Abstract
Enterococcus faecalis is a robust bacterium, which is able to survive in and adapt to hostile environments such as the urinary tract and bladder. In this label-free quantitative proteomic study based on MaxQuant LFQ algorithms, we identified 127 proteins present in the secretome of the clinical vancomycin-resistant isolate E. faecalis V583 and we compared proteins secreted in the initial phase of cultivation in urine with the secretome during cultivation in standard laboratory medium, 2xYT. Of the 54 identified proteins predicted to be secreted, six were exclusively found after cultivation in urine including the virulence factor EfaA (“endocarditis specific antigen”) and its homologue EF0577 (“adhesion lipoprotein”). These two proteins are both involved in manganese transport, known to be an important determinant of colonization and infection, and may additionally function as adhesins. Other detected urine-specific proteins are involved in peptide transport (EF0063 and EF3106) and protease inhibition (EF3054). In addition, we found an uncharacterized protein (EF0764), which had not previously been linked to the adaptation of V583 to a urine environment, and which is unique to E. faecalis. Proteins found in both environments included a histone-like protein, EF1550, that was up-regulated during cultivation in urine and that has a homologue in streptococci (HlpA) known to be involved in bacterial adhesion to host cells. Up-regulated secreted proteins included autolysins. These results from secretome analyses are largely compatible with previously published data from transcriptomics studies. All in all, the present data indicate that transport, in particular metal transport, adhesion, cell wall remodelling and the unknown function carried out by the unique EF0764 are important for enterococcal adaptation to the urine environment. These results provide a basis for a more targeted exploration of novel proteins involved in the adaptability and pathogenicity of E. faecalis.
Introduction
Enterococci are gram-positive bacteria normally found in food, plants, soil and water, as well as in the gastro-intestinal tracts of animals and humans. While enterococci generally are non-virulent, several species are known as antibiotic resistant opportunistic pathogenic bacteria, with Enterococcus faecalis and Enterococcus faecium being the two most important clinical species. E. faecalis is one of the leading causes of hospital-acquired bloodstream and urinary tract infections (UTIs) [1, 2] and may also cause other problems such as abdominal- and wound infections and endocarditis [3]. UTIs are thought to depend on several virulence traits, including biofilm formation, the ability to adhere to host surfaces, the presence of adhesion-promoting cell-wall/cell-membrane components such as glycolipids, and mechanisms for evading the host immune system [4–8]. Interestingly, recent results indicate that E. faecalis can not only adhere to epithelial cells in the urinary tract [4], but also invade cells, leading to formation of intracellular bacterial communities in the bladder [5]. The combination of these multiple virulence mechanisms, the ability to survive in harsh conditions, and both intrinsic and acquired resistance to many antibiotics explains the high frequency of infection by E. faecalis. E. faecalis strains of different origins, including probiotic, pathogenic and laboratory strains, show the same capacity to grow in urine [6], indicating that variation in the ability to exploit urine nutrients is not a pathogenicity-determining factor.
E. faecalis V583 (hereafter referred to as V583) was the first vancomycin-resistant clinical isolate reported in the USA and the first sequenced E. faecalis strain [7, 8]. More than 25% of its genome contains mobile genetic elements, including a large pathogenicity island. V583 has been intensively studied due to its pathogenic significance [7, 8]; however, still, little is known about the proteins and molecular mechanisms that endow this bacterium with the ability to survive and grow within an infected patient. Enterococcal responses in in vivo infection models and under infection-relevant growth conditions such as the presence of urine, blood, bile, or antibiotics have been studied quite extensively using transcriptomics [1, 6, 9–13]. As to cultivation in urine, four studies have been published. Vebø et al. [6] studied the response of enterococcal strains with varying virulence to growth in urine and found that transcriptional responses were quite similar. Some differential regulation was observed for genes related to stress responses, energy metabolism, acquisition of trace metals, and cell envelope modification, but it was not possible to detect pathogen-specific E. faecalis genes. Hanin et al. [12] combined quantitative reverse transcription PCR with recombination-based in vivo expression technology (R-IVET) screening to study the E. faecalis V19 strain (a V583 derivative) and found 18 genes induced after growth in urine, the majority of which were (putatively) involved in transport and protein-protein interactions, or had an unknown function. Furthermore, using real-time quantitative PCR, Shepard and Gilmore [1] studied the response of targeted genes suspected to play a role in pathogenesis of E. faecalis strain MMH594. Studies with urine revealed several genes whose expression was induced with the E. faecalis endocarditis antigen (efaA) demonstrating the largest change in expression. Finally, Carlos et al. [14] used reverse transcription PCR to study the response of selected virulence genes in enterococcal strains of various origins and found that growth in urine resulted in strain-specific regulation of these genes.
The proteomic approach has the advantage that it includes information from the regulatory steps between transcription and the final protein product, including translation, protein translocation and protein turnover. Furthermore, proteins from different cellular locations (intracellular, extracellular, membrane) can be identified and quantified separately, providing an extra dimension to the analysis. Extracellular proteins (the secretome) are of particular interest because they are likely to play a major role in most aspects of the host-pathogen interaction. The secretome of E. faecalis under standard conditions has been analysed by Benachour et al. in 2009 [15] and Bøhle et al. in 2011 [16], leading to the identification of 38 and 69 secreted and surface-located proteins, respectively. A comparative study of the secretomes of a food and a clinical strain of E. faecalis grown on laboratory medium led to the identification of 16 and 33 extracellular proteins, respectively [17]. The membrane proteome of E. faecalis has also been mapped, identifying 102 proteins [18]. Stress responses have so far only been studied looking at the intracellular proteome, including responses to starvation [19], the presence of bovine bile [20], vancomycin treatment [21, 22], and other stress conditions such as heat, H2O2, salt, and pH [23]. Responses of the intracellular proteome to different growth conditions (aerobic, anaerobic) and exposure to mouse intestinal cells have also been studied [24].
Here, we have used a proteomics approach to study the immediate response of E. faecalis to urine, with focus on the secretome. In an attempt to identify proteins possibly involved in UTIs, we have compared proteins in culture supernatants of the clinical isolate E. faecalis V583 when cultivated in urine or in a rich laboratory medium, 2xYT. We used recently developed label-free methods for protein quantification implemented in the MaxQuant software tool [25, 26]. Using this approach allowed identification of proteins that are clearly overproduced during incubation in urine. Of the 54 detected proteins that are predicted to be secreted, 27 were found in urine, but only six of these showed quantitative differences that warrant them being qualified as urine-specific. The present approach provides a basis for more targeted exploration of proteins that govern the behaviour of E. faecalis.
Material and Methods
Culture conditions and harvesting for proteomics analysis
The strain used in this study, Enterococcus faecalis V583 (ATCC 700802) [8] was grown overnight in 2xYT medium [1% (w/v) yeast extract, 1.6% (w/v) tryptone and 1% (w/v) NaCl] at 37°C without agitation. Growth conditions used for proteome sampling were similar to those used by Vebø et al. [6] in a previously published transcriptome study. The overnight cultures were diluted in 250 ml pre-warmed 2xYT medium to an optical density (OD600) of ~0.01 and incubated further at 37°C without agitation until the OD600 reached 0.1. The culture was then divided into two and cells were collected by centrifugation (10.000 x g, 3 min, 37°C). The cell pellets were re-suspended to an OD600 of 0.02 in pre-warmed 2xYT medium or pre-warmed sterile urine, followed by incubation at 37°C, without agitation. For incubation in urine, fresh human urine was collected from four healthy individuals, pooled and sterilized as described by Vebø et al [6]. The experiment was performed in triplicate, on three different days. Fresh urine was collected on each of these days from the same donors, and similarly, fresh 2xYT was prepared every day. This approach was chosen to account for batch-to-batch variation. Samples of each culture were collected 1.5 hours after the re-suspension in 2xYT or urine. Prior to harvesting, phenylmethylsulfonyl fluoride (PMSF) to 0.1 mM final concentration was added to the cultures to prevent proteolysis during sample preparation. The bacteria were harvested by centrifugation (6.500 x g, 10 min, 4°C) and the supernatant was sterilized by filtration (0.2 μm) and stored at -20°C, until use.
Preparation of secreted proteins
To collect proteins from the supernatants, 1.6 mL of the culture supernatant was thawed at room temperature and the pH was adjusted to a pH between 7 and 8 by adding 2 μl 6 M NaOH. Proteins were then precipitated using trichloroacetic acid (TCA). First, Na-deoxycholate was added to the samples to a final concentration of 0.2 mg/mL followed by 30 minutes incubation on ice. Protein precipitation was induced by addition of TCA to a final concentration of 16% (v/v), followed by incubation on ice for 20 minutes and subsequent overnight incubation at -20 °C. Precipitated proteins were collected by centrifugation (16.000 x g, 30 min, 4°C) and the pellets were washed twice with ice-cold acetone, before air-drying the pellets at 60°C for ~5 minutes.
SDS-PAGE and in-gel trypsination
The dried protein pellets were dissolved directly in NuPAGE LDS sample buffer (Invitrogen) before SDS-PAGE. Sample loading was normalized so that the amount of protein loaded corresponded to approximately the same amount of colony forming units (approximately 2.8x108 CFU) for all lanes. Proteins were separated by SDS-PAGE using a 12% NuPAGE Novex Bis-Tris gel and MOPS running buffer (Invitrogen), and the gel was stained using Coomassie Brilliant Blue R250 (Bio-Rad Laboratories, Hercules, CA). Each gel lane was cut into 10 pieces and proteins therein were reduced by incubating the gel pieces in 10 mM DTT at 56°C for 30 minutes and further alkylated using 55 mM iodoacetamide at room temperature in the dark for 30 minutes. Proteins were digested overnight with 0.1 μg trypsin (Promega, Mannheim, Germany) in 25 mM ammonium bicarbonate at 37 °C as previously described [27]. Before MS analysis (see below), peptides from 3–4 gel pieces were pooled together resulting in three samples per gel lane. The peptides were dried in a speed-vac, solubilized in 0.1% (v/v) trifluoroacetic acid, desalted using C18 ZipTips (Merck Millipore, Darmstadt, Germany) according to the manufacturer’s instructions, dried again, and then analysed by liquid chromatography combined with mass spectrometry (LC-MS/MS) as described below.
NanoLC-Orbitrap MS/MS analysis of tryptic peptides
Dried peptides were dissolved in 1% (v/v) formic acid, 2% (v/v) acetonitrile in water and injected into an Ultimate 3000 nano ultra high performance liquid chromatography system (Dionex, Sunnyvale CA, USA) connected to a Q-Exactive quadrupole-orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany) equipped with a nano-electrospray ion source. Chromatographic separation was done using a nanoViper Acclaim PepMap 100 column (C18, 3μm, 100 Å) (Dionex, Sunnyvale CA, USA) with 50 cm bed length. The flow rate was 300 nL/min and the solvent gradient was 12% B to 43% B in 93 minutes, followed by further increase to 90% B in 6 minutes. Solvent A was 0.1% (v/v) formic acid in water and solvent B was 80% (v/v) acetonitrile, 0.08% (v/v) formic acid in water. The mass spectrometer was operated in data-dependent mode to switch automatically between orbitrap-MS and higher-energy collisional dissociation (HCD) orbitrap-MS/MS acquisition. The resolution was R = 70.000 and R = 35.000 for MS and MS/MS, respectively. For optimal acquisition of MS/MS spectra, automatic gain control (AGC) target values were set to 1.000.000 charges or a maximum injection time of 128 ms was allowed. Data dependent analysis was applied in order to isolate and fragment the 10 most intense ions at any given time throughout the chromatographic elution. These ion precursors were then excluded for further fragmentation for 20 seconds.
Bioinformatics analysis
Raw files were imported into MaxQuant [26] version 1.4.1.2 for identification and label-free quantification (LFQ) of proteins. For protein identification in MaxQuant, the database search engine Andromeda [28] was used to search MS/MS spectra against the UniProt Complete Proteome Set of E. faecalis V583 (3,240 sequences) with a tolerance level of 6 ppm for MS and 20 ppm for MS/MS. Trypsin was used as enzyme and two missed cleavages were allowed. Carbamidomethylation of cysteines was set as a fixed modification and protein N-terminal acetylation, oxidation of methionines, deamidation of asparagines and glutamines and formation of pyro-glutamic acid at N-terminal glutamines were allowed as variable modifications. The ‘match between runs’ feature of MaxQuant, which enables identification transfer between samples based on accurate mass and retention time [25], was applied with a match time window of one minute and an alignment time window of 20 minutes. All identifications were filtered in order to achieve a protein false discovery rate (FDR) of 1%. Proteins were considered as “present” if they were detected by at least two peptides in at least two of the three biological replicates.
Protein quantification was based on the MaxQuant label-free algorithm [25], using both unique and razor peptides for protein quantification; at least 2 ratio counts were required for a protein quantification to be considered valid. Calculations of Benjamini-Hochberg corrections were done using the Perseus package (available at http://www.maxquant.org/). Protein abundance was calculated as a ratio of protein LFQ intensity and the sum of all LFQ intensities (total protein) in the sample, as previously described by Wisniewski and colleagues [29].
To predict secreted proteins we used a combination of three prediction algorithms. The SignalP server [30] version 4.1 (available at http://www.cbs.dtu.dk/services/SignalP/) was used with default settings for gram-positive bacteria to predict signal peptide cleavage sites. LipoP [31] version 1.0 (available at http://www.cbs.dtu.dk/services/LipoP/) was used to predict lipoproteins and TMHMM [32] version 2.0 (available at http://www.cbs.dtu.dk/services/TMHMM/) was used for prediction of transmembrane helices. Proteins predicted by TMHMM to harbour a single transmembrane helix located within the first 60 N-terminal amino acids where assumed to have a signal peptide. A protein was considered secreted if this was predicted by at least two of three prediction algorithms.
Information on the domain structure and potential function of uncharacterized proteins was extracted from Pfam version 27.0 (available at http://pfam.xfam.org/) and InterPro version 47.0 (available at http://www.ebi.ac.uk/interpro/). Operon structures were retrieved from ProOpDB [33] (available at http://operons.ibt.unam.mx/OperonPredictor/).
Results and Discussion
E. faecalis V583 was pre-cultivated in the standard laboratory medium 2xYT and then diluted into either pre-warmed 2xYT or to pre-warmed urine. The cultures were further allowed to incubate for 1.5 hours before harvesting for proteomics analysis (Fig 1); at this point the growth rate started to differ between cultures with and without urine.
Fig 1. Growth curves.
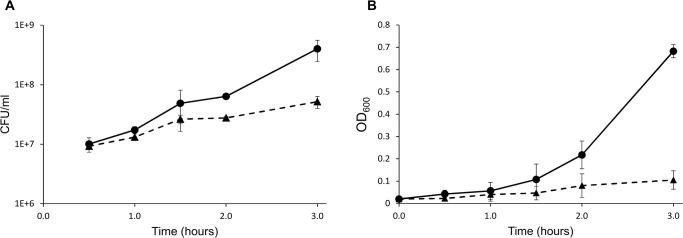
The curves show initial growth of E. faecalis after resuspending cells harvested from an overnight culture in 2xYT to OD600 ~0.02 in fresh medium, either 2xYT (circles on solid lines) or sterile urine (triangles on dashed lines). Growth was monitored by measuring colony forming units per milliliter (CFU/ml) (A) and optical density at 600 nm (OD600) (B). The data points correspond to the mean ± standard deviation of two individual experiments. Samples for proteomic analysis were collected after 1.5 hours.
Quantitative comparison of E. faecalis V583 secretomes
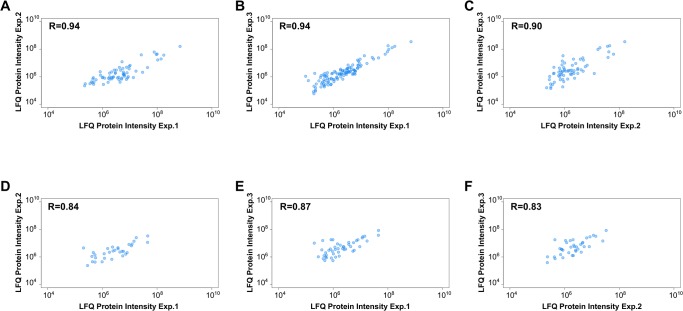
To identify differentially expressed proteins, the secretomes of E. faecalis V583 incubated in control medium (2xYT) or urine were analysed using an MS-based proteomic approach. Precipitated proteins from cell free supernatants were separated by SDS-PAGE and in-gel digested using trypsin. The peptides were analysed by orbitrap LC-MS and identified/quantified using MaxQuant. The relative label-free quantification was highly reproducible between biological replicates, showing Pearson correlations between LFQ intensities varying from 0.83 to 0.94 (Fig 2A–2F), which are values similar to those observed in other LFQ studies [34]. Using a FDR of 1%, and after removal of contaminants, reversed hits and proteins identified with one peptide only, we identified 153 proteins represented by 2.235 peptide sequences. In line with common practice, another filter was applied, requiring a protein to be detected by at least two peptides in at least two of three biological replicates, in order for the detection to be considered significant. This led to a total of 127 proteins, which were distributed over the various samples as shown in Fig 3. The relatively low number of identified proteins reflects the fact that the bacterium was only allowed to grow for 1.5 hours (roughly to an OD600 of 0.1, Fig 1). The fact that fewer proteins were identified in the urine culture may partly be explained by human urine being a complex matrix that may bind proteins and that itself contains proteins in potentially high concentrations [35]. Urine proteins could mask identification of bacterial proteins in the data-dependent acquisition strategy used here, as only the most abundant ions are selected for fragmentation in the mass spectrometer. In addition, urine has been proposed to have limited amounts of iron, manganese and glucose [36] and under these conditions the bacterium may not utilize all pathways functional in a normal environment and thus produce fewer proteins.
Fig 2. Quality of protein quantification.
The figures show the pairwise reproducibility of label-free quantification (LFQ) between the replicates used for secretome quantification for the three 2xYT replicates (A-C) and the three urine replicates (D-F). Pearson correlations are indicated in each graph.
Fig 3. Proteins identified in the secretome of E. faecalis V583 when grown in 2xYT or urine for 1.5 hours.
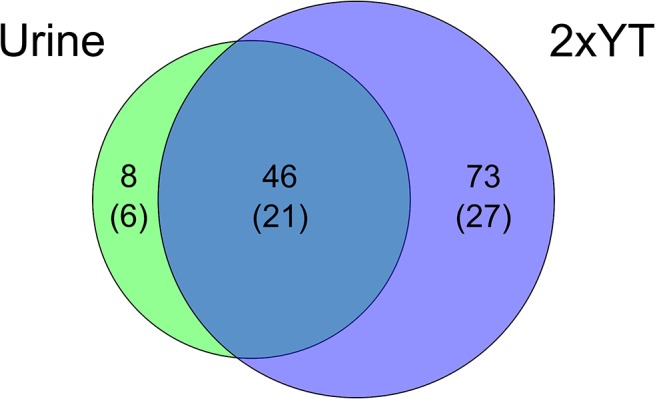
The Venn diagram shows the number of identified proteins in 2xYT, urine and in both conditions. The number of proteins predicted to be secreted is indicated in parenthesis.
The complete genome of V583 codes for 3240 putative proteins (Uniprot complete proteome set) of which 9% (294 proteins) are predicted as secreted when using the combination of SignalP, LipoP and TMHMM, as described in the Methods section. We detected 127 proteins, of which 43% (54 proteins) were predicted to be secreted. Proteomic analysis of the culture supernatant thus led to the expected enrichment of extracellular proteins, although datasets are inevitably polluted with intracellular proteins, as observed in other secretome studies [37, 38]. In total, 18% of all predicted secreted proteins in E. faecalis were detected, which is reasonable considering the results reported in other studies and the fact that one would not expect all predicted extracellular proteins to be secreted at 1.5 hours harvest time.
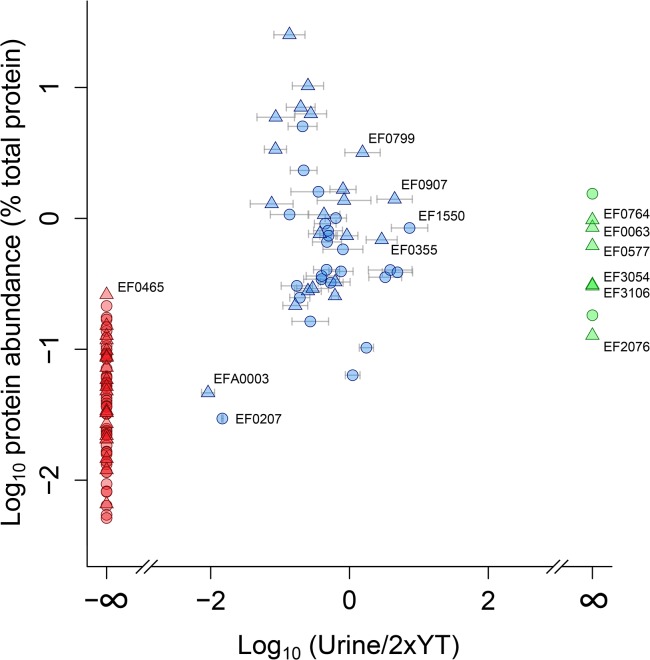
Looking only at the 54 detected proteins that are predicted to be secreted, 6 proteins were found exclusively in the urine culture (Table 1, Fig 4 green points, S1 Table), 27 proteins were exclusively found in the 2xYT culture (Fig 4 red points, S2 Table), and 21 proteins were found in both (Fig 4 blue points, S3 Table). Fig 4 shows that the most abundant proteins were present in both conditions while the least abundant proteins where predominantly those that were only detected in the 2xYT culture.
Table 1. Proteins with a predicted N-terminal signal peptide identified exclusively in the secretome of E. faecalis grown on urine.
| UniProt Accession No. | Gene name | Protein name / Domain structure | Sequence coverage [%] | Protein abundance(% total protein) | Biological process (GO) |
|---|---|---|---|---|---|
| H7C720 | EF0577 | Adhesion lipoprotein | 45.9 | 0.62 | Metal ion transport; cell adhesion |
| Q832Z2 | EF2076 | Endocarditis specific antigen (EfaA) | 9.7 | 0.13 | Metal ion transport; cell adhesion |
| Q839T9 | EF0063 | Pheromone binding protein, putative | 25.6 | 0.85 | Transport |
| Q82ZF4 | EF3106 | Peptide ABC transporter peptide-binding protein | 8.6 | 0.30 | Transport |
| Q837R7 | EF0764 | Uncharacterized protein | 43.1 | 0.97 | |
| Q82ZK1 | EF3054 | Lipoprotein | 15.4 | 0.31 |
Domain structures were identified using Pfam and shown in S1 Table. EF0577 and EF2076 contains TroA domains, which are part of a periplasmic solute binding protein family; part of a family of ABC transporter metal-binding lipoproteins. EF0063 and EF3106 contains SBP_bac_5 domains, which are bacterial extracellular solute-binding proteins, believed to be involved in active transport of solutes across the cytoplasmic membrane. EF3054 contains two PepSY domains, which are thought to have a protease inhibitory function. For EF0764, Pfam does not find any known domains, nor does it annotate a signal peptide as with the other proteins, however both SignalP, LipoP and THMM indicate the presence of such a signal peptide. The functional annotation provided by Gene Ontology (GO) is based on inferences from the domain annotations (InterPro2GO). Additional information on these proteins is available in Table 2 and S1 Table.
Fig 4. Quantitative analysis of proteins identified in the E. faecalis V583 secretome.
The graph shows quantitative analyses of proteins identified exclusively in urine (green), in both urine and 2xYT (blue) and exclusively in 2xYT (red). For finite values, error bars displaying standard deviations of three replicates are shown. The X-axis shows the log ratio of the levels observed in urine and 2xYT (urine/2xYT) and the Y-axis shows protein abundance as a percentage of total protein. Proteins predicted to be secreted are shown as triangles while intracellular proteins are shown as circles. Proteins that are discussed in detail in the text are labeled. All eight proteins reported here as urine specific were identified and quantified with at least two peptides in at least two of three replicates. See S1 Table for details.
Interestingly, while previous transcriptomics studies of enterococcal responses to urine have yielded lists of significantly up-/down-regulated proteins containing up to >700 candidates, the present rigorous proteomic analysis of the immediate response provides a much more limited list of potentially important proteins.
Proteins exclusively detected in the urine cultures
To be able to cause a clinical infection such as UTI, a bacterium needs to colonize the gut and periurethral mucosa and subsequently adhere to the uroepithelium. Indeed, of the six secreted proteins unique to cultivation in urine, two, EF0577 and EF2076, are homologous to known streptococcal adhesins such as SsaB from Streptococcus sanguis [39, 40] and FimA from S. parasanguis [41, 42]. EF0577 is currently annotated as “Adhesion lipoprotein” and is located in the pathogenicity island of V583. EF2076 is annotated as “Endocarditis specific antigen” (EfaA), a well-known virulence factor of E. faecalis originally detected in connection with studies of endocarditis [43, 44]. Using a mouse model system, it has been shown that an EfaA mutant of E. faecalis OG1RF is less virulent than the wild-type strain [44]. Both proteins, which share 60% sequence identity, contain a TroA domain (Pfam accession PF01297) belonging to a family of ABC transporter metal-binding lipoproteins. EF0577 and EF2076 are likely the metal binding surface lipoproteins of ABC transporters encoded by the ef0575-ef0578 and ef2074-ef2076 (efaCBA) operons. The transcriptional regulation of the two proteins is similar, both during incubation in urine (Table 2) and during growth in the presence of excess zinc or excess manganese [45].
Table 2. Previously observed regulation of proteins identified exclusively in the secretome of E. faecalis grown on urine.
| UniProt Accession No. (Gene name) | Previously observed regulatory effects | ||||||
|---|---|---|---|---|---|---|---|
| Urine[1, 6] | Serum[1] | Bile[10] | SDS[10] | Bile+SDS[10] | Blood[11] | Erythromycin[9] | |
| H7C720 (EF0577) | M: 10.6 | V: 16.0 | |||||
| M: 17.1 | |||||||
| Q832Z2 (EF2076) | M: 4.9 | M: 66 (qPCR) | V:3.7 | V: -1.7 | |||
| M: 7.5 | M: 5 (qPCR) | V: -2.1 | |||||
| M: 89 (qPCR) | V: -2.8 | ||||||
| M: 2195 (qPCR) | |||||||
| O: 7.0 | |||||||
| O: 10.6 | |||||||
| S: 8.0 | |||||||
| S: 9.2 | |||||||
| Q839T9 (EF0063) | M: 4.0 | V: 5.7 | |||||
| M: 8.6 | |||||||
| O: 26.0 | |||||||
| O: 16.0 | |||||||
| S: 5.7 | |||||||
| Q82ZF4 (EF3106) | M: 1.5 | V: 68.6 | V: -3.7 | ||||
| M: 34.3 | V: -2.1 | ||||||
| O: 1.7 | |||||||
| O: 10.6 | |||||||
| Q837R7 | V: -3.7 | V: 6.1 | |||||
| Q82ZK1 | M: 1.3 | ||||||
The table summarizes results from previous studies on enterococcal responses involving the proteins listed in Table 1. All data are derived from transcriptomics studies, except if stated otherwise. Only significant changes are included and the numbers are recalculated as fold change; positive numbers indicate up-regulation, negative numbers indicate down-regulation. In addition to the data shown here, regulation data exist for the effects of bivalent metal ions, showing up-regulation (Zn2+) or down-regulation (Cu2+, Mn2+) of EF0577 and EF2076 only [45], and NaCl, showing up-regulation of EF2076 [68]. Blank cells indicate lack of data. The strains used were V583 (V), MMH594 (M), OG1RF (O), and Symbiflor 1 (S). Additional q-PCR-based transcriptome studies, which are cited in the main text [12, 14], did not include quantitative data and are therefore not included in the Table. The results of these studies indicated that of the six proteins in the Table, EF2076 was up-regulated for the food strain LA160 during growth on urine. The other five of these six proteins were not tested/found in these studies.
While homologies with streptococcal adhesion proteins suggest roles of EF0577 and EF2076 in enterococcal adhesion, available experimental evidence for the functionality of these proteins is limited to their role in metal, in particular manganese, transport. Both the efaCBA operon and the ef0575-0578 operon are regulated by the transcriptional regulator (EfaR), in a manganese-dependent manner [46, 47]. In a recent study by Abrantes et al., it was shown that EfaR is a major transcriptional regulator of manganese transporters in E. faecalis and that its mutations reduces the ability of E. faecalis to form biofilms, survive inside macrophages and tolerate oxidative stress, all properties associated with host colonization and infection [47]. The observed down-regulation of the ef0577 and ef2076 genes in response to excess manganese [45] further confirms their role in manganese transport, and could also explain the increased production of EF0577 and EF2076 in urine, where metal concentrations are low and high-affinity transporters thus are required for survival [48]. Interestingly, while the EfaR insertion mutant showed clearly weakened virulence traits, similar effects were not observed for insertion mutants of EF2076 (EfaA) or EF0577 [47], suggesting that the two proteins have overlapping functions. Combined with existing data, the present proteomic data clearly show that the EF0577 and EF2076 (EfaA) are of major importance for enterococcal interactions with its host, including in a urine environment. Further work to verify, characterize and analyse the functional implications of the predicted adhesin function of EF0577 and EF2076 is clearly needed.
In addition to metal ABC transporters, amino acid, peptide and amine ABC transporters are also believed to be linked to bacterial virulence [49]. We identified a lipoprotein (EF3106) annotated as a peptide-binding protein with an extracellular solute-binding domain (Pfam accession PF00496) covering 69% of the protein. EF3106 is part of an ABC transporter involved in peptide transport (the last in an operon of five genes) and the transcription of the ef3106 is known to be up-regulated during growth in urine and blood (Table 2). Another lipoprotein found exclusively in the urine fraction is EF0063, harboring the same domain structure as EF3106, with the extracellular solute-binding domain covering 70% of the protein. However, although EF0063 possess similar domain structure as EF3106, its genetic context is very different. The ef0063 gene is not located in an operon with other ABC transporter components. Interestingly CodY regulatory motifs are located upstream of ef0063 in E. faecalis [50], meaning that expression of the gene might be regulated by CodY (EF1645 in V583), a protein known to be involved in adapting bacteria to stress and starvation and recognized as a global regulator of virulence-associated properties in several Gram-positive bacteria [51].
Lipoproteins in general are involved in many important cellular processes such as substrate-binding and-presenting to ABC transporters, folding of excreted proteins, pheromone production, and antibiotic resistance, as reviewed by Reffuveille et al. [49]. Another lipoprotein identified in the urine cultures only, EF3054, has no gene ontology annotation, but Pfam identifies two PepSY domains together covering 57% of the protein. PepSY domains comprise 60–90 amino acids and proteins harbouring PepSY domains typically contain a signal peptide or a transmembrane helix that would direct the proteins to the exterior of the cell wall [52]. Yeats et al. have shown that PepSY domains serve as inhibitors of members of the M4 family of metallopeptidases [52] and the ability of PepSY domains to regulate protease activity could be linked to pathogenicity [52]. The fact that we find this lipoprotein exclusively after incubation in urine suggests that regulation of protease activity plays a role in the adaptability or pathogenicity of V583. Transcriptomics studies also indicated up-regulation of this protein in response to a urine environment, although the effects were small (up-regulated by 1.3 fold, Table 2).
The sixth secreted protein, EF0764, is a protein of unknown function, giving no hits in Pfam. The protein contains a signal peptide cleavage site and its identification in the supernatants of urine-incubated cells is evident, with six different unique peptides being detected giving 43% combined sequence coverage. Two of the same peptides were also found in one of the three replicates of the 2xYT culture (S1 Table), but in this case, peptide intensities were 14 times lower when compared to the peptides detected in samples from the urine culture. To our knowledge, regulation of this uncharacterized protein in response to a urine environment has not been reported before, but expression of its gene was found to be affected by bile (down, 3.7-fold) and SDS (up, 6.1-fold) (Table 2). Interestingly, EF0764 seems to be unique to E. faecalis, indicating that it could be exploited as a marker. Analysis of the genomic context and synteny using The SEED [53] (Fig 5) showed that ef0764 is located upstream (separated by 72 nucleotides) of another small gene, ef0765, coding for a hypothetical protein of 55 amino acids belonging to the functionally uncharacterized YusW-like protein family, however, the two genes do not appear to be part of an operon structure. Two genes upstream on the same strand, uvrA and uvrB, are both important players in DNA repair [54]. Downstream on the same strand, there is a predicted gene cluster that commonly co-occurs with DNA repair genes. This cluster potentially contains a predicted P-loop-containing kinase (ATPase), a CofD-like protein that may be a 2-phospho-L-lactate transferase [48] and a cytoplasmic hypothetical protein belonging to COG1481, an uncharacterized protein domain. This conserved gene cluster has an operon structure, but its function is not known, nor why it co-occurs with DNA repair genes. DNA damage can result from oxidative stress, and both proteins (UvrA and UvrB) involved in DNA repair mechanisms and antioxidant defense systems are known virulence factors in E. faecalis [55, 56]. Perhaps, the hitherto uncharacterized EF0764 protein is involved in DNA repair or protection from oxidative stress imposed by the host. More targeted experiments on this protein need to be conducted to elucidate its potential function in E. faecalis adaptation and pathogenicity.
Fig 5. Gene context analysis.
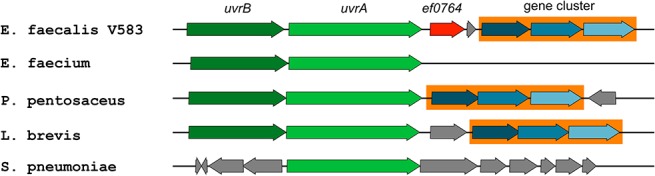
The figure shows the gene context around ef0764 (indicated in red) as well as (potentially) related gene clusters in other gram-positive bacteria. Two genes upstream of ef0764, uvrA and uvrB (green), are involved in DNA repair and the downstream gene cluster of unknown function (orange box) commonly co-occurs with these known DNA repair genes. This cluster potentially encodes a predicted P-loop-containing kinase, a CofD-like protein and a cytoplasmic hypothetical protein. Genes in grey are genes that do not show similarity to other genes in this specific region of the genomes shown; while some have annotated functions, their genomic location is not conserved, nor associated with the location of uvrA and uvrB. The ef0764 gene shows no similarity to the gene located in a similar position in L. brevis, and BLAST analysis could not identify any homologues in any other species.
Proteins exclusively detected in the 2xYT cultures
Most proteins identified exclusively in the 2xYT medium (Fig 4 red points; S2 Table) were present in low amounts and not obviously of interest in relation to pathogenicity and survival. The most abundant secreted protein only found in the 2xYT culture is EF0465, which is a transcriptional regulator belonging to the LytR family. In accordance with this observation, transcriptomics studies by Vebø et al. [6] showed that transcription of ef0465 was down-regulated 1.7–1.9 fold during growth in urine, for all three E. faecalis strains tested, MMH594, OG1RF and Symbiflor.
Proteins detected in both urine and the 2xYT cultures
Proteins occurring in the supernatants of both cultures are quantitatively visualized in Fig 4 (see full list in S3 Table) and include secreted and intracellular proteins present in intermediate to high amounts. The majority of these proteins are hydrolases and many have cell wall/membrane associated functions such as peptidoglycan degradation (S3 Table).
Significance B calculations in Perseus using a Benjamini-Hochberg correction at 5% reveals two proteins standing out as significantly down-regulated in urine. One of these is secreted TraC (EFA0003; traC-1), with a suggested role in pheromone-binding [57], while the other is a ribosomal protein with a predicted intracellular location (EF0207; rplD). The significance of these two identifications is questionable, because imputed intensities had to be used for calculating ratios, due to the low abundance in the urine samples (see S3 Table for details).
The protein with the highest degree of up-regulation on urine was EF1550, a histone-like DNA binding protein, which shares 82% homology with the streptococcal histone-like protein HlpA. Interestingly, in streptococci, HlpA, which is predicted to be cytoplasmic, has been detected on the cell surface, where it forms complexes with lipoteichoid acid (LTA) and mediates bacterial adherence by also interacting with heparan sulfate proteoglycans of host cells [58, 59]. Binding of HlpA-LTA complexes to host cells is known to stimulate production of pro-inflammatory cytokines in cultured murine macrophages, suggesting a role of HlpA in promoting tissue damage at infection sites [60]. To our knowledge, regulation of EF1550 in response to a urine environment has not been reported before.
Three secreted proteins were up-regulated in urine, EF0799 (atlA; Autolysin), EF0355 (atlB; Peptidoglycan hydrolase / Endolysin) and EF0907 (Peptide ABC transporter peptide-binding protein). EF0907 harbours an extracellular solute-binding domain (Pfam accession PF00496) similar to EF0063 and EF3106 (discussed above, see Table 1) and is involved in transport of peptides. The primary function of EF0799 is thought to be peptidoglycan degradation, in particular during digestion of the septum after cell division, and this protein has been linked to enterococcal biofilm formation [61] and antibiotic sensitivity [62]. EF0355 has a similar activity and mutation studies have shown that it can act as a surrogate for EF0799 [63]. A third hydrolytic enzyme related to EF0799 and EF0355, EF1992 (atlC), was not detected in this study.
Concluding remarks
In this proteomics study, we have compared the secretome of E. faecalis during the initial phase of cultivation in urine with the secretome during cultivation in standard laboratory medium, 2xYT. This led to the identification of extracellular proteins and a histone-like “moonlighting” protein, which are all putatively involved in early phases of infection processes in the urinary tract. It must be emphasized that urine is a poor nutrient source [1] and that during development of an UTI, the bacteria are likely to feed on glycoproteins of the UT epithelium. This source of additional energy was not present in our experiments. Thus, while our study reveals interesting proteins likely to be directly or indirectly involved in UTI development, these proteins do not tell the full story.
Known E. faecalis virulence factors include gelatinase (GelE) and serine protease (SprE), two proteins regulated via the Fsr two-component signal transduction system that mediates quorum sensing-based responses [64]. Shankar et al. [65] have shown that the secretomes of Fsr-positive and Fsr-negative strains of E. faecalis grown on BHI medium show distinct differences, beyond the mere presence of GelE and SprE in the Fsr-positive strains. In our study, using an Fsr-positive strain [66], GelE or SprE were detected neither in the urine nor in the 2xYT secretome, indicating that quorum activation had not occurred in any of the two conditions. We detected 17 of the 34 unique proteins detected by Shankar et al. in the Fsr-negative strain (one on urine only, the cytoplasmic EF1167, 7 on 2xYT only and 9 in both). In contrast, we detected only three of the 25 unique proteins detected by Shankar et al. in the Fsr-positive strain (two on 2xYT only, and one on both). Thus, the conditions used in this study may seem to mimic an Fsr-null mutant.
The proteins detected include a known virulence factor, EfaA, and a homologue, EF0577. These two proteins, as well as two of the other detected urine-specific proteins are involved in solute transport, which conceivably is important for survival in the urine environment. Adhesion seems to be important too, since EfaA and EF0577 have putative roles in this respect, whereas streptococcal HlpA, a homologue of up-regulated EF1550, is a known adhesion factor. The detection of EF0764 is of interest because this protein is highly specific for E. faecalis, has a completely unknown function, and had previously not been associated with the urine response, whereas current data show a clear association. The fact that the transcript encoding this protein was not detected in a comprehensive transcriptome analysis of E. faecalis cultivated in urine is remarkable and may indicate unusual regulation justifying further investigations. The rigorous proteomics approach applied here illustrates the power of label-free quantification in combination with bacterial culturing in human urine as a method for finding candidates potentially responsible for UTI by E. faecalis.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [67] with the dataset identifier PXD001345.
The authors would like to thank Helge Holo (Norwegian University of Life Sciences) for the E. faecalis strain used in this study, Jeremy Frank (Norwegian University of Life Sciences) for discussions on gene context analysis and the PRIDE team for support in uploading the proteomics data to ProteomeXchange.
Data Availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001345. [For review, please use this URL: http://www.ebi.ac.uk/pride/archive/login and this login information: Username: reviewer46762@ebi.ac.uk Password: P41ZM2vR]
Funding Statement
This work was funded by the FUGE programme of the Norwegian Research Council, grant numbers 183627 & 183637. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shepard BD, Gilmore MS. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect Immun. 2002;70(8):4344–52. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155(Pt 6):1749–57. 10.1099/mic.0.026385-0 PubMed . [DOI] [PubMed] [Google Scholar]
- 3. Murray BE. Diversity among multidrug-resistant enterococci. Emerging infectious diseases. 1998;4(1):37–47. 10.3201/eid0401.980106 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guzman CA, Pruzzo C, LiPira G, Calegari L. Role of adherence in pathogenesis of Enterococcus faecalis urinary tract infection and endocarditis. Infect Immun. 1989;57(6):1834–8. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Horsley H, Malone-Lee J, Holland D, Tuz M, Hibbert A, Kelsey M, et al. Enterococcus faecalis subverts and invades the host urothelium in patients with chronic urinary tract infection. PLoS One. 2013;8(12):e83637 10.1371/journal.pone.0083637 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vebo HC, Solheim M, Snipen L, Nes IF, Brede DA. Comparative genomic analysis of pathogenic and probiotic Enterococcus faecalis isolates, and their transcriptional responses to growth in human urine. PLoS One. 2010;5(8):e12489 10.1371/journal.pone.0012489 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, et al. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis . Antimicrob Agents Chemother. 1989;33(9):1588–91. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis . Science. 2003;299(5615):2071–4. 10.1126/science.1080613 PubMed . [DOI] [PubMed] [Google Scholar]
- 9. Aakra A, Vebo H, Snipen L, Hirt H, Aastveit A, Kapur V, et al. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob Agents Chemother. 2005;49(6):2246–59. 10.1128/AAC.49.6.2246-2259.2005 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Solheim M, Aakra A, Vebo H, Snipen L, Nes IF. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl Environ Microbiol. 2007;73(18):5767–74. 10.1128/AEM.00651-07 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vebo HC, Snipen L, Nes IF, Brede DA. The transcriptome of the nosocomial pathogen Enterococcus faecalis V583 reveals adaptive responses to growth in blood. PLoS One. 2009;4(11):e7660 10.1371/journal.pone.0007660 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanin A, Sava I, Bao Y, Huebner J, Hartke A, Auffray Y, et al. Screening of in vivo activated genes in Enterococcus faecalis during insect and mouse infections and growth in urine. PLoS One. 2010;5(7):e11879 10.1371/journal.pone.0011879 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abranches J, Tijerina P, Aviles-Reyes A, Gaca AO, Kajfasz JK, Lemos JA. The cell wall-targeting antibiotic stimulon of Enterococcus faecalis . PLoS One. 2013;8(6):e64875 10.1371/journal.pone.0064875 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carlos AR, Semedo-Lemsaddek T, Barreto-Crespo MT, Tenreiro R. Transcriptional analysis of virulence-related genes in enterococci from distinct origins. Journal of applied microbiology. 2010;108(5):1563–75. 10.1111/j.1365-2672.2009.04551.x PubMed . [DOI] [PubMed] [Google Scholar]
- 15. Benachour A, Morin T, Hebert L, Budin-Verneuil A, Le Jeune A, Auffray Y, et al. Identification of secreted and surface proteins from Enterococcus faecalis . Can J Microbiol. 2009;55(8):967–74. 10.1139/w09-052 PubMed . [DOI] [PubMed] [Google Scholar]
- 16. Bohle LA, Riaz T, Egge-Jacobsen W, Skaugen M, Busk OL, Eijsink VG, et al. Identification of surface proteins in Enterococcus faecalis V583. BMC genomics. 2011;12:135 10.1186/1471-2164-12-135 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pessione A, Lamberti C, Cocolin L, Campolongo S, Grunau A, Giubergia S, et al. Different protein expression profiles in cheese and clinical isolates of Enterococcus faecalis revealed by proteomic analysis. Proteomics. 2012;12(3):431–47. 10.1002/pmic.201100468 PubMed . [DOI] [PubMed] [Google Scholar]
- 18. Maddalo G, Chovanec P, Stenberg-Bruzell F, Nielsen HV, Jensen-Seaman MI, Ilag LL, et al. A reference map of the membrane proteome of Enterococcus faecalis . Proteomics. 2011;11(19):3935–41. 10.1002/pmic.201100103 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heim S, Lleo M, Bonato B, Guzman CA, Canepari P. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J Bacteriol. 2002;184(23):6739–45. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bohle LA, Faergestad EM, Veiseth-Kent E, Steinmoen H, Nes IF, Eijsink VG, et al. Identification of proteins related to the stress response in Enterococcus faecalis V583 caused by bovine bile. Proteome science. 2010;8:37 10.1186/1477-5956-8-37 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, He X, Jiang Z, Wang J, Chen X, Liu D, et al. Proteomic analysis of the Enterococcus faecalis V583 strain and clinical isolate V309 under vancomycin treatment. J Proteome Res. 2010;9(4):1772–85. 10.1021/pr901216e PubMed . [DOI] [PubMed] [Google Scholar]
- 22. Eirich J, Orth R, Sieber SA. Unraveling the protein targets of vancomycin in living S. aureus and E. faecalis cells. J Am Chem Soc. 2011;133(31):12144–53. 10.1021/ja2039979 PubMed . [DOI] [PubMed] [Google Scholar]
- 23. Giard JC, Laplace JM, Rince A, Pichereau V, Benachour A, Leboeuf C, et al. The stress proteome of Enterococcus faecalis . Electrophoresis. 2001;22(14):2947–54. PubMed . [DOI] [PubMed] [Google Scholar]
- 24. Lindenstrauss AG, Behr J, Ehrmann MA, Haller D, Vogel RF. Identification of fitness determinants in Enterococcus faecalis by differential proteomics. Arch Microbiol. 2013;195(2):121–30. 10.1007/s00203-012-0857-3 PubMed . [DOI] [PubMed] [Google Scholar]
- 25.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics. 2014. 10.1074/mcp.M113.031591 PubMed . [DOI] [PMC free article] [PubMed]
- 26. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. Epub 2008/11/26. nbt.1511 [pii] 10.1038/nbt.1511 PubMed . [DOI] [PubMed] [Google Scholar]
- 27. Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–60. 10.1038/nprot.2006.468 PubMed . [DOI] [PubMed] [Google Scholar]
- 28. Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–805. 10.1021/pr101065j PubMed . [DOI] [PubMed] [Google Scholar]
- 29. Wisniewski JR, Ostasiewicz P, Dus K, Zielinska DF, Gnad F, Mann M. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol Syst Biol. 2012;8:611 10.1038/msb.2012.44 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6. 10.1038/nmeth.1701 PubMed . [DOI] [PubMed] [Google Scholar]
- 31. Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12(8):1652–62. 10.1110/ps.0303703 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings / International Conference on Intelligent Systems for Molecular Biology; ISMB International Conference on Intelligent Systems for Molecular Biology. 1998;6:175–82. PubMed . [PubMed] [Google Scholar]
- 33. Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. ProOpDB: Prokaryotic Operon DataBase. Nucleic Acids Res. 2012;40(Database issue):D627–31. 10.1093/nar/gkr1020 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32(2):279–89. 10.1016/j.immuni.2010.01.013 PubMed . [DOI] [PubMed] [Google Scholar]
- 35. Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80 10.1186/gb-2006-7-9-R80 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DebRoy S, Gao P, Garsin DA, Harvey BR, Kos V, Nes IF, et al. Transcriptional and Post Transcriptional Control of Enterococcal Gene Regulation. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston2014.
- 37. Huang CJ, Damasceno LM, Anderson KA, Zhang S, Old LJ, Batt CA. A proteomic analysis of the Pichia pastoris secretome in methanol-induced cultures. Appl Microbiol Biotechnol. 2011;90(1):235–47. 10.1007/s00253-011-3118-5 PubMed . [DOI] [PubMed] [Google Scholar]
- 38. Komeil D, Padilla-Reynaud R, Lerat S, Simao-Beaunoir AM, Beaulieu C. Comparative secretome analysis of Streptomyces scabiei during growth in the presence or absence of potato suberin. Proteome science. 2014;12:35 10.1186/1477-5956-12-35 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ganeshkumar N, Song M, McBride BC. Cloning of a Streptococcus sanguis adhesin which mediates binding to saliva-coated hydroxyapatite. Infect Immun. 1988;56(5):1150–7. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ganeshkumar N, Hannam PM, Kolenbrander PE, McBride BC. Nucleotide sequence of a gene coding for a saliva-binding protein (SsaB) from Streptococcus sanguis 12 and possible role of the protein in coaggregation with actinomyces. Infect Immun. 1991;59(3):1093–9. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fachon-Kalweit S, Elder BL, Fives-Taylor P. Antibodies that bind to fimbriae block adhesion of Streptococcus sanguis to saliva-coated hydroxyapatite. Infect Immun. 1985;48(3):617–24. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fenno JC, LeBlanc DJ, Fives-Taylor P. Nucleotide sequence analysis of a type 1 fimbrial gene of Streptococcus sanguis FW213. Infect Immun. 1989;57(11):3527–33. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lowe AM, Lambert PA, Smith AW. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63(2):703–6. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh KV, Coque TM, Weinstock GM, Murray BE. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol Med Microbiol. 1998;21(4):323–31. PubMed . [DOI] [PubMed] [Google Scholar]
- 45. Abrantes MC, Lopes Mde F, Kok J. Impact of manganese, copper and zinc ions on the transcriptome of the nosocomial pathogen Enterococcus faecalis V583. PLoS One. 2011;6(10):e26519 10.1371/journal.pone.0026519 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Low YL, Jakubovics NS, Flatman JC, Jenkinson HF, Smith AW. Manganese-dependent regulation of the endocarditis-associated virulence factor EfaA of Enterococcus faecalis . J Med Microbiol. 2003;52(Pt 2):113–9. PubMed . [DOI] [PubMed] [Google Scholar]
- 47. Abrantes MC, Kok J, Lopes Mde F. EfaR is a major regulator of Enterococcus faecalis manganese transporters and influences processes involved in host colonization and infection. Infect Immun. 2013;81(3):935–44. 10.1128/IAI.06377-11 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kovacs-Simon A, Titball RW, Michell SL. Lipoproteins of bacterial pathogens. Infect Immun. 2011;79(2):548–61. 10.1128/IAI.00682-10 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reffuveille F, Leneveu C, Chevalier S, Auffray Y, Rince A. Lipoproteins of Enterococcus faecalis: bioinformatic identification, expression analysis and relation to virulence. Microbiology. 2011;157(Pt 11):3001–13. 10.1099/mic.0.053314-0 PubMed . [DOI] [PubMed] [Google Scholar]
- 50. Guedon E, Sperandio B, Pons N, Ehrlich SD, Renault P. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes . Microbiology. 2005;151(Pt 12):3895–909. 10.1099/mic.0.28186-0 PubMed . [DOI] [PubMed] [Google Scholar]
- 51. Stenz L, Francois P, Whiteson K, Wolz C, Linder P, Schrenzel J. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol Med Microbiol. 2011;62(2):123–39. 10.1111/j.1574-695X.2011.00812.x PubMed . [DOI] [PubMed] [Google Scholar]
- 52. Yeats C, Rawlings ND, Bateman A. The PepSY domain: a regulator of peptidase activity in the microbial environment? Trends Biochem Sci. 2004;29(4):169–72. 10.1016/j.tibs.2004.02.004 PubMed . [DOI] [PubMed] [Google Scholar]
- 53. Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–702. 10.1093/nar/gki866 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pakotiprapha D, Samuels M, Shen K, Hu JH, Jeruzalmi D. Structure and mechanism of the UvrA-UvrB DNA damage sensor. Nat Struct Mol Biol. 2012;19(3):291–8. 10.1038/nsmb.2240 PubMed . [DOI] [PubMed] [Google Scholar]
- 55. La Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, et al. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis . Mol Microbiol. 2007;66(5):1148–63. 10.1111/j.1365-2958.2007.05987.x PubMed . [DOI] [PubMed] [Google Scholar]
- 56. Peppoloni S, Posteraro B, Colombari B, Manca L, Hartke A, Giard JC, et al. Role of the (Mn)superoxide dismutase of Enterococcus faecalis in the in vitro interaction with microglia. Microbiology. 2011;157(Pt 6):1816–22. 10.1099/mic.0.047381-0 PubMed . [DOI] [PubMed] [Google Scholar]
- 57. Ruhfel RE, Manias DA, Dunny GM. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J Bacteriol. 1993;175(16):5253–9. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stinson MW, McLaughlin R, Choi SH, Juarez ZE, Barnard J. Streptococcal histone-like protein: primary structure of hlpA and protein binding to lipoteichoic acid and epithelial cells. Infect Immun. 1998;66(1):259–65. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boleij A, Schaeps RM, de Kleijn S, Hermans PW, Glaser P, Pancholi V, et al. Surface-exposed histone-like protein a modulates adherence of Streptococcus gallolyticus to colon adenocarcinoma cells. Infect Immun. 2009;77(12):5519–27. 10.1128/IAI.00384-09 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang L, Ignatowski TA, Spengler RN, Noble B, Stinson MW. Streptococcal histone induces murine macrophages To produce interleukin-1 and tumor necrosis factor alpha. Infect Immun. 1999;67(12):6473–7. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, et al. Contribution of autolysin and Sortase a during Enterococcus faecalis DNA-dependent biofilm development. Infect Immun. 2009;77(9):3626–38. 10.1128/IAI.00219-09 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bravetti AL, Mesnage S, Lefort A, Chau F, Eckert C, Garry L, et al. Contribution of the autolysin AtlA to the bactericidal activity of amoxicillin against Enterococcus faecalis JH2-2. Antimicrob Agents Chemother. 2009;53(4):1667–9. 10.1128/AAC.00692-08 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mesnage S, Chau F, Dubost L, Arthur M. Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J Biol Chem. 2008;283(28):19845–53. 10.1074/jbc.M802323200 PubMed . [DOI] [PubMed] [Google Scholar]
- 64. Qin X, Singh KV, Weinstock GM, Murray BE. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol. 2001;183(11):3372–82. 10.1128/JB.183.11.3372-3382.2001 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shankar J, Walker RG, Ward D, Horsburgh MJ. The Enterococcus faecalis exoproteome: identification and temporal regulation by Fsr. PLoS One. 2012;7(3):e33450 10.1371/journal.pone.0033450 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dundar H, Brede DA, La Rosa SL, El-Gendy AO, Diep DB, Nes IF. Fsr quorum sensing system and cognate Gelatinase orchestrate the expression and processing of pro-protein EF1097 into mature antimicrobial peptide enterocin O16. J Bacteriol. 2015. 10.1128/JB.02513-14 PubMed . [DOI] [PMC free article] [PubMed]
- 67. Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41(Database issue):D1063–9. 10.1093/nar/gks1262 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Solheim M, La Rosa SL, Mathisen T, Snipen LG, Nes IF, Brede DA. Transcriptomic and functional analysis of NaCl-induced stress in Enterococcus faecalis . PLoS One. 2014;9(4):e94571 10.1371/journal.pone.0094571 PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001345. [For review, please use this URL: http://www.ebi.ac.uk/pride/archive/login and this login information: Username: reviewer46762@ebi.ac.uk Password: P41ZM2vR]