Abstract
Background
TNF-α and IFN-γ play a role in the development of mucosal damage in celiac disease (CD). Polymorphisms of TNFA and IFNG genes, as well as of the TNFRSF1A gene, encoding the TNF-α receptor 1, might underlie different inter-individual disease susceptibility over a common HLA risk background. The aims of this study were to ascertain whether five SNPs in the TNFA promoter (-1031T>C,-857C>T,-376G>A,-308G>A,-238G>A), sequence variants of the TNFRSF1A gene and IFNG +874A>T polymorphism are associated with CD in a HLA independent manner.
Methods
511 children (244 CD, 267 controls) were genotyped for HLA, TNFA and INFG (Real Time PCR). TNFRSF1A variants were studied (DHPLC and sequence).
Results
Only the rare TNFA-1031C (OR=0.65, 95% CI:0.44-0.95), -857T (OR=0.42, 95% CI:0.27-0.65), -376A (OR=2.25, 95% CI:1.12-4.51) and -308A (OR=4.76, 95% CI:3.12-7.26) alleles were significantly associated with CD. One TNFRSF1A variant was identified (c.625+10A>G, rs1800693), but not associated with CD. The CD-correlated TNFA SNPs resulted in six haplotypes. Two haplotypes were control-associated (CCGG and TTGG) and three were CD-associated (CCAG, TCGA and CCGA). The seventeen inferred haplotype combinations were grouped (A to E) based on their frequencies among CD. Binary logistic regression analysis documented a strong association between CD and HLA (OR for intermediate risk haplotypes=178; 95% CI:24-1317; OR for high risk haplotypes=2752; 95% CI:287-26387), but also an HLA-independent correlation between CD and TNFA haplotype combination groups. The CD risk for patients carrying an intermediate risk HLA haplotype could be sub-stratified by TNFA haplotype combinations.
Conclusion
TNFA promoter haplotypes associate with CD independently from HLA. We suggest that their evaluation might enhance the accuracy in estimating the CD genetic risk.
Introduction
Celiac disease (CD), an autoimmune enteropathy, develops in genetically predisposed individuals after the exposure to gluten-derived peptides [1,2]. CD patients make antibodies specific for gluten peptides, but also for autoantigens, tissue transglutaminase (tTG) being a highly specific autoantigen, and develop enterocytes destruction by CD8+ T cells [2]. Moreover, numerous innate and adaptive immune response alterations occur in both the intestinal epithelial layer and the lamina propria, resulting in the infiltration of leukocytes, the activation of dendritic cells and CD4+ T cells with an increased expression, and the release of IL-15, interferon (IFN)-γ and possibly IL-21 [3]. In the mucosa of CD patients, macrophages are also activated, these cells being the main producers of the tumor necrosis factor (TNF)-α, another relevant cytokine in CD pathogenesis [4–7].
The maintenance of the immune alterations seen in CD intestinal mucosa is strictly dependent on gluten ingestion, which was demonstrated to increase TNF-α, not IFN-γ expression in CD patients, and to increase IFN-γ, not TNF-α in “non-celiac gluten sensitivity” patients [7]. Gluten derived peptides, which undergo enzymatic deamidation by tTG, are transformed into deamidated gluten derived peptides, which are preferentially recognized by lamina propria CD4+ T cells in the presence of disease-associated HLA molecules [2]. The HLA locus, the most relevant genetic factor for CD, accounts for 40 to 50% of the genetic variance occurring in patients with the disease. The HLA-DQ2 haplotype (DQA1*05/DQB1*02, also known as DQ2.5) is expressed in 90% of CD cases, but also in one third of the general population, while HLA-DQ8 haplotype (DQA1*03/DQB1*0302) or another variant of HLA-DQ2 (DQA1*02/DQB1*02, also known as DQ2.2) is expressed in about 5%. The influence of HLA on CD susceptibility shows a dose effect. Individuals can be classified as being at a high or intermediate risk of CD on the basis of the HLA-DQ haplotype and on the number of DQB1*02 carrying alleles [8–11].
Several non-HLA genes, mainly derived from GWAS, have recently been identified as new susceptibility factors for CD. So far, 39 loci with 57 independent association signals have been described and overall they have been estimated to account for about 14% of the genetic variance of the disease [2]. Many of the newly discovered CD associated loci harbor genes that are related to the immune response, particularly B and T cell function [12–14] and some polymorphisms that have emerged from GWAS are indirectly related to the physiology of IFN-γ and/or TNF-α [13,15]. IFN-γ and TNF-α production might be dependent also on IFNG and TNFA genes polymorphisms, and in particular on IFNG +874A>T and TNFA -308G>A which have a functional significance [16–18]. Few and contrastive studies are available in literature focusing on IFNG +874A>T polymorphism in CD [19,20], while some studies support the existence of an association between CD and the TNFA -308 A allele [21–25]. Notably, it has been observed that this latter association is independent of the HLA-DQ2 haplotype despite the linkage between TNFΑ gene and HLA class II genes, both located in close proximity on the human chromosome 6p21.3 region [26]. Furthermore, although the TNFA -308 A allele is reported to be correlated with a higher transcription than the G allele [26], this finding might depend solely on the fact that the allele belongs to specific extended TNFA haplotypes [27].
TNF-α acts by binding to TNF-α receptors, mainly TNFΑ-R1 and TNFΑ-R2. Mutations in the TNFΑ-R1 extracellular domain are associated with the auto-inflammatory TNF receptor-associated periodic syndrome (TRAPS) but may also be involved in CD, which is included in the autoinflammatory-autoimmune continuum [28]. The hypothesis that autoinflammation may play a pathogenic role in CD is supported by the recent findings of Palova-Jelinkova et al. [29], who demonstrated that the breakdown of wheat gliadin by pepsin induces a robust IL-1β and IL-1α secretion by peripheral blood mononuclear cells through the activation of innate immune pathways, including the NLRP3 inflammasome. To our knowledge, no study in the literature has investigated whether sequence variations of the TNF-α receptor gene TNFRSF1A are associated with CD.
We hypothesized that a combined analysis of TNFA promoter haplotypes, TNFRSF1A sequence and of IFNG +874A>T polymorphism will allow to identify HLA-DQ2/DQ8 independent genetic risk factors for CD. To verify this hypothesis, we thought to study the IFNG +874A>T and five polymorphisms of the TNFA promoter (-1031T>C, -857C>T, -376G>A, -308G>A, -238G>A) and to inspect for variants of the TNFRSF1A sequence in a large retrospective cohort of CD and control children.
Materials and Methods
Ethics statement
The parents of all the children gave their fully informed written consent to the study for their offspring. The study was approved by the Local Institutional Ethic Committee (“Comitato Etico per la Sperimentazione, Azienda Ospedaliera di Padova”, ce.sperimentazione@sanita.padova.it).
Patients
In the present retrospective cohort study, we investigated a total of 511 young patients coming from the North East of Italy (Table 1): 198 boys (mean and median age 9 [range 4–12] years) and 313 girls (mean and median age 9 [range 4–12]) years. All children consecutively underwent upper gastrointestinal endoscopy for persistent abdominal symptoms. In all cases, fasting blood for DNA and IgA tTG analyses (S1 Materials and Methods), duodenal and gastric biopsies for histology were taken. CD was histologically diagnosed in 244 (cases) and ruled out in the remaining 267 children (controls). The endoscopic findings and intestinal type lesions classified following theMarsh–Oberhuber criteria for celiac lesions [30], are detailed in Table 1.
Table 1. Patients’ details and clinicopathological characteristics.
| CD | Controls | |
|---|---|---|
| Patients, n | 244 | 267 |
| Females, n (%) a | 167 (68%) | 146 (55%) |
| Mean age ±s.d. (years) b | 7±4 | 10±4 |
| Age range (years) | 1–23 | 0.5–19 |
| Endoscopic findings: | ||
| Normal | 19 | 167 |
| Esophagitis / Hiatus hernia | 0 / 0 | 17 / 6 |
| Antral hyperemia / Diffuse gastric hyperemia / Duodenitis | 3 / 0 / 1 | 24 / 11 / 1 |
| Nodularity of the antral mucosa / Antrum and corpus nodularity | 4 / 0 | 19 / 12 |
| Duodenal nodular mucosa with mosaic pattern / Duodenal nodular mucosa with mosaic pattern and scalloping | 149 / 68 | 9 / 1 |
| Histology (Marsh-Oberhuber classification): | ||
| No Lesions | 0 | 262 |
| Type 1 | 9 | 5 |
| Type 2 | 16 | 0 |
| Type 3 | 219 (3a = 34, 3b = 51, 3c = 134) | 0 |
| H. pylori infection c | 12 | 50 |
| Mean tTG±s.d. (Units) d | 153±79 | 5±9 |
CD, celiac disease
a Chi-square test: 10.17, p = 0.001
b Student’s t test for unpaired data: t = 7.13, p<0.0001
c Chi-square test: 20.09, p<0.0001
d Student’s t test for unpaired data: t = 30.14, p<0.0001
TNFA and IFNG genotyping
Genomic DNA, extracted from EDTA-K2 peripheral blood by the Qiamp DNA blood maxi kit (QIAGEN S.p.A., Milan, Italy), was used to study IFNG +874A>T (rs2430561) and TNFA (-1031T>C, rs1799964; -857C>T, rs1799724; -376G>A, rs1800750; -308G>A, rs1800629; -238G>A, rs361525) polymorphisms, to detect TNFRSF1A variants and to genotype HLA. Single nucleotide polymorphisms of IFNG and TNFA genes were assayed by real-time PCR (ABI Prism 7900, Applied Biosystem, Foster City, CA, USA) as previously described in details [31,32].
TNFRSF1A analysis
Exons 2, 3, 4 and 6 of the TNFΑRSF1A gene were studied using denaturing high-performance liquid chromatography (DHPLC; Wave 2100 Fragment Analysis, Transgenomic, Omaha, NE, USA) and gene sequencing (ABI PRISM 3130 Genetic Analyzer, Applied Biosystem) according to D’Osualdo et al. [33]. Details are reported in S1 File and S1 Table.
HLA genotyping
A real-time polymerase chain reaction was employed by means of Taqman probes using an ABI Prism 7900 HT sequence detection system. Primers and probes for HLA-DQA1*0201, HLA-DQA1*03 and HLA-DQA1*05 detection were described by Fernandez et al. [34]. For HLA-DQB1*02 and HLA-DQB1*0302 alleles typing, primers were those described by Ferstlet al. [35]. Homozygous status was evaluated for the HLA-DQB1*02 allele by the set up of RT-PCR reactions to detect most of the already known HLA-DQB1 alleles different from *02 [35]. Primers and probes were aligned with sequences reported in the IMGT/HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/) and were purchased by Invitrogen Life Technologies (Monza, Italy). The analyses were made starting with about 100 ng DNA in a final volume of 25 μl containing 1X Taqman Universal PCR Master Mix (Applied Biosystems), 900 nM primers and 250 nM probes. The thermocycling conditions were 50°C for 2 minutes, 94°C for 7 minutes, followed by 42 cycles at 92°C for 20 seconds, 60°C for 1 minute and 20 seconds.
Statistics
The statistical analysis of data was made by using the chi-square test, binary logistic regression analysis, Student’s t test for unpaired data, Shapiro-Wilk W test for normal data, one way Anova (Stata 13.1, StataCorp, Lakeway Drive, TX, USA). The Hardy-Weinberg equilibrium test and pairwise linkage disequilibrium calculation analysis were done using Arlequin, version 2.000 (http://cmpg.unibe.ch/software/arlequin3/). Haplotypes with frequency and odds ratios were estimated by the retrospective profile-likelihood approach (dominant model; haplologit package; Stata 13.1) [36].
Results
In the retrospective cohort of 511 children and adolescents evaluated in this study, CD was diagnosed in 244 patients (cases) and ruled out in the remaining 267 (controls). Among controls mean age was higher, female gender lower and H. pylori infection more frequent than in CD (Table 1). Accordingly, age, gender and H. pylori infection were considered as potential confounding factors in all subsequent statistical analyses.
Dose effect of HLA-DQA1 and HLA-DQB1 genetics on CD susceptibility
HLA-DQ haplotypes were classified in agreement with Megiorni et al. [10] on the basis of the combination between HLA-DQA1 and of HLA-DQB1 alleles. Table 2 shows the HLA-DQ haplotypes, the corresponding HLA-DQA1 and HLA-DQB1 alleles, the number of patients bearing each individual haplotype and its frequency in CD cases and controls. In Table 2, the Odds Ratio (OR) and the 95% Confidence Interval (95% CI) for any HLA-DQ haplotype, calculated with respect to HLA-DQ negative patients, are also reported together with the estimated post-test probability of CD, considering that its reported prevalence among the general population of children and adolescents from the same geographic area is 1:184 [37]. The HLA-DQ2.5 homozygous haplotype was associated with the highest risk of CD (1:7). Interestingly, the homozygous HLA-DQB1*02 allele alone increased CD risk to levels comparable with those of HLA-DQ2.5 heterozygotes, while HLA-DQ8 had only a modest effect on CD risk (1:450).
Table 2. HLA haplotype and CD risk.
| HLA | HLA-DQA1 alleles | HLA-DQB1 alleles a | Total, Nr. | Cases, Nr.(frequency) | Controls, Nr.(frequency) | OR (95%CI) b | p-value | Adjusted p-value | Risk of CD c |
|---|---|---|---|---|---|---|---|---|---|
| Neg | Any | X/X | 159 | 1 (0.01) | 158 (0.99) | Ref. | - | - | 1:18216 |
| DQ8 | *03/any | *0302/X | 24 | 7 (0.29) | 17 (0.71) | 83 (9–742) | <0.0001 | <0.0001 | 1:450 |
| B2, hetero | *05 neg | *02/X | 27 | 8 (0.30) | 19 (0.70) | 63 (7–547) | <0.0001 | <0.0001 | 1:429 |
| DQ8/B1*02 pos | *03/*05 neg | *0302/*02 | 8 | 3 (0.37) | 5 (0.63) | 104 (8–1375) | <0.0001 | <0.0001 | 1:313 |
| DQ2.5 hetero | *05/any | *02/X | 162 | 104 (0.64) | 58 (0.36) | 284 (38–2107) | <0.0001 | <0.0001 | 1:103 |
| DQ2/DQ8 | *03/*05 | *0302/*02 | 14 | 10 (0.71) | 4 (0.29) | 585 (52–6523) | <0.0001 | <0.0001 | 1:75 |
| B2, homo | *05 neg | *02/*02 | 9 | 7 (0.78) | 2 (0.22) | 664 (50–8790) | <0.0001 | <0.0001 | 1:52 |
| DQ2.5 homo | *05/any | *02/*02 | 108 | 104 (0.96) | 4 (0.04) | 4422 (474–41281) | <0.0001 | <0.0001 | 1:7 |
p values in boldface are significant
aX = any HLA-DQB1 allele different from *02 or *0302
bOR = Odds Ratio; 95%CI = 95% Confidence Intervals. Odds Ratios were calculated by binary logistic regression analysis adjusted for age, gender and H. pylori infection
c Risk of CD is expressed as 1:N, where N is the number of individuals among which one case is present. Considering a disease prevalence of 1:184 in the general population [37], for each HLA-DQ category, N is calculated as a percentage of controls with that particular HLA-DQ status multiplied by 184 and divided by percentage of patients with the same DQ typing [10].
TNFA, not IFNG and TNFRSF1A polymorphisms, are associated with CD
Table 3 reports the results of the studied polymorphisms of TNFA and IFNG genes. Considering each individual single nucleotide polymorphism (SNP), the minor allele frequency (MAF), the number and frequency of genotypes in cases and controls are shown. To ascertain whether minor alleles exert any effect on CD risk, we estimated the OR with 95% CI associated with dominant and recessive models (Table 3). With the exception of the TNFA -238G>A, all the studied TNFA SNPs were correlated with a diagnosis of CD, and all the rare alleles had a dominant effect on CD risk. The IFNG+874A>T polymorphism was not correlated with CD. Interestingly H. pylori infection was associated with IFNG +874A>T (dominant model: chi-square = 4.73, p = 0.030), not with the other polymorphisms.
Table 3. TNFA, IFNG and TNFRSF1A gene polymorphisms in CD.
| Gene | dbSNP | MAF a | Genotypes | Cases b | Controls c | Dominant model | Recessive model | ||
|---|---|---|---|---|---|---|---|---|---|
| n (freq) | n (freq) | p-value (2df) | OR d (95%CI) | p-value (2df) | OR d (95%CI) | ||||
| TNFA | -1031T>C | C (0.027) | C/C | 9 (0.04) | 22 (0.08) | 0.027 | 0.65(0.44–0.95) | 0.042 | 0.41 (0.17–0.97) |
| T/C | 73 (0.30) | 97 (0.36) | |||||||
| T/T | 162 (0.66) | 148 (0.56) | |||||||
| -857C>T | T (0.153) | T/T | 3 (0.01) | 8 (0.03) | <0.0001 | 0.42 (0.27–0.65) | 0.316 | 0.48 (0.12–2.01) | |
| C/T | 45 (0.19) | 89 (0.33) | |||||||
| C/C | 196 (0.8) | 170 (0.64) | |||||||
| -376G>A | A (0.047) | A/A | 2 (0.01) | 0 (-) | 0.023 | 2.25 (1.12–4.51) | - | - | |
| G/A | 28 (0.11) | 16 (0.06) | |||||||
| G/G | 214 (0.88) | 251 (0.94) | |||||||
| -308G>A | A (0.216) | A/A | 20 (0.08) | 3 (0.01) | <0.0001 | 4.76 (3.12–7.26) | 0.001 | 10.09 (2.71–37.55) | |
| G/A | 117 (0.48) | 58 (0.22) | |||||||
| G/G | 107 (0.44) | 206 (0.77) | |||||||
| -238G>A | A (0.068) | A/A | 2 (0.01) | 1 (0.01) | 0.241 | 1.41 (0.80–2.46) | 0.841 | 1.28 (0.11–14.67) | |
| G/A | 34 (0.14) | 29 (0.11) | |||||||
| G/G | 208 (0.85) | 237 (0.88) | |||||||
| IFNG | +874A>T | T (0.458) | T/T | 52 (0.21) | 56 (0.21) | 0.929 | 0.98 (0.65–1.48) | 0.810 | 1.06 (0.67–1.68) |
| A/T | 116 (0.48) | 136 (0.51) | |||||||
| A/A | 76 (0.31) | 75 (0.28) | |||||||
| TNFRSF1A | c.625+10A>G | A (0.363) | G/G | 17 (0.33) | 26 (0.39) | 0.739 | 1.16 (0.47–2.76) | 0.061 | 0.21 (0.04–1.07) |
| G/A | 32 (0.63) | 31 (0.47) | |||||||
| A/A | 2 (0.04) | 9 (0.14) | |||||||
p values in boldface are significant
aMAF = minor allele frequency
bCases: n = 244; n = 51 for TNFRSF1A
cControls: n = 267; n = 66 for TNFRSF1A
dOR = Odds Ratio; 95%CI = 95% Confidence Intervals. Odds Ratios were calculated by binary logistic regression analysis adjusted for age, gender and H. pylori infection.
In a first series of 16 CD and 14 controls, exons 2-3-4-6 and the flanking intronic DNA sequences of the TNFRSF1A gene were screened by DHPLC to identify the presence of DNA sequence variants (heteroduplex). No variants were found on considering exons 2-3-4 and the adjacent intronic regions. Since in 9 CD and in 4 controls DHPLC analysis revealed the presence of a variant within amplicons spanning exon 6 and adjacent intronic regions, these regions were analyzed by DHPLC in a further series of 35 cases and 52 controls. At DNA sequence analysis, all variant samples were found to carry intronic c.625+10A>G (rs1800693) polymorphism. In our series of patients, differently from what reported in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=1800693) (last accessed: 30th January 2015), the minor allele was the c.625+10G, not the c.625+10A. This TNFRSF1A polymorphism was not correlated with a diagnosis of CD (Table 3).
The findings made on ascertaining whether any of the studied TNFΑ, IFNG and TNFRSF1A polymorphisms were associated with the HLA-DQ haplotypes are shown in S2 Table. Three polymorphisms (-857C>T, -376G>A and -308G>A) of TNFΑ gene were associated with HLA-DQ haplotypes, whereas the remaining two TNFA(-1031C>T and -238G>A), IFNG +874A>T and TNFRSF1A c.625+10A>G polymorphisms were not. Multivariate binary logistic regression analyses were then made considering CD diagnosis as outcome variable, while HLA-DQ haplotypes, the carriage of TNFA and IFNG rare alleles in homozygosis (recessive effect) or in heterozygosis (dominant effect) were predictor variables (S3 Table). The reduction in the risk of CD by the TNFA -1031 A rare allele and the increase by the TNFA-308 Α rare allele occurred independent of HLA-DQ haplotype effects.
TNFA haplotypes are HLA-independent risk factors for CD
The studied polymorphisms of the TNFA, all closely located in the gene promoter region, are carried as different haplotypes. We therefore determined the haplotypes resulting from the combination of TNFA-1031T>C,-857C>T,-376G>A and -308G>A polymorphisms, which were singly correlated with CD diagnosis. A total of six haplotypes with an overall frequency above 10-5 were found: their relative frequencies in CD cases and controls and the OR with 95% CI with respect to the most frequent haplotype (CCGG) are reported in S4 Table. For a clearer reading of subsequent results, the six haplotypes were coded as H1 to H6. TNFA haplotypes combinations were then estimated for any studied subject: a total of 17 haplotype combinations were inferred and are shown together with their frequencies in CD and controls in S5 Table (chi-square = 103, p<0.0001). The H1/H1 and H1/H2 combinations were rare among CD cases (<6%) and frequent among controls (>94%), while the inverse was observed for the H4/H5 and H5/H5 combinations (>80% among CD and <14% among controls). The rare H4/H4, H6/H6 and H3/H6 were exclusive to CD. The frequency of the remaining haplotype combinations ranged from 27 to 72% among CD and 31 to 73% among controls. On the basis of the frequency recorded among CD cases, haplotypes combinations were grouped as follows: group A (frequency among CD<10%); group B (10–40%); group C (40–60%); group D (60–80%); group E (>80%). TNFA haplotype combinations groups were correlated with the HLA-DQ haplotypes (chi-square = 190, p<0.0001). In particular, group A was strongly associated with HLA-DQ negativity, whereas group E strongly associated with HLA-DQ2.5 homozygotes (S1 Fig).
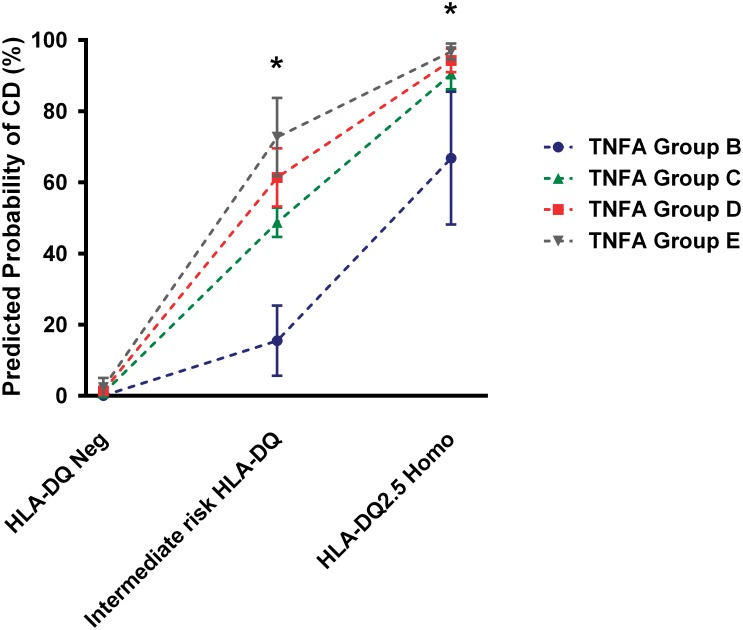
Table 4 shows the results obtained on ascertaining whether TNFA haplotype combinations exerted an independent effect on CD risk by performing multivariate binary logistic regression analysis adjusted for age, gender and H. pylori infection and considering CD diagnosis as an outcome variable, HLA-DQ haplotypes and TNFΑ haplotype combinations as predictors. TNFΑ haplotype combinations play an independent role in enhancing CD risk, although the ROC curve area obtained using the logistic regression model including as predictors HLA-DQ haplotypes, age, gender and H. pylori infection was not significantly different from the model that also included the TNFA haplotype combinations (p = 0.104) (S2 Fig). The multivariate binary logistic regression model presented in Table 4 was used to estimate the marginal probability of CD on the basis of HLA-DQ and TNFA haplotype combinations (Fig 1). The probability of CD was confirmed to be dependent on TNFA haplotype combinations mainly in the subset of patients carrying HLA-DQ intermediate risk haplotypes, which included HLA-DQ8, HLA-B2 hetero, HLA-DQ8/B1*02 pos, HLA-DQ2.5 hetero, HLA-DQ2/DQ8 and HLA-B2 homo.
Table 4. Binary logistic regression analysis.
| OR a | 95%CI | p-value | Adjusted p-value b | |
|---|---|---|---|---|
| HLA-DQ haplotype: | ||||
| Neg | Ref | |||
| Intermediate risk c haplotypes c | 178 | 24–1317 | <0.0001 | <0.0001 |
| DQ2.5 homo | 2752 | 287–26387 | <0.0001 | <0.0001 |
| TNFA haplotype: combinations | ||||
| Group A | Ref | |||
| Group B | 5 | 1–28 | 0.049 | 0.490 |
| Group C | 11 | 2–65 | 0.011 | 0.110 |
| Group D | 13 | 2–70 | 0.003 | 0.030 |
| Group E | 13 | 2–104 | 0.016 | 0.160 |
| Constant | 0.0055 | 0.0004–0.07 | <0.0001 | - |
Outcome variable: CD diagnosis; Predictor variables: HLA-DQ haplotypes, TNFA haplotype combinations.
p values in boldface are significant
a OR = Odds Ratio; Odds Ratios were calculated by binary logistic regression analysis adjusted for age, gender and H. pylori infection
bBonferroni’s adjusted p-values for multiple testing
cHLA-Q8, HLA-B2 hetero, HLA-DQ8/B1*02 pos, HLA-DQ2.5 hetero, HLA-DQ2/DQ8, HLA-B2 homo.
Fig 1. HLA-DQ and TNFA haplotype combinations predict CD risk.
The marginal probability of CD was estimated on the basis of the multivariate binary logistic regression model reported in Table 4 adjusted for age, gender and H. pylori infection. Reference was the group of patients with negative HLA-DQ and TNFA group A haplotype combination. Dots and bars represent the estimated CD probability with their respective Bonferroni’s adjusted standard errors.
*: p<0.05 for all groups with respect to TNFA Group A and p<0.05 for TNFA Group D with respect to TNFA Group B (pairwise comparisons of marginal linear predictions).
TNFA haplotypes and CD histology
We verified whether HLA-DQ and TNFA genetics correlate with CD histology. Patients were classified on the basis of the presence or absence of total atrophy [38]. HLA-DQ haplotype was not significantly correlated with total atrophy, which was found in 64/104 (61.5%) HLA-DQ2.5 homozygotes and in 70/140 (50.0%) remaining CD patients (Odds ratio = 1.6, 95%CI = 0.92–2.77; p-value = 0.073). The risk of total atrophy was stratified for TNFA haplotype combinations groups A to E considering CD patients carrying a high (HLA-DQ2.5 homozygous) or intermediate risk HLA-DQ haplotypes. A significant decrease of the risk of total atrophy was found in the intermediate risk HLA-DQ group (OR = 0.72, 95% CI = 0.52–0.99, p = 0.048), but not in HLADQ2.5 homozygotes (OR = 0.99, 95% CI = 0.67–1.44, p = 0.941).
Serum TNF-α increases in CD and associates with TNFRSF1A, not with TNFA genetics
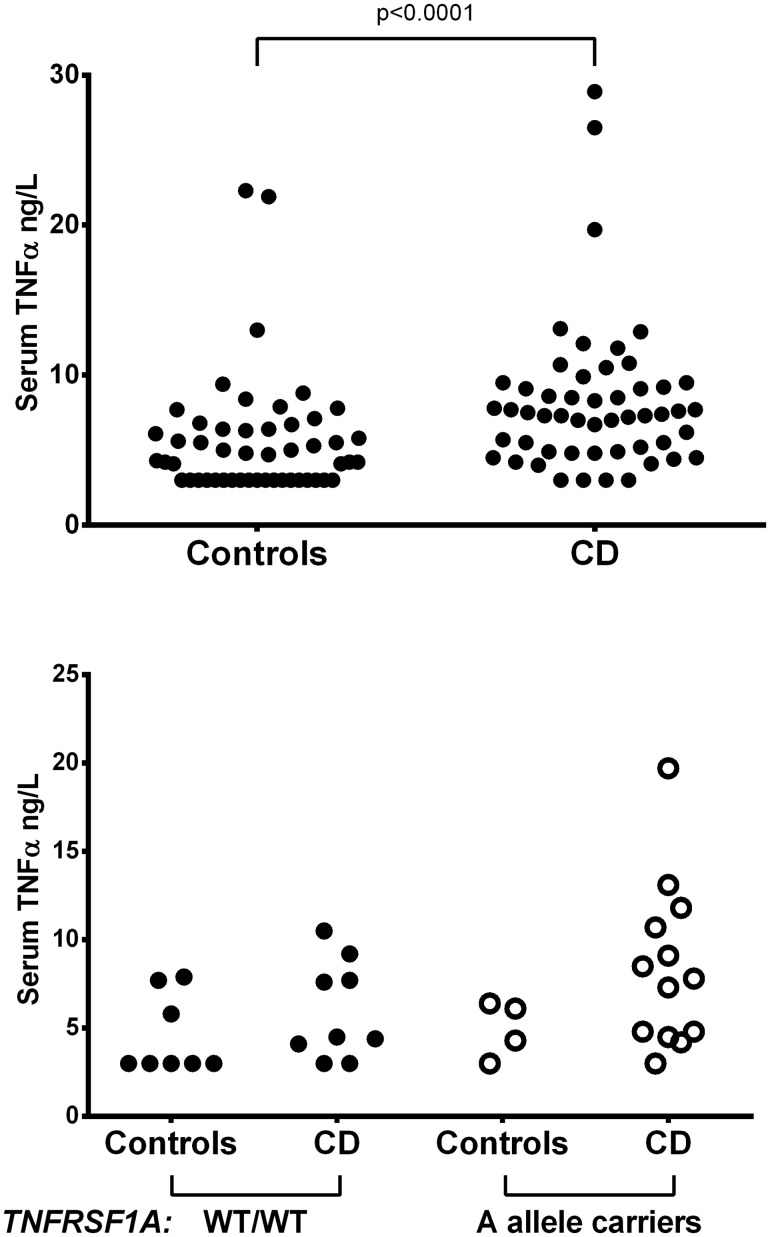
We then verified whether serum TNF-α levels are influenced by CD presence and/or TNFA/TNFRSF1Α genetics. Serum TNF-α was measured in 52 cases and in 50 controls. Due to asymmetrical distribution (Shapiro-Wilk W test for normal data: p<0.0001), the variable was transformed to 1/square root. Although serum TNF-α values were significantly higher in CD patients than in controls (Student’s t test: t = 4.32, p<0.0001) (Fig 2, upper panel), they were not associated with any TNFA studied polymorphisms considered singly or as haplotypes (F = 0.98, p = 0.422). Interestingly, the circulating levels of TNF-α tended to be higher in CD patients carrying the TNFRSF1AA allele (one way Anova: F = 2.55, p = 0.0742; Fig 2, lower panel).
Fig 2. TNFRSF1A polymorphism affects serum TNF-α levels.
Serum TNF-α in controls and CD patients (upper panel) and in patients subdivided according to the TNFRSF1Ac.625+10A>G (rs1800693) polymorphism (lower panel).
Discussion
In the year 2012, the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition formulated new guidelines for the diagnosis of CD in the pediatric age [39]. One algorithm was developed for symptomatic, and another for asymptomatic children and adolescents at an increased risk of developing the disease. In the former algorithm, positive findings for HLA-DQ2 or HLA-DQ8 are confirmatory for CD diagnosis in symptomatic children and adolescents with positive anti-tTG2 (10x normal) and EMA, while in the latter algorithm HLA-DQ2 or HLA-DQ8 testing is proposed as the initial action to identify negative patients who can be excluded from further studies. However, in patients positive at HLA-DQA1 and HLA-DQB1 typing, the CD risk depends on the resulting haplotype, spanning from very high (about 1:10 for HLA-DQ2.5) to low risk values (about 1:200 for B1*02/X), the latter being similar to that of the general population [10,37]. The findings made in the present study confirm the presence of a risk gradient for CD associated with the HLA-DQ haplotype resulting from HLA-DQA1 and HLA-DQB1 allele combinations: the HLA-DQ8 haplotype or the presence of HLA-DQB1*02 allele in single dose and not associated with HLA-DQA1*05 allele represent modest risk factors for CD, while the probability of developing the disease progressively increases as the number of HLA-DQB1*02 and HLA-DQA1*05 alleles increases, the highest risk (1:7) being found among HLA-DQ2.5 homozygotes. This high risk HLA-DQ haplotype was recently demonstrated to be the only factor that was significantly associated with the development of CD autoimmunity and overt CD in children who had at least one first-degree affected relative [40]. Since in our series of patients only few cases carried a single or double copy of the DQB1*02 allele associated with DQA1 alleles different from DQA1*05 or carried the HLA-DQ8, in agreement with Lionetti et al. [40], our studied children were classified as having no HLA-DQ risk alleles, a high risk (HLA-DQ2.5 Homo) and an intermediate risk HLA-DQ haplotype, the latter including all together DQ8, B2 hetero, DQ8/B1*02 pos, DQ2.5 hetero, DQ2/DQ8 and B2 homo. From a pathophysiological viewpoint, the diversity in genetic susceptibility associated with different HLA-DQ haplotypes is probably due, at least in part, to the recognition of different gluten peptide repertoires affecting T cell activation [9]. From a practical clinical viewpoint this diversity raises the question of whether a different management should be proposed for asymptomatic patients carrying high risk or intermediate risk HLA-DQ haplotypes. More specifically, the identification of HLA-DQ independent genetic risk or protective factors for CD might improve our understanding of the disease and ameliorate risk stratification of patients. We focused our study on one HLA-associated gene, TNFA [26], and on two non-HLA-associated genes, IFNG and TNRFSF1A [32,33], whose protein products play an active role in the pathophysiology of CD. TNF-α and IFN-γ are known to play an active role in CD, but also in refractory CD [41], and sequence variations of the main TNF-α receptor, TNF-R1, caused by mutations of the encoding TNRFSF1A gene, may determine an altered cellular response to TNF-α stimulation [42].
IFNG +874A>T polymorphism was not associated with CD, but it was associated with H. pylori, thus further supporting the role of IFN-γ genetics in infectious diseases [32]. Four of the five TNFA promoter polymorphisms were found to be correlated with a diagnosis of CD: the rare alleles of the -1031T>C and -857C>T had a protective dominant effect, while the rare alleles of the -376G>A and -308G>A enhanced the risk of CD. The TNFA-308 A allele had an additional effect on CD risk, unlike the -376A allele; this difference is probably due to the fact that the latter minor allele frequency is very low (0.047) while the former is not (0.216). Since the TNFA gene is located in close proximity of the HLA class II region, we ascertained whether TNFA genotypes were associated with HLA-DQ haplotypes, and this occurred for the -857C>T, -376G>A and -308G>A, not for the -1031T>C, polymorphisms. To ascertain whether the TNFA-associated risk for CD is dependent or independent from its linkage with the HLA-DQA1 and—B1 genes, multivariate statistical analysis was performed, including as cofactors the potential confounders age, gender and H. pylori infection which was less frequently found among CD patients than in controls. The rare TNFA-1031 A and -308 A alleles were confirmed to be HLA-DQ independent CD-associated variants: carriers of the -1031 A allele in homozygosis have a reduced risk, while carriers of the -308 A allele have about a 3 fold increased risk of CD, this finding being in agreement with previously reported data in the literature [21,22,24].
To obtain further insight on the association between TNFA genetics and CD, our subsequent analysis of data was focused on TNFA haplotypes. Although any single polymorphism might influence gene transcription to different extents, in the presence of a string of SNPs located in close proximity with each other, their different combinations in haplotypes might reinforce or otherwise revert the effects on gene transcription of any single SNP. We considered the four TNFA SNPs significantly associated with CD (-1031T>C, -857C>T, -376G>A and -308G>A) and, using the Arlequin statistical software, estimated the haplotypes resulting from their combinations. Two of the six possible obtained haplotypes were more frequent among controls, the CCGG (H1) and TTGG (H2), three were more frequent among CD cases, the CCAG (H4), TCGA (H5) and CCGA (H6) haplotypes, while the frequency of the TCGG (H3) haplotype was similar in CD and controls. Since diploid cells carry pairs of haplotypes, we estimated the possible combinations resulting from H1–H6 haplotypesin our patients, and found 17 combinations. To reduce data dispersion five classes of haplotype combinations (groups A to D) were identified on the basis of their frequency among CD: very low (<10%), low (10–40%), intermediate (40–60%), high (60–80%) and very high (>80%). Haplotype combinations were confirmed to be correlated with CD diagnosis; although all TNFA haplotype combinations groups B to E increased CD risk with respect to group A, an HLA-DQ independent statistically significant effect was found for Group D with respect to group A. In the attempt to translate these findings in clinical practice, we verified whether TNFA haplotype combinations might improve risk estimation among the three HLA-DQ identified categories: HLA-DQ negative, HLA-DQ2.5 homozygotes and intermediate risk HLA-DQ. In the absence of HLA-DQ risk alleles, the TNFA haplotype did not modify the predicted CD risk. Vice versa in HLA-DQ high risk, but mainly in intermediate HLA-DQ risk subjects, carriage of any TNFA haplotype different from group A increased the predicted probability of disease and this increase appeared to be progressively higher in groups from B to E. Therefore TNFA genetic testing might be suggested as being of some help in improving CD risk assessment and in further studying patients. Interestingly, while HLA-DQ was not significantly associated with the histopathological lesion severity, in agreement with Ruiz-Ortiz et al. [43], TNFA haplotype A to E showed a negative trend of association with severity of lesions. In other words, the progressive increase in CD risk due to TNFA haplotypes is associated with a progressive less severe histopathology. If confirmed, this finding indicates that patients carrying high risk TNFA haplotypes are at risk of a less severe mucosal damage with a consequent uncertainty in the histological diagnosis [44].
In order to ascertain whether TNFA haplotypes influence TNF-α protein levels, we measured this cytokine in a series of case and control sera. Serum TNF-α values were higher in CD patients than in controls, supporting the role of TNF-α in CD, but they were not correlated with TNFA haplotypes. Although these findings argue against the hypothesis that TNFA haplotypes increase CD risk by enhancing TNF-α production, we cannot rule out that they affect the amount of TNF-α produced in the affected duodenal mucosa microenvironment.
One of the main TNF-α receptors mediating its the inflammatory effect is TNF-R1, encoded by the TNFRSF1A gene [45]. The R92Q polymorphism of this gene has been associated with an increased risk of TRAPS [46]. Due to the relevant impact of TNF-α on CD pathogenesis, we verified whether the TNFRSF1A gene had any variant correlated with CD. Only one polymorphism of this gene was identified in our series of cases and controls, the c.625+10A>G (rs1800693), which is reportedly a risk factor for multiple sclerosis [47]. The lack of any association between this polymorphism and CD allows us to rule out the hypothesis that it takes a part in causing duodenal atrophy upon exposure to gluten peptides. However, in CD patients this SNP was associated with slightly increased serum TNF-α levels. This might be due to an altered TNF-α/TNF-R1 balance consequent to the synthesis of a truncated TNF-R1 in patients carrying this polymorphism, as previously demonstrated by Ottoboniet al. [48].
In this study we have demonstrated that extended TNFA promoter haplotypes are HLA-DQ independent risk factors for CD and that their analysis might enhance the reliability in estimating the risk of CD especially in patients carrying an intermediate risk HLA-DQ haplotype. The strength of our principal findings rely upon the large number of patients with the same ethnicity entered the study. This renders our results transferrable within European populations, but require confirmation in populations that have a different LD structure, like Africans, Japanese or Chinese. The extended analysis of other genetic loci reportedly associated with CD by GWAS, which will be fast and cost-effective by the incoming high-throughput technology, represents a focus for future research.
Supporting Information
Predictors blu line: HLA-DQ haplotypes, age and gender; predictors red line: HLA-DQ haplotypes, TNFA haplotype combinations, age and gender.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
The OR with 95% CI were calculated with respect to the most frequent haplotype (TCGG)[36].
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1. Fasano A, Catassi C. Clinical practice. Celiac disease. N Engl J Med. 2012; 367:2419–26. 10.1056/NEJMcp1113994 [DOI] [PubMed] [Google Scholar]
- 2. Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol. 2013; 13:294–302. 10.1038/nri3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kupfer SS, Jabri B. Pathophysiology of celiac disease. Gastrointest Endosc Clin N Am. 2012; 22:639–60. 10.1016/j.giec.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nilsen EM, Jahnsen FL, Lundin KE, Johansen FE, Fausa O, Sollid LM, et al. Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology 1998; 115:551–63. [DOI] [PubMed] [Google Scholar]
- 5. Westerholm-Ormio M, Garioch J, Ketola I, Savilahti E. Inflammatory cytokines in small intestinal mucosa of patients with potential coeliac disease. Clin Exp Immunol. 2002; 128:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Westerholm-Ormio M, Vaarala O, Pihkala P, Ilonen J, Savilahti E. Immunologic activity in the small intestinal mucosa of pediatric patients with type 1 diabetes. Diabetes. 2003; 52:2287–95. [DOI] [PubMed] [Google Scholar]
- 7. Brottveit M, Beitnes AC, Tollefsen S, Bratlie JE, Jahnsen FL, Johansen FE, et al. Mucosal cytokine response after short-term gluten challenge in celiac disease and non-celiac gluten sensitivity. Am J Gastroenterol. 2013; 108:842–50. 10.1038/ajg.2013.91 [DOI] [PubMed] [Google Scholar]
- 8. Ahn R, Ding YC, Murray J, Fasano A, Green PH, Neuhausen SL, et al. Association analysis of the extended MHC region in celiac disease implicates multiple independent susceptibility loci. PLoS One. 2012; 7:e36926 10.1371/journal.pone.0036926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koning F. Celiac disease: quantity matters. Semin Immunopathol. 2012; 34:541–9. 10.1007/s00281-012-0321-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Megiorni F, Mora B, Bonamico M, Barbato M, Nenna R, Maiella G, et al. HLA-DQ and risk gradient for celiac disease. Hum Immunol. 2009; 70:55–9. 10.1016/j.humimm.2008.10.018 [DOI] [PubMed] [Google Scholar]
- 11. Romanos J, Rosén A, Kumar V, Trynka G, Franke L, Szperl A, et al. Improving coeliac disease risk prediction by testing non-HLA variants additional to HLA variants. Gut. 2014; 63:415–22. 10.1136/gutjnl-2012-304110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007; 39:827–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008; 40:395–402. 10.1038/ng.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010; 42:295–302. 10.1038/ng.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PloS Genet. 2011; 7:e1002004 10.1371/journal.pgen.1002004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mourão AF, Caetano-Lopes J, Costa P, Canhão H, Santos MJ, Pinto P, et al. Tumor necrosis factor-alpha -308 genotypes influence inflammatory activity and TNF-alpha serum concentrations in children with juvenile idiopathic arthritis. J Rheumatol. 2009; 36:837–42. 10.3899/jrheum.080615 [DOI] [PubMed] [Google Scholar]
- 17. PrabhuAnand S, Harishankar M, Selvaraj P. Interferon gamma gene +874A/T polymorphism and intracellular interferon gamma expression in pulmonary tuberculosis. Cytokine. 2010; 49:130–3. 10.1016/j.cyto.2009.11.009 [DOI] [PubMed] [Google Scholar]
- 18. Huang HR, Zhong YQ, Wu JF. The association between IFN-γ and IL-4 genetic polymorphisms and childhood susceptibility to bronchial asthma. Gene. 2012; 494:96–101. 10.1016/j.gene.2011.09.027 [DOI] [PubMed] [Google Scholar]
- 19. Cataldo F, Scola L, Piccione M, Giuffrè M, Crivello A, Forte GI, et al. Evaluation of cytokine polymorphisms (TNFalpha, IFNgamma and IL-10) in Down patients with coeliac disease. Dig Liver Dis. 2005; 37:923–7. [DOI] [PubMed] [Google Scholar]
- 20. Lio D, Scola L, Forte GI, Accomando S, Giacalone A, Crivello A, et al. TNFalpha, IFNgamma and IL-10 gene polymorphisms in a sample of Sicilian patients with coeliac disease. Dig Liver Dis. 2005; 37:756–60. [DOI] [PubMed] [Google Scholar]
- 21. de la Concha EG, Fernández-Arquero M, Vigil P, Rubio A, Maluenda C, Polanco I, et al. Celiac disease and TNF promoter polymorphisms. Hum Immunol. 2000; 61:513–7. [DOI] [PubMed] [Google Scholar]
- 22. Garrote JA, Arranz E, Tellería JJ, Castro J, Calvo C, Blanco-Quirós A. TNF alpha and LT alpha gene polymorphisms as additional markers of celiac disease susceptibility in a DQ2-positive population. Immunogenetics. 2002; 54:551–5. [DOI] [PubMed] [Google Scholar]
- 23. Louka AS, Lie BA, Talseth B, Ascher H, Ek J, Gudjónsdóttir AH, et al. Coeliac disease patients carry conserved HLA-DR3-DQ2 haplotypes revealed by association of TNF alleles. Immunogenetics. 2003; 55:339–43. [DOI] [PubMed] [Google Scholar]
- 24. Sumnik Z, Cinek O, Bratanic N, Kordonouri O, Kulich M, Roszai B, et al. Risk of celiac disease in children with type 1 diabetes is modified by positivity for HLA-DQB1*02-DQA1*05 and TNF -308A. Diabetes Care. 2006; 29:858–63. [DOI] [PubMed] [Google Scholar]
- 25. Cherñavsky AC, Páez MC, Periolo N, Correa P, Guillén L, Niveloni SI, et al. The simultaneous presence of IL-1B and TNFA two-positions risk haplotypes enhances the susceptibility for celiac disease. Cytokine. 2008; 42:48–54. 10.1016/j.cyto.2008.01.015 [DOI] [PubMed] [Google Scholar]
- 26. Hajeer AH, Hutchinson IV. TNF-alpha gene polymorphism:clinical and biological implications. Microsc Res Tech. 2000; 50:216–28. [DOI] [PubMed] [Google Scholar]
- 27. O'Dwyer MJ, Mankan AK, Ryan AW, Lawless MW, Stordeur P, Kelleher D, et al. Characterization of tumour necrosis factor-alpha genetic variants and mRNA expression in patients with severe sepsis. Int J Immunogenet. 2008; 35:279–85. 10.1111/j.1744-313X.2008.00773.x [DOI] [PubMed] [Google Scholar]
- 28. McGonagle D, Aziz A, Dickie LJ, McDermott MF. An integrated classification of pediatric inflammatory diseases, based on the concepts of autoinflammation and the immunological disease continuum. Pediatr Res. 2009; 65:38R–45R. 10.1203/PDR.0b013e31819dbd0a [DOI] [PubMed] [Google Scholar]
- 29. Palová-Jelínková L, Dáňová K, Drašarová H, Dvořák M, Funda DP, Fundová P, et al. Pepsin digest of wheat gliadin fraction increases production of IL-1β via TLR4/MyD88/TRIF/MAPK/NF-κB signaling pathway and an NLRP3 inflammasome activation. PLoS One. 2013; 8:e62426 10.1371/journal.pone.0062426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oberhuber G. Histopathology of celiac disease. Biomed Pharmacother. 2000; 54:368–72. [DOI] [PubMed] [Google Scholar]
- 31. Zambon C-F, Basso D, Navaglia F, Belluco C, Falda A, Fogar P, et al. Pro- and antiinflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005; 29:141–52. [DOI] [PubMed] [Google Scholar]
- 32. Huang Y, Yang H, Borg BB, Su X, Rhodes SL, Yang K, et al. A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection. Proc Natl Acad Sci USA. 2007; 104:985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. D'Osualdo A, Ferlito F, Prigione I, Obici L, Meini A, Zulian F, et al. Neutrophils from patients with TNFRSF1A mutations display resistance to tumor necrosis factor-induced apoptosis: pathogenetic and clinical implications. Arthritis Rheum. 2006; 54:998–1008. [DOI] [PubMed] [Google Scholar]
- 34. Fernandez S, Wassmuth R, Knerr I, Frank C, Haas JP. Relative quantification of HLA-DRA1 and -DQA1 expression by real-time reverse transcriptase-polymerase chain reaction (RT-PCR). Eur J Immunogenet. 2003; 30:141–8. [DOI] [PubMed] [Google Scholar]
- 35. Ferstl B, Zacher T, Lauer B, Blagitko-Dorfs N, Carl A, Wassmuth R. Allele-specific quantification of HLA-DQB1 gene expression by real-time reverse transcriptase-polymerase chain reaction. Genes Immun. 2004; 5:405–16. [DOI] [PubMed] [Google Scholar]
- 36. Marchenko YV, Carroll RJ, Lin DY, Amos CI, Gutierrez RG. Semiparametric analysis of case–control genetic data in the presence of environmental factors. Stata Journal Volume 8 Number 3; 2008. [Google Scholar]
- 37. Catassi C, Fabiani E, Rätsch IM, Coppa GV, Giorgi PL, Pierdomenico R, et al. The coeliac iceberg in Italy. A multicentreantigliadin antibodies screening for coeliac disease in school-age subjects. Acta Paediatr. 1996; Suppl 412:29–35. [DOI] [PubMed] [Google Scholar]
- 38. Villanacci V, Ceppa P, Tavani E, Vindigni C, Volta U. Coeliac disease: the histology report. Dig Liver Dis. 2011; 43 Suppl 4:S385–95. 10.1016/S1590-8658(11)60594-X [DOI] [PubMed] [Google Scholar]
- 39. Husby S, Koletzko S, Korponay-Szabó IR, Mearin ML, Phillips A, Shamir R, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012; 54:136–60. 10.1097/MPG.0b013e31821a23d0 [DOI] [PubMed] [Google Scholar]
- 40. Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S, et al. Introduction of gluten, HLA status, and the risk ofceliac disease in children. N Engl J Med. 2014; 371:1295–303. 10.1056/NEJMoa1400697 [DOI] [PubMed] [Google Scholar]
- 41. Caruso R, Marafini I, Sedda S, Del Vecchio Blanco G, Giuffrida P, MacDonald TT, et al. Analysis of the cytokine profile in the duodenal mucosa of refractory coeliac disease patients. Clin Sci (Lond). 2014; 126:451–8. 10.1042/CS20130478 [DOI] [PubMed] [Google Scholar]
- 42. Negm OH, Mannsperger HA, McDermott EM, Drewe E, Powell RJ, Todd I, et al. A pro-inflammatory signalome is constitutively activated by C33Y mutant TNF receptor 1 in TNF receptor-associated periodic syndrome (TRAPS). Eur J Immunol. 2014; 44:2096–110. 10.1002/eji.201344328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ruiz-Ortiz E, Montraveta M, Cabré E, Herrero-Mata MJ, Pujol-Borrell R, Palou E, et al. HLA-DQ2/DQ8 and HLA-DQB1*02 homozygosity typing by real-time polymerase chain reaction for the assessment of celiac disease genetic risk: evaluation of a Spanish celiac population. Tissue Antigens. 2014; 84:545–53. 10.1111/tan.12472 [DOI] [PubMed] [Google Scholar]
- 44. Villanacci V. The histological classification of biopsy in celiac disease: Time for a change? Dig Liver Dis. 2015; 47:2–3. 10.1016/j.dld.2014.09.022 [DOI] [PubMed] [Google Scholar]
- 45. MacEwan DJ. TNF receptor subtype signalling: differences and cellular consequences. Cell Signal. 2002; 14:477–92. [DOI] [PubMed] [Google Scholar]
- 46. Caso F, Rigante D, Vitale A, Lucherini OM, Costa L, Atteno M, et al. Monogenic autoinflammatory syndromes: state of the art on genetic, clinical, and therapeutic issues. Int J Rheumatol. 2013; 2013:513782 10.1155/2013/513782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Comabella M, Caminero AB, Malhotra S, Agulló L, Fernández O, Reverter F, et al. TNFRSF1A polymorphisms rs1800693 and rs4149584 in patients with multiple sclerosis. Neurology. 2013; 80:2010–6. 10.1212/WNL.0b013e318294b2d6 [DOI] [PubMed] [Google Scholar]
- 48. Ottoboni L, Frohlich IY, Lee M, Healy BC, Keenan BT, Xia Z, et al. Clinical relevance and functional consequences of the TNFRSF1A multiple sclerosis locus. Neurology. 2013; 81:1891–9. 10.1212/01.wnl.0000436612.66328.8a [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predictors blu line: HLA-DQ haplotypes, age and gender; predictors red line: HLA-DQ haplotypes, TNFA haplotype combinations, age and gender.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
The OR with 95% CI were calculated with respect to the most frequent haplotype (TCGG)[36].
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.