Abstract
DNA extracted from formalin-fixed, paraffin-embedded (FFPE) tissues has been used in the past to analyze genetic polymorphisms. We evaluated the technical reproducibility of different types of assays for gene polymorphisms using DNA extracted from FFPE material. By using the MassARRAY iPLEX system, we investigated polymorphisms in DPYD (rs1801159 and rs3918290), UMPS (rs1801019), ERCC1 (rs11615), ERCC1 (rs3212986), and ERCC2 (rs13181) in 56 FFPE DNA samples. By using PCR, followed by size-based gel electrophoresis, we also examined TYMS 5′ untranslated region 2R/3R repeats and GSTT1 deletions in 50 FFPE DNA samples and 34 DNAs extracted from fresh-frozen tissues and cell lines. Each polymorphism was analyzed by two independent runs. We found that iPLEX biomarker assays measuring single-nucleotide polymorphisms provided consistent concordant results. However, by using FFPE DNA, size-based PCR biomarkers (GSTT1 and TYMS 5′ untranslated region) were discrepant in 32.7% (16/49, with exact 95% CI, 19.9%–47.5%; exact binomial confidence limit test) and 4.2% (2/48, with exact 95% CI, 0.5%–14.3%) of cases, respectively, whereas no discrepancies were observed using intact genomic DNA. Our findings suggest that DNA from FFPE material can be used to reliably test single-nucleotide polymorphisms. However, results based on size-based PCR biomarkers, and particularly GSTT1 deletions, using FFPE DNA need to be interpreted with caution. Independent repeated assays should be performed on all cases to assess potential discrepancies.
Genetic and epigenetic variations, which govern acquisition and progression of cancer as well as treatment-related responses and toxicities, are hallmarks of all human cancers.1 Numerous studies have described individual profiles of tumors at the somatic mutation and germline polymorphism level.2–4 Previous research has suggested that specific germline polymorphisms may influence several important cancer-relevant traits. For example, dihydropyrimidine dehydrogenase (DPYD), uridine monophosphate synthetase (UMPS), and thymidylate synthase (TYMS) are involved in the metabolism of 5-fluorouracil, a common chemotherapy agent used to treat solid tumors, and polymorphisms in these genes have been associated with 5-fluorouracil resistance.5–8 ERCC1 and ERCC2 are involved in DNA repair, and single-nucleotide polymorphisms (SNPs) in these genes have been associated with response to chemotherapies, such as cisplatin.9,10 GSTT1 is a member of the glutathione-S-transferase (GST) family, and 20% to 60% of individuals do not express this gene because they carry homozygous GSTT1 deletions (GSTT1*0/0 allele). Individuals who carry GSTT1*0/0 may have an impaired ability to metabolize carcinogenic compounds and may exhibit worse responses to platinum-/5-fluorouracil–based chemotherapies.11 Accurate determination of germline polymorphisms are thus important for predicting drug response, susceptibility to environmental factors, and risk of cancer development.
The type of biological material that can be used for measuring germline polymorphisms in cancer patients is often limited by practical considerations. In particular for retrospective studies, blood samples from patients are rarely available. In contrast, routine archival formalin-fixed, paraffin-embedded (FFPE) tissue samples are readily available from all resected specimens or from diagnostic biopsy specimens, providing large and valuable collections for genetic studies. Thus, it is important to determine the accuracy and potential limitations of genetic polymorphism studies using this type of material.
Several factors should be considered when measuring genetic polymorphisms in DNA from FFPE samples. Specifically, tissue fixation in formaldehyde and long-term storage at room temperature can damage nucleic acids through denaturation, fragmentation, and cross-linking. For example, A:T base pairs within archival tissues may experience greater degradation and increased transition-type mutations.12–14 The Taq polymerase error rate for FFPE DNA has been reported to be 1 in 500 bp, compared to 1 in 105 bp for fresh tissues.14 Agell et al15 reported high rates of sequencing artifacts present in FFPE DNA but not found in DNA extracted from fresh-frozen samples. However, despite this high rate of artifacts, it was still possible to identify true sequence variants through a second confirmatory PCR, because the probability of finding the same artifactual mutation in a second independent PCR was extremely low. This second confirmation approach was used by Marchetti et al,16 who initially observed several uncommon EGFR mutations in FFPE DNA from lung cancer samples. However, these mutations were nonreproducible across multiple PCR amplifications and were eventually discarded.16
Germline polymorphisms can be divided into two major categories: single-nucleotide variants/polymorphisms (SNVs/SNPs) and copy number polymorphisms, such as GSTT1*0/0. Regarding the former, previous analyses have shown that with careful experimental planning, concordant germline SNP genotyping results can be generated between FFPE DNA and fresh-frozen DNA.17–20 However, many SNP genotyping studies have been performed using quantitative real-time PCR, which is expensive, labor intensive, and not suitable or scalable to the simultaneous genotyping of many SNPs. An alternative platform for SNV/SNP genotyping is the MassARRAY iPLEX system (Sequenom, San Diego, CA), which can genotype as many as 40 SNPs in one single reaction.21,22 Briefly, the iPLEX system exploits oligonucleotide PCR extension over a SNP to produce different-sized products that are allele specific, which are then detected using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. However, to date, relatively few studies have evaluated the performance of the iPLEX platform using FFPE DNA.23–25
Our aim was to evaluate whether FFPE DNA can be used to reliably test germline SNPs using the iPLEX system. We also evaluated whether FFPE DNA can produce reliable results for copy number/size-based PCR biomarkers, such as TYMS 5′ untranslated region (UTR) 2R/3R repeats or GSTT1 deletion. For the latter, we compared results between FPPE DNA and DNA from fresh-frozen tissues and cell lines.
Materials and Methods
Primary Tissue and Cell Line Samples
FFPE blocks of nonmalignant tissues were selected from three clinical cohorts: OEO2, MAGIC (Medical Research Council adjuvant gastric infusional chemotherapy trial), and JUST (Japan/UK/Singapore translation study). The OEO2 and MAGIC cohorts were derived from patients enrolled in randomized, multicenter, phase 3 clinical trials of neoadjuvant chemotherapy versus surgery alone from the United Kingdom (OEO2 patients recruited between 1992 and 1998, average sample storage length, 16 years 2 months; MAGIC patients recruited between 1994 and 2002, average sample storage length, 12 years 3 months).26,27 The JUST cohort comprises Japanese gastric cancer patients who underwent curative resection followed by treatment with adjuvant S1 between 2001 and 2010 at Kanagawa Cancer Center (Yokohama, Japan),28 with an average storage length of 5 years 4 months. Normal fresh-frozen colon tissues were obtained from the Singapore General Hospital (Singapore) Tissue Repository. Gastric cancer cell lines were obtained from ATCC (Manassas, VA) or from collaborating institutions. This study was approved by the respective Institutional Research Ethics Review committees.
Extraction of DNA from FFPE Samples
All hematoxylin and eosin–stained tissue sections from resection specimens were reviewed by a histopathologist (H.I.G., OEO2 and JUST cohorts; MAGIC cohort was not reviewed by an author). DNA was extracted from nonmalignant tissues, either lymph nodes without evidence of metastatic disease or nonneoplastic normal esophagus or stomach. Five sections (10 μm thick) were cut and deparaffinized using a standard protocol, and the area of interest was dissected using a sterile scalpel blade. Genomic DNA was extracted using a protocol based on the QIAmp DNA Micro Kit and QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany), following the manufacturer's instructions. Briefly, after dewaxing and rehydrating the slides, the area of interest was microdissected and placed into a 1.5-mL Eppendorf tube with buffer ATL and proteinase K for digestion (Qiagen). DNA was eluted in buffer ATE (Qiagen) with an elution volume of 30 μL for OEO2 and JUST cohort samples and 60 μL for MAGIC cohort samples. Quality control of the DNA was performed on the basis of 260:230 and 260:280 ratio values and visual inspection of the wavelength spectral pattern provided by the NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE). A 260:230 ratio of approximately 2.0, together with a 260:280 ratio of approximately 1.8 and the presence of a peak at 260 nm with a steep decrease toward 280 nm in the wavelength spectrum, was considered sufficiently good quality DNA.
Extraction of DNA from Normal Colon Tissues and Cell Lines
Genomic DNA was extracted from frozen tissues and cell lines using a blood and cell culture DNA extraction kit, according to the manufacturer's instructions (Qiagen, Valencia, CA). Briefly, 10 to 15 mg frozen tissue was homogenized and incubated in buffer G2 containing RNase A and Qiagen Protease K, then incubated at 50°C for 4 hours. DNA was resuspended in 100 to 200 μL of nuclease-free water (Life Technologies, Carlsbad, CA) and dissolved overnight on a shaker. Human genomic DNA was purchased from Promega (Madison, WI).
Polymorphism Selection
For this study, we chose eight polymorphisms that have been proposed as predictive biomarkers of chemotherapy and prognosis in the literature.5–11 These included six SNPs: DPYD (rs1801159), DPYD (rs3918290), UMPS (rs1801019), ERCC1 (rs11615), ERCC1 (rs3212986), ERCC2 (rs13181), and two non-SNV/SNP polymorphisms (GSTT1-null and TYMS 5′-UTR 2R/3R repeats), which we defined as copy number/size-based polymorphisms.
Genotype Analysis
SNP Genotyping by MassARRAY iPLEX System
Genotype analysis was performed using the MassARRAY iPLEX system (Sequenom), according to the manufacturer's instructions. The assay used was a six-plex assay designed using MassARRAY Online Design Tools (Sequenom). Briefly, multiplexed PCR amplification was performed using 20 ng of DNA in a 5-μL reaction containing 0.5 U of Taq polymerase (Sequenom), 1× PCR buffer, 4 mmol/L MgCl2, 500 μmol/L deoxynucleotide triphosphates, and 0.1 μmol/L of primers (Table 1). RKO colon cell line DNA was used for interrun controls and nuclease-free water as a nontemplate control (Supplemental Table S1). The following program was used for PCR amplification: 95°C for 2 minutes, followed by 45 cycles of 95°C for 30 seconds, 56°C for 30 seconds, 72°C for 1 minute, and a final extension step of 72°C for 5 minutes. Unincorporated deoxynucleotide triphosphates were removed using 0.3 U of shrimp alkaline phosphatase (Sequenom). Single-base extension was performed in a 9-μL reaction containing iPLEX GOLD buffer, iPLEX termination mix, extend primer mix, and iPLEX enzyme (Sequenom). The reactions were performed using the following two cycling loop programs: 94°C for 30 seconds, followed by 40 cycles of 94°C for 5 seconds, 52°C for 5 seconds, and 80°C for 5 seconds. Within the 40 cycles, the annealing and extension step was repeated five times (ie, 40 × 5 = 200 short cycles), before a final extension step of 3 minutes at 72°C. Reactions were desalted using 6 mg of clean resin (Sequenom) and diluted with 16 μL water. A total of 10 nL of each reaction was spotted onto the 384-spot SpectroChipII using MassARRAY Nanodispenser (Sequenom). This was followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis using the MassARRAY Compact system (Sequenom). The mass spectra analysis and genotype calls were generated using Sequenom TYPER software version 4.0.22
Table 1.
Sequences of PCR Primers Used for SNP Genotyping
| Gene | First PCR primer | Second PCR primer | PCR size, bp | Extension primer |
|---|---|---|---|---|
| DPYD (rs1801159) | F: 5′-ACGTTGGATGTGCGC-3′ R: 5′-TAGCAAGACCAAAAG-3′ |
F: 5′-ACGTTGGATGCTCCTAT-3′ R: 5′-TGATCTGGTGGAC-3′ |
97 | 5′-ATGGCCGGATTGAAGTTT-3′ |
| DPYD (rs3918290) | F: 5′-ACGTTGGATGTCACT-3′ R: 5′-GAACTAAAGGCTGAC-3′ |
F: 5′-ACGTTGGATGCACCAA-3′ R: 5′-CTTATGCCAATTCTC-3′ |
100 | 5′-GTTTTAGATGTTAAATCACACTTA-3′ |
| UMPS (rs1801019) | F: 5′-ACGTTGGATGAGAAT-3′ R: 5′-GTCTTTGTGGCAGCG-3′ |
F: 5′-ACGTTGGATGAAGCTG-3′ R: 5′-AGTTCTTTGGGTGC-3′ |
93 | 5′-TCCTTTATAGAAAGGGGAGAA-3′ |
| ERCC1 (rs11615) | F: 5′-ACGTTGGATGGCACA-3′ R: 5′-TAGTCGGGAATTACG-3′ |
F: 5′-ACGTTGGATGGGCAAT-3′ R: 5′-CCCGTACTGAAGTT-3′ |
90 | 5′-CTGAAGTTCGTGCGCAA-3′ |
| ERCC1 (rs3212986) | F: 5′-ACGTTGGATGTTTAG-3′ R: 5′-TTCCTCAGTTTCCCG-3′ |
F: 5′-ACGTTGGATGCACAGG-3′ R: 5′-CCGGGACAAGAAG-3′ |
97 | 5′-CCGGGACAAGAAGCGGAAG-3′ |
| ERCC2 (rs13181) | F: 5′-ACGTTGGATGCTCAG-3′ R: 5′-AGCTGCTGAGCAATC-3′ |
F: 5′-ACGTTGGATGAGCCTG-3′ R: 5′-GAGCAGCTAGAATC-3′ |
87 | 5′-TAGAATCAGAGGAGACGCTG-3′ |
F, forward; R, reverse; SNP, single-nucleotide polymorphism.
Size-Based Polymorphism Genotyping by Gel Electrophoresis
TYMS 5′-UTR 2R/3R repeats and GSTT1 size-based polymorphisms were examined by PCR DNA amplification, followed by analysis in 4% agarose/1× Tris-borate-EDTA gel electrophoresis.29,30 Intact gastric cell line DNA was used as positive controls and nuclease-free water as negative controls (Supplemental Table S1). For TYMS 5′-UTR genotyping, 50 ng of genomic DNA was amplified in a 20-μL PCR mixture containing 10 μL of GoTaq hot start Taq colorless master mix (Promega) and 0.3 μmol/L of primers (Table 2).29 PCR conditions were as follows: 95°C for 5 minutes, followed by 40 cycles of 95°C for 50 seconds, 58°C for 50 seconds, 72°C for 60 seconds, and a final extension at 72°C for 10 minutes. After PCR amplification, PCR products were directly electrophoresed in 4% agarose/1× Tris-borate-EDTA gel stained with GelRed Nucleic Acid Stain (Biotium Inc., Hayward, CA) for 50 minutes at 100 V, and visualized under UV light. For the TYMS 5′-UTR, the predominant alleles expected at TYMS 5′-UTR are the 2R and 3R alleles. PCR products from 2R and 3R alleles differ by a single 28-bp repeat, which can be resolved using gel electrophoresis.
Table 2.
Sequences of PCR Primers Used for Size-Based Polymorphisms
| Gene | PCR primer | Amplicon length (bp) |
|---|---|---|
| TYMS29 | F: 5′-AGGCGCGCGGAAGGGGTCCT-3′ R: 5′-CCGAGCCGGCCACAGGCAT-3′ |
140 |
| GSTT131 | F: 5′-GTGCAAACACCTCCTGGAGAT-3′ R: 5′-AGTCCTTGGCCTTCAGAATGA-3′ |
229 |
| ACTB | F: 5′-CAGTAGGTCTGAACAGACTCCCCA-3′ R: 5′-CTGGATAGCAACGTACATGGCTG-3′ |
187 |
F, forward; R, reverse.
GSTT1 genotyping was performed in a multiplex PCR format with ACTB as the internal control gene.30–32 Briefly, multiplex PCR was performed in a 25-μL reaction mixture including 100 ng of DNA and 12.5 μL of GoTaq hot start Taq colorless master mix (Promega) and 0.3 μmol/L of primers (Table 2). PCR conditions were the same as those used for TYMS analysis. Complete absence of the GSTT1 product in the presence of an amplicon in the control sample was interpreted as a homozygous deletion (GSTT1*0/0 null genotype). The presence of a GSTT1 product was interpreted as GSTT1 positive. In this assay, we are unable to identify whether a GSTT1-positive sample is diploid or heterozygous.30 All of the above biomarkers were tested in two independent assays, and the two assays were done within 1 week.
Statistical Analysis
Exact binomial confidence limits were calculated for the proportion of concordant genotypes obtained on two independent runs.33 The software used to compute confidence intervals was Stata version 13 (StataCorp LP, College Station, TX).
Results
MassARRAY iPLEX SNP Genotyping Provides Reproducible Genotypes Using FFPE DNA
We chose six SNPs that have been associated previously with differential chemotherapy response and disease prognosis in cancer.5–11 These SNPs were DPYD (rs1801159), DPYD (rs3918290), UMPS (rs1801019), ERCC1 (rs11615), ERCC1 (rs3212986), and ERCC2 (rs13181). We sought to test the reproducibility of measuring these SNPs using FFPE DNA, by comparing genotyping results across two independent runs.
By using the iPlex system, we designed a custom-multiplex assay allowing us to genotype the DPYD, UMPS, ERCC1, and ERCC2 SNPs in a single reaction (six SNPs). This multiplexed assay was then tested on 56 randomly selected FFPE DNAs (23 from MAGIC and 33 from OEO2). In the first run, only one case (from OEO2) experienced a genotyping failure. In the second run, all samples were successfully genotyped. Results from the two independent runs revealed 100% concordance (55/55, with exact 95% CI, 93.5%–100%; exact binomial confidence limit test), indicating that the SNP genotyping result is reproducible using the MassARRAY iPlex system. As a benchmark, genotyping of GSTP1 (lle105Val, rs1695), another SNP involved in chemotherapy response, in 30 randomly selected MAGIC cases by direct Sanger sequencing also confirmed concordant genotypes in two independent runs (data not shown).
Copy Number/Size-Based Polymorphism Genotyping Using Gel Electrophoresis Provides Discordant Genotypes Using FFPE DNA
In addition to the SNV/SNP polymorphisms, we also evaluated two copy number/size-based polymorphisms by size-based PCR gel electrophoresis—the GSTT1 deletion and the TYMS 5′-UTR 2R/3R repeats,8,11 in 50 randomly selected cases (20 JUST and 30 MAGIC). Cases from OEO2 were not evaluated in this exercise, because of sample availability and access. However, because sample ages, fixation conditions, storage conditions, collection, and DNA extraction protocols for MAGIC and OEO2 are similar because they are from the same hospitals in the United Kingdom,26,27 it is reasonable to assume that findings for the MAGIC samples will also apply to OEO2 cases.
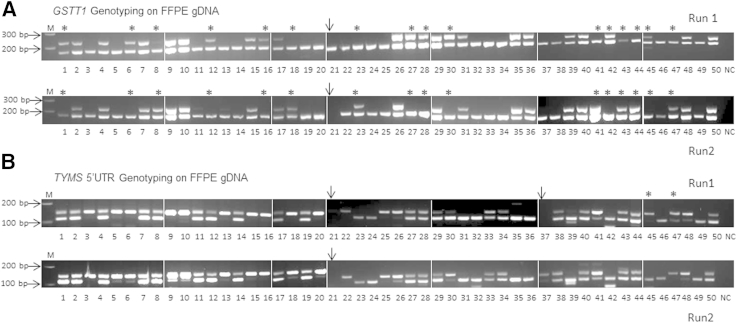
For the GSTT1 copy number polymorphism, 98% of samples were successfully genotyped. All samples were successfully analyzed in the first experiment, and one MAGIC sample failed in the second experiment. In the first experiment, there were 25 GSTT1-positive samples (50%) and 25 GSTT1 *0/0 null samples (50%). In the second experiment, there were 23 GSTT1-positive samples (46%) and 26 GSTT1 *0/0 null samples (52%). Notably, although the distribution of genotypes was similar between experiments, 16 (32.7%) of 49 samples (16/49, with exact 95% CI, 19.9%–47.5%) had a different genotype in the two independent experiments. Seven samples (14.3%) were GSTT1 *0/0 null in the first experiment and GSTT1 positive in the second experiment, whereas nine samples (18.4%) were GSTT1 *0/0 positive in the first experiment and GSTT1 null in the second experiment (Table 3). The GSTT1 genotype of 33 samples (66%) (33/49, with exact 95% CI, 52.5%–80.1%) was the same in both experiments. The concordance rate between two runs was thus 67.3% using FFPE DNA (Figure 1A).
Table 3.
Samples Exhibiting Discordant GSTT1 Genotyping Results between Two Independent Runs
| GSTT1 first run |
GSTT1 second run |
|
|---|---|---|
| No. of null samples | No. of positive samples | |
| No. of null samples | 17 | 7 |
| No. of positive samples | 9 | 16 |
Total number of samples analyzed = 49 (excluding one failed sample).
Figure 1.
GSTT1 and TYMS 5′ untranslated region (UTR) genotyping on formalin-fixed, paraffin-embedded (FFPE) samples. A:GSTT1 genotyping on 50 FFPE samples. Samples 1 to 30 are from the MAGIC cohort and 31 to 50 from the JUST cohort. Complete absence of the GSTT1 product (top panel) in the presence of an ACTB amplicon (bottom panel) was interpreted as a homozygous deletion (GSTT1 *0/0 null genotype). The presence of a GSTT1 product was interpreted as GSTT1 positive. Samples labeled with stars exhibit discrepant results between two independent runs. Sample 21 (arrow) failed in the second run and was excluded from the analysis. The concordance rate between the two runs was 69.4% for FFPE DNA. B:TYMS 5′-UTR genotyping on 50 FFPE samples. Sequence repeats in the TYMS 5′-UTR enhancer region can be classified into 2R/2R, 2R/3R, and 3R/3R genotypes. Samples labeled with stars exhibited discrepant results between the two independent runs. Sample 21 (arrow) failed in both runs, and sample 37 failed in the first run. The concordance rate between the two runs was 95.8% for FFPE DNA. gDNA, genomic DNA; M, 100-bp DNA ladder; NC, negative control.
To assess if this relatively low level of concordance might also be encountered for other copy number/size-based polymorphisms, we performed TYMS 5′-UTR 2R/3R enhancer genotyping in 50 samples. Two samples (MAGIC) failed in the first experiment, and of these, one also failed in the second experiment. In total, 96% of samples were successfully genotyped in both experiments. For the first run, there were 10 (20.8%) 2R/2R samples, 24 (50%) 2R/3R samples, and 14 (29.2%) 3R/3R samples. In the second run, there were 11 (22.9%) 2R/2R samples, 23 (47.9%) 2R/3R samples, and 14 (29.2%) 3R/3R samples. Two (4.2%) of 48 samples (2/48, with exact 95% CI, 0.5%–14.3%) demonstrated different genotypes in the two independent runs—one case changed from 3R/3R in the first run to 2R/2R in the second run, and another case changed from 2R/3R in the first run to 3R/3R in the second run (Table 4). Forty-six cases (95.8%) (46/48, with exact 95% CI, 85.7%–99.5%) remained unchanged. The concordance rate between two assays was thus 95.8% (Figure 1B). The overall GSTT1 and TYMS 5′-UTR genotyping results are summarized in Table 5.
Table 4.
Samples Exhibiting Discordant TYMS 5′-UTR Genotyping Results between Two Independent Runs
| TYMS 5′-UTR first run |
TYMS 5′-UTR second run |
||
|---|---|---|---|
| 2R/2R samples | 2R/3R samples | 3R/3R samples | |
| No. of 2R/2R samples | 10 | 0 | 0 |
| No. of 2R/3R samples | 0 | 23 | 1 |
| No. of 3R/3R samples | 1 | 0 | 13 |
Total number of samples analyzed = 48 (excluding two failed samples).
UTR, untranslated region.
Table 5.
Summary of GSTT1 and TYMS 5′-UTR Genotyping Results
| Gene polymorphism | Genotype frequency (n = 50) |
Genotype concordance in runs 1 and 2 (%) | |
|---|---|---|---|
| Run 1, No. (%) of samples | Run 2, No. (%) of samples | ||
| GSTT1* 0/0 null | 25 (50) | 26 (53)∗ | 66.70 |
| GSTT1 positive | 25 (50) | 23 (47)∗ | |
| TYMS 5′-UTR 2R/2R | 10 (20.8)† | 11 (22.9)† | 95.80 |
| TYMS 5′-UTR 2R/3R | 24 (50)† | 23 (47.9)† | |
| TYMS 5′-UTR 3R/3R | 14 (29.2)† | 14 (29.2)† | |
UTR, untranslated region.
Excluding one failed case.
Excluding two failed cases.
Copy Number/Size-Based Polymorphism Genotyping Using Gel Electrophoresis Provides Concordant Genotypes Using Fresh-Frozen Extracted DNA
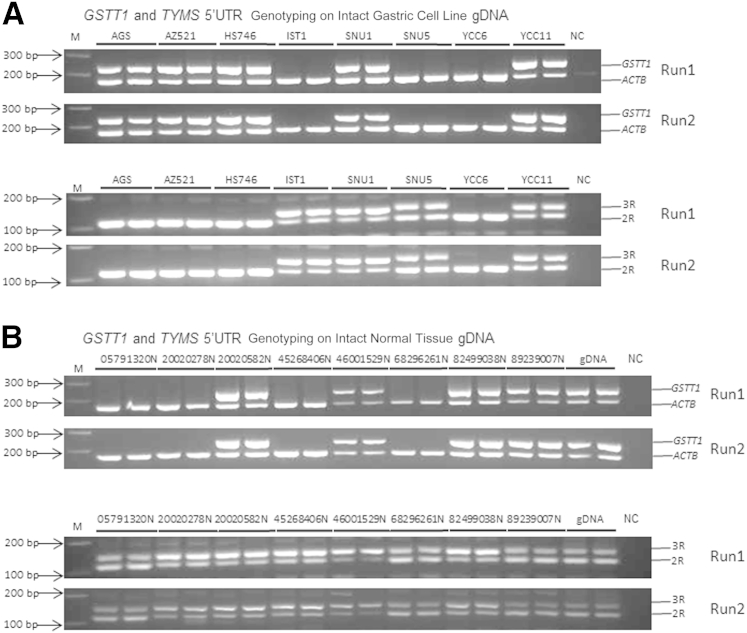
We considered whether discrepancies encountered in the copy number/size-based polymorphism analysis might be due to either the nature of the DNA material (FFPE) or intrinsic to the PCR primers used. To distinguish between these possibilities, we decided to evaluate the GSTT1 and TYMS 5′-UTR polymorphisms in DNA isolated from fresh-frozen samples. We randomly selected eight gastric cell lines, eight fresh-frozen normal colon tissues, and a commercially available human genomic DNA sample. We successfully genotyped the GSTT1 and TYMS 5′-UTR 2R/3R polymorphisms in all cell lines and fresh tissue samples. Three cell lines showed GSTT1 *0/0 null (37.5%), and five cell lines showed GSTT1 positivity (62.5%). Among the colon tissues and commercial human DNA samples, four primary tissues showed GSTT1 *0/0 null (44.4%) and five primary tissues showed GSTT1 positivity (55.6%). For TYMS 5′-UTR, four cell lines showed 2R/2R (50%) and four cell lines showed 2R/3R repeats (50%). All nine primary tissues showed 2R/3R (100%) (17/17, with exact 95% CI, 80.5%–100%). Most important, both GSTT1 and TYMS 5′-UTR genotyping results showed 100% concordance in the two independent runs (Figure 2) using DNA extracted from cell lines or from fresh-frozen tissue samples.
Figure 2.
GSTT1 and TYMS 5′ untranslated region (UTR) genotyping on fresh-frozen gastric cell lines and normal tissues. A:GSTT1 (top panel) and TYMS 5′-UTR (bottom panel) genotyping on gastric cell lines. B:GSTT1 (top panel) and TYMS 5′-UTR (bottom panel) genotyping on normal tissues. There is no discrepancy found between the two independent runs on both fresh-frozen gastric cell lines and normal tissues. gDNA, genomic DNA; M, 100-bp DNA ladder; NC, negative control.
Discussion
PCR approaches targeting well-conserved genomic sequences are widely used for SNP genotyping. However, most techniques currently used for SNP genotyping, such as quantitative real-time PCR, are not scalable, in both time and cost, for high-throughput measurements of multiple SNPs in large series of samples.22 As an alternative, the MassARRAY iPLEX System may facilitate high-throughput SNP analysis, because it permits multiplexing of up to 40 SNPs in a single reaction and can process up to 384 samples in parallel. Input DNA requirements of the iPLEX system are also as little as 10 to 20 ng DNA per sample,22,34 which is another technical advantage.
To date, few reports have investigated the ability of the MassARRAY iPLEX System to accurately genotype SNVs/SNPs in FFPE DNA.35,36 However, determining the applicability of the iPLEX system on FFPE DNA is important, given that many retrospective clinical series may not have blood or buccal epithelial cell samples available, thereby necessitating the use of FFPE samples for molecular analysis. Compared to DNA extracted from frozen samples, FFPE DNA is highly fragmented, with an average fragment size of 200 to 300 bp, and with wide variation in lengths between samples.37 To increase the efficacy of PCR, in our study, each amplicon was approximately 100 bp for iPLEX genotyping. By using the iPLEX system, we tested the reproducibility of six separate SNPs in a cohort of FFPE DNAs. These SNPs were chosen for the previous association with cancer chemotherapy response and disease prognosis. Our investigation demonstrated that germline SNVs/SNPs were associated with reproducible results when measured by iPLEX. A potential caveat of this conclusion is that the SNPs we evaluated are known SNPs common to the general population, and our overall frequencies were generally concordant with previous population studies (Supplemental Table S2).38–43 Formally, it still remains unknown if similar levels of reproducibility will be observed if such assays are extended to rare or private germline variants observed in single individuals.
In addition to SNVs/SNPs, we also tested copy number/size-based polymorphisms, specifically GSTT1 null and TYMS 5′-UTR 2R/3R, using previously described PCR-based methods, followed by agarose gel electrophoresis.29,30 Our result revealed that in contrast to SNVs/SNPs, copy number/size-based polymorphisms showed greater variability between two independent runs when performed using the same FFPE DNA. Specifically, GSTT1 showed significant discordance (33.30%), whereas TYMS 5′-UTR 2R/3R showed a minor 5% discordance. Compared to the SNP genotypes, size-based polymorphism measurements, such as those for TYMS, were also less concordant with previous population studies, particularly for MAGIC samples (Supplemental Tables S3 and S4).44–47 The latter may be due to the extended storage type of the MAGIC samples compared to Japanese samples (12 versus 5 years) and our small sample size.
In the case of GSTT1, similar discordant rates have recently been reported in a study genotyping pediatric brain tumors using FFPE tumor tissue as a DNA source.31 In that study comparing 50 frozen tissue samples to their matched FFPE counterparts, GSTM1 and GSTT1 deletion polymorphisms had a relatively low genotype concordance (77% and 82%, respectively), and subsequent retesting of the FFPE DNA for GSTM1 and GSTT1 demonstrated irreproducible genotype results.31 Our results independently confirm that GSTT1 deletion polymorphism results cannot be reproduced with confidence when FFPE DNA is used. One potential explanation for the lower reproducibility of the FFPE DNA, compared to frozen DNA, is the low level at which FFPE detection is occurring, which can cause stochastic sampling effects despite using more DNA (50 ng for TYMS 5′ UTR and 100 ng for GSTT1 assay) compared to 20 ng of DNA for the SNP genotyping assay.48 Unfortunately, the low levels at which FFPE DNA analysis usually occurs is not revealed by standard spectrophotometric analysis because many elements in FFPE DNA extracts may provide absorbance at 260 and 280 nm. Also, spectrophotometric analysis counts both fragments long enough for amplification and those that are not.
For TYMS 5′ UTR, the discordance rate was minor (5%) but still more than that experienced for SNPs. We have since repeated the TYMS 5′-UTR 2R/3R genotyping on 241 FFPE DNAs on independent runs spaced apart by 1.5 years, and in this setting the TYMS 5′-UTR 2R/3R discordance rate was 10.37% (data not shown). These results show that TYMS 5′-UTR 2R/3R genotype results can be discordant, albeit at a minor level, when applied to FFPE DNA.
Several reasons may explain the differences in discordance rates between the GSTT1 and TYMS 5′-UTR 2R/3R results. First, for example, differences in the sequences of the targeting primers may have resulted in subtle effects on primer binding, leading to differences in PCR amplification rates (eg, melting temperature for 5′ TYMS primers were 63°C, but 58°C for GSTT1 primers). Second, alternatively, differences in the target amplicon size (5′ TYMS versus GSTT1: 140 versus 229 bp) may also contribute to the discordance, particularly when applied to FFPE DNA that is highly fragmented. Third, the use of a multiplex PCR involving a control gene (ACTB) in the GSTT1 may have resulted in within-reaction competition between PCR primers and template DNA, exacerbating the ability of certain fragments to be amplified and not others. Further research should be directed to exploring these possibilities, such that important polymorphisms, such as GSTT1 deletion, can be reliably determined from FFPE DNA. Another possibility is to explore the use of other technology platforms for measuring such size-based polymorphisms, such as real-time PCR.31,32
In conclusion, our data suggest that DNA from FFPE material can be used to reliably test germline SNVs/SNPs by the MassARRAY iPLEX system, particularly when assessing common variants. However, our results based on GSTT1 and TYMS 5′-UTR 2R/3R suggest that results using FFPE DNA to measure copy number/size-based biomarkers may need to be interpreted with caution. On a practical level, our results suggest that when measuring size-based polymorphisms using FFPE DNA, independent repeated assays should routinely be performed on all samples, to determine the potential existence and rate of discrepancies.
Acknowledgment
We thank all of the study participants and OEO2, JUST, and MAGIC groups for providing the formalin-fixed, paraffin-embedded samples.
Footnotes
Supported by the National University of Singapore Graduate School for Integrative Sciences and Engineering Scholarship (I.B.T.), The National Institute for Health Research (NIHR), Royal Marsden (RM)/ The Institute of Cancer Research (ICR) Biomedical Research Centre (E.C.S., D.C., and A.W.), and grants NMRC/TCR/009-NUHS/2013 and BMRC 10/1/24/19/655 (P.T.). Collection and analysis of samples from the OEO2 and MAGIC trials was funded by Cancer Research UK grants C26441/A8944 and C20023/A7217, respectively, with support from the Medical Research Council (MRC) through the MRC Clinical Trials Unit, which sponsored and coordinated the MAGIC trial. JUST study was funded by a non-profit organization, Kanagawa Standard Anti-Cancer Therapy Support System.
Disclosures: None declared.
Current address of H.I.G., Department of Pathology, GROW School for Oncology and Development Biology, Maastricht University Medical Center, Maastricht, the Netherlands.
Supplemental Data
References
- 1.Hahn W.C., Weinberg R.A. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 2.Relling M.V., Dervieux T. Pharmacogenetics and cancer therapy. Nat Rev Cancer. 2001;1:99–108. doi: 10.1038/35101056. [DOI] [PubMed] [Google Scholar]
- 3.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig J.A., Weinstein J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Hahnvajanawong C., Chaiyagool J., Seubwai W., Bhudhisawasdi V., Namwat N., Khuntikeo N., Sripa B., Pugkhem A., Tassaneeyakul W. Orotate phosphoribosyl transferase mRNA expression and the response of cholangiocarcinoma to 5-fluorouracil. World J Gastroenterol. 2012;18:3955–3961. doi: 10.3748/wjg.v18.i30.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwab M., Zanger U.M., Marx C., Schaeffeler E., Klein K., Dippon J., Kerb R., Blievernicht J., Fischer J., Hofmann U., Bokemeyer C., Eichelbaum M., German 5-FU Toxicity Study Group Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU toxicity study group. J Clin Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H., Li Y.M., Zhang H., Jin X. DPYD*5 gene mutation contributes to the reduced DPYD enzyme activity and chemotherapeutic toxicity of 5-FU. Med Oncol. 2007;24:251–258. doi: 10.1007/BF02698048. [DOI] [PubMed] [Google Scholar]
- 8.Lecomte T., Ferraz J.M., Zinzindohoué F., Loriot M.A., Tregouet D.A., Landi B., Berger A., Cugnenc P.H., Jian R., Beaune P., Laurent-Puig P. Thymidylate synthase gene polymorphism predicts toxicity in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. Clin Cancer Res. 2004;10:5880–5888. doi: 10.1158/1078-0432.CCR-04-0169. [DOI] [PubMed] [Google Scholar]
- 9.Cheng J., Ha M., Wang Y., Sun J., Chen J., Wang Y., Tong C. A C118T polymorphism of ERCC1 and response to cisplatin chemotherapy in patients with late-stage non-small cell lung cancer. J Cancer Res Clin Oncol. 2012;138:231–238. doi: 10.1007/s00432-011-1090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiraishi K., Kohno T., Tanai C., Goto Y., Kuchiba A., Yamamoto S., Tsuta K., Nokihara H., Yamamoto N., Sekine I., Ohe Y., Tamura T., Yokota J., Kunitoh H. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol. 2010;28:4945–4952. doi: 10.1200/JCO.2010.30.5334. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z., Chen J.Q., Liu J.L., Qin X.G., Huang Y. Polymorphisms in ERCC1, GSTs, TS and MTHFR predict clinical outcomes of gastric cancer patients treated with platinum/5-Fu-based chemotherapy: a systematic review. BMC Gastroenterol. 2012;12:137. doi: 10.1186/1471-230X-12-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quach N., Goodman M.F., Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol. 2004;4:1. doi: 10.1186/1472-6890-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pääbo S., Irwin D.M., Wilson A.C. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem. 1999;265:4718–4721. [PubMed] [Google Scholar]
- 14.Williams C., Pontén F., Moberg C., Söderkvist P., Uhlén M., Pontén J., Sitbon G., Lundeberg J. A high frequency of sequence alterations is due to formalin fixation of achival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agell L., Hernández S., de Muga S., Lorente J.A., Juanpere N., Esgueva R., Serrano S., Gelabert A., Lloreta J. KLF6 and TP53 mutations are a rare event in prostate cancer: distinguishing between Taq polymerase artifacts and true mutations. Mod Pathol. 2008;21:1470–1478. doi: 10.1038/modpathol.2008.145. [DOI] [PubMed] [Google Scholar]
- 16.Marchetti A., Felicioni L., Buttitta F. Assessing EGFR mutations. N Engl J Med. 2006;354:526–528. doi: 10.1056/NEJMc052564. [DOI] [PubMed] [Google Scholar]
- 17.Ahern T.P., Christensen M., Cronin-Fenton D.P., Lunetta K.L., Rosenberg C.L., Sørensen H.T., Lash T.L., Hamilton-Dutoit S. Concordance of metabolic enzyme genotypes assayed from paraffin-embedded, formalin-fixed breast tumors and normal lymphatic tissue. Clin Epidemiol. 2010;2:241–246. doi: 10.2147/CLEP.S13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rae J.M., Regan M.M., Thibert J.N., Gersch C., Thomas D., Leyland-Jones B., Viale G., Pusztai L., Hayes D.F., Skaar T., Van Poznak C. Concordance between CYP2D6 genotypes obtained from tumor-derived and germline DNA. J Natl Cancer Inst. 2013;105:1332–1334. doi: 10.1093/jnci/djt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonacopoulou A.G., Floratou K., Bravou V., Kottorou A., Dimitrakopoulos F.I., Marousi S., Stavropoulos M., Koutras A.K., Scopa C.D., Kalofonos H.P. The survivin -31 snp in human colorectal cancer correlates with survivin splice variant expression and improved overall survival. Cell Oncol (Dordr) 2011;34:381–391. doi: 10.1007/s13402-011-0038-4. [DOI] [PubMed] [Google Scholar]
- 20.Burchard P.R., Malhotra S., Kaur P., Tsongalis G.J. Detection of the FCGR3a polymorphism using a real-time polymerase chain reaction assay. Cancer Genet. 2013;206:130–134. doi: 10.1016/j.cancergen.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Wright W.T., Heggarty S.V., Young I.S., Nicholls D.P., Whittall R., Humphries S.E., Graham C.A. Multiplex MassArray spectrometry (iPLEX) produces a fast and economical test for 56 familial hypercholesterolaemia-causing mutations. Clin Genet. 2008;74:463–468. doi: 10.1111/j.1399-0004.2008.01071.x. [DOI] [PubMed] [Google Scholar]
- 22.Syrmis M.W., Moser R.J., Whiley D.M., Vaska V., Coombs G.W., Nissen M.D., Sloots T.P., Nimmo G.R. Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus. Clin Microbiol Infect. 2011;17:1804–1810. doi: 10.1111/j.1469-0691.2011.03521.x. [DOI] [PubMed] [Google Scholar]
- 23.Perkins G., Yap T.A., Pope L., Cassidy A.M., Dukes J.P., Riisnaes R., Massard C., Cassier P.A., Miranda S., Clark J., Denholm K.A., Thway K., Gonzalez De Castro D., Attard G., Molife L.R., Kaye S.B., Banerji U., de Bono J.S. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS One. 2012;7:e47020. doi: 10.1371/journal.pone.0047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cote M.L., Atikukke G., Ruterbusch J.J., Olson S.H., Sealy-Jefferson S., Rybicki B.A., Alford S.H., Elshaikh M.A., Gaba A.R., Schultz D., Haddad R., Munkarah A.R., Ali-Fehmi R. Racial differences in oncogene mutations detected in early-stage low-grade endometrial cancers. Int J Gynecol Cancer. 2012;22:1367–1372. doi: 10.1097/IGC.0b013e31826b1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng J., Gao L., Guo J., Wang T., Wang L., Yao Q., Zhu H., Jin Q. Type-specific detection of 30 oncogenic human papillomaviruses by genotyping both E6 and L1 genes. J Clin Microbiol. 2013;51:402–408. doi: 10.1128/JCM.01170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allum W.H., Stenning S.P., Bancewicz J., Clark P.I., Langley R.E. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. 2009;27:5062–5067. doi: 10.1200/JCO.2009.22.2083. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham D., Allum W.H., Stenning S.P., Thompson J.N., Van de Velde C.J., Nicolson M., Scarffe J.H., Lofts F.J., Falk S.J., Iveson T.J., Smith D.B., Langley R.E., Verma M., Weeden S., Chua Y.J., MAGIC Trial Participants Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355:11–20. doi: 10.1056/NEJMoa055531. [DOI] [PubMed] [Google Scholar]
- 28.Sakuramoto S., Sasako M., Yamaguchi T., Kinoshita T., Fujii M., Nashimoto A., Furukawa H., Nakajima T., Ohashi Y., Imamura H., Higashino M., Yamamura Y., Kurita A., Arai K., ACTS-GC Group Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007;357:1810–1820. doi: 10.1056/NEJMoa072252. [DOI] [PubMed] [Google Scholar]
- 29.Kawakami K., Watanabe G. Identification and functional analysis of single nucleotide polymorphism in the tandem repeat sequence of thymidylate synthase gene. Cancer Res. 2003;63:6004–6007. [PubMed] [Google Scholar]
- 30.Singh S., Kumar V., Thakur S., Banerjee B.D., Grover S.S., Rawat D.S., Pasha S.T., Jain S.K., Lal S., Rai A. Genetic polymorphism of glutathione S-transferase M1 and T1 in Delhi population of Northern India. Environ Toxicol Pharmacol. 2009;28:25–29. doi: 10.1016/j.etap.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Koh W.P., Nelson H.H., Yuan J.M., Van den Berg D., Jin A., Wang R., Yu M.C. Glutathione S-transferase (GST) gene polymorphisms, cigarette smoking and colorectal cancer risk among Chinese in Singapore. Carcinogenesis. 2011;32:1507–1511. doi: 10.1093/carcin/bgr175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson A.E., Cohn R.J., Ashton L.J. Use of formalin-fixed paraffin-embedded tumor tissue as a DNA source in molecular epidemiological studies of pediatric CNS tumors. Diagn Mol Pathol. 2012;21:105–113. doi: 10.1097/PDM.0b013e3182340a78. [DOI] [PubMed] [Google Scholar]
- 33.Armitage P., Berry G. ed 2. Blackwell; Oxford: 1987. Statistical Methods in Medical Research; pp. 117–120. [Google Scholar]
- 34.Bouakaze C., Keyser C., Gonzalez A., Sougakoff W., Veziris N., Dabernat H., Jaulhac B., Ludes B. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based single nucleotide polymorphism genotyping assay using iPLEX gold technology for identification of Mycobacterium tuberculosis complex species and lineages. J Clin Microbiol. 2011;49:3292–3299. doi: 10.1128/JCM.00744-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaremko M., Justenhoven C., Abraham B.K., Schroth W., Fritz P., Brod S., Vollmert C., Illig T., Brauch H. MALDI-TOF MS and TaqMan assisted SNP genotyping of DNA isolated from formalin-fixed and paraffin-embedded tissues (FFPET) Hum Mutat. 2005;25:232–238. doi: 10.1002/humu.20141. [DOI] [PubMed] [Google Scholar]
- 36.Horn H., Pott C., Kalla J., Dreyling M., Rosenwald A., Ott G., Schwab M., Schaeffeler E. A multiplex MALDI-TOF MS approach facilitates genotyping of DNA from formalin-fixed paraffin-embedded tumour specimens. Pharmacogenet Genomics. 2010;20:598–604. doi: 10.1097/FPC.0b013e32833deb16. [DOI] [PubMed] [Google Scholar]
- 37.Imyanitov E.N., Grigoriev M.Y., Gorodinskaya V.M., Kuligina E.S., Pozharisski K.M., Togo A.V., Hanson K.P. Partial restoration of degraded DNA from archival paraffin-embedded tissues. Biotechniques. 2001;31:1000–1002. doi: 10.2144/01315bm04. [DOI] [PubMed] [Google Scholar]
- 38.Fariña-Sarasqueta A., Van Lijnschoten G., Rutten H.J., Van den Brule A.J. Value of gene polymorphisms as markers of 5-FU therapy response in stage III colon carcinoma: a pilot study. Cancer Chemother Pharmacol. 2010;66:1167–1171. doi: 10.1007/s00280-010-1403-0. [DOI] [PubMed] [Google Scholar]
- 39.Salgado J., Zabalegui N., Gil C., Monreal I., Rodríguez J., García-Foncillas J. Polymorphisms in the thymidylate synthase and dihydropyrimidine dehydrogenase genes predict response and toxicity to capecitabine-raltitrexed in colorectal cancer. Oncol Rep. 2007;17:325–328. [PubMed] [Google Scholar]
- 40.Jennings B.A., Loke Y.K., Skinner J., Keane M., Chu G.S., Turner R., Epurescu D., Barrett A., Willis G. Evaluating predictive pharmacogenetic signatures of adverse events in colorectal cancer patients treated with fluoropyrimidines. PLoS One. 2013;8:e78053. doi: 10.1371/journal.pone.0078053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palli D., Polidoro S., D'Errico M., Saieva C., Guarrera S., Calcagnile A.S., Sera F., Allione A., Gemma S., Zanna I., Filomena A., Testai E., Caini S., Moretti R., Gomez-Miguel M.J., Nesi G., Luzzi I., Ottini L., Masala G., Matullo G., Dogliotti E. Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis. 2010;25:569–575. doi: 10.1093/mutage/geq042. [DOI] [PubMed] [Google Scholar]
- 42.Santos L.S., Gomes B.C., Gouveia R., Silva S.N., Azevedo A.P., Camacho V., Manita I., Gil O.M., Ferreira T.C., Limbert E., Rueff J., Gaspar J.F. The role of CCNH Val270Ala (rs2230641) and other nucleotide excision repair polymorphisms in individual susceptibility to well-differentiated thyroid cancer. Oncol Rep. 2013;30:2458–2466. doi: 10.3892/or.2013.2702. [DOI] [PubMed] [Google Scholar]
- 43.Moreno V., Gemignani F., Landi S., Gioia-Patricola L., Chabrier A., Blanco I., González S., Guino E., Capellà G., Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 44.Katoh T., Nagata N., Kuroda Y., Itoh H., Kawahara A., Kuroki N., Ookuma R., Bell D.A. Glutathione S-transferase M1 (GSTM1) and T1 (GSTT1) genetic polymorphism and susceptibility to gastric and colorectal adenocarcinoma. Carcinogenesis. 1996;17:1855–1859. doi: 10.1093/carcin/17.9.1855. [DOI] [PubMed] [Google Scholar]
- 45.Deakin M., Elder J., Hendrickse C., Peckham D., Baldwin D., Pantin C., Wild N., Leopard P., Bell D.A., Jones P., Duncan H., Brannigan K., Alldersea J., Fryer A.A., Strange R.C. Glutathione S-transferase GSTT1 genotypes and susceptibility to cancer: studies of interactions with GSTM1 in lung, oral, gastric and colorectal cancers. Carcinogenesis. 1996;17:881–884. doi: 10.1093/carcin/17.4.881. [DOI] [PubMed] [Google Scholar]
- 46.Shitara K., Muro K., Ito S., Sawaki A., Tajika M., Kawai H., Yokota T., Takahari D., Shibata T., Ura T., Ito H., Hosono S., Kawase T., Watanabe M., Tajima K., Yatabe Y., Tanaka H., Matsuo K. Folate intake along with genetic polymorphisms in methylenetetrahydrofolate reductase and thymidylate synthase in patients with advanced gastric cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:1311–1319. doi: 10.1158/1055-9965.EPI-09-1257. [DOI] [PubMed] [Google Scholar]
- 47.Graziano F., Kawakami K., Watanabe G., Ruzzo A., Humar B., Santini D., Catalano V., Ficarelli R., Merriman T., Panunzi S., Testa E., Cascinu S., Bearzi I., Tonini G., Magnani M. Association of thymidylate synthase polymorphisms with gastric cancer susceptibility. Int J Cancer. 2004;112:1010–1014. doi: 10.1002/ijc.20489. [DOI] [PubMed] [Google Scholar]
- 48.Soong R., Ladányi A. Improved indicators for assessing the reliability of detection and quantification by kinetic PCR. Clin Chem. 2003;49:973–976. doi: 10.1373/49.6.973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.