Abstract
Recent work on blind compressed sensing (BCS) has shown that exploiting sparsity in dictionaries that are learnt directly from the data at hand can outperform compressed sensing (CS) that uses fixed dictionaries. A challenge with BCS however is the large computational complexity during its optimization, which limits its practical use in several MRI applications. In this paper, we propose a novel optimization algorithm that utilize variable splitting strategies to significantly improve the convergence speed of the BCS optimization. The splitting allows us to efficiently decouple the sparse coefficient, and dictionary update steps from the data fidelity term, resulting in subproblems that take closed form analytical solutions, which otherwise require slower iterative conjugate gradient algorithms. Through experiments on multi coil parametric MRI data, we demonstrate the superior performance of BCS, while achieving convergence speed up factors of over 15 fold over the previously proposed implementation of the BCS algorithm.
I. INTRODUCTION
Over the recent years, compressed sensing (CS) schemes have shown considerable potential to accelerate MRI acquisition. CS exploits sparse representation of data in a known dictionary bases. For instance, wavelet bases in [1] and temporal Fourier bases in [2] have been used in static and dynamic MRI applications. A challenge in using such predetermined dictionaries often lies with the misfit between the representation and the data; many coefficients are often required for an accurate representation. For instance, in free breathing perfusion MRI, many temporal Fourier bases are required to represent the temporal dynamics of the data, thereby restricting the maximum achievable acceleration factor.
Recently, several researchers have proposed to jointly estimate the sparse representations and the dictionaries from the under sampled data at hand. Dictionaries containing atoms of one-dimensional non-orthogonal temporal bases [3], two-dimensional spatial patches [4], or three-dimensional spatio-temporal cubes [5], [6] have been proposed for dynamic and static applications. These schemes termed as blind compressed sensing (BCS) have shown considerable promise over conventional CS schemes in several MRI applications such as dynamic contrast enhanced MRI [3], cardiac cine MRI [5], [6], functional lung [7], parametric MRI, and high resolution static MRI [4].
The BCS scheme is formulated as a constrained optimization problem consisting of linear combination of data fidelity l2 norm, and a sparsity promoting norm on the coefficients subject to a Frobenius norm constraint on the dictionary. The single coil BCS algorithmic implementation in [3] relies on iterations between quadratic update steps of the coefficient and dictionary update steps. These steps were solved using slow iterative conjugate gradient algorithms due to the complexity in constructing the inverses of the matrices resulting from the data fidelity. The large computational complexity encountered during the optimization pose a challenge in dealing with large practical datasets such as multi-coil and three-dimensional applications.
In this work, we propose to employ variable splitting strategies [8] to efficiently decouple the coefficient update and dictionary update steps from the data fidelity term. Through the decoupling, we develop a computational efficient algorithm that efficiently cycles between different steps that have analytical closed form solutions. We demonstrate through experiments on multi coil parametric MRI data, the proposed algorithm obtains a significant speed up factor of ten fold over the previous implementation.
II. Background
A. BCS Model representation
In parametric imaging, the k-space corresponds to several images acquired at different values of encoding parameters (such as echo time, spin lock time, flip angle) denoted by p. We model the multi-coil undersampled measurement as:
| (1) |
where b(k, p) represents the concatenated vector of the k − p measurements from all the coils. γ(x, p); (x = (x1, y1)) denote the underlying images pertaining to different contrasts; n is additive noise. 𝒜 is the operator that models coil sensitivity C and Fourier encoding F on a specified k − p sampling trajectory S. The dataset is represented as M × N Casoriti matrix [9] ΓM × N
| (2) |
where M is the number of voxels in the image and N is the number of encoding parameters.
B. Image reconstruction
We model Γ as a product of spatial coefficients UM × R and dictionary VR × N. The joint recovery of U, V is formulated as a constrained optimization problem:
| (3) |
The first term ensures data consistency. The second term promotes sparsity on the spatial coefficients by using a nonconvex ℓp; p < 1 semi-norm prior on U. A unit Frobenius norm is imposed on the dictionary V making the recovery problem well posed.
C. Algorithm 1: Without using variable splitting
We majorize an approximation of the ℓp penalty on U in Eq. (3) as , where L is an auxiliary variable. This approximation becomes exact as β → ∞. We use augmented Lagrangian optimization scheme to enforce the constraint V = Q, where Q is the auxiliary variable for V. Thus, the optimization problem is given by
| (4) |
We use an alternating strategy to solve for the variables U, V, Q and L. The L and Q subproblems are solved analytically. Eq. 4 is quadratic in U and V. Iterative methods like conjugate gradient are required to solve these subproblems due to the enormous size of A. In addition, for the majorization to well approximate the ℓp penalty, β needs to be a high value. At higher values of β, the condition number of these subproblems is significantly high resulting in slow convergence as many iterations of CG are required.
III. Proposed Algorithm : Using variable splitting
To improve convergence speed, Ramani and Fessler proposed the use of the technique of variable splitting to decouple the effect of coil sensitivities C and the regularization [8]. We introduce a novel optimization algortihm using variable splitting technique to accelerate the convergence of Eq. 4. First, we decouple the data fidelity term from sparse coefficients U and dictionary V by introducing a constraint X = UV where X is the auxiliary variable for UV. The optimization problem is of the form
| (5) |
We further decouple the coil sensitivities from X by introducing another constraint Z=CX where Z is the auxiliary variable. The constrained optimization problem can be written as
| (6) |
We majorize an approximation of the ℓp penalty on U in (3) as , where L is an auxiliary variable. We enforce the constraints in Eq. 6 by using an augmented Lagrangian (AL) framework [?]. The associated AL function is written as
| (7) |
Here Q is the auxiliary variable for V, ΛX, ΛV and ΛZ are the Lagrange multipliers. βX, βV, βU and βZ are the penalty parameters. We use an alternating strategy to solve for the variables U, V, Q, L, X and Z. All of these subproblems are solved analytically as described below, by minimizing the Eq. 7 with respect to these variables one at a time assuming the other variables to be fixed.
L subproblem
Ignoring all the terms independent of L, Eq. 7 can be written as
| (8) |
The L subproblem is solved using shrinkage rule as
| (9) |
where ‘+’ represents the soft thresholding operator defined as (τ)+ = max{0, τ} and βU is the penalty parameter.
U subproblem
The minimization of Eq. 7 with respect to U results in a quadratic subproblem which has an closed form solution given by
| (10) |
The quadratic subproblem can be solved analytically as shown below.
| (11) |
| (12) |
Q subproblem
The Q subproblem is obtained by minimizing Eq. 7 with respect to Q
| (13) |
The above problem is solved using a projection scheme as specified in Eq. 14. If Frobenius norm of Q is less than unity, we set Q = V, else we scale V to have a unit Frobenius norm
| (14) |
Note that Qn is obtained by scaling Vn so that the Frobenius norm is unity.
V subproblem
The V subproblem is a quadratic subproblem as shown below.
| (15) |
Minimization of the above equation with respect to V yields the following closed form solution
| (16) |
| (17) |
X subproblem
Dropping all the terms independent of X we get
| (18) |
The closed form solution to the above minimization problem is given by
| (19) |
| (20) |
Z subproblem
Writing the Eq. 7 with respect to Z (ignoring constants independent of Z) we get,
| (21) |
This problem is a Fourier domain replacement problem which can be solved analytically as shown below.
| (22) |
We update all the Lagrange multipliers using a steepest ascent method at each iteration as shown below.
| (23) |
| (24) |
| (25) |
The optimization algorithm in Eq. 6 is solved by cycling between the above subproblems. The matrix HU and HV are R × R and can be easily inverted. Since C′C is a diagonal, HX matrix is also diagonal and is therefore easily inverted. Splitting the k-space and coil sensitivities (F and C components) from UV in the data fidelity term has led to separate matrix inverses involving F′F and C′C which are easier to compute. Since, all the steps can be solved analytically, the convergence is much faster than conventional iterative conjugate gradient steps. Although βX and βZ parameters do not affect the final solution, they can affect the convergence rate. These parameters were chosen empirically. Since we use the augmented Lagrangian framework for enforcing the constraint on the dictionary, it is not necessary for βV to tend to 1 for the constraint to hold, allowing faster convergence. The quality of reconstruction is affected by βU parameters as the non-convex penalty is enforced using majorization. As discussed earlier, the majorization is only exact when βU → ∞. We initialize βU to a small value and gradually increment it when the cost in Eq. 3 stagnates to a threshold level of 10−2.
The pseudo-code of the algorithm is shown below.
Algorithm III.1.
FastBCS(S,F,C,b,λ)
| Input : b, βX > 0, βZ > 0, βU > 0 | ||||
| while |𝒞n − 𝒞n−1| > 10−5 𝒞n | ||||
| do | Initialize βV > 0 | |||
| while |V − Q|2 > 10−5 | ||||
| do | Update L :← Eq:[9] | |||
| Update U :← Eq:[11] | ||||
| Update Q :← Eq:[14] | ||||
| Update V :← Eq:[16] | ||||
| Update Z :← Eq:[22] | ||||
| Update X :← Eq:[19] | ||||
| Update ΛV :← Eq:[23] | ||||
| Update ΛX :← Eq:[24] | ||||
| Update ΛZ :← Eq:[25] | ||||
| βV = 5 * βV | ||||
| if |𝒞n − 𝒞n−1| < 10−2 𝒞n | ||||
| then {βU = 50 * βU | ||||
| return (U, V) | ||||
IV. Experimental Evaluation
To study the convergence rate of both the algorithms, we acquired a single slice fully sampled 2D dataset on a Siemens 3T Trio scanner using a turbo spin echo (TSE) sequence with turbo factor (TF) of 8, matrix size = 128×128, FOV= 22×22cm2, TR=2500ms, slice thickness =5mm, B1 spin lock frequency=400Hz, bandwidth= 130Hz/pixel, and echo spacing of 12.2ms. A 12-channel phased array coil was used. T1ρ preparation pulse [10] and T2 preparation pulse [11] were used prior to readout. The data was collected for 12 equispaced spin lock times (TSLs) and 12 equispaced echo times (TEs) values, both ranging from 10 ms to 120 ms. This provided a total of 24 parametric measurements. The dataset was undersampled using a Cartesian hybrid sampling pattern (acceleration of 1.5 using pseudo-random variable density sampling and an acceleration factor of 4 from uniform sampling pattern) giving a net acceleration of 6. The coil sensitivity maps were obtained using Walsh method for coil map estimation [12]. Both the algorithms were implemented in MATLAB on a quad core linux machine.
We compare the performance and convergence speed of both the algorithms using a Mean square error (MSE) metric given by
| (26) |
The regularization parameter λ of both the algorithms was chosen such that the error between reconstructions and the fully sampled data given by MSE was minimized. Comparisons were done for the optimal λ value of 0.05.
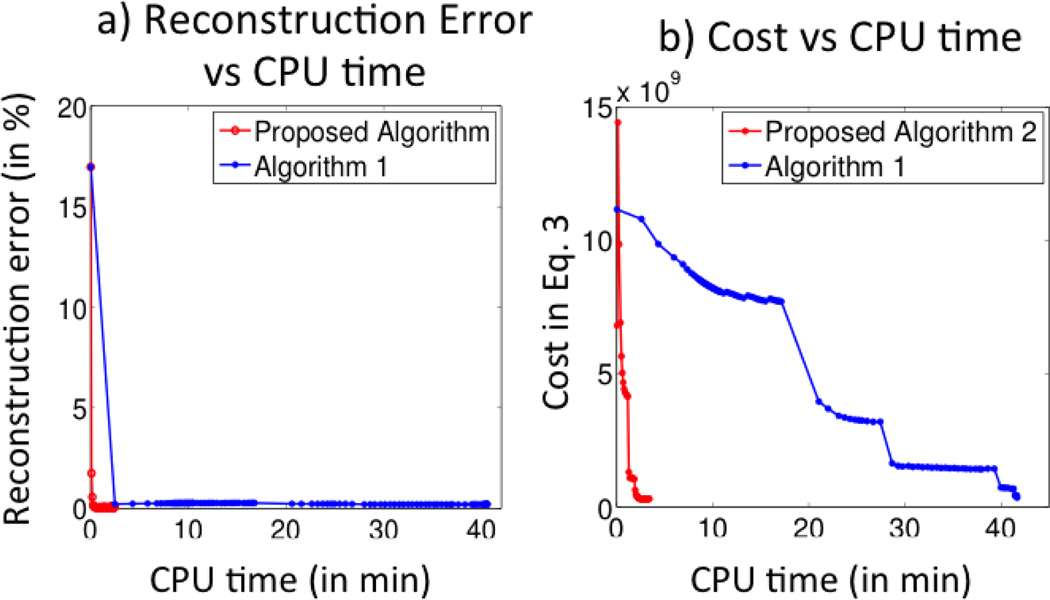
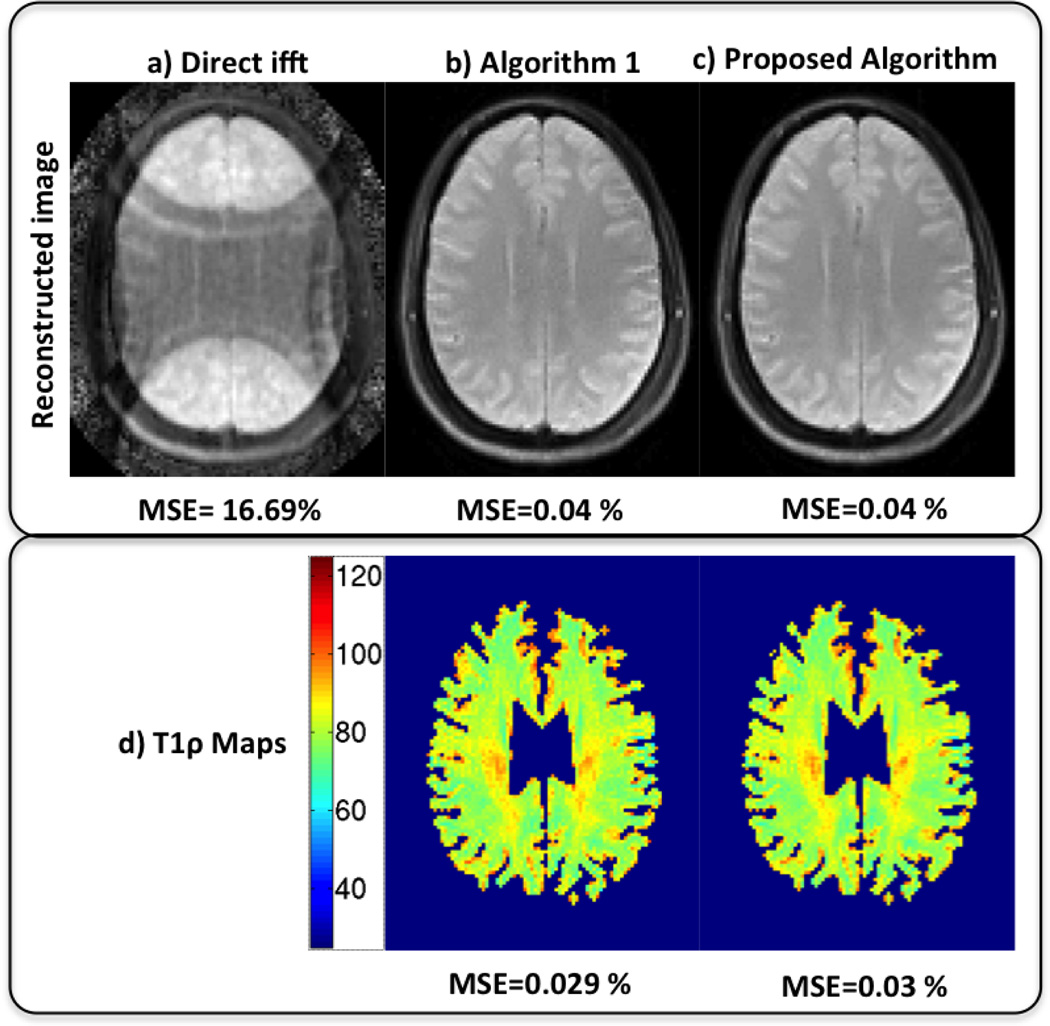
The reconstruction error vs CPU time is shown in Fig. 1(a). It is observed that proposed algorithm converges to almost the same solution in just 3 min. This is also demonstrated by the reconstructed images and T1ρ parameter maps shown in Fig. 2. Every time, when the β in Eq. 4 is incremented, the condition number of the U subproblem increases and the CG algorithm needs many iterations to converge thus increasing the reconstruction time considerably. This behavior can be seen in Fig. 1(b). In contrast the proposed algorithm takes much lesser time as it solves the subproblems analytically. The proposed algorithm converges in about 146 secs (≈ 2.5 min) while the Algorithm 1 takes about 2500 secs (≈ 41 min) resulting in 16 fold acceleration without degradation in image quality.
Fig. 1.
Convergence plots: Reconstruction error vs CPU time and the cost vs CPU time plots are shown in a) and b) respectively. It is observed that proposed algorithm converges to same solution in about 2.5 min where as the Algorithm 1 takes roughly 41 min to converge. Note the threshold in both algorithms was set to 10−6
Fig. 2.
Qualitative results: The first row shows reconstructed images using Direct ifft, Algorithm 1 and the Proposed algorithm in a–c. The second row shows T1ρ parameter maps for Algorithm 1 and proposed Algorithm. It is seen that proposed algorithm converges 10 times faster without degradation in quality as compared to the Algorithm 1.
V. Conclusion
In this work, we have introduced a novel variable splitting based algorithm to improve the convergence rate of blind compressed sensing MRI reconstruction. We have derived analytical closed form update rules by splitting data fidelity term from the dictionary and the coefficients terms. These analytical rules lead to simpler subproblems which were solved by fast operations. Through experiments on multi-coil parametric MRI data, we have observed the new optimization algorithm to provide a speed up factor of about 16 fold in comparison to our previous BCS implementation. The speed up factors provided show potential in handling large scale datasets such those in multi-coil multidimensional MRI applications.
Acknowledgments
This work is supported by grants NSF CCF-0844812, NSF CCF-1116067, NIH 1R21HL109710-01A1, ACS RSG-11-267-01-CCE, and ONR N00014-13-1-0202.
References
- 1.Lustig M, Donoho D, Pauly JM. Sparse mri: The application of compressed sensing for rapid mr imaging. Magnetic resonance in medicine. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 2.Jung H, Park J, Yoo J, Ye JC. Radial k-t focuss for high-resolution cardiac cine mri. Magnetic Resonance in Medicine. 2010;63(1):68–78. doi: 10.1002/mrm.22172. [DOI] [PubMed] [Google Scholar]
- 3.Lingala S, Jacob M. Blind compressive sensing dynamic mri. IEEE transactions on medical imaging. 2013;32(6):1132. doi: 10.1109/TMI.2013.2255133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravishankar S, Bresler Y. Mr image reconstruction from highly undersampled k-space data by dictionary learning. Medical Imaging, IEEE Transactions on. 2011;30(5):1028–1041. doi: 10.1109/TMI.2010.2090538. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Ying L. Compressed sensing dynamic cardiac cine mri using learned spatiotemporal dictionary. IEEE transactions on Biomedical Engineering. 2014 Apr;61(4):1109–1120. doi: 10.1109/TBME.2013.2294939. [DOI] [PubMed] [Google Scholar]
- 6.Caballero J, Price AN, Rueckert D, Hajnal J. Dictionary learning and time sparsity for dynamic mr data reconstruction. IEEE transactions on medical imaging. 2014;33(4):979–994. doi: 10.1109/TMI.2014.2301271. [DOI] [PubMed] [Google Scholar]
- 7.Lingala SG, Mohsin Y, Newell JD, Sieren JC, Thedens D, Kollasch P, Jacob M. Accelerating dynamic imaging of the lung using blind compressed sensing. Journal of Cardiovascular Magnetic Resonance. 2014;16(Suppl 1):W27. [Google Scholar]
- 8.Ramani S, Fessler JA. Parallel mr image reconstruction using augmented lagrangian methods. Medical imaging, iEEE Transactions on. 2011;30(3):694–706. doi: 10.1109/TMI.2010.2093536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Z-P. Noninvasive Functional Source Imaging of the Brain and Heart and the International Conference on Functional Biomedical Imaging, 2007. NFSI-ICFBI 2007. Joint Meeting of the 6th International Symposium on. IEEE; 2007. Spatiotemporal imaging with partially separable functions; pp. 181–182. [Google Scholar]
- 10.Charagundla SR, Borthakur A, Leigh JS, Reddy R. Artifacts in T1ρ-weighted imaging: correction with a self-compensating spinlocking pulse. Journal of Magnetic Resonance. 2003;162(1):113–121. doi: 10.1016/s1090-7807(02)00197-0. [DOI] [PubMed] [Google Scholar]
- 11.Brittain JH, Hu BS, Wright GA, Meyer CH, Macovski A, Nishimura DG. Coronary angiography with magnetization-prepared T2 contrast. Magnetic resonance in medicine. 1995;33(5):689–696. doi: 10.1002/mrm.1910330515. [DOI] [PubMed] [Google Scholar]
- 12.Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magnetic Resonance in Medicine. 2000;43(5):682–690. doi: 10.1002/(sici)1522-2594(200005)43:5<682::aid-mrm10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]