Coarse-grained modeling and all-atom molecular dynamics simulation provide insight into the mechanism for heat activation of TRPV1 gating.
Abstract
The transient receptor potential (TRP) channels act as key sensors of various chemical and physical stimuli in eukaryotic cells. Despite years of study, the molecular mechanisms of TRP channel activation remain unclear. To elucidate the structural, dynamic, and energetic basis of gating in TRPV1 (a founding member of the TRPV subfamily), we performed coarse-grained modeling and all-atom molecular dynamics (MD) simulation based on the recently solved high resolution structures of the open and closed form of TRPV1. Our coarse-grained normal mode analysis captures two key modes of collective motions involved in the TRPV1 gating transition, featuring a quaternary twist motion of the transmembrane domains (TMDs) relative to the intracellular domains (ICDs). Our transition pathway modeling predicts a sequence of structural movements that propagate from the ICDs to the TMDs via key interface domains (including the membrane proximal domain and the C-terminal domain), leading to sequential opening of the selectivity filter followed by the lower gate in the channel pore (confirmed by modeling conformational changes induced by the activation of ICDs). The above findings of coarse-grained modeling are robust to perturbation by lipids. Finally, our MD simulation of the ICD identifies key residues that contribute differently to the nonpolar energy of the open and closed state, and these residues are predicted to control the temperature sensitivity of TRPV1 gating. These computational predictions offer new insights to the mechanism for heat activation of TRPV1 gating, and will guide our future electrophysiology and mutagenesis studies.
INTRODUCTION
The mammalian transient receptor potential (TRP) channels are a superfamily of polymodal cation-permeable channels divided into six subfamilies (Montell, 2005) based on sequence homology (TRPC, TRPV, TRPM, TRPP, TRPML, and TRPA). TRP channels are versatile cellular sensors (Clapham, 2003; Voets et al., 2005) activated and regulated by various physical and chemical stimuli such as heat (Caterina et al., 1997, 1999), cold (Story et al., 2003; Karashima et al., 2009; Nilius et al., 2012), voltage (Voets et al., 2004), acid (Tominaga et al., 1998; Jordt et al., 2000), force (Howard and Bechstedt, 2004; Sotomayor et al., 2005; Yin and Kuebler, 2010), exogenous ligands (e.g., capsaicin; Caterina et al., 1997), endogenous agents (e.g., ATP; Lishko et al., 2007), and phosphatidylinositol-4,5-bisphosphate (PIP2; Qin, 2007; Rohacs and Nilius, 2007). TRP channels are among the most actively pursued drug targets (Gunthorpe and Szallasi, 2008; Nilius, 2013) in recent years thanks to their extensive involvements in many intracellular signaling pathways and pathophysiology linked to various diseases (Nilius, 2007; Nilius et al., 2005b).
As a founding member of the TRPV subfamily, TRPV1 forms a homotetramer. Each of its four subunits consists of a six-helix (S1–S6) transmembrane domain (TMD) and an intracellular domain (ICD; see Fig. 1 A). The TMD consists of two functional modules: the S1–S4 voltage-sensing module (Voets et al., 2007) on the channel periphery and the S5–S6 pore module enclosing a central pore (Fig. 1, A and B). The N-terminal part of ICD is an ankyrin repeats domain (ARD; Fig. 1 A) comprised of six ankyrin repeats, which are ubiquitous motifs involved in protein–ligand interactions (Gaudet, 2008a). The C-terminal domain (CTD) of ICD (Fig. 1 A) contains a highly conserved TRP box helix (Montell, 2001), which is implicated in coupling to TRPV1 gating (García-Sanz et al., 2007) and interactions with other proteins/ligands (Numazaki et al., 2003; Prescott and Julius, 2003). At the TMD–ICD interface is a membrane proximal domain (MPD; Fig. 1 A), which was found to harbor a heat-sensing module in the TRPV subfamily (Yao et al., 2011). Alternative heating-sensing sites were found in CTD (Vlachová et al., 2003; Brauchi et al., 2006, 2007) and outer pore (Grandl et al., 2010; Yang et al., 2010; Cui et al., 2012; Kim et al., 2013). It remains uncertain which specific residues are involved in heat activation of TRPV1 and how they communicate with the channel gate to trigger its opening (e.g., via allosteric coupling; Latorre et al., 2007).
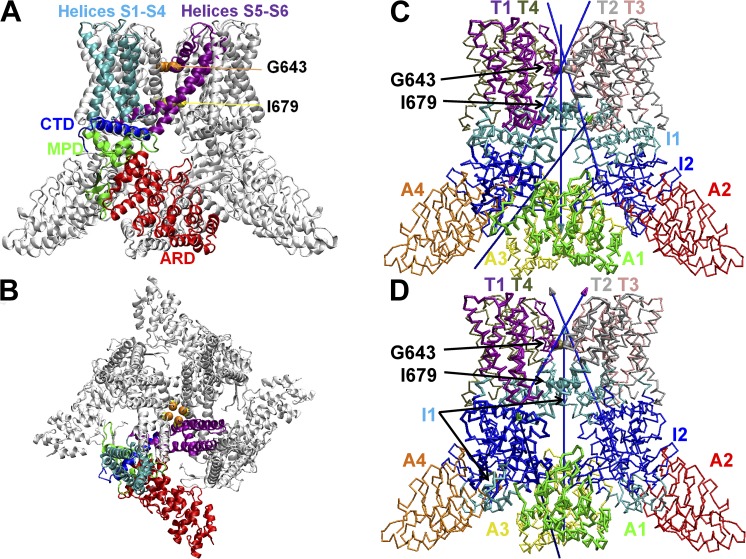
Figure 1.
Structural architecture of TRPV1. (A) The side view showing ARD (red), MPD (green), helices S1–S4 (cyan), helices S5–S6 (purple), and CTD (blue) for a representative subunit, with the remaining subunits colored in light gray, and residues G643 and I679 (at the selectivity filter and lower gate) shown as spheres (colored in orange and yellow, respectively). (B) The top view with the same color scheme as A. (C) Dynamic domain partition for mode 6, where domains T1–T4 are colored purple, silver, pink, and tan; domains I1 and I2 are colored cyan and blue; domains A1–A4 are colored green, red, yellow, and orange; the arrows represent the rotational axes for interdomain movements using the following color scheme: the axis of rotation of domain X relative to domain I2 is represented by an arrow with the tip colored the same as domain X; residues G643 and I679 are shown as spheres. (D) Dynamic domain partition for mode 13 with the same color scheme as C.
To elucidate the molecular mechanisms of TRP channels, tremendous efforts in structural biology (Gaudet, 2008b; Hellmich and Gaudet, 2014) have resulted in low resolution (>13-Å) structures of TRP channels visualized by electron microscopy (Mio et al., 2005, 2007; Maruyama et al., 2007; Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010; Cvetkov et al., 2011; Huynh et al., 2014), and high resolution structures for various fragments of TRP channels solved by x-ray crystallography, including the isolated ARD of TRPV channels (Jin et al., 2006; McCleverty et al., 2006; Lishko et al., 2007; Phelps et al., 2008; Landouré et al., 2010; Inada et al., 2012; Shi et al., 2013). In two landmark papers published in 2013, high resolution (3–4-Å) structures were solved by cryo-electron microscopy (Cao et al., 2013; Liao et al., 2013) for a minimal functional construct of rat TRPV1 in three distinct forms: a closed-channel apo form, an open-channel form bound with DkTx (a peptide toxin) and RTX (a small vanilloid agonist), and an intermediate form bound with capsaicin (an agonist). The apo structure is thought to capture the closed (C) state of TRPV1 channel, and the DkTx–RTX-bound structure likely captures the open (O) state of TRPV1, although other possibilities like a desensitized state cannot yet be ruled out. These structures revealed a dual gate with two constrictions in the channel pore (Fig. 1, A and B): a selectivity filter or upper gate (near residue G643) and a lower gate (near residue I679). Both are closed (open) in the closed-channel (open-channel) structure, whereas the selectivity filter is closed and the lower gate is partially open in the capsaicin-bound structure (Cao et al., 2013; Liao et al., 2013; see Fig. 3 C). Despite great functional insights offered by these structures, to fully understand the activation and regulation mechanisms of TRP channels, we need quantitative knowledge of the underlying dynamics and energetics, which cannot be directly deduced from static structures. It is imperative to obtain and comprehend high resolution structural and dynamic information for key functional states (i.e., C and O state) and the gating transition between them in TRP channels.
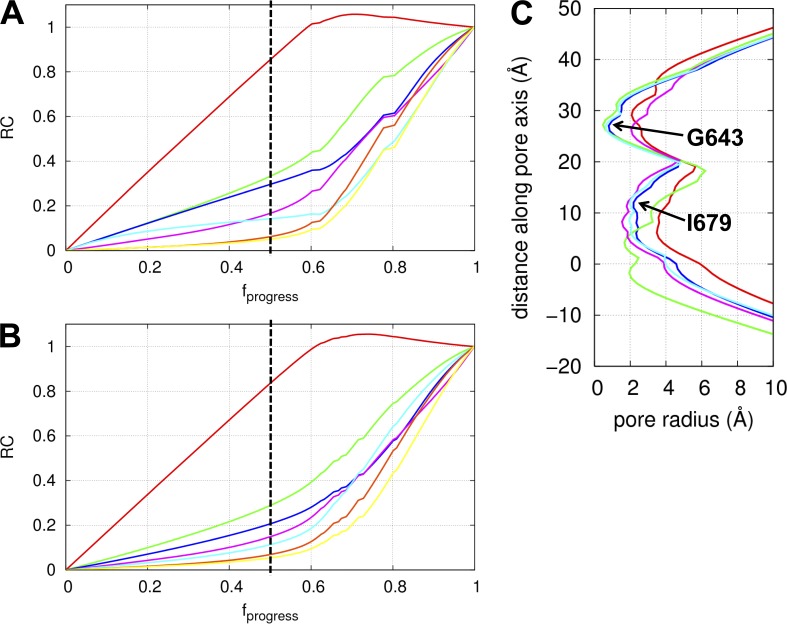
Figure 3.
Results of the transition pathway modeling in the absence or presence of lipids. (A) Reaction coordinates along the closed-to-open transition pathway (calculated in the absence of lipids) for the following functional parts of TRPV1: ARD (red), MPD (green), CTD (blue), helices S1–S4 (cyan), helices S5–S6 (purple), and residues G643 (orange) and I679 (yellow). (B) Reaction coordinates along the closed-to-open transition pathway (calculated in the presence of lipids; same color scheme as A). (C) Pore radius as a function of distance along the pore axis for the C-state structure (blue; PDB accession no. 3J5P), O-state structure (red; PDB accession no. 3J5Q), capsaicin-bound structure (green; PDB accession no. 3J5R), and the ICD-activated model of TRPV1 calculated in the absence of lipids (purple) and presence of lipids (cyan). The two constrictions at residues G643 and I679 are shown. This calculation was done using the HOLE program (Smart et al., 1996).
Molecular dynamics (MD) simulation is the method of choice for exploring protein dynamics and energetics under physiological conditions with atomic resolution (Karplus and McCammon, 2002). MD was used previously to study various ion channels (Lindahl and Sansom, 2008; Nury et al., 2010, 2011; Zhu and Hummer, 2010; Dong and Zhou, 2011; Delemotte et al., 2012; Du et al., 2012; Vargas et al., 2012; Calimet et al., 2013), including truncated domains (Sotomayor et al., 2005; Susankova et al., 2007) and homology models (Brauchi et al., 2007; Susankova et al., 2007; Pedretti et al., 2011) of TRP channels. However, MD simulation is computationally expensive. Until recently, it was impractical to simulate an ion channel beyond tens of nanoseconds (ns) without using a massively parallelized or special-purpose supercomputer (Jensen et al., 2012). Thanks to rapid progress in computational hardware and software (e.g., use of graphics-processing unit to accelerate MD simulation; Stone et al., 2010), one can now routinely run MD simulation of a large biomolecular system (with >105 atoms) at a speed of >1 ns/day on a single computer node. However, it remains difficult for MD to access microsecond–millisecond time scales relevant to many functionally important conformational transitions of protein complexes (e.g., the gating of a TRP channel).
To overcome the timescale limit of MD simulation, coarse-grained modeling has been vigorously pursued using reduced protein representations (e.g., one bead per amino acid residue) and simplified force fields (Tozzini, 2005, 2010). As a popular example of coarse-grained models, the elastic network model (ENM) is constructed by connecting nearby Cα atoms with harmonic springs (Atilgan et al., 2001; Tama and Sanejouand, 2001; Zheng and Doniach, 2003). Despite its simplicity, the normal mode analysis (NMA) of ENM can be used to predict a few low frequency modes of collective motions, which often compare well with conformational changes observed between experimentally solved protein structures in different functional states (e.g., the gating conformational changes in a pentameric ligand-gated ion channel; Krebs et al., 2002; Taly et al., 2005; Sauguet et al., 2014). Numerous studies have established ENM as a useful and efficient means to probe dynamic mechanisms of protein complexes (including membrane proteins; Bahar et al., 2010) with virtually no limit in timescale or system size (Bahar and Rader, 2005; Tama and Brooks, 2006). ENM has formed the basis for our coarse-grained modeling of various conformational transitions between two given protein structures (Zheng et al., 2007a; Tekpinar and Zheng, 2010, 2013; Zheng, 2010, 2011, 2012; Zheng and Auerbach, 2011; Zheng and Tekpinar, 2012), such as the closed and open conformations of a pentameric ligand-gated ion channel (Zheng and Auerbach, 2011).
In this study, to elucidate the molecular mechanism of TRPV1 gating, we have performed a comprehensive coarse-grained modeling and all-atom MD simulation based on the recently solved high resolution structures of open and closed form of TRPV1 (Cao et al., 2013; Liao et al., 2013). Our coarse-grained NMA has captured two key modes of collective motions involved in the gating transition of TRPV1, featuring a quaternary twist motion of the TMDs relative to the ICDs. Our transition pathway modeling has predicted a sequence of structural motions propagating from the ICDs to the TMDs via the MPDs and the CTDs, leading to sequential opening of the selectivity filter followed by the lower gate in the channel pore (confirmed by our modeling of conformational changes induced by the activation of ICDs). The above findings of coarse-grained modeling are shown to be robust to the perturbation of lipids. Finally, our MD simulation of the ICD has identified key residues that contribute differently to the nonpolar energy in the open and closed state, which are predicted to control the temperature sensitivity of TRPV1 gating. This study is, to our knowledge, the first detailed modeling/simulation study of full-length TRPV1 after the publication of the TRPV1 structures (Cao et al., 2013; Liao et al., 2013). The novel computational predictions from this study will guide our future electrophysiology and mutagenesis studies to investigate the heat-activation mechanism of TRPV1 gating.
MATERIALS AND METHODS
ENM and NMA
In an ENM, a protein structure is represented as a network of Cα atoms of amino acid residues. Harmonic springs link all pairs of Cα atoms within a cutoff distance Rc chosen to be 10 Å (following Hafner and Zheng, 2010). The ENM potential energy is:
| (1) |
where N is the number of Cα atoms, θ(x) is the Heaviside function, dij is the distance between the Cα atom i and j, and is the value of dij as given by a reference structure. The spring constant kij is 1 for nonbonded interactions and 10 for bonded interactions (in arbitrary units).
In NMA, the following eigenproblem is solved for the Hessian matrix H, which is obtained by calculating the second derivatives of ENM potential energy (see Zheng and Auerbach, 2011):
| (2) |
where λm and Wm represent the eigenvalue and eigenvector of mode m. After excluding six zero modes corresponding to three rotations and three translations, we kept 3N-6 nonzero modes, which are numbered from 1 to 3N-6 in order of ascending eigenvalue.
To validate ENM-based NMA, we compared each mode (i.e., mode m) with the observed structural changes Xobs between two superimposed experimental structures by calculating an overlap value Im (Zheng, 2012). Im varies between 0 and 1, with higher value meaning greater similarity. gives the fractional contribution of mode m to Xobs. The sum of over the lowest M modes (named cumulative squared overlap) gives the fractional contribution of these modes to Xobs (Zheng, 2012). To assess the local flexibility at individual residue positions as described by the lowest M modes, we define the following cumulative flexibility:
where Wm,nx, Wm,ny, and Wm,nz are the x, y, and z component of mode m’s eigenvector at residue n. We have analyzed the lowest M = 20 and 100 modes (see Fig. 2 and Fig. S1 for results).
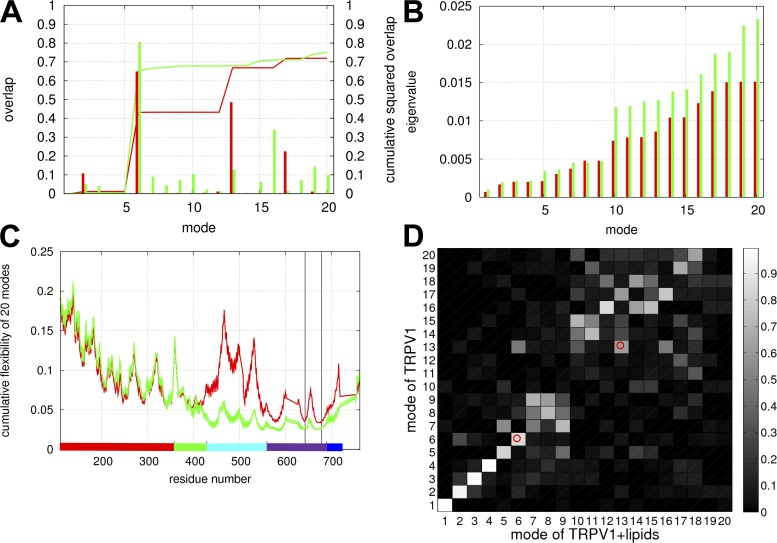
Figure 2.
Results of NMA for the lowest 20 modes calculated in the absence (red) or presence (green) of lipids. (A) The overlap and cumulative squared overlap as a function of mode number (shown as impulses and lines, respectively). (B) The eigenvalue as a function of mode number. (C) The cumulative flexibility as a function of residue number. (D) The all-to-all pairwise overlaps between the modes calculated in the absence or presence of lipids (with high/low overlap corresponding to light/dark gray). In C, the ranges of residue numbers are colored as follows: red, ARD; green, MPD; cyan, S1–S4 helices; purple, S5–S6 helices; blue, CTD. The positions of residue G643 and I679 are marked by vertical lines. In D, the two cells corresponding to the overlaps of modes 6 and 13 are marked by small circles.
We performed the above ENM-based NMA using the AD-ENM webserver (http://enm.lobos.nih.gov/start.html).
ENM of TRPV1 in the presence of lipids
We used the CHARMM-GUI PACE CG Builder (Qi et al., 2014) to embed the C-state structure of TRPV1 (Protein Data Bank [PDB] accession no. 3J5P) in a coarse-grained bilayer of POPC lipids and keep 102 POPC molecules within 10 Å of the Cα atoms of TRPV1. Each POPC molecule contains 13 coarse-grained beads as defined in the Martini force field (Marrink et al., 2007). Then we built an ENM of 2,368 Cα beads and 1,326 POPC beads with springs connecting any two beads within a cutoff distance of 10 Å.
Coarse-grained transition pathway modeling by interpolated ENM (iENM)
We previously developed an iENM protocol to construct a transition pathway (i.e., a series of intermediate conformations) between two given protein conformations by solving the saddle points of a double-well potential built from these two conformations (Tekpinar and Zheng, 2010). Here we applied this method to the closed and open conformation of TRPV1 using the iENM webserver (http://enm.lobos.nih.gov/start_ienm.html).
The iENM can be used to model an unknown activated conformation from a known inactivated conformation together with a target conformation of the activation domain. The idea is to progressively transform the activation domain toward its target conformation and let the rest of the protein relax to a series of minimal-energy conformations leading to a final model for the unknown activated conformation. We have recently used this method to build the pre-powerstroke conformation of dynein motor domain from its post-powerstroke conformation after ATP binding (Zheng, 2012). Here, we used this method to model an intermediate conformation of TRPV1 upon activation of the ICDs toward channel opening.
Using the iENM-predicted transition pathway, we can determine the motional order of various protein parts by calculating a reaction coordinate for each part S (denoted RCS; for details see Zheng, 2010). RCS varies from 0 to 1 as the transition proceeds from the beginning to the end conformation. For two protein parts S and S′, if RCS < RCS′ along the pathway, we infer that the motion of S′ precedes that of S. Here, we plotted RCS as a function of fprogress (a fractional progress parameter as defined in Zhu and Hummer, 2009) to track the evolution of RCS along the transition pathway.
Coarse-grained transition pathway modeling of TRPV1 in the presence of lipids
We first used iENM to construct a coarse-grained O-state model of TRPV1 in the presence of lipids. Starting from the C-state model of the TRPV1–lipids system (see above), we used iENM to progressively transform TRPV1 toward the O-state conformation (PDB accession no. 3J5Q) and let the surrounding lipids relax to a minimal-energy conformation. Then we used iENM to construct a transition pathway from the C-state model to the O-state model of the TRPV1–lipids system, and calculated reaction coordinates for various functional parts of TRPV1 (see above). We also used iENM to model an intermediate conformation of the TRPV1–lipids system upon activation of the ICDs toward channel opening (see above).
All-atom MD simulation of ICD
Based on two experimental structures of TRPV1 (PDB accession nos. 3J5P and 3J5Q), we prepared two systems (i.e., C and O state) for the truncated ICD of TRPV1 (residues 111–429 and 690–719). The protonation states of residues with ionizable side chains were determined by the PDB2PQR server (Dolinsky et al., 2007). The hydrogen atoms were added with the visual MD (VMD) program (Humphrey et al., 1996). Both systems were immersed into a rectangular box of water molecules extending 10 Å from the protein in each direction by using VMD. To ensure a physiological ionic concentration of 0.15 M and zero net charge, Na+ and Cl− ions were added to the systems by VMD. The systems were refined with two rounds of energy minimization using the steepest descent method: first, a 5,000-step energy minimization with harmonic constraints (force constant = 10 kcal/mol/Å2) applied to all backbone heavy atoms, then a 5,000-step energy minimization with harmonic constraints (force constant = 1 kcal/mol/Å2) applied to all Cα atoms. The systems were then heated to 300 K over 300 ps by MD with the same constraints as the second minimization. Then the systems were equilibrated for 500 ps with MD performed in the NVT ensemble with the same constraints as in heating. Finally, the systems were subject to production MD runs performed in the NPT ensemble. The Nosé–Hoover method (Martyna et al., 1999) was used with temperature T = 300 K and pressure P = 1 atm. The periodic boundary conditions were applied to the systems. A 10-Å switching distance and a 12-Å cutoff distance were used for nonbonded interactions. The particle mesh Ewald method (Deserno and Holm, 1998) was used to calculate long-range electrostatic interactions. The SHAKE algorithm (Hoover, 1985) was used to constrain the bond lengths of hydrogen-containing bonds, which allowed a time step of 2 fs for MD simulation. The energy minimization and MD simulation were performed with the NAMD program (Phillips et al., 2005) using the CHARMM27 force field (MacKerell et al., 1998) and TIP3P water model (Mackerell et al., 2004). For production MD runs in the C/O state, we applied harmonic constraints (force constant = 1 kcal/mol/Å2) to all Cα atoms to restrict conformational sampling near the experimentally solved backbone structures of TRPV1 (allowing side chains to relax freely), and ran five 20-ns-long MD trajectories (total 100 ns in simulation time).
Energetic analysis of ICD
We calculated the nonbonded energy in C/O state for 5 × 500 snapshots of the ICD during the last 10 ns of five MD trajectories. For each snapshot, we separately calculated the van der Waals (vdW) contribution EvdW based on CHARMM27 force field (MacKerell et al., 1998) and the electrostatic contribution Eelec using the Poisson–Boltzmann method (Gilson and Honig, 1988; Im et al., 1998). We used the CHARMM program (Brooks et al., 2009) to partition EvdW to contributions from individual ICD residues (denoted EvdW,n for residue n, following Li and Zheng, 2013, and Zheng et al., 2013). For the Poisson–Boltzmann calculation, a dielectric constant of 4 was used for the protein interior (Gilson and Honig, 1986; Sharp and Honig, 1990a,b; Olson and Reinke, 2000). A dielectric constant of 80 was used for the exterior aqueous environment. A probe radius of 1.4 Å was used to define the molecular surface corresponding to the dielectric boundary. The salt concentration was set to 0.15 M. All the Poisson–Boltzmann calculations were performed using the PBEQ module (Nina et al., 1997; Roux, 1997; Im et al., 1998) of the CHARMM program (Brooks et al., 1983). The atomic Born radii used here were previously calibrated and optimized to reproduce the electrostatic free energy of the 20 amino acids in MD simulations with explicit water molecules (Nina et al., 1997).
Conservation analysis
We used the ConSurf web server (http://consurf.tau.ac.il/) to assess the degree of residue conservation within the TRPV subfamily. Using the rat TRPV1 sequence as query, we obtained 194 homologous sequences of TRPV proteins from the UNIREF90 database with CS-BLAST (cutoff E-value = 0.0001; maximum/minimum sequence identity = 95/35%). Based on the multiple sequence alignment of these sequences, each residue position is assigned a grade of 1–9 (see supplemental material for details). If the grade is 8 or 9, then the residue is considered to be well conserved.
Online supplemental material
The supplemental material contains three figures, six videos, two PDB coordinate files for MD trajectories in the C and O state, and results of residue conservation analysis from the ConSurf server. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201411335/DC1.
RESULTS and DISCUSSION
ENM-based NMA predicts key collective motions involved in TRPV1 gating
Starting from the C-state structure of TRPV1 (PDB accession no. 3J5P), we constructed a Cα-only ENM, and then performed NMA, resulting in a spectrum of a total 7,098 normal modes (see Materials and methods). Here, we focus on the lowest 20 modes (for expanded results of the lowest 100 modes, see Fig. S1), each describing a specific pattern of collective motion energetically favored by the given structure (see below). For validation of these modes, we assessed how well they capture the experimentally observed conformational change from the C-state structure to the O-state structure of TRPV1 (PDB accession no. 3J5Q). To this end, we calculated the overlap between each mode and the observed closed-to-open conformational change and the cumulative squared overlap of the lowest 20 (100) modes (see Materials and methods). Encouragingly, the lowest 20 (100) modes have captured 72% (82%) of the observed conformational change, with modes 6 and 13 having the highest overlap (Fig. 2 A). Therefore, as found in many protein complexes (Tama and Sanejouand, 2001; Krebs et al., 2002), the ENM-based NMA offers a good description of the functional conformational change involved in TRPV1 gating.
We also compared the lowest 20 modes with the experimentally observed conformational change from the C-state structure to the capsaicin-bound structure of TRPV1 (PDB accession no. 3J5R). In contrast to the closed-to-open conformational change (see above), only 21% of the capsaicin-induced conformational change was captured by the lowest 20 modes, suggesting that high energy modes are involved in driving this conformational change. Collectively, the above findings imply that different gating transition pathways are induced by capsaicin and DkTx/RTX. Future study will be needed to explore the physiological significance of such differences. Here, we will focus on the closed-to-open conformational transition as induced by DkTx/RTX.
Next, we focused on the two modes that contribute most significantly to the closed-to-open conformational change in TRPV1: Mode 6 (with 0.65 overlap and 42% contribution to the observed conformational change) predicts a quaternary twist motion between the TMDs and the ICDs (i.e., a clockwise rotation of TMDs and a counterclockwise rotation of ICDs in the top view; see Video 1). Based on a dynamic domain partition analysis (Zheng et al., 2007b), the collective motion of this mode can be described in terms of rigid-body rotations between the following dynamic domains (Fig. 1 C):
(a) Four dynamic domains in the TMDs (named T1–T4; Fig. 1 C), with each consisting of the S1–S4 helices of one subunit and the S5–S6 helices of an adjacent subunit (with residue G643 of the selectivity filter at the interface of domains T1–T4; Fig. 1 C).
(b) One joint dynamic domain at the TMD–ICD interface (named I1; Fig. 1 C) comprised of residues 400–415 of the MPDs; residues 690–710 of the CTDs; and the bottom of S1, S5, and S6 helices (with residue I679 of the lower gate located inside domain I1; Fig. 1 C).
(c) Another joint dynamic domain at the TMD–ICD interface (named I2; Fig. 1 C) involving the rest of MPDs and the C-terminal residues 752–762.
(d) Four dynamic domains in the ICDs involving primarily four ARDs (named A1–A4; Fig. 1 C).
The above dynamic domain partition roughly agrees with the functional domain assignment (i.e., ARD, MPD, CTD, S1–S4 helices, and S5–S6 pore helices; Fig. 1, A and B), supporting the functional relevance of the collective motions between these domains described by this mode. Strategically located at the hinge between TMDs and ICDs, domains I1 and I2 may be sensitive to chemical/physical perturbations that affect this twist motion and thereby the gating transition of TRPV1. Interestingly, a similar quaternary twist motion was observed between the TMDs and the extracellular domains of a pentameric ligand-gated ion channel, and was proposed to drive its channel opening (Taly et al., 2005, 2006; Cheng et al., 2006; Zheng and Auerbach, 2011).
Mode 13 (with 0.49 overlap and 24% contribution to the observed conformational change) predicts a counterclockwise rotation of the TMDs and a hinge bending motion of the ICDs (see Video 2). According to the dynamic domain partition analysis (Zheng et al., 2007b), this mode can be described in terms of rigid-body rotations between the following dynamic domains (Fig. 1 D):
(a) Four dynamic domains in the TMDs (named T1–T4; Fig. 1 D), with each consisting of the S1–S4 helices of one subunit and the S5–S6 helices of an adjacent subunit (with residue G643 at the interface of domains T1–T4; Fig. 1 D).
(b) One joint dynamic domain at the TMD–ICD interface (named I1; Fig. 1 D) comprised of residues 310–330 of the ARDs; residues 355–370 of the MPDs; and the bottom of S1, S4, S5, and S6 helices (with residue I679 located inside domain I1; Fig. 1 D).
(c) Another joint dynamic domain at the TMD–ICD interface (named I2; Fig. 1 D) involving the rest of MPDs, the CTDs, and residues 280–310 of the ARDs.
(d) Four dynamic domains in the ICDs involving the rest of four ARDs (named A1–A4; Fig. 1 D).
Compared with mode 6, mode 13 describes a similar twist motion of domains T1–T4 and I1 relative to domain I2 (Fig. 1 D), but a different hinge bending motion of domains A1–A4 (Fig. 1 D). Unlike mode 6, domain I1 of mode 13 includes two disjoint parts, and domain I2 of mode 13 is larger (Fig. 1 D). Together, domains I1 and I2 encompass a larger hinge region in mode 13, involving more residues to sense external perturbations that affect the collective motion of this mode. Similar to mode 6, mode 13 shows no opening in the central pore (see Videos 1 and 2).
Besides the above two modes, other low frequency modes describe more collective motions (for detailed visualization of all the lowest 20 modes, check out the NMA results at the AD-ENM webserver: http://enm.lobos.nih.gov/runoutput/TRPV1/run.output). Together, these modes predict high flexibility toward the N terminus of ARDs, at the N terminus of MPDs, and in parts of S1–S4 helices, and low flexibility in the S5–S6 pore helices near residues G643 and I679 (Fig. 2 C). Consistent with our finding, functional data indicated that the ARD is structurally flexible (Liao et al., 2013) and may undergo large conformational change (Salazar et al., 2008), or even unfold in the absence of ligand (Croy et al., 2004).
In sum, our ENM-based NMA predicted two key modes of collective motions involved in the gating transition from the C state toward the O state. However, both modes show no sign of channel pore opening, which is not favored energetically by the C-state conformation of TRPV1.
NMA of TRPV1–lipid system supports the robustness of TRPV1 dynamics to perturbation of lipids
To assess how lipids around TRPV1 affect its collective motions, we repeated NMA for an ENM of TRPV1 in the presence of lipids (see Materials and methods), and compared it with the results of NMA in the absence of lipids (see above). As expected, the introduction of lipid–TMD interactions increases the energy of collective motions in TRPV1, as indicated by greater eigenvalues of the lowest 20 modes (Fig. 2 B). The local flexibility of TRPV1 is significantly reduced in the TMD (particularly in S1–S4 helices) but changes little in the ICD (Fig. 2 C). Then we calculated the overlap between each mode and the observed closed-to-open conformational change. Encouragingly, the lowest 20 (100) modes have captured 75% (83%) of the observed conformational change, with mode 6 having the highest overlap (Fig. 2 A). Next, we compared the lowest 20 modes solved in the presence and absence of lipids by calculating all-to-all pairwise overlaps between them (Fig. 2 D). Although some modes (such as modes 1–5) are essentially invariant to the perturbation of lipids, some (such as modes 10–12) are strongly perturbed by lipids (Fig. 2 D). Among the two key modes involved in the gating of TRPV1, the old mode 6 (solved in the absence of lipids) is highly similar to the new mode 6 (solved in the presence of lipids) with overlap of 0.84 (Fig. 2 D), whereas mode 13 is less invariant but still relatively robust to lipids (with overlap of 0.62; Fig. 2 D). In sum, despite the perturbation of lipids, the low frequency normal modes remain highly effective in capturing the gating conformational change in TRPV1. In particular, the key modes involved in the gating transition (especially mode 6) are robust to the perturbation of lipids. Our finding is consistent with the notion that the structural dynamics of a protein (including membrane protein) depends, to a large extent, on its native structure. However, our finding does not downplay the key role of a protein’s environment (such as lipids and solvent) in tuning its dynamics.
Transition pathway modeling reveals a sequence of structural events during TRPV1 gating
To determine the structural basis of TRPV1 gating, it is critical to probe the entire conformational transition from the C-state conformation to the O-state conformation (Cao et al., 2013; Liao et al., 2013). To this end, we have used the iENM protocol (see Materials and methods) to generate a transition pathway (consisting of a series of intermediate conformations; see Videos 3 and 4) between the two conformations. To assess the robustness of the predicted pathway, we repeated the C-to-O transition pathway modeling in the presence of more complete CTDs, which yielded similar results (see Fig. S3). By performing a reaction-coordinate (RC) analysis of the predicted transition pathway (see Materials and methods), we determined the motional order of the following numerically labeled functional parts of TRPV1 (Fig. 1, A and B):
(a) ARD (residues 110–357, corresponding to RC1): sensing chemical signals from binding of various ligands such as ATP (Lishko et al., 2007) and calmodulin (Rosenbaum et al., 2004; Lishko et al., 2007);
(b) MPD (residues 358–429, corresponding to RC2): sensing heat that activates TRPV1 gating (Yao et al., 2011);
(c) CTD (residues 691–719, corresponding to RC3): sensing chemical signals from binding of ligands such as calmodulin (Numazaki et al., 2003) and PIP2 (Kwon et al., 2007);
(d) S5–S6 helices of TMD (residues 560–690, corresponding to RC4): forming the channel pore featuring a dual gate (at residues G643 and I679);
(e) S1–S4 helices of TMD (residues 430–559, corresponding to RC5): corresponding to the voltage-sensing module well studied in a related family of voltage-gated ion channels (Catterall, 2012).
By comparing the RCs of the above parts (RC1–RC5) near the middle of the transition pathway, we found RC1 > RC2 > RC3 > RC4, which implies the following motional order: ARD→MPD→CTD→S5–S6 helices (Fig. 3 A). The early motion of ARD is consistent with the finding of highly flexible ARD by NMA (see above), which allows ligand binding at the ARD to affect early events of the gating transition. The motions of MPD and CTD occur at a very similar pace during the first half of the transition pathway, which is consistent with them belonging to the same dynamic domains as found by NMA (Fig. 1, C and D). The above order is consistent with the general model of TRPV1 gating in which the S5–S6 pore helices open in response to physical/chemical perturbations at the ARD, MPD, or CTD. Interestingly, during the first half of the transition pathway, the motion of S1–S4 helices precedes that of S5–S6 helices (i.e., RC5 > RC4; Fig. 3 A), which features a quaternary twist motion (Video 4) also captured by NMA (Video 1). However, the motion of S1–S4 helices lags behind that of S5–S6 helices later during the transition. Therefore, the twisting S1–S4 helices may drive some of the early motion of S5–S6 helices in the gating transition, which may allow voltage change to activate the opening of TRPV1 channel (Voets et al., 2004; Nilius et al., 2005a).
Next we used the RC analysis to compare the motional order of the selectivity filter (represented by residues G643 from four subunits) versus the lower gate (represented by residues I679 from four subunits). Although both remain closed during the first half of the transition pathway, G643 precedes I679 in opening during the second half of the transition pathway (Fig. 3 A). We note that the capsaicin-bound structure of TRPV1 (featuring a closed selectivity filter and a partially open lower gate; Fig. 3 C) does not obey the above order and is therefore not on the closed-to-open transition pathway.
In sum, our transition pathway modeling has predicted a sequence of structural motions propagating from the ARDs to the TMDs via the MPDs and the CTDs, resulting in sequential opening of the selectivity filter followed by the lower gate.
Transition pathway modeling of TRPV1–lipid system supports the robustness of the gating transition pathway to perturbation of lipids
To assess how lipids affect the gating transition of TRPV1, we performed the transition pathway modeling for TRPV1 in the presence of lipids (see Materials and methods), and compared that with the results obtained in the absence of lipids (see above). Despite moderate changes to the RCs, the introduction of lipid–TMD interactions did not change the motional order ARD→MPD→CTD→G643→I679 as found in the absence of lipids (Fig. 3 B). Among all functional parts, the motion of S1–S4 helices is affected the most by lipids (Fig. 3 B): although leading the S5–S6 helices in the absence of lipids (Fig. 3 A), the S1–S4 helices lag behind the S5–S6 helices during the first half of the transition in the presence of lipids (Fig. 3 B). This may be attributed to a larger drag of the S1–S4 helices than the S5–S6 helices by lipids because the former forms more extensive contact with lipids than the latter. Therefore, our transition pathway modeling of TRPV1 gating is largely insensitive to the perturbation of lipids. This finding is consistent with our previous modeling study of the gating transition pathway of a pentameric ligand-gated ion channel, which found good agreement with the experimental Φ values without including lipids in the modeling (Zheng and Auerbach, 2011). However, our finding does not exclude a likely role of lipids in tuning the gating transition of an ion channel like TRPV1.
Modeling of allosteric conformational changes in TMDs induced by activating conformational changes in ICDs
To probe the structural mechanism of allosteric coupling from the ICDs to the TMDs during TRPV1 gating, we attempted to construct an intermediate structural model of TRPV1 after the activation of ICDs. To this end, we started from the C-state structure and used the iENM protocol (see Materials and methods) to progressively transform the ICDs toward the activated conformation given by the O-state structure, then we let the TMDs relax to a series of minimal-energy conformations (Videos 5 and 6). To enable unhindered opening/closing of the channel pore formed by the S5–S6 helices, we turned off the intersubunit elastic interactions between the S5–S6 helices of the TRPV1 tetramer.
The above modeling predicts a quaternary twist motion of the TMDs relative to the ICDs (Video 6), followed by opening of the selectivity filter (at residue G643) while the lower gate (at residue I679) remains closed (Fig. 3 C). This is consistent with our finding that the selectivity filter opens before the lower gate during the gating transition (see above). Therefore, the activated ICDs are allosterically coupled to the more distant selectivity filter instead of the nearer lower gate, highlighting the key role of collective motions in transmitting intramolecular signals over a large distance. Although the activating conformational changes in ICDs can trigger partial opening of the selectivity filter (upper gate), a complete opening of the channel pore requires additional conformational changes in the rest of TRPV1 (i.e., the TMDs).
To further assess the effect of lipids, we repeated the above modeling for the TRPV1–lipid system (see Materials and methods). Unlike the result for TRPV1 alone (see above), we found that neither gate opens after the activation of ICDs in the presence of lipids (Fig. 3 C). Therefore, the lipids seemed to alter the conformational changes in TMDs induced by the ICD activation. This does not necessarily invalidate our above finding (i.e., opening of the upper gate induced by the activation of ICDs). It is possible that our treatment of the TRPV1–lipid system as a homogeneous ENM with uniform force constant may be inaccurate (e.g., the lipids may be less solid-like and more liquid-like than protein, and protein–lipid interactions may be weaker than intra-protein interactions). Indeed, after removing either lipid–lipid or protein–lipid springs while keeping the short-range repulsive interactions between them to prevent collisions (Tekpinar and Zheng, 2010), we were able to recover the finding of upper-gate opening. Therefore, the ENM description of the TRPV1–lipid system may need to be further refined to accurately describe the allosteric conformational changes in TRPV1 induced by the activation of ICDs.
All-atom MD simulation of ICD in C/O state
To further probe how the heat activation of ICD drives the gating transition in TRPV1, we used all-atom MD simulation to analyze the energetics of ICD in the C/O state. Our goal is to identify hotspot residues in ICD that undergo significant change in energy between the C and O state, which may contribute to the temperature sensitivity of TRPV1 gating. We acknowledge that the contributions from residues outside the ICD will not be considered in this simulation, although they may be important to the temperature sensitivity.
To simulate TRPV1 with limited computing resources, we constructed two all-atom systems for a truncated ICD (in the C and O state) submerged in a box of solvent and ions (see Materials and methods), and then conducted extensive MD simulations (i.e., five 20-ns-long trajectories per system; see Materials and methods). To prevent the ICD from being overly flexible (because of the removal of the rest of TRPV1), we applied positional restraints to the Cα atoms (see Materials and methods) during the MD simulation. The MD trajectories were found to stabilize rapidly (within <10 ns; see Fig. S2 A). The simulation of truncated ICD is warranted because: (a) the ICD is known to sense various signals (including heat) for activation and regulation of TRP channels (Gees et al., 2012); and (b) the TRPV1 structures (Cao et al., 2013; Liao et al., 2013) show only a few contacts between the ICDs of adjacent subunits, and previous studies found the ARD forms a structurally independent functional module (Jin et al., 2006; Zheng, 2013). We acknowledge that the bulk of CTD (after residue 719) is unresolved in the cryo-EM structures of TRPV1 (but could be probed using fluorescence resonance energy transfer measurement; De-la-Rosa et al., 2013), which may contribute additional intra- and intersubunit interactions involving the ICDs.
Energetic analysis of ICD in C/O state
The temperature sensitivity of TRPV1 gating requires a large enthalpy difference between the C and O state (denoted ΔH = HO − HC). To probe the energetic basis of heat activation of TRPV1, we assessed contributions to ΔH from polar (electrostatic) energy and nonpolar vdW energy (see Materials and methods). The two contributions were separately evaluated and averaged based on the last 10 ns of five 20-ns-long MD simulations in the C/O state (see Materials and methods). We repeated the calculation for the last 5 ns of MD trajectories to make sure the results are insensitive to truncation of trajectories.
The difference in total vdW energy between the C and O state is 35.3 kcal/mol per ICD (Table 1), which is statistically significant despite large fluctuations in the vdW energy (see Fig. S2 B). This energy difference is sufficient to account for the large enthalpy difference in TRPV1 gating (ΔH of ∼100 kcal/mol for the tetramer or ∼25 kcal/mol per subunit) as determined from the patch-clamp measurement of temperature dependence of single-channel current (Yao et al., 2010). The contribution to vdW energy difference by the MPD is 18.3 kcal/mol (∼52% of the total vdW energy difference; Table 1). In contrast, the difference in electrostatic energy between the C and O state is only −2.7 kcal/mol (Table 1). Therefore, the temperature sensitivity of TRPV1 gating can be explained by a large change in nonpolar energy within the ICD (especially the MPD), which does not require large conformational change in the ICD (root mean squared deviation of ∼1.3 Å). Limited by the truncated ICD simulation and lack of lipids, we are unable to assess the contributions from TMD, ICD–TMD, intersubunit, and protein–lipid interactions. These contributions will be evaluated in the future based on MD simulations of a full-length TRPV1 in the presence of a lipid bilayer.
Table 1.
Results of energetic analysis (mean ± SD (SE)) of the ICD in the C and O state
| Energy term | C state | O state | O state–C state |
| kcal/mol | |||
| vdW | −1,229.0 ± 28.5 (0.57) | −1,193.7 ± 28.7 (0.57) | 35.3 |
| vdW MPD | −287.1 ± 14.3 (0.29) | −268.8 ± 13.9 (0.28) | 18.3 |
| Electrostatic | 5,8172.7 ± 342.8 (6.9) | 5,8170.0 ± 340.7 (6.8) | −2.7 |
To identify key residues of ICD that differentially stabilize the C and O state, we used the CHARMM program (Brooks et al., 2009) to partition the vdW energy to individual residues (see Materials and methods), and calculated the difference (denoted as ΔEvdW,n; Fig. 4 A) between the C and O state for each residue (the distribution of ΔEvdW,n is 0.10 ± 0.496 kcal/mol). Using a 2σ cutoff, we identified the following residues of ICD with significant change in vdW energy between the C and O state (Fig. 4 B):
Figure 4.
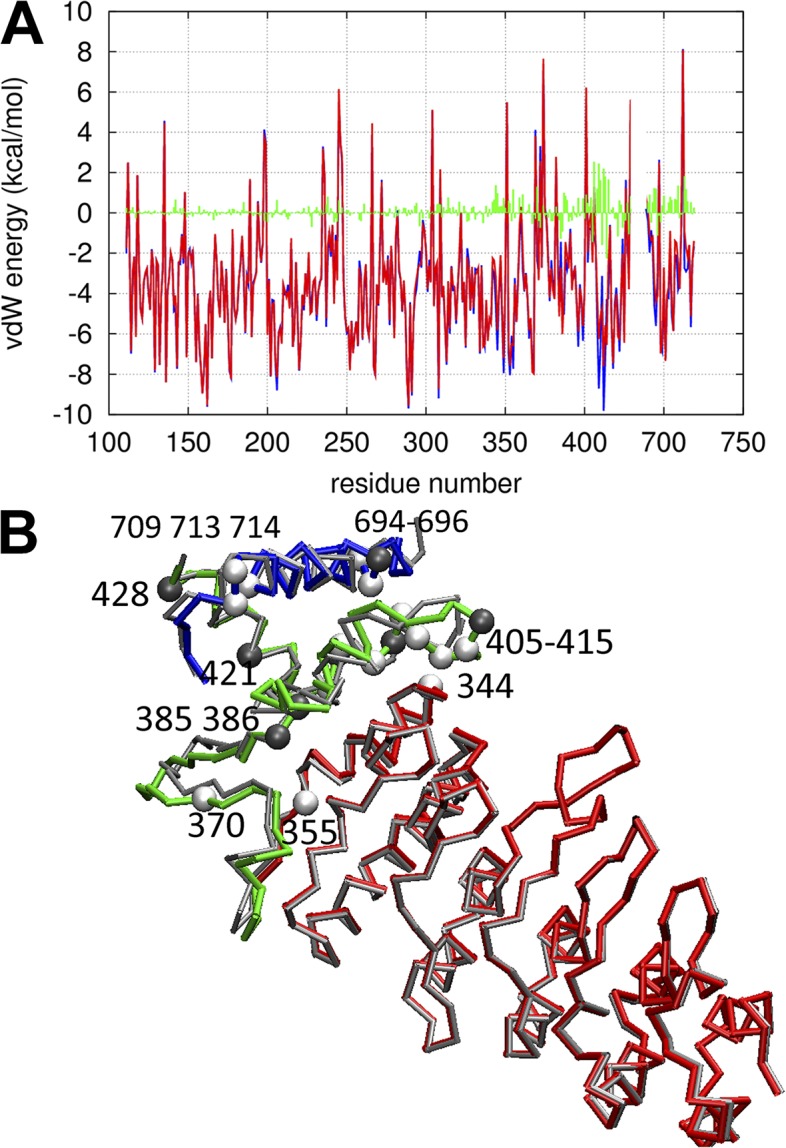
Results of MD-based energetic analysis. (A) vdW energy as a function of residue position for: C state (blue), O sate (red), and difference between C and O state (green). (B) Key residues in the ICD identified by energetic analysis: the backbone trace of the C-state structure is colored as follows: red, ARD; green, MPD; blue, CTD. The backbone trace of the O-state structure is colored in gray; residues with significantly higher (lower) vdW energy in the O state than the C state are shown as spheres colored white (dark gray).
(a) Residues 344, 355, 370, 406, 408, 409, 410, 412, 413, 415, 694, 695, 709, 713, and 714 have higher vdW energy in the O state than the C state. These residues may contribute significantly to the positive enthalpy difference between these states.
(b) Residues 385, 386, 405, 411, 414, 421, 428, and 696 have lower vdW energy in the O state than the C state. These residues may contribute negatively to the enthalpy difference between these states.
The majority of the above residues are in the MPD, supporting the importance of MPD in controlling the temperature sensitivity of TRPV1 gating as found by one of us (Yao et al., 2011). The three top-contributing residues to the vdW energy difference (T406, R409, and M412) are located at a strategic position interfacing between the MPD, the CTD, and the TMD (Figs. 1 A and 4 B). It will be interesting to investigate the functional importance of these residues with mutagenesis and electrophysiology experiments.
Comparison of predicted residues with literature
Many of the above predicted residues in ICD are well conserved in the TRPV subfamily, including G344, R355, T370, R409, M412, L413, L421, I696, and E709 (see Materials and methods). Some of the predicted residues were at (or in proximity to) the targets of past functional and mutational studies of TRPV1. Gain-of-function mutations S343G/R, I352N/T, I689V, and K710A caused weak or strong toxic phenotypes in yeast (Myers et al., 2008). T370 is known to be phosphorylated by protein kinase A and involved in the sensitization of heat-evoked TRPV1 responses (Winter et al., 2013). Mutants I696A, W697A, and R701A showed a severe effect on voltage- and heat-dependent activation of TRPV1 gating (Valente et al., 2008). The K694A/K698A/K710A triple mutant was found to lose its binding affinity to PIP2 (Grycova et al., 2012). In our future mutagenesis and functional studies, we will focus on those untested residues (especially the ones in MPD). In the future, we will also perform MD simulations of the TRPV1 tetramer (in the presence of solvent, ions, and lipids), which will allow us to improve the energy calculations by accounting for contributions from lipids, TMDs, and intersubunit interactions, etc. Before ending, we would like to point out that the cryo-EM structures used for this computational study have several truncated or unresolved regions (such as the outer-pore turret and part of the CTD), which are potentially important to the TRPV1 channel function. We await a future solution of a more complete TRPV1 structure to computationally probe the roles of these missing parts.
Note: while this manuscript was under review, Darré et al. (2015) published an MD simulation study of the permeation and dynamics of the open pore domain of TRPV1 channel, which nicely complements our MD simulation of the ICD of TRPV1.
Supplementary Material
Acknowledgments
This study was funded by grants from American Heart Association (14GRNT18980033) and National Science Foundation (0952736) to W. Zheng, and a National Institutes of Health grant (GM104521) to F. Qin. Computational support was provided by the Center for Computational Research at the University at Buffalo and the Biowulf system at the National Institutes of Health.
The authors declare no competing financial interests.
Kenton J. Swartz served as editor.
Footnotes
Abbreviations used in this paper:
- ARD
- ankyrin repeats domain
- CTD
- C-terminal domain
- ENM
- elastic network model
- ICD
- intracellular domain
- iENM
- interpolated ENM
- MD
- molecular dynamics
- MPD
- membrane proximal domain
- NMA
- normal mode analysis
- PIP2
- phosphatidylinositol-4,5-bisphosphate
- RC
- reaction-coordinate
- TMD
- transmembrane domain
- TRP
- transient receptor potential
- vdW
- van der Waals
References
- Atilgan A.R., Durell S.R., Jernigan R.L., Demirel M.C., Keskin O., and Bahar I.. 2001. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 80:505–515 10.1016/S0006-3495(01)76033-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I., and Rader A.J.. 2005. Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct. Biol. 15:586–592 10.1016/j.sbi.2005.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I., Lezon T.R., Bakan A., and Shrivastava I.H.. 2010. Normal mode analysis of biomolecular structures: Functional mechanisms of membrane proteins. Chem. Rev. 110:1463–1497 10.1021/cr900095e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauchi S., Orta G., Salazar M., Rosenmann E., and Latorre R.. 2006. A hot-sensing cold receptor: C-terminal domain determines thermosensation in transient receptor potential channels. J. Neurosci. 26:4835–4840 10.1523/JNEUROSCI.5080-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauchi S., Orta G., Mascayano C., Salazar M., Raddatz N., Urbina H., Rosenmann E., Gonzalez-Nilo F., and Latorre R.. 2007. Dissection of the components for PIP2 activation and thermosensation in TRP channels. Proc. Natl. Acad. Sci. USA. 104:10246–10251 10.1073/pnas.0703420104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., and Karplus M.. 1983. Charmm: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217 10.1002/jcc.540040211 [DOI] [Google Scholar]
- Brooks B.R., Brooks C.L. III, Mackerell A.D. Jr, Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. 2009. CHARMM: The biomolecular simulation program. J. Comput. Chem. 30:1545–1614 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calimet N., Simoes M., Changeux J.P., Karplus M., Taly A., and Cecchini M.. 2013. A gating mechanism of pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. USA. 110:E3987–E3996 10.1073/pnas.1313785110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E., Liao M., Cheng Y., and Julius D.. 2013. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 504:113–118 10.1038/nature12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., and Julius D.. 1997. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 389:816–824 10.1038/39807 [DOI] [PubMed] [Google Scholar]
- Caterina M.J., Rosen T.A., Tominaga M., Brake A.J., and Julius D.. 1999. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature. 398:436–441 10.1038/18906 [DOI] [PubMed] [Google Scholar]
- Catterall W.A.2012. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J. Physiol. 590:2577–2589 10.1113/jphysiol.2011.224204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Lu B., Grant B., Law R.J., and McCammon J.A.. 2006. Channel opening motion of alpha7 nicotinic acetylcholine receptor as suggested by normal mode analysis. J. Mol. Biol. 355:310–324 10.1016/j.jmb.2005.10.039 [DOI] [PubMed] [Google Scholar]
- Clapham D.E.2003. TRP channels as cellular sensors. Nature. 426:517–524 10.1038/nature02196 [DOI] [PubMed] [Google Scholar]
- Croy C.H., Bergqvist S., Huxford T., Ghosh G., and Komives E.A.. 2004. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 13:1767–1777 10.1110/ps.04731004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Yang F., Cao X., Yarov-Yarovoy V., Wang K., and Zheng J.. 2012. Selective disruption of high sensitivity heat activation but not capsaicin activation of TRPV1 channels by pore turret mutations. J. Gen. Physiol. 139:273–283 10.1085/jgp.201110724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkov T.L., Huynh K.W., Cohen M.R., and Moiseenkova-Bell V.Y.. 2011. Molecular architecture and subunit organization of TRPA1 ion channel revealed by electron microscopy. J. Biol. Chem. 286:38168–38176 10.1074/jbc.M111.288993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darré L., Furini S., and Domene C.. 2015. Permeation and dynamics of an open-activated TRPV1 channel. J. Mol. Biol. 427:537–549 10.1016/j.jmb.2014.11.016 [DOI] [PubMed] [Google Scholar]
- De-la-Rosa V., Rangel-Yescas G.E., Ladrón-de-Guevara E., Rosenbaum T., and Islas L.D.. 2013. Coarse architecture of the transient receptor potential vanilloid 1 (TRPV1) ion channel determined by fluorescence resonance energy transfer. J. Biol. Chem. 288:29506–29517 10.1074/jbc.M113.479618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delemotte L., Klein M.L., and Tarek M.. 2012. Molecular dynamics simulations of voltage-gated cation channels: insights on voltage-sensor domain function and modulation. Front Pharmacol. 3:97 10.3389/fphar.2012.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deserno M., and Holm C.. 1998. How to mesh up Ewald sums. I. A theoretical and numerical comparison of various particle mesh routines. J. Chem. Phys. 109:7678–7693 10.1063/1.477414 [DOI] [Google Scholar]
- Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., and Baker N.A.. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35:W522–W525 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., and Zhou H.X.. 2011. Atomistic mechanism for the activation and desensitization of an AMPA-subtype glutamate receptor. Nat. Commun. 2:354 10.1038/ncomms1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Dong H., and Zhou H.X.. 2012. Gating mechanism of a P2X4 receptor developed from normal mode analysis and molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 109:4140–4145 10.1073/pnas.1119546109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sanz N., Valente P., Gomis A., Fernández-Carvajal A., Fernández-Ballester G., Viana F., Belmonte C., and Ferrer-Montiel A.. 2007. A role of the transient receptor potential domain of vanilloid receptor I in channel gating. J. Neurosci. 27:11641–11650 10.1523/JNEUROSCI.2457-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R.2008a. A primer on ankyrin repeat function in TRP channels and beyond. Mol. Biosyst. 4:372–379 10.1039/b801481g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R.2008b. TRP channels entering the structural era. J. Physiol. 586:3565–3575 10.1113/jphysiol.2008.155812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gees M., Owsianik G., Nilius B., and Voets T.. 2012. TRP channels. Compr Physiol. 2:563–608. [DOI] [PubMed] [Google Scholar]
- Gilson M.K., and Honig B.H.. 1986. The dielectric constant of a folded protein. Biopolymers. 25:2097–2119 10.1002/bip.360251106 [DOI] [PubMed] [Google Scholar]
- Gilson M.K., and Honig B.H.. 1988. Energetics of charge-charge interactions in proteins. Proteins. 3:32–52 10.1002/prot.340030104 [DOI] [PubMed] [Google Scholar]
- Grandl J., Kim S.E., Uzzell V., Bursulaya B., Petrus M., Bandell M., and Patapoutian A.. 2010. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 13:708–714 10.1038/nn.2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grycova L., Holendova B., Bumba L., Bily J., Jirku M., Lansky Z., and Teisinger J.. 2012. Integrative binding sites within intracellular termini of TRPV1 receptor. PLoS ONE. 7:e48437 10.1371/journal.pone.0048437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunthorpe M.J., and Szallasi A.. 2008. Peripheral TRPV1 receptors as targets for drug development: New molecules and mechanisms. Curr. Pharm. Des. 14:32–41 10.2174/138161208783330754 [DOI] [PubMed] [Google Scholar]
- Hafner J., and Zheng W.. 2010. Optimal modeling of atomic fluctuations in protein crystal structures for weak crystal contact interactions. J. Chem. Phys. 132:014111 10.1063/1.3288503 [DOI] [PubMed] [Google Scholar]
- Hellmich U.A., and Gaudet R.. 2014. Structural biology of TRP channels. Handbook Exp. Pharmacol. 223:963–990 10.1007/978-3-319-05161-1_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover W.G.1985. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A. 31:1695–1697 10.1103/PhysRevA.31.1695 [DOI] [PubMed] [Google Scholar]
- Howard J., and Bechstedt S.. 2004. Hypothesis: A helix of ankyrin repeats of the NOMPC-TRP ion channel is the gating spring of mechanoreceptors. Curr. Biol. 14:R224–R226 10.1016/j.cub.2004.02.050 [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Huynh K.W., Cohen M.R., Chakrapani S., Holdaway H.A., Stewart P.L., and Moiseenkova-Bell V.Y.. 2014. Structural insight into the assembly of TRPV channels. Structure. 22:260–268 10.1016/j.str.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im W., Beglov D., and Roux B.. 1998. Continuum solvation model: Computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comput. Phys. Commun. 111:59–75 10.1016/S0010-4655(98)00016-2 [DOI] [Google Scholar]
- Inada H., Procko E., Sotomayor M., and Gaudet R.. 2012. Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel. Biochemistry. 51:6195–6206 10.1021/bi300279b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.O., Jogini V., Borhani D.W., Leffler A.E., Dror R.O., and Shaw D.E.. 2012. Mechanism of voltage gating in potassium channels. Science. 336:229–233 10.1126/science.1216533 [DOI] [PubMed] [Google Scholar]
- Jin X., Touhey J., and Gaudet R.. 2006. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J. Biol. Chem. 281:25006–25010 10.1074/jbc.C600153200 [DOI] [PubMed] [Google Scholar]
- Jordt S.E., Tominaga M., and Julius D.. 2000. Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc. Natl. Acad. Sci. USA. 97:8134–8139 10.1073/pnas.100129497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karashima Y., Talavera K., Everaerts W., Janssens A., Kwan K.Y., Vennekens R., Nilius B., and Voets T.. 2009. TRPA1 acts as a cold sensor in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 106:1273–1278 10.1073/pnas.0808487106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M., and McCammon J.A.. 2002. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 9:646–652 10.1038/nsb0902-646 [DOI] [PubMed] [Google Scholar]
- Kim S.E., Patapoutian A., and Grandl J.. 2013. Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations. PLoS ONE. 8:e59593 10.1371/journal.pone.0059593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs W.G., Alexandrov V., Wilson C.A., Echols N., Yu H., and Gerstein M.. 2002. Normal mode analysis of macromolecular motions in a database framework: Developing mode concentration as a useful classifying statistic. Proteins. 48:682–695 10.1002/prot.10168 [DOI] [PubMed] [Google Scholar]
- Kwon Y., Hofmann T., and Montell C.. 2007. Integration of phosphoinositide- and calmodulin-mediated regulation of TRPC6. Mol. Cell. 25:491–503 10.1016/j.molcel.2007.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landouré G., Zdebik A.A., Martinez T.L., Burnett B.G., Stanescu H.C., Inada H., Shi Y., Taye A.A., Kong L., Munns C.H., et al. 2010. Mutations in TRPV4 cause Charcot-Marie-Tooth disease type 2C. Nat. Genet. 42:170–174 10.1038/ng.512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre R., Brauchi S., Orta G., Zaelzer C., and Vargas G.. 2007. ThermoTRP channels as modular proteins with allosteric gating. Cell Calcium. 42:427–438 10.1016/j.ceca.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Li M., and Zheng W.. 2013. All-atom molecular dynamics simulations of actin-myosin interactions: A comparative study of cardiac α myosin, β myosin, and fast skeletal muscle myosin. Biochemistry. 52:8393–8405 10.1021/bi4006896 [DOI] [PubMed] [Google Scholar]
- Liao M., Cao E., Julius D., and Cheng Y.. 2013. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 504:107–112 10.1038/nature12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E., and Sansom M.S.. 2008. Membrane proteins: molecular dynamics simulations. Curr. Opin. Struct. Biol. 18:425–431 10.1016/j.sbi.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Lishko P.V., Procko E., Jin X., Phelps C.B., and Gaudet R.. 2007. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 54:905–918 10.1016/j.neuron.2007.05.027 [DOI] [PubMed] [Google Scholar]
- MacKerell A.D. Jr, Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Mackerell A.D. Jr, Feig M., and Brooks C.L. III. 2004. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25:1400–1415 10.1002/jcc.20065 [DOI] [PubMed] [Google Scholar]
- Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., and de Vries A.H.. 2007. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B. 111:7812–7824 10.1021/jp071097f [DOI] [PubMed] [Google Scholar]
- Martyna G.J., Hughes A., and Tuckerman M.E.. 1999. Molecular dynamics algorithms for path integrals at constant pressure. J. Chem. Phys. 110:3275–3290 10.1063/1.478193 [DOI] [Google Scholar]
- Maruyama Y., Ogura T., Mio K., Kiyonaka S., Kato K., Mori Y., and Sato C.. 2007. Three-dimensional reconstruction using transmission electron microscopy reveals a swollen, bell-shaped structure of transient receptor potential melastatin type 2 cation channel. J. Biol. Chem. 282:36961–36970 10.1074/jbc.M705694200 [DOI] [PubMed] [Google Scholar]
- McCleverty C.J., Koesema E., Patapoutian A., Lesley S.A., and Kreusch A.. 2006. Crystal structure of the human TRPV2 channel ankyrin repeat domain. Protein Sci. 15:2201–2206 10.1110/ps.062357206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio K., Ogura T., Hara Y., Mori Y., and Sato C.. 2005. The non-selective cation-permeable channel TRPC3 is a tetrahedron with a cap on the large cytoplasmic end. Biochem. Biophys. Res. Commun. 333:768–777 10.1016/j.bbrc.2005.05.181 [DOI] [PubMed] [Google Scholar]
- Mio K., Ogura T., Kiyonaka S., Hiroaki Y., Tanimura Y., Fujiyoshi Y., Mori Y., and Sato C.. 2007. The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. J. Mol. Biol. 367:373–383 10.1016/j.jmb.2006.12.043 [DOI] [PubMed] [Google Scholar]
- Moiseenkova-Bell V.Y., Stanciu L.A., Serysheva I.I., Tobe B.J., and Wensel T.G.. 2008. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA. 105:7451–7455 10.1073/pnas.0711835105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C.2001. Physiology, phylogeny, and functions of the TRP superfamily of cation channels. Sci. STKE. 2001:re1. [DOI] [PubMed] [Google Scholar]
- Montell C.2005. The TRP superfamily of cation channels. Sci. STKE. 2005:re3. [DOI] [PubMed] [Google Scholar]
- Myers B.R., Bohlen C.J., and Julius D.. 2008. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 58:362–373 10.1016/j.neuron.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B.2007. TRP channels in disease. Biochim. Biophys. Acta. 1772:805–812 10.1016/j.bbadis.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Nilius B.2013. Transient receptor potential TRP channels as therapeutic drug targets: Next round! Curr. Top. Med. Chem. 13:244–246 10.2174/1568026611313030002 [DOI] [PubMed] [Google Scholar]
- Nilius B., Talavera K., Owsianik G., Prenen J., Droogmans G., and Voets T.. 2005a. Gating of TRP channels: a voltage connection? J. Physiol. 567:35–44 10.1113/jphysiol.2005.088377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B., Voets T., and Peters J.. 2005b. TRP channels in disease. Sci. STKE. 2005:re8. [DOI] [PubMed] [Google Scholar]
- Nilius B., Appendino G., and Owsianik G.. 2012. The transient receptor potential channel TRPA1: from gene to pathophysiology. Pflugers Arch. 464:425–458 10.1007/s00424-012-1158-z [DOI] [PubMed] [Google Scholar]
- Nina M., Beglov D., and Roux B.. 1997. Atomic radii for continuum electrostatics calculations based on molecular dynamics free energy simulations. J. Phys. Chem. B. 101:5239–5248 10.1021/jp970736r [DOI] [Google Scholar]
- Numazaki M., Tominaga T., Takeuchi K., Murayama N., Toyooka H., and Tominaga M.. 2003. Structural determinant of TRPV1 desensitization interacts with calmodulin. Proc. Natl. Acad. Sci. USA. 100:8002–8006 10.1073/pnas.1337252100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nury H., Poitevin F., Van Renterghem C., Changeux J.P., Corringer P.J., Delarue M., and Baaden M.. 2010. One-microsecond molecular dynamics simulation of channel gating in a nicotinic receptor homologue. Proc. Natl. Acad. Sci. USA. 107:6275–6280 10.1073/pnas.1001832107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nury H., Van Renterghem C., Weng Y., Tran A., Baaden M., Dufresne V., Changeux J.P., Sonner J.M., Delarue M., and Corringer P.J.. 2011. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 469:428–431 10.1038/nature09647 [DOI] [PubMed] [Google Scholar]
- Olson M.A., and Reinke L.T.. 2000. Modeling implicit reorganization in continuum descriptions of protein-protein interactions. Proteins. 38:115–119 [DOI] [PubMed] [Google Scholar]
- Pedretti A., Labozzetta A., Lo Monte M., Beccari A.R., Moriconi A., and Vistoli G.. 2011. Exploring the activation mechanism of TRPM8 channel by targeted MD simulations. Biochem. Biophys. Res. Commun. 414:14–19 10.1016/j.bbrc.2011.08.134 [DOI] [PubMed] [Google Scholar]
- Phelps C.B., Huang R.J., Lishko P.V., Wang R.R., and Gaudet R.. 2008. Structural analyses of the ankyrin repeat domain of TRPV6 and related TRPV ion channels. Biochemistry. 47:2476–2484 10.1021/bi702109w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., and Schulten K.. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott E.D., and Julius D.. 2003. A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science. 300:1284–1288 10.1126/science.1083646 [DOI] [PubMed] [Google Scholar]
- Qi Y., Cheng X., Han W., Jo S., Schulten K., and Im W.. 2014. CHARMM-GUI PACE CG Builder for solution, micelle, and bilayer coarse-grained simulations. J. Chem. Inf. Model. 54:1003–1009 10.1021/ci500007n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F.2007. Regulation of TRP ion channels by phosphatidylinositol-4,5-bisphosphate. Handbook Exp. Pharmacol. 179:509–525 10.1007/978-3-540-34891-7_30 [DOI] [PubMed] [Google Scholar]
- Rohacs T., and Nilius B.. 2007. Regulation of transient receptor potential (TRP) channels by phosphoinositides. Pflugers Arch. 455:157–168 10.1007/s00424-007-0275-6 [DOI] [PubMed] [Google Scholar]
- Rosenbaum T., Gordon-Shaag A., Munari M., and Gordon S.E.. 2004. Ca2+/calmodulin modulates TRPV1 activation by capsaicin. J. Gen. Physiol. 123:53–62 10.1085/jgp.200308906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B.1997. Influence of the membrane potential on the free energy of an intrinsic protein. Biophys. J. 73:2980–2989 10.1016/S0006-3495(97)78327-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar H., Llorente I., Jara-Oseguera A., García-Villegas R., Munari M., Gordon S.E., Islas L.D., and Rosenbaum T.. 2008. A single N-terminal cysteine in TRPV1 determines activation by pungent compounds from onion and garlic. Nat. Neurosci. 11:255–261 10.1038/nn2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauguet L., Shahsavar A., Poitevin F., Huon C., Menny A., Nemecz À., Haouz A., Changeux J.P., Corringer P.J., and Delarue M.. 2014. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. USA. 111:966–971 10.1073/pnas.1314997111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp K.A., and Honig B.. 1990a. Calculating total electrostatic energies with the nonlinear Poisson-Boltzmann equation. J. Phys. Chem. 94:7684–7692 10.1021/j100382a068 [DOI] [Google Scholar]
- Sharp K.A., and Honig B.. 1990b. Electrostatic interactions in macromolecules: Theory and applications. Annu. Rev. Biophys. Biophys. Chem. 19:301–332 10.1146/annurev.bb.19.060190.001505 [DOI] [PubMed] [Google Scholar]
- Shi D.J., Ye S., Cao X., Zhang R., and Wang K.. 2013. Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 reveals unique conformation of finger 3 loop critical for channel function. Protein Cell. 4:942–950 10.1007/s13238-013-3091-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H., Sokabe T., Danev R., Tominaga M., and Nagayama K.. 2010. A 3.5-nm structure of rat TRPV4 cation channel revealed by Zernike phase-contrast cryoelectron microscopy. J. Biol. Chem. 285:11210–11218 10.1074/jbc.M109.090712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O.S., Neduvelil J.G., Wang X., Wallace B.A., and Sansom M.S.. 1996. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14:354–360 10.1016/S0263-7855(97)00009-X [DOI] [PubMed] [Google Scholar]
- Sotomayor M., Corey D.P., and Schulten K.. 2005. In search of the hair-cell gating spring elastic properties of ankyrin and cadherin repeats. Structure. 13:669–682 10.1016/j.str.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Stone J.E., Hardy D.J., Ufimtsev I.S., and Schulten K.. 2010. GPU-accelerated molecular modeling coming of age. J. Mol. Graph. Model. 29:116–125 10.1016/j.jmgm.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story G.M., Peier A.M., Reeve A.J., Eid S.R., Mosbacher J., Hricik T.R., Earley T.J., Hergarden A.C., Andersson D.A., Hwang S.W., et al. 2003. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 112:819–829 10.1016/S0092-8674(03)00158-2 [DOI] [PubMed] [Google Scholar]
- Susankova K., Ettrich R., Vyklicky L., Teisinger J., and Vlachova V.. 2007. Contribution of the putative inner-pore region to the gating of the transient receptor potential vanilloid subtype 1 channel (TRPV1). J. Neurosci. 27:7578–7585 10.1523/JNEUROSCI.1956-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly A., Delarue M., Grutter T., Nilges M., Le Novère N., Corringer P.J., and Changeux J.P.. 2005. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys. J. 88:3954–3965 10.1529/biophysj.104.050229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly A., Corringer P.J., Grutter T., Prado de Carvalho L., Karplus M., and Changeux J.P.. 2006. Implications of the quaternary twist allosteric model for the physiology and pathology of nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. USA. 103:16965–16970 10.1073/pnas.0607477103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F., and Brooks C.L.. 2006. Symmetry, form, and shape: Guiding principles for robustness in macromolecular machines. Annu. Rev. Biophys. Biomol. Struct. 35:115–133 10.1146/annurev.biophys.35.040405.102010 [DOI] [PubMed] [Google Scholar]
- Tama F., and Sanejouand Y.H.. 2001. Conformational change of proteins arising from normal mode calculations. Protein Eng. 14:1–6 10.1093/protein/14.1.1 [DOI] [PubMed] [Google Scholar]
- Tekpinar M., and Zheng W.. 2010. Predicting order of conformational changes during protein conformational transitions using an interpolated elastic network model. Proteins. 78:2469–2481. [DOI] [PubMed] [Google Scholar]
- Tekpinar M., and Zheng W.. 2013. Coarse-grained and all-atom modeling of structural states and transitions in hemoglobin. Proteins. 81:240–252 10.1002/prot.24180 [DOI] [PubMed] [Google Scholar]
- Tominaga M., Caterina M.J., Malmberg A.B., Rosen T.A., Gilbert H., Skinner K., Raumann B.E., Basbaum A.I., and Julius D.. 1998. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 21:531–543 10.1016/S0896-6273(00)80564-4 [DOI] [PubMed] [Google Scholar]
- Tozzini V.2005. Coarse-grained models for proteins. Curr. Opin. Struct. Biol. 15:144–150 10.1016/j.sbi.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Tozzini V.2010. Minimalist models for proteins: a comparative analysis. Q. Rev. Biophys. 43:333–371 10.1017/S0033583510000132 [DOI] [PubMed] [Google Scholar]
- Valente P., García-Sanz N., Gomis A., Fernández-Carvajal A., Fernández-Ballester G., Viana F., Belmonte C., and Ferrer-Montiel A.. 2008. Identification of molecular determinants of channel gating in the transient receptor potential box of vanilloid receptor I. FASEB J. 22:3298–3309 10.1096/fj.08-107425 [DOI] [PubMed] [Google Scholar]
- Vargas E., Yarov-Yarovoy V., Khalili-Araghi F., Catterall W.A., Klein M.L., Tarek M., Lindahl E., Schulten K., Perozo E., Bezanilla F., and Roux B.. 2012. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J. Gen. Physiol. 140:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachová V., Teisinger J., Susánková K., Lyfenko A., Ettrich R., and Vyklický L.. 2003. Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J. Neurosci. 23:1340–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voets T., Droogmans G., Wissenbach U., Janssens A., Flockerzi V., and Nilius B.. 2004. The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature. 430:748–754 10.1038/nature02732 [DOI] [PubMed] [Google Scholar]
- Voets T., Talavera K., Owsianik G., and Nilius B.. 2005. Sensing with TRP channels. Nat. Chem. Biol. 1:85–92 10.1038/nchembio0705-85 [DOI] [PubMed] [Google Scholar]
- Voets T., Owsianik G., Janssens A., Talavera K., and Nilius B.. 2007. TRPM8 voltage sensor mutants reveal a mechanism for integrating thermal and chemical stimuli. Nat. Chem. Biol. 3:174–182 10.1038/nchembio862 [DOI] [PubMed] [Google Scholar]
- Winter Z., Buhala A., Ötvös F., Jósvay K., Vizler C., Dombi G., Szakonyi G., and Oláh Z.. 2013. Functionally important amino acid residues in the transient receptor potential vanilloid 1 (TRPV1) ion channel—an overview of the current mutational data. Mol. Pain. 9:30 10.1186/1744-8069-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Cui Y., Wang K., and Zheng J.. 2010. Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proc. Natl. Acad. Sci. USA. 107:7083–7088 10.1073/pnas.1000357107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Liu B., and Qin F.. 2010. Kinetic and energetic analysis of thermally activated TRPV1 channels. Biophys. J. 99:1743–1753 10.1016/j.bpj.2010.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Liu B., and Qin F.. 2011. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. USA. 108:11109–11114 10.1073/pnas.1105196108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., and Kuebler W.M.. 2010. Mechanotransduction by TRP channels: General concepts and specific role in the vasculature. Cell Biochem. Biophys. 56:1–18 10.1007/s12013-009-9067-2 [DOI] [PubMed] [Google Scholar]
- Zheng J.2013. Molecular mechanism of TRP channels. Compr Physiol. 3:221–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W.2010. Multiscale modeling of structural dynamics underlying force generation and product release in actomyosin complex. Proteins. 78:638–660. [DOI] [PubMed] [Google Scholar]
- Zheng W.2011. Coarse-grained modeling of conformational transitions underlying the processive stepping of myosin V dimer along filamentous actin. Proteins. 79:2291–2305 10.1002/prot.23055 [DOI] [PubMed] [Google Scholar]
- Zheng W.2012. Coarse-grained modeling of the structural states and transition underlying the powerstroke of dynein motor domain. J. Chem. Phys. 136:155103 10.1063/1.4704661 [DOI] [PubMed] [Google Scholar]
- Zheng W., and Auerbach A.. 2011. Decrypting the sequence of structural events during the gating transition of pentameric ligand-gated ion channels based on an interpolated elastic network model. PLOS Comput. Biol. 7:e1001046 10.1371/journal.pcbi.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., and Doniach S.. 2003. A comparative study of motor-protein motions by using a simple elastic-network model. Proc. Natl. Acad. Sci. USA. 100:13253–13258 10.1073/pnas.2235686100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., and Tekpinar M.. 2012. Structure-based simulations of the translocation mechanism of the hepatitis C virus NS3 helicase along single-stranded nucleic acid. Biophys. J. 103:1343–1353 10.1016/j.bpj.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Brooks B.R., and Hummer G.. 2007a. Protein conformational transitions explored by mixed elastic network models. Proteins. 69:43–57 10.1002/prot.21465 [DOI] [PubMed] [Google Scholar]
- Zheng W., Liao J.C., Brooks B.R., and Doniach S.. 2007b. Toward the mechanism of dynamical couplings and translocation in hepatitis C virus NS3 helicase using elastic network model. Proteins. 67:886–896 10.1002/prot.21326 [DOI] [PubMed] [Google Scholar]
- Zheng W., Barua B., and Hitchcock-DeGregori S.E.. 2013. Probing the flexibility of tropomyosin and its binding to filamentous actin using molecular dynamics simulations. Biophys. J. 105:1882–1892 10.1016/j.bpj.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., and Hummer G.. 2009. Gating transition of pentameric ligand-gated ion channels. Biophys. J. 97:2456–2463 10.1016/j.bpj.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., and Hummer G.. 2010. Pore opening and closing of a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. USA. 107:19814–19819 10.1073/pnas.1009313107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.