Abstract
Nanoparticles hold great promise for delivering medical cargos to cancerous tissues to enhance contrast and sensitivity of imaging agents or to increase specificity and efficacy of therapeutics. A growing body of data suggests that nanoparticle shape, in combination with surface chemistry, affects their in vivo fates, with elongated filaments showing enhanced tumor targeting and tissue penetration, while promoting immune evasion. The synthesis of high aspect ratio filamentous materials at the nanoscale remains challenging using synthetic routes; therefore we turned toward nature’s materials, developing and studying the filamentous structures formed by the plant virus potato virus X (PVX). We recently demonstrated that PVX shows enhanced tumor homing in various preclinical models. Like other nanoparticle systems, the proteinaceous platform is cleared from circulation and tissues by the mononuclear phagocyte system (MPS). To increase bioavailability we set out to develop PEGylated stealth filaments and evaluate the effects of PEG chain length and conformation on pharmacokinetics, biodistribution, as well as potential immune and inflammatory responses. We demonstrate that PEGylation effectively reduces immune recognition while increasing pharmacokinetic profiles. Stealth filaments show biodistribution consistent with MPS clearance mechanisms; the protein:polymer hybrids are cleared from the body indicating biodegradability and biocompatibility. Tissue compatibility is indicated with no apparent inflammatory signaling in vivo. Tailoring PEG chain length and conformation (brush vs. mushroom) allows tuning of the pharmacokinetics, yielding long-circulating stealth filaments for applications in nanomedicine.
Introduction
The development of nanoparticles for drug delivery and imaging is a rapidly growing area with many novel platform technologies in the pipeline. For example, nanoparticles engineered with appropriate surface chemistries and equipped with therapeutics allow targeting of toxic payloads to cancerous tissues through passive or active targeting mechanisms. This partitioning toward the diseased tissues increases payload delivery and reduces systemic side effects often associated with potent chemotherapies or toxins. Commonly used nanoparticles include lipid-based micelles, polymeric capsules, iron oxide nanoparticles, gold nanoparticles, carbon nanotubes, and protein-based particles and nanocages; each system has its advantages and disadvantages in terms of nanomanufacturing, engineerability, and in vivo properties. Surface chemistries, such as modification with stealth coatings or targeting ligands, and nanoparticle shape are handles to tailor tissue specificity and bioavailability.
Mounting evidence suggests advantageous behaviors of elongated, filamentous nanomaterials: i) non-spherical materials show increased margination toward the vessel wall and increasing tumor homing;[1–7] ii) elongated materials present ligands more effectively to the larger and flat vessel wall or target cells compared to their spherical counterparts;[8–10] and iii) elongated materials have increased immune evasion and reduced macrophage uptake, therefore further contributing to synergistic target enhancement.[11, 12] Most platform technologies currently under development are spherical or elongated low aspect ratio materials (AR < 5). Exemptions are carbon nanotubes and filomicelles; however carbon-based materials have low biocompatibility[13] and filomicelles are in the micron-size regime.[14] Synthetic approaches to high aspect ratio materials remain challenging because of polydispersity. Synthetic chemistry and nanotechnology seek to mimic what nature has achieved, i.e. self-assembly and programmability at the atomic level. Therefore, we turned toward a bio-inspired approach and are studying and developing filamentous plant viruses for nanomedical applications. Mammalian virus-based nanoparticles for gene therapy and oncolytic virotherapy are in clinical investigations,[15–17] so the potential of virus-based materials for medical applications has clearly been recognized. There are many novel viruses in the development pipeline including bacteriophages and plant viruses; these non-mammalian pathogens may be advantageous because they are non-infectious toward humans.
Specifically, we turned toward the filamentous plant virus potato virus X (PVX), which measures 515 nm in length and 13 nm in width. The filaments can be obtained in gram scales through farming in plants using N. benthamiana plants as the production species. The proteinaceous scaffold is amenable to chemical modification and genetic engineering. For example, we recently demonstrated expression of green fluorescent protein (GFP) and other fluorescent proteins as genetic coat protein fusions.[18] Furthermore, solvent-exposed lysine side chains offer a convenient means of modification with non-peptide-based ligands (e.g. therapeutics or contrast agents) via chemical bioconjugation.[19]
We have shown that plant virus-based materials accumulate in tumors; targeting is achieved based on passive accumulation via the enhanced permeability and retention (EPR) effect[20] or active receptor targeting of cancer signatures.[21–25] Data indicate that filaments show more efficient passive tumor partitioning compared to spherical nanoparticles; this shape-mediated enhanced tumor homing and penetration is reproducible in a variety of models, including human tumor xenografts of fibrosarcoma, squamous cell sarcoma, colon cancer, and breast cancer.[20, 26] Together, these data provide strong support for the further development and investigation of filamentous plant viruses for biomedical applications.
Like other nanomaterials, the proteinaceous carriers are cleared by the mononuclear phagocyte system (MPS).[26] Conjugation of stealth polymers to coat the nanocarriers allows to reduce interaction with the MPS. The most extensively studied stealth polymer is polyethylene glycol (PEG).[27–31] PEG is a non-charged, hydrophilic polymer with low toxicity and immunogenicity; a wide-variety of functionalized PEG monomers and chains are available for nanoparticle modification. The hydrophilic shield provided by the PEG coating of nanoparticles decreases serum protein adsorption, resulting in the “stealth” properties commonly reported for PEGylated nanoparticles (e.g. increased circulation time, decreased accumulation in liver and spleen).[27–31]
In the present studies, we set out to develop and study stealth filaments using PVX-PEG hybrids. While we previously reported the in vivo properties of PEGylated PVX modified with linear PEG chains of 5,000 Da [20], this study set out to determine whether the pharmacokinetic profiles could be further optimized to generate long-circulating stealth filaments with favorable properties for tumor targeting and payload delivery. To do this, we considered PEG chains of various molecular weights and conformations using linear and branched PEGs with molecular weights of 5,000 Da and 20,000 Da. The biological fate of stealth filaments as a function of surface coating was analyzed in vitro and in vivo to study pharmacokinetics, biodistribution, immunogenicity, and immune cell interactions.
Materials and Methods
PVX propagation
PVX was propagated in Nicotiana benthamiana and purified using previously described procedures.[19] Purified PVX was stored in 0.1 M potassium phosphate buffer pH 7.0 at 4°C. PVX concentration was determined by UV/visible spectroscopy (εPVX= 2.97 mL mg−1 cm−1 at 260 nm).
Bioconjugation of PVX with fluorophores and PEG
PVX was modified with Alexa Fluor 647 (A647) or VivoTag-S 750 and/or polyethylene glycol (PEG) using N-hydroxysuccinimide (NHS)-activated esters. First, PVX (at 1–2 mg mL−1 in 0.1 M potassium phosphate buffer, pH 7.0) was reacted with a 2,000 molar excess of NHS-A647 (Life Technologies) or VivoTag-S 750 (Perkin-Elmer) at a 10% (v/v) final concentration of DMSO. PVX has a molar mass of 35x106 g mol−1. The reaction was allowed to proceed for 2 hours at room temperature, with agitation, and purified using 10-kDa cut-off centrifugal filters (Millipore). Second, A647-modified PVX (A-PVX) or VivoTag-S 750-modified PVX (V-PVX) was reacted with linear or branched PEG-NHS with molecular weights of 5,000 Da (P5L and P5B) or linear PEG-NHS with a molecular weight of 20,000 Da (P20) (Nanocs). PEG was added at a molar excess of 10,000 in a 10% (v/v) final concentration of DMSO and incubated overnight at room temperature, with agitation. PVX-A647-PEG (A-PVX-PEG) and PVX-VivoTag-PEG (V-PVX-PEG) were purified using 10- and 100-kDa-cut off centrifugal filter units (Millipore). A-PVX-PEG and V-PVX-PEG filaments were stored in 0.1 M potassium phosphate buffer, pH 7.0 at 4°C and characterized using a combination of UV/visible spectroscopy, denaturing gel electrophoresis, and transmission electron microscopy (TEM).
UV/visible spectroscopy
The number of A647 or VivoTag molecules per PVX filament was determined by UV/visible spectroscopy using a NanoDrop 2000 spectrophotometer (Thermo Fisher). Dye number was determined by the ratio of fluorophore-to-PVX concentration, using the Beer-Lambert law and the fluorophore and PVX-specific extinction coefficients: εPVX = 2.97 mL mg−1 cm−1 at 260 nm, εA647 = 270,000 M−1 cm−1 at 650 nm, and εVivoTag = 240,000 M−1 cm−1 at 775 nm.
Denaturing gel electrophoresis
SDS-PAGE was used to determine the number of PEG molecules per PVX filament. 15 µg of denatured protein samples were loaded and run on 4–12% NuPage gels (Life Technologies) in 1x MOPS SDS running buffer (Life Technologies). Protein bands were visualized under white light before and after staining with Coomassie blue (0.25% w/v).
TEM
A-PVX-PEG filaments (20 µL, 0.1 mg mL−1) were negatively stained with 2% (w/v) uranyl acetate for 5 min on a carbon-coated copper grid (Ted Pella). Samples were analyzed using a Zeiss Libra 200FE transmission electron microscope operated at 200 kV.
Animals
All experiments were carried out in accordance with Case Western Reserve University’s Institutional Animal Care and Use Committee. Balb/C mice (Charles River) and C57/BL6 mice (Taconic) were used.
Pharmacokinetics
Pharmacokinetics and biodistribution (see below) were evaluated in healthy Balb/C mice (Charles River) (n=3). Animals were maintained on an alfalfa-free diet (Teklad) for two weeks prior to the administration of fluorescently labeled PVX filaments to reduce tissue autofluorescence. A-PVX, A-PVX-P5B, or A-PVX-P20 formulations were injected via the tail vein (at 100 µg in 100 µL sterile PBS) and blood was collected into heparin-coated tubes (Fisher) using retro-orbital bleeding; time course studies were conducted and samples were collected up to 36 hours post-administration. Serum was isolated from samples by centrifuging at 14,500 g for 10 minutes. Fluorescence was read (λEx = 600 nm, λEm = 660 nm) using a Tecan microplate reader, and the fluorescence reading was correlated to a standard curve normalized for each particle formulation to determine the amount of particle (in µg) at each time point. Percent injected dose was determined based on the fluorescence reading and amounts of PVX per 50 µL of serum from each time point and assuming a total blood volume of 1.5 mL.
Biodistribution using fluorescence molecular tomography (FMT)
Biodistribution and clearance were evaluated using healthy Balb/C mice with FMT imaging as well as immunofluorescence microscopy (see below). Animals were maintained on an alfalfa-free diet (Teklad) for two weeks prior to the study to reduce tissue autofluorescence. For FMT imaging, mice were injected with V-PVX (n=2, one animal died prior to the start of the experiment), V-PVX-P5B (n=3), or V-PVX-P20 (n=3) via the tail vein (at 100 µg in 100 µL sterile PBS) and imaged using a FMT 2500 quantitative tomography in vivo imaging system (Perkin-Elmer) at 0 minutes (pre-scan), 10 minutes, 30 minutes, 1 hour, 2 hours, 4 hours, 6 hours, 8 hours, 24 hours, 32 hours, and 52 hours post-administration. For each image, regions of interest (ROIs) were chosen for the whole body, liver and spleen, and bladder. Fluorescence intensity was calculated using the normalized total fluorescence within each ROI (based on the number of dyes per particle).
Immunofluorescence microscopy
Biodistribution and clearance were evaluated using immunofluorescence microscopy. Mice were sacrificed at 0 hours, 6 hours, 24 hours, 52 hours, 72 hours, 96 hours, and 7 days post-administration of A-PVX-P20 (via the tail vein at 100 µg in 100 µL sterile PVX), and livers and spleens were harvested. Ten µm thick tissue sections were prepared using a Leica CM1850 cryostat. Slides were fixed in 95% (v/v) ethanol for 20 minutes on ice and permeabilized using 0.2% (v/v) Triton X-100 (EMD Chemicals) in PBS for 2 minutes. Slides were blocked using 10% (v/v) goat serum (GS) (Life Technologies) in PBS. PVX staining was carried out using a rabbit anti-PVX antibody (Pacific Immunology, at 1:250 in 1% (v/v) GS in PBS), followed by Alexa Fluor 488-labeled goat anti-rabbit secondary antibody (Life Technologies, 1:500 in 1% (v/v) GS in PBS); each step was carried out for 1 hour at room temperature and slides were rinsed three times in PBS in between each step. Slides were mounted with Fluoroshield with DAPI (Sigma Aldrich) and stored at −20°C until imaged. Analysis of sections was performed on an Olympus Fluoview FV1000 confocal microscope.
Immunogenicity
Antibody titers were evaluated in healthy male Balb/C mice after repeated administration of PVX, PVX-P5L, PVX-P5B, or PVX-P20. PVX-based formulations were administered intravenously via tail vein injections at 100 µg protein in 100 µL sterile PBS at days 0, 5, 10, and 66. Blood was collected on days 0 (pre-bleed), 5, 10, 18, 24, 66, 74, 85, and 97 using heparin-coated tubes (Fisher Scientific) and retro-orbital bleeding. On days on which both blood collection and particle injection were carried out, blood was collected first. Serum was isolated by centrifuging samples at 10,000 g for 10 minutes and analyzed using enzyme-linked immunosorbent assay (ELISA) as follows: 96-well Nunc Polysorp Immuno plates (Thermo Scientific) were coated with 10 µg/well of either PVX in coating buffer (0.05 M Na2CO3, 0.05 M NaHCO3, 0.015 M NaN3 in dH2O, pH 9.6) and incubated overnight at 4°C. After coating, wells were blocked using 200 µL/well blocking buffer (2.5% (w/v) milk, 25% (v/v) FBS in 1x PBS, pH 7.4) at 37°C for 1 hour. After blocking, 100 µL sera (at various dilutions in blocking buffer) were added to the wells and incubated at 37°C for 2 hours. After serum incubation, 100 µL of alkaline phosphatase-labeled goat anti-mouse IgG (Life Technologies, at 1:3,000 in blocking buffer) was added and incubated at 37°C for 1 hour. In between each step, plates were washed four times with washing buffer (0.1% (v/v) Tween 20 in PBS, 200 µL/well). Wells were developed by adding 100 µL of 1-step PNPP substrate (Fisher) for 10 minutes at 4°C. The reaction was stopped using 100 µL of 2M NaOH. Absorbance was read at 405 nm using a Tecan microplate reader.
Sandwich ELISA
Sandwich ELISAs were performed using sera collected from mice treated with various PVX formulations on day 85 (see above). 96-well Nunc Polysorp Immuno plates were coated overnight at 4°C with a rabbit anti-PVX antibody (Pacific Immunology, 1:500 in 150 µL coating buffer (15 mM Na2CO3, 35 mM NaHCO3 in dH2O, pH 5.6)). After coating, wells were blocked using 150 µL/well blocking buffer (1.5% (w/v) BSA in coating buffer) for 1 hour at 37°C. After blocking, 5 µg of PVX, PVX-P5B, or PVX-P20 in 150 µL incubation buffer (0.5% (w/v) BSA and 0.5% (v/v) Tween 20 in PBS, pH 7.4) was added and incubated for 1 hour at 37°C. Next, 150 µL of PVX serum were added at a dilution of 1:25,000 in incubation buffer and incubated for 1 hour at 37°C. Then, 150 µL of alkaline phosphatase-labeled goat anti-mouse IgG (Life Technologies, 1:3,000 in incubation buffer) was added and incubated for 1 hour at 37°C. After each incubation step, plates were washed four times with washing buffer (0.5% (v/v) Tween 20 in PBS, pH 7.4, 200 µL/well). Wells were developed by adding 100 µL of 1-step PNPP substrate (Fisher) for 10 minutes at 4°C. The reaction was stopped using 100 µL of 2 M NaOH. Absorbance was read at 405 nm using a Tecan microplate reader.
Tissue Culture
RAW264.7 macrophages (ATCC) were cultured in Dulbecco’s Minimum Essential Media (DMEM, Life Technologies), supplemented with 10% (v/v) FBS, 1% (v/v) penicillin-streptomycin (PenStrep), and 1% (v/v) L-glutamine and cultured at 37°C and 5% CO2. Bone marrow derived dendritic cells (BMDC) were isolated from wild-type C57/BL6 mice (Taconic) as described previously.[32] Briefly, mice were euthanized by CO2 inhalation and femurs and tibiae were collected and sterilized in 70% (v/v) ethanol. Marrow cavity plugs were washed out using PBS. BMDC were resuspended in DMEM (Sigma Aldrich) and supplemented with 10% (w/v) bovine calf serum, 100 unit/mL penicillin, 100 µg/mL streptomycin, 2 mM L-glutamine, and 15 ng/mL granulocyte macrophage colony-stimulating factor (GM-CSF), plated onto 150-mm dishes and cultured at 37°C and 10% CO2. Three days post-isolation, 80% of the non-adherent cells were removed and centrifuged at 300 g for 5 minutes at room temperature and fresh medium was added. Five days post-isolation, loosely adherent BMDC were collected and resuspended at 1x106 cells/mL. Cells were used between 7 and 10 days post-isolation.
Flow Cytometry
RAW264.7 were grown to confluency, washed three times with PBS, and collected using enzyme-free Hank’s based cell dissociation buffer (Fisher). Cells were added to 96-well v-bottom plates (1x106 cells/200 µL/well) and incubated with 10,000 or 100,000 PVX particles/cell, in triplicate, using A-PVX, A-PVX-P5L, A-PVX-P5B, or A-PVX-P20 for either 20 minutes or 2 hours at 37°C and 5% CO2. BMDC were removed from plates using Lidocaine-EDTA (40 mg/mL Lidocaine, 10 mM EDTA in PBS, pH 7.4), added to 96-well v-bottom plates (1x106 cells/200 µL/well) and incubated for 2 hours with 10,000 particles/cell of A-PVX, A-PVX-P5L, A-PVX-P5B, or A-PVX-P20 at 37°C and 5% CO2. Following incubation, cells were washed twice in FACS buffer (1 mM EDTA, 25 mM HEPES, 1% (v/v) FBS in PBS, pH 7.0) and fixed in 2% (v/v) paraformaldehyde (Electron Microscopy Sciences) in FACS buffer for 10 minutes at room temperature. Cells were washed twice after fixation, resuspended in 400 µL FACS buffer, and stored at 4°C until analysis. Cells were analyzed using a BD LSRII Flow Cytometer and 10,000 gated events were recorded. Data were analyzed using FlowJo 8.6.3 software.
Cytokine Activation
BMDC were seeded onto treated plates at a concentration of 1x106 cells/mL and allowed to grow for 7 days. Before the start of the assay, plates were centrifuged at 400 g for 5 minutes. Culture medium was aspirated and fresh complete high glucose DMEM (HG-DMEM) was added. PVX, PVX-P5B, or PVX-P20 was added to the cells at a concentration of 100,000 particles/cell for 6 or 24 hours. Media was collected by centrifugation and analyzed for TNFα, IL-6, or IL-12 using ELISA kits (Biolegend mouse ELISA MAX Standard (TNFα and IL-6); R&D Systems mouse IL-12/IL-23 p40 Non Allele-specific DuoSet (IL-12)). IL-6 activation was analyzed following 6-hour treatment with cytochalasin D (CytoD, Sigma Aldrich). CytoD (10 µg/mL final concentration) was added 10 minutes prior to addition of PVX formulations. Lipopolysaccharide (LPS) (30 ng/mL final concentration), Pam3Cys (200 ng/mL final concentration), and PolyI:C (25 µg/mL final concentration) were used as positive controls. Untreated media was used as a negative control. All methods were performed as described by the manufacturer’s instructions. Concentration of cytokine for each sample was determined using a standard curve of TNFα, IL-6, or IL-12 of known concentration supplied with each kit.
Inflammasome Activation
BMDC were prepared as described above. Following 8 days of growth, cells were divided into two groups. All groups were treated with PVX, PVX-P5B, or PVX-P20 (100,000 particles/cell) for 6 hours. Group 1 was untreated, except for PVX stimuli. Group 2 was primed with LPS (1 µg/mL final concentration) for 4 hours prior to addition of stimuli. Alum (480 µg/mL final concentration), LPS (1 µg/mL final concentration), Nigericin (10 µM final concentration), and untreated media were used as controls. Media was collected as described above and analyzed for IL-1β using mouse IL-1β/ILF2 DuoSet ELISA kit (R&D Systems). All methods were performed as described by the manufacturer’s instructions. Concentration of IL-1β for each sample was determined using a standard curve of IL-1β of known concentration supplied with the kit.
Chronic Inflammation
Sera were collected from mice on days 66 and 87 post treatment with PVX formulations (see immunogenicity study) and subsequently analyzed for IL-6 content using mouse IL-6 ELISA MAX standard (Biolegend), as described above. Pooled sera were analyzed and analysis was performed in triplicate.
Statistical analysis
Statistical analysis was calculated in Microsoft Excel using a two-tailed Student’s t-Test, assuming unequal variances between the two data sets. Each data set contained a minimum of n=3. Specific n are indicated for each experiment.
Results
Synthesis and characterization of fluorescently labeled and PEGylated PVX filaments
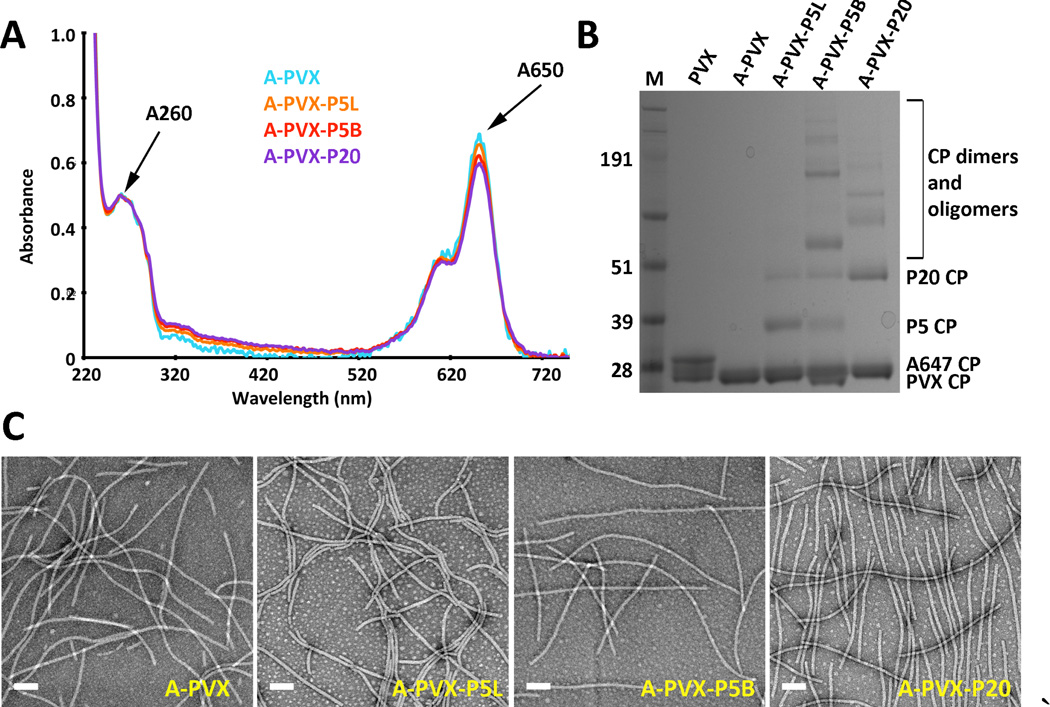
PVX was produced through farming in N. benthamiana plants using previously established protocols and extracted at yields of 20 mg of pure PVX from 100 grams of infected leaf material.[19] PVX filaments were chemically modified with fluorescent dyes and PEG at solvent-exposed lysine side chains.[33] Chemical conjugation was achieved using N-hydroxysuccinimide (NHS)-activated esters of Alexa Fluor 647 (A647), VivoTag-S 750, and/or PEG (conjugated particles denoted A-PVX or V-PVX depending on dye). Specifically, we set out to determine the impact of PEG chain length and conformation on the biological fate of PVX; therefore we considered coating PVX using PEG with a molecular weight of 5,000 Da in linear or branched conformation (P5L and P5B) or linear PEG with a molecular weight of 20,000 Da (P20). Three particle configurations were prepared: PVX-P5L, PVX-P5B, and PVX-P20; for some assays fluorophores were added to allow direct detection of the nanoparticles in cells, blood, or tissues. PEG was added using 10,000 molar excess per PVX and incubated overnight to force maximum coverage. The reaction mixtures were purified by dialysis and the final products were characterized by UV/visible spectroscopy, SDS-PAGE, and TEM (Figure 1).
Figure 1.
Characterization of A-PVX-PEG. A) UV/visible spectroscopy of A-PVX-PEG particles. B) SDS-PAGE after staining with Coomassie blue. Ladder = SeeBlue Plus2 protein standard, numbers represent MW in kDa. C) Negatively stained TEM images of A-PVX-PEG particles. Scale bar = 100 nm. (Double bands for the PVX CPs at ~ 25 kDa can be explained by a proteolytic cleavage that can occur during purification – this cleavage does not effect the reactivity of the particles nor does it compromise the structural integrity of the samples.)
UV/visible spectroscopy was used to determine the degree of fluorescent labeling. Beer-Lambert law and the fluorophore- and PVX-specific extinction coefficients were used determine the concentration of fluorophores and PVX in solution. (The PVX-specific extinction coefficient is εPVX = 2.97 mL mg−1 cm−1 at 260 nm with PVX molecular weight MW = 35 MDa; and the fluorophore-specific extinction coefficients are εA647 = 270,000 M−1 cm−1 at 650 nm, and εVivoTag = 240,000 M−1 cm−1 at 775 nm). The ratio of fluorophore:PVX concentration was used to determine the number of dyes per particle formulation (Figure 1A). On average 20–50% of the surface lysine side chains were modified with fluorophores; the dye concentration was kept constant or was normalized between particle formulations for various assays (see Supporting Information Table S1). For dual-labeled PVX, i.e. PVX filaments with dyes and PEG, dyes were conjugated prior to addition of PEG. We tested the conjugation of PEG with and without fluorophores and found that the presence of dyes did not reduce the PEG conjugation efficiency (see below).
SDS-PAGE and lane analysis tool using ImageJ software was used to determine the number of PEG chains per particle (Figure 1B). Higher molecular weight bands indicated successful PEGylation; in addition to the PVX coat protein band at ~25 kDa, bands corresponding to coat proteins modified with PEG of molecular weights of 5 or 20 kDa were detected (i.e., ~30 kDa and ~45 kDa). Dimers and oligomers of PEGylated PVX coat proteins were also apparent. The degree of coat protein oligomerization for each sample was calculated using band density analysis tool and ImageJ software to determine the percentage of PVX coat proteins in dimers and/or oligomers versus coat protein monomers. Unmodified coat proteins (MW ~25 kDa) and coat proteins modified with a single PEG (MW ~30 kDa or ~45 kDa) were considered coat protein monomers. Any additional higher molecular weight bands were considered dimers or oligomers of PVX-PEG. We determined that 6%, 17%, and 35% of the coat proteins were present as dimers or higher order oligomers for the A-PVX-P5L, A-PVX-P20, and A-PVX-P5B, respectively. Formation of oligomers may be explained by the entangling of the PEGylated coat proteins during the process of denaturing and electrophoresis. A significantly increased number of oligomers was observed for the A-PVX-P5B formulation; in this case, it is possible that multiple coat proteins are entangled or covalently linked together since the branched PEG presents four NHS groups, one at each distal end, therefore possibly interlinking adjacent coat proteins (see Figure 2). Interparticle crosslinking or aggregation was not observed for any of the formulations (Figure 1C).
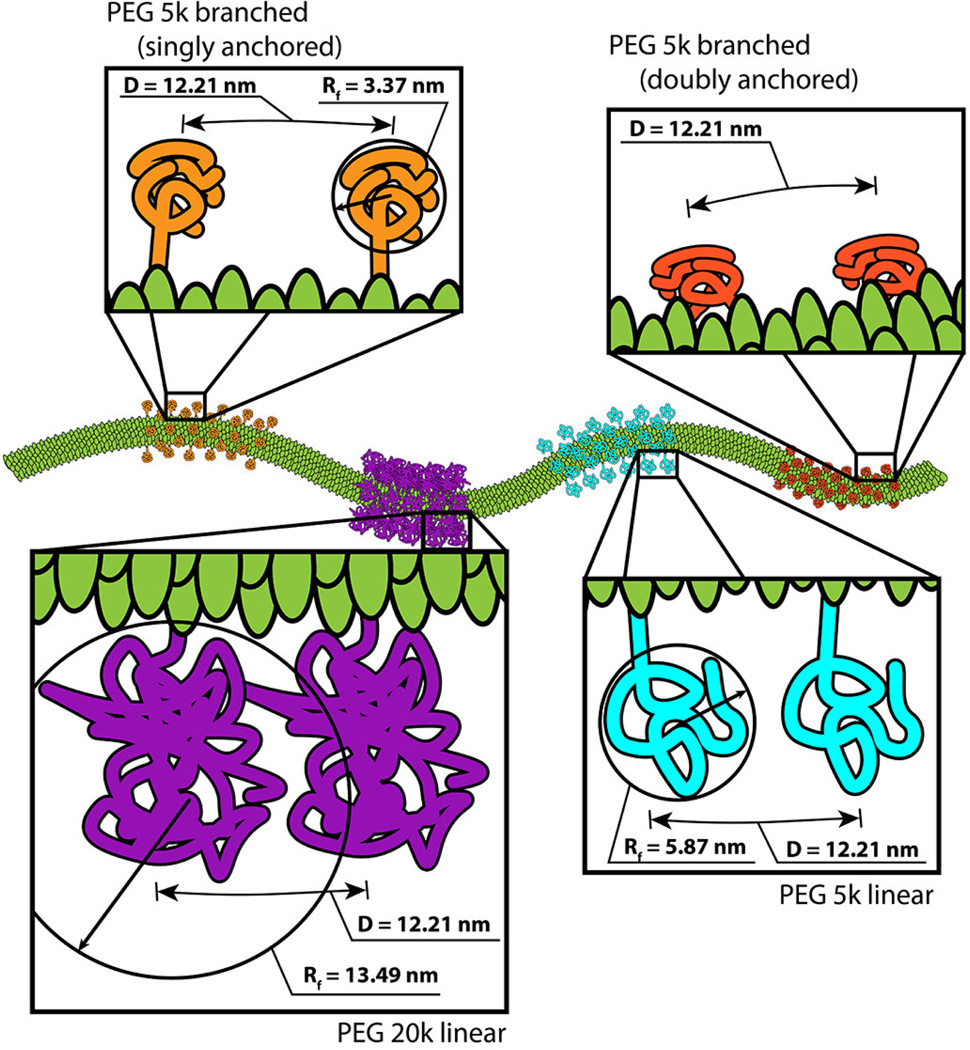
Figure 2.
Schematic (drawn to scale) showing the various PEG conformations presented on the PVX filament. The distance between PEG molecules was estimated based on the grafting density and the dimensions of PVX. The conformation and inter-PEG spacing (D) was determined based on the Flory dimension (RF) of the various PEGs under consideration.
To estimate the degree of PEGylation (number of PEG chains per PVX particle), band density analysis was performed and the number of modified PEG coat proteins was compared to total number of coat proteins. Data indicate that on average 25–50% of the coat proteins (i.e., up to ~600 coat proteins per PVX filament) were modified with PEG, independent of molecular weight or conformation (see Supporting Information Table S1 for a detailed analysis of PEG-per-particle ratios). The batch-to-batch variation may be explained with the inherent instability of the NHS-functional group, which is prone to hydrolysis in aqueous buffer conditions; fresh aliquots were prepared and data were normalized to keep the formulations as consistent as possible.
TEM imaging confirmed that all particle formulation were structurally sound post-chemical modification. Elongated filaments of approximately 515x13 nm were observed in TEM imaging (Figure 1C). While the addition of PEG would increase the hydrodynamic radius of PVX, this was not detectable in TEM imaging using negatively-stained samples.
PEG conformation
Immobilized PEG molecules exist in different conformations based on their grafting density and chain length; one differentiates the brush versus mushroom conformation (as well as various intermediates).[34] We estimated the conformation of the PEG chains grafted to PVX based on the grafting density (PEG chains per PVX particle), the resulting distance between conjugated PEG chains (D), and the Flory dimension (RF) of the various PEG chains. If RF < D, PEG is assumed to exist in a mushroom conformation, and if RF > D, PEG is assumed to exist in a brush conformation.[34]
RF for each PEG was calculated using the following equations:
| (1) |
| (2) |
where a = length of one monomer (0.35 nm for PEG), N = fn (total number of monomers), n = number of monomers (for linear polymers) or number of monomers per branch (for branched polymers), and f = number of branches. For P5L and P20, n is 110 and 440, respectively. For P5B, f is 4 and N is 110. Based on these equations, we calculated RF for P5L, P5B, and P20 to be 5.87 nm, 3.37 nm, and 13.49 nm.
Distance (D) was calculated considering the available surface area of PVX and numbers of PEGs per PVX; we assumed a statistically random distribution and homogenous distribution of PEGs on the PVX surface and considered a ratio of CP:CP-PEG of 3:1, or one PEG for every 3 coat protein residues (Supporting Information, Table S1). The 515x13 nm PVX filament has a surface area of APVX = 21,033 nm2 with each of the 1270 identical coat proteins (CP) providing a surface area of ACP =16.6 nm2 of space. If the PVX CP is assumed to be a square, we determine the distance between two lysines to be 4.1 nm. Since one PEG is found per every three lysine residues, each PEG is 12.2 nm apart (therefore D=12.2 nm).
From these calculations, it appears that for the PVX formulations with short PEG chains, i.e. PVX-P5B and PVX-P5L, the PEG chains are likely to exist in a mushroom conformation (D > RF) and a brush conformation is achieved for the PVX formulations with higher molecular weight PEG, PVX-P20 (Figure 2), however it is also possible that the PEG chains exist in a more complex conformation. For the branched PEG formulation PVX-P5B, as discussed above, it is possible some PEG chains are attached to multiple adjacent coat proteins due to the presence of an NHS group on each arm. In a fully extended conformation, the distance between NHS groups is 19 nm, while considering a more relaxed configuration, the estimated distance would be on the order of 6 nm. The helical pitch and the center-to-center distance of PVX is 2.3 nm. Therefore, in either configuration, it would be feasible for the branched P5B to interlink PVX coat proteins within a single particle. If multiple coat proteins are interlinked, the PEG chains may be presented closer to the PVX surface (Figure 2).
It should be noted that the proposed conformations are models based on theory and mathematical assumptions; there is currently no direct measurement available to probe, measure, or visualize the conformation of PEG chains attached to the (virus-based) nanoparticle.
Pharmacokinetics of PEGylated PVX
The conjugation of PEG to nanoparticle carriers is known to reduce non-specific interactions with tissues, cells, and blood components, therefore increasing nanoparticle bioavailability. We thus set out to investigate how the different PEG configurations affect the PVX pharmacokinetic properties. In this study, we determined the plasma half-life of A-PVX, A-PVX-P5B, and A-PVX-P20. We previously reported the pharmacokinetics of A-PVX-P5L;[35] for comparison these data are shown as a grey line in Figure 3.
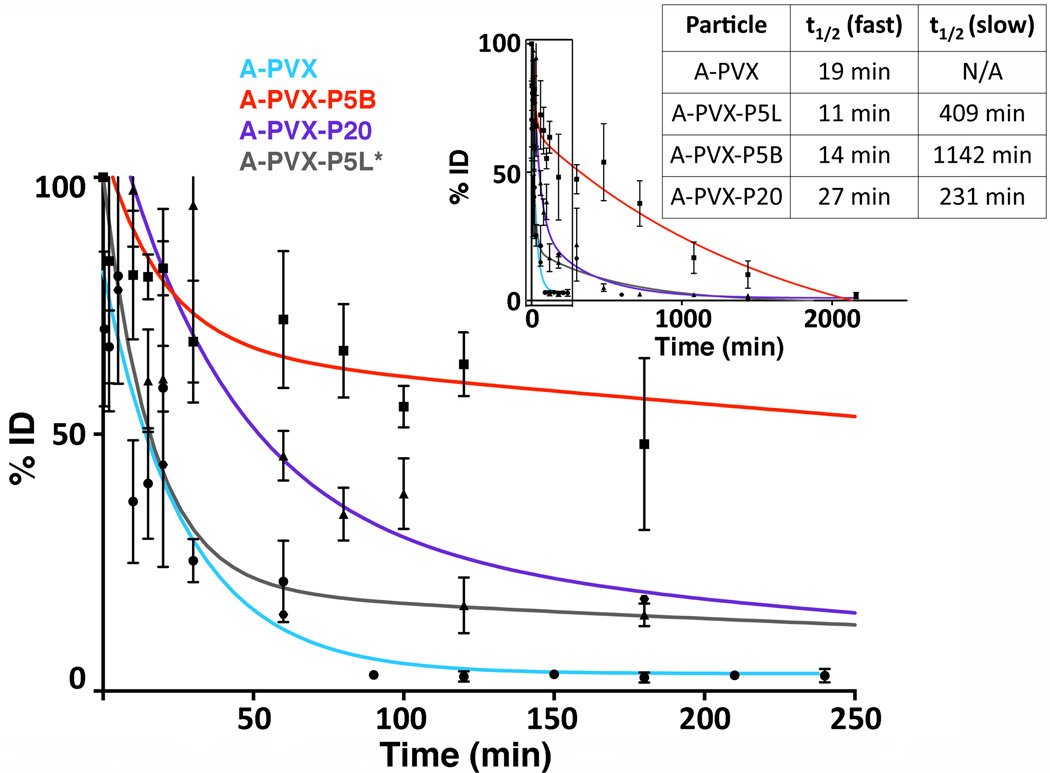
Figure 3.
Pharmacokinetics of A-PVX-PEG. A-PVX-PEG particles were administered intravenously into Balb/C mice (100 µg/mouse); plasma was collected and analyzed at various time points over 2160 minutes (inset). Percent injected dose (%ID) was determined based on a standard curve of known PVX concentration. GraphPad Prism software was used to analyze and fit the data; plasma half-lives are shown in the Table. PVX = light blue line, data points as circles, A-PVX-P5L = grey line, previously reported in the literature,[35] A-PVX-P5B = red line, data points as squares, A-PVX-P20 = purple line, data points as triangles, A-PVX-P5 = red line, data points as squares.
PVX-based formulations were administered intravenously in the tail vein of Balb/C mice, and blood was collected prior to injection and up to 36 hours post-administration. Sera were collected and the percent injected dose (%ID) was determined based on fluorescence obtained in each sample (normalized against a standard curve based on A-PVX samples of known concentration). Data were fitted for one-phase decay or two-phase decay using GraphPad Prism to determine the circulation time and half-life (Figure 3). Data indicate that non-PEGylated PVX exhibited a one-phase decay, while PEGylated PVX formulations follow a two-phase decay. Data indicate that PVX has a half-life of t1/2 = 19 minutes. A-PVX-P5L, A-PVX-P5B, and A-PVX-P20 all showed two-phase decay, which is characterized by an initial fast clearance rate, up to 100 minutes, and a slower clearance for the remaining time: A-PVX-P5B had a fast initial half-life of tI1/2 = 14 minutes and a slower second half-life of tII1/2 = 1142 minutes, while A-PVX-P20 had a fast half-life of tI1/2 = 27 minutes, and a slower second half-life of tII1/2 = 231 minutes. Previously reported,[35] A-PVX-P5L had a tI1/2 = 11 minutes and a tII1/2 = 409 minutes.
Overall, the half-life of PVX is significantly improved upon PEGylation. After a fast initial clearance rate (~10–30 minutes), particles remained in circulation for several hours with tII1/2 of reaching up to 19 hours for the A-PVX-P5B formulation.
Biodistribution and clearance
Fluorescence molecular tomography (FMT) was used to observe biodistribution of V-PVX, V-PVX-P5B, and V-PVX-P20 up to 52 hours post intravenous administration into the tail vein of Balb/C animals. (We previously reported the biodistribution of PVX-P5L[26] and therefore evaluated only the new formulations. The PVX-P5L biodistribution and clearance is discussed in the context of this new data; see Discussion section). For all PVX formulations, fluorescence signals were no longer detectable at 52 hours post-administration using FMT imaging, indicating clearance from the body (Figure 4A). Images were analyzed and the following regions of interest (ROIs) were considered: whole body, liver and spleen, and bladder. Data were normalized to number of dyes per particle for each formulation (Figure 4B).
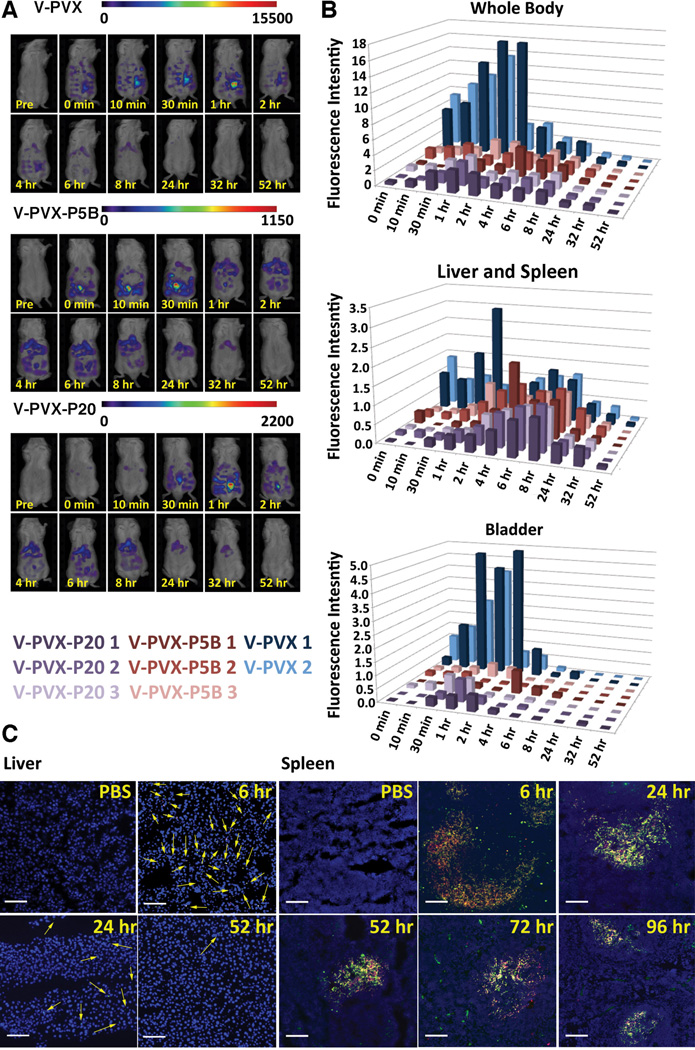
Figure 4.
Biodistribution of native and PEGylated PVX. A) FMT imaging of Balb/C mice following intravenous tail vein injection of V-PVX, V-PVX-P5B, or V-PVX-P20. B) Quantification of FMT signal for the regions of interest (ROIs): whole body (top), liver/spleen (middle), or bladder (bottom); V-PVX (n=2, one animal died prior to start of this study, in blue), V-PVX-P5B (n=3, in red), or V-PVX-P20 (n=3, in purple). C) Fluorescence microscopy tissue sections of liver (left) and spleen (right) from mice post A-PVX-P20 administration (intravenous in the tail vein). Sections were stained with a rabbit anti-PVX antibody and secondary Alexa Fluor 488-labeled antibody (green); the antibody staining colocalized with A-PVX-P20 (red, signals derived form conjugated A647 dye). Nuclei were stained with DAPI (blue). Arrows indicate A-PVX-P20 in liver. Scale bar is 100 µm.
V-PVX (shown in blue) showed maximum fluorescence intensity at one hour post-injection; by 24 hours the fluorescence intensity had dropped below 10% of that maximum. V-PVX showed relatively consistent levels of accumulation in the liver and spleen until 8 hours, at which point the fluorescent signals dropped significantly (which is in agreement with the whole body ROI). At early time points, up to 2 hours post-administration, V-PVX showed increased fluorescence intensity in the bladder compared to the liver and spleen. These data are in good agreement with the pharmacokinetics study, with the one-phase decay for PVX indicative of renal clearance.
It was apparent that V-PVX-P5B and V-PVX-P20 showed much lower tissue accumulation (as indicated by lower fluorescence intensities) overall, with whole body fluorescence intensity levels being about 3-fold lower compared to signals observed for the non-PEGylated PVX formulation. This reduced tissue accumulation is a desired outcome consistent with conjugation of PEG. PEGylation of nanocarriers reduces interaction with blood components, cells, and tissues, and therefore results in a longer circulation time for PEGylated versus native PVX. It should be noted that the threshold of the FMT instrument was set to measure particles deposited in tissues; signals from particles diluted in circulation are not detected. Overall, V-PVX-P5B and V-PVX-P20 showed similar trends. Both formulations accumulated in the liver and spleen until approximately 6 to 8 hours post-administration, and neither showed significant accumulation in the bladder. These data also are in good agreement with pharmacokinetics studies, which indicated a two-phase decay for the PEGylated formulations. The increased diameter of the PEGylated filaments (see Figure 2) may reduce renal filtration, leading to the two-phase pharmacokinetics inherent with combined renal and mononuclear phagocyte system (MPS) clearance.
Immunofluorescence imaging was used to confirm that fluorescence signals detected in FMT correlated with the presence of PVX particles. The possibility exists that the fluorophores would be cleaved in vivo, resulting in imaging of free dye and not PVX particles. To evaluate co-distribution of fluorescent cargo and PVX carriers, confocal microscopy studies were carried out using the PVX-P20 formulation. FMT imaging indicated maximum accumulation of PVX-P20 in liver and spleen sections at 6 hours post-administration and clearance at 52 hours post-administration. Imaging of tissue sections using fluorescence microscopy indicated colocalization of PVX-specific antibodies with the conjugated A647 dye (Figure 4C), which suggests that the fluorescent cargo remains attached to PVX in vivo.
We have previously shown that PVX accumulated in liver macrophages and in the white pulp of the spleen, where it colocalizes with the B cell marker B220;[26] this general pattern is consistent with the present study. While PVX-derived signals were no longer detectable in liver sections 24 hours post-administration, PVX remained detectable in the spleen for up to 96 hours, indicating that imaging of tissue sections is more sensitive than FMT imaging.
Immunogenic properties of PEGylated PVX
Like other viruses, protein-based formulations, or other nanoparticles, plant viruses interact with immune cells and are cleared from the body via the MPS (see Figure 4). In addition, a recent study suggested prevalence of anti-tobacco mosaic virus (TMV) antibodies in humans.[36] Therefore, we set out to determine the IgG titers after repeat administration of various PVX-based formulations and to test whether produced antibodies would recognize the stealth PVX-PEG filaments.
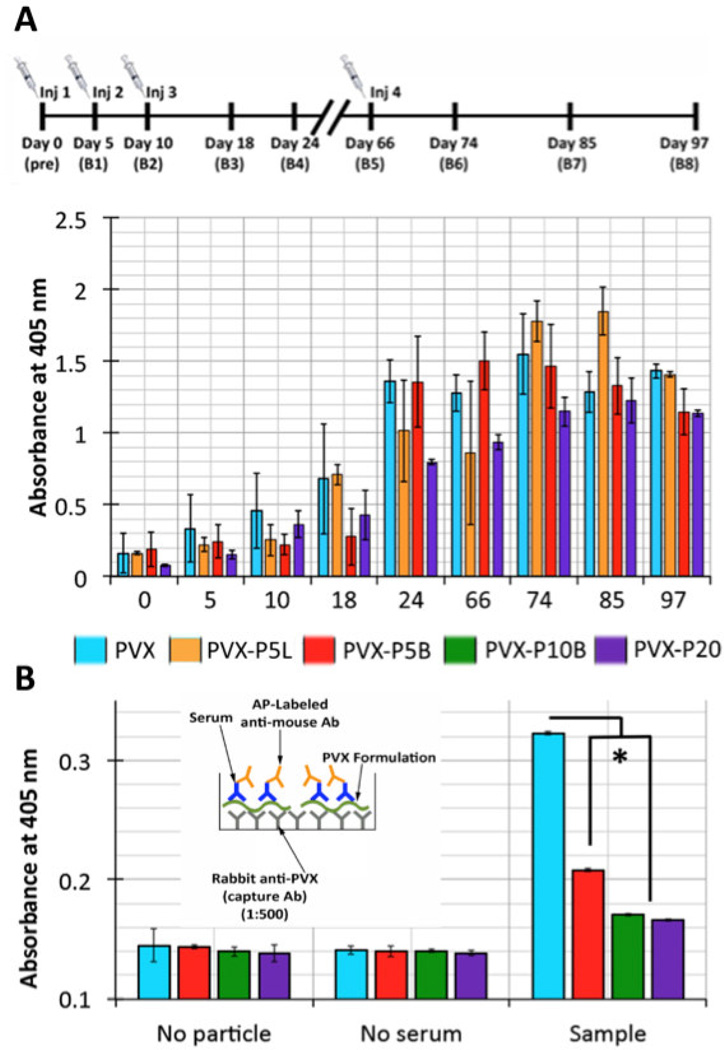
Balb/C mice were treated with PVX-based carriers on days 0, 5, 10, and 66. The initial 5-day interval was chosen because this would follow a typical chemotherapy schedule (the long-term goal is to develop PVX as a drug delivery vehicle), while the late time point (66 days) was chosen as a secondary booster. Blood and sera were collected over a 3-month time frame (see immunization schedule, Figure 5A). The sera were analyzed for IgG content specific for PVX using ELISA in which sera were probed against PVX-coated plates. Data indicate that for each formulation analyzed, PVX-specific IgG antibodies were raised. It was interesting to note that the overall IgG titers were lower for animals treated with PVX-P20 compared to all other PVX-based formulations (Figure 5A). Future studies will set out to determine the antibody titers to make quantitative comparisons between the formulations.
Figure 5.
Anti-PVX IgG titers and antibody-to-PVX binding. A) Timeline of treatment schedule; time points of PVX administration and blood collection are indicated. Anti-PVX IgG titers of sera from mice treated with PVX, PVX-P5L, PVX-P5B, and PVX-P20. B) Sandwich ELISA testing the reactivity of anti-PVX antibodies against PVX, PVX-P5B, PVX-10B, and PVX-P20. All data were analyzed using Excel software and Student’s t-test * p < 0.001; Ab = antibody; AP = alkaline phosphatase.
It was found that PVX-specific antibodies were raised in mice regardless of shielding. Therefore we sought to address whether these antibodies would recognize the stealth filaments. A sandwich ELISA was performed, in which plates were coated with an anti-PVX antibody produced in rabbits immunized with native PVX, and native and PEGylated PVX particles were subsequently captured. The captured PVX samples were then probed with the sera from immunized mice and detection was carried out using an anti-mouse secondary antibody. Data indicate a significant decrease in particle recognition for both PEGylated samples studied, PVX-P5B and PVX-P20, compared to non-PEGylated PVX (Figure 5B), with PVX-P20 being more efficiently shielded compared to PVX-P5B. This data indicates that when covering a sufficiently large surface area with stealth coatings, effective shielding from antibodies is indeed achieved.
To further test this, we synthesized a PVX modified with 4-arm branched PEG but with a molecular weight of 10,000 Da to increase surface coverage and therefore minimize antibody recognition. PVX-P10B was synthesized with 609 PEG chains; based on the RF = 5.11 nm for P10B, and the number of PEG chains attached per PVX, the PEG chains are likely displayed in a mushroom conformation (D = 8.14 nm, and therefore D > RF) covering the entire surface area of the filamentous particle (Supporting Information Figure S3 and Table S1). Sandwich ELISA to probe for antibody recognition indicated that PVX-P10B was indeed effectively shielded from antibody recognition: immune recognition of all PEGylated samples was significantly reduced with PVX-P10B and PVX-P20 only showing 5% of the signal while PVX-P5B showed 40% of signal compared to non-shielded PVX (Figure 5B).
PVX–cell interactions
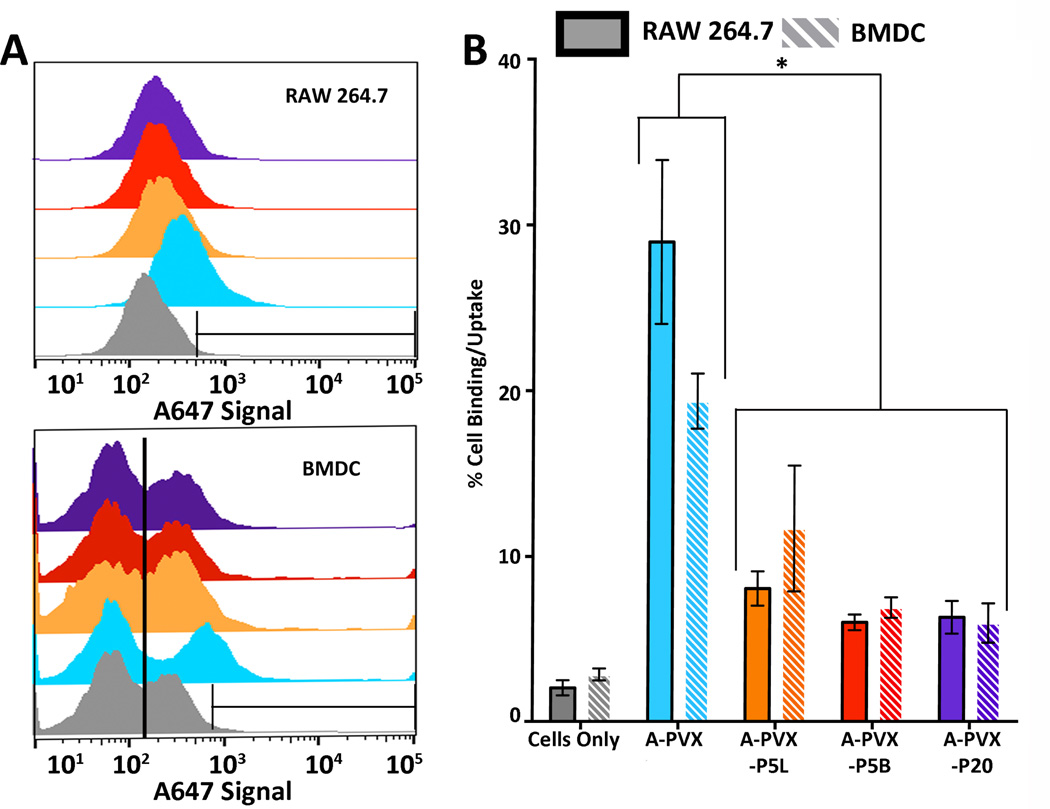
To correlate the in vivo properties, PVX uptake by immune cells was studied. RAW264.7 macrophages (ATCC) and primary bone marrow derived dendritic cells (BMDC) were used. Concentration- (10,000 vs. 100,000 particles per cell) and time-dependent studies (20 minutes vs. 2 hour incubation) were performed and cell interactions were quantified using flow cytometry (Figure 6 and Supporting Information Figure S1).
Figure 6.
A-PVX-PEG–cell interactions measured by flow cytometry. A-PVX-PEG formulations were incubated with RAW264.7 and BMDC at a concentration of 10,000 particles/cell for 2 hours. There are approximately 1x106 circulating monocytes (macrophage precursors) in a mouse. For in vivo studies, we injected 100 µg of PVX per mouse, resulting in approximately 1,000,000 particles/cell immediately after injection. 10,000 and 100,000 particles/cell (see Figure S1) therefore reflects in vivo conditions. A) Histograms of A647 signal versus count for A-PVX-PEG particles in RAW264.7 (top) and BMDC (bottom). Gray = cells only, light blue = A-PVX, orange = A-PVX-P5L, red = A-PVX-P5B, purple = A-PVX-P20. Any counts within the indicated gate represent positive cells. At least 10,000 events were considered; all studies were done in triplicate and data were analyzed using FlowJo software. B) Statistical analysis and quantitative data showing percent cell uptake of A-PVX-PEG particles in RAW264.7 or BMDC. * p<0.05.
Data indicate that immune cell uptake of PVX is concentration- and time-dependent. Overall PEGylated PVX particles are less prone to macrophage and BMDC uptake; i.e. uptake of PEGylated PVX formulations are delayed and reduced as a result of PEG coating and shielding (Figure 6). While 29.0% of macrophages and 19.4 of BMDC stained positive for A-PVX, only 8.1%, 6.0%, and 6.3% of macrophages and 11.7%, 6.9%, and 6.0% of BMDC stained positive for A-PVX-P5L, A-PVX-P5B, and A-PVX-P20, respectively. No significant differences in cell uptake were observed between any particles (native and PEGylated) when incubated for 20 minutes at a concentration of 10,000 particles/cell; at early time points and low particle concentrations, PVX–cell interactions are negligible (Supporting Information Figure S1). Similarly, no significant differences were observed when particles were incubated with cells for 2 hours at a concentration of 100,000 particles/cell; at longer time points and high concentration all PVX formulations are taken up by macrophages (Supporting Information Figure S1). Differences between PEGylated and native PVX became apparent when particles were incubated for 20 minutes at a concentration of 100,000 particles/cell and when particles were incubated for 2 hours at a concentration of 10,000 particles/cell. Under these conditions, statistically significant decreases in percent cell uptake for PEGylated particles were detected compared to native PVX. However, there were no differences in percent cell uptake comparing the different PEGylated formulations, A-PVX-P5L, A-PVX-P5B, and A-PVX-P20. PVX–cell interactions in macrophages and BMDC showed similar trends (Figure 6). (The two populations observed using BMDC maybe explained by a mixture of non-activated and activated cells; cells were not sorted for the uptake studies). Overall, these data indicate that PEGylation results in a decreased rate of cell uptake in immune cells.
Cytokine activation in vitro and in vivo
Knowing that PVX interacts with macrophages and BMDC, we studied whether PVX–cell interactions would trigger inflammatory cytokine signaling. To do this, we incubated PVX, PVX-P5B, or PVX-P20 with BMDC for 6 or 24 hours and analyzed the media for TNFα, IL-6, or IL-12 using ELISA kits.
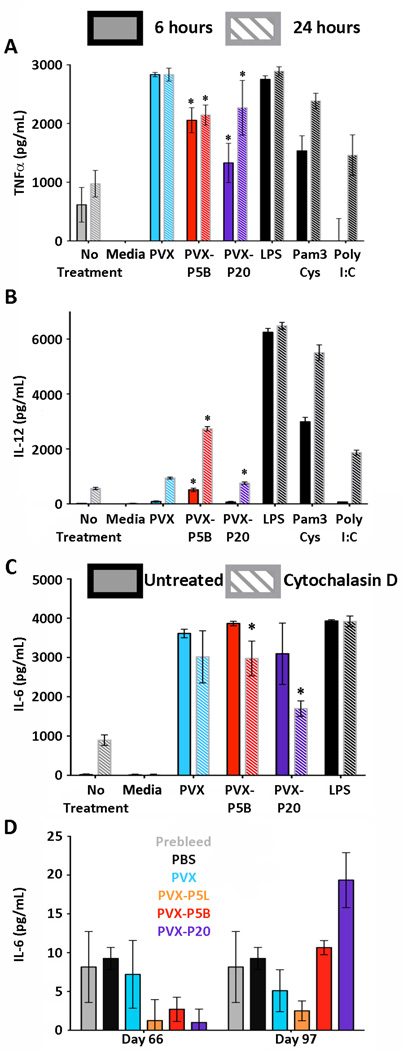
Treatment with non-PEGylated PVX resulted in the largest amount of TNFα activation, with detectable levels of 2,834 pg/mL at 6 and 24 hours post-treatment. PEGylated PVX-P5B and PVX-P20 showed significantly reduced amounts of TNFα activation compared to PVX after both 6 and 24 hours of incubation (2,057 and 1,330 pg/mL, respectively at 6 hours and 2,146 and 2,269 pg/mL, respectively at 24 hours). There were no differences comparing the two PEGylated formulations (Figure 7A). Reduction of cytokine activation is in agreement with reduced cell interactions of PEGylated PVX.
Figure 7.
Cytokine activation in BMDC induced by PVX-PEG particles. Induction of A) TNFα or B) IL-12 after 6 (solid) or 24 hour (striped) incubation with PVX-PEG formulations. C) IL-6 production after 6 hours in the absence (solid) or presence (striped) of cytochalasin D. D) Analysis of mouse sera for IL-6 after repeat intravenous administration of the PVX-PEG particles. * p<0.05.
Treatment of BMDC with PVX, PVX-P5B, and PVX-P20 induced IL-12 production. However, modest levels of 939 pg/mL and 756 pg/mL of IL-12 were reached when treated with PVX and PVX-P20, respectively (Figure 7B). Treatment of PVX-P5B resulted in elevated levels of IL-12 production, reaching levels of 2734 pg/mL at 24 hours post-treatment, which is a threefold increase compared to IL-12 levels from cells treated with PVX or PVX-P20. These data indicate that the branched PEG may be processed differently, which may be a result of interparticle crosslinking (see Figures 1 and 2).
PVX, PVX-P5B, and PVX-P20 triggered production of IL-6, reaching levels of up to 4,000 pg/mL (Figure 7C). To determine whether cytokine activation is correlated with cell uptake, we studied different cell uptake mechanisms. BMDC were treated with cytochalasin D (CytoD) to block actin polymerization, therefore preventing phagocytosis.[37] In all cases, IL-6 production was reduced upon CytoD treatment, with statistically significant differences observed for the PEGylated PVX formulations: IL-6 production was reduced by 23% and 45% when cells were treated with CytoD prior to treatment with PVX-P5B and PVX-P20, respectively (Figure 7C).
Although both PVX-P5B and PVX-P20 showed statistically significant decreases in IL-6 production after treatment with CytoD, a complete knockdown of IL-6 production was not achieved. This indicates that multiple cell entry mechanisms may play a role and that IL-6 production is not solely linked to phagocytosis. PVX-induced IL-6 activation may be a result of either particle interaction with cell surface receptors or non-phagocytosis cellular uptake.
In addition to studying inflammatory cytokine signaling, we also set out to determine whether PVX-treatment would result in inflammasome-mediated cytokine expression, since activation of the inflammasome has been linked to nanoparticles;[38–40] specifically we studied IL-1β activation (Supporting Information Figure S2). Prior to incubation with PVX formulations, samples were primed with lipopolysaccharide (LPS) for 4 hours to increase the amount of pro-IL-1β available; alum treatment was used as a positive control for inflammasome activation. Data indicate that IL-1β is not produced in BMDC after PVX treatment. No significant differences were observed comparing non-PEGylated and PEGylated PVX formulations, and there was no difference comparing non-primed and LPS-primed BMDCs. Overall, these results indicate that PVX, PVX-P5B, and PVX-P20 do not activate inflammasome-mediated cytokine, IL-1β.
Data from in vitro studies suggest that PVX-based formulations are taken up by macrophages and dendritic cells, with PEGylated formulations showing reduced cell uptake rates. Further data suggest that PVX–BMDC interactions trigger expression of pro-inflammatory cytokines when incubated with BMDCs in vitro (Figure 7A–C). Therefore, we studied whether repeated treatment of mice resulted in chronic inflammation; this was assessed by analyzing IL-6 levels from sera collected from immunized mice during the immunogenicity studies on days 66 and 97 (Figure 7D). By day 66, the mice had received three treatments of PVX (on days 0, 5, and 10), and by day 95, the mice were treated with the initial three injections followed by an additional booster on day 66 (see Figure 5A). Elevated levels of IL-6 were not detectable for any of the sera analyzed (Figure 7D), therefore indicating that chronic inflammation is not induced.
Discussion
In summary, we synthesized stealth filaments using the PVX platform modified with PEG. Pharmacokinetics, biodistribution, as well as potential immune and inflammatory responses were evaluated in response to stealth filaments coated with PEG chains of varying length and conformation. PEGylation effectively reduces (but does not prevent) interaction with immune cells in vitro and in vivo. While stealth coatings do not eliminate production of antibodies against the carrier (future studies will set out to determine antibody titers as a function of PEG coating), immune recognition and antibody neutralization can be overcome upon polymer coating. Stealth filaments show rapid tissue clearance through combined renal and MPS clearance mechanisms. Further, tissue compatibility was indicated, with no apparent inflammatory signaling in vivo. Furthermore, stark differences were noted in the pharmacokinetic properties; tailoring PEG chains length and conformation (brush vs. mushroom) allows tuning of the pharmacokinetics, yielding long-circulating stealth filaments for applications in nanomedicine.
While the development pipeline of nanoparticle-based nanomedicines is moving rapidly, a fundamental understanding of the in vivo biology of various systems is lacking. Recent research has shown cell target specificity results from a combined contribution by particle morphology and surface chemistry.[11, 12] Stealth coating is an accepted technology to increase the bioavailability of inorganic and organic nanoparticle formulations. Increased plasma circulation is a result of PEG conjugation to nanoparticles and has been observed for various synthetic platforms,[41, 42] and this in agreement with the data presented in this study (Figure 3). Previous studies have shown that protein adsorption is impacted by PEG density and molecular weight on both 2D surfaces and 3D nanoparticle surfaces and increased pharmacokinetics is explained by the fact that the hydrophilic PEG shell reduces the rate of formation of the so-called ‘protein corona’, which consists of plasma proteins (opsonins, complement, albumin, fibrinogen, immunoglobulins, etc.[43–46]) surrounding the nanocarrier, which in turn results in reduced uptake by immune cells and diminished deposition in non-target organs.[42, 47–49] Indeed, we show that PEGylation of PVX results in decreased liver and spleen deposition in vivo (Figure 4) and reduced uptake in mononuclear phagocytotic cells in vitro (Figure 6). These findings are in good agreement with reports on other PEGylated plant viruses, such as cowpea mosaic virus (CPMV),[50] mammalian viruses, including adenoviruses under development for gene therapy applications,[51] as well as synthetic nanoparticles.[52–55]
Pharmacokinetic data indicate that the addition of PEG induced a switch from one-phase to two-phase plasma clearance kinetics: native PVX exhibited a one-phase clearance, which is indicative of clearance by primarily renal filtration. Upon PEGylation, PVX exhibited a two-phase clearance pattern, which is due to combined renal filtration and MPS clearance (Figure 3); these findings are in agreement with other reports.[56] Biodistribution profiles of PVX-PEG stealth filaments reflect the pharmacokinetic studies. As predicted by pharmacokinetics, injection of native PVX is followed by rapid renal clearance. The size cut-off for glomerular filtration is sub-20 nm;[57] therefore, PVX aligned in the flow is theoretically thin enough to pass through the renal filtration system. Indeed, others have reported that carbon nanotubes are cleared through renal filtration,[58, 59] supporting the hypothesis that elongated, rod-shaped nanomaterials with diameters less than 20 nm can be cleared by renal filtration. PEGylation of PVX increases its hydrodynamic radius (Figure 2), thereby reducing but not completely eliminating renal filtration. Reduction in renal clearance rates leads to MPS clearance; however, it should be noted that liver and spleen deposition of stealth PVX-PEG particles is generally reduced compared to naked PVX (Figure 4).
Whole mouse imaging using FMT indicates rapid tissue clearance of stealth filaments (within 3 days, Figure 4). Sectioning and immunofluorescence also confirm rapid tissue clearance of the filamentous nanoparticles from the liver, while small quantities of particles were still detectable in the spleen at 96 hours post-administration, most likely as a result of less metabolic activity in the spleen. Within the spleen, PVX-PEG particles travel through the red pulp (~6 hours post-administration) and then accumulate within the white pulp of the spleen (at ~24 hours post-administration), where the B cells reside. This pattern is in agreement with our previous observation of PVX[26] and TMV,[60] and it is also consistent with the deposition of gold nanoparticles within the spleen.[61]
Overall, biodistribution of native and PEGylated PVX is consistent with trends seen for a wide range of other nanoparticles, including various viral nanoparticle systems.[60, 62–65] For example, 30 nm-sized icosahedrons CPMV and phage Qβ predominantly accumulate in the liver, while 300x18 nm rod TMV, 30 nm-sized phage MS2, and 900x6 nm filamentous phage M13 were found in both the liver and spleen.[60, 62–65] Of note, a different biodistribution pattern was observed for the 30 nm-sized plant virus cowpea chlorotic mottle virus (CCMV) with much of the administered dose accumulating in the thyroid.[66]
While the overall biodistribution of PVX-PEG stealth filaments is consistent with synthetic nanocarriers, it is of importance to note that compared to synthetic nanoparticles, several of which persist within the body for weeks and longer, virus-based materials are biodegradable and thus are removed from the body within days (Figure 4). For some nanoparticles, such as gold, silica, and carbon nanostructures, clearance by the MPS, and ultimately from the body, can be slow. Synthetic materials, such as gold,[67, 68] polymeric materials,[69] iron oxide nanoparticles,[70] and carbon nanotubes,[71] may persist in the liver for months after injection, which may lead to long-term adverse effects. Fast breakdown and clearance of the protein-based stealth filaments may be explained by the fact that the cellular machinery such as proteases, hydrolyases, and other enzymes are naturally trained to break down proteinaceous materials. In addition, some synthetic nanoparticles have previously been reported to accumulate in lungs, heart, and skin.[68, 72–74] PVX-based stealth filaments were not detected in any of these tissues. It is especially interesting to note that lung deposition did not occur, because other elongated, synthetic systems commonly accumulate in the lungs.[72, 74] A main difference is the flexible nature of the soft, PVX-based nanomaterial vs. rigid hard materials such as carbon nanotubes or silica-based nanorods.
Our data show that PEGylation successfully reduces the immunogenic properties of filamentous PVX, including reduced uptake in immune cells and overcoming antibody recognition (Figures 5 and 6). Since most foreign nanomaterials, including synthetic nanoparticles, are immunogenic, leading to production of carrier-specific antibodies,[66, 75–79] it is most important to understand whether stealth particles are recognized and potentially neutralized upon repeated administration. PEGylation indeed significantly reduced immune recognition of PEGylated stealth filaments (Figure 5). This is consistent with reports on adeno-associated viruses and adenoviruses, both of which are protected from neutralizing antibodies in vitro and in vivo upon PEGylation.[80–83] The stealth filaments therefore provide an attractive platform for drug delivery applications, overcoming immune recognition opens the door for repeat administration, a requirement for treatment of oncological diseases.
To gain insight into the potential inflammatory response, we investigated in vitro uptake by immune cells and cytokine activation, both in vitro and in vivo. As with other stealth nanomaterials,[50–55] PEGylated PVX showed a decreased rate of uptake into immune cells, and this correlated to reduced production of inflammatory cytokines, such as TNFα, IL-12, and IL-6, which also is in agreement with previous reports investigating the in vivo biology of other virus-based platforms.[51, 84] Most interesting, neither native nor PEGylated PVX induced activation of the inflammasome-mediated cytokine, IL-1β; this is in contrast to many synthetic nanoparticles.[72, 74, 76, 85–87] It is also important to note that while in vitro induction of inflammatory signaling was observed, no chronic inflammation was observed in vivo after repeat administration of naked or stealth filaments, as monitored by IL-6 levels (Figure 6); this is in contrast to mammalian virus-based gene delivery vectors, where elevated IL-6 levels have been reported.[51] The plant pathogen is less likely to trigger adverse side effects because it is less likely to interact with the mammalian cell machinery or receptors triggering signal transduction events.
A common challenge for nanoparticle-based delivery systems is to determine the correct balance between circulation time, targeting, and clearance. Tailoring PVX with various PEG coatings, we were able to tune the pharmacokinetic properties of stealth PVX. For the linear PEG formulations we found that higher molecular weight PEG increases circulation time more significantly[48, 88] than lower molecular weight PEG. Biochemical data (Figure 1) in combination with modeling (Figure 2) indicates that PVX-P20 presents the PEGs in a brush-like conformation, while PVX-P5L presents PEG chains in a mushroom conformation. More efficient surface coverage in combination with the brush conformation of the higher molecular weight PEGs results in a more efficient stealth effect. This is consistent with other reports that demonstrate that a brush-like conformation of PEG is most effective to decrease clearance by immune cells.[89]
Furthermore, our data indicate that branched PEG (PVX-P5B) proved more effective than linear PEG (PVX-P5L) in increasing circulation time. Based on Flory Dimension and grafting distance along PVX, both P5L and P5B present in a mushroom conformation. However, P5B is more densely packed and closer to the surface of PVX due to simultaneous binding of multiple coat proteins, therefore potentially shielding the particle more efficiently. While modeling indicates that the PEG arms of the P5B formulations are folded into a mushroom-based conformation, it is also possible that the non-conjugated PEG arms are presented as a brush-like structure, extending off the surface and leading to the highly increased pharmacokinetic profiles observed, with a phase II half-life of almost 20 hours. In ongoing studies we are developing novel imaging methods to directly visualize the PEG chains immobilized on the particle surface to validate or refine the proposed structural models.
The ability to tailor circulation times based on surface chemistry is interesting and allows customization of PVX-PEG filaments for specific desired applications. For example, longer circulation times may be desirable for therapeutic applications and passive tumor targeting (Figure 9),[90] while shorter circulation times are advantageous for receptor-targeted contrast agents or to reduce adverse systemic effects when highly toxic payloads are delivered.[91, 92]
Understanding the in vivo properties of nanoparticles is important for translation, and it is equally important to understand efficacy and sensitivity, as well as clearance profiles. In addition to safety and biocompatibility considerations, another important point to consider for a nanoparticle formulation’s foray into the clinical world is its reproducibility and the completeness of the material’s characterization. In particular, it is important that each compound/nanoparticle is the same. The nano-manufacturability and engineerability of plant virus-based materials is a strength of the material, as size and monodispersity impact in vivo distribution. Therefore, the goal must be to make protocols for the synthesis of uniform nanoparticle formulations. Since they are genetically controlled, to a first approximation, every particle is identical. Furthermore, genetic engineering provides a means of controlling the formulation with atomic precision. Most chemical modifications are stochastic, but it is relatively straightforward to determine concentrations of the protein-based carrier along with how many sites are conjugated per nanoparticle through the use of colorimetric assays, gel electrophoresis, UV/visible spectroscopy, and/or other methods. The same level of quantification is difficult to achieve with liposomes or any other nanoparticle, as it is challenging to determine the number of nanoparticles in solution.
Conclusions
Filamentous plant viruses, such as PVX, present a novel class of nanom aterials with potential applications in drug delivery and tissue-specific imaging. We previously showed that filamentous plant virus-based nanoparticles home to tumor tissues, extravasate, and penetrate into the cancerous tissue; data indicate that this passive tumor accumulation is shape-mediated.[35] Recent data indicating advantageous behaviors of elongated nanomaterials for in vivo medical applications have instigated the search and development for novel chemistries and synthetic routes to yield high aspect ratio nanomaterials. Produced by nature, the highly symmetrical and proteinaceous filaments offer an ideal scaffold for the manufacturing of such materials. In this paper, we discuss the development of stealth filaments by grafting PEG chains to the 515x13 nm PVX-based protein scaffold. We demonstrate increased bioavailability with pharmacokinetics tunable through PEG chain length and conformation. Polymer coating using PEG effectively reduced interaction with immune cells and limited immune recognition by antibodies. The stealth filaments were found to be compatible with tissues and did not induce extended inflammatory responses upon repeat administration in animals. Further improvements may be achieved through incorporation of alternative polymers, such as zwitterionic species. Furthermore, higher polymer densities and favorable brush conformations may be achieved using higher-order branched PEG molecules (e.g. 8-arm PEGs vs. 4-arm PEGs; as an alternative one might consider grafting polymers from the protein scaffold rather than grafting-to).[93] Further surface chemistry optimization and a more comprehensive understanding of the biocompatibility of this filamentous platform technology will pave the way for potential translational research in the future.
Supplementary Material
Acknowledgements
This work was supported by the following grants: NSF CHEM MSN 1306447 (to N.F.S. and J.K.P.) for synthesis and characterization of PVX-polymer hybrids, Ohio Cancer Research Associates (to N.F.S.) and Mt. Sinai Foundation (N.F.S.) for in vivo evaluation of PVX-polymer hybrids, NIH R01 GM36387 (to G.R.D.), and NIH grant NCI R25 CA148052 Cancer Pharmacology training grant (to K.L.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cai S, Vijayan K, Cheng D, Lima EM, Discher DE. Micelles of different morphologies--advantages of worm-like filomicelles of PEO-PCL in paclitaxel delivery. Pharm Res. 2007;24:2099–2109. doi: 10.1007/s11095-007-9335-z. [DOI] [PubMed] [Google Scholar]
- 2.Chauhan VP, Popovic Z, Chen O, Cui J, Fukumura D, Bawendi MG, Jain RK. Fluorescent nanorods and nanospheres for real-time in vivo probing of nanoparticle shape-dependent tumor penetration. Angewandte Chemie (International ed. 2011;50:11417–11420. doi: 10.1002/anie.201104449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christian DA, Cai S, Garbuzenko OB, Harada T, Zajac AL, Minko T, Discher DE. Flexible filaments for in vivo imaging and delivery: persistent circulation of filomicelles opens the dosage window for sustained tumor shrinkage. Molecular pharmaceutics. 2009;6:1343–1352. doi: 10.1021/mp900022m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, Ferrari M. Size and shape effects in the biodistribution of intravascularly injected particles. Journal of controlled release : official journal of the Controlled Release Society. 2010;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentile F, Chiappini C, Fine D, Bhavane RC, Peluccio MS, Cheng MM, Liu X, Ferrari M, Decuzzi P. The effect of shape on the margination dynamics of non-neutrally buoyant particles in two-dimensional shear flows. J Biomech. 2008;41:2312–2318. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Lee SY, Ferrari M, Decuzzi P. Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 8.Doshi N, Prabhakarpandian B, Rea-Ramsey A, Pant K, Sundaram S, Mitragotri S. Flow and adhesion of drug carriers in blood vessels depend on their shape: a study using model synthetic microvascular networks. Journal of controlled release : official journal of the Controlled Release Society. 2010;146:196–200. doi: 10.1016/j.jconrel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S-Y, Ferrari M, Decuzzi P. Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 10.Tan J, Shah S, Thomas A, Ou-Yang HD, Liu Y. The influence of size, shape and vessel geometry on nanoparticle distribution. Microfluid Nanofluidics. 2013;14:77–87. doi: 10.1007/s10404-012-1024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vacha R, Martinez-Veracoechea FJ, Frenkel D. Receptor-mediated endocytosis of nanoparticles of various shapes. Nano letters. 2011;11:5391–5395. doi: 10.1021/nl2030213. [DOI] [PubMed] [Google Scholar]
- 12.Arnida, Janát-Amsbury MM, Ray A, Peterson CM, Ghandehari H. Geometry and surface characteristics of gold nanoparticles influence their biodistribution and uptake by macrophages. European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft für Pharmazeutische Verfahrenstechnik eV. 2011;77:417–423. doi: 10.1016/j.ejpb.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firme CP, 3rd, Bandaru PR. Toxicity issues in the application of carbon nanotubes to biological systems. Nanomedicine : nanotechnology, biology, and medicine. 2010;6:245–256. doi: 10.1016/j.nano.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chitale R. Merck hopes to extend gardasil vaccine to men. J Natl Cancer Inst. 2009;101:222–223. doi: 10.1093/jnci/djp014. [DOI] [PubMed] [Google Scholar]
- 16.Liu TC, Galanis E, Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat Clin Pract Oncol. 2007;4:101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- 17.Shirakawa T. Clinical trial design for adenoviral gene therapy products. Drug News Perspect. 2009;22:140–145. doi: 10.1358/dnp.2009.22.3.1354090. [DOI] [PubMed] [Google Scholar]
- 18.Shukla S, Dickmeis C, Nagarajan AS, Fischer R, Commandeur U, Steinmetz NF. Molecular farming of fluorescent virus-based nanoparticles for optical imaging in plants, human cells and mouse models. Biomaterials science. 2014;2:784–797. doi: 10.1039/c3bm60277j. [DOI] [PubMed] [Google Scholar]
- 19.Lee KL, Uhde-Holzem K, Fischer R, Commandeur U, Steinmetz NF. Genetic engineering and chemical conjugation of potato virus X. Methods Mol Biol. 2014;1108:3–21. doi: 10.1007/978-1-62703-751-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shukla S, Ablack AL, Wen AM, Lee KL, Lewis JD, Steinmetz NF. Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle Potato virus X. Molecular pharmaceutics. 2013;10:33–42. doi: 10.1021/mp300240m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinmetz NF, Cho CF, Ablack A, Lewis JD, Manchester M. Cowpea mosaic virus nanoparticles target surface vimentin on cancer cells. Nanomedicine (Lond) 2011;6:351–364. doi: 10.2217/nnm.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinmetz NF, Ablack A, Hickey JL, Ablack J, Manocha B, Mymryk JS, Luyt LG, Lewis JD. Intravital Imaging of Human Prostate Cancer using Viral Nanoparticles Targeted to Gastrin-Releasing Peptide Receptors. Small. 2011 doi: 10.1002/smll.201000435. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang RK, Steinmetz NF, Fu CY, Manchester M, Johnson JE. Transferrin-mediated targeting of bacteriophage HK97 nanoparticles into tumor cells. Nanomedicine (Lond) 2011;6:55–68. doi: 10.2217/nnm.10.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovlid ML, Steinmetz NF, Laufer B, Lau JL, Kuzelka J, Wang Q, Hyypia T, Nemerow GR, Kessler H, Manchester M, Finn MG. Guiding plant virus particles to integrin-displaying cells. Nanoscale. 2012;4:3698–3705. doi: 10.1039/c2nr30571b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokorski JK, Hovlid ML, Finn MG. Cell targeting with hybrid qbeta virus-like particles displaying epidermal growth factor. Chembiochem. 2011;12:2441–2447. doi: 10.1002/cbic.201100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla S, Wen AM, Ayat NR, Commandeur U, Gopalkrishnan R, Broome AM, Lozada KW, Keri RA, Steinmetz NF. Biodistribution and clearance of a filamentous plant virus in healthy and tumor-bearing mice. Nanomedicine (Lond) 2014;9:221–235. doi: 10.2217/nnm.13.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 28.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond) 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts MJ, Bentley MD, Harris JM. Chemistry for peptide and protein PEGylation. Adv Drug Deliv Rev. 2002;54:459–476. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 30.Veronese FM, Pasut G. PEGylation, successful approach to drug delivery. Drug Discov Today. 2005;10:1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 31.Wattendorf U, Merkle HP. PEGylation as a tool for the biomedical engineering of surface modified microparticles. J Pharm Sci. 2008 doi: 10.1002/jps.21350. [DOI] [PubMed] [Google Scholar]
- 32.Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science. 2002;296:1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 33.Steinmetz NF, Mertens ME, Taurog RE, Johnson JE, Commandeur U, Fischer R, Manchester M. Potato virus X as a novel platform for potential biomedical applications. Nano letters. 2010;10:305–312. doi: 10.1021/nl9035753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Gennes PG. Polymers at an interface: a simplified view. Adv Colloid Interface Sci. 1987;27:189–209. [Google Scholar]
- 35.Shukla S, Ablack AL, Wen AM, Lee KL, Lewis JD, Steinmetz NF. Increased Tumor Homing and Tissue Penetration of the Filamentous Plant Viral Nanoparticle Potato virus X. Mol Pharm. 2013;10:33–42. doi: 10.1021/mp300240m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Vaishnav RA, Roberts AM, Friedland RP. Humans have antibodies against a plant virus: evidence from tobacco mosaic virus. PLoS One. 2013;8:e60621. doi: 10.1371/journal.pone.0060621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elliott JA, Winn WC., Jr Treatment of alveolar macrophages with cytochalasin D inhibits uptake and subsequent growth of Legionella pneumophila. Infection and immunity. 1986;51:31–36. doi: 10.1128/iai.51.1.31-36.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunov O, Syrovets T, Loos C, Nienhaus GU, Mailander V, Landfester K, Rouis M, Simmet T. Amino-functionalized polystyrene nanoparticles activate the NLRP3 inflammasome in human macrophages. ACS Nano. 2011;5:9648–9657. doi: 10.1021/nn203596e. [DOI] [PubMed] [Google Scholar]
- 39.Vaine CA, Patel MK, Zhu J, Lee E, Finberg RW, Hayward RC, Kurt-Jones EA. Tuning innate immune activation by surface texturing of polymer microparticles: the role of shape in inflammasome activation. J Immunol. 2013;190:3525–3532. doi: 10.4049/jimmunol.1200492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yazdi AS, Guarda G, Riteau N, Drexler SK, Tardivel A, Couillin I, Tschopp J. Nanoparticles activate the NLR pyrin domain containing 3 (Nlrp3) inflammasome and cause pulmonary inflammation through release of IL-1alpha and IL-1beta. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19449–19454. doi: 10.1073/pnas.1008155107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond) 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry JL, Reuter KG, Kai MP, Herlihy KP, Jones SW, Luft JC, Napier M, Bear JE, DeSimone JM. PEGylated PRINT Nanoparticles: The Impact of PEG Density on Protein Binding, Macrophage Association, Biodistribution, and Pharmacokinetics. Nano letters. 2012;12:5304–5310. doi: 10.1021/nl302638g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfram J, Yang Y, Shen J, Moten A, Chen C, Shen H, Ferrari M, Zhao Y. The nano-plasma interface: Implications of the protein corona. Colloids Surf B Biointerfaces. 2014 doi: 10.1016/j.colsurfb.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundqvist M. Nanoparticles: Tracking protein corona over time. Nat Nanotechnol. 2013;8:701–702. doi: 10.1038/nnano.2013.196. [DOI] [PubMed] [Google Scholar]
- 45.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 46.Dell’Orco D, Lundqvist M, Cedervall T, Linse S. Delivery success rate of engineered nanoparticles in the presence of the protein corona: a systems-level screening. Nanomedicine. 2012;8:1271–1281. doi: 10.1016/j.nano.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Gunawan C, Lim M, Marquis CP, Amal R. Nanoparticle-protein corona complexes govern the biological fates and functions of nanoparticles. Journal of Materials Chemistry B. 2014;2:2060–2083. doi: 10.1039/c3tb21526a. [DOI] [PubMed] [Google Scholar]
- 48.Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. International journal of pharmaceutics. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Michel R, Pasche S, Textor M, Castner DG. Influence of PEG architecture on protein adsorption and conformation. Langmuir : the ACS journal of surfaces and colloids. 2005;21:12327–12332. doi: 10.1021/la051726h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agrawal A, Manchester M. Differential uptake of chemically modified cowpea mosaic virus nanoparticles in macrophage subpopulations present in inflammatory and tumor microenvironments. Biomacromolecules. 2012;13:3320–3326. doi: 10.1021/bm3010885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mok H, Palmer DJ, Ng P, Barry MA. Evaluation of polyethylene glycol modification of first-generation and helper-dependent adenoviral vectors to reduce innate immune responses. Mol Ther. 2005;11:66–79. doi: 10.1016/j.ymthe.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 52.Nakano K, Bando Y, Tozuka Y, Takeuchi H. Cellular interaction of PEGylated PLGA nanospheres with macrophage J774 cells using flow cytometry. Asian Journal of Pharmaceutical Sciences. 2007;2:220–226. [Google Scholar]
- 53.Ni F, Jiang L, Yang R, Chen Z, Qi X, Wang J. Effects of PEG length and iron oxide nanoparticles size on reduced protein adsorption and non-specific uptake by macrophage cells. J Nanosci Nanotechnol. 2012;12:2094–2100. doi: 10.1166/jnn.2012.5753. [DOI] [PubMed] [Google Scholar]
- 54.Sheng Y, Yuan Y, Liu CS, Tao XY, Shan XQ, Xu F. In vitro macrophage uptake and in vivo biodistribution of PLA-PEG nanoparticles loaded with hemoglobin as blood substitutes: effect of PEG content. Journal of Materials Science-Materials in Medicine. 2009;20:1881–1891. doi: 10.1007/s10856-009-3746-9. [DOI] [PubMed] [Google Scholar]
- 55.Xie J, Xu C, Kohler N, Hou Y, Sun S. Controlled PEGylation of monodisperse Fe3O4 nanoparticles for reduced non-specific uptake by macrophage cells. Advanced Materials. 2007;19 3163-+. [Google Scholar]
- 56.Quan Q, Xie J, Gao H, Yang M, Zhang F, Liu G, Lin X, Wang A, Eden HS, Lee S, Zhang G, Chen X. HSA coated iron oxide nanoparticles as drug delivery vehicles for cancer therapy. Molecular pharmaceutics. 2011;8:1669–1676. doi: 10.1021/mp200006f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostarelos K. Carbon nanotubes: Fibrillar pharmacology. Nature materials. 2010;9:793–795. doi: 10.1038/nmat2871. [DOI] [PubMed] [Google Scholar]
- 58.Ruggiero A, Villa CH, Bander E, Rey DA, Bergkvist M, Batt CA, Manova-Todorova K, Deen WM, Scheinberg DA, McDevitt MR. Paradoxical glomerular filtration of carbon nanotubes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12369–12374. doi: 10.1073/pnas.0913667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh R, Pantarotto D, Lacerda L, Pastorin G, Klumpp C, Prato M, Bianco A, Kostarelos K. Tissue biodistribution and blood clearance rates of intravenously administered carbon nanotube radiotracers. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3357–3362. doi: 10.1073/pnas.0509009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, Steinmetz NF. Biodistribution, pharmacokinetics, and blood compatibility of native and PEGylated tobacco mosaic virus nano-rods and -spheres in mice. Virology. 2014;449:163–173. doi: 10.1016/j.virol.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Almeida JP, Lin AY, Langsner RJ, Eckels P, Foster AE, Drezek RA. In vivo immune cell distribution of gold nanoparticles in naive and tumor bearing mice. Small. 2014;10:812–819. doi: 10.1002/smll.201301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farkas ME, Aanei IL, Behrens CR, Tong GJ, Murphy ST, O’Neil JP, Francis MB. PET Imaging and biodistribution of chemically modified bacteriophage MS2. Molecular pharmaceutics. 2013;10:69–76. doi: 10.1021/mp3003754. [DOI] [PubMed] [Google Scholar]
- 63.Molenaar TJ, Michon I, de Haas SA, van Berkel TJ, Kuiper J, Biessen EA. Uptake and processing of modified bacteriophage M13 in mice: implications for phage display. Virology. 2002;293:182–191. doi: 10.1006/viro.2001.1254. [DOI] [PubMed] [Google Scholar]
- 64.Prasuhn DE, Jr, Singh P, Strable E, Brown S, Manchester M, Finn MG. Plasma clearance of bacteriophage Qbeta particles as a function of surface charge. J Am Chem Soc. 2008;130:1328–1334. doi: 10.1021/ja075937f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. Bio-distribution, toxicity and pathology of cowpea mosaic virus nanoparticles in vivo. J Control Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaiser CR, Flenniken ML, Gillitzer E, Harmsen AL, Harmsen AG, Jutila MA, Douglas T, Young MJ. Biodistribution studies of protein cage nanoparticles demonstrate broad tissue distribution and rapid clearance in vivo. Int J Nanomedicine. 2007;2:715–733. [PMC free article] [PubMed] [Google Scholar]
- 67.Arnida, Janat-Amsbury MM, Ray A, Peterson CM, Ghandehari H. Geometry and surface characteristics of gold nanoparticles influence their biodistribution and uptake by macrophages. Eur J Pharm Biopharm. 2011;77:417–423. doi: 10.1016/j.ejpb.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang XD, Wu D, Shen X, Liu PX, Yang N, Zhao B, Zhang H, Sun YM, Zhang LA, Fan FY. Size-dependent in vivo toxicity of PEG-coated gold nanoparticles. International journal of nanomedicine. 2011;6:2071–2081. doi: 10.2147/IJN.S21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mohammad AK, Reineke JJ. Quantitative Detection of PLGA Nanoparticle Degradation in Tissues following Intravenous Administration. Mol Pharm. 2013;10:2183–2189. doi: 10.1021/mp300559v. [DOI] [PubMed] [Google Scholar]
- 70.Park EJ, Umh HN, Kim SW, Cho MH, Kim JH, Kim Y. ERK pathway is activated in bare-FeNPs-induced autophagy. Archives of Toxicology. 2014;88:323–336. doi: 10.1007/s00204-013-1134-1. [DOI] [PubMed] [Google Scholar]
- 71.Bottini M, Rosato N, Bottini N. PEG-Modified Carbon Nanotubes in Biomedicine: Current Status and Challenges Ahead. Biomacromolecules. 2011;12:3381–3393. doi: 10.1021/bm201020h. [DOI] [PubMed] [Google Scholar]
- 72.Hamilton RF, Jr, Wu Z, Mitra S, Shaw PK, Holian A. Effect of MWCNT size, carboxylation, and purification on in vitro and in vivo toxicity, inflammation and lung pathology. Part Fibre Toxicol. 2013;10:57. doi: 10.1186/1743-8977-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kraft JC, Freeling JP, Wang Z, Ho RJ. Emerging research and clinical development trends of liposome and lipid nanoparticle drug delivery systems. J Pharm Sci. 2014;103:29–52. doi: 10.1002/jps.23773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin S, Wang X, Ji Z, Chang CH, Dong Y, Meng H, Liao YP, Wang M, Song TB, Kohan S, Xia T, Zink JI, Lin S, Nel AE. Aspect Ratio Plays a Role in the Hazard Potential of CeO Nanoparticles in Mouse Lung and Zebrafish Gastrointestinal Tract. ACS nano. 2014 doi: 10.1021/nn5012754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baiu DC, Brazel CS, Bao Y, Otto M. Interactions of iron oxide nanoparticles with the immune system: challenges and opportunities for their use in nano-oncology. Curr Pharm Des. 2013;19:6606–6621. doi: 10.2174/13816128113199990409. [DOI] [PubMed] [Google Scholar]
- 76.Meng J, Yang M, Jia F, Xu Z, Kong H, Xu H. Immune responses of BALB/c mice to subcutaneously injected multi-walled carbon nanotubes. Nanotoxicology. 2011;5:583–591. doi: 10.3109/17435390.2010.523483. [DOI] [PubMed] [Google Scholar]
- 77.Raja KS, Wang Q, Gonzalez MJ, Manchester M, Johnson JE, Finn MG. Hybrid virus-polymer materials. 1. Synthesis and properties of PEG-decorated cowpea mosaic virus. Biomacromolecules. 2003;4:472–476. doi: 10.1021/bm025740+. [DOI] [PubMed] [Google Scholar]
- 78.Semple SC, Harasym TO, Clow KA, Ansell SM, Klimuk SK, Hope MJ. Immunogenicity and rapid blood clearance of liposomes containing polyethylene glycol-lipid conjugates and nucleic Acid. J Pharmacol Exp Ther. 2005;312:1020–1026. doi: 10.1124/jpet.104.078113. [DOI] [PubMed] [Google Scholar]
- 79.Shimizu T, Ishida T, Kiwada H. Transport of PEGylated liposomes from the splenic marginal zone to the follicle in the induction phase of the accelerated blood clearance phenomenon. Immunobiology. 2013;218:725–732. doi: 10.1016/j.imbio.2012.08.274. [DOI] [PubMed] [Google Scholar]
- 80.Croyle MA, Chirmule N, Zhang Y, Wilson JM. “Stealth” adenoviruses blunt cell-mediated and humoral immune responses against the virus and allow for significant gene expression upon readministration in the lung. Journal of Virology. 2001;75:4792–4801. doi: 10.1128/JVI.75.10.4792-4801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee GK, Maheshri N, Kaspar B, Schaffer DV. PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization. Biotechnology and Bioengineering. 2005;92:24–34. doi: 10.1002/bit.20562. [DOI] [PubMed] [Google Scholar]
- 82.O’Riordan CR, Lachapelle A, Delgado C, Parkes V, Wadsworth SC, Smith AE, Francis GE. PEGylation of adenovirus with retention of infectivity and protection from neutralizing antibody in vitro and in vivo. Hum Gene Ther. 1999;10:1349–1358. doi: 10.1089/10430349950018021. [DOI] [PubMed] [Google Scholar]
- 83.Wortmann A, Vohringer S, Engler T, Corjon S, Schirmbeck R, Reimann J, Kochanek S, Kreppel F. Fully detargeted polyethylene glycol-coated adenovirus vectors are potent genetic vaccines and escape from pre-existing anti-adenovirus antibodies. Mol Ther. 2008;16:154–162. doi: 10.1038/sj.mt.6300306. [DOI] [PubMed] [Google Scholar]
- 84.De Geest B, Snoeys J, Van Linthout S, Lievens J, Collen D. Elimination of innate immune responses and liver inflammation by PEGylation of adenoviral vectors and methylprednisolone. Hum Gene Ther. 2005;16:1439–1451. doi: 10.1089/hum.2005.16.1439. [DOI] [PubMed] [Google Scholar]
- 85.Brown DM, Johnston H, Gubbins E, Stone V. Serum enhanced cytokine responses of macrophages to silica and iron oxide particles and nanomaterials: a comparison of serum to lung lining fluid and albumin dispersions. J Appl Toxicol. 2014 doi: 10.1002/jat.2998. [DOI] [PubMed] [Google Scholar]
- 86.Khan HA, Abdelhalim MA, Alhomida AS, Al Ayed MS. Transient increase in IL-1beta, IL-6 and TNF-alpha gene expression in rat liver exposed to gold nanoparticles. Genet Mol Res. 2013;12:5851–5857. doi: 10.4238/2013.November.22.12. [DOI] [PubMed] [Google Scholar]
- 87.Meunier E, Coste A, Olagnier D, Authier H, Lefevre L, Dardenne C, Bernad J, Beraud M, Flahaut E, Pipy B. Double-walled carbon nanotubes trigger IL-1beta release in human monocytes through Nlrp3 inflammasome activation. Nanomedicine (Lond) 2012;8:987–995. doi: 10.1016/j.nano.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 88.Maruyama K, Yuda T, Okamoto A, Kojima S, Suginaka A, Iwatsuru M. Prolonged circulation time in vivo of large unilamellar liposomes composed of distearoyl phosphatidylcholine and cholesterol containing amphipathic poly(ethylene glycol) Biochim Biophys Acta. 1992;1128:44–49. doi: 10.1016/0005-2760(92)90255-t. [DOI] [PubMed] [Google Scholar]
- 89.Yang Q, Jones SW, Parker CL, Zamboni WC, Bear JE, Lai SK. Evading Immune Cell Uptake and Clearance Requires PEG Grafting at Densities Substantially Exceeding the Minimum for Brush Conformation. Molecular pharmaceutics. 2014 doi: 10.1021/mp400703d. [DOI] [PubMed] [Google Scholar]
- 90.Fang J, Nakamura H, Maeda H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv Drug Deliv Rev. 2011;63:136–151. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 91.Murahari MS, Yergeri MC. Identification and usage of fluorescent probes as nanoparticle contrast agents in detecting cancer. Curr Pharm Des. 2013;19:4622–4640. doi: 10.2174/1381612811319250009. [DOI] [PubMed] [Google Scholar]
- 92.Sosnovik DE, Nahrendorf M, Weissleder R. Magnetic nanoparticles for MR imaging: agents, techniques and cardiovascular applications. Basic Res Cardiol. 2008;103:122–130. doi: 10.1007/s00395-008-0710-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pokorski JK, Breitenkamp K, Liepold LO, Qazi S, Finn MG. Functional virus-based polymer-protein nanoparticles by atom transfer radical polymerization. J Am Chem Soc. 2011;133:9242–9245. doi: 10.1021/ja203286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.