Abstract
MondoA is a basic helix–loop–helix (bHLH)/leucine zipper (ZIP) transcription factor that is expressed predominantly in skeletal muscle. Studies in vitro suggest that the Max-like protein X (MondoA:Mlx) heterodimer senses the intracellular energy status and directly targets the promoter region of thioredoxin interacting protein (Txnip) and possibly glycolytic enzymes. We generated MondoA-inactivated (MondoA−/−) mice by gene targeting. MondoA−/− mice had normal body weight at birth, exhibited normal growth and appeared to be healthy. However, they exhibited unique metabolic characteristics. MondoA−/− mice built up serum lactate and alanine levels and utilized fatty acids for fuel during exercise. Gene expression and promoter analysis suggested that MondoA functionally represses peroxisome-proliferator-activated receptor γ co-activator-1α (PGC-1α)–mediated activation of pyruvate dehydrogenase kinase 4 (PDK4) transcription. PDK4 normally down-regulates the activity of pyruvate dehydrogenase, an enzyme complex that catalyses the decarboxylation of pyruvate to acetyl-CoA for entry into the Krebs cycle; in the absence of MondoA, pyruvate is diverted towards lactate and alanine, both products of glycolysis. Dynamic testing revealed that MondoA−/− mice excel in sprinting as their skeletal muscles display an enhanced glycolytic capacity. Our studies uncover a hitherto unappreciated function of MondoA in fuel selection in vivo. Lack of MondoA results in enhanced exercise capacity with sprinting.
Keywords: energy metabolism, exercise, fuel selection, glycolysis, MondoA
INTRODUCTION
Fuel selection is a highly regulated process which ensures that an adequate energy supply is met for cellular function in different physiological environments. Glucose, absorbed from the gastrointestinal tract after a meal, stimulates insulin secretion from pancreatic β-cells. Insulin, in turn, stimulates glucose uptake in skeletal muscle and adipose tissue, whereas it suppresses hepatic glucose production. In addition to stimulating insulin production, hyperglycaemia directly regulates the expression of different target genes, including those involved in glycolysis and lipogenesis [1]. Many of the genes that respond to high glucose share a common molecular signature in their promoter region, known as the carbohydrate-response element (ChoRE), which is composed of two E-boxes separated by five nucleotides. These genes are regulated by two basic helix–loop–helix (bHLH) leucine zipper (ZIP) transcription factors, carbohydrate-response element-binding protein (ChREBP, also known as MondoB) and MondoA. Both bind to the ChoRE in the form of heterodimeric complexes with another factor, Max-like protein X (Mlx). MondoA is expressed predominantly in skeletal muscle, whereas ChREBP is expressed at a high level in the liver [2,3]. Both factors are also expressed in many other tissues.
In vitro experiments suggest that MondoA binds to the CACGTG E-box and/or ChoRE in the promoter regions of a number of putative target genes, including hexokinase II (HKII), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3), lactate dehydrogenase A (LDH-A) and thioredoxin interacting protein (Txnip) [4,5]. C2C12 cells, overexpressing an active form of MondoA, exhibit activated glycolysis because of increased expression of HKII, PFKFB3 and LDH-A, rate-limiting enzymes in glycolysis [6]. Txnip is implicated as a negative regulator of glucose uptake in skeletal muscle in vivo [7] and MondoA negatively regulates glucose uptake via up-regulation of Txnip in C2C12 and HA1E cells [5].
ChREBP and MondoA sense intracellular nutrient states [6,8]. The two Mondo-family transcription factors [9] contain a glucose-sensing module, encompassing a low-glucose inhibitory domain (LID) and a glucose-responsive activation conserved domain (GRACE). Glucose responsiveness to the Mondo transcription factors is mediated by inhibition of the transactivation activity of GRACE by LID and release of the inhibition by high glucose [9]. The MondoA:Mlx heterodimer complex shuttles between cytosol and nucleus and when it is not targeted to the nucleus, MondoA is also found at the outer mitochondrial membrane (OMM) by protein–protein interaction with OMM proteins [6]. It has been suggested that MondoA plays a key role in sensing the intracellular energy state as well as controlling the transcription of glycolytic enzymes, making it a pivotal regulator of energy homoeostasis [6].
In the present study, we generated MondoA-inactivated (MondoA−/−) mice to examine the function of MondoA in vivo. Intriguingly, the expression level of many of the putative MondoA target genes was not affected by loss of MondoA. A major phenotype of the MondoA−/− mice was hyperlactataemia at rest and after exercise. Pyruvate dehydrogenase kinase 4 (PDK4) expression was increased, which accounts for decreased conversion of pyruvate to acetyl-CoA. Moreover, we found that MondoA regulates PDK4 expression indirectly via peroxisome-proliferator-activated receptor γ co-activator-1 (PGC-1α), placing MondoA as a master regulator of fuel selection in skeletal muscle as part of Randle’s ‘glucose-fatty acid cycle’ [10]. The skeletal muscle of MondoA−/− mice also displayed increased transcript levels for enzymes involved in β-oxidation, suggesting up-regulation of fatty acid oxidation. Interestingly, enhanced glycolysis in the MondoA−/− mice was associated with an enhanced capacity for short-term sprint exercise.
MATERIALS AND METHODS
Animals
Using a mouse 129-strain λ genomic library (Stratagene), we isolated clones containing MondoA genomic DNA. Two overlapping clones encompassing exons 2–4 were used to generate a replacement targeting construct. The recombination arms were comprised of a 5.1 kb intron 1 DNA fragment and a 2.3 kb DNA fragment containing exons 3 and 4 and intron 3. A 700 bp exon 2-containing DNA fragment was amplified by PCR as the targeted region and inserted between two loxP sites of the neo cassette in the targeting vector (Supplementary Figure S1). A thymidine kinase cassette was ligated to the 5′ end of the construct for selection against random insertion events.
We used an R1 mouse embryonic stem (ES) cell line [11] to generate the gene targeted ES cell clones as described previously [12]. We used Southern blotting to screen and to select targeted ES cell clones. Cre-recombinase expression vector was introduced into targeted ES cell clones using Lipofectamine 2000 (Invitrogen) to remove the neo cassette only or both the neo cassette and the floxed exon 2-containing genomic region. This gave rise to cells that contained either MondoA-floxed allele or MondoA-deleted allele, separately. ES cells were screened by PCR and injected into blastocysts of C57BL/6J mice to generate chimaeric mice. After germ-line transmission was confirmed by PCR, the targeted mice were back-crossed to C57BL/6J mice for four generations before being used in the study.
Mice were maintained in a temperature-controlled facility with a fixed 12-h-light and 12-h-dark cycle and free access to regular chow and water. In selected experiments, we used a high-sucrose fat-free diet (MP Biomedicals, catalogue no. 901683). Age- and gender-matched mice were used throughout, unless otherwise indicated. Txnip global knockout (KO) mice were provided by Dr Simon Hu (University of California at Los Angeles). All animal experiments were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
Plasma chemistry measurements
We measured blood glucose level using a One touch Ultra (Lifescan) glucometer and blood lactate levels by a Lactate-pro kit (Arkray). Plasma non-esterified fatty acid (NEFA), total cholesterol, triacylglycerol (Waco) and glycerol (Sigma) were measured by using enzymatic kits provided by the manufacturers whose names are given in parenthesis.
Dichloroacetate administration
We diluted dichloroacetate (DCA) in 0.9% NaCl and administrated the solution (200 mg/kg of body weight) by intraperitoneal (i.p.) injection in a single bolus.
Quantitative real-time PCR
Total RNA was isolated from tissues or cultured cells with TRIzol (Invitrogen) and reverse transcribed using SuperScript III reverse transcriptase with random primers (Invitrogen). The cDNA generated was used for quantitative real-time PCR (qRT–PCR) on a MX3000 System (Stratagene) using iQ SYBR Green PCR reagent kit (Bio-Rad Laboratories).
Glucose tolerance test and insulin tolerance test
We performed glucose tolerance test (GTT) and insulin tolerance test (ITT) on mice fasted for 4–6 h. D-Glucose (1–2g/kg of body weight; Sigma) or Humulin R (1 unit/kg of body weight; Novo Nordisk) were injected i.p. and plasma glucose levels were measured before as well as 15, 30, 60 and 120 min after injection.
Histology
We fixed tissues in formalin overnight, dehydrated them overnight in a series of ethanol solutions and performed H&E staining and immunohistochemistry on them afterwards. Samples were then embedded in paraffin, sectioned and stained.
Antibodies used for immunostaining were anti-Myosin Slow (clone NOQ7.5.4D, Sigma) and monoclonal anti-Myosin Fast (clone MY-32, Sigma), for slow- and fast-twitch myosin heavy-chain isoforms respectively. Antigen retrieval was attained by incubating the sectioned tissue in citric acid (pH 6.0) for 20 min in a pressure cooker.
NADH and succinic dehydrogenase (SDH) staining were performed as described by Sheehan and Hrapchak [13]. We performed EM of muscle tissues at the Integrated Microscopy Core Facility of Baylor College of Medicine.
Treadmill exercises
Mice were placed on either an Exer-3/6 rodent treadmill or a 1012M-2 Modular Enclosed Metabolic Treadmill for Mice (Columbus Instruments). The treadmill was equipped with a rear electrical stimulus grid set to deliver 0.2 mA electric shock. Mice were considered to have reached the point of exhaustion when they made contact with the grid for a period of greater than 5 s.
Three different exercise regimens were tested. (1) Mild exercise: the standardized non-intensive exercise regimen consisted of a 10 min ‘walk’ at 6 m/min at a 0° incline followed by 1 min rest. The speed was then increased stepwise from 6 m/min to 10 m/min with an increment of 2 m/min and each step for 2 min at a 15° incline. This regimen was followed by increasing the speed to 12 m/min and 14 m/min, each for 5 min and finally to 16 m/min for 45 s at a 15° incline. The total running time was therefore 16 min 45 s. We confirmed that every mouse was able to accomplish the standardized exercise regimen. (2) Endurance exercise: the endurance exercise regimen consisted of mice warming up at 8 m/min for 6 min at a 0° incline. The speed was then increased to 12, 14 and 16 m/min for 6 min each and increased the speed by 2 m/min every 3 min at a 15° incline until the mice were exhausted and gave up. O2 consumption and CO2 production were monitored during exercise by computer-controlled open-circuit indirect calorimetry (Oxymax, Colombus Instruments). (3) Sprint exercise regimen: the sprint regimen consisted of mice warming up for 3 min at 6 m/min at a 0° incline followed by a 2-min run at the same speed with a 15° incline (see Figure 2E). Subsequent runs were performed at a 15° incline. Speed was first increased to 14 m/min for 30 s, followed by a short 1.5 min of low speed running at 6 m/min and the speed was increased to 18 m/min for 30 s. Afterwards, we increased the speed from 18 m/min to 24 m/min with an increment of 2 m/min and each increment lasting 15 s (total 15 × 4 = 60 s). After that, the mice were kept under interval running at 6 m/min for 3 min, followed by an increase in the speed from 20 m/min to 32 m/min with an increment of 4 m/min and each increment for 15 s. This pattern of exercise continued with a subsequent interval running at 6 m/min for 3 min followed by a sharp increase in speed from 22 m/min to 34 m/min with an increment of 4 m and each for 15 s. The pattern was repeated with increasing speed until the mice could no longer keep up with the speed and gave up at the grid.
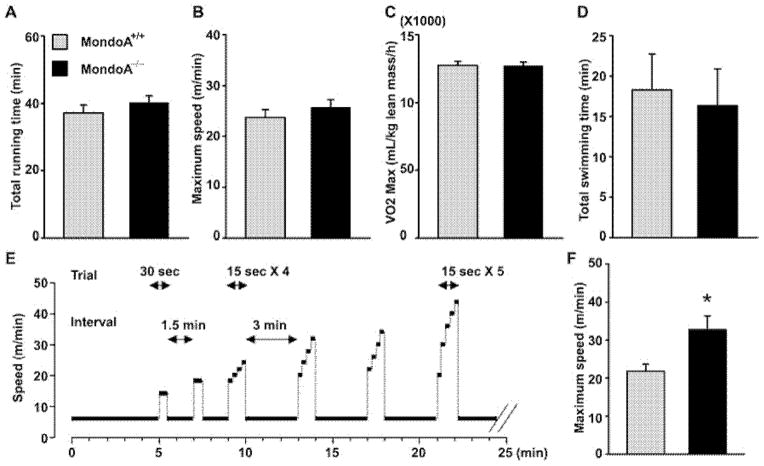
Figure 2. MondoA−/− mice preserve endurance capacity (A–E) while performing better at sprint running (F and G).
Four-month-old male MondoA+/+ (n = 8) and MondoA−/− mice (n = 8) were acclimated for treadmill running 5 min a day for 2 days. On the day of the trial, mice were committed to treadmill running and oxygen consumption was monitored during the running bout. Mice were forced to run until they were exhausted and stayed at the edge of grid with electric shock for 5 s. The figures indicate time period until exhaustion (A), running speed at exhaustion (B) and V̇o2 max (C). All data were expressed as means ± S.E.M. For the swimming endurance test, 5-month-old male MondoA+/+ (n = 13) and MondoA−/− mice (n = 14) were acclimated for swimming 5 min a day for 2 days. On the day of test, mice were attached a weight (3% of body weight) and released into a water bucket. Mice were rescued when they showed signs of fatigue and failed to keep their noses above the water; the total time mice stayed in the water is recorded in (D). Sprinting exercise regime (E) and maximal sprinting speed (F) of 5-month-old MondoA+/+ mice (n = 17) and MondoA−/− mice (n = 14). *P < 0.05 comparing MondoA+/+ and MondoA−/− mice. See text for details.
Swimming endurance
To test the exercise endurance capacity, we also placed the mice in a clear plastic bucket filled with water at a temperature of 31 °C. A weight equivalent to 3% of mouse body weight was tied to its tail during swimming. Exhaustion was deemed when a mouse could not keep its nose above the water surface, at which time the animal was immediately pulled out of the water.
Metabolomics analysis
Serum from MondoA−/− and control mice were harvested before or after standardized exercise using a treadmill, as described above, and the amino acids and acylcarnitines were analysed by MS/MS as described by others [14,15].
Western blot analysis
Tissues (muscles) were freshly isolated from mice and immediately frozen in liquid nitrogen. We homogenized the frozen tissues in protein extraction buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate and 1% NP40) and separated the protein mixture by SDS/PAGE (4–15% gel). The proteins were immobilized on to a nylon membrane and probed with specific antibodies for visualization by enhanced chemiluminescence (SuperSignal ECL kit). The polyclonal antibody against mouse PDK4 (1:1000 dilution) has been used in previous studies for Western blot analysis [16]. Specificity for PDK4 was confirmed with tissue extracts from PDK4 KO mice. The anti-β-actin (1:2000 dilution) monoclonal antibody was purchased from Cell Signaling Technology and served as normalizers.
In vivo tissue glucose transport during a glucose tolerance test and swimming test
Per mouse, 10 μCi of 2-deoxy-D-[1, 2-3H]-glucose ([2-3H]2DG) (MP Biomedicals) in 200 μl of PBS (for swimming test) or 10% dextrose (for GTT) was injected i.p. (1 g/kg of body weight) to weight-matched 6–8-month-old female mice. We obtained blood samples (10 μl each) via the tail vein at 0, 10, 20, 30 and 40 min after injection and determined blood glucose levels.
To determine glucose-specific activity, 10 μl of plasma was deproteinized with Ba(OH)2 (0.15 M) and ZnSO4 (0.15 M) and the radioactivity was then measured in a liquid scintillation counter (LS 6500; Beckman Instruments). The glucose-specific activity (in d.p.m. per μmol of glucose) was calculated by dividing the radioactivity of the plasma sample by the glucose concentration. The area under the curve (AUC) was integrated for the duration of the experiment (40 min). At 40 min after glucose challenge, we killed the mice and snap-froze the muscle tissues in liquid nitrogen for determination of [2-3H]2DG-6-phosphate accumulation. We incubated tissues in 0.5 M NaOH at 60 °C for 60 min and neutralized it with 0.5 M HC1. One aliquot was counted directly to determine the total radioactivity of [2-3H]2DG + [2-3H]2DG-6-phosphate. A second aliquot was treated with Ba(OH)2 and ZnSO4 to remove [2-3H]2DG-6-phosphate and any tracer incorporated into the glycogen and then counted to determine [2-3H]deoxy-glucose radioactivity. The amount of [2-3H]2DG-6-phosphate is the difference between the two aliquots. The accumulation of [2-3H]2DG-6-phosphate was normalized to tissue weight. To calculate the 2DG uptake, we divided the counts (d.p.m.) by the integrated glucose-specific activity AUC and the tissue weight.
Rates of glucose and fatty acid oxidation ex vivo
We isolated skeletal muscle [extensor digitorum longus (EDL)] from anaesthetized mice and pre-incubated them for 15 min with oxygenated Krebs–Henseleit (K–H) bicarbonate buffer (118 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 1.75 mM CaCl2, 0.5 mM Na2EDTA, 25 mM NaHCO3 and 1.2 mM KH2PO4, pH 7.4) containing 4% BSA, glucose (5.5 mM) and bovine insulin (100 mM). We then transferred the muscle to a glass tube with oxygenated K–H buffer and [14C]glucose or [14C]palmitic acid (0.5 μCi/ml or 1 μCi per sample), sealed the tube with a rubber cap and incubated the mixture at 37 °C for 45 min. CO2 was trapped with a 3M paper soaked with hyamine hydroxide. The 3M paper was then transferred into a scintillation vial and the total radioactivity was then measured in a scintillation counter (LS 6500; Beckman Instruments).
Pyruvate dehydrogenase complex assay
We isolated mouse soleus muscles and snap-froze them in liquid nitrogen immediately. Total and actual pyruvate dehydrogenase complex (PDC) were analysed as described in [17,18].
Microarray
Skeletal muscles were isolated from 4-h fasted 6-month-old MondoA+/+ and MondoA−/− mice (n = 3 with each of RNA samples derived from pools of soleus muscle from three mice) and total RNA was extracted using TRIzol (Invitrogen) and after DNaseI treatment (Qiagen). We purified RNA by RNase Easy kit (Qiagen) following the manufacturer’s instruction. Microarray analysis was performed by the Microarray Core Facility, Baylor College of Medicine using Mouse Genome 430 2.0 Array (Affymetrix). Normalizations of gene expression were performed by using d-Chip software (http://www.hsph.harvard.edu/complab/dchip/) and gene set enrichment analysis (GSEA) was performed using GSEA software (http://www.broadinstitute.org/gsea/index.jsp).
Luciferase assay
Luciferase reporter, constitutively active and dominant-negative MondoA-expression constructs were generated by PCR cloning. Luciferase assays were performed with the Gaussia Luciferase Assay Kit (New England Biolabs). The results were normalized to the activity of the co-transfected internal control plasmid pSR-secreted alkaline phosphate (SEAP) Reporter measured with Phospha-Light™ SEAP Reporter Gene Assay System (Applied Biosystems).
The pSR-secreted alkaline phosphate construct was kindly provided by Dr David Spencer (Department of Immunology, Baylor College of Medicine).
Cell lines
C2C12 and BOSC-23 cells (A.T.C.C., Manassas, VA, U.S.A.) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS, penicillin (100 units/ml) and streptomycin (100 μg/ml) (Invitrogen).
Statistical analysis
We used Student’s t-test throughout the studies to analyse the difference between the means and a difference was considered significant when P < 0.05. Data were expressed as means ± S.E.M., unless specified otherwise.
RESULTS
Generation and baseline characterization of MondoA−/− mice
We used a replacement targeting strategy to create a conditional targeting construct for the mouse MondoA gene (Supplementary Figure S1). Floxed mice were generated using standard methods in which exon-2 of the MondoA gene was flanked by two loxP sites in the targeted allele to achieve Cre-recombinase-mediated deletion either globally or in a muscle-specific manner, through transfection of Cre-expression construct at ES cell stage or breeding the mutant F1 to muscle-specific Cre (Mck-Cre)-transgenic mice respectively.
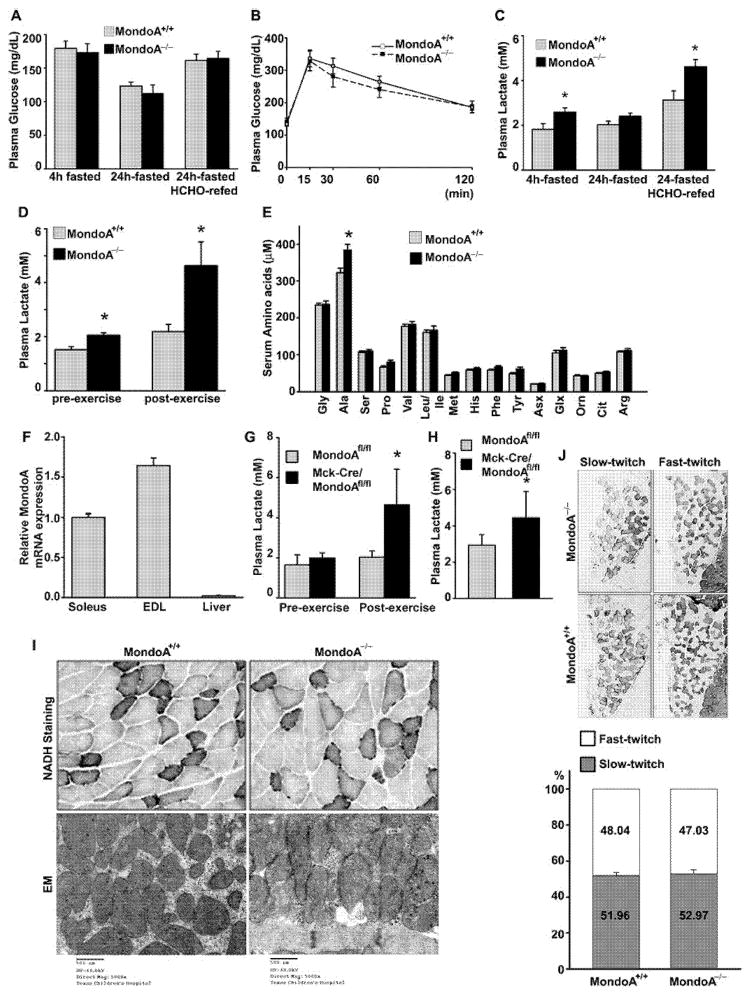
MondoA global KO (MondoA−/−) mice were viable and did not differ from wild-type (WT) mice in their body weight, growth curves or gross appearance (results not shown). They had normal fasting glucose levels (Figure 1A; Supplementary Figure S3) and normal GTTs (Figure 1B), as well as normal plasma lipid profiles (Supplementary Table S1) under basal chow-fed conditions as well as after 4 h and 24 h fasting or after 24 h fasting followed by high-sucrose fat-free diet re-feeding for 18 h. Plasma lactate levels were, however, higher in the 4 h fasted MondoA−/− mice compared with WT mice (Figure 1C). The difference disappeared in mice that were fasted for 24 h with a trend of plasma lactate in MondoA−/− mice decreasing at 24 h of fasting, suggesting that a continued food supply is required to sustain high lactate levels in the MondoA−/− mice. We, next, measured the plasma lactate levels after first fasting the mice for 24 h and then re-feeding them with high-sucrose diet for 18 h. This dietary manipulation increased the plasma lactate levels in both WT and MondoA−/− mice but much more so in the latter, which went up to a level that was significantly higher than that in WT (Figure 1C).
Figure 1. MondoA−/− mice exhibit higher levels of plasma lactate and alanine than MondoA+/+ mice.
(A) Plasma glucose levels of MondoA−/− and MondoA+/+ mice measured at 4-h-, 24-h-fasted and 24-h-fasted followed by 18 h high sucrose fat-free (HCHO) diet re-feeding 12-week-old male mice (n = 6–7 each). (B) GTT of MondoA+/+ (n = 11) and MondoA−/− (n = 12) mice. Mice were fasted for 4 h before i.p. injection of D-glucose (2 g/kg of body weight). Blood glucose was measured at 0, 15, 30, 60 and 120 min after glucose challenge. (C) Plasma lactate was measured in 4-h-, 24-h-fasted without or with HCHO re-fed mice (n = 6–7 each). *P < 0.05 comparing MondoA+/+ and MondoA−/− mice. (D) Plasma lactate level of MondoA+/+ and MondoA−/− was measured before and after non-intensive treadmill exercise as described in the Materials and methods section (4-month-old male, n = 6 each). *P < 0.05 comparing MondoA+/+ and MondoA−/− mice. (E) Serum amino acids profile after non-intensive treadmill exercise. *P < 0.05 comparing MondoA+/+ and MondoA−/− 6-month-old male mice. (F) MondoA mRNA expression from skeletal muscle (soleus and EDL muscles) and liver was determined by qRT-PCR. MondoA mRNA expression was normalized to three house-keeping genes, B2m, Hmbs and Ppia, using GeNorm algorithm [42] and expressed as fold changes relative to the expression level in soleus muscle in eight-week-old female mice (n = 5–6 each) (G) Plasma lactate level of muscle-specific MondoA−/− KO mice (MCK-Cre MondoA−/−) and MondoA floxed control (MondoAflox/flox) mice measured before and after non-intensive treadmill exercise (n = 6). *P < 0.05 comparing muscle-specific MondoA−/− and floxed control 4-month-old mice. (H) Plasma lactate levels in HCHO re-fed MCK-Cre MondoA−/− (n = 7) and MondoAflox/flox mice (n = 6). *P < 0.05 comparing MondoAflox/flox and MCK-Cre MondoA−/− mice. (I) Evaluation of mitochondrial function in skeletal muscle isolated from 12-month-old female MondoA+/+and MondoA−/− mice. Upper panels, NADH staining (gastrocnemius); lower panels, EM (soleus). (J) Muscle fibre type identification. Type I (slow twitch) and type II (fast twitch) fibres of soleus muscle of MondoA+/+ and MondoA−/− 12-month-old female mice were determined by immunohistochemisty against fibre type-specific antibodies (upper panel) and the proportion of each fibre type determined (lower panel, MondoA+/+ n = 4, MondoA−/− n = 3). Data in (G) and (H) were expressed as means ± S.D.
Short-term non-intensive exercise is known to increase glucose uptake and metabolism [20]. This manoeuvre stimulated plasma lactate levels to a moderate degree (~20 %) in WT mice but more than doubled the plasma lactate in MondoA−/− mice (Figure 1D). Presumably, high rates of glycogenolysis and glycolysis led to accumulation of pyruvate which was not disposed in the Krebs cycle and converted into lactate.
We also measured the serum amino acid concentrations in WT and MondoA−/− mice. Under basal conditions there was no difference between the two types of mice. However, after mild exercise we found that alanine level was significantly increased in MondoA−/− mice, although the levels of the other amino acids were very similar (Figure 1E). The elevated plasma alanine is consistent with part of the pyruvate being shunted to alanine through the action of alanine aminotransferase [21].
MondoA is normally expressed at the highest level in skeletal muscle [4]. By qRT-PCR analysis, we confirmed a much higher level of MondoA mRNA in the soleus and EDL as compared with the liver (Figure 1F). To determine whether the exaggerated rise in plasma lactate levels in MondoA−/− mice came predominantly from skeletal muscle, we repeated the analysis in skeletal muscle-specific MondoA-KO mice (MCK-Cre/MondoA−/− mice that we obtained by breeding the MondoAflox/flox mice with Mck-Cre transgenic mice). We found that MCK-Cre/MondoA−/− mice recapitulated the phenotypes of the global MondoA−/− mice with respect to the plasma lactate levels observed under a 4-h fast or 4-h fast followed by exercise (Figure 1G) and a 24-h fast followed by high sucrose re-feeding (Figure 1H). The results suggest that the loss of function of muscle MondoA expression is the principal cause of the plasma lactate accumulation in MondoA−/− mice.
The elevation of plasma lactate and alanine, induced by mild exercise, recapitulates some of the features of mitochondrial myopathy [22], which prompted us to scrutinize the skeletal muscle of MondoA−/− mice by histological examination (H&E and NADH staining) and EM. However, morphological analysis at the light and electron microscopic levels failed to reveal any morphological evidence of mitochondrial abnormalities (Figure 1I; Supplementary Figure S4). There was also no difference in the total amount of mitochondria in skeletal muscle as estimated by mitochondrial DNA content between the WT and MondoA−/− mice (Supplementary Figure S5). Furthermore, the fibre-type distribution and ratio between slow twitch (type I, represented by soleus muscle) and fast twitch (type II, represented by gastrocnemius muscle) fibres (Figure 1J, top and bottom; Supplementary Figure S6) were similar in the two genotypes.
MondoA−/− and wild-type mice perform equally well in endurance exercise
The elevated basal and post-exercise levels of lactate in the MondoA−/− mice suggest a possible defect in glucose metabolism. We therefore subjected the MondoA−/− and WT mice to different exercise protocols.
We used treadmill running to examine the capacity of the WT and MondoA−/− mice for endurance exercise. Despite the elevated plasma lactate, MondoA−/− mice and WT mice performed equally well on the treadmill as judged by total running time and maximal speed attained (Figures 2A and 2B). The two genotypes also exhibited similar maximal oxygen consumption during treadmill exercise (Figure 2C). We, next, subjected the mice to swimming until exhaustion, another form of endurance exercise, and found that the average swimming endurance time before exhaustion was also not different between MondoA−/− and WT mice (Figure 2D).
MondoA−/− mice out-perform their wild-type littermates at sprint exercise
During burst exercise, such as sprinting, skeletal muscle preferentially utilizes glycolysis rather than oxidative phosphorylation for ATP production. We, therefore, developed a protocol in which we placed the mice on the treadmill at a very low speed and introduced short step-wise bursts of increasing speed building up to 20–45 m/min for up to 75 s, as shown in Figure 2(E) (see also the Materials and methods section). The vast majority of the animals would abort their sprints on the treadmill at or before the speed reached 40 m/min. Comparison with the performance of WT and MondoA−/− mice showed that, before exhaustion, MondoA−/− mice had accomplished a terminal speed that is ~30% higher than that reached by their WT littermate controls (P < 0.05) (Figure 2F).
Lowered respiratory exchange ratio in the muscle of MondoA−/− mice under exercise
To test whether glucose uptake in excess of its oxidation by skeletal muscle underlies the lactate build-up, we quantified in vivo skeletal muscle glucose uptake in mice using tritium-labelled 2-DG as a tracer. The rate of 2-DG uptake at the basal resting condition was the same in the two groups (Supplementary Figure S7A) in the two different skeletal muscle groups (EDL and gastrocnemius) that were tested. We, next, subjected the mice to swimming exercise and measured 2DG uptake immediately afterwards. We found that 2DG uptake increased in response to swimming in EDL and gastrocnemius muscles. There was, however, no difference between the two genotypes in the uptake value after swim exercise (Supplementary Figure S7B).
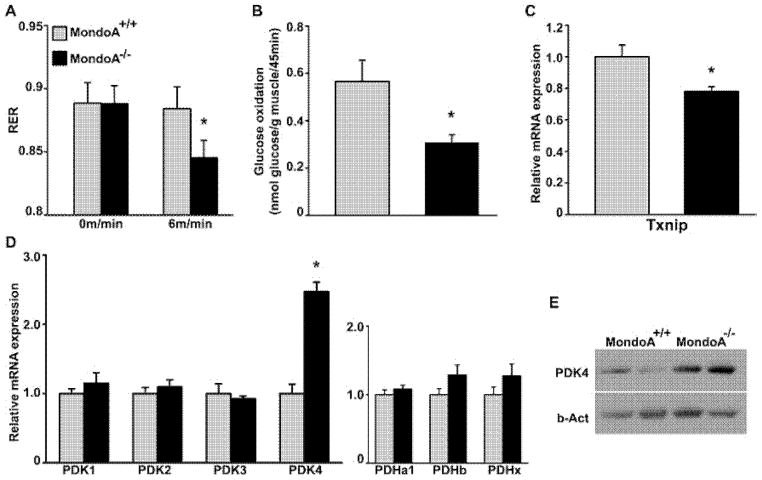
The respiratory exchange ratio (RER) is an indicator of fuel source in energy expenditure. The MondoA−/− and WT mice had a similar RER at the resting state (Figure 3A), but under mild exercise (6 m/min on treadmill for 15 min), the MondoA−/− mice had a lower RER compared with WT mice, suggesting a suboptimal glucose oxidation rate in these mice (Figure 3A). Therefore, we measured rates of glucose oxidation in isolated skeletal muscle (at resting state). Indeed, rates of glucose oxidation were lower in MondoA−/− muscle compared with WT muscle (Figure 3B). In contrast, the rates of palmitate oxidation were similar in the MondoA−/− and WT mice (Supplementary Figure S8). This indicates that MondoA plays a role in the coupling of glucose uptake and oxidation by skeletal muscle.
Figure 3. Reduced glucose oxidation and increased PDK4 expression in the muscles of MondoA−/− mice.
(A) RER under mild exercise (6 m/min treadmill running) was measured. Four-month-old male mice were used (n = 8). *P < 0.05 comparing MondoA+/+ and MondoA−/− mice. (B) Ex vivo glucose oxidation rate of isolated EDL from MondoA+/+ and MondoA−/− 6-month-old female mice (n = 7). *P < 0.05 comparing MondoA+/+ and MondoA−/− mice. (C and D) Gene expression of representative glycolytic genes in skeletal muscle (EDL) was determined by qRT-PCR. Relative expression levels of gene of interest were normalized to Alas, Eef1g and Hmbs using GeNorm algorithm [42]. Eight-week-old female mice were used (n = 5–6). *P < 0.05 comparing MondoA+/+ and MondoA−/− mice. (E) Western blot analysis of PDK4 in skeletal muscle (soleus) isolated from 6-month-old female MondoA+/+ and MondoA−/− mice determined by immunoblotting.
Pyruvate dehydrogenase kinase-4 expression is up-regulated in skeletal muscle of MondoA−/− mice
To explore the mechanism for impaired glucose oxidation and excess lactate production in MondoA−/− mice, we performed qRT-PCR analysis of the known MondoA target genes in the EDL. Txnip mRNA was down-regulated (~25%; Figure 3C), whereas that for other putative MondoA target genes, such as HKII, PFKFB3 and LDH-A, were the same as WT control (Supplementary Figure S9). Complete oxidation of glucose to H2O and CO2 requires that pyruvate enters mitochondria and, through the action of PDC, a multi-enzyme complex, be converted into acetyl-CoA. Pyruvate dehydrogenase kinase (PDK) phosphorylates pyruvate dehydrogenase (or E1) and, subsequently, the entire PDC [23]. There are four PDK isoforms in mice, PDK1–4. We examined the level of the mRNA for these four genes in the skeletal muscles of WT and MondoA−/− mice and found that the level of the mRNA for PDK4 was increased ~100% in MondoA−/− mice as compared with WT (Figure 3D, left panel). The level of the other three PDK isoforms was similar in the two genotypes. The mRNA expression of the other components of the PDC (Pdha1, -b and -x) was also not different between the two genotypes (Figure 3D, right panel). Western blot analysis showed that PDK4 expression was also up-regulated at the protein level in the skeletal muscle of MondoA−/− mice (Figure 3E).
Impaired PDH activity is responsible for lactate build-up in MondoA−/− mice
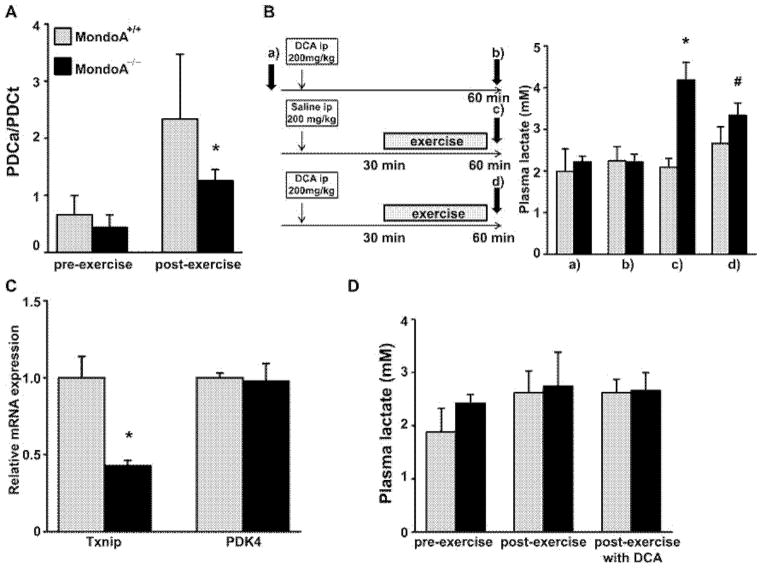
The up-regulation of PDK4 would inhibit the PDC activity and thus the oxidative decarboxylation of pyruvate to acetyl-CoA, preventing the pyruvate from entering the Krebs cycle. We measured the PDC activity in the EDL muscle and found that, under resting condition, the MondoA−/− mice showed a trend of having lower PDC activity; after exercise, they exhibited significantly lower PDC activity than that in WT mice (Figure 4A).
Figure 4. Impaired PDH activity is responsible for lactate build-up in MondoA−/− mice.
(A) Down-regulation of PDH)activity in skeletal muscle of MondoA−/− mice (6-month-old males; n = 5–7). Actual (PDCa) and total (PDCt) PDH activity of skeletal muscle (soleus) was determined as described in the Materials and methods section. The ratio of actual:total PDH of skeletal muscle isolated before or after treadmill exercise is shown. (B) DCA ameliorates the elevation of lactate in MondoA−/− mice (n = 6–7; 5-month-old males). Plasma lactate was measured in MondoA+/+ and MondoA−/− mice before DCA injection (a), 60 min after DCA injection (b), 60 min after saline injection with a 30 min of exercise on treadmill (c) and 60 min after DCA injection and 30 min of exercise on treadmill (d). *P < 0.0001 comparing MondoA+/+ and MondoA−/− mice treated with saline and exercise [under (c)]. #P < 0.01 comparing MondoA−/− mice injected with saline and DCA [between (c) and (d)]. (C) PDK4 expression in EDL muscle isolated from 5-month-old male Txnip heterozygous mice (Txnip+/− was determined by qRT-PCR. Relative expression was normalized by Alas, β-actin and B2m using GeNorm algorithm [42] (n = 3). (D) Plasma lactate level of 4-month-old Txnip+/+ (n = 5) and Txnip+/− (n = 5) mice before exercise, after treadmill exercise with DCA administration. *P < 0.05 comparing Txnip+/+ and Txnip+/− mice.
We reasoned that we could reverse the elevated plasma lactate level in the MondoA−/− mice by suppressing the activity of PDK4 using a pharmacological approach. DCA is a pyruvate analogue and a well-known inhibitor of PDK activity [24]. By inhibiting PDK, it potently stimulates PDC activity to ameliorate lactic acidosis in mitochondrial myopathy and PDC deficiency [25,26]. We devised a trial of DCA therapy using the protocol shown in Figure 4(B). After obtaining basal plasma lactate in WT and MondoA−/− mice, we treated these animals with i.p. DCA (200 mg/kg of body weight), waited 30 min and then subjected a group of the treated animals to mild exercise for 30 min, at the end of which we re-measured lactate levels. Before exercise, treatment of WT or MondoA−/− mice with DCA did not affect plasma lactate levels [Figures 4B, (a) and (b)]. Although exercise had no effect on plasma lactate levels in WT mice, it markedly increased plasma lactate in MondoA−/− mice [Figure 4B, (c)]. In contrast, the exercise-induced rise of lactate in the MondoA−/− mice was significantly blunted when animals were treated with DCA [Figure 4B, (d)]. These data suggest that the elevation of plasma lactate in MondoA−/− mice is, at least partly, the result of elevated PDK activity associated with loss of MondoA function [27].
We showed above (Figure 3C) that MondoA deficiency was associated with a ~25% lower Txnip transcript level in EDL. It is known that skeletal muscle mitochondrial fuel oxidation is impaired and glycolytic formation of lactate is enhanced in Txnip global KO mice [27]. We, next, determined whether the increased blood lactate levels in MondoA−/− mice was a consequence of down-regulated Txnip expression in these animals by studying Txnip-inactivated mice. Heterozygous Txnip-KO (Txnip+/−) mice exhibit a ~50% reduction in the skeletal muscle expression of Txnip mRNA (Figure 4C), a level similar to that in MondoA−/− mice (Figure 3C). First, we examined PDK4 mRNA expression in the skeletal muscle of these mice and found that it was not affected by Txnip haplo-insufficiency (Figure 4C). Treadmill exercise together with or without DCA treatment also failed to induce blood lactate elevation in Txnip+/− mice (Figure 4D). These data suggest that the ~25% down-regulation of Txnip expression in the skeletal muscle of MondoA−/− mice is unlikely to be the cause of the hyperlactataemia.
MondoA negatively regulates PDK4 promoter activity
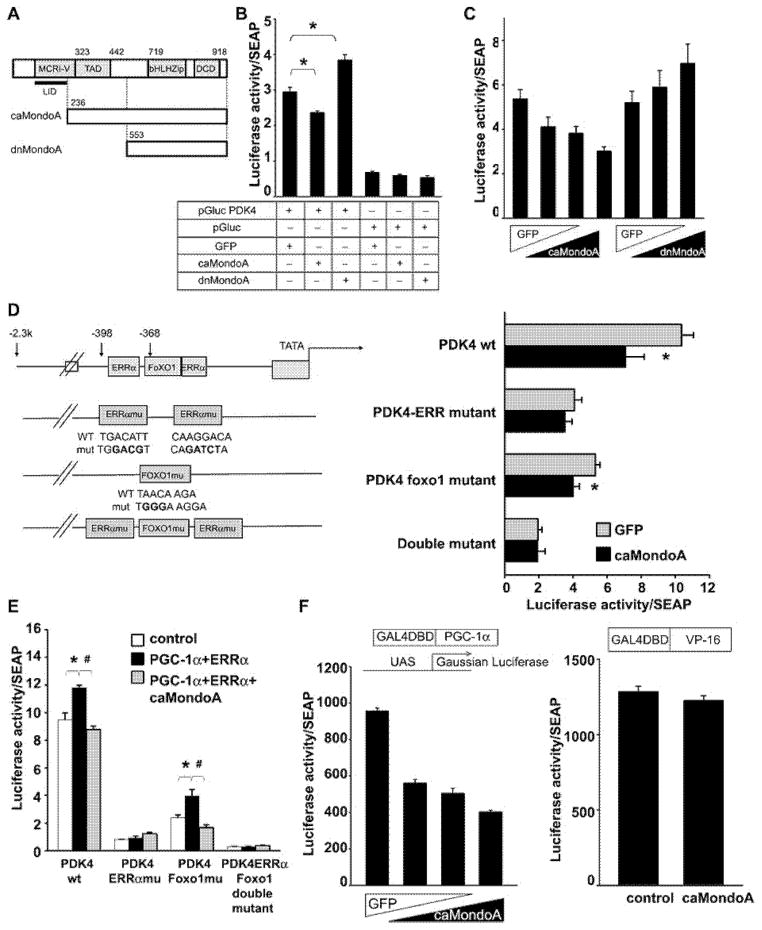
We examined whether MondoA regulates the mouse PDK4 promoter activity in vitro using C2C12 myoblasts transfected with a Gaussia luciferase construct (pGluc) driven by the mouse PDK4 promoter (from −2312 to +147). We co-transfected MondoA constructs in the form of a constitutively active (caMondoA) or a dominant-negative (dnMondoA) protein [9] (Figure 5A). As shown in Figure 5(B), the PDK4 promoter activity was down-regulated by the co-expression of caMondoA and up-regulated by dnMondoA. The effects of ca- and dnMondoA were dose-dependent (Figure 5C). MondoA is known to bind to a single E-box (CACGTG) or ChoRE, neither of which is identified within the promoter region assessed. We therefore examined the effect of caMondoA on the luciferase activity of serial deletion mutants of the PDK4 reporters (Supplementary Figure S10). Deletion mutant 5 (−352/+147), which had lost its consensus oestrogen-related receptor α (ERRα) and forkhead box O1 (FOXO1)-binding sites, as well as other more extensive deletion mutants, no longer responded to caMondoA suppression (Supplementary Figure S10), suggesting that ERRα- and FOXO1-binding sites may be important to MondoA regulation of the PDK4 promoter. Next, we mutated the nucleotide sequence of the putative ERRα- and FOXO1-binding sites [28–30] and analysed the effect of caMondoA on the PDK promoter activity in these luciferase reporter mutants. As shown in Figure 5(D), caMondoA suppressed the activity of the WT, as well as the FOXO1-binding site mutants, but had no effect on the PDK4 promoter that carried mutations in the ERRα-binding sites. The ERRα and FOXO1 double-mutant had lost the PDK4 promoter activity, as well as caMondoA responsiveness.
Figure 5. MondoA negatively regulates PDK4 promoter activity.
(A) Schematic representation of the full-length as well as caMondoA and dnMondoA expression constructs. (B) Luciferase reporter plasmid (pGluc PDK4 or pGluc) and an internal control construct, pSRa-SEAP, which express secreted alkaline phosphatase were co-transfected with vector-expressing caMondoA or dnMondoA or GFP in C2C12 cells. Forty-eight hours after transfection, medium was collected and luciferase activity was analysed. *P < 0.05 between groups indicated by bracket. (C) pGluc PDK4 and pSRa-SEAP were co-transfected with increasing amount caMondoA or dnMondoA into BOSC23 cells. The total amounts of transfected DNA for each group were held the same by adding GFP expression vector. Forty-eight hours after transfection, medium was collected and luciferase activity was analysed. (D) Schematic representation of ERRα- and FOXO1-binding sites of PDK4 promoter and mutant constructs (left panel). pGluc PDK4 or pGluc PDK4 mutants and pSRa-SEAP were co-transfected with caMondoA or GFP control vector into BOSC23 cells. Forty-eight hours after transfection, medium was collected and luciferase activity was analysed. *P < 0.05 between GFP control and caMondoA transfected cells. (E) pGluc PDK4 or pGluc PDK4 mutants (as shown in D) and pSRa-SEAP were co-transfected with vectors expressing PGC-1α and ERRα with or without caMondoA into BOSC23 cells. The total amounts of transfected DNA for each group were held the same with either pcDNA3.1 empty vector or GFP expression vector. Forty-eight hours after transfection, medium was collected and luciferase activity was analysed. (F) Plasmids encoding either GAL4-PGC-1α (left panel) or GAL4 VP-16 (right panel) and pUAS-Gluc and pSEAP were co-transfected with indicated amount of vector-expressing caMondoA into BOSC23 cells. Twenty-four hours after transfection, medium was collected and luciferase activity was analysed.
PGC-1α is known to co-activate PDK4 gene expression by first inducing ERRα expression and the ERRα protein binds to the ERRα site of the PDK4 promoter to which PGC-1α also binds and together they activate PDK4 expression [31]. The PGC-1α-mediated PDK4 co-activation is ERRα-dependent as it does not occur in ERRα-null cells [31]. We examined the effect of PGC-1α and ERRα co-activation on the activities of WT and PDK4 promoter mutants in response to caMondoA suppression. The WT PDK4 promoter responded to PGC1α and ERRα co-activation and this co-activation was suppressed by caMondoA (Figure 5E). The ERRα-binding site mutant PDK4 promoter did not respond to PGC-1α and ERRα co-expression and caMondoA did not change the promoter activity any further. The FOXO1-binding site mutant PDK4 promoter activities were up-regulated by PGC-1α and ERRα co-transfection and this up-regulation was suppressed by the simultaneous expression of caMondoA (Figure 5E). This suggests that MondoA negatively regulates PDK4 promoter activity via modulating PGC-1α-ERRα binding to the PDK4 promoter.
To further analyse the regulation of PGC-1α transcriptional activity by caMondoA, we adopted a GAL4-UAS driven luciferase reporter system. PGC-1α was fused with the GAL4-DNA-binding domain and co-transfected with UAS-driven-luciferase into BOSC23 cells. The fusion GAL4-DNA-binding domain-PGC-1α construct turned on the UAS-Luc robustly, but caMondoA suppresses this activity in a dose-dependent manner (Figure 5F, left panel). caMondoA does not suppress GAL4-VP16 transcriptional activity (Figure 5F, right panel), indicating that the regulation of PGC-1α activity by caMondoA was not a non-specific action of caMondoA.
Metabolic shift from glucose oxidation to fatty acid oxidation
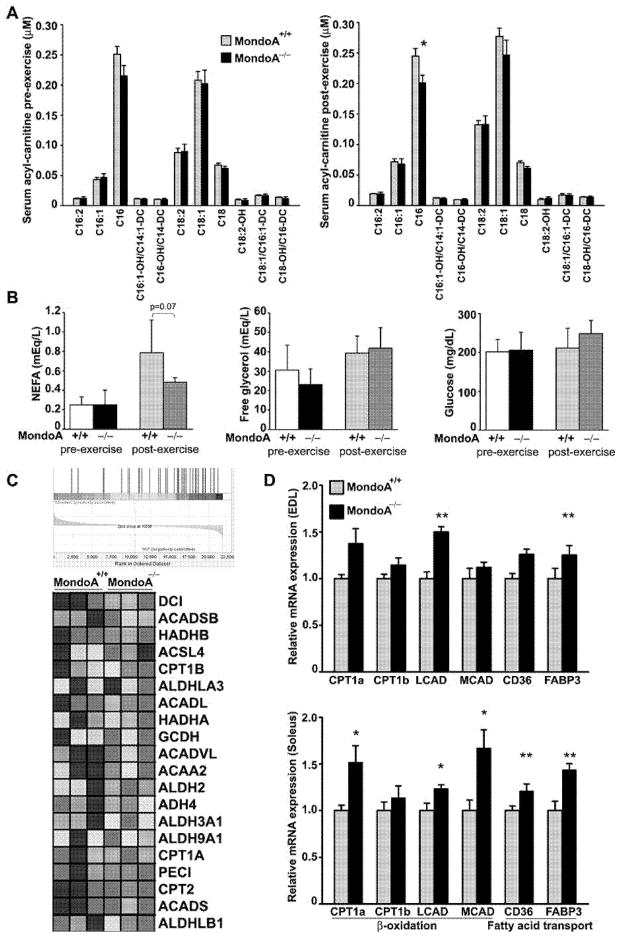
Despite lactate build-up, MondoA−/− mice performed as well as WT mice in endurance exercise and out-performed the latter in sprint exercise. The yield of ATP from glycolysis is lower than that from a complete oxidation of glucose to CO2 and H2O. It is plausible that skeletal muscles in MondoA−/− mice utilize fuel source(s) other than glucose to meet the energy demands. The reduction in the RER indicates that fatty acid oxidation is probably the other source of energy. As part of the metabolomic analysis, we therefore measured the serum acylcarnitine profiles in WT and MondoA−/− mice before and after 15-min of treadmill exercise. In the resting state, there was no significant difference in the level of long-chain acylcarnitine in the serum of WT and MondoA−/− mice (Figure 6A, left panel), although the serum palmitoylcarnitine (C16) tended to be lower in the MondoA−/− mice. After standard non-intensive exercise (see the Materials and methods section), the serum palmitoylcarnitine was significantly lower in MondoA−/− mice (Figure 6A, right panel) and serum NEFAs tended to be lower after exercise (Figure 6B, left panel) consistent with less restriction of fatty acid oxidation at the level of the rate-limiting enzyme carnitine palmitoyltransferase 1. The serum-free glycerol level, an indicator of lipolysis in adipose tissue, remained the same in control and MondoA−/− mice (Figure 6B, middle panel) suggesting that the reduction in palmitoylcarnitine was not due to a reduction in lipolysis. Serum glucose levels were similar in the two groups of mice (Figure 6B, right panel).
Figure 6. Metabolic shift from glucose oxidation to fatty acid oxidation in MondoA−/− mice.
(A) Serum long-chain acylcarnitine profile. Serum harvested from 6-month-old male MondoA+/+ (n = 6) and MondoA−/− mice (n = 6), before and after treadmill exercise, were analysed. (B) Plasma glucose, NEFA and free glycerol before and after treadmill exercise. Four-month-old male (n = 6 each genotype). (C) Heat map of GSEA of fatty acid metabolism gene expressions in soleus muscles of 6-month-old male MondoA−/− and MondoA+/+ mice (n = 3 each). (D) qRT-PCR analysis of lipid-metabolism-associated gene-expression in skeletal muscle (EDL and soleus). Relative expression levels of gene of interest were normalized by Alas, Eef1g and Hmbs using GeNorm algorithm [42]. Eight-week-old female mice were used (n = 5–6). *P < 0.05, **P < 0.01 comparing MondoA+/+ and MondoA−/− mice.
We performed microarray analysis of skeletal muscle mRNA isolated from WT and MondoA−/− mice. Gene-set enrichment analysis showed that the transcripts for three sets of gene that pertain to extracellular matrix (ECM) receptor interaction (HSA04512), fatty acid metabolism (HSA00071) and focal adhesion (HSA04510) were significantly enriched with a false discovery rate of 25%. As shown in the heat map (Figure 6C), top-ranked genes within the gene set for fatty acid metabolism (HSA00071) involved in β-oxidation were uniformly up-regulated in MondoA−/− mice. When we used qRT-PCR to validate results from the microarray analysis, we found that in both EDL and soleus the level of gene transcripts involved in fatty-acid transport and β-oxidation were significantly up-regulated in MondoA−/− compared with WT mice (Figure 6D).
DISCUSSION
We had set out to test the hypothesis that disruption of the gene encoding MondoA would induce major effects on glucose homoeostasis. In contrast with expectations, MondoA deficient mice were normal with respect to fed and fasting blood glucose levels, glucose tolerance, appearance, growth and blood analyte levels. These findings suggest MondoA plays a relatively minor role in fuel homoeostasis of mice in the resting, unstressed state. Unexpectedly, mice lacking MondoA showed an enhanced sprinting performance coupled to enhanced rates of glycolysis as suggested by the build-up of lactate and alanine.
ChREBP and MondoA respond to glucose because they have highly conserved domains in common that render them inactive when glucose levels are low but active when glucose levels are high [9]. Although related in structure and regulation, ChREBP and MondoA differ greatly in tissue distribution and the genes they regulate. ChREBP is expressed primarily in the liver. In contrast, MondoA is expressed primarily in the skeletal muscle. In vitro studies using C2C12 cells suggest that MondoA may promote the transcription of HKII, PFKFB3, LDH-A and Txnip [4–6]. Because skeletal muscle plays an important role in the clearance of blood glucose, it would seem that MondoA deletion should reduce the transcription of these genes, which in turn would result in a reduced rate of glycolysis and elevated blood glucose levels. This was not found to be the case and may simply reflect the fact that the rate of blood glucose clearance by resting skeletal muscle depends more upon conversion of glucose to glycogen than oxidative disposal of glucose by oxidation of pyruvate [32]. The finding in the present study that MondoA deletion had no effect on the expression levels of established target genes (HKII, PFKFB3, LDH-A; Supplementary Figure S9) and only a modest effect on the expression of Txnip (Figure 3C) may simply reflect a low activity state of MondoA in the WT mice. It is noteworthy that the effects of MondoA in vitro on HKII, PFKB3 and LDH-A are cell-line-dependent; the same laboratory that found effects in C2C12 cells [4,6] failed to see any effect of MondoA knockdown on these putative target genes in HA1ER cells [5].
Serum lactate levels of MondoA−/− mice were induced to higher levels than WT mice by short-term fasting, re-feeding following fasting and exercise (Figures 1C and 1D). The accumulation of lactate can lead to fatal lactic acidosis [33]. Therefore, it is important to understand the molecular mechanism by which MondoA controls blood levels of lactic acid. Lactic acidosis can be the result of overproduction by the muscle or under-utilization by the liver. Mice lacking MondoA, specifically in the skeletal muscle, were used to establish that it was due to overproduction of lactate by the muscle (Figures 1G and 1H).
Although there are several possible reasons for greater lactate production by muscle during exercise, the finding that serum alanine levels were also increased was informative. Of the four PDKs expressed in tissues, PDK4 was found to be up-regulated at both the mRNA and the protein level in the skeletal muscle of MondoA KO mice (Figures 3D and 3E). As predicted, greater PDK4 expression resulted in a lower activity state of the PDC (Figure 4A). Reduced PDC activity conserves pyruvate for conversion to alanine and lactate, resulting in higher serum levels of these compounds in MondoA−/− mice.
The finding that PDK4 expression is up-regulated in MondoA−/− mice resulted in the elucidation of a novel mechanism that plays an important role in regulation of PDK4 expression in skeletal muscle. Building on the evidence that the promoter for PDK4 is stimulated by the transcription factors ERRα, FOXO1 and PGC1α, we now found that MondoA exerts a negative effect upon the expression of PGC1α. Because PGC1α regulates expression of ERRα and it also binds to ERRα as a co-activator, down-regulation of PGC1α by MondoA inhibits expression of PDK4. That down-regulation of PDK4 by this mechanism is physiologically important as evidenced by greater PDK4 expression, lower pyruvate dehydrogenase activity and higher lactate and alanine levels in MondoA−/− mice. Although this is the first report that a mechanism exists by which MondoA inhibits PDK4 expression, suggestive evidence for a link between MondoA and PDK4 was noted previously in a study on the effects of the thiazolidinedione (TZD) on insulin resistant subjects [34]. The effectiveness of TZD in improving the sensitivity to insulin, as measured by down-regulation of the expression of PDK4 during a hyperinsulinaemic–euglycaemic clamp, was dependent upon a high level of MondoA expression [34]. These findings suggest that reversible regulation of PDK4 expression, which is clearly important for good metabolic flexibility [34], may require the concerted actions of insulin and MondoA.
At first glance, it seems puzzling to find that serum lactate levels were higher in MondoA−/− mice than WT mice after 4 h of fasting but not after 24 h of starvation (Figure 1C). However, it can be rationalized on the basis of what is known about regulation of the expression of PDK4. Starvation for 24 h results in a large increase in PDK4 expression in skeletal muscle of WT mice [35]. This is due to activation by NEFAs of peroxisome-proliferator-activated receptor (PPAR)α and/or PPARδ, which are known to induce PDK4 expression. Because insulin is a potent inhibitor of PDK4 expression [36–38], it also reflects the low insulin levels that were obtained after 24 h of fasting. Under these conditions of maximum PDK4 expression, an effect of lack of MondoA on PDK4 expression would not register and, therefore, higher serum lactate levels are not observed in the MondoA−/− mice. In contrast, fasting for 4 h is not long enough to induce PDK4 expression in WT mice [38]. NEFA levels are not sufficiently elevated and insulin levels are still very high. Because conservation of three carbon compound (lactate, pyruvate and alanine) for glucose synthesis by the liver is the primary reason for induction of PDK4 in skeletal muscle during starvation [39], PDK4 expression should be and is normally low in skeletal muscle after 4 h of fasting. As a result, three carbon compounds produced in skeletal muscle are largely oxidized as a source of energy. The finding that 4 h of fasting induces greater plasma lactate levels in MondoA−/− mice suggests that the activity of the PDC has been reduced by increased expression of PDK4. From this finding, we can conclude that suppression of PDK4 expression by MondoA plays an important role in maintaining the capacity for complete oxidation of glucose to CO2 and water by skeletal muscle. The importance of this mechanism is further corroborated by higher lactate levels in mice fed a high-sucrose, no-fat diet for 18 h after a 24-h fast.
The activity state of the PDC increases in skeletal muscle during aerobic exercise [40]. Greater capacity for pyruvate oxidation promotes complete glucose oxidation to meet the need for ATP and to minimize lactate and alanine production. Normally, however, the activity of the PDC is kept partially in check during exercise by phosphorylation of the complex by PDK4. Evidence for this was obtained with PDK4 KO mice [41]. Forced muscle contraction in these mice results in significantly greater activation of the PDC, establishing an important role for PDK4 in controlling the capacity for pyruvate oxidation during exercise. Consistent with these findings, exercise induced a larger increase in serum lactate in MondoA−/− mice compared with WT mice. This follows from greater expression of PDK4 and lower activity of the PDC of the MondoA−/− mice. Although this might have been expected to decrease the exercise endurance, this was not observed, perhaps because of compensatory changes in the capacity for fatty acid oxidation.
Furthermore, it is likely that the increased capacity for glycolysis explains the enhanced capacity of the mice for sprinting. Since NAD+ is required for the step of glycolysis catalysed by glyceraldehyde-3-phosphate dehydrogenase, anaerobic glycolysis is totally dependent upon the regeneration of NAD+ from NADH by the reaction catalysed by lactate dehydrogenase, which in turn is dependent upon stoichiometric availability of pyruvate. Inhibition of the PDC by increased expression of PDK4 promotes glycolysis by minimizing the competition between the PDC and lactate dehydrogenase. Although the yield of ATP per molecule of glucose is low, for a short period of time glycolysis can produce ATP at the high rate required for sprinting.
In conclusion, studies of MondoA-inactivated mice in vivo indicate that MondoA promotes skeletal muscle glucose oxidation. In the absence of MondoA, pyruvate is shunted into lactate and the bulk of energy is derived from increased fatty acid oxidation. The transcriptional regulation of PDK4 by MondoA appears to be mediated indirectly via PGC-1α. Interestingly, MondoA−/− mice out performed their WT littermates in the sprinting protocol, highlighting the pivotal role of MondoA in skeletal muscle energy metabolism and performance.
Acknowledgments
We thank Dr Naravat Poungvarin for assistance with the luciferase assay system, the Digestive Disease Center (P30 DK56338) at Baylor College of Medicine for providing EM support and Dr Simon Hu at the University of California at Los Angeles for providing Txnip−/− mice. We also thank Dr Christopher Newgard, Dr Robert Stevens, Dr James Bain, and Dr Olga Ilkayeva of the Duke University Medical Center for metabolomic analyses.
FUNDING
This work was supported by the National Institutes of Health [grant numbers R01 HL51586 (to L.C.), R01 DK084495 (to B.H.-J.C.), R01 HL061483 (to H.T.) and P30 DK079638 (for the Diabetes Research Center at Baylor College of Medicine)]; a VA Merit Review Award (to R.A.H.)]; and the Betty Rutherford Chair for Diabetes Research (to L.C.).
Abbreviations
- [2-3H]2DG
2-deoxy-D-[1,2-3H]-glucose
- AUC
area under the curve
- caMondoA
constitutively active MondoA
- ChoRE
carbohydrate-response element
- ChREBP
carbohydrate-response element-binding protein
- DCA
dichloroacetate
- dnMondoA
dominant-negative MondoA
- EDL
extensor digitorum longus
- ERRα
oestrogen-related receptor α
- ES
embryonic stem
- FOXO
forkhead box O
- GRACE
glucose-responsive activation conserved domain
- GSEA
gene set enrichment analysis
- GTT
glucose tolerance test
- HKII
hexokinase II
- i.p
intraperitoneal
- KO
knockout
- LDH-A
lactate dehydrogenase A
- LID
low-glucose inhibitory domain
- Mck-Cre
muscle creatine kinase Cre
- Mix
Max-like protein X
- NEFA
non-esterified fatty acid
- OMM
outer mitochondrial membrane
- PDC
pyruvate dehydrogenase complex
- PDK
pyruvate dehydrogenase kinase
- PFKFB3
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3
- PGC-1α
peroxisome-proliferator-activated receptor γ co-activator-α
- PPAR
peroxisome-proliferator-activated receptor
- qRT-PCR
quantitative real-time PCR
- RER
respiratory exchange ratio
- SEAP
secreted alkaline phosphatase
- Txnip
thioredoxin interacting protein
- TZD
thiazolidinedione
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
Minako Imamura and Lawrence Chan conceived and designed the experiments. Minako Imamura and Motoyuki Kohjima performed the experiments. Minako Imamura, Ming Li and Benny Hung-Junn Chang analysed the data. Ming Li, Byounghoon Hwang, Robert Harris and Heinrich Taegtmeyer contributed reagents/materials/analysis tools. Minako Imamura, Benny Hung-Junn Chang, Heinrich Taegtmeyer, Robert Harris and Lawrence Chan wrote the paper.
References
- 1.Girard J, Ferré P, Foufelle F. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu Rev Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 2.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uyeda K, Yamashita H, Kawaguchi T. Carbohydrate responsive element-binding protein (ChREBP): a key regulator of glucose metabolism and fat storage. Biochem Pharmacol. 2002;63:2075–2080. doi: 10.1016/s0006-2952(02)01012-2. [DOI] [PubMed] [Google Scholar]
- 4.Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. MondoA, a novel basic helix–loop–helix–leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol. 2000;20:8845–8854. doi: 10.1128/mcb.20.23.8845-8854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci USA. 2008;105:6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA–Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muoio DM. TXNIP links redox circuitry to glucose control. Cell Metab. 2007;5:412–414. doi: 10.1016/j.cmet.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Li MV, Chang B, Imamura M, Poungvarin N, Chan L. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes. 2006;55:1179–1189. doi: 10.2337/db05-0822. [DOI] [PubMed] [Google Scholar]
- 10.Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 11.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang BH, Liao W, Li L, Nakamuta M, Mack D, Chan L. Liver-specific inactivation of the abetalipoproteinemia gene completely abrogates very low density lipoprotein/low density lipoprotein production in a viable conditional knockout mouse. J Biol Chem. 1999;274:6051–6055. doi: 10.1074/jbc.274.10.6051. [DOI] [PubMed] [Google Scholar]
- 13.Sheehan DC, Hrapchak BB. Theory and Practice of Histotechnology. 2. Battelle Press; 1987. [Google Scholar]
- 14.An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- 15.Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu P, Inskeep K, Bowker-Kinley MM, Popov KM, Harris RA. Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes. 1999;48:1593–1599. doi: 10.2337/diabetes.48.8.1593. [DOI] [PubMed] [Google Scholar]
- 17.Jeoung NH, Sanghani PC, Zhai L, Harris RA. Assay of the pyruvate dehydrogenase complex by coupling with recombinant chicken liver arylamine N-acetyltransferase. Anal Biochem. 2006;356:44–50. doi: 10.1016/j.ab.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Jeoung NH, Wu P, Joshi MA, Jaskiewicz J, Bock CB, Depaoli-Roach AA, Harris RA. Role of pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) in glucose homoeostasis during starvation. Biochem J. 2006;397:417–425. doi: 10.1042/BJ20060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted
- 20.Rose AJ, Richter EA. Skeletal muscle glucose uptake during exercise: how is it regulated? Physiology. 2005;20:260–270. doi: 10.1152/physiol.00012.2005. [DOI] [PubMed] [Google Scholar]
- 21.Taegtmeyer H, Peterson MB, Ragavan VV, Ferguson AG, Lesch M. De novo alanine synthesis in isolated oxygen-deprived rabbit myocardium. J Biol Chem. 1977;252:5010–5018. [PubMed] [Google Scholar]
- 22.Behbehani AW, Goebel H, Osse G, Gabriel M, Langenbeck U, Berden J, Berger R, Schutgens RB. Mitochondrial myopathy with lactic acidosis and deficient activity of muscle succinate cytochrome-c-oxidoreductase. Eur J Pediatr. 1984;143:67–71. doi: 10.1007/BF00442753. [DOI] [PubMed] [Google Scholar]
- 23.Harris RA, Popov KM, Zhao Y, Kedishvili NY, Shimomura Y, Crabb DW. A new family of protein kinases–the mitochondrial protein kinases. Adv Enzyme Regul. 1995;35:147–162. doi: 10.1016/0065-2571(94)00020-4. [DOI] [PubMed] [Google Scholar]
- 24.Kato M, Li J, Chuang JL, Chuang DT. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure. 2007;15:992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morten KJ, Beattie P, Brown GK, Matthews PM. Dichloroacetate stabilizes the mutant E1alpha subunit in pyruvate dehydrogenase deficiency. Neurology. 1999;53:612–616. doi: 10.1212/wnl.53.3.612. [DOI] [PubMed] [Google Scholar]
- 26.Stacpoole PW, Kurtz TL, Han Z, Langaee T. Role of dichloroacetate in the treatment of genetic mitochondrial diseases. Adv Drug Deliv Rev. 2008;60:1478–1487. doi: 10.1016/j.addr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hui ST, Andres AM, Miller AK, Spann NJ, Potter DW, Post NM, Chen AZ, Sachithanantham S, Jung DY, Kim JK, Davis RA. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci USA. 2008;105:3921–3926. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki M, Motojima K. Identification of ERRalpha as a specific partner of PGC-1alpha for the activation of PDK4 gene expression in muscle. FEBS J. 2006;273:1669–1680. doi: 10.1111/j.1742-4658.2006.05183.x. [DOI] [PubMed] [Google Scholar]
- 29.Nishiyama A, Matsui M, Iwata S, Hirota K, Masutani H, Nakamura H, Takagi Y, Sono H, Gon Y, Yodoi J. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Ma K, Sadana P, Chowdhury F, Gaillard S, Wang F, McDonnell DP, Unterman TG, Elam MB, Park EA. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J Biol Chem. 2006;281:39897–39906. doi: 10.1074/jbc.M608657200. [DOI] [PubMed] [Google Scholar]
- 31.Wende AR, Huss JM, Schaeffer PJ, Giguère V, Kelly DP. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jue T, Rothman DL, Shulman GI, Tavitian BA, DeFronzo RA, Shulman RG. Direct observation of glycogen synthesis in human muscle with 13C NMR. Proc Natl Acad Sci USA. 1989;86:4489–4491. doi: 10.1073/pnas.86.12.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson BH. Lactic acidemia and mitochondrial disease. Mol Genet Metab. 2006;89:3–13. doi: 10.1016/j.ymgme.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Sears DD, Hsiao G, Hsiao A, Yu JG, Courtney CH, Ofrecio JM, Chapman J, Subramaniam S. Mechanisms of human insulin resistance and thiazolidinedione-mediated insulin sensitization. Proc Natl Acad Sci USA. 2009;106:18745–18750. doi: 10.1073/pnas.0903032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P, Peters JM, Harris RA. Adaptive increase in pyruvate dehydrogenase kinase 4 during starvation is mediated by peroxisome proliferator-activated receptor alpha. Biochem Biophys Res Commun. 2001;287:391–396. doi: 10.1006/bbrc.2001.5608. [DOI] [PubMed] [Google Scholar]
- 36.Huang B, Wu P, Bowker-Kinley MM, Harris RA. Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor-alpha ligands, glucocorticoids, and insulin. Diabetes. 2002;51:276–283. doi: 10.2337/diabetes.51.2.276. [DOI] [PubMed] [Google Scholar]
- 37.Kwon HS, Harris RA. Mechanisms responsible for regulation of pyruvate dehydrogenase kinase 4 gene expression. Adv Enzyme Regul. 2004;44:109–121. doi: 10.1016/j.advenzreg.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 38.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor F0X01 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeoung NH, Wu P, Joshi MA, Jaskiewicz J, Bock CB, Depaoli-Roach AA, Harris RA. Role of pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) in glucose homoeostasis during starvation. Biochem J. 2006;397:417–425. doi: 10.1042/BJ20060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watt MJ, Heigenhauser GJ, Dyck DJ, Spriet LL. Intramuscular triacylglycerol, glycogen and acetyl group metabolism during 4 h of moderate exercise in man. J Physiol. 2002;541:969–978. doi: 10.1113/jphysiol.2002.018820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbst EA, Dunford EC, Harris RA, Vandenboom R, Leblanc PJ, Roy BD, Jeoung NH, Peters SJ. Role of pyruvate dehydrogenase kinase 4 in regulating PDH activation during acute muscle contraction. Appl Physiol Nutr Metab. 2012;37:48–52. doi: 10.1139/h11-136. [DOI] [PubMed] [Google Scholar]
- 42.Pattyn F, Robbrecht P, De Paepe A, Speleman F, Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database, major update 2006. Nucleic Acids Res. 2006;34:D684–D688. doi: 10.1093/nar/gkj155. [DOI] [PMC free article] [PubMed] [Google Scholar]