Abstract
Glycosylation has significant effects on protein function and cell metastasis, which are important in cancer progression. It is of great interest to identify site-specific glycosylation in search of potential cancer biomarkers. However, the abundance of glycopeptides is low compared to that of nonglycopeptides after trypsin digestion of serum samples, and the mass spectrometric signals of glycopeptides are often masked by coeluting nonglycopeptides due to low ionization efficiency. Selective enrichment of glycopeptides from complex serum samples is essential for mass spectrometry (MS)-based analysis. Herein, a strategy has been optimized using LCA enrichment to improve the identification of core-fucosylation (CF) sites in serum of pancreatic cancer patients. The optimized strategy was then applied to analyze CF glycopeptide sites in 13 sets of serum samples from pancreatic cancer, chronic pancreatitis, healthy controls, and a standard reference. In total, 630 core-fucosylation sites were identified from 322 CF proteins in pancreatic cancer patient serum using an Orbitrap Elite mass spectrometer. Further data analysis revealed that 8 CF peptides exhibited a significant difference between pancreatic cancer and other controls, which may be potential diagnostic biomarkers for pancreatic cancer.
Keywords: Pancreatic cancer, lectin, core fucosylation, glycoproteomics, iTRAQ, serum
INTRODUCTION
Glycosylation of proteins is one of the most common post-translational modifications (PTMs) involved in biological processes such as cell adhesion, receptor activation, and inflammatory response.1,2 N-Glycosylation occurs on the asparagine amino acid in the sequence of N-X-S/T/C, with X being any amino acid with the exception of proline. Core fucosylation (CF), which is one type of N-linked glycosylation modification, consists of an α-1,6 fucose substitution on the innermost N-acetylglucosamine (GlcNAc) of the common core pentascaccharide, triple mannose glycans and biacetylglucosamine (Man3-GlcNAc-GlcNAc-(Asn))3.
Aberrant glycosylation may contribute to various aspects of cancer development and progression, where aberrant CF glycoproteins have been reported to undergo significant changes in various tumors and cancers.4,5 Accumulated studies have revealed a series of different CF levels in patient serum samples among normal, benign disease, and cancer.6,7 For example, it has been shown that 25 fucosylated proteins in serum show significant differences between pancreatic cancer and other conditions using a TMT label strategy.8 Increased fucosylation and branching of N-linked oligosaccharides were also observed in pancreatic cancer during glycan profiling using hydrophilic interaction separation.9 Alpha-2-macroglobulin (A2MG) CF levels at sites N396 and N1424 were found to decrease in both chronic pancreatitis and pancreatic cancer compared to that in healthy controls.10 These results imply that site-specific CF aberrations are highly associated with cancer and that the ability to identify and quantify aberrant CF proteins and CF sites in patient serum may provide a means for marker identification for pancreatic cancer.
The detection of low-abundance glycopeptides in complex mixtures is difficult due to the suppression of the glycopeptide mass spectral signals in the presence of nonglycopeptides.11 In order to markedly increase the number of CF glycopeptides identified, highly efficient enrichment of CF glycopeptides from complex serum samples is crucial. One method to achieve this goal is to use lectin affinity enrichment.12 LCA lectin, which is isolated from legume plant Lens culinaris, recognizes glycopeptide sequences containing α-linked mannose residues. LCA shows high binding affinity to glycan moieties, where the corefucose attaches to bi- and triantennary complex type N-acetylglucosamine (GlcNAc).13,14 Addition of Ca2+ and Mn2+ to the binding buffer further enhances LCA affinity activity.15 Previous studies have demostrated that LCA lectin affinity enrichment has a high specificity in the CF glycopeptides enrichment from tissues and serum.5,16
In this study, we have optimized a protocol to identify CF proteins in 13 sets of serum samples from pancreatic cancer, chronic pancreatitis, and healthy controls and characterized CF sites in all CF proteins. Our improved strategy using an optimized elution buffer enabled the large-scale enrichment of CF proteins. A yield of 193 CF proteins was achieved in a single analytical run using our optimized enrichment method. A total of 630 sites were ultimately identified, and 8 CF peptides were found to discriminate pancreatic cancer from other controls. In addition, an overview of cellular location, type, and annotation of these CF proteins was analyzed using Gene Ontology (GO).
MATERIALS AND METHODS
Chemical Materials
Trifluoroacetic acid (TFA), tris (2-carboxyethyl) phosphine (TCEP), iodoacetamide (IAA), α-methylmannoside, α-methylglucoside, TEAB, Bradford reagents, water (HPLC grade), formic acid (HPLC grade), and acetonitrile (HPLC grade) were purchased from Sigma-Aldrich (St. Louis, MO). The Pierce centrifuge column (2 mL), C18 spin column, and low binding tips were purchased from Thermo Fisher Scientific (Fair Lawn, NJ). Low protein binding tubes were purchased from Eppendorf (Hauppauge, NY). Agarose bound Lens culinaris agglutinin (LCA) was purchased from Vector Laboratories (Burlingame, CA). Amicon Ultra 3K centrifugal filters (15 and 4 mL) were purchased from Millipore (Billerica, MA). The Bradford assay kit was purchased from BioRad (Hercules, CA). Isobaric tags for relative and absolute quantitation (iTRAQ) 4-plex reagent kits were purchased from AB Sciex (Framingham, MA). Sequencing grade modified trypsin enzyme was obtained from Promega (Madison, WI). Endoglycosidase F3 (Endo F3) was purchased from BA-Bio (San Mateo, CA).
Serum Sample Preparation
Human healthy controls, chronic pancreatitis, and pancreatic cancer sera were provided by the University Hospital, Ann Arbor, Michigan, according to Institutional Review Board (IRB) approval. Analysis on 13 sample sets was performed in this study, where each sample set includes pancreatic cancer, chronic pancreatitis, healthy control, and a standard reference. A standard reference was employed to estimate protein relative abundances in human serum samples where it was used as an internal standard to compare the 13 sample sets by iTRAQ. The healthy controls were used to compare CF differences with pancreatic cancer or chronic pancreatitis. All samples in different groups were matched in gender, age, and population. Chronic pancreatitis samples were included as benign disease controls due to the typically desmoplastic response observed in pancreatic cancer samples. The samples were aliquoted and stored at −80 °C until use. Two-hundred fifty microliters of serum sample was thawed at 4 °C and filtered through a 0.45 μm filter to remove cell particles or cell debris. Following the strategy shown in Supporting Information Figure 1, the top 14 high-abundance proteins of the serum were depleted first with an IgY14 LC10 column (Sigma, St. Louis, MO) according to previous work.17 In brief, the serum sample was diluted with 1× depletion buffer and loaded onto the IgY14 LC10 column on a HPLC system (Beckman Coulter, Brea, CA). The flow-through fraction was collected from 0 to 30 min and then transferred into a 15 mL YM-3 centrifugal device and centrifuged at 5000g at 4 °C for 1 h, followed by buffer exchange three times with 5 mL of deionized water. The buffer exchange sample was transferred to an Eppendorf tube. Protein concentration was measured using a Bradford assay kit. Each sample was aliquoted as 250 μg of protein per tube, of which 100 μg was pooled to create a mixture sample. A standard reference sample for quantification of mass spectrometry analyses was collected from a mixture of each sample used in this study, including healthy controls, chronic pancreatitis, and pancreatic cancer. The mixed reference sample was aliquoted as 100 μg of protein per tube. All protein aliquots were stored at −80 °C for further use.
Trypsin Digestion and iTRAQ Labeling
One-hundred micrograms of protein from each serum sample was used for one labeling reaction. The protein samples were denatured by adding 8 M urea to a final concentration of 4 M urea. Then the protein samples were reduced by adding 0.5 M TCEP to a final concentration of 0.01 M and kept at room temperature for 30 min. Subsequently the protein sample was alkylated by adding IAA to a final concentration of 22 mM and kept in the dark for 30 min at room temperature. The protein samples were diluted 5-fold with 50 mM TEAB solution to reduce the urea concentration and digested by sequencing grade trypsin using a 1:30 (w/w) ratio at 37 °C overnight. Digestion was stopped by adding 1 μL of trifluoroacetic acid. The resulting peptide mixtures were dried using a SpeedVac (Labconco, Kansas City, MO). Four samples, including a standard reference, healthy control, chronic pancreatitis, and pancreatic cancer were labeled with iTRAQ 4-plex isobaric reagents, according to the manufacturer’s instructions. iTRAQ tag 114 was assigned to the standard reference, and iTRAQ tags 115, 116, and 117 were assigned randomly to healthy controls, chronic pancreatitis, and pancreatic cancer in 13 sample sets. The labeling reaction was quenched using 30 μL of 5% NH3· H2O. The four samples were then pooled together and dried using a SpeedVac.
CF Peptides Enriched by Lens culinaris Agglutinin (LCA)
Prior to LCA enrichment, buffer exchange was performed for the sample labeled with iTRAQ reagents using an Amicon Ultra 3K centrifugal filter (4 mL). After buffer exchange, approximately 40 μL of peptide solution sample was obtained. Two milliliters of LCA binding buffer (20 mM TrisHCl, 0.1 M NaCl, pH 7.4) was added into the tube and centrifuged at 7500g at 4 °C for 50 min, where this process was repeated twice. One milliliter of agarose-bound LCA slurry (1:1, v/v slurry) was transferred into a 2 mL disposable gravity-flow centrifuge column using a 1 mL pipet, where 4–5 mm of the tip was cut off. The resin slurry was washed by LCA binding buffer three times. Peptide solution sample was transferred to the centrifuge spin column. Four-hundred sixty microliters of binding buffer was added into the centrifuge column. The mixture of agarose-bound LCA and peptide sample solution was incubated at room temperature with gentle shaking for 20 min. The flow through was collected and reloaded into the original centrifuge column followed by another 20 min incubation. The flow through was discarded, and then the sample mixture was washed three times by approximately 4 mL of binding buffer, which filled the centrifuge column. During the first washing, binding buffer was applied to wash the cover and tip of the centrifuge column. The washing buffer was collected and transferred into the centrifuge column using a 200 μL pipet, where 3 to 4 mm of the tip was cut off. Bound peptides were then eluted with 800 μL of elution buffer (LCA binding buffer with 200 mM α-methylglucoside and 200 mM α-methylmannoside) after a 20 min incubation with gentle shaking, where this process was repeated twice. Further elution of tightly bound peptides was performed using 800 μL of elution buffer without NaCl, Ca2+, and Mn2+ ions three times (20 mM Tris buffer with 200 mM α-methylglucoside and 200 mM α-methylmannoside). All collected samples were desalted and buffer exchanged to 50 mM sodium acetate, pH 4.5, using a 3K centrifugal filter (4 mL).
Partial Deglycosylation from CF Peptides by Endo F3, Desalted by C18 Spin Column
The volume of enriched glycopeptides sample was adjusted to 50 μL using 50 mM sodium acetate solution (pH 4.5). Three units of Endo F3 (0.5 unit/μL) were added, and samples were incubated at 37 °C overnight. The partially deglycosylated peptides were desalted by a C18 spin column according to the manufacture’s guidance with some modification. The resin was activated with 200 μL of 50% ACN and then equilibrated with 200 μL of 0.1% TFA. The pH of the deglycosylated peptide sample was adjusted to 3 with 1% TFA solution. The sample was loaded into a C18 spin column and centrifuged at 1500g for 1 min. The flow through was reloaded and centrifuged for another four times. Nonbinding peptides were removed by washing with 0.1% TFA and centrifuged at 1500g for 0.5 min three times. Peptides binding to C18 reversed-phase resins were eluted by adding 25 μL of 70% ACN solution with centrifugation at 1500g for 1 min. The elution step was repeated three times, and approximately 100 μL of flow through was collected. The sample was dried in a SpeedVac at room temperature.
NanoLC–MS/MS Analysis
The desalted glycopeptide mixtures were analyzed on a Proxeon Easy-nLC II system (Thermo), directing the eluent to the nanospray source of a linear ion trap Oribitrap Elite mass spectrometer (Thermo) operating in positive mode. One-fourth of each sample was injected for one run. Each sample was run in duplicate. The peptides were separated on a 25 cm column with 75 μm inner diameter, packed in-house with the reverse-phase material Magic C18 AQ 100, 5 μm particle size. Mobile phase A (0.1% formic acid in water) and mobile phase B (99.9% acetonitrile and 0.1% formic acid) were used with a linear gradient of mobile phase B, 5 to 35% for 74 min and 35 to 95% for 7 min, at a flow rate 400 nL/min. MS data acquisition was carried out using the Xcalibur software (Thermo Fisher Scientific, San Jose, CA) in data-dependent mode with the following settings: the resolution of full-scan was 120 000 and the mass window for precursor ion was m/z 400.0–1800.0, automated peak recognition, multiply charged top 15 most intense precursor ions were chosen for tandem MS, and normalized collision energy was set at 35% for higher-energy collision dissociation (HCD) analysis. The dynamic exclusion was enabled at a repeat count 1, repeat duration 30 s, exclusion list size 500, and exclusion duration 30 s.
Data Analysis
All spectra were analyzed with Proteome Discoverer 1.4 (Thermo Fisher Scientific, San Jose, CA) software using SEQUEST HT search engine with the following parameters: (1) static modifications, N-terminus labeling iTRAQ 4-plex (+144.102 Da), cysteine carbamidomethylation (+57.021 Da), lysine iTRAQ labeling (+144.102 Da); (2) dynamic modifications, methionine oxidation (+15.995 Da), asparagines glycosylation GlcNac + fucose (+349.137 Da); (3) 2 maximum missed cleavage sites were allowed; (4) Swiss-Prot Homo sapiens database (reviewed, downloaded in April, 2014) was used; (5) trypsin was selected as digestion enzyme; (6) precursor mass tolerance 10 ppm and fragment mass tolerance 0.05 Da; (7) high peptide confidence filter; (8) the peptide FDR was set at 1% and the shortest peptide length was set at 6 amino acids. The peptide identification data were exported into the XML-formatted file from Proteome Discoverer software followed by manual assessment of the spectra. Protein function was annotated using Gene Ontology (GO) with the DAVID bioinformatics online tool (http://david.abcc.ncifcrf.gov/). The significantly differentially expressed CF levels between each of two groups among pancreatic cancer, healthy controls, and chronic pancreatitis were evaluated using Student’s t test at confidence intervals of 95% and two-tailed p values. The scatterplots of CF peptide ratio were generated using ANOVA with Prism 6 (GraphPad, La Jolla, CA). The receiver operator characteristic (ROC) curve of CF ratio among pancreatic cancer, healthy controls, and chronic pancreatitis was generated with SPSS 16 (IBM, Armonk, NY).
RESULTS AND DISCUSSION
Optimization of the LCA Lectin Enrichment Method
An important aspect of this work involved optimizing the elution efficiency for CF peptides for biomarker discovery. One-hundred micrograms of protein in commercial normal serum was applied using the optimized procedure, where the top 14 high-abundance proteins in commercial normal serum were depleted. Thirty CF peptides were identified using standard elution buffer, whereas 49 CF peptides were identified using an additional elution buffer without NaCl, Ca2+, and Mn2+ after standard elution buffer using an LTQ mass spectrometer for analysis (Supporting Information Figure 2). Increasing the binding efficiency of CF glycopeptides to LCA lectin and efficient release of the bound CF glycopeptides from the LCA lectin column are two essential factors for CF glycopeptides enrichment. In previous studies, NaCl, Ca2+, and Mn2+ were added to increase the lectin affinity.16,18 In this work, we applied an additional elution step using the elution buffer without NaCl, Ca2+, and Mn2+, which resulted in a greater number of CF glycopeptides identified compared to that using standard elution buffer only. The additional eluent was collected separately to detect the additional CF peptides released by nonionic elution buffer. An additional 35.4% of CF peptides were obtained by using the nonionic elution buffer after regular elution.
The metal ions not only serve to maintain the integrity of the subunits of the lectins but also help to position amino acid residues for carbohydrate binding.19,20 Elution buffer has been prepared by adding the appropriate sugar to compete for binding glycoprotein in previous reports.21–23 These reports applied various lectins to enrich glycoproteins routinely according to the manufacture’s guidance without consideration of the factors involved in the elution step, where the effect of NaCl and the divalent metal ions Ca2+ and Mn2+ were neglected in the elution process. In our study, the use of elution buffer without NaCl, Ca2+, and Mn2+ increased the elution efficiency of CF peptides from the LCA lectin. More than one-third of the CF peptides was recovered from the lectin beads. In previous reports of CF proteins or CF peptide enrichment approaches, HILIC or hydrazine has been combined with lectin affinity enrichment to increase the purity of glycopeptides; however, the additional step may also increase the potential sample loss.23,24 In this study, the enrichment at the glycopeptide level was applied in order to quantify the differential CF levels at the peptide level in various disease samples.
It is expected that only those glycopeptides with LCA binding motifs would be eluted using elution buffer. However, we found that more non-CF peptides than CF peptides were coeluted (data not shown) and that a small portion of resin was retained in the cover, tip, and on the tube wall of the spin column. To minimize sample loss and to avoid contamination of non-CF peptides, binding buffer was added into the cover and tip to collect the peptides retained in the cover. In addition, the centrifuge column was filled with binding buffer in order to wash all of the slurry to the bottom so that the nonglycopeptides remaining on the wall of the spin column were reduced to a minimum. With nonglycopeptides efficiently washed, more glycopeptides were obtained due to the increased mass spectrometric signals of glycopeptides by decreasing the interference from nonglycopeptides, where the latter have a much higher ionization efficiency.25,26 To maximize CF elution efficiency, two elution steps were applied: first using regular elution buffer and then by elution buffer without NaCl and the divalent cations Ca2+ and Mn2+.
Criteria for Determination of the CF Site
All candidate CF sites were initially identified by the SEQUEST searching algorithm with precursor ions plus 349.137 Da (GlcNAc-fucose) and confirmed by manual checking for the presence of oxonium ions at m/z 126.055, m/z 138.055, m/z 186.066, and m/z 204.087.23,24 Only precursor ions that were identified with high confidence (1% FDR) by SEQUEST and generated oxonium ions were considered as true CF peptides. Diagnostic ions were not observed in 2 of 274 peptides derived from one mass spectrometry run (Supporting Information Table 1). We further checked the frequency and ratio of diagnostic ions. Diagnostic ions at m/z 126.055 and m/z 204.087 were found in 99.3 and 93.4% of all assigned HCD spectra, respectively. All four oxonium ions were found in 62.0% of all assigned HCD spectra in one analytical run (Supporting Information Table 2). We combined all peptides with CF modification assigned by SEQUEST and the N-X-S/T/C motif to further screen the potential CF peptides by manual inspection. Most potential CF peptides were consistent with the consensus motif; however, 28 candidate CF peptides assigned by the SEQUEST algorithm did not have the consensus motif.
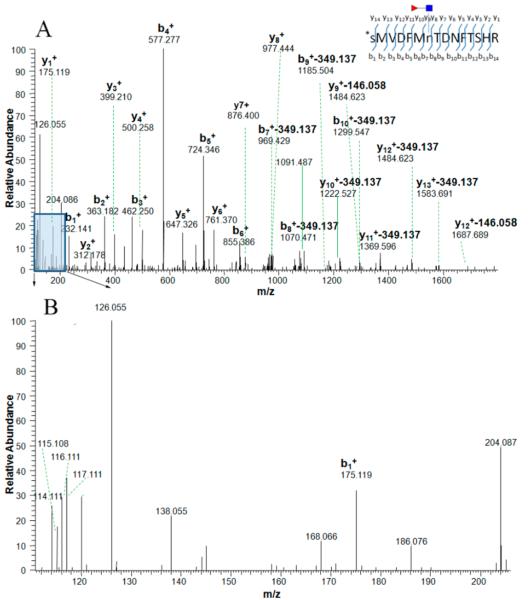
In previous studies, candidate N-linked peptides without an N-X-S/T/C motif were excluded.24,27 This type of N-linked peptide is often ignored due to its low presence in the total N-linked peptide cohort. However, some peptides without the consensus motif are also N-linked peptides.28 A similar phenomenon was also found in mouse tissues and blood plasma, where 2.2% of all identified N-linked peptides did not match the consensus motif.11 The diagnostic ions of all these peptides without the consensus motif were checked manually. For example, the mass spectrum of peptide SMVDFMnTDNFTSHR (n means the CF modification at asparagine acid residue) has diagnostic ions, but it does not match with the consensus motif, N-X-S/T/C (Figure 1A,B). Other potential CF peptides assigned by the SEQUEST searching algorithm without the N-X-S/T/C consensus motif and without characteristic diagnostic ions were excluded. Due to the presence of trypsin missed cleavage, especially the presence of attached carbohydrates where a K or R residue was located immediately after the glycosylated site,29,30 numerous CF peptides could be calculated redundantly without excluding the duplication. In the current study, in order to calculate the number of CF sites, all peptides were calculated based on the number of K and R sites recognized by trypsin enzyme and checked for the presence of trypsin missed cleavage sites, which ensures that each CF peptide is unique when the number of CF peptides is calculated. In a previous report, the missed cleavages were not excluded when the number of glycosites was counted.16 Thus, herein, not only were the duplicated CF peptides due to the trypsin missed cleavage removed but also real CF peptides without the consensus motif were retrieved by checking the oxonium ions (Figure 1B). In the present study, only CF glycopeptides were assigned, where the non-CF and falsely identified CF glycopeptides were removed after screening as described above.
Figure 1.
MS/MS spectra of a tryptic CF peptide, SMVDFMnTDNFTSHR, labeled by iTRAQ (N-terminal amino acid residue) without the N-X-S/T/C consensus motif from protein afamin, where the CF modification was confirmed by diagnostic ions. The ion at m/z 765.763 (2+) was isolated and fragmented by high-energy collision-induced dissociation (HCD) (A). The typical N-linked glycan (NGlcNAc) backbone fragmentation can be identified by the oxonium ions in the lower mass range (B); the signal from the GlcNAc oxonium ion at m/z 126.055 was prominent in the spectrum and oxonium ions at m/z 138.055, 168.088, 186.066, 204.087 were also present. In the higher mass range, the fragment peaks exhibit specific mass differences corresponding to neutral loss product ions and provide information on the CF position on the CF peptide. Product ions with a loss of 349.137 Da (GlcNAcFuc) or 146.058 Da (fucose) are present. Red triangle stands for the fucosyl-GlcNAc glycan residues retained on glycoside.
Evaluation of the Coverage of CF Proteins and CF Peptides in Serial Sample Sets
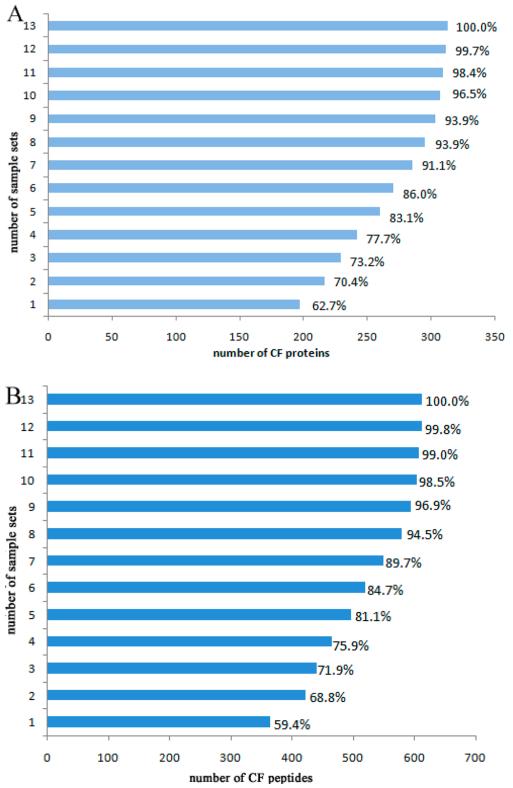
Thirteen sample sets were evaluated to test the coverage of CF glycopeptides and CF glycoproteins in serial sample sets including pancreatic cancer, chronic pancreatitis, healthy control, and a standard reference. Each sample set was run in technical duplicates in the Orbitrap Elite mass spectrometer. The average number of CF glycopeptides and CF glycoproteins identified in a single set was 295 and 174, respectively, whereas the maximum was 350 and 193, respectively (Table 1). To our knowledge, this is the largest data set of CF glycoproteins identified from a single analytical run of human serum samples to date. The results illustrate that the optimized method was very efficient for extraction of CF peptides. In addition, the results also showed that both the number of CF peptides and CF proteins increased with the number of samples run (Figure 2A,B). The number of CF peptides identified grew rapidly at the beginning, but when the number of sample sets run was larger than 7, this number increased only slightly. Thus, the number of CF peptides and CF proteins did not significantly increase along with the increase of sample size above 7 sets. The 7 sample sets covered approximately 90% of total CF peptides and CF proteins obtained from the 13 sample sets.
Table 1. CF Peptides and CF Proteins Identified among 13 Sample Sets.
| no. of CF peptides | no. of CF proteins | |
|---|---|---|
| average of 13 sample sets | 295 ± 76 | 174 ± 39 |
| maximum of single run | 350 | 193 |
Figure 2.
Coverage of CF proteins (A) and CF peptides (B) identification in 13 sample sets. Total CF peptides or proteins derived from 13 sample sets were considered as 100%. Comparison of total CF peptides or proteins derived from 13 sample sets, where the coverage was based on accumulation of sample set 1 to 13. Each sample set includes pancreatic cancer, chronic pancreatitis, healthy control, and a standard reference.
Large-Scale CF Glycosylation Sites Analysis in Pancreatic Cancer Serum
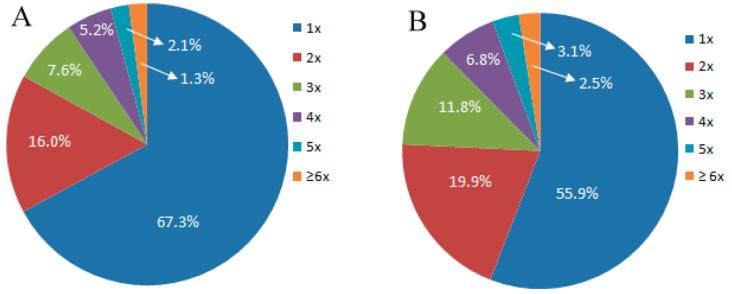
By optimizing the elution buffer in the LCA enrichment procedure and increasing coverage in a sufficient number of pancreatic cancer sample sets, 630 CF sites from 613 CF peptides and 322 CF proteins were identified using 13 sample sets (Supporting Information Table 3). Among all identified CF proteins, 58% CF proteins contain one CF site and 22% CF proteins contain 2 CF sites, whereas 11.2, 6.9, and 3.1% CF proteins have 3, 4, and 5 CF sites, respectively. However, more than 5 CF sites in the same CF proteins are very rare, where only 1.3% CF proteins were identified with 6 CF peptides (Figure 3A). Our results revealed that 97.22 and 2.78% of the peptides have one and two CF sites, respectively (Supporting Information Figure 3). The distribution of CF sites in proteins derived from pancreatic cancer serum (Figure 3B) is also similar to that of N-glycosylation sites in proteins derived from human pancreas tissue.31 Due to the importance of glycoproteins in disease detection and clinical diagnosis, numerous studies have focused on glycoprotein identification and quantification in human serum, urine, and tissue.8,32 Efforts have been devoted to CF protein identification by improvements in experimental methodology.16,23
Figure 3.
Frequency of CF peptides in CF proteins (A) and the distribution of single and multiple CF sites per protein (B).
Improved MS technology has also increased the identification of CF peptide sites and CF proteins. Seventy nine CF proteins were identified using an LTQ-FT mass spectrometer in a mixture of plasma sample from 8 HCC patients where IgG was depleted in one study.16 Ma et al. further identified 357 CF sites from 209 CF proteins recently from a mixture of 6 healthy human plasma samples using a low- and high-normalized collision energy strategy in an Orbitrap Elite mass spectrometer.33 The data were derived from two MS analytical runs under the same conditions except that different normalized collision energy in the fragmentation of HCD was applied. The searching algorithm pFind was applied to the Ma et al. study, where pFind identified more peptides than SEQUEST.34 It should be noted that Cao et al. recently also reported that 702 and 449 CF proteins were identified from mouse liver tissue and HeLa cell samples, respectively.23 A nearly 7-fold increase in the identification efficiency of CF peptides has been obtained using the glycan diagnostic ion-based spectrum refinement method in the same RAW data.23 The pFind search engine was also applied to search the candidate spectra against human Refseq database.23 In general, the number of global glycoproteins in tissue or cell lines is much higher than that in serum. The spectrum refinement method is also potentially useful in our future work, where, compared to other reports, we have identified the largest number of CF proteins in human serum thus far.
Characterization of CF-Glycosylation Motif in CF-Glycosylation Proteins from Pancreatic Cancer Serum
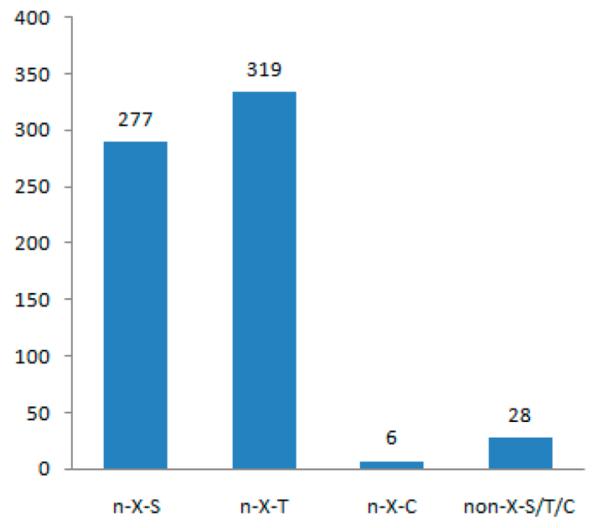
Of the 630 CF sites in 613 CF peptides identified, the number of CF sites with the N-X-S and N-X-T consensus motifs are 277 and 319, respectively, where 95.56% of identified CF peptides match the consensus motif N-X-S/T/C and the threonine residue occurs a little more frequently than the serine residue at the second position of this motif (Figure 4). The results are consistent with previous reports on N-linked glycoproteins in cell surface N-glycoproteomes of T and B cells35 or N-linked glycosylation in four mouse tissues and blood plasma.11 It should be noted that the total number of 33 CF sites were retrieved by manually checking whether the amino acid residue is S or T after lysine (K) and arginine (R) amino acid residues that were cleaved by trypsin. Furthermore, there were 6 CF sites (approximately 1% total CF sites) with the N-X-C consensus motif. This result is consistent with other previous reports in mouse tissue and plasma.11 However, there are still 28 CF identified sites (approximately 4.4% of total CF sites) that do not contain an N-X-S/T/C sequon, but the existence of CF sites is supported by searching the results and diagnostic ions. Some previous reports included these N-linked peptide sites, whereas others excluded these peptides without the consensus motif.36,37 It is interesting to note that in the identified 613 CF peptides, besides the 630 CF sites, there are still 72 nonglycopeptide sites containing the consensus motif N-X-S/T/C but without CF modification, which means that the presence of this consensus peptide or the presence of nearby CF modification does not always lead to CF modification of these sites. These results are consistent with previous studies,38,39 illustrating that the consensus motif N-XS/T/C is required but not sufficient for N-linked glycopeptides.
Figure 4.
Peptide motifs of CF sites identified in this study.
Gene Ontology (GO) Analysis of CF Proteins
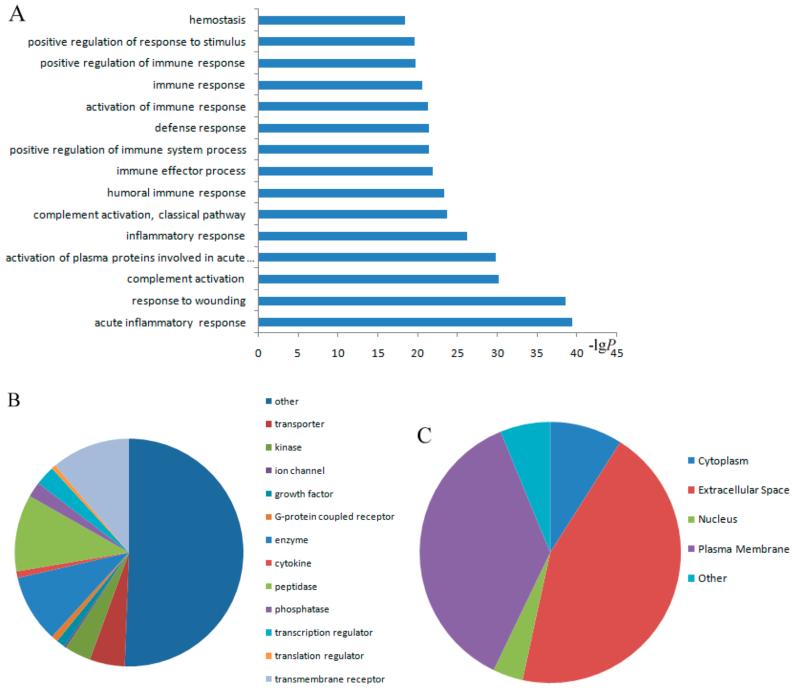
GO analysis was conducted by submitting the list of the UniProt IDs of the 322 CF proteins to the DAVID bioinformatics online tool. The results from the GO analysis revealed that the CF proteins were preferentially found in acute inflammatory response, response to wounding, activation of plasma proteins involved in acute inflammatory response, and other immune responses. Figure 5A highlights the list of significant CF protein p values in biological processes. Half of the CF proteins were assigned to transporter, kinase, ion channel, growth factor, G-protein coupled receptor, enzyme, cytokine, peptidase, etc. (Figure 5B). The dominant types of CF proteins were enzyme, peptidase, and transmembrane receptors. In addition, most identified CF proteins were located in the extracellular space and plasma membrane (Figure 5C). These results are consistent with other studies in which N-linked glycoproteins were found to be involved in a series of biological processes such as protein folding, host–pathogen interactions, immune response, and pathogenesis of many cancers.40,41 All CF protein p values derived from GO analysis are shown in Supporting Information Table 4.
Figure 5.
GO analysis of CF proteins. Significance of the p values of CF proteins that are responsible for different biological processes compared to total proteins from a Homo sapiens database (A). Types of CF proteins (B). Subcellular distribution of CF proteins (C).
Discovery of Potential CF Protein Markers
On the basis of the reference standard sample, the ratio of three different samples in the same set can be calculated since they were run together using iTRAQ-based labeling. The ratio of healthy controls, chronic pancreatitis, and pancreatic cancer over the standard reference was obtained by the SEQUEST algorithm during database searching. A comparative iTRAQ-based analysis of identified CF peptides between groups was performed. CF peptides that were present at significantly different levels between each of two groups among pancreatic cancer, healthy controls, and chronic pancreatitis are listed in Tables 2 and 3. In general, most peptides with CF modifications among pancreatic cancer, chronic pancreatitis, and healthy controls are quite stable without significant change. However, of these 613 CF unique peptides, 55 showed differences between pancreatic cancer and healthy controls. Also, 33 out of 55 of these CF peptides showed a significant difference between chronic pancreatitis and healthy controls (Table 2). Most CF proteins have only one peptide that shows such differences between pancreatic cancer and the control sample. Only 10 CF proteins have two or three unique CF peptides differentially expressed between groups. For example, three unique CF peptides of antithrombin-III, beta-2-glycoprotein 1, and IgGFc-binding were significantly different between groups. Thirteen unique CF peptides from 13 proteins showed significant differences between pancreatic cancer and chronic pancreatitis (Table 3). Each CF protein has only one peptide with CF modification. In addition, CF peptides from the proteins voltage-dependent calcium, aggrecan, and lysyl oxidase homologue 3 were not able to distinguish chronic pancreatitis from healthy controls.
Table 2. List of Differentially Expressed CF Peptides between Healthy Control Samples and Pancreatic Cancer or Chronic Pancreatitisa.
| accession no. | protein name | CF peptideb | N/SR | CP/SR | PC/SR |
|---|---|---|---|---|---|
| Q86TH1 | ADAMTS-like protein 2 | DFTLn*ETVNSIFAQGAPR | 0.71 ± 0.26 | 1.15 ± 0.37c | 1.01 ± 0.28c |
| P43652 | Afamin | DIENFn*STQK | 0.54 ± 0.33 | 1.20 ± 0.67c | 1.28 ± 0.85c |
| YAEDKFn*ETTEK | 0.40 ± 0.23 | 1.29 ± 0.85c | 1.42 ± 1.09c | ||
| P02765 | Alpha-2-HS-glycoprotein | AALAAFNAQNn*GSNFQLEEISR | 0.82 ± 0.26 | 1.43 ± 0.53c | 1.14 ± 0.16c |
| P01008 | Antithrombin-III | SLTFn*ETYQDISELVYGAK | 0.77 ± 0.28 | 1.30 ± 0.75 | 1.23 ± 0.41c |
| AAINKWVSn*KTEGR | 0.68 ± 0.40 | 1.41 ± 0.69c | 1.17 ± 0.52c | ||
| LGACn*DTLQQLMEVFK | 0.82 ± 0.43 | 1.19 ± 0.58 | 1.14 ± 0.33c | ||
| P02749 | Beta-2-glycoprotein 1 | DTAVFECLPQHAMFGn*DTITCTTHGn*WTK | 0.60 ± 0.33 | 1.42 ± 0.87c | 1.19 ± 0.67c |
| LGn*WSAMPSCK | 0.68 ± 0.44 | 1.16 ± 0.47c | 1.22 ± 0.67c | ||
| VYKPSAGn*NSLYR | 0.67 ± 0.34 | 1.55 ± 1.21c | 1.18 ± 0.71c | ||
| P20851 | C4b-binding protein beta chain | EWDn*TTTECR | 0.59 ± 0.26 | 1.13 ± 0.54c | 1.12 ± 0.63c |
| P07339 | Cathepsin D light chain | GSLSYLn*VTR | 0.81 ± 0.24 | 1.06 ± 0.23c | 1.10 ± 0.21c |
| P02760 | cDNA FLJ51445, highly similar to AMBP protein |
SKWn*ITMESYVVHTNYDEYAIFLTK | 0.83 ± 0.46 | 1.52 ± 0.88c | 1.71 ± 0.88c |
| P00450 | Ceruloplasmin | EHEGAIYPDn*TTDFQR | 0.75 ± 0.32 | 1.10 ± 0.37c | 1.05 ± 0.30c |
| En*LTAPGSDSAVFFEQGTTR | 0.78 ± 0.34 | 1.14 ± 0.42c | 1.12 ± 0.38c | ||
| Q9NZP8 | Complement C1r subcomponent-like protein |
n*QSVNVFLGHTAIDEMLK | 0.90 ± 0.41 | 1.55 ± 0.64c | 1.38 ± 0.50c |
| Q03591 | Complement factor H-related protein 2 | LQNNENn*ISCVER | 0.53 ± 0.27 | 1.40 ± 0.79c | 1.26 ± 0.97c |
| P05156 | Complement factor I light chain | n*GTAVCATNR | 0.83 ± 0.26 | 1.36 ± 0.60c | 1.16 ± 0.45c |
| Contactin-4 | SDVGn*YTCVVTNTVTNHK | 0.86 ± 0.25 | 1.06 ± 0.22 | 1.14 ± 0.18c | |
| Q02487 | Desmocollin-2 | An*YTILK | 0.71 ± 0.33 | 0.91 ± 0.33 | 1.08 ± 0.22c |
| Q9H4A9 | Dipeptidase 2 | GVHSFYNn*ISGLTDFGEK | 0.97 ± 0.19 | 1.25 ± 0.22c | 1.15 ± 0.20c |
| Q14118 | Dystroglycan | LGAn* GSHIPQTSSVFSIEVYPEDHSELQSVR | 0.80 ± 0.23 | 1.19 ± 0.27c | 1.03 ± 0.17c |
| P02790 | Hemopexin | ALPQPQn*VTSLLGCTH | 0.62 ± 0.25 | 1.18 ± 0.56c | 1.13 ± 0.34c |
| SWPAVGn*CSSALR | 0.70 ± 0.38 | 1.04 ± 0.44c | 1.21 ± 0.55c | ||
| P05546 | Heparin cofactor 2 | DFVn*ASSK | 0.71 ± 0.35 | 1.30 ± 0.50c | 1.16 ± 0.50c |
| n*LSMPLLPADFHK | 0.71 ± 0.35 | 1.15 ± 0.40c | 1.07 ± 0.44c | ||
| P26927 | Hepatocyte growth factor-like protein alpha chain |
GTGn*DTVLNVALLNVISNQECNIK | 0.86 ± 0.22 | 1.14 ± 0.56 | 1.24 ± 0.35c |
| Q9Y6R7 | IgGFc-binding protein | VITVQVAn*FTLR | 0.74 ± 0.34 | 1.14 ± 0.89 | 1.39 ± 0.73c |
| FDFQGTCEYLLSAPCHGPPLGAEn*FTVTVANEHR | 0.72 ± 0.35 | 1.25 ± 0.34c | 1.12 ± 0.39c | ||
| YLPVn*SSLLTSDCSER | 0.79 ± 0.28 | 1.03 ± 0.60 | 1.23 ± 0.67c | ||
| O14498 | Immunoglobulin superfamily containing leucine-rich repeat protein |
FQAFAn*GSLLIPDFGK | 0.78 ± 0.19 | 1.08 ± 0.26c | 1.04 ± 0.30c |
| Q06033 | Interalpha-trypsin inhibitor heavy chain H3 |
NAHGEEKEn*LTAR | 0.54 ± 0.18 | 1.25 ± 0.74c | 1.10 ± 0.64c |
| P40189 | Interleukin-6 receptor subunit beta | EQYTIIn*R(TA) | 0.82 ± 0.22 | 1.03 ± 0.20 | 1.13 ± 0.31c |
| P01042 | Kininogen-1 | YNSQn*QSNNQFVLYR | 0.48 ± 0.37 | 2.12 ± 3.39c | 1.39 ± 0.84c |
| P02750 | Leucine-rich alpha-2-glycoprotein | KLPPGLLAn*FTLLR | 0.68 ± 0.40 | 1.65 ± 1.32c | 1.49 ± 0.85a |
| MFSQn*DTR | 0.66 ± 0.14 | 1.50 ± 1.08c | 1.02 ± 0.50c | ||
| P51884 | Lumican | LSHNELADSGIPGNSFn*VSSLVELDLSYNK | 0.78 ± 0.21 | 1.04 ± 0.15c | 1.00 ± 0.17c |
| P22897 | Macrophage mannose receptor 1 | WECKn*DTLLGIK | 0.88 ± 0.45 | 0.99 ± 0.32 | 1.37 ± 0.35c |
| P01033 | Metalloproteinase inhibitor 1 | FVGTPEVn* QTTLYQR | 0.82 ± 0.25 | 1.14 ± 0.52 | 1.19 ± 0.38c |
| Q92859 | Neogenin | TLSDVPSAAPQn*LSLEVR | 0.73 ± 0.15 | 1.09 ± 0.31c | 1.02 ± 0.16c |
| P55058 | Phospholipid transfer protein | IYSn*HSALESLALIPLQAPLK | 0.82 ± 0.24 | 1.11 ± 0.40 | 1.05 ± 0.24c |
| P05155 | Plasma protease C1 inhibitor | VLSn*NSDANLELINTWVAK | 0.90 ± 0.34 | 1.43 ± 0.69c | 1.33 ± 0.57c |
| P01833 | Polymeric immunoglobulin receptor | VPGn*VTAVLGETLK | 0.62 ± 0.29 | 1.34 ± 0.84c | 1.15 ± 0.74c |
| P41222 | Prostaglandin-H2 d-isomerase | WFSAGLASn*SSWLR | 0.88 ± 0.34 | 1.20 ± 0.93 | 1.58 ± 0.85c |
| SVVAPATDGGLn*LTSTFLR | 0.82 ± 0.38 | 1.08 ± 0.87 | 1.29 ± 0.66c | ||
| Q86VB7 | Scavenger receptor cysteine-rich type 1 protein M130 |
CKGn*ESSLWDCPAR | 0.74 ± 0.35 | 1.07 ± 0.49 | 1.10 ± 0.26c |
| P01009 | Short peptide from AAT | ADTHDEILEGLNFn*LTEIPEAQIH | 0.50 ± 0.17 | 0.80 ± 0.55 | 0.90 ± 0.34c |
| Thyroxine-binding globulin | VTACHSSQPn*ATLYK | 0.71 ± 0.33 | 1.08 ± 0.32c | 1.09 ± 0.58c | |
| P00751 | Uncharacterized protein | SPYYn*VSDEISFHCYDGYTLR | 0.75 ± 0.39 | 1.22 ± 0.93 | 1.28 ± 0.53c |
| P04004 | Vitronectin | Nn*ATVHEQVGGPSLTSDLQAQSK | 0.59 ± 0.46 | 2.70 ± 6.67 | 1.26 ± 0.79c |
| P04275 | von Willebrand factor | GLQPTLTNPGECRPn*FTCACR | 0.71 ± 0.40 | 1.02 ± 0.98 | 1.44 ± 0.84c |
| von Willebrand factor | MEACMLn*GTVIGPGK | 0.78 ± 0.22 | 1.03 ± 0.37 | 1.30 ± 0.79c | |
| P25311 | Zinc-alpha-2-glycoprotein | FGCEIENn*R(SS) | 0.69 ± 0.28 | 1.48 ± 1.08 | 1.29 ± 0.43c |
| DIYEYYn*DSNGSHVLQGR | 0.78 ± 0.21 | 1.02 ± 0.38 | 1.04 ± 0.37c | ||
| P05154 | Plasma serine protease inhibitor | VLPSLGISNVFTSHADLSGISn*HSNIQVSEMVHK | 0.78 ± 0.42 | 1.02 ± 0.54 | 1.23 ± 0.39c |
Data represent the mean and SD value based on the reporter ion ratios for CF proteins quantified in at least 5 sample sets. Significant difference level P ≤ 0.05; N, healthy controls; CP, chronic pancreatitis; PC, pancreatic cancer; SR, standard reference.
Asterisk (*) indicates CF modification at asparagine (n) residue.
Difference between healthy controls and pancreatic cancer or chronic pancreatitis.
Table 3. CF Peptides Presented in Significantly Different Levels between Pancreatic Cancer and Chronic Pancreatitisa.
| accession no. | protein name | CF peptideb | N/SR | CP/SR | PC/SR |
|---|---|---|---|---|---|
| P54289 | Voltage-dependent calcium channel subunit alpha-2/delta-1 |
DDLDPEKn*DSEPGSQR | 1.13 ± 1.01 | 1.16 ± 0.42 | 0.70 ± 0.15d |
| Q9BXJ4 | Complement C1q tumor necrosis factor |
TGTVDn*NTSTDLK | 9.88 ± 0.42 | 1.45 ± 0.55c | 0.83 ± 0.25d |
| Q12913 | Receptor-type tyrosine-protein phosphatase eta |
Sn*DTAASEYK | 0.85 ± 0.25 | 1.26 ± 0.26c | 0.85 ± 0.26d |
| P01023 | Alpha-2-macroglobulin | GNEANYYSn*ATTDEHGLVQFSIn*TTNVMGTSLTVR | 1.14 ± 0.49 | 1.88 ± 0.34c | 1.04 ± 0.39d |
| Q7Z7G0 | Target of Nesh-SH3 | En*GSFSGK | 0.80 ± 0.27 | 1.24 ± 0.20c | 0.85 ± 0.19d |
| P16112 | Aggrecan | TVYLYPn*QTGLPDPLSR | 0.91 ± 0.29 | 1.25 ± 0.39 | 0.83 ± 0.14d |
| P58215 | Lysyl oxidase homologue 3 | n*ITAEDCSHSQDAGVR | 0.88 ± 0.41 | 0.75 ± 0.30 | 1.19 ± 0.19d |
| P12111 | Collagen alpha-3(VI) chain | VAVVQHAPSESVDn*ASMPPVK | 0.93 ± 0.24 | 1.34 ± 0.31c | 0.99 ± 0.20d |
| P20023 | Complement receptor type 2 | TPNGn*HTGGNIAR | 0.87 ± 0.30 | 1.10 ± 0.30c | 0.57 ± 0.34d |
| P13591 | Neural cell adhesion molecule 1 |
n*ISSEEK | 1.05 ± 0.43 | 1.26 ± 0.44c | 0.89 ± 0.32d |
| Q13477 | Mucosal addressin cell adhesion molecule 1 | n*ASLSAAGTR | 1.13 ± 0.50 | 1.76 ± 0.72c | 0.94 ± 0.20d |
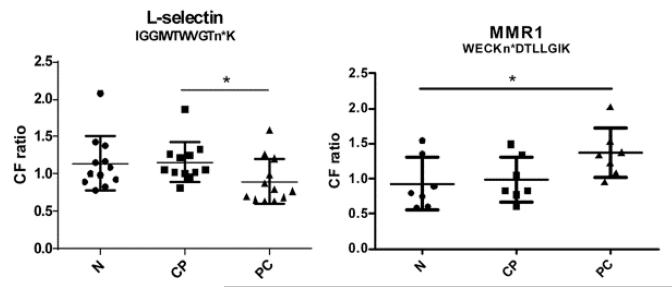
| P14151 | L-selectin | IGGIWTWVGTn*K(SL) | 1.09 ± 0.41 | 1.15 ± 0.27c | 0.90 ± 0.29d |
| P01011 | Alpha-1-antichymotrypsin | LINDYVKn*GTR | 0.53 ± 0.38 | 1.41 ± 0.64c | 0.91 ± 0.39cd |
Data represent the mean and SD value based on the reporter ion ratios for CF proteins quantified in at least 5 sample sets. Significant difference level P ≤ 0.05; N, healthy controls; CP, chronic pancreatitis; PC, pancreatic cancer; SR, standard reference.
Asterisk (*) indicates CF modification at asparagine (n) residue.
Difference between healthy controls and pancreatic cancer or chronic pancreatitis.
Difference between pancreatic cancer and chronic pancreatitis.
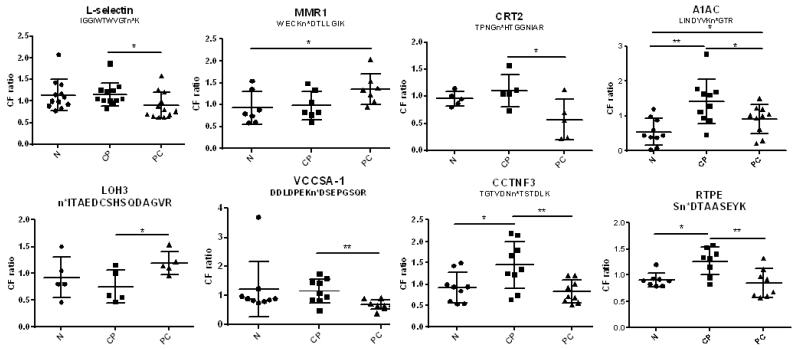
From the comparative analysis of the CF proteome among pancreatic cancer, chronic pancreatitis, and healthy controls based on iTRAQ analysis, 8 CF peptides were differentially expressed between chronic pancreatitis and pancreatic cancer, where the CF peptides from VCCSA1 (voltage-dependent calcium channel subunit alpha-2/delta-1), CCTNFP3 (complement C1q tumor necrosis factor-related protein 3), RTPE (receptor-type tyrosine-protein phosphatase eta), CRT2 (complement receptor type 2), A1AC (alpha-1-antichymotrypsin), complement receptor type 2, or L-selectin decreased in pancreatic cancer compared to chronic pancreatitis, whereas the CF peptide from LOH3 (lysyl oxidase homologue 3) increased in pancreatic cancer relative to chronic pancreatitis. The CF ratio of MMR1 (macrophage mannose receptor 1) was higher in pancreatic cancer than normal samples, but it was not significantly different from chronic pancreatitis. Relative quantitative analysis showed that 8 CF peptides from 8 different CF proteins could be potential biomarkers to distinguish individuals with pancreatic cancer from chronic pancreatitis or healthy controls (Figure 6). Among these CF proteins, MMR1 participates in pathogen recognition, clearance of endogenous serum glycoproteins to maintain the homeostatic process, and antigen presentation.42
Figure 6.
Differentially expressed CF peptides between each of two groups among pancreatic cancer, chronic panceatitis, and healthy control samples. The ratios (PC/SR, CP/SR, or N/SR) of each peptide in individual patient serum samples were subjected to nonparametric analysis of the Student’s t test. VCCSA-1, voltage-dependent calcium channel subunit alpha-2/delta-1; CCTNF3, complement C1q tumor necrosis factor-related protein 3; RTPE, receptor-type tyrosine-protein phosphatase eta; MMR1, macrophage mannose receptor 1; CRT2, complement receptor type 2; LOH3, lysyl oxidase homologue 3; A1AC, alpha-1-antichymotrypsin; n* in different peptide sequences means CF modification at asparagine (n). N, healthy controls; CP, chronic pancreatitis; PC, pancreatic cancer; SR, standard reference; * and ** are significant differences at p < 0.05 and p < 0.01, respectively.
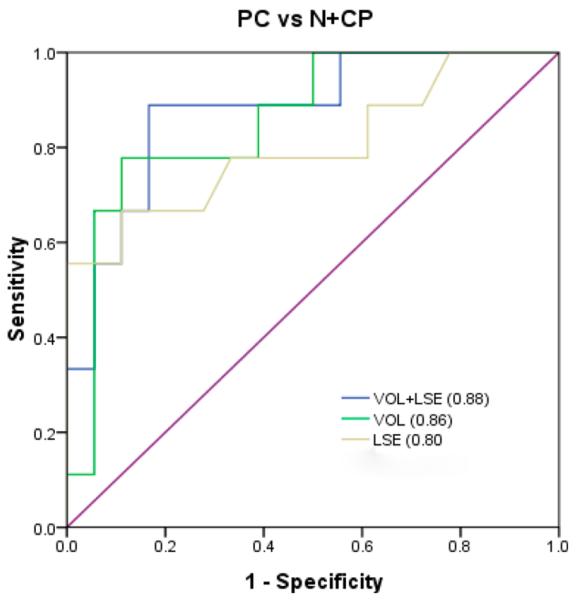
In order to investigate the CF peptide marker, potential candidates were combined to analyze the area-under-the-curve (AUC) value in various combinations of 2 CF peptides among the 8 CF peptide candidates. Chronic pancreatitis and healthy controls were combined as noncases to perform the ROC analysis. The ROC curve analysis between pancreatic cancer and noncases for the combination of VOL and LSE resulted in an AUC value of 0.88 with a specificity of 0.83 at a sensitivity of 0.89 (Figure 7). The biomarker panel was improved compared with VOL alone (AUC value 0.86) or LSE alone (AUC value 0.77). Both sensitivity and specificity increased compared with VOL or LSE alone. Thus, a biomarker panel combining VOL and LSE showed a high diagnostic potential in distinguishing pancreatic cancer from pancreatitis or healthy controls.
Figure 7.
Combination analysis of the CF ratio of voltage-dependent calcium channel subunit alpha-2/delta-1 (VOL) and L-selectin (LSE) among pancreatic cancer, healthy controls, and chronic pancreatitis.
CONCLUSIONS
The results presented herein show that the optimization of the LCA elution conditions can increase the efficiency of CF peptide enrichment. Single LCA column enrichment without any additional serial enrichment may decrease the loss of candidate CF peptides. A maximum of 350 CF sites among 193 CF proteins from pancreatic cancer serum can be identified using the optimized LCA enrichment method in a single run. In total, 630 CF sites were identified among 322 CF proteins from 13 sample sets of human serum. The coverage of CF sites can be improved significantly by increasing the sample sets up to 7, which also showed that three biological repeats are not sufficient to detect the global CF sites in human serum. In addition, CF proteins are involved in a series of immune and inflammatory responses based on GO analysis in the present study. Eight CF peptides could potentially distinguish pancreatic cancer from healthy controls or chronic pancreatitis. A biomarker panel combining VOL and LSE showed high diagnostic potential. Further work will be needed to determine the quantitative content of the identified CF proteins in different samples and to validate these 8 CF candidate peptides as potential markers for pancreatic cancer in a large cohort study.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for support from the National Cancer Institute through SPORE Program Grant 1 P50CA130810 (M.A.A., D.M.L., and M.T.R.), grants 1RO1 CA160254 (D.M.L.) and 1RO1 CA154455 (D.M.L.), and the National Institutes of Health through grant RO1 GM 49500 (D.M.L.).
Footnotes
ASSOCIATED CONTENT
Figure S1: Workflow diagram showing experimental process used in large-scale identification of CF sites in pancreatic cancer serum. Figure S2: Comparison of the number of CF peptides resulting from different elution buffer strategies. Figure S3: Distribution of CF sites in glycopeptides identified in pancreatic cancer serum with single and double CF sites. Table S1: Diagnostic ions frequency and percent in 274 candidate CF peptides from one run mass spectra. Table S2: Different patterns of diagnostic ions in candidate CF peptides from one run mass spectra. Table S3: Identified 630 CF sites in 613 CF peptides. Table S4: All CF proteins p values derived from GO analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Osumi D, Takahashi M, Miyoshi E, Yokoe S, Lee SH, Noda K, Nakamori S, Gu J, Ikeda Y, Kuroki Y, Sengoku K, Ishikawa M, Taniguchi N. Core fucosylation of E-cadherin enhances cell–cell adhesion in human colon carcinoma WiDr cells. Cancer Sci. 2009;100:888–895. doi: 10.1111/j.1349-7006.2009.01125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dennis JW, Laferte S, Waghorne C, Breitman ML, Kerbel RS. Beta-1-6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science. 1987;236:582–585. doi: 10.1126/science.2953071. [DOI] [PubMed] [Google Scholar]
- (3).Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158–184. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- (4).Hakomori S. Tumor malignancy defined by aberrant glycosylation and sphingo(glyco) lipid metabolism. Cancer Res. 1996;56:5309–5318. [PubMed] [Google Scholar]
- (5).Muinelo-Romay L, Villar-Portela S, Cuevas E, Gil-Martin E, Fernandez-Briera A. Identification of alpha(1,6)fucosylated proteins differentially expressed in human colorectal cancer. BMC Cancer. 2011;11:508. doi: 10.1186/1471-2407-11-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM. Core fucosylation and alpha2-3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology. 2011;21:195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- (7).Yin H, Lin Z, Nie S, Wu J, Tan Z, Zhu J, Dai J, Feng Z, Marrero J, Lubman DM. Mass-selected site-specific core-fucosylation of ceruloplasmin in alcohol-related hepatocellular carcinoma. J. Proteome Res. 2014;13:2887–2896. doi: 10.1021/pr500043k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nie S, Lo A, Wu J, Zhu J, Tan Z, Simeone DM, Anderson MA, Shedden KA, Ruffin MT, Lubman DM. Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis. J. Proteome Res. 2014;13:1873–1884. doi: 10.1021/pr400967x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhao J, Qiu WL, Simeone DM, Lubman DM. N-Linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J. Proteome Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- (10).Lin ZX, Yin HD, Lo A, Ruffin MT, Anderson MA, Simeone DM, Lubman DM. Label-free relative quantification of alpha-2-macroglobulin site-specific core-fucosylation in pancreatic cancer by LC-MS/MS. Electrophoresis. 2014;35:2108–2115. doi: 10.1002/elps.201300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- (12).Fanayan S, Hincapie M, Hancock WS. Using lectins to harvest the plasma/serum glycoproteome. Electrophoresis. 2012;33:1746–1754. doi: 10.1002/elps.201100567. [DOI] [PubMed] [Google Scholar]
- (13).Kornfeld K, Reitman ML, Kornfeld R. The carbohydrate-binding specificity of pea and lentil lectins. Fucose is an important determinant. J. Biol. Chem. 1981;256:6633–6640. [PubMed] [Google Scholar]
- (14).Tateno H, Nakamura-Tsuruta S, Hirabayashi J. Comparative analysis of core-fucose-binding lectins from Lens culinaris and Pisum sativum using frontal affinity chromatography. Glycobiology. 2009;19:527–536. doi: 10.1093/glycob/cwp016. [DOI] [PubMed] [Google Scholar]
- (15).Sharon N, Lis H. Legume lectins—a large family of homologous proteins. FASEB J. 1990;4:3198–3208. doi: 10.1096/fasebj.4.14.2227211. [DOI] [PubMed] [Google Scholar]
- (16).Jia W, Lu Z, Fu Y, Wang HP, Wang LH, Chi H, Yuan ZF, Zheng ZB, Song LN, Han HH, Liang YM, Wang JL, Cai Y, Zhang YK, Deng YL, Ying WT, He SM, Qian XH. A strategy for precise and large scale identification of core fucosylated glycoproteins. Mol. Cell. Proteomics. 2009;8:913–923. doi: 10.1074/mcp.M800504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Nie S, Lo A, Zhu JH, Wu J, Ruffin MT, Lubman DM. Isobaric protein-level labeling strategy for serum glycoprotein quantification analysis by liquid chromatography-tandem mass spectrometry. Anal. Chem. 2013;85:5353–5357. doi: 10.1021/ac400838s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liu Y, He J, Li C, Benitez R, Fu S, Marrero J, Lubman DM. Identification and confirmation of biomarkers using an integrated platform for quantitative analysis of glycoproteins and their glycosylations. J. Proteome Res. 2010;9:798–805. doi: 10.1021/pr900715p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sharon N. Lectin carbohydrate complexes of plants and animals—an atomic view. Trends Biochem. Sci. 1993;18:221–226. doi: 10.1016/0968-0004(93)90193-q. [DOI] [PubMed] [Google Scholar]
- (20).Loris R, Hamelryck T, Bouckaert J, Wyns L. Legume lectin structure. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1998;1383:9–36. doi: 10.1016/s0167-4838(97)00182-9. [DOI] [PubMed] [Google Scholar]
- (21).Lewandrowski U, Moebius J, Walter U, Sickmann A. Elucidation of N-glycosylation sites on human platelet proteins—a glycoproteomic approach. Mol. Cell. Proteomics. 2006;5:226–233. doi: 10.1074/mcp.M500324-MCP200. [DOI] [PubMed] [Google Scholar]
- (22).Kaji H, Yamauchi Y, Takahashi N, Isobe T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat. Protoc. 2006;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- (23).Cao Q, Zhao X, Zhao Q, Lv X, Ma C, Li X, Zhao Y, Peng B, Ying W, Qian X. Strategy integrating stepped fragmentation and glycan diagnostic ion-based spectrum refinement for the identification of core fucosylated glycoproteome using mass spectrometry. Anal. Chem. 2014;86:6804–6811. doi: 10.1021/ac501154a. [DOI] [PubMed] [Google Scholar]
- (24).Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- (25).Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291:2351–2356. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- (26).Peterman SM, Mulholland JJ. A novel approach for identification and characterization of glycoproteins using a hybrid linear ion trap/FT-ICR mass spectrometer. J. Am. Soc. Mass Spectrom. 2006;17:168–179. doi: 10.1016/j.jasms.2005.10.008. [DOI] [PubMed] [Google Scholar]
- (27).Zhang W, Wang H, Zhang L, Yao J, Yang PY. Large-scale assignment of N-glycosylation sites using complementary enzymatic deglycosylation. Talanta. 2011;85:499–505. doi: 10.1016/j.talanta.2011.04.019. [DOI] [PubMed] [Google Scholar]
- (28).Valliere-Douglass JF, Eakin CM, Wallace A, Ketchem RR, Wang W, Treuheit MJ, Balland A. Glutamine-linked and non-consensus asparagine-linked oligosaccharides present in human recombinant antibodies define novel protein glycosylation motifs. J. Biol. Chem. 2010;285:16012–16022. doi: 10.1074/jbc.M109.096412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lee JY, Kim JY, Park GW, Cheon MH, Kwon KH, Ahn YH, Moon MH, Lee HJ, Paik YK, Yoo JS. Targeted mass spectrometric approach for biomarker discovery and validation with nonglycosylated tryptic peptides from N-linked glycoproteins in human plasma. Mol. Cell. Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- (31).Pan S, Tamura Y, Chen R, May D, McIntosh MW, Brentnall TA. Large-scale quantitative glycoproteomics analysis of site-specific glycosylation occupancy. Mol. Biosyst. 2012;8:2850–2856. doi: 10.1039/c2mb25268f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ferreira JA, Daniel-da-Silva AL, Alves RMP, Duarte D, Vieira I, Santos LL, Vitorino R, Amado F. Synthesis and optimization of lectin functionalized nanoprobes for the selective recovery of glycoproteins from human body fluids. Anal. Chem. 2011;83:7035–7043. doi: 10.1021/ac200916j. [DOI] [PubMed] [Google Scholar]
- (33).Ma C, Zhang Q, Qu J, Zhao X, Li X, Liu Y, Wang PG. A precise approach in large scale core-fucosylated glycoproteins identification with low- and high-normalized collision energy. J. Proteomics. 2014;114:61–70. doi: 10.1016/j.jprot.2014.09.001. [DOI] [PubMed] [Google Scholar]
- (34).Wang LH, Li DQ, Fu Y, Wang HP, Zhang JF, Yuan ZF, Sun RX, Zeng R, He SM, Gao W. PFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:2985–2991. doi: 10.1002/rcm.3173. [DOI] [PubMed] [Google Scholar]
- (35).Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lei Z, Beuerman RW, Chew AP, Koh SK, Cafaro TA, Urrets-Zavalia EA, Urrets-Zavalia JA, Li SF, Serra HM. Quantitative analysis of N-linked glycoproteins in tear fluid of climatic droplet keratopathy by glycopeptide capture and iTRAQ. J. Proteome Res. 2009;8:1992–2003. doi: 10.1021/pr800962q. [DOI] [PubMed] [Google Scholar]
- (37).Zielinska DF, Gnad F, Schropp K, Wisniewski JR, Mann M. Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common rore machinery. Mol. Cell. 2012;46:542–548. doi: 10.1016/j.molcel.2012.04.031. [DOI] [PubMed] [Google Scholar]
- (38).Marshall RD. Glycoproteins. Annu. Rev. Biochem. 1972;41:673–702. doi: 10.1146/annurev.bi.41.070172.003325. [DOI] [PubMed] [Google Scholar]
- (39).Gavel Y1 v. H. G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Helenius A. How N-Linked oligosaccharides affect glycoprotein folding in the endoplasmic-reticulum. Mol. Biol. Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Saldova R, Wormald MR, Dwek RA, Rudd PM. Glycosylation changes on serum glycoproteins in ovarian cancer may contribute to disease pathogenesis. Dis. Markers. 2008;25:219–232. doi: 10.1155/2008/601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Marttila-Ichihara F, Turja R, Miiluniemi M, Karikoski M, Maksimow M, Niemela J, Martinez-Pomares L, Salmi M, Jalkanen S. Macrophage mannose receptor on lymphatics controls cell trafficking. Blood. 2008;112:64–72. doi: 10.1182/blood-2007-10-118984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.