity, but are resistant to TNF-induced cell death. Subcutaneous injections of TNF into homozygotes elicit much less tissue necrosis. |
| Mouse | ppp1r14c: B6;129S6-Ppp1r14ctm1Uhl/J (stock no. 013041) | No gene product (mRNA) is detected by RT-PCR analysis of brain tissue from homozygotes. Homozygotes exhibit increased phosphatase 1 activity in the thalamus. |
| Mouse | Tnfrsf1a/1b: B6.129S7-Tnfrsf1btm1Imx/J (stock no. 003243) | Double homozygous mutant mice fail to bind TNF. |
| Mouse | PLAT: PLAT−/− (stock no. 002508) | Pulmonary clot lysis is 21% that of normal wildtype siblings. Endotoxin induced venous thrombosis is increased over that seen in normal wildtype siblings. Fibrin dissolution by PLAT-deficient macrophages is unaffected. |
| Mouse | Serpine1 (PAI1): Serpine1−/− (stock no. 002507) | Compared to wildtype mice, pulmonary clot lysis is increased in the heterozygote and further increased in the homozygote. Endotoxin induced venous thrombosis is decreased compared to wildtype mice. Thus, disruption of the PAI-1 (Serpine1) gene induces a mild hyperfibrinolytic state. |
| Mouse | TIMP1: B6.129S4-Timp1tm1Pds/J (stock no. 006243) | No endogenous transcript is detected in lung tissue from affected mice by Northern blot analysis. Homozygous mice have increased resistance to corneal and pulmonary infection with P. aeruginosa, and have altered immune, hematopoietic, and vascular permeability in bleomycin-induced lung injury trials. |
| Mouse | BALB/c | General purpose model used to study numerous biological processes including infectious disease |
| Mouse | CXCR3: B6.129P2-Cxcr3tm1Dgen/J (stock no. 005796) | ‘Changes that may be related to genotype: Glucose level, elevated, hemizygous mutant males.’ (This targeted mutant was created and characterized by Deltagen, Inc) |
| Human | Human Airway Epithelial (HAE) cells | Human airway epithelium cultures (HAE) resemble in vivo pseudo- stratified mucociliary epithelium |
| Human | Calu-3 cells | Human lung adenocarcinoma derived from a caucasian male |
| Human | Calu-3 cells (clone 2B5) | Sub-population of Calu3 cells that express high levels of SARS-CoV cellular receptor, angiotensin converting enzyme 2 (ACE2) |
To prevent RNA degradation, immediately after dissection, lung tissues were directly submerged in RNAlater stabilization solution (Ambion, Catalog number AM7021) following manufacturer’s recommendations. Specifically, tissue was cut into small chunks (<0.5 cm in any single dimension) and placed immediately into 10-20 volumes (w/v) of RNAlater. Samples were thoroughly immersed in RNAlater solution and incubated overnight at 4° C, followed by freezing at −80° C.
Homogenization for later RNA isolation was performed as follows. Lung tissues were thawed, removed from RNAlater, dabbed on a kimwipe to remove excess liquid, transferred to 1 ml TRIzol and homogenized using a TissueLyser or MagnaLyser using several 30 second bursts to prevent overheating of the sample. Samples were transferred to a clean microfuge tube and stored at −80° C until RNA isolation. (CA04M001, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, IM009, IM010, SM001, SM003, SM004, SM007, SM009, SM012, SM014, SM015, SM019, SM020).
Homogenization of tissue for protein isolation was performed as follows: lung tissue were rinsed in 50 mM ammonium bicarbonate to remove blood and other fluid, then frozen at −80 °C and stored until the next step. The tissue was homogenized in 2 ml of 8 M urea dissolved in 50 mM ammonium bicarbonate buffer [pH 7.8]. The lung homogenates were then centrifuged at low speed (e.g. 5000xg) to remove large pieces of debris. The supernatants were transferred to new 2 mL siliconized microcentrifuge tubes, and incubated at room temperature for 1 hour to inactivate viruses. Samples were then frozen in liquid nitrogen or dry ice/ethanol bath and stored at −80 °C until further processing. (CA04M001, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, SM001).
Alternatively, mice were anesthetized with 350 μL of Avertin (2, 2, 2-Tribromoethanol) administered intraperitoneally. The virus inoculum was administered intranasally in a total volume of 30 μL (105 PFU of virus in 1x PBS). After infection, mice were kept on a heating pad until they regained consciousness. At 12 hours following infection, mice were sacrificed by CO2 inhalation. A hole was clipped in the trachea and bronchoalveolar fluid (BALF) collected. Lungs were perfused three times with 1 mL cold 1x PBS, for total of 3 mL. Whole lungs were then removed and homogenized in 1 mL TRIzol. (SBRI_AA_E1).
Viral titers and infection confirmation
Influenza viral titers were determined by standard plaque assay34. Briefly, MDCK cells were plated in 6-well plates, washed twice and then inoculated with 100 μL of 10-fold diluted virus in minimum essential medium supplemented with 0.3% BSA (MEM-BSA). Cells were incubated at room temperature for 1 hour, after which the inoculation solution was then removed. Cells were washed three times with PBS and overlaid with 1% Seaplaque agarose in MEM-BSA, supplemented with TPCK-treated trypsin (final 0.5 μg/mL). After the agarose solidified, plates were incubated upside down at 37° C and 5% CO2 for 48 hours. To visualize and count plaques, all wells were fixed with 10% phosphate-buffered formalin overnight, after which the agarose plugs were removed and plates were dried. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, IM009, IM010, SBRI_AA_E1).
Plaque assays for members of the Betacoronavirus genus were performed on supernatants collected at various times post-infection. Briefly, 5x105 Vero E6/Vero 81 cells were plated in 6-well plates. 10-fold dilutions of harvested viruses were generated in 1 mL of PBS. Then 200 μL of diluted virus was added to each well. Inoculated cells were incubated at 37° C for 1 hour (rocking gently every 15 minutes). Then 3 mL per well of agarose, consisting of 2x DMEM, 1.6% Seaplaque low melting point agarose, 10% final concentration fetal clone II for SARS-CoV and 4% for MERS-CoV, and 1x Antibiotic/Antimycotic was added to the cells. After solidification, plates were incubated face up at 37° C and 5% CO2 for ~48 hours. 1x neutral red solution was added to the top of the agar and incubated for at least 2 hours. Neutral red was then removed and the plaques were counted. (ECL001, SCL005, SCL006, SHAE002, SHAE003, SHAE004, SM001, SM003, SM004, SM007, SM009, SM012, SM014, SM015, SM019, SM020).
In addition to viral titers, real-time PCR quantification of viral genomic and viral mRNA species was performed to confirm viral infection and for comparisons between time points both within an infection and across studies. For Betacoronavirus genus, first-strand cDNA synthesis was performed using 500 ng of total RNA and Thermo-Script reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. For influenza, first-strand cDNA synthesis was performed similar to above except 15 pmol of RNA-specific primers for VN1203, CA04 or NL602 nucleoprotein genomic RNA and mRNA and endogenous control were used to prime the reactions. Primers for NP RNAs were designed with unique 5′ sequences, thereby ensuring differentiation between genomic RNA and mRNA species. The reverse transcriptase reaction primer sequences used were: NP genomic RNA (VN1203, CA04, and NL602) - 5′GGCCGTCATGGTGGCGAATAGCAAAAGCAGGGTAGATAATCACTC3′. NP mRNA (VN1203) - 5′CCAGATCGTTCGAGTCGTTTTTTTTTTTTTTTTTCTTTAATTGTC3′. NP mRNA (NL602 and CA04) - 5′CCAGATCGTTCGAGTCGTTTTTTTTTTTTTTTTTCAACTGTCATACTC3′. RPL14 (human endogenous control) - 5′TTCAATCTTCTTGGCCCATC3′. RPL10 (mouse endogenous control) - 5′GAACGATTTGGTAGGGTATAGGAG3′.
The qPCR assay was performed using a SYBR green kit (Applied Biosystems, Carlsbad, CA, USA) with specific primers for the different RNA species, according to the manufacturer’s standard protocol. Relative RNA quantities were determined using the comparative threshold cycle (CT) method, with human RPL14 or mouse RPL10 as endogenous. The qPCR primer sequences for endogenous controls used were:
RPL14 forward - 5′TTCATCCTCAAGTTTCCGC3′
RPL14 reverse - 5′TTCAATCTTCTTGGCCCATC3′
RPL10 forward - 5′TGAAGACATGGTTGCTGAGAAG3′
RPL10 reverse - 5′GAACGATTTGGTAGGGTATAGGAG3′.
The SARS-CoV qPCR primer sequences used were:
Forward primer for genomic RNA and mRNAs - 5′CCTACCCAGGAAAAGCCAAC3′.
Genomic RNA reverse primer - 5′GGACGAAACCGTAAGCAGTC3′
S mRNA reverse primer - 5′ACCGGTCAAGGTCACTAC3′
E mRNA reverse primer - 5′GCAAGAATACCACGAAAGCA3′.
The MERS-CoV qPCR primer sequences used were:
Forward primer for genomic RNA and mRNAs - 5′GAATAGCTTGGCTATCTCAC3′
Genomic RNA reverse primer - 5′CACAATCCCACCAGACAA3′
ORF2 reverse primer - 5′AGTGTATCATTGTCACGGATAAG3′
ORF5 reverse primer - 5′ACTAGCTGGACGGGTTTA3′
The Influenza qPCR primers sequences used were:
Genomic forward primer (VN1203, NL602 and CA04) - 5′GGCCGTCATGGTGGCGAAT3′
Genomic reverse primer (VN1203) - 5′GTTCCCCACCAGTTTCCATC3′
Genomic reverse primer (NL602 and CA04) - 5′GCTCCCCACCAGTCTCCATT3′
Forward mRNA primer (VN1203, NL602 and CA04) - 5′CCAGATCGTTCGAGTCGT3′
Reverse mRNA primer (VN1203, NL602 and CA04) - 5′CGAACCCGATCGTGCCTTCC3′
Isolation of total RNA and quality control
Isolation of RNA from Trizol lysates was performed using Qiagen RNeasy Mini kit (Qiagen Inc., Valencia, CA). The steps of the protocol were performed as follows: lysates were mixed by vortexing followed by a 5-minute incubation at RT. To each sample, 200 μL of chloroform per mL Trizol was added and the tubes shook vigorously for 15 seconds. In many cases only a portion of the 1 ml Trizol lysate was used and the volume of chloroform was adjusted accordingly. After incubation for 10 minutes at RT, samples were spun at 12,000 g for 15 minutes at 4° C. The aqueous phase was transferred to a clean tube and an equal volume of RLT +BME added. The volume was then doubled by adding 70% ethanol and mixed well by pipetting. This solution was applied to a RNeasy mini column and the RNA was isolated from the column per the manufacturer’s instructions. Appropriate amounts of RNA for quality control were removed and the remaining RNA was frozen on dry ice and store at −80° C. Determination of RNA quality and quantity was done using an Agilent Bioanalyzer 2100 and a Nanodrop spectrophotometer following the manufacturers’ recommendations. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, IM009, IM010, ECL001, SCL005, SCL006, SHAE002, SHAE003, SHAE004, SM001, SM003, SM004, SM007, SM009, SM012, SM014, SM015, SM019, SM020).
Alternatively, chloroform (0.2 mL per 1 mL TRIzol) was added to samples, tubes were inverted 20 times, and incubated at room temperature for 3 minutes. Samples were centrifuged at 12,000 g for 15 minutes at 4° C. The aqueous phase (top phase) was transferred to a new tube and an equal volume of isopropyl alcohol was added and samples incubated for 10 minutes. Tubes were centrifuged at 12,000 g for 10 minutes at 4° C. The RNA pellet was washed once with 70% ethanol and re-suspended in 100 μL of H20, 1 μL of glycogen and 1/10 volume of 3 M NaAc. RNA was re-precipitated with an equal volume of 100% ethanol and washed 2 times with 70% ethanol. The dried pellet was resuspended in 30 μL water. The RNA concentration and purity was determined by absorbance at 260 nm, 270 nm, and 280 nm using a plate reader. RNA integrity was determined using a BioAnalyzer (Agilent, Santa Clara, CA). (SBRI_AA_E1).
Labeling and quantification of total RNA for agilent microarray
Fluorescently labeled probes were generated from each total RNA sample using Agilent one-color Low Input Quick Amp Labeling Kit (Agilent Technologies) following the manufacturer’s instructions. The cRNA probe synthesis began with adding 100 ng total intact RNA to 3.25 μL water, combining RNA and Master Mix (consisting of 1.0 μL H2O, 0.25 μL Spike-in (dilution 2), 0.8 μL T7 Promoter Primer, 2.05 μL Total Volume per reaction), denaturing for 10 minutes at 65° C, and incubating at 4° C for 5 minutes. Next, double-stranded cDNA was synthesized by combining the above cRNA with 4.7 μL per reaction of First Strand mix (consisting of 2.0 μL 5X first stand Buffer1, 1.0 μL 0.1 M DTT, 0.5 μL 10 mM dNTP mix, and 1.2 μL AffinityScript RNase Block Mix) at room temperature followed by incubations at 40° C for 2 hours, 70° C for 15 minutes, and 4° C for 5 minutes.
Labeling of each sample was done by adding 6.0 μL of transcription master mix (consisting of 0.75 μL H2O, 3.2 μL 5X Transcription Buffer, 0.6 μL 0.1 M DTT, 1.0 μL NTP Mix, 0.21 μL T7 RNA Polymerase Blend, and 0.24 μL of Cy3 per reaction), mixing by pipette, and incubating at 40° C for 2 hours in the dark. Next, probes were purified with the Qiagen Rneasy kit (RNA cleanup protocol). After 84 μL of nuclease free water was added (to 100 μL total volume), samples were transferred to 1.5 mL microfuge tubes. Next, 350 μL of RLT, and 250 μL of 100% ethanol were added and mixed thoroughly by pipetting. 700 μL of this mixture was then applied to a column, centrifuged 30 seconds at 13,000 rpm at 4° C (discard flow through and collection tube). The column was then transferred to a new collection tube, 500 μL of RPE washing buffer was added, and centrifuged for 30 seconds at 13,000 rpm at 4° C (discard flow through, reuse tube). Another 500 μL of RPE was added and centrifuged at 60 seconds at 13,000 rpm at 4° C, then transferred to a clean microfuge tube and spun for 30 seconds on max to dry column.
To elute labelled RNA, the column was transferred to a clean labeled collection microfuge tube and 30μL of nuclease free H2O was added to the center of the column for 60 seconds followed by a 60 second centrifugation at 13,000 rpm at 4° C. A small aliquot (~4 μL) was removed for quantification (see below). Samples were quick frozen on dry ice and stored at −80° C while protected from light. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, IM009, IM010, ECL001, SCL005, SCL006, SHAE002, SHAE003, SHAE004, SM001, SM003, SM004, SM007, SM009, SM012, SM014, SM015, SM019, SM020).
Quantification of cRNA probes was done by the Nanodrop spectrophotometry. For each sample 1.2 μL of purified probe was measured and cRNA concentration (ng/μL) and dye incorporation (pmol/μL) recorded. Specific activity was calculated by: (dye incorporation/ cRNA concentration)*1000. Results greater than 8.0 were used for hybridization.
Agilent microarray hybridization and scanning
In summary, 250 ng of each RNA sample was hybridized to either an Agilent 4x44K mouse whole-genome oligonucleotide microarray G4122F (design ID 014868) or 4X44K human HG (design ID 014850) array according to the manufacturer’s instructions (details below).
Prior to hybridization, cRNA was first fragmented by mixing Cy3-cRNA per reaction volume of 55.0 μL (consisting of Cy3-cRNA 1650 ng, 11 μL of 10X Blocking Reagent, and Nuclease free H2O; Bring volume to 52.8 μL), with fragmentation buffer (consisting of 2.2 μL 25X fragmentation buffer) and then incubating at 60° C for 30 minutes in the dark. 55 μL of 2X Gene Expression Hybridization buffer HI-RPM was then added (Total volume=110 μL). Samples were mixed well by pipetting, and spun for 1 minute at room temperature at 13,000 rpm.
Subsequent to fragmentation, hybridization of 100 μL of the fragmented cRNA samples was transferred from above onto the center of each of the wells (4x44K format has 4 wells per slide with one sample per well). A slide was carefully lowered, bar code facing up, onto the bead of liquid, followed by the metal chamber cover and clamp. The chamber was rotated vertically to wet the gasket and make sure that all bubbles were mobile. Two chambers were load at a time into the hybridization oven, set at 65° C and level 10-rotation speed (10 rpm) for 17 hours.
Slides were then washed once with slide cover on in Gene Expression Wash Buffer 1, then cover was pried off and the slide washed in Gene Expression Wash Buffer 1 for a second time. Next, slides were washed in Gene Expression Wash Buffer 1 on a stir plate for ~1 minute and placed into pre-warmed Gene Expression Wash buffer 2 on a stir plate for ~1 minute. The excess liquid was blotted onto a Kimwipe and the dry slides placed into a light tight slide box to be scanned at once.
Dry slides were scanned on an Agilent DNA microarray scanner (model G2505B) using the XDR setting. Raw images were analyzed using the Agilent Feature Extraction software (version 9.5.3.1) and the GE1-v5_95_Feb07 extraction protocol. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, IM009, IM010, ECL001, SCL005, SCL006, SHAE002, SHAE003, SHAE004, SM001, SM003, SM004, SM007, SM009, SM012, SM014, SM015, SM019, SM020).
Agilent microarray processing and expression analysis
Normalization and QA/QC: Raw data consisted of the output from the Agilent Feature Extraction (AFE) software. Data was read into the R/Bioconductor Framework and visualization of raw intensity level was performed using boxplots, histograms, and correlation plots. All intensities were background corrected using the norm-exp method and quantile normalized using Agi4x44PreProcess and RMA Bioconductor packages. Distribution of control probes was examined and then those probes were removed. Descriptive statistics were reported for non-control replicate probes and the intensity distributions were examined via histograms. All non-control replicate probes were then summarized by taking the mean intensity measurement. To assess the quality of the reading for each probe, five Agilent QC flags were considered. The first is a test for minimal spot diameter and signal greater than 1.5 times background noise (gIsFound); the second requires positive intensity and background-subtracted intensity greater than 2.6 times background noise (gIsWellAboveBG); the third is true if more than 50% of spot pixels are greater than the saturation threshold (gIsSaturated); the fourth examines variance of spot pixels (gIsFeatNonUnifOL); the fifth checks if replicate probe is greater than 1.42 times the interquartile range (gIsFeatPopnOL).
A given probe must pass all 5 QC flags for all treatment (i.e., infected) replicates of at least one experimental time point. This allowed a conservative criteria regarding probe quality, but was moderate enough to detect transcripts that may be expressed only briefly (i.e., as little as one time point) during an infection time course. It was imperative that this filtering occurred before differential expression analysis so that low intensity (below background) and low quality probes were not considered in the analysis. Final QA/QC involved CV plots, heatmaps, cluster plots, scatter plots and PCA as needed in order to assess if there were sample outliers or additional batch effects.
Differential expression analysis was performed by comparing infected samples to time-matched mock-infected controls, based on a linear model fit for each transcript/probe using the R package Limma35. Fold changes were calculated by taking the average log2 probe intensity of infected replicates and subtracting the average log2 probe intensity of mock-infected replicates from the same time point. Significant differential expression of a probe was defined using the following criteria: log2 fold change >|1.5| and an adjusted false discovery rate (FDR) P value of <0.05 for a given time point.
Software Utilized: Custom R Bioconductor workflow that includes the Agi4x44 Pre-Process and RMA Bioconductor packages36 plus in house scripts for QA/QC visualization and differential expression analysis. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, IM009, IM010, ECL001, SCL005, SCL006, SHAE002, SHAE003, SHAE004, SM001, SM003, SM004, SM007, SM009, SM012, SM014, SM015, SM019, SM020).
Positive-strand cDNA synthesis, fragmentation, and single-stranded cDNA for affymetrix microarray analysis
cDNA was synthesized from isolated RNA (250 ng from each sample) using the Ambion WT kit (Life Technologies, Grand Island, NY, USA - http://tools.lifetechnologies.com/content/sfs/manuals/cms_064619.pdf) following the manufacturers protocol. Briefly: First strand cDNA was prepared from total RNA using reverse transcriptase and specific primers (5 μL of First-Strand master mix) resulting in single stranded cDNA which contains a T7 promoter sequence. Second strand cDNA was synthesized using Second-Strand master mix and the newly created double stranded DNA was isolated. Antisense cRNA was synthesized from the second strand DNA using T7 RNA polymerase then stored at 4 °C overnight. In the next step, cRNA was purified using cRNA Binding Mix and the yield was determined by absorbance at 260 nm. Purified cRNA was used to synthesize positive sense strand cDNA using reverse transcriptase and random primers. In this reaction, dUTP was added at a fixed ratio relative to dTTP. cRNA was then hydrolyzed using RNase H and cDNA purified using cDNA Binding Mix and cDNA yield determined by absorbance at 260 nm.
Single stranded cDNA was prepared for chip hybridization using the Affymetrix GeneChip WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA, http://www.umich.edu/~caparray/Files/protocols/affymetrix/Ambion%20WT%20Labelling%20and%20Hyb.pdf) following the manufacturer’s protocol. Briefly: Single-stranded cDNA was fragmented using Fragmentation master mix from the kit, then labeled by terminal deoxynucleotidyl transferase (TdT) using the Affymetrix proprietary DNA Labeling Reagent. The labeled mix was then hybridized to GeneChip Mouse Exon 1.0 ST Arrays in a hybridization oven for 17 hours. (SBRI_AA_E1).
Affymetrix microarray processing and expression analysis
Probe intensities were measured using the Affymetrix GeneChip Scanner 3000 and processed into CEL files using the Affymetrix GeneChip Operating Software. Microarrays were normalized at the gene level using the BrainArray custom CDF (Entrez Gene, Version 14) for probeset definitions and RMA as implemented in the justRMA function of the Bioconductor package affy for background adjustment, quantile normalization, and summarization. Linearized data was imported into Genedata Analyst software (Genedata AG, Basel, Switzerland). ANOVA was performed on all samples (100 balanced permutations) and genes with a permutation Q-value of 0.01 or less and fold change of at least 2 (between any two groups) were exported. (SBRI_AA_E1).
Preparation of protein extracts for proteomic analysis
Protein concentrations of cell lysates or lung tissue homogenates were determined by BCA protein assay. The range of final protein amount was between 100-226 μg with an average of 150 μg per sample. Each sample was then diluted to uniform volume in 50 mM ammonium bicarbonate, pH 7.8. Proteins were reduced with 10 mM dithiothreitol, followed by alkylation of free sulfhydryl groups with 40 mM iodoacetamide at 37° C in the dark; each reaction was performed for 1 hour at 37° C with constant shaking at 550 rpm. Denatured and reduced samples were diluted 10-fold with 50 mM ammonium bicarbonate, pH 7.8, and CaCl2 was added to a final concentration of 1 mM prior to enzymatic digestion. Sequencing-grade modified trypsin was activated by adding 20 μL of 50 mM ammonium bicarbonate, pH 7.8, to 20 μg lyophilized trypsin and incubating for 10 min at 37° C. Activated trypsin was added to the samples at 1:50 (w/w) trypsin-to-protein ratio, and samples were digested at 37° C for 3 hours with constant shaking at 800 rpm; reactions were quenched by rapid freezing in liquid nitrogen. Digested samples were desalted using solid phase extraction columns (Discovery C18, Supelco, Bellefonte, PA, USA), which were conditioned with 3 mL of methanol and rinsed with 2 mL of 0.1% trifluoroacetic acid (TFA) in water. Digest-loaded columns were washed with 4 mL of H2O/acetonitrile (95:5, v/v) containing 0.1% TFA, and peptides were eluted with 1 mL of acetonitrile/H2O (80:20, v/v) containing 0.1% TFA. Samples were concentrated to 100 μL in vacuo (Speed-Vac SC 250 Express, Thermo Savant, Holbrook, NY, USA), and a BCA protein assay was performed to verify final peptide concentrations. Samples were stored at −80° C until either strong cation exchange fractionation followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) or quantitative LC-MS analyses. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, SCL005, SCL006, SM001).
Strong cation exchange fractionation
Strong cation exchange fractionation was performed on pooled cell or lung protein digests using an Agilent 1100 HPLC System (Agilent, Palo Alto, CA, USA) equipped with a quaternary pump, degasser, diode array detector, peltier-cooled autosampler, and fraction collector (set at 4° C). Peptides (350 μg per injection) were separated with a PolySulfoethyl A (PolyLC Inc., Columbia, MD, USA) column (200 mm × 2.1 mm; 5 μm particles with 300-Å pores) with a 10 mm×2.1 mm guard column packed with the same material at a flow rate of 0.2 mL/min. The solvents consisted of (a) 10 mM ammonium formate, pH 3.0, and 25% acetonitrile, and (b) 500 mM ammonium formate [pH 6.8] and 25% acetonitrile. The following linear gradient was used: 100% solvent (a) for 10 min; ramp to 50% solvent (b) in 40 min; ramp to 100% solvent (b) in the next 10 min; followed by a 10 min hold in 100% solvent (b). Routinely, 24 fractions were collected from minute 30 to minute 65 of the gradient, and they were subsequently dried in vacuo and stored at −80° C until LC-MS/MS analysis. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, SCL005, SCL006, SM001).
Reversed-phase capillary LC-MS-MS and LC-MS analyses
Capillary LC-MS/MS analysis was used to generate separate accurate mass and time (AMT) tag databases for virus-infected cells and lung homogenates (see below). For this, dried peptide fractions from pooled samples were reconstituted in 30 μL of 25 mM ammonium bicarbonate, pH 7.8, and analyzed using a 4-column custom-built capillary LC system coupled online to a linear ion trap mass spectrometer (LTQ; Thermo Scientific, San Jose, CA, USA) by way of an in-house manufactured electrospray ionization interface. Electrospray emitters were custom made using 150 μm outer diameter (o.d.) x 20 μm inner diameter (i.d.) chemically etched fused silica. Reversed-phase capillary columns were prepared by slurry packing 3-μm Jupiter C18 bonded particles (Phenomenex, Torrence, CA, USA) into a 75 μm x 65 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ, USA) using 0.5 cm sol-gel plugs for particle retention. Mobile phases consisted of (a) 0.1% formic acid in water and (b) 0.1% formic acid in acetonitrile, and they were degassed on-line using a Degasys Model DG-2410 vacuum degasser (Dionex, Germany); the HPLC system was equilibrated at 10,000 psi with 100% mobile phase (a) for initial starting conditions. After loading 2.5 μg of peptides onto the column, the mobile phase was held at 100% mobile phase (a) for 50 min. Exponential gradient elution was initiated 50 min after sample loading with a column flow rate of 300 nL/min, and the mobile phase was ramped from 0% to 55% mobile phase (b) over 100 min using a 2.5 mL stainless steel mixing chamber, followed by a rapid increase to ~100% (b) for 10 min to wash the column. To identify the eluting peptides, the LTQ was operated in a data-dependent MS/MS mode (400-2,000 m/z), in which a full MS scan was followed by ten MS/MS scans using a normalized collision energy of 35%. A dynamic exclusion window of 1 min was used to discriminate against previously analyzed ions. The temperature of the heated capillary and the electrospray ionization (ESI) voltage were 200° C and 2.2 kV, respectively.
Following AMT tag database generation, capillary LC-MS analyses were performed on all virus-infected and mock-infected samples to generate quantitative data using the AMT tag approach. For this, individual dried peptide samples (see above) were reconstituted in 30 μL of 25 mM ammonium bicarbonate, pH 7.8, and analyzed in either duplicate or triplicate and in random order using identical chromatographic and electrospray conditions as for capillary LC-MS/MS analyses. The LC system was interfaced to an Exactive Orbitrap mass spectrometer (Thermo Scientific), and the temperature of the heated capillary and the ESI voltage were 250° C and 2.2 kV, respectively. Data were collected over the mass range 400-2,000 m/z. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, SCL005, SCL006, SM001).
Development of the AMT tag database for virus-infected model systems
A novel AMT database was generated for peptides within the specific model system, using mock-infected and virus-infected samples. To generate the AMT tag database, aliquots of the virus infected or mock-infected samples were combined to make the appropriate sample pools. Each pool was subjected to strong cation exchange fractionation as described above, and each fraction was analyzed by capillary LC- MS/MS. The SEQUEST analysis software was used to match the MS/MS fragmentation spectra with sequences from the appropriate UniProt/Swiss-Prot protein database. When searching, SEQUEST used a dynamic mass modification on methionine residues corresponding to oxidation (15.9949 Da) and a static mass modification on cysteinyl residues to account for alkylation by iodoacetamide (57.0215 Da). Peptides passing the following filter criteria were stored as AMT tags in a Microsoft SQL Server database: 1) SEQUEST DelCn2 value≥0.10 (normalized Xcorr difference between the top scoring peptide and the second highest scoring peptide in each MS/MS spectrum) and 2) for the mouse lung tissue database, a SEQUEST correlation score (Xcorr)≥1.6, 2.4, and 3.2 for fully tryptic peptides with 1+, 2+, and 3+ charge states, respectively, and Xcorr≥4.3, and 4.7 for partially tryptic or non-tryptic protein terminal peptides with 2+, and 3+ charge states, respectively. While for the human cell line database, a SEQUEST correlation score (Xcorr)≥2, 2.6, and 3.5 for fully tryptic peptides with 1+, 2+, and 3+ charge states, respectively, and Xcorr≥2.5, 3.6, and 4.1 for partially tryptic or non-tryptic protein terminal peptides with 1+, 2+, and 3+ charge states, respectively. These filter criteria resulted in estimated peptide false discovery rates of <2% based on target-decoy database searches for both lung and cell samples. Non-tryptic peptides were excluded, and a minimum peptide length of 6 amino acid residues was required. The elution times for these peptides were normalized to a range of 0 to 1 using a predictive peptide LC normalized elution time (NET) model and linear regression. A NET average and standard deviation are assigned to each identified peptide if the same peptide is observed in multiple analyses. Both calculated monoisotopic masses and observed NETs of identified peptides were included in the AMT tag database. From the primary data records corresponding to the data used to generate the AMT tag database (PASS00416 and PASS00417), the user can download the raw data, the assembled database, and parameter files necessary to recreate the AMT-tag approach. In addition to the data files, a detailed tutorial has been made available for download. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, SCL005, SCL006, SM001).
Processing of quantitative LC-MS datasets
Quantitative LC-MS datasets were processed using the PRISM Data Analysis system37, which is a series of software tools developed in-house (e.g. Decon2LS38 and VIPER39; freely available at http://ncrr.pnl.gov/software/). Individual steps in this data processing approach are reviewed here40. The peptide identities for detected features in each dataset (i.e. a single LC-MS analysis) were determined for features matched to AMT tags with high confidence based upon the accurate measured monoisotopic masses and NETs for each of the peptides in the filtered AMT tag databases within initial search tolerances of ±6 ppm and ±0.025 NET for monoisotopic mass and elution time, respectively. The peptides identified from this matching process were retained as matrices for subsequent data analysis. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, SCL005, SCL006, SM001).
Proteomic data statistical processing
The integrated LC-MS peak intensity values (i.e. abundances) for the final peptide identifications were processed in a series of steps using MatLab R2010b that included quality control, normalization, protein quantification, and comparative statistical analyses. Quality control processing was performed to identify and remove peptides with an insufficient amount of data across the set of samples41, and LC-MS runs that showed significant deviation from the standard behavior of all LC-MS analyses42, using a significance level of 0.0001 (ref. 43). The peptide abundance values were normalized across the technical replicates using a rank invariant subset of peptides (p-value threshold of 0.1) followed by median absolute deviation centering of the data. Normalized log10 abundance values were averaged across the technical replicates within each biological sample. Proteins were quantified using a standard R-Rollup method44 using the most abundant reference peptide--after filtering the peptides that were redundant, had low data content, or were outside the dominant significance pattern. Comparative statistical analyses of time-matched mock with viral infected samples were performed using a Dunnett adjusted t-test to assess differences in protein average abundance. (CA04M001, ICL004, ICL006, ICL010, ICL011, ICL012, IM001, IM002, IM004, IM005, IM006A, IM006B, IM007, SCL005, SCL006, SM001).
Data Records
Study designs for all experiments, including relevant repository identifiers, are summarized in Table 1 (Available online only), with individual RNA experiment samples detailed in Table 2 (Available online only) and individual protein experiment samples detailed in Table 3 (Available online only). Sample tracking from animal subjects to experiment samples is given in Table 4 (Available online only). Primary assay results (txt and mzXML files) are archived in public repositories as listed below (Data Record 1 and 2). Derived data resulting from processing and hypothesis testing accompanied by structured metadata describing both the experimental and data analysis methods are available at the Influenza Research Database and the Viral Pathogen Resource (Data Record 3).
Data Record 1
Primary data for Agilent and Affymetrix microarray experiments are available at the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE37569 (Data Citation 1)8, GSE45042 (Data Citation 2)5, GSE28166 (Data Citation 3)4,45, GSE37571 (Data Citation 4), GSE43203 (Data Citation 5), GSE43204 (Data Citation 6), GSE33263 (Data Citation 7)8,45, GSE36328 (Data Citation 8)6, GSE37572 (Data Citation 9)8, GSE43301 (Data Citation 10)8, GSE43302 (Data Citation 11)8, GSE44441 (Data Citation 12), GSE44445 (Data Citation 13)8, GSE40792 (Data Citation 14), GSE37245 (Data Citation 15)9, GSE33267 (Data Citation 16)5,12, GSE37827 (Data Citation 17), GSE47960 (Data Citation 18)11, GSE47961 (Data Citation 19)11, GSE47962 (Data Citation 20)11, GSE33266 (Data Citation 21), GSE50000 (Data Citation 22), GSE51386 (Data Citation 23), GSE50878 (Data Citation 24), GSE51387 (Data Citation 25), GSE49262 (Data Citation 26), GSE49263 (Data Citation 27), GSE40824 (Data Citation 28), GSE40827 (Data Citation 29).
Data Record 2
Raw data files and associated parameter files (.raw,.mzXML) for all proteomics experiments including those used for AMT-TAG database generation have been uploaded to PeptideAtlas (http://www.peptideatlas.org). Experiment accession numbers are as follows: PASS00416 (Data Citation 30), PASS00417 (Data Citation 31), PASS00418 (Data Citation 32), PASS00419 (Data Citation 33), PASS00420 (Data Citation 34), PASS00421 (Data Citation 35), PASS00422 (Data Citation 36), PASS00423 (Data Citation 37), PASS00424 (Data Citation 38), PASS00425 (Data Citation 39), PASS00426 (Data Citation 40), PASS00427 (Data Citation 41), PASS00428 (Data Citation 42), PASS00429 (Data Citation 43), PASS00430 (Data Citation 44), PASS00431 (Data Citation 45), PASS00432 (Data Citation 46), PASS00433 (Data Citation 47).
Data Record 3
Structured experiment metadata and derived data (.xls,.txt) including fold change values and p-values are available in the Host Factor Component of the Influenza Research Database (IRD, www.fludb.org) and the Virus Pathogen Resource (ViPR, www.viprbrc.org) under accession numbers: IRD_SV_CA04M001-P (Data Citation 48), IRD_SV_ICL004-P (Data Citation 49), IRD_SV_ICL006-P (Data Citation 50), IRD_SV_ICL010-P (Data Citation 51), IRD_SV_ICL011-P (Data Citation 52), IRD_SV_ICL012-P (Data Citation 53), IRD_SV_IM001-P (Data Citation 54), IRD_SV_IM004-P (Data Citation 55), IRD_SV_IM005-P (Data Citation 56), IRD_SV_IM006A-P (Data Citation 57), IRD_SV_IM006B-P (Data Citation 58), IRD_SV_IM007-P (Data Citation 59), IRD_SV_SCL005-P (Data Citation 60), IRD_SV_SCL006-P (Data Citation 61), IRD_SV_SM001-P (Data Citation 62), IRD_SV_CA04M001-R (Data Citation 63), IRD_SV_ECL001-R (Data Citation 64), IRD_SV_ICL004-R (Data Citation 65), IRD_SV_ICL006-R (Data Citation 66), IRD_SV_ICL010-R (Data Citation 67), IRD_SV_ICL011-R (Data Citation 68), IRD_SV_ICL012-R (Data Citation 69), IRD_SV_IM001-R (Data Citation 70), IRD_SV_IM002-R (Data Citation 71), IRD_SV_IM004-R (Data Citation 72), IRD_SV_IM005-R (Data Citation 73), IRD_SV_IM006A-R (Data Citation 74), IRD_SV_IM006B-R (Data Citation 75), IRD_SV_IM007-R (Data Citation 76), IRD_SV_IM009-R (Data Citation 77), IRD_SV_IM010-R (Data Citation 78), IRD_SI_SBRI_AA_E1 (Data Citation 79), IRD_SV_SCL005-R (Data Citation 80), IRD_SV_SCL006-R (Data Citation 81), IRD_SV_SHAE002-R (Data Citation 82), IRD_SV_SHAE003-R (Data Citation 83), IRD_SV_SHAE004-R (Data Citation 84), IRD_SV_SM001-R (Data Citation 85), IRD_SV_SM003-R (Data Citation 86), IRD_SV_SM004-R (Data Citation 87), IRD_SV_SM007-R (Data Citation 88), IRD_SV_SM009-R (Data Citation 89), IRD_SV_SM012-R (Data Citation 90), IRD_SV_SM014-R (Data Citation 91), IRD_SV_SM015-R (Data Citation 92), IRD_SV_SM019-R (Data Citation 93), IRD_SV_SM020-R (Data Citation 94).
Technical Validation
Two approaches were used to establish the technical validity of the datasets described. First, the biological samples used to generate the transcriptomic and proteomic assay results reported here have also been assessed using various criteria to ensure technical validity. For example, productive viral infections following inoculation of animals used for in vivo studies was confirmed by examining viral titers, viral mRNA levels, and body weight loss to quantify virus replication and host response to infection as detailed below. For in vitro studies, viral genomic RNA, viral mRNA, and viral titers were quantified to ensure that the virus was actively replicating and transcribing its genome in cell culture. Second, the processed data produced from in vivo and in vitro studies were compared, using Boolean logic, to quantify the number of overlapping significant genes identified in multiple studies performed under similar experimental conditions as an assessment of general reproducibility of the derived results between experiments.
In vivo validation
To validate the in vivo studies, mean viral mRNA levels, mean % body weight, and mean viral titers were examined as surrogates for viral infection in the animal subjects. The CA04M001 and IM001 experiments, in which mice were infected with influenza A virus strains A/California/04/2009 (pandemic H1N1) and A/Viet Nam/1203/2004 (H5N1) respectively, were used as representative datasets for this validation process. For both experiments, the viral mRNA levels indicate that the virus was actively transcribing mRNA within the host organism with mean viral mRNA levels peaking at either 2 or 4 days post infection (dpi), when either a high or low initial inoculum was used, respectively8. Similarly, the mice that were exposed to virus in either experiment showed a dose-dependent decrease in mean % body weight. The viral growth kinetics for A/California/04/2009 in the CA04M001 study showed mean viral titers peaking at 2 dpi regardless of the viral concentration present in the initial inoculation. In contrast, the growth kinetics for A/Viet Nam/1203/2004 used in the IM001 study showed mean viral titers peaking at 2 days post infection when the initial inoculum contained either 103 or 104 plaque forming units (PFU) of virus; whereas mean viral titers were highest at 4 dpi when the initial inoculum contained 102 PFU of virus.
In vitro validation
For in vitro studies, measurements from study ICL004, performed in Calu-3 cells infected with the A/Viet Nam/1203/2004 influenza virus strain, were used as representative of all studies. Mean viral genomic RNA levels increased logarithmically between 0 and 12 hours post infection (hpi), eventually peaking at 18 hpi before decreasing at 24 hpi. Similarly, mean viral mRNA levels increased exponentially between 0 and 7 hpi, where they peaked before decreasing through the 24-hour time point. Mean viral titers were observed to increase logarithmically between the 7 and 12-hour time points, and then continued to rise through 24 hpi8.
Overall, the metrics used to validate the in vivo and in vitro studies indicate that the biological samples that were taken from these sample sources were productively infected with the intended virus. Consequently, any downstream assays that were performed on these biological samples should give rise to biologically valid results.
Reproducibility of derived results
The primary assay results obtained from the transcriptomic and proteomic experiments were processed to identify specific RNAs or peptides that were differentially expressed under a particular set of conditions (e.g. virus amount, time post infection, virus strain, etc.) in comparison with relevant control samples to derive so called ‘Host Factor Biosets’. Another measure of technical validity of the combined laboratory and analytical workflow is that the Host Factor Biosets derived from similar experiment samples treated in a similar way should have similar membership, thereby indicating experimental reproducibility. To examine experimental reproducibility, we determined the overlap that was observed between all experiments that used the same virus strains in the same infection system. In the evaluation reported here, we used datasets reporting results from wild-type A/California/04/2009 (H1N1), A/Viet Nam/1203/04 (H5N1), and SARS-CoV as being representatives of all experiments. In order to avoid complications related to slight differences in infection kinetics, we took the union of all biosets across the entire time course from each experiment (i.e. the non-redundant set of differentially expressed genes) for comparison. This approach can lead to a discrepancy between the total number of genes in each experiment due to differences in number of time points sampled. As an alternative strategy, the data could also be more strictly paired using specific time points and dosages.
Seven situations in which the same treatment conditions (viral strain and viral amount) were reproduced in separate experiments were identified. Since the magnitude of the responses and the number of time points differ between experiments, we calculated the percentage overlap of the number of host factors using the smallest set as the denominator for the sake of comparison. The smallest set was chosen as the denominator to calculate percentage overlap, which may not be an ideal comparison since this would tend to result in higher reproducibility values than if the largest set was used as the denominator. However, since we are primarily interested in measuring the commonality between similar experiments that have slight variations in study designs and are inherently noisy, we chose to focus on determining if the smaller set is largely a subset of the larger sets, which would demonstrate that the these significant hits are reproducible even in the presence of confounding variables such as biological and technical variation.
For transcriptomic experiments the following overlap was observed: 83% for A/California/04/2009 in Calu-3 (Fig. 2a), 91% for A/Viet Nam/1203/2004 in Calu-3 (Fig. 2b, 3-way comparison), 62% for A/California/04/2009 in C57BL/6 (Fig. 2c), 77% for A/Viet Nam/1203/2004 in C57BL/6 (Fig. 2d), 72% for A/California/04/2009 in HAE cells (Fig. 2e), and 40% for SARS-CoV in HAE cells (Fig. 2f). For the one replicated proteomic experiment, in which A/Viet Nam/1203/2004 was used to infect Calu-3, 43% overlap was observed (Fig. 2g). The reproducibility of host responses to both pandemic and highly pathogenic avian influenza viruses was high in both the Calu-3 cell line and mouse infection systems. Reproducibility using HAE was somewhat lower both in terms of the absolute number of host factor responding (Fig. 2e) and the percentage overlap (only 39% in Fig. 2f), perhaps due to the variability in establishing similar cell culture conditions using primary epithelial cells. Reproducibility of proteomic results was also somewhat lower with an overlap of 43% following infection of Calu-3 with A/Viet Nam/1203/2004 (Fig. 2g).
Figure 2. Computational validation of results.
(a) Shows the comparison performed between all significant host factors identified by transcriptomics in the ICL006-R and ICL010-R experiments, both of which involved infection of Calu-3 cells with A/California/04/2009 at 3 multiplicity of infection (MOI). (b) Compares all significant host factors identified by transcriptomics in the ICL004-R, ICL011-R and ICL012-R experiments, which involved infection of Calu-3 cells with the A/Viet Nam/1203/2004 strain at 1 MOI. (c) A comparison between all significant host factors identified by transcriptomics in the CA04M001-R and IM009-R experiments, both of which involved infection of C57BL/6 mice with 106 plaque forming units (PFU) of A/California/04/2009. (d) Compares all significant host factors identified by transcriptomics in the IM001-R and IM010-R experiments, both of which involved infection of C57BL/6 mice with 103 PFU of A/Viet Nam/1203/2004. (e) Compares all significant host factors identified by transcriptomics in the SHAE002-R and SHAE003-R experiments, which involved infection of HAE cells with A/California/04/2009 at 2 MOI. (f) Compares all significant host factors identified by transcriptomics in the SHAE002-R and SHAE003-R experiments, which involved infection of HAE cells with the SARS-CoV Urbani strain at 2 MOI. (g) Compares all significant host factors identified by proteomics in the ICL011-P and ICL012-P experiments, which involved infection of Calu-3 cells with A/Viet Nam/1203/2004 at 1 MOI.
In general, the reproducibility of this collection of experiments using well-controlled standard operating procedures was quite high. In other microarray studies it has been shown that technical variability can result in 10% difference between replicate RNA samples46,47, while biological variability can cause anywhere from 10% to 30% differences depending on the biological system sampled48. In addition, variability in stimulation conditions has been found to have an even more profound impact than either technical or biological variability47. Thus, the studies compared here demonstrated considerable reproducibility in results in spite of the number of sources of variability inherent in their designs. Furthermore, reduction in the number of false positive results in any individual experiment (high specificity) could be achieved by focusing on the host factors that were reproducibly identified in similar experiments. Reduction in the number of false negative results (high sensitivity) could be achieved by using the union of host factors identified among similar experiments.
Overall, the results from the independent in vitro and in vivo validation experiments and the high degree of reproducibility of the derived host factor lists show that the datasets described here are of high quality and can be useful for in-depth investigation and hypothesis generation in follow-up analyses.
Usage Notes
The systems biology studies described herein were designed to characterize how the host responds to viral infection. Separate experiments were performed to measure the response to multiple independent variables that include sampling at multiple time points after infection, at different viral dosages, in various model systems, and in systems with various genetic backgrounds (both viral and host). A number of publications have reported the use of these datasets to address questions concerning pathogenesis. For instance, comparisons between the similarities and dissimilarities of the host response at early and late stages of infection with genetically modified viral strains have been performed to identify possible correlates of disease severity9. Likewise, the host response was measured in different model systems to identify host factors that have a role in disease progression45. Lastly, all of these variables were combined to conduct a detailed evaluation of HPAI H5N1 infection based on time, dosage, and genetically-modified viruses8.
Various analytical and statistical tools can be used to interpret the results from systems biology experiments. The commonly-used methods fall into three categories: enrichment analysis, co-expression network analysis, and transcription factor predictions. Enrichment analysis tools, such as DAVID or GSEA49–51, use diverse statistical methods to identify annotations that are over-represented in a list of significant genes or proteins. Co-expression network analysis methods, such as Weighted Correlation Network Analysis (WGCNA)52, find correlated patterns across multiple microarray samples. Lastly, transcription factor prediction tools, such as PSCAN and PRISM53,54, examine the promoter regions of co-expressed genes to identify over-represented transcription factor binding sites. All of these tools allow researchers to generate hypotheses concerning how different components of the biological system being studied are modulated.
The host factor component within IRD and ViPR contains the processed data derived from these studies focusing primarily on the host response to virus infection. Here we present several scientific use cases that highlight some of the exploration and data mining uses that are currently possible using these data in the IRD/ViPR systems.
Host responses to related viruses
Comparisons of host responses to infection by viruses that are phylogenetically related but associated with subtle or dramatic differences in disease symptoms or severity can be used to help identify the genetic determinants of virulence.
The IM002 experiment was designed to interrogate the different host responses that occurred when mice were infected with more distantly related H1N1 influenza A viruses: A/Brisbane/59/2007, A/New Jersey/8/1976, A/Mexico/4482/2009, and a reconstructed ‘Spanish flu’ strain r1918 at 1, 3, and 5 days post infection. Although all four viruses are of the H1N1 subtype, they differ in their apparent virulence characteristics in humans, with the original isolates of the A/New Jersey/8/1976, A/Mexico/4482/2009, and the r1918 strain all described as causing severe respiratory illness, whereas the A/Brisbane/59/2007 strain was isolated from a relatively mild case, and was included in the trivalent vaccine from 2008-2010. Previous analyses of the pathways and functions that were over-represented in the list of differentially expressed genes revealed that inflammatory response, cell-to-cell signaling, immune cell trafficking, lipid metabolism, amino acid metabolism, cardiovascular disease, and cancer gene responses were associated with virulence differences6.
To illustrate one potential way to compare datasets in ViPR/IRD, we quantified the overlap between differentially expressed genes identified from samples infected with this collection of ‘pre-pandemic’ H1N1 influenza A virus. As before, we compared the non-redundant union of significant genes derived from the transcriptomic results reported in the IM002 experiment to determine differences detected in mice infected with these viruses. Although the pairwise comparisons of the host response to virus infection were somewhat variable, the differences appeared to correlate with the virulence of the infecting virus. No overlap was observed between all three virulent viruses and the A/Brisbane/59/2007 strain. In contrast, 262 host factors (66%) were shared in mice infected with the three virulent viruses (Fig. 3a). Performing a functional annotation analysis for this list of 262 genes identified the same list of GO terms as reported in the original study6 except for the exclusion of cardiovascular disease and cancer.
Figure 3. Comparative analyses usage examples.

(a) Compares the significant host factors identified in the IM002-R experiment by transcriptomics at all time points by infecting C57BL/6 mice with 106 PFU of H1N1 influenza strains A/Mexico/4482/2009, A/New Jersey/8/1976, or r1918. (b) Shows the comparison of significant factors identified by transcriptomics in the SCL005-R experiment that infected Calu-3 cells at 5 MOI with either wild-type SARS-CoV or a mutant containing a functional ORF6 knockout. (c) Results from the ICL004-R and ICL004-P experiments show the overlapping and unique host factors identified by transcriptomics and proteomics respectively for time points after (and including) 12 hpi in Calu-3 cells infected at 1 MOI with influenza strain A/Viet Nam/1203/2004.
Similar analyses designed to compare the host response to other related viruses could be performed using the datasets that are currently available in IRD/ViPR. For example, the ICL010-R experiment examines the host transcriptional response to 2 different 2009 influenza A (H1N1) pandemic strains in Calu-3 cells at 12-, 24, and 48 hpi. The ECL001-R and SCL005-R experiments could be analysed to better understand the similarities and differences in the host response during infection with either MERS or SARS-CoV in Calu-3 cells across multiple time points.
Host responses to mutant viruses
In order to investigate how specific viral proteins might function to regulate host intracellular processes, experiments were performed to identify shared and unique host genes that are differentially expressed in hosts infected with either wild-type or mutant viruses5,11,13. For example, analysis of experimental results involving variants of SARS-CoV with a deletion in the ORF6 coding region performed in human cell lines (SCL005) or mice (SM012) determined that antagonism of karyopherin-based transport was an important function of the ORF6 gene product in SARS-CoV. As nuclear signaling disruption is a strategy used by many viral pathogens, identifying trends in these data presents a possible area for general therapeutic intervention.
Using Host Factor Bioset data in ViPR/IRD comparing human cell lines infected with either wild-type or ORF6 mutant viruses, 74% of significant host factors were shared between the two infection conditions (Fig. 3b). When an enrichment analysis was performed on the non-overlapping host factors (i.e. unique to infection with the ORF6 mutant strain), we found enrichment for terms relating to cell proliferation and immune response (P<0.05), which matches what had been reported previously12.
Similar analyses can be performed with data from other studies in IRD/ViPR to compare infection with wild-type versus genetically-engineered mutant strains. For instance, there are multiple studies that characterize the host responses to SARS-CoV variant strains in a human cell line (SCL006) and a mouse model (SM003, and SM012), and host responses to four influenza mutant variants of VN1203 in mice (IM004, IM005, IM006A, IM006B, and IM007) and a human cell line (ICL011 and ICL012).
Differences between RNA and protein responses
While proteins carry the functional workload in cells, it is still challenging to achieve the levels of dynamic range necessary to measure their abundances and activities at a comprehensive genome-wide level to support systems-level analysis, given practical constraints upon measurement throughput and sensitivity. In contrast, while gene expression arrays allow for the comprehensive measurement of gene expression levels across the entire genome, these levels do not directly reflect functional physiology mediated by the encoded proteins. Therefore, it is interesting to compare quantitative proteomic and transcriptomic experiments using similar samples to estimate how often transcriptional profiles are reflected in protein profiles.
Prior reports have compared transcriptomic and proteomic data as a way of validating experimental results12. In these cases, proteomic data was integrated with the corresponding transcriptomic data to improve the biological networks produced and to support the associated statistical analysis of SARS-CoV host responses in general, and the karyopherin transport hubs in particular.
We performed a similar comparison of RNA and protein Host Factor Biosets in ViPR/IRD from the same tissue/cell samples using either microarray or mass spectrometry technologies (ICL004). In this case, transcriptomic data from the 12 h time point of human Calu-3 cells infected with A/Viet Nam/1203/2004 influenza virus was chosen because this time point contained the highest number of significant probes. These transcriptomic data were compared with a pooled list from the time-matched and all subsequent time points from the related proteomics data in order to control for the ‘lag’ that occurs between when the changes in mRNA expression are translated into corresponding changes in the translated protein product. We found approximately 56% overlap between the statistically significant RNA at 12 hpi and the statistically significant protein at the combined 12 and 18/24 hpi time points (Fig. 3c), similar to what has been reported previously55,56.
This type of analysis can be conducted on a number of other studies hosted by ViPR/IRD that evaluated both the transcript and protein host responses. These experiments included influenza infection in human cell lines (ICL006, ICL010, ICL011, ICL012), mice (CA04M001, IM001, IM004, IM005, IM006A, IM006B, IM007), and SARS-CoV infections in human cell lines (SCL005, SCL006) and mice (SM001).
Meta-analysis
While many of the usage examples provided involve comparative analysis of results obtained in the same experiment, in some cases similar comparisons can be performed between experiments. Indeed, the Validation section focused on the reproducibility of derived results and involved the comparison of similar experiment conditions performed in separate experiments. Combining and contrasting results from different studies is often referred to as meta-analysis. One of the goals behind making these datasets freely available through public database resources like GEO, ViPR and IRD is to support their incorporation into meta-analysis approaches that include new experiments being performed by the research community. To facilitate this kind of meta-analysis, considerable effort has been dedicated to providing complete descriptions of all experimental and data analysis methodologies, combined with structured, standardized metadata describing selected key aspects of the experiment design, especially regarding the experiment factors that serve as the independent (manipulated) variables in the study in ViPR and IRD. Clear representations of the experiment metadata will make it relatively simple to appropriately combine these primary and derived results with data from future related experiments for meta-analysis.
Re-analysis
Although we have emphasized the use of the derived Host Factor Biosets, in every case a close linkage is maintained between the processed data and the primary assay results from which they were derived-even though they may be stored in distinct resources. Thus, the final usage example is related to the re-processing and re-analysis of primary assay results.
Different processing algorithm(s) chosen to perform a specific processing or analysis step in a multistep workflow can yield different results. Many bioinformatics data processing workflows currently implement a ‘one size fits all’ approach with a single algorithm being applied to all datasets without regard for the specific nuances of each individual dataset. The problem with this approach is that the processing algorithms and parameters that are ideal for one dataset may not be optimal for other datasets—especially when dealing with different experimental systems. Hence, processing of the data using a single universally-applied algorithm may not sufficiently address the individual requirements of each dataset. To truly solve this underlying problem, an analytical workflow that explores the relevant parameter space for a diverse set of algorithms would need to be implemented. This approach would provide an objective and automated process to intelligently ‘choose’ the optimal combination of parameters and algorithms that produce the best set of results for each individual dataset. Such an approach could be implemented in analytical workflow suites like Galaxy or GenePattern57,58 to objectively and confidently reprocess the raw data from each ‘-omics’ dataset in a data-driven fashion to further improve the quality of the derived data. Until such a system is developed, we advocate for using the processed data that was provided by the original submitters and subjected to the peer-review process when performing meta-analyses given its overall high quality.
Additional information
Tables 1–6 are only available in the online version of this paper.
How to cite: Aevermann, B. D. et al. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci. Data 1:140033 doi: 10.1038/sdata.2014.33 (2014).
Supplementary Material
Acknowledgments
Data described here was funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services. Data generation was performed under the Systems Virology Contract HHSN272200800060C and Systems Influenza contract HHSN272200800058C. The Influenza Research Database (IRD, www.fludb.org) and the Virus Pathogen Resource (ViPR, www.viprbrc.org) also received generous support from the NIAID under N01AI40041. We also wish to thank Valentina Di Francesco and Alison Yao for their programmatic support of the Systems Biology and the Bioinformatics Resource Centers Programs at the NIAID. Additional support for methodology development was provided by NIH/NCATS (5UL1RR024140). Proteomics data were obtained using capabilities developed under the support of the National Institute of General Medical Sciences (8 P41 GM103493-11), National Institutes of Health, and analyses were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy’s (DOE) Office of Biological and Environmental Research and located at PNNL. PNNL is operated by the Battelle Memorial Institute for the DOE under contract number DE-AC05-76RLO1830. We would also like to acknowledge Drew Trevor Scobey and Alexandra Schaefer for their help performing experiments at University of North Carolina Chapel Hill.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Baric R., Katze M., Chang J., Gralinski L., Law L. 2012. Gene Expression Omnibus. GSE37569
- Josset L. 2013. Gene Expression Omnibus. GSE45042
- Li C. 2011. Gene Expression Omnibus. GSE28166
- Li C. 2012. Gene Expression Omnibus. GSE37571
- Katze M. G. 2013. Gene Expression Omnibus. GSE43203
- Katze M. G. 2013. Gene Expression Omnibus. GSE43204
- Li C. 2011. Gene Expression Omnibus. GSE33263
- Josset L. 2011. Gene Expression Omnibus. GSE36328
- Kawaoka Y. 2012. Gene Expression Omnibus. GSE37572
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE43301
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE43302
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE44441
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE44445
- Kawaoka Y. 2012. Gene Expression Omnibus. GSE40792
- Aderem A. 2011. Gene Expression Omnibus. GSE37245
- Bankhead A. 2011. Gene Expression Omnibus. GSE33267
- Bankhead A. , III2012. Gene Expression Omnibus. GSE37827
- Baric R., Katze M., Sims A., Law G. L., Chang J. 2013. Gene Expression Omnibus. GSE47960
- Baric R., Katze M., Sims A., Law G. L., Chang J. 2013. Gene Expression Omnibus. GSE47961
- Baric R., Katze M., Sims A., Law G. L., Chang J. 2013. Gene Expression Omnibus. GSE47962
- Gralinski L. 2011. Gene Expression Omnibus. GSE33266
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE50000
- Katze M., Baric R., Gralinski L., Law L., Chang J. 2014. Gene Expression Omnibus. GSE51386
- Baric R., Katze M., Gralinski L., Chang J., Law L. 2014. Gene Expression Omnibus. GSE50878
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE51387
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE49262
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE49263
- Baric R., Katze M., Gralinski L., Law L., Chang J. 2012. Gene Expression Omnibus. GSE40824
- Baric R., Katze M., Gralinski L., Law L., Chang J. 2012. Gene Expression Omnibus. GSE40827
- Metz T. 2014. PeptideAtlas. PASS00416
- Metz T. 2014. PeptideAtlas. PASS00417
- Metz T. 2014. PeptideAtlas. PASS00418
- Metz T. 2014. PeptideAtlas. PASS00419
- Metz T. 2014. PeptideAtlas. PASS00420
- Metz T. 2014. PeptideAtlas. PASS00421
- Metz T. 2014. PeptideAtlas. PASS00422
- Metz T. 2014. PeptideAtlas. PASS00423
- Metz T. 2014. PeptideAtlas. PASS00424
- Metz T. 2014. PeptideAtlas. PASS00425
- Metz T. 2014. PeptideAtlas. PASS00426
- Metz T. 2014. PeptideAtlas. PASS00427
- Metz T. 2014. PeptideAtlas. PASS00428
- Metz T. 2014. PeptideAtlas. PASS00429
- Metz T. 2014. PeptideAtlas. PASS00430
- Metz T. 2014. PeptideAtlas. PASS00431
- Metz T. 2014. PeptideAtlas. PASS00432
- Metz T. 2014. PeptideAtlas. PASS00433
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_CA04M001-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL004-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL006-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL010-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL011-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL012-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM001-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM004-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM005-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM006A-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM006B-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database . IRD_SV_IM007-P
- Metz T., Smith R., Baric R., Katze M. 2013. Influenza Research Database. IRD_SV_SCL005-P
- Metz T., Smith R., Baric R., Katze M. 2013. Influenza Research Database. IRD_SV_SCL006-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_SM001-P
- Gralinski L., Baric R., Katze M. 2013. Influenza Research Database. IRD_SV_CA04M001-R
- Josset L. 2013. Influenza Research Database. IRD_SV_ECL001-R
- Kawaoka Y., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_ICL004-R
- Eisfeld A., Katze M., Kawaoka Y. 2012. Influenza Research Database. IRD_SV_ICL006-R
- Ellis A., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_ICL010-R
- Fan S., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_ICL011-R
- Fan S., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_ICL012-R
- Kawaoka Y., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_IM001-R
- Carter V., Katze M., Tumpey M.T. 2013. Influenza Research Database. IRD_SV_IM002-R
- Li C., Eisfeld A., Katze M., Kawaoka Y. 2012. Influenza Research Database. IRD_SV_IM004-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM005-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM006A-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM006B-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM007-R
- Chang J., Katze M., Tumpey T. 2013. Influenza Research Database. IRD_SV_IM009-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM010-R
- Aderem A., Thomas P. 2013. Influenza Research Database. IRD_SI_SBRI_AA_E1
- Baric R., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_SCL005-R
- Baric R., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_SCL006-R
- Sims A., Burkett S., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SHAE002-R
- Sims A., Burkett S., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SHAE003-R
- Sims A., Burkett S., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SHAE004-R
- Baric R., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_SM001-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM003-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM004-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM007-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM009-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM012-R
- Menachery V., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM014-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM015-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM019-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM020-R
References
- Peng X. et al. Virus-host interactions: from systems biology to translational research. Curr. Opin. Microbiol. 12, 432–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak D. E. & Aderem A. Systems biology of innate immunity. Immunol. Rev. 227, 264–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aderem A. et al. A systems biology approach to infectious disease research: innovating the pathogen-host research paradigm. MBio 2, e00325–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J. Virol. 85, 10955–10967 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L. et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 4, e00165–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L. et al. Implication of inflammatory macrophages, nuclear receptors, and interferon regulatory factors in increased virulence of pandemic 2009 H1N1 influenza A virus after host adaptation. J. Virol. 86, 7192–7206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L. E. et al. Mechanisms of severe acute respiratory syndrome coronavirus-induced acute lung injury. MBio. 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchitchek N. et al. Specific mutations in H5N1 mainly impact the magnitude and velocity of the host response in mice. BMC Syst. Biol. 7, 69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askovich P.S. et al. Differential host response, rather than early viral replication efficiency, correlates with pathogenicity caused by influenza viruses. PLoS ONE 8, e74863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V. C. et al. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell 154, 213–227(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell H. D. et al. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS ONE 8, e69374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A. C. et al. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 87, 3885–3902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V. D. et al. Attenuation and restoration of SARS-CoV mutant lacking 2' O methyltransferase activity. J. Virol. 88, 4251–4264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires R. B. et al. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir. Viruses 6, 404–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B. E. et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 40, D593–D598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J. S., Yuen K. Y., Osterhaus A. D. & Stohr K. The severe acute respiratory syndrome. N. Engl. J. Med. 349, 2431–2441 (2003). [DOI] [PubMed] [Google Scholar]
- Neumann G. et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl Acad. Sci. USA 96, 9345–9350 1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Watanabe S., Kim J. H., Hatta M. & Kawaoka Y. Novel approach to the development of effective H5N1 influenza A virus vaccines: use of M2 cytoplasmic tail mutants. J. Virol. 82, 2486–2492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M. et al. Impact of amino acid mutations in PB2, PB1-F2, and NS1 on the replication and pathogenicity of pandemic (H1N1) 2009 influenza viruses. J. Virol. 85, 4596–4601 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M. et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486, 420–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M., Gao P., Halfmann P. & Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293, 1840–1842 (2001). [DOI] [PubMed] [Google Scholar]
- Manicassamy B. et al. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 6, e1000745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash J. C. et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443, 578–581 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G. & Kawaoka Y. Reverse genetics systems for the generation of segmented negative-sense RNA viruses entirely from cloned cDNA. Curr. Top. Microbiol. Immunol. 283, 43–60 (2004). [DOI] [PubMed] [Google Scholar]
- Sanders C. J. et al. Compromised respiratory function in lethal influenza infection is characterized by the depletion of type I alveolar epithelial cells beyond threshold levels. Am. J. Physiol. Lung Cell. Mol. Physiol. 304, L481–L488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutigliano J. A. et al. Protective memory responses are modulated by priming events prior to challenge. J. Virol. 84, 1047–1056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyushina N. A. et al. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 84, 8607–8616 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M. M. et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl Acad. Sci. USA 105, 19944–19949 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B. et al. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79, 14909–14922 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B. et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA 100, 12995–13000 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3, e5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T. et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS ONE 5, e8729 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher M. L., Gabriel S., Burns K. A., Yankaskas J. R. & Randell S. H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107, 183–206 (2005). [DOI] [PubMed] [Google Scholar]
- Zeng H. et al. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J. Virol. 81, 12439–12449 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. in Bioinformatics and Computational Biology Solutions Using R and Bioconductor 397–420 (Springer: New York, NY, 2005). [Google Scholar]
- Gentleman R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebel G. R. et al. PRISM: a data management system for high-throughput proteomics. Proteomics 6, 1783–1790 (2006). [DOI] [PubMed] [Google Scholar]
- Jaitly N. et al. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics 10, 87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe M. E. et al. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23, 2021–2023 (2007). [DOI] [PubMed] [Google Scholar]
- Zimmer J. S., Monroe M. E., Qian W. J. & Smith R. D. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom. Rev. 25, 450–482 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson B. J. et al. Combined statistical analyses of peptide intensities and peptide occurrences improves identification of significant peptides from MS-based proteomics data. J. Proteome Res. 9, 5748–5756 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. M. et al. Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics 27, 2866–2872 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson B. J., Matzke M. M., Jacobs J. M., Pounds J. G. & Waters K. M. A statistical selection strategy for normalization procedures in LC-MS proteomics experiments through dataset-dependent ranking of normalization scaling factors. Proteomics 11, 4736–4741 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polpitiya A. D. et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24, 1556–1558 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J. E. et al. Conserved host response to highly pathogenic avian influenza virus infection in human cell culture, mouse and macaque model systems. BMC Syst. Biol. 5, 190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L. et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics 9(Suppl 9): S10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant P. A., Smyth G. K., Robins-Browne R. & Curtis N. Technical variability is greater than biological variability in a microarray experiment but both are outweighed by changes induced by stimulation. PLoS ONE 6, e19556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. Variability of DNA microarray gene expression profiles in cultured rat primary hepatocytes. Gene Regul. Syst. Bio. 1, 235–249 (2007). [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger A. M. et al. PRISM offers a comprehensive genomic approach to transcription factor function prediction. Genome Res. 23, 889–904 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F., Pesole G. & Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37, W247–W252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters K. M. et al. Network analysis of epidermal growth factor signaling using integrated genomic, proteomic and phosphorylation data. PLoS ONE 7, e34515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters K. M., Pounds J. G. & Thrall B. D. Data merging for integrated microarray and proteomic analysis. Brief Funct. Genomic Proteomic 5, 261–272 (2006). [DOI] [PubMed] [Google Scholar]
- Reich M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006). [DOI] [PubMed] [Google Scholar]
- Giardine B. et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Baric R., Katze M., Chang J., Gralinski L., Law L. 2012. Gene Expression Omnibus. GSE37569
- Josset L. 2013. Gene Expression Omnibus. GSE45042
- Li C. 2011. Gene Expression Omnibus. GSE28166
- Li C. 2012. Gene Expression Omnibus. GSE37571
- Katze M. G. 2013. Gene Expression Omnibus. GSE43203
- Katze M. G. 2013. Gene Expression Omnibus. GSE43204
- Li C. 2011. Gene Expression Omnibus. GSE33263
- Josset L. 2011. Gene Expression Omnibus. GSE36328
- Kawaoka Y. 2012. Gene Expression Omnibus. GSE37572
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE43301
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE43302
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE44441
- Kawoaka Y. 2013. Gene Expression Omnibus. GSE44445
- Kawaoka Y. 2012. Gene Expression Omnibus. GSE40792
- Aderem A. 2011. Gene Expression Omnibus. GSE37245
- Bankhead A. 2011. Gene Expression Omnibus. GSE33267
- Bankhead A. , III2012. Gene Expression Omnibus. GSE37827
- Baric R., Katze M., Sims A., Law G. L., Chang J. 2013. Gene Expression Omnibus. GSE47960
- Baric R., Katze M., Sims A., Law G. L., Chang J. 2013. Gene Expression Omnibus. GSE47961
- Baric R., Katze M., Sims A., Law G. L., Chang J. 2013. Gene Expression Omnibus. GSE47962
- Gralinski L. 2011. Gene Expression Omnibus. GSE33266
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE50000
- Katze M., Baric R., Gralinski L., Law L., Chang J. 2014. Gene Expression Omnibus. GSE51386
- Baric R., Katze M., Gralinski L., Chang J., Law L. 2014. Gene Expression Omnibus. GSE50878
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE51387
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE49262
- Katze M., Baric R., Law L., Gralinski L., Chang J. 2014. Gene Expression Omnibus. GSE49263
- Baric R., Katze M., Gralinski L., Law L., Chang J. 2012. Gene Expression Omnibus. GSE40824
- Baric R., Katze M., Gralinski L., Law L., Chang J. 2012. Gene Expression Omnibus. GSE40827
- Metz T. 2014. PeptideAtlas. PASS00416
- Metz T. 2014. PeptideAtlas. PASS00417
- Metz T. 2014. PeptideAtlas. PASS00418
- Metz T. 2014. PeptideAtlas. PASS00419
- Metz T. 2014. PeptideAtlas. PASS00420
- Metz T. 2014. PeptideAtlas. PASS00421
- Metz T. 2014. PeptideAtlas. PASS00422
- Metz T. 2014. PeptideAtlas. PASS00423
- Metz T. 2014. PeptideAtlas. PASS00424
- Metz T. 2014. PeptideAtlas. PASS00425
- Metz T. 2014. PeptideAtlas. PASS00426
- Metz T. 2014. PeptideAtlas. PASS00427
- Metz T. 2014. PeptideAtlas. PASS00428
- Metz T. 2014. PeptideAtlas. PASS00429
- Metz T. 2014. PeptideAtlas. PASS00430
- Metz T. 2014. PeptideAtlas. PASS00431
- Metz T. 2014. PeptideAtlas. PASS00432
- Metz T. 2014. PeptideAtlas. PASS00433
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_CA04M001-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL004-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL006-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL010-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL011-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_ICL012-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM001-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM004-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM005-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM006A-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_IM006B-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database . IRD_SV_IM007-P
- Metz T., Smith R., Baric R., Katze M. 2013. Influenza Research Database. IRD_SV_SCL005-P
- Metz T., Smith R., Baric R., Katze M. 2013. Influenza Research Database. IRD_SV_SCL006-P
- Metz T., Smith R., Kawaoka Y., Katze M. 2013. Influenza Research Database. IRD_SV_SM001-P
- Gralinski L., Baric R., Katze M. 2013. Influenza Research Database. IRD_SV_CA04M001-R
- Josset L. 2013. Influenza Research Database. IRD_SV_ECL001-R
- Kawaoka Y., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_ICL004-R
- Eisfeld A., Katze M., Kawaoka Y. 2012. Influenza Research Database. IRD_SV_ICL006-R
- Ellis A., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_ICL010-R
- Fan S., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_ICL011-R
- Fan S., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_ICL012-R
- Kawaoka Y., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_IM001-R
- Carter V., Katze M., Tumpey M.T. 2013. Influenza Research Database. IRD_SV_IM002-R
- Li C., Eisfeld A., Katze M., Kawaoka Y. 2012. Influenza Research Database. IRD_SV_IM004-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM005-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM006A-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM006B-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM007-R
- Chang J., Katze M., Tumpey T. 2013. Influenza Research Database. IRD_SV_IM009-R
- Li C., Eisfeld A., Chang J., Katze M., Kawaoka Y. 2013. Influenza Research Database. IRD_SV_IM010-R
- Aderem A., Thomas P. 2013. Influenza Research Database. IRD_SI_SBRI_AA_E1
- Baric R., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_SCL005-R
- Baric R., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_SCL006-R
- Sims A., Burkett S., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SHAE002-R
- Sims A., Burkett S., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SHAE003-R
- Sims A., Burkett S., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SHAE004-R
- Baric R., Smith D., Katze M., McWeeney S., Waters K. 2012. Influenza Research Database. IRD_SV_SM001-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM003-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM004-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM007-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM009-R
- Gralinski L., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM012-R
- Menachery V., Chang J., Katze M., Baric R. 2013. Influenza Research Database. IRD_SV_SM014-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM015-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM019-R
- Gralinski L., Chang J., Katze M., Yoshihiro K. 2013. Influenza Research Database. IRD_SV_SM020-R