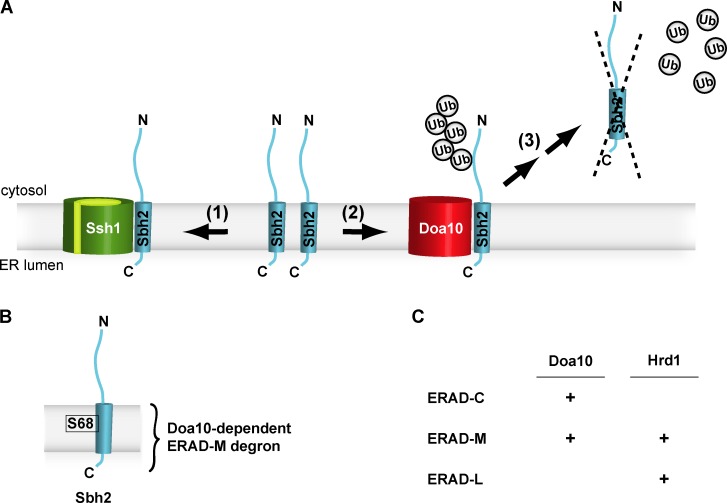
In Saccharomyces cerevisiae, surprisingly, the transmembrane protein Sbh2, which harbors an intramembrane degron, is a substrate of the ubiquitin-protein ligase Doa10.
Abstract
Aberrant endoplasmic reticulum (ER) proteins are eliminated by ER-associated degradation (ERAD). This process involves protein retrotranslocation into the cytosol, ubiquitylation, and proteasomal degradation. ERAD substrates are classified into three categories based on the location of their degradation signal/degron: ERAD-L (lumen), ERAD-M (membrane), and ERAD-C (cytosol) substrates. In Saccharomyces cerevisiae, the membrane proteins Hrd1 and Doa10 are the predominant ERAD ubiquitin-protein ligases (E3s). The current notion is that ERAD-L and ERAD-M substrates are exclusively handled by Hrd1, whereas ERAD-C substrates are recognized by Doa10. In this paper, we identify the transmembrane (TM) protein Sec61 β-subunit homologue 2 (Sbh2) as a Doa10 substrate. Sbh2 is part of the trimeric Ssh1 complex involved in protein translocation. Unassembled Sbh2 is rapidly degraded in a Doa10-dependent manner. Intriguingly, the degron maps to the Sbh2 TM region. Thus, in contrast to the prevailing view, Doa10 (and presumably its human orthologue) has the capacity for recognizing intramembrane degrons, expanding its spectrum of substrates.
Introduction
In a eukaryotic cell, most membrane and secretory proteins are synthesized at the ER. A sophisticated quality-control machinery at the ER ensures that only properly folded and assembled proteins continue along the secretory pathway. Misfolded or unassembled proteins are eliminated by ER-associated degradation (ERAD; Vembar and Brodsky, 2008; Hirsch et al., 2009; Hegde and Ploegh, 2010; Christianson and Ye, 2014; Ruggiano et al., 2014; Zattas and Hochstrasser, 2015). ERAD substrates are retrotranslocated into the cytosol and undergo proteasomal degradation. In most cases, substrates are ubiquitylated concomitantly with their retrotranslocation. Ubiquitylation involves the covalent attachment of ubiquitin monomers or polymers to a substrate protein and is accomplished by a series of enzymatic reactions featuring three classes of enzymes: ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes (Hochstrasser, 2009). E3 ligases thereby facilitate the transfer of ubiquitin from an E2 to a substrate protein. Within ERAD, the attached ubiquitin chain promotes substrate retrotranslocation and proteasomal degradation. Substrate recognition and membrane extraction are coordinated by ER-embedded E3s, which together with membrane-associated E2s catalyze substrate ubiquitylation on the cytosolic face of the ER membrane. The two major ERAD E3 complexes in yeast are the Hrd1 complex and the Doa10 complex, centered around the polytopic E3 ligases Hrd1/Der3 (Bays et al., 2001; Deak and Wolf, 2001) and Doa10 (Swanson et al., 2001), respectively. A third E3 ligase, the cytosolic Ubr1 protein, has recently been implicated in ERAD of some substrates (Stolz et al., 2013). ERAD substrates are classified into three categories: ERAD-L (lumen), ERAD-M (membrane), and ERAD-C (cytosol), depending on the localization of the folding lesion or degradation signal (degron; Vashist and Ng, 2004; Carvalho et al., 2006). Recently, a fourth class of ERAD substrates was described; ERAD-T (translocon) substrates are proteins that stall co- or posttranslationally in the translocon (Rubenstein et al., 2012).
For Saccharomyces cerevisiae, the prevailing notion, which is based on a limited number of investigated ERAD substrates, is that ERAD-L and ERAD-M (and ERAD-T) substrates are exclusively handled by the Hrd1 complex, whereas ERAD-C substrates are recognized by the Doa10 complex (Hirsch et al., 2009; Brodsky and Skach, 2011; Finley et al., 2012; Rubenstein et al., 2012; Christianson and Ye, 2014; Ruggiano et al., 2014; Zattas and Hochstrasser, 2015).
Little is known about the exact nature/structure of intramembrane degrons. In some cases, unassembled subunits of membrane protein complexes are targeted for degradation by exposing a charged amino acid within a transmembrane (TM) domain that would normally interact with and be masked by a binding partner as has been shown for unassembled T cell receptor α-chain (TCR-α; Bonifacino et al., 1989, 1990). In other cases, the overall conformation of a TM region, rather than its sequence per se, is recognized by the ERAD machinery. For example, the TM region of the Hmg2 protein, a key enzyme in the sterol pathway, becomes misfolded when sterol pathway products are high, resulting in recognition by the Hrd1 complex and degradation of Hmg2 (Gardner and Hampton, 1999; Sato et al., 2009).
The ERAD E3 Doa10 is a large membrane protein with a cytosolic N-terminal RING (really interesting new gene)-CH domain and a total of 14 TM domains (Swanson et al., 2001; Kreft et al., 2006). Doa10 orthologues are present in most sequenced eukaryotic genomes, and the human Doa10 orthologue is MARCH6/TEB4 (Swanson et al., 2001; Kreft and Hochstrasser, 2011; Stuerner et al., 2012). Doa10 localizes to the ER membrane as well as to the inner nuclear membrane (Swanson et al., 2001; Deng and Hochstrasser, 2006). It has a remarkable substrate range comprising soluble nuclear and cytoplasmic substrates as well as ER and nuclear envelope integral membrane proteins with cytosolic (or nuclear) degrons (Huyer et al., 2004; Ravid et al., 2006; Deng and Hochstrasser, 2006).
All eukaryotes contain a trimeric Sec61 complex in the ER membrane consisting of the polytopic membrane protein Sec61, the Sec61 γ subunit (Sss1 in yeast), and the Sec61 β subunit (Sbh1 in yeast; Shao and Hegde, 2011; Park and Rapoport, 2012; Mandon et al., 2013). The heterotrimeric Sec61 complex is implicated in co- and posttranslational protein translocation into the ER. S. cerevisiae contains a second, structurally related translocon complex for cotranslational translocation, the Ssh1 complex, consisting of the polytopic membrane protein Ssh1 (Sec61 homologue 1), Sss1 and the β-subunit Sbh2 (Fig. 1 A; Finke et al., 1996). Ssh1 and Sbh2 share homology to Sec61 and Sbh1, respectively (Finke et al., 1996; Toikkanen et al., 1996). The β-subunits Sbh1 and Sbh2 are tail-anchored (TA) proteins (Hegde and Keenan, 2011), and their TM domains bind Sec61 and Ssh1, respectively (Van den Berg et al., 2004; Becker et al., 2009).
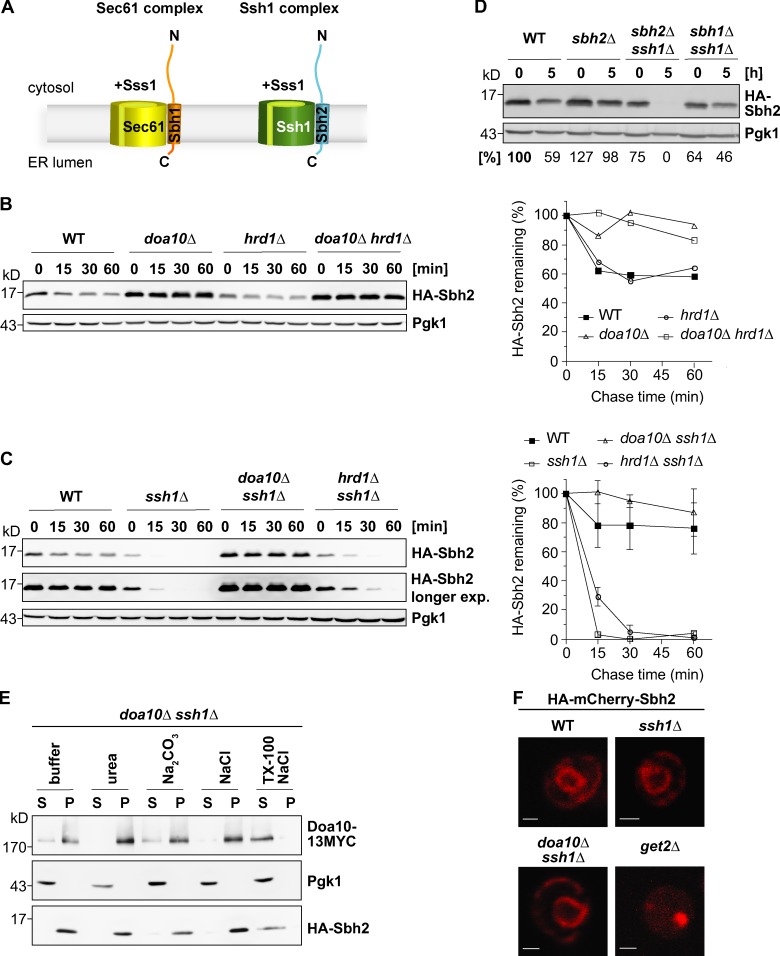
Figure 1.
Sbh2 is a Doa10 substrate and association with Ssh1 protects it from degradation. (A) Schematic of heterotrimeric yeast Sec61 and Ssh1 translocon complexes. The integral membrane protein Sss1, which is part of both complexes, is not depicted in the illustration. N, N terminus; C, C terminus. (B) Cycloheximide (chx) chase analysis of ectopically expressed (low-copy plasmid; MET25 promoter) HA-Sbh2 (in the presence of endogenous Sbh2) in WT, doa10Δ, hrd1Δ, and doa10Δ hrd1Δ cells. Pgk1 served as a loading control. The experiment shown is representative of n = 3 experiments. (right) Quantification of the gel on the left. HA-Sbh2 levels at t = 0 min were set to 100%. (C) Ssh1 protects Sbh2 from Doa10-dependent degradation. chx chase analysis of ectopically expressed HA-Sbh2 (as in B) in WT, ssh1Δ, doa10Δ ssh1Δ, and hrd1Δ ssh1Δ cells. Two different exposures of the anti-HA immunodetection are shown. The graph at right shows the mean degradation rates observed from three independent experiments. HA-Sbh2 levels at t = 0 h were set to 100%. Error bars represent ± SD. exp., exposure. (D) Degradation of unassembled Sbh2. chx chase analysis (time points t1 = 0 h and t2 = 5 h) of ectopically expressed HA-Sbh2 (as in B) in WT, sbh2Δ, sbh2Δ ssh1Δ, and sbh1Δ ssh1Δ cells. Relative protein levels listed below the blots were determined by quantification of pixel densities of HA-Sbh2 bands relative to those of Pgk1. HA-Sbh2 levels of WT cells at t1 = 0 h were set to 100%. (E) HA-Sbh2 is an integral membrane protein in ssh1Δ cells. Subcellular fractionation of doa10Δ ssh1Δ cells expressing HA-Sbh2 from a plasmid (as in B). HA-Sbh2–expressing ssh1Δ doa10Δ cells and Doa10-13MYC–expressing cells were mixed at a 5:1 ratio before lysis. Lysates were treated with buffer alone or buffer containing 2.5 M urea, 0.1 M Na2CO3, pH 11.5, and 0.5 M NaCl or 1% Triton X-100 (TX-100) and 0.5 M NaCl, and divided into microsomal pellet (P) and supernatant (S) fraction by centrifugation. Fractions were examined by immunoblotting with appropriate antibodies. (F) Fluorescence microscopy of WT, ssh1Δ, ssh1Δ doa10Δ, and get2Δ cells overexpressing HA-mCherry-Sbh2 from a low-copy plasmid under the strong TEF2 promoter. Bars, 1 µm.
Here, we show that Sbh2 is rapidly turned over in cells lacking its binding partner Ssh1. We identify the ERAD E3 ligase Doa10 to be responsible for the fast degradation of unassembled Sbh2. Unexpectedly, and in contradiction to the accepted view, the degron is located within the membrane-spanning TA sequence of Sbh2. This establishes unassembled Sbh2 as the first ERAD-M substrate. It can be expected that other Doa10 ERAD-M substrates exist as well.
Results
Sbh2 is a Doa10 substrate and association with Ssh1 protects it from degradation
Sbh2 is a subunit of the trimeric Ssh1 complex in S. cerevisiae (Fig. 1 A). It was reported previously (Finke et al., 1996) that upon deletion of SSH1, Shb2 levels are strongly reduced, suggesting rapid Sbh2 turnover in the absence of Ssh1. It therefore seemed likely that association with Ssh1 protects Sbh2 from degradation. To investigate whether unassembled Sbh2 is indeed short lived, we first analyzed the stability of ectopically expressed N-terminally HA-tagged Sbh2 in wild-type (WT) cells by cycloheximide (chx) chase analysis (Fig. 1 B). In WT cells, HA-Sbh2 levels dropped in the first 15 min after chx addition and remained constant thereafter (Fig. 1, B and C). This is consistent with a scenario in which Ssh1-bound Sbh2 is protected from degradation, whereas surplus Sbh2 is rapidly degraded. As Sbh2 is an ER-localized TA protein, we asked whether Sbh2 is degraded via one of the ERAD pathways. Deletion of the gene encoding Doa10, one of the two major ERAD E3 ligases in S. cerevisiae, led to an increase in HA-Sbh2 steady-state levels (Fig. 1 B, compare 0 min time points of WT and doa10Δ cells) accompanied by an almost complete stabilization of the protein (Fig. 1 B). In contrast, deletion of the HRD1 gene, which encodes the second ERAD E3 Hrd1/Der3, had no detectable effect on HA-Sbh2 stability (Fig. 1 B). The combined deletion of DOA10 and HRD1 had no additional stabilizing effect beyond that of the DOA10 knockout alone (Fig. 1 B).
We next investigated Sbh2 stability in cells lacking its binding partner, the translocon α-subunit Ssh1. As previously observed (Finke et al., 1996), steady-state levels of ectopically expressed HA-Sbh2 were strongly reduced in ssh1Δ cells (approximately fourfold as compared with WT yeast; Fig. 1 C). Moreover, the entire HA-Sbh2 pool was degraded in these cells in <30 min (Fig. 1 C). Codeletion of SSH1 and DOA10 resulted in a pronounced increase in HA-Sbh2 steady-state levels and an almost complete stabilization of the entire HA-Sbh2 pool (Fig. 1 C). HA-Sbh2 was only very moderately stabilized upon deletion of the second yeast ERAD E3, Hrd1, in cells lacking Ssh1 (Fig. 1 C), supporting the notion that HA-Sbh2 is almost exclusively recognized by the Doa10 complex.
The aforementioned findings indicated that Sbh2 stability and consequently Sbh2 quantity are regulated by Doa10. To test whether association with Ssh1 protects Sbh2 from degradation, we compared the amounts of metabolically stable (ectopically expressed) HA-Sbh2 in WT and sbh2Δ cells (Fig. 1 D). In WT cells, the ectopically expressed HA-Sbh2 has to compete with endogenous Sbh2 for incorporation into the Ssh1 complex, whereas in sbh2Δ cells it represents the only source of Sbh2. The amount of stable HA-Sbh2 was determined 5 h after translational inhibition by chx. After such treatment, HA-Sbh2 levels were ∼65% higher in sbh2Δ cells (where the ectopically expressed HA-Sbh2 does not compete with endogenous Sbh2 for Ssh1 binding) as in WT cells (Fig. 1 D). In cells lacking both endogenous Sbh2 and Ssh1, the entire HA-Sbh2 pool was degraded (Fig. 1 D). It was also previously reported that Sbh2 can bind to Sec61, but only in the absence of both Ssh1 and Sbh1, the β subunit of the Sec61 complex (Finke et al., 1996). We found a significant fraction of ectopically expressed HA-Sbh2 to be stable in sbh1Δ ssh1Δ cells, consistent with association with and protection by Sec61 (Fig. 1 D). We conclude that unassembled Sbh2 is an endogenous Doa10 substrate.
Proper membrane insertion of HA-Sbh2 in ssh1Δ cells
To exclude that degradation of HA-Sbh2 in ssh1Δ cells is caused by an incomplete membrane insertion of HA-Sbh2, its membrane association was analyzed biochemically and microscopically. HA-Sbh2 was expressed in doa10Δ ssh1Δ cells, and a crude microsomal fraction was prepared and subjected to various treatments before separation into pellet and supernatant fractions by centrifugation (Fig. 1 E). In this assay, HA-Sbh2 behaved similarly to the polytopic ER-membrane protein Doa10-13MYC, which served as a control (Fig. 1 E). HA-Sbh2 was efficiently extracted from membrane pellets upon solubilization of membranes with detergent and salt together (Triton X-100 and NaCl), but not by salt or urea alone, treatments that strip off peripheral membrane proteins (Fig. 1 E). Neither did an alkaline solution, sodium carbonate (pH 11.5), which liberates peripheral membrane proteins as well as ER-luminal proteins, lead to extraction of HA-Sbh2 (Fig. 1 E). As expected, the soluble protein Pgk1 was found in all cases in the supernatant.
We also investigated the subcellular localization of Sbh2 by confocal microscopy. Sbh2 with an N-terminal HA-mCherry tag was overexpressed ectopically from the strong TEF2 promoter. The HA-mCherry-Sbh2 protein displayed proper ER localization in WT, ssh1Δ, and doa10Δ ssh1Δ cells, respectively, (Fig. 1 F). (Note, HA-mCherry-Sbh2 is detectable even in ssh1Δ cells under these strong overexpression conditions.) Consistent with an earlier study (Schuldiner et al., 2008), deletion of Get2, which is required for insertion of TA proteins into the ER membrane, resulted in accumulation of HA-mCherry-Sbh2 in a single cytosolic aggregate (Fig. 1 F). Together, these results establish that HA-Sbh2/HA-mCherry-Sbh2 is correctly inserted into the ER membrane even in the absence of Ssh1, suggesting that Sbh2 recognition and degradation occurs after membrane insertion.
Degradation of Sbh2 requires Doa10 E3 ligase activity and occurs via the 26S proteasome
Doa10 usually decorates its substrates with a polyubiquitin chain, which is a signal for proteasomal degradation (Nakatsukasa et al., 2008). To test whether Sbh2 degradation depends on the E3 ligase activity of Doa10, Sbh2 stability was investigated in cells expressing an inactive Doa10 variant, doa10-C39S, which contains a mutated RING-CH domain (Swanson et al., 2001). HA-Sbh2 steady-state levels were strongly increased, and the protein was completely stabilized in doa10-C39S cells (Fig. 2 A), indicating that Sbh2 degradation is strictly dependent on Doa10 ligase activity. Next, we analyzed whether Sbh2 is ubiquitylated in cells. Overexpressed HA-Sbh2 was immunoprecipitated from cell lysates prepared from ssh1Δ and doa10Δ ssh1Δ cells expressing MYC-tagged ubiquitin (Fig. 2 B). After precipitation from ssh1Δ cells, a ladder of ubiquitylated HA-Sbh2 was detected. In contrast, a much weaker and shorter ubiquitin ladder was detected when HA-Sbh2 was precipitated from doa10Δ ssh1Δ cells, demonstrating that Doa10 is the major E3 ligase for Sbh2. The residual Sbh2 ubiquitylation observed in the absence of Doa10 is likely performed by Hrd1, given its minor role in HA-Sbh2 turnover (Fig. 1 B). Importantly, no ubiquitylation was observed after anti-HA immunoprecipitation (IP) from ssh1Δ cells overexpressing FLAG-Sbh2 (Fig. 2 B). Next, we investigated whether Doa10-dependent Sbh2 degradation proceeds via the 26S proteasome. To that end, HA-Sbh2 stability was studied upon proteasome inhibition with MG132 (Lee and Goldberg, 1998). HA-Sbh2–expressing cells lacking Ssh1 and the multidrug exporting ATP-binding cassette–transporter Pdr5 were treated with MG132 or DMSO (mock), and HA-Sbh2 levels were determined immediately before and 3 h after drug addition (Fig. 2 C). MG132 treatment led to a strong (>20-fold) increase in HA-Sbh2 levels, indicating that Sbh2 is degraded via the 26S proteasome.
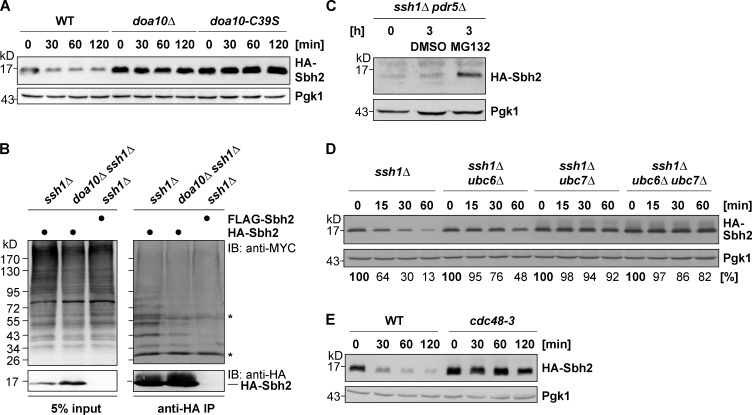
Figure 2.
Sbh2 degradation requires Doa10 E3 ligase activity and proceeds via the 26S proteasome. (A) Doa10 E3 ligase activity is required for Sbh2 degradation. chx chase assay of ectopically expressed (low-copy plasmid; MET25 promoter) HA-Sbh2 in WT, doa10Δ, and doa10-C39S cells (DF5 strain background). (B) Sbh2 is ubiquitylated in cells. In vivo ubiquitylation of HA-Sbh2: HA- or FLAG-tagged Sbh2 was ectopically expressed (low-copy plasmid; GPD promoter) in ssh1Δ and doa10Δ ssh1Δ cells together with MYC-ubiquitin. HA-Sbh2 was precipitated from the cell lysates with anti-HA agarose beads. Precipitates were analyzed by immunoblotting with anti-HA and anti-MYC. Asterisks indicate cross-reactive bands (IgG heavy and light chain, respectively) recognized by the secondary antibody (anti–rabbit peroxidase). IB, immunoblotting. (C) Proteasomal degradation of Sbh2. ssh1Δ pdr5Δ cells ectopically expressing HA-Sbh2 (low-copy plasmid; MET25 promoter) were grown to log phase (0 h time point), divided into two cultures, and treated for 3 h with either the proteasome inhibitor MG132 (50 µM) or the solvent DMSO. Samples were normalized for equal amounts of the stable protein Pgk1 before gel loading. (D) Ubc6 and Ubc7 are required for efficient Sbh2 degradation. Degradation of ectopically overexpressed HA-Sbh2 (low-copy plasmid; GPD promoter) in WT, ubc6Δ, ubc7Δ, and ubc6Δ ubc7Δ cells (DF5 strain background). chx chase analysis was performed as in Fig. 1 B. Relative protein levels listed below the blots were determined by quantification of pixel densities of HA-Sbh2 bands relative to those of Pgk1. 0 h time point was in each case set to 100%. (E) The AAA-ATPase Cdc48 is required for Sbh2 degradation. Degradation of ectopically overexpressed HA-Sbh2 (low-copy plasmid; GPD promoter) in WT and cdc48-3 cells (W303-1A strain background). Cells were grown to log phase at 25°C and shifted to the nonpermissive temperature of 37°C 30 min before addition of chx.
Accessory factors required for efficient Sbh2 degradation
Ubiquitylation of most Doa10 substrates depends on the two E2s Ubc6 and Ubc7 (Swanson et al., 2001). To test the E2 requirements for efficient Sbh2 degradation, we analyzed Sbh2 stability in ssh1Δ cells lacking either one or both E2s. Deletion of UBC6 or UBC7 alone resulted in a detectable stabilization of HA-Sbh2, whereby deletion of UBC7 had the stronger stabilizing effect (Fig. 2 D). No further stabilization was observed in cells lacking both E2s as compared with the ubc7Δ single deletion. Therefore, as for the majority of Doa10 substrates (Zattas and Hochstrasser, 2015), Doa10-dependent degradation of HA-Sbh2 is dependent on both Ubc6 and Ubc7 with Ubc7 playing the major role.
Efficient ER extraction and degradation of Doa10 membrane protein substrates requires the Cdc48 AAA-ATPase in complex with Npl4 and Ufd1 (Ravid et al., 2006). To determine whether degradation of Sbh2 is dependent on a functional Cdc48 complex, we analyzed Sbh2 stability in a cdc48-3 strain expressing a temperature-sensitive (ts) Cdc48 protein (Ye et al., 2001). At nonpermissive temperature, overexpressed HA-Sbh2 was significantly stabilized in cdc48-3 cells (Fig. 2 E), consistent with a role for Cdc48 in Sbh2 degradation.
The cytosolic domain of Sbh2 is dispensable for Sbh2 degradation
Next, we aimed to localize the degron within Sbh2. We first tested whether the degron is contained within the C-terminal half of the 88-residue Sbh2 polypeptide by analyzing the stability of a fusion protein consisting of the soluble Ura3 protein, a HA epitope tag, and the last 48 residues of Sbh2 (Ura3-HA-Sbh2(aa 41–88); Fig. 3 A). The Sbh2 moiety included the TA sequence (Sbh2 residues 62–88) and the 21 preceding residues (residues 41–61). This Sbh2 segment was sufficient to render the otherwise stable Ura3 protein short lived, with a half-life of ∼30 min in ssh1Δ cells (Fig. 3 B). Degradation of the Ura3-HA-Sbh2(aa 41–88) fusion depended on Doa10 in otherwise WT or ssh1Δ cells (Fig. 3 B).
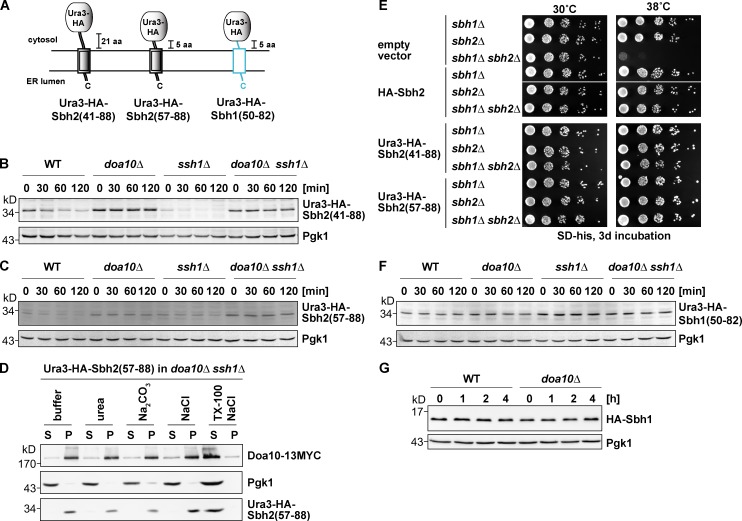
Figure 3.
Mapping of the degron within Sbh2. Analysis of Ura3-HA-Sbh2 fusion proteins. (A) Schematic depiction of Ura3-HA-Sbh2 and Ura3-HA-Sbh1 fusion proteins used in this study. Sequence regions derived from Sbh1 are in light blue, and Sbh2 derived sequences are depicted as double black lines. (B) The last 48 residues of Sbh2 promote degradation of a stable protein. Degradation of ectopically expressed Ura3-HA-Sbh2(aa 41–88) (low-copy plasmid; MET25 promoter) in WT, doa10Δ, ssh1Δ, and doa10Δ ssh1Δ cells. chx chase was performed as in Fig. 1 B. (C) The last 32 residues of Sbh2 promote degradation of a stable protein. Degradation of ectopically expressed Ura3-HA-Sbh2(aa 57–88) as in B. (D) Ura3-HA-Sbh2(aa 57–88) is an integral membrane protein in ssh1Δ cells. Subcellular fractionation of doa10Δ ssh1Δ cells expressing Ura3-HA-Sbh2(aa 57–88) as in Fig. 1 E. S, supernatant; P, pellet; TX-100, Triton X-100. (E) Suppression of growth defect of sbh1Δ sbh2Δ cells at a high temperature by HA-Sbh2 and Ura3-HA-Sbh2 fusion proteins. Cells were transformed with an empty vector (p413MET25) or a p413MET25-based plasmid encoding HA-Sbh2 or the indicated Ura3-HA-Sbh2 protein. Serial dilutions (sixfold) of cultures were spotted onto plates, and plates were incubated as indicated. Empty vector and HA-Sbh2 lanes are from one plate (one plate for 30°C and a second one for 38°C). Both Ura3-HA-Sbh2(aa 41–88) and Ura3-HA-Sbh2(aa 57–88) lanes are from plates that were incubated parallel to the empty vector and HA-Sbh2 plates. (F) The Ura3-HA-Sbh1(aa 50–82) fusion protein is stable. chx chase with ectopically expressed Ura3-HA-Sbh1(aa 50–82) as in B. (G) Sbh1 is a stable protein. chx chase with ectopically expressed HA-Sbh1 (as in B) in WT and doa10Δ cells.
The destabilizing effect on Ura3 is a specific property of the Sbh2 sequence, as a similar fusion protein containing the last 42 amino acids of the TA protein Prm3, was previously shown to be stable (Kreft and Hochstrasser, 2011). We next analyzed a fusion protein that contained only five Sbh2 residues upstream of the Sbh2 TM domain (Ura3-HA-Sbh2(aa 57–88); Fig. 3 A). This fusion protein was also degraded in a Doa10-dependent manner in both WT and ssh1Δ cells (Fig. 3 C), albeit more slowly than HA-Sbh2 and Ura3-HA-Sbh2(aa 41–88). Importantly, the Sbh2 moiety of the Ura3-HA-Sbh2(aa 57–88) fusion protein was sufficient to confer proper ER membrane insertion as judged by subcellular fractionation (Fig. 3 D). Cells lacking SBH1 and SBH2 display a strong growth defect at an elevated temperature (Finke et al., 1996), which is suppressed by expression of the TM domain of Sbh1 or Sbh2 (Feng et al., 2007; Leroux and Rokeach, 2008). Ura3-HA-Sbh2(aa 57–88) was able to suppress the ts growth phenotype of sbh1Δ sbh2Δ cells (Fig. 3 E), providing additional evidence for correct insertion into the ER membrane.
A similar fusion in which the Ura3-HA sequence was fused to the last 33 residues of the other yeast Sec61 β-subunit Sbh1, Ura3-HA-Sbh1(aa 50–82) (Fig. 3 A), was stable in WT, doa10Δ, and ssh1Δ as well as doa10Δ ssh1Δ cells (Fig. 3 F). Consistent with an earlier study (Biederer et al., 1996), full-length HA-Sbh1 was also stable in WT cells (Fig. 3 G). Together, these results demonstrated that the degron is contained within the tail anchor (residues 57–88) of Sbh2.
TM and ER-luminal segments of Sbh2 are required for degradation
To provide additional evidence that the degron is contained within the C-terminal membrane anchor of Sbh2, a chimeric protein consisting of the cytosolic domain of the stable Sbh1 (aa 1–53) and the last 28 residues of Sbh2 (aa 61–88), HA–Sbh-122 (for nomenclature, see legend of Fig. 4), was analyzed (Fig. 4 A). In the absence of Ssh1, the HA–Sbh-122 chimera was degraded with comparable kinetics as the HA-Sbh2 protein (compare t1/2 = ∼7 min for HA-Sbh2 vs. t1/2 = ∼10 min for HA–Sbh-122; Figs. 1 C and 4 B). Degradation of the fusion protein was also evident in WT cells (t1/2 = ∼13 min), but unlike full-length HA-Sbh2, nearly the entire HA–Sbh-122 pool was degraded (Fig. 4 B). In each case, turnover of the fusion protein was dependent on Doa10 (Fig. 4 B). This demonstrated that the Sbh2 TA region consisting of the TM domain and the short ER-luminal domain can target an otherwise stable protein for Doa10-dependent degradation. This is remarkable as all as-of-yet described Doa10 substrates harbor a cytosolic degron (Hirsch et al., 2009; Brodsky and Skach, 2011; Finley et al., 2012; Ruggiano et al., 2014; Christianson and Ye, 2014; Zattas and Hochstrasser, 2015).
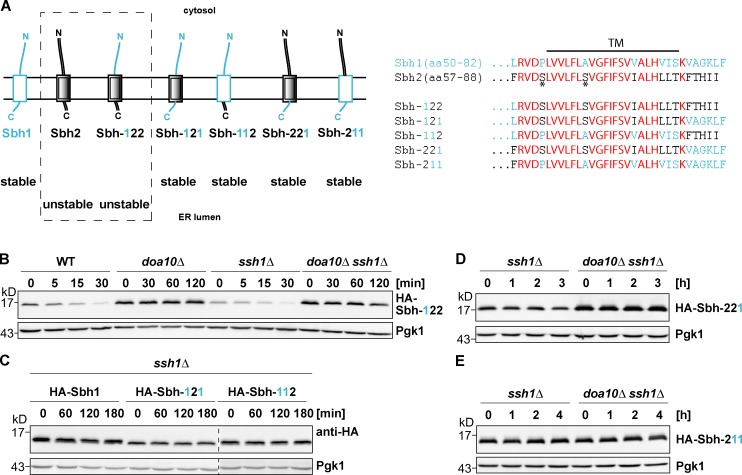
Figure 4.
Investigation of Sbh1/Sbh2 chimeric proteins. Both Sbh2 TM and ER-luminal regions contribute to the degron. (A) Schematic representation of Sbh1, Sbh2, and Sbh1/Sbh2 chimeric proteins used in this study. Sbh1-derived regions are depicted in light blue, and Sbh2-derived regions are shown in black. The nomenclature for Sbh1/Sbh2 chimeric proteins is as follows: “Sbh-XYZ” designates a chimeric protein in which the number at position X indicates the source of the cytosolic domain (“1”: Sbh1; or “2”: Sbh2), the number at position Y indicates the source of the TM helix, and the number at position Z indicates the source of the ER-luminal domain, respectively. (right) Comparison of Sbh1, Sbh2, and Sbh1/Sbh2 chimera TA sequences. Residues depicted bold in red represent identical residues shared between Sbh1 and Sbh2; residues depicted in light blue are Sbh1-specific residues, and residues depicted in black indicate residues specific for Sbh2; the black horizontal line indicates the TM helix, and the asterisks indicate Sbh2 residues Ser61 and Ser68, respectively. N, N terminal; C, C terminal. (B) Degradation of HA–Sbh-122. chx chase as in Fig. 1 B. Note: There are different chase times for WT and ssh1Δ strains (30-min chase) and doa10Δ and doa10Δ ssh1Δ strains (120-min chase). (C) Degradation of HA-Sbh1, HA–Sbh-121, and HA–Sbh-112. All blots are from same experiment/gel but were cropped for clarity (dashed line). (D) Degradation of HA-Sbh-221. (E) Degradation of HA–Sbh-211.
To investigate the individual contribution of the Sbh2 TM and ER-luminal regions to the degron, they were exchanged with the corresponding Sbh1 sequences (Fig. 4 A). Sbh1 and Sbh2 TA sequences exhibit strong homology over the TM helix (76% identity and 24% homology) but differ in primary sequence in the short ER-luminal tail (albeit both tails share an overall hydrophobic character; Fig. 4 A). Swapping of the TM helix or the ER-luminal domain alone, resulting in chimeras HA–Sbh-121 and HA–Sbh-112 (Fig. 4 A), respectively, failed to destabilize the Sbh1 protein in ssh1Δ cells (Fig. 4 C), indicating that both contribute to the functional degron. In support of this notion, replacement of only the ER-luminal part of Sbh2 with the ER-luminal Sbh1 sequence, HA–Sbh-221, resulted in nearly complete stabilization in ssh1Δ cells (Fig. 4 D). Furthermore, replacement of both TM and ER-luminal domains of Sbh2 with the Sbh1 sequence also resulted in a stable protein (HA–Sbh-211; Fig. 4 E). Collectively, these data strongly implied that the Doa10-specific degron is contained in the TA sequence of Sbh2 and that both the TM domain and the short ER-luminal domain contribute to the degron. It is noteworthy that each of the aforementioned Sbh1/Sbh2 chimeric constructs suppresses the ts phenotype of sbh1Δ sbh2Δ cells and behaves like a membrane protein in subcellular fractionation experiments (Fig. S1, A–C), indicating that each chimera is correctly inserted into the ER membrane.
Sbh2 TM residue Ser68 is part of the degron
Having mapped the degron to the TA region (residues 61–88) of Sbh2, we wanted to investigate the degron at a higher resolution. Strong sequence conservation exists in the TM helices of Sbh1, Sbh2, and Sec61 β subunits in other species (Fig. S2 A; Kinch et al., 2002). A highly conserved proline is present at the cytosol/ER membrane interface (e.g., S. cerevisiae Sbh1 Pro54; Fig. 4 A). Strikingly, unlike the translocon β subunit from most other species, S. cerevisiae Sbh2 has a serine at this position (Sbh2 Ser61). Moreover, most eukaryotic Sec61 β subunits contain a strictly conserved serine residue within the TM helix (Sbh2 Ser68), except for S. cerevisiae Sbh1, where an alanine is present at this position (Sbh1 Ala61). Sbh2 residues Ser61 and Ser68 are both predicted to face Ssh1 (Fig. 5 A; Becker et al., 2009). To investigate the role of Ser61 and Ser68 in Sbh2 degradation, they were mutated to the corresponding residues present in Sbh1 (S61P and S68A, respectively), and turnover of each Sbh2 single point mutant was analyzed in ssh1Δ cells (Fig. 5 B). Mutation of either of the two residues resulted in a partial stabilization (Fig. 5 B). The observed residual degradation was in each case dependent on Doa10 (Fig. 5, C and D). The combination of the two mutations (Sbh2(S61P,S68A)) led to a complete stabilization (presumably through binding to Sec61—see following paragraph; Fig. 5 E). The introduced point mutations are not likely to have interfered with proper membrane insertion because the mutant proteins all suppressed the ts growth defect of sbh1Δ sbh2Δ cells (Fig. 5 F).
Figure 5.
TM-residue Ser68 of the Sbh2 tail anchor is a part of the degron. (A) Sbh2 TA sequence in complex with Ssh1 TM helices 1 and 4. Top view of the interface of Sbh2 and Ssh1. Sbh2 residues 58–79 and Ssh1 residues 26–53 (TM1) and 148–179 (TM4) are shown. Sbh2 Ser61 and Ser68 are depicted in red in ball and stick mode. Picture was generated with PyMOL using the atomic coordinates from Protein Data Bank accession no. 2WWA (Becker et al., 2009). (B) Degradation of HA-tagged Sbh2 WT and S61P and S68A point mutants in ssh1Δ cells. A representative blot is shown. chx chase was performed as in Fig. 1 B. The graph at right shows the mean degradation rates observed from at least three independent experiments. Error bars represent ± SD (C and D) HA-Sbh2(S61P) and HA-Sbh2(S68A) are stable in doa10Δ ssh1Δ cells. chx chase analysis of HA-Sbh2(S61P) and HA-Sbh2(S68A) stability. (E) HA-Sbh2(S61P,S68A) is stable even in ssh1Δ cells. chx chase analysis of HA-Sbh2(S61P,S68A) stability. (F) Suppression of growth defect of sbh1Δ sbh2Δ cells at high temperature by mutant Sbh2 variants (S61P), (S68A), and (S61P,S68A), respectively. Growth assay as in Fig. 3 E. Empty vector and HA-Sbh2 lanes are from one plate. (G) Co-IP analysis of digitonin-solubilized microsomes to investigate the interaction between Sec61 β subunits and Sec61. WT or mutant HA-Sbh2 was ectopically expressed in doa10Δ ssh1Δ cells and was precipitated with anti-HA agarose beads. Precipitates were analyzed by immunoblotting with the indicated antibodies. The TA Ubc6 protein served as a negative control. (H) Co-IP analysis to investigate the interaction between Sec61 β subunits and Sec61. WT HA-Sbh1 or HA-Sbh2 or mutant HA-Sbh2 was ectopically expressed in SEC61-9MYC ssh1Δ cells (W303-1B strain background), and Sec61-9MYC was precipitated with an anti-MYC antibody (rabbit polyclonal). Precipitates were analyzed by immunoblotting with indicated antibodies. IB, immunoblot.
These data demonstrated that a stepwise conversion of the Sbh2 TM region into the Sbh1 sequence results in progressive stabilization of the Sbh2 mutant protein. However, neither Sbh1 nor Sbh2 are “free”/unbound inside the cell. Sbh1 binds to Sec61, and Sbh2 binds to Ssh1. Binding of Sbh2 to Ssh1 protects Sbh2 from degradation as does its association with Sec61 (Fig. 1 C; Finke et al., 1996). This made it difficult to judge whether the observed stabilization of the analyzed Sbh2 mutants was caused by an inactivation of the degron itself or whether it resulted from an acquired capacity to associate with Sec61. To examine to what extent each Sbh2 mutant binds Sec61, a series of co-IP experiments were conducted. The three HA-tagged Sbh2 point mutants as well as HA-Sbh1 and HA-Sbh2 were expressed in doa10Δ ssh1Δ cells, and corresponding digitonin-solubilized microsomes were subjected to anti-HA precipitation followed by analysis of the amount of coprecipitating Sec61 protein (Fig. 5 G). Both, the HA-Sbh2(S61P) single mutant and the HA-Sbh2(S61P,S68A) double mutant interacted strongly (comparable to Sbh1) with Sec61 as judged by the amounts of coprecipitated Sec61 (Fig. 5 G). In stark contrast, HA-Sbh2(S68A), showed only a weak interaction with Sec61, and the HA-Sbh2 control showed an even weaker interaction with Sec61 (Fig. 5 G). The specificity of the co-IP protocol is demonstrated by the fact that the endogenous TA protein Ubc6 did not copurify with any of the Sbh2/Sbh1 proteins (Fig. 5 G). A similar outcome was observed when the co-IP was reversed in ssh1Δ SEC61-9MYC cells (Fig. 5 H). Moreover, consistent with the observed individual stabilities of the different Sbh2 variants, WT Sbh2 interacted most strongly with a ligase-inactive Doa10 variant (doa10(C39S)-13MYC) and the S61P,S68A double mutant interacted the weakest with Doa10 (Fig. S3, A and B). The two single mutants, S61P and S68A, displayed an intermediate interaction. Furthermore, the observed interaction for each Sbh2 variant with Doa10 correlated with their ubiquitylation efficiency (Fig. S3 C).
In summary, the very weak interaction of Sbh2(S68A) with Sec61 (Fig. 5, G and H) indicated that the stabilization of the Sbh2(S68A) mutant in ssh1Δ cells is caused by the (partial) inactivation of the degron per se and not to a novel, protective association with Sec61. The TM residue Ser68 of Sbh2 is a critical part of the Doa10-dependent degron. Therefore, Doa10 can function in ERAD-M.
Discussion
Here, we establish the TA protein Sbh2, which is a subunit of the heterotrimeric Ssh1 translocon complex in S. cerevisiae, as the first ERAD-M substrate that is recognized by the ERAD E3 ligase Doa10. The identification of a Doa10-dependent ERAD-M degron shows that Doa10 has the capacity for recognizing intramembrane degrons and allows for the first time to place Doa10 in the ERAD-M pathway (Carvalho et al., 2006; Hirsch et al., 2009; Brodsky and Skach, 2011; Finley et al., 2012; Christianson and Ye, 2014; Ruggiano et al., 2014; Zattas and Hochstrasser, 2015).
Sbh2 is a quantity control substrate
It was previously observed that Sbh2 levels are drastically reduced in cells lacking Ssh1, the primary binding partner of Sbh2 (Finke et al., 1996). Here, we show that, under normal conditions, Sbh2 levels are dictated by the quantity of its binding partner Ssh1 (Fig. 1). Sbh2 can also be stabilized by association with Sec61, which, however, occurs only in the absence of SSH1 and SBH1 (Fig. 1; Finke et al., 1996). Hence, Sbh2 represents a quantity control substrate, which is rapidly degraded if it fails to associate with Ssh1 (Fig. 6).
Figure 6.
Model for Sbh2 quantity control via a Doa10-dependent intramembrane degron. (A) The TA Sbh2 protein associates with the Ssh1 protein in the ER membrane (1). Together with a third integral membrane protein, Sss1 (not depicted), Ssh1, and Sbh2 form the heterotrimeric Ssh1 complex implicated in cotranslational protein translocation in S. cerevisiae (besides the heterotrimeric Sec61 complex). Assembled Sbh2 is stable/protected from degradation. In contrast, unassembled Sbh2 (e.g., in ssh1Δ cells or surplus Sbh2 in WT cells) is readily degraded via a Doa10-dependent ERAD pathway involving ubiquitylation (2), retrotranslocation, and proteasomal degradation of Sbh2 (3). Ub, ubiquitin. (B) Schematic depiction of the Doa10-dependent intramembrane (ERAD-M) degron of Sbh2. The degron encompasses the Sbh2 TM domain and the short ER-luminal domain. The serine residue at position 68 (S68) located within the Sbh2 TM helix is a crucial part of the degron. (C) Assignment of the two major S. cerevisiae ERAD E3 ligases to different ERAD substrate classes. The identification of a Doa10-dependent ERAD-M substrate, Sbh2, allows assigning Doa10 to the ERAD-M pathway.
Other unassembled membrane proteins are destroyed via ERAD. For example, the TCR-α in mammals is degraded in the absence of the TCR-β chain (Bonifacino et al., 1989). In yeast, the polytopic Vph1 protein, a subunit of the vacuolar H+-ATPase, is degraded in the absence of the ER resident protein Vma22 (Hill and Cooper, 2000). Likewise, the yeast 4-TM Der1 protein is turned over if it fails to assemble with the Hrd1 ligase complex (Mehnert et al., 2014). The existence of a quantity control pathway for Sbh2 suggests that elevated Sbh2 levels (i.e., in excess over Ssh1) might be deleterious under some conditions. Although strong overexpression of Sbh2 did not result in a detectable growth phenotype under standard conditions (unpublished data), there could be other circumstances under which excess unassembled Sbh2 negatively impacts cell growth.
Sbh2 degradation is Doa10 dependent
HA-Sbh2 degradation is strongly dependent on Doa10 (Fig. 1). In ssh1Δ cells, HA-Sbh2 steady-state levels are strongly decreased, and HA-Sbh2 is rapidly degraded. Deletion of DOA10 in ssh1Δ cells results in an almost complete stabilization of HA-Sbh2. The second yeast ERAD E3, Hrd1, plays only a minor role in Sbh2 turnover (Fig. 1). It is not known whether Doa10 and Hrd1 target different pools or conformations of Sbh2. Recently, a third E3, Ubr1, was found to play an ancillary role in ERAD in yeast (Stolz et al., 2013), but it is unlikely to play a prominent role in Sbh2 degradation given the nearly complete stabilization of Sbh2 in doa10Δ ssh1Δ cells.
Sbh2 contains an intramembrane degron
Analysis of truncated Sbh2 variants fused to Ura3-HA (Fig. 3) and chimeric proteins in which regions or individual residues of Sbh2 were replaced by the corresponding Sbh1 sequence (Figs. 4 and 5) allowed us to place the degradation signal within the TA region of Sbh2. Both the predicted TM helix and the short intraluminal stretch of Sbh2 are required for formation of the functional degron (Fig. 4). Several independent lines of evidence further indicate that the degron is recognized by Doa10 after membrane insertion of Sbh2, i.e., in context of the membrane. First, subcellular fractionation experiments demonstrated that the investigated Sbh2 proteins insert properly into the membrane even in the absence of Ssh1 (Figs. 1, 3, and S1), indicating that the turnover of the various Sbh2 variants analyzed is not caused by defects in membrane insertion. Second, the analyzed Sbh2 variants are all able to suppress the ts growth phenotype of sbh1Δ sbh2Δ cells (Figs. 3, 5, and S1), which requires proper insertion into the ER membrane and interaction with Sec61 or Ssh1 (Feng et al., 2007; Zhao and Jäntti, 2009). Third, efficient Sbh2 turnover is strongly dependent on the Cdc48 complex (Fig. 2), which is a general requirement for turnover of membrane-embedded (but not soluble) Doa10 substrates (Ravid et al., 2006).
It was recently reported that the C-terminal hydrophobic motifs of a subset of GPI (glycosylphosphatidylinositol)-anchored proteins are sometimes recognized in the cytosol before their insertion/translocation and are targeted for degradation via a Doa10-dependent, ERAD-C–like pathway (Ast et al., 2014). It was also shown that TM segments of low hydrophobicity, such as the TCR-α TM helix, often fail to stay embedded/inserted in the ER membrane and translocate into the ER lumen where they are recognized and subjected to ERAD (Feige and Hendershot, 2013). Although the degron is formally contained within the TM segment in the aforementioned cases, it is recognized by the ERAD machinery in the ER lumen or cytoplasm. In contrast, the Sbh2 degron is very likely to be recognized in the context of the membrane. Turnover of what would normally be GPI-anchored proteins occurs before their membrane insertion and, in contrast to Sbh2, does not require Cdc48 (Ast et al., 2014). Furthermore, the fact that the Sbh2 homologue Sbh1, whose ER membrane insertion can be expected to be identical as that of Sbh2, is a stable protein (Fig. 3; Biederer et al., 1996) further argues against preinsertional degradation of Sbh2. Importantly, unlike the TCR-α TM helix, the TM segments of Sbh1, Sbh2, and Sbh2(S68A) all share a high hydrophobicity (Fig. S2 B), making mislocalization as described for unassembled TCR-α subunits unlikely. It can thus be concluded that the Sbh2 TM degron is recognized by Doa10 in context of the membrane.
Additional evidence for a direct contribution of the intramembrane region to the degron comes from an Sbh2 variant, Sbh2(S68A), which bears a single point mutation in the center of the TM helix at the face of the helix predicted to be in contact with Ssh1 (Fig. 5). Like WT Sbh2, the Sbh2(S68A) protein showed only a weak interaction with Sec61 (in ssh1Δ cells), yet it was significantly more stable than WT Sbh2 (Fig. 5). It is tempting to speculate that Ser68 is part of the degron/interface recognized by Doa10. As Ser68 is predicted to interact with Ssh1, it seems likely that the Doa10-dependent degron at least partially overlaps with the Ssh1 binding interface of the Sbh2 TM helix. Such a scenario would readily explain why an association with Ssh1 protects Sbh2 from being recognized by Doa10.
Our analysis of the Sbh2(S61P) mutant (Fig. 5) indicates that the amino acid at position 61 of Sbh2, which is located at the cytosol/ER membrane boundary, is a decisive factor determining which translocon α subunit is bound. WT Sbh2, with serine at this position, preferentially associates with Ssh1 and only weakly interacts with Sec61 (Fig. 5; Finke et al., 1996). Exchange of this serine to proline, the corresponding residue in Sbh1, led to a strong increase in Sec61 binding (in ssh1Δ cells). Molecular modeling of the Sec61–Sbh2 complex and prediction of binding affinity changes by single point mutations in Sbh2 supports the hypothesis that the residue at the cytosol/ER membrane boundary dictates which α subunit Sbh2 binds to (Fig. S4). In silico, mutating Sbh2 Ser61 to proline increases the predicted affinity to Sec61 (ΔΔGBind = −0.5 kcal/mol). In contrast, mutating Sbh1 Pro54 into serine (residue 54 in Sbh1 is equivalent to residue 61 of Sbh2) results in a decreased calculated affinity for Sec61 (ΔΔGBind = +1.17 kcal/mol).
An obvious question is how the Sbh2 ERAD-M degron is recognized by Doa10. Recently, it has been shown that Hrd1 directly recognizes ERAD-M degrons via its TM region (Sato et al., 2009). ERAD-M degrons are believed to display some hydrophilicity in the TM region, and selected hydrophilic TM residues of the Hrd1 polypeptide have been shown to be crucial for substrate recognition (Sato et al., 2009). Like Hrd1, Doa10 also contains several hydrophilic TM residues. It is therefore tempting to speculate that the Sbh2 intramembrane degron is directly recognized by Doa10 TM residues. Future work will show whether or not this is true.
Our data reveal that the short ER-luminal stretch of Sbh2 is part of the degron (Fig. 4). Replacement of the last five residues of Sbh2 by the last six residues of Sbh1 resulted in a stable chimera (Fig. 4). Because the ER-luminal tail of Sbh2 is presumably too short (and too close to the membrane) to be bound by luminal factors such as the Hsp70 chaperone Kar2, it seems likely that the short luminal sequence of Sbh2 is also recognized directly by Doa10. Considering the hydrophobic nature of the last five residues of Sbh2 (FTHII), it is possible that this region remains largely within the membrane bilayer.
The human orthologue of Doa10, MARCH6/TEB4, is a member of the mammalian membrane-associated RING-CH (MARCH) protein family (Bartee et al., 2004). Most viral and mammalian MARCH proteins are characterized by an N-terminal RING-CH domain and only two TM domains, and many of these are involved in ubiquitylation of surface receptors to trigger their endocytosis or function as ERAD E3s (Wang et al., 2008; Nathan and Lehner, 2009). For several MARCH proteins, TM interactions between the MARCH protein and its substrate are essential for substrate down-regulation (e.g., for the viral kk3 and mk3 [Coscoy and Ganem, 2001; Wang et al., 2004] and the mammalian MARCH1 protein [Corcoran et al., 2011]). Our data suggest that MARCH6 may also be able to recognize some of its substrates via intramembrane interactions.
Comparison of Sbh2 and Ubc6 TA sequences
Sbh2 is not the only TA protein known to interact with Doa10. The TA E2 enzyme Ubc6 is one of two E2s functioning with Doa10 and is also a Doa10 substrate (Swanson et al., 2001). Doa10-dependent degradation of Ubc6 requires both the tail anchor and the E2 activity of Ubc6 (Walter et al., 2001). However, the Ubc6 tail anchor does not represent a degron as the Ubc6 TA sequence alone is not sufficient to confer a short half-life to Ubc6 or an otherwise stable reporter (Fig. S5; Walter et al., 2001; Kreft and Hochstrasser, 2011; a degron is defined as a minimal element within a protein that is sufficient for recognition and degradation by a proteolytic apparatus [Varshavsky, 1991]). In contrast, the TA sequence of Sbh2 is sufficient to confer a short half-life to an otherwise stable reporter (Figs. 3 and S5) and hence represents a de facto (ERAD-M) degron (making Sbh2 the first ERAD-M substrate of Doa10). The differential behavior of Doa10 toward the Sbh2 and Ubc6 TA sequences further highlights the multifaceted interactions of Doa10 with TA sequences and intramembrane degrons.
This study describes the first intramembrane degron that is recognized by the ERAD E3 ligase Doa10. The Doa10-dependent degradation of an ERAD-M substrate challenges the notion that ERAD-M substrates are exclusively recognized by Hrd1 (Fig. 6). Although Hrd1 might represent the major ERAD-M E3 ligase (based on the number of ERAD-M substrates already identified), it is likely that Doa10 recognizes additional, unidentified ERAD-M substrates. Recently, the squalene monooxygenase Erg1 has been identified as a Doa10 substrate (Foresti et al., 2013). The stability of the 2-TM Erg1 polypeptide is regulated by lanosterol. In analogy to the Hrd1 ERAD-M substrate Hmg2, Erg1 could represent a Doa10-dependent ERAD-M substrate. Future work is needed to test this possibility and to identify additional Doa10 ERAD-M substrates.
Materials and methods
Yeast and bacterial methods
Yeast rich (YPD [yeast, peptone, dextrose]) and minimal (SD [synthetic dextrose]) medium/plates were prepared as described previously (Guthrie and Fink, 1991); yeast strains and cultures were grown at 30°C unless indicated otherwise. All plasmids and yeast strains used in this study are summarized in Tables S1 and S2. Unless indicated otherwise, yeast strain background was BY4741.
Antibodies
The following antibodies were used: anti-HA mouse monoclonal antibody HA.11 (Covance), anti-MYC mouse monoclonal antibody 9E10 (Covance), anti-MYC rabbit polyclonal antibody sc-789 (Santa Cruz Biotechnology, Inc.), anti-Pgk1 mouse monoclonal antibody 22C5 (Molecular Probes), anti-Ubc6 rabbit polyclonal antiserum (raised against an bacterially expressed N-terminally His6-tagged version of S. cerevisiae Ubc6ha without TM domain; Walter et al., 2001), and anti-Sec61 rabbit polyclonal antiserum raised against a peptide corresponding to the C terminus of S. cerevisiae Sec61 plus an additional N-terminal cysteine (CLVPGFSDLM; Panzner et al., 1995) were gifts from T. Sommer (Max-Delbrück Center, Berlin, Germany). Secondary antibodies were peroxidase-conjugated goat anti–rabbit (Dianova) and peroxidase-conjugated goat anti–mouse (IgG1 specific; Jackson ImmunoResearch Laboratories, Inc.).
Preparation of cell extracts
Cell extracts were prepared as previously described (Loayza et al., 1998). In brief, 2.5 OD600 of log-phase yeast cultures were lysed by β-mercaptoethanol/NaOH and vigorous vortexing, and proteins were precipitated in 5% TCA. The pellet was resuspended in SDS gel loading buffer.
chx chase/immunoblot analyses
0.25 mg/ml chx was added to log-phase yeast cultures, and cell aliquots were removed at the indicated times after addition. Cells were harvested by centrifugation and resuspended in cold STOP mix (0.5× SD and 10 mM NaN3). After preparation of lysates (see Preparation of cell extracts), proteins were separated by SDS-PAGE and electrotransferred onto a polyvinylidene fluoride membrane (EMD Millipore). Immunodetection was performed with appropriate primary antibodies and horseradish peroxidase–conjugated secondary antibodies. Immunoreactive species were visualized using the ECL system (Western Lightning Plus-ECL; PerkinElmer). Protein degradation rates were determined using an imaging system (LAS-3000; Fujifilm) and AIDA image analyzer software (Bio Imaging). Values for each time point were normalized using an anti-Pgk1 loading control.
Ubiquitylation assay
Substrate ubiquitylation was examined essentially as described previously (Loayza et al., 1998). In brief, cells coexpressing HA-Sbh2 (or FLAG-Sbh2) together with a copper-induced N-terminally MYC-tagged ubiquitin were incubated in media containing 100 µM CuSO4. Where indicated (i.e., for pdr5Δ strains; Fig. S3 C) proteasome inhibitor MG132 (Sigma-Aldrich) dissolved in DMSO was added to a final concentration of 50 µM 2 h before cells were harvested. Cell lysates were prepared using the β-mercaptoethanol/NaOH extraction procedure as described (Preparation of cell extracts) with 5 mM N-ethylmaleimide (Sigma-Aldrich) being present throughout the preparation. After TCA precipitation, protein pellets were resuspended in TCA sample buffer (3.5% SDS, 0.5 M DTT, 80 mM Tris, 8 mM EDTA, 15% glycerol, and 4 mg/ml bromophenol blue) and boiled. Extracts were diluted 30-fold with cold dilution buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100) containing protease inhibitors (1 µg/ml aprotinin/leupeptin, 2 µM pepstatin, and 1 mM Pefabloc) and 5 mM N-ethylmaleimide. The extract was precleared with protein A–Sepharose beads (GE Healthcare) before HA-Sbh2 was precipitated with anti-HA agarose beads (Sigma-Aldrich). Beads were collected and washed with cold dilution buffer. After addition of sample buffer containing 8 M urea and 50 mg/ml DTT, samples were boiled before gel loading. Proteins were visualized by immunoblotting (see chx chase/immunoblot analyses).
Co-IP
Cell lysis and IP were performed as previously described (Stuerner et al., 2012). After cell lysis, the membrane fraction was solubilized in 1% digitonin (Serva); for anti-HA IP, anti-HA agarose beads (Sigma-Aldrich) were used, and for anti-MYC IP, polyclonal rabbit anti-MYC antibody was first added to the lysate, and antigen–antibody conjugates were coupled to preequilibrated protein A–Sepharose beads (GE Healthcare). Proteins were visualized by immunoblotting as described (chx chase/immunoblot analyses).
Spot growth assay
Overnight precultured cells grown at 30°C were diluted to an OD of A600 = 0.2 and spotted in a sixfold dilution series (2.5 µl/spot) in parallel onto SD-His plates. Plates were incubated at 30°C or 38°C for 2–3 d.
Proteasome inhibition
Cells were grown in selective media to mid–log phase. After an aliquot was taken, the culture was divided into two and proteasome inhibitor MG132 (dissolved in DMSO; final concentration 50 µM) was added to one culture, and an equivalent amount of DMSO was added to the second culture. Cultures were incubated for 3 h before cells were harvested. Lysates were prepared as described (Preparation of cell extracts).
Fluorescence microscopy
Yeast cells were grown to log phase in minimal medium, and 1 OD600 of cells was harvested. Cells were fixed with 4% (vol/vol) paraformaldehyde in PBS for 30 min at room temperature. Cells were washed twice with PBS and resuspended in 50 µl PBS. 2.5 µl of the resulting suspension was spotted on a slide and sealed under a coverslip. Confocal microscopy was performed at the Bioimaging Center at the University of Konstanz, Germany, using a point laser scanning confocal microscope (LSM 510 Meta; Carl Zeiss) with a 100× (NA = 1.4) Plan Apochromatic correction Carl Zeiss objective. Images were captured using the internal detectors of the microscope with Efficient Navigation (ZEN 2008; Carl Zeiss) software at room temperature and were transferred to ImageJ (National Institutes of Health) for background subtraction and Photoshop CS4 (Adobe) for contrast and brightness adjustments.
Subcellular fractionation
Yeast subcellular fractionation was performed as previously described (Swanson et al., 2001) with minor alterations. Log-phase doa10Δ ssh1Δ yeast cells expressing the indicated HA-tagged Sec61 β subunit from a plasmid were mixed at a 5:1 ratio with log-phase yeast cells expressing Doa10-13MYC from the DOA10 locus directly before cell harvest (Doa10-13MYC served as an integral membrane protein control). 25 OD600 of cells were harvested, and cell pellets were washed with H2O before cells were resuspended in 500 µl cold fractionation buffer (FB; 200 mM d-mannitol, 150 mM NaCl, 20 mM sodium phosphate, and 1 mM MgCl2) plus protease inhibitors. After glass bead lysis, the beads were washed three times with 200 µl FB plus protease inhibitors. The initial supernatant and supernatants from washing steps were combined and cleared by centrifugation (600 g for 5 min at 4°C). The resulting supernatant was split into five aliquots, and each aliquot was subjected to one of the following treatments (1-h incubation on ice with occasional mixing): FB alone, 0.5 M NaCl, 0.1 M Na2CO3, pH 11.5, and 2.5 M urea or 1% Triton X-100 and 0.5 M NaCl. After incubation, the lysates were separated into pellet (P) and supernatant (S) fractions by centrifugation (16,100 g for 30 min at 4°C). Pellet fractions were washed with FB containing the respective supplements and centrifuged again (16,100 g for 10 min at 4°C). The resulting supernatant fractions were discarded, and pellet fractions were resuspended in FB containing the appropriate supplements. Both the initial supernatant and the “washed” and resuspended pellet were subjected to TCA precipitation (5% TCA). After centrifugation (16,100 g for 10 min at 4°C), the protein pellets were resuspended in 35 µl TCA sample buffer (3.5% SDS, 0.5 M DTT, 80 mM Tris, 8 mM EDTA, 15% glycerol, and 4 mg/ml bromophenol blue) plus 15 µl of 4× sample buffer. Samples were incubated for 15 min at 37°C before gel loading.
In some cases, it was only determined whether a given Sec61 β subunit is present in the crude microsome fraction. As described in the previous paragraph, log-phase ssh1Δ doa10Δ cells expressing the indicated tagged Sec61 β subunit were mixed with log-phase cells expressing Doa10-13MYC. Cells were harvested and resuspended in 300 µl cold FB plus protease inhibitors. After glass bead lysis, the lysate was cleared, and the resulting supernatant was divided into two (input and fractionation sample). The fractionation sample was centrifuged (16,100 g for 20 min at 4°C), the resulting supernatant contained the soluble proteins, and the pellet represented the crude microsome fraction. The pellet was washed and resuspended in FB plus protease inhibitors. Proteins in the input, supernatant, and resuspended pellet (crude microsomes) fractions were precipitated in 5% TCA, resuspended, and evaluated by immunoblotting.
Online supplemental material
Fig. S1 shows analysis of Sbh1/Sbh2 chimeric proteins. Fig. S2 shows a sequence alignment of C-terminal regions of Sec61 β subunits and hydrophobicity of Sbh1, Sbh2, and Sbh2(S68A) TM domains. Fig. S3 shows Doa10 interaction with Sec61 β subunits and ubiquitylation of WT and mutant Sec61 β subunits. Fig. S4 shows modeled Sec61 complex with Sbh2. Fig. S5 shows degradations assays with Ura3-HA-Ubc6TM and Ura3-HA-Sbh2(aa 41–88). Table S1 lists plasmids used in this study. Table S2 lists yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408088/DC1.
Supplementary Material
Acknowledgments
We thank Mark Hochstrasser, Thomas Sommer, Jonathan Weissman, and Dieter H. Wolf for yeast strains, plasmids, and antibodies. We also thank the Bioimaging Center at the University of Konstanz and Elisabeth Stuerner for help with fluorescence microscopy and Daria Bova for generating yeast strains SKY386 and 388. We thank Mark Hochstrasser and Eric M. Rubenstein for critical reading of the manuscript. S.G. Kreft thanks Martin Scheffner for stimulating discussions and continuous support.
This work was supported by intramural funds of the University of Konstanz.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- chx
- cycloheximide
- ERAD
- ER-associated degradation
- FB
- fractionation buffer
- IP
- immunoprecipitation
- MARCH
- membrane-associated RING-CH
- TA
- tail anchored
- TCR-α
- T cell receptor α chain
- TM
- transmembrane
- ts
- temperature sensitive
- WT
- wild type
References
- Ast T., Aviram N., Chuartzman S.G., and Schuldiner M.. 2014. A cytosolic degradation pathway, prERAD, monitors pre-inserted secretory pathway proteins. J. Cell Sci. 127:3017–3023 10.1242/jcs.144386 [DOI] [PubMed] [Google Scholar]
- Bartee E., Mansouri M., Hovey Nerenberg B.T., Gouveia K., and Früh K.. 2004. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78:1109–1120 10.1128/JVI.78.3.1109-1120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays N.W., Gardner R.G., Seelig L.P., Joazeiro C.A., and Hampton R.Y.. 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3:24–29 10.1038/35050524 [DOI] [PubMed] [Google Scholar]
- Becker T., Bhushan S., Jarasch A., Armache J.P., Funes S., Jossinet F., Gumbart J., Mielke T., Berninghausen O., Schulten K., et al. 2009. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 326:1369–1373 10.1126/science.1178535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T., Volkwein C., and Sommer T.. 1996. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Suzuki C.K., Lippincott-Schwartz J., Weissman A.M., and Klausner R.D.. 1989. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J. Cell Biol. 109:73–83 10.1083/jcb.109.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Suzuki C.K., and Klausner R.D.. 1990. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science. 247:79–82 10.1126/science.2294595 [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., and Skach W.R.. 2011. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23:464–475 10.1016/j.ceb.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., and Rapoport T.A.. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Christianson J.C., and Ye Y.. 2014. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat. Struct. Mol. Biol. 21:325–335 10.1038/nsmb.2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran K., Jabbour M., Bhagwandin C., Deymier M.J., Theisen D.L., and Lybarger L.. 2011. Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1). J. Biol. Chem. 286:37168–37180 10.1074/jbc.M110.204040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L., and Ganem D.. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Invest. 107:1599–1606 10.1172/JCI12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak P.M., and Wolf D.H.. 2001. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 276:10663–10669 10.1074/jbc.M008608200 [DOI] [PubMed] [Google Scholar]
- Deng M., and Hochstrasser M.. 2006. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 443:827–831 10.1038/nature05170 [DOI] [PubMed] [Google Scholar]
- Feige M.J., and Hendershot L.M.. 2013. Quality control of integral membrane proteins by assembly-dependent membrane integration. Mol. Cell. 51:297–309 10.1016/j.molcel.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Zhao X., Soromani C., Toikkanen J., Römisch K., Vembar S.S., Brodsky J.L., Keränen S., and Jäntti J.. 2007. The transmembrane domain is sufficient for Sbh1p function, its association with the Sec61 complex, and interaction with Rtn1p. J. Biol. Chem. 282:30618–30628 10.1074/jbc.M701840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke K., Plath K., Panzner S., Prehn S., Rapoport T.A., Hartmann E., and Sommer T.. 1996. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Finley D., Ulrich H.D., Sommer T., and Kaiser P.. 2012. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 192:319–360 10.1534/genetics.112.140467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O., Ruggiano A., Hannibal-Bach H.K., Ejsing C.S., and Carvalho P.. 2013. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife. 2:e00953 10.7554/eLife.00953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.G., and Hampton R.Y.. 1999. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. EMBO J. 18:5994–6004 10.1093/emboj/18.21.5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., and Fink G., 1991. Guide to yeast genetics and molecular biology. Methods in Enzymology Vol. 194 Academic Press, San Diego. [Google Scholar]
- Hegde R.S., and Keenan R.J.. 2011. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 12:787–798 10.1038/nrm3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R.S., and Ploegh H.L.. 2010. Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22:437–446 10.1016/j.ceb.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., and Cooper A.A.. 2000. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 19:550–561 10.1093/emboj/19.4.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C., Gauss R., Horn S.C., Neuber O., and Sommer T.. 2009. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 458:453–460 10.1038/nature07962 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M.2009. Origin and function of ubiquitin-like proteins. Nature. 458:422–429 10.1038/nature07958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G., Piluek W.F., Fansler Z., Kreft S.G., Hochstrasser M., Brodsky J.L., and Michaelis S.. 2004. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279:38369–38378 10.1074/jbc.M402468200 [DOI] [PubMed] [Google Scholar]
- Kinch L.N., Saier M.H. Jr, and Grishin N.V.. 2002. Sec61β—a component of the archaeal protein secretory system. Trends Biochem. Sci. 27:170–171 10.1016/S0968-0004(01)02055-2 [DOI] [PubMed] [Google Scholar]
- Kreft S.G., and Hochstrasser M.. 2011. An unusual transmembrane helix in the endoplasmic reticulum ubiquitin ligase Doa10 modulates degradation of its cognate E2 enzyme. J. Biol. Chem. 286:20163–20174 10.1074/jbc.M110.196360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft S.G., Wang L., and Hochstrasser M.. 2006. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J. Biol. Chem. 281:4646–4653 10.1074/jbc.M512215200 [DOI] [PubMed] [Google Scholar]
- Lee D.H., and Goldberg A.L.. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397–403 10.1016/S0962-8924(98)01346-4 [DOI] [PubMed] [Google Scholar]
- Leroux A., and Rokeach L.A.. 2008. Inter-species complementation of the translocon beta subunit requires only its transmembrane domain. PLoS ONE. 3:e3880 10.1371/journal.pone.0003880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D., Tam A., Schmidt W.K., and Michaelis S.. 1998. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 9:2767–2784 10.1091/mbc.9.10.2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon E.C., Trueman S.F., and Gilmore R.. 2013. Protein translocation across the rough endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5:a013342 10.1101/cshperspect.a013342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert M., Sommer T., and Jarosch E.. 2014. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat. Cell Biol. 16:77–86 10.1038/ncb2882 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., and Brodsky J.L.. 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 132:101–112 10.1016/j.cell.2007.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan J.A., and Lehner P.J.. 2009. The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp. Cell Res. 315:1593–1600 10.1016/j.yexcr.2008.10.026 [DOI] [PubMed] [Google Scholar]
- Panzner S., Dreier L., Hartmann E., Kostka S., and Rapoport T.A.. 1995. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 81:561–570 10.1016/0092-8674(95)90077-2 [DOI] [PubMed] [Google Scholar]
- Park E., and Rapoport T.A.. 2012. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 41:21–40 10.1146/annurev-biophys-050511-102312 [DOI] [PubMed] [Google Scholar]
- Ravid T., Kreft S.G., and Hochstrasser M.. 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25:533–543 10.1038/sj.emboj.7600946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein E.M., Kreft S.G., Greenblatt W., Swanson R., and Hochstrasser M.. 2012. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J. Cell Biol. 197:761–773 10.1083/jcb.201203061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A., Foresti O., and Carvalho P.. 2014. Quality control: ER-associated degradation: protein quality control and beyond. J. Cell Biol. 204:869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato B.K., Schulz D., Do P.H., and Hampton R.Y.. 2009. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol. Cell. 34:212–222 10.1016/j.molcel.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H.D., Schwappach B., and Weissman J.S.. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 134:634–645 10.1016/j.cell.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., and Hegde R.S.. 2011. Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 27:25–56 10.1146/annurev-cellbio-092910-154125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Besser S., Hottmann H., and Wolf D.H.. 2013. Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum-associated protein degradation. Proc. Natl. Acad. Sci. USA. 110:15271–15276 10.1073/pnas.1304928110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuerner E., Kuraku S., Hochstrasser M., and Kreft S.G.. 2012. Split-Doa10: a naturally split polytopic eukaryotic membrane protein generated by fission of a nuclear gene. PLoS ONE. 7:e45194 10.1371/journal.pone.0045194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R., Locher M., and Hochstrasser M.. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15:2660–2674 10.1101/gad.933301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toikkanen J., Gatti E., Takei K., Saloheimo M., Olkkonen V.M., Söderlund H., De Camilli P., and Keränen S.. 1996. Yeast protein translocation complex: isolation of two genes SEB1 and SEB2 encoding proteins homologous to the Sec61 β subunit. Yeast. 12:425–438 [DOI] [PubMed] [Google Scholar]
- Van den Berg B., Clemons W.M. Jr, Collinson I., Modis Y., Hartmann E., Harrison S.C., and Rapoport T.A.. 2004. X-ray structure of a protein-conducting channel. Nature. 427:36–44 10.1038/nature02218 [DOI] [PubMed] [Google Scholar]
- Varshavsky A.1991. Naming a targeting signal. Cell. 64:13–15 10.1016/0092-8674(91)90202-A [DOI] [PubMed] [Google Scholar]
- Vashist S., and Ng D.T.. 2004. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165:41–52 10.1083/jcb.200309132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S.S., and Brodsky J.L.. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Urban J., Volkwein C., and Sommer T.. 2001. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 20:3124–3131 10.1093/emboj/20.12.3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lybarger L., Connors R., Harris M.R., and Hansen T.H.. 2004. Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex. J. Virol. 78:8673–8686 10.1128/JVI.78.16.8673-8686.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Herr R.A., and Hansen T.. 2008. Viral and cellular MARCH ubiquitin ligases and cancer. Semin. Cancer Biol. 18:441–450 10.1016/j.semcancer.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., and Rapoport T.A.. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656 10.1038/414652a [DOI] [PubMed] [Google Scholar]
- Zattas D., and Hochstrasser M.. 2015. Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit. Rev. Biochem. Mol. Biol. 50:1–17 10.3109/10409238.2014.959889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., and Jäntti J.. 2009. Functional characterization of the trans-membrane domain interactions of the Sec61 protein translocation complex β-subunit. BMC Cell Biol. 10:76 10.1186/1471-2121-10-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.