Abstract
Matrix degradation is central to tumor pathogenesis. Enzymes that degrade extracellular matrix are abundant in tumors. But which out of the complex mixture of cells that form a tumor produces them? Surprisingly, several hundred studies devoted to this question have provided confusion rather than clarity. Our analysis of these studies identifies likely reasons as to why this may be the case, which has implications for the broader issue of research reproducibility.
Tumor invasion—that is, the infiltration, dissolution, and eventual substitution of normal tissues with tumor tissue—is a hallmark of human cancer and is key to the morbidity and mortality of the disease. The process involves the complete degradation of the preexisting extracellular matrix scaffolds of the invaded tissue concomitant with the formation of a new tumor-derived extracellular matrix that supports the continued expansion of the tumor mass (Jones and De Clerck, 1982; Liotta et al., 1983; Danø et al., 1985; Liotta, 1985; Tryggvason et al., 1987; Lu et al., 2012). Tumors consist of malignant cells and an assortment of nonmalignant cells, termed stromal cells (Fig. 1). The most abundant stromal cells are fibroblasts and macrophages, but other cell types, such as lymphocytes, neutrophils, mast cells, myoepithelial cells, endothelial cells, lymphendothelial cells, and platelets may also be present (Ehrlich, 1907; Borst, 1924; Engels et al., 2012; Gajewski et al., 2013; Galdiero et al., 2013; Ribatti, 2013; Noy and Pollard, 2014; Öhlund et al., 2014; Sharma et al., 2014). It is a widely held notion that matrix dissolution in human cancer is initiated by the release from the tumor of a limited number of hydrolytic enzymes that are unique in that they display potent enzymatic activity toward intact extracellular matrices (Liotta et al., 1982; Sloane and Honn, 1984; Danø et al., 1985). But which cells, among the diverse population of cell types that constitute a human tumor, produce these enzymes? Despite intense research, there is no clear answer to this question, which is not merely an academic one. Rather, it is central to being able to productively model human tumor invasion in animals and in cell-based ex vivo assays of extracellular matrix dissolution, which is key to the successful development of much-needed novel cancer therapeutics. Moreover, serious problems with reproducing preclinical cancer research have recently been uncovered (Prinz et al., 2011; Begley and Ellis, 2012). These problems are a major concern and may jeopardize both successful cancer therapy development and the integrity of the research field. Here, we set out to critically evaluate the published cancer research literature in order to identify the cellular sources of extracellular matrix–degrading enzymes in human cancer. In the process, we identified possible reasons for the lack of consensus in the field, which may be important to other fields as well and are relevant to the current dialogue on research reproducibility.
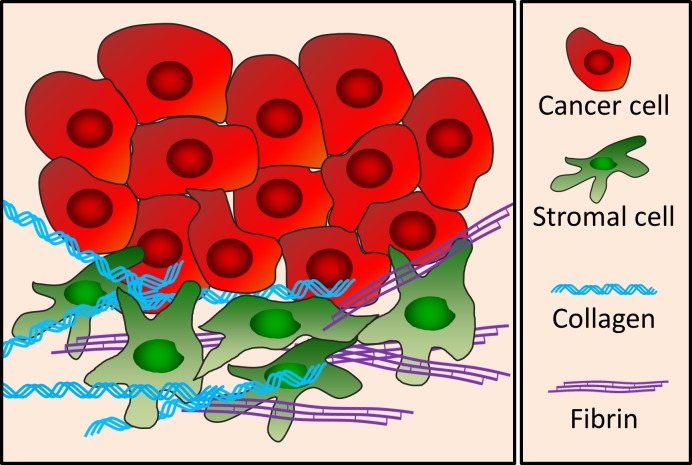
Figure 1.
Human tumors consist of a mixture of cell types associated with a tumor-derived extracellular matrix. Schematic showing cancer cells (red) and an assortment of nonmalignant stromal cells (green) embedded in an extracellular matrix rich in cross-linked interstitial collagen (blue) and fibrin (purple).
A widely discrepant literature
To obtain insights into the cellular sources of matrix-degrading enzymes in human cancer, we performed a literature analysis spanning the last two-and-a-half decades. Due to the vastness of the existing literature, which was evident from our preliminary search, we limited our analysis to the study of four human cancers—breast, colon, lung, and prostate—because they are estimated to account for about half of all newly reported cancer cases and cancer deaths in the United States in 2014 (American Cancer Society, 2014). We focused our analysis on proteases with the capacity to degrade nondenatured interstitial collagen and fibrin, which are the two principal cross-linked protein matrices that are encountered by the expanding tumor mass (Dvorak, 1986; Hiraoka et al., 1998; Hotary et al., 2003; Palumbo et al., 2003; Rowe and Weiss, 2009). These enzymes were matrix metalloproteinase (MMP)-1 (interstitial collagenase, collagenase-1), MMP-2 (gelatinase A, Mr 72,000 type IV collagenase), MMP-13 (collagenase-3), MMP-14 (membrane-type-1 MMP), and urokinase-type plasminogen activator (uPA), which activates the ubiquitous and abundant fibrinolytic protease zymogen, plasminogen (Danø et al., 1985; Aimes and Quigley, 1995; Gill and Parks, 2011). We excluded MMP-8 because of the very low number of studies addressing the expression of MMP-8 in cancer. We compiled 452 datasets from 248 published studies in which the cellular expression of either of these five proteolytic enzymes was analyzed in a manner that provided spatial resolution (immunohistochemistry [IHC] or immunofluorescence [IF]) for protein detection or in situ hybridization for detection of mRNA). We were unable to evaluate protease localization in 161 of these datasets (from 109 research papers), because the cellular localization was not described by the authors or because no figure example of the IHC, IF, or in situ hybridization was included. This left a total of 291 datasets, which included 105 studies of breast cancer, 100 studies of colon cancer, 52 studies of lung cancer, and 34 studies of prostate cancer, with 41 studies analyzing the location of MMP-1, 119 studies of MMP-2, 17 studies of MMP-13, 45 studies of MMP-14, and 69 studies of uPA. The complete list of studies can be found in the supplemental material.
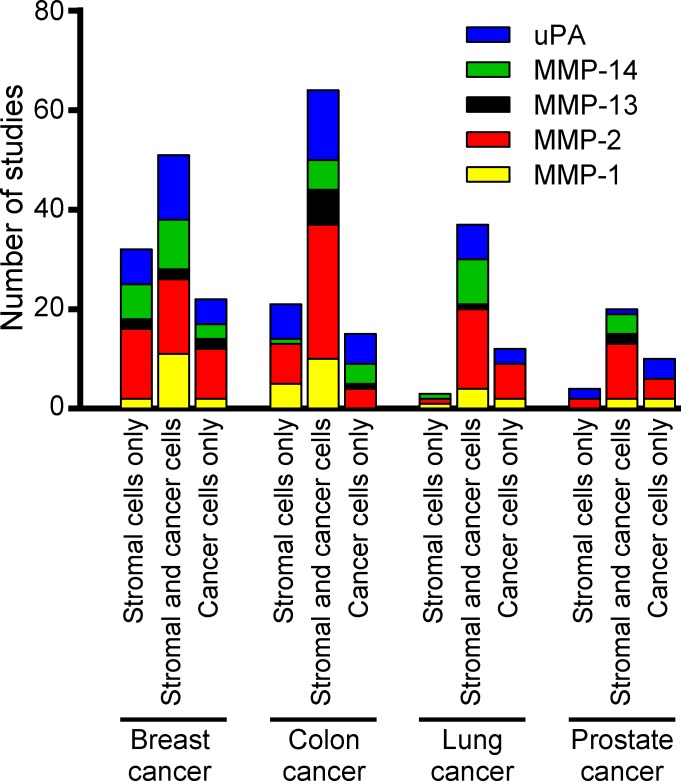
To our surprise, no consensus emerged from these studies as to the cellular source of any of these five proteases in any of the four human tumors. Some studies reported expression of the proteases exclusively by cancer cells, whereas other studies found expression exclusively in nonmalignant stromal cells, and yet other studies found expression of these proteases in both cancer cells and stromal cells (Fig. 2). Furthermore, the reported frequency with which the expression of these proteases was found in individual human tumors, in studies where this was reported, varied from infrequently to ubiquitously. Likewise, the reported fraction of cells within each individual tumor that expressed a given protease varied from a small percentage to all cells within the tumor. Also, some studies found expression of matrix-degrading proteases to be restricted to tumor tissue, whereas other studies also found expression in corresponding normal tissues, again with widely varying cellular locations and frequencies. In studies that found protease expression in stromal cells, incongruity in the reported identity of the nonmalignant protease-expressing cells was frequent. These striking discrepancies could not easily be explained by sampling bias, as the average number of tumors analyzed in each individual study was 108 (with a range of 4–1,420). Furthermore, a comparison of studies of histologically stage-matched tumors, such as ductal breast carcinoma in situ, matched Gleason score for prostate cancer, or matched Dukes classification for colon cancer, did not reveal a consistent pattern in the reported localization of the five proteases.
Figure 2.
Localization of extracellular matrix–degrading proteases in human cancer. Compilation of expression localization data for MMP-1, MMP-2, MMP-13, MMP-14, and uPA obtained from 291 published studies of human breast, colon, lung, and prostate cancer (supplemental text). The bar graphs show the number of studies reporting the expression of any of the proteases in stromal cells only, both cancer and stromal cells, and cancer cells only.
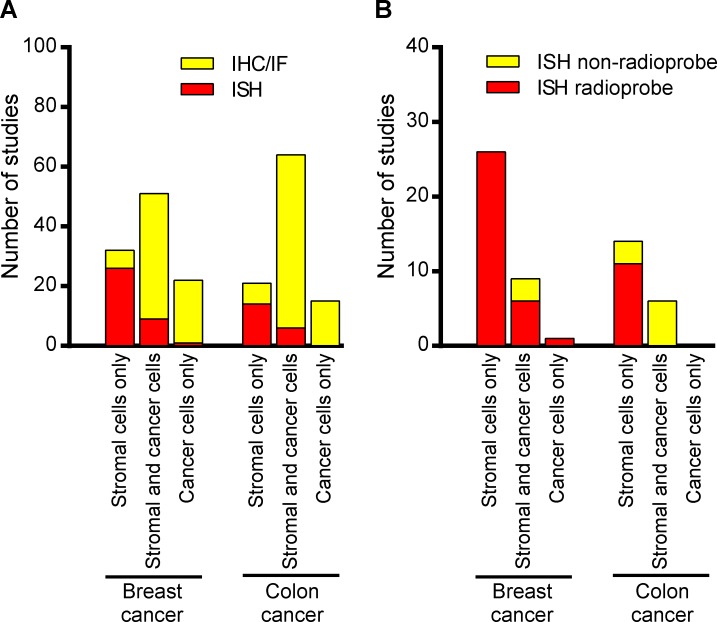
The remarkable absence of an inter-study consensus as to matrix-degrading protease location in human breast, colon, lung, and prostate cancer led us to investigate whether the detection method affected the reported protease localization. Studies on breast and colon cancer constituted the majority (70%) of all the evaluated studies, and we therefore focused on these two types of cancer for this analysis. We first divided the studies into those using IHC or IF, and those using in situ hybridization as the method of protease detection. We found a striking difference in the expression pattern reported based on IHC or IF and the expression pattern reported based on in situ hybridization. The majority of the IHC- or IF-based studies found that both cancer and stromal cells expressed the proteases (61% in breast cancer and 73% in colon cancer), with the remaining 39% and 27% of studies finding expression exclusively in either cancer cells or in stromal cells (Fig. 3 A). Within the in situ hybridization studies, 72% and 70% of the breast cancer and colon cancer studies, respectively, identified stromal cells as the sole source of the five proteases (Fig. 3 A). The remaining studies reported the proteases to be expressed in both stromal and cancer cells or in the cancer cells only. We found no consistency in terms of which of the five proteases were observed in cancer cells, or in both stromal and cancer cells. If we focused entirely on the studies that used radiolabeled probes for in situ hybridization, which made up the majority of the studies, we saw an even stronger consensus, wherein 79% (26 out of 33 studies) and 100% (11 out of 11 studies) of the studies of breast cancer and colon cancer, respectively, found the matrix-degrading proteases to be expressed exclusively by stromal cells (Fig. 3 B). The majority (68%) of the relatively few in situ hybridization studies that used nonradioactive DNA or oligonucleotide probes, however, found the proteases to be expressed in both cancer and stromal cells, frequently in 100% of the cells. This relatively high inter-study consensus suggests that tumor stromal cells, and not tumor cells per se, may be the primary source of matrix-degrading protease in human breast and colon carcinomas. This finding is at odds with the high-level expression of matrix-degrading enzymes found in many established human breast and colon cancer cell lines. However, the degree to which these tumor cell lines, often established multiple decades ago, are representative of the tumors from which they derive in terms of expression of matrix-degrading proteases, is unclear. These cell lines may derive from rare subpopulations of tumor cells endowed with the capacity to be propagated ex vivo. Furthermore, the continuous growth under 2D culture conditions may have led to a phenotypic drift.
Figure 3.
The reported location of extracellular matrix-degrading proteases in human cancer depends on detection method. (A) Compilation of expression localization data for MMP-1, MMP-2, MMP-13, MMP-14, and uPA in breast and colon cancer divided into in situ hybridization studies (ISH) and IHC or IF studies (IHC/IF). The bar graphs show the number of studies reporting the expression of any of the proteases in stromal cells only, both cancer and stromal cells, and cancer cells only. (B) Compilation of expression localization data for MMP-1, MMP-2, MMP-13, MMP-14, and uPA in breast and colon cancer divided into radioprobe in situ hybridization studies and nonradioprobe in situ studies. The bar graphs show the number of studies reporting the expression of any of the proteases in stromal cells only, both cancer and stromal cells, and cancer cells only.
Why such inconsistency?
In summary, a large body of work, spanning more than two decades, has failed to provide a consensus as to the cellular source of extracellular matrix–degrading enzymes in four of the most clinically important human cancers. Why do different researchers, to this extraordinary degree, get different answers when studying the same question? The reason for this will have to remain a matter of speculation. However, our in-depth analysis of the literature would suggest that some of the following issues alone or in combination have contributed to this:
•Inherently low expression levels of extracellular matrix–degrading enzymes, as compared with, for example, structural components of the tumor stroma, making their detection technically challenging, particularly by IHC of IF. This problem may not be restricted to protease detection, but may extend to the detection of other low-abundance secreted proteins in malignant as well as nonmalignant tissues.
•Inconsistent fixation conditions for excised human tumor tissues, resulting in variable epitope and mRNA preservation, and an associated inter-study variability.
•Extensive reliance on manufacturer specifications or the use of antibodies in prior publications as validation of antibody specificity. In this regard, significant problems with quality control of commercial antibodies recently were highlighted by Bradbury and Plückthun (2015).
•The use of excessively long in situ hybridization probes with a propensity for cross-hybridization or, conversely, very short in situ hybridization probes with insufficient ability to visualize nonabundant mRNAs.
•Lack of markers to positively identify antibody-stained or hybridization signal–positive cell populations. Only 10% of the studies analyzed here used cell type–specific markers to identify protease-expressing cell populations.
•The failure to use multiple complementary approaches to validate findings: 87% of the published studies relied on a single antibody or in situ probe to identify protease-expressing cell populations in the four human cancers.
It should be mentioned, however, that contradictory findings were reported even for studies that were seemingly well controlled. In a very thorough study of breast cancer by Nielsen et al. (2001), they used seven different uPA antibodies and an in situ probe, combined with markers for myofibroblasts and myoepithelial cells (α-smooth muscle actin), macrophages (CD68), and endothelial cells (CD31). This study led to the conclusion that uPA is found almost exclusively in stromal cells. In contrast, Carriero et al. (1994), using six different uPA antibodies, reported localization of the protease predominantly to breast cancer cells.
Concluding remarks and recommendations
Knowledge of the cellular sources of matrix-degrading protease production is much needed to assess the validity of current models of human cancer invasion, to generate new models that faithfully replicate human cancer invasion, and to provide a more solid basis for the development of new cancer therapeutics aimed at interfering with matrix degradation. As revealed in our analysis, the identification of the cellular source of matrix-degrading protease expression in human cancer clearly has proven problematic. Therefore, to resolve this issue, future studies would need to apply more highly quality-controlled IHC/IF and in situ hybridization studies as well as complementary approaches to determine the localization of protease protein and mRNA. These may also be useful to other fields where similar controversies may exist.
These include:
•Routine application on serial sections of several extensively validated antibodies for IHC and IF studies and multiple nonoverlapping probes for in situ hybridization studies. In both cases, identical staining/hybridization patterns should be used as the principal evidence for signal specificity, rather than, respectively, absence of staining after primary antibody omission or lack of a hybridization signal from complementary sense probes.
•Critical evaluation of whether the subcellular staining pattern corresponds to the expected localization of the protease.
•mRNA and protease expression profiling of rigorously quality-controlled laser capture microdissected tumor cell populations.
•mRNA and protease expression profiling of immunologically isolated cell populations from human tumor tissues.
Future studies should also integrate the recent revolutionary advances in molecular classification of human cancers to tease out distinct protease expression profiles (if any) linked to each molecular subclass of human cancer. Likewise, these studies should take into consideration the remarkable complexity of the cellular tumor stroma revealed by recent research (Gajewski et al., 2013; Galdiero et al., 2013; Ribatti, 2013; Noy and Pollard, 2014; Öhlund et al., 2014; Sharma et al., 2014).
Journal publication and funding agency policies historically have not stimulated the execution of such time-, labor-, and cost-consuming, yet critically important studies. Encouragingly, however, the heightened awareness of problems with reproducibility of preclinical cancer research (Prinz et al., 2011; Begley and Ellis, 2012; Bradbury and Plückthun, 2015) has triggered new initiatives involving representatives from major funding agencies, academic researchers, reviewers, journal editors, the pharmaceutical industry, and patient advocacy groups to provide improved guidelines for the execution and reporting of science (Landis et al., 2012; Collins and Tabak, 2014).
Online supplemental material
A complete list of studies is available as supplemental text. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501034/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Silvio Gutkind, Alfredo Molinolo, Daniel Lawrence, Karin List, and Mary Jo Danton for critically reviewing this manuscript.
This work was supported by the NIDCR Intramural Research Program.
The authors declare no competing financial interests in relation to the work described.
References
- Aimes R.T., and Quigley J.P.. 1995. Matrix metalloproteinase-2 is an interstitial collagenase. Inhibitor-free enzyme catalyzes the cleavage of collagen fibrils and soluble native type I collagen generating the specific 3/4- and 1/4-length fragments. J. Biol. Chem. 270:5872–5876. [DOI] [PubMed] [Google Scholar]
- American Cancer Society. 2014. Cancer Facts & Figures 2014. American Cancer Society, Atlanta: 72 pp. [Google Scholar]
- Begley C.G., and Ellis L.M.. 2012. Drug development: Raise standards for preclinical cancer research. Nature. 483:531–533 10.1038/483531a [DOI] [PubMed] [Google Scholar]
- Borst M.1924. Allgemeine Pathologie der malignen Geschwülste. S. Hirzel, Leipzig: 322 pp. [Google Scholar]
- Bradbury A., and Plückthun A.. 2015. Reproducibility: Standardize antibodies used in research. Nature. 518:27–29 10.1038/518027a [DOI] [PubMed] [Google Scholar]
- Carriero M.V., Franco P., Del Vecchio S., Massa O., Botti G., D’Aiuto G., Stoppelli M.P., and Salvatore M.. 1994. Tissue distribution of soluble and receptor-bound urokinase in human breast cancer using a panel of monoclonal antibodies. Cancer Res. 54:5445–5454. [PubMed] [Google Scholar]
- Collins F.S., and Tabak L.A.. 2014. Policy: NIH plans to enhance reproducibility. Nature. 505:612–613 10.1038/505612a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danø K., Andreasen P.A., Grøndahl-Hansen J., Kristensen P., Nielsen L.S., and Skriver L.. 1985. Plasminogen activators, tissue degradation, and cancer. Adv. Cancer Res. 44:139–266 10.1016/S0065-230X(08)60028-7 [DOI] [PubMed] [Google Scholar]
- Dvorak H.F.1986. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 315:1650–1659 10.1056/NEJM198612253152606 [DOI] [PubMed] [Google Scholar]
- Ehrlich P.1907. Experimentelle Studien an Mäusetumoren. Experimentelle Studien an Mäusetumoren. 5:59–81. [Google Scholar]
- Engels B., Rowley D.A., and Schreiber H.. 2012. Targeting stroma to treat cancers. Semin. Cancer Biol. 22:41–49 10.1016/j.semcancer.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski T.F., Schreiber H., and Fu Y.X.. 2013. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14:1014–1022 10.1038/ni.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero M.R., Bonavita E., Barajon I., Garlanda C., Mantovani A., and Jaillon S.. 2013. Tumor associated macrophages and neutrophils in cancer. Immunobiology. 218:1402–1410 10.1016/j.imbio.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Gill S.E., and Parks W.C.. 2011. Matrix metalloproteinases and their inhibitors in turnover and degradation of extracellular matrix. Extracellular Matrix Degradation. Parks W.C. and Mecham R.P., Springer, New York: 1–22 10.1007/978-3-642-16861-1_1 [DOI] [Google Scholar]
- Hiraoka N., Allen E., Apel I.J., Gyetko M.R., and Weiss S.J.. 1998. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell. 95:365–377 10.1016/S0092-8674(00)81768-7 [DOI] [PubMed] [Google Scholar]
- Hotary K.B., Allen E.D., Brooks P.C., Datta N.S., Long M.W., and Weiss S.J.. 2003. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 114:33–45 10.1016/S0092-8674(03)00513-0 [DOI] [PubMed] [Google Scholar]
- Jones P.A., and De Clerck Y.A.. 1982. Extracellular matrix destruction by invasive tumor cells. Cancer Metastasis Rev. 1:289–317 10.1007/BF00124214 [DOI] [PubMed] [Google Scholar]
- Landis S.C., Amara S.G., Asadullah K., Austin C.P., Blumenstein R., Bradley E.W., Crystal R.G., Darnell R.B., Ferrante R.J., Fillit H., et al. 2012. A call for transparent reporting to optimize the predictive value of preclinical research. Nature. 490:187–191 10.1038/nature11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta L.A. 1985. Mechanisms of cancer invasion and metastasis. Important Adv. Oncol. 1985:28–41. [PubMed] [Google Scholar]
- Liotta L.A., Thorgeirsson U.P., and Garbisa S.. 1982. Role of collagenases in tumor cell invasion. Cancer Metastasis Rev. 1:277–288 10.1007/BF00124213 [DOI] [PubMed] [Google Scholar]
- Liotta L.A., Rao C.N., and Barsky S.H.. 1983. Tumor invasion and the extracellular matrix. Lab. Invest. 49:636–649. [PubMed] [Google Scholar]
- Lu P., Weaver V.M., and Werb Z.. 2012. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196:395–406 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen B.S., Sehested M., Duun S., Rank F., Timshel S., Rygaard J., Johnsen M., and Danø K.. 2001. Urokinase plasminogen activator is localized in stromal cells in ductal breast cancer. Lab. Invest. 81:1485–1501 10.1038/labinvest.3780363 [DOI] [PubMed] [Google Scholar]
- Noy R., and Pollard J.W.. 2014. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 41:49–61 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhlund D., Elyada E., and Tuveson D.. 2014. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 211:1503–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo J.S., Talmage K.E., Liu H., La Jeunesse C.M., Witte D.P., and Degen J.L.. 2003. Plasminogen supports tumor growth through a fibrinogen-dependent mechanism linked to vascular patency. Blood. 102:2819–2827 10.1182/blood-2003-03-0881 [DOI] [PubMed] [Google Scholar]
- Prinz F., Schlange T., and Asadullah K.. 2011. Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov. 10:712 10.1038/nrd3439-c1 [DOI] [PubMed] [Google Scholar]
- Ribatti D.2013. Mast cells and macrophages exert beneficial and detrimental effects on tumor progression and angiogenesis. Immunol. Lett. 152:83–88 10.1016/j.imlet.2013.05.003 [DOI] [PubMed] [Google Scholar]
- Rowe R.G., and Weiss S.J.. 2009. Navigating ECM barriers at the invasive front: the cancer cell-stroma interface. Annu. Rev. Cell Dev. Biol. 25:567–595 10.1146/annurev.cellbio.24.110707.175315 [DOI] [PubMed] [Google Scholar]
- Sharma D., Brummel-Ziedins K.E., Bouchard B.A., and Holmes C.E.. 2014. Platelets in tumor progression: a host factor that offers multiple potential targets in the treatment of cancer. J. Cell. Physiol. 229:1005–1015 10.1002/jcp.24539 [DOI] [PubMed] [Google Scholar]
- Sloane B.F., and Honn K.V.. 1984. Cysteine proteinases and metastasis. Cancer Metastasis Rev. 3:249–263 10.1007/BF00048388 [DOI] [PubMed] [Google Scholar]
- Tryggvason K., Höyhtyä M., and Salo T.. 1987. Proteolytic degradation of extracellular matrix in tumor invasion. Biochim. Biophys. Acta. 907:191–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.